Abstract
Nucleation of microtubules is central to assembly of the mitotic spindle, which is required for each cell division. γ-Tubulin is a universal component essential for microtubule nucleation from centrosomes. To elucidate the mechanism of microtubule nucleation in budding yeast we reconstituted and characterized the yeast γ-tubulin complex (Tub4p complex) produced in insect cells. The recombinant complex has the same sedimentation coefficient (11.6 S) as the native complex in yeast cell extracts and contains one molecule of Spc97p, one molecule of Spc98p, and two molecules of Tub4p. The reconstituted Tub4p complex binds preformed microtubules and has a low nucleating activity, allowing us to begin a detailed analysis of conditions that enhance this nucleating activity. We tested whether binding of the recombinant Tub4p complex to the spindle pole body docking protein Spc110p affects its nucleating activity. The solubility of recombinant Spc110p in insect cells is improved by coexpression with yeast calmodulin (Cmd1p). The Spc110p/Cmd1p complex has a small sedimentation coefficient (4.2 S) and a large Stokes radius (14.3 nm), indicative of an elongated structure. The Tub4p complex binds Spc110p/Cmd1p via Spc98p and the Kd for binding is 150 nM. The low nucleation activity of the Tub4p complex is not enhanced when it is bound to Spc110p/Cmd1p, suggesting that it requires additional components or modifications to achieve robust activity. Finally, we report the identification of a large 22 S Tub4p complex in yeast extract that contains multimers of Spc97p similar to γ-tubulin ring complexes found in higher eukaryotic cells.
INTRODUCTION
The microtubule cytoskeleton plays essential roles in chromosome segregation, cellular organization, and vesicle trafficking in eukaryotic cells. Effective activities of the microtubule network require precise spatial and temporal control of assembly. In most cells, the centrosome is the primary microtubule-organizing center that nucleates and organizes microtubules. γ-Tubulin is a conserved protein that localizes to all centrosomes and is required for microtubule nucleation (Oakley et al., 1990; Horio et al., 1991; Stearns et al., 1991; Zheng et al., 1991; Joshi et al., 1992). The majority of cytoplasmic γ-tubulin is found in soluble complexes with a ring-like structure (γ-tubulin ring complex, γ-TuRC) as examined by electron microscopy (Zheng et al., 1995; Oegema et al., 1999; Murphy et al., 2001). Ring-like structures labeled with γ-tubulin are also observed in the pericentriolar material of centrosomes (Moritz et al., 1995; Vogel et al., 1997), suggesting a role for the γ-TuRC in microtubule nucleation from centrosomes.
In Saccharomyces cerevisiae, γ-tubulin or Tub4p exists in a stable complex with Spc97p and Spc98p (Knop et al., 1997; Knop and Schiebel, 1997). The γ-tubulin small complex (γ-TuSC) in higher eukaryotes contains orthologs of Tub4p, Spc97p, and Spc98p and forms the core subunit in the γ-TuRC (Martin et al., 1998; Murphy et al., 1998; Tassin et al., 1998; Oegema et al., 1999). Vertebrate γ-TuRCs have sedimentation coefficients ranging from 25 to 32 S (Stearns and Kirschner, 1994; Meads and Schroer, 1995; Moritz et al., 1998; Murphy et al., 1998, 2001; Oegema et al., 1999). In addition to multimers of γ-TuSC, at least five other components are found in each γ-TuRC (Fava et al., 1999; Gunawardane et al., 2000; Zhang et al., 2000; Murphy et al., 2001). These additional components are required for assembly of the γ-TuRC and/or recruitment of the γ-TuRC to the centrosomes. Recent electron microscopic tomography reconstruction studies propose a structural model for the Drosophila γ-TuRC in which a ring of repeating V-shaped units of γ-TuSCs are capped at one end with the remaining components (Moritz et al., 2000).
In vitro, γ-TuRCs can promote microtubule polymerization and cap microtubule minus ends (Zheng et al., 1995; Oegema et al., 1999; Wiese and Zheng, 2000). However, γ-tubulin alone has also been found to bind, cap, and promote microtubule polymerization (Li and Joshi, 1995; Leguy et al., 2000). Two models are currently proposed for the mechanism of microtubule nucleation (reviewed in Erickson, 2000). In the template model, the γ-TuRC acts as a template with a ring of 13 γ-tubulins on which the α/β protofilaments grow longitudinally and the microtubule end becomes flushed with the γ-TuRC surface. In the protofilament model, a single molecule or short filament of γ-tubulins stacked longitudinally to each other, unwinds from the γ-TuRC and acts as the seed from which α/β protofilaments grow both laterally and longitudinally. Recent electron microscopic studies provide evidence for the template model to explain how the γ-TuRC is structurally incorporated into the microtubule. Microtubules polymerized in the presence of γ-TuRCs or derived from animal centrosomes and yeast spindle pole bodies contain pointed ends that are capped similarly to the structure of purified γ-TuRC as analyzed by electron microscopic tomography (Byers et al., 1978; Moritz et al., 2000). Consistent with these findings, components of the γ-TuRC are also localized to the capped ends of microtubules (Keating and Borisy, 2000; Zhang et al., 2000). However, the biochemical mechanism of nucleation remains unclear.
In S. cerevisiae, microtubules are only found to emanate from the spindle pole body (SPB), which is the sole microtubule organizing center. The substructures of the SPB include the outer plaque, which organizes cytoplasmic microtubules; the central plaque, which is embedded in the nuclear envelope; and the inner plaque, which organizes nuclear microtubules. All three components of the Tub4p complex (Tub4p, Spc97p, and Spc98p) are localized to the outer and inner plaques and play key roles in microtubule nucleation and spindle assembly (Rout and Kilmartin, 1990; Sobel and Snyder, 1995; Marschall et al., 1996; Spang et al., 1996a; Knop et al., 1997). Yeast exhibits a closed mitosis in which the nuclear membrane remains intact throughout the cell cycle, resulting in independent regulation of cytoplasmic and nuclear microtubule polymerization. This simpler regulatory network together with the extremely well-defined SPB offers many advantages for the studies of microtubule nucleation in budding yeast. A combination of genetic and two-hybrid analyses shows that the Tub4p complex is anchored to the inner plaque by interaction with Spc110p (Knop and Schiebel, 1997; Nguyen et al., 1998). Spc110p is a major SPB protein that has a calmodulin binding domain at the C terminus embedded in the SPB core, and a Tub4p complex binding domain at the N terminus pointed toward the microtubule ends (Geiser et al., 1993; Kilmartin et al., 1993; Kilmartin and Goh, 1996; Spang et al., 1996b; Sundberg et al., 1996). The two domains are separated by a coiled coil rod that acts as a spacer between the central and inner plaques (Kilmartin et al., 1993). The human ortholog of Spc110p is kendrin, an alternative splice variant of pericentrin that contains a C-terminal extension, including a calmodulin-binding region (Flory et al., 2000). Pericentrin has previously been shown to play a role in recruiting the γ-TuRC to centrosomes (Dictenberg et al., 1998), suggesting that the mechanism of assembling the nucleation machinery at the centrosome may be conserved between yeast and human.
Centrosome-dependent microtubule polymerization in higher eukaryotes involves recruitment of the γ-TuRC to the centrosome and activation of the nucleation machinery that results in aster formation (Felix et al., 1994; Stearns and Kirschner, 1994; Moritz et al., 1998; Schnackenberg et al., 1998). Studies in Drosophila indicate that although the large γ-TuRC has robust nucleating activity in vitro, the γ-TuSC has weak activity (Oegema et al., 1999; Gunawardane et al., 2000). The fact that a γ-TuRC–like complex has not been found in yeast raised the question as to whether the Tub4p complex has nucleating activity. To address these questions, we have reconstituted the Tub4p complex and measured its in vitro microtubule nucleating activity. Because microtubules are only found at the SPB and Spc110p is the only component known to directly link the Tub4p complex to the nuclear side of the SPB, we further postulate that anchoring the Tub4p complex to the SPB may regulate its nucleating activity. We therefore characterized the assembly and function of the Tub4p complex when bound to Spc110p. Our results indicate that linking to Spc110p does not enhance the nucleation activity of the Tub4p complex. This result led to a careful examination of Tub4p complexes in yeast extracts and identification of a large complex that may be the yeast version of the higher eukaryotic γ-TuRC.
MATERIALS AND METHODS
Baculovirus Construction
Recombinant proteins were produced using the Bac-to-Bac baculovirus/insect cell expression system (Invitrogen, Carlsbad, CA). Yeast genes were inserted into the baculovirus vectors pFastbac or pFastbacDual (Table 1). Clones of SPC97, SPC98, TUB4, SPC110, or CMD1 were derived from plasmids previously described (Nguyen et al., 1998). Plasmids carrying SPC29 and SPC42 were from S. Francis (University of Washington, Seattle, WA). A plasmid carrying tub4-34 was a gift from R. Jeng and T. Stearns (Stanford University, Palo Alto, CA).
Table 1.
Expression vectors
| Protein | Parent vector | Method of construction |
|---|---|---|
| Cmd1p | pFastBac | Subclone |
| Spc29p-Flag | pFastBac | PCR with epitope taga |
| Spc42p-2PY | pFastBac | PCR with epitope taga |
| Spc97p | pFastBac | Subclone |
| Spc97p-GST | pFastBac | PCR and subcloneb |
| 10XHis-Spc97p | pFBNHis10c | Subclone |
| Spc98p | pFastBac | Subclone |
| GST-Spc98p551–846 | pGEX2T | Subclone |
| Spc110p-GST | pFastBac | PCR and subcloneb |
| GST-Spc110p/Cmd1p | pFastBac Dual | Subclone and PCRd |
| GST-Spc110p1–220 | pMIT77c | PCRe |
| Tub4p | pFastBac | Subclone |
| Tub4-34p | pFastBac | Subclone |
Gene was amplified by PCR with primers encoding the epitope tag.
