Abstract
At least six rust resistance specificities (P and P1 to P5) map to the complex P locus in flax. The P2 resistance gene was identified by transposon tagging and transgenic expression. P2 is a member of a small multigene family and encodes a protein with nucleotide binding site (NBS) and leucine-rich repeat (LRR) domains and an N-terminal Toll/interleukin-1 receptor (TIR) homology domain, as well as a C-terminal non-LRR (CNL) domain of ∼150 amino acids. A related CNL domain was detected in almost half of the predicted Arabidopsis TIR-NBS-LRR sequences, including the RPS4 and RPP1 resistance proteins, and in the tobacco N protein, but not in the flax L and M proteins. Presence or absence of this domain defines two subclasses of TIR-NBS-LRR resistance genes. Truncations of the P2 CNL domain cause loss of function, and evidence for diversifying selection was detected in this domain, suggesting a possible role in specificity determination. A spontaneous rust-susceptible mutant of P2 contained a G→E amino acid substitution in the GLPL motif, which is conserved in the NBS domains of plant resistance proteins and the animal cell death control proteins APAF-1 and CED4, providing direct evidence for the importance of this motif in resistance gene function. A P2 homologous gene isolated from a flax line expressing the P resistance specificity encodes a protein with only 10 amino acid differences from the P2 protein. Chimeric gene constructs indicate that just six of these amino acid changes, all located within the predicted β-strand/β-turn motif of four LRR units, are sufficient to alter P2 to the P specificity.
INTRODUCTION
Plants contain a large number of genes that provide resistance to pests and pathogens. Many of these are characterized by a “gene-for-gene” interaction in which each resistance gene provides resistance to only those pathogen strains carrying a corresponding avirulence gene. This relationship was first elucidated in the flax–flax rust system (Flor, 1971), but it has been observed in many other disease systems. A simple model to explain the gene-for-gene observation is that resistance genes encode receptors that recognize the direct or indirect products of pathogen avirulence genes (van der Biezen and Jones, 1998a). Specific recognition of invading pathogens results in the subsequent triggering of an arsenal of defense responses.
The majority of resistance genes cloned to date encode proteins classified as NBS-LRR proteins because they contain a nucleotide binding site (NBS) domain and a leucine-rich repeat (LRR) domain (Ellis and Jones, 1998). Different genes in this group confer host resistance to viruses, bacteria, oomycetes, fungi, nematodes, and sucking insects. This class of resistance proteins can be further divided into two subgroups. The first group consists of proteins that have a distinct N-terminal region (TIR domain) that resembles the cytoplasmic domains of the Drosophila protein Toll and the mammalian interleukin-1 receptor protein. Proteins in the second group lack this region but may have a leucine zipper or coiled-coil domain in the N-terminal region (Pan et al., 2000).
Most variation between resistance genes and their closely related homologs occurs within the LRR, particularly within the xxLxLxx (where L indicates a conserved leucine or other aliphatic residue and x represents any amino acid) motif of the repeat units, and evidence suggests that this region has been subject to diversifying selection (Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Ellis et al., 1999; Noël et al., 1999; Dodds et al., 2000). On the basis of comparisons with the human and porcine ribonuclease inhibitor proteins (Kobe and Diesenhofer, 1994; Papageorgiou et al., 1997), the xxLxLxx motif is predicted to form a β-strand/β-turn structure in which the x residues are exposed to solvent and are available for interactions with potential ligands (Jones and Jones, 1997). These observations suggest a model of resistance protein function in which NBS and TIR domains have a conserved signaling function and LRR domains are involved in pathogen recognition. Because LRR domains are generally involved in protein–protein interactions (Kobe and Diesenhofer, 1995), they may interact with avirulence proteins either directly or through binding to protein complexes formed between avirulence proteins and other host proteins.
Direct evidence for the role of the LRR domain in resistance specificity has come from analysis of the L locus in flax. The L locus contains a single gene of the TIR-NBS-LRR class, and 11 alleles with different specificities of resistance to rust fungus isolates have been cloned (Lawrence et al., 1995; Ellis et al., 1999). The L6 and L11 proteins differ only in the LRR domain and recognize distinct avirulence products from the flax rust fungus. Furthermore, chimeric genes encoding the L2 LRR domain fused to the L6 or L10 TIR-NBS domain express the L2 specificity. However, the L6 and L7 proteins differ only in the TIR domain (Ellis et al., 1999), and some chimeric genes with exchanges in this region show altered specificities (Luck et al., 2000). Thus, the TIR domain can also influence specificity.
Five multiple-allele loci (K, L, M, N, and P) have been described in flax that confer gene-for-gene resistance to flax rust. In addition to the 11 cloned L alleles, we have isolated the M gene (Anderson et al., 1997). This gene is closely related to L but occurs in a complex locus with at least 15 related genes. These two genes probably represent homeologous loci, because flax is an ancient tetraploid. To isolate additional rust resistance genes in flax, we have extended the transposon tagging approach with the maize Ac element that was successful in isolating L6 and M (Lawrence et al., 1995; Anderson et al., 1997), and we now describe the isolation of the P2 rust resistance gene by Ac tagging. The P2 gene is part of a complex locus containing a small multigene family. P2 encodes a protein of the TIR-NBS-LRR class, but with an additional domain at the C terminus that is related to the C-terminal domain of other resistance proteins, including RPS4 (Gassmann et al., 1999); RPP1-WsA, RPP1-WsB, and RPP1-WsC (Botella et al., 1998) from Arabidopsis; and the N resistance protein from tobacco. P2 homologous genes were isolated from other flax lines expressing different P locus resistance specificities, and domain swap experiments between P2 and a gene from the P haplotype identified six amino acid differences that are sufficient to distinguish the P2 and P resistance specificities. These differences all occur in the β-strand/β-turn motif of the LRR domain and thus provide direct evidence of the role of this motif in determining the specificity of resistance genes.
RESULTS
Isolation of the Ac-Tagged P2 Mutant Allele p2-X157
We targeted the P2 rust resistance gene of flax in a transposon tagging experiment with the maize Ac element. A line of flax homozygous for P2 (Forge) and containing multiple copies of Ac, including a cluster of three Ac elements linked to P2, was crossed to the susceptible cultivar Hoshangabad. Among 38,400 progeny from this cross, we identified several plants that were susceptible to the rust strain CH5F2(133), which is avirulent to P2. One of these plants, X157, contained a newly transposed Ac element as determined by gel blot analysis (data not shown). X157 was crossed to the cultivar Abyssinian (homozygous for P2), and one progeny plant containing the newly transposed Ac was testcrossed to Hoshangabad. Of 50 testcross progeny, 10 expressed P2 resistance and did not contain this Ac element, whereas the remaining 40 susceptible plants all contained the Ac element. This segregation result indicated that the Ac element was closely linked to the P2 mutant allele p2-X157, and although it does not fit the expected 1:1 ratio, it is consistent with the disturbed segregation observed previously for P2 (G.J. Lawrence, unpublished data).
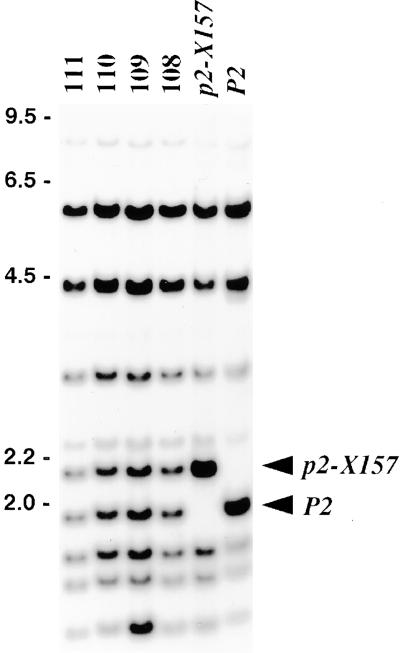
Because p2-X157 was potentially an Ac-tagged P2 gene, we used polymerase chain reaction (PCR)–based DNA walking (Siebert et al., 1995) to isolate a DNA fragment spanning the 3′ end of Ac and including 1.4 kb of flax genomic DNA. This 3′ Ac-flanking DNA hybridized to an altered restriction fragment in plants containing p2-X157 (Figure 1), which was the same size as the linked Ac fragment (data not shown). A restriction fragment length polymorphism (RFLP) marker detected by the Ac-flanking DNA probe cosegregated with the wild-type P2 among 116 testcross progeny, confirming its linkage to the P locus. To further test whether the p2-X157 mutation was due to Ac insertion, we analyzed the reversion of this allele to wild-type resistance. Five lines homozygous for p2-X157 were grown in isolation to prevent cross-pollination, and 2017 selfed progeny were tested for P2 rust resistance. Four individuals were identified that had reverted to the resistant phenotype. Gel blot analysis of these individuals showed that reversion of p2-X157 to wild-type P2 was associated with the excision of Ac from the P2-linked site (Figure 1). Together, these data provide strong evidence that the p2-X157 allele is an Ac insertion mutant of P2.
