Abstract
The mutation rate used in the previous analyses of pig evolution and demographics was cursory and hence invited potential bias in inferring evolutionary history. Herein, we estimated the de novo mutation rate of pigs as 3.6 × 10−9 per base per generation using high-quality whole-genome sequencing data from nine individuals in a three-generation pedigree through stringent filtering and validation. Using this mutation rate, we re-investigated the evolutionary history of pigs. The estimated divergence time of ∼ 10 kiloyears ago (KYA) between European wild and domesticated pigs was consistent with the domestication time of European pigs based on archaeological evidence. However, other divergence events inferred here were not as ancient as previously described. Our estimates suggest that Sus speciation occurred ∼ 1.36 million years ago (MYA); European wild pigs split from Asian wild pigs only ∼ 219 KYA; and south and north Chinese wild pigs split ∼ 25 KYA. Meanwhile, our results showed that the most recent divergence event between Chinese wild and domesticated pigs occurred in the Hetao Plain, northern China, approximately 20 KYA, supporting the possibly independent domestication in northern China along the middle Yellow River. We also found that the maximum effective population size of pigs was ∼ 6 times larger than estimated before. An archaic migration from other Sus species originating ∼ 2 MYA to European pigs was detected during western colonization of pigs, which may affect the accuracy of previous demographic inference. Our de novo mutation rate estimation and its consequences for demographic history inference reasonably provide a new vision regarding the evolutionary history of pigs.
Keywords: Pig, De novo mutation rate, Three-generation pedigree, Evolutionary history, Archaic migration
Introduction
Sus scrofa (wild boars and domestic pigs) belongs to a subfamily of Suidae, a widespread pig species group of Cetartiodactyla that originated in the Oligocene at least 20 million years ago (MYA). Larson et al. [1], Groenen et al. [2], and Frantz et al. [3] made significant contributions to and systemically illustrated the evolutionary history of pigs. Sus scrofa originated on the island south east Asia (ISEA) during the early Pliocene climatic fluctuations about 3–4 MYA [2]. The oldest diverging lineage of pigs found to date is of a wild boar population from the North of Sumatra that split from the Eurasian wild boars around 1.6–2.4 MYA [3]. Over the past one million years, Sus scrofa spread into and colonized almost the entire Eurasian continent [3], [4]. North and south Chinese Sus scrofa populations separated from each other during the Ionian stage, approximately 0.6 MYA [3]. The domestication of pigs is one of the critical events in the history of human agricultural civilization. Pigs were domesticated in at least two locations: Anatolia (Near East) and China. Pig domestication in Anatolia has been well documented, indicating domestication ∼ 10 kiloyears ago (KYA) based on archaeological evidence [1], [5], [6], while pig domestication in China happened at least 8 KYA based on zooarchaeological analyses from middle China [7], [8]. However, studies on the domestication of Chinese wild boars based on genomic analyses remain limited.
An accurate estimate of the mutation rate plays an essential role in understanding many critical questions in evolutionary biology and population genetics, including effective population size (Ne), divergence time, and migration between populations [9]. The two conventional methods used to estimate the mutation rate are 1) phylogenetic approaches, in which the rate of neutral sequence divergence is equal to the rate of mutation [10]; and 2) direct detection of the spontaneous germline mutations in a known pedigree [11], [12], [13], [14], which was used in this study. The latter method, benefiting from the popularity of high-throughput sequencing technologies, has many advantages over the former [11], [12]. A directed per-generation mutation rate derived from a known pedigree has taken an essential part in effectively revising human history [15] and the evolutionary history of dogs [12].
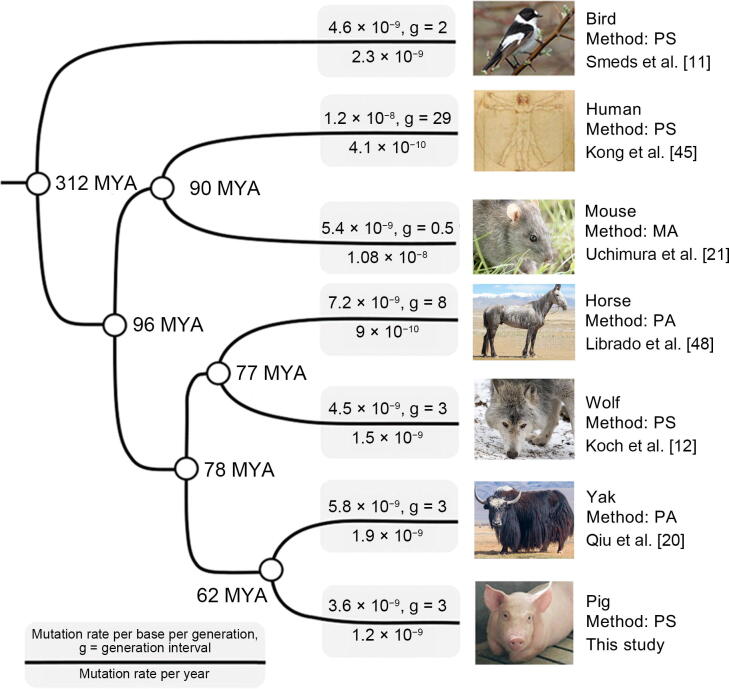
However, at present, there is still no research targeting the accurate estimation of the mutation rate of pigs. The mutation rate used in almost all previous demographics of pigs was set as 2.5 × 10−8 per base per generation, the same as the default value for humans, and five years was used as the generation time of pigs [2], [3], [16], [17], [18]. This resulted in an abnormally large annual mutation rate (5 × 10−9) of pigs, which was twice the average value of mammals [19] and 3.3- and 2.6-fold those of wolves [12] and yaks [20], respectively (Figure 1). The inaccuracy of mutation rate estimation may result in bias in the inference of pig demographic parameters and evolutionary history. This study aimed to estimate the mutation rate of pigs directly using the genomes of nine individuals from a three-generation pig pedigree. Based on this mutation rate, we re-investigated the evolutionary and domestication history of pigs through genomic analyses.
Figure 1.
The mutation rate and generation interval used in the demographic inference in birds and several mammals
Different methods were used to estimate mutation rates: PS, MA, and PA. Photo credit via wikimedia commons: mouse from Zeynel Cebeci (CC BY-SA 4.0); collared flycatcher from Andrej Chudy (CC BY-SA 2.0); horse and yak from Alexandr Frolov (CC BY-SA 4.0); wolf from Christian Mehlführer (CC BY 2.5). PS, pedigree sequencing; MA, sequencing of mutation accumulation lines [21]; PA, phylogenetic approach; MYA, million years ago.
Results
Identification and validation of de novo mutations
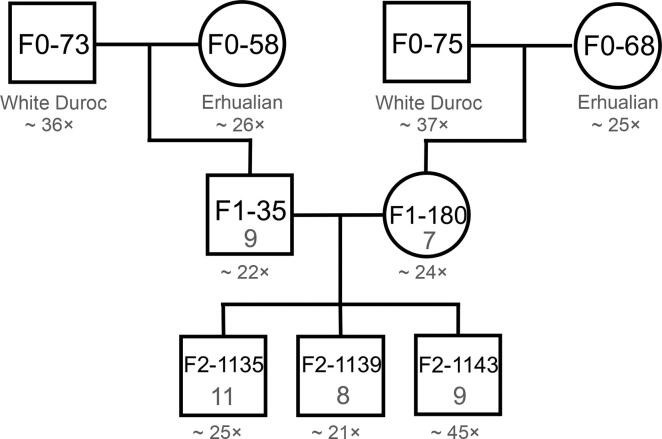
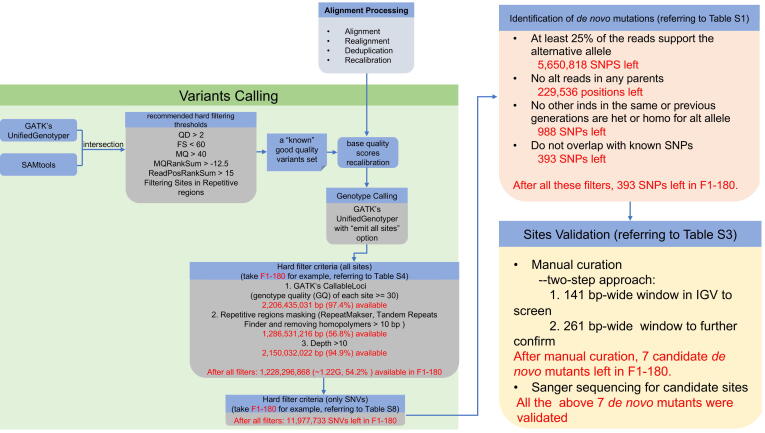
A complete three-generation pedigree consisting of nine pigs (4 parents, 2 children, and 3 grandchildren; Figure 2) was resequenced with depth more than 20×. In the pedigree, two boars in the parent generation (F0) were White Duroc, and two F0 sows were Erhualian. We applied highly stringent filtering criteria as previously described [11] to carefully screen for de novo mutations (DNMs). After all these filters for DNMs, the numbers of single nucleotide polymorphisms (SNPs) left in F1-180, F1-35, F2-1135, F2-1139, and F2-1143 were 393, 360, 180, 213, and 227, respectively (Table S1). Additionally, we took the individual F1-180 as an example to digitize the whole process of DNM screening in more detail (Figure S1). After the stringent hard filter in the variant calling step, we detected 11,977,733 single nucleotide variants (SNVs) in an effective sequence with a length of 1,228,196,868 bp in F1-180. Next, we screened DNMs in these SNVs, and 393 SNVs remained as candidate DNMs. After manual curation, 386 loci were filtered out, leaving only seven candidate DNMs. For these seven DNMs, we designed primers and validated them by Sanger sequencing.
Figure 2.
Whole-genome sequences from nine pigs ofaknown pedigree were analyzed to detect DNMs
The characters and numbers in the squares (male) and circles (female) indicate pig IDs and the numbers of DNMs, respectively. Below each individual is its breed and the average sequencing depth of coverage (see Table S4 for more details of per individual). DNM, de novo mutation.
In total, 44 DNMs (Tables S2 and S3) were identified in the child (F1) and grandchild (F2) generation pigs (7–11 DNMs per individual) that were homozygous for the reference allele in all F0 individuals. None of these DNM sites were known to segregate when searching in the Sus scrofa dbSNPs 150.
