Abstract
The Schizosaccharomyces pombe septation initiation network (SIN) signals the onset of cell division from the spindle pole body (SPB) and is regulated by the small GTPase Spg1p. The localization of SIN components including Spg1p to the SPB is required for cytokinesis and is dependent on Sid4p, a constitutive resident of SPBs. However, a direct interaction between Sid4p and other members of the SIN has not been detected. To understand how Sid4p is linked to other SIN components, we have begun to characterize an S. pombe homolog of the Saccharomyces cerevisiae SPB protein Nud1p. We have determined that this S. pombe Nud1p homolog corresponds to Cdc11p, a previously uncharacterized SIN element. We report that Cdc11p is present constitutively at SPBs and that its function appears to be required for the localization of all other SIN components to SPBs with the exception of Sid4p. The Cdc11p C terminus localizes the protein to SPBs in a Sid4p-dependent manner, and we demonstrate a direct Cdc11p-Sid4p interaction. The N-terminus of Cdc11p is required for Spg1p binding to SPBs. Our studies indicate that Cdc11p provides a physical link between Sid4p and the Spg1p signaling pathway.
INTRODUCTION
To ensure proper segregation of genetic material and organelles to daughter cells during cell division, the onset of cytokinesis must be coordinated with the completion of mitosis. The yeast Schizosaccharomyces pombe has proven to be a valuable organism for the study of cytokinesis and its regulation because it is amenable to both genetic and biochemical studies. Furthermore, S. pombe divides using a medial actomyosin contractile ring, a process similar to cell division in vertebrate cells (Marks et al., 1986; Kitayama et al., 1997).
The key to S. pombe cytokinesis is the activity of a signaling cascade termed the septation initiation network (SIN; reviewed in Cerutti and Simanis, 2000; McCollum and Gould, 2001). The SIN is required for the final steps in cell division including contraction of the actomyosin ring and formation of the septum. Mutants in the SIN give rise to the septation initiation defective (sid) phenotype in which cells become highly elongated and multinucleate. The SIN is triggered by the activity of Spg1p (Schmidt et al., 1997), a small Ras-superfamily GTPase that resides at the SPB(s) throughout the cell cycle (Sohrmann et al., 1998). During interphase, Spg1p is in an inactive GDP-bound form. During metaphase, it becomes activated at both SPBs (GTP-bound form), but during anaphase B, it becomes inactivated at only one pole giving rise to a poorly understood asymmetric state (Sohrmann et al., 1998). The GTP-bound form of Spg1p recruits the Cdc7p protein kinase resulting in Cdc7p localization to both SPBs during metaphase and just one SPB during anaphase B (Sohrmann et al., 1998). The Sid1p protein kinase, in a complex with Cdc14p, is then recruited to the SPB that contains Cdc7p and activated Spg1p at this time (Guertin et al., 2000). The Sid2p protein kinase is also found constitutively at SPBs (Sparks et al., 1999; Hou et al., 2000; Salimova et al., 2000). After recruitment of the Sid1p-Cdc14p complex to a single SPB during anaphase, Sid2p is activated and recruited to the medial ring (Sparks et al., 1999). The recruitment of Sid2p to either the SPB or the medial ring depends on the function of its binding partner, Mob1p (Hou et al., 2000; Salimova et al., 2000).
Sid4p functions upstream of all the aforementioned SIN components. It resides at the SPB throughout the cell cycle and is required for the SPB localization of all other tested SIN components (Sparks et al., 1999; Chang and Gould, 2000; Hou et al., 2000; Salimova et al., 2000), including the two-component GAP for Spg1p that is comprised of Cdc16p and Byr4p (Furge et al., 1998; Li et al., 2000). Although the dependency on Sid4p is consistent with its functioning as a SPB scaffold for the SIN, a direct interaction between Sid4p and other SIN components has not been detected.
In Saccharomyces cerevisiae, a pathway analogous to the SIN is termed the mitotic exit network (MEN; reviewed in McCollum and Gould, 2001). No counterpart of Sid4p is obvious among known MEN components, but a SPB protein termed Nud1p is essential for the localization of at least some MEN proteins to the SPB (Bardin et al., 2000; Gruneberg et al., 2000). Further, Nud1p was found to interact physically with the S. cerevisiae counterparts of Byr4p and Cdc16p (Bfa1p and Bub2p; Gruneberg et al., 2000). In this study, we examined a potential S. pombe homolog of Nud1p identified in the S. pombe database to determine whether it functions in the SIN. We found that the Nud1p-homolog is a constitutive SPB protein, and interestingly, it represents the previously unidentified SIN component Cdc11p. Analysis of protein interactions among Sid4p, Cdc11p, and Spg1p provides evidence that Cdc11p links Sid4p to the Spg1p signaling cascade.
MATERIALS AND METHODS
Strains, Media, and Genetic Methods
S. pombe strains used in this study (Table 1) were grown in yeast extract (YE) or minimal medium with appropriate supplements (Moreno et al., 1991). The parental sid2-250 and spg1-106 strains were isolated in (Balasubramanian et al., 1998) and the parental cdc16-116, cdc11-119, cdc11-136, cdc11-123, and cdc7-24 strains were from Dr. Paul Nurse. Crosses were performed on glutamate medium, and double-mutant strains were constructed by tetrad analysis. S. pombe transformations were performed by electroporation (Prentice, 1992). Regulated expression of genes from various strengths of the nmt1 promoter (Basi et al., 1993; Maundrell, 1993) was achieved by growth in the presence of thiamine (promoter repressed) and then washing into medium lacking thiamine (promoter nonrepressed). S. cerevisiae strain PJ69-4A was used for two-hybrid analysis (James et al., 1996) and was transformed by a lithium acetate method (Gietz et al., 1995). Leu+ Trp+ transformants were scored for positive interactions by plating on synthetic dextrose medium lacking adenine and histidine. β-Galactosidase reporter enzyme activity in the two-hybrid strains was measured using the Galacto-Star chemiluminescent reporter assay system according to the manufacturer's instructions (Tropix Inc., Bedford, MA), with the exception that cells were lysed by glass bead disruption. Each sample was measured in triplicate.
Table 1.
