Abstract
The myb-homologous p1 gene regulates the synthesis of flavonoid pigments in maize kernel pericarp and cob; these floral organs are greatly modified in size and shape compared with their counterparts in teosinte, the progenitor of maize. To elucidate the molecular evolution of the p1 gene in relation to its expression and possible functions in maize and teosinte, we have isolated a second maize gene (p2) that is highly homologous with the p1 gene, and a related gene (p2-t) from Zea mays subsp parviglumis. We present evidence that the maize p1 and p2 genes were generated by duplication of an ancestral p gene (ppre) and its downstream sequences; the duplicated 3′ flanking sequences were inserted upstream of the ppre gene, thereby changing its transcription pattern. This model accounts for the structural organization and the observed differential expression of the p1 and p2 genes: p1 transcripts accumulate in kernel pericarp, cob, tassel glumes, and silk, whereas p2 transcripts are found in developing anther and silk. The duplication is estimated to have occurred 2.75 million years ago; subsequently, multiple retroelements have been inserted between the p1 and p2 genes. Our results demonstrate the evolution of a single gene into a compound locus containing two component genes with different tissue specificities. Expression of the p1 gene in the kernel pericarp may have provided a selective advantage during the evolution of maize kernel morphology.
INTRODUCTION
Eukaryotic genomes have been shaped to a large extent by gene duplications. Changes in ploidy are common in plants and give rise to immediate whole-genome duplications. Local gene duplications are commonly observed in genomic sequence analysis; for example, a 1.9-Mb stretch of Arabidopsis genomic sequence contains eight pairs of related genes, located adjacent to each other and in the same orientation (Bevan et al., 1998). These local sequence repeats apparently are produced by segmental duplications of discreet chromosome intervals. By whatever mechanism they occur, duplications can have a fundamentally important role in evolution. A complete gene duplication produces two identical copies, which then may undergo one of several alternative fates. Both gene copies may retain their original function, enabling the organism to produce a greater quantity of RNA or proteins. Or one of the copies may retain the original function, whereas the other copy may mutate to a functionless state (pseudogene) or acquire mutations that generate a new function (neomorph) (Li and Graur, 1991). Finally, one copy may retain the same coding sequence function but acquire new regulatory elements that specify a different pattern of expression. The latter process is best exemplified in the evolution of genes that regulate biosynthesis of flavonoid pigments in plants. In maize, anthocyanin biosynthesis is controlled by the combined function of two groups of regulatory genes: the c1/pl genes, which encode Myb-homologous transcriptional activators, and the r/b gene family, which encodes helix-loop-helix coactivators. Expression of these regulatory genes in different tissues produces distinct patterns of anthocyanin pigmentation in maize (Dooner et al., 1991). The c1/pl and r/b gene families have each been proposed to be derived from ancient gene duplications (Chandler et al., 1989; Cone et al., 1993), as evidenced by the conserved coding sequences and the exchangeable functions of different gene family members (Goff et al., 1990; Ludwig et al., 1990). Furthermore, the r gene family includes other linked loci, such as the sn and lc genes, that confer additional distinct pigmentation patterns (Dooner and Kermicle, 1976; Ludwig et al., 1989; Tonelli et al., 1991)—diverse expression patterns that may reflect differences in expression elicited by different 5′ regulatory sequences. This idea has been confirmed in two alleles of the b gene by transformation analysis (Radicella et al., 1992; Selinger et al., 1998).
In contrast to the multiple regulators of the anthocyanin pathway, the p1 gene is the only known regulatory gene required for biosynthesis of phlobaphene pigment in maize. The p1 gene encodes a Myb-like transcriptional activator (Grotewold et al., 1991a, 1994) of the structural genes c2, chi, and a1, which encode chalcone synthase, chalcone isomerase, and dihydroflavonol reductase, respectively. The sequential function of these enzymes converts simple organic compounds to polyphenolic flavonoids, including red phlobaphene pigment (Styles and Ceska, 1977; Byrne et al., 1996). The most notable pigmentation conferred by the p1 gene is in the kernel pericarp and cob; indeed, different p1 alleles are identified according to their pigmentation patterns in these two tissues. The P1-rr allele specifies red pericarp and red cob, P1-wr specifies white (colorless) pericarp and red cob, and P1-ww specifies white (colorless) pericarp and cob (Figure 1). Unlike the R-r gene, which has two independently mutable and separable components for seed color and plant color (Stadler and Neuffer, 1953), a single coding sequence of P1-rr controls both pericarp and cob color (Athma and Peterson, 1991). Although the P1-rr coding sequence is simplex, it is flanked on both the 5′ and 3′ sides by 5.2-kb direct repeat sequences (Athma and Peterson, 1991; Xiao et al., 2000). These repeats do not resemble transposons in structure and are not bordered by the target site duplications that are normally generated by transposon insertion (Athma and Peterson, 1991).
Figure 1.

Phlobaphene Pigmentation of Floral Organs Controlled by the p Gene.
(A) Maize ears with pericarp and cob pigmentation conferred by the P1-rr, P1-wr, P1-rw, and P1-ww alleles (left to right).
(B) Mature tassel inflorescence of maize, showing the central spike and four lateral branches with attached spikelets (some lateral branches have been removed for clarity). Teosinte tassel morphology is similar.
(C) Close-up of tassel spikelet of teosinte (Zea mays subsp parviglumis, accession BK1). Note the light brown pigmentation of the spikelet glume margins (arrow).
(D) Maize ear shoot with emerged silks ready for pollination.
(E) Teosinte parviglumis female inflorescence with emerged silks.
(F) Teosinte parviglumis inflorescence at the same stage as in (E), but with husk and silks removed.  .
.
(G) Seeds of teosinte (Z. m. parviglumis Iltis 81-5; upper left), maize (P1-ww-1112; upper right), and their F1 hybrid (lower). Note that the teosinte seed is enclosed within a cupulate fruitcase, whereas the kernels produced by the F1 hybrid are partially exposed.
To clarify the origin of the unusual P1-rr gene structure, we isolated and characterized a highly homologous gene (p2) located upstream of the P1-rr allele. We propose that the P1-rr and the p2 genes were formed by tandem (head to tail) duplication of an ancestral p gene (ppre) and its 3′ flanking sequence. As a result, the incipient P1-rr gene coding region was flanked by directly duplicated sequences. After this duplication event, multiple retroelement insertions separated the p1 and p2 genes. This model is supported by analysis of the sequences of the maize p1 and p2 genes, and an orthologous gene (p2-t) from teosinte, the progenitor of maize. In addition to duplicating the p coding sequences, the duplication event placed new regulatory sequences 5′ to the p1 gene. Thus, differential regulation of duplicated genes can arise as a direct consequence of their duplication. We show that consistent with this model, the p1 and p2 genes are differentially expressed in maize floral organs. The possible implication of p gene duplication in maize evolution is discussed.
