Abstract
Plants respond to day/night cycling in a number of physiological ways. At the mRNA level, the expression of some genes changes during the 24-hr period. To identify novel genes regulated in this way, we used microarrays containing 11,521 Arabidopsis expressed sequence tags, representing an estimated 7800 unique genes, to determine gene expression levels at 6-hr intervals throughout the day. Eleven percent of the genes, encompassing genes expressed at both high and low levels, showed a diurnal expression pattern. Approximately 2% cycled with a circadian rhythm. By clustering microarray data from 47 additional nonrelated experiments, we identified groups of genes regulated only by the circadian clock. These groups contained the already characterized clock-associated genes LHY, CCA1, and GI, suggesting that other key circadian clock genes might be found within these clusters.
INTRODUCTION
Plants have adapted their growth and development to use the diurnal cycling of light and dark. This is manifested at both the physiological level, with leaf movement, growth, and stomatal opening, and the molecular level, with expression of some genes occurring only at certain times of the day. The day/night cycling of gene expression is called a diurnal rhythm and is achieved primarily by two mechanisms: first, by light, and second, by a free-running internal circadian clock. Circadian clocks have been well characterized in animals, fungi, and bacteria, and in all cases they have a central oscillator that measures time with a molecular feedback loop that cycles over a 24-hr period (Dunlap, 1999). Although a growing number of genes either regulated by the clock or affecting clock function have been identified in plants, a full picture has yet to emerge.
The ability of plants to respond to light is achieved through photoreceptors. In Arabidopsis, two classes of photoreceptors are known: the red/far-red receptors, phytochrome A to E (Sharrock and Quail, 1989; Clack et al., 1994), and the blue light receptors, CRY1 (Ahmad and Cashmore, 1993), CRY2 (Guo et al., 1998), and NPH1 (Liscum and Briggs, 1995). Using these photoreceptors, a plant can detect a range of light intensities and wavelengths, with which it senses not only whether light is present but also from which direction the light is coming and whether there is competing vegetation (reviewed in Ballare, 1999). The best characterized of the photoreceptors are the phytochromes, for which the events that convert the light signal into transcriptional regulation have been described. Phytochrome is transported into the nucleus in a light-dependent manner (Sakamoto and Nagatani, 1996; Kircher et al., 1999). In the nucleus, it interacts with a basic helix-loop-helix transcription factor, PIF3 (Ni et al., 1998, 1999), which has been shown to bind to the G box element found in the promoters of many light-activated genes (Giuliano et al., 1988; Martinez-Garcia et al., 2000). This chain of events allows the plants to respond to light after germination, by stopping hypocotyl elongation and allowing cotyledon expansion, and in recurring diurnal cycles with the light and dark of the day.
For convenience, the circadian clock can be divided into three components: input, oscillator, and output (reviewed in Somers, 1999). In plants, these components have been best described in Arabidopsis, whose main input pathway is by light through the photoreceptors. Light is not the only regulator, however, because the clock can be initiated independently of light at imbibition (Zhong et al., 1998) and also can be altered by temperature changes (Kreps and Simon, 1997). The input pathway signal is transmitted to the oscillator, at least in part, through the ELF3 protein, because elf3 mutants show no circadian rhythm in the light, whereas in constant dark a rhythm is maintained (Hicks et al., 1996). Another potential input component is GI, because gi mutants affect expression patterns of clock-regulated genes, with one allele (gi-1) altering light signaling to the clock (Fowler et al., 1999; Park et al., 1999). The cycling of the input pathways trains the clock to a certain period that is maintained by the oscillator.
For the oscillator, a number of genes have been described that, when mutated, alter the period length of the clock under free-running conditions. These genes are of interest because the altered period lengths show that they are necessary to maintain true timing of the clock and therefore are part of either the clock or clock regulators. The toc1 mutant has a shorter period length and encodes a protein similar to a response regulator. It also contains a common domain with the flowering time gene product CO (Millar et al., 1995; Putterill et al., 1995; Strayer et al., 2000). The mutants ztl and fkf (Nelson et al., 2000; Somers et al., 2000) show a lengthening of period. Both of these genes have a similar predicted protein sequence with LOV domains (Christie et al., 1998) and ubiquitination binding sites. FKF is expressed in a circadian fashion, whereas ZTL is not. Because both affect the period of the clock, modulation of the protein might occur with the selective degradation of the protein by the ubiquitin pathway. Two other genes, LHY and CCA1, encode myb-related DNA binding proteins, are regulated in a circadian pattern, and show high homology with each other (Schaffer et al., 1998; Wang and Tobin, 1998). When these genes are overexpressed, no circadian rhythms (including their own) are detectable, implying that they are involved either in the oscillator or in the immediate output pathway. If CCA1 is part of the clock, then it is also involved in the output pathway because it binds to sequences in the promoter region of the photosynthetic gene CAB1 (Wang et al., 1997). Circadian-regulated genes in cca1 null mutants still cycle but with an altered period length, suggesting that CCA1 most likely acts redundantly with LHY (Green and Tobin, 1999).
Genes regulated by the circadian output pathway have been shown to peak in expression at different times of the day. Some examples include genes involved in photosynthesis, oxidative stress, cold response, and cell wall production. The photosynthetic genes CAB and RUBISCO anticipate the dawn by having a high level of expression in the morning (Ernst et al., 1990), whereas the cold-induced, glycine-rich RNA binding genes CCR1 and CCR2 (ATGRP7) have a high level of expression in the afternoon (Carpenter et al., 1994). ATGER3, a germin-like cell wall gene, has a high level of expression that peaks in the night (Staiger et al., 1999). Members of the catalase gene family are expressed at different times: CAT2 peaks in the morning, and CAT3 peaks in the afternoon (Zhong and McClung, 1996). These examples suggest that there are multiple regulatory mechanisms required to regulate circadian expression profiles.