Strains used in this study
| Strain | Genotype | Source/Reference |
|---|---|---|
| KGY246 | h− ade6-M210 ura4-D18 leu1-32 | Lab stock |
| KGY653 | h− cdc11-123 ade6-M210 ura4-D18 leu1-32 | Paul Nurse |
| KGY1234 | h− sid4-SA1 ade6-M210 ura4-D18 leu1-32 | Chang and Gould (2000) |
| KGY1340 | h− sid4-myc∷KanR ade6-M210 ura4-D18 leu1-32 | Chang and Gould (2000) |
| KGY2627 | h− spg1-GFP∷KanR ade6-M210 ura4-D18 leu1-32 | This study |
| KGY2628 | h− sid4-GFP∷KanR ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3197 | h− spg1-GFP∷KanR ade6-M210 cdc11-123 | This study |
| KGY3198 | h− cdc11-GFP∷KanR sid4-SA1 ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3202 | h− cdc11-HA∷KanR ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3218 | h− cdc11-GFP∷KanR spg1-106 ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3340 | h− cdc11-GFP∷KanR sid2-250 ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3341 | h− cdc11-GFP∷KanR ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3342 | h− cdc11-GFP∷KanR cdc16-116 ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3395 | h+ spg1-GFP∷KanR cdc11-119 | This study |
| KGY3526 | h− cdc11-YFP∷KanR sid4-CFP∷KanR ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3538 | h− cdc11-GFP∷KanR cdc7-24 ade6-M210 ura4-D18 leu1-32 | This study |
| KGY3561 | h− spg1-GFP∷KanR cdc11-136 | This study |
| KGY3713 | h− cdc11-HA∷KanR ade6-M210 ura4-D18 leu1-32 sid4-myc∷KanR | This study |
Epitope tagging of cdc11+, sid4+, and spg1+
The cdc11+ chromosomal locus was tagged at its 3′ end with sequences encoding green fluorescent protein (GFP), three copies of the HA epitope, or yellow fluorescent protein (YFP) by a PCR-mediated system as described previously (Bähler et al., 1998). The sid4+ and spg1+ loci were tagged by the same method to encode Sid4p-GFP, Sid4p-cyan fluorescent protein (CFP), and Spg1p-GFP fusion proteins. The sid4-myc strain was constructed previously (Chang and Gould, 2000).
Cloning of cdc11+
The SPCC1739.11c open reading frame (ORF) encoding CAA20785 with 386 base pairs 5′ of the predicted start codon and 163 base pairs 3′ of the predicted stop codon was amplified by PCR from S. pombe genomic DNA and cloned into the S. pombe shuttle vector pUR18 (Barbet et al., 1992).
Plasmids
Pieces of the sid4+ ORF were amplified by PCR from pKG1354 (Chang and Gould, 2000). In each case, a NdeI site was added to the 5′ end of the fragment and a BamHI or BglII site to the 3′ end. After restriction enzyme digestion, the PCR fragments were cloned into pREP1 (Maundrell, 1993) for overproduction studies or pREP81GFP (Basi et al., 1993) to examine the localization of proteins produced at low level. Pieces of the sid4+ ORF indicated in the text were also cloned after PCR amplification into the two hybrid vectors pGAD424 and pGBT9 (James et al., 1996).
The cdc11+ ORF was amplified by PCR from genomic DNA and cloned into the “prey” vector pGAD424 (James et al., 1996). It was also cloned into pSK(+) to make pKG2268. Pieces of the cdc11+ ORF indicated in the text were amplified from pKG2268 and cloned into pGAD424 (James et al., 1996) using the BamHI and PstI sites. For expression in S. pombe, pieces of the cdc11+ ORF were amplified by PCR with a NdeI site at the 5′ end of the fragment and a BamHI site at the 3′ end and cloned into pREP41 (Maundrell, 1993) for overproduction studies or pREP41GFP (Basi et al., 1993) to examine the localization of proteins produced at low level. Sequences encoding Cdc11p residues 1–630 and 631-1045 were also amplified and cloned into pSK(+) for in vitro binding studies.
In Vitro Binding Assays
GST and GST-Sid4p(1–191) were produced in Escherichia coli from pGEX-2T and purified on glutathione agarose beads. pSK(+)cdc11+ (1–630) (pKG2589) and 631-1045 (pKG2590) were translated in vitro in the presence of [35S]-Trans label (ICN Pharmaceuticals, Irvine, CA) with the use of the TNT-coupled reticulocyte lysate system (Promega, Madison, WI). Purified GST or GST-Sid4p bound to glutathione-agarose beads were mixed with 35S-labeled Cdc11p fragments in binding buffer (20 mM Tris-HCl, pH 7.0, 150 mM NaCl, 2 mM EDTA, 0.1% NP-40) and incubated for 1 h at 4°C. The beads were washed five times in binding buffer, and the proteins were resolved by SDS-PAGE, treated with Amplify (Amersham Pharmacia Biotech, Piscataway, NJ), and exposed to film.
Protein Lysates, Immunoprecipitations, and Immunoblots
Protein lysates were prepared in NP-40 buffer as detailed by Gould et al. (1991). Immunoprecipitations with anti-HA (12CA5) or anti-myc (9E10) antibodies were performed as described by McDonald et al. (1999). Proteins were resolved by SDS-PAGE on a 10% gel. For immunoblotting, proteins were transferred by electroblotting to a PVDF membrane (Immobilon P; Millipore Corp., Bedford, MA). Anti-HA (12CA5) and anti-myc (9E10) antibodies were used as described (McDonald et al., 1999). Antibodies were detected using horseradish-peroxidase–conjugated goat anti-mouse secondary antibodies (0.8 mg/ml; Jackson ImmunoResearch Laboratories, West Grove, PA) at a dilution of 1:50,000. Immunoblots were visualized using ECL reagents (Amersham Pharmacia Biotech).
Microscopy
Strains producing GFP-tagged proteins were grown in YE medium, fixed with methanol, and processed as described by Balasubramanian et al. (1997) or visualized in live cells that were stained with Hoechst 33258. Briefly, Hoechst 33258 stock solution was prepared at 10 mg/ml in PBS. Cells from 1 ml of culture were collected by centrifugation, resuspended in 1 ml of a 1:1000 dilution of Hoechst stock, incubated for 30–60 s, collected by centrifugation, and resuspended in 20–50 μl PBS. For temperature-shift experiments, cells were grown overnight at 25°C and shifted to 36°C for 4 h. The restrictive temperature of the sample was maintained while visualizing GFP-tagged proteins in live cells using an objective heater (Bioptechs, Butler, PA). Images were acquired digitally using an Orca-II CCD camera (Hamamatsu Photonics, Hamamatsu City, Japan) mounted on a Zeiss microscope (Axioskop 2; Carl Zeiss, Thornwood, NY) equipped with a GFP, YFP, CFP, and DAPI filter set (Chroma Technology Corp., Brattleboro, VT) on an automated filter wheel (Ludl Electronic Products Ltd., Hawthorne, NY). Z-series optical sections of live cells were taken at 0.5-μm spacing. In all cases, the excitation shutter, filter wheel, Z-motor, and Orca-II CCD camera were controlled by OpenLab software (Improvision, Lexington, MA). For colocalization studies, images from each filter set were deconvolved individually, merged, and rendered into a single plane using OpenLab software.
