Abstract
Pancreatic ductal adenocarcinoma (PDAC) is one of the main aggressive types of cancer, characterized by late prognosis and drug resistance. Among the main factors sustaining PDAC progression, the alteration of cell metabolism has emerged to have a key role in PDAC cell proliferation, invasion, and resistance to standard chemotherapeutic agents. Taking into account all these factors and the urgency in evaluating novel options to treat PDAC, in the present work we reported the synthesis of a new series of indolyl-7-azaindolyl triazine compounds inspired by marine bis-indolyl alkaloids. We first assessed the ability of the new triazine compounds to inhibit the enzymatic activity of pyruvate dehydrogenase kinases (PDKs). The results showed that most of derivatives totally inhibit PDK1 and PDK4. Molecular docking analysis was executed to predict the possible binding mode of these derivatives using ligand-based homology modeling technique. Evaluation of the capability of new triazines to inhibit the cell growth in 2D and 3D KRAS-wild-type (BxPC-3) and KRAS-mutant (PSN-1) PDAC cell line, was carried out. The results showed the capacity of the new derivatives to reduce cell growth with a major selectivity against KRAS-mutant PDAC PSN-1 on both cell models. These data demonstrated that the new triazine derivatives target PDK1 enzymatic activity and exhibit cytotoxic effects on 2D and 3D PDAC cell models, thus encouraging further structure manipulation for analogs development against PDAC.
Keywords: pancreatic ductal adenocarcinoma (PDAC), nortopsentin analogues, antitumor activity, pyruvate dehydrogenase kinases (PDKs), cytotoxic activity, metabolic alterations, ligand-based homology modeling, KRAS
1. Introduction
Cancer cells rewire many metabolic pathways to sustain their survival, growth, invasiveness and resistance to cancer treatments [1].
Pyruvate dehydrogenase complex (PDC) has emerged as a key enzyme in the regulation of metabolic pathway of glucose. In normal human cells, PDC catalyzes the decarboxylation of the pyruvate in acetyl CoA and carbon dioxide—a process often deregulated in cancer cells, in which aerobic glycolysis is used to produce ATP instead of oxidative phosphorylation (OXPHOS), even in presence of adequate oxygen supply [2]. This phenomenon is well known as the “Warburg effect” and provides some tangible advantages to cancer survival, such as a high rate of ATP production, low immunity microenvironment and enhanced cancer cell invasion [3].
The pyruvate dehydrogenase kinases (PDKs) have been identified as the leading players of this metabolic swing since they are involved in PDC inactivation [4,5].
Four PDK isoforms were isolated in humans (PDK1, PDK2, PDK3, PDK4) [6] and their overexpression was frequently associated with the KRAS-mutated cancer types, such as pancreatic ductal adenocarcinoma (PDAC) and colorectal cancer [7,8].
PDAC adopts a distinct metabolic process to meet the energetic requests of growing cancer cells [9] which is clearly associated with chemo-, radio- and immunotherapy resistance [10,11,12].
Therefore, there is an urgent need to develop novel therapeutic strategies to treat PDAC, and the enzymes involved in metabolic alterations, such as PDKs, can be considered valuable targets to develop efficacious anticancer strategies [13,14].
Considering this aim and that approximately 80% of the approved chemotherapeutic drugs are based on bioactive natural products [15,16], the marine microenvironment has been exploited as a huge resource of novel-drug-lead compounds.
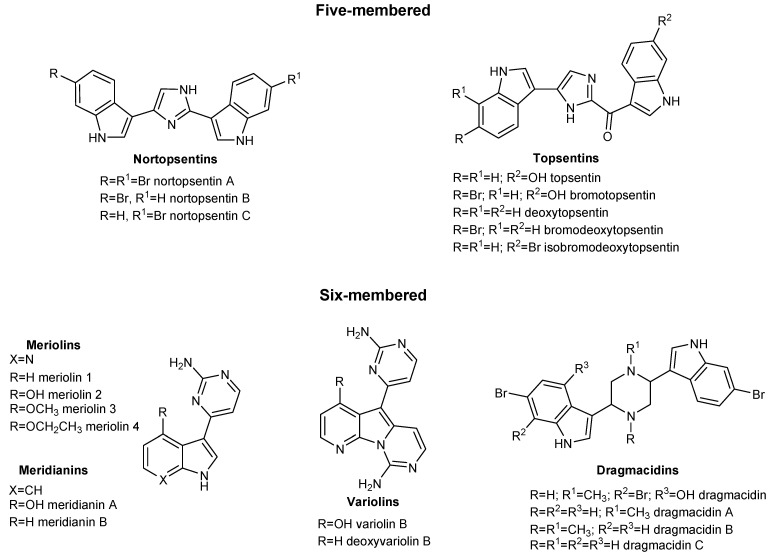
Many marine alkaloids, including nortopsentin, topsentin [17,18], meridianins, variolins, meriolins and dragmacidins [19,20], reported in Figure 1, have been described for their antiproliferative activity often due to their ability in inhibiting different kinases.
Figure 1.
Five- and six-membered natural compounds isolated from marine sponges.
We have previously synthetized a library of marine analogs, replacing the central imidazole ring of nortopsentin and topsentin with several five-membered heterocycles or by manipulation of their indolyl portion [21,22,23,24,25,26].
Many of these derivatives have been proven to be extremely potent against a wide range of pancreatic cancer cells, hampering the activity of different protein kinases heavily involved in cancer progression [27,28].
Specifically, the series of the indolyl thiazolyl 7-azaindole nortopsentin analogs showed a good level of antiproliferative activity in the micro-submicromolar range and cyclin-dependent kinase 1 (CDK1) inhibition with IC50 values of 0.64–0.89 µM [28]. Similarly, structural manipulations of topsentin, replacing the imidazole central core with 1,2,4-oxadiazole moiety led to a series of derivatives which showed good antiproliferative activity in pancreatic cancer cell lines, associated with glycogen synthase kinase 3 β (GSK3-β) or CDK1 inhibition [27,28].
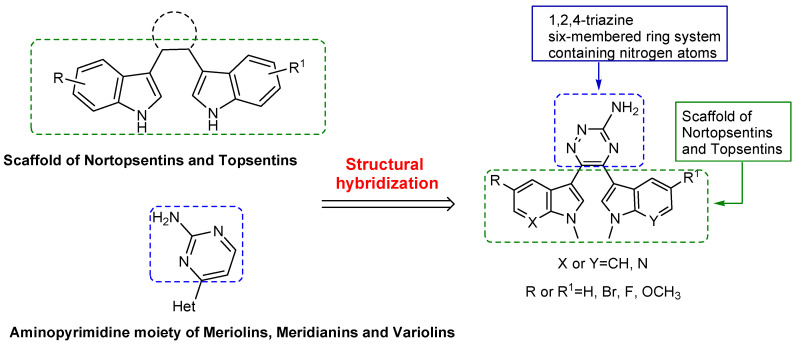
In order to identify novel agents for pancreatic cancer treatment targeting protein kinases involved in the cancer metabolism and still unexplored, we mainly focus our attention on reviewing the progress made in the development of PDKs inhibitors. Since for the latter, including dichloroacetate (DCA), AZD7545, radicicol and M77976, no specific chemical features responsible for the PDKs inhibition were recognized [4], we chose to adopt a molecular hybridization approach that combines different pharmacophoric sub-units in a single structure, to design the novel PDK inhibitors. Among the molecules isolated from natural sources, compounds bearing a six-membered ring system, such as the aminopyrimidine, showed potent kinases inhibition [19,20]. We decided to investigate the effect of the substitution of the aminopyrimidine scaffold with a 3-amino-1,2,4-triazine central core, which was widely described for its antitumor activity [29,30,31,32,33]. Meanwhile, the bioactive bis-indolyl scaffold of nortopsentin and topsentin was maintained, resulting in the novel 1,2,4-triazine-based marine analogs reported in Figure 2.
Figure 2.
Drug design of azaindole triazine marine analogs using a structural hybridization approach.
The antiproliferative activity of the novel triazine compounds was evaluated in pancreatic cancer cell lines, demonstrating their ability to hamper cancer cell growth. Moreover, in order to elucidate the mechanism of action and the ability of the new compounds to revert altered pancreatic cancer cells metabolism, a specific enzymatic assay against the four isoforms of PDKs was carried out. The results showed a selective inhibition of PDK1 and PDK4 isoforms, which was further confirmed by molecular docking analysis.
2. Results and Discussion
2.1. Synthesis
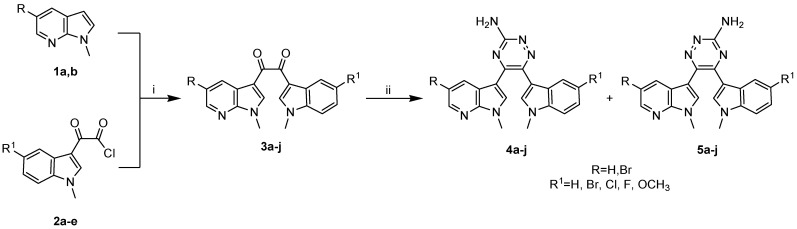
The synthesis of 1,2,4-triazines 4–5 was accomplished as depicted in Scheme 1. The key intermediate diketones 3a–j were synthesized by Friedel–Crafts acylation of methylindole precursor of type 1a,b with indolyl-oxo-acetyl chlorides 2a–e, synthesized as previously described [28,34,35]. This reaction, performed in a mixture of DCM/heptane and in the presence of AlCl3 as a strong Lewis acid catalyst, was carried out via electrophilic aromatic substitution to form the 1,2- diones 3a–j in good yields (76–92%).
Scheme 1.
Synthesis of azaindole triazine derivatives 4a–j, 5a–j. Reagents and conditions: (i) AlCl3, DCM/Heptane 2:1, RT, 2 h, 76–92%; (ii) aminoguanidine bicarbonate, n-butanol, reflux, 3 h, overall yields: 79–96%, 4a–j: 40–49%, 5a–j: 37–48%.
The subsequent reaction of α-diketones 3a–j with aminoguanidine bicarbonate produced new specimens of 3-amino-substituted 1,2,4-triazines 4a–j, together with the corresponding structural isomers 5a–j. All the synthesized derivatives 4a–j and 5a–j and their relative yields are summarized in Table 1.
Table 1.
3-Amino-1,2,4-triazine derivatives 4a–j, 5a–j and their relative yields.
| Cpd | R | R1 | Overall Yield% | 4 (%) 5 (%) |
Cpd | R | R1 | Overall Yield% | 4 (%) 5 (%) |
|---|---|---|---|---|---|---|---|---|---|
| 4a | H | H | 80 | 42 | 4f | Br | Cl | 79 | 40 |
| 5a | 38 | 5f | 39 | ||||||
| 4b | Br | H | 86 | 44 | 4g | H | F | 81 | 41 |
| 5b | 42 | 5g | 40 | ||||||
| 4c | H | Br | 94 | 49 | 4h | Br | F | 85 | 43 |
| 5c | 45 | 5h | 42 | ||||||
| 4d | Br | Br | 92 | 48 | 4i | H | OCH3 | 84 | 47 |
| 5d | 44 | 5i | 37 | ||||||
| 4e | H | Cl | 87 | 46 | 4j | Br | OCH3 | 96 | 48 |
| 5e | 41 | 5j | 48 |
2.2. Enzyme Inhibition Activity
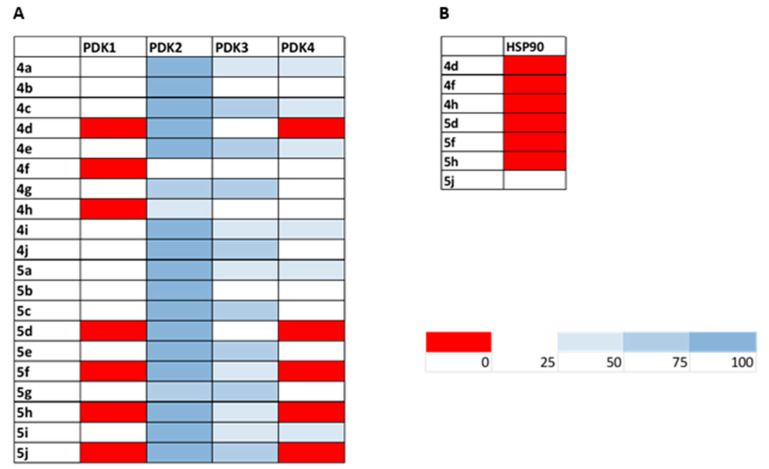
The ability of the tested compounds to hamper the activity of PDK1-4 isoforms was firstly evaluated by means of a chemioluminescent assay, the ADPGlo™ Kinase Assay kit (Promega Corporation, Madison, WI, USA). The results obtained by incubating PDK1-4 isoforms for 30 min at room temperature with 1.5 µM of tested compounds are depicted in Figure 3 panel A as a heat-map summary of the inhibition percentage on PDK1-4 enzymatic activity. All the newly synthesized compounds were able to inhibit the pyruvate dehydrogenase kinases, although to a different extent and showing a preferential activity towards the PDK1 and PDK4 isoforms. In particular, compounds 4d, 4f, 4h, 5d, 5f, 5h and 5j totally hampered the enzymatic activity of PDK1 at 1.5 µM. Derivatives 4d, 5d, 5f, 5h and 5j were also able to completely suppress the PDK4 activity. Conversely, all tested 1,2,4-triazin-3-amines were barely effective in blocking PDK2 activity at 1.5 µM concentration. In addition, IC50 values on the PDK1 isoform were also calculated for all synthesized derivatives and the results are shown in Table 2. All compounds were extremely efficient in decreasing PDK1 catalytic activity at sub-micromolar/nanomolar concentrations, with IC50 values ranging from 0.04 to 0.33 µM. Noticeably, all 1,2,4-triazin-3-amines prominently outperformed the well-known PDK reference inhibitors, DAP and DCA.
