Abstract
A major quantitative trait locus (QTL) controlling response to photoperiod, Hd1, was identified by means of a map-based cloning strategy. High-resolution mapping using 1505 segregants enabled us to define a genomic region of ∼12 kb as a candidate for Hd1. Further analysis revealed that the Hd1 QTL corresponds to a gene that is a homolog of CONSTANS in Arabidopsis. Sequencing analysis revealed a 43-bp deletion in the first exon of the photoperiod sensitivity 1 (se1) mutant HS66 and a 433-bp insertion in the intron in mutant HS110. Se1 is allelic to the Hd1 QTL, as determined by analysis of two se1 mutants, HS66 and HS110. Genetic complementation analysis proved the function of the candidate gene. The amount of Hd1 mRNA was not greatly affected by a change in length of the photoperiod. We suggest that Hd1 functions in the promotion of heading under short-day conditions and in inhibition under long-day conditions.
INTRODUCTION
The transition of the apical meristem from vegetative to reproductive growth is a critical event in the life cycle of a plant. In rice, the timing of this transition affects the timing of heading. This timing, or heading date, is one of the critical traits considered for adapting rice to different cultivation areas and cropping seasons. Rice is a short-day (SD) plant; its heading is promoted by short photoperiods. The response of the plant to length of day (referred to as photoperiod sensitivity [PS]) and its basic vegetative growth determine the heading date of rice. Many genetic studies of heading date have been performed, and several genes controlling PS in rice have been genetically identified, including Se1 (Lm), Se3 to Se7, and E1 to E3 (Yokoo et al., 1980; Yamagata et al., 1986; Poonyarit et al., 1989; Sano, 1992; Yokoo and Okuno, 1993; Tsai, 1995; Kinoshita, 1998). However, only one gene involving photoperiod response in rice has been cloned, Se5 (Izawa et al., 2000). Little is known about the structure and function of PS genes in rice at the molecular level. In contrast, several genes involved in flowering time have been isolated, allowing clarification of part of the genetic control pathway in Arabidopsis (reviewed by Levy and Dean, 1998; Fowler et al., 1999; Kobayashi et al., 1999; Sheldon et al., 1999). Identification of the genes involved in flowering time has made it possible to determine the genetic control pathways for the response to photoperiod and vernalization in Arabidopsis (reviewed by Levy and Dean, 1998; Samach and Coupland, 2000). In addition, homologs of Arabidopsis genes for flowering time also function in Brassica napus (Robert et al., 1998).
The major genes or quantitative trait loci (QTLs) for heading date have been mapped by using DNA markers in rice (Mackill et al., 1993; Li et al., 1995; Xiao et al., 1996; Yano et al., 1997; Lin et al., 1998; Tamura et al., 1998). Four QTLs for heading date involved in PS were mapped precisely as single Mendelian factors by using advanced backcross progeny (Yamamoto et al., 1998, 2000; Lin et al., 2000). A major gene controlling PS, Se1, first was identified as a naturally occurring variant (Yokoo et al., 1980) and was thought to lie at the same locus as the PS QTL Hd1, based on a comparison of their map locations (Yano et al., 1997; Tamura et al., 1998; Yamamoto et al., 1998). Mutant lines with less PS were induced, and some were caused by mutations in the Se1 locus (Inoue et al., 1992). However, no direct evidence was available to prove their allelic relationship, and isolation of the QTL Hd1 or Se1 gene might be an effective way to prove allelism. Linkage mapping for Se1 with restriction fragment length polymorphism (RFLP) markers has been conducted, but the resolution was insufficient for map-based cloning of Se1. To analyze the genetic control of rice heading at the molecular level, we attempted to isolate the genes involved by using naturally occurring variations and a quantitative genetic approach (Yano and Sasaki, 1997). One QTL for PS, Hd6, identified in a recent study, has been isolated by map-based cloning (Y. Takahashi and M. Yano, unpublished data).
In this article, we report the isolation of a major rice PS QTL, Hd1, by map-based cloning. We found that Hd1 is a homolog of CONSTANS from Arabidopsis and encodes a protein with a zinc finger domain. We also confirmed by structural and expression analysis that the major gene controlling PS, Se1, is allelic to Hd1. The nucleotide sequence data reported in this article have been deposited in the DNA Data Bank of Japan nucleotide sequence databases with the accession numbers AB041837 (Nipponbare Hd1; genomic), AB041838 (Nipponbare Hd1; cDNA), AB041839 (Kasalath Hd1), AB041840 (Ginbouzu Hd1), AB041841 (HS66 Hd1), and AB041842 (HS110 Hd1).
RESULTS
Fine-Scale and High-Resolution Mapping
Of >9000 BC3F3 plants grown under natural field conditions, nearly 2000 exhibited early heading, indicating that those plants were homozygous for the recessive Kasalath allele at the Hd1 locus. Yamamoto et al. (1998) reported that early heading plants were homozygous for the Kasalath allele at Hd1, in contrast with plants that were homozygous or heterozygous for the Nipponbare allele, which were later heading under natural field conditions. To avoid contamination by plants heterozygous at Hd1, we carefully selected 1505 plants that showed extreme early heading. Two RFLP markers that flank Hd1, R1679 and P130 (Yamamoto et al., 1998), were used to detect recombination events between Hd1 and markers in the pooled DNAs. Nine and two recombinant chromosomes were detected among 301 pools for R1679 and P130, respectively (Figure 1). Plants containing the recombinant chromosome in each pool were identified from their progeny. The total DNA of these selected plants was used to develop a fine-scale map of the Hd1 region. Eleven plants were selected and analyzed with additional RFLP markers (C235 and S2539) and a cleaved amplified polymorphic sequence (CAPS) marker (S20481) that had been developed from random expressed sequence tag (EST) clones (Yamamoto and Sasaki, 1997). The results suggest that Hd1 lies in the interval between S20481 and P130 (Figure 1).