RESULTS
An S. pombe Homolog of Nud1p is a SPB Protein
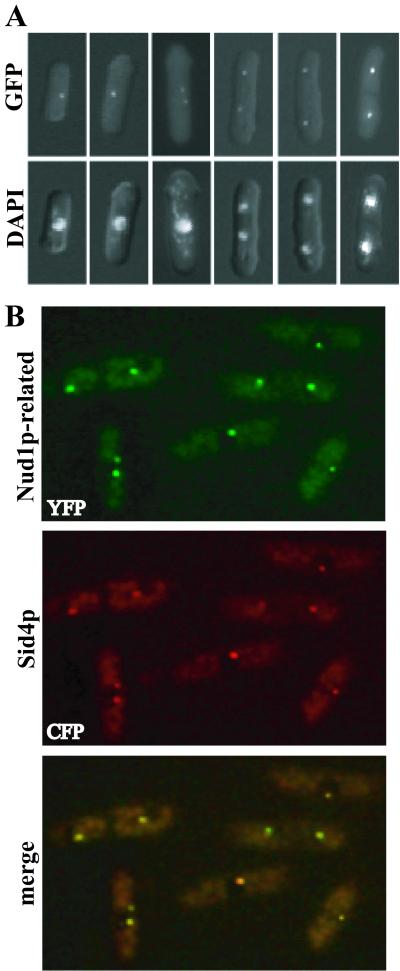
Examination of the S. pombe genome sequence data in the Sanger Center database revealed an uncharacterized protein related to Nud1p (GenBank accession no. CAA20785; BLAST score 1e–12), which we show below corresponds to Cdc11p. Like Nud1p, CAA20785 is predicted to contain a number of leucine-rich repeats (Figure 1A). To determine whether this Nud1p homolog was a SPB component, the chromosomal locus encoding it was modified to produce a C-terminal GFP or YFP fusion protein. The GFP fusion protein was localized to one or two dots at the periphery of nuclei throughout the cell cycle (Figure 2A), a staining pattern indicative of SPB localization. To confirm that these dots corresponded to SPBs, we examined the localization of the YFP-tagged Nud1p homolog in cells also producing a CFP-tagged version of Sid4p. Sid4p is a known SPB and SIN component (Chang and Gould, 2000). The single and merged images indicate that CAA20785 colocalizes with Sid4p to SPBs throughout the cell cycle (Figure 2B).
Figure 1.
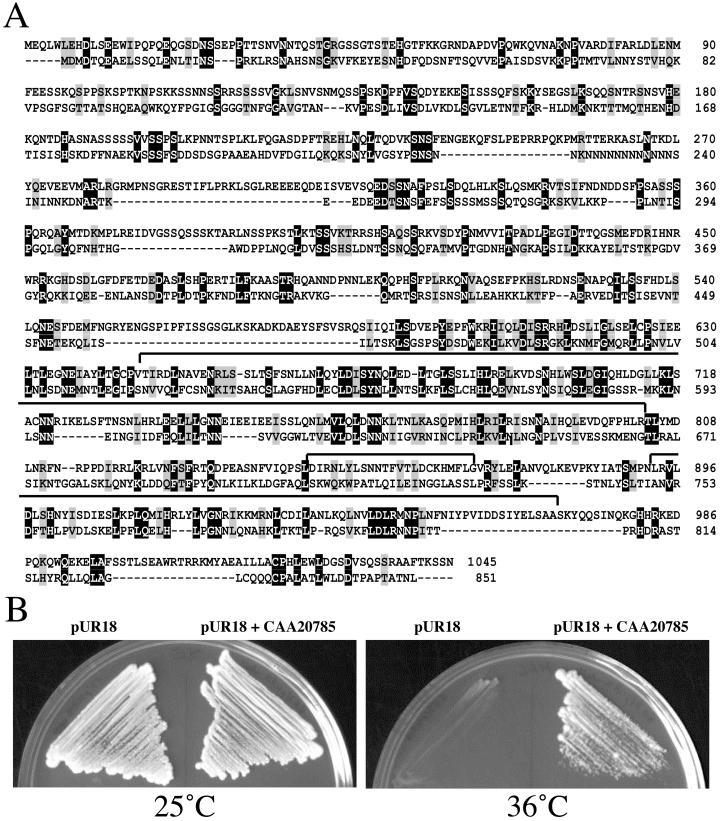
The S. pombe Nud1p homolog is Cdc11p. (A) Sequence alignment of S. pombe CAA20785 (upper lines) with S. cerevisiae Nud1p (lower lines). The leucine-rich repeats are indicated with lines above the sequences. (B) The cdc11-123 strain (KGY653) was transformed with pUR18 or pUR18 containing a CAA20785 gene fragment. Transformants isolated at 25°C were struck to selective plates and incubated at 25 or 36°C.
Figure 2.
The S. pombe Cdc11p is a SPB component. (A) Cells producing the Nud1p homolog tagged with GFP at its endogenous locus (KGY3341) were fixed and stained with DAPI at various cell cycle stages. (B) Cells producing Sid4p-CFP and the Nud1p homolog tagged with YFP (KGY3526) were imaged separately and the images were also merged.
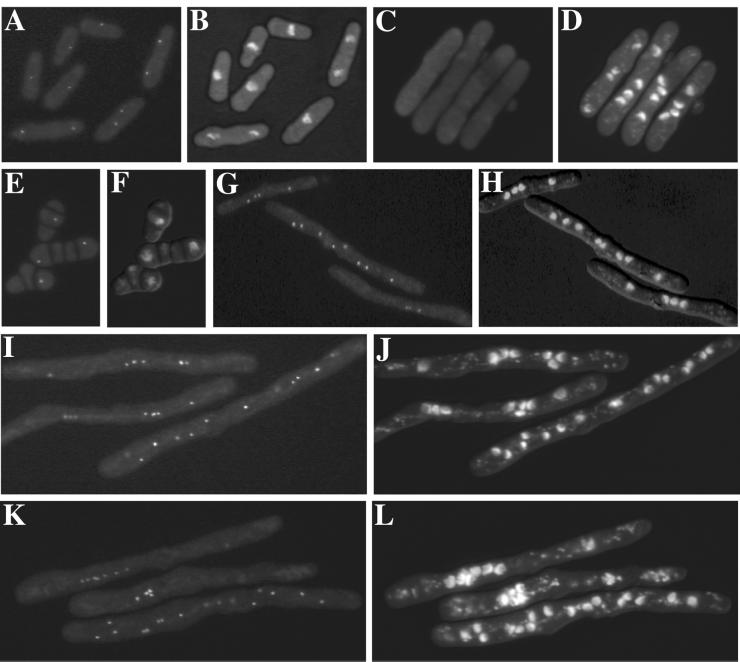
To test whether CAA20785 might play a role in the SIN analogous to that of Nud1p in the MEN and also whether it might interact with Sid4p, we asked whether its SPB localization was altered in SIN mutants. The CAA20785 GFP fusion protein localized normally in cdc16-116, spg1-106, cdc7-24, and sid2-250 temperature-sensitive mutants. However, its localization to the SPB was lost in the sid4-SA1 mutant at restrictive temperature (Figure 3). This indicated that the Nud1p homolog might play a role in the SIN downstream of Sid4p but upstream of Spg1p, Cdc7p, and Sid2p.
Figure 3.
Sid4p function is required for SPB localization of Cdc11p. The intracellular location of the GFP-tagged Nud1p homolog (A, C, E, G, I, and K) was determined after fixation and DAPI staining (B, D, F, H, J, and L) of sid4-SA1 cells (KGY3198) grown at 25°C (A and B) or sid4-SA1 (C and D), cdc16-116 (KGY3342) (E and F), spg1-106 (KGY3218) (G and H), cdc7-24 (KGY3538) (I and J) and sid2-250 (KGY3340) (K and L) cells that had been shifted to 36°C for 4–8 h.
The Nud1p Homolog Is Cdc11p
Of all SIN pathway components described, the molecular identity of one, cdc11+, has remained unknown. Therefore, the possibility that CAA20785 corresponded to Cdc11p was tested. We found that in crosses of the tagged ORF to the cdc11-123 mutation, not a single temperature-sensitive colony (containing the cdc11 mutation) that was also G418 resistant (containing the tagged locus) was isolated from 44 complete tetrads; all 44 tetrads were parental ditypes. This result indicated that CAA20785 was very tightly linked to cdc11+. We next determined that a plasmid containing the CAA20785 ORF and flanking DNA, but not empty plasmid, was able to suppress the cdc11-123 mutation at 36°C (Figure 1B). We conclude that CAA20785 represents Cdc11p.