Figure 1.
Gel Blot Analysis of p2-X157 and Its Revertants.
Hybridization of a DNA probe from the NBS-encoding region of P2 to a gel blot filter of HindIII-digested genomic DNA from lines homozygous for P2 or the p2-X157 mutant allele and four resistant revertant lines (108 to 111) isolated among selfed progeny of p2-X157 homozygotes. Bands corresponding to the wild-type (P2) and mutant (p2-X157) genes are indicated by arrowheads. The four revertant lines are heterozygous for the P2 and p2-X157 fragments, indicating excision of the Ac element from one copy of the p2-X157 allele. The numbers at left indicate the size of DNA markers in kilobases.
The P2 Locus Contains Multiple Genes of the TIR-NBS-LRR Class
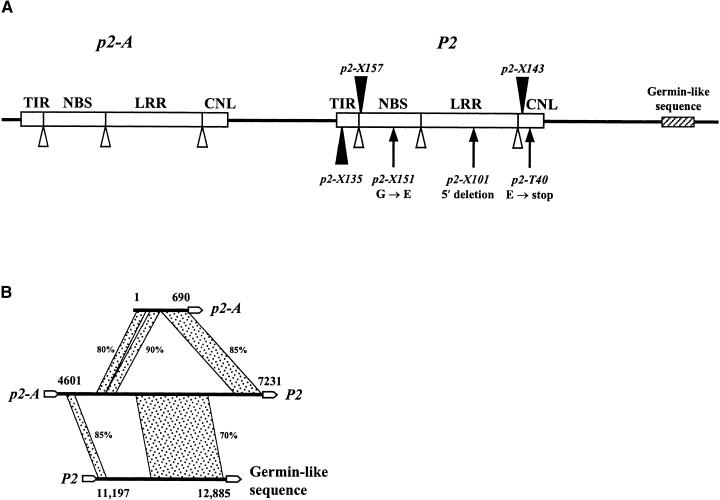
The DNA sequence flanking the 3′ end of Ac in p2-X157 encoded an amino acid sequence related to the NBS domain of plant resistance proteins, and we used this probe to screen a genomic lambda library constructed from a flax line carrying the wild-type P2 gene. Lambda clones containing sequences identical to those of the DNA probe were identified, and a 14-kb region surrounding the Ac insertion site was sequenced. The Ac insertion occurred within a gene predicted to encode a protein with TIR, NBS, and LRR domains characteristic of many resistance proteins. PCR amplification of the 5′ Ac junction using primers directed to the cloned genomic sequence confirmed the site of Ac insertion in p2-X157, and as expected for Ac transposition, an 8-bp target site duplication was present. We also amplified and sequenced the region flanking the Ac insertion site in the four revertant alleles derived from p2-X157 (Figure 1), and all four were identical to the wild-type P2 sequence. In addition to the putative P2 gene, two other potential genes were identified in the 14-kb sequenced region (Figure 2A). One of these, p2-A, is a closely related (95% DNA identity) homolog of P2 and is predicted to encode a protein with only single amino acid differences and small deletions/insertions relative to P2. It is likely that several additional homologs occur at this locus because probes derived from P2 hybridized to at least nine related sequences in the flax genome (Figure 1). RFLP mapping revealed at least two fragments, not derived from P2 or p2-A, that also cosegregated with P2 (data not shown). Only one RFLP was observed that segregated independently of P2. Thus, there may be between four and eight related sequences linked to P2. In addition to P2 and p2-A, a third predicted gene in this region (Figure 2A) encodes a protein closely related to the germin family. Although initially characterized by a germination-related protein in wheat (Lane et al., 1993), members of this family have been shown to be induced by pathogen infection (Zhou et al., 1998) and to provide effective resistance to powdery mildew infection when transiently expressed in wheat (Schweizer et al., 1999). Some but not all of these proteins possess a hydrogen peroxide–generating oxalate oxidase activity. Despite the high level of sequence similarity between the P2 and p2-A coding regions, there was little conservation in the intergenic sequences, with several large deletions/insertions apparent (Figure 2B).
Figure 2.
Features of the Flax Genomic Sequence Containing the P2 Rust Resistance Gene.
(A) Scheme of a 14-kb genomic DNA fragment containing the P2 gene. Open boxes represent the coding regions of P2 and an adjacent related gene, p2-A, with intron positions (predicted for p2-A by comparison with P2) marked by open triangles. The relative positions of the TIR, NBS, LRR, and CNL domains are indicated. The insertion sites of the Ac elements in p2-X157 and p2-X143 are shown by closed triangles, as is the site of the native flax transposable element insertion in p2-X135. The sites of the point mutations in p2-X151 and p2-T40 and the 3′ deletion break point in p2-X101 are also indicated. An open reading frame encoding a germin-like protein is indicated by the hatched box.
(B) Scheme of the intergenic sequences of the P2 region. The start and end of the open reading frames are indicated by the open arrows and are numbered according to their nucleotide positions within the 14-kb sequenced region. Regions of conserved sequence are connected by shaded boxes, with the percentage of sequence identity indicated. No significant sequence similarity was observed between region 1 to 690 bp and region 11,197 to 12,885 bp.
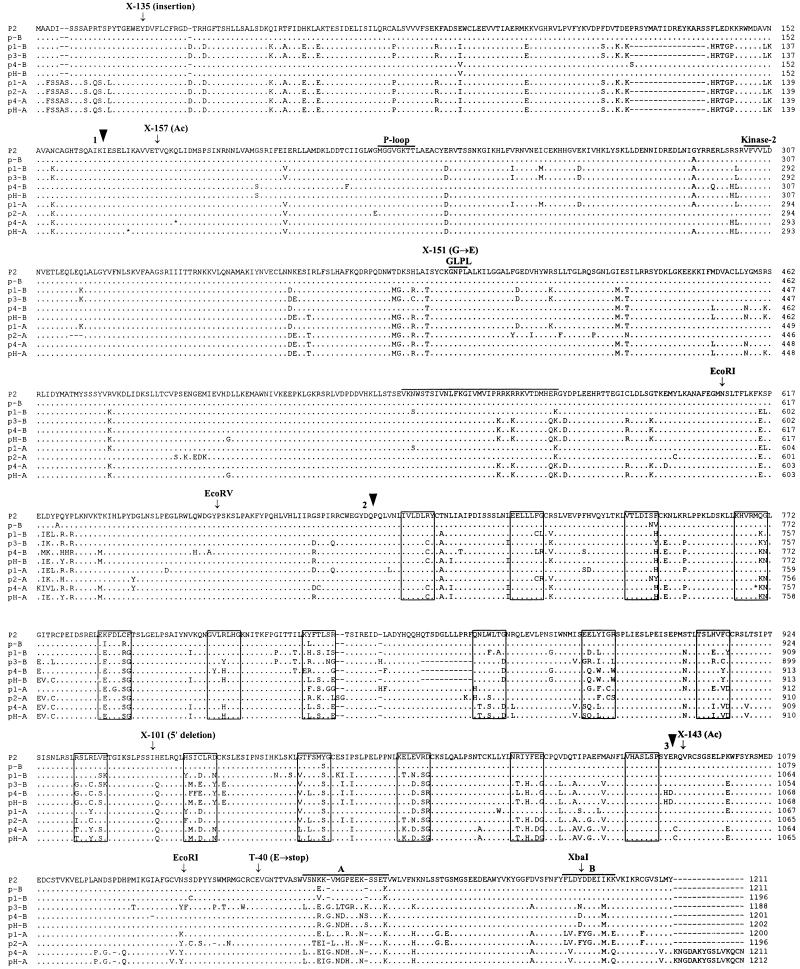
The L6 and M rust resistance genes of flax each contain three introns (Lawrence et al., 1995; Anderson et al., 1997), and the positions of these introns are conserved in the other cloned TIR-NBS-LRR resistance genes N (Whitham et al., 1994), RPP5 (Parker et al., 1997), RPP1 (Botella et al., 1998), and RPS4 (Gassmann et al., 1999). Analysis of the P2 genomic sequence revealed potential intron splice junctions at positions corresponding to these three sites. Reverse transcription (RT)–PCR with primers spanning the intron 1 site confirmed the correct splicing of this sequence. However, RT-PCR with a primer pair spanning the predicted intron 2 and 3 sites gave products in which the third predicted intron was spliced but the sequence corresponding to intron 2 was not. This 99-bp sequence encodes an uninterrupted reading frame continuous with the rest of the P2 coding sequence, and its retention in P2 transcripts results in the insertion of a 33–amino acid peptide. This peptide is highly hydrophilic (10 positively charged and two negatively charged residues) but is not related to other resistance gene sequences, and BLAST searches (Altschul et al., 1997) failed to detect any matching sequences. An additional intron was spliced from P2 transcripts and occurred after the LRR-encoding region. The last exon of P2 encodes a 153–amino acid sequence that we have designated the C-terminal non-LRR (CNL) domain. The predicted 1211–amino acid protein product of the P2 gene after removal of the three confirmed introns is shown in Figure 3. The P2 protein is most closely related to the RPP1-WsA, RPP1-WsB, and RPP1-WsC (Botella et al., 1998) and RPS4 (Gassmann et al., 1999) resistance proteins from Arabidopsis (45% amino acid identity in TIR-NBS domains) but is only distantly related to the L and M rust resistance proteins of flax (30% amino acid identity in TIR-NBS) (Lawrence et al., 1995; Anderson et al., 1997).
