Abstract
Auxins are growth regulators involved in virtually all aspects of plant development. However, little is known about how plants synthesize these essential compounds. We propose that the level of indole-3-acetic acid is regulated by the flux of indole-3-acetaldoxime through a cytochrome P450, CYP83B1, to the glucosinolate pathway. A T-DNA insertion in the CYP83B1 gene leads to plants with a phenotype that suggests severe auxin overproduction, whereas CYP83B1 overexpression leads to loss of apical dominance typical of auxin deficit. CYP83B1 N-hydroxylates indole-3-acetaldoxime to the corresponding aci-nitro compound, 1-aci-nitro-2-indolyl-ethane, with a Km of 3 μM and a turnover number of 53 min−1. The aci-nitro compound formed reacts non-enzymatically with thiol compounds to produce an N-alkyl-thiohydroximate adduct, the committed precursor of glucosinolates. Thus, indole-3-acetaldoxime is the metabolic branch point between the primary auxin indole-3-acetic acid and indole glucosinolate biosynthesis in Arabidopsis.
INTRODUCTION
Apical dominance, cell expansion, vascular differentiation, lateral root and root hair formation, phototropism, and root gravitropism are among the many processes in plants controlled by auxins (Davies, 1995). The level of auxin is regulated by both de novo biosynthesis and reversible and irreversible conjugation to sugars, amino acids, and peptides as well as by degradation. Although the chemical structure of the primary auxin, indole-3-acetic acid (IAA), has been known since the 1930s (Wildman, 1997), not much is known about how plants actually synthesize this essential compound. Plants appear to be capable of synthesizing IAA by both tryptophan-dependent and tryptophan-independent pathways. Classical incorporation studies with radiolabeled compounds have not unambiguously identified the precursors or elucidated the biosynthetic pathway for IAA. (For a recent review of IAA metabolism, see Normanly and Bartel [1999].)
Although a number of mutants in IAA metabolic pathways and perception have been described, the genes involved and their biochemical function and physiological relevance have not all been elucidated (reviewed in Bartel, 1997; Normanly and Bartel, 1999). For example, both the rty/sur1/hls3/alf1 (Boerjan et al., 1995; Celenza et al., 1995; King et al., 1995; Lehman et al., 1996) and sur2 (Delarue et al., 1998) mutants are known to accumulate increased levels of free auxin. Identification of the gene products affected and elucidation of the biochemical roles of these proteins should increase our limited knowledge of IAA biosynthesis and regulation.
A link between indole glucosinolates and auxin has often been suggested in the literature. Glucosinolates are sulfur-containing bioactive natural products derived from amino acids and sequestered in vacuoles of cruciferous plants (Halkier, 1999). It has recently been shown that the cytochromes CYP79B2 and CYP79B3 of Arabidopsis metabolize tryptophan to indole-3-acetaldoxime. This metabolite is often suggested to be the precursor of indole-3-acetonitrile (IAN) in IAA biosynthesis as well as the precursor of thiohydroximates in glucosinolate biosynthesis (Normanly and Bartel, 1999; Hull et al., 2000; Mikkelsen et al., 2000), although neither step has been characterized biochemically. Nitrilases that catalyze the conversion of IAN to IAA are well characterized in Arabidopsis (Bartel and Fink, 1994). In this species, four differentially regulated nitrilases have been identified, although their physiological role is not clear (Normanly and Bartel, 1999). A mutation for one of the nitrilase genes, nit1, renders Arabidopsis seedlings insensitive to exogenously applied IAN, yet this mutant does not have an apparent physiological IAA phenotype under normal conditions (Normanly et al., 1997).
In this article, we demonstrate by genetic and biochemical analyses that a cytochrome P450, CYP83B1, is a regulator of auxin production in Arabidopsis. Knockout of CYP83B1 leads to plants characterized by phenotypes that include severe apical dominance. Conversely, overexpression of CYP83B1 leads to plants with decreased apical dominance. In addition, we show that CYP83B1 catalyzes the first committed step in indole glucosinolate biosynthesis by metabolizing indole 3-acetaldoxime to the corresponding aci-nitro compound, which leads to S-alkyl-thiohydroximates, the committed precursors of glucosinolates. Our data demonstrate not only that the first part of IAA and of indole glucosinolate biosynthesis is shared but also that indole glucosinolates may function as a regulatory sink for IAA. Furthermore, the phylogenetic relationship between CYP83B1 and CYP71E1, the cytochrome P450 involved in the oxime-metabolizing step in cyanogenic glucoside biosynthesis, argues for an evolutionary relationship between IAA, glucosinolate, and cyanogenic glucoside biosynthesis.
RESULTS
CYP83B1 Is Essential for Normal Seedling Development
A null mutation of CYP83B1 was identified by a systematic reverse genetics approach for T-DNA–mediated gene disruptions of cytochromes P450 in Arabidopsis (Winkler et al., 1998). We called this mutation rnt1-1 because of its runt phenotype. The T-DNA is inserted in the first exon of the CYP83B1 gene between positions 316 and 325 relative to the start codon. Eight base pairs of CYP83B1 were deleted at the insertion site. The entire sequence of chromosome 4 obtained recently (Mayer et al., 1999) revealed that among the ∼50 P450 genes of chromosome 4 (http://ag.arizona/p450/), CYP83B1 maps most closely to the prha marker, which is within 1.3 centimorgans of sur2 (Delarue et al., 1998). This strongly suggested that rnt1-1 is an allele of sur2, a mutant known to accumulate elevated levels of free IAA (Delarue et al., 1998). Recently, sur2 was indeed demonstrated to be a CYP83B1 mutant (Barlier et al., 2000). As described for sur2 seedlings, rnt1-1 seedlings are characterized by increased hypocotyl length, epinastic cotyledons, exfoliation of the hypocotyl, adventitious root formation from the hypocotyl, enhanced secondary root and root hair formation, and eventually callus formation and increasing disintegration of the seedling (Table 1 and Figures 1 and 2). The majority of rnt1-1 seedlings never develop more than a few leaves before the organization of the tissue is lost. In soil, a minority of seedlings are able to overcome the initial defects and develop into plants with strong apical dominance, characterized by reduced height, an increased number of epinastic rosette leaves, and a single inflorescence (Table 1 and Figure 2), a phenotype historically associated with auxin overproduction (Davies, 1995). rnt1-1 differs from sur2 in two major aspects, however. Exfoliation is observed at the root–hypocotyl junction in rnt1-1, whereas in sur2, exfoliation starts in the middle of the hypocotyl. Therefore, with respect to exfoliation, rnt1-1 is more similar to sur1 (for a comparison of sur1 and sur2, see Delarue et al. [1998]). rnt1-1 plants grown in soil did not display a wild-type appearance (Table 1), whereas sur2 grown in soil did (Delarue et al., 1998). The phenotype of rnt1-1 therefore appears to be stronger than that observed for sur2.
Table 1.
