Abstract
Most indole-3-acetic acid (IAA) in higher plants is conjugated to amino acids, sugars, or peptides, and these conjugates are implicated in regulating the concentration of the free hormone. We identified iar1 as an Arabidopsis mutant that is resistant to the inhibitory effects of several IAA–amino acid conjugates but remains sensitive to free IAA. iar1 partially suppresses phenotypes of a mutant that overproduces IAA, suggesting that IAR1 participates in auxin metabolism or response. We used positional information to clone IAR1, which encodes a novel protein with seven predicted transmembrane domains and several His-rich regions. IAR1 has homologs in other multicellular organisms, including Drosophila, nematodes, and mammals; in addition, the mouse homolog KE4 can functionally substitute for IAR1 in vivo. IAR1 also structurally resembles and has detectable sequence similarity to a family of metal transporters. We discuss several possible roles for IAR1 in auxin homeostasis.
INTRODUCTION
Indole-3-acetic acid (IAA) is the major endogenous auxin and participates in many plant developmental processes, including cell enlargement and division, differentiation of vascular tissue, initiation of lateral roots, apical dominance, and responses to environmental stimuli such as gravity and light (reviewed by Estelle and Klee, 1994; Bennett et al., 1998). Plants contain little free IAA; most IAA is found conjugated to amino acids, peptides, sugars, or high molecular weight glycans. These conjugates have been implicated in such processes as storage, transport, and protection from oxidative degradation (reviewed in Cohen and Bandurski, 1982; Bandurski et al., 1995). Plants apparently also permanently inactivate excess IAA by conjugation (reviewed in Normanly, 1997). For example, many plants form IAA-Asp as an intermediate in IAA catabolism (Tsurumi and Wada, 1986; Monteiro et al., 1988; Tuominen et al., 1994; Östin et al., 1998).
Conjugation and hydrolysis of conjugates are probable mechanisms used to regulate the concentrations of free IAA (reviewed in Normanly and Bartel, 1999), and characterization of the genes involved in these processes is a prerequisite to understanding this regulation. Although diverse land plants conjugate IAA to glucose and other molecules (Sztein et al., 1995, 1999), the only plant gene so involved that has been identified is the maize iaglu gene, which encodes an enzyme esterifying IAA to glucose (Szerszen et al., 1994). Various plants hydrolyze IAA conjugates, and IAA-glucose hydrolases have been identified in maize, potato, oat, and bean (Kowalczyk and Bandurski, 1990; Jakubowska et al., 1993). IAA-Ala hydrolases have been partially purified from bean and carrot (Cohen et al., 1988; Kuleck and Cohen, 1993), and Chinese cabbage extracts contain isoenzymes that hydrolyze IAA-Ala, IAA-Asp, and IAA-Phe (Ludwig-Müller et al., 1996).
The ability of several IAA conjugates to mimic the effects of free IAA on plant growth (reviewed by Bartel, 1997) has been exploited to identify Arabidopsis mutants that respond abnormally to IAA conjugates. The ilr1 (IAA-Leu–resistant) mutant elongates roots at concentrations of IAA-Leu that inhibit wild-type root growth. The gene defective in ilr1 encodes an amidohydrolase with high affinity for IAA-Leu and IAA-Phe and lesser affinity for IAA-Ala, IAA-Gly, and IAA-Val (Bartel and Fink, 1995). Four ILR1-like genes, ILL1, ILL2 (Bartel and Fink, 1995), ILL3, and ILL5 (Davies et al., 1999), have been identified on the basis of their similarity to ILR1. A sixth member of this gene family is defective in the iar3 (IAA-Ala–resistant) mutant and encodes an amidohydrolase specific for IAA-Ala (Davies et al., 1999). Two other mutants, icr1 and icr2, are resistant to growth inhibition by IAA-Phe, IAA-Ala, and IAA-Gly (Campanella et al., 1996), but the genes defective in these mutants remain unidentified.
We have been identifying and characterizing Arabidopsis mutants with altered responses to IAA–amino acid conjugates to elucidate the role of conjugate metabolism in plant development. Here, we report the isolation of the iar1 auxin conjugate–resistant mutant and the positional cloning of IAR1, which encodes a polytopic membrane protein with several His-rich regions. IAR1 represents a novel component of the auxin conjugate metabolic machinery in plants, and several possible models for its function are discussed.
RESULTS
Isolation and Characterization of iar1 Mutants
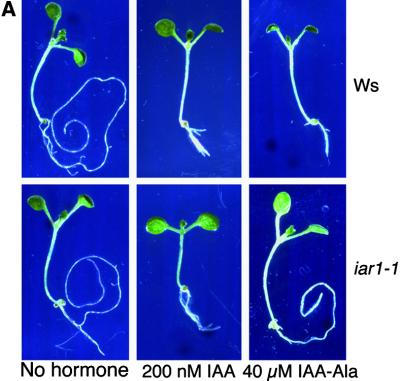
Certain IAA–amino acid conjugates mimic free IAA in bioassays (reviewed in Bartel, 1997), and IAA conjugates that inhibit Arabidopsis root elongation have been used in mutant screens to identify genes involved in hydrolysis of IAA conjugates (Bartel and Fink, 1995; Davies et al., 1999). We initially identified the iar1 mutant because its roots elongate at concentrations of IAA-Ala that are normally inhibitory, as shown in Figure 1. Table 1 lists the seven iar1 alleles, four of which were isolated in screens for IAA-Ala resistance. Three additional iar1 alleles were identified as enhancers of the ilr1 mutant, which is defective in an IAA conjugate hydrolase (Bartel and Fink, 1995). These alleles were isolated based on the increased resistance of ilr1 iar1 double mutants to IAA-Leu or IAA-Phe. The recessive nature of the iar1 mutations (data not shown) suggests that they are loss-of-function alleles.
Figure 1.
iar1 Roots Have Decreased Sensitivity to IAA–Amino Acid Conjugates and Normal Sensitivity to IAA.
(A) Eight-day-old iar1-1 mutant and Wassilewskija (Ws) wild-type seedlings were removed from the agar and photographed after growth on medium containing no hormone, 200 nM IAA, or 40 μM IAA-Ala.
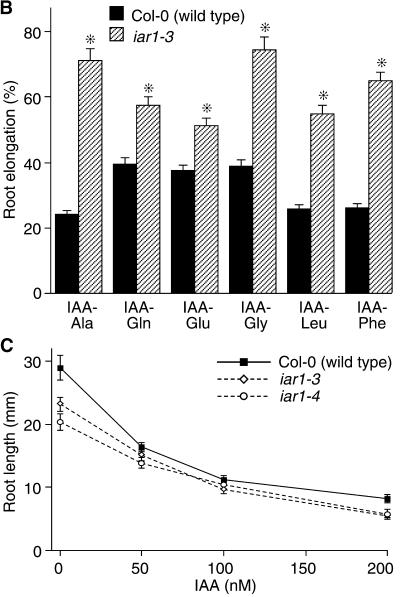
(B) Wild-type Columbia (Col-0) and iar1 mutant seed were plated on medium containing 20 μM of the indicated IAA–amino acid conjugates. After 8 days, the primary root of each seedling was measured and normalized against growth on unsupplemented medium. Error bars indicate standard errors of the means (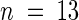 ), and asterisks indicate significant differences from the wild type (Student's t test, P < 0.0001).
), and asterisks indicate significant differences from the wild type (Student's t test, P < 0.0001).
(C) Wild-type (Col-0) and iar1 mutant seed were plated on medium containing the indicated concentrations of IAA. After 8 days, the primary root of each seedling was measured. Error bars indicate standard errors of the means (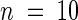 ).
).
Table 1.
iar1 Mutant Alleles
| Allele | Accession | Screen | Mutagen | Codon Changea | Amino Acid Change | Allele Detection |
|---|---|---|---|---|---|---|
| iar1-1 | Ws | IAA-AlaR | EMS | GGA→ΑGA | Gly400→Arg | Destroys HpaII site |
| iar1-2 | Ws | IAA-AlaR | EMS | GGA→ΑGA | Gly423→Arg | Destroys AciI site |
| iar1-3 | Col-0 | IAA-AlaR | Fast neutron | GGAATG→GGGCCTG | Frameshift at codon 458 | Creates HaeIII site |
| iar1-4 | Col-0 | IAA-AlaR | Fast neutron | 65 to 80-kb deletion including entire IAR1 coding region |
Late-flowering due to FKF1 deletionb |
|
| iar1-5 | Ws | ilr1 enhancer IAA-LeuR | EMS | AGA→AAA | Arg357→Lys | Sequencing |
| iar1-6 | Ws | ilr1 enhancer IAA-LeuR | EMS | CAA→ΤAA | Gln162→stop | Destroys HphI site |
| iar1-7 | Ws | ilr1 enhancer IAA-PheR | EMS | tagCT→taaCT (3′ splice site of first intron)c |
Predicted to disrupt splicing |
Creates AluI site |
Nucleotide changes are underlined.