In the past, identification of novel plant genes that are regulated by different rhythms has been achieved a few genes at a time. DNA microarrays can measure large numbers of gene expression patterns simultaneously (reviewed in Schaffer et al., 2000). In plants, large arrays have been used to identify novel genes involved in nutrient response and seed formation (Girke et al., 2000; Wang et al., 2000). As part of the Arabidopsis Functional Genomics Consortium, we have constructed a cDNA microarray containing 11,521 clones and used it to characterize gene expression patterns that are regulated in a diurnal and a circadian manner.
RESULTS
Identification of Genes with a Diurnal Rhythm
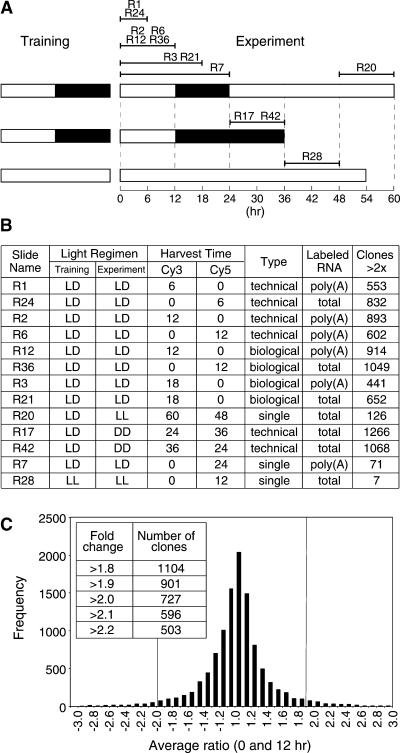
To identify genes with transcripts regulated in a diurnal cycle, we harvested tissue four times during the day: at 0, 6, 12, and 18 hr after dawn. The 0-hr time point was used as a reference sample with which the other time points were compared. In each case, the experiment was repeated, reversing the fluorescent dye. In addition, two biological repetitions of the 0- and 12-hr time points were compared (Figure 1A). For each experiment, the ratios of the fluorescence intensities of the two probes were calculated, and the number of clones showing a ratio greater than twofold was determined. Choosing cutoff values for ratios slightly different from 2.0 greatly altered the number of clones selected (Figures 1B and 1C). To identify a useful cutoff value for differentially expressed clones, expressed sequence tags (ESTs) representing known cycling genes were first examined (Figure 2A). These included genes expressed in the morning (LHY, CCA1, and CAB) and genes expressed later in the day (GI, CCR1, CCR2, and CAT3). Although difference of expression of these genes covered a range of ratios that occasionally were just less than twofold, the average of the experiments was consistently more than twofold (Figure 2B).
Figure 1.
Harvesting Regimen.
(A) Plants were trained in 12-hr-light/12-hr-dark cycles and then harvested at different times (except for R28 plants, which were grown in continuous light). Black bars represent dark, and white bars represent light. Slide names of the microarrays comparing different time points are shown and can be accessed via the Stanford Microarray Database (SMD) (http://genome-www.stanford.edu/microarray).
(B) Table of microarray results shows the slide name, light regimen of the training and experimental periods, Cy dye allocation, type of repetition conducted, RNA labeling method, and the number of clones showing greater than twofold ratios. DD, continuous dark; LD, 12-hr light/dark cycles; LL, continuous light; technical, a repetition that used the same RNA; biological, a repetition that used RNA from different plants; single, an experiment that was not repeated.
(C) Distribution of the average ratios from experiments comparing 0- and 12-hr time points. Small variations in the cutoff value greatly changed the number of clones selected, as shown by the inset table.
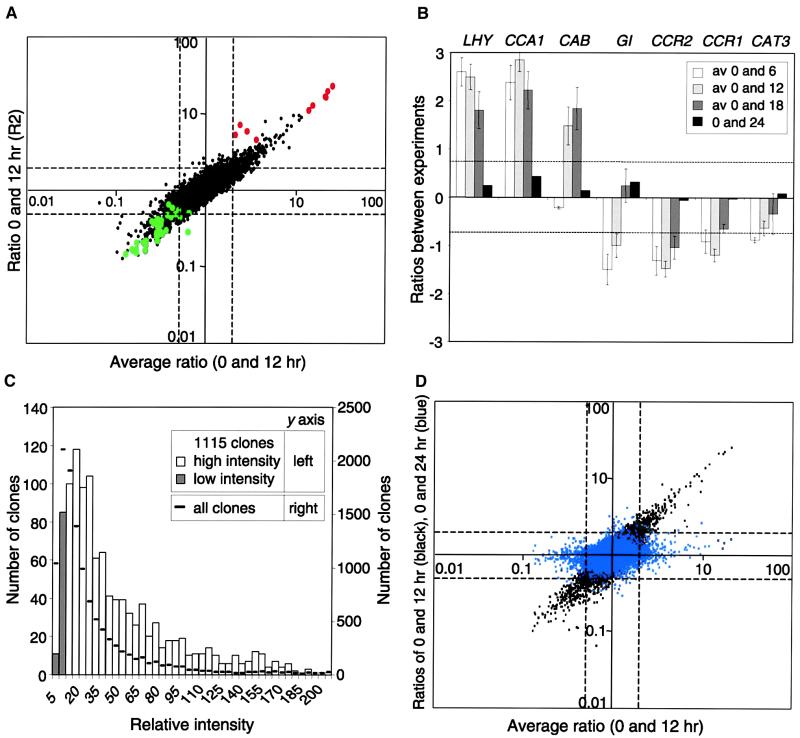
Figure 2.
Identification of Cycling Genes on the Array.
