Abstract
Tandem-duplication mutations of the UBTF gene (UBTF-TDs) coding for the upstream binding transcription factor have recently been described in pediatric patients with acute myeloid leukemia (AML) and were found to be associated with particular genetics (trisomy 8 (+8), FLT3-internal tandem duplications (FLT3-ITD), WT1-mutations) and inferior outcome. Due to limited knowledge on UBTF-TDs in adult AML, we screened 4247 newly diagnosed adult AML and higher-risk myelodysplastic syndrome (MDS) patients using high-resolution fragment analysis. UBTF-TDs were overall rare (n = 52/4247; 1.2%), but significantly enriched in younger patients (median age 41 years) and associated with MDS-related morphology as well as significantly lower hemoglobin and platelet levels. Patients with UBTF-TDs had significantly higher rates of +8 (34% vs. 9%), WT1 (52% vs. 7%) and FLT3-ITD (50% vs. 20.8%) co-mutations, whereas UBTF-TDs were mutually exclusive with several class-defining lesions such as mutant NPM1, in-frame CEBPAbZIP mutations as well as t(8;21). Based on the high-variant allele frequency found and the fact that all relapsed patients analyzed (n = 5) retained the UBTF-TD mutation, UBTF-TDs represent early clonal events and are stable over the disease course. In univariate analysis, UBTF-TDs did not represent a significant factor for overall or relapse-free survival in the entire cohort. However, in patients under 50 years of age, who represent the majority of UBTF-mutant patients, UBTF-TDs were an independent prognostic factor for inferior event-free (EFS), relapse-free (RFS) and overall survival (OS), which was confirmed by multivariable analyses including established risk factors such as age and ELN2022 genetic risk groups (EFS [HR: 2.20; 95% CI 1.52–3.17, p < 0.001], RFS [HR: 1.59; 95% CI 1.02–2.46, p = 0.039] and OS [HR: 1.64; 95% CI 1.08–2.49, p = 0.020]). In summary, UBTF-TDs appear to represent a novel class-defining lesion not only in pediatric AML but also younger adults and are associated with myelodysplasia and inferior outcome in these patients.
Subject terms: Genetics research, Acute myeloid leukaemia
Introduction
The upstream binding transcription factor (UBF or UBTF), encoded by the UBTF gene located on chromosome 17, is a member of the high mobility group (HMG)‑box protein family, a group of ubiquitously expressed non-histone architectural proteins (reviewed in ref. [1]). UBTF is a key regulator of ribosomal RNA transcription, mediating the recruitment of RNA polymerase I to rDNA promoter regions through the formation of nucleosome free regions [2] and the assembly of the preinitiation complex [3, 4]. UBTF is expressed in two variant isoforms as the result of differential splicing, with the shorter variant UBTF2 lacking a segment of HMG-Box 2 [5]. Recent data suggest that the UBTF1/2 ratio regulates the rate of rRNA synthesis and determines the sensitivity of rRNA genes to growth factor stimulation in different cell types [6]. Both Germline and somatic genomic aberrations of UBTF have been linked to several diseases, including childhood neurodegenerative disorders [7, 8], solid tumors such as melanoma [9] or colorectal cancer [10], and hematopoietic malignancies such as precursor acute lymphoblastic leukemia [11]. Recently, tandem duplications in exon 13 of UBTF (UBTF-TDs) were identified as novel, recurrent alterations in newly diagnosed and relapsed cases of pediatric acute myeloid leukemia (AML) by a whole genome sequencing approach [12–14]. In children, UBTF-TDs were associated with poor outcome and a distinct genetic profile, including high rates of trisomy 8 (+8), FLT3-internal tandem duplications (FLT3-ITDs) and WT1-mutations. To date, data on the role of UBTF-TDs in adults are limited, prompting us to investigate the prevalence and prognostic impact of UBTF-TDs in a large and well characterized cohort of adult AML patients.
Patients, materials, and methods
Patients
A total of 4247 adult patients with newly diagnosed AML (n = 3300 de novo AML, n = 796 secondary or therapy-related AML) or higher-risk myelodysplastic syndrome (HR-MDS, n = 151) were retrospectively screened for the presence of UBTF-TDs. Patients with appropriate genetic data (n = 1456) were re-stratified according to ELN2022 [15] and included in the outcome analyses. Most of these patients (n = 1265) were treated in prospective studies involving risk stratified post induction therapy according to cytogenetic risk groups, i. e. the AML96, AML2003, AML60+ and SORAML protocols of the Study Alliance Leukemia (SAL); the remaining patients (n = 191) were recruited to the SAL registry. Detailed treatment protocols have been published previously [16–19] and are summarized in the supplement, including the number of patients treated in each protocol (Table S1). This study was approved by the ethical board of the Medical Faculty TU Dresden. Each patient gave written informed consent to participate in the respective studies.
Patient samples
All materials investigated were obtained at the time of diagnosis. Bone marrow was used whenever available, in all other cases, peripheral blood samples were examined. Genomic DNA was extracted from mononuclear cells using standard procedures (DNA Blood mini kit, Qiagen, Hilden, Germany).
Mutational analysis of UBTF
UBTF-TD screening was done by PCR covering exon 13 of the UBTF gene with 6-FAM labeled primers followed by high-resolution fragment analysis. PCR amplified mutant samples were purified and sequenced on an ABI3130xl instrument (Life Technologies, Darmstadt, Germany). Details of the PCR primers and cycling conditions are given in the supplement (Table S2 and S3).
