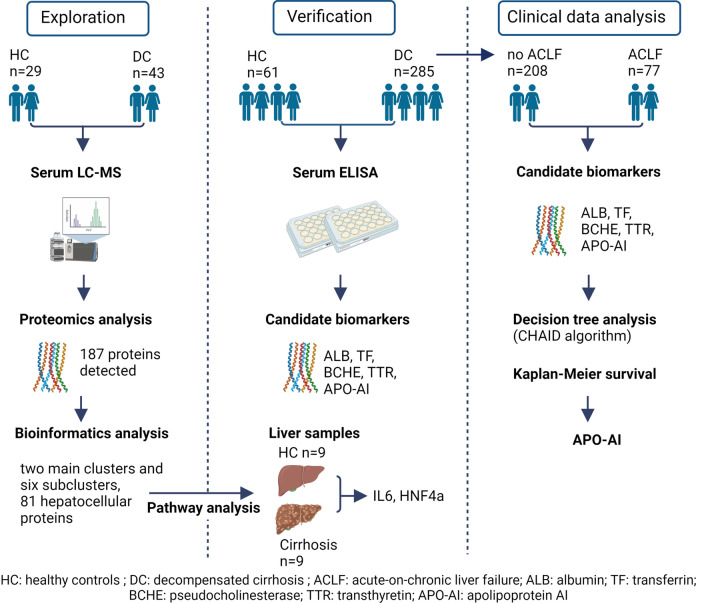
Abstract
Background and aim
Since hepatocytes produce majority of serum proteins, patients with cirrhosis display substantial alterations in the serum proteome. The aim of the current study was to characterize these changes and to study the prognostic utility of hepatocellular proteins available in routine clinical testing.
Methods
Sera from 29 healthy controls and 43 patients with cirrhosis were subjected to untargeted proteomic analysis. Unsupervised hierarchical clustering was performed with Perseus software and R. Ingenuity pathway analysis (IPA) suggested upstream regulators that were validated in liver tissues. The behavior and prognostic usefulness of selected biomarkers was investigated in 61 controls and 285 subjects with decompensated cirrhosis.
Results
Proteomics uncovered 65 and 16 hepatocellular serum proteins that are significantly downregulated or upregulated in patients with cirrhosis vs. controls. Hierarchical clustering revealed two main clusters and six sub-clusters. IPA identified HNF4α and IL-6 as the two major upstream regulators that were confirmed by hepatic gene expression analyses. Among pseudocholinesterase, transferrin, transthyretin, albumin, and apolipoprotein AI (Apo-AI), Apo-AI was the best predictor of 90-days transplant-free survival (AUROC 0.678; p = 0.0001) and remained an independent predictor in multivariable Cox independently of the presence of acute-on-chronic liver failure.
Conclusion
Our study reveals cirrhosis-associated changes in hepatocellular serum proteins and underlying transcription factors. Serum apolipoprotein AI may constitute a useful prognostic adjunct in patients with decompensated cirrhosis.
Graphical abstract
Supplementary Information
The online version contains supplementary material available at 10.1007/s12072-022-10473-x.
Keywords: Mass spectrometry, Fibrosis, Interleukin, Hepatocyte nuclear factor, Decompensated cirrhosis
Introduction
Liver constitutes a central metabolic hub of the human body [1]. It receives nutrients from the intestine as well as compounds from peripheral tissues that are either stored or used for synthesis of new products [1]. Hepatocytes, the parenchymal cells of the liver, are responsible for these processes. They constitute highly active secretory cells and produce majority of proteins found in the serum including hormones, carrier and homeostatic proteins [2]. Protein synthesis is directed by a concerted action of liver-enriched transcription factors [3]. This teamwork enables the generation of multiple carrier and homeostatic proteins that are essential for organismal wellbeing. At the same time, it is responsible for adjustment to stress situations and because of that it responds to inflammatory stimuli [3, 4] It leads to a decreased generation of compounds that are deemed dispensable (so called anti-acute-phase proteins) and increased manufacturing of mediators supposed to help with the stress (termed acute-phase proteins, APPs) [2, 5] APPs are further subdivided into type I and type II, depending on whether they are regulated by interleukin 1-like cytokines such as interleukin-1α/β and tumor necrosis factor or IL-6 like cytokines such as IL-6, IL-11, oncostatin M and leukemia inhibitory factor.
Cirrhosis is the result of an evolutionarily conserved wound-healing response of the liver to tissue injury, usually triggered by inflammatory or immune-mediated mechanisms resulting in a loss of hepatocytes and the remodeling of the tissue architecture [6]. Capillarization of the sinusoids promotes hypoxia, which further impairs the synthetic ability of hepatocytes perpetuating injury and the release of damage-associated molecular patterns (DAMPs) [7]. In parallel, portal hypertension, intestinal dysbiosis, and impaired gut barrier function promote translocation of pathogen-associated molecular patterns (PAMPs) into the circulation [8].
In summary, cirrhosis is characterized by a hepatocellular loss together with hepatocellular re-programming triggered by hepatocellular de-differentiation, hypoxia, and innate immune activation [7, 8]. Since production of secreted proteins constitutes one of the key functions of the liver and changes in serum proteome can be easily measured, serum levels of hepatocellular proteins are potentially useful biomarkers reflecting the functional state of the liver. Notably, some of them, such as albumin or coagulation factors, became well-established components of prognostic scores [9]. Despite that, the exact alterations in hepatocellular serum proteome occurring in advanced liver disease still remain to be systematically characterized. Therefore, we performed an unbiased proteomic analysis of sera from cirrhotic patients as well as corresponding controls and used a bioinformatics approach to elucidate the biological pathways responsible for the observed changes. To validate the bioinformatics predictions, the results were corroborated via RNA expression analyses and the prognostic ability of selected hepatocellular products was studied in an independent cohort of patients with decompensated cirrhosis.
