Abstract
Mapping genes by chromosome walking is a widely used technique applicable to cloning virtually any gene that is identifiable by mutagenesis. We isolated the gene responsible for the recessive mutation rsf1 (for reduced sensitivity to far-red light) in the Arabidopsis Columbia accession by using classical genetic analysis and two recently developed technologies: genotyping high-density oligonucleotide DNA array and denaturing high-performance liquid chromatography (DHPLC). The Arabidopsis AT412 genotyping array and 32 F2 plants were used to map the rsf1 mutation close to the top of chromosome 1 to an interval of ∼500 kb. Using DHPLC, we found and genotyped additional markers for fine mapping, shortening the interval to ∼50 kb. The mutant gene was directly identified by DHPLC by comparing amplicons generated separately from the rsf1 mutant and the parent strain Columbia. DHPLC analysis yielded polymorphic profiles in two overlapping polymorphic amplicons attributable to a 13-bp deletion in the third of five exons of a gene encoding a 292–amino acid protein with a basic helix-loop-helix (bHLH) domain. The mutation in rsf1 results in a truncated protein consisting of the first 129 amino acids but lacking the bHLH domain. Cloning the RSF1 gene strongly suggests that numerous phytochrome A–mediated responses require a bHLH class transcription factor.
INTRODUCTION
Now that the genome of Arabidopsis has been sequenced, this cruciferous plant has emerged as the preferred reference organism for understanding the function of unknown genes in higher plants. The most common method for elucidating the function of unknown genes is to use various mutagenesis procedures, such as insertional mutagenesis (Feldmann, 1991; Koncz et al., 1992; Bancroft and Dean, 1993; Aarts et al., 1995), gene silencing (Baulcombe, 1996; Kooter et al., 1999), and physical or chemical mutagenesis (Redei and Koncz, 1992). Gene disruption by insertional mutagenesis can be accomplished by either transposon mutagenesis (Bancroft and Dean, 1993; Aarts et al., 1995) or Agrobacterium-mediated T-DNA transformation (Feldmann, 1991; Koncz et al., 1992). The advantage of insertional mutagenesis is that the gene affected can be identified easily. However, with few exceptions, such as the recently reported overexpression of PIF3 (for phytochrome-interacting factor) after T DNA insertion in its promoter region (Halliday et al., 1999), insertional mutagenesis lacks the versatility to create partial loss-of-function or gain-of-function alleles, which are often very informative in elucidating the biological function of genes. In contrast, chemical or physical mutagenesis can generate simple sequence mutations, such as point mutations or small deletions and insertions, and could thus be used to dissect all functional domains of proteins by producing a variety of different phenotypes associated with a specific genetic locus. The major drawback in creating mutations by chemical or physical means is the ability to identify the locus responsible for the mutant phenotype because locus identification, unless limited to a few candidate genes (McCallum et al., 2000), entails a laborious process involving map-based cloning or chromosome walking.
Sequence variation between individuals includes single nucleotide polymorphisms (SNPs), deletions, and insertions. These make up the backbone for the mapping and positional cloning of genes responsible for phenotypic traits. By tracking DNA sequence variants that cosegregate with heritable traits, the genes giving rise to these traits can be localized to specific chromosomal locations. The majority of known DNA sequence variants in use over the past 20 years have been polymorphisms affecting the recognition sequence of a restriction endonuclease, commonly referred to as restriction fragment length polymorphisms (RFLPs) (Botstein et al., 1980). With the development of the polymerase chain reaction (PCR), a modified version of RFLPs was developed, cleaved amplified polymorphic sequences (CAPS) (Weining and Langridge, 1991; Konieczny and Ausubel, 1993). The CAPS method uses amplified DNA fragments that are digested with a restriction endonuclease to display any polymorphic restriction sites, and the products are analyzed by agarose gel electrophoresis. In a more sophisticated use of restriction endonucleases as tools to generate molecular markers, genomic DNA is digested with a restriction enzyme followed by linker addition and amplification with random sequence-tagged primers to yield amplified fragment length polymorphisms (AFLPs) (Thomas et al., 1995; Alonso-Blanco et al., 1998). Multiple markers can be analyzed in a single lane of a sequencing gel. The disadvantage of relying on restriction endonucleases to generate molecular markers, however, is that only 28 to 30% of polymorphisms affect the recognition sequence of a restriction endonuclease.
In this study, we describe the use of two novel technologies, high-density oligonucleotide array and denaturing HPLC (DHPLC), to clone an Arabidopsis gene. High-density DNA arrays, which are well known for their application to genomewide expression analysis (Cho et al., 1998; Lockhart and Winzeler, 2000), have also been applied to the large-scale discovery and genotyping of biallelic single nucleotide polymorphisms by direct hybridization with complementary oligonucleotide probes on the array (Wang et al., 1998; Lindblad-Toh et al., 2000). This approach has led to the successful mapping of genes in both yeast (Winzeler et al., 1998) and Arabidopsis (Cho et al., 1999).
DHPLC, in contrast, is an efficient tool for both the discovery and genotyping of polymorphisms in the fine-structure mapping of genes (Giordano et al., 1999; Schriml et al., 2000) and the eventual identification of mutations responsible for particular phenotypes (Ophoff et al., 1996). The technique is based on the detection of heteroduplexes in PCR products of <1000 bp by ion-pair reversed-phase HPLC (Oefner and Underhill, 1998). Under conditions of partial heat denaturation, heteroduplexes are retained for shorter times than are the corresponding perfectly matched homoduplexes, resulting in the appearance of one or more additional peaks in the chromatographic profile. The sensitivity and specificity of DHPLC have been reported to be consistently >96%, clearly surpassing other mutation detection methods, such as single-strand conformation analysis and denaturing gradient gel electrophoresis (O'Donovan et al., 1998; Jones et al., 1999; Wagner et al., 1999; Spiegelman et al., 2000). Currently, both DHPLC and high-density oligonucleotide arrays require the amplification of the target locus before examination; however, both are amenable to high-throughput automation, which is an essential prerequisite in the mapping of genes.
As a test case to demonstrate the validity and complementary nature of high-density oligonucleotide genotyping arrays and DHPLC, we cloned the RSF1 locus (defined by a mutant with reduced sensitivity to far-red light). Phenotypic analysis of this mutant demonstrated that the RSF1 gene encodes an important mediator of phytochrome A (phyA) signaling (Fankhauser and Chory, 2000). The rsf1 mutation identifies one of several loci that have been implicated specifically in signaling downstream of phyA (Bolle et al., 2000; Buche et al., 2000; Hsieh et al., 2000; reviewed in Neff et al., 2000). We show here that RSF1 encodes a basic helix-loop-helix (bHLH) transcription factor with high identity to PIF3, a bHLH protein that interacts with phyA and phyB in vitro and plays a major role in phyB signaling in vivo (Ni et al., 1998; Halliday et al., 1999). The cloning of the RSF1 gene demonstrates that multiple phyA-mediated responses also require the activity of a bHLH class transcription factor.
RESULTS
General Strategy
Genes for Mendelian segregating mutations can be cloned in Arabidopsis by using the AT412 array and DHPLC in three successive steps. The AT412 array is a custom-made high-density oligonucleotide probe array for determining the allelic state of 412 sites known to be polymorphic in Arabidopsis, particularly between the Columbia and Landsberg erecta ecotypes (Cho et al., 1999). The AT412 array is used first to evaluate a limited number (in this case, 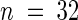 ) of F2 segregating plants that carry the mutant phenotype. This allows the mutation to be mapped onto one of the five Arabidopsis chromosomes to a region within a few centimorgans (cM) of the nearest marker on the chip. Next is the fine-mapping of the gene, which is accomplished by developing new markers by DHPLC and chromosome walking until all or almost all of the recombinants between the gene and the accession genotype used for the genetic cross have been eliminated. Assuming an average recombination rate during the F1 meiosis, this requires the examination of ∼1500 chromosomes carrying the mutant gene. This allows successive narrowing of the interval between the gene and the nearest recombination event to a distance of typically 15 to 30 kb. Ultimately, the entire remaining interval is scanned by tiling across it in overlapping 550-bp sections that are amplified separately from mutant and wild-type genomic DNA before they are mixed and subjected to DHPLC analysis. Sections that yield more than one peak in the chromatographic profile are considered polymorphic, and the exact position and chemical nature of the causative mutation are determined by dye-terminator sequencing.
