Abstract
Certain mutations isolated in the 5′ untranslated region (5′UTR) of the chloroplast rps7 gene in Chlamydomonas reduce expression of reporter genes. Second site suppressors in this 5′UTR sequence restore reporter expression. 5′UTR sequences with the original mutations fail to bind a 20-kD protein, one of five proteins that bind to leaders of several chloroplast genes. However, 5′UTRs from suppressed mutants restore binding to this protein but do not bind a 47-kD protein present on the wild type and the original mutant 5′UTRs. The 20-kD protein was shown to be the S7 protein of the chloroplast ribosomal small subunit encoded by rps7, whereas the 47-kD protein was shown to be RB47, a poly(A) binding protein. Our data are consistent with the hypothesis that the S7 protein plays either a general or a specific regulatory role in translation initiation in the chloroplast.
INTRODUCTION
Regulation of translation is an important determinant of chloroplast gene expression (reviewed in Danon, 1997; Goldschmidt- Clermont, 1998; Hauser et al., 1998; Sugiura et al., 1998). Complex interactions between the mRNA, the ribosomal preinitiation complex, and other trans-acting factors (many of which are encoded by nuclear genes) are essential for translation initiation in chloroplasts (reviewed in Rochaix, 1996; Hauser et al., 1998). Although the chloroplast translational machinery is prokaryotic in many respects (Harris et al., 1994), differences in the mechanisms for translation initiation are emerging between the two systems. In Escherichia coli, translation initiation is predominantly dependent on interactions between the anti-Shine-Dalgarno (SD) sequence (CCUCC) near the 3′ end of the 16S small subunit rRNA and the SD sequence in the mRNA (GGAGG) located 5 to 9 ± 2 nucleotides upstream of the initiation AUG codon. In contrast, translation initiation in chloroplasts (Fargo et al., 1998; reviewed in Hauser et al., 1998) as well as in mitochondria (Dunstan et al., 1997; reviewed in Fox, 1996) seems not to require an SD-based association of the mRNA leader and the small subunit rRNA. Instead, initiation is likely contingent on interactions that include the association of the higher order structure of the 5′ untranslated region (5′UTR) with trans-acting factors. Such mRNA–protein complexes may then bind to the preinitiation complex by forming an association with ribosomal proteins and/or the small subunit rRNA.
To examine the cis-acting sequences required for initiation of chloroplast translation, we generated random mutations in the 5′UTR from the Chlamydomonas chloroplast rps7 mRNA encoding the S7 ribosomal protein. These leaders were linked to the lacZ′ reporter gene and assayed for activity in E. coli using a blue-white colony screen (Fargo et al., 1999). These analyses revealed that transformants with seven of 12 mutant leaders accumulated abundant reporter mRNA that failed to be translated. These leader sequences were then individually linked upstream of an aadA reporter gene encoding aminoglycosyl adenyltransferase and assayed in both E. coli and Chlamydomonas chloroplasts. The reporter genes exhibited a spectrum of responses ranging from zero to wild-type levels of activity, with the response of each mutant being similar in E. coli and the Chlamydomonas chloroplast. Each of the original 5′UTR mutations greatly altered the predicted secondary structure of the rps7 5′UTR. Therefore, strains with compensating second site cis-acting mutations that restored the predicted secondary structure of the rps7 5′UTR were generated to determine whether RNA structural changes were responsible for the diminished translational activity, as shown in Figure 1. UV melting and RNase T1 protection analyses of the wild-type, original mutant, and suppressor double mutant 5′UTRs indicated that the original mutants disrupted the predicted wild-type RNA folding pattern of the first two stem-loops. However, the suppressors restored the secondary structure. These data indicate that the wild-type folding pattern is likely essential for rps7 translation initiation (Fargo et al., 1999).
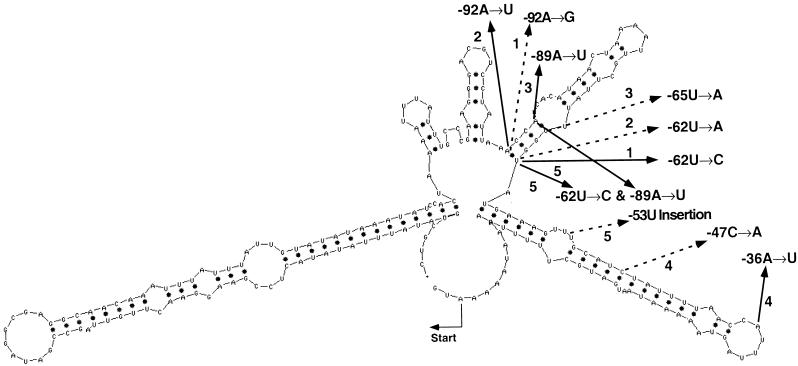
Figure 1.
MFOLD Predicted Secondary Structure at 25°C of the 225 Nucleotides Upstream of the Initiation Codon of the rps7 Gene from the Chlamydomonas Chloroplast.
The locations of four randomly induced mutations, and one site-directed double mutation, that block the expression of the lacZ′ or aadA reporter (Fargo et al., 1999) are indicated with solid arrows. Site-directed and in vivo–selected second site suppressor mutations are indicated with dashed arrows. Original mutant–suppressor mutant combinations include pair (1), −36A→U and −47C→A; pair (2), −62U→C and −92A→G; pair (3), −89A→U and −65U→A; pair (4), −92A→U and −62U→A; and pair (5) −62U→C + −89A→U and −53U insertion.
In this article, we examine whether translation initiation on the Chlamydomonas chloroplast-encoded rps7 5′UTR correlates with the presence or absence of specific trans-acting proteins. We identify five RNA binding proteins ranging from 16 to 80 kD that UV cross-linked to the wild-type rps7 5′UTR and show that each of the original mutants specifically failed to bind a 20-kD protein. The suppressor mutant 5′UTRs, which possess wild-type or near wild-type levels of the AAD reporter activity, restored binding of the 20-kD protein but were unable to bind a 47-kD protein associated with the 5′UTRs of the wild type and the original mutants. Immunoblot analyses identified the 20-kD protein as the S7 protein of the chloroplast ribosomal small subunit, encoded by the rps7 gene, and the 47-kD protein as RB47, a nuclear-encoded poly(A) binding protein reported to be specific for the psbA leader (Yohn et al., 1998). The S7 protein also bound to the rps12, rbcL, atpB, and psbA 5′UTRs, whereas the RB47 protein only bound to the rps7, rps12, and psbA leaders.
