Abstract
Mitogen-activated protein kinase (MPK) cascades play vital roles in plant innate immunity, growth, and development. Here, we report that the rice (Oryza sativa) transcription factor gene OsWRKY31 is a key component in a MPK signaling pathway involved in plant disease resistance in rice. We found that the activation of OsMKK10-2 enhances resistance against the rice blast pathogen Magnaporthe oryzae and suppresses growth through an increase in jasmonic acid and salicylic acid accumulation and a decrease of indole-3-acetic acid levels. Knockout of OsWRKY31 compromises the defense responses mediated by OsMKK10-2. OsMKK10-2 and OsWRKY31 physically interact, and OsWRKY31 is phosphorylated by OsMPK3, OsMPK4, and OsMPK6. Phosphomimetic OsWRKY31 has elevated DNA-binding activity and confers enhanced resistance to M. oryzae. In addition, OsWRKY31 stability is regulated by phosphorylation and ubiquitination via RING-finger E3 ubiquitin ligases interacting with WRKY 1 (OsREIW1). Taken together, our findings indicate that modification of OsWRKY31 by phosphorylation and ubiquitination functions in the OsMKK10-2-mediated defense signaling pathway.
Phosphorylation and ubiquitination of OsWRKY31 in OsMKK10-2-mediated defense signaling.
Introduction
Plants have developed sophisticated defense mechanisms during the arms race with pathogens. Preformed defenses include physical barriers and constitutive chemicals with antimicrobial activities. Inducible defenses are triggered during plant–pathogen interactions and include pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI) and effector-triggered immunity (ETI), which are activated to restrict pathogen infection (Jones and Dangl 2006). In general, both PTI and ETI induce similar signal transduction and defense responses, although the ETI response is more pronounced. However, the exact signal transduction that leads to global transcriptional reprogramming remains largely unknown.
PAMPs, such as bacterial flagellin or peptidoglycan and fungal chitin, are perceived by pattern-recognition receptors, activating a signaling cascade. Upon perception of a chitin elicitor, rice (Oryza sativa) chitin elicitor-binding proteins dimerize and then form a receptor complex with CHITIN ELICITOR RECEPTOR KINASE 1 (OsCERK1) to initiate the signaling process (Hayafune et al. 2014). OsCERK1 directly phosphorylates RECEPTOR-LIKE CYTOPLASMIC KINASE 185 (OsRLCK185) in response to chitin and is required to activate a mitogen-activated protein kinase (MPK) cascade (Yamaguchi et al. 2013). MPKs are important players in many biological processes, including responses to abiotic and biotic stresses in plants. In each MPK cascade, a MPK is phosphorylated by a MPK kinase (MKK), which may be activated by an MKK kinase (MKKK). The activated MPK can phosphorylate target proteins to confer their responses to environmental cues and developmental requirements. The rice genome contains 74 MKKKs, eight MKKs, and 17 MPKs (Hamel et al. 2006; Reyna and Yang 2006; Yang et al. 2015); MKKs are particularly important because they are considered the convergence points in the MPK signaling modules (Andreasson and Ellis 2010).
In rice, OsMPK3 interacts with SUBMERGENCE1A1 (SUB1A1), an ethylene response factor-like protein, regulating submergence tolerance in rice (Singh and Sinha 2016). Activation of OsMPK4 can enhance rice resistance against Xanthomonas oryzae pv. oryzae (Xoo), the causal pathogen of bacterial blight disease (Shen et al. 2010). Chitin induces the OsMKK4–OsMPK3/OsMPK6 cascade for cell death and biosynthesis of diterpenoid phytoalexins and lignin (Kishi-Kaboshi et al. 2010). OsMKK4 can be phosphorylated by OsMKKK18 and OsMKKKε, and in turn, OsMKKKε is phosphorylated by OsRLCK185 (Wang et al. 2017; Yamada et al. 2017). Interestingly, the OsMKKK10–OsMKK4–OsMPK6 signaling pathway positively controls grain size and weight (Guo et al. 2018; Xu et al. 2018). Moreover, OsMKK10-2 has been reported to activate OsMPK3 and OsMPK6 for enhanced resistance to a bacterial streak pathogen, X. oryzae pv. oryzicola (Xoc), and tolerance to drought stress (Ma et al. 2017). OsMKK10-2–OsMPK6 pathway is required for OsWRKY45-mediated resistance against Magnaporthe oryzae (Ueno et al. 2013). Recently, OsMPK6 has been shown to phosphorylate a Raf-like kinase ENHANCED DISEASE RESISTANCE 1 (OsEDR1), leading to increased OsMKK10-2 activity and resistance to Xoc (Ma et al. 2021).
Auxins regulate many processes of growth and development in plants, including petiole elongation, root architecture, and gravity perception (Kazan 2013), and implicated in promoting disease susceptibility in Arabidopsis thaliana (Chen et al. 2007; Kidd et al. 2011). In this context, flg22 induces accumulation of Arabidopsis microRNA393 (miR393), which targets the auxin F-box receptor genes TRANSPORT INHIBITOR 1 (TIR1), AUXIN SIGNALING F-BOX 2 (AFB2), and AFB3, resulting in repression of auxin signaling and restriction of Pseudomonas syringae growth (Navarro et al. 2006). Overexpression of NONEXPRESSOR OF PATHOGENESIS-RELATED 1 (OsNPR1) increases resistance to Xoo but attenuates rice growth (Yuan et al. 2007). A later study suggested that the indole-3-acetic acid (IAA) content and the auxin distribution pattern are altered in OsNPR1-overexpressing plants, partially owing to the upregulation of IAA–amido synthase gene GRETCHEN HAGEN 3.8 (OsGH3.8) (Li et al. 2016). Constitutive expression of OsGH3.8 in rice reduces free IAA content, suppresses plant growth, and enhances resistance to bacterial pathogen Xoo (Ding et al. 2008). In Arabidopsis, the activation-tagged bushy and dwarf 1 (bud1) mutant, which results from the increased expression of the MKK7 gene, inhibits auxin transport indicating that MKK7 negatively regulates polar auxin transport (PAT) (Dai et al. 2006). Conversely, bud1 mutant accumulates elevated amounts of salicylic acid (SA) and displays enhanced resistance to pathogens. In this context, MKK7 phosphorylates MPK3 and MPK6 in planta and positively modulates plant basal and systemic acquired resistance (Zhang et al. 2007; Jia et al. 2016).
WRKY transcription factors (TFs) participate in various plant processes, including growth, development, and responses to abiotic and biotic stresses (Chen et al. 2019). In previous studies, we showed that OsWRKY31 (renamed as OsWRKY55, Rice WRKY Working Group 2012) and OsWRKY89 are positive regulators of rice resistance against M. oryzae (Wang et al. 2007; Zhang et al. 2008). Both OsWRKY31 and OsWRKY89 belong to the WRKY IIIb subclade, which formed after the divergence of Arabidopsis and rice (Wu et al. 2005). In a yeast two-hybrid (Y2H) screen using OsWRKY89 as a bait, OsMKK10-2 and OsREIW1 were obtained as interactors, and these proteins also interact with OsWRKY31. Here, we revealed that OsWRKY31 acts as a key regulator downstream of OsMKK10-2 and OsMPK3 is also a downstream component of the kinase. Moreover, OsWRKY31 stability was affected by its phosphorylation and ubiquitination statuses. Our study sheds light on post-transcriptional regulation of plant immunity.
Results
OsWRKY31 physically interacts with OsMKK10-2 and OsMPKs
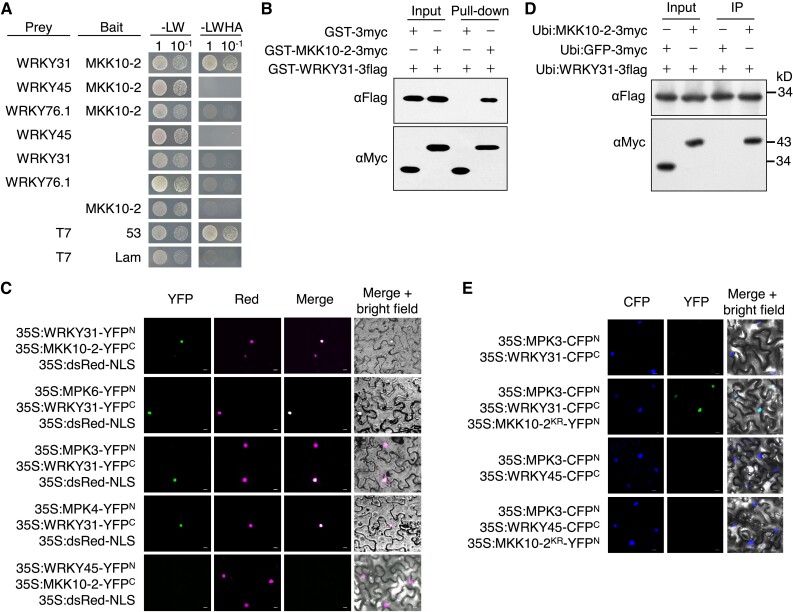
The interaction between OsMKK10-2 and OsWRKY31 was tested in yeast cells with OsMKK10-2 and OsWRKY31 fused with the Gal4 DNA-binding domain (in pBD bait vector) and activation domain (in pAD prey vector), respectively. The interaction between OsMKK10-2 and OsWRKY31 was confirmed by the growth of yeast cells harboring BD-OsMKK10-2 and AD-OsWRKY31 plasmids on the selection media deficient in Leu, Trp, His, and adenine (-LWHA; Fig. 1A). However, no interaction was observed between OsMKK10-2 and OsWRKY76.1 or OsWRKY45, suggesting a specific interaction between OsMKK10-2 and OsWRKY31. For protein pull-down assays, OsMKK10-2 and OsWRKY31 were expressed as GST-MKK10-2-3myc with glutathione transferase (GST) and 3×Myc epitope tags and GST-WRKY31-3flag, respectively, in Escherichia coli; GST-WRKY31-3flag was found to interact specifically with GST-MKK10-2-3myc (Fig. 1B).
Figure 1.
OsMKK10-2 and OsMPKs interact with OsWRKY31. A) OsWRKY31, OsWRKY45, OsWRKY76.1, and OsMKK10-2 were fused to the Gal4 activation domain (prey) or DNA-binding domain (bait). The resulted plasmids in appropriate combinations were introduced into yeast cells and then incubated in synthetic dropout medium without Leu and Trp (-LW) or lacking Leu, Trp, His, and adenine (-LWHA) and photographed 3 d after plating. Yeast cells containing AD-T7 plus BD-53, and AD-T7 plus BD-Lam plasmids were used as the positive and negative controls, respectively. B) OsMKK10-2 and OsWRKY31 were sandwiched with GST and 3×Myc (GST-MKK10-2-3myc) or GST and 3×Flag (GST-WRKY31-3flag) tags. Purified protein (each about 1 μg) was combined accordingly and incubated at 4 °C for 2 h in immunoprecipitation buffer. The protein mixture was precipitated with anti-c-Myc agarose affinity gels, separated on 10% SDS-PAGE gels, and detected by immunoblots with αMyc and αFlag antibodies. C) OsWRKY31, OsWRKY45, OsMKK10-2, OsMPK3, OsMPK4, and OsMPK6 were fused in frame with the YFP N-terminal region (YFPN) or the YFP C-terminal region (YFPC). The Agrobacteria with combined plasmids were infiltrated into the leaves of N. benthamiana. Confocal images were taken at 2 d after the infiltrations. Red fluorescence signals indicative of the nuclei were from co-infiltrated 35S:dsRed-NLS (NLS: nuclear localization signal). D) Plasmids of Ubi:MKK10-2-3myc, Ubi:WRKY31-3flag, and Ubi:GFP-3flag control were introduced separately in A. tumefaciens and co-expressed in appropriate combinations in leaves of N. benthamiana. Total proteins were extracted and immunoprecipitated with anti-c-Flag affinity gels. The precipitates were subjected for immunoblots with αMyc and αFlag antibodies. E) OsWRKY31 or OsWRKY45 and OsMPK3 were fused with CFPC and CFPN, respectively. OsMKK10-2KR, a kinase-dead mutant with Lys81 to Arg81 substitution, was fused with YFPN. Images photographed at 2 d after the infiltrations are CFP (left), YFP (middle), and merged with bright field (right). Bar = 10 μm.
To validate the interaction between OsWRKY31 and OsMKK10-2 in planta, OsMKK10-2 and OsWRKY31 were fused with N- or C-terminal parts of a split yellow fluorescent protein (YFP) for bimolecular fluorescence complementation (BiFC) assays. The 35S:MKK10-2-YFPC and 35S:WRKY31-YFPN plasmids were co-expressed with nuclear localization signal-targeted dsRED (35S:dsRED-NLS) in Nicotiana benthamiana leaves via Agrobacterium tumefaciens-mediated infiltration. The detection of YFP fluorescence indicated the interaction of MKK10-2-YFPC with WRKY31-YFPN; the co-localization of YFP and RED fluorescence signals indicated that the interaction occurred exclusively in the nuclei of N. benthamiana epidermal cells (Fig. 1C). OsWRKY45, belonging to the WRKY IIIa subclade, functions in the OsMKK10-2–OsMPK6 cascade (Ueno et al. 2015). Interaction between WRKY45-YFPN and MKK10-2-YFPC was not detected. The interaction of OsMKK10-2 with OsWRKY31 was also confirmed by the transient expression of Ubi:MKK10-2-3myc and Ubi:WRKY31-3flag proteins in N. benthamiana leaves using co-immunoprecipitation (CoIP) (Fig. 1D).
OsMKK10-2 has been found to interact with OsMPK3, OsMPK4, and OsMPK6 but not with OsMPK17-1 (Zhou et al. 2017). Thus, we further tested the interactions between OsWRKY31 with these OsMPKs and found that OsMPK3, OsMPK4, and OsMPK6 were able to interact with OsWRKY31 in yeast cells (Supplemental Fig. S1). Physical interactions of OsWRKY31 with the three OsMPKs were further revealed by BiFC assays using 35S:WRKY31-YFPN co-transformed with 35S:MPK3-YFPN, 35S:MPK4-YFPN, or 35S:MPK6-YFPN in epidermal cells of N. benthamiana leaves with a dsRed-fused nuclear marker. The co-localization of YFP and red fluorescence signals indicated that the interactions occurred in the nuclear compartment (Fig. 1C).
Furthermore, multicolor BiFC analysis was used to test the complex formation among OsWRKY31, OsMPK3, and OsMKK10-2KR (the kinase-dead mutant generated by changing the amino acid Lys81 to Arg), as OsMKK10-2 caused cell death in N. benthamiana leaves, especially in the presence of OsMPKs (Kamigaki et al. 2016; Zhou et al. 2017). Two types of fluorescence signals were visualized among WRKY31-CFPC, MPK3-CFPN, and MKK10-2KR-YFPN in the N. benthamiana cells but not with WRKY45-CFPC substitution of WRKY31-CFPC (Fig. 1E), revealing the formation of OsWRKY31–OsMPK3–OsMKK10-2KR nuclear complex. These results indicate that OsWRKY31 can physically interact with OsMPK3 and its upstream OsMKK10-2, forming a ternary complex.