Morphometric Analysis of Wild-Type and rnt1-1 Plants Grown in Soila
| Characteristics | Wild Type | rnt1-1 |
|---|---|---|
| Time of bolting | ∼1 month | ∼2 months |
| Plant height | 35.1 ± 0.60 cm (n = 16) | 17.65 ± 1.63 cm (n = 20) |
| No. of inflorescences | 3.28 ± 0.30 (n = 15) | 1.125 ± 0.07 (n = 24) |
| No. of rosette leaves | 7.45 ± 0.28 (n = 11) | 26.1 ± 2.4 (n = 10) |
| Rosette leaf width | 1.47 ± 0.05 cm (n = 15) | 0.69 ± 0.02 cm (n = 40) |
| Rosette leaf length | 2.93 ± 0.15 cm (n = 16) | 1.40 ± 0.05 cm (n = 44) |
| Pedicel length | 1.01 ± 0.02 cm (n = 21) | 0.75 ± 0.02 cm (n = 19) |
| Distance between siliques on main inflorescence | 1.01 ± 0.11 cm (n = 20) | 0.76 ± 0.05 cm (n = 31) |
Although analysis was conducted on the wild type at 7.5 weeks of age and on the rnt1-1 plants at 18 weeks, the plants were analyzed at the same developmental stage.
Figure 1.
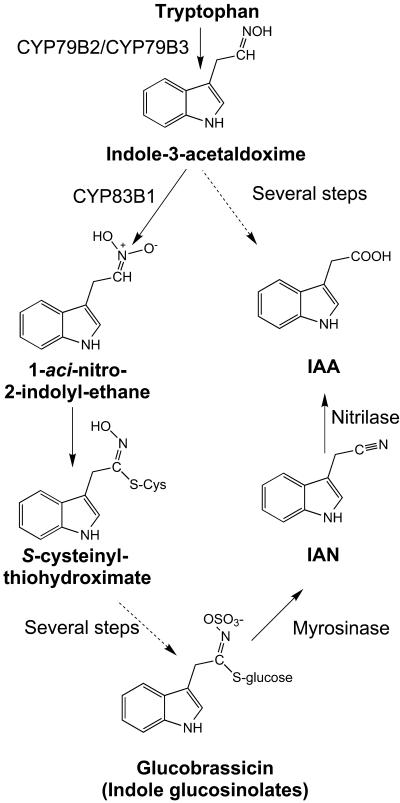
Phenotypes of rnt1-1 Seedlings.
Arrows indicate the approximate position of the root–hypocotyl junction.
(A) Wild type at 2 weeks.
(B) Adventitious roots are formed from the hypocotyl of rnt1-1 at 2 weeks. The scale is the same as in (A).
(C) Peeling of tissue from the root–hypocotyl junction in rnt1-1 seedling at 1 week.
(D) Disintegration of the hypocotyl, adventitious root formation and callus formation, and development of callus from secondary roots in rnt1-1 plant at 6 weeks.
Figure 2.
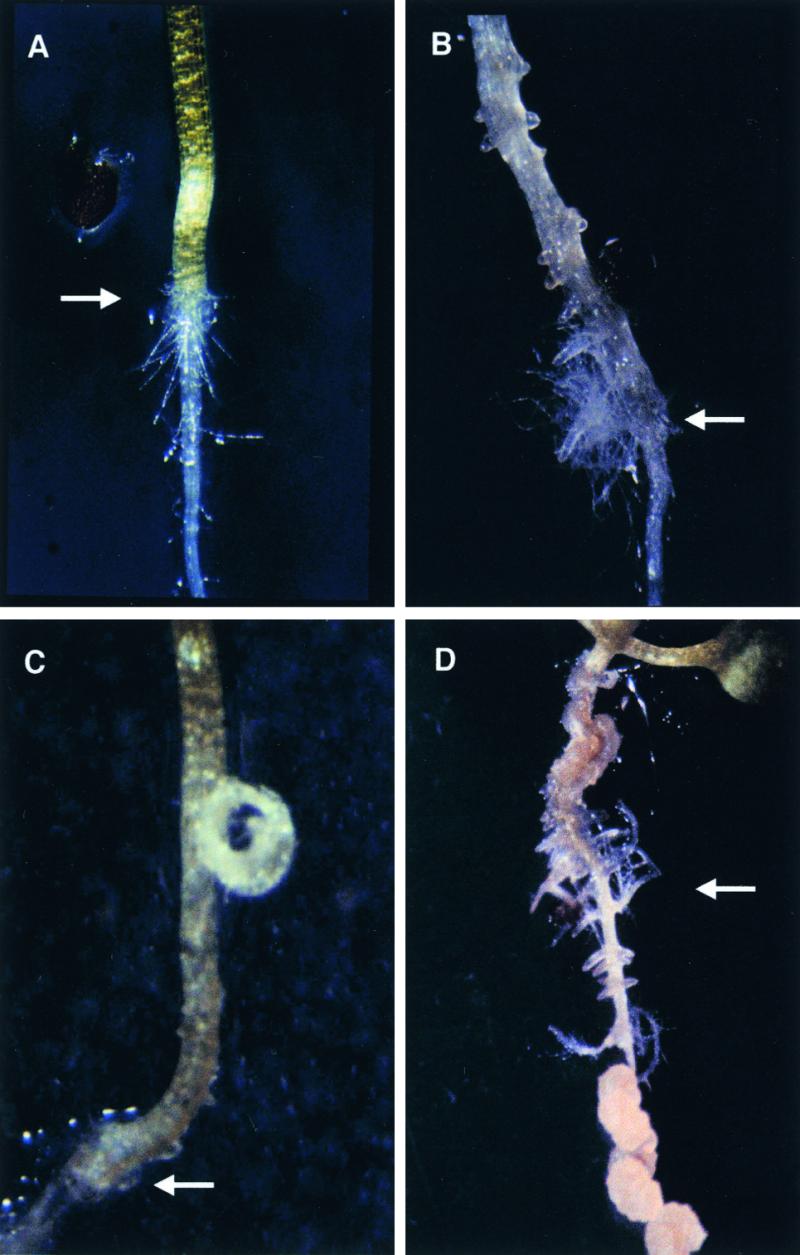
Molecular Complementation of rnt1-1.
(A) Seven-day-old seedlings of the wild type (3.65 ± 0.14 mm), rnt1-1 (5.06 ± 0.10 mm), and the molecularly complemented line 3.25.11 (3.00 ± 0.14 mm). The hypocotyl lengths of rnt1-1 and 3.25.11 differ significantly from that of the wild type at a 1% confidence level (t test). Bar = 5 mm.
(B) Phenotypes of 6-week-old wild type, rnt1-1, molecularly complemented line 3.25.11, and overexpression line 1.4.7 (35S::CYP83B1).
That the increased auxin phenotype is due to knockout of the CYP83B1 gene is demonstrated by molecular complementation of rnt1-1 with a 5.5-kb genomic fragment that comprises the CYP83B1 gene (Figure 2). Of 18 independent lines, none displayed the hypocotyl disintegration phenotype at the seedling stage. Overexpression of a CYP83B1 cDNA under control of the constitutive cauliflower mosaic virus 35S promoter generated lines with reduced hypocotyl length and increased number of inflorescences, as in, for example, the 1.4.7 line (Figure 2). Whereas the wild type has typically three inflorescences and rnt1-1 has one, the molecularly complemented line 3.25.11 and the 1.4.7 overexpression line have between three and six inflorescences. More severe overexpression phenotypes include reduced height, a bushy appearance due to extensive branching, and reduced seed set (data not shown). These bushy plants with decreased height and seed set are phenotypically similar to strong alleles of axr1, which are characterized by showing decreased apical dominance due to reduced sensing of auxin (Lincoln et al., 1990).
