Abstract
Orally administered Lacticaseibacillus rhamnosus CRL1505 enhances respiratory immunity, providing protection against respiratory viruses and Streptococcus pneumoniae. However, the capacity of the CRL1505 strain to improve respiratory immunity against Gram-negative bacterial infections has not been evaluated before. The aim of this work was to evaluate whether the Lcb. rhamnosus CRL1505 was able to beneficially regulate the respiratory innate immune response and enhance the resistance to hypermucoviscous KPC-2-producing Klebsiella pneumoniae of the sequence type 25 (ST25). BALB/c mice were treated with the CRL1505 strain via the oral route and then nasally challenged with K. pneumoniae ST25 strains LABACER 01 or LABACER 27. Bacterial cell counts, lung injuries and the respiratory and systemic innate immune responses were evaluated after the bacterial infection. The results showed that K. pneumoniae ST25 strains increased the levels of TNF-α, IL-1β, IL-6, IFN-γ, IL-17, KC and MPC-1 in the respiratory tract and blood, as well as the numbers of BAL neutrophils and macrophages. Mice treated with Lcb. rhamnosus CRL1505 had significantly lower K. pneumoniae counts in their lungs, as well as reduced levels of inflammatory cells, cytokines and chemokines in the respiratory tract and blood when compared to infected controls. Furthermore, higher levels of the regulatory cytokines IL-10 and IL-27 were found in the respiratory tract and blood of CRL1505-treated mice than controls. These results suggest that the ability of Lcb. rhamnosus CRL1505 to help with the control of detrimental inflammation in lungs during K. pneumoniae infection would be a key feature to improve the resistance to this pathogen. Although further mechanistic studies are necessary, Lcb. rhamnosus CRL1505 can be proposed as a candidate to improve patients’ protection against hypermucoviscous KPC-2-producing strains belonging to the ST25, which is endemic in the hospitals of our region.
Keywords: Lcb. rhamnosus CRL1505, K. pneumoniae, innate immunity, probiotics
1. Introduction
Hypervirulent Klebsiella pneumoniae has become one of the most relevant pathogens in the world. The alarming emergence of this pathogen is due to the wide dissemination of different variants with the increasing ability to resist several antibiotic treatments, including carbapenems. In fact, carbapenemase-producing K. pneumoniae strains have been associated with a wide range of hospital acquired-infections and are associated with high morbidity and mortality rates [1,2,3]. In recent years, an increase in the prevalence of hypermucoviscous carbapenem-resistant K. pneumoniae of the sequence type (ST) 25 was detected in patients hospitalized in Northwest Argentina. This hypermucoviscous phenotype has been associated with hypervirulent strains, leading this bacterium to cause different infectious diseases including liver abscesses, meningitis, endophthalmitis, prostate abscesses and respiratory infections [4,5]. In this sense, our previous studies have shown that K. pneumoniae carbapenemase-2 (KPC-2)-producing ST25 strains are endemic in hospitals in the province of Tucuman (Argentina) and are associated with respiratory and systemic infections [5,6,7]. Notably, hypermucoviscous KPC-2-producing K. pneumoniae ST25 strains are resistant to almost all available antibiotics, making the search for strategies to prevent their infections a high priority.
Probiotic microorganisms have been proved to improve the resistance to pathogens. Using in vitro approaches or mice models of multiresistant K. pneumoniae intestinal colonization, it was demonstrated that some Lactobacillus spp. strains diminished the pathogen colonization and biofilm formation [8,9,10,11,12]. The ability of lactobacilli to inhibit the colonization of K. pneumoniae was associated with organic acid production [10,11] and the secretion of proteins with antimicrobial activity [13,14]. In addition, few studies reported that the oral administration of probiotic microorganisms to mice significantly reduced the lung inflammatory damage induced by K. pneumoniae nasal challenge [15,16], demonstrating that immunomodulatory bacteria are an interesting alternative to enhance resistance to this pathogen.
Lacticaseibacillus rhamnosus CRL1505 has been widely studied in our laboratory because of its remarkable immunomodulatory properties. Our previous reports demonstrated that the nasal priming with Lcb. rhamnosus CRL1505 enhanced respiratory immunity, improving protection against respiratory syncytial virus (RSV), influenza virus (IFV) and Streptococcus pneumoniae [17,18]. We also demonstrated that orally administered CRL1505 strain is able to beneficially modulate innate and adaptive immune responses in the respiratory tract, allowing a reduction of the susceptibility to viral and Gram-positive bacterial infections [19,20,21,22]. However, the capacity of orally administered Lcb. rhamnosus CRL1505 to modulate respiratory immunity in the context of Gram-negative bacterial infections in general, and multiresistant pathogens in particular, has not been evaluated before. Thus, the aim of this work was to evaluate whether the immunomodulatory strain Lbc. rhamnosus CRL1505 was able to beneficially regulate the respiratory innate immune response and enhance the resistance to hypermucoviscous KPC-2-producing K. pneumoniae ST25.
2. Materials and Methods
2.1. Microorganisms
Lacticaseibacillus rhamnosus CRL1505 belong to the CERELA (Reference Center for Lactobacilli, CONICET, Tucuman, Argentina) Culture Collection. The CRL1505 strain was isolated from goat milk [17,18]. Lcb. rhamnosus was cultured for 18 h at 37 °C, the late log phase, in MRS broth (Man–Rogosa–Sharpe, Britannia). After growth, bacteria were harvested by centrifugation at 3000× g for 10 min, washed three times with sterile 0.01 M phosphate-buffered saline (PBS, pH 7.2) and suspended in sterile PBS.
LABACER 01 and LABACER 27, hypermucoviscous carbapenem resistant K. pneumoniae ST25 strains, were isolated at the “Angel Cruz Padilla” hospital (San Miguel de Tucuman, Tucuman, Argentina). ST25 K. pneumoniae strains were identified by MALDI-TOF (matrix-assisted laser desorption/ionization) (Microflex LT; Bruker Daltonik GmbH, Bremen, Germany) and kept in the Culture Collection of the Certified Bacteriology Laboratory (LABACER, National University of Tucuman, Tucuman, Argentina) [4,5,6].
2.2. Animals
Six-week-old male BALB/c mice were obtained from the closed colony maintained in the CERELA-CONICET (Tucuman, Argentina) Animal Facility. All procedures for handling animals were performed according with the guide for the care and use of laboratory animals and approved by the CERELA-CONICET Animal Care and Ethics Committee under the BIOT-CRL/19 protocol. Eighteen mice were divided into three groups: basal, K. pneumoniae LABACER 01 (Kp01) and K. pneumoniae LABACER 27 (Kp27). Each of these groups was further subdivided into two groups. Half were treated with Lcb. rhamnosus CRL1505 while the other half received no treatment. Thus, each subgroup contained 3 mice and the experiments were repeated three times (n = 9 for each parameter evaluated). During the experiments, mice were housed in plastic cages at room temperature and fed a conventional balanced diet ad libitum.