OsWRKY31 is phosphorylated by the OsMKK10-2–OsMPK module
To examine whether OsMKK10-2 could activate the phosphorylation of OsMPKs and OsWRKY31, we first analyzed the autophosphorylation activity of OsMKK10-2. The amino acids Arg203 and Ser209 of OsMKK10-2 were mutated into Asp (OsMKK10-2DD) for constitutive activation, although OsMKK10-2 did not contain the typical S/T-X5-S/T motif for a MKKK phosphorylation (Hamel et al. 2006). Autophosphorylation activities were observed for the recombinant GST-MKK10-2DD-3myc protein and, to a lesser extent, for the wild-type GST-MKK10-2-3myc, but not for the kinase-dead mutant GST-MKK10-2KR-3myc (Supplemental Fig. S2A).
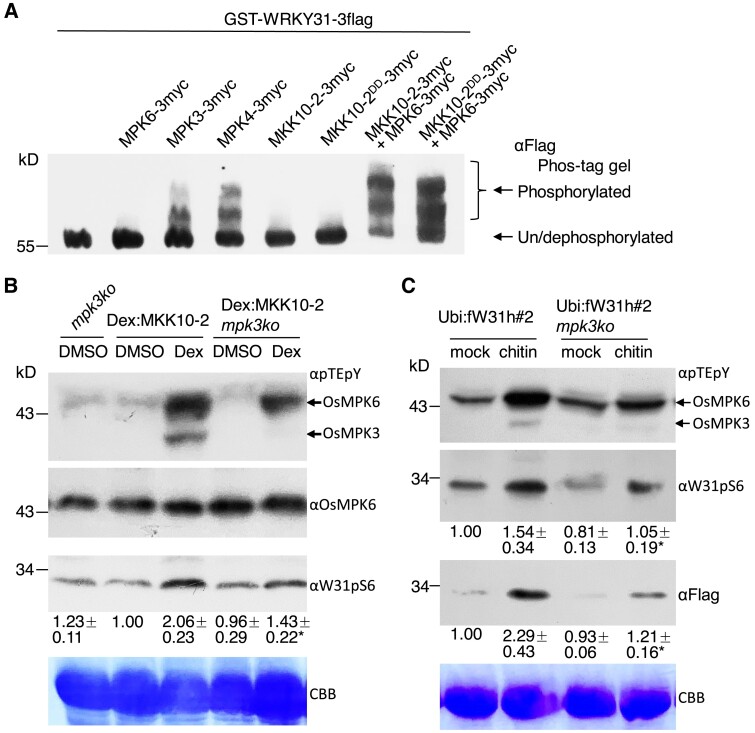
Second, we investigated the phosphorylation of OsWRKY31 by OsMPK proteins in vitro using Phos-tag gel assays. The migration of phosphorylated GST-WRKY31-3flag proteins was clearly retarded for samples treated with GST-MPK3-3myc and GST-MPK4-3myc, while no significant retarded bands were found for those treated with only GST-MPK6-3myc, GST-MKK10-2-3myc, or GST-MKK10-2DD-3myc. However, GST-WRKY31-3flag was markedly phosphorylated when GST-MPK6-3myc was added in combination with GST-MKK10-2-3myc or GST-MKK10-2DD-3myc (Fig. 2A), suggesting that the activation of OsMPK6 via OsMKK10-2 was required for OsWRKY31 phosphorylation in vitro.
Figure 2.
OsMKK10-2 activates phosphorylation of OsMPKs and OsWRKY31. A) The recombinant proteins were expressed with GST plus 3×Myc or 3×Flag tag. GST-WRKY31-3flag with appropriate combination of kinase(s) were subjected to phosphorylation at 30 °C for 2 h. Proteins were separated on Phos-tag gels and blotted with αFlag antibody. The retarded bands indicated the phosphorylated GST-WRKY31-3flag. OsMKK10-2DD: mutations of Arg203 and Ser209 to Asp for constitutive activation. B) Seven-day-old seedlings cultured hydroponically were treated with 10 µM dexamethasone (Dex) or DMSO as the mock. Proteins extracted were separated on 10% SDS-PAGE gels and blotted with αpTEpY, αOsMPK6, or αW31pS6 (generated for detection of OsWRKY31 S6 phosphorylation) antibody. Representative data from four independent experiments. C) Ubi:fW31h#2 mpk3ko and Ubi:fW31h#2 seedlings were grown hydroponically for 7 d, and then treated with 200 µg mL−1 chitin. The proteins extracted were analyzed by immunoblots with αpTEpY, αW31pS6, and αFlag. CBB, Coomassie brilliant blue staining for loading control. Representative data from three independent experiments. Relative intensity (numbers below the gels) of bands was analyzed using ImageJ software. The values below indicate the relative gray density of three experiments. The value marked with * indicates statistically significant difference between Dex treatment B) and chitin C), analyzed by the Student's t-test (*, P < 0.05). Dex:MKK10-2 for Dex:MKK10-2-myc; mpk3ko for OsMPK3 knockout; Ubi:fW31h for Ubi:Flag-WRKY31-6×His.
Third, we examined whether OsMKK10-2 activated phosphorylation of OsMPKs and OsWRKY31 in planta. Dexamethasone (Dex)-inducible MKK10-2-myc plants and knockout plants of OsMKK10-2 (mkk10-2ko), OsMPK3 (mpk3ko), and OsWRKY31 (w31ko) were obtained through the CRISPR/Cas9 method (Supplemental Fig. S3). Rice seedlings harboring the Dex:MKK10-2-myc construct were treated with Dex or dimethyl sulfoxide (DMSO) solvent as control, and sampled for immuno-blot analysis. The prominent phosphorylation of OsMPK6 and OsMPK3 was detected after Dex treatment using anti-pTEpY antibody, while in Dex:MKK10-2 mpk3ko plants, the DEX treatment failed to activate OsMPK3 phosphorylation (Fig. 2B, Supplemental Fig. S2B), suggesting that OsMPK3 was also a component downstream of OsMKK10-2 phosporylation cascade. Moreover, using an antibody for detection of specific phosphorylation of OsWRKY31 at Ser6 (anti-W31pS6) (Supplemental Fig. S4), an increase of OsWRKY31 phosphorylation levels was observed in Dex-induced plants compared with the mock treatment (Fig. 2B). Furthermore, chitin-induced OsWRKY31 accumulation was reduced in the mpk3ko background, although the relative amounts of phosphorylated OsWRKY31 vs. total OsWRKY31 were found to be somewhat variable in different experiments (Fig. 2C).
OsWRKY31 is a key player in OsMKK10-2-mediated resistance against M. oryzae
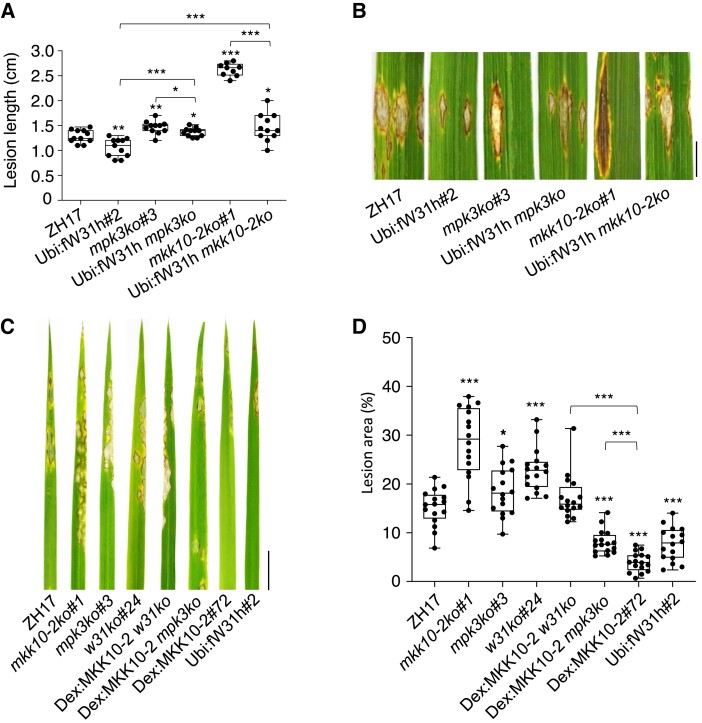
To examine whether OsWRKY31 acts downstream of OsMKK10-2 during defense responses, the transgenic and wild-type plants were inoculated with the rice blast fungus M. oryzae. Spores of M. oryzae SZ strain were injected into the leaf sheaths of rice plants (about 3-mo-old), and the newly grown leaves were collected for disease evaluation. The mkk10-2ko and w31ko plants were more susceptible to M. oryzae SZ than those of Zhonghua 17 (ZH17), whereas the OsWRKY31-overexpressing plants showed reduced susceptibility to the SZ strain compared with the ZH17 controls (Supplemental Fig. S5). However, introduction of Ubi:fW31h into the mkk10-2ko and mpk3ko background significantly decreased the susceptibility to M. oryzae (Fig. 3, A and B). To activate OsMKK10-2, Dex:MKK10-2-myc and Dex:MKK10-2 w31ko plants were sprayed with Dex 12 h before the pathogen inoculation. Dex-induced expression of OsMKK10-2 enhanced plant resistance to M. oryzae, but the resistance was eliminated when the OsWRKY31 gene was disrupted in the Dex:MKK10-2 w31ko lines (Fig. 3, C and D). Conversely, knocking out OsMPK3 only partially attenuated OsMKK10-2-conferred resistance against rice blast fungus. Taken together, these data strongly indicate that OsWRKY31 is a key player in OsMKK10-2-mediated resistance against M. oryzae at a wide period of vegetative stage.
Figure 3.
OsWRKY31 and OsMPK3 are involved in OsMKK10-2 activated disease resistance. Lesion length A) and disease symptoms B) of the transgenic and wild-type ZH17 plants after infiltration with M. oryzae SZ strain. Plants of tillering stage (about 3-mo-old) in paddy field were injected with SZ spores (1 × 105 conidia mL−1) in year 2020. The segments of leaves were photographed and quantified at 9 d after the infiltrations. The median was the crossline in each boxplot showing the lesion length distributions (n ≥ 9). Disease symptoms C) and lesion areas D) of the transgenic and ZH17 plants infected with SZ strain by spraying. Eighteen-day-old plants were inoculated with SZ spores (5 × 105 conidia mL−1) by foliar spraying; the Dex-inducible lines were treated with 20 µM Dex for 12 h before the pathogen inoculation and the other plants were received with DMSO as the mock. Evaluation of disease severity and photography taken were conducted at 7 d after the inoculation. Ubi:fW31h mkk10-2ko for Ubi:fW31h and mkk10-2ko genetic crossing progeny; other plants described in Fig. 2. The median was the crossline in each boxplot showing the lesion area distributions (n = 16). Significance was evaluated by comparing with ZH17 or each other as bracketed using the Student's t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bar = 1 cm.
OsMKK10-2 alters auxin distribution and accumulation
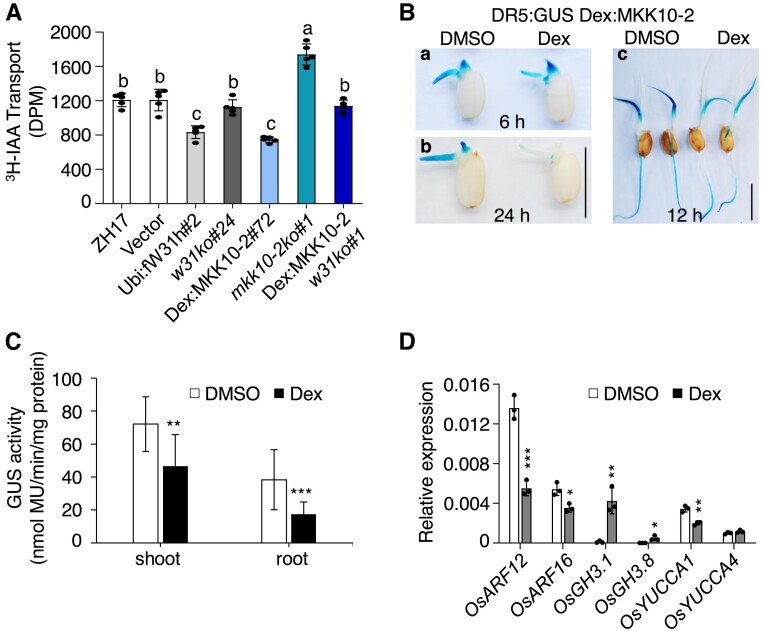
Phylogenetic analysis revealed that OsMKK10-2 is a homolog of Arabidopsis MKK10, belonging to the same D group as MKK7 (Supplemental Fig. S6) (Hamel et al. 2006). The gain-of-function bud1 mutant with increased MKK7 expression exhibits PAT deficiency (Dai et al. 2006). To know whether OsMKK10-2 is involved in auxin transportation, we measured basipetal movement of tritium-labeled IAA (3H-IAA) in the etiolated hypocotyls of the transgenic plants overexpressing OsMKK10-2. The 3H-IAA transport in hypocotyl segments of Dex:MKK10-2-myc plants was reduced to ∼60% of that in the control plants 12 h after Dex treatment, whereas the levels of auxin transport were similar between Dex:MKK10-2 w31ko and ZH17-control plants (Fig. 4A). The basipetal PAT was significantly inhibited in OsWRKY31-overexpressing hypocotyls and enhanced in mkk10-2ko segments when compared with the control plants (Fig. 4A).
Figure 4.
