Abstract
HS3ST1 is a genetic risk gene associated with Alzheimer’s disease (AD) and overexpressed in patients, but how it contributes to the disease progression is unknown. We report the analysis of brain heparan sulfate (HS) from AD and other tauopathies using a LC-MS/MS method. A specific 3-O-sulfated HS displayed sevenfold increase in the AD group (n = 14, P < 0.0005). Analysis of the HS modified by recombinant sulfotransferases and HS from genetic knockout mice revealed that the specific 3-O-sulfated HS is made by 3-O-sulfotransferase isoform 1 (3-OST-1), which is encoded by the HS3ST1 gene. A synthetic tetradecasaccharide (14-mer) carrying the specific 3-O-sulfated domain displayed stronger inhibition for tau internalization than a 14-mer without the domain, suggesting that the 3-O-sulfated HS is used in tau cellular uptake. Our findings suggest that the overexpression of HS3ST1 gene may enhance the spread of tau pathology, uncovering a previously unidentified therapeutic target for AD.
A specific heparan sulfate subpopulation is a glycan-based risk marker for Alzheimer’s disease.
INTRODUCTION
Alzheimer’s disease (AD) is the most common form of dementia that affects 5.8 million Americans aged 65 and older, and this number is projected to grow to 13.8 million by 2050 (1). One of the hallmarks of AD is the presence of neurofibrillary tangles composed of hyperphosphorylated and aggregated protein tau (2). A process that transforms unfolded monomeric protein tau into highly structured aggregates is the primary contributing factor for tauopathies. The roles of heparan sulfate (HS) in the progression of AD are gaining interest. As a sulfated polysaccharide, HS is colocalized with neurofibrillary tangles in AD brains (3), promotes amyloid pathology by modulating brain β amyloid clearance and aggregation (4), and contributes to tau cellular internalization and spreading (5, 6). Recent studies show that a subpopulation of HS, known as 3-O-sulfated HS, binds to tau (7, 8). The 3-O-sulfated HS displays higher binding affinity to tau to increase the cellular internalization (8). 3-O-sulfated HS also causes hyperphosphorylation of tau (7) and induces the aggregation of tau (9). The evidence for the involvement of HS in patients with AD is also emerging from etiology studies (10). Genome-wide association study (GWAS) involving 9751 patients demonstrates that the HS3ST1 gene is a risk locus associated with AD (11). Furthermore, the expression of HS3ST1 is elevated in patients with AD (11). The purpose of this investigation is to connect the expression of the HS3ST1 gene with the biosynthesis of the specific 3-O-sulfated HS that is elevated in AD brains.
HS is biosynthesized in a form of HS proteoglycan (HSPG), which consists of a core protein and HS sugar chains (12). HSPG can be found on the surface of neuronal cells and in the surrounding matrix. The biosynthesis of HS is initiated from the synthesis of nonsulfated polysaccharide with a repeating unit of disaccharide of glucuronic acid (GlcA) and N-acetyl glucosamine (GlcNAc). The polysaccharide is then subjected to a series of modifications by HS biosynthetic enzymes, including N-deacetylase/N-sulfotransferase (NST), C5-epimerase (C5-epi), 2-O-sulfotransferase (2-OST), 6-OST, and 3-OST. The modifications install sulfations at the different positions of glucosamine and iduronic acid (IdoA) residues as well as convert a GlcA to an IdoA residue. The sulfation patterns dictate the biological functions of HS (13).
3-OSTs are a family of HS biosynthetic enzyme that transfers sulfo groups to the 3-hydroxyl (3-OH) positions of glucosamine residues to biosynthesize 3-O-sulfated HS. The 3-O-sulfation coexists with other sulfation types on a single sugar chain, including N-sulfation, 6-O-sulfation, and 2-O-sulfation, to form a special subpopulation designated as 3-O-sulfated HS. The HS isolated from biological sources is a mixture of 3-O-sulfated HS and non–3-O-sulfated HS. Although it is a rare modification in HS, the 3-O-sulfation is closely related to the biological functions of HS (14, 15). There are seven 3-OST isoforms in the human genome encoded by HS3ST1, HS3ST2, HS3ST3A, HS3ST3B, HS3ST4, HS3ST5, and HS3ST6 (16). Each isoform has unique substrate specificity and produces the HS with distinct 3-O-sulfated domains. Connecting a 3-OST isoform to a specific 3-O-sulfated HS product is a central issue in studying the biology and biochemistry of 3-O-sulfated HS (17). Because of its low abundance, lack of a method in measuring the levels of 3-O-sulfated HS has hampered the effort to correlate the expression of HS3ST1 gene and the HS subpopulation with AD.
Here, we used a liquid chromatography–tandem mass spectrometry (LC-MS/MS) method to measure the level of 3-O-sulfated HS from AD and other neurodegenerative disease brains. Five 13C-labeled 3-O-sulfated HS calibrants were used to achieve high sensitivity. We discovered a substantial increase of a specific 3-O-sulfated HS domain, designated as Tetra-1, from AD brain. Our results demonstrate that HS3ST1 gene is responsible for the synthesis of the specific 3-O-sulfated HS carrying the Tetra-1 domain. Our findings suggest that the 3-O-sulfated HS may serve as a new target for modifying AD and early diagnosis.
RESULTS
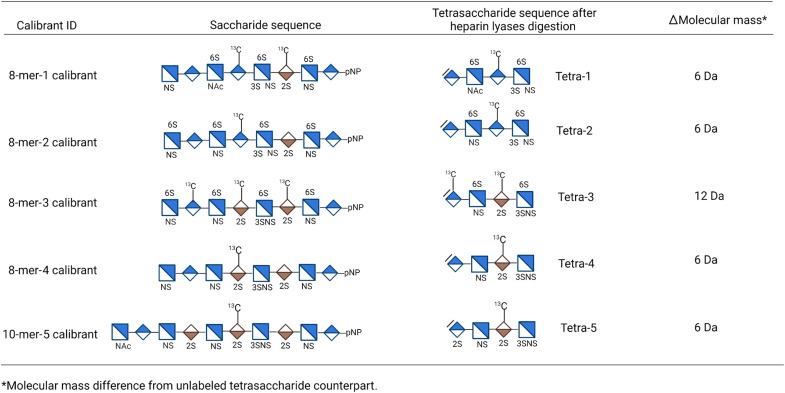
To devise the method for the analysis of 3-O-sulfated HS, we synthesized five 13C-labeled 3-O-sulfated oligosaccharide calibrants, including four 8-mers and one 10-mers, using the chemoenzymatic approach (Fig. 1) (18). Single or multiple 13C-labeled GlcA residues were introduced in the oligosaccharide backbones using the glycosyltransferase and uridine diphosphate (UDP)–[13C]GlcA. The 13C-labeled oligosaccharide backbones were then modified by sulfotransferases and C5-epi to yield the desired 13C-labeled oligosaccharide calibrants as described previously (19). The structures and purities of the calibrants were confirmed by MS and high-performance LC (HPLC) (figs. S1 to S5). The calibrants were converted to the corresponding 13C-labeled 3-O-sulfated tetrasaccharides (Tetra-1 to Tetra-5) after heparin lyases digestion (Fig. 1). The 8-mer-1 and 8-mer-3 calibrant have more than one13C-labeled saccharide residues. The extra 13C labels in the calibrants were used to determine the extent of saccharide chains digestion.
Fig. 1. List of 13C-labeled oligosaccharide calibrants and the anticipated 3-O-sulfated products after heparin lyase digestion.
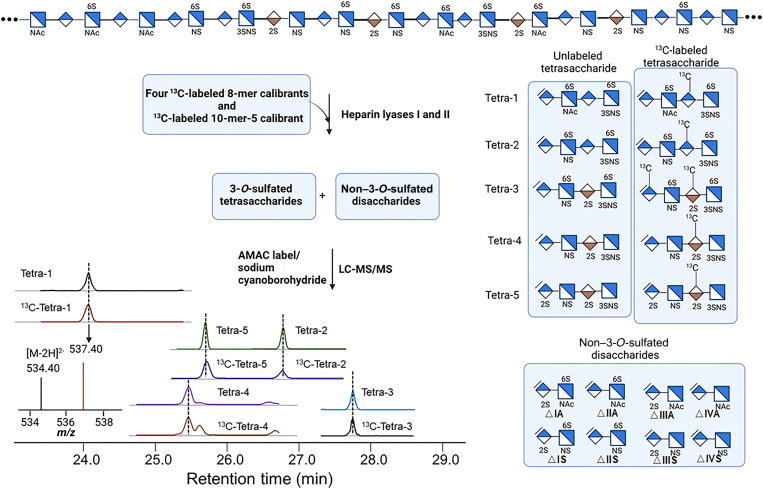
The analysis covers a two-step process, including the heparin lyases degradation followed by the LC-MS/MS analysis (Fig. 2). The 13C-labeled calibrants were mixed with HS polysaccharides and subjected to heparin lyases digestion, resulting in both 3-O-sulfated tetrasaccharides and non–3-O-sulfated disaccharides (Fig. 2). The unlabeled 3-O-sulfated tetrasaccharides can be identified, because the tetrasaccharides are coeluted with 13C-labeled tetrasaccharide counterparts on the LC before the MS/MS analysis. The quantification was achieved by measuring the ratio of the MS/MS signal peak areas for the13C-labeled tetrasaccharide and the unlabeled tetrasaccharide (Fig. 2). The molecular mass of 13C-labeled tetrasaccharide is 6 or 12 Da higher than its unlabeled counterpart (Fig. 1). The13C-labeled disaccharide calibrants were also added to the mixture during the analysis to determine the amount and composition of non–3-O-sulfated disaccharides. A total of 13 structural domains covering five 3-O-sulfated tetrasaccharides and eight non–3-O-sulfated disaccharides were detected from a single analytical process.
Fig. 2. Schematic presentation of the analysis of 3-O-sulfated HS.
Heparan sulfate (HS) was mixed with 13C-labeled oligosaccharide calibrants, and the mixture was subjected to digestion with heparin lyases I and II. The products containing both 3-O-sulfated tetrasaccharides and non–3-O-sulfated disaccharides were chemically modified by AMAC (2-aminoacridone) and sodium cyanoborohydride followed by the liquid chromatography–tandem mass spectrometry (LC-MS/MS) analysis. Five 3-O-sulfated tetrasaccharides were resolved by LC as shown on the bottom left. Both 13C-labeled tetrasaccharides and unlabeled tetrasaccharides were detected. Shorthand structures of five unlabeled and 13C-labeled 3-O-sulfated tetrasaccharides are shown. The shorthand structures of eight unlabeled non–3-O-sulfated disaccharides are also shown. From a single process, the LC-MS/MS method yields the results for the composition of five 3-O-sulfated tetrasaccharides and eight non–3-O-sulfated disaccharides. m/z, mass/charge ratio.
Elevated levels of 3-O-sulfated HS were detected in the HS from AD brain
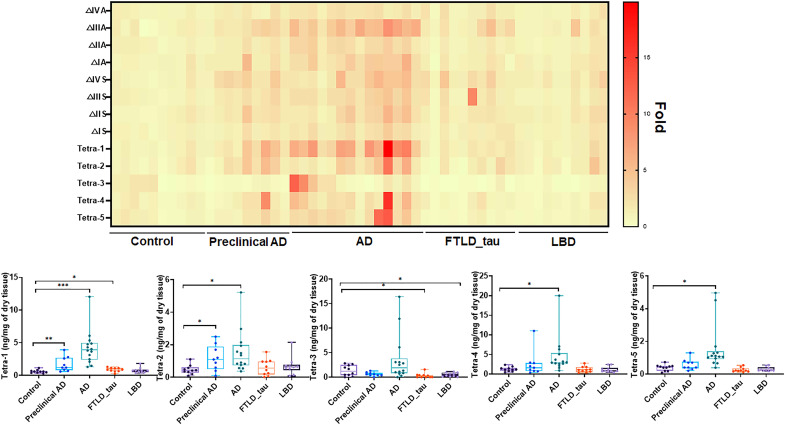
We used our method to analyze the levels of 3-O-sulfated HS from the frontal cortex of the brain from four neurodegenerative disease groups and the healthy control group (Table 1). The disease groups include preclinical AD, end-stage AD, frontotemporal lobar degeneration with tau pathology (FTLD_tau), and Lewy body dementia (LBD), which were diagnosed following the criteria described in Table 1. We discovered changes in the amount of specific 3-O-sulfated tetrasaccharide markers between disease groups and the control group (Fig. 3). In particular, the amount of Tetra-1 showed a significant increase in AD (4.24 ± 2.56 ng/mg) compared to the control group (0.60 ± 0.23 ng/mg), representing a seven-fold increase from the control group (P < 0.0005) (Fig. 3 and tables S1 to S13). Accordingly, the mass percentage of Tetra-1 in the HS from AD brain (1.5 ± 0.66%) was determined to be 2.8-fold higher than the control group (0.54 ± 0.15%) (table S9), suggesting that the concentration of the Tetra-1 domain in HS from AD brain was elevated. A significant, but a smaller, increase in the amount of Tetra-1, was observed in preclinical AD (1.72 ± 1.07 ng/mg) (P < 0.01) group. Statistically significant increases of other 3-O-sulfated tetrasaccharides were observed in AD and other neurodegenerative disease patient groups (Fig. 3 and tables S9 to S12), but the levels of increases are less than Tetra-1 in AD. The total amounts of HS from brain tissues were also measured. We observed a statistically significant increase in the total amount of HS in preclinical AD and AD groups (fig. S6). The quantities of eight non–3-O-sulfated disaccharides of the HS from each patient group are shown in fig. S7 and tables S1 to S8.
Table 1. Information for the patients’ samples used in this study.
AD, Alzheimer’s disease; FTLD_tau, frontotemporal lobar degeneration with tau pathology; LBD, Lewy body dementia; PMI, postmortem interval; M, male; F, female; NIA-AA, National Institute on Aging–Alzheimer's Association.
| Control | Preclinical AD | AD | FTLD_tau | LBD | |
|---|---|---|---|---|---|
| Description | Healthy individuals/normal controls–frontal cortex | Asymptomatic/preclinical AD with mild AD pathology–frontal cortex | End-stage AD | FTLD_tau–frontal cortex | LBD–frontal cortex (with minimal neurodegeneration) |
| n | 10 | 9 | 14 | 10 | 10 |
| Age at death (means ± SD) | 69.3 ± 9.7 | 81.4 ± 9.5 | 69.7 ± 11.0 | 66.4 ± 11.9 | 71.3 ± 9.3 |
| Sex | 5 (M) and 5 (F) | 5 (M) and 4 (F) | 8 (M) and 6 (F) | 7 (M) and 3 (F) | 6 (M) and 4 (F) |
| Race | 7 (white) and 3 (Black) | 8 (white) and 1 (Black) | 13 (white) and 1 (Black) | 9 (white) and 1 (Hispanic) | 10 (white) |
| PMI | 8.2 ± 3.5 | 8.4 ± 5.3 | 8.2 ± 4.8 | 5.8 ± 4.4 | 13.7 ± 5.3 |
| Diagnosis | The normal control group was composed of individuals with no history of neurologic disease and no obvious neurodegenerative pathology at autopsy. | The asymptomatic AD group was composed of individuals with normal cognition who exhibit obvious AD pathology at death (ABC scores of “low” and “intermediate”) (49). | “ABC” scores were determined per NIA-AA guidelines (49) | FTLD_tau was assessed according to consensus guidelines (50, 51). | The diagnosis of Lewy body disease was established using consensus criteria (51, 52). |
Fig. 3. Compositional analysis of HS from neurodegenerative disease brain tissues.