Lcb. rhamnosus CRL1505 was administered via the oral route in a dose of 108 cells per mouse per day during 5 consecutive days. The dose was selected as optimal for immunomodulation in previous studies [17,18]. On day 6, the administration of the CRL1505 strain was terminated and mice were challenged with K. pneumoniae strains, as described below. During the following 2 days post-infection mice did not receive the CRL1505 strain.
2.3. Respiratory Infection with K. pneumoniae LABACER 01 and LABACER 27
K. pneumoniae LABACER 01 or LABACER 27 were administered to lightly anesthetized mice dropwise, through the nostrils. For this purpose, 100 uL of sterile PBS containing 107 CFU of bacteria were used. Mice in the control group were administered only 100 µL from PBS. The infective dose selected for this work was determined previously [23,24].
2.4. K. pneumoniae Counts in Lungs and Blood
Bacterial cell counts in the lungs and blood were performed in mice sacrificed on day 2 post-infection. The lungs of animals were excised, weighed, and homogenized in 5 mL of sterile peptone water. Homogenates were appropriately diluted in BHI broth, plated on blood agar and cultured at 37 °C for 18 h. K. pneumoniae LABACER 01 or LABACER 27 colonies were counted, and results were expressed as log10 CFU per gram of lung. Hemocultures were performed similarly using 100 µL of blood, and results were expressed as log10 CFU per mL of blood.
2.5. Lung Damage
Broncho alveolar lavages (BAL) were used for the determination of albumin and lactate dehydrogenase (LDH) [17,20]. BAL samples were obtained according to the technique described previously [13]. The trachea was exposed and intubated with a catheter, then two sequential bronchoalveolar lavages were performed in each mouse by injecting sterile PBS. The recovered fluid was centrifuged for 10 min at 900× g and the fluid frozen at −70 °C for subsequent determinations. Albumin, as a measure to quantify the increased permeability of the alveolar–capillary barrier, was determined colorimetrically based on albumin binding to bromocresol green using a Wiener Lab albumin diagnostic kit. LDH activity, an indicator of cellular cytotoxicity, was determined by measuring the formation of the reduced form of NAD+ using Wiener’s reagents and procedures. Results were expressed as units per liter of BAL.
2.6. Leukocytes Counts
The total number of leukocytes and differential cell counts in BAL samples were determined as described previously [13,14]. A hemocytometer was used to determine the total number of BAL and blood leukocytes. Differential cell counts in BAL and blood were assessed by microscopically counting cells in smears stained with May–Grunwald–Giemsa.
2.7. Serum Cytokines and Bronchoalveolar Lavages
BAL samples were obtained as described above. Blood samples were obtained via cardiac puncture and collected in heparinized tubes [14,15]. Cytokines determinations were performed in serum and BAL samples using commercial ELISA kits: IFN-γ (Mouse IFN-gamma Quantikine ELISA Kit, sensitivity: 2 pg/mL), and IL-10 (Mouse IL-10 Quantikine ELISA Kit, sensitivity: 5.2 pg/mL) from R&D Systems (Minneapolis, MN, USA), TNF-α (Mouse TNF alpha ELISA Kit, sensitivity: 9.1 pg/mL), IL-27 (Mouse IL-27 p28/IL30 Quantikine ELISA Kit, sensitivity: 4.7 pg/mL), IL-17 (Mouse IL-17 Quantikine ELISA Kit, sensitivity 5 pg/mL), IL-6 (Mouse IL-6 Quantikine ELISA Kit, sensitivity: 1.8 pg/mL), IL-8 mouse homolog chemokine KC (Mouse CXCL1/KC DuoSet ELISA, sensitivity 2.3 pg/mL), IL-1β (Mouse IL-1 beta ELISA Kit (ab197742), sensitivity: 1 pg/mL) and MCP-1 (Mouse MCP1 ELISA Kit (ab208979), sensitivity: 0.487 pg/mL) from Abcam (Cambridge, UK).
2.8. Statistical Analysis
The experiments were performed in triplicate and the results were expressed as mean ± SD. Statistical analyses were performed using Prism 8.0 (GraphPad software). Comparisons among multiple groups across multiple time points were performed using a two-way ANOVA with Tukey’s multiple comparison post hoc test. Comparisons between two groups were performed using unpaired Student’s t-tests. Differences were considered significant at p < 0.05.
3. Results
3.1. Lcb. rhamnosus CRL1505 Improves Resistance to K. pneumoniae ST25 Infection
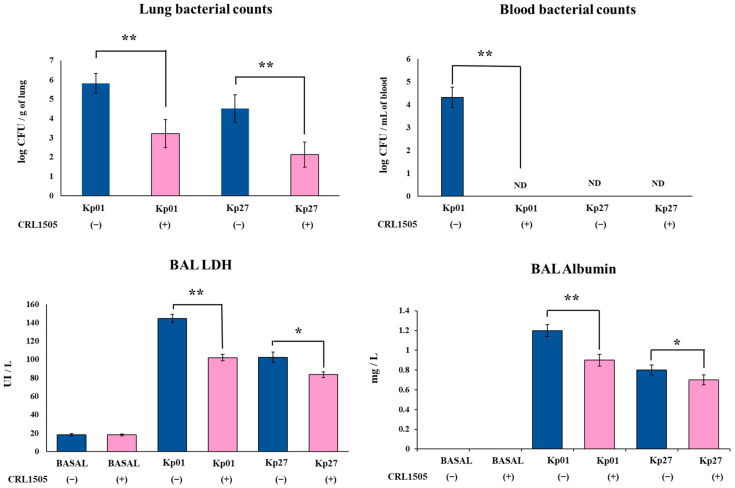
K. pneumoniae strains were able to colonize the respiratory tract of mice, with the LABACER 01 strain (5.8 ± 0.5 log CFU/g of lung) being more efficient than LABACER 27 strain (4.5 ± 0.6 log CFU/g of lung) (Figure 1). In addition, blood bacterial cultures showed that only K. pneumoniae LABACER 01 was able to spread from the lungs (4.3 ± 0.4 log CFU/mL of blood), since the blood cultures in the LABACER 27 group were negative (Figure 1). The oral treatment of mice with Lcb. rhamnosus CRL1505 before the challenge with the K. pneumoniae ST25 strains significantly reduced lung bacterial cell counts (3.2 ± 0.6 and 2.1 ± 0.6 log CFU/g of lung for LABACER 01 and LABACER 27, respectively) (Figure 1). The treatment with the CRL1505 strain was also capable of avoiding the dissemination of K. pneumoniae LABACER 01 into blood (Figure 1).
Figure 1.