GST was first subcloned into pFastBac to make pFastBac-GST. The 3′ end of the yeast gene was amplified using primers that removed the stop codon and added a PstI site. The PCR fragment was subcloned into the pFastBac-GST vector at the PstI site. The remainder of the gene was then added.
The vector pFBNHis10 is pFastBac containing a 10×His tag at the N terminus, and pMIT77 is pFastBac containing an N-terminal GST (both were gifts of K. Kaplan and P. Sorger, Massachusetts Institute of Technology, MA).
Both CMD1 and SPC110 were cloned into pFastBacDual. GST was generated by PCR and inserted at the 5′ end of SPC110.
Nucleotides 1–660 of the SPC110 open reading frame were amplified using a 3′ primer encoding a stop codon.
Antibodies
Anti-Spc98p antibodies were generated against a glutathione S-transferase (GST)-Spc98p fusion protein containing residues 551–846 of Spc98p. The fusion protein was expressed in Escherichia coli, enriched on the basis of its insolubility in 6 M urea, and gel purified before injection into chickens (Aves Labs, Tigard, OR). Anti-Spc97p antibodies were produced in chickens by using a 10XHis-Spc97p (full-length) fusion protein expressed in insect cells. The insoluble fusion protein was enriched on the basis of its insolubility in 250 mM KCl and gel purified. Anti-PY antibodies were generated from tissue culture supernatant of mouse monoclonal cell line AK1310 (gift of R. Deshaies, California Institute of Technology, Pasadena, CA). Anti-Spc110p and anti-Cmd1p antibodies have been described elsewhere (Geiser et al., 1991, 1993). Anti-Tub4p was a gift of T. Stearns (Stanford University), monoclonal anti-FLAG was a gift of G. Zhu (University of Washington, Seattle, WA), and rabbit polyclonal anti-GST, mouse monoclonal anti-MYC, and mouse monoclonal anti-hemagglutinin (HA) antibodies were from Santa Cruz Biotechnology (Santa Cruz, CA).
Baculovirus Expression
We used Sf9 or Hi5 insect cell lines for baculovirus expression. Recombinant baculoviruses were produced from Sf9 cells as recommended by the manufacturer's instructions (Invitrogen). For recombinant proteins, Hi5 cells grown on plates were used for small-scale studies (5–10 × 106 cells), and Sf9 cells grown in shaking flasks were used for large purification preps (>2 × 108 cells). Cells were coinfected with the appropriate baculoviruses for 48–72 h. The amount of virus was optimized for protein expression. Infected cells were harvested and spun down at 500 × g for 10 min. Cell pellets were gently washed once with phosphate-buffered saline, repelleted, and then either processed for protein purification or frozen in liquid nitrogen and stored at −80°C.
Protein Purification and GST-Copurification Assay
We purified the Tub4p complex by using two methods. In method 1, Tub4p and Spc98p were copurified with Spc97p-GST from cells coinfected with the three viruses and the GST domain subsequently removed by cleavage. Two to three volumes of HB100 (40 mM K-HEPES, pH 7.5, 100 mM KCl, 1 mM EGTA, 1 mM MgCl2, 0.1 mM GTP) + 1 mM dithiothreitol (DTT) + 1 mM phenylmethylsulfonyl fluoride (PMSF) + 10 μg/ml pepstatin A + a 1:1000 dilution of a stock of three protease inhibitors (10 mg/ml each of chymostatin, leupeptin, and aprotinin [CLA]) were added to frozen cell pellets from 1 liter of infected culture. The pellets were then quickly thawed at room temperature (RT) and the cell suspension was sonicated twice with a tip sonicator. NP-40 (1% vol/vol) was then added to the suspension and incubated on ice for 30–45 min. The lysate was centrifuged at 40,000 × g for 40 min in a Beckman Coulter JA25.50 rotor, and the cleared supernatant loaded onto a column (2 ml) of glutathione-Sepharose (Amersham Biosciences, Piscataway, NJ) equilibrated in HB100. The column was washed with at least 20 column volumes of HB100 + 1 mM DTT, but minus protease inhibitors. The Spc97p/Spc98p/Tub4p complex was released by incubation for 4 h at 4°C with GST-PreScission protease (Amersham Biosciences) suspended in HB100 + 1 mM DTT. Protease cleavage results in the addition of six amino acid residues (LEVLFQ) after the last codon of Spc97p. Samples were eluted in HB100 + 1 mM DTT + 1/10,000 dilution of CLA, and fractions containing protein were identified by dot blotting on nitrocellulose. Peak fractions were pooled and concentrated with Microcon or Centricon filters (Millipore, Bedford, MA). Sucrose was added to a final concentration of 10% (wt/vol), the protein solution was frozen in liquid nitrogen and stored at −80°.
In method 2, Spc97p, Spc98p, and Tub4p were copurified with GST-Spc110p1–220 from cells coinfected with the four viruses. The large complex [GST-Spc110p1–220 + Tub4p complex] was first affinity purified with glutathione-Sepharose beads as described above. Then the entire large complex was eluted with 10 mM glutathione in 25 mM Tris buffer, pH 7.8, 50 mM KCl, 1 mM MgCl2. Fractions were pooled and applied to a 1.7-ml POROS Q/M anion exchange column run on the BioCAD Sprint Perfusion Chromatography System (Applied Biosystems, Foster City, CA) to separate GST-Spc110p1–220 from the Tub4p complex. Samples were eluted with a 0–1 M KCl gradient in 25 mM Tris buffer, pH 7.8, 1 mM MgCl2 and analyzed by SDS-PAGE. Peak fractions were pooled, concentrated, and dialyzed into HB100 + 10% glycerol + 1 mM DTT + 1 × 10−5 dilution of CLA protease inhibitors, followed by freezing in liquid nitrogen and storage at −80°C. The usual yield of method 1 is 900 μg of Tub4p complex per 1 liter of infected cell culture, whereas method 2 can produce up to 3 mg of Tub4p complex per 1 liter of culture. Protein concentration was determined either using bicinchoninic acid (Sigma Chemical, St. Louis, MO), or by comparison with known amounts of bovine serum albumin (BSA) after separation on SDS-PAGE and staining with Coomassie blue. Protein bands were scanned using a Wratten filter (#0.2) on an ArcusII, Agfa scanner. NIH Image software was used to quantify band intensities.
To purify an Spc110p-GST/Cmd1p complex, frozen cell pellets were quickly lysed at RT in HB300 (HB100, except with 300 mM KCl) plus 4 mM DTT, 1 mM PMSF, 20 μg/ml pepstatin, and a 1/500 dilution of CLA. NP-40 was added to a final concentration of 1% and the sample incubated on ice for 30 min. The lysate was initially spun at 40,000 × g for 40 min, and then the supernatant was respun at 80,000 × g for 10 min in a Sorvall F28/36 rotor. The cleared supernatant was loaded onto a glutathione-Sepharose column and washed with five column volumes of HB300, five column volumes of HB200, and at least 10 column volumes of HB100. Samples were either eluted with glutathione (producing Spc110p-GST/Cmd1p) or by protease treatment (producing Spc110p/Cmd1p) as described above.
In the GST-copurification assay, 5 × 106 infected cells were processed per sample. Different combinations of baculoviruses were coinfected with a virus expressing GST-tagged protein and cell lysates prepared in HB100 and spun at 14,000 × g in a microfuge. Incubation of the cleared supernatant with glutathione beads and elution with glutathione in HB100 were both done in microfuge tubes on a nutator for 30 min each at 4°C. The eluted samples were analyzed by Western blot to look for their copurification with the GST-tagged protein.
Estimating Kd for Spc110p Binding to Tub4p Complex
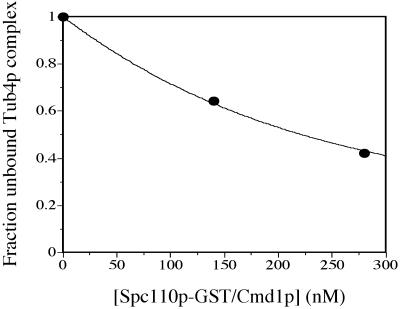
A fixed concentration of 138 nM purified Tub4p complex was incubated at RT for 30 min with various concentrations of purified Spc110p-GST/Cmd1p in a constant volume of HB100 containing 10% glycerol, 2 mM DTT, 0.1% NP-40, and 0.3% casamino acids. Each sample was then added to an equal volume of glutathione-Sepharose beads equilibrated in HB100 with 2 mM DTT, 0.1% NP-40, and 0.3% casamino acids, and incubated for another 20 min at RT. Samples were centrifuged for 1 min at 2000 × g; the supernatant was carefully removed and analyzed by SDS-PAGE. The relative amount of unbound Tub4p complex in the supernatant was determined by quantifying the intensity of the Tub4p bands on a Coomassie-blue stained gel. The fraction of unbound [Tub4p complex] shown in Figure 5 = (intensity of unbound Tub4p at a given [Spc110p]/intensity of unbound Tub4p in the absence of Spc110p).
Figure 5.
Spc110p interacts with the Tub4p complex with modest affinity. Fitted curve for binding of purified Spc110p/Cmd1p to the Tub4p complex. Fraction of [unbound Tub4p complex] is drawn as a function of total [Spc110p-GST/Cmd1p]. Total [Tub4p complex] is 138 nM at all concentrations of Spc110p-GST. The Kd estimated by a linear regression fit of the data is 150 nM (see MATERIALS AND METHODS).