Nelson et al., 2000.
Intron bases are in lowercase letters; exon bases are in uppercase letters.
We quantitated the IAA conjugate resistance of the iar1 alleles by comparing root lengths of seedlings grown on medium containing various conjugates. All iar1 alleles are resistant to IAA-Ala, iar1-4 and iar1-5 being the strongest and weakest alleles, respectively (data not shown). Because ilr1 iar1 double mutants are highly resistant to IAA-Leu and IAA-Phe (data not shown), we tested the response of iar1 single mutants to these conjugates in root elongation assays. iar1-3 is resistant to IAA-Phe at all concentrations tested (10 to 40 μM) but is resistant to IAA-Leu only at lower concentrations (10 to 20 μM; Figure 1B and data not shown). iar1 roots are also resistant to elongation inhibition by IAA-Gly, IAA-Gln, and IAA-Glu (Figure 1B), indicating that IAR1 is required for IAA conjugate sensitivity in general rather than being conjugate specific. In contrast, iar1 plants respond normally to a range of concentrations of free IAA (Figures 1A and 1C), indicating that mutations in IAR1 alter conjugate sensitivity rather than general auxin responsiveness.
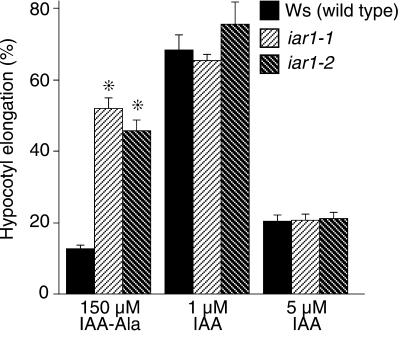
As shown in Figure 2, exogenous IAA or IAA conjugates can inhibit Arabidopsis hypocotyl elongation in the light; accordingly, we examined the effects of IAA conjugates on iar1 hypocotyl elongation. iar1 mutant hypocotyls respond normally to free IAA but are resistant to the inhibitory effects of IAA-Ala on elongation (Figure 2). These results suggest that IAR1 functions in the hypocotyls as well as the roots of Arabidopsis seedlings.
Figure 2.
iar1 Hypocotyls Have Decreased Sensitivity to IAA-Ala.
Wild-type and iar1 seed were plated on medium containing the indicated concentrations of IAA or IAA-Ala. After 8 days in yellow-filtered light at 22°C, hypocotyls were measured and normalized against growth on unsupplemented medium. Error bars indicate standard errors of the means (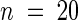 for Ws and iar1-1,
for Ws and iar1-1, 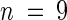 for iar1-2), and asterisks indicate significant differences from the wild type (Student's t test, P < 0.0001).
for iar1-2), and asterisks indicate significant differences from the wild type (Student's t test, P < 0.0001).
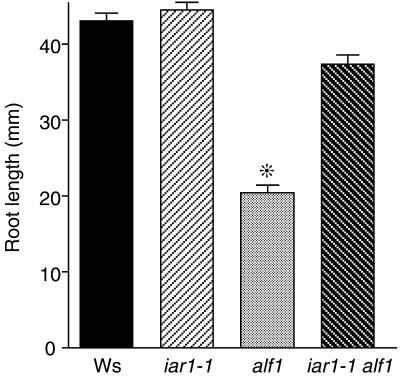
The iar1 Mutation Partially Suppresses alf1 Phenotypes
The Arabidopsis aberrant lateral root formation (alf1) mutant (Celenza et al., 1995), also isolated as rooty (King et al., 1995), superroot1 (Boerjan et al., 1995), and hookless3 (Lehman et al., 1996), overproduces both free and conjugated IAA (Boerjan et al., 1995; King et al., 1995; Lehman et al., 1996). alf1 plants have short primary roots, display increased numbers of adventitious and lateral roots, fail to make an apical hook when germinated in the dark, and are completely sterile under normal growth conditions (Celenza et al., 1995). These phenotypes can be copied by applying auxin to wild-type plants (Boerjan et al., 1995; Celenza et al., 1995; King et al., 1995; Lehman et al., 1996). To investigate whether IAR1 influences IAA responses in vivo, we generated homozygous iar1/iar1 alf1/alf1 plants (see Methods). iar1 partially rescues the fertility defect of alf1, in that double mutants yield some seed. In addition, iar1 alf1 plants are able to form a partial apical hook when germinated in the dark (data not shown). We quantified the degree to which iar1 suppresses alf1 by monitoring root elongation on unsupplemented medium. As shown in Figure 3, the iar1 mutation suppresses the alf1 root elongation defect. The observation that loss of IAR1 function can partially alleviate defects in the alf1 auxin-overproducing mutant suggests that IAR1 normally acts to increase the concentrations of free IAA or the sensitivity to IAA.
Figure 3.
iar1 Suppresses the alf1 Root Elongation Defect.
Seed of Ws (wild type), iar1-1 and alf1 single mutants, and iar1-1 alf1 double mutants were plated on unsupplemented medium. After 10 days under yellow-filtered light at 22°C, the primary root of each seedling was measured. Homozygous alf1 mutants were identified among the progeny of an alf1/ALF1 parent based on the epinastic cotyledon phenotype. Error bars indicate standard errors of the means (n ⩾ 18), and the asterisk indicates a significant difference from the wild type (Student's t test, P < 0.0001).
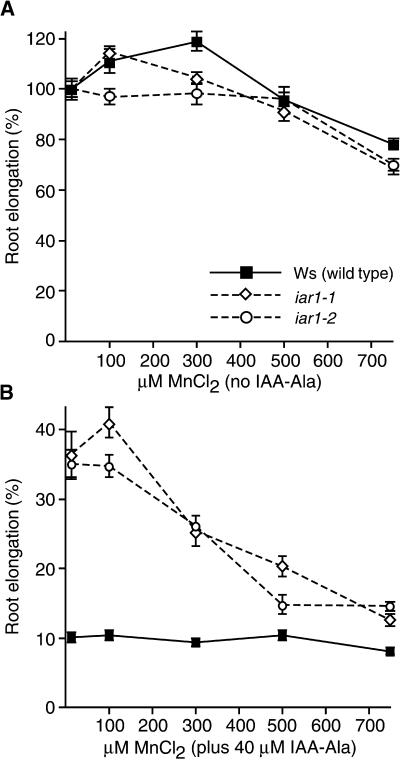
Suppression of the iar1 Phenotype by Manganese
The Arabidopsis amidohydrolases characterized to date require a metal cofactor. Mn2+ and Co2+ are the most effective cofactors identified for heterologously expressed Arabidopsis IAA–amino acid conjugate hydrolases purified from Escherichia coli (Bartel and Fink, 1995; Davies et al., 1999; R. Tellez and B. Bartel, unpublished data). Because mutations in these amidohydrolases can cause IAA–amino acid conjugate resistance (Bartel and Fink, 1995; Davies et al., 1999), mutations that prevent the hydrolases from assembling with or retaining the proper cofactor also might cause this phenotype. We examined the response of iar1 to Mn2+ alone or to IAA-Ala and Mn2+. As shown in Figure 4A, Mn2+ alone did not dramatically inhibit root growth at the concentrations tested. However, high Mn2+ concentrations partially suppressed the IAA-Ala resistance of iar1 (Figure 4B). This finding suggests that IAR1 might be necessary to provide the correct metal cofactor to the IAA–amino acid conjugate hydrolases.
Figure 4.
Manganese Suppresses iar1 IAA-Ala Resistance.
Wild-type (Ws) and iar1 mutant seed were plated on medium containing 14 to 750 μM manganese either with or without 40 μM IAA-Ala. After 8 days under yellow-filtered light at 22°C, the primary root of each seedling was measured and normalized to growth on unsupplemented medium (which contained 14 μM manganese). Error bars indicate standard errors of the means (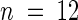 ).
).
(A) Without IAA-Ala.
(B) With 40 μM IAA-Ala.
