Abstract
The tomato resistance genes Cf-4 and Cf-9 confer specific, hypersensitive response–associated recognition of Cladosporium carrying the avirulence genes Avr4 and Avr9, respectively. Cf-4 and Cf-9 encode type I transmembrane proteins with extracellular leucine-rich repeats (LRRs). Compared with Cf-9, Cf-4 lacks two LRRs and differs in 78 amino acid residues. To investigate the relevance of these differences for specificity, we exchanged domains between Cf-4 and Cf-9, and mutant constructs were tested for mediating the hypersensitive response by transient coexpression with either Avr4 or Avr9. We show that the number of LRRs is essential for both Cf-4 and Cf-9 function. In addition, Cf-9 specificity resides entirely in the LRR domain and appears to be distributed over several distant LRRs. In contrast, Cf-4 specificity determinants reside in the N-terminal LRR-flanking domain and three amino acid residues in LRRs 13, 14, and 16. These residues are present at putative solvent-exposed positions, and all are required for full Cf-4 function. Finally, we show that Cf-9 carrying the specificity determinants of Cf-4 has recognitional specificity for AVR4. The data indicate that diversifying selection of solvent-exposed residues has been a more important factor in the generation of Cf-4 specificity than has sequence exchange between Cf-4 progenitor genes. The fact that most variant residues in Cf-4 are not essential for Cf-4 specificity indicates that the diverse decoration of R proteins is not fully adapted to confer recognition of a certain avirulence determinant but likely provides a basis for a versatile, adaptive recognition system.
INTRODUCTION
Because plants are subjected to environments that change continuously, they are armed with recognition systems that can sense diverse biotic and abiotic stresses and subsequently mediate the induction of appropriate responses. A recognition system that can deal with various pathogens is especially crucial for the survival of the plant. Specific recognition of invading pathogens frequently is mediated by resistance (R) genes. Upon recognition of the matching pathogen-derived avirulence determinant (AVR), various defense responses are triggered, often including a hypersensitive response. The hypersensitive response involves the death of the tissue surrounding the primary infection site and thereby restricts further growth of the invading pathogen (Hammond-Kosack and Jones, 1996).
Surprisingly, R genes that confer resistance to different types of pathogens encode very similar proteins, indicating that in plants, flexible recognition systems are used to monitor attacks by a diverse array of pathogens. The largest class of R genes encodes proteins that are likely located in the cytoplasm and contain leucine-rich repeats (LRRs) and a nucleotide binding site (NBS). Members of this NBS-LRR class have been cloned from various plant species and confer race-specific resistance against viruses, bacteria, fungi, oomycetes, nematodes, or insects (reviewed in Van der Biezen and Jones, 1998). Some R genes encode proteins that are likely located in the plasma membrane and carry extracellular LRRs. The Xa21 gene from rice encodes a receptor-like kinase containing a cytoplasmic kinase and an extracellular LRR domain (Song et al., 1995). The products that are encoded by HSPro-1 from sugar beet and the Cf genes from tomato lack the cytoplasmic kinase domain and predominantly consist of extracellular LRRs (Jones et al., 1994; Dixon et al., 1996, 1998; Cai et al., 1997; Thomas et al., 1997; Takken et al., 1999).
Because pathogens continuously attempt to circumvent recognition by the host, the plant recognition system must be able to generate new specificities. Mechanisms by which R genes with new specificities evolve include sequence exchange between homologous genes and selective mutations of solvent-exposed amino acid residues (Parniske et al., 1997; Song et al., 1997; Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Ellis et al., 1999; Noël et al., 1999; Parniske and Jones, 1999; Bittner-Eddy et al., 2000).
Specific recognition mediated by LRR proteins also plays a role in other plant processes. For example, many different receptor-like kinases with extracellular LRRs are involved in plant development. ERECTA is required for proper organ elongation, CLAVATA-1 determines cell fate in shoot and floral meristems, and BRI1 encodes a putative brassinosteroid receptor (Torii et al., 1996; Clark et al., 1997; Li and Chory, 1997). Polygalacturonase-inhibiting proteins have a role in plant defense and are extracellular LRR proteins that bind specifically, and consequently inhibit, fungal endopolygalacturonases (De Lorenzo and Cervone, 1997).
LRRs are highly specialized protein binding motifs that are also present in various proteins of animals and bacteria (reviewed in Buchanan and Gay, 1996). Within a single LRR module, the xxLxLxx motif folds as a β-sheet in which the leucine residues (L) form a hydrophobic core, whereas the side chains of the flanking amino acid residues (x) are solvent exposed. In multiple LRRs, the β-sheets are aligned in parallel and form a surface decorated with solvent-exposed residues that can interact specifically with a ligand. Significant progress in the understanding of protein–protein interactions involving LRR proteins comes from crystallographic studies of ribonucleases and LRR-containing ribonuclease inhibitors (RIs). Cocrystallization of porcine RI and human RI (hRI) with the enzymes RNaseA and angiogenin, respectively, revealed that of the 28 amino acid residues of porcine RI that interact with RNaseA, 26 are at solvent-exposed positions (Kobe and Deisenhofer, 1996). Similarly, of the 26 amino acid residues of hRI that interact with angiogenin, 25 are at solvent-exposed positions (Papageorgiou et al., 1997). Likewise, it can be expected that solvent-exposed amino acid residues of LRRs in R proteins are involved in interactions with matching AVR proteins. Consistent with this theory, it was found that R proteins with specificity for different avirulence determinants differ predominantly in amino acid residues at putative solvent-exposed positions (Parniske et al., 1997; Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Noël et al., 1999; Bittner-Eddy et al., 2000). This finding indicates that these variant amino acid residues are present to adapt the decoration of R proteins to optimal recognition of the matching AVR determinants.
To understand or even modify the specificity of R proteins, studies on domains that determine their specificity are of great interest. For such studies, two functional homologous R proteins with different specificity are required. The R genes Cf-4 and Cf-9 from tomato confer resistance to strains of the biotrophic leaf mold fungus Cladosporium that carry the avirulence genes Avr4 and Avr9, respectively (reviewed in Joosten and De Wit, 1999). Both Avr genes encode stable elicitor proteins that are secreted into the apoplast of tomato leaves. Apart from being small and cysteine rich, the avirulence determinants AVR4 and AVR9 have no sequence similarities. In contrast, the matching R genes encode proteins that share 91% amino acid residue identity (Thomas et al., 1997). The primary structures and the alignment of Cf-4 and Cf-9 are shown in Figure 1. Compared with Cf-9, Cf-4 lacks 10 amino acid residues in the B-domain, one residue in LRR 4, and two complete LRRs at the position where Cf-9 has LRRs 11 and 12. Cf-4 further differs from Cf-9 by 67 amino acid residues that are confined to the N-terminal half of the proteins. Six and four variant residues reside in the signal peptide (A-domain) and B-domain, respectively, whereas the remaining 57 variant amino acid residues are present in the LRRs, of which 32 are located at putative solvent-exposed positions of the β-sheets (Figure 1).
Figure 1.
Primary Structure and Alignment of Cf-4 and Cf-9 Resistance Proteins.
