Abstract
The irregular xylem 1 (irx1) mutant of Arabidopsis has a severe deficiency in the deposition of cellulose in secondary cell walls, which results in collapsed xylem cells. This mutation has been mapped to a 140-kb region of chromosome 4. A cellulose synthase catalytic subunit was found to be located in this region, and genomic clones containing this gene complemented the irx1 mutation. IRX1 shows homology to a previously described cellulose synthase (IRX3). Analysis of the irx1 and irx3 mutant phenotypes demonstrates that both IRX1 and IRX3 are essential for the production of cellulose in the same cell. Thus, IRX1 and IRX3 define distinct classes of catalytic subunits that are both essential for cellulose synthesis in plants. This finding is supported by coprecipitation of IRX1 with IRX3, suggesting that IRX1 and IRX3 are part of the same complex.
INTRODUCTION
Cellulose is a polymer of β(1,4)-linked glucose, with each glucose residue oriented 180° to its neighbor such that the polymeric repeating unit is cellobiose. This allows the chain to adopt a flat, ribbonlike structure (Brown et al., 1996). In the plant cell wall, ∼36 individual β(1,4)-glucose chains crystallize to form microfibrils (Delmer and Amor, 1995).
Cellulose is an essential component of both the primary and secondary cell walls of higher plants. In expanding plant cells, the cellulose–xyloglucan network is considered the main load-bearing network that controls the extent of cell expansion, whereas the orientation of cellulose microfibril deposition has a major role in controlling the direction of cell expansion (Fisher and Cyr, 1998). After a period of expansion, certain cell types lay down a thick secondary cell wall inside the primary wall. Cellulose can make up to 90% of the dry weight of these secondary walls. In some cells, such as those in the xylem, lignin may also be incorporated into these walls, contributing to their mechanical strength.
Cellulose synthesis is relatively well understood in two bacterial species, Acetobacter xylinum and Agrobacterium tumefaciens, which require four and five genes, respectively, for cellulose synthesis. Only one of these genes shows homology between those species, however, and that is the catalytic subunit (Saxena et al., 1990; Matthysse et al., 1995). Analysis of expressed sequence tags from developing cotton fibers led to the identification of plant sequences showing homology with the bacterial gene. This observation, along with the ability of the products to bind UDP glucose, the substrate for cellulose biosynthesis, indicated that those sequences were likely to be the catalytic subunit of the higher plant cellulose synthase complex (Pear et al., 1996). This was confirmed by genetic analysis of Arabidopsis mutants deficient in cellulose deposition in the primary cell wall (rsw1) or secondary cell wall (irx3) (Arioli et al., 1998a; Taylor et al., 1999). Both of these mutations are the result of a mutation in different members of a large family of Arabidopsis genes showing homology to bacterial cellulose synthases.
Cellulose synthesis in higher plants occurs at the plasma membrane. Freeze-fracture studies of the plasma membrane have shown unique “rosette” structures associated with the ends of microfibrils, suggesting that these rosettes are the site of cellulose synthesis (reviewed in Brown, 1996). Confirmation of these results has come from Arabidopsis (rsw1) and barley (brittle culm) mutants that have less cellulose than their respective wild types (Kokubo et al., 1991; Arioli et al., 1998a). Plasma membranes from the cells affected in these mutants exhibit greatly decreased numbers of rosettes. Kimura et al. (1999) recently showed that antibodies recognizing the catalytic central region of a cotton cellulose synthase specifically label rosette structures in freeze-fracture studies, demonstrating that these are indeed the site of cellulose synthesis. The rosette structures are a characteristic feature of cellulose synthesis in higher plants. In view of the importance of the orientation of cellulose microfibrils in determining cell shape, how cellulose is synthesized by these rosettes and how the orientation of cellulose deposition by these rosettes is controlled by the cytoskeleton are central to a further understanding of plant morphology.
The nearing completion of the Arabidopsis genome sequence has made clear that most genes are not single-copy genes but rather are members of small multigene families (Lin et al., 1999). Understanding the role of different family members is an essential part of exploiting this genome information. At least 40 Arabidopsis genes show substantial homology with bacterial cellulose synthases (http://cellwall.stanford.edu/cellwall/index.shtml). Genes known to be involved in cellulose synthesis, such as IRX3 and RSW1, appear to define a subset of this family (Arioli et al., 1998a; Taylor et al., 1999). This subgroup, described as CesA genes, still contains at least 10 family members. To enable use of a common nomenclature, a new naming system has been adopted (Delmer, 1999). Under this system, IRX3 corresponds to AtCesA7 and RSW1 corresponds to AtCesA1 (see above [cellwall] website). The function of most of these genes remains unknown. IRX3 affects secondary cell walls and has little affect on primary cell walls, whereas RSW1 is essential for cellulose synthesis in the primary cell wall (Turner and Somerville, 1997; Arioli et al., 1998a). Although different family members may be required for cellulose synthesis in different tissues under various different conditions, whether more than one family member is essential for cellulose synthesis in the same cell type has remained an open question.
Two other complementation groups, irx1 and irx2, exhibiting the same phenotype of decreased cellulose and irregular xylem have also been described (Turner and Somerville, 1997). In the present study, we describe the isolation of the gene responsible for the irx1 phenotype. The IRX1 gene also appears to encode a cellulose synthase catalytic subunit. Thus, IRX1 and IRX3 define two distinct classes of catalytic subunit, both of which are required for cellulose synthesis in the same cell type. This provides a novel insight into the synthesis of cellulose in plants and raises several interesting questions about the composition and complexity of the rosette structures.
RESULTS
Determination of the Map Position of irx1
Preliminary linkage studies involving 12 F2 irx1 mutant plants suggested that the irx1 locus was linked to two published cleaved amplified polymorphic sequences (CAPS) markers on chromosome 4—g4539 (http://genome-www.stanford.edu/Arabidopsis/maps/CAPS.Chr4.html) and AG (Konieczny and Ausubel, 1993)—and that it was located between the two (data not shown). These markers are located at 54.83 (g4539) and 60.35 centimorgans (AG) on the recombinant inbred (RI) map for Arabidopsis chromosome 4 (Lister and Dean, 1993; http://www.nasc.nott.ac.uk/RI.data/gifs/chrom4.gif).
