Abstract
Yeast protein insertion orientation (PIO) mutants were isolated by selecting for growth on sucrose in cells in which the only source of invertase is a C-terminal fusion to a transmembrane protein. Only the fraction with an exocellular C terminus can be processed to secreted invertase and this fraction is constrained to 2–3% by a strong charge difference signal. Identified pio mutants increased this to 9–12%. PIO1 is SPF1, encoding a P-type ATPase located in the endoplasmic reticulum (ER) or Golgi. spf1-null mutants are modestly sensitive to EGTA. Sensitivity is considerably greater in an spf1 pmr1 double mutant, although PIO is not further disturbed. Pmr1p is the Golgi Ca2+ ATPase and Spf1p may be the equivalent ER pump. PIO2 is STE24, a metalloprotease anchored in the ER membrane. Like Spf1p, Ste24p is expressed in all yeast cell types and belongs to a highly conserved protein family. The effects of ste24- and spf1-null mutations on invertase secretion are additive, cell generation time is increased 60%, and cells become sensitive to cold and to heat shock. Ste24p and Rce1p cleave the C-AAX bond of farnesylated CAAX box proteins. The closest paralog of SPF1 is YOR291w. Neither rce1-null nor yor291w-null mutations affected PIO or the phenotype of spf1- or ste24-null mutants. Mutations in PIO3 (unidentified) cause a weaker Pio phenotype, enhanced by a null mutation in BMH1, one of two yeast 14-3-3 proteins.
INTRODUCTION
The topology of an integral membrane protein is defined by the orientation of its transmembrane (TM) segments. We designate these orientations as Nexo and Cexo, indicating whether the N or C terminus is “exocellular” because of translocation from the cytoplasm. In a eukaryotic cell, protein secretion and translocation of exocellular components of most TM proteins occur at the endoplasmic reticulum (ER), initially to the lumen of the secretory pathway, independent of final cellular location of the protein. The protein must incorporate signals for insertion at the ER, topogenic signals determining the orientation of insertion of TM segments and signals for translocation to its functional site. Assembly of plastids and peroxisomes and delayed membrane insertion events, such as occur in viral entry, the action of pore-forming toxins, and insertion of the effector proteins of pathogenic bacteria, may use different insertion signals and mechanisms. Results, however, are similar: a structure in which energy is minimized because hydrogen-bonding requirements of TM segments are satisfied, whereas surfaces exposed to the internal membrane environment are hydrophobic.
Apart from the β-barrel TM proteins of Gram negative bacterial outer membranes, most TM segments are α-helices of 20 or more predominantly hydrophobic amino acids. Although topogenic signals in TM proteins have been analyzed in considerable detail (Hartmann et al., 1989; Nilsson and von Heijne, 1990; Beltzer et al., 1991; von Heijne, 1992; Gafvelin et al., 1997; Wahlberg and Spiess, 1997), the cellular mechanisms for response to these signals in eukaryotes are poorly understood. Moreover, the extent to which TM proteins in general and polytopic proteins in particular can adopt transient and reversible topologies at the ribosome-translocon interface during translocation at the ER remains to be established (Goder et al., 1999; Heinrich et al., 2000). Dynamic interactions between potential TM segments and between these segments and a signal peptide, when present, may influence final topology, resulting in interdependent insertion (Monne et al., 1999; Rutkowski et al., 2001). In addition, the extent to which the apparent unique insertion orientations usually observed in vivo result from proteolytic destruction of alternate topological forms by the quality control machinery of the ER (Schubert et al., 2000; Travers et al., 2000) also remains to be better defined. Disturbance of this pattern may have pathogenic consequences (Hegde et al., 1998).
The major topogenic signals determining protein insertion orientation (PIO) of a TM segment are the charge difference across it and its total hydrophobicity. Although responses to charge-independent topogenic signals such as hydrophobicity are likely to involve direct contact with proteins or lipids at the translocation site (Prinz et al., 1998; Heinrich et al., 2000), effects of the often-predominant charge difference signal appear to be purely electrostatic. This has been shown most clearly by demonstrating, in yeast (Saccharomyces cerevisiae), that an N-terminal positive charge has the same effect on PIO as a C-terminal negative charge, and vice versa (Harley et al., 1998). Sequence-independent interaction with an electrostatic field is implied. In most prokaryotes this field is provided by the positive-outside potential gradient at the cytoplasmic membrane (Andersson and von Heijne, 1994; Delgado-Partin and Dalbey, 1998). However, charges on anionic phospholipids may also play a significant role (van Klompenburg et al., 1997; van Klompenburg and de Kruijff, 1998). Topological responses to this field are codified in the “positive inside rule” (von Heijne, 1992). Although the ER apparently lacks a TM potential gradient, insertion at the ER follows the related, statistically derived charge difference rule (Hartmann et al., 1989), which gives equal weight to positive and negative charges. We have confirmed the validity of this rule in yeast (Harley et al., 1998) by using fusions of a C-terminal β-lactamase reporter to an N-terminal fragment of the Ste2p α-factor receptor. The product is a model type III TM protein, i.e., one that is predominantly inserted with its single TM segment Nexo. We also determined the relative strength of the charge difference and hydrophobicity signals (Harley and Tipper, 1996; Harley et al., 1998). A similar invertase fusion has now been used to isolate yeast pio mutants defective in response to a strong charge difference signal. Yeast, apparently, easily tolerate a significant increase in the error rate of TM protein insertion. Surprisingly, only two nonessential genes were identified in this search. Although both encode highly conserved polytopic TM proteins resident in or near the ER and present in all yeast cell types, their functions are apparently unrelated to each other or to translocation. Double mutants, however, are highly sensitive to heat shock and die slowly at 4°C, implying redundant functions important to cell survival.
MATERIALS AND METHODS
Yeast Strains
All S. cerevisiae strains used are listed in Table 1. The SUC2 strain CRY2 was provided by Dr. Robert Fuller (Komano and Fuller, 1995). The suc2-null strain SEY6210 was provided by Dr. Scott Emr (Robinson et al., 1988). The MATa strain CHYY100, isogenic to SEY6210, was obtained by transforming SEY6210 with a URA3 plasmid that expresses the HO endonulclease under the GAL1 promoter. Strain CHYY100 was recovered after growth on 5-fluoroorotic acid (5-FOA) plates to select for Ura− derivatives that had lost the HO plasmid. Strains CHYY255 and 256 were constructed by transforming strains SEY6210 and CHYY100, respectively, with the integrating plasmid YIp can1-S+5Inv-URA3 that had been cut at SpeI within the can1 fragment to target integration at CAN1, resulting in insertion flanked by C- and N-terminally truncated fragments of CAN1. Transformants grew on Ura− plates containing 50 μg/ml canavanine, demonstrating disruption of CAN1 and integration of URA3. The integration site was confirmed by reversion to canavanine sensitivity in Ura− loop-out segregants selected on 5-fluoroorotic acid plates. These strains occasionally lost the S+5Inv reporter, presumably due to recombination with the adjacent ura3-52 locus. Stable strains carrying LEU2, TRP1, or HIS3 markers at the same locus were constructed using appropriate variants of the YIp can1-S+5Inv integrating vector.
Table 1.
Yeast strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| SEY6210 | MATα; GAL; suc2-Δ9; ura3-52; leu2-3,112; trp1-Δ901; his3-Δ200; lys2-801 | Rose et al., 1987 |
| CRY2 | MATα ura3-1 leu2-3,112 trp1-1 his3-11 can1-100 ade2 GAL SUC | Maudoux et al., 2000 |
| CRY2A | CRY2 with pPGK-ERKEX2[HIS3] integrated at his3 | Harley et al., 1998 |
| CHY100 | MATa: derivative of SEY6210 | This study |
| CHY255/6 | MATa/α canR [S+5Inv-URA3] derivatives of SEY6210 | This study |
| CHY298/9 | MATa/α canR [S+5Inv-LEU2] derivatives of SEY6210 | This study |
| CHY300/1 | MATa/α canR [S+5Inv-TRP1] derivatives of SEY6210 | This study |
| CHY302/3 | MATa/α canR [S+5Inv-HIS3] derivatives of SEY6210 | This study |
| DK4 | MATa canR [S+5Inv-TRP1] pio1.1 | This study |
Vectors for Expression of SPF1 and STE24 (Table 2)
Table 2.
Plasmids used for expression or deletion of SPF1 and STE24
| Plasmid | Type | Copy no. | Selection marker | Genes expressed |
|---|---|---|---|---|
| pRS314-SPF1 | YCp | 1 | TRP1 | SPF1 |
| pRS314-SPF1-STE24 | YCp | 1 | TRP1 | SPF1 STE24 |
| YEp351-SPF1 | YEp | 12–15 | LEU2 | SPF1 |
| pSM1069 | YCp | 1 | LEU2 | STE24 |
| pSM1104 | YCp | 1 | URA3 | ste24-E269A |
| pSM1072 | Disruption | Replaces STE24 with LEU2 | ||
| pCS202 | Disruption | Replaces SPF1 with TRP1 | ||
All except the disruption plasmids are single copy centromere (YCp) or multicopy yeast episomal (YEp) plasmids, maintained in yeast by URA3, TRP1, or LEU2 markers. Fragments excised from the disruption plasmids were used to replace the indicated genes.
All DNA manipulations were performed using the Escherichia coli strain DH5α [supE44D lacU169(f80 lacZDM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1]. The BamHI to SpeI fragment of the genomic clones encompassing the SPF1 gene isolated by complementation of pio1.1 was cloned into pRS314 (YCp-TRP1) (Sikorski and Hieter, 1989), producing pRS314-SPF1 and into YEp351 (LEU2) (Hill et al., 1986), producing YEp351-SPF1. pSM1069 (YCp LEU2) encoding Ste24p and pSM1104 (YCp URA3) that expresses an E269A mutant of Ste24p were provided by Dr. Susan Michaelis, Johns Hopkins School of Medicine, Baltimore, MD (Fujimura-Kamada et al., 1997). Ste24 was cloned from pSM1069 as a SacI to SpeI fragment by using synthetic primers and inserted into pRS314-SPF1, producing pRS314-SPF1-STE24.
Disruption of SPF1, STE24, YOR291w, RCE1, PMR1, UBC7, and BMH1
Plasmid pCS202 (Table 2) (Suzuki and Shimma, 1999) was used to replace the C-terminal 75% of SPF1 with the TRP1 marker. pSM1072 (Table 2) used for constructing ste24Δ::LEU2 disruptants was provided by Dr. Susan Michaelis (Fujimura-Kamada et al., 1997). To delete YOR291w, primers were synthesized that encompassed bases −45 to −1 and 4277–4318 of this gene. The former was linked to 18 bp homologous to the Sun1-SalI sites in the multicloning site of pFA-HIS3-MX6 (Wach et al., 1997) and the latter to 19 bp complementary to the SacI-ClaI sites of the same vector. The Schizosaccharomyces pombe his5 gene in this vector complements S. cerevisiae his3 mutants. Use of these primers for polymerase chain reaction (PCR) with pFA-HIS3-MX6 as template cloned the his5 gene flanked by the fragments of YOR291w. Transformation into a his3 yeast strain and selection for His+ resulted in replacement of all but the last 33 codons of YOR291w by the his5 gene. Disruption was proven by isolation of genomic DNA and PCR by using primers internal to his5 and flanking the primers used for disruption. Disruption of RCE1 was achieved by the same technique with primers provided by Dr. Jasper Rine, University of California, Berkeley, CA, resulting in replacement of RCE1 by the TRP1 gene (Trueblood et al., 2000). Plasmid pL119, used for constructing pmr1Δ::LEU2 disruptants, was provided by Dr. Kyle Cunningham, Johns Hopkins University, Baltimore, MD (Rudolph et al., 1989). The plasmid used for constructing ubc7::LEU2 disruptants was constructed by replacing the URA3 marker in p ubc7::URA3, provided by Dr. Randy Hampton, University of California, San Diego, CA (Cronin et al., 2000) with the LEU2 marker. Plasmid pB3455 encoding BMH1 (YCpURA3) and pB3453 used for constructing bmh1Δ::HIS3 disruptants and the bmh1 bmh2 double disruptant strain were provided by Dr. Bing Guo, Whitehead Institution of Biomedical Research, Cambridge, MA (Roberts et al., 1997). The URA3 marker in pB3455 was exchanged for HIS3 to allow selection in mTn3 (URA3) mutants. pNS3.8 encoding ADA2 was provided by Dr. Craig Peterson, University of Massachusetts Medical School, Worcester, MA.
Construction of YEp-S+5αHAβla and YEp-S+2αHA βla Reporters of Topology (Table 3)
Table 3.