Spectral Characterization of Heterologously Expressed CYP83B1 Enzyme
To elucidate the biochemical function of CYP83B1, we produced the enzyme in yeast. Yeast microsomes containing CYP83B1 were screened with auxin-related molecules to identify potential ligands by using spectral analysis. Tryptamine was found to cause a typical type IIa binding spectrum (Jefcoate, 1978) with a trough at 390 nm and a peak at 427 nm and with a spectral dissociation constant Ks of 24 μM. The type IIa spectrum indicates access of the amine to the vicinity of the heme active site. In addition to tryptamine, other aromatic primary amines also produced similar spectra changes, but none with the same high amplitude or low dissociation constant (data not shown).
Indole-3-acetaldoxime caused a weak reverse type I spectrum (Jefcoate, 1978) with a trough at 385 nm and a peak at 400 nm (data not shown), which is indicative of binding to the active site. To quantify the affinity of indole-3-acetaldoxime to CYP83B1 properly, we first saturated the enzyme with 100 μM tryptamine. We subsequently displaced tryptamine from the active site by titration with indole-3-acetaldoxime, causing a gradual appearance of a reverse type IIa spectrum, from which a Ks of 0.2 μM for indole-3-acetaldoxime was estimated (Figure 3). Indole-3-acetaldoxime is therefore a high-affinity ligand for CYP83B1, and the ability of indole-3-acetaldoxime to displace tryptamine argues that tryptamine is a competitive inhibitor that binds to the active site of CYP83B1.
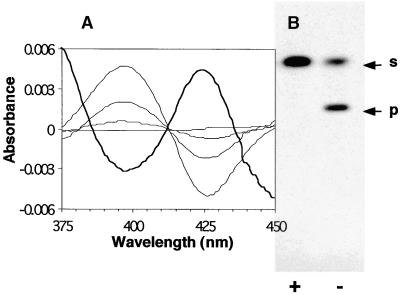
Figure 3.
Characterization of CYP83B1.
(A) Analysis of CYP83B1 by optical difference spectroscopy. A saturated type IIa spectrum was obtained with 100 μM tryptamine (indicated by the thick line, with the trough at 390 nm and the peak at 425 nm) in the sample cuvette. The addition of 100 μM tryptamine to both cuvettes gave a baseline. The increasing concentrations of indole-3-acetaldoxime in the sample cuvette (0.2, 0.8, and 3.0 μM) then displaced tryptamine, giving the reverse type IIa spectrum.
(B) Tryptamine is an inhibitor of CYP83B1 catalysis. We incubated 22 nM CYP83B1 with 35 μM 5-3H-indole-3-acetaldoxime in the absence (−) or presence (+) of 17.5 mM tryptamine. After incubation for 10 min at 28°C, reaction mixtures were extracted with ethyl acetate and analyzed by TLC. p, product; s, substrate.
CYP83B1 Catalyzes the Oxime Metabolizing Step in Indole Glucosinolate Biosynthesis
Isolated yeast microsomes containing CYP83B1 and Arabidopsis NADPH cytochrome P450 reductase catalyzed the metabolism of indole-3-acetaldoxime to a single product that was identified by mass spectrometry (MS) as a covalent adduct with the major nucleophile present in the reaction mixture (Figure 4). These S-alkyl-thiohydroximate adducts were observed with a variety of nucleophiles, showing that formation of the adducts was independent of the structure of the nucleophile and was not limiting under our experimental conditions. Nucleophiles were tested by thin layer chromatography (TLC), and the RF of the adducts formed were as follows: β-mercaptoethanol (RF 0.37), l-cysteine (RF 0.04), ethanethiol (RF 0.54), 1-thio-β-d-glucose (RF 0.02), and reduced glutathione (RF 0.01). In the absence of added nucleophile, the product of the enzymatic reaction inactivated the enzyme. In the presence of cysteine, the Michaelis-Menten constant Km for indole-3-acetaldoxime was estimated to be 3 μM and the turnover 53 min−1. In accordance with the spectral data, tryptamine was found to be an inhibitor of indole-3-acetaldoxime N-oxidation (Figure 3B).
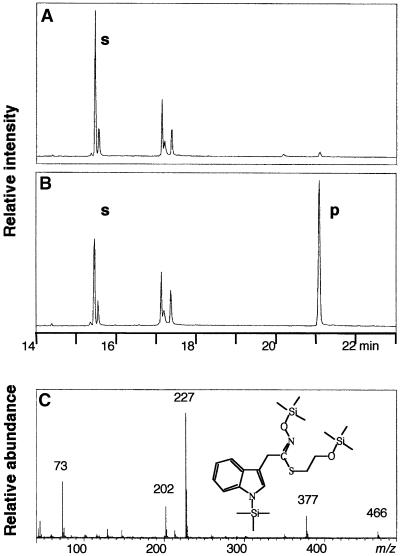
Figure 4.
Analysis of Reaction Mixtures using Gas Chromatography–Electron Impact Mode.
After incubation for 0 min (A) or 15 min (B) at 28°C, reaction mixtures were subjected to gas chromatography–electron impact mode analysis (C). The new peak (p) at 21.083 min shows a molecular ion at m/z 466 and a fragmentation pattern consistent with the structure shown in the insert, with m/z 377, loss of the oxime O-trimethylsilyl (TMS); m/z 228, further loss of S-C2H5-O-TMS; m/z 202, further loss of nitrile; and m/z 73, TMS. The structure was verified by electrospray (ES)-MS of underivatized ethyl acetate–extracted reaction mixtures. p, TMS derivative of the β-mercaptoethanol adduct of the reaction mixture; s, TMS derivative of indole-3-acetaldoxime (15.467 min).
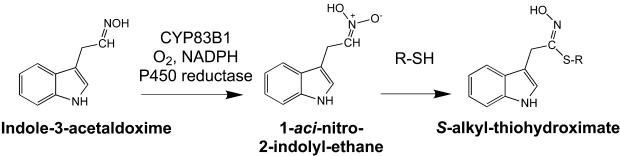
We deduce that the S-alkyl-thiohydroximate is formed by N-hydroxylation of the nitrogen atom at the oxime function to generate an electrophilic aci-nitro compound, 1-aci-nitro-2-indolyl-ethane, that non-enzymatically forms an adduct with nucleophiles at the α-carbon (Figure 5).
Figure 5.
Proposed Reaction Scheme of CYP83B1.
CYP83B1 catalyzes the first committed step of indole glucosinolate biosynthesis, the N-hydroxylation of indole-3-acetaldoxime to a highly reactive electrophile aci-nitro compound, 1-aci-nitro-2-indolyl-ethane, that non-enzymatically forms an adduct with a nucleophile (R-SH).
