Abstract
Plants produce a wide array of natural products, many of which are likely to be useful bioactive structures. Unfortunately, these complex natural products usually occur at very low abundance and with restricted tissue distribution, thereby hindering their evaluation. Here, we report a novel approach for enhancing the accumulation of natural products based on activation tagging by Agrobacterium-mediated transformation with a T-DNA that carries cauliflower mosaic virus 35S enhancer sequences at its right border. Among ∼5000 Arabidopsis activation-tagged lines, we found a plant that exhibited intense purple pigmentation in many vegetative organs throughout development. This upregulation of pigmentation reflected a dominant mutation that resulted in massive activation of phenylpropanoid biosynthetic genes and enhanced accumulation of lignin, hydroxycinnamic acid esters, and flavonoids, including various anthocyanins that were responsible for the purple color. These phenotypes, caused by insertion of the viral enhancer sequences adjacent to an MYB transcription factor gene, indicate that activation tagging can overcome the stringent genetic controls regulating the accumulation of specific natural products during plant development. Our findings suggest a functional genomics approach to the biotechnological evaluation of phytochemical biodiversity through the generation of massively enriched tissue sources for drug screening and for isolating underlying regulatory and biosynthetic genes.
INTRODUCTION
Ethnobotany and limited screens of medicinal plants indicate that the huge repertoire of chemical diversity in plants contains many potentially useful bioactive structural principles for developing novel drugs, flavors, fragrances, and other specialty chemicals. Unfortunately, these complex natural products usually occur in very low abundance and with a restricted tissue distribution. Moreover, almost all of this phytochemical biodiversity resides in exotic, uncultivated species. Whereas drugs such as taxol, vinblastine, and vincristine illustrate the potential of plants as sources of new drugs, the development of rational approaches for the generation of useful amounts of complex natural products for industrial evaluation remains an unsolved problem. In particular, an intense 30-year effort using cell and tissue cultures from medicinal plants has failed to generate useful quantities of complex products for the commercial production of established drugs in vitro or for high-throughput, multiplex screening of phytochemicals (Facchini and Deluca, 1995; McCaskill and Croteau, 1998). This failure probably reflects the stringent spatial and temporal transcriptional controls governing the biosynthesis of specific natural products during plant development (Fowler, 1983; Robins, 1994; Facchini and Deluca, 1995). Transgenic manipulation to override these genetic controls thus may provide the key to enhancing natural product biosynthesis for industrial evaluation and exploitation.
Activation tagging with the enhancer from the cauliflower mosaic virus 35S transcript promoter (35Se) is an emerging technology in plant functional genomics (Weigel et al., 2000). This approach, based on Agrobacterium-mediated transformation, can create transgenic plants in which the T-DNA carrying 35Se at its right border is spliced into the plant genome at random sites. In each independent transgenic line, 35Se strongly activates the plant gene to which, by chance, it lies adjacent. Activation of a gene in this fashion may lead to observable effects on the modified plant, providing important clues about the function of the activated gene. Screening large collections of independent, activation-tagged lines thus represents a powerful way of surveying the genome and isolating genes that affect traits of interest.
Using activation tagging, we have isolated a bright-purple mutant, production of anthocyanin pigment 1-Dominant (pap1-D), in which genes encoding enzymes involved in the biosynthesis of phenylpropanoid natural products exhibit massive and widespread activation throughout plant development. The pap1-D phenotype, which is caused by overexpression of a gene encoding an MYB transcription factor, indicates that activation tagging can be used to overcome the stringent genetic controls regulating the developmental accumulation of specific natural products. These findings suggest a new approach for the systematic biotechnological evaluation of phytochemical biodiversity through the generation of massively enriched tissue sources for drug screening and for isolation of the underlying regulatory and biosynthetic genes.
RESULTS
Mutant Characterization
Approximately 5000 activation-tagged primary lines of Arabidopsis ecotype Columbia (Col-0) were generated by using pSKI015, which contains four copies of 35Se at the right border of the T-DNA, pBluescript KS+ for plasmid rescue, and the BAR gene for Basta resistance as a selectable marker (Kardailsky et al., 1999; Weigel et al., 2000). A single bright-purple plant was observed in this collection, and its seed was collected for progeny analysis. T2 plants segregated for the purple coloration characteristic of anthocyanins in a 3:1 ratio, which is consistent with this trait being determined by a single dominant allele, an allele we named pap1-D (see above). The whole plant, including the roots, stems, leaves, primary and secondary branches, and cauline leaves as well as sepals, anthers, and carpels, exhibited purple pigmentation (Figures 1B and 1C). The purple coloration was more pronounced when plants were grown under high-intensity light or other stress conditions, such as drought and pathogen infection (data not shown). Under these conditions, leaves and stems of wild-type plants also show a slight pigmentation, suggesting that the pap1-D phenotype might in part reflect enhancement of an endogenous stress response. However, we never observed pigmentation in the roots of wild-type plants—in marked contrast to the strong pigmentation at the base of pap1-D roots (Figure 1B). Except for very weak pigmentation in flower petals (Figure 1A), enhanced pigmentation in pap1-D was observed throughout development. No other morphological phenotypes were observed.
Figure 1.
pap1-D Phenotypes.
(A) pap1-D (left) and Col-0 (right) flowers.
