Abstract
This review is an attempt to establish concepts of splicing and alternative splicing giving proper relevance to introns, the key actors in this mechanism. It might also work as a guide for those who found their favorite gene undergoes alternative splicing and could benefit from gaining a theoretical framework to understand the possible impacts of this process. This is not a thorough review of all the work in the field, but rather a critical review of some of the most relevant work done to understand the underlying mechanisms of splicing and the key questions that remain unanswered such as: What is the physiological relevance of alternative splicing? What are the functions of the different outcomes? To what extent do different alternative splicing types contribute to the proteome? Intron retention is the most frequent alternative splicing event in plants and, although scientifically neglected, it is also common in animals. This is a heterogeneous type of alternative splicing that includes different sub-types with features that have distinctive consequences in the resulting transcripts. Remarkably, intron retention can be a dead end for a transcript, but it could also be a stable intermediate whose processing is resumed upon a particular signal or change in the cell status. New sequencing technologies combined with the study of intron lariats in different conditions might help to answer key questions and could help us to understand the actual relevance of introns in gene expression regulation.
IN A NUTSHELL.
Introns are the key actors in splicing. Alternative splicing can be defined as the differential selection of introns in different transcripts, leading to the generation of distinct RNA isoforms from a single gene. Intron retention is the most frequent alternative splicing event in plants. Transcripts with introns retained may have completely different fates, from being kept in the nucleus to being actively translated. This review explores the mechanisms underlying these different outcomes of alternative splicing.
Introduction
Eukaryotic genomes contain large amounts of so-called junk DNA interrupting informative and/or coding regions (Fagundes et al., 2022). Among this junk DNA, eukaryotic genes contain introns that separate the regions that constitute mature RNAs, the exons. Hence, introns are often acknowledged as noncoding and/or noninformative sequences that interrupt the coding messages and/or separate informative regions.
Though all eukaryotes have genes with introns, there is a significant variation in their frequency, length, base composition, and distribution between different phyla and species. Humans, similar to other mammals, have long introns of 5,850 nucleotides (nt) on average, with 45% Guanine and cytosine (GC) richness and an average frequency of eight introns per gene. These numbers are somewhat smaller in Drosophila melanogaster, where the average length of an intron is about 1,531 nt, and they are less frequent and have lower GC richness (36.5%). By contrast, plants introns are much smaller, for example averaging about 164 nt in Arabidopsis (Arabidopsis thaliana) and 396 nt in rice (Oryza sativa). Their frequency in plant genomes and GC content is also lower than mammalian genomes (e.g. GC contents of about 33% and 38%, in Arabidopsis and rice, respectively; see the complete dataset in Zhu et al., 2009). The correct identification and removal of introns by the spliceosome is a central conserved step during gene expression in all eukaryotes. One of the key consequences of this noncontinuous exon–intron structure is the formation of alternative splicing isoforms (Gehring and Roignant, 2021). This process allows the generation of different mature RNAs or messenger RNAs (mRNAs) from a single gene, expanding the transcriptome and the proteome of an organism (Keren et al., 2010).
Splicing: finding the introns to eliminate
The term splicing means to join or to connect two strands by their ends. When considering the splicing of RNA molecules, the mechanism involves the removal of an internal region of the RNA, defined as the intron, and the consequent connection of the neighbor regions, the exons. Since introns are the units of splicing, it is relevant to note that there are four types of introns: (1) transfer RNA introns are found in eukaryotes and archaea and their splicing is catalyzed by protein enzymes, (2) group I introns, which are autocatalytic and found in organelles and ribosomal RNA genes of eukaryotes, (3) group II introns, which are found in bacteria and are self-spliced in a two-step process, and (4) spliceosomal introns, which are found in eukaryotes and are processed by a complex cellular machinery known as the spliceosome (Irimia and Roy, 2014). In this review, I will focus on this latter group. Splicing and alternative splicing were discovered at the same time (Chow et al., 1977; Berget et al., 1977; Kornblihtt et al., 2013). However, it is easier to understand alternative splicing if we first comprehend splicing.
