Abstract
Both physiological and genetic evidence indicate interconnections among plant responses to different hormones. We describe a pleiotropic recessive Arabidopsis transposon insertion mutation, designated hyponastic leaves (hyl1), that alters the plant's responses to several hormones. The mutant is characterized by shorter stature, delayed flowering, leaf hyponasty, reduced fertility, decreased rate of root growth, and an altered root gravitropic response. It also exhibits less sensitivity to auxin and cytokinin and hypersensitivity to abscisic acid (ABA). The auxin transport inhibitor 2,3,5-triiodobenzoic acid normalizes the mutant phenotype somewhat, whereas another auxin transport inhibitor, N-(1-naph-thyl)phthalamic acid, exacerbates the phenotype. The gene, designated HYL1, encodes a 419–amino acid protein that contains two double-stranded RNA (dsRNA) binding motifs, a nuclear localization motif, and a C-terminal repeat structure suggestive of a protein–protein interaction domain. We present evidence that the HYL1 gene is ABA-regulated and encodes a nuclear dsRNA binding protein. We hypothesize that the HYL1 protein is a regulatory protein functioning at the transcriptional or post-transcriptional level.
INTRODUCTION
The development, growth, and survival of plants under a wide range of environmental conditions reflect an intricate interplay of physical and chemical conditions with the highly integrated sensing and response networks in plants. The growth habit and physiological properties of plants can differ markedly under different regimes of light, gravity, temperature, humidity, and salinity, among others. Hormones have long been known to be important internal mediating signals in plants, but the components of the underlying cellular machinery are just beginning to be identified and characterized (Trewavas and Malho, 1997; Grill and Himmelbach, 1998; Solano and Ecker, 1998; D'Agostino and Kieber, 1999). The range of proteins involved in receiving, transmitting, and responding to external signals includes receptor-like and other kinds of protein kinases, phosphatases, and transcription factors, as well as enzymes such as thioredoxin and farnesyltransferase, which influence protein structure or localization through mechanisms other than phosphorylation (Mulligan et al., 1997; Becraft, 1998; Bonetta and McCourt, 1998; Hooley, 1998; Bleecker, 1999; Thornton et al., 1999; Hirt, 2000; Urao et al., 2000).
Ample physiological evidence supports the presence of interconnections among plant responses to different environmental stimuli; moreover, evidence is accumulating that certain mutations can simultaneously influence the response to more than one hormone or altered physical parameter (Wilson et al., 1990; Clouse et al., 1996; Nemeth et al., 1998; Ephritikhine et al., 1999a; Beaudoin et al., 2000; Ghassemian et al., 2000). The implication is that individual proteins can be responsible for such interconnections, either transmitting multiple signals or participating in distinct complexes that transmit different signals (Elion, 1998). Proteins can receive information from a small molecule, such as a hormone, and transmit it to a macromolecule, such as another protein, commonly through either a covalent modification, such as phosphorylation, or protein binding (Mulligan et al., 1997; Trewavas and Malho, 1997; Moller and Chua, 1999; Nambara and McCourt, 1999). Proteins can also receive and transmit information at the macromolecular level through protein–protein interactions, phosphorylation or other structural modifications, and protein–nucleic acid interactions. Thus, the concept of a signal transduction “pathway” representing a linear succession of molecules transmitting information is giving way to the concept of an interconnected signaling “network,” the protein “nodes” of which are characterized by the number and nature of their interconnections (Trewavas and Malho, 1997; Elion, 1998; Bhalla and Iyengar, 1999; Weng et al., 1999).
In the present report, we describe an Arabidopsis insertion mutation that alters the plant's responses to several exogenous hormones and present evidence that the mutated gene encodes an abscisic acid (ABA)–regulated nuclear dsRNA binding protein. We develop the hypothesis that the HYPONASTIC LEAVES1 (HYL1) protein is a regulatory protein with transcriptional or post-transcriptional functions.
RESULTS
The hyl1 Mutation Has Pleiotropic Effects on Growth and Development
The hyl1 mutant was identified in an Arabidopsis Dissociation (Ds) insertion mutant collection generated by using a previously described transposon tagging system (Fedoroff and Smith, 1993). Germination of hyl1 mutant seed is normal, as is cotyledon structure. Seedling, rosette, and cauline leaves of the hyl1 mutant are narrower than are wild-type leaves and exhibit hyponasty (Figure 1). The hypocotyl elongation rate is reduced in both dark- and light-grown plants, and hypocotyl cells are shorter than those of wild-type plants (Figure 1E). Mature plants attain a stature of <30 cm, whereas wild-type plants reach ∼45 cm (Figure 1D). The leaves and flowers of hyl1 plants are smaller than those of wild-type plants (Figures 1B and 1C), and hyl1 siliques are both shorter and twisted (not shown). Flowering of the hyl1 mutant is delayed by ∼10 days under short-day conditions, and mutant plants produce more rosette leaves than do wild-type plants. The markedly reduced fertility of the hyl1 mutant appears to be attributable to the shorter stamen filaments, because fertility is restored upon manual pollination. Mutant plants also exhibit more lateral branching than do wild-type plants, suggesting decreased apical dominance.
Figure 1.
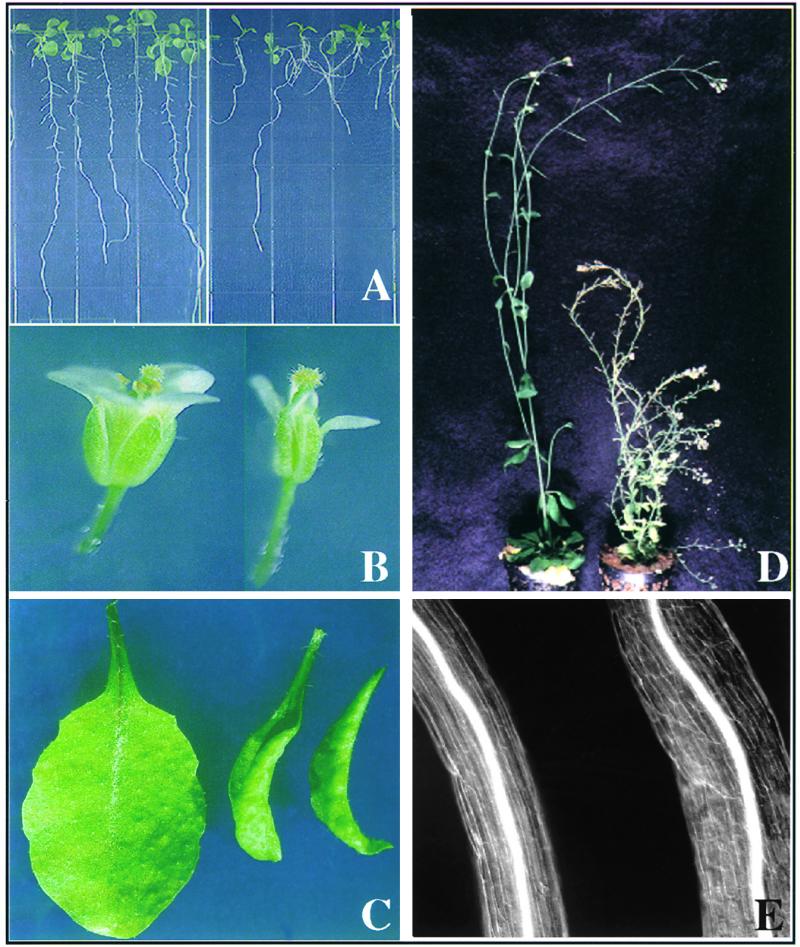
Pleiotropic Phenotype of the Arabidopsis hyl1 Mutant.
(A) Wild-type (left) and hyl1 mutant (right) seedlings grown on Murashige and Skoog (MS) plates.
(B) Flowers of the wild-type (left) and hyl1 mutant (right) plants.
(C) Rosette leaves from three-week-old wild-type (left) and hyl1 mutant (right two) plants.
(D) Mature wild-type and hyl1 plants.
(E) Hypocotyls of wild-type (left) and hyl1 (right) plants.
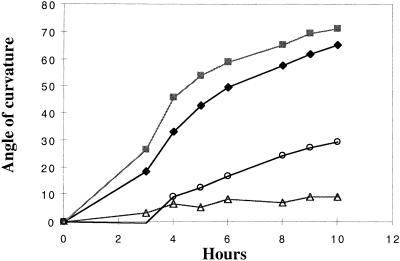
The hyl1 seedling root elongation rate is somewhat slower than that of wild-type plants in both light and dark conditions, and mutant roots exhibit both a reduced gravitropic response and plagiotropic growth (Figure 1A). The roots of wild-type seedlings growing on agar medium showed a curvature of ∼70° toward the gravity vector within 10 hr of gravistimulation, whereas the curvature of hyl1 seedling roots was only ∼10° (Figure 2). The root elongation rates were quite similar (5 mm/day for hyl1 seedlings and 5.5 mm/day for wild-type plants) at the stage at which the measurement was made, indicating that the difference in gravitropic response is not attributable simply to the decreased rate of root elongation in the mutant. In contrast, the hyl1 mutation does not affect shoot gravitropism. When wild-type Arabidopsis plants are placed in a horizontal position, they curve 90° upward within 90 min in the dark. The curvature rate of hyl1 mutant stems placed horizontally was similar; thus, the hyl1 mutation markedly reduces the gravitropic response of the root but not of the shoot.
