Abstract
To assess the extent to which a nuclear gene for a chloroplast protein retained the ability to be expressed in its presumed preendosymbiotic location, we relocated the RbcS gene for the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) to the tobacco plastid genome. Plastid RbcS transgenes, both with and without the transit presequence, were equipped with 3′ hepta-histidine–encoding sequences and psbA promoter and terminator elements. Both transgenes were transcribed abundantly, and their products were translated into small subunit polypeptides that folded correctly and assembled into the Rubisco hexadecamer. When present, either the transit presequence was not translated or the transit peptide was cleaved completely. After assembly into Rubisco, transplastomic small subunits were relatively stable. The hepta-histidine sequence fused to the C terminus of a single small subunit was sufficient for isolation of the whole Rubisco hexadecamer by Ni2+ chelation. Small subunits produced by the plastid transgenes were not abundant, never exceeding ∼1% of the total small subunits, and they differed from cytoplasmically synthesized small subunits in their N-terminal modifications. The scarcity of transplastomic small subunits might be caused by inefficient translation or assembly.
INTRODUCTION
The process of symbiogenesis that has given rise to organelles has involved the progressive migration of most of the genes of the original prokaryotic endosymbiont's chromosome to the nucleus of the host (Gray, 1992; Whatley, 1993). This has occurred to differing extents in different organelles and in different hosts. The RbcS gene for the small subunit of the chloroplast photosynthetic enzyme ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) is a much studied example of a gene that has so migrated. In green algae and higher plants, RbcS has been transferred from the ancestral plastid's genome to become a nuclear multigene family (Rodermel, 1999). On the other hand, in nongreen eukaryotic algae, rbcS has remained in its presumed ancestral location as the second gene in the plastid rbcLS operon (except in the case of some dinoflagellates that apparently lack RbcS altogether) (Delwiche and Palmer, 1996).
A transferred gene for a plastid protein, or a subunit thereof, must become equipped with regulatory and transcriptional elements suitable to its new nuclear location and, in addition, requires a sequence that encodes a transit peptide to target the polypeptide to the plastid. Furthermore, the precursor polypeptide must be able to be synthesized on cytoplasmic ribosomes, folded with the aid of cytoplasmic chaperones, transported through the envelope membrane system of the plastid, relieved of its transit peptide, refolded with the aid of plastid chaperones, and finally assembled in the plastid into the appropriate protein complex, which also may contain other nucleus- and/or plastid-encoded subunits. These several requirements, which have been studied in varying degrees of detail for the RbcS genes of higher plants and green algae, must coexist with the requirements imposed by the function of the subunit in its ultimate complex (Rodermel, 1999; Roy and Andrews, 2000).
Genetic transformation of the genomes of organelles provides a means of artificially reversing the endosymbiotic migration of genes for organelle proteins to the nucleus. This enables analysis of the extent to which nuclear genes and their products, now acclimated to the eukaryotic expression system and the cytoplasmic folding and transport machineries, have retained the ability to be expressed in their presumed original prokaryotic compartment. As an example, we describe here the relocation of the RbcS gene from the nucleus to the plastid genome of tobacco. Two forms of the RbcS gene, one with and one without the presequence that encodes the transit peptide, were equipped with plastid regulatory elements and inserted in the tobacco plastome. Both functioned in the plastid environment, and their products were assembled correctly into Rubisco. However, the amounts of small subunits synthesized in the chloroplast were small compared with the products of the nuclear RbcS gene family. This indicates that acclimation to a nuclear location has compromised, in ways not understood, the ability of this gene to be expressed abundantly in the chloroplast.
RESULTS
Relocation of RbcS to the Plastid Genome
A tobacco cDNA encoding RbcS, both with and without its transit peptide–encoding sequence and with a hepta-His–encoding sequence inserted immediately 5′ to the terminator codon, was equipped with promoter and terminator sequences derived from the plastid psbA gene. The construct lacking the transit sequence also had an Ala codon inserted between codons 1 and 2 of the mature peptide. These cassettes were inserted into a vector derived from pRV112a (Zoubenko et al., 1994) between flanking regions that direct insertion of the cassette into a specific site in the inverted repeat regions of the tobacco plastid genome (Figure 1). These plasmids (ptpSSuH with the transit sequence and pSSuH without) were introduced into tobacco chloroplasts biolistically (see Methods).
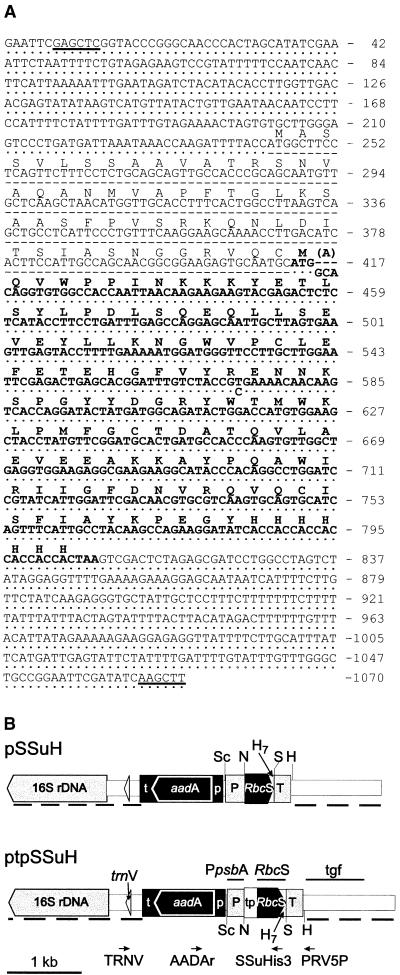
Figure 1.
Plasmids Used for Insertion of RbcS Genes into the Inverted Repeat Region of the Tobacco Plastid Genome.
(A) Sequences of the SSu cassettes of ptpSSuH (upper nucleotide sequence), which directs the synthesis of the SSu precursor, and pSSuH (lower sequence), which directs the synthesis of the mature SSu with an alanyl insertion at residue 2 (in parentheses). The translated amino acid sequence is shown above the upper nucleotide sequence. The region corresponding to the mature SSu is shown in boldface. Deletions are shown as dashes; dots represent identity. The nucleotide sequence includes the psbA promoter and 5′ UTR (nucleotides 17 to 243), the psbA terminator (nucleotides 822 to 1043), and rbcS (with and without the transit peptide–encoding region) with a C-terminal hepta-His–encoding sequence (nucleotides 244 to 807).
(B) Plasmids pSSuH and ptpSSuH. To generate these plasmids, we substituted the SacI-HindIII (sites underlined in [A]) fragments of the cassettes for the SacI-HindIII fragment of pRV112a (Zoubenko et al., 1994) by using the multistep procedure described in Methods. Long dashed lines indicate the homologous flanking regions that direct introduction of the RbcS and aadA genes into the inverted repeats. These flanking regions constitute nucleotides 101,145 (141,481) to 102,309 (140,317) and 102,316 (140,310) to 104,085 (138,541) of inverted repeat A (inverted repeat B in parentheses) of the tobacco plastid genome (GenBank accession number Z00044). The tgf, RbcS, and PpsbA probes used in DNA gel blot and RNA gel blot analyses and the annealing sites of the primers (TRNV, AADAr, SSuHis3, and PRV5P) used to amplify and sequence the inserted genes are indicated. H, HindIII; H7, hepta-His–encoding sequence; N, NcoI; p, rrn promoter and 5′ UTR; P, psbA promoter and 5′ UTR; S, SalI; Sc, SacI; t, rps16 terminator; T, psbA terminator; tp, RbcS transit presequence.
Ten leaves were bombarded with each plasmid, and five and 12 spectinomycin-resistant plantlets were obtained for pSSuH and ptpSSuH, respectively. After tissue from all plantlets was subjected to another round of regeneration on spectinomycin-containing medium, polymerase chain reaction (PCR) analysis (see Methods) was used to identify two SSuH and four tpSSuH plantlets as genuine transformants. Both SSuH transformants (SSuH2 and SSuH5) and two of the tpSSuH transformants (tpSSuH4 and tpSSuH7) were cloned and regenerated twice more on spectinomycin-containing medium before growing to maturity in soil.