RESULTS
Structural Comparison of p Gene Homologs
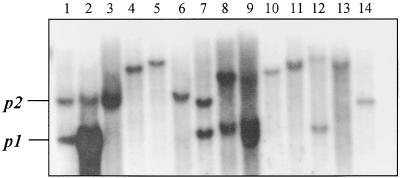
Maize contains a sequence highly homologous with, and tightly linked to, the P1-rr gene (Athma and Peterson, 1991; Das and Messing, 1994). For example, a DNA gel blot of genomic P1-rr DNA probed with a fragment of the second intron of the maize P1-rr gene (probe 8B; Grotewold et al., 1991a) detects a band of 3.7 kb, as expected, from the P1-rr gene and an additional 6-kb band (Figure 2, lane 1). On the same blot, genomic DNA from a P1-wr plant (lane 2) also gives a 6-kb band, in addition to an intensely hybridizing 3.7-kb band reflecting the sixfold tandem repeat of the p1 coding sequences in this allele (Chopra et al., 1998). Moreover, the 6-kb band also is observed in an allele (P1-ww-1112; Figure 2, lane 3) in which the p1 coding sequence has been deleted as a result of recombination between the 5.2-kb repeats flanking the P1-rr gene (Athma and Peterson, 1991). We cloned this p-homologous gene (p2) from a maize stock homozygous for P1-ww-1112 (see Methods). The location and orientation of the p2 gene were deduced by analyzing the structure of an interstitial deletion, p-del2, generated by a nonlinear transposition involving complete and partial Ac transposable elements inserted in the P1-rr gene (Zhang and Peterson, 1999). In the p-del2 mutant, the 5′ region of the p2 gene is joined to the 3′ portion of the P1-rr gene, indicating that the p2 gene is upstream of p1 and in the same transcriptional orientation.
Figure 2.
Detection of p1-Homologous Genes in Maize and Related Species.
DNA gel blot analysis of genomic DNAs digested with XbaI and hybridized with a maize P1-rr genomic fragment (probe 8B; Grotewold et al., 1991a). Lane 1, maize P1-rr; lane 2, maize P1-wr; lane 3, maize P1-ww-1112; lane 4, Z. m. parviglumis (Iltis 81); lane 5, Z. m. parviglumis (BK1); lane 6, Z. m. mexicana (PI 384,060); lane 7, Z. m. mexicana (PI 566,687); lane 8, Z. diploperennis (PI 462,368); lane 9, Z. diploperennis (Ames 21,884); lane 10, Z. luxurians (PI 441,933); lane 11, Z. luxurians (Ames 21,893); lane 12, Z. m. huehuetenangensis (Ames 21,880); lane 13, Z. m. huehuetenangensis (PI 441,934); lane 14, sorghum (y1). In lane 1, maize P1-rr contains bands of 6 and 3.7 kb, derived from the p2 and p1 genes, respectively. In lane 2, maize P1-wr gives an intense 3.7-kb band resulting from the sixfold tandem repeat of p sequences in the P1-wr allele (Chopra et al., 1998). In lane 3, maize P1-ww-1112 has a deletion of the 3.7-kb p1 gene band but retains the 6-kb p2 band.
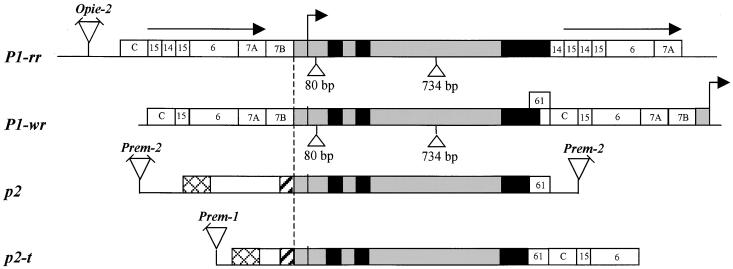
To elucidate the origin of the maize p1/p2 complex, we surveyed by DNA gel blot hybridization (Figure 2) the structures of p1 gene homologs in 10 diverse teosinte accessions, including Zea mays subsp parviglumis (lanes 4 and 5), Z. mays subsp mexicana (lanes 6 and 7), Z. diploperennis (lanes 8 and 9), Z. luxurians (lanes 10 and 11), and Z. mays subsp huehuetenangensis (lanes 12 and 13). The results indicate that the teosinte accessions tested here contain examples of both simple and complex sequences homologous with p1, and that this genic diversity can be seen even within a single teosinte subspecies, for example, mexicana (lanes 6 and 7). We then screened the same teosinte accessions for the presence of red/brown tassel glume margins (Figures 1B and 1C). In maize, tassel glume pigmentation is conferred by functional p1 alleles such as P1-rr and P1-wr. Tassel glume pigmentation was observed in several accessions of Z. m. parviglumis and Z. m. mexicana. One Z. m. parviglumis plant (Iltis 81-5) with pigmented tassel glume margins and a relatively simple p gene structure (Figure 2, lane 4) was selected for further study. Genetic tests indicated that the maize p2 gene and the p-homologous gene in Z. m. parviglumis segregate as alleles in repulsion (see Methods). We cloned this teosinte p gene homolog (termed p2-t) and compared its sequence with that of the maize p2 gene and with two alleles of the maize p1 gene, P1-rr and P1-wr (Figure 3). All four genes have a similar exon/intron structure; moreover, the sequences of the 5′ untranslated region (UTR) and the two introns of the four genes are >91% similar. The P1-rr and P1-wr alleles contain two insertions that are absent in both the maize p2 gene and the teosinte p2-t gene: a 80-bp insertion in the 5′ UTR and a 734-bp insertion in intron 2 (Figure 3).
Figure 3.
Structural Comparison among p-Homologous Genes in Maize and Teosinte: Maize P1-rr, Maize P1-wr, Maize p2, and Teosinte p2-t.
The bent arrow indicates the transcription start site of the P1-rr allele (Grotewold et al., 1991a). Black boxes indicate exons, and gray boxes indicate two introns, 5′ UTRs, and 90-bp conserved promoter sequences. Triangles indicate 80- and 734-bp insertions in the P1-rr and P1-wr leader and intron 2, respectively. The P1-rr gene coding region is flanked by 5.2-kb direct repeats indicated by horizontal arrows. Numbered boxes indicate the locations of p1 gene fragments described previously (Lechelt et al., 1989; Athma and Peterson, 1991; Chopra et al., 1996) and used here for homologous sequence comparison. Hatched and cross-hatched boxes indicate homologous sequences (>90%) in the p2 and p2-t genes. Sequences homologous with retroelements are indicated by triangles; angled lines on triangles indicate the absence of an LTR. In the P1-wr allele, a bent arrow at the 3′ end indicates the transcription start of the downstream tandem p1 gene copy. To show structure, the drawing is not to scale. GenBank accession numbers for maize p2 and teosinte p2-t are AF210616 and AF210617, respectively.
The nucleotide sequence similarity in the open reading frame is 98% or greater among the four genes, up to a point of divergence near the 3′ ends of the genes. In the P1-wr allele, a 210-bp fragment (fragment wr61) replaces a 660-bp fragment (fragment 14) that is present in the P1-rr allele. Following the wr61 fragment, P1-wr contains a 1-kb fragment (fragment C in Figure 3) that is absent from the P1-rr 3′ end but present at the P1-rr 5′ end ∼8 kb from the P1-rr transcription start site (Figure 3; Chopra et al., 1996). In the 3′ direction, the P1-rr and P1-wr alleles are 99% identical over a 3.5-kb region that comprises fragments 15, 6, and 7A (Figure 3). Chopra et al. (1996) previously proposed that the wr61 fragment of the P1-wr allele was replaced by fragment 14 during the generation of the P1-rr allele. The 3′ flanking regions of both the teosinte p2-t gene and the maize p2 gene also contain the wr61 fragment in place of fragment 14. Moreover, the similarity of the teosinte p2-t gene to the maize P1-wr allele extends ∼3.5 kb further in the 3′ direction, including fragments C, 15, and 6. These results support the hypothesis that the 3′ region of the ancestral p gene was more similar to that of the maize P1-wr and teosinte p2-t genes, whereas the P1-rr allele was derived by later modifications (Chopra et al., 1996).