In the F1 generation, seven and nine variants in F1-180 and F1-35, respectively, passed manual curation (Table S2). F1-180 transmitted all seven mutations to the F2 offspring, while one out of the nine mutations in F1-35 was not transmitted to any of the three offspring (Table S2). We detected a total of 28 mutations that passed all the bioinformatic filtering criteria in the F2 generation, and these mutations also passed manual curation (Figure 2; Tables S2 and S3).
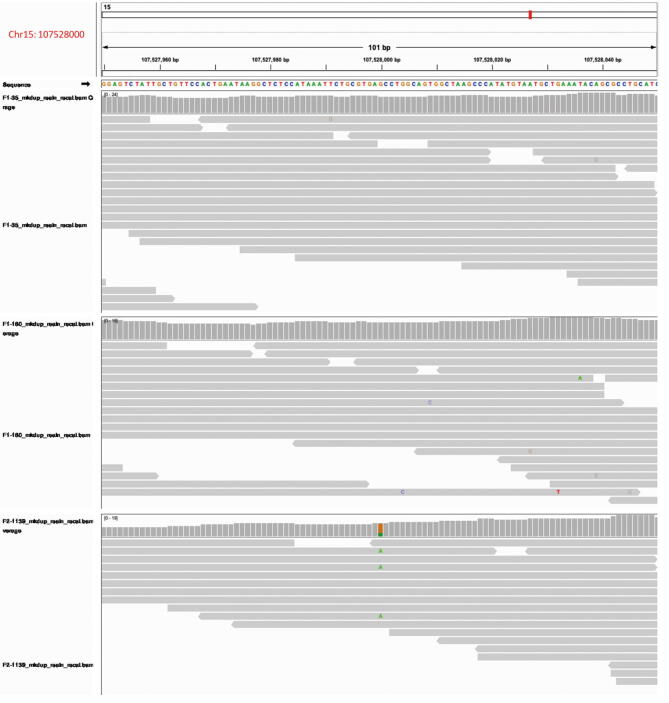
We applied Sanger sequencing to further check the DNMs further. Forty out of the 44 mutants were validated by Sanger sequencing, including the mutation in F1-35 that was not transmitted to any of three offspring (Tables S2 and S3). The remaining four mutations (1, 1, and 2 in F1-35, F2-1135, and F2-1139, respectively) were invalidated and detected as homozygotes for the reference allele by Sanger sequencing. In the mapping results of resequencing data, the ratios of the mapped reads supporting alternative alleles to all the reads at these four sites were 6/17, 10/30, 4/14, and 3/11, respectively (Figures S2–S5). Among these, the mutation at chr7:46553490 was supported by stable inheritance in the F2 generation (Figure S2). We found that the mutation at chr3:9293354 with the ratio of 3/11, the same as the smallest ratio value of the invalidated mutation at chr15:107528000, was detected as real (Table S3). Thus, we did not exclude these four mutations. One possible explanation might be the bias in the sequencing results caused by polymerase chain reaction (PCR) errors before Sanger sequencing [22].
Furthermore, we explored the characteristics of the 44 DNMs. There were 14 mutations in intergenic regions, 25 in introns, one in a 3′-UTR region, one in a splicing region, and three in the coding sequence. Among the three exonic sites (Table S2), one mutation was non-synonymous in the gene encoding piccolo presynaptic cytomatrix protein (PCLO), a part of the presynaptic cytoskeletal matrix. Moreover, there were 13 A:T > G:C and 28 G:C > A:T mutations (Table S2), confirming mutation pressure in the direction of A + T which has been previously observed in both eukaryotes [11], [23] and prokaryotes [24].
The mutation rate in pigs
A total of 44 DNMs were observed in 10 transmissions, with an average of 4.4 mutations per transmission. Among the five offspring, the effective sequences for screening after filtering ranged between 1.17 Gb and 1.28 Gb, with an average size of 1.23 Gb, representing approximately 54.6% of the pig autosomal genome (see details in Materials and methods and Table S4). The parts containing repetitive sequences and not meeting filter criteria for coverage and quality were excluded. Finally, the mutation rate was calculated to be 3.6 × 10−9 [95% confidence interval (CI) = 2.8 × 10−9–4.4 × 10−9] per base pair per generation.
Pigs are typically social animals, breeding in the form of polygamy [25]. The ages of estrus in sows and boars in the wild are different. The age at first pregnancy varies in the wild from about 10 to 20 months [26], while boars begin rut when they are three to five years old. The first rut age of 4–5 years was documented in Russian wild boars [27], and 3–4 years was recorded in Chinese wild boars [28]. Pigs are multiparous animals, and the gestation period lasts about 114–130 days [29]. Comparing to the animals with single birth, such as cattle and yaks, we believe that the generation transmission of pigs is of good continuity. Therefore, we set the generation interval of pigs as 3 years, which is roughly equivalent to the average age of the first pregnancy in sows and the beginning of rut in boars plus the pregnant gestation period of sows. We noticed that evolutionary studies of dogs (wolves) and yaks, which are close in phylogenic distance to pigs, also adopted 3 years as their generation interval [20], [30], suggesting the reasonableness of 3 years as the generation interval of pigs. Based on this generation interval, we obtained an annual mutation rate of 1.2 × 10−9, close to the annual mutation rate of wolves (1.5 × 10−9; the mutation rate was estimated via a known pedigree) [12]. The annual mutation rate of pigs was in the same order of magnitude as those of other mammals (Figure 1) but lower than the mean annual mutation rate (2.2 × 10−9) of mammals [19].
Additionally, we employed the phylogenetic approach to estimate the mutation rate of pigs, following the procedure of mutation rate estimation used in dogs [31]. The annual mutation rate of pigs was estimated to be 1.53 × 10−9 (Table S5). In humans, the annual mutation rate obtained from whole-genome pedigree data is lower than those obtained from the phylogenetic approaches [32], coinciding with the results in dogs and wolves [12], [31]. Here, we obtained a slightly lower annual mutation rate from whole-genome pedigree data than the estimate using the phylogenetic approach, in line with those previous studies in humans and dogs (wolves) and also suggesting the high accuracy of the DNM rate in pigs.
Demographic history of Sus species
We took the DNM rate as a parameter in the pairwise sequentially Markovian coalescent model (PSMC) [33] and the multiple sequential Markovian coalescent model (MSMC2) [34] to reconstruct the population history of pigs. Sumatran wild boars (SMWs), European wild boars (EUWs), north Chinese wild boars (NCWs), and south Chinese wild boars (SCWs) represented Sus scrofa of different geographic distributions in this study. Sus cebifrons, as an outgroup, was involved to date the speciation of Sus scrofa from the Sus genus. Each breed contained two individuals (Table S6).
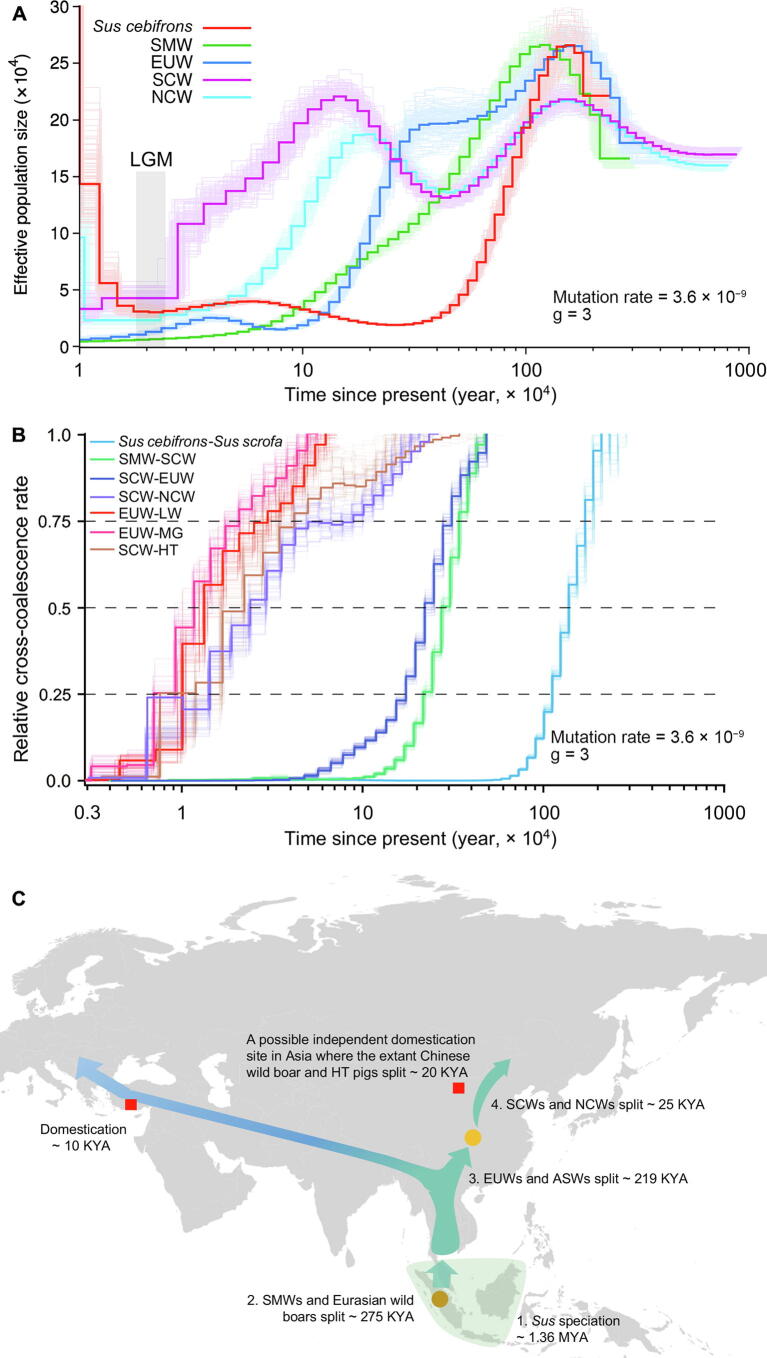
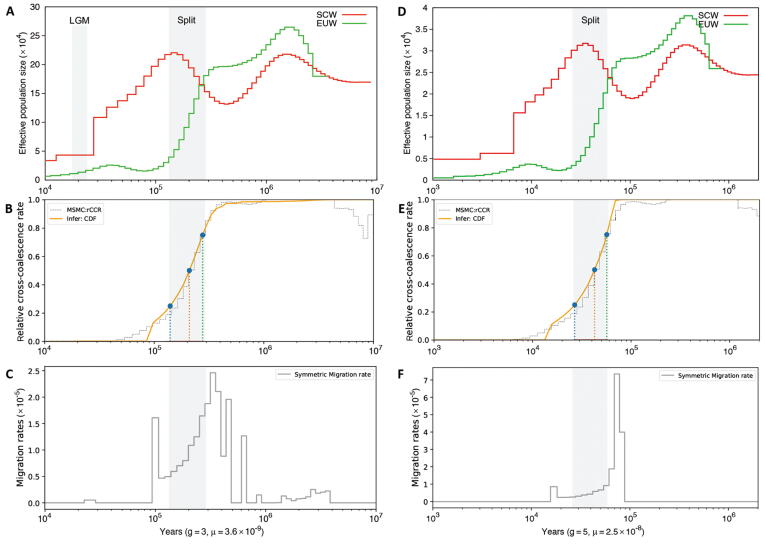
PSMC exhibited messy-looking demographic trajectories: demographic trajectories of different pigs began to separate ∼ 2 MYA, except for those of NCWs and SCWs, which began to separate from each other ∼ 200 KYA (Figure 3A). Such messy curves possibly suggested a short common history among different breeds of pigs and indicated that they diverged from each other million years ago. However, we could find common points, such as all pigs peaked in Ne during 1–2 MYA. Sus cebifrons, SMWs, and EUWs had a similar Ne of ∼ 2.7 × 105 during the peak period, while NCWs and SCWs had a lower Ne in the same period. The maximum estimation of Ne here was ∼ 6 times larger than that estimated previously [2], [3]. Thereafter, Sus cebifrons and SMWs experienced a rapid population decline. EUWs and SCWs/NCWs experienced a lighter decline and then stayed stable or increased 400 KYA. Notably, the trajectory of EUW was similar in trend with that of SCWs and NCWs ∼ 300 KYA but showed relatively higher Ne. All pigs suffered a bottleneck during the Last Glacial Maximum (LGM; 20 KYA; Figure 3A).
