Abstract
Numerous epidemic lung diseases such as COVID-19, tuberculosis (TB), and pneumonia have spread over the world, killing millions of people. Medical specialists have experienced challenges in correctly identifying these diseases due to their subtle differences in Chest X-ray images (CXR). To assist the medical experts, this study proposed a computer-aided lung illness identification method based on the CXR images. For the first time, 17 different forms of lung disorders were considered and the study was divided into six trials with each containing two, two, three, four, fourteen, and seventeen different forms of lung disorders. The proposed framework combined robust feature extraction capabilities of a lightweight parallel convolutional neural network (CNN) with the classification abilities of the extreme learning machine algorithm named CNN-ELM. An optimistic accuracy of 90.92% and an area under the curve (AUC) of 96.93% was achieved when 17 classes were classified side by side. It also accurately identified COVID-19 and TB with 99.37% and 99.98% accuracy, respectively, in 0.996 microseconds for a single image. Additionally, the current results also demonstrated that the framework could outperform the existing state-of-the-art (SOTA) models. On top of that, a secondary conclusion drawn from this study was that the prospective framework retained its effectiveness over a range of real-world environments, including balanced-unbalanced or large-small datasets, large multiclass or simple binary class, and high- or low-resolution images. A prototype Android App was also developed to establish the potential of the framework in real-life implementation.
Keywords: COVID19, Convolutional neural network, Extreme learning machine, Mobile apps, Pneumonia, Tuberculosis
1. Introduction
With the advancement in medical science and technology, a large number of previously incurable diseases now can be completely treatable (Vandenberg et al., 2021). Despite significant advances, emerging new diseases such as COVID-19 continue to present new challenges of quick and accurate identification and finding relevant treatment solutions. COVID-19 has spread in every corner of the world and affected millions of people, and many have lost their lives. It has been estimated that at the beginning of March 2022 nearly six million people have died and over 450 million have been afflicted by COVID-19 in the past three years (World Health Organization, 2022). Flu-like symptoms such as fever, dry cough, fatigue, and difficulty in breathing are generally experienced by the patients. In the severe cases, the COVID-19 frequently results in life-threatening pneumonia (Huff & Singh, 2020), which is a respiratory infection that incapacitates the lungs. When a healthy individual breathes, the lungs' tiny sacs called alveoli get filled with air. However, if a person contracts pneumonia, the alveoli get loaded with pus and fluid, obstructing breathing, and limiting oxygen absorption (Ruuskanen et al., 2011). Around 7% of the global population (450 million people) is impacted by pneumonia alone, and around 2 million people die from pneumonia each year (Ruuskanen et al., 2011).
The popular reverse transcription-polymerase chain reaction (RT-PCR) takes up to forty-eight hours to confirm the presence of coronavirus (Zhu et al., 2020). Since this technique is highly time-consuming and owing to the lack of other resources, a COVID-19 infected person can continue to spread the virus to their close contacts (Vandenberg et al., 2021). However, COVID-19 can be detected using chest X-ray analysis, which is comparatively a faster technique to speed up the diagnosis. However, the X-ray image has to be analyzed by a radiologist manually. Every year in the United States alone, more than 35 million CXR images are collected as part of medical treatment (Kamel et al., 2017). Increasing workload and exhaustion, are already a commonplace among the radiologists who must routinely review in excess of 100 CXR images each day (Kamel et al., 2017). Furthermore, radiologists' diagnosis can differ because of human judgment increasing the prospect of incorrect diagnosis. Numerous other lung diseases such as tuberculosis, cardiomegaly, opacity, and pleural that can also be detected from the X-ray image analysis, make it even more challenging for the radiologists to correctly identify the diseases. Therefore, it requires an automated and intelligent system such as artificial intelligence (AI) that can quickly and accurately identify any lung diseases with high classification accuracy and without consuming a lot of time or resource so that the system can be applied to real-world disease identification while significantly reducing the workload of the radiologists.
Machine learning (ML) and Deep learning (DL) have been applied effectively and efficiently in a wide variety of medical applications. However, for image classification, traditional machine learning models often rely on hand-crafted features, which necessitate multiple steps for extraction. In contrast, DL models have the advantage of automatically learning and extracting relevant features from the data, bypassing the need for manual feature extraction step. The key advantage of the DL in this context is the elimination of the complex feature engineering process typically associated with traditional machine learning techniques (Molina et al., 2021). Due to the recent availability of large-scale data sets, various attempts have been made to automatically identify lung-related anomalies using CXR images.
In 2017, Wang et al. developed a larger database of CXR images that consisted of eight thoracic diseases with over 100,000 images (Wang et al., 2017). Later, this database was expanded by adding images of additional six thoracic diseases to create a ChestX-ray14 (CXR14) dataset containing fourteen thoracic diseases. In the CXR14 dataset, radiologists provided a small number of CXR images with hand-labeled bounding boxes (B-Boxes) to reveal the affected area of the disease in the CXR images. They used several pre-trained transfer learning (TL) models (AlexNet, GoogleNet, VGGNet-16, and ResNet-50) for detecting the diseases from the CXR images and achieved the highest average area under the curve (AUC) of 74.51% using ResNet-50. Yao et al. handled multilabel classification of thoracic diseases from CXR14 images by using a DenseNet-121 as an encoder (Yao et al. 2017). Subsequently, a decoder was utilized to optimize interdependencies among the target labels in predicting 14 pathogenic abnormalities using a long short-term memory (LSTM) network. The proposed framework surpassed the results of Wang et al. with an average AUC of 0.798. Likewise, Rajpurkar et al. proposed a CNN architecture containing 121 layers named CheXNet to detect pneumonia from the CXR14 dataset (Rajpurkar et al., 2017). They employed class activation mappings to display the characteristics region associated with the disease from CXR images (CAMs). In comparison to the prior research (Wang et al., 2017, Yao et al., 2017), the CheXNet produced higher AUC for each condition. Their model’s F1 metric was 0.435, compared to the average F1-score of 0.387 given by four radiologists. On the other hand, Kumar et al. developed a boosted cascaded CNN (BCCNN) for multilabel classification from the CXR14 dataset (Kumar et al., 2018). For calculating the loss for the multilabel classification, they used binary relevance and a pairwise error loss function. The BCCNN achieved a higher AUC for only the cardiomegaly disease with 91.33% than the previous state-of-the-art (SOTA) models (Rajpurkar et al., 2017). The CXR14 database consisted of multiple diseases and was imbalanced. In 2018, Ge et al. addressed these two problems by utilizing a novel error function named multilabel softmax loss (Ge et al., 2018). Various TL models (DenseNet-121, ResNet18, and VGG) were used for predicting the 14 diseases and achieved the highest AUC of 85.37% while using the ensembling model (DenseNet121-VGG) that surpassed the previous SOTA models (Kumar et al., 2018, Rajpurkar et al., 2017, Wang et al., 2017, Yao et al., 2017). In contrast, Gundel et al. developed a location-aware dense network (DNetLoc) based on DenseNet-121 to identify abnormalities in the CXR images from two well-known datasets: CXR14 and PLCO (Guendel et al., 2018). The authors optimally leveraged high-resolution CXR data by comprising spatial knowledge from CXR abnormalities. The proposed DNetLoc (AUC 80.7%) surpassed the AUC performance of Wang et al. (AUC 74.51%). In 2019, Baltruscha et al. employed two TL models named ResNet-38 and ResNet-101 to detect the thoracic diseases from the CXR14 dataset (Baltruschat et al., 2019). The classification was carried out by blending non-image data, for instance, patient age, gender, etc., with CXR images. ResNet-38 achieved the highest Receiver Operating Characteristic (ROC) for discriminating against 14 lung diseases when compared to the SOTA models. In order to minimize the negative effects of imbalance present in the CXR14 dataset, Wang et al. introduced an adaptive sampling strategy that automatically increases the weight of the comparatively poorly performed classes (Wang et al., 2020). For disease detection, they employed a TL model called DenseNet-121, which was trained adaptively and yielded a promising outcome. Ouyang et al. localized and diagnosed the abnormalities from the CXR14 and CheXpert databases while using a new attention-driven weakly supervised algorithm (Ouyang et al., 2020). They used gradient-base visual attention in a holistic way and achieved a more favorable mean AUC of 81.9% which was higher than the AUCs obtained from the SOTA models. Guan and Huang developed a category-wise residual attention learning model for multi-label CXR14 images classification (Guan & Huang, 2020). They considered two methods: feature embedding and attention learning. ResNet-50 and DenseNet-121 models were used for embedding the features and achieved a mean AUC of 81.6% that outperformed the SOTA models.
Oh et al. proposed a patch-based CNN that was trained with a small dataset consisting of three diseases: bacterial pneumonia, tuberculosis (TB), and viral/COVID-19 (Oh et al., 2020). The authors segmented the lung contour using the fully conventional DenseNet103 and then performed classification by ResNet-18. To detect COVID-19 and pneumonia, Khan et al. constructed a CoroNet based on the pre-trained Xception model (Khan et al. 2020). For three-class (Normal vs. Bacterial Pneumonia vs. COVID-19) and four-class (Normal vs. COVID- 19 vs. Bacterial vs. Viral Pneumonia) classifications, the proposed model achieved a precision and recall score of 93.2% and 98.2%, respectively. Pandit et al. employed VGG19 TL model to detect the COVID-19 (Pandit et al., 2021). The authors trained their model with 1,428 CXR images and obtained an accuracy of 92.53 % for binary class (COVID19 vs Normal) and 96 % for three-class (Normal vs Bacterial Pneumonia vs COVID19) classifications. To detect COVID19, viral pneumonia, and bacterial pneumonia from CXR images, Yamac et al. built a convolutional sparse support estimator network based on a neural network (Yamac et al., 2021). After training their model with a total of 6,200 CXR images, an accuracy of 0.8707 for four-class classification and 0.959 for binary classification (COVID-19 and normal). Gour and Jain employed two TL models, VGG19 and Xception, and utilized a softmax classifier to detect COVID-19 from both CXR and CT images (Gour & Jain, 2022). A sensitivity of 97.62% for three-class classification (COVID-19 vs. Pneumonia vs. Normal) was achieved. To detect viral and COVID19 pneumonia from CXR images, Chowdhury et al. used several pre-trained TL models (Chowdhury et al., 2020). For training their models, numerous datasets were merged and accuracies of 99.7% and 97.9% were attained for two- and three-class (normal vs viral vs COVID-19) classifications respectively. Rahman et al. achieved a promising accuracy of 93.3% while using pre-trained DenseNet-201 model for a three-class classification (normal vs bacterial vs viral pneumonia) (Rahman et al., 2020a).
Akter et al. used a variety of TL models, including VGG19 and GoogLeNet to identify COVID-19 (Akter et al., 2021). To balance the datasets, they employed data augmentation and trained the TL models using a total of 52,000 CXR images. Using MobileNetV2, they achieved a high accuracy of 98% for binary classification in 2 hours, 50 minutes, and 21 seconds of compilation time. To diagnose COVID-19, Rasheed et al. developed two classifiers: logistic regression (LR) and CNN (Rasheed et al., 2021). For data augmentation, a generative adversarial network was deployed, and principal component analysis (PCA) was used to identify the most significant features. A model was developed by training 308 CXR images and PCA facilitated to attain an accuracy of 97.6%. Chandra et al. created a computer-aided (CAD) system to detect TB from CXR images (Chandra et al, 2020). The authors employed a guided image filter to de-noise the image before performing lung segmentation and classification. After extracting features, a support vector machine was used to attain accuracies of 95.60% and 99.40% based on the Montgomery and Shenzhen (SZ) datasets, respectively. Sahlol et al. retrieved 50,000 important features from CXR images using a pre-trained MobileNet model. The authors selected relevant features by employing an artificial ecosystem-based optimization algorithm. TB was detected from SZ and Dataset 2 with accuracies of 90.2% and 94.1%, respectively. Rahman et al. used nine TL and two U-net models to diagnose TB, whereas they achieved the highest accuracy of 98.6% using DenseNet201 (Rahman et al., 2020b).
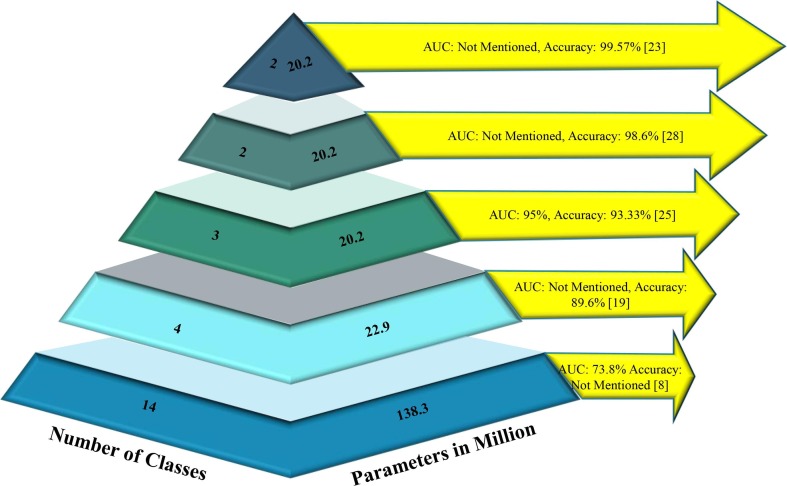
Fig. 1 illustrates the classification performance of selected state-of-the-art (SOTA) deep learning models in relation to the number of classes and the number of parameters involved when classifying lung diseases. It is evident that numerous models are available for multiclass lung disease classification; however, a discernible trend emerges as the number of classes increases. With the growth in the number of classes, the number of parameters escalates, subsequently leading to a decline in classification performance. This demonstrates that model complexity is directly proportional to the number of classes, rendering simple models insufficient for effectively distinguishing between a large number of lung diseases.
