Abstract
Root hair initiation involves the formation of a bulge at the basal end of the trichoblast by localized diffuse growth. Tip growth occurs subsequently at this initiation site and is accompanied by the establishment of a polarized cytoplasmic organization. Arabidopsis plants homozygous for a complete loss-of-function tiny root hair 1 (trh1) mutation were generated by means of the T-DNA–tagging method. Trichoblasts of trh1 plants form initiation sites but fail to undergo tip growth. A predicted primary structure of TRH1 indicates that it belongs to the AtKT/AtKUP/HAK K+ transporter family. The proposed function of TRH1 as a K+ transporter was confirmed in 86Rb uptake experiments, which demonstrated that trh1 plants are partially impaired in K+ transport. In line with these results, TRH1 was able to complement the trk1 potassium transporter mutant of Saccharomyces, which is defective in high-affinity K+ uptake. Surprisingly, the trh1 phenotype was not restored when mutant seedlings were grown at high external potassium concentrations. These data demonstrate that TRH1 mediates K+ transport in Arabidopsis roots and is responsible for specific K+ translocation, which is essential for root hair elongation.
INTRODUCTION
Two fundamental parameters underlie development in higher plants: cell proliferation and differentiation. The coordinated action of these two parameters depends on the perception of and the response to an array of intracellular and extracellular developmental cues. To study cell expansion, we have chosen the epithelial layer of the Arabidopsis root. The outer cell layer of the seedling root, the epidermis, is composed of two distinct cell types that are arranged in files: hair-bearing and non-hair-bearing cells (Dolan et al., 1994; Scheres et al., 1994). Root hairs are tip-growing projections that emerge from specialized epidermal cells, the trichoblasts (Leavitt, 1904).
The development of root hairs in Arabidopsis can be divided into two phases: the early diffuse growth phase (initiation) and the later phase (tip growth); growth rates during these phases also differ (Dolan et al., 1994; Duckett et al., 1994). Tip growth is a form of polarized cell expansion found in fungi and in a number of cell types in plants (pollen tube and root hairs). This polarized growth of the root hair is due to the highly localized exocytosis of Golgi-derived vesicles and the deposition of cell membrane and wall material at a restricted area of the plasma membrane, the tip (Sievers and Schnepf, 1981). Genetic analysis of root hair growth has defined a number of genes involved in both the early and later phases of hair development. RHD1 and RHD6 are required for the earliest phases of root hair outgrowth (Schiefelbein and Somerville, 1990; Masucci and Schiefelbein, 1994), whereas COW1, TIP1, and RHD3 are prerequisites for tip growth (Schiefelbein et al., 1993; Galway et al., 1997; Grierson et al., 1997; Ryan et al., 1997).
In Arabidopsis, initiation is sensitive to both Ca2+ and H+ concentrations in the medium (Schiefelbein et al., 1992; Bibikova et al., 1998), suggesting a role for cytosol-free calcium concentration ([Ca2+]i) and pH in the transition to this developmental stage. Indeed, localized increases in [Ca2+]i have been shown to precede the initiation of apical growth in some systems (Jaffe et al., 1974), although such increases in [Ca2+]i have yet to be shown in root hairs (Wymer et al., 1997). Coordinated changes in cytosolic and apoplastic pH are clearly important for the initiation of root hair growth (Bibikova et al., 1998). Elongation is characterized by rapid tip growth (∼2 μm/min), which becomes evident once the hair outgrowth is ∼20 μm in length. In this final stage, root hair growth is directed by a profound [Ca2+]i gradient along the root hair axis (Wymer et al., 1997).
Root-hair growth assists the acquisition of mineral nutrients not only by increasing the surface of the root but also by exploring new undepleted soil layers. Among the mineral nutrients acquired by plants, potassium is the most abundant. Plant roots are able to accumulate K+ to a level exceeding 100 mM from different types of soil. To adapt to the broad range of K+ concentrations in soil, plants have evolved a “biphasic” mechanism of K+ incorporation (Epstein et al., 1963). Low-affinity transport has been shown to provide a major transport pathway when the K+ concentration in the soil is at the millimolar level, whereas high-affinity transport is vital for plants to sustain growth when external K+ concentrations decrease to the micromolar range (Epstein et al., 1963; Epstein, 1966).
K+ channel proteins have been demonstrated to be a molecular determinant of the low-affinity uptake system (Grabov and Böttger, 1994; Maathuis et al., 1997). In Arabidopsis, AKT1, a channel belonging to the Shaker family, is expressed predominantly in the root cell plasma membrane and has been shown to be responsible for K+ uptake (Lagarde et al., 1996; Hirsch et al., 1998). Surprisingly, the AKT1 channel was also able to mediate potassium uptake from solutions that contained as little as 10 μM K+ (Hirsch et al., 1998).
Electrophysiological studies indicate that the H+-coupled high-affinity transport system operates in Arabidopsis roots (Maathuis and Sanders, 1994). However, no molecular details of K+/H+ transporters are available. Previous indications that the HKT1 transporter from wheat operates as a K+/H+ symporter (Schachtman and Schroeder, 1994) have not been confirmed in later experiments (Rubio et al., 1995). Alternatively, the sodium gradient across the plasma membrane was suggested to drive the high-affinity K+ uptake system (Smith and Walker, 1989). It has been shown that free energy from the Na+ gradient is used by the HKT1 transporter in wheat roots (Rubio et al., 1995). This mode of K+ transport has not been confirmed, however, for AtHKT1, an HKT1 counterpart in Arabidopsis (Uozumi et al., 2000). Novel K+ transporters belonging to the gene family described previously from Schwanniomyces occidentalis and Escherichia coli have been reported recently to mediate low-affinity, high-affinity, and dual-affinity K+ transport (Quintero and Blatt, 1997; Fu and Luan, 1998; Kim et al., 1998; Rubio et al., 2000).
A key physiological role for K+ lies in the osmotic balance of the plant cell, and K+ transport across the vacuolar and plasma membranes contributes directly to turgor regulation. Because cell elongation is driven by turgor pressure, the operation of K+ translocators is crucial for growth. This idea is strongly supported by recent studies of maize coleoptiles, in which auxin failed to stimulate cell elongation in the absence of K+ in the medium or after the addition of K+ channel blockers (Claussen et al., 1997). Moreover, a strong correlation was observed between K+ channel expression and cell elongation after maize coleoptiles were stimulated by auxin (Philippar et al., 1999). The role of K+ transport in root hair growth has yet to be analyzed.
To understand the molecular mechanism that determines root hair elongation, we have isolated a mutant impaired in root hair tip growth. This phenotype was designated tiny root hair 1 (trh1). The mutant was generated by the T-DNA–tagging method, and the disrupted gene was demonstrated to encode a potassium transporter belonging to the AtKT/AtKUP/HAK family. In this study, we show that the TRH1 potassium uptake system is essential for root hair tip growth.