Amino acid residues of Cf-4 and Cf-9 that are identical are shown in black. Cf-4– and Cf-9–specific residues are shown in blue and red, respectively. The numbering (left) corresponds to the Cf-9 protein sequence. Potential N-glycosylation sites (NxS/T) in Cf-4 and Cf-9 are overlined and underlined, respectively. The stippled box indicates the various β-sheets (consensus xxLxLxx), each of which contains five solvent-exposed amino acid residues (x). Residues encoded at restriction sites are indicated with green boxes. Introduction of a ClaI site resulted in two residue changes, whereas the SalI and BamHI sites were introduced as silent mutations. The EcoRI site is endogenous for Cf-4 and Cf-9. Domains, indicated at left, are as follows: SP, signal peptide (A-domain); B, cysteine-rich domain; 1 to 27, LRRs; D, domain without conspicuous features; E, acidic domain; TM, putative transmembrane domain; G, basic domain, representing the putative cytoplasmic tail.
It must be stressed that no evidence exists for a direct interaction between AVR and Cf proteins. It is likely that AVR9 is perceived by plants carrying Cf-9 through a high-affinity binding site for AVR9 that was identified in plasma membranes of solanaceous plants (Kooman-Gersmann et al., 1996, 1998). A similar situation is possible for the perception of AVR4. Although the exact nature of the “ligands” of Cf proteins is unknown, Cf proteins are expected to be involved in specific interactions, possibly through the solvent-exposed amino acid residues in the LRR domain.
The high homology between Cf-4 and Cf-9 proteins provides an excellent opportunity to determine which domains of the Cf proteins confer specificity. Because Cf and AVR function are retained upon Agrobacterium-mediated transient expression in tobacco (Van der Hoorn et al., 2000), we used this technique as a quick and versatile tool to study Cf specificity. In this study, we show that the functions of both Cf-9 and Cf-4 are strongly affected when the number of LRRs is changed. Moreover, the specificity of Cf-9 resides entirely in the LRRs and appears to be distributed over several distant LRRs. In contrast, the specificity of Cf-4 was found to reside in the B-domain and three Cf-4–specific amino acid residues. These residues are present at solvent-exposed positions within LRRs 13, 14, and 16 and collectively contribute to Cf-4 specificity. Introduction of these specificity determinants into the LRRs of Cf-9 results in recognitional specificity for AVR4 instead of AVR9. Our results indicate that diversifying selection of solvent-exposed amino acid residues was a more important factor in the generation of Cf-4 specificity than was sequence exchange between Cf-4 progenitor genes. The fact that most variant amino acid residues in Cf-4 are not essential for Cf-4 specificity indicates that decoration of R proteins is not fully adapted to confer recognition of a certain avirulence determinant but likely provides a basis for a versatile, adaptive recognition system.
RESULTS
The LRR Domain of Cf Proteins Is Required for Specificity
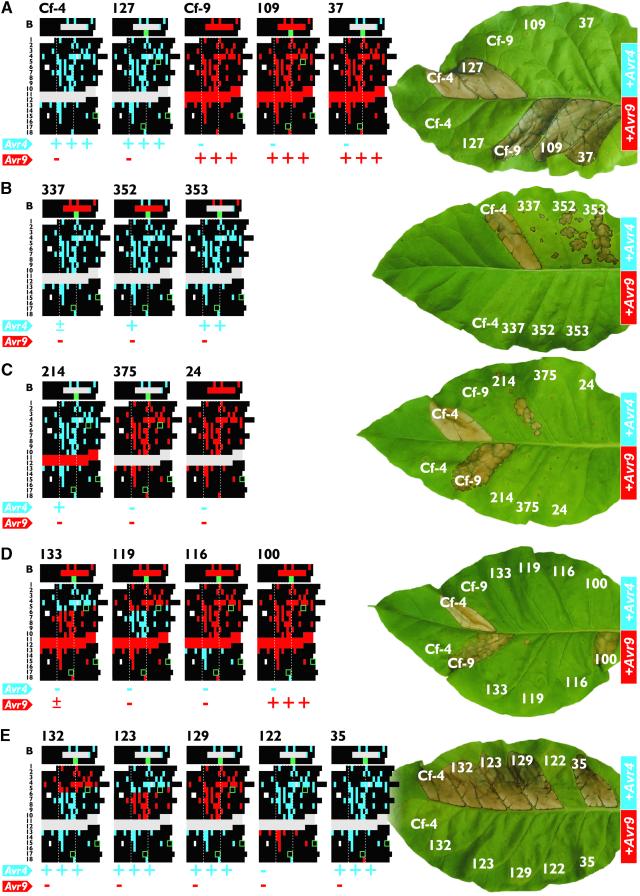
The differences between Cf-4 and Cf-9 suggest that the LRR domain determines specificity for mediating recognition of AVR4 and AVR9, respectively. To test this notion, we exchanged the LRRs of Cf-4 with those of Cf-9. This exchange was facilitated by the introduction of ClaI, SalI, and BamHI restriction sites into the Cf-4 and Cf-9 genes (Figure 1). Only introduction of the ClaI site resulted in a substitution of two amino acid residues (V83I and H84D) in the B-domain of both Cf-4 and Cf-9. Mutants were assayed through transient coexpression with Avr4 or Avr9 by agroinfiltration of tobacco leaves (see Methods). This assay demonstrated that introduction of the restriction sites did not affect Cf gene function (Figure 2A, mutants 127 and 109). Exchange of all LRRs of Cf-4 with those of Cf-9 abolished Cf-4 function in our assays (Figure 2A, mutant 37), indicating that the LRRs of Cf-4 are required for Cf-4 specificity. Because mutant 37 had gained Cf-9 function, we concluded that specificity for AVR9 recognition resides only in the LRRs of Cf-9.
Figure 2.
Domain Swap Analysis Reveals Distinct Specificity Determinants in Cf-4.
Amino acid residues of the Cf-4 and Cf-9 proteins are represented as blocks that make up the different protein domains (indicated at left; see also Figure 1). Only parts of the mature proteins that contain variant residues are shown. Amino acid residues that are identical are shown in black, whereas Cf-4– and Cf-9–specific residues are shown in blue and red, respectively. Cf-4–specific deletions are shown in gray. Positions corresponding to restriction sites are indicated with green blocks and squares. The numbers of the mutants are indicated above the blocks. Below the blocks, the Avr responsiveness of the mutant is indicated compared with wild-type Cf-4 (blue, top) or Cf-9 (red, bottom): −, no necrosis above background; +, incomplete necrosis of infiltrated area (10 to 50%); ++, almost complete necrosis of infiltrated area (50 to 100%); +++, as active as wild-type Cf. The tobacco leaves shown at right illustrate responses observed upon transient coexpression of (mutated) Cf genes with Avr4 (top half of the leaf) or Avr9 (bottom half of the leaf). Photographs were taken 7 days after infiltration and are representative of at least three independent experiments. Color differences of necrotic sectors are due to differences in leaf age and result from the fact that Avr9 is less active than Avr4 when expressed in planta (Van der Hoorn et al., 2000).
(A) The LRR domain of Cf proteins is required for specificity.
(B) Cf-4 specificity also resides in the B-domain.
(C) The number or LRRs strongly affects Cf-4 and Cf-9 function.
(D) Cf-9 specificity appears to be distributed over several distant LRRs.
(E) Cf-4–specific residues in LRRs 13 to 16 are essential for Cf-4 function.