Figure 2.
Gravitropic Responses of Wild-Type and hyl1 Mutant Seedlings.
Seedlings growing on MS medium or MS containing 0.05 μM TIBA were rotated 90° at time 0. Seedlings were aligned on the plates so that all the root tips were perpendicular to the bottom line of the Petri dish. The angle of curvature of the growing root tips was measured at 1- to 2-hr intervals. Filled diamonds, wild-type seedlings on MS medium; filled squares, wild-type seedlings on MS medium + 0.05 μM TIBA; open triangles, hyl1 seedlings on MS medium; open circles, hyl1 seedlings on MS medium + 0.05 mM TIBA.
hyl1 Is a Ds Insertion Mutation
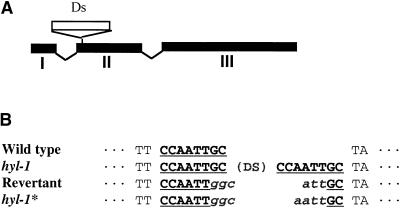
To determine whether the Ds insertion is responsible for the hyl1 mutation, we selected and analyzed phenotypic revertants. The progeny of mutant plants homozygous for the Ds insertion and containing Activator (Ac) transposase were examined for wild-type revertants. We obtained 25 putative wild-type revertants and extracted DNA from 10 plants to examine the Ds insertion site for a transposon footprint compromising all or part of the 8-bp duplication created upon Ds insertion (Pohlman et al., 1984; Schwarz-Sommer et al., 1985). The DNA sequences adjacent to the 5′ and 3′ ends of the Ds were amplified by using the thermal asymmetric interlaced (TAIL) polymerase chain reaction (PCR) technique and then were sequenced directly (Liu and Whittier, 1995; Tsugeki et al., 1996). PCR primers corresponding to the genomic DNA sequences adjacent to the 5′ and 3′ ends of the Ds were tested for their ability to amplify a fragment of the length expected if the Ds had been excised (Tsugeki et al., 1996). A short fragment of the expected size was amplified from eight of the 10 DNA samples. When sequenced, all eight were found to contain a 6-bp footprint at the former insertion site, indicating that phenotypic reversion was caused by excision of Ds (Figure 3B). One of the mutant control plant DNAs also supported amplification of a DNA fragment of the length expected for an empty donor site. However, sequencing revealed that this DNA had a 7-bp footprint at the former insertion site, indicating that another mutant allele had been created by a frame-shift when the Ds excised. These observations show that the complex mutant phenotype is attributable to mutation in a single gene and that the mutation was caused by insertion of the Ds element.
Figure 3.
Ds Insertion Site in the HYL1 Gene.
(A) The exons of the HYL1 gene are numbered I to III. The Ds element insertion site is in the second exon.
(B) The sequence around the Ds insertion site is shown for the wild type, the hyl1 insertion mutant, a wild-type revertant, and the hyl1* mutant allele created by Ds excision. The underlined 8-bp sequence is the target site duplication. The 6- and 7-bp transposon footprints are indicated in italics.
The hyl1 Mutant Is Hypersensitive to ABA
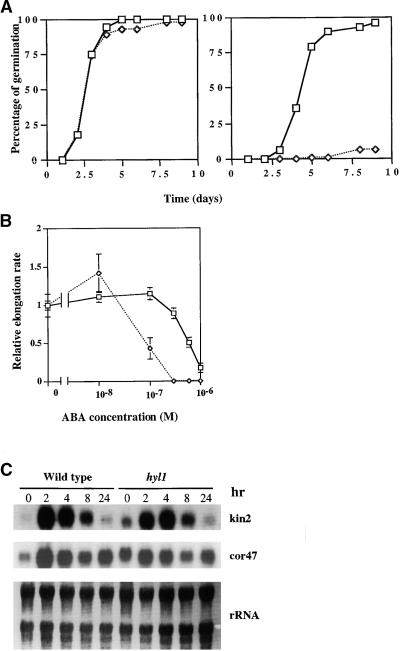
ABA has an inhibitory effect on seed germination, and Arabidopsis mutations have been identified that both increase and decrease sensitivity to such inhibition (Koornneef et al., 1984; Cutler et al., 1996). We tested the hyl1 mutant for the effect of exogenous ABA on seed germination. The hyl1 mutation has no effect on seed germination in the absence of ABA (Figure 4A, left). ABA at 0.5 μM slightly delays germination of wild-type seeds but has a negligible effect on the germination frequency. In contrast, germination of hyl1 mutant seeds is completely inhibited by 0.5 μM ABA (Figure 4A, right). Moreover, growth of hyl1 mutant roots is more sensitive to ABA inhibition than are those of wild-type plants (Figure 4B). Several mutants identified by virtue of their altered response to another hormone also show hypersensitivity of seed germination to exogenous ABA. These include era1-3 (Cutler et al., 1996), jar1 (Staswick et al., 1992), and jin4 (Berger et al., 1996). However, root growth of the era1-3 mutant is unaffected by ABA (P. McCourt, personal communication), suggesting that the hyl1 mutant is more sensitive to exogenous ABA than is the era 1-3 mutant.
Figure 4.
Effects of Exogenous ABA on Wild-Type (open squares) and hyl1 (open diamonds) Mutant Arabidopsis.
(A) hyl1 and wild-type seeds were placed on filter paper saturated with either water (left) or 0.5 μM ABA (right), incubated at 4°C for 48 hr, and then transferred to room temperature for germination.
(B) Relative root elongation rates of wild-type Nossen (No-0) and hyl1 mutant 10-day-old seedlings. Mean values for 100% root elongation were determined on MS medium containing no ABA. Error bars indicate sd.
(C) RNA gel blot analysis of ABA-induced mRNAs in hyl1 and wild-type plants. Arabidopsis plants were grown on MS plates for 2 weeks, then transferred into 0.1 × MS medium containing 50 μM ABA. Ten micrograms of total cellular RNA isolated from the wild-type and hyl1 seedlings at the indicated times after treatment with ABA was used in each lane. Probes are indicated for each panel.
To examine the transcriptional effects of the hyl1 mutation on ABA-induced genes, we monitored expression of two ABA- responsive genes, COR47 and KIN2, in mutant and wild-type plants (Gilmour et al., 1992; Kurkela and Borg-Franck, 1992). The steady state amounts of transcripts of both genes increased in response to exogenous ABA in both mutant and wild-type plants (Figure 4C). However, the amounts of uninduced transcripts of both genes were at least threefold greater in hyl1 plants than in wild-type plants. Thus, both genes tested appear to be deregulated in the mutant in the absence of exogenous ABA, although they remain ABA-inducible.
The hyl1 Mutant Exhibits Altered Sensitivity to Auxin and Inhibitors of Auxin Transport
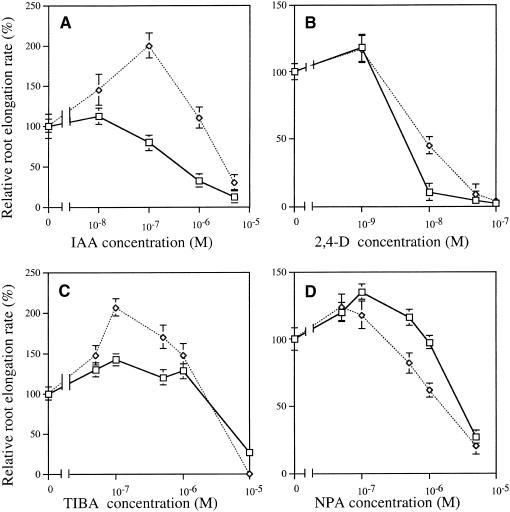
Exogenous auxin inhibits elongation of the primary root in wild-type Arabidopsis plants, and mutants defective in auxin responsiveness do not show normal inhibition (Lincoln et al., 1990). We analyzed the effect of auxin on the growth of hyl1 and wild-type roots. Exogenous indoleacetic acid (IAA) inhibited the growth of wild-type roots by >50% at 1 μM (Figure 5A). In contrast, exogenous IAA stimulated growth of hyl1 mutant roots at concentrations up to 1 μM, inhibiting elongation only at 5 μM, the highest concentration tested. Similarly, wild-type root growth between 5 and 9 days after germination was reduced by 90% by 10 nM 2,4-dichlorophenoxyacetic acid (2,4-D). As noted earlier, hyl1 mutant roots grow more slowly on agar medium in the absence of 2,4-D than do wild-type roots. However, in the presence of 10 nM 2,4-D, mutant root elongation was reduced by only 50%, indicating that the mutant roots are less sensitive to auxin than are wild-type roots (Figure 5B).
Figure 5.
