Abstract
Biofilms are highly tolerant to antimicrobials and host immune defense, enabling pathogens to thrive in hostile environments. The diversity of microbial biofilm infections requires alternative and complex treatment strategies. In a previous work we demonstrated that the human Atrial Natriuretic Peptide (hANP) displays a strong anti-biofilm activity toward Pseudomonas aeruginosa and that the binding of hANP by the AmiC protein supports this effect. This AmiC sensor has been identified as an analog of the human natriuretic peptide receptor subtype C (h-NPRC). In the present study, we evaluated the anti-biofilm activity of the h-NPRC agonist, osteocrin (OSTN), a hormone that displays a strong affinity for the AmiC sensor at least in vitro. Using molecular docking, we identified a pocket in the AmiC sensor that OSTN reproducibly docks into, suggesting that OSTN might possess an anti-biofilm activity as well as hANP. This hypothesis was validated since we observed that OSTN dispersed established biofilm of P. aeruginosa PA14 strain at the same concentrations as hANP. However, the OSTN dispersal effect is less marked than that observed for the hANP (−61% versus −73%). We demonstrated that the co-exposure of P. aeruginosa preformed biofilm to hANP and OSTN induced a biofilm dispersion with a similar effect to that observed with hANP alone suggesting a similar mechanism of action of these two peptides. This was confirmed by the observation that OSTN anti-biofilm activity requires the activation of the complex composed by the sensor AmiC and the regulator AmiR of the ami pathway. Using a panel of both P. aeruginosa laboratory reference strains and clinical isolates, we observed that the OSTN capacity to disperse established biofilms is highly variable from one strain to another. Taken together, these results show that similarly to the hANP hormone, OSTN has a strong potential to be used as a tool to disperse P. aeruginosa biofilms.
Keywords: Biofilm, Natriuretic peptides, Osteocrin, h-NPR-C, Bacterial adaptation, Bacterial sensor
1. Introduction
Biofilms represent major economic and sociological challenges to human and animal health, in addition to their effects on water installations [1]. Unlike biofilms found in the environment that can be treated with varying degrees of success, reducing and especially eradicating a biofilm installed in the tissues of an infected host represents an enormous challenge [2]. Biofilms adopt a specific architecture with a complex matrix trapping and protecting bacteria that merges with human tissues and secretions [3]. Bacteria in a biofilm display high tolerance to antimicrobials and host immune defense, enabling pathogens to survive in hostile environments and to disperse and colonize new tissues [4,5]. In addition, a small percentage of cells in biofilms adopt the persister phenotype. These are highly tolerant to antibiotics, favoring the relapse of infections [[6], [7], [8]]. Among bacteria able to establish biofilms in patient tissue, the opportunistic pathogen Pseudomonas aeruginosa is a major cause of cystic fibrosis (CF) mortality [9,10]. During the infection the bacteria build a biofilm structure embedded in the overproduced viscous mucus in the CF lungs [[11], [12], [13]]. Current treatment strategies include inhalation of enzymes (e.g., Dornase alfa, Pulmozyme®) that reduce the mucus viscosity of CF patients [14]. A very promising new triple therapy combines three CFTR modulators (elexacaftor/tezacaftor/ivacaftor), resulting in a thinner mucus [15,16]. However, this treatment does not eradicate the pathogen responsible for regular recurrences. In this context, new agents or strategies focused on the dispersion and/or eradication of established lung biofilms could improve the overall prognosis.
Over the last 20 years, antimicrobial peptides (AMP) have emerged as an interesting alternative to antibiotics. The prevailing concept is that their antibacterial activities are challenging bacteria exposed to these peptides to deploy resistance mechanisms [17,18]. Therefore, AMP alone or in association with antibiotics [19] have been proposed as one anti-biofilm weapon among the numerous strategies developed to disperse or eradicate biofilm [20]. AMP are classically characterized by their membranolytic activities, likely provoking a physiological stress response by the bacteria. Moreover, reaching all bacteria protected by a biofilm embedded in the middle of the infected host's secretion is challenging. Two novel strategies have been recently proposed to accelerate the discovery of human host produced molecules that act as antimicrobial or antibiofilm compounds. One approach is to search for peptide molecules present in human blood or organs that have been characterized for various metabolic or physiological functions, and that contain partial or complete signatures of potential antimicrobial activities [[21], [22], [23], [24]]. The second concept was inspired by microbial endocrinology [25], and proposes testing the potential antibacterial or antibiofilm activity of human communication molecules, such as hormones, neurotransmitters, or cytokines [[26], [27], [28], [29]].
This latter notion of affecting biofilms has the advantage of having a non-lethal action on bacteria, suggesting a reduction of the risks of bacteria developing resistance to these molecules, even if this is not an absolute rule [30,31]. The mode of action of these eukaryotic communication molecules on bacteria is often similar to their effect on human cells. The signaling molecules bind to a bacterial sensor/receptor, which often has another main function in bacteria, to trigger a cascade of events in the bacteria. This will cause bacterial metabolism and physiology to be disrupted in a manner that does not support continued infection [[26], [27], [28], [29],[32], [33], [34], [35]]. Among hormone peptides able to act on P. aeruginosa, the most studied are the natriuretic peptide family. This family comprises the atrial natriuretic peptide (ANP), the brain natriuretic peptide (BNP) and the c-type natriuretic peptide (CNP). The P. aeruginosa sensor for natriuretic peptides (NP) has been identified as AmiC [[36], [37], [38]]. Noticeably, the AmiC bacterial sensor discriminates between different NPs [36,38], as do the human NP subtype C (hNPR-C) [39,40]. AmiC therefore acts an hNPR-C receptor analog. It has been recently shown that the human ANP (hANP) strongly disperses established P. aeruginosa biofilm and that this effect is consecutive to binding to the AmiC sensor [38], whereas BNP and CNP are unable to disperse P. aeruginosa biofilm. Osteocrin (OSTN) is a peptide that belongs to the family of NP hormones [41], that is mainly expressed in bone [42] and skeletal muscle [43]. OSTN possesses a cardioprotective function after binding to the hNPR-C receptor [44]. Since OSTN is identified as an hNPR-C subtype receptor agonist [45], we speculate that this peptide also has the potential to function as an anti-biofilm agent as well as the hANP [38].
This study aims to investigate the impact of OSTN on P. aeruginosa biofilm. Using molecular docking, we showed that OSTN binds to the P. aeruginosa sensor AmiC. We demonstrated that OSTN strongly disperses established P. aeruginosa biofilms and that AmiC and AmiR, members of the ami operon, are essential to achieve this biofilm dispersal effect. In addition, we evaluated the biofilm dispersal effect of OSTN against P. aeruginosa clinical strains, revealing a high heterogeneity in the sensitivity of clinical strains to the peptide. Finally, the results showed that OSTN has no cytotoxicity or cardio toxicity. We therefore propose this peptide as a potential weapon to disperse P. aeruginosa biofilms in chronically infected patients.
2. Material and methods
2.1. Bacterial strains, media, and growth conditions
The bacterial strains used in this study are listed in Supplementary Table 1. Pseudomonas aeruginosa PA14 strain was obtained from the Biomerit Research Center (Univ. Cork, Ireland) [46]. The PA14ΔamiR mutant contains a clean deletion of the amiR gene following a cross-over performed with a plasmid carrying the flanking regions of the gene of interest [38]. The PA14ΔamiC mutant was obtained by insertion of a transposon [47]. The complemented strain PA14ΔamiC-AmiC + harbors the pBBR-MCS4 plasmid carrying the amiC gene [36]. P. aeruginosa H103 is a prototroph of the wild-type strain PAO1 [48]. P. aeruginosa PAK strain is a non-mucoid clinical strain [49]. Strains MUC-N1, MUC-N2, MUC-P4, MUC-P5 [50] are clinical isolates collected from sputum of CF patients (Nantes hospital) as well as CF6.14 (CNR Besançon, France). Finally, the clinical strains PAL 0.1 and PAL 1.1 [51] were isolated from the respiratory tract of a patient in intensive care unit (Lille hospital, France).
All bacterial strains were cultivated in Luria Bertani (LB) medium (10 g/L tryptone, 5 g/L yeast extract, and 5 g/L NaCl) at 37 °C with agitation (180 rpm). For the mutant strains P. aeruginosa PA14ΔamiC and PA14ΔamiC-AmiC+, the LB medium was supplemented with gentamycin (50 μg/mL) and carbenicillin (400 μg/mL), respectively.