) of F2 segregating plants that carry the mutant phenotype. This allows the mutation to be mapped onto one of the five Arabidopsis chromosomes to a region within a few centimorgans (cM) of the nearest marker on the chip. Next is the fine-mapping of the gene, which is accomplished by developing new markers by DHPLC and chromosome walking until all or almost all of the recombinants between the gene and the accession genotype used for the genetic cross have been eliminated. Assuming an average recombination rate during the F1 meiosis, this requires the examination of ∼1500 chromosomes carrying the mutant gene. This allows successive narrowing of the interval between the gene and the nearest recombination event to a distance of typically 15 to 30 kb. Ultimately, the entire remaining interval is scanned by tiling across it in overlapping 550-bp sections that are amplified separately from mutant and wild-type genomic DNA before they are mixed and subjected to DHPLC analysis. Sections that yield more than one peak in the chromatographic profile are considered polymorphic, and the exact position and chemical nature of the causative mutation are determined by dye-terminator sequencing.
Allelic Discrimination by the AT412 Array
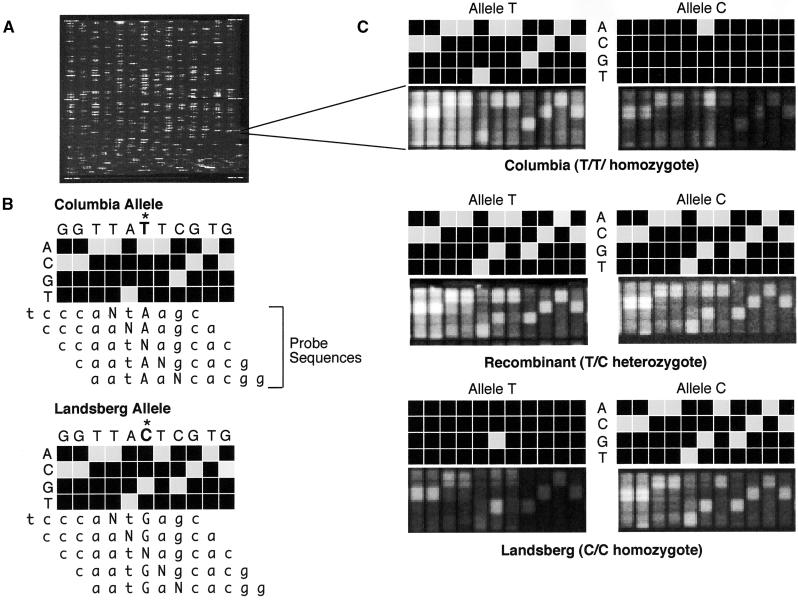
The AT412 array uses the variable detector array (VDA) design to genotype single nucleotide substitutions as well as small deletions or insertions (Wang et al., 1998). Figure 1A shows a fluorescence image of the entire array that was prepared by attaching >72,500 different oligonucleotide probes to the surface of an impermeable glass substrate by photolithographic synthesis (Fodor et al., 1991). These probes represent a total of 1648 VDAs, each consisting of 44 25-mer probes. Four VDAs are required to genotype a simple sequence polymorphism, two for each allele (one for the reverse strand and one for the forward). For each of the 11 positions examined, including the polymorphic site itself and five bases on each side, the VDA has a set of four 25-mer oligonucleotide probes (Figure 1B). These probes are complementary to the reference sequence, except at the central, examined position, for which each of the four nucleotides is substituted in turn. Usually the reference sequence can be read from the hybridization pattern, because the perfectly matching probe yields a much stronger hybridization signal than do the three mismatching oligonucleotide probes in the same column (Figure 1C). This scheme, which tests not only the polymorphic site but also a variety of contexts around the polymorphic site, with mismatches as controls, provides a much more sensitive and reliable assay for genotyping. Ideally, the forward and reverse strand hybridization patterns should corroborate each other. However, quite frequently only one of the two strands yields a hybridization pattern of sufficient quality for accurate allele calling. That is the reason for the redundant nature of the array design.
Figure 1.
Genotyping by Array Hybridization.
(A) Fluorescence image of an entire oligonucleotide probe array after hybridization.
(B) Scheme of the genotyping of the forward strand of SGCSNP4 in Columbia and Landsberg erecta ecotypes on a variant detector high-density oligonucleotide probe array. VDAs using four 25-mer probes that have an A, C, G, or T at the center position (N) are designed to interrogate not only the polymorphic site (marked with an asterisk) but also the flanking five bases on either side. This design allows determination of the sequence context in which the polymorphic site is embedded and adds to the robustness and accuracy of the genotyping assay. The target DNA hybridizes most strongly to the probe that complements its sequence most closely. Therefore, the probe with the correct base at each center position will produce the strongest hybridization signal.
(C) Scans showing the actual and schematic hybridization patterns for homozygous Columbia (top), homozygous Landsberg erecta (bottom), and a heterozygous recombinant (center). In this example, the variant bases are T and C. Hybridization of the T allele to the C allele VDA, and vice versa, result in only one strong hybridization signal in the column that interrogates the polymorphic site itself. Interrogation of the flanking bases yields either no signals or only weak ones because the target sequence does not match perfectly the corresponding probe sequences, given the different allelic states at the polymorphic site.
To enable the amplification of several polymorphic loci in a single PCR, we designed primers with similar calculated melting temperatures that flank the polymorphic site by a few bases on either side. In addition, to accommodate the constraints of hybridization, the PCR target did not exceed a total length of 120 bp. At the 5′ end of the forward primers, an additional 23 bases of T7 sequence were incorporated, and at the 5′ end of the reverse primers, an additional 23 bases of T3 sequence were added. In a secondary reaction, these “tailed” PCR products can be labeled with 5′ biotinylated T7 and T3 primers, which can then be visualized with a streptavidin conjugate.
Individual PCRs were performed to validate the quality of each amplicon. To facilitate the laborious process of amplifying hundreds of PCR products separately, we developed a multiplex PCR strategy in which as many as 55 polymorphic loci were amplified in a single PCR. Our goal with the multiplex strategy was to combine in each multiplex set amplicons having similar amplification efficiencies. In assembling such sets, each marker was amplified individually, and the approximate PCR product yield was determined on a standard ethidium bromide–stained agarose gel. Subsequently, amplicons were grouped according to fluorescence intensities. Each group contained between 45 and 55 markers.
Whole-Genome Mapping of the rsf1 Mutation
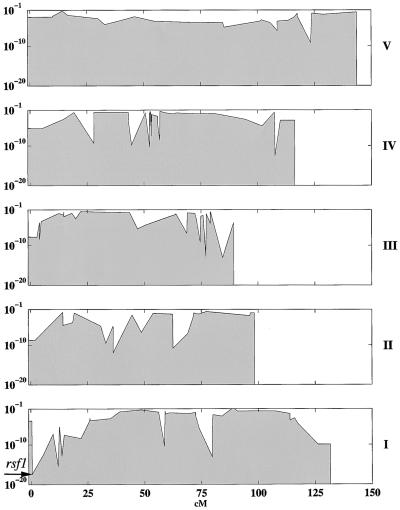
As described previously (Cho et al., 1999), only 235 of the 412 arrayed markers could discriminate accurately between both Columbia and Landsberg erecta homozygous plants and against the heterozygote. Excluding 25 markers from a 150-kb region on chromosome 4 left 210 markers grouped in four pools, which were used to localize the rsf1 mutation in 32 F2 plants from a Columbia rsf1 long hypocotyl in far-red light × Landsberg erecta (Fankhauser and Chory, 2000). Of the 210 SNPs, 182 showed good hybridization. The 28 unexpected failures might have been related to the changes made to the original multiplexing scheme (Cho et al., 1999). No attempt was made to rescue these sites by repooling or changing the protocol because the remaining biallelic sites proved sufficient for the unequivocal mapping of the rsf1 mutation to the top of chromosome 1 between markers SNP5 and SNP247 (Figure 2). This interval encompasses 488,921 bp as determined from publicly available sequence information (http://sequence-www.stanford.edu/ara/chromosome1.html).
Figure 2.
Calculated Probability of Random Segregation in the Arabidopsis Genome for 32 F2 Plants from the Columbia rsf1 × Landsberg erecta Cross That Display Reduced Sensitivity to Far-Red Light.