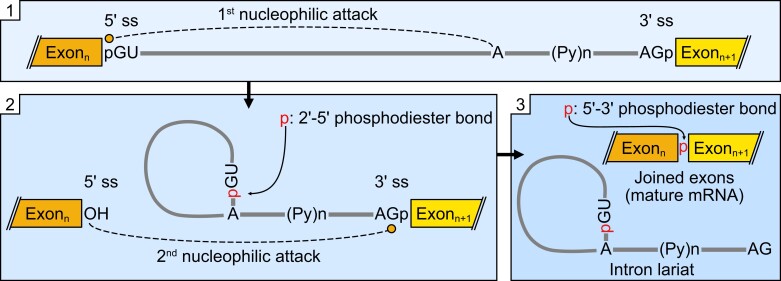
The splicing process requires two consecutive reactions, first a nucleophilic attack of the adenine at the branching point on the 5′ splice site (ss) upstream in the intron, to form a branched 2′–5′ phosphodiester bond; second, the 3′ OH of the upstream exon carries out a nucleophilic attack at the 3′ ss of the intron to ligate the exons in a 5′ –3′ phosphodiester bond. As a consequence, the intron is released as a lariat and the exons are joined together, giving rise to the mature RNA molecules (Figure 1; see also Black, 2003; Smathers and Robart, 2019). These reactions are similar in group II introns and spliceosomal introns with a key difference, while group II introns are self-spliced the spliceosomal introns are processed, as the name implies, by the spliceosome. Importantly, there are two different spliceosomes that are characterized by their respective protein subunits. The major U2-dependent spliceosome catalyzes the splicing of most introns (>99%) while the minor U12-dependent spliceosome is responsible for a small remnant, though there are some exceptions (Akinyi and Frilander, 2021; Larue et al., 2021). In this framework, if a region is recognized by the spliceosome and is excised, then this region is, without question, an intron. However, introns are often defined a priori as “noncoding sequences” interspersed in coding regions of the precursor mRNAs that are then excised by the spliceosome (William Roy and Gilbert, 2006; Rogozin et al., 2012). As Alberto Kornblihtt, a renowned expert in the splicing field, might say “Nope, this is not right. This is wrong!” (Kornblihtt, 2021): introns cannot be defined by their lack of coding capacities, as there are also exons that lack coding capacity and there are introns that can be part of the translated message. Introns can be found interrupting coding exons, but they are also part of the untranslated regions (UTRs) along with exons that are not coding, and exons and introns are also present in long noncoding RNAs (yes, completely noncoding). In addition, some introns that are retained in mature mRNAs actually have coding capacities. So, how can we define an intron? Introns (and exons) are defined by the spliceosome. As introduced above, introns are RNA sequences excised during splicing, while exons are regions that are joined together to form the mature RNA molecule. Gilbert proposed the term intron in 1978: “The notion of the cistron, the genetic unit of function that one thought corresponded to a polypeptide chain, now must be replaced by that of a transcription unit containing regions which will be lost from the mature messenger—which I suggest we call introns (for intragenic regions)—alternating with regions which will be expressed—exons” (Gilbert, 1978). If we consider that “expressed” means to be part of the mature mRNA, then this is, to my knowledge, the most comprehensive and correct definition of splicing. Alternative splicing experts might argue that this definition fails to account for alternative splicing isoforms, since a region that remains in the mRNA in one isoform can be part of an intron, or a whole intron, in another isoform. Although this could mean that introns are not defined unequivocally, if we consider introns and exons to be isoform-specific, the definition given by Gilbert is appropriate even when considering alternative splicing.
Figure 1.
The splicing mechanism. The splicing process follows two consecutive reactions, first a nucleophilic attack of the adenine at the branching point on the upstream 5′ ss generating a branched 2′–5′ phosphodiester bond; second, a nucleophilic attack of the 3′ OH of the upstream exon to the 3′ ss that ligate the exons in a 5′–3′ phosphodiester bond. The excised intron is released as a lariat and the exons are joined together giving rise to the mature RNA molecules. Nucleophilic attacks are schematized as dashed lines.