2.2. Test substances
Osteocrin (SFSGFGSPLDRLSAGSVDHKGKQRKVVDHPKRRFGIPMDRIGRNRLSNSR-NH2) was synthesized by Fmoc solid phase methodology on a liberty microwave assisted automated peptide synthesizer (CEM, Saclay, France) using the standard manufacturer's procedure at 0.1 mmol scale on a Rink amide MBHA resin [52]. The pseudoproline Fmoc-Gly-Ser(Ψ(Me,Me)Pro)-OH was used for coupling of Gly15 and Ser16 to minimize aggregation of the peptide during the synthesis. After completion of the chain assembly, the peptide was deprotected and cleaved from the resin by adding 10 ml of an ice-cold mixture trifluoroacetic acid (TFA)/phenol/H2O/thioanisole/ethane-dithiol (82.5:5:5:5:2.5, v/v/v/v/v) for 120 min at room temperature. After filtration, the crude peptide was washed thrice by precipitation in tert-butyl methyl ether followed by centrifugation (3,260 g, 15 min). The synthetic peptide was purified by reversed phase (RP) HPLC on a 2.2 × 25 cm Vydac 218TP1022C18 column (Grace, Epernon, France) using a linear gradient (20–40% over 45 min) of acetonitrile/TFA (99.9:0.1) at a flow rate of 10 mL/min. The purified peptide was then characterized by MALDI-TOF mass spectrometry on an UltrafleXtreme (Bruker, Strasbourg, France) in the linear mode using sinapinic acid as a matrix. Analytical RP-HPLC, performed on a 0.46 × 25 cm Vydac 218TP54C18 column (Grace), indicated that the purity of the peptide was >99.9%.
The human Atrial Natriuretic Peptide (hANP) was purchased from Calbiochem Merck (United States). Stock solutions at 1 mg/mL were prepared in ultrapure water and stored at −20 °C until use.
2.3. Flow cell biofilm assays under dynamic conditions
The flow cell system, which allows continuous bacterial biofilm formation, was assembled, prepared, and sterilized as described by Tolker-Nielsen and Sternberg [53]. Bacterial cells from an overnight bacterial culture, were recovered by centrifugation (10 min, 7500 g) and washed with sterile physiological water (0.9% w/v NaCl). Each channel of the flow cell (1 mm × 4 mm x 40 mm Bio centrum, DTU, Denmark) was inoculated with 300 μL of bacterial suspension prepared at an OD580 value of 0.1. Bacterial adhesion was performed without any flow for 2 h at 37 °C. After 2 h of adhesion, LB medium was pumped with a flow rate of 2.5 mL/h at 37 °C for 24 h. After 24 h, the different treatments with the compounds of interest were injected onto the formed biofilms using ultra-pure water for the control. The treatment was performed for 2 h at 37 °C and then the bacterial cells were stained with SYTO9 green-fluorescent dye (Invitrogen, Carlsbad, CA) and observed with confocal microscopy.
2.4. Confocal laser scanning microscopy (CLSM)
The CLSM observations of biofilms were performed using a Zeiss LSM710 microscope (Carl Zeiss Microscopy, Oberkochen, Germany) using a x40 oil immersion objective. Bacteria into the biofilm were stained with 5 μM of SYTO9 green-fluorescent dye (Invitrogen, Carlsbad, CA). Images were taken every micrometre throughout the whole biofilm depth. For visualization and processing of three-dimensional (3D) image, the Zen 2.1 SP1 software (Carl Zeiss Microscopy, Oberkochen, Germany) was used. Quantitative analyses of images stacks were performed using the COMSTAT software (http://www.imageanalysis.dk/) [54]. At least three image stacks from at least three independent experiments were used for each analysis.
2.5. Molecular docking
An initial OSTN model was prepared using AlphaFold v.2.2.0 [55], running on a local server using the 2022-03-03 database. This model was docked to the AmiC dimer (PDB ID: 1PEA) using the HADDOCK v. 2.4–2022.08 web server (https://wenmr.science.uu.nl/haddock2.4/) [56,57] and AutoDock Vina v. 1.2.0 [58]. AutoDock Vina was run using the largest box size of 120 and exhaustiveness of 80. The Phyre2 web server [59] was used to predict another fold of the OSTN peptide from the amino acid sequence. The output structures were analyzed using PyMOL v. 2.5.2 (Schrödinger) and LigPlot+ v.2.2 [60]. Surface electrostatics were calculated using the APBS [61] PyMOL plugin.
2.6. Statistical analyses
Statistical analyses were performed using Prism GraphPad software version 9.0. The data were statistically analyzed using unpaired (two samples) two-tailed t-test, or ordinary one-way analysis of variance (ANOVA) followed by Dunnett's multiple comparison test to calculate p values. Experiments were performed with at least three biological independent replicates and results were displayed as means ± SEMs (standard error of the means).
3. Results
3.1. OSTN likely forms a complex with a dimer of the P. aeruginosa sensor protein AmiC
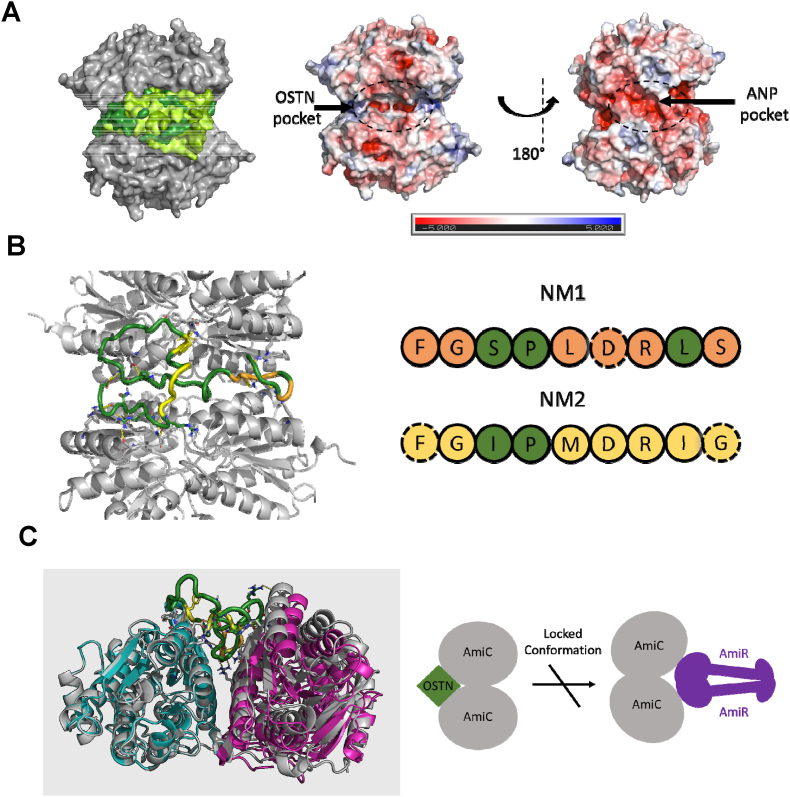
Using microscale thermophoresis, we previously observed that osteocrin (OSTN), like other natriuteric peptide hormones, interacts with AmiC [36]. We therefore evaluated the OSTN-AmiC interaction in silico. As there is no experimental structure of OSTN, we prepared an AlphaFold2 model [55]. The model showed an extended conformation (Supplementary Fig. 1), and AlphaFold could not predict a complex between AmiC and OSTN. We then docked the AlphaFold model into AmiC using HADDOCK [62] and AutoDock Vina [58]. We docked into the AmiC dimer as this is the biologically relevant free AmiC form [63]. Both programs consistently docked OSTN into one pocket at the interface of the AmiC dimer (Fig. 1A/B, Supplementary Fig. 2). OSTN binds to its physiological receptor NPR-1 through two oligopeptides (NM1 and NM2) with similar sequences to the binding regions of ANP, BNP, and CNP [41,64] (Supplementary Fig. 3). HADDOCK showed the NM2 region forming a core hydrophobic interaction with AmiC, whilst the highest energy AutoDock solution showed interactions of both regions with AmiC. AutoDock performed molecular dynamics to optimize the protein-ligand interaction. This produced a series of solutions with similar energy (Fig. 1A, Supplementary Fig. 2; Supplementary Table 2), as expected given the extended native conformation of OSTN. The interactions between and OSTN and AmiC are similar in all the solutions (example shown in Supplementary Fig. 4).
