Abstract
Light perception by the plant photoreceptor phytochrome requires the tetrapyrrole chromophore phytochromobilin (PΦB), which is covalently attached to a large apoprotein. Arabidopsis mutants hy1 and hy2, which are defective in PΦB biosynthesis, display altered responses to light due to a deficiency in photoactive phytochrome. Here, we describe the isolation of the HY2 gene by map-based cloning. hy2 mutant alleles possess alterations within this locus, some of which affect the expression of the HY2 transcript. HY2 encodes a soluble protein precursor of 38 kD with a putative N-terminal plastid transit peptide. The HY2 transit peptide is sufficient to localize the reporter green fluorescent protein to plastids. Purified mature recombinant HY2 protein exhibits PΦB synthase activity (i.e., ferredoxin-dependent reduction of biliverdin IXα to PΦB), as confirmed by HPLC and by the ability of the bilin reaction products to combine with apophytochrome to yield photoactive holophytochrome. Database searches and hybridization studies suggest that HY2 is a unique gene in the Arabidopsis genome that is related to a family of proteins found in oxygenic photosynthetic bacteria.
INTRODUCTION
Plants are exquisitely sensitive to their environment. Because they are sessile and use light as the energy source for photosynthesis, plants have developed well-refined photoreception and signaling systems to modulate their growth and development. The family of phytochromes, which are sensory photoreceptors for red and far-red lights, plays a key role in mediating responses to light quality, quantity, direction, and duration throughout plant development (Kendrick and Kronenberg, 1994; Quail et al., 1995; Furuya and Schäfer, 1996; Neff et al., 2000). Plant phytochromes are homodimers composed of ∼125-kD subunits each with a thioether-linked phytochromobilin (PΦB) prosthetic group (Lagarias and Rapoport, 1980). Phytochrome action depends on its ability to photointerconvert between the red light–absorbing form and the far-red-light–absorbing form, a property conferred by covalently bound PΦB in holophytochrome.
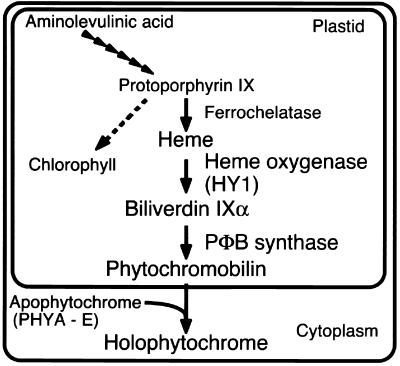
Two pathways are involved in the biosynthesis of holophytochrome, one for the apoprotein, which is encoded by a small multigene family (e.g., PHYA-E in Arabidopsis) (Sharrock and Quail, 1989; Clack et al., 1994), and another for the synthesis of the PΦB (Terry et al., 1993). Apophytochrome is synthesized in the cytosol, whereas PΦB is synthesized entirely within the plastid compartment, followed by its release to the cytosol, where holophytochrome assembly occurs (Figure 1). Based on spectroscopic studies of purified phytochromes, in vitro bilin assembly studies with recombinant apophytochromes, and physiological analyses of chromophore-deficient mutants, PΦB appears to be the immediate chromophore precursor of all higher plant and cryptophyte phytochromes (Terry et al., 1993; Terry, 1997).
Figure 1.
Phytochrome Biosynthesis in Arabidopsis.
PΦB is synthesized from 5-aminolevulinic acid and shares many intermediates with the pathways of chlorophyll and heme biosynthesis (Elich and Lagarias, 1987; Elich et al., 1989). These analyses established that biliverdin IXα (BV) is a PΦB precursor, suggesting the intermediacy of heme in the phytochrome chromophore biosynthetic pathway. Indeed, the first committed step of PΦB biosynthesis is catalyzed by a ferredoxin-dependent heme oxygenase, which is encoded by the HY1 gene in Arabidopsis and by its ortholog in rice (Davis et al., 1999; Muramoto et al., 1999; Izawa et al., 2000). Ferredoxin-dependent heme oxygenases were first identified in red algae and cyanobacteria, in which they catalyze the oxygen-dependent conversion of heme to BV (Beale and Cornejo, 1984; Cornejo and Beale, 1988, 1997; Cornejo et al., 1998). BV, therefore, is the first committed intermediate in the biosynthetic pathways of PΦB as well as those of the phycobilins phycocyanobilin and phycoerythrobilin, which are precursors of the light-harvesting prosthetic groups of the phycobiliproteins in cyanobacteria, red algae, and cryptomonads (Beale, 1993). In plants, BV is subsequently reduced to 3Z-PΦB by the ferredoxin-dependent bilin reductase PΦB synthase, which has not yet been cloned (Terry and Lagarias, 1991; M.T. McDowell and J.C. Lagarias, unpublished data). Although 3Z-PΦB can serve as a functional precursor of the phytochrome chromophore, its facile isomerization to 3E-PΦB, which is also a precursor of the phytochrome chromophore, likely occurs in plants (Terry et al., 1995). Ferredoxin-dependent bilin reductases are also present in cyanobacteria and red algae, where they catalyze the conversion of BV to the phycobilins (reviewed in Beale, 1993). None of these bilin reductases has been cloned.
Our understanding of photomorphogenesis in plants has been aided greatly by the isolation of five classic photomorphogenic Arabidopsis mutants (hy1 to hy5) that are impaired in response to light (Koornneef et al., 1980). Photoreceptor-deficient mutants have proven to be powerful tools to analyze which photoreceptors mediate specific photomorphogenetic responses (Koornneef and Kendrick, 1994; Whitelam and Devlin, 1997). Phytochrome chromophore–deficient mutants, including hy1 and hy2 in Arabidopsis, yg-2 and aurea in tomato, pcd1 and pcd2 in pea, and pew1 and pew2 in Nicotiana plumbaginifolia, have often been used as phytochrome-deficient mutants (reviewed in Terry, 1997). The aurea mutant of tomato has been used widely for physiological studies of phytochrome, for the study of other photoreceptors, and to study phytochrome signaling (Becker et al., 1992; Bowler and Chua, 1994). Knowledge of the molecular basis of these mutations will help in the interpretation of physiological experiments with these mutants. Biochemical analyses have established that the hy1, pcd1, and yg-2 mutants are deficient at the step at which BV is synthesized from heme, whereas pcd2 and aurea mutants are unable to synthesize PΦB from BV (Terry and Kendrick, 1996; van Tuinen et al., 1996; Weller et al., 1996, 1997). The recent cloning of HY1 has provided valuable insight into the first committed enzyme of phytochrome chromophore biosynthesis, heme oxygenase (Davis et al., 1999; Muramoto et al., 1999).
Of the five classic photomorphogenetic mutants, only hy2 remains to be cloned. It is widely believed that HY2 encodes PΦB synthase. However, the observation that a hy2 mutant is partially “rescued” by BV treatment suggests other possibilities (Parks and Quail, 1991). Although it is similar to hy1 mutants, the chlorophyll-deficient phenotype of hy2 mutants is typically less severe (Koornneef et al., 1980; Chory et al., 1989). The gene identification of HY2 in Arabidopsis should help to resolve these paradoxes. In this study, we describe the molecular basis for the phytochrome-deficient phenotype in the hy2 mutant of Arabidopsis. We show that the HY2 gene encodes PΦB synthase, a ferredoxin-dependent BV reductase that is responsible for the final step in phytochrome chromophore biosynthesis in plastids. This work has enabled us to identify other members of the HY2-related, ferredoxin-dependent bilin reductase family in phycobiliprotein-producing photosynthetic organisms (N. Frankenberg, K. Mukougawa, T. Kohchi, and J.C. Lagarias, unpublished data).
