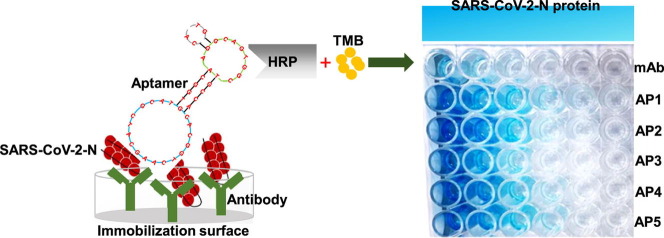
Graphical abstract
Keywords: SARS-CoV-2, COVID-19, Antigen, Nucleocapsid, Diagnostic, Aptamer
Abstract
The severe acute respiratory syndrome coronavirus (SARS-CoV-2) has infected millions of individuals and continues to be a major health concern worldwide. While reverse transcription-polymerase chain reaction remains a reliable method for detecting infections, limitations of this technology, particularly cost and the requirement of a dedicated laboratory, prevent rapid viral monitoring. Antigen tests filled this need to some extent but with limitations including sensitivity and specificity, particularly against emerging variants of concern. Here, we developed aptamers against the SARS-CoV-2 Nucleocapsid protein to complement or replace antibodies in antigen detection assays. As detection reagents in ELISA-like assays, our DNA aptamers were able to detect as low as 150 pg/mL of the protein and under 150 k copies of inactivated SARS-CoV-2 Wuhan Alpha strain in viral transport medium with little cross-reactivity to other human coronaviruses (HCoVs). Further, our aptamers were reselected against the SARS-CoV-2 Omicron variant of concern, and we found two sequences that had a more than two-fold increase in signal compared to our original aptamers when used as detection reagents against protein from the Omicron strain. These findings illustrate the use of aptamers as promising alternative detection reagents that may translate for use in current tests and our findings validate the method for the reselection of aptamers against emerging viral strains.
Introduction
Severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) has infected millions of individuals and caused morbidity and mortality in hundreds of thousands of people worldwide. The coronavirus disease that started in 2019 (COVID-19) ranges in presentation from asymptomatic to fatal with some death rates estimated to be as high as 18 million just in the first two years of the pandemic (Wang, et al. 2022). SARS-CoV-2 infections were primarily monitored through reverse transcription polymerase chain reaction (RT-PCR) (Ji, et al. 2020). While RT-PCR detection has proven to be a reliable method for viral surveillance (Ai, et al. 2020) and remains the gold standard for SARS-CoV-2 testing (Weissleder, et al. 2020), limitations of these tests persist including cost, availability, and in most cases requiring a laboratory to perform the assay and analyze the results (Ji, et al. 2020). In addition, RT-PCR testing requires several hours to days to obtain a result (Long et al., 2020, Weissleder et al., 2020) which translates to increasing the likeliness of not testing at peak viral load during infection (Jones et al., 2021, Stankiewicz Karita et al., 2022) that can create a higher rate of false-negative results and further increase transmission through the population. Alternatively, enzyme-linked immunosorbent assay (ELISA) based antigen tests have been essential as diagnostic tools for the COVID-19 pandemic (Ravi, et al. 2020). Antigen tests offer several advantages over genomic detection methods such as rapid, selective, and low-cost detection (Mahase 2020) which allows for testing multiple timepoints overcoming the caveats of PCR-based testing. However, the use of antibodies (Ab) used in these tests has downsides. In addition to batch-to-batch variations in the production of the Ab (Voskuil 2014), the time to develop a Ab specific to a viral protein is extensive, a point especially emphasized in the context of ever evolving viral strains. This is evidenced by a variant of concern, Omicron, which has 34 to 37 amino acid substitutions in the spike (S) protein rendering many anti-S protein mAbs ineffectual (Cameroni, et al. 2022). While compared to the S protein, the Omicron nucleocapsid (N) protein has fewer mutations including a nine-nucleotide deletion in positions 28370–38462 of the N protein gene and a point mutation, R203M (Syed et al., 2021, Syed et al., 2022), resulting in a 10–20% reduction in the limit of detection for some mAb-based antigen tests (Leuzinger, et al. 2022). Taken together, this evidence suggests the need to improve critical reagents to advance diagnostic testing.
Aptamers are short synthetic oligos selected to bind specific targets(Ellington and Szostak 1990) and provide an alternative or a complement to antibodies (Bunka and Stockley, 2006, Nimjee et al., 2005). Aptamers share similar characteristics to antibodies in that they bind specifically to their targets (Tuerk and Gold 1990) but can be synthesized in a fraction of the time and cost (Baker, 2015, Lakhin et al., 2013) and are temperature and pH resistant(Chen and Yang 2015) suggestive of them being very stable. They are also much smaller than antibodies, ∼2 nm compared to ∼ 14 nm in diameter (Abrego-Martinez, et al. 2022), which allows more aptamer to be used on a capturing surface and for detection, and have less steric hindrance allowing for more binding of targets leading to enhanced sensitivity (Shereen, et al. 2020). Aptamers have been successfully used in several ELISA-like assays (Nguyen et al., 2016, Nikam et al., 2022, Zhang et al., 2022), including LFAs (Chen and Yang 2015). Thus, aptamers have distinct utility for COVID-19 antigen testing as they can be direct replacements of antibodies in ELISA formats, termed Enzyme-linked aptamer sorbent assay (ELASA), by improving sensing performances, provide longer and more robust shelf life (Chen and Yang 2015) and reduced cost (Yang et al., 2022a) that is essential for testing multiple timepoints during viral infection to ensure an accurate positive result.
Already, several groups have had success in selecting aptamers that bind to the SARS-CoV-2 S protein (Avni and Schulman, 1987, Li et al., 2021, Valero et al., 2021, Zhang et al., 2021, AminiLi et al., 2022), including S protein from the Omicron variant (Yang et al., 2022a). The S protein of SARS-CoV-2 is essential for viral infection (Huang, Yang et al. 2020) making it a key target for vaccine development, but it is also one of the most mutated proteins reported in all the emerging strains of SARS-CoV-2 (Magazine, et al. 2022), rendering targeting antibodies and aptamers ineffective. The N protein of SARS-CoV-2 plays a variety of roles in RNA transcription and viral replication (Cubuk, et al. 2021) and is the most abundantly expressed of the structural proteins (He, et al. 2004). Additionally, a standard virion on an average has as few as 30 spike trimers (Sender, et al. 2021), in contrast the N protein is much more abundant and estimated to be between 730 and 2200 copies per virion (Bar-On, et al. 2020) and, as highlighted above, is not under as much evolutionary selection pressure as S protein providing a more conserved target across viral strains. Aptamers against N protein have also been developed (Chen et al., 2020, Cho et al., 2011, Kang et al., 2021, Zhang et al., 2020), but are far less common than those against S and to our knowledge none have been specifically selected against the Omicron variant. In this work, we selected DNA aptamers against SARS-CoV-2 N-protein and successfully used these aptamers as detection reagents in place of antibodies in an ELASA for the detection of SARS-CoV-2 N protein and inactivated virus. Our results showed increased sensitivity towards the SARS-CoV-2 N-protein from the Alpha Wuhan strain and showed low cross reactivity to other common cold coronaviruses. Further, we performed a reselection of our aptamers against SARS-CoV-2 N protein from the Omicron variant and identified novel aptamers that strongly bind to this variant of concern. Our Omicron specific aptamers in an ELASA had a 2-fold increase in the detection of the Omicron variant compared to the Alpha Wuhan strain, thus illustrating this reselection process as a promising method for the replacement or complement to antibodies in COVID-19 antigen detection assays particularly for the detection of emerging variants.