Cf-4 Specificity Also Resides in the B-Domain
To examine whether the LRR domain of Cf-4 is sufficient to confer AVR4 responsiveness, we exchanged all LRRs of Cf-9 with those of Cf-4. Surprisingly, this mutant showed significantly reduced Cf-4 activity (Figure 2B, mutant 337), indicating that the A- and/or B-domains of Cf-4 are required for Cf-4 specificity. The A-domain, which has been suggested to function as a signal peptide for extracellular targeting (Jones et al., 1994), is cleaved off in the mature Cf protein (R. Luderer, unpublished data). Thus, in mature proteins, mutant 337 differs from fully functional Cf-4 by only three amino acid residues and a deletion of 10 amino acid residues in the B-domain (compare mutants 127 and 337 in Figures 2A and 2B, respectively). To determine which of these features are required for Cf-4 function, we constructed mutants that contained either the Cf-4–specific amino acid residues or the Cf-4–specific deletion in the B-domain (Figure 2B, mutants 352 and 353, respectively). Both mutants showed reduced activity compared with wild-type Cf-4, indicating that both features contribute significantly to Cf-4 specificity. In addition, in five replicate experiments, we observed that mutant 353 was more active than mutant 352, indicating that within the B-domain of Cf-4 the deletion of 10 amino acid residues is more important for Cf-4 specificity than the three Cf-4–specific amino acid residues.
The Number of LRRs Strongly Affects Cf-4 and Cf-9 Function
The LRR domain of Cf-4 lacks LRRs 11 and 12 compared with Cf-9 (Figure 1). To examine the role of this deletion in Cf-4 function, we inserted LRRs 11 and 12 of Cf-9 into Cf-4. This mutant showed a significant reduction in activity compared with wild-type Cf-4 (Figure 2C, mutant 214), indicating that this Cf-4–specific deletion is important for Cf-4 function. To determine whether deletion of LRRs 11 and 12 in the LRR domain of Cf-9 is sufficient to confer Cf-4 specificity, we deleted LRRs 11 and 12 of Cf-9 from mutant 37, which already contained the B-domain of Cf-4 (Figure 2A). This mutant was not responsive to AVR4 (Figure 2C, mutant 375), suggesting that in addition to the B-domain and the deletion of LRRs 11 and 12, Cf-4–specific amino acid residues within other LRRs are required for Cf-4 function. Interestingly, mutant 375 also was not active in AVR9 recognition, indicating that deletion of LRRs 11 and 12 from Cf-9 abolishes Cf-9 function (compare with mutant 37, Figure 2A). Indeed, deletion of LRRs 11 and 12 from wild-type Cf-9 protein was sufficient to abolish Cf-9 function in our assays (Figure 2C, mutant 24).
Cf-9 Specificity Is Likely Distributed over Several Distant LRRs
In addition to the presence of LRRs 11 and 12, the LRRs of Cf-9 differ from those of Cf-4 in 57 amino acid residues. To define the region within the LRR domain that specifies Cf-9 function, we exchanged the LRRs of Cf-9 with those of Cf-4 in blocks of five LRRs. Replacement of LRRs 1 to 5 significantly reduced Cf-9 function, whereas exchange of LRRs 6 to 10 or LRRs 13 to 16 abolished Cf-9 function in our assays (Figure 2D, mutants 133, 119, and 116, respectively). This finding indicates that Cf-9–specific amino acid residues that are required for Cf-9 function are distributed over several distant LRRs. The presence of one Cf-4–specific amino acid, N511, in LRR 18 of Cf-9 did not affect Cf-9 function (Figure 2D, mutant 100).
Cf-4–Specific Amino Acid Residues Residing in LRRs 13 to 16 Are Essential for Cf-4 Function
To define the region within the LRR domain that specifies Cf-4 function, the LRRs of Cf-4 were exchanged with those of Cf-9 in blocks of five LRRs. Surprisingly, replacement of LRRs 1 to 5 or LRRs 6 to 10 of Cf-4 with those of Cf-9 did not affect Cf-4 function (Figure 2E, mutants 132 and 123, respectively), indicating that the Cf-4–specific amino acid residues in LRRs 1 to 10 of Cf-4 are not essential for Cf-4 function. Indeed, a Cf-4 mutant containing LRRs 1 to 10 of Cf-9 retained complete Cf-4 function (Figure 2E, mutant 129). In contrast, replacement of LRRs 13 to 16 of Cf-4 with those of Cf-9 abolished Cf-4 function completely in our assays (Figure 2E, mutant 122), indicating that Cf-4–specific amino acid residues within LRRs 13 to 16 are required for Cf-4 function. Finally, the presence of one Cf-9–specific amino acid residue, K511, in LRR 18 of Cf-4 did not affect Cf-4 function (Figure 2E, mutant 35).
Within LRRs 13 to 16, Residues W389, G411, and F457 All Are Required for Full Cf-4 Function
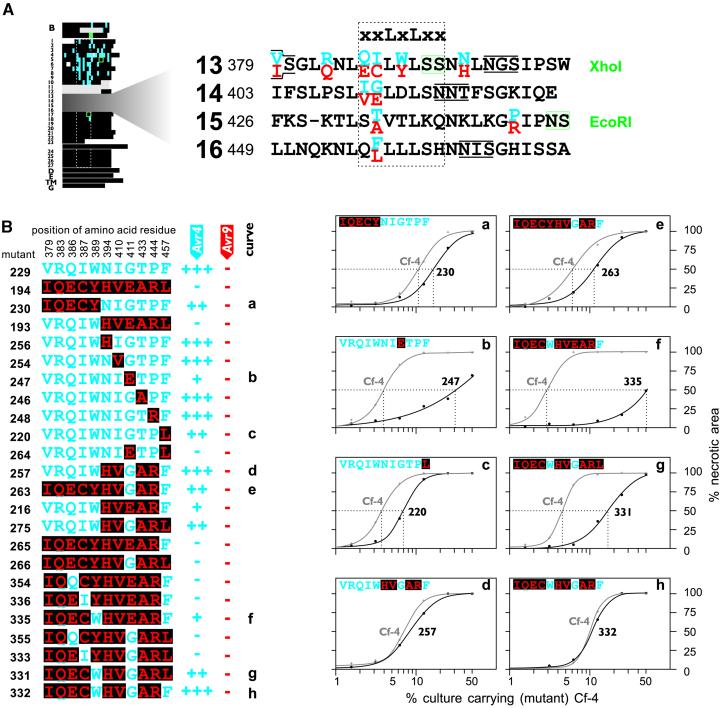
Having shown that only replacement of LRRs 13 to 16 of Cf-4 with those of Cf-9 abolishes Cf-4 function in our assays, we studied the roles of the remaining 11 variant amino acid residues within these LRRs (Figure 3A) in Cf-4 function in more detail. To facilitate the exchange of individual amino acid residues within LRRs 13 to 16, we introduced an XhoI restriction site as a silent mutation at a position in the open reading frame corresponding to LRR 13 (Figure 3A). Introduction of this restriction site did not alter Cf-4 function (Figure 3B, mutant 229). Again, substitution of all variant amino acid residues with Cf-9–specific residues abolished Cf-4 function in our assays (Figure 3B, mutant 194). Exchanging the first five Cf-4–specific amino acid residues in LRR 13 with those of Cf-9 reduced Cf-4 function slightly (Figure 3B, mutant 230). Quantitative comparison of this mutant with wild-type Cf-4 revealed only a minor difference in Cf-4 activity (Figure 3B, curve a). This finding suggests that Cf-4 specificity resides mainly in the remaining six Cf-4–specific residues: N394, I410, G411, T433, P444, and F457. Indeed, a Cf-4 mutant in which these six residues were replaced with those of Cf-9 was inactive in our assays (Figure 3B, mutant 193).
Figure 3.
Three Amino Acid Residues within LRRs 13 to 16 Contribute to Cf-4 Function.
(A) Detailed representation of LRRs 13 to 16. All mutations shown in Figure 3B were introduced into the Cf-4 backbone (left). The enlargement shows an alignment of LRRs 13 to 16 of Cf-4 and Cf-9. Variant amino acid residues between Cf-4 and Cf-9 are shown in blue and red, respectively. Additional details are described in Figure 1.