Growth of hyl1 and Wild-Type Seedlings on Medium Containing Auxin and Auxin Transport Inhibitors.
hyl1 (open diamonds) and wild-type (open squares) seedlings were grown on MS medium supplemented with different concentrations of chemicals; root length was measured 10 days after germination. Root growth is expressed relative to growth on unsupplemented MS medium. Each point represents a test of 20 seedlings.
(A) IAA.
(B) 2,4-D.
(C) TIBA.
(D) NPA.
Error bars indicate sd.
The hyl1 mutant shows a paradoxical response to inhibitors of auxin transport. The auxin transport inhibitors N-(1-naphthyl)phthalamic acid (NPA) and 2,3,5-triiodobenzoic acid (TIBA) are known to inhibit the elongation of wild-type Arabidopsis roots (Katekar and Geissler, 1977). The growth of the hyl1 mutant roots was inhibited by NPA more than was the growth of wild-type roots were (Figure 5D). At low concentrations, TIBA partially corrected the mutant phenotype of hyl1 plants with respect to both root growth rate and gravitropic response (Figures 2 and 5C). The chemical structure of TIBA is markedly different from that of NPA, and TIBA and NPA have been suggested to interact with different target proteins (Katekar and Geissler, 1977; Brunn et al., 1992; Fujita and Syono, 1997). The difference between the responses to NPA and to TIBA in the hyl1 mutant further suggests that the modes of action of the two inhibitors are genetically separable.
The hyl1 Mutant Shows Decreased Sensitivity to Cytokinin but Not to Other Growth Hormones
We examined the effect of exogenous 6-benzylaminopurine (BA) on the growth of hyl1 and wild-type seedlings. The growth of hyl1 shoots was markedly less inhibited than that of wild-type shoots, whereas the sensitivity of hyl1 and wild-type roots to increasing concentrations of BA were similar (Figures 6A and 6B). The fresh weight of untreated 10-day-old hyl1 seedlings is much less than that of wild-type seedlings. However, at BA concentrations >0.01 μM, the fresh weight of hyl1 shoots substantially exceeded that of wild-type shoots (Figure 6D).
Figure 6.
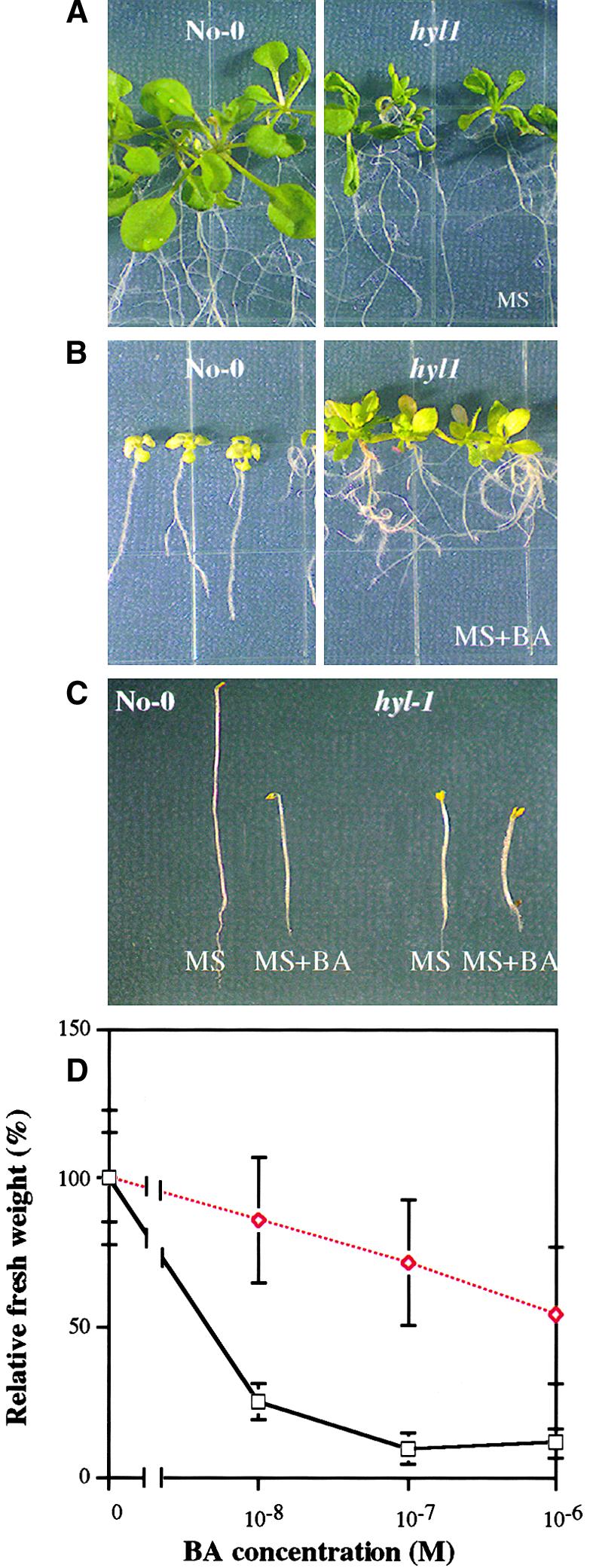
Responses of hyl1 and Wild-Type Seedlings to Exogenous Cytokinin.
(A) Wild-type (left) and hyl1 (right) seedlings grown on MS medium.
(B) Wild-type (left) and hyl1 (right) seedlings grown on MS medium containing 1 μM BA.
(C) Wild-type (left) and hyl1 (right) seedlings grown in the dark for 3 days on MS medium with or without 0.5 μM BA.
(D) The fresh weight of wild-type (open squares) and hyl1 (open diamonds) mutant seedlings at 14 days after germination on MS medium containing the indicated concentrations of BA. Shown is the dose–response curve for one representative experiment of three performed. Error bars indicate sd.
Cytokinins induce ethylene biosynthesis in Arabidopsis, and the ethylene produced in response to low doses of cytokinins is sufficient to induce the triple response in etiolated Arabidopsis seedlings (Vogel et al., 1998). As expected, higher concentrations of cytokinin were required to induce the triple response in etiolated hyl1 seedlings than in wild-type etiolated seedlings (Figure 6C). In contrast to the results obtained with auxin, cytokinin, and ABA, the growth responses of the hyl1 mutant to gibberellins, ethylene, brassinosteroids, methyl jasmonate, and salicylic acid were all similar to those of wild-type plants.
Isolation of the HYL1 cDNA and Characterization of the HYL1 Gene
As noted earlier, the DNA sequences adjacent to the 5′ and 3′ ends of Ds were amplified by TAIL PCR and sequenced directly. The flanking sequences are identical to that of part of an Arabidopsis bacterial artificial chromosome clone (GenBank accession number AC000132, BAC clone F21M12) that has been sequenced and anchored in chromosome 1s, close to marker mi443. The sequence in the immediate vicinity of the Ds insertion site showed extensive homology with a rice expressed sequence tag cDNA clone (c50188), suggesting that the Ds was inserted into an exon. To determine the structure of the HYL1 gene, we isolated an HYL1 cDNA clone. A 400-bp TAIL PCR–amplified fragment adjacent to the Ds insertion site was used to screen a cDNA library (CD4-7 lambda PRL2, a cDNA library prepared from different tissues and developmental stages). One cDNA of ∼1.5 kb was identified among the 400,000 plaques screened, and the cDNA insert was sequenced. The 5′ end of the cDNA has one or more stop codons in reading frames preceding an ATG start codon that initiates an uninterrupted 419–amino acid open reading frame (Figure 7A). RNA gel blot hybridization analysis of RNA isolated from wild-type plants revealed that the HYL1 cDNA hybridizes to a single major transcript of ∼1.5 kb, similar in size to the cDNA insert, suggesting that the cDNA is full-length or nearly full-length (data not shown). Comparing the genomic and cDNA sequences revealed that the HYL1 gene contains three exons of 25, 263, and 972 bp and two introns of 496 and 403 bp. A comparison of the cDNA sequences with that of the TAIL PCR fragment amplified from the 3′ end of Ds showed that the transposon had inserted into the second exon, 31 bp downstream of the putative translation start site (Figure 3A).
Figure 7.
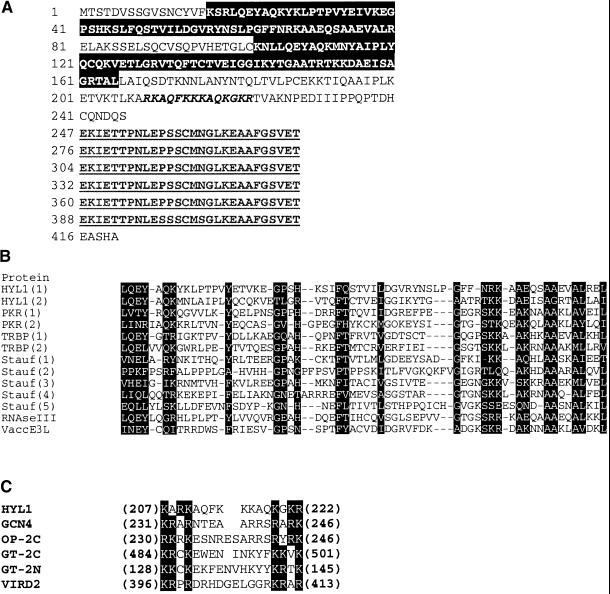
Motifs in the HYL1 Protein Sequence.