Methods
Materials
Biotinylated detection aptamers, scramble aptamer and thiolated aptamers were synthesized by Integrated DNA Technologies (Coralville, IA) and underwent desalting purification. All sequences of aptamers are reported in Supplemental table 1. The scramble used the same base as the detection aptamers but with a random nucleotide (N) repeats: CGA GGC TCT CGG GAC GAC NNN NNN NNN NNN NNN NNN NNN NNN NNN NNN GTC GTC CCG CCT TTA GGA TT ACA G. MyOne T1 Dynabeads® (Life Technologies; Carlsbad, CA) streptavidin-coated magnetic beads were employed for partitioning steps. Purified recombinant SARS-CoV-2 nucleocapsid protein, as well as recombinant SARS-CoV, HuCoV-NL63, HuCoV229E, and HuCoV-OC43 proteins and detergent (0.05% Tween-20) neutralized virus samples were previously described (Terry, et al. 2021). Viral transport medium (VTM) was made by modifying Hanks Balanced Salt Solution (HBSS) by adding 2% heat-inactivated fetal bovine serum, 100 ug/mL gentamicin and 0.5 ug/mL amphotericin B following the CDC specifications (https://www.cdc.gov/coronavirus/2019-ncov/downloads/Viral-Transport-Medium.pdf). Additional recombinant SARS-CoV-2 N protein was purchased from RaybioTech (Peachtree Corners, GA, Code: 230–01104-100,). Anti-SARS-CoV-2 N protein polyclonal antibody (pAb) was purchased from RaybioTech (CODE: 130-10760B-50), used as the capture antibody, and biotinylated SARS-CoV-2 N protein monoclonal antibody (mAb) was purchased from Sino Biological (Houston, TX, Cat: 40143-MM08), used as the detection reagent control. PCR quantified SARS-CoV-2 heat inactivated virus as well as other detergent inactivated common cold corona viruses were purchased from Zeptometrix (Buffalo, NY). The Aptamer Folding Buffer composed of 137 mM Sodium Chloride, 2.7 mM Potassium Chloride, 8 mM Sodium Phosphate dibasic, 1.5 mM Potassium Phosphate monobasic, 0.9 mM Calcium Chloride, and 2 mM Magnesium Chloride. The ELISA/ELASA wash buffer (10x) was made with 320 g NaCl, 46.4 g Na2HP04, 8 g KH2PO4, 8 g KCl, 360 g MgCl2, and 2.0 mL Tween-20.
Melting-Off SELEX for aptamer selection
Aptamers were selected against SARS-CoV-2 nucleocapsid protein under the guidance and in collaboration with Aptagen (Jacobus, PA) using their modified Melting-Off SELEX method. Briefly, DNA aptamers were selected against SARS-CoV-2 nucleocapsid protein (target) and counter selected against pooled nucleocapsid protein from other human coronaviruses (counter targets) used to reduce the enrichment of nonspecific aptamers. Melting-Off SELEX uses a complementary capture probe to immobilize a library to magnetic beads (Supplemental Fig. 1). Introduction of target/counter-target/matrix can then be used to induce conformational changes in the library on binding, allowing for the partitioning of responsive sequences from non-responsive sequences. Selection began with pre-incubating the library with the capture oligo at twice the library amount for refolding in 1X SELEX buffer (1X PBS, pH 7.4) by heating the sample to 90 °C for 1 min, cooling to 60 °C for 5 min, and then cooling to 23 °C for 5 min. While the library was refolding, Dynabeads® were washed three times with 1X PBS containing 0.01% Tween-20 and 5 mM MgCl. High MgCl concentrations at or above 5 mM was used to insure the highest binding to the N protein by preventing oligomerization of N protein. After refolding was completed, the library was captured on the washed Dynabeads®. The Dynabeads® were then washed twice with 1X SELEX buffer to remove non-specifically bound library members. After the initial wash, the library was incubated with counter-targets at 23 °C. Non-binding library was partitioned from counter-binding or buffer-responsive library by magnetic separation, after which the supernatant was discarded. The Dynabeads® were then washed repeatedly (number of washes and concentration of N protein described in Fig. 1 A) to remove any remaining non-specific library members. The remaining Dynabead®-bound-library was then incubated with N protein at 23 °C for 30 min. The recovered library was purified on 10% denaturing PAGE (with 8 M urea and 1X TBE running buffer) for PCR amplification and propagation to the next round. Library response was defined as the ratio of recovered material (determined by spectrophotometric analysis at A260) to the amount of input library. The initial round of selection used omitted counter-targets to maximize recovery of rare sequences from the starting library. Selection continued using target and counter-target at 1 µM until Round 8, where the library response rose to over 10%. At this point, the concentration of target was reduced to 0.5 µM. Once the library response appeared to recover, the target concentration was further reduced to 1 nM. The enriched library was divided into four fractions to perform a parallel assessment against the buffer only (negative condition), pooled nucleocapsid protein from other human coronaviruses (counter condition), purified detergent treated SARS-CoV-2 nucleocapsid protein in VTM (positive condition), and detergent neutralized virus in VTM (Supplemental Fig. 1). The Surface Plasmon Resonance (SPR) measurements were carried out with a Biacore X100 instrument. The Biotin CAPture Kit (GE Healthcare Europe GmbH, Germany) was used for the interaction studies according to manufacturer’s instructions. It enables reversible capture of biotinylated ligands and includes the sensor chip CAP, the Biotin CAPture Reagent, and a two-component regeneration solution. The binding affinity of the aptamers for the N-protein of Wuhan Alpha strain and Omicron variants were performed using a modified protocol developed by Aptagen.
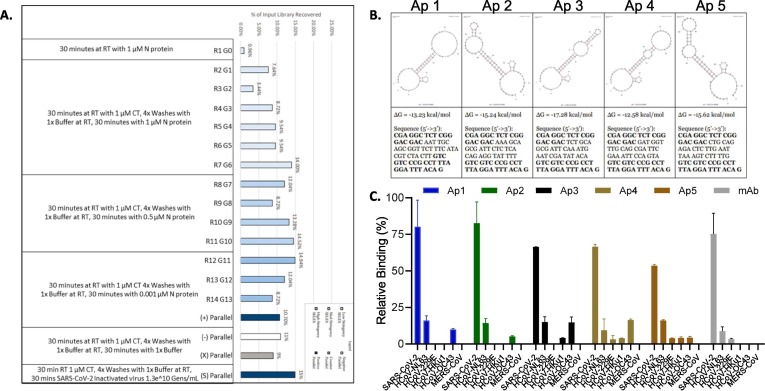
Fig. 1.
Selection of SARS-CoV-2 Nucleocapsid Protein Aptamers using Melting-Off Based Library Enrichment. A. Library recovery following a melting-off based library enrichment determined as a ratio between material recovered from the selection or parallel assessment step and the input amount of material. Specific selection conditions are indicated below the bar graph. B. Bioinformatics based analysis predicted secondary structures (top panels) and sequence data (bottom panels) for the five aptamer candidates. The secondary structure and free energy for each aptamer was computed by Quikfold 2.3 (Zuker 2003) at 23 °C, 150 mM Na+ (combined monovalent cations in 1X SELEX Buffer). Sequences displayed below each structure. PCR primer annealing regions (5′- CGA GGC TCT CGG GAC GAC -[sequence]- GTC GTC CCG CCT TTA GGA TTT ACA G −3′) are highlighted in bold. C. Aptamers identified through SELEX were evaluated for their binding to recombinant nucleocapsid protein from HCoVs (200 ng/mL) using a direct ELASA format and compared to a commercially available monoclonal antibody. Background was subtracted and error bars indicate standard deviation. Statistical significance was reached for all conditions of Alpha variant compared to other HCoV.
Aptamer sequencing
The initial library was subjected to nine rounds of Melting-Off selection followed by parallel assessment. The SELEX process was designed to enrich sequences over multiple rounds of selection that bind to SARS-CoV-2 Nucleocapsid Protein. A counter selection process was used to remove sequences that respond to components of negative patient samples as well as general epitopes associated with human IgG. As a result, the population to be sequenced was expected to contain multiple copies of aptamer candidates. The Illumina (San Diego, CA) MiniSeq system was implemented to sequence the aptamer libraries after the post-parallel selection using a single-end read technique. Deep sequencing and subsequent data analysis reduces the traditional approach of performing many screening rounds, which may introduce error and bias due to the screening process (Schutze, et al. 2011). Hundreds of thousands of sequences were analyzed from the parallel-exposed final libraries. From these sets of data, the library sequence families were constructed at 90% homology (sequence similarity considering mutations, deletions, and insertions). The top 200 sequences are available in Supplemental Table 1.
Direct ELASA
Recombinant nucleocapsid protein (100 µL of 200 ng/mL in 5 mM MgCl- PBS) from SARS-CoV-2 or other human coronaviruses was coated on the wells of a 96-well plate overnight. Protein was blocked with blocking buffer (2% BSA in PBS, pH 7.2) for 30 min and washed with 1x ELISA wash buffer (diluted with pure water). Biotin-modified aptamers (200 nM concentration in folding buffer) were heated to 95 °C for 5 mins and placed on ice. Protein was incubated with 100 µL of aptamers at a 200 nM concentration for 2 h and then washed with 300 µL of 1x ELISA wash buffer. For detection, 100 µL of streptavidin-poly-HRP (50 ng/mL) was added for 30 min followed by 5 washes with 300 µL of 1x ELISA wash buffer. TMB substrate (50 µL) was then added to each well for 30 min and the reaction was stopped with 1 M sulfuric acid and absorbance read at 450 nm. All steps were performed at ambient temperature under mild shaking conditions.