Figure 3.
Population history of Sus species
A. The changes in effective population size of Sus cebifrons and the wild pigs over past years inferred by PSMC. The LGM is highlighted in gray. B. Split time for population pairs estimated by MSMC2. A relative cross-coalescence rate of 0.5 was artificially defined as the divergence time. The cold color lines indicate the split time of wild pig breeds in different regions; the warm color lines could somehow reflect the domestication time, even though the extant wild pigs may not be the direct ancestors of the domesticated breeds. See Table S7 for more details concerning breeds. C. A map depicting the hypothetical spread of wild pigs across Eurasia and the domestication events of pigs which happened in the Middle East and China. The shaded area covered in Southeast Asia indicates that Sus originated here. The circles represent the “node” groups, connecting two different groups. The brown circle depicts SMWs. The yellow circle depicts SCWs. The red squares refer to the domestication events. PSMC, pairwise sequentially Markovian coalescent model; MSMC2, multiple sequential Markovian coalescent model; LGM, Last Glacial Maximum; SMW, Sumatran wild boar; EUW, European wild boar; NCW, north Chinese wild boar; SCW, south Chinese wild boar; LW, Large White; MG, Mangalica; HT, Hetao; ASW, Asian wild boar; KYA, kiloyears ago.
MSMC2 allowed us to study the genetic separation between two populations as a function of time based on relative cross-coalescence rates (RCCRs). The RCCR curve reached a value of 0.5 at ∼ 1.36 MYA (95% CI: 1,335,388–1,461,116 years ago) for the comparison of Sus cebifrons and Sus scrofa (SCW; Figure 3A; Table S7), indicating that Sus speciation occurred during this period on ISEA. According to the RCCR cutoff of 0.5 defined as the divergence time, we could also judge that the divergence time between SMWs and SCWs was ∼ 275 KYA (95% CI: 262,988–283,540 years ago), between EUWs and SCWs was ∼ 219 KYA (95% CI: 212,002–231,589 years ago), between NCWs and SCWs split ∼ 25 KYA (95% CI: 21,908–29,605 years ago), and between EUWs and European domesticated pigs [EUDs; Large White (LW) pigs] was ∼ 1.3 KYA (95% CI: 11,320–15,985 years ago) (Figure 3B; Table S7).
We noticed that the divergence time indicated by MSMC2 was generally more recent than that implied by trajectories from PSMC. To further explore this issue, we took the comparison of EUWs and SCWs as an example. Although the results of PSMC showed that the demographic curves of EUWs and SCWs separated ∼ 2 MYA (Figure S6A), RCCR in MSMC2 did not reach 0.5 until ∼ 219 KYA, indicating that Eurasian pigs did not really split ∼ 2 MYA, but rather split ∼ 219 KYA (Figure S6B). We applied MSMC-IM software [35], fitting a continuous model to coalescence rates to estimate gene flow within and across pairs of populations, to confirm the time of the divergence event. This analysis still showed a peak at ∼ 200 KYA rather than 2 MYA, indicating that Eurasian pigs’ split time was ∼ 200 KYA (Figure S6C).
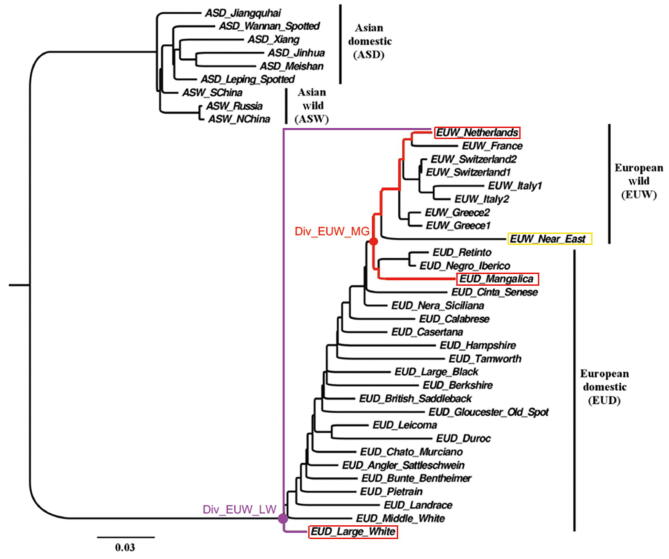
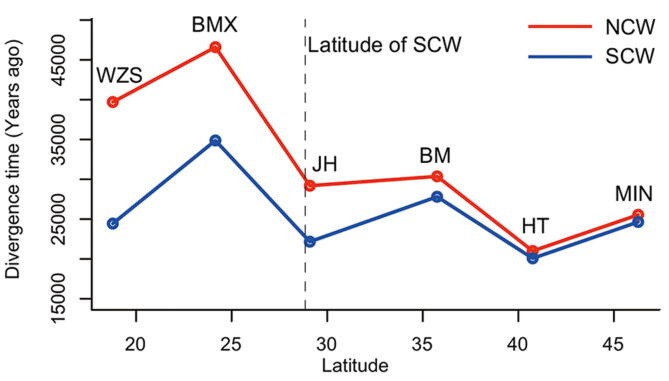
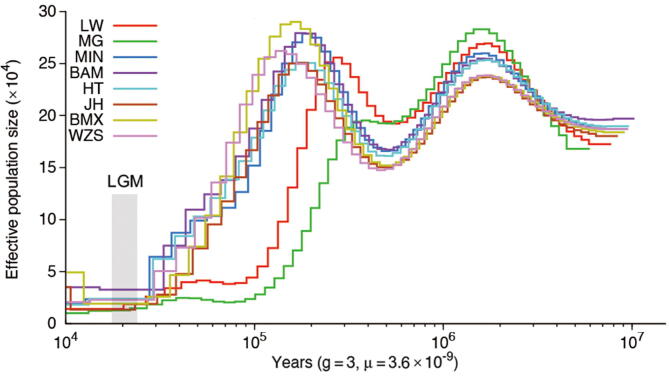
We estimated the divergence time between wild boars and domesticated pigs in Europe and China, respectively. We used EUWs (Netherlands wild boars) paired with two different domesticated pig breeds [LW and Mangalica (MG) pigs] that are located at the most proximal and distal branches relative to the cluster of EUWs, respectively (Figure S7), and found that they diverged ∼ 13 KYA (95% CI: 11,320–15,985 years ago) and ∼ 11 KYA (95% CI: 10,203–13,671 years ago), respectively, coinciding with the generally accepted domestication time of ∼ 10 KYA [2], [3], [5]. However, the direct ancestors of EUDs are not the existing EUWs but the extinct wild boars from the Middle East [1]. The divergence time was expected to be older than the domestication period of 10 KYA, in line with the estimated date of LW and MG pigs splitting from EUWs. We also tested the divergence time between the different geographically distributed Chinese domesticated pig breeds and Chinese wild boars, including those from North China and South China (Figure S8; Table S6). We found that the domesticated pig breed on Hetao plain, located at the intersection of the middle Yellow River and Inner Mongolia, most recently split from Chinese wild pigs ∼ 20 KYA (Figure 3B, Figure S8; Table S7). Interestingly, several studies have addressed the possibility of domestication along the middle Yellow River ∼ 8 KYA based on the ancient mitochondrial DNA [36] and archaeological evidence [37]. Our results further confirmed that north China along the middle Yellow River could be a domestication site in Asia. However, here we cannot decide domestication time according to the divergence time for the same reason as in European pigs, i.e., the ancestor of domesticated pigs might not be the extant wild boars. We also detected a severe bottleneck during the LGM in all domesticated pigs (Figure S9).
To summarize, we can revisit the evolutionary history of the Sus genus (Figure 3C) as follows: Sus speciation occurred on ISEA ∼ 1.36 MYA (95% CI: 1,335,388–1,461,116 years ago), leading to the emergence of the oldest Sus scrofa; then pigs arrived in Eurasia from ISEA and colonized Southeast Asia ∼ 275 KYA (95% CI: 262,988–283,540 years ago); the spread of wild boars into Europe was ∼ 219 KYA (95% CI: 212,002–231,589 years ago); the pigs in South China did not migrate to North China until ∼ 25 KYA (95% CI: 21,908–29,605 years ago). Additionally, North China along the middle Yellow River could be an independent domestication site in Asia where the wild boars and domesticated pigs split ∼ 20 KYA (95% CI: 17,142–25,258 years ago). The divergence time between EUWs and EUDs was first estimated to be ∼ 10 KYA using genomic data.