Next generation sequencing (NGS)-based characterization of co-mutations in UBTF mutant patients
Profiling of mutations in UBTF mutant samples was done by targeted NGS-based resequencing using the Archer VariantPlex Myeloid panel (Illumina, Chesterford, UK) covering 75 genes frequently mutated in AML as described recently [20]. Samples were sequenced paired-end (150 bp PE) on NextSeq- (Illumina) or (300 bp PE) MiSeq-NGS platforms. Sequence data alignment of demultiplexed FastQ files, variant calling and filtering was done using the Sequence Pilot software package (JSI medical systems GmbH, Ettenheim, Germany) with default settings and a 5% variant allele frequency (VAF) mutation calling cut-off.
RNA-sequencing
RNA sequencing was performed on total RNA isolated at diagnosis from 7 AML samples from UBTF-TDpos patients and 42 samples from other well established AML subgroups (t(8;21), t(6;9); inv16, NPM1 and in-frame CEBPAbZIP mutant patients, and patients with a NUP98-NSD1 fusion) using strand-specific RNA-Seq library preparation (Ultra II Directional RNA Library Prep, NEB) and sequenced on an Illumina NovaSeq 6000 instrument. The complete workflow as well as the bioinformatic analysis are detailed in the supplement.
Statistical analysis
All statistical analyses were performed using R version 4.2.0 (https://www.R-project.org/.) and STATA BE 17.0 (Stata Corp, College Station, TX, USA). All analyses were carried out as two‑sided tests. Statistical significance was determined using a significance level α of 0.05. Clinical variables across groups were compared using the Fisher’s exact test for categorial variables, the nonparametric Mann-Whitney U test was applied for continuous variables. p-values of association analyses of UBTF mutations with clinical variables and other molecular abnormalities were adjusted for multiplicity using the Bonferroni-Holm-procedure. With regard to outcome analysis, univariate analysis was carried out using logistic regression to obtain odds ratios (OR). Time-to-event analysis was performed using Cox-proportional hazard models to obtain hazard ratios (HR) as well as the Kaplan–Meier method and the log-rank-test. Multivariable models were adjusted for ELN2022 risk categories [15] and age. Median follow-up time was calculated using the reverse Kaplan–Meier method.
Results
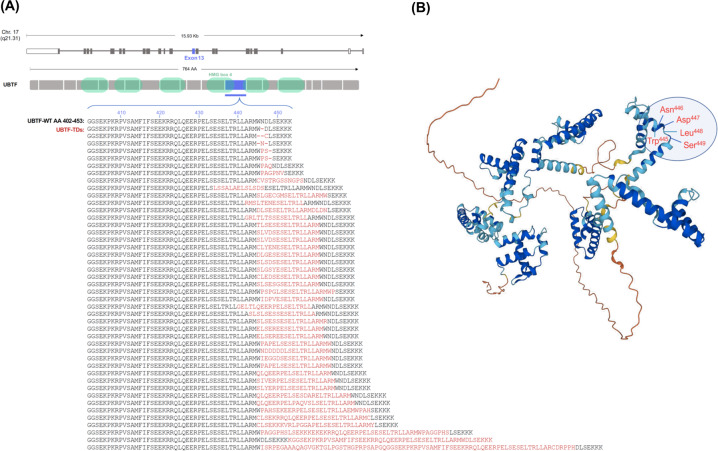
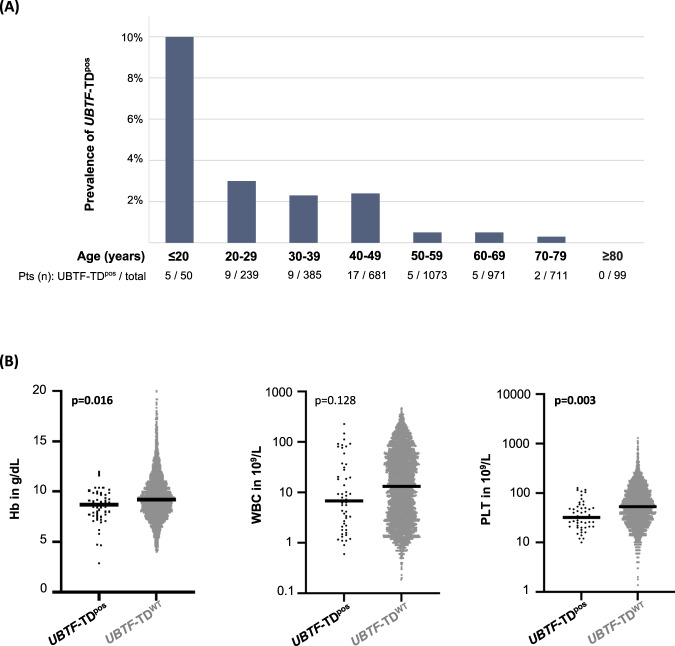
UBTF-TDs were identified in 52 (1.2%) of 4247 patients analyzed. All mutations yielded in-frame insertions, duplications and/or deletions in UBTF exon 13 (median length 51 base pairs; range –6 to +312 bp), specifically affecting the link between the second and third a-helix of HMG-box 4 (Fig. 1). Although UBTF-TDs were rare across the overall cohort, they were considerably more common in younger patients and showed an inverse correlation with age, ranging from 10% in patients below 20 years to 0.25% in patients over 70 years (Fig. 2). Accordingly, the median age was significantly lower in patients with UBTF-TDs (41 years; IQR 28–48.5 years) than in UBTF-TDWT patients (57 years; IQR 46–67 years; p < .001). Based on local evaluation, a high percentage of cases were initially classified as AML M6 at diagnosis, that would qualify as MDS since 2016 [21]. Central cytomorphologic re-assessment of available slides (n = 41) in UBTF-TDpos patients confirmed a strong association with myelodysplastic changes (n = 25/41; 61%; Fig. S1), supporting data in pediatric MDS [22], where UBTF-TDs were found in 25% of patients. This might indicate that in UBTF mutant AML, the disease frequently evolves from preexisting MDS. In line with this, laboratory parameters revealed significantly lower hemoglobin levels and platelet counts at diagnosis for UBTF-TDpos patients compared to UBTF-TDWT patients (median Hb 8.7 vs. 9.2 g/dL; p = 0.02; median PLT 31.5 vs. 53 × 109/L; p = 0.003; Table 1, Fig. 2).