Plasmids for selection and analysis of topology mutants
| New name | Charge difference | Reporter |
|---|---|---|
| YEp-S+5β1a | +5 | P-β-lactamase |
| YEp-S+2β1a | +2 | P-β-lactamase |
| YEp-S−4β1a | −4 | P-β-lactamase |
| YEp-S+5Inv | +5 | P-invertase |
| YCp-S+5Inv | +5 | P-invertase |
| YCp-S+2Inv | +2 | P-invertase |
| YCp-S−4Inv | −4 | P-invertase |
| YEp-S+5αHAβ1a | +5 | α-HA-h9-β1a |
| YEp-S+2αHAβ1a | +2 | α-HA-h9-β1a |
| YIp-can-S+5Inv | +5 | P-invertase |
The first three were previously called YEp-S79a-PB, YEp-S79g-PB, and YEp-S42u-PB, respectively (Harley and Tipper, 1996) where S79 refers to the 79 residue amino terminal fragment of Ste2p. The normal sequence (S79a) has a +5 charge difference, Δ(C-N), across the TM segment. Altered forms produce the modified charge differences indicated. New names emphasize the characteristics relevant to this study, these charge differences and the C-terminal reporter. The β-lactamase (β1a) and invertase (Inv) reporters are susceptible to release by Kex2p cleavage within the P-fragment of P-β-lactamase and P-invertase fusions, but not in the α-HA-h9-β1a fusions. YIp-can-S+5Inv integrates at CAN1 under selection for the coupled marker. The HIS3 derivative, for example, integrated in strain SEY6210, produces strain CHY302 (Tables 1 and 4).
YEp-S79a-αB (pCH10, URA3; Harley and Tipper, 1996) is an episomal, PGK-promoter driven fusion of S79a (S+5), the first 79 residues of Ste2p, including its single efficiently used N-glycosylation site, to an αB reporter where B is the mature sequence of β-lactamase and α is a 30-residue fragment of preproα-factor, including two efficiently used sites for N-glycosylation and a C-terminal LysArg site cleaved by the Kex2p endoprotease (Harley and Tipper, 1996). PCR was used with the primers 5′ G GGC GTG GCC AAG CAG AAT TCT GCA TCG TAC C and 5′ C TCT TTC CCC ATC CTT TAC GC and A3V-3HA-h9-βla (Martinez and Tipper, unpublished data) as template to clone β-lactamase preceded by the triple hemagglutinin (HA) epitope (3HA) and a His9 tag. The product was cleaved with MscI (underlined) and ClaI (within the tPGK transcription terminator) and inserted between the MscI and ClaI sites YEp-S79a-αB. Mutation of the LysArg site to LysGln resulted, precluding cleavage by Kex2p but retaining sensitivity to trypsin. The HA epitopes and His tag are inserted between α- and β-lactamase, producing YEp-S+5αHAβla (Table 3). YEp-S+2αHAβla was constructed in the same way starting with YEp-S+2-αB (pCH16; Harley and Tipper, 1996).
Construction of YEp-S+5Inv (Table 3)
Suc2p, the product of the yeast SUC2 invertase gene, was cloned by PCR by using pSSE14kr (Boehm et al., 1994) as template and the primers 5′ GG GTG GCC AAG AGA GAA GCT GAA GCT TTT ACA AAC GAA and 3′ AAG GTT CAT TCC CTT CAT TTT ATCTAGAGGG. The product includes mature Suc2p from codons 3 to the C terminus at 511 preceded by an MscI site and terminating in a BglII site. Cleavage with MscI and BglII and insertion into YEp-S+5βla (Table 3) (pCH03; Harley and Tipper, 1996) cleaved with the same enzymes produced YEp-S+5Inv, an in-frame fusion of the S+5 (S79a) fragment of Ste2p to invertase separated by the P fragment with its two Kex2p sites, expressed from the PGK promoter. After insertion into the ER membrane in Cexo orientation, cleavage by Kex2p would produce secreted invertase preceded by the peptide Glu Ala Glu Ala Phe. The Ste13p carboxypeptidase should leave a single Phe residue. Invertase activity was not detectably affected.
Construction of YCp-S+5Inv and S+2 and S−4 Derivatives (Table 3)
S+5Inv flanked by the PGK promoter and transcription terminator fragments was cloned by PCR with YEp-S+5Inv as template and the primers 5′ GGG GAA TTC AAG CTT GAA AGA TGC CG and 3′ CGA CCA GCT TTA AGC ATT CAG CTG GGG. The product was cleaved with EcoRI and SalI and cloned into YCp33lac (URA3) (Gietz and Sugino, 1988) cleaved with the same enzymes, producing YCp-S+5Inv. The XhoI-PstI fragment of YCp-S+5Inv, encompassing the S+5 fragment, was replaced with the similar fragments from pCH09 and pCH29 encompassing S+2 (S79g) and S−4 (S42u) fragments, respectively (Harley and Tipper, 1996) to produce the equivalent invertase fusions.
Construction YIp-can-S+5Inv-URA3 and LEU2, TRP1, and HIS3 Derivatives
The S+5Inv fusion, flanked by the PGK promoter and transcription terminator fragments, was excised from YCp-S+5Inv by cutting with EcoRI and SalI and cloned into the URA3 integrating vector pRS306 (Sikorski and Hieter, 1989), producing YIp-S+5Inv. An internal fragment of the CAN1 gene from codons 91–410 was cloned by PCR with yeast genomic DNA as template and primers 5′ GGG GTC GAC ATA TTG GTA TGA TTGC and 3′ GAT AGT TTC TTG TTC AAC CGT ACG TGGG. The product was cut with SalI and SphI and cloned into YIp-S+5Inv, producing YIp-can-S+5Inv-URA3 (Table 3). The LEU2, TRP1 and HIS3 versions of this vector were produced by excising the URA3 fragment from YIp-can-S+5Inv-URA and replacing it with appropriate fragments. To target integration at the CAN1 locus, these constructs were linearized by cutting at the unique SpeI or BstEII sites within the can1 fragment and transformed into yeast strain SEY6210 or CHY100, producing strains CHY255-303 (Table 1).
Construction of pZ-his5 for Rescue of Integrated mTn3 Mutant Sequences
pRSQ2-LEU2 (AmpR) was designed for integration at the unique lacZ fragment in mTnURA3 yeast mutants, allowing rescue of plasmids containing adjacent genomic DNA, as described in the Yale Genome Analysis Center web site (http://ycm1.med.yale.edu/YGAC/3xHAlacZlib.html) (Burns et al., 1994). pZ-his5 was constructed as an equivalent vector containing the S. pombe his5 gene, allowing selection in his3 strains. pFA6a-HIS3MX6 (Wach et al., 1997) was cut with BamHI and BglII and ligated, eliminating both sites. The product was cut with HindIII and AatII and the HindIII to AatII fragment from pRSQ2 encompassing the lacZ fragments was inserted, producing pZ-his5. Cutting pZ-his5 at the unique BamHI site separating the two lacZ fragments targets integration at the lacZ fragment of mTnURA3 or mTnLEU2 transposons in mTn3 yeast mutants. Genomic clones may be isolated from integrants by cutting with EcoRI, SacI, ClaI, SpeI, EcoRV, or SacII; ligation; transformation into E. coli; and selection for AmpR. Recovered clones were sequenced using a primer reading outward from the lacZ sequence into the adjacent genomic sequence.
Media
Media for selection of Suc+ yeast consisted of yeast extract-peptone (YEP) or appropriate drop-out 2% agar media (SC-Ura, SC-Leu, etc.), with filter-sterilized 2% sucrose as the fermentable carbon source, to which antimycin A had been freshly added (100 μl of a 2-mg/ml solution in 100% ethanol/25-ml plate; final concentration 8 μg/ml: YEPSA medium, USA medium, etc.). Cultures were incubated in the dark to protect antimycin from light-induced inactivation. Antimycin A suppresses the respiration that would otherwise allow growth on noncarbohydrate carbon sources (Robinson et al., 1988). Growth of Suc− cells was almost completely suppressed for 4 d after which growth slowly resumed, presumably resulting from hydrolysis of sucrose. YEPD is YEP with 2% glucose. YM-1 broth is YEPD with the addition of Bacto yeast nitrogen base (6.9 g/l) and 170 mM Na succinate pH 5.6.
Enzyme Assays
β-Lactamase activity was assayed as previously described (Harley and Tipper, 1996). Cell wall-bound invertase was assayed using a simplified version of a published procedure (Klionsky et al., 1988), scaled down for use in a Molecular Dynamics microtiter plate reader. Glucose, released by invertase hydrolysis of sucrose, is quantitated using glucostat, that is glucose oxidase coupled to oxidation of o-dianisidine by peroxidase. The signal is proportional to glucose content and invertase activity over a wide range. Overnight 30°C yeast cultures in YEPD were diluted to A600 = 0.4 (∼4 × 106 cells/ml) in fresh YEPD at 30°C and grown to A600 = 1.5–2 (3–4 h). Cells from 0.25 ml of culture were isolated by centrifugation (5 s), suspended in 1 ml of 10 mM azide, pelleted again, and suspended in 0.25 ml of 10 mM azide. Cell density of these washed cells was determined after 10-fold dilution. Duplicate samples (10 μl) of this cell suspension were mixed with 10 μl/l sucrose reagent (50 mg/ml sucrose freshly dissolved in 0.2 M NaOAc pH 5) in wells of a 96-well microtiter plate and incubated for 30 min at 30°C. Standards were 0–20 μl of 2 mM glucose in 20 μl of 0.1 M NaOAc pH 5. After addition of 150 μl of freshly made glucostat reagent (6.6 mg of o-dianisidine dissolved in 9.4 ml water + 0.6 ml of 1.0 M Na PO4 pH 7.0, 0.2 ml of glucose oxidase, 2000 U/ml [G-6891; Sigma Chemical, St Louis, MO], and 0.2 ml of 1 mg/ml horseradish peroxidase), incubation at 30°C was continued for 15 min. The reaction was stopped by the addition of 6 M HCl (100 μl). Absorbance was read at 540 nM.
The plate stain for invertase is a modified version of a similar assay (Klionsky et al., 1988). Fructose agar plates (fructose does not react with the glucostat reagent) were inoculated in patches with 10 μl of saturated broth cultures and grown overnight. Plates were overlaid with Whatman #1 filter paper disks (marked to identify orientation) previously soaked in the glucostat reagent and lightly blotted to a consistently very damp state. Using gloves (to avoid contact with o-dianisidine), light finger pressure was applied to ensure even contact of filter and patches of growth, after which the filters were immediately placed between a folded sheet of plastic cling film to prevent drying, and watched as the color developed at 23°C. The orange-brown color of oxidized o-dianisidine intensifies over time (10–20 min). The reaction can be stopped, providing a reasonably permanent record, by soaking the filter in 2 M Tris base and allowing it to dry in air.
Ethylmethanesulfonate (EMS) Mutagenesis
In a preliminary experiment, two pools of 2 × 108 cells of strain CHY255 cells were each treated with EMS in 1 ml of 0.1 M KPO4 pH 6.8 for 60 or 90 min, after which EMS was inactivated with 4 ml of 5% Na thiosulfate. The four independent pools of survivors (averaging 2 × 107 viable cells) were washed and then allowed to recover in 5 ml of YEPD for 18 h at 30°C, allowing 3–4 generations of growth. Each pool was then plated on five YEPSA plates. After 3–5 d at 30°C, 10 Suc+ clones were recovered from each plate and streaked for single colonies on YEPSA plates.
In a second experiment, six independent pools of strain CHY255 cells (2 ml, 108/ml in 0.1 M KPO4 pH 7.0) were mixed with 50 μl of EMS at 22°C for 70 min, at which time 52–61% of the cells in each pool remained viable. After addition of 8 ml of 5% Na thiosulfate to destroy residual EMS, cells were washed three times in water (12 ml) and suspended in 60 ml of YEPD. Each pool was distributed as 150-μl aliquots into four round-bottomed 96-well microtiter plates, which were sealed and rotated at an angle of 60o for 18 h at 30°C. Then 0.1 ml of each well contents was plated on USA medium (SC-Ura, 2% sucrose, 8 μg/ml antimycin A) and incubated in the dark at 23°C for 5 d to recover Suc+ clones.
Mutagenesis by Using mTn3 Libraries
The procedure described in the Yale Genome Analysis Center web site (http://ycm1.med.yale.edu/YGAC/3xHAlacZ_lib.html) was followed (Burns et al., 1994). The mTn-3xHA/lacZ library (Ross-Macdonald et al., 1999) was received in 18 pools. The 12 pools giving large numbers of transformants in E. coli were amplified. Pooled library DNA's were cut with NotI and transformed into strain CHY298. Overall efficiency was estimated by plating 1% of the total on Ura− glucose plates and the rest was plated on USA plates to select for Suc+ mutants. Candidates were picked for cloning after 3–4 d at 30°C in the dark.
Mutagenesis by Using pGTy1-H3mHIS3A1
Strain CHY301(TRP1) was transformed with pGTy1-H3 mHIS3A1(URA3) (Curcio and Garfinkel, 1991). Ura+ transformants were inoculated at 2 × 106/ml into 10 ml of Ura− galactose medium and grown at 20°C for 4–5 d. Plating on His− glucose medium indicated ∼106 His+ progeny/ml. All cells from each 10-ml culture were then plated on His−Trp− sucrose + antimycin (HTSA) plates and grown at 30°C for 3–4 d for the isolation of Suc+ mutants. These were subsequently cloned on HTSA plates, cured of the Ty1 plasmid by growth in 5-FOA medium, and retested for growth on HTSA plates.
