Abstract
Molybdenum (Mo) plays an essential role in the active site of all eukaryotic Mo-containing enzymes. In plants, Mo enzymes are important for nitrate assimilation, phytohormone synthesis, and purine catabolism. Mo is bound to a unique metal binding pterin (molybdopterin [MPT]), thereby forming the active Mo cofactor (Moco), which is highly conserved in eukaryotes, eubacteria, and archaebacteria. Here, we describe the function of the two-domain protein Cnx1 from Arabidopsis in the final step of Moco biosynthesis. Cnx1 is constitutively expressed in all organs and in plants grown on different nitrogen sources. Mo-repairable cnxA mutants from Nicotiana plumbaginifolia accumulate MPT and show altered Cnx1 expression. Transformation of cnxA mutants and the corresponding Arabidopsis chl-6 mutant with cnx1 cDNA resulted in functional reconstitution of their Moco deficiency. We also identified a point mutation in the Cnx1 E domain of Arabidopsis chl-6 that causes the molybdate-repairable phenotype. Recombinant Cnx1 protein is capable of synthesizing Moco. The G domain binds and activates MPT, whereas the E domain is essential for activating Mo. In addition, Cnx1 binds to the cytoskeleton in the same way that its mammalian homolog gephyrin does in neuronal cells, which suggests a hypothetical model for anchoring the Moco-synthetic machinery by Cnx1 in plant cells.
INTRODUCTION
Molybdenum (Mo) plays an important role as the active center in Mo-containing enzymes catalyzing redox reactions in the global C, N, and S cycles (Hille, 1996). Mo enzymes are essential for diverse metabolic processes such as nitrate assimilation and phytohormone synthesis in plants (Mendel and Schwarz, 1999) and sulfur detoxification and purine catabolism in mammals (Kisker et al., 1998). Except for the nitrogenase, in all the Mo enzymes studied thus far Mo is activated and chelated by a prosthetic group, the so-called molybdenum cofactor (Moco). Moco consists of Mo covalently bound to the unique metal binding molybdopterin (MPT; Rajagopalan and Johnson, 1992), which is highly conserved in eukaryotes, eubacteria, and archaebacteria. Biosynthesis of Moco requires the multiple-step synthesis of the MPT moiety, followed by the subsequent transfer of Mo (Rajagopalan and Johnson, 1992; Mendel, 1997).
Mutants in Moco biosynthesis are known in many organisms, among them the chl mutants in Arabidopsis (Braaksma and Feenstra, 1982; LaBrie et al., 1992) and the cnx mutants in Nicotiana species (summarized in Müller and Mendel, 1989). A mutational block in Moco biosynthesis leads to the combined loss of function of all four of the Mo enzymes known in plants: nitrate reductase (NR; Crawford and Arst, 1993; Campbell, 1999), aldehyde oxidase (Koshiba et al., 1996), xanthine dehydrogenase (Mendel and Müller, 1976), and sulfite oxidase (R.R. Mendel, unpublished result); consequently, affected plant cells can no longer assimilate inorganic nitrogen. However, when grown in culture containing sources of reduced nitrogen, these mutant plants remain alive but develop a peculiar phenotype (retarded growth and chlorotic, small, narrow, crinkled leaves), as has been described for Nicotiana tabacum and N. plumbaginifolia (Müller and Mendel, 1989). The morphology of these cnx mutants probably results from the impaired ability of the plants to synthesize abscisic acid because of the loss of the Moco-dependent aldehyde oxidase activities (Mendel and Schwarz, 1999). The Mo enzyme aldehyde oxidase also is involved in the synthesis of the phytohormone indoleacetic acid (Seo et al., 1998).
At least six gene products are necessary for the biosynthesis of Moco in plants (Hoff et al., 1995; Stallmeyer et al., 1995), humans (Reiss et al., 1998, 1999; Stallmeyer et al., 1999a, 1999b), and bacteria (Rajagopalan, 1996). A guanine derivative, the proposed starting compound, is converted by way of precursor Z (Wuebbens and Rajagopalan, 1993, 1995) to the pyranopterin MPT (Figure 1). The final step of cofactor maturation in eukaryotes, the incorporation of Mo into MPT, is catalyzed by the two-domain protein Cnx1 in plants (Stallmeyer et al., 1995) and by the neuroreceptor anchor protein (Prior et al., 1992) gephyrin in mammals (Stallmeyer et al., 1999b). Both domains are highly homologous with the separately expressed Escherichia coli proteins MoeA and MogA (Figure 2A). The functional integrity and homology of the G domains in Cnx1 and gephyrin were demonstrated by heterologous complementation of the E. coli mogA mutant (Stallmeyer et al., 1995, 1999b). A mutation in proteins catalyzing the last step of Moco synthesis leads to a molybdate-repairable phenotype (MacDonald and Cove, 1974; Tomsett and Garrett, 1980; Mendel et al., 1981; Stewart and MacGregor, 1982; Falciani et al., 1994). Both domains in Cnx1 are able to bind MPT, although with different affinities (Schwarz et al., 1997). Alignment of the amino acid sequences of proteins homologous with Cnx1 E and G domains derived from pro- and eukaryotic origins shows three distinct highly conserved amino acid motifs present in both domains (Figure 2B). For the G domain, mutations in amino acid residues located within or near these motifs completely abolish the function of the G domain (Kuper et al., 2000). The invariant residue 515 aspartate plays a role in the Mo insertion into MPT and is also invariant in all aligned E domains (Figure 2B). A hypothetical active site that is based on the atomic structure of E. coli MogA and is formed by residues of motif 1 and motif 2 has been proposed (Liu et al., 2000).
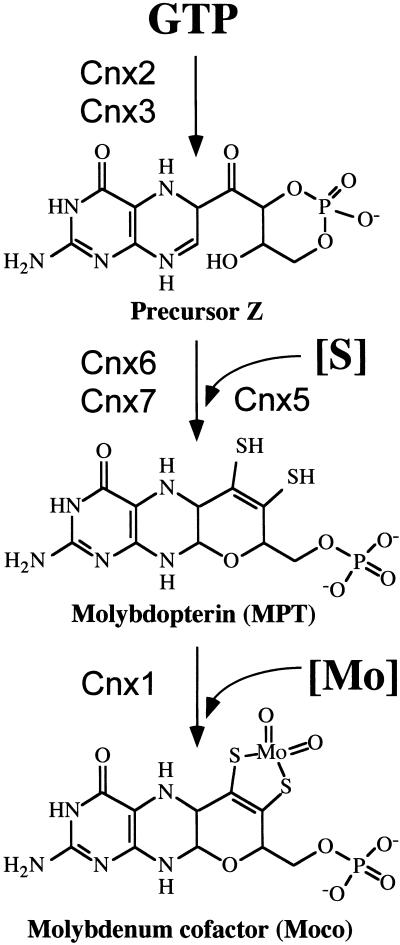
Figure 1.
Pathway of Moco Biosynthesis in Plants.
The proposed starting compound GTP, the intermediates precursor Z, and the metal binding MPT as well as the end product Moco are shown. Cnx2 and Cnx3 (Hoff et al., 1995) are involved in opening the pyrazine ring of guanine in a cyclohydrolase-like reaction (Wuebbens and Rajagopalan, 1995), forming the sulfur-free precursor Z (Wuebbens and Rajagopalan, 1993). In the second step, sulfur has to be introduced to form the dithiolene group. This step is catalyzed by MPT synthase, a complex of Cnx6 and Cnx7 (Mendel and Schwarz, 1999; R.R. Mendel, unpublished result) that is resulfurated by the sulfurase Cnx5 (Nieder et al., 1997). Finally, Mo is activated and incorporated into MPT by the activity of Cnx1 (Stallmeyer et al., 1995).
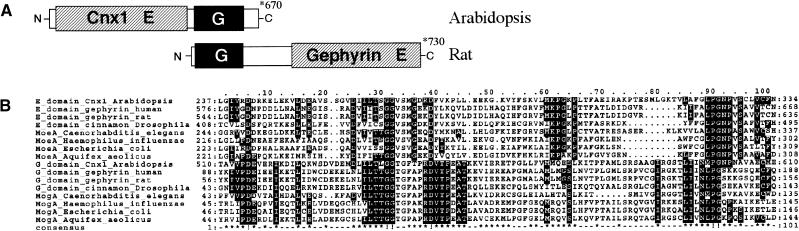
Figure 2.
Two-Domain Structure of Cnx1 and Gephyrin.
(A) Schematic representation of the two highly conserved domains in Cnx1 that are fused in an inversed orientation in gephyrin. The domains are linked by an interdomain region that is enlarged in gephyrin.
(B) Multiple sequence alignment of domains and proteins homologous with the G and E domains of Cnx1, performed with ClustalW (www2.ebi.ac.uk/clustalw/). The aligned protein sequences are marked with the starting and the ending amino acid in each row. Protein sequences and the organism from which they are derived are given; GenBank accession numbers are given in the order as listed for E domain homologs (Q39054, AF272663, Q03555, P39205, T20638, P45210, P12281, E70302) and G domain homologs (Q39054, AF272663, Q03555, P39205, T29649, P44645, P28694, and E703050). The consensus sequences have been calculated with a threshold of 50% (which highlights conservation within one domain). Dots were used to optimize alignment. Three motifs of homology between the E and G domains can be seen. (!), Completely conserved amino acids; (*), highly conserved residues in the consensus sequence; both are shown as white letters on a black background.
Here, we report the characterization of the Arabidopsis Cnx1 protein. We analyze its expression in wild-type and mutant plants, locate the mutation in the chl-6 mutant, demonstrate the functional reconstitution of molybdate-repairable mutants, and describe the functional characterization and biochemical dissection of the purified protein and its domains. Finally, we have found that Cnx1 binds to actin filaments the same way as its mammalian homolog gephyrin binds in neuronal cells to anchor neuroreceptors on the plasma membrane.