Contradictions in previous evolutionary histories of pigs
Frantz et al. [3] applied an approximate likelihood method as implemented in MCMCtree [38] to estimate divergence time between Sus species, in which they set the splitting time between Phacochoerus africanusa and Sus as a root age at 10.5 MYA based on phylogenetic research on mitochondrial DNA of extant sub-Saharan African suids [39]. This ancient root age was used to adjust the prior of the mutation rate, which was set to obey a gamma distribution as G (1125) in Bayesian clock dating. Their divergence time estimates suggested that populations of Sus scrofa from Asia migrated west approximately 1.2 MYA. Groenen et al. [2] and Frantz et al. [3] found that the population sizes of European and Asian lineages started to diverge ∼ 1 MYA (Figure S6D) in PSMC [33], which was considered as a vital line of evidence for the distinct Asian and European pig lineages splitting ∼ 1.2 MYA. Additionally, the aforementioned two studies illustrated an increase in the European population after pigs arrived from Asia.
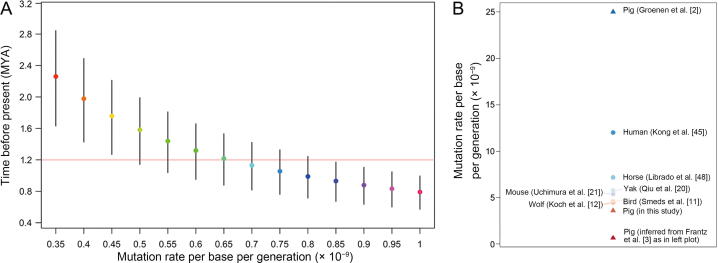
In this study, we repeated PSMC analyses on Eurasian wild pigs and further applied MSMC2 software and MSMC-IM to make forward and backward arguments for the aforementioned studies. First, we used the divergence time between EUWs and SCWs (1.2 MYA) estimated by Frantz et al. [3] to infer the mutation rate for pigs. This resulted in a mutation rate estimation of ∼ 6.5 × 10−10 per base per generation (Figure 4A), which is an order of magnitude less than the mutation rate of other mammals (Figure 1). We also tried to use this mutation rate to estimate divergence time between EUWs and EUDs (i.e., LW pigs). This resulted in a more ancient divergence time (∼ 70 KYA) than the domestication time (∼ 10 KYA) indicated by archaeological evidence. We noticed that the mutation rate applied in PMSC by Groenen et al. [2] was 2.5 × 10−8 per base per generation. This means that two significantly different mutation rates (Figure 4B) reflect a similar divergence history of Eurasian pigs. All of these results suggested that there were some biases in the previously estimated pig history.
Figure 4.
The abnormal mutation rate in the past population studies of pigs
A. The pig mutation rate was inferred using MSMC2 when fixing the divergence time between SCWs and EUWs at 1.2 MYA. SCW–EUW divergence time was inferred by MSMC2 using various mutation rates with 3 years fixed as the generation interval. Dots, lower bars, and upper bars represent the time at which cross-coalescence rate dropped below 50%, 25%, and 75%, respectively. The red horizontal line represents the SCW–EUW divergence time from Frantz and colleagues [3]. B. Scatter plot showing the mutation rates of pigs (triangles) estimated in our study and used in the past research, with the mutation rates of other species (circles) shown in Figure 1.
Then, we ran the PSMC, MSMC2, and MSMC-IM with the mutation rate set as 2.5 × 10−8 per base per generation. In order to compare with the previous results, we only used EUWs and SCWs. Surprisingly, we identified four possible contradictions between the previous estimations and the results of MSMC2 and MSMC-IM. First, the results of both MSMC2 and MSMC-IM indicated a totally different divergence time between EUWs and SCWs as previously discussed [2], i.e., the divergence event appeared at the first crossover of lines corresponding to the time of ∼ 40 KYA (Figure S6D–F) rather than the corresponding intersection point at ∼ 1 MYA. Second, if the divergence event actually happened at the first crossover, the date was only 40 KYA, an illogical split time compared to the declared 1 MYA [2], [3]. Third, the divergence time of EUWs and EUDs was estimated to only ∼ 2 KYA, an unreasonable recent date compared to the domestication time around 10 KYA via MSMC2 and MSMC-IM. Last but not least, compared to other mammals, the Ne of pigs (the maximum Ne was ∼ 4 × 104; see Figure S6D) was much lower than those of dogs (the maximum Ne of ∼ 1.5 × 105) [40] and yaks (the maximum Ne of ∼ 1.6 × 105) [20]. The ratio of non-synonymous to synonymous heterozygosity (πN/πS), as a measure of the mutation load, was applied to the Ne comparisons among distantly related species and was found to be negatively correlated to population size [41]. The πN/πS ratio of 0.62–0.80 in pigs was lower than that of 1.04–1.19 in dogs [42], meaning that pigs had a larger Ne compared to dogs. This is also in contrast to the small Ne estimation of pigs using the previously accepted, commonly used mutation rate of 2.5 × 10−8 per base per generation. We thought that the reason why these contradictions occurred could be the use of abnormally large mutation rate estimates in the previous studies.
Validation of archaic admixture in European Sus scrofa by simulations
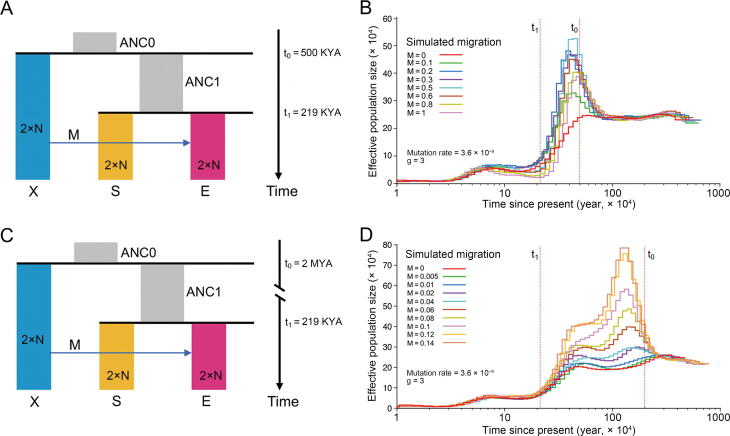
When we applied the mutation rate of 3.6 × 10−9 per base per generation to infer pig demographic history, the MSMC-IM results displayed two pulses (Figure S6C): one pulse corresponding to the place where RCCR was equal to 0.5, and the other pulse appearing between 1 MYA and 4 MYA, indicating a migration into SCWs or EUWs from an archaic population. Correspondingly, recent evidence showed that pygmy hogs and a now-extinct Sus species interbred with Sus scrofa, suggesting that inter-species admixture accompanied the rapid spread of wild boars across mainland Eurasia and North Africa [43]. Thus, wild boars had greater chances of encountering and temporally co-existing with local species during the expansion into Europe, enabling possible inter-species hybridization. The phenomenon that the Ne of EUWs at the peak of the PMSC curve was similar to those of pigs on ISEA (Figure 3A) further suggested a possible migration from an archaic population or another Sus species to EUWs, instead of to SCWs, during the colonization across the Eurasian mainland. We suspected that this introgression from the archaic population possibly contributed to the difference in the curve of Ne between EUWs and SCWs before the point of their separation. To test this hypothesis, we performed a series of simulations in which two populations S and E separated 219 KYA, with population E receiving a varying level of gene flow from an archaic population X (Figure 5). We assumed that the third population (i.e., the ancestral population, ANC1) diverged from the archaic population X 500 KYA (Figure 5A) and 2 MYA (Figure 5C), respectively, to check how the different ancestral donors could affect the trajectories of Ne of the receptor. Using simulations under this split-with-archaic-admixture model (Figure 5), we found that an archaic admixture did lead to the uplift of Ne of a period when varying the extent of migration (Figure 5B and D). Specifically, the receptor with archaic admixture from the ancestral donor that separated from the archaic population 2 MYA (Figure 5D), instead of 500 KYA (Figure 5B) as in our simulations, exhibited a similar trajectory with that of EUWs. Thus, it is most likely that another Sus species, diverging ∼ 2 MYA, introgressed into EUWs. The uplift of the Ne curve attributed to the admixture from an archaic population explained the variation trend of the curves of EUWs and SCWs being nearly same, but Ne of EUWs before the divergence at the first crossover of the PSMC curves was relatively larger (Figure S6A and D). This uplift of the regional Ne curve of EUWs gave us the illusion of deep divergence between EUWs and Asian wild boars (ASWs), and this possibly caused the bias in the previous inference of pig evolutionary history.
Figure 5.
A split-with-archaic-admixturemodel used for simulation testing of whether migration from anarchaicpopulationaffectsPSMC curves
A. The model where the ancestral population ANC1 split from an archaic population X 500 KYA and populations S and E split from each other 219 KYA. B. PSMC was performed using simulated data of population E under the model shown in (A). C. The model where the ancestral population ANC1 split from an archaic population X 2 MYA and populations S and E split from each other 219 KYA. D. PSMC was performed using simulated data of population E under the model shown in (C). M indicates migration strength, which was computed in the form of 4N × m, where N is the initial population size used to scale parameters in the simulations, and m is migration rate. We set M to different values, and N was set to 10,000 (the default value of the ms program).
Discussion
The accuracy of DNMs detected in a three-generation pedigree by high-quality resequencing
Previous studies have shown that the bioinformatic pipeline we refer to can guarantee the high accuracy of DNMs [11], [13], [14], [44]. All of the candidate mutations passed a stringent two-step manual curation by the integrated genomics viewer (IGV) [45]. Several procedures were applied to validate mutations during the DNM identification, including following the stable inheritance of new mutations to subsequent generations and Sanger sequencing. Pfeifer [14] validated the manually curated candidate mutations in the F1 generation using their stable Mendelian inheritance to the next generation (F2) to exclude the false-positive mutations, even though there was only one individual in F2 of that pedigree. Our study had three grandchildren with high depths of coverage (21×–45×) to exclude the false-positive mutations. Additionally, we used another individual, F1-43 (with coverage of 40×) only sharing the same father (F0-73) with F1-35, to check the independent genotyping status of the other two individuals (F1-35 and F1-180) in F1, following the criteria that no other individuals in the same generation are heterozygous or homozygous for the alternative allele. We could judge that the false-positive rate of two individuals in the F1 generation was very low based on the following five reasons: 1) the use of a stringent bioinformatic pipeline; 2) the final manual inspection by a two-step approach to avoid interference of the insertions and deletions around candidate mutations; 3) validation by independent genotyping in the same or the previous generation(s); 4) the fact that sites of mutation events were monomorphic in large population samples (dbSNPs 150); and 5) the fact that stable inheritance was confirmed for mutations appearing in the F2 generation. Notably, there was one mutation in F1 that was not transmitted to any of the three offspring. We detected a total of 28 mutations that passed the manual curation in three F2 generation individuals. The proportions of mutations identified in the F1 (16/44 = 36.4%) and F2 (63.6%) generations were approximately in agreement with the proportions of meiosis scored in the F0 (4/10 = 40%) and F1 (6/10 = 60%) generations (Table S2). The slightly higher ratio (63.6%) in the F2 generation than expected could be attributed to all male individuals, while there was one female containing seven mutations, the lowest number of mutations among all the individuals, in the F1 generation. The mutation rate was reported to have a pronounced male bias in humans and chimpanzees [46], [47]; this possibly explained the slightly lower ratio (36.4%) in the F1 generation. Wang and Zhu [48] also adopted a similar pedigree design to solve the false-positive problem. In this study, we identified 44 DNMs following the pipeline, of which 40 mutations were validated by Sanger sequencing. Even though validation of four mutations failed using Sanger sequencing, we determined to retain those four mutations for the subsequent analysis after balancing the possible unknown errors in Sanger sequencing (e.g., PCR errors) and the low false-positive rate following the pipeline.