RESULTS
Fine Mapping Localizes the HY2 Gene to Two Overlapping Bacterial Artificial Chromosome Clones
We used a positional cloning strategy to isolate the HY2 gene, which previously had been mapped to chromosome 3. Because the hy2 long hypocotyl phenotype is easy to score in seedlings, the HY2 locus has served as a useful landmark for classic mapping. For fine mapping, we crossed the hy2-1 mutant of Landsberg erecta (Ler) ecotype to the wild-type Columbia (Col) ecotype, and segregating F2 populations with the hy2 phenotype were used for DNA preparation. First, we prepared DNA from ∼400 plants to perform genetic mapping of hy2 using cleaved amplified polymorphic sequence (CAPS) markers (Konieczny and Ausubel, 1993) that we developed and that are available in the database at the Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/maps/CAPS_Chr3.html). With ∼400 plants, HY2 was mapped to an interval of ∼360 kb between positional markers cMLP3E-1 and cF3L24 (Figure 2), indicating that recombination frequency in this region was much lower than expected. Therefore, we increased the size of the mapping population to ∼2000 plants. This approach enabled us to map the HY2 locus to an interval of ∼66 kb between the markers cMZB10 and cF3L24 (Figure 2).
Figure 2.
The HY2 Locus of Arabidopsis.
(A) Map of the region of chromosome 3 containing HY2. Two distinct mapping populations were screened, and mapping results with molecular markers are summarized schematically, indicating that HY2 lies in a region 66 kb in length. Markers starting with the letter c are CAPS markers developed during this study. DNA sequence information for bacterial artificial chromosomes (BACs) MZB10 and F3L24 is available in GenBank/EMBL/DDBJ. The HY2 gene structure with mutations is illustrated at the bottom. Exons are depicted as dark boxes and thick lines, which reflect coding regions and 5′/3′ untranslated regions, respectively. Dotted lines indicate introns.
(B) Genomic sequence of HY2 and the deduced protein sequence from the Columbia (Col) ecotype. Uppercase letters represent exons determined by sequence analysis of HY2 cDNAs. Introns and spacer sequences are indicated with lowercase letters. The stop codon is double underlined. Mutations in hy2 alleles are shown in boldface letters. Single nucleotide polymorphisms in both Ler and Wassilewskija (Ws) ecotypes include the following: inserted T (at nucleotide 234), G364T conversion with amino acid change to Asn, and G1182A conversion (silent). Single nucleotide polymorphisms in the Ler ecotype only include the following: C515A (in intron), G884A (silent), C1145T (in intron), and G1717A (in intron). The single nucleotide polymorphism in Ws ecotype only is C1910T (silent).
During these mapping studies, the sequences of two bacterial artificial chromosome clones, MZB10 and F3L24, spanning the HY2 locus genetically defined above, were deposited in the GenBank database (accession numbers AC009326 and AC011436, respectively). There are at least 21 putative genes in the region between the closest recombinations. We screened HY2 candidate genes based on the following expectations. First, HY2 should be categorized as an unknown or putative gene, because neither gene nor protein sequences of any ferredoxin-dependent bilin reductase were known. Second, HY2 should possess a plastid transit peptide, because enzymatic activity for PΦB synthase was detected in plastids (Terry and Lagarias, 1991). Third, weak sequence similarity between HY2 and an unidentified open reading frame(s) (ORF) in fully sequenced cyanobacterial genomes might be detectable, because HY2-related bilin reductase activities have been reported in cyanobacteria (Cornejo and Beale, 1997). The predicted amino acid sequences for all 22 genes in the HY2 region were used for TBLASTN (Altschul et al., 1990) and CHLOROP (Emanuelsson et al., 1999; http://www.cbs.dtu.dk/services/ChloroP/) analyses. By these criteria, one of these genes with two distinct annotations, MZB10.18 (GenBank accession number AC009326-18) or F3L24.1 (GenBank accession number AC0011436-1), appeared to be a strong candidate for HY2.
The HY2 Gene Is Identified by DNA Sequences of Wild-Type and Mutant Alleles
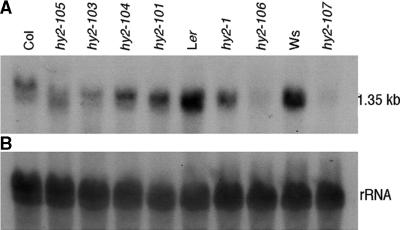
To help identify the HY2 gene, RNA gel blot analysis of wild-type and hy2 mutant seedlings was performed using the cDNA for MZB10.18/F3L24.1 as a probe. Because the hy2 phenotype is readily observed in seedlings, we analyzed the accumulation of transcripts in Arabidopsis seedlings (Figure 3). Transcripts were detected in the wild type of three ecotypes tested. The slow migration of mRNA of Col was verified as a gel artifact (data not shown). RNA gel blotting showed that the transcript levels were decreased severely in the hy2-1, hy2-106, and hy2-107 mutants and were decreased slightly in other mutant lines. Consequently, we focused our attention on the MZB10.18/F3L24.1 gene. To determine if mutations were present in the MZB10.18/F3L24.1 gene in hy2 mutants, DNA fragments corresponding to the region from the end of the upstream gene to the beginning of the downstream gene from various hy2 alleles were amplified by polymerase chain reaction (PCR). The nucleotide sequences were determined directly from the PCR products. In all hy2 alleles tested, nucleotide substitutions or deletions were detected (Figure 2). Complementation of hy2-1 was observed by introducing the construct containing the WT genomic fragment of the MZB10.18/F3L24.1 region (data not shown). Based on these data and biochemical data presented below, we conclude that locus MZB10.18/F3L24.1 corresponds to the HY2 gene.
Figure 3.
RNA Gel Blot Hybridization of MZB10.18/F3L24.1.
(A) Total RNA (10 μg) from 1-week-old seedlings was analyzed by RNA gel blotting using the MZB10.18/F3L24.1 cDNA as a probe. RNA was prepared from seedlings of the hy2 mutants and corresponding wild-type plants.
(B) The same RNA gel blot was probed with rDNA as a loading control of RNA.
As a result of the conflict in annotation of the HY2 gene in MZB10.18 and F3L24.1 (i.e., the former encodes a protein of 273 amino acids, and the latter encodes a protein of 329 amino acids), we sought to verify experimentally the structure of the HY2 gene. To do so, seven cDNA clones prepared from Col seedling mRNA were isolated from ∼300,000 clones examined. The nucleotide sequences of independent cDNA clones were determined, and they revealed a single reading frame that matched that of the annotation for F3L24.1. The HY2 gene contains eight small exons ranging from 51 to 222 nucleotides separated by seven introns ranging from 74 to 183 nucleotides. The longest cDNA insert contained a full-length 990-bp ORF, a 95-bp 5′ untranslated region, a 231-bp 3′ untranslated region, and a poly(A)+ stretch, as shown in Figure 4 (DNA Data Bank of Japan [DDBJ] accession number AB045112).
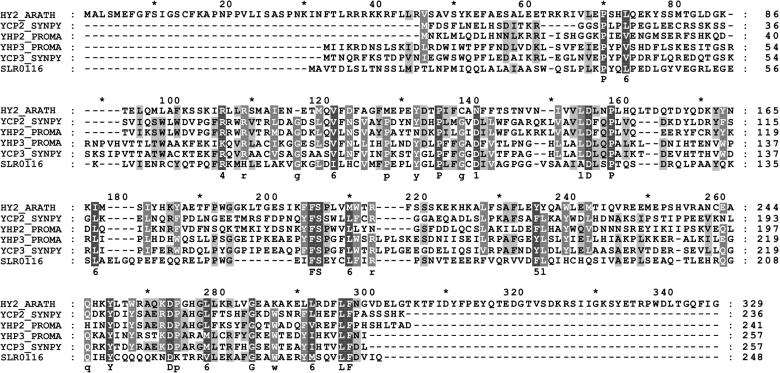
Figure 4.
