Abstract
The Saccharomyces cerevisiae FAB1 gene encodes the sole phosphatidylinositol 3-phosphate [PtdIns(3)P] 5-kinase responsible for synthesis of the polyphosphoinositide PtdIns(3,5)P2. VAC7 encodes a 128-kDa transmembrane protein that localizes to vacuolar membranes. Both vac7 and fab1 null mutants have dramatically enlarged vacuoles and cannot grow at elevated temperatures. Additionally, vac7Δ mutants have nearly undetectable levels of PtdIns(3,5)P2, suggesting that Vac7 functions to regulate Fab1 kinase activity. To test this hypothesis, we isolated a fab1 mutant allele that bypasses the requirement for Vac7 in PtdIns(3,5)P2 production. Expression of this fab1 allele in vac7Δ mutant cells suppresses the temperature sensitivity, vacuolar morphology, and PtdIns(3,5)P2 defects normally exhibited by vac7Δ mutants. We also identified a mutant allele of FIG4, whose gene product contains a Sac1 polyphosphoinositide phosphatase domain, which suppresses vac7Δ mutant phenotypes. Deletion of FIG4 in vac7Δ mutant cells suppresses the temperature sensitivity and vacuolar morphology defects, and dramatically restores PtdIns(3,5)P2 levels. These results suggest that generation of PtdIns(3,5)P2 by the Fab1 lipid kinase is regulated by Vac7, whereas turnover of PtdIns(3,5)P2 is mediated in part by the Sac1 polyphosphoinositide phosphatase family member Fig4.
INTRODUCTION
The various intracellular compartments located along the secretory and endocytic pathways in eukaryotic cells are maintained by highly efficient and accurate sorting systems. Cargo proteins are first selected and packaged into vesicular carriers that bud and ultimately fuse with the proper target membrane (reviewed in Jahn and Sudhof, 1999). These sorting mechanisms maintain the unique composition of proteins and lipids present in each organelle. Besides the protein machinery that regulates selective sorting and trafficking of cargo, several phospholipids, particularly the phosphorylated phosphatidylinositol (PtdIns) derivatives, also have critical roles in this process (reviewed in De Camilli et al., 1996; Odorizzi et al., 2000). The ability of this class of lipids to serve as a regulator in these complex pathways derives from the differential and combinatorial phosphorylation of the inositol headgroup.
Members of both the D-3 and D-4 phosphorylated PtdIns derivatives have been implicated in the regulation of membrane trafficking events (De Camilli et al., 1996). In yeast, PtdIns(4)P synthesis is required for protein secretion (Hama et al., 1999; Walch-Solimena and Novick, 1999; Audhya et al., 2000). The observation that D-3 PtdIns derivatives play distinct roles in intracellular trafficking events was determined by the requirement for the PtdIns 3-kinase Vps34 in yeast vacuolar protein sorting (Schu et al., 1993). vps34 mutants do not produce PtdIns(3)P and have defects in protein transport from the trans-Golgi to the vacuole. Additional phosphorylation of PtdIns(3)P by the yeast PtdIns(3)P 5-kinase Fab1 results in the generation of PtdIns(3,5)P2 (Cooke et al., 1998; Gary et al., 1998). This lipid, although not required for general anterograde transport to the vacuole, is involved in regulating vacuolar homeostasis in yeast (Gary et al., 1998), perhaps through its roles in two novel sorting pathways. Evidence suggests that PtdIns(3,5)P2 is required for inclusion of the vacuolar hydrolase carboxypeptidase S into vesicles that invaginate into the lumen of an endosomal compartment, forming multivesicular bodies (MVBs) (Odorizzi et al., 1998). Additionally, data also suggest that PtdIns(3,5)P2 is required for the recycling of membrane proteins through retrograde transport from the vacuole to earlier compartments (Bryant et al., 1998). The inability to generate PtdIns(3,5)P2 may prevent MVB formation, leading to increased endosomal membrane delivered to the limiting membrane of the vacuole. Likewise, a block in membrane recycling from the vacuole could also result in formation of the grossly enlarged vacuoles in fab1 mutants (Gary et al., 1998).
Downstream of the lipid kinases, effector proteins bind specific PtdIns derivates to mediate downstream biological signaling events. The PH, PX, FYVE, and ENTH domains are four well-characterized lipid-binding domains commonly present in these effectors (reviewed in Hurley and Meyer, 2001; Sato et al., 2001; Wishart et al., 2001). PH domains have been predicted in >200 human proteins and have been found to interact with a number of phosphoinositides (Toker and Cantley, 1997; Schultz et al., 2000). Proteins containing FYVE (Wurmser et al., 1999) and PX (Cheever et al., 2001; Xu et al., 2001; Yu and Lemmon, 2001) domains have been directly implicated in protein trafficking by specifically binding to PtdIns(3)P. Last, ENTH domains specifically bind PtdIns(4,5)P2 to facilitate clathrin-mediated endocytosis (Ford et al., 2001; Itoh et al., 2001). Through protein–lipid interactions, these domains function in targeting proteins to membrane compartments.
The determination that PtdIns derivatives act as signaling molecules to maintain proper membrane trafficking events in yeast suggests that their synthesis and turnover are highly regulated. Indeed, genetic and biochemical studies in yeast have identified upstream activators for some of these PtdIns kinases. The calmodulin-related calcium-binding protein Frq1 interacts with the PtdIns 4-kinase Pik1 to stimulate synthesis of PtdIns(4)P (Hendricks et al., 1999). The recruitment and activation of the Vps34 PtdIns 3-kinase to membranes is dependent upon direct interactions with the membrane-associated protein kinase Vps15 (Stack et al., 1993, 1995). Last, overexpression of Fab1 does not result in increased generation of PtdIns(3,5)P2 (Gary et al., 1998), suggesting that Fab1 PtdIns(3)P 5-kinase activity is regulated by a limiting activator. Both fab1 and vac7 mutants share similar phenotypes, including temperature-sensitive growth, an enlarged vacuole morphology, and severely reduced levels of PtdIns(3,5)P2 (Bonangelino et al., 1997; Gary et al., 1998). Furthermore, both Fab1 and the integral membrane protein Vac7 localize to vacuolar membranes. These similarities suggest that Vac7 may function in the upstream regulation of Fab1 kinase activity.
There are several turnover pathways for the degradation of PtdIns derivatives in yeast. Phospholipase C hydrolyzes PtdIns(4,5)P2 into the secondary signaling molecules diacylglycerol and the soluble Ins(1,4,5)P3 (Rebecchi and Pentyala, 2000). PtdIns(4,5)P2 can also be dephosphorylated by one of four inositol polyphosphate 5-phosphatatases: Inp51, Inp52, Inp53, and Inp54 (Stolz et al., 1998a,b; Guo et al., 1999; Wiradjaja et al., 2001). Inp51–53, also known as the synaptojanin-like proteins Sjl1, Sjl2, and Sjl3, respectively, and Inp54 all contain a PtdIns(4,5)P2 5-phosphatase domain at their carboxy termini (Srinivasan et al., 1997; Guo et al., 1999). Sjl2/Inp52 and Sjl3/Inp53, as well as two additional proteins Sac1 and Fig4, also contain Sac1 domains at their amino termini. The Sac1 domains of Sac1 and Inp53/Sjl3 have been shown to catalyze the dephosphorylation of PtdIns(3)P, PtdIns(4)P, and PtdIns(3,5)P2 in vitro (Guo et al., 1999; Hughes et al., 2000). Furthermore, sac1 mutations result in large accumulations of PtdIns(4)P, as well as increased levels of PtdIns(3)P and PtdIns(3,5)P2 in vivo (Guo et al., 1999). Last, PtdIns(3)P turnover in vivo occurs through multiple mechanisms. PtdIns(3)P on endosomal membranes can be internalized into MVBs, which are degraded by hydrolases upon delivery to the vacuole (Wurmser and Emr, 1998; Gillooly et al., 2000). In addition, PtdIns(3)P that remains on the limiting membrane can be degraded by the myotubularin homolog (Taylor et al., 2000) Ymr1, or phosphorylated by Fab1 (Cooke et al., 1998; Gary et al., 1998).
In this report, we identify genes in yeast that are required for the maintenance of PtdIns(3,5)P2 levels. We isolated a mutant fab1 allele (fab1-5) that bypasses the requirement for Vac7 function. The identification of a fab1 mutant that can bypass the requirement for Vac7 is consistent with Vac7 functioning as a positive regulator of Fab1 kinase activity. Additionally, a genetic screen to isolate mutants that bypass the vac7Δ temperature sensitivity (bvs) identified a mutant allele of FIG4, which contains a Sac1-like polyphosphoinositide phosphatase domain. Expression of the fig4-1 mutant allele or deletion of FIG4 in vac7Δ mutant cells rescued the temperature-sensitive growth defects, aberrant vacuolar morphology, and restored PtdIns(3,5)P2 levels. Together, these results suggest that Vac7 functions as an upstream regulator of the Fab1 lipid kinase activity, whereas the Sac1 lipid phosphatase family member Fig4 mediates turnover of PtdIns(3,5)P2.
MATERIALS AND METHODS
Strains and Media
The Escherichia coli strain used for cloning and plasmid propagation was XL1-Blue (supE44 thi-1 lac endA1 gyrA96 hsdR17 relA1 F′ proAB LacIq ZΔM15). This bacterial strain was grown in standard LB media. The Saccharomyces cerevisiae strains used in this study (Table 1) were grown in standard YPD or SD minimal media with the addition of necessary auxotrophic supplements.