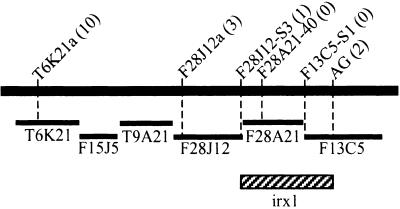
Plants from a test cross between irx1 and Columbia were screened for linkage between the irx1 locus and the g4539 or AG markers. From the 663 plants screened, a total population of 24 recombinant individuals was identified: 22 for g4539 and two for AG. Further novel simple sequence length polymorphism and CAPS markers were identified in the region between AG and g4539. Figure 1 shows that analysis of the recombinants with these novel markers placed irx1 between the markers F28J12-S3 and AG. Consequently, the gene must reside within the region spanned by the bacterial artificial chromosome (BAC) clone F28A21 (94 kb) and the first part (44 kb) of BAC clone F13C5 (Figure 1). While we were refining this map position, the release of annotated sequence from BAC F28A21 (GenBank accession number AL035526) revealed the presence of a homolog of the cotton cellulose synthase gene CelA1 (GhCesA1) (Pear et al., 1996). This Arabidopsis homolog gene was denoted F28A21.190. Because irx1 plants show a specific defect in cellulose content, it was considered that F28A21.190 might be a good candidate for the gene defective in irx1.
Figure 1.
Map Position of irx1 on Chromosome 4 Relative to Several Molecular Markers.
Black bar represents a portion of chromosome 4. Markers are shown above, with the number of recombinants with irx1 given in parentheses. Bars underneath represent the positions of BACs spanning this region. Hatched box denotes region in which irx1 falls.
Cloning a cDNA Corresponding to F28A21.190
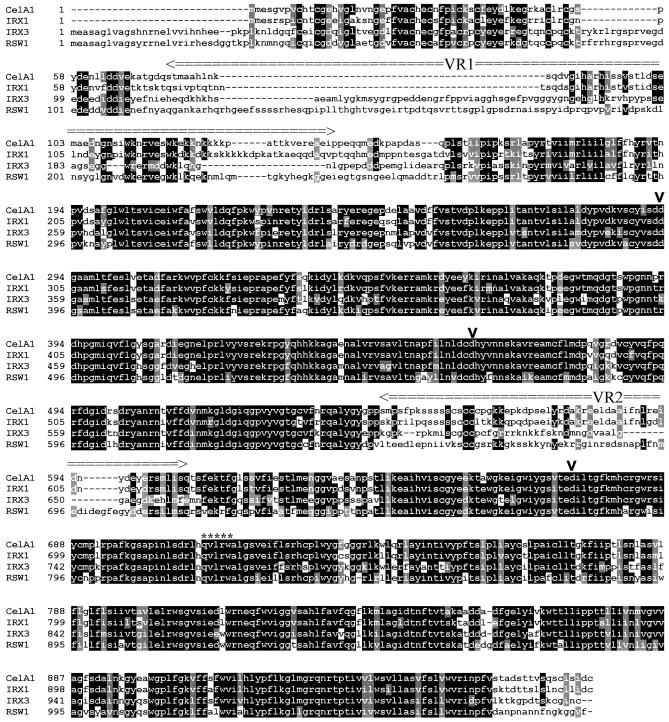
To identify a cDNA that corresponded to the F28A21.190 gene, we constructed a cDNA library produced from stem material. This library was screened with a probe derived from the conserved central region of the IRX3 gene, which has been shown to be a region of conservation between plant cellulose synthase genes (Taylor et al., 1999). One of the clones isolated from this screen contained the entire coding region of the F28A21.190 gene. This sequence has been deposited in GenBank (accession number AF267742) and corresponds to AtCesA8 (see cellwall website, above). This cDNA encodes a predicted protein of 985 amino acids with a predicted molecular mass of 111 kD and a pI of 6.75. Comparison of the cDNA and genomic sequences shows that, in common with IRX3 (Taylor et al., 1999), IRX1 contains 12 exons and 11 introns. In common with other plant cellulose synthase genes, IRX1 encodes a predicted membrane protein having a cytosolic N terminus followed by two membrane-spanning domains (Pear et al., 1996; Arioli et al., 1998a; Taylor et al., 1999). The central portion of the protein is cytosolic with six predicted transmembrane domains at the C terminus. The central cytosolic region also contains, in common with all other plant cellulose synthases, four motifs that have been identified as being conserved in cellulose synthases and in all processive glucosyl transferases (Saxena et al., 1995). The first three of these motifs are centered around aspartate residues, whereas the fourth consists of a QxxRW motif (where x denotes any amino acid). Figure 2 shows the marked sequence similarity between the cellulose synthases whose identity has been confirmed by genetic analysis or partial biochemical characterization. IRX1 shows more similarity to cotton CelA1 (83.5% identity) than to any of the currently identified Arabidopsis genes. This is particularly apparent at the N terminus, where both proteins lack ∼30 residues that are present in the other proteins. Nonetheless, both proteins still contain a cysteine-rich region at their N terminus. This cysteine-rich region has been previously suggested to form a LIM-like zinc finger motif (Delmer, 1998); however, it more resembles the consensus for a RING finger motif (Saurin et al., 1996). These domains have been suggested to be involved in protein–protein interactions (Saurin et al., 1996). The similarity of IRX1 and CelA1 is also apparent in the regions of the protein that have been identified as “variable” regions, where other cellulose synthase genes show very limited or no homology. This close homology in the variable region may mean that IRX1 is the Arabidopsis ortholog of cotton CelA1.
Figure 2.
Alignment of the Amino Acid Sequences of Several Plant Cellulose Synthase Genes.
Solid boxes indicate regions in which more than half of the residues are identical; gray boxes indicate conserved residues. The positions of three aspartic acid (D) residues and QxxRW motifs are indicated by vertical arrowheads and asterisks, respectively. Variable regions VR1 and VR2 are also indicated. Dashes were introduced to optimize alignment. Antibodies were raised against the VR1 from IRX1 (amino acids 75 to 149) and against the VR1 from IRX3 (amino acids 114 to 203).