Fig. 1. Illustration of the effect of UBTF tandem duplications on the amino acid level.
A All patients showed typical inframe insertions and/or deletions leading to alterations in the coding sequence of exon 13. B 3D protein structure of UBTF (Source: Alphafold.ebi.ac.uk Sequence AF-P17480-F1). The highlighted amino acids represent the localization hotspots AA 445-449 of the observed InDel mutations.
Fig. 2. Clinical variables of UBTF-TDpos patients.
A Age distribution of the 52 UBTF-TDpos patients. Prevalence of UBTF-TDs shows a strong negative correlation with age. B Peripheral blood counts in UBTF-TDpos vs. UBTF-TDWT patients. Despite their young age, laboratory parameters revealed significantly lower hemoglobin levels and platelet counts at diagnosis for UBTF-TDpos patients.
Table 1.
Clinical and genetic variables in UBTF-TDpos and UBTF-TDWT patients.
| UBTF-TDWT n = 4195 | UBTF-TDpos n = 52 | p-value (adj.) | |
|---|---|---|---|
| Age, years, median (IQR) | 57 (46–67) | 41 (28–48.5) | <0.001 |
| Sex, n (%) | |||
| Female | 2038 (49) | 22 (42) | 0.368 |
| Male | 2157 (51) | 30 (58) | |
| AML type, n (%) | |||
| de novo | 3261 (78) | 39 (75) | 0.080 |
| sAML | 548 (13) | 11 (22) | |
| tAML | 237 (6) | - | |
| HR-MDS | 149 (3) | 2 (3) | |
| Laboratory, median (IQR) | |||
| BM blasts, % | 61 (38–80) | 46.5 (30.5–71) | 0.057 |
| WBC, 109/L | 13.3 (3–41.6) | 6.3 (1.7–20) | 0.128 |
| PLT, 109/L | 53 (25–98) | 32 (21–50) | 0.003 |
| Hb, g/dL | 9.2 (8.0–10.6) | 8.7 (7.5–9.6) | 0.016 |
| FAB subtype, n (%) | - | ||
| M0 | 446 (11) | 4 (8) | <0.001 |
| M1 | 846 (21) | 8 (15) | |
| M2 | 1143 (28) | 14 (27) | |
| M4 | 702 (17) | 6 (12) | |
| M5 | 559 (14) | 4 (8) | |
| M6 | 117 (3) | 12 (23) | |
| M7 | 27 (1) | 1 (2) | |
| RAEB | 88 (2) | 2 (3) | |
| RAEB-T | 138 (3) | 1 (2) | |
| n-miss | 129 | - | |
| Cytogenetic risk ELN 2022, n (%) | |||
| Favorable | 322 (8) | - | 0.070 |
| Intermediate | 2802 (70) | 47 (90) | |
| Adverse | 902 (22) | 3 (10) | |
| n-miss | 169 | 2 | |
| Trisomy 8 | 372 (9) | 19 (36.5) | <0.001 |
| NPM1mut, n (%) | 1176 (28) | - | <0.001 |
| FLT3-ITDpos, n (%) | 872 (20.8) | 26 (50) | <0.001 |
| CEBPAbZIP-inf, n (%) | 157 (3.7) | - | <0.001 |
IQR interquartile range, AML acute myeloid leukemia, sAML secondary AML, tAML therapy-related AML, HR-MDS higher-risk myelodysplastic syndrome, BM bone marrow, WBC white blood cell count, PLT platelet counts, Hb hemoglobin levels, FAB French-American-British, RAEB refractory anemia with excess blasts, RAEB-T refractory anemia with excess blasts in transformation, ELN European Leukemia Network, CR complete remission, OS overall survival, RFS relapse-free survival, alloHSCT allogeneic hematopoietic stem cell transplantation.
Bold values indicates statisically significant p values.