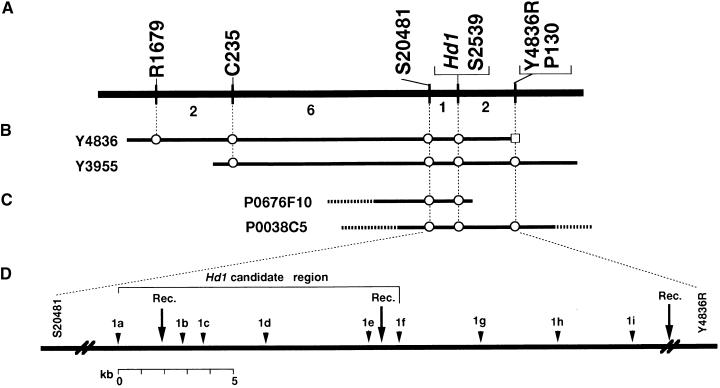
Figure 1.
A Fine-Scale, High-Resolution Genetic and Physical Map of the Hd1 Region on Chromosome 6.
(A) Genetic linkage map showing the relative position of Hd1 with RFLP markers on chromosome 6. Numbers under the horizontal line are numbers of plants with a recombinant chromosome in the adjacent marker intervals.
(B) and (C) Yeast artificial chromosome (B) and PAC clones (C) spanning the Hd1 region. A circle indicates the existence of a sequence corresponding to the RFLP markers. Entire insert sequencing was performed on PAC clone P0038C5.
(D) Detailed genetic and physical map showing the relative positions of the candidate regions of Hd1 and CAPS markers developed based on sequence data. Rec., approximate positions of recombination events that occurred near Hd1.
Yeast artificial chromosome clones containing markers flanking Hd1 have been screened in the physical mapping project of the Rice Genome Research Program (RGP; Kurata et al., 1997). Two yeast artificial chromosome clones, Y4836 and Y3955, were found to contain three flanking markers, C235, S20481, and S2539. The end DNA sequence of Y4836 was cloned; RFLP analysis showed that the resulting clone, Y4836R, cosegregated with P130 (Figure 1). Moreover, two P1-derived artificial chromosome (PAC) clones, P0676F10 and P0038C5, were selected with the markers S20481, S2539, and Y4836R from the PAC genomic library of Nipponbare developed at RGP (Baba et al., 2000). P0038C5 contained sequences for S20481, S2539, and Y4836R. This suggested that P0038C5 encompassed the Hd1 locus (Figure 1).
P0038C5 was sequenced by means of a shotgun strategy. A region of ∼26 kb was defined as an Hd1 candidate region, based on the position of the flanking markers S20481 and Y4836R. To determine the location of Hd1 more precisely, we developed nine CAPS markers (1a to 1i), by using sequence data within the 26-kb candidate genomic region. These CAPS markers detected recombination events between markers 1a and 1b, 1e and 1f, and 1i and Y4836R; four markers—1b, 1c, 1d, and 1e—cosegregated with Hd1 (Figure 1). As a result, a sequence of ∼12 kb was defined as the Hd1-containing region.
Identification and Analysis of the Hd1 Sequence
The candidate genomic sequence was analyzed with the Genscan program (http://genes.mit.edu/GENSCAN.html). Two putative genes were predicted in this region (data not shown). A BLAST search of nonredundant DNA databases revealed that one putative gene showed considerable similarity to the Arabidopsis CONSTANS (CO) gene. Another probable candidate was a sequence identical to that for peroxidase S2539, which was found in random EST sequencing at RGP. At this stage, there was no evidence to exclude S2539 as a candidate for Hd1. However, the sequence showing similarity to CO was further analyzed as a candidate because of the known function of CO in photoperiod response in Arabidopsis (Putterill et al., 1995).
A cosmid clone containing the candidate genomic region was obtained by screening a genomic library of Kasalath. We determined the sequence for a candidate 12-kb region of Kasalath. Comparison of the sequences of Nipponbare and Kasalath revealed many sequence variations. We found four single-base substitutions, one two-base substitution, a 36-bp insertion, and a 33-bp deletion in the putative first exon as well as two single-base substitutions and a two-base deletion in the putative second exon (Figure 2).
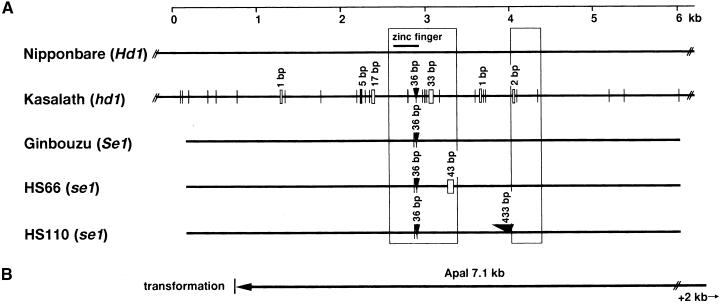
Figure 2.
Scheme of the Structural Differences in the Candidate Region of Hd1 in Nipponbare and Kasalath and the Corresponding Genomic Region of se1 Mutants HS66 and HS110 and Their Original Variety, Ginbouzu.
(A) Comparison of genomic sequences of Nipponbare and Kasalath Hd1 alleles. Boxes show the predicted open reading frames based on the Genscan software maize model. Vertical lines without labels represent single-base substitutions between Nipponbare and Kasalath. Small rectangular boxes and arrowheads represent deletions and insertions, respectively.
(B) A 7.1-kb ApaI genomic fragment containing the entire Hd1 candidate sequence used in the complementation analysis. This fragment does not contain another predicted gene, encoding peroxidase, which was found in the candidate genomic region of Hd1.
Using several kinds of primer pairs that could amplify genomic sequences of the Hd1 candidate, we analyzed the sequences of the se1 mutants HS66 and HS110 and their progenitor variety, Ginbouzu. On the basis of sequence comparison with the Ginbouzu Hd1, we found a 43-bp deletion in the putative first exon of se1 mutant HS66 and a 433-bp insertion in the putative intron of HS110. These results clearly suggested that Hd1 was allelic with Se1. Moreover, we found one single-base substitution and a 36-bp insertion in the first exon of the Ginbouzu Hd1 sequence, compared with that of Nipponbare Hd1 (Figure 2), although both alleles were functional for photoperiod response. The insertion was located at the terminal region of the zinc finger domain (Figures 2A and 3B). Although many variations occurred between the Hd1 sequences of Nipponbare and Kasalath outside of the coding region as well as in the coding regions, no difference was found between Nipponbare and Ginbouzu outside of the coding region. Both Nipponbare and Ginbouzu belong to the japonica subgroup of Oryza sativa, which shows very small genetic variation.