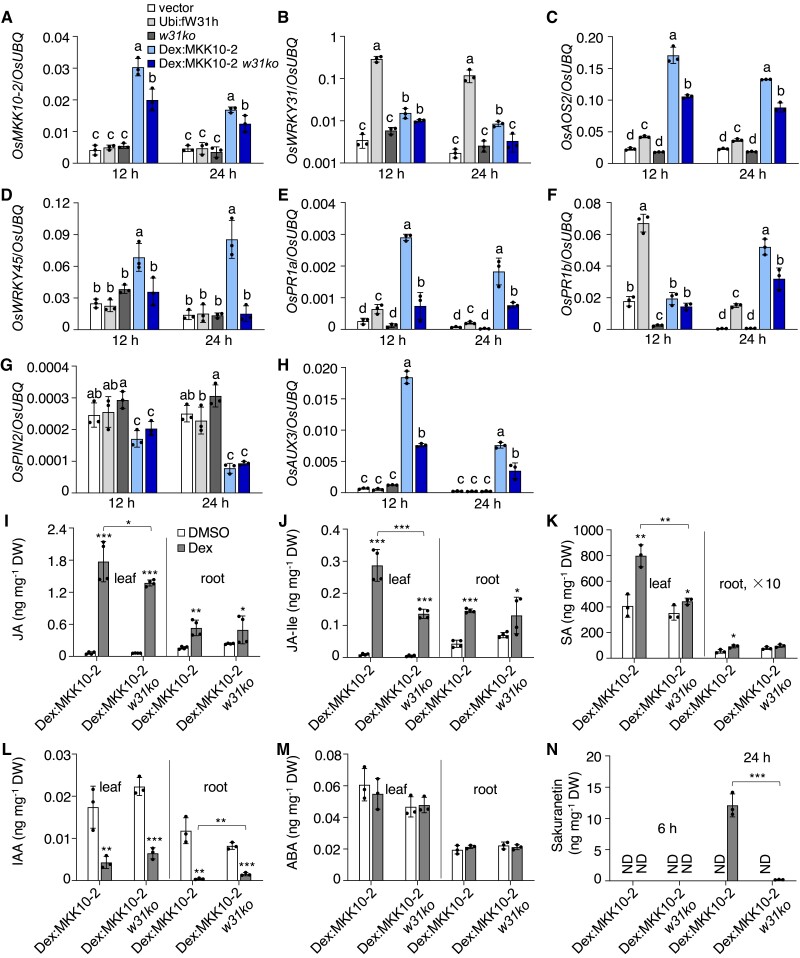
Activation OsMKK10-2 suppresses auxin transport and homeostasis. A) Polar auxin transport (PAT) assay of coleoptiles. Seven-day-old seedlings growing in the dark were treated with 10 µM Dex for 12 h and then segmented for PAT assay. Values shown are means ± SD of five independent replicates. Values marked with different letters indicate statistically significant differences as analyzed by Duncan's multiple range test, α = 0.05. GUS staining B) and activity C) of DR5:GUS Dex:MKK10-2 (genetic crossing of DR5:GUS and Dex:MKK10-2) plants. Seedlings of 2 d after germination were treated with 10 µM Dex or DMSO for 6 (a) and 24 h (b). DR5:GUS Dex:MKK10-2 seedlings of 5 d old were treated with DMSO solvent or Dex for 12 h for GUS staining (c) and activity determination (n = 14) C). DR5:GUS for auxin responsive elements (DR5) controlled GUS reporter gene; Bar = 1 cm. D) Expression of auxin related genes. Dex:MKK10-2 seedlings of 12 d old were treated with 10 µM Dex for 6 h and DMSO as the mock, and the leaves were sampled for total RNA isolation. Gene expression was determined by RT-qPCR using OsUBQ as the reference gene. Results from representative experiments are shown. Experiments were repeated three times with similar results. Values are means ± SD with significant difference compared with DMSO mock using Student's t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
To further illustrate the impact of OsMKK10-2 on auxin levels, an auxin responsive reporter line was crossed with the Dex:MKK10-2-myc plant to generate the DR5:GUS Dex:MKK10-2 progeny. To visualize the distribution of auxin in vivo, GUS staining was performed from the early stage of seed germination on the resulting seedlings treated with Dex or DMSO solvent. A marked reduction of GUS staining was observed on germinating seeds 24 h after Dex treatment (Fig. 4Ba, b), suggesting the decrease of free auxin levels during seed germination when OsMKK10-2 protein was induced. In the 5-d-old seedlings, the Dex treatment also reduced the GUS activity in the leaves and upper part of the primary roots but enhanced the staining in the lower part of the primary roots (Fig. 4, Bc and C). These observations suggested a potential perturbation of auxin distribution in plant tissues upon activation of OsMKK10-2 expression.
We then investigated the expression of genes related to auxin metabolism and signaling in Dex:MKK10-2-myc plants. OsGH3.1 and OsGH3.8 genes, encoding members of the GH3 family of IAA–amido synthetases, are implicated in auxin homeostasis by conjugating free IAA (Ding et al. 2008; Domingo et al. 2009; Li et al. 2016). Levels of OsGH3.1 and OsGH3.8 expression were remarkably increased by the activation of OsMKK10-2 transcription (Fig. 4D). Other genes of the OsGH3 family such as OsGH3.2, OsGH3.6, and OsGH3.9 were upregulated, whereas OsGH3.4 was downregulated in Dex-treated Dex:MKK10-2-myc plants at the time point examined (Supplemental Fig. S7). The YUCCA (YUC) family of flavin monooxygenase genes encodes the rate-limiting enzymes in IAA biosynthesis via the tryptophan-dependent pathway (Yamamoto et al. 2007). OsARF12 and OsARF16 are transcriptional activators in regulating auxin response genes and affect auxin transport and root elongation (Qi et al. 2012; Shen et al. 2015). The levels of OsYUCCA1, OsARF12, and OsARF16 expression were decreased in Dex-induced Dex:MKK10-2-myc plants (Fig. 4D). Collectively, the results indicate that OsMKK10-2 regulates auxin homeostasis and signaling.
To examine possible growth phenotypes of OsMKK10-2 expression, the germinated Dex:MKK10-2-myc seeds were transferred onto half strength Murashige and Skoog (MS) medium containing different concentrations of Dex. Root and coleoptile growth of Dex:MKK10-2-myc seedlings was highly inhibited with increased Dex concentration (Supplemental Fig. S8, A to C), suggesting that the elevation of OsMKK10-2 expression was responsible for these phenotypes. Furthermore, the inhibition of root growth in Dex:MKK10-2-myc plants was attenuated under w31ko background in Dex:MKK10-2 w31ko (Supplemental Fig. S8, D and E), indicating that OsWRKY31 is an important regulator downstream of OsMKK10-2.
Activation of OsMKK10-2 prioritizes defense responses over growth
As the activation of OsMKK10-2 promoted disease resistance and suppressed auxin transport and metabolism, we analyzed transcript levels of genes related to these phenotypes. Transcript levels of OsMKK10-2 increased in different Dex:MKK10-2-myc plants by Dex treatment; however, the increased transcript levels of OsMKK10-2 was slightly suppressed in Dex:MKK10-2 w31ko lines (Fig. 5A, Supplemental Fig. S9A). Activation of OsMKK10-2 increased OsWRKY31 expression in Dex:MKK10-2-myc. However, the level of OsMKK10-2 transcript was not changed significantly in Ubi:fW31h plants, in which the level of OsWRKY31 transcript was very high (Fig. 5B).
Figure 5.
OsWRKY31 is required for OsMKK10-2-activated immune response. The transgenic and control plants were hydroponically cultured for 10 d, treated with 10 µM Dex, and sampled at designated time points for RNA and chemical isolations. Transcript level of each gene A to H) was determined by RT-qPCR using OsUBQ as the internal reference. Values represent means ± SD of three indepent treatments, each including three seedling leaves. Values marked with different letters indicate statistically significant differences as analyzed by the SPSS software (Duncan's multiple range test, α = 0.05). I to N) Contents of compounds were determined by liquid chromatography–tandem mass spectrometry using D6ABA as the internal standard. I to M) sampled at 6 h after the Dex treatment. N) Leaf samples at 6 and 24 h after the Dex treatment; ND, no detection; root × 10, indicating 10 times upscale the value in root. Data are means ± SD of three or four biological repeats. Significance test was evaluated using the Student's t-test by comparing with DMSO treatment or each other as bracketed (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
The jasmonic acid (JA)-inducible allene oxide synthase OsAOS2, a JA biosynthetic gene, was significantly induced by the activation of OsMKK10-2 (Fig. 5C). OsWRKY45 has been reported to play an important role in SA-mediated defense responses in rice (Shimono et al. 2007). The level of OsWRKY45 transcript was upregulated approximately 3-fold in Dex:MKK10-2-myc plants, and this increase required functional OsWRKY31 (Fig. 5D). Moreover, the expression of OsPR1a and OsPR1b was elevated in the Dex-induced OsMKK10-2 and Ubi:fW31h plants (Supplemental Fig. S5, E and F).
Overexpression of OsPIN2, which encodes an auxin efflux transporter of the PIN family, enhances auxin transport from shoots to roots (Chen et al. 2012). In our study, OsPIN2 expression was strongly suppressed in Dex-induced Dex:MKK10-2-myc plants (Fig. 5G). Conversely, the expression of OsAUX3, belonging to the auxin influx carrier family, was elevated in Dex:MKK10-2-myc plants compared with the controls (Fig. 5H). Furthermore, the expression of OsKS1, leading to the synthesis of gibberellin (GA), was downregulated in Ubi:fW31h and Dex-induced Dex:MKK10-2-myc plants, whereas OsKS4, leading to synthesis of phytoalexin momilactones, was upregulated (Supplemental Fig. S9B). Increased OsWRKY11 and OsGH3.8 expression led to growth retardation and enhanced disease resistance (Ding et al. 2008; Cai et al. 2014). Transcript levels of both OsWRKY11 and OsGH3.8 genes were also upregulated in Ubi:fW31h and Dex-treated Dex:MKK10-2-myc plants (Supplemental Fig. S9B).
Hormonal profiling revealed that the accumulation of JA, jasmonoyl isoleucine (JA-Ile), and SA was remarkably elevated in both leaves and roots of Dex-treated Dex:MKK10-2-myc and, to a lesser extent, in Dex:MKK10-2 w31ko plants (Fig. 5, I to K). In contrast, IAA content in both roots and leaves decreased significantly by the induction of OsMKK10-2 expression (Fig. 5L) but without noticeable changes in abscisic acid (ABA) accumulation (Supplemental Fig. S5M). The levels of the phytoalexin sakuranetin were highly increased in Dex:MKK10-2-myc leaves 24 h after Dex treatment; however, this induction was insignificant in Dex:MKK10-2 w31ko plants (Fig. 5N). We also examined phytohormone accumulation in mkk10-2ko, w31ko, and Ubi:fW31h plants with or without the inoculation of rice blast fungus. Contents of SA, JA, and JA-Ile were lower in the leaves of mkk10-2ko and w31ko mutants compared with Ubi:fW31h#2 and ZH17, although induction of JA and JA-Ile was still observed (Supplemental Fig. S10). SA content was not changed largely by M. oryzae challenge, consistent with the previous report (Yang et al. 2004). Collectively, the results suggest that activation of OsMKK10-2 promoted defense responses with a synergistic increase of SA and JA accumulation and suppressed auxin-related plant growth.
OsMKK10-2 and OsWRKY31 participate in PAMP regulation of defense and auxin pathways
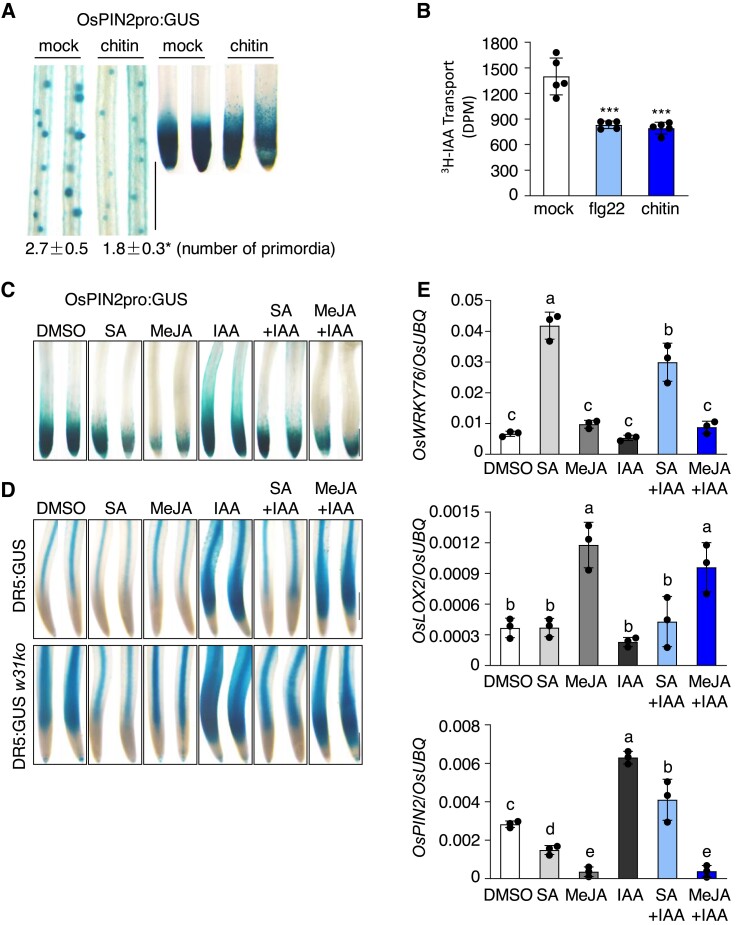
OsPIN2 mediates auxin distribution and plays an important role in root development and system architecture in rice (Wang et al. 2018). We examined the response of OsPIN2pro:GUS to chitin and verified that GUS staining was weakened at the lateral root primordia and root tips upon chitin treatment (Fig. 6A). Meanwhile, the number of the lateral root primordia was reduced by chitin treatment. Tests on basipetal PAT revealed a significant inhibition of PAT in rice seedlings in the presence of chitin or flg22 (Fig. 6B). Gene expression analysis revealed a downregulation of OsPIN2 and upregulation of OsGH3.8 in rice seedlings by chitin (Supplemental Fig. S11, C and D), consistent with the decrease of free IAA and suppression of its translocation by chitin. Expressions of OsMKK10-2 and OsWRKY31 were induced by chitin treatments, and notably, the chitin-induced defense-related OsPR1b and OsKS4 expressions were substantially compromised in the mkk10-2ko and w31ko lines (Supplemental Fig. S11, A, B, E, and F).
Figure 6.
OsMKK10-2 and OsWRKY31 are involved in PAMP regulation of defense and auxin pathways. A) Changes of OsPIN2pro:GUS expression pattern and primordia formation by chitin treatment. Five-day-old OsPIN2pro:GUS seedlings were treated with 200 µg mL−1 chitin for 12 h and stained at 37 °C for 4 h. Values are means ± SD of three biological replicates. B) Chitin and flg22 suppress auxin transport. Seven-day-old ZH17 seedlings grown in the dark were treated with 1 µM flg22 or 200 µg mL−1 chitin for 12 h, and then the coleoptiles were collected for PAT assay as described in Fig. 4. Error bars represent standard deviation of five replicate experiments. Significance test was evaluated using the Student's t-test (***, P < 0.001). Suppression of OsPIN2C) and DR5D) promoter activities by MeJA and SA. Five-day-old OsPIN2pro:GUS, DR5:GUS, and DR5:GUS w31ko seedlings were treated with MeJA (200 µM), SA (500 µM), or in combination with IAA (1 µM), and the mock received the same amount of DMSO at 28 °C for 12 h. Representative GUS staining images of OsPIN2pro:GUS primary roots stained at 37 °C for 4 h, and DR5:GUS and DR5:GUS w31ko staining at 37 °C for 1 h. Bar = 0.5 mm C and D). E) Expression of SA (OsWRKY76), JA (OsLOX2), and IAA (OsPIN2) marker genes in roots. Five-day-old DR5:GUS seedlings were treated with phytohormones as described in D). Gene expression was determined by RT-qPCR analysis using OsUBQ as the reference. Data are means ± SD of three independent treatments. Values marked with different letters indicate statistically significant differences as analyzed by the SPSS software (Duncan's multiple range test, α = 0.05).