Figure 3.
Predicted Amino Acid Sequences of P2 and Its Homologs.
The complete amino acid sequence of P2 is shown in one-letter code. The sites of three introns in the corresponding genomic DNA sequence are indicated by numbered arrowheads. The sites of Ac and flax transposon insertion in the p2-X135, p2-X143, and p2-X157 mutant alleles are indicated, as are the 3′ deletion break point site in p2-X101, the altered amino acid in p2-X151, and the stop codon introduced in p2-T40. The P-loop, kinase-2, and GLPL motifs of the NBS are labeled, and the xxLxLxx motifs of the LRR domain are boxed. A 33–amino acid region encoded by a DNA sequence corresponding to the position of the second intron in L6 and M is overlined. The predicted amino acid sequences of p2-A and of P2/p2-A homologs from the P, P1, P3, and P4 resistance haplotypes and the susceptible variety Hoshangabad (pH-A and pH-B) are also shown. Only amino acid differences from P2 are shown, with identical residues indicated by dots. The p-A and p3-A sequences are not shown because they are identical to the p2-A sequence except for one unique frameshift mutation each at codons 880 and 1116, respectively. Two frameshift mutations occur in the p4-A gene and one in pH-A (asterisks), and the amino acid sequences shown are continued as translated from the shifted reading frame. The sites of EcoRI, EcoRV, and XbaI sites used to construct chimeric P2/p-B genes are also indicated. Two hypervariable regions in the CNL domain are labeled A and B.
Sequence Analysis of P2 Mutant Alleles
To confirm the identity of the putative P2 gene, we examined six independent p2 mutants that were isolated in a separate screen from p2-X157. This screen analyzed 280,000 Forge × Hoshangabad progeny in which the Forge parent did not contain the three P2-linked Ac elements. These six mutations did not appear to result from Ac insertion, because we did not detect newly transposed Ac elements using a 3′ Ac probe. DNA gel blot and sequence analysis revealed that all six of these mutants contained alterations in the P2 gene (Figures 2A and 3). The p2-X46 and p2-X101 mutants contained large deletions of either the entire 14-kb sequenced region (p2-X46) or the 5′ half of P2 and all of p2-A (p2-X101). The p2-X135 allele contained a 3.5-kb insertion of a native flax transposon-like sequence related to the Ac/Ds family (data not shown) and with end sequences similar to those present on dLute, a 314-bp nonautonomous transposable element from flax (Luck et al., 1998). The p2-X143 allele contained an Ac insertion that went undetected earlier because it gives rise to a 3′ Ac restriction fragment of similar size to a fragment present in the parental line. Two other mutants contained single nucleotide substitutions. In p2-T40, a G→T substitution introduces a stop at codon 1126. A G→A nucleotide substitution in p2-X151 causes a glycine-to-glutamate change (395G→E). This change occurs in the conserved GLPL motif (GNPL in the P2 protein), which is common to NBS-containing resistance proteins and also to the animal cell death proteins APAF1 and CED4 (van der Biezen and Jones, 1998b). This mutation demonstrates that this motif is important in resistance protein function.
Transgenic Expression of the P2 Gene Confers P2-Specific Rust Resistance
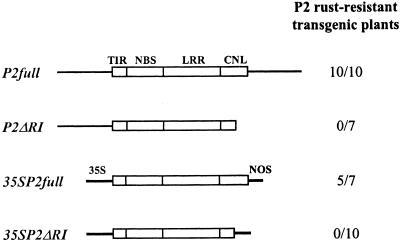
To test whether the cloned P2 gene was sufficient to confer P2 resistance, we introduced this gene into the flax line Ward, which is susceptible to rust strains recognized by P2 (Table 1). We used two transformation constructs (Figure 4): an 8-kb genomic fragment (P2full) with a 1.8-kb upstream promoter region and a 2-kb downstream sequence, and a construct containing the P2 coding sequence (including introns) driven by the cauliflower mosaic virus 35S promoter with the nopaline synthase (NOS) terminator (35SP2full). Transgenic plants were tested for resistance to the rust strain CH5F2(83), which is avirulent to P2 but virulent to Ward. Of 10 plants containing the P2full construct, all 10 expressed resistance to CH5F2(83). Similarly, five of 10 plants containing 35SP2full expressed resistance to this rust. To confirm that the resistance specificity expressed by these plants corresponded to P2, we tested two transgenic lines containing P2full with six rusts of diverse origin that together represent all of the original and unrelated rust strains available in this laboratory. In each case, the reaction of the transgenic plants was identical to that of the flax differential line Abyssinian, the source of the P2 gene (Table 1). This finding indicates that the cloned gene is sufficient to account for all of the rust resistance reactions associated with P2.
Table 1.
Rust Reactions on the P2 and P Flax Differential Lines and Transgenic Lines
| Rust Straina
|
||||||
|---|---|---|---|---|---|---|
| Flax Line | C | Fi | I | H | Bs1 | J |
| Abyssinian (P2) | + | + | + | − | − | − |
| Koto (P) | − | − | + | + | + | − |
| Ward | + | + | + | + | + | +/− |
| Ward::P2full | + | + | + | − | − | − |
| Ward::P2/P-RI | − | − | + | + | + | − |
(+), unrestricted growth of the rust; (−), complete resistance; (+/−), intermediate reaction.
Figure 4.
P2 Transformation Constructs.
Scheme showing transformation constructs containing the full-length genomic P2 gene (P2full), the P2 coding region with the 35S promoter and NOS terminator sequences (35SP2full), and the C-terminal truncated versions of these constructs (P2ΔRI and 35SP2ΔRI, respectively). The coding sequence is indicated by the open box, with intron positions marked by vertical lines; nontranslated sequences are shown as solid lines. The number of transformants expressing P2 resistance out of the total number of transformed plants is indicated to the right of each construct. The relative positions of the TIR, NBS, LRR, and CNL domains are shown.
The P2 Protein Contains a C-Terminal Non-LRR Domain That Is Conserved in a Subclass of TIR-NBS-LRR Resistance Proteins and Is Required for P2 Resistance
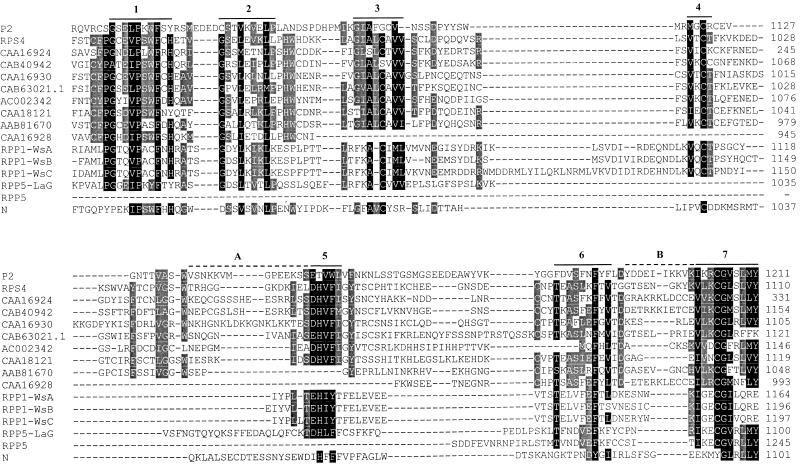
A PSI-BLAST search (Altschul et al., 1997) with the P2 CNL-domain amino acid sequence found several predicted TIR-NBS-LRR proteins with similar C-terminal domains. These included the Pseudomonas syringae resistance protein RPS4 (Gassmann et al., 1999) and eight predicted proteins from the Arabidopsis genome. Alignment of these sequences revealed seven short conserved regions separated by more variable sequences (Figure 5). A second PSI-BLAST search using the RPS4 CNL domain as a query sequence identified 63 predicted TIR-NBS-LRR proteins from the Arabidopsis genome that contained similar sequence motifs in a C-terminal domain. In comparison, a PSI-BLAST search with the RPS4 TIR domain identified 141 predicted TIR-NBS-LRR proteins from Arabidopsis (data not shown), suggesting that almost half (45%) of this class of genes in Arabidopsis encode a CNL domain related to P2/RPS4. These include the Peronospora resistance genes RPP1-WsA, RPP1-WsB, and RPP1-WsC (Botella et al., 1998), which encode CNL domains of 160 to 180 amino acids, including sequences related to all of the motifs conserved between P2 and RPS4 (Figure 5). In addition, several genes from the RPP5 complex locus (Noël et al., 1999) encode related protein sequences. For instance, RPP5-LaG encodes an amino acid sequence containing six of the conserved CNL motifs, although RPP5 itself contains a deletion of most of this region and only encodes the final two motifs (Figure 5). The N gene for tobacco mosaic virus resistance in tobacco (Whitham et al., 1994) was also detected in this database search. The N protein contains a CNL domain of 216 amino acids with homology to the first half of the P2/RPS4 CNL domain, but it has little similarity to the rest of this region (Figure 5). Each of the protein sequences shown in Figure 5 extends between 24 and 170 amino acids farther than P2, but there were no conserved motifs evident beyond the region shown in Figure 5.