Hybrid sandwich ELASA
To determine the efficiency of the aptamers for detection in a sandwich ELISA-like format, a hybrid sandwich ELASA was developed in which an antibody (Ab) was used as the capturing agent. Briefly, 10 µg/mL Ab was coated on a high binding ELISA 96-well plate (Corning Cat. No. 9018) overnight. Plates were washed with 1x ELASA wash buffer and protein at various concentrations (150 pg to 500 ng) was added for 4 h. Plates were blocked with blocking buffer (2% BSA in PBS, pH 7.2) for 30 min and washed with 1x ELASA wash buffer. Biotin-modified aptamers (200 nM in folding buffer) were heated to 95 °C for 5 mins and placed on ice. Protein was incubated with 100 µL biotin-modified aptamers at 200 nM concentration or mAb (1 µg/ml) for 2 h and then washed with 300 µL of 1x ELASA wash buffer. For detection, 100 µL of streptavidin-polyHRP (50 ng/mL) was added for 30 min followed by 5 washes with 300 µL of 1x ELISA wash buffer. TMB substrate (50 µL) was then added to each well for 30 min and the reaction was stopped with 1 M sulfuric acid and absorbance read at 450 nm. All steps were performed at ambient temperature under mild shaking conditions.
Sandwich ELASA using Thiol-modified aptamers as capture reagents
To develop a sandwich ELASA in which aptamers were used for both the capture and detection we followed the protocol described by Svobodova et al.(Svobodova, et al. 2021). Briefly, thiol-modified aptamers were immobilized on maleimide-activated microtiter plates as capture and, as above, biotinylated aptamers were used as reporters with SA-poly-HRP/TMB for the detection. Thiolated aptamers (50 µL of 500 nM in PBS) were coated on a maleimide-activated microtiter plate (ThermoFisher Scientific, Waltham, MA) overnight. The plates were blocked with 200 μL of 5% skimmed milk (w/v) in PBS for 1 h at ambient temperature and then 200 ng/mL SARS-CoV-2 nucleocapsid protein were added to the wells. Biotin-modified aptamers (200 nM in folding buffer) were heated to 95 °C for 5 mins and placed on ice. Protein was incubated with biotin-modified aptamers for 2 h and then washed with 300 µL of 1x ELISA wash buffer. For detection, 100 µL of streptavidin-polyHRP (50 ng/mL) was added for 30 min followed by 5 washes with 300 µL of 1x ELISA wash buffer. TMB substrate (50 µL) was then added to each well for 30 min and the reaction was stopped with 1 M sulfuric acid and absorbance read at 450 nm. All steps were performed at ambient temperature under mild shaking conditions.
Microarray synthesis against SARS-CoV-2 variants Delta and Omicron
A Cy5-labeled reporter oligo complimentary to a constant region of the library (5′-GTC GTC CCG AGA GCC TCG - Cy5 −3′) was synthesized by IDT (Coralville, IA) and purified by HPLC. This oligo would be displaced during target binding to provide an indication of binding ability. The selected full-length aptamer candidates were arranged randomly by name before being synthesized by LC Sciences (Houston, TX) on duplicate 4 K microarray chips using standard phosphoramidite chemistry and masking techniques proprietary to LC Sciences. Each candidate cluster occupied 18 positions (3 × 6 colonies), with control sequences distributed semi-regularly throughout each chip (Fig. 2 ). Heat-inactivated Omicron (hCoV-19/USA/GA-EHC-2811C/2021, Lineage B.1.1.529, NR56495) and Delta (hCoV-19/USA/MD-HP05285/2021, Lineage B.1.617.2, NR-56128). SARS-CoV (pB700 NP-1), HuCoV-NL63 N (pB702 NP), HuCoV-229E (pB705 NP-1), and HuCoV-OC43 (pB701 NP-2) nucleocapsid protein samples were used as counter-target material. 1X SELEX Buffer was prepared as 1X PBS with 1% Tween-20, pH 7.4.
Fig. 2.
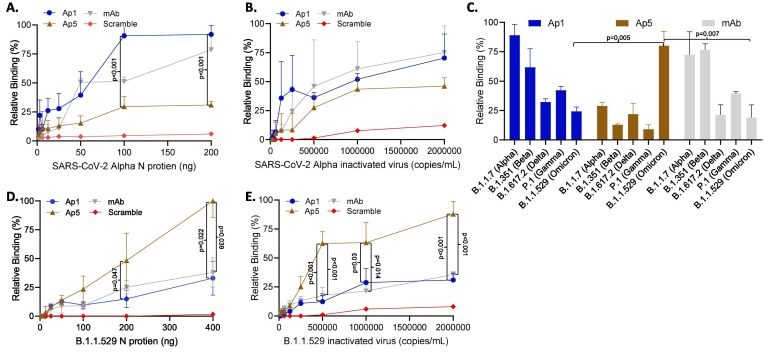
Aptamers as detection reagents for SARS-CoV-2 N protein and inactivated virus in a hybrid ELASA format. Aptamers 1 and 5 were evaluated as detection reagents in a hybrid ELASA format where an antibody was used for capture and aptamers were used as detection reagents. A. Detection of SARS-CoV-2 Alpha recombinant nucleocapsid protein or B. inactivated virus. C. Aptamers were evaluated for their ability to detect N protein from SARS-CoV-2 viral strains of interest (200 ng). D. Detection of SARS-CoV-2 Omicron recombinant nucleocapsid protein or E. inactivated virus. The error bars indicate standard deviation and statistical significance is indicated when p < 0.05 and was calculated using one-way ANOVA with Šidák multiple comparisons test (Figures A, B, D and E) and Welches T Test (Figure C).
Monoclonal colonies of each candidate were synthesized 3′ to 5′, with the 3′-end anchoring each individual candidate to the colony region on the chip. Colonies are defined as aptamer replicates in a single well, while clusters are defined as the group of wells that contain the same candidate. Candidate colonies and clusters are needed to significantly enhance the change in signal when responding to target or counter target, as an individual molecule’s change in fluorescence can be drowned out by the signal of its neighbors. Additional sequences were synthesized as a quality control measure.
Before use, the microarray chip was blocked with 0.1% BSA in 1X PBS solution to prevent nonspecific reporter binding to the chip. Candidates were refolded using a modified protocol, as the microarray setup cannot be heated above 60 °C without compromising the microfluidics. A solution of 100 nM Cy5-labeled Reporter oligo in the 1X SELEX Buffer was introduced to the chip with an oscillating peristaltic pump to ensure even distribution across colonies. During this process, the candidates were refolded by heating the chip and solution to 60 °C for 20 min and then cooling to 23 °C for 20 min. Unbound probe was removed by washing with 1 mL of 1X SELEX Buffer. Fluorescence readings were then taken by a GenePix 4000B Microarray Scanner (Molecular Devices, LLC.; San Jose, CA) with 1X SELEX Buffer in place to establish baseline responses of each candidate. While the microarrays were being prepared, solutions of heat-inactivated nucleocapsid protein from Omicron, Delta, and pooled Control samples were prepared by diluting stock solutions to 1 × 10^8 genome equivalents/mL (Omicron and Delta) or 33 ng/mL (pooled Controls). After the fluorescence probe had been annealed and the pre-incubation baseline image taken, 500 µL of prepared N-protein solution was circulated through the microarray for 16 h at 37 °C. After this incubation step was completed, the microarray was washed with 1 mL of 1X SELEX Buffer and a post-incubation image was taken with 1X SELEX Buffer remaining in the array. Omicron sample was assessed initially, and the same array was washed with 5 mL of 1X SELEX Buffer before undergoing the full preparation process for Delta analysis. Control analysis used a separate microarray chip. Data analysis was conducted as follows. The mean background fluorescence value (fbackground) was subtracted from the mean fluorescence value of each candidate prior to the addition of sample (fpre-treatment) as well as each candidate after the addition of sample (fpost-treatment). The amount of signal loss from the pre-treatment condition to the post-treatment condition was then expressed as a percentage. This process was repeated for the target and counter-target assessment. The percent signal loss for the target assessment was then divided by the percent signal loss for the counter assessment to generate a score. A score greater than one indicates increased response to the target condition over the counter condition. The percent signal loss from the target assessment minus the percent signal loss from the counter assessment is also recorded.
| (1) |
| (2) |
Statistics
Statistical comparisons of aptamer activity relied on two-sided t-tests with Welch’s correction and one-way ANOVA using Šidák multiple comparisons test was used to identify statistical significance between different aptamers or controls. Where each test was used is indicated in the figure legends.
