Abstract
Higher-order chromatin has been implicated in epigenetic gene control and in the functional organization of chromosomes. We have recently discovered mouse (Suv39h1) and human (SUV39H1) histone H3 lysine 9-selective methyltransferases (Suv39h HMTases) and shown that they modulate chromatin dynamics in somatic cells. We describe here the isolation, chromosomal assignment, and characterization of a second murine gene, Suv39h2. Like Suv39h1, Suv39h2 encodes an H3 HMTase that shares 59% identity with Suv39h1 but which differs by the presence of a highly basic N terminus. Using fluorescent in situ hybridization and haplotype analysis, the Suv39h2 locus was mapped to the subcentromeric region of mouse chromosome 2, whereas the Suv39h1 locus resides at the tip of the mouse X chromosome. Notably, although both Suv39h loci display overlapping expression profiles during mouse embryogenesis, Suv39h2 transcripts remain specifically expressed in adult testes. Immunolocalization of Suv39h2 protein during spermatogenesis indicates enriched distribution at the heterochromatin from the leptotene to the round spermatid stage. Moreover, Suv39h2 specifically accumulates with chromatin of the sex chromosomes (XY body) which undergo transcriptional silencing during the first meiotic prophase. These data are consistent with redundant enzymatic roles for Suv39h1 and Suv39h2 during mouse development and suggest an additional function of the Suv39h2 HMTase in organizing meiotic heterochromatin that may even impart an epigenetic imprint to the male germ line.
In eukaryotes, control of gene expression and the functional organization of chromosomes depends on higher-order chromatin, which has been proposed to be nucleated by the covalent modification of histone amino termini (45). In addition to its role in somatic cells, dynamic transitions in the organization of higher-order chromatin are also important during meiosis (15). Although condensation and pairing of meiotic chromosomes is evolutionarily highly conserved, meiosis in male mammals is exceptional because the heteromorphic X and Y chromosomes undergo facultative heterochromatinization that is accompanied by transcriptional silencing (21). This selective inactivation of the sex chromosomes, which is cytologically defined by the appearance of the so-called XY body or sex vesicle (44), has been suggested to restrict promiscuous pairing or recombination between nonhomologous chromosomes, thereby reducing the risk for aneuploidy (21).
Despite the apparent resemblance of the XY body to the Barr body (9) in female somatic cells, it is currently unresolved whether similar mechanism(s) operate in inducing chromosome-specific heterochromatinization in meiotic and somatic cells. For example, although Xist RNA also localizes to the XY body (5), spermatogenesis is unaffected in Xist-deficient mice (29). Moreover, only a few proteins that associate with the XY body have been described (12, 23, 26, 43) of which M31 (HP1β) represents the first bona fide heterochromatic component (32, 49). Because M31 (HP1β) is a mammalian member of the Su(var) gene family, this result suggested a possible link between heterochromatin-induced gene repression in somatic tissues and transcriptional silencing of the sex chromosomes during male meiosis.
Su(var) genes were initially identified by genetic screens on centromeric position effects in Drosophila melanogaster (37) and Schizosaccharomyces pombe (3). Since Su(var) genes suppress position effect variegation (PEV), their gene products have been implicated in the organization of repressive chromatin domains. Indeed, characterized family members represent either chromosomal proteins or enzymes that can modify chromatin (47). Recently, we isolated mouse (Suv39h1) and human (SUV39H1) homologs (1) of the dominant Drosophila PEV modifier Su(var)3-9 (46) and demonstrated that they encode histone methyltransferases which selectively methylate lysine 9 (Lys9) in the histone H3 N terminus (36). Immunolocalization of endogenous Suv39h1 or SUV39H1 proteins in mammalian cells indicated enriched distribution at heterochromatic foci during interphase and transient accumulation at centromeric positions during mitosis (2). In addition, Suv39h1 or SUV39H1 associate with M31 (HP1β), indicating the existence of a mammalian SU(VAR) protein complex (1). Moreover, deregulated SUV39H1 can induce ectopic heterochromatin and redistribute endogeneous M31 (HP1β) (30). Together, our data defined Suv39h1 or SUV39H1 as novel heterochromatin-associated HMTases that are intrinsically involved in the structural organization of mammalian higher-order chromatin in somatic cells.
We describe here the isolation and characterization of a second murine Suv39h gene, Suv39h2. Over the entire length of the 477-amino-acid protein, Suv39h2 shares 59% identity with Suv39h1 and also displays an H3-Lys9-selective HMTase activity. However, Suv39h2 differs from Suv39h1 by the presence of a very basic, histone H1-like N terminus. Moreover, whereas both murine Suv39h loci show overlapping expression profiles during embryogenesis, Suv39h2 transcripts are restricted to adult testes. Immunolocalization of endogenous Suv39h2 protein reveals enriched distributions at heterochromatin during the first meiotic prophase and in the early stages of spermiogenesis. During mid-pachytene, Suv39h2 specifically accumulates with chromatin of the silenced sex chromosomes present in the XY body. These data are consistent with redundant gene functions for both Suv39h loci during mouse development but, in addition, suggest a role for the Suv39h2 HMTase in regulating higher-order chromatin dynamics during male meiosis.
MATERIALS AND METHODS
Molecular cloning of murine Suv39h2.
A 210-bp EST DNA probe (encoding amino acids 219 to 289 of Suv39h2, see Fig. 1) was PCR amplified from murine B-cell-specific (J558L and S194) cDNA libraries and screened against a day 11.5 mouse embryonic λgt11 cDNA library (Clontech) and a λ129/Sv genomic library (Stratagene), resulting in the isolation of six cDNA and three genomic clones. The longest cDNA (1 kb; λ4-Suv39h2) and genomic (14-kb) isolates were sequenced by primer walking on an automated sequencer (Applied Biosystems). Sequence analysis indicated that the λ4-Suv39h2 cDNA encoded amino acids 132 to 477 and that the genomic sequence comprised exons 1 to 3, as predicted by GENE-Finder. Missing 5′ sequences of the Suv39h2 cDNA were extended by nested rapid amplification of cDNA ends (RACE) (Marathon cDNA Amplification Kit; Clontech) from the J558L and S194 cDNA libraries using the exon 3 specific primers 5′-GCCCTCCAAGTCAACAGTG and 5′-GTGTTGAGGTAATCTTGCCATC. The RACE amplifications identified exon 2 (amino acids 83 to 131). Exon 1, including the starting ATG, was deduced from an EST (accession no. AA959164) which correctly spliced into exon 2 and whose sequence information was confirmed by comparison with genomic sequences.
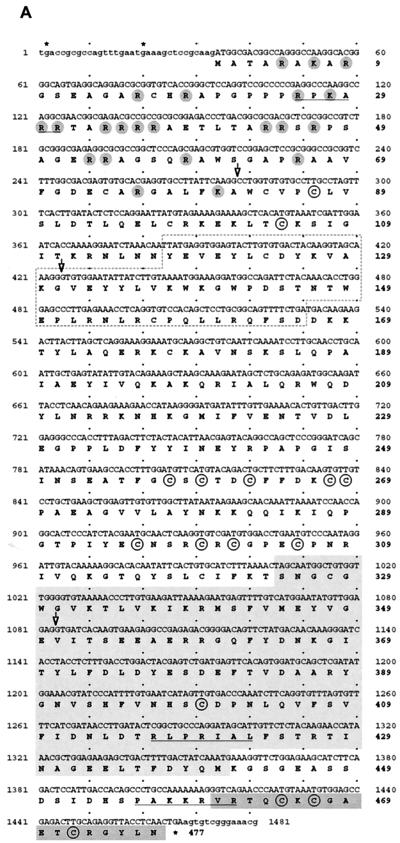
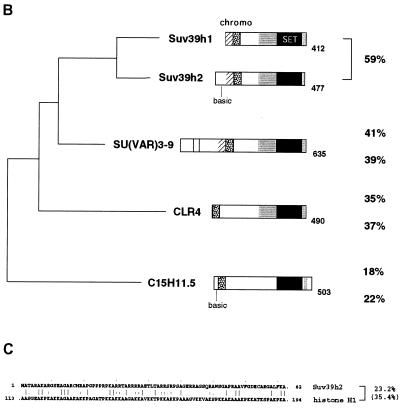
FIG. 1.