Figure 5.
The P2 CNL Domain Is Shared by Several TIR-NBS-LRR Resistance Proteins.
An alignment is shown of the P2 CNL domain with similar sequences from the C termini of the RPS4 resistance protein of Arabidopsis (Gassmann et al., 1999); several putative TIR-NBS-LRR proteins predicted from the Arabidopsis genome sequence (labeled with their GenBank accession numbers); RPP1-WsA, RPP1-WsB, and RPP1-WsC (Botella et al., 1998); RPP5 and RPP5-LaG (Noël et al., 1999); and the N resistance protein from tobacco (Whitham et al., 1994). Positions with identical or conservatively exchanged amino acids in at least 12 of the 16 sequences are shown in black, whereas sites conserved in at least eight sequences are shaded gray. Seven conserved regions are overlined and numbered, and the hypervariable regions A and B of P2 are indicated by dashed lines. The alignment was initially constructed using the Pileup program of GCG and then optimized manually.
To address the requirement for the CNL domain in P2 resistance, we transformed Ward plants with constructs truncated at an EcoRI site within the last exon (Figure 3), resulting in loss of the last 104 amino acids of the protein. The P2ΔRI construct was derived from the P2full construct and thus contained a 1.8-kb genomic promoter sequence but no 3′ transcription termination sequence. The 35SP2ΔRI construct contained the 35S promoter and NOS terminator sequences (Figure 3). Of seven transgenic plants containing P2ΔRI, all were susceptible to CH5F2(83), and all 10 35SP2ΔRI transformants were also susceptible (Figure 4). RT-PCR analysis of three 35SP2ΔRI transgenic lines, using primers specific to the introduced construct, showed that each contained mRNA derived from the truncated P2 gene. Thus, the lack of resistance was not due to lack of expression of the transgene but was likely the result of the truncation of the protein product. The p2-X143 and p2-T40 mutant alleles provide further evidence for the importance of the CNL domain (Figures 2A and 3). In p2-X143, an Ac element is inserted near the beginning of exon 4 and leads to a truncation of most of this domain. In p2-T40, a single base change introduces a stop codon at position 1126 and leads to the loss of the last 85 amino acids from the predicted protein.
P Locus Genes from the P, P1, P3, and P4 Resistance Haplotypes
At least six rust resistance specificities occur at the P locus (Islam and Mayo, 1990). We examined flax lines expressing the P, P1, P3, and P4 resistance phenotypes as well as the Hoshangabad line (which expresses no detectable resistance) by gel blot hybridization with P2-derived probes. Each line contained a similarly complex pattern of hybridizing fragments, with eight or nine homologous sequences, and analysis of backcrossed lines showed that at least some of these fragments are linked to the P locus in each haplotype. We used long-range PCR to isolate sequences corresponding to P2 and p2-A from these flax lines. PCR reactions used a 5′ primer specific to either P2 or p2-A together with a 3′ primer common to both genes. In each case, a single PCR product was observed by gel electrophoresis, and direct sequencing of this product yielded a uniform sequence, indicating that only one gene with the corresponding flanking primer sequences was present in each haplotype. Those sequences amplified with the p2-A–specific primer were designated p-A, p1-A, p3-A, p4-A, and pH-A, whereas those amplified with the P2-specific primer were designated p-B, p1-B, and so forth. Three of the -A class genes (p-A, p3-A, pH-A) contained a single frameshift mutation (different in each case) leading to premature termination of the predicted translation products, whereas p4-A contained two frameshifts. All of the -B class genes encoded potentially full-length proteins. The predicted amino acid sequences encoded by these genes are shown in Figure 3 aligned with the P2 protein sequence. The protein sequences were highly similar (>90% amino acid identity) but showed most variation in the LRR domain. As observed for alleles of the flax L rust resistance gene and other resistance genes, this variation was clustered around the xxLxLxx β-strand/β-turn motif of the LRR (boxed in Figure 3). In addition, we observed two hypervariable regions within the CNL domain, regions A and B (Figure 3). These regions are not conserved between the related Arabidopsis proteins (Figure 5).
Evidence for Diversifying Selection Acting on the CNL Domain and on the xxLxLxx Motif of P2 Homologs
We performed an analysis of synonymous and nonsynonymous nucleotide substitution rates (Ks and Ka, respectively) among the P2 homologs (Table 2). No evidence of diversifying selection (as revealed by higher Ka than Ks values) acting on the TIR or NBS regions was found; in fact, Ka was significantly less than Ks in the NBS domain (in a t test comparison of average Ka and Ks values, 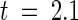 , P < 5%; Kumar et al., 1993). Although average Ka was greater than Ks for comparisons between sequences encoding the entire LRR domain, this difference was significant only at the 10% level. However, when comparisons were confined to the sequences encoding the variant positions in the xxLxLxx motif, average Ka was significantly greater than Ks (
, P < 5%; Kumar et al., 1993). Although average Ka was greater than Ks for comparisons between sequences encoding the entire LRR domain, this difference was significant only at the 10% level. However, when comparisons were confined to the sequences encoding the variant positions in the xxLxLxx motif, average Ka was significantly greater than Ks (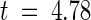 , P < 0.01%). Of the 55 individual pairwise comparisons for this region, 33 showed a statistically significant excess of nonsynonymous substitutions at the 5% probability level and seven showed a statistically significant excess at the 0.5% level (2 × 2 contingency χ2 test). These results indicate that diversifying selection has acted on this region during the evolution of P locus genes. Interestingly, many pairwise comparisons between CNL domains also showed Ka > Ks, and eight of these differences were significant at P < 5%. However, all comparisons involving this region of p4-A or pH-A (which are identical) showed equal rates of synonymous and nonsynonymous substitution, which may indicate neutral selection on these two nonfunctional genes, even though diversifying selection was detected in the LRR domains of these genes. If these two genes are excluded from the overall comparison, average Ka is significantly greater than Ks (
, P < 0.01%). Of the 55 individual pairwise comparisons for this region, 33 showed a statistically significant excess of nonsynonymous substitutions at the 5% probability level and seven showed a statistically significant excess at the 0.5% level (2 × 2 contingency χ2 test). These results indicate that diversifying selection has acted on this region during the evolution of P locus genes. Interestingly, many pairwise comparisons between CNL domains also showed Ka > Ks, and eight of these differences were significant at P < 5%. However, all comparisons involving this region of p4-A or pH-A (which are identical) showed equal rates of synonymous and nonsynonymous substitution, which may indicate neutral selection on these two nonfunctional genes, even though diversifying selection was detected in the LRR domains of these genes. If these two genes are excluded from the overall comparison, average Ka is significantly greater than Ks (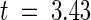 , P < 0.05%) in this domain. We also analyzed two 25-codon windows surrounding the hypervariable regions A and B of the CNL domain (Figure 3 and Table 2). In region A, there was a high rate of nonsynonymous substitutions but no synonymous substitutions. This difference was highly significant (
, P < 0.05%) in this domain. We also analyzed two 25-codon windows surrounding the hypervariable regions A and B of the CNL domain (Figure 3 and Table 2). In region A, there was a high rate of nonsynonymous substitutions but no synonymous substitutions. This difference was highly significant (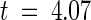 , P < 0.01%). However, in region B, no significant difference was observed between Ka and Ks.
, P < 0.01%). However, in region B, no significant difference was observed between Ka and Ks.
Table 2.
Average Number of Nucleotide Substitutions per 100 Nonsynonymous (Ka) or Synonymous (Ks) Sites between P Locus Genesa
| Region | Ka | Ks |
|---|---|---|
| Whole gene | 4.3 (0.2) | 4.8 (0.5) |
| Exon 1/TIR | 4.4 (0.8) | 7.9 (2.1) |
| Exon 2/NBS | 2.4 (0.3) | 4.0 (0.7)b |
| Exon 3/LRR | 7.0 (0.5) | 5.4 (0.9) |
| xxLxLxx | 22.6 (2.2) | 6.9 (3.4)c |
| Exon 4/CNL | 5.4 (0.8) | 3.5 (1.2) |
| Minus p4-A/pH-A | 5.1 (0.8) | 1.3 (0.8)d |
| Region A | 11.9 (2.9) | 0.0 (0.0)c |
| Region B | 8.1 (2.6) | 7.6 (5.5) |
Standard errors of average Ka and Ks values are shown in parentheses.
Ks > Ka significant at P < 5%.
Ka > Ks significant at P < 0.01%.
Ka > Ks significant at P < 0.05%.
