Abstract
Mutants at the PROCUSTE1 (PRC1) locus show decreased cell elongation, specifically in roots and dark-grown hypocotyls. Cell elongation defects are correlated with a cellulose deficiency and the presence of gapped walls. Map-based cloning of PRC1 reveals that it encodes a member (CesA6) of the cellulose synthase catalytic subunit family, of which at least nine other members exist in Arabidopsis. Mutations in another family member, RSW1 (CesA1), cause similar cell wall defects in all cell types, including those in hypocotyls and roots, suggesting that cellulose synthesis in these organs requires the coordinated expression of at least two distinct cellulose synthase isoforms.
INTRODUCTION
Cellulose, the most abundant biopolymer, is produced by plants, bacteria, and certain animal species; it consists of simple linear (1→4)-β–linked glucan chains assembled in parallel arrays in semicrystalline microfibrils (Delmer, 1999). In primary walls of most cotyledonous species, cellulose microfibrils constitute an extensible network cross-linked by hydrogen-bonded xyloglucans embedded in a hydrophilic pectin matrix (Carpita and Gibeaut, 1993; McCann and Roberts, 1994). In growing cells, cellulose microfibrils are deposited transversely to the elongation axis and are thus thought to constrain growth in one direction. Cortical microtubule arrays often parallel the orientation of the microfibrils and appear to play a key role in controlling the orientation of the microfibrils in the primary wall (Giddings and Staehelin, 1991; Wymer and Lloyd, 1996). Not only do microfibrils control cell shape, but also their orientation in meristematic tissues controls the formation of organ primordia (Green, 1994) and thus plays a key role in the control of plant architecture.
When cell expansion is arrested, a secondary wall is deposited within the bounds of the primary wall. The matrix polysaccharide composition of the secondary wall differs from that of the primary wall and frequently contains a high proportion of lignin. In addition, microfibrils are mostly deposited in the secondary wall in helical arrays in successive layers of alternating pitch (Vian and Reis, 1991). As a result, this wall is much more rigid and provides strength and resistance against compressive forces, in contrast to the flexible primary walls of organs, which maintain their shape as a result of turgor pressure within their tissues. In summary, these observations show that a thorough understanding of the molecular mechanism and the regulation of cellulose synthesis is indispensable for understanding multiple aspects of plant development.
To date, cellulose synthesis remains poorly understood (Kawagoe and Delmer, 1997; Delmer, 1999). UDP-glucose is the direct substrate added to the nonreducing end of the growing (1→4)-β-glucan chain. In plants, a membrane-bound sucrose synthase, the enzyme that converts sucrose to UDP-glucose, may be linked physically to the cellulose synthase complex and may play an important role in channeling sucrose into the cellulose synthase complex (Amor et al., 1995). The biochemistry of cellulose synthesis is extremely complex, and thus far no efficient procedure to assay this activity in vitro has been described (Delmer, 1999). In contrast to other cell wall polysaccharides, which are synthesized in the Golgi apparatus and transported to the cell surface by vesicles, cellulose is thought to be synthesized by plasma membrane–bound complexes. These complexes, which can be visualized in higher plants as hexameric clusters of particles on freeze-fractured plasma membranes, have been named rosette terminal complexes (TCs) or simply rosettes (Emons, 1985).
Genes encoding homologs of the catalytic subunit of cellulose synthase in bacteria have been identified in higher plants (Pear et al., 1996). The roles of at least two family members (CesA1 and CesA7) in the synthesis of cellulose were further demonstrated by the identification of mutants in Arabidopsis. rsw1 carries a mutation in CesA1 that causes a temperature-sensitive radial cell expansion defect and a cellulose deficiency in all cell types investigated (Arioli et al., 1998). In this mutant, moreover, the TCs disappear on images of freeze-fractured plasma membranes. irx3 is mutated in CesA7 and shows a collapsed xylem phenotype and a greatly decreased cellulose content specifically in secondary cell walls (Taylor et al., 1999). These genes thus appear to encode functionally specialized isoforms required for cellulose synthesis in primary or secondary walls. Recently, Kimura et al. (1999) showed that antibodies generated against a member of the cellulose synthase catalytic subunit family preferentially labeled TCs in freeze-fractured plasma membranes of Vigna angularis; thus, TCs were definitively identified as the site of cellulose synthesis.
Besides the polymerization of (1→4)-β-glucan, the assembly of the newly synthesized polymer into microfibrils remains poorly understood. The analysis of rsw1 suggested that the synthesis and formation of microfibrils are two independent processes. Indeed, at the restrictive temperature and in the absence of functional rosettes, an ammonium oxalate–soluble, presumably noncrystalline, (1→4)-β-glucan accumulated (Arioli et al., 1998). Similarly, cotton fibers grown in vitro in the presence of the herbicide Novartis CGA 325′615, which also inhibits the formation of rosettes, produced (1→4)-β-glucan, but also produced it in an alkali or ammonium oxalate–extractable form (Delmer, 1999). These observations are consistent with the view that RSW1 and related proteins polymerize (1→4)-β-glucans but mediate crystallization into microfibrils only when they are associated in TCs.
Cellulose synthase catalytic subunits are encoded by a large gene family. In Arabidopsis, at least 10 isoforms are recognized (http://cellwall.stanford.edu/cellwall and http://www-plb.ucdavis.edu/labs/delmer/genes.html). The strong mutant phenotypes for RSW1 (CesA1) and IRX3 (CesA7) suggest a functional specialization for these isoforms. The functions of the other family members remain to be determined. Besides the catalytic subunits, presumably other genes are involved in cellulose synthesis. In the bacteria Acetobacter xylinum and Agrobacterium tumefaciens, several genes besides the cellulose synthase catalytic subunit are essential for cellulose synthesis, although their exact functions remain to be determined (Saxena et al., 1990; Matthysse et al., 1995a, 1995b). Interestingly, one of the essential genes in Agrobacterium encodes an endo-(1→4)-β-glucanase, which has been proposed to play a controversial role in the cleavage of lipid-linked intermediates of the biosynthetic pathway. In plants, a membrane-bound endo-(1→4)-β-glucanase, KOR, also may play a role in cellulose synthesis, as suggested by the cellulose deficiency of kor mutants (Nicol et al., 1998; M. Fagard, G. Mouille, F. Goubet, and H. Höfte, unpublished data).
In this article, further insight is provided regarding the cellulose synthesis machinery of plants. We report the cloning of PROCUSTE1 (PRC1), which was identified previously by mutations causing a growth defect in roots and dark-grown hypocotyls (Desnos et al., 1996). Here, we show that the decreased cell expansion in the dark-grown hypocotyl is correlated with a deficiency in cellulose and the presence of gaps in internal cortical and epidermal cell walls. PRC1 was cloned and found to encode a novel member of the CesA family, CesA6. Interestingly, both rsw1 and prc1 mutants caused a cell elongation defect and cellulose deficiency in roots and dark-grown hypocotyls; however, prc1 affected only a subset of the cell types that were affected in rsw1. This suggests that cellulose synthesis in this subset of cell types requires the coordinated activity of at least two CesA isoforms.
RESULTS
Identification of Additional prc1 Alleles
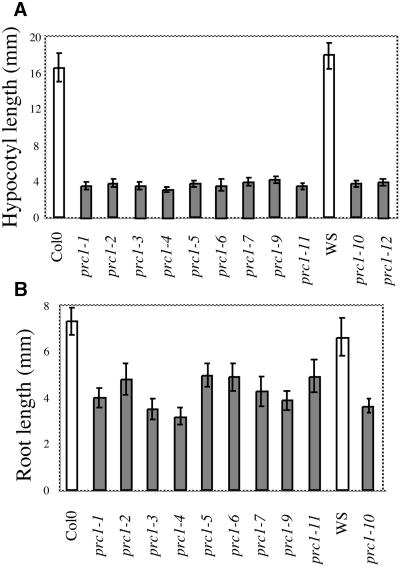
Fourteen new alleles of the prc1 locus were identified from Arabidopsis populations mutagenized with ethyl methanesulfonate, T-DNA, or x-rays. Among them were the previously described quill (qui) mutants (Hauser et al., 1995), which were shown to be allelic to prc1 (data not shown; see below). qui1, qui2, and qui3 were renamed prc1-9, prc1-10, and prc1-11, respectively. Figure 1A shows the hypocotyl lengths of seedlings for nine prc1 alleles grown for 7 days in complete darkness. Consistent with previous results (Desnos et al., 1996), all alleles (regardless of ecotype) showed a four- to fivefold decrease in hypocotyl length compared with that of wild-type seedlings, whereas no large variations in allele strength were observed (Figure 1A). Root length was also determined for seedlings grown for 7 days in white light on agar medium containing 4.5% sucrose (Figure 1B), conditions under which roots of qui alleles exhibit a strong radial expansion phenotype (Hauser et al., 1995). As expected, prc1 roots were 1.5- to twofold shorter than were wild-type roots, with greater variations between alleles than those observed for dark-grown hypocotyls. On the basis of these root lengths, prc1-3 and prc1-4 appeared to be the strongest alleles.
Figure 1.
prc1 Mutants Show Reduced Elongation of Roots and Dark-Grown Hypocotyls.
(A) Dark-grown hypocotyl lengths of wild-type (ecotypes Columbia [Col-0] and Wassilewskija [WS]) seedlings and 11 prc1 alleles. Seedlings were grown for 7 days in total darkness on sucrose-free agar medium.