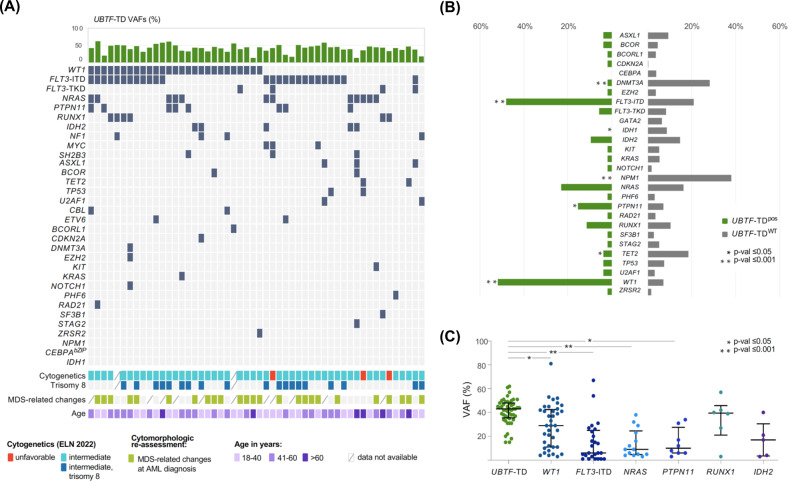
As in pediatric patients [13, 14], there was a significant association with +8 (38% in UBTF-TDpos vs. 9.8% UBTF-TDWT; p < 0.001), 3 patients (6%) showed an adverse karyotype. Similar to findings in children, UBTF-TDs were significantly associated with WT1-mutations (52% vs. 8.6, p < 0.001), FLT3-ITDs (50% vs. 20.8%, p < 0.001) as well as PTPN11-mutations (15.4% vs. 7%; p = 0.022), while DNMT3A (1.9% vs. 28.1%; p < 0.001), TET2 (3.8% vs. 18.4%; p = 0.007) and IDH1-mutations (0% vs. 8.6%; p = 0.027) were significantly less common (Fig. 3A/B, Table S4). Furthermore, UBTF-TDs and several class-defining lesions [23], i.e., reciprocal translocations such as t(8;21), t(6;9) or mutations in CEBPAbZIP or NPM1, were mutually exclusive (Fig. 3A/B). As outlined in Fig. 3C, the variant allele frequency (VAF) of UBTF-TDs (median 43%; range 13-62%) was significantly higher than the VAFs of frequent co-mutations, indicating that the UBTF-TD may represented the earliest clonal event.
Fig. 3. Landscape of co-mutations in UBTF mutant AML and survival analysis according to UBTF mutant status.
A Alignment of additional gene mutations in 52 UBTF-TDpos patients. B Frequency distribution of additional gene mutations identified in UBTF-TDpos vs. UBTF-TDWT patients. C Variant allele frequencies (VAFs) of UBTF-TDs and frequent co-mutations (frequency of at least 10% in UBTF-mut patients). Solid bars indicate VAF median and IQR.
In order to further assess the persistence of UBTF-TDs in patients in complete morphologic remission (CR) we screened DNA from available remission samples in 9 patients using the fragment analysis procedure, which has a sensitivity of about 1%. In 5 of these 9 samples, residual UBTF-TD mutations were still detectable (median VAF 2.7%; range 0.9–4.3%), which is in line with the overall rather low CR rate of 58.8% for these patients.
Relapse samples were available in five patients. In all patients, the initially detected UBTF-TD mutation was also present at the time of relapse. In contrast, the comutational profile showed evidence of profound changes with losses of mutations in NRAS, PTPN11 and FLT3 and acquisition of mutations in NF1, BCOR, WT1, GATA2 and U2AF1 (Fig. S2).
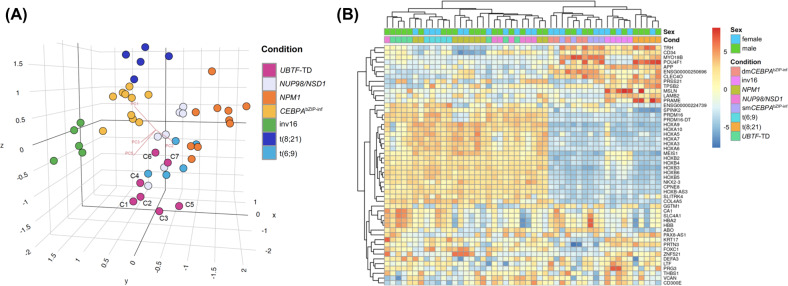
To gain additional insights into the biology of UBTF-TD mutant AML, we performed an RNA-sequencing analysis of 7 AML samples from UBTF-TDpos patients and 42 samples from other well established AML subgroups, i.e., t(8;21), inv16, NPM1 and in-frame CEBPAbZIP mutant patients (single and double), t(6;9) and patients with a NUP98-NSD1 fusion. As shown in Fig. 4A, the principle component analysis (PCA) revealed a clustering of the UBTF-TDpos samples which showed a partial overlap with those of NUP98-NSD1 patients and patients with t(6;9), which is consistent with previous results [13]. A heatmap built on unsupervised clustering based on the top 50 differentially expressed genes showed two major clusters containing the UBTF-TDpos samples, one of which again showed an overlap between the UBTF-TDs and NUP98-NSD1 and t(6;9) patients (Fig. 4B). In line with previous reports [13, 14], UBTF-TDpos samples showed a strong upregulation of several HOX-genes (HOX-A and HOX-B; Fig. 4B, Fig. S3). Interestingly, four of the seven UBTF-TDpos samples showed a strong upregulation of several genes associated with erythroid differentiation (HBA2, HBB, ABO), which might explain the association with the erythroid lineage/FAB M6. Pairwise comparison between UBTF-TD and the other subgroups for the most differentially expressed genes again showed the smallest difference compared with patients with t(6;9) (396 differentially expressed genes (DEGs)) and NUP98-NSD1 fusion (455 DEGs), whereas the largest differences were observed compared with patients with in-frame mutations in CEBPAbZIP (1278 DEGs) and t(8;21) (1424 DEGs) (Fig. S3).