Keightley et al. [13] addressed the rate of false negatives by adding synthetic mutations to read data from a Drosophila melanogaster pedigree containing 14 individuals when using a very similar bioinformatic pipeline for mutation identification as in our study. They detected 99.4% of all callable synthetic mutations following the bioinformatic pipeline, suggesting that the rate of false negatives was negligible. Smeds et al. [11] considered the filtering of both heterozygous sites in the parental generation and candidate mutations in the F1 or F2 generations that corresponded to known segregating alleles in the known SNP datasets (e.g., Sus scrofa dbSNP dataset) as the reason for the low false-negative rate in the aforementioned filter criteria. Similarly, Pfeifer [14] also found a low false-negative rate after the highly stringent computational filters, particularly for those mutations stably inherited to subsequent generations. Strictly following the mutation-detection procedure of Smeds et al. [11], we find no reason to expect that the false-negative rate should be significantly different in our study. Given the relatively high depth of coverage, we also consider the rate of false negatives as very low.
In general, high-quality resequencing, a stringent bioinformatic pipeline, IGV manual curation, and validation via Sanger sequencing ensured the accuracy of DNMs we detected.
The evolutionary history of Sus scrofa revisited through the DNM rate
We considered the results of PSMC and two other methods (MSMC2 and MSMC-IM) and found some contradictions in previous studies. First, two extremely different mutation rates (Figure 4B) were used in the different methods, but the results of divergence time of EUWs from ASWs were similar. Next, the mutation rates in previous studies led to an incredibly advanced or delayed split of EUDs from EUWs. Finally, the Ne of pigs was an order of magnitude less than those of the other domesticated animals such as dogs [40], yaks [20], and horses [49]. Additionally, MSMC-IM exhibited evidence of archaic gene flow. Correspondingly, it was reported that at least two events of inter-species admixture occurred when wild boars rapidly spread into Europe, including the migration from pygmy hogs and a wave of gene flow contributed by an unknown ancient ghost population [43]. Based on our simulations, we found that the complex gene flow from another Sus species into EUWs might have lifted the Ne curve and have interfered with the previous judgment of divergence events based on PSMC. In our case, the date estimation suggested that EUWs and ASWs actually shared a long history, but the complex migration from an archaic population into EUWs misled us to accept the deep divergence between EUWs and ASWs. The phenomenon that introgression influences the Ne seems to be common. Hawks [50] found that the Ne inferred for a particular interval of time in the past is strongly affected by the history of introgression or gene flow, such as the gene flow between non-Africans and Neandertals or Denisovans. Inside Africa, introgression was also detected, and was suspected from other archaic human groups, with approximately the same inferred divergence date as Neandertals [51]. Therefore, all modern humans have a clear “wave” of larger inferred Ne with a “crest”, including Africans [50]. A simulation study in dogs also concluded that imported gene flow lifted the estimated evolutionary trajectory of the target population in PSMC [40]. In another recent study, a hump in the historical Ne estimated by PSMC was attributed to admixture events occurring in donkeys [52]. Hawks [50] found that the longer the introgression donor diverged from its ancestor, the greater amplitude of the “wave” in Ne occurred for the target population. This is consistent with our simulation results. When M = 0.1, a larger Ne rose in the target population resulting from introgression from the donor population separating from the archaic population 2 MYA (∼ 6 × 105, Figure 5D) than 500 KYA (∼ 3 × 105, Figure 5B). The Sus species introgression into EUWs can be traced to at least 1 MYA based on this study (1–4 MYA suggested by MSMC-IM, Figure S6C) and a previous study [43]. The receptor with archaic admixture from the donor that separated from the archaic population 2 MYA in our simulations exhibited a similar trajectory with that of EUWs. This suggested that the origin of the introgression donor Sus population can be traced to 2 MYA, and introgression from this population into EUWs further caused the demographic curves of EUWs and SCWs to separate ∼ 2 MYA (Figure S6A). This independent introgression from an archaic population into EUWs resulted in a unique phenomenon of PSMC in pigs, i.e., the Ne trajectories of SCWs and EUWs separated significantly before their populations separated. This reminds us to make demographic inferences based on multiple lines of evidence to avoid the interference of introgression or gene flow.
The DNM rate directly estimated from the pedigree can be used to reliably characterize the demographic history of Sus scrofa [12], [15]. Herein, we used the DNM rate to reconstruct the population history of pigs. Sus speciation occurred on ISEA ∼ 1.36 MYA (95% CI: 1,335,388–1,461,116 years ago), leading to the emergence of the oldest Sus scrofa; then, pigs arrived in Eurasia from ISEA and colonized Southeast Asia ∼ 275 KYA (95% CI: 262,988–283,540 years ago). Next, the spread of wild boars into Europe occurred ∼ 219 KYA (95% CI: 212,002–231,589 years ago). The colonization in North China, where the climate was cold, happened ∼ 25 KYA (95% CI: 21,908–29,605 years ago). These estimated population histories were much more recent than the generally accepted history [3], but they were consistent with some lines of evidence, including documentary records of domestication. The new date estimation could yield new thinking regarding the population genetics of pigs.
The divergence time between EUWs and EUDs was estimated at around 10 KYA (95% CI: 10,203–13,671 years ago for EUW–MG pair; 95% CI: 11,320–15,985 years ago for EUW–LW pair) using the DNM rate, a result that perfectly coincided with the generally accepted domestication time of ∼ 10 KYA based on documentary records [2], [3], [5]. We first validated the domestication of pigs ∼ 10 KYA by genomic data. The ASWs and EUWs split ∼ 219 KYA (95% CI: 212,002–231,589 years ago) (Figure 3B and C; Table S7), far more recent than ∼ 1.2 MYA [2], [3] and suggesting that EUWs shared a considerably longer history than previously thought. SCWs (Nanchang, Jiangxi Province) and NCWs diverged ∼ 25 KYA (95% CI: 21,908–29,605 years ago), less than 600 KYA [3]. Based on this more recent split time, we proposed that human hunting and domestication activities might have accelerated the divergence of SCWs and NCWs and forced the pigs to migrate into a cold environment that was a severe challenge to their survival.
The new estimated mutation rate also revealed a maximum Ne of 2.7 × 105 in pigs, ∼ 6 times larger than that estimated previously [2], [3] (Figure 3A, Figure S9) and similar to the Ne values of other mammals such as dogs [40], yak [20], and horses [49]. Our results also revealed a bottleneck in the EUWs after colonizing Europe (Figure 3A) rather than a population expansion [2]. Similar bottlenecks observed in non-African human populations [33] and western Eurasian dogs [53] have been interpreted as signs of migration to a new living environment. The penultimate glacial period (PGP, 135–194 KYA) followed the western diffusion of pigs. The cold climate exacerbated the bottleneck of the western migrating pigs. Instead, during this period, pigs in Southeast Asia had not yet started to spread northward, and the cold concentrated the pigs more in warm areas, leading to a temporary increase of Ne. We have previously found that an ∼ 50 Mb segment on the X chromosome of European pigs from another genus of pigs may have led to their cold adaptability [54]. Combined with the results of this study, we speculated that after pigs dispersed out of Southeast Asia, due to the new environment and the following PGP, the western-spreading pigs experienced a severe population bottleneck. During this period, the pig hybridized with other pig genera and received the “gift” of adaptation to the cold to a certain extent, and then the beneficial introgressed fragment further fixed in the population during their colonization in the European continent. All Sus populations investigated here, even Sus cebifrons, suffered bottlenecks during LGM (20 KYA) (Figure 3A, Figure S9). The bottleneck of Sus cebifrons was not found during LGM in the previous study [3]. The bottlenecks observed here were more severe than those reported previously [2]. Notably, the wild sow is the only ungulate that must build a nest to provide the litter with a warm microenvironment [55]. This is due to an essential gene, UCP1, which participates in brown adipose tissue-mediated adaptive non-shivering thermogenesis having been lost ∼ 20 MYA [56]. The cold climate during the glacial period was fatal to pigs. As expected, we found the lowest Ne of pigs during LGM.
Conclusion
Altogether, we found some irrationalities and contradictions in the previously estimated evolutionary history of pigs. To address these incompatibilities, we estimated the DNM rate of pigs via a whole-genome three-generation pedigree containing nine individuals. The pig is another non-primate mammalian species for which the mutation rate has been directly obtained after wolves (dogs) and mice. The estimated mutation rate of pigs using a pedigree ultimately did not show an abnormally large or small value, rather being at the same order of magnitude as the mutation rate of other common mammals such as wolves (dogs) and yaks (Figure 1). This directed mutation rate provides a new vision regarding the origin and evolutionary history of pigs. In addition, complex archaic admixture could have led to misjudgment of the population history, and thus we advise taking the results of demographic history with caution in the research on human beings and animals, and demographic inferences should be based on multiple lines of evidence. Our results advance the understanding of the population history of pigs.
Materials and methods
Samples and sequencing
We used whole genomes to construct a known pedigree of nine pigs from a three-generation pedigree (Figure 2; Table S4). Two boars in the parent generation (F0) were White Duroc, and two sows in the F0 generation were Erhualian. The two boars F0-73 and F0-75 were sequenced [57] using the HiSeq 2000 platform. The other seven individuals were sequenced in the same way. Briefly, genomic DNA was extracted from ear tissues using a standard phenol–chloroform method and then sheared into fragments of 200–800 bp according to the Illumina DNA sample preparation protocol. These treated fragments were end-repaired, A-tailed, ligated to paired-end adaptors, and PCR amplified with 500 bp (or 350 bp) inserts for library construction. Sequencing was performed to generate 100 bp (or 150 bp) paired-end reads on a HiSeq 2000 (or 2500) platform (Illumina) according to the manufacturer’s standard protocols.