In the 5′ flanking regions, the maize P1-rr, P1-wr, and p2 genes and the teosinte p2-t gene all contain a 90-bp segment of 98% similarity located immediately upstream of the transcription start site and including the TATA box. Further in the 5′ direction of this 90-bp segment, the P1-rr and P1-wr 5′ flanking sequences are nearly identical (99.3% similarity over 5.2 kb). The P1-rr gene 5′ flanking sequence is directly repeated 3′ of the coding sequence (Figure 3). In contrast, the maize p2 and teosinte p2-t genes are not flanked by direct repeats. The 5′ flanking sequences of the maize p2 and teosinte p2-t genes share a 170-bp region of similarity (93%) and an additional 1-kb block of similarity (96%) located further in the 5′ direction. The 170-bp and 1-kb regions of similarity are separated by nonhomologous sequence blocks of 2.6 and 120 bp in the maize p2 and teosinte p2-t genes, respectively. The maize p2 clone contains a region similar to retroelement Prem-2 (Turcich et al., 1996) located 3.9 kb 5′ of the presumptive p2 transcription start site. In contrast, the teosinte p2-t gene contains a 3′ long terminal repeat (LTR) of retroelement Prem-1 (Turcich and Mascarenhas, 1994), located ∼2 kb in the 5′ direction of the transcription start site.
Maize p1, p2, and Teosinte p2-t Genes Have Distinct Expression Patterns
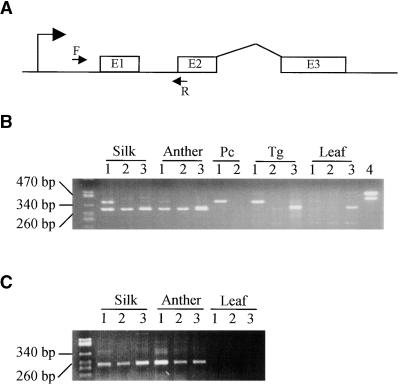
The maize p1 gene controls phlobaphene pigment accumulation in kernel pericarp, cob, and tassel glume, and the presence of P1-rr and P1-wr transcripts in pericarp and cob has been confirmed by RNA gel blot and reverse transcription–polymerase chain reaction (RT-PCR) analysis (Grotewold et al., 1991a; Chopra et al., 1996). In contrast, the p2 gene does not induce detectable phlobaphene pigmentation, as evidenced by the absence of phlobaphene pigments in plants that contain p2 but lack p1 (Athma and Peterson, 1991; Athma et al., 1992). As with the maize p2 gene, the teosinte p2-t gene does not induce pigmentation of maize kernel pericarp or cob, as shown by a test cross of a teosinte parviglumis p2-t line (Iltis 81-5) with a maize P1-ww line (Figure 1G). However, the tassel glume margin of the original teosinte Iltis 81-5 plant was pigmented slightly reddish brown, and this tassel glume pigmentation cosegregates with the teosinte p2-t gene in the test-cross progeny (see Methods). To examine the expression pattern of p1 and p2 transcripts, we used RT-PCR to amplify transcripts in kernel pericarp, tassel glume, silk, anther, and leaf. RNA samples from these tissues were reverse-transcribed, and the resulting first-strand cDNA preparations were PCR-amplified by using primers EP5-8 and EP3-13 (see Methods). This primer pair can amplify transcripts of both p1 and p2, producing a different size for each: The p1 gene product is 380 bp, whereas the p2 gene product is 300 bp because of an 80-bp deletion in the p2 5′ UTR. Additionally, p2 gene transcripts were specifically amplified with primer P2-5, which matches the 5′ UTR sequence of p2 but not p1. Figure 4 clearly shows that the maize p1 and p2 genes have distinct patterns of transcript accumulation. With RNAs isolated from P1-rr (Figure 4B, lanes 1) pericarp or tassel glume, the primer pair EP 5-8 and EP 3-13 amplified only a 380-bp band, derived from P1-rr transcripts. In contrast, with RNA isolated from young anther, the same primer pair amplified only a 300-bp band, which corresponds to p2 transcripts. In silk, both p1 and p2 transcripts were detected (Figure 4B). Expression of the p2 gene was confirmed by using RNAs isolated from a maize line carrying the P1-ww-1112 allele (Figure 4B, lanes 2), in which p1 is deleted but p2 is present. Additionally, RNAs from silk and young anther were amplified with the p2-specific primer pair P2-5 and EP 3-13 to produce a 240-bp p2 product in both the P1-rr and P1-ww-1112 genotypes (Figure 4C). As with the maize p2 gene, the teosinte p2-t gene also is expressed in silk and young anther, as indicated by RT-PCR of RNAs isolated from teosinte Iltis81-5: primers EP5-8/EP3-13 yield a 300-bp product (Figure 4B, lanes 3), and primers P2-5/EP3-13 yield a 240-bp product (Figure 4C, lanes 3). Additionally, RT-PCR analysis of RNA from teosinte tassel glume, performed with primer pair EP5-8 plus EP3-13, produces a 300-bp band (Figure 4B, lane 3), which represents the p2-t transcript.
Figure 4.
RT-PCR Detection of p Gene Transcripts in Maize and Teosinte Floral Organs.
(A) Locations of primers in the p gene. The bent arrow at left indicates the transcription start site, and boxes E1, E2, and E3 indicate p gene open reading frame sequences (not to scale). The arrow F indicates forward primers EP5-8 (p1) or P2-5 (p2); the arrow R indicates the reverse primer EP3-13 (p1 and p2).
(B) RT-PCR analysis with primers EP5-8 and EP3-13, which amplify both p1 and p2 transcripts to generate products of 380 and 300 bp, respectively. Lanes 1, 2, and 3 use RNAs from maize P1-rr, maize P1-ww-1112, and teosinte parviglumis Iltis81-5, respectively. Lane 4 uses genomic DNA from P1-rr as a PCR control. RNA samples were prepared from the indicated organs. Neither p1 nor p2 transcripts were detected in the seedling leaf tissues tested here; however, more recent RT-PCR experiments have de-tected small amounts of p1 transcripts in leaf tissues (S.M. Cocciolone and T. Peterson, unpublished results). The band in the teosinte leaf RNA lane visible on the ethidium-stained gel appeared to be a nonspecific RT-PCR product, given its very weak hybridization to a p-specific probe (not shown). The numbers at left denote the sizes of DNA molecular weight standards. Pc, pericarp; Tg, tassel glume.
(C) RT-PCR analysis with primers P2-5 and EP3-13, which specifically amplify a 240-bp product derived from the p2 transcript. Lanes 1, 2, and 3 use RNAs prepared from the indicated organs of maize P1-rr, maize P1-ww-1112, and teosinte parviglumis Iltis81-5, respectively. The numbers at left denote the sizes of DNA molecular weight standards. The agarose gel containing RT-PCR products was blotted and hybridized with probe PMD, which detects the region amplified by PCR; the results (not shown) confirmed that the 240-bp band visible in the ethidium-stained gel is a p-specific PCR product.