Because DNA gel blots of total leaf DNA from the T0 plants digested with HindIII showed no trace of the 7.7-kb fragment observed with the wild type (Figure 2A), we concluded that the transformants were homoplasmic. For DNA digested with HindIII (Figure 2A) and other restriction enzymes (data not shown), the sizes of the fragments detected by the tgf probe (which anneals to a sequence adjacent to the point of insertion of the transgenes [Figure 1B]) corresponded to those expected for correct insertion of the transgenes into the target site in the inverted repeat regions. Correct insertion also was confirmed by PCR (Figure 2B; see Methods).
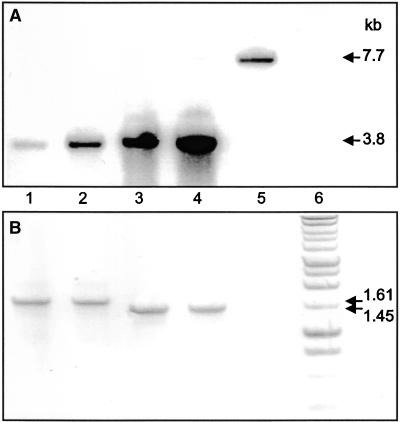
Figure 2.
Analysis of DNA from Homoplasmic Transformants and a Nontransformed Control.
The mature T0 transformants, tpSSuH4 (lane 1), tpSSuH7 (lane 2), SSuH2 (lane 3), and SSuH5 (lane 4), and a nontransformed control (lane 5) were analyzed.
(A) DNA gel blot of HindIII-digested leaf DNA probed with the 32P-labeled 787-bp tgf fragment (see Methods). The sizes of the bands detected were assigned by comparison with an unlabeled DNA ladder. The point of insertion of the transgenes occurs within a 7.7-kb HindIII fragment of the tobacco plastome. These restriction sites lie outside of the flanking regions incorporated into the transforming plasmids shown in Figure 1B. Insertion of either transgene introduces a new HindIII site that reduces the size of the fragment detected by the tgf probe (Figure 1B) to 3.8 kb.
(B) PCR amplification of the 1613-bp (with transit presequence) and 1446-bp (without presequence) DNA fragments by using the primers AADAr and PRV5P (see Figure 1B). A 1-kb DNA ladder (Promega) is shown in lane 6.
Both Plastid RbcS Transgenes Were Strongly Transcribed
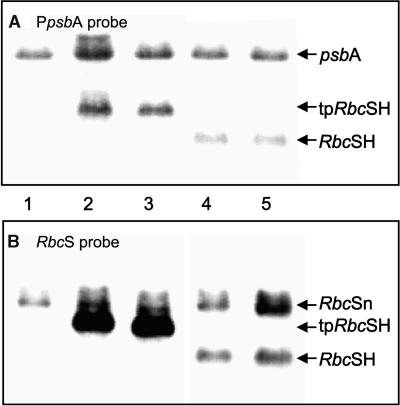
Because both transgenes contained the 5′ untranslated region (UTR) derived from the plastid psbA gene, the PpsbA probe (Figure 1B) hybridized to the mRNA produced by the endogenous psbA gene and to that produced by the transgenes (Figure 3A). This provided a means of comparing the amount of message produced by the transgenes with the abundant psbA message. In young leaves from the transformants tpSSuH4 and tpSSuH7, the message produced by the RbcS transgene with the transit sequence was approximately two-thirds as abundant as that of the psbA mRNA. In similar leaves of the transformants SSuH2 and SSuH5, in which the transit sequence was missing, the transgene produced approximately one-third as much message as the psbA gene in both cases. The greater abundance of the message with the transit sequence was reproduced in RNA gel blots probed with the RbcS probe (Figure 3B). This probe also recognizes the message produced by the nuclear RbcS gene family, which acts as the internal control in this case. For both tpSSuH transformants, the chloroplast RbcS message exceeded the nuclear RbcS message by at least fivefold (Figure 3B). In contrast, the SSuH2 and SSuH5 transformants had approximately one-half as much chloroplast message as nuclear message (Figure 3B).
Figure 3.
Detection of mRNA Produced by the Transgenes.
(A) RNA (10 μg) from nontransformed tobacco (lane 1) and the T1 transformants tpSSuH4 (lane 2), tpSSuH7 (lane 3), SSuH2 (lane 4), and SSuH5 (lane 5) was hybridized with a 239-bp SacI-NcoI fragment corresponding to the psbA promoter and 5′ UTR (PpsbA [Figure 1B]).
(B) Duplicate blot hybridized with a 377-bp NcoI-SalI fragment corresponding to the coding sequence of the mature small subunit (RbcS [Figure 1B]).
The positions of the mRNAs for psbA, nucleus-encoded RbcS (RbcSn), and plastid-encoded His-tagged RbcS with (tpRbcSH) and without (RbcSH) the transit presequence are indicated.
The Plastid RbcS Transgenes Directed the Synthesis of Small Subunits, and These Were Assembled into Hexadecameric Rubisco
Fusion of hepta-His to the C terminus of the tobacco small subunit increases its molecular mass from 14.6 to 15.6 kD. The molecular mass of the His-tagged precursor of the small subunit is 21.3 kD. Overexpression of these constructs in Escherichia produced His-tagged polypeptides of these expected sizes in the soluble fractions of the cell extracts (Figure 4A). Although it has been reported that tobacco small subunits and precursors expressed in Escherichia were located predominantly in inclusion bodies (Klein and Salvucci, 1992), we observed that substantial amounts of the His-tagged versions of these polypeptides could be isolated from the soluble fraction by Ni2+-nitrilotriacetic acid (Ni-NTA) agarose chromatography (Figure 4A).
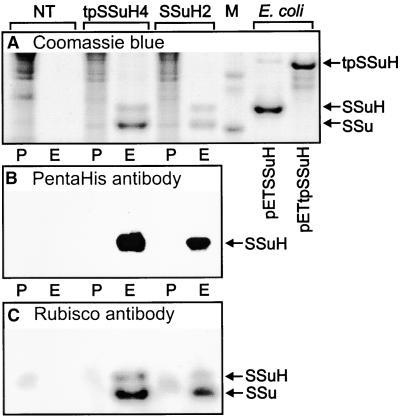
Figure 4.
Expression of Plastid-Encoded RbcS Transgenes and Assembly of the Products into the Rubisco Holoenzyme.
(A) Coomassie blue–stained SDS gel of protein extracted from young leaves of nontransformed tobacco (NT), the T1 transformants tpSSuH4 and SSuH2, and Escherichia (E. coli) BL21 (DE3) expressing the plasmids pETSSuH and pETtpSSuH. The insoluble protein (P) was pelleted, and His-tagged protein complexes (E) were isolated from the supernatant fraction as described in Methods. For the leaf extracts, each lane was loaded with protein (pelleted or His tagged) that had been derived from 0.5 cm2 of leaf. For the Escherichia extracts, the His-tagged polypeptides in the soluble fractions constituted 5.4% (pETtpSSuH) and 2% (pETSSuH) of the total soluble protein as determined by the dye binding method (Pierce Coomassie Plus).
(B) Immunoblot of an analogous gel, loaded with leaf samples only, probed with PentaHis antibody that recognized the His tag.
(C) Immunoblot probed with tobacco Rubisco antibody.
The positions corresponding to the 14.6-kD mature small subunit (SSu), the 15.6-kD hepta-His–tagged mature small subunit (SSuH), and the 21.3-kD hepta-His–tagged small subunit precursor with the N-terminal transit peptide (tpSSuH) are indicated. M, molecular mass marker lane showing soybean trypsin inhibitor (20.1 kD) and α-lactalbumin (14.4 kD).
