Abstract
The amino- and carboxy-terminal domains of mitochondrially encoded cytochrome c oxidase subunit II (Cox2p) are translocated out of the matrix to the intermembrane space. We have carried out a genetic screen to identify components required to export the biosynthetic enzyme Arg8p, tethered to the Cox2p C terminus by a translational gene fusion inserted into mtDNA. We obtained multiple alleles of COX18, PNT1, and MSS2, as well as mutations in CBP1 and PET309. Focusing on Cox18p, we found that its activity is required to export the C-tail of Cox2p bearing a short C-terminal epitope tag. This is not a consequence of reduced membrane potential due to loss of cytochrome oxidase activity because Cox2p C-tail export was not blocked in mitochondria lacking Cox4p. Cox18p is not required to export the Cox2p N-tail, indicating that these two domains of Cox2p are translocated by genetically distinct mechanisms. Cox18p is a mitochondrial integral inner membrane protein. The inner membrane proteins Mss2p and Pnt1p both coimmunoprecipitate with Cox18p, suggesting that they work together in translocation of Cox2p domains, an inference supported by functional interactions among the three genes.
INTRODUCTION
The production and assembly of subunits of the cytochrome c oxidase complex requires a large number of genes in Saccharomyces cerevisiae and appears to be a highly regulated process (Tzagoloff and Dieckmann, 1990; Pel et al., 1992; Dieckmann and Staples, 1994; Fox, 1996). The three mitochondrially encoded subunits are synthesized in the matrix and inserted into the inner membrane where they become tightly associated in the core of the enzyme, surrounded by imported subunits encoded by nuclear genes (Tsukihara et al., 1996).
Cytochrome oxidase subunit II (Cox2p) has substantial hydrophilic domains comprising both its N- and C-terminal portions, which are exported through the inner membrane to the intermembrane space (IMS; Poyton et al., 1992; Iwata et al., 1995; Tsukihara et al., 1996). Association of Cox2p with the membrane appears to begin with localized translation activated by the membrane-bound mRNA-specific translational activator Pet111p, which recognizes the 5′-untranslated leader of the COX2 mRNA (Mulero and Fox, 1993; Sanchirico et al., 1998). Export of the Cox2p N-terminal hydrophilic domain depends on Oxa1p, a translocase component embedded in the inner membrane (Bonnefoy et al., 1994a; Altamura et al., 1996; He and Fox, 1997; Hell et al., 1997; Kermorgant et al., 1997; Hell et al., 1998). Another inner membrane protein, Mba1p (Rep and Grivell, 1996), is also involved in N-tail export (Preuss et al., 2001). After export, a 15-amino acid leader peptide is removed from the N terminus by a protease comprised of Imp1p, Imp2p, and Som1p (Schneider et al., 1991; Nunnari et al., 1993; Jan et al., 2000), in a reaction facilitated by the interaction of preCox2p with the inner membrane protein Cox20p (Hell et al., 2000).
Export of the Cox2p C-tail depends on the inner membrane potential, whereas export of the N-tail does not (He and Fox, 1997). Thus, these two processes appear to be mechanistically distinct despite the fact that they both depend on Oxa1p (He and Fox, 1997; Hell et al., 1997). To study further the export of the Cox2p C-tail, we have used a genetic screen to identify S. cerevisiae mutants that are defective in exporting a Cox2p-Arg8p fusion protein across the mitochondrial inner membrane (He and Fox, 1999). Arg8p is normally a nuclearly encoded protein that is imported into the mitochondrial matrix where it participates in arginine biosynthesis (Jauniaux et al., 1978). We have synthesized a gene, ARG8m, that specifies Arg8p in the mitochondrial genetic code and complements a nuclear arg8 deletion when expressed within mitochondria (Steele et al., 1996). When ARG8m was translationally fused to COX2, it directed the synthesis of a fusion protein whose Arg8p moiety was largely exported to the IMS (He and Fox, 1997). Synthesis of the fusion protein supports respiratory growth (Pet+) but, in certain genetic backgrounds, fails to support Arg+ growth due to export of Arg8p from the matrix (He and Fox, 1999). Thus, mutants that fail to export the Arg8p moiety of the fusion protein can be selected from the Arg−,Pet+ parent strain by isolating Arg+,Pet− colonies (He and Fox, 1999). To date, this screen has identified two nuclear genes required for export of the C-tail of a Cox2p-Arg8p fusion protein: PNT1 (He and Fox, 1999) and MSS2 (Broadley et al., 2001), both of which encode mitochondrial inner membrane proteins.
Here we report the isolation and analysis of 35 new mutants that have defects in export of the Cox2p-Arg8p fusion protein. Most of the mutations affected the genes COX18, MSS2, and PNT1. Cox18p was previously shown to be required for the accumulation of Cox2p and was proposed to play a role in assembly of cytochrome oxidase (Souza et al., 2000). We have studied further the phenotypes and genetic interactions of cox18 mutations, as well as the topology of Cox18p and its interactions with other proteins involved in Cox2p C-tail export.
MATERIALS AND METHODS
Yeast Strains and Genetic Methods
S. cerevisiae strains utilized in this study are listed in Table 1. Genetic manipulations and standard growth media (YPD, containing 2% glucose; YPEG, containing 3% ethanol and 3% glycerol; YPRaf containing 2% raffinose) were as previously described (Rose et al., 1988; Fox et al., 1991; Guthrie and Fink, 1991). Synthetic complete medium (SC) was purchased from Bio 101 (Vista, CA). Null cox18 mutations were generated using a cox18::URA3 disruption construct (Souza et al., 2000) or polymerase chain reaction (PCR) products specifying G418 resistance (Wach, 1996). The cox4Δ strain was generated using a PCR product encoding a cox4Δ::LEU2 construct (Dowhan et al., 1985).
Table 1.