Table 1.
S. cerevisiae strains used in this study
| Strain | Genotype | Reference or source |
|---|---|---|
| SEY6210 | MATα leu2-3, 112 ura3-52 his3-Δ 200 trp1-Δ 901 lys2-801 suc2-Δ 9 | Robinson et al., 1988 |
| SEY6211 | MATa leu2-3, 112 ura3-52 his3-Δ 200 trp1-Δ 901 ade2-101 suc2-Δ 9 | Robinson et al., 1988 |
| JGY132 | SEY6210; sjl2Δ ∷HIS3 sjl3Δ ∷TRP1 | Foti et al., 2001 |
| JGY133 | SEY6210; fab1Δ ∷LEU2 | Gary et al., 1998 |
| JGY134 | SEY6210; vac7Δ ∷HIS3 | This study |
| JGY135 | SEY6210; fab1Δ ∷LEU2 vac7Δ ∷HIS3 | This study |
| JGY136 | SEY6210; vac7Δ ∷HIS3 bvs16 | This study |
| JGY137 | SEY6210; fig4Δ ∷LEU2 vac7Δ ∷HIS3 | This study |
| JGY138 | SEY6210; fig4Δ ∷LEU2 | This study |
| JGY139 | SEY6210; vac7Δ ∷HIS3 sac1Δ ∷TRP1 | This study |
| JGY140 | SEY6210; vac7Δ ∷HIS3 sjl2Δ ∷HIS3 | This study |
| JGY141 | SEY6210; vac7Δ ∷HIS3 sjl3Δ ∷TRP1 | This study |
| JGY142 | SEY6210; FIG4-HA ∷HIS3MX6 | This study |
| JGY143 | SEY6211; vac7Δ ∷HIS3 | This study |
| JGY144 | SEY6211; vac7Δ ∷HIS3 bvs16 | This study |
| YCS191 | SEY6210; fig4Δ ∷LEU2 sjl2Δ ∷HIS3 sjl3Δ ∷TRP1 | This study |
Genetic and DNA Manipulations
Restriction enzymes (Roche Applied Sciences, Indianapolis, IN), T4 DNA ligase, synthetic oligonucleotides, and dNTPs (Invitrogen, Carlsbad, CA) were used according to company specifications. Standard molecular biology techniques were used for all other DNA-based protocols (Maniatis et al., 1982). Yeast transformations and isolation of yeast genomic DNA are described in Gary et al. (1998). The chromosomal deletion of VAC7 in SEY6210 and JGY133 strains was accomplished using the constructs and protocols previously described (Bonangelino et al., 1997). FIG4 was polymerase chain reaction (PCR) amplified from genomic DNA by using the primers 500 base pairs 5′ and 3′ from the open reading frame (ORF). The resulting PCR product was digested with BamHI and KpnI ∼370 base pairs 5′ and 460 base pairs 3′ of the FIG4, respectively, and cloned into similarly digested pRS416 vector (Sikorski and Hieter, 1989). An identical method was used to clone the bvs16 allele (fig4-1) from strain JGY136.
The FIG4 ORF was PCR cloned from genomic DNA by using the primers incorporating a unique SalI and EagI restriction sites 300 base pairs 5′ from the start codon and EagI 200 base pairs 3′ from the stop codon, respectively. This PCR product was digested with SalI/EagI and cloned into SalI/NotI digested pBluescriptII (KS−) (Stratagene, La Jolla, CA), generating pBS-FIG4. The construct used for deletion of the FIG4 gene pBS-FIG4::LEU2 was made by digesting pBS-FIG4 with XbaI/Nru I (removing 1.8 kb of FIG4) and ligating to a similarly digested 2.2-kb fragment that included the LEU2 gene. The chromosomal FIG4 gene was replaced with the fig4Δ::LEU2 construct by linearizing pBS-FIG4::LEU2 with BglI and transforming it into SEY6210 or JGY134. Chromosomal insertion of the fig4Δ::LEU2 disruption was confirmed by PCR analysis. The genomic integration construct was PCR amplified from the pFA6a-3HA-His3MX6 template described by Longtine et al. (1998). The resulting 1.7-kb PCR product was then transformed into SEY6210. Chromosomal integration was confirmed by PCR analysis and expression of Fig4-HA was verified by Western blotting.
To mutagenize FAB1, a 4.8-kb fragment of FAB1 was PCR amplified from the plasmid pAY60 (Yamamoto et al., 1995) by using 0.3× dATP and 150 μM MnCl2. The primers corresponded to sites 550 base pairs upstream of the 5′ Sfo I site and 550 base pairs downstream of the Nru I site, respectively. Mutagenized PCR products were then cotransformed into JGY135 with a pAY60 fragment that had been gapped by a Sfo I/Nru I digestion. The transformed cells were then plated on selective media and incubated at 38°C. Plasmids from mutants viable at 38°C were isolated and retested for plasmid linkage.
FM4-64 Labeling of Yeast Vacuoles
Yeast were harvested at an OD600 reading of 0.6–0.8. Approximately 1 OD600 of cells was labeled with FM4-64 (Molecular Probes, Eugene, OR) as previously described (Vida and Emr, 1995). Labeled cells were then observed by Nomarski optics and fluorescence (rhodamine channel) as described previously (Wendland et al., 1996).
Steady-State In Vivo Analysis of Phosphoinositides
The labeling of cells with myo-[2-3H]inositol (Amersham Biosciences, Piscataway, NJ) was done in SD-inositol as described (Gary et al., 1998; Bonangelino and Weisman, unpublished data). For this study however, the 10-min 0.9 M NaCl shock was omitted from the labeling protocol and lipid extraction was conducted by a perchloric acid, precipitation-based procedure (Whiteford et al., 1996, 1997). The subsequent high-performance liquid chromatography (HPLC) analysis of deacylated phosphoinositides was done as previously described (Gary et al., 1998). A total of 3.5 million cpm of deacylated lipid extract was injected for each analysis. To more accurately compare gPtdIns(3,5)P2 levels between experiments, the levels of each of the lipids from a single HPLC run were normalized based on the integration of the gPtdIns(4,5)P2 peak being 30,000 cpm. Despite the variation in the raw cpm data from many wild-type labelings, the ratio of gPtdIns(4,5)P2 to gPtdIns(3,5)P2 always remained constant.
Mutagenic Screen for vac7Δ Bypass Mutants
Ethyl methanesulfonate mutagenesis of the JGY134 strain was performed as described (Wendland et al., 1996). The treatment was titrated to allow ∼40% viability when grown at the permissive temperature of 26°C. After mutagenesis, cells were diluted and plated at 38°C for selection of suppressors. The original isolate, JGY136, was backcrossed to strain JGY143 three times. The final isolate, JGY144, was then transformed with a LEU2-CEN S. cerevisiae genomic library (Rose and Broach, 1991). Transformants were plated onto SD-LEU-ADE plates at 26°C supplemented with 5.4 μg/ml adenine. After 6 d, white colonies were selected and rescreened on identical plates. The library plasmid (pBVS16-107) was isolated and retransformed into JGY136 to verify for plasmid-dependent temperature sensitivity.
Analysis of Fig4-HA Membrane Association
Subcellular fractionation and immunoblot blot analyses were performed as previously described (Gary et al., 1998). Monoclonal antibody against the hemagglutinin (HA) epitope (Roche Applied Sciences) was used at a 1:1500 dilution. The sucrose density gradients were done as previously described (Babst et al., 1998).
RESULTS
Isolation of a fab1 Mutant That Suppresses vac7Δ
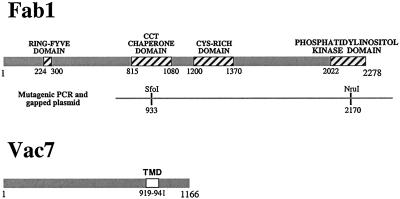
VAC7 and FAB1 were each isolated in a screen for mutants defective in vacuolar inheritance (Weisman et al., 1990). Both vac7 and fab1 mutants share a common set of phenotypes, including temperature-sensitive growth, dramatically enlarged vacuoles (Wang et al., 1996), and a severe reduction in steady-state PtdIns(3,5)P2 levels (Gary et al., 1998). These data raise the possibility that Vac7 and Fab1 function in a common pathway, with Vac7 playing an important regulatory role in activation of the Fab1 kinase. A prediction from this hypothesis is that mutant fab1 alleles may be capable of suppressing the defects associated with the loss of Vac7 function. We tested this hypothesis by identifying fab1 mutant alleles that bypassed the requirement for VAC7 function. Accordingly, a 4.8-kb fragment of FAB1 was randomly mutagenized by error-prone PCR (Figure 1). This region of Fab1 includes the lipid kinase domain as well as the CCT chaperone and cysteine-rich domains conserved in Fab1 homologs (Schultz et al., 2000). Mutagenized fab1 was then transformed into the fab1Δ vac7Δ double mutant strain and transformants were screened for growth at 38°C, the restrictive temperature for the vac7Δ strain.
Figure 1.
Schematic diagram of the S. cerevisiae Fab1 and Vac7 proteins. Four domains identified within Fab1 are indicated by the hatched boxes and their amino acid positions are noted. The PCR-mutagenized region of FAB1 is denoted by the gray line and restriction sites used in gapped plasmid repair are also indicated. Vac7 contains a transmembrane domain (TMD) at its carboxy terminus (amino acids 919–941).