Fig. 1.
Summary of ML model’s classification performance in identifying lung diseases against the number of classes and parameters employed.
Although there exist many models, it has been aimed here to showcase the best five SOTA models where the selected models represent the highest performance levels achieved in their respective categories. For instance, the best AUC for 14-class classification was achieved by the model presented by Wang et al. (2017), while the highest accuracy of 99.57% for TB classification was obtained by Chowdhury et al. (2020). By choosing the top-performing models in each category, it is intended to provide a concise overview of the current advancements in this research area, illustrating the benchmark performances for various classification tasks ranging from 14-class to binary class.
In recent years, various SOTA models have demonstrated promising results in detecting multiple lung diseases, particularly in two-, three-, and four-class classifications. However, the accuracy of these models in classifying 14 distinct lung-related disorders using the CXR14 dataset was notably low. Consequently, researchers have predominantly relied on the AUC value for performance comparison. The majority of SOTA models that focused on the CXR14 dataset solely compared the AUC, neglecting other essential performance metrics such as precision, recall, specificity, and accuracy, as their values were deemed inconsequential. Remarkably, Rajpurkar et al. achieved an f1-metric of 0.435, the highest value in the last decade using the CXR14 dataset (Rajpurkar et al., 2017). However, earlier SOTA models struggled to deliver satisfactory classification performance, especially when taking into account the heightened complexity of the models, characterized by a larger number of parameters and layers. These observations underscore the limitations of existing research in handling the classification of a more extensive range of lung diseases, highlighting the need for further advancements in deep learning models to address these challenges. The development of novel methodologies capable of maintaining high classification performance, even with increased complexity, remains a crucial research objective in the field of lung disease classification.
Upon reviewing the relevant SOTA studies, the following challenges for detecting the lung diseases using the CXR images are identified.
-
•
Challenge 1: Binary classification between normal and one specific lung disease refers to a highly idealized situation. Real-world CXR images would contain features associated with a variety of lung diseases creating a large multiclass classification problem. Furthermore, this raises the level of complexity in classification owing to larger number of parameters and layers.
-
•
Challenge 2: The CXR image datasets are highly imbalanced in nature as some diseases are more frequently identified than the others. For instance, in the CXR14 dataset, the number of hernia images appeared 227 times, whereas infiltration was found in 19,871 images. One of the primary issues is that each class must contribute substantially equal to the final categorization.
-
•
Challenge 3: Many of the lung diseases such as COVID-19 are life threatening, therefore detecting each lung-related disease with high classification accuracy and faster processing time is of utmost importance.
The main goal of this study was to develop a computer-aided framework that could detect multi-class (17 classes: atelectasis, cardiomegaly, effusion, infiltration, mass, nodule, pneumothorax, consolidation, edema, emphysema, bacterial pneumonia, viral pneumonia, COVID-19, pleural thickening, fibrosis, hernia, tuberculosis) lung-related diseases fast and accurately with a relatively small number of parameters and layers in any practical environment (larger or small number of classes, larger or smaller dataset, balanced or unbalanced dataset, higher or lower resolution CXR images), in order to demonstrate the proposed framework's suitability in real-world applications without requiring huge computational resources. The classification of 17 types of lung diseases has been attempted for the first time in this study with simpler processing to achieve a more optimistic result than the SOTA models while cutting down on processing time and the number parameters, layers, and size. Aside from that, a hybrid parallel CNN-ELM model, combining both DL and ML models, was developed. It is challenging to integrate the ELM with parallel CNN to perform Grad-CAM visualization. Up until now, according to the authors’ best knowledge, limited study has shown Grad-CAM or any other visualization by combining the DL and ML models. Therefore, in this study, a hybrid framework was designed that combined ELM with parallel CNN to calculate the gradient from the last layer of the ELM to the first layer of the CNN, which makes the model used for explaining the decision-making of the black box CNN-ELM model. The novel hybrid model’s interpretability was demonstrated by Grad-CAM visualization to explain which part of the model focused more on images during classification than the other. Furthermore, a prototype mobile app was developed to simulate the real-life application of the proposed framework.
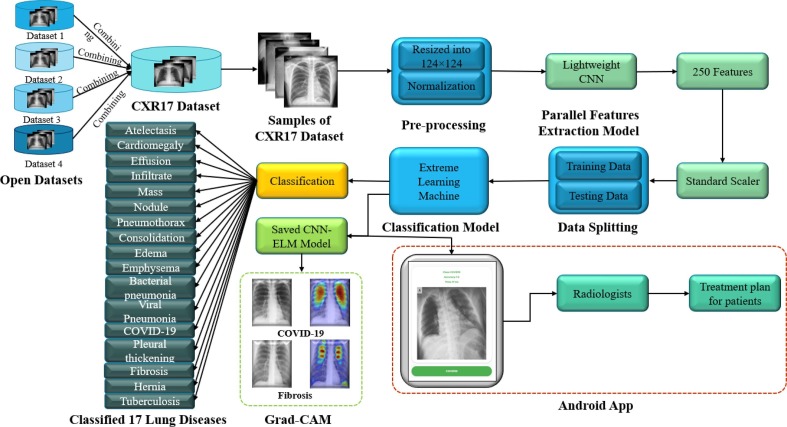
2. Prospective framework
Fig. 2 depicts the prospective framework for detecting multi-class lung disorders. It was crucial to merge different publicly available datasets that contained lung-related disorders in order to create a competitive dataset that represented a scenario much closer to the real world. In this study, CXR images only related to seventeen most well-known lung-related diseases including COVID-19 were combined. CXR images are easy to obtain from the patients and comparatively cheaper than other imaging methods (CT scan or MRI). No consideration was given to other less-known diseases or other sources of images in this approach. The CXR images were reshaped and normalized, then inputted into a lightweight CNN for extracting the most discriminating features. The preference was given to the lightweight CNN over other TL models due to its lower number of layers and parameters. After extracting the features, standardization was applied to the features. Finally, ELM was proposed as a classifier to detect 17 classes lung diseases from 250 extracted features. Apart from these, a heatmap from Grad-CAM was used to explore the black-box approach of the proposed parallel CNN-ELM model. From the author's best knowledge, this is the first time that a DL model was combined with a ML model to explain the proposed hybrid parallel CNN-ELM with Grad-CAM visualization. The step required for calculating the gradient from the last layer (output) of the ELM to the first layer is shown in Algorithm 1. This is carried out by replacing the final layer of CNN with the layers (input, hidden, and output layers) of ELM and integrating the trained weights and biases of the ELM hidden layers with the CNN model.
Algorithm 1
CNN-ELM algorithm for lung disease classification and for Grad-CAM visualization
| 1: | ModelCNN: parameter set-up CNN model | |
| 2: | Train the ModelCNN (70 epochs) | |
| 3: | FE: feature extraction from last dense layer before classification layer | |
| 4: | ModelELM: parameter set-up | |
| 5: | Train the ModelELM using extracted features (1 epoch) | |
| 6: | InputWeight (W): generates randomly | |
| 7: | OutputWeight (β): ReLU (X.W)-1.Y, where X and Y be the input and output | |
| 8: | Merging CNN-ELM | |
| 9: | Drop last 2 dense layers from the ModelCNN | |
| 10: | Add 2 new dense layers in the ModelCNN with same parameter of the ModelELM | |
| 11: | Set weight for newly added 2 layers using layers weight (W, β) of the ModelELM | |
| 12: | Classification using CNN-ELM | |
Fig. 2.
The prospective CNN-ELM framework for detecting seventeen lung diseases.
2.1. Coupling of data sets
The maximum number of images related to 13 lung diseases (Atelectasis, Cardiomegaly, Effusion, Infiltrate, Mass, Nodule, Pneumothorax, Consolidation, Edema, Emphysema, Fibrosis, Pleural Thickening, and Hernia) were collected from the ChestX-Ray14 dataset (Mooney, 2017). The CXR images of pneumonia from the ChestX-Ray14 were not considered since the difference between viral and bacterial pneumonia was not distinctive enough. Therefore, the CXR images of viral and bacterial pneumonia were collected from the Kaggle pneumonia dataset (JtiptJ, 2021). Since COVID-19 is also influenced by a virus, the viral pneumonia was also taken into account. Hence, the proposed framework was designed to test its performance in distinguishing COVID-19 from the viral pneumonia CXR images. It should be noted that large number of COVID-19 CXR images were not available in a single publicly open dataset, hence numerous datasets were integrated to construct a bespoke database with 4,192 COVID-19 CXR images (BIMCV Medical Imaging Databank of the Valencia Region, 2020, Chowdhury et al., 2020, Cohen, 2021, Haghanifar, 2019, JtiptJ, “Chest x-ray (pneumonia, covid-19,tuberculosis),”, 2021, ml-workgroup, “covid-19-image-repository,”, 2019). Finally, 1,037 CXR images of tuberculosis (TB) were gathered from three separate datasets to complete the construction of a unique dataset that contains a total of seventeen lung diseases with 40,490 CXR images (CXR17) (Health, 2020, Jaeger et al., 2014, Mader, 2021). In this study, only multi-class classification was considered, whereas multi-label was not considered due to a lack of multi-label CXR images in the public domain. Again, the number of patients affected by multiple lung diseases at a time is a very rare case. In Table 1 , the train and test split of the dataset are revealed. About 80% of the dataset was utilized to train the model, with the remaining 20% being used for the testing. The samples of each disease are depicted in Fig. 3 . Since the datasets were compiled from a variety of distinct CXR images, their resolutions varied widely. This issue was resolved by resizing all the CXR images to a same size of 124 × 124 pixels for promoting easier processing by the network. A multitude of pixel values ranging from 0 to 255 were used to depict an image. Image rescaling was used to make the range smaller by dividing each image by 255 and transforming the range from 0 to 1 making them ready for feature extraction using the CNN model.
Table 1.
Train and test split of the CXR17 dataset.
| Diseases Name | No of Training Images | No of Testing Images |
|---|---|---|
| Atelectasis | 3,724 | 843 |
| Cardiomegaly | 874 | 219 |
| Effusion | 3,164 | 791 |
| Infiltration | 7,637 | 1,910 |
| Mass | 1,711 | 428 |
| Nodule | 2,164 | 541 |
| Pneumothorax | 1,755 | 439 |
| Consolidation | 1,048 | 262 |
| Edema | 502 | 126 |
| Emphysema | 714 | 178 |
| Bacterial Pneumonia | 2,222 | 555 |
| Viral Pneumonia | 1,194 | 299 |
| COVID-19 | 3,354 | 838 |
| Pleural Thickening | 901 | 225 |
| Fibrosis | 582 | 145 |
| Hernia | 88 | 22 |
| Tuberculosis | 829 | 207 |
| Total | 32,463 | 8,028 |
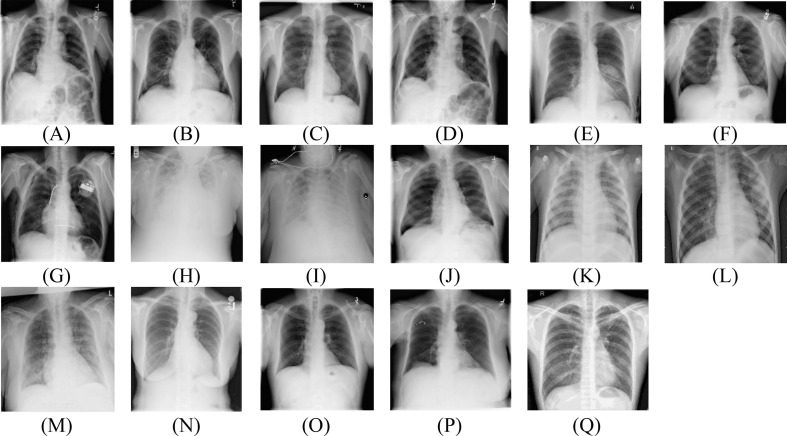
Fig. 3.
Sample CXR images of (A) Atelectasis, (B) Cardiomegaly (C) Effusion, (D) Infiltration, (E) Mass, (F) Nodule, (G) Pneumothorax, (H) Consolidation, (I) Edema, (J) Emphysema, (K) Bacterial pneumonia, (L) Viral pneumonia, (M) COVID-19, (N) Pleural thickening, (O) Fibrosis, (P) Hernia, and (Q) Tuberculosis.
2.2. Features extraction
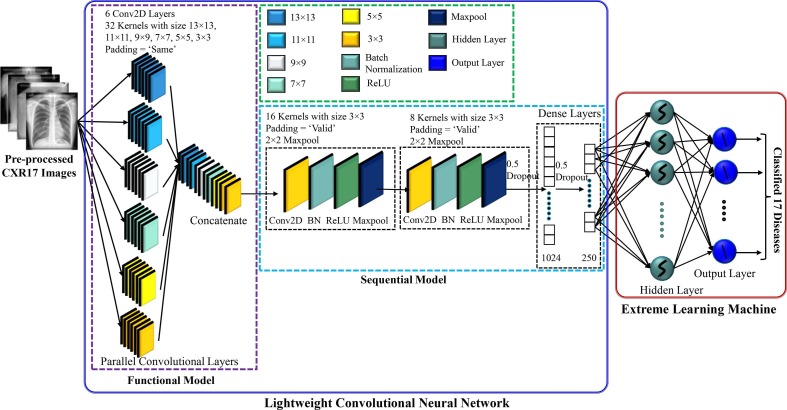
The main challenging task was to design a CNN model that can effectively extract the most prominent features with a relatively small number of parameters and layers suitable for easy implementation in real-world applications. However, too few parameters and layers might not be able to adequately capture the discriminating features, while too many parameters and layers might overfit the model requiring a longer processing time as well as increased computing resources. Fig. 4 depicts architecture of the lightweight parallel CNN model employed to extract the features.
