Abstract
The barley Mla locus encodes 28 characterized resistance specificities to the biotrophic fungal pathogen barley powdery mildew. We describe a single-cell transient expression assay using entire cosmid DNAs to pinpoint Mla1 within the complex 240-kb Mla locus. The MLA1 cDNA encodes a 108-kD protein containing an N-terminal coiled-coil structure, a central nucleotide binding domain, and a C-terminal leucine-rich repeat region; it also contains a second short open reading frame at the 5′ end that has a possible regulatory function. Although most Mla-encoded resistance specificities require Rar1 for their function, we used the single-cell expression system to demonstrate that Mla1 triggers full resistance in the presence of the severely defective rar1-2 mutant allele. Wheat contains an ortholog of barley Mla, designated TaMla, that is tightly linked to (0.7 centimorgan) but distinct from a tested resistance specificity at the complex Pm3 locus to wheat powdery mildew. Thus, the most polymorphic powdery mildew resistance loci in barley and wheat may have evolved in parallel at two closely linked homeoloci. Barley Mla1 expressed in wheat using the single-cell transformation system failed to trigger a response to any of the wheat powdery mildew Avr genes tested, indicating that AvrMla1 is not genetically fixed in wheat mildew strains.
INTRODUCTION
Powdery mildew fungi are one of the most widespread pathogens of plants and infect thousands of dicot and monocot species. These obligate biotrophic fungi are ectoparasites and attack epidermal tissue exclusively (Jørgensen, 1988). Growth of the fungus is almost entirely external, and infection is limited to the formation of haustoria within epidermal cells for the purpose of nutrient retrieval. Blumeria graminis f sp hordei is one of the grass powdery mildew pathogens, which colonize only species of a single genus, Hordeum (barley), a feature shared by other grass powdery mildews (Jørgensen, 1988). Resistance to this fungus can be mediated by one of at least three genetically separable pathways in Hordeum. Two of these pathways involve recognition of isolate-specific fungal determinants, whereas the third pathway is thought to function by potentiating a broad-spectrum defense response (reviewed in Schulze-Lefert and Vogel, 2000). Isolate-specific resistance in the former two pathways is triggered by a multitude of powdery mildew resistance genes (Mlx) and is almost invariably associated with the activation of a rapid host cell death (hypersensitive response [HR]) at sites of attempted fungal ingress.
Many of the powdery mildew R genes require for their function two additional genes, Rar1 and Rar2 (Freialdenhoven et al., 1994; Jørgensen, 1996). Rar1 has been shown to encode a small, highly conserved cytoplasmic Zn2+ binding protein and to function upstream of a whole-cell H2O2 burst that precedes HR, thus demonstrating a role for Rar1 in R gene signaling (Shirasu et al., 1999a).
The Mla locus encodes an exceptionally large number of characterized resistance specificities, each recognizing unique fungal determinants that are encoded by cognate fungal avirulence (Avr) genes (Jørgensen, 1994). Curiously, although many tested Mla resistance genes require Rar1 and Rar2 for their function, some appear to have different signaling requirements (Jørgensen, 1996). The Mla locus is also of interest because of the diversity of resistant phenotypes that are conferred by different Mla resistance specificities. These phenotypes can range from near immunity, associated with a rapid single-cell epidermal HR and early growth arrest of the fungus, to a late and spatially extended HR consuming mesophyll cells, allowing the development of some fungal mycelium (Boyd et al., 1995).
Recently, the Mla locus was mapped physically to a 240-kb region on barley chromosome 1HS that exhibits suppressed recombination (Wei et al., 1999). Sequencing of bacterial artificial chromosome DNA clones from a cultivar lacking a known Mla resistance specificity indicated the presence of eight genes with products similar to those of nucleotide binding leucine-rich repeat (NB-LRR)–type R genes, the predominant class of known plant R genes. On the basis of sequence similarity, the R gene homologs (RGH) at Mla were classified into three families, RGH1, RGH2, and RGH3.
In this study, we identify molecularly the gene encoding Mla1 specificity using a single-cell transient expression assay. Unique mutational events identified in susceptible Mla1 mutant lines corroborate the functional identification of the candidate gene. We examine Mla1 signaling requirements by taking advantage of its cell-autonomous activity. Comparative mapping of barley Mla-derived probes in wheat provides insight into the evolution of the most polymorphic resistance loci in barley and wheat, Mla and Pm3, respectively. Functional analysis of barley Mla1 in wheat implies that powdery mildew R–Avr gene interactions within the Triticeae tribe have evolved in a genus-specific manner.
RESULTS
Molecular Analysis of Susceptible Mla1 Mutants
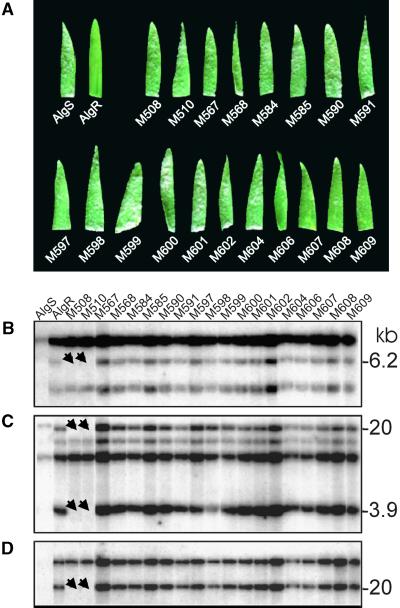
Approximately 39,000 M2 seedlings derived from γ-irradiated M1 seed of the Mla1-containing barley line CI-16137 (AlgR) were screened for altered infection phenotypes upon challenge with the AvrMla1-containing powdery mildew isolate CR3 (see Methods). A majority of the mutants (47 of 54) were fully susceptible to race CR3. The remaining mutants were only partially susceptible. Of the fully susceptible mutants, 19 were used in this study. Challenge with another AvrMla1-containing isolate (K1) showed the same susceptibility in M3 families derived by selfing of M2 candidates (Figure 1A). These data suggest that susceptibility in the 19 candidate mutants was due to mutations either in Mla1 or in genes required for its function.
Figure 1.
Phenotypic and Molecular Characteristics of Susceptible Mla1 Mutants.
(A) Primary leaf tip fragments were collected from the infected parent lines, AlgR (resistant) and AlgS (susceptible), and the Mla1 mutants at 7 days after inoculation with the K1 (AvrMla1) powdery mildew isolate. Powdery mildew hyphae are visible as white spots.
(B) to (D) Results of sequential hybridization to a single blot (HindIII digestion) with probes RGH1a (B), MWG2083 (C), and MWG2197 (D) using normal stringency wash conditions (2 × SSC, 0.5% SDS for 30 min and 0.1 × SSC, 0.5% SDS for 30 min at 65°C; 1× SSC is 0.15 M NaCl and 0.015 M sodium citrate). Arrows indicate the deletions in M508 and M510.
Because many of the candidate mutants originated from γ-ray–treated seed, we reasoned that some of them may contain easily detectable DNA rearrangements at the Mla locus (such as large insertions or deletions). We surveyed gel blots containing DNA of the 19 susceptible mutants with DNA probes representing the Mla-encoded RGH1, RGH2, and RGH3 families (see Methods; Wei et al., 1999). Only the probe RGH1a detected DNA polymorphisms among the mutants. A 6.2-kb HindIII restriction fragment, detected by the RGH1a probe in AlgR and most of the mutants, was missing in mutant lines M508 and M510 (Figure 1B). When the same blots were probed with the restriction fragment length polymorphism (RFLP) markers MWG2083 and MWG2197, which are known to cosegregate with and map 0.05 centimorgan (cM) distal to Mla, respectively, we again noted the absence of hybridization signals in mutant lines M508 and M510 (Figures 1C and 1D). This suggested a large deletion in these two mutants at Mla of at least 140 kb based on the estimated physical distance between RFLPs MWG2083 and MWG2197 (Wei et al., 1999).
To obtain a genomic copy of the region containing the Mla1 gene, we constructed a cosmid library from the Mla1-containing cultivar AlgR (see Methods). Twelve cosmid clones were then isolated using various RGH and RFLP probes known to map at the Mla locus (Table 1; Wei et al., 1999). Four different cosmid clones were isolated with probe RGH1a, which was shown to detect mutation-induced polymorphisms in the susceptible mutant lines M508 and M510. However, only one of the isolated cosmids, p6-49-2, contained an RGH1a-homologous HindIII fragment of the same size as that deleted in mutants M508 and M510 (data not shown). Thus, cosmid p6-49-2 was identified as a candidate to contain the Mla1 gene and was used in the first round of functional tests.
Table 1.