RESULTS
The Genomic Structure of Arabidopsis cnx1
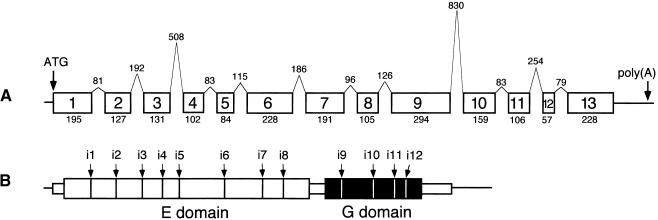
The genomic structure of Arabidopsis cnx1 is shown in Figure 3. The cnx1 cDNA is encoded by 13 exons covering 4646 bp of chromosomal DNA (GenBank accession number AJ236870). Interestingly, the two-domain structure of Cnx1 is not reflected by the organization of the exons. Whereas exons 1 to 8 and 10 to 13 encode for the E and G domain, respectively, exon 9 is actually the largest exon and encodes for the end of the E domain (50 amino acid residues), the interdomain region (22 amino acid residues), and the beginning of the G domain (22 amino acid residues; Figure 3C). In the 2685-bp region upstream of the transcription start, we have identified two putative TATA box elements (−20 and −35 bp), three CAAT boxes (between −50 and −150 bp), and two repeats (between −200 and −700 bp; data not shown). Within the 2-kb promoter sequence upstream of −700 bp, no open reading frame was found that was homologous with any expressed cDNA. Promoter–β-glucuronidase (GUS) fusions showed the presence of essential elements between −2685 and −700 bp (data not shown).
Figure 3.
Genomic Structure of the cnx1 Gene from Arabidopsis.
(A) The relative sizes of the exons (boxes) and introns (peaks) are shown; their lengths are indicated as numbers of base pairs.
(B) The domain structure of Cnx1, encoded by 13 exons, is shown on the protein level.
Expression of Cnx1 in Wild-Type Arabidopsis and the Moco-Deficient Mutant chl-6
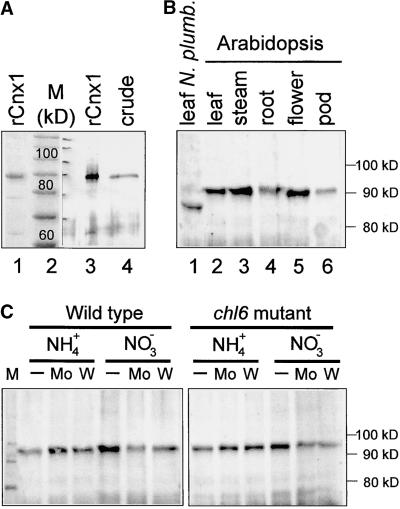
Cnx1 expression was analyzed by immunoblotting crude leaf extracts of wild-type Arabidopsis with a polyclonal antibody raised against recombinant Cnx1 (Figures 4A and 4B). A major post-translational modification of Cnx1 can be excluded because there was no detectable difference between the size of recombinantly expressed Cnx1 and the protein present in Arabidopsis crude extracts (Figure 4A). Cnx1 was expressed in all organs (Figure 4B) but slightly less in seedpods and roots than in other parts of the plant. Cnx1 expression also was assayed in the Arabidopsis chl-6 mutant (B73) because cnx1 had been mapped previously to the molybdate-repairable chl-6 locus of Arabidopsis (Stallmeyer et al., 1995). Expression of Cnx1 in the chl-6 mutant was essentially the same as in wild-type plants; moreover, neither expression was substantially altered when cultured on media supplemented either with additional molybdate or with tungstate as a molybdate antagonist (Figure 4C). Nitrate is known to induce the synthesis of the Mo enzyme NR. Growth of wild-type and chl-6 mutant plants on nitrate-containing medium did not markedly influence the expression Cnx1 in comparison with that in ammonium-grown plants (Figure 4C). Expression of cnx1 RNA was detectable only when the very sensitive RNase protection assay was used, thus indicating a weak expression of cnx1 (Figure 5A). Furthermore, analysis of plants transformed by a fusion of the cnx1 promoter with the reporter gene GUS showed GUS activity in all tissues, regardless of the nitrogen source (ammonia or nitrate) used for growing the plants. GUS activity, and hence cnx1 expression, was greatest in 2-week-old plants, thereafter decreasing with increasing age (data not shown). Together, these data suggest a constitutive expression of Cnx1.
Figure 4.
Expression of Cnx1 in Arabidopsis and N. plumbaginifolia.
(A) Expression of recombinant and endogenous Arabidopsis Cnx1. His-tagged Cnx1 (rCnx1) was separated on a 10% SDS–polyacrylamide gel and visualized by staining with Coomassie blue (lane 1, 2 μg) or by immunoblotting (lane 3, 0.1 μg). Fifty micrograms of crude protein extract from Arabidopsis plants was loaded in lane 4 (crude) and immunoblotted with polyclonal Cnx1 antibodies (Ak90cnx1; 1:2000 dilution). All other Cnx1 protein gel blots were performed under the same conditions. The discrepancy between the calculated molecular mass of Cnx1 (73 kD) and the observed migration in the gel (90 kD) reflects the greater mobility of the standard used (lane 2) and a lower SDS/protein ratio for denatured Cnx1.
(B) Cnx1 protein gel blot analysis of crude protein extracts (50 μg) from different Arabidopsis organs (lanes 2 to 6) and from a N. plumbaginifolia plant (lane 1).
(C) Cnx1 protein gel blot analysis of crude protein extracts from Arabidopsis wild-type and chl-6 mutant plants (B73) cultured on ammonium (NH4+)- or nitrate (NO3−)-containing culture medium either otherwise unsupplemented (−) or supplemented with 0.1 mM sodium molybdate (Mo) or 0.1 mM sodium tungstate (W).
Figure 5.
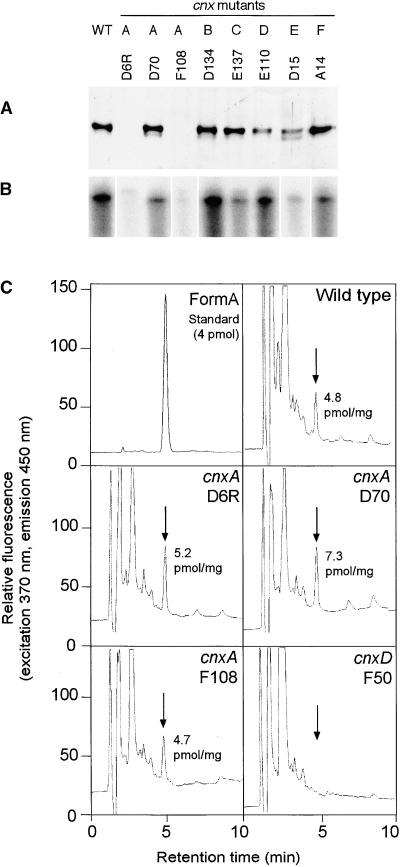
Biochemical Characterization of Moco-Deficient Mutants from N. plumbaginifolia.
(A) Expression of Cnx1 in Moco-deficient mutants cnxA to cnxF. Shown is protein gel blot analysis of crude protein extracts from callus cultures of wild-type (WT) and mutant lines cnxA to cnxF (A to F). Equivalent amounts of total protein (50 μg) were loaded onto a 7.5% SDS–polyacrylamide gel, immunoblotted, and detected with polyclonal Cnx1 antibodies (Ak90cnx1; 1:2000 dilution).
(B) RNase protection assay of total RNA preparations (50 μg) of wild-type and mutant lines cnxA to cnxF. A 0.5-kb fragment encoding for the Cnx1 G domain was used as labeled antisense RNA probe (106 cpm of 32P), hybridized with total RNA, digested, separated on a 5% acrylamide gel, and exposed to radiographic film.
(C) Analysis of MPT/Moco in N. plumbaginifolia wild-type and mutant lines of cnxA and cnxD. Crude protein extracts of N. plumbaginifolia callus were oxidized with I2/KI to generate FormA-phospho from Moco and MPT. After dephosphorylation, FormA was purified further on QAE–Sephadex columns and analyzed by C18 reversed-phase HPLC with fluorescence detection. Shown is the elution profile of 4 pmol of FormA standard prepared from bovine xanthine oxidase. Arrows indicate the elution time of FormA in each chromatogram. The amount of MPT (per milligram of total protein) in the extract is given in each chromatogram.
Expression of Cnx1 in N. plumbaginifolia Moco Mutants
For further analysis of Cnx1 expression and Moco biosynthesis, a few Arabidopsis Moco mutants derived from five Moco-specific loci are available (Braaksma and Feenstra, 1982; LaBrie et al., 1992). However, all these mutants are leaky and exhibit a relatively high background activity of NR (Braaksma and Feenstra, 1982; LaBrie et al., 1992), which hampers their detailed biochemical characterization. Therefore, we analyzed the expression of Cnx1 protein in the N. plumbaginifolia system, which includes nonleaky Moco mutants (Mendel et al., 1986; Gabard et al., 1988; Müller and Mendel, 1989; Marion-Poll et al., 1991). In crude protein extracts of N. plumbaginifolia wild-type plants and cell cultures, the Arabidopsis Cnx1 antiserum detected a single band migrating at 85 kD in SDS-PAGE gels, indicating a lower molecular mass for Cnx1 in this species than in Arabidopsis (Figure 4B, lane 1). The corresponding N. plumbaginifolia cnx1 cDNA recently cloned (J. Schulze, unpublished result) encodes for a protein of 668 amino acids with a molecular mass of 70.3 kD. In both Arabidopsis and N. plumbaginifolia, Cnx1 was detected in the cytoplasmic fraction without prior solubilization of organelle membranes (data not shown).
N. plumbaginifolia Moco mutants fall into six complementation groups (cnxA to cnxF). Only cnxA mutants are partially repairable by supplementing the growth medium with 1 mM molybdate (Marton et al., 1982; Gabard et al., 1988). Analysis of Cnx1 expression by immunoblotting crude cell extracts from the wild type, from three different cnxA mutants, and from one representative mutant of each of the loci for cnxB to cnxF showed that Cnx1 was expressed in the wild type as well as in cnxB to cnxF mutant lines (Figure 5A). Among cnxA mutants, only D70 showed unaffected Cnx1 expression, whereas the other two mutants (D6R and F108) failed to express detectable amounts of Cnx1 (Figure 5A). Similar expression patterns were observed for the RNAs: cnx1 mRNA was not detectable in the cnxA mutants D6R and F108 (Figure 5B).
Analysis of MPT in N. plumbaginifolia cnxA Mutants
Based on the molybdate-repairable phenotype of N. plumbaginifolia cnxA mutants, a defect in the transfer of Mo to MPT can be supposed (Mendel and Schwarz, 1999). Hence, such mutants should be able to synthesize the Moco precursor MPT, whereas plants with mutations in earlier steps of Moco biosynthesis should lack MPT. Therefore, we analyzed and quantified by HPLC FormA analysis (Schwarz et al., 1997) the MPT content in wild-type and cnxA mutants and in the cnxD F50 mutant as negative control (Figure 5C). In agreement with the assumption that MPT synthesis is an essential prerequisite for molybdate repairability, all three cnxA mutants contained amounts of MPT comparable to that of the wild-type plants (Figure 5C).