Conceptional reading frame and domain conservation of the Suv39h2 protein. (A) The ∼1.5-kb nucleotide sequence and conceptional reading frame of the coding part of the Suv39h2 cDNA is shown. Exon 1, including the starting ATG preceded by in-frame stop codons (asterisks), has been derived from genomic Suv39h2 sequences and from an EST that correctly spliced into exon 2. From the available genomic sequences, exons 1 to 3 could be identified, and their respective exon-intron boundaries are indicated by open arrowheads at nucleotide positions 278, 424, and 1083. The 477-amino-acid Suv39h2 protein contains several conserved sequence motifs, including a chromo domain (dashed box), the SET domain (light gray shading), and a C-terminal tail (dark gray shading). Basic amino acids in the N terminus are highlighted by gray circles. In addition, cysteine residues that are also conserved in Suv39h1 are circled. Putative nuclear localization signals are underlined. (B) Phylogenetic relationships of murine Suv39h1 (412 amino acids) (1), murine Suv39h2 (477 amino acids), Drosophila SU(VAR)3-9 (635 amino acids) (46), S. pombe CLR4 (490 amino acids) (25), and a C. elegans ORF C15H11.5 (503 amino acids) (accession no. Z81035). Over the entire length of the protein, Suv39h1 shares 59% identity with Suv39h2, 41% identity with SU(VAR)3-9, 35% identity with CLR4, and 18% identity with C15H11.5. Similarly, Suv39h2 shares 59% identity with Suv39h1, 39% identity with SU(VAR)3-9, 37% identity with CLR4, and 22% identity with C15H11.5. Highly conserved sequence motifs are indicated and comprise the chromo (stippled) and SET (black) domains and the SET-associated cysteine-rich clusters (gray), which are only in part present in C15H11.5. In addition, an N-terminal region (hatched) shared by the murine and fly proteins (1), a putative GTP-binding domain (light-shaded box) in SU(VAR)3-9 (46), and the basic N termini (basic) in Suv39h2 and C15H11.5 are also highlighted. (C) Amino acid identity and similarity (in brackets) between the Suv39h2 N terminus and the C-terminal portion of histone H1.
Chromosomal assignment and fine mapping of the Suv39h1 and Suv39h2 loci.
To obtain genomic Suv39h1 sequences, we hybridized a Suv39h1-specific DNA probe encoding amino acids 19 to 223 of Suv39h1 (1) against a λ129/Sv genomic library (Stratagene). Among several isolates, a 13-kb partial genomic Suv39h1 fragment comprising exons 1 to 3 was sequenced by primer walking. Both Suv39h1 and Suv39h2 genomic sequences were subcloned into pBluescript (see Fig. 2, top) and used for fluorescence in situ hybridization (FISH) analysis on Robertsonian mouse chromosomes as described previously (28). Fine mapping by single-stranded conformation polymorphism (SSCP) was performed with a 170-bp exon 3-, intron 3-specific DNA probe for Suv39h1 (primers 5′-CACTAATGATGGCCGAGG and 5′-GCAGAAGAGTTTGAGGTACAG) and with the 210-bp exon 3-specific EST-Suv39h2 probe (primers 5′-GGGGATGATATTTGTTGAAAACAC and 5′-GGTTGGATTTTAATTTGTTGCTTC). The SSCP analysis and calculation of genetic distances was as recently reported (28).
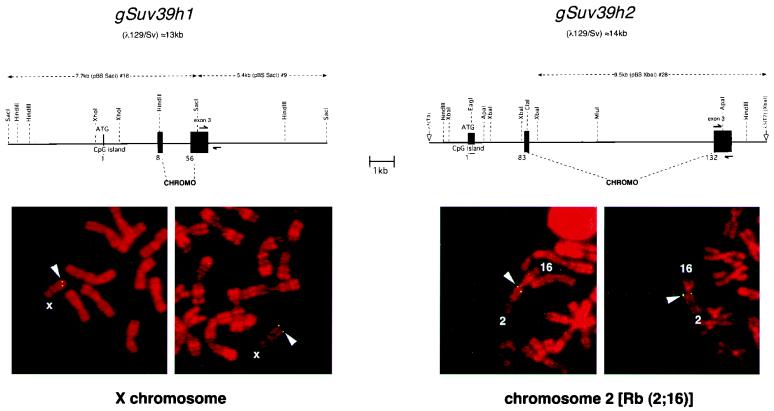
FIG. 2.
Genomic organization and chromosomal localization of the Suv39h1 and Suv39h2 loci. (Top) Organization of partial genomic clones comprising the 5′ regions of murine Suv39h1 and Suv39h2. Exons are shown by black boxes, and numbers indicate the starting amino acid positions of the respective exons. Demarcated by the dashed lines are exon regions that encode the conserved chromo domain. Both loci are drawn to scale, and the respective Suv39h1 and Suv39h2 subclones (dashed lines above the loci) that were hybridized to metaphase spreads (below) are shown, as are the specific primer pairs (split arrows) used for the haplotype analysis (see Fig. 3). (Bottom) FISH analysis of Suv39h1 and Suv39h2 localization on metaphase chromosomes prepared from lymphocytes of WMP male mice. Chromosomes were counterstained and R banded with propidium iodide. Arrowheads indicate symmetrical signals on the X chromosome for Suv39h1 (left panel) and on the Rb(2;16) chromosome for Suv39h2 (right panel). Based on the R banding, localization of Suv39h1 is assigned to regions A1 and A2 of chromosome X and that of Suv39h2 to region A of chromosome 2.
RNA isolation and blot analysis.
RNA isolation and analysis was done as described previously (1). Membranes were sequentially hybridized under stringent Church conditions (41) with a 1.6-kb EcoRI cDNA fragment comprising nearly full-length Suv39h1 (1) or with a 980-bp cDNA PCR amplicon which codes for amino acids 143 to 477 of Suv39h2. To control for the quality of the RNA preparations, blots were rehybridized with a DNA probe that is specific for Gadph sequences (17).
In situ hybridization for Suv39h1 and Suv39h2 expression.
To obtain Suv39h1- and Suv39h2-specific riboprobes, PCR-converted SalI/BamHI DNA fragments were subcloned into the polylinker of pGEM-3Zf (Promega). Similar to an internal 395-bp DNA fragment encoding amino acids 113 to 237 of Suv39h1 (1), a 325-bp internal DNA fragment encoding amino acids 186 to 290 of Suv39h2 was used. Within this region, Suv39h1 and Suv39h2 nucleotide sequences are only ∼53% identical and do not cross-hybridize. In situ RNA probes were internally labeled with DIG-UTP (Boehringer Mannheim) by transcription with SP6 (antisense probes of EcoRI-linearized plasmid) or T7 RNA polymerase (sense probes of BamHI-linearized plasmid).
In situ hybridizations of whole-mount embryos or of 5-μm sections of paraffin-embedded testes were performed at 65 to 70°C overnight, washed under high stringency, and processed for detection after incubation with α-DIG alkaline phosphatase-conjugated antibodies and BM purple as the chromogenic substrate (Boehringer Mannheim).
Generation and purification of rabbit polyclonal α-Suv39h2-specific antibodies.
Suv39h2 coding sequences comprising amino acids 157 to 477 were converted into a BamHI-EcoRI DNA fragment by PCR amplification and combined in-frame with N-terminal glutathione S-transferase (GST) in the bacterial expression vector pGEX-2T (Pharmacia). Purification of recombinant protein and immunization of rabbits with the GST-Suv39h2 antigen was done as described elsewhere (1). An immunoglobulin G fraction was prepared from the crude serum of rabbit 2218, batch preabsorbed against GST-Suv39h1 glutathione-Sepharose beads (1), and α-Suv39h2 antibodies were affinity purified over a glutathione-Sepharose (Pharmacia) column that had been loaded with GST-Suv39h2. After elution with 100 mM glycine (pH 2.5), antibodies were neutralized with a 1/10 volume of 2 M HEPES (pH 7.9). These affinity-purified α-Suv39h2 antibodies (concentration, ∼0.5 mg/ml) were used at 1:250 or 1:500 dilutions for protein blot analysis or at 1:10 to 1:20 dilutions for indirect immunofluorescence.
Epitope-tagged Suv39h2 protein in HeLa cells.
To generate a (myc)3-tagged Suv39h2 protein that would resemble the shorter Suv39h1 or SUV39H1 gene products, the portion of the Suv39h2 cDNA comprising amino acids 83 to 477 was converted into a NotI/XhoI DNA fragment by PCR amplification and transferred into a NotI/XhoI-digested pKW2T-(myc)3H6SUV39H1 derivative which contains a unique in-frame NotI cloning site immediately following the N-terminal tag (1). The (myc)3-Suv2(83–477) construct was confirmed by sequencing and “stably” cotransfected into HeLa cells as described earlier (1). One clone (HeLa-S2/5) with significant overexpression of the ectopic protein in ∼65% of clonal cells was characterized further and, together with HeLa-B55 cells which overexpress (myc)3-SUV1(3–412) (30), used to analyze specificity of the α-Suv39h2 antibodies.
Isolation of PMEFs.
Primary mouse fibroblasts (PMEFs) were derived from day E12.5 C57BL6/129SvJ fetuses after removing the head and inner organs. Trunk tissues were partially homogenized in 5 ml of medium by repeated passage through a 20-gauge hypodermic needle, reseeded onto a 10-cm-diameter dish, and cultivated in high-glucose Dulbecco modified Eagle medium supplemented with 15% fetal calf serum–2 mM glutamine–1% nonessential amino acids–0.1 mM β-mercaptoethanol–1% penicillin-streptomycin (all Gibco-BRL). This primary culture was trypsinized after 3 days, and the floating single cell suspension was expanded into passage 1, which already contained a highly enriched population of PMEFs.