Cdc11p Is Required for Correct Spg1p Localization
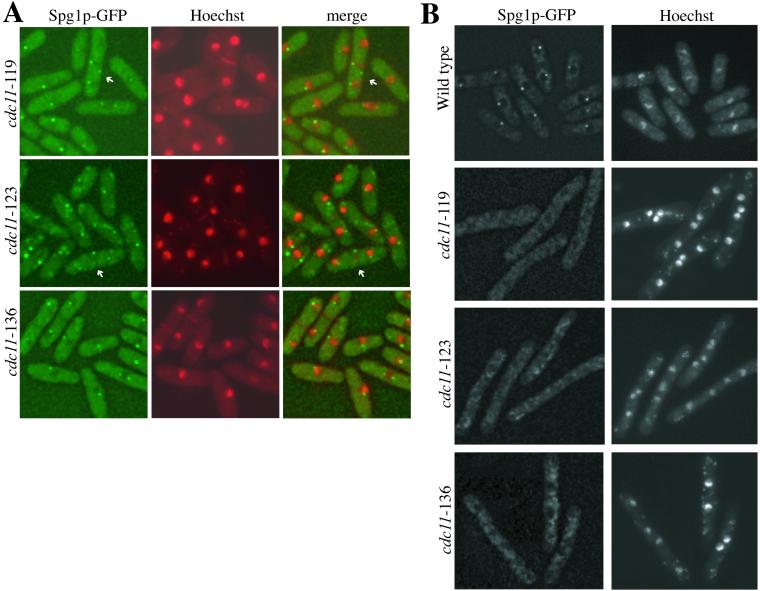
In S. cerevisiae, it has been demonstrated that the Spg1p homolog, Tem1p, does not localize to the SPB in two NUD1 mutant strains, nud1-2 (Gruneberg et al., 2000) and nud1-44 (Bardin et al., 2000). Hence, we examined whether Spg1p localized correctly in cdc11 mutants. At 25°C, Spg1p-GFP could be detected at SPBs in cdc11-123, cdc11-119, and cdc11-136 mutant strains (Figure 4A). Interestingly, in 5 and 15% of cdc11-123 and cdc11-119, respectively, Spg1p-GFP was observed in additional dots that were not adjacent to the nuclear periphery. In live cells, these “extra” dots were observed to move freely (our unpublished results). At 36°C, Spg1p-GFP was detected at SPBs in wild-type cells but was absent from SPBs in the cdc11 mutant strains (Figure 4B). We conclude that Cdc11p function is required for the normal localization of Spg1p.
Figure 4.
Spg1p SPB localization requires Cdc11p function. (A) The cdc11-119 spg1-GFP (KGY3395), cdc11-123 spg1-GFP (KGY3197), and cdc11-136 spg1-GFP (KGY3561) strains were grown at 25°C and stained with Hoechst. Single and merged images are shown. “Extra” Spg1p-containing dots are indicated by arrows. (B) KGY3395, KGY3197, KGY3561, and the spg1-GFP strain (KGY2627) were grown at 25°C, then shifted to 36°C for 4 h, and stained with Hoechst.
The Cdc11p C Terminus Directs Its SPB Localization
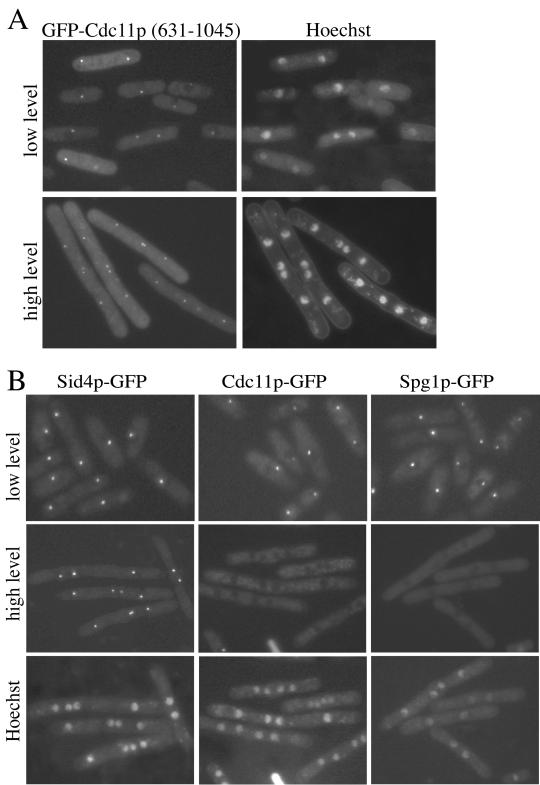
We next tested whether the N or C terminus of Cdc11p contained the SPB binding domain of the protein. Cdc11p residues 1–630 and 631-1045 were fused to GFP under control of the medium strength nmt41 promoter. GFP-Cdc11p(1–630) was distributed throughout the cytoplasm (our unpublished results), but GFP-Cdc11p(631–1045) localized to SPBs (Figure 5A). This fragment of the protein contains the majority of the leucine repeats. Interestingly, we found that overproduction of GFP-Cdc11p(631–1045) generated a sid phenotype (Figure 5A). Thus, we examined whether this fragment interfered with the localization of Sid4p, full-length Cdc11p, and/or Spg1p to SPBs. For this purpose Cdc11p(631–1045) was overproduced without a tag. Overproduction of Cdc11p(631–1045) had no effect on the localization of Sid4p-GFP but caused the loss of Cdc11p-GFP and Spg1p-GFP from SPBs (Figure 5B). This is consistent with the idea that Cdc11p(631–1045) saturates the SPB binding site for Cdc11p, thus eliminating the opportunity for the full-length protein to localize to the SPB.
Figure 5.
The Cdc11p C terminus directs its SPB localization. (A) Wild-type cells (KGY246) expressing GFP-Cdc11p(631–1045) under control of the nmt41 promoter in pREP41 were grown to midlog phase at 32°C in the presence (top panels) or absence (bottom panels) of thiamine. Images of live cells that had been stained with Hoechst to visualize nuclei were captured. (B) The sid4-GFP (KGY2628), cdc11-GFP (KGY3341), and spg1-GFP (KGY2627) strains were transformed with pREP41cdc11(631–1045). Live cell images were captured after cells had been grown in the presence of thiamine (low level, top row) and after derepression of the nmt1-T41 promoter and Hoechst staining (high level, middle and bottom rows).