The binding equation for Spc110p and Tub4p complex is as follows:
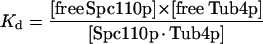 |
where [free Spc110p] = [total Spc110p] − [Spc110p · Tub4p], and [Spc110p · Tub4p] = [total Tub4p] − [free Tub4p]. By solving for [free Tub4p] as a function of [total Spc110p] and [total Tub4p] and using linear regression (SigmaPlot) to fit the data, we estimated the Kd for binding to be 150 nM.
Sucrose Gradient Sedimentation and Gel Filtration Chromatography
Sedimentation coefficients of proteins were determined by sucrose gradient centrifugation. To generate a larger complex containing Spc110p/Cmd1p and the Tub4p complex, 200 nM purified Spc110p was incubated with 700 nM purified Tub4p complex for 30 min at RT before loading on the gradient. Sucrose gradients (10–40%) were generated by allowing five steps of equal volume to diffuse into continuous gradients at RT and chilled at 4°C before use. Gradient buffers were either HB100 or TB100 (see “Yeast Extract” below) with 1 mM DTT, 0.1 mM PMSF, 1 μg/ml pepstatin A, and a 1 × 10−4 dilution of CLA. Each experimental run was calibrated against protein standards that were loaded onto the same gradient as the tested sample. Protein standards BSA (4.4 S), aldolase (7.4 S), catalase (11.3 S), and thyroglobulin (19.4 S) (all from Sigma Chemical) were dissolved in either HB100 or TB100. Between 100 and 200 μg of total protein was loaded onto each gradient and centrifuged at 210,000 × g for 5 h at 4°C in a TLS55 rotor. Thirteen to fourteen fractions (150 μl) were collected from the top with cut off pipette tips. Recombinant proteins and standards were analyzed directly by SDS-PAGE followed by staining with Coomassie blue. When yeast extracts were analyzed, protein fractions were first precipitated with 10% trichloroacetic acid (TCA) and then analyzed by Western blot. Protein gel bands were quantified and peak fractions identified.
The Stokes radii of protein complexes were estimated by gel filtration chromatography on a Superose-6 column (Amersham Biosciences) by using the Biocad sprint perfusion chromatography system. Each experimental run was calibrated against standards: dextran (void volume), thyroglobulin (8.6 nm), ferritin (6.1 nm), catalase (5.2 nm), aldolase (4.6 nm), and BSA (3.5 nm). The column buffer was HB100 containing 1 mM DTT and 0.1 mM GTP. Peak fractions (300 μl) identified by absorbance at 230 nm were precipitated with 10% TCA and analyzed by Western blots. The molecular mass of each protein complex was estimated using the method of Siegel and Monty (1966).
Nucleation Assay
Reagents.
BRB80 is 80 mM K-piperazine-N,N′-bis(2-ethanesulfonic acid) buffer, pH 6.9, 1 mM EGTA, and 1 mM MgCl2; GTP stock (Roche Applied Sciences, Indianapolis, IN) is 100 mM in 100 mM MgCl2; HB100gly is HB100 containing 10% glycerol. All assay buffers were made at one time, distributed to small aliquots, frozen, and stored at −20°C. The background microtubule polymerization activity was similar when the same buffer was assayed within a year of storage. Tubulin was purified from bovine brain (Mitchison and Kirschner, 1984) and labeled with tetramethylrhodamine as described (Hyman et al., 1991).
Assay.
The nucleation assay was optimized using short rhodamine microtubule seeds (<2 μm) generated by polymerizing microtubules in the presence of guanylyl-(α,β)-methylene-diphosphonate (Hyman et al., 1991). The concentration of rhodamine tubulin used in the assay was chosen because it elongated >80% of guanylyl-(α,β)-methylene-diphosphonate seeds in 4 min and gave the lowest background of polymerization in the absence of seeds. A 5-μl total reaction included 2.5 μl of HB100gly alone or purified yeast proteins diluted in HB100gly, plus a 2.5-μl solution of 2.0 mg/ml rhodamine tubulin, 1 mM GTP, and 1× BRB80 (from a 5× stock). Polymerization was achieved by incubation for 4 min at 37°C, and stopped by adding 50 μl of prewarmed 1% gluteraldehyde (Ted Pella, Redding, CA) in BRB80. Samples were gently mixed with a cut off pipette tip, incubated for 3 min at RT, and then 1 ml of ice-cold BRB80 was added to each sample, the tubes gently inverted, and transferred to ice. Various microtubule dilutions (50 μl) were each pelleted in TLS55 centrifuge tubes at 173,000 × g for 8 min, 4°C, through 0.5 ml of BRB80 and a cushion of 0.75 ml of 15% glycerol in BRB80 onto poly-lysine–coated 5-mm coverslips placed on Teflon adaptors. After centrifugation, coverslips were carefully removed, postfixed in methanol at −20°C, briefly air-dried, mounted on slides with Citifluor (Ted Pella), and sealed with nail polish. Rhodamine microtubules were viewed with a 100× objective on a fluorescence microscope (Diastar, Leica, NY). Magnified images were projected onto a silicon-intensified target camera (Hamamatsu C2400-8, Bartels and Stout, WA), displayed on a high-resolution video monitor, and recorded on VHS tapes. Microtubules were counted in 20–40 random video fields, and specific images were captured on the computer by using NIH Image software.
Microtubule Copelleting Assay
Taxol-stabilized microtubules were made by incubating bovine tubulin in 1× BRB80 plus 10% dimethyl sulfoxide, 1 mM GTP, and 1 mM DTT for 30 min at 37°C, and stopping the reaction with 9 volumes of 20 μM taxol (Sigma Chemical) in BRB80. The solution was cleared by centrifugation in a microfuge at 5000 × g to remove aggregates. The remaining microtubules were collected by pelleting through a 33% glycerol/taxol/DTT/BRB80 cushion in a TLA100-2 rotor for 20 min at 200,000 × g, RT. The pellet was rinsed once and resuspended in 10 μM taxol in BRB80 + 1 mM DTT. Tubulin concentration was quantified using the Bradford assay (Bio-Rad reagent; Bio-Rad, Hercules, CA).
For the copelleting assay, all tested samples were subjected to a prespin of 120,000 × g for 2 min, at 2°C in a TLA45 rotor. Various concentrations of taxol stabilized microtubules were incubated for 30 min at RT with either 10 μM taxol/DTT/BRB80 buffer, purified Tub4p complex, or purified bacterial GST in the presence of 0.5 mg/ml BSA prepared in taxol/BRB80. The sample was then pelleted through a 15% glycerol/taxol/BRB80 cushion at 109,000 × g for 10 min in a TLA100 rotor at 25°C. The supernatant was removed from the microtubule pellet and then both supernatant and pellet were analyzed by Western blotting.
Yeast Strains, Cell Extracts, and Immunoprecipitation
SPC97 was tagged in either CRY1 (MATa) or CRY2 (MATα), which are isogenic yeast strains with the genotype ade2–1oc can1-100 his3-11,15 leu2-3,112 trp1-1 ura3-1. We used the polymerase chain reaction (PCR)-based gene targeting method originally described in Wach et al. (1997) and as modified in http://depts.washington.edu/∼yeastrc/fm_home3.htm. pFA6a-13MYC-kanMX6 was used as the PCR template to create SPC97-13MYC cells (DVY52) and pFA6a-3HA-kanMX6 was used to create SPC97–3HA cells (DVY51) (Bahler et al., 1998). To make the SPC97-13MYC/SPC97-3HA diploid (DVY53), DVY52 (MATα), and DVY51 (MATa) cells were mated to each other and the resulting diploids tested for their ability to sporulate. Heterozygous SPC97-13MYC/SPC97 (DVY54) and SPC97-3HA/SPC97 (DVY55) strains were generated by crossing DVY52 or DVY51 to CRY1 or CRY2, respectively. In all cases, correct protein expression was confirmed by Western blots with anti-MYC and/or anti-HA antibodies. All yeast strains appear to grow normally on plates and in culture at 30°C.
To make cell extracts, all strains were grown in YPD at 30°C to a density of 2 × 107 cells/ml. Cells were pelleted and washed once with water, stored at −80°C, or resuspended in the appropriate lysis buffer. For extracts in TB100 (20 mM Tris buffer, pH 7.5, 100 mM NaCl, 10 mM EDTA, 2 mM EGTA), buffer and lysis conditions of a 40-ml culture cell pellet were essentially as described in Knop et al. (1997), except 5% glycerol was omitted in the buffer and the final protease inhibitors included 1 mM PMSF and a 1/1000 dilution of CLA. An extract in HB100 buffer was made by first resuspending a cell pellet from a 40-ml culture in 500 μl of HB (HB100 without KCl) plus 1 mM EDTA, 2 mM DTT, 1 mM PMSF, and 1/1000 dilution of CLA. Cells were vortexed with glass beads until >90% of cells were lysed. Cell lysate was retrieved to a fresh tube, and an additional 250 μl of HB was added to the glass beads. The entire sample was spun for 10 s in a microfuge to remove cell debris, and then for another 20 min at 13,000 × g, 4°C. The pellet was resuspended in 100–200 μl of HB100 plus 1 mM EDTA, 2 mM DTT, 1 mM PMSF, 1/1000 dilution CLA, and 1% NP-40 and incubated for 30 min on ice. Insoluble material was removed by centrifugation at 100,000 × g for 15 min at 4°C. The final supernatant was saved for sucrose gradient centrifugation or immunoprecipitation. Protein concentrations were measured using bicinchoninic acid.