IAR1 Cloning
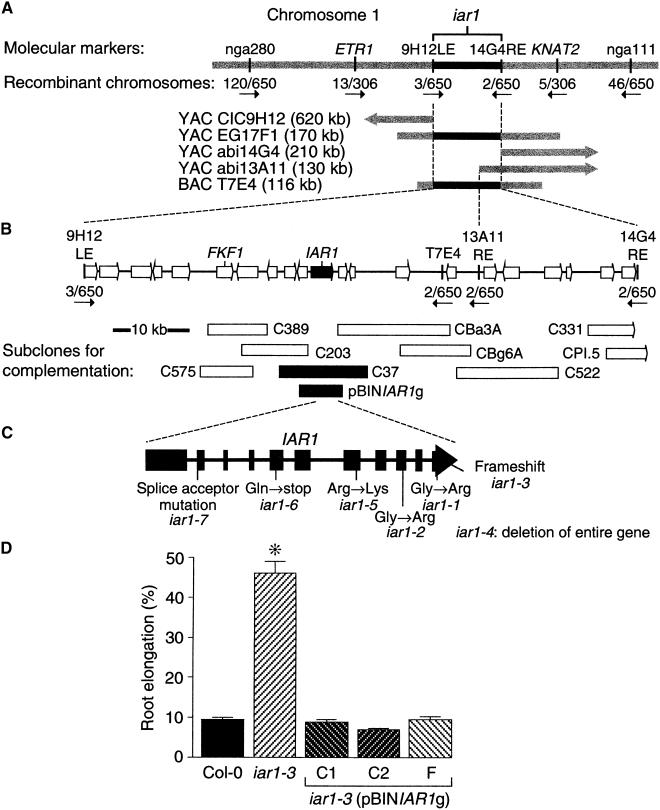
To elucidate the role of IAR1 in conjugate metabolism, we cloned IAR1 by using a map-based positional approach. A recombination mapping population was prepared by outcrossing iar1 plants in the Wassilewskija (Ws) accession to wild-type Columbia (Col-0) plants. DNA isolated from IAA-Ala–resistant F2 progeny was analyzed with polymerase chain reaction (PCR)–based polymorphic markers (Konieczny and Ausubel, 1993; Bell and Ecker, 1994). The iar1 mutation was localized to an unsequenced (at that time) region at the bottom of chromosome 1, between simple sequence length polymorphism markers nga280 and nga111 (Bell and Ecker, 1994). An 800-kb yeast artificial chromosome (YAC) contig had been described in this interval (Vijayraghavan et al., 1995), and we used subclones from these and other nearby YACs to develop additional PCR-based markers for mapping (see Methods). We localized the iar1 mutation between the markers 9H12LE and 13A11RE, as shown in Figure 5. Using PCR and DNA hybridization analysis, we determined that this interval is completely contained on the 116-kb bacterial artificial chromosome (BAC) T7E4 (Figure 5A).
Figure 5.
Positional Cloning of IAR1.
(A) Recombination mapping with PCR-based markers nga111 and nga280 (Bell and Ecker, 1994) localized iar1 to the bottom of chromosome 1. This position was refined to between ETR1 (Chang et al., 1993) and KNAT2 (Lincoln et al., 1994). Additional markers from nearby YAC clones included 9H12LE and 14G4RE (Nelson et al., 2000) and a marker made from the right end of YAC abi13A11 (see Methods). These three markers hybridized to BAC T7E4 (Choi et al., 1995).
(B) A complementation library was constructed (see Methods) from BAC T7E4, and subclones that were tested for iar1 complementation are indicated by rectangles below the predicted genes (arrows) in the region. Subclones shown in black rescued the iar1 IAA-Ala resistance, and those shown in white did not.
(C) Positions of the seven iar1 mutations are shown below a scheme of the IAR1 coding region. Exons are indicated by closed rectangles, and introns are represented by lines.
(D) A genomic construct containing the predicted IAR1 open reading frame controlled by its own promoter rescued the iar1 mutant phenotype. Seed from the wild type (Col-0), the iar1-3 mutant, and three iar1-3 transgenic lines homozygous for the pBINIAR1g construct, shown in (B), were plated on medium containing 40 μM IAA-Ala. After 8 days of growth under yellow-filtered light at 22°C, the primary root of each seedling was measured and standardized against growth on unsupplemented medium. Error bars indicate standard errors of the means (n ⩾ 17), and the asterisk indicates a significant difference from the wild type (Student's t test, P < 0.0001).
We constructed a complementation library from BAC T7E4 (see Methods) and used Agrobacterium to transform (Clough and Bent, 1998) these subclones into the iar1 mutant. T2 progeny of iar1-3 plants transformed with the various constructs were tested for conjugate resistance. Approximately 75% of the T2 progeny of C37 (Figure 5B) transformants had wild-type IAA-Ala sensitivity, as expected for a rescuing construct. We sequenced the 12-kb insert of C37 and identified several potential coding regions. One of these genes was altered in all seven iar1 alleles. Three alleles (iar1-1, iar1-2, and iar1-5) had mutations that caused amino acid substitutions; iar1-6 had a nonsense mutation; the mutation in iar1-7 probably disrupted splicing because it changed the final G of the first intron to an A; iar1-3 had a frameshift mutation predicted to replace the last 11 amino acids of IAR1 with an alternative 14–amino acid stretch (Table 1; Figure 5C); and iar1-4 had an ∼65- to 80-kb deletion that included the entire C37 region (Nelson et al., 2000). Finally, the 6.5-kb genomic clone pBINIAR1g (see Methods), which contains the IAR1 open reading frame with 2.1 kb of DNA upstream of the initiator ATG, completely restored IAA-Ala sensitivity to the iar1 mutant (Figure 5D), confirming that we had identified IAR1.
IAR1 Encodes a Membrane Protein with Multiple His-Rich Regions
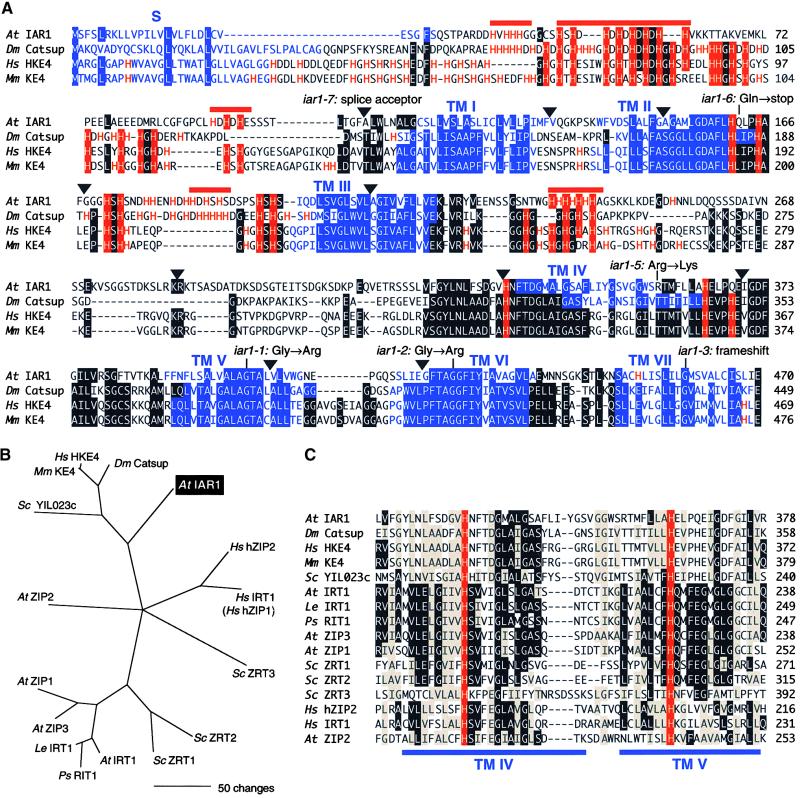
We isolated a full-length IAR1 cDNA (GenBank accession number AF216524) by hybridization to a cDNA library (see Methods). Comparing this cDNA with various genomic sequences revealed that the IAR1 coding region has 10 introns (Figure 5C) and potentially encodes a 469–amino acid protein with seven or eight transmembrane domains, as shown in Figures 6A and 7. IAR1 is 28% identical to the Drosophila Catsup (catecholamines up) protein (GenBank accession number AAF37226) and 26% identical to the predicted mouse KE4 and human HKE4 proteins (Abe et al., 1988; St.-Jaques et al., 1990; Janatipour et al., 1992; Ando et al., 1996). Uncharacterized IAR1 homologs are found in other multicellular organisms, including Drosophila (GenBank accession numbers AAF49687, AAF50401, and AAF56960), Caenorhabditis elegans (GenBank accession numbers CAB17070, CAB05297.1, and CAB02806.1), and zebrafish (GenBank accession number AAF05821). Figure 6A shows an alignment of IAR1 with similar proteins from Drosophila and mammals.
Figure 6.
Alignment of IAR1 and Similar Proteins from Other Organisms.
(A) IAR1, Drosophila (Dm) Catsup, human (Hs) HKE4 (Janatipour et al., 1992; Ando et al., 1996), and mouse (Mm) KE4 (Abe et al., 1988; St.-Jaques et al., 1990) predicted proteins were aligned with the Megalign program (DNAStar, Madison, WI) by using the Clustal method (Higgins and Sharp, 1989) with PAM250 residue weights. Residues conserved in at least three proteins are shaded. Triangles indicate the positions of introns in the IAR1 coding sequence. Positions of iar1 mutations are shown above the IAR1 sequence, except for iar1-4, which is a deletion of the entire IAR1 gene. The His residues are highlighted in red, and IAR1 potential metal-binding sequences of the type (HX)3-6 are indicated by red bars above the sequence. Regions predicted by the SMART program (Schultz et al., 2000) to be transmembrane domains (TM I to VII) are indicated in blue.