The C Terminus of Sid4p Binds the SPB
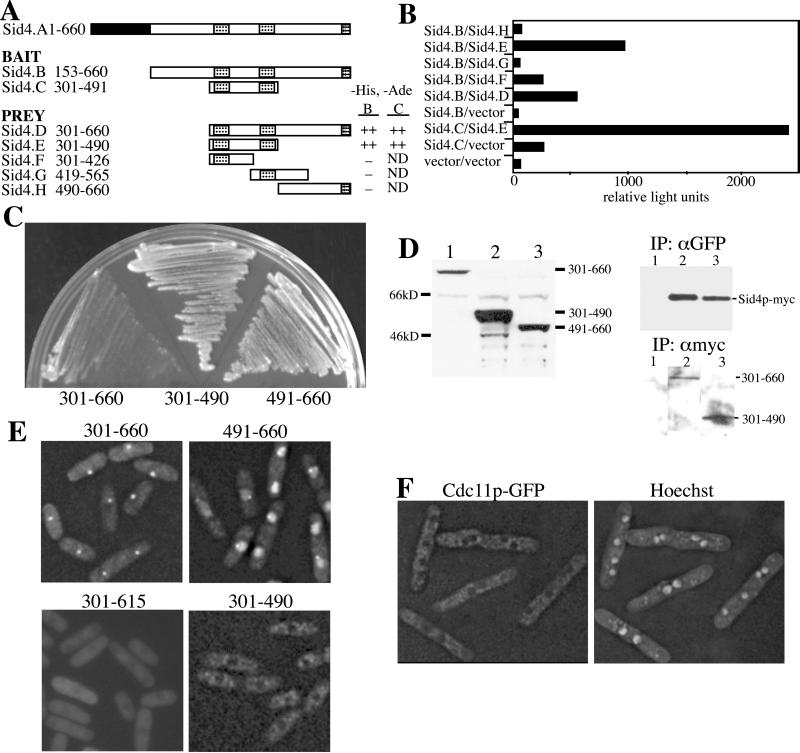
We next wanted to determine whether Cdc11p bound Sid4p directly and thus might serve to link Sid4p to the Spg1p signaling cascade. First, however, the regions of Sid4p most likely to interact with Cdc11p and the SPB were better defined. We had found previously that overproduction of N-terminally truncated Sid4p (Sid4ΔNp; residues 153–660) gave rise to a dominant negative phenotype (Chang and Gould, 2000). This phenotype resulted from the displacement of full-length Sid4p and Spg1p from the SPB (Chang and Gould, 2000). Because we did not detect Sid4ΔNp at SPBs, we hypothesized that the dominant negative effect produced by Sid4ΔNp was due to dimerization of Sid4ΔNp with full-length Sid4p and the consequent titration of full-length Sid4p from the SPB (Chang and Gould, 2000). Sid4ΔNp contains three putative coiled-coil domains that could be involved in dimerization. Although we did not previously define the domain responsible for the possible dimerization more precisely than amino acids 300–660 (Chang and Gould, 2000), new two-hybrid constructs indicated that the central two coiled-coil regions (residues 301–490) were sufficient to support robust Sid4p-Sid4p interaction (Figure 6, A and B). Inconsistent with our original hypothesis, however, was the finding that overproduction of Sid4p(301–490) was unable to produce a dominant negative phenotype (Figure 6C) despite the ability of a GFP-tagged version of it to bind endogenous Sid4p-myc (Figure 6D). Thus, we reexamined our previous conclusion that Sid4p lacking its N-terminus could not localize to the SPB.
Figure 6.
The Sid4p C terminus binds SPBs and displaces Cdc11p. The indicated regions of Sid4p in either bait or prey plasmids were cotransformed into S. cerevisiae strain PJ69–4A. Leu+Trp+ transformants with either bait “B” or “C” fragments were scored for growth on selective medium (A) or for β-galactosidase activity (B) that was measured in relative light units. The three coiled-coil domains of Sid4p are indicated in the diagram by hatched boxes. (C) Wild-type cells (KGY246) expressing the indicated GFP-Sid4p fusion proteins from the full-strength nmt1 promoter were struck to medium lacking thiamine at 32°C. Colonies were allowed to form for 3 days. (D and E) The sid4-myc (KGY1340) (D) and wild-type (KGY246) (E) strains were transformed with pREP81 vectors directing production of the indicated GFP-Sid4p fusion proteins. Transformants were grown in the absence of thiamine and either protein lysates were prepared (D) or live cell images captured (E). A sample of each lysate was resolved by SDS-PAGE and immunoblotted with antibodies to GFP (D, left panel). The remaining lysates were divided in half and subjected to immunoprecipitation with either polyclonal anti-GFP serum or the 9E10 monoclonal anti-myc antibody. Immunoprecipitates were resolved by SDS-PAGE and immunoblotted with 9E10 (D, right upper panel) or anti-GFP serum (right lower panel). (F) The cdc11-GFP strain (KGY3341) was transformed with pREP1sid4ΔNp (residues 153–660) and grown in the absence of thiamine for 20 h. Images of live cells stained with Hoechst were captured.
For this purpose, GFP fusions of Sid4p residues 301–660, 301–490, and 491–660 were created in pREP81, a vector that contains a low-strength thiamine-regulatable promoter. All three fusion proteins were stable when produced in sid4-myc cells (Figure 6D). GFP-Sid4p(301–660) was detected readily at SPBs (Figure 6E), and it could interact well with endogenous Sid4p-myc as determined by coimmunoprecipitation (Figure 6D). Further, this portion of Sid4p induced a septation initiation defective (sid) phenotype when overproduced from the full-strength nmt1 promoter (Figure 6C and our unpublished results). Thus, Sid4p residues 301–660 contain both SPB and self-interaction domains. Our previous inability to detect Sid4p(153–660) at the SPB we now attribute to a technical difficulty, and we have localized this larger fragment to the SPB as well (our unpublished results).
GFP-Sid4p(491–660), which does not contain the self-interaction domain (Figure 6, A and B), localized to SPBs, but a strong nuclear localization predominated (Figure 6E). It was unable to induce a dominant negative phenotype when overproduced (Figure 6C). The third fragment, GFP-Sid4p(301–490), was observed throughout the cytoplasm and not at SPBs (Figure 6E). It accumulated to high levels and interacted well with endogenous Sid4p-myc (Figure 6D). Taken in combination, these results suggest that overproduced Sid4p(301–660) can displace full-length Sid4p from the SPB because it saturates the Sid4p binding site at the SPB and not simply because it titrates the full-length Sid4p protein from SPBs. We also conclude that residues 491–660 contain a minimal SPB recognition domain that is enhanced by the ability of Sid4p to dimerize through its central coiled-coil regions.
To determine the section of Sid4p within residues 301–660 involved in
SPB localization, a GFP fusion to residues 301–615 was created in
pREP81. This Sid4p fragment was not detected at SPBs (Figure 6E),
suggesting that the extreme C terminus of Sid4p, possibly the third
coiled-coil domain, is essential for interacting with a SPB component.
Consistent with this interpretation, a GFP fusion of Sid4p residues
554–660 gave a localization pattern similar to GFP-Sid4p(491–660) in
that it was predominantly nuclear but could be detected at SPBs (our
unpublished results). These Sid4p localization results are summarized
in Table 2.
![]()
We next asked whether Cdc11p-GFP remained at SPBs in cells overproducing Sid4ΔNp. This was of particular interest because we had shown previously that Spg1p and full-length Sid4p were driven off SPBs overproducing Sid4ΔNp (Chang and Gould, 2000). Cdc11p-GFP could not be detected at SPBs in these cells (Figure 6F) or in cells overproducing Sid4p(301–660) (our unpublished results). Thus, the presence of the Sid4p N-terminus is required for docking of Cdc11p to SPBs as well as the SPB localization of other downstream components of the SIN.