Fig. 4.
A lightweight parallel CNN architecture for extracting features from CXR images.
The CNN contained eight convolutional layers (CLs), the first six of which were executed in parallel to speed up the processing and they were adopted through a trial-and-error method. Szegedy et al. were the ones who came up with the idea of parallel CLs first (Szegedy et al., 2015). Longer processing time would be required if the six CLs were run separately. The size of each CL was 32, whereas the kernel size of the first, second, third, fourth, fifth, and six was 13 × 13, 11 × 11, 9 × 9, 7 × 7, 5 × 5, and 3 × 3, respectively. In fact, for selecting the kernel size, this study followed the architecture provided by Krizhevsky et al., who used large kernel sizes like 11 × 11, for instance and achieved a better classification performance (Krizhevsky et al., 2017, Nahiduzzaman et al., 2023a). Since different kernels generate different feature maps, therefore, in this study, different kernels were considered and concatenated to capture the relevant features, which would assist in obtaining high classification performance. As the size of the features could vary depending on the disease type, variable kernel size was also maintained for each CL. This arrangement would allow to easily capture the key features even if the size of the features varied from small to large. The padding size was kept ‘SAME’ in the first six CLs in order to extract any crucial details in the border element of the CXR images. The output of the simultaneous CLs was then concatenated and fed into a sequential CNN. The rest of the CLs were followed by a batch normalization and a max-pooling layer with 2 × 2 kernel. The sizes of the two CLs were set to 16 and 8, each with 3 × 3 kernel and the padding sizes were kept “valid”. The ReLU activation function was employed for all the CLs. A total of two fully connected (FC) layers were present, with the features being extracted from the last FC layer. By ignoring 50% of all nodes at random, dropout was utilized to reduce overfitting and speed up the training process. One dropout (0.5 probability) was used after the final CL and another was employed after the first FC layer. Moreover, the CNN model was trained for 70 epochs with a batch size of 64. While taking into account the learning rate of 0.0001 with the ADAM optimizer, the model loss was dealt with the sparse categorical cross entropy loss function - for the purpose of extracting the features. Using a trial-and-error method, 250 features were selected from the final FC layer.
The extracted features need to be standardized (standard scaler) before feeding into the ELM. As the ELM is a type of ML algorithm, standardizing the features will improve its performance (Nahiduzzaman et al., 2019). If one of the features has a vast range of values, this feature will govern the distance. For this purpose, the range of each feature should be normalized in the same scale. All features’ ranges were normalized (Szegedy et al., 2015) using Eq. (1).
| (1) |
where represents the output of standard scaler, represents the input sample, is the mean of the samples and σ represents the standard deviation of the samples.
2.3. Extreme Learning Machine (ELM)
Using a forward feed network, Huang developed ELM (Huang et al. 2006), a type of neural network (NN) based on supervised learning. In order to classify the extracted features from the lung CXR images, a single hidden layer was applied. As there was no backpropagation involved in ELM, the training time was a thousand times faster than that of a typical NN, and the model possessed superior generalization power and classification performance than the traditional NN. When it comes to large multi-class classification, the ELM produces higher classification performance (Nahiduzzaman et al., 2021a, Nahiduzzaman et al., 2021b). The parameters between the input layer and the hidden layer were determined randomly, but the parameters between the hidden layer and the output layer were determined by adopting pseudoinverse. Table 2 shows the number of hidden and output nodes used in this study for different trials. Table 3 reveals the summary of proposed CNN-ELM model to detect multiple diseases.
Table 2.
Parameters of ELM for different trials.
| Scheme | Total Nodes in Input Layer | Total Nodes in Hidden Layer | Total Nodes in Output Layer |
|---|---|---|---|
| Trial 1–17 class | 250 | 1500 | 17 |
| Trial 2–14 class | 250 | 1500 | 14 |
| Trial 3–4 class | 250 | 500 | 4 |
| Trial 4–3 class | 250 | 500 | 3 |
| Trial 5–2 class | 250 | 200 | 1 |
| Trial 6–2 class | 250 | 200 | 1 |
| Activation Function | ReLU | ||
Table 3.
Summary of proposed CNN-ELM model to detect multiple lung diseases.
| Layer (Type) | Output Shape | Parameters |
|---|---|---|
| model (Functional) | (None, 124, 124, 192) | 43,776 |
| Conv7 (Conv2D) | (None, 122, 122, 16) | 27,664 |
| bn1 (BatchNormalization) | (None, 122, 122, 16) | 64 |
| Av7 (Activation) | (None, 122, 122, 16) | 0 |
| mp1 (MaxPooling2D) | (None, 61, 61, 16) | 0 |
| Conv8 (Conv2D) | (None, 59, 59, 8) | 1,160 |
| bn2 (BatchNormalization) | (None, 59, 59, 8) | 32 |
| av2 (Activation) | (None, 59, 59, 8) | 0 |
| mp2 (MaxPooling2D) | (None, 29, 29, 8) | 0 |
| dp1 (Dropout) | (None, 29, 29, 8) | 0 |
| ft (Flatten) | (None, 6,728) | 0 |
| dense (Dense) | (None, 1,024) | 6,890,496 |
| bn4 (BatchNormalization) | (None, 1,024) | 4,096 |
| dp2 (Dropout) | (None, 1,024) | 0 |
| Feature Extraction (Dense) | (None, 250) | 256,250 |
| Hidden Layer (Dense) | (None, 1,500) | 376,500 |
| av3 (Activation) | (None, 1,500) | 0 |
| Output (Dense) | (None, 17) | 25,500 |
| Total Parameters | 7,625,538 | |
| Trainable Parameters | 7,623,442 | |
| Non-trainable Parameters | 2,096 | |
3. Experimental procedure
Both binary and multiclass classifications with six trials were considered in this study to determine the suggested framework's performance. The first trial was carried out using the mixed dataset that included 17 different lung illnesses as shown in Table 1. The rest of the trials (Table 4 ) were utilized to validate that the suggested framework’s ability to provide optimistic classification performance in terms of reduction in the number of model parameters and layers, and processing time compared to the SOTA models. The second experiment was carried out using the CXR14 dataset. Trial 3 and Trial 4 were conducted to detect COVID-19 from normal and pneumonia. Trial 5 and Trial 6 were used for identifying COVID-19 or TB from normal. A variety of classification trials were conducted to ensure that the framework under consideration could adapt to any situation (e.g., binary or multiclass classifications, balanced or imbalanced datasets (Mukherjee et al. 2020), small or large datasets, and high or low-resolution images) and address the challenges mentioned in the Introduction section. All of the trials were assessed using an 80/20 split for training and testing schemes.
Table 4.
Description of Trial 2 to Trial 6 for detecting multiple lung diseases.
| Trial Number | Number of Classes | Disease Names | Training Images | Testing Images |
|---|---|---|---|---|
| Trial 2 | 14 | Atelectasis | 3,372 | 843 |
| Cardiomegaly | 874 | 219 | ||
| Effusion | 3,164 | 791 | ||
| Infiltration | 7,637 | 1,910 | ||
| Mass | 1,711 | 428 | ||
| Nodule | 2,164 | 541 | ||
| Pneumonia | 258 | 64 | ||
| Pneumothorax | 1,755 | 439 | ||
| Consolidation | 1,048 | 262 | ||
| Edema | 502 | 126 | ||
| Emphysema | 714 | 178 | ||
| Fibrosis | 582 | 145 | ||
| Pleural Thickening | 901 | 225 | ||
| Hernia | 88 | 22 | ||
| Total | 24,770 | 6,139 | ||
| Trial 3 | 4 | Normal | 8,153 | 2,039 |
| COVID-19 | 3,354 | 838 | ||
| Bacterial Pneumonia | 2,222 | 555 | ||
| Viral Pneumonia | 1,194 | 299 | ||
| Total | 14,923 | 3,731 | ||
| Trial 4 | 3 | Normal | 8,153 | 2,039 |
| Pneumonia | 1,194 | 299 | ||
| COVID-19 | 3,354 | 838 | ||
| Total | 12,701 | 3,176 | ||
| Trial 5 | 2 | Normal | 8,153 | 2,039 |
| COVID-19 | 3,354 | 838 | ||
| Total | 11,507 | 2,877 | ||
| Trial 6 | 2 | Normal | 8,153 | 2,039 |
| Tuberculosis | 829 | 207 | ||
| Total | 8,982 | 2,246 | ||
All the experiments were conducted using a Python-based framework operated by a system with an Intel Core i9 processor and graphics processing unit (GPU). The parameters for the proposed framework are mentioned in Table 5 . A confusion matrix (CM) was used to assess the ELM model's performance. The accuracy, precision, recall, f1-score, and area under the curve (AUC) were evaluated from the CM using the following equation (Powers, 2010, Swets, 1988).
| (2) |
| (3) |
| (4) |
| (5) |
| (6) |
where , , and , denote true positives, true negatives, false positives, and false negatives, respectively.
Table 5.
The parameters for the proposed framework.
| Name | Parameters |
|---|---|
| Programming Language | Python |
| Environment | PyCharm Community Edition (2021.2.3) |
| Backend | Keras with TensorFlow |
| Processor | 11th generation Intel(R) Core (TM) i9-11900 CPU @2.50 GHz |
| Installed RAM | 32 GB |
| GPU | NVIDIA GeForce, RTX 3090 24 GB |
| Operating system | Windows 10 Pro |
| Input | Chest X-Ray Images |
| Input Size | 124 × 124 |
| App Development Platform | Android Studio 2021.1.1 (Bumblebee) |
| Tensorflow Lite | tensorflow-lite-support:0.3.0 |
| Metadata Extractor | tensorflow-lite-metadata:0.3.0 |
| Tensorflow Lite GPU Acceleration | tensorflow-lite-gpu:0.3.0 |
4. Results and discussions
4.1. Trial 1: Multiclass-17 lung diseases
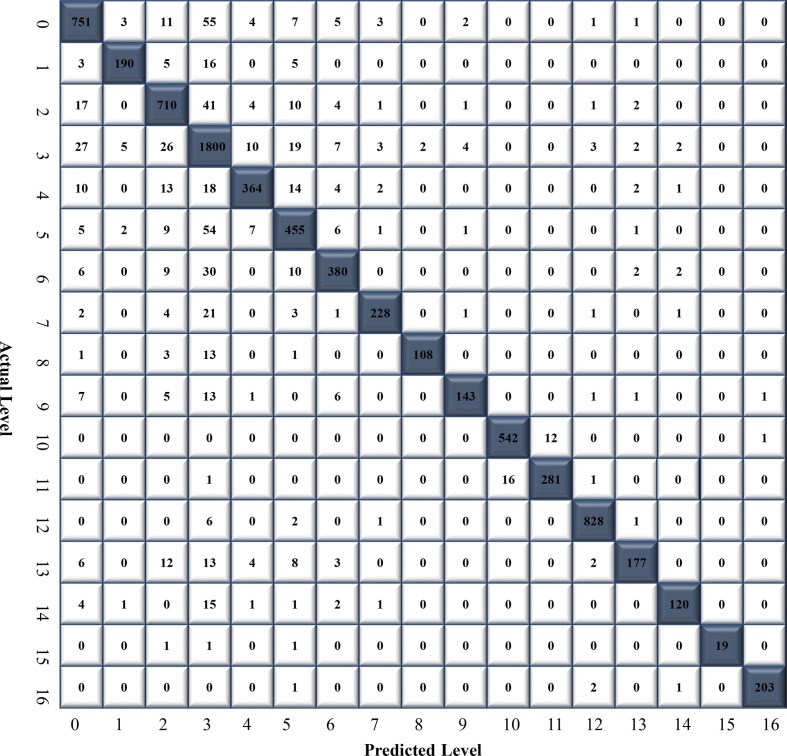
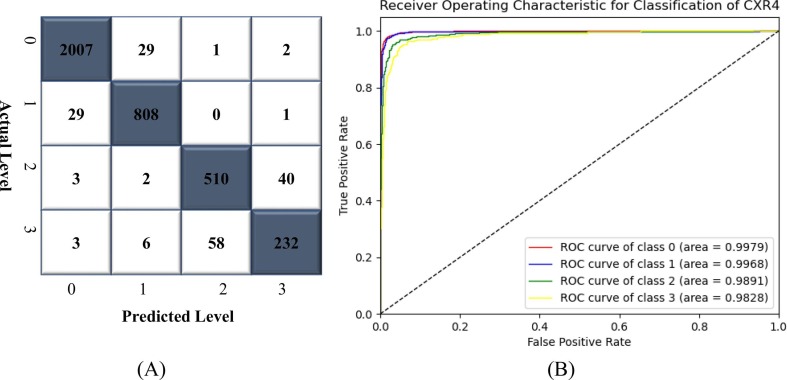
In this trial, a total of 32,463 CXR images were used to train the ELM, with 3,724, 874, 3,164, 7,637, 1,711, 2,164, 1,755, 1,048, 502, 714, 2,222, 1,194, 3,354, 901, 582, 88 and 829 images for atelectasis, cardiomegaly, effusion, infiltrate, mass, nodule, pneumothorax, consolidation, edema, emphysema, bacterial pneumonia, viral pneumonia, COVID-19, pleural thickening, fibrosis, hernia, and tuberculosis, respectively. The classification performance of the CNN-ELM models was tested using 8,028 CXR images (atelectasis: 843, cardiomegaly: 219, effusion: 791, infiltrate: 1,910, mass: 428, nodule: 541, pneumothorax: 439, consolidation: 262, edema: 126, emphysema: 178, bacterial pneumonia: 555, viral pneumonia: 299, COVID-19: 838, pleural thickening: 225, fibrosis: 145, hernia: 22, and tuberculosis: 207). The CM of 17 diseases is illustrated in Fig. 5 .