Alignment of HY2 and HY2-Related Proteins.
Alignment of the HY2 protein with proteins of unknown function from oxygenic photosynthetic bacteria identified by PSI-BLAST. Conserved residues in 100 or 80% of the aligned sequences are depicted in the consensus sequence with uppercase or lowercase letters, respectively. Sequence similarity groups shown in the consensus sequence reflect conservation in >100% of the sequences. These are labeled as follows: 1,  ; 4,
; 4, 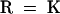 ; 5,
; 5, 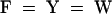 ; and 6,
; and 6, 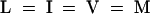 . Dark shading with white letters, gray shading with white letters, and gray shading with black letters reflect 100, 80, and 60% sequence conservation, respectively. Sequence identifiers correspond to hypothetical proteins from Synechococcus sp WH8020 (YCP2_SYNPY and YCP3_SYNPY), from Prochlorococcus (YHP2_PROMA and YHP3_PROMA), and from Synechocystis sp PCC 6803 gene (cyanobase locus slr0116; see http://www.kazusa.or.jp/cyano/cyano.html). Database accession numbers are AB045112 for HY2 (DDBJ), Q02189 for YCP2_SYNPY (SWISS-PROT), Q02190 for YCP3_SYNPY (SWISS-PROT), CAB95700.1 for YHP2_PROMA (EMBL), CAB95701.1 for YHP3_PROMA (EMBL), and S76709 for slr0116 (Protein Information Resource). Asterisks are indicated every 20 residues.
. Dark shading with white letters, gray shading with white letters, and gray shading with black letters reflect 100, 80, and 60% sequence conservation, respectively. Sequence identifiers correspond to hypothetical proteins from Synechococcus sp WH8020 (YCP2_SYNPY and YCP3_SYNPY), from Prochlorococcus (YHP2_PROMA and YHP3_PROMA), and from Synechocystis sp PCC 6803 gene (cyanobase locus slr0116; see http://www.kazusa.or.jp/cyano/cyano.html). Database accession numbers are AB045112 for HY2 (DDBJ), Q02189 for YCP2_SYNPY (SWISS-PROT), Q02190 for YCP3_SYNPY (SWISS-PROT), CAB95700.1 for YHP2_PROMA (EMBL), CAB95701.1 for YHP3_PROMA (EMBL), and S76709 for slr0116 (Protein Information Resource). Asterisks are indicated every 20 residues.
Figure 2 shows the genomic structure of the HY2 gene with positions of the mutations in hy2 alleles. Two hy2 alleles, hy2-102 and hy2-107, were found to have point mutations at 3′ splice sites in the seventh and fifth introns, respectively. Such mutations in the G of the essential AG dinucleotide at the 3′ splice site have been reported to lead to missplicing with a downstream AG, resulting in a frameshift in the protein (Brown, 1996). hy2-105 was another possible splicing mutant, with a 25-bp deletion in the second intron. This mutation truncates the second intron to 57 nucleotides, much smaller than the average size of Arabidopsis introns (240 nucleotides). The efficiency of intron splicing might be reduced because of a minimum intron size requirement (Deutsch and Long, 1999), although we have not checked the significance of defects in pre-mRNA splicing experimentally. A fast neutron–generated allele, hy2-106, carries a 5-bp deletion in the first exon, making an immediate stop codon. Four ethyl methanesulfonate–generated alleles, hy2-1, hy2-101, hy2-103, and hy2-104, have single nucleotide changes to produce amino acid substitutions compared with the corresponding wild-type allele. Two of these alleles, hy2-1 and hy2-104, have the same mutation (P128L), whereas hy2-101 and hy2-103 possess G181R and R252Q substitutions, respectively.
The HY2 Protein Is Related to a Family of Cyanobacterial Proteins
Based on cDNA sequence analysis, the HY2 protein contains 329 residues with a calculated molecular mass of 38.1 kD. At its N terminus, the HY2 protein sequence is rich in serine, with few acidic residues (six serine and one aspartic acid among 45 residues), which suggests a possible transit peptide for localization to plastids (Gravel and von Heijne, 1990). The second amino acid after the initiation methionine is alanine, which is often observed in plastid transit peptides. The program CHLOROP was also used to predict the transit peptide of HY2, and it indicated that the first 45 amino acid residues of the HY2 protein form a chloroplast transit peptide (Emanuelsson et al., 1999; http://www.cbs.dtu.dk/services/ChloroP/). The calculated molecular mass of the mature HY2 protein is 33.0 kD and its predicted pI is 5.66, which are similar to those of PΦB synthase purified from oat seedlings (M.T. McDowell and J.C. Lagarias, unpublished data). The HY2 protein has no predicted transmembrane helices, which is also consistent with the observation that oat PΦB synthase is a soluble protein (M.T. McDowell and J.C. Lagarias, unpublished data).
Using the HY2 protein sequence as a query sequence, we performed an iterative PSI-BLAST search of the nonredundant GenBank/EMBL database (http://www.ncbi.nlm.nihgov/blast/psiblast.cgi) by using default search parameters (Altschul et al., 1997). Surprisingly, no HY2-related gene was identified by this search in the nearly complete Arabidopsis genome. In contrast, this search identified HY2-related sequences from two marine cyanobacteria, Prochlorococcus marinus sp SS120 (EMBL accession numbers CAB95700.1 and CAB95701.1) and Synechococcus sp WH8020 (SWISS-PROT accession numbers Q02189 and Q02190), and a related protein sequence from the cyanobacterium Synechocystis sp PCC 6803 (cyanobase locus slr0116; Protein Information Resource accession number S76709). Both marine cyanobacteria possess two HY2-related ORFs that appear to be part of multigene operons. Interestingly, the Synechococcus ORFs, ycp2_synpy and ycp3_synpy, are located within a cluster of genes involved in phycobiliprotein biosynthesis (Wilbanks and Glazer, 1993), whereas the Prochlorococcus ORFs, which we term yhp2_proma and yhp3_proma, are located immediately downstream of a gene related to heme oxygenase (GB:AJ278499.1). These observations strongly support the hypothesis that these genes are involved in phycobilin biosynthesis.
Figure 4 shows an optimized multiple sequence alignment of HY2 and HY2-related cyanobacterial sequences using the programs CLUSTALW (Higgins et al., 1996), MEME to guide hand alignments (http://meme.sdsc.edu/meme/website/), and GENEDOC for highlighting (http://www.psc.edu/biomed/genedoc). As expected, the HY2-related cyanobacterial proteins lack the putative plastid transit peptide sequence found at the N terminus of HY2. Pairwise sequence identities between HY2 and the cyanobacterial ORFs are quite low (<20%), although the similarities between YCP2_SYNPY and YHP2_PROMA and between YCP3_SYNPY and YHP3_PROMA suggest that these pairs of proteins have similar functions. That the mutation in the hy2-1 and hy2-104 alleles (P128L) lies in a conserved proline residue is consistent with a critical role of this residue in the enzyme's structure. Proline residues are typically involved in cis-peptide bonds, which occur at β-turns in proteins. Examination of the amino acid alterations in the two other missense alleles, G181R in hy2-101 and R252Q in hy2-103, reveals that neither mutation corresponds to a strongly conserved residue in this protein family.