Cdc11p and Sid4p Interact Directly
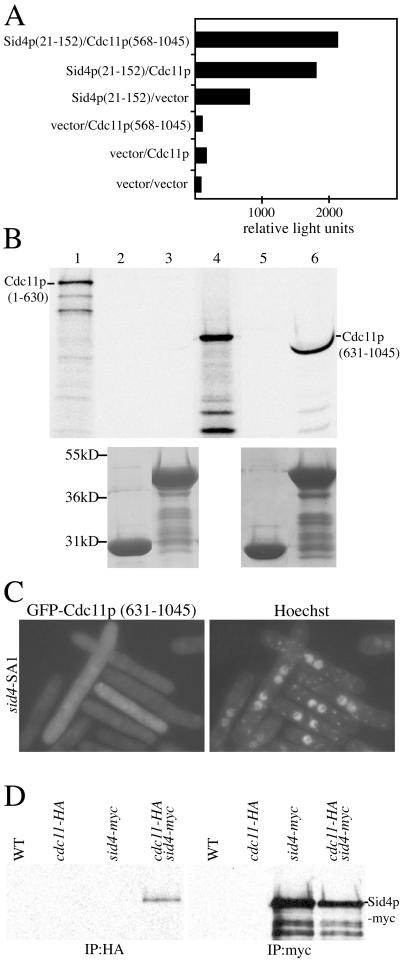
Because Cdc11p was not present at SPBs when Sid4p(153–660) was overproduced, we tested whether the N-terminal region of Sid4p (amino acids 1–152) might contain a Cdc11p binding site. This question was examined first by two-hybrid analysis, and in this assay, Sid4p residues 21–152 as bait transactivated to some extent but showed an enhanced interaction with full-length Cdc11p and Cdc11p(568–1045) as prey (Figure 7A). When the N-terminal region was extended to include the self-interaction domain, we observed a very robust two-hybrid interaction with Cdc11p (our unpublished results). To confirm this interaction, a GST-fusion protein of Sid4p(1–191) was produced in E. coli and tested for its ability to bind Cdc11p fragments produced in a coupled transcription/translation in vitro system. This system presumably lacks yeast spindle pole body constituents that could act to bridge a Sid4p-Cdc11p interaction. As predicted from the two-hybrid results, GST-Sid4p(1–191) bound Cdc11p(631–1045) but not Cdc11p(1–630) (Figure 7B). GST alone was unable to bind either fragment of Cdc11p (Figure 7B). These results indicate that Cdc11p binds directly to Sid4p through its C-terminal domain, the same domain required for its SPB localization. As expected from these results, GFP-Cdc11p(631–1045) is unable to bind SPBs in the sid4-SA1 mutant at its restrictive temperature (Figure 7C).
Figure 7.
Cdc11p binds Sid4p directly. (A) The Sid4p(21–152) bait plasmid was cotransformed with either Cdc11p full-length or Cdc11p(568–1045) prey plasmids into the S. cerevisiae strain PJ69–4A. Leu+Trp+ transformants were scored for β-galactosidase activity in relative light units. (B) GST-Sid4p(1–191) binds directly to Cdc11p(631–1045). Approximately equal amounts of GST (lanes 2 and 5) and GST-Sid4p(1–191) (lanes 3 and 6) bound to glutathione beads were mixed with in vitro translated Cdc11p(1–630) (lanes 2 and 3) or Cdc11p(631–1045) (lanes 5 and 6). After extensive washes, the proteins were resolved by SDS-PAGE and detected by autoradiography (upper panel) or Coomassie staining (lower panel). Lanes 1 and 4 represent samples of in vitro translated Cdc11p(1–630) and Cdc11p(631–1045), respectively, before the binding reaction. Lane 6 shows a compression due to GST-Sid4p(1–191) and Cdc11p(631–1045) having similar molecular weights. (C) Cdc11p SPB localization requires Sid4p. sid4-SA1 (KGY1234) cells expressing pREP41GFP Cdc11p(631–1045) were grown in the absence of thiamine to induce GFP-Cdc11p (631–1045) and then shifted to 36°C for 4 h to inactivate Sid4p. (D) Cdc11p-HA and Sid4p-myc interact in vivo. Lysates from wild-type (KGY246), cdc11-HA (KGY3202), sid4-myc (KGY1340), and the double-tag strain (KGY3713) were immunoprecipitated using anti-HA (12CA5) or anti-myc (9E10) antibodies. After SDS-PAGE, samples were blotted with anti-myc (9E10) antibodies.
We also tested whether Cdc11p and Sid4p could coimmunoprecipitate from S. pombe protein lysates. To this end, a cdc11-HA sid4-myc strain was produced. An anti-HA immunoprecipitate from this double-tag strain, but not from the sid4-myc or cdc11-HA individually tagged strains, contained Sid4p-myc (Figure 7D), confirming the ability of these proteins to associate in vivo.
DISCUSSION
Through genetic and biochemical studies in S. pombe, a signal transduction pathway involving Spg1p and downstream protein kinases has been shown to regulate the onset of cytokinesis. All known components of this signaling cascade, termed the SIN, localize to SPBs in a Sid4p-dependent manner. However, we were unable previously to demonstrate a direct interaction between Sid4p and other SIN elements. Here, we have found that the previously uncharacterized Cdc11p is a homolog of S. cerevisiae Nud1p that bridges an interaction between Sid4p and other SIN components at the SPB. A similar conclusion has been reached recently by another group (Krapp et al., 2001).
Strains containing mutations in the cdc11+ gene were first identified as part of a screen for cell cycle mutants (Nurse et al., 1976). They were members of a family of mutants that failed to undergo septation but continued to elongate and formed multiple nuclei. This phenotype has since been given the name of sid. Subsequent genetic analyses indicated that cdc11 mutants displayed numerous genetic interactions with other sid mutants, indicating that Cdc11p was likely to participate in the SIN pathway (Marks et al., 1992). However, although all other genetically defined members of the pathway have been cloned and studied (for review, see Cerutti and Simanis, 2000; McCollum and Gould, 2001), throughout this time the identity of cdc11+ has eluded discovery. In retrospect, this may be due to its relatively large ORF (>3 kb), making it poorly represented in most gene libraries. The genetic studies mentioned above had previously placed Cdc11p toward the top of the SIN pathway. cdc11 mutants rescue a mutation in cdc16+, one of the GAP components of the system (Marks et al., 1992). In addition, increased expression of Cdc7p or Spg1p could rescue mutations in cdc11 (Fankhauser and Simanis, 1994; Schmidt et al., 1997). Our localization data support placing Cdc11p function near the top of the SIN pathway and indicate that Cdc11p most likely functions with Sid4p to provide a SPB platform on which the SIN pathway might organize. This interpretation is consistent with the recent characterization of S. cerevisiae Nud1p, a homolog of Cdc11p (Bardin et al., 2000; Gruneberg et al., 2000). Nud1p was found to interact with Bfa1p and Bub2p (Gruneberg et al., 2000), the S. cerevisiae homologues of Byr4p and Cdc16p, respectively. It was also reported that nud1 mutants mislocalized Tem1p, the S. cerevisiae equivalent of Spg1p, and failed to exit mitosis (Bardin et al., 2000; Gruneberg et al., 2000).