Fig. 5.
Confusion matrix (CM) of CNN-ELM to detect 17 diseases.
The CNN-ELM model's average precision, recall, F1-score, and accuracy were 0.94, 0.89, 0.91, and 90.92% respectively. Table 6 reflects the effectiveness of the model to detect 17 diseases in terms of class-wise classification.
Table 6.
Class-wise classification performance of CNN-ELM to detect 17 diseases.
| Diseases Name | Precision | Recall | F1-Score | Accuracy (%) |
|---|---|---|---|---|
| Atelectasis (0) | 0.90 | 0.89 | 0.89 | – |
| Cardiomegaly (1) | 0.95 | 0.87 | 0.90 | – |
| Effusion (2) | 0.88 | 0.90 | 0.89 | – |
| Infiltrate (3) | 0.86 | 0.94 | 0.90 | – |
| Mass (4) | 0.92 | 0.85 | 0.88 | – |
| Nodule (5) | 0.85 | 0.84 | 0.84 | – |
| Pneumothorax (6) | 0.91 | 0.87 | 0.89 | – |
| Consolidation (7) | 0.95 | 0.87 | 0.89 | – |
| Edema (8) | 0.98 | 0.86 | 0.92 | – |
| Emphysema (9) | 0.94 | 0.80 | 0.87 | – |
| Bacterial Pneumonia (10) | 0.97 | 0.98 | 0.97 | – |
| Viral Pneumonia (11) | 0.96 | 0.94 | 0.95 | – |
| COVID-19 (12) | 0.99 | 1.00 | 0.99 | – |
| Pleural Thickening (13) | 0.94 | 0.83 | 0.88 | – |
| Fibrosis (14) | 0.94 | 0.79 | 0.86 | – |
| Hernia (15) | 1.00 | 0.86 | 0.93 | – |
| Tuberculosis (16) | 0.99 | 0.98 | 0.99 | – |
| Average | 0.94 | 0.89 | 0.91 | 90.92 |
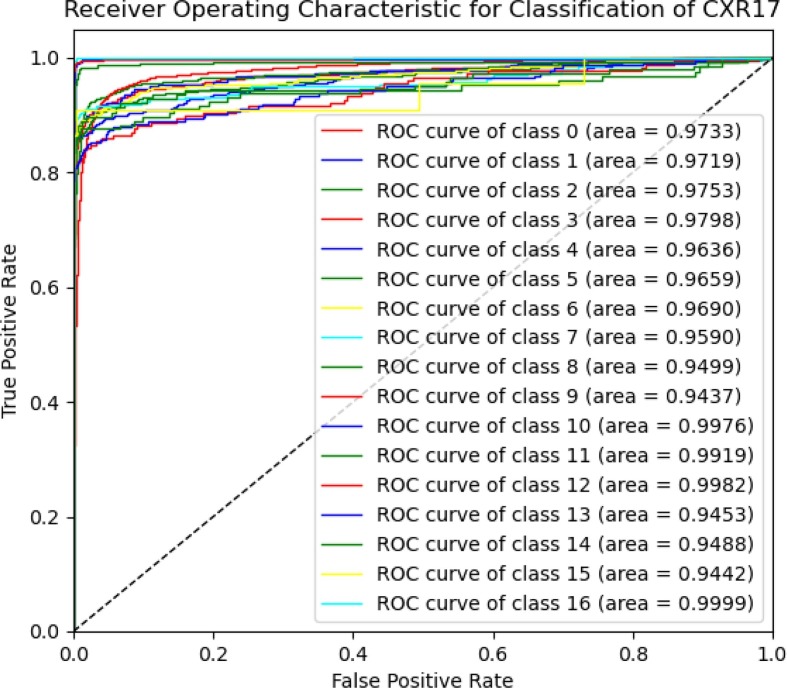
Fig. 6 illustrates a class-wise ROC to show how well the CNN-ELM model distinguishes 17 classes of lung diseases. The models' robustness was reflected in the fact that the ROC values for all classes appeared higher than 94%. While the average ROC of the proposed framework was 96.93%, the ROC value of COVID-19 was 99.82%, which indicated the model's ability to achieve high classification accuracy. The CNN-ELM model took 0.1904 seconds (s) for testing 8,028 CXR images in contrast to only 0.9 miliseconds (ms) for testing a single CXR image.
Fig. 6.
The ROC of CNN-ELM to detect 17 diseases.
It should be noted that the model has been developed with an imbalanced dataset mainly due to the unavailability of data for each category in the public domain. Indeed, one of our primary objectives in designing the model was to effectively handle the data imbalance problem. Although the number of images in the classes considered in this work was unbalanced, each class contributed equally to the final score. For example, the F1-score for all classes ranged between 0.89 and 0.99 indicating that even with unbalanced datasets a range of F1-score was obtained. For COVID-19, a recall of 100% was achieved, demonstrating the suggested framework's optimistic performance in distinguishing COVID-19 from all other lung diseases with superior classification performance. Recently, there has been an urgent requirement for the radiologists to reliably detect COVID-19. Therefore, the suggested model would reduce radiologist's liability and increase confidence. Further success even with the imbalanced data can be demonstrated by the results presented in Fig. 6. For instance, the infiltration class, with 7,637 images, achieved an AUC of 97.98%. Similarly, the TB class, consisting of 829 images, obtained an AUC of 99.9%, and the hernia class, with 88 images, reached an AUC of 94.42%. These results indicated that despite the presence of data imbalance across different classes, the model ensured that all classes contribute almost equally to the final classification outcomes. This approach effectively addresses the performance disparities that may arise due to the variations in the number of samples available for different lung disease classes, demonstrating the model's potential for real-world applications.
Due to the fact that the CXR17 is a combined dataset containing 17 diseases, the outcome could not be compared to any SOTA models in an effort to clarify whether the model perform well or not. Though the results of the large class classification (17 diseases) were highly promising, additional trials were conducted to demonstrate the proposed framework's consistency and robustness.
4.2. Trial 2: Multiclass-14 lung diseases
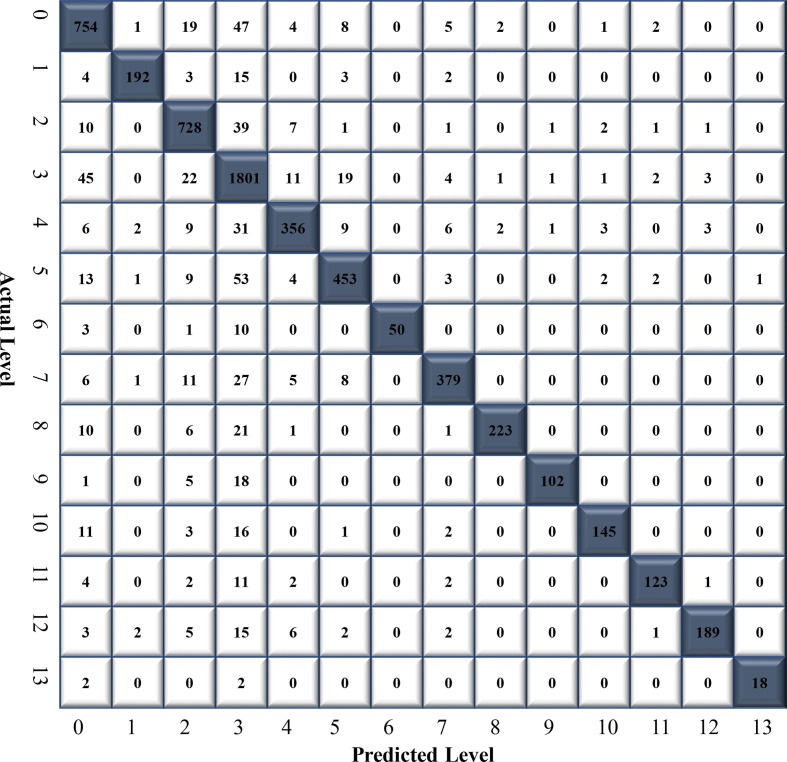
This trial with ChestX-Ray14 dataset was conducted to compare CNN-ELM model’s performance with other SOTA models. A total of 24,770 CXR images (atelectasis: 3,372, cardiomegaly: 874, effusion: 3,164, infiltrate: 7,637, mass: 1,711, nodule: 2,164, pneumonia: 258, pneumothorax: 1,755, consolidation: 1,048, edema: 502, emphysema: 714, fibrosis: 582, pleural thickening: 901, and hernia: 88) were used to train the CNN-ELM model. The remaining of 6,193 CXR images (atelectasis: 843, cardiomegaly: 219, effusion: 791, infiltrate: 1,910, mass: 428, nodule: 541, pneumonia: 64, pneumothorax: 439, consolidation: 262, edema: 126, emphysema: 178, fibrosis: 145, pleural thickening: 225, and hernia: 22) were employed to test the model's performance. The precision, recall, F1-socre and accuracy were calculated from the CM illustrated in Fig. 7 .
Fig. 7.
The confusion matrix of CNN-ELM to detect 14 diseases in Trial 2.
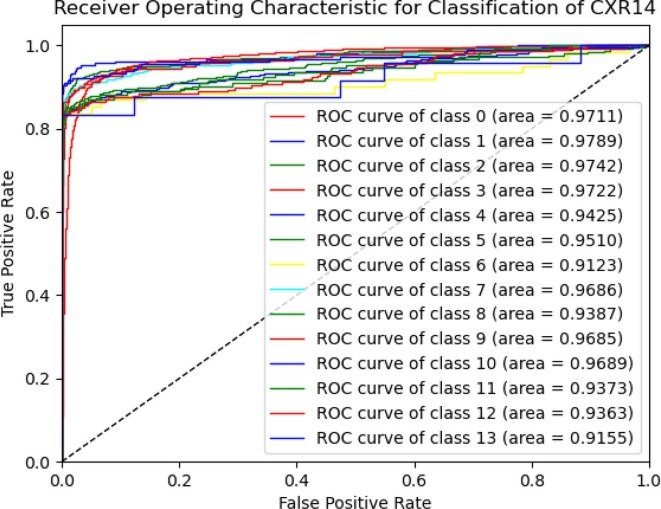
Table 7 presents the class-wise CNN-ELM's precision, recall, and F1-score for detecting 14 lung diseases. Even though the dataset was imbalanced, the outcome was relatively stable, indicating that each class has a significant impact on the final classification performance. The proposed framework attained an average accuracy of 89.10% and a receiver operating characteristic (ROC) value of 95.62%.
Table 7.
Class-wise classification performance of CNN-ELM to detect 14 diseases.
| Diseases Name | Precision | Recall | F1-Score | Accuracy (%) |
|---|---|---|---|---|
| Atelectasis (0) | 0.86 | 0.89 | 0.88 | – |
| Cardiomegaly (1) | 0.96 | 0.88 | 0.92 | – |
| Effusion (2) | 0.88 | 0.92 | 0.90 | – |
| Infiltrate (3) | 0.86 | 0.94 | 0.90 | – |
| Mass (4) | 0.90 | 0.83 | 0.86 | – |
| Nodule (5) | 0.90 | 0.83 | 0.87 | – |
| Pneumonia (6) | 1.00 | 0.78 | 0.88 | |
| Pneumothorax (7) | 0.93 | 0.86 | 0.90 | – |
| Consolidation (8) | 0.97 | 0.85 | 0.91 | – |
| Edema (9) | 0.97 | 0.81 | 0.88 | – |
| Emphysema (10) | 0.94 | 0.81 | 0.87 | – |
| Fibrosis (11) | 0.96 | 0.84 | 0.90 | – |
| Pleural Thickening (12) | 0.93 | 0.85 | 0.89 | – |
| Hernia (13) | 0.95 | 0.82 | 0.88 | – |
| Average | 0.93 | 0.85 | 0.91 | 89.10 |
As illustrated in Fig. 8 , the class-wise ROC curves for each class affirmed the model's efficiency in discriminating different diseases.
Fig. 8.
The ROC of CNN-ELM to detect 14 diseases in Trial 2.