Figure 3.
Inhibition of isolated enzyme isoforms. Heat-map showing the relative inhibition of tested compounds (at 1.5 µM) on PDK1-4 (panel A) and HSP90 (panel B) enzymes. Results are expressed as enzyme residual activity (range 0–100%). Data are the means of three independent experiments.
Table 2.
Inhibition of isolated PDK1 isoform or HSP90. IC50 values were calculated by a four-parameter analyzed logistic model (p < 0.05). DCA and DAP were reported as reference compounds. Data represent mean values from 3 independent experiments. S.D. = standard deviation.
| Cpd | IC50 (µM) ± S.D. PDK1 | IC50 (µM) ± S.D. HSP90 | Cpd | IC50 (µM) ± S.D. PDK1 | IC50 (µM) ± S.D. HSP90 |
|---|---|---|---|---|---|
| 4a | 0.30 ± 0.10 | - | 5a | 0.32 ± 0.10 | - |
| 4b | 0.33 ± 0.05 | - | 5b | 0.21 ± 0.07 | - |
| 4c | 0.26 ± 0.07 | - | 5c | 0.27 ± 0.08 | - |
| 4d | 0.04 ± 0.02 | 0.06 ± 0.02 | 5d | 0.05 ± 0.02 | 0.07 ± 0.01 |
| 4e | 0.29 ± 0.06 | - | 5e | 0.32 ± 0.10 | - |
| 4f | 0.12 ± 0.04 | 0.10 ± 0.03 | 5f | 0.08 ± 0.02 | 0.11 ± 0.04 |
| 4g | 0.19 ± 0.04 | - | 5g | 0.29 ± 0.10 | - |
| 4h | 0.06 ± 0.02 | - | 5h | 0.05 ± 0.03 | 0.09 ± 0.03 |
| 4i | 0.33 ± 0.10 | - | 5i | 0.31 ± 0.07 | - |
| 4j | 0.18 ± 0.06 | - | 5j | 0.04 ± 0.02 | 0.18 ± 0.04 |
| DCA | 15,890 ± 256 | 29,740 ± 558 | DAP | 212 ± 48 | 698 ± 83 |
Since the ATP binding pockets in PDKs and HSP90 are similar and taking into consideration that these 1,2,4-triazine derivatives conserved a chemical scaffold resembling that of the well-known Hsp90 inhibitor Ganetespib, the most performant PDK inhibitors 4d, 4f, 4h, 5d, 5f, 5h and 5j were also screened for their ability to target HSP90. As reported in Figure 3 panel B, derivatives 4d, 4f, 4h, 5d, 5f, and 5h were highly effective against HSP90, being able to completely abolish the enzyme activity at 1.5 µM. Conversely, derivative 5j was able to decrease HSP90 catalytic activity by only 60–80% at equi-doses, thus attesting to its ability to act as a more selective PDK1 inhibitor. While derivatives 4d, 4f, 4h, 5d, 5f, and 5h retained a similar inhibitory profile against HSP90, compound 5j showed an IC50 value about two times higher towards isolated HSP90 compared with PDK-1 (Table 2).
2.3. Molecular Modeling Studies
The most appropriate protein–ligand complex of both HSP90 and PDK1 was selected on the basis of their crystallographic resolution, paying particular attention to the structures in which the co-crystallized ligand showed the highest structural similarity to compounds 4 and 5. The crystal structures with PDB codes 2Q8F (method: X-ray diffraction, resolution: 2.03 Å) [36] and 5J64 (method: X-ray diffraction, resolution: 1.38 Å) [37] were chosen for PDK1 and HSP90, respectively. The PDB files were prepared for the computational analysis using MOE suite; first, the small missing loops in the structures were rebuilt trough the Structure Preparation program, then the proper protonation state was conferred employing Protonate 3D application, and finally, the added hydrogen atoms were minimized with the AMBER10:EHT [38] force field implemented in MOE.
The ligands, as well as the protein structures, underwent a proper preparation procedure, exploiting the tools of the QUACPAC package of the OpenEye suite. First, the tautomers tool was used for selecting the dominant tautomeric state for each compound, then the three-dimensional conformations for each molecule were generated with the OMEGA program, and the MolCharge tool was used to assign the proper partial charges (with the AM1BCC [39] method). Finally, the FixpKa application was exploited to determine the dominant protonation state for each ligand at pH 7.4.
For both HSP90 and PDK1, the ligands were docked in the orthosteric ATP-binding site with the program PLANTS which is based on an Ant-Colony-Optimization algorithm, which was developed by the University of Tübingen [40,41,42]. For each compound, 50 poses were generated. All the final conformations presenting steric clashes with the receptor were discarded, as well as the ones showing unfavorable electrostatic interactions within the binding pocket. In the case of HSP90, the remaining poses were filtered by passing them through a three-dimensional pharmacophoric model created by consensus on the different orthosteric HSP90 crystallographic ligand binding modes. For each of the remaining compounds, the final poses were inspected and the best one was selected.
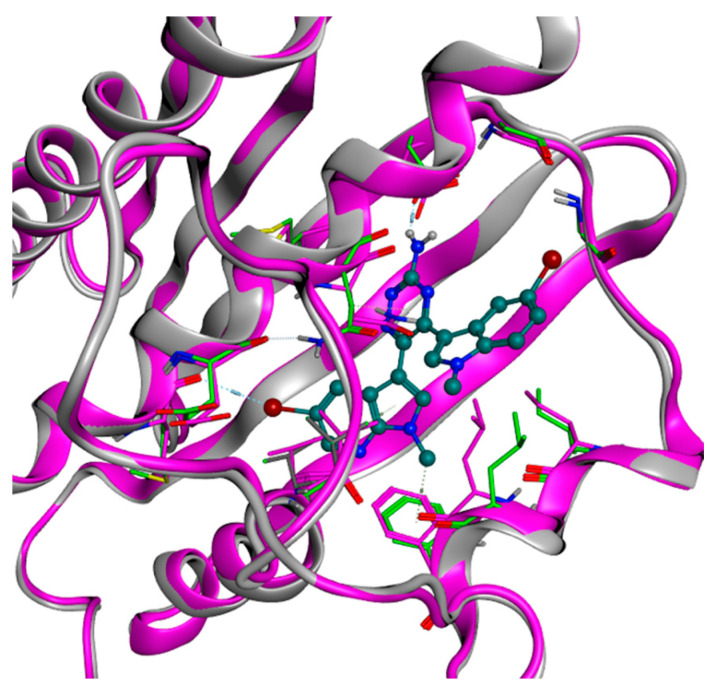
In the case of PDK1, molecular docking was not able to generate reasonable binding modes for the tested molecules, which displayed important clashes with the binding pocket. Knowing that the binding of these compounds is experimentally validated, we employed a technique called ligand-based homology modeling [43]. This method consists of the execution of a homology modeling operation, but with a defined ligand placed in a desired location (the ATP-binding site, in our case). The sequence to model is the same as the receptor of interest, indeed the main focus of the technique is to “reorganize” the amino acid side chain around the ligand, allowing it to get a reasonable placement in the pocket. All the ligand-based homology modeling passages were executed in MOE. The ligand chosen as the reference for the modeling was compound 5d, one of the most potent derivatives of the series examined with respect to PDK1. Specifically, this compound was manually placed in the binding site, following our structure-activity relationship (SAR) hypothesis, which was also already reported in a previous study [43]. In this model, the triazine binds the residue Asp318, while one of the two bicyclic aromatic moieties is placed in the inner part of the pocket, leaving the other bicycle more exposed to the solvent. Figure 4 shows the clean superimposition of the protein backbones of the crystal structure and the model of PDK1, in which the side chains have moved to create the proper room for the ligand placement.
Figure 4.
Representation of the superimposition between the original structure of PDK1 (PDB code: 2Q8F, colored in magenta) and our ligand-based homology model (depicted in grey), created on the manually placed conformation of compound 5d (pine green in the figure). As can be seen, the side chains of the amino acids in the binding pocket have adapted to the presence of the ligand, while the backbone structure did not show significant changes (RMSD between the crystal and the model backbones: 0.87 Å). The shifted side chains of interest in the homology model have been highlighted in lime color. The image was created and rendered with MOE.
The molecular docking calculation was executed again on PDK1 using the model created instead of the crystallographic structure. The results were much more promising, with many clash-free conformations which also displayed a proficient electrostatic interaction with the binding site. It is important to highlight that the created poses for the ligands resemble the three-dimensional placement of compound 5d in the pocket, giving credit to our 3D SAR hypothesis (see Figure 5 as an example). Indeed, even if the best docking pose displays a 180 degrees rotation of the ligand through the longitudinal axis, the interaction pattern with the protein is retained. Lacking several PDK1-ligand complexes in public databases, the best conformation for each ligand was selected based on the main literature information about the interaction with this target.
Figure 5.
Image representing the best pose obtained for 5d (highlighted in gold) and the originally manually placed conformation (colored in pine green), upon which the creation of the ligand-based homology model was run. As can be seen, the molecule orientation in the pocket is shifted of 180 degrees, but the interactive pattern is maintained. The image was created and rendered with MOE.
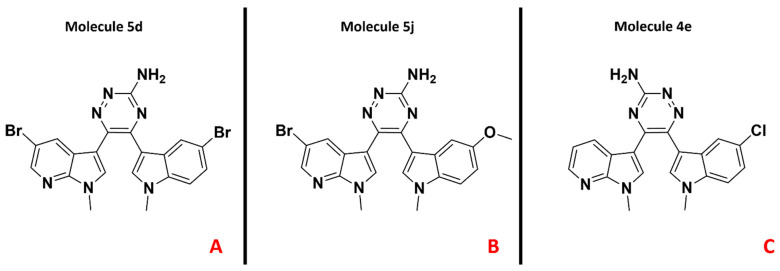
In order to confirm the experimental data presented in this study from a molecular modeling perspective, we decided to focus our analysis on three specific compounds. The first, compound 5d, shows a very high-potency profile on both PDK1 (IC50 = 0.05 µM) and HSP90 (IC50 = 0.07 µM). The second, compound 5j, has shown a significant inhibitory activity on PDK1 (IC50 = 0.04 µM) and a slightly lower potency against HSP90 (IC50 = 0.18 µM on HSP90), while the third, derivative 4e, displays an even lower activity on PDK1 (IC50 = 0.29 µM), while it was not tested on HSP90. The selected ligands are represented in Figure 6, while their binding modes with the targets under examination are depicted in Figure 7 and Figure 8.
Figure 6.
Representation of the chemical structures of the molecules taken as reference for the present study. Compound 5d (PDK1 IC50= 0.05 µM, HSP90 IC50= 0.07 µM), 5j (PDK1 IC50= 0.04 µM, HSP90 IC50= 0.18 µM), and 4e (PDK1 IC50= 0.29 µM, not tested on HSP90) are depicted in (panel A), (panel B), and (panel C), respectively.
Figure 7.
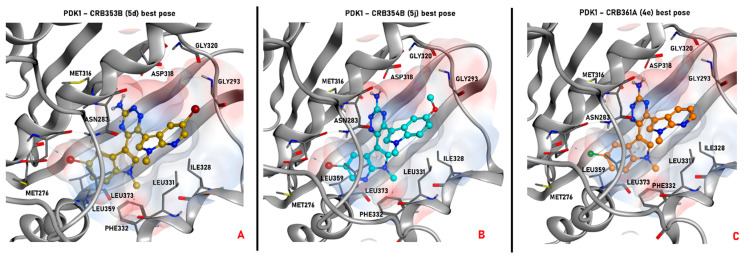
Representation of the selected pose produced by molecular docking with the program PLANTS for the compound 5d (colored in gold, panel A), 5j (depicted in cyan, panel B), and 4e (colored in orange, panel C) in the ATP-binding site of the ligand-based homology model created for PDK1. All the selected conformations passed the steric and electrostatic filtering processes. In each panel, also the electrostatic surface around the ligand in the binding site is represented. The images were created and rendered with MOE.