Cosmids Isolated from AlgR (Mla1) Library
|
Mla-Linked Probes
| ||||
|---|---|---|---|---|
| RGH1a | RGH1b | 80 H14-R1.1 | MWG2083 | MWG2197 |
| p5-33-1 | p7-35-1 | p3-9-1 | p7-35-2 | p5-3-1 |
| p5-42-2 | p4-42-1 | p7-24-1 | ||
| p6-49-2 | p4-25-4 | |||
| p7-36-2 | p6-16-1 | |||
A Single-Cell Expression System to Identify Mla1-Containing Cosmids
Although susceptibility in mutants M508 and M510 is likely to be the result of the detected deletion events at Mla, the large size of the deletions prevented us from using these mutants to determine the gene encoding Mla1. Therefore, we devised an alternative experimental route to identify candidates for Mla1. This route was based on the transient single-cell expression of candidate genes that are introduced into epidermal cells of detached leaf segments by particle bombardment (Shirasu et al., 1999b). This approach takes advantage of the recessive nature of mlo resistance and capitalizes on the fact that mlo- and R gene–mediated resistance signal through separate pathways in barley (Peterhänsel et al., 1997). Figure 2 shows a scheme of the assay (designated hereafter the “three-component test”) aimed at identifying candidate genomic clones containing Mla1. Biolistic delivery of DNA of the construct pUGLUM (see Methods), which harbors the genes Mlo and GFP, into leaf epidermal cells of an mlo-resistant genotype and subsequent challenge inoculation with powdery mildew spores results in single-cell complementation of mlo resistance in the leaf epidermis (Figure 2; Shirasu et al., 1999b). This complementation can be quantified by counting the number of green fluorescent protein (GFP)–expressing epidermal cells that support the growth of a sporulating fungal colony 5 days after spore inoculation. Importantly, single Mlo-expressing and GFP-marked cells become susceptible, although all neighboring nontransformed cells respond to pathogen challenge with mlo-mediated resistance. We expected that cotransformation of genomic DNA clones harboring Mla1 together with the pUGLUM construct would lead to R gene–triggered resistance in GFP-expressing cells after challenge inoculation with an AvrMla1-containing isolate only (Figure 2).
Figure 2.
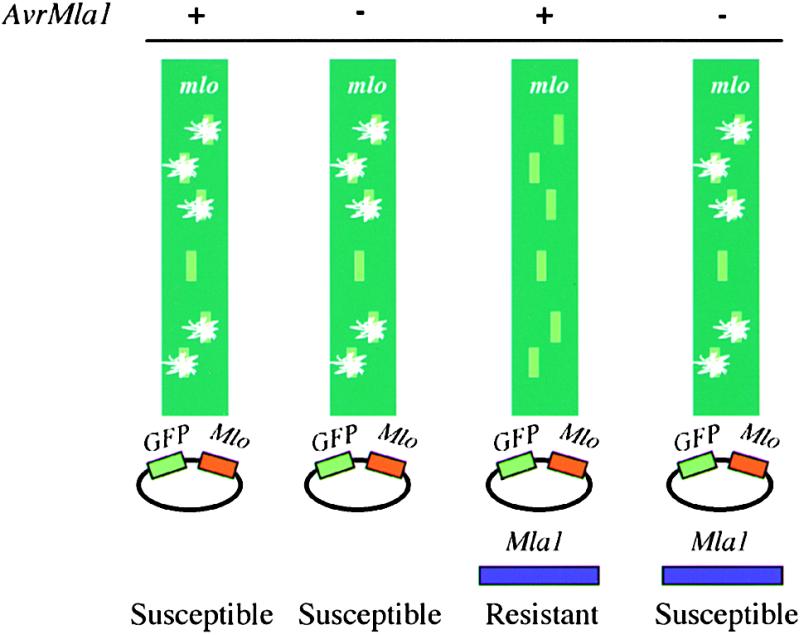
Scheme of the Three-Component Single-Cell Transient Expression Assay.
The mlo-mediated broad-spectrum resistant barley plants were used for the transient expression assay. After bombardment with pUGLUM (expression of GFP and Mlo driven by the Ubi promoter) and inoculation with either A6 (lack of AvrMla1) or K1 (containing AvrMla1), colony growth was observed by UV light microscopy only in the transformed GFP- and Mlo-expressing cells. The percentage of the GFP cells with colonies was dependent on inoculation density (to determine the proportion of challenged GFP cells) and particle-coating quality. In this study, >90% of GFP-expressing cells were usually challenged by a germinated spore. When mlo leaves were bombarded with pUGLUM together with an Mla1-containing cosmid and inoculated with either A6 or K1, fungal colonies growing from the transformed epidermal cells were observed in the case of the virulent isolate, whereas no fungal colony growth was observed in the case of the avirulent isolate (i.e., the cosmid-conferred resistance was race specific).
Table 2 summarizes transient expression data obtained upon transformation with the pUGLUM construct with various cosmid clones from the Mla1 locus and challenge with fungal isolates K1 (containing AvrMla1) or A6 (lacking AvrMla1). Isolates K1 and A6 gave comparable numbers of colonies on GFP-expressing epidermal cells (38 and 35%, respectively) when bombardment was performed with the pUGLUM construct only. The proportion of GFP cells with colonies was also similar with isolates K1 and A6 when pUGLUM was cobombarded with either cosmid p7-35-1 or p7-35-2 (46 and 43% for K1 and 46 and 37% for A6, respectively). However, the results of inoculation with K1 but not A6 were altered in two respects when pUGLUM was introduced together with cosmid p6-49-2. First, the percentage of GFP-expressing cells supporting K1 growth (12.2%) was clearly lower compared with that of GFP cells supporting A6 colony formation (44.1%). Second, the number of detectable GFP-expressing cells after inoculation was lower with K1 than with A6 (148 versus 263). We interpreted the former effect as evidence of the presence of Mla1 in cosmid p6-49-2 conferring race-specific resistance to isolate K1. The latter effect could be explained by GFP protein inactivation resulting from frequent activation of the Mla1-triggered HR operating in the single transformed epidermal cells (Boyd et al., 1995).
Table 2.
Identification of Candidate Cosmids Containing Mla1 by Transient Expression in mlo Leavesa
| pUGLUM
|
pUGLUM + p7-35-1 |
pUGLUM + p7-35-2 |
pUGLUM + p6-49-2 |
pUGLUM + p6-49-2-15 |
pUGLUM + p6-49-2-7 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Fungal Isolates | GFP + Spore |
GFP + Colony |
GFP + Spore |
GFP + Colony |
GFP + Spore |
GFP + Colony |
GFP + Spore |
GFP + Colony |
GFP + Spore |
GFP + Colony |
GFP + Spore |
GFP + Colony |
| −AvrMla1 | 412 | 156 | 147 | 67 | 97 | 42 | 263 | 116 | 136 | 56 | 148 | 67 |
| 37.9% | 45.6% | 43.3% | 44.1% | 41.2% | 45.3% | |||||||
| +AvrMla1 | 495 | 175 | 140 | 57 | 85 | 31 | 148 | 18 | 79 | 2 | 139 | 53 |
| 35.4% | 40.7% | 36.5% | 12.2%b | 2.5%b | 38.1% | |||||||
GFP + Spore indicates number of GFP-expressing epidermal cells attacked by a germinated fungal spore; GFP + Colony indicates number of GFP-expressing epidermal cells supporting a sporulating fungal colony. Data in the table were pooled from at least two independent experiments showing similar trends.
Indicates the significant effect of candidate cosmids on the reduction of colony growth of K1 (AvrMla1) from transformed cells.
Molecular Characterization of Mla1
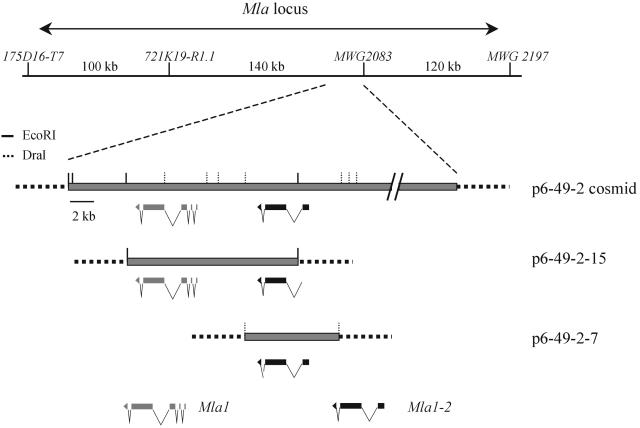
Next, we determined a contiguous stretch of 26.5 kb of DNA sequence from cosmid p6-49-2. We initiated a search for candidate genes in the 26.5-kb contig using the BLAST2 algorithm (Altschul and Gish, 1996) and available databases. We also tested for regions exhibiting high coding probabilities. This analysis revealed only two conceptual genes within the contig (Figure 3), both of which encode proteins with similarity to NB-LRR–type R genes (see below). To determine whether one or both of these candidates mediates AvrMla1-dependent resistance, we isolated two subclones of cosmid p6-49-2. Subclone p6-49-2-15 contains a predicted full-length copy of one of the two NB-LRR genes and a truncated copy of the other (Figure 3). Subclone p6-49-2-7 contains a predicted full-length copy of the gene that is truncated in p6-49-2-15 (Figure 3). Subclone p6-49-2-15 but not p6-49-2-7 showed a dramatic decrease in the proportion of GFP cells supporting growth of the K1 isolate relative to the A6 isolate (2.5 and 41.2%; Table 2). These data corroborate the conclusion that cosmid p6-49-2 may contain Mla1 and show that only one of the two candidate genes within this cosmid mediates AvrMla1-dependent activity.