Complementation of Molybdate-Repairable Moco Mutants by cnx1 cDNA
Our data support the view that cnx1 is defective or absent (Figure 5A) in those mutants of N. plumbaginifolia and Arabidopsis that are molybdate repairable. To prove this assumption, we transformed N. plumbaginifolia cnxA mutants and the Arabidopsis chl-6 mutant with cnx1 cDNA to complement the mutant phenotype. For the cnxA mutants, full-length cnx1 cDNA was expressed under control of the cauliflower mosaic virus (CaMV) 35S promoter. Because of the very sick phenotype of cnx mutants that would not survive Agrobacterium-mediated transformation, we chose to use direct DNA transfer into protoplasts (Negrutiu et al., 1987) isolated from leaves of in vitro–grown mutant plants.
Three different cnxA mutants (D6R, D70, and F108) and one representative mutant of each of the loci cnxB to cnxF were transformed with cnx1 in several independent experiments (Table 1). Among all kanamycin-selected colonies, only cnxA mutants continued to grow on nitrate-containing media, because of their regained ability to assimilate inorganic nitrogen, as confirmed by the presence of NR activity. In contrast, all other mutants (cnxB to cnxF) failed to grow on nitrate-containing media and had no measurable NR enzyme activity (Table 1). In these experiments, we were able to restore NR activity in all three cnxA mutants (D6R, D70, and F108) by heterologous expression of Arabidopsis Cnx1. The successful incorporation of the transgene into the plant genome was confirmed by DNA gel blot analysis (data not shown) of 10 and six randomly chosen NR-positive colonies of D6R and D70, respectively. All plants regenerated from NR-positive clones regained their ability to grow on nitrate as the sole nitrogen source.
Table 1.
Transformation of pRT101cnx1 into N. plumbaginifolia Mutants cnxA-cnxF
| No. of Clones
|
|||||
|---|---|---|---|---|---|
| Locus | Mutant | Independent Transformation Experiments | Selected on Kanamycin | Tested for NR Activity | NR Positive |
| cnxA | D6R | 4 | 101 | 66 | 18 |
| D70 | 4 | 301 | 185 | 21 | |
| F108 | 2 | 27 | 23 | 2 | |
| cnxB | D134 | 3 | 110 | 31 | 0 |
| cnxC | E137 | 4 | 126 | 42 | 0 |
| cnxD | E110 | 3 | 61 | 26 | 0 |
| cnxE | D15 | 6 | 158 | 86 | 0 |
| cnxF | A14 | 1 | 35 | 21 | 0 |
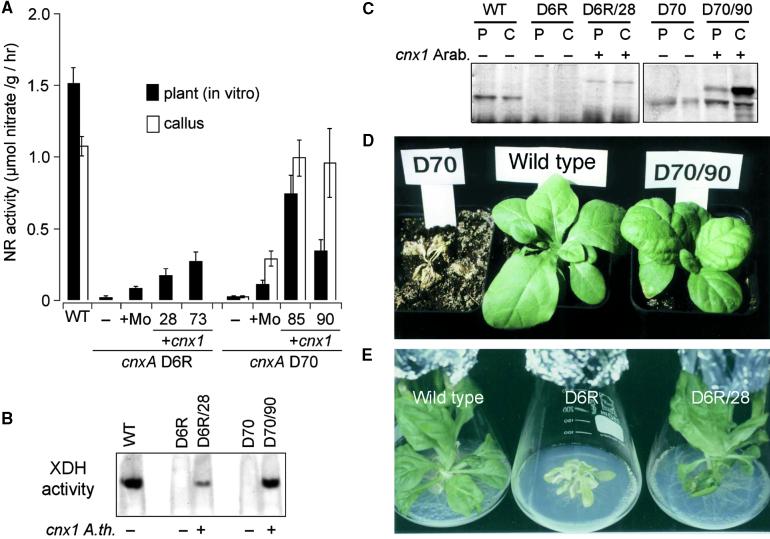
Two representative transformants chosen from each of the D6R and D70 lines displayed NR activities of 11.5 to 17.9% and of 22.7 to 49.1%, respectively, those of N. plumbaginifolia wild-type plants (Figure 6A); in contrast, untransformed cnxA mutants D70 and D6R had only 1.1 to 1.5% of the wild-type activity, respectively. NR activities after complementation by the cnx1 cDNA were noticeably higher than after molybdate repair of cnxA mutants, the latter ranging between 7.3 and 9.7% of wild-type activity. Molybdate repair is due to a noncatalyzed insertion of Mo. At callus stage, NR activities in some of the complemented lines were even greater than at the plant stage. In lines D70/85 and D70/90 (Figure 7A), these activities reached wild-type values (89.4 to 92.5%). The activity of another Moco-containing enzyme, xanthine dehydrogenase, also was restored after expression of Cnx1 (Figure 6B). Although no xanthine dehydrogenase activity was detectable in either cnxA mutant (D6R and D70), callus cultures expressing Arabidopsis Cnx1 exhibited xanthine dehydrogenase activity in the native gel, reflecting a complementation rate comparable to that determined for the amount of NR activity. Hence, expression of the cnx1 cDNA in molybdate-repairable Moco mutants of N. plumbaginifolia results in restored Moco biosynthesis that reverses the pleiotropic loss of Mo enzyme activities.
Figure 6.
Functional Complementation of Moco Biosynthesis in N. plumbaginifolia cnxA Mutants by Heterologous Expression of Arabidopsis cnx1 cDNA.
(A) NR activity of the wild-type (WT) and cnxA mutant plants (WT, D6R and D70; black bars) and callus cultures (WT and D70 [line D6R was not determined]; white bars). The mutants were either untransformed (−) or grown in the presence of 1 mM sodium molybdate in the growth media (+Mo) or transformed with Arabidopsis cnx1 cDNA (+cnx1). Two representative clones of transformed D6 (D6R/28 and D6R/73) and D70 (D70/85 and D70/90) were analyzed. Before harvesting, all plants and callus cultures were grown for 6 days on nitrate-containing media. The bars represent mean values of three to eight independent measurements with the standard errors shown.
(B) Xanthine dehydrogenase activity (XDH) of wild-type (WT, −) and mutant lines cnxA D6R (−) and D70 (−) as well as D6R and D70 complemented (+) with the Arabidopsis cnx1 cDNA (D6R/28 and D70/90). Crude protein extracts of callus cultures (25 μg per lane) were separated in a 5% discontinuous native gel electrophoresis system and stained for in situ enzyme activity according to Mendel and Müller (1976).
(C) Protein gel blot analysis of crude protein extracts shown in (B) that were derived from plants (P) and callus cultures (C). The same amounts of total protein (50 μg) were loaded onto a 7.5% SDS–polyacrylamide gel, blotted onto a polyvinylpolypyrrolidone membrane, and detected by using polyclonal Cnx1 antibodies (Ak90cnx1; 1:2000 dilution). Samples transformed with cnx1 are indicated (+).
(D) and (E) Normalization of the mutant phenotype. In (D), all plants shown were grown in sterile culture and transferred on the same day into soil fertilized with potassium nitrate as the nitrogen source. At left is the Moco-deficient cnxA mutant D70; it is unable to grow in soil and dies. At center is the wild-type N. plumbaginifolia. At right is the D70 transformant (D70/90) expressing Arabidopsis Cnx1. The wild-type–like phenotype of the D70/90 plant is representative of 21 cnx1-transformed and regenerated mutant plants. (E) In vitro culture of the wild-type N. plumbaginifolia (left), the cnxA D6R plant with small, crinkled leaves (middle), and a representative D6R transformant (D6R/28) expressing Arabidopsis Cnx1 (right).
Figure 7.
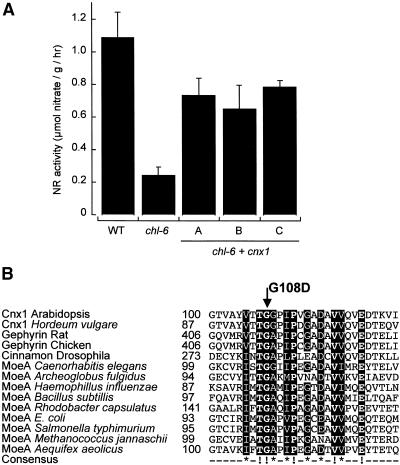
Functional Complementation and Molecular Characterization of the Arabidopsis chl-6 Mutant.
(A) NR activity of the wild-type (WT) and chl-6 mutant plants (B73). Shown are the untransformed mutant (chl-6) and three representative chl-6 plants (A to C) transformed with Arabidopsis cnx1 cDNA (chl-6 + cnx1) under control of the CaMV 35S core promoter (Benfey et al., 1989). Before harvest, plants were grown for 5 days on nitrate-containing media. The bars represent mean values of three to five independent measurements with the standard errors shown.
(B) Alignment of the mutation surrounding the Arabidopsis chl-6 mutation in cnx1. The mutation in the Arabidopsis chl-6 mutant reflects a point mutation at the cDNA position 361 (G → A), resulting in the substitution of aspartate for the glycine 108 (G108D) located in the E domain. Various E domain–homologous sequences were aligned by using ClustalW (www2.ebi.ac.uk/clustalw/); the region surrounding G108D is shown. The numbers shown indicate the first amino acid in each sequence. Protein sequences and the organism from which they are derived are given; GenBank accession numbers are given in the order listed: Q39054, AF268595, Q03555, AF174130, P39205, T20638, A69270, P45210, E69659, AJ23848, P12281, Q56066, Q58296 and E70302. The consensus sequence has been calculated with a threshold of 70%; completely conserved amino acids (boldface letters) are marked (!), and highly conserved residues show an asterisk in the consensus sequence. Both kinds of conserved regions are given in white letters over a black background.
Heterologous expression of Arabidopsis Cnx1 was demonstrated by protein gel blot analysis: Arabidopsis Cnx1 is easily distinguished from the endogenous N. plumbaginifolia Cnx1 protein by the difference in molecular weight (Figure 6C). D6R mutants showed no Cnx1 signals, whereas a single band of Arabidopsis Cnx1 could be seen in transgenic plants and callus cultures. D70 transformants showed a second band for a protein larger (greater molecular weight) than the endogenous Cnx1 protein.
Furthermore, the phenotype of the cnxA mutant plants, characterized by chlorotic, small, crinkled leaves (Gabard et al., 1988), was completely reversed to wild-type–like green plants by Cnx1 expression (Figures 6D and 6E). These experiments demonstrate that cnx1 is the defective gene in N. plumbaginifolia cnxA mutants.