Of the ∼10,000 colonies screened, seven were found to grow at 38°C. Next, the vacuole morphology of these fab1 mutants was assessed using the vital, lipophilic dye FM4-64 (Vida and Emr, 1995). One allele, fab1-5, completely suppressed the temperature sensitivity and the enlarged vacuole morphology of the vac7Δ fab1Δ double mutant (Table 2 and Figure 2A). The vacuoles in vac7Δ fab1-5 double mutant cells appear similar in size and number to the vacuoles found in wild-type cells, despite the absence of the VAC7 gene product. With pulse-chase labeling followed by immunoprecipitation experiments, we did not observe any difference in protein amount or stability between wild-type Fab1 and the Fab1-5 mutant protein (our unpublished data), suggesting that suppression of the mutant phenotypes by the fab1-5 allele was not due to an increase in Fab1 stability or expression.
Table 2.
Summary of growth phenotypes
| Strain | Growth at 38°C |
|---|---|
| Wild-type | 214 |
| vac7Δ | − |
| vac7Δ fab1-5 | 214 |
| vac7Δ bvs16 | 214 |
| vac7Δ bvs16 213 FIG4 | − |
| vac7Δ fig4Δ | 213 |
| vac7Δ fig4-1 | 214 |
| vac7Δ sac1Δ | 214 |
| vac7Δ sjl2Δ | − |
| vac7Δ sjl3Δ | − |
, strong growth.
, moderate growth.
−, no growth.
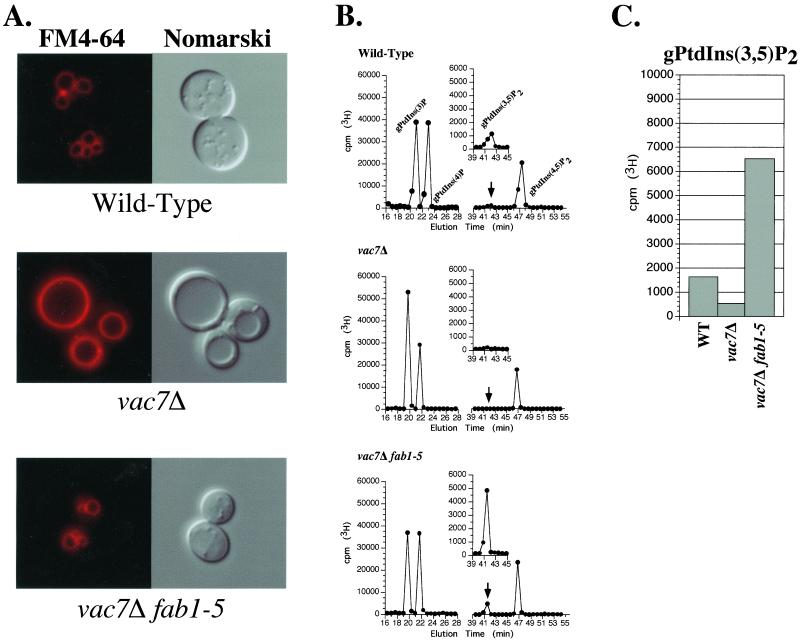
Figure 2.
Expression of the fab1-5 mutant allele suppresses the vacuolar morphology defects and restores PtdIns(3,5)P2 levels in fab1Δ vac7Δ double mutant cells. (A) Visualization of vacuolar morphology in wild-type, vac7Δ, and vac7Δ fab1-5 cells. Vacuoles of the indicated strains were visualized by staining with FM 4-64 (left) or by Nomarski optics (right). Cells grown to mid-log phase were labeled with FM 4-64 for 10 min and chased with media for 1 h at 26°C as described in MATERIALS AND METHODS. (B) Representative HPLC profile of the four known PtdIns derivatives from wild-type, vac7Δ, and vac7Δ fab1-5 cells. Cells were labeled with myo-[2-3H]inositol for 14 h at 26°C. After precipitation of crude cell lysates with perchloric acid, the total cellular lipids were deacylated as described in MATERIALS AND METHODS. Deacylated phosphoinositides were separated by HPLC analysis and measured by radiolabel detection. The positions of the deacylated PtdIns derivatives are indicated. The peak corresponding to gPtdIns(3,5)P2 is highlighted by an arrow and shown in the inset. (C) Bar graph representing the quantified levels of gPtdIns(3,5)P2 from the indicated strains. The height of each bar represents the normalized average of three or more independent labeling and HPLC experiments with SD <10%.
fab1-5 Mutation Results in Dramatic Increase in PtdIns(3,5)P2 Levels
We determined whether suppression of the vac7Δ mutant phenotypes by the fab1-5 mutation was due to a restoration of PtdIns(3,5)P2 levels by measuring in vivo steady-state phosphoinositide levels. PtdIns derivatives were isolated from myo-[2-3H]inositol-labeled cells by lipid precipitation (Whiteford et al., 1996, 1997; Bonangelino and Weisman, unpublished data) and chemical deacylation. Deacylated phosphoinositides were separated by HPLC and quantified by 3H detection. Wild-type yeast cells produced detectable quantities of four glycero-phosphoinositide derivatives corresponding to PtdIns(3)P, PtdIns(4)P, PtdIns(3,5)P2, and PtdIns(4,5)P2 (Figure 2B). Compared with wild-type cells, vac7Δ cells contained nearly undetectable levels of PtdIns(3,5)P2 (100–200 cpm above background) and an increased amount of PtdIns(3)P, which is consistent with this lipid being a precursor of PtdIns(3,5)P2 (Figure 2, B and C). Furthermore, vac7Δ mutant cells hyperosmotically shocked in 0.9 M NaCl did not display detectable levels of PtdIns(3,5)P2 by this analysis. This is similar to results from a previous study in which fab1Δ mutants also lacked detectable levels of PtdIns(3,5)P2 under hyperosmotic conditions (Cooke et al., 1998).
In striking contrast, we found that expression of the fab1-5 allele in vac7Δ fab1Δ double mutant cells resulted in a 40-fold increase in the steady-state levels of PtdIns(3,5)P2 compared with vac7Δ cells expressing wild-type Fab1 (Figure 2, B and C). In addition, vac7Δ fab1-5 double mutant cells produced approximately fourfold more PtdIns(3,5)P2 than even wild-type cells. Expression of the fab1-5 mutation, however, did not significantly alter the levels of the other PtdIns derivatives (Figure 2, B and C). Furthermore, expression of the fab1-5 allele in wild-type cells also results in a similar increase in PtdIns(3,5)P2 levels (our unpublished data), indicating that the fab1-5 mutation is dominant. This dramatic increase in PtdIns(3,5)P2 indicates that the fab1-5 mutation restores PtdIns(3,5)P2 levels in the absence of VAC7, suggesting that this mutation allows the bypass of Vac7 function required for the generation of PtdIns(3,5)P2. We have attempted to map the mutation within the fab1-5 allele that confers the suppression of the vac7Δ phenotype and have found that this effect is dependent upon multiple mutations in distinct regions of the FAB1 gene (our unpublished data).
Isolation of Other Mutants That Bypass the Requirement for Vac7
Previously, we proposed that PtdIns(3)P 5-kinase activity was regulated, because overexpression of Fab1 alone did not lead to an increase in PtdIns(3,5)P2 levels (Gary et al., 1998). Furthermore, the identification of the gain-of-function fab1-5 allele that suppresses the mutant phenotypes associated with the vac7Δ strain suggests that Fab1 activity is indeed regulated, possibly through an interaction with Vac7. We attempted to identify additional genes involved in regulating cellular PtdIns(3,5)P2 levels by screening ethyl methanesulfonate-mutagenized vac7Δ cells for the restoration of growth at 38°C. We reasoned that mutations in at least three classes of genes might be identified: 1) positive and negative regulators of the Fab1 kinase activity; 2) downstream factors responsible for PtdIns(3,5)P2 degradation, such as PtdIns phosphatases; and 3) genes encoding PtdIns(3,5)P2 binding effectors, in which mutations would suppress the temperature-sensitivity phenotype but not affect PtdIns(3,5)P2 synthesis.
