Abstract
Full-length cDNAs are essential for functional analysis of plant genes. Using the biotinylated CAP trapper method, we constructed full-length Arabidopsis cDNA libraries from plants in different conditions, such as drought-treated, cold-treated, or unstressed plants, and at various developmental stages from germination to mature seed. We prepared a cDNA microarray using ∼1300 full-length Arabidopsis cDNAs to identify drought- and cold-inducible genes and target genes of DREB1A/CBF3, a transcription factor that controls stress-inducible gene expression. In total, 44 and 19 cDNAs for drought- and cold-inducible genes, respectively, were isolated, 30 and 10 of which were novel stress-inducible genes that have not been reported as drought- or cold-inducible genes previously. Twelve stress-inducible genes were identified as target stress-inducible genes of DREB1A, and six of them were novel. On the basis of RNA gel blot and microarray analyses, the six genes were identified as novel drought- and cold-inducible genes that are controlled by DREB1A. Eleven DREB1A target genes whose genomic sequences have been registered in the GenBank database contained the dehydration-responsive element (DRE) or DRE-related CCGAC core motif in their promoter regions. These results show that our full-length cDNA microarray is a useful material with which to analyze the expression pattern of Arabidopsis genes under drought and cold stresses, to identify target genes of stress-related transcription factors, and to identify potential cis-acting DNA elements by combining the expression data with the genomic sequence data.
INTRODUCTION
Sequencing projects are producing large quantities of genomic and cDNA sequences for a number of organisms. In the model plant Arabidopsis, the complete genomic sequences of two chromosomes have been determined (Lin et al., 1999; Mayer et al., 1999), and the entire genomic sequence was completed by the end of 2000. Expressed sequence tag (EST) projects have also provided a major contribution with the discovery of expressed genes (Höfte et al., 1993; Newman et al., 1994; Cooke et al., 1996; Asamizu et al., 2000). A recent release of dbEST (the EST database of the National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov) contained partial cDNA sequences in which more than half of the total gene complement (i.e., ∼28,000 genes) is represented (as estimated from the gene content of the entirely sequenced chromosome 2 in Arabidopsis [Lin et al., 1999]). A major challenge for the coming decade is the functional analysis of this large set of genes.
Recently, microarray technology has become a useful tool for the analysis of genome-scale gene expression (Schena et al., 1995; Eisen and Brown, 1999). This DNA chip-based technology arrays cDNA sequences on a glass slide at a density >1000 genes/cm2. These arrayed sequences are hybridized simultaneously to a two-color fluorescently labeled cDNA probe pair prepared from RNA samples of different cell or tissue types, allowing direct and large-scale comparative analysis of gene expression. This technology was first demonstrated by analyzing 48 Arabidopsis genes for differential expression in roots and shoots (Schena et al., 1995). Microarrays were used to study 1000 randomly chosen clones from a human cDNA library for identification of novel genes responding to heat shock and protein kinase C activation (Schena et al., 1996). In another study, expression profiles of inflammatory disease-related genes were analyzed under various induction conditions by this chip-based method (Heller et al., 1997). Furthermore, the yeast genome of >6000 coding sequences has been analyzed for dynamic expression by the use of microarrays (DeRisi et al., 1997; Wodicka et al., 1997). However, in plant science, only several reports of microarray analyses have been published (Schena et al., 1995; Ruan et al., 1998; Aharoni et al., 2000; Reymond et al., 2000). We constructed Arabidopsis full-length cDNA libraries (Seki et al., 1998) from plants in different conditions, such as drought-treated and cold-treated plants, by using the biotinylated CAP trapper method (Carninci et al., 1996). In this study, we prepared an Arabidopsis full-length cDNA microarray using ∼1300 full-length cDNAs, including stress-inducible genes, to monitor the expression patterns of genes under drought and cold stresses.
Plant growth is greatly affected by environmental abiotic stresses, such as drought, high salinity, and low temperature. Plants respond and adapt to these stresses to survive under stress conditions. Among these abiotic stresses, drought or water deficit is the most severe limiting factor of plant growth and crop production. Drought stress induces various biochemical and physiological responses in plants. Recently, a number of genes have been described that respond to drought at the transcriptional level (Bohnert et al., 1995; Ingram and Bartels, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 1997, 1999, 2000). Stress-inducible genes have been used to improve the stress tolerance of plants by gene transfer (Holmberg and Bülow, 1998; Bajaj et al., 1999). It is important to analyze the functions of stress-inducible genes not only to understand the molecular mechanisms of stress tolerance and the responses of higher plants but also to improve the stress tolerance of crops by gene manipulation. Hundreds of genes are thought to be involved in abiotic stress responses. In the present study, we applied cDNA microarray analysis to identify new drought- or cold-inducible genes.
Dehydration-responsive element/C-repeat (DRE/CRT) has been identified as an important cis-acting element in drought-, high salt–, and cold stress–responsive gene expression in an abscisic acid (ABA)–independent manner (Yamaguchi-Shinozaki and Shinozaki, 1994; Thomashow, 1999; Shinozaki and Yamaguchi-Shinozaki, 2000). Transcription factors (DREB/CBF) involved in DRE/CRT-responsive gene expression have been cloned (Stockinger et al., 1997; Gilmour et al., 1998; Liu et al., 1998; Shinwari et al., 1998). DREB1/CBFs are thought to function in cold-responsive gene expression, whereas DREB2s are involved in drought-responsive gene expression. Strong tolerance to freezing stress was observed in transgenic Arabidopsis plants that overexpress CBF1 (DREB1B) cDNA under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Jaglo-Ottosen et al., 1998). We reported that overexpression of the DREB1A (CBF3) cDNA under the control of the CaMV 35S promoter or the stress-inducible rd29A promoter in transgenic plants gave rise to strong constitutive expression of the stress-inducible DREB1A target genes and increased tolerance to freezing, drought, and salt stresses (Liu et al., 1998; Kasuga et al., 1999). Previously (Kasuga et al., 1999), we identified six DREB1A target genes: rd29A/lti78/cor78, kin1, kin2/cor6.6, cor15a, rd17/cor47, and erd10. However, it is not well understood how overexpression of the DREB1A cDNA in transgenic plants increases stress tolerance to freezing, drought, and salt. To study the molecular mechanisms of drought and freezing tolerance, it is important to identify and analyze more genes that are controlled by DREB1A. Therefore, in the present study, we used cDNA microarray analysis to identify novel DREB1A target genes.
A complete list of ∼1300 Arabidopsis cDNA clones and the expression data from this study are available at http://rtcweb.rtc.riken.go.jp/lab/pmb.
RESULTS
Arabidopsis Full-Length cDNA Microarray
Using the biotinylated CAP trapper method, we constructed full-length cDNA libraries from Arabidopsis plants in different conditions, such as drought-treated, cold-treated, and unstressed plants, at various developmental stages from germination to mature seed (Seki et al., 1998). From the full-length cDNA libraries, we isolated ∼1300 independent Arabidopsis full-length cDNAs. We used a method described previously (Eisen and Brown, 1999) to array polymerase chain reaction (PCR)–amplified cDNA fragments onto glass slides. We prepared a full-length cDNA microarray containing 1300 Arabidopsis full-length cDNAs, including drought-inducible genes responsive to dehydration (rd) and early responsive to dehydration (erd) (Taji et al., 1999), as positive controls, the Arabidopsis α-tubulin gene (Ludwig et al., 1987) with same expression level in our experimental conditions as an internal control, and the mouse nicotinic acetylcholine receptor epsilon-subunit (nAChRE) gene and the mouse glucocorticoid receptor homolog gene, which have no substantial homology to any sequences in the Arabidopsis database, to assess for nonspecific hybridization as negative controls. To assess the reproducibility of the microarray technique, we arrayed the PCR products from each cDNA clone twice on each slide.