Fig. 1.
Osteocrin (OSTN) likely binds to a hydrophobic pocket in the AmiC dimer. (A) Docking of OSTN to AmiC with AutoDock Vina consistently targets one pocket. AmiC is shown as a gray surface; the top five OSTN poses are shown as a surface representation in shades of green. The core interaction with AmiC is consistent with the folding of OSTN around this core differing. Right: the AmiC dimer surface colored by charge (red: negative charge; blue: positive charge). OSTN binds to a largely hydrophobic pocket with limited charge density, whilst the positively charged ANP binds to a highly negatively charged pocket on the opposite face of AmiC. (B) Highlight of the highest energy predicted OSTN pose. OSTN is shown as a colored ribbon, and AmiC as cartoon. The conserved elements of the NM1 and NM2 regions are shown in orange and yellow respectively. Conserved residues interacting with AmiC are shown with dashed outlines (right). (C) OSTN binding causes a flex of the AmiC dimer. The OSTN-bound AmiC is shown in gray, with the acetamide bound AmiC shown in cyan (protomer superimposed with OSTN bound structure) and magenta. OSTN binding draws the two AmiC protomers towards OSTN. This interaction is antithetical to the AmiC movement required to accommodate AmiR binding (Supplementary Fig. 5). (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
The optimized OSTN-AmiC complex requires a flexing in the AmiC dimer (Fig. 1C). The extensive OSTN-AmiC interaction is likely to stabilize this AmiC conformation. This conformation would prevent the opening of the AmiC dimer that is required for AmiR binding [65]. The flexing of the AmiC dimer would also likely prevent the interaction of hANP with AmiC, as hANP is predicted to bind to a negatively charged pocket on AmiC that is disrupted in the optimized AmiC-OSTN complex (Fig. 1A, right).
3.2. OSTN disperses P. aeruginosa pre-formed biofilms
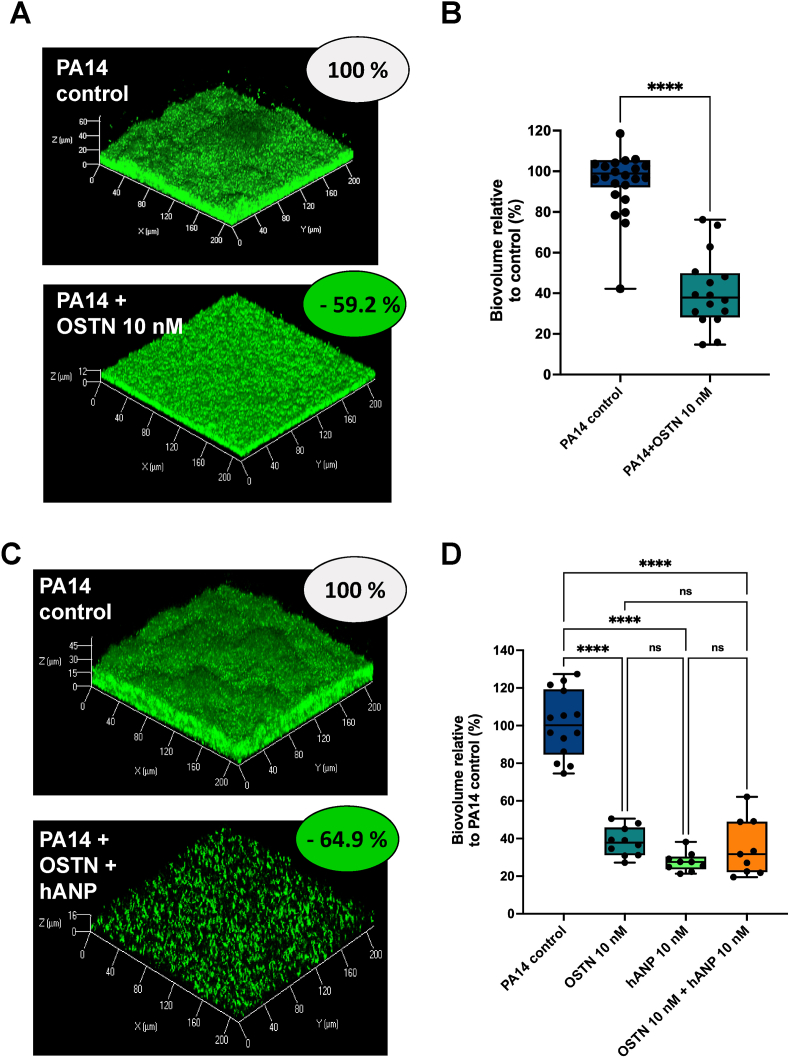
In a previous work, we showed that the AmiC-binding peptide hANP can strongly disperse established P. aeruginosa biofilms [38]. We therefore evaluated the potential biofilm dispersal activity of OSTN. Biofilms of P. aeruginosa PA14 strain formed over 24 h were exposed to OSTN at 10 nM for 2 h. As a control condition, we used established biofilms exposed to pure water which was used as a solvent to dilute OSTN. The results indicated that the exposure to OSTN induced a significant dispersion of established biofilms of P. aeruginosa notably with a loss of mushroom-like structures (Fig. 2A). COMSTAT analyses of CLSM images revealed a significant reduction (59.2 ± 4.5%) of the biofilm biovolume after exposure to OSTN as compared to the control condition (Fig. 2B). Since the in silico molecular docking predicted that the binding of OSTN and hANP to P. aeruginosa AmiC is mutually exclusive, we exposed established biofilms of P. aeruginosa to a mix of OSTN and hANP (10 nM each peptide) (Fig. 2C). The biofilm dispersal was only 64.9 ± 5.0% upon exposure to the OSTN-hANP cocktail, whereas in the same conditions OSTN alone and hANP alone resulted in 61.7 ± 2.4% and 72.4 ± 1.7% dispersion of preformed biofilm, respectively, showing no significant difference (ns) in the antibiofilm effect when biofilms were exposed to these three conditions (Fig. 2D). These data demonstrated that there is no additional or synergistic effect of the cocktail OSTN-hANP compared with the two natriuretic peptides when used independently, suggesting a common mechanism of action for these two hormone peptides to disperse established biofilms.
Fig. 2.
Effect of osteocrin (OSTN) on established biofilm of P. aeruginosa. (A) 3D-shadow representations of 24 h pre-formed P. aeruginosa PA14 biofilm in control condition (upper part) and 24 h pre-formed P. aeruginosa PA14 biofilm exposed to OSTN (10 nM) for 2 h at 37 °C (lower part). (B) COMSTAT image analyses of biofilms structures of P. aeruginosa PA14 control or exposed to OSTN (10 nM). Data are the result of the analysis of nineteen views from five independent biological experiments (control) and sixteen measurements from five independent experiments (OSTN 10 nM). (C) 3D-shadow representations of 24 h pre-formed P. aeruginosa PA14 biofilm structures untreated (control condition) or exposed to a cocktail of OSTN (10 nM) and hANP (10 nM) for 2 h at 37 °C. (D) COMSTAT image analyses of biofilms structures of P. aeruginosa PA14 control or exposed to OSTN (10 nM) alone, hANP (10 nM) alone or exposed to a cocktail of OSTN (10 nM) and hANP (10 nM) for 2 h at 37 °C. Data are the result of the analysis of fifteen views from three independent biological experiments (control condition), ten measurements from three independent biological experiments (OSTN 10 nM) and nine measurements from three independent biological experiments (hANP10 nM; and OSTN 10 nM + hANP 10 nM). All biofilms were stained with the SYTO9 and observed by CLSM. Statistics were achieved using unpaired (two samples) two-tailed t-test, or ordinary one-way ANOVA followed by Dunnett's multiple-comparison test. Values that are significantly different are indicated by asterisks as follows: ****, p < 0.0001.