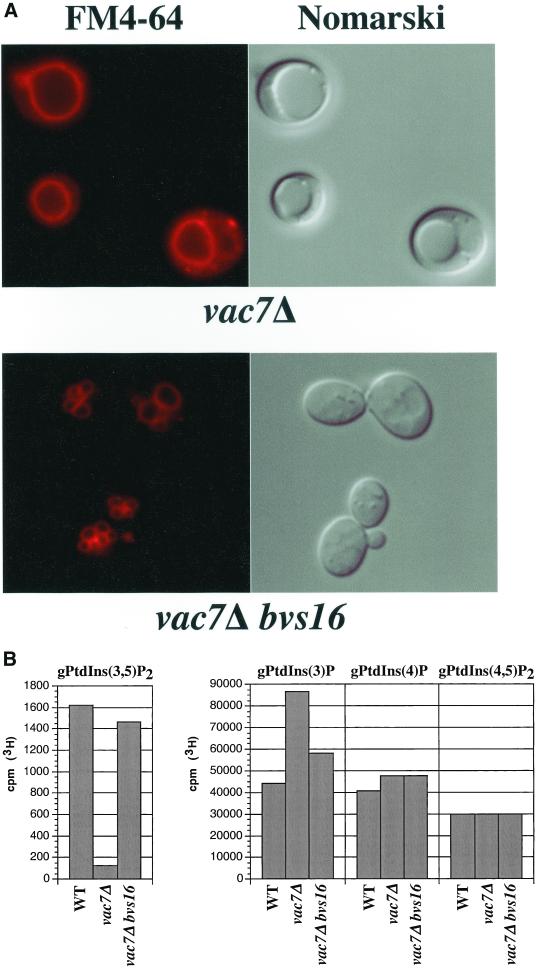
Of the 50,000 colonies screened, we isolated 17 mutants that were able to bypass the vac7Δ temperature sensitivity (bvs) phenotype. These bvs mutants were then assessed for suppression of the enlarged vacuole morphology phenotype by FM4-64. All of the bvs isolates had morphologies that differed to some degree from the parental vac7Δ strain. We focused our attention on the vac7Δ bvs16 double mutant, which grew at 38°C (Table 2) and displayed a near wild-type vacuolar morphology (Figure 3A). Lipid labeling of the vac7Δ bvs16 double mutant with myo-[2-3H]inositol and subsequent HPLC analysis revealed that this double mutant had restored the steady-state level of PtdIns(3,5)P2 (Figure 3B). The vac7Δ bvs16 double mutant strain maintained near wild-type levels of PtdIns(3,5)P2, >18-fold more PtdIns(3,5)P2 than the vac7Δ strain. The relative levels of other PtdIns derivatives were slightly altered in this mutant. PtdIns(4)P and PtdIns(3)P levels were 1.2- and 1.3-fold, respectively, above wild-type levels. Thus, the bvs16 mutation restored the steady-state PtdIns(3,5)P2 level and suppressed the mutant phenotypes in the vac7Δ mutant strain.
Figure 3.
bvs16 mutation suppresses the mutant phenotypes associated with vac7Δ mutants. (A) Vacuolar morphology of vac7Δ bvs16 double mutants. As in Figure 2, vacuoles from vac7Δ or vac7Δ bvs16 mutant cells were visualized by fluorescent staining with FM 4-64 (left) or Nomarski optics (right). (B) Quantification of PtdIns derivatives in vac7Δ bvs16 mutants. The levels of radiolabeled PtdIns derivatives from the indicated strains were isolated, separated, and measured as described in MATERIALS AND METHODS. The values of each bar represents the normalized average of three or more experiments with SD <15%.
The bvs16 Mutation Is Allelic to FIG4
The diploid strain resulting from crossing the vac7Δ bvs16 isolate to the parental vac7Δ strain displayed the identical mutant phenotypes of the vac7Δ/vac7Δ diploid strain (our unpublished data). In addition, suppression of the mutant phenotypes segregated with a 2:2 ratio in the haploid progeny from this cross, indicating that the bvs16 allele results from a single recessive mutation (our unpublished data). To clone the gene allelic to bvs16, we generated a vac7Δ bvs16 ade2 triple mutant that, due to the ade2 mutation, accumulated a red pigment in its vacuoles that cause colonies to appear red (Wada et al., 1992). The color change is dependent upon vacuolar acidification. Thus, vac7Δ ade2 double mutant cells, which do not contain acidified vacuoles, appear white. Therefore, we reasoned that we would be able to identify the gene allelic to bvs16 by transforming the vac7Δ bvs16 ade2 triple mutant with a S. cerevisiae genomic library and screening for white colonies.
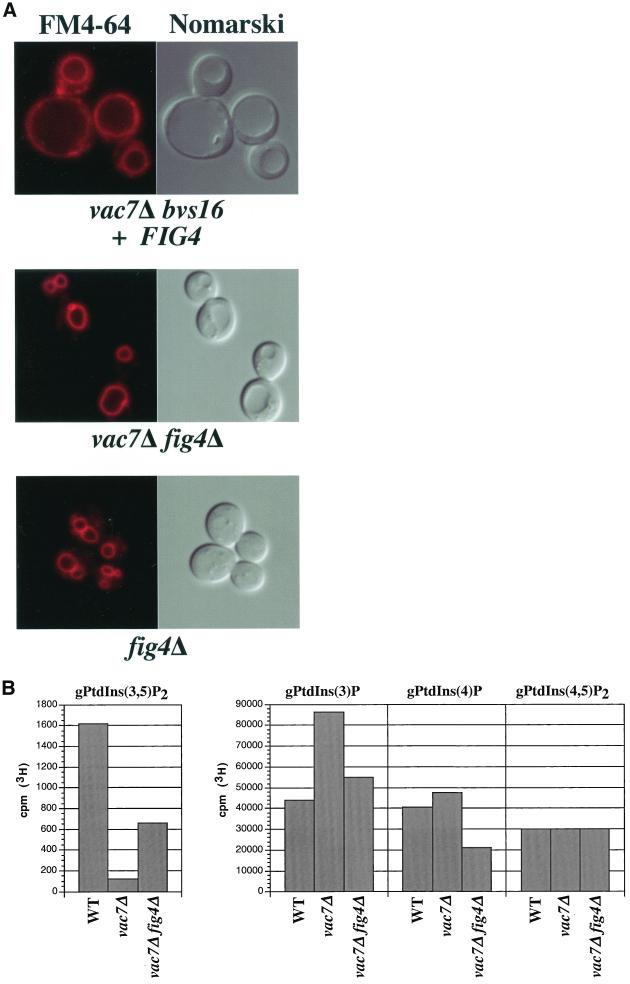
From this screen, we isolated one transformant that displayed the enlarged vacuole phenotype of vac7Δ mutants and was no longer able to grow at 38°C. The library plasmid pBVS16-107 from this transformant was then rescued and sequenced, revealing a 10.8-kb genomic DNA insert containing six ORFs, which included the FIG4 gene. Additional complementation analysis determined that transformation of the FIG4 gene alone into the vac7Δ bvs16 double mutant strain was sufficient for reversion to vac7Δ mutant phenotypes (Figure 4A and Table 2). Furthermore, we found that deletion of the FIG4 gene in vac7Δ cells suppressed the vac7Δ temperature sensitivity and vacuole morphology defects (Table 2 and Figure 4A). Quantification of the PtdIns derivatives in the vac7Δ Fig4Δ double mutant strain showed that steady-state PtdIns(3,5)P2 levels increased sevenfold compared with the vac7Δ alone (Figure 4B). Slight changes in the other phosphorylated forms were also observed relative to vac7Δ (Figure 4B); PtdIns(3)P and PtdIns(4)P levels decreased 40 and 55%, respectively.
Figure 4.
Deletion of FIG4 suppresses the mutant phenotypes of vac7Δ mutants. (A) Vacuole morphology of the indicated strains was visualized by labeling with FM 4-64 (left) or Nomarski optics (right). (B) Quantification of radiolabeled PtdIns derivatives from wild-type, vac7Δ, and vac7Δ fig4Δ mutant strains. The normalized average of deacylated PtdIns derivatives from the indicated strains were quantified as described in MATERIALS AND METHODS. The values of each bar represents the average of three or more independent labeling experiments with SD <10%.
Fig4 Contains a Polyphosphoinositide Phosphatase Domain
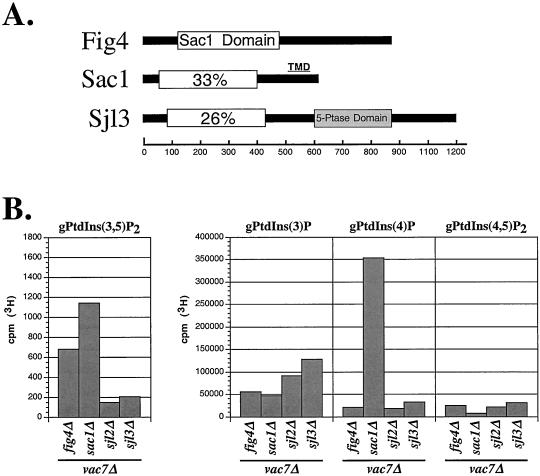
Fig4 is one of four proteins in yeast that contains a polyphosphoinositide phosphatase domain called the Sac1 domain (Figure 5A; Guo et al., 1999). Other representatives include Sac1 and the inositide polyphosphate 5-phosphatases Sjl2/Inp52 and Sjl3/Inp53 (Srinivasan et al., 1997; Stolz et al., 1998a,b; Guo et al., 1999). In vitro, the Sac1 domains from Sac1 and Sjl3 are able to dephosphorylate PtdIns(3,5)P2, PtdIns(3)P, and PtdIns(4)P but do not appear to recognize PtdIns(4,5)P2 (Guo et al., 1999; Hughes et al., 2000). Furthermore, sac1 mutants accumulate ∼2.5-fold higher levels of PtdIns(3,5)P2 than wild-type cells (Guo et al., 1999). Because evidence suggests that PtdIns(3,5)P2 may be a substrate for the Sac1 domains of Sac1 and Sj13, we addressed the specificity of fig4 inactivation to mediate bypass of VAC7 function by assessing whether the loss of other Sac1 domain-containing proteins similarly affected vac7Δ mutants. The sac1Δ vac7Δ double mutant strain was able to grow at 38°C, whereas sjl2Δ vac7Δ and sil3Δ vac7Δ double mutant strains were not able to grow (Table 2). As expected, neither deletion of SJL2 nor SJL3 was able to rescue PtdIns(3,5)P2 levels in vac7Δ mutant cells. In contrast, the PtdIns(3,5)P2 levels in the vac7Δ sac1Δ double mutant strain were higher than in vac7Δ fig4Δ double mutant cells (Figure 5B). However, in addition to the elevation in the level of PtdIns(3,5)P2, the level of PtdIns(4)P also was dramatically elevated, ∼16-fold more than in wild-type or vac7Δ cells (Figures 4B and 5B). Furthermore, transformation of the SAC1 gene in the remaining bvs mutants did not reverse the vac7Δ suppression, suggesting that mutations in SAC1 were not isolated. Sac1 is an integral membrane protein localized to the endoplasmic reticulum in yeast that primarily dephosphorylates PtdIns(4)P in vivo (Guo et al., 1999; Foti et al., 2001). These results suggest that inhibition of PtdIns(3,5)P2 turnover by inactivation of Fig4 or Sac1 can restore PtdIns(3,5)P2 levels to allow the bypass of Vac7 function.