The heatmap shows the relative fold changes of non–3-O-sulfated disaccharides and 3-O-sulfated tetrasaccharides between disease groups and the control group. The amounts of five 3-O-sulfated tetrasaccharides from individual subject of disease groups and the control are presented on the bottom. P values were determined by two tailed unpaired t test, *P < 0.05; **P < 0.01; ***P < 0.001. AD, Alzheimer’s disease; FTLD_tau, frontotemporal lobar degeneration with tau pathology; LBD, Lewy body dementia. The data for the 3-O-sulfated tetrasaccharides and non–3-O-sulfated disaccharides from analyses are presented in tables S1 to S13.
3-OST-1 is the key isoform to biosynthesize Tetra-1 domain
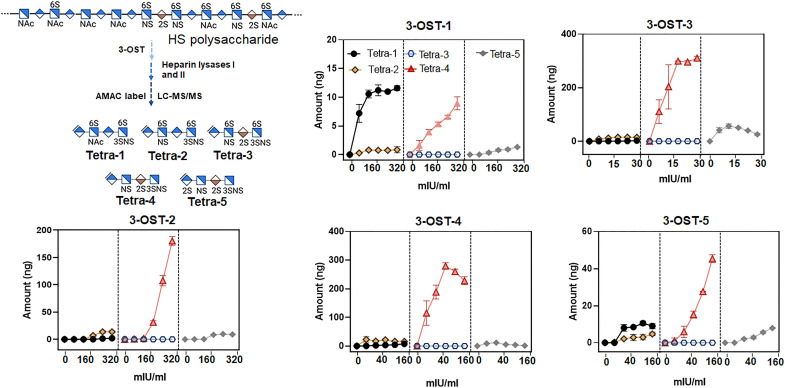
We then examined which isoform is responsible for the synthesis of the HS that contains the Tetra-1 domain. First, we analyzed the levels of 3-O-sulfated tetrasaccharides from HS after being modified by different 3-OST isoforms under in vitro conditions. Here, HS was incubated with different recombinant 3-OST isoforms, including 3-O-sulfotransferase isoform 1 (3-OST-1) to 3-OST-5 to yield 3-O-sulfated HS polysaccharide products as illustrated in Fig. 4. Notably, 3-OST-3A and 3-OST-3B isoforms contain an identical amino acid sequence in the sulfotransferase domain, and both isoforms have same substrate specificity (16). The recombinant 3-OST-3 used in this study contains only the sulfotransferase domain; therefore, it represents the activities of both 3-OST-3A and 3-OST-3B. The preparation of recombinant 3-OST-6 is still unsuccessful at present time, limiting our ability to synthesize 3-OST-6–modified HS; however, results from the analysis of HS from mammalian cell lines that 3-OST-6 was transiently expressed suggested that this isoform is very similar to 3-OST-2 to 3-OST-4 (20).
Fig. 4. Amounts of 3-O-sulfated tetrasaccharides from HS after being modified by 3-OST isoforms.
The schematic presentation of the experiment is shown in the top left corner. HS polysaccharide (from wild-type Chinese hamster ovary cells) was individually incubated with five different 3-O-sulfotransferase (3-OST) isoforms. The amount of 3-O-sulfated tetrasaccharides was determined by the LC-MS/MS. Each data point is the average of three independent determinations ± SD. mIU, milli-international units.
The LC-MS/MS analysis was next performed to determine the amounts of 3-O-sulfated domains made by different isoforms (Fig. 4). Our results show that 3-OST-1 is the isomer that produces Tetra-1, although it also produced Tetra-4 under higher enzyme concentrations. Other isoforms, including 3-OST-2 to 3-OST-4, primarily generated Tetra-4. The 3-OST-3 enzyme also generated Tetra-5 and so did 3-OST-2 to a lesser extent. 3-OST-5 is the least selective enzyme, as it generated Tetra-1, Tetra-2, Tetra-4, and Tetra-5. The level of Tetra-4 in 3-OST-5–modified HS was about fivefold higher than the level of Tetra-1 (Fig. 4). Together, there is no clear distinction in the 3-O-sulfated domains from 3-OST-2– to 3-OST-4–modified HS with an exception that a low level of Tetra-2 was detected in 3-OST-2–modified HS under high enzyme concentrations. Note that 3-OST-1 is reportedly capable of synthesizing the 3-O-sulfated Tetra-2 domain, and the enzyme is unable to synthesize Tetra-4 domain when 6- and 12-mer substrates are used (18, 21). Our data suggest that the substrate specificity of 3-OST-1 using 6- or 12-mer oligosaccharide substrates may not completely reflect the specificity toward the full-length polysaccharide substrates (60- to 200-mers in length) isolated from cells and tissues.
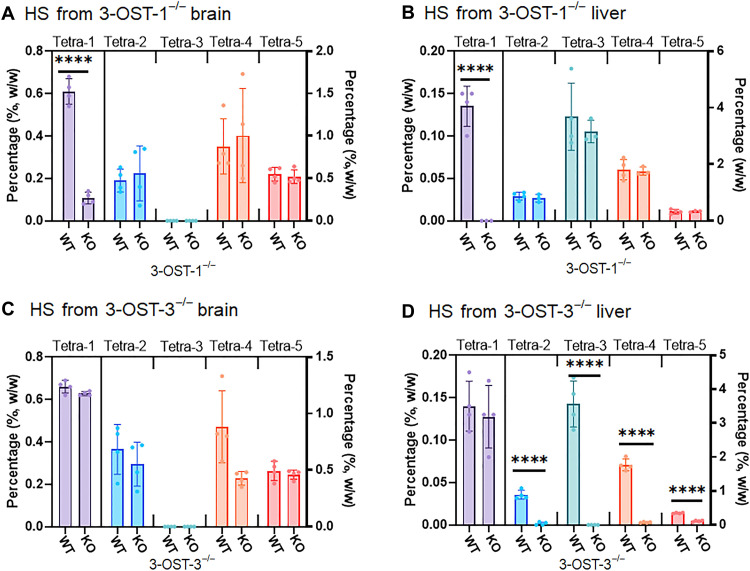
To investigate in vivo 3-OST-1– and 3-OST-3–modified HS structures, we compared HS from animal tissues using wild-type, 3-OST-1−/− mice (22), and 3-OST-3−/− mice. Because 3-OST-3 exists in both 3-OST-3A and 3-OST-3B, we chose to use Hs3st3a/Hs3st3b double-knockout mouse in our study, referred to as 3-OST-3−/−. Compared to the mass percentages of 3-O-sulfated domains in the brain HS from wild-type and knockout mice, there was a significant decrease in the level of Tetra-1 from 3-OST-1−/− mice, but not from 3-OST-3−/− mice (Fig. 5, A and B), suggesting that 3-OST-1 is the key isoform to synthesize Tetra-1 domain in brain HS. Furthermore, we did not find any significant difference in the levels of 3-O-sulfated HS from the brain comparing wild-type and 3-OST-3−/− mice (Fig. 5B). Such finding is not unanticipated, as neither 3-OST-3A nor 3-OST-3B is expressed in the wild-type brain (16). The Tetra-2, Tetra-4, and Tetra-5 in the brain HS are likely the products of the isoforms of 3-OST-2, 3-OST-4, or 3-OST-5. We also analyzed the HS from liver from different mice (Fig. 5, C and D). The level of Tetra-1 from the liver in 3-OST-1−/− mice was also significantly decreased, but not in HS from 3-OST-3−/− mice (Fig. 5, C and D). The data demonstrate that 3-OST-1 is also responsible for the biosynthesis of the Tetra-1 domain in liver HS. The levels of Tetra-2 to Tetra-5 were all decreased in the liver in 3-OST-3−/− mice, suggesting that 3-OST-3 enzymes are responsible for generating various 3-O-sulfated HS, with the exception of Tetra-1. It is of interest to note that we discovered Tetra-3 in the liver, but not in the brain from wild-type mice, demonstrating that the 3-O-sulfated HS saccharide sequence has unique tissue distribution. Together, our data suggest that the elevated level of Tetra-1 found in the brain HS is attributed to the up-regulation of the enzymatic activity of 3-OST-1 in patients with AD.
Fig. 5. Levels of 3-O-sulfated tetrasaccharides in HS from wild-type, 3-OST-1−/−, and 3-OST-3−/− mice.
(A) shows the composition of 3-O-sulfated tetrasaccharides in brain HS from 3-OST-1−/− mice. WT, wild type; KO, knockout. (B) shows the composition of 3-O-sulfated tetrasaccharides in liver HS from 3-OST-1−/− mice. (C) shows the composition of 3-O-sulfated tetrasaccharides in brain HS from 3-OST-3−/− mice. (D) shows the composition of 3-O-sulfated tetrasaccharides in liver HS from 3-OST-3−/− mice. Data are presented as average (n = 3 or 4) ± SD. The P value was determined by two-tailed unpaired t test, ****P < 0.0001. The complete dataset for the analysis is shown in tables S14 and S15.
3-O-sulfated 14-mer inhibits cellular internalization of tau
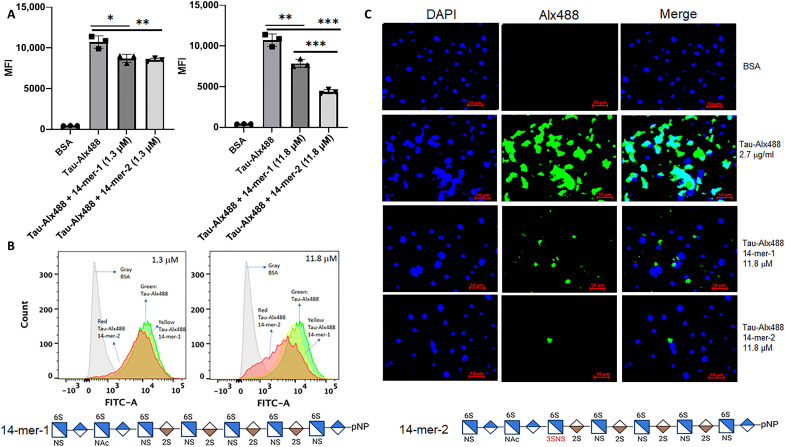
One pathological impact from the elevated level of the 3-O-sulfated HS in AD is that the specific HS epitopes enhances the cellular internalization of tau to enhance cell-to-cell spread of tau pathology (7, 8). To test this hypothesis, we determined whether exogenous 3-O-sulfated HS blocks the cellular internalization of tau. To this end, we synthesized two tetradecasaccharides (14-mers) to investigate the contribution of 3-O-sulfated HS to the inhibition of tau internalization in a mouse lung endothelial cell line that expresses a high level of 3-OST-1 (23). The two 14-mers are (i) 14-mer-1 without the Tetra-1 domain and (ii) 14-mer-2 with the domain (figs. S8 and S9). The 14-mer-2 mimics the structure of the 3-O-sulfated HS in AD brain as Tetra-1 domain was released after the heparin lyases degradation. In contrast, the Tetra-1 domain was absent in 14-mer-1 after the digestion (fig. S10). In support of our hypothesis, the 14-mer-2, at 11.8 μM, displayed about twofold stronger potency, inhibiting the internalization of tau to MLE (mouse lung epithelial) cells (Fig. 6A). The binding affinities of 14-mer-2 to tau were measured to be 14.4 nM by surface plasmon resonance (SPR), whereas the binding affinity for 14-mer-1 was determined to be 137 nM, nearly 10-fold weaker than 14-mer-2 (fig. S12). Higher binding affinity from 14-mer-2 corroborates the observation that it displays stronger inhibition to inhibit tau internalization to MLE cells. Furthermore, we demonstrate that the HS from MLE cells contains the domain of Tetra-1 (table S16). Our data suggest that HS carrying the Tetra-1 domain competes binding to the endogenous cell surface and therefore likely displays stronger binding to tau and is important for tau internalization.
Fig. 6. The inhibition of the cellular internalization of tau by 14-mers.
(A) shows the internalization of Alexa Fluor 488–labeled tau (Tau-Alx488) to the cells in the absence or the presence of 14-mer-1 (1.3 or 11.8 μM) and 14-mer-2 (1.3 or 11.8 μM). “MFI” represents mean fluorescence intensity measured in flow cytometry analysis. (B) shows plots from flow cytometry analysis after the cells incubated with bovine serum albumin (BSA; gray line), with Tau-Alx488 (green line), Tau-Alx488 and 14-mer-1 (1.3 or 11.8 μM; yellow line), and Tau-Alx488 and 14-mer-2 (1.3 or 11.8 μM; red line). FITC-A, fluorescein isothiocyanate A. (C) shows the microscopic images of the cells after incubation with BSA or Tau-Alx488 with or without 14-mers (11.8 μM). Incubation with 14-mer-2 reduced the internalization of tau to a greater extent than 14-mer-1. A wild-type mouse lung endothelial cell line that expresses 3-OST-1 enzyme was used in the experiment (23). The cells in 12-well plates were incubated with Tau-Alx488 [2.7 μg/ml (56 nM), 0.3 ml per well] at 37°C for 3 hours. Saccharide structures for 14-mer-1 and 14-mer-2 are shown at the bottom of the figure. DAPI, 4′,6-diamidino-2-phenylindole. The inhibition plot (A) was calculated using MFI of internalized Tau-Alx488 in the cells. The fluorescence intensity from the images (C) appears that the 14-mer-1 and 14-mer-2 were more inhibitory than what was observed in the flow cytometry analysis (A) and (B) due to the image signal includes both the cell surface bound and the internalized Tau-Alx488, and both assays gave the same conclusion. Two controls were carried out for the tau internalization experiment: (i) The treatment of the cells with heparin lyases reduced the internalization of tau, and (ii) incubation with heparin reduced the internalization of tau (fig. S11). *P < 0.05, **P < 0.01, ***P < 0.001.