(B) Roots of pap1-D (left) and Col-0 (right) plants.
(C) Six-week-old adult pap1-D (front) and Col-0 (back) plants.
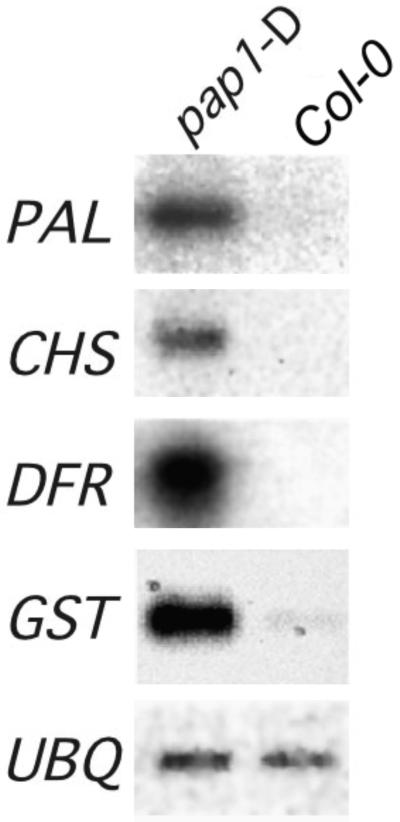
Because anthocyanins are a subclass of flavonoid natural products derived from the phenylpropanoid skeleton, we examined the expression of phenylpropanoid biosynthetic genes and the accumulation of natural products. RNA gel blot analysis showed massive enhancement of the expression of phenylpropanoid biosynthetic genes in pap1-D plants (Figure 2). The amounts of transcripts encoding chalcone synthase (CHS), the entry point enzyme into the flavonoid branch pathway, and dihydroflavonol reductase, an enzyme of flavonoid biosynthesis specific for anthocyanins, were greater in pap1-D plants than in wild-type Col-0 plants. Transcripts encoding glutathione S-transferase, which has been implicated in the transport of anthocyanins into the vacuole (Alfenito et al., 1998), also were expressed in increased amounts. Moreover, the accumulation of transcripts that encode phenylalanine ammonia-lyase (PAL), the first enzyme of the overall phenylpropanoid biosynthetic pathway, also was markedly enhanced, indicating that transcriptional activation was not confined to the flavonoid branch.
Figure 2.
Enhanced Expression of Phenylpropanoid Biosynthetic Genes in pap1-D.
RNA gel blot hybridization was conducted with total RNA isolated from 6-week-old pap1-D and Col-0 wild-type plants. DFR, dihydroflavonol reductase; GST, glutathione S-transferase; UBQ, ubiquitin.
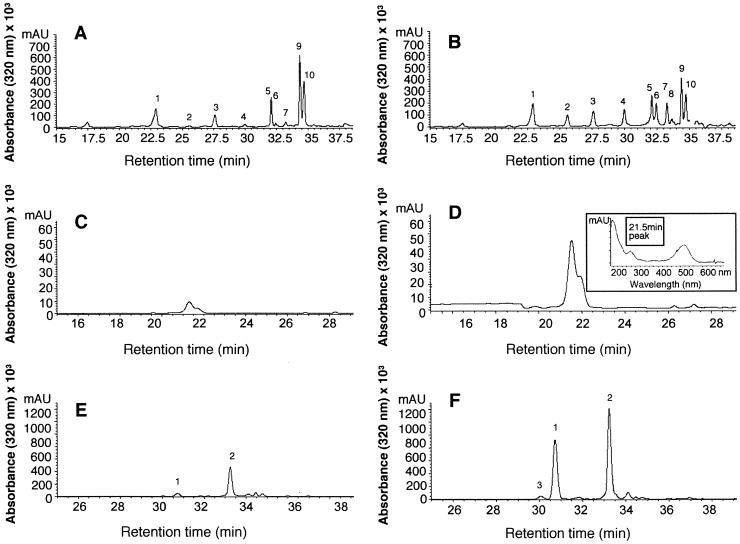
To determine the extent of changes in anthocyanins and other phenylpropanoid-derived compounds in pap1-D, we extracted and analyzed soluble and cell wall–bound phenolic compounds by HPLC. Analysis of the soluble fraction, which was obtained by extraction in acetone, revealed several quantitative differences between pap1-D and wild-type Col-0 plants—in particular, increased concentrations of certain flavonol glycosides, including Glc-rhamnose (Rha)-quercetin, Glc-Rha-kaempferol, and unidentified conjugates of kaempferol and quercetin (Figures 3A and 3B). After alkaline hydrolysis of the residue that was obtained after acetone extraction, one portion was freeze-dried for analysis of anthocyanidins (anthocyanin aglycones); the remainder was partitioned into ethyl acetate for determination of cell wall–bound hydroxycinnamic acids. Anthocyanidins were present in greater concentrations in pap1-D than in Col-0 (Figures 3C and 3D), as were coumaric and sinapic acids measured in alkaline hydrolysates of the wall-bound phenolic fraction (Figures 3E and 3F). Thus, pap1-D is characterized by strongly increased concentrations of glycosylated anthocyanins, flavonols, and cell wall–esterified hydroxycinnamic acids in comparison with wild-type Col-0.
Figure 3.