(A) Deduced amino acid sequence of the HYL1 gene. The two putative dsRNA binding motifs are shown in white letters on a black background, the putative nuclear localization motif is indicated in italics, and the six consecutive C-terminal repeats are underlined.
(B) Alignment of the dsRNA binding motifs of HYL1 protein with those of other proteins with dsRNA binding domains. The conserved regions are in white letters on a black background. Dashes were introduced to optimize alignments.
(C) Alignment of the putative bipartite nuclear localization sequence of HYL1 with similar motifs in other proteins. The basic residues at the ends of the motif are shown in white letters on a black background.
The predicted sequence of the HYL1 protein was used to search the GenBank database. The search results showed that the HYL1 protein contains two regions with marked similarities to dsRNA binding motifs (St. Johnston et al., 1992; Bass et al., 1994). The highest scoring segment pairs were those matching the dsRNA binding domains of the human interferon-induced RNA-dependent protein kinase P68, Escherichia coli ribonuclease III, and Xenopus dsRNA adenosine deaminase (Burd and Dreyfuss, 1994; Kharrat et al., 1995; Brooks et al., 1998). The two dsRNA binding motifs of the HYL1 protein are shown aligned with those of other proteins in Figure 7B. We identified three plant homologs of the HYL1 protein, one of which was the protein encoded by the rice cDNA c26837, which shows 69% identity and 80% similarity with the first dsRNA binding domain in the HYL1 protein. Based on the cDNA sequence, the rice protein contains only one dsRNA binding motif, but whether the cDNA is full length is not known. No homology was found outside of the dsRNA binding domains. We identified two additional putative Arabidopsis genes on chromosome 5 encoding proteins having considerable homology with HYL1 in the dsRNA binding domain (P1 clones MEE6 and AB010072).
As do other dsRNA binding proteins, the predicted HYL1 protein appears to have additional domains. The C-terminal end of the molecule includes six almost perfect repeats of a 28–amino acid sequence. A recent search of the Protein Sequence Databases identified a protein with sequence similarity to the HYL1 protein. Protein F, a fibronectin binding protein from Streptococcus pyogenes, contains five repeats of a 37–amino acid sequence having 30% identity and 48% similarity to the repeat in the HYL1 protein (Hanski and Caparon, 1992; Sela et al., 1993). The HYL1 protein also contains a positively charged region between residues 209 and 222 that resembles a bipartite nuclear localization signal (Varagona et al., 1992). Plant nuclear localization signals generally comprise two regions of basic amino acids separated by ∼10 amino acids (Figure 7C). This finding therefore suggests that HYL1 is a nuclear protein.
To determine whether HYL1 has homologs other than those found in the Arabidopsis sequence database, we performed low-stringency DNA gel blot hybridization experiments, using a cDNA probe. Genomic DNA from the Columbia, Nossen (No-0), and Landsberg erecta ecotypes was digested with EcoRI, HindIII, XbaI, and BglII and then probed with the full-length HYL1 cDNA. A single, strong band was seen in all lanes, as we expected from the absence of internal sites in the gene for these enzymes (data not shown). A single fragment was detected at both high and low stringencies, indicating that the HYL1 gene is unique in the Arabidopsis genome.
Expression of the HYL1 Gene
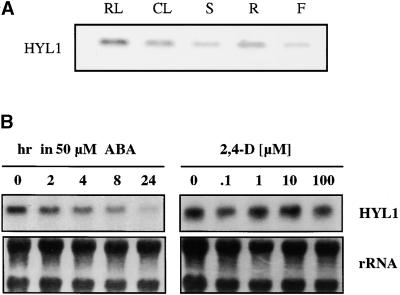
The phenotype of the hyl1 mutant suggests that the HYL1 protein plays a key role in Arabidopsis development throughout the life cycle of the plant. That is, the mutant has abnormal roots, leaves, and flowers, indicating that this gene has a function in most or all tissues. We used gel blot analysis of total RNA isolated from various Arabidopsis tissues to investigate the pattern of HYL1 gene expression directly. As expected, HYL1 transcripts were present in all of the tested tissues, including rosette leaves, cauline leaves, stems, roots, and flowers (Figure 8A). Moreover, the abundance of the HYL1 transcript was similar in different tissues. No hybridization was detected to RNA from hyl1 plants, indicating that expression of the HYL1 gene is considerably decreased or even abolished in the hyl1 mutant (data not shown). Given the site of insertion within the gene, hyl1 is probably a null mutation.
Figure 8.
Expression of the HYL1 Gene.
(A) RNA was isolated from rosette leaves (RL), cauline leaves (CL), inflorescent stems (S), roots (R), and flowers (F); fractionated on a 1.2% formaldehyde–agarose gel; blotted onto nitrocellulose; and probed with a full-length HYL1 cDNA probe.
(B) RNA was isolated from seedlings 0, 2, 4, 8, or 24 hr after exposure to 50 μM ABA (left) or after 1.5-hr exposure to different concentrations of 2,4-D, ranging from 0.1 to 100 μM. RNA was fractionated and analyzed as described in (A).
Because the hyl1 mutation impairs plant hormone responses, we wanted to determine whether the hyl1 gene itself is responsive to exogenous hormones. Accordingly, we analyzed HYL1 gene expression in Arabidopsis seedlings treated with auxin and ABA. Exposure of 2-week-old seedlings to 50 μM ABA for 24 hr decreased the HYL1 transcript levels to approximately one-third of those in control seedlings, whereas treatment with 2,4-D (0 to 100 μM) had no marked effect on HYL1 expression (Figure 8B). Thus we conclude that the HYL1 gene is downregulated by ABA.
Nuclear Localization of HYL1 Protein
To gain further insight into the function of the HYL1 protein, we investigated its subcellular location. As noted earlier, no transit or signal sequences have been identified in the HYL1 protein, and it lacks the structural features of a membrane-associated domain. However, the sequence contains a putative nuclear localization signal (Varagona et al., 1992). To determine whether the HYL1 protein is targeted to the nucleus, we fused the coding sequence of the HYL1 protein to that of the GUS gene (which encodes β-glucuronidase) expressed from a cauliflower mosaic virus (CaMV) 35S promoter. We introduced the resulting 35S-HYL1-GUS construct into onion epidermal cells by particle bombardment (Shieh et al., 1993). For a control, the onion epidermal cells were bombarded with an unmodified GUS gene expressed from the same promoter. In cells bombarded with the 35S-GUS gene, GUS activity was observed throughout the cell. In contrast, GUS activity was concentrated in the nuclei of all cells bombarded with the 35S-HYL1-GUS construct (Figure 9). This result confirms the inference from sequence homology that the HYL1 protein contains a nuclear localization sequence and suggests that the protein functions in the nucleus.
Figure 9.
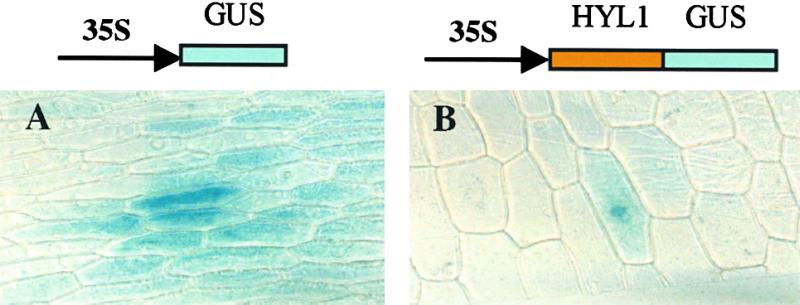
Subcellular Localization of an HYL1–GUS Fusion Protein.
(A) Structure of the GUS reporter construct.
(B) Structure of the HYL1 cDNA–GUS fusion construct.
The constructs were introduced into onion epidermal cells by particle bombardment. The cells were stained for GUS activity after 20 hr of incubation at room temperature. The two cells shown are representative of the results obtained in two independent experiments.
HYL1 Protein Binds dsRNA
The results of sequence analyses revealed that the HYL1 protein contains two tandem repeats of a motif characteristic of proteins that bind dsRNA, but not dsDNA. We used the protein encoded by the HYL1 cDNA to study the nucleic acid–binding properties of the HYL1 protein. To obtain large quantities of the protein, the cDNA was cloned in a pQE vector, which adds a histidine tag to the N terminus of the protein (Makrides, 1996). The fusion protein was purified from E. coli by two rounds of ion-exchange chromatography on a nickel–nitrilo triacetic acid (Ni-NTA) column. Protein gel blots showed that in the protein purified under native conditions, the main product was a 30-kD degradation product (data not shown). Because this fragment contains >200 amino acids of the HYL1 N terminus and because the two dsRNA binding motifs are located between amino acids 17 and 165, we reasoned that the truncated protein should contain all of the amino acids necessary for binding to dsRNA.