RESULTS
Loss of Translation Associated with Mutant rps7 5′UTRs Was Correlated with Diminished Polysome Association
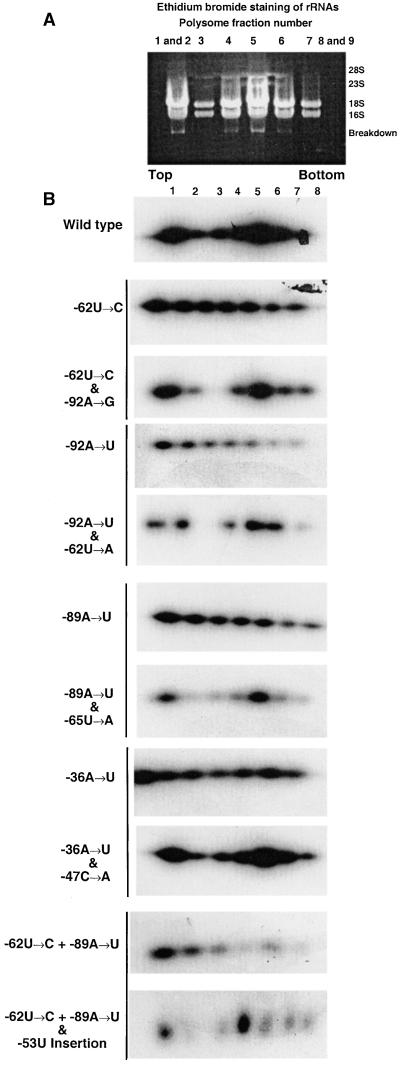
Because the original mutant rps7 5′UTRs were presumed to be unable to initiate translation of the aadA reporter mRNA in contrast to the suppressed mutants (see Figure 1; Fargo et al., 1999), we examined the polysome association profile of the wild type, the original mutants, and each original mutant in combination with its suppressor mutant. Ethidium bromide staining of rRNAs was conducted on a Chlamydomonas strain transformed with the wild-type rps7::reporter construct to identify the pattern of mRNA distribution on polysomes of various sizes throughout the sucrose gradient, as seen in Figure 2A. Polysomes of individual strains transformed with the wild-type, original mutant, and suppressor mutant rps7::reporter constructs were also characterized for aadA mRNA association, as illustrated in Figure 2B. A consistent pattern with two peaks of hybridization to the aadA probe, the first in lanes 1 and 2 and the second close to lanes 4 and 5, was found with the wild type and suppressor mutants. In contrast, the original mutants displayed decreasing hybridization in the fractions toward the bottom of the gradient. The observed decreasing hybridization pattern did not correspond to the pattern of rRNA staining shown in Figure 2A. The original mutant fractions were heavily overexposed in this figure to make any peaks detectable. The reporter activity for a given rps7 5′UTR correlated well with the polysome profile, as indicated in Table 1, suggesting that AAD reporter enzyme activity and growth in selective antibiotics was directly related to the loading of reporter mRNAs with rps7 5′UTRs onto chloroplast polysomes.
Figure 2.
Separation of rps7::aadA Polysomes with Wild-Type, Original Mutant, and Suppressor Mutant 5′UTRs on Linear Sucrose Density Gradients.
(A) Ethidium bromide staining of rRNA from sucrose density gradient fractions of wild-type cells to identify the rRNA distribution pattern.
(B) Gel blots of total RNA from fractions 1 through 8 were probed with a 0.81-kb aadA fragment. The original mutant and corresponding suppressor mutant 5′UTRs are grouped in pairs designated by the horizontal line to the left of the blots. Exposures of the individual blots varied to determine relative amounts of reporter mRNA within a gradient profile. Therefore, no comparison of the amount of reporter mRNA between the gradients is possible. In general, the original mutant 5′UTRs were exposed longer than the wild-type and suppressor mutant 5′UTRs. The pattern of message accumulation on the original mutant gradients suggests a general decrease in accumulation along the gradient but does not correspond with the rRNA pattern shown in (A).
Table 1.
Relationship between Reporter Activity and Polysome Association for rps7 5′UTR Reporter Constructs in Chlamydomonas
| rps7 5′UTR | Origin | Spr Growth and AAD (%) Activitya | Polysome Associationb |
|---|---|---|---|
| Wild type | — | 100 | + |
| −62U→C | Random mutagenesis | 55 | − |
| −62U→C and −92A→G | Random mutagenesis, site-directed | 84 | + |
| −92A→U | Random mutagenesis | 24 | − |
| −92A→U and −62U→A | Random mutagenesis, site-directed | 89 | + |
| −89A→U | Random mutagenesis | 28 | − |
| −89A→U and −65U→A | Random mutagenesis, site-directed | 101 | + |
| −36A→U | Random mutagenesis | 13 | − |
| −36A→U and −47C→A | Random mutagenesis, in vivo selection | 79 | + |
| −62U→C + −89A→U | Random mutagenesis, site-directed | 7 | − |
| −62U→C + −89A→U and | Random mutagenesis, site-directed | 87 | + |
| −53U insertion | in vivo selection |
All values are standardized to those of Chlamydomonas chloroplast transformants with the wild-type rps7 reporter construct as 100%. Spectinomycin-resistant (Spr) growth and AAD activity is the average value of Spr growth and AAD activity, as described in Fargo et al. (1999).
(+) and (−) indicate the presence or absence, respectively, of an aadA mRNA hybridization pattern consistent with polysome association, as illustrated in Figure 2.
UV Cross-Linking of Chloroplast-Enriched Heparin-Actigel–Purified Proteins to Wild-Type, Mutant, and Suppressor Mutant rps7 5′UTRs
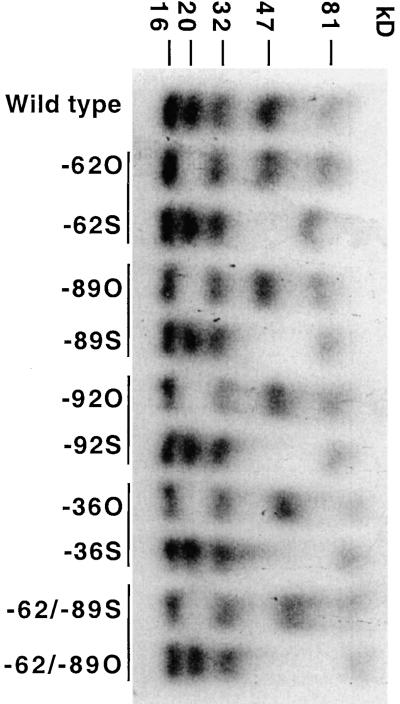
Each of the rps7 5′UTRs was transcribed in vitro with 32P-labeled UTP and UV cross-linked with chloroplast-enriched heparin-Actigel–purified proteins. As shown in Figure 3, a set of five proteins (81, 47, 32, 20, and 16 kD) was found to cross-link to the wild-type 5′UTR. 5′UTRs from each of the randomly selected or site-directed original mutants (−62U→C, −89A→U, −92A→U, −36A→U, −62U→C + −89A→U) failed to cross-link the 20-kD protein. In contrast, each of the site-directed or in vivo–selected suppressor mutant 5′UTRs (−62U→C and −92A→G, −89A→U and −65U→A, −92A→U and −62U→A, −36A→U and −89A→U, −62U→C + −89A→U and −53U insertion) restored the ability to cross-link the 20-kD protein but lost the ability to cross-link the 47-kD protein. All five mutants and their five suppressors had nucleotide substitutions that fall within the first two predicted stem-loop structures of the rps7 5′UTR, thus implicating this region of the leader in the binding of these two proteins, as seen in Figure 1.