Figure 3.
Scheme of Mla1-Containing Cosmids and the Subclones.
Cosmid p6-49-2 was isolated by screening a library with the RGH1a probe. RGH1a was derived from the bacterial artificial chromosome clone 80H14, which was mapped genetically and physically to the Mla locus (Wei et al., 1999). Two full-length NB-LRR genes are predicted to be encoded by the 26.5-kb contiguous sequence contained in the p6-49-2 cosmid. Both of them were deleted in the Mla1 mutants M508 and M510 (see Figure 5). The two NB-LRR genes were subcloned into p6-49-2-15 and p6-49-2-7 using EcoRI and DraI digestion, respectively. Each subclone contains one full-length NB-LRR gene.
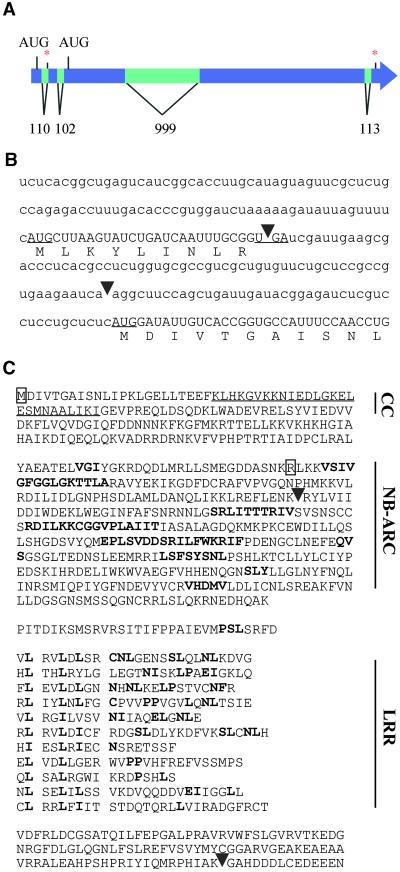
Primers derived from the candidate gene in subclone p6-49-2-15 were used to obtain the full-length MLA1 candidate cDNA sequence from leaf RNA of cultivar AlgR (see Methods). A comparison of genomic and cDNA sequences revealed five exons, each flanked by splice site consensus sequences (Figure 4A). Two open reading frames were found upon inspection of the full-length cDNA: a long one encoding a predicted protein of 108.6 kD and a short one, located 5′ to the other, encoding a predicted peptide of 1.1 kD (Figures 4B and 4C). Sequence analysis of the deduced 108.6-kD protein revealed sequence similarities to known plant R gene products and a tripartite arrangement of modules comprising a predicted N-terminal coiled-coil structure, a central ATP/GTP binding domain (NB-ARC; van der Biezen and Jones, 1998), and 11 imperfect C-terminal LRRs. No sequence similarities to characterized proteins were found for the predicted small 1.1-kD peptide.
Figure 4.
Mla1 Is a Coiled-Coil NB-LRR Protein.
(A) Scheme of the open reading frames, introns, and exons in the Mla1 gene. Introns are represented by lighter bars. Asterisks mark stop codon positions.
(B) 5′-end sequence of Mla1 mRNA and corresponding translation. Arrowheads indicate the intron positions of introns 1 and 2 Mla1 at the 5′ end of mRNA. Start and stop codons of the upstream 1.1-kD open reading frame are underlined. The start codon of the 108.6-kD MLA1 protein is also underlined. Lowercase letters indicate untranslated 5′-end mRNA; uppercase letters indicate coding 5′-end mRNA sequences. Uppercase letters below the RNA sequence indicate deduced protein sequences of the 1.1-kD upstream open reading frame and of the N terminus of MLA1.
(C) Amino acid sequences and the conserved motifs of MLA1. The coiled-coil domain (underlined, CC) was identified with the COILS modeling server described in Methods. Boldface letters in the NB-ARC and LRR regions indicate conserved amino acid motifs to known NB-LRR proteins. The outlined M indicates the point mutation (M→V) in mutants M567, M568, M584, M585, M590, M591, M597, M599, M600, M601, M602, M604, M606, M607, M608, and M609, and the outlined R indicates the point mutation (R→STOP codon) in mutant M598. Arrowheads indicate the intron positions in the corresponding genomic Mla1 sequence. The MLA1 cDNA sequence GenBank accession number is AY009939, and the Mla1 sequence GenBank accession number is AY009938.
Mla1 Mutants Reveal Unique Sequence Alterations in the Mla1 Candidate Gene
To obtain complementary evidence that the candidate gene in subclone p6-49-2-15 is Mla1, we sequenced DNA from 17 susceptible Mla1 mutants (as shown in Figure 1A except for M508 and M510) and compared these sequences with the gene sequence derived from the resistant line AlgR. Mutant M598 was found to contain an A→T change at position 806 of the cDNA, thus generating a stop codon early in the nucleotide binding domain. Sixteen other mutants each contain an A→G change at position 230, substituting a valine for the first methionine of the predicted NB-LRR protein. The identification of identical mutational changes in these mutants suggests that they are likely to have derived from only two independent mutational events. Because mutants M508 and M510 (described above) had been shown to contain a deletion affecting the Mla1 candidate gene, we have identified three different mutational events in the candidate gene that are correlated with the loss of Mla1-specified resistance within the mutant collection. Together with the race-specific resistance resulting from the transient expression of subclone p6-49-2-15 in single leaf epidermal cells, these mutants demonstrate that we have isolated the Mla1 gene. We have designated the sequence-related but functionally inactive homolog of Mla1 that is present in the p6-49-2 clone Mla1-2.
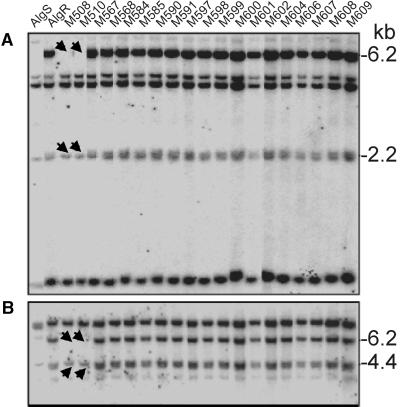
Genomic DNA gel blot hybridization of the congenic lines AlgS and AlgR and the susceptible mutants with probes from the MLA1 NB-ARC or LRR domains generated complex hybridization signals (Figure 5). This finding suggests the presence of multiple sequence-related Mla1 homologs in both resistant AlgR and susceptible AlgS lines. Both probes detected restriction fragments missing in DNA of mutants M508 and M510. The deleted fragments are identical in length to two genomic fragments that are predicted from the 26.5-kb sequence contig of cosmid p6-49-2 and that encompass Mla1 and Mla1-2. This suggests that the large deletion in mutants M508 and M510 includes, in addition to Mla1, only a single Mla1 homolog, Mla1-2, that was shown to be juxtaposed physically with Mla1 in cosmid p6-49-2.
Figure 5.
DNA Hybridization Analysis of Mla1 Mutants Using Mla1 Probes.
(A) A genomic DNA gel blot of the parents and Mla1 mutant lines digested with HindIII hybridized with MLA1-NB probe.
(B) The same DNA gel blot hybridized with MLA1-LRR probe.
The arrows indicate the deletions in M508 and M510. Normal stringency wash conditions (2 × SSC, 0.5% SDS for 30 min and 0.1 × SSC, 0.5% SDS for 30 min at 65°C) were used for both probes.
Mla1 Function Is Independent of Rar1
Previous genetic data provided evidence that Mla1, unlike most other resistance specificities encoded at Mla, does not require Rar1 for its function (Jørgensen, 1996). To further test the signaling requirements of Mla1, we took advantage of the cell-autonomous function of Mla1 resistance in the transient three-component expression assay. We compared Mla1 activity in mlo-resistant lines containing either wild-type Rar1 or a rar1 mutant allele (genotypes Rar1 mlo and rar1-2 mlo; see Methods). These two genetic lines gave similar proportions of GFP cells, supporting K1 growth after bombardment with pUGLUM alone containing the GFP reporter gene and wild-type Mlo (50 versus 48% in genotypes Rar1 mlo and rar1-2 mlo; Table 3). Similarly, comparable levels of single-cell susceptibility were recorded in the Rar1 mlo and rar1-2 mlo lines when pUGLUM was cobombarded with p6-49-2-7, encoding Mla1-2 (41 and 42%, respectively). In contrast, complete resistance was observed in both wild-type Rar1 and mutant rar1 cells upon cobombardment of pUGLUM with p6-49-2-15, encoding Mla1 (Table 3). These findings provide evidence that Rar1 is not required for the function of Mla1.
Table 3.
