Abstract
Multiple morphological abnormalities of the sperm flagella (MMAF) is a severe form of asthenozoospermia categorized by immotile spermatozoa with abnormal flagella in ejaculate. Whole-exome sequencing (WES) is used to detect pathogenic variants in patients with MMAF. In this study, a novel homozygous frameshift variant (c.6158_6159insT) in dynein axonemal heavy chain 8 (DNAH8) from two infertile brothers with MMAF in a consanguineous Pakistani family was identified by WES. Reverse transcription-polymerase chain reaction (RT-PCR) confirmed DNAH8 mRNA decay in these patients with the DNAH8 mutation. Hematoxylin–eosin staining and transmission electron microscopy revealed highly divergent morphology and ultrastructure of sperm flagella in these patients. Furthermore, an immunofluorescence assay showed the absence of DNAH8 and a reduction in its associated protein DNAH17 in the patients’ spermatozoa. Collectively, our study expands the phenotypic spectrum of patients with DNAH8-related MMAF worldwide.
Keywords: DNAH8, gene mutation, male infertility, multiple morphological abnormalities of the sperm flagella
INTRODUCTION
Infertility affects approximately 8%–12% of couples worldwide, and infertility is attributed to the male factor in 20%–25% of these couples.1 Poor semen quality, including low sperm count and motility and high abnormal sperm morphology, is the main factor contributing to male infertility. Both intrinsic and extrinsic factors affect normal semen parameters, among which genetic variants may cause severe spermatogenic impairment. Although more than 1000 genes are predominantly expressed in the human testis, only a few genes have been shown to contribute to male fertility.2 Accordingly, more genetic determinants need to be identified.
Multiple morphological abnormalities of the sperm flagella (MMAF) is an infertility phenotype with severe flagellar morphological defects, such as short, coiled, bent, absent, and/or irregular flagella.3 The ultrastructure of the sperm flagellum in patients with MMAF shows the absence of the axonemal central pair complex (CPC) and/or microtubule doublet, outer dense fiber, mitochondrial sheath, and dynein arm disorganization. A large percentage of sperm morphological abnormalities have been found in infertile men with testicular disorders (i.e., toxic substances and varicocele).4 Given that genetic defect is one of the main etiological causes of severe asthenoteratospermia, more than 20 genes have been reported to be related to MMAF (e.g., cilia- and flagella-associated protein 43 [CFAP43], cilia- and flagella-associated protein 44 [CFAP44], armadillo repeat-containing 2 [ARMC2], DAZ-interacting zinc finger protein 1 [DZIP1], dynein axonemal heavy chain 17 [DNAH17], and serine/threonine kinase 33 [STK33]).5,6,7,8,9,10 However, these genetic discoveries account for only approximately 35%–60% of MMAF cases, indicating that the causes of many MMAF remain unknown.11,12
The human dynein axonemal heavy chain 8 (DNAH8) gene located at chromosome 6p21.2 encodes 92 exons, producing a 4695-amino acid protein. According to the Human Protein Atlas (HPA) database, DNAH8 encodes an outer dynein arm protein that is specifically expressed in the testis.13 Mutation in DNAH8 was first reported in a man with primary ciliary dyskinesia (PCD), a hereditary disorder characterized by aberrant ciliary structure causing persistent respiratory tract infections and asthenozoospermia.14 Subsequently, biallelic pathogenic variants in DNAH8 were identified in infertile men with MMAF.15,16 In line with studies in humans, DNAH8 mutant mice also exhibit a typical MMAF phenotype.15 In addition, one study by Zhou et al.17 reported that approximately 60% of the spermatozoa of two affected men expressing a DNAH8 splicing variant exhibited abnormal sperm heads. All of these studies indicate that there is a causal association between DNAH8 variants and male infertility.
This study involved two primary infertile males with the MMAF phenotype in a consanguineous Pakistani family. Through whole-exome sequencing (WES) and Sanger sequencing, we identified a novel homozygous frameshift variant (c.6158_6159insT, p.Gly2054Trpfs*50) of the DNAH8 gene in these patients that resulted in a reduction in the expression levels of DNAH8 and its associated protein DNAH17. Our findings expand the genetic cause of the human MMAF phenotype.
PARTICIPANTS AND METHODS
Participants, clinical examination, and semen analysis
This study was approved by the Ethical Committee of the University of Science and Technology of China (USTC; Hefei, China) with approval No. USTCEC202000003. We recruited two infertile brothers from a consanguineous Pakistani family (PK-INF-379) who were diagnosed with asthenozoospermia. Written informed consent was obtained from all participants. A detailed questionnaire on PCD and a clinical history of infertility was obtained from all available family members. A detailed physical examination, including testicular size, secondary sexual characteristics, genital surgery, and varicocele, was performed. After 3–5 days of abstinence, semen samples were collected by masturbation, and semen analysis was carried out after liquefaction at 37°C for 30 min according to the World Health Organization (WHO) guidelines.18 For each ejaculate, sperm motility was assessed by counting at least 200 sperm across ten randomly selected individual fields. Sperm concentration was measured using a hemocytometer. Semen parameters were examined at least twice for both patients (IV:1 and IV:3). Sperm morphological analyses were performed in patients IV:1 and IV:3, and at least 200 sperm per sample were observed using a light microscope at 100× (Nikon Eclipse 80i, Nikon, Tokyo, Japan). Liquefied semen was smeared onto a glass slide, fixed in 4% paraformaldehyde, washed twice with 1× phosphate-buffered saline (PBS) at room temperature for 5 min each time, and stored at −20°C until assay.