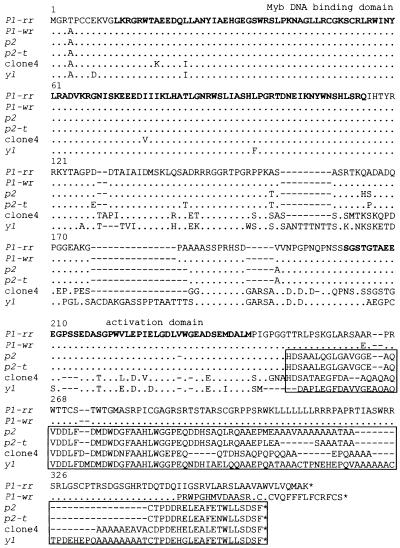
Conserved Functional Domains and Diverged C Termini of P Proteins
The deduced amino acid sequences of the maize p2 gene and the teosinte p2-t gene products were aligned with those of the P1-rr and P1-wr alleles of the maize p1 gene (Figure 5). The Myb-homologous DNA binding domain and the putative transcriptional activation domain (Grotewold et al., 1991a, 1994) are highly conserved in all four proteins. Outside of these two functional domains are several amino acid substitutions and insertion/deletion mutations. The most striking differences are found at the C termini of the P1-rr and P1-wr proteins compared with the p2 and p2-t proteins. The maize p2 and teosinte p2-t gene translation products diverge from that of the maize p1 gene shortly after the presumptive activation domain. However, their nucleotide sequences are 98 to 99% identical in the same region. The divergence in amino acid sequences reflects a reading frame difference caused by a single-base insertion/deletion between the p1 and p2 nucleotide sequences. The deduced p2 and teosinte p2-t proteins are shorter than both the P1-rr and P1-wr encoded proteins. Further C-terminal, beyond the region of comparison with p2, the P1-rr– and P1-wr–encoded proteins differ from each other as reported previously (Chopra et al., 1996).
Figure 5.
Alignment of Amino Acid Sequences of Maize, Teosinte, and Sorghum P Proteins.
Amino acid sequences were deduced from the maize P1-rr, maize P1-wr, maize p2, teosinte parviglumis p2-t, teosinte parviglumis clone 4, and sorghum y1 nucleotide sequences. Amino acids of the Myb DNA binding domain and putative transcription activation domain are shown in boldface. Dots indicate identical residues; dashes indicate gaps. The C-terminal boxed regions contain blocks of amino acids conserved among the predicted protein products of the maize p2, teosinte p2-t, teosinte clone 4, and sorghum y1 genes. The P1-rr sequence reported here is derived from the P1-rr-4B2 allele (Grotewold et al., 1991b) and differs at the C-terminal region from the previously reported P1-rr (Bloody Butcher) allele (Grotewold et al., 1991b). The Y1 protein sequence is derived from sorghum genomic (Chopra et al., 1999) and cDNA (S. Chopra and T. Peterson, unpublished data) sequences. The alignments were created by using the Pileup program of Genetics Computer Group software, with subsequent adjustments by inspection to optimize alignments in the C-terminal regions. GenBank accession numbers are as follows: p2, AF210616; p2-t, AF210617; and clone 4, AF210618 and AF210619.
An additional p gene homolog (teosinte clone 4) also was isolated during screening of the genomic library of Z. m. parviglumis. Genetic tests (see Methods) indicate that the clone 4 locus segregates independently of p2-t. Although the map position and function, if any, of the clone 4 gene are unknown, analysis of its nucleotide sequence provides some insights into p gene evolution. Teosinte clone 4 has greatest homology with p gene sequences in the exons (81 to 94%) and lower homology in the adjacent noncoding sequences (39 to 60% identity over sequenced regions); these results indicate that the teosinte clone 4 gene diverged before the duplication of the p1 and p2 genes. Interestingly, the teosinte clone 4 gene encodes a protein with a C-terminal sequence very similar to that of the maize p2 and teosinte p2-t genes but unlike that of the P1-rr and P1-wr genes (Figure 5, boxed sequences). A similar result is obtained by analysis of the y1 gene, a p gene homolog that controls phlobaphene pigmentation in sorghum pericarp (Chopra et al., 1999); the deduced Y1 protein C-terminal region is unlike that of P1 but contains blocks of amino acid sequences with high homology with the maize p2 gene product (Figure 5, boxed sequences). The simplest explanation for the similar C-terminal sequences of the proteins encoded by the maize p2, teosinte p2-t and clone 4, and sorghum y1 genes is that they share identity by descent from a common progenitor.
DISCUSSION
Duplication and Divergence of the Maize p1/p2 Genes: Evidence from Gene Structure and Sequence Comparisons
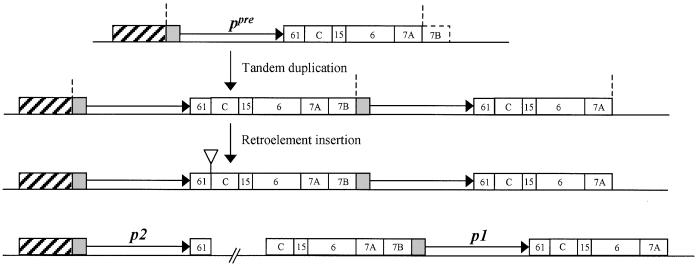
A current challenge in plant biology is to understand the generation of diversity in expression and coding potential associated with the presence of multiple copies of related genes in plant genomes. A favorable system for the analysis of functional diversity among repeated gene copies is found in the regulation of nonessential plant pigments. In maize, the p1 gene controls the synthesis of a red flavonoid pigment in floral organs, including kernel pericarp, the floral bracts of the cob, and tassel glumes (Figure 1). We have isolated a second p gene (p2) from maize that is tightly linked to the p1 gene, as well as a single-copy p gene from teosinte (p2-t) that segregates as an allele of the maize p1/p2 complex. Interestingly, the teosinte p2-t gene is more closely related to the maize p2 gene throughout 5′ and 3′ flanking regions as well as coding sequences. These results suggest that the maize p1 and p2 genes were generated by duplication of the coding sequence and 3′ flanking region of an ancestral p gene. As shown in Figure 6, the structure of the preduplication p gene (ppre) most probably resembled that of the teosinte p2-t gene. We propose that the ppre gene contained the two 5′ flanking sequence fragments (1 kp and 170 bp) common to the teosinte p2-t gene and the maize p2 gene (hatched box). The ppre gene also would have the conserved 90-bp promoter sequence (gray box) common among the teosinte p2-t gene, maize p2 gene, and P1-rr and P1-wr alleles of the maize p1 gene. The ppre gene 3′ flanking region would resemble the 3′ region of the P1-wr and p2-t genes and would contain fragment wr61, followed by fragment C and fragments 15, 6, 7A, and possibly 7B (see below). We propose that ∼10 kb of the progenitor ppre gene sequence, extending from the 5′ end of the conserved 90-bp promoter sequence to the 3′ end of fragment 7A where the 5.2-kb direct repeat sequence ends, was duplicated in a tandem head-to-tail arrangement. Once duplicated, the formerly 3′ flanking sequences were inserted at the 5′ side of the downstream gene, thus forming the p2 gene and p1 gene backbones. After this duplication, sequential retroelement insertions truncated the p2 3′ flanking region and enlarged the region between the two duplicated genes (see below). Subsequent to the formation of the p1 backbone, some further rearrangements occurred in the P1-rr allele, most notably in the C-terminal region (Chopra et al., 1996). Additionally, the extremely high sequence homology (99% at the nucleotide level) between the 5.2-kb repeats flanking the P1-rr gene suggests recent copy correction of these sequences. The P1-wr allele retains the basic p1 gene backbone with six tandem direct repeats of a 12.6-kb sequence; the 12.6-kb repeats are nearly identical, and each contains a single p1 gene coding sequence and its associated upstream regulatory sequences (Chopra et al., 1998). The sixfold repetition of the P1-wr allele produces an intensely hybridizing band in genomic DNA gel blots (Figure 2, lane 2). We proposed previously that the P1-wr amplification occurred after the domestication of modern maize, and possibly within the last 500 years (Chopra et al., 1998). Interestingly, however, one accession of Z. diploperennis appears to contain a similar repetition of p1-homologous sequences (Figure 2, lane 9), which may indicate that the P1-wr amplification occurred much earlier than previously thought. Alternatively, the p1 repetition in Z. diploperennis may result from an independent gene amplification event.
Figure 6.
Model for Origin of the Maize p1/p2 Gene Complex.