Isolation of Mutant Alleles of IRX1
To confirm that F28A21.190 corresponded to the gene causing the defect in cellulose production in irx1 plants, it was necessary to identify the mutation causing this defect. Reverse transcription–polymerase chain reaction (RT-PCR) was used to clone the alleles from irx1-1 and irx1-2. The mutant alleles were amplified in two halves, with two independent clones being sequenced for each allele to control for the possibility of nucleotide misincorporation by Taq polymerase. irx1-1 presented a G-to-A nucleotide substitution in both clones. The corresponding substitution was also demonstrated in two independently amplified clones from irx1-1 genomic DNA. This G-to-A substitution results in replacing an aspartate residue with an asparagine residue (D683N). The aspartate residue replaced in irx1-1 is the third of the conserved aspartate residues thought to be essential for cellulose synthase activity (Pear et al., 1996). irx1-2 had a C-to-T nucleotide substitution in both clones and also in the corresponding region of genomic DNA. This substitution, which results in the alteration of a serine residue to a leucine residue (S679L), occurs within four residues of the third conserved aspartate residue described above and is within a region conserved among all CesA genes.
Complementation of the irx1 Phenotype with the Wild-Type Gene
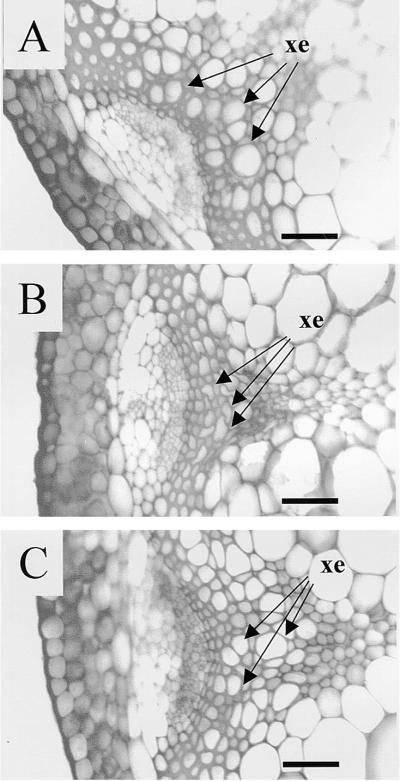
To prove definitively that the F28A21.190 gene corresponded to the IRX1 gene, we identified an 8.1-kb SalI fragment from F28A21 that contained the entire coding region of the gene, along with 2555 bp of the 5′ sequence and 1669 bp of the 3′ sequence. Analysis of this fragment of DNA predicts the presence of portions of two other genes in addition to the CesA gene. The 5′ region contains most of the coding sequence of a predicted protein that shows similarity to a rice integral membrane protein, and the 3′ region of this fragment contains most of the coding sequence for a predicted protein showing similarity to a human transforming protein myb. Neither of these genes is complete, however, and both are unlikely to affect the outcome of the complementation experiments. The 8.1-kb fragment was cloned into a binary vector (pCB2300). The resulting construct (pCS52) was transformed into Agrobacterium and used to transform irx1 plants. Figure 3 shows the phenotype of the resulting transformants. In wild-type plants transformed with an unrelated β-glucuronidase (GUS) reporter gene (pP8GUS), the xylem elements are seen to be open and regular in shape (Figure 3A). In comparison, irx1-1 plants transformed with the same construct show the characteristic inward collapse of the xylem elements (Figure 3B). irx1-1 plants transformed with pCS52 showed a wild-type phenotype (Figure 3C).
Figure 3.
Toluidine Blue–Stained Sections of Arabidopsis Vascular Bundles Showing Complementation of irx1-1.
(A) Wild type transformed with pP8GUS.
(B) irx1-1 transformed with pP8GUS.
(C) irx1-1 transformed with pCS52.
xe, xylem elements.  .
.
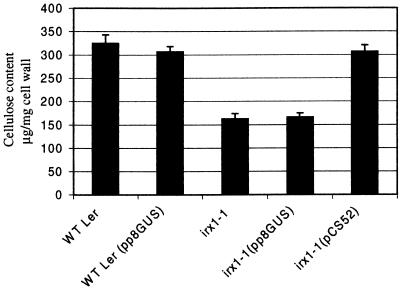
Figure 4 shows biochemical analysis of the stems of these plants, demonstrating that irx1-1 plants have approximately half of the cellulose that the wild type does, but irx1-1 plants transformed with pCS52 show a return to wild-type values. The controls in which irx1-1 or wild-type Landsberg erecta were transformed with pP8GUS exhibited no differences from the corresponding untransformed irx1-1 or wild-type plants. Similar results were obtained with irx1-2 transformants (data not shown). Hence, the 8.1-kb SalI fragment contains the gene able to complement both the collapsed xylem and cellulose-deficient phenotype of irx1 plants.
Figure 4.
Cellulose Measurements Showing Complementation of irx1-1.
Error bars represent standard error. Values are the mean of measurements from five independent transformants. WT Ler, wild-type Landsberg erecta.
IRX1 and IRX3 Are Expressed in the Same Tissues
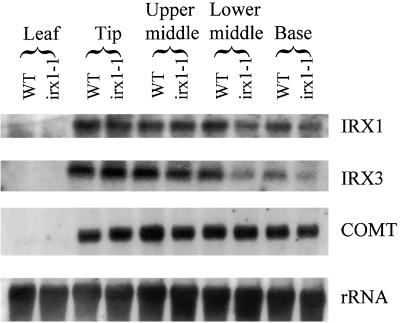
IRX3 has previously been demonstrated to be the catalytic domain of cellulose synthase and is required for secondary rather than primary cell wall synthesis. Xylem elements of irx1 and irx3 exhibit a very similar characteristic collapsed xylem phenotype (Turner and Somerville, 1997). To examine the expression pattern of these genes, we extracted RNA from leaves and four discrete stem sections (the tip, upper middle, lower middle, and base) from mature wild-type and irx1-1 plants. Figure 5 shows the results of probing this RNA with probes derived from IRX1 and IRX3. Neither gene is expressed in leaves, but both are highly expressed in the stems, showing identical expression patterns for IRX1 and IRX3. This is consistent with both genes being involved only in secondary cell wall synthesis. The lower two segments of the irx1-1 stems exhibit a slight decrease in the amount of transcript for both IRX1 and IRX3, but this probably reflects that the mutant plants grow slightly more slowly than the wild type (Turner and Somerville, 1997), and so the mutant plants may be at a slightly different developmental stage than the wild type. For controls, these blots were also probed with COMT, a component of the lignin biosynthesis pathway; the results showed that the irx1-1 mutation has no effect on a gene involved in the lignin biosynthetic pathway.