The x axis represents genetic positions; the y axis (log base 10) indicates the probability of random segregation. The approximate position of rsf1, as determined by the genotyping of 182 biallelic markers on an oligonucleotide array, is indicated by an arrow. The roman numerals at the right side of the plot indicate the five chromosomes of Arabidopsis.
Fine Mapping of the rsf1 Mutation by DHPLC
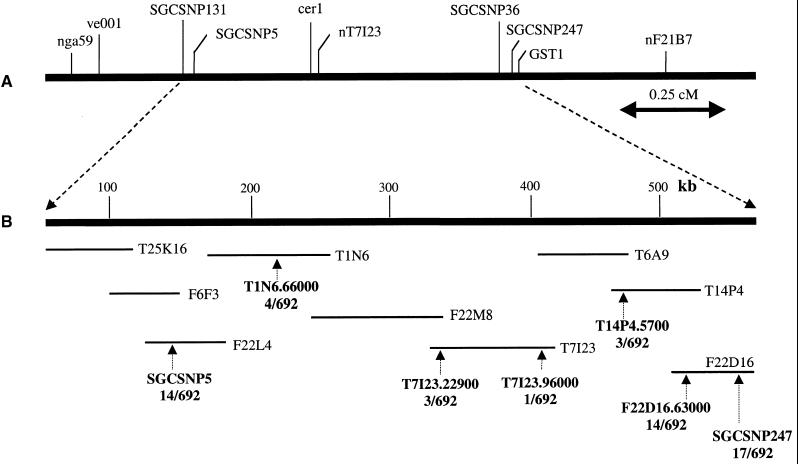
To shorten the interval between the two chip markers, we used DHPLC to determine and genotype additional polymorphic markers. Using primarily intergenic regions as templates, PCR primers were designed approximately every 10,000 bp to yield amplicons with an average size of 600 nucleotides. Mixtures of corresponding fragments amplified from genomic DNA of the Columbia and Landsberg erecta accessions were screened by DHPLC at temperatures recommended by the results of computer simulation of the DNA melting behavior of the amplicons (Jones et al., 1999). In the presence of a mismatch, the characteristic heteroduplex profiles obtained contain two or more clearly resolved peaks, whereas monomorphic samples yield only one peak. The heteroduplexes are generally eluted first. As shown previously, the sensitivity of DHPLC in detecting a single nucleotide polymorphism exceeds 96% (Jones et al., 1999; Spiegelman et al., 2000). Only DHPLC profiles displaying unambiguously resolved heteroduplexes were used for further genotyping. The primers and the temperatures used for genotyping seven markers in 692 F2 plants that had not been prescreened for recombination in this interval are listed in Table 1. Figure 3A shows a genetic map of known phenotypic and molecular markers in the area of interest. Figure 3B shows the contig of bacterial artificial chromosomes (BACs) sequenced between markers SGCSNP5 and SGCSNP247 and the physical positions of the seven markers used to shorten the interval by progressive elimination of recombinants. The gene was fine-mapped to a 55,331-bp interval on the BAC T6A9, between markers T7I23.96000 and T14P4.5700. Overall, the number of polymorphic sites observed at the top of chromosome 1 was ∼10% as many as in other regions of chromosome 1 as well as of chromosomes 2 and 4. From a total of 134,230 bp screened in BAC clones T25K16, T1N6, T7I23, and T14P4, only 45 polymorphic sites were detected, corresponding to one in 2983 nucleotides screened. When we started with BAC F22D16, the polymorphism rate increased to one in 361 nucleotides. This compared well with findings for other regions on chromosomes 1, 2, and 4, in which the polymorphism rate ranged from one in 164 to one in 417. Subsequent comparisons with other ecotypes (Lagostera, Nossen, and Argentat) showed that low nucleotide diversity at the top of chromosome 1 was limited to Landsberg erecta. Given the scarcity of polymorphisms in this region of the Columbia rsf1 × Landsberg erecta cross, we opted to forego further mapping and rely on the proven sensitivity of DHPLC in detecting mutations by screening ∼100 overlapping PCR products that covered the entire interval.
Table 1.
Primers, Amplicon Sizes, and Temperatures Used for DPHLC-Based Genotyping of Polymorphic Fragments in Fine Mapping of the rsf1 Mutation
| Marker Name | Forward Primer (5′ to 3′) | Reverse Primer (5′ to 3′) | Size (bp) | Temperature (°C) |
|---|---|---|---|---|
| SNP5 | CATGCTTTGACTGGTCAATAAG | AACTGGACTATGATCACTGTGA | 259 | 58 |
| T1N6.66000 | GGTTTCGTAAACACAGAAATCA | CTTCTCTTAGTTTCCGGCTTC | 520 | 54 |
| T7I23.22900 | TGAGCTTGATTAGTATCATTCCA | ATTATTCGTCACACCATCCATA | 569 | 53 |
| T7I23.96000 | GTTCAAACCCCAAAATTCAATG | AATATTCAAAGGTATAGCAGG | 371 | 48 |
| T14P4.5700 | TGTGTGAATACTGATTGAAGCC | ATCAAATTTGCCTCCTTAAGTT | 54 | 54 |
| F22D16.63000 | CAGATTCAAAAATCCAAACATG | TCTGGATGGAAATTGATTGTTG | 448 | 56 |
| SNP247 | CTACTTCAATAAATCTCCACC | GTGTTTCTGCTTTCTATTATTGC | 429 | 53 |
Figure 3.
Chromosome Walking toward the RSF1 Gene.
(A) Genetic map positions, in relation to other known phenotypic and molecular markers, of SGCSNP5 and SGCSNP247, the two SNPs that mapped the rsf1 mutation to the top of chromosome 1 by array hybridization.
(B) Physical map of the BACs sequenced between SGCSNP5 and SGCSNP247 as well as the positions of the seven markers (see Table 1) used to shorten the interval by DHPLC-based genotyping of 692 F2 plants. The 14 recombinants that were heterozygous at SGCSNP5 were gradually reduced to one recombinant at T7I23.96000. Correspondingly, the 17 recombinants heterozygous at SGCSNP247 were reduced to three at T14P4.5700. The remaining gap between markers T7I23.96000 and T14P4.5700 occupied 55,331 bp.
Identification of the RSF1 Locus
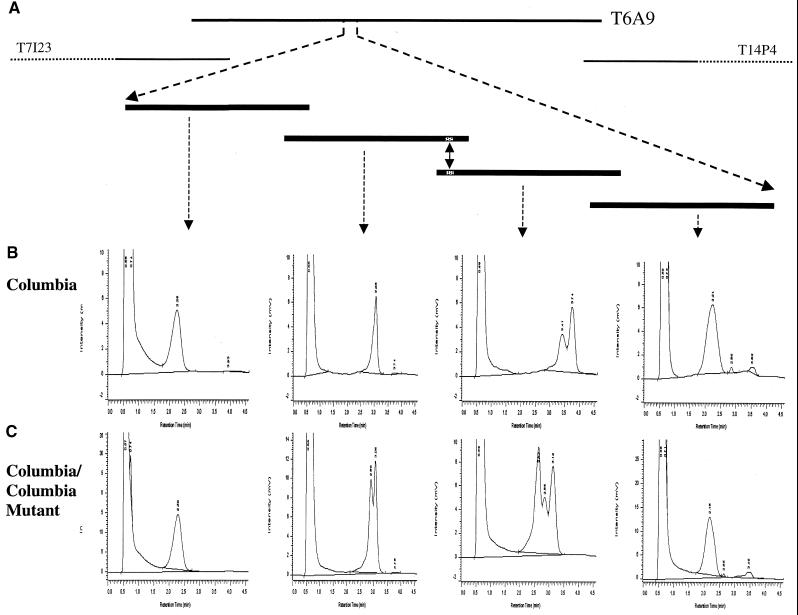
To identify the rsf1 mutation, we mixed corresponding amplicons from the rsf1 mutant and the Columbia wild type in an equimolar ratio and subjected them to DHPLC analysis at the temperatures recommended by the melting algorithm. For this purpose, the entire interval between markers T7I23.22900 and T14P4.5700 was amplified by PCR in fragments of ∼550 bp with a minimal overlap of 30 bp. This is depicted schematically in Figure 4A for the four amplicons that cover the immediate vicinity of the mutation. The genomic sequence of BAC T6A9 was used as a template for the design of primers. Typically, the appearance of one or more additional peaks in a chromatographic profile indicates the presence of a mutation. However, because enzyme slippage during amplification or the presence of a second locus amplified with the same set of primers sometimes may cause the appearance of additional peaks aside from the homoduplex peak (Spiegelman et al., 2000), we recommended analyzing the wild-type DNA alone (Figure 4B) as a control against which the chromatographic profile obtained for the mixture of wild-type and mutant is then compared (Figure 4C). Of the ∼100 amplicons analyzed, two overlapping fragments showed characteristic changes in the chromatographic profiles when the rsf1 mutant and the parent strain Columbia were mixed (the second and third chromatogram in Figure 4C, respectively), indicative of a mismatch. Subsequent dye-terminator sequencing showed that the polymorphic DHPLC profiles were the result of a 13-bp deletion in the rsf1 mutant.