The HY2 Protein Is Localized to the Plastid
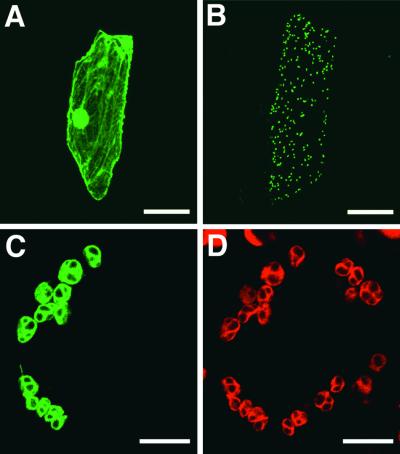
The N terminus of HY2 has a stretch of 45 amino acids with features of a chloroplast transit peptide. To determine whether this peptide is a functional plastid-targeting sequence, we fused the transit peptide–coding region of HY2 to a modified gene of green fluorescent protein (GFP) from jellyfish under the control of modified cauliflower mosaic virus 35S promoter (Chiu et al., 1996). The construct was introduced into onion skin cells and tobacco leaves by bombardment with DNA-coated particles, and transient expression was analyzed using confocal laser scanning microscopy. Although a control construct without the putative transit peptide showed GFP fluorescence throughout the cytoplasm and the nucleus of onion cells (Figure 5A), clear localization of GFP fluorescence to small dots, most likely plastids, was observed when the putative transit peptide was fused to GFP (Figure 5B). For better visualization, we also introduced the construct into tobacco leaves, where the chloroplasts are well developed in guard cells. GFP fluorescence was localized exclusively in oval structures (Figure 5C) that match the red autofluorescence from the chlorophyll of the chloroplasts (Figure 5D), demonstrating that the fusion protein is efficiently targeted to chloroplasts. This finding confirms the presence of a functional transit peptide and implies that the HY2 gene product is localized in the chloroplast.
Figure 5.
Transient Expression of GFP Fusion in Onion Cells and Tobacco Cells.
(A) to (C) Cells expressing GFP (onion) (A), the HY2 transit peptide fused to GFP (HY2TP-GFP) (onion) (B), and HY2TP-GFP (tobacco) (C) were analyzed by fluorescence microscopy using the green channel for GFP.
(D) The same sample as in (C) imaged using the red channel for chlorophyll.
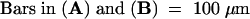 ;
; 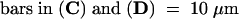 .
.
Recombinant HY2 Exhibits PΦB Synthase Activity
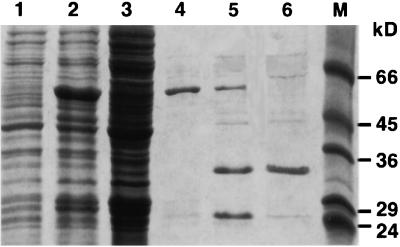
The HY2 protein lacking the transit peptide mHY2 was synthesized in Escherichia coli as a fusion protein with glutathione S-transferase (GST) and purified by affinity chromatography, as described in Methods. The GST tag was removed by site-specific protease digestion. A second round of affinity chromatography yielded protein at >90% homogeneity. Figure 6 shows SDS-PAGE results of the purification and processing of the protein. One liter of bacterial culture yielded ∼1 mg of recombinant protein. The molecular mass of the Arabidopsis mHY2 deduced from the cDNA is 33 kD. However, the cloning and expression strategy for the mHY2 cDNA using pGEX-6P-1 were responsible for an additional five N-terminal amino acids (GPLGS) after protease treatment.
Figure 6.
SDS-PAGE of the Purification of Arabidopsis mHY2.
Lane 1, cell-free extract from uninduced Escherichia DH5α carrying pGEX-mHY2; lane 2, cell-free extract from isopropylthio-β-galactoside−induced Escherichia DH5α carrying pGEX-mHY2; lane 3, soluble fraction of the induction; lane 4, GST-mHY2 after glutathione-agarose affinity chromatography; lane 5, GST-mHY2 after PreScission protease treatment; lane 6, purified recombinant mHY2 after a second round of glutathione-agarose affinity chromatography; lane M, molecular mass standards. Numbers at right indicate positions of molecular weight markers (Sigma, SDS-7) in kilodaltons.
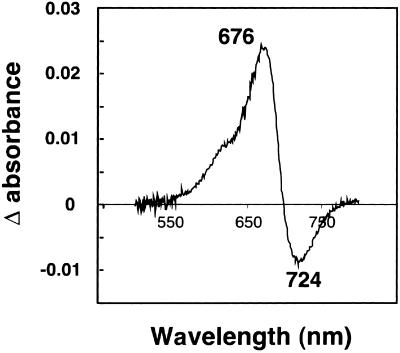
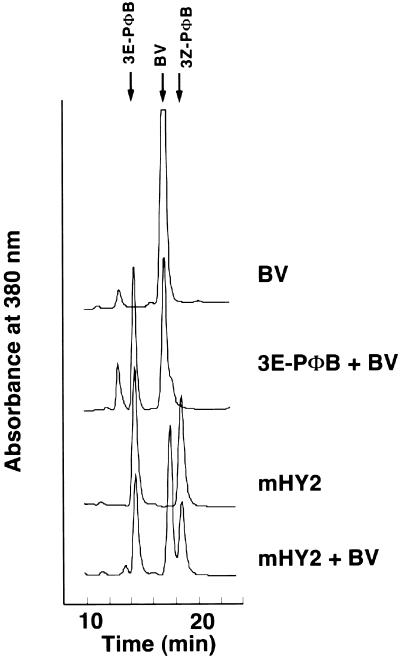
To determine whether mHY2 has PΦB synthase activity, its ability to reduce BV to PΦB was first assessed with a “coupled” holophytochrome assembly assay in which the reaction products were incubated with recombinant cyanobacterial phytochrome 1 (Cph1) apoprotein (Yeh et al., 1997). Figure 7 shows a phytochrome difference spectrum obtained after incubation of apoCph1 with the bilin products from a PΦB synthase assay of a crude cell-free bacterial extract expressing GST-mHY2. The difference spectrum has a peak at 676 nm and a valley at 724 nm, which is consistent with a PΦB-Cph1 adduct (Yeh et al., 1997). To ensure that this activity was not due to a component of the crude Escherichia lysate, the ability of purified mHY2 to reduce BV to PΦB was analyzed using the coupled assembly assay as well as an HPLC assay. A phytochrome difference spectrum identical to that shown in Figure 7 was obtained (data not shown). The HPLC results of the PΦB synthase assay mixture are shown in Figure 8. After incubation of HY2 for 30 min under standard PΦB synthase assay conditions, all of the BV was converted to PΦB. Interestingly, both 3Z- and 3E-PΦB isomers were produced, although the relative amount of the 3E-PΦB isomer varied for different HY2 samples and may be an artifact of the presence of residual glutathione (N. Frankenberg and J.C. Lagarias, unpublished data).
Figure 7.
Phytochrome Difference Spectra of Cph1 Incubated with BV Metabolites.
A soluble protein extract of isopropylthio-β-galactoside−induced E. coli DH5α carrying pGEX-mHY2 was assayed for PΦB synthase activity as described in Methods. Recombinant Cph1 was added to the bilin reaction mixture, which was incubated for another 30 min at room temperature under green safe light, and a phytochrome difference spectrum was obtained. The absorption maximum and minimum are indicated in nanometers.
Figure 8.
HY2 Converts BV to PΦB as Detected by HPLC.
Purified recombinant HY2 protein (40 μg) was assayed for PΦB synthase activity as described in Methods. Bilins were extracted from the incubation mixture using a SepPak C18 reversed phase cartridge and analyzed by reversed phase HPLC as described in Methods. The HPLC solvent was acetone:20 mM formic acid (50:50 [v/v]), and the eluate was monitored at 380 nm. The top traces represent the standard bilins BV and 3E-PΦB. The third trace shows the bilin metabolites obtained after incubation of BV with HY2. The bottom trace has, in addition, BV as an internal standard.