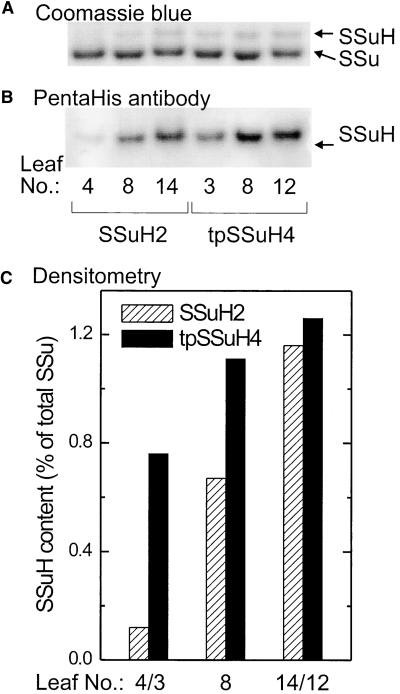
His-tagged small subunits produced by the tobacco plastid transformants were soluble and assembled into Rubisco hexadecamers (Figure 4). No small subunits were detected in the particulate fraction by either the PentaHis antibody (Figure 4B) or the anti-Rubisco antibody (Figure 4C). (The traces of immunoreactive material detected in the pellet lanes of all three samples by the anti-Rubisco antibody [Figure 4C] did not correspond to His-tagged or untagged small subunits or precursors and are likely to result from trace contamination of the tobacco Rubisco preparation used to raise the antibody.) His-tagged Rubisco was affinity purified and concentrated from the soluble fractions of the leaf extracts by using Ni-NTA agarose. SDS-PAGE of these preparations revealed three bands with molecular masses of 53, 15.6, and 14.6 kD, corresponding to the Rubisco large subunit, His-tagged small subunit, and untagged small subunit, respectively (Figures 4A, 4B, and 4C; large subunit region of the gels not shown). All three bands were recognized by the anti-Rubisco antibody. Because no trace of a 21.3-kD band was detected by Coomassie blue or either antibody in the sample derived from the tpSSuH transformant, we conclude that either the transit peptide was not incorporated into the initial translation product or it had been cleaved completely and apparently normally. For both types of transformants, the intensities of the 15.6- and 14.6-kD bands when stained with Coomassie blue appeared to be approximately in the ratio 1:7, implying that only a single small subunit in the hexadecamer bore a His tag. Immunoblotting with the anti-Rubisco antibody (Figure 4C) followed by densitometry also showed an approximate 1:7 ratio between His-tagged and untagged small subunits. Sedimentation of the affinity purified preparations through a sucrose gradient, as described by Whitney et al. (1999), confirmed that all of the His-tagged small subunits were assembled in hexadecameric Rubisco complexes. A single, rapidly sedimenting, UV light–absorbing peak corresponding to Rubisco hexadecamers was observed. SDS-PAGE followed by immunoblotting with the PentaHis antibody showed no traces of unassembled His-tagged small subunits in fractions higher in the gradient than this peak, even after 30-fold concentration by absorption onto Ni-NTA agarose (data not shown).
Plastid-Synthesized Small Subunits Were Not Abundant but Accumulated as the Leaves Aged
In both types of transformants, the numbers of His-tagged small subunits present were too low for each Rubisco hexadecamer to contain one His-tagged small subunit. Therefore, most hexadecamers were not bound by the affinity resin. The proportion of His-tagged small subunits to total small subunits was measured by purifying the total Rubisco content of leaves of different ages by conventional procedures (Whitney et al., 1999) and measuring its content of His tags by immunoblotting with the PentaHis antibody. (The antibody to tobacco Rubisco was not well suited to this purpose because the two small subunit bands it recognized were too closely spaced for accurate densitometry when the band corresponding to the untagged small subunit was in vast excess.) Calibration was achieved by separating a range of concentrations of an affinity-purified Rubisco standard on the same gel and assuming that one-eighth of the small subunits in the standard were His tagged (Figure 5). In younger leaves, Rubisco in the tpSSuH transformant had a much higher proportion of its small subunits His tagged than did Rubisco in the SSuH transformant. However, His-tagged small subunits increased in abundance more rapidly in the latter transformant than in the former as the leaves aged. Eventually, in older leaves, both transformants had similar contents of His-tagged small subunits. Even so, only ∼1.2% of the small subunits were the products of the transgenes (Figure 5C).
Figure 5.
Extent of Assembly of Plastid-Encoded Small Subunits into Hexadecameric Rubisco.
(A) Coomassie blue–stained SDS gel loaded with equal amounts (11.5 pmol) of total Rubisco (His tagged plus untagged) purified from leaves of the tobacco transformants SSuH2 and tpSSuH4 by a combination of polyethylene glycol precipitation and Suc gradient centrifugation, as described previously (Whitney et al., 1999). Separate purifications were performed for leaves at different positions on the plants (counting downward from the apical meristem). At the time of sampling, the SSuH2 plants had 18 leaves and the tpSSuH4 plants had 16 leaves. Total Rubisco content of these preparations was measured by binding of 14C-2′-carboxyarabinitol-1,5-bisphosphate (Butz and Sharkey, 1989; Ruuska et al., 1998).
(B) Immunoblot of an analogous gel probed with the PentaHis antibody. The content of His-tagged small subunits was measured by densitometry of the immunoblots by comparison with a series of standards containing 0.25 to 1.2 pmol of His-tagged small subunits separated on the same gel. These standards were derived by separating His-tagged Rubisco holoenzyme from the purified Rubisco preparation from the tpSSuH transformant by using Ni-NTA agarose, as described in Methods. The Rubisco content of this standard preparation was measured by binding of 14C-2′-carboxyarabinitol-1,5-bisphosphate. Because each hexadecamer in this preparation must have at least one His-tagged small subunit and the preparation was derived from a total Rubisco preparation with, on average, much less than one His-tagged small subunit per hexadecamer, we assumed that His-tagged hexadecamers contained only one His-tagged small subunit. Densitometry of immunoblots (anti-Rubisco antibody) of SDS-PAGE gels run with the standard preparation confirmed the assumed 1:7 stoichiometry between His-tagged and untagged small subunits (see Figure 4C). Densitometry of these standards after immunoblotting with the PentaHis antibody produced a linear calibration ( ) passing through the origin.
) passing through the origin.
(C) The content of His-tagged small subunits expressed as a fraction of the total small subunits calculated from the total Rubisco content.
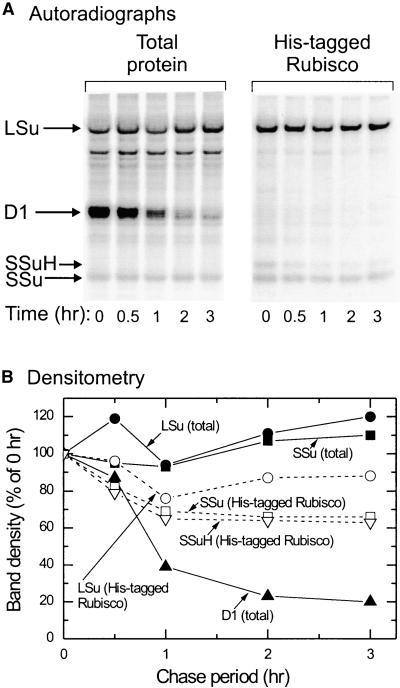
Turnover of Plastid-Synthesized Small Subunits after Assembly into Rubisco
Pulse–chase labeling with 35S was used to investigate how quickly newly synthesized small subunits were turned over. Autoradiography of SDS-PAGE gels loaded with total protein (insoluble and soluble) from leaf extracts of a tpSSuH4 plant showed an intensely labeled band near 30 kD whose label decreased rapidly during the chase period. Although we have no other evidence regarding the identity of this protein, its size and rapid turnover are consistent with it being the D1 protein of photosystem II, and we have labeled it as such (Figure 6). Other prominently labeled bands correspond to the large and untagged small subunits of Rubisco, and the label in these remained constant during the chase period, consistent with little or no turnover. In the total protein gel, no discrete band of label corresponded to the His-tagged small subunit. However, isolation and concentration of His-tagged Rubisco from the soluble fraction of the labeled extracts by using Ni-NTA agarose, followed by SDS-PAGE and autoradiography, allowed ready identification of the His-tagged small subunit band, together with more intense bands for the large and untagged small subunits (Figure 6A). Loss of label from all subunits derived from these His-tagged hexadecamers was noticeably faster than from total Rubisco but much slower than from the D1 protein. Within this fraction, the His-tagged and untagged small subunits turned over at the same rate and more rapidly than the large subunits (Figure 6B).
Figure 6.
Turnover of Rubisco Subunits and the D1 Subunit of Photosystem II Measured by 35S Pulse–Chase Labeling.
(A) Autoradiographs of SDS gels loaded with total leaf protein and isolated His-tagged Rubisco (see Methods), equivalent to 1.8 and 14.5 cm2 of leaf, respectively. Bands corresponding to the D1 protein and the Rubisco large (LSu), small (SSu), and His-tagged small (SSuH) subunits are indicated.
(B) Densitometric measurement of 35S labeling in the various subunits during the chase period. LSu (closed circles), SSu (closed squares), and D1 (closed triangles) band densities were measured in the total protein lanes. Because the SSuH bands in these lanes were too faint for accurate measurement, they were measured in the His-tagged Rubisco lanes, in which a lower background allowed a 10-fold greater amplification of the intensities of the labeled bands. The LSu (open circles) and SSu (open squares) bands also were measured in these lanes.