Figure 5.
RNA Gel Blots Showing Expression of the IRX1 Gene.
Blots containing RNA from developing stems and leaves from wild-type (WT) and irx1-1 plants were probed with IRX1, IRX3, COMT, and rRNA.
irx1 and irx3 Are Phenotypically Indistinguishable in the Xylem
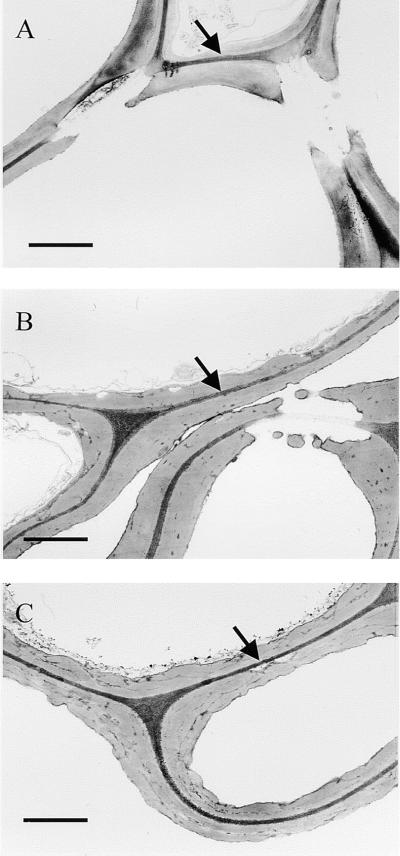
Although the patterns of expression of IRX1 and IRX3 transcripts are similar, that does not necessarily demonstrate a requirement for the expression of both transcripts in the same cell. Evidence that IRX1 and IRX3 are both required in the same cell comes from careful examination of the irx1 and irx3 phenotypes. In vascular bundles of the stem, the irx1 and irx3 mutations affect both early-forming protoxylem and later-forming metaxylem, causing the collapse or distortion of all, or the large majority of, xylem elements within a vascular bundle. To further confirm the similarity of the irx1 and irx3 phenotypes, we examined xylem cell walls by transmission electron microscopy. Figure 6A shows that cell walls of xylem elements from wild-type plants have a characteristic appearance; they tend to stain poorly and have a well-defined and smooth edge. In the interfascicular region of irx3 plants, however, the secondary wall has a very irregular appearance (Turner and Somerville, 1997). This irregular appearance of the cell wall is also seen in the cell walls of the xylem elements (Figure 6B). The walls of irx1 plants (Figure 6C) look apparently identical to those of irx3 cell walls (Figure 6B), having the same characteristically dark-staining cell wall material deposited irregularly in comparison with the pattern in the wild type. Thus, even at the ultrastructural level, the same cell types exhibit an identical phenotype when either IRX1 or IRX3 is mutated. Clearly, the same cell types are affected by the irx1 and irx3 mutations, and the products of both of these genes are required within the same cell to give normal levels of cellulose deposition in the secondary cell wall.
Figure 6.
Ultrastructure of Xylem Element Cell Walls.
Transmission electron microscopy was used to study cell wall structure.
(A) Wild type.
(B) irx3.
(C) irx1-1.
Arrows indicate middle lamellae.  .
.
Interaction between IRX3 and IRX1
Because both IRX1 and IRX3 appear to be required in the same cell types, it is important to determine whether they interact directly and are part of the same protein complex. To address this question, we placed an epitope tag at the N terminus of the IRX3 protein. This tag was inserted between amino acids 3 and 4 of IRX3 and contained the amino acid sequence RGSHHHHHH. The RGSHHHH residues form the epitope for recognition by a highly specific monoclonal antibody, and the hexahistidine tag allows purification by conventional methods of immobilized metal affinity chromatography. This epitope-tagged IRX3 was transformed into irx3 mutant plants (NHisIRX3), and was found to complement the mutation fully (data not shown).
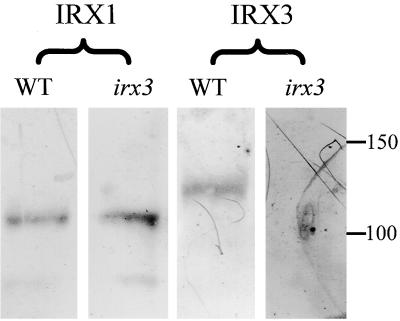
To detect IRX1 and IRX3, we raised polyclonal antibodies against variable region 1 from each of these proteins (Figure 2). These antibodies specifically recognize IRX1 and IRX3, respectively. Figure 7 shows protein gel blots demonstrating this specificity. Each antibody recognized a single band in wild-type extracts that show a difference in mobility between IRX1 and IRX3. In addition, the anti-IRX3 antibody did not recognize a band in irx3 extracts, indicating that this is a null mutation. Importantly, no other bands were detected, demonstrating the specificity of this antibody. The anti-IRX1 antibody recognizes a band of the correct size, however, demonstrating that there is no cross-reaction between the antibodies. These isoform-specific antibodies also demonstrate that IRX1 and IRX3 are regulated independently.
Figure 7.
Specificity of IRX1 and IRX3 Antibodies.
Protein gel blots of wild-type and irx3 extracts probed with anti-IRX1 antibody and anti-IRX3 antibody. Molecular mass markers are given at right in kilodaltons. WT, wild type.
