Abstract
Cyclin F (CCNF) variants have been found to be associated with amyotrophic lateral sclerosis (ALS)/frontotemporal dementia (FTD). However, the genetic and clinical characteristics of ALS patients who carry CCNF variants are largely unknown. Genetic analysis was performed for 1587 Chinese ALS patients, and missense variants were predicted by software analyses. Additionally, we searched PubMed, Embase, and Web of Science for relevant literature and conducted a meta-analysis of the frequency of variants. In our ALS cohort, we identified 29 nonsynonymous variants in 41 ALS patients. Among these ALS patients, 18 (1.1%) were carriers of 15 rare missense variants that were considered probably pathogenic variants, and 11 of 15 variants were novel. Seven relevant studies were identified, and a total of 43 CCNF variants in 59 ALS patients with a frequency of 0.8% were reported. The ratio of males to females in our cohort (10/8) was similar to that in Caucasian populations (4/7) and significantly higher than that in Asian populations (10/1). The proportion of bulbar onset in Caucasian CCNF carriers was similar to our cohort (25.0 vs. 27.8%); however, bulbar onset had never been reported in previous Asian studies (0/11). FTD was not found in CCNF carriers in previous Asian studies and our cohort, but it has been reported in a FALS cohort (1/75) of Caucasian individuals. There were some differences in the clinical characteristics among different ethnic ALS populations. More basic scientific studies are needed to elucidate the pathogenic mechanisms and genotype-phenotype associations of CCNF variants.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12035-023-03380-1.
Keywords: Amyotrophic lateral sclerosis, CCNF gene variant, Clinical characteristics, Genotype
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating and rapidly progressive neurodegenerative disease. There are clinical heterogeneities among different patients; these include heterogeneities in age of onset, initial symptoms, clinical subtypes and disease progression. Approximately 10–15% of ALS patients are also diagnosed with frontotemporal dementia (FTD), and particularly, these patients carry chromosome 9 open reading frame 72 gene (C9orf72) mutations [1, 2]. Most patients die from respiratory failure within 3–5 years after onset, and the number of ALS cases is estimated to grow to approximately 380,000 by 2040 due to an aging population [3]. However, the etiology of ALS is still poorly understood. Genetic studies have shown that more than 50 genes are involved in the pathogenesis of ALS, and approximately 70% of familial cases can be explained by known gene mutations [4].
Recently, a single cyclin F (CCNF) variant that segregated with seven affected family members was identified by whole-genome linkage analysis and whole-exome sequencing (WES) [5]. Furthermore, screening for CCNF variants in ALS/FTD cohorts from diverse geographic populations (mostly European ancestry) revealed that CCNF variants in different ethnic ALS cohorts and novel and rare protein-altering CCNF variants were significantly enriched in sporadic ALS (SALS) patients [5]. CCNF encodes cyclin F, which is a component of the E3 ubiquitin-protein ligase complex (SCFcyclin F) that mediates protein ubiquitination and proteasomal degradation [6]. Functional analysis using neuronal cells revealed that mutant cyclin F leads to the abnormal accumulation of ubiquitinated proteins, including TAR DNA-binding protein of 43 kDa (TDP-43), and dysfunction in protein homeostasis [5].
However, the genetic spectrum and clinical phenotypes of CCNF variants in ALS patients are not fully understood due to a lack of sufficient samples and studies. Therefore, to further understand the role of CCNF variants in ALS, we screened 1587 ALS patients from southwestern China using WES and investigated the clinical characteristics of the patients carrying CCNF variants.
Materials and Methods
Study Population and Assessment
A total of 1587 definite or probable ALS patients diagnosed according to El Escorial ALS criteria [7] were included in the current study, all of whom were from the Department of Neurology, West China Hospital, Sichuan University, China. Familial ALS (FALS) is defined as a patient's first-, second-, or third-degree relative with ALS. The rest of the cases are classified as SALS. This study was approved by the Ethics Committee of West China Hospital of Sichuan University, and all recruited patients signed an informed consent form before participating in the study.
Demographic and clinical information was collected for all patients, and the patients were followed up in person or by telephone every three or six months. The severity of disease was assessed by the ALS function rating scale-revised (ALSFRS-R) at baseline and visits. The disease progression rate at baseline was calculated as (48 - “baseline” ALSFRS-R score) / the time from onset to “baseline” (months), and the follow-up disease progression rate was calculated as (“baseline” ALSFRS-R score - “last follow-up” ALSFRS-R score) / the time between “last follow-up” and “baseline” (months). Rapid progression was defined when the disease progression rate was greater than 0.5 [8]. The frontal assessment battery (FAB) [9] was used to assess frontal lobe function. The Montreal Cognitive Assessment (MoCA) was used to evaluate global cognitive function [10]. In our study, impairment of the frontal lobe and cognitive function was defined as a score of FAB ≤ 14 and MoCA ≤ 22, respectively [11, 12]. The Hamilton Depression Scale (HAMD, 24 items) [13], Hamilton Anxiety Scale (HAMA) [14], and Beck Depression Scale (BDI) [15] were used to evaluate mood. When the HAMD/HAMA score was > 7 or the BDI score was ≥ 5, the patient was considered to have symptoms of anxiety or depression.
Genetic Analysis
The genetic analysis of patients was performed using WES and repeat-primed polymerase chain reaction (RP-PCR), in which RP-PCR was used to detect G4C2 repeats in C9orf72. Genomic DNA was collected from peripheral blood leukocytes using standard phenol–chloroform procedures. We choose 41 known ALS-related genes based on our previous study [16]. Rare missense variants were defined as having a minor allele frequency (MAF) < 0.1% in the Exome Aggregation Consortium-East Asian (ExAC_EAS) and Genome Aggregation Database-East Asian (GnomAD_EAS). In addition, we found the RefSeq ID of the transcript sequence of each CCNF variant. The novel variant is defined as a variant that has not been reported in ALS patients in previous studies. We enrolled ALS patients with the CCNF variant, and excluded the ALS patients with ALS gene classified as definitive on the ALSoD website (https://alsod.ac.uk/).
