Abstract
Arabidopsis COP1 acts within the nucleus to repress photomorphogenesis, and its nuclear abundance is negatively regulated by light. Here, we report the identification of a COP1-interactive partner, CIP4. CIP4 is a nuclear protein and a potent transcription coactivator. Conditional suppression of CIP4 expression resulted in an elongated hypocotyl and reduced chlorophyll content in the light, indicating that CIP4 is required for the promotion of photomorphogenesis. Furthermore, CIP4 was revealed to act downstream of multiple photoreceptors as well as COP1 in mediating light control of development. CIP4 expression is light inducible and regulated by COP1. However, CIP4 does not play a role in mediating the light induction of anthocyanin accumulation. Together with our previous studies of CIP7 and HY5, our data suggest that COP1 interacts directly with and regulates multiple physiological targets, which in turn regulate distinct sets of light-regulated responses.
INTRODUCTION
Like other higher plants, developing Arabidopsis seedlings exhibit dramatic adaptation to the surrounding light environment to optimize their chance of survival. Light-grown seedlings have open cotyledons, short hypocotyls, and extensive cell differentiation and accumulate chlorophyll and anthocyanin. Within the cells, chloroplasts are well developed and fully capable of photosynthesis. On the other hand, dark-grown seedlings have closed cotyledons and an elongated hypocotyl without chlorophyll or anthocyanin. The plastid of the etiolated seedlings (named the etioplast) is morphologically distinct from the chloroplast (Kendrick and Kronenberg, 1994; von Arnim and Deng, 1996). Environmental light signals are perceived by several families of photoreceptors. In Arabidopsis, there are five phytochromes, phyA to phyE, recognizing red/far-red light; two cryptochromes, CRY1 and CRY2, as blue light receptors; and NPH1, which is a blue light receptor for phototropic response (Fankhauser and Chory, 1997; Christie et al., 1998). The quality, quantity, duration, and direction of environmental light are perceived by these color-specific photoreceptors, and the information is transmitted to regulate transcription and other cellular responses (Kendrick and Kronenberg, 1994; von Arnim and Deng, 1996; Fankhauser and Chory, 1997).
Complementary approaches have been used to reveal the molecular mechanisms of the light regulation of development. A pharmacological approach using microinjection with tomato phytochrome–deficient mutant cells and a soybean cell culture line revealed the involvement of cyclic GMP, trimeric G proteins, and calcium/calmodulin in phyA signaling (Neuhaus et al., 1993; Bowler et al., 1994). Genetic approaches to identify photoreceptor-specific signaling components revealed several genetic loci for phytochrome-specific signaling in Arabidopsis, including FHY1, FHY3 (Whitelam et al., 1993), FIN2 (Soh et al., 1998), PSI2 (Genoud et al., 1998), SPA1 (Hoecker et al., 1999), FAR1 (Hudson et al., 1999), PAT1 (Bolle et al., 2000), and FIN219 (Hsieh et al., 2000). Isolation of phytochrome-interactive proteins identified nuclear and cytoplasmic factors that play a role in both phyA- and phyB-dependent signals (Ni et al., 1998; Choi et al., 1999; Fankhauser et al., 1999). Genetic screens for mutants with photomorphogenic development in darkness have defined another group of negative regulators, including COP1, DET1, COP10, and components of the COP9 signalosome (von Arnim and Deng, 1996; Fankhauser and Chory, 1997; Serino et al., 1999; Wei and Deng, 1999). Loss-of-function mutants of these genes show hypersensitivity to at least phyA-, phyB-, and CRY1-mediated signals (von Arnim and Deng, 1996; Fankhauser and Chory, 1997; Osterlund and Deng, 1998). This group of genes is highly conserved among all higher eukaryotes, including mammals (Wang et al., 1999; Wei and Deng, 1999). However, how this group of regulators acts at the molecular and biochemical levels is not clear and remains a major focus of current research.
Among the group of negative regulators, COP1 is a rate-limiting factor and possibly a regulatory step (McNellis et al., 1994). COP1 is a protein containing a RING finger, a coiled-coil region (COIL), and WD40 repeats, all of which are known protein–protein interaction modules (Deng et al., 1992). Analysis of the intracellular localization of COP1 using β-glucuronidase (GUS) as a fused reporter revealed that GUS-COP1 localizes to the nucleus in the dark and is excluded from the nucleus in the light (von Arnim and Deng, 1994; Osterlund and Deng, 1998). This finding indicates that COP1 acts within the nucleus to suppress photomorphogenic development. Recent analyses of COP1-interacting protein CIP7 and the bZIP protein HY5 suggest that COP1 regulates positive regulators of photomorphogenesis through direct physical interaction within the nucleus (Ang et al., 1998; Yamamoto et al., 1998). Furthermore, in the case of HY5, this negative regulation by the nuclear COP1 may involve targeted degradation of HY5 via the 26S proteasome (Hardtke et al., 2000; Osterlund et al., 2000). Here, we report a new COP1-interacting protein, CIP4. CIP4 is a novel type of nuclear protein and a positive regulator of photomorphogenesis. Analysis of this new COP1 target indicates that COP1 regulates multiple physiological responses by directly modulating distinct regulators for those branched pathways.
RESULTS
Isolation and Characterization of CIP4
To isolate COP1-interacting proteins, we screened an unamplified Arabidopsis cDNA expression library from dark-grown seedlings with a 32P-labeled COP1 protein as a probe (Yamamoto et al., 1998). From the screening of 3 × 106 independent cDNA clones, three positive clones were isolated, including CIP7 (Yamamoto et al., 1998). The second clone, designated CIP4, represents a partial cDNA as determined by RNA gel blot and sequencing analyses (data not shown). This CIP4 cDNA was used as a probe to screen for longer cDNAs and genomic clones. Sequence analyses of a combination of genomic and cDNA clones as well as reverse transcription–polymerase chain reaction (RT-PCR) revealed an open reading frame encoding 915 amino acid residues (Figure 1; DNA Data Bank of Japan accession number AB036832). The sequence analyses also revealed another gene highly homologous with CIP4. Database searches and DNA gel blot hybridization confirmed that there are only two sequences related to the CIP4 gene in the Arabidopsis genome (data not shown). However, the second gene, designated CIP4.1 (DNA Data Bank of Japan accession number AB036833), contains a stop codon (TAA) in the middle of the coding region (after 371 amino acid residues), in spite of an otherwise highly homologous amino acid sequence corresponding to the entire coding region of CIP4 (78.8%). Therefore, although CIP4.1 is transcribed and spliced (data not shown), this gene might be a pseudogene that was recently duplicated. Higher homology between CIP4 and CIP4.1 at the DNA level (85.0%) than at the amino acid level (78.8%) may support this notion. On the basis of the released genome sequence, CIP4 is located in the middle of chromosome 5 and CIP4.1 is located at the top of chromosome 4.
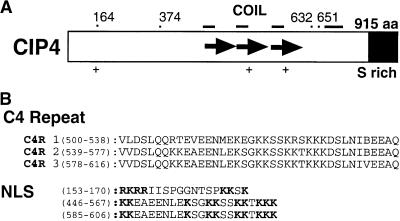
Figure 1.
Structure of CIP4.
(A) Diagram showing CIP4 protein structure. The small bars at the top indicate putative COILs. (+) indicates the location of a putative bipartite NLS. Tandem repeats consisting of 39 amino acid (aa) residues (C4 repeats) are shown as arrows. The amino acid residue positions of several key CIP4 deletion mutant breakpoints are indicated by dots at the top. The position of the serine-rich C terminus is marked.
(B) Sequences of the C4 repeats (C4Rs) and putative NLS. The basic charged residues in the NLS sequences are in boldface. The positions of the first and last amino acids in those sequences are in parentheses.