Figure 3.
UV Cross-Linking of Chloroplast-Enriched Heparin-Actigel–Purified Proteins to 32P-Labeled rps7 5′UTRs Synthesized in Vitro from the Wild Type, Mutants, and Suppressor Mutants.
The original mutant and corresponding suppressor mutant 5′UTRs are grouped in pairs designated by the horizontal line on top of the lanes. Abbreviations in lanes for the original and suppressor mutants include −62U→C (−62O), −62U→C and −92A→G (−62S), −89A→U (−89O), −89A→U and −65U→A (−89S), −92A→U (−92O), −92A→U and −62U→A (−92S), −36A→U (−36O), −36A→U and −47C→A (−36S), −62U→C plus −89A→U (−62/−89O), and −62U→C plus −89A→U and −53U insertion (−62/−89S). Apparent molecular masses of the individual bound proteins are indicated at top in kilodaltons compared with prestained molecular mass standards.
Immunoblot Analyses of RNA Affinity-Purified Proteins
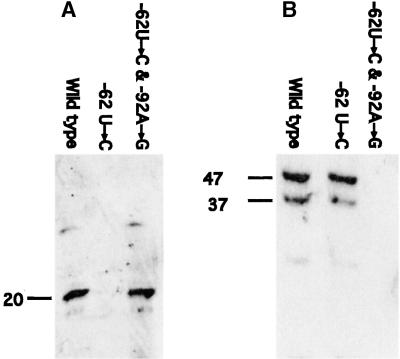
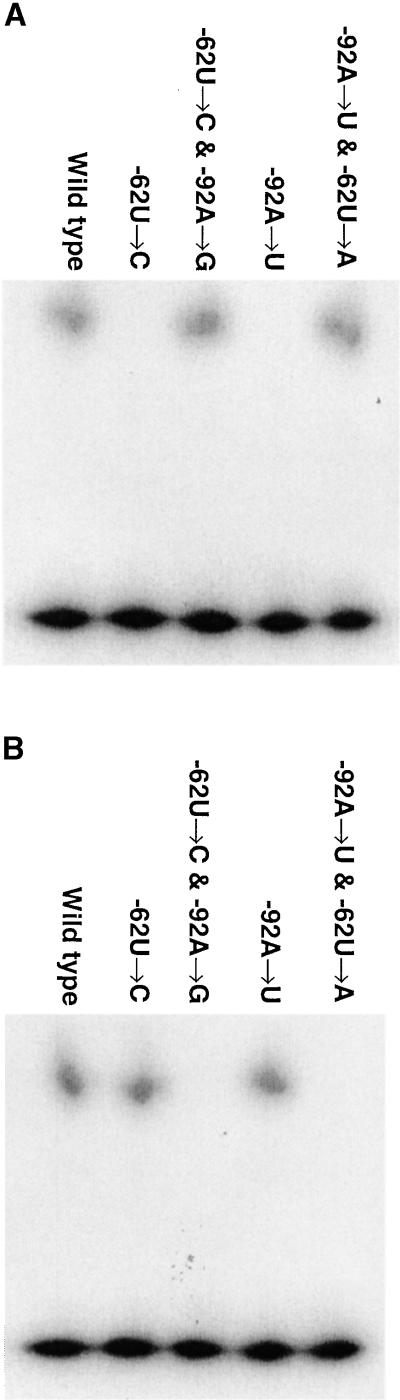
Chloroplast-enriched proteins were RNA affinity purified on columns containing rps7 5′UTRs from either the wild type, the −62U→C, and −92A→U original mutants or the suppressor mutants −62U→C and −92A→G as well as −92A→U and −62U→A. Figure 4A, showing immunoblot analyses using antisera raised against either the S7 or RB47 proteins, indicates that the 20-kD protein was affinity purified by the wild-type and the suppressor 5′UTRs but not by the mutant 5′UTRs. In contrast, Figure 4B reveals that the RB47 protein was affinity purified by the wild type and the original mutant but not by the suppressor mutant 5′UTRs. These data confirm the pattern seen in the UV cross-linking analyses and indicate that the 20-kD protein is the S7 protein and the 47-kD protein is the RB47 poly(A) binding protein described by Yohn et al. (1998) as specific for the psbA leader. Additional immunoblot analyses using antisera raised against other ribosomal proteins of ∼20 kD, including antisera against the 18-kD S19 and 17-kD S21 (Randolph-Anderson et al., 1989), showed no reaction with any of the proteins affinity purified by the rps7 5′UTR (data not shown).
Figure 4.
Immunoblot Analyses of RNA Affinity-Purified Proteins.
(A) RNA affinity-purified proteins were separated by gel electrophoresis and probed with the antisera raised against the recombinant S7 protein.
(B) RNA affinity-purified proteins were probed with the antisera raised against the recombinant RB47 protein. The lower molecular mass band at 37 kD is likely a breakdown product of RB47 (Yohn et al., 1998).
Apparent molecular masses are indicated at left in kilodaltons compared with prestained molecular mass standards.