In line with the above results, Arabidopsis chl-6 mutants also were complemented by the expression of cnx1 cDNA. Under control of the 35S core promoter mediating a moderate expression of Cnx1, 60 to 72% of wild-type NR activity was regained in the transformants. In contrast with N. plumbaginifolia cnxA mutants, chl-6 mutants exhibited a background (no complementation) NR activity of 22% of the wild-type expression (Figure 7A). The cnx1 cDNA was amplified by reverse transcription–polymerase chain reaction, subcloned, and sequenced to verify the mutation in the chl-6 mutant. We found the point mutation G→A at position 361 of the cnx1 cDNA (GenBank accession number L47323). The mutation leads to the substitution of an aspartate residue for glycine 108, that is, a negatively charged amino acid replacing a noncharged one (Figure 7B). Within the Cnx1 E domain and among other E domain–homologous proteins of pro- and eukaryotic origin, G108 belongs to the group of the most conserved residues. Furthermore, G108 is part of a motif of highly conserved residues located between residues 105 and 122 (Figure 7B) such that G108 presumably is essential for the function of the protein. Importantly, this motif of the E domain differs from the motifs (Figure 2) that are conserved between the E and G domains. Sequence data of the chl-6 mutant also show that the G domain of Cnx1 is not mutated. To verify the functionality of the G domain, we subcloned the chl-6 cnx1 cDNA into a bacterial expression vector and used it for complementation of E. coli mogA mutants that are defective in the bacterial homologue of the G domain. Expression of chl-6 cnx1 resulted in a complementation of E. coli mogA mutant RK5206 (data not shown), demonstrating that the G domain is active and is not affected by the mutation found in the E domain. Furthermore, we conclude that a mutation in the E domain of Cnx1 results in a molybdate-repairable phenotype, whereas in E. coli, only G domain–homologous mutants exhibit such a phenotype. In addition, the chl-6 mutation does not affect the expression of Cnx1 (cf. the protein gel blots shown in Figure 4C).
Binding and Activation of MPT by Cnx1 and Its G Domain in Vitro and in Vivo
Our results show that Cnx1-defective plant cells accumulate the final precursor MPT that cannot be further converted to active Moco. The molybdate-repairable phenotype of cnx1 mutants suggests a defect in processing or inserting the molybdate anion, the supposed source of Mo. Previously, we found that Cnx1 and both of its domains are able to bind MPT, although with different affinities (Schwarz et al., 1997). To understand how Cnx1 catalyzes the conversion of bound MPT into active Moco, we expressed holo-Cnx1 and its E and G domains separately in E. coli, purified them by affinity chromatography (data not shown), and tested them for the ability to bind MPT and convert it into active Moco. For this experiment, MPT was prepared as described in Methods and was coincubated with the protein to be tested. The binding mixture was separated by ultrafiltration (Schwarz et al., 1997) into protein-bound MPT, which was retained by the filter, and unbound MPT, which went through the filter into the salt fraction. Aliquots of both the retained and the flowthrough fraction were used for chemical quantification of MPT. Both fractions also were analyzed for biologically active Moco as follows: To distinguish between (1) bound but not processed MPT and (2) active Moco, we used the nit-1 reconstitution assay, which is based on the transfer of Moco to the NR apoprotein in crude extracts of the Moco-deficient Neurospora crassa nit-1 mutant and the detection of reconstituted NADPH-NR activity (Nason et al., 1971).
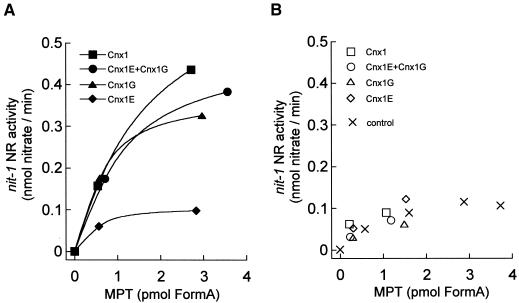
In the presence of molybdate, Cnx1-bound MPT showed markedly more NADPH-NR activity in the nit-1 assay than did unbound MPT or MPT not incubated with Cnx1 (Figures 8A and 8B). The same was found for the separated G domain as well as for an equimolar mixture of E domain and G domain. Interestingly, the E domain, although able to bind MPT, could not activate bound MPT the way that Cnx1 and the G domain did; E domain–bound MPT behaved the same as unbound (free) MPT. The results show a positive effect of Cnx1 and the G domain on the stabilization of Moco previously bound or formed in the binding mixture.
Figure 8.
In Vitro Binding of Moco and MPT to Cnx1 and Its Domains.
Shown is the nit-1 activity of MPT bound (A) to 100 nM Cnx1, G domain (Cnx1G), E domain (Cnx1E), or an equimolar mixture of both domains or of unbound MPT (B) in the presence of 5 mM sodium molybdate (50 μL reconstitution volume) (Cnx1E+Cnx1G). MPT was isolated from xanthine oxidase, and the binding mixture (500 μL) was separated by ultrafiltration (Schwarz et al., 1997). The protein-bound MPT was retained by the filter (∼250 μL), and the unbound MPT was in the flowthrough fraction (∼250 μL). Bound and unbound MPT were determined by FormA analysis, and aliquots of different dilutions were used for nit-1 reconstitution. The control consisted of free MPT not incubated with protein. NADPH-NR activities in the nit-1 assay were plotted against the amount of MPT in each sample.
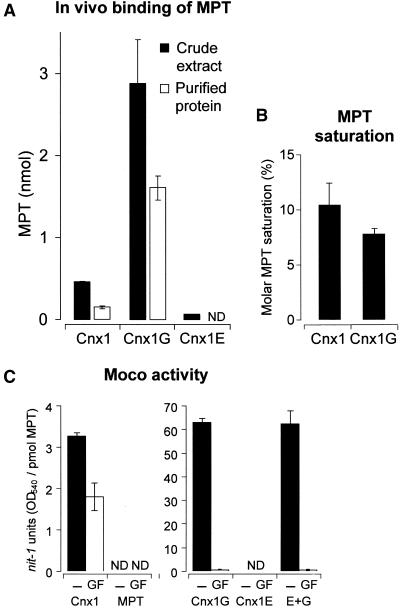
To determine to what extent Cnx1 and its domains can bind MPT under in vivo conditions, we investigated the process in E. coli mogA mutants. As are the cnx1 mutants, these bacterial mutants are defective in the last step of Moco biosynthesis; that is, they accumulate MPT. When we overexpressed Cnx1 in an E. coli mogA mutant, endogenous MPT was found to bind to Cnx1, and after purifying the overexpressed protein, the amount of protein-bound MPT could be detected and quantified (Figure 9A). After purification of Cnx1 and its domains from E. coli mogA cells, the molar saturations of Cnx1 (10.4%) and the separately expressed G domain (7.8%) with MPT were comparable (Figure 9B), whereas the E domain had no MPT bound to the protein after purification (Figure 9A). Obviously, the lesser affinity of the E domain to MPT (Kd = 1.6 μM) and the cooperative type of binding (Schwarz et al., 1997) are not sufficient for copurification of MPT from E. coli crude extracts.
Figure 9.
Cnx1 Generates Active Moco from Prebound MPT in the Absence of External Molybdate.
(A) Copurification of MPT with Cnx1 and its domains after recombinant expression in E. coli mogA RK5206 mutant. Cnx1 and the E and G domains were purified on small columns of nickel–trinitriloacetic acid matrix; minimal volumes of washing buffers were used to reduce dissociation of the bound MPT/Moco from the proteins. Shown are the total MPT content in crude extracts (black bars) and the total amount of MPT present in the purified protein fraction (white bars); the latter is called copurified MPT. Average values for MPT crude extracts are derived from triplicate purifications. The MPT values and standard deviations for copurified MPT were calculated from the percentage of MPT values for each purification in relation to the corresponding crude extract.
(B) Molar saturation of purified Cnx1 and G domains with copurified MPT. The data shown in (A) were correlated with the amount of purified protein (data not shown) and expressed as the percentage ratio of picomoles of MPT bound per picomole of protein.
(C) nit-1 reconstitutions of either the nit-1 crude extract (−) or the protein fraction of gel-filtrated nit-1 extract (GF) in the absence of external molybdate, as done with MPT bound to Cnx1, to G and E domains, or to an equimolar mixture of E and G domain (E+G), or with free MPT isolated from xanthine oxidase. The activity is given in nit-1-NR units per picomole of MPT. Standard errors were calculated from three different reconstitutions with at least three different MPT concentrations chosen from the linear range of the reconstitution assay.
ND, not detectable.
Catalyzing the Conversion of MPT to Moco
Cnx1 and its domains, after overexpression in E. coli mogA mutants and purification along with prebound MPT, were further analyzed in the nit-1 reconstitution assay, but this time in the absence of molybdate. Under these conditions, only active Moco but not MPT was detectable. Cnx1 and the G domain but not the E domain showed Moco activity. Cnx1 exhibited only 5% of the activity of the G domain (Figure 9C, black bars), whereas the total amounts of MPT bound to both proteins (Figure 9B) were comparable, indicating less dissociation of bound MPT from Cnx1 to nit-1 apo-NR. An equimolar addition of E domain to the G domain did not affect the activity of the G domain (Figure 9C, black bars).
To dissect whether Cnx1 and the G domain recruit a Mo species from the nit-1 crude extract, we treated the extract by gel filtration, which removes all of the low molecular weight compounds that might serve as a source of Mo. If NADPH-NR activity is detected under these conditions in the nit-1 assay, then all of the Mo inserted into MPT must have come from the protein to be tested; that is, Mo must have been already bound to the protein before the assay, and the protein inserts Mo into MPT during the experiment, generating active Moco. Alternatively, NADPH-NR activity in the gel-filtrated nit-1 extract could indicate that the protein to be tested already contains prebound Moco and then stabilizes it in a biologically active form. The results of this experiment showed that gel filtration of the nit-1 extract did not strongly affect the activity of Cnx1 (Figure 9C; cf. activities before [black bars] and after [white bars] gel filtration). However, the nit-1 reconstituting activity of the G domain decreased steeply, to only 1% of its prefiltration value. Moreover, adding an equimolar amount of E domain to the G domain did not increase the low activity of the G domain.
These results can be interpreted as follows: Holo-Cnx1 protein, copurified with MPT from E. coli mogA cells, is able to donate active Moco in a molybdate-independent way to the nit-1 apo-NR. Because no low molecular weight compound from the nit-1 crude extract was found to be essential to this outcome, we propose that the protein itself possesses the intrinsic activity for the synthesis of Moco. The G domain alone, however, is not able to catalyze this reaction; a low molecular weight Mo compound must be present in the nit-1 extract for the G domain to be able to activate prebound MPT.