Sorting Intolerant from Tolerant (SIFT), Polymorphism Phenotyping v2 (PPH2), Functional Analysis through Hidden Markov Models (FATHMM), Combined Annotation Dependent Depletion (CADD), Mutation Assessor (MA), Deleterious Annotations of genetic variants using Neural Networks (DANN), Protein Variation Effect Analyzer (PROVEAN), and Likelihood Ratio Test (LRT) were used to predict the harmfulness of CCNF variants. Genomic Evolutionary Rate Profiling++ (GERP++) was used to predict nucleic acid conservation. If more than four of the eight software programs predicted the variant to be deleterious (prediction ≥ 4/8), the variant was considered probably pathogenic (PP) in our study.
Search of the Literature
We searched articles on CCNF variants in ALS patients using the keywords (“amyotrophic lateral sclerosis” or “ALS” or “motor neuron disease”) and (“CCNF” or “Cyclin F”) in PubMed, Embase, and Web of Science (from inception until June 2022).
Statistical Analysis
Statistical analysis was performed using the statistical software SPSS (IBM SPSS Statistics for Windows, version 26, IBM Corporation, Inc., Chicago, IL, USA, 2019). Wilcoxon's T test or rank-sum test was used to compare the clinical characteristics of carriers of the CCNF missense variants. The Kaplan–Meier method was used for survival analysis. We performed a meta-analysis of variant frequency using STATA 16.0 software (version 16.0; Stata Corp., College Station, TX, USA) to calculate effect sizes and 95% confidence intervals (CIs) for each previously reported study.
Results
Genetic Analyses
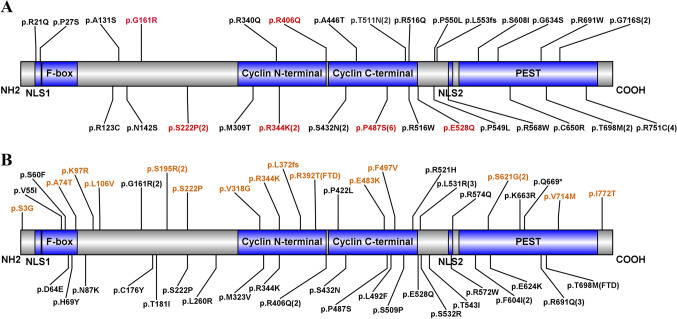
Among the 1587 recruited ALS patients, 29 nonsynonymous variants in the CCNF gene, including 28 rare missense variants and one frameshift variant, were identified in 41 ALS patients (Supplementary Table 1). Among the 29 variants, six variants (p.Gly161Arg, p.Arg344Lys, p.Arg406Gln, p.Glu528Gln, p.Pro487Ser, and p.Ser222Pro) have been reported in previous studies [5, 17–22]. Ten variants were located in the cyclin domains, seven variants were located in the PEST sequence, three variants were located in the nuclear localization signal (NLS), and nine variants were not located in any of the above domains (Fig. 1A). Using eight software programs to predict protein structure or function, 15 rare missense variants detected in 18 ALS patients were considered PP variants, of which 11 variants detected in 13 ALS patients were novel (Supplementary Table 2).
Fig. 1.
The distribution of CCNF variants at the protein level in our ALS cohort and previously reported cohorts
Interestingly, seven variants (p.Arg751Cys, p.Gly716Ser, p.Thr698Met, p.Arg691Trp, p.Cys650Arg, p.Gly634Ser, and p.Ser608Ile) located in the PEST sequence were all predicted to be nonpathogenic by software (prediction < 4/8) in our study.
Demographics and Clinical Characteristics of CCNF Carriers
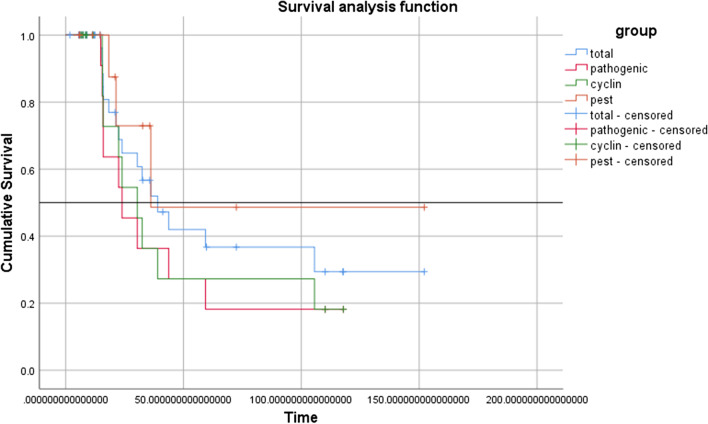
The demographic and clinical characteristics of the patients with CCNF variants are shown in Table 1. In our cohort, no patients carrying CCNF variants had a positive family history. The mean age of onset of all patients with CCNF variants and those with PP variants were 57.13 ± 10.12 and 59.67 ± 8.79 years, respectively. Approximately one-third of patients carrying CCNF variants had bulbar onset. The proportion of CCNF variant carriers with a rapidly progressive disease rate decreased from baseline to the last visit (58.5 to 25.0%). At the last follow-up visit, 20 patients were lost to follow-up, five patients were still alive, and 16 patients died from respiratory failure or nutritional problems. The median survival times of all CCNF variant carriers and PP missense variant carriers were 39.00 ± 7.83 months and 23.97 ± 7.96 months, respectively (Fig. 2). There were no significant differences in the clinical characteristics between CCNF PP carriers and ALS patients who did not carry the CCNF gene variant and other identified pathogenic gene mutations. In addition, there were no significant differences in the clinical features, including nonmotor symptoms, between the two patient groups with variants in the cyclin domains and PEST sequence (Table 1).
Table 1.