(A) The distribution of known cycling genes on the array is shown. The averages of the four 0- and 12-hr experiments (x axis) were plotted against the ratio from the first 0- and 12-hr experiment (R2) (y axis). Ratios >1 represent genes highly expressed in the morning (top right quadrant), and ratios <1 represent genes highly expressed in the afternoon (bottom left quadrant). Known circadian genes with high expression in the morning or afternoon are shown in red or green, respectively.
(B) The average ratios for LHY, CCA1, CAB, GI, CCR2, CCR1, and CAT3 plotted in log space are shown. Positive values represent high expression at 0 hr; negative values represent low expression at 0 hr and high expression at the other times. A twofold difference in expression equals 0.69 and −0.69, as shown by the dashed lines. Error bars represent standard deviation values for each time point. av, average.
(C) Distribution of signal intensities of the differentially expressed genes compared with the complete clone set is shown. The sums of the normalized intensities for all of the experiments were plotted. Intensities of clones showing values at three- to eightfold those of the background are shown as shaded bars.
(D) As a negative control, ratios of the 0- and 24-hr experiment were plotted against the average ratios of 0- and 12-hr time points. Black spots represent ratios of ESTs in the first 0- and 12-hr experiment (R2) (as in [A]). Blue spots represent the ratios of expression between 0 and 24 hr (R7). Clones showing a difference of expression between 0 and 12 hr do not show a difference between the 0- and 24-hr time points.
Using this twofold average, 1115 ESTs showed a difference of expression during the day/night period, with LHY and CCA1 having the highest ratios (Figure 2B). To establish the expression range of these selected clones, we calculated the distribution of total signal intensities from all of the experiments. The results show a range of representation from genes expressed at a low level (threefold that of the background signal) to the most highly expressed genes, implying that there is no intensity-dependent bias in their selection. Genes expressed at a low level are more sensitive to variation in background signal, so 96 clones with expression levels three- to eightfold that of background level (Figure 2C) are shown in subsequent figures marked with asterisks. Clones with values less than threefold that of the background value were exwcluded from data analysis.
To identify ESTs representing the same gene, we mapped the 1115 ESTs to bacterial artificial chromosome (BAC) locations by using a BLAST search comparison with Arabidopsis genomic DNA. Using a cutoff value of 10−50 (the probability that alignment would be generated randomly is <1 in 1050), we mapped 75% of the ESTs to a BAC. ESTs that mapped within 5 kb of each other and had similar expression patterns were identified as the same gene. ESTs that did not map to BACs were compared using a BLAST search against the nonredundant protein database, and those with a BLAST probability cutoff value of 10−30 for the same gene and showing similar expression patterns were assumed to be the same gene. When duplicates were identified, the first spot (numerically) was chosen to represent that gene. Using these criteria, the 1115 ESTs represented 831 genes showing a 25% redundancy on the array: 446 of these genes had high expression in the morning, and 385 had high expression in the afternoon.
As a control, tissue harvested at 0 hr was also compared with tissue harvested at 24 hr. These time points are identical in both diurnal and circadian cycles, so all of the ratios for these samples should be close to 1 (representing no differences in gene expression between the time points). Only a few ESTs showed expression ratios greater than a twofold difference (Figure 2D), and of the 831 genes selected as described above, only 14 had expression ratios greater than twofold. This finding supports the hypothesis that differences seen between the time points are the result of varying expression patterns throughout the day. For an additional control, plants were grown in continuous light for 3 weeks, and two samples were harvested 12 hr apart. It was found that all but seven genes of the total gene set had expression ratios less than twofold. All but one of the 831 genes that show a different expression through the day had a ratio less than twofold, showing that the expression pattern of these genes had dampened to a constant level of expression in continuous light. The 15 genes that showed different expression between the 0- and 24-hr time points and the continuous light were removed from further analysis because they were likely to be genes that had variable expression rather than light-regulated genes.
To identify the potential biological functions of the 816 ESTs, we translated them into all open reading frames and compared them with the nonredundant protein database using BLASTX. Using a probability cutoff of 10−30, we found that 594 ESTs showed similarity to known proteins and that 222 showed no similarity. Of the 594 genes that showed similarity, 413 showed similarity to proteins of known function and 181 to proteins of unknown function. Two or more ESTs that mapped to different BAC locations but were similar to the same protein were taken to represent genes from a gene family. Of the 590 genes with similarity, 37 were found to represent gene families. A list of the 816 genes is available on the Internet (http://www.prl.msu.edu/circl).
Identification of Genes Regulated by the Circadian Clock
To identify genes controlled by a circadian clock, we transferred plants that had been trained in 12-hr-light/12-hr-dark cycles to continuous darkness. Plants were harvested 12 and 24 hr after transfer (Figure 1A). When hybridized to an array, 792 ESTs showed consistent changes in expression between the two time points. Of the 816 diurnal cycling genes, 206 showed differential expression patterns. The 586 genes that showed changes of expression but did not cycle with a diurnal rhythm were not investigated further because they were likely to be induced or repressed with longer periods of darkness (a list of these genes can be viewed at http://www.prl.msu.edu/circl). To further study genes regulated by circadian rhythm, we repeated this experiment, but rather than transferring the plants to the dark, we transferred them to continuous light. These plants were harvested 24 and 36 hr after transfer (Figure 1A). When data from these two time points were compared, 126 ESTs showed a greater than twofold change of expression. Of the 816 diurnal cycling genes, only 35 showed a difference of expression. The difference in the numbers of genes identified between continuous light and dark might be due to an overall increase of cycling gene expression in the light that would create a smaller difference of expression.