Nuclear extracts and protein blot analysis.
Isolation of nuclei from mouse testes was performed according to described protocols (11, 32). Approximately 30 μg of nuclear extracts from testes, the HeLa cell clones, or from PMEFs were analyzed on protein blots with α-myc, α-M31 (HP1β) (49), α-Suv39h1, and α-Suv39h2 antibodies as reported elsewhere (1).
HMTase assays with recombinant Suv39h2.
Methyltransferase assays with free histones (Boehringer Mannheim) or histone H3, CENP-A, and macroH2A N-terminal peptides and the recombinant GST-Suv2(157–477) product were done as recently described (36).
Immunofluorescence analysis of testes sections.
Testes were surgically removed from adult C57BL6/129SvJ mice, embedded in O.C.T. 4853 (Tissue-Tek), and frozen in liquid nitrogen using precooled isopentane. Then, 10-μm sections were fixed in 2% paraformaldehyde in phosphate-buffered saline (PBS; pH 7.4) for 10 min on ice, washed in cold PBS, and treated with 0.1% sodium citrate buffer (pH 6.0) containing 0.1% Triton X-100 for 5 min on ice. Subsequently, the sections were washed and blocked with PBS–2.5% bovine serum albumin (BSA)–0.1% Tween 20–10% goat serum for 30 min at room temperature.
For indirect immunofluorescence of M31 (HP1β) and Scp3 epitopes, sections were simultaneously incubated with rat monoclonal α-M31 antibodies (49) and mouse monoclonal α-Scp3 antibodies (22) overnight at 4°C. After several washes (PBS, 0.2% BSA, 0.1% Tween 20), samples were incubated for 1 h at room temperature with secondary CY3-conjugated goat anti-rat and CY5-conjugated goat anti-mouse antibodies (both from Dianova). After three final washes with PBS containing 0.1% Tween 20, sections were mounted in Vectashield antifade solution (Vector Laboratories) containing 4′,6′-diamidino-2-phenylindole (DAPI).
For double labeling of Suv39h2 and Scp3 epitopes, sections were incubated with rabbit polyclonal α-Suv39h2 antibodies and mouse monoclonal α-Scp3 antibodies. Suv39h2 staining was visualized by immunoamplification, using biotinylated goat anti-rabbit antibodies, Alexa488-conjugated avidin, biotinylated goat anti-avidin antibodies, and Alexa488-conjugated avidin (Vector Laboratories and Molecular Probes). Scp3 epitopes were visualized by secondary Alexa568-conjugated goat anti-mouse antibodies (Molecular Probes). Processed samples were evaluated using a Zeiss Axiophot epifluorescence microscope. Digital black-and-white images were recorded with a cooled charge-coupled device camera (Princeton Instruments), merged to RGB images using the Metamorph Imaging System (Universal Imaging Corporation), and processed in Adobe Photoshop 5.1.
Immunofluorescense analysis of testis suspension cells.
Testes were minced with scalpel blades in cold minimal essential medium (Gibco) containing protease inhibitors (Roche Biochemicals). Structurally preserved suspension cells were prepared by cross-linking fixation as described elsewhere (35). Testis suspension cells were mixed with equal volumes of PBS-buffered (pH 7.2) 3.7% formaldehyde–0.1 M sucrose, placed on silanized glass slides, and allowed to dry down until they were coated by a thin layer of sucrose.
For indirect immunofluorescence of Suv39h2 epitopes, sucrose-embedded cells were briefly washed with PBS, extracted for 30 min with 0.2% Triton X-100 in PBS, and incubated overnight at 4°C with rabbit polyclonal α-Suv39h2 antibodies. After three 3-min washes in PBS–0.1% Tween 20–0.2% BSA–0.1% gelatin, samples were either incubated for 45 min at 37°C with secondary, CY3-conjugated goat anti-rabbit antibodies (Vector Laboratories) or with secondary goat anti-rabbit biotinylated antibodies (Dianova) that were visualized after a third incubation by using avidin-fluorescein isothiocyanate (FITC) (Sigma). After three final washes in PBS–0.1% Tween 20, preparations were mounted in Antifade solution containing DAPI. Staging of individual mouse spermatogenic cells was determined by the specific distribution of heterochromatin (42).
For double-labeling experiments, samples were first incubated with α-Suv39h2 antibodies, followed by sandwich detection with anti-rabbit biotinylated antibodies and avidin-CY3. After a brief fixation with 1% formaldehyde in PBS, Xmr epitopes were then detected with mouse monoclonal α-Xmr antibodies (12) that were visualized with secondary goat anti-mouse FITC-conjugated antibodies (Dianova).
GenBank accession numbers.
The murine Suv39h2 cDNA (accession no. AF149205) and the murine genomic Suv39h2 (accession no. AF149204) and murine genomic Suv39h1 (accession no. AF149203) sequences have been deposited in GenBank.
RESULTS
Isolation of a second murine Suv39h gene: Suv39h2.
Sequence similarity searches (4, 7) with the murine Suv39h1 or human SUV39H1 cDNAs (1) revealed the presence of related, yet distinct expressed sequence tags (ESTs) in the DDBJ, EMBL, and GenBank databases. In particular, the mouse ESTs fall into two categories that are either homologous to Suv39h1 or indicative of a second gene. Using oligonucleotides specific for this second class of Suv39h ESTs, an internal (lacking the conserved chromo and SET domain sequences) DNA probe was PCR amplified from murine cDNAs and screened against a mouse embryonic day 11.5 (E11.5) cDNA library (see Materials and Methods). Of six positive isolates, the longest insert was subcloned and sequenced, revealing an open reading frame (ORF) which comprises the chromo and the C-terminal SET domain. RACE amplifications with cDNA templates from the murine B-cell-specific cell lines J558L and S194 extended the missing 5′ end; however, they did not detect a starting ATG.
To obtain more sequence information, a partial Suv39h2 genomic clone of approximately 14 kb was isolated (see Materials and Methods). In addition, we also identified a partial genomic clone of approximately 13 kb for the mouse Suv39h1 locus. Overall, the genomic organization, including the exon-intron structure and the presence of a predicted CpG-island across the first exon, is very similar for both loci (see Fig. 2 and below). Comparison of the available genomic, cDNA, and EST sequences for the Suv39h1-related gene allowed the definition of exon 1 (see Materials and Methods) that contains a consensus ATG preceded by in-frame stop codons and which can correctly splice into exon 2. In analogy to Suv39h1, we designated this novel gene Suv39h2 [for Su(var)3-9 homolog 2]. The nucleotide sequence (∼1.5 kb) and conceptional reading frame (477 amino acids) of the composite coding Suv39h2 cDNA are shown in Fig. 1A.
Sequence conservation of Suv39h2 within the SU(VAR)3-9 protein family.
Over the length of the 477-amino-acid protein, Suv39h2 is 59% identical to Suv39h1 (412 amino acids) (1). Cross-species comparison of Suv39h1 or Suv39h2 with other representative members of the SU(VAR)3-9 protein family, such as Drosophila SU(VAR)3-9 (46), S. pombe CLR4 (25), and a putative ORF in Caenorhabditis elegans (C15H11.5; accession no. Z81035) indicate very similar sequence identities and phylogenetic relationships (Fig. 1B).
Interestingly, however, Suv39h2 contains a highly basic (20.7%) N-terminal extension of 82 amino acids that is not present in Suv39h1, although a very basic, yet distinct N terminus is also found in the C15H11.5 ORF. In addition to its resemblance to arginine-rich protamines, the Suv39h2 N terminus shows moderate sequence identity (23.2%) with the C-terminal portion of the linker histone H1 that is not restricted to basic residues (Fig. 1C). With the exception of this extended N terminus, Suv39h2 maintains all other conserved domains outlined previously for Suv39h1 (1). For example, both proteins display highest identity in the 130-amino-acid SET domain core (75.2%) and in the conspicuous C-terminal tail (69.6%) with its three conserved cysteine residues. The 60-amino-acid chromo domain (62.7%), the SET-associated cysteine-rich region (54.9%), and the SU(VAR)3-9-specific N-terminal region (45.0%) are also highly identical. Like Suv39h1, Suv39h2 is also significantly shorter than the 635-amino-acid fly protein (46).
Chromosomal assignment of the murine Suv39h1 and Suv39h2 loci.
To investigate the chromosomal assignment of the two Suv39h loci, metaphase spreads prepared from stimulated lymphocytes of WMP mice, which are homozygous for metacentric Robertsonian (Rb) translocations in all autosomes except chromosome 19, were analyzed by FISH with genomic Suv39h1 and Suv39h2 DNA probes (Fig. 2, top, and Materials and Methods). The genomic Suv39h1 probe indicated a symmetrical, two-dotted signal at the tip of the mouse X chromosome, a localization consistent with the previous assignment of a SUV39H1 homologous sequence (MG-44) to the Xp11.2 region on the human X chromosome (20). In contrast, the genomic Suv39h2 probe specifically hybridized with the A region of mouse chromosome 2 present in the Rb(2;16) translocation (Fig. 2, bottom).