4.3. Trial 3: Multiclass- COVID-19, bacterial and viral pneumonia
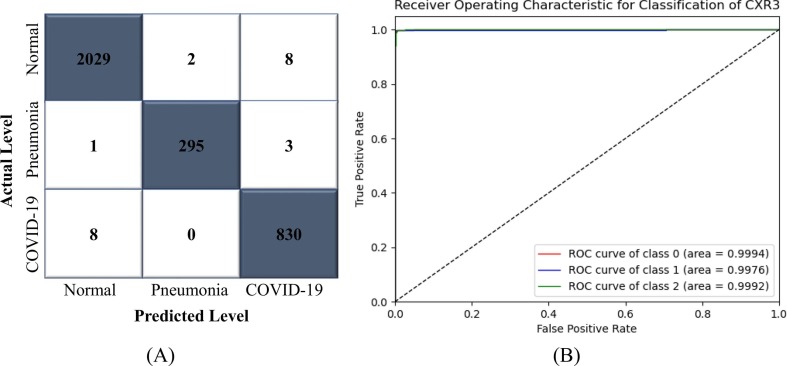
Since the proposed methodology produced favorable outcomes in the cases of 17- and 14-class classifications, the framework's ability to handle small class classifications were also tested with 4-, 3- and 2-class classifications. The CNN-ELM model was trained using 14,923 CXR images (normal: 8,153, COVID-19: 3,354, bacterial pneumonia: 2,222, and viral pneumonia: 1,194) in Trial 3. 3,731 images (normal: 2,039, COVID-19: 838, bacterial pneumonia: 555, and viral pneumonia: 299) were utilized to assess the model's categorization performance. Fig. 9 shows that the proposed approach was able to discriminate COVID-19 from the other lung disorders with an optimistic ROC of 99.68%.
Fig. 9.
(A) CM and (B) ROC of CNN-ELM to detect 4 classes in Trial 3.
Table 8 shows how well the CNN-ELM model can classify different types of diseases in a 4-class including COVID-19. The average precision, recall, F1-score, and accuracy obtained were 0.92, 0.91, 0.91, and 95.33%, respectively. Since viral and bacterial pneumonias are difficult to differentiate, hence the model discriminant ability was not so high as shown in CM of Fig. 9.
Table 8.
Class-wise classification performance of CNN-ELM to detect 4 classes in Trial 3.
| Disease Name | Precision | Recall | F1-Score | Accuracy (%) |
|---|---|---|---|---|
| Normal (0) | 0.98 | 0.98 | 0.98 | – |
| COVID-19 (1) | 0.96 | 0.96 | 0.96 | – |
| Bacterial Pneumonia (2) | 0.90 | 0.92 | 0.91 | – |
| Viral Pneumonia (3) | 0.84 | 0.78 | 0.81 | – |
| Average | 0.92 | 0.91 | 0.91 | 95.33 |
4.4. Trial 4: Multiclass-COVID-19 and pneumonia
The primary objective of this trial is to investigate how well the model distinguishes COVID-19 from viral pneumonia, as both of these disorders are caused by viruses. Therefore, the CNN-ELM model was trained using 12,701 CXR images (normal: 8,153, pneumonia: 2,222, and COVID-19: 3,354). A total of 3,176 images (normal: 2,039, pneumonia: 299, and COVID-19: 838) were utilized to calculate the model's precision, recall, F1-score, and accuracy from the confusion matrix illustrated in Fig. 10 (A). From Table 9 , it was observed that the results associated with each class's precision, recall, and F1-score were higher than 99% with an accuracy of 99.30%. The optimistic discriminant capability of the proposed framework was verified by the receiver operating characteristic (ROC) value of 99.88%. The class-wise ROC value (99.92%) demonstrated in Fig. 10(B) also revealed the model's discriminant capability to detect COVID-19.
Fig. 10.
(A) CM and (B) ROC of CNN-ELM to detect 3 classes in Trial 4.
Table 9.
Class-wise classification performance of CNN-ELM to detect 3 classes in Trial 4.
| Disease Name | Precision | Recall | F1-Score | Accuracy (%) |
|---|---|---|---|---|
| Normal (0) | 1.00 | 1.00 | 1.00 | – |
| Pneumonia (1) | 0.99 | 0.99 | 0.99 | – |
| COVID-19 (2) | 0.99 | 0.99 | 0.99 | – |
| Average | 0.99 | 0.99 | 0.99 | 99.30 |
4.5. Trial 5: Binary-Normal and COVID-19
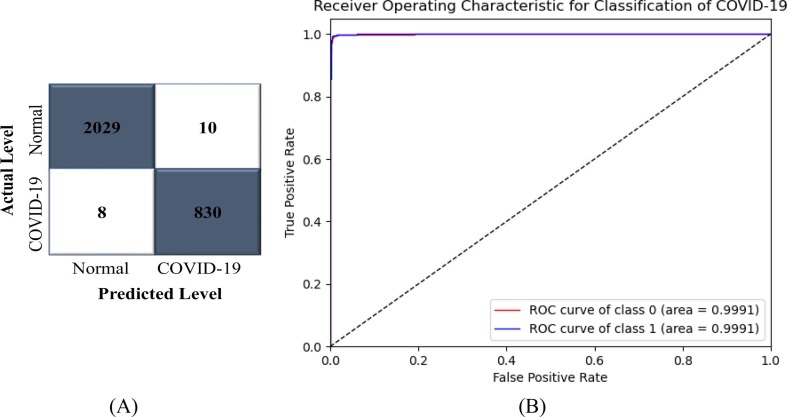
A significant percentage of research in the field of medical image analysis over the last few decades has focused on two-class classification rather than multi-class classification (Akter et al., 2021, Chandra et al., 2020, Chowdhury et al., 2020, Khan et al., 2020, Rahman et al., 2020b, Yamac et al., 2021). As a result, the model's performance was also evaluated in two-class environment. A total of 11,507 CXR images (normal: 8,153 and COVID-19: 3,354) were used for training, and 2,877 images (normal: 2,039 and COVID-19: 838) were used to evaluate the CNN-ELM's performance in detecting COVID-19. Fig. 11 (A) illustrates the confusion matrix (CM) that was used to determine the precision, recall, and F1-score. The accuracy of the proposed framework was 99.37% and the average precision, recall, and F1-score were all over 99%, as shown in Table 10 . The class-wise ROC of two classes was 99.91% in this trial as illustrated in Fig. 11(B), validating the framework's highest discriminant competence.
Fig. 11.
(A) CM and (B) ROC of CNN-ELM to detect COVID-19.
Table 10.
Class-wise classification performance of CNN-ELM to detect COVID-19.
| Disease Name | Precision | Recall | F1-Score | Accuracy (%) |
|---|---|---|---|---|
| Normal (0) | 1.00 | 1.00 | 1.00 | – |
| COVID-19 (1) | 0.99 | 0.99 | 0.99 | – |
| Average | 0.99 | 0.99 | 0.99 | 99.37 |
4.6. Trial 6: Binary-Normal and TB
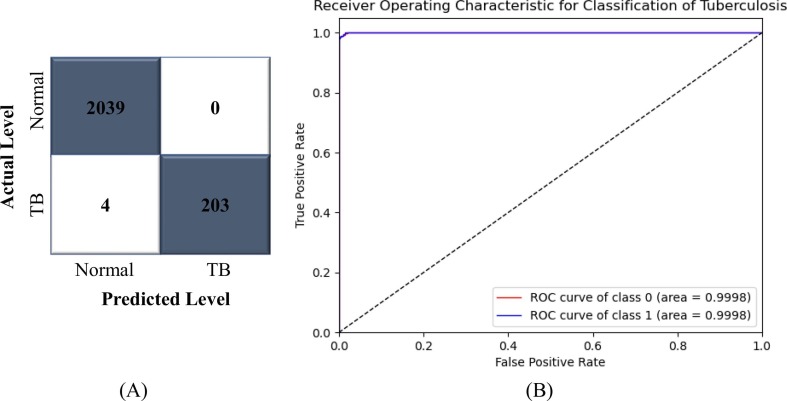
In this trial, the CNN-ELM model was trained with a total of 8,982 CXR images (normal: 8,153 and tuberculosis: 829) and evaluated its performance on 2,246 images (normal: 2,039 and tuberculosis: 207). The confusion matrix (CM) used to calculate the precision, recall, and F1-score is depicted in Fig. 12 (A). As shown in Table 11 , the proposed framework was 99.82% accurate, with an average precision, recall, and F1-score of 100%, 99%, and 99%, respectively. The CM demonstrated that the CNN-ELM model correctly identified all normal patients. The class-wise ROC for two classes was 99.98%, as shown in Fig. 12 (B), proving the framework's strongest classifier skill.
Fig. 12.
(A) CM and (B) ROC of CNN-ELM to detect Tuberculosis (TB).
Table 11.
Class-wise classification performance of CNN-ELM to detect TB.
| Disease Name | Precision | Recall | F1-Score | Accuracy (%) |
|---|---|---|---|---|
| Normal (0) | 1.00 | 1.00 | 1.00 | – |
| TB (1) | 1.00 | 0.98 | 0.99 | – |
| Average | 1.00 | 0.99 | 0.99 | 99.82 |
4.7. Performance comparison with SOTA models
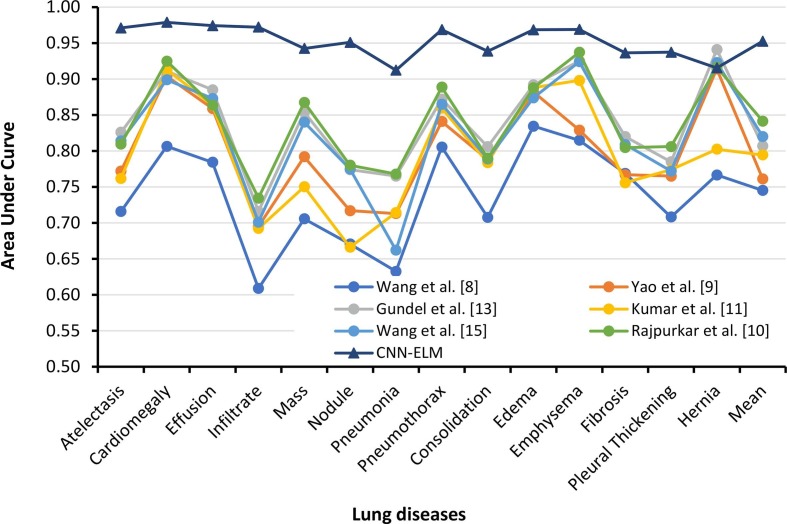
The proposed CNN-ELM model was compared to the SOTA models based on classification performance as well as model parameters, layers, and processing time. As Trial 1 used a completely new merged dataset, it could not be compared. The previously used SOTA models simply cited the AUC values, as no acceptable level of precision, recall, F1-score, or accuracy could not be achieved. Table 12 compares class wise AUC values of the earlier works with the present model developed in Trial 2. Only Rajpurkar et al. demonstrated an F1-score of 0.435, but the proposed CNN-ELM demonstrated an encouraging F1-score of 0.89, over 46% higher than the prior study. On top of that, Rajpurkar et al. achieved the highest AUC values for almost each class compared to the prior SOTA models. However, the proposed CNN-ELM surpassed Rajpurkar et al.'s AUC values except for the AUC value for “Hernia” (only 0.0009 higher). The suggested CNN-ELM model achieved an AUC of 95.26%, which was 11% higher than best mean AUC of the SOTA models (84.14%) validating the proposed framework's resilience. Fig. 13 also clearly indicates the similar observations in a pictorial view.
Table 12.
Class-wise comparison Area under the curves of ROC for diseases classification in the CXR14 dataset (Trial 2).
| Diseases | Area Under Curve (AUC) |
||||||
|---|---|---|---|---|---|---|---|
| Wang et al., 2017 | Yao et al., 2017 | Guendel et al., 2018 | Kumar et al., 2018 | Wang et al., 2020 | Rajpurkar et al., 2017 | CNN-ELM | |
| Atelectasis | 0.7158 | 0.772 | 0.826 | 0.7618 | 0.814 | 0.8094 | 0.9711 |
| Cardiomegaly | 0.8065 | 0.904 | 0.911 | 0.9133 | 0.899 | 0.9248 | 0.9789 |
| Effusion | 0.7843 | 0.859 | 0.885 | 0.8635 | 0.873 | 0.8638 | 0.9742 |
| Infiltrate | 0.6089 | 0.695 | 0.716 | 0.6923 | 0.701 | 0.7345 | 0.9722 |
| Mass | 0.7057 | 0.792 | 0.854 | 0.7502 | 0.840 | 0.8676 | 0.9425 |
| Nodule | 0.6706 | 0.717 | 0.774 | 0.6662 | 0.775 | 0.7802 | 0.9510 |
| Pneumonia | 0.6326 | 0.713 | 0.765 | 0.7145 | 0.662 | 0.7680 | 0.9123 |
| Pneumothorax | 0.8055 | 0.841 | 0.872 | 0.8594 | 0.865 | 0.8887 | 0.9686 |
| Consolidation | 0.7078 | 0.788 | 0.806 | 0.7838 | 0.789 | 0.7901 | 0.9387 |
| Edema | 0.8345 | 0.882 | 0.892 | 0.8880 | 0.874 | 0.8878 | 0.9685 |
| Emphysema | 0.8149 | 0.829 | 0.925 | 0.8982 | 0.924 | 0.9371 | 0.9689 |
| Fibrosis | 0.7688 | 0.767 | 0.820 | 0.7559 | 0.809 | 0.8047 | 0.9363 |
| Pleural Thickening | 0.7082 | 0.765 | 0.785 | 0.7739 | 0.772 | 0.8062 | 0.9373 |
| Hernia | 0.7667 | 0.914 | 0.941 | 0.8024 | 0.923 | 0.9164 | 0.9155 |
| Mean | 0.7451 | 0.761 | 0.807 | 0.7945 | 0.82 | 0.8414 | 0.9526 |
Bold values indicate the best values.