Experimental procedures
Patient cohorts
In total, frozen serum samples from 285 patients with acute decompensation (AD) of cirrhosis and from 61 self-identified healthy individuals were used for this study. Of these, sera from 21 patients with acute chronic liver failure (ACLF) or pre-ACLF, 22 patients with stable decompensated cirrhosis (SDC) according to the PREDICT study [10], and 29 healthy individuals were randomly selected for proteomic analysis (Table 1). Patient samples were derived from patients hospitalized for acute decompensation of cirrhosis treated at the Jena University Hospital between 09/2010 and 07/2015 as described previously [11] and frozen at − 80 °C until analysis. Sera from self-declared healthy individuals were collected at the University Hospital Aachen, Germany between the years 2016 and 2019 as described previously [12].
Table 1.
Baseline characteristics
| Proteome analysis | Candidate biomarker analysis | |||
|---|---|---|---|---|
| Decompensated cirrhosis (n = 43) | Healthy subjects (n = 29) | Decompensated cirrhosis (n = 285) | Healthy subjects (n = 61) | |
| Age (yrs) | 53 (47–62) | 57 (53–61) | 59 (53–68) | 61 (56–67) |
| Male sex (%) | 38 (88) | 21 (72) | 210 (74) | 45 (74) |
| Alcohol-related liver disease (%) | 39 (91) | 0 | 227 (80) | 0 |
| Ascites | 43 (100) | 0 | 285 (100) | 0 |
| ACLF | 13 (30) | 0 | 77 (27) | 0 |
| Pre-ACLF | 9 (21) | 0 | 66 (23) | 0 |
| Hepatocellular carcinoma | 7 (16) | 0 | 40 (14) | 0 |
| MELD score | 16 (12–25) | N/A | 17 (12–22) | N/A |
| Bilirubin (µmol/L) | 58 (23–184) | 10 (6–13) | 46 (24–109) | 8 (6–13) |
| ALT (µmol/[L × s]) | 0.5 (0.4–1.3) | 0.5 (0.4–0.6) | 0.6 (0.4–0.9) | 0.4 (0.3–0.5) |
| AST (µmol/[L × s]) | 1.0 (0.4–1.3) | 0.4 (0.4–0.5) | 1.1 (0.7–1.9) | 0.4 (0.4–0.5) |
| INR | 1.5 (1.3–1.9) | 0.9 (0.9–1) | 1.4 (1.2–1.7) | 0.9 (0.9–1) |
| Platelets (/nL) | 137 (96–178) | 244 (206–285) | 124 (80–177) | 224 (187–284) |
| Creatinine (µmol/L) | 72 (61–148) | 80 (63–96) | 91 (63–142) | 76 (61–94) |
| Albumin (g/L) | 23 (20–27) | 48 (46–50) | 24 (20–29) | 48 (44–50) |
| BCHE (ng/mL) | 720 (358–1286) | 3861 (3308–4553) | 691 (424–949) | 3966 (3198–4791 |
| Transferrin (mg/dL) | 86 (57–151) | 268 (230–292) | 104 (65–162) | 263 (231–284) |
| Transthyretin (mg/dL) | 51 (35–95) | 215 (157–250) | 60 (39–85) | 222 (177–270) |
| Apolipoprotein AI (g/L) | 0.77 (0.30–1.28) | 2.6 (2.4–3.0) | 0.83 (0.50–1.27) | 2.8 (2.5–3.2) |
Baseline characteristics are shown as frequencies or medians with interquartile ranges
Human liver tissue
Liver tissues from nine patients who underwent liver surgery at the University of Aachen between the years 2006 and 2018 were analyzed. Unaffected surrounding portions of liver tissue from nine patients collected during oncological surgery for exclusion of liver malignancy were used as controls (Supplementary Table S1). RNA was isolated using the RNeasy tissue mini isolation kit (Qiagen, Hilden, Germany). RNA was translated to cDNA using the M-MLV reverse transcriptase kit (Promega, Madison, WI, USA) with random hexamer primers (Thermo Scientific, Waltham, MA, USA). The relative expression of genes of interest was determined using qPCR using specific primers (Supplementary Table S2). The human ribosomal gene RPLPO was used as an internal loading control.
Further experimental procedures (e.g., proteomics, bioinformatics, and statistical analysis) are given in the supplementary materials.