(B) Identification of single amino acid residues W389, G411, and F457 that contribute to Cf-4 function. Only amino acid residues that differ between Cf-4 and Cf-9 within LRRs 13 to 16 are shown. All mutations were present in the Cf-4 backbone (Figure 3A). Cf-4–specific amino acid residues are shown in blue, and Cf-9–specific amino acid residues are boxed and shown in red. The activity of the various Cf-4 mutants, upon coexpression with either Avr4 or Avr9, is indicated at right: −, no necrosis; +, incomplete necrosis; ++ and +++, complete necrosis. Discrimination between ++ and +++ was determined by quantitative comparisons (curves a to h). For quantitative comparisons, Agrobacterium cultures that carry a plasmid that encodes a (mutant) Cf-4 protein were mixed in different ratios with a culture of equal density carrying an AVR4-encoding plasmid and infiltrated into opposite tobacco leaf halves. At 7 days after infiltration, the percentage of infiltrated area that had become necrotic was measured and plotted against the percentage of culture carrying (mutant) Cf-4.
To identify which of the six Cf-4–specific amino acid residues are essential for Cf-4 function, Cf-4 mutants were generated in which each of the six amino acid residues was replaced individually with a Cf-9–specific amino acid residue (Figure 3B, mutants 256, 254, 247, 246, 248, and 220). Surprisingly, all single mutants retained Cf-4 function, albeit not all at the same level. Mutants carrying N394H, I410V, T433A, or P444R substitutions fully retained Cf-4 function (Figure 3B, mutants 256, 254, 246, and 248, respectively), whereas mutants carrying G411E or F457L substitutions showed reduced Cf-4 activity (Figure 3B, mutants 247 [curve b] and 220 [curve c], respectively). Significantly, a Cf-4 double mutant with both G411E and F457L substitutions was inactive in our assays (Figure 3B, mutant 264), suggesting that the G411E and F457L substitutions collectively disrupt Cf-4 function. Moreover, simultaneous introduction of the N394H, I410V, T433A, and P444R substitutions did not affect Cf-4 function (Figure 3B, mutant 257, curve d). Thus, within the last six Cf-4–specific amino acid residues present in LRRs 13 to 16, only G411 and F457 are essential for full Cf-4 function.
Having identified the two Cf-4–specific amino acid residues that are required in the last six variant positions in LRRs 13 to 16, we focused on the importance for Cf-4 function of the residues at the first five variant positions of LRRs 13 to 16. Again, a mutant of Cf-4 carrying Cf-9–specific amino acid residues except G411 and F457 showed reduced Cf-4 activity (Figure 3B, mutant 263, curve e), consistent with the reduction in activity of mutant 230 (Figure 3B, curve a). The presence of an essential Cf-4–specific residue in the first five positions became more evident from a comparison based on Cf-4 mutants that carried either F457 or G411 in the last six variant positions (Figure 3B, mutants 216 and 275, respectively). In both cases, Cf-4 function was abolished completely in our assays by introducing Cf-9–specific residues in the first five positions (Figure 3B, mutants 265 and 266, respectively). Therefore, the inactive mutants carrying only F457 (mutant 265) or G411 (mutant 266) were used in a gain-of-function approach to determine which of the first five variant amino acid residues in LRR 13 is/are important for Cf-4 function.
Because we had shown that within the last six variant amino acid residues, only residues at putative solvent-exposed positions are important for Cf-4 function, we focused on amino acid residues Q386, I387, and W389, which are present at putative solvent-exposed positions of the β-sheets of LRR 13 (Figure 3A). We inserted each of these residues separately into the inactive mutants 265 and 266. Combination of amino acid residue F457 with residue Q386 or I387 did not restore Cf-4 function (Figure 3B, mutants 354 and 336, respectively), whereas combination with residue W389 restored Cf-4 activity (Figure 3B, mutant 335, curve f) to a level comparable to that of mutant 216, which carries all Cf-4–specific amino acid residues in the first five variant positions of LRR 13. Similarly, combination of amino acid residue G411 with residue Q386 or I387 did not restore Cf-4 function (Figure 3B, mutants 355 and 333, respectively), whereas combination with residue W389 restored Cf-4 activity (Figure 3B, mutant 331, curve g) to a level comparable to that of mutant 275. These data demonstrate that within LRR 13, only the Cf-4–specific amino acid residue W389 contributes to Cf-4 function.
To establish whether amino acid residues W389, G411, and F457 are sufficient to confer Cf-4 function, they were introduced simultaneously into Cf-4 mutant 194, which carries LRRs 13 to 16 of Cf-9 (Figure 3B). Introduction of the three Cf-4–specific amino acid residues into these LRRs restored Cf-4 function completely (Figure 3B, mutant 332, curve h), indicating that within LRRs 13 to 16 of Cf-4, presence of amino acid residues W389, G411, and F457 are required and sufficient for full Cf-4 function.
Quantitative analysis of the gain-of-function mutants (Figure 3B, curves f and g) also confirmed the relative importance of the substitutions observed in the loss-of-function approach (Figure 3B, curves a to c). The W389Y substitution resulted in a twofold reduction in Cf-4 activity compared with wild-type Cf-4 (Figure 3B, curves a and e), whereas the G411E substitution resulted in an eight- to 16-fold reduction in Cf-4 activity (Figure 3B, curves b and f). The F457L substitution resulted in two- to threefold reduced Cf-4 activity (Figure 3B, curves c and g). Thus, the relative importance of these substitutions for the attenuation of Cf-4 function is G411E > F457L ⩾ W389Y.
Within the LRRs of Cf-9, Deletion of LRRs 11 and 12 and Introduction of W389, G411, and F457 Are Sufficient to Confer Cf-4 Function
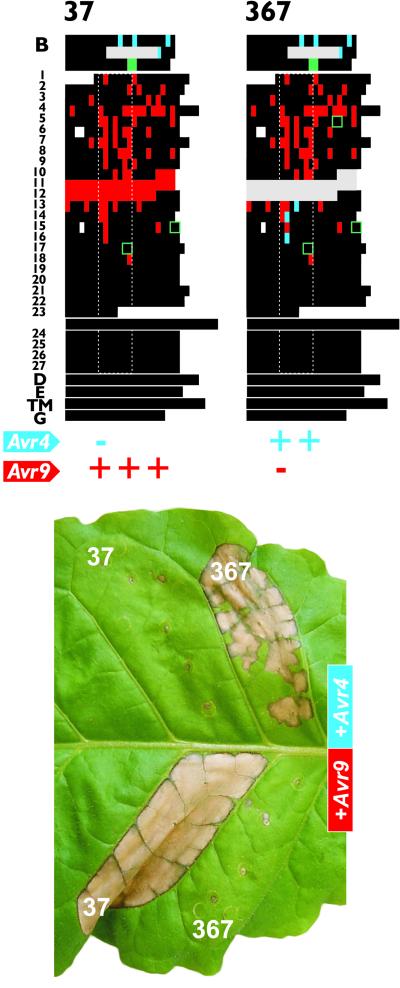
To determine whether deletion of LRRs 11 and 12, together with introduction of the amino acid residues W389, G411, and F457 into the LRRs of Cf-9, is sufficient to confer Cf-4 function, such a mutant was generated from mutant 37, which was fully responsive to AVR9 (Figures 2A and 4). Although slightly reduced compared with that of wild-type Cf-4, this mutant clearly showed specificity for AVR4, whereas Cf-9 function was lost completely (Figure 4, mutant 367). Thus, within the LRRs of Cf-9, deletion of LRRs 11 and 12 and introduction of amino acid residues W389, G411, and F457 are sufficient to confer Cf-4 function.