The reads were aligned to the Sus scrofa reference genome (build 11.1) using BWA [58] with default options. The mapped reads were subsequently processed by sorting, indel realignment, duplicate marking, and low-quality filtering using Picard (http://picard.sourceforge.net) and GATK v3.5.0 [59], [60]. To generate an initial trial set of variants for recalibrating base quality scores, we used GATK UnifiedGenotyper and SAMtools [61] to call variant sites separately and then used the intersection of variant sites from these two methods. Next, we kept SNP sites if they passed the recommended hard filtering thresholds (QD > 2, FS < 60, MQ > 40, MQRankSum > −12.5, and ReadPosRankSum > 15) as described previously [12] and filtered sites in repetitive regions from this set using RepeatMasker v4.0.6 (http://repeatmasker.org/). Finally, we treat the remaining SNPs as a “known” good-quality variant set to recalibrate base quality scores using GATK v3.5.0.
After recalibrating base quality scores, the genotypes of all sites were called with GATK UnifiedGenotyper with the “emit all sites” option. We did not perform variant quality recalibration (VQSR) suggested by GATK’s best practices since the fact that DNMs should only occur in a single individual and are therefore more likely to be filtered out as low-quality variants. Instead, we applied a set of highly stringent hard filter criteria to weed out potential false positives (Table S4). Following Smeds et al. [11], repetitive regions were masked with a combination of RepeatMasker v4.0.6 (http://repeatmasker.org/), Tandem Repeats Finder v4.07 [62], and a custom Shell script to remove any homopolymers longer than 10 bp that were not already masked (this criterion excluded ∼ 43.4% of the autosomal genome). Then sites passing GATK’s CallableLoci level were kept, and genotype quality (GQ) of each site had to be at least 30 (96.2%–97.5% of the autosome genome met this criterion). A hard coverage threshold of 10 was used to minimize false variant calls due to insufficient read data (90.6%–99.1% of the autosome genome met this criterion) following Keightley and colleagues [13]. These procedures ensured sufficient data to obtain accurate DNMs and filter false positives. A total of 1.17–1.28 Gb sequence per individual (51.8%–56.7% of the autosome genome) was kept for further quality control of DNMs. Finally, we excluded nonvariant sites and indels, only keeping SNVs in this study (Table S8).
The procedure of DNM identification
We applied extremely stringent bioinformatic filtering in an attempt to have high confidence on the DNMs following previous studies [46], [11], [12], [13], [14]. Before filtering, we detected a total of 24.3 million SNPs that segregated in the pedigree, concordant with expectations based on previously reported nucleotide levels [63]. For each individual in the F1 and F2 generations, heterozygous positions were extracted from the background and had to meet the following criteria to be considered as potential DNMs: 1) both parents were required to be homozygous for the reference allele with no reads supporting the alternative allele (discard the possibility of potential parental mosaicism); 2) no other individuals in the same or the previous generation(s) are heterozygous or homozygous for the alternative allele; 3) at least 25% of the reads support the alternative allele; and 4) the allele does not overlap with the known SNPs from Build 150 of the Sus scrofa dbSNP dataset [64] from the NCBI database (since DNMs are rare events, candidates also detected as variation segregating in unrelated individuals are likely false positives).
The filtering criteria in the F2 generation were stricter than that in the F1 generation; this helped reduce background noise. The DNMs in the F2 generation must simultaneously be homozygous for the reference allele with no reads supporting the alternative allele in the F0 and F1 generations. For the mutation candidates in the F1 generation, only F0 individuals were required to be homozygous for the reference allele with no potential parental mosaicism.
Manual curation and annotation of DNMs
Candidate mutations were manually curated using IGV [45], similar to the procedure described by Keightley et al. [13], to visually detect false positives caused by misaligned reads, sequencing errors, insertions, and deletions. The types of false positives detected were excluded as described in the examples shown in Figures S1–S4 in the study by Keightley and colleagues [13]. In the curation process, we applied a two-step approach: first, we used a 141 bp-wide window in IGV to screen for the reliable DNMs; second, we enlarged the window to 261 bp to further confirm whether the mutation was robustly true based on the status of linked loci on the same read of the candidate mutation, and further to determine whether the mutation originated from the father or the mother (Table S2). Sites were annotated using ANNOVAR v2020Apr28 [65] with the annotation of the Sus scrofa genome (build 11.1).
PCR for the detection of DNMs
In this study, we designed a pair of specific primers for each of 44 DNMs identified in a family composed of nine individuals (Table S9). The PCR comprised 2.5 µl of 10× buffer (Mg2+ plus) (TaKaRa, Japan), 1.5 µl of 25 mM MgCl2, 2.0 µl of 2.5 mM dNTP, 1.0 µl of each primer (10 µM), 0.4 µl of taq DNA polymerase (5 U/µl), and 50 ng of the genomic DNA, and the final volume was set to 25 µl with ddH2O. The mixture was then run in a thermocycler under the following conditions: 94 °C for 5 min; 26 cycles of 94 °C for 30 s, 68 °C (−0.5 °C/cycle) for 30 s, 72 °C for 45 s; 14 cycles of 94 °C for 30 s, 55 °C for 30 s, 72 °C for 45 s; and 72 °C for 10 min. The PCR products were sequenced on the 3130XL Genetic Analyzer ThermoFisher Scientific, Waltham, MA.
Evolutionary history inference methods and models
We used PSMC [33] and MSMC2 [34] to infer population sizes and split time for Sus populations. The genomic data of Sus populations, including Sus cebifrons and Sus scrofa (Table S6), were downloaded from the NCBI Sequence Read Archive (SRA: SRA096093 [54], SRP039012 [66], and SRP115801 [67]) and European Nucleotide Archive (ENA: PRJEB9326 [18], PRJEB9922 [68], and ERP001813 [2]). Among the data of Sus cebifrons in the public database, there were two good-quality individuals, allowing us to estimate the split time. Sus scrofa consists of wild pigs [SMWs, SCWs (Nanchang), NCWs, EUWs (Netherlands)], and domesticated pigs [Europe: LW and MG pigs; China: HT, Bamei (BM), Min (MIN), Jinhua (JH), Bamaxiang (BMX), and Wuzhishan (WZS) pigs]. The reads from all the aforementioned individuals were aligned to the Sus scrofa reference genome (build 11.1) using BWA [58]. The subsequent steps, including sorting, indel realignment, and deduplication, were processed via GATK v3.5.0 [59], [60].
For Ne inference, PSMC requires diploid consensus sequences. The consensus was generated from the ‘mpileup’ command of SAMtools software package [61] using the “-C 50, -O, -D 2*reads_depth, -d 1/3*reads_depth” option, which was set as recommended by PSMC’s manual. Then we used the tool ‘fq2psmcfa’ from the PSMC package to create the input file. We used Tmax = 20, n = 64 (4 + 50*1 + 4 + 6) following the study by Groenen and colleagues [2]. To validate the variance of Ne, we performed 100 bootstrap replications for each subspecies’ representative samples. Here we adopted the mutation rate and generation time updated in this study.
Split time estimated by MSMC2 requires two-phased genomes for each population. SNP calling and low-quality filtering were conducted as previously described [54]. We phased the samples using SHAPEIT [69]. In addition, there were two masks applied: one was derived by the tool ‘bamCaller.py’ from the MSMC-tools package; the other included the sites that were masked using Heng Li’s SNPable mask (http://lh3lh3.users.sourceforge.net/snpable.shtml), in which mappability was taken into account, and non-unique sequence positions were not used for calculations. We performed 100 bootstrap replications for each breed to determine the variance in Ne estimates. Then, MSMC2 was run on four haplotypes (two from each of the two populations) to calculate RCCR with the “--skipAmbiguous” argument to skip unphased segments of the genome. The time segment parameters were set to 64 (4 + 50*1 + 4 + 6) with 20 iterations. The RCCR variable ranges between 0 and 1 (occasionally, the calculation is unavoidably greater than 1) [34]. A value close to 1 indicates that two populations were one population at the point of time, and when RCCR = 0.5, the corresponding time is estimated as the timing of the separation of the two populations. The confidence intervals around RCCR estimates were obtained using block-bootstrapping. We used a script called “multihetsep_bootstrap.py” in the MSMC-tools repository to generate artificial “bootstrapped” datasets from the original input data by chopping up the input data into blocks (5 Mb long by default) and randomly sampling with replacement to create artificial 3 Gb long genomes out of these blocks. The confidence intervals were calculated on a total of 100 of these artificially created datasets (100 bootstrap replicates). The results were scaled to real-time by applying a mutation rate of 3.6 × 10−9 per base per generation and a generation time of 3 years derived in this study. MSMC-IM, fitting a continuous isolation-migration model to coalescence rates to obtain a time-dependent estimate of gene flow within and across pairs of populations based on the results of MSMC2, was also used to decide the time of a split event, presented by a signal of strong gene flow [35].
A split-with-archaic-admixture model, in which two populations (S and E) of varying sizes were assumed to have separated 219 KYA, with population E receiving a varying level of gene flow from an archaic population X, was built to check how the archaic admixture would affect the shape of PSMC. The ms software [70] was adopted to perform a series of simulations under the split-with-archaic-admixture model. We ran the simulations under two different models where the third population ANC1 diverged from the archaic population X 500 KYA (Figure 5A) and 2 MYA (Figure 5C), respectively. The initial population size was set as 10,000 to scale the parameters in the simulation.
Ethical statement
All procedures involving pigs were based on the care and use guidelines of experimental animals established by the Ministry of Science and Technology of China. The Ethics Committee of Jiangxi Agricultural University approved this study (Approval No. JXAULL-2003003).
Data availability
The raw sequence reads of nine individuals from the three-generation pedigree have been deposited in the Genome Sequence Archive [71] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: CRA005031), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa.
CRediT author statement
Mingpeng Zhang: Investigation, Methodology, Writing - original draft. Qiang Yang: Methodology, Validation. Huashui Ai: Conceptualization, Supervision, Writing - review & editing. Lusheng Huang: Conceptualization, Supervision, Resources, Writing - review & editing. All authors have read and approved the final manuscript.
Competing interests
The authors have declared no competing interests.