Histological analyses
For hematoxylin–eosin (HE) staining, semen smears were stained with hematoxylin for 30 min, washed with ddH2O for 1 min, dipped in 1% acid alcohol for 30 s, and placed in tap water for 5 min to remove excess dye. The slides were then stained with eosin for 8 min and dehydrated in graded ethanol (80%, 90%, and 100%, respectively) for 1 min each, followed by immersion in xylene for 10 min.
Chromosomal analysis, WES, and variant filtering
Karyotyping was carried out on cultured peripheral blood lymphocytes from the two patients using standard G-banding. Genomic DNA was extracted from peripheral blood samples from five family members (the two patients [IV:1 and IV:3], their father [III:1], and two fertile brothers [IV:5 and IV:7]) using a FlexiGene DNA kit according to the manufacturer’s protocol (51206, QIAGEN, Hilden, Germany). For WES screening, DNA was captured with an AIExome Enrichment Kit V1 (iGeneTech, Beijing, China) and sequenced using the HiSeq2000 platform (Illumina, San Diego, CA, USA). Raw WES data processing was performed as previously reported.19 The bioinformatic pipeline of the filtering process is provided in Supplementary Figure 1 (217KB, tif)
The variants were retained for further screening according to their recessive inheritance pattern.20 In addition, Sanger sequencing was performed to confirm the DNAH8 variant in all participants (III:1, IV:1, IV:3, IV:5, and IV:7) using the following primer sequences: forward 5’-TTTATGTGGGCTACTCAGAC-3’ and reverse 5’-GCAATATCATACGGATCAAC-3’ (438 bp).
In silico study of DNAH8
The genomic sequence of DNAH8 was retrieved from the GenBank NCBI database (https://ncbi.nlm.nih.gov). The functional pathogenicity of the mutation was assessed using the MutationTaster prediction tool.21
RNA extraction and reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted from blood samples of all participants using TRIzol reagent (9109, TAKARA, Otsu, Japan). The concentration and integrity of the RNA samples were determined based on optical density (OD) at 260 nm and the 260/280 nm ratio using a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE, USA). Then, 1 μg of RNA was reverse transcribed to cDNA using a PrimeScript RT Reagent Kit (TAKARA). The cDNA was diluted and used for RT-PCR with primer pairs for position 6022–6254 covering mutation site 6158–6159. The sequences of the primers used are as follows: DNAH8, forward 5’-GATGTTGATTTTATTTACCA-3’ and reverse 5’-GATCAAATGGATTTCAGAGG-3’ (233 bp); and β-actin (ACTB), forward 5’-AATGAGCTGCGTGTGGCTC-3’ and reverse 5’-ATAGCACAGCCTGGATAGCAAC-3’ (214 bp). All amplified DNA fragments were analyzed by 1% agarose gel electrophoresis containing GelRed (APExBIO, Houston, TX, USA). To check the quality of the primers, both a positive control (fertile individual DNA sample as template) and a negative control (ddH2O as template) were used for PCR. A 100-bp DNA ladder (BM311, Trans, Beijing, China) was used as a molecular marker.
Immunofluorescence (IF) staining
Human sperm smears and IF staining were carried out as previously described.9 Briefly, semen smears were permeabilized for 30 min with 0.2% Triton X-100 in 1× PBS (pH = 7.4), followed by incubation in blocking buffer (PBS containing 3% skim milk and 0.1% Triton X-100). The samples were then incubated with the primary antibodies diluted in blocking buffer overnight at 4°C, followed by incubation with Alexa Fluor 488-conjugated goat anti-mouse antibodies (1:100; A21121, Molecular Probes, Eugene, OR, USA) or Alexa Fluor 555-conjugated donkey anti-rabbit antibodies (1:200; A31572, Molecular Probes) for 1 h at 37°C. Anti-tubulin (1:200; T6074, Sigma-Aldrich, St. Louis, MO, USA), anti-DNAH8 (1:50; HPA028447, Sigma-Aldrich), and anti-DNAH17 (1:100; ABclonal Biotechnology, Wuhan, China) antibodies were also used. A Nikon Eclipse 80i microscope with a color charge-coupled device (CCD) camera (Hamamatsu Photonics, Hamamatsu, Japan) was utilized to capture images.
