Abstract
Actin assembly on membrane surfaces is an elusive process in which several phosphoinositides (PIPs) have been implicated. We have reconstituted actin assembly using a defined membrane surface, the latex bead phagosome (LBP), and shown that the PI(4,5)P2-binding proteins ezrin and/or moesin were essential for this process (Defacque et al., 2000b). Here, we provide several lines of evidence that both preexisting and newly synthesized PI(4,5)P2, and probably PI(4)P, are essential for phagosomal actin assembly; only these PIPs were routinely synthesized from ATP during in vitro actin assembly. Treatment of LBP with phospholipase C or with adenosine, an inhibitor of type II PI 4-kinase, as well as preincubation with anti-PI(4)P or anti-PI(4,5)P2 antibodies all inhibited this process. Incorporation of extra PI(4)P or PI(4,5)P2 into the LBP membrane led to a fivefold increase in the number of phagosomes that assemble actin. An ezrin mutant mutated in the PI(4,5)P2-binding sites was less efficient in binding to LBPs and in reconstituting actin assembly than wild-type ezrin. Our data show that PI 4- and PI 5-kinase, and under some conditions also PI 3-kinase, activities are present on LBPs and can be activated by ATP, even in the absence of GTP or cytosolic components. However, PI 3-kinase activity is not required for actin assembly, because the process was not affected by PI 3-kinase inhibitors. We suggest that the ezrin-dependent actin assembly on the LBP membrane may require active turnover of D4 and D5 PIPs on the organelle membrane.
INTRODUCTION
A significant fraction of the de novo nucleation of actin in cells occurs on the cytoplasmic surface of eukaryotic cell membranes, especially the plasma membrane (Tilney, 1976; Carraway and Carraway, 1989; Small et al., 1995; Mitchison and Cramer, 1996), and a role for phosphoinositides in this elusive process has been widely discussed (Divecha and Irvine, 1995; Martin, 1998; Caroni, 2001). However, the precise function of these lipids is still not clear and is likely to be quite complicated. In several cellular systems that show rapid actin assembly in response to extracellular ligands, synthesis of phosphoinositides, especially phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], and in some cases phosphatidylinositol-3,4,5-trisphosphate [PI(3,4,5)P3], coincides precisely with the transient burst of actin assembly (Eberle et al., 1990; Dobos et al., 1992; Apgar, 1995; Hartwig et al., 1995; Gachet et al., 1997). In addition, overexpression of phosphatidylinositol-4-phosphate [PI(4)P] 5-kinase in cells leads to a significant polymerization of actin (Shibasaki et al., 1997). However, in other systems, the synthesis of PI(4,5)P2 as well as PI(3,4,5)P3 coincides more with actin depolymerization, after a transient assembly of F-actin (Apgar, 1995; Gratacap et al., 1998).
One important clue to the functions of phosphoinositides in actin assembly/disassembly is that these lipids can bind in vitro to an increasing number of actin-binding proteins (ABPs). Interestingly, two different behaviors of these ABPs have been described. First, many ABPs, such as profilin, gelsolin, and cofilin, lose their affinity for actin when bound to PI(4,5)P2 (Lassing and Lindberg, 1985; Janmey et al., 1987; Yonezawa et al., 1990; Janmey et al., 1999). The second class, which includes vinculin, talin, and the ezrin/radixin/moesin (ERM) family, appear to be able to bind PI(4,5)P2 and actin simultaneously (Niggli et al., 1995; Gilmore and Burridge, 1996; Isenberg and Niggli, 1998; Nakamura et al., 1999). Lipid vesicles containing PI(4,5)P2 have been shown to recruit ABPs and other regulatory factors (e.g., N-WASP, which recruits Arp2/3 and Cdc42) from cytosolic extracts. When bound to the vesicles, these proteins somehow coordinate the insertion of actin monomers into filaments such that the vesicles are propelled by actin comets (Ma et al., 1998; Rozelle et al., 2000; Taunton et al., 2000). To the best of our knowledge, no systematic attempt using a defined biological membrane system in vitro has been made to address the role of phosphoinositides in membrane-bound actin assembly.
Phagosomes from J774 macrophages can be prepared by internalizing 1-μm latex beads for up to 1 h followed by various times of chase up to 36 h. During this intracellular period, the latex-bead phagosomes (LBPs), which are de novo–assembled membrane organelles, mature, and as they age, show significant changes in their protein and lipid composition (Desjardins et al., 1994a; Desjardins et al., 1994b; Diakonova et al., 1997; Claus et al., 1998; Jahraus et al., 1998), as well as in their ability to interact with microtubules (Blocker et al., 1996; Blocker et al., 1997), to fuse with endocytic organelles (Desjardins et al., 1994a; Desjardins et al., 1994b; Jahraus et al., 1998, 2001), and to assemble actin de novo (Defacque et al., 2000b). LBPs also bind in vitro to F-actin (Al-Haddad et al., 2001) in a process that is distinct from actin nucleation. A recent proteomic analysis of purified LBPs has identified ∼150 of the estimated maximum 500 proteins present on this organelle (Garin et al., 2001).
We have recently established two related assays to monitor in vitro actin assembly on this membrane surface, using either fluorescence microscopy (Defacque et al., 2000b) or flow cytometry analysis (Defacque et al., 2000a). Importantly, neither GTP nor cytosolic proteins are added to this system, which depends on the intrinsic capacity of the phagosomal membrane. As the phagosomes mature in the cell, they fluctuate in their ability to assemble actin, both in vivo and in vitro (Defacque et al., 2000b), and this cyclical pattern of assembly activity correlates strongly with the phosphorylation state of many (still to be identified) phagosomal proteins (Emans et al., 1996). The PI(4,5)P2-binding proteins ezrin and moesin were shown to be essential for the actin assembly process on LBPs (Defacque et al., 2000b).
Here, we provide many lines of evidence that both preexisting and newly synthesized D4 and D5, but not D3 PIPs are essential for the ezrin-dependent process of de novo actin assembly by the phagosomal membrane. This implies that phosphoinositide turnover may be essential for this process to occur.
MATERIALS AND METHODS
Reagents
Phospholipids were obtained from Sigma (St. Louis, MO), except phosphatidylinositol-3,4-bisphosphate [PI(3,4)P2] and PI(3,4,5)P3, which were from Matreya Inc., State College, PA; PI(4)P and PI(4,5)P2 were from Calbiochem (San Diego, Ca). Recombinant wild-type and mutant ezrins were expressed in bacteria as previously described (Roy et al., 1997). The ezrin mutant K63N, K64N, K253N, K254N, K262N, K263N is described in Barret et al. (2000).
J774 Cell Culture, Phagosome Purification, and Treatment
Phagosomes containing 1-μm latex beads were prepared in J774A.1 mouse macrophages as described previously (Jahraus et al., 1998). “Salt stripping” of the phagosomes with 1.3 M NaCl and recovery of the peripheral proteins were performed as described (Defacque et al., 2000b). The protein concentration of the salt-stripped extracts was typically ∼160 μg/ml as determined by use of BCA Protein Assay Reagent (Pierce Chemical Company, Rockford, IL). Of this, ∼1.5 μg/ml (22 nM) is ezrin (Defacque et al., 2000b).
For phospholipase C treatment, phosphatidylinositol-specific phospholipase C from Bacillus cereus (PI-PLC; Sigma) was reconstituted in PLC buffer (10 mM PIPES, pH 6.8, 200 mM sorbitol, 150 mM KCl, 0.5 mM MgCl2), stored at −20°C, and used within 2 wk. Purified phagosomes were pretreated for 15 min at 37°C with 0, 0.1, or 0.6 U/ml PLC in a minimal volume of PLC buffer supplemented with protease inhibitors and 0.5 mM dithiothreitol. They were then immediately diluted 1:20 in the actin/Tβ4 mix and assayed for their actin assembly activity.
Actin Assembly (Nucleation) Assay by Fluorescence Microscopy
This assay was described in detail by Defacque et al. (2000b). Briefly, glass slides were coated with 0.5% fish-skin gelatin in water and air-dried before the experiment. A constant number of phagosomes (Blocker et al., 1997) was incubated between a slide and a coverslip in P buffer (20 mM HEPES, pH 7.0, 50 mM KCl, 4 mM MgCl2, 0.2 mM CaCl2, 0.2 mM ATP, 0.03% fish-skin gelatin, and protease inhibitors) with 2 μM rhodamine G-actin, 6 μM thymosin β4, and an antifade reagent (Blocker et al., 1997) at room temperature for 15 min. The percentage of positive phagosomes was determined with a Zeiss Axioscope microscope (Zeiss, Oberkochen, Germany). In all experiments described, the errors reported are the SDs from counts from at least three different microscope slides.
For lipid treatments, the PI(4)P and PI(4,5)P2 stock lipids were dissolved in water or chloroform (1 mg/ml). For the preincubations, the phagosomes were mixed for 15–30 min with the respective lipid (or antibody) at concentrations indicated for each experiment and then mixed with sucrose to a final concentration of 35%, placed at the bottom of a tube, and overlaid with a 25%/8% sucrose step gradient. After ultracentrifugation, the phagosomes with the bound lipid float up to the 25%/8% interface, whereas unbound lipid floats to the top. The refloated phagosomes are used immediately in the actin assembly assay.