The N Termini of Small Subunits Synthesized in the Plastid Were Blocked
The SDS-PAGE bands corresponding to His-tagged and untagged small subunits in affinity-purified Rubisco preparations from the transformants were subjected to N-terminal Edman microsequencing (see Methods). The untagged small subunit bands from both tpSSuH4 and SSuH2 plants yielded the sequence X-Q-V-W-P-P-I/Y predicted by the nucleotide sequence of the tobacco RbcS gene family (Muller et al., 1983). The residue released in the first cycle, X, was not the predicted Met, however, but gave rise to a peak that chromatographed between the phenylthiohydantoin derivatives of Phe and Glu. This is consistent with it being N-methyl-Met, as has been reported for the small subunits of several higher plants (Grimm et al., 1997), including tobacco (Ying et al., 1999). A smaller additional peak that was released during the first cycle and eluted after the Glu derivative may be the sulfoxide form of N-methyl-Met.
The His-tagged small subunit bands from the same gels were subjected to similar N-terminal sequencing. No sequence was returned for either transformant, except for a trace level of the same sequence returned for the untagged small subunit that may be attributed to cross-contamination arising from the adjacent untagged band on the gel. The result was the same when the samples were treated with formic acid to cleave a formyl group, if present, from an N-terminal Met residue (see Methods). Presumably, the N termini of the products of both plastid transgenes were blocked with a group(s) other than formyl.
Attempts to obtain information about the nature of the N-terminal blockage of the tpSSuH small subunits by using electrospray ionization mass spectrometry (ESI-MS; see Methods) were unsuccessful. Wild-type small subunits yielded two prominent molecular ions with similar abundances corresponding to subunit masses of 14,530 and 14,573 D. The latter corresponds to an N-terminally methylated polypeptide with the mature sequence shown in Figure 1A (lacking the hepta-His peptide and the inserted Ala-2). The former presumably corresponds to an unresolved mixture of small subunits with closely similar masses encoded by other members of the RbcS gene family (Muller et al., 1983). The small subunit fraction from tpSSuH plants yielded spectra identical to the wild type. Larger ions consistent with the presence of the His tag could not be detected, presumably because of their low abundance. Prior concentration of the His-tagged Rubisco from the transformants by Ni-NTA agarose chromatography before isolation of the small subunits resulted in unreliable mass spectra, despite extensive dialysis. Poor ESI-MS results with Ni-NTA– purified proteins have been reported previously (Neylon et al., 2000).
Plastid-Synthesized Small Subunits Lacking the C-Terminal Hepta-His Sequence Also Were Not Abundant
Preliminary transformations with the pC3 plasmid (which encodes a mature small subunit with an Ala insertion at residue 2 but without a His tag [see Methods]) yielded homoplasmic transformants that resembled the SSuH transformants in having approximately one-third as much transgene message as psbA message (data not shown). Sequencing of the N terminus of the small subunit band from SDS-PAGE gels of Rubisco isolated from these transformants yielded only the wild-type sequence with no trace of Ala in the second position. However, blockage of the N terminus of the plastid-synthesized small subunit (see above) would prevent detection of the transgene's product in this way. ESI-MS of the small subunit fraction also detected only the same prominent pair of molecular ions as observed with the wild-type small subunits (see above) and no larger or smaller species (<2%) consistent with the Ala-2 insertion or the removal of residues from the N terminus. We conclude that, even in the absence of the His-tag, plastid-synthesized small subunits constitute <2% of the total small subunits.
DISCUSSION
Relocation of Genes for Rubisco Subunits between Nuclear and Plastid Genomes
The experiments we report here complement those in which the rbcL gene was deleted from the tobacco plastome, equipped with a transit presequence, and inserted into the nuclear genome (Kanevski and Maliga, 1994). Relocation of the RbcS gene from the nucleus to the plastid is somewhat more complicated because the RbcS gene family cannot be deleted from the nucleus. However, use of a C-terminal hepta-His sequence allowed us to distinguish between the products of the plastid RbcS transgenes and the nuclear RbcS genes. The results of the two types of relocation experiments were similar. The transferred gene functioned in its new location, and its products were assembled correctly into Rubisco in the chloroplast. This finding suggests that both of the Rubisco subunits are physically capable of being synthesized in both cytoplasmic and plastidic compartments. However, in both cases, the products of the transgenes were not very abundant.
The two relocation experiments differ in an important detail: synthesis of the large subunit (nearly 80% of the Rubisco protein by weight) makes much larger demands on the capacity for protein synthesis, folding, transport, and assembly than does synthesis of the small subunit. Because Rubisco is by far the most abundant leaf protein, relocation of the synthesis of its large subunit to the cytoplasm places an extraordinary demand on the protein-synthesizing and protein-folding machineries of this compartment and on the mechanisms by which the precursor protein is transported into the chloroplast. These processes are not naturally adapted to a flux of this magnitude; therefore, it is not surprising that the nuclear rbcL transgene directed the synthesis of only 3 to 10% of the normal Rubisco complement of the wild-type chloroplast (Kanevski and Maliga, 1994). Relocation of RbcS to the plastid genome is not likely to involve capacity problems of this kind. No transport of the product is required, and the protein synthesis capacity needed to produce small subunits in amounts stoichiometric with large subunits would be little more than 20% of that required for the large subunits themselves. Therefore, the reasons for the scarcity of the products of the plastid RbcS transgenes in assembled Rubisco (never more than 1.3% of the total small subunit content; Figure 5) must be sought elsewhere.
Transcript Abundance
It seems unlikely that the scarcity of the mRNA transcripts of the plastid RbcS transgenes can be the cause of the low expression level of the plastid-synthesized small subunits. The abundance of the transcript with the transit presequence rivaled that of the psbA message (one of the most abundant mRNAs in the chloroplast [Gruissem et al., 1988]) and was manyfold greater than that of the nuclear RbcS transcript (Figure 3). In young leaves, the chloroplast RbcS message lacking the transit presequence was two- to 10-fold less abundant than its homolog with the presequence (Figure 3). This could be caused by either slower transcription of the shorter transgene or greater instability of its transcript. Nevertheless, this transcript was still quite abundant. Therefore, the limitation(s) that impaired accumulation of the plastid-encoded small subunits is not likely to be related directly to the transcription rate of the transgenes or the stability of their mRNA products. Translational or post-translational limitations seem to be indicated.
Translational Efficiency
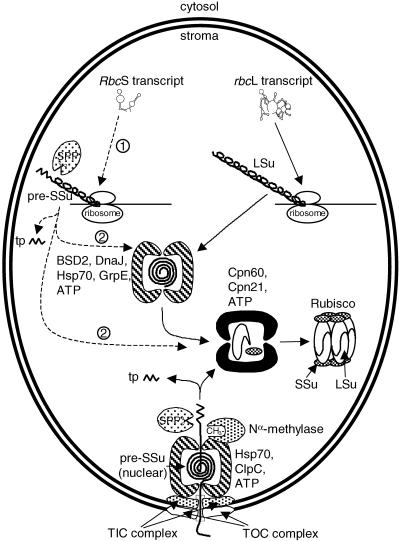
Inefficient translation of the transcripts of the plastid transgenes would be a simple explanation for the scarcity of plastid-encoded small subunits. A restriction at the level of translation (indicated by dashed arrow 1 in Figure 7) could be caused by an inappropriate structure of the mRNAs or unfavorable codon use and would accord with suggestions that translational control is a dominant means of regulating the expression of plastid genes (Staub and Maliga, 1994; Mayfield et al., 1995; Rodermel, 1999).
Figure 7.
Scheme of the Synthesis and Assembly of Rubisco in Tobacco Transformants with a Plastid Transgene Expressing the Small Subunit Precursor (pre-SSu).