Fig. 4. RNA-sequencing analysis in UBTF mutant AML and other well established AML subgroups.
A Principal component analysis (PCA) of the RNA-Sequencing data. B Heat map of RNA-Sequencing analysis indicates the top 50 differentially expressed genes ranked based on FDR between UBTF-TDpos patients and references, with high levels of expression shown in red and low levels shown in blue. Color coding is based on standardized and normalized read counts accounting for the library size.
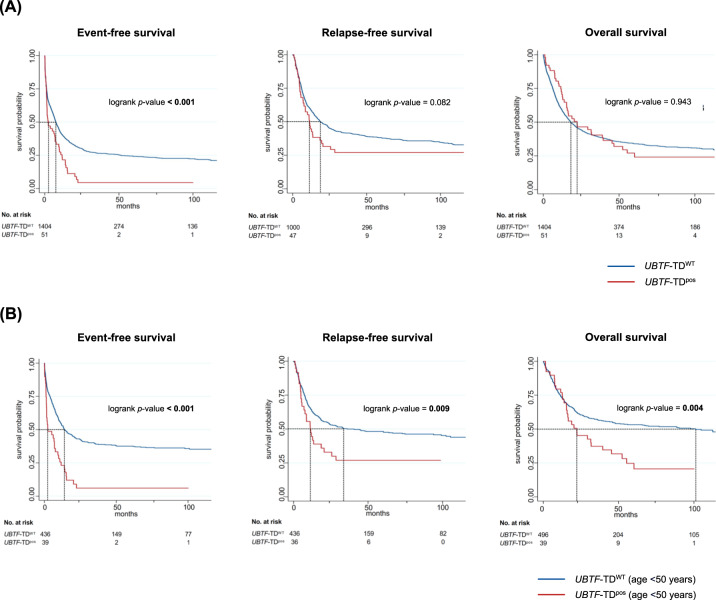
The prognostic relevance of UBTF-TD mutations was analyzed in 1455 intensively treated patients, which had available NGS data allowing to reclassify them according to the ELN2022 risk groups (1404 UBTF-TDWT/51 UBTF-TDpos; median follow-up time of patients alive 61 months; IQR 36–96 months). Individual clinical courses of all UBTF-TDpos patients are summarized in Table S5. In the entire cohort, UBTF-TDs were associated with a significantly shorter event-free survival (EFS, median UBTF-TDpos 2.4 months vs. UBTF-TDWT 7.5 months; p < 0.001), no differences in relapse-free survival (RFS) or overall survival (OS) were observed (Fig. 5A). In a subgroup analysis in patients <50 years of age, who represent the majority of UBTF mutant patients (40/52 = 77%; 3% UBTF-TD prevalence in pts <50 years vs. 0.4% in pts ≥50 years), UBTF-TDs were associated with significantly shorter EFS (median EFS UBTF-TDpos 2.4 [1.5–7.7] months vs. UBTF-TDWT 14.0 [11.6–19.3] months; p < 0.001), RFS (median RFS UBTF-TDpos 11.2 [5.6-20.4] months vs. UBTF-TDWT 35.9 [23.0-102.4] months; p = 0.009) and OS (median OS UBTF-TDpos 22.3 [15.4–39.5] months vs. UBTF-TDWT 101.1 [46.9-n.r.] months; p = 0.005) (Fig. 5B). Multivariable analyses (including age and ELN2022 risk group), confirmed UBTF-TDs as an independent risk factor for dismal EFS (HR: 2.20; 95% CI 1.52–3.17, p < 0.001), RFS (HR: 1.59; 95% CI 1.02–2.46, p = 0.039) and OS (HR: 1.64; 95% CI 1.08–2.49, p = 0.020) in patients <50 years of age (Table S6). The lack of this effect in the overall cohort (Table S7) can probably be explained by the generally poorer outcome of AML patients at higher ages and the low prevalence of UBTF-TD mutations in older patients. To address whether the observed survival differences in younger adults merely result from the absence of favorable risk features such as NPM1 or in-frame CEBPAbZIP mutations, which appear to be mutually exclusive with the presence of UBTF-TDs, we performed an additional subgroup analysis in patients stratified as intermediate risk according to ELN2022 guidelines [15]. Although also in these patients UBTF-TDs were associated with an inferior overall (median OS UBTF-TDpos 22.4 [15.8-55.4] months vs. UBTF-TDWT 97.7 [20.6-n.r.] months; p = 0.204) and relapse-free survival (median RFS UBTF-TDpos 11.5 [5.6–25.6] months vs. UBTF-TDWT 27.9 [12.1-n.r.] months; p = 0.189), only the event-free survival showed a significant difference between UBTF-TDpos and UBTF-TDWT patients (median EFS UBTF-TDpos 2.1 [1.4-7.7] months vs. UBTF-TDWT 11.1 [7.8-24.5] months; p < .001), presumably due to the limited number of patients in this analysis (Fig. S4). For the two most common co-mutations, FLT3-ITD and WT1, there was no evidence of a relevant additional effect on outcome (Fig. S5A). Clearly, these data have to be interpreted with caution, given these even smaller subsets of patients.