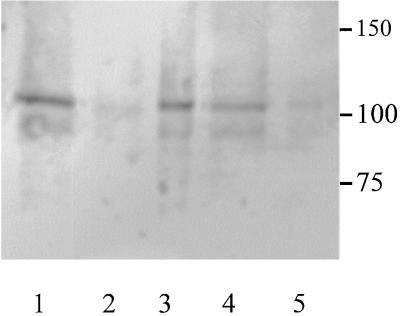
Because both IRX1 and IRX3 are predicted to be membrane proteins, extracts were solubilized in various detergents. Figure 8 shows the result of solubilization in 2% Triton X-100. When no detergent was present, all of the IRX1 protein was found in the pellet after centrifugation at 100,000g, indicative of a membrane protein. After solubilization in 2% Triton X-100, however, >90% of the IRX1 was found in the supernatant after 100,000g centrifugation, indicating that the protein had been solubilized. Under the same conditions, IRX3 showed a similar pattern of solubilization (data not shown). The presence of a faint band at ∼90 kD is thought to be the result of proteolysis of IRX1. This band appears only during manipulation of samples and is not seen when samples are processed immediately (Figure 7).
Figure 8.
Solubilization of IRX1 in Triton X-100.
Protein gel blot probed with the anti-IRX1 antibody. Lane 1 contains total protein after clarification; lane 2, supernatant after centrifugation at 100,000g; lane 3, pellet after 100,000g centrifugation; lane 4, supernatant of solubilized extract after 100,000g centrifugation; lane 5, pellet from solubilized extract after 100,000g centrifugation. Molecular mass markers are given at right in kilodaltons.
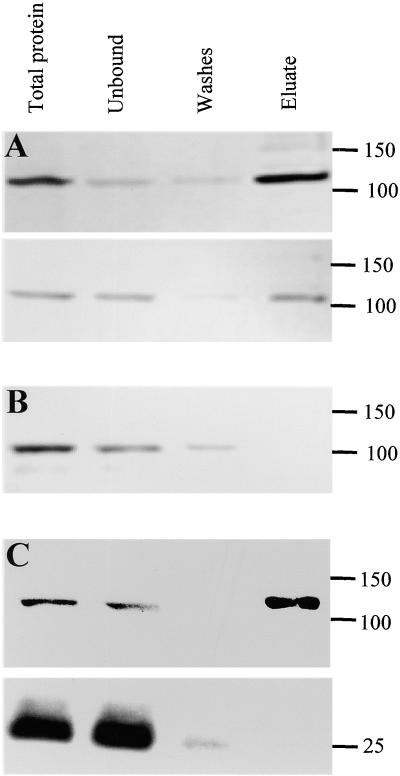
The solubilized extract was then bound to nickel resin, which was used to purify the solubilized NHisIRX3. The top panel in Figure 9A shows a protein gel blot probed with anti- RGSHHHH antibody to determine the purification of NHisIRX3. This antibody recognizes a single band and does not react with extracts from plants not containing the epitope tag (data not shown). As Figure 9A shows, a large proportion of the NHisIRX3 protein binds to the nickel matrix. In addition, this bound protein may be eluted with imidazole, which is consistent with the binding being due to the hexahistidine sequence. Using an identical blot probed with the IRX1-specific polyclonal antibody demonstrated that IRX1 shows a very similar pattern to IRX3, remaining bound to the resin until eluted with imidazole, indicating that IRX1 and IRX3 are associated (Figure 9A). The results are not the result of a nonspecific interaction of IRX1 with the nickel matrix because performing the same experiment with wild-type plants (i.e., plants in which IRX3 was not modified) did not retain IRX1 on the resin (Figure 9B). Thus, the binding of IRX1 to the nickel matrix is dependent on the epitope-tagged IRX3. To determine that the association of IRX1 with IRX3 was not the result of the presence of other unrelated proteins, we used another abundant plasma membrane protein, aquaporin, as a control. Figure 9C shows that aquaporin does not copurify with IRX3, as was detected by using a specific anti-aquaporin polyclonal antibody (Daniels et al., 1994). Similar experiments in which the IRX3-specific antibody was used to coimmunoprecipitate IRX1 have given identical results (data not shown).
Figure 9.
Copurification of IRX3 and IRX1 as Shown by Protein Gel Blots.
(A) NHisIRX3 probed with anti-RGSHHHH antibody (top) and anti-IRX1 antibody (bottom).
(B) Wild type probed with anti-IRX1 antibody.
(C) NHisIRX3 probed with anti-RGSHHHH antibody (top) and anti-aquaporin antibody (bottom).
Molecular mass markers are shown at right in kilodaltons.
DISCUSSION
irx1 plants show a reduction in stem cellulose content to 40% of wild-type amounts. This leads to an alteration of the physical properties of the stems, with decreases in both stiffness and strength (Turner and Somerville, 1997). irx1 plants also show a collapsed xylem phenotype because of the inability of the xylem elements to withstand the negative pressure generated by water transport.
The position of this mutation was mapped to a 140-kb region of chromosome 4 containing BAC clone F28A21. Analysis of the sequence of this BAC revealed the presence of a cellulose synthase gene. RT-PCR was used to isolate the mutant allele from two lines, irx1-1 and irx1-2, from which we found that each allele contained a single mutation in this cellulose synthase gene. In irx1-1, this mutation resulted in the substitution of asparagine for the third conserved aspartate residue. This conserved aspartate has been found in all enzymes that require nucleotide sugars. The mutation in irx1-2 resulted in a serine-to-leucine change within four residues of this third aspartate, and analysis of all known CesA genes has revealed that this serine is also a conserved residue. irx1 plants were complemented with the F28A21.190 gene, confirming that a defect in this gene was responsible for the irx1 mutation.
It was found by RNA gel blot analysis that IRX1 and IRX3, a previously identified secondary cell wall–specific cellulose synthase (Taylor et al., 1999), were expressed in the same tissues. Analysis of the collapsed xylem phenotype and ultrastructure of the cell walls of irx1 and irx3 plants revealed that the mutant phenotypes were indistinguishable in xylem cells. Mutations in either IRX1 or IRX3 result in a decrease in cellulose of >70% in stems and hypocotyls (Turner and Somerville, 1997). However, that is probably an underestimate of the effect of the mutations because the considerable amounts of cellulose in primary cell walls of stems and hypocotyls will be unaffected in these mutants. In the xylem, both mutations also affect the same cell type, showing that IRX1 and IRX3 are both required to make cellulose in the same cells. Thus, IRX1 and IRX3 define distinct classes of catalytic subunits that are both essential for cellulose synthesis.