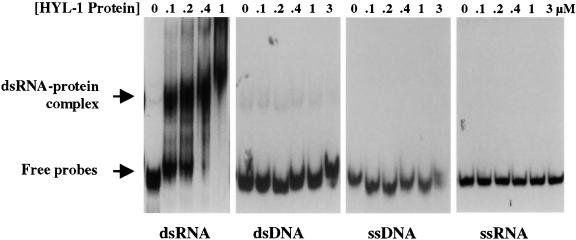
Mobility shift assays have been used extensively to study the properties of dsRNA binding proteins (Bass et al., 1994; Bevilacqua and Cech, 1996; Bevilacqua et al., 1998). The first part of Figure 10 shows the results of a gel mobility shift experiment performed under nondenaturing conditions with the truncated HYL1 protein and a 32P-labeled dsRNA fragment of ∼100 bp derived from the pBluescript II KS vector. A dsRNA–protein complex was first observed at a protein concentration of 0.1 μM. As the protein concentration increased, all of the labeled dsRNA shifted to a protein-bound form. However, at the same or greater concentrations of protein, no shift was detected when a ssRNA, dsDNA, or ssDNA of the same sequence was used (Figure 10). These results show that HYL1 binds dsRNA preferentially, as suggested by sequence homology with the dsRNA binding motifs of other proteins.
Figure 10.
Native Gel Mobility Shift Analysis of HYL1 Protein Binding to Nucleic Acids.
An RNA duplex of ∼100 bp was prepared by end-labeling hybridized T7 and T3 transcripts of the pBluescript II KS plasmid. A PCR fragment that includes both T7 and T3 promoters and intervening sequences of the same plasmid was used as dsDNA substrate. ssDNA was produced by denaturing and gel-purifying the same dsDNA fragment. A T7 transcript of the plasmid was used for ssRNA. Approximately the same amount of each test molecule (10 ng) was used for each experiment; the amount of the His-tagged HYL1 protein fragment added is indicated.
DISCUSSION
The pleiotropic recessive hyl1 mutation identifies an Arabidopsis gene that is both hormonally regulated itself and involved in auxin, ABA, and cytokinin responses. The pleiotropic developmental phenotype of the mutation suggests defects in the ability to perceive or transmit hormonal information. Thus, the mutant homozygote grows more slowly than does the wild type and shows less ability to respond to gravistimulation. Several of its defects, including reduced fertility, are attributable to decreased relative growth rates. Moreover, the plant shows excessive branching, implying reduced apical dominance. Direct analysis of the responses of the hyl1 mutant plant to exogenous hormones revealed that the mutation decreases the sensitivity of the plant to inhibition of growth by auxin and cytokinin but also increases its sensitivity to the inhibition of seed germination and root growth by ABA.
The protein encoded by the HYL1 gene contains a sequence resembling the plant consensus nuclear localization sequence, and an HYL1–GUS fusion protein accumulates in the nucleus, implying that the native protein is targeted to the nucleus. The HYL1 protein has two domains homologous with the dsRNA binding domains of other proteins. We have clearly demonstrated that an N-terminal fragment of the protein expressed in and purified from E. coli binds to dsRNA but not to ssRNA or to ss- or dsDNA of the same sequence. Although it remains to be determined whether the dsRNA binding domain of the HYL1 protein is essential for its function, the presence of canonical RNA binding domains and the binding properties of the recombinant protein strongly support the view that the HYL1 protein is a dsRNA binding protein.
Two genes that have previously been shown to be ABA-inducible, the KIN2 gene coding for a protein similar to type 1 fish antifreeze proteins (Kurkela and Borg-Franck, 1992) and the ABA- and cold-regulated COR47 gene (Gilmour et al., 1992), exhibit increased basal amounts of transcripts in hyl1 mutant homozygotes. This suggests that the HYL1 protein is itself a negative transcriptional regulator or is part of a negative regulatory complex. The observation that the HYL1 gene is itself downregulated by ABA is consistent with such an interpretation. The diminished sensitivity of the hyl1 mutant to inhibition by high exogenous concentrations of auxins and cytokinins, and the significant stimulatory effects of low concentrations of auxin and of auxin transport inhibitors, suggest that the protein mediates a stimulatory effect of these hormones, either directly or indirectly. Although we do not understand the differential response of the hyl1 mutant to the auxin transport inhibitors, we note that both NPA and TIBA show a slight but significant stimulation of root growth at very low concentrations, suggesting that inhibition of auxin transport partially compensates for the loss of HYL1 function.
Growing genetic evidence indicates that a single mutation can affect responses to more than one hormone. Interactions between ABA and such environmental signals as salt, cold, or drought are well documented in Arabidopsis (Ishitani et al., 1997; Zhu et al., 1997). Moreover, antagonisms have been observed between ABA and gibberellic acid (GA) in dormancy and germination (Steber et al., 1998; Wobus and Weber, 1999). ABA was originally identified as an inhibitor of auxin-induced growth of coleoptiles and stems (Davies and Jones, 1991; Taiz and Zeiger, 1991). ABA-deficient and ABA-insensitive mutants rescue the germination of the ga1 auxotroph and of seeds treated with an inhibitor of GA biosynthesis (Nambara et al., 1992; Steber et al., 1998). Genetic evidence also supports interconnections between ABA and other plant hormones. Thus, for example, the axr2 mutant was isolated on the basis of its decreased auxin sensitivity, but it is also resistant to ABA (Wilson et al., 1990). The brassinosteroid bri1 mutant is hypersensitive to ABA (Clouse et al., 1996). The phenotype of the sax1 mutant in brassinosteroid biosynthesis suggests that brassinosteroids have a negative regulatory effect on ABA and auxin responses and a positive regulatory effect on GA and ethylene responses (Ephritikhine et al., 1999a, 1999b). Two recent reports describe the interactions between ABA and ethylene signaling cascades (Beaudoin et al., 2000; Ghassemian et al., 2000).
Some of the foregoing examples of interconnections among hormonal responses are probably mediated by small intermediary molecules, such as a brassinosteroid or Ca2+. In contrast, the properties of the HYL1 protein suggest that it functions at the macromolecular level. In this regard, it resembles the nuclear WD protein encoded by the Arabidopsis PRL1 locus (Nemeth et al., 1998). A mutation in that locus increases the sensitivity of the plant to cytokinin, ethylene, ABA, and auxin and confers hypersensitivity to glucose. The observed increase in the basal abundance of transcripts of ABA-inducible genes in the hyl1 mutant suggests that HYL1 is involved in either the regulation of transcription itself or of mRNA stability, conjectures consonant with its nuclear localization. The HYL1 protein lacks an identifiable DNA binding motif and does not resemble known transcription factors. Although that does not prove that the HYL1 protein is not a transcriptional regulator, it does suggest that the protein is more likely to exert its regulatory effect indirectly, through interactions with other proteins. One possibility is that the HYL1 protein is a coactivator for certain auxin- and cytokinin-inducible genes and a corepressor for certain ABA-inducible genes. Also, it is possible that its molecular effects are similar, despite the apparently opposite phenotypic effects, or that some of its hormonal effects are primary and others secondary.
dsRNA binding proteins have been identified in many organisms, including humans, Drosophila, yeast, E. coli, and viruses (Burd and Dreyfuss, 1994). One of the best-characterized dsRNA binding proteins is PKR, a dsRNA-dependent protein kinase. PKR phosphorylates eukaryotic initiation factor 2 when activated by dsRNA. Induced by interferon treatment, PKR mediates the antiviral and antiproliferative effects of interferon; it also has a role in regulating cellular differentiation, stress response, and apoptosis (Clemens, 1997). Staufen, a Drosophila dsRNA binding protein with five dsRNA binding motifs, is required to properly localize bicoid and oskar mRNAs in the developing oocyte (St. Johnston et al., 1991). A plant protein with an RNA helicase domain and an RNase III–like domain has recently been identified through analysis of an Arabidopsis floral mutant (Jacobsen et al., 1999). These observations suggest that dsRNA binding proteins play several cellular roles.
Consistent with the diverse functions of proteins with dsRNA binding domains, most have a second functional or catalytic domain. PKR contains a conserved protein kinase domain at its C terminus. dsRNA adenosine deaminase has an adenosine deaminating domain required for conversion of adenosine to inosine. The Drosophila maleless protein, on the other hand, carries an RNA helicase domain (Hitti et al., 1998). The HYL1 protein also appears to contain an additional domain. The C terminus of this protein contains six consecutive copies of a 28–amino acid repeat. Using the Smith–Waterman algorithm, we found a weakly homologous repeat in protein F from S. pyogenes (24.7% identity and 55% similarity in the 190–amino acid repetitive region). In protein F, however, the domain comprises five repeats compared with six repeats in the HYL1 protein. The repeats are at the C-terminal end of both proteins, with no gaps between the repeated units, and the repeats of both proteins are rich in acidic amino acids (Asp and Glu). Protein F is a fibronectin binding protein, and its repeat region has been shown to be necessary for binding (Hanski and Caparon, 1992; Sela et al., 1993). Using the C terminus of the protein as bait in a yeast two-hybrid system, we were able to identify several interacting proteins (C. Lu and N.V. Fedoroff, unpublished data), which suggests that HYL1 contains a protein–protein interaction domain.