(B) Phylogenetic tree of IAR1 and its relatives. The tree reconstructs the evolutionary relationship between IAR1 family members from (A) and ZIP proteins, including Arabidopsis (At; Korshunova et al., 1999) and tomato (Le) IRT1 (GenBank accession number AF136579); pea (Ps) RIT1 (GenBank accession number AF065444); Arabidopsis ZIP1, ZIP2, and ZIP3 (Grotz et al., 1998); yeast (Sc) ZRT1 (Zhao and Eide, 1996a), ZRT2 (Zhao and Eide, 1996b), ZRT3 (MacDiarmid et al., 2000), and YIL023c (GenBank accession number P40544); and human (Hs) IRT1 (hZIP1) and hZIP2 (Lioumi et al., 1999; Gaither and Eide, 2000). Sequences (from predicted transmembrane domain IV to the end of the proteins) were aligned as described in (A), and the phylogenetic tree was generated by using PAUP 4.0b3a (Swofford, 2000). The bootstrap method was performed for 100 replicates with a maximum parsimony optimality criterion. All characters were weighted equally. Starting trees for the heuristic search were obtained by random stepwise addition, and tree-bisection-reconnection was the branch-swapping algorithm.
(C) Alignment of predicted transmembrane domains IV and V from the ZIP family members described in (B) with the similar region from IAR1 family members. Identical residues and conservative changes seen in at least seven proteins are shown in black and gray boxes, respectively. Transmembrane domains in the ZIP family described by MacDiarmid et al. (2000) are indicated by blue bars below the alignment. The His residues conserved with those in Arabidopsis IRT1 that are required for zinc transport (Rogers et al., 2000) are indicated in red.
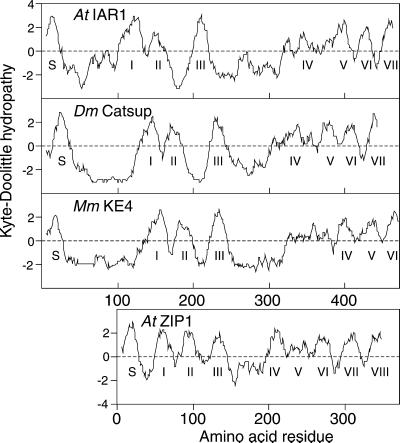
Figure 7.
IAR1 Encodes a Potential Transmembrane Protein.
Hydropathy was calculated for IAR1, Drosophila (Dm) Catsup, mouse (Mm) KE4 (Abe et al., 1988; St.-Jaques et al., 1990), and Arabidopsis (At) ZIP1 (Grotz et al., 1998) with a window size of 13 and a linear weight variation model (Kyte and Doolittle, 1982). Positive and negative values indicate hydrophobic and hydrophilic regions, respectively. Predicted signal sequences are indicated by S, and the transmembrane domains predicted by the SMART program (Schultz et al., 2000) are numbered.
The closest IAR1 homologs of known function are members of the ZIP (ZRT, IRT-like protein) family of metal transporters (Eng et al., 1998; Guerinot and Eide, 1999; Guerinot, 2000), which are ∼10 to 13% identical to IAR1 (Figure 6B). These widespread transporters have been characterized from yeast (Zhao and Eide, 1996a, 1996b; MacDiarmid et al., 2000), Arabidopsis (Grotz et al., 1998), and mammals (Lioumi et al., 1999; Gaither and Eide, 2000).
The Mouse KE4 Gene Can Functionally Substitute for IAR1 in Arabidopsis
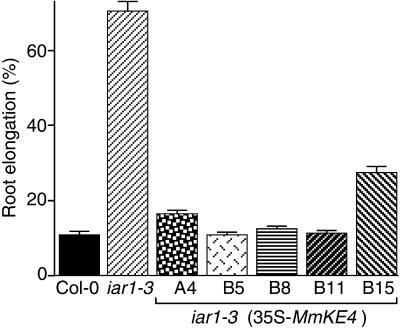
The IAR1 homologs KE4 and HKE4 are mouse and human genes of unknown function (Abe et al., 1988; St.-Jaques et al., 1990; Janatipour et al., 1992; Ando et al., 1996). In addition to 26% amino acid identity (Figure 6A), IAR1 and KE4 share very similar hydropathy plots, suggesting comparable membrane-spanning topologies (Figure 7). To determine whether the Arabidopsis IAR1 and mammalian KE4 proteins function analogously, we transformed iar1-3 mutant plants with a construct expressing the KE4 cDNA from the constitutive cauliflower mosaic virus 35S promoter (35S-MmKE4). As shown in Figure 8, the 35S-MmKE4 construct substantially restores IAA-Ala sensitivity to the iar1 mutant, indicating that the mouse KE4 gene can substitute for IAR1 in vivo. This finding suggests that IAR1 and KE4 have similar functions in Arabidopsis and mammals.
Figure 8.
The Mouse KE4 Gene Can Functionally Substitute for IAR1.
Wild-type (Col-0), iar1-3, and homozygous T3 progeny of five independent lines of iar1-3 plants transformed with 35S-MmKE4, a construct containing the mouse KE4 cDNA (Abe et al., 1988; St.-Jaques et al. 1990) under the control of the cauliflower mosaic virus 35S promoter, were grown for 8 days under yellow-filtered light at 22°C on medium containing 40 μM IAA-Ala. The primary root of each seedling was measured and standardized against growth on unsupplemented medium. Error bars indicate standard errors of the means (n ⩾ 20).
Several Arabidopsis ZIP genes have been isolated (Grotz et al., 1998) that restore growth on low-zinc medium to a S. cerevisiae zrt1 zrt2 double mutant that is defective in high- and low-affinity plasma membrane–localized zinc transporters (Zhao and Eide, 1996a, 1996b). We tested whether IAR1 could rescue this mutant because these transporters are apparently related to IAR1 (Figure 6B) and are structurally similar (Figure 7). In contrast to the expression of several Arabidopsis ZIP genes (Grotz et al., 1998), IAR1 expression does not rescue the zrt1 zrt2 strain in the presence of limiting zinc (data not shown), suggesting that IAR1 does not encode a plasma membrane–localized zinc transporter. However, perhaps IAR1 does not fold or localize properly in yeast, because the human hZIP2 zinc transporter also fails to rescue zrt1 zrt2 mutants (Gaither and Eide, 2000).
DISCUSSION
Requirement of IAR1 for IAA–Amino Acid Conjugate Sensitivity
Phenotypic analysis of iar1 mutants indicates that IAR1 participates in IAA–amino acid conjugate metabolism or sensing. iar1 mutant roots and hypocotyls respond normally to free IAA but are less sensitive than wild-type roots and hypocotyls to several IAA–amino acid conjugates (Figures 1 and 2). In contrast, two other IAA–amino acid–resistant mutants, ilr1 and iar3, are defective in IAA conjugate hydrolases that have distinct substrate specificities and display correspondingly restricted conjugate resistance profiles (Bartel and Fink, 1995; Davies et al., 1999). The broad iar1 conjugate resistance profile suggests a general role for IAR1 in IAA–amino acid conjugate metabolism rather than an IAA-Ala–specific role.
Mutation of IAR1 partially alleviates several auxin overproduction phenotypes of the alf1 mutant, indicating that IAR1 normally increases free IAA or sensitivity to IAA. The alf1 mutant has shortened primary roots, has more adventitious and lateral roots, and is completely sterile (Celenza et al., 1995), probably because of increased concentrations of free IAA (Boerjan et al., 1995; King et al., 1995; Lehman et al., 1996). The iar1 mutation partially suppresses the alf1 phenotype, restoring normal root elongation (Figure 3) and partially restoring fertility and the ability to form an apical hook when germinated in the dark (data not shown).
IAR1 Homologs in Other Multicellular Organisms
The IAR1 gene potentially encodes a 469–amino acid protein that is 26 to 28% identical to the Drosophila Catsup protein and to mammalian KE4 proteins encoded in the major histocompatibility complex (MHC) class II locus (Abe et al., 1988; St.-Jaques et al., 1990; Janatipour et al., 1992; Ando et al., 1996). Interestingly, these IAR1-like genes (Figure 6A) probably play similar or identical roles in plants and animals, because expression of the mouse KE4 cDNA restores normal IAA-Ala sensitivity to iar1 plants (Figure 8). These proteins all have predicted cleavable N-terminal signal sequences, several regions rich in His residues, and six or seven transmembrane domains (Figure 6A). Their almost identical hydropathy plots (Figure 7) suggest common membrane topologies. IAR1 is most similar to its homologs in and near potential transmembrane domains, and two of the iar1 point mutations have Arg residues substituted for conserved Glys in predicted transmembrane domains (Figure 6A), an indication that these regions are functionally important.