Results
Cirrhosis is associated with a profound alteration of hepatocellular serum proteins
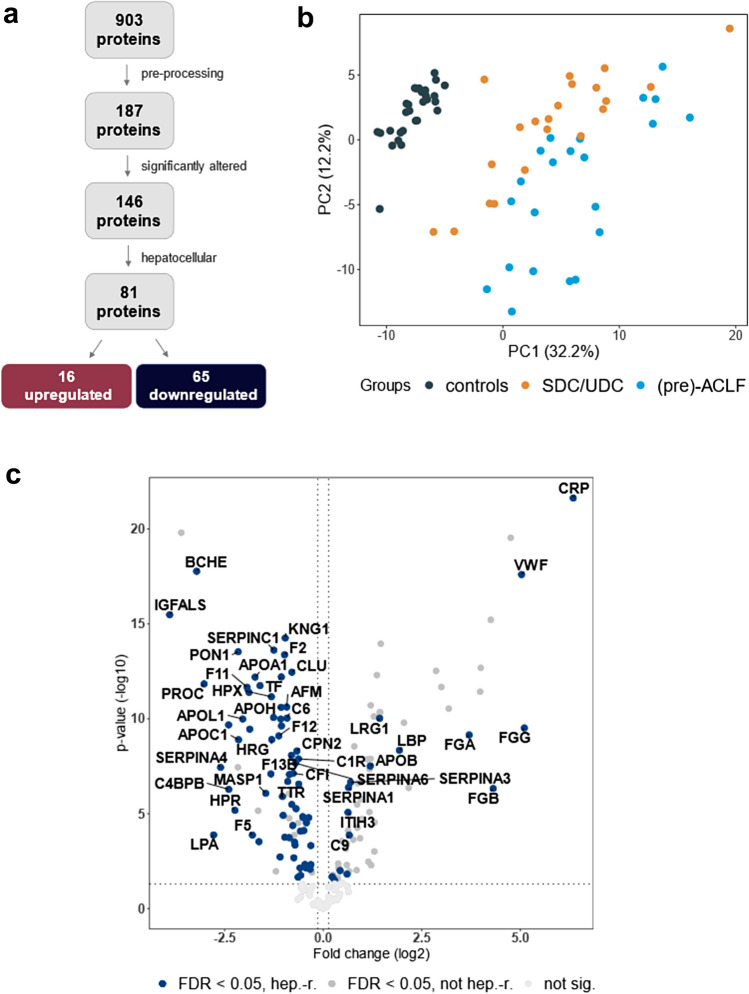
To identify biomarkers of hepatocellular function altered in patients with cirrhosis, sera from 29 healthy individuals and 43 patients with acute decompensation of cirrhosis were subjected to untargeted proteomic analysis (Table 1). Out of 903 identified proteins, 187 were detected in at least 50% of the samples (Fig. 1A, Supplementary Table S3). Serum levels of 146 proteins differed significantly (false discovery rate (FDR) < 0.05) between groups and both groups were clearly separated by principal component analysis (Fig. 1B, Supplementary Table S4). Eighty-one of the proteins were of hepatocellular origin, and of these, sixty-five were significantly lower in sera from patients with cirrhosis as compared to healthy controls applying FDR of less than 0.05. Notably, C-reactive protein (CRP) and pseudocholinesterase (or butyrylcholinesterase; BCHE) were among the most significantly upregulated and downregulated proteins, respectively, demonstrating both loss of hepatocellular synthesis and activation of acute-phase response (Fig. 1C). On the other hand, albumin (ALB), a well-established surrogate of hepatocellular synthesis, displayed only a moderate difference between patients with cirrhosis and controls (Supplementary Table S4).
Fig. 1.
Serum proteome analysis. a Flow chart shows the number of detected proteins (903) and the number of proteins that were identified in a sufficient number of participants (187). One hundred and forty-six of them differentially regulated in a group of patients with decompensated cirrhosis, comprising patients with stable (SDC) and unstable decompensated cirrhosis (UDC) and (pre-)acute-on-chronic liver failure (ALCF) compared to controls (FDR < 0.05). Among those, 81 were of hepatocellular origin (16 upregulated, 65 downregulated). b The first two dimensions of the principal component analysis present a clear discrimination between patients with decompensated cirrhosis and healthy controls. c Volcano plot illustrates differentially abundant hepatocellular proteins. The − log10 (false discovery-adjusted p value) is plotted against the log2-fold change (resembling the differences between means of log2-transformed LFQ intensities for patients with cirrhosis vs. those of healthy controls). A log2-fold change > 0 indicates proteins upregulated in patients with cirrhosis, while a value < 0 identifies downregulated species. The non-axial vertical lines denote ± 0.05-fold change (the smallest log2-fold change observed for any significantly altered protein), while the non-axial horizontal line denotes p = 0.05 as the significance threshold. hep-r hepatocyte-related
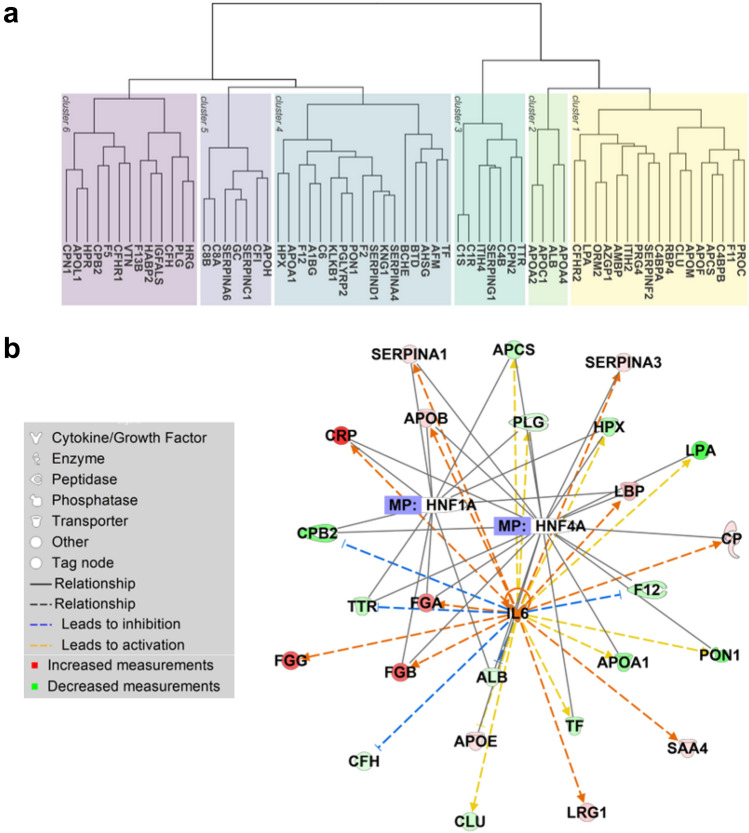
Unsupervised hierarchical cluster analysis revealed that these 65 proteins of interest could be categorized into 2 main clusters and 6 sub-clusters (Fig. 2A, Supplementary Table S5). The number of proteins per cluster ranged from 4 to 20. Overall, the analysis demonstrated profound differences in the hepatocellular serum proteome of patients with decompensated cirrhosis when compared to controls, as shown in Supplementary Tables S5 and S6, but also the fact that hepatocellular proteins exhibit unique behavior during the development of end-stage liver disease.