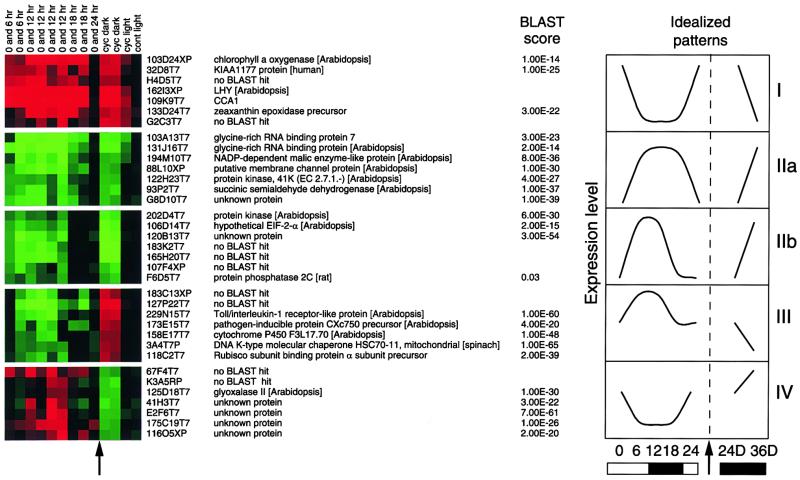
When the expression patterns of known circadian cycling genes from the microarrays were examined under light/dark cycles, different types of patterns could be seen. For example, LHY and CCA1 had high expression at 0 hr and low expression for the rest of the day. CAB had a longer expression pattern, showing a high level of transcript at 0 and 6 hr. GI expression was low at dawn, peaking at 6 and 12 hr, and then decreasing at 18 hr, whereas CCR1 and CCR2 were expressed at 6, 12, and 18 hr (Figure 2B). This can be seen more clearly by clustering the experiments. Cluster analysis mathematically arranges genes according to similarity of gene expression (Eisen et al., 1998). When the 206 genes showing differential expression in the circadian experiment were clustered, a number of distinct patterns emerged, including those consistent with being regulated by a circadian clock (Figure 3). Of the 139 genes showing patterns consistent with circadian regulation, 59 genes had high expression in the morning and 80 genes had high expression in the afternoon. The complete clusters can be viewed at the Internet site mentioned above. Most of the genes that peaked in the morning have a limited expression period, with high expression at dawn and low expression at all other times. The genes expressed in the afternoon show two patterns, both of which have longer periods of expression: some genes have high expression at 6 and 12 hr, whereas others have high expression at 6, 12, and 18 hr after dawn. In addition to circadian patterns, two other types of differentially regulated genes were also identified. Forty-four genes showed intermediate expression at dawn that was then downregulated during the day; when the plants were transferred to continuous dark, the expression of these 44 genes increased. These genes would fall into the class of dark-induced/light-inhibited genes. Twenty-three genes showed the opposite pattern and would be classified as light-induced/dark-inhibited genes.
Figure 3.
Comparison of Different Cycling Patterns.
The 206 clones with both diurnal cycling and differential expression in the dark were clustered. Five representative patterns, each with an idealized graph representing patterns of expression, are shown. Arrows indicate similar times. For the clusters, green represents expression that is greater in the afternoon, and red represents expression that is greater in the morning. cyc dark, differences in darkness; cyc light, differences in continuous light (of trained plants); cont light, differences in plants grown in continuous light. The protein with the most similar BLAST score is shown. Numbers above the clusters represent time points. Graph I represents 61 circadian genes with high expression in the morning. Graph II represents 78 circadian genes with high expression in the afternoon with (a) longer time of expression and (b) shorter time of expression. Graph III represents 23 genes induced in light and repressed in dark, and graph IV represents 44 genes repressed in light and induced in dark.
When the gene functions of these circadian-regulated clones were examined in more detail (Table 1), 25% of the genes were found to be similar to genes with unknown function and 28% showed no significant similarity to any proteins in the protein databases. The genes that were similar to known proteins could be separated into potential regulators, for example, kinases and potential transcription factors, and response elements. Response elements that had previously been shown to be regulated by cold, stress, pathogens, and auxins were identified. The previous categorization of these circadian-regulated response elements show that many circadian-regulated genes are co-regulated by other factors.
Table 1.
Categories of Genes That Cyclea
| Category | a.m. | p.m. |
|---|---|---|
| Unknown | 18 | 17 |
| No similarity | 16 | 23 |
| Transcription factors | 5 | 3 |
| Kinase/phosphatase | 4 | 3 |
| Photosynthesis/carbon metabolism | 7 | 4 |
| Auxin response | 1 | 3 |
| Stress (cold/pathogen/water) | 0 | 9 |
| Membrane proteins | 0 | 2 |
| Glycine-rich RNA binding | 0 | 3 |
| Other | 8 | 14 |
ESTs that show an expression pattern consistent with circadian rhythms were compared with all proteins in the protein database, using BLASTX with a probability cutoff of 10−30. Genes were divided into classes, depending on the most similar protein in the databases.
Clustering Data from Different Experiments
Data from the 816 clones showing diurnal cycling were compared with data from 47 Arabidopsis microarray experiments available in the Stanford Microarray Database (SMD) (http://genome-www.stanford.edu/microarray). These 47 experiments, conducted with the same clone set, compared different tissue types, light conditions, hormone treatments, and stress and pathogen responses. A selection of different clusters is shown in Figure 4. The whole cluster can be viewed on the Internet (http://www.prl.msu.edu/circl).
Figure 4.
Clustering Different Experiments.
Data from the 831 genes showing a diurnal expression pattern were clustered with data from 47 experiments in the SMD. A selection of clusters is shown here along with the name of each EST, the protein showing the highest homology, and the P score. Asterisks denote genes with lower expression levels.
(A) Genes with predominantly circadian-regulated expression.
(B) Photosynthetic genes that cluster into one highly expressed group in the morning and show differential tissue expression.