Reconstitution Assay
For ezrin binding to phagosomes, the indicated amount of recombinant ezrin was incubated with previously salt-stripped phagosomes in D buffer (10 mM HEPES, pH 7.5, 150 mM KCl, 1 mM MgCl2, 1 mM EGTA, 1 mM dithiothreitol, protease inhibitors) with 0.3% fish-skin gelatin for 15 min at 25°C. For each condition, 100 μl (binding experiment) or 6 μl (microscopy assay) of a phagosome preparation (OD600 ∼0.4) (Blocker et al., 1997) was used. The phagosomes were then repurified on a sucrose gradient as previously described (Jahraus et al., 1998) before the fluorescence microscopy actin assembly assay was performed. Alternatively, for the ezrin-binding experiments, recovered phagosomes were diluted with 4 volumes of S/J buffer (25 mM HEPES-KOH, 115 mM potassium acetate, 25 mM magnesium acetate, protease inhibitors, pH 7.4) and pelleted by centrifugation at 12,000 rpm for 10 min in a TLS55 rotor (Beckman TL100 ultracentrifuge). The pellets were resuspended in Laemmli buffer (Laemmli, 1970), heated at 95°C for 5 min, and separated by SDS-PAGE. Western blotting onto polyvinylidene difluoride membranes was performed with a polyclonal anti-ezrin antibody (Andreoli et al., 1994). Before solubilization of proteins in Laemmli buffer, we checked that the amounts of phagosomes recovered in all the pellets were constant by measuring the OD at 600 nm.
Fluorometric Assay of Actin Polymerization
Polymerization of G-actin (10% pyrenyl-labeled) was carried out exactly as described by Cooper (1992) in the presence of 10 nM unlabeled F-actin seeds when indicated in the legend to Figure 1. Nucleation and polymerization of pyrene G-actin (in the absence of F-actin) was performed in P buffer at 25°C for 10 min immediately after addition of salts (50 mM KCl, 1 mM MgCl2). An increase of fluorescence was followed in an Aminco-Bowman Series 2 Luminescence Spectrometer (SLM-Aminco Inc., Northampton, MA). Excitation and emission wavelengths were 365 and 407 nm, respectively.
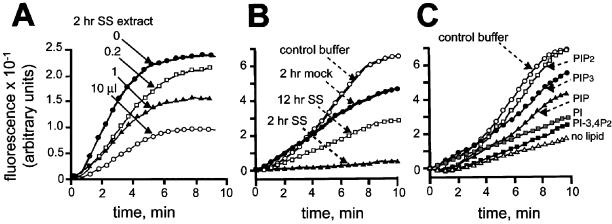
Figure 1.
Inhibition of actin polymerization by a salt-stripped (SS) extract of 2-h phagosomes can be restored by PI(4,5)P2 and PI(3,4,5)P3. (A) Unlabeled F-actin seeds were preincubated in a total final volume of 50 μl with a control buffer (●), 0.2 μl (□), 1 μl (▴), or 10 μl (○) of a 2-h salt-stripped extract (containing ∼160 μg/ml total protein) for 5 min at 25°C. After preincubation, 1 μM pyrene G-actin was added. (B) 1 μM pyrene G-actin was preincubated for 5 min at 25°C with a 2-h salt extract fraction (▴), a 2-h mock extract (●), a 12-h salt extract (□), or control buffer (○). (C) In the absence of membranes, 1 μM pyrene G-actin was preincubated for 5 min at 25°C with a control buffer alone (○) or a 2-h salt extract without (●) or with 50 μM PI(4,5)P2 (□), PI(3,4,5)P3 (●), PI (░⃞), PI(3,4)P2 (▪), or PI(4)P (▴). PI(4,5)P2 itself had no effect on pyrene actin polymerization (data not shown). In all experiments, after preincubation, actin polymerization was triggered with 50 mM KCl and 2 mM MgCl2 and followed by spectrofluorometry. Fluorescence increase was expressed in arbitrary units. All extracts, corresponding to ∼30 ng/ml protein, were prepared with same numbers of phagosomes (see Blocker et al., 1997).
Specificity of the KT10 Anti-PI(4,5)P2 Antibody
The highly specific mAb KT10 against PI(4,5)P2 has been shown to be effective in blocking PI(4,5)P2-regulated functions in many systems (Fukami et al., 1988; Matuoka et al., 1988; Uno et al., 1988; Gilmore and Burridge, 1996; Mayer et al., 2000). According to the manufacturer's data (Assay Designs, Inc., Ann Arbor, MI; Fukami et al., 1988; Matuoka et al., 1988), the KT10 anti-PI(4,5)P2 antibody gives <0.2% cross-reactivity for phosphatidylinositol, phosphatidylcholine, phosphatidylserine, phosphatidylglycerol, cardiolipin, cholesterol, or diacylglycerol. It cross-reacts with PI(4)P at similar low levels but has slightly higher cross-reactivity with phosphatidic acid (<5%). We also set up an ELISA assay by coating each lipid on ELISA plates. A strong signal was seen with PI(4,5)P2, but there was no significant detection of antibody binding to PI(4)P. A low degree of cross-reactivity was obtained with PI(3,4)P2 (results not shown). As additional evidence of antibody specificity in the fluorescence-activated cell sorter (FACS) analysis (see below), the phagosome labeling was abolished when the antibody was preincubated with PI(4,5)P2. A similar preincubation with PI(4)P or PI showed no difference in the signal relative to control phagosomes (results not shown). Collectively, these data indicate that the KT10 antibody is highly specific for PI(4,5)P2.
Immunofluorescence Labeling and FACS Analysis of Phosphoinositides on Phagosomes
Phagosome preparations were incubated for 5 min at room temperature with monoclonal anti-PI(4,5)P2 antibody KT10 (1:25 diluted; Assay Designs, Inc., Ann Arbor, MI) in a minimal volume of PBS, 0.03% fish-skin gelatin, and protease inhibitors, followed by 5 min of incubation with a fluorescein-labeled antimouse IgG (Dianova, Hamburg, Germany) in the same buffer. In parallel, a control sample was prepared by incubating phagosomes under the same conditions but without the primary antibody. The samples were then gently fixed in the same tube with 1% paraformaldehyde/PBS, and FACS analysis (Becton Dickinson, San Jose, CA) was performed by acquisition of 10,000 events. Quantification of positive phagosomes corresponded to the percentage of individual phagosomes incubated with both antibodies and having a fluorescence intensity higher than that of phagosomes incubated with the secondary antibody alone. The errors reported are the population SDs from at least three separate reactions. PLC-treated and mock-treated phagosomes were also assessed for their PI(4,5)P2 content by indirect immunofluorescence microscopy as described above, but without fixation.
32P-Labeling of Phospholipids
Phagosomes were incubated as in the actin assembly assay, except that the ATP concentration in the P buffer was lowered to 0–20 μM and 10 μCi of γ-32P[ATP] (10 mCi/ml, Amersham) was added. After the incubation, the volume was adjusted up to 50 μl with buffer before addition of 50 μl 1N HCl and 200 μl methanol/chloroform (1:1, vol/vol). The organic phase was collected and washed with an equal volume of 1 M HCl/chloroform (1:1, vol/vol). The sample was dried under vacuum and dissolved in chloroform/methanol/water (75:25:2, vol/vol). Subsequently, 32P incorporation into the lipids was quantified in a scintillation counter, or the lipids were separated by TLC on Silica Gel G60 plates [pretreated with 1% potassium oxalate/2 mM EDTA in methanol/water (1:1, vol/vol)] using a solvent mixture of chloroform/acetone/methanol/glacial acetic acid/water (80:30:26:24:14, vol/vol) (Norris and Majerus, 1994). Phospholipid standards [PI, PI(4)P, PI(4,5)P2, PI(3,4,5)P3, and PA] were stained with iodine. The quantification of 32P-labeled phospholipids separated by TLC was performed with a Fujifilm Imaging Plate and Fujifilm Fluorescent Image Analyzer FLA-2000 equipment (Fujifilm, Elmsford, NY).
Deacylation of Phosphoinositides and High-Pressure Liquid Chromatography Analysis
Spots scraped from TLC plates were incubated with 1.5 ml methylamine reagent (5.77 ml 25% methylamine in water, 6.16 ml methanol, and 1.54 ml 1-butanol) at 53°C for 50 min. The samples were subsequently dried under vacuum, and the lipids were redissolved in 1 ml water and redried. The residue was subsequently dissolved in 600 μl water and extracted with 700 μl 1-butanol/petroleum ether/ethyl formate (20:4:1, vol/vol). The upper phase, containing the fatty acids, was discarded, and the lower phase was washed twice in 700 μl of the above solvent mixture, dried under vacuum, and dissolved in water for SAX high-pressure liquid chromatography (HPLC) analysis.
RESULTS
Membrane-associated Proteins from Salt-Stripped Phagosomes Inhibit Actin Assembly, Which Can Be Rescued by PI(4,5)P2
We previously showed both in vitro and in vivo that 2-h phagosomes (1-h pulse of beads, followed by another 1-h chase) were active in the process of in vitro actin assembly, but 12-h LBPs (1-h pulse, followed by an 11-h chase) were inactive, whereas 24-h phagosomes regain a high activity (Defacque et al., 2000b). The 2-h, active LBPs lost most of their actin-assembling capacity when treated with 1.3 M NaCl. When we added the ensuing phagosome-derived salt extract to the 2-h salt-stripped phagosomes, actin assembly was restored (Defacque et al., 2000b). Here, we show that when the salt extract of 2-h phagosomes was mixed with pyrene G-actin and F-actin seeds in the absence of membranes, actin polymerization was inhibited in a dose-dependent manner (Figure 1A). As shown in Figure 1B, the extract from 2-h phagosomes could completely inhibit the polymerization of actin from seeds, whereas the extract from 12-h phagosomes, which is inactive in rescuing actin assembly on salt-stripped 2-h LBPs (Defacque et al., 2000b), inhibited actin polymerization only partially. Thus, the 2-h salt extract, whose components behave as a positive effector of actin assembly in the presence of membranes, inhibits actin polymerization when free in solution.
The LBP membrane is known to contain many ABPs (Desjardins et al., 1994a; Dermine et al., 2001; Garin et al., 2001), and a number of these ABPs, such as ezrin, moesin, gelsolin, capping proteins, profilin, and cofilin, are known to bind PI(4,5)P2 and other PIPs (Isenberg et al., 1996). We therefore tested the effects of different phosphoinositides in combination with the 2-h salt-stripped extract on pyrene actin assembly (in the absence of membranes). As seen in Figure 1C, micelles of some pure phosphoinositide lipids could completely rescue the ability of pyrene actin to elongate from seeds. In the presence of PI(4,5)P2 or PI(3,4,5)P3, the rate of polymerization and relative levels of total polymerized actin were similar to those measured with actin alone (Figure 1C). PI(3,4)P2, PI(4)P, and PI were much less efficient in the restoration of actin polymerization. The above data indirectly suggest that ABPs that can bind PI(4,5)P2 or PI(3,4,5)P3 must be present on phagosomal membranes.