Demographic and clinical characteristics of ALS patients carrying CCNF gene variants in the study
| Variables | All (n=41) |
Pathogenic (n=18) |
Cyclin (n=17) |
Pest (n=12) |
Control (n=90) |
p value |
|---|---|---|---|---|---|---|
| Sex (F/M) | 17/24 | 8/10 | 7/10 | 5/7 | 40/50 | P1=1,P2=0.98 |
| Years of education, n | 34, 6.53 ± 4.00a | 17, 5.47 ± 3.56a | 14, 6.36 ± 5.37a | 11, 7.55 ± 2.70a | 6.77 ± 4.97a | P1=0.57,P2=0.51 |
| BMI, kg/m2, n | 33, 21.61 ± 2.99a | 16, 22.95 ± 2.81a | 14, 21.83 ± 2.41a | 10, 20.55 ± 3.26a | 21.78 ± 2.78a | P1=0.09,P2=0.31 |
| Age at onset, years | 57.13 ± 10.12a | 59.67 ± 8.79a | 58.25 ± 9.67a | 56.76 ± 8.64a | 60.39 ± 1.01a | P1=0.58,P2=0.67 |
| Site of onset, n (%) | ||||||
| U, 15 (36.6) | U, 6 (33.3) | U, 8 (47.1) | U, 4 (33.3) | U, 48 (53.3) | P1=0.37,P2=0.62 | |
| L, 12 (29.3) | L, 7 (38.9) | L, 4 (23.5) | L, 2 (16.7) | L, 27 (30.0) | ||
| B, 14 (34.1) | B, 5 (27.8) | B, 5 (29.4) | B, 6 (50.0) | B, 15 (16.7) | ||
| Family history, n | 0 | 0 | 0 | 0 | 0 | NA |
| Disease duration, months | 12.17 (14.15)b | 10.22 (8.41)b | 8.90 (10.22)b | 15.42 (21.62)b | 12.52 (15.33)b | P1=0.09,P2=0.08 |
| Diagnosis delay, months | 11.20 (10.13)b | 10.12 (7.92)b | 8.10 (9.62)b | 14.22 (18.03)b | 12.17 (11.71)b | P1=0.09,P2=0.18 |
| ALSFRS-R | 41.00 (7.50)b | 42.50 (8.50)b | 39.00 (8.50)b | 40.00 (8.25)b | 39.00 (8.25)b | P1=0.10,P2=0.90 |
| Progression rate | ||||||
| Baseline | 0.63 (0.88)b | 0.58 (1.00)b | 0.92 (1.33)b | 0.49 (0.92)b | 0.71 (0.69)b | P1=0.52,P2=0.16 |
| RP/total (%) | 24/41 (58.5) | 10/18 (55.5) | 12/17(70.6) | 5/12(41.7) | 56/90 (62.2) | |
| Follow-up | 0.30 (0.34)b | 0.31 (0.51)b | 0.20 (0.35)b | 0.36 (0.50)c | NA | P2=0.91 |
| RP/total (%) | 4/16 (25.0) | 2/8 (25.0) | 1/7 (14.3) | 1/3(33.3) | NA | |
| Survival time, months | 39.00 ±7.83 (23.66-54.34)d | 23.97 ± 7.96 (8.36-39.58)d | 30.40 ± 5.50 (19.61-41.19)d | 36.13 (median) | 34.00 ± 3.09 (27.95-40.05)d | P1=0.79,P2=0.23 |
| FAB, A/T (%) | 7/23 (30.4), 16.00 (3.00)b | 3/10(30.0), 17.00 (4.00)b | 2/8(25.0), 16.50 (3.25)b | 2/8(25.0), 15.50 (1.75)b | 19/59(32.2), 16.00 (4.00)b | P1=0.42,P2=0.31 |
| MOCA, A/T (%) | 12/22(54.5), 22.00 (5.00)b | 5/9(55.5), 22.00 (4.50)b | 5/8(62.5), 20.88 ± .06a | 3/8(37.5), 22.75 ± 3.77a | 25/52(48.1), 23.50 (8.50)b | P1=0.75,P2=0.56 |
| FBI | 0/20(0), 0 (3.75)b | 0/10 (0), 0 (0)b | 0/9(0), 0 (5.50)b | 0/5(0), 0 (9.00)b | NA | P2=0.88 |
| BDI, A/T (%) | 13/26(50.0), 4.00 (8.50)b | 4/12(33.3), 2.00 (8.75)b | 6/12(50.0), 4.00 (9.25)b | 5/8(62.5), 6.50 (9.75)b | NA | P2=0.35 |
| HAMD, A/T (%) | 15/31(48.4), 7.00 (7.00)b | 5/14(35.7), 5.50 (6.50)b | 5/14(35.7), 6.50 (5.50)b | 7/10(70.0), 10.50 (11.75)b | NA | P2=0.06 |
| HAMA, A/T (%) | 12/31(38.7), 5.00 (8.00)b | 3/14(57.1), 2.00 (4.50)b | 4/14(28.6), 2.00 (8.25)b | 7/10(70.0), 9.00 (5.00)b | NA | P2=0.02 |
| PSQI, A/T (%) | 1/10(10.0), 2.00 (4.00)b | 1/6(21.4), 1.00 (3.50)b | 1/5(20.0), 3.00 (7.00)b | NA | NA | NA |
| RBD, A/T (%) | 1/10(10.0), 1.00 (2.00)b | 1/6(16.7), 1.00 (2.00)b | 1/5(20.0), 1.00 (3.00)b | NA | NA | NA |
| ESS, A/T (%) | 0/9(0), 3.00 (4.00)b | 0/6(0), 3.50 (3.50)b | 0/4(0), 4.00 (4.25)b | NA | NA | NA |
a: mean ± SD, b: median (IQR), c: median (range), d: median ± SE (95% CI), RP rapid progression, Control: age- and sex-matched ALS patients without CCNF variants and any other identified ALS-related pathogenic gene mutations. A/T abnormal/total, p1 value represents the comparison between pathogenic group and control group, p2 value represents the comparison between cyclin group and pest group, IQR interquartile range, SD standard deviation, SE standard error, CI confidence interval, F female, M male, PEST PEST sequence (short stretch of amino acids enriched in proline, glutamic acid, serine and threonine, cyclin: cyclin domains, U upper limbs, L lower limbs, B bulbar, ALSFRS-R ALS function rating scale-revised, FAB frontal assessment battery, MOCA Montreal cognitive assessment, FBI frontal behavioral inventory, BDI Beck Depression Inventory, HAMD Hamilton depression scale, HAMA Hamilton anxiety scale, PSQI Pittsburgh sleep quality index, RBD rapid eye movement sleep behavior disorder, ESS Epworth sleepiness scale
Fig. 2.
Survival analysis of ALS patients with CCNF variants in our cohort
In addition to motor symptoms, we assessed nonmotor symptoms of patients and found that approximately 54% of patients had cognitive impairment assessed by MoCA. None of the patients met the diagnosis criteria of FTD according to the FTDC criteria [23]. Half of the patients had depressive symptoms, and 38.7% had anxiety symptoms.