Figure 5.
Fig4 contains a Sac1 polyphosphoinositide phosphatase domain. (A) Schematic diagram indicating the positions of Sac1 domains (open rectangle) within Fig4, Sac1, and Sjl3/Inp53. The Sac1 domain of Fig4 is 33 and 25% identical to the Sac1 domains of Sac1 and Sjl3, respectively. In addition, Sac1 contains a predicted transmembrane domain (TMD) at its carboxy terminus, whereas Sjl3 also contains an inositol polyphosphate 5-phosphatase domain. (B) Quantification of radiolabeled PtdIns derivatives from vac7Δ fig 4Δ, vac7Δ sac1Δ, vac7Δ sjl2Δ, and vac7Δ sjl3Δ double mutants. Radiolabeled PtdIns derivatives were isolated and quantified from the indicated mutant strains as described in MATERIALS AND METHODS. The values of each bar represents the average of three or more independent labeling experiments with SD <15%.
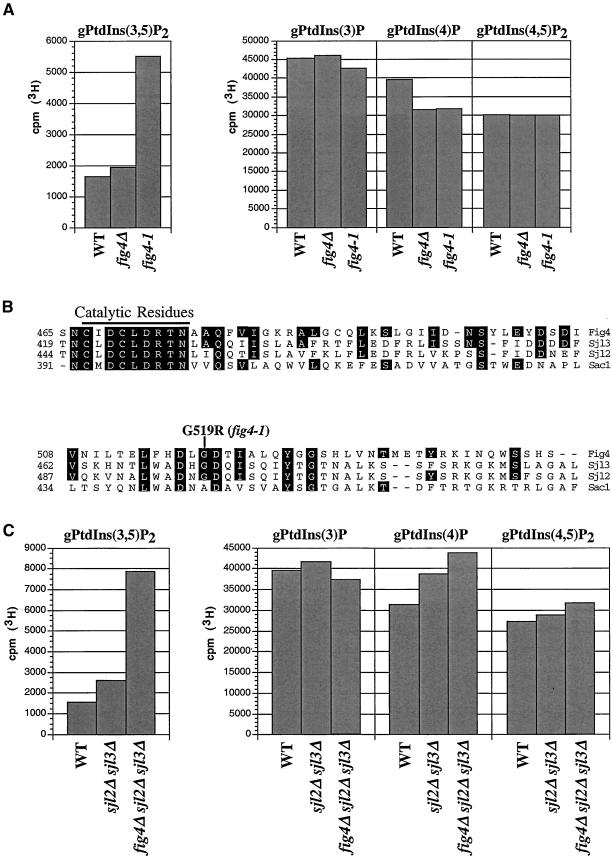
Identification of fig4-1 Mutation
Our genetic evidence and the presence of a Sac1 domain suggests that Fig4 is a lipid phosphatase that regulates the turnover of PtdIns(3,5)P2. Surprisingly, however, analysis of steady-state lipid levels revealed that fig4Δ cells did not contain significantly higher levels of PtdIns(3,5)P2 compared with wild-type cells (Figure 6A). In sharp contrast, fig4-1 mutant cells had almost threefold more PtdIns(3,5)P2 than wild-type and fig4Δ cells. Similarly, the level of PtdIns(3,5)P2 in vac7Δ fig4-1 double mutants was almost 2.5-fold greater than in vac7Δ fig4Δ double mutant cells (compare Figure 3B with 4B). These observations suggest that the fig4-1 mutation has a greater effect on the restoration of PtdIns(3,5)P2 levels than does deletion of FIG4 itself. To understand the nature of the fig4-1 mutation, we sequenced the fig4-1 allele and identified a single mutation that resulted in the exchange of glycine at position 519 for arginine (G519R; Figure 6B). This amino acid change occurred ∼50 amino acids outside of the Sac1 catalytic domain (RXNCXDCLDRTN) and is conserved in both Sjl2/Inp52 and Sjl3/Inp53 (Figure 6B). Sac1 does not have a conserved glycine at this position, however, a mutation that changes the corresponding alanine inactivates Sac1 phosphatase activity (Whitters et al., 1993).
Figure 6.
fig4-1 mutants accumulate high levels of PtdIns(3,5)P2. (A) Steady-state quantification of PtdIns derivatives from wild-type, fig4Δ, and fig4-1 cells. Radiolabeled PtdIns derivatives were isolated and quantified from the indicated strains as described in MATERIALS AND METHODS. The height of each bar represents the normalized average of three or more experiments with a SD <15%. (B) Quantification of radiolabeled PtdIns derivatives from wild-type, sjl2Δ sjl3Δ, and fig4Δ sjl2Δ sjl3Δ mutant cells. Radiolabeled PtdIns derivatives were isolated and quantified from the indicated strains as described in MATERIALS AND METHODS. The height of each bar represents the normalized average of three or more experiments with SD <10%. (C) Protein sequence alignment of a region of the Sac1 domain in Fig4, Sjl3/Inp53, Sjl2/Inp52, and Sac1. The lipid phosphatase catalytic motif is denoted by the black line. Boxed amino acids identify residues that are conserved between Fig4 and other Sac1 domain family members. The glycine at position 519 changed to an arginine in the fig4-1 mutant is indicated.
A possible explanation for the lack of elevated PtdIns(3,5)P2 levels in fig4Δ cells may be due to other Sac1 domain-containing lipid phosphatases, which compensate for the loss in Fig4 activity. Consistent with earlier work (Guo et al., 1999), we found that sjl2Δ sjl3Δ double mutant cells maintain increased cellular levels of PtdIns(3,5)P2 compared with wild-type cells (Figure 6C). This suggests that both Sjl2 and Sjl3 may be responsible for dephosphorylating PtdIns(3,5)P2 in fig4Δ mutant cells. Indeed, in fig4Δ sjl2Δ sjl3Δ triple mutant cells, the PtdIns(3,5)P2 level was more than fivefold greater than in wild-type cells with modest increases in both PtdIns(4)P and PtdIns(4,5)P2 (Figure 6C). These results indicate that Sjl2, Sjl3, and Fig4 may have overlapping functions in regulating PtdIns(3,5)P2 homeostasis in vivo.
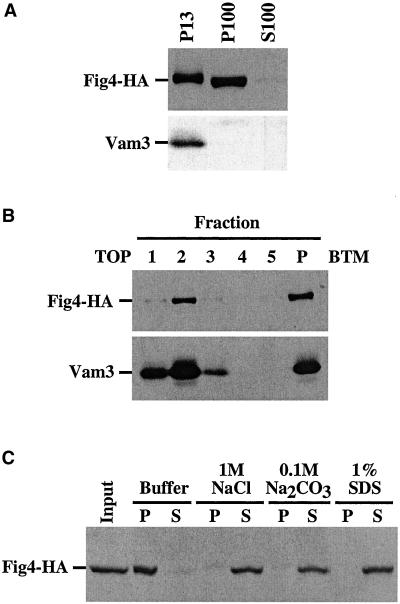
Fig4 Is a Peripheral Membrane Protein
Because our data suggest that Fig4 is a lipid phosphatase, we expected that Fig4 would associate with membrane compartments. To determine whether Fig4 associates with membranes, we constructed a strain that expressed a Fig4-HA tag fusion and used differential centrifugation to generate two membrane-enriched fractions. The Fig4-HA fusion protein was equally enriched in both the 13,000 × g (P13) and 100,000 × g (P100) pelletable fractions (Figure 7A), indicating that Fig4 may associate with distinct compartments. Furthermore, the vacuolar transmembrane protein Vam3 was found exclusively in the P13 fraction (Figure 7A), suggesting that a fraction of Fig4 associates with vacuolar membranes. Alternatively, the P13 fraction of Fig4-HA may result from an insoluble aggregation or association with cytoskeletal elements. We addressed these possibilities by loading the P13 fraction at the bottom (fraction 5) of a sucrose density step gradient. During a 20-h centrifugation at 200,000 × g, the low-density membranes, such as vacuolar membranes, migrate up toward the top of the gradient into fractions 1–3, whereas insoluble proteins are found at the bottom (fraction 5 and pellet). After centrifugation, Fig4-HA appeared primarily in the second fraction of the gradient, as did Vam3 (Figure 7B). These results are consistent with an association of Fig4 with membrane compartments.
Figure 7.