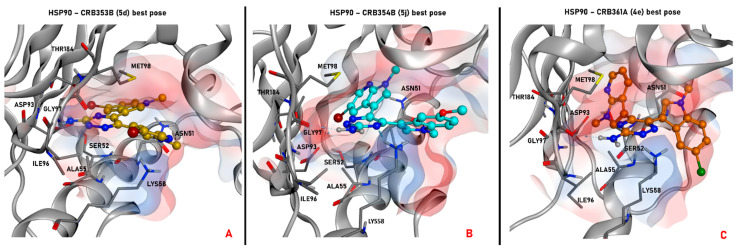
Figure 8.
Representation of the selected pose produced by molecular docking with the program PLANTS for the compound 5d (colored in gold, panel A), 5j (depicted in cyan, panel B), and 4e (colored in orange, panel C) in the ATP-binding site of HSP90. All the conformations passed the steric, electrostatic, and pharmacophoric filtering procedures. In each panel, also the electrostatic surface around the ligand in the binding site is represented. The images were created and rendered with MOE.
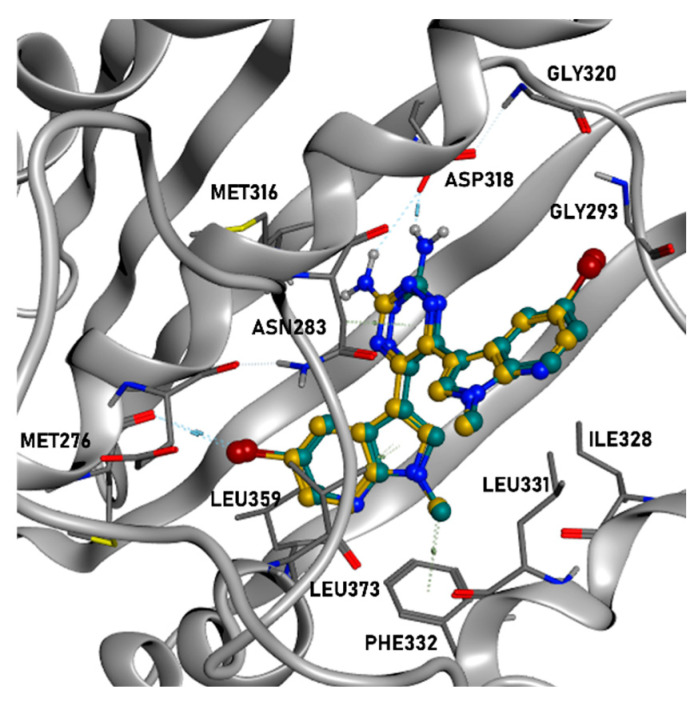
As depicted, all compounds interact through a hydrogen bond with Asp318 in the PDK1 ATP-binding site using their aromatic amine moiety. Moreover, all the molecules place the halogenated bicyclic portion in the inner part of the binding site, establishing a halogen bond with Met276. No significant differences between the binding poses for azaindole or indole insertion in the enzymatic pocket (both proficiently occy the volume of the region) were observed, as the only feature which seems to be relevant is the presence of a halogen atom (bromine or chlorine) for the stabilization of the cited molecular contact. Additionally, the methyl group of the indole (in the case of 5j) and azaindole (for 5d and 4e) establishes a hydrophobic contact with the phenyl side chain of Phe332.
Focusing on the difference between the binding modes of 5d and 5j, it seems that the pocket occupancy and the halogen bond with Met276 play a more important role in ligand stabilization in respect to the hydrogen bond with Asp318. Moreover, the bromine seems to guide such interaction in a more proficient way in respect to chlorine, and this can be appreciated considering that 4e retains the same interaction pattern of the other two examined ligands, but showing a 6-fold decrease in potency. Nevertheless, looking at the differences in potencies between the regioisomers of series 4 and 5, most probably the hydrogen bond with Asp318 “guides” the binding event, favoring the placement of the ligand with an halogen directed towards Met276, proficiently occupying the pocket. This can be hypothesized taking into account that regioisomers with the same substitions in R and R1 (e.g, 4a vs. 5a, 4d vs. 5d) do not show noticeable differences in potencies, while there are some cases in which, when R and R1 are different (e.g., 4b vs. 5b, 4g vs. 5g, 4j vs. 5j), there is the tendency for a shift in favor to one of the two isomers. The case of 4j vs. 5j is representative because, being the compounds characterized by the bulky and rotatable methoxy substituent, it could be important for a binding perspective to place this moiety in the outer part of the pocket, and this event could be “guided” more proficiently by H-bonding with Asp318 for 5j in respect to 4j.
A slightly different situation is retrievable with HSP90. Indeed, while the binding modes of compounds 5j and 4e are very similar to each other, with the aromatic amine in contact with Gly97 and the azaindole in the inner portion of the pocket, the conformational bound form for 5d is much more “flat”, with the ligand core directed to the left part of the ATP-binding site (the hydrogen bond is established with Asp93).
Differently from what was observed for the previously described series of analogues [44,45], the potencies of compounds 4 and 5 are similar for both PDK1 and HSP90.
Most probably, the key differences in the activity of these molecules are due to binding or unbinding kinetics rather than their interaction pattern in the pocket. Indeed, it is very likely that some dynamic processes affect the way in which the compounds approach the binding site, allowing some molecules to enter and place in the pocket more easily than others. This kind of behavior can be analyzed with molecular dynamics (MD) simulations, and more specifically with methodologies able to sample the protein–ligand recognition pathway. One of the techniques which have shown success in this field is Supervised Molecular Dynamics (SuMD) [46], an unbiased enhanced sampling approach able to proficiently describe the recognition event between a ligand and a biological target in the nanosecond timescale. This method has been already applied to several targets and scenarios, giving very promising results [47,48,49,50]
On the other side, if the main discriminant in the potency discrepancy is related to the unbinding process, some other MD methodologies can be applied. This is the case for some enhanced sampling methods, such as steered MD [51] or Thermal Titration Molecular Dynamics (TTMD) [52,53]. Specifically, this last technique has been recently developed, and its main advantage is that it is not based on the introduction of energetical bias in the system. Indeed, the system temperature is the only parameter that is changed in the simulation in order to allow for more proficient discrimination of the ligands based on their residence time, which is progressively increased with simulation time, in the binding pocket.
Currently, a computational work combining SuMD and TTMD is taking place in our laboratory, with the specific goal to rationalize the potency discrepancies in this and other series of ligands, with a specific focus on PDK1 and HSP90. Indeed, the very high similarities of these proteins, in particular in their ATP-binding site [44], suggest that the binding/unbinding events are not extremely important for understanding potency discrepancies within the series of ligands considered. We are confident that the combination of SuMD and TTMD will help to elucidate this very impactful aspect, also allowing to guide further efforts in drug design of new PDK inhibitors.
2.4. 2D and 3D Cytotoxicity Studies
All newly developed 1,2,4-amino-triazine compounds were screened for their cytotoxic activity against two human PDAC cell lines, namely, a KRAS-wild-type PDAC cell line (BxPC-3 cells) and a KRAS-mutant PDAC cell line (PSN-1 cells). For comparison purposes, the efficacy of the PDAC gold-standard chemotherapeutic gemcitabine, as well as of the well-known PDK inhibitors DCA and DAP were assessed under the same experimental conditions. The cytotoxicity parameters, expressed in terms of IC50 and obtained after 72 h of drug exposure by MTT assay, are reported in Table 3.
Table 3.
Antiproliferative activity of tested compounds against PSN-1 and BxPC3 cancer cells evaluated in 2D and 3D cytotoxicity assay.
| Cpd | IC50 (µM) ± S.D. BxPC-3 a |
IC50 (µM) ± S.D. PSN-1 a |
IC50 (µM) ± S.D. BxPC-3 b | IC50 (µM) ± S.D. PSN-1 b |
|---|---|---|---|---|
| 4a | 15.7 ± 2.2 | 11.3 ± 2.0 | 141.3 ± 0.1 | 97.5 ± 2.8 |
| 4b | 23.5 ± 3.2 | 19.5 ± 2.9 | 223.7 ± 14.8 | 188.5 ± 13.8 |
| 4c | 9.5 ± 1.7 | 7.8 ± 1.5 | 76.8 ± 5.1 | 55.8 ± 3.5 |
| 4d | 30.2 ± 6.2 | 23.4 ± 3.7 | 175.5 ± 15.7 | 102.8 ± 9.1 |
| 4e | 32.3 ± 8.2 | 26.5 ± 4.3 | 159.5 ± 11.4 | 59.6 ± 4.5 |
| 4f | 18.4 ± 1.4 | 11.3 ± 1.8 | 168.9 ± 12.4 | 162.2 ± 6.8 |
| 4g | 6.5 ± 1.1 | 4.5 ± 1.3 | 71.5 ± 5.2 | 65.5 ± 6.8 |
| 4h | 22.3 ± 2.4 | 17.5 ± 2.8 | 187.5 ± 14.4 | 77.8 ± 3.8 |
| 4i | 31.2 ± 5.5 | 26.5 ± 4.0 | 189.4 ± 10.9 | 139.5 ± 7.5 |
| 4j | 30.5 ± 3.4 | 27.8 ± 3.8 | 159.7 ± 10.5 | 102.8 ± 8.7 |
| 5a | 27.2 ± 3.7 | 22.2 ± 2.7 | 169.5 ± 9.4 | 78.8 ± 8.5 |
| 5b | 9.4 ± 1.3 | 5.2 ± 0.9 | 68.5 ± 2.5 | 73.6 ± 6.8 |
| 5c | 13.7 ± 2.9 | 9.2 ± 1.9 | 156.5 ± 11.7 | 78.4 ± 5.8 |
| 5d | 26.7 ± 6.1 | 21.3 ± 3.5 | 198.5 ± 14.2 | 215.6 ± 13.6 |
| 5e | 16.5 ± 3.8 | 14.2 ± 3.1 | 199.7 ± 16.3 | 197.5 ± 12.4 |
| 5f | 22.4 ± 3.2 | 21.3 ± 4.4 | 229.4 ± 17.2 | 212.8 ± 14.4 |
| 5g | 19.8 ± 3.2 | 15.2 ± 4.6 | 79.7 ± 11.4 | 129.5 ± 11.8 |
| 5h | 0.05 ± 0.03 | 27.4 ± 5.1 | 208.3 ± 12.7 | 132.5 ± 5.9 |
| 5i | 0.31 ± 0.07 | 14.9 ± 3.2 | 153.4 ± 10.3 | 112.5 ± 9.1 |
| 5j | 0.04 ± 0.02 | 0.1 ± 0.03 | 79.5 ± 5.4 | 89.7 ± 5.7 |
| DCA | >1000 | >1000 | 36200 ± 125 | 29800 ± 357 |
| DAP | 16.5 ± 0.9 | 10.20 ± 1.6 | 78.2 ± 6.2 | 87.4 ± 8.9 |
| Gemcitabine | 0.020 ± 0.003 | 0.10 ± 0.04 | 159.5 ± 5.6 | 102.6 ± 6.3 |
a Cells (5 × 103·well) were treated for 72 h with increasing concentrations of tested compounds. Cytotoxicity was assessed by MTT test. b Spheroids (2.5 × 103 cells/well) were treated for 72 h with increasing concentrations of tested compounds. The growth-inhibitory effect was evaluated by means of the acid phosphatase (APH) test. IC50 values were calculated by a four-parameter logistic model (p < 0.05). Gemcitabine, DCA and DAP were reported as reference compounds. Data represent mean values from at least 3 independent experiments. S.D. = standard deviation.
As expected, DCA was effective at millimolar concentrations in determining a reduction in cancer cell viability. On the contrary, all tested compounds showed a promising cytotoxic activity, with IC50 values in the micromolar range. Remarkably, all derivatives were much more effective against KRAS mutant PSN1 cancer cells compared with wild-type BxPC-3 pancreatic cancer cells. On the other hand, all derivatives were less cytotoxic compared with the reference chemotherapeutic drug gemcitabine and some compounds were also less effective than DAP in decreasing cancer cell proliferation. Among all, compounds 4c, 4g, 5b and 5j were the most effective, being able to inhibit cell growth more potently than DAP. In particular, 5j was extremely potent towards both considered PDAC cells, eliciting IC50 values in the sub-micromolar range.