Interestingly, in our ALS cohort, there were two patients (Case No. 1901 and Case No. 1120) carrying two different missense variants in CCNF. Case No. 1901, who carried the p.Pro487Ser and p.Arg516Gln variants of CCNF (predictions were 2/8 and 5/8, respectively; the latter located in the cyclin domain was considered to be a PP missense variant), developed dysarthria at the age of 65 with a disease progression rate of 1.17 and died of nutritional problems 22.5 months after onset. Another patient, Case No. 1120, carried the CCNF p.Ala131Ser and p.Arg751Cys variants (predictions were 6/8 and 3/8, respectively, and the former was considered to be a PP missense variant). This patient presented with dysarthria at the age of 64 with a disease progression rate of 0.26 and was lost to follow-up at 11 months of the disease course. A novel frameshift variant (p.Leu553fs) was also identified in our cohort in a patient with bulbar onset at the age of 41, with baseline and follow-up disease progression rates of 0.34 and 0.20, respectively. The survival time was more than 117 months.
In addition, 13 patients in our cohort were carriers of additional ALS-related gene variants in addition to CCNF variants. The clinical characteristics of the 13 individual patients are shown in Table 2. There were no significant differences in the clinical features of the 13 ALS patients who carried additional ALS-related gene variants compared with those who did not carry the CCNF gene variant and other identified pathogenic gene mutations (Supplementary Table 5). Case No. 3457 carried the CCNF p.Arg516Trp variant (which was considered PP) and ALS2 p.Pro955Arg variant (which was considered PP), neither of which has been reported previously. This patient had extremely rapid disease progression with a progression rate of 2.32 and died from choking approximately 16 months after disease onset. Two female patients (Case Nos. 1149 and 1168) in our cohort carried additional variants besides CCNF p.Gly716Ser variant which was predicted to be benign. Case No. 1168 was a carrier of an additional TIA1 variant (p.Phe214Tyr, prediction 2/6) and present with a fast progression rate of 0.66 at baseline. This patient survived for more than 152 months and was still alive at the last follow-up. Case No. 1149 was a carrier of an additional CHMP2B variant (p.Asp151Glu, prediction 4/8) but was lost to follow-up.
Table 2.
Clinical features of CCNF variant carriers who had other ALS-related gene variants
| Case No. | CCNF variant | Age at onset, y | Disease duration, m | Sex | Site of onset | ALSFRS-R | Progression rate | Survival time, m | Outcome | Other gene variant |
|---|---|---|---|---|---|---|---|---|---|---|
| 1046 | p.Ser222Pro | 56.49 | 12.17 | M | L | 46 | 0.16 | 12.17 | 2 | FIG4(p.Pro851Leu) |
| 1149 | p.Gly716Ser | 43.44 | 35.70 | F | B | 45 | 0.08 | 35.70 | 2 | CHMP2B(p.Asp151Glu) |
| 1168 | p.Gly716Ser | 46.97 | 42.37 | F | B | 20 | 0.66 | 152.13 | 1 | TIA1(p.Phe214Tyr) |
| 1228 | p.Arg340Gln | 44.49 | 8.90 | M | B | 36 | 1.35 | 8.90 | 2 | FIG4(c.1389-2A>G) |
| 1363 | p.Pro487Ser | 69.81 | 11.60 | M | B | 39 | 0.78 | 11.60 | 2 | EWSR1(p.Gly589Ser) |
| 2330 | p.Ala446Thr | 45.26 | 6.30 | M | B | 44 | 0.63 | 39.00 | 0 | SETX(p.Asn1654Ser) |
| 3017 | p.Arg568Trp | 74.25 | 5.47 | M | B | 41 | 1.28 | 14.90 | 0 | CHMP2B(p.Lys8Asn),ALS2(p.Asp298Tyr) |
| 3199 | p.Thr511Asn | 69.55 | 18.30 | F | U | 37 | 0.60 | 23.97 | 0 | PNPLA6(p.Ala344Thr) |
| 3327 | p.Pro550Leu | 44.44 | 22.20 | M | U | 41 | 0.32 | 59.77 | 1 | DAO(p.Gly129Ser) |
| 3355 | p.Pro27Ser | 71.11 | 23.53 | F | L | 43 | 0.21 | 59.33 | 0 | SETX(p.Thr1188Asn) |
| 3457 | p.Arg516Trp | 61.18 | 6.03 | F | L | 34 | 2.32 | 15.90 | 0 | ALS2(p.Pro955Arg) |
| 3474 | p.Pro487Ser | 71.20 | 5.53 | M | U | 36 | 2.17 | 32.47 | 0 | FIG4(p.Thr270Ile) |
| 3493 | p.Arg344Lys | 63.44 | 14.93 | M | U | 41 | 0.47 | 15.67 | 0 | NEFH(p.Glu700Ala) |
y years, m months, 0: death, 1: alive, 2: loss, M male, F female, U upper limbs, L lower limbs, B bulbar
Literature Overview
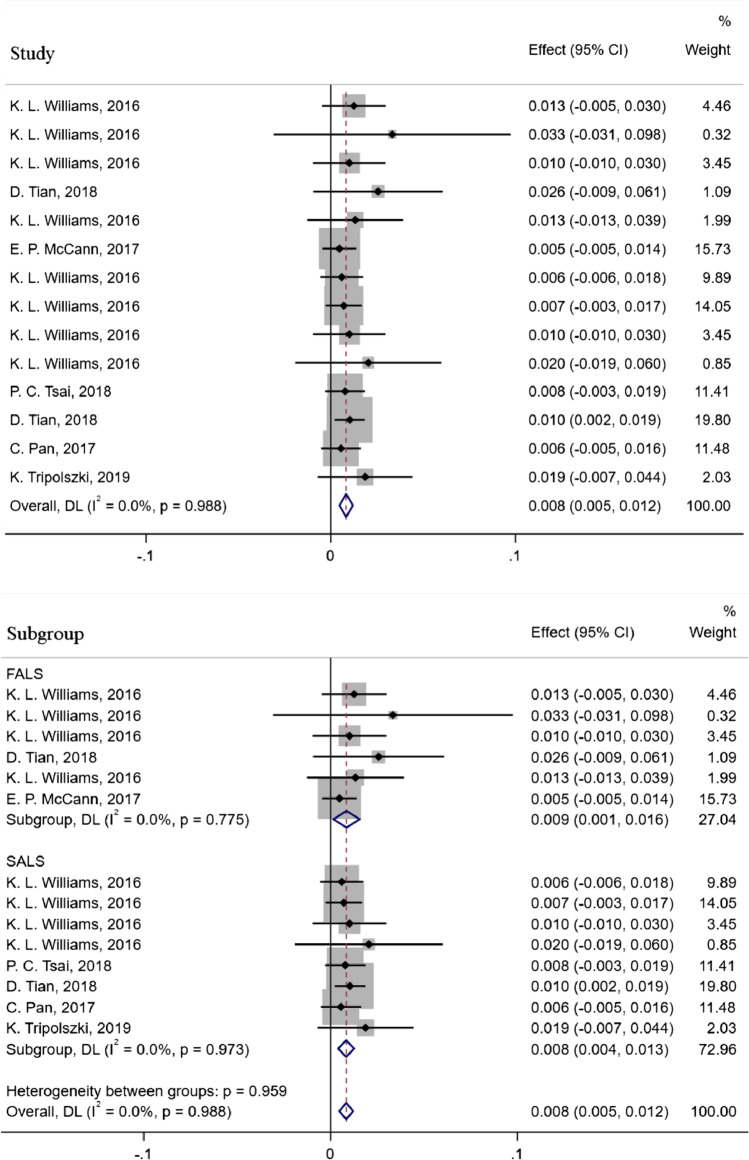