3.3. Effect of OSTN on various P. aeruginosa clinical isolates pre-formed biofilms
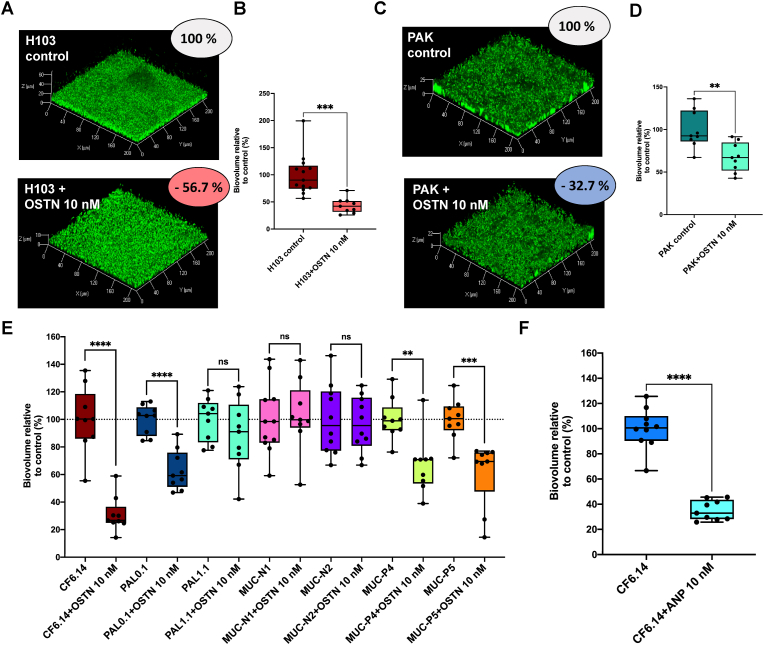
Next, we investigated whether the dispersal effect of OSTN is specific to PA14 strain. The impact of OSTN on established biofilms was first assessed on two other laboratory reference strains (H103 and PAK). The results showed that the biofilm established by H103 strain (a prototroph derivative of PAO1) has the same sensitivity as the PA14 strain to OSTN exposure (10 nM) displaying a reduction of biofilm biovolume of 56.7 ± 4.7% (H103 and OSTN) (Fig. 3A–B) and 59.2 ± 4.5% (PA14 and OSTN), respectively (Fig. 2B). In contrast, the biofilm formed by the PAK strain is less sensitive since OSTN exposure triggers 32.7 ± 5.7% reduction of the biofilm biovolume (Fig. 3C–D). To further evaluate the sensitivity of different P. aeruginosa biofilms to OSTN we tested its dispersal activity on various clinical isolates that formed different type of biofilms in term of global architecture and matrix organization. The results showed substantial heterogeneity of the clinical isolates response to OSTN. Interestingly, the CF 6.14 clinical strain collected from a CF patient appeared to be significantly sensitive to 10 nM of OSTN (69.0 ± 4.2% of biofilm dispersion) (Fig. 3E). The clinical isolates MUC-P4, MUC-P5, and PAL 0.1 showed a moderate sensitivity to OSTN with a biofilm dispersion of approximatively 40% (Fig. 3E). In contrast, PAL 1.1 clinical isolate showed weak sensitivity to OSTN (11.7 ± 8.5% of biofilm dispersion), and MUC-N1 and MUC-N2 were totally insensitive (Fig. 3E). Taken together, these results indicate that the dispersal effect of OSTN on established biofilms is not restricted to laboratory reference strains, however, OSTN showed an heterogenous dispersal effect on clinical isolates.
Fig. 3.
Effect of osteocrin (OSTN) on established biofilm of both P. aeruginosa lab strains and clinical isolates. (A) 3D-shadow representations of 24 h pre-formed P. aeruginosa H103 biofilm in control condition (top image) and 24 h pre-formed P. aeruginosa H103 biofilm exposed to OSTN (10 nM) for 2 h at 37 °C (bottom image). (B) COMSTAT image analyses of biofilms structures of P. aeruginosa H103 control or exposed to OSTN (10 nM). Data are the result of the analysis of thirteen views from three independent biological experiments (control) and nine measurements from three independent experiments (exposed to OSTN 10 nM). (C) 3D-shadow representations of 24 h pre-formed P. aeruginosa PAK biofilm in control condition (top image) and 24 h pre-formed P. aeruginosa PAK biofilm exposed to OSTN (10 nM) for 2 h at 37 °C (bottom image). (D) COMSTAT image analyses of biofilms structures of P. aeruginosa PAK control or exposed to OSTN (10 nM). Data are the result of the analysis of nine views from three independent biological experiments (control condition and exposed to OSTN 10 nM condition). (E) Effect of osteocrin (OSTN) on established biofilm from a couple of P. aeruginosa clinical isolate. The results of the COMSTAT image analyses of biofilms structures of P. aeruginosa strain, in control condition, are shown in left bar chart whereas the results of the COMSTAT image analyses of biofilms of the same strain, exposed to OSTN (10 nM; 2 h) are presented in the right bar chart right next to it control condition. For CF6.14 strain, PAL 0.1 strain, PAL 1.1 strain, P4 strain and P5 strain, data are the result of the analysis of nine views from three independent biological experiments (both in control condition and exposed to OSTN 10 nM condition). For N1 strain data are the result of the analysis of eleven views from three independent biological experiments (control condition) and nine views from three independent biological experiments (OSTN exposed condition). For N2 strain data are the result of the analysis of ten views from three independent biological experiments (control condition) and nine views from three independent biological experiments (OSTN exposed condition). (F) COMSTAT image analyses of biofilms structures of P. aeruginosa CF6.14 strain control or exposed to OSTN (10 nM). Data are the result of the analysis of ten views from three independent biological experiments and nine views from three independent biological experiments for respectively control condition and exposed to OSTN 10 nM condition. Statistics were achieved using unpaired (two samples) two-tailed t-test. Asterisks indicate values that are significantly different as follows: **, p < 0.01; ***, p < 0.001; ****, p < 0.0001.
3.4. OSTN biofilm dispersal mechanism of action: involvement of the ami operon
Our in silico data (Fig. 1) and our previous work [36] strongly suggest that the P. aeruginosa ami operon could be involved in the OSTN biofilm dispersion activity. The primary ami operon function is to allow hydrolysis of short-chain aliphatic amides to their corresponding organic acids through the final product of the operon, the aliphatic amidase AmiE. The genomic organization of the ami operon differs among P. aeruginosa strain. In strain PA14, the ami operon consists of four genes whereas in PAO1, the operon comprises six genes (www.pseudomonas.com) (Supplementary Fig. 6). The role of the three main proteins encoded by this operon (i.e., AmiC, AmiR and AmiE) is well defined. The AmiC sensor protein is the negative regulator that sequesters AmiR. After binding of acetamide or other agonists to the AmiC sensor, the AmiR regulator is released. This antiterminator regulator binds to ami mRNA sequences allowing the transcription of the full ami operon, including the amidase enzyme AmiE. Notably, despite the differential genomic architecture of the ami operon, the AmiC sensor amino-acid sequence is 100% identical between PAO1 and PA14 strains. The AmiR protein sequences show 98% identity between PAO1 and PA14 strains (4 mismatches among the 196 amino-acids constituting AmiR), strongly suggesting that the AmiC-AmiR pathway is activated similarly across P. aeruginosa strains.