The structure of the hypothetical p progenitor coding region (ppre) is shown at the top, and the diagrams below denote the structures generated by tandem duplication and retroelement insertion. The structure of the extant p2/p1 complex is shown at the bottom, with a broken line indicating the uncloned region between fragments 61 and C. The hatched boxes denote the 5′ regulatory sequences of the progenitor ppre gene and the extant p2 gene; the gray boxes indicate the 90-bp conserved promoter sequences; the horizontal arrows indicate the p gene coding sequences; and the numbered boxes indicate the identities of flanking sequences (cf. Figure 3). The open triangle denotes the initial retroelement insertion between fragments 61 and C (cf. Figure 7). The vertical dashed lines delimit the duplicated region of ∼10 kb. To show structure, the drawing is not to scale.
To estimate when the p1/p2 gene duplication occurred, we compared the p1 and p2 coding sequences, using the formula R = K/2T (Li and Graur, 1991), where K is the number of substitutions at synonymous sites (Nei and Gojobori, 1986), R is the rate of substitution per synonymous site per year (estimated at 6 × 10−9 for grass nuclear genes; Gaut, 1998), and T is time (years). According to this formula, p gene duplication occurred ∼2.75 million years ago, that is, long before maize was domesticated from Z. m. parviglumis ∼7500 years ago (Iltis, 1983; Doebley et al., 1984). The single-copy p gene we isolated from teosinte parviglumis Iltis81-5 may have retained the original nonduplicated gene structure and evolved independently. Our genomic DNA gel blot data (Figure 2) indicate that both single-copy and duplicated p genes are present in various teosinte accessions. It will be interesting to determine whether the duplicated genes in the other wild taxa have the 3′ 1-bp frameshifting insertion that produces the distinct C-terminal region conserved among the p2-like genes (Figure 5).
How the initial step of p gene duplication occurred is unclear. Transposable elements may have been involved in inducing rearrangements of gene flanking regions (Habu et al., 1997). Tandem duplication also can occur by way of an unequal crossover in repeated sequences flanking a single-copy gene. For example, an unequal crossover between two copies of a transposable element inserted on either side of the Drosophila Bar gene generates a tandem duplication associated with the Bar-B allele (Tsubota et al., 1989). In maize, the formation of inverted duplicates of the S component (for seed pigmentation) of the R-r complex was associated with a dopia transposable element (Walker et al., 1995). As shown in Figure 6, the break point between the p1 and p2 duplications is expected to lie in the region delimited by the 3′ end of the 5.2-kb direct repeat at position −1034 bp upstream of the p1 transcription start site and by the beginning of the 90-bp conserved promoter sequence at position −92 upstream of the p1 transcription start site. This 923-bp sequence (fragment 7B in Figure 6) contains a tRNA-like gene as well as several inverted and direct repeats that may be involved in the transcriptional regulation of the P1-rr promoter (Sidorenko et al., 1999, 2000). However, no transposon-like sequences were detected in this sequence, and thus there is no evidence for involvement of transposable elements in the initial p gene duplication. Fragment 7B sequences may have been derived from the 3′ flanking region of the ancestral p gene, as proposed in Figure 6. However, because no sequences homologous with fragment 7B were identified downstream of the teosinte p2-t gene, the origin of the fragment 7B sequences is unclear at this time.
Retroelement Insertions in the p2/p1 Interval
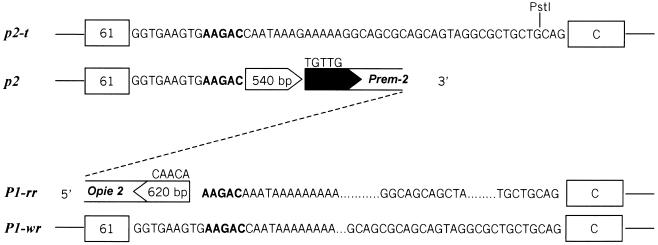
The p2 gene 3′ flanking sequence is similar to the p2-t and P1-wr sequences at 250 bp 3′ of the translational stop (Figure 3); the homology ends 35 bp before the PstI site in fragment wr61 (Figure 7). The homology is interrupted by a 540-bp sequence of unknown origin, followed further in the 3′ direction by a Prem-2 element, a highly repetitive retroelement in maize (Turcich et al., 1996). The Prem-2 insertion continues for 2.2 kb, at which point the p2 genomic clone ends. Interestingly, the homology with p2-t and P1-wr resumes upstream of the 5.2-kb 5′ flanking sequence of P1-rr (Figure 7). In P1-rr, 5′ upstream of the homology break point is a 620-bp sequence of unknown origin (Figure 7), followed further 5′ by sequences homologous with the maize opie-2 retroelement (SanMiguel et al., 1996). Thus, the p2 and P1-rr genes appear to have been separated by insertions of retroelements. The 540- and 620-bp sequences at the p2/p1 rearrangement break points are not homologous with known retroelements, but they may represent remnants of LTRs of an undefined retroelement. Retroelement LTRs commonly have short (4 to 12 bp) inverted repeats at their ends. For example, the yeast Ty retroelement family (Boeke, 1989), Drosophila copia retroelement (Bingham and Zachar, 1989), and maize retroelements (SanMiguel et al., 1996) all contain the sequences TGTTG and CAACA at the LTR ends. The 620-bp presumptive sequence next to the P1-rr gene 5′ end terminates in CAACA, suggesting that this 620-bp sequence is derived from a retroelement LTR. More importantly, a 5-bp direct duplication (AAGAC) is found adjacent to the 540- and 620-bp segments at the 3′ end of the p2 gene and the 5′ end of the P1-rr gene, respectively (Figure 7). Because retroelement insertions in maize are commonly flanked by 5-bp target site sequence duplications (SanMiguel et al., 1996), the presence of a 5-bp duplication at the p2/p1 break point is a strong indication that retroelement insertions separated the two copies of the original tandemly (head-to-tail) duplicated p genes. In maize, retroelements are commonly found inserted into other retroelements, and occasionally within LTRs (SanMiguel et al., 1996). Similarly, an unidentified retroelement could have inserted between the duplicated p2 and p1 genes, followed by subsequent insertions of other retroelements, including Prem-2 and Opie-2. The physical distance between the 3′ end of p2 and the 5′ end of P1-rr is not known. However, this interval does not contain any essential gene, as shown by the fact that plants homozygous for an interstitial deletion (p-del2) that removes this region are viable (J. Zhang and P. Zhang, unpublished observations). Possibly, the chromosome region between p2 and p1 may be composed entirely of retroelement sequences. These retroelement insertions would have necessarily occurred subsequent to the p2/p1 duplication ∼2.75 million years ago. This time range is consistent with a reported burst of retroelement activity in the maize genome within the last 3 million years (SanMiguel et al., 1998).
Figure 7.
Sequences at the Maize p2/p1 Break Point.
The p2/P1-rr break point sequences are aligned with the teosinte p2-t sequence (top) and the maize P1-wr sequence (bottom). The open arrowhead at the p2 gene 3′ end indicates a 540-bp sequence that may be a partial 5′ LTR of a undefined retroelement. The adjacent black arrowhead is a 5′ LTR sequence of retroelement Prem-2 with the typical TGTTG terminal sequence. The open arrowhead at the 5′ region of P1-rr indicates a presumptive partial retroelement 3′ LTR, ending with the typical CAACA terminal sequence. The 5-bp sequence in boldface (AAGAC) is present once in the p2-t and P1-wr genes and is duplicated in the p2/P1-rr interval at the retroelement insertion end points.