After systematically reviewing the literature, seven relevant studies consisting of 59 ALS/FTD patients with CCNF variants, including 15 Asian patients [17–19] and 34 Caucasian patients [5, 20–22], were analyzed. A total of 43 CCNF variants in ALS/FTD patients were reported [5, 17–22], including one stop-gain variant, one frameshift variant, and 41 missense variants (Supplementary Table 4). There were no significant differences in the frequency of CCNF variants in FALS and SALS patients between Caucasian and Asian populations (1.9% (2/107) vs. 2.6% (2/78), p=0.26; 1.0% (6/575) vs. 0.8% (14/1616), p=0.30). The overall variant frequency was approximately 0.8% (95% CI 0.5–1.2%), with 0.9% in the FALS and 0.8% in the SALS (Fig. 3). In addition, one FTD patient carrying the p.S621G variant of CCNF (1.3%, 1/75) was identified in a FALS cohort (Supplementary Table 4) [5].
Fig. 3.
The frequency of CCNF variants
The mean age of onset of previously reported CCNF carriers was 52.62 ± 13.59 years, the proportion of bulbar onset accounted for 10%, and the median survival time was approximately 43.00 ± 6.12 months. There were no statistically significant differences in the age of onset and site of onset between Caucasian and Asian CCNF carriers. The ratio of males to females was significantly higher in Asian populations than in Caucasian populations (10/1 vs. 4/7, p=0.02) (Table 3).
Table 3.
Previously reported clinical features of ALS and/or FTD patients with CCNF gene variants
| All (n=22) |
Asians (n=11) |
Caucasians (n=11) |
p value | |
|---|---|---|---|---|
| Mutation frequency | 0.8% | 0.8% | 0.9% | 0.62 |
| FALS:0.9%, SALS: 0.8% | FALS: 2.6% (2/78), SALS: 0.8% (14/1616) | FALS: 1.0% (6/575), SALS: 1.9% (2/107) | 0.26,0.30 | |
| Sex, F/M | 8/14 | 1/10 | 7/4 | 0.02 |
| Age at onset, years, n | 21, 52.62 ±13.59a | 11, 54.07 ± 6.52a | 10, 51.03 ± 18.91a | 0.61 |
| Site of onset, n (%) | ||||
| S, 17 (89.5) | S, 11 (100.0) | S, 6(75.0) | 0.16 | |
| B, 2 (10.5) | B, 0(0) | B, 2(25.0) | ||
| Cognition, A/T (%) | 2/20 (10.0) | 0/9(0) | 2/11(18.2) | 0.25 |
| Survival time, months, n | 13 | 11 | 2 | |
| Median ± SE (95% CI) | 43.00 ± 6.12 (31.01 – 54.99)d | 43.00 ± 5.91 (31.41 – 54.99)d | 24.00 (median) | NA |
a: mean ± SD, A/T abnormal/total, F female, M male, d: median ± SE (95% CI), SD standard deviation, SE standard error, CI confidence interval, U upper limbs, L lower limbs, B bulbar, p value represents the comparison between LoF group and missense variant group
Based on the protein level, seven variants in 16 reported ALS patients were located in the PEST sequence, 13 variants in 16 reported ALS patients were located in the cyclin domains, and five variants in five patients were located in the F-box domain (Fig. 1B). Due to insufficient clinical data, it was not possible to compare whether there were differences in the clinical characteristics among ALS patients with variants in the three domains of CCNF.
Discussion
We investigated the genetic spectrum and clinical phenotype of CCNF variants through our large Chinese ALS cohort and literature review. First, the PP variant frequency of CCNF in our cohort (1.1%) was comparable to that in previous studies (0.8%). Second, the 11 novel PP missense variants identified in our cohort expand the genetic variant spectrum of CCNF. Third, our study found that nearly one-third of CCNF variant carriers had additional variants in other ALS-related genes in addition to CCNF variants. Finally, more than half of CCNF carriers in our cohort had cognitive impairment, but no patients met the diagnosis criteria of FTD.
The CCNF gene encodes cyclin F, which is a founding member of the F-box protein family that mainly includes three functional modules: a pseudocatalytic module containing the F-box domain and NLS1, a substrate complement module containing two cyclin domains, and a regulatory module containing NLS2 and PEST (a short amino acid rich in proline, glutamic acid, serine and threonine) [24]. Cyclin F is essential for genome stability and regulates deoxyribonucleotide triphosphate levels, centrosome duplication and spindle formation by targeting and regulating the levels of SCFcyclin F substrates [25, 26]. Recently, it was also found that the expression of CCNF in the motor neuron-like cell line NSC-34 changes after exposure to oxidative stress [27]. CCNF can also affect the activity of valosin-containing protein (VCP) in the cytoplasm [28].