Prompted by these encouraging results, we also screened the newly developed 1,2,4-triazine compounds against 3D spheroids of PDAC KRAS wild-type and mutant cells, to further estimate their anticancer potential in a more predictive environment. Actually, 3D cell cultures possess several features that closely resembles the in vivo tumor architecture complexity and pathophysiology, being more predictive for in vivo effectiveness with respect to bidimensional cancer cell models [54]. In particular, 3D spheroids are significantly more representative of the in vivo tumor metabolic conditions [55], thus emphasizing the need to use 3D cell models for investigating new putative drugs acting on tumor targets implicated in cancer metabolism, such as PDKs. The PDAC cancer spheroids were treated with tested compounds for 72 h, and cell viability was assessed by means of the acid phosphatase (APH) assay (Table 3). The efficacy of gemcitabine as well as of DCA and DAP was assessed under the same experimental conditions. Tested on 3D models, all compounds were much more effective than DCA, whereas only few derivatives (4c, 4g, and 5b) were more active than DAP. Interestingly, some derivatives showed an in vitro antitumor profile greater than gemcitabine on 3D pancreatic models. As for 2D PDAC models, all 1,2,4-triazine compounds were, on average, much more effective against KRAS mutant PSN-1 spheroids compared with KRAS wild-type. 3D assays confirmed derivatives 4c, 4g, 5b and 5j as the most effective, with IC50 values resembling those detected with the well-known PDK inhibitor DAP.
3. Materials and Methods
3.1. Chemistry
The anhydrous solvents used for organic synthesis (n-butanol and heptane) and the reagents were purchased from Sigma-Aldrich Co, Alfa Aesar, VWR International, and Acros Organics. Dichloromethane was purified and dried using calcium hydride and stored over 4 Å molecular sieves. All air- or moisture-sensitive reactions were performed using oven-dried glassware under an inert dry nitrogen atmosphere. Analytical thin-layer chromatography (TLC) was performed on silica gel 60 F254 plates (0.25 mm thickness) and the developed plates were examined under ultraviolet (UV) light. All melting points were taken on a Buchi-Tottoly capillary apparatus and were uncorrected. IR spectra were determined in bromoform with a Shimadzu FT/IR 8400S spectrophotometer and peaks were reported in wavenumber (cm−1). 1H and 13C NMR spectra were measured at 200 and 50 MHz, respectively, on DMSO-d6 solution, using a Bruker Avance II series 200 MHz spectrometer. Chemical shifts were described in parts per million (δ), coupling constants (J) were expressed in Hertz (Hz), and splitting patterns were reported as singlet (s), doublet (d), triplet (t), quartet (q), multiplet (m), doublet of doublets (dd) and triplet of doublets (td). The chromatography column was performed with MERK silica gel 230–400 mesh ASTM or FLASH40i Biotage chromatography or with Buchi Sepacore chromatography module (prepacked cartridge reference). We state that all compounds are >95% pure by LC-MS analysis. MS spectra of all final synthesized compounds are reported in the Supporting Information (Figures S1–S20). The LC–MS was performed using an Agilent 1260 Infinity system (Agilent Technologies, Waldbronn, Germany), quaternary pump and autosampler in association with a Varian MS 500 Ion Trap Mass Spectrometer (Agilent Technologies, Waldbronn, Germany). The analysis was performed using an eclipse plus C18, 4.6 × 150 mm column using the following gradient: 0.0 min 80% A, 20% B; 10 min 20% A, 80% B, 14 min 20% A, 80% B, 15 min 80% A, 20% A (A: MilliQ water 1% formic acid, B: acetonitrile). The analysis lasted 15 min with a 0.750 mL/min flow. The compounds were detected by electrospray ionization ion trap mass spectrometry source under positive-ion conditions in turbo TDDS in the acquisition range m/z 300–550. The following parameters were used: capillary voltage 95.0 V, Needle Voltage +/− 5500 V, RF loading 81%.
3.1.1. General procedure for the preparation of 1-(1-Methyl-1H-indol-3-yl)-2-(1-methyl-1H-pyrrolo [2,3-b]pyridin-3-yl)-ethane-1,2-dione (3a–j)
To a suspension of acyl chloride 2a–e (2.41 mmol) in a mixture of DCM/heptane 2:1 (7.2 mL), under a nitrogen atmosphere, aluminum trichloride (10.84 mmol) was added in small portions. The reaction mixture was left to stir at room temperature for a few minutes and then a solution of the proper 7-azaindole 1a,b (3.98 mmol) in DCM (2.4 mL) was added dropwise. When the reaction reached completion, monitored by TLC, the crude mixture was poured into crushed ice, until the destruction of the aluminum trichloride, which is manifested by the cessation of effervescence. The resulting solid precipitate was collected by vacuum filtration, washed with water, dried under vacuum for 24 h, and purified by column chromatography using DCM/ethyl acetate 7:3 as eluent to give the desired products 3a–j.
1-(1-Methyl-1H-indol-3-yl)-2-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-ethane-1,2-dione (3a) Yield: 78%; light yellow solid; m.p.: 242.4 °C; IR (cm−1): 1697 (CO), 1601 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.57 (dd, J = 7.8, 1.6 Hz), 8.52 (s, 1H), 8.45 (dd, J = 4.7, 1.6 Hz, 1H), 8.32 (s, 1H), 8.31–8.29 (m, 1H), 7.62–7.59 (m, 1H), 7.40–7.32 (m, 4H), 3.98 (s, 3H), 3.90 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 188.6; 187.9, 148.7, 145.1, 141.5, 141.4, 138.0, 130.3, 126.5, 124.1, 123.4, 121.9, 119.4, 118.9, 111.6, 111.5, 110.3, 33.8, 32.1.
1-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-2-(1-methyl-1H-indol-3-yl)-ethane-1,2-dione (3b) Yield: 83%; ochre solid; m.p.: 217.0 °C; IR (cm−1): 1620 (CO), 1616 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.68 (d, J = 2.3 Hz, 1H), 8.60 (s, 1H), 8.54 (d, J = 2.3 Hz, 1H), 8.35 (s, 1H), 8.30–8.28 (m, 1H), 7.62–7.60 (m, 1H), 7.40–7.32 (m, 2H), 3.89 (s, 3H), 3.88 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 188.2, 187.2, 147.2, 145.2, 142.6, 141.7, 138.0, 132.0, 126.5, 124.1, 123.5, 121.9, 120.5, 114.9, 111.6, 111.5, 109.7, 33.8, 32.4.
1-(5-Bromo-1-methyl-1H-indol-3-yl)-2-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-ethane-1,2-dione (3c) Yield: 90%; light yellow solid; m.p.: 278.1 °C; IR (cm−1): 1625 (CO), 1605 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.57 (d, J = 1.5 Hz, 1H), 8.55 (s, 1H), 8.46–8.44 (m, 2H), 8.39 (s, 1H), 7.62 (d, J = 8.7 Hz, 1H), 7.52 (dd J = 8.7, 2.0 Hz, 1H), 7.40 (dd, J = 7.9, 4.7 Hz, 1H), 3.90 (s, 3H), 3.89 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 188.0; 187.6, 148.7, 145.1, 142.4, 141.6, 136.8, 130.3, 128.3, 126.6, 124.1, 119.5, 118.9, 116.3, 113.8, 111.1, 110.1, 34.0, 32.2.
1-(5-Bromo-1-methyl-1H-indol-3-yl)-2-(5-bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-ethane-1,2-dione (3d) Yield: 86%; cream solid; m.p.: 287.6 °C; IR (cm−1): 1624 (CO), 1616 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.68 (d, J = 2.2 Hz, 1H), 8.63 (s, 1H), 8.55 (d, J = 2.2 Hz, 1H), 8.43 (d, J = 2.0 Hz, 1H), 8.42 (s, 1H), 7.62 (d, J = 8.7 Hz, 1H), 7.53 (d, J = 2.0 Hz, 1H), 3.89 (s, 6H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 187.6, 187.0, 147.2, 145.3, 142.9, 142.6, 136.8, 132.0, 128.3, 126.7, 124.0, 120.5, 116.4, 114.9, 113.9, 110.9, 109.6, 34.0, 32.4.
1-(5-Chloro-1-methyl-1H-indol-3-yl)-2-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-ethane-1,2-dione (3e) Yield: 89%; wheat solid; m.p.: 263.0 °C; IR (cm−1): 1624 (CO), 1616 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.56 (dd, J = 7.8, 1.6 Hz, 1H), 8.55 (s, 1H), 8.45 (dd, J = 4.7, 1.6 Hz, 1H), 8.41 (s, 1H), 8.28 (d, J = 2.1 Hz, 1H), 7.68– 7.66 (m, 1H), 7.41- 7.38 (m, 2H), 3.90 (s, 3H), 3.89 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 188.0, 187.6, 148.7, 145.1, 142.5, 141.6, 136.5, 130.3, 128.2, 127.7, 124.0, 121.0, 119.5, 118.9, 113.4, 111.2, 110.1, 34.0, 32.1.
1-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-2-(5-chloro-1-methyl-1H-indol-3-yl)-ethane-1,2-dione (3f) Yield: 76%; light yellow solid; m.p.: 271.0 °C; IR (cm−1): 1643 (CO), 1630 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.68 (d, J = 2.2 Hz, 1H); 8.63 (s, 1H), 8.55 (d, J = 2.2 Hz, 1H), 8.43 (s, 1H), 8.27 (d, J = 2.1 Hz, 1H), 7.67 (d, J = 8.7 Hz, 1H), 7.40 (dd, J = 8.7, 2.1 Hz, 1H), 3.90 (s, 3H), 3.89 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 187.6, 187.0, 145.3, 142.8, 142.7, 136.5, 132.1, 128.4, 128.3, 128.2, 124.1, 121.0, 120.5, 114.9, 113.5, 111.0, 109.6, 34.1, 32.4.
1-(5-Fluoro-1-methyl-1H-indol-3-yl)-2-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-ethane-1,2-dione (3g) Yield: 91%; ochre solid; m.p.: 238.6 °C; IR (cm−1): 1628 (CO), 1616 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.57 (dd, J = 7.8, 1.6 Hz, 1H), 8.55 (s, 1H), 8.45 (dd, J = 4.7, 1.6 Hz, 1H), 8.40 (s, 1H), 7.96 (dd, J = 9.6, 2.6 Hz, 1H), 7.65 (dd, J = 9.0, 4.4 Hz, 1H), 7.39 (dd, J = 7.8, 4.7 Hz, 1H), 7.23 (td, J = 9.2, 9.0, 2.6 Hz, 1H), 3.90 (s, 3H), 3.89 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 188.1, 187.6, 159.8 (d, J= 236.0.3 Hz), 148.7, 145.1, 142.7, 141.5, 134.6, 130.3, 127.3 (d, J = 11.4 Hz), 119.5, 118.9, 113.1 (d, J= 9.9 Hz), 112.1 (d, J = 26.0 Hz), 111.5 (d, J = 3.8 Hz), 110.1, 106.8 (d, J = 25.1 Hz), 34.1, 32.1.
1-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-2-(5-fluoro-1-methyl-1H-indol-3-yl)-ethane-1,2-dione (3h) Yield: 87%; yellow solid; m.p.: 249.7 °C; IR (cm−1): 1636 (CO), 1611 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.68 (d, J = 2.2 Hz, 1H), 8.62 (s, 1H), 8.54 (d, J = 2.2 Hz, 1H), 8.43 (s, 1H), 7.95 (dd, J = 9.6, 2.6 Hz, 1H), 7.66 (dd, J = 9.0, 4.4 Hz, 1H), 7.23 (td, J = 9.2, 9.0, 2.6 Hz, 1H), 3.89 (s, 3H), 3.88 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 187.8, 186.9, 159.8 (d, J = 236.0.3 Hz), 147.2, 145.3, 142.9, 142.7, 134.6, 132.0, 127.3 (d, J = 11.1 Hz), 120.5, 114.9, 113.2 (d, J = 9.5 Hz), 112.1 (d, J= 25.8 Hz), 111.4 (d, J = 3.8 Hz), 109.6, 106.8 (d, J= 25.1 Hz), 34.1, 32.4.