S. cerevisiae strains used in this study
| Strain name | Nuclear (mitochondrial) genotypea | Source or reference |
|---|---|---|
| DAU1ρ0b | MATα ura3Δ ade2 (ρ0) | Marykwas and Fox, 1989 |
| DFS188b | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8∷hisG (ρ+) | Steele et al., 1996 |
| NB75c | MATα ura3-52 leu2Δ ade2-101 arg8Δ∷URA3 kar1-1 (ρ+ COX2(1-91)∷ARG8m) | Bonnefoy and Fox, 2000 |
| NSG132ρ0b | MATα ura3Δ leu2 ade2-101 lys2 arg8∷hisG pet111-14 (ρ+) | N. S. Green-Willms |
| RCB1 | MATa ura3-52 leu2-3,12 his3-11,15 arg8Δ∷hisG cox18Δ∷KanR (ρ+) | R. C. Burgess |
| SB19b | MATα ura3ΔNS leu2-3,112 his4-519 MSS2∷3xHA (ρ+) | Broadley et al., 2001 |
| SH36b | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8∷hisG (ρ+ COX2(1-67)∷ARG8m) | He and Fox, 1997 |
| SH326c | MATa ura3-52 ade2-101 trp1∷hisG arg8Δ∷URA3 (ρ0) | S. He |
| SH345c | MATa ura3-52 leu2Δ ade2-101 trp1∷hisG arg8Δ∷URA3 (ρ+ COX2(1-251)∷ARG8m) | S. He |
| SH404b | MATα ura3Δ ade2-101 PNT1∷3xHA (ρ+) | He and Fox, 1999 |
| TF249b | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8∷hisG (ρ+ COX2(1-251)∷ARG8m) | T. D. Fox |
| TMD28c | MATa ura3-52 leu2-3,12 his3-11,15 arg8Δ∷hisG (ρ+) | C. M. Demlow |
| SCS15ac | MATα ura3-52 leu2Δ ade2-101 arg8Δ∷hisG (ρ+ COX2(1-251)∷ARG8m) | This study |
| SCS32c | MATα ura3-52 leu2Δ ade2-101 arg8Δ∷hisG cox18∷URA3 (ρ+ COX2(1-251)∷ARG8m) | This study |
| SCS43c | MATa ura3-52 leu2 his3-11,15 arg8Δ∷hisG cox18-12 (ρ+) | This study |
| SCS44c | MATa ura3-52 leu2 his3-11,15 arg8Δ∷hisG cox18-80 (ρ+) | This study |
| SCS46c | MATa ura3-52 leu2 his3-11,15 arg8Δ∷hisG cox18-52 (ρ+) | This study |
| SCS61b | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8∷hisG COX18∷3xHA (ρ+) | This study |
| SCS62b | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8∷hisG cox18∷URA3 (ρ+ COX2[1-67]∷ARG8m) | This study |
| SCS63b | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8∷hisG cox18∷URA3 (ρ+ COX2(1-251)∷ARG8m) | This study |
| SCS68b | MATα ura3ΔNS leu2-3,112 his4-519 MSS2∷3xHA COX18∷3xMYC (ρ+) | This study |
| SCS81b | MATα ura3Δ ade2-101 PNT1∷3xHA COX18∷3xMYC (ρ+) | This study |
| SCS95c | MATα ura3-52 leu2Δ ade2-101 arg8Δ∷URA3 kar1-1 (ρ+ COX2∷3xHA) | This study |
| SCS100c | MATα ura3-52 leu2Δ ade2-101 arg8Δ∷URA3 cox18Δ∷KanR kar1-1 (ρ+ COX2∷3xHA) | This study |
| SCS197b | MATa ura3-52 leu2-3,112 lys2 his3Δ arg8∷hisG cox4Δ∷LEU2 (ρ+ COX2∷3xHA) | This study |
All strains with DBY947 or D273-10B nuclear backgrounds have their mtDNA sequences derived from D273-10B strains.
Isogenic or congenic to D273-10B.
Isogenic or congenic to DBY947.
Cox18p was tagged at its C terminus with three hemagglutinin (HA) or three MYC epitopes by transformation of DFS188 with a PCR product (Schneider et al., 1995) generated by primers SS-40 (5′-C T C A C A G G C T C C A T T C C T T C T T T C G C T C T A T T G G A T A T C A T C A C A G C T A T T C T C C C T G G T G C A A A A T A T C A T A T T A A A T T G G A T T T A T C C T T A C C A A C G A A G G G A A C A A A A G C T G G-3′) and SS-41 (5′-G C A G G A A A G A G T C A G G A A C G G T T G A A G G A T A T T G A A A G T C A T T T T T G T T T A T T T A C A A G C T G A T G T A G A A T T A C A T A T C C T A T C T A T G C G T C A G C T T C A C C T A T A G G G C G A A T T G G-3′). Integrants were confirmed using PCR.
Cox2p was tagged at its C terminus with three HA epitopes by amplifying the 3′-end of COX2 with the 5′-primer 1058 (5′-ACAGCTGCTGATGTT-3′), and the 3′-primer SS-79 (5′-GGCCGG A T C C T T A C G C A T A G T C A G G A A C A T C G T A T G G G T A G G A G C C C G C A T A G T C A G G A A C A T C G T A T G G G T A G C C C G C A T A G T C A G G A A C A T C G T A T G G G T A T T G T T C A T T T A A T C A T T C C A-3′) to fuse the last 20 base pairs of the COX2 open reading frame (italics) to a triple HA cassette from the pMPY3x-HA plasmid (Schneider et al., 1995) and produce a BamHI site (underlined) immediately after the stop codon. This PCR fragment was cut with BamHI and BbsI and cloned into the similarly cut vector pSH05 (He and Fox, 1997). The resulting plasmid, pSCS16, was introduced by microprojectile bombardment (Bonnefoy and Fox, 2001) into a ρ+ strain lacking this region of COX2 (NB75) and screening for respiratory growth. Correct integration was confirmed by DNA sequencing.
Mutant Isolation and Characterization
Arg+ mutants were selected from the parent strain SCS15a and screened for respiratory defects as described by He and Fox (1999). After organization into complementation groups (see RESULTS), DNA fragments complementing the Pet− phenotype of nuclear recessive mutations were selected from a CEN plasmid library and sequenced to identify candidate genes on the fragments by comparison to the genomic sequence (Goffeau et al., 1996) through the SGD (http://genome-www.stanford.edu/Saccharomyces/). Overlapping plasmids were analyzed to identify complementing genes, and each mutant allele was determined by sequence analysis of PCR-amplified genomic DNA from the mutant strains. This analysis yielded the following alleles: cox18-9 K146(frame-shift [f.s.]); cox18-12 S34I; cox18-32 M1I; cox18-45 K154(f.s.); cox18-52 L176R; cox18-72 L163(f.s.); cox18-78 L23stop; cox18-80 R69I; mss2-19 K309stop; mss2-31 L266stop; mss2-35 N59(f.s.); mss2-47 K64(f.s.); mss2-53 W316stop; mss2-62 E294stop; mss2-64 E167stop; mss2-67 L251R; mss2-71 Y238stop; pnt1-26 I249L, K350I; pnt1-27 N353(f.s.); pnt1-43 L145(f.s.); pnt1-75 A172(f.s.); pnt1-76 T142(f.s.); pnt1-141 R377K; pet309-65 L138stop; pet309-66 S35stop; cbp1-29 G454(f.s.).
Analysis of Mitochondrial Proteins
Mitochondrial isolation and purification, mitoplasting, proteinase K treatment, and Western blotting were carried out as previously described (Glick and Pon, 1995; He and Fox, 1997, 1999). Analysis of mitochondrial proteins was carried out with cells grow in YPRaf. Coimmunoprecipitations were carried out as previously described (Hell et al., 2000). anti-HA-horseradish peroxidase was from Roche Biochemicals (Indianapolis, IN), and anti-MYC agarose was from Santa Cruz Biotechnology (Santa Cruz. CA).
RESULTS
Isolation and Identification of Mutants Defective for Export of the Cox2p-Arg8p Fusion Protein
Using the previously described genetic screen (He and Fox, 1999), we have identified 35 new Arg+,Pet− mutants from the parent strain SCS15a (Table 2). Western blot analysis of total protein extracts from the mutant strains indicated they all lacked a characteristic Arg8p degradation product that accumulates after export to the IMS (unpublished results; He and Fox, 1997, 1999), suggesting a defect in export of the fusion protein. All of the mutants were mated to an otherwise wild-type rho+ strain containing the COX2::ARG8m fusion gene in its mtDNA (SH345) and to an otherwise wild-type rho0 strain, lacking mtDNA (SH326). Thirty of the mutants produced respiring diploids when crossed to both the rho+ and rho0 tester strains, indicating the presence of nuclear recessive mutations, although in eight cases the heterozygous diploids' respiratory growth was clearly reduced relative to wild type. Five of the mutants produced respiring diploids when mated to the rho+ [COX2::ARG8m] tester but not when mated to the rho0 tester, indicating the presence of mutations in their mtDNAs.
Table 2.
Mutants identified in export defective screen
| Gene | Number of alleles isolated in screen |
|---|---|
| COX18 | 8 |
| MSS2 | 9 |
| PNT1 | 7 |
| CBP1 | 1 |
| PET309 | 2 |
Analysis of the 35 mutants obtained from the screen resulted in the identification of the 27 mutants listed above. Three of the nuclear mutants were not characterized further, and the remaining five contain mutations in their mtDNA.