Effects of Inhibitors of PI 3- and 4-Kinases
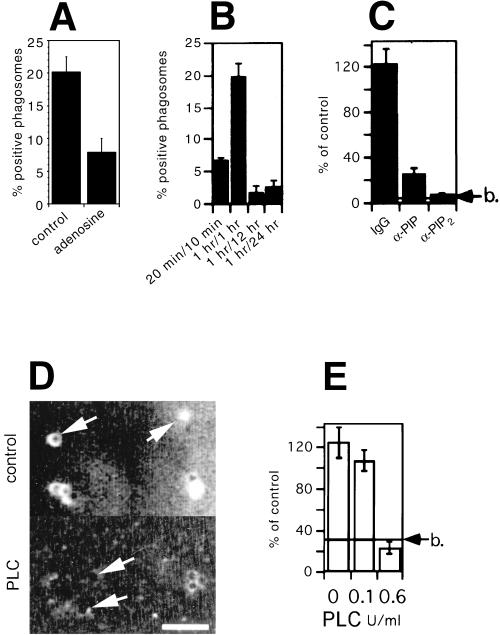
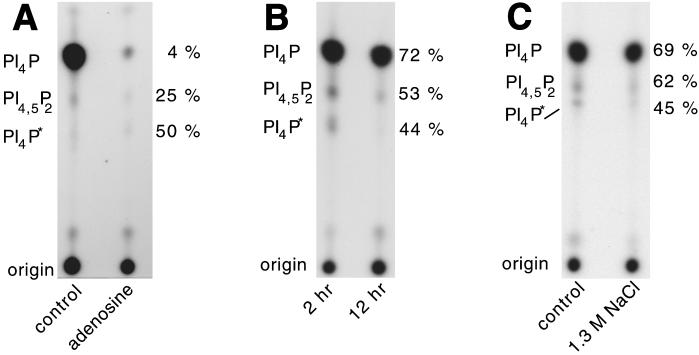
Because PI(3,4,5)P3 rescued the salt-stripped extract–induced inhibition of pyrene actin assembly (Figure 1C), we tested the effects of wortmannin and LY294002, two inhibitors of PI 3-kinases. As determined by fluorescence microscopy, preincubation of 2-h phagosomes with such inhibitors (at concentrations up to 1 μM), or adding them during the assay, had no effect on actin assembly on LBPs (our unpublished results). This argues that the synthesis of the products of PI 3-kinases is not necessary for actin assembly on phagosomal membranes. However, the addition of 200 μM adenosine, an inhibitor of type II PI 4-kinase (Fruman et al., 1998; Barylko et al., 2001), to the LBP actin assembly assay led to an ∼60% inhibition in actin assembly (Figure 2A). This suggests that the synthesis of PI(4)P or its downstream product PI(4,5)P2 is necessary for actin assembly on the phagosomal membrane. That adenosine had the expected inhibitory effect on PIP synthesis is shown below by TLC analysis.
Figure 2.
Involvement of PI(4,5)P2 in actin assembly. (A) Actin assembly on 2-h phagosomes in the absence (control) or presence of 200 μM adenosine. (B) Phagosomes purified after different pulse/chase times of beads in cells were analyzed for their PI(4,5)P2 content, via antibody labeling, followed by flow cytometry analysis. The same treatment of noninternalized beads, Triton X-100–pretreated phagosomes, or incubation of phagosomes with the secondary antibody alone gave no detectable signal (data not shown). (C) 2-h phagosomes were preincubated for 5 min at 25°C with an irrelevant antibody (IgG), anti-PI(4)P antibodies (α-PIP), or anti-PI(4,5)P2 antibodies (α-PIP2). Actin assembly on phagosomes after antibody preincubation was assayed by microscopy. Results show the percentage of positive phagosomes for each sample relative to control phagosomes. (D) 2-h phagosomes were pretreated with 0 (control) or 0.6 U/ml bacterial PI-PLC and immediately labeled for PI(4,5)P2 by indirect immunofluorescence microscopy. Arrows show individual phagosomes seen by phase-contrast microscopy. Bar, 5 μm. (E) In parallel experiments, 2-h phagosomes were pretreated in the absence or presence of 0.1 or 0.6 U/ml bacterial PI-PLC. Actin assembly on subsequently reisolated phagosomes was then assayed by microscopy. RESULTS show the percentage of positive phagosomes for each sample relative to control (untreated) phagosomes. b indicates the value obtained with fish-skin gelatin–coated beads.
Labeling of Phagosomes with Antibodies against PI(4,5)P2 and PI(4)P
We next investigated the presence of PI(4,5)P2 on LPB using a well-characterized monoclonal anti-PI(4,5)P2 antibody (see MATERIALS AND METHODS). We also compared the labeling of LBPs of different ages, because the ability of phagosomes to assemble actin fluctuates with their maturation state in the cell (see INTRODUCTION) (Defacque et al., 2000b). As determined by flow cytometry, there was a relatively low percentage of PI(4,5)P2-labeled phagosomes at the 30-min time point, with a significant rise at the 2-h stage (Figure 2B). Both the 12-h and 24-h LBPs were poorly labeled (Figure 2B). The rise and fall over the first 12 h coincides well with the ability of phagosomes to assemble actin (Defacque et al., 2000b). The absence of labeling on 24-h LBPs may be a result of epitope inaccessibility (see below). Flow cytometry analysis of 2-h phagosomes also showed that the treatment of phagosomes with 1.3 M NaCl had no effect on their labeling with anti PI(4,5)P2, arguing that the lipid is not extracted by the salt treatment (our unpublished results).
Blocking PI(4)P or PI(4,5)P2 with Antibodies Inhibits Phagosomal Actin Assembly
We next attempted to functionally inhibit phosphoinositide function, or synthesis, in the context of actin assembly. The phagosome system has the advantage that reagents can be preincubated with the organelles, followed by a reisolation via flotation to remove unbound reagent (Defacque et al., 2000a,b). We took advantage of this approach, in combination with antibodies against PI(4)P and PI(4,5)P2.
Preincubation of 2-h phagosomes with the highly specific monoclonal anti-PI(4,5)P2 antibody (see MATERIALS AND METHODS) followed by flotation of the organelles gave a significant inhibition of phagosomal actin assembly, whereas the anti-PI(4)P antibody gave a lesser degree of inhibition (Figure 2C). A similar inhibition was obtained with a rabbit anti-PI(4,5)P2 antibody (a gift of Dr. T. Yoshioka; our unpublished results). This result suggests that a preexisting pool of PI(4,5)P2, and/or its more abundant precursor PI(4)P (see below), is involved in LBP actin assembly.
PI-PLC (Divecha and Irvine, 1995) is known to significantly decrease total PI(4,5)P2 levels in mammalian cells (Eberhard et al., 1990; Ross et al., 1992) and to block both PI(4,5)P2 synthesis and vacuole fusion in yeast (Mayer et al., 2000). Pretreatment of phagosomes with this phospholipase drastically lowered their PI(4,5)P2 content, as seen by immunofluorescence microscopy (Figure 2D). After their reisolation, PLC-treated 2-h phagosomes (Figure 2E) as well as 24-h phagosomes (our unpublished results) completely lost their ability to polymerize actin. These data provide further evidence for a role for PI(4)P and/or PI(4,5)P2 in the actin assembly process.
In Vitro Synthesis of PI(4)P and PI(4,5)P2
To investigate whether active synthesis of phospholipids, and in particular PI(4,5)P2, accompanied phagosomal actin assembly, isolated phagosomes or latex beads were incubated with 10 μCi γ-32P–labeled ATP in P buffer (see MATERIALS AND METHODS) and up to 20 μM unlabeled ATP. The standard actin assembly assay contains 0.2 mM ATP. To achieve efficient incorporation of 32P, we routinely reduced the concentration of unlabeled ATP to 10–20 μM. At this concentration, the level of phagosomal actin assembly was modestly reduced relative to the standard 200 μM (∼30%; our unpublished results). At 5 μM, however, no polymerization was observed (our unpublished results).
The incubation of latex beads alone at room temperature or phagosomes at 4°C with [γ-32P]ATP resulted in a low nonspecific binding of label, whereas phagosomes incubated at room temperature for 15 min incorporated much higher amounts of 32P into the lipid fraction (Figure 3A), suggesting that lipids in the phagosomal membrane were indeed phosphorylated during the assay. This phosphorylation of lipids was further shown to be time-dependent and formed a plateau at 15 min, a time corresponding to the end of the actin assembly assay (our unpublished results). When the actin/Tβ4 mixture was omitted from the assay, no significant change in the amount of 32P incorporated into lipids was observed (Figure 3A). Most of the subsequent experiments were carried out without the actin/Tβ4 mixture.
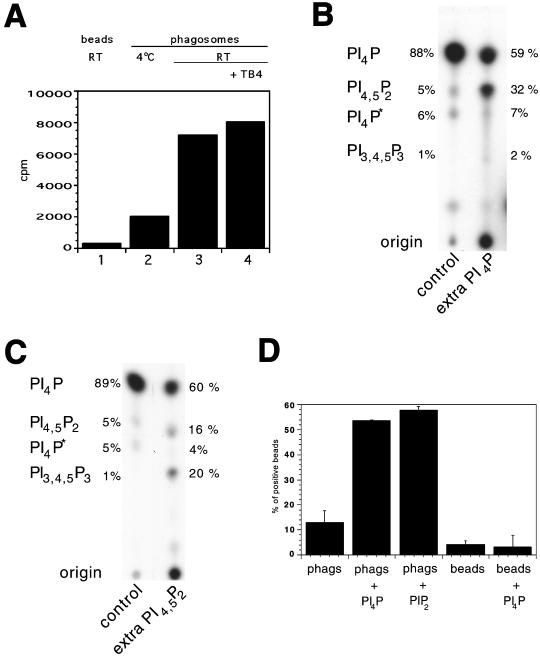
Figure 3.