(C) Genes that peak in expression in the afternoon and exhibit differential tissue expression.
Information from clustering not only allows the identification of related expression patterns of different genes but also shows expression patterns of individual genes over a number of different experiments. Groups of genes with desirable patterns can be identified. One pattern of interest is genes that show few changes apart from being regulated in a circadian fashion. These genes might play a role in clock function because circadian genes would be expected to be expressed in all tissues under different environmental conditions. A single cluster of 33 genes with high expression in the morning falls under this category (Figure 4A). Within this cluster, potential regulators can be identified by similarity searches to predicted protein sequences. The DNA binding myb-related transcription factors LHY and CCA1 were found, as were other potential transcriptional regulators—two similar myb-like transcription factors (137A5T7 and 165H8T7) and a gene similar to CO (166C8T7). At the protein regulation level, the translation initiation factor EIF4B (218O11T7) was identified, along with clone G11D11T7, which shows similarity to a protein kinase. No single group was detected showing circadian-regulated expression in the afternoon and little differences in the other experiments. However, three smaller clusters of 24 genes showed less difference of expression in other experiments. One of these clusters included the flowering time gene GI, whereas another group contained the glycine-rich RNA binding genes CCR1 and CCR2. Other genes in these clusters include a MAP kinase (139A7T7), dehydration response genes, and a late embryo-abundant gene (153B6T7).
In addition to the genes showing predominantly circadian expression groups, other co-ordinately expressed genes were identified. One example is a cluster of genes with a long period of expression in the morning and a high level of expression in leaves and a low level in roots and in tissue culture cells grown in the dark (Figure 4B). Fourteen of the 27 genes in this cluster play a role in the photosynthesis. Along with the photosynthetic genes, there is a gene with a predicted product showing similarity to leucine zipper proteins. Leucine zippers have been shown to be involved in dimerization and are often associated with DNA binding domains. A second example (Figure 4C) is a cluster that is circadian regulated with high expression in the afternoon. It also has high expression in leaves and low expression in roots. This contains the already characterized CAT3 gene as well as an α amalase and a hexose sugar transporter. In addition, this group has several potential regulators, including an elongation initiation factor EIF2a (106D14T7), a protein kinase (202D4T7), and a protein phosphatase (F6D5T7).
DISCUSSION
Using a microarray containing 11,521 Arabidopsis clones, we have identified 816 genes that are regulated in a diurnal pattern and 139 genes with an expression pattern consistent with a circadian rhythm. Arabidopsis is totally reliant on light for growth and development, and it is therefore surprising that only a small proportion of the genes that were measured cycle. The genes that were found to cycle with a circadian rhythm were predicted to encode proteins ranging from those affecting photosynthesis and carbon and nitrogen metabolism to transcription factors and protein kinases. Many of these genes have not previously been shown to be regulated as such, showing that microarrays are a powerful tool for gene discovery. With this information, new insights can be generated about biological processes by identifying genes involved in the same biological pathway and by clustering genes with similar transcription profiles. This is especially relevant because many of the genes characterized in this article have no assigned function.
By estimating the number of unique genes on the array to be 7800, a number reached by subtracting 8% for failed polymerase chain reaction (PCR) amplifications and allowing for 25% redundancy, 11% of genes show diurnal regulation of expression and 2% show circadian regulation. This is likely to be an underestimate of the proportion of genes that cycle in the plant. The ESTs represented on this microarray were generated from multiple tissues harvested at different times and then randomly sequenced. These would be expected to include the most commonly expressed genes in the Arabidopsis genome. Genes that cycle with a high peak of expression would therefore likely be represented in this library, whereas genes with a lower peak might be missed. If this were to happen, it is possible that a number of low expressed cycling genes had not been present on the microarray. In addition, by using an artificially generated cutoff value of twofold, we might exclude genes that cycle with low amplitude from the group. Decreasing the cutoff value by small amounts significantly increased the number of clones that were included (Figure 1C). However, using a twofold cutoff value, which would include previously characterized cycling genes, we are confident that the genes selected have a difference of expression between the time points measured.
The differences measured on the microarrays are likely to be consistent with true expression patterns because the results consistently agree with published data of northern expression patterns of previously characterized genes. In addition, it has been shown that the microarray data produce consistent results with RNA gel blot analysis (Wang et al., 2000). However, one of the main sources of error from microarrays is the identity of the spots found on the array. All of the 94 good sequences obtained from resequencing clones from the spotting plate were as expected, suggesting that there is a <1% annotation error rate on our array.
Clustering gene expression patterns generated by microarray analysis can greatly enhance our understanding of coordinately expressed genes that are involved in similar processes. This is demonstrated in Figure 4B, which shows a group of genes involved with photosynthesis clustered together. Several of these genes are unknown, perhaps pointing to novel photosynthetic genes. In addition, there is a potential transcription factor, which may play a role in regulating these genes. This type of data mining from clusters has been recently shown in yeast, where functions of unknown genes were assigned by clustering gene expression patterns (Hughes et al., 2000). In addition to identifying genes expected to be regulated in a common process, we can gain insight into processes in which gene products might act sequentially. An example of this is found in Figure 4C, where an α amylase (135M15T7) is coregulated with a hexose sugar transporter (181G24T7). In Arabidopsis, starch is synthesized in the plastids during the day and broken down during the night, thus providing plants with sugars (Casper, 1994). From this pattern of accumulation, one would expect the amylase to be expressed in the leaves late in the day. Our array shows that this is the case, along with a sugar transporter that would allow sugar to be moved out of the plastids and into other areas of the plant. A third example of genes found on the array is the identification of genes encoding proteins involved in nitrogen metabolism. On the array, the nitrate reductase clone (216H23T7) shows strong circadian regulation, whereas nitrite reductase (177N14T7) is light regulated; this is consistent with previous observations (Cheng et al., 1991; Vincentz et al., 1993). These examples show how global metabolic processes can be viewed simultaneously to gain insight into the biology of the plant.