Mla1 Function Does Not Require Rar1a
| pUGLUM
|
pUGLUM + p6-49-7
|
pUGLUM + p6-49-15
|
||||
|---|---|---|---|---|---|---|
| Genotypes | GFP + Spore | GFP + Colony | GFP + Spore | GFP + Colony | GFP + Spore | GFP + Colony |
| 102 | 50 | 109 | 45 | 41 | 0 | |
| Rar1 mlob | ||||||
| 49.0% | 41.3% | 0.0% | ||||
| 161 | 76 | 186 | 80 | 68 | 0 | |
| rar1 mlob | ||||||
| 47.2% | 43.0% | 0.0% | ||||
GFP + Spore indicates number of GFP-expressing epidermal cells attacked by a germinated fungal spore; GFP + Colony indicates number of GFP-expressing epidermal cells supporting a sporulating fungal colony. The data in the table were pooled from three independent experiments showing similar trends.
Inoculated with K1 (AvrMla1).
Barley Mla and Wheat Pm3 May Represent Distinct Homeoloci
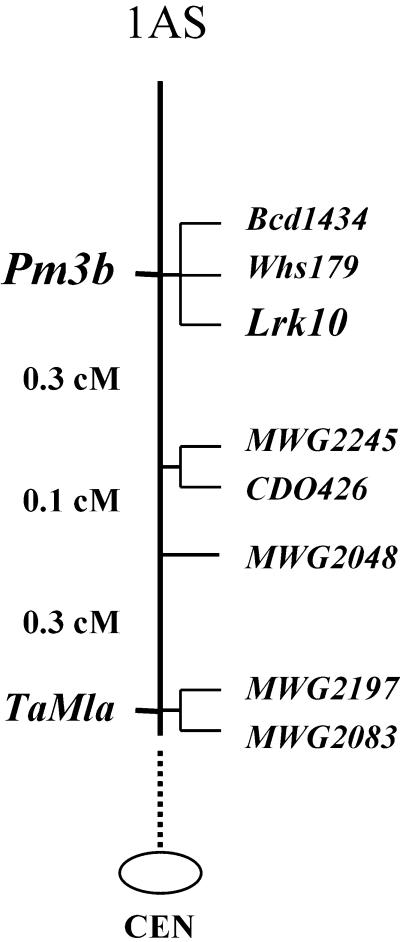
In wheat, the Pm3 locus encodes a large number of resistance specificities against wheat powdery mildew (Zeller et al., 1993). It has been suggested that wheat Pm3 may represent an ortholog of barley Mla, because both resistance loci are known to map on homologous chromosome arms and within orthologous marker intervals (Hartl et al., 1993). To test this hypothesis, a cDNA fragment encoding the MLA1-LRR and probes MWG2197 and MWG2083, which are known to cosegregate with and map 0.05 cM distal to barley Mla, respectively, were used to map RFLP loci in an F2 population of 476 individuals (Figure 6) derived from a cross between wheat lines nearly isogenic for the Pm3b specificity (see Methods). All three barley probes detected orthologs at a single locus on chromosome 1AS. However, seven recombinant plants were found that placed the Mla1 ortholog (designated TaMla) 0.7 cM proximal to Pm3b.
Figure 6.
Genetic Map of Pm3b and TaMla on Chromosome 1AS of Wheat.
The wheat powdery mildew resistance locus Pm3b cosegregated with RFLP markers Bcd1434 and Whs179 and a receptor-like kinase gene, Lrk10, in an F2 population derived from the cross between Chul/8*Chancellor, carrying Pm3b, and the susceptible line Chancellor. The ortholog TaMla of the barley powdery mildew resistance locus Mla was mapped 0.7 cM proximal to Pm3b using Mla1 as a hybridization probe. The Mla1, MWG2197, and MWG2083 probes detected RFLP loci that cosegregated in wheat. CEN, centromere.
Functional Analysis of Mla1 in Wheat
Next, we wanted to determine whether barley Mla1 can function in wheat. However, when the single-cell transformation system is used in an Mlo-susceptible line (i.e., a line not containing mlo resistance), single epidermal GFP cells transformed using the particle bombardment method cannot be assessed for external fungal growth because of the presence of hyphae arising from neighboring cells. Furthermore, fungal growth at the earlier stage of haustorium establishment cannot be assessed because these internal structures cannot be observed under the UV light used to visualize GFP. The absence of mlo resistance in wheat therefore precludes the use of the Mlo/GFP-facilitated functional assay in wheat. Instead, we used a GUS reporter–based functional assay developed for the study of cereal–powdery mildew interactions (Schweizer et al., 1999). In this system, haustoria and β-glucuronidase (GUS) expression within individual transformed epidermal cells can be visualized concurrently. Because Mla1 acts at an early stage, during cell wall penetration and before haustorium formation (Boyd et al., 1995), we anticipated that Mla1 activity, if present in wheat, could be assessed on the basis of haustorium formation (and the appearance of secondary hyphae) 66 hr after inoculation.
Race-specific recognition of AvrMla1 from barley powdery mildews could not be tested in wheat, because all available AvrMla1-containing isolates evoked a nonhost response in wheat, preventing initial penetration of the epidermal cell wall by the fungus (data not shown). Therefore, we tested potential resistance responses to three wheat powdery mildew isolates (JIW2, JIW48, and FZ1) containing 10 characterized avirulence and nine virulence genes and each one virulent on wheat cultivar Cerco (Table 4; see Meth-ods). Each of the tested fungal isolates was capable of establishing differentiated haustoria in 30 to 38% of challenged GUS-expressing epidermal host cells transformed using the construct containing the GUS gene alone (pUGUS; Table 4). Comparable frequencies of successful haustorium formation were also observed after cobombardment with genomic DNA encoding the barley genes Mla1-2 and Mla1 (Table 4), indicating the absence of detectable Mla1 and Mla1-2 activity in wheat. This finding suggests either that an Avr gene recognized by Mla1 is not present in the tested wheat fungal isolates or that other host components required for Mla1 resistance are missing in wheat.
Table 4.
Mla1 Expression in Wheat Does Not Confer Resistance to the Wheat Powdery Mildew Fungusa
| pUGUS
|
pUGUS + p6-49-7
|
pUGUS + p6-49-15
|
||||
|---|---|---|---|---|---|---|
| Isolates | GUS + Spore | GUS + Haustoria | GUS + Spore | GUS + Haustoria | GUS + Spore | GUS + Haustoria |
| 169 | 52 | 232 | 73 | 360 | 105 | |
| JIW48 | ||||||
| 30.8% | 31.5% | 29.2% | ||||
| 84 | 32 | 78 | 28 | 75 | 32 | |
| JIW2 | ||||||
| 38.1% | 35.9% | 42.7% | ||||
| 75 | 23 | 99 | 41 | 92 | 26 | |
| FZ1 | ||||||
| 30.7% | 41.4% | 28.3% | ||||
GUS + Spore indicates number of GUS-expressing epidermal cells attacked by a germinated fungal spore; GUS + Haustoria indicates number of GFP-expressing epidermal cells containinging a fungal haustorium. The data in the table were pooled from two independent experiments showing similar trends.
DISCUSSION
The Three-Component Transient Expression Assay, a Potentially Versatile Tool
The three-component assay developed in this study was instrumental in the molecular isolation and functional analysis of barley Mla1. The simultaneous expression of GFP, Mlo, and Mla1 enabled us to reveal Mla1 activity in single transformed leaf epidermal cells because neighboring, untransformed cells respond with broad-spectrum mlo-type resistance to fungal attack. This makes it possible to detect, even late after spore inoculation, the few single-cell transformation events that otherwise would become masked by spreading fungal mycelium that originates from the majority of nontransformed susceptible cells. Although transformation of both cosmid DNA from p6-49-2 and DNA from subclone p6-49-2-15 indicated that Mla1 activity was detectable in the transient expression assay, a significant proportion of GFP-expressing cells retained susceptibility upon challenge with the AvrMla1 isolate (12.2 and 2.5% of GFP-expressing cells, respectively). In general, we observed that the number of these “escape events” in the single-cell assay is positively correlated with the complexity of cobombarded DNA molecules (note that a 1:2 molar ratio of plasmid to cosmid DNA was used for the cotransformation and that p6-49-2 [45 kb] is threefold larger than p6-49-2-15 [15 kb]). The greater number of escape events recorded in the presence of the entire cosmid p6-49-2 therefore may reflect difficulties in coating carrier particles homogeneously with DNA from each of the three genes. Nevertheless, it is notable that the three-component assay is sufficiently robust to detect Mla1 activity upon transformation of the entire cosmid DNA. Apart from its use in the identification of other powdery mildew R genes (Halterman et al., 2001), this assay should become a broadly applicable tool for facile structure/function analysis of MLA proteins and for studies of Mla signaling, as shown here in the context of Rar1.