Acknowledgments
We thank Mingshan Wang from UC Santa Cruz, for helpful comments and suggestions. Mingpeng Zhang thanks Rasmus Nielsen for useful comments and for hosting him from January 2019 to September 2020 at the Center for Theoretical Evolutionary Genomics, UC Berkeley. We also thank Jiaqi Chen for help on the IGV application. This work was financially supported by the Innovative Research Team of the Ministry of Education of China (Grant No. IRT1136), the National Natural Science Foundation of China (Grant No. 31672383), and the National Swine Industry and Technology System of China (Grant No. nycytx-009).
Handled by Jingfa Xiao
Footnotes
Peer review under responsibility of Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation and Genetics Society of China.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.gpb.2022.02.001.
Contributor Information
Huashui Ai, Email: aihsh@hotmail.com.
Lusheng Huang, Email: Lushenghuang@hotmail.com.
Supplementary material
The following are the Supplementary data to this article:
Supplementary Figure S1.
The pipeline diagram to exhibit the steps of DNMs detection and DNMs validation in detail The individual F1-180 was used as an example in the pipeline, and the specific statistical data in each step was marked. DNM, de novo mutant; SNV, single nucleotide variants; SNP, single nucleotide polymorphism; IGV, integrative genomics viewer.
Supplementary Figure S2.
Screenshot from the IGV showing genotypes at chr7:46553490
Supplementary Figure S3.
Screenshot from the IGV showing genotypes at chr2:1101857
Supplementary Figure S4.
Screenshot from the IGV showing genotypes at chr9:28285405
Supplementary Figure S5.
Screenshot from the IGV showing genotypes at chr15:107528000
Supplementary Figure S6.
Comparison of population histories inferred using different mutation rates by PSMC, MSMC2, and MSMC-IM The split time is identified and highlighted in grey at the first crossover of population size’s curves instead of the second crossover (backward in time). A and D. Demographic history was inferred with two different mutation rates: 3.6 × 10-9 are the mutation rate estimated in this study; 2.5 × 10-8 are the mutation rate of human, which was widely used in pig demography inference. The LGM is highlighted in grey in (A). B and E. RCCR are inferred by MSMC2 with the two different mutation rates, respectively. The split time was indicated by the RCCR (orange solid line). The vertical dotted lines represented the date corresponding to RCCR of 25%, 50%, and 75%, respectively. C and F. Migration rates are inferred by MSMC-IM with the two different mutation rates, respectively. The split time was reflected via signals of strong gene flow across SCW and EUW. SCW, South Chinese wild pigs; EUW, European wild pigs. LGM, Last Glacial Maximum; PSMC, pairwise sequentially Markovian coalescent model; MSMC2, multiple sequential Markovian coalescent model 2; MSMC-IM, multiple sequential Markovian coalescent-isolation and migration model; RCCR, relative cross-coalescent rates.
Supplementary Figure S7.
The pigs sampled for divergence time estimation between European wild and domesticated pigs The phylogenetic tree was constructed by TreeMix using data in Frantz et al. 2015 [68], where the pigs were genotyped by the porcine 60SNP array set. Netherlands wild pigs were selected to be paired with European domesticated pigs, including large white and Mangalica pigs, to estimate the divergence time. Large white and Mangalica pigs located at the most proximal and distal branches relative to the cluster of European wild pigs, respectively.
Supplementary Figure S8.
The relationship between the geographical positions of different Chinese domesticated pig breeds and their split time compared with NCW and SCW, respectively The split time here is derived from MSMC2. The dotted line represents the latitude of SCW. We didn’t know the exact geographic position of NCW (which was used to detect the split time between north and south Chinese pigs [3], and the only sequenced data available in north China in the public database). The split time of all the Chinese domesticated pigs (including ones distributed in North and South) with SCW are closer than that of NCW. Based on this figure, we thought the place where HT is distributed, beside Yellow River, is a possible place of domestication, echoing the views of Larson et al. 2010 [35]. Thus, we set the split time (20,075 years ago) between HT and SCW as the domesticated time of the Chinese pigs. We also noticed that the time of pigs spreading to north China (25,071 years ago) is close to the domestication time. SCW, South Chinese wild pigs; NCW, North Chinese wild pigs; WZS, Wuzhishan pigs; BMX, Bama Xiang pigs; JH, Jinhua pigs; BM, Bamei pigs; HT, Hetao pigs; MIN, Min pigs (see Table S6 for more detail of per breed).
Supplementary Figure S9.
Dynamic changes of effective population size over past year inferred by PSMC The LGM is highlighted in grey. LW, Largewhite pigs; MG, Mangalica pigs; PSMC, pairwise sequentially Markovian coalescent (see Table S6 for more detail of per breed).
The details of individuals and the amount of SNVs available after DNMs filtering for each individual
Details about the identified mutation events
Genotyping of the de novo mutations identified in 9 individuals by Sanger sequencing and next-generation sequencing
The details of individuals and the amount of sequence available for screening after filtering for each individual
Mutation rate along the pig lineage
Sequencing Statistics of samples used for PSMC and MSMC2
MSMC2 results with cross-coalescence rate at 25%, 50%, and 75%.
The details of individuals and the amount of the single nucleotide variants available after filtering for each individual
PCR primers for detecting the de novo mutations
References
- 1.Larson G., Dobney K., Albarella U., Fang M., Matisoo-Smith E., Robins J., et al. Worldwide phylogeography of wild boar reveals multiple centers of pig domestication. Science. 2005;307:1618–1621. doi: 10.1126/science.1106927. [DOI] [PubMed] [Google Scholar]
- 2.Groenen M.A.M., Archibald A.L., Uenishi H., Tuggle C.K., Takeuchi Y., Rothschild M.F., et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature. 2012;491:393–398. doi: 10.1038/nature11622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Frantz L.A.F., Schraiber J.G., Madsen O., Megens H.J., Bosse M., Paudel Y., et al. Genome sequencing reveals fine scale diversification and reticulation history during speciation in Sus. Genome Biol. 2013;14:R107. doi: 10.1186/gb-2013-14-9-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Groenen M.A. A decade of pig genome sequencing: a window on pig domestication and evolution. Genet Sel Evol. 2016;48:23. doi: 10.1186/s12711-016-0204-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frantz L.A.F., Haile J., Lin A.T., Scheu A., Geörg C., Benecke N., et al. Ancient pigs reveal a near-complete genomic turnover following their introduction to Europe. Proc Natl Acad Sci U S A. 2019;116:17231–17238. doi: 10.1073/pnas.1901169116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Giuffra E, Kijas JM, Amarger V, Carlborg O, Jeon JT, Andersson L. The origin of the domestic pig: independent domestication and subsequent introgression. Genetics 2000;154:1785–91. [DOI] [PMC free article] [PubMed]
- 7.Larson G., Cucchi T., Fujita M., Matisoo-Smith E., Robins J., Anderson A., et al. Phylogeny and ancient DNA of Sus provides insights into neolithic expansion in Island Southeast Asia and Oceania. Proc Natl Acad Sci U S A. 2007;104:4834–4839. doi: 10.1073/pnas.0607753104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jing Y., Flad R.K. Pig domestication in ancient China. Antiquity. 2002;76:724–732. [Google Scholar]
- 9.Lynch M. Evolution of the mutation rate. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 11.Smeds L., Qvarnström A., Ellegren H. Direct estimate of the rate of germline mutation in a bird. Genome Res. 2016;26:1211–1218. doi: 10.1101/gr.204669.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koch E.M., Schweizer R.M., Schweizer T.M., Stahler D.R., Smith D.W., Wayne R.K., et al. De novo mutation rate estimation in wolves of known pedigree. Mol Biol Evol. 2019;36:2536–2547. doi: 10.1093/molbev/msz159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keightley P.D., Ness R.W., Halligan D.L., Haddrill P.R. Estimation of the spontaneous mutation rate per nucleotide site in a Drosophila melanogaster full-sib family. Genetics. 2014;196:313–320. doi: 10.1534/genetics.113.158758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pfeifer S.P. Direct estimate of the spontaneous germ line mutation rate in African green monkeys. Evolution. 2017;71:2858–2870. doi: 10.1111/evo.13383. [DOI] [PubMed] [Google Scholar]
- 15.Scally A., Durbin R. Revising the human mutation rate: implications for understanding human evolution. Nat Rev Genet. 2012;13:745–753. doi: 10.1038/nrg3295. [DOI] [PubMed] [Google Scholar]
- 16.Bosse M., Megens H.J., Madsen O., Frantz L.A.F., Paudel Y., Crooijmans R.P.M.A., et al. Untangling the hybrid nature of modern pig genomes: a mosaic derived from biogeographically distinct and highly divergent Sus scrofa populations. Mol Ecol. 2014;23:4089–4102. doi: 10.1111/mec.12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li M., Tian S., Jin L., Zhou G., Li Y., Zhang Y., et al. Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat Genet. 2013;45:1431–1438. doi: 10.1038/ng.2811. [DOI] [PubMed] [Google Scholar]
- 18.Nuijten R.J.M., Bosse M., Crooijmans R.P.M.A., Madsen O., Schaftenaar W., Ryder O.A., et al. The use of genomics in conservation management of the endangered visayan warty pig (Sus cebifrons) Int J Genomics. 2016;2016:1–9. doi: 10.1155/2016/5613862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kumar S., Subramanian S. Mutation rates in mammalian genomes. Proc Natl Acad Sci U S A. 2002;99:803–808. doi: 10.1073/pnas.022629899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qiu Q., Wang L., Wang K., Yang Y., Ma T., Wang Z., et al. Yak whole-genome resequencing reveals domestication signatures and prehistoric population expansions. Nat Commun. 2015;6:1–7. doi: 10.1038/ncomms10283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uchimura A., Higuchi M., Minakuchi Y., Ohno M., Toyoda A., Fujiyama A., et al. Germline mutation rates and the long-term phenotypic effects of mutation accumulation in wild-type laboratory mice and mutator mice. Genome Res. 2015;25:1125–1134. doi: 10.1101/gr.186148.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikegawa S., Mabuchi A., Ogawa M., Ikeda T. Allele-specific PCR amplification due to sequence identity between a PCR primer and an amplicon: is direct sequencing so reliable? Hum Genet. 2002;110:606–608. doi: 10.1007/s00439-002-0735-1. [DOI] [PubMed] [Google Scholar]