Effect of pap1-D Mutation on Accumulation of Phenylpropanoid Products.
(A) to (F) HPLC profiles of phenylpropanoid metabolites in extracts from wild-type Col-0 ([A], [C], and [E]) and pap1-D ([B], [D], [F]) plants.
(A) and (B) Soluble phenolics: peak 1, rhamnose (Rha)-Glc-Rha-quercetin; peak 2, quercetin conjugate; peak 3, Rha-Glc-Rha-kaempferol; peak 4, Glu-Rha-quercetin; peak 5, Rha-Rha-quercetin; peak 6, Glc-Rha-kaempferol; peaks 7 and 8, kaempferol conjugates; peak 9, sinapic acid; peak 10, Rha-Rha-kaempferol.
(C) and (D) Anthocyanidins; the inset in (D) shows the UV light absorption spectrum of the major anthocyanidin eluting at 21.5 min.
(E) and (F) Wall-bound phenolics: peak 1, trans-4-coumaric acid; peak 2, sinapic acid; peak 3, cis-4-coumaric acid.
The observation of increased wall-bound hydroxycinnamic acids in pap1-D prompted us to measure the content and composition of lignin, which is derived from hydroxycinnamic acid precursors. Lignin was analyzed by derivatization followed by reductive cleavage, which helps to determine the absolute amounts of guaiacyl (G, monomethylated) and syringyl (S, dimethylated) lignin monomers (Lu and Ralph, 1997). The results in Table 1 indicate increases in both total G and total S residues in the cell wall fraction of pap1-D compared with those in Col-0, but the S/G ratio varied little. The change in lignin monomers could reflect an increase in lignin content or a change in composition that led to more efficient monomer extractability.
Table 1.
Enhanced PAL Activity and Lignin Levels in pap1-D Plants
| Plant | PAL Activity (mkat/g FW)aStem |
COMTb Activity (μkat/g FW) Stem |
CCOMTc Activity (pkat/g FW) Stem |
Lignin Compostion
|
|||
|---|---|---|---|---|---|---|---|
| Total G (μmol/g) |
Total S (μmol/g) |
Total G and S (μmol/g) |
S/G | ||||
| Wild type | 30.5 | 44.2 | 50.4 | 21.6 | 2.5 | 24.1 | 0.12 |
| pap1-D | 70.3 | 44.4 | 60.6 | 30.6 | 5.0 | 35.6 | 0.16 |
FW, fresh weight; kat, katal.
COMT, caffeic acid O-methyltransferase.
CCOMT, caffeoyl-CoA O-methyltransferase.
Changes in lignin content and composition have been engineered in transgenic plants by downregulation of PAL, caffeic acid O-methyltransferase, and caffeoyl-CoA O-methyltransferase, enzymes of the lignin branch of phenylpropanoid biosynthesis (Atanassova et al., 1995; Zhong et al., 1998). PAL activity in stems of pap1-D plants was approximately twice that found in stems of wild-type plants, whereas the activities of the two O-methyltransferases differed little between the two (Table 1). Thus, the changes in lignin composition and increased concentrations of wall-bound sinapic acid in pap1-D reflect the change in PAL activity but do not appear to be associated with increases in lignin O-methyltransferase activities.
The Arabidopsis transparent testa glabra1-1 (ttg1-1) mutation blocks anthocyanin accumulation and trichome formation (Koornneef, 1981). TTG1 encodes a WD40 repeat protein homologous with an AN11-encoded protein from petunia, which also controls anthocyanin production (de Vetten et al., 1997; Walker et al., 1999). To test the genetic relationship between TTG1 and PAP1, we crossed ttg1-1 with pap1-D. The pap1-D allele was tracked by Basta resistance, and the ttg1-1 mutation was scored visibly. The double mutant F2 plants failed to accumulate anthocyanins, indicating that TTG1 is required for the production of anthocyanins mediated by PAP1 overexpression and acts either downstream from or at the same step as PAP1.
Cloning of PAP1
In a population of >100 segregating T2 plants, each plant that had the pap1-D phenotype showed resistance to Basta, and all plants with a wild-type phenotype (i.e., lacking purple pigmentation) were sensitive to Basta, indicating that the mutation was tightly linked to the T-DNA insert. Moreover, hybridization of DNA gel blots of pap1-D genomic DNA that had been digested with EcoRI or KpnI showed that the 35Se sequences were localized to single fragments of 10 and 12 kb, respectively (data not shown), indicating that the mutant contained a single, simple insertion. The 12-kb KpnI and 10-kb EcoRI fragments were cloned by plasmid rescue (Weigel et al., 2000), generating pPAP1-K1 and pPAP1-E1, respectively. Nucleotide sequencing and restriction analysis showed that the 12-kb KpnI fragment fully overlapped the smaller EcoRI fragment. Probing the Arabidopsis CD4-7 cDNA library at high stringency with pPAP1-K1 resulted in isolation of a single 956-bp cDNA, which defined three exons in the genomic DNA of pPAP1-K1 encoding an MYB transcription factor (Figures 4A and 4B). In the pap1-D line, the 35Se sequences had inserted 5.1 kb 3′ to the start of this gene, designated PAP1, and RNA gel blot hybridization with the PAP1 cDNA revealed a single 1-kb transcript, which was massively overexpressed in the pap1-D mutant (Figure 4C).