Fig. 13.
Class wise AUC values of CNN-ELM model compared to the SOTA models.
Table 13 presents comparative performance between the CNN-ELM model and the SOTA models for Trial 2 to Trail 6. For the detection of four diseases, Khan et al. attained the highest precision, recall, and accuracy of 89.84%, 89.94%, and 89.6% (Khan et al., 2020). In Trial 3, the proposed CNN-ELM model outperformed the SOTA models with an optimistic accuracy of 95.33%, which is 6% higher than the previous SOTA models. Again, Khan et al.'s CoroNet used 71 layers and 33.97 million (M) parameters, whereas CNN-ELM employed only 7 layers with 7.6 M parameters. This meant that CNN_ELM layers were 10 times and the parameters were 6 times lower than the CoroNet model. Jain et al. achieved a maximum accuracy of 97.97% in the SOTA models in Trial 4 by employing the InceptionV3 model, which consisted of 48 layers and 23.9 M parameters. The proposed framework obtained a promising accuracy of 99.30% (2.6% higher) using a relatively modest model with almost 7 times less layers and over 3 times less parameters than those in the InceptionV3 model.
Table 13.
Classification performance comparison with SOTA models for the Trial 3 to Trial 6.
| Scheme No | Reference | No. of Class | Processing Times (seconds) | Precision | Recall | Accuracy | AUC |
|---|---|---|---|---|---|---|---|
| Trial 3 | Khan et al., 2020 | 4 | – | 89.84% | 89.94% | 89.6% | – |
| Yamac et al., 2021 | – | – | 79.79% | 87.07% | – | ||
| Chandra et al., 2020 | – | 80.92% | 85.66% | 76.46% | |||
| CNN-ELM | 0.000997 | 92% | 91% | 95.33% | 99.17% | ||
| Trial 4 | Chandra et al., 2021 | 3 | – | – | – | 93.41% | – |
| Pandit et al., 2021 | – | – | 86.7% | 92.53% | |||
| Sekeroglu and Ozsahin, 2020 | – | 92.70% | 92.70% | 95.99% | – | ||
| Wang et al., 2020 | – | 93.33% | 93.33% | 93.3% | – | ||
| Jain et al., 2021 | – | 96.33% | 93% | 97.97% | – | ||
| Asnaou & Chawki, 2021 | 0.159 | 92.38% | 92.11% | 92.18% | |||
| Ozturk et al., 2020 | <1 | 89.96% | 85.35% | 87.02% | |||
| Chandra et al., 2020 | – | 94.27% | 96.40% | 95.66% | – | ||
| CNN-ELM | 0.000996 | 99% | 99% | 99.30% | 99.88% | ||
| Trial 5 | Pandit et al.,2021 | 2 | – | – | 92.64% | 96% | – |
| Panwar et al., 2020 | – | – | – | 88.10% | 88.10% | ||
| Akter et al., 2021 | – | – | – | 89.6% | – | ||
| Khan et al., 2020 | – | 93% | 98.2% | 89.6% | – | ||
| Sekeroglu and Ozsahin, 2020 | – | – | 93.92% | 98.39% | 96.48% | ||
| Chowdhury et al., 2020 | – | 97% | 98% | 98% | – | ||
| Ozturk et al., 2020 | <1 | 98.03% | 95.13% | 98.08% | |||
| Brunese et al., 2020 | 2.498 | – | 94% | 98% | – | ||
| CNN-ELM | 0.000996 | 99% | 99% | 99.37% | 99.91% | ||
| Trial 6 | Chandra et al., 2020 | 2 | – | 99.42% | 99.40% | 99.40% | 99% |
| Rahman et al., 2020a, Rahman et al., 2020b | – | 98.57% | 98.56% | 98.60% | – | ||
| Sahlol et al., 2020 | – | – | 91.94% | 90.23% | – | ||
| Ayaz et al., 2021 | – | – | – | 97.59% | 99% | ||
| Lopes and Valiati, 2017 | – | – | – | 84.7% | 92.60% | ||
| Duong et al., 2021 | – | – | 97.3% | 98.7% | 99% | ||
| CNN-ELM | 0.000996 | 100% | 99% | 99.82% | 99.98% |
Bold values indicate the best values.
Due to the lower number of parameters, the CNN-ELM analyzed a single image in <1 ms, whereas other models in the literature (Joshi et al., 2021, Ozturk et al., 2020) reported values between 15.9 ms and 100 ms. Trial 5 has been a major focus for the researchers during the past two years. The suggested model performed well in classifying COVID-19 patients from normal patients using the CXR images, with an accuracy and recall of 99.37% and 99%, respectively. Sekeroglu et al. acquired the maximum AUC of 96.48%, whereas the proposed CNN-ELM accomplished an optimistic AUC of 99.91%, demonstrating its potential in diagnosing COVID-19 with high accuracy. CNN-ELM has a processing time of <1 ms, which was faster than that by Ozturk et al. (Joshi et al., 2021) (100 ms) and Brunese et al. (Asnaou & Chawki, 2021) (249 ms) enabling real-time diagnosis. The CNN-ELM model surpassed eight previous SOTA models in Trial 5. In addtion, TB is one of the most severe respiratory disorders, and it was effectively identified in this study as well. The proposed model in Trial 6 recognized TB with 100% precision, 99.82% accuracy and 99.98% AUC, which were greater than six well-known approaches. However, Chandra et al. (2020) achieved slightly better Recall (99.40%) compared to the CNN-ELM model (99.00%).
Table 14 demonstrates that the CNN-ELM model was less complex than the SOTA models for detecting 14 lung diseases. The majority of the SOTA models (Guendel et al., 2018, Rajpurkar et al., 2017, Yao et al., 2017) employed DenseNet-121, which had 121 layers and 8.1 M parameters, whereas the suggested model employed only 7 layers (CNN has three CLs and two FC layers whereas ELM has one hidden and one output layer) and had a total of 7.6 M parameters. The SOTA models that used AlexNet had layer number (8) closes to the model developed in this study but still the number of parameters were much higher.
Table 14.
CNN-ELM simplicity comparison with SOTA models.
| Model Name (Ref.) | Number of Layers | Number of Parameters (million) |
|---|---|---|
| VGG16 (Brunese et al., 2020, Lopes and Valiati, 2017, Pandit et al., 2021, Wang et al., 2017) | 16 | 138.3 |
| DenseNet-121 (Guendel et al., 2018, Rajpurkar et al., 2017, Wang et al., 2020, Yao et al., 2017) | 121 | 8.1 |
| CoroNet (Khan et al., 2020) | 71 | 33.97 |
| DarkNet (Panwar et al., 2020) | 106 | 40.5 |
| Inception Resnet V2 (Asnaou & Chawki, 2021) | 164 | 55.8 |
| DenseNet 201 (Chowdhury et al., 2020, Rahman et al., 2020b) | 201 | 20.2 |
| ResNet 50 (Lopes and Valiati, 2017, Wang et al., 2017) | 50 | 25.6 |
| Inception-V3 (Jain et al., 2021) | 48 | 23.8 |
| Xception (Jain et al., 2021) | 71 | 22.9 |
| AlexNet (Duong et al., 2021, Wang et al., 2017) | 8 | 62.37 |
| Proposed Framework | 7 | 7.6 |
Bold value indicate the best value.
In addition, the earlier SOTA models required high-resolution images, for instance 480 × 480 (Wang et al., 2020), 416 × 416 (Panwar et al., 2020), 299 × 299 (Asnaou & Chawki, 2021), 227 × 227 (Lopes & Valiati, 2017), 224 × 224 (Wang et al., 2017), and 256 × 256 (Yao et al., 2017). The proposed model, on the other hand, was able to correctly recognize lung disorders with low-resolution input images (124 × 124). The CNN-ELM was designed to maintain consistency in any setting, including large- (17 classes) or small-class (two classes) classifications, large (40,139) or small (11,228) datasets, balanced or unbalanced datasets, and low or high resolution CXR images. Therefore, the proposed framework classified accurately 17 different lung diseases with fewer model parameters and layers, low-resolution CXR images, and a shorter processing time. The framework also showed promises to implement in real-world environment with real-time diagnosis outcomes.
4.8. Model’s interpretability capability
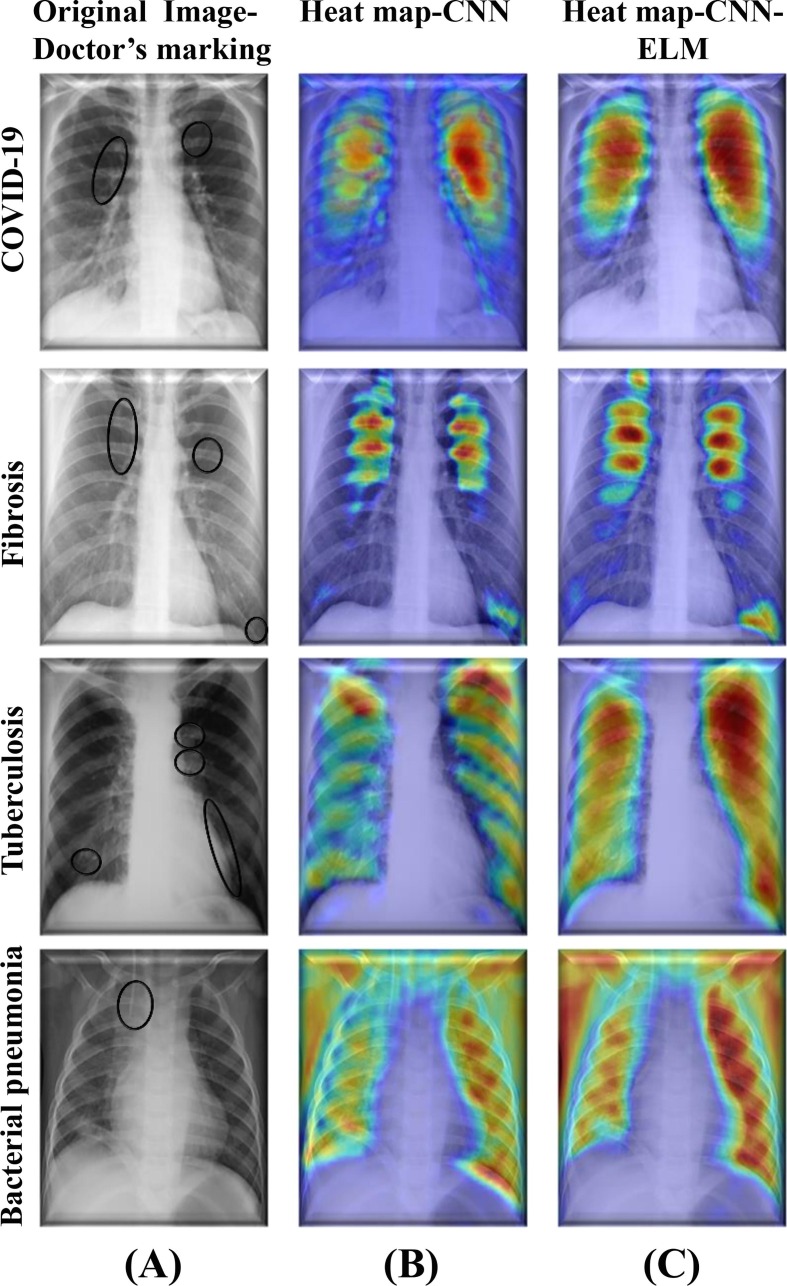
In recent years, to break the black box nature of the DL models, several researchers employed different tools for example gradient weighted class activation mapping (Grad-CAM), Shapley additive explanations (SHAP), and local interpretable model-agnostic explanations (LIME). During classification of different diseases, the tools highlight parts of the images that were identified by the DL models (Arik et al., 2020, Ghoshal and Tucker, 2020, Nahiduzzaman et al., 2023b, Yan et al., 2020). In order to classify various lung-related defects, Grad-CAM was used in this work to determine where exactly in an image the proposed model paid significantly more attention in contrast to the other parts.
According to the author's best knowledge, no study had shown Grad-CAM heatmaps for lung disease classification by combining DL models with the ML models. Fig. 14 shows a comparison of Parallel CNN and Parallel CNN-ELM for Grad-CAM visualization of selected four diseases without any particular preference. In the first row, the CXR image of the COVID-19 patient was marked by an expert radiologist. In general, it was clear that heatmaps generated on the images classified by CNN-ELM was much stronger than that generated by the CNN. For example, in the fibrosis CXR image, the parallel CNN produced the heatmap highlighting the areas marked by the doctor. However, the heatmap for the hybrid parallel CNN-ELM model was even more pronounced with intense color aligned with the areas marked by the doctor. Similar trend was also found in the heatmaps for other diseases. The main challenging task was merging the ELM hidden layers' weight with the CNN model, which was done by dropping the last layer of CNN and updating by adding three layers (input, hidden, and output layers) of ELM with corresponding weights and biases. Consequently, both Parallel CNN and ELM were engaged in classifying a particular lung related disease with high classification performance. Therefore, the proposed hybrid lightweight parallel CNN-ELM model demonstrated a higher capability of interpretability than the parallel CNN model only.
Fig. 14.
(A) CXR images marked by an expert radiologist, (B) Grad-CAM heatmap with Parallel CNN, and (C) Grad-CAM heatmap with hybrid Parallel CNN-ELM.