Characterization of Fig4-HA membrane association. (A) Subcellular fractionation of Fig4-HA. Lysate from wild-type yeast cells expressing Fig4-HA was fractionated by centrifugation at 13,000 × g to generate the P13 pellet and S13 soluble fractions. The S13 soluble fraction was further centrifuged at 100,000 × g to generate the P100 pellet and the S100 soluble fractions. Proteins in these fractions were precipitated, resolved by SDS-PAGE, and detected by Western blotting with anti-HA or anti-Vam3 antibodies, respectively. Each lane represents lysate from 1 OD600 equivalent of cells. (B) Sucrose gradient analysis of Fig4-HA is described in MATERIALS AND METHODS. Briefly, the P13 pellet from the FIG4-HA strain was generated as explained in A and resuspended in 60% sucrose. The resuspended pellet was then loaded beneath a two-step gradient containing 55 and 35% sucrose. After 20-h centrifugation at 200,000 × g, fractions from the gradient were collected, and proteins were precipitated and resolved by SDS-PAGE. Fig4-HA and Vam3 were visualized by Western blotting with anti-HA and anti-Vam3 antibodies, respectively. Fractions are numbered from the top of the gradient (1) down to the pellet material (P). Fraction 5 corresponds to the initial position of the loaded P13 material. (C) Membrane association of Fig4-HA. Lysate from cells expressing Fig4-HA was centrifuged at 3000 × g. The soluble fraction (S3) was then incubated on ice in the indicated buffer conditions for 30 min. After the incubation, samples were then centrifuged at 100,000 × g to generate the pelletable (P) and soluble (S) fractions. Both fractions were precipitated and Fig4-HA was visualized by Western blotting with anti-HA antibody.
Although Fig4 appears to associate with membranes, sequence analysis failed to detect any membrane-spanning domains. We attempted to define the nature of the Fig4 membrane association by determining the conditions in which Fig4 could be extracted from membrane fractions. Lysates from yeast cells expressing Fig4-HA were incubated with buffer alone, 1 M NaCl, 0.1 M Na2CO3 pH 11, or 1% SDS. After incubation and centrifugation at 100,000 × g, Fig4-HA was found to redistribute into the soluble S100 fraction after treatment with NaCl, Na2CO3, or SDS (Figure 7C). As expected, the transmembrane protein Vam3 shifted into the S100 fraction only upon treatment with the detergent SDS (our unpublished data). Consistent with the absence of any identifiable transmembrane domains, these results indicate that Fig4 is a peripheral membrane protein.
DISCUSSION
Regulation of Fab1 Lipid Kinase by Vac7
Both vac7 and fab1 mutants contain dramatically enlarged vacuoles and are temperature sensitive for growth at 38°C (Bonangelino et al., 1997; Gary et al., 1998). Mutations that inactivate the PtdIns(3)P 5-kinase Fab1 result in an inability to generate the polyphosphoinositide PtdIns(3,5)P2 (Gary et al., 1998). Like fab1 mutants, vac7 mutants also lack PtdIns(3,5)P2. Based on these observations, it was proposed that Vac7 functions upstream of Fab1, possibly through direct activation of the lipid kinase. Consistent with Vac7 functioning as an activator of Fab1 activity, we found that Vac7 is necessary for the generation of high levels of PtdIns(3,5)P2 upon hyperosmotic shock. Fab1 has been previously shown to generate the large amounts PtdIns(3,5)P2 within yeast cells in hyperosmotic conditions (Cooke et al., 1998; Gary et al., 1998). Similarly, vac7Δ mutants did not generate detectable amounts of PtdIns(3,5)P2 in 0.9 M NaCl. This type of relationship between positive regulators and lipid kinases has previously been observed in yeast. The PtdIns 4-kinase Pik1 is activated by the calcium-binding protein Frq1 (Hendricks et al., 1999), whereas activation of the PtdIns 3-kinase Vps34 requires the protein kinase Vps15 (Stack et al., 1993, 1995). We have attempted to detect physical interactions between Vac7 and Fab1 by native coimmunoprecipitation experiments; however, because the Vac7 protein is unstable in cell extracts (our unpublished data), it was not possible to perform these experiments.
In a genetic approach to identify regulatory interactions between Vac7 and Fab1, we isolated a mutant fab1 allele that can bypass the requirement for VAC7. Expression of the mutant fab1-5 gene product in vac7Δ fab1Δ double mutant cells results in suppression of the vac7Δ growth and vacuolar morphology phenotypes (Table 2 and Figure 2A) and resulted in ∼40-fold higher PtdIns(3,5)P2 levels than in vac7Δ cells (Figure 2, B and C). This suggests that the fab1-5 mutation results in Vac7-independent activation of the Fab1 kinase. Furthermore, these results are consistent with Vac7 functioning as an upstream regulator of Fab1 kinase activity. However, additional experiments are necessary to determine a role for Vac7 in directly activating Fab1.
Putative Polyphosphoinositide Phosphatase Fig4 Regulates PtdIns(3,5)P2 Levels
In a genetic screen to identify additional regulators of PtdIns(3,5)P2 levels, we identified a mutant allele of FIG4 whose gene product contains a conserved Sac1 lipid phosphatase domain, as a suppressor of the vac7Δ temperature sensitivity. Expression of the fig4-1 mutant allele in vac7Δ cells restored growth at 38°C and suppressed the enlarged vacuole morphology (Table 2 and Figure 3A). Bypass of vac7Δ phenotype was due to a restoration of PtdIns(3,5)P2 levels. In vac7Δ fig4-1 double mutants, PtdIns(3,5)P2 levels were almost equal to that in wild-type cells (18-fold greater than in vac7Δ cells; Figure 3C). Similarly, deletion of the FIG4 gene also bypassed the vac7Δ mutant phenotypes (Figure 4, A and B, and Table 2). Thus, even in the absence of Fab1 activation by Vac7, mutations in Fig4 may prevent the turnover of residual PtdIns(3,5)P2 and allow for the accumulation of PtdIns(3,5)P2 to levels that restore vacuolar homeostasis and function.
Sac1 domains were initially identified in the yeast proteins Sac1 and Fig4, and the synaptojanin-like inositol polyphosphate 5-phosphatases Sjl2/Inp52 and Sjl3/Inp53 (Guo et al., 1999). The Sac1 domains from Sac1 and Sjl3 have been determined to dephosphorylate PtdIns(3)P, PtdIns(4)P, and PtdIns(3,5)P2 in vitro (Guo et al., 1999; Hughes et al., 2000). We are currently attempting to purify and test the Sac1 domain of Fig4 for phosphatase activity in vitro. In addition, we found that deletion of SAC1 suppressed the phenotypes of vac7Δ mutants, whereas deletion of SJL2 or SJL3 did not (Table 2 and Figure 5B). In vac7Δ sac1Δ double mutant cells, PtdIns(3,5)P2 levels were ∼1.7-fold greater than vac7Δ fig4Δ cells (Figure 5B). However, as previously reported for sac1Δ mutants (Guo et al., 1999), the levels of PtdIns(4)P in vac7Δ sac1Δ double mutants were dramatically elevated, >17-fold greater than vac7Δ fig4Δ mutant cells. Recently, characterization of a temperature-conditional sac1tsf mutant determined that PtdIns(4)P is the primary substrate of Sac1 in vivo (Foti et al., 2001). Inactivation of the sac1tsf mutant resulted in a sevenfold increase in PtdIns(4)P levels, with only modest changes in other phosphoinositides.
Our biochemical data indicate that Fig4 is a peripheral membrane protein (Figure 7, A–C). A portion of Fig4 cofractionates with membranes enriched in vacuoles, which is consistent with the previously observed vacuolar localization of both Vac7 and Fab1 (Bonangelino et al., 1997; Gary et al., 1998). These results suggest the possibility that PtdIns(3,5)P2 levels on the vacuolar membrane may be regulated through Fab1-mediated synthesis and Fig4-dependent degradation. In contrast, Sac1 has been shown to primarily localize to the endoplasmic reticulum (Whitters et al., 1993; Foti et al., 2001), raising the possibility that the restoration of PtdIns(3,5)P2 levels in vac7Δ sac1Δ mutants may be indirect. The large excess pool of PtdIns(4)P present in sac1Δ mutant cells may compete with PtdIns(3,5)P2 for Fig4 activity. However, we cannot rule out the possibility that Sac1 may also play a role in the dephosphorylation of a pool of PtdIns(3,5)P2 in endoplasmic reticulum membranes or an additional compensatory role in PtdIns(3,5)P2 dephosphorylation on other membranes.
fig4-1 Mutation Dramatically Stabilizes PtdIns(3,5)P2 Levels
Surprisingly, deletion of the FIG4 gene did not result in a dramatic increase in PtdIns(3,5)P2 levels; the PtdIns(3,5)P2 levels in fig4Δ mutant cells were ∼20% greater than in wild-type cells (Figure 6A). This suggests that Fig4 is not the only PtdIns(3,5)P2 lipid phosphatase in yeast. Consistent with this hypothesis, we found that PtdIns(3,5)P2 levels were significantly greater in fig4Δ sjl2Δ sjl3Δ triple mutant cells (Figure 6C) over fig4Δ (Figure 6A) and sjl2Δ sjl3Δ mutant cells (Figure 6C). Thus, Sjl2 and Sjl3 may also play some roles in PtdIns(3,5)P2 turnover and, in the absence of Fig4 activity, they may compensate by dephosphorylating the majority of the excess PtdIns(3,5)P2.