Interaction of Cnx1 Protein with the Cytoskeleton
Given the homology of Cnx1 with the mammalian neuroreceptor anchor protein gephyrin (Stallmeyer et al., 1995), this latter protein was shown to possess a second function, namely, to catalyze the last step of Moco biosynthesis (Stallmeyer et al., 1999b). Because the association of gephyrin with the cytoskeleton had been demonstrated (Kirsch et al., 1993), we wondered whether Arabidopsis Cnx1 also could bind to the cytoskeleton, like gephyrin.
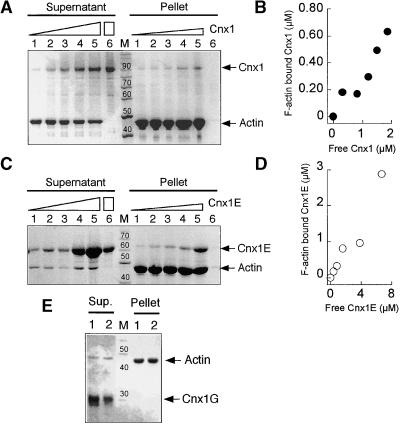
Recombinant Cnx1 was tested for its ability to bind actin filaments (F-actin) by using quantitative cosedimentation assays, which are based on the sedimentation of polymerized F-actin during ultracentrifugation. Proteins that interact with F-actin will cosediment and can be detected in the pellet by SDS-PAGE analysis. Proteins that do not interact will remain in the supernatant. Rabbit actin was used instead of plant actin because it is much easier to prepare in large amounts. Rabbit actin is 94% homologous with plant actin and has been used previously for analyzing plant proteins in this kind of binding assay (McLean et al., 1995; Calvert et al., 1996). Adding increasing amounts of Cnx1 (0.5 to 2.5 μM) to the binding mixture containing 4.1 μM actin resulted in a protein-dependent sedimentation of Cnx1 with F-actin, whereas Cnx1 alone did not sediment under control conditions (Figure 10A, lanes 6). After densitometric quantification of the protein bands in the SDS-PAGE gel, a kinetic analysis of the binding of Cnx1 to F-actin was performed (Figure 10B), yielding a Kd of ∼2 μM. Repeating the same experiment with constant amounts of Cnx1 and increasing amounts of actin resulted in the actin-dependent sedimentation of Cnx1, leaving decreasing amounts of free Cnx1 in the supernatant (data not shown).
Figure 10.
Binding of Cnx1 to Actin Filaments.
(A) Cosedimentation of actin filaments with increasing amounts of Cnx1. SDS-PAGE analysis of pellet and supernatant fractions from 4.1 μM actin and 0.5, 1.0, 1.5, 2.0, and 2.5 μM Cnx1 (lanes 1 to 5, respectively) and 2.5 μM Cnx1 without actin (lane 6). The concentration of sedimented F-actin, determined densitometrically, was 2.3 μM. Triangles denote increasing amounts of Cnx1, whereas the rectangle denotes the greatest amount of Cnx1 used in the control.
(B) Cnx1 binding to F-actin. The protein concentrations of bound (pellet) and unbound (supernatant) Cnx1 were determined by densitometric scanning of the gel shown in (A).
(C) Cosedimentation of actin filaments with increasing amounts of Cnx1 E domain. SDS-PAGE analysis of pellet and supernatant fractions of cosedimentation experiments with 4.2 μM actin and 0.6, 1.2, 2.4, 4.8, and 9.6 μM Cnx1 E domain (lanes 1 to 5, respectively) and 2.4 μM Cnx1 without actin (lane 6). The concentration of sedimented F-actin was determined by densitometry as 3.6 μM. Triangles denote increasing amounts of Cnx1E, whereas the rectangle denotes the greatest amount of Cnx1E used in the control.
(D) Binding of Cnx1 E domain to F-actin. The protein concentrations of bound (pellet) and unbound (supernatant) E domain were determined by densitometric scanning of the gel shown in (C).
(E) Cosedimentation of actin filaments with the Cnx1 G domain. SDS-PAGE analysis of pellet and supernatant fractions of cosedimentation experiments with 4 μM actin and 1 and 2 μM G domain (lanes 1 and 2, respectively).
The length markers in (A), (C), and (E) = 10 kD; the numbers above the bands indicate the molecular weight of the marker bands; M, protein standard for molecular weight.
To determine which domain of Cnx1 mediates the observed F-actin binding, recombinant E and G domains were used for binding assays. Although the G domain showed no sedimentation with F-actin (Figure 10E), the E domain (0.6 to 9.6 μM) cosedimented with actin filaments the same way as the entire Cnx1 protein did (Figure 10C), with an estimated Kd of 5 μM.
DISCUSSION
The structure of the metal binding pterin MPT is highly conserved in all organisms, which leads to the assumption that its biosynthetic pathway also might be conserved (Mendel et al., 1997). Among organisms of diverse phylogenetic origin, Moco mutants have been described that pleiotropically lack all Mo enzyme activities. These mutants have been classified into several complementation groups, and one of the mutant loci was shown to have a molybdate-repairable phenotype (MacDonald and Cove, 1974; Tomsett and Garrett, 1980; Mendel et al., 1981; Stewart and MacGregor, 1982; Falciani et al., 1994). Mutants in this locus were supposed to be defective in the final step of Moco biosynthesis, namely, the activation of molybdate and the incorporation of Mo into MPT to form the active Moco. Here, we have described the functional characterization of the Arabidopsis Cnx1 protein catalyzing the final step of Moco synthesis.
The primary structure of the Cnx1 protein shows two highly conserved domains. In E. coli, mogA mutants homologous with the Cnx1 G domain are molybdate repairable, whereas mutants in MoeA (homologous with the E domain) are not. Interestingly, we have shown that the loss of Cnx1 in the Arabidopsis chl-6 mutant results from a point mutation in the E domain, a mutation that also causes a molybdate-repairable phenotype, suggesting that both domains are essential for conversion of MPT to Moco. In Rhodobacter capsulatus, a moeA mutant was described that showed molybdate repair only for those enzymes that contained the MPT-based cofactor (such as that of eukaryotic enzymes), whereas activities of Mo enzymes with a dinucleotide-based cofactor, as occurs in eubacteria and archaebacteria, were not restored by molybdate (Leimkühler et al., 1999). Those findings and our results lead us to surmise that the function of the E domain might differ between eukaryotes and prokaryotes. Our suggestion is underlined by the observation that Arabidopsis Cnx1 (Stallmeyer et al., 1995) and its G domain can reconstitute Moco biosynthesis in E. coli mogA mutants, but E. coli moeA mutants cannot be reconstituted by Cnx1 (G. Schwarz, unpublished result).
Cnx1 expression was found in all organs of Arabidopsis plants. Whether the plants were grown on nitrate- or ammonium-containing medium, Cnx1 expression did not change markedly. The promoter–GUS fusions also underlined the ubiquitous housekeeping character of Cnx1 expression. During the analysis of Cnx1 expression in molybdate-repairable cnxA mutants of N. plumbaginifolia, we detected mutant lines with complete loss of Cnx1, thus pointing to a mutation in cnx1. These N. plumbaginifolia Moco mutants were generated by mutagenesis with nitroso-urea compounds (Gabard et al., 1988) known preferentially to cause point mutations. Therefore, the observed loss of Cnx1 expression in lines D6R and F108 could indicate a mutation in the promoter region or a mutation generating a stop truncation or a splicing error, whereas in mutant line D70, only single amino acid changes that do not affect overall protein expression should be responsible for the mutant phenotype. Furthermore, all cnxA mutants analyzed contained amounts of MPT that were similar to that in the wild type, making obvious the presence of a defect in the final step of Moco synthesis. Finally, the restoration of Moco biosynthesis in cnxA mutants after transformation with Arabidopsis cnx1 demonstrates that cnxA mutants are defective in Cnx1. In summary, our findings that a mutation in the E domain (chl-6 mutant) or the total loss of Cnx1 (cnxA mutant lines D6R and F108) is molybdate repairable let us conclude that both domains of Cnx1 are essential for catalyzing the insertion of Mo into MPT.
In the next step, we showed that Cnx1 is capable of converting MPT into active Moco by inserting Mo into MPT. Although in vitro binding of MPT to Cnx1 and its domains already had been demonstrated (Schwarz et al., 1997), the in vivo binding of MPT was observed only in the G domain and Cnx1. The molar saturations of both Cnx1 and G domain with MPT after copurification from E. coli mogA cells were similar. Therefore, we assume the G domain is the major site of MPT binding within Cnx1. Recently, we found that MPT bound to the G domain of Cnx1 can be converted to active Moco only by utilizing an unknown Mo compound from nit-1 crude extracts (Kuper et al., 2000). Interestingly, Cnx1 containing both domains exhibited a Moco synthetic activity independent from the low molecular weight compounds provided in the nit-1 extract. These findings suggest a role of the E domain in generating a Mo species that can be used by the neighboring G domain for converting MPT into Moco. When separately expressed E and G domains were coincubated, the activity of holo-Cnx1 was not regained, thereby suggesting the need of a proper (i.e., not just random) interaction between the E and G domains. Our data fit with the proposal of Hasona et al. (1998) that E. coli MoeA activates Mo by forming a thiomolybdate species. In eukaryotes, however, this mechanism of activation might be different because prokaryotic MoeA and the eukaryotic E domain are not functionally interchangeable (G. Schwarz, unpublished result).