As the activation of OsMKK10-2 promoted the accumulation of JA and SA and decreased IAA content (Fig. 5), we investigated the effects of methyl jasmonate (MeJA) and SA on auxin responses using OsPIN2pro:GUS and DR5:GUS plants. Histochemical assays of OsPIN2pro:GUS revealed that the GUS signal was significantly suppressed at the tips of primary roots by SA and MeJA treatments with or without IAA (Fig. 6C). Similarly, GUS activities of DR5:GUS and DR5:GUS w31ko were inhibited by SA and MeJA treatments (Fig. 6D), suggesting that SA and MeJA antagonize auxin effects. Analysis of gene expression revealed that OsPIN2 expression in both roots and leaves is suppressed by the exogenous application of MeJA and SA, even in the presence of IAA (Fig. 6E, Supplemental Fig. S12). Conversely, IAA treatment inhibited the expressions of the MeJA-inducible marker gene OsLOX2 and SA-inducible gene OsWRKY76. These findings suggest that PAMPs triggered immunity recruits OsMKK10-2 and OsWRKY31 to enhance defense reactions, and auxin pathway antagonizes with JA and SA pathways in rice plants.
Phosphomimetic OsWRKY31 enhances cis-element-binding activity and disease resistance
We adopted an in vitro mutation approach to localize potential phosphorylation sites of OsWRKY31. Some of the Ser/Thr residuals of Ser/Thr-Pro loci—potential MAP kinase phosphorylation sites—in OsWRKY31 were mutated to Ala to produce non-phosphorylatable variants of the sites. OsWRKY31 and its mutants were subjected to phosphorylation assays by the interacting MPKs, and Ser6 and Ser101 residues were found as the major phosphorylation sites of OsMPK3, OsMPK4, or OsMKK10-2-activated OsMPK6 (Supplemental Fig. S13).
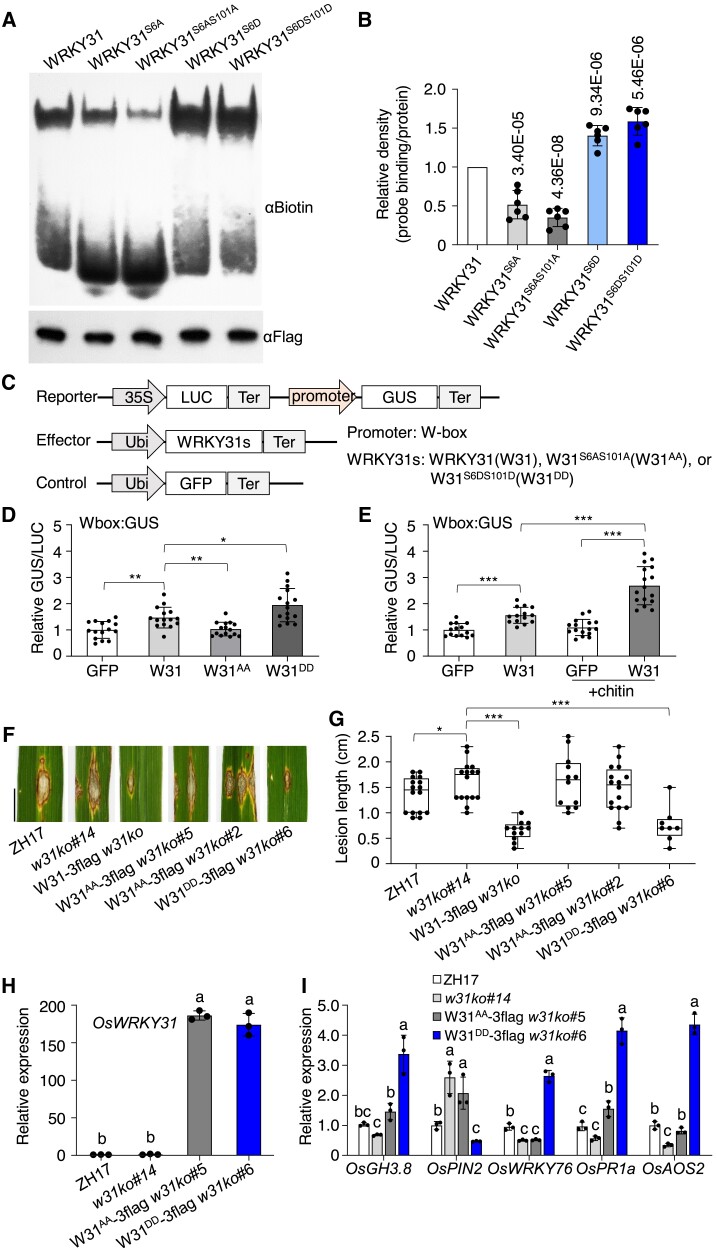
To investigate whether OsWRKY31 phosphorylation could affect its W-box-binding capability, we tested OsWRKY31 binding with probes of various combinations of W-box elements by electrophoretic mobility shift assays (EMSA). The recombinant OsWRKY31 protein interacted specifically with the probes from the promoter of OsPR1b with different affinity; however, OsWRKY31 contained a hepta WRKYGEK instead of the more conserved WRKYGQK motif in most of WRKY TFs (Supplemental Fig. S14A) (Chen et al. 2019). OsWRKY31 showed weaker binding to BP22-f1 (containing the core sequence TGAC of W-box) than BP22-f3 (containing TTGAC) probe, suggesting that OsWRKY31 might prefer to interact with the full W-box elements. Moreover, the phosphomimetic OsWRKY31S6DS101D protein showed stronger intensity of the retarded bands compared with those of OsWRKY31, whereas the phosphonull OsWRKY31S6A and OsWRKY31S6AS101A diminished markedly in the binding activity compared with the wild-type protein (Fig. 7, A and B, Supplemental Fig. S14B). Transactivation activity of OsWRKY31 and its phosphomutants was tested by using Wbox:GUS reporter in N. benthamiana leaves. A steady increase of relative activity was observed using OsWRKY31 and OsWRKY31S6DS101D effectors compared with OsWRKY31S6AS101A or the GFP control (Fig. 7, C and D), and the activity was increased by additional chitin treatment (Fig. 7E), which might increase OsWRKY31 and its phosphorylation levels with chitin application (see Fig. 8, A and B). The data indicated that OsWRKY31 is a transactivator.
Figure 7.
Phosphomimetic OsWRKY31 enhances DNA-binding activity and disease resistance. A) Phosphomimetic (GST-WRKY31S6D-3flag and GST-WRKY31S6DS101D-3flag), phosphonull (GST-WRKY31S6A-3flag and GST-WRKY31S6AS101A-3flag), and GST-WRKY31-3flag protein were expressed in E. coli. and purified for electrophoretic mobility shift assay using BP22-f3 probe (biotin-labeled P22-f3 probe, Supplemental Fig. S14A). The mixture of protein (about 2 µg) with probe (0.03 μM) was separated on native PAGE gels and detected with αBiotin antibody. The presence of retarded band indicating the formation of protein–DNA complex. Protein loading was analyzed using αFlag antibody. B) The relative density is the retarded band in related to the loading protein and normalized with the wild-type OsWRKY31 protein. Values are means ± SD of six repeats. P-values are shown in the graph. C) Schematic diagrams of the reporter and effector constructs. The promoter was fused with GUS reporter gene. LUC gene driven by the cauliflower mosaic virus (CaMV) 35S was used as an internal control. The W-box DNA was put ahead of 35S minimal promoter with TATA-box to generate Wbox:GUS construct. D and E) Effector of OsWRKY31 (W31), phosphomimetic OsWRKY31S6DS101D (W31DD), phosphonull OsWRKY31S6AS101A (W31AA), or GFP control was co-infiltrated with the reporter of Wbox:GUS in the leaves of N. benthamiana. The treated tobacco plants were grown 25 °C for 3 d and sampled for GUS activity. In case of chitin treatment, the leaves were sprayed with chitin solution (200 µg mL−1) 5 h before the sample collection. Values are means ± SD of different leaf discs (n ≥ 14). Overexpression of phosphomimetic OsWRKY31DD elevated resistance to M. oryzae SZ strain. F and G) The phosphomimetic W31DD-3flag, the phosphonull W31AA-3flag, and W31-3flag genes (all controlled by ubiquitin promoter) were transformed into w31ko background. The hygromycin-resistant transgenics of T2 progenies and the ZH17 control were grown to the tillering stage in the paddy filed and inoculated with spores of M. oryzae SZ (1 × 105 conidia mL−1) in year 2020. The leaf segments were photographed and quantified at 9 d after the infiltrations. The median was the crossline in each boxplot showing the lesion length distributions (n ≥ 8). Significance was evaluated by comparing with each other as bracketed using the Student's t-test (*, P < 0.05; **, P < 0.01; ***, P < 0.001). Bar = 1 cm. H and I) Gene expression. Leaves of 10-d-old seedlings cultured hydroponically were used for total RNA isolation. Gene expression was determined by RT-qPCR using OsUBQ as the reference gene. Values are means ± SD of three independent expereiments, each including three seedling leaves. Values marked with different letters indicate statistically significant differences as analyzed by the SPSS software (Duncan's multiple range test, α = 0.05).
Figure 8.
Rapid induction of OsWRKY31 protein by chitin and M. oryzae. A and B) Induction of OsWRKY31 accumulation by chitin and proteasome inhibitor MG132. Leaves of 7-d-old Ubi:fW31h#2 were treated with 200 µg mL−1 chitin, 100 µM MG132 or DMSO as the mock for 1 h B) or with 200 µg mL−1 chitin at various time points A). The extracted proteins were analyzed by immunoblots with antibodies of αFlag, αW31pS6, and αActin as the loading control. C) Induction of OsWRKY31 protein by M. oryzae. Eighteen-day-old rice plants (Ubi:W31DD-3flag w31ko and Ubi:W31-3flag w31ko) growing in soil were inoculated with spores of M. oryzae SZ (5 × 105 conidia mL−1) by spraying, and the leaves were sampled for protein analysis at 3 h after or before (0 h) the challenge. The proteins extracted were analyzed by immunoblots with αFlag antibody. Results from representative experiments are shown. Experiments were performed in three replicates. D) Increase OsWRKY31 accumulation by phosphorylation. Agrobacteria harboring Ubi:W31-3flag plasmid were co-infiltrated with agrobacteria containing 35S:MPK3-myc, 35S:MPK3KR-myc, or 35S:GFP-myc, into the leaves of N. benthamiana. Proteins were extracted from the leaves of 2 d after the infiltrations and analyzed by immunoblots with αMyc and αFlag antibodies. CBB, Coomassie brilliant blue staining for loading control. Representative data from three independent experiments.
To investigate the effects of OsWRKY31 phophosites in planta, we generated OsWRKY31 phosphomimetic and phosphonull plants in the w31ko#14 genetic background (Ubi:W31S6DS101D-3flag w31ko and Ubi:W31S6AS101A-3flag w31ko; abbreviated W31DD-3flag w31ko and W31AA-3flag w31ko, respectively). The phosphomimetic W31DD-3flag w31ko plants presented a markedly increased resistance against M. oryzae in comparison with W31AA-3flag w31ko, w31ko, and ZH17 (Fig. 7, F and G, Supplemental Fig. S14, C and D). Expression of defense-related genes, OsGH3.8, OsPIN2, OsWRKY76, OsPR1a, and OsAOS2, was upregulated in W31DD-3flag w31ko, whereas OsPIN2 expression was suppressed (Fig. 7I). The transcript levels of defense-related genes were similarly much lower in phosphonull W31AA-3flag w31ko and w31ko plants than in W31DD-3flag w31ko lines, although both of the transgene W31AA-3flag and W31DD-3flag were highly expressed (Fig. 7, H and I). The observations indicated that the two phosphosites of OsWRKY31 are essential for its role in activation of disease resistance.
Rapid induction of OsWRKY31 protein by chitin and M. oryzae
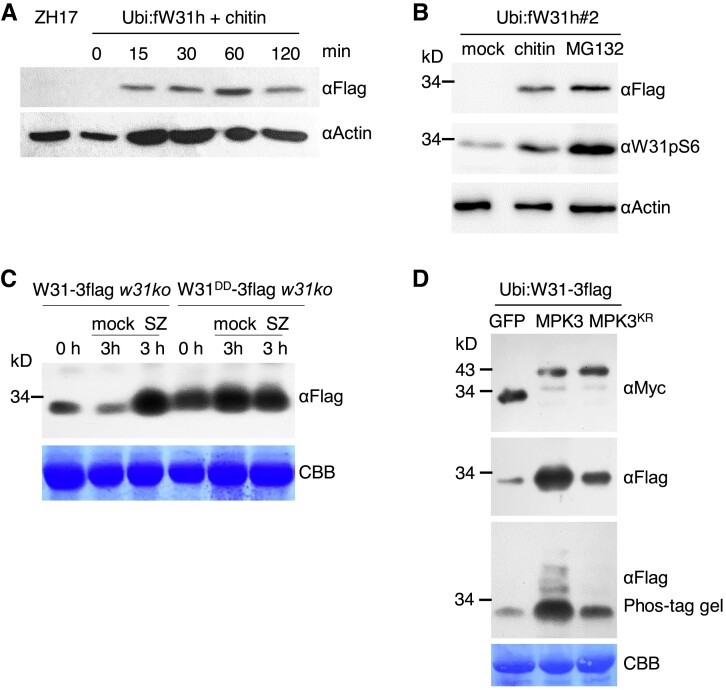
When the abundance of OsWRKY31 protein was examined, we found that the level of OsWRKY31 accumulation was low in Ubi:fW31h plants compared to the very high transcript levels of OsWRKY31, especially in hydroponic culture conditions (Fig. 5B). Rapid and transient induction of OsWRKY31 protein was observed in Ubi:fW31h plants treated with chitin (Fig. 8A). Similar to chitin treatment, the levels of OsWRKY31 protein and its phosphorylation were simultaneously elevated in Ubi:fW31h#2 plants treated with MG132, an inhibitor of proteasome-mediated protein degradation (Fig. 8B), suggesting that OsWRKY31 protein is regulated by the ubiquitin–proteasome system.
To address why W31DD-3flag w31ko and W31-3flag w31ko lines showed similar levels of resistance against M. oryzae (Fig. 7, F and G), we analyzed the levels of the proteins encoded by the respective transgenes. The abundance of the phosphomimetic mutant OsWRKY31DD was higher than wild-type OsWRKY31 revealed by comparing the samples (0 h) before the pathogen infection (Fig. 8C) and among different transgenic lines (Supplemental Fig. S14D). OsWRKY31 abundance was markedly elevated after inoculation of M. oryzae SZ strain in 3 h, a very early infection period for the pathogen infection, and reached to the level of OsWRKY31DD (Fig. 8C). The results imply that the rapid induction of OsWRKY31 protein is able to compensate for the lower level at the beginning than OsWRKY31DD, and also the phosphorylated OsWRKY31 is more stable than its wild type (Figs. 2C and 8B).