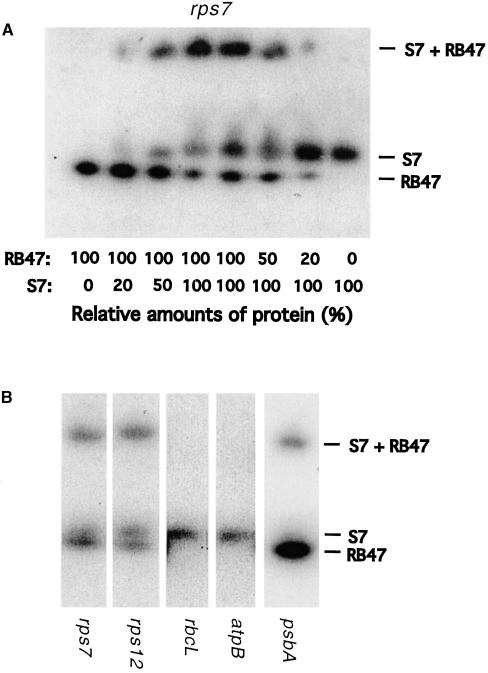
RNA Gel Mobility Shift with Purified Recombinant S7 and RB47 Proteins
DNA corresponding to the rps7 coding region was polymerase chain reaction amplified from Chlamydomonas and expressed in E. coli in a glutathione S-transferase (GST) fusion vector. The recombinant protein was purified on glutathione–Sepharose and isolated from the GST fusion by thrombin digestion. Recombinant RB47 protein (Yohn et al., 1998) was obtained from Stephen Mayfield. The purified recombinant proteins were individually used in RNA gel mobility shift assays to determine if they were able to bind to the rps7 5′UTRs from the wild type, original mutants, and suppressor mutants. The 5′UTRs from the wild type and the −62U→C and −92A→G or −92A→U and −62U→A suppressor mutants were shifted by the purified S7 protein, whereas 5′UTRs from the original mutants −62U→C and −92A→U were not, as shown in Figure 5A. The opposite is shown in Figure 5B for the RB47 protein with the wild type and original mutant 5′UTRs being shifted but not the suppressor mutant 5′UTRs. These data were consistent with the UV cross-linking and RNA affinity purification immunoblot assays regarding the binding of these two proteins to wild-type and the mutant rps7 leaders. In addition, both recombinant proteins were used at a range of concentrations in a gel mobility shift assay with the wild-type rps7 5′UTR. When equal amounts of the two proteins were used, three upshifted bands were seen on the native gel (Figure 6A). This suggested that the RB47 and S7 proteins bound and labeled the wild-type rps7 5′UTR at unique sites and that they bound cooperatively to form the slower moving band. Additionally, a decrease in either the S7 or the RB47 bands and in the S7 + RB47 slower moving band was observed when the amount of one of the individual proteins was reduced. The rps12, rbcL, atpB, and psbA 5′UTRs were also investigated for their ability to bind the recombinant S7 and RB47 proteins. The RB47 protein bound only the rps7, rps12, and psbA 5′UTRs, whereas the S7 protein was able to associate with all of the leaders, as illustrated in Figure 6B.
Figure 5.
rps7 5′UTR RNA Gel Mobility Shift Conducted with the Purified Recombinant S7 or RB47 Proteins.
rps7 5′UTRs from the wild type, the random mutagenesis–isolated −62U→C and −92A→U mutant, and the site-directed mutagenesis–generated −62U→C and −92A→G or −92A→U and −62U→A suppressor mutants were individually incubated with the purified recombinant proteins.
(A) Recombinant S7 protein.
(B) Recombinant RB47 protein.
Figure 6.
T1 RNase Digestion RNA Gel Mobility Shift Assays Conducted Using Both the Purified Recombinant S7 and RB47 Proteins Together.
(A) A wild-type rps7 5′UTR gel mobility shift was conducted using varying concentrations of the S7 and RB47 proteins ranging from 0 to 100% to determine if different binding sites were present.
(B) A gel mobility shift was conducted with equal amounts of the S7 and RB47 proteins using the wild-type rps7, rps12, rbcL, atpB, and psbA 5′UTRs.
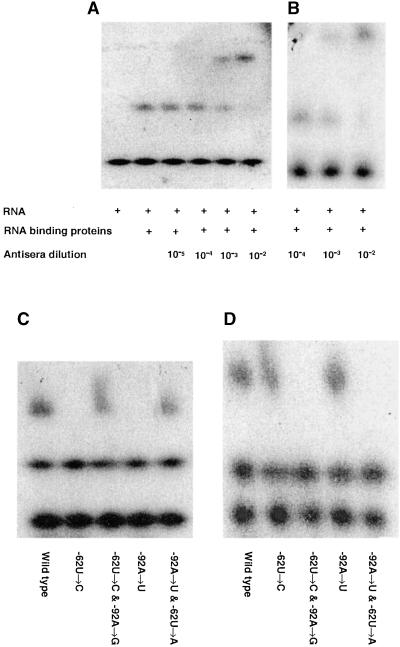
Gel Mobility Supershift Assays with RNA-Purified Proteins Using Antisera Raised against the S7 and RB47 Proteins
A wild-type rps7 5′UTR RNA-heparin-Actigel–purified protein complex was generated in a gel mobility shift assay, and the RB47 or S7 antisera were added at dilutions ranging from 10−5 to 10−2 to determine the minimum concentration at which a specific supershift would occur. The 10−3 and 10−2 dilutions of the S7 and RB47 antisera were able to generate supershifts as illustrated in Figures 7A and 7B, respectively. Chloroplast-enriched heparin-Actigel–purified proteins and rps7 5′UTRs from the wild type, the −62U→C and −92A→U mutants, and the −62U→C and −92A→G and −92A→U and −62U→A suppressor mutants were incubated individually with the RB47 and S7 antisera at a 10−3 dilution, as determined above. The S7 antiserum was able to shift the mobility of the rps7 5′UTR RNA–protein complexes from the wild type and the suppressor mutants but not that of the original mutants, as seen in Figure 7C. In contrast, the RB47 antiserum shifted the mobility of the wild-type and original mutant rps7 5′UTR RNA–protein complexes but not the suppressor mutant RNA–protein complexes, as illustrated in Figure 7D. This indicated an absence of the RB47 protein in the affinity-purified protein complexes from the suppressor mutant. These data agreed with the UV cross-linking and immunoblot analyses demonstrating a differential association of the wild-type, original mutant, and suppressor mutant rps7 5′UTRs with the S7 and RB47 RNA binding proteins.
Figure 7.
RNA Protein Gel Mobility Supershift Assay Conducted with Antisera Raised against the S7 or RB47 Proteins.
(A) and (B) Binding reaction with the wild-type rps7 5′UTR RNA and heparin-Actigel–purified proteins was conducted, and this complex was incubated with S7 or RB47 antisera at concentrations ranging from 10−5 to 10−2 to determine the optimum concentration for the gel mobility retardation assay. The (+) sign indicates the presence of the RNA and the RNA binding proteins in the individual gel lanes.