Several recent observations suggest that dsRNAs have regulatory and signaling functions. For example, in a variety of organisms, including nematodes, fruit flies, trypanosomes, hydra, zebrafish, and mice, homologous dsRNA can suppress gene expression selectively and specifically (Fire et al., 1998; Kennerdell and Carthew, 1998; Ngo et al., 1998; Lohmann et al., 1999; Wargelius et al., 1999). And growing, although still circumstantial, evidence suggests that dsRNA mediates both transcriptional and post-transcriptional gene silencing in plants (Vaucheret et al., 1998; Waterhouse et al., 1998; Fire, 1999; Hamilton and Baulcombe, 1999; Jones et al., 1999). Gene silencing can spread systemically in plants and requires an RNA-dependent RNA polymerase (Vaucheret et al., 1998; Voinnet et al., 1998; Dalmay et al., 2000; Mourrain et al., 2000). The recent identification of a plant protein that can carry RNA systemically is consistent with the notion that RNA is a signaling molecule (Xoconostle-Cazares et al., 1999).
Nothing is known about the role of dsRNA in the normal developmental and physiological processes of plants. Although sequence-specificity is not inherent in dsRNA binding proteins (Bevilacqua and Cech, 1996; Bevilacqua et al., 1998), gene silencing is sequence-specific. The properties of the hyl1 mutant and the protein encoded by the HYL1 locus suggest the involvement of dsRNA in the hormonal signaling networks that control growth and development in Arabidopsis. Recent studies on the mechanism of dsRNA-induced mRNA destabilization suggest that a multiprotein complex may mediate the sequence-specific destabilization of mRNA by short dsRNAs, which in turn might be enzymatically amplified (Hamilton and Baulcombe, 1999; Tuschl et al., 1999; Grishok et al., 2000; Zamore et al., 2000). Thus, an alternative hypothesis for the role of the HYL1 protein would be its participation in a regulatory mechanism that acts to destabilize transcripts by a dsRNA-dependent mechanism. Further investigation of the molecular interactions between the HYL1 protein and dsRNA, as well as the proteins that interact with HYL1, should provide insight into the role of this gene in development and of dsRNA in hormone signaling.
METHODS
Plant Lines and Growth Conditions
All experiments were performed with Arabidopsis thaliana ecotype Nossen. The transposon lines were described by Smith et al. (1997). Plants were grown on Murashige and Skoog (MS) medium (Gibco BRL, Grand Island, NY) containing 1% sucrose. In experiments on sterile medium, seeds were sterilized and grown as described in Lincoln et al. (1990). Indoleacetic acid (IAA), the synthetic auxin 2,4-dichlorophen-oxyacetic acid (2,4-D), abscisic acid (ABA), and the cytokinin 6-benzylaminopurine (BA) were added to the autoclaved medium where indicated. To examine the sensitivity to plant hormones, seedlings were measured ∼5 to 10 days after germination. To assess the root gravitropic response, wild-type and mutant seedlings were grown for 5 days on minimal medium (Lincoln et al., 1990). Plants were placed vertically. After 5 days, seedlings were transferred to square Petri plates containing minimal medium. Gravitropic stimulation was achieved by rotating the square Petri plates by 90°. The angle of curvature of the growing root tips was measured at 1-hr intervals.
TAIL-PCR and Footprint Analysis
Total DNA was extracted from leaves of mutant plants for thermal asymmetric interlaced polymerase chain reaction (TAIL PCR) amplification reactions (Dellaporta et al., 1983). TAIL PCR was performed as described by Tsugeki et al. (1996). After tertiary PCR, fragments were separated by gel electrophoresis and purified from gels by using the QIAquick gel extraction kit (QIAGEN, Valencia, CA). Purified fragments were directly sequenced. DNA was extracted from leaves of putative revertants for detection of a transposon footprint. PCR was performed with primers corresponding to the 5′ and 3′ flanking region of the Ds insertion site (5′-ATTGGCTTAGCTCACTGGATTTTG and 5′-GGTTTAAAACTGTCTCTCC). Amplified fragments were purified and directly sequenced.
Isolation of Full-Length HYL1 cDNA
A full-length cDNA was isolated from the Arabidopsis CD4-7 lambda PRL2 cDNA library (obtained from the Arabidopsis Biological Resource Center) by using the λZIPLOX selection system (Life Technologies, Inc., Bethesda, MD). Briefly, a genomic fragment was amplified by PCR with the primer pair 5′-ATTGGCTTAGCTCACTGGATTTTG and 5′-GGTTTAAAACTGTCTCTCC and then labeled with phosphorus-32 by using random primers and following standard procedures. This was used as a probe to screen the cDNA library. The cDNA clones were sequenced by using shotgun cloning and automated sequencing. Database searches for homologous sequences were performed by using the NCBI BLAST programs and the European Molecular Biology Network FDF (Fast Data Finder) tool, which is an implementation of the original Smith–Waterman dynamic programming algorithm.
DNA and RNA Gel Blot Analysis
DNA was extracted from Arabidopsis leaves as described by Dellaporta et al. (1983). DNA digests (500 ng) were fractionated on an 0.8% agarose gel, then transferred to Hybond-N+ membrane (Amersham). The full-length HYL1 cDNA was labeled with [32P]dCTP by random priming and used as a probe.
Total RNA was isolated by using the RNeasy Plant Mini Kit (QIAGEN) from liquid-grown root cultures, rosette leaves, cauline leaves, stems, and flowers of all stages from soil-grown plants ∼4 to 6 weeks after germination. To analyze the effects of ABA, seedlings were treated as described previously (Wang et al., 1998). Briefly, 2-week-old seedlings grown in pots were cleared of soil with water, then put into 0.1 × MS medium with 50 μM ABA. Seedling samples were collected after various treatment periods and total RNA was isolated. Approximately 10 μg of total RNA was fractioned in a formaldehyde gel. The HYL1-specific probe was from the cDNA coding region. KIN2 cDNA was provided by Dr. M.F. Thomashow. DNA probes for COR47 were amplified by PCR from expressed sequence tag 242B8T7 (Arabidopsis expressed sequence tag library obtained from the Arabidopsis Biological Resource Center). All the DNA probes were labeled with [32P]dCTP. Hybridization was performed at 65°C as previously described (Tsugeki et al., 1996).
Transient Assay in Onion Epidermal Cells
The 35S-HYL1 transcriptional fusion was produced by amplifying the HYL1 coding sequence from the HYL1 cDNA clone with a 5′ T7 primer and the primer 5′-CCATGCCATGGTTGCGTGGCTTGCTTCTGTCTC-3′ to create a 3′ Ncol site at the 3′ end. The amplified fragment was digested with EcoRI and NcoI and cloned into EcoRI/NcoI-digested plasmid pRTL-GUS, which contains the β-glucuronidase (GUS) coding sequence expressed from a cauliflower mosaic virus (CaMV) 35S promoter. The 35S-HYL1-GUS fusion construct was made by fusing the GUS-encoding gene carried by the plasmid pRTL-GUS to the 3′ end of the HYL1 gene by inserting the full-length HYL1 coding sequence into the EcoRI/NcoI site of the 35S-HYL1 plasmid.
Onion epidermal layers were transformed by using biolistic bombardment as described by Varagona et al. (1992). After bombardment, the layers were incubated for 20 hr at room temperature in darkness. GUS activity was detected by staining with 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid (Carrington, 1995). The subcellular location of the blue precipitate was visualized with a Zeiss Axioplan microscope (Carl Zeiss, Inc., Thornwood, NY).
In Vitro Protein Binding Activity Assay
The HYL1 cDNA was subcloned into the vector pQE-32 (QIAGEN), designed for overexpression of proteins fused to RGS–His tag. The fusion protein, a 441–amino acid protein, contained all 419 amino acids encoded by the HYL1 cDNA. The tagged protein was purified by using a Ni–NTA (where NTA is nitrilo triacetic acid) resin according to the manufacturer's protocol under nondenaturing conditions.
Proteins were fractionated by 8% SDS-PAGE and blotted onto PROTRAN membranes (Schleicher and Schuell). The blot was blocked with 3% BSA in Tris-buffered saline (TBS: 10 mM Tris-Cl, pH 7.5, and 150 mM NaCl) and incubated with anti-RGS–His antibody (1:1500 dilution). The blot was then incubated with the secondary antibody (sheep anti–mouse IgG; Amersham-Pharmacia Biotech) conjugated with horseradish peroxidase (1:6000 dilution). After each incubation step with antibodies, the membrane was washed with TBS and TBS-Tween/Triton buffer (20 mM Tris-Cl, pH 7.5, 500 mM NaCl, 0.05% [v/v] Tween 20, and 0.2% [v/v] Triton X-100). The ECL-Plus System (Amersham-Pharmacia Biotech) was used for immunodetection according to the manufacturer's instructions.
The binding activity assays were performed as previously described (Bass et al., 1994). A pBluescript II KS vector (Stratagene) containing a short fragment (multiple cloning sites) between the T7 and T3 promoters was used to prepare substrates. The T7 transcript labeled with T4 polynucleotide kinase was used as the ssRNA substrate. To make the dsRNA substrate, T3 transcripts were first kinase-labeled, then excess T7 transcripts were hybridized with labeled T3 transcripts to form dsRNA. The T7/T3 PCR products, native and denatured, were used as dsDNA and ssDNA substrates, respectively.