1-(5-Methoxy-1-methyl-1H-indol-3-yl)-2-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-ethane-1,2-dione (3i) Yield: 84%; light yellow solid; m.p.: 204.8 °C; IR (cm−1): 1636 (NH), 1616 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.57 (dd, J = 7.8, 1.6 Hz, 1H), 8.54 (s, 1H), 8.45 (dd, J = 4.7, 1.6 Hz, 1H), 8.26 (s, 1H), 7.81 (d, J = 2.5 Hz, 1H), 7.51 (d, J = 8.9 Hz, 1H), 7.39 (dd, J = 7.8, 4.7 Hz, 1H), 6.98 (dd, J = 8.9, 2.5 Hz, 1H), 3.90 (s, 3H), 3.85 (s, 3H), 3.84 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 188.6, 187.7, 156.8, 148.7, 145.1, 141.4, 141.3, 132.8, 130.3, 127.5, 119.4, 118.9, 113.6, 112.5, 111.3, 110.3, 103.8, 55.8, 33.9, 32.1.
1-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-2-(5-methoxy-1-methyl-1H-indol-3-yl)-ethane-1,2-dione (3j) Yield: 92%; ochre solid; m.p.: 264.6 °C; IR (cm−1): 1636 (CO), 1612 (CO); 1H NMR (DMSO-d6, 200 MHz) δ: 8.71–8.56 (m, 3H), 8.36–8.24 (m, 1H), 7.89–7.78 (m, 1H), 7.60–7.48 (m, 1H), 7.03–7.00 (m, 1H), 3.91–3.80 (m, 9H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 187.0, 186.9, 156.9, 147.9, 145.3, 142.6, 141.6, 132.9, 132.1, 127.5, 120.7, 114.8, 113.7, 112.6, 111.2, 103.8, 55.9, 34.0, 32.4.
3.1.2. General procedure for the preparation of 5,6-di(1H-indol-3-yl)-1,2,4-triazin-3-amines (4–5)
To a suspension of the appropriate derivatives 3a–j (5 mmol) in anhydrous n- butanol (20 mL), aminoguanidine bicarbonate (10 mmol) was added in portions. The resulting mixture was heated under reflux for 3 h. The precipitate, which formed upon cooling, was collected by vacuum filtration, washed with n-butanol, and dried under vacuum for 24 h. The crude residue, obtained as a mixture of two diastereomers, was purified by column chromatography (CC) using dichloromethane/methanol 97:3 and 96:4 as eluent, affording the single enantiomers 4 and 5, respectively.
6-(1-Methyl-1H-indol-3-yl)-5-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (4a) Yield: 42%; yellow solid; m.p.: 268.1 °C; IR (cm−1): 3458, 3271 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.81 (dd, J = 8.0, 1.6 Hz, 1H), 8.32 (dd, J = 4.7, 1.6 Hz, 1H), 7.63 (s, 1H), 7.54 (d, J = 8.2 Hz, 1H), 7.24 (s, 1H), 7.22- 7.16 (m, 3H), 6.99- 6.93 (m, 3H), 3.88 (s, 3H), 3.55 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.5, 148.1, 144.1, 143.4, 137.2, 134.5, 131.9, 130.1, 126.3, 122.0, 120.4, 119.9, 119.6, 117.6, 111.6, 110.7, 108.8, 33.1, 31.7; LC-MS: m/z [M + H]+ calcd for C20H18N7+ 356.40; found, 356.41.
5-(1-Methyl-1H-indol-3-yl)-6-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (5a) Yield: 38%; yellow solid; m.p.: 235.5 °C; IR (cm−1): 3397, 3298 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.46 (d, J = 7.9 Hz, 1H) 8.28 (dd, J = 4.6, 1.6 Hz, 1H), 7.80 (s, 1H), 7.59 (dd, J = 7.9, 1.6 Hz, 1H), 7.44 (d, J = 8.1 Hz, 1H), 7.28–7.12 (m, 3H), 6.99 (dd, J = 7.9, 4.6 Hz, 1H), 6.95 (s, 2H), 3.88 (s, 3H), 3.56 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 153.1, 148.0, 143.2, 143.1, 137.4, 134.5, 130.1, 128.9, 127.1, 123.3, 122.9, 121.4, 118.6, 116.2, 110.8, 110.6, 110.0, 33.3, 31.5; LC-MS: m/z [M + H]+ calcd for C20H18N7+ 356.40; found, 356.38.
5-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-6-(1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-amine (4b) Yield: 44%; yellow solid; m.p.: 260.0 °C; IR (cm−1): 3466, 3447 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.92 (d, J = 2.2 Hz, 1H), 8.39 (d, J = 2.2 Hz, 1H), 7.65 (s, 1H), 7.54 (d, J = 8.2 Hz, 1H), 7.29 (s, 1H), 7.20–7.16 (m, 2H), 7.10 (s, 2H), 6.96- 6.92 (m, 1H), 3.89 (s, 3H), 3.53 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 151.9, 146.5, 144.2, 143.2, 137.2, 135.9, 133.6, 130.1, 126.2, 122.0, 121.1, 120.3, 120.0, 113.4, 111.4, 110.7, 108.5, 33.1, 31.9; LC-MS: m/z [M + H]+ calcd for C20H17BrN7+ 434.07; found, 436.30.
6-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-(1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-ylamine (5b) Yield: 42%; yellow solid; m.p.: 275.5.0 °C; IR (cm−1): 3472, 3447 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.37–8.25 (m, 2H), 7.87 (d, J = 2.2 Hz, 1H), 7.82 (s, 1H), 7.47 (d, J = 8.2 Hz, 1H), 7.35 (s, 1H), 7.28–7.20 (m, 1H), 7.14 (t, J = 7.5 Hz, 1H), 6.99 (s, 2H), 3.84 (s, 3H), 3.64 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 146.5, 143.2, 142.8, 137.4, 137.2, 134.4, 131.9, 130.9, 127.0, 123.1, 122.9, 121.3, 120.5, 111.6, 110.8, 110.1, 110.0, 33.3, 31.7; LC-MS: m/z [M + H]+ calcd for C20H17BrN7+ 434.07; found, 434.42.
6-(5-Bromo-1-methyl-1H-indol-3-yl)-5-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (4c) Yield: 49%; yellow solid; m.p.: 176.3 °C; IR (cm−1): 3481, 3402 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.68 (dd, J = 8.0, 1.5 Hz, 1H), 8.33 (dd, J = 4.6, 1.4 Hz, 1H), 7.63 (s, 1H), 7.57–7.47 (m, 2H), 7.40 (s, 1H), 7.31 (dd, J = 8.7, 1.9 Hz, 1H), 7.19 (dd, J = 8.0, 4.7 Hz, 1H), 7.02 (s, 2H), 3.84 (s, 3H), 3.64 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.1, 152.6, 144.1, 143.0, 136.0, 135.4, 134.4, 131.7, 131.6, 128.2, 124.5, 122.7, 119.5, 117.6, 112.8, 112.7, 111.0, 108.9, 33.3, 31.7; LC-MS: m/z [M + H]+ calcd for C20H17BrN7+ 434.07; found, 434.39.
5-(5-bromo-1-methyl-1H-indol-3-yl)-6-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (5c) Yield: 45%; yellow solid; m.p.: 304.5 °C; IR (cm−1): 3304, 3127 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.56 (d, J = 1.7 Hz, 1H), 8.28 (dd, J = 4.6, 1.2 Hz, 1H), 7.82 (s, 1H), 7.55 (dd, J = 7.9, 1.4 Hz, 1H), 7.43 (d, J = 8.7 Hz, 1H), 7.36 (dd, J = 8.7, 1.9 Hz, 1H), 7.24 (s, 1H), 7.05 (s, 2H), 6.98 (dd, J = 7.9, 4.6 Hz, 1H), 3.89 (s, 3H), 3.55 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.6, 148.0, 143.3, 136.8, 136.1, 135.6, 130.2, 128.8, 128.7, 125.5, 125.2, 118.5, 116.2, 114.6, 112.9, 110.4, 109.6, 33.5, 31.5; LC-MS: m/z [M + H]+ calcd for C20H17BrN7+ 434.07; found, 434.40.
6-(5-Bromo-1-methyl-1H-indol-3-yl)-5-(5-bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (4d) Yield: 48%; yellow solid; m.p.: 266.5.0 °C; IR (cm−1): 3481, 3285 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.46 (d, J = 1.9 Hz, 1H), 8.35 (d, J = 2.2 Hz, 1H), 7.84–7.83 (m, 2H), 7.46 (d, J = 8.7 Hz, 1H), 7.38–7.35 (m, 2H), 7.08 (s, 2H), 3.85 (s, 3H), 3.62 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.6, 146.5, 143.3, 142.7, 136.2, 135.5, 131.9, 130.8, 128.6, 125.5, 125.0, 120.4, 114.5, 112.9, 111.6, 110.0, 109.6, 33.5, 31.7; LC-MS: m/z [M + H]+ calcd for C20H16Br2N7+ 513.97; found, 514.30.
5-(5-Bromo-1-methyl-1H-indol-3-yl)-6-(5-bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (5d) Yield: 44%; yellow solid; m.p.: 333.7 °C; IR (cm−1): 3426, 3315 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.82 (d, J = 2.3 Hz, 1H), 8.40 (d, J = 2.2 Hz, 1H), 7.65 (s, 1H), 7.53 (d, J = 8.7 Hz, 1H), 7.48 (d, J = 1.8 Hz, 1H), 7.44 (s, 1H), 7.31 (dd, J = 8.7, 2.0 Hz, 1H), 7.13 (s, 2H), 3.85 (s, 3H), 3.62 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.0, 146.6, 144.3, 142.8, 136.0, 135.7, 133.4, 131.6, 128.2, 124.5, 122.6, 121.0, 113.3, 112.9, 112.8, 110.9, 108.5, 33.3, 31.9; LC-MS: m/z [M + H]+ calcd for C20H16Br2N7+ 513.97; found, 514.28.
6-(5-Chloro-1-methyl-1H-indol-3-yl)-5-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (4e) Yield: 46%; yellow solid; m.p.: 178.0 °C; IR (cm−1): 3481, 3408 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.70 (dd, J = 8.0, 1.6 Hz, 1H), 8.33 (dd, J = 4.7, 1.6 Hz, 1H), 7.60–7.56 (m, 1H), 7.65 (s, 1H), 7.38 (s, 1H), 7.37–7.28 (m, 1H), 7.25–7.10 (m, 2H), 7.02 (s, 2H), 3.85 (s, 3H), 3.63 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.5, 148.2, 144.1, 143.0, 135.7, 134.3, 131.7 (×2), 127.6, 124.8, 122.0, 119.6, 119.5, 117.6, 112.4, 111.2, 108.9, 33.3, 31.7; LC-MS: m/z [M + H]+ calcd for C20H17ClN7+ 390.12; found, 390.40.
5-(5-Chloro-1-methyl-1H-indol-3-yl)-6-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (5e) Yield: 41%; yellow solid; m.p.: 310.3 °C; IR (cm−1): 3454, 3319 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.47 (d, J = 2.0 Hz, 1H), 8.28 (dd, J = 4.6, 1.5 Hz, 1H), 7.83 (s, 1H), 7.55 (dd, J = 7.9, 1.6 Hz, 1H), 7.48 (d, J = 8.6 Hz, 1H), 7.32–7.17 (m, 2H), 7.04 (s, 2H), 6.99 (dd, J = 7.9, 4.6 Hz, 1H), 3.90 (s, 3H), 3.55 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.6, 148.0, 143.3, 143.0, 135.9, 135.8, 130.2, 128.8, 128.1, 126.4, 122.9, 122.3, 118.6, 116.2, 112.5, 110.4, 109.6, 33.5, 31.5; LC-MS: m/z [M + H]+ calcd for C20H17ClN7+ 390.12; found, 390.41.
5-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-6-(5-chloro-1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-amine (4f) Yield: 40%; yellow solid; m.p.: 262.1 °C; IR (cm−1): 3481, 3285 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.84 (d, J = 2.3 Hz, 1H), 8.40 (d, J = 2.3 Hz, 1H), 7.68 (s, 1H), 7.58 (d, J = 8.7 Hz, 1H), 7.43 (s, 1H), 7.32 (d, J = 2.1 Hz, 1H), 7.19 (dd, J = 8.7, 2.1 Hz, 1H), 7.14 (s, 2H), 3.86 (s, 3H), 3.61 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.0, 146.6, 144.2, 142.8, 135.8, 135.7, 133.5, 131.8, 127.5, 124.8, 122.0, 121.0, 119.6, 113.4, 112.4, 111.0, 108.5, 33.4, 31.9; LC-MS: m/z [M + H]+ calcd for C20H16BrClN7+ 470.02; found, 470.30.