Figure 4.
Detection of the rsf1 Mutation by Using DHPLC.
(A) Approximate physical position of the rsf1 mutation on BAC T6A9 and a schematic representation of the four overlapping PCR products that covered the mutation (indicated by double-headed arrow) and its immediate vicinity.
(B) DHPLC profiles for the four overlapping DNA fragments that had been amplified from parent strain Columbia only.
(C) DHPLC profiles obtained after mixing the corresponding amplicons from Columbia wild type and Columbia mutant. The additional peak seen in the second and third profiles represents the heteroduplex created by the 13-bp deletion in the mutant.
The x axis under the chromatographic profiles in (B) and (C) shows retention time in minutes; and the y axis shows intensity in millivolts (mV).
Several expressed sequence tag (EST) clones matching this part of BAC T6A9 (GenBank accession number AC064879) 100% were found in the Arabidopsis database. Sequencing the EST clones 173B11T7 and 209K19T7 showed the first one to be incomplete at the 5′ end, whereas the second one appeared to be full length because it was the same size as the RSF1 mRNA, as determined by RNA gel blot analysis (data not shown). The RSF1 gene has five exons and is predicted to encode a 292–amino acid protein (Figure 5). Database searches with BLAST programs showed that the gene product in the region between amino acids 140 and 200 was highly homologous with bHLH-containing proteins. The closest homolog to RSF1 is the gene encoding bHLH transcription factor PIF3 (a phytochrome-interacting protein), which plays an important role in phytochrome-mediated light signaling (Ni et al., 1998; Halliday et al., 1999). The homology is very obvious in the bHLH region but also extends ∼30 amino acids toward the N terminus and 30 amino acids toward the C terminus of this domain. Over the region of these 120 amino acids, RSF1 and PIF3 are 35% identical and 50% similar (Figure 6). The 13-bp deletion in exon 3 results in a frameshift, such that the rsf1 mutant is predicted to code for a protein with the same 129 first amino acids but followed by 16 novel amino acids and a stop codon (Figure 7). Interestingly, the bHLH domain is lost in this mutant form. However, we have detected no significant reduction in RSF1 mRNA quantities by RNA gel blotting of rsf1 mutants (data not shown).
Figure 5.
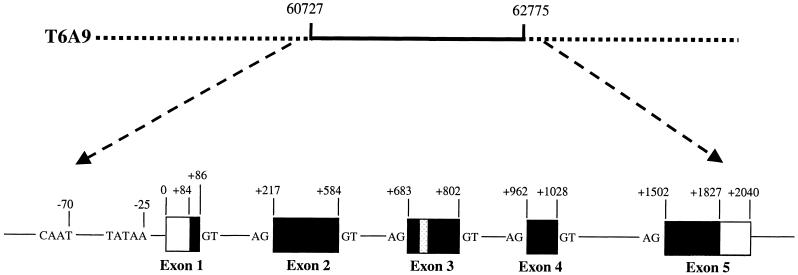
Genomic Structure of RSF1 and Location of the 13-bp Deletion.
The genomic sequence was derived from the BAC clone T6A9, which contained the entire gene between nucleotides 60,727 and 62,775. RSF1 has five exons. A TATA box and a CAAT box were found 25 and 70 nucleotides upstream of exon 1, respectively.
Figure 6.
Comparative Amino Acid Sequence Analysis of Proteins Encoded by RSF1 (Top, Amino Acids 105 to 224) and PIF3 (Bottom, Amino Acids 302 to 431) in the Region Containing the bHLH Domain.
The homology is apparent in the bHLH region but also extends ∼30 amino acids to the N terminus and 30 amino acids to the C terminus of this domain. Over these 120 amino acids, RSF1 and PIF3 are 35% identical and 50% similar. The 13-bp deletion in the rsf1 mutant causes a truncated protein of 145 amino acids that lacks the bHLH domain. Gray regions represent amino acids that are identical between RSF1 and PIF3. White type represents amino acids that are similar. The underlined region denotes the bHLH domain of the PIF3 gene. Dashes indicate amino acids absent from either the RSF1- or PIF3-encoded sequences.
Figure 7.
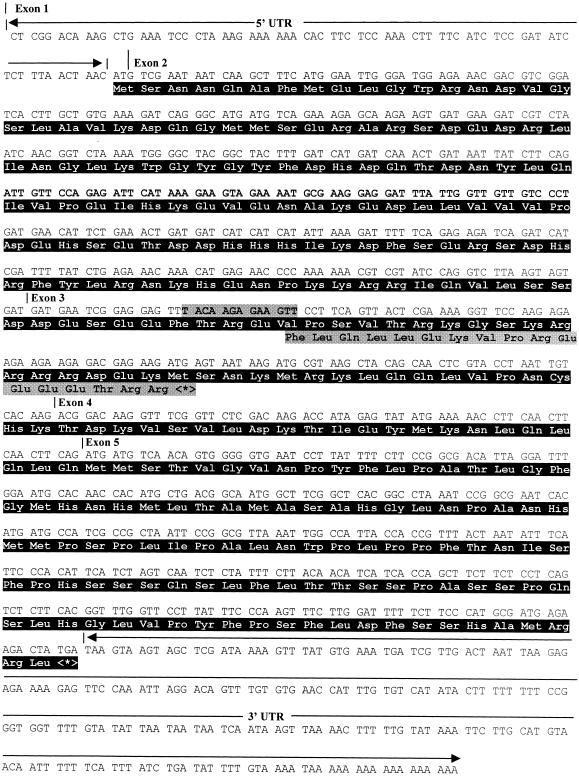
The cDNA Sequence of RSF1.
The cDNA sequence was derived from EST clone 209K19T7. Exons are numbered consecutively, and the positions of introns 1 to 4 are marked by small vertical lines next to the exon numbers. The complete coding sequence and the deduced amino acid sequence (in black area) of RSF1 are shown. The stippled area in the nucleic acid sequence of exon 3 indicates the 13 bp deleted. The consequence of the mutation, a frameshift, on amino acid sequence is also depicted in a stippled box. UTR, untranslated region.
Rescue of rsf1 with the Wild-Type Gene
To demonstrate that this 13-bp deletion is responsible for the phenotype of the rsf1 mutants, we sequenced the entire 50 kb between our closest recombinants and found no other mutation. Moreover, we transformed rsf1 mutants with a construct encompassing 2.8 kb of genomic DNA, including 0.7 kb of sequence 5′ of the transcription start site. The rsf1 mutants have hypocotyl phenotypes in both blue and far-red light (Fankhauser and Chory, 2000). Because it is easier to score for both kanamycin resistance and the RSF1 phenotype in blue light than in far-red light, we scored T1 transformants in blue light 5 days after germination. The majority of transformants (24 of 29) were rescued by this genomic construct, confirming that the 13-bp deletion in the RSF1 gene causes the observed phenotype. This was also confirmed in recent independent studies by Fairchild et al. (2000) and Soh et al. (2000).
DISCUSSION
In this study, we describe the successful genomewide and subsequent fine-structure mapping of the rsf1 mutation by oligonucleotide array hybridization and DHPLC analysis with simple sequence polymorphisms as genetic markers. In general, this highly efficient strategy can be used to clone genes in any organism for which controlled crosses of inbred strains are feasible. In the future, extending this approach to mapping quantitative trait loci (QTL) will allow the determination of genes involved in complex traits.