We intended to measure the mRNA level of 3-OST-1 from the brain samples using reverse transcription polymerase chain reaction (RT-PCR) analysis but did not observe a significant increase in the mRNA of 3-OST-1 in the AD group compared to the controls (fig. S13). However, considerable scatter was observed in all of the experimental groups, likely due at least in part to the inability to control conditions in harvesting human tissues and the effects of postmortem variables on the quality of RNA quality isolated from human tissues (24, 25). Thus, we did not feel that we could draw any reliable conclusions from these data regarding the expression of 3-OST-1 in AD compared to control. Our study further demonstrates the importance of directly analyzing 3-O-sulfated HS in addition to the expression level of 3-OST-1 mRNA.
DISCUSSION
Genetic factors contribute substantially to the pathological process of AD as demonstrated by the fact that the genetic heritability in AD is in the range of between 60 and 80% (10). Delineating the connection between genetic variants in AD and structural changes in HS glycans provides insights into the disease mechanism as well as searching for potential new therapeutic targets. As a risk locus identified from GWAS analysis, HS3ST1 gene displays variants among patients with AD; however, whether the variants alter the level of 3-OST-1 enzyme activity to change the structure of HS remains unknown (11). Here, we provide evidence to link a specific 3-O-sulfated HS structural domain in patients with AD to 3-OST-1 enzyme encoded by HS3ST1 gene. We show that in vivo deletion of Hs3st1 in mice abolishes the synthesis of the specific 3-O-sulfate HS tetrasaccharide in tissues. In a cell-based assay, we demonstrate that a synthetic 3-O-sulfated 14-mer displayed stronger inhibition to the cellular internalization of tau than that for a 14-mer without 3-O-sulfation (Fig. 6). Our data support the conclusion that tau uses 3-O-sulfated HS on the surface to enter cells, suggesting that overexpression of 3-O-sulfated HS may lead to enhancement of tau internalization (Fig. 6). To further support this claim, Ferreira and colleagues (26) recently demonstrated that the up- or down-regulated expression of the 3-OST-1 in HCT-116 cells (a human colorectal carcinoma cell line) substantially affects tau uptake. Our findings offer a causal connection between variants in the genomic region of HS3ST1 gene and structural changes in HS in patients with AD, demonstrating that the specific 3-O-sulfated HS is a glycan risk marker for AD.
We have developed a protocol using 13C-labeled calibrants coupled with LC-MS/MS and carried out an extensive compositional analysis of HS. Our method covers only part of 3-O-sulfated sequences present in HS from biological sources (14). At the current time, it is unknown how many different 3-O-sulfated tetrasaccharide sequences are present. Our rationale to start with five 3-O-sulfated tetrasaccharides is based on the fact that (i) the calibrants can be used to distinguish the products between 3-OST-1–modified HS and other 3-OST isoform-modified HS and (ii) these tetrasaccharides can be resolved with the current LC conditions. We noticed that three other 3-O-sulfated tetrasaccharides were reported from heparin (27). The synthesis of additional 13C-labeled calibrants to cover these three reported 3-O-sulfated tetrasaccharides will be the subject for future study.
The involvement of the13C-labeled calibrants as internal reference standards in the analysis is crucial to allow quantifying HS from AD brains with adequate sensitivity. The inclusion of 13C-labeled calibrants also confers “self-correction” capability to address a question associated with the analysis of HS: For example, if the results are negative, then how do we know whether it is due to the sensitivity, not due to the contaminants that suppress the ionization of the analytes to reduce signals? In our experimental design, failed analyses should give very low signals from the 13C-labeled calibrants. In this case, purification steps can be added to remove contaminants as we reported previously (19). Such capability is essential for the analysis of the HS subpopulation that is present in low abundance, i.e., 3-O-sulfated HS. During the current study, the 13C-labeled calibrants showed comparable signal strengths in all samples, and thereby no additional purification steps were added to individual analysis.
Our findings demonstrate that a specific 3-O-sulfated HS domain that we refer to as Tetra-1 is increased in patients with AD. The increase of the 3-O-sulfated HS in patients with AD is the result of two combined factors: the elevation of total HS and the percentage of the 3-O-sulfated domain in HS. Notably, the increase in Tetra-1 domain alone does not account for all elevated HS. HS contains other sulfation domains, including N-sulfation, 6-O-sulfation, and 2-O-sulfation. To synthesize these structural domains involves N-deacetylase/NST, 6-OST, and 2-OST, respectively, although none of these enzymes has been identified as a risk locus for AD from the available GWAS data (10). Compositional analysis revealed a statistically significant increase of ΔUA-GlcNS (ΔIVS) disaccharide (from 10.75 ± 4.58 to 34.5 ± 16.99 ng/mg; table S4 and fig. S7) representing a building block from the N-sulfated domains and an increase of ΔUA-GlcNAc6S (ΔIIA) disaccharide (from 10.88 ± 3.49 to 30.2 ± 7.64 ng/mg; table S6 and fig. S7) representing a building block from the 6-O-sulfated domain. The elevations of ΔUA-GlcNS and ΔUA-GlcNAc6S in HS may not require substantial overexpression of both N-deacetylase/NST and 6-OST, as both enzymes are constitutively expressed in most cell types. In contrast, the expression level of 3-OST-1 in the healthy human brain is low (11). Because N-sulfation and 6-O-sulfation are the intermediates for the biosynthesis of Tetra-1 domain (28), elevated N-sulfation and 6-O-sulfation may be contributing factors for up-regulating the biosynthesis of 3-O-sulfated HS. The presence of 6-O-sulfation and N-sulfation, but not 2-O-sulfation, has been shown to be required for the binding to tau and its cellular uptake (29, 30). An increase in the amount of ΔUA2S-GlcNAc (ΔIIIA) was also observed in AD (2.12 ± 0.67 ng/mg) and preclinical AD (0.93 ± 0.34 ng/mg) groups from the control group (0.41 ± 0.22 ng/mg) (table S7 and fig. S7). The disaccharide is biosynthesized by 2-OST, and the amount is low compared with ΔIVS and ΔIIA.
The results from the NMR study of the interaction between 3-O-sulfated HS and tau revealed that the sugar mainly interacts with the proline-rich region 2, a repeat domain 2 in tau protein (8). These regions are known to contribute to tau aggregation and microtubule binding as well as serve as hotspots for phosphorylation. In addition to tau, the investigation on whether pathologic effect of 3-O-sulfated HS involves the binding to apolipoprotein E is underway.
Our conclusion is that 3-OST-1 is the primary isoform that carries out the synthesis of Tetra-1 domain based on the structural analysis of in vitro 3-OST-1–modified HS and the HS from 3-OST-1 knockout mice (Figs. 4 and 5). Other isoforms of 3-OST have been implicated in AD in a previous study. The mRNA levels of 3-OST-2 and 3-OST-4 were reportedly elevated in hippocampus of AD, the area relevant to AD pathology (7). Although the 3-O-sulfated HS structures from AD hippocampus have not been reported, we found that 3-OST-2 and 3-OST-4 likely synthesize HS similar to those made by 3-OST-3, not by 3-OST-1. Both 3-OST-2 and 3-OST-4 synthesize the 3-O-sulfated HS carrying the Tetra-2 and Tetra-4 domains. The reason that we were unable to detect an increase of 3-OST-2– and 3-OST-4–modified products in AD brains could be multifactorial. One possibility is that we analyzed the HS from frontal cortex, not from hippocampus, where 3-OST-2 and/or 3-OST-4 expression are up-regulated (31). HS3ST1 is reportedly overexpressed in amygdala, claustrum (31), and cerebellar cortex in patients with AD (11). Further analysis of the distribution of 3-O-sulfated HS from different areas of AD brains will be of interest and the subject for a subsequent study. The dynamic changes of Hs3st1 mRNA expression in AD have been reported by Pérez-López et al. (31). Hs3st1 mRNA expression was down-regulated at the mild and moderate AD stages and increased in the severe stage. It is known that tau cell-cell spreading and cellular internalization are found in many parts of brain, including the amygdala, claustrum, and cerebellar and frontal cortex in the late-stage AD (32, 33). Elevated expression of the 3-O-sulfated HS synthesized by 3-OST-1 in these areas may facilitate the tau spreading and internalization, exacerbating the pathogenic process. Although we proved that 3-OST-1 plays an essential role in controlling the synthesis in mice, cautions should be applied to extrapolate the data from mice to humans. Nevertheless, a substantial similarity to control the biosynthesis of HS between humans and mice exists. A recent study shows that 3-OST-1 is responsible for Tetra-1 expression in HCT 116 cells, a human colon cell line (26), supporting the assertion that both human and mouse 3-OST-1 is responsible for the synthesis of Tetra-1.
The structural changes in HS among different neurodegenerative diseases are not uniform, which is reflected in the total amount of HS and the level of 3-O-sulfated HS. In addition to Tetra-1, we also observed a significant increase in the level of Tetra-2 in both AD and preclinical AD groups as well as Tetra-4 in AD group, but the extent of the increase is smaller (Fig. 3). The increases in Tetra-2 and Tetra-4 can be attributed to the actions of 3-OST-2, 3-OST-4, and 3-OST-5. An increase in the total amount of HS was observed among preclinical AD and AD groups, but not from FTLD_tau and LBD groups, suggesting that 3-O-sulfated HS has distinct roles in contributing to the neuropathology under different disease conditions (fig. S6).
Our findings provide a body of evidence to justify further in-depth studies investigating the contribution of HS to the progression of AD and for the development of therapeutic as well as biomarkers for AD. The 3-O-sulfated HS oligosaccharides could serve as decoy receptors to reduce the cellular internalization and cell-cell spreading of tau (8). Although demonstrated in a cell-based experiment, the delivery of highly sulfated oligosaccharides to neurons perhaps requires penetrating blood-brain barrier, which can be challenging as a therapeutic approach. An alternative approach is to design small-molecule inhibitors that can penetrate the barrier to inhibit the activity of 3-OST-1. A small-molecule scaffold has been reported to inhibit the activity of 3-OST-1, which potentially blocks the synthesis of Tetra-1 domain (34). Another aspect of our findings is to exploit the possibility of using Tetra-1 as a diagnostic marker for AD, since a significant elevation of Tetra-1 was detected in the preclinical AD group. To develop the diagnostic application, our future goal will be to detect Tetra-1 domain from plasma or urine samples of patients with AD, where the samples can be collected from patients noninvasively or with minimum invasion.
In summary, we report an LC-MS/MS method to profile the structural composition of HS from patients with AD and other neurodegenerative diseases. Our data have successfully built the connection between the elevated level of a specific 3-O-sulfated HS domain and HS3ST1 gene. Our findings provide a new molecular target to investigate the mechanism of the progression of AD.
MATERIALS AND METHODS
Experimental design
This study was aimed to establish the connection between a specific 3-O-sulfated tetrasaccharide domain and genetic risk gene HS3ST1. The HS isolated from human brain tissues with different neurodegenerative diseases were analyzed. To develop a method for the analysis of 3-O-sulfated HS, 13C-labeled 3-O-sulfated oligosaccharide calibrants were synthesized using the chemoenzymatic approach. The identification and quantification of 3-O-sulfated HS domains were carried out using LC-MS/MS technique with 13C-labeled internal standards. We used this method to analyze a total of 44 human brain samples. A specific 3-O-sulfated tetrasaccharide domain referred as to Tetra-1 is substantially increased in AD group and pre-AD groups. The HS3ST1 gene encodes 3-OST-1 enzyme. To correlate the activity of 3-OST-1 with the elevated level of Tetra-1 domain, we conducted the analysis of HS modified by HS3ST isoforms in vitro. The structures of HS from 3-OST-1 and 3-OST-3 knockout mice were also analyzed to confirm the role of 3-OST-1 in synthesizing the Tetra-1 domain in vivo. The function of 3-O-sulfated HS on the tau cellular internalization was investigated using the mouse lung endothelial cell line using two synthetic 14-mers.
A key technology in this study is to develop a method to analyze 3-O-sulfated HS. Unlike the disaccharide compositional analysis, our method is designed to analyze tetrasaccharides from HS after being digested with heparin lyases. The glycosidic linkages that are adjacent to the nonreducing end of a 3-O-sulfated glucosamine residue in HS are known to be resistant to the digestion. Consequently, the HS polysaccharides are degraded to 3-O-sulfated tetrasaccharides after heparin lyases digestion rather than disaccharides (35–37). Some 3-O-sulfated HS domains are reportedly depolymerized into disaccharides (38, 39); however, the actual efficiency for cleaving a 3-O-sulfated domain in context of HS polysaccharide into the disaccharides is unknown. In the current study, we only used two heparin lyases, I and II, to depolymerize 3-O-sulfated HS to minimize the formation of 3-O-sulfated disaccharides. Notably, the use of heparin two lyases effectively completely depolymerizes the non–3-O-sulfated domains in HS into disaccharides.
Materials
2-Aminoacridone (AMAC) and sodium cyanoborohydride were obtained from Sigma-Aldrich (St. Louis, MO, USA). Four 13C-labeled 8-mer calibrants and one 13C-labeled 10-mer calibrant were synthesized by chemoenzymatic synthesis method (19). Recombinant heparan lyase I and II were expressed in Escherichia coli and purified by a Ni-agarose column. Diethylaminoethyl (DEAE) Sepharose Fast Flow resin was purchased from GE Healthcare (Chicago, IL, USA). All reagents and chemicals were HPLC grade or LC-MS grade.
Chemoenzymatic synthesis of 13C-labeled 8- and 10-mer calibrants
The use of the 13C-labeled 8- and 10-mer calibrants was the crucial innovation in this study. The calibrants were degraded into five different 13C-labeled 3-O-sulfated tetrasaccharides, including Tetra-1 to Tetra-5. These tetrasaccharides served as tracers to follow the target analytes during LC analysis. Identification of the analytes from LC allows us to perform the multiple reaction monitoring (MRM) analysis to gain sensitivity. Furthermore, 8- and 10-mer calibrants are chemically stable, while 3-O-sulfated tetrasaccharides are labile due to the presence of a 3-O-sulfated glucosamine residue at the reducing end (36, 40).
The synthesis of calibrants 8-mer-1 to 8-mer-3 has been reported previously (39). The synthesis of calibrants 8-mer-4 and 10-mer-5 started from the 6-mer (GlcNS-GlcA-GlcNS-IdoA2S-GlcNS-GlcA-pNP). The 13C-labeled GlcA was inserted in the designed position of oligosaccharides. After heparin lyases digestion, the 13C-labeled 3-O-sulfated tetrasaccharide targets carrying 13C-labeled GlcA or IdoA (IdoA2S) were obtained.