(B) Root lengths of wild-type seedlings and 10 prc1 alleles. Seedlings were grown for 7 days in 16-hr-light/8-hr-dark cycles on agar medium containing 4.5% sucrose.
prc1-1 through prc1-9 and prc1-11 are Col-0 alleles; prc1-10 and prc1-12 are Ws ecotype.
Error bars indicate ±sd.
Alterations in Cell Wall Structure in Dark-Grown prc1 Hypocotyls
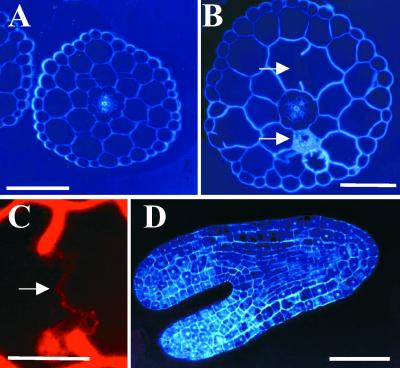
The prc1 phenotype can be phenocopied by treating wild-type seedlings with cellulose biosynthesis inhibitors such as 2,6-dichlorobenzonitrile (DCB) or isoxaben (data not shown). To look for potential cell wall defects, we studied transverse sections halfway through hypocotyls of 4-day-old dark-grown seedlings after staining with Calcofluor or Congo Red (Figure 2). Calcofluor stains cellulose, callose, and other nonsubstituted or weakly substituted β-glucans (Maeda and Ishida, 1967), whereas Congo Red more generally stains reducing sugars. As shown in Figure 2, no differences in the numbers of cells or tissue layers were observed. However, similar to the DCB-treated seedlings, prc1-1 hypocotyls were larger than were wild-type hypocotyls, with more radially expanded cells in epidermis, cortex, and endodermis. Interestingly, in almost every cross-section of dark-grown prc1-1 hypocotyls, incomplete cell walls were found (Figure 2B, top arrow). Incomplete walls were found in epidermal, cortical, and endodermal cell layers. The protruding wall ends were frequently found to be linked by a membranous structure not stained by Calcofluor but observable with Congo Red (Figure 2C) or other stains. This structure presumably corresponds to cell membranes surrounding polysaccharidic material. The study of two other alleles (prc1-3 and prc1-7; data not shown) confirmed the presence of such incomplete cell walls. In some sections (both transverse and longitudinal), cells filled with granular Calcofluor- and Congo Red–stained material were observed (Figure 2B, bottom arrow); these were never observed in the wild type, and the exact nature of the material remains to be determined.
Figure 2.
Dark-Grown prc1 Hypocotyls Contain Incomplete Cell Walls.
(A) and (B) Calcofluor-stained hypocotyl cross-sections of 4-day-old dark-grown wild-type (A) and prc1-1 (B) seedlings. The top arrow indicates an incomplete wall and the bottom arrow indicates an abnormally filled cell in a prc1-1 hypocotyl section.
(C) Congo Red–stained section of a prc1-1 hypocotyl cell in which the incomplete cell wall stubs are connected by a thin structure (arrow).
(D) Calcofluor-stained section through a prc1-1 immature embryo. No incomplete wall or cell filled with stained material was observed.
 ;
;  ;
; 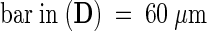 .
.
Incomplete cell walls, referred to as “stubs,” have been described in various mutants that are defective in cytokinesis (Liu et al., 1995a; Assaad et al., 1996; Lukowitz et al., 1996). Because cortical and most epidermal cells in Arabidopsis hypocotyls have an embryonic origin and do not divide during postembryonic development, we examined sections through prc1-1 embryos (illustrated for a torpedo-stage embryo in Figure 2D). Cell wall stubs were never observed. The same was true for mature embryos, or even for transverse sections through mature hypocotyls grown in the light (data not shown). Together, these observations indicate that cytokinesis is unaffected in prc1 and that the observed gapped walls in dark-grown prc1 hypocotyls arise during cell expansion.
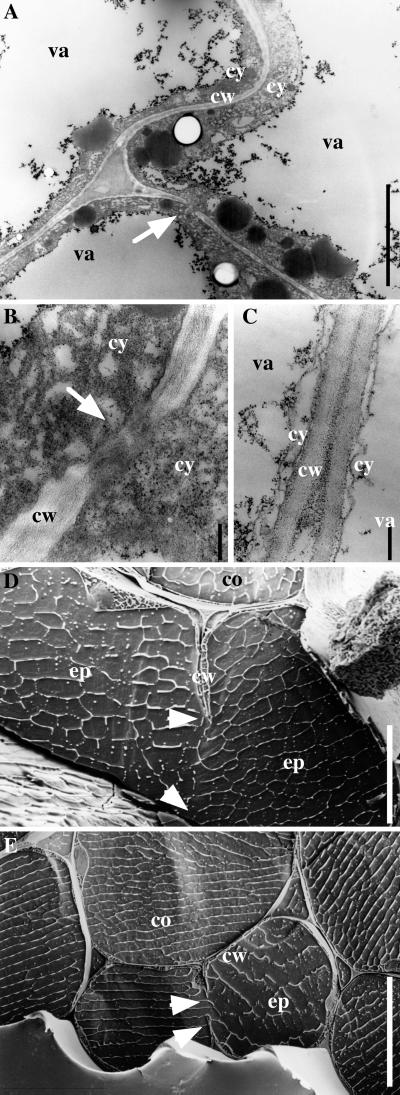
Incomplete walls were also observed in cortical and epidermal walls in dark-grown prc1-1 hypocotyls by scanning electron microscopy on freeze-fractured material or by transmission electron microscopy, as shown in Figure 3; incomplete walls were observed as frequently as on sections prepared for light microscopy. This finding strongly suggests that the discontinuous walls were present in living tissue and were not artifacts of the sample preparation. Furthermore, transmission electron microscopy revealed the presence of fibrillar material in the wall gaps (Figure 3B).
Figure 3.
prc1 Dark-Grown Hypocotyls Display Abnormal Cell Wall Structures.
(A) to (C) Transmission electron micrographs of cross-sections from dark-grown hypocotyls (4 days old) of prc1-1 mutant ([A] and [B]) and wild-type (C) seedlings in the cortical cell layer. Arrow in (A) indicates an abnormal wall portion shown at a higher magnification in (B).
(D) and (E) Scanning electron micrographs of prc1-1 dark-grown hypocotyls. Arrowheads indicate protruding stubs of incomplete cell walls in the epidermal cell layer.
co, cortex cell; cw, cell wall; cy, cytoplasm; ep, epidermal cell; va, vacuole. 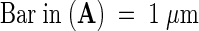 ;
; 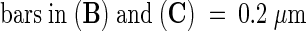 ;
; 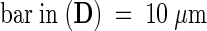 ;
; 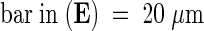 .
.
Mutations at the PRC1 Locus and Cellulose Deficiency
prc1 cell walls were analyzed by using Fourier transform infrared (FTIR) microspectroscopy. This technique allows rapid analysis of the cell wall composition of areas of the tissue as small as one or a few cells (McCann et al., 1997). FTIR absorption spectra of 4-day-old dark-grown hypocotyls of wild-type and two mutant alleles of prc1 were collected from a region of ∼60 × 40 μm midway along the hypocotyl and avoiding the central stele. The spectra correspond to the absorption in the mid-IR region (between wave numbers 600 and 4000 cm−1) of essentially longitudinal and transverse cell walls of epidermal and cortical cells (two layers of epidermis and four layers of cortex cells), both of which are cell types that are affected by the prc1 mutations.
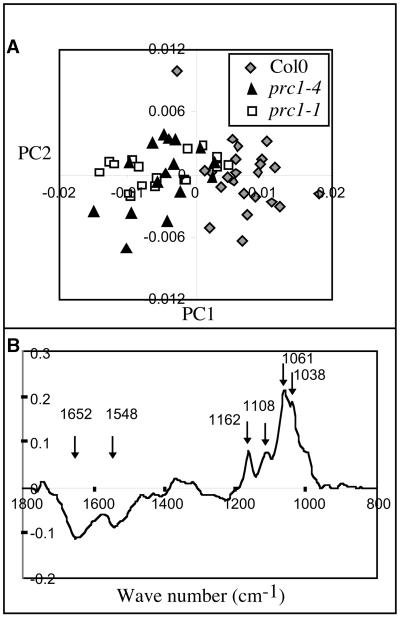
The spectra (20 for each condition) were analyzed by principal components (PCs) analysis (Kemsley, 1998), a statistical method that reduces the dimensionality of the data from >100 variates (one every 8 cm−1 for the region from 1800 to 850 cm−1) to only a few, the PCs. The PCs are ordered in terms of decreasing variance. Each observation (spectrum) has a corresponding set of PC scores describing the variance of that spectrum relative to the mean of the population for each PC. The PC scores of the spectra can then be plotted against one another to reveal patterns or structures in the data (Kemsley, 1998). One can then mathematically derive a “spectrum” (a PC loading) from a PC to identify the molecular factors responsible for the separation of groups of spectra (Kemsley, 1998). As shown in Figure 4A, the analysis showed that wild-type spectra can be separated from prc1 mutant spectra by using a combination of two PC scores. PC1 explained 72% of the variance. The loading for PC1 (Figure 4B) showed characteristics of purified cellulose in the fingerprint region (peaks at 1038, 1064, 1100, and 1162 cm−1, respectively) and of protein (peaks at 1650 and 1550 cm−1). Peaks at 1650 and 1550 cm−1 were negatively correlated with the cellulose fingerprint peaks. Because the PC scores of the spectra of prc1 seedlings were negative relative to the mean (Figure 4A), the data suggest that the cell walls of prc1 seedlings are relatively richer in protein and poorer in cellulose than the cell walls of wild-type seedlings. These results show that the changes in prc1 cell walls are complex but that a major deficiency in crystalline cellulose is a part of the phenotype.