Isolation of Drought- and Cold-Inducible Genes by Use of the cDNA Microarray
The cDNA microarray hybridized with Cy3 and Cy5 fluorescently labeled probe pairs of drought-treated plants plus unstressed plants and cold-treated plants plus unstressed plants prepared as described in Methods. Figure 1 shows an image of a portion of the cDNA microarray. Dual labeling of cDNA probe pairs with Cy3-dUTP for one mRNA sample and Cy5-dUTP for the other sample allows simultaneous hybridization to DNA elements on microarrays and facilitates direct quantitative measurements of gene expression between two different conditions, stressed and unstressed. Hybridized microarrays were scanned by two separate laser channels for Cy3 and Cy5 emissions from each DNA element. The ratio of the two fluorescent signal intensities of each DNA element was then measured as a relative measure to determine changes in the differential expression of genes represented by cDNA spots on the microarrays. In this study, we used the α-tubulin gene as an internal control gene because its expression level is almost the same in the two experimental conditions we analyzed.
Figure 1.
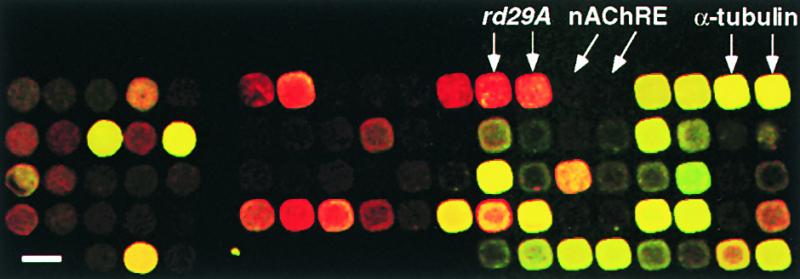
cDNA Microarray Analysis of Gene Expression under Cold Stress.
A fluorescently labeled cDNA was prepared from mRNA isolated from unstressed Arabidopsis plants by reverse transcription in the presence of Cy5-dUTP. A second probe, labeled with Cy3-dUTP, was prepared from cold-treated (2 hr) plants. After simultaneous hybridization of both probes with a cDNA microarray containing ∼1300 Arabidopsis cDNA clones, a pseudocolor image was generated. Genes induced and genes repressed after cold stress are represented as red and green signals, respectively. Genes expressed at approximately equal levels between treatments appear as yellow signals. The intensity of each spot corresponds to the absolute amount of expression of each gene. Cold-inducible genes (rd29A) are shown as red signals. α-Tubulin genes (internal control) are shown as yellow signals. No signals are shown for nAChRE.  .
.
Figure 2 shows the strategy for identification of drought- or cold-inducible genes. mRNAs from drought- or cold-treated plants and wild-type unstressed plants were used for the preparation of Cy3-labeled and Cy5-labeled cDNA probes, respectively. These cDNA probes were mixed and hybridized with the cDNA microarray. To assess the reproducibility of the microarray analysis, we repeated the same experiment five times (see Table 2). Hybridization of different microarrays with the same mRNA samples indicated good correlation. We regarded genes with an expression ratio (drought/unstressed or cold/unstressed) greater than twofold that of the α-tubulin gene as drought- or cold-inducible genes.
Figure 2.
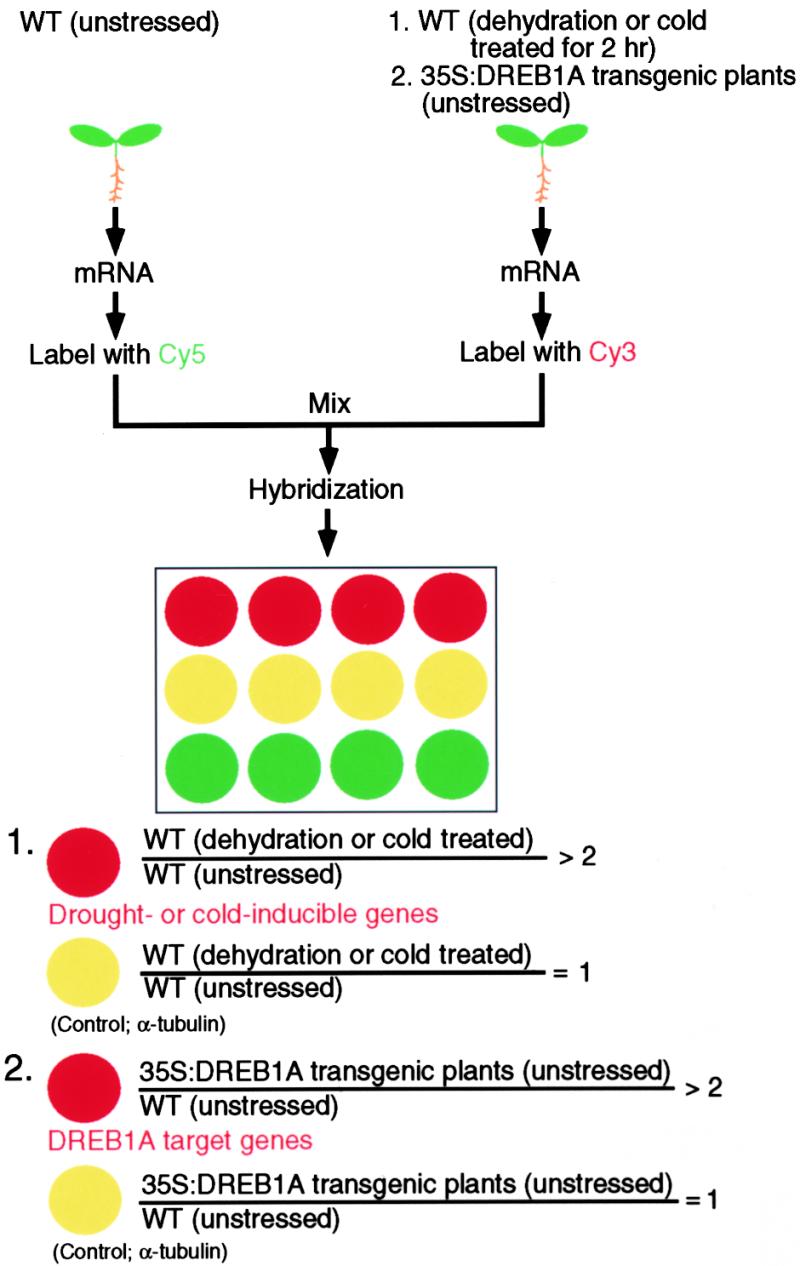
Strategy for the Identification of Drought- or Cold-Inducible Genes and DREB1A Target Genes.
(1) mRNAs from dehydrated or cold-treated plants and wild-type (WT) unstressed plants were used for preparation of Cy3-labeled and Cy5-labeled cDNA probes, respectively. These cDNA probes were mixed and hybridized with the cDNA microarray. In this study, we used the α-tubulin gene as an internal control gene because its expression level is almost the same in the two conditions. We regarded the genes with expression ratios (drought/unstressed or cold/unstressed) greater than twice that of α-tubulin as drought- or cold-inducible genes.
(2) mRNAs from 35S:DREB1A transgenic plants and wild-type (WT) unstressed plants were used for preparation of Cy3-labeled and Cy5-labeled cDNA probes, respectively. These cDNA probes were mixed and hybridized with the cDNA microarray. In this study, we used the α-tubulin gene as an internal control gene because its expression level is almost the same in the two conditions. We regarded the genes with expression levels more than two times greater in 35S:DREB1A transgenic plants than in wild-type unstressed plants as DREB1A target genes.
Table 2.