The elongation and sulfation steps were involved in the synthesis of calibrant 8-mer-4 and 10-mer-5. The elongation from GlcNS-GlcA-GlcNS-IdoA2S-GlcNS-GlcA-pNP (6-mers) to [13C]GlcA-GlcNS-GlcA-GlcNS-IdoA2S-GlcNS-GlcA-pNP (7-mers) was implemented by incubating 6-mers (1.2 mg/ml), 0.2 mM UDP-[13C]GlcA, and PmHS2 (heparosan synthase from Pasteurella multocida; 290 μg/ml) in a buffer containing 25 mM sodium acetate (NaOAc) (pH 5.0) and 15 mM MnCl2 in a total volume of 100 ml at 37°C overnight (where [13C]GlcA indicates universally 13C-labeled GlcA residue). In the elongation of 7-to 8-mers (GlcNTFA-[13C]GlcA-GlcNS-GlcA-GlcNS-IdoA2S-GlcNS-GlcA-pNP), the substrate (0.3 mg/ml) was incubated with 0.2 mM UDP-GlcNTFA (N-trifluoroacetylated glucosamine), PmHS2 (290 μg/ml), 25 mM Mops (pH 6.8), and 15 mM MnCl2 in a total volume of 400 ml at 37°C overnight. The de–N-trifluoroacetylation of 8-mers was implemented by suspending the oligosaccharide in 0.1 M LiOH and incubation on ice for 30 min. The pH was then adjusted to neutral using HCl. The N-sulfation of 8-mers (0.2 mg/ml) was followed by incubating with NST (30 μg/ml), 50 mM Mops (pH 6.8), and 0.2 mM PAPS (3′-phosphoadenosine 5′-phosphosulfate) in a total volume of 500 ml at 37°C overnight. Product formation was monitored by strong anion exchange chromatography on a ProPac PA1 column (9 by 250 mm; Thermo Fisher Scientific) by measuring the absorbance at 310 and 260 nm. The purification was performed using Q Sepharose (GE Healthcare). The epimerization and 2-O-sulfation was performed in the solution containing start material (0.3 mg/ml), C5-epi (3 μg/ml), 2-OST (6.5 μg/ml), 0.2 mM PAPS, and 50 mM Mops (pH 6.8) and 5 mM MnCl2 in the total volume of 400 ml. The mixture was incubated at 37°C overnight. For the 8-mer-4, the 3-O-sulfation was introduced by incubating the 8-mers (0.25 mg/ml) with 3-OST-3 (0.11 mg/ml), 50 mM Mops (pH 7.0), and 0.2 mM PAPS in a total volume of 400 ml at 37°C overnight. The 10-mer-5 calibrant was obtained by two additional elongations with UDP-GlcA and UDP-GlcNAc with 8-mer-4 as start material. From 8- to 9-mers, substrate (0.1 mg/ml) was incubated with 0.1 mM UDP-[13C]GlcA, PmHS2 (290 μg/ml), 25 mM NaOAc (pH 5.0), and 15 mM MnCl2 in a total volume of 400 ml at 37°C overnight. The 9-mer (0.1 mg/ml) was subjected to epimerization and 2-O-sulfation by incubation with C5-epi (3 μg/ml), 2-OST (6.5 μg/ml), and 0.1 mM PAPS in 50 mM Mops (pH 6.8) and 5 mM MnCl2 in a total volume of 400 ml at 37°C overnight. The additional GlcNAc residue was introduced by incubating start material (0.095 mg/ml), 0.05 mM UDP-GlcNAc, and PmHS2 (290 μg/ml) in a buffer containing 25 mM Mops (pH 6.8) and 15 mM MnCl2 at 37°C overnight to obtain the 10-mer-5 calibrant. The chemoenzymatic synthesis involves the elongation, epimerization, and sulfation steps. The recovery yield of each step was in the range of 73 to 92%.
Chemoenzymatic synthesis of 14-mer-1 and 14-mer-2
The synthesis of 14-mer-1 and 14-mer-2 was started from 11-mer with a structure of GlcA-GlcNS-GlcA-GlcNS-IdoA2S-GlcNS-IdoA2S-GlcNS-IdoA2S-GlcNS-GlcA-pNP. In the elongation of 11- to 12-mers with UDP-GlcNAc, the substrate (1 mg/ml) was incubated with 0.6 mM UDP-GlcNAc, PmHS2 (290 μg/ml), 25 mM Mops (pH 6.8), and 15 mM MnCl2 in a total volume of 50 ml at 37°C overnight. The elongation from 12- to 13-mers was performed by incubating start material (0.25 mg/ml), 0.3 mM UDP-GlcA, and PmHS2 (290 μg/ml) in 25 mM NaOAc (pH 5.0) and 15 mM MnCl2 in a total volume of 200 ml at 37°C overnight. The 13-mer (0.2 mg/ml) was subjected to epimerization and 2-O-sulfation by incubating C5-epi (3 μg/ml), 2-OST (6.5 μg/ml), and 0.2 mM PAPS in 50 mM Mops (pH 6.8) and 5 mM MnCl2 in a total volume of 200 ml at 37°C overnight. The elongation step was finished by introducing the UDP-GlcNTFA in the solution containing 0.2 mM UDP-GlcNTFA and PmHS2 (290 μg/ml) in 25 mM Mops and 15 mM MnCl2 at 37°C overnight. The de–N-trifluoroacetylation of 14-mers was implemented by suspending the oligosaccharide (0.19 mg/ml) in 0.1 M LiOH and incubation on ice for 30 min. The pH was then adjusted to neutral using HCl. The N-sulfation of 14-mers was followed by incubating with NST (30 μg/ml) in 50 mM Mops (pH 6.8) and 0.2 mM PAPS in a total volume of 200 ml at 37°C overnight. The 6-O-sulfation of 14-mers was carried out by incubating 14-mers (0.23 mg/ml) with 6-OST-3 (0.9 mg/ml) and 6 mM PAPS in 100 mM tris (pH 8.5) in a total volume of 100 ml at 37°C overnight to get the 14-mer-1. The 14-mer-2 was synthesized by incubating the 14-mer-1 (0.25 mg/ml) with 3-OST-1 (0.15 mg/ml) and 0.7 mM PAPS in 50 mM Mops and 5 mM MgCl2 in a total volume of 40 ml at 37°C overnight. Product formation was monitored by strong anion exchange chromatography on a ProPac PA1 column (9 by 250 mm; Thermo Fisher Scientific) by measuring the absorbance at 310 and 260 nm. The purification was performed using Q Sepharose (GE Healthcare). The recovery yield of each step was in the range of 67 to 91%.
The pNP (p-nitrophenyl)-tagged 14-mers were converted to biotinylated counterparts. 14-mers with a pNP tag (5 to 10 mg) and 0.5 mg of palladium on carbon (Pd/C) were dissolved in 20 mM NaOAc (pH 5.0) in a total volume of 4 ml. Reaction mixture was vacuumed and refilled with H2 three times. The reaction was then incubated at room temperature for 4 hours. After that, it was filtered to remove charcoal. The filtered solution was adjusted to pH 8.5 using 500 mM Na2HPO4. Succinimidyl 6-azidohexanoate (20 molar equivalents of starting oligosaccharides) was added and incubated at 37°C overnight. Reaction was purified by DEAE HPLC column to generate azido-tagged oligosaccharides. Phosphate-buffered saline (PBS; pH 7.4) buffer was bubbled with N2 for 5 min to prepare the sample solution of 0.1 M CuSO4, 0.1 M tris(3-hydroxypropyl-triazolylmethyl)amine (THPTA; Sigma-Aldrich), 0.15 M sodium ascorbate, 0.01 M azido-tagged oligosaccharides, and 0.02 M biotin-PEG4-alkyne (Sigma-Aldrich). The mixture of 400 μl of THPTA and 80 μl of CuSO4 was vortexed, and then 160 μl of sodium ascorbate, 200 μl of azido-tagged oligomers, and 200 μl of biotin-PEG4-alkyne was added and bubbled with N2 for 2 min and then incubated at 37°C overnight. The reaction was purified by DEAE HPLC column to generate biotinylated products. The reactions were monitored using HPLC and MS. The weight-average molecular weight (Mw) of biotinylated 14-mer-1 was determined to be 4468.0, which is very close to the calculated Mw of 4469.9. The Mw of biotinylated 14-mer-2 was determined to be 4547.4, which is very close to the calculated Mw of 4549.9.
Preparation of 3-O-sulfated HS using different 3-OST isoforms
The HS polysaccharides modified by 3-OST isoforms were performed with rHS01 (HS isolated from wild type Chinese hamster ovary cells; TEGA Therapeutics) as a substrate. The modification reaction constituted 10 μg of rHS01, 0.4 mM PAPs, 50 mM Mops, and 5 mM MgCl2 and a series of concentrations of 3-OST isoforms and incubated at 37°C for 5 hours. After incubation, the reaction was boiled at 100°C for 10 min and centrifuged to get the supernatant. The recovered solution was freeze-dried and reconstituted with 100 μl of heparin lyases buffer containing enzymatic buffer [100 mM sodium acetate/2 mM calcium acetate buffer (pH 7.0) containing 0.1 g/liter bovine serum albumin (BSA)] and heparin lyase cocktails containing 5 mg/ml each of heparin lyase I and II. 13C-labeled five 3-O-sulfated oligosaccharides calibrants (500 ng each) were added to the digest solution. The reaction solution was incubated at 37°C overnight. After incubation, the reaction was boiled at 100°C for 10 min and centrifuged to get the supernatant. The recovered solution was freeze-dried and stored at −20°C for AMAC labeling and LC-MS/MS analysis.
Labeling disaccharides and tetrasaccharides with AMAC
The AMAC derivatization was performed with 5 μl of 0.1 M AMAC in the solution in dimethyl sulfoxide/glacial acetic acid (17:3, v/v) and incubated at room temperature for 15 min. Then, 5 μl of 1 M aqueous sodium cyanoborohydride (freshly prepared) was added. The mixture was incubated at 45°C for an additional 2 hours and then centrifuged to obtain the supernatant for the LC-MS/MS analysis.
Patient samples from neurodegenerative diseases
Frozen postmortem frontal cortex samples were provided by the Neuropathology Core Brain Bank at the Goizueta Alzheimer’s Disease Center at Emory University (Institutional Review Board approved no. AM4_IRB00045782). Tissues were provided from 10 control participants, 9 early or preclinical AD samples, 14 end-stage patients with AD, 10 FTLD_tau, and 10 LBD. The neuropathological diagnosis of neurodegenerative diseases was established following current diagnostic criteria. Control participants consisted of individuals with no known history of neurological disease and no obvious neurodegenerative pathology at autopsy. Detailed information for patients is shown in Table 1.
3-OST-1−/− and 3-OST-3−/− knockout mice
All mice used were housed and maintained according to guidelines approved by National Institute of Dental and Craniofacial Research (NIDCR) and National Institutes of Health (NIH). 3-OST-1−/− mice were provided by J. Esko (41) and genotyped as described previously (22). The 3-OST-3−/− mice were generated by the Gene Transfer Core at NIDCR, NIH. The Hs3st3b1−/− mice used were as described previously (42). A zinc finger nuclease (ZFN) mRNA construct targeted to exon 1 of Hs3st3a1 gene (NM_178870.5 AGCTACCACAAGGGCCTCgcctggTACCGGTGAGCGCGGGGA) was designed, assembled, and validated by Sigma-Aldrich. The ZFN mRNA were microinjected into single-cell Hs3st3b1+/− mouse embryos, which were then transferred to the oviduct of day 0.5 pseudo-pregnant females. Offspring were screened for ZFN-induced mutations at the target site of the Hs3st3a1 gene. Hs3st3 mice were confirmed to have cosegregation of both Hs3st3a and Hs3st3b deletion on the same allele. Brain and liver tissues were harvested from 8- to 12-week-old male mice, weighed, and frozen for HS analysis. Samples were blinded before HS analysis.
The expression of 3-OST isoforms
The expression of (His)6-tagged 3-OST-1 and 3-OST-3 was performed in E. coli using BL21 cells (43). The transformed cells were grown in LB medium containing kanamycin (50 μg/liter) for 3-OST-1 and 3-OST-3 and incubated at 37°C until the optical density at 600 nm (OD600nm) reached 0.6 to 0.8. 0.2 mM isopropyl-β-d-thiogalactopyranoside (IPTG) was added to induce the expression of 3-OST-1 and 3-OST-3. The bacteria culture was kept shaking at 22°C overnight. The bacterial cells were harvested by spinning at 8000g for 15 min. The cells were resuspended in 25 ml of buffer solution containing 25 mM tris (pH 7.5), 30 mM imidazole, and 500 mM NaCl. The suspension was sonicated and centrifuged at 15,000g for 30 min. The supernatant was filtered through a 0.45-μm membrane before purification. Nickel-agarose (GE Healthcare) column was used to purify the proteins. Buffer A contained 25 mM tris (pH 7.5), 30 mM imidazole, and 500 mM NaCl; buffer B contained 25 mM tris (pH 7.5), 300 mM imidazole, and 500 mM NaCl. After loading the medium, buffer A was used to wash the column until ultraviolet absorbance at 280 nm reached baseline at a flow rate of 2 ml/min. Buffer B was then applied to elute the protein. The expression of (His)6-tagged 3-OST-5 was performed in the CHAP-OriB cell line (44). The cells were grown at 37°C in terrific broth containing ampicillin (75 μg/ml), kanamycin (37.5 μg/ml), chloramphenicol (26.3 μg/ml), and tetracycline (9.4 μg/ml) to an OD600nm of 0.8 to 0.9. Chaperone GroEL/GroES expression was induced by addition of (l)-arabinose to a final concentration of 1 mg/ml, and the temperature was decreased to 18°C for 30 min. The expression was then induced by addition of IPTG to a final concentration of 0.4 mM IPTG and allowed to proceed overnight at 18°C, with shaking at 275 revolutions per minute (rpm). The cells were lysed by sonication in buffer A containing 25 mM tris (pH 8), 750 mM NaCl, and 10 mM imidazole (pH 8) supplemented with 1 mM phenylmethylsulfonyl fluoride and complete EDTA-free protease inhibitor tablets (one tablet per 40-ml volume of lysis buffer), and the lysate was centrifuged at 18,000 rpm at 4°C. The supernatant was loaded to Ni–nitrilotriacetic acid (QIAGEN) resin at 4°C and washed thoroughly with buffer A; and protein was eluted with buffer B containing 45 mM tris (pH 8), 775 mM NaCl, and 200 mM imidazole (pH 8).