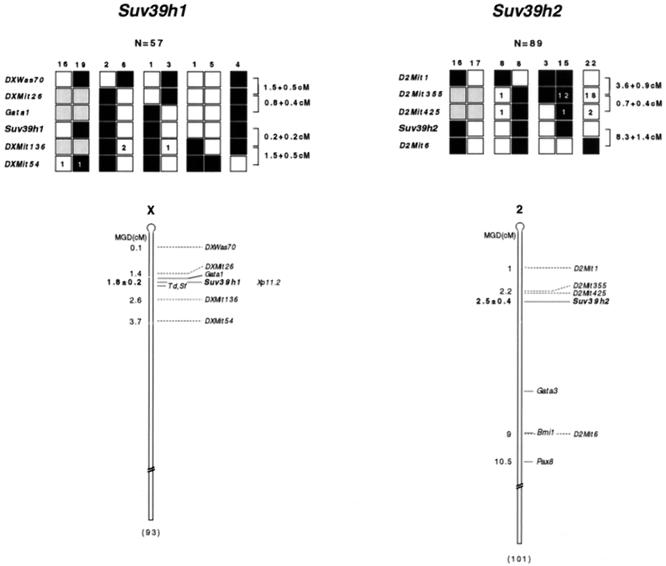
The chromosomal position of the two murine Suv39h loci was further defined by haplotype analysis of recombinant progeny (Mus spretus × C57BL/6) from the European collaborative interspecific backcross (EUCIB) (8, 38). Allelic variants were identified on a series of PCR-amplified DNA fragments by SSCP (see Materials and Methods). SSCP polymorphisms were detected with an exon 3-intron 3 primer pair for Suv39h1 and with an exon 3-specific primer pair for Suv39h2 (see Fig. 3, top). Each of these specific primer pairs was scored through a random panel of EUCIB backcross mice, indicating linkage of Suv39h1 to mouse chromosome X markers and of Suv39h2 to mouse chromosome 2 markers (data not shown). Haplotype analysis localized Suv39h1 to map position 1.8 cM of the mouse chromosome X, in an interval that also harbors the mouse mutations tattered (Td) (13) and scurfy (Sf) (39) (Fig. 3, left panel). In the parallel fine mapping of Suv39h2, haplotype analysis of a panel of 89 mice carrying recombinants in the D2Mit1/D2Mit6 region localized Suv39h2 to this interval. The Suv39h2 locus lies 0.7 ± 0.4 centimorgans (cM) distal of D2Mit355. Genetic distances taken from the Mouse Genome Database (http://www.jax.org) would indicate that Suv39h2 resides at map position 2.5 cM of mouse chromosome 2 (Fig. 3, right panel).
FIG. 3.
Linkage mapping of Suv39h1 and Suv39h2 on mouse chromosomes X and 2. (Top) Haplotype analysis. The segregation of C57BL/6 or M. spretus parental alleles for Suv39h1 (left panel), Suv39h2 (right panel), and for the respective flanking microsatellite markers that are specific for chromosome X or chromosome 2 are shown. Each backcross progeny has inherited the indicated allele from the F1 female parent. A white box indicates an M. spretus allele, and a black box indicates a C57BL/6 allele. Gray boxes represent markers that have not been scored. Numerals inside boxes reflect the number of selected recombinant progeny for which typings are missing in the EUCIB database. The numbers above each column indicate the number of progeny inheriting each type of chromosome. The calculation of genetic distances and standard error, correcting for recombinants not typed, was done as described previously (28). (Bottom) Consensus maps for Suv39h1 and Suv39h2 loci. Since genetic distances are calculated from multiple crosses, they reflect the relative rather than the precise order of markers. All distances (in centimorgans) are according to MGD chromosome committee maps. The position of the corresponding SUV39H1 locus (Xp11.2) on the human X chromosome (20) is also indicated.
Temporal and spatial expression of Suv39h1 and Suv39h2 during mouse development.
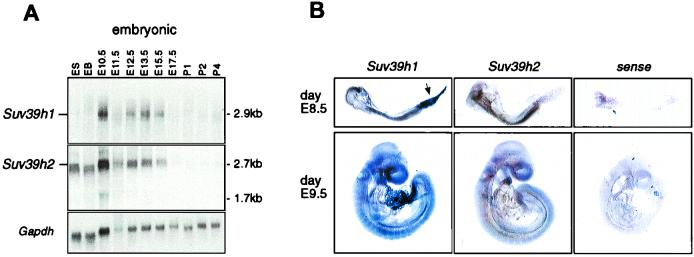
To determine the expression and size of Suv39h2 mRNAs, RNA blots containing total RNA from embryonic stem (ES) cells and mouse fetuses from various embryonic stages (days E10.5 to E17.5) and on postnatal days 1 to 4 (P1 to P4) were hybridized with a 980-bp cDNA probe comprising Suv39h2 coding sequences (amino acids 143 to 477). Within this region, the Suv39h2 cDNA is approximately 60% identical to the Suv39h1 nucleotide sequence and does not cross-hybridize with Suv39h1 transcripts (see Fig. 5A; also data not shown). This Suv39h2-specific cDNA probe recognized a prominent mRNA of approximately 2.7 kb in most RNA preparations of the analyzed stages (Fig. 4A, middle panel), whereas at day E10.5, smaller-sized (1.7-kb) transcripts were also detected. Abundant Suv39h2-specific transcripts are present in ES cells, in in vitro-differentiated embryoid bodies (EB), and appear to peak at ca. day E10.5. In contrast, Suv39h2-specific transcripts are substantially downregulated at day E17.5 and are nearly absent during postnatal development. A very similar dynamic expression profile was also observed for Suv39h1, except that the relative abundance of Suv39h1 transcripts in ES cells and EB is reduced compared to Suv39h2 transcripts (Fig. 4A, top panel). With a more sensitive RNase protection assay, however, Suv39h1 has been shown to be expressed in ES cells (1).
FIG. 5.
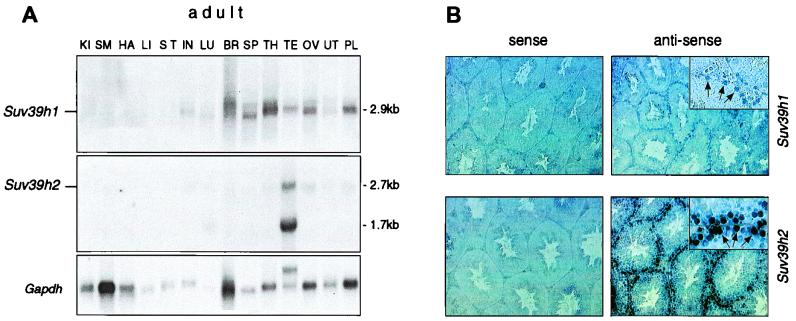
Testis-specific expression of Suv39h2. (A) RNA blot analysis to detect Suv39h1 and Suv39h2 transcripts in 15 μg of total RNA prepared from adult 129/Sv tissues, including kidney (KI), skeletal muscle (SM), heart (HA), liver (LI), stomach (ST), intestine (IN), lung (LU), brain (BR), spleen (SP), thymus (TH), testis (TE), ovaries (OV), uterus (UT), and placenta (PL). As a loading control, the RNA blot was rehybridized with a probe that is specific for Gapdh sequences. (B) RNA in situ hybridizations on 5-μm sections of adult testis with Suv39h1- and Suv39h2-specific riboprobes. As a control, sections were also hybridized with the corresponding Suv39h1- and Suv39h2-specific riboprobes. Enlarged insets show specific staining of type B spermatogonia and preleptotene spermatocytes (indicated by arrows).
FIG. 4.
Temporal and spatial expression of Suv39h1 and Suv39h2 during mouse development. (A) RNA blot analysis to detect Suv39h1 and Suv39h2 transcripts in 15 μg of total RNA prepared from undifferentiated CCE ES cells, EB derived after retinoic-acid-induced in vitro differentiation of CCE cells, and whole fetuses at various stages of embryonic (E10.5 to E17.5) and postnatal (P1 to P4) development. As a control for the quality of the RNA, the RNA blot was rehybridized with a probe that is specific for Gapdh sequences. (B) Whole-mount RNA in situ hybridizations of E8.5 and E9.5 mouse fetuses with Suv39h1- and Suv39h2-specific riboprobes. The arrow indicates the presence of Suv39h1 transcripts in the allantois. As a control, fetuses were also hybridized with a Suv39h2-specific sense probe.