Drought- and Cold-Inducible Genes, and DREB1A Target Genes Identified by cDNA Microarray Analysis
Drought- and Cold-Inducible Genes Identified with the Full-Length cDNA Microarray
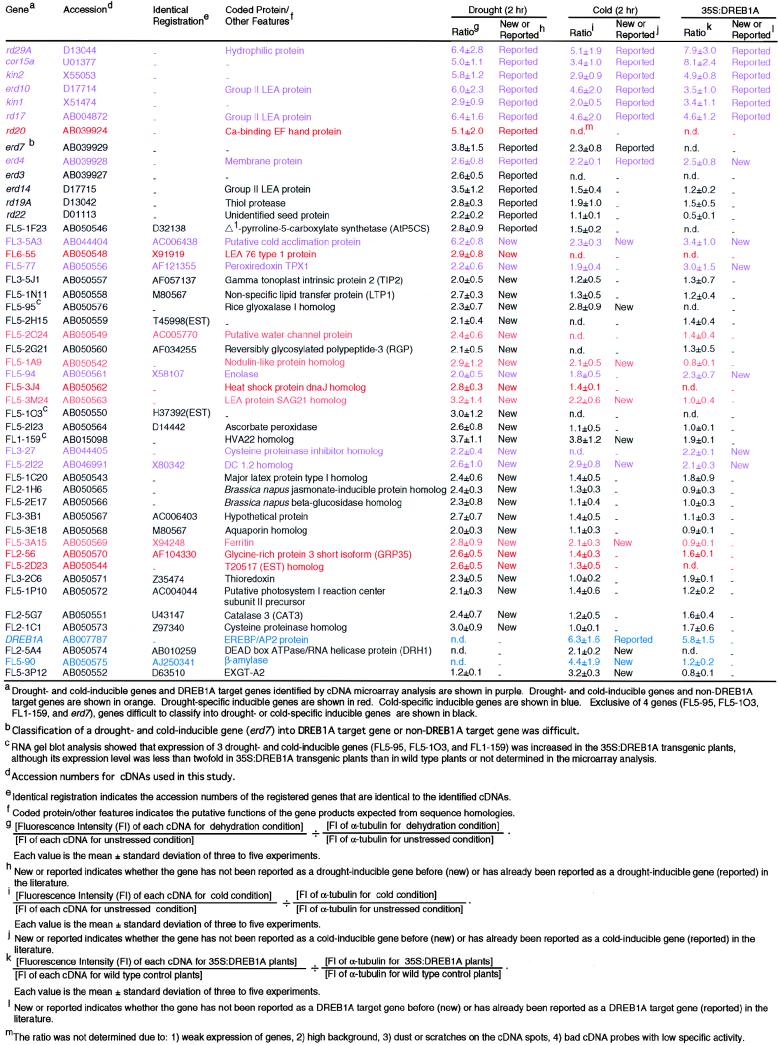
In total, 44 drought-inducible genes were identified by cDNA microarray analysis (Tables 1 and 2). Fourteen of these genes were reported to be drought-inducible genes, such as rd29A/cor78, cor15a, kin1, kin2, rd17/cor47, erd10, and rd20 (Kiyosue et al., 1994a; Bohnert et al., 1995; Ingram and Bartels, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 1997, 1999, 2000; Taji et al., 1999; Takahashi et al., 2000). These results showed that our cDNA microarray system functioned properly to find stress-inducible genes. Among the 30 new drought-inducible genes that have not been reported previously as drought inducible, we found cDNAs (FL3-5A3, FL6-55, FL5-1N11, FL5-2O24, FL5-2H15, and FL1-159) that are identical at the nucleotide level with putative cold acclimation protein (accession number AC006438), LEA 76 type 1 protein (accession number X91919), nonspecific lipid transfer protein (LTP1; accession number M80567), putative water channel protein (accession number AC005770), T45998 EST, and HVA22 homolog (accession number AB015098).
Table 1.
Drought- and Cold-Inducible Genes, and DREB1A Target Genes Identified by cDNA Microarray Analysis
| Genes | No. of Genes |
|---|---|
| Drought-inducible genes | 44 |
| New drought-inducible genesa | 30 |
| Cold-inducible genes | 19 |
| New cold-inducible genesb | 10 |
| DREB1A target genes | 12 |
| New DREB1A target genesc | 6 |
Drought-inducible genes that have not been reported as drought-inducible genes previously.
Cold-inducible genes that have not been reported as cold-inducible genes previously.
DREB1A target genes that have not been reported as DREB1A target genes previously.
Also, 19 cold-inducible genes were identified by the cDNA microarray analysis (Tables 1 and 2). Among them, nine were reported to be cold-inducible genes: rd29A, cor15a, kin1, kin2, rd17, erd10, erd7, erd4 (Kiyosue et al., 1994b; Shinozaki and Yamaguchi-Shinozaki, 1997, 1999, 2000; Taji et al., 1999; Thomashow, 1999), and DREB1A (Liu et al., 1998). Among the 10 new cold-inducible genes that have not been reported as cold inducible previously, we found cDNAs (FL3-5A3, FL5-3A15, FL5-3P12, FL5-90, FL5-2I22, and FL1-159) that are identical with putative cold acclimation protein (accession number AC006438), ferritin (accession number X94248), EXGT-A2 (accession number D63510), β-amylase (accession number AJ250341), DC 1.2 homolog (accession number X80342), and HVA22 homolog (accession number AB015098) and cDNAs (FL5-1A9, FL5-95, and FL5-3M24) showing sequence similarity with nodulin-like protein (accession number CAA22576), rice glyoxalase I (accession number AB017042), and LEA protein homolog (SAG21; accession number AF053065).
Stress-Inducible Genes That Are Controlled by the DREB1A Transcription Factor
We used the full-length cDNA microarray to identify stress-inducible genes that are controlled by the DREB1A transcription factor. The strategy for identification of DREB1A target genes is shown in Figure 2. mRNAs prepared from transgenic Arabidopsis plants that overexpress the DREB1A cDNA under the control of the CaMV 35S promoter (35S:DREB1A transgenic plants) and wild-type control plants were used for the preparation of Cy3-labeled and Cy5-labeled cDNA probes, respectively. These cDNA probes were mixed and hybridized with the cDNA microarray. We regarded genes with expression levels more than two times greater in the 35S:DREB1A transgenic plants than in wild-type control plants as DREB1A target genes.
In total, 12 DREB1A target genes were identified by cDNA microarray analysis (Tables 1 and 2). Among them, six were reported to be DREB1A target genes: rd29A/cor78, cor15a, kin1, kin2, rd17/cor47, and erd10 (Kasuga et al., 1999). Also, among the six novel DREB1A target genes that have not been reported as DREB1A target genes previously, we found cDNAs (FL3-5A3, FL5-2I22, FL5-94, and FL5-77) showing sequence identity with putative cold acclimation protein (accession number AC006438), DC 1.2 homolog (accession number X80342), enolase (accession number X58107), and peroxiredoxin TPX1 (accession number AF121355) and a cDNA (FL3-27) showing sequence similarity with cowpea cysteine proteinase inhibitor (accession number Z21954) and erd4 cDNA (Kiyosue et al., 1994b; Taji et al., 1999). The relationship between the six novel DREB1A target genes and stress tolerance is unelucidated and should be analyzed in the future.
RNA Gel Blot Analysis
In the cDNA microarray analysis, it is important to extract correct data with minimal effort. We evaluated the validity of the cDNA microarray analysis by the following methods. Initially, we identified 80 genes whose expression ratios (dehydration for 2 hr/unstressed) were more than twice that of α-tubulin. We performed RNA gel blot analysis of 80 putative drought-inducible genes and identified 44 as real drought-inducible genes. The inconsistency between the microarray results and the RNA gel blot results on several genes was due to (1) weak expression of genes, (2) high background, (3) dust or scratches on the cDNA spots, and (4) bad cDNA probes with low specific activity. Therefore, we flagged the experimental data mentioned above and excluded half of them from the subsequent analysis. After this data treatment, we identified 44 drought-inducible genes, 19 cold-inducible genes, and 12 DREB1A target genes based on cDNA microarray analysis. Then we performed RNA gel blot analysis to confirm the results obtained with the cDNA microarray. The results of expression data obtained by microarray analysis were in agreement with those obtained by RNA gel blot analysis in 44 drought-inducible genes, 19 cold-inducible genes, and 12 DREB1A target genes identified (data not shown). In Figure 3, the results of the microarray analysis are compared with those of RNA gel blot analysis for the six new DREB1A target genes (FL3-5A3, FL3-27, FL5-2I22, FL5-94, FL5-77, and erd4). All six genes were induced by dehydration and cold treatment and overexpressed in the 35S:DREB1A plants under unstressed conditions.