Effect of Lacticaseibacillus rhamnosus CRL1505 on lung colonization and damage induced by KPC-2-producing hypermucoviscous Klebsiella pneumoniae ST25 strains. BALB/c mice (6 weeks old) were orally treated with the CRL1505 strain for 5 days and then challenged nasally with K. pneumoniae LABACER 01 (Kp01) or LABACER 27 (Kp27). Before (basal) and two days after challenge, bacteria cell counts in lung homogenates, broncho–alveolar lavages (BAL) lactate dehydrogenase (LDH) activity and albumin concentration were determined. Results represent data from three independent experiments. Asterisks indicate significant differences between the indicated groups, (*) p < 0.05, (**) p < 0.01. Basal levels of BAL albumin were below the detection limit. ND: not detected. Pink bars represent mice given Lcb. rhamnosus CRL1505 and blue bars represent control mice.
The biochemical markers albumin and LDH were studied in BAL samples to evaluate the lung damage induced by the KPC-2-producing hypermucoviscous K. pneumoniae ST25 strains. In normal conditions, albumin is not detected in BAL samples, hence the increase in this parameter indicates an enhanced permeability of the bronchoalveolar–capillarity barrier. In addition, the increase in the intracellular enzyme LDH in BAL samples is an indicator of cytotoxicity. As shown in Figure 1, both LABACER 01 and LABACER 27 strains significantly increased BAL albumin (1.21 ± 0.06 and 0.87 ± 0.05 mg/L, respectively) and LDH levels (144.5 ± 4.5 and 102.5 ± 5.6 UI/L, respectively) compared to non-infected controls. It was also observed that mice orally treated with Lcb. rhamnosus CRL1505 prior the infection with K. pneumoniae ST25 strains had significantly lower levels of BAL albumin (0.91 ± 0.06 and 0.71 ± 0.04 mg/L for LABACER 01 and LABACER 27, respectively) and LDH (102.1 ± 3.5 and 83.7 ± 3.1 UI/L for LABACER 01 and LABACER 27, respectively) than those found in their respective control groups (Figure 1).
3.2. Lcb. rhamnosus CRL1505 Modulates Innate Immunity against K. pneumoniae ST25 Infection
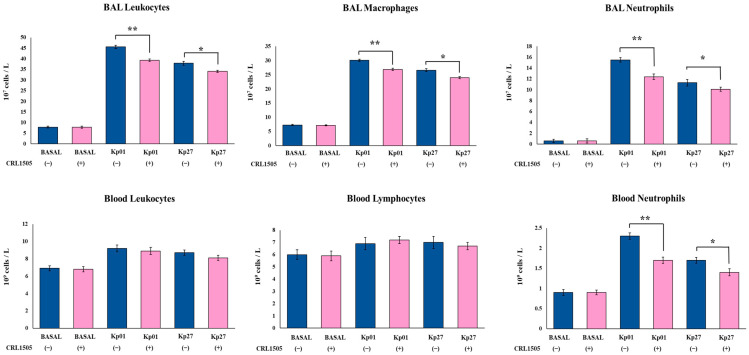
We further studied the influence of the CRL1505 strain on immune cells in both respiratory and systemic compartments (Figure 2). The oral treatment with Lcb. rhamnosus CRL1505 did not modify the numbers either of BAL or blood leucocytes in the steady state. The respiratory challenge with K. pneumoniae ST25 strains significantly increased the numbers of BAL leucocytes, macrophages and neutrophils in both control and lactobacilli-treated mice (Figure 2). However, mice treated with the CRL1505 strain showed statistically reduced levels of BAL leucocytes, macrophages, and neutrophils compared to controls. In fact, BAL leucocyte counts in control mice infected with LABACER 01 and LABACER 27 strains were 45.6 ± 0.8 and 37.9 ± 0.9 107 cells/L while in mice treated with the CRL1505 strain they were 39.3 ± 0.6 and 34.1 ± 0.5, respectively.
Figure 2.
Effect of Lacticaseibacillus rhamnosus CRL1505 on respiratory and blood leukocytes numbers induced by KPC-2-producing hypermucoviscous Klebsiella pneumoniae ST25 strains. BALB/c mice (6 weeks old) were orally treated with the CRL1505 strain for 5 days and then challenged nasally with K. pneumoniae LABACER 01 (Kp01) or LABACER 27 (Kp27). Before (basal) and two days after challenge, total and differential leukocytes counts were determined in broncho–alveolar lavages (BAL) and blood. Results represent data from three independent experiments. Asterisks indicate significant differences between the indicated groups, (*) p < 0.05, (**) p < 0.01. Pink bars represent mice given Lcb. rhamnosus CRL1505 and blue bars represent control mice.
Similarly, the respiratory infection with LABACER 01 and LABACER 27 enhanced the levels of blood leukocytes, neutrophils, and lymphocytes (Figure 2). No significant differences were found between control and CRL1505-treated mice after the challenge with K. pneumoniae LABACER 01 or LABACER 27 when blood leukocytes and lymphocytes were compared. In contrast, significantly lower numbers of blood neutrophils were observed in mice orally treated with Lcb. rhamnosus CRL1505 after infection with both LABACER 01 and LABACER 27 strains (Figure 2). Blood neutrophil counts in control mice infected with LABACER 01 and LABACER 27 strains were 2.31 ± 0.08 and 1.72 ± 0.07 107 cells/L, while in mice treated with the CRL1505 strain they were 1.71 ± 0.06 and 1.43 ± 0.06, respectively.