Fig. 2.
Serum proteins of hepatocellular origin decreased in decompensated cirrhosis. a The dendrogram indicates the hierarchical clustering of 65 serum proteins of hepatocellular origin that were significantly downregulated (log2-fold change < 0 and FDR < 0.05) in patients with cirrhosis as compared to healthy controls. Proteins are labeled via the according gene symbol. b Network of three most strongly enriched upstream regulators (HNF1A, HNF4A, and IL-6) and their corresponding downstream regulators as predicted by the Ingenuity Pathway Core Analysis. The analysis was based on hepatocyte-related proteins that were significantly altered (FDR < 0.05) between healthy controls and individuals with liver cirrhosis
Upstream regulator analyses revealed two major upstream regulators associated with analyzed hepatocellular proteins
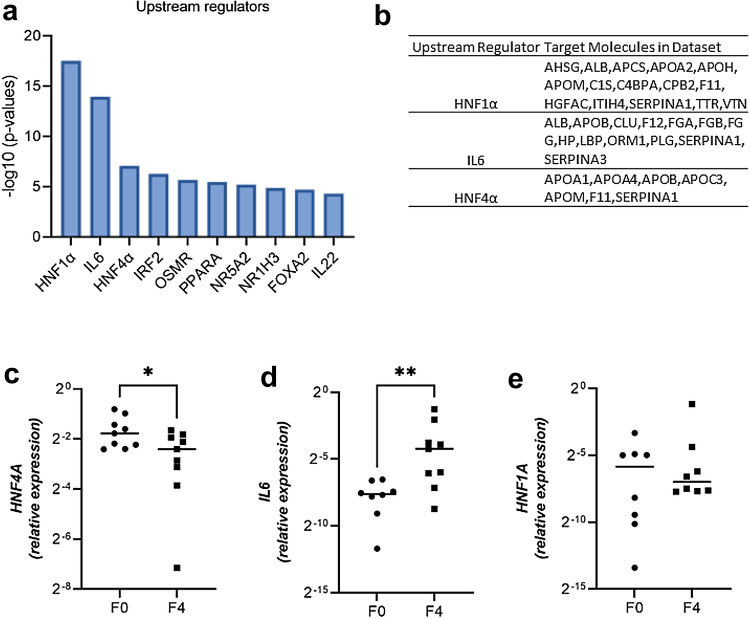
To obtain an insight into the molecular mechanisms underlying serum proteome changes in decompensated cirrhosis, proteomics data were submitted to QIAGEN’s Ingenuity Pathway Analysis (IPA, QIAGEN Redwood City, www.qiagen.com/ingenuity). IPA core analysis revealed 71 upstream regulators to be significantly altered (threshold for overlap p value < 0.05). Hepatocyte nuclear factors 1α, 4α (HNF1α, HNF4α), and interleukin 6 (IL6) were the top three predicted transcriptional regulators that may explain the changes occurring in decompensated cirrhosis (Fig. 3A) and the predicted target proteins included albumin, apolipoproteins, transport proteins, coagulation factors, and acute-phase proteins (Fig. 3B). The suggested expression changes in HNF4A and IL6 mRNA, but not HNF1A were validated in liver tissue of patients with cirrhosis as compared to nine controls without (Fig. 3C–E). Hepatic HNF4A expression positively correlated with hepatic albumin (ALB), transthyretin (TTR), transferrin (TF), and apolipoprotein AI (Apo-AI) expression, whereas hepatic IL6 expression negatively correlated with hepatic ALB and BCHE expression (Fig. 4), but the correlations were not particularly strong. To further visualize the interplay between the key regulators (i.e., HNF1A, HNF4A and IL6) and the serum proteins that were altered in our proteomic dataset, we performed an IPA-driven network visualization (Fig. 2B). It demonstrated that most serum proteins are co-regulated by more of the players and that the observed changes are likely a result of a complex regulation involving several different players.
Fig. 3.
Analysis of upstream regulators altered in control vs. cirrhotic livers. a IPA analysis delineates the pathways altered in patients with decompensated cirrhosis vs. controls. Minus log10 (p values) of predicted upstream regulators are shown. b Target genes that are associated with top three upstream regulators obtained from IPA analysis. C–E Relative mRNA expression of selected genes was assessed in livers from patients with cirrhosis and non-fibrotic livers and was normalized to human ribosomal protein (RPLPO) as a housekeeping gene. Medians and individual values are shown. *p < 0.05; **p < 0.01 in non-parametric t test. F0 no fibrosis, F4 fibrosis stage 4
Fig. 4.