Fig. 5. Survival analysis according to UBTF mutant status.
Kaplan–Meier survival curves showing event-free, relapse-free and overall survival of UBTF-TDpos and UBTF-TDWT patients for A all patients and B for patients <50 years of age. p-values were calculated using the log-rank test. Numbers of patients at risk are provided below the x-axis.
Allogeneic hematopoietic stem cell transplantation (alloHSCT) performed in CR1 in 11 patients (22% vs. 15% in UBTF-TDWT patients) did not improve outcome in this limited data set (Fig. S6), but alloHSCT performed as salvage treatment (30/51 patients, 59%) was the only intervention associated with long-term cure, thus the impact of alloHSCT on outcome cannot be finally addressed in this study due to small numbers.
Discussion
Summarizing these results, UBTF-TDs appear to characterize a novel class-defining lesion not only in children but also younger adult AML patients. In vitro studies confirmed that UBTF-TDs confer a proliferative advantage to cord-blood derived CD34+-cells [13]. However, the functional implications of these mutations are unknown. All UBTF-TDs reported so far are in-frame alterations and affect the HMG-box 4 of UBTF, indicating that these changes are not loss- but gain- or shift-of-function lesions. Among the 6 HMG-boxes of UBTF, HMG-box 4 is unique to mammalian UBTF and appears to regulate species specificity [24]. Since the UBTF-wt protein is predominantly located in the nucleolus and interacts with numerous other nucleosomal proteins, including NPM1, an important regulator of the p14ARF/HDM2/TP53 axis [25], one might speculate that altered protein binding of mutant UBTF and sequestration of other essential proteins are involved in the transformation process.
An interesting observation not previously described is the intriguing association of UBTF-mutations with myelodysplastic features seen in our cohort. This observation and the particularly high rate of UBTF-TDs in pediatric MDS patients [22] together point to the fact that the leukemia in these patients might originate from a prior MDS clone. The high association with the former AML M6 subtype also suggests a defect in erythroid differentiation. Interestingly, a recent comprehensive molecular study of erythroleukemia patients reported 3 patients with 6 bp-deletions in UBTF, similar to those which we observed in our cohort [26].
Transcriptomic profiling via RNA-sequencing revealed several interesting aspects of UBTF-TD mutant AML. As reported previously, samples from UBTF-TDpos patients clustered together with NUP98/NSD1, t(6;9) and NPM1mut. All these subgroups show a strong upregulation of HOX-genes, most importantly HOXA as well HOXB. Another gene highly upregulated in this subgroup is PRDM16. PRDM16 codes for a histone H3K4 methyltransferase which is involved in adipose tissue differentiation, neural stem cell maintenance and represents an important regulator of normal hematopoietic differentiation [27]. In mouse models, forced overexpression of murine PRDM16, especially the short isoform, is able to transform hematopoietic stem cells and induce a fatal, transplantable leukemia [28]. In human AML, overexpression of PRDM16 has been linked to NUP98-NSD1 fusions as well as NPM1 mutant AML and was found to be associated with poor prognosis [29]. Interestingly and in line with the observed association with AML M6 discussed above, samples from UBTF-TD patients also showed an upregulation of several genes associated with erythroid differentiation, e.g., HBB, HBA2, and ABO.
The poor prognosis found in our study as well as in the pediatric patients, despite treatment with alloHSCT, indicates that novel treatment algorithms need to be evaluated in these patients. Interestingly, overexpression of HOXA and HOXB genes has recently been described as a predictive biomarker for sensitivity to treatment with BCL2-inhibitors such as Venetoclax [30], which is in line with the favorable response to Venetoclax treatment observed especially in NPM1 mutant patients [31]. However, more recent data suggest that in patients with erythroid differentiation, the BCX-L pathway might be more relevant, which would suggest that other inhibitors, such as Navitoclax [32] should be taken into account.
Taken together, these results as well as previously published data indicate that UBTF-TDs characterize a novel and specific subtype of AML, predominantly affecting adolescents and younger adults. The fact that UBTF-TDs appear to represent early clonal lesions, which have a very specific pattern of cytogenetic alterations (+8) and co-mutations (FLT3-ITD and WT1) as well as the absence of other subtype-specific lesions such as mutations in NPM1, CEBPAbZIP or CBF-translocations, point to UBTF-TD as a novel class-defining lesion in AML. Based on the significantly higher prevalence in children and adolescents as well as the fact that the gene is not covered in most clinically used diagnostic assays, dedicated screening for this mutation should be considered in patients below the age of 50 years. Due to the overall poor response to chemotherapy and alloHSCT, further understanding of the causative mechanisms appears crucial to improve the treatment and prognosis of these patients.
Supplementary information
Acknowledgements
We thank all centers and participating physicians of the Study Alliance Leukemia who entered their patients into the study (see Appendix). This study was supported in part through funding by the “DKTK JF-Excellence Project RiskY-AML” to CT.