For 3-OST-2 and 3-OST-4 expression, the regions encoding the sulfotransferase domains of 3-OST-2 (Lys108-Glu367) and 3-OST-4 (Glu163-Lys456) were cloned into the pMALX(TEV) vector, generating a maltose binding protein N-terminal fusion protein. The resulting vector was transformed into the CHAP-OriB cells for expression (44). The cells were grown in LB medium supplemented with ampicillin (75 μg/ml), kanamycin (25 μg/ml), chloramphenicol (26.25 μg/ml), and tetracycline (9.4 μg/ml) to an OD600nm of ~0.8. (l)-arabinose was added to a final concentration of 1 mg/ml (to induce expression of the GroES/GroEL chaperone), and the temperature was decreased to 16°C for 35 min. Target protein induction was achieved by addition of IPTG to a final concentration of 0.4 mM, and expression was allowed to proceed overnight at 16°C. The cells were pelleted and lysed by sonication in 25 mM tris (pH 8.0) and 500 mM NaCl. The lysate was cleared by centrifugation, and the soluble proteins were bound in-batch to amylose resin. 3-OST-2 and 3-OST-4 were released from the resin by overnight cleavage with tobacco etch virus (TEV) protease in sonication buffer supplemented with 20 μM PAPS at 4°C. The cleaved proteins were further purified by size exclusion chromatography, using a Superdex 200 10/300 Increase column equilibrated with sonication buffer.
Measurement of the sulfotransferase activities of 3-OSTs
For measuring the activities of 3-OST-1 and 3-OST-5, a 6-mer oligosaccharide (0.1 mg/ml; GlcNS6S-GlcA-GlcNS6S-IdoA2S-GlcNS6S-GlcA-pNP) was incubated with either 3-OST-1 (10 μg/ml) or 3-OST-5 (10 μg/ml) in the reaction buffer containing MOPS (50 mM), MgSO4 (5 mM), MnCl2 (5 mM), and PAPS (248 μM). For measuring the activities of 3-OST-2, 3-OST-3, and 3-OST-4, a 8-mer oligosaccharide (0.1 mg/ml; GlcNS-GlcA-GlcNS-IdoA2S-GlcNS-IdoA2S-GlcNS-GlcA-pNP) was incubated with 3-OST-2 (10 μg/ml), 3-OST-3 (22 μg/ml), or 3-OST-4 (6.7 μg/ml) in the reaction buffer containing Mops (50 mM), MgSO4 (5 mM), MnCl2 (5 mM), and PAPS (248 μM). The reaction mixture was incubated at 37°C for 5 hours and then was quenched by placing on a 100°C heat block for 10 min. The mixture was then spun down at 14,000g for 10 min. The supernatant was collected and analyzed by HPLC on a ProPac PA1 anion exchange column at a flow rate of 1 ml/min buffer A (20 mM NaOAc, pH 5.0). A linear gradient from 35 to 100% buffer B (20 mM NaOAc and 2 M NaCl, pH 5.0) over 60 min with a flow rate of 1 ml/min was used for elution with monitoring the absorbance at 310 nm. The enzyme activity was determined on the basis of the ratio of peak areas of oligosaccharide substrate and the 3-O-sulfated oligosaccharide product. One unit of the sulfotransferase activity was defined as the amount of enzyme that transfers 1 nmol of sulfo group to the substrate under the conditions described above.
Tau internalization assay
The overexpression and purification of full-length tau protein were performed as previously described (45, 46), and the tau protein was conjugated with Alexa Fluor 488 (8). The mouse lung endothelial cell line was developed in our lab and expresses a high level of HS3ST1 (23). The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS), penicillin (100 U/ml) and streptomycin (100 μg/ml) at 37°C, and 5% CO2.
Mouse lung endothelial cells (2.5 × 105) were seeded at DMEM (600 μl per well) containing 10% FBS and penicillin (100 U/ml) and streptomycin (100 μg/ml) in a 12-well plate. After culturing overnight, the cells were washed with Dulbecco's phosphate-buffered saline (DPBS) twice and then incubated with DMEM (500 μl per well) containing BSA (1.6 μg/ml or 24 nM), Alexa Fluor 488–labeled tau (Tau-Alx488; 2.7 μg/ml, 56 nM), Tau-Alx488 (2.7 μg/ml, 56 nM) mixed with heparin (20 or 200 μg/ml, 1.67 or 16.7 μM), or Tau- Alx488 (2.7 μg/ml, 56 nM) mixed with 14-mer-1 or 14-mer-2 (5 or 40 μg/ml, 1.3 or 11.8 μM) at 37°C for 3 hours. Some of the wells were pretreated with heparin lyases I to III (0.3 or 0.4 U/0.3 ml of DMEM per well) at 37°C for 1 hour before the Tau-Alx488 incubation. The cells were then processed for image or flow cytometry analyses. For image analysis, the cells were covered with mounting medium 4b,6-diamidino-2-phenylindole (DAPI) and examined for internalized tau-Alx488 using Keyence BZ-Z800 microscope. The image data were processed using ImageJ. For flow cytometry analysis, the cells were trypsinized and resuspended in DPBS containing 2 mM EDTA, 1% BSA, and propidium iodide (PI), which stain dead cells, and then analyzed by BD FACSCanto II flow cytometer. The flow cytometry data were processed using FlowJo v10.8. GraphPad Prism 9.0 was applied for statistical analysis and graphing of the collected data.
Extraction and quantitation analysis of HS from the AD human brain tissue and mice brain and liver tissues
The HS extraction from tissues was carried out on frozen tissues with homogenization and defatting as described previously (19). Briefly, the tissue (from 800 to 1300 mg) was excised and homogenized in the PBS. Then, the homogenized tissue was subjected to defat by suspension and vortex in the chloroform/methanol mixtures [2:1, 1:1, and 1:2 (v/v)]. The defatted tissues were dried and weighed to obtain the dry weight (from 30 to 80 mg). Then, the dried tissue (20 mg) was subjected to pronase E digest [10 mg:1 g (w/w), tissue/pronase E] at 55°C for 24 hours to degrade the proteins. Twenty nanograms of recovery calibrant 13C-labeled NSK5P (N-sulfo heparosan from E. coli K5 strain) was added into the digestion solution before subjecting to DEAE column purification. The DEAE column mobile phase A consisted of 20 mM tris (pH 7.5) and 50 mM NaCl, and mobile phase B consisted of 20 mM tris (pH 7.5) and 1 M NaCl. After loading the digested solution, the column was washed with 1.5 ml of mobile phase A, followed by 1.5 ml of mobile phase B to elute the HS fraction. The YM-3KDa spin column was applied to desalt the elution, and the retentate was subjected to heparin lyases digest. Before the digestion, 13C-labeled 3-O-sulfated calibrants (500 ng each) were added to the retentate. A total of 100 μl of enzymatic buffer [100 mM sodium acetate/2 mM calcium acetate buffer (pH 7.0) containing BSA (0.1 g/liter)] and the 20 μl of enzyme cocktails containing heparin lyases I and II (5 mg/ml each) was added to degrade the retentate on the filter unit of the YM-3KDa column. The reaction solution was incubated at 4°C for 1 hour. Before recovering the disaccharides from the digest solution, a known amount 13C-labeled non–3-O-sulfated disaccharide calibrants (ΔIS = 80 ng, ΔIIS = 80 ng, ΔIIIS = 40 ng, ΔIVS = 80 ng, ΔIA = 40 ng, ΔIIA = 80 ng, ΔIIIA = 40 ng, and ΔIVA = 250 ng) were added to the digestion solution. The HS disaccharides and tetrasaccharides were recovered by centrifugation, and the filter unit was washed twice with 200 μl of deionized water. The collected filtrates were freeze-dried before the AMAC derivatization. The AMAC label and LC-MS/MS analysis of the collected disaccharides and tetrasaccharides of tissues was performed as described below. The amount of tissue HS was determined by comparing the peak area of native di/tetrasaccharide to each corresponding 13C-labeled internal standard, and the recovery yield was calculated on the basis of a comparison of the amount of 13C-labeled disaccharide (Δ[13C]UA-[13C]GlcNS from heparin lyases degraded 13C-labeled NSK5P) in the tissue samples and control, respectively.
LC-MS/MS analysis
The analysis of AMAC-labeled di/tetrasaccharides was implemented on a Vanquish Flex ultra HPLC System (Thermo Fisher Scientific) coupled with TSQ Fortis triple-quadrupole MS as the detector. The ACQUITY Glycan BEH Amide column (1.7 μm, 2.1 by 150 mm; Waters, Ireland, UK) was used to separate di/tetrasaccharides at 60°C. Mobile phase A was 50 mM ammonium formate in water (pH 4.4). Mobile phase B is acetonitrile. The elution gradient was as follows: 0 to 20 min, 83 to 60% gradient B; 20 to 25 min, 5% B; and 25 to 30 min 83% B. The flow rate was 0.3 ml/min. On-line triple quadrupole MS operating in the MRM mode was used as the detector. The electrospray ionization MS analysis was operated in the negative ion mode using the following parameters: negative ion spray voltage at 3.0 kV, sheath gas at 55 arbitrary units, aux gas at 25 arbitrary units, ion transfer tube temperature at 250°C, and vaporizer temperature at 400°C. TraceFinder software was applied for data processing.
Determination of the binding affinity between 14-mers and tau
SPR measurements were performed on a Biacore 3000 SPR (GE Healthcare, Uppsala, Sweden). The biotinylated 14-mers (14-mer-1 and 14-mer-2) were immobilized to streptavidin (SA) sensor chips (Biacore, GE Healthcare, Uppsala, Sweden) based on the manufacturer’s protocol. In brief, 20 μl of biotinylated 14-mers (0.2 mg/ml) was injected over flow cells 2 and 3 (FC2 and FC3) of the SA chip at a flow rate of 10 μl/min. The successful immobilization of biotinylated 14-mers was confirmed by the observation of 811– and 789–resonance unit increase. The control flow cell (FC1) was prepared by 1-min injection with saturated biotin. The running buffer contained 0.01 M Hepes, 0.15 M NaCl, 3 mM EDTA, and 0.05% surfactant P20 (pH 7.4).
Recombinant tau protein at concentrations of 500, 250, 125, 63, and 32 nM were injected at a flow rate of 40 μl/min. At the end of the recombinant tau injection, the same buffer flowed over the SA surface to facilitate dissociation. After a 3-min dissociation time, the SA surface was regenerated by injecting 40 μl of 2 M NaCl. The response was monitored as a function of time (sensogram) at 25°C. The sensograms of various tau concentrations were globally fitted with 1:1 Langmuir model. SPR measurements were performed on a Biacore 3000 operated using Biacore 3000 control and BIAevaluation software (version 4.0.1).
qPCR analysis of mRNA expression
Total RNA was isolated from human brain tissues with TRIzol reagent (Invitrogen; Thermo Fisher Scientific Inc.) following RNeasy Mini Kit (QIAGEN Inc.). Before RNA extraction, brain samples were homogenized using BeadBlaster homogenizer (Benchmark Scientific Inc.) with beads at 6.5 m/s, 15 s for three cycles. The quality of RNA and concentration were assessed using the NanoDrop one/one Microvolume ultraviolet-visible Spectrophotometer (Thermo Fisher Scientific Inc.). RNA concentrations ranged from 0.1 to 0.6 μg/μl, and an optical density at a wavelength of 260/280 nm with a range of 1.8 to 2.2 were chosen for this study. One microgram of total RNA was reverse-transcribed into cDNA using SuperScript II Reverse Transcriptase (Thermo Fisher Scientific Inc.) with RNaseOUT RNase inhibitors (Invitrogen). mRNA expression of 3OST-1 (sense: 5′-ACCACATGCAGAAGCACAAG-3′ and anti-sense: 5′-TCCAAAAGGTCGAGAGGTTCCT-3′) and internal control glyceraldehyde-3-phosphate dehydrogenase (GAPDH; sense: 5′-ACCACATGCAGAAGCACAAG-3′ and anti-sense: 5′-TCCAAAAGGTCGAGAGGTTCCT-3′) and cytochrome C1 (CYC1; sense: 5′-AGCCTACAAGAAAGTTTGCCTAT-3′ and anti-sense: 5′-TCTTCTTCCGGTAGTGGATCTTGGC-3′) were evaluated by quantitative PCR (qPCR) using the PowerUp SYBR Green Master Mix (Applied Biosystems). Primer pair sequences are designed on the basis of literature (47, 48) and ordered from Eton Bioscience. The amplification condition was as follows: 50° (2 min) and 95° (2 min), followed by 40 cycles at 95° (15 s), 57° (20 s), and 72°C (1 min). The melting curve was tested after the amplification, with the condition as follows: 95° (15 s), 60° (1 min), and 95°C (15 s). The experiment was done using the ViiA 7 Flex Real-Time PCR System (Thermo Fisher Scientific). The range of Ct of 3-OST-1, GAPDH, and CYC1 is 22.8 to 32.3, 16.3 to 23.9, and 17.4 to 26.9. mRNA levels of 3-OST1 were quantified with the 2−∆∆Ct method and normalized to the internal control gene GAPDH and CYC1. The results are shown relative to the expression level of the control group.
The statistics and reproducibility
For the quantification of HS from brain tissues in the different neurodegenerative diseases, two replicates of each sample were performed on LC-MS/MS analysis. For the evaluation of substrate specificity of different 3-OST isoforms using polysaccharide, three separate samples were performed on LC-MS/MS analysis. For the quantification of HS isolated from brain and liver tissues in the 3-OST-1−/− and 3-OST-3−/− mice, two replicates of each sample were performed on LC-MS/MS analysis. The P value was determined by two-tailed unpaired t test. **P < 0.01; ***P < 0.001; ****P < 0.0001. The statistical significance was defined as P < 0.05.
Acknowledgments
J.L. lab at UNC has received a gift from Glycan Therapeutics to support research in glycosciences.
Funding: This work is funded by NIH grants (HL094463, HL144970, GM142304, GM140693, AG069039, AG066511, AG074289, and CA225784) and 1Z1A-ES102645 (L.C.P.) in the Division of Intramural Research Program of the National Institute of Environmental Health Sciences, NIH. This work was also supported by Glycan innovation grants from Eshelman Institute for Innovation and by GlycoMIP, a National Science Foundation Materials Innovation Platform funded through cooperative agreement DMR-1933525. The research was supported by the Intramural Research Program of the National Institute of Dental and Craniofacial Research at NIH (1Z1ADE000707). The lab of C.W. also received grants from NIH (R21GM143507 and R01 CA206592) to develop a method for protein NMR analysis and investigate the mechanisms of hedgehog autoprocessing, respectively.
Author contributions: Conceptualization: Z.W. and J.L. Formal analysis: Z.W., X.S., and V.U.D. Funding acquisition: J.L., V.P., G.S., and C.W. Investigation: Z.W., J.L., Y.X., V.N.P., C.W., L.C.P., L.W., M.P.H., M.G., X.S., V.U.D., A.M.K., V.P., G.S., Y.L., D.M., and F.Z. Methodology: Z.W., X.S., V.N.P., and V.U.D. Supervision: J.L. Writing (original draft): Z.W. and J.L. Writing (review and editing): V.N.P., C.W., L.C.P., L.W., M.P.H., and M.G.