One explanation of the requirement for two family members for proper cellulose production relates to the hypothesis that the initial stage in cellulose production may require the formation of a short primer. Although no direct evidence supporting this idea has been found, such an arrangement could explain the requirement for two similar polypeptides: one to synthesize a short primer and one to synthesize cellulose microfibrils as part of the complex. This idea is not supported by sequence analysis, however, because IRX1 and IRX3 are highly homologous with each other and with other known cellulose synthases (Figure 2). Such a relationship would seem extremely unlikely if IRX1 and IRX3 had different activities.
Another possibility is that the IRX1 and IRX3 polypeptides are randomly assembled into the cellulose synthase complex. In this case, it would be possible that the inclusion of one defective subunit could prevent the production of a single β(1,4), glucose chain, which could in some way stall microfibril synthesis from the entire complex. In that case, however, the mutants would be expected to be dominant or semidominant, whereas the irx1 mutation behaves like a recessive mutation (Turner and Somerville, 1997). Similarly, complementation experiments in which a single IRX1 or IRX3 gene is inserted into the mutant background, which contains two defective copies of the gene, restores cellulose to wild-type values (Figure 4). In addition, the irx3 mutation causes a truncation of the protein, and drastically less mRNA is present (Taylor et al., 1999); indeed, irx3 plants have been shown to contain no detectable IRX3 protein (Figure 7). The inability of IRX1 and other components of the cellulose synthase complex to synthesize cellulose in the absence of IRX3 further suggests that this hypothesis is incorrect.
In view of the similarity of IRX1 and IRX3 to one another, the simplest explanation is that IRX1 and IRX3 both have an essential role in the cellulose synthase complex. Although IRX1 and IRX3 are similar and are both involved in cellulose synthesis in the secondary cell wall, a mutation in either gene results in decreased production of cellulose. That both IRX1 and IRX3 are essential is supported by the interaction of IRX1 and IRX3 in detergent-solubilized extracts, which im-plies that both IRX1 and IRX3 are present in the same complex. Although the possibility that IRX1 and IRX3 associate in posthomogenization steps cannot be discounted, together these data all suggest that IRX1 and IRX3 are both essential components of the cellulose synthase complex responsible for cellulose deposition in the secondary cell walls of xylem elements. These genes are not functionally identical and represent two different classes of catalytic subunits. The exact basis of this difference between IRX1 and IRX3 remains to be defined.
The large number of cellulose synthase genes found in plants (>13 CesA genes in Arabidopsis) has been a subject of much speculation (Arioli et al., 1998b; Delmer, 1998). The possibility that more than one catalytic subunit is required in every cell type may help explain the large number of apparently similar CesA genes found in Arabidopsis.
Clearly, more than one catalytic subunit is not required for the synthesis of cellulose in all organisms; bacteria, for example, appear to require only a single catalytic subunit (Saxena et al., 1990; Matthysse et al., 1995). Most models of cellulose synthesis, however, predict the presence of two catalytic sites, which would allow the simultaneous addition of two glucose residues, each orientated 180° to its neighbor (Carpita and Vergara, 1998). Given that IRX1 and IRX3 have been shown to be present in the same complex, perhaps in plants different subunits combine to provide these multiple catalytic sites. This could help explain the apparently large number of genes encoding these proteins.
It is also possible that the formation of rosettes may require more than one CesA family member. Bacteria do not have a requirement to control the orientation of cellulose microfibril biosynthesis because cellulose is not part of the cell wall and is synthesized from stationary structures in the plasma membrane. In contrast, higher plants need to control strictly the orientation of deposition of cellulose in the cell wall. This control of deposition of microfibrils operates through the cellulose synthase rosettes that move through the plasma membrane. These rosettes, elaborate structures that make as many as 36 chains simultaneously, presumably contain at least 36 copies of the catalytic domain and possibly a variety of other proteins. Perhaps plants have evolved a requirement for at least two different classes of catalytic subunits for the structure and/or assembly of these rosettes. This hypothesis is supported by experiments with the temperature-sensitive mutant rsw1 demonstrating the importance of the catalytic subunit in assembly of these rosettes. At the restrictive temperature, a comparatively minor mutation in the catalytic subunit results in dissociation of the rosette structure (Arioli et al., 1998a).
At present, the possibility that other CesA family members may also be required cannot be discounted. In addition, by analogy with bacterial systems, it is likely that other non-CesA components may be required. Defining the stoichiometry of different CesA family members within the rosette and the way in which they interact is, however, clearly essential to a proper understanding of cellulose synthesis in plants. The identification of the IRX1 gene is an important step forward in attaining these goals.
METHODS
Mapping IRX1
Plants (Arabidopsis thaliana) used were the result of a test cross between the mutant line SRT123-4 (irx1-1) isolated in a Landsberg erecta background (Turner and Somerville, 1997) and Columbia carrying the gl1 mutation. Plants ∼21 days old were scored phenotypically by visual assessment, which was confirmed by using light microscopy, as previously described (Turner and Somerville, 1997). DNA was prepared from these plants as described previously (Turner and Somerville, 1997).
For each bacterial artificial chromosome (BAC), a search was performed for all possible dinucleotide and mononucleotide repeat sequences. Primers were designed that would amplify an 80- to 240-bp fragment of DNA around any repeat >18 bp long, and polymerase chain reaction (PCR) products from parental DNA samples were assayed for polymorphisms. When no suitable microsatellite sequences were found in a BAC of interest, primers were designed that amplified a 1500- to 2000-bp region of the BAC to generate a cleaved amplified polymorphic sequences marker (Konieczny and Ausubel, 1993). The resulting PCR product was screened for restriction endonuclease polymorphisms between the two parental ecotypes by using a selection of 11 restriction endonucleases with 4-bp recognition sites. Where polymorphisms were found, the loci were scored against our initial 24 recombinant individuals as well as at least 20 randomly selected samples as controls to test for proper segregation in each case.