Phosphorylation of phagosomal lipids. (A) Phagosomes (lanes 2–4) or beads (lane 1) were incubated with [γ-32P]ATP. The reactions were either kept on ice (lane 2) or kept at room temperature (lanes 1, 3, and 4) for 15 min. Phagosomes were incubated either without (lanes 1–3) or with actin/thymosin β4 (lane 4). Lipids were extracted as described in MATERIALS AND METHODS, and 32P incorporation was quantified in a scintillation counter. Although the absolute values we observed varied greatly between experiments, the pattern shown in this experiment was seen consistently (5 experiments). In B and C, the 32P-labeled lipids were separated by TLC on Silica Gel G60 plates, followed by autoradiography. Unlabeled PI(4)P (0.2 mg/ml) (B) or PI(4,5)P2 (0.2 mg/ml) (C) were coincubated with phagosomes. The identity of the spots, as determined by HPLC, is indicated on the left. The relative amounts of radioactivity in each lipid spot were quantified by use of a Fujifilm Imaging Plate and Fluorescent Image Analyzer and expressed as a percentage of the total counts in the PIP species (indicated next to the lanes). PI4P* indicates the second form of PI(4)P. The total counts for the signal detected in the PIPs are (B) control, 7800; extra PI4P, 5100; (C) control, 47,700; extra PI4,5P2, 32,700. (D) Phagosomes (phags) or beads were coincubated with PI(4)P or PI(4,5)P2 (0.2 mg/ml) and tested, along with untreated phagosomes/beads, in the actin assembly assay. The signal per phagosome was also distinctly higher in the LBPs preincubated with PI(4)P or PI(4,5)P2.
Analysis of the 32P-labeled phosphoinositides using TLC revealed several labeled species, including an abundant spot that comigrated with PI(4)P and a less dense spot that comigrated with PI(4,5)P2 (Figure 3B). Of the 32P-labeled phospholipids that were detected in the total lipid fraction, ∼90% were PIPs, and of this pool, ∼90% were in PI(4)P, whereas ∼5% comigrated with a PI(4,5)P2 standard. The remaining 5% of total label in the PI(4)P fraction went into a spot that migrated more slowly than the PI(4,5)P2 spot [subsequently identified by HPLC as being another variant of PI(4)P; see below]. A low signal in a species comigrating with phosphatidic acid was usually detected. In a few experiments, we also detected a minor amount of label in a species that comigrated with PI(3,4,5)P3; in most experiments however, this species was not observed. When the labeled ATP was mixed with 0.2 mM ATP (the concentration used in the actin assembly assay), after a longer exposure, again the only PIPs detected were PI(4)P and PI(4,5)P2, at ratios similar to those seen at low ATP (our unpublished results).
To identify more definitively the species of phosphoinositides that were synthesized in the phagosomal membrane, the relevant lipids were scraped from the TLC plates, deacylated, and subjected to HPLC analysis (Figure 4A). Only two phosphoinositides were observed under standard conditions. The major species synthesized after 10 min was PI(4)P (∼90%). The two different TLC spots [indicated as PI(4)P and PI(4)P* in Figure 3B] were resolved as a single species with HPLC. These two TLC species must therefore differ in their fatty acid tails, which affect the migration on TLC but are removed for the HPLC analysis. Importantly, no significant label was incorporated into PI(3)P, a lipid that is essential for endosome (and probably phagosome) fusion (Simonsen et al., 1998).
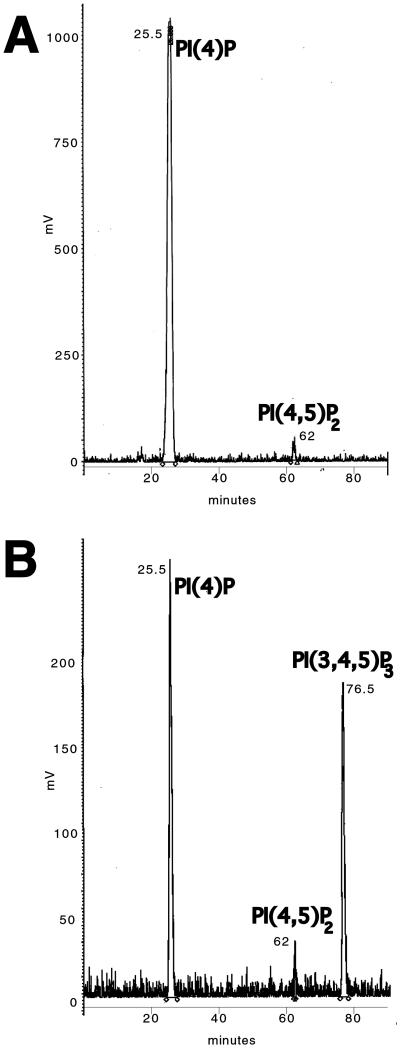
Figure 4.
HPLC analysis of 32P-labeled lipids. All the spots comigrating with phosphoinositide standards were scraped together from TLC plates of 2-h phagosomes (A) and 2-h phagosomes with the addition of unlabeled PI(4,5)P2 (B), and after deacylation, they were analyzed by HPLC. The peak elution times of the different phosphoinositides [PI(4)P, PI(4,5)P2 and PI(3,4,5)P3] are indicated at the peaks.
The second major synthesized species seen by TLC was confirmed by HPLC to be PI(4,5)P2 (Figure 4A), representing ∼6% of the total label, again consistent with the TLC results. PI(3,4)P2 and PI(3,5)P2 were not detected. The triple phosphorylated species PI(3,4,5)P3 was again detected occasionally in a few experiments as a minor species (results not shown).
In conclusion, these data show that under actin assembly conditions, only two phosphoinositides were routinely synthesized by phagosomal membranes, with ∼94% being incorporated into the two isoforms of PI(4)P and ∼6% into PI(4,5)P2. These three species represented the bulk of total phospholipid synthesized from ATP in vitro. The routine absence of newly synthesized PI(3,4)P2 is significant, because a transient rise in this lipid has been correlated with actin assembly in some systems (Apgar, 1995; Gratacap et al., 1998). Our experiments show clearly that PI kinases are present and active on the LBP membrane in the absence of GTP or cytosolic components.
Coincubation of Phagosomes with PI(4)P or PI(4,5)P2 During Actin Assembly: Effects on Actin Assembly and on PI(4)P Synthesis
Because phagosomes synthesized both PI(4)P and PI(4,5)P2 during the actin assembly assay, we next investigated the effects of adding extra amounts of these PIPs to the assay. Strikingly, the addition of PI(4)P resulted in a dramatic increase (approximately fivefold) in the percentage of phagosomes that assembled actin (Figure 3D). When LBPs that had been preincubated with PI(4)P were labeled with [γ-32P]ATP, TLC analysis revealed a significant incorporation of the γ phosphate from ATP into PI(4,5)P2 (Figure 3B). When the LBPs were preincubated with PI(4,5)P2, the levels of actin assembly were even higher than with the PI(4)P preincubation (Figure 3D); under this condition, TLC analysis showed a significant increase in the synthesis of PI(3,4,5)P3. Thus, incorporation of PI(4,5)P2 into the LBP membrane must have activated a PI 3-kinase. The increase in PI(4,5)P2 after preincubation with PI(4)P (data not shown) and of PI(3,4,5)P3 after preincubation with PI(4,5)P2 (Figure 4B) was confirmed by HPLC analyses.
Effects of PI 3- and 4-Kinase Inhibitors on Phosphoinositide Synthesis
To gain information on the type of PI 4-kinases that are active on the phagosomal membrane, we tested the effects of wortmannin and adenosine on PI(4)P and PI(4,5)P2 synthesis. At a concentration of 1 μM, wortmannin inhibits not only PI 3-kinases but also certain type III PI 4-kinases (Downing et al., 1996; Gehrmann and Heilmeyer, 1998). At this concentration of the drug, we observed only a minor inhibition of the level of PI(4)P and PI(4,5)P2 synthesis (<10%; our unpublished results), suggesting that type III PI 4-kinases are not significantly active in our assay.
The other class of PI 4-kinases, type II PI 4-kinases, are generally inhibited by adenosine (Barylko et al., 2001; Endemann et al., 1987). As described above, this drug inhibits actin assembly on phagosomal membranes by 60% (Figure 2A). The addition of 200 μM adenosine to our TLC assay inhibited the synthesis of PI(4)P > 95% (Figure 5A), suggesting that a type II PI 4-kinase is present and active on the LBP membrane. The synthesis of PI(4,5)P2 was also significantly reduced after this treatment (∼75%). Because adenosine inhibits both actin assembly and PIP synthesis, it provides further evidence for a correlation between these two processes on phagosomal membranes.
Figure 5.
Correlation between actin assembly and PI(4,5)P2 synthesis. (A) 2-h phagosomes were 32P-labeled in the presence of 200 μM adenosine. Lipids were analyzed by TLC. (B) Equal amounts of 2-h and 12-h phagosomes were analyzed and quantified. (C) 2-h phagosomes were salt-stripped and subsequently repurified on a sucrose gradient. Equal amounts of control phagosomes and salt-stripped phagosomes were analyzed. In all panels, the lipid species (as determined by HLPC) are indicated on the left. Shown on the right is the level of 32P incorporation into each lipid species compared with control (2-h) phagosomes, quantified as in Figure 3. The total counts for the signal detected in the PIPs are (A) control, 76,000; adenosine, 4100; (B) 2 h, 33,900; 12 h, 23,100; (C) control, 90,200; 1.3 M NaCl, 60,900.