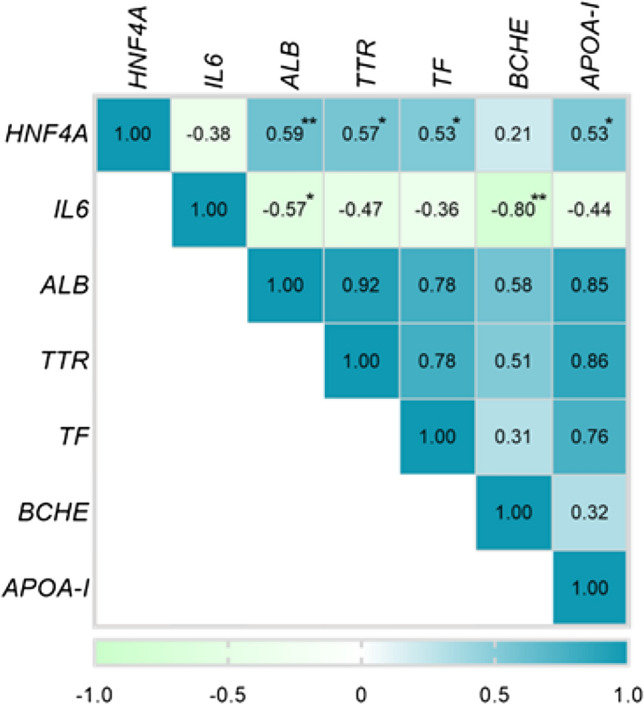
Correlation between hepatic mRNA expression of upstream regulators and their putative target genes. Spearman correlation coefficients based on RT-PCR analysis of cirrhotic and non-fibrotic livers are shown (n = 9 each). *p < 0.05; **p < 0.01. Proteins are labeled via the according gene symbol
Biomarkers of hepatocellular synthesis correlate with the trajectory of acute decompensation in cirrhosis
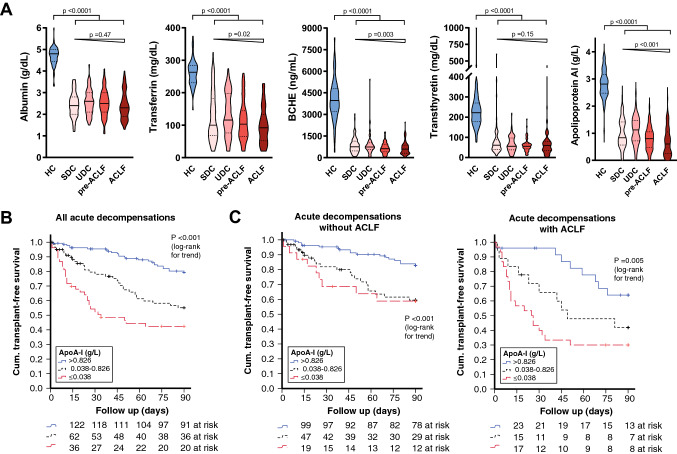
Based on hierarchical cluster analysis (Supplementary Table S5) and the availability in routine clinical testing, we selected five indicator proteins, which were decreased in sera from patients with cirrhosis for further analysis. Their concentrations were analyzed in sera from 61 healthy individuals and from 285 patients with acute decompensation of cirrhosis. As expected, patients with cirrhosis had significantly lower serum concentrations of albumin, transferrin, transthyretin, pseudocholinesterase, and apolipoprotein AI compared to healthy individuals (Fig. 5A). In addition, transferrin, BCHE, and apolipoprotein AI but not albumin and transthyretin display a stage-dependent decrease of serum concentrations from stable (SDC) and unstable decompensated cirrhosis (UDC) to pre-ACLF and ACLF as indicated by a significant Jonckheere-Terpstra test (Fig. 5A).
Fig. 5.
Regulated hepatocellular biomarkers and outcome in acute decompensation of cirrhosis. a Concentrations of albumin, transferrin, pseudocholinesterase (BCHE), transthyretin, and apolipoprotein AI in sera from healthy controls (HC) and patients with stable decompensated cirrhosis (SDC), unstable decompensated cirrhosis (UDC), pre-ACLF and ACLF depicted by violin plots. b Kaplan–Meier analysis of 90-days transplant-free survival stratified for apolipoprotein AI (Apo-AI). Cutoffs were derived from decision tree analysis using the Chi-square automatic interaction detectors (CHAID) algorithm with death/liver transplantation (LTX) within 90 days as events and albumin, pseudocholinesterase, transferrin, transthyretin, and apolipoprotein AI as variables. c Kaplan–Meier analysis of 90-days transplant-free survival stratified for Apo-AI if shown for patients with acute decompensation without (left panel) and with ACLF at baseline (right panel). p values from log-rank test for linear trends are indicated
Higher serum levels of transthyretin are associated with better prognosis in patients with ACLF
Out of 285 patients with acute decompensation of cirrhosis (Table 1), 82 (29%) died within 90 days and 10 (4%) underwent liver transplantation. Diagnostic accuracy in predicting mortality or transplant at 90 days was compared using areas under the receiver operating characteristic curves (AUROC, supplementary table S7). Among the investigated hepatocellular biomarkers, apolipoprotein AI (AUROC 0.678; p = 0.0001), BCHE (AUROC 0.626; p = 0.001), transferrin (AUROC 0.602; p = 0.009), and transthyretin (AUROC 0.593; p = 0.01) were able to discriminate between the two groups, whereas serum albumin was not (AUROC 0.496; p = 0.923). The prognostic abilities of apolipoprotein AI were numerically comparable to those of the MELD score (AUROC = 0.685; p < 0.001). In Chi-square automatic interaction detectors (CHAID) analysis, apolipoprotein AI outperformed BCHE, transferrin, transthyretin, and albumin as predictors of death or transplant within 90 days and suggested stratification of apolipoprotein AI based on two optimized cutoffs, namely 0.380 g/L and 0.826 g/L. In Kaplan–Meier analysis, higher apolipoprotein AI serum concentrations were associated with better 90-days transplant-free survival (Fig. 5B), which remained true in the subgroups of patients with and without ACLF (Fig. 5C). In time-to-event analysis, apolipoprotein AI strata remained significant in two different multivariable Cox regression models adjusting for demographic factors and for severity of liver disease, i.e., MELD score or ACLF (Table 2).
Table 2.