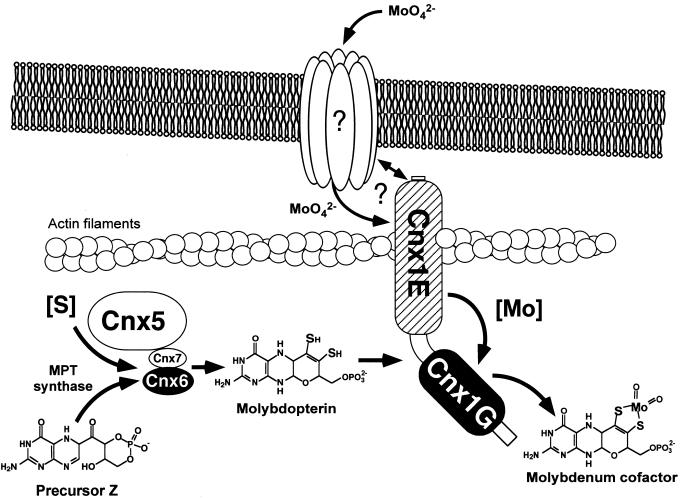
In view of the homology of Cnx1 with the neuroreceptor anchor protein gephyrin, we have shown that Cnx1 is capable of binding to rabbit actin. Gephyrin showed a direct interaction with microtubules (Kirsch et al., 1991), but both microtubules and actin filaments are important for the subcellular localization of gephyrin in cultured neurons (Kirsch and Betz, 1995). The binding of Cnx1 to actin filaments was mediated by the E domain, which corresponds to the microtubule binding C terminus of gephyrin. The hypothetical model shown in Figure 11 summarizes the functions of Cnx1. Our results suggest a function of Cnx1 similar to gephyrin in linking an integral membrane protein to the subcellular cytoskeleton. Such a protein could be an unknown molybdate transporter or an equivalent anion transporter that channels molybdate to Cnx1. Furthermore, we postulate a role for Cnx1 in anchoring a Moco-synthetic multienzyme complex of MPT synthase (Cnx6/7), sulfurase (Cnx5), and Cnx1 (Figure 11). A protein–protein interaction has been demonstrated recently for Cnx5 and Cnx6/7 (R.R. Mendel, unpublished results), but the interaction between Cnx1 and the other Cnx proteins has not been shown. Because MPT is highly sensitive to oxidation, it must be protected by a surrounding protein, which could result in a subcellular joining of the proteins that produce MPT with the protein that further converts MPT into Moco (i.e., Cnx1) and could lead to compartmentalization of Moco biosynthesis. In our speculative model, Cnx1 links MPT synthesis with molybdate uptake to facilitate the subsequent activation and incorporation of Mo into MPT.
Figure 11.
Proposed Model for the Function of Cnx1 in Moco Biosynthesis in Plant Cells.
Cnx1 is located under the plasmalemma and is bound to an actin filament. The interaction of Cnx1 with an integral membrane protein is based on the function described for the Cnx1-homologous animal protein gephyrin in neuroreceptor anchoring and needs to be verified. We propose an unidentified molybdate transport system that interacts with Cnx1 to facilitate substrate channeling to the E domain, given that a mutation in this part of the protein results in a molybdate-repairable phenotype. The conversion of precursor Z to MPT by the MPT synthase (Cnx6 and Cnx7) and the sulfurase (Cnx5) is shown. Because MPT is highly sensitive to oxidation, we suggest that the rapid conversion of precursor Z to Moco occurs in a multienzyme complex anchored by Cnx1 on the cytoskeleton.
METHODS
Materials
All chemicals used were the highest grade available. Xanthine oxidase (EC 1.1.3.22) from buttermilk grade I (0.69 units/mg) was obtained from Sigma (Deissenhofen, Germany). Nickel–nitrilotriacetic acid Superflow matrix was obtained from Qiagen (Hilden, Germany). Prepacked gel-filtration PD10 and Nick columns were used as recommended by the manufacturer (Amersham/Pharmacia, Freiburg, Germany).
Bacterial Strains and Growth Conditions
Escherichia coli strains were grown on Luria-Bertani medium, supplemented with 50 μg/mL kanamycin, 125 μg/mL ampicillin, or both, where appropriate. For subcloning E. coli and plant expression vectors, E. coli DH5α was used. The mogA mutant RK5206 (F− ara D139 Δ[argF-lac] U 169 deoC1 flb B5201 gyr A219 rel A1 rps150 non-9 pts F2 chlG206::Mu cts mogA; Stewart and MacGregor, 1982) was cultured at 30°C and used for recombinant expression of Cnx1 and its domains. Aerobic growth was achieved by vigorous shaking (>200 rpm) on an orbital shaker.
Identification and Sequence Analysis of the cnx1 Gene from Arabidopsis
Approximately 50,000 recombinant phages of the Arabidopsis genomic λ-GEM11 library (originally assembled by J.T. Mulligan and R.W. Davis and distributed by the German Arabidopsis Stock Center, Max Planck Institute, Cologne, Germany) have been grown in the bacterial host LE392 and blotted onto nitrocellulose filters (Pal Gelman Science, Ann Arbor, MI). Subsequent hybridization was performed under stringent conditions as described (Sambrook et al., 1989), with Arabidopsis cnx1 cDNA (GenBank accession number Q39054; Stallmeyer et al., 1995) as a probe. Twelve hybridizing plaques were identified, of which four were purified to homogeneity. Using DNA gel blot analysis, we identified 4.8- and 5.2-kb EcoRI fragments carrying the cnx1 gene. We subcloned these two EcoRI fragments from one clone (λ-GEM11-4) into pBSSKII+ and performed a restriction enzyme mapping. The SacI-SacI and SacI-XbaI fragments have been sequenced on both strands by subsequent subcloning and primer extension.
Plasmids
The plant expression vector pRT101cnx1 was generated by subcloning a 2.3-kb EcoRI fragment, derived from the construct pBSK+cnx1 (Stallmeyer et al., 1995) and containing the total coding region of the cnx1 cDNA clone, into the EcoRI-restricted pRT101 (Töpfer et al., 1988) polylinker. The expression vector pRT101core-cnx1 was generated by EcoRV and HincII restriction and subsequent religation of the plasmid to remove a 328-bp fragment from the cauliflower mosaic virus (CaMV) 35S promoter, generating a core promoter containing domain A (−90 to +8) of the CaMV 35S promoter (Benfey et al., 1989). For transformation of the Arabidopsis chl-6 mutants, the HindIII cassette of pRT101core-cnx1 was subcloned into pBIN19 (Bevan, 1984) and designated pBIN19cnx1. For expression analysis of the cnx1 promoter, the HindIII cassette of pRT103GUS (Töpfer et al., 1988) was subcloned into pBSK+ (pBSK+ β-glucuronidase [GUS]), followed by EcoRI and NcoI restriction of the resulting plasmid to remove the CaMV 35S promoter. The entire cnx1 promoter as well as a 5′ truncated cnx1 promoter were subcloned by polymerase chain reaction into the pBSK+GUS EcoRI-NcoI, resulting in pBSK+prom3GUS and pBSK+prom1GUS. Finally, the EcoRI-HindIII fragment of pBSK+GUS, pBSK+prom3GUS, and pBSK+prom1GUS were cloned into EcoRI-HindIII–digested pBIN19. For recombinant expression in E. coli, we used the previously described expression constructs of Cnx1 and Cnx1 E and G domains (pQE60cnx1, pQE60cnx1E, and pQE60cnx1G; Schwarz et al., 1997).
Sequence Analysis, RNase Protection Assay, Protein Gel Blot Analysis, and Recombinant Protein Expression
Sequence analysis was performed with the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit on an ABI Prism 310 cycle sequencer (PE Applied Biosystems, Warrington, UK) with a pop 6 polymer.
RNase protection assays were performed by using the Maxiscript in vitro transcription kit (Ambion, Austin, TX) and the RNase Protection Assay Kit (Ambion) according to the manufacturer's instructions. The antisense mRNA probes used in the protection assay correspond to 500 nucleotides of the cnx1 cDNA encoding for the G domain.
Protein expression analysis was done by gel-blotting crude protein extracts from whole-plant tissues or cell cultures (suspension or callus) of Arabidopsis and Nicotiana plumbaginifolia. Cells were ground in liquid nitrogen, immediately homogenized in two volumes of extraction buffer (100 mM Hepes, 10% glycerol, 1 mM EDTA, 1% polyvinylpolypyrrolidone, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, 1 mM benzamidine, 20 μM chymostatin, and 20 μM leupeptin, pH 6.8), and centrifugated, and the supernatant was subjected to 7.5% SDS-PAGE (Laemmli, 1970). After the separated proteins were blotted semidry (on polyvinylpolypyrrolidone membrane; Amersham/Pharmacia), Cnx1 was detected with a 1:2000 dilution of an affinity-purified polyclonal antibody (Ak90cnx1) generated against purified recombinant holo-Cnx1 protein (Eurogentec, Seraing, Belgium).
Plant Material and Plant Growth
Arabidopsis plants were grown in vitro on 0.5 × MS medium (Murashige and Skoog, 1962) in the presence of 10 mM KNO3 and 10 mM NH4NO3. N. plumbaginifolia plants were grown on MS medium containing 20 mM KNO3, 20 mM NH4NO3, and 12.5 mM ammonium succinate at 20°C under a 14-hr-light/10-hr-dark regimen. Callus and suspension cultures of N. plumbaginifolia were grown on MS medium containing 20 mM KCl and 12.5 mM ammonium succinate plus 0.9% agar where appropriate. For induction of nitrate reductase (NR), cultures previously grown on ammonium succinate–containing media were shifted to MS medium containing 20 mM KNO3 and 20 mM NH4NO3.
Stable Transformation and Regeneration of Plant Mutants
Plants that were maintained in vitro on solid medium, according to Müller (1983), containing 20 mM ammonium succinate and 9.5 mM KNO3 as the nitrogen source and vitamins according to Gamborg et al. (1968), at 23°C and 25 μmol of photons m−2 sec−1 under a 16-hr-light/8-hr-dark regimen, showed the following for the Moco-deficient phenotype: no typical rosette morphology as in the wild type; small, crinkled, chlorotic leaves; and decreased rooting ability. Mesophyll protoplasts were isolated from 2- to 3-month-old plants after overnight digestion of the leaves in 0.6% (w/v) Onozuka Cellulase R10 (Serva) and 0.2% (w/v) Macerocyme (Serva) dissolved in T0 (Crepy et al., 1982) but with Tween 80 omitted. Transformation was performed according to Negrutiu et al. (1987) by using plasmids pRT101cnx1 and pRT103neo (Töpfer et al., 1988) harboring the nptII gene for antibiotic selection. After transformation, protoplasts were cultured in the dark at 25°C at a density of 8 × 104 mL−1 in liquid T0 supplemented with 5 mM glutamine. After 2 weeks, protoplast-derived microcolonies were plated onto modified solid T8 medium (Crepy et al., 1982) containing macrosalts from the plant medium described above, but with 20 mM ammonium succinate, 220 mM mannitol, and 50 mg/L kanamycin. After 4 to 6 weeks, kanamycin-resistant colonies were transferred to the same medium but with mannitol omitted. Three weeks later, well-grown calli were subcultured on the same medium to which 1.1 μM 6-benzylaminopurine had been added for shoot development and were incubated in the light. After another 3 weeks, growth on nitrate was assayed by cutting calli into two halves and placing them on the same medium containing either ammonium succinate or nitrate as the sole source of nitrogen. We observed a 7% coexpression frequency of the nonlinked marker neomycin phosphotransferase II and the cnx1 cDNA. Plants from calli able to grow and to regenerate on the nitrate-containing medium were transferred and maintained on MS medium without growth regulators. After root development, these were planted in soil and grown in the phytotron.