Sequence Exchange Has Occurred between Duplicated Genes at the P Locus
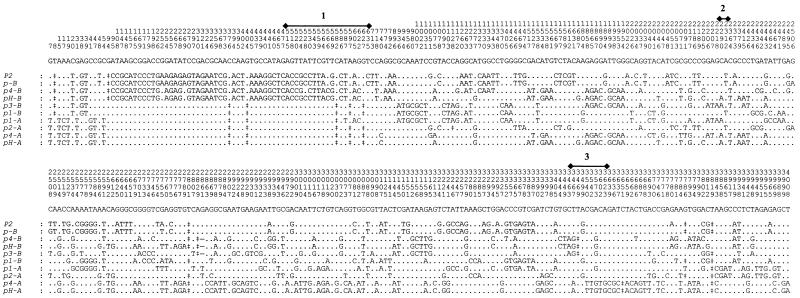
Sequence exchange between duplicated genes (paralogs) in complex resistance loci has been suggested to be a source of new variation and to contribute to the evolution of novel resistance specificities (Pryor and Ellis, 1993; Parniske et al., 1997). Therefore, we examined the evidence for past sequence exchanges between members of the P locus gene family. Figure 6 shows the informative polymorphic sites in an alignment of the P locus gene sequences. It is apparent from this alignment that multiple sequence exchange events have occurred during the evolution of these genes. Statistical analysis of these data with the Geneconv program (Sawyer, 1989) detected strong evidence for sequence exchanges between these genes (global simulated P < 0.01%). In comparisons between individual pairs of sequences, 38 regions were identified in which two sequences shared a long run of identical polymorphic sites flanked by nonconserved polymorphisms (significant at P < 1% for both simulated and Bonferroni-corrected probability values). Of the 38 highly significant runs of matching polymorphisms, 19 occurred between genes of the -A and -B classes. For instance, pH-A and pH-B are identical in a 2032-nucleotide region between positions 793 and 2824 relative to the start codon, but they have 152 nucleotide differences outside this region (P < 0.001%). Thus, sequence exchange events at the P locus have occurred between both orthologous (allelic) and paralogous sequences.
Figure 6.
Alignment of Informative Polymorphic Sites in the DNA Sequences of P2 Homologs.
The nucleotides present at each informative polymorphic site of 10 sequenced P2 homologs from flax are shown. The p-A and p3-A sequences are not shown because these are identical to the p2-A sequence with the exception of two unique and thus noninformative differences. The consensus nucleotide at each site is shown above the gene sequences, and the nucleotide positions relative to the start codon are indicated above the consensus line. The intron positions are indicated by the numbered lines. Deletions of one or more nucleotides are indicated by double daggers and numbered as the first missing nucleotide.
Six Amino Acid Changes Convert P2 to P Specificity
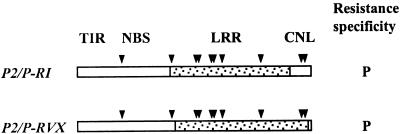
One striking observation from the comparison of P locus sequences is that the p-B gene from the P haplotype is almost identical to P2. Only 10 amino acid differences distinguish the predicted P2 and p-B proteins: one in the NBS domain, one in the region between the NBS and LRR domains, six in the LRR domain, and two in the CNL domain (Figure 3). This raised the question Does p-B express the P specificity and, if so, which of the 10 amino acid differences from P2 confer this change in specificity? To address this question, we constructed two chimeric genes (Figure 7) derived from the full-length P2 genomic construct P2full (Figure 4) by insertion of DNA fragments derived from p-B. P2/P-RI encoded a chimeric protein with seven amino acid changes from P2 (one pre-LRR and six in LRR) between two EcoRI sites in the genomic DNA sequence (Figure 3), whereas P2/P-RVX encoded eight p-B–derived amino acid changes (six in LRR and two in CNL) between an EcoRV and an XbaI site (Figure 3). These constructs were transformed into the flax line Ward and tested with rust strains to distinguish the P2 and P resistance phenotypes. Of 13 transgenic lines containing the P2/P-RI chimeric gene, 12 showed resistance to rust strain CH5F2(84), which is avirulent to P but virulent to P2 and to the untransformed Ward plants. However, these plants were all fully susceptible to rust strain CH5F2(106), which is virulent to P but avirulent to P2. This result is consistent with the chimeric gene expressing the P resistance specificity but not the P2 specificity. We further tested two transgenic lines with the six independent rust isolates described above. In each case, the resistance reaction was identical to that observed on the flax differential line Koto, which was the source of the P specificity (Table 1). Thus, the P2/P-RI construct expresses a resistance specificity indistinguishable from the natural P resistance and is sufficient to account for the full spectrum of resistance associated with P. Six transgenic lines containing P2/P-RVX also showed resistance to the rust strain CH5F2(84) but not to CH5F2(106), indicating that this construct also expresses P but not P2 resistance specificity. Importantly, the P and P2 specificities are clearly distinguished, with some rust strains virulent to P and avirulent to P2 and others avirulent to P and virulent to P2 (Table 1), indicating that different avirulence products are recognized by these genes. Six amino acid changes are common to the two chimeric constructs, and all occur within the xxLxLxx β-strand/β-turn motifs of four individual LRR repeat units (Figure 3).
Figure 7.
Chimeric P2/p-B Transformation Constructs.
Scheme showing chimeric transformation constructs P2/P-RI and P2/P-RVX. The unshaded region represents the P2-derived sequence, and the shaded region shows sequence derived from p-B. The positions of the TIR, NBS, LRR, and CNL domains are indicated above the diagram, and the sites of the 10 amino acid changes between the P2 and p-B proteins are shown by arrowheads. Rust resistance specificities conferred by each construct are shown to the right of the diagram.
DISCUSSION
At least six rust resistance specificities (P and P1 to P5) have been described that segregate as alleles of the P locus in flax (Islam and Mayo, 1990). Here, we describe the identification of a gene that confers the P2 specificity. This gene is part of a family of genes at the P locus that encode TIR-NBS-LRR class proteins. The P2 gene was initially identified by transposon tagging, and subsequently a number of independent mutants were shown to have alterations in the same gene (Figures 2 and 3). Finally, transformation of a susceptible flax variety confirmed that this gene was sufficient to confer resistance to all of the rust strains recognized by the P2 differential line Abyssinian (Figure 4 and Table 1). Because the P2 haplotype contains at least four related sequences, it is possible that other genes present in this haplotype may also confer rust resistance. Certainly, the cloned P2 gene is the only gene active against the A-P2 avirulence gene derived from rust strain H, because the p2 mutant alleles confer susceptibility to rusts containing this gene, such as CH5F2(133). However, we have not tested the mutant lines with other rusts recognized by P2 because of the presence of the L6, M, and N resistance genes in these mutants. The p2-A gene that we have sequenced from this haplotype contains no obvious mutations that would render it incapable of function, so we cannot rule out the possibility that it, or other unsequenced members of this complex, may also have some resistance activity. Significantly, the RPP1-WsA gene from Arabidopsis confers resistance to all of the downy mildew strains recognized by the Wassilewskija ecotype, but the RPP1-WsB and RPP1-WsC genes, which are part of this same haplotype, each provides resistance to a subset of these strains, possibly interacting with different avirulence determinants (Botella et al., 1998).
The Conserved GLPL Motif in the NB-ARC Domain Is Crucial for P2 Resistance
Plant resistance genes of the NBS-LRR class share some common features around the NBS domain with the animal apoptosis-regulating proteins APAF1 and CED4 (van der Biezen and Jones, 1998b). In addition to the P-loop, kinase 2, and kinase 3a motifs present in ATP/GTP binding proteins, several other shared motifs are present in these proteins, and these define the larger NB-ARC (for nucleotide binding APAF1–resistance protein CED4) domain. However, the functions of these additional motifs are not known. The most strongly conserved motif in this domain is characterized by the amino acid sequence GLPL (GNPL in the P2 protein). The G→E amino acid substitution in the p2-X151 mutant allele occurs at the conserved glycine residue of this motif and completely abolishes P2 resistance. This mutation provides direct evidence of the importance of this motif in the function of NB-ARC proteins. On a similar note, Botella et al. (1998) described one mutant of RPP1-WsA that contains an 11–amino acid deletion immediately before the GLPL motif. This region also shows some conservation among the NB-ARC family (van der Biezen and Jones, 1998b). To our knowledge, no other mutations in motifs specific to the NB-ARC family have been described.