Transmission electron microscopy (TEM) analysis
Spermatozoa from patients and fertile men were washed with 1×PBS (pH = 7.4), fixed in 0.1 mol l−1 cacodylate buffer (pH = 7.4) containing 4% paraformaldehyde, 0.2% picric acid, and 8% glutaraldehyde at 4°C and incubated for 12 h. The fixed spermatozoa were washed again four times with 0.1 mol l−1 cacodylate buffer (pH = 7.4), followed by postfixation in 1% osmium tetroxide (OsO4). Subsequently, the samples were dehydrated using alcohol gradients (30%, 50%, 75%, 95%, and 100%, respectively), followed by infiltration with a mixture of acetone and Epon resin. The samples were cut into sections of 70 nm using an ultrathin slicer, followed by staining with lead citrate and uranyl acetate. A transmission electron microscope (Hitachi, Tokyo, Japan) was used to capture images and analyze the ultrastructure of the sperm flagella at 100 kV.
RESULTS
Clinical characteristics of patients with asthenozoospermia in a consanguineous Pakistani family
Two infertile brothers from a consanguineous Pakistani family were recruited for the current study (Figure 1a). The two patients were born to first-cousin parents and were married at 23 (IV:1) and 20 (IV:3) years of age, respectively. Physical examination revealed normal testicular sizes, male external genitalia development, and secondary sexual characteristics. Moreover, the affected individuals exhibited a normal visceral position (Supplementary Figure 2 (78.4KB, tif) ), without any clinical symptoms of PCD. Chromosomal analysis revealed a normal karyotype (46;XY), with no large scale deletions of the Y chromosome. The semen parameters and physical features of the two infertile patients (IV:1 and IV:3) are summarized in Table 1. According to the WHO recommendation standards, the two patients had low sperm motility at 6.5% ± 1.5% for IV:1 and 11.0% ± 1.0% for IV:3; the sperm vitality of IV:3 was normal. Morphologically, the abnormal sperm were characterized by coiled, absent, short, and angulation, with irregular sperm flagellar phenotypes (Figure 1b and Table 1).22 Based on the analysis of sperm morphometric parameters of the patients, the maximal length and width of the sperm head and the ratio of the surface of the acrosomal region were similar to those of fertile controls.23 However, there was a significant difference between the length of the midpiece and the principal piece of the sperm tail of the patients and control individuals (both P < 0.001; Table 1). These results indicate that IV:1 and IV:3 exhibited severe asthenozoospermia due to MMAF.
Figure 1.
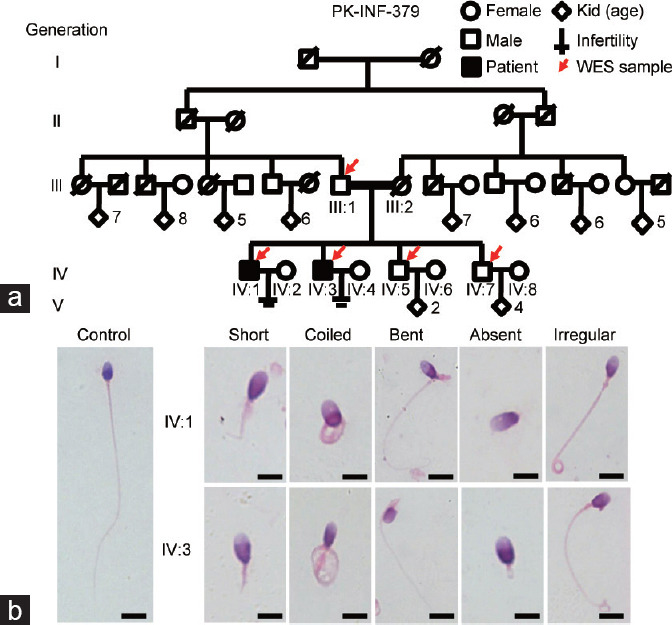
A consanguineous family with two infertile males showing morphological abnormalities of the sperm flagella. (a) According to the family pedigree, the patients (IV:1 and IV:3) were born to a first-cousin marriage. Circles and squares denote females and males, respectively. Red arrows mark the individuals for whom WES was performed. The parents of the two patients (filled black symbols) with severe asthenozoospermia had a first-degree consanguineous marriage (marked as parallel horizontal lines). Slash symbols indicate a deceased family member. (b) Morphological analyses of the patients’ spermatozoa under light microscopy. Representative spermatozoa from a fertile control and two affected individuals (IV:1 and IV:3). Scale bars = 10 µm. WES: whole-exome sequencing.
Table 1.