Results
Aptamer selection against SARS-CoV-2 nucleocapsid protein
Multiple aptamers were selected to target the SARS-CoV-2 nucleocapsid protein as described in materials and methods. The results for nine rounds of pooled-split Melting-Off screening and parallel assessment are summarized in Fig. 1A. The low library recovery seen in Round 1 is typical, a result of collecting sequences that bind to SARS-CoV-2 Nucleocapsid Protein from over 1014 possible species. Once library response significantly increased by Round 7, concentration of the protein target was reduced to increase the stringency of selection. The library recovered after four rounds of exposure to the moderate stringency condition. At this point, target concentration was again decreased. Under these conditions library response did not recover, so the Round 13 (G12) library was taken to parallel assessment against detergent treated N protein and detergent inactivated SARS-CoV-2 virions in viral transport medium (VTM) to evaluate suitability for sequencing (Fig. 1A). Library response to the detergent inactivated SARS-CoV-2 in VTM was at least 4% higher than any other sample, representing a large quantity of sequences that could be elucidated. Thus, the library was deemed sufficiently enriched to proceed to sequencing and bioinformatics.
Bioinformatics and aptamer candidate selection
Five libraries were collected for sequencing: 1) the post-parallel assessment library recovered after incubation with detergent inactivated SARS-CoV-2 in 1X SELEX Buffer (positive population G12(S)); 2) post-parallel assessment library that had been recovered from incubation with positive target in SELEX Buffer (positive population G12(P)); 3) post-parallel assessment library recovered after incubation with counter-target in SELEX Buffer (counter population G12(C)); 4) post-parallel assessment library recovered after incubation with only SELEX Buffer (negative population G12(N)); and 5) the pre-parallel assessment library that had been recovered from incubation with positive target in SELEX Buffer prior to the round of parallel assessment (pre-positive population G11(P)). The positive population was compared against the counter population to identify any sequences that were not removed during the counter selection steps, but still had some affinity for both the target and counter-target (Supplemental Fig. 3). As the positive G12(P) population was the target for screening, candidates are chosen from this population’s sequences (Supplemental Figures 4 and 5). Those sequences that appeared at comparable rates in both positive and counter populations were ranked lower than ones that could predominantly be found in the positive populations. Conversely, families and sequences that appeared with significantly higher frequency in the positive populations over the counter population were considered for aptamer candidacy (Supplemental Fig. 6). Finally, the stability of a candidate’s secondary structure was used as a “tiebreaker” parameter, as stronger aptamers have relatively stable secondary structures. After considering these factors, 200 candidates were chosen for microarray synthesis and high throughput assessment. Five sequences were specifically identified as potential top candidates of the selection process based on the previously described criteria (Fig. 1B).
Since the goal was to identify aptamers that bind to SARS-CoV-2 Nucleocapsid Protein over proteins from other human coronaviruses, the candidates were selected based on greater proportional representation in the G12(P) and G12(S) positive populations over the G12(C) counter population that contained recombinant nucleocapsid protein from three common cold human coronaviruses, HCoV-NL63, HCoV-229E and HCoV-HKu1. Aptamer 1 (Ap1) is the strongest sequence calculated under this criterion – there are other candidates that appear exclusively in the G12(P) and G12(S) positive populations, though some may not be entirely dependable due to low read counts. Sequences that appear in the G12(S) population that do not appear in the G12(P) population are not considered at this point, as they may be responding to other molecules in the processed samples. Although based on sequence enrichment (frequency in generation over generation), the most strongly enriching sequences tended to coincide with sequences chosen considering counter-target response. Finally, the top five candidate sequences exhibited sufficient stability based on m fold secondary structure prediction (Zuker 2003) and were subjected to a standard Surface Plasmon Resonance (SPR) assay and were determined to have high binding affinities with affinity dissociation constant values (KD) below 20 nM, thus are preferential candidates (Fig. 1B).
Detection of SARS-CoV-2 N protein using a direct ELASA
With the aim to develop an aptamer-based SARS-CoV-2 antigen reagent, selected aptamers were challenged to detect SARS-CoV-2 N protein in a direct ELASA assay while maintaining specificity against other human coronaviruses (HCoV) sharing a 65–82.5% genome sequence homology with SARS-CoV-2. In this regard, 200 ng/mL of recombinant protein from SARS-CoV-2, HCoVs NL63, 229E, HKu1 and OC43 and MERS-CoV was plated overnight and detected with the top 5 aptamer candidates and a commercially available mAb (Sino Biological, Cat: 40143-MM08) done in triplicate. In all cases, except a small reaction to NL63 and OC43 (which was not used in the counter selection step during the production of the aptamers), the aptamers had little cross reactivity to the other HCoVs and MERS but showed a strong signal for SARS-CoV-2 (Fig. 1C). The cross reactivity to NL63 and OC43 was not observed at more physiological concentrations, such as under 1 ng/mL of recombinant protein (data not shown) illustrating the aptamers specificity to SARS-CoV-2.
Aptamer as detection reagents for SARS-CoV-2
To further evaluate the aptamers as detection reagents, we used a hybrid sandwich ELISA/ELASA test where an Ab was used for capture while aptamers were used for the detection of soluble viral protein. Using this hybrid format, the Ab was coated on a 96-well plate and contrived negative or positive samples containing either recombinant protein or known concentrations of inactivated virus (determined by qRT-PCR from the manufacture) was incubated for four hours. The top 5 identified aptamers candidates were then used to detect captured recombinant protein (Fig. 2A and Supplemental Fig. 7A) or detergent inactivated virus (Fig. 2B and Supplemental Fig. 7B) and the aptamers were compared for relative binding. For simplicity only aptamers 1 and 5, which had the lowest limits of detection against the Alpha and Omicron variants (described below in detail), respectively, are shown. A scrambled aptamer (scramble) and a detection mAb were used as negative and positive controls, respectively. Ap1 and 5 both had a detectable signal even at the lowest concentration tested of<6.5 ng/mL recombinant protein (Fig. 2A) but Ap1 had a higher relative binding signal compared to Ap5 at 100 ng (p < 0.001) and 200 ng of protein (p < 0.001). Both aptamers had detectable signals at<250 k inactivated SARS-CoV-2 alpha strain viral copies, and while Ap1 had a higher relative binding signal at the highest concentrations compared to Ap5, the difference did not reach statistical significance (Fig. 2B). Aptamers 2, 3 and 4 performed similarly to Ap1 for both recombinant protein and inactivated virus (Supplemental Table 7). Taken together, these data illustrate the ability of the aptamers to act as detection reagents for SARS-CoV-2 N recombinant protein and inactivated virus.
Aptamer detection of other SARS-CoV-2 viral strains
Since viral strains can have changes in their proteins due to evolutionary selection processes, we next sought to evaluate the ability of the aptamers in detecting all viral strains of interest as of June of 2022. Using a hybrid sandwich ELASA in which an Ab was used as capture with aptamers as detection, aptamers 1–5 were evaluated on their ability to detect a consistent amount of 100 ng/mL of recombinant protein from SARS-CoV-2 viral strains B.1.1.7 (Alpha), B.1.351 (Beta), B.1.617.2 (Delta), P.1 and B.1.1.529 (Omicron). Since Ap1-4 had similar results, for simplicity only Ap1 is shown in the main figure (Fig. 2C) while Ap2-4 are shown in Supplemental Fig. 7C. Aptamers 1–4 were able to detect all viral strains in a similar fashion and showed a higher signal (higher relative binding) to the alpha strain compared to the subsequent viral strains, with protein from the Omicron variant having the poorest signal. Interestingly, Ap5 had a higher preference to the Omicron variant than the other viral strains which gave a statistically significant higher relative binding signal compared to Ap1 (p = 0.005) and the mAb control (p = 0.007, Fig. 2C). We followed up this finding by comparing Ap1 and Ap5 for their binding affinity for the Omicron variant at several concentrations and found that Ap5 had a detectable signal at concentrations as low as 0.50 ng/mL of recombinant N protein from the Omicron variant (B.1.1.52) compared to Ap1 at ∼ 50 ng/mL (Fig. 2D). Ap5 also gave a statistically significant higher relative binding signal compared to Ap1 at 100 ng/mL (p = 0.047) and to both Ap1 (p = 0.22) and the mAb (p = 0.039) at 200 ng/mL. This pattern was also true when comparing Ap1 and 5 for detection of inactivated SARS-CoV-2 Omicron variant (B.1.1.529), with Ap5 having a detectable signal at<125 k viral copies and having statistically higher relative binding signal compared to Ap1 and the mAb at all viral titers higher than 500 k/mL (Fig. 2E). These data suggest that identified aptamers differentially bind to N protein from different viral strains indicating preferential binding of different epitopes of the N protein. These preferences could thus provide an improved reagent for the detection of SARS-CoV-2 variants that have specific N protein changes, such as those found in the Omicron variant (Hossain, Akter et al. 2022).