Figure 4.
Molecular Characterization of PAP1.
(A) Genomic context of T-DNA insertion in pap1-D and structure of pMN-PAP1. Bastar, Basta resistance; Kanr, Kanamycin resistance; LB, left border; pBS, pBluescript KS+ plasmid; RB, right border; 4 × 35S denotes four copies of 35Se.
(B) Sequence homology of R2R3 MYB. Proteins were aligned using the ClustalW software program. Red shading denotes 100% conserved residues, and yellow shading denotes matching residues with PAP1. R2 and R3 MYB domains are shown. GenBank accession numbers PAP1 (AF325123), PAP2 (AF325124), AN2 (AAF66727), C1 (AAA33482), P1 (AAB67720), P (AAC49394), GL1 (P27900), Mixta (CAA55725), and c-myb (AAA52031).
(C) Gel blot hybridization of total RNA from 4-week-old pap1-D and Col-0 plants.1.0 denotes transcript size in kilobases.
(D) Neighbor-joining phylogenetic tree built by using full-length proteins, showing the anthocyanin MYB family branch. The human gene c-myb is used as an outgroup.
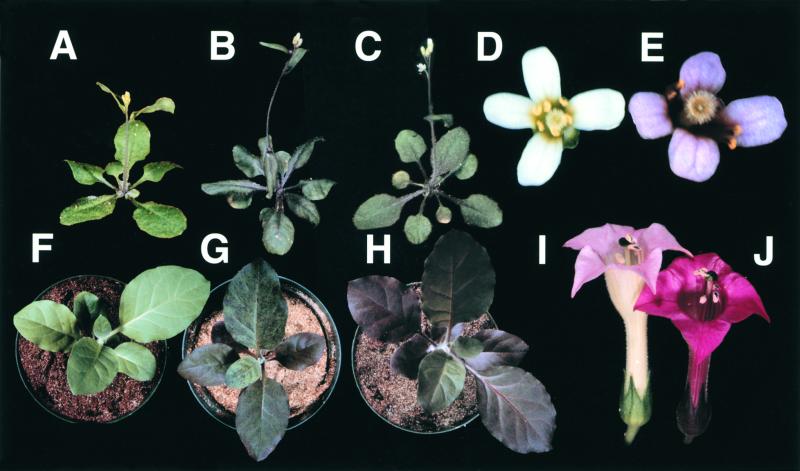
To confirm that overexpression of PAP1 caused the pap1-D phenotype, we cloned a 3-kb genomic fragment spanning the three PAP1 exons and flanking sequences into pMN20-2, which contains two copies of 35Se (Weigel et al., 2000), thereby creating pMN-PAP1. Transformation of wild-type Col-0 with this construct, which mimics the genomic context of the pap1-D allele, generated multiple transgenic lines with the characteristic purple phenotype (Figure 5B). As would be expected from position effects, these transgenic lines represented an allelic series ranging from the wild type to an even more intense phenotype than pap1-D, in some cases having strong purple pigmentation in the petals (Figure 5E). In contrast, transformation with pMN20-2 as an empty vector control gave no enhanced pigmentation phenotype (Figures 5A and 5D).
Figure 5.
Overexpression of PAP1 or PAP2 Enhances Pigmentation in Arabidopsis and Tobacco.
(A) to (E) Arabidopsis plants transformed with pMN20-2 ([A] and [D]), pMN-PAP1 ([B] and [E]), and pCHF3:PAP2 (C). (A) to (C) show six-week-old plants. (D) and (E) show flowers on 12-week-old plants.
(F) to (J) Tobacco plants transformed with pCHF3 ([F] and [I]), pCHF3:PAP1 (G), and pCHF3:PAP2 ([H] and [J]). Plantlets in (F) to (H) were photographed at age 4 weeks, and flowers in (I) and (J) at 10 weeks after transfer to soil. pCHF3-PAP1 plants had brilliant flower pigmentation, identical to that of pCHF3-PAP2 (data not shown).
Sequence alignments with the Arabidopsis databases showed that PAP1 is a member of the R2, R3 MYB family, which is estimated to have >100 members in Arabidopsis (Kranz et al., 1998; Romero et al., 1998) and >80 members in maize (Rabinowicz et al., 1999). PAP1 is identical to ATMYB75 (Kranz et al., 1998) except that ATMYB75 contains a sequencing error (1-bp deletion at position 695), creating an early stop codon. The PAP1 protein shares high homology with other MYB-like transcription factors that regulate anthocyanin production (Figures 4B and 4D). PAP1 is closely related to the product of the petunia AN2 gene, showing 82% identity through the MYB region and 50% identity overall. The products of the maize anthocyanin MYB genes C1 and pl are 64% identical through the MYB region, with 38% identity overall. The MYB transcription factor GLABAROUS1 from Arabidopsis and MIXTA from snapdragon both control trichome development (Oppenheimer et al., 1991; Glover et al., 1998) and are each 58% identical to PAP1 in the MYB domain and 33% identical overall. A phylogenetic tree constructed with these full-length MYB proteins shows that PAP1 belongs to a branch involved in anthocyanin biosynthesis (Figure 4D).