Both Phagosome Maturation and Salt Stripping Affect the Levels of Phagosomal PI(4)P and PI(4,5)P2 Synthesis
We next investigated PIP synthesis under conditions in which actin assembly on phagosomal membranes is poor. As mentioned, phagosomes fluctuate in their ability to assemble actin during maturation. We therefore compared the amount of PIPs produced during the actin assembly assay by 2-h (active) and 12-h (inactive) LBPs. Equal amounts of phagosomes (determined by OD600; Blocker et al., 1997) of the two different time points were included in the assay. TLC analysis showed that the synthesis of PI(4)P and PI(4,5)P2 was significantly reduced on the 12-h phagosomes (Figure 5B).
As previously shown (Defacque et al., 2000b), salt stripping of phagosomal membranes with 1.3 M NaCl results in a 70% reduction of actin assembly. When 2-h phagosomes were salt-stripped, incubated with [γ-32P]ATP, and subsequently analyzed by TLC, we observed a slight (∼30%) reduction in the amount of both PI(4)P and PI(4,5)P2 synthesized (Figure 5C). This result has two implications. First, a large fraction of the phagosomal membrane-bound PI 4- and 5-kinases are not removed by the salt treatment (which removes the bulk of ezrin and moesin). Second, the finding that two conditions (salt stripping and ageing) that strongly inhibit actin assembly only modestly inhibit PI(4)P and PI(4,5)P2 synthesis suggests that a relatively high threshold concentration of these lipids may be required (perhaps as a local patch) on the LBP membrane for the actin assembly process to be switched on. An alternative possibility is that there may be distinct PIP-dependent and -independent processes that assemble actin on the membrane.
An Ezrin Mutant Defective in PI(4,5)P2 Binding Nucleates and Binds Poorly to Phagosomal Membranes
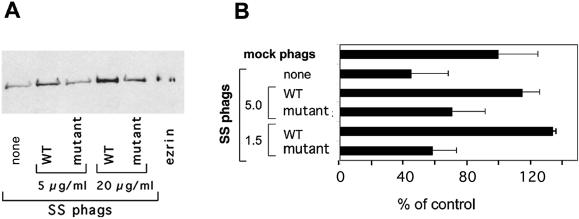
We recently reported that ezrin and moesin are essential for actin assembly on phagosomes (Defacque et al., 2000b). Ezrin binds in vitro to PI(4,5)P2 via its N-terminal domain (Niggli et al., 1995), and it can also bind to protein receptors in the membrane, as well to adaptors, which themselves are associated with membrane components (Hirao et al., 1996; Tsukita and Yonemura, 1997; Heiska et al., 1998; for review, see Bretscher, 1999; Mangeat et al., 1999). In this study, an ezrin mutant with a significantly reduced affinity for PI(4,5)P2 (Barret et al., 2000) bound much less to salt-stripped phagosomes than did wt ezrin (Figure 6A). The ezrin mutant was also much less efficient at reconstituting actin assembly on the salt-stripped LBPs (Figure 6B), presumably a consequence of its lower affinity for phagosomes. These results suggest that in the standard LBP actin assembly assay, ezrin/moesin needs to interact with PI(4,5)P2 for the membrane-dependent actin assembly process to proceed optimally.
Figure 6.
Actin assembly on phagosomes requires PI(4,5)P2/ezrin interactions. (A) 2-h salt-stripped phagosomes were preincubated with a control buffer (none), or with wild-type (WT) or a mutant ezrin defective in PI(4,5)P2-binding at the indicated concentrations. After preincubation the phagosomes were repurified and the amount of ezrin bound to phagosomes was analyzed by Western blotting. Densitometric quantitation of the ezrin signal on phagosomes gave the following values for the different lanes (in percent, from left to right): 100%, 143%, 87%, 199%, 125%. (B) Actin assembly activity was assayed by fluorescence microscopy. The results show the percentage of positive phagosomes for each sample relative to control (mock) phagosomes (phags). The addition of 5 or 1.5 μg of WT or mutant ezrin to the salt-stripped phagosomes is indicated. The errors reported are the SDs from at least three independent experiments. Higher amounts of ezrin and phagosomes were used in (A) [100 μl vs. 6 μl in (B)], because ezrin could not be detected by Western blot below 5 μg/ml.
DISCUSSION
We show here, using a defined in vitro membrane system, that the presence of a preexisting pool, as well as active synthesis of PI(4,5)P2 and probably also PI(4)P, is essential for efficient actin assembly induced by phagosomal membranes, thus extending a large body of evidence that has strongly implicated these phosphoinositides in actin assembly (Lassing and Lindberg, 1985; Eberle et al., 1990; Gilmore and Burridge, 1996; Gachet et al., 1997; DiNubile, 1998; Isenberg and Niggli, 1998; Ma et al., 1998; Janmey et al., 1999; Rozelle et al., 2000). Collectively, our data argue that an active turnover of D4 and D5 PIPs may be required for the ezrin/moesin–facilitated process of actin assembly to proceed on the LBP membrane. An unexpected finding in our studies was that PI 3-, PI 4-, and PI 5-kinases are present on the LBP membrane and can be activated by a low level (0.2 mM) of ATP, even in the absence of GTP or cytosolic factors. However, the PI 3-kinases are not essential for LBP actin assembly, because inhibitors of these enzymes had no effect on the process.
The actin assembly by LBPs that we analyzed is a specific process that probably requires many components besides ezrin/moesin and PIPs to assemble the machinery on the membrane. We recently found that the sphingolipids, sphingomyelin, ceramide, sphingosine, and sphingosine-1-phosphate in the LBP membrane are also major regulators of this process (Bos et al.; manuscript submitted). That the membrane environment is important for actin assembly to occur on the LBP membrane is further supported here by our findings that the salt extract of active 2-h LBPs, which allows actin to assemble on salt-stripped phagosomes, strongly inhibited the polymerization of pure actin free in solution. We suggest that this inhibition is caused by phagosome-derived PI(4,5)P2-binding, actin-capping proteins, two of which, CapG and CapZ, are detected on LBPs (Garin et al., 2001); such a proposal would be in agreement with a similar scenario put forward by DiNubile (1998). Consistent with this notion, the addition of PI(4,5)P2 to the extract allowed pyrene actin polymerization to proceed at the same rate as that of actin alone, presumably by inactivating the actin barbed-end capping function of proteins present in the extract.
As pointed out in the INTRODUCTION, one can classify two groups of ABPs that bind PIPs and could be important for phagosomal actin assembly. Gelsolin is a good candidate among the first category of ABPs that do not bind PIPs and actin simultaneously. The N-terminal three domains of gelsolin (G1–3) can sever and cap, but not nucleate, actin filaments in the absence of calcium (Way et al., 1989). When phagosomes were preincubated with G1–3 and then reisolated on gradients, their subsequent ability to polymerize actin was significantly enhanced (Defacque et al., 2000a). It is possible that gelsolin can bind to and influence the activity of signaling molecules, such as phospholipases C and D (Steed et al., 1996; Baldassare et al., 1997; Sun et al., 1997), that regulate the actin assembly process; pharmacological evidence suggests that these enzymes are also present and active on isolated phagosomes (our unpublished data). However, preincubation or coincubation of LBPs with gelsolin G1–3 had no effects on the synthesis of PI(4)P, P(4,5)P2, or phosphatidic acid by phagosomes (results not shown). Gelsolin has also been found to be phosphorylated by Src in the presence of PI(4,5)P2 (De Corte et al., 1997).
Of the second category of PI(4,5)P2-binding proteins (which can bind PIPs and actin simultaneously), ezrin and moesin are clearly essential for actin assembly by phagosomes (Defacque et al., 2000b). The ERM proteins, as well as talin, bind in vitro to PI(4,5)P2 via their N-terminal domains (Niggli et al., 1995), and this interaction, along with phosphorylation, has been proposed to induce an open conformation of the molecule, as is the case for vinculin (Gilmore and Burridge, 1996). In addition, talin has been shown to nucleate actin when bound to PI(4,5)P2 vesicles (Isenberg et al., 1996). Both PI(4,5)P2 binding and phosphorylation have been proposed to facilitate stabilization of these proteins or a more efficient binding to their various transmembrane receptors, such as CD44 (which by immune electron microscopy is present in J774 cells and in small amounts on LBPs) or intercellular adhesion molecule-ICAM-1 and -2 (Hirao et al., 1996; Heiska et al., 1998; Legg and Isacke, 1998; Nakamura et al., 1999). It also remains to be established whether ezrin and/or moesin phosphorylation plays any role in the LBP system. However, neither genistein (an inhibitor of tyrosine kinases) nor staurosporine (an inhibitor of protein kinase C and other kinases) had any effect on the standard phagosomal actin assay (our unpublished data).
An ezrin mutant unable to bind PI(4,5)P2 binds poorly to phagosomes and is a less potent stimulator of actin assembly on phagosomal membranes. This PI(4,5)P2-independent binding of the mutant ezrin to LBP is probably a result of interactions with membrane receptors, and it seems logical to suggest that binding of newly synthesized PI(4,5)P2 to already receptor-bound ezrin/moesin may help to transiently stabilize an active conformation of these proteins on the phagosomal membrane, a process essential for the membrane-dependent actin assembly process. That binding of proteins to PI(4,5)P2 can change their structure is well established (Raghunathan et al., 1992; Lu and Chen, 1997; Tuominen et al., 1999; for review, see Janmey et al., 1999). It should be noted that in vitro ERM proteins bind significantly better to PI(4,5)P2 than to PI(4)P (Niggli et al., 1995). This fact induces us to believe that in our system, the PI(4,5)P2 that is synthesized may be more crucial for the ezrin-dependent actin assembly than is PI(4)P. Nevertheless, because of the many interacting components available and the relatively large amounts of PI(4)P synthesized, we consider it likely that this lipid is also an important player in our system, perhaps bound to different ABPs relative to PI(4,5)P2.