To investigate the spatial expression profiles of Suv39h2 and Suv39h1, we performed whole-mount in situ hybridizations with Suv39h2- and Suv39h1-specific riboprobes (see Materials and Methods) on day E8.5 and day E9.5 mouse fetuses. Whereas only residual staining is observed with a Suv39h2 control sense probe, the Suv39h2 antisense probe reveals a rather uniform expression throughout the entire fetus (Fig. 4B, middle panel). Similarly, the Suv39h1 antisense probe detects a broad distribution of transcripts, a finding consistent with the ubiquitous expression of Suv39h1 in previous in situ hybridizations on sagittal sections of day E12.5 fetuses (1). In addition to embryonic tissues, the mesenchyme-derived allantois is also prominently stained by the Suv39h1 antisense probe (Fig. 4B, arrow in left panel). Together with the RNA blot shown above, this comparative analysis indicates significant coexpression of Suv39h1 and Suv39h2 during mouse development.
Suv39h2 transcripts are highly expressed in mouse testes.
In contrast to embryonic expression profiles, the abundance of Suv39h1 and Suv39h2 transcripts greatly differs in adult tissues. Whereas Suv39h1 displays broad expression in a panel of RNA preparations comprising 14 adult tissues, the expression of Suv39h2 remains largely restricted to the testes, with mRNAs being present as 2.7- and 1.7-kb transcripts (Fig. 5A, middle panel). In addition to other tissues, Suv39h2 transcripts are also significantly downregulated in ovaries.
To analyze this testis-specific expression in more detail, we performed in situ hybridizations on sagittal sections of adult testes. The Suv39h2 and Suv39h1 antisense probes revealed specific expression in the outermost cell layer of the seminiferous tubules (Fig. 5B, right panels), whereas the corresponding control sense probes proved negative. Suv39h2-specific transcripts appear at elevated levels compared to Suv39h1. Higher magnification (see insets in Fig. 5B) shows predominant staining of type B spermatogonia and preleptotene spermatocytes. Suv39h2-specific transcripts are also detected at reduced levels in several pachytene-stage cells and in mitotically inactive Sertoli cells (data not shown). Together, these data indicate prominent expression of Suv39h2 transcripts in male germ cells during the early stages of spermatogenesis.
Detection and size of the endogenous Suv39h2 protein.
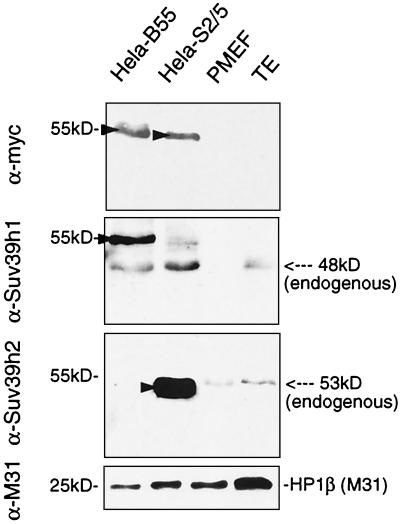
To characterize the Suv39h2 protein at a biochemical level, we generated a Suv39h2-specific polyclonal rabbit antiserum (see Materials and Methods) and probed protein blots containing nuclear extracts from PMEFs and from adult testes with affinity-purified α-Suv39h2 antibodies. As a specificity and size control, we included nuclear extracts from HeLa cell lines that “stably” overexpress (myc)3-SUV39H1 (HeLa-B55) (30) or a corresponding (myc)3-Suv39h2 construct which encodes amino acids 83 to 477 of the Suv39h2 cDNA (HeLa-S2/5) (see Materials and Methods). Immunoblotting with α-Suv39h1 antibodies indicated the presence of ectopic (myc)3-SUV39H1 (55 kDa) and of endogenous SUV39H1 (48 kDa) in HeLa-B55 nuclear extracts. However, endogenous Suv39h1 was undetectable in PMEFs and detectable only at low-abundant levels in testes (Fig. 6, middle panel). By contrast, the α-Suv39h2 antibodies recognize an endogenous protein of ∼53 kDa in both PMEFs and testes (Fig. 6, lower panel), which comigrates with ectopic (myc)3-Suv2(83–477) in HeLa-S2/5 nuclear extracts. We conclude that Suv39h2 is more highly expressed in PMEFs and in testes than is Suv39h1 and that the size of the endogenous Suv39h2 protein is in good agreement with the gene product predicted from the coding sequence of the Suv39h2 cDNA (see Fig. 1).
FIG. 6.
Detection and size of endogenous Suv39h2 protein. Approximately 30 μg of nuclear extracts from HeLa-B55, HeLa-S2/5, PMEFs, and adult testis (TE) were immunoblotted with α-myc, α-Suv39h1, α-Suv39h2, and α-M31 (as a loading control) antibodies. HeLa-B55 cells overexpress (myc)3-SUV1(3–412), and HeLa-S2/5 cells overexpress (myc)3-Suv2(83–477). Ectopic proteins are indicated by arrowheads. Endogenous Suv39h2 (53 kDa) almost comigrates with (myc)3-Suv2(83–477).
Suv39h2 is a second H3 Lys9 HMTase.
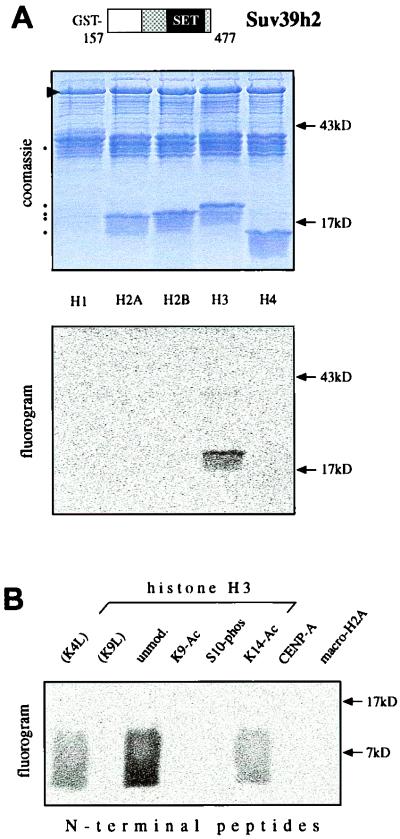
SU(VAR)3-9 related proteins were recently shown to be novel histone H3 methyltransferases. Although Suv39h1 selectively methylates Lys9 of the histone H3 N terminus, a weak signal was also detected if histone H1 was used as a substrate (36). To compare the HMTase specificity of Suv39h2, we performed in vitro methylation assays with free histones and a recombinant GST-Suv39h2 product that comprises amino acids 157 to 477. Purified GST-Suv2(157–477) only methylated histone H3, but not H2A, H2B, or H4 (Fig. 7A). Notably, histone H1 was also not a substrate. To investigate the methylation site profile of Suv39h2, we extended the in vitro methylation assays with unmodified, modified, or mutated H3 N-terminal peptides. Whereas the unmodified H3 peptide was strongly methylated, mutation of Lys9 (K9L) abolished substrate specificity (Fig. 7B). Further, preexisting acetylation of Lys9 (K9-Ac) or phosphorylation of serine 10 (S10-phos) prevented Suv39h2-dependent methylation, and acetylation of lysine 14 (K14-Ac), like mutation of lysine 4 (K4L), significantly reduced the H3 substrate quality. The H3 variant CENP-A has been shown to be retained in sperm chromatin (34), and macroH2A is a new component of the XY body (24). However, the Suv39h2 HMTase did not react with the CENP-A and macroH2A N-terminal peptides (Fig. 7B). Together, these data define Suv39h2 as a second H3 (Lys9) selective HMTase, whose substrate specificity and sensitivity to preexisting H3 tail modifications appears even more stringent than that of Suv39h1.
FIG. 7.
Recombinant Suv39h2 is an H3 Lys9-selective HMTase. (A) In vitro HMTase assays with 10 μg of GST-Suv2(157–477) (murine Suv39h2) and free histones, using S-adenosyl-[methyl-14C]-l-methionine as a methyl donor. Coomassie blue staining (top panel) shows purified proteins (arrowhead) and free histones (dots). Fluorography (bottom panel) indicates H3 HMTase activity of GST-Suv2(157–477). (B) In vitro methylation assays using GST-Suv2(157–477) as enzyme and the indicated N-terminal peptides of H3, CENP-A, and macroH2A as substrates.
In situ localization of Suv39h2 and M31 (HP1β) in testes sections.