DISCUSSION
The hy2 mutant of Arabidopsis is one of five classic long hypocotyl mutants first identified by Koornneef et al. (1980). That the hy2 mutant is photomorphogenetically impaired due to a phytochrome deficiency has been well documented by physiological and photobiological analyses (Koornneef et al., 1980; Chory et al., 1989; Parks and Quail, 1991; Goto et al., 1993). Parks and Quail (1991) showed that the long hypocotyl phenotype of the hy1 and hy2 mutants was in part “rescued” by BV feeding and suggested that these mutants have lesions in the phytochrome chromophore biosynthetic pathway. Indeed, HY1 encodes a plastid-localized heme oxygenase that catalyzes the cleavage of heme to form BV (Davis et al., 1999; Muramoto et al., 1999). The present studies establish that HY2 encodes PΦB synthase, a plastid-localized enzyme responsible for the ferredoxin-dependent conversion of BV to PΦB, the immediate precursor of the phytochrome chromophore.
Based on the presence of a functional plastid-targeting sequence in the HY2 protein, we can confidently conclude that the entire pathway of PΦB biosynthesis occurs within plastids. Nevertheless, the possibility of an alternative pathway in other subcellular compartments cannot be dismissed entirely. In this regard, there are three other heme oxygenase genes besides HY1 in the Arabidopsis genome whose products may play a role in an alternative pathway (M. Masuda, T. Muramoto, and T. Kohchi, unpublished data). However, our database searches revealed no other gene in the Arabidopsis genome that shows statistically significant similarity to HY2. Although a weak similarity between HY2 and a ferredoxin-dependent bilin reductase involved in chlorophyll catabolism, red chlorophyll catabolite reductase, was revealed by profile analysis (N. Frankenberg, K. Mukougawa, T. Kohchi, and J.C. Lagarias, unpublished data), red chlorophyll catabolite reductase does not catalyze the reduction of BV to PΦB (Wüthrich et al., 2000; N. Frankenberg and J.C. Lagarias, unpublished results). Therefore, it appears that HY2 is the only PΦB synthase gene in Arabidopsis.
Physiological comparisons of the hy1 and hy2 mutants indicate that hy1 plants display more severe phytochrome-deficient phenotypes (Koornneef et al., 1980; Chory et al., 1989). These observations are somewhat surprising in view of the apparent uniqueness of the HY2 gene and the existence of multiple HY1-related proteins in the Arabidopsis genome (M. Masuda, T. Muramoto, and T. Kohchi, unpublished data). However, this may reflect the strength of the hy1 and hy2 alleles examined. In this regard, the partial rescue of the hy2-1 mutant treated with BV (Parks and Quail, 1991) can be explained by the hypothesis that the P128L missense mutation affords a partially active enzyme with a lower affinity for BV. Alternately, it is possible that BV might be converted to PΦB by an enzyme unrelated to HY2 in Arabidopsis. Phytochrome chromophore biosynthetic mutants have been identified in other plant species (Terry, 1997). In all cases, two classes of mutants have been identified: those that are deficient in heme oxygenase and those that are deficient in PΦB synthase. Based on biochemical analyses, the aurea mutant of tomato and the pcd2 mutant of pea are deficient in PΦB synthase activity (van Tuinen et al., 1996; Weller et al., 1997). The observations that the corresponding heme oxygenase mutants in these plant species (i.e., yg-2 and pcd1, respectively) exhibit less severe phenotypes further support the hypothesis that the relative allele strength of the two loci determines the phenotype. A phenotypic comparison of null alleles of hy1 and hy2 (e.g., hy2-106 and hy2-107) should help resolve this question.
The cloning of the Arabidopsis HY2 gene will help to identify PΦB synthase genes from other plant species and to confirm that the mutations in aurea and pcd2 occur in homologous genes. The aurea mutant of tomato has been used extensively to analyze phytochrome signal transduction (Bowler et al., 1994), and knowledge of the molecular basis of this mutation is of considerable interest. The molecular basis of such mutations should provide insight into residues critical for substrate and/or potential cofactor (i.e., metal ions or organic single electron carriers) interactions as well as those necessary for protein–protein interactions (i.e., between HY2 and ferredoxin or between HY1 and HY2). The availability of HY1- and HY2-specific cDNA probes and specific antibodies to both enzymes will facilitate experiments to study the regulation of phytochrome chromophore biosynthesis. With such probes, several key questions can be addressed. Are the two enzymes expressed coordinately in all tissues? Is their expression spatially and temporally regulated? Do HY1 and HY2 proteins form a dual enzyme complex in the plastid that channels the conversion of heme to PΦB? Does the expression of HY1 affect HY2 expression and vice versa?
The molecular cloning of HY2 has provided a breakthrough in our knowledge of bilin biosynthesis in general. Our bioinformatic analyses reveal that HY2 is related to a number of cyanobacterial genes of unknown function (Figure 4). Indeed, we hypothesize that these HY2-related proteins are enzymes involved in the biosynthesis of the chromophore precursors of the light-harvesting phycobiliproteins phycocyanobilin and phycoerythrobilin. As might be expected for enzymes with different substrate/product specificities, these proteins are highly diverged from HY2 (<20% sequence identity). The levels of identity between these proteins and HY2, which are highlighted in Figure 4, likely reflect residues involved in overall protein folding and/or ferredoxin interaction that are common to the entire family of enzymes. We demonstrate that these HY2-related proteins are members of a growing family of ferredoxin-dependent bilin reductases with different double bond specificities (N. Frankenberg, K. Mukougawa, T. Kohchi, and J.C. Lagarias, unpublished data).
The pathway for phytochrome chromophore biosynthesis shown in Figure 1 has been clearly documented. Now that the two key genes of the phytochrome chromophore biosynthetic pathway have been cloned, we intend to elucidate how bilin biosynthesis is regulated throughout the plant, a process that is critical to the plant's ability to respond to light. The possible role of bilins as second messengers, which was raised by recent studies of transgenic plants expressing mammalian biliverdin reductase (Montgomery et al., 1999), also will be addressed by manipulating the expression of HY1 and HY2 genes within different cells and tissues of the plant. Finally, it will be of particular interest to address the relationship of phytochrome chromophore biosynthesis and chlorophyll biosynthesis, not only because they share common biosynthetic intermediates but to determine how each pathway influences the other.
METHODS
Plant Materials
Arabidopsis thaliana ecotypes Columbia (Col), Landsberg erecta (Ler), and Wassilewskija (Ws) were obtained from our laboratory stocks. Mutant strains used in this work were obtained from Maarten Koornneef for hy2-1 (distributed as CS68 by the Arabidopsis Biological Stock Center, Columbus, OH; in Ler ecotype); from Jason Reed for hy2-101 (EMS89S73S-E isolated originally by J. Reed; in Col ecotype), hy2-102 (EMS195 isolated by J. Reed; in Col ecotype), hy2-103 (IAAR-7 isolated by Allison Wilson; in Col ecotype), hy2-104 (IAAR-12 isolated by A. Wilson; in Col ecotype), hy2-105 (γ10-9 isolated by J. Reed; in Col ecotype), and hy2-106 (FN16-3 isolated by Aron Silverstone; in Ler ecotype); and from Nam-Hai Chua for hy2-107 (segregated hy2 from T-DNA lines in his laboratory; in Ws ecotype). Plants were grown under long-day conditions at 22°C in a growth chamber.