Semen parameters of the patients carrying the DNAH8 mutation
| Parameter | Patient IV:1 | Patient IV:3 | Reference values |
|---|---|---|---|
| Fertility | Infertile | Infertile | - |
| Infertility phenotype | Asthenozoospermia MMAF | Asthenozoospermia MMAF | - |
| Mutation | MT/MT | MT/MT | - |
| Physical information | |||
| Age (year) | 45 | 43 | - |
| Marriage (year) | 23 | 20 | - |
| Height/weight (cm/kg) | 170.2/61.0 | 167.6/62.0 | - |
| Semen parameters | |||
| Semen volume (ml), mean±s.e.m. | 2.0±0.5 | 1.6±0.2 | >1.5 |
| Semen PH | Alkaline | Alkaline | Alkaline |
| Sperm concentration (×106 ml−1), mean±s.e.m. | 22.0±4.0 | 27.0±2.5 | >15.0 |
| Total sperm count (×106), mean±s.e.m. | 42.0±3.0 | 44.0±9.5 | >39.0 |
| Progressive motility (%), mean±s.e.m. | 6.5±1.5* | 11.0±1.0* | >32.0 |
| Nonprogressive motility (%), mean±s.e.m. | 8.0±2.0* | 15.0±2.0* | - |
| Total motility (%), mean±s.e.m. | 14.5±0.5* | 26.0±1.0* | >40.0 |
| Immotile (%), mean±s.e.m. | 85.5±0.5 | 74.0±1.0 | - |
| Vitality (%), mean±s.e.m. | - | 69.5±4.8 | >58.0 |
| Sperm morphology | |||
| Normal (%) | 8.4 | 4.2 | >4 |
| Abnormal head (%) | 1.3 | 4.5 | - |
| Abnormal flagella (%) | 85.0 | 76.2 | - |
| Abnormal head and tail (%) | 5.3 | 15.1 | - |
| Sperm flagellar morphology | |||
| Absent flagella (%) | 5.3* | 7.1* | <5.0 |
| Short flagella (%) | 13.3* | 6.0* | <1.0 |
| Coiled flagella (%) | 29.2* | 35.2* | <17.0 |
| Angulation (%) | 20.8* | 13.4* | <13.0 |
| Irregular caliber (%) | 21.7* | 29.7* | <2.0 |
| Normal flagella (%) | 9.7* | 8.7* | >23.0 |
| Morphometric parameters of sperm | |||
| Maximal length of head (µm), mean±s.e.m. | 4.8±0.2 | 4.8±0.3 | 5.4±0.2 |
| Maximal width of head (µm), mean±s.e.m. | 2.7±0.3 | 2.9±0.2 | 2.9±0.2 |
| Length of midpiece (µm), mean±s.e.m. | 4.0±0.9 | 3.7±0.6 | 4.7±0.1 |
| Length of principal piece (µm), mean±s.e.m. | 33.9±10.8 | 26.3±12.6 | 50.0±5.0 |
| Acrosomal region of sperm head: total surface of sperm head (×100), mean±s.e.m. | 50.4±0.5 | 49.7±0.7 | 50.5±0.5 |
*Abnormal values. Lower and upper reference limits are shown according to the World Health Organization standards.18 For IV:1 and IV:3, semen volume, sperm concentration, and sperm motility were examined at least twice (n = 2). Sperm morphology was assessed using at least 200 spermatozoa examined for IV:1 and IV:3. Distribution ranges of morphologically abnormal spermatozoa were observed in fertile individuals.22 A total of 60 sperm were counted for each patient to quantify morphometric parameters.23 DNAH8: dynein axonemal heavy chain 8; MMAF: multiple morphological abnormalities of the sperm flagella; s.e.m.: standard error of the mean; MT: mutant allele; -: no value
Identification of the DNAH8 variant
To identify the genetic cause of the abnormal sperm flagella in this family, WES was performed for the two affected individuals (IV:1 and IV:3) and three unaffected individuals (III:1, IV:5, and IV:7). After multiple filtration steps, a homozygous DNAH8 frameshift mutation (c.6158_6159insT, p.Gly2054Trpfs*50) was identified in the patients (Figure 2a and Supplementary Figure 1 (217KB, tif) ). Sanger sequencing confirmed the patients’ father and fertile brothers to be heterozygous carriers (Figure 2b). The allele frequency of the identified mutation is zero in public population databases (1000 Genomes and GnomAD). As DNAH8 variants have been reported in infertile men with an MMAF phenotype, the identified frameshift mutation in DNAH8 is considered a likely pathogenic variant in this family.
Figure 2.
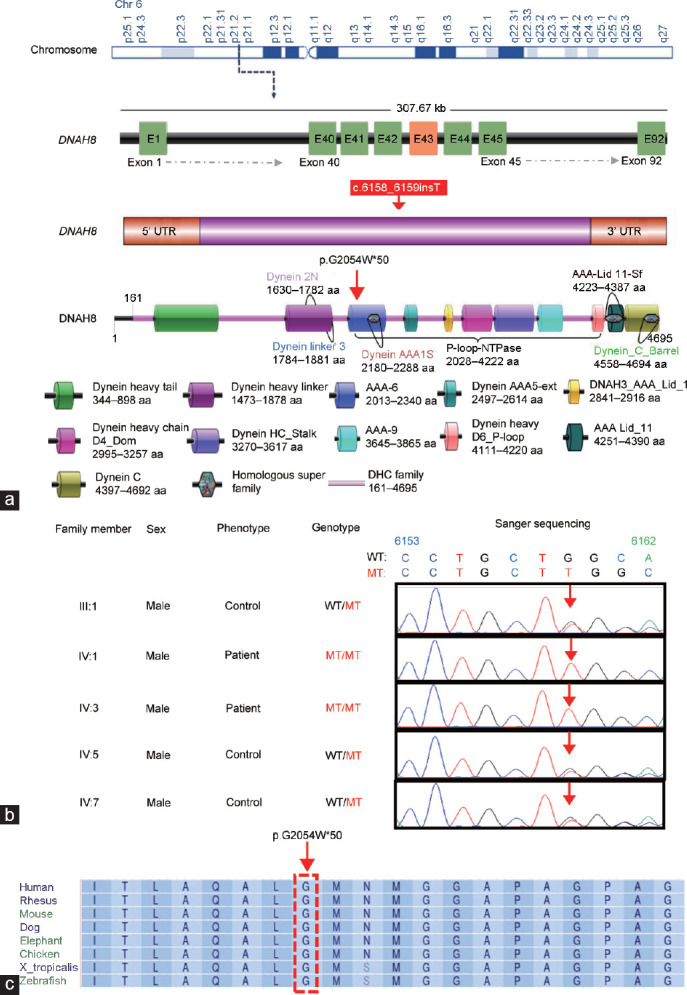
Identification of a homozygous frameshift mutation in DNAH8. (a) The position of the identified DNAH8 mutation at the genomic, transcript, and protein levels. DNAH8 is located at chromosome (chr) 6, g.38862342–38862343T, comprises 92 exons, and encodes a predicted 4695-amino acid protein (NCBI: NP_001193856). The red box indicates the location of the mutation site. (b) The DNAH8 mutation, c.6158_6159insT, in genomic DNA from all available family members. (c) Multiple sequence alignment shows conservation of the affected amino acid (glycine) across different organisms. Arrowheads refer to the mutation sites; WT: wild-type allele; MT: mutant allele; A: adenine; C: cytosine; G: guanine; T: thymine; DNAH8: dynein axonemal heavy chain 8; UTR: untranslated region.