Cox regression analysis of death or transplant within 90 days
| Univariable analysis | Multivariable analysis (model 1) | Multivariable analysis (model 2) | ||||
|---|---|---|---|---|---|---|
| Unadjusted hazard ratio (95% CI) | p value | Adjusted hazard ratio (95% CI) |
p value | Adjusted hazard ratio (95% CI) |
p value | |
| Age | 1.05 (1.03–1.07) | < 0.001 | 1.06 (1.04–1.09) | < 0.001 | 1.07 (1.04–1.09) | < 0.001 |
| MELD score (per 1-point increase) | 1.10 (1.07–1.13) | < 0.001 | Not included | 1.09 (1.05–1.12) | < 0.001 | |
| ACLF | 2.88 (1.91–4.35) | < 0.001 | 2.53 (1.61–3.96) | < 0.001 | Not included | |
| HCC at baseline | 1.96 (1.18–3.24) | 0.009 | 1.88 (1.07–3.30) | 0.029 | 1.67 (0.96–2.91) | 0.071 |
| Apolipoprotein AI* | ||||||
| > 0.826 g/L | 1.00 (reference) | 1.00 (reference) | 1.00 (reference) | |||
| 0.381–0.826 g/L | 2.67 (1.58–4.53) | < 0.001 | 2.72 (1.60–4.63) | < 0.001 | 2.27 (1.33–3.88) | 0.003 |
| ≤ 0.381 g/L | 4.58 (2.69–7.78) | <0.001 | 4.33 (2.49–7.51) | < 0.001 | 3.29 (1.85–5.85) | <0.001 |
*cutoff based on Chi-square automatic interaction detectors (CHAID) algorithm
Discussion
Using untargeted serum proteomics, in silico upstream regulator analysis and validation ELISAs alongside with hepatic gene expression analysis, we herein show that serum concentrations of proteins regulated by HNF-4α-and IL-6 are reduced in cirrhosis, associated with more advanced stages of cirrhosis, and indicate poor transplant-free survival. Low serum levels of apolipoprotein AI identified patients with very low likelihood of transplant-free survival in a large cohort of patients with acute decompensation of cirrhosis. The prognostic usefulness of apolipoprotein AI seen in our study further extends and corroborates previous findings [13, 14].
The prognosis in patients with advanced chronic liver disease is mainly determined by liver function, portal hypertension, the presence of extrahepatic organ failure, and systemic inflammation [15–17]. In advanced cirrhosis, inflammation is a major driver of complications and mortality, and pro-inflammatory cytokines, acute-phase proteins, and immune activation markers have been employed to improve risk prediction in decompensated liver disease [18–21]. The identified prognostic relevant hepatocellular proteins, albumin, apolipoprotein AI, transthyretin, and transferrin, are negative acute-phase proteins, and low concentrations of negative acute-phase proteins can serve as surrogates of reduced hepatic synthesis and impaired hepatocellular reserve but also indicate the presence of systemic inflammation. Upstream regulator analysis identified HNF-4α and IL-6 as the most important regulators underlying the observed serum proteome changes in patients with decompensated cirrhosis. Whereas the hepatic mRNA expression of the principal inducer of acute-phase response, IL6, was increased in cirrhotic livers, hepatic mRNA expression of HNF4A was reduced and positively correlated with the expression of ALB, TTR, TF and Apo-AI. The promoter regions of TTR and other negative APP contain HNF-4α binding sites, and this correlation has been observed in patients with alcoholic hepatitis [22–24].
Although BCHE expression did not correlate with HNF4A expression, its strong negative correlation with IL6 expression alongside the positive correlation of BCHE with transferrin and transthyretin on the protein level suggest a similar role as a negative APP and likely explains its marked decrease in patients with cirrhosis.
Despite a comparable hepatic regulation of the indicator proteins investigated in more detail, i.e., albumin, apolipoprotein AI, transthyretin, transferrin, and BCHE, their role as a prognostic biomarker somewhat differed. This may be due to different serum half-lives ranging from < 1 day (apolipoprotein AI), 2–4 days (transthyretin) across 8–12 days for transferrin and BCHE to 3 weeks (albumin). As higher levels of apolipoprotein AI identified a subgroup of patients with ACLF with better prognosis, apolipoprotein AI may be interpreted as the most dynamic marker of the hepatic reserve after liver injury and organ failure.
Hepatic HNF-4α-dependent gene expression is altered across the spectrum from fibrogenesis to decompensation and liver failure. Mechanistically, in early liver disease with fibrosis, liver matrix stiffness and cytoskeletal tension inhibit the hepatocellular HNF4-α transcriptional network [25]. In advanced liver disease, such as alcohol-related liver failure, hepatic activity of the liver-enriched transcription factor HNF-4α is severely inhibited, reducing hepatic expression of cytochrome P450 enzymes, apolipoproteins, and aldolases [26]. This process is driven by growth factors such as TGF-β, HGF, and EGF, cytokines such as TNF, IL-1β, IFN-γ, and inflammatory mediators such as PGE2 and LPS, and hepatocyte de-differentiation may play an additional role [26]. Because these upstream regulators play a prognostic role for complications of decompensated cirrhosis as well, the association of HNF-4α-regulated proteins with outcome in patients with acute decompensation of cirrhosis is plausible.
In a study on liver tissue from patients at different stages of decompensation, HNF-4α expression was downregulated and correlated with liver dysfunction, fibrosis stage, and prognostically relevant serum parameters bilirubin, albumin, and prothrombin time [27]. As a result, lower serum concentrations of transthyretin and BCHE correlated with the Child–Pugh stage and with the presence of complications such as hepatic encephalopathy in a study on patients with predominantly viral cirrhosis [28]. Plasma proteome of 459 patients with compensated alcohol-related chronic liver disease demonstrated that lower levels of plasma albumin, BCHE, and transthyretin were associated with the presence of significant liver fibrosis and hepatic inflammation (vs. no/minimal fibrosis/inflammation), but not with the degree of hepatic steatosis [29].
In this study, we demonstrate remarkable differences in serum levels of bona fide hepatocellular proteins as well as an association between lower apolipoprotein AI levels and transplant-free survival after adjusting for confounding factors such as age, MELD score, and the presence of hepatocellular carcinoma. Notably, the prognostic ability of apolipoprotein AI was comparable to the routinely used MELD score consisting of three different biomarkers. Moreover, apolipoprotein AI remained a significant predictor of mortality in two different multivariable models including MELD or ACLF.