To immunoprecipitate Spc97p-13MYC, 4 μg of anti-MYC antibodies was added per 100 μg of cell extract and incubated at 4°C for 1 h, followed with another hour of incubation in protein-G Sepharose (Amersham Biosciences) at 4°C. Similar protocols were used for immunoprecipitating Spc97p-3HA, except 8 μg of anti-HA antibodies was used per 100 μg of cell extract. Precipitates on beads were pelleted at 2000 × g for 1 min and washed three times with HB100 containing 1 mM DTT, 1/1000 dilution of CLA, and 0.1% NP-40. Precipitates were resuspended in 2× sample buffer and analyzed by Western blots.
RESULTS
Interactions of Tub4p Complex with Spc110p
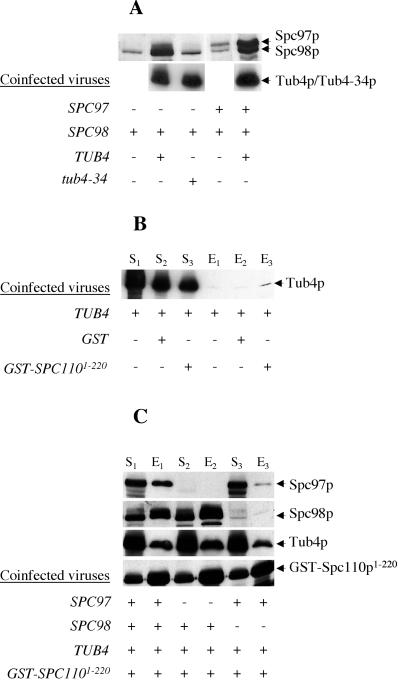
Previous studies by Knop et al. (1997) indicated that Tub4p exists predominantly in complex with Spc97p and Spc98p in yeast cells. We and others have subsequently shown that these three components are anchored to the nuclear side of the SPB via interactions with the core component Spc110p (Knop and Schiebel, 1997; Nguyen et al., 1998). Genetic analyses and two-hybrid assays indicated that the interactions between Spc98p and the N terminus of Spc110p are direct and robust, whereas Spc97p and Tub4p depend on Spc98p for their interactions with Spc110p (Nguyen et al., 1998). To better understand these diverse interactions, we tested the ability of components of the Tub4p complex to bind directly to the first 220 residues of Spc110p fused to GST. We infected insect cells with different combinations of recombinant baculoviruses and tested which components copurify with GST-Spc110p1–220. Initial experiments showed that coexpression of Tub4p greatly increases the production of soluble Spc98p and soluble Spc97p (Figure 1A), so we included Tub4p in all subsequent experiments. Tub4p itself associates very poorly with GST-Spc110p1–220 (Figure 1B). When Tub4p, Spc98p, and Spc97p are expressed in the starting supernatant, all three proteins copurified with GST-Spc110p1–220 (Figure 1C). In the absence of Spc98p, a barely detectable amount of Spc97p and a low level of Tub4p copurify with GST-Spc110p1–220. In contrast, the expression of only Spc98p and Tub4p allows efficient copurification of both components with GST-Spc110p1–220. We conclude that Spc98p is the primary component interacting with Spc110p.
Figure 1.
Spc98p binds most robustly to the N-terminal domain of Spc110p. Insect cells were infected with different combinations of viruses as indicated. (A) Production of soluble Spc97p and Spc98p is greatly enhanced upon coexpression with wild-type Tub4p. Western blots show the cleared lysates derived from insect cells expressing Spc98p or Spc98p and Spc97p with and without the coexpression of Tub4p or Tub4–34p. (B) Tub4p interacts poorly with GST or GST-Spc110p1–220. Tub4p was tested for its ability to bind glutathione-Sepharose beads, GST, or GST-Spc110p1–220. The Western blot shows the starting cleared lysates (S) (0.25% of total) and the eluates (E) (5% of total) from the glutathione-Sepharose beads. (C) An Spc98p/Tub4p complex binds GST-Spc110p1–220 as efficiently as an Spc98p/Tub4p/Spc97p complex. Herein, we loaded 0.5% of S and 10% of E. Antibodies used in all Westerns include anti-Spc97p, anti-Spc98p, anti-Tub4p, and anti-GST.
We also characterized a mutant form of Tub4p, Tub4–34p, with the mutations F244S and Y247C (Jeng and Stearns, personal communication). tub4-34 cells are defective in microtubule nucleation and the SPB is deficient in both Tub4-34p and Spc98p (Marschall et al., 1996). Consistent with the in vivo phenotypes, we observed that even although expressed in insect cells at the same level as wild-type Tub4p, Tub4-34p fails to enhance the soluble expression of Spc98p, suggesting a defective interaction between the two components (Figure 1A). Finally, when coexpressed with GST-Spc110p1–220, only the combination of Spc98p and Tub4p, but not Spc98p and Tub4-34p, copurifies with GST-Spc110p1–220 (our unpublished data).
Reconstitution and Purification of Tub4p Complex
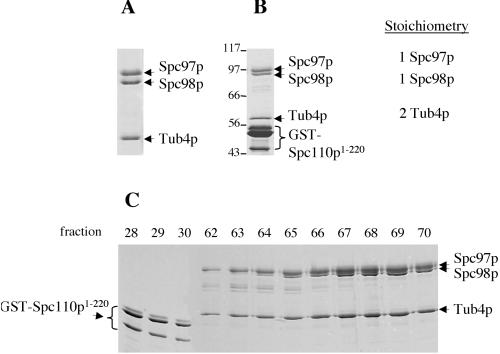
To further understand the assembly and function of the three components in the Tub4p complex, we reconstituted and purified the Tub4p/Spc97p/Spc98p complex expressed from insect cells by using two methods. In method 1, the complex was purified using Spc97p-GST (Figure 2A); in method 2, the complex was purified on the basis of its ability to bind to GST-Spc110p1–220 (Figure 2B). GST-Spc110p1–220 was subsequently removed by anion exchange chromatography (Figure 2C). Both methods yield complexes containing the three proteins in a stoichiometry of 1 Spc97p:1 Spc98p:2 Tub4p. The minor bands detected by Coomassie blue staining (Figure 2B) are breakdown products of Spc98p. Tubulin was not detected in either purified preparation by Western blot with DM1α antibodies, which recognize insect α-tubulin (our unpublished data). We also observed that excess Spc97p was not purified from method 1, consistent with the fact that stable expression of Spc97p-GST is limited by the expression of Tub4p. Generally, method 2 produces about three times more Tub4p complex and has a larger purification capacity, but method 1 is quicker.
Figure 2.
Two methods of generating purified Tub4p complex reconstituted from insect cells. (A) Method 1: the Tub4p complex was isolated by copurification with Spc97p-GST followed by cleavage of GST. (B) Method 2: the Tub4p complex was copurified with GST-Spc110p1–220. Numbers indicate positions of molecular weight standards. In both methods, a similar stoichiometry of 1 Spc98p:1 Spc97p:2 Tub4p was observed. (C) GST-Spc1101–220 is purified away from the Tub4p complex by anion exchange chromatography in method 2. Numbers above panel indicate fractions collected from the anion exchange column. (A–C) SDS-PAGE separated protein samples stained with Coomassie blue.
The molecular mass of the Tub4p complex was determined by sucrose gradient centrifugation and gel filtration chromatography. Purified recombinant Tub4p complex migrates predominantly at a peak of 11.6 S in a sucrose gradient (Figure 3B). By gel filtration, the complex has a Stokes radius of 7.1 nm (Table 2). Based on these values, we estimate the molecular mass of the reconstituted Tub4p complex to be 340 kDa. This number closely matches the predicted molecular mass (300 kDa) of a Tub4p complex based on the amino acid sequences and stoichiometry of the components.
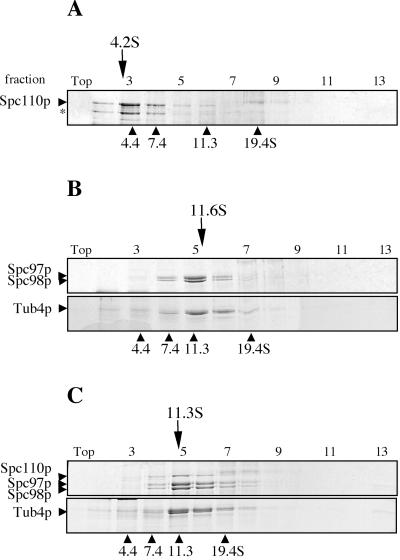
Figure 3.
Sucrose gradients of reconstituted Tub4p complex and Spc110p/Cmd1p purified from insect cells. (A) Spc110p/Cmd1p alone. Asterisk (∗) indicates a breakdown product of Spc110p that was confirmed with anti-Spc110p Western blot. (B) Tub4p complex. (C) Tub4p complex + Spc110p/Cmd1p. In A–C, fractions were analyzed by SDS-PAGE and the gels stained with Coomassie blue. Positions of the peak of the tested samples are indicated above each panel, and positions of internal protein standards are indicated beneath.
Table 2.
Physical characterization of Tub4p complex ± Spc110p/Cmd1p
| Sedimentation coefficient (S) | Stokes radius (nm) | Estimated molecular mass (Da) | |
|---|---|---|---|
| Spc110p/Cmd1p | 4.2 | 14.3 | 250,000 |
| Tub4p complex | 11.6 | 7.1 | 340,000 |
| Spc110p/Cmd1p + Tub4p complex | 11.3 | >14.4 | >620,000 |
Reconstitution of Tub4p Complex with Full-Length Spc110p
Because we were interested in what effects Spc110p has on the organization and nucleation activity of the Tub4p complex, we reconstituted the Tub4p complex together with full-length Spc110p. However, unlike GST-Spc110p1–220, the soluble expression of Spc110p-GST or Spc110p is much lower making copurification with the Tub4p complex inefficient. We separately purified Spc110p and the Tub4p complex and reconstituted them just before biochemical analyses. One way to increase the solubility of Spc110p-GST without disrupting the GST/glutathione-Sepharose binding was to prepare insect cell extract at a higher salt concentration of 300 mM KCl. During purification, the concentration of salt was reduced to 100 mM KCl to allow association with the Tub4p complex. Furthermore, we found that coexpressing yeast calmodulin (Cmd1p) with Spc110p-GST greatly enhances its soluble expression and yield (Figure 4), perhaps by preventing aggregation. Calmodulin binds the C terminus of Spc110p and in yeast cells, calmodulin is required for proper assembly of Spc110p multimers (Sundberg et al., 1996).