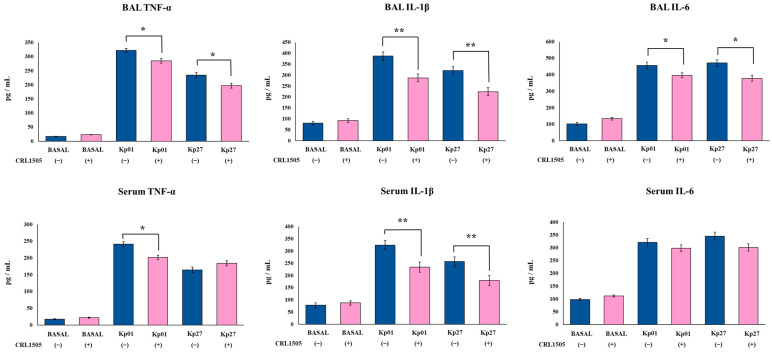
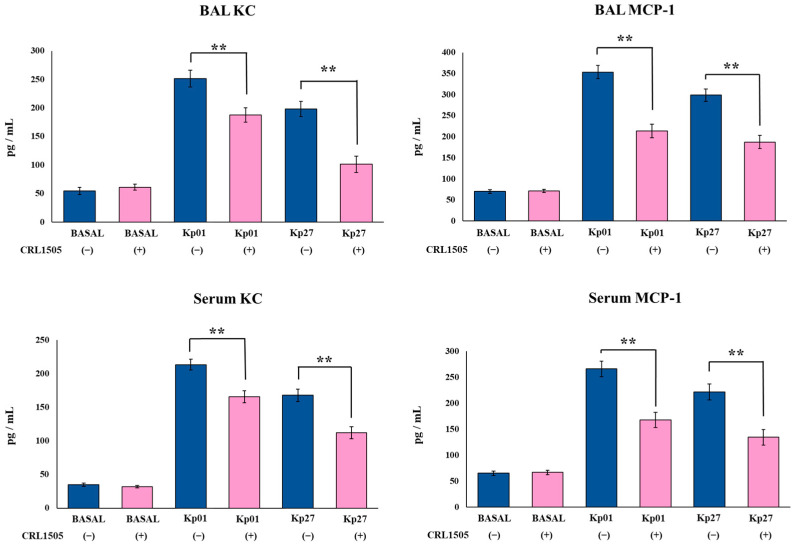
The levels of the inflammatory cytokines TNF-α, IL-1β, and IL-6, as well as the inflammatory chemokines KC and MCP-1, were determined in BAL and serum samples before and after the challenge with the K. pneumoniae ST25 strains. The oral treatment of mice with the CRL1505 strain did not induce changes in the basal levels of BAL and serum TNF-α, IL-1β, IL-6 (Figure 3), KC and MPC-1 (Figure 4). The nasal challenge with LABACER 01 or LABACER 27 increased the levels of all the inflammatory factors compared to basal levels, in both control and lactobacilli-treated mice. However, the levels of BAL TNF-α, IL-1β, IL-6 (Figure 3), KC and MPC-1 (Figure 4) were lower in mice orally treated with Lcb. rhamnosus CRL1505 than in their respective control groups. The most remarkable effects were observed for BAL IL-1β and MCP-1. The levels of IL-1β in control mice infected with LABACER 01 and LABACER 27 strains were 324.5 ± 18.1 and 256.8 ± 19.2 pg/mL while in mice treated with the CRL1505 strain they were 234.23 ± 21.2 and 179.4 ± 20.3, respectively. The concentrations of MCP-1 in LABACER 01 and LABACER 27 control groups were 353.4 ± 15.8 and 298.6 ± 14.5 pg/mL, while in Lcb. rhamnosus CRL1505-treated mice they were 213.4 ± 16.2 and 187.5 ± 15.9, respectively. Similarly, serum TNF-α, IL-1β (Figure 3), KC and MPC-1 (Figure 4) were diminished in lactobacilli-treated mice compared to controls. Notably, no significant differences were found in serum IL-6 levels between mice receiving Lcb. rhamnosus CRL1505 and controls (Figure 3).
Figure 3.
Effect of Lacticaseibacillus rhamnosus CRL1505 on respiratory and blood inflammatory cytokines induced by KPC-2-producing hypermucoviscous Klebsiella pneumoniae ST25 strains. BALB/c mice (6 weeks old) were orally treated with the CRL1505 strain for 5 days and then challenged nasally with K. pneumoniae LABACER 01 (Kp01) or LABACER 27 (Kp27). Before (basal) and two days after challenge, TNF-α, IL-1β and IL-6 levels were determined in broncho–alveolar lavages (BAL) and serum. Results represent data from three independent experiments. Asterisks indicate significant differences between the indicated groups, (*) p < 0.05, (**) p < 0.01. Pink bars represent mice given Lcb. rhamnosus CRL1505 and blue bars represent control mice.
Figure 4.
Effect of Lacticaseibacillus rhamnosus CRL1505 on respiratory and blood inflammatory chemokines induced by KPC-2-producing hypermucoviscous Klebsiella pneumoniae ST25 strains. BALB/c mice (6 weeks old) were orally treated with the CRL1505 strain for 5 days and then challenged nasally with K. pneumoniae LABACER 01 (Kp01) or LABACER 27 (Kp27). Before (basal) and two days after challenge, KC and MCP-1 levels were determined in broncho–alveolar lavages (BAL) and serum. Results represent data from three independent experiments. Asterisks indicate significant differences between the indicated groups, (**) p < 0.01. Pink bars represent mice given Lcb. rhamnosus CRL1505 and blue bars represent control mice.
3.3. Lcb. rhamnosus CRL1505 Modulates Effector and Regulatory Cytokines Produced against K. pneumoniae ST25 Infection
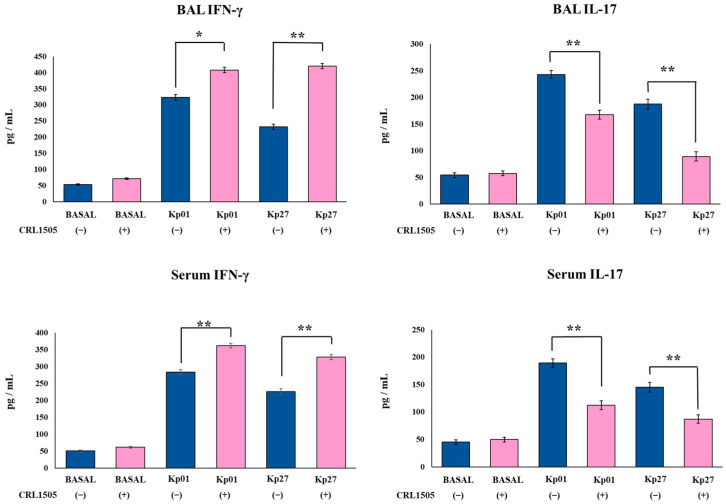
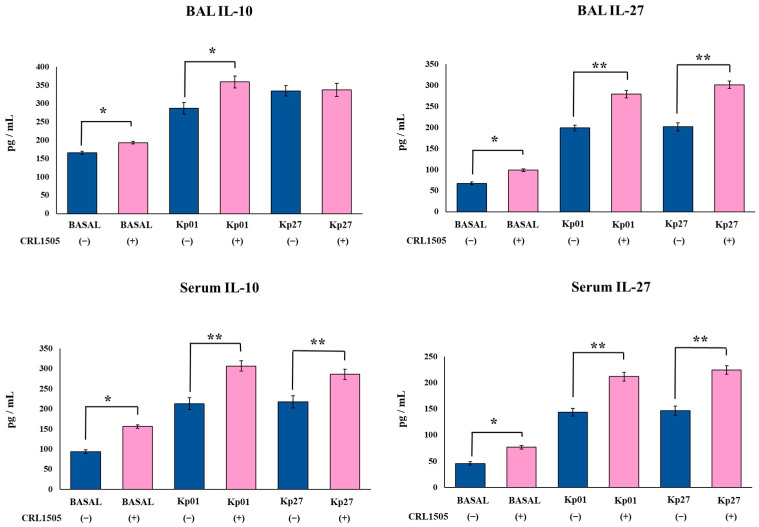
Finally, the levels of the effector cytokines IFN-γ and IL-17 as well as the regulatory cytokines IL-10 and IL-27 were evaluated in BAL and serum samples before and after the challenge with the LABACER strains. The oral treatment of mice with the CRL1505 strain did not induce changes in the basal levels of BAL and serum IFN-γ and IL-17 (Figure 5). However, the oral treatment of mice with Lcb. rhamnosus CRL1505 increased the levels of BAL and serum IL-10 and IL-27 in the steady state (Figure 6). The levels of BAL IL-27 and IL-10 in control mice were 67.4 ± 2.9 and 165.4 ± 4.2 pg/mL, respectively, while in mice treated with the CRL1505 strain they were 98.7 ± 3.1 and 193.3 ± 3.9. The nasal challenge with the K. pneumoniae ST25 strains significantly increased the levels of IFN-γ, IL-17, IL-10, and IL-27 in both the respiratory and systemic compartments in all the experimental groups. However, mice treated with the CRL1505 strain showed higher levels of BAL and serum IFN-γ (Figure 5), IL-10 and IL-27 (Figure 6) than controls. The levels of BAL IFN-γ in control mice infected with LABACER 01 and LABACER 27 strains were 323.9 ± 9.2 and 232.5 ± 8.3 pg/mL, while in mice treated with the CRL1505 strain they were 408.3 ± 8.6 and 420.3 ± 7.9, respectively. On the other hand, the concentrations of BAL IL-27 in LABACER 01 and LABACER 27 control groups were 198.6 ± 7.1 and 201.3 ± 9.2 pg/mL, while in Lcb. rhamnosus CRL1505-treated mice they were 278.9 ± 8.5 and 301.2 ± 8.7, respectively.