- 23.Lynch M. Rate, molecular spectrum, and consequences of human mutation. Proc Natl Acad Sci U S A. 2010;107:961–968. doi: 10.1073/pnas.0912629107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hershberg R., Petrov D.A., Nachman M.W. Evidence that mutation is universally biased towards AT in bacteria. PLoS Genet. 2010;6:e1001115. doi: 10.1371/journal.pgen.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Canu A., Scandura M., Merli E., Chirichella R., Bottero E., Chianucci F., et al. Reproductive phenology and conception synchrony in a natural wild boar population. Hystrix. 2015;26:1–8. [Google Scholar]
- 26.Singer F.J. Wild pig populations in the national parks. Environ Manage. 1981;5:263–270. [Google Scholar]
- 27.Heptner V.G., Nasimovich A.A., Bannikov A.Ge, Hoffmann R.S. Smithsonian Institution Libraries and National Science Foundation; Washington DC: 1988. Mammals of the Soviet Union. [Google Scholar]
- 28.Wang X. A breeding method of Wannan wild boar in China. China Patent Application. 2012 CN102630633A. [Google Scholar]
- 29.Comer C.E., Mayer J.J. In: Wild pigs: biology, damage, control techniques, and management. Mayer J.J., Brisbin I.L., editors. Savannah River National Laboratory; Aiken: 2009. Wild pig reproductive biology; pp. 51–75. [Google Scholar]
- 30.Freedman A.H., Gronau I., Schweizer R.M., Ortega-Del Vecchyo D., Han E., Silva P.M., et al. Genome sequencing highlights the dynamic early history of dogs. PLoS Genet. 2014;10:e1004016. doi: 10.1371/journal.pgen.1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang G.D., Zhai W., Yang H.C., Wang L.u., Zhong L., Liu Y.H., et al. Out of southern East Asia: the natural history of domestic dogs across the world. Cell Res. 2016;26:21–33. doi: 10.1038/cr.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nachman MW, Crowell SL. Estimate of the mutation rate per nucleotide in humans. Genetics 2000;156:297–304. [DOI] [PMC free article] [PubMed]
- 33.Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schiffels S., Durbin R. Inferring human population size and separation history from multiple genome sequences. Nat Genet. 2014;46:919–925. doi: 10.1038/ng.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang K., Mathieson I., O’Connell J., Schiffels S., Schierup M.H. Tracking human population structure through time from whole genome sequences. PLoS Genet. 2020;16:e1008552. doi: 10.1371/journal.pgen.1008552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiang H., Gao J., Cai D., Luo Y., Yu B., Liu L., et al. Origin and dispersal of early domestic pigs in northern China. Sci Rep. 2017;7:1–9. doi: 10.1038/s41598-017-06056-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Larson G., Liu R., Zhao X., Yuan J., Fuller D., Barton L., et al. Patterns of East Asian pig domestication, migration, and turnover revealed by modern and ancient DNA. Proc Natl Acad Sci U S A. 2010;107:7686–7691. doi: 10.1073/pnas.0912264107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 39.Gongora J., Cuddahee R.E., do Nascimento F.F., Palgrave C.J., Lowden S., Ho S.Y.W., et al. Rethinking the evolution of extant sub-Saharan African suids (Suidae, Artiodactyla) Zool Scr. 2011;40:327–335. [Google Scholar]
- 40.Wang MS, Wang S, Li Y, Jhala Y, Thakur M, Otecko NO, et al. Ancient hybridization with an unknown population facilitated high-altitude adaptation of canids. Mol Biol Evol 2020;37:2616–29. [DOI] [PubMed]
- 41.Galtier N., Rousselle M. How much does Ne vary among species? Genetics. 2020;216:559–572. doi: 10.1534/genetics.120.303622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takashi M, Carl-Johan R, Miguel C, Erik A, Leif A, Webster MT. Elevated proportions of deleterious genetic variation in domestic animals and plants. Genome Biol Evol 2018;10:276–90. [DOI] [PMC free article] [PubMed]
- 43.Liu L., Bosse M., Megens H.J., Frantz L.A.F., Lee Y.L., Irving-Pease E.K., et al. Genomic analysis on pygmy hog reveals extensive interbreeding during wild boar expansion. Nat Commun. 2019;10:1992. doi: 10.1038/s41467-019-10017-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Keightley P.D., Pinharanda A., Ness R.W., Simpson F., Dasmahapatra K.K., Mallet J., et al. Estimation of the spontaneous mutation rate in Heliconius melpomene. Mol Biol Evol. 2015;32:239–243. doi: 10.1093/molbev/msu302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., et al. Integrative genomics viewer. Nat Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kong A., Frigge M.L., Masson G., Besenbacher S., Sulem P., Magnusson G., et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature. 2012;488:471–475. doi: 10.1038/nature11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Francioli L.C., Polak P.P., Koren A., Menelaou A., Chun S., Renkens I., et al. Genome-wide patterns and properties of de novo mutations in humans. Nat Genet. 2015;47:822–826. doi: 10.1038/ng.3292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang H., Zhu X. De novo mutations discovered in 8 Mexican American families through whole genome sequencing. BMC Proc. 2014;8:S24. doi: 10.1186/1753-6561-8-S1-S24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Librado P., Der Sarkissian C., Ermini L., Schubert M., Jónsson H., Albrechtsen A., et al. Tracking the origins of Yakutian horses and the genetic basis for their fast adaptation to subarctic environments. Proc Natl Acad Sci U S A. 2015;112:E6889–E6897. doi: 10.1073/pnas.1513696112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hawks J. Introgression makes waves in inferred histories of effective population size. Hum Biol. 2017;89:67–80. doi: 10.13110/humanbiology.89.1.04. [DOI] [PubMed] [Google Scholar]
- 51.Lachance J., Vernot B., Elbers C., Ferwerda B., Froment A., Bodo J.M., et al. Evolutionary history and adaptation from high-coverage whole-genome sequences of diverse African hunter-gatherers. Cell. 2012;150:457–469. doi: 10.1016/j.cell.2012.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C., Li H., Guo Y.u., Huang J., Sun Y., Min J., et al. Donkey genomes provide new insights into domestication and selection for coat color. Nat Commun. 2020;11:1–15. doi: 10.1038/s41467-020-19813-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Frantz L.A.F., Mullin V.E., Pionnier-Capitan M., Lebrasseur O., Ollivier M., Perri A., et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science. 2016;352:1228–1231. doi: 10.1126/science.aaf3161. [DOI] [PubMed] [Google Scholar]
- 54.Ai H., Fang X., Yang B., Huang Z., Chen H., Mao L., et al. Adaptation and possible ancient interspecies introgression in pigs identified by whole-genome sequencing. Nat Genet. 2015;47:217–225. doi: 10.1038/ng.3199. [DOI] [PubMed] [Google Scholar]
- 55.Algers B., Jensen P. Thermal microclimate in winter farrowing nests of free-ranging domestic pigs. Livest Prod Sci. 1990;25:177–181. [Google Scholar]
- 56.Berg F., Gustafson U., Andersson L., Barsh G.S. The uncoupling protein 1 gene (UCP1) is disrupted in the pig lineage: a genetic explanation for poor thermoregulation in piglets. PLoS Genet. 2006;2:e129. doi: 10.1371/journal.pgen.0020129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ai H, Zhang M, Yang B, Goldberg A, Li W, Ma J, et al. Human-mediated admixture and selection shape the diversity on the modern swine (Sus scrofa) Y chromosomes. Mol Biol Evol 2021;38:5051–65. [DOI] [PMC free article] [PubMed]
- 58.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.DePristo M.A., Banks E., Poplin R., Garimella K.V., Maguire J.R., Hartl C., et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., et al. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Benson G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27:573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Choi J.W., Chung W.H., Lee K.T., Cho E.S., Lee S.W., Choi B.H., et al. Whole-genome resequencing analyses of five pig breeds, including Korean wild and native, and three European origin breeds. DNA Res. 2015;22:259–267. doi: 10.1093/dnares/dsv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sherry S.T., Ward M.H., Kholodov M., Baker J., Phan L., Smigielski E.M., et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wang K., Li M., Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Molnár J., Nagy T., Stéger V., Tóth G., Marincs F., Barta E. Genome sequencing and analysis of Mangalica, a fatty local pig of Hungary. BMC Genomics. 2014;15:1–12. doi: 10.1186/1471-2164-15-761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu Y., Li W., Yang B., Zhang Z., Ai H., Ren J., et al. Signatures of selection and interspecies introgression in the genome of Chinese domestic pigs. Genome Biol Evol. 2017;9:2592–2603. doi: 10.1093/gbe/evx186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frantz L.A.F., Schraiber J.G., Madsen O., Megens H.J., Cagan A., Bosse M., et al. Evidence of long-term gene flow and selection during domestication from analyses of Eurasian wild and domestic pig genomes. Nat Genet. 2015;47:1141–1148. doi: 10.1038/ng.3394. [DOI] [PubMed] [Google Scholar]
- 69.Delaneau O., Zagury J.F., Marchini J. Improved whole-chromosome phasing for disease and population genetic studies. Nat Methods. 2013;10:5–6. doi: 10.1038/nmeth.2307. [DOI] [PubMed] [Google Scholar]
- 70.Hudson R.R. Generating samples under a Wright-Fisher neutral model of genetic variation. Bioinformatics. 2002;18:337–338. doi: 10.1093/bioinformatics/18.2.337. [DOI] [PubMed] [Google Scholar]
- 71.Chen T., Chen X., Zhang S., Zhu J., Tang B., Wang A., et al. The Genome Sequence Archive Family: toward explosive data growth and diverse data types. Genomics Proteomics Bioinformatics. 2021;19:578–583. doi: 10.1016/j.gpb.2021.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The details of individuals and the amount of SNVs available after DNMs filtering for each individual
Details about the identified mutation events
Genotyping of the de novo mutations identified in 9 individuals by Sanger sequencing and next-generation sequencing
The details of individuals and the amount of sequence available for screening after filtering for each individual
Mutation rate along the pig lineage
Sequencing Statistics of samples used for PSMC and MSMC2
MSMC2 results with cross-coalescence rate at 25%, 50%, and 75%.
The details of individuals and the amount of the single nucleotide variants available after filtering for each individual
PCR primers for detecting the de novo mutations
Data Availability Statement
The raw sequence reads of nine individuals from the three-generation pedigree have been deposited in the Genome Sequence Archive [71] at the National Genomics Data Center, Beijing Institute of Genomics, Chinese Academy of Sciences / China National Center for Bioinformation (GSA: CRA005031), and are publicly accessible at https://ngdc.cncb.ac.cn/gsa.