Arabidopsis wild-type and chl-6 mutant plants were transformed by vacuum infiltration (Bechtold et al., 1993) of 3-week-old plants with Agrobacterium host strain C58C1 (pGV2260; Chilton and Chilton, 1984) carrying pBIN19 vectors.
Chemical Detection of Metal Binding Pterin
Metal binding pterin (MPT) was detected and quantified by converting it to the stable oxidation product FormA-dephospho, according to Johnson and Rajagopalan (1982). Oxidation, dephosphorylation, QAE chromatography, and HPLC analysis were performed as described in detail previously (Schwarz et al., 1997). FormA-dephospho was quantified by comparison with a standard isolated from xanthine oxidase for which the absorptivity was ɛ380 = 13,200 M−1 cm−1 (Johnson and Rajagopalan, 1982). Crude protein extracts were prepared by solubilization of 100 mg of ground cells in 500 μL of 100 mM Tris-HCl, pH 7.2, centrifugation, and subsequent oxidation of 400 μL of supernatant as described previously (Schwarz et al., 1997). MPT concentrations were expressed in picomoles of FormA per milligram of total protein.
Enzyme Assays
NR activity in N. plumbaginifolia plants was determined in crude extracts exactly as described by Scheible et al. (1997). Xanthine dehydrogenase activity was determined after electrophoretic separation in a nondenaturing polyacrylamide gel by an in situ assay as described by Mendel and Müller (1976). Histochemical localization and in vitro determination of GUS activity were performed according to Jefferson et al. (1987).
Purification of Recombinant Proteins and MPT Copurification
Large-scale purifications of Cnx1 and its E and G domains were performed by metal–chelate affinity chromatography after expression in E. coli strain M15 (Qiagen), according to Schwarz et al. (1997). MPT copurification with Cnx1 and its domains from E. coli mogA cells was performed as described (Kuper et al., 2000). Crude protein extracts from 250 mL of E. coli RK5206 cells were prepared and purified on 0.5 mL of nickel–nitrilotriacetic acid matrix (Qiagen). The eluted purified proteins were immediately desalted on PD10 columns and either used directly for further experiments or shock-frozen in liquid nitrogen and stored at −70°C. The purity of the proteins was checked in discontinuous SDS–polyacrylamide gels with a 12.5% separating gel. The concentrations of the purified proteins were determined by UV light absorption measurements and the calculated extinction coefficient for each protein (Schwarz et al., 1997). The amount of MPT bound to the purified proteins was determined by HPLC FormA analysis. The molar saturation of protein with bound MPT was calculated as picomoles of MPT per picomole of protein.
MPT Binding and nit-1 Reconstitution
MPT binding experiments were performed with protein-free MPT isolated from heat-treated xanthine oxidase by ultrafiltration as described previously (Schwarz et al., 1997). Freshly isolated MPT (120 to 140 nM) was coincubated with 100 nM protein in the presence of 5 mM sodium molybdate for 10 min. Protein-bound and unbound (free) MPT was separated by ultrafiltration, and the amount of MPT in each fraction was determined by FormA analysis. Aliquots (5 to 25 μL) of each fraction then were used for reconstitution of NADPH-NR activity in 25 μL of nit-1 crude extract.
The Neurospora crassa nit-1 extract was prepared as described by Nason et al. (1971) and stored in aliquots at −70°C. All reconstitutions were performed with nit-1 extraction buffer (50 mM sodium phosphate, 200 mM NaCl, and 5 mM EDTA, pH 7.2) in the presence of 4 mM reduced glutathione and molybdate where appropriate. nit-1 reconstitution with copurified MPT was performed with various amounts of protein (0.5 to 10 μL) according to the previously determined linear range of the reconstitution assay in the absence of molybdate. Under these conditions, only Moco can be detected. The reconstitution assay was performed in a 60-μL reaction volume containing 40 μL of non-gel-filtrated nit-1 extract or 50 μL of gel-filtrated extract. Just before use, nit-1 extract was gel-filtrated on Nick columns previously equilibrated with nit-1 extraction buffer by loading 400 μL of extract and eluting 500 μL of sample from the column. Complementation was performed anaerobically for 2 hr at room temperature. After addition of 20 mM NADPH and incubating for at least 10 min, reconstituted NADPH-NR activity was determined as described (Nason et al., 1971).
Cytoskeleton Binding
Rabbit actin was prepared according to Spudich and Watt (1971) and stored in 2 mM Tris-HCl, 0.1 mM CaCl2, 1 mM NaN3, 0.4 mM ATP, and 1 mM DTT, pH 8.3. Polymerization was achieved by adding a solution of 1 mM ATP, 10 mM imidazole, 10 mM KCl, and 2 mM MgCl2 to 0.5 to 3.0 mg/mL actin and incubating for 30 to 60 min at 25°C. Binding assays were performed in a total volume of 150 μL, coincubating 2 to 15 μM polymerized F-actin with 0.5 to 15 μM of the protein of interest (precentrifugated for 30 min at 100,000g) for 1 hr at 25°C. Bound and unbound protein was separated by ultracentrifugation of F-actin at 100,000g (Airfuge; Beckman Instruments, Fullerton, CA). The supernatants containing the unbound protein were concentrated by trichloracetic acid precipitation, and the complete pellet and supernatant fractions were analyzed by SDS-PAGE with Coomassie Brilliant Blue R250 staining; the protein bands were densitometrically quantified.
Acknowledgments
We thank Christian Meyer (Institut National de la Recherche Agronomique, Versailles, France) for providing plant material of N. plumbaginifolia cnx mutants, Birgit Stallmeyer for providing the pRT101cnx1 vector, and the German Arabidopsis Stock Center (Max Planck Institute, Cologne, Germany) for providing the Arabidopsis genomic λ-GEM11 library. Furthermore, we thank Brigitte Jockusch (Technical University of Braunschweig) for providing rabbit muscle powder and for advice about the preparation of actin and the cytoskeleton binding assays. The financial support of the Deutsche Forschungsgemeinschaft (G.S. and R.R.M.) and the Fritz Thyssen Stiftung (R.R.M.) is gratefully acknowledged.
References
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C.R. Acad. Sci. Paris Sci. Vie 316 1194–1199. [Google Scholar]
- Benfey, P., Ren, L., and Chua, N. (1989). The CaMV 35S enhancer contains at least two domains which can confer different developmental and tissue-specific expression patterns. EMBO J. 8 2195–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan, M. (1984). Binary Agrobacterium vectors for plant transformation. Nucleic Acids Res. 12 8711–8721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braaksma, F.J., and Feenstra, W.J. (1982). Isolation and characterization of nitrate reductase–deficient mutants of Arabidopsis thaliana. Theor. Appl. Genet. 64 83–90. [DOI] [PubMed] [Google Scholar]
- Calvert, C.M., Gant, S.J., and Bowles, D.J. (1996). Tomato annexins p34 and p35 bind to F-actin and display nucleotide phosphodiesterase activity inhibited by phospholipid binding. Plant Cell 8 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell, W.H. (1999). Nitrate reductase structure, function and regulation: Bridging the gap between biochemistry and physiology. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 277–303. [DOI] [PubMed] [Google Scholar]
- Chilton, W.S., and Chilton, M.D. (1984). Mannityl opine analogs allow isolation of catabolic pathway regulatory mutants. J. Bacteriol. 158 650–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford, N.M., and Arst, H.N., Jr. (1993). The molecular genetics of nitrate assimilation in fungi and plants. Annu. Rev. Genet. 27 115–146. [DOI] [PubMed] [Google Scholar]
- Crepy, L., Chupeau, M.-C., and Chupeau, Y. (1982). The isolation and culture of leaf protoplasts of Cichorium intybus and their regeneration into plants. Z. Pflanzenphysiol. 107 123–131. [Google Scholar]
- Falciani, F., Terao, M., Goldwurm, S., Ronchi, A., Gatti, A., Minoia, C., Li Calzi, M., Salmona, M., Cazzaniga, G., and Garattini, E. (1994). Molybdenum(VI) salts convert the xanthine oxidoreductase apoprotein into the active enzyme in mouse L929 fibroblastic cells. Biochem. J. 298 69–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabard, J., Pelsy, F., Marion-Poll, A., Caboche, M., Saalbach, I., Grafe, R., and Müller, A.J. (1988). Genetic analysis of nitrate reductase deficient mutants of Nicotiana plumbaginifolia: Evidence for six complementation groups among 70 classified molybdenum cofactor deficient mutants. Mol. Gen. Genet. 213 275–281. [Google Scholar]
- Gamborg, O.L., Miller, R.A., and Ojima, K. (1968). Nutrient requirements of suspension culture of soybean root cells. Exp. Cell Res. 50 151–158. [DOI] [PubMed] [Google Scholar]
- Hasona, A., Ray, R.M., and Shanmugam, K.T. (1998). Physiological and genetic analyses leading to identification of a biochemical role for the moeA (molybdate metabolism) gene product in Escherichia coli. J. Bacteriol. 180 1466–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hille, R. (1996). The mononuclear molybdenum enzymes. Chem. Rev. 96 2757–2816. [DOI] [PubMed] [Google Scholar]
- Hoff, T., Schnorr, K.M., Meyer, C., and Caboche, M. (1995). Isolation of two Arabidopsis cDNAs involved in early steps of molybdenum cofactor biosynthesis by functional complementation of Escherichia coli mutants. J. Biol. Chem. 270 6100–6107. [DOI] [PubMed] [Google Scholar]
- Jefferson, R.A., Kavanagh, T.A., and Bevan, M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6 3901–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, J.L., and Rajagopalan, K.V. (1982). Structural and metabolic relationship between the molybdenum cofactor and urothione. Proc. Natl. Acad. Sci. USA 79 6856–6860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch, J., and Betz, H. (1995). The postsynaptic localization of the glycine receptor-associated protein gephyrin is regulated by the cytoskeleton. J. Neurosci. 15 4148–4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirsch, J., Langosch, D., Prior, P., Littauer, U.Z., Schmitt, B., and Betz, H. (1991). The 93-kDa glycine receptor-associated protein binds to tubulin. J. Biol. Chem. 266 22242–22245. [PubMed] [Google Scholar]