The PAP1 gene was mapped to 0.2 centimorgan (cM) up from mi303 at 83.7 cM on chromosome 1 by using an Xba1 restriction fragment length polymorphism and the Col/Ler recombinant inbred lines (Nottingham University Stock Centre, Nottingham, UK). The sequencing project recently came to PAP1 on bacterial artificial chromosome F25P12 just below mi303 at 85 cM.
PAP2
Also discovered in the Arabidopsis database was 193M15, an expressed sequence tag with very high homology with PAP1. 193M15 encodes a protein with 93% identity to PAP1 in the MYB domain and 77% identity overall. To test whether overexpression of 193M15 also could cause the production of anthocyanin pigments, we made a 35S promoter::cDNA fusion construct, pCHF3-PAP2. Transgenic plants containing pCHF3-PAP2 had purple leaves and stems (Figure 5C), although the pigmentation was somewhat weaker than that of the pap1-D mutant or some pMN-PAP1 lines. In view of the sequence similarity between the PAP1 protein and that encoded by 193M15 (Figures 4B and 4D) and similar overexpression phenotypes (Figure 5C), we named the 193M15 cDNA PAP2. PAP2 is on bacterial artificial chromosome T27F4 near mi424 on chromosome 1, ∼9 cM below PAP1. PAP2 is also a member of the Arabidopsis MYB family reported as ATMYB90 (Kranz et al., 1998).
Overexpression of PAP1 and PAP2 in Tobacco
To test whether PAP1 and PAP2 could enhance pigmentation in other plants, we transformed tobacco cv xanthi with pCHF3-PAP1 and pCHFS-PAP2. Both constructs generated purple tobacco plants, indicating that the Arabidopsis PAP1 and PAP2 genes could activate production of anthocyanin pigments in another species (Figures 5G and 5H). These transgenic tobacco plants also produced flowers with much more pigmentation than did the pCHF3 transgenic control plants (Figures 5I and 5J). Tobacco transformed with pCHF3 as an empty vector control did not have an increased pigmentation phenotype (Figures 5F and 5I).
DISCUSSION
Accumulation of phenylpropanoid products during development is under tight transcriptional regulation, and the pap1-D phenotype represents a striking override of these genetic controls. Thus, specific tranches of the overall pathway appear to be controlled by separate sets of transcription factors. For example, the maize myb genes C1 and pl are involved in the regulation of anthocyanin synthesis from CHS onward but do not regulate PAL and other genes of the upstream central pathway (Cone et al., 1993a, 1993b; Mol et al., 1996), whereas P independently controls the 3-deoxy flavonoid branch pathway (Grotewold et al., 1994). In contrast, the pap1-D phenotype, which results from overexpression of the PAP1 MYB transcription factor, reflects massively enhanced expression of PAL as well as CHS, the gene encoding dihydroflavonol reductase, and the glutathione S-transferase gene. This broad transcriptional activation of the overall pathway is accompanied by a correspondingly broad pattern of enhanced product accumulation with increases in lignin, wall-bound hydroxycinnamic acid esters, flavonols, and anthocyanins. Moreover, pathway activation in pap1-D was observed in all vegetative organs and maintained throughout development, in marked contrast to activation in wild-type plants of individual branch pathways at defined developmental stages and with characteristic cell-type, tissue-type, and organ specificities (Graham, 1991; Grotewold et al., 1994). The relatively modest increase in lignin content probably reflects a major control point at the polymerization stage (Bate et al., 1994) with consequent spillover of lignin monomers and their precursors, which contributes to the marked accumulation of wall-bound hydroxycinnamic acid esters in pap1-D.
MYB genes contribute to the control of flavonoid biosynthesis in a wide range of plant species, often in combination with other regulatory genes. The extensive sequence similarity with AN2, c1, and pl, together with the overexpression phenotypes, suggests that PAP1 and PAP2 may be the Arabidopsis orthologs of these petunia and maize myb genes, with genetically defined functions in phenylpropanoid regulation. In maize, c1 and pl but not P require R and B, encoding basic helix-loop-helix proteins, to activate transcription of maize flavonoid biosynthetic genes (Mol et al., 1996). Basic helix-loop-helix proteins and MYB proteins also function together in the control of flower pigmentation in snapdragon (Goodrich et al., 1992) and petunia (Quattrocchio et al., 1998). Moreover, the WD40 proteins TTG1 and AN11 are required for MYB control of flavonoid biosynthesis in both Arabidopsis and petunia (de Vetten et al., 1997; Larkin et al., 1999; Walker et al., 1999), and the pap1-D phenotypes require the WD40 gene TTG1. Despite the stringent and often complex genetic control of phenylpropanoid biosynthesis, strong overexpression of PAP1 in the pap1-D line was sufficient to hyperactivate the pathway, which is reminiscent of the enhancement of flavonoid biosynthesis by deliberate ectopic expression of P in suspension cultures of maize cells (Grotewold et al., 1999). The pap1-D phenotypes may reflect involvement of PAP1 as the limiting factor in a novel regulatory circuit with atypically broad control functions in phenylpropanoid biosynthesis or may indicate functional spillover from one regulatory circuit to related circuits when PAP1 is massively overexpressed.