RESULTS
Mutant Phenotype of trh1
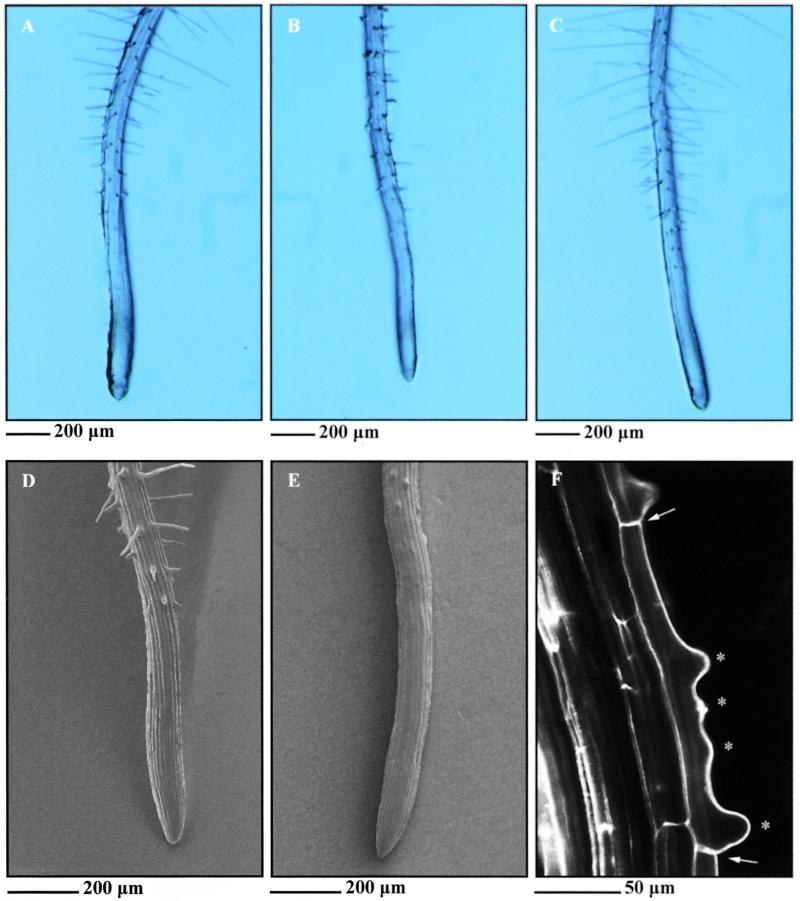
Root hairs grow in the differentiation zone of the root perpendicular to the root axis and can be characterized as polarized outgrowths from the basal end of trichoblasts (Figure 1). The trh1 mutant was isolated from a population of 4000 mutagenized families containing T-DNA insertions (Feldmann, 1991). Consistent with the presence of a closely linked T-DNA insertion, each kanamycin-resistant plant segregated for the trh1 mutation. A complete loss-of-function mutation in the TRH1 gene results in the arrest of hair growth soon after initiation (Figure 1). The fact that TRH1 is required for the establishment of tip growth from the initiation site indicates that TRH1 is a positive genetic regulator of tip growth. However, trh1 trichoblasts occasionally develop multiple initiation sites (Figure 1), showing that the development of root hair growth from the basal end of the trichoblast may negatively regulate the initiation of other hairs in the same cell in a TRH1-dependent process. Segregation data from the F2 progeny of a cross between homozygous mutants and the wild type indicate that TRH1 represents a single Mendelian recessive allele (542 wild-type seedlings and 190 mutant seedlings; ratio 3:1, 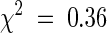 , P > 0.5). Pollen tube growth is not defective in trh1 plants, as might be expected, because this cell type also undergoes tip growth. Further detailed microscopic examination of trh1 plants failed to identify any other cell growth defects, as were observed previously in tip1 and rhd3 mutants (Schiefelbein and Somerville, 1990; Schiefelbein et al., 1993; Galway et al., 1997; Grierson et al., 1997; Ryan et al., 1997).
, P > 0.5). Pollen tube growth is not defective in trh1 plants, as might be expected, because this cell type also undergoes tip growth. Further detailed microscopic examination of trh1 plants failed to identify any other cell growth defects, as were observed previously in tip1 and rhd3 mutants (Schiefelbein and Somerville, 1990; Schiefelbein et al., 1993; Galway et al., 1997; Grierson et al., 1997; Ryan et al., 1997).
Figure 1.
Phenotypes of Wassilewskija Wild Type, trh1 Mutant, and trh1 Transformed with the TRH1 Gene.
(A) and (B) Wild-type (A) and trh1-1 (B) primary roots from 1-week-old seedlings.
(C) trh1 mutant transformed with the 8.5-kb BamHI-SalI genomic fragment containing the TRH1 gene. The same phenotype was observed when trh1 mutant plants were transformed with the 7.1- and 6.6-kb fragments.
(D) and (E) Scanning electron micrographs of wild-type (D) and trh1 (E) primary root hair cells.
(F) Confocal micrograph of trh1 primary root hair cells. Asterisks show multiple sites of root hair initiation in a single root hair cell that is indicated by the arrows.
Isolation of the TRH1 Gene
To determine the copy number of T-DNA inserts, we hybridized T-DNA sequences to genomic DNA from the Arabidopsis mutant. The DNA gel blot showed two hybridizing bands, indicating that two T-DNAs were present (Figure 2). By plasmid rescue, it was possible to isolate two plant DNA fragments flanking the T-DNA insertions: one 200 bp in length and the other 1.3 kb in length. Both fragments hybridized to a single 1.5-kb EcoRI genomic fragment of wild-type plants, indicating that both T-DNA copies were introduced into the same locus. The 1.3-kb fragment isolated from plasmid rescue was used to screen an Arabidopsis genomic library. Eight overlapping genomic clones were identified that spanned the 35-kb region shown in Figure 3. All clones were also positive when counterscreened with the 200-bp rescued fragment. Four identical cDNA clones were isolated from the cDNA library when screened with the 1.5-kb EcoRI fragment, indicating that it contained transcribed sequences.
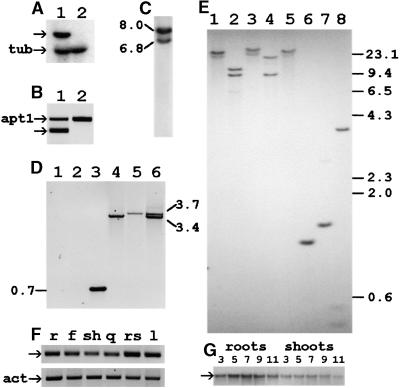
Figure 2.
Expression and Copy Number of the TRH1 Gene.
(A) RNA gel blot analysis. Approximately 30 μg of total RNA was loaded from wild-type (lane 1) and trh1 (lane 2) 1-week-old roots. The TRH1 (arrow) and tubulin (tub) gene transcripts detected are shown.
(B) Reverse transcription–polymerase chain reaction (RT-PCR) analysis of RNA isolated from wild-type (lane 1) and mutant (lane 2) plants. The specific primers used for the TRH1 gene (exon2 and st2rev; arrow) generated a fragment of 250 bp. Primers specific to the APT1 gene generated a fragment of 478 bp (apt1) and were used as a control.
(C) T-DNA copy number. Genomic DNA from trh1 mutant plants was digested with EcoRI, and the resulting DNA gel blot was hybridized to T-DNA. The sizes of the two hybridizing bands are in kilobases.
(D) Two T-DNA copies were inserted into the first intron of the TRH1 gene as an inverted dimer. PCR analysis of the genomic DNA isolated from wild-type (lanes 1 to 3) or trh1 mutant (lanes 4 to 6) plants was performed. The combinations of primers used are as follows: lanes 1 and 4, st2rev/#1242; lanes 2 and 5, exon2/#1242; lanes 3 and 6, exon2/#1242/st2rev. Numbers denote the lengths of the PCR products in kilobases.
(E) Arabidopsis genomic DNA was isolated from ecotype Columbia and digested with XhoI, BamHI, SalI, EcoRI, HindIII, and PstI (lanes 3 to 8, respectively) and XhoI-SalI (lane 1) or BamHI-SalI (lane 2). The numbers at right are in kilobases.
(F) Quantitative RT-PCR analysis of the TRH1 gene performed with RNA isolated from roots (r), flowers (f), shoots (sh), siliques (q), rosette leaves (rs), or leaves (l). The RT-PCR products for TRH1 (arrow) and actin (act) are shown. Actin was used as an internal control.
(G) RNA gel blot analysis of the TRH1 gene transcripts during seedling development. Wild-type Arabidopsis seedlings at 3, 5, 7, 9, and 11 days after germination were dissected, and RNA was isolated from roots and shoots. Each lane contains 20 μg of total RNA. The arrow denotes the TRH1 transcript.
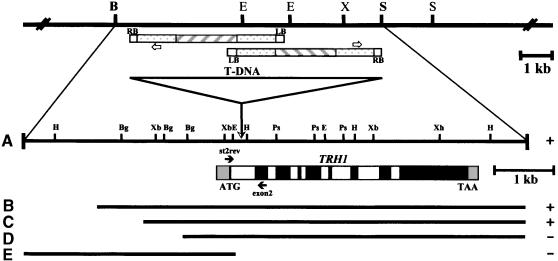
Figure 3.