Figure 4.
prc1 Dark-Grown Hypocotyl Cell Walls Are Cellulose Deficient.
(A) FTIR analysis of 4-day-old dark-grown hypocotyls. PC analysis was performed by using 20 FTIR spectra from wild-type and two different prc1 alleles. PC1 explained 72% of the variance between spectra from wild-type alleles and spectra from prc1 alleles. Col0, Columbia.
(B) The PC1 loading showed positive peaks characteristic of cellulose in the fingerprint region (1162, 1108, 1061, and 1038 cm−1, respectively), indicating that prc1 dark-grown hypocotyls were deficient in cellulose relative to the wild type. Two other major peaks (1652, and 1548 cm−1, respectively) appeared negatively in this PC-loading. These peaks corresponded to amide groups of protein, suggesting an enrichment for protein in prc1 walls relative to the wild type.
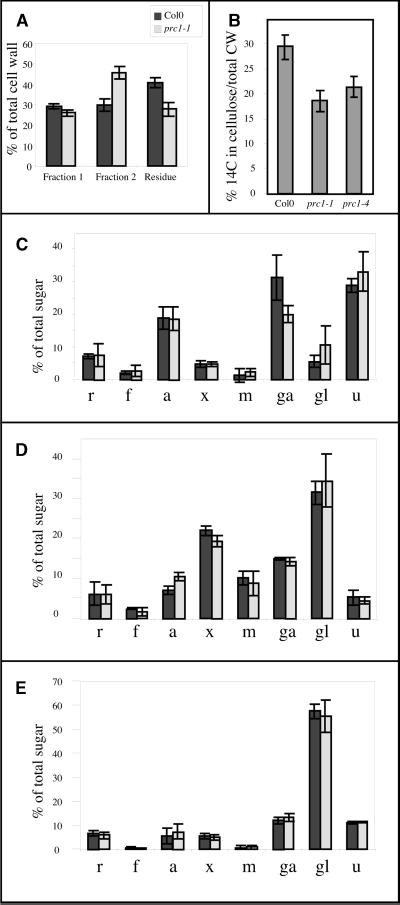
To confirm the observed changes chemically, walls of 4-day-old dark-grown wild-type and prc1-1 seedlings were isolated and fractionated, and the neutral sugar and uronic acid contents were determined for each fraction. In both wild-type and prc1-1 seedlings, ∼2% of the fresh weight consisted of cell walls. Wall material was extracted consecutively with 0.1 and 4 M KOH, yielding two fractions that were enriched for pectins (fraction 1) and hemicellulose (fraction 2), respectively, as shown by the neutral sugar and uronic acid contents (Figures 5C and 5D). The residual fraction contained 60% glucose (Figure 5E), which indicates an enrichment for cellulose, because as shown below, in the culture conditions used, neither wild-type nor prc1-1 accumulated significant levels of (1→3)-β-glucan (callose). Also, staining with iodine failed to detect starch in wild-type or prc1-1 hypocotyls (data not shown). The presence of minor amounts of xylosyl (5%), rhamnosyl (6%), uronic acid (10%), galactosyl (10%), and arabinosyl (6%) residues indicates that a proportion of the noncellulosic polysaccharides also resisted the alkaline treatment, suggesting an intimate association of these polymers with the cellulose microfibrils. Similar observations were reported for cell walls from Arabidopsis leaves (Zablackis et al., 1995).
Figure 5.
Chemical Analysis Confirms Cellulose Deficiency in Dark-Grown prc1 Seedlings.
(A) Walls were purified and extracted successively with 0.1 M KOH, yielding a pectin-rich fraction (fraction 1), and 4 M KOH, yielding a hemicellulose-rich fraction (fraction 2). The residual fraction was enriched for a glucan corresponding to cellulose (see text). prc1-1 walls were deficient for the residual fraction relative to the wild type, indicating a deficiency in cellulose for this mutant. Col0, Columbia.
(B) Incorporation of 14C-glucose (14C) in the acid-resistant cellulosic fraction in wild-type and prc1 alleles. Each value represents the mean of five independent measurements. CW, cell wall.
(C) to (E) Sugar composition of fraction 1 (C), fraction 2 (D), and the residue (E). Only fraction 1 reproducibly showed qualitative differences in sugar composition. r, rhamnose; f, fucose; a, arabinose; x, xylose; m, mannose; ga, galactose; gl, glucose; u, uronic acid.
Error bars indicate ±sd.
Strikingly, in mutant walls, the proportions of the three fractions were altered markedly (Figure 5A). Compared with wild-type walls, prc1-1 walls showed a relative increase in fraction 2, a relative decrease in the residual fraction, and a very minor decrease in fraction 1. Interestingly, in prc1-1, the decrease in the residual fraction was not accompanied by a change in the proportions of neutral sugars and uronic acid, suggesting that with less crystalline cellulose present as a substrate, less xyloglucan and pectin resisted the extraction procedure. The neutral sugar and uronic acid contents also remained unchanged for fractions 1 and 2, except for an important decrease of galactosyl residues in fraction 1. This may be the result of a reduction in the number of galactan side chains on the pectins.
To confirm the reduced cellulose production in prc1, we measured the incorporation of 14C-glucose into the nitric/acetic acid–insoluble cell wall fraction of actively growing 3-day-old seedlings (Updegraff, 1969). This fraction is 97% (1→4)-β–linked glucose (Peng et al., 2000; data not shown) and is generally considered to be crystalline cellulose. Figure 5B shows that in the wild type, 30% of the label in the cell wall was incorporated in the cellulosic fraction. This proportion was reduced to 17 and 19% in prc1-1 and prc1-4, respectively.
In conclusion, chemical analysis of the cell walls of mutant seedlings confirms the deficiency in cellulose observed by FTIR microspectroscopy. Other changes detected included a relative increase in hemicellulosic polysaccharides and qualitative changes in pectin composition.
The inhibition of cellulose by the cellulose synthesis inhibitor DCB in Arabidopsis seedlings causes an accumulation of callose (Nickle and Meinke, 1998). Callose accumulation was also observed in embryos of the cytokinesis-defective mutant cyt1 (Nickle and Meinke, 1998). We investigated whether also in prc1 the cellulose deficiency was accompanied by increased callose production. Staining hypocotyl sections as well as permeabilized seedlings with aniline blue or its fluorescent derivative (Stone et al., 1984) revealed a slight accumulation of callose in DCB-treated seedlings. In prc1-1 hypocotyl sections, we occasionally observed a weak staining, however much weaker than that observed in cyt1 (data not shown).
Map-Based Cloning of PRC1
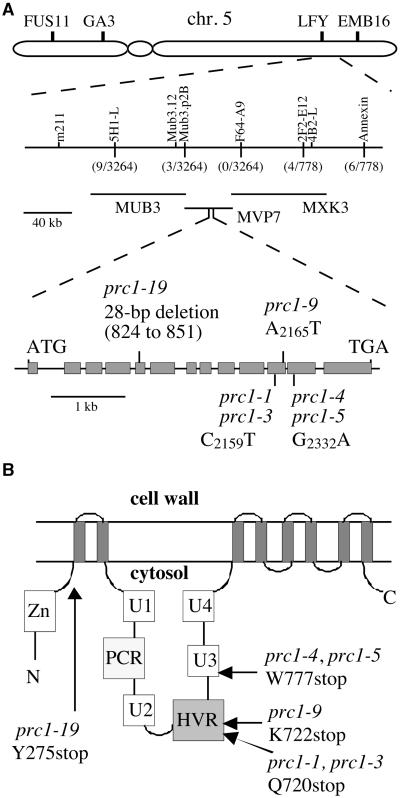
The prc1-1 mutation was mapped previously to the bottom of chromosome 5, 1.7 centimorgans (cM) distal to marker m211, in a small mapping population (Desnos, 1996; Desnos et al., 1996). New mapping populations were established by using a cross between prc1-1 and Landsberg erecta (Ler) as well as by crossing prc1-1 to a line (in the C24 ecotype) carrying a T-DNA insertion close to the LFY3 restriction fragment length polymorphism marker (Lister and Dean, 1993; Van Lijsebettens et al., 1996). By use of restriction fragment length polymorphism and cleaved amplified polymorphic sequence (Konieczny and Ausubel, 1993) markers, the prc1-1 mutation was mapped to a region of ∼120 kb covered by P1 clones (Liu et al., 1995b) MVP7 and MXK3. As shown in Figure 6A, prc1-1 is positioned 0.1 cM distal to MUB3.p2B and 1 cM proximal to 4B2-L/2F2-E12.
Figure 6.
The PRC1 Gene Is Isolated Through Map-Based Cloning.