A search for motifs in the Cdc11p sequence revealed that the C terminus contains a total of 11 leucine-rich repeats arranged into three sets (Figure 1A). These structures are known to act as favorable surfaces for protein interactions and to be particularly common in those involved in signal transduction pathways (for review, see Kobe and Deisenhofer, 1995). No motif involved in catalytic activity was identified in these searches. Based on this information, it seems reasonable to suggest that Cdc11p may act in the capacity of a scaffolding protein for the SIN pathway. Our finding that the C-terminal region is responsible for binding to Sid4p will probably prove to be but one of a number of interactions in which Cdc11p participates. On this note, we found that Spg1p could not localize to SPBs in cells overproducing the C terminus of Cdc11p. It is likely, therefore, that residues 631-1045 lack an intact binding site for Spg1p or another protein that tethers Spg1p to Cdc11p.
The findings of ourselves and others (Sparks et al., 1999; Guertin et al., 2000; Hou et al., 2000; Li et al., 2000) that members of the SIN including its negative regulators depend upon Cdc11p, which in turn depends upon Sid4p, for localization to the SPB allow us to conclude that Sid4p is the SIN pathway component most proximal to the center of the SPB. Its role in the SIN may be purely a structural one, linking the “active” members of the SIN to the SPB. The previously reported ability of Sid4p to dimerize (Chang and Gould, 2000) has now been found to be the function of the two central coiled-coil domains within the protein, and this dimerization is essential for proper Sid4p SPB localization. We reported previously that the Sid4p N-terminus was responsible for SPB localization. This conclusion was based on the observations that Sid4ΔNp overexpression prevented assembly of other pathway components at the SPB, that Sid4ΔNp was not detected at the SPB (this was apparently due to a technical problem that we have since corrected), and that the Sid4p N-terminal portion was detected at the SPB when it was highly overproduced (Chang and Gould, 2000). However, the more detailed analysis reported here has shown that the C terminus of Sid4p, not the N terminus, is responsible for SPB localization. The Sid4p C terminus readily localizes to SPBs, and it is most likely the last coiled-coil region of the protein that interacts with another SPB component. When the Sid4p C terminus is overproduced, it blocks the ability of full-length Sid4p, Cdc11p, and Spg1p to localize to SPBs and hence produces a sid phenotype. This indicates that the C terminus binds efficiently to the Sid4p docking site on SPBs, and it will be interesting to learn what other SPB protein(s) the C terminus of Sid4p interacts with. In contrast, the Sid4p N-terminus, even when highly overproduced, does not disturb the localization of other SIN components (our unpublished results and Chang and Gould, 2000). Our explanation of why the Sid4p N terminus was previously detected at SPBs, when overproduced, is that this fragment, as we have demonstrated here, is able to bind Cdc11p. Presumably, the weak localization we observed was due to its ability to interact with some Cdc11p at SPBs (Chang and Gould, 2000).
Sid4p does not have any immediately obvious homolog in S. cerevisiae. However, it is worth considering the possibility that Cnm67p may be the S. cerevisiae counterpart of Sid4p. Although sequence similarity between the two proteins is undetectable, Cnm67p is of similar size to Sid4p and contains central coiled-coil domains (Brachat et al., 1998). The N-terminal region of Cnm67p interacts with the C terminus of Nud1p (Elliott et al., 1999), these proteins can be found in a cytoplasmic complex together (Elliott et al., 1999), and Cnm67p is required for the localization of Nud1p to the outer plaque of the SPB (Adams and Kilmartin, 1999). Further, it is the C terminus of Cnm67p, like the C terminus of Sid4p, that tethers it to the SPB, and the loss of Cnm67p leads to the formation of multinucleate cells (Schaerer et al., 2001). Importantly, loss of Cnm67p has been shown to result in spindle and/or astral microtubule defects resulting from the loss of the outer plaque of the SPB (Hoepfner et al., 2000). Although we have not detected any obvious microtubule or nuclear positioning defects in sid4-SA1 (our unpublished results; Chang and Gould, 2000), it will be important to examine sid4 null mutants by electron microscopy to determine whether they exhibit SPB defects.
While this article was under review, Krapp et al. (2001) reached similar conclusions regarding the identity and role of Cdc11p in the SIN. Furthermore, these authors reported that cdc11::ura4+ cells displayed defects in astral microtubule attachment to the SPB (Krapp et al., 2001) similar to those observed in S. cerevisiae nud1 mutants (Gruneberg et al., 2000). Although we detected extra Spg1p “dots” in certain cdc11 mutants at permissive temperature, indicating a potential abnormality in SPB structure, both interphase and mitotic microtubule structures in cdc11 temperature-sensitive mutants appeared normal as determined by indirect immunofluoresence microcopy (our unpublished results). Thus, some SPB functions are likely to be retained in the temperature-sensitive cdc11 mutants relative to the null.
In conclusion, Cdc11p now appears to be the previously missing link between Sid4p and the other SIN pathway components. The structure of Cdc11p indicates that it may function as a scaffold for mediating interactions of other SIN pathway members. It will be interesting to learn the full range of Cdc11p-associated proteins and determine how their interactions are regulated.
ACKNOWLEDGMENTS
We are grateful to Anna Feoktistova for excellent technical assistance and to Louise Chang and Hayes McDonald for providing the sid4-gfp and spg1-gfp strains and GFP PCR tagging cassette, respectively. We also thank Fred Chang for providing the PCR CFP and YFP tagging cassettes. This work was supported by the Howard Hughes Medical Institute of which K.L.G. is an associate investigator.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–09–0455. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–09–0455.