Database searches using CIP4 did not identify any other related protein in the database except for CIP4.1, indicating that CIP4 is a novel protein. CIP4 has three tandem repeats consisting of 39 amino acid residues (designated C4 repeat; Figure 1). Overlapping with the C4 repeat, CIP4 has four predicted COILs (Figure 1A) and three typical bipartite nuclear localization signals (NLS), revealed by the Multicoil (http://nightingale.lcs.mit.edu/cgi-bin/multicoil) and PSORT (http://psort.nibb.ac.jp/form.html) programs, respectively. The C-terminal domain is rich in Ser residues (20% Ser in the C-terminal 175 amino acid residues) (see SFigure 1A, S rich).
Determination of the CIP4 Binding Domain of COP1
COP1 contains a RING finger, a COIL, and WD40 repeats (Deng et al., 1992) (Figure 2A), all of which can participate in protein–protein interactions. To determine which domain of COP1 interacts with CIP4, a glutathione S-transferase (GST) pull-down assay was performed. Because the full-length CIP4 cDNA caused a subcloning problem, possibly due to the toxicity of the full-length protein, only partial cDNA clones that can be expressed in Escherichia were used for the assay. The glutathione–Sepharose resin was charged with GST–CIP4 fusion proteins and then incubated with the 32P-labeled COP1 protein. After extensive washing, the bound COP1 was analyzed by SDS-PAGE and then visualized by autoradiography (Figure 2). As shown in Figure 2B, the full-length COP1 bound strongly to GST–CIP4(374-915), but not to GST, demonstrating specific and direct binding of COP1 to CIP4 in vitro. Deletion analysis indicated that the two smaller fragments of CIP4 [CIP4(374-651) and CIP4(632-915)] can interact with COP1. However, the CIP4(374-651) fragment, which contains all three C4 repeats (Figure 1), binds more strongly to COP1 than CIP4(632-915). Of the three constructs, the longest version, GST–CIP4(374-915), binds most strongly to COP1. Therefore, CIP4 may have more than one contact motif for binding to COP1.
Figure 2.
Determination of the CIP4 Binding Domain of COP1.
(A) Diagrams of wild-type and COP1 deletion constructs used for the assay. All of the deletion proteins were fused with a Flag tag and an HMK recognition site sequence (Yamamoto et al., 1998). The three protein–protein interaction domains and domain deletions are marked.
(B) The binding capacities of three versions of GST–CIP4 fusion proteins, and GST itself with full-length 32P-labeled COP1.
(C) The binding capacities of the 32P-labeled COP1 deletion proteins toward the long version of GST–CIP4 in the in vitro binding assay. The input COP1 probes are shown at left and the bound COP1 proteins are shown at right. Calculated molecular masses of the COP1 deletion proteins are shown at right in kilodaltons, and the deletion proteins are indicated by asterisks.
(D) The radioactivity of each band in (C) was quantified, and the ratios of the bound fractions relative to the total input were calculated.
A series of COP1 deletion proteins like those shown in Figure 2A was subjected to the GST pull-down assay using the long GST–CIP4(374-915) (Figures 2C and 2D). Deletion of either the RING (ΔRING) or the WD40 (N282) domain from the full-length COP1 resulted in the reduction of binding activity. Further deletion of RING from N282 (NΔRING) or deletion of WD40 from ΔRING (NΔRING) did not reduce binding. Therefore, the RING and WD40 domains can strengthen binding but are not essential. However, deletion of the COIL from N282 (NΔCOIL) resulted in the loss of binding activity. NΔRING, which is the smallest protein with CIP4 binding activity, bound to the shorter CIP4 fusion protein (GST–CIP4[374-651]) as well (data not shown). It appears that although the full-length COP1 exhibited its strongest binding to CIP4, the COIL motif of COP1 defines a critical binding domain toward CIP4, but the RING and WD40 domains are dispensable. As reported previously, overexpression of the COP1 deletion proteins N282 and NΔRING in transgenic Arabidopsis caused a dominant negative phenotype, whereas overexpression of NΔCOIL did not (McNellis et al., 1996; Torii et al., 1998). Consistent with those past studies and with CIP7 data reported by Yamamoto et al. (1998), our results suggest that the COIL is a critical interface for some interactive partners of COP1.
CIP4 Is a Nuclear Protein
The presence of typical bipartite NLS sequences in CIP4 (Figure 1B) suggests that CIP4 might be a nuclear protein. To examine the intracellular localization of CIP4, a modified green fluorescent protein (GFP) from Aequorea victoria (soluble, modified, red-shifted GFP [smRS-GFP]) (Davis and Vierstra, 1998) was fused with CIP4, and the fusion protein was expressed in transgenic Arabidopsis. To overcome the apparent toxicity of the full-length cDNA during the subcloning process, the first two introns from the CIP4 gene were introduced into the GFP–CIP4 chimeric construct. Although smRS-GFP by itself localized all over the cell (data not shown) (Davis and Vierstra, 1998), the GFP–CIP4 fusion protein was observed exclusively within the nucleus (Figure 3; see Methods). The GFP–CIP4 full-length fusion protein localized to the nuclei in cotyledons, hypocotyls, roots, and leaves (Figure 3 and data not shown). Alteration of the light environment did not affect this nuclear localization, indicating a constitutive nuclear localization of CIP4 (data not shown).
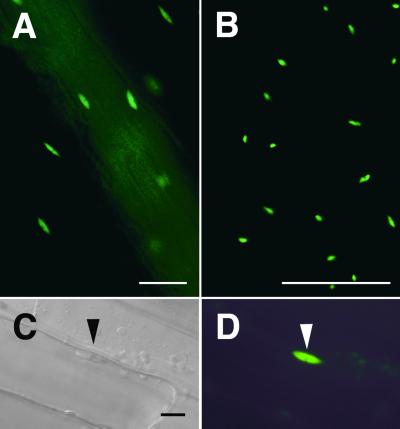
Figure 3.
Nuclear Localization of GFP–CIP4.
Transgenic Arabidopsis expressing GFP–CIP4 was examined under a light microscope.
(A) Fluorescence microscope image of a hypocotyl.
(B) Fluorescence microscope image of a leaf.
(C) Differential interference contrast microscope image of hypocotyl cells.
(D) Florescence microscope image of the same hypocotyl cells shown in (C).
The large track of diffused fluorescence signal from top left to bottom right in (A) is due to a vascular bundle that was out of focus. The positions of GFP fluorescence were confirmed to be nuclei by differential interference contrast imaging or 4′,6-diamidino-2-phenylindole staining (data not shown). The triangles in (C) and (D) point to the nucleus.  ;
; 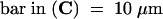 for (C) and (D).
for (C) and (D).
Transcriptional Activation by CIP4
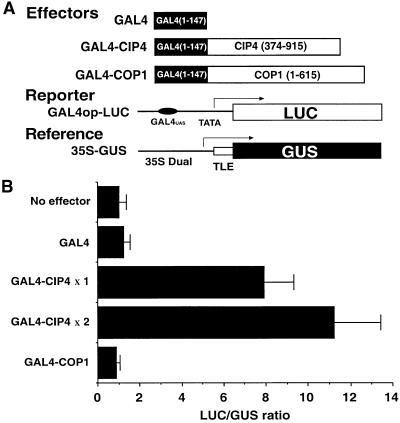
To determine if the nucleus-localized CIP4 is a transcription factor, we examined the ability of CIP4 to activate transcription in plant cells. Due to the apparent toxicity of the full-length CIP4 in Escherichia, which prevents its cloning, a partial CIP4 cDNA (residues 374 to 915) was fused with the DNA binding domain of GAL4. The fusion gene (Figure 4A, GAL4–CIP4) was introduced into tobacco leaves by a microprojectile bombardment method together with a luciferase (LUC) reporter gene under the control of the GAL4 operator (Figure 4A, GAL4op-LUC) (Yamamoto and Deng, 1998). To monitor transfection efficiency, another reporter driven by a constitutive promoter (Figure 4A, 35S-GUS) was also cotransfected and used as an internal control to normalize LUC expression. Although the DNA binding domain of GAL4 and a GAL4–COP1 fusion protein did not activate the LUC reporter gene, the GAL4–CIP4 fusion exhibited ∼10-fold transactivation of reporter gene expression in a dose-dependent manner (Figure 4). Thus, the transcription activation ability of CIP4 in plant cells is even stronger than that of a previously reported COP1 target, CIP7. Like CIP7, CIP4 does not contain any recognizable DNA binding motif (Yamamoto et al., 1998). Therefore, CIP4 is likely a transcriptional coactivator that does not bind directly to DNA.