Whereas deletion of FIG4 did not have a pronounced effect, we found that the PtdIns(3,5)P2 levels in fig4-1 mutant cells were strikingly higher, almost threefold greater than in wild-type and fig4Δ cells (Figure 6A). Mapping of the fig4-1 mutation identified a substitution of amino acid residue 519 from glycine to arginine just outside of the Sac1 catalytic domain (Figure 6B). The glycine at this corresponding position is conserved in Sjl2, Sjl3, and Fig4 homologs, whereas Sac1 contains an alanine. However, mutations that change this alanine in Sac1 also cause the loss of lipid phosphatase activity (Whitters et al., 1993). Thus, this region of the Sac1 domain may have a role in regulating the lipid phosphatase activity. The analogous Fig4 mutant may bind and stabilize a pool of PtdIns(3,5)P2 that is inaccessible to Sjl2 and Sjl3. Alternatively, the subcellular localization of Fig4 may play a critical role in the determining Fig4 substrate specificity. Thus, the fig4-1 mutation may alter the localization of Fig4 to membranes that do not contain PtdIns(3,5)P2. Further biochemical analysis of the fig4-1 mutant protein will be necessary to discern its effect on lipid phosphatase activity.
Vac7 and Fig4 Function in Regulating PtdIns(3,5)P2 Levels
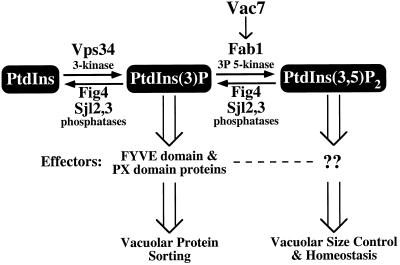
Together, our results suggest that both Vac7 and Fig4 are key regulators of PtdIns(3,5)P2 levels in yeast (Figure 8). The synthesis of PtdIns(3,5)P2 is dependent upon two known lipid kinases. Vps34 is a PtdIns 3-kinase that phosphorylates PtdIns to PtdIns(3)P (Schu et al., 1993), a lipid that has been shown to be required for Golgi-to-vacuole protein sorting through its interactions with proteins containing FYVE (Wurmser et al., 1999) and PX domains (Cheever et al., 2001). Finally, phosphorylation of PtdIns(3)P by the PtdIns(3)P 5-kinase Fab1 results in the generation of PtdIns(3,5)P2 (Gary et al., 1998). The identification of a mutant fab1 allele that bypasses the requirement for Vac7 function, along with the similarities in phenotypes between vac7 and fab1 mutants and their localization on the vacuole (Gary et al., 1998), suggest that Vac7 positively regulates Fab1 kinase activity. The generation of PtdIns(3,5)P2 is necessary for the recruitment of unknown effector molecules for vacuolar membrane homeostasis functions, such as protein sorting at the multivesicular body (Odorizzi et al., 1998) and retrograde transport from the vacuole (Bryant et al., 1998). Furthermore, our data suggest that the Sac1 domain containing protein Fig4 functions, along with Sjl2 and Sjl3, to mediate the turnover of PtdIns(3,5)P2. Sac1 may also play a role in PtdIns(3,5)P2 dephosphorylation. The turnover of PtdIns(3,5)P2 is likely to be essential in attenuating downstream signaling events mediated by PtdIns(3,5)P2 and its downstream effectors.
Figure 8.
Model for regulation of PtdIns(3,5)P2 synthesis and degradation. PtdIns is phosphorylated by the PtdIns 3-kinase Vps34 to generate PtdIns(3)P, which is essential for vacuolar protein sorting through interactions with FYVE and PX domain-containing proteins. PtdIns(3)P can be further phosphorylated by the PtdIns(3)P 5-kinase Fab1 to produce PtdIns(3,5)P2. Fab1 kinase activity is regulated by the upstream activator Vac7. The generation of PtdIns(3,5)P2 is essential for the maintenance of vacuolar size control and homeostasis through interactions with unknown effectors. Fig4, through its Sac1 polyphosphoinositide phosphatase domain, functions with Sj12 and Sjl3, in regulating PtdIns(3,5)P2 levels by mediating the turnover of this lipid.
Although it is apparent that an equilibrium between PtdIns(3,5)P2 production and degradation is essential for maintaining normal growth and membrane trafficking in yeast, the mechanism by which PtdIns(3,5)P2 triggers these downstream signaling events is unknown. Although FYVE and PX domains have been shown to specifically bind to PtdIns(3)P, no PtdIns(3,5)P2 effectors have been identified. In addition to the fig4-1 mutation that restores PtdIns(3,5)P2 levels in vac7Δ mutants, we isolated additional mutants (bvs) that restored growth at 38°C and vacuolar morphology, but had no effect on steady-state PtdIns(3,5)P2 levels, suggesting that these mutations occur in genes that function downstream of PtdIns(3,5)P2. Further characterization of these mutants may identify novel PtdIns(3,5)P2 effectors and/or provide insight as to how PtdIns(3,5)P2 functions in maintaining vacuolar membrane homeostasis.
ACKNOWLEDGMENTS
We thank A. Wurmser and D. Anderson for critical reading of this manuscript. This work was supported by a grant from the National Institutes of Health (CA-58689 to S.D.E). S.D.E. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0498. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0498.
REFERENCES
- Audhya A, Foti M, Emr SD. Distinct roles for the yeast phosphatidylinositol 4-kinases, Stt4p and Pik1p, in secretion, cell growth, and organelle membrane dynamics. Mol Biol Cell. 2000;11:2673–2689. doi: 10.1091/mbc.11.8.2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babst M, Wendland B, Estepa EJ, Emr SD. The Vps4p AAA ATPase regulates membrane association of a Vps protein complex required for normal endosome function. EMBO J. 1998;17:2982–2993. doi: 10.1093/emboj/17.11.2982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonangelino CJ, Catlett NL, Weisman LS. Vac7p, a novel vacuolar protein, is required for normal vacuole inheritance and morphology. Mol Cell Biol. 1997;17:6847–6858. doi: 10.1128/mcb.17.12.6847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Piper RC, Weisman LS, Stevens TH. Retrograde traffic out of the yeast vacuole to the TGN occurs via the prevacuolar/endosomal compartment. J Cell Biol. 1998;142:651–663. doi: 10.1083/jcb.142.3.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheever ML, Sato TK, de Beer T, Kutateladze TG, Emr SD, Overduin M. Phox domain interaction with PtdIns(3)P targets the Vam7 t-SNARE to vacuole membranes. Nat Cell Biol. 2001;3:613–618. doi: 10.1038/35083000. [DOI] [PubMed] [Google Scholar]
- Cooke FT, Dove SK, McEwen RK, Painter G, Holmes AB, Hall MN, Michell RH, Parker PJ. The stress-activated phosphatidylinositol 3-phosphate 5-kinase Fab1p is essential for vacuole function in S. cerevisiae. Curr Biol. 1998;8:1219–1222. doi: 10.1016/s0960-9822(07)00513-1. [DOI] [PubMed] [Google Scholar]
- De Camilli P, Emr SD, McPherson PS, Novick P. Phosphoinositides as regulators in membrane traffic. Science. 1996;271:1533–1539. doi: 10.1126/science.271.5255.1533. [DOI] [PubMed] [Google Scholar]
- Ford MG, Pearse BM, Higgins MK, Vallis Y, Owen DJ, Gibson A, Hopkins CR, Evans PR, McMahon HT. Simultaneous binding of PtdIns(4,5)P2 and clathrin by AP180 in the nucleation of clathrin lattices on membranes. Science. 2001;291:1051–1055. doi: 10.1126/science.291.5506.1051. [DOI] [PubMed] [Google Scholar]
- Foti M, Audhya A, Emr SD. Sac1 lipid phosphatase and Stt4 PtdIns 4-kinase regulate a pool of PtdIns(4)P that functions in the control of the actin cytoskeleton and vacuole morphology. Mol Biol Cell. 2001;12:2396–2411. doi: 10.1091/mbc.12.8.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gary JD, Wurmser AE, Bonangelino CJ, Weisman LS, Emr SD. Fab1p Is Essential for PtdIns(3)P 5-kinase activity and the maintenance of vacuolar size and membrane homeostasis. J Cell Biol. 1998;143:65–79. doi: 10.1083/jcb.143.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillooly DJ, Morrow IC, Lindsay M, Gould R, Bryant NJ, Gaullier JM, Parton RG, Stenmark H. Localization of phosphatidylinositol 3-phosphate in yeast and mammalian cells. EMBO J. 2000;19:4577–4588. doi: 10.1093/emboj/19.17.4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Stolz LE, Lemrow SM, York JD. SAC1-like domains of yeast SAC1, INP52, and INP53 and of human synaptojanin encode polyphosphoinositide phosphatases. J Biol Chem. 1999;274:12990–12995. doi: 10.1074/jbc.274.19.12990. [DOI] [PubMed] [Google Scholar]
- Hama H, Schnieders EA, Thorner J, Takemoto JY, DeWald DB. Direct involvement of phosphatidylinositol 4-phosphate in secretion in the yeast Saccharomyces cerevisiae. J Biol Chem. 1999;274:34294–34300. doi: 10.1074/jbc.274.48.34294. [DOI] [PubMed] [Google Scholar]
- Hendricks KB, Wang BQ, Schnieders EA, Thorner J. Yeast homologue of neuronal frequenin is a regulator of phosphatidylinositol-4-OH kinase. Nat Cell Biol. 1999;1:234–241. doi: 10.1038/12058. [DOI] [PubMed] [Google Scholar]