Pulse Labeling and Immunoprecipitation
Cells transformed with YEp-S+5αHAβla or YEp-S+2αHAβla were grown to mid-log phase (107/ml) in 10 ml Ura− Met− medium at 30°C then pelleted and suspended in 1 ml of fresh prewarmed Ura− Met− medium. After 15 min at 30°C, 0.4 mCi of 35S-L Met was added followed, 20 min later, by cold 10 mM L-Met. For analysis of protease sensitivity, cells were washed in buffer A (50 mM Tris pH 7.5, 150 mM NaCl, 10 mM NaN3, 10 mM KF, 2.5 mM EDTA) then suspended in 0.3 ml of buffer A1 (buffer A plus protease inhibitors: 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml pepstatin A) containing 1.4 M sorbitol and 0.5% β-mercaptoethanol and treated with 0.2 mg/ml zymolyase 100T. After 15 min, spheroplasts were washed twice with buffer A1 plus sorbitol and then lysed in 0.2 ml of 12% sucrose in 100 mM Tris pH 7.5, 1 M EDTA plus protease inhibitors by freezing in liquid nitrogen and thawing in water eight times. For all other analyses, cells were suspended in 0.2 ml of buffer A1 and lysed by vortexing eight times 45 s with 0.5-mm glass beads with intervening cooling in ice water (1 min). After lysis, cell debris was removed by centrifugation for 1 min at 1000 × g and the low-speed supernatant was separated by airfuge (20 min at 28 psi) into membrane (pellet) and cytosolic fractions.
For immediate immunoprecipitation, the membrane pellet was dissolved in 1% SDS in buffer A1 (30 μl, 15 min at 37°C) and then diluted to 0.1% SDS with 270 μl of immunoprecipitation buffer (50 mM Tris pH 8.0, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 0.5% deoxycholate). This solution was mixed with 4 μl of Ultralink protein A beads (Pierce Chemical) for 30 min at 4°C to remove nonspecifically absorbing species, and incubated for 2 h at 4°C with 4 μl of anti-HA monoclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA) and then for an additional 2 h at 4°C with 4 μl of protein A beads. Beads were washed four times with immunoprecipitation buffer + 0.1% SDS. Bound proteins were eluted by heating in 35 μl of gel loading buffer for 15 min at 37°C and immediately fractionated by SDS-PAGE. For trypsin hydrolysis, the airfuge pellet was dispersed in 0.3 ml of buffer A and treated with N-tosyl-l-phenylalanine chloromethyl ketone-treated trypsin (5–50 ng/ml). After 20 min at 16°C, protease inhibitors were added and the membrane pellet isolated again by airfuge, dissolved, and immunoprecipitated as described above. Dried gels were visualized by autoradiography or using a Bio-Rad GS525 imager.
Assays of Growth Rate and Sensitivity to Starvation and Heat Shock
Growth rates were measured in YM-1 medium at 30°C by following absorbance at 600 nM. To test sensitivity to heat shock, 5 μl of late exponential phase cultures was patched on YEPD plates, grown overnight, and replicated to duplicate YEPD plates. One was incubated at 30°C; the duplicate was floated on a water bath at 57°C for 20 min and then cooled on ice water before incubation at 30°C. The effects of starvation were tested by incubating cultures continuously in YM-1 medium at 30°C, or at 4°C after overnight growth at 30°C, and measuring viable counts at 48-h intervals.
RESULTS
Selection for Protein Insertion Orientation (pio) Mutants
To select for pio mutants that fail to respond faithfully to a strong charge difference signal we used a modified form of S79a-PB, the model type III TM protein we previously used to analyze topogenic signals in yeast (Harley and Tipper, 1996; Harley et al., 1998). The yeast Ste2p α-factor receptor (Figure 1A) is a typical seven TM segment G protein-coupled receptor with an Nexo N-terminus. S79a (Figure 1B) is a 79-residue N-terminal fragment of Ste2p, including its normally translocated 51-residue N terminus, its first TM segment (with its central Arg58 replaced by Ile), and, at its C terminus, the eight-residue first cytoplasmic loop. Charged residues closely flanking the TM segment in S79a include three C-terminal positive charges in this loop and two N-terminal negative charges, producing a net charge difference Δ(C-N) = +5 (Hartmann et al., 1989). To simplify plasmid names while emphasizing topogenic determinants, S79a has been renamed S+5 in this article (Table 3). S79g and S42u derivatives of S79a in which the charge difference is altered to +2 and −4, respectively, are called S+2 and S−4.
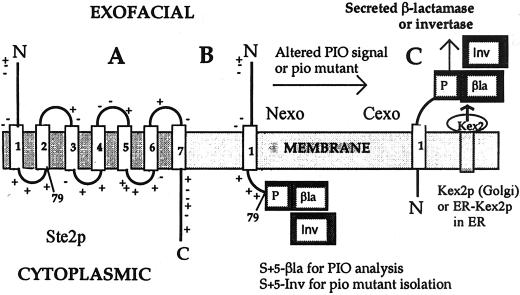
Figure 1.
S79a-PB (S+5-βla) and S79a-PI (S+5-Inv) model type III TM proteins. (A) Schematic representation of Ste2p, the α-factor receptor, showing the seven TM segments and the adjacent charged residues principally responsible for determining insertion orientation. The Arg 58 residues within TM1 is mutated to Ile in S79a (S+5), which terminates at the end of the first cytoplasmic loop, as indicated. (B) S+5-βla and S+5Inv in Nexo orientation. (C) S+5-βla and S+5Inv in Cexo orientation, indicating cleavage by Kex2p in the Golgi lumen or by ERKex2p in the ER lumen.
For topological analysis, S+5 is fused to a β-lactamase (βla) reporter in YEp-S+5βla (Table 3). β-Lactamase is preceded by P, a 58-residue fragment of the yeast secreted K1 killer protoxin that includes two sites for cleavage by Kex2p (Harley and Tipper, 1996). Kex2p is a TM protease normally located in the Golgi with its subtilisin-like active site in the lumen (Redding et al., 1991). Any fraction of S+5βla inserted Cexo and translocated to the Golgi will be processed by Kex2p, resulting in the secretion of β-lactamase (Figure 1C). If translocation and processing are efficient, therefore, topology (the Cexo/Nexo ratio) can be determined from the ratio of secreted to cell-associated β-lactamase activities, after due correction for half-lives of the different species. Retention of fusions in the ER, however, would lead to an underestimate of the Cexo fraction and to avoid this, strain CRY2, routinely used in these analyses, was modified to express high levels of a soluble, secreted form of Kex2p carrying an ER retention signal (Chaudhuri and Stephan, 1992) from a chromosomally integrated PGK promoter, producing strain CRY2A (Harley et al., 1998). Although processing of some PB fusions with longer Ste2p fragments was improved, processing of S+5βla was unaffected (Harley et al., 1998), implying efficient translocation of this fusion to the Golgi in strain CRY2.
Because of the +5 charge difference across its TM segment, S+5βla is inserted 97% Nexo in strains CRY2 and CRY2A. Analysis in strain SEY6210, the parent of the pio mutants, gave the same result. Increasing inversion of PIO is observed as the charge difference is decreased and then reversed (Harley et al., 1998). These fusions, therefore, have been shown to faithfully follow the “charge difference rule” (Hartmann et al., 1989). β-Lactamase in S+5βla was replaced by the mature sequence of invertase, producing S+5Inv (Table 3; Figure 1); this fusion is predicted to have the same 97% Nexo insertion orientation and is designed for the selection of pio mutants that fail to follow this rule faithfully. Fermentative growth of yeast in the presence of antimycin A, which inhibits respiration, requires the use of sugars such as glucose or sucrose as primary carbon and energy sources. S. cerevisiae cannot import sucrose and must secrete invertase to use sucrose for growth. If S+5Inv is expressed at an appropriate level as the only source of invertase, the ∼3% inserted Cexo and therefore processed by Kex2p to produce secreted invertase will be insufficient to support growth on sucrose. A major class of mutants selected for growth on sucrose, having a Suc+ Pio phenotype, should then have a distinct increase in the fraction of S+5Inv inserted Cexo. This phenotype can be confirmed by quantitative analysis of secreted invertase and of the topology of S+5βla and related fusions.
Expression of S+5Inv Allows Growth of suc2-Null Strain on Sucrose, but Only When Basal Levels Are Increased Four- to Sixfold
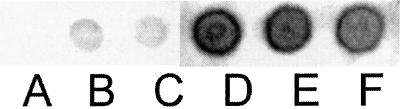
S+5Inv was expressed from the PGK promoter, which is constitutive in the presence of a fermentable carbon source. The suc2Δ strain SEY6210 lacks any sources of invertase and so fails to stain for invertase activity (Figure 2A). When S+5Inv is expressed in this strain from the single copy centromere plasmid YCp-S+5Inv (Table 3), invertase production was only 3.2% of the level in the wild-type SUC2 strain CRY2 (Table 4), giving a weak but readily detectable stain (Figure 2B). This transformant was unable to grow on sucrose media. In strains CHY255, 298, 300, and 302, S+5Inv and the URA3, LEU2, TRP1, and HIS3 markers, respectively, are integrated at the CAN1 locus of strain SEY6210, ensuring a copy number of 1. These strains were also unable to grow on sucrose media (e.g., CHY298; Figure 3A) and expressed essentially the same level of secreted invertase as the YCp-S+5Inv transformant (Table 4; Figure 2C), as did diploids made by crossing these strains.
Figure 2.
Invertase stain of pio mutants. Samples (5 μl) of saturated cultures of the indicated strains were spotted on Ura− fructose plates, grown overnight, and then stained for invertase as described in MATERIALS AND METHODS. Only external, secreted activity is detected. (A) SEY6210 (suc2Δ). (B) SEY6210 [YCp-S+5Inv]. (C) CHY255 (SEY6210 can1::S+5Inv-URA3). (D–F) Representative EMS mutants of CHY255, pio1.1, pio2.1, and pio3.1, respectively.
Table 4.
Relative secreted invertase activities of the SUC2 strain CRY2 (grown on fructose medium, control) and of transformants of the suc2 strain SEY6210, grown on glucose media
| Strain | Invertase source | % Cexo | Invertase activity |
|---|---|---|---|
| CRY2 | SUC2 | secreted | 100 |
| SEY6210 | 0 | ||
| SEY6210 | YCp-S+5Inv | 3 | 3.2 |
| SEY6210 | YEp-S+5Inv | 3 | 16 |
| SEY6210 | YCp-S+2Inv | 8 | 11 |
| SEY6210 | YCp-S−4Inv | 95 | 120 |
| CHY302 | [can1-S+5Inv] | 3 | 2.8 |
All plasmids express the invertase fusions from the PGK promoter. YCp centromere plasmids have a copy number near unity. The copy number of YEp plasmids is 10–15-fold higher. Strain CHY302 has a single copy of S+5Inv integrated at the can1 locus. S+5Inv, S+2Inv, and S−4Inv have charge differences of +5, +2, and −4, respectively, and insert as TM proteins with the indicated approximate %Cexo orientation (Harley and Tipper, 1996).
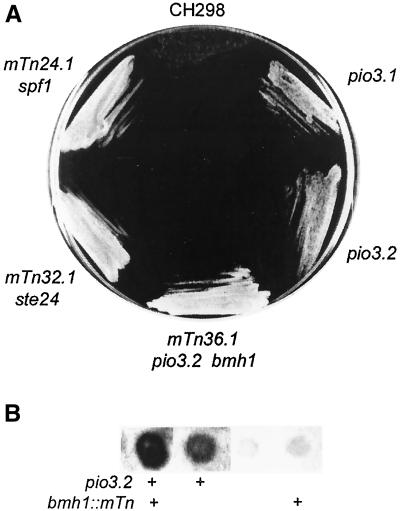
Figure 3.
(A) Growth of strain CHY298 (Suc−, top) and of representative mTn3-URA3 pio mutant derivatives on sucrose-antimycin media. Strains shown are mutant 24.1 (spf1::mTn3), 32.1 (ste24::mTn3), 36.1 (bmh1::mTn3 pio3.2), a Ura− BMH1 pio3.2 segregant from the cross of mutant 32.1 to strain CHY301, and the pio3.1 EMS mutant of strain CHY255. (B) Invertase stain of a tetrad from cross of mutant 36.1 to strain CHY301. The bmh1 mutation is marked by URA3. The pio3.2 mutation is recognized by stain intensity.
Replacement of the three positive charges following the TM segment in S+5Inv with neutral residues (Harley and Tipper, 1996) reduced the charge difference from +5 to +2, producing the YCp-S+2I fusion. Transformants had three- to fourfold higher invertase activity (Table 4) and were able to grow on sucrose, although significantly more slowly than strain CRY2. Episomal YEp plasmids typically have a copy number of 10–15 in yeast. YEp-S+5Inv transformants of strain SEY6210 produced only about fivefold higher secreted invertase activity than the YCp transformant (Table 4), presumably because transcription factors become limiting, consistent with data on β-lactamase fusions (Cartwright et al., 1994). Transformants were able to grow normally on sucrose. A form of S+5βla modified so that the charge difference is reversed to −4 (S−4βla) is inserted 95% Cexo (Harley and Tipper, 1996). SEY6210 transformed by YCp-S−4Inv produced 120% of the invertase activity of strain CRY2 (Table 4). Growth was not further enhanced. These data indicate that 15–20% of wild-type invertase activity is required for growth at the normal rate on sucrose. An approximately fivefold increase in the level of invertase secreted by a strain with a single, integrated copy of S+5Inv should, therefore, allow normal growth.