In our cohort, a total of 41 ALS patients were carriers of 29 variants in the CCNF gene, but only 15 missense variants were identified as deleterious by software analyses. Therefore, the variant frequency of CCNF in our ALS cohort was 1.1% (15/1587), which was similar to the results of previous studies [5, 17–20]. A previous study found an enrichment for rare protein alterations in CCNF (MAF < 0.0001) among SALS patients compared to control individuals [5]. However, this study also identified a CCNF frameshift variant (p.L372fs) that did not segregate with disease, suggesting that a dominant gain in toxic function may be required for the pathogenicity of mutant CCNF [5]. One patient with the frameshift variant (p.Leu553fs) was also found in our cohort. However, because of the lack of genetic information on relatives, it was not possible to determine whether the variant segregated with the disease. Therefore, whether the functional consequences of CCNF variants lead to increased functional toxicity or loss of dominant negative function or haploinsufficiency remains to be determined by more studies.
Currently, studies have not found a clear relationship between the location of CCNF variants and the clinical phenotype. Two variants (p.Pro27Ser and p.Arg21Gln) were located in the NLS1 domain, which mediates interactions with other components of the SCF ubiquitin-protein ligase complex to achieve ubiquitination of target substrates [24]. This is the first report of a variant in the NLS1 domain. Both variants were considered deleterious by software (predictions of 4/8 and 5/8, respectively). In our cohort, there was also a newly discovered variant (p.Arg568Trp) located in the NLS2 domain, which was considered to be harmful with a prediction of 4/8. The patient carrying this variant had extremely rapid disease progression, with a progression rate of 1.28, and died of sputum asphyxia less than 15 months after onset. The p.Arg568Trp variant has not been reported previously. However, two variants (p.R574Q and p.R572W) located in the NLS2 domain have been reported in European populations and are also considered to be pathogenic variants [5, 20].
Ten variants were located in the cyclin domains in our cohort, eight of which (all except the p.Pro487Ser and p.Arg344Lys variants) were predicted to be PP (prediction ≥ 4/8). Interestingly, the benign CCNF p.Pro487Ser variant (prediction 2/8) was present in six patients in our ALS cohort, and this variant was previously reported in an Asian patient [17]. In our cohort, a benign p. Arg344Lys variant (prediction 2/8) identified in two patients has been previously reported once in both European and Asian populations [5, 19].
Seven variants located in the PEST sequence were all predicted to be benign by software analyses (prediction<4/8) in our study, and none have been reported previously. However, the p.S621G variant located in the PEST sequence has been reported repeatedly in Caucasian FALS/FTD patients [5, 22]. The p.S621G variant prevents the phosphorylation of casein kinase II (CK2) and increases the lys48-ubiquitin activity, resulting in dysfunction of the autophagy degradation pathway [5, 29, 30]. It was also found that zebrafish with the p.S621G variant had disrupted axonal growth, suggesting a toxic gain-of-function mechanism in the pathogenesis of ALS [31]. The p.S621G variant was not found in our cohort, and all variants identified in our cohort were not located at the identified phosphorylation sites [29]. Therefore, based on the results predicted by software and the current literature review, we considered these seven variants located in the PEST sequence in our cohort to be likely benign, but more basic experimental studies are needed to explore the pathogenicity and pathogenesis of CCNF variants located in the PEST sequence.
Four CCNF variants (p.Gly161Arg, p.Ser222Pro, p.Ala131Ser, and p.Arg123Cys) predicted to be PP in our study were not located in any functional domain. Among them, the p.Gly161Arg variant of CCNF has been reported previously in both Caucasian and Asian populations. In the Asian populations, the p.Gly161Arg variant was found in a 43-year-old man who presented with rapidly progressive left-hand weakness and died of respiratory failure two years after onset [5, 20]. In our cohort, the patient carrying the p.Gly161Arg variant also had a rapid progression rate of 1.5, but the survival time of this patient could not be obtained due to loss of follow-up. A previously reported Asian carrier of the p.Ser222Pro variant of CCNF had a relatively long survival time of more than 60 months [17]. The in vitro functional study found that the p.Ser222Pro variant of CCNF had a deleterious effect on cyclin F-mediated proteasome degradation, and UPS damage may occur upstream of the proteasome through abnormal ubiquitination or transport to the proteasome [5, 17]. The p.Ala131Ser and p.Arg123Cys variants of CCNF have not been reported previously. Although they are not located in any functional domain, they may also be related to the folding and aggregation of abnormal proteins. Further basic studies are needed to explore the structural or functional changes of these variants.
Currently, this study is the largest sample size of CCNF variant carriers and clinical characteristics analysis. The frequency of PP variants in CCNF in our study was similar to that in previous studies, but there were differences in the results of sex ratio, site of onset, survival, and cognitive function. The CCNF carriers in our ALS cohort had a late age of onset (59.67 years vs. 52.62 years) and a large proportion of bulbar onset (27.8 vs. 10.5%), especially among Asian patients, where bulbar onset has not been reported before. In addition, patients with PP variants of CCNF in our study had a shorter median survival than previously reported results (24.0 months vs. 43.0 months). The proportion of male patients was similar between our cohort and Caucasian patients. In addition to the ALS phenotype, a previous study on FALS and FTD patient cohorts found CCNF carriers with primary lateral sclerosis [5], which was not found in our cohort. This may be because the current cohort only enrolled ALS patients. No significant differences in clinical manifestations were found in patients carrying CCNF variants in different domains in our cohort, but CCNF variant carriers with variants located in cyclin domains tended to have a faster disease progression rate than carriers with variants located in PEST domains (0.92 vs. 0.49). However, the difference was not statistically significant. This was consistent with our finding that all variants located in the PEST sequence were likely benign, while the majority of variants located in the cyclin domains were probably pathogenic.
Our study has the following limitations. First, we did not have genetic information from the relatives of carriers of CCNF variants, so we cannot estimate whether these variants segregate with disease. Second, there are few studies on CCNF variants in ALS, and the correlation between CCNF carriers and clinical phenotypes cannot be clarified. Third, we assessed the pathogenicity of missense variants only by in silico tools, without further basic experiments, which could lead to misjudgments of pathogenicity.