Divergent Expression and Possible Functional Domains of the Maize P1, P2, and Teosinte P2-t Proteins
The proteins encoded by the maize p2 and teosinte p2-t genes are nearly identical to the P1 protein over the Myb DNA binding domain and the putative acidic transcriptional activation domain. This sequence conservation suggests that the functionality of the maize p2 and teosinte p2-t genes has been maintained by selection. However, the p2 gene does not confer phlobaphene pigmentation, as evidenced by the colorless pericarp, cob, and tassel glume phenotype of plants that lack p1 but retain p2. These phenotypes could reflect differential expression of the p2 and p1 genes. A direct outcome of the gene duplication postulated in Figure 6 is that the p1 gene acquired new 5′ regulatory sequences. Indeed, regulatory elements in the P1-rr 5′ flanking sequences have been identified by Ac insertional mutagenesis (Moreno et al., 1992) and promoter analysis in transiently and stably transformed maize (Sidorenko et al., 1999, 2000; Cocciolone et al., 2000). The transcript expression profiles reported here confirm that the maize p1 and p2 genes have different patterns of expression: p1 transcripts are detected in pericarp and silk, whereas the maize p2 and teosinte p2-t gene transcripts are detected in anther and silk. Moreover, genetic cosegregation tests (see Methods) suggest that the teosinte p2-t gene probably regulates the production of reddish brown pigments in tassel glume margins (Figure 1C). This suggests that the protein encoded by p2-t is capable of conferring pigmentation as long as the gene is expressed in a tissue competent for phlobaphene accumulation. The lack of pigmentation in maize pericarp and cob glume is probably the result of the different tissue-specific expression pattern of the p2-t gene. Together, these results suggest that the p2-t protein, the p2 protein, and by extension the protein encoded by the progenitor p gene all have the potential to activate biosynthesis of phlobaphene pigment. Thus, the effective changes in the evolution of the p gene have been in the regulatory sequences, not in the coding sequences. A similar conclusion was reached in a previous study of the evolution of the anthocyanin regulatory gene c1 (Hanson et al., 1996). The results here provide additional support for the hypothesis that key changes in plant evolution have occurred through changes in expression of transcriptional regulators (Doebley and Lukens, 1998).
The characterization of p gene homologs in maize and teosinte raises the question of the possible function or functions of these genes in each plant. Tassel morphology (Figures 1B and 1C) is similar in maize and teosinte, whereas the morphology of the female inflorescence (ear) differs (Figures 1D to 1F). In maize, the pericarp forms the tough outermost layer of the kernel and provides a protective barrier against pathogen infection (Johann, 1935). Among 36 maize cultivars examined, pericarp thickness ranges from 5 to 22 cell layers (Tracy and Galinat, 1987). In contrast, the teosinte seed is enclosed within a cupulate fruitcase formed from the rachis internode and a thick, indurated outer glume (Dorweiler et al., 1993). The teosinte pericarp is only two or three cells thick (Tracy and Galinat, 1987) and is completely enclosed within the shell-like fruitcase (Figure 1G). Evolution of the maize kernel from teosinte was accompanied by marked kernel expansion, resulting in disruption of the fruitcase and exposure of the kernel on the ear (Dorweiler et al., 1993). Kernel exposure also is observed in the F1 hybrid of a cross of Z. m. parviglumis and maize (Figure 1G). The changes in floral structure in the conversion of teosinte to maize probably necessitated the conversion of the pericarp from a rudimentary internal layer into a tough protective seed coat by way of increases in cell layer number, density, and cell wall thickness. Although the p1/p2 duplication described here occurred long before the evolution of maize from teosinte, activation of the p1 gene in the pericarp may have provided some important selective advantages at the time of conversion from encased to exposed kernels. First, synthesis of the deep-red phlobaphene pigments in the exposed pericarp would present an obvious subject for human selection of colored kernels. Second, production of flavonoids in the pericarp may have helped to protect the kernel from damage by UV irradiation (Stapleton 1992), fungal infection (Snyder and Nicholson, 1990; Esele et al., 1993), and insect feeding (Byrne et al., 1996).
The p gene homologs analyzed here have highly conserved N-terminal regions, but the C-terminal regions are not equally conserved. The C-terminal regions of the proteins encoded by the maize p2, teosinte p2-t and clone 4, and the sorghum y1 genes share a conserved region of ∼100 amino acid residues (77 to 96% similarity at the amino acid level), and all four proteins end with an identical seven-residue motif sequence (WLLSDSF; Figure 5). However, this conserved C-terminal region is absent from the p1 alleles (P1-rr and P1-wr) because of a frameshifting insertion/deletion mutation. The fact that the sorghum Y1 protein contains the conserved C-terminal domain strongly supports the idea that the maize p2 and teosinte p2-t genes, and not the P1-rr allele, more closely resemble the ancestral p gene. The extensive conservation of the C-terminal region of these proteins implies that this domain has a functional role. Analysis of the ability of the p2-encoded protein to activate transcription of flavonoid biosynthetic genes in transformed maize cell cultures (Grotewold et al., 1998) may provide clues to its function in the plant.
METHODS
Maize and Teosinte Genetic Stocks
The maize lines used in this study contained the following alleles: P1-rr-4B2, derived from P1-vv by excision of Ac (Grotewold et al., 1991b); P1-ww-1112, derived from P1-vv by Ac-induced deletion of the P1-rr coding sequence (Athma and Peterson, 1991); and P1-wr in the W23 inbred background (Chopra et al., 1996). The maize p2 gene was cloned from a stock homozygous for the P1-ww-1112 allele. Teosinte stocks were obtained from Dr. John Doebley (University of Minnesota). The teosinte p2-t gene was isolated from Zea mays subsp parviglumis (accession Iltis81). In one plant from this line (30:81-5), genomic DNA gel blot hybridization with a maize P1-rr probe corresponding to the Myb-coding sequence (PMD [for P1-rr Myb domain]), or an intron 2 probe (fragment 8B; Grotewold et al., 1991a), detected a single strongly hybridizing band in five different enzyme digestions. Additionally, 22 progeny plants from a single self-pollinated plant gave the same banding pattern as the parental plant (data not shown). These results indicate that the Iltis81-5 stock is homozygous for a single-copy p gene homolog. The Iltis81-5 stock was crossed with a maize P1-ww line containing the p2 gene (P1-ww-1112), and the F1 plants were subsequently crossed to a second, structurally distinct, P1-ww line (P1-ww-4Co63). Among 46 progeny plants, 25 contained only the teosinte p gene band and 21 contained only the maize p2 gene band (data not shown). This ∼1:1 ratio, and the fact that no plants contained both the maize p2 and the teosinte p gene bands, indicates that the teosinte p gene (termed p2-t) and the maize p2 gene segregate as alleles in repulsion.
Genomic Library Construction and Screening
For isolation of the maize p2 gene, genomic DNA was prepared from leaves of plants homozygous for the P1-ww-1112 allele by using cetyltrimethylammonium bromide (CTAB) reagent (Saghai-Maroof et al., 1984). Genomic DNA was partially digested with Sau3AI and then ligated to a partially filled-in XhoI site of λ FixII vector (Stratagene). Approximately 500,000 independent phage clones were screened with two probes of P1-rr gene coding regions, PMD and fragment 12. PMD is a PCR fragment amplified from genomic P1-rr DNA using primers EP5-8 and EP3-13 located at the 5′ end of P1-rr (Grotewold et al., 1991a). P1-rr fragment 12 is a 1.2-kb BglII-SacI fragment covering parts of intron 2 and exon 3 of the P1-rr gene (Lechelt et al., 1989). Clones that hybridized to both probes were selected; NotI fragments of the largest clone containing an ∼20-kb insert were subcloned into pBluescript SK+ for sequencing. For isolation of the teosinte p2-t gene, a genomic library was prepared from a single plant of Z. m. parviglumis (accession Iltis81, plant 30:81-5) as described above; ∼800,000 clones were screened with P1-rr–derived probe PMD and P1-rr genomic fragments 13 (exon 3) and 15 (3′ flanking region; Lechelt et al., 1989). Clones that hybridized to all three probes were selected; SalI fragments from the largest clone containing an ∼19-kb insert were subcloned in pBluescript for sequencing. Plasmid subclones were sequenced with gene-specific primers and the Applied Biosystems (Foster City, CA) fluorescent sequencing system at the Iowa State University Nucleic Acid Facility.