- Hughes WE, Woscholski R, Cooke FT, Patrick RS, Dove SK, McDonald NQ, Parker PJ. SAC1 encodes a regulated lipid phosphoinositide phosphatase, defects in which can be suppressed by the homologous Inp52p and Inp53p phosphatases. J Biol Chem. 2000;275:801–808. doi: 10.1074/jbc.275.2.801. [DOI] [PubMed] [Google Scholar]
- Hurley JH, Meyer T. Subcellular targeting by membrane lipids. Curr Opin Cell Biol. 2001;13:146–152. doi: 10.1016/s0955-0674(00)00191-5. [DOI] [PubMed] [Google Scholar]
- Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Koshiba S, Kigawa T, Kikuchi A, Yokoyama S, Takenawa T. Role of the ENTH domain in phosphatidylinositol-4,5-bisphosphate binding and endocytosis. Science. 2001;291:1047–1051. doi: 10.1126/science.291.5506.1047. [DOI] [PubMed] [Google Scholar]
- Jahn R, Sudhof TC. Membrane fusion and exocytosis. Annu Rev Biochem. 1999;68:863–911. doi: 10.1146/annurev.biochem.68.1.863. [DOI] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, 3rd, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Maniatis T, Fritsch EF, Sambrook J. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Fab1p PtdIns(3)P 5-kinase function essential for protein sorting in the multivesicular body. Cell. 1998;95:847–858. doi: 10.1016/s0092-8674(00)81707-9. [DOI] [PubMed] [Google Scholar]
- Odorizzi G, Babst M, Emr SD. Phosphoinositide signaling and the regulation of membrane trafficking in yeast. Trends Biochem Sci. 2000;25:229–235. doi: 10.1016/s0968-0004(00)01543-7. [DOI] [PubMed] [Google Scholar]
- Rebecchi MJ, Pentyala SN. Structure, function, and control of phosphoinositide-specific phospholipase C. Physiol Rev. 2000;80:1291–1335. doi: 10.1152/physrev.2000.80.4.1291. [DOI] [PubMed] [Google Scholar]
- Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Broach JR. Cloning genes by complementation in yeast. Methods Enzymol. 1991;194:195–230. doi: 10.1016/0076-6879(91)94017-7. [DOI] [PubMed] [Google Scholar]
- Sato TK, Overduin M, Emr SD. Location, location, location: membrane targeting directed by PX domains. Science. 2001;294:1881–1885. doi: 10.1126/science.1065763. [DOI] [PubMed] [Google Scholar]
- Schu PV, Takegawa K, Fry MJ, Stack JH, Waterfield MD, Emr SD. Phosphatidylinositol 3-kinase encoded by yeast VPS34 gene essential for protein sorting. Science. 1993;260:88–91. doi: 10.1126/science.8385367. [DOI] [PubMed] [Google Scholar]
- Schultz J, Copley RR, Doerks T, Ponting CP, Bork P. SMART: a web-based tool for the study of genetically mobile domains. Nucleic Acids Res. 2000;28:231–234. doi: 10.1093/nar/28.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Seaman M, Nemoto Y, Daniell L, Suchy SF, Emr S, De Camilli P, Nussbaum R. Disruption of three phosphatidylinositol-polyphosphate 5-phosphatase genes from Saccharomyces cerevisiae results in pleiotropic abnormalities of vacuole morphology, cell shape, and osmohomeostasis. Eur J Cell Biol. 1997;74:350–360. [PubMed] [Google Scholar]
- Stack JH, DeWald DB, Takegawa K, Emr SD. Vesicle-mediated protein transport: regulatory interactions between the Vps15 protein kinase and the Vps34 PtdIns 3-kinase essential for protein sorting to the vacuole in yeast. J Cell Biol. 1995;129:321–334. doi: 10.1083/jcb.129.2.321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack JH, Herman PK, Schu PV, Emr SD. A membrane-associated complex containing the Vps15 protein kinase and the Vps34 PI 3-kinase is essential for protein sorting to the yeast lysosome-like vacuole. EMBO J. 1993;12:2195–2204. doi: 10.1002/j.1460-2075.1993.tb05867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz LE, Kuo WJ, Longchamps J, Sekhon MK, York JD. INP51, a yeast inositol polyphosphate 5-phosphatase required for phosphatidylinositol 4,5-bisphosphate homeostasis and whose absence confers a cold-resistant phenotype. J Biol Chem. 1998a;273:11852–11861. doi: 10.1074/jbc.273.19.11852. [DOI] [PubMed] [Google Scholar]
- Stolz LE, Huynh CV, Thorner J, York JD. Identification and characterization of an essential family of inositol polyphosphate 5-phosphatases (INP51, INP52 and INP53 gene products) in the yeast Saccharomyces cerevisiae. Genetics. 1998b;148:1715–1729. doi: 10.1093/genetics/148.4.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GS, Maehama T, Dixon JE. Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc Natl Acad Sci USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toker A, Cantley LC. Signaling through the lipid products of phosphoinositide-3-OH kinase. Nature. 1997;387:673–676. doi: 10.1038/42648. [DOI] [PubMed] [Google Scholar]
- Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:779–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada Y, Ohsumi Y, Anraku Y. Genes for directing vacuolar morphogenesis in Saccharomyces cerevisiae. I. Isolation and characterization of two classes of vam mutants. J Biol Chem. 1992;267:18665–18670. [PubMed] [Google Scholar]
- Walch-Solimena C, Novick P. The yeast phosphatidylinositol-4-OH kinase pik1 regulates secretion at the Golgi. Nat Cell Biol. 1999;1:523–525. doi: 10.1038/70319. [DOI] [PubMed] [Google Scholar]
- Wang YX, Zhao H, Harding TM, Gomes de Mesquita DS, Woldringh CL, Klionsky DJ, Munn AL, Weisman LS. Multiple classes of yeast mutants are defective in vacuole partitioning yet target vacuole proteins correctly. Mol Biol Cell. 1996;7:1375–1389. doi: 10.1091/mbc.7.9.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisman LS, Emr SD, Wickner WT. Mutants of Saccharomyces cerevisiae that block intervacuole vesicular traffic and vacuole division and segregation. Proc Natl Acad Sci USA. 1990;87:1076–1080. doi: 10.1073/pnas.87.3.1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendland B, McCaffery JM, Xiao Q, Emr SD. A novel fluorescence-activated cell sorter-based screen for yeast endocytosis mutants identifies a yeast homologue of mammalian eps15. J Cell Biol. 1996;135:1485–1500. doi: 10.1083/jcb.135.6.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford CC, Best C, Kazlauskas A, Ulug ET. D-3 phosphoinositide metabolism in cells treated with platelet-derived growth factor. Biochem J. 1996;319:851–860. doi: 10.1042/bj3190851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whiteford CC, Brearley CA, Ulug ET. Phosphatidylinositol 3,5-bisphosphate defines a novel PI 3-kinase pathway in resting mouse fibroblasts. Biochem J. 1997;323:597–601. doi: 10.1042/bj3230597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitters EA, Cleves AE, McGee TP, Skinner HB, Bankaitis VA. SAC1p is an integral membrane protein that influences the cellular requirement for phospholipid transfer protein function and inositol in yeast. J Cell Biol. 1993;122:79–94. doi: 10.1083/jcb.122.1.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiradjaja F, Ooms LM, Whisstock JC, McColl B, Helfenbaum L, Sambrook JF, Gething MJ, Mitchell CA. The yeast inositol polyphosphate 5-phosphatase Inp54p localizes to the endoplasmic reticulum via a C-terminal hydrophobic anchoring tail: regulation of secretion from the endoplasmic reticulum. J Biol Chem. 2001;276:7643–7653. doi: 10.1074/jbc.M010471200. [DOI] [PubMed] [Google Scholar]
- Wishart MJ, Taylor GS, Dixon JE. Phoxy lipids. revealing px domains as phosphoinositide binding modules. Cell. 2001;105:817–820. doi: 10.1016/s0092-8674(01)00414-7. [DOI] [PubMed] [Google Scholar]
- Wurmser AE, Emr SD. Phosphoinositide signaling and turnover: PtdIns(3)P, a regulator of membrane traffic, is transported to the vacuole and degraded by a process that requires lumenal vacuolar hydrolase activities. EMBO J. 1998;17:4930–4942. doi: 10.1093/emboj/17.17.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurmser AE, Gary JD, Emr SD. Phosphoinositide 3-kinases and their FYVE domain-containing effectors as regulators of vacuolar/lysosomal membrane trafficking pathways. J Biol Chem. 1999;274:9129–9132. doi: 10.1074/jbc.274.14.9129. [DOI] [PubMed] [Google Scholar]
- Xu Y, Hortsman H, Seet L, Wong SH, Hong W. SNX3 regulates endosomal function through its PX-domain-mediated interaction with PtdIns(3)P. Nat Cell Biol. 2001;3:658–666. doi: 10.1038/35083051. [DOI] [PubMed] [Google Scholar]
- Yamamoto A, DeWald DB, Boronenkov IV, Anderson RA, Emr SD, Koshland D. Novel PI(4)P 5-kinase homologue, Fab1p, essential for normal vacuole function and morphology in yeast. Mol Biol Cell. 1995;6:525–539. doi: 10.1091/mbc.6.5.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Lemmon MA. All Phox homology (PX) domains from Saccharomyces cerevisiae specifically recognize phosphatidylinositol 3-phosphate. J Biol Chem. 2001;276:44179–44184. doi: 10.1074/jbc.M108811200. [DOI] [PubMed] [Google Scholar]