Conclusion
In conclusion, we screened Chinese ALS patients for CCNF variants and found that such variants were common, especially in SALS patients. Our study expands the CCNF genetic variant spectrum in ALS patients; however, the pathogenic mechanisms and pathogenicity of many novel variants are poorly understood. There are differences in clinical characteristics between ALS patients in our cohort and those of previous studies. More follow-up observations and basic studies are needed to elucidate the pathogenic mechanisms and genotype-phenotype associations of CCNF variants.
Supplementary Information
Supplementary Table 1 Information on CCNF variants in ALS patients. Supplementary Table 2 Protein function, structure and nucleic acid conservation prediction of CCNF variants in ALS. Supplementary Table 3 Information on additional ALS-related gene variants in ALS patients with CCNF variants. Supplementary Table 4 Gene analysis and clinical characteristics of CCNF variants in previously reported ALS patients. Supplementary table 5 Clinical features of CCNF variants carriers who had other ALS-related gene variants. Supplementary table 6 The ALS-related genes in the study (XLSX 41 kb)
Acknowledgements
The authors thank all the participants of this study.
Author Contribution
BZ and QRJ searched and selected the studies, analyzed the data, and drafted and revised the article. JYL, QQW, CYL, YBH, BC, LYZ, RWO, KCL, TMY, RH and YX prepared the samples and gave suggestions on study design. BZ, QRJ, RH, and HFS designed the study and provided suggestions for revising the article. All authors read and approved the final manuscript.
Funding
This article was supported by the Sichuan Science and Technology Program (Grant No. 2022ZDZX0023), the National Natural Science Foundation of China (Grant No. 82101485), the Sichuan Science and Technology Program (No. 2020YJ0457), and the Science and Technology Commission Foundation of Chengdu City (Grant No. 2021-YF05-00242-SN).
Data Availability
Anonymized data will be shared upon request with any qualified investigator.
Declarations
Ethical Approval
This study was approved by the Ethics Committee of West China Hospital of Sichuan University, and all patients in the study signed informed consent before participating in the study.
Informed Consent
This article does not contain any previously unpublished studies that would require informed consent.
Consent for Publication
All authors have consented to publication.
Conflict of Interest
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Bi Zhao and Qirui Jiang contributed equally to this work.
References
- 1.Strong MJ, Abrahams S, Goldstein LH, Woolley S, McLaughlin P, Snowden J, et al. Amyotrophic lateral sclerosis - frontotemporal spectrum disorder (ALS-FTSD): revised diagnostic criteria. Amyotroph Lateral Scler Frontotemporal Degener. 2017;18(3-4):153–174. doi: 10.1080/21678421.2016.1267768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72(2):245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arthur KC, Calvo A, Price TR, Geiger JT, Chiò A, Traynor BJ. Projected increase in amyotrophic lateral sclerosis from 2015 to 2040. Nat Commun. 2016;7:12408. doi: 10.1038/ncomms12408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chia R, Chiò A, Traynor BJ. Novel genes associated with amyotrophic lateral sclerosis: diagnostic and clinical implications. Lancet Neurol. 2018;17(1):94–102. doi: 10.1016/s1474-4422(17)30401-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams KL, Topp S, Yang S, Smith B, Fifita JA, Warraich ST, et al. CCNF mutations in amyotrophic lateral sclerosis and frontotemporal dementia. Nat Commun. 2016;7:11253. doi: 10.1038/ncomms11253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bai C, Sen P, Hofmann K, Ma L, Goebl M, Harper JW, et al. SKP1 connects cell cycle regulators to the ubiquitin proteolysis machinery through a novel motif, the F-box. Cell. 1996;86(2):263–274. doi: 10.1016/s0092-8674(00)80098-7. [DOI] [PubMed] [Google Scholar]
- 7.Brooks BR, Miller RG, Swash M, Munsat TL. El Escorial revisited: revised criteria for the diagnosis of amyotrophic lateral sclerosis. Amyotroph Lateral Scler Other Motor Neuron Disord. 2000;1(5):293–299. doi: 10.1080/146608200300079536. [DOI] [PubMed] [Google Scholar]
- 8.Cedarbaum JM, Stambler N, Malta E, Fuller C, Hilt D, Thurmond B, et al. The ALSFRS-R: a revised ALS functional rating scale that incorporates assessments of respiratory function. BDNF ALS Study Group (Phase III) J Neurol Sci. 1999;169(1-2):13–21. doi: 10.1016/s0022-510x(99)00210-5. [DOI] [PubMed] [Google Scholar]
- 9.Dubois B, Slachevsky A, Litvan I, Pillon B. The FAB: a Frontal Assessment Battery at bedside. Neurology. 2000;55(11):1621–1626. doi: 10.1212/wnl.55.11.1621. [DOI] [PubMed] [Google Scholar]
- 10.Nasreddine ZS, Phillips NA, Bédirian V, Charbonneau S, Whitehead V, Collin I, et al. The Montreal Cognitive Assessment, MoCA: a brief screening tool for mild cognitive impairment. J Am Geriatr Soc. 2005;53(4):695–699. doi: 10.1111/j.1532-5415.2005.53221.x. [DOI] [PubMed] [Google Scholar]
- 11.Chong MS, Lim WS, Chan SP, Feng L, Niti M, Yap P, et al. Diagnostic performance of the Chinese Frontal Assessment Battery in early cognitive impairment in an Asian population. Dement Geriatr Cogn Disord. 2010;30(6):525–532. doi: 10.1159/000321665. [DOI] [PubMed] [Google Scholar]
- 12.Lu J, Li D, Li F, Zhou A, Wang F, Zuo X, et al. Montreal cognitive assessment in detecting cognitive impairment in Chinese elderly individuals: a population-based study. J Geriatr Psychiatry Neurol. 2011;24(4):184–190. doi: 10.1177/0891988711422528. [DOI] [PubMed] [Google Scholar]