Map-Based Cloning
The hy2-1 mutant was outcrossed with wild-type Col ecotype, and the mapping population was selected from F2 families with the long hypocotyl phenotype. Genomic DNA was prepared using a protocol described by Edwards et al. (1991). We used cleaved amplified polymorphic sequence (CAPS) markers between Col and Ler (Konieczny and Ausubel, 1993), two CAPS markers (C6 and manganese–superoxide dismutase) in the Arabidopsis database, and seven new CAPS markers developed during this study. Primer sequences for polymerase chain reaction (PCR) amplification are listed here with the enzymes used for digestion indicated in parentheses: c4523, 5′-ACAGCGAGATTCAAAGGTCCATTAACCGGA-3′ and 5′-GGGCTTACAGTGATATCTGCAAGACTTCTA-3′ (HpaII); cMLP3E1, 5′-TAATGCTTGCGACAAACAGG-3′ and 5′-GTTCATCTCAGGGCCAAAAA-3′ (RsaI); cMXK7, 5′-GCTTTCAGAAATCAGACCTCAA-3′ and 5′-CTGGTGTGGTTGATCGAATCT-3′ (DdeI); cMZB10, 5′-CTGCCAAGCTTCATTTGGTT-3′ and 5′-GCAGGAGCTGCAGACAATCT-3′ (BsrI); cMZB10.18, 5′-CAATGCAGGTTTAACTTCAGCA-3′ and 5′-CCATGG-GAAAGTCTGCAAAT-3′ (DdeI); cF3L24, 5′-TCAAGCCCTTTTCCAACATC-3′ and 5′-TTCCCCATCTGAACTCAACC-3′ (HinfI); and cF8A24, 5′-AATGATGCATGGTGTTGGTG-3′ and 5′-GCTCGAGGAAAAGTC-ATCCA-3′ (MboI).
Sequence Analysis of the HY2 Locus
A pair of primers (5′-CGTTTGTCTCACTGAAACTG-3′ and 5′-CAATCATCTTGAAATGCAGA-3′) was used to amplify 1.98-kb fragments of the MZB10.18 region from mutants and their corresponding wild-type plants. The PCR products were subjected directly to a cycle-sequencing protocol with several primers, and reactions were analyzed on an ABI373S sequencing apparatus (Applied Biosystems, Foster City, CA).
Isolation of HY2 cDNA
A cDNA library was constructed by K. Ando (Nara Institute of Science and Technology) from Col seedlings in λZAPII (Stratagene) according to the manufacturer's instructions. The DNA fragment containing MZB10.18 described above was used as a probe to screen ∼300,000 cDNA clones by plaque hybridization. Several cDNA plasmids were recovered by in vivo excision according to the manufacturer's instructions.
RNA Isolation and Analysis
RNA was isolated from 1-week-old whole Arabidopsis seedlings by the acid guanidinium thiocyanate-phenol-chloroform extraction method using Isogen (Nippon Gene, Tokyo, Japan). Total RNA (10 μg/lane) was electrophoresed on a 1.2% formaldehyde/agarose gel and transferred to a nylon membrane (Hybond-N; Amersham Corp.). Prehybridization and hybridization were then performed in Church hybridization solution (Church and Gilbert, 1984) using radioactive probes (3 × 106 to 5 × 106 cpm/mL). A fragment of cDNA produced by EcoRI and XhoI digestion was used as a hybridization probe. Filters were washed under highly stringent conditions three times with 1 × SSC (1 × SSC is 0.15 M NaCl and 0.015 M sodium citrate), 0.1% SDS at room temperature, and twice with 0.2 × SSC, 0.1% SDS at 65°C for 15 min. To show equal loading of RNA, an rRNA probe was used for hybridization.
Subcellular Localization Experiment with Green Fluorescent Protein Fusion
The coding region of HY2 for the putative transit peptide and flanking amino acid residues (amino acids 1 to 62) isolated by PCR was cloned into pTH2XA, a modified green fluorescent protein (GFP) vector derived from 35SΩ-sGFP-S65T (Chiu et al., 1996). In pTH2XA, five glycine residues were included at the fusion junction to GFP (M. Takemura, unpublished data). The construct, which can express the HY2 transit peptide fused to the N terminus of a modified GFP gene under the control of the cauliflower mosaic virus 35S promoter, was introduced into onion bulbs and tobacco leaves. The conditions of bombardment were the same as those described by Muramoto et al. (1999). Transient expression was observed after overnight incubation using confocal laser scanning microscopy (LSM510; Carl Zeiss, Jena, Germany).
Construction of the pGEX-mHY2 Expression Vector
mHY2, the mature HY2 gene without the predicted chloroplast transit peptide, was subcloned into the Escherichia coli expression vector pGEX-6-P1 (Amersham Pharmacia Biotech, Piscataway, NJ) to produce pGEX-mHY2. mHY2 was amplified using the primers mHY2BglIIfwd. (5′-GAAGATCTGTCTCTGCTGTGTCGTATAAGG-3′) and HY2SmaIrev. (5′-TCCCCCGGGTTAGCCGATAAATTGTCCTGTTAAATC-3′), which contained BglII and SmaI sites (underlined), respectively, and was cloned into BamHI-SmaI–digested pGEX-6-P1 to give pGEX-mHY2. The integrity of the construct was verified by restriction analysis and complete DNA sequencing of the insert (Davis Sequencing, Inc., Davis, CA). The constructed vector contains the mHY2 sequence placed 3′ to the glutathione S-transferase (GST) gene of Schistosoma japonicum under the control of a Ptac promotor. A recognition sequence for PreScission protease, which is also a GST fusion protein, is located upstream of mHY2. Proteolytic cleavage yields the native Arabidopsis mHY2 with the five–amino acid N-terminal extension GPLGS.
Expression and Purification of Recombinant mHY2
The Escherichia coli strain DH5α containing pGEX-mHY2 was grown at 37°C in 500-mL batches of Luria-Bertani medium containing ampicillin (100 μg/mL) to an OD578 of 0.6. Cultures were induced by the addition of 1 mM isopropylthio-β-galactoside and incubated for an additional 3 hr, and bacteria were harvested subsequently by centrifugation. The bacterial pellet from 3 liters of culture was resuspended in 20 mL of lysis buffer (50 mM Tris-HCl, pH 8.0, 100 mM NaCl, 0.05% Triton X-100, 1 mM DTT, 2 mM benzamidine, 2 mM PMSF, 2.0 μg/mL leupeptin, and 3 μg/mL pepstatin A) and disrupted with a French press (3 × 20,000 p.s.i.). Cell debris were removed by centrifugation for 30 min at 100,000g. The resulting supernatant was loaded directly onto a glutathione-agarose (Sigma) column (1 × 3 cm) that had been equilibrated with 5 column volumes of PBS. Unbound protein was removed by washing the column with 5 column volumes of PBS. GST-mHY2 fusion protein was eluted with 50 mM Tris-HCl, pH 8.0, containing 10 mM reduced glutathione. GST-mHY2–containing fractions were pooled and dialyzed overnight against cleavage buffer (50 mM Tris-HCl, pH 7.0, 100 mM NaCl, 1 mM EDTA, and 1 mM DTT). Digestion of the fusion protein was performed by adding 2 units of PreScission protease (Amersham Pharmacia Biotech) per 100 μg of fusion protein and incubating at 4°C for 5 hr. Removal of uncleaved fusion protein and excised GST tag was achieved by loading the digestion mixture onto a second glutathione-agarose column (1 × 3 cm). Recombinant mHY2 was detected in the flow through, analyzed by SDS-PAGE, and concentrated using Centriprep-10 concentrator devices (Amicon, Beverly, MA). One liter of bacterial culture yielded 1 mg of purified protein.
Determination of Protein Concentration
Protein concentration was determined using the method of Bradford (1976) or by absorption at 280 nm for purified mHY2, where 1 absorption unit represents 0.64 mg/mL mHY2 (Gill and von Hippel, 1989).