The TRH1 Locus.
The genomic region of ∼35 kb of contiguous DNA was isolated from a Columbia genomic library using as a probe the genomic fragment (1.3 kb) that was identified by plasmid rescue of the trh1 mutant plants. Five fragments were used for transformation of mutant plants. The 8.5-kb (A), 7.1-kb (B), and 6.6-kb (C) fragments converted the mutant phenotype to wild type (+). The 5.7-kb (D) and the upstream 3.5-kb (E) fragments did not restore the phenotype (−). The T-DNA insertion site is depicted by the triangle within the first intron. Two T-DNA copies were inserted in the 1.5-kb EcoRI fragment in an inverted orientation. Black boxes represent exons, white boxes represent introns, and gray boxes represent 5′ or 3′ untranslated cDNA sequences. First and last codons are shown. Solid arrows st2rev and exon2 denote the specific primers for the TRH1 gene in the first and second exons, respectively. The open arrows denote the specific primer #1242 from pBR322. RB and LB, left and right border, respectively. The restriction map of the region TRH1 is also shown. B, BamHI; Bg, BglII; E, EcoRI; H, HindIII; Ps, PstI; S, SalI; X or Xh, XhoI; Xb, XbaI. Hatched boxes represent T-DNA structure. Stippled boxes represent pBR322 sequences.
DNA gel blot analysis of the isolated genomic clones showed that the isolated cDNA hybridized to an 8.5-kb BamHI-SalI fragment from the positive clone λ-72 (Figure 3). The cDNA and ∼5 kb from the genomic clone were sequenced and shown to represent the same sequences. The cDNA was truncated in the 5′ coding region because no initiation codon was present. An expressed sequence tag (EST) entry in the Arabidopsis database (H10G7) was completely identical at the 3′ end to the isolated cDNA. This EST is a full-length cDNA that contained the initiation codon and the 5′ untranslated region. Alignment of the sequences from the EST, the isolated cDNA, and the genomic region showed that the TRH1 gene consists of nine exons separated by eight introns ranging from 71 to 430 bp in length. The longer exon is located at the 3′ end of the gene. The first exon consists of only the initiation codon.
Sequence and polymerase chain reaction (PCR) analysis of the genomic DNA from the trh1 plant showed that both T-DNAs had been inserted in the same position within the first intron of the gene in a tail-to-tail orientation. This result indicates that only the isolated gene corresponds to the trh1 phenotype. One of the T-DNAs is complete and the other is truncated (Figures 2 and 3). The gene is localized on chromosome IV (Arabidopsis thaliana ESSAII Sequencing Project, bacterial artificial chromosome clone F9D16; GenBank accession number AL035394). The TRH1 cDNA consists of a single 2325-nucleotide open reading frame encoding a predicted protein of 775 amino acid residues with a molecular mass of 86,842 D (Figure 4). Sequence analysis identified a region similar to a putative cAMP-dependent protein kinase phosphorylation site at amino acid 730 and some other phosphorylation sites.
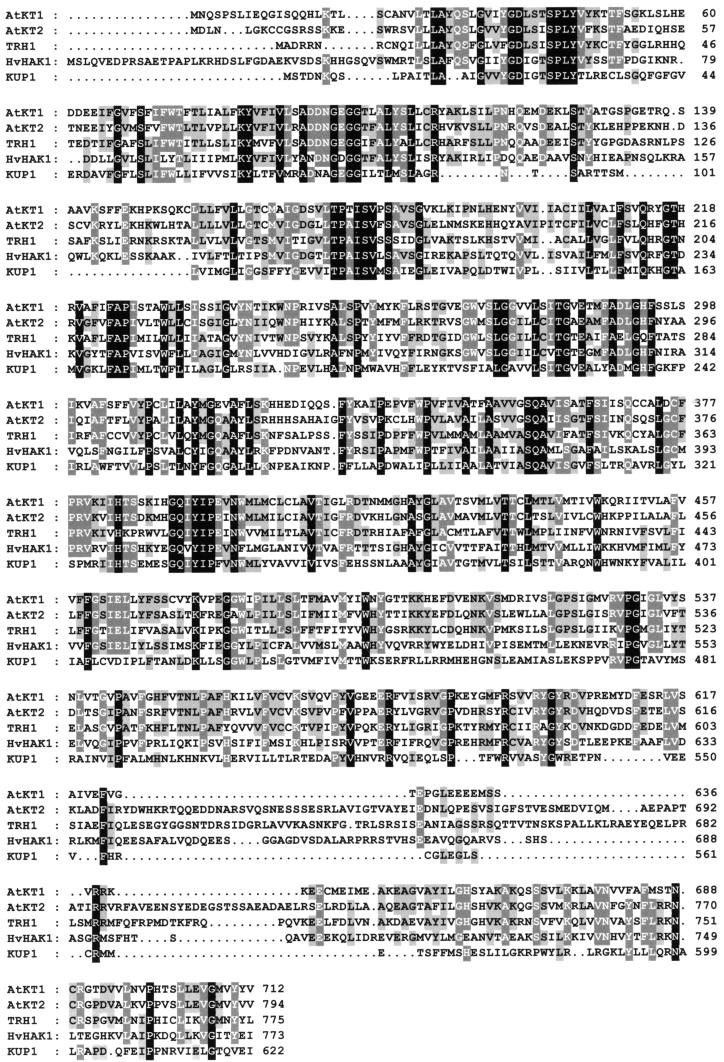
Figure 4.
Alignment of K+ Transporters.
Alignment of the deduced amino acid sequences of TRH1 with bacterial KUP1 and plant HvHAK1 from barley (Schleyer and Bakker, 1993; Santa-Maria et al., 1997) and AtKT1 and AtKT2 from Arabidopsis (Quintero and Blatt, 1997; Kim et al., 1998). One-letter amino acid code is used. Dots have been introduced to optimize sequence alignment. Identical or similar amino acids are shaded black or gray, respectively. The GenBank accession numbers of the complete TRH1 cDNA and genomic sequences are AJ296155 and AJ296156, respectively.
The TRH1 Gene Complements the trh1 Mutant Phenotype
To establish whether the disrupted gene is responsible for the trh1 mutant phenotype, we hybridized the TRH1 gene to total RNA isolated from mutant and wild-type plants. RNA gel blot analysis showed that the mRNA transcribed from the TRH1 gene was not detected in the trh1 plants (Figure 2). A more detailed analysis with reverse transcription (RT)–PCR verified this result (Figure 2). Furthermore, we transformed mutant plants with genomic DNAs spanning the TRH1 gene. Five different wild-type genomic fragments were used: the 8.5-kb BamHI-SalI fragment and the 7.1-, 6.6-, and 5.7-kb fragments containing ∼2.7-kb, 1800-bp, 1300-bp, and 300-bp upstream sequences, respectively (Figure 3). The upstream fragment, from −3.1 kb to +310 bp, was also used for complementation analysis (Figure 3). The 8.5-kb BamHI-SalI fragment and the 7.1- and 6.6-kb fragments (Figure 3) complemented the trh1 phenotype, producing plants with wild-type root hairs (Figure 1). These results clearly demonstrate that the TRH1 gene is required for the establishment of tip growth from the initiation site of the root hair cell. The 5.7-kb fragment (Figure 3) containing ∼300-bp upstream sequences failed to complement the mutant phenotype. The 300 nucleotides of the promoter region therefore were insufficient for proper expression of the TRH1 gene. The fragment containing exclusively upstream sequences was also unable to complement the mutant plant (Figure 3).
TRH1 Gene Expression
Based on high-stringency DNA gel blot analysis, we determined that only one copy of the TRH1 gene is present in the Arabidopsis genome, because one major hybridizing band was generated by the restriction enzymes EcoRI, HindIII, SalI, and PstI (Figure 2). Identical hybridization patterns of genomic DNA isolated from Columbia or Wassilewskija ecotypes were observed. To determine the expression pattern of the TRH1 gene, we isolated RNA from the aerial parts (leaves, stems, flowers, rosette leaves) and from the root of the plant. Quantitative RT-PCR analysis showed that the TRH1 gene is expressed almost equally in all parts of the plant (Figure 2). To investigate the temporal expression and developmental accumulation of the TRH1 transcripts during the early stages of plant development, we isolated RNA from roots and shoots of developing seedlings. RNA gel blot analysis showed that during seedling development, the levels of TRH1 gene transcripts were higher at the early stages of seedling growth and that root tissues accumulated higher levels of these transcripts compared with the aerial parts (Figure 2).