Author contributions
JAG and CT designed the study; performed the research; collected, assembled, analyzed, interpreted the data; and wrote the manuscript; S Stasik and MH performed the molecular analyses and analyzed the data; FK performed the cytomorphologic analyses; UÖ performed the flow cytometric analyses; J-NE, SZ, and JS performed the statistical analyses; CR, JMM, U Krug, TS, S Scholl, AH, THB, R Naumann, BS, HE, MS, AB, AN, KS-E, CS, SWK, MH, R Noppeney, U Kaiser, CDB, MK, CM-T, UP, WEB, HS, GE, MB, and JS treated the patients and collected the clinical data; and all authors approved the final version of the manuscript.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets generated and/or analyzed during the current study are available in the Kaggle repository, 10.34740/kaggle/dsv/5550865.
Competing interests
CT is CEO and co-owner of AgenDix GmbH.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
A list of authors and their affiliations appears at the end of the paper.
A full list of members and their affiliations appears in the Supplementary Information.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-023-00858-y.
References
- 1.McStay B. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev. 2016;30:1598–610. doi: 10.1101/gad.283838.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ranieri R, Pianigiani G, Sciabolacci S, Perriello VM, Marra A, Cardinali V, et al. Current status and future perspectives in targeted therapy of NPM1-mutated AML. Leukemia. 2022;36:2351–67. doi: 10.1038/s41375-022-01666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Panov KI, Friedrich JK, Russell J, Zomerdijk JC. UBF activates RNA polymerase I transcription by stimulating promoter escape. Embo J. 2006;25:3310–22. doi: 10.1038/sj.emboj.7601221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tremblay MG, Sibai DS, Valère M, Mars JC, Lessard F, Hori RT, et al. Ribosomal DNA promoter recognition is determined in vivo by cooperation between UBTF1 and SL1 and is compromised in the UBTF-E210K neuroregression syndrome. PLoS Genet. 2022;18:e1009644. doi: 10.1371/journal.pgen.1009644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.O’Mahony DJ, Rothblum LI. Identification of two forms of the RNA polymerase I transcription factor UBF. Proc Natl Acad Sci USA. 1991;88:3180–4. doi: 10.1073/pnas.88.8.3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stefanovsky VY, Moss T. The splice variants of UBF differentially regulate RNA polymerase I transcription elongation in response to ERK phosphorylation. Nucl Acids Res. 2008;36:5093–101. doi: 10.1093/nar/gkn484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edvardson S, Nicolae CM, Agrawal PB, Mignot C, Payne K, Prasad AN, et al. Heterozygous de novo UBTF gain-of-function variant is associated with neurodegeneration in childhood. Am J Hum Genet. 2017;101:267–73. doi: 10.1016/j.ajhg.2017.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sedláčková L, Laššuthová P, Štěrbová K, Haberlová J, Vyhnálková E, Neupauerová J, et al. UBTF mutation causes complex phenotype of neurodegeneration and severe epilepsy in childhood. Neuropediatrics. 2019;50:57–60. doi: 10.1055/s-0038-1676288. [DOI] [PubMed] [Google Scholar]
- 9.Zhang J, Zhang J, Liu W, Ge R, Gao T, Tian Q, et al. UBTF facilitates melanoma progression via modulating MEK1/2-ERK1/2 signalling pathways by promoting GIT1 transcription. Cancer Cell Int. 2021;21:543. doi: 10.1186/s12935-021-02237-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tsoi H, Lam KC, Dong Y, Zhang X, Lee CK, Zhang J, et al. Pre-45s rRNA promotes colon cancer and is associated with poor survival of CRC patients. Oncogene. 2017;36:6109–18. doi: 10.1038/onc.2017.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bastian L, Hartmann AM, Beder T, Hänzelmann S, Kässens J, Bultmann M, et al. UBTF::ATXN7L3 gene fusion defines novel B cell precursor ALL subtype with CDX2 expression and need for intensified treatment. Leukemia. 2022;36:1676–80. doi: 10.1038/s41375-022-01557-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stratmann S, Yones SA, Mayrhofer M, Norgren N, Skaftason A, Sun J, et al. Genomic characterization of relapsed acute myeloid leukemia reveals novel putative therapeutic targets. Blood Adv. 2021;5:900–12. doi: 10.1182/bloodadvances.2020003709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Umeda M, Ma J, Huang BJ, Hagiwara K, Westover T, Abdelhamed S, et al. Integrated genomic analysis identifies UBTF tandem duplications as a recurrent lesion in pediatric acute myeloid leukemia. Blood Cancer Discov. 2022;3:194–207. doi: 10.1158/2643-3230.BCD-21-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaburagi T, Shiba N, Yamato G, Yoshida K, Tabuchi K, Ohki K, et al. UBTF-internal tandem duplication as a novel poor prognostic factor in pediatric acute myeloid leukemia. Genes Chromosom Cancer. 2023;62:202–9. doi: 10.1002/gcc.23110. [DOI] [PubMed] [Google Scholar]