A recent report describes the use of activation tagging in Catharanthus cell cultures to isolate ORCA3, a jasmonate-responsive transcriptional regulator of primary and secondary metabolism, the upregulation of which promotes biosynthesis of indole alkaloids (Van der Fits and Memelink, 2000). These data, along with the present findings, indicate that activation tagging can be used to generate novel gain- of-function mutations that affect complex biosynthetic pathways under polygenic control; as such, this presents a potentially powerful new approach for isolating the genes that regulate biosynthesis of plant natural products. Loss-of-function screens for transparent testa have been saturated, and no mutations map to the PAP1 or PAP2 loci (Shirley et al., 1995). Moreover, examination of >100 PAP1 antisense lines showed no visible phenotype (data not shown). The similar overexpression phenotypes of PAP1 and PAP2 suggest that these genes may be functionally redundant, such that only activation tagging or some other gain-of-function screen could have readily revealed their key attributes.
Activation tagging as a gene discovery tool based on gain-of-function is intrinsically oriented toward biotechnological utility, and the ability to activate a biosynthetic pathway that will lead to the enhanced accumulation of several distinct subclasses of natural products has several important potential applications. Thus, hyperactivated tissues or organs provide massively enriched sources for passage through multiplex drug screens with the potential for discovery of novel activities based on what combinatorial effects might arise from complex mixtures as well as allowing convenient isolation and characterization of individual bioactive components. This approach in principle could be augmented by feeding studies using pathway intermediates or synthetic derivatives. Moreover, activation of phenylpropanoid biosynthesis in pap1-D reflects massively enhanced expression of genes encoding pathway enzymes; hence, these tissues provide a correspondingly enriched source for isolating the cDNAs that encode key biosynthetic enzymes not readily identified by biochemical approaches.
Although the plant kingdom has a remarkable diversity of natural products, the underlying pathway regulatory mechanisms appear to be at least partially conserved between species (Mol et al., 1996; Quattrocchio et al., 1998). For example, the maize anthocyanin regulatory gene R functions appropriately when expressed in Arabidopsis or tobacco (Lloyd et al., 1992). Likewise, ectopic expression of PAP1 or PAP2 in transgenic tobacco caused phenotypes similar to those observed in Arabidopsis. Therefore, convenient, readily transformed genetic model species, such as Arabidopsis, can be used to isolate candidate regulatory genes for direct evaluation in medicinal plants and other exotic species or as a platform for the identification of orthologs and potentially useful, related genes in target species.
The serendipitous discovery of pap1-D among a large collection of activation-tagged lines was possible because activation of PAP1 enhanced the accumulation of anthocyanin pigments, which was easily scored. Several other plant natural products, such as the isoprenoids lycopene and carotene and the alkaloid sanguinarine, also are colored. Hence, genetic activation of these tranches of plant metabolism also could be scored by visual inspection, but this is not a generally applicable approach. In principle, activation-tagged lines with enhanced accumulation of natural products of interest could be identified by high-throughput metabolic profiling. However, a more promising general strategy may be to make transgenic plants that express easily screened marker genes under the control of promoters from genes encoding enzymes involved in the biosynthesis of natural products of interest.
METHODS
Plant Growth and Transformation
Arabidopsis thaliana ecotype Columbia (Col-0) plants were grown in growth rooms at 22°C in long days (16 hr of light) or short days (9 hr of light) and received 250 μE from three 35-W cool white bulbs and one 35-W Sylvania GrowLux bulb (Osram Sylvania, Danvers, MA). Nicotiana tabacum cv xanthi plants were regenerated under 24-hr-light conditions at 25°C and then transferred to the greenhouse. Tobacco and Arabidopsis transformation was performed as previously described (Neff et al., 1999; Weigel et al., 2000) except that 0.02% Silwet-L77 (Lehle Seeds, Round Rock, TX) was used for the latter. Basta was obtained from AgrEvo (Montvale, NJ).
Gel Blot Hybridization
DNA and RNA gel blot hybridizations were performed according to standard procedures (Sambrook et al., 1989). RNA samples used in the gel blot analysis shown in Figure 2 were from vegetative leaves of pap1-D and Col-0 plants grown under short-day conditions for 4 weeks and long-day conditions for 2 weeks. Probes were full-length cDNA fragments of PAP1, the glutathione S-transferase gene, and the gene encoding ubiquitin. The chalcone synthase (CHS) probe was a polymerase chain reaction product amplified by using the primers 5′-TGGTCTCCGTCCTTCCGTCAA and 5′-CCCTCAAATGTCCGTCTATGGAA. The phenylalanine ammonia-lyase (PAL) probe was amplified by using the primers 5′-CTATACGCTTACCTACCAACAAAC and 5′-TCTCCGATGAGAAGTAGCACCAA, and the dihydroflavonol reductase probe was amplified with primers 5′-AAAAAGATGACAGGATGGGT-3′ and 5′-CCCCTGTTTCTGTCTTGTTA-3′.
Enzyme Assays
PAL activity was measured by using a microcuvette spectrophotometric assay (Blount et al., 2000). Caffeic acid O-methyltransferase and caffeoyl-CoA O-methyltransferase activities were assayed by standard methods (Inoue et al., 1998). Protein concentrations were determined by the procedure of Bradford (1976).