Figure 3.
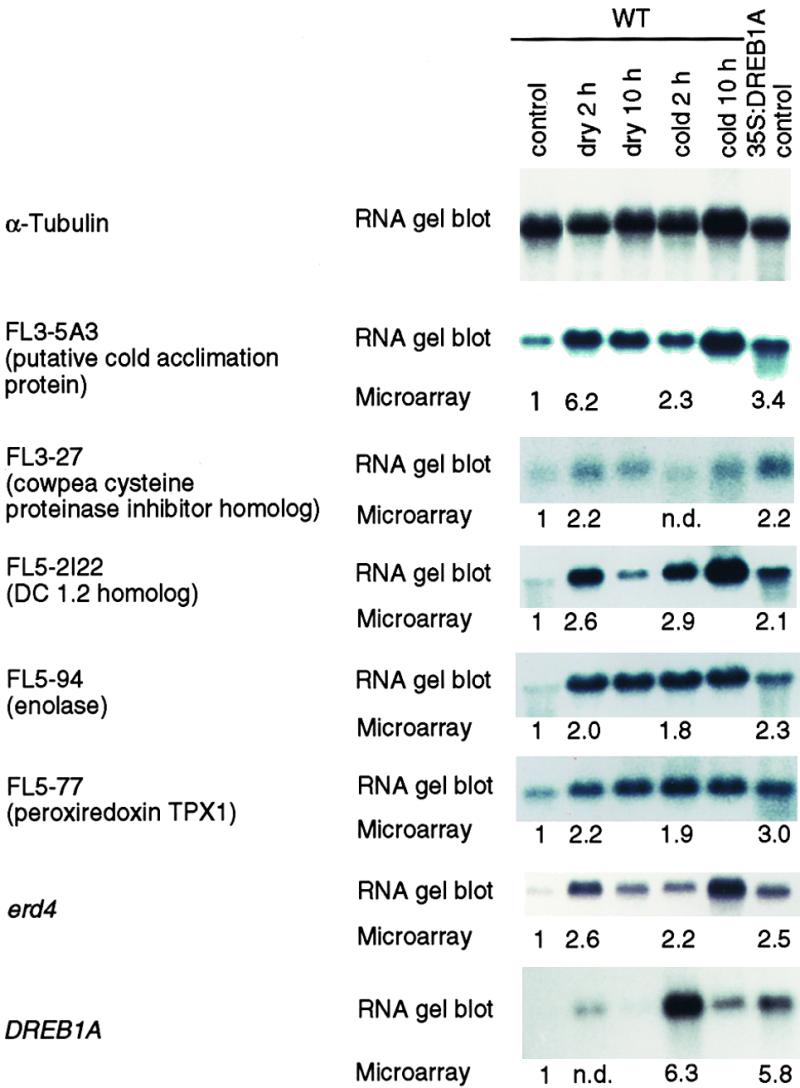
Comparison of cDNA Microarray and RNA Gel Blot Analysis for New DREB1A Target Genes and the DREB1A Gene.
Samples from dehydrated (dry 2 hr or dry 10 hr), cold-treated (cold 2 hr or cold 10 hr), or untreated 35S:DREB1A transgenic plants (35S:DREB1A control) were fluorescently labeled with Cy3-dUTP, and samples from untreated wild-type (WT) plants (control) were labeled with Cy5-dUTP. After hybridization with a cDNA microarray and scanning, relative expression ratios were calculated and are indicated below the RNA gel blots. The full-length cDNA sequences of three DREB1A target genes (FL3-5A3, FL3-27, and FL5-2I22) have been submitted to the GenBank, EMBL, and DDBJ databases with accession numbers AB044404, AB044405, and AB046991 respectively. h, hour; n.d., not determined.
DISCUSSION
Many Novel Drought- or Cold-Inducible Genes Could Be Identified with the Full-Length cDNA Microarray
Hundreds of plant genes are thought to be induced by stress, such as drought, high salinity, and low temperature, and function in the stress tolerance and responses of plants. However, only ∼50 drought- or cold-inducible genes have been reported to date (Shinozaki and Yamaguchi-Shinozaki, 1999; Thomashow, 1999). On the basis of the microarray and RNA gel blot analyses (drought 2 hr, 10 hr and cold 2 hr, 10 hr) of the drought- or cold-inducible genes identified, they were grouped as follows: (1) drought- and cold-inducible genes, (2) drought-specific inducible genes, and (3) cold-specific inducible genes. As a result, 20 of them are drought- and cold-inducible genes, five of them are drought-specific inducible genes, including four novel genes (FL6-55, FL5-2D23, FL2-56, and FL5-3J4), and two of them are cold-specific inducible genes, including one novel gene (FL5-90). However, 21 of the drought- or cold-inducible genes were difficult to classify into the three groups because of weak expression of genes or high background (Table 2 and Figure 4). On the basis of a search for homology in the GenBank database using the BLAST program, two cDNAs (FL6-55 and FL2-56) for drought-specific inducible genes were found to have sequence identity with LEA 76 type I protein (accession number X91919) and glycine-rich protein 3 short isoform (GRP3S; accession number AF104330); two drought-specific cDNAs (FL5-3J4 and FL5-2D23) were found to have sequence similarity to Borrelia burgdorferi heat shock protein dnaJ (accession number M96847) and T20517 EST; and one cDNA (FL5-90) for cold-specific inducible genes was found to have sequence identity with the gene for β-amylase (accession number AJ250341). Analyses of these novel drought- or cold-specific inducible genes will provide information about genes involved in stress tolerance and cis-acting promoter elements that function in drought- or cold-specific gene expression.
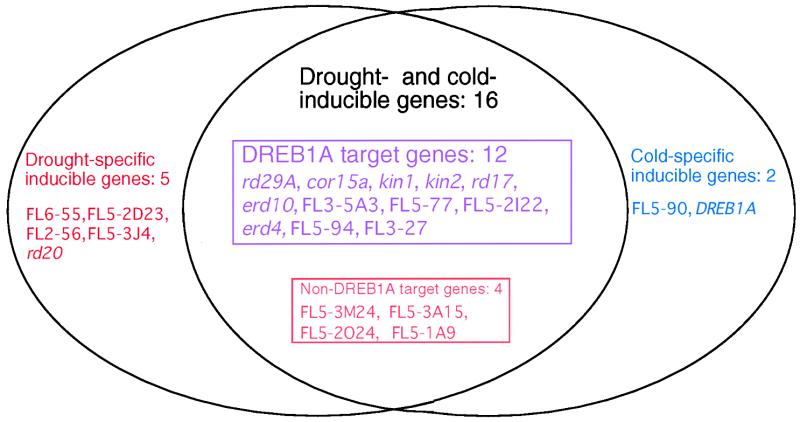
Figure 4.
Classification of the Drought- or Cold-Inducible Genes Identified into Four Groups on the Basis of RNA Gel Blot and Microarray Analyses.