Indole-3-Acetaldoxime Is the Metabolic Branch Point between IAA and Indole Glucosinolate Biosynthesis
The rnt1-1 mutant and the CYP83B1 overexpression lines clearly show auxin-related phenotypes. In addition, the biochemical results shown above imply that the substrate of CYP83B1, tryptophan-derived indole-3-acetaldoxime, is an intermediate in the biosynthesis of indole glucosinolates. In accordance with the in vitro–deduced biochemical function of CYP83B1, seedlings of rnt1-1 contained reduced levels of indole glucosinolates compared with the levels of wild-type seedlings, whereas the molecularly complemented line 3.25.11 contained levels comparable to those of the wild type, and the overexpression line 1.4.7 contained elevated levels of indole glucosinolates compared with the levels in the wild type (Figure 6). In the present study, tryptamine was identified as an inhibitor of CYP83B1 in vitro. Wild-type seedlings germinated on Murashige and Skoog media (1962) supplemented with 100 μM tryptamine, the CYP83B1 inhibitor, contained reduced levels of indole glucosinolates compared with those of the wild type germinated without tryptamine, indicating that tryptamine is also an inhibitor of CYP83B1 in planta. Molecular complementation with the wild-type gene thus eliminates the strong auxin overproduction phenotype of rnt1-1 and at the same time restores the level of indole glucosinolate back to that of wild-type seedlings. Therefore, we conclude that enzymes in glucosinolate and IAA biosynthesis use the same indole-3-acetaldoxime pool.
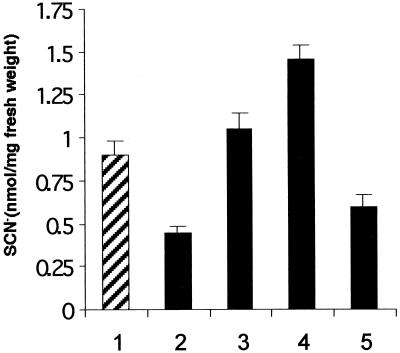
Figure 6.
Indole Glucosinolate Levels Are Affected by CYP83B1 Expression.
Indole glucosinolates in individual 2-week-old seedlings grown on Murashige and Skoog agar were quantified colorimetrically as thiocyanite (SCN−), as described by Bak et al. (1999). Lane 1, wild type; lane 2, rnt1-1; lane 3, molecularly complemented line 3.25.11; lane 4, overexpression line 1.4.7; and lane 5, wild type grown on 100 μM tryptamine. Data are represented as the mean ±se; n = 10 seedlings. Except for the molecularly complemented line 3.25.11, mean indole glucosinolate values all differ from wild-type seedling values at a 1% confidence value (t test).
IAN Is Not a Direct Product of Indole-3-Acetaldoxime Metabolism in IAA Biosynthesis
IAN has often been suggested to be the product of an indole-3-acetaldoxime metabolizing enzyme in IAA biosynthesis (e.g., Normanly and Bartel, 1999). Accordingly, wild-type Arabidopsis seedlings grown on media supplemented with 50 μM IAN phenocopy rnt1-1 seedlings by showing exfoliation at the root–hypocotyl junction and the development of adventitious lateral root primordia on the hypocotyl and epinastic cotyledons (Normanly et al., 1997; S. Bak, unpublished results). The observations that rnt1-1 seedling growth on 50 μM IAN (i) enhances the IAA phenotype of rnt1-1 by almost completely inhibiting germination and (ii) exacerbates the primary/lateral root imbalance (data not shown) further support the involvement of IAN in IAA biosynthesis. Four nitrilases, differentially expressed during development and responding to environmental cues, catalyze hydrolysis of IAN to IAA in Arabidopsis (Bartel and Fink, 1994). The nit1 mutant is insensitive to the auxin effects of exogenously applied IAN at the seedling stage (Normanly et al., 1997). In accordance with the observed insensitivity of nit1 seedlings to IAN, NIT1 is most strongly expressed in the hypocotyl near the junction with the root in young seedlings as well as in the cotyledons. In contrast, NIT2 and NIT3 are expressed primarily in the cotyledons, and NIT4 is expressed in the root tip (Bartel and Fink, 1994). If IAN is a direct metabolite of indole-3-acetaldoxime in IAA biosynthesis, then the rnt1-1 phenotype should be suppressed in the nit1 background. Surprisingly, the rnt1-1 phenotype is not suppressed in the nit1-1 background (Figure 7). This strongly suggests that IAN is not a direct metabolite of indole-3-acetaldoxime in IAA biosynthesis.
Figure 7.
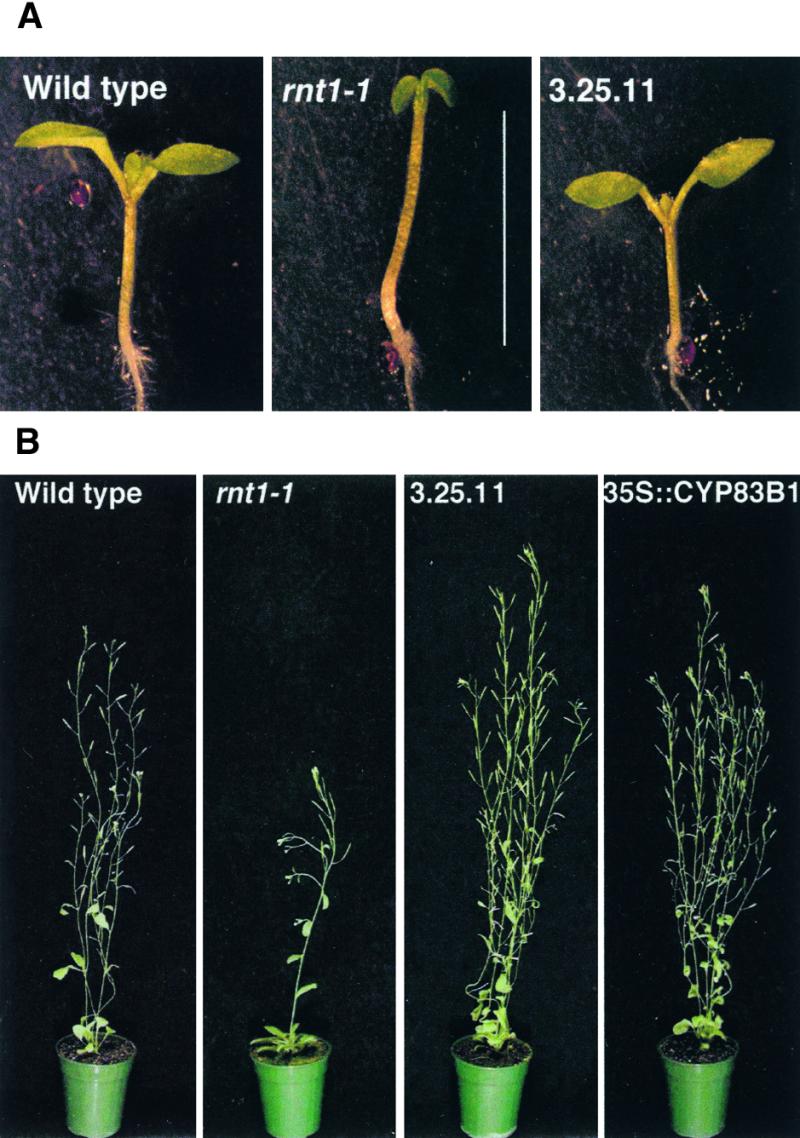
rnt1-1 Is Not Suppressed in a nit1-1 Background.