SARS-COV-2 Omicron microarray reselection assay
Based on our finding that our aptamers preferentially bind N protein from a particular viral strain, to further improve our aptamers, we challenged the top 200 aptamers against N protein from the SARS-CoV-2 Omicron strain to develop a SARS-CoV-2 Omicron variant specific aptamer. Candidates were first tested against 1 × 10^8 genome equivalents/mL of Omicron N-protein target in 1X SELEX Buffer. Supplemental Fig. 8 (left), depicts the result of blocking candidates with Reporter oligo. Based on the image, most candidates interacted well with the Reporter, although there is a strong gradient in probe annealing. Despite this, assessment continued as candidate responses are normalized to pre-incubation fluorescence baselines. After this reading was taken, candidates were incubated with target for 16 h at 37 °C. The solution at the inlet of the peristaltic pump was then replaced with 1X SELEX Buffer to wash the target sample from the microarray (Supplemental Fig. 8, (right)). Fluorescence readings of each pixel were collected and averaged to produce a Mean Fluorescence value and Standard Deviation per candidate. Candidate response to target was then calculated as described in Equation (1). As a comparison similar approaches were used to analyze candidate response to 1 × 10^8 genome equivalents/mL of Delta N-protein in 1X SELEX Buffer (Supplemental Fig. 9) and 33 ng/mL of pooled counter-targets in 1X SELEX Buffer (Supplemental Fig. 10). Candidate percent responses to target samples and pooled counter-targets were calculated as the mean of the 18 replicate positions pre- and post-incubation, omitting colonies that did not successfully incorporate the fluorescent probe. Pre-incubation fluorescence was then divided by post-incubation fluorescence to produce a Signal Ratio value, which was then used to rank how responsive a sequence was to the assessment condition; elevated aptamer response would result in greater displacement of the fluorescent Reporter, and therefore represent a larger difference in pre- and post-incubation readings. The percent responses were then compared to determine candidates that specifically responded to the Omicron variant. Candidates were ranked according to the ratio of signal loss to either the Omicron Variant or to counter targets. The greater this score, the more response to the target condition relative to the counter condition, with scores > 2 being favorable. Only a handful of the strongly counter-target-responsive sequences were the same as those that responded strongly to Omicron (Supplemental Fig. 11). Two aptamer candidates (Fig. 3 A) were then synthesized for qualitative assessment.
Fig. 3.
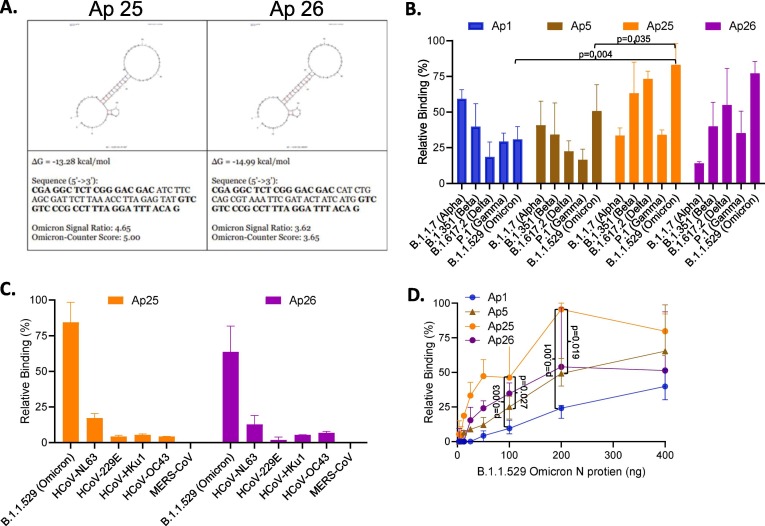
Identification of aptamers reselected against N protein from Omicron SARS-CoV-2 variant. The top 200 aptamers selected against the alpha variant were reselected against the omicron variant using a microarray assay. A. Top 2 Aptamer candidates based on highest binding affinity against SARS-CoV-2 Omicron Nucleocapsid Protein. B. Aptamers were evaluated for their binding to N protein from SARS-CoV-2 viral strains of interest (200 ng) using a hybrid ELASA format. C. Reselected aptamers were evaluated for their binding to recombinant nucleocapsid protein from HCoVs (200 ng/mL) using a direct ELASA format. Background was subtracted and error bars indicate standard deviation. D. Relative binding of aptamers as detection reagents for SARS-CoV-2 Omicron recombinant nucleocapsid protein. The error bars indicate standard deviation and statistical significance is indicated when p < 0.05 and was calculated using one-way ANOVA with Šidák multiple comparisons test (Figure D) with and two-sided t-tests with Welch’s correction (Figure B).
SARS-CoV-2 Omicron variant aptamers as detection reagents
We evaluated the top two aptamers, Ap25 and Ap26, discovered in the microarray assay for their efficiency as detection reagents for SARS-CoV-2 N protein from the Omicron variant of concern. As with Ap1 and Ap5, Ap25 and Ap26 were able to detect recombinant N protein (200 ng) from all variants of concern in a hybrid ELASA but had an improved signal to the Omicron and Delta variants (Fig. 3B). Ap 25 also had an increased signal for the Beta variant while Ap26 had a decreased signal of the Alpha variant compared to Ap1 and Ap5. Ap25 and Ap26 were then compared to Ap1 and Ap5 for their ability to detect various concentrations of recombinant N protein from the Omicron variant. Ap1 had the highest (poorest) limit of detection at ∼ 50 ng, followed by Ap5 and Ap26 at ∼ 12.5 ng with Ap25 reaching a limit of detection at less than ∼ 6.25 ng. Supportive of our reselection, Ap25 had an increase signal at all concentrations compared to any other aptamer, but Ap26 was only slightly better than Ap5 at concentrations at 200 ng and under but was lower at 400 ng of N protein. These data show that reselection of aptamers against emerging variants of concern can provide further improved detection reagents.
Identifying suitable aptamer pairs for a sandwich ELASA
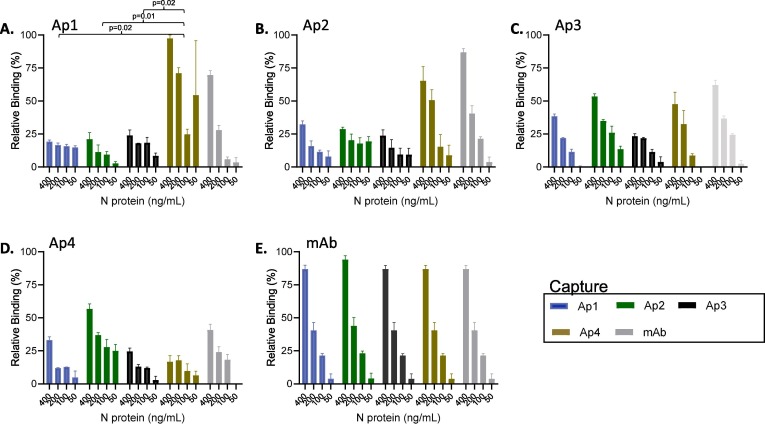
To pursue additional utility of the aptamers we sought to develop an ELASA sandwich assay in which aptamers were used as both capture and detections reagents. Testing several formats including binding directly to plastic, and biotinylating the aptamers and then binding to a streptavidin coated plate, but we found that aptamers in these formats were not well suited as capture reagents. However, following a protocol outlined by Svobodova et al. (Svobodova, et al. 2021) in which thiolated aptamers bound to a maleimide plate, aptamers were able to capture N portion from SARS-CoV-2 Alpha Variant. Specifically, thiolated aptamers 1–4 or a capture Ab were coated on a maleimide plate. SARS-CoV-2 N protein form the Alpha strain was added at various concentrations and incubated overnight. Biotinylated aptamers 1–4 and a detection Ab were used as detection reagents (Fig. 4 ) to identify suitable aptamer pairs. When using Ap1 as the detection and with Ap4 as capture a strong signal was observed that exceeded the Ab as capture control (Fig. 4A). This signal produced with the Ap1 detection and Ap4 capture pair reached statistical significance when compared to Ap1 detection pair with either itself (Ap1, p = 0.02) or the other two tested aptamers (Ap2, p = 0.01 and 3, p = 0.02) as capture, highlighting the Ap1 detection/Ap4 capture as a strong suitable pair. Ap4 as capture also had a signal when combined with Ap2 as detection but did not reach statistical difference to the other aptamers and was lower than the Ap2 detection with Ab capture (Fig. 2B) and to the Ap1 detection with Ap4 capture pair. Ap3 as detection with the capture aptamers produced a titred results correlating with the concentration of the protein, likely indicating a real result however the signal was not as high as other pairs (Fig. 4C). Interestingly, when Ap4 was used as detection with the other aptamers as capture there was only a mild signal produced with Ap2 (Fig. 4D). These findings are particularly interesting when comparing Ap1 detection and Ap4 capture compared to its inverse (Ap4 detection, Ap1 capture) as it indicates not all aptamers hold universal utility for being both capture and detection reagents, however, further work is required to fully understand this effect. While further work is required to develop aptamers as efficient capture reagents, these findings provide support that aptamers can be used as capture reagents for SARS-CoV-2 N protein providing potential future options for novel capture and detection pairs.