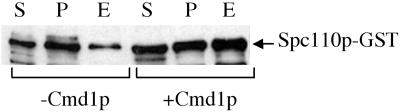
Figure 4.
Yield of Spc110p-GST purified from insect cells is increased by the coexpression of yeast calmodulin, Cmd1p. Western blot with anti-GST shows expression of Spc110p-GST in the starting cleared lysates (S), in the insoluble pellets (P), and collected in the eluates after glutathione-Sepharose beads (E). Loading was equal in S and P samples and 10-fold more in E. Quantitation of the intensity of Spc110p-GST bands shows that fivefold more Spc110p-GST is isolated (E) in the presence of calmodulin.
Finally, we used a modified GST-copurification assay to estimate the Kd for binding of purified Spc110p/Cmd1p to the Tub4p complex. A fixed amount of Tub4p complex was incubated with different concentrations of Spc110p-GST/Cmd1p and then glutathione-Sepharose beads were added. The unbound Tub4p complex was separated from that bound to Spc110p-GST/Cmd1p by centrifugation and analyzed by SDS-PAGE. We measured the amount of unbound Tub4p complex that remains in the supernatant at increasing concentrations of Spc110p-GST/Cmd1p (Figure 5). Using this assay, the Kd is estimated to be 150 nM.
Characterization of Complex Containing [Spc110p/Cmd1p + Tub4p Complex]
To determine whether Spc110p can organize the Tub4p complex into a higher order structure such as the γ-TuRC, we estimated the size of purified Spc110p/Cmd1p alone and a reconstituted complex containing Spc110p/Cmd1p and the Tub4p complex. Spc110p/Cmd1p migrates at 4.2 S in a sucrose gradient, but has a large Stokes radius of 14.3 nm by gel filtration (Figure 3A and Table 2). Based on these values, Spc110p/Cmd1p has a molecular mass of 250 kDa and is a dimer of the Spc110p/Cmd1p complex (predicted molecular mass based on amino acid sequence is 256 kDa).
To confirm reconstitution of a large complex containing Spc110p/Cmd1p and Tub4p complex, we tested for comigration of all components in sucrose gradient sedimentation and gel filtration chromatography. After a short preincubation of purified Spc110p/Cmd1p with the Tub4p complex, all of Spc110p/Cmd1p was observed to shift from the 4 S position and cosediment with the Tub4p complex to a peak of 11.3 S (Figure 3C). This sedimentation coefficient is less than half that of the γ-TuRC in higher eukaryotes. Unfortunately, we could not determine the Stokes radius of this large complex as it fractionates in the void volume (>14.4 nm) of the gel filtration column (Table 2).
Interactions of Spc110p with Other SPB Core Components
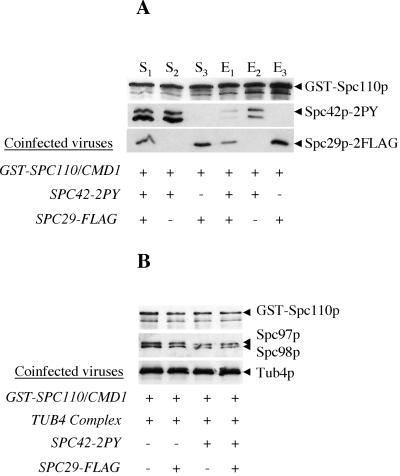
Electron microscopy studies have indicated that the N- and C-terminal domains of Spc110p are separated by a coiled coil rod that acts as a spacer between the central and inner plaques of the SPB. The C terminus of Spc110p is tightly attached to the core of the SPB as its N terminus anchors the Tub4p complex (Kilmartin and Goh, 1996; Sundberg et al., 1996; Knop and Schiebel, 1997; Nguyen et al., 1998). Current models propose that the SPB core, consisting of a crystalline array of Spc42p, acts as a template on which docking proteins such as Spc110p are recruited and assembled (Adams and Kilmartin, 1999). Extraction of the SPB generates a subcomplex of Spc110p/Cmd1p and two other proteins, Spc29p and Spc42p (Elliott et al., 1999). However, there have been conflicting thoughts about whether Spc110p can interact with both Spc29p and Spc42p or whether Spc29p mediates the interactions of Spc42p with Spc110p (Adams and Kilmartin, 1999; Elliott et al., 1999). We are interested in determining whether the interaction of Spc110p with these two other core components at the C terminus can affect its self-assembly and its organization of the Tub4p complex at the N terminus. We first tested whether Spc110p can bind both Spc29p and Spc42p by using the GST-copurification assay with recombinant proteins expressed in insect cells. For these experiments, we tagged the N terminus of Spc110p with GST to allow unhindered interactions at the C terminus. GST-Spc110p/Cmd1p interacts independently with either Spc29p-Flag or Spc42p-2PY (Figure 6A). However, the coexpression of Spc29p-Flag and Spc42p-2PY appears to hinder the binding of Spc42p-2PY to Spc110p (Figure 6A). One interpretation of these results is that Spc29p and Spc42p have an overlapping binding site on Spc110p and neither mediates the binding of the other to Spc110p. Alternatively, Spc29p binds to Spc42p and masks the binding site of Spc42p for Spc110p. However, we have not been able to detect any significant interaction between Spc29p and Spc42p when similar copurification experiments were tested using GST-Spc29p and Spc42p-2PY (GST-copurification) or Spc29p-Flag and Spc42p-2PY (anti-PY immunoprecipitation) (our unpublished data).
Figure 6.
Spc110p/Cmd1p can interact with Spc42p or Spc29p independently of each other. (A) GST-Spc110p/Cmd1p was coexpressed in insect cells with either Spc42p-2PY or Spc29p-FLAG or both. S is the starting cleared lysate and E is the eluate from glutathione-Sepharose beads. Protein samples were analyzed by Western blot with anti-GST, anti-PY, and anti-FLAG antibodies. The lower migrating band detected by anti-GST is a GST-Spc110p breakdown. (B) Interactions at the C terminus of Spc110p with Spc42p or Spc29p do not seem to affect its ability to interact with the Tub4p complex. Insect cells were infected with different combinations of viruses as indicated, and the Tub4p complex was tested for its binding to GST-Spc110p/Cmd1p. Note that the Tub4p complex is present at a 10-fold molar excess to GST-Spc110p/Cmd1p in each cleared crude lysate (our unpublished data). Eluates were analyzed by Western blot.
We next tested whether the binding of GST-Spc110p/Cmd1p to either Spc29p-Flag and/or Spc42p-2PY affects its ability to bind the Tub4p complex. In a coinfection/copurification experiment, we found that the interaction of Spc29p-Flag or Spc42p-2PY to GST-Spc110p/Cmd1p has little effect on its ability to bind to the Tub4p complex (Figure 6B).
In Vitro Microtubule Nucleation Activity of Tub4p Complex
Although soluble Tub4p exists predominantly in stable Tub4p complexes, microtubules are only observed to emanate from the SPB. One hypothesis is that the microtubule nucleation capacity of the cytoplasmic Tub4p complex is minimal until it is recruited to the SPB by a protein such as Spc110p. We therefore tested whether the reconstituted Tub4p complex has microtubule nucleation activity and whether interacting with purified Spc110p enhances this activity. To measure nucleation, we used a modified solution assay adapted from Oegema et al. (1999). Rhodamine-labeled bovine tubulin was incubated with recombinant Tub4p complex or buffer alone, and the resulting polymerized microtubules were pelleted onto coverslips and counted using fluorescence microscopy (Figure 7A). Because there is some variability, we report the average of all experiments and also show the highest and lowest values. An average of six independent experiments shows that incubating 2.0 mg/ml tubulin with 50 nM Tub4p complex promotes microtubule polymerization about threefold above the background level of a buffer control (Figure 7B) (a one-sided Wilcoxon rank sum test yielded a P value of 0.021). A fourfold increase in the concentration of Tub4p complex increased activity by 40% (our unpublished data). Tub4p complexes obtained by either purification method 1 (Spc97p-GST) or method 2 (GST-Spc110p1–220), which has an additional purification step, gave similar results. Thus, the microtubule-nucleating activity observed is likely due to the reconstituted Tub4p complex. The addition of Spc110p has no effect because 50 nM Tub4p complex preincubated with 350 nM Spc110p/Cmd1p produced a similar level of activity as Tub4p complex alone (Figure 7B). Finally, the nucleation activity of the reconstituted Tub4p complex was the same whether purified yeast tubulin or bovine tubulin was used (our unpublished data).