In silico analyses of DNAH8
The glycine residue (G) at position 2054 of DNAH8 is highly conserved among different species (Figure 2c). The homozygous mutation (c.6158_6159insT) causes a frameshift of the coding sequence and amino acid alteration at the Gly2054 site, resulting in a stop codon after a 50-amino acid frameshift sequence (Figure 2a). Furthermore, in silico analysis by MutationTaster predicted that the frameshift mutation in the DNAH8 gene is potentially disease causing.
Decreased DNAH8 transcripts in the two patients
To determine the effect of homozygous frameshift mutation on the expression level of DNAH8 mutant transcripts, we isolated RNA from blood samples from both patients and fertile men and performed RT-PCR assays. Endogenous DNAH8 transcripts were not detected in either patient, but a clear band was obtained for the fertile control (Figure 3). Because the mutation is predicted to cause a premature stop codon, the combined data from RT-PCR and in silico analyses (Figure 2a) suggest that the absence of DNAH8 transcripts in the infertile brothers is due to nonsense-mediated decay machinery.
Figure 3.
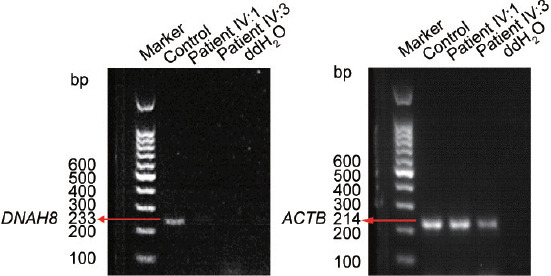
Reduced level of DNAH8 mRNA expression in the patients. Levels of DNAH8 mRNA from both patients (IV:1 and IV:3) and fertile controls were determined by RT-PCR. ACTB (β-actin) served as the loading control. DNAH8: dynein axonemal heavy chain 8; RT-PCR: reverse transcription-polymerase chain reaction; mRNA: messenger RNA.
Loss of the DNAH8 protein in the patients’ spermatozoa
To further assess the influence of the frameshift variant on DNAH8 expression, we performed an immunofluorescence assay using an anti-DNAH8 antibody. In contrast to the strong DNAH8 protein signals in the sperm flagella of fertile men, we did not observe DNAH8 signals in the patients’ spermatozoa (Figure 4a). In addition, expression of DNAH17 was dramatically reduced in the spermatozoa of the patients compared to healthy individuals (Figure 4b).24,25 Together, these results show that the novel DNAH8 variant disrupts expression of the proteins DNAH8 and DNAH17.
Figure 4.
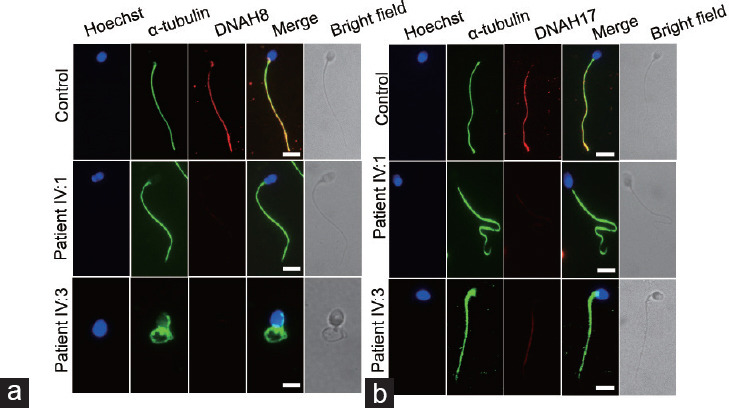
Expression and localization of DNAH8 and DNAH17 in the patients’ spermatozoa. (a) Representative images of spermatozoa from fertile controls and patients carrying the DNAH8 variant stained with an anti-DNAH8 antibody (red), an anti-α-tubulin antibody (green), and Hoechst (blue, nuclear marker). The DNAH8 signal was not detected in the sperm flagella from the two patients. (b) Representative images of spermatozoa from fertile controls and the patients stained with an anti-DNAH17 antibody (red), an anti-α-tubulin antibody (green), and Hoechst (blue). The signal intensity of DNAH17 was decreased in the spermatozoa from the patients with the DNAH8 mutant compared to healthy individuals. Scale bars = 5 µm. DNAH8: dynein axonemal heavy chain 8.