Fig. 4.
Screening of aptamer pairs for the detection of SARS-CoV-2 N protein. Thiolated capture aptamer was analyzed in combination with each of the biotinylated detection aptamers using 400 to 50 ng/mL of SARS-CoV-2 N recombinant protein. The graph title refers to the biotinylated aptamer used as detection (or detection monoclonal antibody), and the colors indicate the thiolated capture aptamer (or capture antibody). The error bars indicate standard deviation and statistical significance is indicated when p < 0.05 and was calculated using one-way ANOVA.
Discussion
Despite the availability of vaccines, SARS-CoV-2 continues to spread worldwide in part due to the challenges of testing to understand infectious status when interacting with the public. While genomic testing remains the most reliable for detection of viral infection, the limitations of this technology have prevented continued effective screening and viral monitoring, leading to increased viral spread. To this extent, antigen diagnostic assays provide alluring alternatives with rapid testing that is affordable and applicable for at home use. However, using antibodies as reagents in these tests comes with limitations, particularly adaption to emerging viral strains. In this regard, we developed aptamers specific to the N protein of SARS-CoV-2 that when used as detection reagents in ELISA based assays were comparable to antibodies regarding detection limits, which was in low picomolar range, and specificity exhibiting only negligible cross reactivity to the N protein from other HCoVs. Furthermore, reselection for aptamers that bind to N protein from the Omicron variant of concern produced sequences with specific responses to Omicron N-Protein that translated to an improved limit of detection when used as detection reagents.
The purpose of this study was to select aptamers that could substitute or support antibodies in antigen-based diagnostic assays, particularly with the reselection of our aptamers to quickly adapt to the detection of emerging variants. Others have been successful in using aptamers in place of antibodies for detecting SARS-CoV-2(Avni and Schulman, 1987, Chen et al., 2020, Cho et al., 2011, Kang et al., 2021, Li et al., 2021, Valero et al., 2021, Zhang et al., 2020, Zhang et al., 2021, AminiLi et al., 2022) in several formats including in a SARS-CoV-2 S protein LFA(Yang et al., 2022b). However, many of these assays used aptamers selected against recombinant protein produced in other cell types and were not treated with detergent which reasonably expected to result in confirmational differences that may prevent optimal binding and reduce effectiveness in tests that require deactivation of the virus. Here, to ensure the target protein was similar to what would be detected in a point of care assay, i.e., detergent inactivated virus, the aptamer selection was performed on N protein that was grown in human cells and was treated with detergent and kept in a high salt concentration to prevent oligomerization. To produce the best aptamers, the SELEX procedure here used high-stringency conditions including low target concentrations (<1 nM) of purified target, detergent included in the selection buffer, and included a parallel assessment using counter targets comprised of protein from other HCoVs and two positive controls: detergent treated N protein in viral transport medium (VTM) and detergent inactivated virus in VTM. Aptamer sequences that were identified in both positive control samples but not in the counter targets provided a highly enriched library specific to SARS-CoV-2.
To confirm our aptamers were viable in an ELASA, we first utilized a direct assay in which protein was bound to a plastic plate and aptamers were used for detection. We found that our top aptamers were comparable to a commercially available antibody in detecting SARS-COV-2 N protein from the Alpha strain, thus illustrating the ability of our aptamers to perform as designed. To take our aptamers a step further, we next evaluated the aptamers in assays against inactivated virus using contrived samples with virus in VTM. In this regard, we utilized a hybrid ELASA format in which a previously validated antibody was used for capture of SARS-CoV-2. This hybrid format allowed soluble virus to be captured, expediting the evaluation of the aptamers as detection reagents, and bypassing the need to determine aptamers pairs for capture and detection. Using this format, we found that our aptamers worked as well as a validated antibody for detection of SARS-CoV-2 Alpha Strain. While our goal was to improve detection limits, this finding of non-inferiority compared to an antibody shows great promise for the aptamers as they are cheaper to produce and are more stable than antibodies among other advantages. We also tested our aptamers in contrived samples mixed with nasopharyngeal swab samples and with saliva and found that nasopharyngeal swabs did not impact our detection limits, but saliva had an inhibitory effect. However, others have had success using aptamers in saliva based tests (Zhang et al. 2022), thus it is possible that with some additional modifications our assay would be suitable for other bodily fluids. Feasibly, these findings suggest aptamers could replace antibodies as the detection portion of the assay.
We next challenged our aptamers against all viral strains of concern as of June 2022 and found that all our aptamers were able to detect all strains, and interestingly Aptamer 5 produced a higher signal against the Omicron variant. This finding was particularly noteworthy as several other antibody tests had been rendered unable to detect, or had a decreased sensitivity, of the Omicron variant and this variant is known to have changes in the N protein (Hossain, et al. 2022). Having specificity to viral strains, particularly of those that evade antibody detection, and can be selected and substituted quickly is one of the distinct advantages of aptamers compared to antibodies. To further pursue this idea, we performed a microarray reselection assay with our aptamers against Omicron inactivated virus to determine which of our aptamers had a higher preference, indicating a better reagent to use in antigen tests to detect the Omicron variant. Considering both affinity and specificity, we were able to identify two novel sequences as strong, specific aptamers for Omicron N-Protein. Identified aptamers were then tested for sensitivity for Omicron N-Protein and found to produce a high signal that indicated increased detection limits by more than two-fold and they maintained their specificity for SARS-CoV-2 compared to other HCoVs. The preference of these new aptamers and Aptamer 5 to Omicron compared to the Alpha strain may indicate a unique binding sequence, which to fully elucidate would present an interesting avenue for a future study. In a cursory attempt to explain this effect we found that each of these aptamers with a preference for the Omicron variant had a sequence of TAAA that the other tested aptamers did not, potentially indicating this sequence as the cause for the higher affinity to the Omicron N protein, however further testing would be required to confirm this.This success highlights the feasibility and provides proof of concept of this approach to quickly adapt aptamers against emerging variants of concern.
Here we illustrate the selection of aptamers for SARS-CoV-2 that can replace antibodies in antigen-based assays. Further, we identified specific aptamers with a higher prefrence to the Omicron variant, illustrating its use as a promising reagent in current tests and validating the method for the selection of aptamers against emerging viral strains. While we did have some success in using aptamers as capture reagents, our aptamers in most cases were not able to perform as well as antibodies, thus the utility of aptamers as capture reagents requires further investigation. Additionally, while our process identified aptamers that could perform as well as antibodies as detection reagents, further research may focus on optimizing aptamer functionality through truncation to a core binding domain, re-selection to further explore sequence space with a biased library based on the successful candidates, and/or the incorporation of modified nucleotides for enhanced affinity and specificity, which may particularly improve aptamers as detection or even capture reagents. Nevertheless, our results here illustrate the utility of aptamers in viral diagnostic assays which could decrease cost, improve detection limits, and be quickly adapted for emerging viral strains of SARS-CoV-2 or even novel viruses yet to be discovered.
Author Contributions
C.P.N. designed research; C.P.N., B.M. and R.S. performed research; C.P.N., B.M and R.S. analyzed data and C.P.N., G.T.C., S.M.A. and A.L. wrote the manuscript. B.J.G. provided key reagents and B.J.G., S.M.A. and M.C. provided critical review of the manuscript.
Sources of Support
This work was supported by NIH Grant U01HL152405.