Mla1 Encodes a CC-NB-LRR Protein
The deduced protein sequence of Mla1 reveals a modular domain architecture and significant sequence similarities to plant NB-LRR proteins. NB-LRR proteins are the predominant class of known plant R proteins (Ellis and Jones, 1998). These are further subdivided on the basis of sequences close to the N termini. One subclass shows similarities to the cytoplasmic domains of the Drosophila Toll and mammalian interleukin-1 receptors, the TIR-NB-LRR proteins. The other subclass has N-terminal sequences with the potential to form coiled-coil structures, the CC-NB-LRR proteins. Because a few members of the latter subclass have sequence signatures in common with leucine zippers, a subtype of coiled-coil domains, CC-NB-LRR proteins have occasionally been called LZ-NB-LRR proteins. A single stretch of 26 amino acids (K24 to I50) of the MLA1 peptide is predicted to adopt a coiled-coil structure ( ; Lupas et al., 1991) similar in location to the predicted coiled-coil segments of other CC-NB-LRR proteins. This enables us to assign MLA1 to the CC-NB-LRR subclass of NB-LRR proteins.
; Lupas et al., 1991) similar in location to the predicted coiled-coil segments of other CC-NB-LRR proteins. This enables us to assign MLA1 to the CC-NB-LRR subclass of NB-LRR proteins.
Successful detection of Mla1 activity in single epidermal cells demonstrates a cell-autonomous function for this R gene and identifies the first attacked host cell as being critical in mediating the growth arrest of the fungus. The lack of detectable transmembrane helices and transit or leader peptide sequences in MLA1 suggests an intracellular localization, consistent with other NB-LRR plant R proteins. Thus, it is likely that the recognition event in MLA1-specified resistance occurs intracellularly. This is notable given the behavior of powdery mildew fungi: they do not show invasive growth in their hosts but merely invaginate the plasma membrane of attacked epidermal cells for haustorium differentiation and nutrient uptake. This suggests that the fungus transports molecules, such as Avr proteins, across the fungal and plant plasma membranes before the MLA1 protein can mediate recognition of the intruder.
Although we have not tested the functional significance of the upstream open reading frame (uORF) in the 5′ leader of the MLA1 mRNA, the presence of a similar uORF in the mRNA of MLA6 suggests that this is a common feature of MLA mRNAs (Halterman et al., 2001). The presence of uORFs in 5′ mRNA leaders is known to suppress the translation of specific mRNA molecules and is in many cases subject to regulation (Hershey, 1991; Geballe and Morris, 1994). Putative uORF-mediated post-transcriptional regulation has not been reported for other CC-NB-LRR proteins. However, it is known that several TIR-NB-LRR–type plant R genes encode multiple transcripts, which may be involved in another form of post-transcriptional control of R gene function (Whitham et al., 1994; Ayliffe et al., 1999). The tobacco N gene, for example, encodes two alternatively spliced transcripts that are both necessary for full resistance to the tobacco mosaic virus, and the splicing process itself appears to be subject to tobacco mosaic virus–induced signals (Dinesh-Kumar and Baker, 2000).
The RGH1a probe, used here to detect the Mla1 deletion in the AlgR-derived mutant lines M508 and M510, encodes one of several NB-LRR homologs that were identified previously at the Mla locus in cultivar Morex, which lacks any known Mla resistance specificity (Wei et al., 1999; F. Wei and R.P. Wise, unpublished data). A comparison of predicted MLA1, MLA1-2, and RGH1a proteins reveals sequence identities ranging from 77 to 85%, suggesting that these proteins may have diverged recently. This interpretation is corroborated by their common gene structure, as demonstrated by the same number of introns and exons as well as conserved intron/exon junctions (data not shown). A comparison of the deduced MLA1 and MLA6 proteins, however, reveals that both are more closely related to each other (91% identity) than to the MLA1-2 homolog or to any of the Mla-encoded NB-LRR genes that were identified in a cultivar lacking Mla resistance specificity (Wei et al., 1999; Halterman et al., 2001). This exceptional sequence similarity may indicate that different Mla resistance specificities represent alleles of the same gene at the complex Mla locus, an interpretation that is supported by preliminary sequence information obtained from Mla12- and Mla13-containing cultivars (Halterman et al., 2001).
Mla1 Signaling Requirements
A previous study provided evidence that the majority of tested Mla resistance specificities require for their function at least two additional genes, Rar1 and Rar2 (Jørgensen, 1996). These data were inferred from complex segregation ratios of infection phenotypes in F2 progeny derived from crosses between various Mla-resistant lines and mutants in Rar1 and Rar2. Because the previous study involved crosses between different cultivars, it was not possible to exclude genetic background effects on the infection phenotypes. Here, we used the single-cell assay to demonstrate that Mla1 expression triggers a full resistance response in both rar1-2 mutant and Rar1 wild-type plants, thereby corroborating the earlier genetic data that Mla1 function does not require Rar1. Intriguingly, Mla6 function, contrary to that of Mla1, is fully dependent on Rar1, although the two deduced proteins are 91% identical in sequence (Halterman et al., 2001).
The cause of the different signaling requirements of Mla-encoded R genes is open to speculation. Neither of the two available Rar1-defective alleles, rar1-1 or rar1-2, is a transcriptional null mutant, suggesting that residual wild-type Rar1 activity even in the severely defective rar1-2 allele may be sufficient for some but not all Mla resistance functions. (Mutant rar1-2 plants are defective in Rar1 transcript splicing, minimal amounts of wild-type mRNA are still detectable using polymerase chain reaction [PCR] amplification, and the rar1-1 mutant allele compromises resistance to a lesser extent than rar1-2 [Shirasu et al., 1999a].) This scenario is unlikely because a highly reactive Rar1 antiserum detects the Rar1 protein in both Rar1 and rar1-1 leaf extracts and fails to detect a signal in the rar1-2 mutant (A. Sandanandom and P. Schulze-Lefert, unpublished data), suggesting that rar1-2 may be a null mutant at the protein level. The differential signaling requirements of Mla1 and Mla6 are also unexpected, because resistance triggered by both genes is associated with a rapid HR of attacked epidermal cells coincident with haustorium differentiation and at an early time during attempted infection (Boyd et al., 1995). This contrasts with many other Mla genes, which trigger an HR at a later stage of the infection process, and makes it unlikely that the different Mla1 and Mla6 signaling requirements reflect grossly different temporal or spatial Avr gene activities during pathogen invasion. Moreover, the single-cell assay demonstrates directly that both Mla1 and Mla6 act in a cell-autonomous fashion in the first attacked epidermal cell. It is possible that few amino acid differences between the MLA1 and MLA6 proteins determine directly interactions with distinct downstream signaling components. Alternatively, each MLA1 and MLA6 may form distinct protein complexes involving cognate Avr gene products and other host factors. In this model, signaling specificity may be determined by unrelated proteins that are present in MLA1 or MLA6 recognition complexes.
Parallel Evolution of Mla and Pm3 R Loci in Barley and Wheat?
The genera Hordeum and Triticum are thought to have evolved recently (∼12 million years ago) and belong to the same tribe of grasses, the Triticeae (Wolfe et al., 1989). Three barley probes, derived from the Mla locus and including an MLA1-LRR cDNA fragment, detected orthologs in wheat that cosegregated on chromosome arm 1AS. Because the tested barley probes encompass a physical interval of >100 kb at Mla (Wei et al., 1999), their cosegregation in wheat is strong evidence for an orthologous locus, designated TaMla. Although previous studies predicted that Pm3, the most polymorphic R locus in wheat to wheat powdery mildew, could be an ortholog of barley Mla (Hartl et al., 1993), our data show that at least Pm3b and Mla orthologs in wheat define separate but tightly linked loci. Although crosses between wheat lines nearly isogenic for different Pm3 resistance specificities did not identify recombinants among several hundred F2 progeny (Zeller et al., 1993), the confidence limits of these genetic tests are insufficient to rule out the possibility that other Pm3 resistance specificities reside at TaMla. Thus, further experiments involving F2 populations that segregate for other Pm3 resistance specificities will be needed to determine whether none of the known Pm3 resistance specificities has a common ancestry with TaMla.
AvrMla1 Is Not Genetically Fixed in Wheat Powdery Mildew
Unlike powdery mildews of dicots, the grass powdery mildews exhibit an exceptionally narrow host range (Jørgensen, 1988). For example, the wheat powdery mildew fungus successfully infects only wheat but not other genera of the Triticeae tribe such as Hordeum or Avena. Similarly, the barley powdery mildew fungus is pathogenic only on Hordeum plants, a phenomenon that led to the classification of different formae specialis among grass mildews (e.g., Blumeria graminis f sp hordei). A number of studies have addressed the genetic basis of formae specialis by analyzing fungal progeny that are derived from crosses between different formae specialis (Tosa, 1989a, 1989b, 1992; Matsumura and Tosa, 1995). What these studies found was that single R loci that control race-specific resistance to the compatible formae specialis of a particular grass species can also recognize individual powdery mildew avirulence genes in other formae specialis. This implies that the host range among grass species is the result of active defense responses and not missing host compatibility or fungal virulence factors. It also suggests that at least some Avr genes of powdery mildews were present in ancestral fungi before their genetic isolation in formae specialis and that these genes may have become genetically fixed in different formae specialis.