IAR1 Similarity to the ZIP Family of Metal Transporters
Intriguingly, members of the ZIP family of metal transporters (reviewed by Guerinot and Eide, 1999; Guerinot, 2000) also have N-terminal signal sequences, seven or eight transmembrane domains (Figure 7), and His-rich regions. Although most ZIP family members were isolated as zinc transporters (Zhao and Eide, 1996a, 1996b; Grotz et al., 1998; Gaither and Eide, 2000), results of both direct uptake assays (Eide et al., 1996; Korshunova et al., 1999; Pence et al., 2000) and competition experiments (Grotz et al., 1998; Gaither and Eide, 2000) suggest that ZIPs can have varied substrate specificities. IAR1, KE4, and Catsup have weak identity to the predicted transmembrane domains of ZIP family members (Figure 6C and data not shown), including most of the consensus residues noted previously (Eng et al., 1998). The conserved His residues in transmembrane domains IV and V of the ZIPs that are essential for metal transport in Arabidopsis IRT1 (Rogers et al., 2000) are conserved in IAR1, Catsup, and KE4 (Figure 6C). However, the IAR1 region corresponding to ZIP transmembrane domains IV and V contains only one transmembrane domain, as predicted by the SMART (Schultz et al., 2000) and PSORT (Nakai and Kanehisa, 1992) motif prediction programs. Although these predicted helices are amphipathic in the ZIPs (Eng et al., 1998), this region in IAR1, Catsup, and KE4 contains even more polar residues. Moreover, IAR1 family members share a conserved Pro residue in the region corresponding to transmembrane domain V of the ZIPs (Figure 6C). This region of IAR1 is probably important because the conservative substitution of a Lys for an Arg at position 357 results in the IAA–amino acid conjugate resistance of the iar1-5 allele and because this region is very similar in IAR1, KE4, and Catsup (Figure 6A).
A striking feature of IAR1 is the presence of several extremely His-rich regions. His represents 17% of the 88 residues between the predicted signal sequence and the first predicted transmembrane domain and 32% of the 40 residues between the second and third transmembrane domains. In addition, the loop between the third and fourth predicted transmembrane domains has seven consecutive His residues (Figure 6A). The Drosophila and mammalian IAR1 homologs are also His-rich in these areas (Figure 6A), suggesting that these residues are important for IAR1 function. Although the membrane topologies of IAR1 and its homologs are unknown, two of these areas flank the third predicted transmembrane domain, indicating that both faces of the membrane should have His-rich regions. Five of these regions in IAR1 are potential metal-binding motifs with the sequence (HX)3-6 (Eng et al., 1998; Guerinot and Eide, 1999). Most ZIP family members contain these motifs between their third and fourth predicted transmembrane domains (Eng et al., 1998), and one of the IAR1 repeats is in this position (Figure 6A). Although IAR1 and the ZIPs share only ∼10% amino acid identity, this similarity between IAR1 family members and ZIP family members suggests a common evolutionary ancestor (Figure 6B) and also that IAR1 may transport or bind metals.
Possible Roles for IAR1 in IAA Conjugate Metabolism
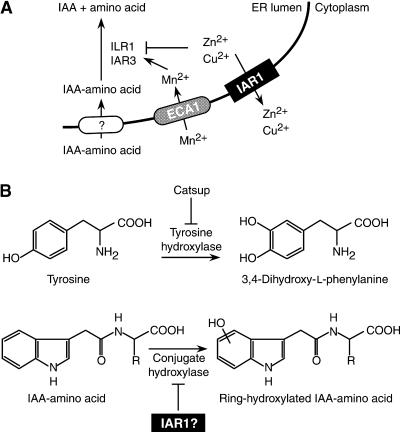
IAR1 encodes a putative polytopic membrane protein weakly similar to metal transporters. This single-copy gene lacks close Arabidopsis relatives, at least in the sequenced portion of the genome (∼98% complete in September 2000; http://arabidopsis.org/agi.html). However, even iar1 null alleles lack striking morphological phenotypes in the absence of exogenous auxin conjugates, indicating that the plant can compensate for IAR1 deficiency in laboratory growth conditions. Figure 9 presents two models that could explain how the loss of IAR1 might result in IAA conjugate resistance.
Figure 9.
Possible Models for IAR1 Function.
(A) IAR1 may transport inhibitory metals out of the ER. This model is suggested by the weak similarity of IAR1 to ZIP family metal transporters, the suppression of the iar1 phenotype by the conjugate hydrolase cofactor Mn2+, and the inhibition of conjugate hydrolases by Cu2+ or Zn2+.
(B) IAR1 may inhibit an auxin conjugate hydroxylase. This model is suggested by the similarity of IAR1 to the Drosophila tyrosine hydroxylase inhibitor Catsup (Stathakis et al., 1999) and is consistent with the presence of ring-hydroxylated IAA conjugates in plants. Flat-tipped arrows indicate inhibitory interactions. See text for details.
Arabidopsis encodes a family of amidohydrolases, some of which hydrolyze IAA–amino acid conjugates and contain sequence motifs that suggest an endoplasmic reticulum (ER) lumen localization (Bartel and Fink, 1995; Davies et al., 1999). If IAR1 encodes an ER-localized IAA conjugate transporter, then loss of IAR1 function would prevent conjugates from reaching the hydrolases and would thus cause conjugate resistance. However, we did not detect conjugate transport in microsomes prepared from yeast expressing IAR1 (data not shown). Additionally, iar1 rescue by the mouse KE4 cDNA (Figure 8) indicates that IAR1 and KE4 probably function similarly in plants and mammals. The lack of an obvious need for such a transporter in mammals argues against a direct role for IAR1 in IAA–amino acid conjugate transport.
The shared structural features with the ZIP family of transporters (Figures 6 and 7) suggest that IAR1 might transport a metal. The characterized Arabidopsis amidohydrolases require a metal cofactor, and mutations in these genes can result in IAA–amino acid conjugate resistance (Bartel and Fink, 1995; Davies et al., 1999). Therefore, it is possible that mutations that prevent hydrolases from assembling with or retaining the proper metal might also cause this phenotype. Supplying more of the required cofactor might alleviate IAA–amino acid resistance resulting from such a mutation. This model has precedent in the copper requirement of Arabidopsis ethylene receptors (Rodríguez et al., 1999). Certain copper transporter mutants respond to an ethylene receptor antagonist as if it were ethylene, a phenotype that can be suppressed by providing exogenous copper (Hirayama et al., 1999). Manganese is an effective amidohydrolase cofactor, and the partial suppression of the IAA-Ala resistance of iar1 by exogenous manganese (Figure 4B) suggests that IAR1 might be an ER-localized manganese transporter. However, IAR1 expression does not rescue the manganese hypersensitivity of the yeast pmr1 mutant (data not shown), which is defective in an ER-localized calcium and manganese transporter (Rudolph et al., 1989; Dürr et al., 1998). In addition, Arabidopsis ECA1 transports manganese into the ER and rescues the pmr1 mutant (Liang et al., 1997). Finally, this model would require that the topology of IAR1 in the membrane be reversed compared with that of the ZIPs, which transport metals into the cytoplasm (Eng et al., 1998; Guerinot and Eide, 1999; MacDiarmid et al., 2000). Therefore, we think it unlikely that IAR1 transports manganese into the ER.
A related possibility is that IAR1 exports an inhibitory metal out of the ER. Zinc and copper inhibit the IAR3 amidohydrolase in vitro, even when stoichiometric manganese is present (Lasswell, 2000). Yeast ZRT1 and human hZIP2 are plasma membrane localized (Gitan et al., 1998; Gaither and Eide, 2000), whereas yeast ZRT3 is in the vacuolar membrane, where it appears to mobilize stored zinc in zinc-limited cells (MacDiarmid et al., 2000). Therefore, one could speculate that IAR1 transports zinc, copper, or another inhibitory metal out of the ER and away from the hydrolases (Figure 9A). This model is consistent with the suppression of the iar1 mutant phenotype by manganese (Figure 4B); excess manganese may outcompete the inhibitory metal from the amidohydrolase and restore activity. It will be interesting to identify the membrane to which IAR1 localizes, because an ER localization would be consistent with this model.
A recent report on the Drosophila Catsup protein (Stathakis et al., 1999), which is ∼28% identical to IAR1 (Figure 6A), suggests an alternative role for IAR1 in IAA conjugate responses. The rate-limiting step in catecholamine biosynthesis is the tyrosine hydroxylase (TH)–mediated conversion of l-tyrosine to 3,4-dihydroxy-l-phenylalanine (Kumer and Vrana, 1996). TH is transcriptionally, translationally, and post-translationally regulated (Kumer and Vrana, 1996). Catsup mutant flies have normal amounts of TH protein but increased TH activity, which leads to a lethal increase in catecholamine synthesis and suggests that Catsup is a post-transcriptional negative regulator of TH (Stathakis et al., 1999) (Figure 9B). Interestingly, Arabidopsis can hydroxylate IAA conjugates (Östin et al., 1998), perhaps as a mechanism to detoxify excess auxin. Thus, if IAR1 regulates an auxin conjugate hydroxylase the way Catsup regulates TH, increased conjugate hydroxylase activity could explain the IAA–amino acid conjugate resistance of the iar1 mutant (Figure 9B). A preliminary report suggests that Catsup interacts directly with TH (Burton et al., 2000), so it will be interesting to isolate proteins that physically or genetically interact with IAR1; these may include the targets of IAR1 action in conjugate metabolism.