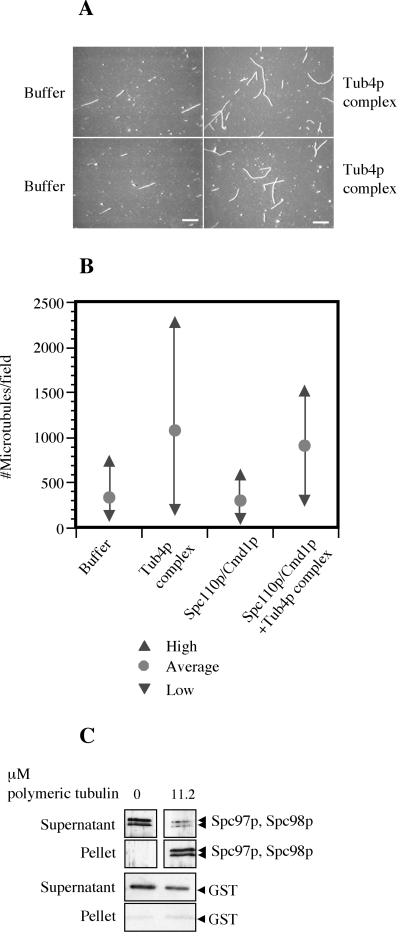
Figure 7.
Reconstituted Tub4p complex can promote microtubule polymerization at a low level. (A) Representative video fields of rhodamine-labeled microtubules from the nucleation assay polymerized in the presence of buffer or recombinant Tub4p complex. Bar, 10 μm. (B) Summary of the nucleation assay. For each experiment, the number of microtubules is counted from 20 fields and an average number of microtubules/field is taken. The indicated #Microtubules/field is normalized as the total number of microtubules per field that would have been counted in a 5-μl reaction. For each sample, an average (of 3–6 independent experiments), a high and a low result are indicated. For Tub4p complex alone, the average was taken of results from two experiments by using complexes purified by method 1 and 4 experiments with complexes purified by method 2. We estimate that the concentration of microtubules produced in the presence of 50 nM Tub4p complex is 1.8 pM. (C) Reconstituted Tub4p complex was tested for its ability to bind taxol-stabilized microtubules in a copelleting assay. After pelleting the microtubules, the supernatant and pellet were separated and both analyzed by Western blot to look for components of the Tub4p complex or the negative control, bacterial GST. Tubulin pellets were also analyzed by Ponceau S staining of the Western blot. In this experiment, 15 nM Tub4p complex was incubated with 11.2 μM polymeric tubulin containing 3 nM microtubule ends.
Because of the low nucleating activity, we tested the binding of the Tub4p complex to microtubules by using a microtubule copelleting assay. The Tub4p complex cosediments with taxol-stabilized microtubules (Figure 7C). This binding is specific because incubating the negative control bacterial GST with taxol-microtubules does not pellet a significant amount of GST (Figure 7C).
Sedimentation of Tub4p Complex from Yeast Whole Cell Extract
The γ-tubulin complex of higher eukaryotic cells contains additional components that enhance the nucleating activity of the small complex (Oegema et al., 1999). No evidence exists for such a large complex in yeast. Furthermore, results from our studies of recombinant Tub4p complex and Spc110p/Cmd1p indicate a low microtubule-nucleating activity. We sought to compare the recombinant Tub4p complex to the native complex in yeast cells. The sedimentation coefficient of the recombinant Tub4p complex is identical to that isolated from yeast (11.6 S) when sedimented in a Tris-based buffer (TB100) developed by Knop and coworkers (Figure 8A; our unpublished data). However, because previous experiments have been performed with a buffer that may interfere with microtubule polymerization, we sedimented yeast extracts by using buffer HB100 known to support microtubule nucleation. Interestingly, we found that under these conditions, a significant portion of Tub4p migrates at 22 S (Figure 8, B and C). High salt does not dissociate this large complex because a cell extract prepared in HB plus 400 mM KCl still retains the 22 S peak (our unpublished data). The 22 S Tub4p complex does not migrate with endogenous Spc110p, which has a sedimentation coefficient of 4.4 S under all conditions (Figure 8C). This result is consistent with the fact that most Spc110p (≥75%; our unpublished data) is found in the SPB pellet, and soluble Spc110p is not associated with Tub4p complexes, perhaps due to the modest affinity of Spc110p for the Tub4p complex. Note that the recombinant form of Spc110p/Cmd1p has the same sedimentation coefficient as that derived from yeast (compare Figure 3A with 8C).
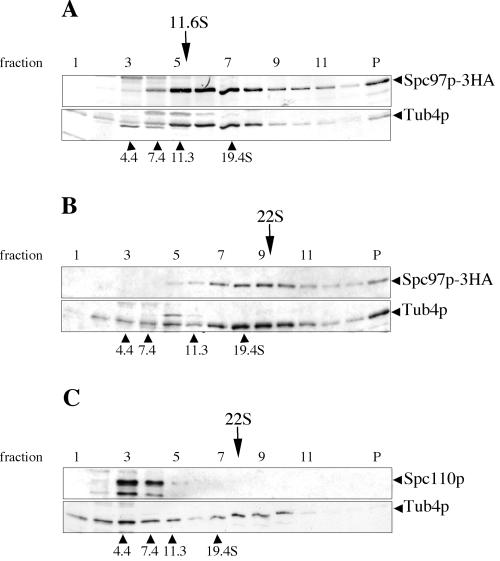
Figure 8.
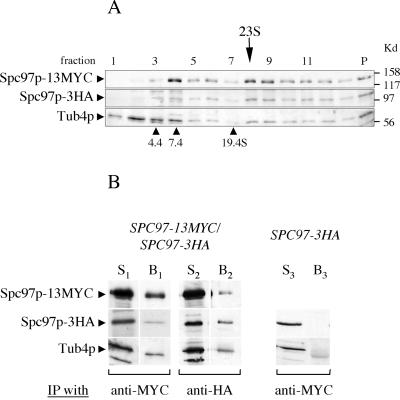
Identification of the 22 S Tub4p complex from yeast extract. (A and B) A logarithmic culture of cells containing SPC97-3HA cells was divided into two and lysed with either TB100 (A) or HB100 buffer (B). (C) Wild-type yeast cell extract prepared in HB100. Cleared cell extracts were loaded onto 10–40% sucrose gradients prepared in the same buffer as the cell extract. Fractions (P is the pellet) were TCA precipitated and analyzed by Western blot with anti-HA, anti-Tub4p, or anti-Spc110p antibodies. Internal protein standards were included in each gradient and were analyzed directly by Coomassie stained SDS-PAGE without TCA precipitation. The estimated sedimentation coefficients of the Spc97-3HA/Tub4p complexes are marked above each gradient.
We determined whether the 22 S Tub4p complex includes other components of the small Tub4p complex by analyzing cell extract from a yeast strain harboring a single genomic copy of SPC97-3HA. The majority of Spc97p-3HA migrates with Tub4p at 22 S in a sucrose gradient (Figure 8B), suggesting that the large Tub4p complex contains Spc97p. The fact that Spc97p-3HA sediments predominantly at 22 S indicates that the fraction of Tub4p that sediments at smaller S values are not associated with Spc97p. α-Tubulin (analyzed by Western blot with YOL/34 antibody) was not associated with the 22 S peak (our unpublished data).
The size of the large Tub4p complex resembles that of vertebrate γ-TuRCs (25–35 S). The γ-TuRC is composed of many small γ-TuSCs and various accessory proteins that are believed to link multimers of γ-TuSCs and enhance their nucleating activity (Oegema et al., 1999; Moritz et al., 2000; Zhang et al., 2000). To determine whether the 22 S Tub4p complex contains more than one small Tub4p complex, we tested for physical interactions between two different tagged versions of Spc97p. A heterozygous yeast strain was constructed to contain a genomic copy of SPC97-3HA and a genomic copy of SPC97-13MYC. Cell extract was prepared in HB100 and analyzed by sucrose gradient centrifugation. All three Spc97p-13MYC, Spc97p-3HA, and Tub4p cosediment at a peak of ∼23 S (Figure 9A). A fraction of Spc97p-13MYC and Spc97p-3HA also sediments at lower S values, suggesting the presence of smaller tagged Spc97p complexes. To demonstrate that the two tagged versions of Spc97p were in the same complex, we immunoprecipitated the same cell extract with anti-HA or anti-MYC antibodies. Anti-MYC antibodies precipitate both Spc97p-13MYC and Spc97p-3HA and anti-HA antibodies precipitate both Spc97p-3HA and Spc97p-13MYC (Figure 9B). In both cases, Tub4p was also coprecipitated, suggesting association between complexes containing Spc97p and Tub4p. These coprecipitations are specific because subjecting an extract from SPC97-3HA cells with anti-MYC antibody does not bring down Spc97p-3HA or Tub4p (Figure 9B). We conclude that at least a subset of the 23 S Tub4p complexes found in SPC97-3HA/SPC97-13MYC cells contain more than one Spc97p.
Figure 9.
The large Tub4p-complex contains more than one Spc97p. (A) Sucrose gradient of cell extract prepared from strain SPC97-13MYC/SPC97-3HA in HB100 buffer. Fractions were TCA precipitated and analyzed by Western blots with anti-MYC, anti-HA, or anti-Tub4p antibodies. Note the comigration of Spc97p-13MYC, Spc97p-3HA, and Tub4p at 23 S. (B) Immunoprecipitations of cell extracts from strain SPC97-13MYC/SPC97-3HA or SPC97-3HA by using anti-MYC or anti-HA antibodies. Specific proteins were identified by Western blot. For each immunoprecipitation, S is the starting cleared lysate, and B is the fraction solubilized from the immunoprecipitate–protein-G beads. S1 and S2 are different sources of cell extracts.
DISCUSSION
Orthologs of components of the yeast Tub4p complex make up the γ-TuSC, which is the core unit of vertebrate γ-TuRCs. The fact that no complex similar to the γ-TuRC had been identified in S. cerevisiae raised the question of whether the mechanism of microtubule nucleation and its regulation in yeast are simplified versions of those in higher eukaryotes. To further understand the assembly and function of the yeast γ-TuSC, we characterized the Tub4p complex alone and when bound to the SPB docking protein Spc110p by using recombinant proteins expressed in insect cells. The strong similarities we found between the small Tub4p complex and the γ-TuSC in other organisms led us to reexamine the presence of a large Tub4p complex in yeast. We present herein evidence for a large Tub4p complex from yeast extracts that may contain multimers of small Tub4p complexes.