High-density DNA arrays, which are well known for their use in studying genomewide patterns of gene expression (Cho et al., 1998; Lockhart and Winzeler, 2000), also represent powerful tools for detection of DNA variation and subsequent parallel genotyping of large numbers of genetic markers (Wang et al., 1998; Steinmetz and Davis, 2000). Arrays have been already applied successfully to the genomewide mapping of phenotypic traits in yeast (Winzeler et al., 1998) and Arabidopsis (Cho et al., 1999). For the smaller and less complex yeast genome, total genomic DNA, after limited digestion with DNase I and end-labeling with biotin, can be hybridized to an oligonucleotide array designed for gene expression monitoring that represents ∼20% of the yeast genome (Winzeler et al., 1998). An imperfect match in the mutant strain results in a loss of hybridization signal, which essentially acts as a marker. This scheme has allowed construction of an inheritance map for the entire yeast genome with an average resolution of 3.5 kb.
For more complex organisms, such as Arabidopsis, the size and complexity of the genome do not allow for direct genomic DNA hybridizations to the array. To increase target concentration and decrease sample complexity, PCR amplification is required before array hybridization. By combining ⩾50 markers with similar PCR amplification efficiencies, as determined by singleplex reactions, hundreds of polymorphic sequences can be amplified in a few PCRs. This allows the array hybridization to be much more efficient in terms of both costs and labor. Critical multiplexing strategies include minimizing the concentration of each primer in the primer cocktail and using a higher than normal magnesium concentration to compensate for the high molarity of nucleic acids and oligonucleotides and for the number of amplifications in the reaction.
The high-density DNA array is very useful because it enables one to analyze many markers at the same time. The use of many probes is successful because of the specificity in detecting individual target sequences within complex mixtures. To maximize array hybridization, no more than seven cytosines in a 20-mer probe or three C residues in a window of eight bases can be used; otherwise, the result will be indiscriminate hybridization. Additional sequence contexts known to cause poor hybridization, in a 20-mer probe, include more than eight A residues; more than nine T residues; 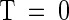 ; more than four consecutive A, G, or T residues; >10 repeats of the same pair of bases; palindromes longer than six bases; >13 bases A and T; and >14 bases A and G. Another challenge is that primer pairs with similar melting characteristics are needed for amplification of polymorphic fragments ⩽120 bp long. Considering that not all SNPs are amenable to genotyping on the array, approximately twice as many polymorphisms must be identified than can be genotyped. However, the usefulness and simplicity of the genotyping protocol outweigh the efforts required to discover which SNPs are abundant between Arabidopsis accessions.
; more than four consecutive A, G, or T residues; >10 repeats of the same pair of bases; palindromes longer than six bases; >13 bases A and T; and >14 bases A and G. Another challenge is that primer pairs with similar melting characteristics are needed for amplification of polymorphic fragments ⩽120 bp long. Considering that not all SNPs are amenable to genotyping on the array, approximately twice as many polymorphisms must be identified than can be genotyped. However, the usefulness and simplicity of the genotyping protocol outweigh the efforts required to discover which SNPs are abundant between Arabidopsis accessions.
Using the AT412 array for genomewide mapping of the rsf1 mutation to the top of Arabidopsis chromosome 1 in 32 F2 individuals homozygous for the rsf1 allele took less than a week. In its current configuration, AT412 is a prototype high-density array used to validate the efficiency of tiling arrays as a tool for genomewide mapping; it is by no means optimized for genetic studies with Arabidopsis. The AT412 array used here was designed to examine 412 biallelic markers between the accessions Columbia and Landsberg erecta. As described by Cho et al. (1999), however, only 56% of the markers can discriminate homozygous and heterozygous genotypes. Although the average genetic distance between the useful markers is 3.5 cM, some areas in the genome have gaps of at least 10 cM. But with the entire genome of Arabidopsis sequenced, we can now create a genotyping array that contains markers at predetermined intervals, for example, every 50 or 100 kb. Even though a genotyping array of such high marker density is unnecessary for mapping monogenic traits, high-density marker arrays will be valuable tools for mapping QTL.
Whereas DNA arrays are able to genotype known polymorphisms between given ecotypes, they will probably be most useful for accessions for which the SNP has been identified. For instance, the AT412 array was designed to interrogate DNA variations between Columbia and Landsberg erecta. If another ecotype is crossed with one of the reference accessions, approximately half of the markers on the array will be useful. The proportion of useful markers can be as low as 30% when two random accessions are analyzed for polymorphisms with the AT412 array (data not shown).
Previously, DHPLC was used to identify several hundred simple sequence polymorphisms that distinguish the Arabidopsis ecotypes Columbia and Landsberg erecta. This approach was economical and efficient as long as ESTs and cDNA sequences were the primary source of sequence for determining DNA sequence variants. With the completion of the Arabidopsis sequencing project, however, direct sequencing of intergenic regions is a far more efficient strategy for the discovery of polymorphisms. Simple sequence polymorphisms, including single-nucleotide substitutions and small insertions and deletions, can be expected approximately every 200 to 400 nucleotides. In the fine-structure mapping of genes, however, DHPLC may still play an important role, as shown in this study, because it can be used for both the discovery and genotyping of sequence variants without prior knowledge of their chemical nature. Because markers in fine-structure mapping are rarely revisited, the cost of genotyping increasingly fewer F2 plants cannot be matched by any other genotyping technology that relies on primary sequence information. Moreover, further improvements in throughput of DHPLC are imminent, in light of the recent description of capillary HPLC columns that can be arrayed and combined with laser-induced fluorescence detection in an arrangement analogous to that of a capillary array sequencer (Premstaller et al., 2000).
DHPLC is useful in detecting and genotyping simple sequence polymorphisms, though it provides only dominant marker scoring unless the samples are mixed with a known reference (Steinmetz et al., 2000). The greatest advantages of DHPLC, however, are its high sensitivity and specificity, which consistently approach 100% (O'Donovan et al., 1998; Jones et al., 1999; Wagner et al., 1999; Spiegelman et al., 2000). These attributes, combined with its low operating cost, make DHPLC an effective and practical method in the automated mutational analysis of inherently monomorphic regions of the genome, such as the human Y chromosome (Shen et al., 2000), and in the targeted screening of chemically induced mutations (McCallum et al., 2000). With a throughput of as many as 300 samples per day, 50 kb of sequence can be scanned in <1 day of analysis. By including an inbred monomorphic control in the analysis, the mutation can be detected by simple visual comparison of the DHPLC profiles.
The recessive mutant rsf1 identified in this study exhibits reduced sensitivity to far-red inhibition of hypocotyl elongation. rsf1 mutants are also affected in other phyA-mediated responses such as anthocyanin accumulation and the opening and expansion of cotyledons in far-red light (Fankhauser and Chory, 2000). The quantities of phyA photoreceptor protein are not affected in rsf1 mutants, strongly suggesting that RSF1 is a major phyA signaling component (Fankhauser and Chory, 2000). Thus, cloning the RSF1 gene provides us with valuable information concerning the mechanism of phyA signaling. Recently, several genes involved in phyA signaling have been cloned (Hoecker et al., 1999; Hudson et al., 1999; Bolle et al., 2000; Hsieh et al., 2000). RSF1 provides us with immediate information concerning the biochemical mechanisms involved; the other three, SPA1 (for suppressor of phyA-105), FAR1 (for far-red–impaired response), and PAT1 (for phyA signal transduction), encode proteins with known protein domains but without assigned biochemical functions.
The currently identified phytochrome signaling components fall into two classes: cytoplasmic and nuclear proteins (Bolle et al., 2000; reviewed in Neff et al., 2000). This probably reflects the complexity of phytochrome responses and suggests that light-activated phytochromes initiate a signaling cascade with several branches. A recent series of publications strongly suggest that one phytochrome signaling route involves light-regulated import of the photoreceptor into the nucleus, followed by interaction of phytochrome with transcription factors such as PIF3 (Martinez-Garcia et al., 2000; Nagatani, 2000). RSF1 most likely belongs to the nuclear signaling branch. While the present study was under review, Fairchild et al. (2000) reported the isolation of HFR1 (for long hypocotyl in far-red), which is allelic to RSF1. Their data suggest that no direct interaction takes place between HFR1/RSF1 and the phytochromes but HFR1/RSF1 can heterodimerize with PIF3.