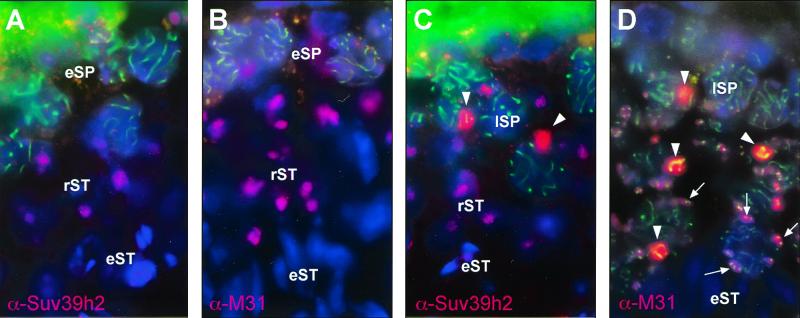
To analyze the distribution and nuclear localization of the Suv39h2 HMTase during spermatogenesis, we performed double-labeling immunofluorescence for Suv39h2 and Scp3 epitopes in testes sections. Scp3 is a major component of the lateral elements of the synaptonemal complex which is formed between homologous chromosomes during meiotic prophase (22). Prominent Suv39h2 signals were visualized in late, but not early, spermatocytes in a subnuclear region that displays a characteristic DAPI staining, reminiscent of the XY body (arrowheads in Fig. 8C). This staining indicated that Suv39h2 has a preference to localize to the sex chromosomes (see below). In addition, Suv39h2 is present at DAPI-dense blocks of heterochromatin (clustered centromeres) in round spermatids but was no longer detectable in elongating spermatids (Fig. 8A and C). To evaluate this staining pattern, we repeated the immunofluorescence with α-Scp3 and α-M31 antibodies (49) that are specific for HP1β, a known interacting partner for SUV39H1 or Suv39h1 proteins in somatic cells (1). As recently described (32), HP1β was detected at the XY body and at clustered centromeres in the nuclear periphery of mid and late spermatocytes (arrows in Fig. 8D). Moreover, HP1β also decorated the heterochromatic blocks of round spermatids. Thus, this comparative analysis indicates significant overlap in meiotic chromatin association between Suv39h2 and HP1β in different stages of mouse spermato- and spermiogenesis.
FIG. 8.
In situ localization of Suv39h2 and M31 (HP1β) in testis sections. Double-labeling indirect immunofluorescence for Suv39h2 (pink) and Scp3 (green) (A and C) or for M31 (pink) and Scp3 (green) (B and D) in testis sections representing different stages of spermatogenesis. DNA was counterstained with DAPI. The positions of early and late spermatocytes (eSP and lSP), round spermatids (rST), and elongating spermatids (eST) are shown. Staining of the XY body in lSP is indicated by arrowheads. Also highlighted by arrows in panel D is the concentration of M31 around clustered centromeres in the nuclear periphery of lSP and rST.
Dynamic heterochromatin association of Suv39h2 in male germ cells.
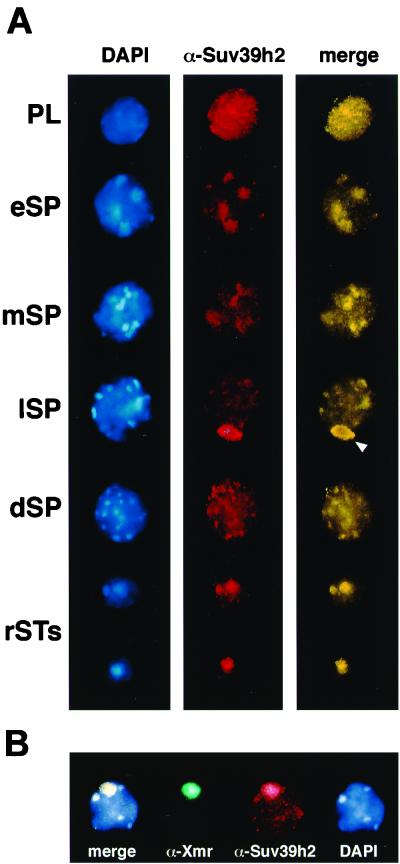
To investigate chromosomal associations of Suv39h2 in more detail, we analyzed its distribution in structurally preserved testis suspension cells (see Materials and Methods). Endogenous Suv39h2 is found in a dispersed distribution in some premeiotic nuclei (data not shown) and as a granular stain in all preleptotene nuclei (Fig. 9A, PL). During the development of leptotene to diplotene spermatocytes (eSP to dSP), Suv39h2 staining is weakly but distinctly apparent at blocks of heterochromatin, as visualized by the bright DAPI counterstaining. Prominent Suv39h2 signals accumulate at the sex chromosomes present in the XY body during the mid to late pachytene stage (lSP, arrowhead). After the meiotic divisions, Suv39h2 remains enriched at the condensing heterochromatic foci and chromocenters of haploid spermatids (rSTs) but is no longer detectable in mature sperm (data not shown).
FIG. 9.
Dynamic heterochromatin association of Suv39h2 in male germ cells. (A) Indirect immunofluorescence of testis suspension cells with α-Suv39h2 antibodies (red). DNA was counterstained with DAPI (blue). Staging of individual mouse spermatogenic cells was determined as described in Materials and Methods and comprised preleptotene spermatogonia (PL); early, middle, and late spermatocytes (eSP, mSP, and lSP); diplotene spermatocytes (dSP); and round spermatids (rST). Merged images are artificially colored yellow. (B) Double-labeling indirect immunofluorescence for Suv39h2 (red) and Xmr (green) in late-pachytene spermatocytes of adult testis suspension cells. DNA was counterstained with DAPI (blue). The XY body (sex vesicle) in panel A is indicated by an arrowhead.
To demonstrate the specific accumulation of Suv39h2 with the XY body, we performed double immunofluorescence analyses for Suv39h2 and Xmr, which selectively associates with the axes and chromatin of sex chromosomes (12). The results of these analyses show that Suv39h2 colocalizes with Xmr at the XY body in 48% of evaluated spermatocytes (n = 194) containing a prominent Xmr staining of the sex chromosomes (Fig. 9B). Whereas immunolocalization of Suv39h2 at the XY body and at the chromocenters of haploid spermatids can be observed by several staining techniques, the visualization of Suv39h2 at heterochromatin in developing spermatocytes requires triple labeling (see Materials and Methods). Despite these variations in the detection sensitivity, our data indicate a dynamic heterochromatin distribution for Suv39h2 during most stages of spermato- and spermiogenesis. By contrast, in parallel analyses of testes swab preparations with α-Suv39h1 antibodies, immunolocalization of Suv39h1 only revealed very weak labeling that did not show a preference for heterochromatin (data not shown).
DISCUSSION
Murine Suv39h genes are encoded by two loci.
Using sequence information from isolated cDNAs, ESTs, and genomic sequences, we defined Suv39h2 as a novel gene encoding a protein of 477 amino acids (Fig. 1). The size and authenticity of the Suv39h2 gene product has been confirmed by immunodetection of a 53-kDa endogenous protein in nuclear extracts of PMEFs and testis (see Fig. 6). RNA blot analyses (see Fig. 4A and data not shown) indicate that the most abundant Suv39h2 transcripts are 2.7 kb, suggesting that the Suv39h2 mRNA is composed of 1.5-kb coding and 1.2-kb 3′-untranslated sequences. In addition, smaller-sized transcripts of 1.7 kb are also present at day E10.5 of mouse embryogenesis and in adult testis. Although an alternative short exon (preceding the starting ATG by ∼260 bp and encoding the amino acids MASDLRT-) can be predicted from the genomic sequence, both the 1.7- and the 2.7-kb transcripts hybridize with exon 1 sequences (data not shown). Since we failed to detect endogenous proteins distinct from the 53-kDa Suv39h2 gene product, the smaller-sized transcripts do not appear to give rise to a largely different Suv39h2 isoform.
From all available sequence information, similarity searches against EST databases and reduced stringency hybridizations (data not shown), murine Suv39h proteins appear to be encoded by no more than two gene loci. Using FISH and haplotype analysis, we mapped the Suv39h2 locus to the subcentromeric region of mouse chromosome 2 (Fig. 2 and 3). Our localization data characterizes Suv39h2 as one of the most proximal gene markers on mouse chromosome 2 and would predict a syntenic position for SUV39H2 on human chromosome 10p13-p15. Within this region, loss of heterozygosity has been correlated with human gliomas (26). In contrast, Suv39h1 resides at the tip of the X chromosome in the immediate vicinity of the mouse mutations Td (13) and Sf (39). However, the recent correlation of mutations in a gene encoding a sterol isomerase with the Td phenotype (16) and the absence of apparent alterations for Suv39h1 in Sf/Y mice (10) would indicate that Suv39h1 is nonallelic to Td or Sf.
Chromatin association of Suv39h2 in somatic versus meiotic cells.
In our previous studies, we identified Suv39h1 and SUV39H1 as heterochromatic proteins that associate with centromeric positions of metaphase chromosomes (1, 2). Although Suv39h2 can be detected in some mammalian cell lines (data not shown) and in PMEFs (see Fig. 6), we failed to visualize endogenous Suv39h2 at heterochromatic foci or at mitotic chromosomes in these cells. By contrast, transient expression of (myc)3-Suv2(83–477) in murine Cop8 cells results in preferred heterochromatic localizations at a low abundance of ectopic protein (data not shown). This observation suggests that the highly basic N terminus is not required for the intrinsic chromatin affinity of Suv39h2 but may modulate chromosomal associations in somatic versus meiotic chromatin. This interpretation is supported by the apparent preference of Suv39h2 for heterochromatin in pachytene spermatocytes and round spermatids (see Fig. 9), whereas Suv39h2 displays a rather broad nuclear staining in Sertoli cells (data not shown). Because SUV39H1 has been shown to be a phosphoprotein with mitosis-specific isoforms (2) which appears further regulated by the anti-phosphatase Sbf1 (18), it is likely that phosphorylation-dependent modifications could also contribute to the distinct distributions of Suv39h2 in mitotic and meiotic chromatin.