Figure 5.
Effect of Lacticaseibacillus rhamnosus CRL1505 on respiratory and blood effector cytokines chemokines induced by KPC-2-producing hypermucoviscous Klebsiella pneumoniae ST25 strains. BALB/c mice (6 weeks old) were orally treated with the CRL1505 strain for 5 days and then challenged nasally with K. pneumoniae LABACER 01 (Kp01) or LABACER 27 (Kp27). Before (basal) and two days after challenge, IFN-γ and IL-17 levels were determined in broncho–alveolar lavages (BAL) and serum. Results represent data from three independent experiments. Asterisks indicate significant differences between the indicated groups, (*) p < 0.05, (**) p < 0.01. Pink bars represent mice given Lcb. rhamnosus CRL1505 and blue bars represent control mice.
Figure 6.
Effect of Lacticaseibacillus rhamnosus CRL1505 on respiratory and blood regulatory cytokines chemokines induced by KPC-2-producing hypermucoviscous Klebsiella pneumoniae ST25 strains. BALB/c mice (6 weeks old) were orally treated with the CRL1505 strain for 5 days and then challenged nasally with K. pneumoniae LABACER 01 (Kp01) or LABACER 27 (Kp27). Before (basal) and two days after challenge, IL-10 and IL-27 levels were determined in broncho–alveolar lavages (BAL) and serum. Results represent data from three independent experiments. Asterisks indicate significant differences between the indicated groups, (*) p < 0.05, (**) p < 0.01. Pink bars represent mice given Lcb. rhamnosus CRL1505 and blue bars represent control mice.
In contrast, mice treated with Lcb. rhamnosus CRL1505 had significantly lower levels of BAL and serum IL-17 than their respective control groups (Figure 5). In fact, the LABACER 01 and LABACER 27 control groups have values of BAL IL-17 of 243.4 ± 7.1 and 187.6 ± 9.2 pg/mL while mice receiving the CRL1505 strain showed values of 167.8 ± 8.2 and 89.5 ± 8.8, respectively.
4. Discussion
K. pneumoniae is one of the most prevalent pathogens causing nosocomial pneumonia [25]. The interest in the study of respiratory infections caused by this bacterium has been renewed in recent years because K. pneumoniae was identified as a major cause of secondary bacterial pneumonia in patients with COVID-19 [26]. Furthermore, the increase in hypervirulent K. pneumoniae strains capable of causing community-acquired infections in otherwise healthy hosts [27] and the enhanced antibiotic resistance among clinical isolates [28] necessitate more in-depth studies of the pathogenesis and immune response, as well as preventive and therapeutic alternatives for these bacteria. Murine models of respiratory infections caused by classical and hypervirulent K. pneumoniae strains have been useful in gaining deeper knowledge of the immunobiology of these infections [29,30]. In this regard, we have used mice as a model to characterize respiratory infections caused by multiresistant K. pneumoniae isolates from the ST25 considering that among the KPC-2-producing strains, this sequence type has emerged as a persistent and overrepresented cause of hospital-associated infections in the Northwest of Argentina [5,6,7]. We demonstrated that K. pneumoniae ST25 strains were able to infect the respiratory tract of adult immunocompetent mice after the nasal challenge, inducing a potent innate immune response characterized by the increase in inflammatory cytokines and chemokines and the recruitment of inflammatory cells into the lung [23,24]. In fact, our previous and present results demonstrated that the nasal challenge of mice with the ST25 K. pneumoniae strains LABACER 01 and LABACER 27 increased the levels of TNF-α, IL-1β, IL-6, IFN-γ, IL-17, KC and MPC-1 in the respiratory tract and blood, as well as the numbers of BAL neutrophils and macrophages.
Inflammatory cytokines and chemokines have been shown to exert a protective role against respiratory K. pneumoniae infection. Deficiency or impairment of IFN-γ, IL-17 [31] and TNF-α [32] significantly reduce the resistance against K. pneumoniae infection. Recent transcriptomic studies performed in the lungs of mice infected with the clinical strain isolated from a patient with severe pneumonia, K. pneumoniae YBQ, demonstrated a remarkable activation of TNF and IL-17 signaling pathways and suggested that neutrophils and CCR2+ monocytes are the key to protection against the infection [33]. Similarly, a time course transcriptomic study of the lungs of mice infected with hypervirulent K. pneumoniae revealed a significant upregulation of inflammatory cytokines and chemokines genes [34]. Notably, the work suggested that the activation of TNF, IL-17, MAPK and NF-kB signaling pathways and the decrease in the expression of genes involved in the structural integrity of lung tissue may play key roles in the immunopathogenesis of K. pneumoniae infection. Then, acute inflammation in the lung is necessary to control K. pneumoniae infection but if it is not tightly regulated, it can cause damage to lung structures, impairing the lung’s normal functions.