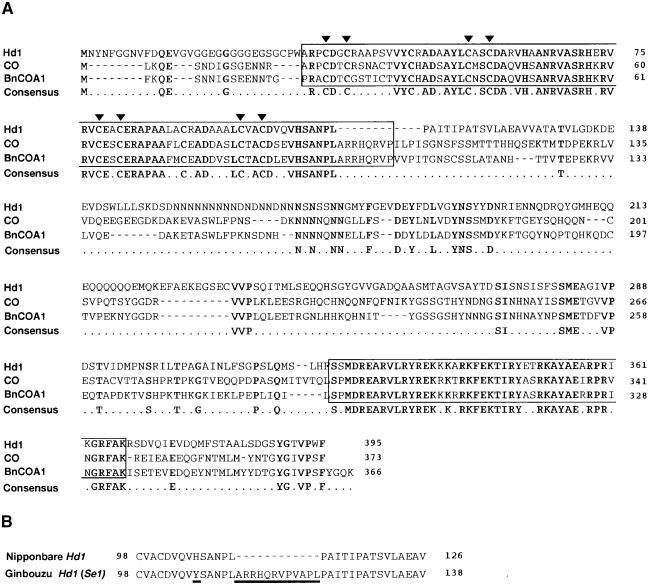
Figure 3.
Deduced Amino Acid Sequence of Hd1 Protein, Amino Acid Alignment with Arabidopsis CO and B. napus BnCOA1, and Comparison of Amino Acid Sequences of C-Terminal Regions of the Zinc Finger Domain between Nipponbare and Ginbouzu Hd1.
(A) Comparison of Nipponbare Hd1, CO, and BnCOA1 alleles. Boxes in N and C termini are conserved domains of the zinc finger motif (N terminus) and nuclear localization signals (C terminus). Arrowheads indicate cysteine residues in the zinc finger domain. Boldface letters represent identical amino acid residues among the three proteins.
(B) Comparison of the C-terminal zinc finger domain of Nipponbare Hd1 and Ginbouzu Se1 (Hd1) proteins. Amino acid substitutions and deletions are indicated by underlines.
In addition to genomic sequencing of Hd1, using the rapid amplification of cDNA ends method, we determined the cDNA sequence of Hd1 by sequencing a product amplified by reverse transcription–polymerase chain reaction (RT-PCR) and 3′ and 5′ primer extension. We thus verified the putative exons and intron based on a prediction from the Genscan maize model. When we compared the sequence of Hd1 with that of CO from Arabidopsis, we found 59% identity in the zinc finger domain and 79% identity in the C-terminal region. The sequence obtained indicates that rice Hd1 is composed of two exons that encode a 395–amino acid protein and is a member of the Arabidopsis CO family with a zinc finger domain (Figure 3A). The deduced amino acid sequence of the Hd1 protein was compared with that of CO from Arabidopsis and BnCOA1 from B. napus. Comparison of two domains conserved in most members of the CO family revealed substantial similarity among them (Figure 3A). The region containing the zinc finger motif showed 65% identity and a consensus structure of CX2CX16CX2C in the CO family (Putterill et al., 1995; Robert et al., 1998). The region near the C terminus showed 83% identity and is thought to be a nuclear localization signal (Putterill et al., 1995; Robert et al., 1998). Downstream from the zinc finger, the Nipponbare Hd1 sequence had little similarity with the sequences of the Arabidopsis CO and B. napus BnCOA1 proteins (Figure 3A). The basic amino acid motif (RRHQR) that is a common feature between the Arabidopsis and B. napus CO family (Robert et al., 1998) in the C terminus of the zinc finger domain was present in the Ginbouzu Hd1 protein but not in Nipponbare (Figure 3B).
Functional Complementation with Candidate Gene in Transgenic Rice
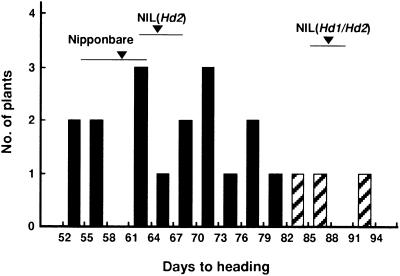
A 7.1-kb ApaI fragment of Nipponbare (Figure 2B) containing the candidate Hd1 region was transferred into a nearly isogenic line (NIL) of Nipponbare, NIL(Hd1/Hd2). The recipient line exhibited no photoperiod response, because Nipponbare functional alleles of Hd1 and Hd2 were replaced with Kasalath nonfunctional alleles (Lin et al., 2000). Thus, the transgenic plants with the Nipponbare Hd1 sequence were expected to exhibit a more sensitive photoperiod response, such as a promotion of heading in SD conditions. Fifty plants with a hygromycin resistance gene and containing the target gene region were obtained by Agrobacterium-mediated transformation with a 7.1-kb ApaI fragment (Figure 2). The ApaI fragment contains only one gene highly similar to Arabidopsis CO. Some transgenic plants showed earlier heading in a growth chamber under SD conditions than did those with only the vector sequence and the nearly isogenic line NIL(Hd1/Hd2) control (data not shown). We selected one transgenic plant that showed early heading and had one copy of the 7.1-kb fragment for further progeny analysis. Self-pollinated progeny of the selected plant showed wide variation in days to heading (53 to 93 days) under SD conditions (Figure 4). We used CAPS marker 1e to determine the presence or absence of the gene in each plant. Plants without the transgene and NIL(Hd1/Hd2) plants showed later heading than did plants homozygous or heterozygous for the transgene. All transgenic plants showed no growth abnormalities. Thus, the 7.1-kb candidate genomic region promoted heading under SD conditions. This is consistent with results comparing NIL(Hd2) and NIL(Hd1/Hd2) plants (Lin et al., 2000). These results clearly suggest that the Hd1 sequence in the 7.1-kb candidate genomic region retains the function of photoperiod response.
Figure 4.
Frequency Distribution for Days to Heading in Self-Pollinated Progeny (T1) of One 7.1-kb Transformant.