(C) and (D) rps7 5′UTRs from the wild-type, the −62U→C and −92A→U original mutants generated by random mutagenesis as well as suppressor mutants −62U→C and −92A→G or −92A→U and −62U→A created by site-directed mutagenesis were incubated individually with chloroplast-enriched heparin-Actigel–purified proteins and antisera at a 10−3 dilution.
(A) and (C) show antisera raised against S7 protein. (B) and (D) show antisera raised against RB47 protein.
DISCUSSION
In E. coli, association of r-proteins with the 5′UTRs of their mRNAs typically leads to the downregulation of the entire operon in which the genes encoding those mRNAs are located (Zengel and Lindahl, 1994; Spiridonova et al., 1998). These interactions have been well defined and are highly conserved throughout the prokaryotes (Nowotny and Nierhaus, 1988; Chiaruttini et al., 1989; Mandiyan et al., 1991; Dragon and Brakier-Gingras, 1993; Samaha et al., 1994; Spiridonova et al., 1998; Miyamoto et al., 1999). The S7 protein has been crystallized from Thermus thermophilus (Wimberly et al., 1997) and Bacillus stearothermophilus (Hosaka et al., 1997), and the active site structures needed for formation of the ribosomal small subunit and to bind the rpsG mRNA 5′UTR (rpsG is the functional equivalent of rps7 in Chlamydomonas chloroplasts) have been determined at a resolution of 1.9 Å (T. thermophilus) and 2.5 Å (B. stearothermophilus). These analyses have identified two separate potential sites for S7 binding to RNAs (Wimberly et al., 1997).
In many prokaryotes and in the majority of chloroplast genomes, the rps7 gene is located in the str operon and is the second gene downstream of the rps12 gene (Harris et al., 1994). However, in the chloroplast of Chlamydomonas, the rps7 gene is not located downstream of rps12 but is instead transcribed as part of a dicistronic message upstream of the atpE gene (Fargo et al., 1999). In E. coli, free S7 protein modulates its own synthesis by binding to rpsG at the 5′UTR. This sequesters the SD sequence in a conformation that prevents association with the preinitiation complex and downregulates translation of the rpsG mRNA (Dragon and Brakier-Gingras, 1993; Spiridonova et al., 1998). In contrast, the S7 protein in Chlamydomonas chloroplasts, which is 56% similar and 37% identical to the E. coli S7 protein (Harris et al., 1994), is clearly not used to sequester the SD sequence and block initiation. The rps7 5′UTR does not possess a putative SD sequence within the first 100 nucleotides of the initiation AUG codon (Fargo et al., 1998), and binding of S7 to the rps7 leader correlates with activation rather than repression of initiation. Therefore, the S7 protein likely plays a different regulatory role in chloroplasts than it does in E. coli.
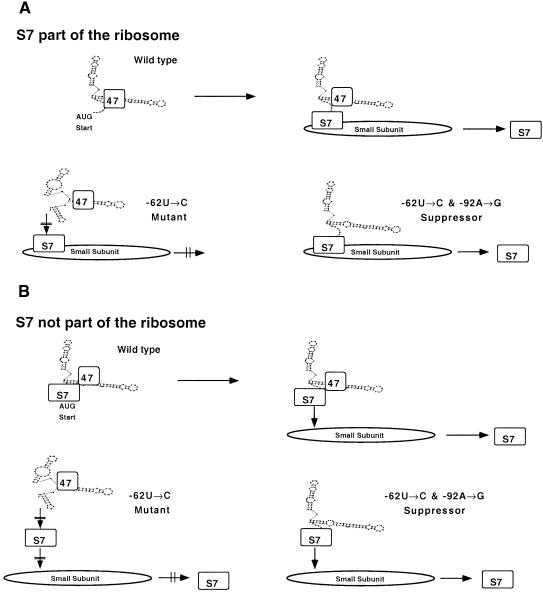
In both chloroplasts (reviewed by Hauser et al., 1998) and mitochondria (reviewed by Fox, 1996), a number of trans-acting factors have been identified that regulate translation via the 5′UTR. We have found that binding of the S7 protein is positively correlated with translation initiation from the rps7 leader, whereas binding of the RB47 protein is dispensable. Although a specific role for S7 in regulating only the expression of rps7 cannot be ruled out, S7 binds to several other 5′UTRs tested (rps12, rbcL, atpB, and psbA). One possible explanation is that S7 acts as a ribosomal docking site for messages like rps7 lacking an SD sequence, as illustrated in Figure 8A. This model predicts that 5′UTRs of mRNAs from the wild-type and suppressor mutant rps7 5′UTRs, as well as 5′UTRs from other genes, dock with the S7 protein in the chloroplast ribosomal small subunit and initiate translation. In contrast, the original mutant rps7 5′UTRs are unable to associate with S7 and to dock with the chloroplast ribosomal small subunit and therefore do not initiate translation. Given that affinity purification of free S7 might be easier than purification of S7 assembled into a ribosomal subunit due to better accessibility to the RNA ligand, an alternative model in which free S7 acts as an upregulator of translation initiation, should also be considered, as shown in Figure 8B. In this model, free S7 protein would bind to the rps7 5′UTR and leaders of several other chloroplast mRNAs (see Figure 6) to activate their translation. The model suggests that S7 would act as a positive facilitator of translation as the RB47 protein does for psbA (Yohn et al., 1998). Preliminary results (data not shown) in which purified small subunits of the chloroplast ribosome were incubated with the in vitro–transcribed wild-type rps7 5′UTR show that several r-proteins ranging from 15 to 61 kD cross-link to this leader and suggest that the 20-kD protein may fail to bind the original mutant leaders.
Figure 8.
Models for the Role of r-Protein S7 in Regulating Translation Initiation.
(A) Docking of chloroplast mRNAs occurs on the small subunit of the chloroplast ribosome. The S7 protein exposed on the surface of the small subunit of the chloroplast ribosome associates with the 5′UTR to facilitate translation initiation in the absence of an SD sequence.
(B) The S7 protein binds to the 5′UTR to permit association with the ribosome in the absence of an SD sequence.
The regulation of translation initiation of chloroplast-encoded mRNAs is likely dependent on several factors. First, the mRNA must fold into the appropriate shape to bind trans-acting factors. Some of these factors likely associate with 5′UTRs in a message-specific manner, whereas others may provide a general housekeeping function and bind to several or all chloroplast-encoded mRNAs (Hauser et al., 1998). After the folded mRNA and trans-acting factors have assembled, the complex is able to initiate translation. This may occur using ribosomal protein S7 as a docking site, whether or not an SD sequence is present in the leader. Binding of RB47 to the 5′UTR of psbA appears to regulate initiation (Yohn et al., 1998), whereas binding of this protein to the 5′UTR of rps7, and other chloroplast leaders, occurs in vitro, but it does not appear to be necessary for translation of their mRNAs in vivo.