CRediT authorship contribution statement
C.P.N.: Conceptualization, Data curation, Formal analysis, Funding acquisition, Methodology, Project administration, Supervision, Writing – original draft; M.C.: Conceptualization, Writing – review & editing; B.J.G.: Resources, Methodology, Writing – review & editing; G.T.C.: Methodology, Writing – original draft; A.L.: Data curation, Methodology, Writing – original draft; S.M.A.: Writing – original draft, Writing – review & editing; B.M.: Data curation, Formal analysis; R.S.: Data curation, Formal analysis
Declaration of Competing Interest
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests: [G. Thomas Caltagirone and Albert Liao reports a relationship with Aptagen that includes: employment.].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crbiot.2023.100132.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- Abrego-Martinez J.C., Jafari M., Chergui S., Pavel C., Che D., Siaj M. Aptamer-based electrochemical biosensor for rapid detection of SARS-CoV-2: Nanoscale electrode-aptamer-SARS-CoV-2 imaging by photo-induced force microscopy. Biosens. Bioelectron. 2022;195 doi: 10.1016/j.bios.2021.113595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ai J.W., Zhang Y., Zhang H.C., Xu T., Zhang W.H. Era of molecular diagnosis for pathogen identification of unexplained pneumonia, lessons to be learned. Emerg Microbes Infect. 2020;9(1):597–600. doi: 10.1080/22221751.2020.1738905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AminiLi, R., Z. ZhangLi, J. LiLi, J. Gu, J. D. Brennan and Y. Li (2022). “Aptamers for SARS-CoV-2: Isolation, Characterization, and Diagnostic and Therapeutic Developments.” Anal Sens: e202200035. [DOI] [PMC free article] [PubMed]
- Avni E.F., Schulman C.C. The status of medical imagery in pediatric urology. Acta Urol. Belg. 1987;55(1):43–46. [PubMed] [Google Scholar]
- Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521(7552):274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- Bar-On Y.M., Flamholz A., Phillips R., Milo R. SARS-CoV-2 (COVID-19) by the numbers. Elife. 2020;9 doi: 10.7554/eLife.57309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunka D.H., Stockley P.G. Aptamers come of age - at last. Nat. Rev. Microbiol. 2006;4(8):588–596. doi: 10.1038/nrmicro1458. [DOI] [PubMed] [Google Scholar]
- Cameroni E., Bowen J.E., Rosen L.E., Saliba C., Zepeda S.K., Culap K., Pinto D., VanBlargan L.A., De Marco A., di Iulio J., Zatta F., Kaiser H., Noack J., Farhat N., Czudnochowski N., Havenar-Daughton C., Sprouse K.R., Dillen J.R., Powell A.E., Chen A., Maher C., Yin L., Sun D., Soriaga L., Bassi J., Silacci-Fregni C., Gustafsson C., Franko N.M., Logue J., Iqbal N.T., Mazzitelli I., Geffner J., Grifantini R., Chu H., Gori A., Riva A., Giannini O., Ceschi A., Ferrari P., Cippa P.E., Franzetti-Pellanda A., Garzoni C., Halfmann P.J., Kawaoka Y., Hebner C., Purcell L.A., Piccoli L., Pizzuto M.S., Walls A.C., Diamond M.S., Telenti A., Virgin H.W., Lanzavecchia A., Snell G., Veesler D., Corti D. Broadly neutralizing antibodies overcome SARS-CoV-2 Omicron antigenic shift. Nature. 2022;602(7898):664–670. doi: 10.1038/s41586-021-04386-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Wu Q., Chen J., Ni X., Dai J. A DNA Aptamer Based Method for Detection of SARS-CoV-2 Nucleocapsid Protein. Virol. Sin. 2020;35(3):351–354. doi: 10.1007/s12250-020-00236-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen A., Yang S. Replacing antibodies with aptamers in lateral flow immunoassay. Biosens. Bioelectron. 2015;71:230–242. doi: 10.1016/j.bios.2015.04.041. [DOI] [PubMed] [Google Scholar]
- Cho S.J., Woo H.M., Kim K.S., Oh J.W., Jeong Y.J. Novel system for detecting SARS coronavirus nucleocapsid protein using an ssDNA aptamer. J. Biosci. Bioeng. 2011;112(6):535–540. doi: 10.1016/j.jbiosc.2011.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubuk J., Alston J.J., Incicco J.J., Singh S., Stuchell-Brereton M.D., Ward M.D., Zimmerman M.I., Vithani N., Griffith D., Wagoner J.A., Bowman G.R., Hall K.B., Soranno A., Holehouse A.S. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat. Commun. 2021;12(1):1936. doi: 10.1038/s41467-021-21953-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- He Y., Zhou Y., Wu H., Kou Z., Liu S., Jiang S. Mapping of antigenic sites on the nucleocapsid protein of the severe acute respiratory syndrome coronavirus. J. Clin. Microbiol. 2004;42(11):5309–5314. doi: 10.1128/JCM.42.11.5309-5314.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hossain A., Akter S., Rashid A.A., Khair S., Alam A. Unique mutations in SARS-CoV-2 Omicron subvariants' non-spike proteins: Potential impacts on viral pathogenesis and host immune evasion. Microb. Pathog. 2022;170 doi: 10.1016/j.micpath.2022.105699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y., Yang C., Xu X.F., Xu W., Liu S.W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol. Sin. 2020;41(9):1141–1149. doi: 10.1038/s41401-020-0485-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji T., Liu Z., Wang G., Guo X., Akbar Khan S., Lai C., Chen H., Huang S., Xia S., Chen B., Jia H., Chen Y., Zhou Q. Detection of COVID-19: A review of the current literature and future perspectives. Biosens. Bioelectron. 2020;166 doi: 10.1016/j.bios.2020.112455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T.C., Biele G., Muhlemann B., Veith T., Schneider J., Beheim-Schwarzbach J., Bleicker T., Tesch J., Schmidt M.L., Sander L.E., Kurth F., Menzel P., Schwarzer R., Zuchowski M., Hofmann J., Krumbholz A., Stein A., Edelmann A., Corman V.M., Drosten C. Estimating infectiousness throughout SARS-CoV-2 infection course. Science. 2021;373(6551) doi: 10.1126/science.abi5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Jang H., Yeom G., Kim M.G. Ultrasensitive Detection Platform of Disease Biomarkers Based on Recombinase Polymerase Amplification with H-Sandwich Aptamers. Anal. Chem. 2021;93(2):992–1000. doi: 10.1021/acs.analchem.0c03822. [DOI] [PubMed] [Google Scholar]
- Lakhin A.V., Tarantul V.Z., Gening L.V. Aptamers: problems, solutions and prospects. Acta Nat. 2013;5(4):34–43. [PMC free article] [PubMed] [Google Scholar]
- Leuzinger K., Roloff T., Egli A., Hirsch H.H. Impact of SARS-CoV-2 Omicron on Rapid Antigen Testing Developed for Early-Pandemic SARS-CoV-2 Variants. Microbiol Spectr. 2022;10(4):e0200622. doi: 10.1128/spectrum.02006-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhang Z., Gu J., Stacey H.D., Ang J.C., Capretta A., Filipe C.D.M., Mossman K.L., Balion C., Salena B.J., Yamamura D., Soleymani L., Miller M.S., Brennan J.D., Li Y. Diverse high-affinity DNA aptamers for wild-type and B.1.1.7 SARS-CoV-2 spike proteins from a pre-structured DNA library. Nucleic Acids Res. 2021;49(13):7267–7279. doi: 10.1093/nar/gkab574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long C., Xu H., Shen Q., Zhang X., Fan B., Wang C., Zeng B., Li Z., Li X., Li H. Diagnosis of the Coronavirus disease (COVID-19): rRT-PCR or CT? Eur. J. Radiol. 2020;126 doi: 10.1016/j.ejrad.2020.108961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magazine N., Zhang T., Wu Y., McGee M.C., Veggiani G., Huang W. Mutations and Evolution of the SARS-CoV-2 Spike Protein. Viruses. 2022;14(3) doi: 10.3390/v14030640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahase E. Covid-19: 120 million rapid tests pledged to low and middle income countries. BMJ. 2020;371 doi: 10.1136/bmj.m3857. [DOI] [PubMed] [Google Scholar]
- Nguyen V.T., Seo H.B., Kim B.C., Kim S.K., Song C.S., Gu M.B. Highly sensitive sandwich-type SPR based detection of whole H5Nx viruses using a pair of aptamers. Biosens. Bioelectron. 2016;86:293–300. doi: 10.1016/j.bios.2016.06.064. [DOI] [PubMed] [Google Scholar]