Because pnt1 (He and Fox, 1999) and mss2 (Broadley et al., 2001) mutations were already known to block fusion protein export, the nuclear recessive mutants were mated to pnt1Δ and mss2Δ strains, and the resulting diploids were tested for respiratory growth to score complementation. By this test, seven of the mutants failed to complement pnt1 and nine failed to complement mss2 mutations. Mutations in these genes were identified by DNA sequence analysis (see MATERIALS AND METHODS), except for one pnt1 mutant. The remaining 14 nuclear mutants complemented both testers, indicating that their mutations were in unknown genes. These 14 mutants, and strains derived from them by meiotic crosses to a rho0, were then mated among each other and scored for respiratory growth to sort them into complementation groups. We found six groups. The major group contained eight mutants and one contained two. The others were represented by a single mutant each.
We had previously learned that some S. cerevisiae mutations blocking export of the Cox2p-Arg8p fusion protein have little effect on assembly of unmodified Cox2p into functional cytochrome c oxidase (He and Fox, 1999). To focus on those mutations having a strong effect in an otherwise wild-type Cox2p background, we combined mtDNA containing wild-type COX2 with the mutations from the 14 unidentified nuclear mutants (by crossing rho0 derivatives of the mutants to strains containing wild-type COX2 in their mtDNA and isolating meiotic progeny) and scored the resulting respiratory phenotypes. Five mutations from the eight-membered complementation group (cox18 mutants, see below) produced detectable respiratory-defective phenotypes in haploids containing wild-type mtDNA (Figure 1), as did both members of the complementation group containing two mutations. One additional mutant, strain SCS15a-29, was also Pet− in the presence of wild-type mtDNA. Thus, mutations in three complementation groups could block respiration in cells containing wild-type mtDNA. The remaining three mutations caused no phenotype in the presence of wild-type mtDNA and were not further studied.
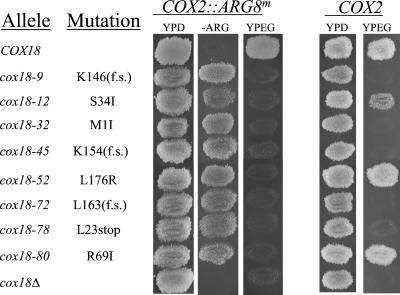
Figure 1.
Growth phenotypes of the eight spontaneous cox18 alleles obtained by selection for Arg+Pet− (retention of the Arg8p moiety of a Cox2p-Arg8p fusion protein within the matrix). Cells with the indicated relevant mitochondrial genotypes were grown on solid complete glucose medium (YPD) then replica plated twice to glucose medium lacking arginine (−ARG) and complete medium containing ethanol and glycerol (YPEG), as indicated. Incubation temperature was 30o. The mtDNA of these strains encoded either the Cox2p-Arg8p fusion protein or wild-type Cox2p, as indicated. All Cox2p-Arg8p–expressing strains are isogenic to strain SCS15a and all Cox2p-expressing strains are isogenic to TMD28 (Table 1). cox18Δ strains are SCS32 and RCB1. f.s. indicates a frame-shift affecting the indicated codon.
To isolate the genes identified by the mutations preventing respiratory growth with wild-type Cox2p, we transformed yeast genomic libraries into one representative each of the three complementation groups (MATERIALS AND METHODS). Plasmids that restored respiratory growth were analyzed to determine the smallest region necessary to restore respiratory growth and then sequenced. The region of overlap among the plasmids complementing the eight-membered group contained SPT4 and COX18. COX18 has previously been shown to be required for cytochrome oxidase function and Cox2p accumulation (Souza et al., 2000). Indeed, all eight strains in this group failed to complement a cox18Δ mutant and contained mutations in COX18, as revealed by DNA sequencing (MATERIALS AND METHODS). Similar analysis of complementing plasmids, failure to complement known chromosomal deletions, and DNA sequencing identified the two-member complementation group as the COX1 translational activator gene PET309 (Manthey and McEwen, 1995; Manthey et al., 1998) and the one-membered group as CBP1, whose product is required to stabilize the mitochondrial mRNA encoding cytochrome b (Dieckmann et al., 1984; Chen and Dieckmann, 1997).
The alleles we selected in the screen for Arg+,Pet− phenotypes exhibited a range of respiratory defects, suggesting that many of the mutated genes were partially functional. We therefore tested the effects of cox18, pet309, and cbp1 null mutations in strains derived from SCS15a, expressing the Cox2p-Arg8p fusion protein (MATERIALS AND METHODS). Interestingly, although deletions of each of these three genes caused a Pet− phenotype, they did not produce Arg+ phenotypes. The same difference in phenotype between selected alleles and complete deletions has also been observed for PNT1 (unpublished results) and MSS2 (Broadley et al., 2001). We conclude that expression of the Arg+ phenotype that we originally selected depends on partial function of the mutated genes.
Cox18p Is Required to Export the C-Tail, but Not the N-Tail, of Cox2p
A previous study had demonstrated that Cox18p was an integral mitochondrial membrane protein required for normal Cox2p accumulation and cytochrome oxidase assembly but did not further define its role (Souza et al., 2000). Because we isolated cox18 mutations in our screen for export defects, we next explored the possibility that Cox18p was required for export of the unmodified Cox2p C-tail domain. We found that deletion of COX18 lowered the steady-state level of Cox2p approximately 10-fold, but the remaining Cox2p could be easily detected by Western analysis using a mAb (Pinkham et al., 1994) that recognizes an epitope in the C-tail (He and Fox, 1997).
Studies of in vivo Cox2p export from the matrix are complicated by the fact that the exported domains of the protein are assembled into the protease-resistant cytochrome c oxidase complex, making assays of protease sensitivity/resistance in mitoplasts problematic (He and Fox, 1999). As a new approach to detecting Cox2p C-tail export directly by proteolysis, we attached a triple-HA epitope tag (30 residues) to the C terminus of Cox2p by modifying COX2 in mtDNA (MATERIALS AND METHODS). Because the Cox2p C terminus is at the surface of cytochrome oxidase exposed to the IMS (Iwata et al., 1995; Tsukihara et al., 1996), we reasoned that the amino acids of a C-terminal epitope might be accessible to protease despite the resistance of native Cox2p domains. Addition of this C-terminal epitope did not appear to affect Cox2p function because strains containing the tagged protein grew at wild-type rates on nonfermentable medium at 14°, 30°, and 37°C. Mitochondria and mitoplasts were prepared from COX18 and cox18Δ strains expressing this Cox2p-HA fusion protein and treated with added protease. Probing the Western blots with anti-HA antibody revealed that the HA tag on Cox2p-HA was highly sensitive to proteinase K degradation in mitoplasts derived from the wild-type COX18 strain (Figure 2, lane 4). In contrast, the HA tag on Cox2p-HA in mitoplasts lacking Cox18p was protected from proteolysis (Figure 2, lane 8). However, Cox2p-HA was shortened by proteolysis of the cox18Δ mitoplasts, by partial digestion of the 25-residue exported N-tail (Figure 2, lane 8). Disruption of the inner membrane by treatment of cox18Δ mitoplasts with the mild detergent octylglucoside made the epitope accessible to protease (Figure 2, lane 9). Reprobing the blots with the C-tail-specific anti-Cox2p antibody showed that the Cox2p C-tail was protected from protease in both COX18 and cox18Δ mitoplasts (Figure 2, lanes 4 and 8). However, the Cox2p-HA in COX18 mitoplasts was shortened by ∼3 kDa, as a result of the removal of the triple-HA epitope (Figure 2, lane 4). (The epitope was also removed from a fraction of the Cox2p-HA by leakage of protease through the outer membrane of the COX18 mitochondria [Figure 2, lane 2].) Similar experiments with mitochondria derived from a different nuclear background (D273-10B) gave identical results (unpublished results).