Reconstitution of Tub4p Complex
We reconstituted the small Tub4p complex from insect cells to produce the large amount of purified complexes required for biochemical analyses. The molecular mass of the reconstituted Tub4p complex is 340 kDa, which closely matches the molecular mass based on the measured stoichiometry of 1 Spc98p:1 Spc97p:2 Tub4p in our purified complexes. Although previous immunoprecipitation studies identified the three components of the Tub4p complex (Knop et al., 1997; Knop and Schiebel, 1997), this is the first time the stoichiometry has been determined. This stoichiometry is identical to that of the Drosophila γ-TuSC (Oegema et al., 1999), suggesting conserved assembly, in addition to composition, of core components of the microtubule nucleation machinery.
The native Tub4p complex derived from yeast extract sediments at 11.6 or 22 S, depending on the buffers used to prepare the extract (see below). In yeast extract prepared in a Tris buffer, TB100, the 11.6 S complex predominates. The recombinant complex prepared from insect cells that consists of one molecule of Spc98p, one molecule of Spc97p, and two molecules of Tub4p also sediments at 11.6 S. This sedimentation coefficient conflicts with that of Knop et al. (1997) who report that the Tub4p complex isolated from yeast in TB-100 sediments at 6 S. However, because the native Tub4p complex in their studies migrates immediately below the protein standard β-amylase, which has a sedimentation coefficient of 8.9 S (Sober, 1968), the actual value appears to be closer to 11.6 S.
Interactions between Tub4p Complex and Spc110p
We and others have previously identified Spc110p as the docking protein of the Tub4p complex to the nuclear side of the SPB (Knop and Schiebel, 1997; Nguyen et al., 1998). The recent identification of kendrin/pericentrin as the mammalian ortholog of Spc110p suggests a universal mechanism of recruitment or attachment of γ-tubulin complexes to the centrosome (Flory et al., 2000). In these studies, we have reconstituted the Tub4p complex with Spc110p to determine what effects Spc110p has on the organization and nucleation activity of Tub4p complexes.
By the two-hybrid assay, we previously observed diverse interactions between the components of the Tub4p complex and Spc110p (Nguyen et al., 1998). To test these interactions in another way and to avoid the background interference of endogenous yeast proteins in the two-hybrid assay, we characterized the interactions of proteins expressed and purified from insect cells. Confirming our two-hybrid results, we observed that Spc98p is the primary component of the Tub4p complex that binds to the N terminus of Spc110p. The affinity of Spc98p for Spc110p and the fact that Spc98p is the only component of the Tub4p complex with a nuclear localization signal may be important in regulating the recruitment of the Tub4p complex to the SPB nuclear side. Unexpectedly, we found that complexing with Tub4p enhances the expression of either soluble Spc98p or Spc97p. One would predict that free Tub4p, but not free Spc98p or Spc97p, can exist in cells. In fact, yeast extracts typically contain species of Tub4p with small sedimentation coefficients that do not cosediment with Spc97p, whereas all of the Spc97p cosediments with Tub4p.
We were particularly interested in characterizing full-length Spc110p and its interactions with the Tub4p complex. We first discovered that the solubility of Spc110p in insect cells is much improved when yeast calmodulin or Cmd1p is coexpressed. This result is consistent with the phenotypes of mutants harboring an Spc110p defective in binding Cmd1p (Sundberg et al., 1996). At the nonpermissive temperature, spc110-220 cells accumulate a nuclear aggregate of Spc110p that is detached from the SPB and devoid of Cmd1p, suggesting that Cmd1p promotes the proper assembly of Spc110p required for incorporation into the SPB.
From its hydrodynamic characteristics, purified recombinant Spc110p/Cmd1p is estimated to have two molecules each of Spc110p and Cmd1p and a molecular mass of 250 kDa. A small measured sedimentation coefficient in combination with the large Stokes radius suggests an asymmetrical structure consistent with the rod shape of its extensive coiled coil region (Kilmartin et al., 1993). The binding of Spc110p/Cmd1p to the Tub4p complex generates a larger complex with a sedimentation coefficient of 11.3 S, but not as high as the 25–32 S values for vertebrate γ-TuRCs (Stearns and Kirschner, 1994; Meads and Schroer, 1995; Murphy et al., 1998; Oegema et al., 1999). Because Spc110p/Cmd1p plus Tub4p complex together migrate in the void volume of a gel filtration column, we cannot determine the molecular mass of this larger complex. Soluble Spc110p/Cmd1p in yeast extracts sediments at 4.2 S identical to that of the recombinant Spc110p/Cmd1p. The native soluble Spc110p is not associated with the Tub4p complex consistent with the low abundance of these proteins and the moderate Kd (150 nM) for their binding.
We tested whether Spc29p or Spc42p enhances the affinity of Spc110p for the Tub4p complex. Both Spc29p and Spc42p are core components required for SPB duplication (Donaldson and Kilmartin, 1996; Elliott et al., 1999) and are part of a template that recruits and organizes the assembly of Spc110p onto the central plaque of the SPB (Adams and Kilmartin, 1999). We provide evidence herein that Spc110p/Cmd1p binds directly to both Spc42p and Spc29p. However, we found that the binding of Spc110p to either Spc42p and/or Spc29p does not affect its interactions with the Tub4p complex. Interestingly, recombinant Spc42p and Spc29p do not mediate the binding of each other to Spc110p; instead, their binding appears to be competitive. How Spc110p, calmodulin, Spc29p, and Spc42p assemble into the central plaque remains an open question.
Microtubule Nucleation and Binding by Tub4p Complex
Studies in Drosophila indicate that the γ-TuRC has robust microtubule nucleation activity, but the individual γ-TuSC subunit exhibits only weak activity (Oegema et al., 1999). Because yeast cells were not known to possess a large Tub4p complex analogous to the γ-TuRC, we tested whether microtubule polymerization in yeast requires only the small Tub4p complex or depends on a critical interaction between the Tub4p complex and the SPB. Recruitment to the SPB may result in the organization of Tub4p complexes into a structure that has robust nucleation activity similar to the γ-TuRC. Furthermore, although the small Tub4p complex can interact directly with the SPB via a docking protein such as Spc110p, the vertebrate γ-TuSC requires additional components contained in the γ-TuRC (Moritz et al., 1998; Zhang et al., 2000). We measured the nucleation activity of the reconstituted Tub4p complex alone and when it is bound to Spc110p. Our results indicate that the Tub4p complex has a low intrinsic ability to promote microtubule polymerization and interaction with Spc110p does not alter this activity. Although we cannot completely rule out the presence of insect cell contaminants in our preparations, we attribute the low nucleation activity to the reconstituted Tub4p complex for several reasons. First, an additional purification step does not alter the nucleation activity of the Tub4p complex. Second, we did not find any trace of insect tubulin contaminants. Finally, this low nucleation activity is equivalent to the activity reported for Drosophila γ-TuSC extracted from the γ-TuRC (Oegema et al., 1999) and more recently, reconstituted from Sf9 insect cells (Gunawardane et al., 2000).
A Larger Tub4p Complex in Yeast
Our findings that the Tub4p complex with or without Spc110p/Cmd1p only has low nucleation activity suggested to us that components associated with an active Tub4p complex are missing. Because the ends of yeast microtubules are capped similarly to microtubules nucleated in the presence of vertebrate γ-TuRC, it is possible that other as yet unknown yeast proteins may organize the Tub4p complexes into a multimeric ring structure such as the γ-TuRC. Careful analysis of yeast extracts prepared under conditions known to promote microtubule nucleation identified a stable 22 S complex containing Tub4p and Spc97p. The size of this complex is close to the 25–32 S size of vertebrate γ-TuRCs. This complex is not associated with Spc110p or tubulin and contains more than one molecule of Spc97p, similar to the γ-TuRC. Future analyses will identify the components of the complex and its role in microtubule nucleation.
ACKNOWLEDGMENTS
We thank R. Jeng and T. Stearns for antibodies and plasmids; R. Deshaies for cell line AK1310; K. Kaplan and P. Sorger for plasmids and advice; and M. Gupta and R. Himes for yeast tubulin. We thank L. Wordeman and S. Francis for helpful discussions and comments on the manuscript and A. Hunter for technical advice. This work was supported by National Institutes of Health grant R01 GM-40506, National Institute of General Medical Sciences (to T.N.D.). D.V. was supported by Public Health Service National Research Service Award F32 GM-17945, National Institute of General Medical Sciences.
Footnotes
DOI: 10.1091/mbc.02–01–0607.