(A) rnt1-1 at 2 weeks.
(B) rnt1-1 in the nit1-1 background at 2 weeks.
Arrows indicate the approximate position of the root–hypocotyl junction.
DISCUSSION
We have shown that CYP83B1 is a regulator of auxin production in Arabidopsis by controlling the flux of indole-3-acetaldoxime into IAA and indole glucosinolate biosynthesis. In addition, we have shown that CYP83B1 catalyzes the first committed step in indole glucosinolate biosynthesis by metabolizing indole-3-acetaldoxime to its corresponding aci-nitro compound. The recently reported identity of sur2 also as CYP83B1 (Barlier et al., 2000) explains the strong auxin phenotype of the rnt1-1 mutant, which is a knockout for CYP83B1. Increased free auxin levels were previously measured in the sur2 mutant (Delarue et al., 1998). In contrast to rnt1-1, in which the CYP83B1 gene is disrupted by insertion of a T-DNA, the CYP83B1 gene in sur2 likely may not be completely inactivated and therefore causes a less severe phenotype. In accordance with the observed phenotypes of sur2/rnt1-1, CYP83B1 is expressed in all tissues, with the highest expression level observed in the roots (Mizutani et al., 1998; W. Xu and S. Bak, unpublished results).
Oximes are unstable compounds that do not accumulate in the cell. The low Ks and Km of CYP83B1 for indole-3-acetaldoxime would prevent accumulation of indole-3-acetaldoxime. In the CYP83B1 knockout plants, the indole-3-acetaldoxime in excess is channeled into IAA biosynthesis, which leads to elevated IAA levels and thus increased apical dominance and reduced indole glucosinolate levels. Conversely, overexpression of CYP83B1 in Arabidopsis leads to a reduced IAA phenotype and loss of apical dominance and elevated indole glucosinolate levels, indicating that increased N-hydroxylation of indole-3-acetaldoxime results in a net loss of IAA.
Glucosinolate Biosynthesis
We show that CYP83B1 catalyzes the formation of 1-aci-nitro-2-indolyl-ethane that non-enzymatically forms an adduct with a nucleophile in vitro. Based on precursor feeding studies, cysteine but not 1-thio-β-d-glucose has been proposed as the sulfur donor in vivo (Wetter and Chrisholm, 1968). 1-aci-Nitro-2-indolyl-ethane produced by CYP83B1 in vitro can form an adduct with cellular nucleophilic substrates such as glutathione, cysteine, and 1-thio-β-d-glucose, but the preferred thiol donor in vivo is unknown. The S-alkyl-thiohydroximate formed by CYP83B1 catalysis can be cleaved by a C-S lyase to generate thiohydroximates. It is well established that thiohydroximates are glucosylated by a soluble UDPG:thiohydroximate glucosyltransferase to form desulfoglucosinolates that are subsequently sulfated by a soluble 3′-phosphoadenosine 5′-phosphosulphate:desulphoglucosinolate sulphotransferase (PAPS transferase) (reviewed in Halkier and Du, 1997).
Arabidopsis contains at least 23 different glucosinolates derived from tryptophan and from chain-elongated homologs of phenylalanine and methionine (Hogge et al., 1988). In Arabidopsis, at least six related members of the CYP79 family (http://ag.arizona.edu/p450) have been identified. These enzymes probably all catalyze the conversion of amino acid and chain-elongated amino acids to their acetaldoximes (Bak et al., 1998b; Hull et al., 2000; Mikkelsen et al., 2000; Wittstock and Halkier, 2000). The next P450 in the common pathway from tryptophan, CYP83B1, shows 65% amino acid sequence identity to the most closely related P450, CYP83A1 (Paquette et al., 2000). The reduction of indole glucosinolate levels by only 50% in rnt1-1 suggests the presence of a functional homolog that partially complements CYP83B1. CYP83A1 may thus be a functional homolog of CYP83B1 and may also metabolize indole-3-acetaldoxime to some degree. Further comparative analysis of CYP83A1 and CYP83B1, and their substrate specificity toward indole-, aromatic-, or methionine-derived acetaldoximes, and thus a possible involvement in the biosynthesis of other glucosinolates, is currently under way (S. Bak and R. Feyereisen, manuscript in preparation).
The Role of IAN in IAA Biosynthesis
The auxin effects of IAN when supplemented to Arabidopsis are well documented (Normanly et al., 1997). The results of IAN treatment of wild-type and rnt1-1 mutant plants confirm that IAN can serve as a precursor in IAA biosynthesis. However, the inability of nit1-1 (which is resistant to exogenous IAN) to suppress the auxin phenotype of rnt1-1 indicates that IAN may not be the direct product of indole-3-acetaldoxime metabolism in IAA biosynthesis as often suggested. The role of IAN in IAA biosynthesis appears to be restricted to glucosinolate-producing species because they contain myrosinases that hydrolyze indole glucosinolates to IAN as well as nitrilases that can convert IAN to IAA (Thimann and Mahadevan, 1964). Apart from their function as bioactive natural products, indole glucosinolates may thus have a role in IAA biosynthesis as a sink for indole-3-acetaldoxime as well as a source of the IAA precursor IAN by turnover of the indole glucosinolate pool (Figure 8), much in the same way that hydrolyzable IAA conjugates (Normanly and Bartel, 1999). The level of IAN is two orders of magnitude higher than that of IAA in 7-day-old seedlings (Normanly et al., 1997), and the Km values for IAN hydrolysis by NIT1 and NIT2 are in the millimolar range (Bartling et al., 1994). This is unexpected for an intermediate in a highly regulated biosynthetic pathway. Also, the observation that the isotope enrichment of IAN is lower than that for IAA when radiolabeled tryptophan is administered to either Arabidopsis roots or shoots (Normanly et al., 1993; Müller et al., 1998) indicates that IAN may not be a direct product of indole-3-acetaldoxime metabolism in IAA biosynthesis. Ectopic overexpression of the nitrilases does not give rise to an apparent IAA phenotype under normal conditions (Normanly et al., 1997; Grsic et al., 1998).
Figure 8.
Indole-3-Acetaldoxime Is the Metabolic Branch Point between Indole Glucosinolates and IAA Biosynthesis.
In rnt1-1, the pathway into indole glucosinolates through CYP83B1 is blocked, which leads to accumulation of IAA and plants with high IAA phenotype. Conversely, in CYP83B1 overexpression lines, additional indole-3-acetaldoxime is channeled into indole glucosinolate biosynthesis, which leads to plants with a low IAA phenotype and increased indole glucosinolate levels.
In addition to indole-3-acetonitrile, indole-3-acetaldehyde derived from indole-3-acetaldoxime or indole-3-pyruvic acid has also been suggested to be a precursor of IAA biosynthesis. Labeling studies have shown that indole-3-acetaldoxime can be converted to IAA with indole-3-acetaldehyde as an intermediate in higher plants (Rajapogal and Larsen, 1972). Accordingly, it has been shown that in the IAA overproducer sur1, the aldehyde oxidase activity is increased (Seo et al., 1998). As opposed to the nitrilases, oxidases that catalyze conversion of indole-3-acetaldehyde to IAA are widespread in higher plants. This suggests that indole-3-acetaldehyde may be the intermediate in IAA biosynthesis from indole-3-acetaldoxime.