Phenylpropanoid Analysis
Soluble and wall-bound phenolics in whole-plant extracts as well as extracts of individual tissues were analyzed by HPLC (Blount et al., 2000). The aqueous phase, which remained after ethyl acetate extraction of the wall-bound phenolics, was lyophilized and resuspended in 70% methanol for analysis. The HPLC eluates were monitored by absorbance at 270, 310, and 550 nm, and the peaks were identified by comparing their retention times and UV light spectra with those of known standards. Lignin was assayed by derivatization followed by reductive cleavage (Lu and Ralph, 1997).
pMN-PAP1, pCHFS-PAP1, and pCHFS-PAP2 Constructs
A PAP1 genomic fragment was amplified by using 5′-AACTACTGC-AGCTAGAGCGTAGAGG-3′ and 5′-TCAAACTGCAGAAACTAAGCC-CA-3′ to construct 5′ and 3′ Pst sites. This fragment was cloned into pMN20-2 (Weigel et al., 2000), which contains two copies of 35Se to create pMN-PAP1. PCHF3-PAP1 was created by amplifying the PAP1 cDNA with primers 5′-ACTGGTACCTTTTACAATTTGTTTA-3′ and 3′-AAGGGATCCTATACACAAACGCA-5′ and cloning it into the KpnI and BamHI sites of pCHF3, a pPZP211-based plant expression vector carrying the cauliflower mosaic virus 35S promoter and a pea ribulose 1,5-bisphosphate carboxylase/oxygenase terminator (C. Fankhauser, K. Hanson, and J. Chory, unpublished data). The PAP2 cDNA was excised from expressed sequence tag clone 193M15 with KpnI and BamHI and was cloned into pCHF3 to create pCHF3-PAP2.
Acknowledgments
We thank the following (all at Salk Institute unless otherwise noted): Tsegaye Dabi for help with transgenic tobacco; Mary Anderson at the Nottingham University Stock Centre for help with restriction fragment length polymorphism mapping; David Huhman (Noble Foundation) for lignin measurements; Michael Neff, Christian Fankhauser, Kim Hanson, and Joanne Chory for pMN20 and pCHF3 vectors; and Igor Kardailsky, Sioux Christensen, and Detlef Weigel for pSKI015. J.O.B. thanks Joanne Chory for providing laboratory facilities for completion of this work.
References
- Alfenito, M.R., Souer, E., Goodman, C.D., Buell, R., Mol, J., Koes, R., and Walbot, V. (1998). Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell 10 1135–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atanassova, R., Foret, N., Martz, F., Chrabbert, B., Tollier, M.-T., Monties, B., Fritig, B., and Legrand, M. (1995). Altered lignin composition in transgenic tobacco expressing O-methyltransferase sequences in sense and antisense orientation. Plant J. 8 465–477. [Google Scholar]
- Bate, N., Orr, J., Ni, W., Meroni, A., Nadler-Hassan, T., Doerner, P.W., Dixon, R.A., Lamb, C.J., and Elkind, Y. (1994). Quantitative relationship between phenylalanine ammonia-lyase levels and phenylpropanoid accumulation in transgenic tobacco identifies a rate-determining step in natural product synthesis. Proc. Natl. Acad. Sci. USA 91 7608–7612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blount, J.W., Korth, K.L., Masoud, S.A., Rasmussen, S., Lamb, C., and Dixon, R.A. (2000). Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol. 122 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Anal Biochem. 72 248–254. [DOI] [PubMed] [Google Scholar]
- Cone, K.C., Cocciolone, S.M., Burr, F.A., and Burr, B. (1993. a). Maize anthocyanin regulatory gene pl is a duplicate of c1 that functions in the plant. Plant Cell 5 1795–1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cone, K.C., Cocciolone, S.M., Moehlenkamp, C.A., Weber, T., Drummond, B.J., Tagliani, L.A., Bowen, B.A., and Perrot, G.H. (1993. b). Role of the regulatory gene pl in the photocontrol of maize anthocyanin pigmentation. Plant Cell 5 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vetten, N., Quattrocchio, F., Mol, J., and Koes, R. (1997). The an11 locus controlling flower pigmentation in petunia encodes a novel WD-repeat protein conserved in yeast, plants, and animals. Genes Dev. 11 1422–1434. [DOI] [PubMed] [Google Scholar]
- Facchini, D., and Deluca, V. (1995). Phloem-specific expression of tyrosine dopa decarboxylase genes and the biosynthesis of isoquinoline alkaloids in opium poppy. Plant Cell 7 1811–1821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fowler, M.W. (1983). Plant-cell cultures: Fact and fantasy. Biochem. Soc. Trans. 11 23–28. [DOI] [PubMed] [Google Scholar]
- Glover, B.J., Perez-Rodriguez, M., and Martin, C. (1998). Development of several epidermal cell types can be specified by the same MYB-related transcription factor. Development 125 3497–3508. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., Carpenter, R., and Coen, E.S. (1992). A common gene regulates pigmentation in diverse plant species. Cell 68 955–964. [DOI] [PubMed] [Google Scholar]