- 13.Hamilton M. A rating scale for depression. J Neurol Neurosurg Psychiatry. 1960;23(1):56–62. doi: 10.1136/jnnp.23.1.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton M. The assessment of anxiety states by rating. Br J Med Psychol. 1959;32(1):50–55. doi: 10.1111/j.2044-8341.1959.tb00467.x. [DOI] [PubMed] [Google Scholar]
- 15.Beck AT, Beck RW. Screening depressed patients in family practice. A rapid technic. Postgrad Med. 1972;52(6):81–85. doi: 10.1080/00325481.1972.11713319. [DOI] [PubMed] [Google Scholar]
- 16.Chen YP, Yu SH, Wei QQ, Cao B, Gu XJ, Chen XP, et al. Role of genetics in amyotrophic lateral sclerosis: a large cohort study in Chinese mainland population. J Med Genet. 2022;59(9):840–849. doi: 10.1136/jmedgenet-2021-107965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsai PC, Liao YC, Chen PL, Guo YC, Chen YH, Jih KY, et al. Investigating CCNF mutations in a Taiwanese cohort with amyotrophic lateral sclerosis. Neurobiol Aging. 2018;62:243.e1–243.e6. doi: 10.1016/j.neurobiolaging.2017.09.031. [DOI] [PubMed] [Google Scholar]
- 18.Pan C, Jiao B, Xiao T, Hou L, Zhang W, Liu X, et al. Mutations of CCNF gene is rare in patients with amyotrophic lateral sclerosis and frontotemporal dementia from Mainland China. Amyotroph Lateral Scler Frontotemporal Degener. 2017;18(3-4):265–268. doi: 10.1080/21678421.2017.1293111. [DOI] [PubMed] [Google Scholar]
- 19.Tian D, Li J, Tang L, Zhang N, Fan D (2018) Screening for CCNF mutations in a Chinese Amyotrophic Lateral Sclerosis Cohort. Front Aging Neurosci:10185. 10.3389/fnagi.2018.00185 [DOI] [PMC free article] [PubMed]
- 20.Tripolszki K, Gampawar P, Schmidt H, Nagy ZF, Nagy D, Klivényi P, et al. Comprehensive genetic analysis of a Hungarian amyotrophic lateral sclerosis cohort. Front Genet. 2019;10:732. doi: 10.3389/fgene.2019.00732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rayner SL, Yang S, Farrawell NE, Jagaraj CJ, Cheng F, Davidson JM, et al. TDP-43 is a ubiquitylation substrate of the SCF(cyclin F) complex. Neurobiol Dis. 2022;167:105673. doi: 10.1016/j.nbd.2022.105673. [DOI] [PubMed] [Google Scholar]
- 22.McCann EP, Williams KL, Fifita JA, Tarr IS, O'Connor J, Rowe DB, et al. The genotype-phenotype landscape of familial amyotrophic lateral sclerosis in Australia. Clin Genet. 2017;92(3):259–266. doi: 10.1111/cge.12973. [DOI] [PubMed] [Google Scholar]
- 23.Rascovsky K, Hodges JR, Knopman D, Mendez MF, Kramer JH, Neuhaus J, et al. Sensitivity of revised diagnostic criteria for the behavioural variant of frontotemporal dementia. Brain. 2011;134(Pt 9):2456–2477. doi: 10.1093/brain/awr179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.D'Angiolella V, Esencay M, Pagano M. A cyclin without cyclin-dependent kinases: cyclin F controls genome stability through ubiquitin-mediated proteolysis. Trends Cell Biol. 2013;23(3):135–140. doi: 10.1016/j.tcb.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.D'Angiolella V, Donato V, Vijayakumar S, Saraf A, Florens L, Washburn MP, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466(7302):138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.D'Angiolella V, Donato V, Forrester FM, Jeong YT, Pellacani C, Kudo Y, et al. Cyclin F-mediated degradation of ribonucleotide reductase M2 controls genome integrity and DNA repair. Cell. 2012;149(5):1023–1034. doi: 10.1016/j.cell.2012.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dangoumau A, Marouillat S, Coelho R, Wurmser F, Brulard C, Haouari S et al (2021) Dysregulations of expression of genes of the ubiquitin/SUMO pathways in an in vitro model of amyotrophic lateral sclerosis combining oxidative stress and SOD1 gene mutation. Int J Mol Sci 22(4). 10.3390/ijms22041796 [DOI] [PMC free article] [PubMed]
- 28.Yu Y, Nakagawa T, Morohoshi A, Nakagawa M, Ishida N, Suzuki N, et al. Pathogenic mutations in the ALS gene CCNF cause cytoplasmic mislocalization of Cyclin F and elevated VCP ATPase activity. Hum Mol Genet. 2019;28(20):3486–3497. doi: 10.1093/hmg/ddz119. [DOI] [PubMed] [Google Scholar]
- 29.Lee A, Rayner SL, De Luca A, Gwee SSL, Morsch M, Sundaramoorthy V et al (2017) Casein kinase II phosphorylation of cyclin F at serine 621 regulates the Lys48-ubiquitylation E3 ligase activity of the SCF((cyclin F)) complex. Open Biol 7(10). 10.1098/rsob.170058 [DOI] [PMC free article] [PubMed]
- 30.Lee A, Rayner SL, Gwee SSL, De Luca A, Shahheydari H, Sundaramoorthy V, et al. Pathogenic mutation in the ALS/FTD gene, CCNF, causes elevated Lys48-linked ubiquitylation and defective autophagy. Cell Mol Life Sci. 2018;75(2):335–354. doi: 10.1007/s00018-017-2632-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hogan AL, Don EK, Rayner SL, Lee A, Laird AS, Watchon M, et al. Expression of ALS/FTD-linked mutant CCNF in zebrafish leads to increased cell death in the spinal cord and an aberrant motor phenotype. Hum Mol Genet. 2017;26(14):2616–2626. doi: 10.1093/hmg/ddx136. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table 1 Information on CCNF variants in ALS patients. Supplementary Table 2 Protein function, structure and nucleic acid conservation prediction of CCNF variants in ALS. Supplementary Table 3 Information on additional ALS-related gene variants in ALS patients with CCNF variants. Supplementary Table 4 Gene analysis and clinical characteristics of CCNF variants in previously reported ALS patients. Supplementary table 5 Clinical features of CCNF variants carriers who had other ALS-related gene variants. Supplementary table 6 The ALS-related genes in the study (XLSX 41 kb)
Data Availability Statement
Anonymized data will be shared upon request with any qualified investigator.