Six Amino Acid Differences in the xxLxLxx Motif of the LRR Distinguish the P2 and P Specificities
The isolation of P2 homologous genes from differential lines containing the P, P1, P3, and P4 resistance specificities has identified candidates for these resistance genes. We examined whether one of these genes, p-B from the P haplotype, encoded protein sequences with the ability to recognize rust strains to which the P haplotype confers resistance. Interestingly, the candidate P resistance protein differs by only 10 amino acids from P2. By introducing certain of these amino acid changes into the P2 protein by in vitro sequence exchanges, we were able to generate chimeric genes that expressed a resistance specificity identical to that of the P differential line Koto (Table 1 and Figure 7). Thus, it is likely that the p-B gene is responsible for the resistance phenotype observed in the P differential line, although this has not been examined directly by flax transformation with p-B. Importantly, P2 and P are clearly distinguished by rust strains that are reciprocally virulent/avirulent to these specificities; that is, some rusts are avirulent to P2 and virulent to P, whereas others have the reverse reaction, virulent to P2 and avirulent to P (Table 1). This is a necessary condition to show that two resistance genes recognize different avirulence products. For example, although there are rust strains that are avirulent to L6 and virulent to L7, there are no rust strains with the reverse reaction. These resistance genes are distinguished by their interaction with a dominant inhibitor of resistance gene, I, in the rust, but they may recognize the same avirulence gene (Lawrence et al., 1981; Luck et al., 2000). The P2/p-B chimeric genes represent the fewest amino acid changes shown to abolish recognition of one avirulence product and allow recognition of a new avirulence product. This may reflect a close relationship between the A-P2 and A-P avirulence genes, which segregate as alleles of a single locus in crosses between the rust strains H and C (80 F2 lines and 32 testcross lines; Lawrence et al., 1981).
The two chimeric genes with P specificity encode seven and eight amino acid differences from P2, and six of these differences are common to both constructs. Thus, these six common amino acid changes are most likely sufficient to account for the difference in specificity between these proteins. These changes occur in the xxLxLxx motifs of four LRR units. In the canonical LRR proteins, the porcine and human ribonuclease inhibitors (PRI and hRI), these motifs adopt β-strand/β-turn structures, with the leucine residues buried in the protein and the intervening x residues exposed (Kobe and Diesenhofer, 1995; Papageorgiou et al., 1997). Juxtaposition of these structures from adjacent repeat units forms a solvent-exposed surface that interacts with the respective ligands of these proteins. In the complex between PRI and its ligand, ribonuclease A, 17 of 28 contact points involve amino acids in the xxLxLxx motif, whereas in a similar complex formed between hRI and the ribonuclease angiogenin, 19 amino acids within the xxLxLxx motif of hRI make strong contacts with angiogenin and six contact points occur outside this motif. The observation that diversifying selection acts on codons encoding these solvent-exposed residues in many resistance gene families supports the hypothesis that this region may be involved in determining the specificity of resistance proteins (Table 2) (Parniske et al., 1997; Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Noël et al., 1999; Bittner-Eddy et al., 2000; Dodds et al., 2000). Another class of plant proteins that contain LRRs is the polygalacturonase-inhibiting proteins (PGIPs). Recently, Leckie et al. (1999) reported that a single amino acid change in this region was sufficient to explain the different polygalacturonase binding specificity of the bean PGIP-1 and PGIP-2 proteins. Our results demonstrate that amino acid changes confined to the LRR β-strand/β-turn motif can determine specificity differences between plant resistance genes.
Interestingly, tests for allelism between P, P1, and P2 (Flor, 1965) detected no recombinant genotypes (double resistant or double susceptible) in testcross progeny derived from P/P1 heterozygotes (7354 progeny) or P/P2 heterozygotes (2306 progeny). In contrast, recombinant genotypes between the M and M3 genes of the complex M locus occurred at a frequency of 1 in 600 (Flor, 1965). Because P and P2 are distinguished by just six amino acid differences and may occur at allelic positions within the P locus cluster, the recombinant phenotype could arise only by recombination within a very small region or by mispairing and unequal recombination during meiosis. The lack of recombination between P and P1 suggests that these genes may also occur at allelic positions; thus, the p1-B gene is a strong candidate for encoding the P1 resistance specificity. An alternate explanation for the lack of recombination observed between P alleles is that there is a reduced rate of recombination within the P locus region compared with the M locus region.
Role of the CNL Domain in TIR-NBS-LRR Proteins
Although the P2 protein and its homologs are members of the TIR-NBS-LRR class, they contain an additional CNL domain. Mutations within this domain of P2 by transposon insertion, introduction of a stop codon, or truncation of a transgene product all resulted in loss of activity of the P2 resistance protein. Although the Ac insertion in p2-X143 and the 3′ truncation of the genomic P2ΔRI construct may result in instability of the transcripts expressed by these genes, this is unlikely to be the case for the p2-T40 mutant allele, which contains a single base change leading to a stop codon at amino acid position 1126. The 35S-driven truncated transgene 35SP2ΔRI contains a NOS 3′ terminator sequence, and RT-PCR analysis showed that transcripts derived from this construct were present in at least three of the transformed flax lines. Thus, we conclude that an intact CNL domain is required for P2 protein function.
The P2 CNL domain is most closely related to a similar domain in the RPS4 resistance protein, but many other TIR-NBS-LRR proteins also encode related domains. We identified 63 predicted TIR-NBS-LRR proteins from Arabidopsis that contain a related CNL domain, which represented 45% of the predicted proteins of this class represented in the protein databases. Thus, the presence or absence of this domain defines two subclasses of resistance proteins within the TIR-NBS-LRR group. Of the loci with known disease resistance function, the RPS4, RPP1, and RPP5 loci in Arabidopsis, the N locus in tobacco, and the P locus in flax encode proteins with related CNL domains (Figure 5). Proteins encoded by the L and M loci in flax do not contain CNL domains. The conservation of this region between a flax rust resistance protein and resistance proteins in Arabidopsis and tobacco is striking and suggests a conserved function for this domain in the resistance mediated by these proteins. We observed an increased ratio of nonsynonymous to synonymous nucleotide substitutions in the CNL domain of P locus genes and particularly within the hypervariable region A (Table 2 and Figure 3), which is indicative of the operation of diversifying selection. Thus, this region may have a role in specificity determination. However, the two amino acid differences between P2 and p-B in region A were not important for determining the difference in resistance specificity between these two genes. In both the RPP5 and RPP1 gene families, hypervariable sites occur within the CNL domains (Botella et al., 1998; Noël et al., 1999), but these regions have not been examined for the action of diversifying selection.
Interestingly, although some paralogs at the complex RPP5 locus in Arabidopsis (La-B, La-C, La-G, La-H, La-I, Col-A, Col-B, Col-D, Col-E, Col-F, and Col–G; Noël et al., 1999) contain sequences similar to the full CNL domain, others, including RPP5 itself, contain deletions of most of this region. This suggests that variation among members of a resistance gene family can encompass the presence/absence of CNL domains and that sequence exchange among these genes could allow interchange of these regions. To our knowledge, none of the non-TIR class of NBS-LRR resistance genes cloned to date encodes more than a few amino acid residues (<20) after the final LRR unit, although some, such as Prf (Salmeron et al., 1996) and Mi (Milligan et al., 1998) in tomato, do encode large domains at the N terminus.
P Locus Gene Evolution
We detected strong evidence for past sequence exchange events between genes at the P locus by using a statistical test for alternating long stretches of matching polymorphic sites between individual sequences. This analysis suggested that sequence exchange has occurred between paralogous genes at the complex P locus. In the absence of such exchanges, the -A and -B gene classes would represent two different allelic groups with independent lineages. This is clearly not the case because significant stretches of sequence identity were observed between members of the -A and -B classes of P locus genes (Figure 6) as well as between individual sequences from the same haplotype (e.g., pH-A and pH-B). These sequence exchange events could result from unequal recombination (or gene conversion) between mispaired duplicated sequences during meiosis. These results suggest that sequence exchange between paralogs at the P locus has been relatively frequent and has made a significant contribution to the generation of sequence diversity. In their model of the evolution of complex resistance loci by a “birth and death” process, Michelmore and Meyers (1998) suggested that sequence exchange between paralogs is rare compared with exchange between orthologs, that is, between strictly allelic genes. This suggestion is based on observations of the Dm3 and Pto complex resistance loci in lettuce and tomato, respectively. Some genes in these loci are more similar in sequence to genes from the corresponding loci in related species than to paralogs in the same species. This suggested that the frequency of recombination between paralogs was not sufficient to cause sequence convergence (concerted evolution) of the repeated genes within a haplotype, as is observed for rRNA genes in yeast (Gangloff et al., 1996). We do not believe that our data for P locus genes are inconsistent with the birth and death model. The difference in the relative importance of sequence exchange between paralogs at Dm3 and Pto compared with the P locus may reflect the different levels of sequence similarity between genes at these loci. The P locus genes that we have examined are highly similar (>95% DNA identity), whereas some genes at Dm3 and Pto share as little as 55% DNA identity. High sequence identity would increase the likelihood of meiotic exchange between mispaired sequences. Shuffling of polymorphic sites between paralogs by unequal recombination is probably an important process in the evolution of new resistance specificities, but it may be limited to closely related sequences within complex loci. As the duplicated sequences at these loci diverge due to the random accumulation of mutations over time, unequal recombination would become less frequent. Importantly, the action of selection to maintain recombinant genes with new and advantageous phenotypes will also influence the frequency with which recombinant genes are observed at complex loci. Indeed, recombinant genes resulting from unequal exchange within the Rp1D rust resistance haplotype of maize have been derived experimentally, and some of these have novel resistance phenotypes (Collins et al., 1998; T. Pryor, personal communication) that could have differential fitness consequences in a natural population. Selection to maintain allelic diversity at resistance loci may counteract to some degree the sequence convergence that would otherwise result from sequence exchange between paralogs. Sequence alignments suggest that past sequence exchange may have occurred between paralogous genes at the RPP8/HRT (McDowell et al., 1998; Cooley et al., 2000) and RPP5 (Noël et al., 1999) clusters in Arabidopsis and at the Cf-4/9 and Cf-2/5 complexes in tomato (Parniske et al., 1997; Dixon et al., 1998; Parniske and Jones, 1999), although these observations have not been tested statistically.