Acknowledgments
We thank Dr. Ryuji Tsugeki for the help with cDNA screening and phenotypic characterization, Maya Olson for revertant screening, and Dr. Michael F. Thomashow for the Kin2 clone. We are also grateful to Drs. Hong Ma, Sarah Assmann, Simon Gilroy, Philip Bevilacqua, Ramamurti Mahalingam, Fuqiang Chen, and Surhabi Raina and to Hongchang Cui and Philip Day for their helpful discussion and critical review. This work was supported by National Aeronautics and Space Administration Grant NAG5-3970.
References
- Bass, B.L., Hurst, S.R., and Singer, J.D. (1994). Binding properties of newly identified Xenopus proteins containing dsRNA-binding motifs. Current Biol. 4 301–314. [DOI] [PubMed] [Google Scholar]
- Beaudoin, N., Serizet, C., Gosti, F., and Giraudat, J. (2000). Interactions between abscisic acid and ethylene signaling cascades. Plant Cell 12 1103–1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becraft, P.W. (1998). Receptor kinases in plant development. Trends Plant Sci. 3 384–388. [Google Scholar]
- Berger, S., Bell, E., and Mullet, J.E. (1996). Two methyl jasmonate–insensitive mutants show altered expression of AtVsp in response to methyl jasmonate and wounding. Plant Physiol. 111 525–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua, P.C., and Cech, T.R. (1996). Minor-groove recognition of double-stranded RNA by the double-stranded RNA-binding domain from the RNA-activated protein kinase PKR. Biochemistry 35 9983–9994. [DOI] [PubMed] [Google Scholar]
- Bevilacqua, P.C., George, C.X., Samuel, C.E., and Cech, T.R. (1998). Binding of the protein kinase PKR to RNAs with secondary structure defects: Role of the tandem A-G mismatch and noncontiguous helixes. Biochemistry 37 6303–6316. [DOI] [PubMed] [Google Scholar]
- Bhalla, U.S., and Iyengar, R. (1999). Emergent properties of networks of biological signaling pathways. Science 283 381–387. [DOI] [PubMed] [Google Scholar]
- Bleecker, A.B. (1999). Ethylene perception and signaling: An evolutionary perspective. Trends Plant Sci. 4 269–274. [DOI] [PubMed] [Google Scholar]
- Bonetta, D., and McCourt, P. (1998). Genetic analysis of ABA signal transduction pathways. Trends Plant Sci. 3 231–235. [Google Scholar]
- Brooks, R., Eckmann, C.R., and Jantsch, M.F. (1998). The double-stranded RNA-binding domains of Xenopus laevis ADAR1 exhibit different RNA-binding behaviors. FEBS Lett. 434 121–126. [DOI] [PubMed] [Google Scholar]
- Brunn, S.A., Muday, G.K., and Haworth, P. (1992). Auxin transport and the interaction of phytotropins. Probing the properties of a phytotropin binding protein. Plant Physiol. 98 101–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd, C.G., and Dreyfuss, G. (1994). Conserved structures and diversity of functions of RNA-binding proteins. Science 265 615–621. [DOI] [PubMed] [Google Scholar]
- Carrington, J.C. (1995). Targeting of proteins to the nucleus. In Methods in Plant Cell Biology, D.W. Galbraith, D.P. Bourque, and H.J. Bohnert, eds (San Diego: Academic Press), pp. 283–293. [DOI] [PubMed]
- Clemens, M.J. (1997). PKR—A protein kinase regulated by double-stranded RNA. Int. J. Biochem. Cell Biol. 29 945–949. [DOI] [PubMed] [Google Scholar]
- Clouse, S.D., Langford, M., and McMorris, T.C. (1996). A brassinosteroid-insensitive mutant in Arabidopsis thaliana exhibits multiple defects in growth and development. Plant Physiol. 111 671–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler, S., Ghassemian, M., Bonetta, D., Cooney, S., and McCourt, P. (1996). A protein farnesyl transferase involved in abscisic acid signal transduction in Arabidopsis. Science 273 1239–1241. [DOI] [PubMed] [Google Scholar]
- D'Agostino, I.B., and Kieber, J.J. (1999). Molecular mechanisms of cytokinin action. Curr. Opin. Plant Biol. 2 359–364. [DOI] [PubMed] [Google Scholar]
- Dalmay, T., Hamilton, A., Rudd, S., Angell, S., and Baulcombe, D.C. (2000). An RNA-dependent RNA polymerase gene in Arabidopsis is required for posttranscriptional gene silencing mediated by a transgene but not by a virus. Cell 101 543–553. [DOI] [PubMed] [Google Scholar]
- Davies, W.J., and Jones, H.G. (1991). Abscisic Acid: Physiology and Biochemistry. (Oxford, UK: Bios Scientific Publishers).
- Dellaporta, S.L., Wood, J., and Hicks, J.B. (1983). A plant DNA minipreparation; version II. Plant Mol. Biol. Rep. 1 19–21. [Google Scholar]
- Elion, E.A. (1998). Routing MAP kinase cascades. Science 281 1625–1626. [DOI] [PubMed] [Google Scholar]
- Ephritikhine, G., Fellner, M., Vannini, C., Lapous, D., and Barbier-Brygoo, H. (1999. a). The sax1 dwarf mutant of Arabidopsis thaliana shows altered sensitivity of growth responses to abscisic acid, auxin, gibberellins and ethylene and is partially rescued by exogenous brassinosteroid. Plant J. 18 303–314. [DOI] [PubMed] [Google Scholar]
- Ephritikhine, G., Pagant, S., Fujioka, S., Takatsuto, S., Lapous, D., Caboche, M., Kendrick, R.E., and Barbier-Brygoo, H. (1999. b). The sax1 mutation defines a new locus involved in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 18 315–320. [DOI] [PubMed] [Google Scholar]
- Fedoroff, N.V., and Smith, D.L. (1993). A versatile system for detecting transposition in Arabidopsis. Plant J. 3 273–289. [DOI] [PubMed] [Google Scholar]
- Fire, A. (1999). RNA-triggered gene silencing. Trends Genet. 15 358–363. [DOI] [PubMed] [Google Scholar]
- Fire, A., Xu, S., Montgomery, M.K., Kostas, S.A., Driver, S.E., and Mello, C.C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391 806–811. [DOI] [PubMed] [Google Scholar]
- Fujita, H., and Syono, K. (1997). PIS1, a negative regulator of the action of auxin transport inhibitors in Arabidopsis thaliana. Plant J. 12 583–595. [DOI] [PubMed] [Google Scholar]
- Ghassemian, M., Nambara, E., Cutler, S., Kawaide, H., Kamiya, Y., and McCourt, P. (2000). Regulation of abscisic acid signaling by the ethylene response pathway in Arabidopsis. Plant Cell 12 1117–1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmour, S.J., Artus, N.N., and Thomashow, M.F. (1992). cDNA sequence analysis and expression of two cold-regulated genes of Arabidopsis thaliana. Plant Mol. Biol. 18 13–21. [DOI] [PubMed] [Google Scholar]
- Grill, E., and Himmelbach, A. (1998). ABA signal transduction. Curr. Opin. Plant Biol. 1 412–418. [DOI] [PubMed] [Google Scholar]
- Grishok, A., Tabara, H., and Mello, C.C. (2000). Genetic requirements for inheritance of RNAi in C. elegans. Science 287 2494–2497. [DOI] [PubMed] [Google Scholar]
- Hamilton, A.J., and Baulcombe, D.C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 950–952. [DOI] [PubMed] [Google Scholar]
- Hanski, E., and Caparon, M. (1992). Protein F, a fibronectin-binding protein, is an adhesin of the group A streptococcus Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 89 6172–6176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirt, H. (2000). MAP kinases in plant signal transduction. Results Probl. Cell Differ. 27 1–9. [DOI] [PubMed] [Google Scholar]
- Hitti, E., Neunteufl, A., and Jantsch, M.F. (1998). The double-stranded RNA-binding protein X1rbpa promotes RNA strand annealing. Nucleic Acids Res. 26 4382–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hooley, R. (1998). Auxin signaling: Homing in with targeted genetics. Plant Cell 10 1581–1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishitani, M., Xiong, L., Stevenson, B., and Zhu, J.K. (1997). Genetic analysis of osmotic and cold stress signal transduction in Arabidopsis: Interactions and convergence of abscisic acid–dependent and abscisic acid–independent pathways. Plant Cell 9 1935–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsen, S.E., Running, M.P., and Meyerowitz, E.M. (1999). Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development 126 5231–5243. [DOI] [PubMed] [Google Scholar]
- Jones, L., Hamilton, A.J., Voinnet, O., Thomas, C.L., Maule, A.J., and Baulcombe, D.C. (1999). RNA–DNA interactions and DNA methylation in post-transcriptional gene silencing. Plant Cell 11 2291–2302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katekar, G.F., and Geissler, A.E. (1977). Auxin transport inhibitors. III. Chemical requirements of a class of auxin transport inhibitors. Plant Physiol. 60 826–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kennerdell, J.R., and Carthew, R.W. (1998). Use of dsRNA-mediated genetic interference to demonstrate that frizzled and frizzled 2 act in the wingless pathway. Cell 95 1017–1026. [DOI] [PubMed] [Google Scholar]