Severe disorganization of axonemal structures in the patients’ spermatozoa
Because disruption of DNAH8 in humans and mice causes abnormalities in the components of the flagellum,15,24 TEM was carried out to investigate the ultrastructural characteristics of the sperm flagella in our patients (Figure 5). In contrast to the typical axoneme ultrastructure that consists of nine doublets of microtubules and two central pairs (9 + 2 organization) in spermatozoa from a fertile man, ultrastructural defects were observed in more than 80% of the sections of sperm flagella from the two patients, including disorganized or missing peripheral microtubule doublets and outer dense fibers, missing or disassembled outer dense arms (ODAs), and absent central pair complexes (Figure 5). Hence, these findings indicate that the homozygous frameshift mutation in DNAH8 causes severe ultrastructural flagellum defects.
Figure 5.
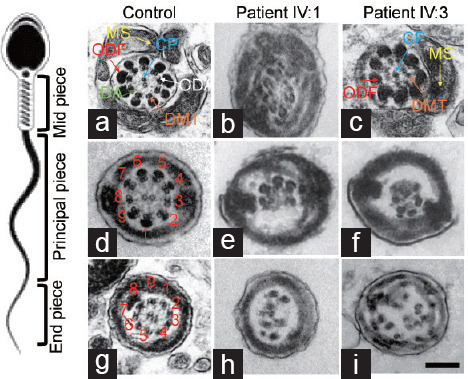
The axonemal structure of sperm flagella in fertile controls and the patients harboring the homozygous frameshift DNAH8 variant. Representative images of midpiece sperm flagella from (a) fertile control and the patients of (b) IV:1 and (c) IV:3, respectively. Both groups show the typical ‘‘9+2’’ microtubule structure, including nine pairs of peripheral microtubule doublets (DMT; orange arrows), nine ODFs (red arrows), ODA (white arrow), IDA (green arrow), and CP (blue arrow). (d) Representative images of the principal piece of sperm flagella from fertile controls. Disarrangement of microtubules and the missing central pair were found in sperm flagella from the patients of (e) IV:1 and (f) IV:3. (g) Representative images of end pieces of sperm flagella from fertile controls. Axonemal anomalies include a lack of CP and ODA, DMT, and ODF missing or disorganized were found in sperm flagella from the patients of (h) IV:1 and (i) IV:3. Scale bars = 500 nm. DNAH8: dynein axonemal heavy chain 8; CP: central pair of microtubules; MS: mitochondria sheath; ODF: outer dense fiber; ODA: outer dynein arm; IDA: inner dynein arm; DMT: doublet microtubule.
DISCUSSION
MMAF, a severe male infertility phenotype, is mainly caused by mutations in axonemal genes. In the present study, we detected a novel homozygous frameshift mutation (c.6158_6159insT) in DNAH8 of two infertile brothers with MMAF in a consanguineous Pakistani family. Multiple sequence alignment and in silico prediction tools showed that the DNAH8 variant is highly conserved among different organisms and predicted to have a high probability of pathogenicity. In our study, the infertile males harboring this homozygous frameshift DNAH8 variant exhibited a typical MMAF phenotype, in line with the results of other studies.15,16,24
As a DNAH component, the DNAH8 protein is highly expressed in spermatozoa,25 suggesting that it plays an important role in spermatogenesis and male fertility. The homozygous variant c.6158_6159insT located in exon 43 causes a frameshift and a premature stop codon after 50 amino acids, resulting in the formation of a truncated DNAH8 protein. There are six conserved AAA+ domains (AAA1–6) in DNAH8: domains AAA1–4 bind/hydrolyze ATP, and domains AAA5 and AAA6 appear to serve a structural role for the adenosinetriphosphatases (ATPase) domain.26 The G2054W mutant is located in the AAA6 domain, resulting in low sperm motility in patients, which is in line with other reports showing that AAA+ domain DNAH8 variants cause asthenozoospermia.15,17 MutationTaster analysis predicted that the c.6158-6159insT mutation causes a premature termination codon. RT-PCR and IF assays showed the absence of DNAH8 transcripts and DNAH8 protein in the patients’ spermatozoa, suggesting that the c.6158-6159insT variant is likely to result in nonsense-mediated decay. In addition, expression of DNAH17, a DNAH8-associated protein, was significantly reduced in the sperm flagella of the patients with the DNAH8 mutation. Disruption of the interaction between DNAH8 and DNAH17 impairs the integrity of the sperm tail, causing asthenozoospermia and male infertility.