As observed in other resistance gene families, we found evidence for diversifying selection acting on DNA sequences encoding the β-strand/β-turn (xxLxLxx) motifs of the LRR repeats of P locus genes (Table 2). The fact that diversifying selection was detected in comparisons involving genes with frameshift mutations suggests either that the frameshifts in these genes occurred relatively recently and subsequent accumulation of random mutations has not yet eliminated the evidence of past selection or that sequence exchange events subsequent to the mutations have allowed reintroduction of actively selected sequences. This latter explanation may be favored by the observation that although increased Ka/Ks ratios were observed in comparisons involving the LRR-encoding regions of p4-A and pH-A, neutral substitution rates were observed in comparisons involving their CNL domains.
METHODS
Plant and Rust Material
Flax (Linum usitatissimum) cultivars Forge (contains L6, M, N, and P2 rust resistance genes), Hoshangabad (contains no active resistance genes), and Ward (L9 and M2) have been described by Lawrence et al. (1995). The flax lines Koto (P), Akmolinsk (P1), Abyssinian (P2), Leona (P3), and CI 1911 Punjab (P4) were from H.H. Flor's original set of flax differential lines and have been described by Islam and Mayo (1990). Flax rust (Melampsora lini) strains I, C, and H and F2 progeny of C × H (prefix CH5F2) were described by Lawrence et al. (1981), and rust strains Fi, Bs1, and J were described by Ellis et al. (1999).
Transposon Tagging with Ac
We have previously described a transposon-tagging approach using the maize transposable element Ac in the flax line Forge (Lawrence et al., 1995). Because Ac preferentially transposes to locations on the same chromosome, we first identified a Forge-Ac line with an Ac element linked to P2. Testcross families of 64 individuals derived from 26 independent Forge-Ac lines were scored for P2 by inoculation with rust strain CH5F2(133) (avirulent to P2 but virulent to L6, M, and N) and for neomycin phosphotransferase (NPT)-II enzyme activity in cotyledons (the NPT-II gene is present on the same T-DNA as Ac). One family showed linkage between P2 and NPT-II (14 map units), and gel blot analysis indicated that three Ac elements cosegregated with the NPT-II marker in 66 testcross progeny. Because Ac is not active in flax lines with few copies, the Forge line containing the three P2-linked Ac elements was crossed to a Forge line containing multiple copies of Ac (Lawrence et al., 1995). Progeny plants, which contained ∼10 copies of Ac, including the P2-linked elements, were crossed extensively to the rust-susceptible line Hoshangabad. Progeny were screened for mutations in the P2 gene by inoculation with the rust strain CH5F2(133). Gel blot hybridization analysis with Ac-derived probes was used to screen for newly transposed Ac elements in mutant lines.
Gel Blot Analysis
Restriction enzyme–digested genomic DNA was separated on 1.0% agarose gels and transferred to Hybond N+ nylon membranes (Amersham, Buckinghamshire, UK). Prehybridization and hybridization with 32P-dCTP–labeled DNA probes were conducted in 7% (w/v) SDS, 1% (w/v) BSA, 0.5 M sodium phosphate, pH 7.2, and 1 mM EDTA at 65°C; washing was performed in 1 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) and 0.1% SDS at 65°C.
Isolation of Ac-Flanking DNA Sequences
DNA flanking an Ac element in p2-X157 was isolated using the polymerase chain reaction (PCR) walking method of Siebert et al. (1995). Blunt end–digested genomic DNA from a line containing the p2-X157–linked Ac but lacking other Ac elements was ligated to an amine-blocked adapter primer and then amplified by nested PCR using the Ap1 and Ap2 primers (Siebert et al., 1995) with two Ac primers, 3′AcA (5′-TGATATGCACAAAAGATTGGGTAGC-3′) and 3′AcB (5′-TGATTGGTGATCTCGAGGTGCTAGAC-3′). The Perkin-Elmer/Applied Biosystems GeneAmp XL PCR kit was used (Roche Molecular Systems, Inc., Branchburg, NJ) with Ampliwax PCR gem wax beads (Roche Molecular Systems) under the following amplification conditions: 94°C for 2 min, 40 cycles of 94°C for 20 sec, 55°C for 30 sec, 72°C for 5 min, and then a final 10 min at 72°C. PCR prod-ucts were isolated using either Wizard PCR purification columns (Promega, Madison, WI) or a Strataprep PCR purification kit (Stratagene, La Jolla, CA); they were then either sequenced directly or subcloned into pBluescript SK+, and a consensus sequence of at least three independent clones was determined. DNA sequences were determined using the ABI dye terminators system and were analyzed on an ABI 377 DNA sequencer (Applied Biosystems, Foster City, CA). Sequences were analyzed using Sequencher (Gene Codes Corp., Ann Arbor, MI) and GCG (Genetics Computer Group, Madison, WI) software.
Isolation of P Locus Genomic DNA Sequences
A lambda DNA library of Sau3A partially digested genomic DNA from the flax line Forge containing the P2 gene (Lawrence et al., 1995) was screened by hybridization with a 32P-labeled DNA probe containing the p2-X157 Ac element flanking sequence. Positive clones were purified, subcloned into pBluescript vectors, and sequenced as described above. Sequences homologous to P2 and p2-A were amplified from other P locus alleles by using the upstream primers P2-18 (5′-AAATACAAACTCTGGATCCTG-3′; P2 specific) or P2-9 (5′-CAGAAGAAGACCACAACATC-3′; p2-A specific) with the downstream primer P2-27 (5′-CTCCCAGTAATGGAAG-3′; common to P2 and p2-A). DNA-flanking known P2 sequence in p2-X101, p2-X135, and p2-X143 alleles was isolated by PCR walking as described above, but with P2-derived primers used in place of the Ac primers. Sequence from the 5′ Ac junction in p2-X157 was obtained by PCR with P2-18 and 5′AcA (5′-CTTGTTCCATGATGACCCTCAGC-3′) primers, whereas the 5′ Ac junction in p2-X143 was amplified with the 5′AcA and P2-16 (5′-ATTGGATGGTCAGGACTATC-3′) primers. The Ac excision site in p2-X157 revertants was amplified with the primers P2-18 and P2-5 (5′-CAAGATTTTGAGTGCCAAAGG-3′). PCR conditions were as described above, and products were sequenced directly.
Preparation of Constructs and Flax Transformation
The P2full construct consisted of an 8-kb genomic DNA fragment containing the P2 gene with a 1.8-kb upstream promoter region and a 2-kb downstream sequence. The P2ΔRI construct contained a similar DNA fragment but was truncated at an EcoRI site in the fourth exon of P2, removing the last 104 codons (Figure 3). The 35SP2 construct contained P2 coding sequence (including introns and 290 bp upstream and 90 bp downstream) inserted between the 35S cauliflower mosaic virus promoter and nopaline synthase (NOS) terminator sequences. The 35SP2ΔRI construct was similar but truncated at the EcoRI site in exon 4. For chimeric constructs, the 5′ half of p-B was amplified with the high-fidelity Pyrococcus furiosus (Pfu) polymerase (Stratagene) and primers P2-1 and P2-27 and cloned into pCRScript (Stratagene). A 1.7-kb EcoRI fragment and a 1.8-kb EcoRV/XbaI fragment from a p-B PCR clone with no sequence errors were inserted into the P2full construct to create the P2/P-RI and P2/P-RVX constructs, respectively. Transformation constructs were inserted as NotI fragments into the binary vector pTNotTReg (derived from pTAB-EPspec and containing the selectable spectinomycin resistance gene between the left and right T-DNA transfer borders) (Anderson et al., 1997; J.G. Ellis, unpublished results) and fully sequenced to confirm their integrity. Transformation of the flax line Ward was as described by Anderson et al. (1997), except that spectinomycin was used at 50 μg/mL.