The dashed arrows indicate steps that may be rate limiting for the production of chloroplast-synthesized small subunits. Arrow 1 indicates the rates of engagement of the RbcS mRNA with the chloroplast ribosomes and its subsequent translation. The arrows labeled 2 indicate possible access routes of the newly synthesized small subunits to the Rubisco assembly pathway. See text for details. Chaperone complexes are indicated by the paired bracket-shaped symbols. BSD2, product of the BUNDLE SHEATH DEFECTIVE 2 gene (Brutnell et al., 1999); Cpn60 and Cpn21, chloroplast chaperonins 60 and 21; DnaJ, Hsp70, GrpE, and ClpC, chaperone proteins thought to be involved in import and assembly of Rubisco (Brutnell et al., 1999; Keegstra and Cline, 1999); LSu and SSu, large and small subunits of Rubisco; Nα-methylase, enzyme responsible for methylating the α-amino group at the N terminus of the mature small subunit after cleavage of the transit peptide (Grimm et al., 1997; Ying et al., 1999); SPP, stromal processing peptidase; TIC and TOC, translocons of the inner and outer chloroplast membranes (Keegstra and Cline, 1999); tp, transit peptide.
We chose to use promoter and 5′ and 3′ UTR elements derived from the plastid psbA gene for our constructs because of previous success in obtaining abundant expression in chloroplasts of a foreign uidA gene flanked by these elements (Staub and Maliga, 1993, 1994). However, the factors that control the translation rate of plastid genes are not fully understood. There is evidence that the 5′ UTR, in particular, has sequences that bind protein factors that regulate the initiation of translation (Mayfield et al., 1995; Somanchi and Mayfield, 1999). It is possible that mismatch between the 5′ UTR and the protein-coding sequence might lead to an mRNA structure that occludes such a binding site. Additionally, an inappropriate secondary structure of the fused mRNA species might directly impair engagement with the ribosome independent of the involvement of trans-acting factors. Similarities between the translational efficiencies in Escherichia and Chlamydomonas chloroplasts of transgenes fused to randomly mutated 5′ UTR elements can be interpreted in this way because these two organisms are not likely to have identical trans-acting factors (Fargo et al., 1999). Therefore, the efficiency of translational initiation might vary unpredictably according to the foreign sequence fused to the psbA 5′ UTR. We will test these possibilities in future studies using 5′ UTR elements derived from other plastid genes.
In young expanding leaves, the plastid RbcS transgene with the transit presequence directed the synthesis of five times as much small subunit as did the transgene without the presequence, but the difference diminished as the leaves aged (Figure 5). If both mRNA species were translated with similar inefficiency, this difference might be explained by the greater abundance of the mRNA with the transit presequence in the young leaves (Figure 3).
The rate of translation of the plastid RbcS genes also might be impaired by nonoptimal codon use. However, the codon bias of RbcS appears to differ from the consensus codon use of plastid genes in tobacco (Nakamura et al., 1999) and dicotyledons generally (Ausubel, 1988) no more significantly than does the codon use of uidA, a gene that is expressed abundantly in tobacco chloroplasts (Staub and Maliga, 1993, 1994). Moreover, Morton and Levin (1997) suggested that the sensitivity of protein synthesis in plastids of higher plants to codon bias has diminished during evolution, perhaps as a result of the increasing copy number of the plastid genome and, consequently, increasing levels of mRNAs. These issues could be addressed experimentally by resynthesis of the RbcS genes to match the codon bias of abundantly expressed plastid genes such as rbcL and psbA.
Processing of the Small Subunit Precursor
No trace of uncleaved small subunit precursor could be detected in tobacco transformants with the transgene incorporating the transit presequence (Figure 4). There could be two explanations for this.
First, it is possible that the presequence might not have been translated, initiation occurring exclusively at the ATG codon corresponding to the Met residue at the N terminus of the mature polypeptide. (There is a GGA sequence 13 to 15 bp upstream that might have served as a ribosome binding site [Figure 1A].) Although our present evidence does not exclude this possibility, it seems unlikely. In Escherichia transformed with a construct that had the same transit presequence and, therefore, the same possibility for translation of the message to commence at the first Met residue of the mature subunit, there was no sign of such a product, initiation at the ATG codon at the 5′ end of the transit presequence being preferred (Figure 4). Because chloroplasts and Escherichia have similar translational machineries, translation might be expected to initiate similarly. Our use of promoter and 5′ UTR sequences identical to those of the psbA gene strengthens the expectation that translation would initiate at the same ATG codon as it does in psbA, that is, at 5′ end of the transit presequence.
Second, if the transit presequence was translated as expected, cleavage of the transit peptide from the small subunit precursor so produced by the stromal processing peptidase must have been complete (Figure 7). This would not be surprising because the translation product would be identical (except for the C-terminal hepta-His tag) to the cytoplasmically synthesized small subunit precursor imported into the chloroplast. However, this scenario does not readily explain why the N terminus of the mature small subunit produced by the nuclear RbcS is methylated whereas its plastid-encoded analog is blocked in a manner not determined (see Results). Either translational initiation is different with the plastid RbcS as discussed above or the plastid-synthesized precursor is denied access to the methylase responsible for modification of the N terminus of the mature small subunit exposed after cleavage of the precursor (Ying et al., 1999). Perhaps the methylase requires a particular context for efficient recognition of the cleaved precursor. This might be provided by docking of the methylase with the translocons of the inner chloroplast membranes complex or its associated chaperones (Figure 7).
The N termini of the mature small subunits produced by both transgenes were blocked to Edman sequencing (see Results). If translation of the message produced by the transit sequence–containing gene initiated at the Met residue corresponding to the mature N terminus (as discussed above), then a similar modification mechanism might have applied to the products of both transgenes. If the N-terminal Met or formyl-Met residue was removed post-translationally, as is sometimes the case with chloroplast gene products (Michel et al., 1988; Houtz et al., 1989), the Ala or Gln residue thus exposed at the N terminus might have conflicted with the specificity of the N-methylase, perhaps triggering another type of N-terminal modification mechanism. For example, acetylation of the N terminus is observed with the Rubisco large subunit (Houtz et al., 1989), the D1 and D2 proteins of photosystem II, and the chlorophyll a binding protein, Cpa-2 (Michel et al., 1988; Sharma et al., 1997). Clearly, the mechanisms by which the N termini of plastid-synthesized proteins are modified, and the generality of such modifications, need further investigation.
Assembly of Small Subunits into Rubisco
Rubisco molecules with at least one (and probably only one) plastid-synthesized small subunit (i.e., L8S7SH) were somewhat less stable than L8S8 molecules. The former lost 35S label slowly during the chase period whereas the latter retained it completely (Figure 6). Perhaps the presence of even one His tag is sufficient to destabilize the whole hexadecamer slightly. However, the turnover rate of L8S7SH was slow compared with that of the D1 protein. Although enhanced turnover might contribute to the scarcity of plastid-synthesized small subunits, it cannot by itself completely explain it.
It is possible, however, that the plastid-synthesized small subunits or their precursors might be subject to rapid turnover before assembly into Rubisco. Although unassembled His-tagged small subunits would be isolated by the Ni2+ chelation procedure, such rapid degradation might keep their steady state pool sizes so small that they would escape detection in pulse-labeling experiments (Figure 6). Rapid degradation of unassembled small subunits or precursors is consistent with their absence from the particulate fraction (Figure 4). Such degradation might be specific to small subunits synthesized in the chloroplast if these subunits were distinguishable in some way from the cytoplasmically derived precursors and subunits.
It is conceivable that the C-terminal hepta-His sequence might slow translation or impair the folding of the nascent small subunits or precursors sufficiently to bring them to the attention of degradative protease(s) thought to be responsible for the disposal of unassembled small subunits that accumulate in excess of those required to assemble with large subunits (Rodermel, 1999). However, even in the C3 transformants in which the RbcS transgene lacked the hepta-His–encoding sequence, the plastid-synthesized small subunits still constituted <2% of the total small subunits (see Results). This finding argues against the hepta-His sequence being the sole reason for the low levels of plastid-synthesized small subunits.
Plastid-synthesized small subunits or precursors might be distinguishable from their cytoplasmic analogs if the two types of subunits were kept separate during the assembly of Rubisco. It is possible that, in the normal assembly pathway, unassembled small subunit precursors and small subunits are always sequestered by chaperones unless the small subunit supply exceeds the supply of large subunits (in which case the unsequestered excess would be rapidly degraded). Ivey et al. (2000) identified domains in the transit peptide of the small subunit with affinity for the Hsp70 chaperone of chloroplasts, consistent with suggestions that this chaperone may function as an ATP-dependent translocase that draws the precursor across the envelope membranes. Once inside the chloroplast, the precursor might be cleaved of its transit peptide, methylated at its newly exposed N terminus, and handed on to other chaperones in the Rubisco assembly sequence without ever existing in significant amounts in an unassociated state in the stroma (Figure 7). Plastid-synthesized small subunit precursors or small subunits might have difficulty joining this “inside track” to assembly, particularly if they had partially misfolded after leaving the plastid ribosomes (dashed arrows labeled 2 in Figure 7). Therefore, they would mostly fall victim to the degradative protease(s). If this is the case, increasing the rate of translational production of small subunits in the chloroplast may not greatly promote the assembly of these subunits into Rubisco.