Figure 4.
A Cf-9 Mutant Carrying the Specificity Determinants of Cf-4 Confers Recognition of AVR4.
The specificity of mutant 37, which confers full AVR9 recognition upon transient expression in tobacco (Figure 2A), was changed into specificity for AVR4 by deleting LRRs 11 and 12 and introducing W389, G411, and F457, resulting in mutant 367. For further details, see legends to Figures 1 and 2.
DISCUSSION
Because many R genes that confer resistance to a variety of pathogens appear to encode homologous proteins, it is of great interest to discover the features of R proteins that determine specificity for avirulence determinants of pathogens. Reports on dissecting the domains that determine specificity in R proteins have been limited, because at least two homologous proteins with different specificities are required to address this question. The distinct specificity and high similarity of Cf-4 and Cf-9, together with the availability of the matching avirulence genes Avr4 and Avr9, respectively, provided a unique opportunity to identify domains within both R proteins that determine specificity. Using agroinfiltration, we found that the number of LRRs is essential for both Cf-4 and Cf-9 function. In addition, we showed that Cf-9 specificity resides entirely in the LRR domain and is likely distributed over several distant LRRs. In contrast, Cf-4 specificity resides in the B-domain and three amino acid residues present at putative solvent-exposed positions of LRRs 13 to 16. Finally, a Cf-9 mutant carrying the features that are required for Cf-4 specificity confers recognition of AVR4.
The domain swap analysis of the tomato Cf resistance genes presented was based on a functional assay in tobacco. We cannot exclude that mutant Cf genes behave somewhat different in tomato and tobacco. Additional proteins that are required for AVR perception and interact with the Cf protein might differ slightly between tomato and tobacco. Thus far, however, we found that AVR9 and AVR4 perception occurs with the same specificity in tomato and tobacco (Van der Hoorn et al., 2000), suggesting that the tomato Cf genes function similarly both in tomato and tobacco. We also cannot exclude that different Cf mutants differ in protein stability, because we did not examine protein accumulation of the mutants. However, because we exchanged domains between functional proteins that occur in nature, destabilization of proteins is not likely to occur.
The B-Domain Contributes to Specificity in Cf Proteins
The presumed binding capacities of the LRR domain, especially the large variation in solvent-exposed amino acid residues in this domain, suggest that the LRRs determine the specificity of Cf proteins. However, we have shown that the B-domain contributes to Cf-4 specificity. This finding suggests that in Cf-4, the B-domain is required for interaction with a component that is not essential for Cf-9 function. Especially the 10–amino acid residue deletion in the B-domain is important for Cf-4 function. It is striking that this region in the B-domain of Cf homologs is hypervariable for deletions and amino acid residue substitutions (Parniske et al., 1997; Parniske and Jones, 1999), suggesting that it generally contributes to specificity in Cf proteins. However, in contrast with Cf-4 function, Cf-9 function is not affected when its B-domain is replaced with that of Cf-4. Thus, at this stage, it is difficult to predict whether the requirement of a particular B-domain for Cf specificity is an exception rather than a rule. Also, for R proteins of the NBS-LRR class, specificity can reside outside the LRR domain, as has been found in flax, in which the L6 and L7 proteins vary only outside the LRR domain and yet have different specificities (Ellis et al., 1999).
Importance of the Number of LRRs
Among the 15 Cf proteins that are homologous with Cf-4 and Cf-9, Cf-4 is exceptional in having 25 rather than 27 LRRs (Parniske et al., 1997; Parniske and Jones, 1999). In this report, we show that this feature is important for Cf-4 function. We also show that Cf-9 function is abolished in our assays when LRRs 11 and 12 are deleted. As a consequence, it is unlikely that a hybrid Cf protein with recognitional specificities for both AVR4 and AVR9 can be constructed.
Variation in the number of LRRs is a general feature of R proteins. Cf proteins that show homology with Cf-2 carry 25, 26, 31, 32, 34, or 38 LRRs (Dixon et al., 1998), whereas NBS-LRR proteins encoded by the RPP5 locus carry 13, 17, 21, 23, or 25 LRRs (Noël et al., 1999). However, the importance of the number of LRRs for R protein function has been reported in only a few cases. The nonfunctional Cf-5 homolog Hcr2-5D encodes a protein that differs from the functional Cf-5 protein by having two additional LRRs (Dixon et al., 1998). Similarly, inactive alleles of the M gene from flax encode NBS-LRR proteins with a reduced number of LRRs (Anderson et al., 1997), and an inactive RPP5 allele from Arabidopsis encodes an NBS-LRR protein with a duplication of four LRRs (Parker et al., 1997).
Studies of the interaction between hRI and angiogenin showed that LRRs of hRI have multiple interaction points with angiogenin (Papageorgiou et al., 1997). The 26 amino acid residues of hRI that interact with angiogenin are scattered throughout the 13 LRRs of hRI. Deletion of one or more LRRs abolished the hRI–angiogenin interaction, an observation that could not be explained solely by the loss of the few interacting amino acid residues within the deleted LRRs. This finding suggests that deletion of LRRs can result in loss of interaction with the remaining LRRs or can significantly change the overall three-dimensional structure of the LRR protein. Similar strong structural effects can explain the requirement of a specific number of LRRs in Cf-4, Cf-9, and other R proteins.
The Role of Putative Solvent-Exposed Amino Acid Residues in Cf Specificity
Based on studies of RIs, solvent-exposed amino acid residues of the β-sheets present in LRR proteins are expected to form a recognition surface at the inner site of the curved LRR domain (Kobe and Deisenhofer, 1995). Thus, variation in the decoration of the recognition surface by substitution of solvent-exposed amino acid residues is likely to affect the recognitional specificity of an LRR protein. Consistent with this theory, it was found that Cf proteins and many R proteins of the NBS-LRR class are hypervariable at solvent-exposed positions in the LRR domain (Parniske et al., 1997; Botella et al., 1998; McDowell et al., 1998; Meyers et al., 1998; Noël et al., 1999; Bittner-Eddy et al., 2000). However, to date, the exact role of solvent-exposed residues in the specificity of R proteins has not been documented. We have now identified three solvent-exposed residues that are essential for full Cf-4 function. Two of these (W389 and G411) reside among the seven positions that have been reported to be hypervariable in Cf proteins encoded by the Cf-4 and Cf-9 gene clusters (Parniske et al., 1997). Both W389 and G411 are unique among homologs of Cf-4 and are clustered in the Cf-4 protein. This finding indicates that diversifying selection of solvent-exposed amino acid residues was more important in the generation of AVR4 recognitional specificity than sequence exchange between Cf-4 progenitor genes.
We also showed that amino acid residues W389, G411, and F457 only partially contribute to Cf-4 specificity. Single W389Y and F457L substitutions reduced Cf-4 function only slightly, whereas Cf-4 function was abolished completely in our assays when W389 and F457 were substituted simultaneously. The slightly reduced activity of mutant 367, which contains the Cf-4–specific features that we have identified, might be explained by the absence of a combination of certain Cf-4–specific residues in the LRRs. When considered individually, these residues contribute only slightly to Cf-4 function, and therefore their relevance was not identified by analysis of mutants 129, 35, and 332 (Figures 2E and 3B). Possibly T433, which is present at a putative solvent-exposed position in LRR 15 of Cf-4, has such a slight contribution to Cf-4 function (Figure 3B, compare curves a and e). We also showed that double mutants can be very useful to identify amino acid residues that contribute only partially to Cf-4 function. In this way, the importance of amino acid residue W389 for Cf-4 function was easily revealed by gain-of-function studies, starting from inactive double mutants.