(A) Physical map of the region of chromosome (chr.) 5 containing the PRC1 gene. The prc1-1 mutation was mapped to the bottom of chromosome 5, south of molecular marker LFY. New markers positioned the prc1-1 mutation between markers Mub3.p2B and 2F2-E12. The number of recombinant chromosomes is indicated in parentheses. Sequence analysis of candidate genes between these two markers showed that six prc1 mutant alleles carried a point mutation or a deletion in the CesA6 gene carried by P1 clone MVP7. The PRC1 (CesA6) gene comprises 13 exons (filled boxes) and 12 introns. The prc1 mutations that were identified are indicated.
(B) Predicted topology of the PRC1 protein. The protein is predicted to contain eight membrane-spanning helices (indicated by the eight gray bars). The putative Zn binding domain, the U1, U2, U3, and U4 domains, the plant conserved region (PCR), and the HVR are predicted to face the cytosol (Delmer, 1999). The positions of the mutations are indicated. N, N terminus; C, C terminus.
From the genomic sequence of the 120-kb region containing the prc1-1 mutation, seven candidate genes were selected. Polymerase chain reaction products for each of these genes were amplified from genomic DNA isolated from several prc1 alleles, sequenced, and compared with the relevant wild-type sequences. Six prc1 alleles showed nucleotide substitutions in the sequence of a predicted gene between positions 18,535 and 23,313 on P1 clone MVP7 (GenBank accession number AB025637). All six mutations caused premature stop codons in the predicted coding region. Interestingly, despite their provenance from independent M2 families, prc1-1 and prc1-3 showed mutations at identical positions, as did prc1-4 and prc1-5, albeit at a different location (Figure 6). The allele prc1-19 generated by T-DNA mutagenesis contained a 28-bp deletion (Figure 6), which may be a hallmark of an aborted T-DNA integration.
Isoform of the Cellulose Synthase Family Encoded by PRC1
The genomic sequence of PRC1 perfectly matches five expressed sequence tag (EST) sequences and a previously reported slightly truncated cDNA (AraxCelA; GenBank accession number AF062485) (Wu et al., 1998). The gene contains 13 exons and encodes a protein of 1084 amino acids with a predicted molecular mass of 123 kD. PRC1 is a member of a large, ancient family of β-glycosyl transferases present in bacteria, plants, and animals. In higher plants, a subfamily of highly conserved proteins can be distinguished, two members of which (RSW1 and IRX3) have been shown to be essential for cellulose synthesis in Arabidopsis. This subfamily, referred to as CesA, probably corresponds to the family of catalytic subunits of cellulose synthase and has at least 10 members in Arabidopsis. According to the classification proposed by Delmer (1999) (see also http://www-plb.ucdavis.edu/labs/delmer/genes.html), PRC1 corresponds to CesA6. The predicted protein structure is shown in Figure 6B.
The PRC1 protein contains the U1, U2, U3, and U4 domains that are characteristic of all β-glycosyl transferases and that form the catalytic domain. A protein fragment of a cotton homolog (CelA1) containing these motifs was shown to bind UDP-glucose in the absence of calcium ions (Kawagoe and Delmer, 1997; Delmer, 1999). All CesA family members from plants contain the so-called plant conserved region and a region that is hypervariable (HVR) between family members. Allele prc1-9 is predicted to produce a protein truncated at the N-terminus of the first transmembrane domain. In the other mutants analyzed, the predicted truncated product contains only the first two transmembrane domains and part of the catalytic domain. All mutants investigated therefore are expected to be null alleles.
Two CesA Isoforms Required for Normal Cell Expansion in Roots and Dark-Grown Hypocotyl Cells
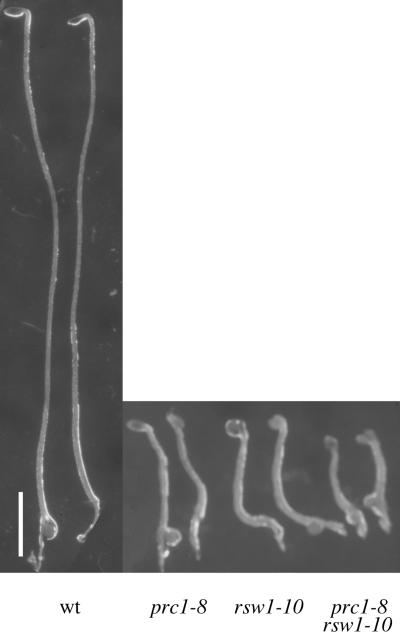
An intriguing observation is that two mutations in members of the same gene family caused very similar phenotypes in the same cell types. Indeed, the leaky allele rsw1-10 (H. Höfte, unpublished data) and the null allele prc1-8 both showed comparable decreases in the length of dark-grown hypocotyls, accompanied by increases in width (Figure 7) in both epidermal and cortical cells (data not shown). FTIR microspectroscopy also indicated a cellulose deficiency in epidermal or cortical cells (or both) in both mutants (Figure 4 and data not shown). The rsw1-10 mutation was additive to prc1-8, as shown by the further shortened hypocotyl length of the double mutant (Figure 7).
Figure 7.
prc1-8 and rsw1-10 Have Similar, and Additive, Hypocotyl Phenotypes.
Seedlings were grown for 4 days in the dark. Left to right: wild type (wt), prc1-8, rsw1-10, and double mutant. The prc1-8 and rsw1-10 mutants showed a similar decrease in hypocotyl length compared with wild-type seedlings, whereas the double mutant showed an even shorter hypocotyl. 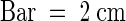 .
.
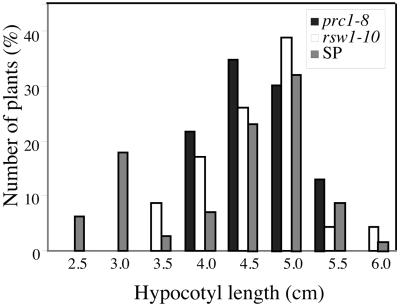
One explanation for these observations is that both gene products have comparable functions and contribute to a pool of enzymes that becomes limiting in a mutant background for one or the other locus. However, that situation is unlikely because both mutations are recessive and no striking phenotype was observed in plants that are transheterozygous for both mutations, whereas if the hypothesis described above were true, then the phenotype should be equivalent to that of a single homozygote. In addition, as shown in Figure 8, even for prc1-8 segregating in a homozygous rsw1-10 mutant background, no gene dosage effects were observed, as shown by the strict 1:3 segregation of the additive phenotype. Therefore, even in a situation in which the RSW1 gene product is growth-limiting, both gene copies of PRC1, rather than a single copy, need to be inactivated to result in a further reduction of hypocotyl length. This strongly suggests that the presence of each protein is critical for normal hypocotyl elongation, rather than the size of the combined pool of the CesA1 and CesA6 proteins.
Figure 8.
No Gene Dosage Effect Is Observed for CesA1 and CesA6.
Hypocotyl lengths of dark-grown prc1-8 seedlings, mutant for CesA6, rsw1-10 seedlings, mutant for CesA1, and the progeny of a plant homozygous for rsw1-10 and heterozygous for prc1-8 were measured. In the segregating population (SP), a discrete 1:3 segregation of the additive phenotype was observed. No gene dosage effect was observed. Indeed, an intermediate hypocotyl length for half of the seedlings, those corresponding to prc1-8 heterozygotes, would be expected if the dosage of the CesA6 gene were critical for hypocotyl growth.
Functional Specialization within the CesA Family
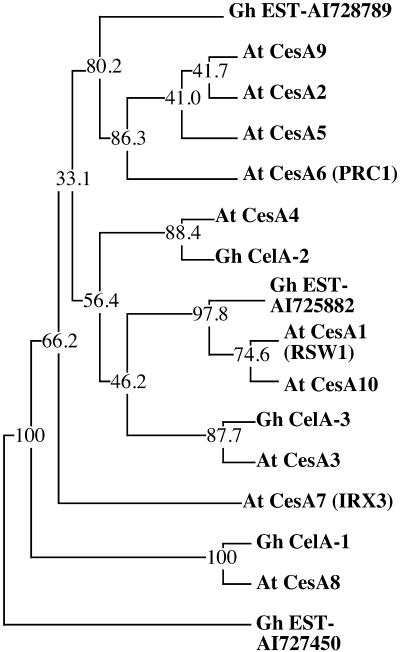
The strong mutant phenotypes for IRX3 (CesA7), RSW1 (CesA1), and PRC1 (CesA6) indicate that the other family members cannot compensate for the absence of their respective gene products. In some cases, this may be simply a result of cell type–specific expression patterns. However, as shown above, this does not seem to be the case, at least not for RSW1 and PRC1. Alternatively, the sequences of different family members may have diverged as adaptations to their specialized cellular roles. To reveal such specialized functions, we investigated the evolutionary relationships between different family members in Arabidopsis and cotton, a species for which a large number of CesA ESTs are available in public databases. The sequences of 10 Arabidopsis CesA family members were compared with those of three full-length cotton cDNAs and two genes identified by ESTs from different cotton libraries. Because the cotton ESTs corresponded to single-pass sequences close to the 3′ end of the mRNA, we chose the amino acid sequence corresponding to the HVR, which is localized between amino acids 690 and 743 close to the C-terminus of PRC1, to construct a similarity tree. The use of this hypervariable protein segment also maximized the chances to distinguish among the different family members, which in many cases are extremely conserved. A sixth, more divergent, cotton sequence (Gh EST-AI727450), presumably corresponding to a cellulose synthase–like gene (http://cellwall.stanford.edu/cellwall), was used as an outlier in the bootstrap procedure. As shown in Figure 9, amino acid sequences from the five cotton genes did not cluster together but in each case clustered with one or more genes from Arabidopsis. This suggests that the HVR is hypervariable among different family members within a species but is conserved between the orthologs in different species. A very similar tree was obtained when we used the complete amino acid sequences of the Arabidopsis genes and the three full-length cotton genes (data not shown). In conclusion, the divergence between the family members, at least those represented by the five cotton ESTs, must have predated the divergence between Brassicaceae and Malvaceae. Interestingly, RSW1 and PRC1 also had their respective orthologs in cotton.