Interestingly, studies performed in germ-free mice demonstrated that these animals had increased susceptibility to a lethal K. pneumoniae respiratory infection because of an impaired innate immune response [35]. This work highlighted the importance of the intestinal microbial population in regulating the respiratory immune response against this Gram-negative pathogen, as was previously described for Gram-positive [36] and viral [37,38] respiratory pathogens. Then, we aimed to evaluate whether the oral administration of an immunomodulatory probiotic strain could beneficially modulate the respiratory immunity and confer some degree of protection against LABACER 01 and LABACER 27 strains. Notably, we demonstrated here that the oral treatment of mice with Lcb. rhamnosus CRL1505 before the challenge with K. pneumoniae ST25 strains significantly improved the mice’s resistance to the pathogens. In our hands, CRL1505-treated mice had lower K. pneumoniae counts in lungs, decreased levels of lung injury markers, reduced levels of BAL macrophages and neutrophils and diminished concentrations of TNF-α, IL-1β, IL-6, IL-17, KC and MPC-1 in the respiratory tract and blood than infected controls. This is the first demonstration of the Lcb. rhamnosus CRL1505’s ability, when orally administered, to beneficially modulate the respiratory immune response against a Gram-negative bacterial pathogen. To the best of our knowledge, only two other studies have demonstrated the capacity of orally administered probiotics to beneficially modulate the immune response to K. pneumoniae respiratory infection. Experiments performed in adult C57BL/6 mice showed that the oral administration of Bifidobacterium longum 51A enhanced their resistance to the nasal challenge with K. pneumoniae ATCC 27,736 [16]. Animals treated with the 51A strain had improved survival and lower lung bacterial counts and injuries than infected controls. These benefits were associated with a differential regulation of the respiratory innate immune response, since B. longum 51A-treated mice had reduced levels of TNF-α, IL-6, CXCL1, and inflammatory cells in lungs [16]. Similarly, the oral administration of Lactiplantibacillus plantarum CIRM653 to C57BL/6 mice before the respiratory challenge with K. pneumoniae LM21 significantly enhanced the resistance to the pathogen by reducing BAL macrophages and neutrophils as well as the levels of BAL TNF-α, IL-6 and KC [15]. Moreover, in vitro studies showed that L. plantarum CIRM653 reduced the activation of the NF-κB pathway in airway epithelial cells induced by K. pneumoniae challenge, reducing the production of IL-8 and IL-6 [15].
In addition to the reduction in inflammatory cells and cytokines, orally administered Lcb. rhamnosus CRL1505 was able to increase the levels of the regulatory cytokines IL-10 and IL-27 in BAL and serum of the mice infected with LABACER 01 and LABACER 27 strains. In line with these results, we demonstrated that orally administered probiotics exert their beneficial effects against Gram-positive and viral respiratory pathogens by modulating the levels of the regulatory cytokines IL-10 and/or IL-27 [17,18,19,20,21,22]. It was also reported that the oral treatment of mice with B. longum 51A significantly increased the levels of BAL IL-10 during K. pneumoniae ATCC 27,736 infection [16]. Furthermore, it was shown that the respiratory challenge of mice with K. pneumoniae significantly increased the expression of T-bet in lungs, while no modifications were detected for ROR-γt or foxp3 [15]. In contrast, mice treated with L. plantarum CIRM653 before K. pneumoniae challenge had reduced T-bet and increased foxp3 and il10 expression in the lung tissue [15]. In agreement, it was reported that the CIRM653 strain increased the numbers of CD4+CD25+Foxp3+ cells in the mediastinal lymph nodes of K. pneumoniae-infected mice. These and our own results suggest that the ability of probiotic microorganisms to help control detrimental inflammation in lungs during K. pneumoniae infection would be a key feature to improve the resistance to this pathogen.
It was shown that IL-10 inhibits immunopathological consequences induced by respiratory pathogens, including K. pneumoniae, particularly during the resolution phase of inflammation [39]. Although the beneficial role of IL-27 in regulating inflammation in the lungs has been described for several respiratory pathogens [40], there is a lack of studies directly investigating the effect of this regulatory cytokine in the context of K. pneumoniae respiratory infection. Considering the improved levels of respiratory and serum IL-27 in CRL1505-treated mice and the lower susceptibility of these mice to the lung bacterial colonization and damage, detailed evaluation of the role of IL-27 in K. pneumoniae infection, as well as in the protective effect of Lcb. rhamnosus CRL1505, is an interesting topic for future research.
A range of recent studies, including from our group, have shown that some orally administered probiotic strains can exert beneficial effects in the immune responses of the respiratory tract and thus increase the protection against pathogens, particularly Gram-positive bacteria and viruses. In contrast, their potential beneficial effects against Gram-negative bacteria such as K. pneumoniae were less explored. In this work, we found a differential regulation of the respiratory and systemic immune responses triggered by the nasal infection with K. pneumoniae ST25 strains in mice preventively treated with Lcb. rhamnosus CRL1505 via the oral route. Although further analysis of immune factors and cells is required to clarify the mechanism involved in the beneficial effect of Lcb. rhamnosus CRL1505 during the infection with K. pneumoniae, this probiotic is an interesting candidate to improve protection of patients against hypermucoviscous KPC-2-producing strains belonging to the ST25, which is endemic in the hospitals of our region.
Author Contributions