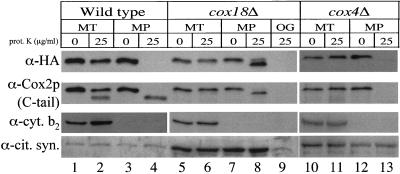
Figure 2.
The C terminus of Cox2p-HA is protected from exogenous protease in mitoplasts from a strain lacking Cox18p. Mitochondria isolated from wild-type (SCS95), cox18Δ (SCS100), and cox4Δ (SCS197) cells expressing Cox2p-HA from their mtDNA were converted to mitoplasts. Intact mitochondria (MT) or mitoplasts (MP; 75 μg of mitochondrial protein) were treated with 25 μg/ml proteinase K (prot. K) or mock treated as controls, as described in MATERIALS AND METHODS. cox18Δ mitochondria were also solubilized with 1% octylglucoside (OG) and treated with protease. The resulting samples were electrophoresed on 12% SDS-PAGE gels, blotted, and probed with anti-HA antibody. The filters were stripped and reprobed with antibodies against Cox2p (CCO6), the IMS marker cytochrome b2 (cyt. b2), and the matrix marker anti-citrate synthase (cit. syn.). Because of the reduced Cox2p accumulation in cox18Δ and cox4Δ mitochondria, 10-fold more mitochondrial proteins from these strains (30 μg) were loaded per lane than from wild-type mitochondria (3 μg). Because of the unequal loadings of COX18 and cox18Δ mitochondria, the anti-cytochrome b2 probing was exposed for different times.
Cox2p C-tail export is dependent on the inner membrane potential (He and Fox, 1997), raising the possibility that the defect caused by the cox18Δ is an indirect effect of the specific loss of cytochrome c oxidase activity (Souza et al., 2000). We therefore asked whether absence of the nuclearly encoded subunit Cox4p, which is known to prevent assembly of cytochrome oxidase (Dowhan et al., 1985), would prevent Cox2p-HA C-tail export. Proteolysis of mitoplasts from a cox4Δ strain degraded the C-terminal HA epitope, as well as the endogenous epitope in the Cox2p C-tail (which is labile because of the assembly defect caused by the lack of Cox4p; Figure 2, lane 13), demonstrating that C-tail export occurs in the absence of cytochrome oxidase activity per se. Thus, decreased membrane potential cannot account for the export defect observed in the cox18Δ strain.
The protease protection experiments presented above indicate that Cox18p does not play a role in exporting the Cox2p N-tail. To test this hypothesis further, we examined the behavior of a fusion protein comprising the first 67 amino acids of Cox2p and Arg8p (Cox2p[1–67]-Arg8p). Previous experiments have shown that in otherwise wild-type strains the Cox2p N-tail of this chimera is exported through the mitochondrial inner membrane while the Arg8p moiety remains in the matrix (He and Fox, 1997). Because cytochrome oxidase is not assembled in these cells, the exported N-tail is sensitive to proteases added to mitoplasts. We carried out protease protection experiments on mitochondria and mitoplasts from COX18 and cox18Δ strains expressing this fusion protein and probed the resulting Western blots with an anti-Arg8p antibody (Figure 3). The Cox2p(1–67)-Arg8p fusion protein in both COX18 and cox18Δ mitoplasts was shortened by proteinase K treatment, confirming that Cox18p is not necessary for Cox2p N-tail export.
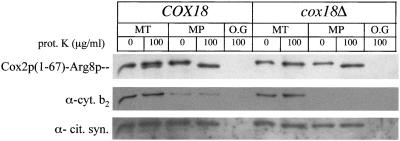
Figure 3.
Export of the Cox2p N-tail, in a Cox2p(1–67)-Arg8p fusion protein, is unaffected in cox18Δ mitochondria. Mitochondria (MT) were isolated from wild-type (SH36) and cox18Δ (SCS62) strains expressing the Cox2p(1–67)-Arg8p fusion protein. Mitoplasting was carried out in the absence or presence of 100 μg/ml proteinase K (prot. K). Mitochondrial proteins (50 μg per lane) were loaded on a 12% SDS-PAGE gel. After separation and blotting, the resulting Western blot was probed with anti-Arg8p antibodies and then stripped and reprobed with anti-cytochrome b2 (cyt. b2) and anti-citrate synthase (cit. syn.) antibodies. MP, mitoplasts; O.G, +1% octylglucoside.
The Cox18p C Terminus Is on the Matrix Side of the Inner Membrane
Previous work has shown Cox18p to be a mitochondrial integral membrane protein (Souza et al., 2000). To examine further the submitochondrial location of Cox18p, we attached a triple-HA epitope tag to its C terminus by altering the chromosomal COX18 gene (MATERIALS AND METHODS). This modification had no effect on respiratory growth (unpublished results). Mitochondria were isolated from the strain containing Cox18p-HA and were treated with protease before and after conversion to mitoplasts. Probing a Western blot of the resulting fractions with anti-HA antibody revealed that Cox18p-HA was present in the mitochondria as two species of approximately 40 kDa (Figure 4A). Although Cox18p was protected from added protease by the outer membrane of whole mitochondria, a significant fraction of it was cleaved by protease treatment of mitoplasts to an approximately 23-kDa C-terminal fragment containing the HA tag (Figure 4A). This C-terminal fragment is protected from protease by the inner membrane because addition of protease to mitoplasts in the presence of 1% octylglucoside resulted in complete degradation of immune-detectable Cox18p-HA. Thus, Cox18p has a domain near the middle that is accessible to protease from the IMS side, and its C terminus appears to be on the matrix side of the inner membrane (Figure 4B).
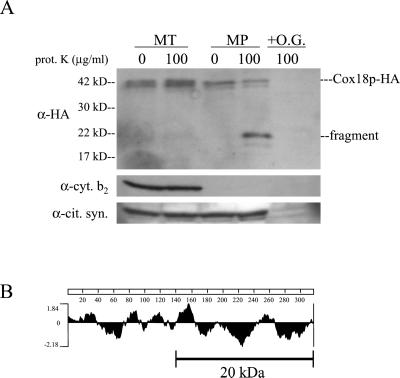
Figure 4.
The C terminus of Cox18p is protected from added protease by the inner membrane. (A) Mitochondria (MT) were isolated from a strain expressing a Cox18p-HA fusion protein (SCS61). The mitochondria were subjected to mitoplasting in the absence or presence of 100 μg/ml proteinase K (prot. K). Protein samples (75 μg per lane) were separated on a 12% SDS-PAGE gel and blotted. The blot was probed with anti-HA antibodies and then stripped and reprobed with anti-cytochrome b2 (cyt. b2) and anti-citrate synthase antibodies (cit. syn.). The C-terminal fragment of Cox18p-HA (fragment) produced in mitoplasts treated with protease is indicated. O.G., +1% octylglucoside. (B) A hydrophilicity plot of the Cox18p protein (Kyte and Doolittle, 1982). The line indicates the approximate length of the C-terminal fragment relative to the plot, taking into account the 3.5-kDa triple-HA tag.