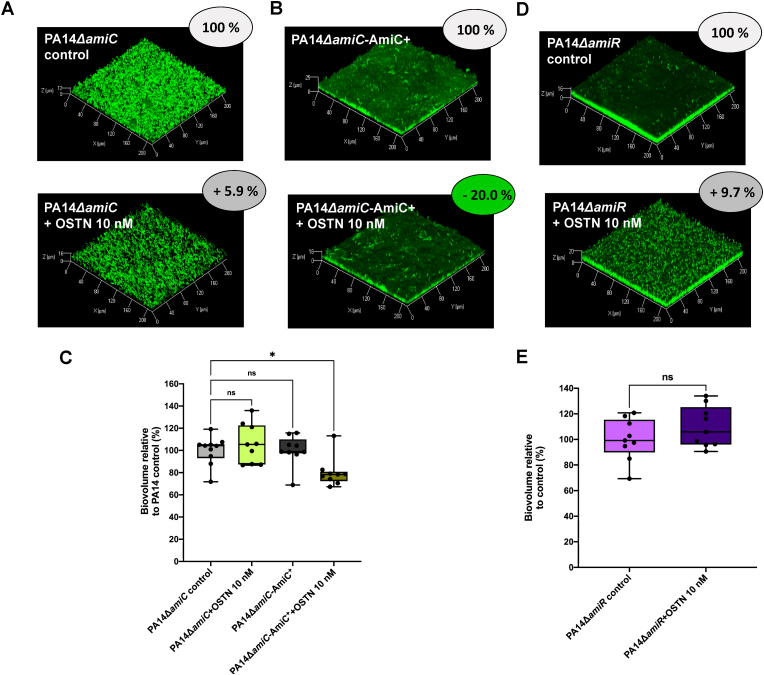
Since OSTN binds to AmiC (Fig. 1), and the dispersal effect of hANP is relayed by the complex AmiC-AmiR of the ami pathway [38], we used PA14ΔamiC and PA14ΔamiR mutant strains to decipher the mechanism of action underlying OSTN biofilm dispersal activity. The results showed that PA14ΔamiC is totally unaffected when treated with OSTN (2 h; 10 nM) (Fig. 4A and C). The PA14ΔamiC-AmiC + complemented with the amiC gene partially recovered sensitivity to OSTN treatment (2 h; 10 nM) (Fig. 4B and C). In parallel, we observed that the biofilm formed by a strain lacking the AmiR regulator (PA14ΔamiR deletion mutant) was also insensitive to OSTN exposure (Fig. 4D and E). However, OSTN treatment resulted in a modification of the global architecture of the biofilm formed by the PA14ΔamiR mutant strain as compared to the control condition (Fig. 4D).
Fig. 4.
Impact of OSTN (10 nM) on biofilm established by PA14-ΔamiC and PA14-ΔamiR strains. (A) 3D-shadow representations of the 24 h-old biofilm structures of the mutant strain PA14-ΔamiC control (above image) or exposed to OSTN (2 h; 10 nM; bottom image). (B) 3D-shadow representations of the 24 h-old biofilm structures of the complemented strain PA14-ΔamiC Comp AmiC (top image) or exposed to OSTN (2 h; 10 nM; above image). (C) COMSTAT image analyses of the biofilm structures of P. aeruginosa PA14-ΔamiC and PA14-ΔamiC Comp AmiC strains in control conditions or exposed to OSTN (10 nM) for 2 h at 37 °C. Data are the result of the analysis of ten views from three independent biological experiments (PA14-ΔamiC control condition), nine measurements from three independent biological experiments (PA14-ΔamiC + OSTN 10 nM), nine measurements from three independent biological experiments (PA14-ΔamiC Comp AmiC control) and nine measurements from three independent biological experiments (PA14-ΔamiC Comp AmiC + OSTN 10 nM). (D) 3D-shadow representations of the 24 h-old biofilm structures of the mutant strain PA14-ΔamiR control (top image) or exposed to OSTN (2 h; 10 nM; above image). (E) COMSTAT image analyses of the biofilm structures of P. aeruginosa PA14-ΔamiR in control conditions or exposed to OSTN (10 nM) for 2 h at 37 °C. Data are the result of the analysis of nine views from three independent biological experiments (PA14-ΔamiR control condition and PA14-ΔamiR exposed to OSTN). Statistics were achieved by ordinary one-way ANOVA followed by Dunnett's multiple-comparison test. Statistics were achieved using unpaired (two samples) two-tailed t-test, or ordinary one-way ANOVA followed by Dunnett's multiple-comparison test. Asterisks indicate values that are significantly different as follows: *, p < 0.05.
4. Discussion
Chronic P. aeruginosa infections result in the long-term establishment of a bacterial colonization of the respiratory tract, which is critical in numerous lung chronic disease such as bronchiectasis, chronic obstructive pulmonary disease (COPD) and in CF patients. Indeed, the ability of P. aeruginosa to form persistent biofilms in which the bacteria are embedded in a matrix, trapped in the mucus overproduced by CF patients, is a major obstacle to the eradication of these chronic infections and requires significant antibiotic treatments in terms of concentration and duration [66]. The repeated treatments trigger the emergence of bacterial resistant strains that necessitate urgent alternative and innovative therapeutics. In this general context emerged the idea of searching for anti-biofilm molecules targeting either the bacteria or the biofilm matrix to allow the dispersion of the biofilm [67], thus favoring the bactericidal action of antibiotics that can then be used at lower concentrations. In a previous study, we showed that hANP is capable of dispersing P. aeruginosa biofilms at very low concentrations [38], opening interesting therapeutic perspectives for NP molecules. In the present study, the impact of the host peptide hormone OSTN on established biofilms of P. aeruginosa was evaluated and its mechanism of action on P. aeruginosa was investigated.
We modelled the interaction between OSTN and AmiC using a suite of docking tools. These revealed that OSTN likely interacts with a hydrophobic pocket on the reverse side of the AmiC dimer to AmiR binding (Fig. 1A). Although OSTN adopts an extended, largely disorder structure in solution, we predicted that it forms a more compact structure in complex with AmiC. This structure interestingly reflects the structure predicted by the Phyre2 threading server (Supplementary Fig. 1A). The predicted AmiC-OSTN complex shows a subtle flexing of the AmiC dimer interface (Fig. 1C). The large stable interface between AmiC and OSTN likely explains the high apparent affinity of OSTN for AmiC and will prevent binding of both AmiR and hANP to AmiC [38,65]. This may explain why hANP and OSTN showed no additive effect when applied to P. aeruginosa. There is sufficient uncertainty in this modelling that it should be considered preliminary until an experimental AmiC-OSTN structure has been determined, which we are currently pursuing. However, using microscale thermophoresis, we previously observed that OSTN binds AmiC with a KD value lower than the detection limit of 100 nM [36].
Given that the biophysically determined interaction between OSTN and AmiC is structurally reasonable, we validated that OSTN disperses established biofilm of P. aeruginosa PA14 strain at the same concentration as was required for hANP (10 nM). This effect is likely to result from a modification of the biofilm organization since OSTN does not alter P. aeruginosa growth (Supplementary Fig. 7). A reduction of 59% of biofilm biovolume was observed upon exposure to OSTN, compared to the 73% biofilm dispersal found with hANP for the same bacterial strain and peptide concentration [38]. We observed no additive effect when 10 nM of both hANP and OSTN were added to the biofilm. Here, 65% of the biofilm was dispersed, representing a slight, but non-significative, reduction of the effect provoked by hANP alone (73% versus 65% of inhibition). These physiological data suggest that OSTN and hANP both bind AmiC in a mutually exclusive fashion. This is consistent with our docking of these peptides to AmiC, which suggested that the AmiC-OSTN interaction causes a small AmiC conformational change that occludes the proposed hANP binding site (Fig. 1C).
To verify whether the different proposed binding modes of OSTN and ANP to AmiC might reflect a difference in the OSTN mechanism of action, we tested OSTN in a mutant strain that did not express the AmiC sensor. This experiment confirmed unequivocally that AmiC is necessary for the dispersal effect of OSTN on P. aeruginosa PA14 biofilm, since PA14ΔamiC mutant strain is totally insensitive to OSTN.
It is well known that AmiC acts as a repressor of the ami operon activation since in the basal condition AmiC sequesters AmiR preventing the transcription of the whole ami operon [68,69]. The agonist (e.g., acetamide) binding to AmiC results in AmiR release and the transcription of the whole ami operon [65,69]. Following this, the final gene product of this operon, the amidase enzyme AmiE, is synthesized. OSTN binding to AmiC seems to prevent the formation of the AmiC-AmiR complex (Fig. 1C) [65]. This logically suggests that exposure of P. aeruginosa to OSTN would enhance the quantity of free AmiR anti-terminator. In this context, AmiR would be the relay of the OSTN effect on biofilm dispersion. We validated by showing that a mutant strain which did not express the AmiR protein (ΔamiR) is also totally insensitive to OSTN.