A cluster of major interest consists of a group of genes showing circadian regulation but few differences of expression in other experiments. These genes are candidates to be components of the central clock, because genes involved in clock function are likely to be unaffected by tissue type and other conditions. When the potential functions of these genes were identified, the genes could be classified into one of two categories: those that are regulated only by the circadian clock and those that may play a role in clock function. Some genes, including APS reductase (73F9T7), formamidase (91H14T7), and catalase (clone 154N18T7), would fall into the first category, whereas LHY, CCA1, and GI have already been shown to play some part in circadian clock function. Among members of this group are many genes of unknown function. However, a number of potential regulators were identified, including putative transcription factors and post-translational modifiers. CCR1, CCR2, and genes involved in dehydration were also identified in this cluster. These genes have been previously characterized and are regulated by other factors (Carpenter et al., 1994; Kiyosue et al., 1994). These examples demonstrate a limitation in clustering a limited number of experiments in the database, because key experiments that would differentiate expression patterns between genes might not be included, thus skewing the picture.
In summary, using microarrays, we have identified a number of genes that cycle with either a diurnal or a circadian rhythm. Many of these genes have not previously been shown to cycle. A large number of genes described here have no homology to proteins in the databases. This is a common problem researchers will have to face now that the genome is sequenced. Microarrays are a useful tool to gain knowledge of potential gene function. Although microarrays offer a powerful method for studying co-ordinate gene expression, much more information can be generated by comparing a large number of different experiments. The ability to mine useful information in this way is continually improving with the increasing number of experiments available in the SMD. Once a representative number of conditions have been examined with microarrays, a fuller picture of gene expression patterns will emerge.
METHODS
Selection of Clones
A total of 11,174 expressed sequence tags (ESTs) were selected for spotting on the microarray from 37,490 Arabidopsis thaliana ESTs in the National Center for Biotechnology Information database dbest. To remove ESTs that represented the same gene, we compared each EST with the others in pairwise comparisons by using a BLAST search (with a P-value cutoff of 10−30) followed by a BLASTX comparison with all available Arabidopsis proteins (using a P-value cutoff of 10−30). A total of 4655 groups of ESTs were identified, and a single EST from each group was chosen; 7809 did not line up with any other EST. When these were combined, a set of 12,464 ESTs was produced. Of these, 11,174 were represented in the Newman EST collection (Newman et al., 1994) and were selected. An additional 257 clones not represented in the library and 45 control clones were added to the set. Included in the control clones were six genes with circadian expression patterns: CAB2, LHY, RUBISCO, CCA1, GI, and CCR2.
Polymerase Chain Reaction Amplification
Ziplox primers 5′-ATTGAATTTAGGTGACACTATAGAAGAGC-3′ and 5′-CGACTCACTATAGGGAAAGCTGG-3′ or pBluescript KS primers 5′-CGACTCACTATAGGGCGAATTGG-3′ and 5′-GGAAACAGCTATGACCATGATTACG-3′ were used to amplify 10 ng of the stock ESTs using polymerase chain reaction (PCR). The PCR products were precipitated in ethanol and resuspended in 25 μL of 3 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and were checked for quality by gel electrophoresis for concentration and multiple bands. Concentration was determined using Quantity1 (Bio-Rad, Hercules, CA) software. Ninety-two percent of the clones showed a single band with a concentration of at least 50 ng/μL. DNA was spotted on sylilated (CEL Associates, Houston, TX) and superaldehyde (Telechem, Sunnydale, CA) glass slides at high density using an Omnigridder robot (Gene Machines, San Carlos, CA) and 16 ArrayIt chipmaker 2 pins (Telechem). Slides were washed and blocked according to the Telechem protocol.
RNA Isolation and Probe Synthesis
Arabidopsis plants (Landsberg erecta) were grown at high density in 12-hr-light/12-hr-dark cycles in growth cabinets on soil until they just showed a floral bolt in the center of the rosette. Due to the sensitivity of microarrays, plant-to-plant variation of gene expression was reduced by bulk harvesting of ∼100 plants. RNA was extracted using a modification of a protocol designed to extract RNA from pine trees (Chang et al., 1993). Instead of using 2 to 3 g of tissue, 1 g was used with 5 mL of extraction buffer. In each experiment, either 1 μg of poly(A)+ RNA or 100 μg of total RNA was labeled. One microgram of poly(A)+ was isolated using the Oligotex (Qiagen, Valencia, CA) poly(A) isolation kit and labeled using Klenow labeling as described in the cDNA protocol of Eisen and Brown (1999) with the following modifications: 1 μg of oligo-dT23V was added to a total volume of 24.5 μL, heated at 70°C for 10 min, reverse transcribed in a total volume of 40 μL (300 units of Superscript II [Gibco BRL, Rockville, MD], 8 μL of buffer, 4 μL of 0.1 M DTT, and 2 μL of 10 mM deoxynucleotide triphosphates) for 2 hr at 42°C, and finally treated with RNase H at 37°C for 30 min. Excess nucleotides and primers were removed by adding 160 μL of TE buffer (TE is 10 mM Tris and 1 mM EDTA, pH 8.0) and centrifugation through a Microcon YM-30 column (Millipore, Bedford, MA), washing with 200 μL of TE buffer, and eluting in 20 μL of TE buffer. When using total RNA, the RNA was further purified according to the RNAeasy kit cleanup protocol (Qiagen). One hundred micrograms of total RNA was labeled as described by Eisen and Brown (1999), except that 2 μL of Cy dye –linked dCTP dye was used instead of 3 μL and the RNA was removed using 0.5 μL of RNase A (10 mg/mL) and 0.2 μL of RNase H (Gibco BRL).