Genetic Interactions among COX18, PNT1, MSS2, and PET111
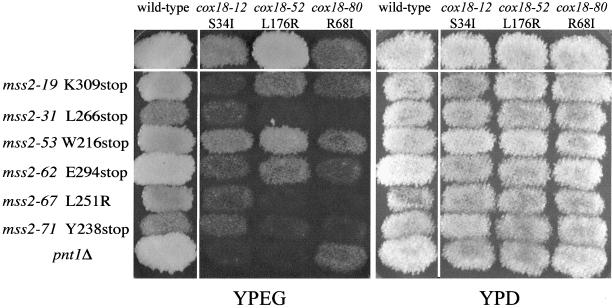
Many of the mutants isolated in our screen for defects in export of the Cox2p-Arg8p fusion protein exhibited leaky or no respiratory growth defects in strains containing wild-type Cox2p (Figures 1 and 5). Thus, we had the opportunity to test for synthetic phenotypes (genetic enhancement) by combining leaky alleles of different genes in double-mutant strains. Three cox18 alleles that had partial or full respiratory ability were mated to six mss2 alleles that also retained at least some respiratory competence with Cox2p. These diploids were sporulated and the resulting tetrads were scored on nonfermentable (YPEG) medium. In many cases, one-quarter of the haploid progeny produced by these crosses exhibited clear synthetic respiratory defects when compared with the parents (Figure 5). DNA sequencing of the cox18 and mss2 genes in these nonrespiring segregants confirmed that they were double mutants. Interestingly, several of these synthetic defects demonstrate allele-specific interactions between COX18 and MSS2 (Figure 5). For example, the cox18-52 mutant is less severe than cox18-12. However, the cox18-52, mss2-67 double mutant has a more severe phenotype than the cox18-12, mss2-67 double mutant.
Figure 5.
Allele-specific synthetic respiratory defects in cox18, mss2, and cox18, pnt1 double mutants. All strains used here contain mtDNA encoding wild-type Cox2p. Haploid strains were patched and replica plated to YPD and YPEG media and incubated at 30o for 3 and 4 d, respectively. Wild type is boxed in the top left corner of each panel. Haploid single mutants with the indicated relevant genotypes are in the left columns and top rows outside the white lines. Double mutants contain the alleles indicated by their positions in the matrix.
Deletion of PNT1 has little effect on respiratory growth of S. cerevisiae strains containing wild-type mtDNA (He and Fox, 1999). However, in combination with two of the leaky cox18 alleles tested, the pnt1Δ caused a clear synthetic defect (Figure 5). Interestingly, no interaction was observed between cox18-80 and the pnt1Δ, providing evidence for allele-specific interactions between these genes as well.
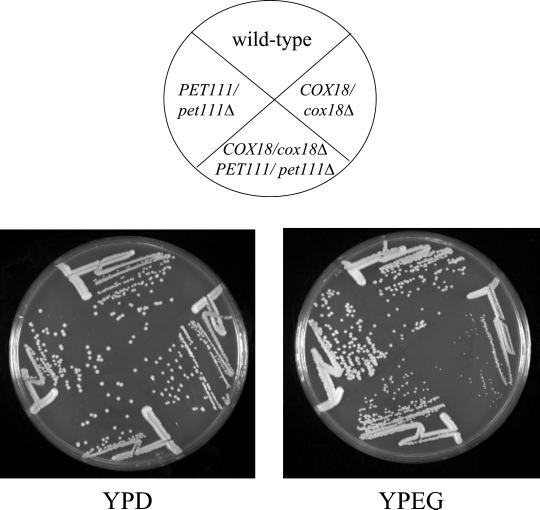
As noted above, when the original set of Arg+,Pet− mutants containing the COX2::ARG8m fusion in mtDNA were crossed to tester strains with wild-type nuclear genes, eight of the mutants produced diploids whose respiratory growth was significantly slower than wild type. All eight of the these strains proved to contain cox18 mutations. To test whether this reduced respiratory growth could be due to COX18 haploinsufficiency in the presence of the Cox2p-Arg8p fusion protein, we created a heterozygous cox18Δ/COX18 diploid strain and found that it indeed had reduced respiratory growth relative to an isogenic COX18/COX18 strain (Figure 6). This haploinsufficiency was not observed in cox18Δ/COX18 strains expressing wild-type Cox2p. Interestingly, when we reduced mitochondrial expression of the COX2::ARG8m mRNA by halving the gene dosage of the rate-limiting COX2 mRNA-specific translational activator PET111 (Green-Willms et al., 2001) in the cox18Δ/COX18 diploid, growth on nonfermentable carbon sources was improved (Figure 6).
Figure 6.
COX18/cox18Δ strains producing the Cox2p-Arg8p fusion protein exhibit respiratory haploinsufficiency that is partially reversed by decreased expression of the fusion protein. Diploid strains with the indicated relevant genotypes were produced by the following matings: (wild type) TF249xDAU1ρ0, (COX18/cox18Δ) SCS63xDAU1ρ0, (COX18/cox18Δ PET111/pet111Δ) SCS63xNSG132ρ0, and (PET111/pet111Δ) TF249xNSG132ρ0. The diploids were streaked to complete glucose (YPD) and ethanol/glycerol (YPEG) media and incubated at 30o for 4 and 5 d, respectively.
Cox18p Interacts with Mss2p and Pnt1p
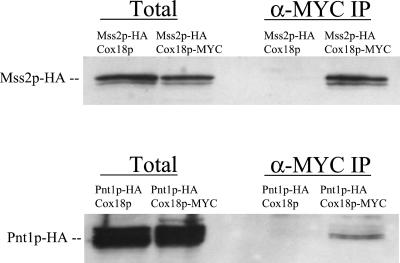
The allele specificity of synthetic defective interactions among cox18, mss2, and pnt1 mutations suggested that their protein products could interact physically. To test this we created strains whose Cox18p was tagged with a triple-MYC epitope and contained a triple-HA epitope tag on either Mss2p or Pnt1p. Cox18p-MYC was immunoprecipitated from solubilized mitochondrial extracts from each strain with anti-MYC antibody bound to agarose beads (MATERIALS AND METHODS). The immunoprecipitates were then analyzed by Western blotting using anti-HA antibody as a probe for coprecipitated proteins. This experiment revealed that Mss2p-HA was coprecipitated with Cox18p-MYC but not with the control Cox18p lacking the MYC epitope (Figure 7). Pnt1p-HA also specifically coprecipitated with Cox18p-MYC, although less efficiently than Mss2p-HA (Figure 7). The integral inner membrane protein Yme1p was not detectable in the immune precipitates (Saracco and Fox, unpublished results), indicating that our results are not due to nonspecific precipitation of membrane fragments. We have not determined whether the incomplete efficiency of coprecipitation reflects dynamic interactions among the proteins or destabilization of a complex by the digitonin used to solubilize membranes (Herrmann et al., 2001).
Figure 7.
Mss2p and Pnt1p coimmunoprecipitate with Cox18p. Mitochondria (500 μg) containing the indicated epitope-tagged proteins were solubilized with 1% digitonin as described by Hell et al. (2000). After a clarifying spin, 250 μg of soluble protein were incubated with anti-MYC agarose beads (MATERIALS AND METHODS). After a 1-h incubation at 4oC, the beads were pelleted and washed three times. Bound proteins were eluted with SDS sample buffer, run on 12% SDS-PAGE gels, blotted, and probed with anti-HA-horseradish peroxidase antibodies (α-MYC IP). Total solubilized mitochondrial proteins (50 μg) were precipitated with trichloroacetic acid and similarly analyzed (Total). Strains were SB19, SCS68, SH404, and SCS81 (Table 1).