The ami operon gene architecture is not identical in different P. aeruginosa strains, especially between PAO1 and PA14. Thus, in P. aeruginosa PA14 strain the ami operon consists of four genes, coding for the AmiE enzyme, the chaperone PA14_20570, the AmiC sensor and the AmiR regulator (www.pseudomonas.com) [70]. In contrast, in P. aeruginosa PAO1 strain, six genes belong to the ami operon: the PA3362 gene (amiS), amiR, amiC, PA3365 gene (coding for the chaperone AmiB), amiE and amiL (a non-coding RNA sequence) (www.pseudomonas.com) [69,70] (Supplementary Fig. 6). To verify if this organization could impact the strain sensitivity to OSTN, we evaluated the impact of OSTN on PAO1 H103 and PAK strains. First, we observed that PAO1 H103 strain is sensitive to OSTN in the same range as PA14 strain, suggesting that the difference of the whole products of the ami operon between PAO1 and PA14 does not alter the sensitivity to OSTN. Since the AmiC sensor is crucial to observe the antibiofilm activity of natriuretic peptides (this study and [38], the fact that the amino-acid sequences of AmiC sensor is strictly identical in PAO1 and PA14 explains the absence of difference in sensitivity to OSTN. Secondly, we have noted that the PAK strain is less sensitive to OSTN than both H103 and PA14. The analysis of the AmiC of amino-acid sequence produced by PAK strain (annotated Y880_RS14905 in www.pseudomonas.com) revealed 99.2% of identity with the PA14 AmiC protein including three mismatches.
An alternative hypothesis is that the architecture of the biofilm, rather than the organization of the ami operon, may explain the strain specific differences in OSTN susceptibility. Indeed, characterization of the clinical strains selected in the panel demonstrated that they form biofilms with significant differences in biofilm spatial organization and matrix composition [50,71]. Moreover, it was recently observed that P. aeruginosa PA14 exposed to hANP modifies the exopolysaccharides composition of the biofilm matrix [38]. The fact that there is no difference in AmiC sequence between PAO1 and PA14 is consistent with this hypothesis, although we cannot exclude that AmiC-AmiR partners encoded by the other genes of the ami operon could play a role. This question is currently under investigation in our laboratory. The OSTN sensitivity of biofilm established by clinical isolates revealed a great heterogeneity and in addition show an important difference when compared with the hANP biofilm sensitivity [38]. First, we observed that the MUC-N1 and MUC-N2 clinical strains are totally insensitive to both OSTN and ANP exposure, confirming that OSTN and ANP antibiofilm activity is triggered by the same mechanism. However, the biofilm formed by the strain MUC-P4 that was highly sensitive to 10 nM ANP (92% biofilm dispersion) [38], is reduced by only 32.9% by OSTN (10 nM). Conversely, the biofilm of the MUC-P5 isolate is more sensitive to OSTN (39% dispersion) than to ANP (22% dispersion) [38]. Lastly, concerning clinical isolates recovered from sputum of CF patient, the biofilm of the strain CF 6.14 is highly sensitive to both OSTN (69% dispersion) and hANP (61% dispersion) (personal data). In parallel, we evaluated the impact of OSTN on biofilm formed by two other P. aeruginosa clinical strains (PAL 0.1 and PAL 1.1) collected from non-CF patients [51]. The biofilm of the PAL 1.1 (PA14 clade) isolate was poorly sensitive to OSTN at 10 nM (11.7% dispersion), whereas this biofilm is clearly more sensitive to hANP (47% dispersion) [38]. Concerning the PAL 0.1 (PAO1 clade) biofilm, we observed that exposure to 10 nM OSTN reduced 37% of the biomass, a biofilm that was also more sensitive to 10 nM ANP (52% dispersion) [38]. Taken together, these data show that strains that are sensitive to hANP are also sensitive to OSTN, with minor differences in effect of the two peptides between strains. This suggests that the precise binding of NPs to AmiC may differentially release AmiR and thus modulate the bacterial response to this exposure. Moreover, the key role of the anti-terminator regulator AmiR on biofilm regulation is currently under investigation.
5. Conclusions
The present study shows that the natriuretic peptide OSTN disperses established biofilms of P. aeruginosa laboratory strains and clinical isolates. The biofilm dispersal effect of OSTN seems to require the activation of the complex AmiC-AmiR of the ami pathway. In support of the development of OSTN as a treatment of P. aeruginosa chronic respiratory infections, OSTN is neither cytotoxic towards cultured lung cells nor cardiotoxic (Supplementary Fig. 8). Overall, our data highlight the potential use of OSTN as a potential antibiofilm agent since it meets several key criteria of a good candidate for therapeutic development.
CRediT authorship contribution statement
ML, AT, CT, TC, JL, BL, EB, MB, FD and OL conceived and designed the experiments. ML, AT and TC performed biofilm experiments. CT, FD and NJH conducted in silico analysis. TC, AT and EB constructed mutant strains. ML, AT and MB realized hERG experiment. ML, ML, AT, EK, TG, PC, MF, NJH, SC and OL analyzed the data. ML, AT, NJH, PC, SC, MF and OL wrote the paper.
Funding
This work was supported by grants from the Region Normandy, the European and Regional Development Fund, InterReg IVA PeReNE project, from the Normandy Valorisation cluster and from the BBSRC.
Declaration of competing interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: Lesouhaitier Olivier reports financial support was provided by University of Rouen Normandy. Lesouhaitier Olivier reports a relationship with University of Rouen Normandy that includes: employment and funding grants. Lesouhaitier Olivier has patent issued to Licensee. No conflict of interest.
Acknowledgments
We wish to thank Olivier Maillot, Lucille Roquigny and Jérémy Enault for technical assistance. We would like to thank William Stuart (University of Exeter) for setting up and maintaining the AlphaFold server. We wish to thank Prof. Dufour Alain (University of South Brittany, Lorient, France) and Prof. Caillon Jocelyne (University of Nantes, Nantes, France) who graciously provided us four clinical strains (MUC-N1, MUC-N2, MUC-P4 and MUC-P5) isolated from sputum samples of adult Cystic Fibrosis patients followed at the Centre de Référence Contre la Mucoviscidose (CRCM), Centre Hospitalier Universitaire (CHU) of Nantes (France). We wish to thank Prof. Jeannot Katy and Prof. Plesiat Patrick (French National Reference Centre for Antibiotic Resistance, Besançon, France) who graciously provided the CF6.14 clinical strain. M. Louis was recipient of a doctoral fellowship from Normandy Region. T. Clamens was recipient of a doctoral fellowship from the French Ministry of Research (MRE). The CBSA Lab is supported by the Région Normandie (France), the European FEDER funds, and Evreux Portes de Normandie (France) and the Living Systems Institute (University of Exeter) by BBSRC.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bioflm.2023.100131.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Camara M., Green W., MacPhee C.E., Rakowska P.D., Raval R., Richardson M.C., et al. Economic significance of biofilms: a multidisciplinary and cross-sectoral challenge. NPJ Biofilms Microbiomes. 2022;8(1):42. doi: 10.1038/s41522-022-00306-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Martin I., Waters V., Grasemann H. Approaches to targeting bacterial biofilms in cystic fibrosis airways. Int J Mol Sci. 2021;22(4) doi: 10.3390/ijms22042155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van den Bossche S., De Broe E., Coenye T., Van Braeckel E., Crabbe A. The cystic fibrosis lung microenvironment alters antibiotic activity: causes and effects. Eur Respir Rev. 2021;30(161) doi: 10.1183/16000617.0055-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciofu O., Tolker-Nielsen T. Tolerance and resistance of Pseudomonas aeruginosa biofilms to antimicrobial agents-how P. aeruginosa can escape antibiotics. Front Microbiol. 2019;10:913. doi: 10.3389/fmicb.2019.00913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cameron D.R., Pitton M., Oberhaensli S., Schlegel K., Prod'hom G., Blanc D.S., et al. Parallel evolution of Pseudomonas aeruginosa during a prolonged ICU-infection outbreak. Microbiol Spectr. 2022 doi: 10.1128/spectrum.02743-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hoiby N., Bjarnsholt T., Moser C., Bassi G.L., Coenye T., Donelli G., et al. ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect. 2015;21(Suppl 1):S1–S25. doi: 10.1016/j.cmi.2014.10.024. [DOI] [PubMed] [Google Scholar]