To further confirm the relationship between OsWRKY31 abundance and its phosphorylation status, we transiently expressed OsWRKY31 with OsMPK3 in N. benthamiana leaves via agroinfiltration. The levels of OsWRKY31 protein and its phosphorylation were higher in plants expressing Ubi:W31-3flag with 35S:MPK3-myc than with 35S:MPK3KR-myc or 35S:GFP-myc (Fig. 8D), supporting the idea that OsWRKY31 phosphorylation favors its protein accumulation and is consistent with that observed in Fig. 2C. The increased OsWRKY31 could be because an increase of phosphorylated active OsWRKY31 triggers a large variation in defense signaling and stimulates a higher level of expression of unphosphorylated OsWRKY31. Collectively, the rapid induction of OsWRKY31 accumulation by chitin and during the early infection of rice blast fungus suggested that OsWRKY31 is highly controlled for efficient defense responses.
OsWRKY31 stability is regulated by ubiquitination
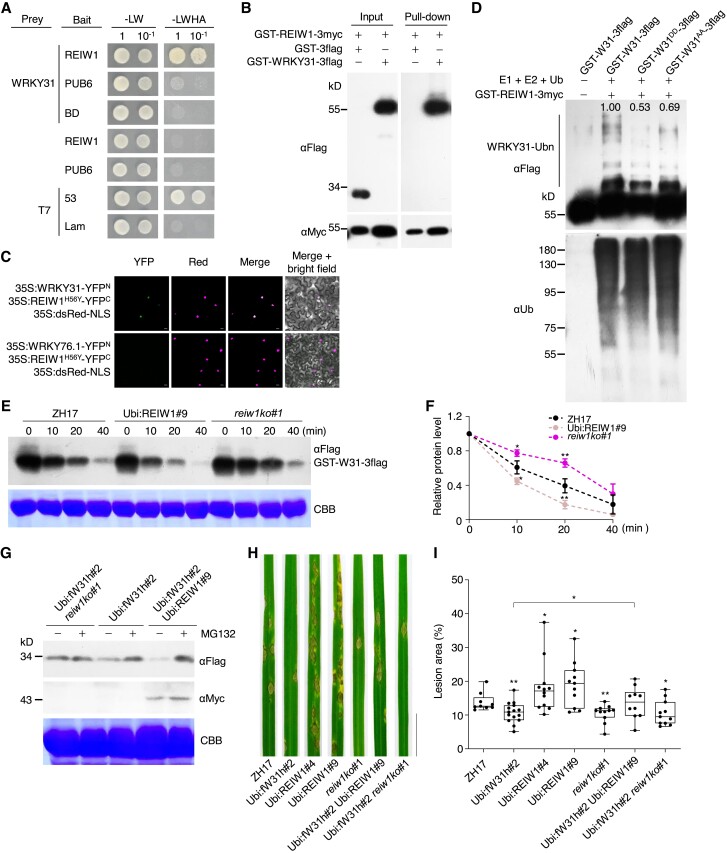
Since OsWRKY31 stability is mediated by the proteasome system, we searched E3 ubiquitin ligases as potential interactors of OsWRKY31. A RING-finger E3 ubiquitin ligase interacting with WRKY, designated as OsREIW1, was previously obtained through a Y2H screen. By transcriptomic analyses of plant responses to a panel of phytohormone treatments (Sato et al. 2013), we found that OsREIW1 is co-regulated with OsWRKY31 (Supplemental Fig. S15A). Further, OsWRKY31 was verified to interact with OsREIW1 protein, but not the E3 ligase PUB6 in yeast cells and by pull-down experiment (Fig. 9, A and B). Interaction of OsWRKY31 with OsREIW1H56Y (His56 to Tyr, to eliminate the enzymatic activity) mutant was mainly in the nucleus of N. benthamiana leaves by BiFC assay (Fig. 9C), supported by the predominant localization of both OsREIW1 and OsWRKY31 in the nuclei (Supplemental Fig. S15B) (Zhang et al. 2008).
Figure 9.
OsREIW1 regulates OsWRKY31 stability. A to C) OsREIW1 interacts with OsWRKY31. A) OsREIW1 and PUB6 as a control were fused to the Gal4 DNA-binding domain (bait). The prey-OsWRKY31 was from Fig. 1. Yeast cells were incubated in SD medium without Leu and Trp (-LW) or lacking Leu, Trp, His, and adenine (-LWHA), and photographed 3 d after plating. Yeast cells containing AD-T7 plus BD-53 and AD-T7 plus BD-Lam plasmids were used as the positive and negative controls, respectively. B) Purified GST-REIW1-3myc and GST-WRKY31-3flag protein (each about 1 μg) were mixed and incubated at 4 °C for 2 h in immunoprecipitation buffer. The protein mixture was precipitated with anti-c-Myc agarose affinity gels, separated on 10% SDS-PAGE gels, and detected by immunoblots with αMyc and αFlag antibodies. C) Agrobacteria harboring 35S:REIW1H56Y-YFPC, 35S:WRKY31-YFPN or 35S:WRKY76.1-YFPN plasmid were combined accordingly and infiltrated into the leaves of N. benthamiana. Confocal images were taken at 2 d after the infiltrations. Red fluorescence signals indicative of the nuclei were from co-infiltrated 35S:dsRed-NLS (NLS: nuclear localization signal). Bar = 10 μm. D) Ubiquitination of OsWRKY31 by OsREIW1. OsWRKY31 recombinant protein GST-W31-3flag and its phosphomimetic GST-W31DD-3flag and phosphonull GST-W31AA-3flag were incubated with E1, E2, and ubiquitin in the presence of GST-REIW1-3myc. The reaction mixture (30 μL) contained 50 ng wheat ubiquitin-activating enzyme E1, 100 ng human ubiquitin-binding enzyme E2 (UBCH5b), 5 µg Arabidopsis ubiquitin (Ub), 500 ng GST-REIW1-3myc, and 2 μg OsWRKY31 or its mutant protein in buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 2 mM DTT, and 5 mM ATP) and incubated at 30 °C for 1 h. The reaction was stopped by adding the 5× SDS loading buffer. Samples were separated by 10% SDS-PAGE gels and analyzed by immunoblots using αUb or αFlag antibody. Relative intensity (numbers in the gel) of ubiquitinated bands was analyzed using ImageJ software. E and F) Cell-free degradation of OsWRKY31. Total proteins were extracted from the leaves of Ubi:REIW1-3myc, reiw1ko, and ZH17 seedlings grown hydroponically for 1 wk. Protein extracts (about 500 μg) were incubated with GST-WRKY31-3flag (2 μg) for various time points at 25 °C. The protein samples were separated and analyzed by immunoblot with αFlag antibody. Representative data from three independent experiments. Quantification of treatments at 0 min was set as 1. Values are mean ± SD (n = 3). G) Segments of leaves from 7-d-old seedlings cultured hydroponically were treated with 100 µM MG132 or DMSO solvent for 1 h and then sampled for protein isolation. The protein mixture was separated on 10% SDS-PAGE gels and detected by immunoblots with αMyc and αFlag antibodies. Representative results from three independent experiments. CBB, Coomassie brilliant blue staining for loading control. H and I) Eighteen-day-old plants were inoculated with M. oryzae SZ spores (5 × 105 conidia mL−1) by foliar spraying. Disease symptoms H) and lesion areas I) were conducted at 7 d after the inoculation. Representative data from three independent experiments. Significance was evaluated by comparing with ZH17 or each other as bracketed using Student's t-test (*, P < 0.05; **, P < 0.01). Bar = 2 cm.
Recombinant GST-REIW1-3myc protein exhibited self-ubiquitination activity, and the OsREIW1H56Y mutant showed no enzymatic activity (Supplemental Fig. S15C). OsWRKY31 was highly ubiquitinated by OsREIW1 (Fig. 9D). In contrast, ubiquitination of the phosphomimetic OsWRKY31DD was weaker than the phosphonull OsWRKY31AA and OsWRKY31 (Fig. 9D), implying that phosphorylation of OsWRKY31 at the two sites potentially attenuates the ubiquitination and increases the protein stability. This note is consistent with the higher level of OsWRKY31DD in its transgenic plants compared to OsWRKY31 plants (Fig. 8C, 0 h).
Moreover, we used a cell-free approach to compare the stability of OsWRKY31 and OsWRKY31DD during incubation with protein extracts from OsREIW1-overexpressing plants. GST-WRKY31DD-3flag showed slower degradation than GST-WRKY31-3flag (Supplemental Fig. S15D). Compared with OsREIW1 knockout (reiw1ko) plants, GST-WRKY31-3flag degradation was faster in the extracts from OsREIW1-overexpressing or wild-type ZH17 plants (Fig. 9, E and F). Lower OsWRKY31 abundance was detected in Ubi:fW31h Ubi:REIW1 plants (Ubi:fW31h and Ubi:REIW1-3myc cross progeny) than in Ubi:fW31h reiw1ko plants (Fig. 9G). Notably, OsWRKY31 levels in Ubi:REIW1-3myc plants but not in reiw1ko plants were increased upon MG132 treatment (Fig. 9G), suggesting that OsWRKY31 is a target of OsREIW1 for degradation via 26S proteasome.
The OsREIW1-related plants were tested for the response to M. oryzae SZ. The knockout mutant of OsREIW1 exhibited enhanced resistance against the fungal pathogen, whereas Ubi:REIW1-3myc plants were more susceptible to the SZ pathogen than ZH17 (Fig. 9, H and I). The elevated resistance in Ubi:fW31h plants was compromised in the presence of Ubi:REIW1-3myc, indicating that OsREIW1 is a negative regulator of OsWRKY31-mediated defense responses. Collectively, these findings demonstrated that OsWRKY31 stability is regulated by phosphorylation and ubiquitination.
Discussion
In this study, we provided evidence that OsWRKY31 is a key regulator of rice defense and growth, acting downstream of OsMKK10-2 in a OsMKK10-2–OsMPK3–OsWRKY31 cascade. In addition, we found that OsWRKY31 stability is regulated by phosphorylation and ubiquitination via a RING-finger E3 ligase, OsREIW1.
OsMKK10-2–OsMPK3/6 activates OsWRKY31 in rice immunity
In the MPK cascades, OsMKK10-2 has been shown to interact directly or indirectly with environmental stress-related OsMPK3, OsMPK4, and OsMPK6 (Ueno et al. 2015; Ma et al. 2017; Zhou et al. 2017). Ueno et al. (2013, 2015) have addressed the OsMKK10-2–OsMPK6–OsWRKY45 pathway in M. oryzae resistance, in which ABA treatment leads to dephosphorylation of OsMPK6 via activation of protein tyrosine phosphatases and improves disease resistance. Thus, OsMKK10-2–OsMPK3–OsWRKY45 appears to mediate SA–ABA signaling crosstalk between disease-resistance and abiotic-stress responses. Similarly, Ma et al. (2017) have shown that overexpressing OsMKK10-2 increases resistance to the bacterial pathogen Xoc and tolerance to drought, whereas its RNAi-knockdown plants are more sensitive to Xoc and drought stress compared with the control plants. The disease-resistance and drought-tolerance pathways are regulated separately by OsMKK10-2–OsMPK6 and OsMKK10-2–OsMPK3 modules, respectively (Ma et al. 2017). In this study, we showed that activation of OsMKK10-2–OsMPK3–OsWRKY31 promotes defensive responses and this particular signaling cascade has a negative effect on auxin-related activities including downregulation of genes for auxin biosynthesis, signaling, and transportation (Figs. 3 to 5). These results point to an involvement of OsMKK10-2–OsMPK3-OsWRKY31 in SA–IAA signaling crosstalk. We find it interesting that both OsMKK10-2–OsMPK6–OsWRKY45 and OsMKK10-2–OsMPK3–OsWRKY31 cascades regulate rice defense but have negative crosstalk with ABA (in the case of OsWRKY45) or auxin (in the case of OsWRKY31) hormonal pathways. Likely, OsWRKY31 is involving defense and auxin, whereas OsWRKY45 participated in defense and ABA signaling. Future research is needed to further examine if OsWRKY31 and OsWRKY45 have nonredundant and/or overlapping roles in mediating other aspects of defense-growth trade-offs. Phylogenetically, OsWRKY45 belongs to the WRKY IIIa subclade, whereas OsWRKY31 is in the IIIb subclade. These two subclades appear to have evolved after the divergence of dicots and monocots (Wu et al. 2005).
Furthermore, we found the formation of OsMKK10-2–OsMPK3–OsWRKY31 ternary complex (Fig. 1E), which potentially increases the efficiency of OsWRKY31 phosphorylation and specificity of the signaling pathway. The protein complex is often observed in the metabolic channeling, which allows an efficient control of metabolic flux (Achnine et al. 2004). The complex of LOX2, AOS2, and AOC2 in the plastid envelope of Arabidopsis chloroplasts provides a channel for JA precursor biosynthesis (Pollmann et al. 2019). In Arabidopsis, MEKK1 interacts with and directly phosphorylates WRKY53 and regulates WRKY53 through binding to its promoter, suggesting that MEKK1 can act as a short cut in MPK signaling (Miao et al. 2007). However, the biochemical function associated with the interaction between OsMKK10-2 and OsWRKY31 is yet to be clarified.
OsMKK10-2 and OsWRKY31 mediate the defense and growth trade-off
PAMPs, such as flg22, have been demonstrated to repress auxin signaling in Arabidopsis (Navarro et al. 2006). Our results showed that chitin inhibited IAA transport in rice seedlings and downregulated OsPIN2 expression (Supplemental Fig. S11C). GUS signal depletion in OsPIN2pro:GUS and DR5:GUS plants by the addition of SA or MeJA revealed the antagonistic effect of SA and MeJA on IAA in rice plants (Fig. 6, C to E). Activation of OsMKK10-2 increased the levels of SA and JA/Ile-JA, whereas knockout of OsWRKY31 compromised the induced phytohormnone accumulation (Fig. 5, I to K). This is consistent with the activation of OsMKK10-2 or overexpressing OsWRKY31 inhibiting root growth and enhancing resistance against M. oryzae (Supplemental Fig. S8, B and C; Zhang et al. 2008). The effects on growth and defense via OsMKK10-2 are remarkably suppressed by knockout of OsWRKY31 (Fig. 3, C and D, Supplemental Fig. S8, D and E), indicating that OsWRKY31 is an important downstream component of OsMKK10-2. Additionally, OsWRKY31 possibly has a feedback effect on OsMKK10-2 expression because OsMKK10-2 expression is slightly suppressed in the w31ko background (Figs. 5A, Supplemental Fig. S9, A and S11A), but we cannot exclude the possibility of partial silencing of the DEX-induced OsMKK10-2 in the w31ko background. Together, our data indicate that the OsMKK10-2–OsMPK3/6–OsWRKY31 module functions at a node for rice defense and growth, even though the study here was focused on the seedling stage, which may not reflect growth effects at the whole-plant scale.