Although PIF3 was originally identified as a protein interacting with both phyA and phyB, more recent biochemical and physiological data suggest that PIF3 plays a more prominent role in phyB signaling than in phyA signaling (Ni et al., 1998; Halliday et al., 1999; Martinez-Garcia et al., 2000). In contrast, based on the rsf1 loss-of-function phenotype, RSF1 is implicated in phyA signaling but not phyB signaling. The identification of transcription factors specifically implicated in phyA or phyB signaling is particularly interesting in view of some recent results demonstrating that signaling initiated by phyA and phyB converges on the same promoter elements (Cerdan et al., 2000). It will be important to test whether these elements are the target sites for transcription factors such as those encoded by PIF3 and RSF1.
Although focusing on the rsf1 gene, the strategy presented here should be taken as an example of how to improve efficiency when cloning genes in Arabidopsis among other organisms. Improvements (e.g., cost and more amenable markers) in the techniques presented here will undoubtedly facilitate the cloning of genes responsible for a given phenotype. For example, the AT412 genotyping chip is currently relatively expensive, and because many of the 412 markers on the array are not ideal for genotyping, it is inefficient. In terms of decreasing these inefficiencies, a generic high-density oligonucleotide array can be designed that contains thousands of preselected 20-mer oligonucleotide tags (Shoemaker et al., 1996) for use with single-nucleotide primer extension reactions (Fan et al., 2000). This approach offers advantages such as increased specificity and resolution of secondary structures because allele discrimination by primer extension is conducted at higher temperatures than in previous methods. However, more steps are required, and the overall efficiency is only slightly higher than that of the AT412 chip, with 29 of 171 SNPs tested failing in multiplex PCRs and only 114 of the remaining 142 SNPs yielding distinct genotype clusters for the homozygote or the heterozygote.
Most of our knowledge of the genetic factors influencing phenotypic traits is based on single genes or alleles. However, in nature, much of the diversity that occurs in a population is attributable to multigenic traits or QTL, which are affected by environmental factors. Here, we looked at a single-gene trait; however, elucidating the complexity of quantitatively inherited phenotypes is one of the main challenges geneticists will face in the future. The limited number of genetic markers has hindered the cloning of QTL and the molecular understanding of quantitative traits. Once such a map is constructed, the use of high-density oligonucleotide arrays and DHPLC can be used to study what genetic factors are involved in quantitative characteristics such as height, seed size, root and stem length, and crop yield.
METHODS
Mutagenesis and Generation of rsf1 Plants
The rsf1 mutant was identified by screening T2 Arabidopsis thaliana seeds of a T-DNA collection of mutants with reduced sensitivity to far-red light. Because the mutation had not been caused by a T-DNA tag, a chromosome walk was undertaken to identify the mutant gene. The isolation of the mutant, the phenotypic characterization, and the identification of mutant F2 plants in a cross between rsf1 (Columbia background) and Landsberg erecta have been described elsewhere (Fankhauser and Chory, 2000). For fine-mapping, we used DNA from 692 F2 recombinants.
Amplification of Biallelic Markers
Genomic DNA from rsf1 mutant plants was extracted as described previously (Neff et al., 1998). We used 210 of the 412 loci present on the AT412 genotyping array (Affymetrix, Santa Clara, CA) for whole-genome mapping. The polymerase chain reaction (PCR) primers used had similar calculated melting temperatures (60°C) and were designed to amplify the polymorphic site in amplicons ⩽120 bp. The T7 sequence was incorporated at the 5′ end of forward primers (5′-TAATACGACTCACTATAGGGAGA…genomic sequence-3′), whereas the T3 sequence was incorporated at the 5′ end of reverse primers (5′-AATTAACCCTCACTAAAGGGAGA…genomic sequence-3′). In this way, the “tailed” PCR products could be labeled in a secondary reaction with 5′-biotinylated T7 and T3 primers. Singleplex PCRs were performed in a 20-μL volume containing 20 ng of Arabidopsis DNA, 0.005 to 0.01 μM each primer, 1 unit of AmpliTaq Gold DNA polymerase (Applied Biosystems, Foster City, CA), 250 μM deoxynucleotide triphosphates, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, and 2.5 mM MgCl2. For thermocycling, we used an Applied Biosystems GeneAmp PCR system 9700, with initial denaturation/activation at 95°C for 10 min, followed by 14 cycles of denaturation at 95°C for 20 sec, annealing at 63°C for 1 min, and primer extension at 72°C for 1 min; in this case, the primer annealing temperature was decreased 0.5°C in each consecutive cycle. Twenty additional cycles were then performed, of 95°C for 20 sec, 56°C for 45 sec, and 72°C for 1 min, with a final extension reaction at 72°C for 10 min. PCR product yields were determined by the semiquantitative assessment of fluorescence intensities on ethidium bromide–stained agarose gels. Amplicons with similar intensities were placed into four groups, averaging 52 amplicons (or 104 primers per group), each having a final concentration of ∼62.5 nM.
A multiplex PCR strategy was then designed to reduce the time and labor expended in amplifying polymorphic loci. After grouping the amplicons into four different groups based on amplification efficacy, PCRs were repeated at various reannealing temperatures. For the two groups with high intensities, 60°C for the first primer annealing stage of the first cycle and 53°C for the second cycle were found to be optimal. For the two groups of intermediate original amplifications, PCR with annealing temperatures of 57 and 52°C for the first and second cycles, respectively, was found to maximize amplification. Assessing the effect of changing MgCl2 concentration between 1.5 and 7.5 mM showed that, overall, the temperature had the greatest effect on amplification efficiency; however, MgCl2 at 7.5 mM was crucial for maximizing the PCR.
The initial multiplexed reaction was secondarily amplified by using 5′-biotinylated T7 and T3 primers. This secondary labeling PCR was performed in a total volume of 30 μL, with 0.6 μL of the multiplex reaction, 0.15 μM each biotinylated primer, 1 unit of AmpliTaq Gold DNA polymerase, 250 μM deoxynucleotide triphosphates, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, and 2.5 mM MgCl2. Thermocycling was performed with an initial denaturation/activation at 95°C for 10 min, followed by 30 cycles at 95°C for 30 sec, 52°C for 45 sec, and 72°C for 1 min. A final extension reaction was performed at 72°C for 10 min. Products of the four 30-μL multiplex reactions for each of the 32 recombinant inbred plants were then pooled, and 76 μL of this sample was used for array hybridization.
Array Hybridization
The AT412 array has been described previously (Cho et al., 1999). The probe array was prewashed with 6 × SSPET (1 × SSPET = 0.15 NaCl, 10 mM NaH2PO4, 1 mM EDTA, pH 7.4, and 0.005% Triton X-100) at 25°C for 10 min and washed once with 3 M tetramethylammonium-chloride (Sigma) for 5 min. The 76-μL biotinylated sample was denatured at 95°C for 5 min, cooled immediately on ice, and brought to a final volume of 200 μL with the addition of hybridization cocktail (3 M tetramethylammonium chloride, 10 mM Tris-HCl, pH 8.0, 1 mM EDTA, 0.01% Triton X-100, 0.1 mg/mL herring sperm DNA [Promega], and 200 pM control oligomer). The 200-μL final volume was applied to the array at 42°C and rotated for 16 to 24 hr on a rotisserie at ∼60 rpm.
Afterward, the probe array was washed on a fluidics station (FS400; Affymetrix) 10 times with 6 × SSPET at 22°C and then three times with 1 × SSPET. Staining was performed at room temperature with 2 mg/mL streptavidin R–phycoerythrin (Molecular Probes, Eugene, OR) and 0.5 mg/mL acetylated BSA (Sigma) in 6 × SSPET. After staining, the probe array was washed another five times with 6 × SSPET at 22°C.
The probe array was scanned on the GeneArray scanner (HP G2500A; Affymetrix) at ∼40 to 80 pixels per probe feature with a 570-nm filter. Affymetrix GeneChip software was used to generate a digitized intensity table for each of the features on the chip. These signal data and the positional information were then processed sequentially by using InterMap analysis package software to compute the probability of observing each segregation pattern (as previously described by Cho et al., 1999). The package is available at ftp://tairpub:tairpub @ftp.arabidopsis.org/home/tair/Software. Marker scores for each genotype from a homozygous recombinant inbred panel were used to compute the expected score for each genotype and its variance. Markers were genotyped as heterozygous when their score was >3 sd from both homozygous control scores. The P value for each marker was directly computed by
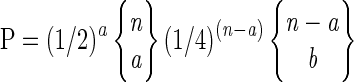 |
where n is the total number of F2 mutant genotypes, a is the number that are heterozygous, and b is the number of homozygous Landsberg genotypes.