All plants were cultivated under SD conditions (10.0 hr) in a controlled growth chamber. Bars indicate plants with (black) and without (hatched) the candidate genomic fragment. NIL(Hd1/Hd2) is the recipient line for transformation, and NIL(Hd2), the nearly isogenic line of Hd2, can be a control of transformation when the functional Hd1 is complemented in NIL(Hd1/Hd2). Means and ranges of days to heading of Nipponbare, NIL(Hd1/Hd2), and NIL(Hd2) are indicated by arrowheads and horizontal bars, respectively.
Expression of Hd1
We could not detect Hd1 mRNA in RNA gel blot analysis. Thus, to determine whether the Hd1 candidate region was expressed and whether the expression of Hd1 was induced by a change of daylength, we performed RT-PCR analysis (Figure 5). The amplified fragment was designed to include a 33-bp deletion in the first exon in Kasalath and a 43-bp deletion in the first exon in HS66 but not a 36-bp insertion in the first exon in Kasalath or Ginbouzu (Figure 2). The fragment also included a 433-bp insertion in the intron in HS110. Hd1 mRNA was detected in Nipponbare, Kasalath, and NIL(Hd1). However, the amplified product was slightly smaller in Kasalath than in Nipponbare. Sequencing the amplified product showed this size difference to be consistent with the 33-bp difference in the genomic sequences (Figure 2). Ginbouzu produced the same amount of mRNA as Nipponbare. The se1 mutant HS66 also produced the same amount of mRNA as Ginbouzu, but its PCR product was slightly smaller than that of Ginbouzu. On the other hand, several different amplified products of the se1 mutant HS110 were seen in the RT-PCR assay (Figure 5). Sequencing these RT-PCR products revealed a 43-bp deletion in the amplified product of HS66 and a 433-bp insertion in the amplified product of HS110. A sequencing product the same as that of Nipponbare also was seen in HS110 (data not shown). These changes are consistent with the results of genomic sequencing of HS66 and HS110 (Figure 2). Thus, the transcription of Hd1 may have been inaccurate in the se1 mutant HS110.
Figure 5.
Detection of mRNA in the Varieties and Lines Used in This Study by RT-PCR Assay.
All plants were raised in LD conditions (16.0 hr) and then subjected to the following treatments: L, additional 10 days of treatment in LD conditions; S0, no additional treatment; S5, additional 5 days of treatment in SD conditions (10.0 hr); S10, additional 10 days of treatment in SD conditions. The actin control is shown at the bottom of figure. Lane M indicates DNA size markers. Lane gDNA is a PCR product of genomic DNA of Nipponbare as a template.
The quantities of Nipponbare mRNA at Hd1 did not change with the transition from long-day (LD) to SD conditions (Figure 5), which was associated with the initiation of transition to heading. We also observed no differences in Hd1 mRNA amounts when the plants were grown solely in SD or LD conditions (data not shown). These results suggest that the expression of Hd1 is not greatly affected by a change of length of photoperiod.
DISCUSSION
Map-Based Cloning of QTLs
Many genes for heading in rice have been genetically identified (Yokoo et al., 1980; Yamagata et al., 1986; Poonyarit et al., 1989; Yokoo and Okuno, 1993; Tsai, 1995; Kinoshita, 1998). Several were suggested to involve response to photoperiod (i.e., PS). However, little is known about the genetic control pathway of PS, owing to a lack of molecular information on the genes involved. In this study, we cloned Hd1, one of the genes that control PS in rice, by map-based cloning with naturally occurring allelic variations. Another QTL, Hd6, which also is involved in photoperiod response, has been isolated by the same map-based strategy (Y. Takahashi and M. Yano, unpublished data). These results imply that QTLs detected based on naturally occurring allelic variation can be isolated with the use of advanced backcross progeny (Yano and Sasaki, 1997; Yamamoto et al., 1998, 2000). In rice as in Arabidopsis (Alonso-Blanco and Koornneef, 2000), naturally occurring variation can provide new gene sources for plant genetics and molecular biology.
Similarity in Structure of Hd1 and CO
The Hd1 protein contains two zinc finger domains of structure CX2CX16CX2C. The zinc finger domain is believed to be involved in DNA binding in the GATA1 gene family (Pevny et al., 1991; Omichinski et al., 1993). The C-terminal region of Hd1, which is thought to be a nuclear localization signal, also showed considerable identity with the CO family at the amino acid level (Robert et al., 1998). Several ESTs in rice exhibited a conserved structure with the CO family (Putterill et al., 1995; Song et al., 1998), and three of them showed strong similarity at the amino acid level to CO in the N-terminal and C-terminal regions. However, Hdl is more closely related to CO than to proteins encoded by the three rice ESTs. Linkage mapping of these ESTs revealed no candidate ESTs for Hd1 (Song et al., 1998), perhaps because of the very low expression of Hd1 as revealed in this study. We have shown here that Hd1, a CO homolog in rice, also has a role in photoperiod response.
Allelic Relationship between Hd1 and Se1 and Loss of Function in Kasalath Hd1 and se1 Mutant Alleles
Genetic analyses of PS in several Japanese rice varieties (Okumoto et al., 1991; Ichitani et al., 1998) indicate that Nipponbare might possess an allele of Se1 with a strong photoperiod response. Genetic linkage mapping revealed that the chromosomal location of Hd1 coincided with that of Se1 (Tamura et al., 1998; Yamamoto et al., 1998), but no direct evidence demonstrated an allelic relationship between Hd1 and Se1. In this study, we identified the allelic relationship by structural and expressional analysis of Hd1.
The Nipponbare and Kasalath Hd1 alleles have many structural differences between them (Figure 2). A 2-bp deletion in the second putative exon in the Kasalath allele results in a premature stop codon. Thus, the Kasalath Hd1 protein could be shorter than the Nipponbare protein because it is missing the C-terminal region (Figure 2). The 43-bp deletion in the se1 mutant HS66 also produces a premature stop codon. Abnormal transcripts with a 433-bp insertion sequence in the intron region were detected in the se1 mutant HS110 by RT-PCR. However, small amounts of mRNA of the same size as in Nipponbare also were detected in this mutant (Figure 5), and the sequence of this product is identical to that of Nipponbare. Days to heading of HS110 was ∼4 days later than that of HS66 under natural field conditions (data not shown). This finding indicates that the function of the se1 allele in HS110 was not completely lost (data not shown). This phenotypic difference might reflect the presence of normal-size transcripts in HS110. Perhaps HS110 might be categorized as a leaky mutant at the Hd1 locus.