We have tested this hypothesis directly by expression of barley Mla1 in single wheat epidermal cells and inoculation with wheat powdery mildew isolates that together harbor most known wheat powdery mildew Avr genes. Lack of detectable Mla1 (and Mla1-2) activity in wheat provides strong evidence against the existence of a genetically fixed AvrMla1 gene in another powdery mildew formae specialis, at least in the tested tritici isolates. However, we cannot rule out the possibility that the failure of Mla1 activity in wheat is due to additional host factors that either are not present or are too much diverged in sequence to permit the assembly of a functional recognition complex. Expression of another specificity, Mla6, in wheat epidermal cells resulted in recognition of the barley fungus in an AvrMla6-dependent manner, but again, this specificity failed to recognize any of 10 tested wheat powdery mildew Avr genes (Halterman et al., 2001). This finding shows that, in principle, Mla proteins can function in wheat, consistent with observations in dicots that NB-LRR–type R gene functions can be transferred to related species of the same plant family (Whitham and McCormick, 1996; Tai et al., 1999). Clearly, molecular isolation of multiple powdery mildew Avr genes will be necessary to examine directly if and how many of these genes are genetically fixed in different powdery mildew formae specialis.
METHODS
Plant and Fungal Material
The Algerian-derived congenic lines C.I. 16,137 (AlgR, Mla1) and C.I. 16,138 (AlgS, mla1) (Moseman, 1972), an mlo-5 backcross line (mlo-5, Rar1), an mlo rar1 double mutant (mlo rar1-2), the susceptible wheat (Triticum aestivum) cultivar Cerco (containing no known powdery mildew resistance genes), and the series of Mla1 mutants were cultivated under conditions described previously (Shirasu et al., 1999b). The barley powdery mildew (Blumeria graminis f sp hordei) isolates A6 (VirMla1) and K1 (AvrMla1) were propagated on the susceptible barley (Hordeum vulgare) cultivar Golden Promise. The wheat powdery mildew (B. graminis f sp tritici) isolates JIW2 (AvrPm1, AvrPm2, AvrPm3a, AvrPm3b, VirPm3c, VirPm3d, VirPm3f, AvrPm4a, AvrPm4b, AvrPm5, AvrPm6, VirPm7, AvrPm8) and JIW48 (AvrPm1, VirPm2, VirPm3a, AvrPm3b, VirPm3c, VirPm3d, VirPm3f, VirPm4a, VirPm4b, VirPm5, AvrPm6, AvrPm7, AvrPm8) (kindly provided by James Brown, John Innes Centre) were propagated on the susceptible wheat cultivar Cerco. A wheat powdery mildew isolate (FZ1) that occurs naturally in the United Kingdom with unknown Avr and Vir spectra was propagated in a greenhouse on cv Cerco (also provided by James Brown). To achieve even inoculation, fresh spores from infected plants were passed through a plastic mesh and onto detached leaves in an inoculation tower.
Mutagenesis and Characterization of the Mla1 Mutants
For γ-irradiation, dry seed of the powdery mildew–resistant line AlgR was irradiated for 3 min at 3300 rad/min (9900 rad), for 4 min at 2200 rad/min (8800 rad), or for 5 min at 2200 rad/min (10,500 rad) using a cobalt 60 source. The mutagenized M1 seed was grown in the field in six plots, and M2 seed was harvested by plot. The M2 seedlings were screened at 9 to 11 days after inoculation for disease response to barley powdery mildew race CR3, which was originally isolated in California (Moseman and Schaller, 1959). Methods for maintaining race CR3 and for performing inoculations have been described (Masri and Ellingboe, 1966; Kerby and Somerville, 1989). Susceptible mutants were advanced to the M3 generation and then retested for their disease reaction to CR3. The hordein banding pattern of all lines was determined and found to be AlgR-like (Doll and Anderson, 1981). All of the lines used in the experiments described here were fully susceptible to CR3. M508 and M510 were derived from the same plot of M1 seed, and M567, M568, M584, M585, M590, M591, M597, M598, M599, M600, M601, M602, M604, M606, M607, M608, and M609 were derived from the same plot of M1 seed. Mutants derived from the same plot may be siblings.
Cosmid Library Construction and Screening
High-molecular-mass genomic DNA was isolated from barley line AlgR using procedures described by Leister et al. (1998) and partially digested with Sau3AI to produce DNA fragments of 30 to 60 kb. After dephosphorylation, the fragments were ligated to the XbaI-BamHI–linearized SuperCos cosmid vector, according to the manufacturer's instructions (Stratagene). The ligations were packaged using Gigapack III XL packaging extract (Stratagene) and then used to transform Escherichia coli strain XL1-Blue MR (Stratagene). A total of 260 pools averaging 3000 clones each were made and kept frozen as glycerol stocks. The library has an average insert size of 35 kb (range 30 to 45 kb) and represents six genome equivalents. Plasmid DNA was made from each pool and was either used for polymerase chain reaction (PCR) screening or spotted onto nylon membranes for hybridization screening. Approximately 10,000 clones from each pool were screened by hybridization to obtain purified clones. The positive clones were fingerprinted by restriction digestion using EcoRI, HindIII, or BamHI and grouped into different classes.
DNA Gel Blot Hybridization Analysis
Plant genomic DNA or cosmid DNA was digested completely using the restriction enzymes indicated in Figures 1 and 5, and the DNA fragments were resolved by agarose gel electrophoresis and blotted onto Hybond-N+ membranes (Amersham Pharmacia Biotech). Following the procedures described by Graner et al. (1990), the blots were hybridized with 32P-labeled probes and washed as indicated in the legends to Figures 1 and 5. Analysis of Mla1 mutants was performed by probing with resistance gene homolog (RGH) PCR products or the cosegregating restriction fragment length polymorphism (RFLP) probes MWG2083 and MWG2197. The RGH PCR products were amplified from bacterial artificial chromosome 80H14 DNA using RGH gene–specific primers (Wei et al., 1999). The MLA1-NB and MLA1-LRR probes were derived from plasmid clones D33 and B76, which contain exon 3 and exon 4 Mla1 sequences, respectively.
Sequencing and Gene Characterization
DNA from cosmid clones was isolated using the Qiagen plasmid midi kit (Qiagen, Ltd., Crawley, UK) and digested completely with HindIII or digested partially with HaeIII, Sau3AI, or Tsp509. The digested DNA fragments (2 to 4 kb) were cloned into the pBluescript II SK− plasmid vector (Stratagene) and propagated in E. coli strain DH10B. DNA from plasmid subclones was prepared using the R.E.A.L. Prep 96 Plasmid Kit (Qiagen, Ltd.) and used as templates for sequencing. Sequencing reactions were performed using a BigDye Terminator sequencing reaction kit (Perkin-Elmer) with T3 or T7 primers and analyzed on an ABI 377 automated sequencer (Applied Biosystems, Foster City, CA). Construction of sequence contigs and estimation of coding probabilities were performed using the GCG9 and STADEN (fourth edition) software packages (University of Wisconsin Genetics Computer Group, Madison, WI). Homology searches were performed using BLAST software (http://www.ncbi.nlm.nih.gov/blast/blast.cgi). Coiled-coil structure prediction was performed with COILS (http://ulrec3.unil.ch/software/COILS_form.html).
Sequencing of Mla1 Mutant Alleles
Mla1-specific primers were designed based on the sequence alignment of Mla1, Mla1-2, and RGH1a. The primers were first tested on two deletion mutants, M508 and M510, and the parent line AlgR. Only primers that worked on AlgR but not on the mutants were used further (Table 5). PCR products amplified from mutant genomic DNA were purified using the Wizard PCR preps kit (Promega) and sequenced directly. The sequences of the PCR products were compared with that of the wild-type Mla1 gene, and mutations were confirmed by sequencing both plus and minus strands.
Table 5.