PCR Amplification to Search for Novel Single Nucleotide Polymorphism Markers
The sequences of the bacterial artificial chromosome (BAC) clones between SGCSNP5 and SGCSNP247, at the bottom of chromosome 1, were downloaded from www.ncbi.nlm.nih.gov. Oligonucleotide primers 18 to 27 bases long were designed, with a bias for intergenic regions, to have a uniform melting temperature of 60°C. These primers yielded amplicons between 500 and 600 bp. PCR amplifications were performed in 50-μL volumes containing ∼50 ng of genomic DNA from both Columbia and Landsberg erecta, 0.15 μM each primer, 1 unit of AmpliTaq Gold DNA polymerase, 250 μM deoxynucleotide triphosphates, 10 mM Tris-HCl, pH 8.3, 50 mM KCl, and 2.5 mM MgCl2. Thermocycling was initiated with a denaturing/activating step at 95°C for 10 min, followed by 20 cycles of 95°C for 20 sec, 60°C (under touchdown conditions) for 1 min, and 72°C for 1 min. Another 25 cycles were performed at 95°C for 20 sec, 52°C for 45 sec, and 72°C for 1 min, followed by a final extension at 72°C for 7 min. Success of amplification was verified by electrophoresis of the products on agarose gel.
Screening for Markers by Denaturing HPLC
Before denaturing HPLC (DHPLC) analysis, corresponding Columbia and Landsberg erecta amplicons were mixed in a roughly equimolar ratio, denatured at 95°C for 3 min, and allowed to renature over 30 min by decreasing the temperature gradually from 95 to 65°C. Each mixture was analyzed, along with the Columbia monomorphic control, with an automated DHPLC instrument (WAVE; Transgenomic, San Jose, CA) equipped with a DNASep column (Transgenomic) at the recommended temperature predicted for each amplicon mixture by the melting algorithm available at http://insertion.stanford.edu/melt.html (Jones et al., 1999). The mobile phase was 0.1 M triethylammonium acetate (Applied Biosystems), pH 7.0, and 0.1 mM Na4EDTA (Sigma). DNA fragments were eluted at a flow rate of 0.9 mL/min in a gradient with acetonitrile (J.T. Baker, Phillipsburg, NJ). The start and end points of the gradient were calculated with the WAVE MAKER (version 3.3.4; Transgenomic) program, and slight adjustments were made after the first run to ensure a time window of 1.5 min between the void and the amplicon peak. Between runs, the column was flushed for 0.5 min with 50% acetonitrile by using the WAVE accelerator and reequilibrated for 1.5 min at the starting conditions of the gradient. Chromatograms were examined visually for differences between the Columbia control only and the Columbia–Landsberg erecta duplex. Only markers with clearly resolved homoduplex and heteroduplex species were used for subsequent genotyping.
Cycle Sequencing
Ten microliters of each PCR amplicon was treated with 1 unit each of exonuclease I and shrimp alkaline phosphatase for 30 min at 37°C and 15 min at 80°C to degrade excess primers and deoxynucleotide triphosphates. The PCR amplicons were then sequenced by using the Applied Biosystems BigDye Deoxy Terminator cycle-sequencing kit according to the manufacturer's instructions. The sequencing reactions were purified with Sephadex G-50 (Amersham Pharmacia, Piscataway, NJ), and analyzed with an Applied Biosystems 377A sequencer.
Complementation of the rsf1 Mutant
For complementation of the rsf1 mutant, we amplified 2.8 kb of genomic DNA encompassing the RSF1 gene from BAC T6A9 (nucleotides 60,634 to 63,431) with primers containing a BamHI linker. We used 20 amplification cycles and DyNAzyme EXT DNA polymerase (Finnzymes, Espoo, Finland) according to the manufacturer's instructions. The PCR product was digested with BamHI and ligated into BamHI-digested and phosphatase-treated pPZP211 vector (Hajdukiewicz et al., 1994). The resulting construct (CF262) was sequenced to ensure for the absence of PCR-generated mutations.
CF262 was introduced into Agrobacterium tumefaciens GV3101. rsf1 plants were transformed with the spray method described by Weigel et al. (2000). T1 seeds were plated on half-strength Murashige and Skoog medium (Gibco BRL; 10632-016) containing 0.7% Phytagar (Gibco BRL; 10675-023) and 30 μg/mL kanamycin. Plated seeds were stratified for 3 days at 4°C, and the plates were then moved into constant 8 μmol m−2 sec−1 blue light (λmax 469 nm) at 22°C in an incubator (E30-LED; Percival, Boone, IA) and scored 5 days after germination.
Acknowledgments
We thank the Arabidopsis stock center (Arabidopsis Biological Resource Center, Columbus, OH) for providing EST clones 173B11T7 and 209K19T7. We are grateful to Consuelo Solomon in Geneva for technical assistance and to Dr. Curt Palm and Ed Allen at Stanford University for sequence annotation and graphics, respectively. This work was supported by National Institutes of Health Grant Nos. R01 GM61973 (to P.J.O.) and 2RO1 GM52413 (to J.C.) and the Swiss National Science Foundation Grant No. 631-58 151.99 (to C.F.). J.C. is an Associate Investigator of the Howard Hughes Medical Institute.
References
- Aarts, M.G.M., Corzaan, P., Stiekema, W.J., and Pereira, A. (1995). A two-element enhancer–inhibitor transposon system in Arabidopsis thaliana. Mol. Gen. Genet. 247 555–564. [DOI] [PubMed] [Google Scholar]
- Alonso-Blanco, C., Peeters, A.J.M., Koornneef, M., Lister, C., Dean, C., van den Bosch, N., Pot, J., and Kuiper, M.T.R. (1998). Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J. 14 259–271. [DOI] [PubMed] [Google Scholar]
- Bancroft, I., and Dean, C. (1993). Heterologous transposon tagging of the DRL1 locus in Arabidopsis. Plant Cell 5 631–638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulcombe, D.C. (1996). RNA as a target and an initiator of post-transcriptional gene silencing in transgenic plants. Plant Mol. Biol. 32 79–88. [DOI] [PubMed] [Google Scholar]
- Bolle, C., Koncz, C., and Chua, N.-H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Botstein, D., White, R.L., Skolnick, M., and Davis, R.W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32 314–331. [PMC free article] [PubMed] [Google Scholar]
- Buche, C., Poppe, C., Schafer, E., and Kretsch, T. (2000). eid1: A new Arabidopsis mutant hypersensitive in phytochrome A–dependent high-irradiance responses. Plant Cell 12 547–558. [PMC free article] [PubMed] [Google Scholar]
- Cerdan, P.D., Staneloni, R.J., Ortega, J., Bunge, M.M., Rodriguez-Batiller, M.J., Sanchez, R.A., and Casal, J.J. (2000). Sustained but not transient phytochrome A signaling targets a region of an Lhcb1*2 promoter not necessary for phytochrome B action. Plant Cell 12 1203–1212. [PMC free article] [PubMed] [Google Scholar]
- Cho, R.J., Campbell, M.J., Winzeler, E.A., Steinmetz, L., Conway, A., Wodicka, L., Wolfsberg, T.G., Gabrielian, A.E., Landsman, D., Lockhart, D.J., and Davis, R.W. (1998). A genome-wide transcriptional analysis of the mitotic cell cycle. Mol. Cell 2 65–73. [DOI] [PubMed] [Google Scholar]
- Cho, R.J., et al. (1999). Genome-wide mapping with biallelic markers in Arabidopsis thaliana. Nat. Genet. 23 203–207. [DOI] [PubMed] [Google Scholar]
- Fairchild, C.D., Schumaker, M.A., and Quail, P.H. (2000). HFR1 encodes an atypical bHLH protein that acts in phytochrome A signal transduction. Genes Dev. 14 2377–2391. [PMC free article] [PubMed] [Google Scholar]
- Fan, J.B., Chen, X., Halushka, M.K., Berno, A., Huang, X., Ryder, T., Lipshutz, R.J., Lockhart, D.J., and Chakravarti, A. (2000). Parallel genotyping of human SNPs using generic high-density oligonucleotide tag arrays. Genome Res. 10 853–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser, C., and Chory, J. (2000). RSF1, an Arabidopsis locus implicated in phytochrome A signaling. Plant Physiol. 124 39–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmann, K.A. (1991). T-DNA insertion mutagenesis in Arabidopsis: Mutational spectrum. Plant J. 1 71–82. [Google Scholar]