We also found a structural difference between the functional Hd1 alleles of Nipponbare and Ginbouzu. A 36-bp nucleotide sequence corresponding to the terminal region of a zinc finger domain and including the basic amino acid motif RRHQR was present in the Ginbouzu Hd1 allele but not in the Nipponbare allele (Figures 2A and 3B). This sequence was also present in Kasalath, which makes it more likely that a 36-bp deletion occurred in the Nipponbare Hd1 allele. This region is thought to be important for DNA–DNA interaction (Omichinski et al., 1993) and is highly conserved between rice and Arabidopsis or B. napus (Robert et al., 1998). At present, no concrete evidence explains the functional difference between the Nipponbare and Ginbouzu Hd1 alleles. Clarifying any functional difference between them by genetic analysis is difficult, because allelic differences in other genes involving heading date are also present. Given the differences between days to heading of Nipponbare (116 ± 1.9) and NIL(Hd1) (97.2 ± 1.3) and between Ginbouzu (125.2 ± 1.5) and HS66 (95.2 ± 2.2), we assume that the inhibition of heading under natural field conditions by the Hd1 allele of Nipponbare is less than that of Ginbouzu. We also observed a difference in days to heading between two se1 mutants, HS66 (95.2 ± 2.2) and HS110 (103.8 ± 1.9). In the RT-PCR assay, we detected a normal-size Hd1 transcript in HS110. These results suggest that the function of the se1 (hd1) allele of HS110 is not completely lost. It will be necessary to confirm the functional difference between Nipponbare and Ginbouzu Hd1 alleles and two mutant alleles of se1(hd1) by transformation analysis.
Function of Hd1 in Photoperiod Response
The function of Hd1 probably is to affect transcription activation because of the presence of a zinc finger domain. Because Hd1 transcription itself was not greatly affected by change in length of day, we speculate that Hd1 affects the transcription of genes for which expression is controlled by photoperiod changes. A nearly isogenic line for Hd1, in which the Kasalath Hd1 chromosomal region was substituted into the Nipponbare background, was developed in a previous study (Lin et al., 2000). NIL(Hd1) exhibited later heading than Nipponbare under SD conditions but earlier heading under LD conditions (Lin et al., 2000). We analyzed naturally occurring allelic variation—whether such genes promote or inhibit heading under certain photoperiod conditions—in contrast with mutants because the functional parental alleles were uncertain. In the case of Hd1, high-resolution linkage mapping clearly suggested that a functional allele of Hd1 inhibited heading under natural field conditions (more like LD conditions in the growth chamber). On the other hand, transformation analysis revealed that Hd1 promoted heading under SD conditions. These results suggest that Hd1 might be bifunctional under SD and LD conditions, promoting heading under SD conditions and inhibiting it under LD conditions. The fact that the same amount of Hd1 mRNA was present under both SD and LD conditions also supports this hypothesis. In Arabidopsis, the CO protein promotes flowering under LD conditions but has no phenotypic effect on flowering time under SD conditions (Putterill et al., 1995). This raises the question of what factors are involved in regulating the marked change in photoperiod response from SD to LD conditions. In Arabidopsis, one possible explanation is derived from expression analysis of CO and GI genes (Samach and Coupland, 2000). The daily GI expression peak occurred 8 to 12 hr after dawn, and the timing and duration of the peak were influenced by length of day (Fowler et al., 1999). This change of expression pattern might be responsible for the photoperiod response in Arabidopsis. In this study, we analyzed mRNA at a single time during daylight (5 hr after dawn). More comprehensive expression analysis of rice Hd1, such as measurement of daily temporal expression pattern, should be performed under conditions of different photoperiod lengths.
Genetic Control of PS in Rice
The genetic control pathway of rice heading has not been modeled successfully. Even though the importance of phytochrome in the photoperiod response has been recognized in other plant species, only recently was it proved to play an important role in rice (Izawa et al., 2000). Other factors involved in photoperiod response in rice are still uncertain. To clarify the genetic control of heading in rice in detail, we have been genetically analyzing naturally occurring variation (Yano and Sasaki, 1997; Yano et al., 1997; Lin et al., 1998; Yamamoto et al., 1998, 2000). Nonfunctional alleles of Hd1 and other genes at QTLs for photoperiod response were combined. Analysis of epistatic interactions revealed that Hd1 is epistatic to other genes that enhance photoperiod response, such as Hd2 and Hd3 (Lin et al., 2000). These results suggest that Hd1 plays a central role in the expression of photoperiod response under both SD and LD conditions. However, this study suggests that the transcription of Hd1 might not be greatly affected by changes in photoperiod length. These observations raise a major question: which factors are involved in the dramatic change in the response to photoperiod in rice? We cannot rule out the hypothesis that Hd1 itself is involved in such a strong response. However, other factors for which expression is affected by changes in photoperiod might be involved downstream of Hd1 in the genetic control pathway. To clarify the real role of Hd1 in the photoperiod response pathway, we need to analyze rhythmic expression under SD and LD conditions. In addition, identification of other QTLs, such as Hd2, Hd3, and Hd6, by map-based cloning is progressing (Yano and Sasaki, 1997; Yamamoto et al., 2000; Y. Takahashi and M. Yano, unpublished data). This simultaneous approach to identifying QTLs that control PS will contribute to our understanding of the genetic control pathway for photoperiod response in rice.
This study proves that genes with the same structure are involved in flowering in the dicot species Arabidopsis and the monocot species rice. A casein kinase IIα subunit also is involved in the photoperiod response of flowering in rice (Y. Takahashi and M. Yano, unpublished data). The casein kinase II activates the CCA1 protein, a circadian oscillator in Arabidopsis (Sugano et al., 1998), and is involved in the control of flowering (Sugano et al., 1999). These results clearly indicate that the same factors (genes) might be involved in photoperiod response in both SD plants (rice) and LD plants (Arabidopsis). However, we do not understand the mechanism that makes each plant species completely opposite in their photoperiod responses. Our efforts to identify genes in rice by using both naturally occurring allelic variation and mutational analysis might provide important clues to the mechanisms of these phenomena.