TRH1 Is Similar to a Family of Potassium Transporters
Alignment of TRH1 showed that the predicted polypeptide shares significant homology with the bacterial KUP1 (Schleyer and Bakker, 1993) and yeast HAK1 (Banuelos et al., 1995) potassium transporters. This alignment yielded 30 and 26% identity overall for KUP1 and HAK1, respectively. The alignment of the predicted amino acids of TRH1 yielded an average 46% identity with the two other isolated members of the potassium transporter gene family from Arabidopsis, AtKT1 and AtKT2 (Quintero and Blatt, 1997; Fu and Luan, 1998; Kim et al., 1998) and 33% identity with HvHAK1 from barley (Santa-Maria et al., 1997). However, the similarity of TRH1 to the other two members of the gene family AtKT1 and AtKT2 is ∼66% (Figure 4).
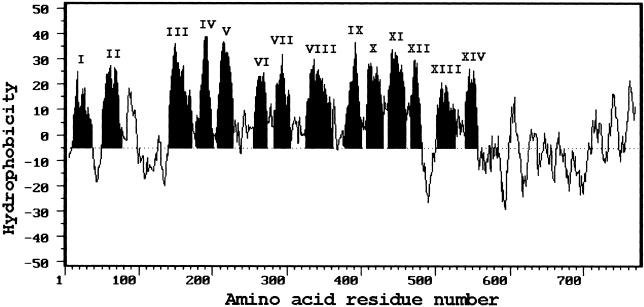
The polypeptide deduced from the TRH1 gene can be broadly categorized into three distinct domains (Figure 5). The N- and C-terminal domains, consisting of 7 and 204 amino acids, respectively, are hydrophilic, and the middle domain, consisting of 564 amino acids, is mostly hydrophobic. By computer analysis of the deduced polypeptide, TRH1 was predicted to contain 14 transmembrane domains. The number of hydrophilic amino acids separating the transmembrane spans ranges from 3 to 73, with an average length of ∼20 amino acids. The largest hydrophilic loop is located between the second and third transmembrane spans (Figure 5). This position is consistent with those predicted for KUP1 from bacteria, HAK1 from yeast, and AtKT1 and AtKT2 from Arabidopsis (Schleyer and Bakker, 1993; Banuelos et al., 1995; Quintero and Blatt, 1997; Kim et al., 1998).
Figure 5.
Hydropathy Profile of Amino Acid Sequences Deduced from the TRH1 Gene.
Hydropathy indices were calculated using an interval of 13 amino acids. Roman numerals denote the number of transmembrane (shaded) spans.
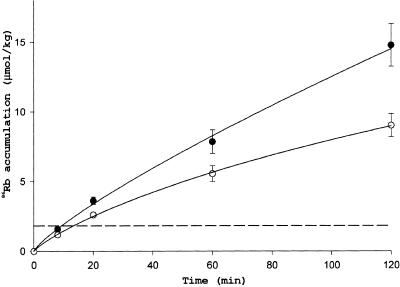
To ascertain the effect of external potassium concentrations on the growth of root hairs, we grew Arabidopsis seedlings on medium containing various concentrations of K+. The trh1 phenotype was prominent from low (0.1 mM) to high (50 mM) external potassium concentrations. Similarly, there was very little effect on root hair growth in wild-type plants grown at various concentrations of external potassium. To determine whether TRH1 functions in potassium uptake, we performed K+ uptake analysis using 86Rb as a radioactive tracer. Wild-type plants accumulated 86Rb at a rate of 3.7 nmol/hr (calculated from data shown in Figure 6 and normalized per 1 g of fresh weight and 1 μmol of Rb in the incubation medium). Similar results (4.5 nmol/hr) were obtained by Hirsch et al. (1998) under similar experimental conditions (10 μmol of alkali cations in the medium). A significantly lower rate of 86Rb accumulation was detectable in the trh1 mutant after only 10 min of incubation. After 2 hr of incubation, the normalized rate of 86Rb uptake by trh1 was 2.2 nmol/hr, which was 40% less than the uptake rate in wild-type plants.
Figure 6.
Time Dependence of 86Rb Uptake by Wild-Type and trh1 Plants.
Each point represents the mean ±se for nine seedlings. The dashed line indicates a concentration of 86Rb in the incubation medium. Solid circles, wild type; open circles, trh1.
TRH1 Complements Yeast Mutants Defective in High-Affinity K+ Transport
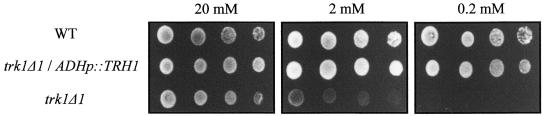
To analyze further TRH1 function as a potassium transporter, we assessed its capacity to rescue yeast mutants defective in K+ transport. Two yeast strains were used: the single mutant strain M398 with the high-affinity potassium transporter TRK1 gene deleted and the double mutant strain CY162 lacking both potassium transporter TRK1 and TRK2 genes (Gaber et al., 1988; Ko et al., 1990; Ko and Gaber, 1991). Both mutant strains carrying the potassium transporter deletions are lethal when grown in medium with low levels of potassium (Anderson et al., 1992). We transformed both strains with the expression vector and the expression vector containing the TRH1 cDNA. The TRH1 cDNA failed to complement the CY162 yeast strain carrying trk1Δtrk2Δ deletions because the transformants were unable to grow in medium containing low potassium concentrations (2 or 0.2 mM). However, the yeast strain M398 transformed with the TRH1 cDNA grew like the wild-type yeast strain in medium containing potassium concentrations as low as 0.2 mM (Figure 7). This finding suggests that the TRH1 gene product possesses high-affinity potassium transporter activity in yeast cells.
Figure 7.
Function of TRH1.
The yeast potassium transport mutant strain M398 lacking the TRK1 gene was transformed with the yeast expression vector pVT100U containing the TRH1 gene under the control of the ADH1 promoter (trk1Δ1/ADHp::TRH1). The wild-type strain R757 (WT) and the mutant strain M398 (trk1Δ1) harbored the plasmid and were used as controls. A fivefold dilution series of cell suspensions were inoculated on ammonium-free solid arginine synthetic medium containing different concentrations of K+.
To determine the functional domains of the TRH1 polypeptide, we deleted the hydrophilic C-terminal end; the remaining truncated TRH1 gene was used for complementation experiments of the M398 yeast strain. This truncated construct, which contained all transmembrane domains but lacked the putative cAMP-dependent kinase phosphorylation site, was unable to complement trk1Δ deletion of the M398 yeast strain (data not shown). This result indicates that the hydrophilic C-terminal end is a key feature of high-affinity potassium transporter activity. Alternatively, this hydrophilic end may be important in the correct deposition and localization of the protein.