Figure 4.
CIP4 Is a Competent Transcriptional Activator in Plant Cells.
(A) The constructs used in the assay. The reporter (GAL4op-LUC) and the reference (35S-GUS), which contains a translational enhancer element (TLE), were mixed together with the effectors (GAL4 DNA binding domain fusion proteins) and introduced into tobacco leaves by the microprojectile bombardment method. Numbers in parentheses indicate the amino acid residue coordinates of the proteins, arrows indicate the direction of transcription, and 35S Dual indicates the 35S promoter with two copies of the enhancer element. The GAL4UAS indicates the GAL4 DNA binding site in the promoter.
(B) Transient activation of the reporter in plant cells. LUC reporter activity was normalized to GUS activity. Average and sd values of the normalized LUC activity (LUC/GUS) are shown. At least six independent tests were performed for each sample. In the case of GAL4–CIP4, two concentrations of the effector plasmids (×2 and ×1) were tested.
Light-Dependent Expression of CIP4
As a first step in examining the role of CIP4 in the light regulation of plant development, we examined the effect of light on CIP4 expression. As shown in Figure 5, the transcript size of CIP4 was ∼3.1 kb, which is in agreement with the cDNA size. Comparison of light-grown seedlings (Figure 5, left, WT L) and dark-grown siblings (WT D) revealed that CIP4 is expressed in a light-dependent manner. In the dark, cop1 seedlings (cop1 D), partially derepressed CIP4 expression compared with the wild type (WT D), indicating the involvement of COP1 in the dark-dependent repression of CIP4 expression. When light-grown seedlings (Figure 5, right, WT L) were transferred to darkness for 2 days (WT LD), the expression of CIP4 decreased considerably, displaying a dark-adaptive decrease. Analysis of CIP4.1 transcripts by quantitative RT-PCR revealed that CIP4.1 also is expressed in the same manner as CIP4, but with an expression level fivefold to tenfold less than that of CIP4 (data not shown). Because CIP4 is expressed at a high level in the light, CIP4 is believed to function primarily in the light.
Figure 5.
CIP4 Expression Is Regulated by Light.
(A) CIP4 RNA gel blot analysis of total RNA levels from 5-day-old wild-type (WT) or mutant (cop1) seedlings grown in darkness (D) or light (L).
(B) Five-day-old wild-type (WT) seedlings grown in light (L) were transferred to dark for 2 days (LD) or left in the light for 2 days (LL). Identical blots were hybridized with the 18S rDNA probe (18S) to confirm equal loading of the RNA samples.
Antisense Inhibition of CIP4 Expression Suppresses Photomorphogenic Development
To obtain direct evidence for the physiological role of CIP4, a reverse genetics approach was used to suppress CIP4 expression. Initially, a constitutive 35S promoter was used for an antisense construct that expresses the complementary strand of a CIP4 fragment (amino acid residues 377 to 653). However, we failed to isolate any transgenic Arabidopsis lines using this construct, possibly due to the lethality of the constitutive suppression of CIP4. Subsequently, we made a conditional antisense construct in which the expression of the antisense gene was induced by dexamethasone (DEX) treatment (Aoyama and Chua, 1997). Using the second construct, we established 50 independent transgenic lines, and of these, 38 lines showed paler leaf color and longer hypocotyl phenotypes in white light with DEX induction (Figures 6A and 6B). These phenotypic features behaved as dominant traits and strictly cosegregated with the antisense transgene (data not shown), indicating that the phenotype was caused by the transgene. At later stages, petioles were also elongated more than in the wild type (Figures 6C and 6D). Because these transgenic plants are less sensitive to light, CIP4 acts as a positive regulator of photomorphogenesis. In our collection of transgenic lines, four of 50 antisense lines and six of 50 vector control transgenic lines showed severe growth arrest at the early seedling stage in the presence of DEX. This growth inhibition was very similar to that of a recently reported artifactual phenotype associated with the glucocorticoid-mediated induction system (Kang et al., 1999). However, the longer hypocotyl and paler leaf phenotypes were clearly distinct and not observed in any of the vector control transgenic lines; thus, these phenotypes of the antisense CIP4 lines are not artifacts of the glucocorticoid induction system but are specific to antisense CIP4 transgenic plants. Two independent antisense CIP4 lines carrying a single T-DNA locus (209-9 and 209-27) and exhibiting a typical antisense phenotype were selected. Homozygous lines in the T3 generation were produced for detailed analysis. Under the induction condition, the accumulation of CIP4 protein in the antisense lines was reduced to <10% of the wild-type level in a DEX-dependent manner, as estimated by protein gel blot analysis (Figure 7).
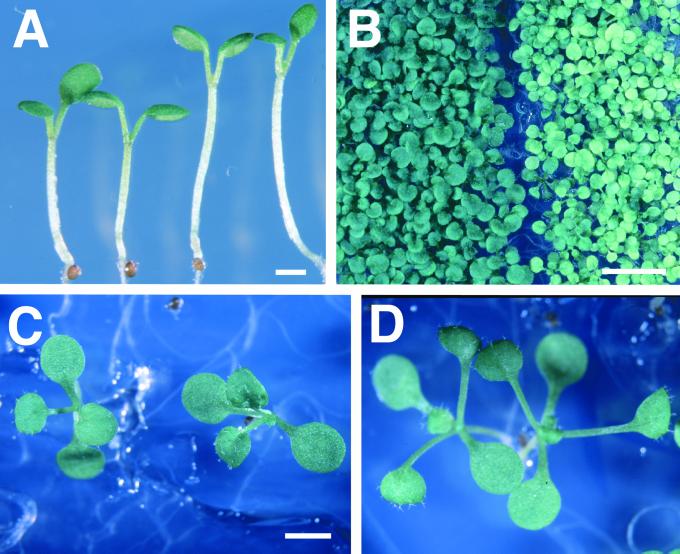
Figure 6.
Antisense CIP4 Transgenic Arabidopsis Seedlings Exhibited Long Hypocotyl and Pale Cotyledon Phenotypes.
(A) Five-day-old wild-type (left) and antisense CIP4 (right) seedlings grown in white light (8 μE m−2 sec−1).
(B) Wild-type (left) and antisense CIP4 (right) seedlings grown in white light (100 μE m−2 sec−1) for 2 weeks.
(C) and (D) Wild-type (left) and antisense CIP4 (right) seedlings were grown under a 12-hr-light/12-hr-dark photoperiod for 2 weeks.
All seedlings were grown in the presence of 10 μM DEX.  ;
;  ;
;  .
.
Figure 7.
Immunoblot Analysis of the Antisense CIP4 Transgenic Lines.