Figure 9.
Members of the CesA Family Exhibit Functional Specialization.
Shown is a phylogenetic tree based on the HVR (see text and Figure 5) of protein sequences of Arabidopsis and cotton CesA family members. Alignment data are from bootstrap values sampled 100 times and used to construct the consensus tree shown. Numbers are bootstrap values and indicate the number of trees in which the proteins to the right of the bootstrap values clustered together. Gh EST-AI727450 was included as an outlier. The five other cotton genes did not cluster together but instead clustered with one or two Arabidopsis sequences, suggesting that the CesA amino acid sequences diverged before the divergence between Brassicaceae and Malvaceae; that, in turn, suggests a functional specialization of most CesA isoforms.
At, Arabidopsis thaliana; Gh, Gossypium hirsutum.
Expression Patterns of PRC1 and RSW1 mRNAs
RNA gel blot assays were performed with probes specific for sequences corresponding to a variable region in the respective N termini of PRC1 and RSW1 (Figure 10). Both PRC1 and RSW1 mRNA were detected in all organs investigated, except in mature flowers. Although the transcript abundance of PRC1 was less than that of RSW1, both mRNAs showed similar expression patterns. In developing seedlings, RSW1 and PRC1 mRNAs were high initially and decreased at later growth stages. Expression was high in elongating stems but dropped in the stem sections in which secondary wall synthesis takes place. Interestingly, despite the light-dependent hypocotyl phenotype of prc1, we did not obtain evidence for a regulation of PRC1 at the transcript level: indeed, transcript quantities did not differ between 4-day-old dark-grown and light-grown seedlings.
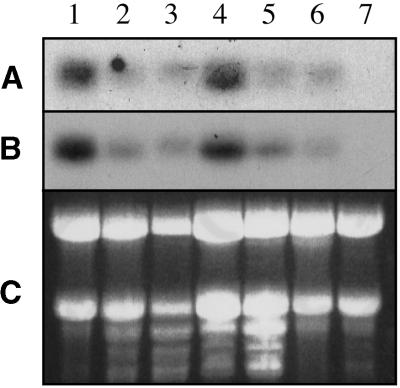
Figure 10.
PRC1 and RSW1 Have Similar Transcript Profiles.
Total RNA of different samples was hybridized with labeled probes corresponding to a variable sequence at the N-terminal end of CesA proteins.
(A) PRC1 probe.
(B) RSW1 probe.
(C) Ethidium bromide–stained agarose gel of the same samples.
Lane 1, 1-day-old germinating seeds; lane 2, 4-day-old dark-grown seedlings; lane 3, 4-day-old light-grown seedlings; lane 4, elongating inflorescence stem sections; lane 5, mature leaves; lane 6, roots; lane 7, mature flowers.
Similar results were obtained by comparing distributions of the EST sequences corresponding to RSW1 and PRC1 obtained from tissue-specific EST libraries (http://cellwall.stanford.edu/cellwall). For instance, in four EST libraries separately covering etiolated seedlings, above-ground tissues, roots, and siliques, we found, respectively, 1, 5, 5, and 1 EST for PRC1 and 4, 13, 18, and 2 ESTs for RSW1. This confirms that PRC1 expresses fewer transcripts than RSW1, that the tissue distribution of each is similar, and that PRC1 mRNA is present in both dark-grown and light-grown tissues.
DISCUSSION
In this report, we describe the cloning of a novel cellulose synthase required for cell elongation in roots and dark-grown hypocotyls in Arabidopsis. A role for PRC1 (CesA6) in the synthesis of cellulose was supported by two main observations: prc1 mutants display a cellulose deficiency, and the PRC1 gene is a member of the cellulose synthase catalytic subunit family.
Prc1 Phenotype a Result of Cellulose Deficiency
Chemical analysis of the wall composition confirmed a cellulose deficiency in mutant dark-grown seedlings. Indeed, after successive extractions with 0.1 and 4 M KOH, the residual fraction in prc1-1 was reproducibly 25% less than in wild-type seedlings. This is probably an underestimate, because not all cell types of the dark-grown seedlings were affected by the mutation. This residual fraction was highly enriched for glucose, which in theory could indicate an enrichment for (1→3)-β-glucan (callose), (1→4)-α-glucan (starch), or (1→4)-β-glucan (cellulose). Specific staining with aniline blue failed to detect a substantial accumulation of callose in wild-type or mutant hypocotyl cells. The resistance to strong alkali and the absence of iodine staining also ruled out the presence of starch in this fraction. The residue therefore must consist primarily of cellulose. This was further confirmed by labeling experiments, which showed that less 14C-glucose was incorporated into the nitric/acetic acid–resistant fraction in prc1 than in the wild type.
A cellulose deficiency was also detected by FTIR microspectroscopy. The advantage of this technique is that the cell wall composition can be assessed at a microscopic level in one or a few cells, whereas chemical methods unavoidably provide an average view of many different cell types. PC analysis of absorption spectra obtained from an area corresponding exclusively to epidermal and cortical cells of dark-grown hypocotyls revealed a cellulose deficiency in mutant cell walls. Similar results were obtained when dark-grown hypocotyls of wild-type seedlings treated with cellulose synthesis inhibitors such as DCB or isoxaben were compared with those of untreated controls, confirming the validity of the method (Fagard, 1999).
PRC1 a Member of the Cellulose Synthase Family
All six mutant alleles sequenced contained premature stop codons in CesA6, which in each case removed a major part or even all of the catalytic domain. The prc1 phenotype is therefore caused by null alleles of the CesA6 gene. This gene belongs to family 2 of glycosyl transferase, which also contains bacterial cellulose synthases. Previous mutant analysis has shown that at least two other family members, CesA1 (Arioli et al., 1998) and CesA7 (Taylor et al., 1999), are required for normal cellulose synthesis in Arabidopsis. Also, the disappearance of rosettes from scanning electron microscopy images of freeze-fractured plasma membranes at the restrictive temperature in rsw1 (mutant for CesA1) and the immunolabeling of V. angularis rosettes with antibodies raised against a CesA family member provide strong evidence for a role of these genes in the synthesis of cellulose. A recent study with virus-induced gene silencing in Nicotiana benthamiana also showed that the silencing of one or several members of the CesA family specifically led to cellulose deficiencies (Burton et al., 2000). The final proof will consist of the in vitro demonstration of cellulose synthesis activity for products of the CesA gene family. In the absence of an in vitro assay, the sequence similarity and the cellulose deficiency of prc1 mutants provide strong evidence that CesA6 encodes another cellulose synthase catalytic subunit.
Other Cell Wall Changes in prc1 Mutants
Not only did chemical analysis reveal a deficiency in cellulose, but also mutant walls showed a 50% increase in the hemicellulose-enriched fraction, the composition of which did not differ between wild-type and mutant plants. Only a slight decrease, if any, was observed for the pectin-enriched fraction. Interestingly, in prc1-1, this fraction showed a decreased galactose content without alterations in the content of uronic acid or other sugars. In primary walls, galactose is found mainly on galactan side chains of RG1 and on the side chains of arabinogalactan proteins. However, given the unaltered arabinose content, the simplest explanation is that the cellulose deficiency in prc1 caused compensatory changes in pectin composition, that is, a decrease in the number or length of the galactan side chains of RG1. We observed similar changes in cell walls of kor, another cellulose-deficient mutant (I. His, A. Driouch, F. Goubet, and H. Höfte, unpublished data). The molecular mechanisms underlying these secondary changes and their functional significance, if any, remain to be determined. Finally, the chemical analysis did not provide evidence for the accumulation of substantial amounts of alkali-extractable nonhemicellulosic glucan in prc1, whereas such accumulation was reported for rsw1.
An intriguing observation was the presence of gapped walls in endodermal, cortical, and epidermal cells of prc1 hypocotyls and roots. This is unlikely to be an artifact of the sample preparation, because the same defect was observed when using three different sample preparation procedures, including scanning electron microscopy, which does not involve tissue fixation. This defect is reminiscent of the stubs observed in cytokinesis mutants. Cortical and epidermal hypocotyl cells essentially have an embryonic origin, and no additional postembryonic divisions take place, except for those that contribute to the formation of stomatal guard cells. Because no gapped walls were observed in immature or mature embryos or in light-grown hypocotyls, the defect must arise during cell expansion. The simplest explanation would be that radial walls, the architecture of which is not compatible with wall expansion, rupture as a result of the increased radial expansion caused by the cellulose deficiency. That would be unlikely, however, because several other radial expansion mutants did not show gapped walls (Nicol et al., 1998; Bichet et al., 2001). An alternative explanation is that wall expansion, presumably mediated by wall-loosening proteins such as expansins, continues in the absence of sufficient cellulose synthesis to maintain a constant wall thickness. This eventually may lead to the rupture of the walls. Interestingly, the space between the wall stubs often contained wall material, composed of at least xyloglucan and pectin, as found with specific antibodies (data not shown). These results suggest that at least in mutant walls, impaired cellulose synthesis does not feed back to the wall-loosening machinery and the synthesis of other wall polysaccharides.