DISCUSSION
TRH1 Gene Disruption Arrests Root Hair Tip Growth
Several genes that disrupt the initiation of root hair development, tip growth, hair morphology, and the spatial arrangement of root hairs have been identified and isolated from Arabidopsis. The TRH1 gene is required for the establishment of tip growth from the initiation site, suggesting that it acts as a positive genetic regulator of tip growth. Trichoblasts of trh1 plants, however, occasionally develop multiple initiation sites, indicating that either the TRH1 gene product itself or the outgrowth from the basal end of the trichoblast may inhibit any initiation of additional hairs from the same cell. Mutants impaired in patterning and cellular differentiation, such as ttg and gl2, cause root hairs to form on nearly every root epidermal cell. This implies that TTG (a MYC transcription factor) and GL2 (a homeobox gene) either promote nonhair differentiation or repress root hair differentiation (Galway et al., 1994; Masucci et al., 1996). A novel gene (WER), which encodes a MYB-type protein, is also required for nonhair fate (Lee and Schiefelbein, 1999). The CPC gene, which encodes a protein with a MYB-like DNA binding domain but no typical transcriptional activation domain, is required for hair/cell differentiation (Wada et al., 1997). Other genes that affect tip growth, such as RHD1 and RHD6, are required for the early stages of root hair diffuse growth and also affect pollen tube formation (Schiefelbein and Somerville, 1990; Masucci and Schiefelbein, 1994; van Den Berg et al., 1998). The COW1, TIP1, and RHD3 genes are important for tip growth but also affect the morphology of root hair outgrowth (Galway et al., 1997; Grierson et al., 1997; Ryan et al., 1997). Plants homozygous for point mutations in the RHD2 gene initiate root hairs, which cease growing almost immediately (Schiefelbein and Somerville, 1990). This phenotype is similar to that of the trh1 mutant. Both trh1 and rhd2 homozygous plants have normal appearance and exhibit no other morphological abnormalities. However, they are not alleles, because rhd2 mapped on chromosome V (Aeschbacher et al., 1994) and trh1 mapped on chromosome IV. The cessation of growth in rhd2 plants may be in part the result of defective Ca2+ gradient formation in the root hairs, because RHD2 is required for calcium gradient formation in the tip of the growing hair. It is possible that RHD2 may be required for the activity of TRH1. Determining the activity of TRH1 in plants homozygous for rhd2 will be instructive.
TRH1 Is a Potassium Transporter
TRH1 shares considerable amino acid sequence homology with the KUP1 and HAK1 genes, which are potassium transporters from Escherichia and Schwanniomyces, respectively (Schleyer and Bakker, 1993; Banuelos et al., 1995). This fact suggests that TRH1 can function as a K+ transporter in Arabidopsis. This hypothesis has been confirmed in uptake experiments in which 86Rb was used as a radioactive tracer. The rate of K+ transport was reduced significantly (40%) in trh1 plants (Figure 6). We cannot exclude, however, that this reduction was partially due to smaller surface area in trh1 roots. The mutant plants did not develop symptoms of K+ starvation. Thus, we conclude that the rate of K+ transport was not a limiting factor for whole-plant growth in our experiments, probably because the total K+ concentration in Murashige and Skoog (1962) medium is as high as 20 mM.
In accordance with the proposed K+ transport function, the TRH1 gene complemented the M398 yeast strain deficient for the high-affinity K+ uptake system (Gaber et al., 1988). These results indicate that the TRH1 gene encodes a high-affinity potassium transporter, at least when expressed in yeast. The TRH1 gene is a member of the AtKT/AtKUP/HAK gene family (Quintero and Blatt, 1997; Fu and Luan, 1998; Kim et al., 1998). Another protein from the same family, AtKT1 (AtKUP1), has been reported to exhibit high-affinity (Kim et al., 1998) or dual-affinity (Fu and Luan, 1998) potassium transport activity. The AtKT2 (AtKUP2) gene was suggested to mediate low-affinity K+ transport, with 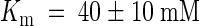 (Quintero and Blatt, 1997).
(Quintero and Blatt, 1997).
Although TRH1 is a member of the AtKUP gene family, DNA gel blot analysis revealed no cross-hybridization with any genomic fragments of the other AtKUP genes, indicating a high degree of divergence within this gene family. TRH1 is constitutively expressed in most organs of Arabidopsis. The developmental pattern of mRNA accumulation during the early stages of postembryonic root growth showed almost constant levels. However, plants homozygous for the loss of function of this gene showed no other phenotype except defective root hair growth. The AtKUP gene family exhibits similar patterns of gene expression (Kim et al., 1998). This result also suggests that the function of each product of the AtKUP gene family may depend on additional factors (i.e., β subunits) or on post-translational modification (i.e., phosphorylation).
The AtKUP1 transporter from Arabidopsis was not expressed in roots (Kim et al., 1998) but it was detected in shoots, implying that the function of TRH1 may be redundant in other plant tissues or organs. We also cannot exclude the possibility that other potassium transporters from the same family are active in roots. This complexity in affinity and redundancy may reflect plant plasticity in adaptation to soil environments with different levels of K+.
Other potassium translocators preferentially present in root tissues are the inward- and outward-rectifying K+ channels AKT1 and SKOR, respectively (Lagarde et al., 1996; Gaymard et al., 1998). AKT1 is localized in the plasma membrane of root cells, and the SKOR gene is expressed in the root pericycle and in stelar parenchyma cells. SKOR is involved in K+ release into the xylem sap, and its disruption results in decreased K+ translocation toward the shoots (Gaymard et al., 1998). Therefore, it is reasonable to believe that potassium is transported from the rhizosphere by the AKT1 localized in the plasma membrane of the root cells and secreted to the xylem sap by the SKOR for transport to other parts of the plant. A place for TRH1 in the K+ transport pathway has yet to be identified, but its significance for root hair development suggests that this transporter may be important for specific localized K+ delivery.
Predicted Protein Structure
Computer analysis showed that the TRH1 gene product is composed of a hydrophobic part consisting of 14 transmembrane domains, in contrast to other members of the AtKT/AtKUP/HAK family, which contain 12 transmembrane regions (Schleyer and Bakker, 1993; Banuelos et al., 1995; Quintero and Blatt, 1997; Fu and Luan, 1998; Kim et al., 1998). All polypeptides from this family have a large hydrophilic loop between the second and third transmembrane spans, an even number of transmembrane domains, and a long, mostly hydrophilic C terminus.
It is well established that the majority of transport proteins function as multimeric proteins. Voltage-gated K+ channels, for instance, are predicted to consist of six transmembrane regions (S1 to S6) and a pore loop (H5) between domains S5 and S6 (Jan and Jan, 1994). The functioning channel is formed by four identical subunits, and a pore is lined by four H5 loops (Jan and Jan, 1994). Fu and Luan (1998) suggested that the AtKUP1 protein may function as a voltage-dependent K+ channel. They also found homology between the signature motif (GYGD) in the K+ channel pore and the IYGD motif in a highly conserved region of the AtKUP1 and HAK1 proteins. We cannot deny the similarity between TRH1 and K+ channels. In TRH1, however, IYGD is substituted with a modified motif, VFGD. At present, we know neither the number of subunits that form the functional transporter nor whether the VFGD site is in fact a determinant of K+ selectivity in TRH1. We will attempt to answer these questions in future investigations.
The structural features necessary for K+ uptake have not been determined for TRH1. However, when the hydrophilic C-terminal end of the protein was removed, the truncated TRH1 polypeptide did not complement the M398 mutant yeast strain, indicating that this domain is essential for potassium translocation. Alternatively, this removal may result in misfolding or misincorporation of the remaining polypeptide in the membrane.
Root Hair Tip Growth
The current paradigm postulates that changes in cell turgor are responsible for the movement of plant cells and organs and for changes in cell shape. A characteristic example of the latter is the opening and closing of stomata, where K+ and anion channel activities are intimately involved in osmoregulation (Schroeder et al., 1984; Grabov and Blatt, 1998). Although mechanisms similar to those that participate in guard cell volume regulation have been implicated in maize coleoptile elongation (Claussen et al., 1997; Philippar et al., 1999), a role for K+ translocators in root hair growth has not been demonstrated. It has been shown that a blocker of K+ channels, tetraethylammonium, inhibits inward-rectifying K+ channels in Arabidopsis, but it caused only a transient cessation in tip growth (Lew, 1991).
In this report, we demonstrate that the potassium transporter TRH1 is involved in root hair outgrowth. The phenotype of the trh1 mutant indicates that the identity of the root hair cells is established in the rhizodermis, but root hairs fail to grow despite the fact that other potassium transporters are known to be present in the root. Moreover, the phenotype of the trh1 mutant plant was not restored even when the plants were grown in a medium containing 50 mM K+. These results indicate that tip growth requires spatially localized and/or temporally coordinated K+ transport activity that cannot be substituted for by increasing the rate of diffuse K+ uptake. This specific transport activity may be important for channelized solute delivery or for the generation of electrochemical gradients around tip-growing cells. These gradients have been shown to accompany the apical growth of root hairs as well as some other cells (Jaffe et al., 1974; Schiefelbein et al., 1992; Jones et al., 1995). This hypothesis is compatible with the fact that trh1 trichoblasts produced multiple initiation sites. More experiments, however, must be performed to prove this theory.