Tryptamine Inhibits Biosynthesis of Indole Glucosinolates
Tryptamine was identified in the present study as an inhibitor of CYP83B1, which suggests that tryptamine may act as a regulator of the flux through the indole-3-acetaldoxime pathway to IAA. Expression of tryptophan decarboxylase from Catharanthus roseus in Brassica napus leads to an accumulation of tryptamine, whereas the level of indole glucosinolates in leaves was reduced (Chavadej et al., 1994). This was interpreted as a redirection of tryptophan into tryptamine rather than into indole glucosinolates. On the basis that tryptamine inhibits the enzymatic reaction catalyzed by CYP83B1 and thereby the production of precursors for indole glucosinolates, the role of tryptamine decarboxylase expression in B. napus should be reevaluated. Compared with expression in, for example, tobacco (Songstad et al., 1989), the B. napus transgenic plants accumulated only 2% of the tryptamine levels found in transgenic tobacco. Tobacco does not produce indole glucosinolates and thus probably does not have a CYP83B1 functional homolog. Therefore, we speculate that expression of high levels of tryptamine in glucosinolate-producing plants may be deleterious to the regeneration of transgenic lines due to elevated IAA levels. This implies that tryptamine may not necessarily be a general regulator of IAA biosynthesis.
Because tryptamine is an inhibitor of CYP83B1, one may wonder whether treating plants with tryptamine can cause an rnt-1–like phenotype. Tryptamine treatment at levels higher than 100 μM inhibited seedling germination. At levels of 50 to 100 μM tryptamine, the only visible phenotype was a slightly more pale appearance than plants grown on 0 or 10 μM (results not shown). The concentration of tryptamine used in our in vivo experiment, 100 μM, thus reflects seedlings that are more or less wild type in appearance but have significant lower indole glucosinolate levels. The level of indole glucosinolates measured in the tryptamine experiments is not as low as in the knockout, thus a visual IAA phenotype may not necessarily be expected.
Has Glucosinolate Biosynthesis Evolved from an IAA Biosynthetic Pathway?
Plants produce a vast array of natural products, often referred to as secondary metabolites, to accommodate their biotic and abiotic environments. These natural products are produced at a high expense of energy, with natural selection as a driving force. Little is known about the evolutionary origin of the biosynthetic pathways behind these versatile compounds. Identification of the cytochrome P450 CYP83B1 as being involved in the regulation of IAA levels as well as in glucosinolate biosynthesis suggests that the glucosinolate biosynthetic pathway may have evolved from an IAA biosynthetic pathway.
The first step in IAA and indole glucosinolate biosynthesis is catalyzed by cytochromes P450 of the CYP79B subfamily (Figure 8) (Hull et al., 2000; Mikkelsen et al., 2000). CYP79 family members also catalyze the conversion of amino acids to their corresponding aldoximes in the biosynthesis of cyanogenic glucosides (Halkier et al., 1995; Andersen et al., 2000; Nielsen and Møller, 2000) and glucosinolates (Hull et al., 2000; Mikkelsen et al., 2000; Wittstock and Halkier, 2000). In contrast to glucosinolates that are found primarily in the order Capparales, cyanogenic glucosides are widespread in nature and represent an evolutionary ancient trait. The oxime-metabolizing step in biosynthesis of the tyrosine-derived cyanogenic glucoside dhurrin is catalyzed by the cytochrome P450 CYP71E1 (Bak et al., 1998a). A phylogenetic analysis of the cytochrome P450 supergene family revealed that CYP83B1 belongs to the larger CYP71 clade (Paquette et al., 2000). It has often been suggested that glucosinolate biosynthesis has evolved from a cyanogenic predisposition (e.g., Poulton and Møller, 1993). The present finding of indole-3-acetaldoxime as a metabolic branch point in IAA and indole glucosinolate biosynthesis implies that the biosynthetic pathways of glucosinolates and cyanogenic glucosides have evolved from an existing IAA biosynthetic pathway and that glucosinolates may not necessarily have evolved from a cyanogenic glucoside pathway.
Cytochromes P450 are heme thiolate enzymes anchored in the endoplasmic reticulum membrane that comprise one of the functionally most versatile supergene families. They are the catalysts of many complex steps in biosynthetic pathways. Currently, 237 P450 genes distributed over 43 families have been identified in the genome of the model plant Arabidopsis (http://ag.arizona.edu/p450). We predict that a total of 250 to 270 individual P450 genes will be identified by the time the genome is fully sequenced. The CYP71 clade is the most expanded cytochrome P450 subfamily, because currently 45 members of the CYP71 family have been identified in Arabidopsis (http://ag.arizona.edu/p450). The significance of the CYP71 clade in the recruitment of enzymes of indole and oxime metabolism also is illustrated by the four CYP71C P450s involved in biosynthesis of indole-derived defense compounds DIBOA (2,4-dihydroxy-1,4-benzoxazin-3-one) and DIMBOA (2,4-dihydroxy-7-methoxy-1,4-benzoxazin-3-one) in grasses (Frey et al., 1997) and by the involvement of CYP71B15 in biosynthesis of the indole-derived phytoalexin camalexin (Zhou et al., 2000).
Overexpression of CYP79B2 and CYP79B3, recently shown to catalyze indole-3-acetaldoxime formation from tryptophan, did not lead to an IAA phenotype (Hull et al., 2000). Similarly, overexpression of any of the four nitrilases in Arabidopsis did not lead to an IAA phenotype under normal conditions (Normanly et al., 1997; Grsic et al., 1998; Grsic-Rausch et al., 2000). CYP83B1 is thus the first enzyme demonstrated to regulate auxin production from tryptophan in Arabidopsis. Identification of CYP83B1 as a regulator of auxin levels opens up the possibilities of regulating plant growth by chemical or molecular intervention on this enzyme. Molecular intervention of CYP83B1 and its homologs also opens up the possibility of genetically engineering novel glucosinolate profiles to increase nutritional value and modulate interactions with pests and pathogens of crop plants.
METHODS
Plants
Arabidopsis thaliana plants were grown at a photosynthetic flux of 100 to 120 μmol photons m−2 sec−1 at 70% humidity and 22°C for a 12-hr photoperiod. Morphometric analyses are shown with their standard errors of the mean.
rnt1-1 was backcrossed once to wild-type Wassilewskija-2 (Ws-2) Arabidopsis. Rnt progeny from the backcross were used for all further analysis. For molecular complementation of rnt1-1, a 5574-bp DNA fragment containing 2276 bp upstream of the start codon and 1703 bp downstream of the stop codon was inserted into the binary vector pPZP221 (Hajdukiewicz et al., 1994) harboring gentamycin resistance and was transformed into rnt1-1/+ plants by the floral dip method. Primary transformants were selected on Murashige and Skoog plates supplemented with 2% sucrose, 0.9% Bacto agar, 50 μg/mL kanamycin, and 200 μg/mL gentamycin. Lines homozygous for the T-DNA insertion in CYP83B1 and harboring the introduced 5.5-kb DNA fragment were identified by co-segregation analysis on selective Murashige and Skoog agar plates. Overexpression constructs comprising the CYP83B1 cDNA under control of a cauliflower mosaic virus 35S promoter and polyadenylation site were made in pPZP221.