cDNA Library Construction and Screening
Total RNA was isolated from 15 g of mature Landsberg erecta stem tissue. Poly(A)+ RNA was isolated from the total RNA by using a polyATract kit (Promega, Madison, WI) according to the manufacturer's instructions. The library was constructed in UniZAP XR (Stratagene, La Jolla, CA) according to the manufacturer's instructions. The amplified library was screened with a probe encoding the central conserved region of IRX3 (amino acids 293 to 566). This fragment was generated by PCR by using primers C1F (5′-GGCCATATGGAACGTGAGACCTATCTAGAT-3′) and C1R (5′-GGCCTCGAGGTTTGTGTCAATGCCATCAAA-3′) (underlined sequences show homology to IRX3) in a standard PCR reaction using 10 ng of IRX3 cDNA 5′ (Taylor et al., 1999) as template. This fragment was then labeled nonradioactively with the Gene Images random prime labeling module (Amersham, Little Chalfont, UK). The library was probed and then developed with the Gene Images CDP-Star detection module (Amersham) according to the manufacturer's instructions, after which the signal was visualized by exposure to Super RX film (Fuji Photo Film Co., Tokyo, Japan). Two rounds of screening were conducted to identify hybridizing clones.
RNA Gel Blot Analysis
Total RNA was isolated as described above. After transfer of 5 μg of electrophoresed RNA to Hybond N+ membranes (Amersham), the membranes were probed with IRX1 (1.4-kb ClaI fragment corresponding to the 3′ end of the gene), 75G11 (IRX3), COMT, and rRNA, as described by Taylor et al. (1999). The probes were labeled with 32P-dCTP, as described by Hodgson and Fisk (1987), and the filters were probed, washed, and developed by the standard techniques described by Sambrook et al. (1989).
PCR and Reverse Transcription–PCR
For reverse transcription(RT)–PCR, total RNA was isolated from mature plants by using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). Five hundred nanograms of this RNA was subjected to RT-PCR by using Reverse-iT One Step (Abgene, Epsom, Surrey, UK). For the 5′ portion of the gene, the primers used were IRX15′F (5′-GGCGAATTCGAAGATGATGGA-3′) and IRX15′R (5′-GTTTACAGAGTCGGGAACACC-3′). For the 3′ portion of the gene, primers IRX13′F (5′-CATTACTTAACTTGTGGCTCA-3′) and IRX13′R (5′-AGACAAGCACTTTACGGATAC-3′) were used, with the reverse primer acting as the gene-specific primer for the RT reaction in each case. After incubation for 60 min at 47°C and inactivation of the RT for 5 min at 94°C, the reactions were subjected to 35 cycles of 94°C for 30 sec, 55°C for 30 sec, and 72°C for 2 min, followed by incubation at 72°C for 5 min. RT-PCR products were gel-purified before being cloned into the vector pGEM-T Easy (Promega) for sequencing.
For PCR amplification from plant genomic DNA, DNA was extracted from leaf tissue according to the method of Konieczny and Ausubel (1993). PCR primers IRX11900+ (5′-CGAGTCTACTCTTATGGAAAA-3′) and IRX12200− (5′-CTAATCTCTGGAGCAACTTGA-3′) were used to amplify the fragments containing the mutation under the following conditions: 30 cycles of 94°C for 30 sec, 50°C for 30 sec, and 72°C for 30 sec. PCR was performed with Taq DNA polymerase (Immunogen International, Sunderland, UK) according to the manufacturer's instructions in a PTC100 thermal cycler (MJ Research Inc., Watertown, MA). Again, the PCR products were gel-purified and cloned into pGEM-T Easy for sequencing.
DNA Sequencing
Plasmid templates purified by Qiagen QIAprep spin miniprep kits (Qiagen) were primed with either universal or gene-specific primers of high-purity salt-free grade (MWG Biotech UK, Milton Keynes, UK) and were sequenced automatically by using ABI PRISM Big Dye Terminators (Applied Biosystems, Inc., Foster City, CA). DNA sequences were analyzed by using programs available for use on the Internet.
Complementation of irx1
irx1-1 and irx1-2 plants were transformed by Agrobacterium tumefaciens (GV3101) carrying the plasmid pCB2300, which contained the 8.1-kb SalI fragment carrying the IRX1 gene, according to Bent and Clough (1998). Transformants were then selected, grown, and analyzed as described previously (Turner and Somerville, 1997; Taylor et al., 1999).
Electron Microscopy
Samples were prepared as described previously (Turner and Somerville, 1997).
Production of IRX1- and IRX3-Specific Polyclonal Antibodies
The region encoding the first variable region of IRX1 was amplified by PCR with primers IRX1VR1FOR (5′-GGCATATGACTCAATCCATTGTTCCAACA-3′) and IRX1VR1REV (5′-GGCTCGAGAGGAACCTGAGCGTCTTGTTG-3′) and cloned into the NdeI and XhoI sites of pET24a (Novagen, Madison, WI). After induction in Escherichia coli BL21(DE3), the overexpressed protein was purified according to the manufacturer's instructions with Ni-NTA Superflow (Qiagen). Antibodies were raised against this purified polypeptide in sheep (Diagnostics Scotland, Carluke, Scotland) and then were affinity-purified with the antigen immobilized on Affigel 10 (Bio-Rad, Hercules CA), according to the manufacturer's instructions. Anti-IRX3 antibodies were produced the same way, using primers IRX3VR1FOR (5′-GGCCATATGATCGAACATGAACAAGATAAGCAT-3′) and IRX3REV (5′-GGCCTCGAGAAGATTTCCATGCTGGAGCTT-3′) to amplify variable region 1.
Interaction between IRX3 and IRX1
An 8.3-kb XhoI-MunI genomic DNA fragment carrying the entire IRX3 coding region and 1.7 kb of promoter sequence cloned into pCB2300 was cut with NheI, and a double-stranded oligonucleotide (the product of annealing His1 [5′-CTAGGGGATCCCATCACCATCACCATCACC-3′] and His2 [5′-CTAGGGTCATGGTGATGGTCATCGGATCCC-3′]) was ligated to insert the epitope. This construct was transformed into irx3 plants by vacuum infiltration (Bent and Clough, 1998).