Because preincubation of LBP with anti-PI(4)P and PI(4,5)P2 antibodies blocked their ability to assemble actin, we conclude that a preexisting pool of PIPs may be necessary for the process to occur. This is also consistent with the less efficient binding to phagosomes of an ezrin mutant defective in PI(4,5)P2 binding (Figure 6). However, significant amounts of PI(4,5)P2 and PI(4)P were also synthesized by the LBPs upon incubation with ATP, and this pool seems to be required for efficient actin assembly, because adenosine, an inhibitor of type II PI 4-kinase, could inhibit LBP actin assembly by 60% and the synthesis of PI(4)P and PI(4,5)P2 by 75–90%. The simplest explanation for these results is that these PIPs need to be dynamically synthesized and degraded for actin assembly on the LBPs to occur. A speculative scenario is that the ezrin would remain bound to the phagosomal membrane mostly via relatively stable interactions with protein receptors. In contrast, the binding to PI(4,5)P2 may be more dynamic; conceivably, it may involve on–off interactions controlled by cycles of alternating PIP synthesis and breakdown via PIP 4- and 5-phosphatase activities. Such a scenario may be linked to the complex process by which actin monomers are inserted into the growing actin filaments that are somehow also attached to the membrane surface. The complexity of this process is evident from the fact that not a single model exists in the literature that can incorporate all the necessary steps in this process.
We speculate that PI(4,5)P2 may exist in raft-like microdomains on the LBP after isolation. On activation of cells with agonists or addition of ATP to the in vitro actin assay, PIPs are rapidly synthesized and may aggregate laterally into larger raft domains that may now become intimately associated with the ezrin/receptor complexes (Oliferenko et al., 1999). The rafts may thus provide a platform for the proteins and lipids necessary for actin assembly to occur locally on the LBP membrane. Rafts are now known to be enriched not only in cholesterol and sphingomyelin but also in PI(4,5)P2 (Pike and Miller, 1998; Toomre et al., 2000), and recent studies are increasingly connecting these domains to dynamic actin processes on membranes (Rozelle et al., 2000; Toomre et al., 2000; Caroni, 2001). Pretreatment of LBP with the cholesterol-depleting reagents methyl-β-cyclodextrin or digitonin led to ∼50% inhibition of the actin assembly process (E.B., unpublished data). Further, Dermine et al. (2001) recently showed that raft subdomains are also present on LBPs prepared identically to the LBPs used in our study; interestingly, in the latter publication, ∼20 ABPs were found to be enriched in these Triton X-100–resistant fractions. Whether ezrin or moesin is in this fraction remains to be determined, but it is interesting to note that the amount of Triton X-100–nonextractable ezrin is higher on cell (ezrin) activation (Berryman et al., 1995; Lamb et al., 1997). Finally, type IIIα PI 4-kinase and a PI-phosphatase have been found in raft fractions (see Payrastre et al., 2001).
The Arp2/3 complex, N-WASP, Cdc42, and partners that undoubtedly nucleate actin under some conditions (Machesky and Gould, 1999; Cooper and Schafer, 2000; Pollard et al., 2000) are present on phagosomes (Garin et al., 2001; our unpublished data), but this whole complex is unlikely to be involved in the LBP actin assay, because no GTP is present. Further, GTPγS, toxin B, and C3 toxin (which cleave or inactivate Rho proteins), as well as the N-WASP WA domain (a potent regulator of the Arp2/3 system, which stimulated cytosolic actin assembly in our hands) had no effect (our unpublished data). We suggest that in the cell, the ezrin/moesin–facilitated process is responsible for the primary membrane nucleation of actin on phagosomes, whereas the Arp2/3 system might drive secondary nucleation (branching) from the sides of these primary actin filaments (see Amann and Pollard, 2001, and references therein).
Actin assembly by the phagosomal membrane is likely to be a highly complex process, even as it occurs in a technically simple, GTP-free in vitro system. The results shown here nevertheless highlight an important role for two phosphoinositides in the regulation of this process. Our more recent data extend this complexity by showing that a large cascade of signaling lipids and enzymes communicate with the PIPs and sphingolipids in the LBP membrane. An eventual understanding of this process will require a more complete deciphering of all the relevant protein–protein, protein–lipid, and lipid–lipid interactions, as well as a detailed structural analysis of these components on their specific membrane subdomains. It will also require signaling network analysis, which is now in progress.
ACKNOWLEDGMENTS
This study was greatly supported by a network grant of the Human Frontier Science Program Organization to G.G. H.D. was supported by a Marie Curie fellowship; E.B. by a Talent Stipendium of the Dutch Organization of Scientific Research; P.M. by a grant from l'Association pour la Recherche sur le Cancer; and C.B. by La Ligue Nationale Française contre le Cancer. We are grateful to Dr. Heinz Faulstich and Dr. Sergei Kuznetsov for their continued support and encouragement. We also thank Dr. Tom Martin for his helpful discussions and suggestions and Dr. Andreas Mayer for providing the phospholipase C protocol. We thank Dr. N. Schmidt of Assay Designs Inc., Dr. K. Fukami, and Dr. T. Yoshioka for their help in deciphering the lineage of the anti-PI(4,5)P2 antibody KT10 (further information can be provided by Dr. N. Schmidt, Assay Designs Inc.), as well as Ann Atzberger, who helped with the FACS analysis. Dr Yoshioka kindly provided anti-PIP2. We also appreciate the generous gift of chemically synthesized thymosin β4 from Dr. W. Voelter and Dr. H. Eichner.
Abbreviations used:
- ABP
actin-binding protein
- ERM
ezrin/radixin/moesin
- HPLC
high-pressure liquid chromatography
- LBP
latex bead phagosome
- N-WASP
neural Wiskott-Aldrich syndrome protein
- PI
phosphatidylinositol
- PIP
phosphoinositide
- PI(4)P
phosphatidylinositol-4-phosphate
- PI(4,5)P2
phosphatidylinositol-4,5-bisphosphate
- PI(3,4,5)P3
phosphatidylinositol-3,4,5-trisphosphate
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–06–0314. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–06–0314.
REFERENCES
- Al-Haddad A, et al. Myosin Va bound to phagosomes binds to F-actin and delays microtubule-dependent motility. Mol Biol Cell. 2001;12:2742–2755. doi: 10.1091/mbc.12.9.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amann KJ, Pollard TD. The Arp2/3 complex nucleates actin filament branches from the sides of pre-existing filaments. Nat Cell Biol. 2001;3:306–310. doi: 10.1038/35060104. [DOI] [PubMed] [Google Scholar]
- Andreoli C, Martin M, Le Borgne R, Reggio H, Mangeat P. Ezrin has properties to self-associate at the plasma membrane. J Cell Sci. 1994;107:2509–2521. doi: 10.1242/jcs.107.9.2509. [DOI] [PubMed] [Google Scholar]
- Apgar JR. Activation of protein kinase C in rat basophilic leukemia cells stimulates increased production of phosphatidylinositol 4-phosphate and phosphatidylinositol 4,5-bisphosphate: correlation with actin polymerization. Mol Biol Cell. 1995;6:97–108. doi: 10.1091/mbc.6.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldassare JJ, Henderson PA, Tarver A, Fisher GJ. Thrombin activation of human platelets dissociates a complex containing gelsolin and actin from phosphatidylinositide-specific phospholipase Cgamma1. Biochem J. 1997;324:283–287. doi: 10.1042/bj3240283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barret C, Roy C, Montcourrier P, Mangeat P, Niggli V. Mutagenesis of the phosphatidylinositol 4,5-bisphosphate (PIP(2)) binding site in the NH(2)-terminal domain of ezrin correlates with its altered cellular distribution. J Cell Biol. 2000;151:1067–1080. doi: 10.1083/jcb.151.5.1067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barylko B, Gerber SH, Binns DD, Grichine N, Khvotchev M, Sudhof TC, Albanesi JP. A novel family of phosphatidylinositol 4-kinases conserved from yeast to humans. J Biol Chem. 2001;276:7705–7708. doi: 10.1074/jbc.C000861200. [DOI] [PubMed] [Google Scholar]
- Berryman M, Gary R, Bretscher A. Ezrin oligomers are major cytoskeletal components of placental microvilli: a proposal for their involvement in cortical morphogenesis. J Cell Biol. 1995;131:1231–1242. doi: 10.1083/jcb.131.5.1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Severin FF, Burkhardt JK, Bingham JB, Yu H, Olivo JC, Schroer TA, Hyman AA, Griffiths G. Molecular requirements for bi-directional movement of phagosomes along microtubules. J Cell Biol. 1997;137:113–129. doi: 10.1083/jcb.137.1.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blocker A, Severin FF, Habermann A, Hyman AA, Griffiths G, Burkhardt JK. Microtubule-associated protein-dependent binding of phagosomes to microtubules. J Biol Chem. 1996;271:3803–3811. doi: 10.1074/jbc.271.7.3803. [DOI] [PubMed] [Google Scholar]