The role of solvent-exposed amino acid residues in LRR proteins also was addressed in reports on mutant LRR proteins that carry single amino acid residue substitutions at putative solvent-exposed positions. These proteins are encoded by alleles of Cf-9 (D509N and S676L; Thomas et al., 1998), clv1-4 and clv1-8 (Clark et al., 1997), rpp8-2 (McDowell et al., 1998), rps5-1 (Warren et al., 1998), rps2-201 (Bent et al., 1994; Mindrinos et al., 1994), and rpm1-4 (Grant et al., 1995). All of these mutants appeared inactive or showed a severely reduced functionality. None of the observed substitutions was present in functional homologs. In the case of Cf-9 and RPS5, the substitutions were found in regions that are conserved among family members. Although these mutations at putative solvent-exposed positions may have a direct effect on interactions with other proteins, the phenotype also can be a result of effects of the mutation on overall protein structure or stability. Only in case of the RPS5 mutation, a role for the mutated amino acid residue can be speculated because this rps5-1 mutation also partially suppresses the function of other homologous R genes in Arabidopsis (Warren et al., 1998).
The putative solvent-exposed positions of W389, G411, and F457 in LRRs of Cf-4 suggest that these residues play an important role at the surface where Cf-4 interacts with a protein or proteins of the AVR4 perception complex. The weak effect of the W389Y and F457L substitutions possibly is due to the high similarity between the side chain of the resident and the introduced amino acid residue. Amino acid substitutions also can affect the orientation of side chains of adjacent solvent-exposed amino acid residues, as was suggested for the V118G substitution in polygalacturonase-inhibiting protein 2 (Leckie et al., 1999). This finding may explain the effect of the G411E substitution, because glycine has no side chain that can participate in interactions.
The Role of Variation in R Proteins
We have shown that of the 57 amino acid residues that differ between the LRRs of Cf-4 and Cf-9, 54 are not essential for Cf-4 specificity. This could be a general feature of LRR proteins. For example, a Cf gene mediating AVR9 recognition in Lycopersicon pimpinellifolium encodes a homolog that differs from Cf-9 by 63 amino acid residues (R.A.L. Van der Hoorn, M. Kruijt, M.H.A.J. Joosten, and P.J.G.M. De Wit, unpublished data). Again, most of the variation between Cf-9 and this functional Cf-9 homolog resides in the N-terminal half of the proteins, particularly at putative solvent-exposed positions in the β-sheets of the LRRs.
These observations raise the question of why R proteins carry so many variant amino acid residues in the LRR domain but only a few residues are required for recognitional specificity. One possibility is that the high number of variant residues at solvent-exposed positions is allowed because diversification changes only the decoration of the protein and not the overall structure. However, the reported diversifying selection on solvent-exposed residues suggests that there must be an additional advantage. We propose four distinct roles for variation in R proteins. First, variation in LRR proteins could be a result of selection in the past. This implies that the existing variation has no current function but remains as a relic from the evolution of new R gene specificities. Second, ongoing variation in LRRs of R proteins is still vital because it provides the basis for the generation of new specificities through recombination and gene conversion. Third, variation in LRRs of R proteins also might give plant populations the ability to recognize a diverse collection of nonself proteins. Finally, the versatile binding capabilities of LRR proteins also suggest that variable amino acid residues can be involved in recognition of multiple ligands, thus generating dual or multiple recognitional specificities on a single R protein.
Thus, R proteins could act as scaffolds that can easily change their decoration as a result of gene shuffling and diversifying selection on solvent-exposed amino acid residues. We have shown that most of the specific decorations found on an R protein are not necessarily involved in the determination of recognitional specificity of that protein. This finding indicates that the decoration of R proteins is not fully adapted to a certain function but rather reflects the fact that R proteins have the versatility to adapt to sense the presence of new “foreign” proteins. For plants, such a flexible recognition system for diverse pathogens is crucial for survival among adapting pathogens. The elucidation of where specificity resides in Cf proteins eventually might allow the design of custom R proteins to provide durable resistance by targeting recognitional specificity to conserved proteins of pathogens that are crucial for their pathogenicity.
METHODS
DNA Manipulations and Plasmids
All DNA manipulations were performed using standard protocols (Sambrook et al., 1989). Polymerase chain reaction (PCR) was performed with Pfu polymerase (Stratagene, La Jolla, CA) or AmpliTaq polymerase (Perkin-Elmer Applied Biosystems, Foster City, CA), according to the manufacturer's instructions. Restriction enzymes, Klenow polymerase, T4 ligase, and Escherichia coli DH5α cells were from Life Technologies (Breda, The Netherlands). Primers were synthesized by Amersham-Pharmacia (Buckinghamshire, UK). The authenticity of all cloned PCR fragments was confirmed by sequencing. The presence of correct Cf-9 and Cf-4 fragments in binary constructs was assayed using PCR on plasmid DNA isolated from Agrobacterium tumefaciens, followed by restriction analysis of the amplified fragments to reveal polymorphic sites.
The following plasmids that were used in this study have been described previously (Van der Hoorn et al., 2000): pRH46 (35S-driven Cf-4), pRH1 (35S-driven Cf-9), pRH48 (binary Cf-4 vector), pRH21 (binary Cf-9 vector), pRH87 (binary Avr4 vector), pMOG978 (binary Avr9 vector), and pMOG800 (binary vector).
Construction of Binary Plasmids
To construct a binary plasmid carrying 35S-driven Cf genes lacking the part encoding leucine-rich repeats (LRRs) 1 to 17, we removed BamHI and ClaI sites from the multiple cloning sites of pRH1, resulting in pRH17. Cf-4 and Cf-9 fragments that lacked the part encoding LRRs 1 to 17 were amplified from pRH46 and pRH1, respectively. This was done with PCR overlap extension by using primers that anneal in the 35S promoter (5′-gttcatttcatttggagagg-3′) and at the conserved HindIII site, which is present in the open reading frame at a position corresponding to LRR 21 of the encoded Cf protein (5′-catgcaacttatttgatctcaagc-3′) and overlap primers 5′-aacaatatcaataggcctgtcgtctcgtcacaatcgatgccatcc-3′ and 5′-gacgacaggcctattgatattgtt-agacttgggatccaataatttgg-3′ (the ClaI and BamHI sites, respectively, are underlined). The XbaI and EcoRI sites in the Cf-4 sequence encoding the B-domain were removed using PCR overlap extension with overlap primers 5′-gctcttcttgagttcaagaac-3′ and 5′-gttcttgaactc-aagaagagc-3′ in such a way that the encoded protein sequence remained the same. The amplified Cf fragments lacking the part encoding LRRs 1 to 17 were cloned into pRH17 by using NcoI and HindIII restriction sites, resulting in pRH26 and pRH18 for Cf-4 and Cf-9, respectively. The promoter–open reading frame–terminator cassettes of pRH26 and pRH18 were transferred subsequently to pMOG800 by using XbaI and EcoRI restriction sites, creating binary vectors pRH94 and pRH22, respectively. Mutants 353 and 352, encoding Cf-4 mutants with exchanges within the B-domain, were generated by PCR overlap extension by using overlap primers 5′-gcttctgattattgttacgac-3′ and 5′-gtcgtaacaataatcagaagc-3′.