Role of OsMPK3 in rice resistance
We demonstrated, using a mpk3ko mutant, that OsMPK3 is an important kinase activated by OsMKK10-2 in defense response (Figs. 2B and 3, A to D), providing updated information on MPK signaling in rice. Knockout of OsMPK3 only partially compromised OsMKK10-2-induced resistance to M. oryzae (Fig. 3D), suggesting that other kinases may have contributed to OsMKK10-2-mediated disease resistance, as OsMKK10-2 was able to phosphorylate several OsMPKs (Fig. 2B) (Zhou et al. 2017). Disease assays on mpk3ko mutants, Dex:MKK10-2 mpk3ko and Ubi:fW31h mpk3ko plants, suggested that OsMPK3 acts as a positive regulator of resistance against rice blast fungus, which differs from previous studies showing OsMPK3 as a negative regulator (Xiong and Yang 2003; named as OsMAPK5 therein). The phenotypic discrepancies between gene knockout and gene knockdown may be caused by a genetic compensation response, as noted in zebrafish (Rossi et al. 2015; Ma et al. 2019) and plants (Gao et al. 2015). Alternatively, the difference is possible due to cultivar variations and pathogens used in the studies.
Phosphomimetic OsWRKY31 facilitates its DNA-binding activity
OsWRKY31 with a hepta WRKYGEK instead of the more conserved WRKYGQK motif shows binding activity to different combinations of W cis-elements (Supplemental Fig. S14A). Phosphorylation analyses in vitro demonstrated that OsWRKY31 could be phosphorylated directly by OsMPK3 and OsMPK4 but not OsMPK6 and that OsWRKY31 S6 and S101 are major phosphosites of the OsMPKs (Supplemental Fig. S13). Other studies have shown that MPKs can directly phosphorylate substrates in vitro. For instance, Arabidopsis MPK6 directly phosphorylates elFiso4G1, a component of the translational preinitiation complex and OsMPK3 phosphorylates SUB1A (Singh and Sinha 2016; Wang et al. 2022). We found that the phosphomimetic OsWRKY31S6DS101D facilitated W-box-binding and enhanced disease resistance against rice blast fungus, whereas the phosphonull OsWRKY31S6AS101A diminished W-box-binding activity (Fig. 7, A and B).
Phosphorylation-mediated effects on WRKY binding to target DNA have been reported previously. For example, Arabidopsis WRKY70 is able to bind the W-box for inhibition and WT-box for activation regulation (Liu et al. 2021). Phosphomimetic WRKY70 can still bind the WT-box but not the W-box. Phosphorylation of OsWRKY53 markedly enhances binding activity to a W-box-containing DNA sequence (Tian et al. 2017). Similarly, phosphomimetic MdWRKY17 elevates W-box binding, and its nonphosphorylatable mutant compromises the DNA-binding activity (Shan et al. 2021). Change of phosphorylation status may affect protein charges, conformation, and formation of homocomplexes, leading to alteration of DNA-binding activity. The results indicate that the selectivity and affinity of a WRKY TF towards variant W-boxes is influenced by its post-translational modification by phosphorylation (Chen et al. 2019; Liu et al. 2021).
Phosphomimetic OsWRKY31 increases its stability
We found that degradation of OsWRKY31 is mediated by OsREIW1, a RING-finger ubiquitin E3 ligase, through the 26S proteasome (Fig. 9), likely explaining the low level of OsWRKY31 protein in OsWRKY31-overexpressing plants (Fig. 2C). Intriguingly, the phosphomimetic OsWRKY31DD seems more stable than the wild-type protein (Fig. 8D, Supplemental Fig. S14D), which is consistent with weaker ubiquitination in in vitro assays (Fig. 9D). Transcription regulators activated via phosphorylations are inclined to degrade in comparison with their native forms (Min et al. 2019; Liu et al. 2021). This on-and-off regulatory process can presumably avoid overdose responses to stimuli. Upregulation of defense-related genes in Ubi:W31DD-3flag w31ko plants is consistent with the relatively high amount of OsWRKY31DD protein; however, levels of the increase in gene transcripts, such as OsPR1a and OsAOS2, are lower than those induced by activation OsMKK10-2 at 12 h (Figs. 5, C and E and 7I), implying a restricted elevation of OsWRKY31DD. Conversely, OsWRKY31 protein can be rapidly induced by chitin and M. oryzae (Figs. 2C and 8C), thereby promptly raising the defenses in OsWRKY31-overexpressing plants. Under chilling stress, OsbHLH002/OsICE1 is phosphorylated by OsMPK3, leading to inhibition of OsbHLH002 ubiquitination by the OsHOS1 E3 ligase and enhanced chilling tolerance (Zhang et al. 2017). Nevertheless, the stability of OsWRKY31DD needs further clarification. Based on our findings and those of previous studies, we propose that OsMKK10-2 activated by biotic and abiotic stimuli phosphorylates OsMPKs that lead to the phosphorylation of TFs including OsWRKY31 to promote immunity; OsWRKY31 protein is controlled by phosphorylation and ubiquitination for fine-tuning the defense and growth balance.
Materials and methods
Plant growth and treatments
The seeds of wild-type (O. sativa L. ZH17) and transgenic plants were surface sterilized and germinated in 1/2 MS medium. The seedlings were transferred to 96-hole plates and cultured in 1/4 MS liquid medium at 28 °C under the white fluorescent light intensity of 200 μmol m−2 s−1 with a 12 h light/12 h dark photoperiod. Seedlings at designated age were treated with 5 mM MES (4-morpholineethanesulfonic acid, pH 5.8) buffer containing 500 μM SA, 200 μM MeJA, 1 μM flg22, or 200 μg mL−1 chitin and used for RNA isolation or GUS activity.
To investigate the effect of OsMKK10-2 on seedling growth, the germinated seeds of Dex:MKK10-2-myc and ZH17 controls were transferred to 1/2 MS solid medium containing 0.24% [w/v] phytagel and various contents of Dex or the same amount volume of DMSO solvent as the mock treatment and grown at 28 °C for 5 d.
Generation of transgenic plants
The coding sequences of OsMKK10-2 and OsREIW1 were amplified from ZH17 cDNA using gene specific primers (Supplemental Table S1, including all primers used in this study). OsMKK10-2DD, OsWRKY31S6AS101A, and OsWRKY31S6DS101D variants were generated by PCR-based site-directed mutagenesis. The sequence of OsMKK10-2 was fused with one Myc tag at the 3′ end and put under the control of a Dex-inducible promoter to generate Dex:MKK10-2-myc. OsWRKY31S6AS101A and OsWRKY31S6DS101D were cloned to fuse with 3×Flag and controlled by the maize (Zea mays) Ubiquitin promoter, ligated into restriction endonuclease sites of BamH I and PmaC I in a modified pCAMBIA1301 vector to generate Ubi:W31AA-3flag and Ubi:W31DD-3flag plasmids, respectively (Liu et al. 2016). Similarly, OsREIW1 was constructed as Ubi:REIW1-3myc. For CRISPR/Cas9 editing, the target sequence of each gene (Supplemental Table S1) was put under the control of the U3 promoter in pOsCas9 vector and verified by sequence. DR5:GUS vector was constructed by substitution of the W-box fragment in Wbox:GUS plasmid (Liu et al. 2016).
All of the plasmids constructed were verified by sequencing and introduced into A. tumefaciens EHA105 for rice transformation. The transgenic plants were generated from the immature seeds of ZH17 by the Agrobacterium-mediated transformation method (Hiei et al. 1994) unless indicated specifically. Ubi:W31AA-3flag and Ubi:W31DD-3flag plasmids were transformed into the immature seeds of OsWRKY31 knockout (w31ko#14) segregant without the transferred CRISPR/Cas9 vector. Transgenic plants were analyzed by PCR amplification and the positive plants of T2 or higher progeny were used in the experiments. Dex:MKK10-2 w31ko, Dex:MKK10-2 mpk3ko, and Ubi:fW31h mkk10-2ko were generated by genetic crossing.
Pathogen inoculation
Eighteen-day-old rice plants, grown in soil containing peat moss and vermiculite (1:1), were inoculated with a virulent M. oryzae SZ strain by spraying the spore suspension (5 × 105 conidia mL−1 containing 0.005% [v/v] Silwet L-77) as described by Liu et al. (2016). In the case of Dex treatment required, 20 μM Dex solution containing 0.002% [v/v] Silwet L-77 was sprayed 12 h before the spore inoculation, whereas the mock treatment received the same amount of DMSO solvent. For injection of rice blast spores, rice plant after tillering (3-mo-old) was injected with the spores (1 × 105 conidia mL−1) to each leaf sheath at the bottom, and the newly grown leaves were collected for disease severity evaluation.
Auxin transport assay
The sterilized seeds were grown in 1/2 MS liquid medium with 5 mM MES (pH 5.8) at 28 °C in the dark. Seven-day-old seedlings were treated with 200 µg mL−1 chitin or 1 µM flg22 (in 5 mM MES, pH 5.8) for 12 h, and the seedlings harboring Dex:MKK10-2-myc construct were treated with 10 µM Dex for 12 h. The seedlings were segmented from 2 mm below the tip (20 mm length) and incubated in 1/2 MS medium with moderate shaking for 3 h to deprive endogenous IAA. A microdroplet of 3H-IAA (30 µL, 500 nM 3H-IAA and 500 nM unlabeled IAA in 1/2 MS medium containing 5 mM MES, pH 5.8) was applied to the apical tips at 28 °C for 3 h. Fifteen-millimeter segments from the other ends were harvested and quickly rinsed with 1/2 MS medium for three times. The samples were incubated in 3 mL scintillation fluid for 18 h and then used for radioactivity measurement on liquid scintillation counter (Perkin-Elmer, 1450 MicroBeta TriLux).
Yeast two-hybrid assay
The DNA fragments of genes and their mutants were inserted into pGBKT7 or pGADT7 vector (Clontech) to generate bait or prey plasmid. Appropriate combinations of bait and prey constructs were transformed into yeast cells and then grown at 28 °C for 3 d on the synthetic dropout medium without Leu and Trp and the selection medium deficiency of Leu, Trp, His, and Ade for interaction test.
Protein expression and purification
OsMKK10-2, OsMPK3, OsMPK4, OsMPK6, OsWRKY31, OsREIW1, and their mutants were cloned into a modified pGEX-tag vector, respectively (Liu et al. 2016), with 3×Myc or 3×Flag at the C-terminus of the recombinant protein. Each plasmid was transformed into E. coli BL21 (DE3), and protein expression was induced by addition of 0.2 mM IPTG followed by incubation at 28 °C for 6 h. The recombinant proteins were purified using Glutathione Sepharose 4B (GE Healthcare).
Pull-down assay
GST-MKK10-2-3myc, GST-MKK10-2DD-3myc (Arg203 and Ser209 mutated to Asp), GST-MKK10-2KR-3myc (Lys81 mutated to Arg), or GST-3myc was combined with GST-WRKY31-3flag (each recombinant protein 2 μg) and incubated with EZview Red anti-c-Myc affinity gels (Sigma-Aldrich, E6654) with moderate shaking at 4 °C for 3 h. The beads were washed five times for 5 min with IP buffer (50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.1% [v/v] Triton X-100). Proteins bound to the affinity gels were eluted by boiling the beads in 100 µL 1× Laemmli SDS loading buffer. The immunocomplexes were separated on 10% polyacrylamide gels, blotted onto PVDF membranes, and detected using horseradish peroxidase (HRP)-conjugated anti-Flag (Sigma-Aldrich, A8592, 1: 10,000 dilution) and HRP-conjugated anti-Myc (Sungene Biotech, LK8003, 1: 10,000 dilution) antibodies.
BiFC assay
BiFC assays were performed as described previously (Kamigaki et al. 2016; Liu et al. 2016). The coding sequences of OsWRKY31, OsMPK3, OsMPK4, OsMPK6, and OsMKK10-2 were constructed in-frame with YFPN (N-terminal YFP) and/or YFPC (C-terminal YFP). For multicolor BiFC analysis, OsMPK3, OsWRKY31, and OsWRKY45 were fused with CFPN or CFPC, whereas OsMKK10-2KR was fused with YFPN. Each plasmid was introduced into A. tumefaciens EHA105, and then appropriate combinations of BiFC constructs with the nuclear localization signal-targeted dsRED (35S:dsRED-NLS) plasmid were co-infiltrated into the abaxial side of 4-wk-old N. benthamiana leaves with a 1 mL needleless syringe. Two days later, fluorescence was visualized with a confocal microscope (Eclipse TE2000, Nikon) using the preset settings for YFP (Ex: 514 nm, Em: 510 to 550 nm), dsRED (Ex: 557 nm, Em: 579 to 586 nm), and CFP (Ex: 458 nm, Em: 470 to 500 nm).
Plant protein extraction and immunoblot
Total plant proteins were extracted with buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10% [v/v] glycerol, 150 mM NaCl, 1% [v/v] Triton X-100, 1 mM DTT, 1 mM PMSF, 0.1 mM MG132, 1 mM cantharidin, and 1:200 protease inhibitor cocktail) on ice for 30 min. The supernatants were collected after centrifugation at 12,000 × g for 10 min at 4 °C, and repeated the centrifugation again for use.
Proteins or protein–DNA complexes after separation on gels were transferred to nylon membranes. Immunoblots were performed by using appropriate dilution of each antibody and then exposed to X-ray films.
Phosphorylation assay
Potential MPK phosphorylation (SP/TP) sites of OsWRKY31 (including S2, S6, S101, S133, and T143) were sequentially mutated to phosphonull (S or T substituted to A) mutants and cloned for protein expression. OsWRKY31 and each mutant protein were subjected to phosphorylation with GST-MPK4-3myc. Phosphorylation mixture was 4:1:2 for OsWRKY31 (2 μg), OsMKK10-2, and OsMPK recombinant protein, in 100 μL volume containing 50 mM Tris-HCl (pH 7.5), 10 mM MgCl2, 10 mM MnCl2, 1 mM DTT, and 2 mM ATP, and reacted at 30 °C for 2 h. The reactions were terminated by adding 5× SDS loading buffer. GST-WRKY31-3flag and its S6A and S101A single and double mutants were also examined for phosphorylation by OsMPK3 or OsMPK6 in combination with OsMKK10-2. Dephosphorylation was carried out by addition of 0.5 μL calf intestinal alkaline phosphatase (CIAP) and appropriate amount of 10× alkaline buffer provided by the supplier into aliquot phosphorylation mixture and incubated at 37 °C for 1 h. The denatured samples were separated on Phos-tag gels and analyzed by immunoblots. To detect phosphorylation of OsMPKs and OsWRKY31 in plants, anti-pTEpY (Cell Signaling Technology, 9101S, 1:2,000 dilution) and anti-W31pS6 (ChinaPeptides, 2286-R4, 1:10,000) were used in immunoblotting.