DISCUSSION
The components of translocases necessary to export mitochondrially coded protein domains from the matrix to the IMS have not been identified by comparisons of the yeast proteome to known translocases in other systems (Glick and von Heijne, 1996). Furthermore, biochemical analysis of these translocation events is hampered by the lack of a true in vitro system for mitochondrial translation. We have therefore exploited a genetic screen for mutants with defects in the export of a mitochondrially encoded Cox2p-Arg8p fusion protein (He and Fox, 1999) to find nuclear genes required to export the Cox2p C-tail domain. This screen depends on the fact that export of the fusion protein removes Arg8p from the matrix, causing an Arg− phenotype, but proteolysis in the IMS generates functional Cox2p that supports respiratory growth. Synthesis and export of this fusion protein appear to stress the mitochondrial system because the organelles have an abnormally light buoyant density, and some mutations that cause nonrespiratory (Pet−) growth in conjunction with the COX2::ARG8m mitochondrial gene do not completely prevent cytochrome oxidase assembly and respiratory growth in conjunction with the wild-type COX2 gene (He and Fox, 1997). Thus, selection for Arg+ mutants is highly sensitive for decreased efficiency of export, and the presence of the fusion protein can enhance export defective phenotypes to produce synthetic respiratory deficiency.
In this study we identified several mutations in the nuclear gene COX18. cox18 mutants had previously been shown to lack cytochrome c oxidase but not ATPase or NADH-cytochrome c reductase (Souza et al., 2000). Thus, Cox18p is not generally required for insertion of mitochondrially coded proteins into the inner membrane and may be specific for Cox2p. Cox18p had previously been shown to encode an integral mitochondrial membrane protein with four predicted transmembrane helices (Souza et al., 2000). We confirmed the location of Cox18p and found that its C terminus appears to be on the inner face of the inner membrane, consistent with a possible role in translocation.
It is difficult to detect export of the Cox2p C-tail using protease sensitivity in mitoplasts because the exported protein is assembled into the protease-resistant cytochrome oxidase complex (He and Fox, 1999). To ask whether cox18 mutants were in fact defective for C-tail export, we utilized a new tool to circumvent this problem. The addition of an HA epitope to the C terminus of Cox2p, which did not detectably affect respiratory growth, allowed us to determine whether the Cox2p C-tail was exported to the IMS in the absence of Cox18p. As expected from the structure of the complex (Iwata et al., 1995; Tsukihara et al., 1996), this epitope was accessible to protease added from the IMS side of the inner membrane of mitoplasts despite the fact that Cox2p was assembled. In the absence of Cox18p, this HA epitope was protected from proteolytic digestion by the inner membrane. This experiment demonstrated that Cox18p is necessary for C-tail export through the inner membrane.
A previous study demonstrated that export of the Cox2p N-tail was independent of the inner membrane potential, whereas export of the C-tail depends on this potential, indicating that these two processes are mechanistically distinct (He and Fox, 1997). Our present results support this conclusion genetically, because Cox2p N-tail export proceeded normally in a cox18 deletion mutant. Mutations in COX18 or the other genes we identified cannot simply destroy the inner membrane potential because that would prevent protein import and result in lethality (Lill et al., 1996; Schatz, 1996). Furthermore, we found that elimination of cytochrome oxidase activity by deletion of COX4 did not prevent C-tail export.
Export of both the N-tail and C-tail is dependent on the conserved inner membrane protein Oxa1p, which appears to function as a translocase (Bauer et al., 1994; Bonnefoy et al., 1994a, b; He and Fox, 1997; Hell et al., 1997, 1998; Herrmann et al., 1997; Kermorgant et al., 1997; Scotti et al., 2000). Interestingly, a PSI-Blast comparison suggests that Cox18p resembles Oxa1p homologues from several species, and both proteins have their C termini in the matrix. This similarity suggests that Cox18p could be directly involved in translocation of the C-tail, a reaction that could be dependent on prior translocation of the N-tail by Oxa1p. However, we found that elevated OXA1 gene dosage does not detectably suppress a cox18Δ, suggesting that Cox18p carries out a distinct function. A Cox18p orthologue (42.8% identity) has been identified in the yeast Kluyveromyces lactis (Hikkel et al., 1997), and a gene coding a homologous protein with 28.5% identity to Cox18p is present in the Candida albicans genome (http://sequence-www.stanford.edu/group/candida/index.html). However, we have found no homologues in more distantly related species. Although Cox18p is clearly required for export of the Cox2p C-tail, we have not demonstrated that Cox18p functions by direct interaction with Cox2p.
In addition to COX18, our selection identified mutations in two other nuclear genes, PNT1 (He and Fox, 1999) and MSS2 (Broadley et al., 2001), that were already known to play a role in Cox2p C-tail export. Pnt1p is an integral mitochondrial inner membrane protein with hydrophilic domains facing the matrix side (He and Fox, 1999), whereas Mss2p is a peripheral inner membrane protein on the matrix side. Thus, it is plausible that Cox18p, Pnt1p, and Mss2p function together in the export of the Cox2p C-tail. Consistent with this idea, we detected synthetic respiratory-defective interactions between certain combinations of cox18 and mss2 alleles, as well as certain cox18 alleles and a pnt1 deletion, in strains containing wild-type Cox2p. In addition, we detected biochemical interactions among Cox18p/Pnt1p and Cox18p/Mss2p through coimmunoprecipitations. Thus, it appears that these three membrane-bound proteins interact on the matrix side of the inner membrane to facilitate Cox2p C-tail translocation. Whether or not they form a stable complex remains to be determined.
The cox18 deletion mutation was fully recessive at the level of respiratory growth in strains containing wild-type Cox2p. However, in diploids containing the Cox2p-Arg8p fusion protein, cox18Δ/COX18 cells grew less well on nonfermentable medium than did COX18/COX18 cells. This partial haploinsufficiency suggests that reduced levels of Cox18p create a bottleneck in translocation that exacerbates the deleterious effect of the fusion protein on mitochondrial biogenesis. If this were the case, then reduced expression of the Cox2p-Arg8p fusion protein might improve respiratory growth of the cox18Δ/COX18 cells. Indeed, deleting one copy of the rate-limiting COX2 mRNA-specific translational activator PET111 (Green-Willms et al., 2001) improved respiratory growth of these diploids. This interaction between PET111 and COX18 suggests that the levels of Cox2p translation and translocation activities may be roughly coordinated.
Several of the cox18 mutations we selected by asking for Arg+ growth due to reduced fusion protein export were found by DNA sequence analysis to be frame-shift or nonsense alleles near the beginning of the open reading frame, suggesting that they were null mutations. Surprisingly however, we found that a complete cox18 deletion in the same strain background exhibited an Arg− growth phenotype. Thus, it appears that the selected alleles retain some residual gene expression that is necessary for the unexported fusion protein to function in arginine biosynthesis. Consistent with this idea, we found that treating some of the selected cox18 mutants with guanidine hydrochloride, to eliminate the PSI+ prion and thereby increase cytoplasmic translational fidelity, produced an Arg− phenotype similar to the deletion allele (unpublished results). This residual gene expression does not appear to increase the steady-state levels of the Cox2p-Arg8p fusion protein over that seen in the deletion mutant (data not shown). Cox18p is not absolutely required to allow the unexported fusion protein to function in arginine biosynthesis because the cox18Δ mutation in the D273-10B strain background, which has more robust mitochondrial gene expression than the DBY947 background used here (He and Fox, 1999), does not cause an Arg− phenotype. Although we do not understand the basis for this phenotypic difference between the selected alleles and deletions, we have obtained similar results with all of the nuclear genes identified in this study. Deletion of OXA1 also prevents fusion protein-dependent Arg+ growth even in the D273-10B background, but in this case the steady-state level of the unexported fusion protein is severely reduced, presumably due to extreme instability (He and Fox, 1997).