Isolation of pio Mutants by Three Mutagenic Strategies
EMS Mutagenesis
Cells of strain CHY255 (expressing S+5Inv and URA3 integrated at the CAN1 locus) were mutagenized with EMS to 55–60% lethality and Suc+ clones were isolated from four independent pools by growth on sucrose + antimycin A plates. Because it was anticipated that a major class of pio mutants would arise from mutations in essential components of the translocation machinery, selection was performed at 23°C to allow recovery of partially defective mutants with a ts phenotype. Of 120 Suc+ mutants tested, none were ts. The frequency of rapidly growing Suc+ clones among survivors was 3–6 × 10−6. Approximately 10 times as many slowly growing clones were present but were not pursued. The low frequency of rapidly growing mutants suggests that the number of loci with a major effect on the Suc phenotype of this strain is small. All Suc+ mutants reverted to Suc− when cured of the S+5Inv insert by growth on 5-FOA plates and regained Suc+ phenotype when retransformed with YCp-S+5Inv, so resulted from mutations independent of this reporter fusion. Colony stain for invertase indicated that all had a distinctly stronger secreted activity than the parent. It should be noted that although this stain can easily detect the 3% of wild-type activity seen in the parent strain 255 (Figure 2C), it is insensitive to variation above ∼30% of wild-type levels, as defined by strains such as CRY2. Nevertheless, invertase stain intensity correlated with the rate of growth on sucrose for all of the Suc+ mutants isolated in this study. This suggests that none arose from acquisition of permease activity for sucrose. Release of intracellular invertase by partial lysis of mutants with fragile cell walls was another potential source of misleading mutants. However, none of these mutants, or the additional pio mutants described below, became Suc− when grown on 1 M sorbitol medium. In addition, several representative pio mutants were tested for release of β-lactamase expressed cytoplasmically from cPB (Harley and Tipper, 1996). None did so, although release of 2% would have been readily detected (our unpublished data).
Twenty EMS mutants with the strongest Suc+ and invertase phenotypes were chosen for genetic analysis. Diploids produced from mating to the Trp+ strain CHY301 were Suc−, indicating that the Suc+ phenotypes were recessive. On sporulation, seven of the diploid derivatives gave 2:2 segregation of Suc+/Suc− phenotypes, consistent with mutations in single loci. All others gave patterns indicating segregation of multiple determinants of Suc+ phenotype. Allelism among the seven mutants was tested by mating of appropriate segregants and analysis of growth on sucrose and invertase stain intensity. Confirmation required analysis of meiotic segregation patterns because some diploids, although Suc−, had an intermediate level of invertase stain intensity. Three complementation groups were found: four mutants belonged to pio1, two to pio2, and one to pio3. Invertase stains of representative segregants are shown in Figure 2, D–F. Segregation patterns showed clear linkage of pio1 to the marker integrated at CAN1, a gene on chromosome V. Additionally, pio1 mutants all showed a modest reduction in growth rate at all temperatures. At 30°C in YEPD medium, doubling time increased from 95 min in the CHY255 parent to 104 min in the mutants.
EMS mutagenesis was repeated under milder conditions to measure the frequency of ts mutants among a much larger group of independent pio Suc+ mutants. Six independent pools of strain CHY255 cells were mutagenized to 35–40% lethality and 384 pools from each were allowed to recover in microtiter plates at 23°C. Suc+ clones were isolated from each well on USA plates at 23°C, resulting in recovery of 2216 independent mutants. The frequency of Suc+ clones among survivors was ∼2 × 10−6. Only 32 of these clones were ts, a frequency of 1.5%, most likely deriving from mutations unrelated to those causing Suc+ phenotype. pio mutations in essential genes are, therefore, very rare or nonexistent. Because the weak phenotype of pio mutants impeded cloning by complementation (see below) and this analysis of EMS mutants suggested that no essential genes may be involved in the Pio phenotype, we decided to isolate additional mutants by using transposon mutagenesis. This greatly facilitates the cloning and genetic analysis of nonessential genes with weak phenotypes, but is essentially limited to the isolation of null mutants.
mTn3 Mutagenesis.
mTn3-URA3 mutant libraries from the Snyder laboratory (Burns et al., 1994) were used to isolate additional pio mutants from strain CHY298. The URA3 libraries used can create in-frame fusions to lacZ, which can be used to analyze expression. Isolation of mutations in any particular gene depends on the completeness of the library in use. Twelve independent pools of mTn-URA3 libraries were amplified in E. coli and transformed into strain CHY298. Although the efficiency of transformation to Ura+ was >105 in each case, only six pools (pools 21, 22, 24, 32, 36, and 38) elicited Suc+ colonies, and subsequent genetic and sequence analysis showed that each of these libraries produced multiple isolates of a single mutation. Growth of representative mTn3 mutants from pools 24, 32, and 36 on sucrose-antimycin medium is compared with the strain CHY298 parent in Figure 3. These amplified libraries were clearly incomplete, suggesting that the pio mutants class may not have been saturated by this technique. We therefore isolated additional pio mutants by random mutagenesis with a version of the yeast Ty retroposon that allows direct selection for transposition, potentially providing both random mutagenesis and the same benefits for cloning as use of the mTn3 libraries.
pGTy1-H3 mHIS3A1 Mutagenesis
Ty1-H3 mHIS3A1 is a modified form of the Ty1, a retroposon that transposes via an RNA intermediate (Curcio and Garfinkel, 1991). Ty1 transcription is driven by the galactose-inducible GAL1 promoter, so that growth on galactose induces a high rate of transposition. The HIS3 gene is inserted in opposite orientation to the Ty1 transcript and is interrupted by an inverted intron that is excised only in Ty1 transcripts, that is during transposition. The Ty1-HIS3A1 element is introduced on a URA3 plasmid and, following growth on galactose medium at 18°C (the transposase is temperature sensitive), ∼1% of the cells became His+, indicating that at least one Ty1 transposition event has occurred. Although 90% of Ty1 transpositions result in insertion in AT-rich intergenic regions, essentially any gene can be interrupted by Ty1 transposition (Smith et al., 1996). Genomic DNA adjacent to the Ty1 insertion site can be characterized as described for mTn3 mutants. In our hands, however, the mutagenesis efficiency was low and the majority of mutants were unstable, reverting to Suc− with high frequency. Fifty independent cultures of Ura+ pGTy1-H3 mHIS3A1 transformants of strain CHY301 were grown on galactose at 18°C for 4 d and His+ transposition products with Suc+ Pio phenotype were selected. Only eight stable, independent Suc+ mutants were isolated.
Expression of Translocon Components Fails to Complement pio Mutants
SEC61, SEC62, SEC72, SSS1, SBH1, and SBH2 genes encode major components of the yeast translocon and SEC65 encodes the major protein of the signal recognition particle. These genes, cloned on YCp vectors, failed to complement any of the seven genetically characterized EMS pio mutants, the six mTn3 mutants, or the eight Ty-HIS3A mutants.
Cloning of SPF1 by Complementation of pio1.1
Strain DK4 (Table 1) is a pio1.1 Suc+ segregant from genetic analysis of EMS mutants (Figure 2D) and has the TRP1 marker integrated with S+5Inv at can1. This strain was transformed with several pools of a YCp50 (URA3) library of random yeast genomic fragments (Rose et al., 1987). Complementation of the Suc+ phenotype was tested by staining for invertase activity after growth on Ura− Trp− fructose plates. Selection for Trp+ prevented isolation of clones from which the S+5Inv-TRP1 reporter had looped out, which were otherwise observed at a frequency of 3 × 10−4. Eleven of 6500 Ura+ transformants stained only weakly for invertase, resembling the pio1.1/WT diploid, and were phenotypically Suc−. In contrast, a control transformed with the YCp50 vector grew normally on sucrose. Plasmids were recovered in E. coli. Transformation back into strain DK4 confirmed complementation of the Suc+ phenotype. Restriction analysis showed that all plasmids had the identical insert of 13.2 kDa. Sequence analysis, using primers complementary to the pBR322 sequences adjacent to the BamHI site used in library construction, confirmed this identity and showed the clone inserts to span the YEL030w, 31w, and 32w genes on chromosome V. This observation is consistent with the observed linkage of pio1 to CANI (YEL063c), from which this segment is separated by ∼45 kDa. Subcloning into the YCp vector pRS314 identified the complementing gene as YEL031w. YEL031w was previously cloned as sensitive to Pichia farinosa killer toxin (SPF1) by selection for resistance to the proteinaceous salt-mediated killer (SMK) toxin produced by strain KK1 of P. farinosa (Suzuki and Shimma, 1999). The SPF1 subclone was transferred from pRS314 to YEp351 (LEU2) (Table 2). This multicopy plasmid also complemented the pio1.1 mutant. The complemented strain and a control strain carrying the functional chromosomal copy as well as these multiple copies of SPF1 secreted normal amounts of invertase and grew normally on sucrose. Overexpression of SPF1, therefore, had no detected phenotype.
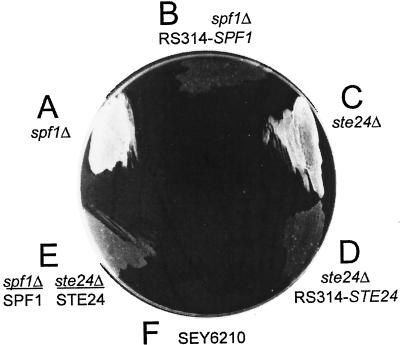
SPF1 is a gene of unknown function that encodes one of the 16 P-type ATPases found in the S. cerevisiae genome (Catty et al., 1997). Saturation Ty1 mutagenesis of chromosome V showed it to be nonessential (Smith et al., 1996); the only phenotype observed was a reduction of ∼10% in growth rate, as observed in our pio1 mutants. However, Dr. Chise Suzuki showed that spf1-null mutants are resistant to SMK toxin (Suzuki and Shimma, 1999) and confirmed that all of our pio1 mutants were also fully resistant (Suzuki, unpublished data). We used plasmid pCS202 (Suzuki and Shimma, 1999) to replace the C-terminal 75% of SPF1 with the TRP1 marker in strains CHY255 and 298. These spf1Δ-TRP1 mutants had the same Suc+ phenotype (Figure 4A) as the EMS pio1 mutants whose recessive phenotypes are consistent with loss of function. All pio1 mutants and the spf1Δ-TRP1 mutant were fully complemented by the pRS314 subclone of SPF1 (Figure 4B). Spf1-null mutants, therefore, have a strong Pio phenotype. Invertase secretion levels were increased sevenfold (Table 5). An spf1-null/SPF1 diploid, although Suc−, showed slightly enhanced growth (Figure 4E) and had a slight increase in invertase activity relative to the SPF1 haploid (Table 5), presumably the effect of a gene dosage of 0.5.
Figure 4.
Growth of null mutants of strain CHY302 on sucrose-antimycin media. Strains shown are spf1Δ::TRP1, with and without complementation by pRS314-SPF1; ste24Δ::LEU2, with and without complementation by pRS314-STE24; the Suc− diploid from cross of the spf1Δ::TRP1 and ste24Δ::LEU2 strains; and the suc2Δ strain SEY6210.
Table 5.
Invertase activities of intact cells of strain CHY302, expressing an integrated copy of S+5Inv, and of its pio mutant derivatives, relative to strain CRY2 (100%; Table 4)
| Mutant | Invertase activity |
|---|---|
| Wild type | 3 |
| spf1-null | 22 |
| ste24-null | 24 |
| spf1-ste24-null | 39 |
| bmh1-TnURA3 | 3 |
| pio3.2 | 12 |
| mTn3 mutant 36 pio3.2 bmh1-TnURA3 | 21 |
| spf1Δ∷TRP1/SPF1 diploid | 5 |
The pio3.2 bmh1-TnURA3 double mutant is the mTn3 mutant 36 (Figure 3).
We were unable to clone either pio2 or the weaker pio3 EMS mutants by complementation using the YCp50 library or a YEp24 library (Carlson and Botstein, 1982), in spite of screening large numbers of transformants. Three distinct YEp24 library clones substantially suppressed the secreted invertase activity of pio3 mutants but only moderately suppressed their growth on sucrose media. Analysis of these multicopy weak suppressors (YDR305c-07w, YMR272c-3c, YOR193w-5w) was not pursued.