Vector transformants (Vector), two antisense CIP4 lines (T3 homozygotes of 209-9 and 209-27), and the wild type (WT) were grown in the light on a medium with (+) or without (−) DEX and subjected to immunoblot analysis for the CIP4 protein. The relative amounts of the CIP4 protein were calculated based on a series of dilutions of the wild-type sample and are indicated at the bottom. Asterisks indicate that the signal intensity was below the quantitative range. Note that the lower level of the CIP4 protein in the antisense lines without DEX may be due to partial cosuppression.
CIP4 Is Required for Photomorphogenic Responses Mediated by Multiple Photoreceptors
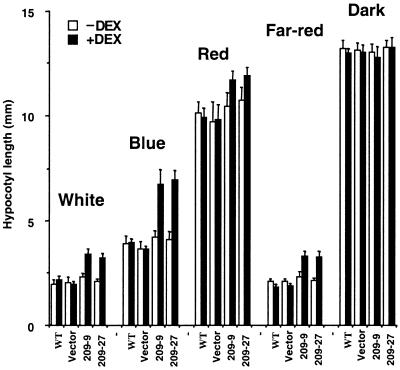
Multiple photoreceptors are involved in mediating the light inhibition of hypocotyl elongation. For example, the inhibition of hypocotyl elongation in Arabidopsis seedlings is mediated primarily by CRY1/CRY2 in continuous blue light, by phyB in continuous red light, and by phyA in continuous far-red light (Fankhauser and Chory, 1997). To identify which pathways CIP4 is involved in, the hypocotyl elongation of the representative antisense CIP4 transformants was examined under different light conditions. As shown in Figure 8, antisense CIP4 lines (209-9 and 209-27) showed longer hypocotyls under white light conditions in a DEX induction–dependent manner. This finding indicates that the phenotypic change is caused by transgene induction. However, the dark-grown seedlings showed the same hypocotyl length as the wild-type seedlings. Therefore, the effect of antisense induction was strictly light dependent. DEX-dependent long hypocotyl phenotypes of the antisense CIP4 lines were most drastic in continuous blue and far-red light. In red light, the effect of the antisense CIP4 was less evident, although the antisense CIP4 seedlings still had longer hypocotyls than did the wild-type seedlings. This phenotype is somewhat similar to that of the hy5 mutants and the COP1-overexpressing strains (Ang and Deng, 1994; McNellis et al., 1994). This indicates that CIP4 is involved in light inhibition of hypocotyl elongation that is mediated by multiple photoreceptors. Because COP1 also acts downstream of the multiple photoreceptors (von Arnim and Deng, 1996; Osterlund and Deng, 1998), these data are consistent with the notion that CIP4 and COP1 are physiological partners in the light regulatory network.
Figure 8.
Antisense CIP4 Seedlings Are Defective in the Inhibition of Hypocotyl Elongation Responses Mediated by White, Blue, Red, and Far-Red Light.
Hypocotyl elongation of wild type (WT), vector transformants (Vector), and two antisense CIP4 lines (209-9 and 209-27) under various light conditions. Average length and sd values are indicated. Approximately 30 to 40 seedlings were examined for each data point. Seedlings grown with (+) and without (−) DEX were compared.
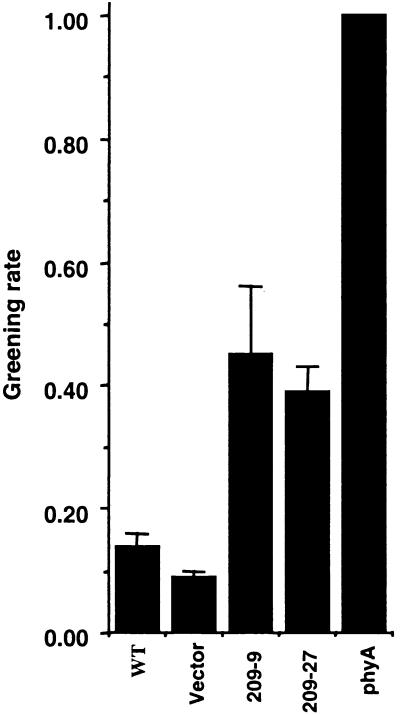
We further examined the role of CIP4 in another phyA-mediated response, the greening of the seedlings after transferring from far-red light-grown condition to white light (Barnes et al., 1996). When Arabidopsis seedlings grown in far-red light were transferred to white light, partially developed chloroplasts in far-red light underwent photobleaching. However, phyA or mutants of its specific signaling components do not develop chloroplasts in far-red light, so they can turn green after the shift from far-red to white light (Barnes et al., 1996). As shown in Figure 9, although wild-type seedlings exhibited the expected photobleaching, the antisense CIP4 lines avoided photobleaching and turned green after transfer to white light. This result indicates that the antisense line partially mimics the phyA phenotype and provides another line of evidence for the involvement of CIP4 in a phyA-mediated response.
Figure 9.
Antisense CIP4 Seedlings Exhibit a Partial Defect in Greening after Growth in Continuous Far-Red Light.
Fractions of 4-day-old seedlings grown in far-red light that turned green after transfer to white light for 1 week are shown. The results are averages of four tests, and sd values are indicated. All seedlings (WT, vector control, two antisense lines, and phyA mutant) were grown in the presence of DEX. WT, wild type.
CIP4 Is Required for Light-Dependent Chloroplast Development but Not Anthocyanin Accumulation
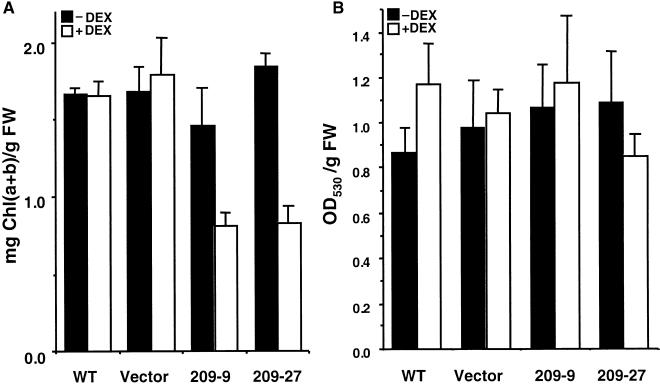
The pale cotyledons of the antisense transgenic seedlings suggest a possible role of CIP4 in light-induced pigment accumulation. Therefore, we examined the effect of CIP4 antisense expression on chlorophyll and anthocyanin accumulation. As shown in Figure 10A, under DEX induction, chlorophyll accumulation in antisense seedlings was reduced to approximately half that seen in the wild type, whereas the vector control transgenic lines showed no detectable effect. However, anthocyanin accumulation was not altered significantly in the antisense lines (Figure 10B). Thus, the induction of antisense CIP4 caused the inhibition of chlorophyll but not anthocyanin accumulation. Because COP1 is involved in hypocotyl elongation, chloroplast development, and anthocyanin accumulation (von Arnim and Deng, 1996), CIP4 may regulate only part of the COP1-mediated responses.
Figure 10.
Pigment Accumulation of Antisense CIP4.
(A) Chlorophyll accumulation.
(B) Anthocyanin accumulation.
Two-week-old seedlings of wild type (WT), vector transformants (Vector), and antisense lines (209-9 and 209-27) were used for chlorophyll (Chl) and anthocyanin quantitation. The effects of the presence (+) or absence (−) of DEX were compared. Average amounts of pigment per fresh weight (FW) of plants and sd values are shown.