Surprisingly, we also isolated mutations in two genes whose known functions are not directly related to membrane translocation: PET309, a COX1 mRNA-specific translational activator gene (Manthey and McEwen, 1995), and CBP1, which is required to stabilize the mitochondrial mRNA encoding cytochrome b and possibly play a role in its translation (Dieckmann et al., 1984; Weber and Dieckmann, 1990; Dieckmann and Staples, 1994; Chen and Dieckmann, 1997). The products of both are present in mitochondria and functionally interact with mRNAs. The submitochondrial location of Cbp1 has not been established, but Pet309p is an integral inner membrane protein (Manthey et al., 1998). One interpretation of the fact that mutations in these genes can cause an Arg+ phenotype in our system is that they might lead to increased expression of COX2::ARG8m by preventing expression of other mitochondrial genes. However, complete pet309 and cbp1 deletions did not lead to the Arg+ phenotype observed with the selected alleles, as would be expected if this mechanism were correct. An alternative possibility is that proteins involved in translation of mitochondrial gene products and their membrane insertion are organized in large complexes. In this case, a mutation in one component of the complex could potentially affect the efficiency with which the Cox2p-Arg8p fusion protein is exported, even if that component were not directly involved in expression of COX2.
ACKNOWLEDGMENTS
We thank S. Broadley for sequencing the mss2 alleles, G. Schatz and T. Mason for gifts of antisera, and E. Williams for critical reading of the manuscript. This work was supported by the U.S. National Institutes of Health in the form of a training grant (GM07617) to S.A.S. and a research grant (GM29362) to T.D.F.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc. 01–12–0580. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–12–0580.
REFERENCES
- Altamura N, Capitanio N, Bonnefoy N, Papa S, Dujardin G. The Saccharomyces cerevisiae OXA1 gene is required for the correct assembly of cytochrome c oxidase and oligomycin-sensitive ATP synthase. FEBS Lett. 1996;382:111–115. doi: 10.1016/0014-5793(96)00165-2. [DOI] [PubMed] [Google Scholar]
- Bauer M, Behrens M, Esser K, Michaelis G, Pratje E. PET1402, a nuclear gene required for proteolytic processing of cytochrome oxidase subunit 2 in yeast. Mol Gen Genet. 1994;245:272–278. doi: 10.1007/BF00290106. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Chalvet F, Hamel P, Slonimski PP, Dujardin G. OXA1, a Saccharomyces cerevisiae nuclear gene whose sequence is conserved from prokaryotes to eukaryotes controls cytochrome oxidase biogenesis. J Mol Biol. 1994a;239:201–212. doi: 10.1006/jmbi.1994.1363. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Fox TD. In vivo analysis of mutated initiation codons in the mitochondrial COX2 gene of Saccharomyces cerevisiae fused to the reporter gene ARG8m reveals lack of downstream reinitiation. Mol Gen Genet. 2000;262:1036–1046. doi: 10.1007/pl00008646. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Fox TD. Genetic transformation of Saccharomyces cerevisiae mitochondria. Methods Cell Biol. 2001;65:381–396. doi: 10.1016/s0091-679x(01)65022-2. [DOI] [PubMed] [Google Scholar]
- Bonnefoy N, Kermorgant M, Groudinsky O, Minet M, Slonimski PP, Dujardin G. Cloning of a human gene involved in cytochrome oxidase assembly by functional complementation of an oxa1− mutation in Saccharomyces cerevisiae. Proc Natl Acad Sci USA. 1994b;91:11978–11982. doi: 10.1073/pnas.91.25.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broadley SA, Demlow CM, Fox TD. A peripheral mitochondrial inner membrane protein, Mss2p, required for export of the mitochondrially coded Cox2p C-tail in Saccharomyces cerevisiae. Mol Cell Biol. 2001;21:7663–7672. doi: 10.1128/MCB.21.22.7663-7672.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Dieckmann CL. Genetic evidence for interaction between Cbp1 and specific nucleotides in the 5′ untranslated region of mitochondrial cytochrome b mRNA in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:6203–6211. doi: 10.1128/mcb.17.11.6203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieckmann CL, Homison G, Tzagoloff A. Assembly of the mitochondrial membrane system: nucleotide sequence of a yeast nuclear gene (CBP1) involved in 5′ end processing of cytochrome b pre-mRNA. J Biol Chem. 1984;259:4732–4738. [PubMed] [Google Scholar]
- Dieckmann CL, Staples RR. Regulation of mitochondrial gene expression in Saccharomyces cerevisiae. Int Rev Cytol. 1994;152:145–181. doi: 10.1016/s0074-7696(08)62556-5. [DOI] [PubMed] [Google Scholar]
- Dowhan W, Bibus CR, Schatz G. The cytoplasmically-made subunit IV is necessary for assembly of cytochrome c oxidase in yeast. EMBO J. 1985;4:179–184. doi: 10.1002/j.1460-2075.1985.tb02334.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox TD. Genetics of mitochondrial translation. In: Hershey JWB, Matthews MB, Sonenberg N, editors. Translational Control. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1996. pp. 733–758. [Google Scholar]
- Fox TD, Folley LS, Mulero JJ, McMullin TW, Thorsness PE, Hedin LO, Costanzo MC. Analysis and manipulation of yeast mitochondrial genes. Methods Enzymol. 1991;194:149–165. doi: 10.1016/0076-6879(91)94013-3. [DOI] [PubMed] [Google Scholar]
- Glick BS, Pon LA. Isolation of highly purified mitochondria from Saccharomyces cerevisiae. Methods Enzymol. 1995;260:213–223. doi: 10.1016/0076-6879(95)60139-2. [DOI] [PubMed] [Google Scholar]
- Glick BS, von Heijne G. Saccharomyces cerevisiae mitochondria lack a bacterial-type Sec machinery. Protein Sci. 1996;5:2651–2652. doi: 10.1002/pro.5560051229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffeau A, Barrell BG, Bussey H, Davis RW, Dujon B, Feldmann H, Galibert F, Hoheisel JD, Jacq C, Johnston M, Louis EJ, Mewes HW, Murakami Y, Philippsen P, Tettelin H, Oliver SG. Life with 6000 genes. Science. 1996;274:546. doi: 10.1126/science.274.5287.546. , 563–567. [DOI] [PubMed] [Google Scholar]
- Green-Willms NS, Butler CA, Dunstan HM, Fox TD. Pet111p, an inner membrane-bound translational activator that limits expression of the Saccharomyces cerevisiae mitochondrial gene. COX2. J Biol Chem. 2001;276:6392–6397. doi: 10.1074/jbc.M009856200. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. In: Abelson JN, Simon MI, editors. Methods in Enzymology. Vol. 194. San Diego: Academic Press; 1991. [PubMed] [Google Scholar]
- He S, Fox TD. Membrane translocation of mitochondrially coded Cox2p: distinct requirements for export of amino- and carboxy-termini, and dependence on the conserved protein Oxa1p. Mol Biol Cell. 1997;8:1449–1460. doi: 10.1091/mbc.8.8.1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, Fox TD. Mutations affecting a yeast mitochondrial inner membrane protein, Pnt1p, block export of a mitochondrially synthesized fusion protein from the matrix. Mol Cell Biol. 1999;19:6598–6607. doi: 10.1128/mcb.19.10.6598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Herrmann J, Pratje E, Neupert W, Stuart RA. Oxa1p mediates the export of the N- and C-termini of pCoxII from the mitochondrial matrix to the intermembrane space. FEBS Lett. 1997;418:367–370. doi: 10.1016/s0014-5793(97)01412-9. [DOI] [PubMed] [Google Scholar]