- 7.Ciofu O., Rojo-Molinero E., Macia M.D., Oliver A. Antibiotic treatment of biofilm infections. APMIS. 2017;125(4):304–319. doi: 10.1111/apm.12673. [DOI] [PubMed] [Google Scholar]
- 8.Balaban N.Q., Helaine S., Lewis K., Ackermann M., Aldridge B., Andersson D.I., et al. Definitions and guidelines for research on antibiotic persistence. Nat Rev Microbiol. 2019;17(7):441–448. doi: 10.1038/s41579-019-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Malhotra S., Hayes D., Jr., Wozniak D.J. Cystic fibrosis and Pseudomonas aeruginosa: the host-microbe interface. Clin Microbiol Rev. 2019;32(3) doi: 10.1128/CMR.00138-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Guillaume O., Butnarasu C., Visentin S., Reimhult E. Interplay between biofilm microenvironment and pathogenicity of Pseudomonas aeruginosa in cystic fibrosis lung chronic infection. Biofilms. 2022;4 doi: 10.1016/j.bioflm.2022.100089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolpen M., Jensen P.O., Faurholt-Jepsen D., Bjarnsholt T. Prevalence of biofilms in acute infections challenges a longstanding paradigm. Biofilms. 2022;4 doi: 10.1016/j.bioflm.2022.100080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolpen M., Kragh K.N., Enciso J.B., Faurholt-Jepsen D., Lindegaard B., Egelund G.B., et al. Bacterial biofilms predominate in both acute and chronic human lung infections. Thorax. 2022;77(10):1015–1022. doi: 10.1136/thoraxjnl-2021-217576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sauer K., Stoodley P., Goeres D.M., Hall-Stoodley L., Burmolle M., Stewart P.S., et al. The biofilm life cycle: expanding the conceptual model of biofilm formation. Nat Rev Microbiol. 2022;20(10):608–620. doi: 10.1038/s41579-022-00767-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang C., Montgomery M. Dornase alfa for cystic fibrosis. Cochrane Database Syst Rev. 2021;3(3):CD001127. doi: 10.1002/14651858.CD001127.pub5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heijerman H.G.M., McKone E.F., Downey D.G., Van Braeckel E., Rowe S.M., Tullis E., et al. Efficacy and safety of the elexacaftor plus tezacaftor plus ivacaftor combination regimen in people with cystic fibrosis homozygous for the F508del mutation: a double-blind, randomised, phase 3 trial. Lancet. 2019;394(10212):1940–1948. doi: 10.1016/S0140-6736(19)32597-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Middleton P.G., Mall M.A., Drevinek P., Lands L.C., McKone E.F., Polineni D., et al. Elexacaftor-Tezacaftor-Ivacaftor for Cystic Fibrosis with a Single Phe508del Allele. N Engl J Med. 2019;381(19):1809–1819. doi: 10.1056/NEJMoa1908639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Somma A., Moretta A., Cane C., Cirillo A., Duilio A. Antimicrobial and antibiofilm peptides. Biomolecules. 2020;10(4) doi: 10.3390/biom10040652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hancock R.E.W., Alford M.A., Haney E.F. Antibiofilm activity of host defence peptides: complexity provides opportunities. Nat Rev Microbiol. 2021;19(12):786–797. doi: 10.1038/s41579-021-00585-w. [DOI] [PubMed] [Google Scholar]
- 19.Reffuveille F., de la Fuente-Nunez C., Mansour S., Hancock R.E. A broad-spectrum antibiofilm peptide enhances antibiotic action against bacterial biofilms. Antimicrob Agents Chemother. 2014;58(9):5363–5371. doi: 10.1128/AAC.03163-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wille J., Coenye T. Biofilm dispersion: the key to biofilm eradication or opening Pandora's box? Biofilms. 2020;2 doi: 10.1016/j.bioflm.2020.100027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pane K., Cafaro V., Avitabile A., Torres M.T., Vollaro A., De Gregorio E., et al. Identification of novel cryptic multifunctional antimicrobial peptides from the human stomach enabled by a computational-experimental platform. ACS Synth Biol. 2018;7(9):2105–2115. doi: 10.1021/acssynbio.8b00084. [DOI] [PubMed] [Google Scholar]
- 22.Torres M.D.T., Cao J., Franco O.L., Lu T.K., de la Fuente-Nunez C. Synthetic biology and computer-based frameworks for antimicrobial peptide discovery. ACS Nano. 2021;15(2):2143–2164. doi: 10.1021/acsnano.0c09509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cesaro A., Torres M.D.T., Gaglione R., Dell'Olmo E., Di Girolamo R., Bosso A., et al. Synthetic antibiotic derived from sequences encrypted in a protein from human plasma. ACS Nano. 2022;16(2):1880–1895. doi: 10.1021/acsnano.1c04496. [DOI] [PubMed] [Google Scholar]
- 24.Torres M.D.T., Melo M.C.R., Flowers L., Crescenzi O., Notomista E., de la Fuente-Nunez C. Mining for encrypted peptide antibiotics in the human proteome. Nat Biomed Eng. 2022;6(1):67–75. doi: 10.1038/s41551-021-00801-1. [DOI] [PubMed] [Google Scholar]
- 25.Lyte M. Microbial endocrinology and infectious disease in the 21st century. Trends Microbiol. 2004;12(1):14–20. doi: 10.1016/j.tim.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Lesouhaitier O., Veron W., Chapalain A., Madi A., Blier A.S., Dagorn A., et al. Gram-negative bacterial sensors for eukaryotic signal molecules. Sensors. 2009;9(9):6967–6990. doi: 10.3390/s90906967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kumar A., Sperandio V. Indole signaling at the host-microbiota-pathogen interface. mBio. 2019;10(3) doi: 10.1128/mBio.01031-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lesouhaitier O., Clamens T., Rosay T., Desriac F., Louis M., Rodrigues S., et al. Host peptidic hormones affecting bacterial biofilm formation and virulence. J Innate Immun. 2019;11(3):227–241. doi: 10.1159/000493926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar A., Russell R.M., Pifer R., Menezes-Garcia Z., Cuesta S., Narayanan S., et al. The serotonin neurotransmitter modulates virulence of enteric pathogens. Cell Host Microbe. 2020;28(1):41–53. doi: 10.1016/j.chom.2020.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garcia-Contreras R., Maeda T., Wood T.K. Resistance to quorum-quenching compounds. Appl Environ Microbiol. 2013;79(22):6840–6846. doi: 10.1128/AEM.02378-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bove M., Bao X., Sass A., Crabbe A., Coenye T. The quorum-sensing inhibitor furanone C-30 rapidly loses its tobramycin-potentiating activity against Pseudomonas aeruginosa biofilms during experimental evolution. Antimicrob Agents Chemother. 2021;65(7) doi: 10.1128/AAC.00413-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wu L., Estrada O., Zaborina O., Bains M., Shen L., Kohler J.E., et al. Recognition of host immune activation by Pseudomonas aeruginosa. Science. 2005;309(5735):774–777. doi: 10.1126/science.1112422. [DOI] [PubMed] [Google Scholar]
- 33.Zaborina O., Lepine F., Xiao G., Valuckaite V., Chen Y., Li T., et al. Dynorphin activates quorum sensing quinolone signaling in Pseudomonas aeruginosa. PLoS Pathog. 2007;3(3):e35. doi: 10.1371/journal.ppat.0030035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chotirmall S.H., Smith S.G., Gunaratnam C., Cosgrove S., Dimitrov B.D., O'Neill S.J., et al. Effect of estrogen on pseudomonas mucoidy and exacerbations in cystic fibrosis. N Engl J Med. 2012;366(21):1978–1986. doi: 10.1056/NEJMoa1106126. [DOI] [PubMed] [Google Scholar]
- 35.Scardaci R., Bietto F., Racine P.J., Boukerb A.M., Lesouhaitier O., Feuilloley M.G.J., et al. Norepinephrine and serotonin can modulate the behavior of the probiotic Enterococcus faecium NCIMB10415 towards the host: is a putative surface sensor involved? Microorganisms. 2022;10(3) doi: 10.3390/microorganisms10030487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rosay T., Bazire A., Diaz S., Clamens T., Blier A.S., Mijouin L., et al. Pseudomonas aeruginosa expresses a functional human natriuretic peptide receptor ortholog: involvement in biofilm formation. mBio. 2015;6(4) doi: 10.1128/mBio.01033-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Desriac F., Clamens T., Rosay T., Rodrigues S., Tahrioui A., Enault J., et al. Different dose-dependent modes of action of C-type natriuretic peptide on Pseudomonas aeruginosa biofilm formation. Pathogens. 2018;7(2) doi: 10.3390/pathogens7020047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Louis M., Clamens T., Tahrioui A., Desriac F., Rodrigues S., Rosay T., et al. Pseudomonas aeruginosa biofilm dispersion by the human atrial natriuretic peptide. Adv Sci. 2022;9(7) doi: 10.1002/advs.202103262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potter L.R., Abbey-Hosch S., Dickey D.M. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr Rev. 2006;27(1):47–72. doi: 10.1210/er.2005-0014. [DOI] [PubMed] [Google Scholar]