Antisense CIP4 Can Partially Suppress the cop1 Short Hypocotyl Phenotype in the Dark
The data described thus far all strongly suggest that CIP4 is a downstream factor of COP1. To further verify this possibility, we first introduced the CIP4 antisense transgene (209-9) into the GUS–COP1 fusion protein background. Analyses of the intracellular localization pattern of GUS–COP1 revealed no observable effect of CIP4 antisense expression (data not shown). This result is consistent with the idea of CIP4 being downstream of COP1. Next, we examined the genetic interaction between the antisense CIP4 and cop1 mutations. We introduced the antisense CIP4 (209-9) into a weak cop1 allele, cop1-6, and the lethal and null allele, cop1-5. The hypocotyl elongation of double mutants and their parental strains in darkness was examined (Table 1). Although antisense CIP4 itself did not affect hypocotyl length in dark-grown wild-type seedlings, antisense CIP4 increased the hypocotyl length of the cop1-6 and cop1-5 mutant seedlings (33.0 and 13.9%, respectively). This result suggests that antisense CIP4 can partially suppress the short hypocotyl phenotype of the cop1 mutations and supports the conclusion that CIP4 acts downstream of COP1. The fact that antisense CIP4 only partially suppressed the cop1 mutations (Table 1) suggests the presence of a downstream pathway that bypasses CIP4. Among the identified downstream targets of COP1 (HY5 and CIP7), only HY5 is known to be involved in the regulation of hypocotyl elongation (Koornneef et al., 1980; Yamamoto et al., 1998). Therefore, HY5 might be involved in the predicted bypass pathway that acts in conjunction with CIP4. Further studies are needed to substantiate this notion.
Table 1.
The Effect of Antisense CIP4 and cop1 Mutations on the Hypocotyl Elongation of Dark-Grown Arabidopsis Seedlingsa
| Parentalb | Antisense CIP4c | Effect of Antisense CIP4d | t Valuee | Significance | |
|---|---|---|---|---|---|
| Wild type | 12.8 ± 2.20 | 12.2 ± 2.52 | −4.7% | 0.98 | No |
| cop1-6 | 2.85 ± 0.32 | 3.79 ± 0.30 | 33.0% | 11.7 | Yes |
| cop1-5 | 2.22 ± 0.44 | 2.53 ± 0.52 | 13.9% | 2.49 | Yes |
All of the seedlings were grown under dark conditions in the presence of 10 μM DEX, and the hypocotyl lengths of 30 seedlings are analyzed here. For hypocotyl length measurements, the seedlings on the plate were photographed, the image was enlarged 400%, and the length was measured in 0.125-mm increments of the actual length.
Average of hypocotyl length (mm ±sd) of the parental lines (wild type, cop1-6, and cop1-5).
Average of hypocotyl length (mm ±sd) of the F3 seedlings carrying 209-9 T-DNA.
Percentage of changes in hypocotyl length due to the presence of the antisense CIP4 transgene.
Student's t test value under the assumption that two corresponding populations have the same average. With this population size, a t value >2.00 precludes the assumption that the two populations have the same average; thus, in this case, the antisense CIP4 has a significant effect on hypocotyl elongation.
DISCUSSION
This report describes a new downstream target of nuclear COP1 in mediating light control of Arabidopsis development. Like two previously identified COP1-interactive downstream targets, HY5 and CIP7 (Yamamoto et al., 1998), CIP4 is a positive regulator of photomorphogenesis and may be a transcription factor. Because COP1 is a negative regulator of photomorphogenesis (von Arnim and Deng, 1996), this study, together with our previous work, begins to suggest that nucleus-localized COP1 interacts directly with and negatively regulates multiple transcriptional regulators that promote photomorphogenesis. Therefore, the role of COP1 in mediating light signaling would be to interact with and regulate transcription factors according to the light environment. These altered gene expression patterns could be responsible for the corresponding developmental and morphological changes.
Our studies also revealed another important feature of the mechanism of COP1-mediated developmental processes. Functional studies of COP1-interacting partners clearly suggest that they promote overlapping but distinct combinations of light-regulated processes. For example, CIP7 plays a role in promoting chloroplast development and anthocyanin accumulation in the light (Yamamoto et al., 1998), whereas CIP4 promotes light-mediated inhibition of hypocotyl elongation and chloroplast development. On the other hand, HY5 seems to affect primarily hypocotyl elongation and anthocyanin accumulation. This notion is illustrated in Figure 11, which shows how COP1 achieves its effect by regulating multiple downstream targets, such as CIP4, HY5, and CIP7. The close relationship between CIP4 and COP1 is further supported by the fact that both COP1 and CIP4 act downstream of multiple photoreceptors, including phyA and phyB, and cryptochromes (von Arnim and Deng, 1996; Osterlund and Deng, 1998). The working model shown in Figure 11 suggests that the light signals perceived by distinct photoreceptors are transduced to inactivate COP1 and negatively regulate its nuclear abundance. This reduced nuclear abundance of COP1 results in relief of its negative regulation of multiple transcription factors. Our recent study suggests that in the case of HY5, this COP1 regulation is achieved by controlling the degradation of HY5 (Osterlund et al., 2000). As a consequence, the altered activity levels of these transcription factors lead to dramatically altered gene expression profiles and thus to new developmental patterns. Additional studies are needed to reveal the biochemical nature of the negative regulation of these transcription factors by COP1 and the specific gene targets for each transcription factor involved.
Figure 11.
A Model for the COP1-Mediated Light Control of Arabidopsis Development.
In this model, COP1 is a key regulator that interacts directly with and negatively regulates multiple targets, including CIP4, HY5, and CIP7. The different COP1 targets specifically regulate distinct sets of light-controlled processes. For example, CIP7 is a positive regulator of chlorophyll (Chl) and anthocyanin (Antho) accumulation but not of the repression of hypocotyl elongation (Hyp) (Yamamoto et al., 1998). CIP4 is a positive regulator of hypocotyl and chlorophyll accumulation but not of anthocyanin accumulation. HY5 regulates both hypocotyl length and anthocyanin accumulation but not chlorophyll accumulation. However, COP1 activity is negatively regulated by multiple light-mediated photoreceptors, including phyA, phyB, and CRY1. T-bars represent negative interactions, and arrows indicate positive interactions. Except in the cases of COP1 and its targets, other interactions are likely indirect. The molecular mechanism that permits COP1 to negatively regulate CIP4 and CIP7 is not known.
CIP4 is a nuclear protein and a potent transcriptional activator without any recognizable DNA binding motif. Therefore, CIP4 could be a transcriptional coactivator that does not recognize DNA directly but rather works together with another specific DNA binding factor. This would put CIP4 and CIP7 in the same class (Yamamoto et al., 1998), whereas HY5 would represent another group of transcription factors that bind promoter elements directly (Ang et al., 1998). Puente et al. (1996) reported that combinatorial interactions of two distinct light-responsive promoter elements constitute an autonomous light-responsive determinant in the promoter. This phenomenon has been termed “combinatorial interplay” of the promoter elements. At present, the molecular basis for this phenomenon is not known. At least two general schemes can be envisioned: cooperative binding of two DNA binding proteins that recognize the individual promoter elements, and the involvement of coactivators that simultaneously recognize both promoter element binding proteins. The emergence of putative coactivators such as CIP4 and CIP7 involved in the light control of development might support the latter scheme. It is possible that CIP4 and CIP7 could activate transcription of the target genes by interacting cooperatively with more than one protein that is bound to distinct promoter elements. The identification of CIP4 and CIP7 thus allows us to test this speculation on the combinatorial interplay of promoter elements.