Sequestration of unassembled small subunits in chaperone complexes also might provide an explanation for the lack of N-terminal methylation of the plastid-synthesized small subunits. Only unassembled small subunits are capable of acting as the substrate for the methylase (Grimm et al., 1997; Ying et al., 1999). If, as suggested above, the most effective substrate for methylation is the small subunit/chaperone/TIC complex (Figure 7), unsequestered small subunits might be methylated inefficiently, leaving them vulnerable to a different kind of N-terminal processing that leads to blockage of Edman sequencing.
Conclusions
Rubisco small subunits can be synthesized within the chloroplast as well as in the cytoplasm. Chloroplast-synthesized small subunits assemble successfully into L8S8 Rubisco complexes. However, the N termini of the plastid-encoded small subunits are modified incorrectly, and the combined synthesis/assembly process is inefficient. Slow translation of the abundant chloroplast RbcS message could be one reason for the inefficiency. Another might be the limited access of chloroplast-synthesized small subunits or precursors to a preferred assembly pathway enjoyed by their cytoplasmically derived analogs. Such a pathway might connect import of the precursor, cleavage, post-translational modification, and assembly via a sequence of chaperone complexes without the release of significant amounts of the precursors or small subunits at any stage (Figure 7).
METHODS
Construction of Transformation Plasmids
A tobacco (Nicotiana tabacum) RbcS gene was amplified from pTSSU2 (Hudson et al., 1992) by using the primers SSu5C (5′- GGGTCTAGAGTCGACGAGGTTAACGCCATGGCACAGGTGTGG-CCACC-3′, which introduces an NcoI site [underlined] and has an Ala codon [italic] inserted between codons 1 and 2 of the mature small subunit) and SSu3B (5′-GGGTCTAGAGTCGACTTAGTAGCCTTCTGGCTT-3′, which introduces XbaI [underlined] and SalI [italic] restriction sites). The uidA gene in pJS25 (Staub and Maliga, 1993) was replaced with the 383-bp NcoI-XbaI RbcS fragment of the amplified RbcS gene to produce pJSTSSu. An 846-bp SacI-DraI cassette from pJSTSSu, comprising the psbA promoter–RbcS-psbA terminator sequence, was ligated into SacI/SmaI-digested pBluescript II KS+ (Stratagene) to give pBlueSSu. The SacI-HindIII cassette from pBlueSSu was inserted into the multiple cloning site of pRV112a (Zoubenko et al., 1994) to give the preliminary plastid-transforming plasmid pC3. This plasmid was used for preliminary transformations only. For most of the present work, variant RbcS genes with 3′ hepta-His–encoding sequences were substituted into it.
To construct the hepta-His variants of RbcS, RNA was purified from leaves of tobacco cultivar Petit Havana (N,N) by using the SV Total RNA isolation system (Promega, Madison, WI). Using avian myeloblastosis virus reverse transcriptase (Promega), we reverse transcribed 0.4 μg of RNA with primer TPRIME (5′-GGGAGCTCGCTA-GCGGATCCTTTTTTTTTTTTTTTTT-3′). A full-length RbcS gene (including the transit peptide–coding sequence) was amplified using primers SSuTP5 (5′-TAACCATGGCTTCCTCAGTTCT-3′ [NcoI site underlined]) and SSuHis3 (5′-TAGTCGACTTAgtggtggtggtggtggtggtgA-TATCCTTCTGGCTTGTAGGCAATA-3′, introducing SalI [underlined] and EcoRV [italic] sites and with a hepta-His–encoding sequence [lowercase] immediately 3′ to the complement of the RbcS stop codon [boldface]) and cloned into pGEM-T Easy (Promega) to give ptpSSuHT. An RbcS gene encoding only the mature small subunit with a C-terminal hepta-His extension and an additional Ala at codon 2 was amplified from ptpSSuHT by using primers SSu5C and SsuHis3 (Figure 1). The amplified product and ptpSSuHT were digested with NcoI and SalI, and the corresponding 398- and 569-bp fragments were ligated into NcoI-SalI–digested pC3 to give the transforming plasmids pSSuH and ptpSSuH, respectively (Figure 1). The plasmids were sequenced by BigDye terminator cycle sequencing (Applied Biosystems, Foster City, CA) using the primers PRV5P (5′-CTTCGGAATCGGTAGTCA-3′) and AADAr (5′-GAATGTCATTGC-GCTGCCATTCTCCA-3′; Figure 1B). The sequence obtained from ptpSSuH (Figure 1A) is identical (except for the hepta-His–encoding sequence) to that inferred from a previously reported RbcS genomic sequence from tobacco (Mazur and Chui, 1985). The sequence from pSSuH had the expected deletion of the transit sequence and insertion of an Ala codon at position 2 and, in addition, one polymerase chain reaction (PCR)–induced silent substitution within the coding sequence (Figure 1A). The amino acid sequence of the small subunit encoded by pC3 is identical to that shown for pSSuH except for the omission of the C-terminal hepta-His peptide and a substitution of Val for Ile at residue 8 (not shown).
Analogous plasmids for transforming Escherichia coli BL21 (DE3), pETtpSSuH and pETSSuH, were constructed by inserting the NcoI-SalI fragments from pSSuH and ptpSSuH, respectively, into pET28a(+) (Novagen, Madison, WI).
Plastid Transformation and Plant Growth
Tobacco (cultivar Petit Havana [N,N]) leaves were transformed using the biolistic method (Svab and Maliga, 1993). Tissue from spectinomycin-resistant plantlets was put through two additional rounds of regeneration on regeneration medium (Carrer et al., 1993) containing 500 μg/mL spectinomycin before transformed plantlets were identified by PCR analysis of leaflet DNA isolated according to Saghai-Maroof et al. (1984) using two different pairs of primers: SSuHis3 with TRNV (5′-CAGGCTCGAACTGATGACTTCCACC-3′) and AADr with PRV5P (Figure 1B). Two independently transformed lines of pSSuH (SSuH2 and SSuH5) and ptpSSuH (tpSSuH4 and tpSSuH7) transformants were maintained through two additional rounds of regeneration. Homoplasmicity then was confirmed by DNA gel blot analysis (Sambrook et al., 1989) of HindIII-digested leaf DNA probed with a 32P-labeled (Amersham Megaprime labeling kit) 787-bp HindIII-BamHI fragment of pRV112a derived from the flanking region adjacent to the insertion site (tgf; Figure 1B). Correct insertion into the plastid genome was confirmed by PCR using the primers AADAr (Figure 1B) and PRV5′ (5′-CCGAAGAGTAACTAGGACCAAT-3′). The latter primer anneals to a sequence in the tobacco plastome just beyond the right flanking region (Figure 1B) of the transforming plasmids (nucleotides 101,000 to 101,021 of inverted repeat A and 141,605 to 141,626 of inverted repeat B in the update of the sequence of Shinozaki et al. [1986] provided in GenBank accession number Z00044). Another transformed line (C3) was obtained using the pC3 plasmid, and its homoplasmicity was confirmed after several rounds of regeneration by the same procedures.
Regenerated transformed plantlets (and nontransformed controls) were transferred to 5-liter pots of soil and grown to maturity in an air-conditioned glasshouse. Pots were watered daily, and a complete nutrient solution was applied three times per week.
RNA Gel Blotting
Total RNA was extracted from young expanding leaves with the TriPure isolation reagent (Roche Molecular Biochemicals, Mannheim, Germany) according to the manufacturer's instructions. RNA pellets were dissolved in water and immediately electrophoresed, blotted, and cross-linked to Hybond N+ membranes (Amersham) as described by Kucharski and Maleska (1998). Duplicate blots were hybridized with two 32P-labeled DNA fragments derived from pBlueSSu (PpsbA and RbcS; Figure 1B). Membranes were exposed to a storage phosphor screen, and computer-generated images were analyzed using ImageQuant software (Molecular Dynamics [Sunnyvale, CA] PhosphorImager 400S).