The drought- or cold-inducible genes identified were grouped into the following three groups: (1) drought- and cold-inducible genes, (2) drought-specific inducible genes, and (3) cold-specific inducible genes. The following 21 genes were not grouped because of the difficulty of grouping them into the three groups: erd3, erd14, rd19A, rd22, FL5-1F23, FL3-5J1, FL5-1N11, FL5-2H15, FL5-2G21, FL5-2I23, FL5-1C20, FL2-1H6, FL5-2E17, FL3-3B1, FL5-3E18, FL3-2C6, FL5-1P10, FL2-5G7, FL2-1C1, FL2-5A4, and FL5-3P12. As a result, they were grouped as 20 drought- and cold-inducible genes, five drought-specific inducible genes, and two cold-specific inducible genes. Drought- and cold-inducible genes were classified into two groups: (1) DREB1A target genes and (2) non-DREB1A target genes. Sixteen drought- and cold-inducible genes were grouped as 12 DREB1A target genes and four non-DREB1A target genes. Four drought- and cold-inducible genes (FL5-95, FL5-1O3, FL1-159, and erd7) are not shown because (1) the expression level of three genes (FL5-95, FL5-1O3, and FL1-159) was less than twofold in 35S:DREB1A transgenic plants than in wild-type control plants or not determined in the cDNA microarray analysis, and (2) it was difficult to group erd7 into DREB1A target genes or non-DREB1A target genes.
Most of the identified genes are induced by both drought and cold stress (Table 2 and Figure 4) and contain the DRE or DRE-related CCGAC core motif in their promoters, as described below. One of the drought-specific inducible genes is rd20, which encodes a Ca2+ binding protein. The rd20 gene is induced by dehydration and high salinity but not by cold stress (Takahashi et al., 2000). The rd20 gene is induced by ABA treatment and contains the ABA-responsive element (ABRE) motif in its promoter, which indicates that part of drought-specific gene expression is regulated by an ABA-dependent pathway (Shinozaki and Yamaguchi-Shinozaki, 2000). In contrast, one of the cold-specific inducible genes encodes DREB1A/CBF3, a cold-inducible transcription factor with the AP2/ERF domain. The DREB1A/CBF3 promoter is thought to contain cis-acting elements involved in cold-specific gene expression (Gilmour et al., 1998; Shinwari et al., 1998). These results support the existence of different regulatory systems in drought- and cold-inducible gene expression (Shinozaki and Yamaguchi-Shinozaki, 2000).
Most of the Drought- and Cold-Inducible Genes Are Target Genes of DREB1A/CBF3
We have reported the presence of at least four signal transduction pathways that function in the activation of stress-inducible genes under drought conditions, two of which are ABA dependent and two of which are ABA independent (Shinozaki and Yamaguchi-Shinozaki, 1997, 1999, 2000). To understand the molecular mechanisms of gene expression in response to water stress, studies of cis- and trans-acting elements are important. A conserved sequence, PyACGTG(G/T)C, has been reported to function as an ABRE in many ABA-responsive genes (Ingram and Bartels, 1996; Bray, 1997; Shinozaki and Yamaguchi-Shinozaki, 1999). Also, a 9-bp conserved sequence, TACCGACAT, named the dehydration-responsive element (DRE), has been reported to be essential for regulation of the induction of rd29A expression under drought, low temperature, and high salt stress conditions, but it does not function as an ABRE (Yamaguchi-Shinozaki and Shinozaki, 1994). DRE and DRE-related core motifs (CCGAC), CRT, and LTRE have also been reported in the promoter regions of drought- and cold-inducible genes such as kin1, kin2, rd17/cor47, and cor15a (Table 3) (Baker et al., 1994; Wang et al., 1995; Iwasaki et al., 1997).
Table 3.
ABRE, DRE, and CCGAC Core Sequences Observed in the Promoter Regions of the DREB1A Target Genes Identified by cDNA Microarray Analysisa
| Gene | ABRE (PyACGTG (T/G) C) | DRE (TACCGACAT) | CCGAC Core Motif (CCGAC) |
|---|---|---|---|
| rd29A | TACGTGTC (−45 to −38)b | TACCGACAT (−148 to −140) | AGCCGACAC (−111 to −103) |
| TACCGACAT (−206 to −198) | GACCGACTA (−256 to −248) | ||
| cor15a | CACGTGGC (−132 to −125) | — | GGCCGACCT (−184 to −176) |
| GGCCGACAT (−361 to −353) | |||
| AACCGACAA (−416 to −424) | |||
| kin1 | — | TACCGACAT (−120 to −112) | ATCCGACAT (−720 to −712) |
| kin2 | CACGTGGC (−68 to −61) | TACCGACAT (−127 to −119) | CCCCGACGC (−403 to −395) |
| rd17 | TACGTGTC (−920 to −913) | — | TACCGACTT (−162 to −154) |
| AGCCGACCA (−967 to −959) | |||
| GACCGACAT (−997 to −989) | |||
| erd10 | CACGTGGC (−838 to −831) | — | GACCGACGT (−198 to −190)c |
| GACCGACCG (−202 to −194)c | |||
| CACCGACCG (−206 to −198)c | |||
| GACCGACAT (−999 to −991) | |||
| FL3-5A3 | CACGTGGC (−74 to −67) | TACCGACAT (−415 to −407) | TGCCGACAT (−806 to −798) |
| FL3-27 | — | TACCGACAT (−89 to −91) | — |
| FL5-2122 | — | — | TACCGACTC (−191 to −183) |
| TACCGACTA (−266 to −258) | |||
| TGCCGACCT (−418 to −410) | |||
| ACCCGACTA (−695 to −703) | |||
| GACCGACGT (−786 to −778) | |||
| FL5-77 | — | — | CCCCGACTA (−315 to −307) |
| FL5-94 | — | — | TACCGACTA (−190 to −198) |
| TTCCGACTA (−260 to −268) | |||
| ATCCGACGC (−630 to −622) |
ABRE, DRE, and CCGAC core sequences observed in 1000-bp upstream regions of the 5′ termini of the longest cDNA clones isolated are listed.
Numbers in parentheses indicate the nucleotide beginning at the 5′ terminus of the longest cDNA clone isolated. Minus signs indicate that the nucleotide exists upstream of the 5′ terminus of the putative transcription start site.
These 9-bp sequences that contain a CCGAC core motif overlap each other.
In this study, we identified 12 DREB1A target genes by cDNA microarray analysis. Among the six new DREB1A target genes, genomic sequences of the five target genes were registered in the GenBank database in October 2000. A 9-bp DRE sequence was observed in the promoter regions of genes corresponding to FL3-5A3 and FL3-27 cDNAs (Table 3). The CCGAC core sequence was observed in the promoter regions of genes corresponding to FL3-5A3, FL5-2I22, FL5-77, and FL5-94 cDNAs (Table 3). Most of the drought- and cold-inducible genes are DREB1A target genes and contain DRE or DRE-related CCGAC core motifs in their promoters (Table 3 and Figure 4). The ABRE sequence, PyACGTG(G/T)C, was observed in the promoter regions of six of the 12 DREB1A target genes identified (Table 3). This finding indicates that many drought- and cold-inducible genes are controlled by both ABA-dependent and ABA-independent pathways. However, expression of several drought- and cold-inducible genes (FL5-3M24, FL5-3A15, FL5-1A9, and FL5-2O24) was not increased in the 35S:DREB1A transgenic plants (Figure 4), indicating that these genes are not DREB1A target genes. The CCGAC core sequence was not observed in the 2000-bp upstream region of the 5′ terminus of the FL5-3M24 cDNA. These results suggest the existence of novel cis-acting elements involved in drought- and cold-inducible gene expression. By comparing the Arabidopsis full-length cDNA sequences with the Arabidopsis genomic sequences (the entire Arabidopsis genomic sequence was completed by the end of 2000), the promoter sequences and cis-acting elements of each gene can be studied on the basis of full-length cDNA sequences. These analyses will provide more information on gene expression and signal transduction in abiotic stress responses.
In the present study, we identified 12 DREB1A target genes containing six new target genes by using cDNA microarray analysis. However, the cDNA microarray analysis does not permit us to distinguish direct targets from indirect targets of the DREB1A gene. Resolving this issue and the complex interrelationships among the induced genes will require another approach, such as analysis for cis-acting elements or gel shift assays by using promoter regions of each stress-inducible gene.