Constructs for Mutagenesis in the LRR Domain
Mutations within the LRR domain were generated as follows. A construct encoding the LRRs of Cf-9, with the Cf-4–specific deletion of LRRs 11 and 12, was made by removing a 138-bp AvaII fragment encoding LRRs 11 and 12 from pRH1, resulting in pRH5. DNA fragments encoding LRRs 1 to 17 were amplified from pRH46, pRH1, and pRH5 by using primers that are described in Table 1. For some fragments, PCR overlap extension was used or cloned PCR products were used as a template. In a few Cf-9 constructs, the EcoRI site present in the DNA encoding LRR 15 was removed by PCR overlap extension by using overlap primers 5′-gctgaaaggtcgtattccgaatttactcctaaaccagaagaacc-3′ and 5′-ggttcttctggtttaggagtgaatttggaa-tacgacctttcagc-3′ without changing the coding sequence. The fragments were cloned subsequently into pBluescript SK− (Stratagene) by using ClaI, BamHI, EcoRI, and XhoI restriction sites. LRR-encoding fragments were cloned subsequently into binary vectors pRH22 and pRH94, encoding Cf-9 and Cf-4 that lack LRRs 1 to 17. For some cloning steps, removal of XhoI and EcoRI restriction sites from the multiple cloning sites of the vectors pRH22 and pRH94 was required.
Table 1.
Primers Used to Generate Fragments Encoding LRRsa
| Primer
|
|
Sequence (5′ to 3′)
|
Cf-4
|
Cf-9
|
|---|---|---|---|---|
| AF | (f) | ccaaaacattaagtgccgttactctaaaac | 1123 | |
| AR | (r) | gttttagagtaacggcacttaatgttttgg | 1142 | |
| BR1 | (r) | attattggatcccaagtctaacaatatc | 1314 | 1485 |
| CF1 | (f) | ggcatcgattgtgacgagacg | 215 | 245 |
| CY | (r) | acgtggatccgaattcgctcgagaggtaaagacattgtaggtt | 1188 | |
| DF1 | (f) | ataaccatcttgaaggaccaatt | 933 | 966 |
| DF2 | (f) | ggbccaattccatccaac | 947 | 1118 |
| DR1 | (r) | aattggtccttcaagatggttat | 955 | 988 |
| DR2 | (r) | gttggatggaattggwcc | 964 | 1135 |
| EC | (r) | acgtggatccgaattcgctcgagagccaaagacattctaggtt | 1188 | |
| EF | (f) | ccatcactgatagagttagacttgagc | 1049 | 1220 |
| ER | (r) | gctcaagtctaactctatycagtgatggaaggg | 1075 | 1246 |
| FY | (r) | acgtggatccgaattcgctcgagaggtaaagtaaaactaggtt | 1188 | |
| GF | (f) | ccatcactggttgggttagacttgagc | 1049 | 1220 |
| GR | (r) | gctcaagtctaacccaaccagtgatggaaggg | 1075 | 1246 |
| QF4 | (f) | acttctcgagtaacaacttgaatggg | 995 | |
| QF9 | (f) | acttctcgagtaaccacttgaatggg | 995 | 1166 |
| QR4 | (r) | ttaaggatcctctcgagagccaaagtatttgtagg | 1018 | |
| QR9 | (r) | tcaaggatcctctcgagaggtagagacattctagg | 1189 | |
| RR | (r) | gagtgaattcggaatacgaccttttagc | 1177 | |
| SF1 | (f) | gccgatcgataacatctcgtcgactattcctt | 568 | 601 |
| SR2 | (r) | ccccggatccaggaatagtcgacgagatgtt | 608 | 641 |
Forward (f) or reverse (r) primers are indicated, and restriction sites are underlined. The position of the 5′ end of the primer in Cf-4 or Cf-9 is indicated relative to the start codon of the open reading frame. Only primer positions that were used are indicated.
Agroinfiltration
Tobacco plants (Nicotiana tabacum cv Petite Havana SR1) were grown under normal greenhouse conditions. Binary plasmids were transferred to Agrobacterium strain MOG101 (Hood et al., 1993) by electroporation. Culture preparation and infiltration of leaves of 4- to 8-week-old tobacco plants were performed as described previously (Van der Hoorn et al., 2000).
For quantitative comparisons, Agrobacterium cultures that carry a plasmid that encodes a (mutant) Cf protein were mixed in different ratios with a culture of equal density carrying an AVR-encoding plasmid and infiltrated into opposite tobacco leaf halves. At 7 days after infiltration, outlines of sectors and necrotic areas were drawn on a sheet. Areas on the scanned sheet were quantified subsequently using the magnetic lasso and histogram options of Adobe Photoshop (version 5.0; Adobe Systems, Mountain View, CA). Each pair of curves represents one leaf. Differences in Cf-4 curves are due to difference in responsiveness of the infiltrated leaves. The dose–response curves that are shown are representative of at least four independent experiments.
Acknowledgments
We thank Matthieu Joosten for critically reading the manuscript; Marco Kruijt for technical assistance; Jacques Vervoort, Harrold van den Burg (Department of Biochemistry, Wageningen University), and Bas Brandwagt for valuable discussions; and Maarten de Kock (Departement of Plant Breeding, Wageningen University) for his excellent suggestions for quantifying necrotic responses. R.A.L.V.d.H. was supported by a grant from the Dutch Foundation for Earth and Life Sciences, project No. ALW 805.33-231; R.R. was supported by grants from the European Molecular Biology Organization and the European Community (ER4001GT97-2132).
References
- Anderson, P.A., Lawrence, G.J., Morrish, B.C., Ayliffe, M.A., Finnegan, E.J., and Ellis, J.G. (1997). Inactivation of the flax rust resistance gene M associated with loss of a repeated unit within the leucine-rich repeat coding region. Plant Cell 9, 641–651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bent, A., Kunkel, B.N., Dahlbeck, D., Brown, K.L., Schidt, R., Giraudat, J., Leung, J., and Staskawicz, B.J. (1994). RPS2 of Arabidopsis thaliana: A leucine-rich repeat class of plant disease resistance genes. Science 265, 1856–1860. [DOI] [PubMed] [Google Scholar]
- Bittner-Eddy, P.D., Crute, I.R., Holub, E.B., and Beynon, J.L. (2000). RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21, 177–188. [DOI] [PubMed] [Google Scholar]
- Botella, M.A., Parker, J.E., Frost, L.N., Bittner-Eddy, P.D., Beynon, J.L., Daniels, M.J., Holub, E.B., and Jones, J.D.G. (1998). Three genes of the Arabidopsis RPP1 complex resistance locus recognize distinct Peronospora parasitica avirulence determinants. Plant Cell 10, 1847–1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchanan, S.G., and Gay, N.J. (1996). Structural and functional diversity in the leucine-rich repeat family of proteins. Prog. Biophys. Mol. Biol. 65, 1–44. [DOI] [PubMed] [Google Scholar]
- Cai, D., Kleine, M., Kifle, S., Harloff, H.J., Sandal, N.N., Marcker, K.A., Klein-Lankhorst, R.M., Salentijn, E.M.J., Lange, W., Stiekema, W.J., Wyss, U., Grundler, F.M.W., Jung, C. (1997). Positional cloning of a gene for nematode resistance in sugar beet. Science 275, 832–834. [DOI] [PubMed] [Google Scholar]
- Clark, S.E., Williams, R.W., and Meyerowitz, E.M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. [DOI] [PubMed] [Google Scholar]
- De Lorenzo, G., and Cervone, F. (1997). Polygalacturonase-inhibiting proteins (PGIPs): Their role in specificity and defense against pathogenic fungi. In Plant-Microbe Interactions, Vol. 3, G. Stacey and N.T. Keen, eds (New York: Chapman & Hall), pp. 76–93.