Functional Specialization within the Cellulose Synthase Gene Family
Large gene families are more the rule than the exception in plants, including Arabidopsis. Reverse genetics frequently has failed to associate clear-cut mutant phenotypes with mutations in individual genes within such families (Gilliland et al., 1998; D. Bouchez, personal communication). This suggests that many gene family members may have at least partially redundant roles. The CesA gene family is atypical in this respect, in that strong mutant phenotypes have been observed for at least three genes. Tissue-specific expression may explain specific phenotypes in some cases, for example, RSW1 in cells producing primary walls and IRX3 in xylem elements producing secondary walls. However, the conservation of specific amino acid sequence motifs, such as the HVR, between species suggests a functional specialization of different isoforms. We think it likely that the HVR domain interacts with other proteins with specific roles in different cell types or environmental conditions.
Another complication involves the observation that in some cell types at least two isoforms are required for normal cellulose synthesis. Indeed, rsw1-1 is affected in all cell types at the restrictive temperature, including in cells in the hypocotyl and root (Arioli et al., 1998). Cell expansion defects in all expanding cell types, including those in the central cylinder, were also observed in roots and hypocotyls of rsw1-10. The phenotype of rsw1-10 is less severe than rsw1-1 at the restrictive temperature (data not shown), which suggests that rsw1-10 is a leaky allele. Null alleles of PRC1 also showed expansion defects in endodermis, cortex, and epidermis of dark-grown hypocotyls and roots, indicating a requirement for both gene products to support normal cellulose synthesis and cell expansion in these cell types. The simplest explanation for this observation is a quantitative one: to sustain the demand for cellulose in the cell, a sufficiently large pool of CesA protein is required, which in turn requires synthesis from both genes. The strictly recessive phenotypes for both genes do not support a strong dependence on dose. An even stronger argument against this hypothesis is the absence of a gene dosage effect in a population segregating for the prc1-8 mutation in a genetic background homozygous for rsw1-10. The results suggest a strict requirement for two distinct isoforms rather than a certain pool size of CesA protein. This implies that different CesA isoforms need to be present at different times in the same cell or at different locations within the cell. Alternatively, CesA isoforms may act in concert or might form heterodimers. Immunolocalization studies with specific antibodies or with epitope-tagged versions of RSW1 (CesA1) and PRC1 (CesA6) should clarify this issue.
A final intriguing observation is the light-dependent conditional phenotype in mutant prc1 hypocotyls. We previously showed that the rescue of the hypocotyl phenotype is mediated by phytochrome (Desnos et al., 1996). The simplest explanation for this phenomenon is that phytochrome controls the expression of one or more other CesA isoforms, which either replace PRC1 (CesA6) in light-grown hypocotyls or have a function redundant with it. These observations indicate that phytochrome acts on the cellulose synthesis machinery and suggest a connection between phytochrome signaling and cell wall–related processes involved in growth control. RNA gel blot experiments thus far do not provide evidence for the regulation of PRC1 mRNA quantities by light. We are currently investigating the control by phytochrome of the expression of other CesA family members.
METHODS
Plant Material and Growth Conditions
Wild-type Arabidopsis thaliana plants were of the Columbia (Col-0), Wassilewskija (Ws), or Landsberg erecta (Ler) ecotype. Alleles prc1-1 through prc1-4 have been described previously (Desnos et al., 1996) and were of the Col-0 ecotype; these lines were all backcrossed four times. Alleles prc1-5 through prc1-7, prc1- 9, and prc1-11 (T. Desnos, F. Nicol, and H. Höfte, unpublished data; P. Benfey and M.-T. Hauser, personal communication) were of the Col-0 ecotype; these lines were backcrossed two, two, one, two, and one time, respectively. Mutant rsw1-1 was of the Col-0 ecotype (Arioli et al., 1998). Mutants rsw1-10, prc1-8, prc1-10, prc1-12, and prc1-19 were isolated from T-DNA–mutagenized populations and were of the Ws ecotype.
For growth in vitro, plants were grown on Petri dishes containing Murashige and Skoog (1962) medium (Estelle and Somerville, 1987) without sucrose at 25°C. For dark growth, seeds were cold-treated for 48 hr and then exposed to fluorescent white light (200 μM.m−2.sec−1) for 2 hr to synchronize germination, and finally the plates were wrapped in three layers of aluminum foil. For culture in the light, seeds were cold-treated for 48 hr and then placed under fluorescent white light (200 μM.m−2.sec−1) in 16-hr-light/8-hr-dark cycles. The time of measurement or collection of tissue was counted as days after the beginning of the light treatment. For cellulose incorporation assays, 200 Arabidopsis seedlings were grown at 20°C in the dark for 3 days in 10 mL of liquid medium containing 0.5% glucose. Seeds were cold-treated for 48 hr and then exposed to fluorescent white light (200 μM.m−2.sec−1) for 5 hr to synchronize germination, after which the flasks were wrapped in three layers of aluminum foil.
Measurement of Hypocotyl and Root Lengths
The growth of the seedlings was arrested by adding an aqueous solution of 0.4% formaldehyde. Hypocotyls and roots were spread on agar plates, and their image was captured with a digital camera. Lengths were measured by using image analysis software (Optimas 5.2; IMASYS, Surennes, France), as described by Gendreau et al. (1997).
Cross-Sections
For light microscopy, seedlings were fixed in PBS buffer containing 4% paraformaldehyde and 0.2% glutaraldehyde and were embedded in historesin (Technovit 7100; Kulzer, Wehrheim, Germany), according to the manufacturer's instructions. Sections 3 μm thick were cut by using a Jung RM2055 microtome (Leica Instruments, Nussloch, Germany). The cross-sections were stained with a 0.005% aqueous solution of Calcofluor (fluorescent brightener 28; Sigma) for 2 min and visualized under UV light with a Microphot FXA microscope (Nikon, Champigny-sur-Marne, France).
For transmission electron microscopy, seedlings were fixed with a 2.5% glutaraldehyde solution in phosphate buffer and then stained with a 2% osmium tetroxide solution. Seedlings were then embedded in Epon resin (TAAB 812, TAAB Laboratories Equipment Ltd., Reading, UK). Ultrathin sections (100 nm thick) were cut with a Reichert (Leica Instruments) Ultracut E ultramicrotome and deposited on 300-mesh copper–palladium grids. Samples were stained a second time with a 5% uranyl acetate solution followed by a 0.5% lead citrate solution and visualized with an EM 420 transmission electron microscope (Philips, Eindhoven, The Netherlands) at 80-kV acceleration.
Fourier Transform Infrared Spectroscopy
The seedlings (10 for each condition) were pressed onto a barium fluoride window and rinsed with water to remove cytoplasmic debris. The samples were dried at 37°C for 20 min. Two spectra of each seedling were collected on a Bio-Rad FTS-40 FT spectrometer in the middle region of the hypocotyl, avoiding the central cylinder, in a 60- × 40-μm window. Because absorbance varies with sample thickness, all data sets were corrected for baseline and normalized for area before statistical methods were applied. Exploratory principal components analysis was performed with Win-Discrim software (E.K. Kemsley, Institute of Food Research, Norwich, UK). Reference infrared absorption spectra of cellulose and other β-glucans were obtained from the literature (Tsuboi, 1957; Liang and Marchessault, 1959; Séné et al., 1994).
Preparation of Cell Wall Material
The plants were incubated for 30 min in 90% ethanol at 65°C to inactivate enzymes. The tissues were then ground in a Tenbroeck glass Potter–Elvehjem homogenizer (Janke and Kunkel IKA Labortechnik, Staufen, Germany). The homogenate was centrifuged at 2300g for 15 min. The pellet was washed with ethanol (three or four times), methanol/chloroform (2:3 [v/v], overnight), and ethanol again. The remaining pellet, which contained the cell wall, was dried overnight at 80°C. The yield of cell walls was defined as the weight (in grams) of dry cell walls per 100 g of fresh plants.
Extraction of Polymers
Pectic and hemicellulosic polymers were solubilized successively from cell walls by treatments with 0.1 M KOH at 20°C (three times for 1 hr) and 4 M KOH at 20°C (three times for 1 hr), respectively. The remaining pellet was washed with water until the pH of the supernatant was equal to that of water. The KOH extracts were neutralized with HCl, dialyzed against water with a molecular weight cutoff of 1000, and lyophilized.
Sugar Composition
Neutral sugar composition was determined as described by Englyst and Cummings (1984). Inositol was added to samples as an internal reference. Uronic acid contents were determined as described by Thibault (1979).