Our identification of CIP4 adds to an increasing list of nuclear regulators involved in the light control of development. Recently, several light-signaling factors of Arabidopsis that mediate signaling events specific to individual or multiple photoreceptors have been characterized and shown to be nuclear factors. They include SPA1 (Hoecker et al., 1999), PIF3 (Ni et al., 1998), FAR1 (Hudson et al., 1999), DET1 (Fankhauser and Chory, 1997), the COP9 signalosome (Serino et al., 1999; Wei and Deng, 1999), COP1 (Fankhauser and Chory, 1997), HY5 (Oyama et al., 1997), and CIP7 (Yamamoto et al., 1998). Furthermore, several photoreceptors, including phyA and phyB (Kircher et al., 1999; Yamaguchi et al., 1999), CRY1 (Cashmore et al., 1999), and CRY2 (Guo et al., 1999; Kleiner et al., 1999), also are able to localize to the nucleus. Therefore, it is possible that a large portion of light-signaling events, from the activation of immediate downstream targets of photoreceptors to transcriptional regulation as an output, may occur within the nucleus. The same nuclear localization of all of these signaling components would provide fertile ground for communication among or coaction by the photoreceptor-specific signaling pathways (Kendrick and Kronenberg, 1994; Ahmad and Cashmore, 1997).
METHODS
Protein Gel Blot Screening
The Flag-tagged COP1 protein was expressed in Escherichia coli BL21 (DE3) pLysS, purified with an M2 anti-Flag affinity resin, labeled with γ-32P-ATP by heart muscle kinase, and used as a probe to screen an unamplified cDNA expression library prepared from etiolated Arabidopsis thaliana seedlings as described previously (Yamamoto et al., 1998). The isolated CIP4 clone (SK-CIP4) was used subsequently as a probe to isolate longer cDNA clones as well as genomic clones. The longest cDNA clone (yy173) isolated from the CD4-16 library (Arabidopsis Biological Resource Center, Columbus, OH) still lacked the region encoding the N terminus of CIP4 as judged from the transcript size of CIP4. The corresponding part of a λ genomic clone (4G5), which contains the CIP4 gene, was sequenced, and the missing N-terminal part of yy173 was synthesized by reverse transcription–polymerase chain reaction (RT-PCR) based on the genomic and cDNA sequences. The RT-PCR product and the cDNA clone together were used to construct a full-length cDNA (yy193). Coding regions of CIP4.1 were amplified by RT-PCR, and the products were subjected to sequence analysis to reveal the open reading frame. Although homology between CIP4 and CIP4.1 spans more than 900 amino acid residues, CIP4.1 was found to have a stop codon in frame at position 372. This stop codon was present in all of the RT-PCR products from six independent PCR reactions (data not shown); therefore, it is not a PCR artifact. Unless mentioned otherwise, all molecular biological procedures were performed according to standard manuals (Sambrook et al., 1989). In most cases, cloned PCR products were subjected to sequence analysis to select the correct clones with no PCR errors.
Glutathione S-Transferase Pull-Down Assay
The cDNA fragments containing partial CIP4 were prepared by PCR and inserted into the EcoRI site of pGEX4T1 (Amersham Pharmacia Biotech, Tokyo, Japan) to make GST–CIP4 fusion proteins. The GST–CIP4 fusion proteins were purified from E. coli, charged to glutathione–Sepharose 4B resin (Amersham Pharmacia Biotech) according to the manufacturer's protocol. Because expression levels were different depending on the construct, the amount of resin relative to the amount of bacterial extract was normalized to give equal amounts of the bound glutathione S-transferase (GST) fusion proteins per resin volume, as judged by SDS-PAGE. The COP1 deletion proteins were expressed in Escherichia BL21 (DE3) pLysS, purified with the M2 resin, and labeled with 32P (Yamamoto et al., 1998). The labeled proteins were analyzed by SDS-PAGE to quantitate the incorporated radioactivity. Twenty microliters of the charged resin were incubated in 300 μL of binding buffer (10 mM Hepes, pH 7.9, 1 mM KCl, 0.4 mM MgCl2, 0.1 mM ZnSO4, 60 mM NaCl, 0.8 mM EDTA, 0.05% BSA, 0.1% Nonidet P-40, and 10% glycerol), with the 32P-labeled COP1 deletion proteins carrying an equal amount of radioactivity, and washed extensively with PBS, as described previously (Yamamoto et al., 1998). Strong and specific binding was observed in a range of 0.05 to 0.3% Nonidet P-40 or Triton X-100 (data not shown). After washing, bound COP1 was analyzed by SDS-PAGE with a 10% polyacrylamide gel. The gel was dried, and the radioactivity of each band was quantitated using a Fujix BAS2500 phosphoimager (Fuji Photo Film, Tokyo, Japan).
Green Fluorescent Protein Fusion Study
The cDNA clone encoding the soluble, modified, red-shifted green fluorescent protein (smRS-GFP) (Davis and Vierstra, 1998) was subcloned into the binary vector pPZPY122 (Yamamoto et al., 1998) with minor modifications to make yy217. The smRS-GFP gene in yy217 contains a translational enhancer from tobacco psaDb (Yamamoto et al., 1995), the plant consensus sequence for translational initiation (Lutcke et al., 1987), and duplicated translational start sites (Yamamoto et al., 1995) to enhance expression. The DNA sequence of the clone is available upon request.
To avoid the toxicity of the full-length CIP4 cDNA, the first four introns, isolated by genomic PCR, were introduced to stabilize the cDNA clone. The genomic/cDNA fusion of CIP4 (yy220) was inserted into the BamHI site of yy217, and the resulting GFP–CIP4 fusion (yy283) was transferred to Arabidopsis by the vacuum infiltration method (Bechtold et al., 1993) together with yy217, which expresses only smRS-GFP, as a control. The T2 seedlings were inspected for intracellular localization of the GFP fusion protein by using a fluorescence microscope with a filter set for fluorescein isothiocyanate (model U-MWBV; Olympus, Tokyo, Japan). The positions of GFP fluorescence were confirmed to be in the nucleus by differential interference contrast imaging or 4′,6-diamidino-2-phenylindole staining (data not shown).
In Planta Transactivation Assay
The EcoRI fragment of yy173 was initially inserted into the EcoRI site of pMA424 (Yamamoto and Deng, 1998), then the GAL4–CIP4 fusion was digested with XhoI-SalI and the fragment was inserted into the XhoI-SalI site of pMA560 (Yamamoto and Deng, 1998). The resulting construct (yy95) had a nopaline synthase promoter, a GAL4–CIP4 fusion, and a polyadenylation signal from nopaline synthase (Yamamoto and Deng, 1998). To make GAL4–COP1 (yy68), pBSCOP1 (von Arnim and Deng, 1993) was digested with BglII and the COP1 fragment was inserted into the BglII site of yy64 (Yamamoto and Deng, 1998). The plasmid DNAs of a reporter (yy96) and a reference 35S::GUS (pRTL2–GUS) (Yamamoto and Deng, 1998) were mixed with the effector plasmids (pMA560, yy95, and yy68) and subjected to transfection of tobacco leaves. Transient activity of the reporters was determined as described (Yamamoto and Deng, 1998).
RNA Analysis
Total RNA was extracted from cotyledons and hypocotyls of seedlings grown on half-strength Murashige and Skoog (1962) salts with 0.8% Bactoagar (Difco Laboratories, Detroit, MI) and subjected to RNA gel blot analysis (Yamamoto et al., 1998). A partial CIP4 clone, SK-CIP4, which contains a region corresponding to the region from amino acids 374 to 915, plus 170 bases of 3′ untranslated region were used to prepare the 32P-labeled antisense riboprobe. The results shown in Figure 5 were confirmed by three independent experiments.
The amount of CIP4.1 mRNA was determined by quantitative RT-PCR. Two primers, 5′-CCTTGAGTTGGTTGAGAAGAAGGA-3′ and 5′-TGCCCATCTACAAGCTCTGAATTA-3′, perfectly match both CIP4 and CIP4.1 and give PCR products of 418 and 382 bp, respectively. Unspliced mRNA species give longer PCR products and thus are distinct from these fragments. After 20 to 30 cycles of PCR, samples were subjected to DNA gel blot analysis for quantitation. A series of diluted RNA samples was also subjected to the same analysis to determine the linearity of the signal. Hybridization signals were analyzed using a Fujix BAS2500 phosphoimager (Fuji Photo Film).