IAR1 is a novel membrane protein that is involved directly or indirectly in auxin homeostasis. The identification of the IAR1 gene demonstrates the capacity of genetic screens to uncover unanticipated players in biochemical processes, and the conservation of IAR1 homologs in metazoans suggests that similar proteins play important roles beyond the plant kingdom.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana seed were surface-sterilized (Last and Fink, 1988) and grown on plant nutrient medium (PNS) containing 0.5% sucrose (Haughn and Somerville, 1986) solidified with 0.6% agar. PNS either contained no supplement or was supplemented with 50 nM to 5 μM indole-3-acetic acid (IAA; from a 10 mM stock in ethanol), 20 to 150 μM IAA–amino acid conjugates (from 100 mM stocks in ethanol), 15 μg/mL kanamycin (Kan; from a 25 mg/mL stock in water), 7.5 μg/mL glufosinate-ammonium (BASTA; from a 15 mg/mL stock in 25% ethanol), or 100 to 750 μM MnCl2 (from a 1 M stock in water). IAA and IAA conjugates were from Aldrich (Milwaukee, WI) or were synthesized (S. LeClere and S.P.T. Matsuda, unpublished data). Plates were wrapped in gas-permeable Leukopor surgical tape (Beierdorf Inc., Norwalk, CT) and grown at 22°C in 24-hr illumination under yellow long-pass filters (25 to 45 μE m−2 sec−1) to prevent breakdown of indolic compounds (Stasinopoulos and Hangarter, 1990). Plants transferred to soil (Metromix 200; Scotts, Marysville, OH) were grown at 22 to 25°C under continuous illumination (∼200 μE m−2 sec−1) with Cool White fluorescent bulbs (Sylvania, Versailles, KY).
Mutant Isolation
The iar1-1 and iar1-2 mutants were isolated from pools of Wassilewskija (Ws) seed mutagenized with ethyl methanesulfonate (EMS) as described previously (Normanly et al., 1997). Approximately 24,000 surface-sterilized M2 seed were grown on PNS supplemented with 50 μM IAA-Ala. Two-week-old putative mutants with long roots were transferred to soil and allowed to set seed. The resulting M3 seed were screened separately for resistance to 50 μM IAA-Ala and for sensitivity to 1 μM IAA. iar1-3 and iar1-4 were isolated similarly from M2 progeny of fast neutron–mutagenized Columbia (Col-0) gl1-1 seed (purchased from Lehle Seeds, Round Rock, TX). The other alleles were identified from M2 pools of EMS-mutagenized ilr1-1 seed on PNS supplemented with 100 μM IAA-Leu (iar1-5 and iar1-6) or 70 μM IAA-Phe (iar1-7), as described previously (Davies et al., 1999). The independence of the seven alleles was confirmed by sequencing polymerase chain reaction (PCR) amplification products of genomic DNA. iar1 mutants were backcrossed at least three times to the parental accession (Col-0 or Ws) before phenotypic analysis.
To obtain the iar1-1 alf1 homozygous double mutant, an alf1/ALF1 heterozygous plant (accession Ws) was crossed to the iar1-1 mutant, and F2 plants with the phenotypes of both single mutants were transferred to soil and allowed to set seed. Some of these plants were partially fertile (unlike alf1/alf1 IAR1/IAR1 plants). These plants yielded 100% IAA-Ala–resistant, alf1-like progeny, indicating that they were homozygous for both mutations. Root lengths were measured in progeny from these iar1-1/iar1-1 alf1/alf1 plants after 10 days at 22°C under continuous yellow-filtered light.
Genetic Mapping of iar1
To map the iar1 mutation, we used segregating populations from crosses between IAR1/IAR1 (accession Col-0) and iar1/iar1 (accession Ws) plants. Genomic DNA prepared (Celenza et al., 1995) from 325 IAA-Ala–resistant F2 plants was scored by using published (Konieczny and Ausubel, 1993; Bell and Ecker, 1994; Nelson et al., 2000) and new PCR-based polymorphic markers. New markers developed for mapping include 13A11RE (amplification with the oligonucleotides 5′-GGTCTTCTGTATTTCACATGGC-3′ and 5′-GTGACA-ATAAGATAATTAGATCCGG-3′ yielded a 144-bp product with one Tsp509I site in Ws and none in Col-0), T7E4 (amplification with the oligonucleotides 5′-CGGTTTCGAGAGTTGCCTCGT-3′ and 5′-GCG-AACTGAATAAAAACCTCCG-3′ yielded a 260-bp product with one MseI site in Ws and none in Col-0), ETR1 (amplification with the oligonucleotides 5′-ATCTCCGTAACCGCTCTTGTCACC-3′ and 5′-ACC-ACCACCATCTTGTTCCTCTAT-3′ yielded a 1200-bp product with one NcoI site in Ws and none in Col-0), and KNAT2 (amplification with the oligonucleotides 5′-GGTAGCCATATCAGTTCATTG-3′ and 5′-ATTTAGTTGACACATCAGCTATC-3′ yielded a 660-bp product with three AluI sites in Ws and two in Col-0).
Complementation Library Construction
DNA (∼50 μg) from the T7E4 bacterial artificial chromosome (BAC) (Choi et al., 1995) was partially digested with Sau3AI and separated on 0.8% agarose gel containing 1 mM guanosine to prevent UV light damage to the DNA (Gründemann and Schömig, 1996). Fragments of ∼10 kb were purified by using the Qiaex II gel extraction kit (Qiagen, Valenica, CA), ligated into a BamHI-digested pBIN19 plant transformation vector (Bevan, 1984), and transformed into Escherichia coli strain DH5α. A contig of overlapping clones covering the region in which iar1 mapped was assembled by using a combination of DNA blot hybridization and sequencing. Selected clones were introduced into Agrobacterium tumefaciens GV3101 (Koncz and Schell, 1986) by electroporation (Ausubel et al., 1995). iar1-1 and iar1-3 mutant plants were transformed with these constructs by the floral dip method (Clough and Bent, 1998), and Kan-resistant transformants were transferred to soil ∼14 days after the T1 seed had been plated on PNS supplemented with 15 μg/mL Kan. T2 plants were tested for rescue of the iar1 IAA-Ala–resistant phenotype by plating on 40 μM IAA-Ala and examining root elongation after 8 days.
pBINIAR1g was made by cloning a 6.5-kb EcoRI fragment from C37 into the EcoRI site of pBIN19. This construct was transformed into iar1-3 as described above, and homozygous lines were identified by examining the pattern of Kan resistance in the T3 generation.
To determine the sequences of the various iar1 mutations, we amplified genomic DNA by PCR with the following pairs of oligonucleotides: 5′-GAGATGCAAAGAGCAACACTC-3′ with 5′-CACTGATGG-AATGGCATTAGG-3′, and 5′-CACTGATGGAATGGCATTAGG-3′ with 5′-CTGCAAGAACTCCAGCAACAGCTA-3′ (40 cycles of 15 sec at 94°C, 15 sec at 55°C, and 30 sec at 72°C); and 5′-GAACCAGGACAATCATCGTTG-3′ with 5′-GGTACTGCTATCCAATCCGG-3′, and 5′-CGTGACGATCACGTGCATCATC-3′ with 5′-GCGAATCCGAATGAGAATGATC-3′ (40 cycles of 30 sec at 95°C, 30 sec at 56°C, and 3 min at 72°C). Products were sequenced directly with the primers used for amplification after sequential precipitations with ethanol, polyethylene glycol, and ethanol (Ausubel et al., 1995). The base changes in most iar1 alleles alter restriction enzyme recognition sites (Table 1).
cDNA Isolation
A full-length IAR1 cDNA was isolated by hybridizing an Arabidopsis accession Landsberg erecta cDNA library (Minet et al., 1992) with a 1.2-kb HindIII/PstI genomic fragment subcloned from the rescuing C37 construct (Figure 5B) containing the last four exons of the IAR1 gene. The NotI insert of the cDNA was subcloned into the NotI site of pBluescript KS+ (Stratagene, La Jolla, CA) to create KSIAR1c, which was sequenced by using vector-derived and internal primers (GenBank accession number AF216524).
Transgenic Lines Expressing the Mouse KE4 cDNA
A full-length cDNA encoding the mouse IAR1 homolog KE4 in the vector pME18S-FL3 was purchased from Research Genetics (Huntsville, AL). The ∼1.6-kb cDNA insert was excised with StuI and XmnI and ligated into the SmaI site of 35SpBARN (S. LeClere and B. Bartel, unpublished data) to create 35S-MmKE4, such that the mouse KE4 cDNA was under the control of the 35S promoter. This construct was transformed into iar1-3 plants as described above, transformants were selected on PNS supplemented with 7.5 μg/mL BASTA, and homozygous lines were identified by examining the pattern of BASTA resistance in the T3 generation.