- Kharrat, A., Macias, M.J., Gibson, T.J., Nilges, M., and Pastore, A. (1995). Structure of the dsRNA binding domain of E. coli RNase III. EMBO J. 14 3572–3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef, M., Reuling, G., and Karssen, C.M. (1984). The isolation and characterization of abscisic acid–insensitive mutants of Arabidopsis thaliana. Physiol. Plant. 61 377–383. [Google Scholar]
- Kurkela, S., and Borg-Franck, M. (1992). Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 19 689–692. [DOI] [PubMed] [Google Scholar]
- Lincoln, C., Britton, J.H., and Estelle, M. (1990). Growth and development of the axr1 mutants of Arabidopsis. Plant Cell 2 1071–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, Y.G., and Whittier, R.F. (1995). Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25 674–681. [DOI] [PubMed] [Google Scholar]
- Lohmann, J.U., Endl, I., and Bosch, T.C. (1999). Silencing of developmental genes in Hydra. Dev. Biol. 214 211–214. [DOI] [PubMed] [Google Scholar]
- Makrides, S.C. (1996). Strategies for achieving high-level expression of genes in Escherichia coli. Microbiol. Rev. 60 512–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller, S.G., and Chua, N.H. (1999). Interactions and intersections of plant signaling pathways. J. Mol. Biol. 293 219–234. [DOI] [PubMed] [Google Scholar]
- Mourrain, P., et al. (2000). Arabidopsis SGS2 and SGS3 genes are required for posttranscriptional gene silencing and natural virus resistance. Cell 101 533–542. [DOI] [PubMed] [Google Scholar]
- Mulligan, R.M., Chory, J., and Ecker, J.R. (1997). Signaling in plants. Proc. Natl. Acad. Sci. USA 94 2793–2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara, E., and McCourt, P. (1999). Protein farnesylation in plants: A greasy tale. Curr. Opin. Plant Biol. 2 388–392. [DOI] [PubMed] [Google Scholar]
- Nambara, E., Naito, S., and McCourt, P. (1992). A mutant of Arabidopsis which is defective in seed development and storage protein accumulation is a new abi3 allele. Plant J. 2 435–441. [Google Scholar]
- Nemeth, K., et al. (1998). Pleiotropic control of glucose and hormone responses by PRL1, a nuclear WD protein, in Arabidopsis. Genes Dev. 12 3059–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngo, H., Tschudi, C., Gull, K., and Ullu, E. (1998). Double-stranded RNA induces mRNA degradation in Trypanosoma brucei. Proc. Natl. Acad. Sci. USA 95 14687–14692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlman, R.F., Fedoroff, N.V., and Messing, J. (1984). The nucleotide sequence of the maize controlling element Activator. Cell 37 635–643. [DOI] [PubMed] [Google Scholar]
- Schwarz-Sommer, Z., Gierl, A., Cuypers, H., Peterson, P.A., and Saedler, H. (1985). Plant tranposable elements generate the DNA sequence diversity needed in evolution. EMBO J. 14 591–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sela, S., Aviv, A., Tovi, A., Burstein, I., Caparon, M.G., and Hanski, E. (1993). Protein F: An adhesin of Streptococcus pyogenes binds fibronectin via two distinct domains. Mol. Microbiol. 10 1049–1055. [DOI] [PubMed] [Google Scholar]
- Shieh, M.W., Wessler, S.R., and Raikhel, N.V. (1993). Nuclear targeting of the maize R protein requires two nuclear localization sequences. Plant Physiol. 101 353–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, J.D., Todd, P., and Staehelin, A.L. (1997). Modulation of statolith mass and grouping in white clover (Trifolium repens) grown in 1-g, microgravity and on the clinostat. Plant J. 12 1361–1373. [DOI] [PubMed] [Google Scholar]
- Solano, R., and Ecker, J.R. (1998). Ethylene gas: Perception, signaling and response. Curr. Opin. Plant Biol. 1 393–398. [DOI] [PubMed] [Google Scholar]
- St. Johnston, D., Beuchle, D., and Nusslein-Volhard, C. (1991). Staufen, a gene required to localize maternal RNAs in the Drosophila egg. Cell 66 51–63. [DOI] [PubMed] [Google Scholar]
- St. Johnston, D., Brown, N.H., Gall, J.G., and Jantsch, M. (1992). A conserved double-stranded RNA-binding domain. Proc. Natl. Acad. Sci. USA 89 10979–10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E., Su, W.P., and Howell, S.H. (1992). Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proc. Natl. Acad. Sci. USA 89 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steber, C.M., Cooney, S.E., and McCourt, P. (1998). Isolation of the GA-response mutant sly1 as a suppressor of ABI1-1 in Arabidopsis thaliana. Genetics 149 509–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taiz, L., and Zeiger, E. (1991). Plant Physiology. (Redwood City: Benjamin/Cummings Publishing), pp. 473–488.
- Thornton, T.M., Swain, S.M., and Olszewski, N.E. (1999). Gibberellin signal transduction presents … the SPY who O-GlcAc'd me. Trends Plant Sci. 4 424–428. [DOI] [PubMed] [Google Scholar]
- Trewavas, A.J., and Malho, R. (1997). Signal perception and transduction: The origin of the phenotype. Plant Cell 9 1181–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsugeki, R., Kochieva, E.Z., and Fedoroff, N.V. (1996). A transposon insertion in the Arabidopsis SSR16 gene causes an embryo-defective lethal mutation. Plant J. 10 479–489. [DOI] [PubMed] [Google Scholar]
- Tuschl, T., Zamore, P.D., Lehmann, R., Bartel, D.P., and Sharp, P.A. (1999). Targeted mRNA degradation by double-stranded RNA in vitro. Genes. Dev. 13 3191–3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urao, T., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2000). Two-component systems in plant signal transduction. Trends Plant Sci. 5 67–74. [DOI] [PubMed] [Google Scholar]
- Varagona, M.J., Schmidt, R.J., and Raikhel, N.V. (1992). Nuclear localization signal(s) required for nuclear targeting of the maize regulatory protein Opaque-2. Plant Cell 4 1213–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J.B., Mourrain, P., Palauqui, J.C., and Vernhettes, S. (1998). Transgene-induced gene silencing in plants. Plant J. 16 651–659. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Woeste, K.E., Theologis, A., and Kieber, J.J. (1998). Recessive and dominant mutations in the ethylene biosynthetic gene ACS5 of Arabidopsis confer cytokinin insensitivity and ethylene overproduction, respectively. Proc. Natl. Acad. Sci. USA 95 4766–4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voinnet, O., Vain, P., Angell, S., and Baulcombe, D.C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95 177–187. [DOI] [PubMed] [Google Scholar]
- Wang, H., Qi, Q., Schorr, P., Cutler, A.J., Crosby, W.L., and Fowke, L.C. (1998). ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. Plant J. 15 501–510. [DOI] [PubMed] [Google Scholar]
- Wargelius, A., Ellingsen, S., and Fjose, A. (1999). Double-stranded RNA induces specific developmental defects in zebrafish embryos. Biochem. Biophys. Res. Commun. 263 156–161. [DOI] [PubMed] [Google Scholar]
- Waterhouse, P.M., Graham, M.W., and Wang, M.B. (1998). Virus resistance and gene silencing in plants can be induced by simultaneous expression of sense and antisense RNA. Proc. Natl. Acad. Sci. USA 95 13959–13964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng, G., Bhalla, U.S., and Iyengar, R. (1999). Complexity in biological signaling systems. Science 284 92–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, A.K., Pickett, F.B., Turner, J.C., and Estelle, M. (1990). A dominant mutation in Arabidopsis confers resistance to auxin, ethylene and abscisic acid. Mol. Gen. Genet. 222 377–383. [DOI] [PubMed] [Google Scholar]
- Wobus, U., and Weber, H. (1999). Seed maturation: Genetic programmes and control signals. Curr. Opin. Plant Biol. 2 33–38. [DOI] [PubMed] [Google Scholar]
- Xoconostle-Cazares, B., Xiang, Y., Ruiz-Medrano, R., Wang, H.L., Monzer, J., Yoo, B.C., McFarland, K.C., Franceschi, V.R., and Lucas, W.J. (1999). Plant paralog to viral movement protein that potentiates transport of mRNA into the phloem. Science 283 94–98. [DOI] [PubMed] [Google Scholar]
- Zamore, P.D., Tuschl, T., Sharp, P.A., and Bartel, D.P. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101 25–33. [DOI] [PubMed] [Google Scholar]
- Zhu, J., Hasegawa, P.M., and Bressan, R.A. (1997). Molecular aspects of osmotic stress in plants. Crit. Rev. Plant Sci. 16 253–277. [Google Scholar]