6-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-(5-chloro-1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-amine (5f) Yield: 39%; yellow solid; m.p.: 334.6 °C; IR (cm−1): 3445, 3420 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.39 (d, J = 2.1 Hz, 1H), 8.35 (d, J = 2.2 Hz, 1H), 7.86 (s, 1H), 7.84 (d, J = 2.2 Hz, 1H), 7.55–7.45 (m, 1H), 7.38 (s, 1H), 7.25 (dd, J = 8.7, 2.1 Hz, 1H), 7.09 (s, 2H), 3.86 (s, 3H), 3.62 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 152.6, 146.5, 143.3, 142.6, 136.0, 135.7, 131.9, 130.8, 128.1, 126.4, 122.9, 122.2, 120.4, 112.5, 111.6, 110.0, 109.6, 33.6, 31.7; LC-MS: m/z [M + H]+ calcd for C20H16BrClN7+ 470.02; found, 470.30.
6-(5-Fluoro-1-methyl-1H-indol-3-yl)-5-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (4g) Yield: 41%; yellow solid; m.p.: 267.4 °C; IR (cm−1): 3402, 3308 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.74 (dd, J = 8.0, 1.6 Hz, 1H), 8.33 (dd, J = 4.7, 1.6 Hz, 1H), 7.69 (s, 1H), 7.55 (dd, J = 8.9, 4.5 Hz, 1H), 7.34 (s, 1H), 7.20 (dd, J = 8.0, 4.7 Hz, 1H), 7.08–6.94 (m, 4H), 3.86 (s, 3H), 3.60 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 156.6 (d, J = 262.7 Hz), 152.4, 148.1, 144.1, 143.1, 134.3, 134.0, 132.0, 131.8, 126.6 (d, J = 10.8 Hz), 119.5, 117.6, 112.0 (d, J= 9.4 Hz), 111.6 (d, J= 4.8 Hz), 110.2 (d, J = 26.6 Hz), 108.9, 105.1 (d, J = 23.5 Hz), 33.4, 31.7; LC-MS: m/z [M + H]+ calcd for C20H17FN7+ 374.15; found, 374.39.
5-(5-Fluoro-1-methyl-1H-indol-3-yl)-6-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (5g) Yield: 40%; yellow solid; m.p.: 281.4 °C; IR (cm−1): 3487, 3410 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.32 (d, J = 2.6 Hz, 1H), 8.29 (dd, J = 4.6, 1.6 Hz, 1H), 7.84 (s, 1H), 7.57 (dd, J = 7.9, 1.6 Hz, 1H), 7.47 (dd, J = 9.0, 4.6 Hz, 1H), 7.21 (s, 1H), 7.10 (td, J = 9.1, 9.0, 2.6 Hz, 2H), 7.03 (s, 2H), 7.00 (dd, J = 7.9, 4.6 Hz, 1H), 3.91 (s, 3H), 3.55 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 160.3, 155.1 (d, J= 237.3 Hz), 152.8, 148.0, 143.3, 142.8, 136.0, 134.1, 128.8, 127.7 (d, J = 10.8 Hz), 118.6, 116.2, 112.1 (d, J = 10.0 Hz), 111.2 (d, J = 4.8 Hz), 110.7 (d, J = 27.0 Hz), 108.6, 108.5 (d, J= 23.5 Hz), 33.7, 31.5; LC-MS: m/z [M + H]+ calcd for C20H17FN7+ 374.15; found, 374.38.
5-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-6-(5-fluoro-1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-amine (4h) Yield: 43%; yellow solid; m.p.: 155.6 °C; IR (cm−1): 3481, 3288 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.86 (d, J = 2.2 Hz, 1H), 8.40 (d, J = 2.2 Hz, 1H), 7.71 (s, 1H), 7.56 (dd, J = 9.1, 4.5 Hz, 1H), 7.38 (s, 1H), 7.13 (s, 2H), 7.03 (td, J = 9.2, 9.1, 2.6 Hz, 1H), 6.96 (dd, J = 10.0, 2.6 Hz, 1H), 3.87 (s, 3H), 3.59 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 156.6 (d, J = 255.8 Hz), 151.9, 146.6, 144.2, 142.9, 135.7, 134.0, 133.5, 132.0, 129.9 (d, J = 12.8 Hz), 121.06, 113.4, 112.0 (d, J = 10.2 Hz), 111.4 (d, J = 4.5 Hz), 110.3 (d, J = 25.7 Hz), 108.5, 105.0 (d, J = 23.5 Hz), 33.4, 31.9; LC-MS: m/z [M + H]+ calcd for C20H16BrFN7+ 452.06; found, 452.45.
6-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-(5-fluoro-1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-amine (5h) Yield: 42%; yellow solid; m.p.: 311.1 °C; IR (cm−1): 3479, 3450 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.36 (d, J = 2.2 Hz, 1H), 8.23 (dd, J = 10.6, 2.6 Hz, 1H), 7.88–7.87 (m, 2H), 7.49 (dd, J = 8.9, 4.6 Hz, 1H), 7.35 (s, 1H), 7.11 (td, J = 9.1, 2.6 Hz, 1H), 7.06 (s, 2H), 3.87 (s, 3H), 3.62 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 156.8 (d, J = 267.1 Hz), 152.8, 146.5, 143.3, 142.4, 135.9, 134.2, 131.9, 130.8, 127.7 (d, J = 11.2 Hz), 120.5, 112.1 (d, J = 10.2 Hz), 111.6, 111.1 (d, J = 26.4 Hz), 110.0, 109.7 (d, J= 4.5 Hz), 108.3 (d, J = 24.8 Hz), 33.7, 31.7; LC-MS: m/z [M + H]+ calcd for C20H16BrFN7+ 452.06; found, 452.45.
6-(5-methoxy-1-methyl-1H-indol-3-yl)-5-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (4i) Yield: 47%; yellow solid; m.p.: 217.0 °C; IR (cm−1): 3466, 3285 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.72 (dd, J = 8.0, 1.6 Hz, 1H), 8.32 (dd, J = 4.7, 1.6 Hz, 1H), 7.58 (s, 1H), 7.49–7.39 (m, 1H), 7.30 (s, 1H), 7.19 (dd, J = 7.9, 4.7 Hz, 1H), 6.97 (s, 2H), 6.81 (dd, J = 8.8, 2.4 Hz, 1H), 6.61 (d, J = 2.4 Hz, 1H), 3.83 (s, 3H), 3.58 (s, 3H), 3.44 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 154.2, 152.4, 150.9, 148.1, 144.0, 134.4, 132.4, 131.6, 130.5, 126.7, 119.5, 117.6, 112.2, 111.5, 111.2, 109.0, 102.0, 55.5, 33.3, 31.7; LC-MS: m/z [M + H]+ calcd for C21H20N7O+ 386.17; found, 386.44.
5-(5-Methoxy-1-methyl-1H-indol-3-yl)-6-(1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,2,4-triazin-3-amine (5i) Yield: 37%; yellow solid; m.p.: 288.2 °C; IR (cm−1): 3421, 3329 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.28 (dd, J = 4.6, 1.5 Hz, 1H), 7.77 (d, J = 2.4 Hz, 2H), 7.62 (dd, J = 7.9, 1.5 Hz, 1H), 7.33 (d, J = 8.9 Hz, 1H), 7.21 (s, 1H), 7.00 (dd, J = 7.9, 4.6 Hz, 1H), 6.94 (s, 2H), 6.84 (dd, J = 8.8, 2.5 Hz, 1H), 3.86 (s, 3H), 3.68 (s, 3H), 3.54 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.2, 155.3, 153.2, 148.0, 143.3, 143.0, 134.8, 132.5, 130.1, 128.9, 127.6, 118.7, 116.2, 112.7, 111.5, 110.7, 109.7, 105.1, 55.8, 33.5, 31.4; LC-MS: m/z [M + H]+ calcd for C21H20N7O+ 386.17; found, 386.43.
5-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-6-(5-methoxy-1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-amine (4j) Yield: 48%; yellow solid; m.p.: 197.6 °C; IR (cm−1): 3481, 3447 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 8.82 (d, J = 2.2 Hz, 1H), 8.39 (d, J = 2.2 Hz, 1H), 7.60 (s, 1H), 7.42 (d, J = 8.8 Hz, 1H), 7.35 (s, 1H), 7.07 (s, 2H), 6.80 (dd, J = 8.9, 2.4 Hz, 1H), 6.59 (d, J = 2.4 Hz, 1H), 3.84 (s, 3H), 3.57 (s, 3H), 3.45 (s, 3H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 161.3, 154.2, 151.8, 146.5, 144.1, 143.5, 135.8, 133.3, 132.4, 130.5, 126.6, 121.0, 113.3, 112.3, 111.5, 111.0, 108.7, 101.9, 55.6, 33.3, 31.9; LC-MS: m/z [M + H]+ calcd for C21H18BrN7O+ 465.32; found, 464.40.
6-(5-Bromo-1-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-(5-methoxy-1-methyl-1H-indol-3-yl)-1,2,4-triazin-3-amine (5j) Yield: 48%; yellow solid; m.p.: 269.8 °C; IR (cm−1): 3481, 3291 (NH2); 1H NMR (DMSO-d6, 200 MHz) δ: 3.61 (s, 3H), 3.68 (s, 3H), 3.83 (s, 3H), 6.84 (dd, J = 8.9, 2.5 Hz, 1H), 6.97 (s, 2H), 7.36–7.33 (m, 2H), 7.68 (d, J = 2.5 Hz, 1H), 7.81 (s, 1H), 7.87 (d, J = 2.2 Hz, 1H), 8.35 (d, J = 2.2 Hz, 1H); 13C{1H} NMR (DMSO-d6, 50 MHz) δ: 31.7, 33.5, 55.8, 105.0, 109.7, 110.3, 111.5, 111.6, 112.8, 120.5, 127.5, 130.8, 131.8, 132.6, 134.7, 142.7, 143.2, 146.4, 153.1, 155.3, 161.2; LC-MS: m/z [M + H]+ calcd for C21H18BrN7O+ 465.32; found, 466.34.
3.2. Software Overview
All the molecular modeling operations were executed within the Molecular Operating Environment (MOE) suite (version 2019.01) [56] [. The proteins were downloaded as PDB files from the Protein Data Bank (PDB) [57], and their preparation for the computational studies was executed by exploiting the dedicated tools available in the MOE package, as well as the generation of the homology models used for the molecular docking studies. The ligands were properly prepared for computational handling with the tools of the QUACPAC package available in the OpenEye suite [58]. The molecular docking calculations were executed with the program PLANTS [59], setting PLANTSCHEMPLP as the scoring function. The hardware exploited for all the computational studies was a 20-CPU Linux Workstation (Intel i9-9820X).
3.3. PDK1-4 Kinase Assay
The in vitro inhibitory activity of tested compounds on recombinant human PDK1-4 isoforms (Abcam, Cambridge, MA, USA) was evaluated by using the ADPGlo™ Kinase Assay kit from Promega (Promega Corporation, Madison, WI, USA) following the instructions of the manufacturer, as previously described [44].
3.4. Heat Shock Protein 90 (HSP90) Inhibition Assay
The in vitro inhibitory activity of tested compounds on recombinant human HSP90 was assessed by The Transcreener™ ADP kit (Bellbrook Labs, Madison, WI, USA) according to the manufacturer’s protocol as previously described [44].
3.5. Experiments with Cultured Human Cancer Cells
All tested compounds were dissolved in DMSO just before the experiment, and a calculated amount of drug solution was added to the cell growth medium to a final solvent concentration of 0.5%, which had no detectable effects on cell viability. Dichloroacetate (DCA), 2,2-dichloroacetophenone (DAP) and gemcitabine hydrochloride were purchased by Merck KGaA (Darmstadt, Germany).
3.6. Cell Cultures
Human pancreatic PSN-1 and BxPC-3 carcinoma cell lines were obtained by American Type Culture Collection (ATCC, Rockville, MD, USA). Cell lines were maintained in the logarithmic phase at 37 °C in a 5% carbon dioxide atmosphere using RPMI-1640 medium (EuroClone) containing 10% fetal calf serum (EuroClone, Milan, Italy), antibiotics (50 units/mL penicillin and 50 μg/mL streptomycin) and 2 mM l-glutamine.
3.7. MTT Assay
The growth inhibitory effect on tumor cells was evaluated by means of MTT assay. Briefly, 5 × 103 cells/well were seeded in 96-well microplates in a growth medium (100 μL). After 24 h, the medium was removed and replaced with a fresh one containing the compound to be studied at the appropriate concentration. Triplicate cultures were established for each treatment. After 72 h, each well was treated with 10 μL of a 5 mg/mL MTT saline solution, and following 5 h of incubation, 100 μL of a sodium dodecyl sulfate (SDS) solution in HCl 0.01 M was added. After overnight incubation, cell growth inhibition was detected by measuring the absorbance of each well at 570 nm using a Bio-Rad 680 microplate reader. The mean absorbance for each drug dose was expressed as a percentage of the control untreated well absorbance and plotted vs. drug concentration. IC50 values, the drug concentrations that reduce the mean absorbance at 570 nm to 50% of those in the untreated control wells, were calculated by the four-parameter logistic (4-PL) model. The evaluation was based on means from at least four independent experiments.