4.9. Key technical contributions and limitations
Although the CNN-ELM architecture and the application of Grad-CAM in deep learning and machine learning models have been previously explored, this work addresses specific challenges and offers unique contributions that differentiate it from the existing research. The key challenges were aimed to overcome in this study include classifying a large number of lung diseases (17 classes) with improved performance compared to the previous works. The best research prior to this work achieved an AUC of 73.8% for 14 lung diseases classification using a model with 138.3 M parameters. Designing a lightweight model with a reduced number of layers and parameters to facilitate implementation on the embedded systems can support medical practitioners in detecting life-threatening lung diseases. This study addresses these challenges by developing a lightweight model with only 7 layers and 7.6 M parameters while running 6 convolution layers in parallel, reduced any overfitting problem and enhanced classification performance across all schemes. It was demonstrated that the proposed model did not require pretraining, allowing for faster classification of lung diseases (<1 ms on a GPU computer and 21 ms on an Android phone). A novel Grad-CAM comparison between parallel CNN and parallel CNN-ELM models was made to again demonstrate a better outcome through combining ML and DL. GRAD-CAM was successfully applied to identify 17 lung disease classes in comparison to limited number of classes available in the literature. Furthermore, GRAD-CAM results were verified by the expert radiologists which can rarely be found in the existing literature. By addressing these challenges and offering novel contributions, this study advances the current understanding of efficient large multiclass lung disease classification using a combined ML and DL approach.
In this study, classifications were conducted with images only containing single label. However, it was not totally unusual that an image could have multiple labels. Only few images in the original databases contained diseases with multiple labels. To train these multi-label CXR images (10 images in classes), the parameters and structure of the DL and ML models must be altered, which could impair the results of the multi-class classification (Kumar et al., 2018). Again, multi-label disorders would confound the model's ability to detect diseases accurately which means reduced the classification performance (Baltruschat et al., 2019, Ge et al., 2018). It is extremely rare for a patient to be affected by many lung diseases concurrently. Therefore, only multi-class categorization sounds a better proposition as it would not be a preferable choice to evaluate only few multi-label diseases with relatively low accuracy rather than achieving promising outcomes in the case of multi-class classification. Again, in Trial 3, the classification performance was not so high compared to the Trials 4 to Trial 6. Since Trial 3 contained both viral and bacterial pneumonia, the model might not be able to detect minute differences in the features associated with these two similar classes. Even though the CXR images are cheaper compared to the computed tomography (CT) scan images, but the latter could produce better classification performance (Baghdadi et al., 2022, Islam and Nahiduzzaman, 2022). Even after combining multiple datasets, number of images in some categories are still limited, which can limit the overall performance of the model developed. However, the model can be further updated with additional images from different categories when available in public.
5. Application of CNN-ELM model in Android App
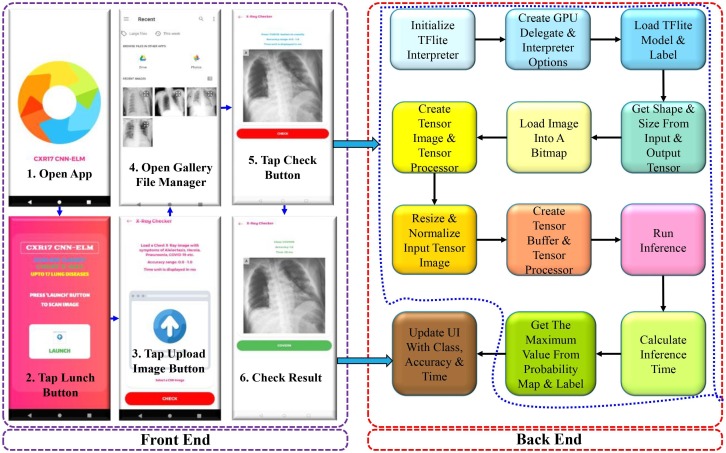
5.1. App design and development
A functional prototype of an Android App was designed and developed to demonstrate the implementation of the developed model in real life application. The trained CNN-ELM model was converted to TFLite model format for developing an Android app with a user interface (UI), which was used to classify sample X-ray images in a live Android device. The front end and back end operational flow charts are illustrated in Fig. 15 . To scan a sample Chest X-ray image, first the affected subject must undergo a chest X-ray in a health centre or hospital. The X-ray film will then be scanned and uploaded to the mobile device, followed by fast results via the mobile app. After a successful classification, the class, accuracy, and processing time of the CXR image will be shown.
Fig. 15.
Steps for scanning and classifying an X-ray image on an Android device.
5.2. App testing
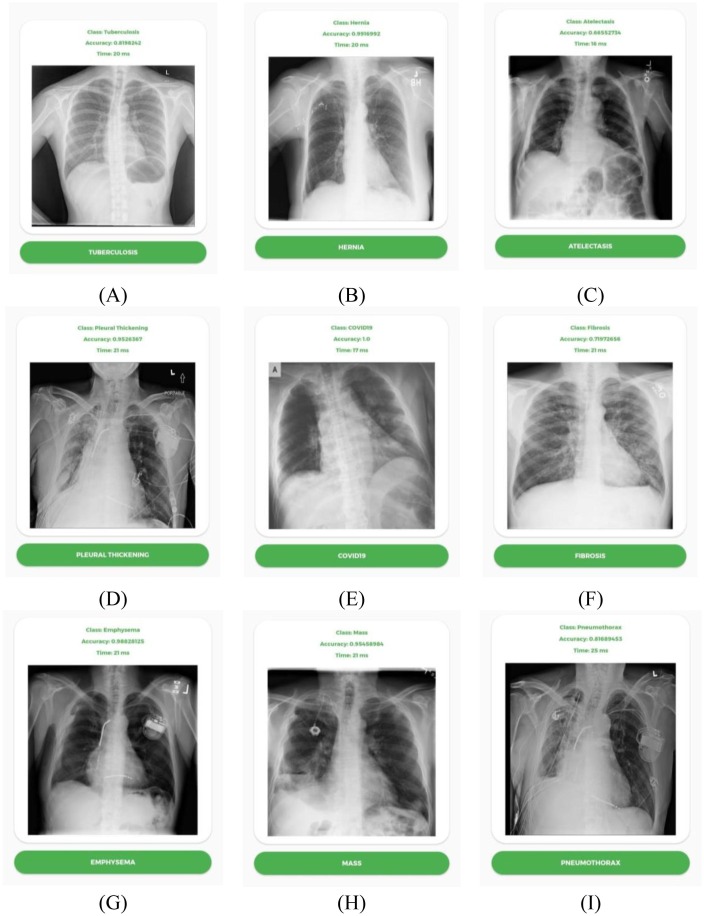
There are in total 17 classes of respiratory diseases that can be detected using the TFLite model. Among the 17 classes, 9 classes with test accuracy and inference runtime are shown in Fig. 16 . All the tests were performed using GPU acceleration and took around 15–21 ms to run inference using the sample CXR images. However, GPU acceleration is not mandatory to classify a sample image. For the 9 classes listed above, all the tests indicate accuracy in the range of 0.7–1.0. All tests are performed using a OnePlus 6T running Android 11 on Snapdragon 835 SoC.
Fig. 16.
TFLite based image classification of (A) Tuberculosis, (B) Hernia, (C) Atelectasis, (D) (E) Pleural Thickening, (F) COVID-19, (G) Fibrosis, (H) Emphysema, (I) Mass and Pneumothorax.
It is the first time an Android app has been able to detect 17 different types of respiratory diseases from the CXR images using the CNN-ELM model developed in this study. The time required (15–21 ms) to detect the lung diseases with high accuracy via the Android app would be much less than a medical professional. Hence, this app can save a lot of time when multiple X-ray images needs to be checked one-by-one. Furthermore, the App will be a guiding tool for the radiologists in the cases where human eyes fail to identify the distinctive features in the CXR image for lung disease identification. The number of disease classes and accuracy of the model can be further improved in the future by collecting additional CXR images. Currently, the app is only compatible with Android devices, but an iOS app can also be developed.
6. Conclusion
The study provided a novel CNN-ELM framework for accurately detecting various life-threatening lung diseases from CXR images with a small number of model parameters and layers and a short processing time. A large dataset containing 17 classes was customized by integrating several publicly available datasets. Following that, a parallel CNN was designed to extract the most discriminant features, which were then entered into a simple ELM for disease classification. For the first time, the proposed CNN-ELM model successfully identified 17 types of lung diseases with an average Precision, Recall, F1-Score and Accuracy of 0.94, 0.89, 0.91 and 90.92% respectively. Even when other SOTA models failed to achieve any reasonable performance metrics other than AUC with 14 diseases, whereas the proposed model achieved performance metric values close to 90% or above with 17 diseases. Significant reduction in number of layers (7) and parameters (7.6 M) compared to other SOTA models indicated the model’s simplicity. Class wise COVID-19 Precision, Recall, and F1-Score of 0.99 or above with CXR17 demonstrated that the model could reliably detect COVID-19. Furthermore, the models outperformed the previously established and well-known SOTA models for other various multiclass and binary classifications with respect to all performance metrics considered here. The development of an Android mobile app using the CNN-ELM model further shows the feasibility to implement the model in real-life scenario for fast detecting the lung diseases precisely and confidently during this COVID-19 pandemic. To detect a single CXR image, the CNN-ELM consumed <1 ms on a computer and 21 ms on an Android mobile device, respectively. This further illustrated the model’s suitability in helping the medical professionals to receive real-time prediction of lung diseases.
CRediT authorship contribution statement
Md. Nahiduzzaman: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Validation, Visualization, Writing – original draft, Writing – review & editing. Md. Omaer Faruq Goni: Formal analysis, Investigation, Methodology, Software, Writing – original draft, Writing – review & editing. Rakibul Hassan: Formal analysis, Investigation, Visualization, Writing – original draft. Md. Robiul Islam: Methodology, Investigation, Formal analysis, Validation, Writing – original draft, Writing – review & editing. Md Khalid Syfullah: Formal analysis, Investigation, Visualization, Writing – original draft. Saleh Mohammed Shahriar: Formal analysis, Investigation, Visualization, Writing – original draft. Md. Shamim Anower: Project administration, Methodology, Supervision, Formal analysis, Writing – review & editing. Mominul Ahsan: Methodology, Visualization, Supervision, Formal analysis, Writing – review & editing. Julfikar Haider: Supervision, Formal analysis, Visualization, Validation, Writing – review & editing. Marcin Kowalski: Methodology, Supervision, Formal analysis, Validation, Writing – review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Data availability
The authors do not have permission to share data.
References
- Akter S., Shamrat F., Chakraborty S., Karim A., Azam S. Covid-19detection using deep learning algorithm on chest x-ray images. Biology. 2021;10(11):1174. doi: 10.3390/biology10111174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arik S., Li C.L., Yoon J., Sinha R., Epshteyn A., Le L., et al. Interpretable sequence learning for COVID-19 forecasting. Advances in Neural Information Processing Systems. 2020 [Google Scholar]
- Asnaou K.E., Chawki Y. Using x-ray images and deep learning for automated detection of coronavirus disease. Journal of Biomolecular Structure and Dynamics. 2021;39(10):3615–3626. doi: 10.1080/07391102.2020.1767212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaz M., Shaukat F., Raja G. Ensemble learning based automatic detection of tuberculosis in chest x-ray images using hybrid feature descriptors. Physical and Engineering Sciences in Medicine. 2021;44(1):183–194. doi: 10.1007/s13246-020-00966-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baghdadi, N. A. , Malki, A., Abdelaliem, S. F., Balaha, H. M., Badawy, M., and Elhosseini, M., “An automated diagnosis and classification of covid-19 from chest ct images using a transfer learning-based convolutional neural network,” Computers in Biology and Medicine, p. 105383, 2022. [DOI] [PMC free article] [PubMed]
- Baltruschat I.M., Nickisch H., Grass M., Knopp T., Saalbach A. Comparison of deep learning approaches for multi-label chest x-ray classification. Scientific Reports. 2019;9(1):1–10. doi: 10.1038/s41598-019-42294-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunese L., Mercaldo F., Reginelli A., Santone A. Explainable deep learning for pulmonary disease and coronavirus covid-19 detection from x-rays. Computer Methods and Programs in Biomedicine. 2020;196 doi: 10.1016/j.cmpb.2020.105608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Automatic detection of tuberculosis related abnormalities in chest x-ray images using hierarchical feature extraction scheme. Expert Systems with Applications. 2020;158 [Google Scholar]
- Chandra T.B., Verma K., Singh B.K., Jain D., Netam S.S. Coronavirus disease (covid-19) detection in chest x-ray images using majority voting-based classifier ensemble. Expert Systems with Applications. 2021;165 doi: 10.1016/j.eswa.2020.113909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chowdhury, M. E., Rahman, T., Khandakar, A., Mazhar, R., Kadir. M. A., Mahbub, Z. B., Islam, K. R., Khan, M. S., Iqbal, A., Al Emadi N. et al., “Can ai help in screening viral and covid-19 pneumonia?” IEEE Access, vol. 8,pp. 132 665–132 676, 2020.
- Duong L.T., Le N.H., Tran T.B., Ngo V.M., Nguyen P.T. Detection of tuberculosis from chest x-ray images: Boosting the performance with vision transformer and transfer learning. Expert Systems with Applications. 2021;184 [Google Scholar]
- Ge, Z., Mahapatra, D., Sedai, S., Garnavi, R., and Chakravorty, R., “Chest x-rays classification: A multi-label and fine-grained problem,” arXiv preprintarXiv:1807.07247, 2018.