METHODS
Strains and Culture Conditions
Chlamydomonas reinhardtii strains carrying the wild-type and mutant rps7 5′ untranslated regions (5′UTRs) ligated upstream of the aadA reporter construct (CC-) and plasmids with the same chimeric constructs (P-) described in this article are available from the Chlamydomonas Genetics Center (c/o Dr. Elizabeth Harris, DCMB Group, Box 91000, Duke University, Durham, NC 27708-1000). Escherichia coli and Chlamydomonas strains were grown and harvested for transformation, biochemical analysis, and molecular analysis, as described previously (Sambrook et al., 1989; Fargo et al., 1998).
Transformation of the aadA Reporter Constructs with Mutant rps7 Leaders into the Chlamydomonas Chloroplast and into E. coli
Transformation of the aadA reporter constructs with the original mutant and suppressor mutant rps7 leaders into the Chlamydomonas chloroplast was conducted using standard biolistic techniques and involved complementation of a partial atpB deletion mutant (CC-373) by a wild-type atpB gene linked to the reporter construct (Boynton et al., 1988; Fargo et al., 1999). E. coli cells (XL1-Blue and BL-21) were transformed by electroporation and transformants selected on Luria-Bertani plates containing 100 μg/mL ampicillin using standard techniques (Sambrook et al., 1989).
Protein Purification and in Vitro RNA Generation
Chloroplast proteins used in UV cross-linking assays were obtained from a cell-wall-free strain of C. reinhardtii (CC-400). Cells were grown in high salt-acetate (HSA) medium, as described elsewhere (Fargo et al., 1999), and gently broken in a Yeda press (Harris, 1989). The preparation was enriched for chloroplasts by centrifugation over a 10 to 80% percoll gradient (Zerges and Rochaix, 1994), and nucleic acid binding proteins were separated by heparin-Actigel chromotography, as described elsewhere (Danon and Mayfield, 1994).
The wild-type and mutant rps7 and wild-type rps12, rbcL, atpB, and psbA 5′UTRs were synthesized in vitro, as described elsewhere (Fargo et al., 1999), from an SKII+ plasmid with the T7 promoter upstream of sequences corresponding to (1) the wild-type 5′UTR (P-833); (2) three of the randomly generated mutant 5′UTRs affecting the second predicted stem-loop (−62U→C [P-848], −89A→U [P-849], and −92A→U [P-850]); (3) three of the 5′UTRs with the corresponding suppressor mutants (−62U→C and −92A→G [P-851], −89A→U and −65U→A [P-852], −92A→U and −62U→A [P-853]) generated by site-directed mutagenesis; (4) the 5′UTR with a randomly generated mutation in a loop (−36A→U [P-856]) and the in vivo–selected suppressor of that mutation (−36A→U and −47C→A [P-866]); (5) the double mutant (−62U→C and −89A→U [P-867]) generated by site-directed mutagenesis and the in vivo–selected suppressor of that mutation (−53U insertion [P-868]); and (6) the wild-type 5′UTRs from the Chlamydomonas chloroplast-encoded genes rps12 (P-877), rbcL (P-878), atpB (P-879), and psbA (P-880).
RNA Gel Blot Analyses
RNA was isolated for gel blot analysis from the transformed cells of E. coli and Chlamydomonas using standard techniques (Chomczynski and Sacchi, 1987; Fargo et al., 1998). Extracted RNA was separated on 1.1% agarose–formaldehyde gels, transferred to nylon membranes, and probed with a cloned fragment corresponding to the 0.81-kB aadA reporter coding region, as described elsewhere (Fargo et al., 1998). For each RNA sample, nucleic acid concentrations were measured spectrophotometrically and standardized to ensure equal loading of lanes. Stringent hybridization and washing conditions were used on all RNA gel blots. The probes were labeled with α-32P-dATP (New England Nuclear) using a random priming kit (Boehringer Mannheim). Blots were exposed to x-ray film (Kodak X OMAT-AR) at −70°C and were also quantified using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
RNA Affinity Purification of Proteins, Protein Electrophoresis, and Immunoblotting
Ten microliters of in vitro–synthesized RNA from wild-type and mutant rps7 5′UTRs was oxidized with 10 μL of 20 mM NaIO4 in dH2O for 30 min in the dark at 4°C. The reaction was quenched with 0.2% glycerol. This solution was mixed with 1 mL of Aminogel-B that had been washed with NaBH4 on a shaker for 2 hr at 25°C. Each matrix was then washed with 15 mL of high-salt buffer (20 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, 0.55 M KOAc) followed by a wash with 15 mL of low-salt buffer (20 mM Tris-HCl, pH 7.5, 3 mM MgCl2, 0.1 mM EDTA, 5 mM 2-mercaptoethanol, 0.1 M KOAc). Chloroplast-enriched, heparin-Actigel–purified Chlamydomonas proteins were then affinity purified over each matrix and eluted with 1.0 M KOAc.
Twenty micrograms of RNA affinity-purified proteins were mixed with a loading buffer (1% SDS, 0.5% mercaptoethanol, 30 mM Tris-HCl, pH 6.8, 0.01% bromophenol blue) and heated to 95°C for 3 min. Proteins were separated by 5% SDS-PAGE and stained with Coomassie Brilliant Blue R 250 or electroblotted to nitrocellulose. Blotted filters were treated as described elsewhere (Fargo et al., 1999), except that antisera specific for S7 or RB47 were used at a 10−3 dilution.
Polysome Fraction Isolation and mRNA Association Profile
Total Chlamydomonas polysomes were isolated as described elsewhere (Rott et al., 1998), with a few modifications. Thirty-milliliter cultures of log phase cells grown in HSA medium (2 × 106 cells/mL) were broken using a French press in a buffer containing 0.2 M Tris-HCl, pH 9.0, 0.2 M KCl, 35 mM MgCl2, 25 mM EGTA, 0.2 M sucrose, 1% Triton X-100, 2% polyoxyethylene-10-tridecyl ether, 0.5 mg/mL heparin, and 0.1 mg/mL chloramphenicol. After centrifugation at 3000g for 5 min at 4°C, the supernatant was adjusted to 0.5% Na-deoxycholate to solubilize microsomal membranes, incubated for 5 min on ice, and centrifuged for 15 min at 10,000g. Aliquots (0.5 mL) were layered onto 4.5-mL 15 to 55% linear sucrose gradients in a buffer containing 40 mM Tris-HCl, pH 8.0, 20 mM KCl, 10 mM MgCl2, 0.5 mg/mL heparin, and 0.1mg/mL chloramphenicol and centrifuged for 65 min at 50,000 rpm in a Beckman SW60 rotor. Fractions (0.5 mL) were collected, and RNA from each fraction was extracted with phenol-chloroform and ethanol precipitated. Electrophoresis of RNA, blotting, hybridization with random-primed 32P-labeled 0.81-kB aadA DNA fragment, and autoradiography were also conducted as described elsewhere (Fargo et al., 1999).