Acknowledgments
We thank Patrick Masson, John Sedbrook, and Daphne Preuss for sharing markers; John Celenza for alf1/ALF1 seed; Mary Lou Guerinot for the zrt1 zrt2 yeast strain; Sherry LeClere and Seiichi Matsuda for IAA-Gln and IAA-Glu; Sherry LeClere for 35SpBARN; and Mindy Cohen, Rosie Tellez, and Melanie Stagger for iar1 alleles. We thank Sherry LeClere, Mónica Magidin, Seiichi Matsuda, and Bethany Zolman for critical comments on the manuscript, and Kevin MacKenzie for helpful discussions. The Arabidopsis Biological Resource Center at The Ohio State University supplied BAC and YAC clones. This research was supported by the National Institutes of Health (Grant No. R29 GM54749), the Robert A. Welch Foundation (Grant No. C-1309), and National Aeronautics and Space Administration/Texas Space Grant Consortium and Houston Livestock Show and Rodeo fellowships (to J.L.).
References
- Abe, K., Wei, J.-F., Wei, F.-S., Hsu, Y.-C., Uehara, H., Artzt, K., and Bennett, D. (1988). Searching for coding sequences in the mammalian genome: The H-2K region of the mouse MHC is replete with genes expressed in embryos. EMBO J. 7 3441–3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando, A., Kikuti, Y.Y., Shigenari, A., Kawata, H., Okamoto, N., Shiina, T., Chen, L., Ikemura, T., Abe, K., Kimura, M., and Inoko, H. (1996). cDNA cloning of the human homologues of the mouse Ke4 and Ke6 genes at the centromeric end of the human MHC region. Genomics 35 600–602. [DOI] [PubMed] [Google Scholar]
- Ausubel, F., Brent, R., Kingston, R.E., Moore, D.D., Seidman, J.G., Smith, J.A., and Struhl, K. (1995). Short Protocols in Molecular Biology. (New York: John Wiley and Sons).
- Bandurski, R.S., Cohen, J.D., Slovin, J.P., and Reinecke, D.M. (1995). Auxin biosynthesis and metabolism. In Plant Hormones, P.J. Davies, ed (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 39–65.
- Bartel, B. (1997). Auxin biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48 51–66. [DOI] [PubMed] [Google Scholar]
- Bartel, B., and Fink, G.R. (1995). ILR1, an amidohydrolase that releases active indole-3-acetic acid from conjugates. Science 268 1745–1748. [DOI] [PubMed] [Google Scholar]
- Bell, C.J., and Ecker, J.R. (1994). Assignment of 30 microsatellite loci to the linkage map of Arabidopsis. Genomics 19 137–144. [DOI] [PubMed] [Google Scholar]
- Bennett, M., Kieber, J., Giraudat, J., and Morris, P. (1998). Hormone regulated development. In Arabidopsis, M. Anderson and J. Roberts, eds (Boca Raton, FL: CRC Press), pp. 107–150.
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boerjan, W., Cervera, M.T., Delarue, M., Beeckman, T., Dewitte, W., Bellini, C., Caboche, M., Van Onckelen, H., Van Montagu, M., and Inzé, D. (1995). superroot, a recessive mutation in Arabidopsis, confers auxin overproduction. Plant Cell 7 1405–1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, D.Y., Stathakis, D.G., and O'Donnell, J.M. (2000). The regulation of tyrosine hydroxylase and guanosine triphosphate cyclohydrolase in Catsup mutants (abstract no. 751C). In 41st Annual Drosophila Research Conference (Pittsburgh, PA: Genetics Society of America), p. a259.
- Campanella, J.J., Ludwig-Müller, J., and Town, C.D. (1996). Isolation and characterization of mutants of Arabidopsis thaliana with increased resistance to growth inhibition by indoleacetic acid–amino acid conjugates. Plant Physiol. 112 735–745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celenza, J.L., Grisafi, P.L., and Fink, G.R. (1995). A pathway for lateral root formation in Arabidopsis thaliana. Genes Dev. 9 2131–2142. [DOI] [PubMed] [Google Scholar]
- Chang, C., Kwok, S.F., Bleecker, A.B., and Meyerowitz, E.M. (1993). Arabidopsis ethylene-response gene ETR1: Similarity of product to two-component regulators. Science 262 539–544. [DOI] [PubMed] [Google Scholar]
- Choi, S., Creelman, R.A., Mullet, J.E., and Wing, R.A. (1995). Construction and characterization of a bacterial artificial chromosome library of Arabidopsis thaliana. Weeds World 2 17–20. [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. [DOI] [PubMed] [Google Scholar]
- Cohen, J.D., and Bandurski, R.S. (1982). Chemistry and physiology of the bound auxins. Annu. Rev. Plant Physiol. 33 403–430. [Google Scholar]
- Cohen, J.D., Slovin, J.P., Bialek, K., Chen, K.H., and Derbyshire, M.K. (1988). Mass spectrometry, genetics, and biochemistry: Understanding the metabolism of indole-3-acetic acid. In Biomechanisms Regulating Growth and Development, G.L. Steffens and T.S. Rumsey, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 229–241.
- Davies, R.T., Goetz, D.H., Lasswell, J., Anderson, M.N., and Bartel, B. (1999). IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11 365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dürr, G., Strayle, J., Plemper, R., Elbs, S., Klee, S.K., Catty, P., Wolf, D.H., and Rudolph, H.K. (1998). The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum–associated protein degradation. Mol. Biol. Cell 9 1149–1162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide, D., Broderius, M., Fett, J., and Guerinot, M.L. (1996). A novel iron-regulated metal transporter from plants identified by functional expression in yeast. Proc. Natl. Acad. Sci. USA 93 5624–5628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng, B.H., Guiernot, M.L., Eide, D., and Saier, M.H. (1998). Sequence analysis and phylogenetic characterization of the ZIP family of metal ion transport proteins. J. Membr. Biol. 166 1–7. [DOI] [PubMed] [Google Scholar]
- Estelle, M., and Klee, H.J. (1994). Auxin and cytokinin in Arabidopsis. In Arabidopsis, E.M. Meyerowitz and C.R. Somerville, eds (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press), pp. 555–578.
- Gaither, L.A., and Eide, D.J. (2000). Functional expression of the human hZIP2 zinc transporter. J. Biol. Chem. 275 5560–5564. [DOI] [PubMed] [Google Scholar]
- Gitan, R.S., Luo, H., Rodgers, J., Broderius, M., and Eide, D. (1998). Zinc-induced inactivation of the yeast ZRT1 zinc transporter occurs through endocytosis and vacuolar degradation. J. Biol. Chem. 273 28617–28624. [DOI] [PubMed] [Google Scholar]
- Grotz, N., Fox, T., Connolly, E., Park, W., Guerinot, M.L., and Eide, D. (1998). Identification of a family of zinc transporter genes from Arabidopsis that respond to zinc deficiency. Proc. Natl. Acad. Sci. USA 95 7220–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gründemann, D., and Schömig, E. (1996). Protection of DNA during preparative agarose gel electrophoresis against damage induced by ultraviolet light. BioTechniques 21 898–903. [DOI] [PubMed] [Google Scholar]
- Guerinot, M.L. (2000). The ZIP family of metal transporters. Biochim. Biophys. Acta 1465 190–198. [DOI] [PubMed] [Google Scholar]
- Guerinot, M.L., and Eide, D. (1999). Zeroing in on zinc uptake in yeast and plants. Curr. Opin. Plant Biol. 2 244–249. [DOI] [PubMed] [Google Scholar]
- Haughn, G.W., and Somerville, C. (1986). Sulfonylurea-resistant mutants of Arabidopsis thaliana. Mol. Gen. Genet. 204 430–434. [Google Scholar]
- Higgins, D.G., and Sharp, P.M. (1989). Fast and sensitive multiple sequence alignments on a microcomputer. Comput. Appl. Biosci. 5 151–153. [DOI] [PubMed] [Google Scholar]
- Hirayama, T., Kieber, J.J., Hirayama, N., Kogan, M., Guzman, P., Nourizadeh, S., Alonso, J.M., Dailey, W.P., Dancis, A., and Ecker, J.R. (1999). RESPONSIVE-TO-ANTAGONIST1, a Menkes/Wilson disease–related copper transporter, is required for ethylene signaling in Arabidopsis. Cell 97 383–393. [DOI] [PubMed] [Google Scholar]
- Jakubowska, A., Kowalczyk, S., and Leznicki, A.J. (1993). Enzymatic hydrolysis of 4-O- and 6-O-indol-3-ylacetyl—d-glucose in plant tissues. J. Plant Physiol. 142 61–66. [Google Scholar]
- Janatipour, M., Naumov, Y., Ando, A., Sugimura, K., Okamoto, N., Tsuji, K., Abe, K., and Inoko, H. (1992). Search for MHC-associated genes in human: Five new genes centromeric to HLA-DP with yet unknown functions. Immunogenetics 35 272–278. [DOI] [PubMed] [Google Scholar]
- King, J.J., Stimart, D.P., Fisher, R.H., and Bleecker, A.B. (1995). A mutation altering auxin homeostasis and plant morphology in Arabidopsis. Plant Cell 7 2023–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koncz, C., and Schell, J. (1986). The promoter of the TL-DNA gene 5 controls the tissue-specific expression of chimaeric genes carried by a novel type of Agrobacterium binary vector. Mol. Gen. Genet. 204 383–396. [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Korshunova, Y.O., Eide, D., Clarke, W.G., Guerinot, M.L., and Pakrasi, H.B. (1999). The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 40 37–40. [DOI] [PubMed] [Google Scholar]
- Kowalczyk, S., and Bandurski, R.S. (1990). Enzymatic hydrolysis of 1-O-, 4-O-, and 6-O-indol-3-ylacetyl—d-glucose and the enzymatic synthesis of indole-3-acetyl glycerol by a hormone metabolizing complex. Plant Physiol. 94 4–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuleck, G.A., and Cohen, J.D. (1993). An indole-3-acetyl amino acid hydrolytic enzyme from carrot cells. Plant Physiol. 102 (suppl.), 60 [Google Scholar]
- Kumer, S.C., and Vrana, K.E. (1996). Intricate regulation of tyrosine hydroxylase activity and gene expression. J. Neurochem. 67 443–462. [DOI] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R.F. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157 105–132. [DOI] [PubMed] [Google Scholar]
- Lasswell, J. (2000). Genetic analyses of auxin metabolism and of the transition to flowering in the model plant Arabidopsis thaliana. Ph.D. Dissertation (Houston, TX: Rice University).