- Bretscher A. Regulation of cortical structure by the ezrin-radixin-moesin protein family. Curr Opin Cell Biol. 1999;11:109–116. doi: 10.1016/s0955-0674(99)80013-1. [DOI] [PubMed] [Google Scholar]
- Caroni P. New EMBO members' review. Actin cytoskeleton regulation through modulation of PI(4,5)P(2) rafts. EMBO J. 2001;20:4332–4336. doi: 10.1093/emboj/20.16.4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraway KL, Carraway CA. Membrane-cytoskeleton interactions in animal cells. Biochim Biophys Acta. 1989;988:147–171. doi: 10.1016/0304-4157(89)90017-8. [DOI] [PubMed] [Google Scholar]
- Claus V, Jahraus A, Tjelle T, Berg T, Kirschke H, Faulstich H, Griffiths G. Lysosomal enzyme trafficking between phagosomes, endosomes, and lysosomes in J774 macrophages: enrichment of cathepsin H in early endosomes. J Biol Chem. 1998;273:9842–9851. doi: 10.1074/jbc.273.16.9842. [DOI] [PubMed] [Google Scholar]
- Cooper JA, Schafer DA. Control of actin assembly and disassembly at filament ends. Curr Opin Cell Biol. 2000;12:97–103. doi: 10.1016/s0955-0674(99)00062-9. [DOI] [PubMed] [Google Scholar]
- Cooper V. Actin filament assembly and organization in vitro. In: Carraway KL, Cooper JA, editors. The Cytoskeleton: A Practical Approach. New York: IRL Press; 1992. pp. 52–60. [Google Scholar]
- De Corte V, Gettemans J, Vandekerckhove J. Phosphatidylinositol 4,5-bisphosphate specifically stimulates PP60(c-src) catalyzed phosphorylation of gelsolin and related actin-binding proteins. FEBS Lett. 1997;401:191–196. doi: 10.1016/s0014-5793(96)01471-8. [DOI] [PubMed] [Google Scholar]
- Defacque H, Egeberg M, Antzberger A, Ansorge W, Way M, Griffiths G. Actin assembly induced by polylysine beads or purified phagosomes: quantification by a new flow cytometry assay. Cytometry. 2000a;41:46–54. [PubMed] [Google Scholar]
- Defacque H, et al. Involvement of ezrin/moesin in de novo actin assembly on phagosomal membranes. EMBO J. 2000b;19:199–212. doi: 10.1093/emboj/19.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dermine JF, Duclos S, Garin J, St-Louis F, Rea S, Parton RG, Desjardins M. Flotillin-1-enriched lipid raft domains accumulate on maturing phagosomes. J Biol Chem. 2001;27:27. doi: 10.1074/jbc.M101113200. [DOI] [PubMed] [Google Scholar]
- Desjardins M, Celis JE, van Meer G, Dieplinger H, Jahraus A, Griffiths G, Huber LA. Molecular characterization of phagosomes. J Biol Chem. 1994a;269:32194–32200. [PubMed] [Google Scholar]
- Desjardins M, Huber LA, Parton RG, Griffiths G. Biogenesis of phagolysosomes proceeds through a sequential series of interactions with the endocytic apparatus. J Cell Biol. 1994b;124:677–688. doi: 10.1083/jcb.124.5.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diakonova M, Gerke V, Ernst J, Liautard JP, van der Vusse G, Griffiths G. Localization of five annexins in J774 macrophages and on isolated phagosomes. J Cell Sci. 1997;110:1199–1213. doi: 10.1242/jcs.110.10.1199. [DOI] [PubMed] [Google Scholar]
- DiNubile MJ. Nucleation and elongation of actin filaments in the presence of high speed supernate from neutrophil lysates: modulating effects of Ca2+ and phosphatidylinositol-4,5-bisphosphate [published erratum appears in Biochim. Biophys. Acta 1999;1450:107] Biochim Biophys Acta. 1998;1405:85–98. doi: 10.1016/s0167-4889(98)00108-6. [DOI] [PubMed] [Google Scholar]
- Divecha N, Irvine RF. Phospholipid signaling. Cell. 1995;80:269–278. doi: 10.1016/0092-8674(95)90409-3. [DOI] [PubMed] [Google Scholar]
- Dobos GJ, Norgauer J, Eberle M, Schollmeyer PJ, Traynor-Kaplan AE. C5a reduces formyl peptide-induced actin polymerization and phosphatidylinositol(3,4,5)trisphosphate formation, but not phosphatidylinositol (4,5) bisphosphate hydrolysis and superoxide production, in human neutrophils. J Immunol. 1992;149:609–614. [PubMed] [Google Scholar]
- Downing GJ, Kim S, Nakanishi S, Catt KJ, Balla T. Characterization of a soluble adrenal phosphatidylinositol 4-kinase reveals wortmannin sensitivity of type III phosphatidylinositol kinases. Biochemistry. 1996;35:3587–3594. doi: 10.1021/bi9517493. [DOI] [PubMed] [Google Scholar]
- Eberhard DA, Cooper CL, Low MG, Holz RW. Evidence that the inositol phospholipids are necessary for exocytosis: loss of inositol phospholipids and inhibition of secretion in permeabilized cells caused by a bacterial phospholipase C and removal of ATP. Biochem J. 1990;268:15–25. doi: 10.1042/bj2680015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberle M, Traynor-Kaplan AE, Sklar LA, Norgauer J. Is there a relationship between phosphatidylinositol trisphosphate and F-actin polymerization in human neutrophils? J Biol Chem. 1990;265:16725–16728. [PubMed] [Google Scholar]
- Emans N, Nzala NN, Desjardins M. Protein phosphorylation during phagosome maturation. FEBS Lett. 1996;398:37–42. doi: 10.1016/s0014-5793(96)01213-6. [DOI] [PubMed] [Google Scholar]
- Endemann G, Dunn SN, Cantley LC. Bovine brain contains two types of phosphatidylinositol kinase. Biochemistry. 1987;26:6845–6852. doi: 10.1021/bi00395a039. [DOI] [PubMed] [Google Scholar]
- Fruman DA, Meyers RE, Cantley LC. Phosphoinositide kinases. Annu Rev Biochem. 1998;67:481–507. doi: 10.1146/annurev.biochem.67.1.481. [DOI] [PubMed] [Google Scholar]
- Fukami K, Matsuoka K, Nakanishi O, Yamakawa A, Kawai S, Takenawa T. Antibody to phosphatidylinositol 4,5-bisphosphate inhibits oncogene-induced mitogenesis. Proc Natl Acad Sci USA. 1988;85:9057–9061. doi: 10.1073/pnas.85.23.9057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gachet C, Payrastre B, Guinebault C, Trumel C, Ohlmann P, Mauco G, Cazenave JP, Plantavid M, Chap H. Reversible translocation of phosphoinositide 3-kinase to the cytoskeleton of ADP-aggregated human platelets occurs independently of Rho A and without synthesis of phosphatidylinositol (3,4)-bisphosphate. J Biol Chem. 1997;272:4850–4854. doi: 10.1074/jbc.272.8.4850. [DOI] [PubMed] [Google Scholar]
- Garin J, Diez R, Kieffer S, Dermine JF, Duclos S, Gagnon E, Sadoul R, Rondeau C, Desjardins M. The phagosome proteome: insight into phagosome functions. J Cell Biol. 2001;152:165–180. doi: 10.1083/jcb.152.1.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrmann T, Heilmeyer LM., Jr Phosphatidylinositol 4-kinases. Eur J Biochem. 1998;253:357–370. doi: 10.1046/j.1432-1327.1998.2530357.x. [DOI] [PubMed] [Google Scholar]
- Gilmore AP, Burridge K. Regulation of vinculin binding to talin and actin by phosphatidyl-inositol-4–5-bisphosphate. Nature. 1996;381:531–535. doi: 10.1038/381531a0. [DOI] [PubMed] [Google Scholar]
- Gratacap MP, Payrastre B, Viala C, Mauco G, Plantavid M, Chap H. Phosphatidylinositol 3,4,5-trisphosphate-dependent stimulation of phospholipase C-gamma2 is an early key event in FcgammaRIIA-mediated activation of human platelets. J Biol Chem. 1998;273:24314–24321. doi: 10.1074/jbc.273.38.24314. [DOI] [PubMed] [Google Scholar]
- Hartwig JH, Bokoch GM, Carpenter CL, Janmey PA, Taylor LA, Toker A, Stossel TP. Thrombin receptor ligation and activated Rac uncap actin filament barbed ends through phosphoinositide synthesis in permeabilized human platelets. Cell. 1995;82:643–653. doi: 10.1016/0092-8674(95)90036-5. [DOI] [PubMed] [Google Scholar]
- Heiska L, Alfthan K, Gronholm M, Vilja P, Vaheri A, Carpen O. Association of ezrin with intercellular adhesion molecule-1 and -2 (ICAM-1 and ICAM-2): regulation by phosphatidylinositol 4, 5-bisphosphate. J Biol Chem. 1998;273:21893–21900. doi: 10.1074/jbc.273.34.21893. [DOI] [PubMed] [Google Scholar]
- Hirao M, Sato N, Kondo T, Yonemura S, Monden M, Sasaki T, Takai Y, Tsukita S. Regulation mechanism of ERM (ezrin/radixin/moesin) protein/plasma membrane association: possible involvement of phosphatidylinositol turnover and Rho-dependent signaling pathway. J Cell Biol. 1996;135:37–51. doi: 10.1083/jcb.135.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg G, Niggli V. Interaction of cytoskeletal proteins with membrane lipids. Int Rev Cytol. 1998;178:73–125. doi: 10.1016/s0074-7696(08)62136-1. [DOI] [PubMed] [Google Scholar]
- Isenberg G, Niggli V, Pieper U, Kaufmann S, Goldmann WH. Probing phosphatidylinositolphosphates and adenosinenucleotides on talin nucleated actin polymerization. FEBS Lett. 1996;397:316–320. doi: 10.1016/s0014-5793(96)01203-3. [DOI] [PubMed] [Google Scholar]
- Jahraus A, Egeberg M, Hinner B, Habermann A, Sackman E, Pralle A, Faulstich H, Rybin V, Defacque H, Griffiths G. ATP-dependent membrane assembly of F-actin facilitates membrane fusion. Mol Biol Cell. 2001;12:155–170. doi: 10.1091/mbc.12.1.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahraus A, Tjelle TE, Berg T, Habermann A, Storrie B, Ullrich O, Griffiths G. In vitro fusion of phagosomes with different endocytic organelles from J774 macrophages. J Biol Chem. 1998;273:30379–30390. doi: 10.1074/jbc.273.46.30379. [DOI] [PubMed] [Google Scholar]