DNA Purification and Sequence Determination
DNA was purified using the Qiagen plasmid mini-purification kit protocol provided by the manufacturer. Individual DNA sequences were determined by using the ABI Prism Big Dye Terminator system. Reactions (10 μL) of each sample contained 4 μL of Terminator Ready Reaction Mix, 400 ng of double-stranded DNA template, and 15 ng of primer corresponding to 5′-TTTGTTGCCTCGCCTAATC-3′ for the SKII+ transformants or 5′-CTGGTTCCGCGTGGGATCCATGCCA-3′ for the pGEX transformants were used.
Amplification of the rps7 Coding Region, Expression of a GST-S7 Fusion Protein, and Purification of Recombinant S7 Protein
Total Chlamydomonas DNA was isolated from the wild type (CC-125), as described elsewhere (Lers et al., 1992). The coding region of the rps7 gene from the chloroplast was amplified by polymerase chain reaction using primers corresponding to the 5′UTR and the N-terminal region of the coding sequence 5′-TAAAATAAAAGGATCCATGCCACGTCGTCCCATTAATA-3′, with the initiation codon in boldface; and to the 3′UTR and the C-terminal region of the coding sequence 5′-TAATAATTGAATTCTTAATCAACTAATAAATTAATCGCA-3′, with BamHI and EcoRI restriction sites underlined. The amplified sequence was then digested with BamHI and EcoRI, placed into a pGEX-4T-1 vector, and transformed into BL-21 electrocompetent E. coli cells for expression of a GST-S7 fusion protein. The DNA sequence of the vector was then determined as described above to verify the transformation. The fusion protein was induced with isopropyl-β-d-galactosidase, the cells were lysed in a French press, and total E. coli protein was extracted. Purification of the fusion protein was conducted on a glutathione–Sepharose column. After purification, the native protein was released from the GST fusion by digestion with thrombin (the thrombin site is immediately upstream of the BamHI site in the vector). The native protein elute was then investigated for purity by polyacrylamide gel electrophoresis at 4°C followed by Coomassie blue staining. Stephen Mayfield supplied the purified recombinant RB47 protein (Yohn et al., 1998).
UV Cross-Linking Analyses with RNA-Purified Proteins or Proteins on Chloroplast Ribosomal Small Subunits
Conditions for UV cross-linking were as described (Fargo et al., 1999). Binding reactions (15 μL) were conducted using 7 μg of chloroplast-enriched heparin-Actigel–purified proteins or 0.5 OD260 of chloroplast ribosomal small subunits. The reactions were preincubated in 3 mM MgCl2 and 0.5 units Rnasin for 10 min at 22°C. E. coli tRNA (2.5 μg) was added as a nonspecific competitor, and 32P-UTP–labeled chloroplast 5′UTR probes (wild type and each mutant) were added. After incubation at 22°C for 15 min, the reactions were placed on ice and cross-linked with 254-nm UV irradiation 1.0 J/cm2 using a Stratalinker (Stratagene). RNA transcripts were then digested with 10 μg of RNase A (Sigma) for 30 min at 55°C and boiled for 1 min in protein loading buffer. SDS 5% polyacrylamide gel electrophoresis was followed by autoradiography with Kodak XAR5 film, and a PhosphorImager was used to visualize cross-linked labeled RNA–protein complexes, as described elsewhere (Fargo et al., 1999).
RNA–Protein Gel Mobility Shift Assays with Purified Recombinant S7 and RB47 Proteins and Gel Mobility Supershift Assays with Antisera Raised against the Proteins
For RNA mobility shift assays, individual in vitro–generated transcripts were incubated with various concentrations ranging to 7 μg of purified recombinant S7 or RB47 proteins with 100-fold excess E. coli tRNA in 3 mM MgCl2 and digested with 10 units of RNase T1, as described elsewhere (Hauser et al., 1998). For supershift assays, individual transcripts were incubated with chloroplast-enriched heparin-Actigel–purified proteins, as described above. After the 5′UTR RNA protein binding reactions (10 min at 25°C), various concentrations of the antisera raised against S7 or RB47 ranging from 10−5 to 10−2 dilution for wild-type and 10−3 for mutant and suppressor 5′UTRs were individually incubated with the RNA–protein solutions for 30 min at 25°C. Native 5% polyacrylamide gel electrophoresis and autoradiography followed (Fargo et al., 1999).
Analyses of Predicted RNA Secondary Structures
Predicted RNA secondary structures were generated using the MFOLD algorithm (Zuker, 1994) available at http://mfold1.wustl.edu/~mfold/rna/form1.cgi using default conditions at 25°C (temperature of Chlamydomonas growth) and 37°C (temperature of E. coli growth), as described elsewhere (Fargo et al., 1999).
Acknowledgments
We thank Stephen Mayfield for the purified RB47 protein as well as the RB47 antisera and Barbara Randolph-Anderson for the antisera raised against the S7 protein. Thanks also to Charles Hauser for incisive discussions leading to the docking model. This work was supported by National Institutes of Health Grant No. GM-19427.