In summary, plants lacking TRH1 gene function are morphologically normal in most respects under standard growth conditions, indicating either that TRH1 activity is not required for whole-plant development or that other genes can compensate for its absence. TRH1-mediated K+ translocation is not crucial for overall plant K+ nutrition, particularly when plants are grown on rich medium. In contrast, there is an absolute requirement for TRH1 activity during root hair elongation, indicating that TRH1 plays a critical role in specific K+ translocation that is essential for the growth of this cell type.
METHODS
T-DNA Tagging and Plasmid Rescue
Germinating seed from the geographic race Wassilewskija of Arabidopsis thaliana were transformed with Agrobacterium tumefaciens strain C58C1 RifR. This plasmid carries the pGV3850:pAK1003 Ti plasmid (Velten and Schell, 1985; Feldmann, 1991), which contains the neomycin phosphotransferase II gene that confers resistance to kanamycin, and duplicated pBR322 sequences from both ends of the T-DNA. trh1 plants were identified in a population transformed with this Ti plasmid.
For the recovery of plant DNA disrupted by the T-DNA insertion, the plasmid rescue technique was applied (Behringer and Medford, 1992). The recovered plasmids from both EcoRI-digested (right border rescue) and SalI-digested (left border rescue) genomic DNA isolated from trh1 homozygous plants were analyzed further.
One plasmid that was rescued from the SalI digest, named pBSalI0.7, consisted of only T-DNA sequences, as if two copies were inserted in a single locus of the genome. Two plasmids were rescued from the EcoRI digest, pBER2.4 and pBER1.3, containing 1.3 and 0.2 kb, respectively, of genomic DNA. T-DNA–plant DNA junction fragments of rescued clones were isolated and either used as hybridization probes or subcloned into pBluescript KS+ or SK− (Stratagene).
Library Screening and Sequencing
The 1.3-kb plant DNA that flanked the T-DNA of the pBER2.4 clone was used as a probe to screen an Arabidopsis genomic library in the λGEM11 vector (Promega). Approximately 100,000 plaques were screened as described by Maniatis et al. (1982). The eight positive overlapping λ clones were subjected to restriction analysis. A 1.5-kb EcoRI genomic fragment was hybridized to a 1.3-kb rescued fragment and identified as being present in all isolated λ clones. This genomic fragment was used as a probe for the screening of a cD4-7 library (Newman et al., 1994). Four positively hybridizing plaques were identified and subjected to plaque purification. The cDNA clones were converted to recombinant pZL1 plasmids. The restriction analysis pattern and sequencing results revealed that all four cDNAs were identical and found to be truncated at the 5′ missing 18 amino acids but homologous with expressed sequence tag (EST) H10G7. Sequencing was performed using a 35S-dATP for double-stranded DNA sequencing (Sanger et al., 1977).
DNA and RNA Gel Blot Analysis
Genomic DNA was isolated from Arabidopsis (Columbia ecotype) by the cetyl-trimethyl-ammonium bromide method (Murray and Thompson, 1980). After digestion with restriction enzymes, DNA (5 μg in each lane) was loaded and separated on a 0.7% agarose gel and blotted onto a nylon membrane (Hybond+; Amersham Corp.). The 32P-labeled probes were prepared using the nick translation protocol. Prehybridization for 1 hr, overnight hybridization, and washes were performed as described by Church and Gilbert (1984).
RNA was isolated from 7-day-old seedlings grown in liquid Murashige and Skoog (1962) (MS) medium. Frozen tissue was ground in liquid nitrogen with a mortar and pestle. Total RNA was isolated using the phenol-SDS extraction method. RNA concentrations were determined spectrophotometrically and verified by ethidium bromide staining of agarose gels. The RNAs were electrophoresed on 1.2% agarose gels containing formaldehyde and blotted to a nitrocellulose membrane. The membrane was hybridized as described for DNA gel blot analysis.
Polymerase Chain Reaction Analysis
The oligonucleotides used for polymerase chain reaction (PCR) analysis were as follows: st2rev, 5′-CCAAGCTACGCCGCATCTTACTCGC-3′, and exon2, 5′-CTTCACCATTGTCATCTGCACTTAAC-3′, corresponding to the 5′ untranslated region and the second exon of the TRH1 gene, respectively; #1242, 5′-AAGTGCGGCGACGATAGTCATGCCCCG-3′, corresponding to the pBR322 sequence; apt1, 5′-TCCCAGAATCGCTAAGATTGCC-3′, and apt2, 5′-CCTTTCCCTTAAGCTCTG-3′, corresponding to the APT1 gene; actin2, 5′-AAGATG-ACCCAAATCATGTTTGAGAC-3′, and actin3, 5′-ACGACCTTGATC-TTCATGCTGC-3′, corresponding to the ACTIN gene. To identify the molecular organization of the T-DNA inserts, PCR reactions were performed using genomic DNA of both wild-type and trh1 plants as templates. Various combinations of st2rev, exon2, and #1242 were selected. The templates were amplified with the Expand High Fidelity PCR Polymerase (Roche Molecular Biochemicals, Mannheim, Germany). Total RNA (0.5 μg) was isolated and treated with RQ1 RNase-free DNase (Promega). Reverse transcription (RT)–PCR was performed using the Titan one-tube RT-PCR system (Roche Molecular Biochemicals), according to the manufacturer's instructions. The APT1 gene is expressed at a low level in all tissues of Arabidopsis and was used as an endogenous control mRNA, producing a 478-bp PCR product (Moffatt et al., 1994).
Complementation of the trh1 Mutation and Medium Preparation
For Arabidopsis transformation, genomic fragments were filled in by Klenow DNA polymerase and inserted into the filled HindIII site of pGPTV-HPT binary vector (Becker et al., 1992). The Agrobacterium strain C58C1 RifR containing the nononcogenic Ti plasmid pGV3101 was transformed with the pGPTV-HPT constructs by electroporation (Gene Pulser II; Bio-Rad). The vacuum infiltration method was used for Arabidopsis transformation, according to Bechtold et al. (1993).
Seed (T1 generation) were selected on 1 × MS medium, 2% (w/v) sucrose, 0.05% Mes, pH 5.7, 0.6% (w/v) agarose plates containing 30 mg/L hygromycin (Sigma) and 50 mg/L Claforan (Hoechst, Frankfurt, Germany). Resistant individuals could be clearly identified 10 to 15 days after germination. Seed of T2 and T3 generations were plated individually on the medium described above, except that Claforan was omitted.
To resolve the effects of various concentrations of K+ on root hair morphogenesis in both wild-type and trh1 plants, artificial plant growth medium was prepared according to Hirsch et al. (1998). The primary roots of 7-day-old seedlings were observed and photographed using an Olympus (Tokyo, Japan) SZX12 stereomicroscope.
86Rb Uptake Experiments
Plants were grown hydroponically for 7 days in sterile conditions on 1 × MS medium, 1% sucrose, and 0.05% Mes, pH 5.7. Seedlings were harvested and preincubated for 5 hr in medium containing 10 μM KNO3, 3 mM Ca(NO3)2, 1.5 mM MgSO4, 1 mM NaH2PO4/Na2HPO4 buffer, pH 5.6, and 7 mM Ca2+-Mes buffer, pH 5.6. Incubation buffer of the same composition was supplemented with 1.8 μM 86Rb-Cl (DuPont–New England Nuclear, Boston, MA) with a specific activity of 21.9 Bq/pmol. After loading with 86Rb, samples (three seedlings in each) were washed with 10 × 5 mL of preincubation buffer supplemented with 50 μM unlabeled Rb. Samples were dried for 15 sec on 55-mm Ø paper filters (Whatman) using a vacuum filtration apparatus and placed in preweighed scintillation vials. After determination of plant weight, vials were filled with Ecolite (ICN, Costa Mesa, CA) scintillation cocktail. Radioactivity was measured using a Rackbeta 1211 (LKB-Wallac, Turku, Finland) scintillation counter.