It should be noted that mutations in DNAH genes may cause MMAF or PCD. The DNAH8 gene was initially reported to be a candidate PCD gene.14,27 However, recent studies have shown that DNAH8 mutation causes MMAF in infertile men.15,16,24 In this study, we confirmed the MMAF phenotype rather than the symptoms of PCD in spermatozoa from two infertile brothers with the DNAH8 mutation, suggesting that this novel pathogenic DNAH8 variant is a cause of MMAF and primary infertility. Although DNAH8 mutations contribute to male infertility by inducing the MMAF phenotype, good clinical outcomes can be achieved by intracytoplasmic sperm injection (ICSI) treatment,24 indicating high-quality sperm nuclei in patients with DNAH8 mutations. Therefore, genetic diagnostics for MMAF patients may provide important information and help predict the success of ICSI.
In summary, we identified a novel homozygous frameshift DNAH8 variant in two infertile brothers with the MMAF phenotype from a consanguineous Pakistani family. The findings of this study improve our understanding of the genetic basis of MMAF and provide knowledge for genetic counseling of infertile men.
AUTHOR CONTRIBUTIONS
SD carried out most of the experiments (histological, immunofluorescence, and TEM analyses) and drafted the manuscript. AK, AU, MLY, IA, AZ, HZ, JTZ, MZ, and KK carried out the semen analysis and performed the WES sequencing and in silico analysis. SB reviewed and edited the manuscript. QHS conceived the study, participated in its design and coordination, and helped to draft the manuscript. All authors read and approved the final manuscript.
COMPETING INTERESTS
All authors declare no competing interests.
WES data analysis. (a) WES data analysis flow chart. A MAF less than 0.01 was considered for filtration of existing mutations in the 1000 Genomes, ESP6500, ExAC, and GenomAD databases. (b) Genome-wide logarithm of the odds ratio (LOD) scores using WES-derived genotypes for the family. WES: whole-exome sequencing; MAF: minor allele frequency.
Chest X-ray of the patients. Chest X-ray showing no PCD symptoms in the patients with the homozygous frameshift DNAH8 mutation. PCD: primary ciliary dyskinesia; DNAH8: dynein axonemal heavy chain 8.
ACKNOWLEDGMENTS
We thank Dr. Bo Xu and Dr. Xiao-Hua Jiang from The First Affiliated Hospital of USTC (Hefei, China) for their valuable contributions to the data analysis. This work was supported by the National Natural Science Foundation of China (No. 31871514, No. 81971333, and No. 82071709) and the National Key Research and Development Program of China (2019YFA0802600 and 2021YFC2700202).
Supplementary Information is linked to the online version of the paper on the Asian Journal of Andrology website.
REFERENCES
- 1.Vander Borght M, Wyns C. Fertility and infertility:definition and epidemiology. Clin Biochem. 2018;62:2–10. doi: 10.1016/j.clinbiochem.2018.03.012. [DOI] [PubMed] [Google Scholar]
- 2.Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 3.Sudhakar DV, Shah R, Gajbhiye RK. Genetics of male infertility - present and future:a narrative review. J Hum Reprod Sci. 2021;14:217–27. doi: 10.4103/jhrs.jhrs_115_21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal A, Sharma R, Harlev A, Esteves SC. Effect of varicocele on semen characteristics according to the new 2010 World Health Organization criteria:a systematic review and meta-analysis. Asian J Androl. 2016;18:163–70. doi: 10.4103/1008-682X.172638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ma H, Zhang B, Khan A, Zhao D, Ma A, et al. Novel frameshift mutation in STK33 is associated with asthenozoospermia and multiple morphological abnormalities of the flagella. Hum Mol Genet. 2021;30:1977–84. doi: 10.1093/hmg/ddab165. [DOI] [PubMed] [Google Scholar]
- 6.Tang S, Wang X, Li W, Yang X, Li Z, et al. Biallelic mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genet. 2017;100:854–64. doi: 10.1016/j.ajhg.2017.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv M, Liu W, Chi W, Ni X, Wang J, et al. Homozygous mutations in DZIP1 can induce asthenoteratospermia with severe MMAF. J Med Genet. 2020;57:445–53. doi: 10.1136/jmedgenet-2019-106479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coutton C, Martinez G, Kherraf ZE, Amiri-Yekta A, Boguenet M, et al. Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am J Hum Genet. 2019;104:331–40. doi: 10.1016/j.ajhg.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang B, Ma H, Khan T, Ma A, Li T, et al. A DNAH17 missense variant causes flagella destabilization and asthenozoospermia. J Exp Med. 2020;217:e20182365. doi: 10.1084/jem.20182365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Toure A, Martinez G, Kherraf ZE, Cazin C, Beurois J, et al. The genetic architecture of morphological abnormalities of the sperm tail. Hum Genet. 2021;140:21–42. doi: 10.1007/s00439-020-02113-x. [DOI] [PubMed] [Google Scholar]