MTn3 Mutants 21 and 24 Are Alleles of SPF1
The mTn3 insertion mutants from libraries 21, 24, and 38 were all complemented by pRS314-SPF1. Transposon mutants 21 and 24 had phenotypes identical to the original EMS pio1.1 mutants: an intermediate level of secreted invertase activity (Table 5), a near-normal growth rate on sucrose, a modest reduction in growth rate on glucose media, and resistance to SMK toxin (Suzuki, personal communication). pRS314-SPF1 transformants of these mutants were Suc− and secreted invertase activity was reduced to that of the strain CHY298 parent. Sequence analysis confirmed insertion of the transposon into SPF1 in both mutants. Neither insertion produced an in-frame fusion to lacZ. Mutant 38 isolates had a weaker phenotype, growing slowly on sucrose, and remained sensitive to SMK toxin. Sequence analysis showed the transposon insertion to be located in rDNA, probably irrelevant to the Pio phenotype. Presumably, transformation had induced a leaky point mutation in SPF1.
MTn3 Mutants 22 and 36 Are Double Mutants
Mutant 22 had a relatively weak phenotype. The transposon insertion site, identified by cloning and sequence analysis, was in the ADA2 gene. However, transformation with the wild-type gene in pNS3.8 failed to complement the phenotype, which again appears to result from a separate adventitious mutation. It was complemented by neither SPF1 nor STE24 (see below). It was not further characterized.
The invertase activity of mTn3 mutant 36 was equivalent to that of an spf1-null (Table 5) and the mutant grew normally on sucrose media (Figure 3A). Genetic analysis showed that this mutation was allelic to the weaker EMS pio3.1 mutation; the diploid from mating of these mutants remained Suc+, as did all haploid segregants. However, these segregants had two levels of secreted invertase activity, the higher cosegregating with the mTn3-URA3 marker and resembling the original mutant 36 isolate, and the lower resembling the pio3.1 mutant. When mutant 36 was crossed to the Suc− strain CHY301, the diploid was Suc− and haploid segregants again demonstrated both strong and weaker invertase activities, the higher activity always segregating with the URA3 mTn3 marker, although some Ura+ segregants were Suc− (Figure 3B). Two unlinked mutations are apparently involved in the phenotype of this mutant. The transposon insertion site was identified as the BMH1 gene by cloning and sequence analysis. However, transformation with the wild-type gene in pB3455 only modestly reduced secreted invertase activity in mTn3 mutant 36. A bmh1-null mutant of strain CHY301, constructed using pB3453, had no significant increase in secreted invertase activity, as in some bmh1::mTn3 Ura+ segregants (Figure 3B; Table 5) and remained Suc−. The strong phenotype of mutant 36 results, therefore, from combination of an mTn3 insertion in bmh1 with a second mutation allelic to pio3.1 that was named pio3.2. The combination results in normal growth on sucrose media, whereas the pio3.1 and pio3.2 mutants grew more slowly (Figure 3A) and have about half of the secreted invertase activity of the double mutant (Table 5). A bmh1 bmh2 double null mutant (Roberts et al., 1997) also had no Pio phenotype: when transformed with the YEp-S+5βla plasmid: β-lactamase activity data indicated normal 97% Nexo insertion (our unpublished data). PIO3 remains unidentified. The high frequency of secondary mutations appears to be a weakness of the mTn3 mutagenesis technique, at least in strain SEY6210.
PIO2 Is STE24. Null Mutants Have a Strong Pio Phenotype
The mTn3 mutant 32 grew normally on sucrose (Figure 3A). The transposon insertion site was identified in STE24 by pZ-his5 insertion, cloning, and sequence analysis. Both Suc+ and invertase stain Pio phenotypes were fully complemented by pSM1069, carrying the wild-type gene in pRS315 (LEU2) (Table 2; Fujimura-Kamada et al., 1997). A ste24ΔLEU2 deletion mutant of strain CHY301, constructed using pSM1072 (Table 2; Fujimura-Kamada et al., 1997), had the same Pio phenotype as mTn3 mutant 32 (Figure 4C) and was fully complemented by STE24 subcloned into pRS314 (Figure 4D). Growth rate was not altered. Ste24p is a polytopic transmembrane protein, located in the ER with its zinc metalloprotease active site located in a cytoplasmic loop between predicted TM segments 4 and 5 (Schmidt et al., 1998). pSM1104 encodes a ste24-null mutant in which Ala replaces Glu298 in the HEXXH sequence required for zinc binding and activity in Ste24p (Fujimura-Kamada et al., 1997). This plasmid failed to complement the Pio phenotypes of mutant 32, indicating that loss of Ste24p protease activity is sufficient to cause these Pio phenotypes. The diploid made by crossing the spf1ΔTRP1 and ste24ΔLEU2 mutants, heterozygous for both markers, was Suc−, further demonstrating that both phenotypes are recessive (Figure 4E).
All pGTy1-H3mHIS3A1 Mutants Are Alleles of SPF1 or STE24
Eight stable mutants were isolated by selection of His+ Ty1-H3 mHIS3A1 transposition products for growth on sucrose, a frequency of ∼10−7/His+ mutant, or ∼10−6/nonsilent transposition event (see MATERIALS AND METHODS), similar to the EMS-induced frequency. Transformation of the Ty-HIS3A insertion mutants with the wild-type genes showed that five were alleles of SPF1 and three were alleles of STE24. Mutagenesis was repeated in strain CHY298 transformed with a derivative of pRS314 carrying both the SPF1 and STE24 genes (Table 2). No stable Suc+ mutants were recovered. We again conclude that the number of nonessential loci potentially responsible for a Pio phenotype is small.
Confirmation of Pio Phenotypes of spf1 and ste24 Mutants by Using β-Lactamase Fusions
The selection used for isolation of pio mutants requires import of sucrose or enhanced secretion of invertase expressed from S+5Inv. The colony stain for invertase activity indicated enhanced secretion, confirmed by assay (Figure 3; Table 5). Both spf1- and ste24-null mutations resulted in a five- to sevenfold increase in invertase secretion, as expected for mutants with a strong Suc+ phenotype (Table 4). This increase could result from an increase in total expression from the PGK promoter, from stabilization and enhanced export from the ER of the S+5Inv fraction normally inserted Cexo, from enhanced cell lysis, or from an increase in Cexo insertion of S+5Inv. We previously demonstrated that PIO in yeast could be determined, for the equivalent S+5βla fusion and its variants, from the ratio of cell-associated β-lactamase activity (derived from Nexo fusions) to secreted activity (derived from Cexo fusions by Kex2p cleavage) (Harley and Tipper, 1996; Harley et al., 1998). PGK promoter-driven expression of the S+5βla fusion showed unaltered total expression in spf1- and ste24-null mutants. Cexo insertion was increased four- to fivefold, similar to the increase observed in invertase secretion (Table 6). Cexo insertion of the S+2βla fusion with a smaller +2 charge difference was increased approximately threefold above a distinctly higher basal level in these mutants. The PIO of an S0βla fusion (S60-PB) with zero charge difference is normally ∼38% Cexo, dictated by the hydrophobicity of its TM segment (Harley et al., 1998). Cexo insertion was unaffected by an spf1 mutation and slightly decreased by a ste24 mutation (Table 6). These results are entirely consistent with a reduced response to charge difference signals being the major effect of the pio mutants, because a complete failure to respond to such a signal should presumably result in 38% Cexo insertion of the S+5 βla fusion.
Table 6.
Pio phenotypes of the pio3.1 and pio null mutant derivatives of strain CHY302
| Reporter Δ(C-N) = | S+5β1a +5 | S+2β1a +2 | S0β1a 0 | S+5αHAh9β1a +5 | S+2αHAh9β1a +2 |
|---|---|---|---|---|---|
| Strain | Secreted β-lactamase activity | 35S-Met incorporation | |||
| CHY302 | 2 | 6 | 37 | 8 | 16 |
| spf1-null | 9 | 18 | 38 | 22 | 29 |
| ste24-null | 8 | 16 | 31 | 25 | 32 |
| spf1-ste24-null | 14 | 26 | 30 | 36 | 43 |
| pio3.1 | 6 | 9 | nd | nd | nd |
| pmr1-null | 2 | 3 | 35 | nd | nd |
| spf1-pmr1-null | 11 | 18 | nd | nd | nd |
Δ (C-N), the charge difference flanking the TM segment, is indicated for each fusion. Data are expressed as %Cexo insertion of the indicated reporter fusions, expressed from the PGK promoter in YEp vectors. Data for β1a fusions (columns 2–5) are derived from the percentage of total β-lactamase activity secreted, as previously described (Harley and Tipper, 1996; Harley et al., 1998). Total activities were similar in all strains. Data for the αHAh9-β1a fusions in columns 5 and 6 are derived from scans of the gels whose autoradiograms are shown in Figure 6.
nd, not determined.
Effects of spf1 and ste24 Mutations on Invertase and β-Lactamase Secretion Are Additive. Double Mutants Are Stress Sensitive
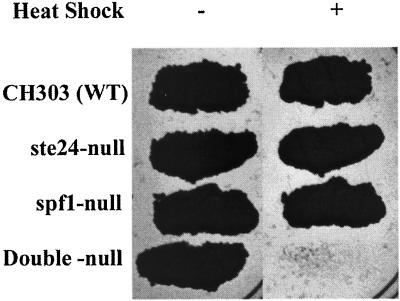
Double mutant meiotic segregants were derived from mating spf1Δ::TRP1 and ste24Δ::LEU2 mutants. Secreted invertase activity was near the sum of activities for the single mutants (Table 5), indicating additivity of mutant effects. Effects on β-lactamase secretion from the S+5βla and S+2βla fusions were also additive in the double mutant (Table 6). Growth on sucrose appeared normal but the growth rate on glucose was markedly decreased relative to that of the spf1Δ::TRP1 mutant, with a doubling time increased by 60% to 152 min. Growth to stationary phase in YM-1 medium allows survival of cultures of normal yeast strains for several months at 4°C. Both spf1- and ste24-null mutants were normal in this respect and all mutants survived normally at 30°C. The double mutant, however, had 10% survival after 15 d at 4°C and <1% after a month. Cold sensitivity suggests sensitivity to stress and this was confirmed by testing sensitivity to heat shock. Patches of the parent strain CHY303 and of single and double spf1 and ste24 mutants were replica plated on YEPD and grown 24 h at 30°C, with and without prior exposure to 57°C for 20 min. No double mutant cells survived this heat shock although the single mutants were unaffected (Figure 5).
Figure 5.
Heat shock sensitivity of pio mutants. YEPD plates were inoculated with 5 μl of fresh saturated cultures of the indicated strains, grown overnight, and then replica plated to fresh YEPD plates. These were grown at 30°C with (+) and without (−) prior exposure to 57°C for 20 min.
Pulse Label Analysis of pio Mutant Phenotypes
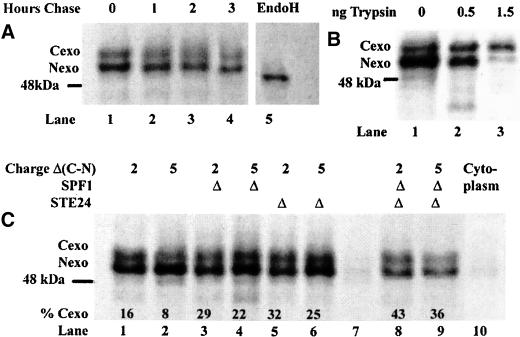
PIO data from the analysis of secreted invertase and β-lactamase activities in pio mutants are directly relevant to the mutant selection used and are internally consistent, so that deduced relative levels of Cexo insertion are significant. However, these assays are indirect and may give a distorted view of the actual PIO because of variations in half-life and processing efficiency of the different fusion species. We therefore used direct, independent assays of PIO based on the analysis of pulse-labeled species isolated by immunoprecipitation. The S+5αHAβla and S+2αHAβla fusions (Table 3) were constructed for this purpose and are based on the βla fusions described above with three modifications. First, insertion of the triple HA epitope allows efficient precipitation with commercially available monoclonal antisera. Second, the preceding 30-residue α segment provides two efficiently used N-glycosylation sites that should be modified only in the Cexo orientation, whereas the S+5 N terminus will be glycosylated, at a single site, only in the Nexo orientation. Third, this α segment terminates in a single trypsin-sensitive Lys residue, derived from the original Kex2p cleavage site of the α-factor precursor. In the absence of a Kex2p sensitive site, both Cexo and Nexo species should be stably incorporated into membranes. Predicted sizes are 52 and 49 kDa, assuming core N-glycosylation. The C-terminal βla sequence provides nine additional Met residues, greatly enhancing the efficiency of labeling with 35S-Met, and was previously shown to be without detectable effect on the insertion orientation of similar fusions in yeast (Harley et al., 1996). If the predicted species are observed exclusively in membranes and have similar half-lives, and if topology assignments are confirmed by analysis of glycosylation and protease sensitivity then PIO can be deduced directly from the ratio of label incorporated into these species.