One gram of stems from transformed plants was ground well in lysis buffer (50 mM NaH2PO4, pH 8.0, 300 mM NaCl) containing 10 mM imidazole. After clarification by centrifugation, Triton X-100 was added to a final concentration of 2%. Next, 100 μL of Ni-NTA Superflow (Qiagen) was added to these solubilized extracts, which were mixed end over end for 60 min. After centrifugation, the resin was washed three times with 250 μL of lysis buffer containing 20 mM imidazole. Proteins were eluted from the resin twice with 30 μL of lysis buffer containing 250 mM imidazole. The entire purification procedure was performed at 4°C in the presence of protease inhibitors (protease inhibitor cocktail for mammalian cell extracts; Sigma, Poole, Dorset, UK). Ten-microliter aliquots were denatured in loading buffer for 60 min at 37°C before electrophoresis through 7.5% SDS–polyacrylamide gels (Laemmli, 1970). After transfer to Immuno-blot polyvinylidene difluoride membrane (Bio-Rad), protein gel blots were performed according to standard protocols (Harlow and Lane, 1988). Epitope-tagged IRX3 was detected by using an anti-RGSHis monoclonal antibody (Qiagen).
Acknowledgments
We are grateful to the Arabidopsis Biological Resource Center at The Ohio State University (Columbus, OH) for providing BAC clones. This work was supported by the Biology and Biotechnology Research Council (Grant No. 34/P06859).
References
- Arioli, T., et al. (1998. a). Molecular analysis of cellulose biosynthesis in Arabidopsis. Science 279 717–720. [DOI] [PubMed] [Google Scholar]
- Arioli, T., Burn, J.E., Betzner, A.S., and Williamson, R.E. (1998. b). Response: How many cellulose synthase–like gene products actually make cellulose? Trends Plant Sci. 3 165–166. [Google Scholar]
- Bent, A.F., and Clough, S.J. (1998). Agrobacterium germ-line transformation: Transformation of Arabidopsis without tissue culture. In: Plant Molecular Biology Manual, 2nd ed, S.B. Gelvin and R.A. Schilperoot, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), Section B7, pp. 1–14.
- Brown, R.M. (1996). The biosynthesis of cellulose. J. Macromol. Sci. Pure Appl. Chem. A33 1345–1373. [Google Scholar]
- Brown, R.M., Saxena, I.M., and Kudlicka, K. (1996). Cellulose biosynthesis in higher plants. Trends Plant Sci. 1 149–156. [Google Scholar]
- Carpita, N., and Vergara, C. (1998). A recipe for cellulose. Science 279 672–673. [DOI] [PubMed] [Google Scholar]
- Daniels, M.J., Mirkov, T.E., and Chrispeels, M.J. (1994). The plasma membrane of Arabidopsis thaliana contains a mercury-insensitive aquaporin that is a homolog of the tonoplast water channel protein TIP. Plant Physiol. 106 1325–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delmer, D.P. (1998). A hot mutant for cellulose synthesis. Trends Plant Sci. 3 164–165. [Google Scholar]
- Delmer, D.P. (1999). Cellulose biosynthesis: Exciting times for a difficult field of study. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 245–276. [DOI] [PubMed] [Google Scholar]
- Delmer, D.P., and Amor, Y. (1995). Cellulose biosynthesis. Plant Cell 7 987–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher, D.D., and Cyr, R.J. (1998). Extending the microtubule/microfibril paradigm. Plant Physiol. 116 1043–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hodgson, C.P., and Fisk, R.Z. (1987). Hybridization probe size control: Optimized ‘oligolabelling.’ Nucleic Acids Res. 15 6295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, S., Laosinchai, W., Itoh, T., Cui, X., Linder, C.R., and Brown, R.M. (1999). Immunogold labeling of rosette terminal cellulose-synthesizing complexes in the vascular plant Vigna angularis. Plant Cell 11 2075–2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokubo, A., Sakurai, N., Kuraishi, S., and Takeda, K. (1991). Culm brittleness of barley (Hordeum vulgare L.) mutants is caused by a smaller number of cellulose molecules in the cell wall. Plant Physiol. 91 509–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using codominant ecotype-specific PCR-based markers. Plant J. 4 403–410. [DOI] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227 680–685. [DOI] [PubMed] [Google Scholar]
- Lin, X., et al. (1999). Sequence and analysis of chromosome 2 of the plant Arabidopsis thaliana. Nature 402 761–769. [DOI] [PubMed] [Google Scholar]
- Lister, C., and Dean, C. (1993). Recombinant inbred lines for mapping RFLP and phenotypic markers in Arabidopsis thaliana. Plant J. 4 745–750. [DOI] [PubMed] [Google Scholar]
- Matthysse, A.G., White, S., and Lightfoot, R. (1995). Genes required for cellulose synthesis in Agrobacterium tumefaciens. J. Bacteriol. 177 1069–1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear, J.P., Kawagoe, Y., Schreckengost, W.E., Delmer, D.P., and Stalker, D.M. (1996). Higher plants contain homologs of the bacterial CelA genes encoding the catalytic subunit of cellulose synthase. Proc. Natl. Acad. Sci. USA 93 12637–12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Saurin, A.J., Borden, K.L.B., Boddy, M.N., and Freemont, P.S. (1996). Does this have a familiar RING? Trends Biochem. Sci. 21 208–214. [PubMed] [Google Scholar]
- Saxena, I.M., Lin, F.C., and Brown, R.M. (1990). Cloning and sequencing of the catalytic subunit of Acetobacter xylinum. Plant Mol. Biol. 15 673–683. [DOI] [PubMed] [Google Scholar]
- Saxena, I.M., Brown, R.M., Fevre, M., Geremia, R.A., and Henrissat, B. (1995). Multidomain architecture of β-glycosyl transferases: Implications for mechanism of action. J. Bacteriol. 177 1419–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, N.G., Scheible, W.-R., Cutler, S., Somerville, C.R., and Turner, S.R. (1999). The irregular xylem3 locus of Arabidopsis encodes a cellulose synthase required for secondary cell wall synthesis. Plant Cell 11 769–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, S.R., and Somerville, C.R. (1997). Collapsed xylem phenotype of Arabidopsis identifies mutants deficient in cellulose deposition in the secondary cell wall. Plant Cell 9 689–701. [DOI] [PMC free article] [PubMed] [Google Scholar]