Suv39h HMTases and histone H3 tail modifications.
Suv39h-dependent methylation of Lys9 in the histone H3 N terminus has been shown to modulate chromatin dynamics in somatic cells, in part by interfering with the phosphorylation of adjacent serine 10 (phosH3) (36), a modification required for the condensation and segregation of chromosomes (48). Moreover, heterochromatin association of HP1 is perturbed upon forced expression of SUV39H1 in HeLa cells (30), and methylation of Lys9 in H3 generates a high-affinity binding site for HP1 proteins in native chromatin of PMEFs (M. Lachner, D. O'Carroll, S. Rea, K. Mechtler, and T. Jenuwein, submitted for publication). Similarly, overexpression of (myc)3-Suv2(83–477) in HeLa cells also redistributes endogenous HP1β (data not shown). Suv39h2 is an H3 Lys9-selective HMTase, whose substrate specificity and sensitivity to preexisting H3 tail modifications appears to be even more stringent than that of Suv39h1 (see Fig. 7). These findings indicate that both Suv39h HMTases can transduce the H3 Lys9 methylation mark into an important epigenetic signal for the induction and assembly of mammalian heterochromatin in somatic cells.
A role for the Suv39h2 HMTase in the male germ line?
Dynamic transitions in chromatin structure are particularly important during male meiosis (see the introduction), where heterochromatinization has been proposed to be involved in the progressive centromere clustering of chromosomes (31). Because phosH3 also defines centromeric heterochromatin in meiosis (14), Suv39h2 could modulate the timing and/or degree of H3 phosphorylation, thereby influencing chromosome alignments during the meiotic divisions. In addition to such a centromeric model, meiotic chromatin associations of the Suv39h2 HMTase extend to the spermatid stage and significantly overlap with Hp1β (see Fig. 8), suggesting that Suv39h2-induced alterations could further contribute to the dense packaging of chromatin in elongating spermatids. HP1β has recently been shown to be retained in mature spermatozoa by protein blot analysis (23). Similarly, ∼15% of human sperm chromatin remains complexed with histones (19). Although the presence of Suv39h2 in mature sperm is currently undefined, its highly basic N terminus could facilitate associations with condensing chromatin in elongating and elongated spermatids, when histones are replaced by protamines (6).
Most interestingly, Suv39h2 and HP1β (32) accumulate with the silenced sex chromosomes present in the XY body (Fig. 8 and 9). This finding is particularly intriguing, since SUV39H1 (18) and HP1β (33, 40) have been shown to repress gene activity in somatic cells (18). Because of the high conservation between mammalian Suv39h proteins (Fig. 1), these data imply that Suv39h2 would represent an important regulator to induce silenced chromatin domains at selective chromosomes during spermatogenesis. Based on the results presented here, we even propose that the Suv39h2 HMTase could impart an epigenetic imprint to the male germ line.
ACKNOWLEDGMENTS
We thank Gotthold Schaffner, Robert Kurzbauer, and Ivan Botto for sequence analysis and oligonucleotide synthesis; Alexander Schleiffer for database searches on the C. elegans C15H11.5 ORF; Meinrad Busslinger for advice on the exon/intron organization of the genomic Suv39h2 clone; Jean-Louis Guénet (Pasteur Institute, Paris, France) for his gift of WMP mice; Cécile Mignon-Ravix for help in the FISH analysis; Prim B. Singh (The Roslin Institute, Roslin, Midlothian, United Kingdom) for α-M31 (HP1β) antibodies; Christa Heyting (Wageningen, The Netherlands) for α-Scp3 antibodies; H.-J. Garchon (Hopital Necker, Paris, France) for α-Xmr antibodies; and Karl Mechtler for Suv39h2 antiserum purification.
This work was supported by the IMP through Boehringer Ingelheim and by grants from by the Austrian Research Promotion Fund to T.J., the Deutsche Forschungsgemeinschaft (grant number DFG #350/8-2) to H.S., the Medical Research Council (United Kingdom) to S.D.M.B., and the ARC (Association pour la Recherche sur le Cancer) to M.G.M.
REFERENCES
- 1.Aagaard L, Laible G, Selenko P, Schmid M, Dorn R, Schotta G, Kuhfittig S, Wolf A, Lebersorger A, Singh P B, Reuter G, Jenuwein T. Functional mammalian homologues of the Drosophila PEV modifier Su(var)3-9 encode centromere-associated proteins which complex with the heterochromatin component M31. EMBO J. 1999;18:1923–1938. doi: 10.1093/emboj/18.7.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aagaard L, Schmid M, Warburton P, Jenuwein T. Mitotic phosphorylation of SUV39H1, a novel component of active centromeres, coincides with transient accumulation at mammalian centromeres. J Cell Sci. 2000;113:817–829. doi: 10.1242/jcs.113.5.817. [DOI] [PubMed] [Google Scholar]
- 3.Allshire R C, Nimmo E R, Ekwall K, Javerzat J P, Granston G. Mutations derepressing silent centromeric domains in fission yeast disrupt chromosome segregation. Genes Dev. 1995;9:218–233. doi: 10.1101/gad.9.2.218. [DOI] [PubMed] [Google Scholar]
- 4.Altschul S F, Madden T L, Schäffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3398–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ayoub N, Richler C, Wahrman J. Xist RNA is associated with the transcriptionally inactive XY body in mammalian male meiosis. Chromosoma. 1997;106:1–10. doi: 10.1007/s004120050218. [DOI] [PubMed] [Google Scholar]
- 6.Ballhorn R, Weston S, Thomas C, Wyrobek A J. DNA packaging in mouse spermatids: synthesis of protamine variants and four transition proteins. Exp Cell Res. 1984;150:298–308. doi: 10.1016/0014-4827(84)90572-x. [DOI] [PubMed] [Google Scholar]
- 7.Basset D R, Jr, Boguski M S, Spencer F, Reeves R, Goebl M, Hieter P. Comparative genomics, genome cross-referencing and XREFdb. Trends Genet. 1995;11:372–373. doi: 10.1016/s0168-9525(00)89109-x. [DOI] [PubMed] [Google Scholar]
- 8.Breen M, Deakin L, Macdonald B, Miller S, Sibson R, Tarttelin E, et al. Towards high resolution maps of the mouse and human genomes—a facility for ordering markers to 0.1 cM resolution. Hum Mol Genet. 1994;3:621–627. [PubMed] [Google Scholar]
- 9.Brown S W. Heterochromatin. Science. 1966;151:417–425. doi: 10.1126/science.151.3709.417. [DOI] [PubMed] [Google Scholar]
- 10.Bultman S, Magnuson T. Molecular and genetic analysis of the mouse homolog of the Drosophila suppressor of position-effect variegation 3-9 gene. Mamm Genome. 2000;11:251–254. doi: 10.1007/s003350010049. [DOI] [PubMed] [Google Scholar]
- 11.Bunick D, Johnson P A, Johnson T R, Hecht N B. Transcription of the testis-specific mouse protamine 2 gene in a homologous in vitro system. Proc Natl Acad Sci USA. 1990;87:891–895. doi: 10.1073/pnas.87.3.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Calenda A B, Allenet D, Escalier D, Bach J F, Garchon H-J. The meiosis-specific Xmr gene product is homologous to the lymphocyte Xlr protein and is a component of the XY body. EMBO J. 1994;13:100–109. doi: 10.1002/j.1460-2075.1994.tb06239.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cattanach B M. A new X-linked mutation, Td. Mouse News Lett. 1982;66:61–62. [Google Scholar]
- 14.Cobb J, Miyaike M, Kikuchi A, Handel M A. Meiotic events at the centromeric heterochromatin: histone H3 phosphorylation, topoisomerase IIa localization and chromosome condensation. Chromosoma. 1999;108:412–425. doi: 10.1007/s004120050393. [DOI] [PubMed] [Google Scholar]
- 15.Dernburg A F, Sedat J W, Hawley R S. Direct evidence for a role of heterochromatin in meiotic chromosome segregation. Cell. 1996;86:135–146. doi: 10.1016/s0092-8674(00)80084-7. [DOI] [PubMed] [Google Scholar]
- 16.Derry J M, Gormally E, Means G D, Zhao W, Meindl A, Kelley R I, et al. Mutations in a delta 8-delta 7 sterol isomerase in the tattered mouse and X-linked dominant chondrodysplasia punctata. Nat Genet. 1999;3:286–290. doi: 10.1038/10350. [DOI] [PubMed] [Google Scholar]