PΦB Synthase Activity Assay
All enzymes used for PΦB synthase assay were obtained from Sigma. For a 1-mL assay of PΦB synthase, the protein fraction to be assayed was diluted into 50 mM Tes-KOH, pH 7.3, containing an NADPH-regenerating system (6.5 mM glucose-6-phosphate, 0.82 mM NADP+, 1.1 units/mL glucose-6-phosphate dehydrogenase type XII from Torula yeast [EC 1.1.1.49]), a ferredoxin-reducing system (4.6 μM spinach ferredoxin, 0.025 units/mL spinach ferredoxin: NADP+ oxidoreductase [EC 1.18.1.2]), and 10 μM BSA (fraction V, heat shock) (M.T. McDowell and J.C. Lagarias, unpublished data). Glucose-6-phosphate and NADP+ were prepared as 100- and 25-mM stocks, respectively, in water; both were stored at 4°C. The glucose-6-phosphate stock was filter sterilized before storage. Glucose-6-phosphate dehydrogenase was prepared as a 500-unit/mL stock in 5 mM sodium citrate, pH 7.4, and stored at 4°C. Spinach ferredoxin:NADP+ oxidoreductase was prepared as a 10-unit/mL stock with sterile water and stored at 4°C. BSA was made up as 100 μM stock in 0.1 M potassium phosphate buffer, pH 7.4, and stored at either 4 or −20°C. The reaction was initiated by the addition of 5 μM (final concentration) purified biliverdin IXα (BV) (McDonagh and Palma, 1980) in 5 μL of DMSO. Assay mixtures were incubated in a 28°C water bath under green safe light or under subdued light for the desired amount of time. The assays were stopped by placing them on ice. Product analysis used a direct HPLC assay or a coupled assay after the addition of recombinant cyanobacterial phytochrome 1 (Cph1) apoprotein and difference spectroscopy (see below).
Direct HPLC Assay
For the quantitative analysis of PΦB synthase activity, assay mixtures (outlined above) were loaded onto a Waters (Milford, MA) C18 Sep-Pak Light (catalog No. WAT023501) preconditioned as follows: 3-mL wash with acetonitrile to wet the Sep-Pak, 3-mL wash with MilliQ water, and 3-mL wash with 50 mM 4-methylmorpholine/glacial acetic acid, pH 7.7. After the sample was loaded onto the Sep-Pak, it was washed with 3 mL of 4-methylmorpholine/glacial acetic acid, pH 7.7, followed by 3 mL of 0.1% (v/v) trifluoroacetic acid. The Sep-Pak was then eluted with 2 mL of 100% acetonitrile. The eluate was dried using a Speed-Vac lyophilizer. The dried samples were analyzed by HPLC. Samples were first dissolved in 10 μL of DMSO and then diluted with 200 μL of the HPLC mobile phase (acetone:20 mM formic acid [50:50, v/v]). After the samples were dissolved, they were centrifuged briefly, passed through a 0.45-μm polytetrafluoroethylene syringe filter, and chromatographed using a Varian (Palo Alto, CA) 5000 liquid chromatograph. The column eluate was monitored at 380 nm using a Varian UV100 flow-through absorbance detector. Peak areas were quantitated using a 3365 Chemstation II (Hewlett-Packard, Waldbronn, Germany). The HPLC column used for all of the analyses was a Phenomenex (Torrance, CA) Ultracarb 5-μm ODS (20) 4.6 × 250-mm analytical column with a 4.6 × 30-mm guard column of the same material. The mobile phase used with this column was acetone:20 mM formic acid (50:50 [v/v]). The flow rate was 0.8 mL/min.
Coupled Difference Spectral Assay
An alternative to the direct analysis of PΦB synthase activity was the coupled, or indirect, assay. This assay was based on the method outlined previously (Terry and Lagarias, 1991). The assay described above for PΦB synthase was performed as before, but instead of working up the sample by Sep-Pak, an aliquot of recombinant apophytochrome (Cph1 from Synechocystis sp PCC 6803) was added to the sample. The sample was incubated for an additional 20 to 30 min at room temperature under green safe light, and then a difference spectrum was taken. The method for difference spectroscopy was described previously (Terry and Lagarias, 1991).
Acknowledgments
We thank the Arabidopsis Biological Resource Center and Drs. M. Koornneef (Wageningen Agricultural University, Wageningen, The Netherlands ), J.W. Reed (University of North Carolina, Chapel Hill), and N.-H. Chua (Rockefeller University, New York) for providing hy2 seed. We also thank Drs. R. Finkelstein (University of California, Santa Barbara), Y. Nakamura, and S. Tabata (Kazusa DNA Research Institute, Chiba, Japan) for sharing mapping information regarding the HY2 locus. We are grateful to Drs. M. Aida, T. Shikanai, and M. Takemura (Nara Institute of Science and Technology) for their technical suggestions and to Ms. E. Harada and T. Iwamae for technical assistance. This work was supported in part by Research for the Future Program Grant 00L01605 and by a Grant-in-Aid for Scientific Research on Priority Areas from the Ministry of Education, Sciences, Sports, and Culture of Japan to T.K. and in part by United States Department of Agriculture Competitive Research Grant Initiative AMD-9801768 to J.C.L. N.F. was supported by a fellowship from the Deutsche Forschungsgemeinschaft (FR 1487/1-1).
References
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Altschul, S.F., Madden, T.L., Schaffer, A.A., Zhang, J.H., Zhang, Z., Miller, W., and Lipman, D.J. (1997). Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale, S.I. (1993). Biosynthesis of phycobilins. Chem. Rev. 93, 785–802. [Google Scholar]
- Beale, S.I., and Cornejo, J. (1984). Enzymatic heme oxygenase activity in soluble extracts of the unicellular red alga, Cyanidium caldarium. Arch. Biochem. Biophys. 235, 371–384. [DOI] [PubMed] [Google Scholar]
- Becker, T.W., Foyer, C., and Caboche, M. (1992). Light-regulated expression of the nitrate reductase and nitrate reductase genes in tomato and in the phytochrome-deficient aurea mutant of tomato. Planta 188, 39–47. [DOI] [PubMed] [Google Scholar]
- Bowler, C., and Chua, N.-H. (1994). Emerging themes of plant signal transduction. Plant Cell 6, 1529–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowler, C., Neuhaus, G., Yamagata, H., and Chua, N.H. (1994). Cyclic GMP and calcium mediate phytochrome phototransduction. Cell 77, 73–81. [DOI] [PubMed] [Google Scholar]
- Bradford, M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein using the principle of protein-dye binding. Anal. Biochem. 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Brown, J.W.S. (1996). Arabidopsis intron mutations and pre-mRNA splicing. Plant J. 10, 771–780. [DOI] [PubMed] [Google Scholar]
- Chiu, W., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Chory, J., Peto, C., Ashbaugh, M., Saganich, R., Pratt, L., and Ausubel, F. (1989). Different roles for phytochrome in etiolated and green plants deduced from characterization of Arabidopsis thaliana mutants. Plant Cell 1, 867–880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G.M., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack, T., Mathews, S., and Sharrock, R.A. (1994). The phytochrome apoprotein family in Arabidopsis is encoded by five genes: The sequences and expression of phyD and phyE. Plant Mol. Biol. 25, 413–427. [DOI] [PubMed] [Google Scholar]
- Cornejo, J., and Beale, S.I. (1988). Algal heme oxygenase from Cyanidium caldarium. J. Biol. Chem. 263, 11915–11921. [PubMed] [Google Scholar]
- Cornejo, J., and Beale, S.I. (1997). Phycobilin biosynthetic reactions in extracts of cyanobacteria. Photosynth. Res. 51, 223–230. [Google Scholar]
- Cornejo, J., Willows, R.D., and Beale, S.I. (1998). Phytobilin biosynthesis: Cloning and expression of a gene encoding soluble ferredoxin-dependent heme oxygenase from Synechocystis sp. PCC 6803. Plant J. 15, 99–107. [DOI] [PubMed] [Google Scholar]