3.8. Spheroid Cultures
Spheroid cultures were obtained by seeding 2.5 × 103 PSN-1 cancer cells/well in a round-bottom non-treated tissue culture 96-well plate (Greiner Bio-one, Kremsmünster, Austria) in phenol red-free RPMI-1640 medium (Sigma Chemical Co, St. Louis, MO, USA) containing 10% fetal calf serum and supplemented with 20% methylcellulose stock solution.
3.9. Acid Phosphatase (APH) Assay
An APH-modified assay was used for determining cell viability in 3D spheroids, as previously described [60]. Briefly, the pre-seeded spheroids were treated with fresh medium containing the compound to be studied at the appropriate concentration (range 2.5–200 μM). Triplicate cultures were established for each treatment. After 72 h, each well was treated with 100 μL of the assay buffer (0.1 M sodium acetate, 0.1% Triton-X-100, supplemented with ImmunoPure p-nitrophenyl phosphate; Sigma Chemical Co.) and, following 3 h of incubation, 10 μL of 1 M NaOH solution were added. The inhibition of the cell growth induced by the tested compounds was detected by measuring the absorbance of each well at 405 nm, using a Bio-Rad 680 microplate reader. The mean absorbance for each drug dose was expressed as a percentage of the control untreated well absorbance (T/C) and plotted vs. drug concentration. IC50 values, the drug concentrations that reduce the mean absorbance at 405 nm 50% of those in the untreated control wells, were calculated by the four-parameter logistic (4-PL) model. The evaluation was based on means from at least four independent experiments.
4. Conclusions
To the best of our knowledge, very few studies reported small molecules as PDK inhibitors employed for the treatment of metabolic altered types of cancer, such as PDAC. PDKs are a family of kinases with a key role of control in cellular energy metabolism, whose deregulation led to cancer formation sustaining tumour cell proliferation, invasion, angiogenesis, apoptosis resistance and metabolic switch. Among the existing therapies, chemotherapy regimens represent the standard treatment for most patients; unfortunately, PDAC tumor metabolism alteration has been proven to be associated with chemoresistance. Therefore, there is an urgent need to develop novel therapeutic strategies to treat PDAC. In the present study, we presented the synthesis and pharmacological evaluation of a new series of 1,2,4-triazine derivatives 4a-j, 5a-j, with promising PDKs enzymatic inhibition, especially with preferential activity towards the PDK1 and PDK4 isoforms. In particular, compounds 4d, 4f, 4h, 5d, 5f, 5h, and 5j totally hampered the enzymatic activity of PDK1 at the tested concentration of 1.5 µM, at the same time compounds 4d, 5d, 5f, 5h and 5j were also able to suppress the enzymatic activity of PDK4. On the other hand, all the tested derivatives were scarcely effective against PDK2 and PDK3. The IC50 values on the PDK1 isoforms were also calculated for all derivatives, confirming their efficiency in decreasing the PDK1 catalytic activity, with IC50 values ranging from 0.04 to 0.33 µM. In addition, considering the high degree of similarity shared by PDK1 and HSP90, the most promising compounds were also screened for their ability to block the enzymatic activity on HSP90. The results showed that the selected derivatives were able to completely suppress the activity of HSP0, except for compound 5j. Considering these results, molecular docking analyses were performed on both, PDK1 and HSP90. All the new synthesized compounds were docked into the orthosteric ATP- binding site. In the case of PDK1, binding mode of these new compounds was experimentally validated, using the ligand-based homology modeling technique, which showed that all derivatives interact through hydrogen bond with Asp318 by nitrogen triazine central core, whereas the bicyclic portions are placed in the inner part of the binding site interacting with solvent exposed area. Moreover, the methyl groups of the indole/azaindole interact with hydrophobic area. A very similar binding mode was also observed for HSP0; therefore, the key differences in the activities could be explained by the dynamic processes, which allows some molecules to better fit in the pocket than others. In vitro antiproliferative activities against KRAS-wild-type and mutant PDAC cells BXPC-3 and PSN-1 were assessed by using an MTT assay. Interestingly, all derivatives were much more effective against KRAS-mutant PSN-1 showing promising cytotoxic activity, with IC50 values in the micromolar range. In order to further confirm the antiproliferative activity found, the compounds were tested on 3D spheroids of PDAC which better simulate the pathophysiological tumor specific microenvironment. Interestingly, also in 3D models, the compounds were much more effective in the KRAS-mutant PSN-1, confirming the cytotoxic activity of the new triazine derivatives.
Acknowledgments
MMS lab is very grateful to Chemical Computing Group, OpenEye, and Acellera for the scientific and technical partnership.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/md21050288/s1, Figures S1–S20: Chromatogram and MS spectrum of compound (4a–j and 5a–j).
Author Contributions
The manuscript was written through the contributions of all authors. The synthetic workflow, D.C. and C.P.; the in vitro experiments, M.D.F.; the molecular modeling studies, D.B. and M.P.; writing—original draft preparation, S.C., B.P., G.C., S.D. and S.S.; study concept, writing—review and editing, S.M., V.G. and P.D.; funding acquisition, P.D. and V.G. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Funding Statement
This work was partially supported by the following grants: PRIN2017, Prot. No. 2017E84AA4 (to P.D. and V.G.) and European Union 2014–2020 PON Ricerca e Innovazione grant from the Italian Ministry of Education, University and Research, entitled “PROGEMA-Processi Green per l’Estrazione di Principi Attivi e la Depurazione di Matrici di Scarto e Non” (ARS01_00432) (to P.D.).
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Phan L.M., Yeung S.C., Lee M.H. Cancer metabolic reprogramming: Importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biol. Med. 2014;11:1–19. doi: 10.7497/j.issn.2095-3941.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stacpoole P.W. Therapeutic Targeting of the Pyruvate Dehydrogenase Complex/Pyruvate Dehydrogenase Kinase (PDC/PDK) Axis in Cancer. J. Natl. Cancer Inst. 2017;109:djx071. doi: 10.1093/jnci/djx071. [DOI] [PubMed] [Google Scholar]
- 3.Liberti M.V., Locasale J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016;41:211–218. doi: 10.1016/j.tibs.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anwar S., Shamsi A., Mohammad T., Islam A., Hassan M.I. Targeting pyruvate dehydrogenase kinase signaling in the development of effective cancer therapy. Biochim. Biophys. Acta Rev. Cancer. 2021;1876:188568. doi: 10.1016/j.bbcan.2021.188568. [DOI] [PubMed] [Google Scholar]
- 5.Harris R.A., Bowker-Kinley M.M., Huang B., Wu P. Regulation of the activity of the pyruvate dehydrogenase complex. Adv. Enzym. Regul. 2002;42:249–259. doi: 10.1016/S0065-2571(01)00061-9. [DOI] [PubMed] [Google Scholar]
- 6.Wynn R.M., Kato M., Chuang J.L., Tso S.C., Li J., Chuang D.T. Pyruvate dehydrogenase kinase-4 structures reveal a metastable open conformation fostering robust core-free basal activity. J. Biol. Chem. 2008;283:25305–25315. doi: 10.1074/jbc.M802249200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tambe Y., Terado T., Kim C.J., Mukaisho K.I., Yoshida S., Sugihara H., Tanaka H., Chida J., Kido H., Yamaji K., et al. Antitumor activity of potent pyruvate dehydrogenase kinase 4 inhibitors from plants in pancreatic cancer. Mol. Carcinog. 2019;58:1726–1737. doi: 10.1002/mc.23045. [DOI] [PubMed] [Google Scholar]
- 8.Trinidad A.G., Whalley N., Rowlinson R., Delpuech O., Dudley P., Rooney C., Critchlow S.E. Pyruvate dehydrogenase kinase 4 exhibits a novel role in the activation of mutant KRAS, regulating cell growth in lung and colorectal tumour cells. Oncogene. 2017;36:6164–6176. doi: 10.1038/onc.2017.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qin C., Yang G., Yang J., Ren B., Wang H., Chen G., Zhao F., You L., Wang W., Zhao Y. Metabolism of pancreatic cancer: Paving the way to better anticancer strategies. Mol. Cancer. 2020;19:50. doi: 10.1186/s12943-020-01169-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grasso C., Jansen G., Giovannetti E. Drug resistance in pancreatic cancer: Impact of altered energy metabolism. Crit. Rev. Oncol. Hematol. 2017;114:139–152. doi: 10.1016/j.critrevonc.2017.03.026. [DOI] [PubMed] [Google Scholar]
- 11.Gunda V., Souchek J., Abrego J., Shukla S.K., Goode G.D., Vernucci E., Dasgupta A., Chaika N.V., King R.J., Li S., et al. MUC1-Mediated Metabolic Alterations Regulate Response to Radiotherapy in Pancreatic Cancer. Clin. Cancer Res. 2017;23:5881–5891. doi: 10.1158/1078-0432.CCR-17-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang C.H., Qiu J., O’Sullivan D., Buck M.D., Noguchi T., Curtis J.D., Chen Q., Gindin M., Gubin M.M., van der Windt G.J., et al. Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression. Cell. 2015;162:1229–1241. doi: 10.1016/j.cell.2015.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sutendra G., Michelakis E.D. Pyruvate dehydrogenase kinase as a novel therapeutic target in oncology. Front. Oncol. 2013;3:38. doi: 10.3389/fonc.2013.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mehla K., Singh P.K. Metabolic Subtyping for Novel Personalized Therapies Against Pancreatic Cancer. Clin. Cancer Res. 2020;26:6–8. doi: 10.1158/1078-0432.CCR-19-2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li T., Wang N., Zhang T., Zhang B., Sajeevan T.P., Joseph V., Armstrong L., He S., Yan X., Naman C.B. A Systematic Review of Recently Reported Marine Derived Natural Product Kinase Inhibitors. Mar. Drugs. 2019;17:493. doi: 10.3390/md17090493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Luo M., Groaz E., De Jonghe S., Snoeck R., Andrei G., Herdewijn P. Amidate Prodrugs of Cyclic 9-( S)-[3-Hydroxy-2-(phosphonomethoxy)propyl]adenine with Potent Anti-Herpesvirus Activity. ACS Med. Chem. Lett. 2018;9:381–385. doi: 10.1021/acsmedchemlett.8b00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sakemi S., Sun H.H. Nortopsentins A., B, and C. Cytotoxic and antifungal imidazolediylbis[indoles] from the sponge Spongosorites ruetzleri. J. Org. Chem. 1991;56:4304–4307. doi: 10.1021/jo00013a044. [DOI] [Google Scholar]
- 18.Bao B., Sun Q., Yao X., Hong J., Lee C.O., Sim C.J., Im K.S., Jung J.H. Cytotoxic bisindole alkaloids from a marine sponge Spongosorites sp. J. Nat. Prod. 2005;68:711–715. doi: 10.1021/np049577a. [DOI] [PubMed] [Google Scholar]
- 19.Bettayeb K., Tirado O.M., Marionneau-Lambot S., Ferandin Y., Lozach O., Morris J.C., Mateo-Lozano S., Drueckes P., Schächtele C., Kubbutat M.H., et al. A new class of cell death-inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases. Cancer Res. 2007;67:8325–8334. doi: 10.1158/0008-5472.CAN-07-1826. [DOI] [PubMed] [Google Scholar]
- 20.Kohmoto S., Kashman Y., McConnell O.J., Rinehart K.L., Wright A., Koehn F. Dragmacidin; a New Cytotoxic Bis (indole) Alkaloid from a Deep Water Marine Sponge, Dragmacidon sp. J. Org. Chem. 1988;53:3116–3118. doi: 10.1021/jo00248a040. [DOI] [Google Scholar]
- 21.Cascioferro S., Parrino B., Petri G.L., Cusimano M.G., Schillaci D., Di Sarno V., Musella S., Giovannetti E., Cirrincione G., Diana P. 2;6-Disubstituted imidazo[2;1-b][1;3;4]thiadiazole derivatives as potent staphylococcal biofilm inhibitors. Eur. J. Med. Chem. 2019;167:200–210. doi: 10.1016/j.ejmech.2019.02.007. [DOI] [PubMed] [Google Scholar]