METHODS
Plant Materials
Rice (Oryza sativa) varieties Nipponbare and Kasalath were used to develop a large mapping population. Self-pollinated progeny (BC3F3), derived from BC3F2 plants heterozygous for the proximal region of chromosome 6 (including the Hd1 target locus), were used as the segregating population. The BC3F2 plants used in small-scale mapping of Hd1 were described previously (Yamamoto et al., 1998). Six varieties or lines were used in genomic DNA gel blot hybridization analysis, reverse transcription–polymerase chain reaction (RT-PCR) assay, and sequencing of the genomic candidate region. A nearly isogenic line for Hd1 (NIL[Hd1]), in which the Kasalath chromosomal region of Hd1 was substituted into the genetic background of Nipponbare, was developed previously (Lin et al., 2000). Artificial mutants for the Se1 locus, HS66 and HS110, were induced by gamma irradiation from the variety Ginbouzu at the Plant Breeding Laboratory at Kyoto University (Inoue et al., 1992). These rice varieties and lines were cultivated in controlled growth chambers or under natural field conditions. Leaves were collected for DNA and RNA extraction at the appropriate stages (see legend to Figure 5). Nipponbare and Ginbouzu showed a strong photoperiod response, but Kasalath showed a weak response. Days to heading of Nipponbare, NIL(Hd1), Ginbouzu, and the se1 mutants HS66 and HS110 under natural field conditions are stated in Results, except for Kasalath (105.2 ± 1.3 days).
Growth Conditions for Expression Analysis
All varieties, mutants, and the nearly isogenic line were grown in growth chambers with different photoperiod conditions at temperatures of 26°C for 12 hr and 22°C for 12 hr. Two photoperiod treatments were used: short day (SD; 10.0 hr) and long day (LD; 16.0 hr). Photon irradiance was ∼700 μmol m−2 sec−1. The plants were grown under LD conditions for 30 days, after which some were transferred to SD conditions. Plants were sampled at 30 and 40 days under LD conditions and at 5 and 10 days under SD conditions. Sampling time was ∼5 hr after dawn.
High-Resolution Mapping
Approximately 9000 BC3F3 plants were grown in a field at the National Institute of Agrobiological Resources. Segregants homozygous for the Kasalath Hd1 allele were selected based on days to heading (early heading). The selected plants were used for recombination detection in the genomic region flanking Hd1 by the pooled sampling method (Churchill et al., 1993). Green leaves of five individuals were pooled for DNA extraction by the cetyltrimethylammonium bromide method (Murray and Thompson, 1980). DNA was digested with the restriction enzymes BamHI, BglII, HindIII, and EcoRI for restriction fragment length polymorphism (RFLP) analysis. DNA gel blotting, probe production, labeling, and detection were performed as described previously (Kurata et al., 1994; Harushima et al., 1998). Two probes, R1679 and P130, which flanked Hd1, were used to detect recombination events between Hd1 and the markers. When a particular pool of five plants was found to contain a recombinant chromosome, the five plants were analyzed individually to identify the plant with the recombinant chromosome. The total DNA of each BC3F3 plant was reconstituted from the bulked DNA of BC3F4 seedlings. Approximately 100 BC3F4 seedlings were grown in a greenhouse, and the leaves were harvested for DNA extraction. RFLP analysis was performed as described above. Plants with a crossover between the flanking markers and Hd1 were used for further fine mapping with additional RFLP and cleaved amplified polymorphic sequence (CAPS) markers.
Screening of Genomic Clones Containing the Hd1 Locus
A rice P1-derived artificial chromosome (PAC) library (inserts averaging 112 kb from ∼70,000 clones) was constructed with genomic DNA of Nipponbare at the Rice Genome Research Program (RGP; Baba et al., 2000). From this library, 18,432 clones were screened by PCR with sequence tag site primers designed for markers S2539, S20481, and Y4836R. To obtain sequences of the corresponding Kasalath candidate region for Hd1, we screened a cosmid library constructed with Kasalath genomic DNA. The genomic library was constructed using the superCos vector (Stratagene, La Jolla, CA). Flanking markers S2539 and S20481 were used to screen the library.
Generation of New DNA Markers Flanking Hd1
To produce additional DNA markers flanking the Hd1 locus, we cloned the end sequence of Y4836R. Yeast artificial chromosome end DNA fragments were amplified by a slightly modified cassette PCR method (Isegawa et al., 1992) and were cloned with the TA vector (Invitrogen, Carlsbad, CA). CAPS markers were generated to narrow the minimum candidate region for Hd1. From the sequence data of the candidate region in Nipponbare, we designed several primer pairs to amplify specific amplified products of the target region. The amplified products were digested with 28 restriction enzymes to search for polymorphisms. When we could not find an appropriate enzyme for polymorphisms, we cloned and sequenced the corresponding genomic region in Kasalath. Using sequence comparison, we then found an appropriate restriction enzyme to produce polymorphisms between Nipponbare and Kasalath.
Sequence Analysis
The shotgun sequencing strategy was used to obtain genomic sequencing data covering the Hd1 locus. The insert of PAC clone P0038C5 was purified by ultracentrifugation and fragmented by ultrasonication, and the ends of each piece were blunt-ended with T4 DNA polymerase. After electrophoresis on agarose gels, fractions corresponding to 2 and 5 kb were cut out, and the eluted DNA fragments were ligated to pUC18 to make sublibraries. Sequencing was performed with an automated fluorescent laser sequencer and a Big-Dye Primer Cycle Sequencing Kit (Perkin-Elmer Applied Biosystems, Foster City, CA). The restriction enzymes PstI and AvrII were used to subclone Kasalath cosmid clone no. 47. If necessary, additional restriction enzymes were used to obtain subclones of the appropriate insert size. Sequencing was performed as described above. The raw sequence data of the PAC insert were scored with phred software (Ewing et al., 1998) and then assembled with phrap/cross-match software to make a contiguous nucleotide sequence (Ewing and Green, 1998).
The sequence of the candidate genomic region was analyzed with Genscan software (Burge and Karlin, 1997). A maize model was used to predict putative open reading frames in the candidate region. The predicted amino acid sequence was used for BLAST searches (Altschul et al., 1990) of nonredundant protein databases at GenBank.