Electrophoretic mobility shift assay
The W-box containing DNA elements were derived from the promoter region of OsPR1b and labeled on 3′ end with biotin. EMSA was performed in a 20 μL reaction volume (10 mM Tris-HCl, pH 7.5, 50 mM NaCl, 1 mM DTT, 1 mM EDTA, and 5% [v/v] glycerol) containing about 2 μg recombinant protein, 0.03 μM biotin-labeled probe, and designated amount of competitor probes if required, at 25 °C for 30 min. The reaction was terminated by addition of 2 μL 10× loading buffer (250 mM Tris-HCl, pH 8.0, and 40% [v/v] glycerol), separated on 6% native polyacrylamide gels, and then transferred onto PVDF membranes for immunoblot using HRP-conjugated anti-Biotin antibody (Thermo Fisher 89880).
Ubiquitination assay
Ubiquitination assay was performed by following the method described previously with slight modification (Chen et al. 2021). Each 30 μL reaction mixture contained 50 ng recombinant wheat (Triticum aestivum) ubiquitin-activating enzyme E1 (GI: 136632), 100 ng human ubiquitin-binding enzyme E2 (UBCH5b), 5 µg ubiquitin protein (UBQ14, At4g02890), 500 ng GST-REIW1-3myc, and 2 μg OsWRKY31 substrate or its mutant in a reaction buffer (50 mM Tris-HCl pH 7.5, 10 mM MgCl2, 2 mM DTT, and 5 mM ATP) and incubated at 30 °C for 1 h. The reaction was stopped by adding 5× SDS loading buffer. Samples were separated by 10% SDS-PAGE gel and analyzed by immunobloting with anti-Ubiquitin (Sigma-Aldrich, U0508, 1:10,000 dilution) and HRP-conjugated anti-Flag (Sigma-Aldrich, A8592, 1: 10,000 dilution) antibodies.
Protein degradation assay
To investigate whether the stability of OsWRKY31 is related with OsREIW1, the leaves of Ubi:REIW1-3myc, reiw1ko, and ZH17 seedlings grown hydroponically for 1 wk were used to extract the total proteins in buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 10% [v/v] glycerol, 150 mM NaCl, 1% [v/v] Triton X-100, and 1 mM DTT). The protein extracts (about 500 μg) were incubated with GST-WRKY31-3flag (2 μg) for designated time points at 25 °C. The protein samples were separated and analyzed by immunobloting with HRP-conjugated anti-Flag (Sigma-Aldrich, A8592, 1: 10,000 dilution).
GUS and LUC assays
Whole seedlings or various tissues were collected for GUS staining in the staining buffer (50 mM sodium phosphate at pH 7.0, 0.5 mM K3Fe[CN]6, 0.5 mM K4Fe[CN]6, 10 mM EDTA, 0.1% [v/v] Triton X-100, and 0.5 mg mL−1 X-gluc), and 70% [v/v] ethanol was used to remove chlorophyll.
For GUS and luciferin (LUC) activities, the samples were homogenized in liquid nitrogen and lysed in 200 µL GUS I buffer (50 mM sodium phosphate, pH 7.0, 10 mM EDTA, 0.1% [v/v] Triton X-100, and 10 mM β-mercaptoethanol). After centrifugation, 5 µL supernatant was incubated with 45 µL GUS II buffer (GUS I buffer/methanol/20 mM 4-methyl umbelliferone glucuronide [MUG] = 7/4/1) for 30 min at 37 °C in the darkness, and the reaction was terminated by addition of 200 µL 0.2 M Na2CO3. Fluorescent product 4-methylumbelliferone (MU) was measured at Ex: 360 nm and Em: 485 nm using Multiscan Spectrum (TECAN F200). The values were calibrated against a standard curve prepared with known concentrations of MU. For LUC activity, 5 µL supernatant was added to 100 µL LR buffer (20 mM Tricine, pH 7.8, 26.7 mM MgSO4, 0.1 mM EDTA, 2 mM DTT, and 5 µM ATP). After addition of 15 µL LUC, chemiluminescence was immediately measured using TECAN F200. The protein contents were detected using BSA method.
Transcriptional activity assay
The Wbox:GUS reporter plasmid was constructed by fusion the W-box and 35S mini-promoter to GUS gene. The reporter and effector plasmids were transferred separately into A. tumefaciens EHA105 and then co-infiltrated into leaves of N. benthamiana with designated combinations. When chitin treatment required, the chitin solution (200 μg mL−1) was smeared on the injection sites 3 h before the samplings. GUS and LUC activities were determined 48 h after the agroinfiltration.
Rt-qPCR analysis
Total RNAs of various tissues was extracted via the TRIzol method and treated with DNase I to remove possible DNA contaminations. Two micrograms of total RNAs were transcribed with random hexamers and oligo(dT)18 primers using Moloney murine leukemia virus (M-MLV) reverse transcriptase (Takara). The relative transcript levels were quantified using SYBR Green PCR MasterMix (Takara) and normalized to the ubiquitin gene (OsUBQ, XM_01587471). The relative expression level of each gene was analyzed using the delta–delta Ct method. Gene-specific primers used in Reverse transcription quantitative PCR (RT-qPCR) are listed in Supplemental Table S1.
Determination of metabolites
Compound extraction and determination were described previously (Liang et al. 2017). Briefly, the leaves or roots freeze-dried (approximate 25 mg for each replicate) were extracted with 90% [v/v] aqueous methanol containing 0.1% [v/v] formic acid and internal standards of 10 ng D6ABA and 0.05 ng lidocaine. After dry of the supernatants by nitrogen gas, the residues were dissolved in 0.1 mL of 90% aqueous ethanol containing 0.1% formic acid for LC-MS/MS. Chemicals were separated by a C18 column (2.1 × 150 mm, 3 μm, Phenomenex) on Agilent 1260 separation module (Agilent).
Statistical analysis
GraphPad Prism 8.0 and Microsoft Office Excel were used for statistical analysis of the numerical data. Significant differences between two groups were analyzed by using the one-tailed or two-tailed Student's t-test. Significant differences between multiple samples were determined by one-way analysis of variance (ANOVA) followed by Duncan's multiple range test with SPSS statistics software. Statistical data are provided in Supplemental Data Set 1.
Phylogenetic analysis
The full-length amino acid sequences of 10 Arabidopsis and one maize MKK proteins combine with eight rice MKK proteins were aligned by ClustalW, and the neighbor-joining tree was constructed using MEGA7.0 with 1,000 bootstrap replicates.
Accession numbers
Sequence data from this article can be found in the GenBank data libraries or Rice Genome Annotation Project Database under accession numbers showed below and also listed in Supplemental Table S1. OsWRKY11 (LOC_Os01g43650), OsWRKY31 (LOC_Os03g20550), OsWRKY45 (LOC_Os05g25770), OsWRKY76 (LOC_Os09g25060), OsMKK10-2 (LOC_Os03g12390), OsMPK3 (LOC_Os03g17700), OsMPK4 (LOC_Os10g38950), OsMPK6 (LOC_Os06g06090), OsAOS2 (LOC_Os03g12500), OsLOX2 (LOC_Os03g08220), OsAUX3 (LOC_Os05g37470), OsPIN2 (LOC_Os06g44970), OsARF12 (LOC_Os04g57610), OsARF16 (LOC_Os06g09660), OsYUCCA1 (LOC_Os01g45760), OsYUCCA4 (LOC_Os01g12490), OsGH3.1 (LOC_Os01g57610), OsGH3.2 (LOC_Os01g55940), OsGH3.3 (LOC_Os01g12160), OsGH3.4 (LOC_Os05g42150), OsGH3.5 (LOC_Os05g50890), OsGH3.6 (LOC_Os05g05180), OsGH3.7 (LOC_Os06g30440), OsGH3.8 (LOC_Os07g40290), OsGH3.9 (LOC_Os07g38890), OsGH3.10 (LOC_Os07g38860), OsGH3.11 (LOC_Os07g47490), OsGH3.12 (LOC_Os11g08340), OsGH3.13 (LOC_Os11g32520), OsKS1 (LOC_Os04g52230), OsKS4 (LOC_Os04g10060), OsPR1a (LOC_Os07g03710), OsPR1b (LOC_Os07g03600), OsPUB6 (LOC_Os06g46770), OsREIW1 (LOC_Os04g44820).
Supplementary Material
Acknowledgments
We thank Prof. Chuanzao Mao and Prof. De An Jiang (Zhejiang University) for providing OsPIN2pro:GUS seeds and DR5-GFP plasmid, respectively. We thank Dr. Qian Chen (China Agricutural University) for help of ubiquitination and providing the plasmids, Ms. Yi Sun for assistance of rice transformation, Mr. Zongliang Chen (Yangzhou University) for help of field management. We thank Prof. Sheng Yang He (Duke University), Prof. Kabin Xie (Huazhong Agricutural University), and Prof. Jun Fan (China Agricutural University) for useful discussion and suggestions.
Contributor Information
Shuai Wang, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Shuying Han, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Xiangui Zhou, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Changjiang Zhao, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Lina Guo, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Junqi Zhang, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Fei Liu, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Qixin Huo, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Wensheng Zhao, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Zejian Guo, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Xujun Chen, Key Laboratory of Pest Monitoring and Green Management, MOA, Joint Laboratory for International Cooperation in Crop Molecular Breeding, Department of Plant Pathology, China Agricultural University, Beijing 100193, China.
Author contributions
X.C. and Z.G. conceptualized and supervised the project. S.W., S.H., and X.Z. performed most of the experiments. L.G., J.Z., F.L., and Q.H. participated in some experiments. C.Z. initiated the project. Z.G. and X.C. drafted the manuscript. All authors contributed to review and editing the manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Interaction of OsWRKY31 with OsMPKs.
Supplemental Figure S2 . Autophosphorylation of OsMKK10-2 and activation of OsMKK10-2 increased phosphorylation of OsMPKs.
Supplemental Figure S3 . Identification of mkk10-2ko, mpk3ko, and w31ko mutants.
Supplemental Figure S4 . Characterization of αW31pS6 antibody and induction of OsWRKY31 accumulation by chitin.
Supplemental Figure S5 . OsWRKY31 and OsMKK10-2 altered rice disease resistance.
Supplemental Figure S6 . Phylogenetic analysis of OsMKK10-2.
Supplemental Figure S7 . Expression of OsGH3 family genes in Dex-induced OsMKK10-2 plants.
Supplemental Figure S8 . OsWRKY31 is implication in OsMKK10-2 inhibition of seedling growth.
Supplemental Figure S9 . Induction of OsMKK10-2 and activation of OsMKK10-2-regulated gene expression in favor of defense over growth.
Supplemental Figure S10 . Analysis of phytohormone contents.
Supplemental Figure S11 . Induction of OsMKK10-2 and OsWRKY31 by chitin and knockout of OsMKK10-2 and OsWRKY31 compromised chitin-induced gene expression.
Supplemental Figure S12 . Antagonistic effect of SA and MeJA to IAA on gene expression in leaves.
Supplemental Figure S13 . Analyses of OsWRKY31 phosphorylation sites.
Supplemental Figure S14 . Phosphomimetic OsWRKY31S6DS101D enhanced DNA-binding and resistance to rice blast fungus.
Supplemental Figure S15 . OsREIW1 expression, localization, and self-ubiquitination activity.
Supplemental Table S1 . Primers sequences used in this study.
Supplemental Data Set 1 . Statistical data.
Funding
This work was supported by the National Natural Science Foundation of China (U22A20463, 31972253, and 31571947), the National Key Research and Development Program of China (2016YFD0100601), and Frontiers Science Center for Molecular Design Breeding.