- Kirsch, J., Wolters, I., Triller, A., and Betz, H. (1993). Gephyrin antisense oligonucleotides prevent glycine receptor clustering in spinal neurons. Nature 366 745–748. [DOI] [PubMed] [Google Scholar]
- Kisker, C., Schindelin, H., and Rees, D.C. (1997). Molybdenum cofactor–containing enzymes: Structure and mechanism. Annu. Rev. Biochem. 66 233–267. [DOI] [PubMed] [Google Scholar]
- Koshiba, T., Saito, E., Ono, N., Yamamoto, N., and Sato, M. (1996). Purification and properties of flavin- and molybdenum-containing aldehyde oxidase from coleoptyles of maize. Plant Physiol. 110 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuper, J., Palmer, T., Mendel, R.R., and Schwarz, G. (2000). Mutations in the molybdenum cofactor biosynthetic protein Cnx1G from Arabidopsis thaliana define functions for molybdopterin binding, molybdenum insertion, and molybdenum cofactor stabilization. Proc. Natl. Acad. Sci. USA 97 6475–6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaBrie, S.T., Wilkinson, J.Q., Tsay, Y.F., Feldmann, K.A., and Crawford, N.M. (1992). Identification of two tungstate-sensitive molybdenum cofactor mutants, chl2 and chl7, of Arabidopsis thaliana. Mol. Gen. Genet. 233 169–176. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Leimkühler, S., Angermüller, S., Schwarz, G., Mendel, R.R., and Klipp, W. (1999). Activity of the molybdopterin-containing xanthine dehydrogenase of Rhodobacter capsulatus can be restored by high molybdenum concentrations in a moeA mutant defective in molybdenum cofactor biosynthesis. J. Bacteriol. 181 5930–5939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, M.T., Wuebbens, M.M., Rajagopalan, K.V., and Schindelin, H. (2000). Crystal structure of the gephyrin-related molybdenum cofactor biosynthesis protein MogA from Escherichia coli. J. Biol. Chem. 275 1814–1822. [DOI] [PubMed] [Google Scholar]
- MacDonald, D.W., and Cove, D.J. (1974). Studies on temperature-sensitive mutants affecting the assimilatory nitrate reductase of Aspergillus nidulans. Eur. J. Biochem. 47 107–110. [DOI] [PubMed] [Google Scholar]
- Marion-Poll, A., Cherel, I., Gonneau, M., and Leydecker, M.T. (1991). Biochemical characterization of cnx nitrate reductase deficient mutants from Nicotiana plumbaginifolia. Plant Sci. 76 201–209. [Google Scholar]
- Marton, L., Dung, T.M., Mendel, R.R., and Maliga, P. (1982). Nitrate reductase deficient cell lines from haploid protoplast cultures of Nicotiana plumbaginifolia. Mol. Gen. Genet. 183 301–304. [Google Scholar]
- McLean, B.G., Zupan, J., and Zambryski, P.C. (1995). Tobacco mosaic virus movement protein associates with the cytoskeleton in tobacco cells. Plant Cell 7 2101–2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendel, R.R. (1997). Molybdenum cofactor of higher plants: Biosynthesis and molecular biology. Planta 203 399–405. [DOI] [PubMed] [Google Scholar]
- Mendel, R.R., and Müller, A.J. (1976). A common genetic determinant of xanthine dehydrogenase and nitrate reductase in Nicotiana tabacum. Biochem. Physiol. Pflanz. 170 538–541. [Google Scholar]
- Mendel, R.R., and Schwarz, G. (1999). Molybdoenzymes and molybdenum cofactor in plants. Crit. Rev. Plant Sci. 18 33–69. [Google Scholar]
- Mendel, R.R., Alikulov, Z.A., Lvov, N.P., and Müller, A.J. (1981). Presence of the molybdenum-cofactor in nitrate reductase–deficient mutant cell lines of Nicotiana tabacum. Mol. Gen. Genet. 181 395–399. [Google Scholar]
- Mendel, R.R., Marton, L., and Müller, A.J. (1986). Comparative biochemical characterization of nitrate reductase/molybdenum cofactor loci cnxA, cnxB and cnxC of Nicotiana plumbaginifolia. Plant Sci. 43 125–129. [Google Scholar]
- Mendel, R.R., Brinkmann, H., Greger, K., Nerlich, A., Nieder, J., Schledzewksi, K., Schulze, J., Schwarz, G., and Stallmeyer, B. (1997). Molybdenum cofactor biosynthesis in plants and humans. In Chemistry and Biology of Pteridines and Folates, W. Pfleiderer and H. Rokos, eds (Berlin: Blackwell Science), pp. 681–692.
- Müller, A. (1983). Genetic analysis of nitrate reductase–deficient tobacco plants regenerated from mutant cells. Mol. Gen. Genet. 192 275–281. [Google Scholar]
- Müller, A.J., and Mendel, R.R. (1989). Biochemical and somatic cell genetics of nitrate reductase in Nicotiana. In Molecular and Genetic Aspects of Nitrate Assimilation, J.L. Wray and J.R. Kinghorn, eds (Oxford: Oxford University Press), pp. 166–185.
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15 473–497. [Google Scholar]
- Nason, A., Lee, K.Y., Pan, S.S., Ketchum, P.A., Lamberti, A., and DeVries, J. (1971). In vitro formation of assimilatory reduced nicotinamide adenine dinucleotide phosphate: Nitrate reductase from a Neurospora mutant and a component of molybdenum-enzymes. Proc. Natl. Acad. Sci. USA 68 3242–3246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negrutiu, I., Shillito, R., Potrykus, I., Biasini, G., and Sala, F. (1987). Hybrid genes in the analysis of transformation conditions. I. Setting up a simple method for direct gene transfer in plant protoplasts. Plant Mol. Biol. 8 363–373. [DOI] [PubMed] [Google Scholar]
- Nieder, J., Stallmeyer, B., Brinkmann, H., and Mendel, R.R. (1997). Molybdenum cofactor biosynthesis: Identification of A. thaliana cDNAs homologous to E. coli sulfotransferase MoeB. In Sulfur Metabolism in Higher Plants: Molecular, Ecophysiological and Nutritional Aspects, W.J. Cram, L.J. De Kork, I. Stulen, C. Brunold, and H. Rennenberg, eds (Amsterdam: SPB Academic Publisher) pp. 275–277.
- Prior, P., Schmitt, B., Grenningloh, G., Pribilla, I., Multhaup, G., Beyreuther, K., Maulet, Y., Werner, P., Langosch, D., Kirsch, J., and Betz, H. (1992). Primary structure and alternative splice variants of gephyrin, a putative glycine receptor–tubulin linker protein. Neuron 8 1161–1170. [DOI] [PubMed] [Google Scholar]
- Rajagopalan, K.V. (1996). Biosynthesis of the molybdenum cofactor. In Escherichia coli and Salmonella typhimurium, F.C. Neidhardt, ed (Washington, DC: ASM Press), pp. 674–679.
- Rajagopalan, K.V., and Johnson, J.L. (1992). The pterin molybdenum cofactors. J. Biol. Chem. 267 10199–10202. [PubMed] [Google Scholar]
- Reiss, J., Cohen, N., Dorche, C., Mandel, H., Mendel, R.R., Stallmeyer, B., Zabot, M.T., and Dierks, T. (1998). Mutations in a polycistronic nuclear gene associated with molybdenum cofactor deficiency. Nat. Genet. 20 51–53. [DOI] [PubMed] [Google Scholar]
- Reiss, J., Dorche, C., Stallmeyer, B., Mendel, R.R., Cohen, N., and Zabot, M.T. (1999). Human molybdopterin synthase gene: Genomic structure and mutations in molybdenum cofactor deficiency type B. Am. J. Hum. Genet. 64 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Scheible, W.R., Gonzalez-Fontes, A., Morcuende, R., Lauerer, M., Geiger, M., Glaab, J., Gojon, A., Schulze, E.D., and Stitt, M. (1997). Tobacco mutants with a decreased number of functional nia genes compensate by modifying the diurnal regulation of transcription, post-translational modification and turnover of nitrate reductase. Planta 203 304–319. [DOI] [PubMed] [Google Scholar]
- Schwarz, G., Boxer, D.H., and Mendel, R.R. (1997). Molybdenum cofactor biosynthesis. The plant protein Cnx1 binds molybdopterin with high affinity. J. Biol. Chem. 272 26811–26814. [DOI] [PubMed] [Google Scholar]
- Seo, M., Akaba, S., Oritani, T., Delarue, M., Bellini, C., Caboche, M., and Koshiba, T. (1998). Higher activity of an aldehyde oxidase in the auxin-overproducing superroot1 mutant of Arabidopsis thaliana. Plant Physiol. 116 687–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich, J.A., and Watt, S. (1971). The regulation of rabbit skeletal muscle concentration. I. Biochemical studies of the interaction of the tropomyosin–troponin complex with actin and the proteolytic fragments of myosin. J. Biol. Chem. 246 4866–4871. [PubMed] [Google Scholar]
- Stallmeyer, B., Nerlich, A., Schiemann, J., Brinkmann, H., and Mendel, R.R. (1995). Molybdenum co-factor biosynthesis: The Arabidopsis thaliana cDNA cnx1 encodes a multifunctional two-domain protein homologous to a mammalian neuroprotein, the insect protein Cinnamon and three Escherichia coli proteins. Plant J. 8 751–762. [DOI] [PubMed] [Google Scholar]
- Stallmeyer, B., Drugeon, G., Reiss, J., Haenni, A.L., and Mendel, R.R. (1999. a). Human molybdopterin synthase gene: Identification of a bicistronic transcript with overlapping reading frames. Am. J. Hum. Genet. 64 698–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallmeyer, B., Schwarz, G., Schulze, J., Nerlich, A., Reiss, J., Kirsch, J., and Mendel, R.R. (1999. b). The neurotransmitter receptor–anchoring protein gephyrin reconstitutes molybdenum cofactor biosynthesis in bacteria, plants, and mammalian cells. Proc. Natl. Acad. Sci. USA 96 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart, V., and MacGregor, C.H. (1982). Nitrate reductase in Escherichia coli K-12: Involvement of chlC, chlE, and chlG loci. J. Bacteriol. 151 788–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomsett, A.B., and Garrett, R.H. (1980). The isolation and characterization of mutants defective in nitrate assimilation in Neurospora crassa. Genetics 95 649–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Töpfer, R., Schell, J., and Steinbiss, H.H. (1988). Versatile cloning vectors for transient gene expression and direct gene transfer in plant cells. Nucleic Acids Res. 16 8725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuebbens, M.M., and Rajagopalan, K.V. (1993). Structural characterization of a molybdopterin precursor. J. Biol. Chem. 268 13493–13498. [PubMed] [Google Scholar]
- Wuebbens, M.M., and Rajagopalan, K.V. (1995). Investigation of the early steps of molybdopterin biosynthesis in Escherichia coli through the use of in vivo labeling studies. J. Biol. Chem. 270 1082–1087. [DOI] [PubMed] [Google Scholar]