- Hell K, Herrmann JM, Pratje E, Neupert W, Stuart RA. Oxa1p, an essential component of the N-tail protein export machinery in mitochondria. Proc Natl Acad Sci USA. 1998;95:2250–2255. doi: 10.1073/pnas.95.5.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hell K, Tzagoloff A, Neupert W, Stuart RA. Identification of Cox20p, a novel protein involved in the maturation and assembly of cytochrome oxidase subunit 2. J Biol Chem. 2000;275:4571–4578. doi: 10.1074/jbc.275.7.4571. [DOI] [PubMed] [Google Scholar]
- Herrmann JM, Neupert W, Stuart RA. Insertion into the mitochondrial inner membrane of a polytopic protein, the nuclear-encoded Oxa1p. EMBO J. 1997;16:2217–2226. doi: 10.1093/emboj/16.9.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann JM, Westermann B, Neupert W. Analysis of protein-protein interactions in mitochondria by coimmunoprecipitation and chemical cross-linking. Methods Cell Biol. 2001;65:217–230. doi: 10.1016/s0091-679x(01)65013-1. [DOI] [PubMed] [Google Scholar]
- Hikkel I, Gbelska Y, van der Aart QJ, Lubecu G, Subik J. Cloning and characterization of KlCOX18, a gene required for activity of cytochrome oxidase in Kluyveromyces lactis. Curr Genet. 1997;32:267–272. doi: 10.1007/s002940050276. [DOI] [PubMed] [Google Scholar]
- Iwata S, Ostermeier C, Ludwig B, Michel H. Structure at 2.8 A resolution of cytochrome c oxidase from Paracoccus denitrificans. Nature. 1995;376:660–669. doi: 10.1038/376660a0. [DOI] [PubMed] [Google Scholar]
- Jan P-S, Esser K, Pratje E, Michaelis G. Som1p, a third component of the yeast mitochondrial inner membrane peptidase complex that contains Imp1p and Imp2p. Mol Gen Genet. 2000;263:483–491. doi: 10.1007/s004380051192. [DOI] [PubMed] [Google Scholar]
- Jauniaux J-C, Urrestarazu LA, Wiame J-M. Arginine metabolism in Saccharomyces cerevisiae: subcellular localization of the enzymes. J Bacteriol. 1978;133:1096–1107. doi: 10.1128/jb.133.3.1096-1107.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kermorgant M, Bonnefoy N, Dujardin G. Oxa1p, which is required for cytochrome c oxidase and ATP synthase complex formation, is embedded in the mitochondrial inner membrane. Curr Genet. 1997;31:302–307. doi: 10.1007/s002940050209. [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lill R, Nargang FE, Neupert W. Biogenesis of mitochondrial proteins. Curr Opin Cell Biol. 1996;4:505–512. doi: 10.1016/s0955-0674(96)80028-7. [DOI] [PubMed] [Google Scholar]
- Manthey GM, McEwen JE. The product of the nuclear gene PET309 is required for translation of mature mRNA and stability or production of intron-containing RNAs derived from the mitochondrial COX1 locus of Saccharomyces cerevisiae. EMBO J. 1995;14:4031–4043. doi: 10.1002/j.1460-2075.1995.tb00074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manthey GM, Przybyla-Zawislak BD, McEwen JE. The Saccharomyces cerevisiae Pet309 protein is embedded in the mitochondrial inner membrane. Eur J Biochem. 1998;255:156–161. doi: 10.1046/j.1432-1327.1998.2550156.x. [DOI] [PubMed] [Google Scholar]
- Marykwas DL, Fox TD. Control of the Saccharomyces cerevisiae regulatory gene PET494: transcriptional repression by glucose and translational induction by oxygen. Mol Cell Biol. 1989;9:484–491. doi: 10.1128/mcb.9.2.484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulero JJ, Fox TD. Alteration of the Saccharomyces cerevisiae COX2 5′-untranslated leader by mitochondrial gene replacement and functional interaction with the translational activator protein PET111p. Mol Biol Cell. 1993;4:1327–1335. doi: 10.1091/mbc.4.12.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunnari J, Fox TD, Walter P. A mitochondrial protease with two catalytic subunits of nonoverlapping specificities. Science. 1993;262:1997–2004. doi: 10.1126/science.8266095. [DOI] [PubMed] [Google Scholar]
- Pel HJ, Tzagoloff A, Grivell LA. The identification of 18 nuclear genes required for the expression of the yeast mitochondrial gene encoding cytochrome c oxidase subunit I. Curr Genet. 1992;21:139–146. doi: 10.1007/BF00318473. [DOI] [PubMed] [Google Scholar]
- Pinkham JL, Dudley AM, Mason TL. T7 RNA polymerase-dependent expression of COXII in yeast mitochondria. Mol Cell Biol. 1994;14:4643–4652. doi: 10.1128/mcb.14.7.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyton RO, Duhl DMJ, Clarkson GHD. Protein export from the mitochondrial matrix. Trends Cell Biol. 1992;2:369–375. doi: 10.1016/0962-8924(92)90049-s. [DOI] [PubMed] [Google Scholar]
- Preuss M, Leonhard K, Hell K, Stuart RA, Neupert W, Herrmann JM. Mba1, a novel component of the mitochondrial protein export machinery of the yeast Saccharomyces cerevisiae. J Cell Biol. 2001;153:1085–1096. doi: 10.1083/jcb.153.5.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rep M, Grivell LA. MBA1 encodes a mitochondrial membrane-associated protein required for biogenesis of the respiratory chain. FEBS Lett. 1996;388:185–188. doi: 10.1016/0014-5793(96)00543-1. [DOI] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]
- Sanchirico ME, Fox TD, Mason TL. Accumulation of mitochondrially synthesized Saccharomyces cerevisiae Cox2p and Cox3p depends on targeting information in untranslated portions of their mRNAs. EMBO J. 1998;17:5796–5804. doi: 10.1093/emboj/17.19.5796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatz G. The protein import system of mitochondria. J Biol Chem. 1996;271:31763–31766. doi: 10.1074/jbc.271.50.31763. [DOI] [PubMed] [Google Scholar]
- Schneider A, Behrens M, Scherer P, Pratje E, Michaelis G, Schatz G. Inner membrane protease I, an enzyme mediating intramitochondrial protein sorting in yeast. EMBO J. 1991;10:247–254. doi: 10.1002/j.1460-2075.1991.tb07944.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Scotti PA, Urbanus ML, Brunner J, de Gier JW, von Heijne G, van der Does C, Driessen AJ, Oudega B, Luirink J. YidC, the Escherichia coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 2000;19:542–549. doi: 10.1093/emboj/19.4.542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Souza RL, Green-Willms NS, Fox TD, Tzagoloff A, Nobrega FG. Cloning and characterization of COX18, a Saccharomyces cerevisiae pet gene required for the assembly of cytochrome oxidase. J Biol Chem. 2000;275:14898–14902. doi: 10.1074/jbc.275.20.14898. [DOI] [PubMed] [Google Scholar]
- Steele DF, Butler CA, Fox TD. Expression of a recoded nuclear gene inserted into yeast mitochondrial DNA is limited by mRNA-specific translational activation. Proc Natl Acad Sci USA. 1996;93:5253–5257. doi: 10.1073/pnas.93.11.5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R, Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 Å. Science. 1996;272:1136–1144. doi: 10.1126/science.272.5265.1136. [DOI] [PubMed] [Google Scholar]
- Tzagoloff A, Dieckmann CL. PET genes of Saccharomyces cerevisiae. Microbiol Rev. 1990;54:211–225. doi: 10.1128/mr.54.3.211-225.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in Saccharomyces cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Weber ER, Dieckmann CL. Identification of the CBP1 polypeptide in mitochondrial extracts from Saccharomyces cerevisiae. J Biol Chem. 1990;265:1594–1600. [PubMed] [Google Scholar]