- 22.Cascioferro S., Petri G.L., Parrino B., Carbone D., Funel N., Bergonzini C., Mantini G., Dekker H., Geerke D., Peters G.J., et al. Imidazo[2;1-b] [1;3;4]thiadiazoles with antiproliferative activity against primary and gemcitabine-resistant pancreatic cancer cells. Eur. J. Med. Chem. 2020;189:112088. doi: 10.1016/j.ejmech.2020.112088. [DOI] [PubMed] [Google Scholar]
- 23.Cascioferro S., Li Petri G., Parrino B., El Hassouni B., Carbone D., Arizza V., Perricone U., Padova A., Funel N., Peters G.J., et al. 3-(6-Phenylimidazo [2;1-b][1;3;4]thiadiazol-2-yl)-1H-Indole Derivatives as New Anticancer Agents in the Treatment of Pancreatic Ductal Adenocarcinoma. Molecules. 2020;25:329. doi: 10.3390/molecules25020329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carbone A., Cascioferro S., Parrino B., Carbone D., Pecoraro C., Schillaci D., Cusimano M.G., Cirrincione G., Diana P. Thiazole Analogues of the Marine Alkaloid Nortopsentin as Inhibitors of Bacterial Biofilm Formation. Molecules. 2020;26:81. doi: 10.3390/molecules26010081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Carbone D., Vestuto V., Ferraro M.R., Ciaglia T., Pecoraro C., Sommella E., Cascioferro S., Salviati E., Novi S., Tecce M.F., et al. Metabolomics-assisted discovery of a new anticancer GLS-1 inhibitor chemotype from a nortopsentin-inspired library: From phenotype screening to target identification. Eur. J. Med. Chem. 2022;234:114233. doi: 10.1016/j.ejmech.2022.114233. [DOI] [PubMed] [Google Scholar]
- 26.Di Franco S., Parrino B., Gaggianesi M., Pantina V.D., Bianca P., Nicotra A., Mangiapane L.R., Lo Iacono M., Ganduscio G., Veschi V., et al. CHK1 inhibitor sensitizes resistant colorectal cancer stem cells to nortopsentin. iScience. 2021;24:102664. doi: 10.1016/j.isci.2021.102664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carbone D., Parrino B., Cascioferro S., Pecoraro C., Giovannetti E., Di Sarno V., Musella S., Auriemma G., Cirrincione G., Diana P. 1,2,4-Oxadiazole Topsentin Analogs with Antiproliferative Activity against Pancreatic Cancer Cells; Targeting GSK3β Kinase. ChemMedChem. 2021;16:537–554. doi: 10.1002/cmdc.202000752. [DOI] [PubMed] [Google Scholar]
- 28.Pecoraro C., Parrino B., Cascioferro S., Puerta A., Avan A., Peters G.J., Diana P., Giovannetti E., Carbone D. A New Oxadiazole-Based Topsentin Derivative Modulates Cyclin-Dependent Kinase 1 Expression and Exerts Cytotoxic Effects on Pancreatic Cancer Cells. Molecules. 2022;27:19. doi: 10.3390/molecules27010019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kumar R., Sirohi T.S., Singh H., Yadav R., Roy R.K., Chaudhary A., Pandeya S.N. 1,2,4-triazine analogs as novel class of therapeutic agents. Mini Rev. Med. Chem. 2014;14:168–207. doi: 10.2174/1389557514666140131111837. [DOI] [PubMed] [Google Scholar]
- 30.El-Wakil M.H., Ashour H.M., Saudi M.N., Hassan A.M., Labouta I.M. Design; synthesis and molecular modeling studies of new series of antitumor 1;2;4-triazines with potential c-Met kinase inhibitory activity. Bioorg Chem. 2018;76:154–165. doi: 10.1016/j.bioorg.2017.11.006. [DOI] [PubMed] [Google Scholar]
- 31.Verma T., Sinha M., Bansal N. Heterocyclic Compounds Bearing Triazine Scaffold and Their Biological Significance: A Review. Anticancer. Agents Med. Chem. 2020;20:4–28. doi: 10.2174/1871520619666191028111351. [DOI] [PubMed] [Google Scholar]
- 32.Song J., Cui X.X., Wu B.W., Li D., Wang S.H., Shi L., Zhu T., Zhang Y.B., Zhang S.Y. Discovery of 1;2;4-triazine-based derivatives as novel neddylation inhibitors and anticancer activity studies against gastric cancer MGC-803 cells. Bioorg Med. Chem. Lett. 2020;30:126791. doi: 10.1016/j.bmcl.2019.126791. [DOI] [PubMed] [Google Scholar]
- 33.Branowska D., Karczmarzyk Z., Wolińska E., Wysocki W., Morawiak M., Urbańczyk-Lipkowska Z., Bielawska A., Bielawski K. 1,2,4-Triazine Sulfonamides: Synthesis by Sulfenamide Intermediates, In Vitro Anticancer Screening, Structural Characterization, and Molecular Docking Study. Molecules. 2020;25:2324. doi: 10.3390/molecules25102324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parrino B., Carbone D., Cascioferro S., Pecoraro C., Giovannetti E., Deng D., Di Sarno V., Musella S., Auriemma G., Cusimano M.G., et al. 1,2,4-Oxadiazole topsentin analogs as staphylococcal biofilm inhibitors targeting the bacterial transpeptidase sortase A. Eur. J. Med. Chem. 2021;209:112892. doi: 10.1016/j.ejmech.2020.112892. [DOI] [PubMed] [Google Scholar]
- 35.Dhingra U.H., Huryn D.M., Keith D.D. Preparation of substituted bisindolylmaleimides for the inhibition of cell proliferation. WO9804551 A1. World Intellectual Property Organization. 1998 February 5;
- 36.Kato M., Li J., Chuang J.L., Chuang D.T. Distinct structural mechanisms for inhibition of pyruvate dehydrogenase kinase isoforms by AZD7545, dichloroacetate, and radicicol. Structure. 2007;15:992–1004. doi: 10.1016/j.str.2007.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Amaral M., Kokh D.B., Bomke J., Wegener A., Buchstaller H.P., Eggenweiler H.M., Matias P., Sirrenberg C., Wade R.C., Frech M. Protein conformational flexibility modulates kinetics and thermodynamics of drug binding. Nat. Comm. 2017;8:2276. doi: 10.1038/s41467-017-02258-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Case D.A., Darden T.A., Cheatham T.E., Simmerling C.L., Wang J., Duke R.E., Luo R., Crowley M., Walker R.C., Zhang W., et al. AMBER 10. University of California; Davis, CA, USA: 2008. [Google Scholar]
- 39.Jakalian A., Jack D.B., Bayly C.I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 40.Bassani D., Pavan M., Bolcato G., Sturlese M., Moro S. Re-Exploring the Ability of Common Docking Programs to Correctly Reproduce the Binding Modes of Non-Covalent Inhibitors of SARS-CoV-2 Protease Mpro. Pharmaceuticals. 2022;15:180. doi: 10.3390/ph15020180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bolcato G., Cescon E., Pavan M., Bissaro M., Bassani D., Federico S., Spalluto G., Sturlese M., Moro S. A Computational Workflow for the Identification of Novel Fragments Acting as Inhibitors of the Activity of Protein Kinase CK1δ. Int. J. Mol. Sci. 2021;22:9741. doi: 10.3390/ijms22189741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bassani D., Pavan M., Sturlese M., Moro S. Sodium or Not Sodium: Should Its Presence Affect the Accuracy of Pose Prediction in Docking GPCR Antagonists? Pharmaceuticals. 2022;15:346. doi: 10.3390/ph15030346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Moro S., Deflorian F., Bacilieri M., Spalluto G. Ligand-based homology modeling as attractive tool to inspect GPCR structural plasticity. Curr Pharm Des. 2006;12:2175–2185. doi: 10.2174/138161206777585265. [DOI] [PubMed] [Google Scholar]
- 44.Pecoraro C., De Franco M., Carbone D., Bassani D., Pavan M., Cascioferro S., Parrino B., Cirrincione G., Dall’Acqua S., Moro S., et al. 1,2,4-Amino-triazine derivatives as pyruvate dehydrogenase kinase inhibitors: Synthesis and pharmacological evaluation. Eur. J. Med. Chem. 2023;249:115134. doi: 10.1016/j.ejmech.2023.115134. [DOI] [PubMed] [Google Scholar]
- 45.Carbone D., De Franco M., Pecoraro C., Bassani D., Pavan M., Cascioferro S., Parrino B., Cirrincione G., Dall’Acqua S., Moro S., et al. Discovery of the 3-Amino-1,2,4-triazine-Based Library as Selective PDK1 Inhibitors with Therapeutic Potential in Highly Aggressive Pancreatic Ductal Adenocarcinoma. Int. J. Mol. Sci. 2023;24:3679. doi: 10.3390/ijms24043679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sabbadin D., Moro S. Supervised molecular dynamics (SuMD) as a helpful tool to depict GPCR-ligand recognition pathway in a nanosecond time scale. J. Chem. Inf. Mod. 2014;54:372–376. doi: 10.1021/ci400766b. [DOI] [PubMed] [Google Scholar]
- 47.Bissaro M., Bolcato G., Pavan M., Bassani D., Sturlese M., Moro S. Inspecting the Mechanism of Fragment Hits Binding on SARS-CoV-2 Mpro by Using Supervised Molecular Dynamics (SuMD) Simulations. ChemMedChem. 2021;16:2075–2081. doi: 10.1002/cmdc.202100156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pavan M., Bolcato G., Bassani D., Sturlese M., Moro S. Supervised Molecular Dynamics (SuMD) Insights into the mechanism of action of SARS-CoV-2 main protease inhibitor PF-07321332. J. Enzyme Inhib. Med. Chem. 2021;36:1646–1650. doi: 10.1080/14756366.2021.1954919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bolcato G., Pavan M., Bassani D., Sturlese M., Moro S. Ribose and Non-Ribose A2A Adenosine Receptor Agonists: Do They Share the Same Receptor Recognition Mechanism? Biomedicines. 2022;10:515. doi: 10.3390/biomedicines10020515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hassankalhori M., Bolcato G., Bissaro M., Sturlese M., Moro S. Shedding Light on the Molecular Recognition of Sub-Kilodalton Macrocyclic Peptides on Thrombin by Supervised Molecular Dynamics. Front. Mol. Biosci. 2021;8:707661. doi: 10.3389/fmolb.2021.707661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Do P.C., Lee E.H., Le L. Steered Molecular Dynamics Simulation in Rational Drug Design. J. Chem. Inf. Model. 2018;58:1473. doi: 10.1021/acs.jcim.8b00261. [DOI] [PubMed] [Google Scholar]
- 52.Pavan M., Menin S., Bassani D., Sturlese M., Moro S. Qualitative Estimation of Protein-Ligand Complex Stability through Thermal Titration Molecular Dynamics Simulations. J. Chem. Inf. Mod. 2022;62:5715–5728. doi: 10.1021/acs.jcim.2c00995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Menin S., Pavan M., Salmaso V., Sturlese M., Moro S. Thermal Titration Molecular Dynamics (TTMD): Not Your Usual Post-Docking Refinement. Int. J. Mol. Sci. 2023;24:3596. doi: 10.3390/ijms24043596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jubelin C., Muñoz-Garcia J., Griscom L., Cochonneau D., Ollivier E., Heymann M.F., Vallette F.M., Oliver L., Heymann D. Three-dimensional in vitro culture models in oncology research. Cell. Biosci. 2022;12:155. doi: 10.1186/s13578-022-00887-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomás-Bort E., Kieler M., Sharma S., Candido J.B., Loessner D. 3D approaches to model the tumor microenvironment of pancreatic cancer. Theranostics. 2020;10:5074–5089. doi: 10.7150/thno.42441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chemical Computing Group, ULC. Molecular Operating Environment (MOE) 2022. [(accessed on 10 January 2023)]. Available online: https://www.chemcomp.com/Products.html.
- 57.Berman H.M., Westbrook J., Feng Z., Gilliland G., Bhat T.N., Weissig H., Shindyalov I.N., Bourne P.E. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.QUACPAC 2.1.3.0: OpenEye Scientific Software, Santa Fe, NM. [(accessed on 10 January 2023)]. Available online: http://www.eyesopen.com.
- 59.Korb O., Stützle T., Exner T.E. Proceedings of the Ant Colony Optimization and Swarm Intelligence: 5th International Workshop, ANTS 2006, Brussels, Belgium, 4–7 September 2006. Springer; Berlin/Heidelberg, Germany: 2006. PLANTS: Application of Ant Colony Optimization to Structure-Based Drug Design. [Google Scholar]
- 60.Pellei M., Bagnarelli L., Luciani L., Del Bello F., Giorgioni G., Piergentili A., Quaglia W., De Franco M., Gandin V., Marzano C., et al. Synthesis and Cytotoxic Activity Evaluation of New Cu(I) Complexes of Bis(pyrazol-1-yl) Acetate Ligands Functionalized with an NMDA Receptor Antagonist. Int. J. Mol. Sci. 2020;21:2616. doi: 10.3390/ijms21072616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Not applicable.