Transcript Analysis by Reverse Transcription–PCR
P2 transcripts were analyzed by reverse transcription (RT)–PCR to confirm the splicing of predicted intron sequences. Flax RNA was reverse transcribed using Superscript Reverse Transcriptase (Gibco BRL, Rockville, MD) with the downstream PCR primer and then amplified by Taq polymerase. The primer pairs P2-8 (5′-TAATTGTGCTGGTCATACTTC-3′) and P2-5 (see above) span intron 1, and primers P2-1 (5′-TCCCTATTAACATGTGTTCCTAGCGAG-3′) and P2-15 (5′-TAGTAATCCATCAGAAGTCTG-3′) span intron 2. For amplification across intron 3 and detection of the polyadenylation site, the SMART PCR cDNA synthesis and Advantage cDNA PCR kits (Clontech, Palo Alto, CA) were used with internal primer P2-21 (5′-GGGCTCGGGATCACTCG-3′). PCR products were cloned into pGEMT-Easy (Promega), and at least four independent clones were sequenced. For detection of transcripts derived from the 35SP2ΔRI construct in transgenic flax plants, the primers Nos1 (5′-ATTGCCAATGTTTGAACGATC-3′) and P2-13 (5′-GAGAACCACCCACCAATGC-3′) were used.
DNA Evolutionary Analysis
Rates of synonymous and nonsynonymous nucleotide substitutions between P locus homologs were determined from alignments of coding sequences using Molecular Evolutionary Genetics Analysis software version 1.02 (Kumar et al., 1993; http://evolgen.biol.metro-u.ac.jp/MEGA/) for global comparisons and the Diverge program of the Wisconsin Package version 10.1 (Genetics Computer Group) for individual pairwise comparisons. The Geneconv program (Sawyer, 1989; http://www.math.wustl.edu/~sawyer/geneconv/index.html) was used to detect evidence for sequence exchanges between P locus genes.
Acknowledgments
This research was supported by grants from the Grains Research and Development Corporation and the Australian Research Grants Scheme. Valerie Ryle and Patricia Atkinson provided excellent technical assistance.
References
- Altschul, S.F., Madden, T.L., Schäffer, A.A., Zhang, J., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bittner-Eddy, P.D., Crute, I.R., Holub, E.B., and Beynon, J.L. (2000). RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21 177–188. [DOI] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Beynon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N.C., Webb, C.A., Seah, S., Ellis, J.G., Hulbert, S.H., and Pryor, A. (1998). The isolation and mapping of disease resistance gene analogs in maize. Mol. Plant-Microbe Interact. 11 968–978. [DOI] [PubMed] [Google Scholar]
- Cooley, M.B., Pathirana, S., Wu, H.-J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and oomycete pathogens. Plant Cell 12 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, M.S., Hatzixanthis, K., Jones, D.A., Harrison, K., and Jones, J.D.G. (1998). The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., Pryor, T., and Ellis, J.G. (2000). Genetic analysis and evolution of plant disease resistance genes. In Molecular Plant Pathology, Annual Plant Reviews, Vol. 4, M. Dickinson and J. Beynon, eds (Sheffield, UK: Academic Press), pp. 88–107.
- Ellis, J., and Jones, D. (1998). Structure and function of proteins controlling strain-specific pathogen resistance in plants. Curr. Opin. Plant Biol. 1 288–293. [DOI] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flor, H.H. (1965). Tests for allelism of rust-resistance genes in flax. Crop Sci. 5 415–418. [Google Scholar]
- Flor, H.H. (1971). Current status of the gene-for-gene concept. Annu. Rev. Phytopathol. 9 275–296. [Google Scholar]
- Gangloff, S., Zou, H., and Rothstein, R. (1996). Gene conversion plays the major role in controlling the stability of large tandem repeats in yeast. EMBO J. 15 1715–1725. [PMC free article] [PubMed] [Google Scholar]
- Gassmann, W., Hinsch, M.E., and Staskawicz, B.J. (1999). The Arabidopsis RPS4 bacterial resistance gene is a member of the TIR-NBS-LRR family of disease resistance genes. Plant J. 20 265–277. [DOI] [PubMed] [Google Scholar]
- Islam, M.R., and Mayo, G.M.E. (1990). A compendium on host genes in flax conferring resistance to flax rust. Plant Breed. 104 89–100. [Google Scholar]
- Jones, D.A., and Jones, J.D.G. (1997). The role of leucine-rich repeat proteins in plant defenses. Adv. Bot. Res. Adv. Plant Pathol. 24 89–167. [Google Scholar]
- Kobe, B., and Diesenhofer, J. (1994). The leucine-rich repeat: A versatile binding motif. Trends Biol. Sci. 19 415–421. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Diesenhofer, J. (1995). A structural basis of the interactions between leucine-rich repeats and protein ligands. Nature 374 183–186. [DOI] [PubMed] [Google Scholar]
- Kumar, S., Tamura, K., and Nei, M. (1993). MEGA: Molecular Evolutionary Genetics Analysis, version 1.0. (University Park, PA: Pennsylvania State University).
- Lane, B.G., Dunwell, J.M., Ray, J.A., Schmitt, M.R., and Cuming, A.C. (1993). Germin, a protein marker of early plant development, is an oxalate oxidase. J. Biol. Chem. 268 12239–12242. [PubMed] [Google Scholar]
- Lawrence, G.J., Mayo, G.M.E., and Shepherd, K.W. (1981). Interactions between genes controlling pathogenicity in the flax rust fungus. Phytopathology 71 12–19. [Google Scholar]
- Lawrence, G.J., Finnegan, E.J., Ayliffe, M.A., and Ellis, J.G. (1995). The L6 gene for flax resistance is related to the Arabidopsis bacterial resistance gene Rps2 and the tobacco viral resistance gene N. Plant Cell 7 1195–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckie, F., Mattei, B., Capodicasa, C., Hemmings, A., Nuss, L., Aracri, B., De Lorenzo, G., and Cervone, F. (1999). The specificity of a polygalacturonase-inhibiting protein (PGIP): A single amino acid substitution in the solvent-exposed β-strand/β-turn region of the leucine-rich repeats (LRRs) confers a new recognition capability. EMBO J. 18 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luck, J.E., Lawrence, G.J., Finnegan, E.J., Jones, D.A., and Ellis, J.G. (1998). A flax transposon identified in two spontaneous mutant alleles of the L6 rust resistance gene. Plant J. 16 365–369. [DOI] [PubMed] [Google Scholar]
- Luck, J.E., Lawrence, G.J., Dodds, P.N., Shepherd, K.W., and Ellis, J.G. (2000). Regions outside of the leucine-rich repeats of flax rust resistance proteins have a role in specificity determination. Plant Cell 12 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G.M., Goff, S., Holub, E.B., and Dangl, J.E. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michelmore, R.W. (1998). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8 1113–1130. [DOI] [PubMed] [Google Scholar]
- Milligan, S.B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P., and Williamson, V.M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël, L., Moores, T.L., van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergences at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Pan, Q., Wendel, J., and Fluhr, R. (2000). Divergent evolution of plant NBS-LRR resistance gene homologues in dicot and cereal genomes. J. Mol. Evol. 50 203–213. [DOI] [PubMed] [Google Scholar]
- Papageorgiou, A.C., Shapiro, R., and Acharya, K.R. (1997). Molecular recognition of human angiogenin by placental ribonuclease inhibitor: An X-ray crystallographic study at 2.0 Å resolution. EMBO J. 16 5162–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabo, V., Frost, L.N., Schmidt, R., van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and interleukin-1 receptors with N and L6. Plant Cell 9 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., and Jones, J.D.G. (1999). Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA 96 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B.H., and Jones, J.D.G. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91 821–832. [DOI] [PubMed] [Google Scholar]
- Pryor, T., and Ellis, J. (1993). The genetic complexity of fungal resistance in plants. Adv. Plant Pathol. 10 282–305. [Google Scholar]
- Salmeron, J.M., Oldroyd, G.E.D., Rommens, C.M.T., Scofield, S.R., Kim, H.-S., Lavelle, D.T., Dahlbeck, D., and Staskawicz, B.J. (1996). Tomato Prf is a member of the leucine-rich repeat class of plant disease resistance genes and lies embedded within the Pto kinase gene cluster. Cell 86 123–133. [DOI] [PubMed] [Google Scholar]
- Sawyer, S.A. (1989). Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6 526–538. [DOI] [PubMed] [Google Scholar]
- Schweizer, P., Christoffel, A., and Dudler, R. (1999). Transient expression of members of the germin-like gene family in epidermal cell of wheat confers disease resistance. Plant J. 20 541–552. [DOI] [PubMed] [Google Scholar]
- Siebert, P.D., Chenchik, A., Kellog, D.E., Lukyanov, K.A., and Lukyanov, S.A. (1995). An improved PCR method for walking in uncloned genomic DNA. Nucleic Acids Res. 23 1087–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D.G. (1998. a). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23 454–456. [DOI] [PubMed] [Google Scholar]
- van der Biezen, E.A., and Jones, J.D.G. (1998. b). The NB-ARC domain: A novel signaling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8 R226–R227. [DOI] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the interleukin-1 receptor. Cell 78 1101–1115. [DOI] [PubMed] [Google Scholar]
- Zhou, F., Zhang, Z., Gregersen, P.L., Mikkelsen, J.D., de Neergaard, E., Collinge, D.B., and Thordal-Christensen, H. (1998). Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus. Plant Physiol. 117 33–41. [DOI] [PMC free article] [PubMed] [Google Scholar]