Cellulose Incorporation Assays
Two hundred Arabidopsis seedlings, grown in liquid culture in the dark, as described above, were washed three times with 15 mL of glucose-free growth medium before being resuspended in 1 mL of growth media containing 14C-glucose (NEN Research, Boston, MA), 1.0 μCi.mL−1 . The seedlings were then incubated for 1 hr in the dark at 15°C in glass tubes. After treatment, the seedlings were washed three times with 6 mL of glucose-free growth medium and then extracted with 5 mL of boiling absolute ethanol for 20 min. This step was repeated three times. Next, seedlings were resuspended in 3 mL of chloroform/methanol (1:1 [v/v]), extracted for 20 min at 45°C, and finally resuspended in 3 mL of acetone for 15 min at room temperature with gentle shaking. The remaining material was resuspended in 500 μL of an acetic acid/nitric acid/water solution (8:1:2 [v/v/v]), as described by Updegraff (1969), for 1 hr in a boiling water bath. Acid-soluble material and acid-insoluble material were separated by vacuum filtration through glass microfiber filters (GF/A; 2.5 cm diameter; Whatman, Maidstone, UK), after which the filters were washed with 1.5 mL of water. The acid solution and water wash constitute the acid-soluble fraction. Finally, the filters were washed with 30 mL of water and 20 mL of ethanol, yielding the acid-insoluble fraction, which is essentially crystalline cellulose. The amount of label in each fraction was determined by scintillation counting in a Betamatic scintillation counter (Kontron Instruments, Montigny le Bretonneux, France) using Ultima-Flo AP (Packard, Groningen, The Netherlands) as the scintillation liquid. The percentage of label incorporation was expressed as 100 × the ratio between the amount of label in the cellulosic fraction and the amount in the acid-soluble plus cellulosic fractions.
Genetic Analysis and Map-Based Cloning
The prc1-1 mutant (ecotype Col-0; backcrossed three times to Col-0) was crossed to line KanR5 (ecotype C24), which bears a T-DNA (conferring resistance to kanamycin) inserted close to marker LFY3 (Van Lijsebettens et al., 1996) at the bottom of chromosome 5 (Lister and Dean, 1993). F2 seedlings were sown on Murashige and Skoog medium supplemented with 50 mM kanamycin and grown for 3 days in total darkness. Of these, 1632 seedlings with short hypocotyls were scored and cultured for 3 more days in white light. Individuals with the prc1 phenotype and resistant to kanamycin were considered recombinant between the prc1-1 mutation and the T-DNA insertion. These plants were transferred to the greenhouse, and genomic DNA was extracted from leaves and flower buds, as described previously (Bouchez and Camilleri, 1998).
To determine the position of the prc1-1 mutation, DNA of the recombinant plants was analyzed with restriction fragment length polymorphism marker 5H1-L; this marker was designed by isolating the left border of yeast artificial chromosome 5H1 using the vectorette technique (Riley et al., 1990) and looking for polymorphism between the Col-0 and C24 ecotypes by DNA gel blotting with different restriction enzymes. The MUB3.12 and MUB3.p2B cleaved amplified polymorphic sequence (CAPS) markers were designed by polymerase chain reaction amplification of introns from genes 12 and p2B of P1 clone MUB3 (GenBank accession number AB010076) and digestion of the polymerase chain reaction products, amplified from genomic DNA of Col-0 and C24 ecotypes, with pools of restriction enzymes (40 were tested in all). The MUB3.12 marker (specific oligonucleotides: forward, CGCCGACGGAGAAATGACCAAG; reverse, TCACCAACACGAATGCGAGTACGA) is polymorphic for an EcoRI restriction site that is present in ecotype Col-0 but absent in C24 and Ler, whereas the MUB3.p2B marker (specific oligonucleotides: forward, CTCTCCCAAAGTAATCGTGCTA; reverse, TACCAGATGAACAGGGCTAAGT) is polymorphic for an AccI site that is present in C24 but not in Col-0.
The prc1-1 mutant was also crossed to a wild-type plant of the Ler ecotype. F2 seedlings were grown in the dark on Murashige and Skoog medium, and individuals homozygous for the prc1-1 mutation were selected on the basis of their short hypocotyls. These plants were transferred to the greenhouse, and genomic DNA was extracted from leaves and flower buds (Edwards et al., 1991). Recombinant individuals were identified by using CAPS markers based on partial sequences of markers ATPK41B (Recombinant inbred marker; Lister and Dean, 1993) and ATHB-5 (GenBank accession number X67033). Specific oligonucleotides (forward, TGTGATCAGAAATGGATCG; reverse, CTTCCTGTGTTGCTCTCGTT) amplified an ∼700-bp fragment at the ATPK41B locus, which was found, by testing with 40 commonly used restriction enzymes, to be cleaved by HinfI in the Ler allele but not in the Col-0 allele. For optimal resolution of the polymorphism, the polymerase chain reaction fragments were also cut with the restriction enzyme DraI. Other specific oligonucleotides (forward, TAACCAGCTCATCAACCAAAC; reverse, AAGATGGAAACAGAGAATTGACTA) amplified an ∼850-bp fragment at the ATHB-5 locus, which is cleaved by the restriction enzyme AvaI in the Col-0 allele but not in the Ler allele. Recombinant inbred marker Annexin (ve028; Lister and Dean, 1993) was converted to a CAPS marker (specific oligonucleotides: forward, TCCACCAAGATGTTCACCAAGA; reverse, CGTTCCATGTCAACTTCAGTTC) by using the restriction enzyme TfiI. Finally, a restriction fragment length polymorphism marker (2F2-E12) was found on the proximal part of P1 clone MXK3, which is polymorphic for an HpaII restriction site between Ler and Col-0.
RNA Gel Blotting
RNAs were extracted as described previously (Brusslan and Tobin, 1992). RNA gel blotting on Hybond N membranes and labeling of the probes by random priming in the presence of 50 μCi of 32P-dCTP was performed according to the manufacturer's instructions (Amersham, Little Chalfont, UK). The membrane was hybridized overnight at 42°C in 10 mL of SDS-rich buffer, as described by Church and Gilbert (1984) with 50% formamide and 100 μg/mL salmon sperm DNA after overnight prehybridization. Washes were performed at 65°C in 2 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate) plus 0.1% SDS for 15 min and again for 15 min in 1 × SSC plus 0.1% SDS (once for the RSW1 probe and twice for the PRC1 probe).
The RSW1 probe was prepared with a 300-bp-long EcoRV-HincII fragment covering the N-terminal variable region, which is specific for each CesA (positions 322 to 622 from ATG). The PRC1 probe was prepared with a 379-bp-long BglII-HindIII fragment covering the same region (positions 237 to 616 from ATG). The specificity of the probes was tested on dot blots of cloned cDNAs corresponding to Arabidopsis cellulose synthase genes, in the same hybridization conditions as those used for the RNA gel blots. The hybridization signal of the RSW1 probe was at least 10-fold greater for RSW1 than for PRC1. The hybridization signal of the PRC1 probe was at least 10-fold greater for PRC1 than for the very similar sequence CesA5 and at least 100-fold greater than for RSW1 (data not shown).
Phylogenetic Analysis
Unrooted trees were built by using an alignment of a variable region corresponding to amino acids 690 and 743 in PRC1 obtained with the program ClustalW (http://www.infobiogen.fr/services/analyseq/cgi-bin/clustalw_in.pl). This alignment was used by the algorithm PROTPARS (also available at the above-mentioned website) to construct a consensus tree from 100 bootstrapped data sets.
Acknowledgments
We thank Brigitte Gelie, Jocelyne Kronenberger, Joël Talbotec, Sigolène Müller, Audrey Hantiu, and Olivier Grandjean for technical assistance; Estelle Aletti for providing the data described in Figure 7; Hervé Vaucheret for enthusiastic discussions and critical reading of the manuscript; Phillip Benfey and Marie-Therese Hauser for providing prc alleles; and Reginald Wilson for insightful advice regarding FTIR. We thank Vincent Chiang for kindly providing the AraxCelA cDNA clone. Jean-François Thibault and Marc Lahaye are thanked for thoughtful advice concerning sugar chemistry and for making their equipment available. The Arabidopsis Biological Resources Center and the Nottingham Stock Centre are thanked for providing seed and DNA stocks. This work was financed in part by Framework 4 Grant PHOTARCH (No. BIO4-CT97-2124) from the European Economic Community; Grant No. A.I.P. 00176 from the Groupement de Recherche, Génomes et Mutants d'Arabidopsis; and Grant No. ACC-SV 9501006 from the Ministère de la Recherche et de la Technologie. T. Desnos and M.F. were bursaries from the Ministère de la Recherche et de la Technologie.
References
- Amor, Y., Haigler, C.H., Johnson, S., Wainscott, M., and Delmer, D.P. (1995). A membrane-associated form of sucrose synthase and its potential role in synthesis of cellulose and callose in plants. Proc. Natl. Acad. Sci. USA 92 9353–9357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arioli, T., Peng, L., Betzner, A.S., Burn, J., Wittke, W., Herth, W., Camilleri, C., Hofte, H., Plazinski, J., Birch, R., Cork, A., Glover, J., Redmond, J., and Williamson, R.E. (1998). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720. [DOI] [PubMed] [Google Scholar]
- Assaad, F.F., Mayer, U., Wanner, G., and Jurgens, G. (1996). The KEULE gene is involved in cytokinesis in Arabidopsis. Mol. Gen. Genet. 253 267–277. [DOI] [PubMed] [Google Scholar]
- Bichet, A., Desnos, T., Turner, S., Grandjean, O., and Höfte, H. (2001). BOTERO1 is required for normal orientation of cortical microtubules and anisotropic cell expansion in Arabidopsis. Plant J., in press. [DOI] [PubMed]
- Bouchez, D., and Camilleri, C. (1998). High molecular weight DNA extraction from Arabidopsis. Methods Mol. Biol. 82 61–70. [DOI] [PubMed] [Google Scholar]
- Brusslan, J.A., and Tobin, E.M. (1992). Light-independent developmental regulation of CAB gene expression in Arabidopsis thaliana seedlings. Proc. Natl. Acad. Sci. USA 89 7791–7795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burton, R.A., Gibeat, D.M., Bacic, A., Findlay, K., Roberts, K., Hamilton, A., Baulcombe, D., and Fincher, G.B. (2000). Virus-induced silencing of a plant cellulose synthase gene. Plant Cell 12 691–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpita, N.C., and Gibeaut, D.M. (1993). Structural models of primary cell walls in flowering plants: Consistency of molecular structure with the physical properties of the walls during growth. Plant J. 3 1–30. [DOI] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer, D. (1999). Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 245–276. [DOI] [PubMed] [Google Scholar]
- Desnos, T. (1996). Etude Génétique et Physiologique de l'Elongation Cellulaire chez Arabidopsis thaliana. (Paris: Sciences de la Vie).