- 17.Dugaiczyk A, Haron J A, Stone E M, Dennison O E, Rothbum K N, Schwartz R J. Cloning and sequencing of a deoxyribonucleic acid copy of glyceraldyhde-3-phosphate dehydrogenase messenger ribonucleic acid isolated from chicken muscle. Biochemistry. 1983;22:1605–1613. doi: 10.1021/bi00276a013. [DOI] [PubMed] [Google Scholar]
- 18.Firestein R, Cui X, Huie P, Cleary M L. SET domain-dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila SU(VAR)3-9. Mol Cell Biol. 2000;20:4900–4909. doi: 10.1128/mcb.20.13.4900-4909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gusse M, Sautiere P, Bélaiche D, Martinage A, Roux C, Dadoune J-P, Chevaillier P. Purification and characterization of nuclear basic proteins of human sperm. Biochim Biophys Acta. 1986;884:124–134. doi: 10.1016/0304-4165(86)90235-7. [DOI] [PubMed] [Google Scholar]
- 20.Geraghty M T, Brody L C, Martin L S, Marble M, Kearns W, Pearson P, Monaco A P, Lehrach H, Valle D. The isolation of cDNAs from OATL1 at Xp11.2 using a 480-kb YAC. Genomics. 1993;16:440–446. doi: 10.1006/geno.1993.1208. [DOI] [PubMed] [Google Scholar]
- 21.Handel M A, Hunt P A. Sex-chromosome pairing and activity during mammalian meiosis. Bioessays. 1992;12:817–822. doi: 10.1002/bies.950141205. [DOI] [PubMed] [Google Scholar]
- 22.Heyting C, Dettmers R J, Dietrich A J J, Redeker E J W, Vink A C G. Two major components of synaptonemal complexes are specific for meiotic prophase nuclei. Chromosoma. 1988;96:325–332. doi: 10.1007/BF00286921. [DOI] [PubMed] [Google Scholar]
- 23.Hoyer-Fender S, Singh P B, Motzkus D. The murine heterochromatin protein M31 is associated with the chromocenter of round spermatids and is a component of mature spermatozoa. Exp Cell Res. 2000;254:72–79. doi: 10.1006/excr.1999.4729. [DOI] [PubMed] [Google Scholar]
- 24.Hoyer-Fender S, Costanzi C, Pehrson J R. Histone macroH2A1.2 is concentrated in the XY-body by the early pachytene stage of spermatogenesis. Exp Cell Res. 2000;258:254–260. doi: 10.1006/excr.2000.4951. [DOI] [PubMed] [Google Scholar]
- 25.Ivanova A V, Bonaduce M J, Ivanov S V, Klar A J S. The chromo and SET domains of the Clr4 protein are essential for silencing in fission yeast. Nat Genet. 1998;19:192–195. doi: 10.1038/566. [DOI] [PubMed] [Google Scholar]
- 26.Kimmelman A C, Ross D A, Liang B C. Loss of heterozygosity of chromosome 10p in human gliomas. Genomics. 1996;34:250–254. doi: 10.1006/geno.1996.0277. [DOI] [PubMed] [Google Scholar]
- 27.Kralewski M, Novello A, Benavente R. A novel Mr 77,000 protein of the XY body of mammalian spermatocytes: its localisation in normal animals and in Searle's translocation carriers. Chromosoma. 1997;106:160–167. doi: 10.1007/s004120050235. [DOI] [PubMed] [Google Scholar]
- 28.Laible G, Haynes A R, Lebersorger A, O'Carroll D, Mattei M-G, Denny P, Brown S D M, Jenuwein T. The murine Polycomb-group genes Ezh1 and Ezh2 map close to Hox gene clusters on mouse chromosomes 11 and 6. Mamm Genome. 1999;10:311–314. doi: 10.1007/s003359900993. [DOI] [PubMed] [Google Scholar]
- 29.Marahrens Y, Panning B, Dausmann J, Strauss W, Jaenisch R. Xist-deficient mice are defective in dosage compensation but not spermatogenesis. Genes Dev. 1997;11:156–166. doi: 10.1101/gad.11.2.156. [DOI] [PubMed] [Google Scholar]
- 30.Melcher M, Schmid M, Aagaard L, Selenko P, Laible G, Jenuwein T. Structure-function analysis of SUV39H1 reveals a dominant role in heterochromatin organization, chromosome segregation and mitotic progression. Mol Cell Biol. 2000;20:3728–3741. doi: 10.1128/mcb.20.10.3728-3741.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer-Ficca M, Müller-Navia J, Scherthan H. Clustering of pericentromeres initiates in step 9 of spermiogenesis of the rat (Rattus norvegicus) and contributes to a defined genome architecture in the sperm nucleus. J Cell Sci. 1998;111:1363–1370. doi: 10.1242/jcs.111.10.1363. [DOI] [PubMed] [Google Scholar]
- 32.Motzkus D, Singh P B, Hoyer-Fender S. M31, a murine homolog of Drosophila HP1, is concentrated in the XY body during spermatogenesis. Cytogenet Cell Genet. 1999;86:83–88. doi: 10.1159/000015418. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen A L, Ortiz J A, You J, Oulad-Abdelghani M, Khechumian R, Gansmuller A, Chambon P, Losson R. Interaction with members of the heterochromatin protein 1 (HP1) family and histone deacetylation are differentially involved in transcriptional silencing by members of the TIF1 family. EMBO J. 1999;18:6385–6395. doi: 10.1093/emboj/18.22.6385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Palmer D K, O'Day K, Margolis R L. The centromere specific histone CENP-A is selectively retained in discrete foci in mammalian sperm nuclei. Chromosoma. 1990;100:32–36. doi: 10.1007/BF00337600. [DOI] [PubMed] [Google Scholar]
- 35.Pandita T K, Westphal C H, Anger M, Sawant S G, Géard C R, Pandita R K, Scherthan H. Atm inactivation results in aberrant telomere clustering during meiotic prophase. Mol Cell Biol. 1999;19:5096–5105. doi: 10.1128/mcb.19.7.5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rea S, Eisenhaber F, O'Carroll D, Strahl B D, Sun Z-W, Schmid M, Opravil S, Mechtler K, Ponting C P, Allis C D, Jenuwein T. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature. 2000;406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 37.Reuter G, Spierer P. Position-effect variegation and chromatin proteins. Bioessays. 1992;14:605–612. doi: 10.1002/bies.950140907. [DOI] [PubMed] [Google Scholar]
- 38.Rhodes M, Straw R, Fernando S, Evans A, Lacey T, Dearlove A, et al. A high resolution microsatellite map of the mouse genome. Genome Res. 1998;8:531–542. doi: 10.1101/gr.8.5.531. [DOI] [PubMed] [Google Scholar]
- 39.Russell W L, Russell L B, Gower J S. Exceptional inheritance of a sex-linked gene in the mouse explained on the basis that the X/O sex-chromosome constitution is female. Proc Natl Acad Sci USA. 1959;45:554–560. doi: 10.1073/pnas.45.4.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryan F R, Schultz D C, Ayyanathan K, Singh P B, Friedman J R, Fredericks W J, Rauscher F J., III KAP-1 corepressor protein interacts and colocalizes with heterochromatic and euchromatic HP1 proteins: a potential role for Krüppel-associated box-zinc finger proteins in heterochromatin-mediated gene silencing. Mol Cell Biol. 1999;19:4366–4378. doi: 10.1128/mcb.19.6.4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory, Press; 1989. [Google Scholar]
- 42.Scherthan H, Weich S, Schwegler H, Härle M, Heyting C, Cremer T. Centromere and telomere movements during early meiotic prophase of mouse and man are associated with the onset of chromosome pairing. J Cell Biol. 1996;134:1109–1125. doi: 10.1083/jcb.134.5.1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Smith A, Benavente R. An Mr 51,000 protein of mammalian spermatogenic cells that is common to the whole XY body and centromeric heterochromatin of autosomes. Chromosoma. 1995;103:591–596. doi: 10.1007/BF00357685. [DOI] [PubMed] [Google Scholar]
- 44.Solari A J. The behaviour of the XY pair in mammals. Int Rev Cytol. 1974;38:273–317. doi: 10.1016/s0074-7696(08)60928-6. [DOI] [PubMed] [Google Scholar]
- 45.Strahl B D, Allis C D. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 46.Tschiersch B, Hofmann A, Krauss V, Dorn R, Korge G, Reuter G. The protein encoded by the Drosophila position-effect variegation suppressor gene Su(var)3-9 combines domains of antagonistic regulators of homeotic gene complexes. EMBO J. 1994;13:3822–3831. doi: 10.1002/j.1460-2075.1994.tb06693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wallrath L L. Unfolding the mysteries of heterochromatin. Curr Opin Genet Dev. 1998;8:147–153. doi: 10.1016/s0959-437x(98)80135-4. [DOI] [PubMed] [Google Scholar]
- 48.Wei Y, Lanlan Y, Bowen J, Gorovsky M A, Allis C D. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- 49.Wreggett K A, Hill F, James P S, Hutchings G W, Singh P B. A mammalian homologue of Drosophila heterochromatin protein 1 (HP1) is a component of constitutive heterochromatin. Cytogenet Cell Genet. 1994;66:99–103. doi: 10.1159/000133676. [DOI] [PubMed] [Google Scholar]