RNA Isolation and Reverse Transcription–Polymerase Chain Reaction
Pericarps were dissected from kernels at 20 days after pollination, tassel glumes were dissected from tassels at anthesis, immature anthers were collected at early pollen development stage (between uninucleate and binucleate stages), and leaf samples were taken from three or four leaf seedlings. Samples were frozen in liquid nitrogen and ground in a mortar and pestle. Total RNA was extracted into Trizol reagent (Gibco BRL) and treated with DNase to remove residual genomic DNA. Aliquots of RNA (1 to 2 μg) were reverse-transcribed by using Superscript reverse transcriptase (Gibco BRL) at 42°C. Reactions were primed with either oligo(dT) or EP3-12 (5′-AAGCTTGAATTCGAGTTCCAGTAGTTCTTGATC-3′), the latter being homologous with p gene exon 3. Reactions were stopped by heating at 95°C for 5 min. The first-strand cDNA pool then was diluted fivefold in dH2O, and one-tenth of this was used in polymerase chain reaction (PCR) amplification. Primers EP5-8 (5′-ACGCGCGACCAGCTGCTA-ACCGTG-3′; homologous with the 5′ untranslated region of the maize p1 gene) and EP3-13 (5′-AGGAATTCCGCCCGAAGGTAGTTGATCC-3′; homologous with p1 exon 2) were used to amplify both the p1 and p2 transcripts. Primers P2-5 (5′-CTCGATTGGCGGGACCAGC-3′; homologous with the maize p2 5′ untranslated region) and EP3-13 were used to specifically amplify the maize p2 and the teosinte p2-t transcripts. PCR amplifications were performed in 1.5 mM MgCl2 and 2% DMSO for the following cycle conditions: 94°C for 4 min, followed by 30 cycles at 94°C for 45 sec, 60°C for 1 min, and 72°C for 1 min, and a final extension for 10 min at 72°C. Reverse transcription (RT)–PCRs were repeated at least twice to verify the results.
Acknowledgments
We thank John Doebley (University of Wisconsin) for providing teosinte stocks and for performing the initial maize by teosinte crosses. We thank Jianbo Zhang for assistance in the isolation of the P1-del2 deletion and Terry Olson for assistance with DNA gel blot hybridization (shown in Figure 2). We thank Phil Becraft, John Imsande, Jonathan Wendel, and Suzy Cocciolone for advice and comments on the manuscript. This research was supported by United States Department of Agriculture National Research Initiative Competitive Grants Program Award No. 9701354. Journal Paper No. J-18498 of the Iowa Agriculture and Home Economics Experiment Station, Ames, Iowa, Project No. 3297, was supported by Hatch Act and State of Iowa funds.
References
- Athma, P., and Peterson, T. (1991). Ac induces homologous recombination at the maize P locus. Genetics 128 163–173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athma, P., Grotewold, E., and Peterson, T. (1992). Insertional mutagenesis of the maize P gene by intragenic transposition of Ac. Genetics 131 199–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M., et al. (1998). Analysis of 1.9 Mb of contiguous sequence from chromosome 4 of Arabidopsis thaliana. Nature 391 485–488. [DOI] [PubMed] [Google Scholar]
- Bingham, P.M., and Zachar, Z. (1989). Retrotransposons and the FB transposon from Drosophila melanogaster. In Mobile DNA, D.E. Berg and M.M. Howe, eds (Washington, DC: American Society of Microbiology), pp. 485–502.
- Boeke, J.D. (1989). Transposable elements in Saccharomyces cerevisiae. In Mobile DNA, D.E. Berg and M.M. Howe, eds (Washington, DC: American Society of Microbiology), pp. 335–374.
- Byrne, P.F., McMullen, M.D., Snook, M.E., Musket, T.A., Theuri, J.M., Widstrom, N.W., Wiseman, B.R., and Coe, E.H. (1996). Quantitative trait loci and metabolic pathways: Genetic control of the concentration of maysin, a corn earworm resistance factor, in maize silks. Proc. Natl. Acad. Sci. USA 93 8820–8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandler, V.L., Radicella, J.P., Robbins, T.P., Chen, J., and Turks, D. (1989). Two regulatory genes of the maize anthocyanin pathway are homologous: Isolation of B utilizing R genomic sequences. Plant Cell 1 1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, S., Athma, P., and Peterson, T. (1996). Alleles of the maize P gene with distinct tissue specificities encode Myb-homologous proteins with C-terminal replacements. Plant Cell 8 1149–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chopra, S., Athma, P., Li, X., and Peterson, T. (1998). A maize myb homolog is encoded by a multicopy gene complex. Mol. Gen. Genet. 260 372–380. [DOI] [PubMed] [Google Scholar]
- Chopra, S., Brendel, V., Zhang, J., Axtell, J.D., and Peterson, T. (1999). Molecular characterization of a mutable pigmentation phenotype and isolation of the first active transposable element from Sorghum bicolor. Proc. Natl. Acad. Sci. USA 96 15330–15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cocciolone, S.M., Sidorenko, L.V., Chopra, S., Dixon, P.M., and Peterson, T. (2000). Hierarchical patterns of transgene expression indicate involvement of developmental mechanisms in the regulation of the maize P1-rr promoter. Genetics 156 839–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K.C., Cocciolone, S.M., Burr, F.A., and Burr, B. (1993). Maize anthocyanin regulatory gene Pl is a duplicate of C1 that functions in the plant. Plant Cell 5 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das, O.P., and Messing, J. (1994). Variegated phenotype and developmental methylation changes of a maize allele originating from epimutation. Genetics 136 1121–1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J., and Lukens, L. (1998). Transcriptional regulators and the evolution of plant form. Plant Cell 10 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doebley, J.F., Goodman, M.M., and Stuber, C.W. (1984). Isoenzymatic variation in Zea. Syst. Bot. 9 203–218. [Google Scholar]
- Dooner, H.K., and Kermicle, J.L. (1976). Displaced and tandem duplications in the long arm of chromosome 10 in maize. Genetics 82 309–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., Robbins, T.P., and Jorgensen, R.A. (1991). Genetic and developmental control of anthocyanin biosynthesis. Annu. Rev. Genet. 25 173–199. [DOI] [PubMed] [Google Scholar]
- Dorweiler, J., Stec, A., Kermicle, J., and Doebley, J. (1993). Teosinte glume architecture 1: A genetic locus controlling a key step in maize evolution. Science 262 233–235. [DOI] [PubMed] [Google Scholar]
- Esele, J.P., Frederiksen, R.A., and Miller, F.R. (1993). The association of genes controlling caryopsis traits with grain mold resistance in sorghum. Phytopathology 83 490–495. [Google Scholar]
- Gaut, B.S. (1998). Molecular clocks and nucleotide substitution rates in higher plants. Evol. Biol. 30 93–120. [Google Scholar]
- Goff, S.A., Klein, T.M., Roth, B.A., Fromm, M.E., Cone, K.C., Radicella, J.P., and Chandler, V.L. (1990). Transactivation of anthocyanin biosynthetic genes following transfer of B regulatory genes into maize tissues. EMBO J. 9 2517–2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold, E., Athma, P., and Peterson, T. (1991. a). Alternatively spliced products of the maize P gene encode proteins with homology to the DNA-binding domain of Myb-like transcription factors. Proc. Natl. Acad. Sci. USA 88 4587–4591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold, E., Athma, P., and Peterson, T. (1991. b). A possible hot spot for Ac insertion in the maize P gene. Mol. Gen. Genet. 230 329–331. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 543–553. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., Maddock, S., Clair, G., and Bowen, B. (1998). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10 721–740. [PMC free article] [PubMed] [Google Scholar]
- Habu, Y., Peyachoknagul, S., Sakata, Y., Fukasawa, K., and Ohno, T. (1997). Evolution of a multigene family that encodes the kunitz chymotrypsin inhibitor in winged bean: A possible intermediate in the generation of a new gene with a distinct pattern of expression. Mol. Gen. Genet. 254 73–80. [DOI] [PubMed] [Google Scholar]
- Hanson, M.A., Gaut, B.S., Stec, A.O., Fuerstenberg, S.I., Goodman, M.M., Coe, E.H., and Doebley, J.F. (1996). Evolution of anthocyanin biosynthesis in maize kernels: The role of regulatory and enzymatic loci. Genetics 143 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iltis, H. (1983). From teosinte to maize: The catastrophic sexual transmutation. Science 222 886–894. [DOI] [PubMed] [Google Scholar]
- Johann, H. (1935). Histology of the caryopsis of yellow dent corn with reference to resistance and susceptibility to kernel rots. J. Agric. Res. 51 855–883. [Google Scholar]
- Lechelt, C., Peterson, T., Laird, A., Chen, J., Dellaporta, S.L., Dennis, E., Peacock, W.J., and Starlinger, P. (1989). Isolation and molecular analysis of the maize P locus. Mol. Gen. Genet. 219 225–234. [DOI] [PubMed] [Google Scholar]
- Li, W., and Graur, D. (1991). Fundamentals of Molecular Evolution. (Sunderland, MA: Sinauer Associates).