Competing interests: Y.X. and J.L. are founders of Glycan Therapeutics and have equity. V.P. and G.S. are employees of Glycan Therapeutics and have the equity of Glycan Therapeutics. Z.W. is presently an employee of Glycan Therapeutics and has the equity option for the company. J.L. lab at UNC has received a gift from Glycan Therapeutics to support research in glycosciences. All other authors declare that they have no competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. The genetically modified mice used in this study can be provided by M.P.H. pending scientific review and a completed material transfer agreement. Requests for the mice should be submitted to: mhoffman@dir.nidcr.nih.gov.
Supplementary Materials
This PDF file includes:
Figs. S1 to S13
Tables S1 to S16
REFERENCES AND NOTES
- 1.2020 Alzheimer’s disease facts and figures. Alzheimers Dement. 16, 391–460 (2020). [Google Scholar]
- 2.I. Grundke-Iqbal, K. Iqbal, Y. C. Tung, M. Quinlan, H. M. Wisniewski, L. I. Binder, Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. U.S.A. 83, 4913–4917 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.A. D. Snow, H. Mar, D. Nochlin, R. T. Sekiguchi, K. Kimata, Y. Koike, T. N. Wight, Early accumulation of heparan sulfate in neurons and in the beta-amyloid protein-containing lesions of Alzheimer’s disease and Down’s syndrome. Am. J. Pathol. 137, 1253–1270 (1990). [PMC free article] [PubMed] [Google Scholar]
- 4.C. C. Liu, N. Zhao, Y. Yamaguchi, J. R. Cirrito, T. Kanekiyo, D. M. Holtzman, G. Bu, Neuronal heparan sulfates prmotes amyloid pathology by modulating brain amyloid-β clearance and aggregation in Alzheimer’s disease. Sci. Tansl. Med. 8, 332ra44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.B. B. Holmes, S. DeVos, N. Kfoury, M. Li, R. Jacks, K. Yanamandra, M. O. Ouidja, F. M. Brodsky, J. Marasa, D. P. Bagchi, P. T. Kotzbauer, T. M. Miller, D. Papy-Garcia, M. I. Diamond, Heparan sulfate proteoglycans mediate internalization and propagation of specific proteopathic seeds. Proc. Natl. Acad. Sci. U.S.A. 110, E3138–E3147 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.J. N. Rauch, J. J. Chen, A. W. Sorum, G. M. Miller, T. Sharf, S. K. See, L. C. Hsieh-Wilson, M. Kampmann, K. S. Kosik, Tau internalization is regulated by 6-O sulfation on heparan sulfate proteoglycans (HSPGs). Sci. Rep. 8, 6382 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.J. E. Sepulveda-Diaz, S. M. Alavi Naini, M. B. Huynh, M. O. Ouidja, C. Yanicostas, S. Chantepie, J. Villares, F. Lamari, E. Jospin, T. H. van Kuppevelt, A. G. Mensah-Nyagan, R. Raisman-Vozari, N. Soussi-Yanicostas, D. Papy-Garcia, HS3ST2 expression is critical for the abnormal phosphorylation of tau in Alzheimer’s disease-related tau pathology. Brain 138, 1339–1354 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.J. Zhao, Y. Zhu, X. Song, Y. Xiao, G. Su, X. Liu, Z. Wang, Y. Xu, J. Liu, D. Eliezer, T. F. Ramlall, G. Lippens, J. Gibson, F. Zhang, R. J. Linhardt, L. Wang, C. Wang, 3-O-sulfation of heparan sulfate enhances tau interaction and cellular uptake. Angew. Chem. Int. Ed. 59, 1818–1827 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.M. B. Huynh, N. Rebergue, H. Merrick, W. Gomez-Henao, E. Jospin, D. S. F. Biard, D. Papy-Garcia, HS3ST2 expression induces the cell autonomous aggregation of tau. Sci. Rep. 12, 10850 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.C. Bellenguez, F. Küçükali, I. E. Jansen, L. Kleineidam, S. Moreno-Grau, N. Amin, A. C. Naj, R. Campos-Martin, B. Grenier-Boley, V. Andrade, P. A. Holmans, A. Boland, V. Damotte, S. J. van der Lee, M. R. Costa, T. Kuulasmaa, Q. Yang, I. de Rojas, J. C. Bis, A. Yaqub, I. Prokic, J. Chapuis, S. Ahmad, V. Giedraitis, D. Aarsland, P. Garcia-Gonzalez, C. Abdelnour, E. Alarcón-Martín, D. Alcolea, M. Alegret, I. Alvarez, V. Álvarez, N. J. Armstrong, A. Tsolaki, C. Antúnez, I. Appollonio, M. Arcaro, S. Archetti, A. A. Pastor, B. Arosio, L. Athanasiu, H. Bailly, N. Banaj, M. Baquero, S. Barral, A. Beiser, A. B. Pastor, J. E. Below, P. Benchek, L. Benussi, C. Berr, C. Besse, V. Bessi, G. Binetti, A. Bizarro, R. Blesa, M. Boada, E. Boerwinkle, B. Borroni, S. Boschi, P. Bossù, G. Bråthen, J. Bressler, C. Bresner, H. Brodaty, K. J. Brookes, L. I. Brusco, D. Buiza-Rueda, K. Bûrger, V. Burholt, W. S. Bush, M. Calero, L. B. Cantwell, G. Chene, J. Chung, M. L. Cuccaro, Á. Carracedo, R. Cecchetti, L. Cervera-Carles, C. Charbonnier, H. H. Chen, C. Chillotti, S. Ciccone, J. A. H. R. Claassen, C. Clark, E. Conti, A. Corma-Gómez, E. Costantini, C. Custodero, D. Daian, M. C. Dalmasso, A. Daniele, E. Dardiotis, J. F. Dartigues, P. P. de Deyn, K. de Paiva Lopes, L. D. de Witte, S. Debette, J. Deckert, T. del Ser, N. Denning, A. DeStefano, M. Dichgans, J. Diehl-Schmid, M. Diez-Fairen, P. D. Rossi, S. Djurovic, E. Duron, E. Düzel, C. Dufouil, G. Eiriksdottir, S. Engelborghs, V. Escott-Price, A. Espinosa, M. Ewers, K. M. Faber, T. Fabrizio, S. F. Nielsen, D. W. Fardo, L. Farotti, C. Fenoglio, M. Fernández-Fuertes, R. Ferrari, C. B. Ferreira, E. Ferri, B. Fin, P. Fischer, T. Fladby, K. Fließbach, B. Fongang, M. Fornage, J. Fortea, T. M. Foroud, S. Fostinelli, N. C. Fox, E. Franco-Macías, M. J. Bullido, A. Frank-García, L. Froelich, B. Fulton-Howard, D. Galimberti, J. M. García-Alberca, P. García-González, S. Garcia-Madrona, G. Garcia-Ribas, R. Ghidoni, I. Giegling, G. Giorgio, A. M. Goate, O. Goldhardt, D. Gomez-Fonseca, A. González-Pérez, C. Graff, G. Grande, E. Green, T. Grimmer, E. Grünblatt, M. Grunin, V. Gudnason, T. Guetta-Baranes, A. Haapasalo, G. Hadjigeorgiou, J. L. Haines, K. L. Hamilton-Nelson, H. Hampel, O. Hanon, J. Hardy, A. M. Hartmann, L. Hausner, J. Harwood, S. Heilmann-Heimbach, S. Helisalmi, M. T. Heneka, I. Hernández, M. J. Herrmann, P. Hoffmann, C. Holmes, H. Holstege, R. H. Vilas, M. Hulsman, J. Humphrey, G. J. Biessels, X. Jian, C. Johansson, G. R. Jun, Y. Kastumata, J. Kauwe, P. G. Kehoe, L. Kilander, A. K. Ståhlbom, M. Kivipelto, A. Koivisto, J. Kornhuber, M. H. Kosmidis, W. A. Kukull, P. P. Kuksa, B. W. Kunkle, A. B. Kuzma, C. Lage, E. J. Laukka, L. Launer, A. Lauria, C. Y. Lee, J. Lehtisalo, O. Lerch, A. Lleó, W. Longstreth Jr., O. Lopez, A. L. de Munain, S. Love, M. Löwemark, L. Luckcuck, K. L. Lunetta, Y. Ma, J. Macías, C. A. MacLeod, W. Maier, F. Mangialasche, M. Spallazzi, M. Marquié, R. Marshall, E. R. Martin, A. M. Montes, C. M. Rodríguez, C. Masullo, R. Mayeux, S. Mead, P. Mecocci, M. Medina, A. Meggy, S. Mehrabian, S. Mendoza, M. Menéndez-González, P. Mir, S. Moebus, M. Mol, L. Molina-Porcel, L. Montrreal, L. Morelli, F. Moreno, K. Morgan, T. Mosley, M. M. Nöthen, C. Muchnik, S. Mukherjee, B. Nacmias, T. Ngandu, G. Nicolas, B. G. Nordestgaard, R. Olaso, A. Orellana, M. Orsini, G. Ortega, A. Padovani, C. Paolo, G. Papenberg, L. Parnetti, F. Pasquier, P. Pastor, G. Peloso, A. Pérez-Cordón, J. Pérez-Tur, P. Pericard, O. Peters, Y. A. L. Pijnenburg, J. A. Pineda, G. Piñol-Ripoll, C. Pisanu, T. Polak, J. Popp, D. Posthuma, J. Priller, R. Puerta, O. Quenez, I. Quintela, J. Q. Thomassen, A. Rábano, I. Rainero, F. Rajabli, I. Ramakers, L. M. Real, M. J. T. Reinders, C. Reitz, D. Reyes-Dumeyer, P. Ridge, S. Riedel-Heller, P. Riederer, N. Roberto, E. Rodriguez-Rodriguez, A. Rongve, I. R. Allende, M. Rosende-Roca, J. L. Royo, E. Rubino, D. Rujescu, M. E. Sáez, P. Sakka, I. Saltvedt, Á. Sanabria, M. B. Sánchez-Arjona, F. Sanchez-Garcia, P. S. Juan, R. Sánchez-Valle, S. B. Sando, C. Sarnowski, C. L. Satizabal, M. Scamosci, N. Scarmeas, E. Scarpini, P. Scheltens, N. Scherbaum, M. Scherer, M. Schmid, A. Schneider, J. M. Schott, G. Selbæk, D. Seripa, M. Serrano, J. Sha, A. A. Shadrin, O. Skrobot, S. Slifer, G. J. L. Snijders, H. Soininen, V. Solfrizzi, A. Solomon, Y. Song, S. Sorbi, O. Sotolongo-Grau, G. Spalletta, A. Spottke, A. Squassina, E. Stordal, J. P. Tartan, L. Tárraga, N. Tesí, A. Thalamuthu, T. Thomas, G. Tosto, L. Traykov, L. Tremolizzo, A. Tybjærg-Hansen, A. Uitterlinden, A. Ullgren, I. Ulstein, S. Valero, O. Valladares, C. V. Broeckhoven, J. Vance, B. N. Vardarajan, A. van der Lugt, J. V. Dongen, J. van Rooij, J. van Swieten, R. Vandenberghe, F. Verhey, J. S. Vidal, J. Vogelgsang, M. Vyhnalek, M. Wagner, D. Wallon, L. S. Wang, R. Wang, L. Weinhold, J. Wiltfang, G. Windle, B. Woods, M. Yannakoulia, H. Zare, Y. Zhao, X. Zhang, C. Zhu, M. Zulaica; EADB, J. Laczo, V. Matoska, M. Serpente, F. Assogna, F. Piras, F. Piras, V. Ciullo, J. Shofany, C. Ferrarese, S. Andreoni, G. Sala, C. P. Zoia, M. D. Zompo, A. Benussi, P. Bastiani, M. Takalo, T. Natunen, T. Laatikainen, J. Tuomilehto, R. Antikainen, T. Strandberg, J. Lindström, M. Peltonen, R. Abraham, A. al-Chalabi, N. J. Bass, C. Brayne, K. S. Brown, J. Collinge, D. Craig, P. Deloukas, N. Fox, A. Gerrish, M. Gill, R. Gwilliam, D. Harold, P. Hollingworth, J. A. Johnston, L. Jones, B. Lawlor, G. Livingston, S. Lovestone, M. Lupton, A. Lynch, D. Mann, B. McGuinness, A. McQuillin, M. C. O’Donovan, M. J. Owen, P. Passmore, J. F. Powell, P. Proitsi, M. Rossor, C. E. Shaw, A. D. Smith, H. Gurling, S. Todd, C. Mummery, N. Ryan, G. Lacidogna, A. Adarmes-Gómez, A. Mauleón, A. Pancho, A. Gailhajenet, A. Lafuente, D. Macias-García, E. Martín, E. Pelejà, F. Carrillo, I. S. Merlín, L. Garrote-Espina, L. Vargas, M. Carrion-Claro, M. Marín, M. Labrador, M. Buendia, M. D. Alonso, M. Guitart, M. Moreno, M. Ibarria, M. Periñán, N. Aguilera, P. Gómez-Garre, P. Cañabate, R. Escuela, R. Pineda-Sánchez, R. Vigo-Ortega, S. Jesús, S. Preckler, S. Rodrigo-Herrero, S. Diego, A. Vacca, F. Roveta, N. Salvadori, E. Chipi, H. Boecker, C. Laske, R. Perneczky, C. Anastasiou, D. Janowitz, R. Malik, A. Anastasiou, K. Parveen, C. Lage, S. López-García, A. Antonell, K. Y. Mihova, D. Belezhanska, H. Weber, S. Kochen, P. Solis, N. Medel, J. Lisso, Z. Sevillano, D. G. Politis, V. Cores, C. Cuesta, C. Ortiz, J. I. Bacha, M. Rios, A. Saenz, M. S. Abalos, E. Kohler, D. L. Palacio, I. Etchepareborda, M. Kohler, G. Novack, F. A. Prestia, P. Galeano, E. M. Castaño, S. Germani, C. R. Toso, M. Rojo, C. Ingino, C. Mangone, D. C. Rubinsztein, S. Teipel, N. Fievet, V. Deramerourt, C. Forsell, H. Thonberg, M. Bjerke, E. D. Roeck, M. T. Martínez-Larrad, N. Olivar; GR@ACE, N. Aguilera, A. Cano, P. Cañabate, J. Macias, O. Maroñas, R. Nuñez-Llaves, C. Olivé, E. Pelejá; DEGESCO, A. D. Adarmes-Gómez, M. D. Alonso, G. Amer-Ferrer, M. Antequera, J. A. Burguera, F. Carrillo, M. Carrión-Claro, M. J. Casajeros, M. Martinez de Pancorbo, R. Escuela, L. Garrote-Espina, P. Gómez-Garre, S. Hevilla, S. Jesús, M. A. L. Espinosa, A. Legaz, S. López-García, D. Macias-García, S. Manzanares, M. Marín, J. Marín-Muñoz, T. Marín, B. Martínez, V. Martínez, P. Martínez-Lage Álvarez, M. M. Iriarte, M. T. Periñán-Tocino, R. Pineda-Sánchez, D. Real de