Conceptualization, M.Á.J., H.K. and J.V.; methodology, S.D.M., Y.I., M.E., L.A. and K.N.; validation, Y.S. and M.Á.J.; formal analysis, S.D.M., Y.I., M.E., L.A., K.N. and S.K.; investigation S.D.M. and J.V.; resources, S.K., H.K. and J.V.; writing—original draft preparation, J.V.; writing—review and editing, H.K. and M.Á.J.; visualization, S.D.M. and L.A.; supervision, H.K. and J.V.; project administration, J.V.; funding acquisition, H.K. and J.V. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All the data related to this project are presented here.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This study was supported by a Grant-in-Aid for Scientific Research (A) (19H00965) from the Japan Society for the Promotion of Science (JSPS), and by the research program on development of innovative technology grants (JPJ007097) from the Project of the Bio-oriented Technology Research Advancement Institution (BRAIN), and by the Japan Racing Association (JRA) Livestock Industry Promotion Project (which supported Haruki Kitazawa). This study was also supported by ANPCyT–FONCyT Grant PICT-2016-0410 to Julio Villena, and by the JSPS Core-to-Core Program, A. Advanced Research Networks, entitled Establishment of international agricultural immunology research core for a quantum improvement in food safety. The study was also supported by the Tohoku University Research Program “Frontier Research in Duo” (FRiD), and by AMED (Moonshot R&D—MILLENNIA Program) Grant Number JP21zf0127001. Kohtaro Fukuyama was supported by JST, the establishment of university fellowships towards the creation of science technology innovation, Grant Number JPMJFS2102.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Gomez S.A., Pasteran F.G., Faccone D., Tijet N., Rapoport M., Lucero C., Lastovetska O., Albornoz E., Galas M., Melano R.G., et al. Clonal dissemination of Klebsiella pneumoniae ST258 harbouring KPC-2 in Argentina. Clin. Microbiol. Infect. 2011;17:1520–1524. doi: 10.1111/j.1469-0691.2011.03600.x. [DOI] [PubMed] [Google Scholar]
- 2.Cejas D., Fernandez Canigia L., Nastro M., Rodríguez C., Tanco A., Rodríguez H., Vay C., Maldonado I., Famiglietti A., Giovanakis M., et al. Hyperendemic clone of KPC producing Klebsiella pneumoniae ST 258 in Buenos Aires hospitals. Infect. Genet. Evol. 2012;12:499–501. doi: 10.1016/j.meegid.2011.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Cejas D., Elena A., Guevara Nuñez D., Sevilla Platero P., De Paulis A., Magariños F., Alfonso C., Berger M.A., Fernández-Canigia L., Gutkind G., et al. Changing epidemiology of KPC-producing Klebsiella pneumoniae in Argentina: Emergence of hypermucoviscous ST25 and high-risk clone ST307. J. Glob. Antimicrob. Resist. 2019;18:238–242. doi: 10.1016/j.jgar.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 4.Lin J., Huang Y., Qian L., Pan X., Song Y. Liver Abscess Combined with Endogenous Endophthalmitis Caused by Genotype ST25 Serotype K2 Hypervirulent Klebsiella pneumoniae: A Case Report. Infect. Drug Resist. 2022;15:4557–4561. doi: 10.2147/IDR.S376443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vargas J.M., Moreno Mochi M.P., Nuñez J.M., Cáceres M., Mochi S., del Campo Moreno R., Jure M.A. Virulence factors and clinical patterns of multiple-clone hypermucoviscous KPC-2 producing K. pneumoniae. Heliyon. 2019;5:e01829. doi: 10.1016/j.heliyon.2019.e01829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jure M.A., Castillo M.E., Musa H.E., López C.G., Cáceres M.A., Mochi S.D., Bousquet A.A., Genel N.A., Arlet G.A., Decré D.C. Novel patterns in the molecular epidemiology of KPC-producing Klebsiella pneumoniae in Tucumán, Argentina. J. Glob. Antimicrob. Resist. 2019;19:183–187. doi: 10.1016/j.jgar.2019.02.015. [DOI] [PubMed] [Google Scholar]
- 7.Jure M.A., Albarracin L., Vargas J.M., Maidana S.D., Zamar J.C., Kitazawa H., Villena J. Draft genome sequences of two hypermucoviscous carbapenem-resistant ST25 Klebsiella pneumoniae strains causing respiratory and systemic infections. J. Glob. Antimicrob. Resist. 2021;26:174–176. doi: 10.1016/j.jgar.2021.05.018. [DOI] [PubMed] [Google Scholar]
- 8.Villena J., Kitazawa H. The Modulation of Mucosal Antiviral Immunity by Immunobiotics: Could They Offer Any Benefit in the SARS-CoV-2 Pandemic? Front. Physiol. 2020;11:699. doi: 10.3389/fphys.2020.00699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Raras T.Y.M., Firdausy A.F., Kinanti I.R., Noorhamdani N. Anti-Biofilm Activity of Lactic Acid Bacteria Isolated from Kefir against Multidrug-Resistant Klebsiella pneumoniae. J. Pure Appl. Microbiol. 2019;13:983–992. doi: 10.22207/JPAM.13.2.35. [DOI] [Google Scholar]
- 10.El-Mokhtar M.A., Hassanein K.M., Ahmed A.S., Gad G.F.M., Amin M.M., Hassanein O.F.E. Antagonistic Activities of Cell-Free Supernatants of Lactobacilli Against Extended-Spectrum β-Lactamase Producing Klebsiella pneumoniae and Pseudomonas aeruginosa. Infect. Drug Resist. 2020;13:543–552. doi: 10.2147/IDR.S235603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yan R., Lu Y., Wu X., Yu P., Lan P., Wu X., Jiang Y., Li Q., Pi X., Liu W., et al. Anticolonization of Carbapenem-Resistant Klebsiella pneumoniae by Lactobacillus plantarum LP1812 through Accumulated Acetic Acid in Mice Intestinal. Front. Cell. Infect. Microbiol. 2021;11:1276. doi: 10.3389/fcimb.2021.804253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lagrafeuille R., Miquel S., Balestrino D., Vareille-Delarbre M., Chain F., Langella P., Forestier C. Opposing effect of Lactobacillus on in vitro Klebsiella pneumoniae in biofilm and in an in vivo intestinal colonisation model. Benef. Microbes. 2017;9:87–100. doi: 10.3920/BM2017.0002. [DOI] [PubMed] [Google Scholar]
- 13.Ermolenko E., Rybalchenko O., Borshev Y., Tarasova E., Kramskaya T., Leontieva G., Kotyleva M., Orlova O., Abdurasulova I., Suvorov A. Influence of monostrain and multistrain probiotics on immunity, intestinal ultrastructure and microbiota in experimental dysbiosis. Benef. Microbes. 2018;9:937–949. doi: 10.3920/BM2017.0117. [DOI] [PubMed] [Google Scholar]
- 14.Savinova O.S., Glazunova O.A., Moiseenko K.V., Begunova A.V., Rozhkova I.V., Fedorova T.V. Exoproteome analysis of antagonistic interactions between the probiotic bacteria Limosilactobacillus reuteri LR1 and Lacticaseibacillus rhamnosus F and multidrug resistant strain of klebsiella pneumonia. Int. J. Mol. Sci. 2021;22:10999. doi: 10.3390/ijms222010999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vareille-Delarbre M., Miquel S., Garcin S., Bertran T., Balestrino D., Evrard B., Forestier C. Immunomodulatory effects of Lactobacillus plantarum on inflammatory response induced by Klebsiella pneumoniae. Infect. Immun. 2019;87:e00570-19. doi: 10.1128/IAI.00570-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vieira A.T., Rocha V.M., Tavares L., Garcia C.C., Teixeira M.M., Oliveira S.C., Cassali G.D., Gamba C., Martins F.S., Nicoli J.R. Control of Klebsiella pneumoniae pulmonary infection and immunomodulation by oral treatment with the commensal probiotic Bifidobacterium longum 51A. Microbes Infect. 2016;18:180–189. doi: 10.1016/j.micinf.2015.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Raya Tonetti F., Clua P., Fukuyama K., Marcial G., Sacur J., Marranzino G., Tomokiyo M., Vizoso-Pinto G., Garcia-Cancino A., Kurata S., et al. The ability of postimmunobiotics from L. rhamnosus CRL1505 to Protect against respiratory syncytial virus and pneumococcal super-infection is a strain-dependent characteristic. Microorganisms. 2022;10:2185. doi: 10.3390/microorganisms10112185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kitazawa H., Villena J. Modulation of respiratory TLR3-anti-viral response by probiotic microorganisms: Lessons learned from Lactobacillus rhamnosus CRL1505. Front. Immunol. 2014;5:201. doi: 10.3389/fimmu.2014.