In preliminary experiments it was shown, by Western blot with anti-HA, that expression of these fusions gave rise to just two bands corresponding to glycosylated species of apparent size 55 and 52 kDa, respectively (based on mobility relative to a prestained standard mix), located >95% in the membrane pellet derived from airfuge fractionation of low-speed supernatants from cell lysis. Immunoprecipitation resulted in complete recovery of these species (our unpublished data). Immunoprecipitated species from expression of S+2αHAβla in normal cells (strain CHY303), labeled for 20 min with 35S-Met, are shown in Figure 6A. The same two species seen by Western blot were observed in the membrane pellets, and sensitivity to endoglycosidase H confirmed that both species were N-glycosylated (lane 5). Less than 3% of either species was present in the airfuge supernatant (cytoplasmic) fraction (Figure 6C, lane 10). Chase in the presence of excess cold methionine for 1–3 h showed that both species had 2–3 h half-lives (lanes 1–4), similar to the 2–4-h half-life deduced for the Nexo species of related fusions by Western blot (Harley et al., 1998). Because the N terminus of the Ste2p fragment in these fusions lacks Lys and Arg residues, the Cexo species should be completely resistant to trypsin in intact microsomes, whereas the Nexo species should be rapidly degraded. Trypsin treatment of gently solubilized membranes gave the expected result, although the Cexo species was only partially protected (Figure 6B). The validity of topology assays based on the ratio of incorporation of label into the 56- and 52-kDa bands was, therefore, confirmed.
Figure 6.
TM topology of S+5αHAh9-βla and of S+2αHAh9-βla, expressed in strain CHY302 and in single and double pio mutant derivatives. The membrane-associated 35S-Met-labeled doubly glycosylated Cexo (top) and singly glycosylated Nexo (bottom) species were isolated by immunoprecipitation and detected by autoradiography after fractionation by SDS-PAGE. In each segment, the position of the nominally 48-kDa prestained marker is indicated. (A) Cells of strain CHY302 expressing S+2αHAh9-βla were labeled for 20 min and then chased with excess cold Met for 1, 2, or 3 h, as indicated (lanes 1–4). The sample in lane 5 is identical to that in lane 1 except that the solubilized membrane fraction was hydrolyzed with endoglycosidase H (EndoH) before immunoprecipitation. (B) Microsomal membrane fraction from strain CHY302 expressing S+2αHAh9-βla and labeled for 20 min was treated with the indicated quantity of trypsin for 15 min before solubilization and immunoprecipitation. (C) Expression of S+2αHAh9-βla (charge +2, lanes 1, 3, 5, and 8) and S+5αHAh9-βla (charge +5, lanes 2, 4, 6, and 9) in strain CHY302 (lanes 1 and 2) and in the indicated single and double mutant derivatives. All are membrane fractions except lane 10, the cytoplasmic fraction corresponding to lane 1 (strain CHY302). The data for percentage of Cexo are derived from scanning with the GS-525 molecular imager. Lane 7 is a negative control of labeled membranes from cells expressing the YEp352 URA3 vector.
Both S+5αHAβla la and S+2αHAβla fusions were expressed in single and double pio mutants and the labeled species were quantitated using a Bio-Rad GS525 molecular imager. The data (Figure 6C; Table 6) confirmed the results obtained by analysis of invertase and β-lactamase secretion, although deduced levels of Cexo insertion, especially in cells of the normal CHY303 strain, were significantly higher. Assays based on secretion may underestimate Cexo insertion if processing or post-Golgi sorting to the plasma membrane is incomplete. Cexo insertion of the S+5 fusion was increased approximately threefold in the single mutants and four- to fivefold in the double mutant. Insertion of the S+2 fusion was increased about twofold in single mutants. The increase in insertion in the double mutant was less pronounced than in the S+5 fusion, probably because of approach to the limit seen in the absence of a charge difference, although this limit has not been determined for related fusions by this technique.
Tests of Mutations in Genes Related to SPF1 and STE24 for Pio Phenotype
Because the known functions of the SPF1 and STE24 gene products are apparently unrelated to each other or to protein translocation, we tested the effects on PIO of mutations in related genes, hoping to clarify the mechanisms of pio mutations.
YOR291w Is a Paralog of SPF1; Mutants Have No Pio or Other Detected Phenotype
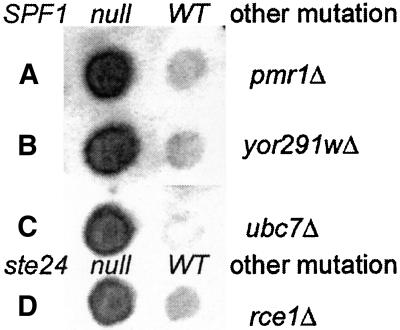
Spf1p is a P-type ATPase, presumably an ATP-driven ion pump. Its closest relative is YOR291wp (26% identity, 44% similarity across all 1124 residues of homology) with which it forms a newly recognized subfamily, highly conserved in metazoans (Axelsen and Palmgren, 1998). Because their unknown function is presumably important, the nonessential nature of SPF1 might result from redundant function with YOR291w. We disrupted YOR291w by replacement with the S. pombe his5 gene in a diploid heterozygous for spf1Δ::TRP1 and ste24Δ::LEU2 mutations. The pombe his5 gene complements S. cerevisiae his3 mutations and its use avoids the possibility of confounding gene conversion or recombination events. Disruption of YOR291w in the resultant His+ diploid was confirmed by PCR analysis. After sporulation, most tetrads had 4:0 viability, and all combinations of markers were viable. Phenotypes are given in Table 7. Deletion of YOR291w was without detectable phenotype in WT or spf1-null cells (Figure 7) and also failed to affect ste24-null cells. The growth rate of the ste24-spf1-double null mutant was unaltered on either glucose or sucrose media by deletion of YOR291w. It appears, therefore, that a yor291w-null mutation has no Pio or other detectable phenotype, consistent with a previous report (Cronin et al., 2000).
Table 7.
Effect of pmr1, yor291w, and ubc7-null mutations on invertase activities of spf1-null mutants of strain CHY302 and of an rce1-null mutation on the ste24-null derivative
| Mutant | Invertase activity |
|---|---|
| Wild type | 3 |
| spf1-null | 22 |
| yor291w-null | 3 |
| yor291w-spf1-null | 21 |
| pmr1-null | 4 |
| pmr1-spf1-null | 25 |
| ubc7-null | 3 |
| ubc7-spf1-null | 23 |
| ste24-null | 24 |
| rce1-null | 3 |
| rce1-ste24-null | 27 |
The data for the single pio mutants are repeated from Table 5.
Figure 7.
Invertase stain of pmr1-, yor291w-, and ubc7-null mutants with and without an spf1Δ::TRP1 mutation and of rce1-null with and without a ste24Δ::TRP1 mutation.
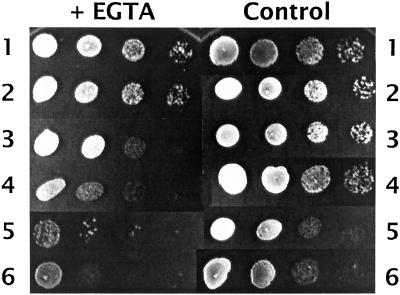
spf1 Mutations Exacerbate EGTA Sensitivity of pmr1 Mutants, but pmr1 Mutants Have No Pio Phenotype
Pmr1p, the major identified Ca2+ pump in S. cerevisiae (Rudolph et al., 1989; Okorokov and Lehle, 1998), is a nonessential P-type ATPase located in the Golgi. It is more distantly related to Spf1p (22% identity, 39% similarity across 733 residues of homology) than is YOR291wp. Because Spf1p is located in the ER or Golgi (Cronin et al., 2000; Suzuki, 2001), it seemed plausible that it might be the unidentified yeast ER Ca2+ pump and that Pio phenotypes might result from disturbance of Ca2+ homeostasis mechanisms. We therefore disrupted PMR1 in a diploid heterozygous for spf1Δ::TRP1 using pmr1Δ::LEU2 (Rudolph et al., 1989). After meiosis, viability in spore tetrads was 4:0. The spf1 pmr1 double mutants grew significantly more slowly than spf1 segregants (Figure 8, controls 3 and 5), but did not show temperature sensitivity. The pmr1 disruptants showed no Pio phenotype, failing to increase invertase secretion (Table 7; Figure 7) or to grow on sucrose. The double spf1 pmr1 mutants retained their Pio phenotype, but did not increase invertase secretion (Table 7; Figure 7). On synthetic media containing 2 mM Ca2+, the pmr1 disruptants were distinctly more sensitive to EGTA (which preferentially chelates Ca2+) than WT cells, as previously reported (Rudolph et al., 1989). The spf1 disruptants had a modest increase in sensitivity, whereas the double mutants had a marked increase in sensitivity (Figure 8). Ca2+ (60–300 mM), however, failed to suppress the Pio phenotype of spf1 disruptants (our unpublished data). These results are consistent with a role for Spf1p in controlling Ca2+ levels in the ER, but show that the Pmr1p Golgi Ca2+ pump fails to share its role in determining Pio responses.
Figure 8.
Sensitivity of pio and pmr1 mutants of strain CHY302 to EGTA. Suspensions containing (5 μl) ∼50,000, 5000, 500, and 50 cells (left to right) were spotted onto complete synthetic medium (2.5 mM Ca2+) containing 15 mM EGTA (left) or no EGTA (right). Strains were wild type (1), ste24-null (2), spf1-null (3), pmr1-null (4), pmr1-spf1-null (5), and pmr1-spf1-ste24-null (6). Note that the pmr1-spf1 double null mutants (strains 5 and 6) grow more slowly, as seen in the controls. Growth is shown after 34 h except for strain 5 + EDTA, which is shown after 48 h, when viability is seen more clearly.
UBC7 Mutants Have No Pio Phenotype
It has recently been observed that SPF1, like the ER-associated degradation (ERAD) pathway, is among the many ER and secretory pathway functions regulated by the unfolded protein response (Travers et al., 2000). Moreover, pmr1-null mutants are defective in the ER export and subsequent proteosomal degradation of mutant forms of carboxypeptidase Y (Duerr et al., 1998). It was therefore necessary to test whether spf1-null Pio phenotypes might result from a defect in ERAD of miss-inserted TM proteins, possibly resulting in a marked increase in the stability of the Cexo TM form. However, a ubc7-null mutation, which inactivates a major branch of the ERAD pathway, failed to affect Pio phenotypes of WT or spf1-null cells (Figure 7; Table 7). Also, the analysis of Pio phenotypes with β-lactamase fusions failed to detect any effects of pio mutations on stability.
RCE1 Encodes a Second CAAX Box Peptidase; rce1 Mutant Have No Pio Phenotype
Ste24p has two distinct roles in the maturation of a-factor and ste24 mutants have a MATa-specific sterile phenotype (Fujimura-Kamada et al., 1997; Boyartchuk and Rine, 1998). The sterility is only partial, because there is some redundancy in processing activities (Tam et al., 1998). The precursor of a-factor, a farnesylated dodecapeptide, has a C-terminal CAAX box and a 21-residue N-terminal extension. Ste24p and Rce1p both induce cleavage of the AAX C-terminal tripeptide from CAAX box family proteins. Double mutants are viable but completely MATa sterile. They cleave the C-AAX bond in the a-factor precursor with similar efficiency (Tam et al., 1998). Ste24p, but not Rce1p, is also specifically required for N-terminal cleavage of the farnesylated a-factor precursor between residues 7 and 8, a sequence unrelated to C-AAX (Schmidt et al., 1998; Tam et al., 1998). The roles of these two ste24p functions in the Pio phenotype should be distinguished by testing the effect of an rce1-null mutation. An rce1-null derivative of strain CHY301 was unaffected in invertase secretion and the double mutant with ste24-null, which grows well, showed the same level of secreted invertase activity as the ste24 mutant itself (Figure 7; Table 7).
DISCUSSION
The function of the signals determining TM PIO and of the mechanisms for response to these signals must be to maximize insertion in functional orientation because of the inherent waste in metabolic energy, potential for cumulative toxicity, and burden on protein turnover and stress response mechanisms resulting from errors. The normal error rate is uncertain, but is certainly significant, probably varying widely between proteins. Overall turnover of nascent proteins in mammalian cells is estimated at 30% (Schubert et al., 2000). The assembly of multicomponent TM protein complexes, such as T-cell receptors, is known to be highly regulated and inherently inefficient, whereas the error rate for simple monotopic TM proteins may be low. Our analysis of model monotopic TM proteins in yeast would suggest a range extending from 3% to at least 10%. A reduction in efficiency resulting from mutagenic damage to the mechanisms for response to PIO signals would place mutants at a significant selective disadvantage in a normal population. However, in pure culture, such mutants can apparently survive, albeit with some reduction in growth rate. Even in a double mutant where the error rate approached 30%, this probably represents no more than 10% of total protein production, suggesting that the observed marked growth inhibition and sensitivity to stresses are largely due to toxic effects of miss-incorporation.
The PIO selection was designed to isolate mutants that respond poorly to the strong +5 charge difference signal in the S+5Inv fusion, a model type III (Nexo) TM protein (Figure 1). An increase in Cexo insertion should cause a corresponding increase in secretion of invertase, allowing growth on sucrose. Only two complementation groups with strong phenotype, SPF1 and STE24, were identified after extensive mutagenesis by three independent techniques. A third group, PIO3, has a weaker phenotype enhanced by loss of function in Bmh1p and has not been identified or extensively characterized. The low frequency of pio mutants among survivors of mutagenesis suggests that few if any additional loci give a strong phenotype in this selection. The phenotypes of spf1 and ste24 mutants were shown to result from the predicted alteration in insertion orientation, apparently resulting in turn from decreased responsiveness to a charge difference signal.