- Ludwig, S.R., Habera, L.F., Dellaporta, S.L., and Wessler, S.R. (1989). Lc, a member of the maize R gene family responsible for tissue-specific anthocyanin production, encodes a protein similar to transcriptional activators and contains the myc-homology region. Proc. Natl. Acad. Sci. USA 86 7092–7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ludwig, S.R., Bowen, B., Beach, L., and Wessler, S.R. (1990). A regulatory gene as a novel visible marker for maize transformation. Science 247 449–450. [DOI] [PubMed] [Google Scholar]
- Moreno, M., Chen, J., Greenblatt, I., and Dellaporta, S. (1992). Reconstitutional mutagenesis of the maize P gene by short-range Ac transpositions. Genetics 131 939–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., and Gojobori, T. (1986). Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3 418–426. [DOI] [PubMed] [Google Scholar]
- Radicella, J.P., Brown, D., Tolar, L.A., and Chandler, V.L. (1992). Allelic diversity of the maize B regulatory gene: Different leader and promoter sequences of two alleles determine distinct tissue specificities of anthocyanin production. Genes Dev. 6 2152–2164. [DOI] [PubMed] [Google Scholar]
- Saghai-Maroof, M.A., Soliman, K.M., Jordensen, R.A., and Allard, R.W. (1984). Ribosomal DNA spacer-length polymorphism in barley: Mendelian inheritance, chromosomal location, and population dynamics. Proc. Natl. Acad. Sci. USA 81 8014–8018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SanMiguel, P., Tikhonov, A., Jin, Y., Motchoulskaia, N., Zakharov, D., Melake-Berhan, A., Springer, P.S., Edwards K.J., Lee, M., Avramova, Z., and Bennetzen, J.L. (1996). Nested retrotransposons in the intergenic regions of the maize genome. Science 274 765–768. [DOI] [PubMed] [Google Scholar]
- SanMiguel, P., Gaut, B.S., Tikhonov, A., Nakajima, Y., and Bennetzen, J.L. (1998). The paleontology of intergene retrotransposons of maize. Nat. Genet. 30 43–45. [DOI] [PubMed] [Google Scholar]
- Selinger, D.A., Lisch, D., and Chandler, V.L. (1998). The maize regulatory gene B-Peru contains a DNA rearrangement that specifies tissue-specific expression through both positive and negative promoter elements. Genetics 149 1125–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidorenko, L.V., Li, X., Tagliani, L., Bowen, B., and Peterson, T. (1999). Characterization of the regulatory elements of the maize P1-rr gene by transient expression assays. Plant Mol. Biol. 39 11–19. [DOI] [PubMed] [Google Scholar]
- Sidorenko, L.V., Li, X., Cocciolone, S.M., Tagliani, L., Chopra, S., Bowen, B., Daniels, M., and Peterson, T. (2000). Complex structure of a maize myb gene promoter: Functional analysis in transgenic plants. Plant J. 22 471–482. [DOI] [PubMed] [Google Scholar]
- Snyder, B.A., and Nicholson, R.L. (1990). Synthesis of phytoalexins in sorghum as a site specific response to fungal ingress. Science 248 1637–1639. [DOI] [PubMed] [Google Scholar]
- Stadler, L.J., and Neuffer, M.G. (1953). Problems of gene structure. II. Separation of R-r elements (S) and (P) by unequal crossing over. Science 117 471–472. [Google Scholar]
- Stapleton, A.E. (1992). Ultraviolet radiation and plants: Burning questions. Plant Cell 4 1353–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Styles, E.D., and Ceska, O. (1977). The genetic control of flavonoid synthesis in maize. Can. J. Genet. Cytol. 19 289–302. [Google Scholar]
- Tonelli, C., Consonni, G., Dolfini, S.F., Dellaporta, S.L., Viotti, A., and Gavazzi, G. (1991). Genetic and molecular analysis of Sn, a light-inducible, tissue-specific regulatory gene in maize. Mol. Gen. Genet. 225 401–410. [DOI] [PubMed] [Google Scholar]
- Tracy, W.F., and Galinat, W.C. (1987). Thickness and cell layer number of the pericarp of sweet corn and some of its relatives. HortScience 22 645–647. [Google Scholar]
- Tsubota, S.I., Rosenberg, D., Szostak, H., Rubin, D., and Schedl, P. (1989). The cloning of the Bar region and the B breaking point in Drosophila melanogaster: Evidence for a transposon-induced rearrangement. Genetics 122 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcich, M.P., and Mascarenhas, J.P. (1994). Prem-1, a putative maize retroelement has LTR sequences that are preferentially transcribed in pollen. Sex. Plant Reprod. 7 2–11. [Google Scholar]
- Turcich, M.P., Bokhari-Riza, A., Hamilton, D.A., He, C., Messier, W., Stewart, C., and Mascarenhas, J.P. (1996). Prem-2, a copia-type retroelement in maize is expressed preferentially in early microspores. Sex. Plant Reprod. 9 65–74. [Google Scholar]
- Walker, E.L., Robbins, T.P., Bureau, T.E., Kermicle, J., and Dellaporta, S.L. (1995). Transposon-mediated chromosome rearrangements and gene duplications in the formation of the maize R-r complex. EMBO J. 14 2350–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao, Y., Li, X., and Peterson, T. (2000). Ac insertion site affects the frequency of transposon-induced homologous recombination at the maize p1 locus. Genetics, in press. [DOI] [PMC free article] [PubMed]
- Zhang, J., and Peterson, T. (1999). Genome rearrangements by non-linear transposons in maize. Genetics 153 1403–1410. [DOI] [PMC free article] [PubMed] [Google Scholar]