- Nikam P.S., Palachandra S., Kingston J.J. In vitro selection and characterization of ssDNA aptamers by cross-over SELEX and its application for detection of S. Typhimurium. Anal. Biochem. 2022;656 doi: 10.1016/j.ab.2022.114884. [DOI] [PubMed] [Google Scholar]
- Nimjee S.M., Rusconi C.P., Sullenger B.A. Aptamers: an emerging class of therapeutics. Annu. Rev. Med. 2005;56:555–583. doi: 10.1146/annurev.med.56.062904.144915. [DOI] [PubMed] [Google Scholar]
- Ravi N., Cortade D.L., Ng E., Wang S.X. Diagnostics for SARS-CoV-2 detection: A comprehensive review of the FDA-EUA COVID-19 testing landscape. Biosens. Bioelectron. 2020;165 doi: 10.1016/j.bios.2020.112454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schutze T., Wilhelm B., Greiner N., Braun H., Peter F., Morl M., Erdmann V.A., Lehrach H., Konthur Z., Menger M., Arndt P.F., Glokler J. Probing the SELEX process with next-generation sequencing. PLoS One. 2011;6(12):e29604. doi: 10.1371/journal.pone.0029604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sender R., Bar-On Y.M., Gleizer S., Bernshtein B., Flamholz A., Phillips R., Milo R. The total number and mass of SARS-CoV-2 virions. PNAS. 2021;118(25) doi: 10.1073/pnas.2024815118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shereen M.A., Khan S., Kazmi A., Bashir N., Siddique R. COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses. J. Adv. Res. 2020;24:91–98. doi: 10.1016/j.jare.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz Karita H.C., Dong T.Q., Johnston C., Neuzil K.M., Paasche-Orlow M.K., Kissinger P.J., Bershteyn A., Thorpe L.E., Deming M., Kottkamp A., Laufer M., Landovitz R.J., Luk A., Hoffman R., Roychoudhury P., Magaret C.A., Greninger A.L., Huang M.L., Jerome K.R., Wener M., Celum C., Chu H.Y., Baeten J.M., Wald A., Barnabas R.V., Brown E.R. Trajectory of Viral RNA Load Among Persons With Incident SARS-CoV-2 G614 Infection (Wuhan Strain) in Association With COVID-19 Symptom Onset and Severity. JAMA Netw. Open. 2022;5(1):e2142796. doi: 10.1001/jamanetworkopen.2021.42796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svobodova M., Skouridou V., Jauset-Rubio M., Vieitez I., Fernandez-Villar A., Cabrera Alvargonzalez J.J., Poveda E., Bofill C.B., Sans T., Bashammakh A., Alyoubi A.O., O'Sullivan C.K. Aptamer Sandwich Assay for the Detection of SARS-CoV-2 Spike Protein Antigen. ACS Omega. 2021;6(51):35657–35666. doi: 10.1021/acsomega.1c05521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed, A. M., A. Ciling, T. Y. Taha, I. P. Chen, M. M. Khalid, B. Sreekumar, P. Y. Chen, G. R. Kumar, R. Suryawanshi, I. Silva, B. Milbes, N. Kojima, V. Hess, M. Shacreaw, L. Lopez, M. Brobeck, F. Turner, L. Spraggon, T. Tabata, M. Ott and J. A. Doudna (2022). “Omicron mutations enhance infectivity and reduce antibody neutralization of SARS-CoV-2 virus-like particles.” Proc Natl Acad Sci U S A119(31): e2200592119. [DOI] [PMC free article] [PubMed]
- Syed A.M., Taha T.Y., Tabata T., Chen I.P., Ciling A., Khalid M.M., Sreekumar B., Chen P.Y., Hayashi J.M., Soczek K.M., Ott M., Doudna J.A. Rapid assessment of SARS-CoV-2-evolved variants using virus-like particles. Science. 2021;374(6575):1626–1632. doi: 10.1126/science.abl6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry J.S., Anderson L.B., Scherman M.S., McAlister C.E., Perera R., Schountz T., Geiss B.J. Development of a SARS-CoV-2 nucleocapsid specific monoclonal antibody. Virology. 2021;558:28–37. doi: 10.1016/j.virol.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249(4968):505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- Valero J., Civit L., Dupont D.M., Selnihhin D., Reinert L.S., Idorn M., Israels B.A., Bednarz A.M., Bus C., Asbach B., Peterhoff D., Pedersen F.S., Birkedal V., Wagner R., Paludan S.R., Kjems J. A serum-stable RNA aptamer specific for SARS-CoV-2 neutralizes viral entry. PNAS. 2021;118(50) doi: 10.1073/pnas.2112942118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voskuil J. Commercial antibodies and their validation. F1000Res. 2014;3:232. doi: 10.12688/f1000research.4966.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, H., K. R. Paulson, S. A. Pease, S. Watson, H. Comfort, P. Zheng, A. Y. Aravkin, C. Bisignano, R. M. Barber, T. Alam, E. A. M. John E Fuller, Darwin Phan Jones, Meghan E Frisch, Cristiana Abbafati, Chri and S. A. P. Katherine R Paulson, Stefanie Watson, Haley Comfort, Peng Zheng, Aleksandr Y Aravkin, Catherine Bisignano, Ryan M Barber, Tahiya Alam, John E Fuller, Erin A May, Darwin Phan Jones, Meghan E Frisch, Cristiana Abbafati, Christopher Adolph, Adrien Allorant, Joanne O Amlag, Bree Bang-Jensen, Gregory J Bertolacci, Sabina S Bloom, Austin Carter, Emma Castro, Suman Chakrabarti, Jhilik Chattopadhyay, Rebecca M Cogen, James K Collins, Kimberly Cooperrider, Xiaochen Dai, William James Dangel, Farah Daoud, Carolyn Dapper, Amanda Deen, Bruce B Duncan, Megan Erickson, Samuel B Ewald, Tatiana Fedosseeva, Alize J Ferrari, Joseph Jon Frostad, Nancy Fullman, John Gallagher, Amiran Gamkrelidze, Gaorui Guo, Jiawei He, Monika Helak, Nathaniel J Henry, Erin N Hulland, Bethany M Huntley, Maia Kereselidze, Alice Lazzar-Atwood, Kate E LeGrand, Akiaja Lindstrom, Emily Linebarger, Paulo A Lotufo, Rafael Lozano, Beatrice Magistro, Deborah Carvalho Malta, Johan Månsson, Ana M Mantilla Herrera, Fatima Marinho, Alemnesh H Mirkuzie, Awoke Temesgen Misganaw, Lorenzo Monasta, Paulami Naik, Shuhei Nomura, Edward G O'Brien, James Kevin O'Halloran, Latera Tesfaye Olana, Samuel M Ostroff, Louise Penberthy, Robert C Reiner Jr, Grace Reinke, Antonio Luiz P Ribeiro, Damian Francesco Santomauro, Maria Inês Schmidt, David H Shaw, Brittney S Sheena, Aleksei Sholokhov, Natia Skhvitaridze, Reed J D Sorensen, Emma Elizabeth Spurlock, Ruri Syailendrawati, Roman Topor-Madry, Christopher E Troeger, Rebecca Walcott, Ally Walker, Charles Shey Wiysonge, Nahom Alemseged Worku, Bethany Zigler, David M Pigott, Mohsen Naghavi, Ali H Mokdad, Stephen S Lim, Simon I Hay, Emmanuela Gakidou, Christopher J L Murray (2022). “Estimating excess mortality due to the COVID-19 pandemic: a systematic analysis of COVID-19-related mortality, 2020-21.” Lancet399(10334): 1513-1536.
- Weissleder R., Lee H., Ko J., Pittet M.J. COVID-19 diagnostics in context. Sci. Transl. Med. 2020;12(546) doi: 10.1126/scitranslmed.abc1931. [DOI] [PubMed] [Google Scholar]
- Yang L.F., Kacherovsky N., Liang J., Salipante S.J., Pun S.H. SCORe: SARS-CoV-2 Omicron Variant RBD-Binding DNA Aptamer for Multiplexed Rapid Detection and Pseudovirus Neutralization. Anal. Chem. 2022 doi: 10.1021/acs.analchem.2c01993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang L.F., Kacherovsky N., Panpradist N., Wan R., Liang J., Zhang B., Salipante S.J., Lutz B.R., Pun S.H. Aptamer Sandwich Lateral Flow Assay (AptaFlow) for Antibody-Free SARS-CoV-2 Detection. Anal. Chem. 2022;94(20):7278–7285. doi: 10.1021/acs.analchem.2c00554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Fang X., Liu X., Ou H., Zhang H., Wang J., Li Q., Cheng H., Zhang W., Luo Z. Discovery of sandwich type COVID-19 nucleocapsid protein DNA aptamers. Chem. Commun. (Camb) 2020;56(70):10235–10238. doi: 10.1039/d0cc03993d. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Pandey R., Li J., Gu J., White D., Stacey H.D., Ang J.C., Steinberg C.J., Capretta A., Filipe C.D.M., Mossman K., Balion C., Miller M.S., Salena B.J., Yamamura D., Soleymani L., Brennan J.D., Li Y. High-Affinity Dimeric Aptamers Enable the Rapid Electrochemical Detection of Wild-Type and B.1.1.7 SARS-CoV-2 in Unprocessed Saliva. Angew. Chem. Int. Ed. Engl. 2021;60(45):24266–24274. doi: 10.1002/anie.202110819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X., Zhang Z., Li J., Huang X., Wei J., Yang J., Guan L., Wen X., Wang S., Qin Q. A Novel Sandwich ELASA Based on Aptamer for Detection of Largemouth Bass Virus (LMBV) Viruses. 2022;14(5) doi: 10.3390/v14050945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.