Double mutants of nit1-1 and rnt1-1 were made by crossing rnt1-1/ RNT1 (Ws-2; the pollen donor) into nit1-1 Columbia (Col-0). Double mutants homozygous for nit1-1 were identified according to Normanly et al. (1997) and verified by sequence analysis. To account for possible ecotypic differences, rnt1-1 was crossed into wild-type Col-0. No ecotypic differences of the rnt1-1 phenotype were observed in the Col-0 ecotype compared with that of Ws-2.
Analysis of Recombinant CYP83B1 Enzyme
Microsomes from yeast WAT11 cells expressing the CYP83B1 cDNA using the pYeDP60 vector were isolated essentially according to Pompon et al. (1996). Indole-3-acetaldoxime was synthesized from indole-3-acetaldehyde according to Rausch et al. (1985). The structure of indole-3-acetaldoxime was confirmed by 1H NMR, electrospray–mass spectrometry (ES-MS), and gas chromatography–mass spectrometry (GC-MS). 5-3H-Indole-3-acetaldoxime was synthesized from 5-3H-l-tryptophan according to Hofmann et al. (1980). Binding spectra were recorded using 0.44 μM CYP83B1 on a Lambda19 spectrophotometer (Perkin Elmer). To determine the spectral dissociation constant Ks of tryptamine, we used nine concentrations ranging from 2.2 to 160 μM. To determine the Ks of indole-3-acetaldoxime, we used 11 concentrations ranging from 0.2 to 7 μM.
For analysis of CYP83B1, catalysis reaction mixtures of 25 μL containing 8.8 nM CYP83B1, 6% glycerol, 50 mM Tris-HCl, pH 7.6, 70 μM 5-3H-indole-3-acetaldoxime (specific activity 350 mCi/mmol), 0.1 mM NADPH, 2 mM glucose-6-phosphate, 0.075 units of glucose-6-phosphate dehydrogenase, and 10 mM cysteine or other nucleophiles as indicated were incubated for 15 min at 28°C. After incubation, reaction mixtures were stopped by adding SDS to 0.4% and were analyzed by thin layer chromatography (TLC). TLC plates were developed in chloroform/methanol/water (85:14:1), and radioactive bands were visualized by autoradiography.
To determine Km and Vmax, reaction mixtures containing 2.2 nM CYP83B1, 0.2 to 60 μM 5-3H-indole-3-acetaldoxime, and 50 mM l-cysteine were incubated for 1 min at 28°C. After incubation, reactions were stopped by addition of ethyl acetate, and product and substrate were partitioned by three extractions with ethyl acetate. Aliquots of the combined substrate-containing ethyl acetate phase and product-containing water phase were subjected to liquid scintillation counting.
For structural analysis, reaction mixtures of 100 μL containing 0.5 μM CYP83B1, 1 mM indole-3-acetaldoxime, 10 mM β-mercaptoethanol, 50 mM MOPS, pH 7.6, 0.1 mM NADPH, 2 mM glucose-6-phosphate, 0.2 units of glucose-6-phosphate dehydrogenase, and 6% glycerol were incubated at 28°C. After incubation, reaction mixtures were extracted with ethyl acetate and dried. ES-MS samples were dissolved in acetonitrile. For GC-MS, analysis samples were derivatized for 20 min at 90°C with a 30-μL 1:1 mixture of bis-trimethylsilyl-trifluoroacetamide:pyridine containing 1% trimethylchlorosilane before analysis. For GC-MS analysis, we used a VA-5MS column (10 m × 0.25 mm × 0.25 μm film thickness). The oven temperature program used was as follows: 100°C for 2 min, 100 to 250°C at 10°C min−1, and 250°C for 10 min. Samples were run in both electron impact mode and chemical ionization mode.
Acknowledgments
We thank Drs. Denis Pompon and Philippe Urban for the gift of pYeD60 and WAT11; Dr. Bonnie Bartel for the kind gift of nit1-1 seeds; Drs. Marat Murataliev, Sunghwa Choe, and HoBang Kim for helpful discussions; Kathryn Byrnes for isolating the CYP83B1 genomic clone; Adria Decker for technical assistance; and Dr. Arpad Somogyi for performing the GC-MS and ES-MS analysis. The Human Frontier Science Program (Grant No. RG0280/1999M) and the U.S. Department of Argiculture (Grant No. NRICGP 97 01472) supported this work. S.B. was supported by the Danish Veterinary and Agricultural Research Council (Grant No. 970265).
References
- Andersen, M.D., Busk, P.K., Svendsen, I., and Møller, B.L. (2000). Cytochromes P450 from cassava (Manihot esculenta Crantz) catalyzing the first steps in the biosynthesis of the cyanogenic glucosides linamarin and lotaustralin. Cloning, functional expression in Pichia pastoris, and substrate specificity of the isolated recombinant enzymes. J. Biol. Chem. 275 1966–1977. [DOI] [PubMed] [Google Scholar]
- Bak, S., Kahn, R.A., Nielsen, H.L., Møller, B.L., and Halkier, B.A. (1998. a). Cloning of three A-type cytochromes P450, CYP71E1, CYP98, and CYP99 from Sorghum bicolor (L.) Moench by a PCR approach and identification by expression in Escherichia coli of CYP71E1 as a multifunctional cytochrome P450 in the biosynthesis of the cyanogenic glucoside dhurrin. Plant Mol. Biol. 36 393–405. [DOI] [PubMed] [Google Scholar]
- Bak, S., Nielsen, H.L., and Halkier, B.A. (1998. b). The presence of CYP79 homologues in glucosinolate-producing plants shows evolutionary conservation of the enzymes in the conversion of amino acids til aldoxime in the biosynthesis of cyanogenic glucosides and glucosinolates. Plant Mol. Biol. 38 725–734. [DOI] [PubMed] [Google Scholar]
- Bak, S., Olsen, C.E., Petersen, B.L., Møller, B.L., and Halkier, B.A. (1999). Metabolic engineering of p-hydroxybenzylglucosinolate in Arabidopsis by expression of the cyanogenic CYP79A1 from Sorghum bicolor. Plant J. 20 663–671. [DOI] [PubMed] [Google Scholar]
- Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., Sandberg, G., and Bellini, C. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. USA 97 14819–14824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 51–66. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Fink, G.E. (1994). Differential regulation of an auxin-producing nitrilase gene family in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 91 6649–6653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartling, D., Seedorf, M., Schmidt, R.C., and Weiler, E.W. (1994). Molecular characterization of two cloned nitrilases from Arabidopsis thaliana: Key enzymes in biosynthesis of the plant hormone indole-3-acetic acid. Proc. Natl. Acad. Sci. USA 91 6021–6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W., Cervera, M.-T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Oncklen, H.V., Montagu, M.V., and Inzé, P. (1995). Superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. [DOI] [PubMed] [Google Scholar]
- Chavadej, S., Brisson, N., McNeil, J.N., and De Luca, V. (1994). Redirection of tryptophan leads to production of low indole glucosinolate canola. Proc. Natl. Acad. Sci. USA 91 2166–2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, P.J. (1995). The plant hormones: Their nature, occurrence and functions. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 1–12.