These data suggest that the activity of the liver-enriched transcription factor HNF-4α as a surrogate for hepatocellular reserve, degree of fibrosis, hepatocyte differentiation, and inflammatory status may provide additional prognostic information in advanced stages of cirrhosis. However, several limitations of our work need to be considered. The observed analyses are based on a single-center cohort, and the clinical cohorts used for mRNA analyses differed from cohorts used for proteomic assessment since liver biopsies are rarely ethically justifiable in subjects with decompensated liver cirrhosis. Furthermore, the single biomarkers assessed in our study have been studied before and in this respect, our results are confirmative. The demonstrated heterogeneity in hepatocellular proteins in subjects with cirrhosis should spur large-scale proteomic studies addressing the prognostic usefulness of the individual protein patterns. Such studies might be able to identify and validate clinically useful prognostic biomarkers as it has been recently demonstrated for alcoholic liver disease [29].
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We thank all patients for their participation in our study.
Abbreviations
- APPs
Acute-phase proteins
- ALB
Albumin
- TF
Transferrin
- TTR
Transthyretin
- BCHE
Pseudocholinesterase
- Apo-AI
Apolipoprotein A1
- CRP
C-reactive protein
- HC
Healthy controls
- SDC
Stable decompensated cirrhosis
- UDC
Unstable decompensated cirrhosis
- ACLF
Acute-on-chronic liver failure
- MELD
Model of end-stage liver disease
- CHAID
Chi-square automatic interaction detectors
- HNF1A and 4A
Hepatocyte nuclear factor 1 or 4 alpha
- RPLPO
Human ribosomal protein
- TGF-b
Transforming growth factor beta
- HGF
Hepatocyte growth factor
- EGF
Epidermal growth factor
- TNF
Tumor necrosis factor
- IL-1b
Interleukin-1 beta
- IFN-g
Interferon gamma
- PGE2
Prostaglandin E2
- IL-6
Interleukin-6
Author contributions
Study concept and design: BG, NG, TB, PS. Acquisition of data: BG, NG, PAR, CP, IK, KR, AS, TB, PS. Analysis and interpretation of data: all authors. Drafting of the manuscript: BG, NG, TB, PS. Critical revision of the manuscript for important intellectual content: all authors. Statistical analysis: NG, BG, CP, KR, TB. Figures and tables: NG, BG, CP, KR, TB. Obtained funding: LF, NG, TB, PS. Study supervision: NG, TB, PS. All authors had full access to all of the data and approved the final version of this manuscript. All authors can take responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was supported by the Deutsche Forschungsgemeinschaft (DFG) grants SFB 1382 (ID 403224013) (to P.S., N.G. T.B., M.v.B), STR 1095/6–1 (Heisenberg professorship to P.S.), UFZ for the ProMetheus platform for proteomics and metabolomics (I.K., K.S., M.v.B.), National Natural Science Foundation of China (81804019, to L.F.) and Guangxi Natural Science Foundation (2018GXNSFBA050041, to L.F.)
Data availability
Proteomic data were deposited in a publicly available database. Further data are available from authors upon reasonable request.
Declarations
Conflict of interest
All authors declare: no support from any organization other than the below mentioned ones for the submitted work; no financial relationships with any organizations that might have an interest in the submitted work in the previous 3 years; no other relationships or activities that could appear to have influenced the submitted work. Hence, all authors declare themselves to be independent from funders with respect to this manuscript.
Ethical approval
The study protocol conformed to the ethical guidelines of the 1975 Declaration of Helsinki and was approved by the Ethics committees of Jena University Hospital (2880–08/10, 3683–02/3, 3150–06/11), Aachen University Hospital (EK 173/15), by the Ethics committees of First Faculty of Medicine and Prague General University Hospital (MZ10-UK1LF-BruhaRadan-B).
Further remarks
We attest that we did not use any copyright protected material in our manuscript. No writing assistance was provided.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The authors Berivan Gurbuz, Nurdan Guldiken, Tony Bruns, and Pavel Strnad share authorship.
References
- 1.Rui L. Energy metabolism in the liver. Compr Physiol. 2014;4:177–197. doi: 10.1002/cphy.c130024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kuscuoglu D, Janciauskiene S, Hamesch K, Haybaeck J, Trautwein C, Strnad P. Liver—master and servant of serum proteome. J Hepatol. 2018;69:512–524. doi: 10.1016/j.jhep.2018.04.018. [DOI] [PubMed] [Google Scholar]
- 3.Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part I: The hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev. 2002;54:129–158. [DOI] [PubMed]
- 4.Schrem H, Klempnauer J, Borlak J. Liver-enriched transcription factors in liver function and development. Part II: The C/EBPs and D site-binding protein in cell cycle control, carcinogenesis, circadian gene regulation, liver regeneration, apoptosis, and liver-specific gene regulation. Pharmacol Rev. 2004;56:291–330. [DOI] [PubMed]
- 5.Strnad P, Tacke F, Koch A, Trautwein C. Liver-guardian, modifier and target of sepsis. Nat Rev Gastroenterol Hepatol. 2017;14:55–66. doi: 10.1038/nrgastro.2016.168. [DOI] [PubMed] [Google Scholar]
- 6.Le Couteur DG, Fraser R, Hilmer S, Rivory LP, McLean AJ. The hepatic sinusoid in aging and cirrhosis: Effects on hepatic substrate disposition and drug clearance. Clin Pharmacokinet. 2005;44:187–200. doi: 10.2165/00003088-200544020-00004. [DOI] [PubMed] [Google Scholar]