Extraction of Rubisco from Tobacco Leaves
Tissue from young mature leaves (5 cm2) was frozen in liquid N2 and then ground in ice-cold glass homogenizers (Wheaton, Millville, NJ) in 1 mL of extraction buffer (0.1 M Tris-HCl, pH 8.0, 0.3 M NaCl, 20 mM imidazole, 5 mM MgCl2, and 1 mM phenylmethylsulfonyl fluoride). The extracts were centrifuged at 13,000g for 2 min at 4°C, and the pellet fraction was washed with three 1-mL volumes of extraction buffer before resuspension in SDS-containing sample buffer (NuPAGE; Novex, San Diego, CA) to a final volume of 0.2 mL and applied to NuPAGE electrophoresis gels (see below). His-tagged ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) complexes were isolated from the supernatant fraction as described below.
Extraction of Rubisco Small Subunits and Precursors from Escherichia Cells
Escherichia BL21 (DE3) cells transformed with plasmids pETtpSSuH and pETSSuH were grown at 28°C on a rotary shaker (150 rpm) in 0.3 liters of Luria-Bertani medium containing 30 μg/mL kanamycin. When cell absorbance (A600) reached 0.6, isopropyl-β-d-thiogalactopyranoside was added to 0.5 mM. After 150 min, the cells (A600 ∼1.3 to 1.4) were harvested by centrifugation at 3300g for 10 min at 4°C and resuspended in 10 mL of ice-cold extraction buffer. The cells were lysed by two passes through a prechilled French pressure cell at 140 MPa. The extract was centrifuged at 33,000g for 10 min at 4°C. His-tagged small subunits or precursors were isolated from the supernatant fraction as described below.
Affinity Purification of His-Tagged Rubisco Hexadecamers or Small Subunits
Total Rubisco (untagged plus His tagged, purified from leaves according to Whitney et al. [1999]) or the soluble protein fraction from extracts of leaves or Escherichia cells was mixed with Ni2+-nitrilotriacetic acid (Ni-NTA) agarose (Qiagen, Valencia, CA) by rotation at 4°C for 2 hr. The resin was collected into a Wizard minicolumn (Promega) and washed with 40 to 100 bed volumes of extraction buffer. Bound protein was collected by successive applications of 0.5 bed volumes of elution buffer (0.1 M Tris-HCl, pH 8.0, 0.3 M NaCl, and 200 mM imidazole). These washes were collected by centrifugation for 10 sec at 13,000g.
Electrophoresis and Immunoblotting Procedures
Proteins were separated by SDS-PAGE using NuPAGE 4 to 12% Bis-Tris gels (Novex) buffered with Mes according to the supplier's instructions. Protein bands were visualized by staining with Coomassie Brilliant Blue G 250 by using the Gelcode Blue reagent (Pierce Chemical Co.). Separated proteins were transferred to nitrocellulose (Hybond C; Amersham) by using an Xcell transfer cell (Novex). After blocking for 45 min with 3% (w/v) BSA (Sigma) in 10 mM Tris-HCl, pH 7.5, 150 mM NaCl (TBS), membranes were probed for 1 hr with antiserum raised in rabbits against Rubisco purified from tobacco according to Servaites (1985) (diluted 1:20,000 in TBS) or the PentaHis antibody preparation (Qiagen; diluted 1:1000 in TBS containing 3% [w/v] BSA). Membranes were then probed with appropriate secondary antibodies conjugated to alkaline phosphatase (Bio-Rad; diluted 1:2000 with TBS). Immunoreactive polypeptides were visualized by fluorescence with AttoPhos (JBL Scientific, San Luis Obispo, CA) by using a FluorImager (Molecular Dynamics). Band densities were measured with ImageQuant software.
35S Pulse–Chase Labeling
Ten leaf discs (0.97 cm2) from young fully expanded leaves of transformant tpSSuH4 growing in natural illumination ( ) were sampled, layered immediately on 5 mL of Murashige and Skoog (1962) nutrient solution, pH 7.2, containing 10 mM NaHCO3, 0.25% (v/v) Silwet-L77 (Lehle Seeds, Round Rock, TX), and 400 μCi of Trans35S-label (ICN, Costa Mesa, CA), and vacuum infiltrated for 90 sec. The leaf discs then were illuminated (600 μmol m−2 sec−1) for 20 min at 25°C, blotted briefly with blotting paper, rinsed by vacuum infiltration with 25 mL of water, blotted again, and vacuum infiltrated with Murashige and Skoog (1962) solution, pH 7.2, containing 10 mM NaHCO3 and 10 mM Met. After further illumination for various chase periods, duplicate leaf discs were frozen in liquid N2 and extracted with 0.4 mL of leaf extraction buffer. For analysis of total protein, 0.4 mL of Tris-glycine-SDS sample buffer (Novex) was added to one of the extracts. His-tagged Rubisco hexadecamers were isolated from the other extract by affinity purification as described above and eluted in a final volume of 50 μL, to which 50 μL of Tris-glycine-SDS buffer was added. Proteins in both samples were separated by SDS-PAGE, and the resulting gels were dried and exposed to a storage phosphor screen for 14 days.
) were sampled, layered immediately on 5 mL of Murashige and Skoog (1962) nutrient solution, pH 7.2, containing 10 mM NaHCO3, 0.25% (v/v) Silwet-L77 (Lehle Seeds, Round Rock, TX), and 400 μCi of Trans35S-label (ICN, Costa Mesa, CA), and vacuum infiltrated for 90 sec. The leaf discs then were illuminated (600 μmol m−2 sec−1) for 20 min at 25°C, blotted briefly with blotting paper, rinsed by vacuum infiltration with 25 mL of water, blotted again, and vacuum infiltrated with Murashige and Skoog (1962) solution, pH 7.2, containing 10 mM NaHCO3 and 10 mM Met. After further illumination for various chase periods, duplicate leaf discs were frozen in liquid N2 and extracted with 0.4 mL of leaf extraction buffer. For analysis of total protein, 0.4 mL of Tris-glycine-SDS sample buffer (Novex) was added to one of the extracts. His-tagged Rubisco hexadecamers were isolated from the other extract by affinity purification as described above and eluted in a final volume of 50 μL, to which 50 μL of Tris-glycine-SDS buffer was added. Proteins in both samples were separated by SDS-PAGE, and the resulting gels were dried and exposed to a storage phosphor screen for 14 days.
N-Terminal Protein Sequencing
N-terminal amino acid sequencing was performed on the small subunit bands obtained by SDS-PAGE from Rubisco preparations that had been purified by Ni-NTA agarose affinity chromatography from transformants tpSSuH4 and SSuH2. The separate bands corresponding to the two types of small subunits (His tagged and untagged) were excised from the gels, washed extensively with water, and freeze dried. The gel fragments were allowed to reswell in 10 mM 3-(cyclohexylamino)-1-propanesulfonic acid, pH 11.0, containing 0.2% SDS before 10 volumes of 0.1 M 3-(cyclohexylamino)-1-propanesulfonic acid was added. The polypeptides were allowed to adsorb passively overnight onto pieces of polyvinylidene difluoride membrane. Half of the membrane pieces were treated overnight with 70% formic acid to cleave any N-terminal formyl groups that might be present. Polypeptides on all membrane pieces were sequenced using an Applied Biosystems Procise-cLC sequencer equipped with a 0.8 × 250-mm phenylthiohydantoin column.
Mass Spectrometric Analyses
Total Rubisco, purified from the wild type and tpSSuH and C3 transformants as described by Whitney et al. (1999), was denatured by dilution with 4 volumes of 0.1 M Tris-HCl buffer, pH 7.2, containing 6 M guanidine-HCl. The large and small subunits were separated by gel filtration on a Beckman (San Ramon, CA) Ultraspherogel SEC 4000 column (7.5 × 300 mm) equilibrated with the same buffer. Samples from the small subunit peaks were prepared for electrospray ionization mass spectrometry (ESI-MS) as described by Neylon et al. (2000). Mass spectra were obtained with a VG Biotech (Altrincham, Cheshire, UK) Quattro II mass spectrometer.
Acknowledgments
We thank G. Hudson for contributing to the construction of plasmid pC3, P. Maliga for plasmids and advice about the biolistic technique, and H. Kane for reading the manuscript.
References
- Ausubel, F.M. (1988). Current Protocols in Molecular Biology. (New York: John Wiley and Sons).