- 15.Döhner H, Wei AH, Appelbaum FR, Craddock C, DiNardo CD, Dombret H, et al. Diagnosis and management of AML in adults: 2022 recommendations from an international expert panel on behalf of the ELN. Blood. 2022;140:1345–77. doi: 10.1182/blood.2022016867. [DOI] [PubMed] [Google Scholar]
- 16.Schaich M, Röllig C, Soucek S, Kramer M, Thiede C, Mohr B, et al. Cytarabine dose of 36 g/m² compared with 12 g/m² within first consolidation in acute myeloid leukemia: results of patients enrolled onto the prospective randomized AML96 study. J Clin Oncol. 2011;29:2696–702. doi: 10.1200/JCO.2010.33.7303. [DOI] [PubMed] [Google Scholar]
- 17.Röllig C, Serve H, Hüttmann A, Noppeney R, Müller-Tidow C, Krug U, et al. Addition of sorafenib versus placebo to standard therapy in patients aged 60 years or younger with newly diagnosed acute myeloid leukaemia (SORAML): a multicentre, phase 2, randomised controlled trial. Lancet Oncol. 2015;16:1691–9. doi: 10.1016/S1470-2045(15)00362-9. [DOI] [PubMed] [Google Scholar]
- 18.Schetelig J, Schaich M, Schäfer-Eckart K, Hänel M, Aulitzky WE, Einsele H, et al. Hematopoietic cell transplantation in patients with intermediate and high-risk AML: results from the randomized Study Alliance Leukemia (SAL) AML 2003 trial. Leukemia. 2015;29:1060–8. doi: 10.1038/leu.2014.335. [DOI] [PubMed] [Google Scholar]
- 19.Röllig C, Kramer M, Gabrecht M, Hänel M, Herbst R, Kaiser U, et al. Intermediate-dose cytarabine plus mitoxantrone versus standard-dose cytarabine plus daunorubicin for acute myeloid leukemia in elderly patients. Ann Oncol. 2018;29:973–8. doi: 10.1093/annonc/mdy030. [DOI] [PubMed] [Google Scholar]
- 20.Stasik S, Middeke JM, Kramer M, Röllig C, Krämer A, Scholl S, et al. EZH2 mutations and impact on clinical outcome: an analysis in 1,604 patients with newly diagnosed acute myeloid leukemia. Haematologica. 2020;105:e228–e31. doi: 10.3324/haematol.2019.222323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arber DA, Orazi A, Hasserjian R, Thiele J, Borowitz MJ, Le Beau MM, et al. The 2016 revision to the World Health Organization classification of myeloid neoplasms and acute leukemia. Blood. 2016;127:2391–405. doi: 10.1182/blood-2016-03-643544. [DOI] [PubMed] [Google Scholar]
- 22.Erlacher M, Stasik S, Yoshimi A, Georgi J-A, Göhring G, Rudelius M, et al. UBTF tandem duplications account for a third of advanced pediatric mds without genetic predisposition to myeloid neoplasia. Blood. 2022;140:1355–6. doi: 10.1182/blood-2022-159002. [DOI] [Google Scholar]
- 23.Khoury JD, Solary E, Abla O, Akkari Y, Alaggio R, Apperley JF, et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia. 2022;36:1703–19. doi: 10.1038/s41375-022-01613-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cairns C, McStay B. HMG box 4 is the principal determinant of species specificity in the RNA polymerase I transcription factor UBF. Nucl Acids Res. 1995;23:4583–90. doi: 10.1093/nar/23.22.4583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mitrea DM, Kriwacki RW. On the relationship status for Arf and NPM1-it’s complicated. Febs J. 2018;285:828–31. doi: 10.1111/febs.14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iacobucci I, Wen J, Meggendorfer M, Choi JK, Shi L, Pounds SB, et al. Genomic subtyping and therapeutic targeting of acute erythroleukemia. Nat Genet. 2019;51:694–704. doi: 10.1038/s41588-019-0375-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou B, Wang J, Lee SY, Xiong J, Bhanu N, Guo Q, et al. PRDM16 Suppresses MLL1r leukemia via intrinsic histone methyltransferase activity. Mol Cell. 2016;62:222–36. doi: 10.1016/j.molcel.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hu T, Morita K, Hill MC, Jiang Y, Kitano A, Saito Y, et al. PRDM16s transforms megakaryocyte-erythroid progenitors into myeloid leukemia-initiating cells. Blood. 2019;134:614–25. doi: 10.1182/blood.2018888255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shiba N, Ohki K, Kobayashi T, Hara Y, Yamato G, Tanoshima R, et al. High PRDM16 expression identifies a prognostic subgroup of pediatric acute myeloid leukaemia correlated to FLT3-ITD, KMT2A-PTD, and NUP98-NSD1: the results of the Japanese Paediatric Leukaemia/Lymphoma Study Group AML-05 trial. Br J Haematol. 2016;172:581–91. doi: 10.1111/bjh.13869. [DOI] [PubMed] [Google Scholar]
- 30.Kontro M, Kumar A, Majumder MM, Eldfors S, Parsons A, Pemovska T, et al. HOX gene expression predicts response to BCL-2 inhibition in acute myeloid leukemia. Leukemia. 2017;31:301–9. doi: 10.1038/leu.2016.222. [DOI] [PubMed] [Google Scholar]
- 31.Issa GC, Bidikian A, Venugopal S, Konopleva M, DiNardo CD, Kadia TM, et al. Clinical outcomes associated with NPM1 mutations in patients with relapsed or refractory AML. Blood Adv. 2023;7:933–42. doi: 10.1182/bloodadvances.2022008316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuusanmäki H, Dufva O, Vähä-Koskela M, Leppä AM, Huuhtanen J, Vänttinen IM, et al. Erythroid/megakaryocytic differentiation confers BCL-XL dependency and venetoclax resistance in acute myeloid leukemia. Blood. 2022;141:1610–25. doi: 10.1182/blood.2021011094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the Kaggle repository, 10.34740/kaggle/dsv/5550865.