Because the insertion of most TM proteins is apparently irreversible, responses to topogenic signals must occur during membrane insertion of the nascent protein, usually during translocation at the ER. It is consistent, therefore, that the products of both SPF1 and STE24 are polytopic TM proteins located in or near the ER. Because the response to charge is independent of sequence and N- or C-terminal location of the charge relative to the TM segment (Harley et al., 1998), the signal appears to be electrostatic and an electrostatic field at the ER is presumably responsible for this response. Plausible contributors are an electrochemical gradient, for which there is no evidence (although the Ca2+ ion gradient is strong); a marked restriction of anionic phospholipids to the cytoplasmic face of the ER, for which there is ample evidence at the plasma membrane; or a highly charged domain on a translocon component in proximity to the nascent cargo. Mutations in translocon components, especially the core trimeric Sec61p complex, which forms the channel into which TM segments are initially inserted (Heinrich et al., 2000), were anticipated. They were not found and the 1.5% frequency of ts mutants among >2000 independent pio mutants derived from EMS mutagenesis suggests that no essential genes are involved in the PIO response. This excludes Sec61p and many of the core and peripheral translocon components from the PIO gene class, although it does not preclude their involvement in the PIO response, because point mutations compatible with function may not have a significant effect. Mutations resulting in fragile cells, in import of sucrose or in a marked increase in total expression or stability of the fusion might also have allowed growth on sucrose, but were also not found among the pio mutants. In fact, this selection turned out to be surprisingly specific, producing multiple alleles of SPF1, STE24, and PIO3.
SPF1 (PIO1) is a nonessential gene whose inactivation by Ty1 insertion caused a modest, ∼10% reduction in growth rate, but had no other detected effect on growth under various nutritional and temperature conditions (Smith et al., 1996). Loss of function mutations in SPF1 have now been identified in three independent selection regimens, including the selection of pio mutants described herein. In each case, all mutants were recessive and equivalent to null mutants, showing that these phenotypes all resulted from loss of function. Overexpression of the normal gene from YEp plasmids had no detectable phenotype. The slight increase in invertase activity detected in SPF1/spf1-null diploids, however, presumably results from reduced gene dosage.
SPF1 was first cloned by selection for resistance to the P. farinosa SMK toxin. This protein toxin forms pores in the plasma membrane of S. cerevisiae cells. Spf1-null mutants are resistant, apparently because changes in cell wall structure are induced, resulting in increased cell wall binding of toxin and reduced access to the unidentified lethal target in the plasma membrane (Suzuki and Shimma, 1999). This is in contrast to better studied yeast killer toxins, for which binding to a cell wall receptor is actually required for access to their membrane targets (Tipper and Schmitt, 1991). spf1-null mutants are hypersensitive to calcofluor white and hygromycin and N-glycosylation of invertase is reduced due to a reduction in the length of outer chains added in the Golgi (Suzuki and Shimma, 1999). All of these phenotypes are characteristic of a disturbance of the normal secretory pathway, because a sizeable number of such mutants, including pmr1 (see below), have a similar phenotype.
SPF1 was next identified in a screen for mutants that fail to stabilize Hmg2p, one of the two hydroxymethylglutaryl-CoA reductases in yeast, in response to its inhibition by lovastatin (Cronin et al., 2000). Hmg2p is a very hydrophobic, polytopic TM protein located in the ER that catalyzes the first committed step in ergosterol synthesis in yeast, identical to the first step in cholesterol synthesis in metazoans. Degradation of this enzyme by export to the cytoplasm, ubiquitination, and destruction by the proteasome (Cronin et al., 2000) are normally inhibited when ergosterol and its precursors become limiting as a consequence of inhibition of Hmg2p by drugs such as lovastatin. Thirty-eight mutants were isolated that had lost this response; all were alleles of a single gene called COD1, later identified as SPF1. The mechanism of this effect is unknown.
Potential Roles of Spf1p, a P-Type ATPase, in Membrane Protein Topogenesis
Spf1p is localized in the ER or Golgi membrane (Cronin et al., 2000; Suzuki, 2001), perhaps shuttling between the two, where it presumably functions as a pump. Together with its relative YOR291wp, it forms a new subfamily among the 16 P-type ATPases in S. cerevisiae (Catty et al., 1997). This family is highly conserved in metazoans, so presumably has important functions (Axelsen and Palmgren, 1998). Deletion of YOR291w was without detectable effect on PIO in WT, spf1-null, or ste24-null cells and also failed to affect Hmg2p stability (Cronin et al., 2000), so its cellular role is unknown and is apparently not redundant with SPF1.
A major family of the yeast P-type ATPases may function as lipid translocases (Tang et al., 1996) and it is plausible to propose that the PIO response to a charge difference could be mediated by restriction of anionic phospholipids to the cytoplasmic leaflet of the ER membrane, so that a defect in an ER-associated phosphatidyl serine translocase might have a Pio phenotype. However, in spite of the absence of motifs in Spf1p characteristic of Ca2+ pumps, several lines of evidence indicate that it may serve that function in the ER, as does Pmr1p in the Golgi. First, as pointed out by Suzuki and Shimma (1999), pmr1-null mutants share several of the phenotypes of spf1-nulls, including effects on N-glycosylation and sensitivity to calcofluor white and hygromycin. Second, we showed that an spf1-null mutant shows modest sensitivity to EGTA, whereas the double mutant with pmr1-null is hypersensitive and is significantly retarded in growth, suggesting some overlap in function. Third, effects of a pmr1-null mutation on N-glycosylation and of spf1-null on Hmg2p stability are partially suppressed by high Ca2+, and EGTA addition inhibits stabilization of Hmg2p by lovastatin in normal cells (Cronin et al., 2000). However, pmr1-null mutations did not affect Hmg2p stability (Cronin et al., 2000) and, as we have shown, had no Pio phenotype. In addition, the Pio phenotype of spf1-null was unaffected by high Ca2+ concentrations. It is possible, nevertheless, that all of the phenotypes observed for spf1 mutants are a secondary consequence of altered ER Ca2+ homeostasis. Because of the role of Spf1p in controlling Hmg2p degradation, it has been suggested as a target for controlling cholesterol synthesis (Cronin et al., 2000). Our data suggest caution, because Spf1p may also play an important general role in controlling the translocation of TM proteins.
PIO3 has not been identified. The phenotype of pio3.2 is enhanced by a null mutation in BMH1, although this has no effect on PIO in isolation. Bmh1p encodes a member of the 14-3-3 family and is 90% identical to Bmh2p. A double null mutant is viable but temperature sensitive. As we have shown, it also lacks Pio phenotype. It may be significant that 14-3-3 proteins are implicated in binding to and activating P-type ATPases (Maudoux et al., 2000).
Potential Roles of Ste24p, a TM Protease, in Membrane Protein Topogenesis
Ste24p and Rce1p are polytopic transmembrane proteins located in the ER (Schmidt et al., 1998). Ste24p is a Zn-protease and Rce1p is either a protease of closely related function or controls such a protease (Tam et al., 1998). These are the only proteases in yeast known to be responsible for the removal of the C-terminal AAX tripeptide from prenylated CAAX box proteins (Fujimura-Kamada et al., 1997; Boyartchuk and Rine, 1998). They have distinct but overlapping specificities (Trueblood et al., 2000). Prenylation of substrates by cytoplasmic transferases is required before they can be cleaved and subsequently carboxymethylated, apparently because this lipid modification is required to bring them to the ER membrane, allowing access to the protease active sites. Double null ste24 rce1 mutants lack AAX processing but are viable, although the function of well-known substrates, such as Ras and Rho proteins, are attenuated (Trueblood et al., 2000) and a-factor cannot be exported, resulting in complete MATa sterility.
An E298A mutation in the HEXXH zinc-binding motif of Ste24p has a null phenotype (Fujimura-Kamada et al., 1997). Because this mutant had the same Pio phenotype as a ste24-deletion, Ste24p functions as a protease in controlling PIO. The substrate is unknown, but the gene is expressed in all yeast cell types and the function of Ste24p is clearly not restricted to processing of a-factor. Because Rce1p plays no detectable role in PIO function, the critical Ste24p substrate may be a C-AAX motif completely resistant to Rce1p. The yeast genome contains 98 open reading frames terminating in a CAAX motif. Analysis of the specificities of Ste24p and Rce1p, by using variants of a-factor, suggested that 11 of these 98 proteins may require Ste24p for efficient processing (Trueblood et al., 2000). One of these may be the substrate critical for PIO function. However, besides sharing responsibility with Rce1p for C-terminal AAX processing, Ste24p alone also trims the a-factor precursor N terminus between residues 7 and 8. Trimming is subsequently completed by cutting at 21/22 by Axl1p (Tam et al., 1998). It seems more likely that the Ste24p substrate critical to PIO is an analog of the a-factor precursor N terminus. Ste24 defines a novel family of related polytopic transmembrane metalloproteases conserved from bacteria to man. When the cloned human analog was used to precisely replace STE24, the a-factor processing defects in an ste24 rce1 double mutant were suppressed, demonstrating ability to perform both roles of Ste24p (Tam et al., 1998). We intend to test its ability to suppress the Pio phenotype of ste24-null.
A tenuous connection between the PIO functions of Ste24p and Spf1p may be provided by the role of proteases in a second mechanism for the feedback control of cholesterol synthesis. In mammalian cells, ER membrane-bound transcription factors called sterol regulatory element binding proteins are proteolytically activated in response to a sensor of lipid composition. Two cleavage events are involved, the first catalyzed by S1P, a membrane-bound serine protease, and the second by S2P, a membrane-bound zinc protease that cleaves the protein within the first TM domain (Nohturfft et al., 2000). Perhaps both Spf1p and Ste24p help, indirectly, to control ER membrane lipid composition.
PIO Genes, ERAD Pathway, and Stress
The selective destruction of incorrectly folded secreted proteins and incorrectly inserted or assembled TM proteins is the responsibility of the ERAD pathway. This pathway is among the select group of ER and secretory functions regulated by the unfolded protein response; SPF1, but neither STE24 nor PMR1, is also regulated by this pathway (Travers et al., 2000). This pathway is induced by unfolded proteins in the ER, so may be induced by the high PIO error rate in the double spf1 ste24 mutant, perhaps contributing to the observed stress sensitivity. This has not been tested. pmr1-null mutants are defective in ERAD degradation of mutant forms of carboxypeptidase Y (Duerr et al., 1998). spf1-null, however, did not affect the stability of Hmg1p, a normally stable isozyme of Hmg2p, so apparently had no general effect on the stability of ER proteins (Cronin et al., 2000). Because it seemed plausible that a defect in the ERAD pathway might produce a Pio phenotype, we tested the effect of a ubc7-null mutation on PIO. This mutation, by inhibiting the ubiquitination of most proteins exported from the ER, inactivates the ERAD pathway. It failed to affect the Pio phenotype of WT or spf1-null cells.
In conclusion, the products of two genes, SPF1 and STE24, have been implicated in the control of TM protein insertion orientation at the ER in yeast. Both are members of highly conserved families, both are polytopic TM proteins present in all yeast cell types, and both are located in the ER, all appropriate for components of a topogenic signal response mechanism. Effects of null mutants on PIO are additive, implying involvement in separate pathways. The consequent high-stress sensitivity, however, implies a common and important function in cell survival. Available information on the function of neither gene product provides an obvious model for this role or for the effects on PIO. Clearly, more remains to be learned about mechanisms for response to topogenic signals in eukaryotic TM proteins.
ACKNOWLEDGMENTS
We thank Jonothan A. Holt, Din Kagalwala, and Erin Hokanson for excellent technical assistance and the Council for Tobacco Research for financial support. This work was also supported by grant A2001-080 from the American Health Assistance Foundation. We thank Drs. Bing Guo, Kyle Cunningham, Susan Michaelis, Craig Peterson Jasper Rine, H.D. Schmitt, and Chise Suzuki for gifts of plasmids and primers used in these studies and are particularly grateful to Dr. Suzuki for performing tests of sensitivity to SMK toxin and for helpful discussions.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0488. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0488.