Asúa, S. Rodrigo, I. Sastre, M. P. Vicente, R. Vigo-Ortega, L. Vivancos; EADI, J. Epelbaum, D. Hannequin, D. campion, V. Deramecourt, C. Tzourio, A. Brice, B. Dubois; GERAD, A. Williams, C. Thomas, C. Davies, W. Nash, K. Dowzell, A. C. Morales, M. Bernardo-Harrington, J. Turton, J. Lord, K. Brown, E. Vardy, E. Fisher, J. D. Warren, M. Rossor, N. S. Ryan, R. Guerreiro, J. Uphill, N. Bass, R. Heun, H. Kölsch, B. Schürmann, A. Lacour, C. Herold, J. A. Johnston, P. Passmore, J. Powell, Y. Patel, A. Hodges, T. Becker, D. Warden, G. Wilcock, R. Clarke, P. Deloukas, Y. Ben-Shlomo, N. M. Hooper, S. Pickering-Brown, R. Sussams, N. Warner, A. Bayer, I. Heuser, D. Drichel, N. Klopp, M. Mayhaus, M. Riemenschneider, S. Pinchler, T. Feulner, W. Gu, H. van den Bussche, M. Hüll, L. Frölich, H. E. Wichmann, K. H. Jöckel, M. O’Donovan, M. Owen; Demgene, S. Bahrami, I. Bosnes, P. Selnes, S. Bergh; FinnGen, A. Palotie, M. Daly, H. Jacob, A. Matakidou, H. Runz, S. John, R. Plenge, M. McCarthy, J. Hunkapiller, M. Ehm, D. Waterworth, C. Fox, A. Malarstig, K. Klinger, K. Call, T. Behrens, P. Loerch, T. Mäkelä, J. Kaprio, P. Virolainen, K. Pulkki, T. Kilpi, M. Perola, J. Partanen, A. Pitkäranta, R. Kaarteenaho, S. Vainio, M. Turpeinen, R. Serpi, T. Laitinen, J. Mäkelä, V. M. Kosma, U. Kujala, O. Tuovila, M. Hendolin, R. Pakkanen, J. Waring, B. Riley-Gillis, J. Liu, S. Biswas, D. Diogo, C. Marshall, X. Hu, M. Gossel, R. Graham, B. Cummings, S. Ripatti, J. Schleutker, M. Arvas, O. Carpén, R. Hinttala, J. Kettunen, A. Mannermaa, J. Laukkanen, V. Julkunen, A. Remes, R. Kälviäinen, J. Peltola, P. Tienari, J. Rinne, A. Ziemann, J. Waring, S. Esmaeeli, N. Smaoui, A. Lehtonen, S. Eaton, S. Lahdenperä, J. van Adelsberg, J. Michon, G. Kerchner, N. Bowers, E. Teng, J. Eicher, V. Mehta, P. Gormley, K. Linden, C. Whelan, F. Xu, D. Pulford, M. Färkkilä, S. Pikkarainen, A. Jussila, T. Blomster, M. Kiviniemi, M. Voutilainen, B. Georgantas, G. Heap, F. Rahimov, K. Usiskin, T. Lu, D. Oh, K. Kalpala, M. Miller, L. McCarthy, K. Eklund, A. Palomäki, P. Isomäki, L. Pirilä, O. Kaipiainen-Seppänen, J. Huhtakangas, A. Lertratanakul, M. Hochfeld, N. Bing, J. E. Gordillo, N. Mars, M. Pelkonen, P. Kauppi, H. Kankaanranta, T. Harju, D. Close, S. Greenberg, H. Chen, J. Betts, S. Ghosh, V. Salomaa, T. Niiranen, M. Juonala, K. Metsärinne, M. Kähönen, J. Junttila, M. Laakso, J. Pihlajamäki, J. Sinisalo, M. R. Taskinen, T. Tuomi, B. Challis, A. Peterson, A. Chu, J. Parkkinen, A. Muslin, H. Joensuu, T. Meretoja, L. Aaltonen, J. Mattson, A. Auranen, P. Karihtala, S. Kauppila, P. Auvinen, K. Elenius, R. Popovic, J. Schutzman, A. Loboda, A. Chhibber, H. Lehtonen, S. McDonough, M. Crohns, D. Kulkarni, K. Kaarniranta, J. A. Turunen, T. Ollila, S. Seitsonen, H. Uusitalo, V. Aaltonen, H. Uusitalo-Järvinen, M. Luodonpää, N. Hautala, S. Loomis, E. Strauss, H. Chen, A. Podgornaia, J. Hoffman, K. Tasanen, L. Huilaja, K. Hannula-Jouppi, T. Salmi, S. Peltonen, L. Koulu, I. Harvima, Y. Wu, D. Choy, P. Pussinen, A. Salminen, T. Salo, D. Rice, P. Nieminen, U. Palotie, M. Siponen, L. Suominen, P. Mäntylä, U. Gursoy, V. Anttonen, K. Sipilä, J. W. Davis, D. Quarless, S. Petrovski, E. Wigmore, C. Y. Chen, P. Bronson, E. Tsai, Y. Huang, J. Maranville, E. Shaikho, E. Mohammed, S. Wadhawan, E. Kvikstad, M. Caliskan, D. Chang, T. Bhangale, S. Pendergrass, E. Holzinger, X. Chen, Å. Hedman, K. S. King, C. Wang, E. Xu, F. Auge, C. Chatelain, D. Rajpal, D. Liu, K. Call, T. H. Xia, M. Brauer, M. Kurki, J. Karjalainen, A. Havulinna, A. Jalanko, P. Palta, P. della Briotta Parolo, W. Zhou, S. Lemmelä, M. Rivas, J. Harju, A. Lehisto, A. Ganna, V. Llorens, H. Laivuori, S. Rüeger, M. E. Niemi, T. Tukiainen, M. P. Reeve, H. Heyne, K. Palin, J. Garcia-Tabuenca, H. Siirtola, T. Kiiskinen, J. Lee, K. Tsuo, A. Elliott, K. Kristiansson, K. Hyvärinen, J. Ritari, M. Koskinen, K. Pylkäs, M. Kalaoja, M. Karjalainen, T. Mantere, E. Kangasniemi, S. Heikkinen, E. Laakkonen, C. Sipeky, S. Heron, A. Karlsson, D. Jambulingam, V. S. Rathinakannan, R. Kajanne, M. Aavikko, M. G. Jiménez, P. della Briotta Parola, A. Lehistö, M. Kanai, M. Kaunisto, E. Kilpeläinen, T. P. Sipilä, G. Brein, G. Awaisa, A. Shcherban, K. Donner, A. Loukola, P. Laiho, T. Sistonen, E. Kaiharju, M. Laukkanen, E. Järvensivu, S. Lähteenmäki, L. Männikkö, R. Wong, H. Mattsson, T. Hiekkalinna, T. Paajanen, K. Pärn, J. Gracia-Tabuenca; ADGC, E. Abner, P. M. Adams, A. Aguirre, M. S. Albert, R. L. Albin, M. Allen, L. Alvarez, L. G. Apostolova, S. E. Arnold, S. Asthana, C. S. Atwood, G. Ayres, C. T. Baldwin, R. C. Barber, L. L. Barnes, S. Barral, T. G. Beach, J. T. Becker, G. W. Beecham, D. Beekly, J. E. Below, P. Benchek, B. A. Benitez, D. Bennett, J. Bertelson, F. E. Margaret, T. D. Bird, D. Blacker, B. F. Boeve, J. D. Bowen, A. Boxer, J. Brewer, J. R. Burke, J. M. Burns, W. S. Bush, J. D. Buxbaum, N. J. Cairns, C. Cao, C. S. Carlson, C. M. Carlsson, R. M. Carney, M. M. Carrasquillo, S. Chasse, M. F. Chesselet, A. Chesi, N. A. Chin, H. C. Chui, J. Chung, S. Craft, P. K. Crane, D. H. Cribbs, E. A. Crocco, C. Cruchaga, M. L. Cuccaro, M. Cullum, E. Darby, B. Davis, P. L. de Jager, C. DeCarli, J. DeToledo, M. Dick, D. W. Dickson, B. A. Dombroski, R. S. Doody, R. Duara, N. Ertekin-Taner, D. A. Evans, T. J. Fairchild, K. B. Fallon, M. R. Farlow, J. J. Farrell, V. Fernandez-Hernandez, S. Ferris, M. P. Frosch, B. Fulton-Howard, D. R. Galasko, A. Gamboa, M. Gearing, D. H. Geschwind, B. Ghetti, J. R. Gilbert, T. J. Grabowski, N. R. Graff-Radford, S. F. A. Grant, R. C. Green, J. H. Growdon, J. L. Haines, H. Hakonarson, J. Hall, R. L. Hamilton, O. Harari, L. E. Harrell, J. Haut, E. Head, V. W. Henderson, M. Hernandez, T. Hohman, L. S. Honig, R. M. Huebinger, M. J. Huentelman, C. M. Hulette, B. T. Hyman, L. S. Hynan, L. Ibanez, G. P. Jarvik, S. Jayadev, L. W. Jin, K. Johnson, L. Johnson, M. I. Kamboh, A. M. Karydas, M. J. Katz, J. A. Kaye, C. D. Keene, A. Khaleeq, R. Kim, J. Knebl, N. W. Kowall, J. H. Kramer, P. P. Kuksa, F. M. LaFerla, J. J. Lah, E. B. Larson, C. Y. Lee, E. B. Lee, A. Lerner, Y. Y. Leung, J. B. Leverenz, A. I. Levey, M. Li, A. P. Lieberman, R. B. Lipton, M. Logue, C. G. Lyketsos, J. Malamon, D. Mains, D. C. Marson, F. Martiniuk, D. C. Mash, E. Masliah, P. Massman, A. Masurkar, W. C. McCormick, S. M. McCurry, A. N. McDavid, S. McDonough, A. C. McKee, M. Mesulam, J. Mez, B. L. Miller, C. A. Miller, J. W. Miller, T. J. Montine, E. S. Monuki, J. C. Morris, A. J. Myers, T. Nguyen, S. O’Bryant, J. M. Olichney, M. Ory, R. Palmer, J. E. Parisi, H. L. Paulson, V. Pavlik, D. Paydarfar, V. Perez, E. Peskind, R. C. Petersen, J. E. Phillips-Cremins, A. Pierce, M. Polk, W. W. Poon, H. Potter, L. Qu, M. Quiceno, J. F. Quinn, A. Raj, M. Raskind, E. M. Reiman, B. Reisberg, J. S. Reisch, J. M. Ringman, E. D. Roberson, M. Rodriguear, E. Rogaeva, H. J. Rosen, R. N. Rosenberg, D. R. Royall, M. A. Sager, M. Sano, A. J. Saykin, J. A. Schneider, L. S. Schneider, W. W. Seeley, S. H. Slifer, S. Small, A. G. Smith, J. P. Smith, Y. E. Song, J. A. Sonnen, S. Spina, P. S. George-Hyslop, R. A. Stern, A. B. Stevens, S. M. Strittmatter, D. Sultzer, R. H. Swerdlow, R. E. Tanzi, J. L. Tilson, J. Q. Trojanowski, J. C. Troncoso, D. W. Tsuang, O. Valladares, V. M. van Deerlin, L. J. van Eldik, R. Vassar, H. V. Vinters, J. P. Vonsattel, S. Weintraub, K. A. Welsh-Bohmer, P. L. Whitehead, E. M. Wijsman, K. C. Wilhelmsen, B. Williams, J. Williamson, H. Wilms, T. S. Wingo, T. Wisniewski, R. L. Woltjer, M. Woon, C. B. Wright, C. K. Wu, S. G. Younkin, C. E. Yu, L. Yu, Y. Zhang, Y. Zhao, X. Zhu; CHARGE, H. Adams, R. O. Akinyemi, M. Ali, N. Armstrong, H. J. Aparicio, M. Bahadori, J. T. Becker, M. Breteler, D. Chasman, G. Chauhan, H. Comic, S. Cox, A. L. Cupples, G. Davies, C. S. DeCarli, M. G. Duperron, J. Dupuis, T. Evans, F. Fan, A. Fitzpatrick, A. E. Fohner, M. Ganguli, M. Geerlings, S. J. Glatt, H. M. Gonzalez, M. Goss, H. Grabe, M. Habes, S. R. Heckbert, E. Hofer, E. Hong, T. Hughes, T. F. Kautz, M. Knol, W. Kremen, P. Lacaze, J. Lahti, Q. L. Grand, E. Litkowski, S. Li, D. Liu, X. Liu, M. Loitfelder, A. Manning, P. Maillard, R. Marioni, B. Mazoyer, D. M. van Lent, H. Mei, A. Mishra, P. Nyquist, J. O’Connell, Y. Patel, T. Paus, Z. Pausova, K. Raikkonen-Talvitie, M. Riaz, S. Rich, J. Rotter, J. Romero, G. Roshchupkin, Y. Saba, M. Sargurupremraj, H. Schmidt, R. Schmidt, J. M. Shulman, J. Smith, H. Sekhar, R. Rajula, J. Shin, J. Simino, E. Sliz, A. Teumer, A. Thomas, A. Tin, E. Tucker-Drob, D. Vojinovic, Y. Wang, G. Weinstein, D. Williams, K. Wittfeld, L. Yanek, Y. Yang, L. A. Farrer, B. M. Psaty, M. Ghanbari, T. Raj, P. Sachdev, K. Mather, F. Jessen, M. A. Ikram, A. de Mendonça, J. Hort, M. Tsolaki, M. A. Pericak-Vance, P. Amouyel, J. Williams, R. Frikke-Schmidt, J. Clarimon, J. F. Deleuze, G. Rossi, S. Seshadri, O. A. Andreassen, M. Ingelsson, M. Hiltunen, K. Sleegers, G. D. Schellenberg, C. M. van Duijn, R. Sims, W. M. van der Flier, A. Ruiz, A. Ramirez, J. C. Lambert, New insights into the genetic etiology of Alzheimer’s disease and related dementias. Nat. Genet. 54, 412–436 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.A. Witoelar, A. Rongve, I. S. Almdahl, I. D. Ulstein, A. Engvig, L. R. White, G. Selbæk, E. Stordal, F. Andersen, A. Brækhus, I. Saltvedt, K. Engedal, T. Hughes, S. Bergh, G. Bråthen, N. Bogdanovic, F. Bettella, Y. Wang, L. Athanasiu, S. Bahrami, S. le Hellard, S. Giddaluru, A. M. Dale, S. B. Sando, S. Steinberg, H. Stefansson, J. Snaedal, R. S. Desikan, K. Stefansson, D. Aarsland, S. Djurovic, T. Fladby, O. A. Andreassen, Meta-analysis of Alzheimer’s disease on 9,751 samples from Norway and IGAP study identifies four risk loci. Sci. Rep. 8, 18088 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.J. Bishop, M. Schuksz, J. D. Esko, Heparan sulphate proteoglycans fine-tune mammalian physiology. Nature 446, 1030–1037 (2007). [DOI] [PubMed] [Google Scholar]
- 13.C. Gama, S. E. Tully, N. Sotogaku, P. M. Clark, M. Rawat, N. Vaidehi, W. A. Goddard III, A. Nishi, L. C. Hsieh-Wilson, Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat. Chem. Biol. 2, 467–473 (2006). [DOI] [PubMed] [Google Scholar]
- 14.J. Liu, L. C. Pedersen, Emerging chemical and biochemical tools for studying 3-O-sulfated heparan sulfate. Am. J. Physiol. Cell Physiol. 322, C1166–C1175 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.B. Thacker, D. Xu, R. Lawrence, J. Esko, Heparan sulfate 3-O-sulfation: A rare modification in search of a function. Matrix Biol. 35, 60–72 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.N. W. Shworak, J. Liu, L. M. Petros, L. Zhang, M. Kobayashi, N. G. Copeland, N. A. Jenkins, R. D. Rosenberg, Multiple Isoforms of heparan sulfate D-glucosaminyl 3-O-sulfotransferase. Isolation, characterization, and expression of human cdnas and identification of distinct genomic loci. J. Biol. Chem. 274, 5170–5184 (1999). [DOI] [PubMed] [Google Scholar]