Establishment of Inducible Antisense CIP4 Transgenic Arabidopsis
A CIP4 region corresponding to amino acids 377 to 653 was amplified by PCR and inserted into the SpeI-XhoI site of pTA7002 (Aoyama and Chua, 1997) to make yy209. Both the vector (pTA7002) and yy209 were used for stable transformation of Arabidopsis by the vacuum infiltration method (Bechtold et al., 1993).
Analysis of Antisense CIP4 Plants
Surface-sterilized seed were sown on GM medium (Ang and Deng, 1994) with 1% sucrose, 0.8% Bactoagar, and 0.1% ethanol, which was used as a solvent for dexamethasone (DEX) (Sigma, Chiba, Japan), with or without 10 μM DEX, cold treated for 4 days, and germinated under dark, white, red, or blue light conditions at 22°C. Hypocotyl length was measured 7 days after transfer to 22°C. For the hypocotyl elongation assay in far-red light, cold-treated seed were germinated in the dark at 22°C for 1 day before transfer to far-red light conditions at 22°C. The white light source has been described elsewhere (Nagatani et al., 1993). Blue light was generated from fluorescent tubes (model FL20SB; National, Tokyo, Japan) wrapped with one layer of blue filter (No. 72; Tokyo Butai Shomei, Tokyo, Japan). Red light was generated by light emission diodes (660 ± 20 nm; EYELA, Tokyo, Japan). Far-red light was provided by light emission diodes (735 ± 25 nm; EYELA), and wavelengths shorter than 705 nm were eliminated with a plastic filter (model PIR705; Seiko, Osaka, Japan).
The fluence rates of white, red, far-red, and blue light were 8, 60, 25, and 20 μE m−2 sec−1, respectively. For the greening experiments, seedlings of wild-type, vector transformant, 209 transgenic, and phyA-201 plants were grown in far-red light as described above for 4 days, transferred to 0.8% agar, and illuminated with white light with a fluence rate of 40 μE m−2 sec−1 for 1 week at 22°C before visual inspection. For chlorophyll and anthocyanin measurements, 2-week-old seedlings grown in 100 μE m−2 sec−1 white light were used according to a published protocol (Yamamoto et al., 1998).
Immunoblot Analysis
Partial CIP4 cDNA clones corresponding to amino acids 374 to 651 or 632 to 915 were inserted into the pET29a vector (Novagen, Tokyo, Japan), and the resulting HIS-tagged proteins were expressed, purified from Escherichia, and used to raise anti-CIP4 antibody in rabbits. The CIP4-specific antibody was purified from the rabbit sera using a Hi-Trap NHS column (Amersham Pharmacia Biotech) that had been coupled with the corresponding CIP4 regions fused with GST instead of a HIS tag (Harlow and Lane, 1999). Total protein was extracted from 10-day-old seedlings grown in continuous white light (100 μmol m−2 sec−1) as described (Martinez-Garcia et al., 1999). Equal amounts of the extracted protein (150 μg) were separated on a 6% polyacrylamide gel, blotted onto a polyvinylidene difluoride membrane, and probed with anti-CIP4 antibody using a chemifluorescence method (ECF protein gel blot system; Amersham Pharmacia Biotech). Detection and quantitation of the bands were performed using FluoroImager SI (Amersham Pharmacia Biotech).
Double Mutant Analysis
An antisense CIP4 line (209-9) (T2 generation) was crossed with cop1-6, cop1-5, (GUS)–COP1, and GUS–NIa strains (von Arnim and Deng, 1994). All of the double homozygous lines except cop1-5/209-9 were identified in the F3 generation. Because cop1-5 homozygotes are lethal, double mutants of cop1-5 and 209-9 seedlings were analyzed using progeny of cop1-5+/−/209(HygR)+/+ plants, which produced one-quarter deetiolated seedlings and three-quarters etiolated seedlings in the absence and presence of DEX (data not shown). Analysis of the intracellular localization of GUS–COP1 and GUS–NIa in the wild-type and antisense CIP4 backgrounds was performed according to a published procedure (von Arnim and Deng, 1994).
Acknowledgments
We thank Dr. N.-H. Chua for providing pTA7002, Dr. T. Aoyama for technical information on the DEX induction system, Drs. S.J. Davis and R. Vierstra for providing smRS-GFP and technical information about the GFP variant, the Arabidopsis Biological Resource Center for providing cDNA and genomic libraries, Dr. M. Nakazawa for technical information regarding affinity purification of antibodies, and Drs. C. Hardtke, M. Holm, H. Okamoto, T. Yoshizumi, and K. Gohda for critical reading of the manuscript. Y.Y.Y. was the recipient of a Long Term Fellowship from the Human Frontier Science Program (at Yale) and a Special Postdoctoral Researcher of RIKEN, and was supported by a Grant-in-Aid for Encouragement of Young Scientists from the Ministry of Education, Science, Sports, and Culture, Japan. This work was supported by a grant from the National Institutes of Health (No. GM47850) to X.-W.D., a grant from the Research for the Future Program from the Japan Society for the Promotion of Science (No. JSPS-RFTF96L00601) to M.M., and a Human Frontier long-term grant (No. RG43/97) to M.M. and X.-W.D.
References
- Ahmad, M., and Cashmore, A.R. (1997). The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J. 11, 421–427. [DOI] [PubMed] [Google Scholar]
- Ang, L.-H., and Deng, X.-W. (1994). Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell 6, 613–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ang, L.-H., Chattopadhyay, S., Wei, N., Oyama, T., Okada, K., Batschauer, A., and Deng, X.-W. (1998). Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol. Cell 1, 213–222. [DOI] [PubMed] [Google Scholar]
- Aoyama, T., and Chua, N.-H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11, 605–612. [DOI] [PubMed] [Google Scholar]
- Barnes, S.A., Nishizawa, N.K., Quaggio, R.B., Whitelam, G.C., and Chua, N.-H. (1996). Far-red light blocks greening of Arabidopsis seedlings via a phytochrome A-mediated change in plastid development. Plant Cell 8, 601–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. 316, 1194–1199. [DOI] [PubMed] [Google Scholar]
- Bolle, C., Koncz, C., and Chua, N.-H. (2000). PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes Dev. 14, 1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Bowler, C., Neuhaus, G., Yamagata, H., and Chua, N.-H. (1994). Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77, 73–81. [DOI] [PubMed] [Google Scholar]
- Cashmore, A.R., Jarillo, J.A., Wu, Y.-J., and Liu, D. (1999). Cryptochromes: Blue light receptors for plants and animals. Science 284, 760–765. [DOI] [PubMed] [Google Scholar]
- Choi, G., Yi, H., Lee, J., Kwon, Y.-K., Soh, M.S., Shin, B., Luka, Z., Hahn, T.R., and Song, P.S. (1999). Phytochrome signaling mediated through nucleoside diphosphate kinase 2. Nature 401, 610–613. [DOI] [PubMed] [Google Scholar]
- Christie, J.M., Reymond, P., Powell, G.K., Bernasconi, P., Raibekas, A.A., Liscum, E., and Briggs, W.R. (1998). Arabidopsis NPH1: A flavoprotein with the properties of a photoreceptor for phototropism. Science 282, 1698–1701. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., and Vierstra, R.D. (1998). Soluble, highly fluorescent variants of green fluorescent protein (GFP) for use in higher plants. Plant Mol. Biol. 36, 521–528. [DOI] [PubMed] [Google Scholar]
- Deng, X.-W., Matsui, M., Wei, N., Wagner, D., Chu, A.M., Feldmann, K.A., and Quail, P.H. (1992). COP1, an Arabidopsis regulatory gene, encodes a protein with both a zinc-binding motif and a G beta homologous domain. Cell 71, 791–801. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., and Chory, J. (1997). Light control of plant development. Annu. Rev. Cell Dev. Biol. 13, 203–229. [DOI] [PubMed] [Google Scholar]
- Fankhauser, C., Yeh, K.C., Lagarias, J.C., Zhang, H., Elich, T.D., and Chory, J. (1999). PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science 284, 1539–1541. [DOI] [PubMed] [Google Scholar]
- Genoud, T., Millar, A.J., Nishizawa, N., Kay, S.A., Schafer, E., Nagatani, A., and Chua, N.H. (1998). An Arabidopsis mutant hypersensitive to red and far-red light signals. Plant Cell 10, 889–904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, H., Duong, H., Ma, N., and Lin, C. (1999). The Arabidopsis blue light receptor cryptochrome 2 is a nuclear protein regulated by a blue light–dependent posttranscriptional mechanism. Plant J. 19, 279–287. [DOI] [PubMed] [Google Scholar]
- Hardtke, C.S., Gohda, K., Osterlund, M.T., Oyama, T., Okada, K., and Deng, X.-W. (2000). HY5 stability and activity in Arabidopsis is regulated by phosphorylation within its COP1 binding domain. EMBO J. 19, 4997–5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1999). Antibodies. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Hoecker, U., Tepperman, J.M., and Quail, P.H. (1999). SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science 284, 496–499. [DOI] [PubMed] [Google Scholar]
- Hsieh, H.L., Okamoto, H., Wang, M.L., Ang, L.H., Matsui, M., Goodman, H., and Deng, X.-W. (2000). FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsis development. Genes Dev. 14, 1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hudson, M., Ringli, C., Boylan, M.T., and Quail, P.H. (1999). The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes Dev. 13, 2017–2027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang, H.-G., Fang, Y., and Singh, K.B. (1999). A glucocorticoid-inducible transcription system causes severe growth defects in Arabidopsis and induces defense-related genes. Plant J. 20, 127–133. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. eds (1994). Photomorphogenesis in Plants, 2nd ed. (Dordrecht, The Netherlands: Kluwer Academic Publishers).