- Ghoshal, B., & Tucker, B. (2020). Estimating uncertainty and interpretability in deep learning for coronavirus (COVID-19) detection, 2020, arXiv preprint arXiv:2003, pp. 10769.
- Gour M., Jain S. Automated covid-19 detection from x-ray and ct im-ages with stacked ensemble convolutional neural network. Biocybernetics and Biomedical Engineering. 2022;42(1):27–41. doi: 10.1016/j.bbe.2021.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan Q., Huang Y. Multi-label chest x-ray image classification via category-wise residual attention learning. Pattern Recognition Letters. 2020;130:259–266. [Google Scholar]
- Guendel S., Grbic S., Georgescu B., Liu S., Maier A., Comaniciu D. Learning to recognize abnormalities in chest x-rays with location-aware dense networks. IBERO American Congress on Pattern Recognition. Springer. 2018:757–765. [Google Scholar]
- Huang G.-B., Zhu Q.-Y., Siew C.-K. Extreme learning machine: Theory and applications. Neurocomputing. 2006;70(1–3):489–501. [Google Scholar]
- Huff H.V., Singh A. Asymptomatic transmission during the coronavirus disease 2019 pandemic and implications for public health strategies. Clinical Infectious Diseases. 2020;71(10):2752–2756. doi: 10.1093/cid/ciaa654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam M.R., Nahiduzzaman M. Complex features extraction with deep learning model for the detection of COVID19 from CT scan images using ensemble based machine learning approach. Expert Systems with Applications. 2022 doi: 10.1016/j.eswa.2022.116554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain R., Gupta M., Taneja S., Hemanth D.J. Deep learning-based detection and analysis of covid-19 on chest x-ray images. Applied Intelligence. 2021;51(3):1690–1700. doi: 10.1007/s10489-020-01902-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi R.C., Yadav S., Pathak V.K., Malhotra H.S., Khokhar H.V.S., Parihar A., et al. A deep learning-based covid-19 automatic diagnostic framework using chest x-ray images. Biocybernetics and Biomedical Engineering. 2021;41(1):239–254. doi: 10.1016/j.bbe.2021.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamel S.I., Levin D.C., Parker L., Rao V.M. Utilization trends in noncardiac thoracic imaging, 2002–2014. Journal of the American College of Radiology. 2017;14(3):337–342. doi: 10.1016/j.jacr.2016.09.039. [DOI] [PubMed] [Google Scholar]
- Khan A.I., Shah J.L., Bhat M.M. Coronet: A deep neural network for detection and diagnosis of covid-19 from chest x-ray images. Computer Methods and Programs in Biomedicine. 2020;196 doi: 10.1016/j.cmpb.2020.105581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhevsky A., Ilya S., Geoffrey E.H. Imagenet classification with deep convolutional neural networks. Communications of the ACM. 2017;60(6):84–90. [Google Scholar]
- Kumar P., Grewal M., Srivastava M.M. International conference image analysis and recognition. 2018. Boosted cascaded convnets for multilabel classification of thoracic diseases in chest radiographs; pp. 546–552. [Google Scholar]
- Lopes U., Valiati J.F. Pre-trained convolutional neural networks as feature extractors for tuberculosis detection. Computers in Biology and Medicine. 2017;89:135–143. doi: 10.1016/j.compbiomed.2017.08.001. [DOI] [PubMed] [Google Scholar]
- Molina A., José R., Laura B., Andrea A., Santiago A., Anna M. Automatic identification of malaria and other red blood cell inclusions using convolutional neural networks. Computers in Biology and Medicine. 2021 doi: 10.1016/j.compbiomed.2021.104680. [DOI] [PubMed] [Google Scholar]
- Mukherjee H., Ghosh S., Dhar A., Obaidullah S.M., Santosh K.C., Roy K. Shallow convolutional neural network for COVID-19 outbreak screening using chest X-rays. Cognitive Computation. 2020:1–14. doi: 10.1007/s12559-020-09775-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nahiduzzaman, M., Islam, M. R., Islam, S. R., Goni, M. O. F., Anower, M. S., and Kwak, K.S., “Hybrid cnn-svd based prominent feature extraction and selection for grading diabetic retinopathy using extreme learning machine algorithm,” IEEE Access, vol. 9, pp. 152 261–152 274, 2021. 4 VOLUME 4, 2016.
- Nahiduzzaman M., Nayeem M.J., Ahmed M.T., Zaman M.S.U. 4th International conference on electrical information and communication technology (EICT) IEEE; 2019. Prediction of heart disease using multi-layer perceptron neural network and support vector machine. [Google Scholar]
- Nahiduzzaman, M., Goni, M. O. F., Anower, M. S., Islam, M. R., Ahsan, M., Haider, J., Gurusamy, S., Hassan, R., and Islam, M. R., “A novel method for multivariant pneumonia classification based on hybrid CNN-PCA based feature extraction using extreme learning machine with CXR images,” IEEE Access, vol. 9, pp. 147 512–147 526, 2021.
- Nahiduzzaman M., Islam M.R., Goni M.O.F., Anower M.S., Ahsan M., Haider J., et al. Diabetic retinopathy identification using parallel convolutional neural network based feature extractor and ELM classifier. Expert Systems with Applications. 2023;9 [Google Scholar]
- Nahiduzzaman M., Islam M.R., Hassan R. ChestX-Ray6: Prediction of multiple diseases including COVID-19 from chest X-ray images using convolutional neural network. Expert Systems with Applications. 2023 doi: 10.1016/j.eswa.2022.118576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh Y., Park S., Ye J.C. Deep learning covid-19 features on cxr using limited training data sets. IEEE Transactions on Medical Imaging. 2020;39(8):2688–2700. doi: 10.1109/TMI.2020.2993291. [DOI] [PubMed] [Google Scholar]
- Ouyang X., Karanam S., Wu Z., Chen T., Huo J., Zhou X.S., et al. Learning hierarchical attention for weakly-supervised chest x-ray abnormality localization and diagnosis. IEEE Transactions on Medical Imaging. 2020 doi: 10.1109/TMI.2020.3042773. [DOI] [PubMed] [Google Scholar]
- Ozturk T., Talo M., Yildirim E.A., Baloglu U.B., Yildirim O., Acharya U.R. Automated detection of covid-19 cases using deep neural networks with x-ray images. Computers in Biology and Medicine. 2020;121 doi: 10.1016/j.compbiomed.2020.103792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandit M.K., Banday S.A., Naaz R., Chishti M.A. Automatic detection of covid-19 from chest radiographs using deep learning. Radiography. 2021;27(2):483–489. doi: 10.1016/j.radi.2020.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panwar H., Gupta P., Siddiqui M.K., Morales-Menendez R., Singh V. Application of deep learning for fast detection of covid-19 in x-rays using ncovnet. Chaos, Solitons & Fractals. 2020;138 doi: 10.1016/j.chaos.2020.109944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers, D. M. (2020). “Evaluation: from precision, recall and f-measure to roc, informedness, markedness and correlation, arXiv preprint arXiv:2010.16061, 2020.
- Rahman T., Chowdhury M.E., Khandakar A., Islam K.R., Islam K.F., Mahbub Z.B., et al. Transfer learning with deep convolutional neural network (cnn) for pneumonia detection using chest x-ray. Applied Sciences. 2020;10(9):3233. [Google Scholar]
- Rahman, T., Khandakar, A., Kadir, M. A., Islam, K. R., Islam, K. F., Mazhar, R., Hamid, T., Islam, M. T., Kashem, S., Mahbub, Z. B. et al., “Reliable tuberculosis detection using chest x-ray with deep learning, seg-mentation and visualization,” IEEE Access, vol. 8, pp. 191 586–191 601, 2020.
- Rajpurkar, P., Irvin, J., Zhu, K., Yang, B., Mehta, H., Duan, T., Ding, D., Bagul, A., Langlotz, C., Shpanskaya K. et al. (2017). “Chexnet: Radiologist-level pneumonia detection on chest x-rays with deep learning,” arXiv preprintarXiv:1711.05225.
- Ruuskanen O., Lahti E., Jennings L.C., Murdoch D.R. Viral pneumonia. The Lancet. 2011;377(9773):1264–1275. doi: 10.1016/S0140-6736(10)61459-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasheed J., Hameed A.A., Djeddi C., Jamil A., Al-Turjman F. A machine learning-based framework for diagnosis of covid-19 from chest x-ray images. Interdisciplinary Sciences: Computational Life Sciences. 2021;13(1):103–117. doi: 10.1007/s12539-020-00403-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahlol A.T., Abd Elaziz M., Tariq Jamal A., Damaševiˇcius R., Hassan O.F. A novel method for detection of tuberculosis in chest radiographs using artificial ecosystem-based optimisation of deep neural network features. Symmetry. 2020;12(7):1146. [Google Scholar]
- Sekeroglu B., Ozsahin I. <? COVID19?> detection of covid-19 from chest x-ray images using convolutional neural networks. SLAS TECHNOLOGY: Translating Life Sciences Innovation. 2020;25(6):553–565. doi: 10.1177/2472630320958376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swets J.A. Measuring the accuracy of diagnostic systems. Science. 1988;240(4857):1285–1293. doi: 10.1126/science.3287615. [DOI] [PubMed] [Google Scholar]
- Szegedy C., Wei L., Yangqing J., Pierre S., Scott R., Dragomir A., et al. Proceedings of the IEEE conference on computer vision and pattern recognition. 2015. Going deeper with convolutions. [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nature Reviews Microbiology. 2021;19(3):171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Yang Y.-Y., Pan Y., Han P., Li Z.-X., Huang H.-G., et al. Detecting thoracic diseases via representation learning with adaptive sampling. Neurocomputing. 2020;406:354–360. [Google Scholar]
- Wang X., Peng Y., Lu L., Lu Z., Bagheri M., Summers R.M. Proceedings of the IEEE conference on computer vision and pattern recognition. 2017. Chestx-ray8: Hospital-scale chest x-ray database and benchmarks on weakly-supervised classification and localization of common thorax diseases. [Google Scholar]
- World Health Organization, “Who coronavirus (covid-19) dashboard,” 2022. Accessed on: March, 12, 2022. [Online]. Available: https://COVID19.who.int/table.
- Yamac M., Ahishali M., Degerli A., Kiranyaz S., Chowdhury M.E., Gabbouj M. Convolutional sparse support estimator-based covid-19recognition from x-ray images. IEEE Transactions on Neural Networks and Learning Systems. 2021;32(5):1810–1820. doi: 10.1109/TNNLS.2021.3070467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan L., Hai-Tao Z., Jorge G., Yang X., Maolin W., Yuqi G., et al. An interpretable mortality prediction model for COVID-19 patients. Nature Machine Intelligence. 2020;2(5):283–288. [Google Scholar]
- Yao, L., Poblenz, E., Dagunts, D., Covington, B., Bernard, D., & Lyman, K. (2017). Learning to diagnose from scratch by exploiting dependencies among labels, arXiv preprint arXiv:1710.10501.
- Zhu, N., Zhang, D., Wang, W., Li, X., Yang, B., Song, J., ... & Tan, W. (2020). A novel coronavirus from patients with pneumonia in China, 2019. New England Journal of Medicine. [DOI] [PMC free article] [PubMed]
- BIMCV Medical Imaging Databank of the Valencia Region, “Bimcv-covid19,” 2020. Accessed on: December, 20, 2021. [Online]. Available: https://bimcv.cipf.es/bimcv-projects/bimcv-covid19/#1590858128006-9e640421-6711.
- Cohen, J. P. “covid-chest xray-dataset,” 2019. Accessed on: December, 20, 2021. [Online]. Available: https://github.com/ieee8023/covid-chestxray-dataset/tree/master/images.
- Haghanifar, A. “Covid-cxnet,” 2019. Accessed on: December, 20,2021. [Online]. Available: https://github.com/armiro/COVID-CXNet/tree/master/chest_xray_images/covid19.
- Health, B. P. “Belarus tuberculosis portal,” 2020. Accessed on: December,20, 2021. [Online]. Available: http://tuberculosis.by.
- Jaeger S., Candemir S., Antani S., Wáng Y.-X.-J., Lu P.-X., Thoma G. Two public chest x-ray datasets for computer-aided screening of pulmonary diseases. Quantitative Imaging in Medicine and Surgery. 2014;4(6):475. doi: 10.3978/j.issn.2223-4292.2014.11.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- JtiptJ, “Chest x-ray (pneumonia, covid-19,tuberculosis),” 2021. Accessed on: December, 20, 2021. [Online]. Available: https://www.kaggle.com/jtiptj/chest-xray-pneumoniacovid19tuberculosis.
- Mader, K. S. “Pulmonary chest x-ray abnormalities,” 2017. Accessed on: December, 20, 2021. [Online]. Available: https://www.kaggle.com/kmader/pulmonary-chest-xray-abnormalities.
- ml-workgroup, “covid-19-image-repository,” 2019. Accessed on: December, 20, 2021. [Online]. Available: https://github.com/ml-workgroup/covid-19-image-repository/tree/master/png.
- Mooney, P. “Chest x-ray images (pneumonia),” 2017. Accessed on: December, 20, 2021. [Online]. Available: https://www.kaggle.com/paultimothymooney/chest-xray-pneumonia.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors do not have permission to share data.