Advantages and Disadvantages of the Full-Length cDNA Microarray
Several reports have been published on the use of plant EST microarrays. In the present study, we used an Arabidopsis full-length cDNA microarray, which has several advantages. Using the full-length cDNA microarray, it is easy to isolate full-length cDNAs for further functional analysis. Also, by comparing the Arabidopsis full-length cDNA sequences with the Arabidopsis genomic sequences, the promoter sequences and cis-acting elements of each gene can be studied on the basis of full-length cDNA sequences. Moreover, there is little cross-hybridization with pseudogenes compared with using the genomic DNA array. However, cross-hybridization between highly related sequences may occur in the full-length cDNA microarray (Richmond and Somerville, 2000). To avoid cross-hybridization problems, the use of oligonucleotide microarrays such as the Affymetrix GeneChip Array (Affymetrix, Inc., Santa Clara, CA) may be appropriate.
cDNA Microarray Technology
One of the major technical merits of microarray analysis is its high sensitivity in the detection of mRNAs. Sensitivity, which is determined by the minimal signal that can be reliably detected above the background, is dependent in part on background levels and on the specific incorporation of label into the probes. EST microarrays have been reported to detect 1:100,000 (w/w) (Ruan et al., 1998) or 1:500,000 (w/w) (Schena et al., 1996) of the total mRNAs. In our system, 3.3 pg out of 1 μg of poly(A)+ RNA (1:300,000) could be detected. These levels are generally considered sufficient to detect an mRNA present at a few copies per cell (Ruan et al., 1998). In this study, we prepared the probes from 1 μg of poly(A)+ RNA. The original methods for preparing a probe required >1 μg of poly(A)+ RNA (Eisen and Brown, 1999); however, it is difficult to obtain much mRNA from specialized cell types. The method for amplification of RNA isolated from a single cell has been reported (Eberwine et al., 1992). This method may extend the utility of microarray analysis of tissue-specific gene expression.
In the present study, we used Imagene version 2.0 (BioDiscovery, Inc., Los Angeles, CA) and QuantArray (GSI Lumonics, Watertown, MA) as microarray data analysis software. Companies that provide microarrays often provide the necessary software and information to examine the experimental data (Richmond and Somerville, 2000). Scanning and image processing currently require human intervention to ensure that grids are properly aligned and that artifacts are flagged and excluded from subsequent analysis. Adoption of standard input/output formats, automation of feature identification, and software identification of common artifacts are important goals for the next generation of microarray analysis software. The ideal image analysis software package should detect automatically each valid spot on the array, flag and exclude bad spots, and subtract the local background (Bassett et al., 1999). Also, genomic information resources can be highly synergistic, and public databases and tools such as GenBank, ENTREZ, and BLAST provide biologists with integrated and linked information. A GenBank-like public database of gene expression measurements, integrated with MEDLINE, ENTREZ, and other data and tools, would be a useful resource for the biological community.
Conclusions and Perspectives
In the present study, we identified 44 drought-inducible genes and 19 cold-inducible genes using the full-length cDNA microarray (Tables 1 and 2). Among them, 30 and 10 genes are novel drought- and cold-inducible genes, respectively. Furthermore, we identified 12 DREB1A target genes, six of which are novel genes. These results indicate that full-length cDNA microarray analysis is a powerful tool for the identification of stress-inducible genes and target genes of transcription factors that control stress-inducible gene expression. Using our full-length cDNA microarray, it is easy to isolate full-length cDNAs for further functional analysis. We are planning to isolate >10,000 Arabidopsis full-length cDNAs and prepare the cDNA microarrays using the cDNA clones for the identification of new stress-inducible genes and new target genes of stress-related transcription factors. Furthermore, we will apply this cDNA microarray analysis to identify plant hormone-inducible genes, tissue-specific genes, and target genes of transcription factors.
Full-length cDNA microarray analysis is not only a method for the systematic analysis of quantitative gene expression but also an extremely powerful tool with which to find novel genes that are expressed in certain conditions or in certain tissues. Hybridization of cDNA microarrays containing complete sets of genes may eventually replace classic differential screening and display procedures. This method can provide new markers for various physiological processes involved in abiotic stress responses, hormonal signal transduction, pathogen attack, and developmental processes. It can also be used for the characterization of the molecular basis of phenotypic changes in various mutants and transgenic plants and for the study of genetic networks through the analysis of epistatic relationships between mutant phenotypes. Functional analysis, by use of parallel expression monitoring, should help researchers to better understand the fundamental mechanisms that underlie plant growth and development. Microarray analysis using full-length cDNAs provides a means to link genomic sequence information and functional analysis. By accumulating data on gene expression by tissue type, developmental stage, hormone and herbicide treatment, genetic background, and environmental conditions, it should be possible to identify the genes involved in many important processes of development and responses to environmental conditions in plants.
METHODS
cDNA Clones
In the cDNA microarray analyses, we used ∼1300 cDNA sequences representing Arabidopsis thaliana full-length cDNA clones isolated from full-length cDNA libraries, the drought- and cold-inducible genes responsive to dehydration (rd) and early responsive to dehydration (erd), kin1, kin2, and cor15a, and the α-tubulin gene as an internal control. As a negative control, two DNAs derived from the mouse nicotinic acetylcholine receptor epsilon-subunit (nAChRE) gene and the mouse glucocorticoid receptor homolog gene were used.
Sequence Analysis
Plasmid DNA was extracted with a Kurabo DNA extraction instrument (model PI-100; Kurabo, Tokyo, Japan) and subjected to sequencing. DNA sequences were determined using the dye terminator cycle sequencing method with a DNA sequencer (model ABI Prism 3700; Perkin-Elmer Applied Biosystems, Foster City, CA). Sequence homologies were examined with the GenBank/EMBL database using the BLAST program.
Amplification of cDNA Inserts
The vector used for cDNA library construction was λZAPII (Carninci et al., 1996). Inserts of cDNA clones were amplified by polymerase chain reaction (PCR) using primers that were complementary to vector sequences flanking both sides of the cDNA insert. The primers were 5′-CGCCAGGGTTTTCCCAGTCACGA (FL forward 1224 primer) and 5′-AGCGGATAACAATTTCACACAGGA (FL reverse 1233 primer). Plasmid templates (1 to 2 ng) were added to 100 μL of a PCR mixture containing 0.25 mM each nucleotide, 0.2 μM each primer, 1 × Ex Taq buffer (Takara Shuzo, Kyoto, Japan), and 1.25 units of Ex Taq polymerase (Takara Shuzo). PCR was performed as follows: at 94°C for 3 min; for 35 cycles at 95°C for 1 min, 60°C for 30 sec, and 72°C for 3 min; and at 72°C for 3 min. To clean up PCR products and prepare the DNA for printing, we precipitated PCR products in ethanol and resuspended the DNA in 25 μL of 3 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate). One microliter of each finished reaction was electrophoresed on a 0.7% agarose gel to confirm amplification quality and quantity.
cDNA Microarray Preparation
PCR products were arrayed from 384-well microtiter plates onto poly-l-lysine–coated micro slide glass (model S7444; Matsunami, Osaka, Japan) using the GTMASS System gene tip microarray stamping machine (Nippon Laser and Electronics Laboratory, Nagoya, Japan). The tip loaded 0.5 μL of PCR products (100 to 500 ng/μL) from 384-well microtiter plates and deposited 5 nL per slide on six slides with spacing of 280 μm. The printed slides were rehydrated in a beaker with hot distilled water and snap dried at 100°C for 3 sec. The slides were placed into a slide rack, and the rack was placed into a glass chamber. The blocking solution, containing 15 mL of 1 M sodium borate, pH 8.0, 5.5 g of succinic anhydride (Wako, Osaka, Japan), and 335 mL of 1-methyl-2-pyrrolidone (Wako), was poured into the glass chamber. The slide racks were plunged up and down five times, shaken gently for 15 min, transferred into a chamber containing boiling water, plunged up and down another five times, and left for 2 min. The slide racks were transferred into a chamber containing 95% ethanol, plunged up and down five times, and centrifuged at 2500g for 30 sec.