REFERENCES
- Andersson H, von Heijne G. Membrane protein topology: effects of Δμ(H)+ on the translocation of charged residues explain the “positive inside” rule. EMBO J. 1994;13:2267–2272. doi: 10.1002/j.1460-2075.1994.tb06508.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J Mol Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- Beltzer JP, Fiedler K, Fuhrer C, Geffen I, Handschin C, Wessels HP, Spiess M. Charged residues are major determinants of the transmembrane orientation of a signal-anchor sequence. J Biol Chem. 1991;266:973–978. [PubMed] [Google Scholar]
- Boehm J, Ulrich HD, Ossig R, Schmitt HD. Kex2-dependent invertase secretion as a tool to study the targeting of transmembrane proteins which are involved in ER→Golgi transport in yeast. EMBO J. 1994;13:3696–3710. doi: 10.1002/j.1460-2075.1994.tb06679.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyartchuk VL, Rine J. Roles of prenyl protein proteases in maturation of Saccharomyces cerevisiae a-factor. Genetics. 1998;150:95–101. doi: 10.1093/genetics/150.1.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns N, Grimwade B, Ross-Macdonald PB, Choi EY, Finberg K, Roeder GS, Snyder M. Large-scale analysis of gene expression, protein localization, and gene disruption in Saccharomyces cerevisiae. Genes Dev. 1994;8:1087–1105. doi: 10.1101/gad.8.9.1087. [DOI] [PubMed] [Google Scholar]
- Carlson M, Botstein D. Two differentially regulated mRNAs with different 5′ ends encode secreted with intracellular forms of yeast invertase. Cell. 1982;28:145–154. doi: 10.1016/0092-8674(82)90384-1. [DOI] [PubMed] [Google Scholar]
- Cartwright CP, Li Y, Zhu YS, Kang YS, Tipper DJ. Use of beta-lactamase as a secreted reporter of promoter function in yeast. Yeast. 1994;10:497–508. doi: 10.1002/yea.320100409. [DOI] [PubMed] [Google Scholar]
- Catty P, de Kerchove d'Exaerde A, Goffeau A. The complete inventory of the yeast Saccharomyces cerevisiae P-type transport ATPases. FEBS Lett. 1997;409:325–332. doi: 10.1016/s0014-5793(97)00446-8. [DOI] [PubMed] [Google Scholar]
- Chaudhuri B, Stephan C. A modified Kex2 enzyme retained in the endoplasmic reticulum prevents disulfide-linked dimerisation of recombinant human insulin-like growth factor-1 secreted from yeast. FEBS Lett. 1992;304:41–45. doi: 10.1016/0014-5793(92)80585-5. [DOI] [PubMed] [Google Scholar]
- Cronin SR, Khoury A, Ferry DK, Hampton RY. Regulation of HMG-CoA reductase degradation requires the P-type ATPase Cod1p/Spf1p. J Cell Biol. 2000;148:915–924. doi: 10.1083/jcb.148.5.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curcio MJ, Garfinkel DJ. Single-step selection for Ty1 element retrotransposition. Proc Natl Acad Sci USA. 1991;88:936–940. doi: 10.1073/pnas.88.3.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado-Partin VM, Dalbey RE. The proton motive force, acting on acidic residues, promotes translocation of amino terminal domains of membrane proteins when the hydrophobicity of the translocation signal is low. J Biol Chem. 1998;273:9927–9934. doi: 10.1074/jbc.273.16.9927. [DOI] [PubMed] [Google Scholar]
- Durr G, Strayle J, Plemper R, Elbs S, Klee SK, Catty P, Wolf DH, Rudolph HK. The medial-Golgi ion pump Pmr1 supplies the yeast secretory pathway with Ca2+ and Mn2+ required for glycosylation, sorting, and endoplasmic reticulum-associated protein degradation. Mol Biol Cell. 1998;9:1149–1162. doi: 10.1091/mbc.9.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujimura-Kamada K, Nouvet FJ, Michaelis S. A novel membrane-associated metalloprotease, Ste24p, is required for the first step of NH2-terminal processing of the yeast a-factor precursor. J Cell Biol. 1997;136:271–285. doi: 10.1083/jcb.136.2.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gafvelin G, Sakaguchi M, Andersson H, von Heijne G. Topological rules for membrane protein assembly in eukaryotic cells. J Biol Chem. 1997;272:6119–6127. doi: 10.1074/jbc.272.10.6119. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Sugino A. New yeast-Escherichia coli shuttle vectors constructed with in vitro mutagenized yeast genes lacking six-base pair restriction sites. Gene. 1988;74:527–534. doi: 10.1016/0378-1119(88)90185-0. [DOI] [PubMed] [Google Scholar]
- Goder V, Bieri C, Spiess M. Glycosylation can influence topogenesis of membrane proteins and reveals dynamic reorientation of nascent polypeptides within the translocon. J Cell Biol. 1999;147:257–266. doi: 10.1083/jcb.147.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley CA, Holt JA, Turner R, Tipper DJ. Transmembrane protein insertion orientation in yeast depends on the charge difference across transmembrane segments, their total hydrophobicity, and its distribution. J Biol Chem. 1998;273:24963–24971. doi: 10.1074/jbc.273.38.24963. [DOI] [PubMed] [Google Scholar]
- Harley CA, Tipper DJ. The role of charged residues in determining transmembrane protein insertion orientation in yeast. J Biol Chem. 1996;271:24625–24633. doi: 10.1074/jbc.271.40.24625. [DOI] [PubMed] [Google Scholar]
- Hartmann E, Rapoport TA, Lodish HF. Predicting the orientation of eukaryotic membrane-spanning proteins. Proc Natl Acad Sci USA. 1989;86:5786–5790. doi: 10.1073/pnas.86.15.5786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde RS, Mastrianni JA, Scott MR, DeFea KA, Tremblay P, Torchia M, DeArmond SJ, Prusiner SB, Lingappa VR. A transmembrane form of the prion protein in neurodegenerative disease. Science. 1998;279:827–834. doi: 10.1126/science.279.5352.827. [DOI] [PubMed] [Google Scholar]
- Heinrich SU, Mothes W, Brunner J, Rapoport TA. The Sec61p complex mediates the integration of a membrane protein by allowing lipid partitioning of the transmembrane domain. Cell. 2000;102:233–244. doi: 10.1016/s0092-8674(00)00028-3. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner TJ, Tzagoloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Banta LM, Emr SD. Intracellular sorting and processing of a yeast vacuolar hydrolase: proteinase A propeptide contains vacuolar targeting information. Mol Cell Biol. 1988;8:2105–2116. doi: 10.1128/mcb.8.5.2105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komano H, Fuller RS. Shared functions in vivo of a glycosyl-PI linked aspartyl protease, Mkc7, and the proprotein processing proteasse Kex2 in yeast. Proc Natl Acad Sci. 1995;92:10752–10756. doi: 10.1073/pnas.92.23.10752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maudoux O, Batoko H, Oecking C, Gevaert K, Vandekerckhove J, Boutry M, Morsomme P. A plant plasma membrane H+-ATPase expressed in yeast is activated by phosphorylation at its penultimate residue and binding of 14-3-3 regulatory proteins in the absence of fusicoccin. J Biol Chem. 2000;275:17762–17770. doi: 10.1074/jbc.M909690199. [DOI] [PubMed] [Google Scholar]
- Monne M, Gafvelin G, Nilsson R, von Heijne G. N-Tail translocation in a eukaryotic polytopic membrane protein: synergy between neighboring transmembrane segments. Eur J Biochem. 1999;263:264–269. doi: 10.1046/j.1432-1327.1999.00498.x. [DOI] [PubMed] [Google Scholar]
- Nilsson I, von Heijne G. Fine-tuning the topology of a polytopic membrane protein: role of positively and negatively charged amino acids. Cell. 1990;62:1135–1141. doi: 10.1016/0092-8674(90)90390-z. [DOI] [PubMed] [Google Scholar]
- Nohturfft A, Yabe D, Goldstein JL, Brown MS, Espenshade PJ. Regulated step in cholesterol feedback localized to budding of SCAP from ER membranes [In Process Citation] Cell. 2000;102:315–323. doi: 10.1016/s0092-8674(00)00037-4. [DOI] [PubMed] [Google Scholar]
- Okorokov LA, Lehle L. Ca(2+)-ATPases of Saccharomyces cerevisiae: diversity and possible role in protein sorting. FEMS Microbiol Lett. 1998;162:83–91. doi: 10.1111/j.1574-6968.1998.tb12982.x. [DOI] [PubMed] [Google Scholar]
- Prinz WA, Boyd DH, Ehrmann M, Beckwith J. The protein translocation apparatus contributes to determining the topology of an integral membrane protein in Escherichia coli. J Biol Chem. 1998;273:8419–8424. doi: 10.1074/jbc.273.14.8419. [DOI] [PubMed] [Google Scholar]
- Redding K, Holcomb C, Fuller RS. Immunolocalization of Kex2 protease identifies a putative late Golgi compartment in the yeast Saccharomyces cerevisiae. J Cell Biol. 1991;113:527–538. doi: 10.1083/jcb.113.3.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RL, Mosch HU, Fink GR. 14–3-3 proteins are essential for RAS/MAPK cascade signaling during pseudohyphal development in S. cerevisiae. Cell. 1997;89:1055–1065. doi: 10.1016/s0092-8674(00)80293-7. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Novick P, Thomas JH, Botstein D, Fink GR. A Saccharomyces cerevisiae genomic plasmid bank based on a centromere-containing shuttle vector. Gene. 1987;60:237–243. doi: 10.1016/0378-1119(87)90232-0. [DOI] [PubMed] [Google Scholar]
- Ross-Macdonald P, et al. Large-scale analysis of the yeast genome by transposon tagging and gene disruption. Nature. 1999;402:413–418. doi: 10.1038/46558. [DOI] [PubMed] [Google Scholar]
- Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao JI, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Rutkowski DY, Lingappa VR, Hegde RS. Substrate-specific regulation of the ribosome-translocon junction by N-terminal signal sequences. Proc Natl Acad Sci USA. 2001;98:7823–7828. doi: 10.1073/pnas.141125098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt WK, Tam A, Fujimura-Kamada K, Michaelis S. Endoplasmic reticulum membrane localization of Rce1p and Ste24p, yeast proteases involved in carboxyl-terminal CAAX protein processing and amino-terminal a-factor cleavage. Proc Natl Acad Sci USA. 1998;95:11175–11180. doi: 10.1073/pnas.95.19.11175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schubert U, Anton LC, Gibbs J, Norbury CC, Yewdell JW, Bennink JR. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith V, Chou KN, Lashkari D, Botstein D, Brown PO. Functional analysis of the genes of yeast chromosome V by genetic footprinting. Science. 1996;274:2069–2074. doi: 10.1126/science.274.5295.2069. [DOI] [PubMed] [Google Scholar]
- Suzuki C. Immunochemical and mutational analysis of p-Type ATPase Spf1p involved in the yeast a secretory pathway. Biosci Biotechnol Biochem. 2001;65:2405–2411. doi: 10.1271/bbb.65.2405. [DOI] [PubMed] [Google Scholar]
- Suzuki C, Shimma YI. P-type ATPase spf1 mutants show a novel resistance mechanism for the killer toxin SMKT. Mol Microbiol. 1999;32:813–823. doi: 10.1046/j.1365-2958.1999.01400.x. [DOI] [PubMed] [Google Scholar]
- Tam A, Nouvet FJ, Fujimura-Kamada K, Slunt H, Sisodia SS, Michaelis S. Dual roles for Ste24p in yeast a-factor maturation: NH2-terminal proteolysis and COOH-terminal CAAX processing. J Cell Biol. 1998;142:635–649. doi: 10.1083/jcb.142.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X, Halleck MS, Schlegel RA, Williamson P. A subfamily of P-type ATPases with aminophospholipid transporting. Science. 1996;272:1495–1497. doi: 10.1126/science.272.5267.1495. [DOI] [PubMed] [Google Scholar]
- Tipper DJ, Schmitt MJ. Yeast dsRNA viruses: replication and killer phenotypes. Mol Microbiol. 1991;5:2331–2338. doi: 10.1111/j.1365-2958.1991.tb02078.x. [DOI] [PubMed] [Google Scholar]
- Travers KJ, Patil CK, Wodicka L, Lockhart DJ, Weissman JS, Walter P. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell. 2000;101:249–258. doi: 10.1016/s0092-8674(00)80835-1. [DOI] [PubMed] [Google Scholar]
- Trueblood CE, Boyartchuk VL, Picologlou EA, Rozema D, Poulter CD, Rine J. The CaaX proteases, Afc1p and Rce1p, have overlapping but distinct substrate specificities. Mol Cell Biol. 2000;20:4381–4392. doi: 10.1128/mcb.20.12.4381-4392.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Klompenburg W, de Kruijff B. The role of anionic lipids in protein insertion and translocation in bacterial membranes [In Process Citation] J Membr Biol. 1998;162:1–7. doi: 10.1007/s002329900336. [DOI] [PubMed] [Google Scholar]
- van Klompenburg W, Nilsson I, von Heijne G, de Kruijff B. Anionic phospholipids are determinants of membrane protein topology. EMBO J. 1997;16:4261–4266. doi: 10.1093/emboj/16.14.4261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Alberti-Segui C, Rebischung C, Philippsen P. Heterologous HIS3 marker and GFP reporter modules for PCR-targeting in Saccharomyces cerevisiae. Yeast. 1997;13:1065–1075. doi: 10.1002/(SICI)1097-0061(19970915)13:11<1065::AID-YEA159>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- Wahlberg JM, Spiess M. Multiple determinants direct the orientation of Signal-Anchor proteins: the topogenic role of the hydrophobic signal domain. J Cell Biol. 1997;137:555–562. doi: 10.1083/jcb.137.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]