The labeling reactions were purified using the QiaQuick PCR cleanup kit (Qiagen). The experimental and reference tissues were labeled with either Cy3 or Cy5 fluorescent dye and hybridized to a microarray in a total volume of 30 μL of hybridization buffer (3.4 × SSC, 0.32% SDS, and 5 μg of yeast tRNA) for 16 hr at 60°C. These tissues were then washed at room temperature in 2 × SSC, 0.1% SDS for 5 min, in 1 × SSC, 0.1% SDS for 5 min, and in 0.1 × SSC for 15 sec. The slides were centrifuged dry and scanned with a Scanarray 4000 (GSI Lumonics, Billerica, MA). Each microarray experiment was repeated twice, either as a technical repetition, in which the same RNA was labeled but reversing the Cy dye, or as a biological replicate, with new tissue grown under the same conditions. Figure 1B shows the type of replication and the labeling method used for each clone.
Data Analysis
The intensities of the spots were measured using Scanalyze 4.24 (available at http://genome-www4.stanford.edu/MicroArray/SMD/restech.html). The two channels were normalized in log space using the z-score normalization on a 95% trimmed data set. To remove unreliable spots, the data were screened using the following criteria. Spots containing clones that had poor amplification or multiple bands, as well as those that were flagged due to a false intensity caused by dust or background on the array, were removed. Spots with <65% of the spot intensity at >1.5-fold that of the background in both channels were ignored. These procedures usually removed ∼10% of the spots. Spots that had an intensity of less than twofold that of the background had their intensity increased to that value. This reduced artificially generated ratios in spots with intensities close to background, and it gave a value by which to divide for spots that had expression in only one channel. Spots that had their normalized intensities increased artificially in one channel to greater than the saturated value due to the normalization factor had this value reduced to the saturated value in the other channel. Comparison of the arrays was achieved using Microsoft Excel and Microsoft Access database. Multiple experiments were analyzed using Cluster and Treeview software (Eisen et al., 1998; available at http://genome-www4.stanford.edu/MicroArray/SMD/restech.html).
Quality Control of the Array
A number of quality controls of the array were performed. To test whether the annotations of the ESTs spotted on the array were correct, we selected 107 clones showing differential expression from the printing plates, which we then reamplified and resequenced. Reliable sequence was obtained for 94 clones, and all represented the expected clones for that location. The sequences from the remaining 13 clones were too poor to resolve and therefore could not be classified as correct or incorrect. Spiking of in vitro–labeled mRNA into the labeling reaction in known concentrations was used to determine the sensitivity of the array. Using these controls, it was estimated that the microarray could reliably detect amounts of RNA as low as 20 pg (data not shown).
Supplementary Material
Acknowledgments
The Arabidopsis Functional Genomics Consortium array was developed in collaboration with Pamela Green and John Ohlrogge at Michigan State University and Shauna Somerville and Mike Cherry at Stanford University. We thank Tom Newman for generously donating EST clones; Curt Wilkerson for help with bioinformatics and EST mapping; Ken Keegstra, Thomas Girke, and Miguel Perez for useful discussions; and Pamela Green, Christoph Benning, Rachel Green, and Alison Murray for critically reading the manuscript. This work was funded by National Science Foundation Grant No. DBI9872638 to Pamela Green, Rick Amasino, Mike Cherry, Steve Delaporta, Ken Keegstra, John Ohlrogge, Shauna Somerville, and Mike Sussman of the Arabidopsis Functional Genomics Consortium.
References
- Ahmad, M., and Cashmore, A.R. (1993). HY4 gene of A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366 162–166. [DOI] [PubMed] [Google Scholar]
- Ballare, C.L. (1999). Keeping up with the neighbours: Phytochrome sensing and other signalling mechanisms. Trends Plant Sci. 4 97–102. [DOI] [PubMed] [Google Scholar]
- Carpenter, C.D., Kreps, J.A., and Simon, A.E. (1994). Genes encoding glycine-rich Arabidopsis thaliana proteins with RNA-binding motifs are influenced by cold treatment and an endogenous circadian rhythm. Plant Physiol. 104 1015–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casper, T. (1994). Genetic dissection of the biosynthesis, degradation, and biological functions of starch. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 913–936.