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garcia-Castillo V., Tomokiyo M., Raya Tonetti F., Islam M.A., Takahashi H., Kitazawa H., Villena J. Alveolar Macrophages Are Key Players in the Modulation of the Respiratory Antiviral Immunity Induced by Orally Administered Lacticaseibacillus rhamnosus CRL1505. Front. Immunol. 2020;11:568636. doi: 10.3389/fimmu.2020.568636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Albarracin L., Garcia-Castillo V., Masumizu Y., Indo Y., Islam M.A., Suda Y., Garcia-Cancino A., Aso H., Takahashi H., Kitazawa H., et al. Efficient Selection of New Immunobiotic Strains With Antiviral Effects in Local and Distal Mucosal Sites by Using Porcine Intestinal Epitheliocytes. Front. Immunol. 2020;11:543. doi: 10.3389/fimmu.2020.00543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chiba E., Tomosada Y., Vizoso-Pinto M.G., Salva S., Takahashi T., Tsukida K., Kitazawa H., Alvarez S., Villena J. Immunobiotic Lactobacillus rhamnosus improves resistance of infant mice against respiratory syncytial virus infection. Int. Immunopharmacol. 2013;17:373–382. doi: 10.1016/j.intimp.2013.06.024. [DOI] [PubMed] [Google Scholar]
- 22.Salva S., Villena J., Alvarez S. Immunomodulatory activity of Lactobacillus rhamnosus strains isolated from goat milk: Impact on intestinal and respiratory infections. Int. J. Food Microbiol. 2010;141:82–89. doi: 10.1016/j.ijfoodmicro.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Dentice Maidana S., Ortiz Moyano R., Vargas J.M., Fukuyama K., Kurata S., Melnikov V., Jure M.Á., Kitazawa H., Villena J. Respiratory Commensal Bacteria Increase Protection against Hypermucoviscous Carbapenem-Resistant Klebsiella pneumoniae ST25 Infection. Pathogens. 2022;11:1063. doi: 10.3390/pathogens11091063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Albarracin L., Ortiz Moyano R., Vargas J.M., Andrade B.G.N., Zamar J.C., Dentice Maidana S., Fukuyama K., Kurata S., Jure M.Á., Kitazawa H., et al. Genomic and Immunological Characterization of Hypermucoviscous Carbapenem-Resistant Klebsiella pneumoniae ST25 Isolates from Northwest Argentina. Int. J. Mol. Sci. 2022;23:7361. doi: 10.3390/ijms23137361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lou W., Venkataraman S., Zhong G., Ding B., Tan J.P.K., Xu L., Fan W., Yang Y.Y. Antimicrobial polymers as therapeutics for treatment of multidrug-resistant Klebsiella pneumoniae lung infection. Acta Biomater. 2018;78:78–88. doi: 10.1016/j.actbio.2018.07.038. [DOI] [PubMed] [Google Scholar]
- 26.Zhu X., Ge Y., Wu T., Zhao K., Chen Y., Wu B., Zhu F., Zhu B., Cui L. Co-infection with respiratory pathogens among COVID-2019 cases. Virus Res. 2020;285:198005. doi: 10.1016/j.virusres.2020.198005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russo T.A., Marr C.M. Hypervirulent Klebsiella pneumoniae. Clin. Microbiol. Rev. 2019;32:e00001-19. doi: 10.1128/CMR.00001-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.CDCP . Antibiotic Resistance Threats in the United States, 2019. CDC; Atlanta, GA, USA: 2019. [DOI] [Google Scholar]
- 29.Wasbotten R.K., Dahler A.A., Mackel J.J., Smith C.M., Rosen D.A. Murine Respiratory Tract Infection with Classical Klebsiella pneumoniae Induces Bronchus-Associated Lymphoid Tissue. Infect. Immun. 2022;90:e00596-21. doi: 10.1128/iai.00596-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ahn D., Bhushan G., McConville T.H., Annavajhala M.K., Soni R.K., Wong Fok Lung T., Hofstaedter C.E., Shah S.S., Chong A.M., Castano V.G., et al. An acquired acyltransferase promotes Klebsiella pneumoniae ST258 respiratory infection. Cell Rep. 2021;35:109196. doi: 10.1016/j.celrep.2021.109196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Happel K.I., Dubin P.J., Zheng M., Ghilardi N., Lockhart C., Quinton L.J., Odden A.R., Shellito J.E., Bagby G.J., Nelson S., et al. Divergent roles of IL-23 and IL-12 in host defense against Klebsiella pneumoniae. J. Exp. Med. 2005;202:761–769. doi: 10.1084/jem.20050193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moore T.A., Lau H.Y., Cogen A.L., Standiford T.J. Defective innate antibacterial host responses during murine Klebsiella pneumoniae bacteremia: Tumor necrosis factor (TNF) receptor 1 deficiency versus therapy with anti-TNF-alpha. Clin. Infect. Dis. 2005;41((Suppl. 3)):S213–S217. doi: 10.1086/430126. [DOI] [PubMed] [Google Scholar]
- 33.Lei L., Zhang X., Yang R., Jing H., Yuan Y., Chen Z., Gou Q., Zhao Z., Zhang J., Wang X. Host Immune Response to Clinical Hypervirulent Klebsiella pneumoniae Pulmonary Infections via Transcriptome Analysis. J. Immunol. Res. 2022;2022:5336931. doi: 10.1155/2022/5336931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zheng X., Guo J., Cao C., Qin T., Zhao Y., Song X., Lv M., Hu L., Zhang L., Zhou D., et al. Time-Course Transcriptome Analysis of Lungs From Mice Infected With Hypervirulent Klebsiella pneumoniae via Aerosolized Intratracheal Inoculation. Front. Cell. Infect. Microbiol. 2022;12:386. doi: 10.3389/fcimb.2022.833080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fagundes C.T., Amaral F.A., Vieira A.T., Soares A.C., Pinho V., Nicoli J.R., Vieira L.Q., Teixeira M.M., Souza D.G. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J. Immunol. 2012;188:1411–1420. doi: 10.4049/jimmunol.1101682. [DOI] [PubMed] [Google Scholar]
- 36.Schuijt T.J., Lankelma J.M., Scicluna B.P., De Sousa E Melo F., Roelofs J.J.T.H., De Boer J.D., Hoogendijk A.J., De Beer R., De Vos A., Belzer C., et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. 2016;65:575–583. doi: 10.1136/gutjnl-2015-309728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ichinohe T., Pang I.K., Kumamoto Y., Peaper D.R., Ho J.H., Murray T.S., Iwasaki A. Microbiota regulates immune defense against respiratory tract influenza a virus infection. Proc. Natl. Acad. Sci. USA. 2011;108:5354–5359. doi: 10.1073/pnas.1019378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abt M.C., Osborne L.C., Monticelli L.A., Doering T.A., Alenghat T., Sonnenberg G.F., Paley M.A., Antenus M., Williams K.L., Erikson J., et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012;37:158–170. doi: 10.1016/j.immuni.2012.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peñaloza H.F., Noguera L.P., Ahn D., Vallejos O.P., Castellanos R.M., Vazquez Y., Salazar-Echegarai F.J., González L., Suazo I., Pardo-Roa C., et al. Interleukin-10 produced by Myeloid-Derived suppressor cells provides protection to Carbapenem-Resistant Klebsiella pneumoniae sequence type 258 by enhancing its clearance in the airways. Infect. Immun. 2019;87:e00665-18. doi: 10.1128/IAI.00665-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Branchett W.J., Lloyd C.M. Regulatory cytokine function in the respiratory tract. Mucosal Immunol. 2019;12:589–600. doi: 10.1038/s41385-019-0158-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data related to this project are presented here.