- Davis, S.J., Kurepa, J., and Vierstra, R.D. (1999). The Arabidopsis thaliana HY1 locus, required for phytochrome-chromophore biosynthesis, encodes a protein related to heme oxygenases. Proc. Natl. Acad. Sci. USA 96, 6541–6546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deutsch, M., and Long, M. (1999). Intron-exon structures of eukaryotic model organisms. Nucleic Acids Res. 27, 3219–3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, K., Johnstone, C., and Thompson, C. (1991). A simple and rapid method for the preparation of plant genomic DNA for PCR analysis. Nucleic Acids Res. 19, 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich, T.D., and Lagarias, J.C. (1987). Phytochrome chromophore biosynthesis: Both 5-aminolevulinic acid and biliverdin overcome inhibition by gabaculine in etiolated Avena sativa L. seedlings. Plant Physiol. 84, 304–310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elich, T.D., McDonagh, A.F., Palma, L.A., and Lagarias, J.C. (1989). Phytochrome chromophore biosynthesis: Treatment of tetrapyrrole-deficient Avena explants with natural and non-natural bilatrienes leads to formation of spectrally active holoproteins. J. Biol. Chem. 264, 183–189. [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., and von Heijne, G. (1999). ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci. 8, 978–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furuya, M., and Schäfer, E. (1996). Photoperception and signaling of induction reaction by different phytochrome. Trends Plant Sci. 1, 301–307. [Google Scholar]
- Gill, S.C., and von Hippel, P.H. (1989). Calculation of protein extinction coefficients from amino acid sequence. Anal. Biochem. 182, 319–326. [DOI] [PubMed] [Google Scholar]
- Goto, N., Yamamoto, K.T., and Watanabe, M. (1993). Action spectra for inhibition of hypocotyl growth of wild-type plants and of the hy2 long-hypocotyl mutant of Arabidopsis thaliana L. Photochem. Photobiol. 57, 867–871. [Google Scholar]
- Gravel, Y., and von Heijne, G. (1990). A conserved cleavage-site motif in chloroplast transit peptides. FEBS Lett. 261, 455–458. [DOI] [PubMed] [Google Scholar]
- Higgins, D.G., Thompson, J.D., and Gibson, T.J. (1996). Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266, 383–402. [DOI] [PubMed] [Google Scholar]
- Izawa, T., Oikawa, T., Tokutomi, S., Okuno, K., and Shimamoto, K. (2000). Phytochromes confer the photoperiodic control of flowering in rice (a short-day plant). Plant J. 22, 391–399. [DOI] [PubMed] [Google Scholar]
- Kendrick, R.E., and Kronenberg, G.H.M. (1994). Photomorphogenesis in Plants. (Dordrecht, The Netherlands: Martinus Nijhoff Publishers).
- Konieczny, A., and Ausubel, F.M. (1993). A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J. 4, 403–410. [DOI] [PubMed] [Google Scholar]
- Koornneef, M., and Kendrick, R.E. (1994). Photomorphogenic mutants of higher plants. In Photomorphogenesis in Plants, R.E. Kendrick and G.H.M. Kronenberg, eds (Dordrecht, The Netherlands: Kluwer Academic Publishers), pp. 601–628.
- Koornneef, M., Rolff, E., and Spruit, C.J.P. (1980). Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana L. Heynh. Z. Pflanzenphysiol. 100, 147–160. [Google Scholar]
- Lagarias, J.C., and Rapoport, H. (1980). Chromopeptides from phytochrome: The structure and linkage of the Pr form of the phytochrome chromophore. J. Am. Chem. Soc. 102, 4821–4828. [Google Scholar]
- McDonagh, A.F., and Palma, L.A. (1980). Preparation and properties of crystalline biliverdin IXα. Biochem. J. 189, 193–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery, B.L., Yeh, K.-C., Crepeau, M.W., and Lagarias, J.C. (1999). Modification of distinct aspects of photomorphogenesis via targeted expression of mammalian biliverdin reductase in transgenic Arabidopsis plants. Plant Physiol. 121, 629–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muramoto, T., Kohchi, T., Yokota, A., Hwang, I., and Goodman, H.M. (1999). The Arabidopsis photomorphogenic mutant hy1 is deficient in phytochrome chromophore biosynthesis as a result of a mutation in a plastid heme oxygenase. Plant Cell 11, 335–347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff, M.M., Frankhauser, C., and Chory, J. (2000). Light: An indicator of time and place. Genes Dev. 14, 257–271. [PubMed] [Google Scholar]
- Parks, B.M., and Quail, P.H. (1991). Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsis are defective in phytochrome chromophore biosynthesis. Plant Cell 3, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wargner, D. (1995). Phytochromes: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Sharrock, R.A., and Quail, P.H. (1989). Novel phytochrome sequences in Arabidopsis thaliana: Structure, evolution and differential expression of a plant regulatory photoreceptor family. Genes Dev. 3, 1745–1757. [DOI] [PubMed] [Google Scholar]
- Terry, M.J. (1997). Phytochrome chromophore-deficient mutants. Plant Cell Environ. 20, 740–745. [Google Scholar]
- Terry, M.J., and Kendrick, R.E. (1996). The aurea and yellow-green-2 mutants of tomato are deficient in phytochrome chromophore synthesis. J. Biol. Chem. 271, 21681–21686. [DOI] [PubMed] [Google Scholar]
- Terry, M.J., and Lagarias, J.C. (1991). Holophytochrome assembly: Coupled assay for phytochromobilin synthase in organello. J. Biol. Chem. 266, 22215–22221. [PubMed] [Google Scholar]
- Terry, M.J., Wahleithner, J.A., and Lagarias, J.C. (1993). Biosynthesis of the plant photoreceptor phytochrome. Arch. Biochem. Biophys. 306, 1–15. [DOI] [PubMed] [Google Scholar]
- Terry, M.J., McDowell, M.T., and Lagarias, J.C. (1995). (3Z)- and (3E)-phytochromobilin are intermediates in the biosynthesis of the phytochrome chromophore. J. Biol. Chem. 270, 11111–11118. [DOI] [PubMed] [Google Scholar]
- van Tuinen, A., Hanhart, C.J., Kerckhoffs, L.H.J., Nagatani, A., Boylan, M.T., Quail, P.H., Kendrick, R.E., and Koornneef, M. (1996). Analysis of phytochrome-deficient yellow-green-2 and aurea mutants of tomato. Plant J. 9, 173–182. [Google Scholar]
- Weller, J.L., Terry, M.J., Rameau, C., Reid, J.B., and Kendrick, R.E. (1996). The phytochrome-deficient pcd1 mutant of pea is unable to convert heme to biliverdin IXα. Plant Cell 8, 55–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller, J.L., Terry, M.J., Reid, J.B., and Kendrick, R.E. (1997). The phytochrome-deficient pcd2 mutant of pea is unable to convert biliverdin IXα to 3(Z)-phytochromobilin. Plant J. 11, 1177–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitelam, G.C., and Devlin, P.F. (1997). Roles of different phytochromes in Arabidopsis photomorphogenesis. Plant Cell Environ. 20, 752–758. [Google Scholar]
- Wilbanks, S.M., and Glazer, A.N. (1993). Rod structure of a phycoerythrin-II–containing phycobilisome 1. Organization and se-quence of the gene cluster encoding the major phycobiliprotein rod components in the genome of marine Synechococcus sp. WH8020. J. Biol. Chem. 268, 1226–1235. [PubMed] [Google Scholar]
- Wüthrich, K.L., Bovet, L., Hunziker, P.E., Donnison, I.S., and Hörtensteiner, S. (2000). Molecular cloning, functional expression and characterisation of RCC reductase involved in chlorophyll catabolism. Plant J. 21, 189–198. [DOI] [PubMed] [Google Scholar]
- Yeh, K.C., Wu, S.H., Murphy, J.T., and Lagarias, J.C. (1997). A cyanobacterial phytochrome two-component light sensory system. Science 277, 1505–1508. [DOI] [PubMed] [Google Scholar]