- Fodor, S.P., Read, L.J., Pirrung, M.C., Stryer, L., Lu, A.T., and Solas, D. (1991). Light-directed, spatially addressable parallel chemical synthesis. Science 251 767–773. [DOI] [PubMed] [Google Scholar]
- Giordano, M., Oefner, P.J., Underhill, P.A., Cavalli-Sforza, L.L., Tosi, R., and Richiardi, P. (1999). Identification by denaturing high-performance liquid chromatography of numerous polymorphisms in a candidate region for multiple sclerosis susceptibility. Genomics 56 247–253. [DOI] [PubMed] [Google Scholar]
- Hajdukiewicz, P., Svab, Z., and Maliga, P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25 989–994. [DOI] [PubMed] [Google Scholar]
- Halliday, K.J., Hudson, M., Ni, M., Qin, M., and Quail, P.H. (1999). poc1: An Arabidopsis mutant perturbed in phytochrome signalling because of a T DNA insertion in the promoter of PIF3, a gene encoding a phytochrome-interacting bHLH protein. Proc. Natl. Acad. Sci. USA 96 5832–5837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284 496–499. [DOI] [PubMed] [Google Scholar]
- Hsieh, H.L., Okamoto, H., Wang, M., Ang, L.H., Matsui, M., Goodman, H., and Deng, X.W. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14 1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hudson, M., Ringli, C., Boylan, M.T., and Quail, P.H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, A.C., Austin, J., Hansen, N., Hoogendoorn, B., Oefner, P.J., Cheadle, J.P., and O'Donovan, M.C. (1999). Optimal temperature selection for mutation detection by denaturing high performance liquid chromatography and comparison to SSCP and heteroduplex analysis. Clin. Chem. 45 1133–1140. [PubMed] [Google Scholar]
- Koncz, C., Nemeth, K., Redei, G.P., and Schell, J. (1992). T-DNA insertional mutagenesis in Arabidopsis. Plant Mol. Biol. 20 963–976. [DOI] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Kooter, J.M., Matzke, M.A., and Meyer, P. (1999). Listening to the silent genes: Transgene silencing, gene regulation and pathogen control. Trends Plant Sci. 4 340–347. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh, K., et al. (2000). Large-scale discovery and genotyping of single-nucleotide polymorphisms in the mouse. Nat. Genet. 24 381–386. [DOI] [PubMed] [Google Scholar]
- Lockhart, D.J., and Winzeler, E.A. (2000). Genomics, gene expression and DNA arrays. Nature 405 827–836. [DOI] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Huq, E., and Quail, P.H. (2000). Direct targeting of light signals to a promoter element–bound transcription factor. Science 288 859–863. [DOI] [PubMed] [Google Scholar]
- McCallum, C.M., Comai, L., Greene, E.A., and Henikoff, S. (2000). Targeting induced local lesions IN genomes (TILLING) for plant functional genomics. Plant Physiol. 123 439–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagatani, A. (2000). Plant biology. Lighting up the nucleus. Science 288 821–822. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Neff, J.D., Chory, J., and Pepper, A.E.. (1998). dCAPS, a simple technique for the genetic analysis of single nucleotide polymorphisms: Experimental applications in Arabidopsis thaliana genetics. Plant J. 14 387–392. [DOI] [PubMed] [Google Scholar]
- Neff, M.M., Fankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14 257–271. [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95 657–667. [DOI] [PubMed] [Google Scholar]
- O'Donovan, M.C., Oefner, P.J., Roberts, S.C., Austin, J., Hoogendoorn, B., Guy, C., Speight, G., Upadhyaya, M., Sommer, S.S., and McGuffin, P. (1998). Blind analysis of denaturing high-performance liquid chromatography as a tool for mutation detection. Genomics 52 44–49. [DOI] [PubMed] [Google Scholar]
- Oefner, P.J., and Underhill, P.A. (1998). DNA mutation detection using denaturing high-performance liquid chromatography. In Current Protocols in Human Genetics, N.C. Dracopoli, J.L. Haines, B.R. Korf, D.T. Moir, C.C. Morton, C.E. Seidman, J.G. Seidman, and D.R. Smith, eds (New York: John Wiley & Sons), pp. 7.10.1–7.10.12. [DOI] [PubMed]
- Ophoff, R.A., et al. (1996). Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4. Cell 87 543–552. [DOI] [PubMed] [Google Scholar]
- Premstaller, A., Oberacher, H., and Huber, C.G. (2000). High-performance liquid chromatography–electrospray ionization mass spectrometry of single- and double-stranded nucleic acids using monolithic capillary columns. Anal. Chem. 72 4386–4393. [DOI] [PubMed] [Google Scholar]
- Redei, P.G., and Koncz, C. (1992). Classical mutagenesis. In Methods in Arabidopsis Research, C. Koncz, N.-H. Chua, and J. Schell, eds (Singapore: World Scientific Publishing Co.), pp. 16–82.
- Schriml, L.M., Peterson, R.J., Gerrard, B., and Dean, M. (2000). Use of denaturing HPLC to map human and murine genes and to validate single-nucleotide polymorphisms. BioTechniques 28 740–745. [DOI] [PubMed] [Google Scholar]
- Shen, P., et al. (2000). Population genetic implications from se-quence variation in four Y chromosome genes. Proc. Natl. Acad. Sci. USA 97 7354–7359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoemaker, D.D., Lashkari, D.A., Morris, D., Mittmann, M., and Davis, R.W. (1996). Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat. Genet. 14 450–456. [DOI] [PubMed] [Google Scholar]
- Soh, M.-S., Kim, Y.-M., Han, S.-J. and Song, P.-S. (2000). REP1, a basic helix-loop-helix pattern, is required for a branch pathway of phytochrome A signaling in Arabidopsis. Plant Cell 12 2061–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiegelman, J.I., Mindrinos, M.N., and Oefner, P.J. (2000). High accuracy DNA sequence variation screening by DHPLC. BioTechniques 29 2–7. [DOI] [PubMed] [Google Scholar]
- Steinmetz, L.M., and Davis, R.W. (2000). High-density arrays and insights into genome function. Biotechnol. Genet. Eng. Rev. 17 109–146. [DOI] [PubMed] [Google Scholar]
- Steinmetz, L.M., Mindrinos, M., and Oefner, P.J. (2000). Combining genome sequences and new technologies for dissecting the genetics of complex phenotypes. Trends Plant Sci. 5 396–400. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M., Vos, P., Zabeau, M., Jones, D.A., Norcott, K.A., Chadwick, B.P., and Jones, J.D.G. (1995). Identification of amplified restriction fragment polymorphism (AFLP) markers tightly linked to the tomato Cf-9 gene for resistance to Cladosporium fulvum. Plant J. 8 785–794. [DOI] [PubMed] [Google Scholar]
- Wagner, T., Stoppa-Lyonnet, D., Fleischmann, E., Muhr, D., Pagès, S., Sandberg, T., Caux, V., Moeslinger, R., Langbauer, G., Borg, A., and Oefner, P. (1999). Denaturing high-performance liquid chromatography (DHPLC) detects reliably BRCA1 and BRCA2 mutations. Genomics 62 369–376. [DOI] [PubMed] [Google Scholar]
- Wang, D.G., et al. (1998). Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280 1077–1082. [DOI] [PubMed] [Google Scholar]
- Weigel, D., et al. (2000). Activation tagging in Arabidopsis. Plant Physiol. 122 1003–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weining, S., and Langridge, P. (1991). Identification and mapping of polymorphisms in cereal based on the polymerase chain reaction. Theor. Appl. Genet. 82 209–216. [DOI] [PubMed] [Google Scholar]
- Winzeler, E.A., Richards, D.R., Conway, A.R., Goldstein, A.L., Kalman, S., McCullough, M.J., McCusker, J.H., Stevens, D.A., Wodicka, L., Lockhart, D.J., and Davis, R.W. (1998). Direct allelic variation scanning of the yeast genome. Science 281 1194–1197. [DOI] [PubMed] [Google Scholar]