- Brutnell, T.P., Sawers, R.J.H., Mant, A., and Langdale, J.A. (1999). BUNDLE SHEATH DEFECTIVE 2, a novel protein required for post-translational regulation of the rbcL gene of maize. Plant Cell 11 849–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butz, N.D., and Sharkey, T.D. (1989). Activity ratios of ribulose-1,5-bisphosphate carboxylase accurately reflect carbamylation ratios. Plant Physiol. 89 735–739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrer, H., Hockenberry, T.N., Svab, Z., and Maliga, P. (1993). Kanamycin resistance as a selectable marker for plastid transformation in tobacco. Mol. Gen. Genet. 241 49–56. [DOI] [PubMed] [Google Scholar]
- Delwiche, C.F., and Palmer, J.D. (1996). Rampant horizontal transfer and duplication of Rubisco genes in eubacteria and plastids. Mol. Biol. Evol. 13 873–882. [DOI] [PubMed] [Google Scholar]
- Fargo, D.C., Boynton, J.E., and Gillham, N.W. (1999). Mutations altering the predicted secondary structure of a chloroplast 5′ untranslated region affect its physical and biochemical properties as well as its ability to promote translation of reporter mRNAs both in the Chlamydomonas reinhardtii chloroplast and in Escherichia coli. Mol. Cell. Biol. 19 6980–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141 233–357. [DOI] [PubMed] [Google Scholar]
- Grimm, R., Grimm, M., Eckerskorn, C., Pohlmeyer, K., Röhl, T., and Soll, J. (1997). Postimport methylation of the small subunit of ribulose-1,5-bisphosphate carboxylase in chloroplasts. FEBS Lett. 408 350–354. [DOI] [PubMed] [Google Scholar]
- Gruissem, W., Barkan, A., Deng, X.-W., and Stern, D. (1988). Transcriptional and post-transcriptional control of plastid mRNA levels in higher plants. Trends Genet. 4 258–263. [DOI] [PubMed] [Google Scholar]
- Houtz, R.L., Stults, J.T., Mulligan, R.M., and Tolbert, N.E. (1989). Post-translational modifications in the large subunit of ribulose bisphosphate carboxylase/oxygenase. Proc. Natl. Acad. Sci. USA 86 1855–1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, G.S., Evans, J.R., von Caemmerer, S., Arvidsson, Y.B.C., and Andrews, T.J. (1992). Reduction of ribulose-1,5-bisphosphate carboxylase/oxygenase content by antisense RNA reduces photosynthesis in transgenic tobacco plants. Plant Physiol. 98 294–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivey, R.A., Subramanian, C., and Bruce, B.D. (2000). Identification of a Hsp70 recognition domain within the Rubisco small subunit transit peptide. Plant Physiol. 122 1289–1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanevski, I., and Maliga, P. (1994). Relocation of the plastid rbcL gene to the nucleus yields functional ribulose-1,5-bisphosphate carboxylase in tobacco chloroplasts. Proc. Natl. Acad. Sci. USA 91 1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegstra, K., and Cline, K. (1999). Protein import and routing systems of chloroplasts. Plant Cell 11 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein, R.R., and Salvucci, M.E. (1992). Photoaffinity labeling of mature and precursor forms of the small subunit of ribulose-1,5-bisphosphate carboxylase/oxygenase after expression in Escherichia coli. Plant Physiol. 98 546–553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski, R., and Maleska, R. (1998). Arginine kinase is highly expressed in the compound eye of the honey bee, Apis mellifera. Gene 211 343–349. [DOI] [PubMed] [Google Scholar]
- Mayfield, S.P., Yohn, C.B., Cohen, A., and Danon, A. (1995). Regulation of chloroplast gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46 147–166. [Google Scholar]
- Mazur, B.J., and Chui, C.-F. (1985). Sequence of a genomic DNA clone for the small subunit of ribulose bisphosphate carboxylase-oxygenase from tobacco. Nucleic Acids Res. 13 2373–2386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel, H., Hunt, D.F., Shabanowitz, J., and Bennett, J. (1988). Tandem mass spectrometry reveals that three photosystem II proteins of spinach chloroplasts contain N-acetyl-O-phosphothreonine at their NH2 termini. J. Biol. Chem. 263 1123–1130. [PubMed] [Google Scholar]
- Morton, B.R., and Levin, J.A. (1997). The atypical codon usage of the plant psbA gene may be the remnant of an ancestral bias. Proc. Natl. Acad. Sci. USA 94 11434–11438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller, K.D., Salnikow, J., and Vater, J. (1983). Amino-acid sequence of the small subunit of d-ribulosebisphosphate carboxylase oxygenase from Nicotiana tabacum. Biochim. Biophys. Acta 742 78–83. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nakamura, Y., Gojobori, T., and Ikemura, T. (1999). Codon usage tabulated from the international DNA sequence databases; its status 1999. Nucleic Acids Res. 27 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neylon, C., Brown, S.E., Kralicek, A.V., Miles, C.S., Love, C.A., and Dixon, N.E. (2000). Interaction of the Escherichia coli replication terminator protein (Tus) with DNA: A model derived from DNA-binding studies of mutant proteins by surface plasmon resonance. Biochemistry 39 11989–11999. [DOI] [PubMed] [Google Scholar]
- Rodermel, S. (1999). Subunit control of Rubisco biosynthesis: A relic of an endosymbiotic past? Photosynth. Res. 59 105–123. [Google Scholar]
- Roy, H., and Andrews, T.J. (2000). Rubisco: Assembly and mechanism. In Photosynthesis: Physiology and Metabolism, R.C. Leegood, T.D. Sharkey, and S. von Caemmerer, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 53–83.
- Ruuska, S., Andrews, T.J., Badger, M.R., Hudson, G.S., Laisk, A., Price, G.D., and von Caemmerer, S. (1998). The interplay between limiting processes in C-3 photosynthesis studied by rapid-response gas exchange using transgenic tobacco impaired in photosynthesis. Aust. J. Plant Physiol. 25 859–870. [Google Scholar]
- Saghai-Maroof, M.A., Soliman, K.M., Jorgensen, R.A., and Allard, R.W. (1984). Ribosomal DNA spacer-length polymorphisms in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Servaites, J.C. (1985). Crystalline ribulose bisphosphate carboxylase/oxygenase of high integrity and catalytic activity from Nicotiana tabacum. Arch. Biochem. Biophys. 238 154–160. [DOI] [PubMed] [Google Scholar]
- Sharma, J., Panico, M., Shipton, C.A., Nilsson, F., Morris, H., and Barber, J. (1997). Primary structure characterization of the photosystem II D1 and D2 subunits. J. Biol. Chem. 272 33158–33166. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome, its gene organization and expression. EMBO J. 5 2043–2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somanchi, A., and Mayfield, S.P. (1999). Nuclear-chloroplast signalling. Curr. Opin. Plant Biol. 2 404–409. [DOI] [PubMed] [Google Scholar]
- Staub, J.M., and Maliga, P. (1993). Accumulation of D1 polypeptide in tobacco plastids is regulated via the untranslated region of the psbA mRNA. EMBO J. 12 601–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staub, J.M., and Maliga, P. (1994). Translation of psbA mRNA is regulated by light via the 5′-untranslated region in tobacco plastids. Plant J. 6 547–553. [DOI] [PubMed] [Google Scholar]
- Svab, Z., and Maliga, P. (1993). High-frequency plastid transformation in tobacco by selection for a chimeric aadA gene. Proc. Natl. Acad. Sci. USA 90 913–917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whatley, J.M. (1993). The endosymbiotic origin of chloroplasts. Int. Rev. Cytol. 144 259–299. [Google Scholar]
- Whitney, S.M., von Caemmerer, S., Hudson, G.S., and Andrews, T.J. (1999). Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiol. 121 579–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying, Z.T., Mulligan, R.M., Janney, N., and Houtz, R.L. (1999). Rubisco small and large subunit N-methyltransferases: Bi- and mono-functional methyltransferases that methylate the small and large subunits of Rubisco. J. Biol. Chem. 274 36750–36756. [DOI] [PubMed] [Google Scholar]
- Zoubenko, O.V., Allison, L.A., Svab, Z., and Maliga, P. (1994). Efficient targeting of foreign genes into the tobacco plastid genome. Nucleic Acids Res. 22 3819–3824. [DOI] [PMC free article] [PubMed] [Google Scholar]