- 7.Luedde T, Kaplowitz N, Schwabe RF. Cell Death and Cell Death Response in Liver Disease: Mechanisms and Clinical Relevance. Gastroenterology. 2014;147:765–783. doi: 10.1053/j.gastro.2014.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szabo G, Petrasek J. Inflammasome activation and function in liver disease. Nat Rev Gastroenterol Hepatol. 2015;12:387–400. doi: 10.1038/nrgastro.2015.94. [DOI] [PubMed] [Google Scholar]
- 9.Durand F, Valla D. Assessment of the prognosis of cirrhosis: child-pugh versus MELD. J Hepatol. 2005;42:100–107. doi: 10.1016/j.jhep.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 10.Trebicka J, Fernandez J, Papp M, et al. The PREDICT study uncovers three clinical courses of acutely decompensated cirrhosis that have distinct pathophysiology. J Hepatol. 2020;73:842–854. doi: 10.1016/j.jhep.2020.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Wirtz TH, Reuken PA, Jansen C, et al. Balance between macrophage migration inhibitory factor and sCD74 predicts outcome in patients with acute decompensation of cirrhosis. JHEP Reports. 2021;3:100221. doi: 10.1016/j.jhepr.2020.100221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hamesch K, Mandorfer M, Pereira VM, et al. Liver Fibrosis and Metabolic Alterations in Adults With alpha-1-antitrypsin Deficiency Caused by the Pi*ZZ Mutation. Gastroenterology. 2019;157:705–719.e18. doi: 10.1053/j.gastro.2019.05.013. [DOI] [PubMed] [Google Scholar]
- 13.Tsai MH, Peng YS, Chen YC, et al. Low serum concentration of apolipoprotein A-I is an indicator of poor prognosis in cirrhotic patients with severe sepsis. J Hepatol. 2009;50:906–915. doi: 10.1016/j.jhep.2008.12.024. [DOI] [PubMed] [Google Scholar]
- 14.Trieb M, Rainer F, Stadlbauer V, et al. HDL-related biomarkers are robust predictors of survival in patients with chronic liver failure. J Hepatol. 2020;73:113–120. doi: 10.1016/j.jhep.2020.01.026. [DOI] [PubMed] [Google Scholar]
- 15.Moreau R, Jalan R, Gines P, et al. Acute-on-chronic liver failure is a distinct syndrome that develops in patients with acute decompensation of cirrhosis. Gastroenterology. 2013;144:1426–1437.e9. doi: 10.1053/j.gastro.2013.02.042. [DOI] [PubMed] [Google Scholar]
- 16.Jalan R, D’Amico G, Trebicka J, Moreau R, Angeli P, Arroyo V. New clinical and pathophysiological perspectives defining the trajectory of cirrhosis. J Hepatol. 2021;75:S14–S26. doi: 10.1016/j.jhep.2021.01.018. [DOI] [PubMed] [Google Scholar]
- 17.D’Amico G, Morabito A, D’Amico M, et al. Clinical states of cirrhosis and competing risks. J Hepatol. 2018;68:563–576. doi: 10.1016/j.jhep.2017.10.020. [DOI] [PubMed] [Google Scholar]
- 18.Costa D, Simbrunner B, Jachs M, et al. Systemic inflammation increases across distinct stages of advanced chronic liver disease and correlates with decompensation and mortality. J Hepatol. 2021;74:819–828. doi: 10.1016/j.jhep.2020.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Jalan R, Saliba F, Pavesi M, et al. Development and validation of a prognostic score to predict mortality in patients with acute-on-chronic liver failure. J Hepatol. 2014;61:1038–1047. doi: 10.1016/j.jhep.2014.06.012. [DOI] [PubMed] [Google Scholar]
- 20.Solé C, Solà E, Morales-Ruiz M, et al. Characterization of inflammatory response in acute-on-chronic liver failure and relationship with prognosis. Sci Rep. 2016;6:1–10. doi: 10.1038/srep32341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grønbæk H, Rødgaard-Hansen S, Aagaard NK, et al. Macrophage activation markers predict mortality in patients with liver cirrhosis without or with acute-on-chronic liver failure (ACLF) J Hepatol. 2016;64:813–822. doi: 10.1016/j.jhep.2015.11.021. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z, Burke PA. Effects of hepatocyte nuclear factor-4α on the regulation of the hepatic acute phase response. J Mole. 2007;371:323–335. doi: 10.1016/j.jmb.2007.05.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guldiken N, Argemi J, Gurbuz B, et al. Serum transferrin as a biomarker of hepatocyte nuclear factor 4 alpha activity and hepatocyte function in liver diseases. BMC Med. 2021;19:1–12. doi: 10.1186/s12916-021-01917-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sladek FM, Zhong W, Lai E, Darnell JE. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990;4:2353–2365. doi: 10.1101/gad.4.12b.2353. [DOI] [PubMed] [Google Scholar]
- 25.Desai SS, Tung JC, Zhou VX, et al. Physiological ranges of matrix rigidity modulate primary mouse hepatocyte function in part through hepatocyte nuclear factor 4 alpha. Hepatology. 2016;64:261–275. doi: 10.1002/hep.28450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Argemi J, Latasa MU, Atkinson SR, et al. Defective HNF4alpha-dependent gene expression as a driver of hepatocellular failure in alcoholic hepatitis. Nat Commun. 2019;10:3126. doi: 10.1038/s41467-019-11004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guzman-Lepe J, Cervantes-Alvarez E, Collin de l’Hortet A, et al. Liver-enriched transcription factor expression relates to chronic hepatic failure in humans. Hepatol Commun. 2018;2:582–594. [DOI] [PMC free article] [PubMed]
- 28.Tan L, Meng Y, Zeng T, et al. Clinical diagnostic significance of prealbumin, cholinesterase and retinol binding protein in liver cirrhosis combined with encephalopathy. Br J Biomed Sci. 2019;76:24–28. doi: 10.1080/09674845.2018.1523673. [DOI] [PubMed] [Google Scholar]
- 29.Niu L, Thiele M, Geyer PE, et al. Noninvasive proteomic biomarkers for alcohol-related liver disease. Nat Med. 2022;28:1277–1287. doi: 10.1038/s41591-022-01850-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Proteomic data were deposited in a publicly available database. Further data are available from authors upon reasonable request.