- 40.Potter L.R. Natriuretic peptide metabolism, clearance and degradation. FEBS J. 2011;278(11):1808–1817. doi: 10.1111/j.1742-4658.2011.08082.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moffatt P., Thomas G.P. Osteocrin--beyond just another bone protein? Cell Mol Life Sci. 2009;66(7):1135–1139. doi: 10.1007/s00018-009-8716-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thomas G., Moffatt P., Salois P., Gaumond M.H., Gingras R., Godin E., et al. Osteocrin, a novel bone-specific secreted protein that modulates the osteoblast phenotype. J Biol Chem. 2003;278(50):50563–50571. doi: 10.1074/jbc.M307310200. [DOI] [PubMed] [Google Scholar]
- 43.Nishizawa H., Matsuda M., Yamada Y., Kawai K., Suzuki E., Makishima M., et al. Musclin, a novel skeletal muscle-derived secretory factor. J Biol Chem. 2004;279(19):19391–19395. doi: 10.1074/jbc.C400066200. [DOI] [PubMed] [Google Scholar]
- 44.Miyazaki T., Otani K., Chiba A., Nishimura H., Tokudome T., Takano-Watanabe H., et al. A new secretory peptide of natriuretic peptide family, osteocrin, suppresses the progression of congestive heart failure after myocardial infarction. Circ Res. 2018;122(5):742–751. doi: 10.1161/CIRCRESAHA.117.312624. [DOI] [PubMed] [Google Scholar]
- 45.Yasoda A. Physiological and pathophysiological effects of C-type natriuretic peptide on the heart. Biology. 2022;11(6) doi: 10.3390/biology11060911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rahme L.G., Stevens E.J., Wolfort S.F., Shao J., Tompkins R.G., Ausubel F.M. Common virulence factors for bacterial pathogenicity in plants and animals. Science. 1995;268(5219):1899–1902. doi: 10.1126/science.7604262. [DOI] [PubMed] [Google Scholar]
- 47.Liberati N.T., Urbach J.M., Miyata S., Lee D.G., Drenkard E., Wu G., et al. An ordered, nonredundant library of Pseudomonas aeruginosa strain PA14 transposon insertion mutants. Proc Natl Acad Sci U S A. 2006;103(8):2833–2838. doi: 10.1073/pnas.0511100103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hancock R.E., Carey A.M. Outer membrane of Pseudomonas aeruginosa: heat- 2-mercaptoethanol-modifiable proteins. J Bacteriol. 1979;140(3):902–910. doi: 10.1128/jb.140.3.902-910.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeya K., Amako K. A rod-shaped Pseudomonas phage. Virology. 1966;28(1):163–165. doi: 10.1016/0042-6822(66)90317-5. [DOI] [PubMed] [Google Scholar]
- 50.Boukerb A.M., Simon M., Pernet E., Jouault A., Portier E., Persyn E., et al. Draft genome sequences of four Pseudomonas aeruginosa clinical strains with various biofilm phenotypes. Microbiol Resour Announc. 2020;9(1) doi: 10.1128/MRA.01286-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grandjean T., Le Guern R., Duployez C., Faure K., Kipnis E., Dessein R. Draft genome sequences of two Pseudomonas aeruginosa multidrug-resistant clinical isolates, PAL0.1 and PAL1.1. Microbiol Resour Announc. 2018;7(17) doi: 10.1128/MRA.00940-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Touchard A., Aili S.R., Tene N., Barasse V., Klopp C., Dejean A., et al. Venom peptide repertoire of the European myrmicine ant manica rubida: identification of insecticidal toxins. J Proteome Res. 2020;19(4):1800–1811. doi: 10.1021/acs.jproteome.0c00048. [DOI] [PubMed] [Google Scholar]
- 53.Tolker-Nielsen T., Sternberg C. Growing and analyzing biofilms in flow chambers. Curr Protoc Microbiol. 2011;1 doi: 10.1002/9780471729259.mc01b02s21. Unit 1B 2. [DOI] [PubMed] [Google Scholar]
- 54.Heydorn A., Nielsen A.T., Hentzer M., Sternberg C., Givskov M., Ersboll B.K., et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology (Read) 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 55.Jumper J., Evans R., Pritzel A., Green T., Figurnov M., Ronneberger O., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583–589. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.van Zundert G.C.P., Rodrigues J., Trellet M., Schmitz C., Kastritis P.L., Karaca E., et al. The HADDOCK2.2 web server: user-friendly integrative modeling of biomolecular complexes. J Mol Biol. 2016;428(4):720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 57.Honorato R.V., Koukos P.I., Jimenez-Garcia B., Tsaregorodtsev A., Verlato M., Giachetti A., et al. Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front Mol Biosci. 2021;8 doi: 10.3389/fmolb.2021.729513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Eberhardt J., Santos-Martins D., Tillack A.F., Forli S. AutoDock Vina 1.2.0: new docking methods, expanded force field, and Python bindings. J Chem Inf Model. 2021;61(8):3891–3898. doi: 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kelley L.A., Mezulis S., Yates C.M., Wass M.N., Sternberg M.J. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10(6):845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wallace A.C., Laskowski R.A., Thornton J.M. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8(2):127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 61.Baker N.A., Sept D., Joseph S., Holst M.J., McCammon J.A. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98(18):10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dominguez C., Boelens R., Bonvin A.M. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125(7):1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 63.Pearl L., O'Hara B., Drew R., Wilson S. Crystal structure of AmiC: the controller of transcription antitermination in the amidase operon of Pseudomonas aeruginosa. EMBO J. 1994;13(24):5810–5817. doi: 10.1002/j.1460-2075.1994.tb06924.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nishida M., Miyamoto K., Abe S., Shimada M., Shimizu Y., Tsuji A., et al. Natriuretic peptide receptor-C releases and activates guanine nucleotide-exchange factor H1 in a ligand-dependent manner. Biochem Biophys Res Commun. 2021;552:9–16. doi: 10.1016/j.bbrc.2021.03.028. [DOI] [PubMed] [Google Scholar]
- 65.O'Hara B.P., Norman R.A., Wan P.T., Roe S.M., Barrett T.E., Drew R.E., et al. Crystal structure and induction mechanism of AmiC-AmiR: a ligand-regulated transcription antitermination complex. EMBO J. 1999;18(19):5175–5186. doi: 10.1093/emboj/18.19.5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dingemans J., Monsieurs P., Yu S.H., Crabbe A., Forstner K.U., Malfroot A., et al. Effect of shear stress on Pseudomonas aeruginosa isolated from the cystic fibrosis lung. mBio. 2016;7(4) doi: 10.1128/mBio.00813-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.King J., Murphy R., Davies J.C. Pseudomonas aeruginosa in the cystic fibrosis lung. Adv Exp Med Biol. 2022;1386:347–369. doi: 10.1007/978-3-031-08491-1_13. [DOI] [PubMed] [Google Scholar]
- 68.Wilson S., Drew R. Cloning and DNA sequence of amiC, a new gene regulating expression of the Pseudomonas aeruginosa aliphatic amidase, and purification of the amiC product. J Bacteriol. 1991;173(16):4914–4921. doi: 10.1128/jb.173.16.4914-4921.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wilson S.A., Wachira S.J., Drew R.E., Jones D., Pearl L.H. Antitermination of amidase expression in Pseudomonas aeruginosa is controlled by a novel cytoplasmic amide-binding protein. EMBO J. 1993;12(9):3637–3642. doi: 10.1002/j.1460-2075.1993.tb06037.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Winsor G.L., Griffiths E.J., Lo R., Dhillon B.K., Shay J.A., Brinkman F.S. Enhanced annotations and features for comparing thousands of Pseudomonas genomes in the Pseudomonas genome database. Nucleic Acids Res. 2016;44(D1):D646–D653. doi: 10.1093/nar/gkv1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Jouault A., Gobet A., Simon M., Portier E., Perennou M., Corre E., et al. Alterocin, an antibiofilm protein secreted by pseudoalteromonas sp. strain 3J6. Appl Environ Microbiol. 2020;86(20) doi: 10.1128/AEM.00893-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.