Gene-Specific Primers for PCR and Sequencing of Mla1 from the Mutants
| Primer Name | Primer Sequences | Annealing Temperature | Region of Mla1 | Size (bp) of Product |
|---|---|---|---|---|
| Mla1S6 | 5′-CTGTCACGCCTATCAGCCACCTTT-3′ | 60°C | 5′UTRa | 1206 |
| Mla1AS6 | 5′-TCTCCTCCACAAACTTTTCTTTCC-3′ | 3rd exon | ||
| Mla146S1 | 5′-TTGGAGTTGCTCTTGGATGTC-3′ | 60°C | 3rd exon | 566 |
| Mla1AS3 | 5′-GTTACAGCTCCCTTTATTCATCA-3′ | 3rd exon | ||
| Mla1S7 | 5′-CGTTTAGTGTGAACTGCTTATGCC-3′ | 60°C | 3rd intron | 481 |
| Mla1AS7 | 5′-CCCTTGCCTTCGAGCTTTGTATGC-3′ | 3rd exon | ||
| Mla1S3 | 5′-ATCTTCCTCTTTCCTTCCTCTC-3′ | 60°C | 3rd intron | 720 |
| Mla1AS7 | 5′-CCCTTGCCTTCGAGCTTTGTATGC-3′ | 3rd exon | ||
| MlaNS1 | 5′-AGGAGAGGAAGGAAAGAGGAAGA-3′ | 60°C | 3rd intron | 313 |
| MlaNAS1 | 5′-AATATATGACAATTAACAAATCTCTTG-3′ | 3rd intron | ||
| MlaGS1 | 5′-TGGTCACCGGCCAAAGCACTAGC-3′ | 60°C | 4th exon | 987 |
| MlaNS1 | 5′-AGGAGAGGAAGGAAAGAGGAAGA-3′ | 3rd intron | ||
| Mla55S1 | 5′-AATCCAAACTACCATCCGTGAA-3′ | 60°C | 4th exon | 1001 |
| Mla1AS5 | 5′-CCAGAAGATGAAACCAAAGTGTGAG-3′ | 4th exon | ||
| MlaLS1 | 5′-CACGGTTACCATCCTCTTTCGTCAC-3′ | 65°C | 4th exon | 450 |
| Mla1AS4 | 5′-GTCTACGTATTGAGTGCAATTCCAG-3′ | 4th exon | ||
| Mla1S1 | 5′-GATAAGAACATACATCAATCCACCC-3′ | 60°C | 3′UTRa | 691 |
| Mla1AS1 | 5′-TAATAACGAGCACCGACCAAAC-3′ | 4th exon |
UTR, untranslated region.
Reporter Constructs
The construct pUGLUM was created by modifying the vector pU-hGFP-C3-N (Shirasu et al., 1999b) to contain the barley Mlo cDNA behind a second maize ubiquitin promoter followed by the nopaline synthase (NOS) terminator sequence. First, a plasmid pUGL was made by the addition of a multiple cloning site containing EcoRV, Asp718, and NotI into the EcoRI site just downstream of the NOS terminator of pU-hGFP-C3-N. The second ubiquitin promoter was amplified by PCR with the primers Ubi1 (5′-TAATGAGCATTGCATGTC-TAAG-3′) and Ubi2 (5′-TGCAGAAGTAACACCAAACAAC-3′), cloned into pGEMT (Promega), and confirmed by sequencing. The promoter was released by digestion with SacII and NotI, and the ends were polished using the Klenow fragment of E. coli DNA polymerase I and cloned into the EcoRV site of pUGL to create pUGLU. The MLO cDNA (Büschges et al., 1997) was cloned into a pBluescript II KS+ plasmid vector containing the NOS terminator, and the MLO-NOS fragment was released with Asp718 and NotI and cloned into pUGLU to create pUGLUM. The β-glucuronidase (GUS) reporter construct was kindly provided by Ken Shirasu (Sainsbury Laboratory, John Innes Centre).
Bombardment and Inspection of Resistance Phenotypes
Gold particles (0.9 μm; Bio-Rad) were coated with reporter plasmid DNA alone or together with the DNA of cosmids or cosmid subclones p6-49-2-15 and p6-49-2-7 at a plasmid:cosmid molar ratio of 1:2. Particles were delivered into epidermal cells of detached barley or wheat primary leaves using a particle inflow gun, basically as described by Shirasu et al. (1999b) and Schweizer et al. (1999). After bombardment, the specimens were kept on a 1% (w/v) Phytoagar (GIBCO) medium containing 10% (w/v) sucrose for 4 hr and then moved to a 1% (w/v) Phytoagar plate containing 1 mM benzimidazole for fungal inoculation. To compare the effects of Mla1 candidate genes on fungal isolates A6 (VirMla1) and K1 (AvrMla1), we inoculated an equal number of the transformed leaves with A6 and K1. After inoculation, the plates were placed in a growth chamber at 15°C with 16 hr of light and 8 hr of darkness. At 66 hr after inoculation, the specimens were infiltrated with GUS staining solution containing 0.1 M Na2HPO4/NaH2PO4, pH 7.0, 10 mM N-EDTA, 5 mM potassium hexacyanoferrat (II) and potassium hexacyanoferrat (III), 1 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, cyclohexylammonium salt, 0.1% (v/v) Triton X-100, and 20% (v/v) methanol and incubated at 37°C overnight. The specimens were rinsed in water and stained in Coomassie Brilliant Blue R 250 solution (0.3% [w/v] Coomassie Brilliant Blue, 7.5% [w/v] trichloroacetic acid, and 30% [v/v] methanol) for 5 min to visualize fungal structures, as described (Schweizer et al., 1999). The challenged GUS-positive epidermal cells were characterized for the presence or absence of fungal haustoria under a light microscope. When green fluorescent protein (GFP) was used as the reporter, the specimens were examined 4 to 5 days after inoculation by UV light incident fluorescence microscopy (excitation filter, 450 to 490 nm; bypass filter, 515 to 565 nm; Leica, Wetzlar, Germany). The challenged GFP-expressing epidermal cells were characterized for the presence or absence of colonies.
Reverse Transcription–PCR and Rapid Amplification of cDNA Ends
Rapid amplification of cDNA ends (RACE) was performed with the Marathon cDNA amplification kit (Clontech, Palo Alto, CA) using 1 μg of poly(A)+ RNA from the leaves of AlgR as template. To obtain the 5′ end, two rounds of PCR were performed using the Mla1-specific primer MLA1GSP1 (5′-TCGGCCACCCACTTCCATATCAGTTC-3′) for the first round and the nested primer MLA1NGSP1 (5′-ACTGAAGGAGAATATCCCACTCACAC-3′) for the second round. Conditions for the first and second rounds of PCR (25 cycles each) were as recommended by the manufacturer (Clontech). Amplification of the 3′ end was performed using the same conditions as in 5′-end RACE but with the single Mla1-specific primer MLA1LAS1 (5′-TCTCTGTTTATA-TGTATTGTGGTGGA-3′). RACE products were cloned into the pGEM-T Easy vector (Promega), and six independent clones for each end were sequenced.
cDNA sequences internal to RACE products were obtained by amplification of two overlapping Mla1 fragments. Advantage cDNA polymerase mix (Clontech) and Expand Long Template enzyme mixture (Boehringer Mannheim) were used for reverse transcription–PCR of the internal fragments with the Mla1-specific primers MLA1S2 (5′-TACAAATCCAAACTACCATCCC–3′) and MLA1AS2 (5′-CAGTGT-CTCTAATTCATGTTGCTCA-3′) for the first fragment and the primers MLA1LS1 (Table 5) and MLA1AS4 (Table 5) for the second fragment. Five independent clones of each cDNA product were obtained using the pGEM-T Easy vector (Promega) and sequenced.
TaMla and Pm3b Mapping
The mapping analysis involved 476 susceptible plants selected from 2000 segregating F2 plants from a cross between the nearly isogenic lines Chul/8*Chancellor (Pm3b) and Chancellor. Two different isolates carrying AvrPm3b were used for the infection of wheat leaf sections (isolates 96,236 and 96244; FAL Reckenholz, Zürich-Reckenholz, Switzerland). F3 seedlings of recombinant plants were retested with the same isolates. Isolation of wheat genomic DNA, DNA gel blotting, and labeling experiments were performed as described by Graner et al. (1990). Seven restriction enzymes were used for genomic digests: EcoRI, HindIII, XbaI, EcoRV, BamHI, DraI, and BglII. Barley probes were mostly from the München-Weihenstephan collection (A. Graner, Institut für Pflanzengenetik und Kulturpflanzenforschung, Gatersleben, Germany). Barley probe BCD1434 and oat probe CDO426 were provided by M. Sorrels (Cornell University, Ithaca, NY). Wheat probe Whs179 was provided by L. Hartl (Technische Universität München, Freising-Weihenstephan, Germany). Lrk10-A is a 1-kb fragment encoding the extracellular domain of the Lrk10 gene (Feuillet et al., 1997). Linkage estimation was based on the maximum likelihood method using Mapmaker (Lander et al., 1987).
Acknowledgments
We thank James Orme for providing the mlo rar1-2 mutant for this study, Evonne Waterman for technical assistance, and our colleague Nicholas Collins for critical reading of the manuscript. Mla research in the R.W. laboratory was supported in part by U.S. Department of Agriculture National Research Initiative Competitive Grants Program Grant No. 98-35300-6170. Pm3 research in the B.K. laboratory was supported by Swiss Priority Program Biotechnology Grant No. 5002-57824. The S.S. laboratory was supported in part by grants from the National Institutes of Health and the U.S. Department of Energy. Work in the P.S.-L. laboratory was supported by a grant from the GATSBY Charitable Organization and the European Union–funded European Gramineae Mapping Project consortium.