REFERENCES
- Adams IR, Kilmartin JV. Localization of core spindle pole body (SPB) components during SPB duplication in Saccharomyces cerevisiae. J Cell Biol. 1999;145:809–823. doi: 10.1083/jcb.145.4.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bähler J, Wu JQ, Longtine MS, Shah NG, McKenzie A, 3rd, Steever AB, Wach A, Philippsen P, Pringle JR. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast. 1998;14:943–951. doi: 10.1002/(SICI)1097-0061(199807)14:10<943::AID-YEA292>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Chang L, Wong KC, Naqvi NI, He X, Sazer S, Gould KL. Isolation and characterization of new fission yeast cytokinesis mutants. Genetics. 1998;149:1265–1275. doi: 10.1093/genetics/149.3.1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balasubramanian MK, McCollum D, Gould KL. Cytokinesis in fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1997;283:494–506. doi: 10.1016/s0076-6879(97)83039-x. [DOI] [PubMed] [Google Scholar]
- Barbet N, Muriel WJ, Carr AM. Versatile shuttle vectors and genomic libraries for use with Schizosaccharomyces pombe. Gene. 1992;114:59–66. doi: 10.1016/0378-1119(92)90707-v. [DOI] [PubMed] [Google Scholar]
- Bardin AJ, Visintin R, Amon A. A mechanism for coupling exit from mitosis to partitioning of the nucleus. Cell. 2000;102:21–31. doi: 10.1016/s0092-8674(00)00007-6. [DOI] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Brachat A, Kilmartin JV, Wach A, Philippsen P. Saccharomyces cerevisiaecells with defective spindle pole body outer plaques accomplish nuclear migration via half-bridge-organized microtubules. Mol Biol Cell. 1998;9:977–991. doi: 10.1091/mbc.9.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti L, Simanis V. Controlling the end of the cell cycle. Curr Opin Genet Dev. 2000;10:65–69. doi: 10.1016/s0959-437x(99)00044-1. [DOI] [PubMed] [Google Scholar]
- Chang L, Gould KL. Sid4p is required to localize components of the septation initiation pathway to the spindle pole body in fission yeast. Proc Natl Acad Sci USA. 2000;97:5249–5254. doi: 10.1073/pnas.97.10.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott S, Knop M, Schlenstedt G, Schiebel E. Spc29p is a component of the Spc110p subcomplex and is essential for spindle pole body duplication. Proc Natl Acad Sci USA. 1999;96:6205–6210. doi: 10.1073/pnas.96.11.6205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fankhauser C, Simanis V. The cdc7 protein kinase is a dosage dependent regulator of septum formation in fission yeast. EMBO J. 1994;13:3011–3019. doi: 10.1002/j.1460-2075.1994.tb06600.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge KA, Wong K, Armstrong J, Balasubramanian M, Albright CF. Byr4 and Cdc16 form a two-component GTPase-activating protein for the Spg1 GTPase that controls septation in fission yeast. Curr Biol. 1998;8:947–954. doi: 10.1016/s0960-9822(98)70394-x. [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–360. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Gould KL, Moreno S, Owen DJ, Sazer S, Nurse P. Phosphorylation at Thr167 is required for Schizosaccharomyces pombep34cdc2 function. EMBO J. 1991;10:3297–3309. doi: 10.1002/j.1460-2075.1991.tb04894.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruneberg U, Campbell K, Simpson C, Grindlay J, Schiebel E. Nud1p links astral microtubule organization and the control of exit from mitosis. EMBO J. 2000;19:6475–6488. doi: 10.1093/emboj/19.23.6475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guertin DA, Chang L, Irshad F, Gould KL, McCollum D. The role of the sid1p kinase and cdc14p in regulating the onset of cytokinesis in fission yeast. EMBO J. 2000;19:1803–1815. doi: 10.1093/emboj/19.8.1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoepfner D, Brachat A, Philippsen P. Time-lapse video microscopy analysis reveals astral microtubule detachment in the yeast spindle pole mutant cnm67. Mol Biol Cell. 2000;11:1197–1211. doi: 10.1091/mbc.11.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou MC, Salek J, McCollum D. Mob1p interacts with the Sid2p kinase and is required for cytokinesis in fission yeast. Curr Biol. 2000;10:619–622. doi: 10.1016/s0960-9822(00)00492-9. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitayama C, Sugimoto A, Yamamoto M. Type II myosin heavy chain encoded by the myo2 gene composes the contractile ring during cytokinesis in Schizosaccharomyces pombe. J Cell Biol. 1997;137:1309–1319. doi: 10.1083/jcb.137.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobe B, Deisenhofer J. Proteins with leucine-rich repeats. Curr Opin Struct Biol. 1995;5:409–416. doi: 10.1016/0959-440x(95)80105-7. [DOI] [PubMed] [Google Scholar]
- Krapp A, Schmidt S, Cano E, Simanis V. S. pombecdc11p, together with sid4p, provides an anchor for septation initiation network proteins on the spindle pole body. Curr Biol. 2001;11:1559–1568. doi: 10.1016/s0960-9822(01)00478-x. [DOI] [PubMed] [Google Scholar]
- Li C, Furge KA, Cheng QC, Albright CF. Byr4 localizes to spindle-pole bodies in a cell cycle-regulated manner to control Cdc7 localization and septation in fission yeast. J Biol Chem. 2000;275:14381–14387. doi: 10.1074/jbc.275.19.14381. [DOI] [PubMed] [Google Scholar]
- Marks J, Fankhauser C, Simanis V. Genetic interactions in the control of septation in Schizosaccharomyces pombe. J Cell Sci. 1992;101:801–808. doi: 10.1242/jcs.101.4.801. [DOI] [PubMed] [Google Scholar]
- Marks J, Hagan IM, Hyams JS. Growth polarity and cytokinesis in fission yeast: the role of the cytoskeleton. J Cell Sci Suppl. 1986;5:229–241. doi: 10.1242/jcs.1986.supplement_5.15. [DOI] [PubMed] [Google Scholar]
- Maundrell K. Thiamine-repressible expression vectors pREP and pRIP for fission yeast. Gene. 1993;123:127–30. doi: 10.1016/0378-1119(93)90551-d. [DOI] [PubMed] [Google Scholar]
- McCollum D, Gould KL. Timing is everything: regulation of mitotic exit and cytokinesis by the MEN and SIN. Trends Cell Biol. 2001;11:89–95. doi: 10.1016/s0962-8924(00)01901-2. [DOI] [PubMed] [Google Scholar]
- McDonald WH, Ohi R, Smelkova N, Frendewey D, Gould KL. Myb-related fission yeast cdc5p is a component of a 40S snRNP-containing complex and is essential for pre-mRNA splicing. Mol Cell Biol. 1999;19:5352–5362. doi: 10.1128/mcb.19.8.5352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno S, Klar A, Nurse P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 1991;194:795–823. doi: 10.1016/0076-6879(91)94059-l. [DOI] [PubMed] [Google Scholar]
- Nurse P, Thuriaux P, Nasmyth K. Genetic control of the cell division cycle in the fission yeast Schizosaccharomyces pombe. Mol Gen Genet. 1976;146:167–178. doi: 10.1007/BF00268085. [DOI] [PubMed] [Google Scholar]
- Prentice HL. High efficiency transformation of Schizosaccharomyces pombeby electroporation. Nucleic Acids Res. 1992;20:621. doi: 10.1093/nar/20.3.621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salimova E, Sohrmann M, Fournier N, Simanis V. The S. pombe orthologue of the S. cerevisiae mob1gene is essential and functions in signaling the onset of septum formation. J Cell Sci. 2000;113:1695–1704. doi: 10.1242/jcs.113.10.1695. [DOI] [PubMed] [Google Scholar]
- Schaerer F, Morgan G, Winey M, Philippsen P. Cnm67p is a spacer protein of the Saccharomyces cerevisiaespindle pole body outer plaque. Mol Biol Cell. 2001;12:2519–2533. doi: 10.1091/mbc.12.8.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt S, Sohrmann M, Hofmann K, Woollard A, Simanis V. The Spg1p GTPase is an essential, dosage-dependent inducer of septum formation in Schizosaccharomyces pombe. Genes Dev. 1997;11:1519–1534. doi: 10.1101/gad.11.12.1519. [DOI] [PubMed] [Google Scholar]
- Sohrmann M, Schmidt S, Hagan I, Simanis V. Asymmetric segregation on spindle poles of the Schizosaccharomyces pombeseptum-inducing protein kinase Cdc7p. Genes Dev. 1998;12:84–94. doi: 10.1101/gad.12.1.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparks CA, Morphew M, McCollum D. Sid2p, a spindle pole body kinase that regulates the onset of cytokinesis. J Cell Biol. 1999;146:777–790. doi: 10.1083/jcb.146.4.777. [DOI] [PMC free article] [PubMed] [Google Scholar]