Plant Materials and RNA Isolation
Wild-type Columbia plants and transgenic plants overexpressing DREB1A cDNA under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Kasuga et al., 1999) were grown on germination medium agar plates for 3 weeks as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). Dehydration and cold stress treatments were performed as described previously (Yamaguchi- Shinozaki and Shinozaki, 1994). The Columbia plants were subjected to stress treatments for 2 or 10 hr and then frozen in liquid nitrogen for further analysis. In the experiments to identify DREB1A target genes, transgenic plants overexpressing DREB1A cDNA and wild-type plants grown on germination medium lacking kanamycin were used. Transgenic plants overexpressing DREB1A cDNA were not subjected to stress treatment. Total RNA was prepared using Isogen (Nippon Gene, Tokyo, Japan), and mRNA was prepared using the Oligotex-dT30 mRNA purification kit (Takara, Tokyo, Japan).
Preparation of Fluorescent Probes
Each mRNA sample was reverse transcribed in the presence of Cy3 dUTP or Cy5 dUTP (Amersham Pharmacia). The reverse transcription reaction was performed in a 30-μL volume containing 1 μg of poly(A)+ RNA with 6 μg of oligo(dT) 18mer, 10 mM DTT, 500 μM each dATP, dCTP, and dGTP, 200 μM dTTP, 100 μM Cy3 dUTP or Cy5 dUTP, and 400 units of SuperScript II reverse transcriptase (Life Technologies, Grand Island, NY) in 1 × Superscript first-strand buffer (50 mM Tris-HCl, pH 8.3, 75 mM KCl, 3 mM MgCl2, and 20 mM DTT) (Life Technologies). After incubation at 42°C for 1 hr, the reaction products of two samples (one with Cy3 labeling and the other with Cy5 labeling) were combined, treated with 15 μL of 0.1 M NaOH and 1.5 μL of 20 mM EDTA, incubated at 70°C for 10 min, and treated with 15 μL of 0.1 M HCl. The samples were placed in a Microcon 30 microconcentrator (Amicon, Beverly, MA). Four hundred microliters of TE buffer (10 mM Tris-HCl, pH 8.0, and 1 mM EDTA) was added and spun for 5 to 10 min in a benchtop microcentrifuge at a high speed to a volume of 10 to 20 μL. The flow-through product was discarded. Four hundred microliters of TE buffer and 20 μL of 1 mg/mL human Cot-1 DNA (Gibco BRL) were added and spun again. After the second spin, the probe retained by the Microcon was significantly brighter than the flow-through product. This is a very strong indicator of a successful labeling reaction. The probes were collected by inverting the filter and spun for 5 min. Several microliters of distilled water was added to the Microcon. The filter was inverted and spun so that the final volume of the collected probes was 12 μL. Two microliters of 10 μg/μL yeast tRNA, 2 μL of 1 μg/μL pd(A)12–18 (Amersham Pharmacia), 3.4 μL of 20 × SSC, and 0.6 μL of 10% SDS were added to the probes. The probe samples were denatured by placing them in a 100°C water bath for 1 min, left at room temperature for 30 min, and then used for hybridization.
Microarray Hybridization and Scanning
The probe samples were spun for 1 min in a benchtop microcentrifuge at high speed to pellet any particulate matter. The probes were placed onto the center of the array to avoid forming bubbles. A cover slip was placed over the entire array surface to avoid the formation of bubbles. Four 5-μL drops of 3 × SSC were placed on a separate part of the slide to provide humidity in the hybridization chamber and thus ensure that the probe mixture did not dehydrate during hybridization. The slides were placed in a sealed hybridization cassette (model THC-1; BM Equipment, Tokyo, Japan) and submerged in a 65°C water bath for 12 to 16 hr. After hybridization, the outside of the slide chamber was dried carefully. The slides were removed and placed in a slide rack submerged in washing solution 1 (2 × SSC, 0.1% SDS), with the array face of the slide tilted down so that when the cover slip dropped off it did not scratch the array surface. After the cover slip came off, the slide racks were plunged up and down three times in washing solution 1 and transferred to washing solution 2 (1 × SSC) carefully to minimize the transfer of washing solution 1 to the second chamber, because SDS can interfere with slide imaging. The slide chamber was rocked gently for 2 min. The slide racks were transferred to washing solution 3 (0.2 × SSC), allowed to stand for 2 min, spun at 2500g for 1 min, and dried.
Microarrays were scanned with a scanning laser microscope (model ScanArray4000; GSI Lumonics). Separate images were acquired for each fluor at a resolution of 10 μm per pixel. To normalize the two channels with respect to signal intensity, we adjusted photomultiplier and laser power settings so that the signal ratio of the α-tubulin genes (internal control) was as close to 1.0 as possible.
For the microarray data analysis, we used Imagene version 2.0 (BioDiscovery) and QuantArray (GSI Lumonics) software.
RNA Gel Blot Analysis
Isolated total RNA was also used for RNA gel blot hybridization. Hybridization was performed as described previously (Yamaguchi-Shinozaki and Shinozaki, 1994). The PCR-amplified fragments prepared from the full-length Arabidopsis cDNAs were used as probes for RNA gel blot hybridization.
Acknowledgments
We thank Suzurei Shimamura, Masahiro Yonezawa, Suguru Okunuki, Setsuko Kawamura, Mihoko Ohsaki, and Shigeko Nagano for their skilled technical assistance. We thank Dr. Yasushi Okazaki and Rika Miki for technical advice. We thank Nippon Laser and Electronics Laboratory, BM Equipment Co., Ltd., General Scanning, Inc., and Dr. Takaaki Sato for technical support. We thank Dr. D.P. Snustad for the gift of the α-tubulin gene. This work was supported in part by a grant for genome research from RIKEN, the Program for the Promotion of Basic Research Activities for Innovative Biosciences, the Special Coordination Fund of the Science and Technology Agency, and a grant-in-aid from the Ministry of Education, Science, and Culture of Japan to K.S. It was also supported by a Grant-in-Aid for Scientific Research on Priority Areas (C) “Genome Science” from the Ministry of Education, Science, and Culture of Japan to M.S. It was also supported in part by the Core Research for Evolutional Science and Technology program of the Japan Science and Technology Corporation, the Special Coordination Fund, a research grant for the Genome Exploration Research Project from the Science and Technology Agency, and a Grant-in-Aid for Scientific Research on Priority Areas and the Human Genome Program from the Ministry of Education and Culture, Japan, to Y.H.