- Dixon, M.S., Jones, D.A., Keddie, J.S., Thomas, C.M., Harrison, K., and Jones, J.D.G. (1996). The tomato Cf-2 disease resistance locus comprises two functional genes encoding leucine-rich repeat proteins. Cell 84, 451–459. [DOI] [PubMed] [Google Scholar]
- Dixon, M.S., Hatzixanthis, K., Jones, D.A., Harrison, K., and Jones, J.D.G. (1998). The tomato Cf-5 disease resistance gene and six homologs show pronounced allelic variation in leucine-rich repeat copy number. Plant Cell 10, 1915–1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant, M.R., Godiard, L., Straube, E., Ashfield, T., Lewald, J., Sattler, A., Innes, R.W., and Dangl, J.L. (1995). Structure of the Arabidopsis RPM1 gene enabling dual specificity disease resistance. Science 269, 843–846. [DOI] [PubMed] [Google Scholar]
- Hammond-Kosack, K.E., and Jones, J.D.G. (1996). Resistance gene dependent plant defense responses. Plant Cell 8, 1773–1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood, E.E., Gelvin, S.B., Melchers, L.S., and Hoekema, A. (1993). New Agrobacterium helper plasmids for gene transfer to plants. Transgenic Res. 2, 208–218. [Google Scholar]
- Jones, D.A., Thomas, C.M., Hammond-Kosack, K.E., Balint-Kurti, P.J., and Jones, J.D.G. (1994). Isolation of the tomato Cf-9 gene for resistance to Cladosporium fulvum by transposon tagging. Science 266, 789–793. [DOI] [PubMed] [Google Scholar]
- Joosten, M.H.A.J., and De Wit, P.J.G.M. (1999). The tomato-Cladosporium fulvum interaction: A versatile experimental system to study plant-pathogen interactions. Annu. Rev. Phytopathol. 37, 335–367. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1995). Proteins with leucine-rich repeats. Curr. Opin. Struct. Biol. 5, 409–416. [DOI] [PubMed] [Google Scholar]
- Kobe, B., and Deisenhofer, J. (1996). Mechanism of ribonuclease inhibition by ribonuclease inhibitor protein based on the crystal structure of its complex with ribonuclease A. J. Mol. Biol. 264, 1028–1043. [DOI] [PubMed] [Google Scholar]
- Kooman-Gersmann, M., Honée, G., Bonnema, G., and De Wit, P.J.G.M. (1996). A high-affinity binding site for the AVR9 peptide elicitor of Cladosporium fulvum is present on plasma membranes of tomato and other solanaceous plants. Plant Cell 8, 929–938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kooman-Gersmann, M., Vogelsang, R., Vossen, P., Van den Hooven, H.W., Mahe, E., Honée, G., and De Wit, P.J.G.M. (1998). Correlation between binding affinity and necrosis-inducing activity of mutant AVR9 peptide elicitors. Plant Physiol. 117, 609–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leckie, F., Mattei, B., Capodicasa, C., Hemmings, A., Nuss, L., Aracri, B., De Lorenzo, G., and Cervone, F. (1999). The specificity of polygalacturonase-inhibiting protein (PGIP): A single amino acid substitution in the solvent-exposed β-strand/β-turn region of the leucine-rich repeats (LRRs) confers new recognition capability. EMBO J. 18, 2352–2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., and Chory, J. (1997). A putative leucine-rich repeat receptor kinase involved in brassinosteriod signal transduction. Cell 90, 929–938. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G.M., Goff, S., Holub, E.B., and Dangl, J.L. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michalmore, R.W. (1998). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 11, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mindrinos, M., Katagiri, F., Yu, G.L., and Ausubel, F.M. (1994). The A. thaliana disease resistance gene RPS2 encodes a protein containing a nucleotide-binding site and leucine-rich repeats. Cell 78, 1089–1099. [DOI] [PubMed] [Google Scholar]
- Noël, L., Moores, T.L., Van der Biezen, E.A., Parniske, M., Daniels, M.J., Parker, J.E., and Jones, J.D.G. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Papageorgiou, A.C., Shapiro, R., and Acharya, K.R. (1997). Molecular recognition of human angiogenin by placental ribonuclease inhibitor: An X-ray crystallographic study at 2.0 Å resolution. EMBO J. 16, 5162–5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker, J.E., Coleman, M.J., Szabò, V., Frost, L.N., Schmidt, R., Van der Biezen, E.A., Moores, T., Dean, C., Daniels, M.J., and Jones, J.D.G. (1997). The Arabidopsis downy mildew resistance gene RPP5 shares similarity to the Toll and interleukin-1 receptors with N and L6. Plant Cell 9, 879–894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., and Jones, J.D.G. (1999). Recombination between divergent clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA 96, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B.H., and Jones, J.D.G. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf4/9 locus of tomato. Cell 91, 821–832. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T.T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Song, W.Y., Wang, G.L., Chen, L., Kim, H.S., Pi, L.Y., Gardner, J., Wang, B., Holsten, T., Zhai, W.X., Zhu, L.H., Fauquet, C., and Ronald, P.C. (1995). A receptor kinase-like protein encoded by the rice disease resistance gene Xa21. Science 270, 1804–1806. [DOI] [PubMed] [Google Scholar]
- Song, W.Y., Pi, L.Y., Wang, G.L., Gardner, J., Holsten, T., and Ronald, P.C. (1997). Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9, 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takken, F.L.W., Thomas, C.M., Joosten, M.H.A.J., Golstein, C., Westerink, N., Hille, J., Nijkamp, H.J.J., De Wit, P.J.G.M., and Jones, J.D.G. (1999). A second gene at the tomato Cf-4 locus confers resistance to Cladosporium fulvum through recognition of a novel avirulence determinant. Plant J. 20, 279–288. [DOI] [PubMed] [Google Scholar]
- Thomas, C.M., Jones, D.A., Parniske, M., Harrison, K., Balint-Kurti, P.J., Hatzixanthis, K., and Jones, J.D.G. (1997). Characterization of the tomato Cf-4 gene for resistance to Cladosporium fulvum identifies sequences that determine recognitional specificity in Cf-4 and Cf-9. Plant Cell 9, 2209–2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas, C.M., Dixon, M.S., Parniske, M., Golstein, C., and Jones, J.D.G. (1998). Genetic and molecular analysis of tomato Cf genes for resistance to Cladosporium fulvum. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 353, 1413–1424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torii, K.U., Mitsukawa, N., Oosumi, T., Matsuura, Y., Yokoyama, R., Whittier, R.F., and Komeda, Y. (1996). The Arabidopsis ERECTA gene encodes a putative receptor protein kinase with extracellular leucine-rich repeats. Plant Cell 8, 735–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Biezen, E.A., and Jones, J.D.G. (1998). Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem. Sci. 23, 454–456. [DOI] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., Laurent, F., Roth, R., and De Wit, P.J.G.M. (2000). Agroinfiltration is a versatile tool that facilitates comparative analysis of Avr9/Cf-9-induced and Avr4/Cf-4-induced necrosis. Mol. Plant-Microbe Interact. 13, 439–446. [DOI] [PubMed] [Google Scholar]
- Warren, R.F., Henk, A., Mowery, P., Holub, E., and Innes, R.W. (1998). A mutation within the leucine-rich repeat domain of the Arabidopsis disease resistance gene RPS5 partially suppresses multiple bacterial and downy mildew resistance genes. Plant Cell 10, 1439–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]