References
- Boynton, J.E., Gillham, N.W., Harris, E., Hosler, A.M., Johnson, A.R., Jones, A., Randolph-Anderson, B., Robertson, D., Klein, K., Shark, K., and Sanford, J. (1988). Chloroplast transformation in C. reinhardtii with high velocity microprojectiles. Science 240 1534–1538. [DOI] [PubMed] [Google Scholar]
- Chiaruttini, C., Milet, M., Hayes, D.H., and Expert-Bezancon, A. (1989). Crosslinking of ribosomal proteins S4, S5, S7, S8, S11, S12 and S18 to domains 1 and 2 of 16S rRNA in the Escherichia coli 30S particle. Biochimie 71 839–852. [DOI] [PubMed] [Google Scholar]
- Chomczynski, P., and Sacchi, H. (1987). Single-step method of RNA isolation by acid guanidium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162 156–159. [DOI] [PubMed] [Google Scholar]
- Danon, A. (1997). Translational regulation in the chloroplast. Plant Physiol. 115 1293–1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danon, A., and Mayfield, S.P. (1994). ADP-dependent phosphorylation regulates RNA-binding in vitro: Implications in light-modulated translation. EMBO J. 13 2227–2235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragon, F., and Brakier-Gingras, L. (1993). Interaction of Escherichia coli ribosomal protein S7 with 16S rRNA. Nucleic Acids Res. 21 1199–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunstan, H.M., Green-Willms, N.S., and Fox, T.D. (1997). In vivo analysis of Saccharomyces cerevisiae COX2 mRNA 5′ untranslated leader functions in mitochondrial translation initiation and translational activation. Genetics 147 87–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fargo, D.C., Zhang, M., Gillham, N.W., and Boynton, J.E. (1998). Shine-Dalgarno-like sequences are not required for translation of chloroplast mRNAs in Chlamydomonas reinhardtii chloroplast or in Escherichia coli. Mol. Gen. Genet. 257 271–282. [DOI] [PubMed] [Google Scholar]
- Fargo, D.C., Gillham, N.W., and Boynton, J.E. (1999). Mutations altering the predicted secondary structure of a chloroplast 5′ untranslated region affect its physical and biochemical properties as well as its ability to promote translation of reporter mRNAs in both the Chlamydomonas reinhardtii chloroplast and in Escherichia coli. Mol. Cell. Biol. 19 6980–6990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox, T.D. (1996). Genetics of mitochondrial translation. In Translational Control, J. Hershey, M. Mathews, and N. Sonenberg, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 733–758.
- Goldschmidt-Clermont, M. (1998). Coordination of nuclear and chloroplast gene expression in plant cells. Int. Rev. Cytol. 177 115–180. [DOI] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook. (San Diego, CA: Academic Press, Inc.).
- Harris, E.H., Boynton, J.E., and Gillham, N.W. (1994). Chloroplast ribosomes and protein synthesis. Microbiol. Rev. 58 700–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser, C.R., Gillham, N.W., and Boynton, J.E. (1998). Regulation of chloroplast translation. In Molecular Biology of Chlamydomonas: Chloroplasts and Mitochondria, J.-D. Rochaix, M. Goldschmidt-Clermont, and S. Merchant, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 197–217.
- Hosaka, H., Nakagawa, A., Tanaka, I., Harada, N., Sano, K., Kimura, M., Yao, M., and Wakatsuki, S. (1997). Ribosomal protein S7: A new RNA-binding motif with structural similarities to a DNA architectural factor. Structure 5 1199–1208. [DOI] [PubMed] [Google Scholar]
- Lers, A., Heifetz, P.B., Boynton, J.E., Gillham, N.W., and Osmond, C.B. (1992). The carboxyl-terminal extension of the D1 protein of photosystem II is not required for optimal photosynthetic performance under CO2- and light saturated growth conditions. J. Biol. Chem. 267 17494–17497. [PubMed] [Google Scholar]
- Mandiyan, V., Tumminia, S.J., Wall, J.S., Hainfeld, J.F., and Boublik, M. (1991). Assembly of the Escherichia coli 30S ribosomal subunit reveals protein-dependent folding of the 16S rRNA domains. Proc. Natl. Acad. Sci. USA 88 8174–8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto, A., Usui, M., Yamasaki, N., Yamada, N., Kuwano, E., Tanaka, I., and Kimura, M. (1999). Role of the N-terminal region of ribosomal protein S7 in its interaction with 16S rRNA which binds to the concavity formed by the beta-ribbon arm and the alpha-helix. Eur. J. Biochem. 266 591–598. [DOI] [PubMed] [Google Scholar]
- Nowotny, V., and Nierhaus, K.H. (1988). Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry 27 7051–7055. [DOI] [PubMed] [Google Scholar]
- Randolph-Anderson, B.L., Gillham, N.W., and Boynton, J.E. (1989). Electrophoretic and immunological comparisons of chloroplast and prokaryotic ribosomal proteins reveal that certain families of large subunit proteins are evolutionarily conserved. J. Mol. Evol. 29 68–88. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.-D. (1996). Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas reinhardtii. Plant Mol. Biol. 32 327–341. [DOI] [PubMed] [Google Scholar]
- Rott, R., Haim, L., Drager, R.G., Stern, D.B., and Schuster, G. (1998). 3′-Processed mRNA is preferentially translated in Chlamydomonas reinhardtii chloroplasts. Mol. Cell. Biol. 18 4605–4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samaha, R.R., O'Brien, B., O'Brien, T.W., and Noller, H.F. (1994). Independent in vitro assembly of a ribonucleoprotein particle containing the 3′ domain of 16S rRNA. Proc. Natl. Acad. Sci. USA 91 7884–7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Spiridonova, V.A., Golovin, A.V., Drygin, D., and Kopylov, A.M. (1998). An extremely high conservation of RNA–protein S7 interactions during prokaryotic ribosomal biogenesis. Biochem. Mol. Biol. Int. 44 1141–1146. [DOI] [PubMed] [Google Scholar]
- Sugiura, M., Hirose, T., and Sugita, M. (1998). Evolution and mechanism of translation in chloroplasts. Annu. Rev. Genet. 32 437–459. [DOI] [PubMed] [Google Scholar]
- Wimberly, B.T., White, S.W., and Ramakrishnan, V. (1997). The structure of ribosomal protein S7 at 1.9 Å resolution reveals a beta-hairpin motif that binds double-stranded nucleic acids. Structure 5 1187–1198. [DOI] [PubMed] [Google Scholar]
- Yohn, C.B., Cohen, A., Danon, A., and Mayfield, S.P. (1998). A poly(A) binding protein functions in the chloroplast as a message-specific translation factor. Proc. Natl. Acad. Sci. USA 95 2238–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zengel, J.M., and Lindahl, L. (1994). Diverse mechanisms for regulation of ribosomal protein synthesis in Escherichia coli. Prog. Nucleic Acids Res. 47 331–370. [DOI] [PubMed] [Google Scholar]
- Zerges, W., and Rochaix, J.-D. (1994). The 5′ leader of a chloroplast mRNA mediates the translational requirements for two nucleus encoded functions in Chlamydomonas reinhardtii. Mol. Cell Biol. 14 5268–5277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. (1994). Prediction of RNA secondary structure by energy minimization. Methods Mol. Biol. 25 267–294. [DOI] [PubMed] [Google Scholar]