- Last, R.L., and Fink, G.R. (1988). Tryptophan-requiring mutants of the plant Arabidopsis thaliana. Science 240 305–310. [DOI] [PubMed] [Google Scholar]
- Lehman, A., Black, R., and Ecker, J.R. (1996). HOOKLESS1, an ethylene response gene, is required for differential cell elongation in the Arabidopsis hypocotyl. Cell 85 183–194. [DOI] [PubMed] [Google Scholar]
- Liang, F., Cunningham, K.W., Harper, J.F., and Sze, H. (1997). ECA1 complements yeast mutants defective in Ca2+ pumps and encodes an endoplasmic reticulum–type Ca2+-ATPase in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 94 8579–8584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln, C., Long, J., Yamaguchi, J., Serikawa, K., and Hake, S. (1994). A knotted1-like homeobox gene in Arabidopsis is expressed in the vegetative meristem and dramatically alters leaf morphology when overexpressed in transgenic plants. Plant Cell 6 1859–1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lioumi, M., Ferguson, C.A., Sharpe, P.T., Freeman, T., Marenholz, I., Mischke, D., Heizmann, C., and Ragoussis, J. (1999). Isolation and characterization of human and mouse ZIRTL, a member of the IRT1 family of transporters, mapping within the epidermal differentiation complex. Genomics 62 272–280. [DOI] [PubMed] [Google Scholar]
- Ludwig-Müller, J., Epstein, E., and Hilgenberg, W. (1996). Auxin-conjugate hydrolysis in Chinese cabbage: Characterization of an amidohydrolase and its role during infection with clubroot disease. Physiol. Plant. 97 627–634. [Google Scholar]
- MacDiarmid, C.W., Gaither, L.A., and Eide, D. (2000). Zinc transporters that regulate vacuolar zinc storage in Saccharomyces cerevisiae. EMBO J. 19 2845–2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minet, M., Dufour, M.-E., and Lacroute, F. (1992). Complementation of Saccharomyces cerevisiae auxotrophic mutants by Arabidopsis thaliana cDNAs. Plant J. 2 417–422. [DOI] [PubMed] [Google Scholar]
- Monteiro, A.M., Crozier, A., and Sandberg, G. (1988). The biosynthesis and conjugation of indole-3-acetic acid in germinating seed and seedlings of Dalbergia dolchipetala. Planta 174 561–568. [DOI] [PubMed] [Google Scholar]
- Nakai, K., and Kanehisa, M. (1992). A knowledge base for predicting protein localization sites in eukaryotic cells. Genomics 14 897–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson, D.C., Lasswell, J., Rogg, L.E., Cohen, M.A., and Bartel, B. (2000). FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101 331–340. [DOI] [PubMed] [Google Scholar]
- Normanly, J. (1997). Auxin metabolism. Physiol. Plant. 100 431–442. [Google Scholar]
- Normanly, J., and Bartel, B. (1999). Redundancy as a way of life: IAA metabolism. Curr. Opin. Plant Biol. 2 207–213. [DOI] [PubMed] [Google Scholar]
- Normanly, J., Grisafi, P., Fink, G.R., and Bartel, B. (1997). Arabidopsis mutants resistant to the auxin effects of indole-3-acetonitrile are defective in the nitrilase encoded by the NIT1 gene. Plant Cell 9 1781–1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Östin, A., Kowalyczk, M., Bhalerao, R.P., and Sandberg, G. (1998). Metabolism of indole-3-acetic acid in Arabidopsis. Plant Physiol. 118 285–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pence, N.S., Larsen, P.B., Ebbs, S.D., Letham, D.L.D., Lasat, M.M., Garvin, D.F., Eide, D., and Kochian, L. (2000). The molecular physiology of heavy metal transport in the Zn/Cd hyperaccumulator Thlaspi caerulescens. Proc. Natl. Acad. Sci. USA 97 4956–4960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodríguez, F.I., Esch, J.J., Hall, A.E., Binder, B.M., Schaller, G.E., and Bleecker, A.B. (1999). A copper cofactor for the ethylene receptor ETR1 from Arabidopsis. Science 283 996–998. [DOI] [PubMed] [Google Scholar]
- Rogers, E.E., Eide, D.J., and Guerinot, M.L. (2000). Altered selectivity in an Arabidopsis metal transporter. Proc. Natl. Acad. Sci. USA 97 12356–12360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph, H.K., Antebi, A., Fink, G.R., Buckley, C.M., Dorman, T.E., LeVitre, J., Davidow, L.S., Mao, J., and Moir, D.T. (1989). The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+-ATPase family. Cell 58 133–145. [DOI] [PubMed] [Google Scholar]
- Schultz, J., Copley, R.R., Doerks, T., Ponting, C.P., and Bork, P. (2000). SMART: A web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 28 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stasinopoulos, T.C., and Hangarter, R.P. (1990). Preventing photochemistry in culture media by long-pass light filters alters growth of cultured tissues. Plant Physiol. 93 1365–1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stathakis, D.G., Burton, D.Y., McIvor, W.E., Krishnakumar, S., Wright, T.R.F., and O'Donnell, J.M. (1999). The catecholamines up (Catsup) protein of Drosophila melanogaster functions as a negative regulator of tyrosine hydroxylase activity. Genetics 153 361–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St.-Jaques, B., Han, T.-H., MacMurray, A., and Shin, H.-S. (1990). A putative transmembrane protein with histidine-rich charge clusters encoded in the H-2K/tw5 region of mice. Mol. Cell. Biol. 10 138–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford, D.L. (2000). PAUP*: Phylogenetic Analysis Using Parsimony (*and Other Methods). (Sunderland, MA: Sinauer Associates).
- Szerszen, J.B., Szczyglowski, K., and Bandurski, R.S. (1994). iaglu, a gene from Zea mays involved in conjugation of growth hormone indole-3-acetic acid. Science 265 1699–1701. [DOI] [PubMed] [Google Scholar]
- Sztein, A.E., Cohen, J.D., Slovin, J.P., and Cooke, T.J. (1995). Auxin metabolism in representative land plants. Am. J. Bot. 82 1514–1521. [Google Scholar]
- Sztein, A.E., Cohen, J.D., García de la Fuente, I., and Cooke, T.J. (1999). Auxin metabolism in mosses and liverworts. Am. J. Bot. 86 1544–1555. [PubMed] [Google Scholar]
- Tsurumi, S., and Wada, S. (1986). Dioxindole-3-acetic acid conjugates formation from indole-3-acetylaspartic acid in Vicia seedlings. Plant Cell Physiol. 27 1513–1522. [Google Scholar]
- Tuominen, H., Östin, A., Sandberg, G., and Sundberg, B. (1994). A novel metabolic pathway for indole-3-acetic acid in apical shoots of Populus tremula (L.) × Populus tremuloides (Michx.). Plant Physiol. 106 1511–1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vijayraghavan, U., Siddiqi, I., and Meyerowitz, E. (1995). Isolation of an 800 kb contiguous DNA fragment encompassing a 3.5-cM region of chromosome 1 in Arabidopsis using YAC clones. Genome 38 817–823. [DOI] [PubMed] [Google Scholar]
- Zhao, H., and Eide, D. (1996. a). The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc. Natl. Acad. Sci. USA 93 2454–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, H., and Eide, D. (1996. b). The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J. Biol. Chem. 271 23203–23210. [DOI] [PubMed] [Google Scholar]