- Delarue, M., Prinsen, E., Onckelen, H.V., Caboche, M., and Bellini, C. (1998). Sur2 mutations of Arabidopsis thaliana define a new locus involved in the control of auxin homeostatis. Plant J. 14 603–611. [DOI] [PubMed] [Google Scholar]
- Frey, M., Chomet, P., Glawischnig, E., Stettner, C., Grun, S., Winklmair, A., Eisenreich, W., Bacher, A., Meeley, R.B., Briggs, S.P., Simcox, K., and Gierl, A. (1997). Analysis of a chemical plant defense mechanism in grasses. Science 277 696–699. [DOI] [PubMed] [Google Scholar]
- Grsic, S., Sauerteig, S., Neuhaus, K., Albrecht, M., Rossiter, J., and Ludwig-Müller, J. (1998). Physiological analysis of transgenic Arabidopsis thaliana plants expressing one nitrilase isoform in the sense or antisense direction. J. Plant Physiol. 153 446–456. [Google Scholar]
- Grsic-Rausch, S., Kobelt, P., Siemens, J.M., Bischoff, M., and Ludwig-Müller, J. (2000). Expression and localization of nitrilase during symptom development of the clubroot disease in Arabidopsis. Plant Physiol. 122 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A. (1999). Glucosinolates. In Natural Occurring Glycosides, R. Ikan, ed (Chichester, UK: John Wiley), pp. 193–223.
- Halkier, B.A., and Du, L. (1997). The biosynthesis of glucosinolates. Trends Plant Sci. 11 425–430. [DOI] [PubMed] [Google Scholar]
- Halkier, B.A., Nielsen, H.L., Koch, B., and Møller, B.L. (1995). Purification and characterization of recombinant cytochrome P450TYR expressed at high levels in Escherichia coli. Arch. Biochem. Biophys. 322 369–377. [DOI] [PubMed] [Google Scholar]
- Hofmann, F., Rausch, T., and Hilgenberg, W. (1980). Preparation of radioactively labelled indole-3-acetic acid precursors. J. Labelled Compd. Radiopharm. 18 1491–1495. [Google Scholar]
- Hogge, R.L., Reed, D.W., and Underhill, E.W. (1988). HPLC separation of glucosinolates from leaves and seeds of Arabidopsis thalina and their identification using termospray liquid chromotography/mass spectrometry. Chromatogr. Sci. 26 551–556. [Google Scholar]
- Hull, A.K., Vij, R., and Celenza, J.L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. USA 97 2379–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jefcoate, C.R. (1978). Measurement of substrate and inhibitor binding to microsomal cytochrome P-450 by optical-difference spectroscopy. Methods Enzymol. 27 258–279. [DOI] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidospis. Plant Cell 7 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehman, A., Black, R., and Ecker, J.R. (1996). HOOKLESS1, an ethylene responsive gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85 183–194. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402 769–777. [DOI] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Hansen, C.H., Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275 33712–33717. [DOI] [PubMed] [Google Scholar]
- Mizutani, M., Ward, E., and Ohta, D. (1998). Cytochrome P450 superfamily in Arabidopsis thaliana: Isolation of cDNA's, differential expression, and RFLP mapping of multiple cytochromes P450. Plant Mol. Biol. 37 39–52. [DOI] [PubMed] [Google Scholar]
- Müller, A., Hillebrand, H., and Weiler, E.W. (1998). Indole-3-acetic acid is synthesized from L-tryptophan in roots of Arabidopsis thaliana. Planta 206 362–369. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, R. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nielsen, J.S., and Møller, B.L. (2000). Cloning and expression of cytochrome P450 enzymes catalysing the conversion of tyrosine to p-hydroxyphenylacetaldoxime in the biosynthesis of cyanogenic glucosides in Triglochin maritima. Plant Physiol. 122 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly, J., and Bartel, B. (1999). Redundancy as a way of life—IAA metabolism. Curr. Opin. Plant Biol. 2 207–213. [DOI] [PubMed] [Google Scholar]
- Normanly, J., Cohen, J.D., and Fink, G.R. (1993). Arabidopsis thaliana auxotrophs reveal a tryptophan-independent biosynthetic pathway for indole-3-acetic acid. Proc. Natl. Acad. Sci. USA 90 10355–10359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Normanly, J., Grisafi, P., Fink, G.R., and Bartel, B. (1997). Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette, S.M., Bak, S., and Feyereisen, R. (2000). Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol. 19 307–317. [DOI] [PubMed] [Google Scholar]
- Pompon, D., Louerat, B., Bronine, A., and Urban, P. (1996). Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272 51–64. [DOI] [PubMed] [Google Scholar]
- Poulton, J.E., and Møller, B.L. (1993). Glucosinolates. Methods Plant Biochem. 9 209–237. [Google Scholar]
- Rajapogal, R., and Larsen, P. (1972). Metabolism of indole-3-acetaldoxime in plants. Planta 103 45–54. [DOI] [PubMed] [Google Scholar]
- Rausch, T., Helmlinger, J., and Hilgenberg, W. (1985). High-performance liquid chromotographic separation and some properties of (E)- and (Z)-3-indoleacetaldoxime. J. Chromatogr. 318 95–102. [Google Scholar]
- Seo, M., Akaba, S., Oritani, T., Delarue, M., Bellini, C., Caboche, M., and Koshiba, T. (1998). Higher activity of an aldehyde oxidase in the auxin-overproducing superroot mutant of Arabidopsis thaliana. Plant Physiol. 116 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Songstad, D.D., De Luca, V., Brisson, N., Kurz, W.G.W., and Nessler, C.L. (1989). High-levels of tryptamine accumulation in transgenic tobacco expressing tryptophan decarboxylase. Plant Physiol. 94 1410–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thimann, K.V., and Mahadevan, S. (1964). Nitrilase. I. Occurrence, preparation, and general properties of the enzyme. Arch. Biochem. Biophys. 105 133–141. [DOI] [PubMed] [Google Scholar]
- Wetter, L.R., and Chrisholm, M.D. (1968). Sources of sulfur in the thioglucosides of various higher plants. Can. J. Biochem. 46 931–935. [DOI] [PubMed] [Google Scholar]
- Wildman, S.G. (1997). The auxin-A, B enigma: Scientific fraud or scientific ineptitude? Plant Growth Reg. 23 37–68. [Google Scholar]
- Winkler, R.G., Frank, M.R., Galbraith, D.W., Feyereisen, R., and Feldmann, K.A. (1998). Systematic reverse genetics of transfer-DNA-tagged lines of Arabidopsis. Plant Physiol. 118 743–750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock, U., and Halkier, B.A. (2000). Cytochrome P450 CYP79A2 from Arabidopsis thaliana L. catalyzes the conversion of L-phenylalanine to phenylacetaldoxime in the biosynthesis of benzylglucosinolates. J. Biol. Chem. 275 14659–14666. [DOI] [PubMed] [Google Scholar]
- Zhou, N., Tootle, T.L., and Glazebrook, J. (2000). Arabidopsis PAD3, a gene required for camalexin biosynthesis, encodes a putative cytochrome P450 monooxygenase. Plant Cell 11 2419–2428. [DOI] [PMC free article] [PubMed] [Google Scholar]