REFERENCES
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahler J, Wu J-Q, Longtine MS, Shah NG, McKenzie III A, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Byers B, Shriver K, Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae. J Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- Dictenberg JB, Zimmerman W, Sparks CA, Young A, Vidair C, Zheng Y, Carrington W, Fay FS, Doxsey SJ. Pericentrin and gamma-tubulin form a protein complex and are organized into a novel lattice at the centrosome. J Cell Biol. 1998;141:163–174. doi: 10.1083/jcb.141.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson AD, Kilmartin JV. Spc42p: a phosphorylated component of the S. cerevisiae spindle pole body (SPD) with an essential function during SPB duplication. J Cell Biol. 1996;132:887–901. doi: 10.1083/jcb.132.5.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S, Knop M, Schlenstedt G, Schiebel E. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc Natl Acad Sci USA. 1999;96:6205–6210. doi: 10.1073/pnas.96.11.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. γ-Tubulin nucleation: template or protofilament? Nat Cell Biol. 2000;2:E93–E96. doi: 10.1038/35014084. [DOI] [PubMed] [Google Scholar]
- Fava F, Raynaud-Messina B, Leung-Tack J, Mazzolini L, Li M, Guillemot JC, Cachot D, Tollon Y, Ferrara P, Wright M. Human 76p: a new member of the γ-tubulin-associated protein family. J Cell Biol. 1999;147:857–868. doi: 10.1083/jcb.147.4.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felix MA, Antony C, Wright M, Maro B. Centrosome assembly in vitro: role of gamma-tubulin recruitment in Xenopus sperm aster formation. J Cell Biol. 1994;124:19–31. doi: 10.1083/jcb.124.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flory MR, Moser MJ, Monnat RJ, Jr, Davis TN. Identification of a human centrosomal calmodulin-binding protein that shares homology with pericentrin. Proc Natl Acad Sci USA. 2000;97:5919–5923. doi: 10.1073/pnas.97.11.5919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, Sundberg HA, Chang BH, Muller EG, Davis TN. The essential mitotic target of calmodulin is the 110-kilodalton component of the spindle pole body in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:7913–7924. doi: 10.1128/mcb.13.12.7913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiser JR, van-Tuinen D, Brockerhoff SE, Neff MM, Davis TN. Can calmodulin function without binding calcium? Cell. 1991;65:949–959. doi: 10.1016/0092-8674(91)90547-c. [DOI] [PubMed] [Google Scholar]
- Gunawardane RN, Martin OC, Cao K, Zhang L, Dej K, Iwamatsu A, Zheng Y. Characterization, and Reconstitution of Drosophila γ-tubulin ring complex subunits. J Cell Biol. 2000;151:1513–1523. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horio T, Uzawa S, Jung MK, Oakley BR, Tanaka K, Yanagida M. The fission yeast gamma-tubulin is essential for mitosis and is localized at microtubule organizing centers. J Cell Sci. 1991;99:693–700. doi: 10.1242/jcs.99.4.693. [DOI] [PubMed] [Google Scholar]
- Hyman A, Drechsel D, Kellogg D, Salser S, Sawin K, Steffen P, Wordeman L, Mitchison T. Preparation of modified tubulins. Methods Enzymol. 1991;196:478–485. doi: 10.1016/0076-6879(91)96041-o. [DOI] [PubMed] [Google Scholar]
- Joshi HC, Palacios MJ, McNamara L, Cleveland DW. Gamma-tubulin is a centrosomal protein required for cell cycle-dependent microtubule nucleation. Nature. 1992;356:80–83. doi: 10.1038/356080a0. [DOI] [PubMed] [Google Scholar]
- Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- Kilmartin JV, Dyos SL, Kershaw D, Finch JT. A spacer protein in the Saccharomyces cerevisiae spindle poly body whose transcript is cell cycle-regulated. J Cell Biol. 1993;123:1175–1184. doi: 10.1083/jcb.123.5.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilmartin JV, Goh PY. Spc110p: assembly properties and role in the connection of nuclear microtubules to the yeast spindle pole body. EMBO J. 1996;15:4592–4602. [PMC free article] [PubMed] [Google Scholar]
- Knop M, Pereira G, Geissler S, Grein K, Schiebel E. The spindle pole body component Spc97p interacts with the gamma-tubulin of Saccharomyces cerevisiae and functions in microtubule organization and spindle pole body duplication. EMBO J. 1997;16:1550–1564. doi: 10.1093/emboj/16.7.1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knop M, Schiebel E. Spc98p and Spc97p of the yeast gamma-tubulin complex mediate binding to the spindle pole body via their interaction with Spc110p. EMBO J. 1997;16:6985–6995. doi: 10.1093/emboj/16.23.6985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leguy R, Melki R, Pantaloni D, Carlier M-F. Monomeric γ-tubulin nucleates microtubules. J Biol Chem. 2000;275:21975–21980. doi: 10.1074/jbc.M000688200. [DOI] [PubMed] [Google Scholar]
- Li Q, Joshi HC. γ-Tubulin is a minus end-specific microtubule binding protein. J Cell Biol. 1995;131:207–214. doi: 10.1083/jcb.131.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschall LG, Jeng RL, Mulholland J, Stearns T. Analysis of Tub4p, a yeast gamma-tubulin-like protein: implications for microtubule-organizing center function. J Cell Biol. 1996;134:443–454. doi: 10.1083/jcb.134.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin OC, Gunawardane RN, Iwamatsu A, Zheng Y. Xgrip109: a gamma tubulin-associated protein with an essential role in gamma tubulin ring complex (γTuRC) assembly and centrosome function. J Cell Biol. 1998;141:675–687. doi: 10.1083/jcb.141.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meads T, Schroer TA. Polarity and nucleation of microtubules in polarized epithelial cells. Cell Motil Cytoskeleton. 1995;32:273–288. doi: 10.1002/cm.970320404. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Microtubule assembly nucleated by isolated centrosomes. Nature. 1984;312:232–237. doi: 10.1038/312232a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the γ-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by γ-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- Moritz M, Zheng Y, Alberts BM, Oegema K. Recruitment of the gamma-tubulin ring complex to Drosophila salt-stripped centrosome scaffolds. J Cell Biol. 1998;142:775–786. doi: 10.1083/jcb.142.3.775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Preble AM, Patel UK, O'Connell KL, Dias DP, Moritz M, Agard D, Stults JT, Stearns T. GCP5 and GCP6: two new members of the human gamma-tubulin complex. Mol Biol Cell. 2001;12:3340–3352. doi: 10.1091/mbc.12.11.3340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen T, Vinh DBN, Crawford DK, Davis TN. A genetic analysis of interactions with Spc110p reveals distinct functions of Spc97p and Spc98p, components of the yeast gamma-tubulin complex. Mol Biol Cell. 1998;9:2201–2216. doi: 10.1091/mbc.9.8.2201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley BR, Oakley CE, Yoon Y, Jung MK. Gamma-tubulin is a component of the spindle pole body that is essential for microtubule function in Aspergillus nidulans. Cell. 1990;61:1289–1301. doi: 10.1016/0092-8674(90)90693-9. [DOI] [PubMed] [Google Scholar]
- Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnackenberg BJ, Khodjakov A, Rieder CL, Palazzo RE. The disassembly and reassembly of functional centrosomes in vitro. Proc Natl Acad Sci USA. 1998;95:9295–300. doi: 10.1073/pnas.95.16.9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel GLM, Monty KJ. Determination of molecular weights and frictional ratios of proteins in impure systems by use of gel filtration and density gradient centrifugation. Application to crude preparations of sulfite and hydroxylamine reductases. Biochim Biophys Acta. 1966;112:346–362. doi: 10.1016/0926-6585(66)90333-5. [DOI] [PubMed] [Google Scholar]
- Sobel SG, Snyder M. A highly divergent gamma-tubulin gene is essential for cell growth and proper microtubule organization in Saccharomyces cerevisiae. J Cell Biol. 1995;131:1775–1788. doi: 10.1083/jcb.131.6.1775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sober HA. Handbook of Biochemistry, Selected Data for Molecular Biology. Cleveland, OH: The Chemical Rubber Co.; 1968. [Google Scholar]
- Spang A, Geissler S, Grein K, Schiebel E. γ-Tubulin-like Tub4p of Saccharomyces cerevisiae is associated with the spindle pole body substructures that organize microtubules and is required for mitotic spindle formation. J Cell Biol. 1996a;134:429–441. doi: 10.1083/jcb.134.2.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A, Grein K, Schiebel E. The spacer protein Spc110p targets calmodulin to the central plaque of the yeast spindle pole body. J Cell Sci. 1996b;109:2229–2237. doi: 10.1242/jcs.109.9.2229. [DOI] [PubMed] [Google Scholar]
- Stearns T, Evans L, Kirschner M. γ-Tubulin is a highly conserved component of the centrosome. Cell. 1991;65:825–836. doi: 10.1016/0092-8674(91)90390-k. [DOI] [PubMed] [Google Scholar]
- Stearns T, Kirschner M. In vitro reconstitution of centrosome assembly and function: the central role of γ-tubulin. Cell. 1994;76:623–637. doi: 10.1016/0092-8674(94)90503-7. [DOI] [PubMed] [Google Scholar]
- Sundberg HA, Goetsch L, Byers B, Davis TN. Role of calmodulin and Spc110p interaction in the proper assembly of spindle pole body components. J Cell Biol. 1996;133:111–124. doi: 10.1083/jcb.133.1.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassin AM, Celati C, Moudjou M, Bornens M. Characterization of the human homologue of the yeast spc98p and its association with γ-tubulin. J Cell Biol. 1998;141:689–701. doi: 10.1083/jcb.141.3.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel JM, Stearns T, Rieder CL, Palazzo RE. Centrosomes isolated from Spisula solidissima oocytes contain rings and an unusual stoichiometric ratio of α/β tubulin. J Cell Biol. 1997;137:193–202. doi: 10.1083/jcb.137.1.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wiese C, Zheng Y. A new function for the γ-tubulinring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- Zhang L, Keating TJ, Wilde A, Borisy GG, Zheng Y. The role of Xgrip210 in γ-tubulin ring complex assembly and centrosome recruitment. J Cell Biol. 2000;151:1525–1535. doi: 10.1083/jcb.151.7.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jung MK, Oakley BR. Gamma-tubulin is present in Drosophila melanogaster and Homo sapiens and is associated with the centrosome. Cell. 1991;65:817–823. doi: 10.1016/0092-8674(91)90389-g. [DOI] [PubMed] [Google Scholar]
- Zheng Y, Wong M-L, Alberts B, Mitchison T. Nucleation of microtubule assembly by a γ-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]