- Janmey PA, Iida K, Yin HL, Stossel TP. Polyphosphoinositide micelles and polyphosphoinositide-containing vesicles dissociate endogenous gelsolin-actin complexes and promote actin assembly from the fast-growing end of actin filaments blocked by gelsolin. J Biol Chem. 1987;262:12228–12236. [PubMed] [Google Scholar]
- Janmey PA, Xian W, Flanagan LA. Controlling cytoskeleton structure by phosphoinositide-protein interactions: phosphoinositide binding protein domains and effects of lipid packing. Chem Phys Lipids. 1999;101:93–107. doi: 10.1016/s0009-3084(99)00058-4. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lamb RF, Ozanne BW, Roy C, McGarry L, Stipp C, Mangeat P, Jay DG. Essential functions of ezrin in maintenance of cell shape and lamellipodial extension in normal and transformed fibroblasts. Curr Biol. 1997;7:682–688. doi: 10.1016/s0960-9822(06)00295-8. [DOI] [PubMed] [Google Scholar]
- Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- Legg JW, Isacke CM. Identification and functional analysis of the ezrin-binding site in the hyaluronan receptor, CD44. Curr Biol. 1998;8:705–708. doi: 10.1016/s0960-9822(98)70277-5. [DOI] [PubMed] [Google Scholar]
- Lu PJ, Chen CS. Selective recognition of phosphatidylinositol 3,4,5-trisphosphate by a synthetic peptide. J Biol Chem. 1997;272:466–472. doi: 10.1074/jbc.272.1.466. [DOI] [PubMed] [Google Scholar]
- Ma L, Cantley LC, Janmey PA, Kirschner MW. Corequirement of specific phosphoinositides and small GTP-binding protein Cdc42 in inducing actin assembly in Xenopusegg extracts. J Cell Biol. 1998;140:1125–1136. doi: 10.1083/jcb.140.5.1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machesky LM, Gould KL. The Arp2/3 complex: a multifunctional actin organizer. Curr Opin Cell Biol. 1999;11:117–121. doi: 10.1016/s0955-0674(99)80014-3. [DOI] [PubMed] [Google Scholar]
- Mangeat P, Roy C, Martin M. ERM proteins in cell adhesion and membrane dynamics. Trends Cell Biol. 1999;9:187–192. doi: 10.1016/s0962-8924(99)01544-5. [DOI] [PubMed] [Google Scholar]
- Martin TF. Phosphoinositide lipids as signaling molecules: common themes for signal transduction, cytoskeletal regulation, and membrane trafficking. Annu Rev Cell Dev Biol. 1998;14:231–264. doi: 10.1146/annurev.cellbio.14.1.231. [DOI] [PubMed] [Google Scholar]
- Matuoka K, Fukami K, Nakanishi O, Kawai S, Takenawa T. Mitogenesis in response to PDGF and bombesin abolished by microinjection of antibody to PIP2. Science. 1988;239:640–643. doi: 10.1126/science.2829356. [DOI] [PubMed] [Google Scholar]
- Mayer A, Scheglmann D, Dove S, Glatz A, Wickner W, Haas A. Phosphatidylinositol 4,5-bisphosphate regulates two steps of homotypic vacuole fusion. Mol Biol Cell. 2000;11:807–817. doi: 10.1091/mbc.11.3.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchison TJ, Cramer LP. Actin-based cell motility and cell locomotion. Cell. 1996;84:371–379. doi: 10.1016/s0092-8674(00)81281-7. [DOI] [PubMed] [Google Scholar]
- Nakamura F, Huang L, Pestonjamasp K, Luna EJ, Furthmayr H. Regulation of F-actin binding to platelet moesin in vitro by both phosphorylation of threonine 558 and polyphosphatidylinositides. Mol Biol Cell. 1999;10:2669–2685. doi: 10.1091/mbc.10.8.2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggli V, Andreoli C, Roy C, Mangeat P. Identification of a phosphatidylinositol-4,5-bisphosphate-binding domain in the N-terminal region of ezrin. FEBS Lett. 1995;376:172–176. doi: 10.1016/0014-5793(95)01270-1. [DOI] [PubMed] [Google Scholar]
- Norris FA, Majerus PW. Hydrolysis of phosphatidylinositol 3,4-bisphosphate by inositol polyphosphate 4-phosphatase isolated by affinity elution chromatography. J Biol Chem. 1994;269:8716–8720. [PubMed] [Google Scholar]
- Oliferenko S, Paiha K, Harder T, Gerke V, Schwarzler C, Schwarz H, Beug H, Gunthert U, Huber LA. Analysis of CD44-containing lipid rafts: recruitment of annexin II and stabilization by the actin cytoskeleton. J Cell Biol. 1999;146:843–854. doi: 10.1083/jcb.146.4.843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payrastre B, Missy K, Giuriato S, Bodin S, Plantavid M, Gratacap M. Phosphoinositides: key players in cell signaling, in time and space. Cell Signal. 2001;13:377–387. doi: 10.1016/s0898-6568(01)00158-9. [DOI] [PubMed] [Google Scholar]
- Pike LJ, Miller JM. Cholesterol depletion delocalizes phosphatidylinositol bisphosphate and inhibits hormone-stimulated phosphatidylinositol turnover. J Biol Chem. 1998;273:22298–22304. doi: 10.1074/jbc.273.35.22298. [DOI] [PubMed] [Google Scholar]
- Pollard TD, Blanchoin L, Mullins RD. Molecular mechanisms controlling actin filament dynamics in nonmuscle cells. Annu Rev Biophys Biomol Struct. 2000;29:545–576. doi: 10.1146/annurev.biophys.29.1.545. [DOI] [PubMed] [Google Scholar]
- Raghunathan V, Mowery P, Rozycki M, Lindberg U, Schutt C. Structural changes in profilin accompany its binding to phosphatidylinositol, 4,5-bisphosphate. FEBS Lett. 1992;297:46–50. doi: 10.1016/0014-5793(92)80324-a. [DOI] [PubMed] [Google Scholar]
- Ross TS, Wang FP, Majerus PW. Mammalian cells that express Bacillus cereusphosphatidylinositol-specific phospholipase C have increased levels of inositol cyclic 1:2-phosphate, inositol 1-phosphate, and inositol 2-phosphate. J Biol Chem. 1992;267:19919–19923. [PubMed] [Google Scholar]
- Roy C, Martin M, Mangeat P. A dual involvement of the amino-terminal domain of ezrin in F- and G-actin binding. J Biol Chem. 1997;272:20088–20095. doi: 10.1074/jbc.272.32.20088. [DOI] [PubMed] [Google Scholar]
- Rozelle AL, Machesky LM, Yamamoto M, Driessens MH, Insall RH, Roth MG, Luby-Phelps K, Marriott G, Hall A, Yin HL. Phosphatidylinositol 4,5-bisphosphate induces actin-based movement of raft-enriched vesicles through WASP-Arp2/3. Curr Biol. 2000;10:311–320. doi: 10.1016/s0960-9822(00)00384-5. [DOI] [PubMed] [Google Scholar]
- Shibasaki Y, Ishihara H, Kizuki N, Asano T, Oka Y, Yazaki Y. Massive actin polymerization induced by phosphatidylinositol-4-phosphate 5-kinase in vivo. J Biol Chem. 1997;272:7578–7581. doi: 10.1074/jbc.272.12.7578. [DOI] [PubMed] [Google Scholar]
- Simonsen A, Lippe R, Christoforidis S, Gaullier JM, Brech A, Callaghan J, Toh BH, Murphy C, Zerial M, Stenmark H. EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature. 1998;394:494–498. doi: 10.1038/28879. [DOI] [PubMed] [Google Scholar]
- Small JV, Herzog M, Anderson K. Actin filament organization in the fish keratocyte lamellipodium. J Cell Biol. 1995;129:1275–1286. doi: 10.1083/jcb.129.5.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steed PM, Nagar S, Wennogle LP. Phospholipase D regulation by a physical interaction with the actin-binding protein gelsolin. Biochemistry. 1996;35:5229–5237. doi: 10.1021/bi952370j. [DOI] [PubMed] [Google Scholar]
- Sun H, Lin K, Yin HL. Gelsolin modulates phospholipase C activity in vivo through phospholipid binding. J Cell Biol. 1997;138:811–820. doi: 10.1083/jcb.138.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taunton J, Rowning BA, Coughlin ML, Wu M, Moon RT, Mitchison TJ, Larabell CA. Actin-dependent propulsion of endosomes and lysosomes by recruitment of N-WASP. J Cell Biol. 2000;148:519–530. doi: 10.1083/jcb.148.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG. In: In: International Cell Biology. Brinkley BR, Porter RK, editors. Boston: Rockefeller University Press/USA; 1976. pp. 388–402. [Google Scholar]
- Toomre D, Steyer JA, Keller P, Almers W, Simons K. Fusion of constitutive membrane traffic with the cell surface observed by evanescent wave microscopy. J Cell Biol. 2000;149:33–40. doi: 10.1083/jcb.149.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukita S, Yonemura S. ERM (ezrin/radixin/moesin) family: from cytoskeleton to signal transduction. Curr Opin Cell Biol. 1997;9:70–75. doi: 10.1016/s0955-0674(97)80154-8. [DOI] [PubMed] [Google Scholar]
- Tuominen EK, Holopainen JM, Chen J, Prestwich GD, Bachiller PR, Kinnunen PK, Janmey PA. Fluorescent phosphoinositide derivatives reveal specific binding of gelsolin and other actin regulatory proteins to mixed lipid bilayers. Eur J Biochem. 1999;263:85–92. doi: 10.1046/j.1432-1327.1999.00464.x. [DOI] [PubMed] [Google Scholar]
- Uno I, Fukami K, Kato H, Takenawa T, Ishikawa T. Essential role for phosphatidylinositol 4,5-bisphosphate in yeast cell proliferation. Nature. 1988;333:188–190. doi: 10.1038/333188a0. [DOI] [PubMed] [Google Scholar]
- Way M, Gooch J, Pope B, Weeds AG. Expression of human plasma gelsolin in Escherichia coliand dissection of actin binding sites by segmental deletion mutagenesis. J Cell Biol. 1989;109:593–605. doi: 10.1083/jcb.109.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonezawa N, Nishida E, Iida K, Yahara I, Sakai H. Inhibition of the interactions of cofilin, destrin, and deoxyribonuclease I with actin by phosphoinositides. J Biol Chem. 1990;265:8382–8386. [PubMed] [Google Scholar]