Complementation Test for the Function of the Candidate Genomic Region
A 7.1-kb ApaI fragment containing the candidate Hd1 region was subcloned into binary vector pPZP2H-lac, a plasmid vector without promoter (T. Fuse and M. Yano, unpublished data). An Agrobacterium-mediated method was used for transformation (Toki, 1997). Subclones of the 7.1-kb fragment were transformed into Agrobacterium strain EHA101 and then infected into callus of a nearly isogenic line of Nipponbare (NIL[Hd1/Hd2]), in which the chromosomal regions of the photoperiod sensitivity (PS) loci Hd1 and Hd2 had been replaced with chromosomal segments of Kasalath; the resulting nearly isogenic line showed no photoperiod response (Lin et al., 2000). Plants regenerated from hygromycin-resistant calluses were grown in the controlled growth chamber under SD conditions (10.0 hr). Self-pollinated progeny of transformants were grown there under the same conditions. Days to heading of individual plants were scored and used for confirmation of cosegregation of both heading date and the integrated gene.
RNA Extraction and RT-PCR Assay
Total RNA was extracted from leaves at several stages under various photoperiod treatments in a single-step method (Chomcznski and Sacchi, 1987) with minor modifications. A first-strand cDNA was synthesized with the First-Strand cDNA Synthesis Kit (Amersham Pharmacia Biotech, Uppsala, Sweden) according to the manufacturer's instructions. The first-strand cDNA was used as a template, and amplification was performed for 30 PCR cycles (1 min at 94°C, 2 min at 55°C, 3 min at 72°C) followed by 7 min at 72°C. The primers for Hd1 were 5′-TTCTCCTCTCCAAAGATTCC-3′ (sense) and 5′-CATACGCCTTTCTTGTTTCA-3′ (antisense). Actin primers were 5′-TCCATCTTGGCATCTCTCAG-3′ (sense) and 5′-GTACCCGCATCAGGC-ATCTG -3′ (antisense). RT-PCR assay was performed at least twice for each sample. Rapid amplification of cDNA ends PCR was performed by using a Marathon cDNA Amplification Kit (Clontech, Palo Alto, CA). Amplification using reduced cycles in the RT-PCR assay was performed to confirm the results obtained.
Acknowledgments
We thank Prof. T. Tanisaka and Dr. Y. Okumoto (Kyoto University, Kyoto, Japan) for providing se1 mutants and for valuable comments. We thank Dr. Ben Burr for critical reading of the manuscript. We also thank Dr. K. Eguchi for his advice and encouragement. This work was supported mainly by funds from the Program for the Promotion of Basic Research Activities for Innovative Biosciences and partly by funds from the Ministry of Agriculture, Forestry, and Fisheries and the Japan Racing Association.
References
- Alonso-Blanco, C., and Koornneef, M. (2000). Naturally occurring variation in Arabidopsis: An underexploited resource for plant genetics. Trends Plant Sci. 5 22–29. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. [DOI] [PubMed] [Google Scholar]
- Baba, T., et al. (2000). Construction and characterization of rice genomic libraries, PAC library of japonica variety Nipponbare, and BAC library of indica variety Kasalath. Bull. Natl. Inst. Agrobiol. Resour. 14 41–49. [Google Scholar]
- Burge, C., and Karlin, S. (1997). Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268 78–94. [DOI] [PubMed] [Google Scholar]
- Chomcznski, P., and Sacchi, N. (1987). Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Anal. Biochem. 162 156–159. [DOI] [PubMed] [Google Scholar]
- Churchill, G.A., Giovannoni, J.J., and Tanksley, S.D. (1993). Pooled-sampling makes high-resolution mapping practical with DNA markers. Proc. Natl. Acad. Sci. USA 90 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing, B., and Green, P. (1998). Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8 186–194. [PubMed] [Google Scholar]
- Ewing, B., Hillier, L., Wendl, M.C., and Green, P. (1998). Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8 175–185. [DOI] [PubMed] [Google Scholar]
- Fowler, S., Lee, K., Onouchi, H., Samach, A., Richardson, K., Morris, B., Coupland, G., and Putterill, J. (1999). GIGANTEA: A circadian clock–controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18 4679–4688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harushima, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148 479–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichitani, K., Okumoto, Y., and Tanisaka, T. (1998). Genetic analyses of low photoperiod sensitivity of rice cultivars from the northernmost regions of Japan. Plant Breed. 117 543–547. [Google Scholar]
- Inoue, H., Tanisaka, T., Okumoto, Y., and Yamagata, H. (1992). An early-heading mutant gene of a mutant line HS66 of rice (in Japanese with English summary). Rep. Soc. Crop Sci. Breed. Kinki 37 47–52. [Google Scholar]
- Isegawa, Y., Sheng, J., Sokawa, Y., Yamanishi, K., Nakagomi, O., and Ueda, S. (1992). Selective amplification of cDNA sequence from total RNA by cassette-ligation mediated polymerase chain reaction (PCR): Application to sequencing 6.5 kb genome segment of hantavirus strain B-1. Mol. Cell. Probes 6 467–475. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Tokutomi, S., Okuno, K., and Shimamoto, K. (2000). Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 22 391–399. [DOI] [PubMed] [Google Scholar]
- Kinoshita, T. (1998). Report of the committee on gene symbolization, nomenclature and linkage groups. II. Linkage mapping using mutant genes in rice. Rice Genet. Newsl. 15 13–74. [Google Scholar]
- Kobayashi, Y., Kaya, H., Goto, K., Iwabuchi, M., and Araki, T. (1999). A pair of related genes with antagonistic roles in mediating flowering signals. Science 286 1960–1962. [DOI] [PubMed] [Google Scholar]
- Kurata, N., et al. (1994). A 300-kilobase-interval genetic map of rice including 883 expressed sequences. Nat. Genet. 8 365–372. [DOI] [PubMed] [Google Scholar]
- Kurata, N., Umehara, Y., Tanoue, H., and Sasaki, T. (1997). Physical mapping of the rice genome with YAC clones. Plant Mol. Biol. 35 101–113. [PubMed] [Google Scholar]