- Desnos, T., Orbovic, V., Bellini, C., Kronenberger, J., Caboche, M., Traas, J., and Höfte, H. (1996). Procuste1 mutants identify two distinct genetic pathways controlling hypocotyl cell elongation, respectively, in dark- and light-grown Arabidopsis seedlings. Development 122 683–693. [DOI] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emons, A. (1985). Role of particle rosettes and terminal globules in cellulose synthesis. In Biosynthesis and Biodegradation of Cellulose, C. Haigler and P. Weimer, eds (New York: Marcel Dekker), pp. 71–98.
- Englyst, H.N., and Cummings, J.H. (1984). Simplified method for the measurement of total non-starch polysaccharides by gas–liquid chromatography of constituent sugars as alditol acetates. Analyst 109 937–942. [DOI] [PubMed] [Google Scholar]
- Estelle, M.A., and Somerville, C. (1987). Auxin-resistant mutants of Arabidopsis thaliana with an altered morphology. Mol. Gen. Genet. 206 200–206. [Google Scholar]
- Fagard, M. (1999). Analyse Physiologique et Moléculaire du Mutant procuste1 d'Arabidopsis thaliana Affecté dans l'Elongation Cellulaire. (Paris: Sciences de la Vie).
- Gendreau, E., Traas, J., Desnos, T., Grandjean, O., Caboche, M., and Höfte, H. (1997). Cellular basis of hypocotyl growth in Arabidopsis thaliana. Plant Physiol. 114 295–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings, T.H., and Staehelin, L.A. (1991). Microtubule-mediated control of microfibril deposition: A re-examination of the hypothesis. In The Cytoskeletal Basis of Plant Growth and Form, C.W. Lloyd, ed (New York: Academic Press), pp. 85–99.
- Gilliland, L., McKinney, E., Asmussen, M., and Meagher, R. (1998). Detection of deleterious genotypes in multigenerational studies. I. Disruptions in individual actin genes. Genetics 2 717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green, P.B. (1994). Connecting gene and hormone action to form, pattern and organogenesis: Biophysical transductions. J. Exp. Bot. 45 1775–1788. [Google Scholar]
- Hauser, M.T., Morikami, A., and Benfey, P.N. (1995). Conditional root expansion mutants of Arabidopsis. Development 121 1237–1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagoe, Y., and Delmer, D. (1997). Pathways and genes involved in cellulose biosynthesis. Genet. Eng. 19 63–87. [DOI] [PubMed] [Google Scholar]
- Kemsley, E.K. (1998). Chemometric Methods for Classification Problems: Discriminant Analysis and Modelling of Spectroscopic Data (Chichester, UK: John Wiley & Sons), pp. 1–47.
- Kimura, S., Laosinchai, W., Itoh, T., Cui, X., Linder, R., and Brown, R. (1999). Immunogold labeling of rosette terminal cellulose-synthase complexes in the vascular plant Vigna angularis. Plant Cell 11 2075–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Liang, C.Y., and Marchessault, R.H. (1959). Infrared spectra of crystalline polysaccharides. II. Native celluloses in the region from 640 to 1700 cm−1. J. Polym. Sci. 31 269–278. [Google Scholar]
- Lister, C., and Dean, C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4 745–750. [DOI] [PubMed] [Google Scholar]
- Liu, C.-M., Johnson, S., and Wang, T.L. (1995. a). cyd, a mutant of pea that alters embryo morphology, is defective in cytokinesis. Dev. Genet. 16 321–331. [Google Scholar]
- Liu, Y.-G., Mitsukawa, N., Vazquez-Tello, A., and Whittier, R.F. (1995. b). Generation of a high-quality P1 library of Arabidopsis suitable for chromosome walking. Plant J. 7 351–358. [Google Scholar]
- Lukowitz, W., Mayer, U., and Jürgens, G. (1996). Cytokinesis in the Arabidopsis embryo involves the syntaxin-related KNOLLE gene product. Cell 84 61–71. [DOI] [PubMed] [Google Scholar]
- Maeda, H., and Ishida, N. (1967). Specificity of binding of hexopyranosyl polysaccharides with fluorescent brightener. J. Biochem. 62 276–278. [DOI] [PubMed] [Google Scholar]
- Matthysse, A.G., Thomas, D.L., and White, A.R. (1995. a). Mechanism of cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177 1076–1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthysse, A.G., White, S., and Lightfoot, R. (1995. b). Genes required for cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann, M.C., and Roberts, K. (1994). Changes in cell wall architecture during cell elongation. J. Exp. Bot. 45 1683–1691. [Google Scholar]
- McCann, M., Chen, L., Roberts, K., Kemsley, E.K., Séné, C., Carpita, N.C., Stacey, N.J., and Wilson, R.H. (1997). Infrared microspectroscopy: Sampling heterogeneity in plant cell wall composition and architecture. Physiol. Plant. 100 729–738. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nickle, T.C., and Meinke, D.W. (1998). A cytokinesis-defective mutant of Arabidopsis (cyt1) characterized by embryonic lethality, incomplete cell walls, and excessive callose accumulation. Plant J. 15 321–332. [DOI] [PubMed] [Google Scholar]
- Nicol, F., His, I., Jauneau, A., Vernhettes, S., Canut, H., and Höfte, H. (1998). A plasma membrane–bound putative endo-1,4-beta-d-glucanase is required for normal wall assembly and cell elongation in Arabidopsis. EMBO J. 17 5563–5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear, J.R., Kawagoe, Y., Schreckengost, W.E., Delmer, D.P., and Stalker, D.M. (1996). Higher plants contain homologs of the bacterial celA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng, L., Hocart, C.H., Redmond, J.W., and Williamson, R.E. (2000). Fractionation of carbohydrates in Arabidopsis root cell walls shows that three radial swelling loci are specifically involved in cellulose production. Planta 211 406–414. [DOI] [PubMed] [Google Scholar]
- Riley, J., Butler, R., Ogilvie, D., Finniear, R., Jenner, D., Powell, S., Anand, R., Smith, J.C., and Markham, A.F. (1990). A novel, rapid method for the isolation of terminal sequences from yeast artificial chromosome (YAC) clones. Nucleic Acids Res. 18 2887–2890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena, I.M., Lin, F.C., and Brown, R.M.J. (1990). Cloning and sequencing of the cellulose synthase catalytic subunit gene of Acetobacter xylinum. Plant Mol. Biol. 16 673–683. [DOI] [PubMed] [Google Scholar]
- Séné, C., McCann, M., Wilson, R.H., and Grinter, R. (1994). FT-Raman and FT-infrared spectroscopy: An investigation of five higher plant cell walls and their components. Plant Physiol. 106 1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone, B.A., Evans, N.A., Bonig, I., and Clarcke, A.E. (1984). The application of Sirofluor, a chemically defined fluorochrome from aniline blue, for the histochemical detection of callose. Protoplasma 122 191–195. [Google Scholar]
- Taylor, N., Scheible, W., Cutler, S., Somerville, C., and Turner, S. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault, J.-F. (1979). Automatisation du dosage des substances pectiques par la méthode du méta-hydroxyphenyl. Lebensm. Wiss. Technol. 12 247–251. [Google Scholar]
- Tsuboi, M. (1957). Infrared spectrum and crystal structure of cellulose. J. Polym. Sci. 25 159–171. [Google Scholar]
- Updegraff, D.M. (1969). Semi-micro determination of cellulose in biological materials. Anal. Biochem. 32 420–424. [DOI] [PubMed] [Google Scholar]
- Van Lijsebettens, M., Wang, X., Cnops, G., Boerjan, W., Desnos, T., Höfte, H., and Van Montagu, M. (1996). Transgenic Arabidopsis tester lines with dominant marker genes. Mol. Gen. Genet. 251 365–372. [DOI] [PubMed] [Google Scholar]
- Vian, B., and Reis, D. (1991). Relationship of cellulose and other cell wall components: Supramolecular organisation. In Biosynthesis and Biodegradation of Cellulose, C. Haigler and P. Weimer, eds (New York: Marcel Dekker), pp. 25–50.
- Wu, L., Joshi, C.P., and Chiang, V.L. (1998). AraxCelA, a new member of cellulose synthase gene family from Arabidopsis thaliana (accession no. AF 062485) (PGR98–114). Plant Physiol. 117 1125. [Google Scholar]
- Wymer, C., and Lloyd, C. (1996). Dynamic microtubules: Implications for cell wall patterns. Trends Plant Sci. 1 222–228. [Google Scholar]
- Zablackis, E., Huang, J., Müller, B., Darvill, A.G., and Albersheim, P. (1995). Characterization of the cell-wall polysaccharides of Arabidopsis thaliana leaves. Plant Physiol. 107 1129–1138. [DOI] [PMC free article] [PubMed] [Google Scholar]