- Graham, T.L. (1991). Flavonoid and isoflavonoid distribution in developing soybean seedling tissues and in seed and root exudates. Plant Physiol. 95 594–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotewold, E., Drummond, B.J., Bowen, B., and Peterson, T. (1994). The myb-homologous P gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76 543–553. [DOI] [PubMed] [Google Scholar]
- Grotewold, E., Chamberlin, M., Snook, M., Siame, B., Butler, L., Swenson, J., Maddock, S., St. Clair, G., and Bowen, B. (1999). Engineering secondary metabolism in maize cells by ectopic expression of transcription factors. Plant Cell 10 721–740. [PMC free article] [PubMed] [Google Scholar]
- Inoue, K., Sewalt, V.J.H., Balance, G.M., Ni, W., Sturzer, C., and Dixon, R.A. (1998). Developmental expression and substrate specificities of alfalfa caffeic acid 3-O-methyltransferase and caffeoyl coenzyme A 3-O-methyltransferase in relation to lignification. Plant Physiol. 117 761–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kardailsky, I., Shukla, V., Ahn, J.H., Dagenais, N., Christensen, S.K., Nguyen, J.T., Chory, J., Harrison, M.J., and Weigel, D. (1999). A pair of related genes with antagonistic roles in floral induction. Science 286 1962–1965. [DOI] [PubMed] [Google Scholar]
- Koornneef, M. (1981). The complex syndrome of ttg mutants. Arabidopsis Inf. Serv. 18 45–51. [Google Scholar]
- Kranz, H.D., et al. (1998). Towards functional characterisation of the members of the R2R3-MYB gene family from Arabidopsis thaliana. Plant J. 16 263–276. [DOI] [PubMed] [Google Scholar]
- Larkin, J.C., Walker, J.D., Bolognesi-Winfield, A.C., Gray, J.C., and Walker, A.R. (1999). Allele-specific interactions between ttg and gl1 during trichome development in Arabidopsis thaliana. Genetics 151 1591–1604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd, A.M., Walbot, V., and Davis, R.W. (1992). Arabidopsis and Nicotiana anthocyanin production activated by maize regulators R and C1. Science 258 1773–1775. [DOI] [PubMed] [Google Scholar]
- Lu, F.C., and Ralph, J. (1997). Derivatization followed by reductive cleavage (DFRC method), a new method for lignin analysis: Protocol for analysis of DFRC monomers. J. Agric. Food Chem. 45 2590–2592. [Google Scholar]
- McCaskill, D., and Croteau, R. (1998). Some caveats for bioengineering terpenoid metabolism in plants. Trends Biotechnol. 16 349–355. [Google Scholar]
- Mol, J., Jenkins, G., Schafer, E., and Weiss, D. (1996). Signal perception, transduction, and gene expression involved in anthocyanin biosynthesis. Crit. Rev. Plant Sci. 15 525–557. [Google Scholar]
- Neff, M.M., Nguyen, S.M., Malancharuvil, E.J., Fujioka, S., Noguchi, T., Seto, H., Tsubuki, M., Honda, T., Takatsuto, S., Yoshida, S., and Chory, J. (1999). BAS1: A gene regulating brassinosteroid levels and light responsiveness in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 15316–15323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheimer, D.G., Herman, P.L., Sivakumaran, S., Esch, J., and Marks, M.D. (1991). A MYB gene required for leaf trichome differentiation in Arabidopsis is expressed in stipules. Cell 67 483–493. [DOI] [PubMed] [Google Scholar]
- Quattrocchio, F., Wing, J.F., van der Woude, K., Mol, J.N.M., and Koes, R. (1998). Analysis of bHLH and MYB domain proteins: Species-specific regulatory differences are caused by divergent evolution of target anthocyanin genes. Plant J. 13 475–488. [DOI] [PubMed] [Google Scholar]
- Rabinowicz, P.D., Braun, E.L., Wolfe, A.D., Bowen, B., and Grotewold, E. (1999). Maize R2R3 myb genes. Sequence analysis reveals amplification in higher plants. Genetics 153 427–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robins, R.J. (1994). Secondary products from cultured cells and organs. I. Molecular and cellular approaches. In Plant Cell Culture: A Practical Approach, R.A. Dixon and R.A. Gonzales, eds (Oxford, UK: IRL Press), pp. 169–198.
- Romero, I., Fuertes, A., Benito, M.J., Malpica, J.M., Leyva, A., and Paz-Ares, J. (1998). More than 80 R2R3-MYB regulatory genes in the genome of Arabidopsis thaliana. Plant J. 14 273–284. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Shirley, B.W., Lubasek, W., Storz, G., Bruggemann, E., Koornneef, M., Ausubel, F.M., and Goodman, H. (1995). Analysis of Arabidopsis mutants deficient in flavonoid biosynthesis. Plant J. 8 659–671. [DOI] [PubMed] [Google Scholar]
- Van der Fits, L., and Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289 295–297. [DOI] [PubMed] [Google Scholar]
- Walker, A.R., Davison, P.A., Bolognesi-Winfield, A.C., James, C.M., Srinivasan, N., Blundell, T.L., Esch, J.J., Marks, M.D., and Gray, J.C. (1999). The TRANSPARENT TESTA GLABRA1 locus, which regulates trichome differentiation and anthocyanin biosynthesis in Arabidopsis, encodes a WD40 repeat protein. Plant Cell 11 1337–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong, R., Morrison III, W.H., Negrel, J., and Ye, Z.-H. (1998). Dual methylation pathways in lignin biosynthesis. Plant Cell 10 2033–2045. [DOI] [PMC free article] [PubMed] [Google Scholar]