- Kircher, S., Kozma-Bongar, L., Kim, L., Adam, E., Harter, K., Schäfer, E., and Nagy, F. (1999). Light quality–dependent nuclear import of the plant photoreceptors phytochrome A and B. Plant Cell 11, 1445–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner, O., Kircher, S., Harter, K., and Batschauer, A. (1999). Nuclear localization of the Arabidopsis blue light receptor cryptochrome 2. Plant J. 19, 289–296. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., Rolff, E., and Spruit, C.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Lutcke, H.A., Chow, K.C., Mickel, F.S., Moss, K.A., Kern, H.F., and Scheele, G.A. (1987). Selection of AUG initiation codons differs in plants and animals. EMBO J. 6, 43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Garcia, J.F., Monte, E., and Quail, P.H. (1999). A simple, rapid and quantitative method for preparing Arabidopsis protein extracts for immunoblot analysis. Plant J. 20, 251–257. [DOI] [PubMed] [Google Scholar]
- McNellis, T.W., von Arnim, A.G., and Deng, X.-W. (1994). Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell 6, 1391–1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNellis, T.W., Torii, K.U., and Deng, X.-W. (1996). Expression of an N-terminal fragment of COP1 confers a dominant-negative effect on light-regulated seedling development in Arabidopsis. Plant Cell 8, 1491–1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473–497. [Google Scholar]
- Nagatani, A., Reed, J.W., and Chory, J. (1993). Isolation and initial characterization of Arabidopsis mutants that are deficient in phytochrome A. Plant Physiol. 102, 269–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus, G., Bowler, C., Kern, R., and Chua, N.-H. (1993). Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell 73, 937–952. [DOI] [PubMed] [Google Scholar]
- Ni, M., Tepperman, J.M., and Quail, P.H. (1998). PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix-loop-helix protein. Cell 95, 657–667. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., and Deng, X.-W. (1998). Multiple photoreceptors mediate the light-induced reduction of GUS-COP1 from Arabidopsis hypocotyl nuclei. Plant J. 16, 201–208. [DOI] [PubMed] [Google Scholar]
- Osterlund, M.T., Hardtke, C.S., Wei, N., and Deng, X.-W. (2000). Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405, 462–466. [DOI] [PubMed] [Google Scholar]
- Oyama, T., Shimura, Y., and Okada, K. (1997). The Arabidopsis HY5 gene encodes a bZIP protein that regulates stimulus-induced development of root and hypocotyl. Genes Dev. 11, 2983–2995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puente, P., Wei, N., and Deng, X.-W. (1996). Combinational interplay of promoter elements constitutes the minimal determinants for light and developmental control of gene expression in Arabidopsis. EMBO J. 15, 3732–3743. [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Serino, G., Tsuge, T., Kwok, S., Matsui, M., Wei, N., and Deng, X.-W. (1999). Arabidopsis cop8 and fus4 mutations define the same gene that encodes subunit 4 of the COP9 signalosome. Plant Cell 11, 1967–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soh, M.S., Hong, S.H., Hanzawa, H., Furuya, M., and Nam, H.G. (1998). Genetic identification of FIN2, a far red light-specific signaling component of Arabidopsis thaliana. Plant J. 16, 411–419. [DOI] [PubMed] [Google Scholar]
- Torii, K.U., McNellis, T.W., and Deng, X.-W. (1998). Functional dissection of Arabidopsis COP1 reveals specific roles of its three structural modules in light control of seedling development. EMBO J. 17, 5577–5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.-W. (1993). Ring finger motif of Arabidopsis thaliana COP1 defines a new class of zinc-binding domains. J. Biol. Chem. 268, 19626–19631. [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.-W. (1994). Light inactivation of Arabidopsis photomorphogenic repressor COP1 involves a cell-specific regulation of its nucleocytoplasmic partitioning. Cell 79, 1035–1045. [DOI] [PubMed] [Google Scholar]
- von Arnim, A.G., and Deng, X.-W. (1996). Light control of seedling development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 215–243. [DOI] [PubMed] [Google Scholar]
- Wang, H., Kang, D., Deng, X.-W., and Wei, N. (1999). Evidence for functional conservation of a mammalian homologue of the light-responsive plant protein COP1. Curr. Biol. 9, 711–714. [DOI] [PubMed] [Google Scholar]
- Wei, N., and Deng, X.-W. (1999). Making sense of the COP9 signalosome: A regulatory protein complex conserved from Arabidopsis to human. Trends Genet. 15, 98–103. [DOI] [PubMed] [Google Scholar]
- Whitelam, G.C., Johnson, E., Peng, J., Carol, P., Anderson, M.L., and Harberd, N. (1993). Phytochrome A null mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5, 757–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, R., Nakamura, M., Mochizuki, N., Kay, S.A., and Nagatani, A. (1999). Light-dependent translocation of a phytochrome B-GFP fusion protein to the nucleus in transgenic Arabidopsis. J. Cell Biol. 3, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, Y.Y., and Deng, X.-W. (1998). A new vector set for GAL4-dependent transactivation assay in plants. Plant Biotechnol. 15, 217–220. [Google Scholar]
- Yamamoto, Y.Y., Tsuji, H., and Obokata, J. (1995). 5′ Leader of a photosystem I gene in Nicotiana sylvstris, psaDb, contains a translational enhancer. J. Biol. Chem. 270, 12466–12470. [DOI] [PubMed] [Google Scholar]
- Yamamoto, Y.Y., Matui, M., Ang, L.-H., and Deng, X.-W. (1998). Role of COP1 interactive protein in mediating light-regulated gene expression in Arabidopsis. Plant Cell 10, 1083–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]