References
- Aharoni, A., et al. (2000). Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 12, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu, E., Nakamura, Y., Sato, S., and Tabata, S. (2000). A large scale analysis of cDNA in Arabidopsis thaliana: Generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res. 7, 175–180. [DOI] [PubMed] [Google Scholar]
- Bajaj, S., Targolli, J., Liu, L.F., Ho, T.H.D., and Wu, R. (1999). Transgenic approaches to increase dehydration-stress tolerance in plants. Mol. Breed. 5, 493–503. [Google Scholar]
- Baker, S.S., Wilhelm, K.S., and Thomashow, M.F. (1994). The 5′-region of Arabidopsis thaliana cor15a has cis-acting elements that confer cold-, drought-, and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. [DOI] [PubMed] [Google Scholar]
- Bassett, D.E., Jr., Eisen, M.B., and Boguski, M.S. (1999). Gene expression informatics: It's all in your mine. Nat. Genet. 21, 51–55. [DOI] [PubMed] [Google Scholar]
- Bohnert, H.J., Nelson, D.E., and Jensen, R.G. (1995). Adaptations to environmental stresses. Plant Cell 7, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray, E.A. (1997). Plant responses to water deficit. Trends Plant Sci. 2, 48–54. [Google Scholar]
- Carninci, P., et al. (1996). High-efficiency full-length cDNA cloning by biotinylated CAP trapper. Genomics 37, 327–336. [DOI] [PubMed] [Google Scholar]
- Cooke, R., et al. (1996). Further progress towards a catalogue of all Arabidopsis genes: Analysis of a set of 5,000 non-redundant ESTs. Plant J. 9, 101–124. [DOI] [PubMed] [Google Scholar]
- DeRisi, J.L., Iyer, V.R., and Brown, P.O. (1997). Exploring the metabolic and genetic control of gene expression on a genomic scale. Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- Eberwine, J., Yeh, H., Miyashiro, K., Cao, Y., Nair, S., Finnell, R., Zattel, M., and Coleman, P. (1992). Analysis of gene expression in single live neurons. Proc. Natl. Acad. Sci. USA 89, 3010–3014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen, M.B., and Brown, P.O. (1999). DNA arrays for analysis of gene expression. Methods Enzymol. 303, 179–205. [DOI] [PubMed] [Google Scholar]
- Gilmour, S.J., Zarka, D.G., Stockinger, E.J., Salazar, M.P., Houghton, J.M., and Thomashow, M.F. (1998). Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 16, 433–443. [DOI] [PubMed] [Google Scholar]
- Heller, R.A., Schena, M., Chai, A., Shalon, D., Bedilion, T., Gilmore, J., Woolley, D.E., and Davis, R.W. (1997). Discovery and analysis of inflammatory disease-related genes using cDNA microarrays. Proc. Natl. Acad. Sci. USA 94, 2150–2155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Höfte, H., et al. (1993). An inventory of 1,152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J. 4, 1051–1061. [DOI] [PubMed] [Google Scholar]
- Holmberg, N., and Bülow, L. (1998). Improving stress tolerance in plants by gene transfer. Trends Plant Sci. 3, 61–66. [Google Scholar]
- Ingram, J., and Bartels, D. (1996). The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 377–403. [DOI] [PubMed] [Google Scholar]
- Iwasaki, T., Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1997). The dehydration-inducible RD17 (Cor47) gene and its promoter region in Arabidopsis thaliana (accession no. AB004872) (PGR 97–156). Plant Physiol. 115, 1287.9390449 [Google Scholar]
- Jaglo-Ottosen, K.R., Gilmour, S.J., Zarka, D.G., Schabenberger, O., and Thomashow, M.F. (1998). Arabidopsis CBF1 overexpression induces cor genes and enhances freezing tolerance. Science 280, 104–106. [DOI] [PubMed] [Google Scholar]
- Kasuga, M., Liu, Q., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1999). Improving plant drought, salt, and freezing tolerance by gene transfer of a single stress-inducible transcription factor. Nat. Biotechnol. 17, 287–291. [DOI] [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994. a). Characterization of two cDNAs (ERD10 and ERD14) corresponding to genes that respond rapidly to dehydration stress in Arabidopsis thaliana. Plant Cell Physiol. 35, 225–231. [PubMed] [Google Scholar]
- Kiyosue, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994. b). Cloning of cDNAs for genes that are early-responsive to dehydration stress (ERDs) in Arabidopsis thaliana L.: Identification of three ERDs as HSP cognate genes. Plant Mol. Biol. 25, 791–798. [DOI] [PubMed] [Google Scholar]
- Lin, X., et al. (1999). Sequence and analysis of chromosome 2 of Arabidopsis thaliana. Nature 402, 761–768. [DOI] [PubMed] [Google Scholar]
- Liu, Q., Kasuga, M., Sakuma, Y., Abe, H., Miura, S., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). The transcription factors, DREB1 and DREB2, with an EREBP/AP2 DNA binding domain separate two cellular signal transduction pathways in drought- and low-temperature-responsive gene expression, respectively, in Arabidopsis. Plant Cell 10, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, S.R., Oppenheimer, D.G., Silflow, C.D., and Snustad, D.P. (1987). Characterization of the alpha-tubulin gene family of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 84, 5833–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayer, K., et al. (1999). Sequence and analysis of chromosome 4 of the plant Arabidopsis thaliana. Nature 402, 769–777. [DOI] [PubMed] [Google Scholar]
- Newman, T., DeBruijn, F.J., Green, P., Keegstra, K., Kende, H., Mcintosh, L., Ohlrogge, J., Raikhel, N., Somerville, S., Thomashow, M., Retzel, E., and Somerville, C. (1994). Genes galore: A summary of methods for accessing results from large-scale partial sequencing of anonymous Arabidopsis cDNA clones. Plant Physiol. 106, 1241–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richmond, T., and Somerville, S. (2000). Chasing the dream: Plant EST microarrays. Curr. Opin. Plant Biol. 3, 108–116. [DOI] [PubMed] [Google Scholar]
- Ruan, Y., Gilmore, J., and Conner, T. (1998). Towards Arabidopsis genome analysis: Monitoring expression profiles of 1400 genes using cDNA microarrays. Plant J. 15, 821–833. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Davis, R.W., and Brown, P.O. (1995). Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470. [DOI] [PubMed] [Google Scholar]
- Schena, M., Shalon, D., Heller, R., Chai, A., Brown, P.O., and Davis, R.W. (1996). Parallel human genome analysis: Microarray-based expression monitoring of 1000 genes. Proc. Natl. Acad. Sci. USA 93, 10614–10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki, M., Carninci, P., Nishiyama, Y., Hayashizaki, Y., and Shinozaki, K. (1998). High-efficiency cloning of Arabidopsis full-length cDNA by biotinylated CAP trapper. Plant J. 15, 707–720. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1997). Gene expression and signal transduction in water-stress response. Plant Physiol. 115, 327–334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (1999). Molecular responses to drought stress. In Molecular Responses to Cold, Drought, Heat and Salt Stress in Higher Plants, K. Shinozaki and K. Yamaguchi-Shinozaki, eds (Austin, TX: R.G. Landes), pp. 11–28.
- Shinozaki, K., and Yamaguchi-Shinozaki, K. (2000). Molecular responses to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. [PubMed] [Google Scholar]
- Shinwari, Z.K., Nakashima, K., Miura, S., Kasuga, M., Seki, M., Yamaguchi-Shinozaki, K., and Shinozaki, K. (1998). An Arabidopsis gene family encoding DRE/CRT binding proteins involved in low-temperature-responsive gene expression. Biochem. Biophys. Res. Commun. 250, 161–170. [DOI] [PubMed] [Google Scholar]
- Stockinger, E.J., Gilmour, S.J., and Thomashow, M.F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcription activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 94, 1035–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taji, T., Seki, M., Yamaguchi-Shinozaki, K., Kamada, H., Giraudat, J., and Shinozaki, K. (1999). Mapping of 25 drought-inducible genes, RD and ERD, in Arabidopsis thaliana. Plant Cell Physiol. 40, 119–123. [DOI] [PubMed] [Google Scholar]
- Takahashi, S., Katagiri, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). An Arabidopsis gene encoding a Ca2+-binding protein is induced by abscisic acid during dehydration. Plant Cell Physiol. 41, 898–903. [DOI] [PubMed] [Google Scholar]
- Thomashow, M.F. (1999). Plant cold acclimation: Freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. [DOI] [PubMed] [Google Scholar]
- Wang, H., Datla, R., Georges, F., Loewen, M., and Cuter, A.J. (1995). Promoters from Kin1 and cor6.6, two homologous Arabidopsis thaliana genes: Transcriptional regulation and gene expression induced by low temperature, ABA osmoticum and dehydration. Plant Mol. Biol. 28, 605–617. [DOI] [PubMed] [Google Scholar]
- Wodicka, L., Dong, H., Mittmann, M., Ho, M.H., and Lockhart, D.J. (1997). Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat. Biotechnol. 15, 1359–1367. [DOI] [PubMed] [Google Scholar]
- Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low-temperature, or high-salt stress. Plant Cell 6, 251–264. [DOI] [PMC free article] [PubMed] [Google Scholar]