- Levy, Y.Y., and Dean, C. (1998). The transition to flowering. Plant Cell 10 1973–1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Z.K., Pinson, S.R.M., Stansel, J.W., and Park, W.D. (1995). Identification of quantitative trait loci (QTLs) for heading date and plant height in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 91 374–381. [DOI] [PubMed] [Google Scholar]
- Lin, H.X., Yamamoto, T., Sasaki, T., and Yano, M. (2000). Characterization and detection of epistatic interactions of three QTLs, Hd1, Hd2 and Hd3, controlling heading date in rice using nearly isogenic lines. Theor. Appl. Genet., 101 1021–1028. [Google Scholar]
- Lin, S.Y., Sasaki, T., and Yano, M. (1998). Mapping quantitative trait loci controlling seed dormancy and heading date in rice, Oryza sativa L., using backcross inbred lines. Theor. Appl. Genet. 96 997–1003. [Google Scholar]
- Mackill, D.J., Salam, M.A., Wang, Z.Y., and Tanksley, S.D. (1993). A major photoperiod-sensitivity gene tagged with RFLP and isozyme markers in rice. Theor. Appl. Genet. 85 536–540. [DOI] [PubMed] [Google Scholar]
- Murray, M.G., and Thompson, W.F. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8 4321–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto, Y., Tanisaka, T., and Yamagata, H. (1991). Heading-time genes of rice varieties grown in south-west warm region in Japan (in Japanese with English summary). Jpn. J. Breed. 41 135–152. [Google Scholar]
- Omichinski, J.G., Clore, G.M., Schaad, O., Felsenfeld, G., Trainor, C., Appella, E., Stahl, S.J., and Gronenborn, A.M. (1993). NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1. Science 261 438–446. [DOI] [PubMed] [Google Scholar]
- Pevny, L., Simon, M.C., Robertson, E., Klein, W.H., Tsai, S.F., D'Agati, V., Orkin, S.H., and Costantini, F. (1991). Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature 349 257–260. [DOI] [PubMed] [Google Scholar]
- Poonyarit, M., Mackill, D.J., and Vergara, B.S. (1989). Genetics of photoperiod sensitivity and critical daylength in rice. Crop Sci. 29 647–652. [Google Scholar]
- Putterill, J., Robson, F., Lee, K., Simon, R., and Coupland, G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80 847–857. [DOI] [PubMed] [Google Scholar]
- Robert, L.S., Robson, F., Sharpe, A., Lydiate, D., and Coupland, G. (1998). Conserved structure and function of the Arabidopsis flowering time gene CONSTANS in Brassica napus. Plant Mol. Biol. 37 763–772. [DOI] [PubMed] [Google Scholar]
- Samach, A., and Coupland, G. (2000). Time measurement and the control of flowering in plants. Bioessays 22 38–47. [DOI] [PubMed] [Google Scholar]
- Sano, Y. (1992). Genetic comparisons of chromosome 6 between wild and cultivated rice. Jpn. J. Breed. 42 561–572. [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, A., Peacock, W.J., and Dennis, E. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, J., Yamamoto, K., Shomura, A., Itadani, H., Zhong, H.S., Yano, M., and Sasaki, T. (1998). Isolation and mapping of a family of putative zinc-finger protein cDNAs from rice. DNA Res. 30 95–101. [DOI] [PubMed] [Google Scholar]
- Sugano, S., Andronis, C., Green, R.M., Wang, Z.Y., and Tobin, E.M. (1998). Protein kinase CK2 interacts with and phosphorylates the Arabidopsis circadian-associated 1 protein. Proc. Natl. Acad. Sci. USA 95 11020–11025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugano, S., Andronis, C., Ong, M.S., Green, R.M., and Tobin, E.M. (1999). The protein kinase CK2 is involved in regulation of circadian rhythms in Arabidopsis. Proc. Natl. Acad. Sci. USA 96 12362–12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura, K., Nomura, K., Oshima, I., Namai, H., Yano, M., Sasaki, T., and Kikuchi, F. (1998). Identification of restriction fragment length polymorphism markers tightly linked to a major photoperiod sensitivity gene, Se-1, and to a blast resistance gene, Pi-zt, in rice. SABRAO J. Breed. Genet. 30 61–67. [Google Scholar]
- Toki, S. (1997). Rapid and efficient Agrobacterium-mediated transformation in rice. Plant Mol. Biol. Rep. 15 16–21. [Google Scholar]
- Tsai, K.H. (1995). Genetic analysis for heading time in wild rice strains. Jpn. J. Genet. 70 555–562. [Google Scholar]
- Xiao, J., Li, J., Yuan, L., and Tanksley, S.D. (1996). Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 92 230–244. [DOI] [PubMed] [Google Scholar]
- Yamagata, H., Okumoto, Y., and Tanisaka, T. (1986). Analysis of genes controlling heading time in Japanese rice. In Rice Genetics, (Los Baños, Philippines: International Rice Research Institute), pp. 351–359.
- Yamamoto, K., and Sasaki, T. (1997). Large-scale EST sequencing in rice. Plant Mol. Biol. 35 135–144. [PubMed] [Google Scholar]
- Yamamoto, T., Kuboki, Y., Lin, S.Y., Sasaki, T., and Yano, M. (1998). Fine mapping of quantitative trait loci Hd-1, Hd-2 and Hd-3, controlling heading date of rice, as single Mendelian factors. Theor. Appl. Genet. 97 37–44. [Google Scholar]
- Yamamoto, T., Lin, H.X., Sasaki, T., and Yano, M. (2000). Identification of heading date quantitative trait locus Hd6 and characterization of its epistatic interactions with Hd2 in rice using advanced backcross progeny. Genetics 154 885–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano, M., and Sasaki, T. (1997). Genetic and molecular dissection of quantitative traits in rice. Plant Mol. Biol. 35 145–153. [PubMed] [Google Scholar]
- Yano, M., Harushima, Y., Nagamura, Y., Kurata, N., Minobe, Y., and Sasaki, T. (1997). Identification of quantitative trait loci controlling heading date in rice using a high-density linkage map. Theor. Appl. Genet. 95 1025–1032. [Google Scholar]
- Yokoo, M., and Okuno, K. (1993). Genetic analysis of earliness mutations induced in the rice cultivar Norin 8. Jpn. J. Breed. 43 1–11. [Google Scholar]
- Yokoo, M., Kikuchi, F., Nakane, A., and Fujimaki, H. (1980). Genetical analysis of heading time by aid of close linkage with blast, Pyricularia oryzae, resistance in rice. Bull. Natl. Inst. Agric. Sci. Ser. D 31 95–126. [Google Scholar]