- Chang, S., Puryear, J., and Cairney, J. (1993). A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11 113–116. [Google Scholar]
- Cheng, C.L., Acedo, G.N., Dewdney, J., Goodman, H.M., and Conkling, M.A. (1991). Differential expression of the two Arabidopsis nitrate reductase genes. Plant Physiol. 96 275–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282 1698–1701. [DOI] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of PHYD and PHYE. Plant Mol. Biol. 25 413–427. [DOI] [PubMed] [Google Scholar]
- Dunlap, J.C. (1999). Molecular bases for circadian clocks. Cell 96 271–290. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., and Brown, P.O. (1999). DNA arrays for analysis of gene expression. Methods Enzymol. 303 179–205. [DOI] [PubMed] [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst, D., Apfelbock, A., Bergmann, A., and Weyrauch, C. (1990). Rhythmic regulation of the light-harvesting chlorophyll a/b protein and the small subunit of ribulose-1,5-bisphosphate carboxylase mRNA in rye seedlings. Photochem. Photobiol. 52 29–33. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock–controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girke, T., Todd, J., Ruuska, S., White, J., Benning, C., and Ohlrogge, J. (2000). Microarray analysis of developing Arabidopsis seeds. Plant Physiol. 124 1570–1581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliano, G., Pichersky, E., Malik, V.S., Timko, M.P., Scolnik, P.A., and Cashmore, A.R. (1988). An evolutionarily conserved protein binding sequence upstream of a plant light-regulated gene. Proc. Natl. Acad. Sci. USA 85 7089–7093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, R.M., and Tobin, E.M. (1999). Loss of the circadian clock–associated protein 1 in Arabidopsis results in altered clock-regulated gene expression. Proc. Natl. Acad. Sci. USA 96 4176–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Yang, H., Mockler, T.C., and Lin, C. (1998). Regulation of flowering time by Arabidopsis photoreceptors. Science 279 1360–1363. [DOI] [PubMed] [Google Scholar]
- Hicks, K.A., Millar, A.J., Carre, I.A., Somers, D.E., Straume, M., Meeks-Wagner, D.R., and Kay, S.A. (1996). Conditional circadian dysfunction of the Arabidopsis early-flowering 3 mutant. Science 274 790–792. [DOI] [PubMed] [Google Scholar]
- Hughes, T.R., et al. (2000). Functional discovery via a compendium of expression profiles. Cell 102 109–126. [DOI] [PubMed] [Google Scholar]
- Kircher, S., Kozma-Bognar, L., Kim, L., Adam, E., Harter, K., Schafer, E., and Nagy, F. (1999). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol. 35 225–231. [PubMed] [Google Scholar]
- Kreps, J.A., and Simon, A.E. (1997). Environmental and genetic effects on circadian clock–regulated gene expression in Arabidopsis. Plant Cell 9 297–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum, E., and Briggs, W.R. (1995). Mutations in the NPH1 locus of Arabidopsis disrupt the perception of phototropic stimuli. Plant Cell 7 473–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element–bound transcription factor. Science 288 859–863. [DOI] [PubMed] [Google Scholar]
- Millar, A.J., Carre, I.A., Strayer, C.A., Chua, N.H., and Kay, S.A. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267 1161–1163. [DOI] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101 331–340. [DOI] [PubMed] [Google Scholar]
- Newman, T., de Bruijn, F.J., Green, P., Keegstra, K., Kende, H., McIntosh, L., Ohlrogge, J., Raikhel, N., Somerville, S., and Thomashow, M. (1994). Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1999). Binding of phytochrome B to its nuclear signalling partner PIF3 is reversibly induced by light. Nature 400 781–784. [DOI] [PubMed] [Google Scholar]
- Park, D.H., Somers, D.E., Kim, Y.S., Choy, Y.H., Lim, H.K., Soh, M.S., Kim, H.J., Kay, S.A., and Nam, H.G. (1999). Control of circadian rhythms and photoperiodic flowering by the Arabidopsis GIGANTEA gene. Science 285 1579–1582. [DOI] [PubMed] [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Sakamoto, K., and Nagatani, A. (1996). Nuclear localization activity of phytochrome B. Plant J. 10 859–868. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Ramsay, N., Samach, A., Corden, S., Putterill, J., Carre, I.A., and Coupland, G. (1998). The late elongated hypocotyl mutation of Arabidopsis disrupts circadian rhythms and the photoperiodic control of flowering. Cell 93 1219–1229. [DOI] [PubMed] [Google Scholar]
- Schaffer, R., Landgraf, J., Perez-Amador, M., and Wisman, E. (2000). Monitoring genome-wide expression in plants. Curr. Opin. Biotechnol. 11 162–167. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution, and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3 1745–1757. [DOI] [PubMed] [Google Scholar]
- Somers, D.E. (1999). The physiology and molecular bases of the plant circadian clock. Plant Physiol. 121 9–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somers, D.E., Schultz, T.F., Milnamow, M., and Kay, S.A. (2000). ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101 319–329. [DOI] [PubMed] [Google Scholar]
- Staiger, D., Apel, K., and Trepp, G. (1999). The Atger3 promoter confers circadian clock–regulated transcription with peak expression at the beginning of the night. Plant Mol. Biol. 40 873–882. [DOI] [PubMed] [Google Scholar]
- Strayer, C., Oyama, T., Schultz, T.F., Raman, R., Somers, D.E., Mas, P., Panda, S., Kreps, J.A., and Kay, S.A. (2000). Cloning of the Arabidopsis clock gene TOC1, an autoregulatory response regulator homolog. Science 289 768–771. [DOI] [PubMed] [Google Scholar]
- Vincentz, M., Moureaux, T., Leydecker, M.T., Vaucheret, H., and Caboche, M. (1993). Regulation of nitrate and nitrite reductase expression in Nicotiana plumbaginifolia leaves by nitrogen and carbon metabolites. Plant J. 3 315–324. [DOI] [PubMed] [Google Scholar]
- Wang, R., Guegler, K., LaBrie, S.T., and Crawford, N.M. (2000). Genomic analysis of a nutrient response in Arabidopsis reveals diverse expression patterns and novel metabolic and potential regulatory genes induced by nitrate. Plant Cell 12 1491–1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, Z.Y., and Tobin, E.M. (1998). Constitutive expression of the CIRCADIAN CLOCK ASSOCIATED 1 (CCA1) gene disrupts circadian rhythms and suppresses its own expression. Cell 93 1207–1217. [DOI] [PubMed] [Google Scholar]
- Wang, Z.Y., Kenigsbuch, D., Sun, L., Harel, E., Ong, M.S., and Tobin, E.M. (1997). A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 9 491–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, H.H., and McClung, C.R. (1996). The circadian clock gates expression of two Arabidopsis catalase genes to distinct and opposite circadian phases. Mol. Gen. Genet. 251 196–203. [DOI] [PubMed] [Google Scholar]
- Zhong, H.H., Painter, J.E., Salome, P.A., Straume, M., and McClung, C.R. (1998). Imbibition, but not release from stratification, sets the circadian clock in Arabidopsis seedlings. Plant Cell 10 2005–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.