References
- Altschul, S., and Gish, W. (1996). Computer methods for macromolecular sequence analysis. Methods Enzymol. 266, 460–480. [DOI] [PubMed] [Google Scholar]
- Ayliffe, M.A., Frost, D.V., Finnegan, E.J., Lawrence, G.J., Anderson, P.A., and Ellis, J.G. (1999). Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J. 17, 287–292. [DOI] [PubMed] [Google Scholar]
- Boyd, L.A., Smith, P.H., Foster, E.M., and Brown, J.K.M. (1995). The effects of allelic variation at the Mla resistance locus in barley on the early development of Erysiphe graminis f. sp. hordei and host responses. Plant J. 7, 959–968. [Google Scholar]
- Büschges, R., et al. (1997). The barley Mlo gene: A novel control element of plant pathogen resistance. Cell 88, 695–705. [DOI] [PubMed] [Google Scholar]
- Dinesh-Kumar, S.P., and Baker, B.J. (2000). Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc. Natl. Acad. Sci. USA 97, 1908–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll, H., and Anderson, B. (1981). Preparation of barley storage protein, Hordein, for analytical sodium dodecyl sulfate–polyacrylamide gel electrophoresis. Anal. Biochem. 115, 61–66. [DOI] [PubMed] [Google Scholar]
- Ellis, J., and Jones, D. (1998). Structure and function of proteins controlling strain-specific pathogen resistance in plants. Curr. Opin. Plant Biol. 1, 288–293. [DOI] [PubMed] [Google Scholar]
- Feuillet, C., Schachermayr, G., and Keller, B. (1997). Molecular cloning of a new receptor-like kinase gene encoded at the Lr10 disease resistance locus of wheat. Plant J. 11, 45–52. [DOI] [PubMed] [Google Scholar]
- Freialdenhoven, A., Scherag, B., Hollricher, K., Collinge, D.B., Thordal-Christensen, H., and Schulze-Lefert, P. (1994). Nar-1 and Nar-2, two loci required for Mla12-specified race-specific resistance to powdery mildew in barley. Plant Cell 6, 983–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geballe, A.P., and Morris, D.R. (1994). Initiation codons within 5′-leaders of messenger-RNAs as regulators of translation. Trends Biochem. Sci. 19, 159–164. [DOI] [PubMed] [Google Scholar]
- Graner, A., Siedler, H., Jahoor, A., Herrmann, R.G., and Wenzen, G. (1990). Assessment of the degree and the type of restriction fragment length polymorphism in barley (Hordeum vulgare). Theor. Appl. Genet. 80, 826–832. [DOI] [PubMed] [Google Scholar]
- Halterman, D., Zhou, F., Wei, F., Wise, R.P., and Schulze-Lefert, P. (2001). The MLA6 coiled-coil, NBS-LRR protein confers AvrMla6-dependent resistance specificity to Blumeria graminis f. sp. hordei in barley and wheat. Plant J. In Press. [DOI] [PubMed]
- Hartl, L., Weiss, H., Zeller, F.J., and Jahoor, A. (1993). Use of RFLP markers for the identification of alleles of the Pm3 locus conferring powdery mildew resistance in wheat (Triticum aestivum L.). Theor. Appl. Genet. 86, 959–963. [DOI] [PubMed] [Google Scholar]
- Hershey, J.W.B. (1991). Translational control in mammalian cells. Annu. Rev. Biochem. 60, 717–755. [DOI] [PubMed] [Google Scholar]
- Jørgensen, J.H. (1988). Erysiphe graminis, powdery mildew of cereals and grasses. Adv. Plant Pathol. 6, 137–157. [Google Scholar]
- Jørgensen, J.H. (1994). Genetics of powdery mildew resistance in barley. Crit. Rev. Plant Sci. 13, 97–119. [Google Scholar]
- Jørgensen, J.H. (1996). Effect of three suppressors on the expression of powdery mildew resistance genes in barley. Genome 39, 492–498. [DOI] [PubMed] [Google Scholar]
- Kerby, K., and Somerville, S. (1989). Enhancement of specific intercellular peroxidases following inoculation of barley with Erysiphe graminis f. sp. hordei. Physiol. Mol. Plant Pathol. 35, 323–337. [Google Scholar]
- Lander, E.S., Green, P., Abrahamson, J., Barlow, A., Daly, M., Lincoln, S., and Newburg, L. (1987). MAPMAKER: An interactive computer package for constructing primary genetic linkage maps of experimental and natural populations. Genomics 1, 174–181. [DOI] [PubMed] [Google Scholar]
- Leister, D., Kurth, J., Laurie, D.A., Yano, M., Sasaki, T., Devos, K., Graner, A., and Schulze-Lefert, P. (1998). Rapid reorganization of resistance gene homologues in cereal genomes. Proc. Natl. Acad. Sci. USA 95, 370–375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupas, A., Vandyke, M., and Stock, J. (1991). Predicting coiled coils from protein sequences. Science 252, 1162–1164. [DOI] [PubMed] [Google Scholar]
- Masri, S., and Ellingboe, A. (1966). Germination of conidia and formation of appressoria and secondary hyphae in Erysiphe graminis f. sp. tritici. Phytopathology 56, 304–308. [PubMed] [Google Scholar]
- Matsumura, K., and Tosa, Y. (1995). The rye mildew fungus carries avirulence genes corresponding to wheat genes for resistance to races of the wheat mildew fungus. Phytopathology 85, 753–756. [Google Scholar]
- Moseman, J., and Schaller, C. (1959). The effect on various cultures of Erysiphe graminis f. sp. hordei of the genes in barley that condition resistance to culture CR3. Phytopathology 49, 207–209.
- Moseman, J.G. (1972). Isogenic barley lines for reaction to Erysiphe graminisi f. sp. hordei. Crop Sci. 12, 681–682.
- Peterhänsel, C., Freialdenhoven, A., Kurth, J., Kolsch, R., and Schulze-Lefert, P. (1997). Interaction analyses of genes required for resistance responses to powdery mildew in barley reveal distinct pathways leading to leaf cell death. Plant Cell 9, 1397–1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulze-Lefert, P., and Vogel, J. (2000). Closing the ranks to attack by powdery mildew. Trends Plant Sci. 5, 343–348. [DOI] [PubMed] [Google Scholar]
- Schweizer, P., Pokorny, J., Abderhalden, O., and Dudler, R. (1999). A transient assay system for the functional assessment of defense-related genes in wheat. Mol. Plant-Microbe Interact. 12, 647–654. [Google Scholar]
- Shirasu, K., Lahaye, T., Tan, M.-W., Zhou, F., Azevedo, C., and Schulze-Lefert, P. (1999. a). A novel class of eukaryotic zinc-binding proteins is required for disease resistance signaling in barley and development in C. elegans. Cell 99, 355–366. [DOI] [PubMed] [Google Scholar]
- Shirasu, K., Nielsen, K., Piffanelli, P., Oliver, R., and Schulze-Lefert, P. (1999. b). Cell-autonomous complementation of mlo resistance using a biolistic transient expression system. Plant J. 17, 293–299. [Google Scholar]
- Tai, T.H., Dahlbeck, D., Clark, E.T., Gajiwala, P., Pasion, R., Whalen, M.C., Stall, R.E., and Staskawicz, B.J. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. USA 96, 14153–14158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosa, Y. (1989. a). Evidence on wheat gene-for-gene relationship between formae speciales of Erysiphe graminis and genera of gramineous plants. Genome 32, 918–924. [Google Scholar]
- Tosa, Y. (1989. b). Genetic analysis of the avirulence of wheatgrass powdery mildew fungus on common wheat. Genome 32, 913–917. [Google Scholar]
- Tosa, Y. (1992). A model for the evolution of formae speciales and races. Phytopathology 82, 728–730. [Google Scholar]
- van der Biezen, E.A., and Jones, J.D.G. (1998). The NB-ARC domain: A novel signalling motif shared by plant resistance gene products and regulators of cell death in animals. Curr. Biol. 8, R226–R227. [DOI] [PubMed] [Google Scholar]
- Wei, F., Gobelman-Werner, K., Morroll, S.M., Kurth, J., Mao, L., Wing, R., Leister, D., Schulze-Lefert, P., and Wise, R.P. (1999). The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153, 1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., and McCormick, S. (1996). The N gene of tobacco confers resistance to tobacco mosaic virus in transgenic tomato. Proc. Natl. Acad. Sci. USA 93, 8776–8781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S., Dinesh-Kumar, S.P., Choi, D., Hehl, R., Corr, C., and Baker, B. (1994). The product of the tobacco mosaic virus resistance gene N: Similarity to Toll and the interleukin-1 receptor. Cell 78, 1101–1115. [DOI] [PubMed] [Google Scholar]
- Wolfe, K.H., Gouy, M.L., Yang, Y.W., Sharp, P.M., and Li, W.H. (1989). Date of the monocot–dicot divergence estimated from chloroplast DNA-sequence data. Proc. Natl. Acad. Sci. USA 86, 6201–6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeller, F.J., Lutz, J., and Stephan, U. (1993). Chromosome location of genes for resistance to powdery mildew in common wheat (Triticum aestivum L.). 1. Mlk and other alleles at the Pm3 locus. Euphytica 68, 223–229. [Google Scholar]