- 11.Lores P, Dacheux D, Kherraf ZE, Nsota Mbango JF, Coutton C, et al. Mutations in TTC29, encoding an evolutionarily conserved axonemal protein, result in asthenozoospermia and male infertility. Am J Hum Genet. 2019;105:1148–67. doi: 10.1016/j.ajhg.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ma A, Zeb A, Ali I, Zhao D, Khan A, et al. Biallelic variants in CFAP61 cause multiple morphological abnormalities of the flagella and male infertility. Front Cell Dev Biol. 2021;9:803818. doi: 10.3389/fcell.2021.803818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Karlsson M, Zhang C, Méar L, Zhong W, Digre A, et al. A single-cell type transcriptomics map of human tissues. Sci Adv. 2021;7:eabh2169. doi: 10.1126/sciadv.abh2169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Watson CM, Crinnion LA, Morgan JE, Harrison SM, Diggle CP, et al. Robust diagnostic genetic testing using solution capture enrichment and a novel variant-filtering interface. Hum Mutat. 2014;35:434–41. doi: 10.1002/humu.22490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu C, Miyata H, Gao Y, Sha Y, Tang S, et al. Bi-allelic DNAH8 variants lead to multiple morphological abnormalities of the sperm fagella and primary male infertility. Am J Hum Genet. 2020;107:330–41. doi: 10.1016/j.ajhg.2020.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Jiang C, Zhang X, Liu X, Li J, et al. Loss-of-function mutation in DNAH8 induces asthenoteratospermia associated with multiple morphological abnormalities of the sperm flagella. Clin Genet. 2020;98:396–401. doi: 10.1111/cge.13815. [DOI] [PubMed] [Google Scholar]
- 17.Zhou Z, Mao X, Chen B, Mu J, Wang W, et al. A novel splicing variant in DNAH8 causes asthenozoospermia. J Assist Reprod Genet. 2021;38:1545–50. doi: 10.1007/s10815-021-02116-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. WHO Laboratory Manual for the Examination and Processing of Human Semen. 5th ed. Geneva: World Health Organization; 2010. p. 271. [Google Scholar]
- 19.Jiao Y, Fan S, Jabeen N, Zhang H, Khan R, et al. A TOP6BL mutation abolishes meiotic DNA double-strand break formation and causes human infertility. Sci Bull. 2020;65:2120–9. doi: 10.1016/j.scib.2020.08.026. [DOI] [PubMed] [Google Scholar]
- 20.Yin H, Ma H, Hussain S, Zhang H, Xie X, et al. A homozygous FANCM frameshift pathogenic variant causes male infertility. Genet Med. 2019;21:62–70. doi: 10.1038/s41436-018-0015-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwarz JM, Cooper DN, Schuelke M, Seelow D. MutationTaster2:mutation prediction for the deep-sequencing age. Nat Methods. 2014;11:361–2. doi: 10.1038/nmeth.2890. [DOI] [PubMed] [Google Scholar]
- 22.Auger J, Jouannet P, Eustache F. Another look at human sperm morphology. Hum Reprod. 2016;31:10–23. doi: 10.1093/humrep/dev251. [DOI] [PubMed] [Google Scholar]
- 23.Sofikitis NV, Miyagawa I, Zavos PM, Toda T, Iino A, et al. Confocal scanning laser microscopy of morphometric human sperm parameters:correlation with acrosin profiles and fertilizing capacity. Fertil Steril. 1994;62:376–86. doi: 10.1016/s0015-0282(16)56894-0. [DOI] [PubMed] [Google Scholar]
- 24.Weng M, Sha Y, Zeng YU, Huang N, Liu W, et al. Mutations in DNAH8 contribute to multiple morphological abnormalities of sperm flagella and male infertility. Acta Biochim Biophys Sin. 2021;53:472–80. doi: 10.1093/abbs/gmab013. [DOI] [PubMed] [Google Scholar]
- 25.Whitfield M, Thomas L, Bequignon E, Schmitt A, Stouvenel L, et al. Mutations in DNAH17, encoding a sperm-specific axonemal outer dynein arm heavy chain, cause isolated male infertility due to asthenozoospermia. Am J Hum Genet. 2019;105:198–12. doi: 10.1016/j.ajhg.2019.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schmidt H, Carter AP. Review:structure and mechanism of the dynein motor ATPase. Biopolymers. 2016;105:557–67. doi: 10.1002/bip.22856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pazour GJ, Agrin N, Walker BL, Witman GB. Identification of predicted human outer dynein arm genes:candidates for primary ciliary dyskinesia genes. J Med Genet. 2006;43:62–73. doi: 10.1136/jmg.2005.033001. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
WES data analysis. (a) WES data analysis flow chart. A MAF less than 0.01 was considered for filtration of existing mutations in the 1000 Genomes, ESP6500, ExAC, and GenomAD databases. (b) Genome-wide logarithm of the odds ratio (LOD) scores using WES-derived genotypes for the family. WES: whole-exome sequencing; MAF: minor allele frequency.
Chest X-ray of the patients. Chest X-ray showing no PCD symptoms in the patients with the homozygous frameshift DNAH8 mutation. PCD: primary ciliary dyskinesia; DNAH8: dynein axonemal heavy chain 8.