- 17.R. D. Rosenberg, N. W. Shworak, J. Liu, J. J. Schwartz, L. Zhang, Heparan sulfate proteoglycans of the cardiovascular system: Specific structures emerge but how is synthesis regulated? J. Clin. Invest. 99, 2062–2070 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Y. Xu, C. Cai, K. Chandarajoti, P. H. Hsieh, L. Li, T. Q. Pham, E. M. Sparkenbaugh, J. Sheng, N. S. Key, R. Pawlinski, E. N. Harris, R. J. Linhardt, J. Liu, Homogeneous low-molecular-weight heparins with reversible anticoagulant activity. Nat. Chem. Biol. 10, 248–250 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Z. Wang, K. Arnold, Y. Xu, V. Pagadala, G. Su, H. Myatt, R. J. Linhardt, J. Liu, Quantitative analysis of heparan sulfate using isotopically labeled calibrants. Commun. Biol. 3, 425 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.D. Xu, V. Tiwari, G. Xia, C. Clement, D. Shukla, J. Liu, Characterization of heparan sulphate 3-O-sulphotransferase isoform 6 and its role in assisting the entry of herpes simplex virus type 1. Biochem. J. 385, 451–459 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Z. Wang, P. H. Hsieh, Y. Xu, D. Thieker, E. J. E. Chai, S. Xie, B. Cooley, R. J. Woods, L. Chi, J. Liu, Synthesis of 3-O-sulfated oligosaccharides to understand the relationship between structures and functions of heparan sulfate. J. Am. Chem. Soc. 139, 5249–5256 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.S. HajMohammadi, K. Enjyoji, M. Princivalle, P. Christi, M. Lech, D. Beeler, H. Rayburn, J. J. Schwartz, S. Barzegar, A. I. de Agostini, M. J. Post, R. D. Rosenberg, N. W. Shworak, Normal levels of anticoagulant heparan sulfate are not essential for normal hemostasis. J. Clin. Invest. 111, 989–999 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.H. Qiu, S. Shi, J. Yue, M. Xin, A. V. Nairn, L. Lin, X. Liu, G. Li, S. A. Archer-Hartmann, M. dela Rosa, M. Galizzi, S. Wang, F. Zhang, P. Azadi, T. H. van Kuppevelt, W. V. Cardoso, K. Kimata, X. Ai, K. W. Moremen, J. D. Esko, R. J. Linhardt, L. Wang, A mutant-cell library for systematic analysis of heparan sulfate structure-function relationships. Nat. Methods 15, 889–899 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.A. C. Birdsill, D. G. Walker, L. F. Lue, L. I. Sue, T. G. Beach, Postmortem interval effect on RNA and gene expression in human brain tissue. Cell Tissue Bank. 12, 311–318 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.B. Highet, R. Parker, R. L. M. Faull, M. A. Curtis, B. Ryan, RNA quality in post-morten human brain tissu is afftected by Alzheimer’s disease. Front. Neurosci. 14, 780352 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.A. Ferreira, I. Royaux, J. Liu, Z. Wang, G. Su, D. Moechars, N. Callewaert, L. de Muynck, The 3-O-sulfation of heparan sulfate proteoglycans contributes to the cellular internalization of tau aggregates. BMC Mol. Cell Biol. 23, 61 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Y. Chen, L. Lin, I. Agyekum, X. Zhang, K. St. Ange, Y. Yu, F. Zhang, J. Liu, I. J. Amster, R. J. Linhardt, Structural analysis of heparin-derived 3-O-sulfated tetrasaccharides: Antithrombin binding site variants. J. Pharm. Sci. 106, 973–981 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.D. Xu, J. Esko, Demystifying heparan sulfate-protein interactions. Annu. Rev. Biochem. 83, 129–157 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.J. Zhao, I. Huvent, G. Lippens, D. Eliezer, A. Zhang, Q. Li, P. Tessier, R. J. Linhardt, F. Zhang, C. Wang, Glycan determinants of heparin-tau interaction. Biophys. J. 112, 921–932 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.B. E. Stopschinski, B. B. Holmes, G. M. Miller, V. A. Manon, J. Vaquer-Alicea, W. L. Prueitt, L. C. Hsieh-Wilson, M. I. Diamond, Specific glycosaminoglycan chain length and sulfation patterns are required for cell uptake of tau versus α-synuclein and β-amyloid aggregates. J. Biol. Chem. 293, 10826–10840 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.N. Pérez-López, C. Martín, B. García, M. P. Solís-Hernández, D. Rodríguez, I. Alcalde, J. Merayo, I. Fernández-Vega, L. M. Quirós, Alterations in the expression of the genes responsible for the synthesis of heparan sulfate in brains with Alzheimer disease. J. Neuropathol. Exp. Neurol. 80, 446–456 (2021). [DOI] [PubMed] [Google Scholar]
- 32.Y. Wang, E. Mandelkow, Tau in physiology and pathology. Nat. Rev. Neurosci. 17, 5–21 (2016). [DOI] [PubMed] [Google Scholar]
- 33.C. A. Brunello, M. Merezhko, R. L. Uronen, H. J. Huttunen, Mechanisms of secretion and spreading of pathological tau protein. Cell. Mol. Life Sci. 77, 1721–1744 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.S. T. Cheung, M. S. Miller, R. Pacoma, J. Roland, J. Liu, A. M. Schumacher, L. C. Hsieh-Wilson, Discovery of a small-molecule modulator of glycosaminoglycan sulfation. ACS Chem. Biol. 12, 3126–3133 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.S. Yamada, K. Yoshida, M. Sugiura, K. Sugahara, K. H. Khoo, H. R. Morris, A. Dell, Structural studies on the bacterial lyase-resistant tetrasaccharides derived from the antithrombin III-binding site of porcine intestinal heparin. J. Biol. Chem. 268, 4780–4787 (1993). [PubMed] [Google Scholar]
- 36.V. M. Dhurandhare, V. Pagadala, A. Ferreira, L. De Muynck, J. Liu, Synthesis of 3-O-sulfated disaccharide and tetrasaccharide standards for compositional analysis of heparan sulfate. Biochemistry 59, 3186–3192 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.K. St Ange, A. Onishi, L. Fu, X. Sun, L. Lin, D. Mori, F. Zhang, J. S. Dordick, J. Fareed, D. Hoppensteadt, W. Jeske, R. J. Linhardt, Analysis of heparins derived from bovine tissues and comparison to porcine intestinal heparins. Clin. Appl. Thromb. Hemost. 22, 520–527 (2016). [DOI] [PubMed] [Google Scholar]
- 38.R. Karlsson, P. Chopra, A. Joshi, Z. Yang, S. Y. Vakhrushev, T. M. Clausen, C. D. Painter, G. P. Szekeres, Y.-H. Chen, D. R. Sandoval, L. Hansen, J. D. Esko, K. Pagel, D. P. Dyer, J. E. Turnbull, H. Clausen, G.-J. Boons, R. L. Miller, Dissecting structure-function of 3-O-sulfated heparin and engineered heparan sulfates. Sci. Adv. 7, eabl6026 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Z. Wang, K. Arnold, V. M. Dhurandahare, Y. Xu, V. Pagadala, E. Labra, W. Jeske, J. Fareed, M. Gearing, J. Liu, Analysis of 3-O-sulfated heparan sulfate using isotopically labeled oligosaccharide calibrants. Anal. Chem. 94, 2950–2957 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Y. Huang, Y. Mao, C. Zong, C. Lin, G. J. Boons, J. Zaia, Discovery of a heparan sulfate 3-O-sulfation specific peeling reaction. Anal. Chem. 87, 592–600 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.B. E. Thacker, E. Seamen, R. Lawrence, M. W. Parker, Y. Xu, J. Liu, C. W. Vander Kooi, J. D. Esko, Expanding the 3-O-sulfate proteome—enhanced binding of neuropilin-1 to 3-O-sulfated heparan sulfate modulates its activity. ACS Chem. Biol. 11, 971–980 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.V. N. Patel, D. L. Pineda, E. Berenstein, B. R. Hauser, S. Choi, M. Prochazkova, C. Zheng, C. M. Goldsmith, T. H. van Kuppevelt, A. Kulkarni, Y. Song, R. J. Linhardt, A. M. Chibly, M. P. Hoffman, Loss of Hs3st3a1 or Hs3st3b1 enzymes alters heparan sulfate to reduce epithelial morphogenesis and adult salivary gland function. Matrix Biol. 103-104, 37–57 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.R. Wander, A. M. Kaminski, Y. Xu, V. Pagadala, J. M. Krahn, T. Q. Pham, J. Liu, L. C. Pedersen, Deciphering the substrate recognition mechanisms of the heparan sulfate 3-O-sulfotransferase-3. RSC ChemBiol. 2, 1239–1248 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D. Xu, A. F. Moon, D. Song, L. C. Pedersen, J. Liu, Engineering sulfotransferases to modify heparan sulfate. Nat. Chem. Biol. 4, 200–202 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.H. Qi, C. Despres, S. Prabakaran, F. X. Cantrelle, B. Chambraud, J. Gunawardena, G. Lippens, C. Smet-Nocca, I. Landrieu, The study of posttranslational modifications of tau protein by nuclear magnetic resonance spectroscopy: Phosphorylation of tau protein by ERK2 recombinant kinase and rat brain extract, and acetylation by recombinant creb-binding protein. Methods Mol. Biol. 1523, 179–213 (2017). [DOI] [PubMed] [Google Scholar]
- 46.P. Barré, D. Eliezer, Structural transitions in tau k18 on micelle binding suggest a hierarchy in the efficacy of individual microtubule-binding repeats in filament nucleation. Protein Sci. 22, 1037–1048 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.R. Rydbirk, J. Folke, K. Winge, S. Aznar, B. Pakkenberg, T. Brudek, Assessment of brain reference genes for RT-qPCR studies in neurodegenerative diseases. Sci. Rep. 6, 37116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.M. B. Huynh, M. O. Ouidja, S. Chantepie, G. Carpentier, A. Maïza, G. Zhang, J. Vilares, R. Raisman-Vozari, D. Papy-Garcia, Glycosaminoglycans from Alzheimer’s disease hippocampus have altered capacities to bind and regulate growth factors activities and to bind tau. PLOS ONE 14, e0209573 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.T. J. Montine, C. H. Phelps, T. G. Beach, E. H. Bigio, N. J. Cairns, D. W. Dickson, C. Duyckaerts, M. P. Frosch, E. Masliah, S. S. Mirra, P. T. Nelson, J. A. Schneider, D. R. Thal, J. Q. Trojanowski, H. V. Vinters, B. T. Hyman; National Institute on Aging; Alzheimer’s Association , National Institute on Aging–Alzheimer’s Association guidelines for the neuropathologic assessment of Alzheimer’s disease: A practical approach. Acta Neuropathol. 123, 1–11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.N. J. Cairns, E. H. Bigio, I. R. Mackenzie, M. Neumann, V. M. Lee, K. J. Hatanpaa, C. L. White III, J. A. Schneider, L. T. Grinberg, G. Halliday, C. Duyckaerts, J. S. Lowe, I. E. Holm, M. Tolnay, K. Okamoto, H. Yokoo, S. Murayama, J. Woulfe, D. G. Munoz, D. W. Dickson, P. G. Ince, J. Q. Trojanowski, D. M. Mann; Consortium for Frontotemporal Lobar Degeneration , Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: Consensus of the consortium for frontotemporal lobar degeneration. Acta Neuropathol. 114, 5–22 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.I. R. A. Mackenzie, M. Neumann, E. H. Bigio, N. J. Cairns, I. Alafuzoff, J. Kril, G. G. Kovacs, B. Ghetti, G. Halliday, I. E. Holm, P. G. Ince, W. Kamphorst, T. Revesz, A. J. M. Rozemuller, S. Kumar-Singh, H. Akiyama, A. Baborie, S. Spina, D. W. Dickson, J. Q. Trojanowski, D. M. A. Mann, Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: Consensus recommendations. Acta Neuropathol. 117, 15–18 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.I. G. McKeith, B. F. Boeve, D. W. Dickson, G. Halliday, J. P. Taylor, D. Weintraub, D. Aarsland, J. Galvin, J. Attems, C. G. Ballard, A. Bayston, T. G. Beach, F. Blanc, N. Bohnen, L. Bonanni, J. Bras, P. Brundin, D. Burn, A. Chen-Plotkin, J. E. Duda, O. el-Agnaf, H. Feldman, T. J. Ferman, D. ffytche, H. Fujishiro, D. Galasko, J. G. Goldman, S. N. Gomperts, N. R. Graff-Radford, L. S. Honig, A. Iranzo, K. Kantarci, D. Kaufer, W. Kukull, V. M. Y. Lee, J. B. Leverenz, S. Lewis, C. Lippa, A. Lunde, M. Masellis, E. Masliah, P. McLean, B. Mollenhauer, T. J. Montine, E. Moreno, E. Mori, M. Murray, J. T. O'Brien, S. Orimo, R. B. Postuma, S. Ramaswamy, O. A. Ross, D. P. Salmon, A. Singleton, A. Taylor, A. Thomas, P. Tiraboschi, J. B. Toledo, J. Q. Trojanowski, D. Tsuang, Z. Walker, M. Yamada, K. Kosaka, Diagnosis and management of dementia with Lewy bodies. Neurology 89, 88–100 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S13
Tables S1 to S16