Yeast Complementation
To determine whether TRH1 can function as a potassium transporter, we tested cDNA for the ability to complement the potassium transport deficiency of the M398 strain of Saccharomyces cerevisiae (Gaber et al., 1988; Sentenac et al., 1992). The H10G7 EST full-length cDNA (GenBank accession number AA042476) was cloned into the XbaI site of the yeast expression vector pVT100U that carries the constitutively expressed system of the yeast alcohol dehydrogenase gene (ADH1) promoter–terminator sequences and a modified URA3 gene for selection in yeast (Vernet et al., 1987). Yeast transformation was performed by the lithium acetate method. The transformed yeast cells were selected on URA-glucose-ammonium–free solid arginine synthetic medium supplemented with 100 mM KCl (Rodriguez-Navarro and Ramos, 1984; Ko et al., 1990). Growth restoration of the M398 mutant strain was performed on the synthetic medium described above supplemented with various concentrations of K+.
Microscopy
Three- and five-day old roots were frozen in nitrogen slush at −190°C. Ice was sublimed at −90°C, and the specimen was sputter coated and examined on an XL 30 FEG (Philips, Eindhoven, The Netherlands) cryoscanning electron microscope fitted with a cold stage.
Acknowledgments
The authors thank the Arabidopsis Biological Resource Center (Ohio State University, Columbus) for providing the cD4-7 cDNA library and the EST H10G7 clone; Dr. J. Schell for providing pGPTV-HPT binary vector; and Dr. R. Gaber for providing the R757, M398, and CY162 yeast strains and the shuttle vector pVT100U. We thank Dr. Ben Scheres (University of Utrecht, Utrecht, The Netherlands) for critical reading of the manuscript and suggestions and Dr. D. Milioni for helpful discussions. S.R. was supported by the Greek State Scholarships Foundation. This research was also supported by grants to L.D. and P.H. from the European Union (Grant No. BIO4-CT96-0217 [DGXII]), to P.H. from the General Secretariat of Research and Technology, Greece (Grant No. 95/1300), and to A.G. and L.D. from the Biotechnology and Biological Science Research Council, UK. We acknowledge the massive input of Scott Poethig (University of Pennsylvania, Philadelphia), in whose laboratory this research was initiated. We are grateful for the help and advice of Dr. Pablo Scolnik (E.I. Du Pont De Nemours Co., Wilmington, DE).
References
- Aeschbacher, R.A., Schiefelbein, J.W., and Benfey, P. (1994). The genetic and molecular basis of root development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 45 25–45. [Google Scholar]
- Anderson, J.A., Huprikar, S.S., Kochian, L.V., Lucas, W.J., and Gaber, R.F. (1992). Functional expression of a probable Arabidopsis thaliana potassium channel in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 89 3736–3740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banuelos, M.A., Klein, R.D., Alexander-Bowman, S.J., and Rodriguez-Navarro, A. (1995). A potassium transporter of the yeast Schwanniomyces occidentalis homologous to the Kup system of Escherichia coli has a high concentrative capacity. EMBO J. 14 3021–3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316 1194–1199. [Google Scholar]
- Becker, D., Kemper, E., Schell, J., and Masterson, R. (1992). New plant binary vectors with selectable markers located proximal to the left T-DNA border. Plant Mol. Biol. 20 1195–1197. [DOI] [PubMed] [Google Scholar]
- Behringer, F.J., and Medford, J.I. (1992). A plasmid rescue technique for the recovery of plant DNA disrupted by T-DNA insertion. Plant Mol. Biol. Rep. 10 190–198. [Google Scholar]
- Bibikova, T.N., Jacob, T., Dahse, I., and Gilroy, S. (1998). Localized changes in apoplastic and cytoplasmic pH are associated with root hair development in Arabidopsis thaliana. Development 125 2925–2934. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussen, M., Lüthen, H., Blatt, M., and Böttger, M. (1997). Auxin-induced growth and its linkage to potassium channels. Planta 201 227–234. [Google Scholar]
- Dolan, L., Duckett, C.M., Grierson, C., Linstead, P., Schneider, K., Lawson, E., Dean, C., Poethig, S., and Roberts, K. (1994). Clonal relationships and cell patterning in the root epidermis of Arabidopsis. Development 120 2465–2474. [Google Scholar]
- Duckett, C.M., Oparka, K.J., Prior, D.A.M., Dolan, L., and Roberts, K. (1994). Dye-coupling in the root epidermis of Arabidopsis is progressively reduced during development. Development 120 3247–3255. [Google Scholar]
- Epstein, E. (1966). Dual pattern of ion absorption by plant cells and by plants. Nature 212 1324–1327. [Google Scholar]
- Epstein, E., Rains, D.W., and Elzam, O.E. (1963). Resolution of dual mechanisms of potassium absorption by barley roots. Proc. Natl. Acad. Sci. USA 49 684–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, K.A. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1 71–82. [Google Scholar]
- Fu, H.H., and Luan, S. (1998). AtKUP1: A dual-affinity K+ transporter from Arabidopsis. Plant Cell 10 63–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaber, R.F., Styles, C.A., and Fink, G.R. (1988). TRK1 encodes a plasma membrane protein required for high-affinity potassium transport in Saccharomyces cerevisiae. Mol. Cell. Biol. 8 2848–2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galway, M.E., Masucci, J.D., Lloyd, A.M., Walbot, V., Davis, R.W., and Schiefelbein, J.W. (1994). The TTG gene is required to specify epidermal cell fate and cell patterning in the Arabidopsis root. Dev. Biol. 166 740–754. [DOI] [PubMed] [Google Scholar]
- Galway, M.E., Heckman, J.W., Jr., and Schiefelbein, J.W. (1997). Growth and ultrastructure of Arabidopsis root hairs: The rhd3 mutation alters vacuole enlargement and tip growth. Planta 201 209–218. [DOI] [PubMed] [Google Scholar]
- Gaymard, F., Pilot, G., Lacombe, B., Bouchez, D., Bruneau, D., Boucherez, J., Michaux-Ferriere, N., Thibaud, J.B., and Sentenac, H. (1998). Identification and disruption of a plant Shaker-like outward channel involved in K+ release into the xylem sap. Cell 94 647–655. [DOI] [PubMed] [Google Scholar]
- Grabov, A., and Blatt, M. (1998). Coordination of signalling elements in guard cell ion channel control. J. Exp. Bot. 49 351–360. [Google Scholar]
- Grabov, A., and Böttger, M. (1994). Are redox reactions involved in regulation of K+ channels in the plasma membrane of Limnobium stoloniferum root hairs? Plant Physiol. 105 927–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grierson, C.S., Roberts, K., Feldmann, K.A., and Dolan, L. (1997). The COW1 locus of Arabidopsis acts after RHD2, and in parallel with RHD3 and TIP1, to determine the shape, rate of elongation, and number of root hairs produced from each site of hair formation. Plant Physiol. 115 981–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsch, R.E., Lewis, B.D., Spalding, E.P., and Sussman, M.R. (1998). A role for AKT1 potassium channel in plant nutrition. Science 280 918–921. [DOI] [PubMed] [Google Scholar]
- Jaffe, L.F., Robinson, K.R., and Nuccitelli, R. (1974). Local cation entry and self-electrophoresis as an intracellular localization mechanism. Ann. N.Y. Acad. Sci. 238 372–389. [DOI] [PubMed] [Google Scholar]
- Jan, L.Y., and Jan, Y.N. (1994). Potassium channels and their evolving gates. Nature 371 119–122. [DOI] [PubMed] [Google Scholar]
- Jones, D.L., Shaff, J.E., and Kochian, L.V. (1995). Role of calcium and other ions in directing root hair tip growth in Limnobium stoloniferum. I. Inhibition of tip growth by aluminium. Planta 197 672–680. [Google Scholar]