References
- Achnine L, Blancaflor EB, Rasmussen S, Dixon RA. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell. 2004:16(11): 3098–3109. 10.1105/tpc.104.024406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson E, Ellis B. Convergence and specificity in the Arabidopsis MAPK nexus. Trends Plant Sci. 2010:15(2): 106–113. 10.1016/j.tplants.2009.12.001 [DOI] [PubMed] [Google Scholar]
- Cai Y, Chen X, Xie K, Xing Q, Wu Y, Li J, Du C, Sun Z, Guo Z. Dlf1, a WRKY transcription factor, is involved in the control of flowering time and plant height in rice. PLoS One. 2014:9(7): e102529. 10.1371/journal.pone.0102529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Liu R, Wu Y, Wei S, Wang Q, Zheng Y, Xia R, Shang X, Yu F, Yang X, et al. . ERAD-related E2 and E3 enzymes modulate the drought response by regulating the stability of PIP2 aquaporins. Plant Cell. 2021:33(8): 2883–2898. 10.1093/plcell/koab141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Li C, Wang H, Guo Z. WRKY transcription factors: evolution, binding, and action. Phytopathol Res. 2019:1(1): 13. 10.1186/s42483-019-0022-x [DOI] [Google Scholar]
- Chen Y, Fan X, Song W, Zhang Y, Xu G. Over-expression of OsPIN2 leads to increased tiller numbers, angle and shorter plant height through suppression of OsLAZY1. Plant Biotechnol J. 2012:10(2): 139–149. 10.1111/j.1467-7652.2011.00637.x [DOI] [PubMed] [Google Scholar]
- Chen Z, Agnew JL, Cohen JD, He P, Shan L, Sheen J, Kunkel BN. Pseudomonas syringae type III effector AvrRpt2 alters Arabidopsis thaliana auxin physiology. Proc Natl Acad Sci USA. 2007:104(50): 20131–20136. 10.1073/pnas.0704901104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Wang H, Li B, Huang J, Liu X, Zhou Y, Mou Z, Li J. Increased expression of MAP KINASE KINASE7 causes deficiency in polar auxin transport and leads to plant architectural abnormality in Arabidopsis. Plant Cell. 2006:18(2): 308–320. 10.1105/tpc.105.037846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding X, Cao Y, Huang L, Zhao J, Xu C, Li X, Wang S. Activation of the indole-3-acetic acid–amido synthetase GH3-8 suppresses expansin expression and promotes salicylate-and jasmonate-independent basal immunity in rice. Plant Cell. 2008:20(1): 228–240. 10.1105/tpc.107.055657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domingo C, Andrés F, Tharreau D, Iglesias DJ, Talón M. Constitutive expression of OsGH3.1 reduces auxin content and enhances defense response and resistance to a fungal pathogen in rice. Mol Plant Microbe Interact. 2009:22(2): 201–210. 10.1094/MPMI-22-2-0201 [DOI] [PubMed] [Google Scholar]
- Gao Y, Zhang Y, Zhang D, Dai X, Estelle M, Zhao Y. Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development. Proc Natl Acad Sci USA. 2015:112(7): 2275–2280. 10.1073/pnas.1500365112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo T, Chen K, Dong N-Q, Shi C-L, Ye W-W, Gao J-P, Shan J-X, Lin H-X. GRAIN SIZE AND NUMBER1 negatively regulates the OsMKKK10-OsMKK4-OsMPK6 cascade to coordinate the trade-off between grain number per panicle and grain size in rice. Plant Cell. 2018:30(4): 871–888. 10.1105/tpc.17.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamel L-P, Nicole M-C, Sritubtim S, Morency M-J, Ellis M, Ehlting J, Beaudoin N, Barbazuk B, Klessig D, Lee J. Ancient signals: comparative genomics of plant MAPK and MAPKK gene families. Trends Plant Sci. 2006:11(4): 192–198. 10.1016/j.tplants.2006.02.007 [DOI] [PubMed] [Google Scholar]
- Hayafune M, Berisio R, Marchetti R, Silipo A, Kayama M, Desaki Y, Arima S, Squeglia F, Ruggiero A, Tokuyasu K. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc Natl Acad Sci USA. 2014:111(3): E404–E413. 10.1073/pnas.1312099111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiei Y, Ohta S, Komari T, Kumashiro T. Effificient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994:6(2): 271–282. 10.1046/j.1365-313X.1994.6020271.x [DOI] [PubMed] [Google Scholar]
- Jia W, Li B, Li S, Liang Y, Wu X, Ma M, Wang J, Gao J, Cai Y, Zhang Y. Mitogen-activated protein kinase cascade MKK7-MPK6 plays important roles in plant development and regulates shoot branching by phosphorylating PIN1 in Arabidopsis. PLoS Biol. 2016:14(9): e1002550. 10.1371/journal.pbio.1002550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JD, Dangl JL. The plant immune system. Nature. 2006:444(7117): 323–329. 10.1038/nature05286 [DOI] [PubMed] [Google Scholar]
- Kamigaki A, Nito K, Hikino K, Goto-Yamada S, Nishimura M, Nakagawa T, Mano S. Gateway vectors for simultaneous detection of multiple protein-protein interactions in plant cells using bimolecular fluorescence complementation. PLoS One. 2016:11(8): e0160717. 10.1371/journal.pone.0160717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. Auxin and the integration of environmental signals into plant root development. Ann Bot. 2013:112(9): 1655–1665. 10.1093/aob/mct229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd BN, Kadoo NY, Dombrecht B, Tekeoglu M, Gardiner DM, Thatcher LF, Aitken EA, Schenk PM, Manners JM, Kazan K. Auxin signaling and transport promote susceptibility to the root-infecting fungal pathogen Fusarium oxysporum in Arabidopsis. Mol Plant Microbe Interact. 2011:24(6): 733–748. 10.1094/MPMI-08-10-0194 [DOI] [PubMed] [Google Scholar]
- Kishi-Kaboshi M, Okada K, Kurimoto L, Murakami S, Umezawa T, Shibuya N, Yamane H, Miyao A, Takatsuji H, Takahashi A. A rice fungal MAMP-responsive MAPK cascade regulates metabolic flow to antimicrobial metabolite synthesis. Plant J. 2010:63(4): 599–612. 10.1111/j.1365-313X.2010.04264.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang D-L, Sun L, Li Q, Mao B, He Z. The systemic acquired resistance regulator OsNPR1 attenuates growth by repressing auxin signaling through promoting IAA-amido synthase expression. Plant Physiol. 2016:172(1): 546–558. 10.1104/pp.16.00129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang X, Chen X, Li C, Fan J, Guo Z. Metabolic and transcriptional alternations for defense by interfering OsWRKY62 and OsWRKY76 transcriptions in rice. Sci Rep. 2017:7(1): 2474. 10.1038/s41598-017-02643-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Liu B, Lou S, Bi H, Tang H, Tong S, Song Y, Chen N, Zhang H, Jiang Y, et al. . CHYR1 Ubiquitinates the phosphorylated WRKY70 for degradation to balance immunity in Arabidopsis thaliana. New Phytol. 2021:230(3): 1095–1109. 10.1111/nph.17231 [DOI] [PubMed] [Google Scholar]
- Liu J, Chen X, Liang X, Zhou X, Yang F, Liu J, He SY, Guo Z. Alternative splicing of rice WRKY62 and WRKY76 transcription factor genes in pathogen defense. Plant Physiol. 2016:171(2): 1427–1442. 10.1104/pp.15.01921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Chen J, Zhang Z, Ma L, Yang Z, Zhang Q, Li X, Xiao J, Wang S. MAPK Kinase 10.2 promotes disease resistance and drought tolerance by activating different MAPKs in rice. Plant J. 2017:92(4): 557–570. 10.1111/tpj.13674 [DOI] [PubMed] [Google Scholar]
- Ma H, Li J, Ma L, Wang P, Xue Y, Yin P, Xiao J, Wang S. Pathogen-inducible OsMPKK10.2–OsMPK6 cascade phosphorylates the Raf-like kinase OsEDR1 and inhibits its scaffold function to promote rice disease resistance. Mol Plant. 2021:14(4): 620–632. 10.1016/j.molp.2021.01.008 [DOI] [PubMed] [Google Scholar]
- Ma Z, Zhu P, Shi H, Guo L, Zhang Q, Chen Y, Chen S, Zhang Z, Peng J, Chen J. PTC-bearing mRNA elicits a genetic compensation response via Upf3a and COMPASS components. Nature. 2019:568(7751): 259–263. 10.1038/s41586-019-1057-y [DOI] [PubMed] [Google Scholar]
- Miao Y, Laun TM, Smykowski A, Zentgraf U. Arabidopsis MEKK1 can take a short cut: it can directly interact with senescence-related WRKY53 transcription factor on the protein level and can bind to its promoter. Plant Mol Biol. 2007:65(1-2): 63–76. 10.1007/s11103-007-9198-z [DOI] [PubMed] [Google Scholar]
- Min HJ, Cui LH, Oh TR, Kim JH, Kim TW, Kim WT. OsBZR1 turnover mediated by OsSK22-regulated U-box E3 ligase OsPUB24 in rice BR response. Plant J. 2019:99(3): 426–438. 10.1111/tpj.14332 [DOI] [PubMed] [Google Scholar]
- Navarro L, Dunoyer P, Jay F, Arnold B, Dharmasiri N, Estelle M, Voinnet O, Jones JD. A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science. 2006:312(5772): 436–439. 10.1126/science.1126088 [DOI] [PubMed] [Google Scholar]
- Pollmann S, Springer A, Rustgi S, von Wettstein D, Kang C, Reinbothe C, Reinbothe S. Substrate channeling in oxylipin biosynthesis through a protein complex in the plastid envelope of Arabidopsis thaliana. J Exp Bot. 2019:70(5): 1483–1495. 10.1093/jxb/erz015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi Y, Wang S, Shen C, Zhang S, Chen Y, Xu Y, Liu Y, Wu Y, Jiang D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012:193(1): 109–120. 10.1111/j.1469-8137.2011.03910.x [DOI] [PubMed] [Google Scholar]
- Reyna NS, Yang Y. Molecular analysis of the rice MAP kinase gene family in relation to Magnaporthe grisea infection. Mol Plant Microbe Interact. 2006:19(5): 530–540. 10.1094/MPMI-19-0530 [DOI] [PubMed] [Google Scholar]
- Rice WRKY Working Group . Nomenclature report on rice WRKY's-Conflict regarding gene names and its solution. Rice. 2012:5(1): 3. 10.1186/1939-8433-5-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi A, Kontarakis Z, Gerri C, Nolte H, Hölper S, Krüger M, Stainier DY. Genetic compensation induced by deleterious mutations but not gene knockdowns. Nature. 2015:524(7564): 230–233. 10.1038/nature14580 [DOI] [PubMed] [Google Scholar]
- Sato Y, Takehisa H, Kamatsuki K, Minami H, Namiki N, Ikawa H, Ohyanagi H, Sugimoto K, Antonio BA, Nagamura Y. RiceXPro Version 3.0: expanding the informatics resource for rice transcriptome. Nucleic Acids Res. 2013:41(D1): D1206–D1213. 10.1093/nar/gks1125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan D, Wang C, Zheng X, Hu Z, Zhu Y, Zhao Y, Jiang A, Zhang H, Shi K, Bai Y, et al. . MKK4-MPK3-WRKY17-mediated salicylic acid degradation increases susceptibility to Glomerella leaf spot in apple. Plant Physiol. 2021:186(2): 1202–1219. 10.1093/plphys/kiab108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen C, Yue R, Sun T, Zhang L, Yang Y, Wang H. OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. 2015:231: 148–158. 10.1016/j.plantsci.2014.12.003 [DOI] [PubMed] [Google Scholar]
- Shen X, Yuan B, Liu H, Li X, Xu C, Wang S. Opposite functions of a rice mitogen-activated protein kinase during the process of resistance against Xanthomonas oryzae. Plant J. 2010:64(1): 86–99. 10.1111/j.1365-313X.2010.04306.x [DOI] [PubMed] [Google Scholar]
- Shimono M, Sugano S, Nakayama A, Jiang CJ, Ono K, Toki S, Takatsuji H. Rice WRKY45 plays a crucial role in benzothiadiazole-inducible blast resistance. Plant Cell. 2007:19(6): 2064–2076. 10.1105/tpc.106.046250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh P, Sinha AK. A positive feedback loop governed by SUB1A1 interaction with MITOGEN-ACTIVATED PROTEIN KINASE3 imparts submergence tolerance in rice. Plant Cell. 2016:28(5): 1127–1143. 10.1105/tpc.15.01001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X, Li X, Zhou W, Ren Y, Wang Z, Liu Z, Tang J, Tong H, Fang J, Bu Q. Transcription factor OsWRKY53 positively regulates brassinosteroid signaling and plant architecture. Plant Physiol. 2017:175(3): 1337–1349. 10.1104/pp.17.00946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Yoshida R, Kishi-Kaboshi M, Matsushita A, Jiang C-J, Goto S, Takahashi A, Hirochika H, Takatsuji H. MAP Kinases phosphorylate rice WRKY45. Plant Signal Behav. 2013:8(6): e24510. 10.4161/psb.24510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueno Y, Yoshida R, Kishi-Kaboshi M, Matsushita A, Jiang C-J, Goto S, Takahashi A, Hirochika H, Takatsuji H. Abiotic stresses antagonize the rice defence pathway through the tyrosine-dephosphorylation of OsMPK6. PLoS Pathog. 2015:11(10): e1005231. 10.1371/journal.ppat.1005231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Wang G, Zhang C, Zhu P, Dai H, Yu N, He Z, Xu L, Wang E. OsCERK1-mediated chitin perception and immune signaling requires RECEPTOR-LIKE CYTOPLASMIC KINASE 185 to activate an MAPK cascade in rice. Mol Plant. 2017:10(4): 619–633. 10.1016/j.molp.2017.01.006 [DOI] [PubMed] [Google Scholar]
- Wang H, Hao J, Chen X, Hao Z, Wang X, Lou Y, Peng Y, Guo Z. Overexpression of rice WRKY89 enhances ultraviolet B tolerance and disease resistance in rice plants. Plant Mol Biol. 2007:65(6): 799–815. 10.1007/s11103-007-9244-x [DOI] [PubMed] [Google Scholar]
- Wang J, Zhang X, Greene GH, Xu G, Dong X. PABP/purine-rich motif as an initiation module for cap-independent translation in pattern-triggered immunity. Cell. 2022:185(17): 3186–3200.e17. 10.1016/j.cell.2022.06.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Guo M, Li Y, Ruan W, Mo X, Wu Z, Sturrock CJ, Yu H, Lu C, Peng J, et al. . LARGE ROOT ANGLE1, encoding OsPIN2, is involved in root system architecture in rice. J Exp Bot. 2018:69(3): 385–397. 10.1093/jxb/erx427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu KL, Guo ZJ, Wang HH, Li J. The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 2005:12(1): 9–26. 10.1093/dnares/12.1.9 [DOI] [PubMed] [Google Scholar]
- Xiong L, Yang Y. Disease resistance and abiotic stress tolerance in rice are inversely modulated by an abscisic acid-inducible mitogen-activated protein kinase. Plant Cell. 2003:15(3): 745–759. 10.1105/tpc.008714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu R, Duan P, Yu H, Zhou Z, Zhang B, Wang R, Li J, Zhang G, Zhuang S, Lyu J. Control of grain size and weight by the OsMKKK10-OsMKK4-OsMAPK6 signaling pathway in rice. Mol Plant. 2018:11(6): 860–873. 10.1016/j.molp.2018.04.004 [DOI] [PubMed] [Google Scholar]
- Yamada K, Yamaguchi K, Yoshimura S, Terauchi A, Kawasaki T. Conservation of chitin-induced MAPK signaling pathways in rice and Arabidopsis. Plant Cell Physiol. 2017:58(6): 993–1002. 10.1093/pcp/pcx042 [DOI] [PubMed] [Google Scholar]
- Yamaguchi K, Yamada K, Ishikawa K, Yoshimura S, Hayashi N, Uchihashi K, Ishihama N, Kishi-Kaboshi M, Takahashi A, Tsuge S. A receptor-like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe. 2013:13(3): 347–357. 10.1016/j.chom.2013.02.007 [DOI] [PubMed] [Google Scholar]
- Yamamoto Y, Kamiya N, Morinaka Y, Matsuoka M, Sazuka T. Auxin biosynthesis by the YUCCA genes in rice. Plant Physiol. 2007:143(3): 1362–1371. 10.1104/pp.106.091561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Qi M, Mei C. Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. 2004:40(6): 909–919. 10.1111/j.1365-313X.2004.02267.x [DOI] [PubMed] [Google Scholar]
- Yang Z, Ma H, Hong H, Yao W, Xie W, Xiao J, Li X, Wang S. Transcriptome-based analysis of mitogen-activated protein kinase cascades in the rice response to Xanthomonas oryzae infection. Rice. 2015:8(1): 4. 10.1186/s12284-014-0038-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Zhong S, Li Q, Zhu Z, Lou Y, Wang L, Wang J, Wang M, Li Q, Yang D. Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biotechnol J. 2007:5(2): 313–324. 10.1111/j.1467-7652.2007.00243.x [DOI] [PubMed] [Google Scholar]
- Zhang J, Peng Y, Guo Z. Constitutive expression of pathogen-inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 2008:18(4): 508–521. 10.1038/cr.2007.104 [DOI] [PubMed] [Google Scholar]
- Zhang X, Dai Y, Xiong Y, DeFraia C, Li J, Dong X, Mou Z. Overexpression of Arabidopsis MAP kinase kinase 7 leads to activation of plant basal and systemic acquired resistance. Plant J. 2007:52(6): 1066–1079. 10.1111/j.1365-313X.2007.03294.x [DOI] [PubMed] [Google Scholar]
- Zhang Z, Li J, Li F, Liu H, Yang W, Chong K, Xu Y. OsMAPK3 phosphorylates OsbHLH002/OsICE1 and inhibits its ubiquitination to activate OsTPP1 and enhances rice chilling tolerance. Dev Cell. 2017:43(6): 731–743.e5. 10.1016/j.devcel.2017.11.016 [DOI] [PubMed] [Google Scholar]
- Zhou X, Zhao C, Guo Z. Rice MAP kinase kinase MKK10-2 triggers cell death and H2O2 production in Nicotiana benthamiana. Acta Phytopathol Sin. 2017:47(6): 767–775. 10.13926/j.cnki.apps.000035 [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.