- Kim, E.J., Kwak, J.M., Uozumi, N., and Schroeder, J.I. (1998). AtKUP1: An Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10 51–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, C.H., and Gaber, R.F. (1991). TRK1 and TRK2 encode structurally related K+ transporters in Saccharomyces cerevisiae. Mol. Cell. Biol. 11 4266–4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko, C.H., Buckley, A.M., and Gaber, R.F. (1990). TRK2 is required for low affinity K+ transport in Saccharomyces cerevisiae. Genetics 125 305–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde, D., Basset, M., Lepetit, M., Conejero, G., Gaymard, F., Astruc, S., and Grignon, C. (1996). Tissue-specific expression of Arabidopsis AKT1 gene is consistent with a role in K+ nutrition. Plant J. 9 195–203. [DOI] [PubMed] [Google Scholar]
- Leavitt, R.G. (1904). Trichomes of the root in vascular cryptograms and angiosperms. Proc. Boston Soc. Natl. Hist. 31 273–313. [Google Scholar]
- Lee, M.M., and Schiefelbein, J. (1999). WEREWOLF, a MYB-related protein in Arabidopsis, is a position-dependent regulator of epidermal cell patterning. Cell 99 473–483. [DOI] [PubMed] [Google Scholar]
- Lew, R.R. (1991). Electrogenic transport properties of growing Arabidopsis root hairs: The plasma membrane proton pump and potassium channels. Plant Physiol. 97 1527–1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis, F.J.M., and Sanders, D. (1994). Mechanism of high-affinity potassium uptake in roots of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 91 9272–9276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maathuis, F.J.M., Ichida, A.M., Sanders, D., and Schroeder, J.I. (1997). Roles of higher plant K+ channels. Plant Physiol. 114 1141–1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maniatis, T., Fritsch, E.F., and Sambrook, J. (1982). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Masucci, J.D., and Schiefelbein, J.W. (1994). The rhd6 mutation of Arabidopsis thaliana alters root hair initiation through an auxin- and ethylene-associated process. Plant Physiol. 106 1335–1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masucci, J.D., Rerie, W.G., Foreman, D.R., Zhang, M., Galway, M.E., Marks, M.D., and Schiefelbein, J.W. (1996). The homeobox gene GLABRA2 is required for position-dependent cell differentiation in the root epidermis of Arabidopsis thaliana. Development 122 1253–1260. [DOI] [PubMed] [Google Scholar]
- Moffatt, B.A., McWhinnie, E.A., Agarwal, S.K., and Schaff, D.A. (1994). The adenine phosphoribosyltransferase–encoding gene of Arabidopsis thaliana. Gene 143 211–216. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Murray, M.G., and Thompson, W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman, T., De Bruijn, F.J., Green, P., Keegstra, K., Kende, H., McIntosh, L., Ohlrogge, J., Raikhel, N., Somerville, S., Thomashow, M., Retzel, E., and Somerville, C. (1994). Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippar, K., Fuchs, I., Lüthen, H., Hoth, S., Bauer, C.S., Haga, K., Thiel, G., Ljung, K., Sandberg, G., Böttger, M., Becker, D., and Hedrich, R. (1999). Auxin-induced K+ channel expression represents an essential step in coleoptile growth and gravitropism. Proc. Natl. Acad. Sci. USA 96 12186–12191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintero, F.J., and Blatt, M.R. (1997). A new family of K+ transporters from Arabidopsis that are conserved across phyla. FEBS Lett. 415 206–211. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Navarro, A., and Ramos, J. (1984). Dual system for potassium transport in Saccharomyces cerevisiae. J. Bacteriol. 159 940–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio, F., Gassmann, W., and Schroeder, J.I. (1995). Sodium-driven potassium uptake by the plant potassium transporter HKT1 and mutations conferring salt tolerance. Science 270 1660–1663. [DOI] [PubMed] [Google Scholar]
- Rubio, F., Santa-Maria, G.E., and Rodriguez-Navarro, A. (2000). Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol. Plant. 109 34–43. [Google Scholar]
- Ryan, E., Grierson, C.S., Cavell, A., Steer, M., and Dolan, L. (1997). TIP1 is required for both tip growth and non-tip growth in Arabidopsis. New Phytol. 138 49–58. [Google Scholar]
- Sanger, F., Nicklen, S., and Coulson, A.R. (1977). DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 74 5463–5467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santa-Maria, G.E., Rubio, F., Dubcovsky, J., and Rodriguez-Navarro, A. (1997). The HAK1 gene of barley is a member of a large gene family and encodes a high affinity potassium transporter. Plant Cell 9 2281–2289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schachtman, D.P., and Schroeder, J.I. (1994). Structure and transport mechanism of a high-affinity potassium uptake transporter from higher plants. Nature 370 655–658. [DOI] [PubMed] [Google Scholar]
- Scheres, B., Wolkenfelt, H., Willemsen, V., Terlouw, M., Lawson, E., Dean, C., and Weisbeek, P. (1994). Embryonic origin of the Arabidopsis primary root and root meristem initials. Development 120 2475–2487. [Google Scholar]
- Schiefelbein, J.W., and Somerville, C. (1990). Genetic control of root hair development in Arabidopsis thaliana. Plant Cell 2 235–243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiefelbein, J.W., Shipley, A., and Rowse, P. (1992). Calcium influx at the tip of growing root-hair cells of Arabidopsis thaliana. Planta 187 455–459. [DOI] [PubMed] [Google Scholar]
- Schiefelbein, J., Galway, M., Masucci, J., and Ford, S. (1993). Pollen tube and root-hair tip growth is disrupted in a mutant of Arabidopsis thaliana. Plant Physiol. 103 979–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schleyer, M., and Bakker, E.P. (1993). Nucleotide sequence and 3′-end deletion studies indicate that the K+ uptake protein Kup from Escherichia coli is composed of a hydrophobic core linked to a large and partially essential hydrophilic C terminus. J. Bacteriol. 175 6925–6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder, J.I., Hedrich, R., and Fernandez, J.M. (1984). Potassium-selective single channels in guard cell protoplasts of Vicia faba. Nature 312 361–362. [Google Scholar]
- Sentenac, H., Bonneaud, N., Minet, M., Lacroute, F., Salmon, J.M., Gaymard, F., and Grignon, C. (1992). Cloning and expression in yeast of a plant potassium ion transport system. Science 256 663–665. [DOI] [PubMed] [Google Scholar]
- Sievers, A., and Schnepf, E. (1981). Morphogenesis and polarity of tubular cells with tip-growth. Cell Biol. Monogr. 8 265–299. [Google Scholar]
- Smith, F.A., and Walker, N.A. (1989). Transport of potassium by Chara australis. I. A symport with sodium. J. Membr. Biol. 108 125–137. [DOI] [PubMed] [Google Scholar]
- Uozumi, N., Kim, E.J., Rubio, F., Yamaguchi, T., Muto, S., Tsuboi, A., Bakker, E.P., Nakamura, T., and Schroeder, J.I. (2000). The Arabidopsis HKT1 gene homolog mediates inward Na+ currents in Xenopus laevis oocytes and Na+ uptake in Saccharomyces cerevisiae. Plant Physiol. 4 1249–1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Den Berg, C., Weisbeek, P., and Scheres, B. (1998). Cell fate and cell differentiation status in the Arabidopsis root. Planta 205 483–491. [DOI] [PubMed] [Google Scholar]
- Velten, J., and Schell, J. (1985). Selection-expression plasmid vectors for use in genetic transformation of higher plants. Nucleic Acids Res. 13 6981–6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vernet, T., Dignard, D., and Thomas, D.Y. (1987). A family of yeast expression vectors containing the phage f1 intergenic region. Gene 52 225–233. [DOI] [PubMed] [Google Scholar]
- Wada, T., Tachibana, T., Shimura, Y., and Okada, K. (1997). Epidermal cell differentiation in Arabidopsis determined by a Myb homolog. CPC Sci. 277 1113–1116. [DOI] [PubMed] [Google Scholar]
- Wymer, C.L., Bibikova, T.N., and Gilroy, S. (1997). Cytoplasmic free calcium distributions during the development of root hairs of Arabidopsis thaliana. Plant J. 12 427–439. [DOI] [PubMed] [Google Scholar]