The splicing machinery is mostly conserved among eukaryotes, and introns are pervasive in their genomes, but why do they exist? This question arose as soon as splicing was discovered, by the already mentioned Gilbert (1978). Because introns generally belong to noncoding regions in DNA that are mostly absent in mature RNAs, they had been considered part of the junk DNA, lacking any relevant information (Poverennaya and Roytberg, 2020). In this sense, introns are often considered as mutational buffers that protect coding sequences from random mutations. This view is not well-sustained, however, as most mutagens generate errors or changes in the DNA sequence with a given frequency (1 base per X bases) that correlates with their dose. Having more nucleotides as mutational substrates does not alter that frequency, it only affects the total number of mutated bases, so coding sequences are expected to be equally affected by mutation with or without extra noncoding sequences. Hence, there must be other possible functions, as having introns is expensive for cells, which must transcribe the intron sequences and then remove them to generate mature mRNA (Jo and Choi, 2015). Relevant putative functions of introns are usually divided into the following categories: (1) functions associated with splicing; (2) generic functions of noncoding DNA sequences; (3) storage of regulatory elements; (4) location of nested protein-coding genes; and (5) hot-spots of intra-genic recombination (Rogozin et al., 2012; Jo and Choi, 2015; Poverennaya and Roytberg, 2020). These have been extensively reviewed (see Jo and Choi (2015) for a complete list with supporting studies), so in this review I focus on those functions that seem to be well supported, and I introduce and discuss others that are commonly ignored, considering the particular features of plant genomes and transcriptomes.
The advantage (?) of having introns
Introns are a hallmark of eukaryotic genomes, as spliceosomes and spliceosomal introns are present in every sequenced eukaryote (Collins and Penny, 2005; Martin and Koonin, 2006) and are absent in prokaryotes. In prokaryotes, where translation is cotranscriptional, introns would be a great burden. In other words, “the coexistence of DNA and functional ribosomes in the same cell compartment would allow ribosomes to translate unspliced premessengers” (Cavalier-Smith, 1991). Ergo, the nuclear compartment is highly relevant when analyzing the existence and function of spliceosomal introns. In this sense, it is important to note that for an mRNA to be translated, introns must be removed in order for the transcript to be exported to the cytosol. This points to a potential key function for introns that is linked to splicing: to regulate the timing and intensity of gene expression. Interestingly, intron retention is a major phenomenon in plants (Ner-Gaon et al., 2004; Marquez et al., 2012, 2015; Zhang et al., 2022). Transcripts that retain introns are mostly kept inside the nucleus (Göhring et al., 2014; Jia et al., 2020; Fuchs et al., 2021), which mostly prevents these molecules from being translated. There are, however, specific subsets of introns that manages to escape this obstacle, and the transcripts that contain them are efficiently exported and translated (Ner-Gaon et al., 2004; Marquez et al., 2015).
Intron retention—the presence of an intron in a processed transcript (i.e. except for that intron perhaps) could be an outcome of alternative splicing, but could also be a way to stall the complete processing of a transcript in order to regulate the timing of gene expression (read expression here as the generation of the protein-coding transcript; see Boothby et al., 2013; Buccitelli and Selbach, 2020). In fact, Jia and collaborators suggested that chromatin-tethered post-transcriptional splicing is a major contributor to the widespread intron retention in plants, and furthermore, they indicate this could be a mechanism to produce functional mRNAs when needed (Boothby et al., 2013; Jia et al., 2020). Such a mechanism was previously described in animal cells and was designated as intron detention (Shalgi et al., 2014; Boutz et al., 2015). Boutz and collaborators identified thousands of detained introns in human and mouse cell lines as well as in the adult mouse liver. The transcripts with detained introns can have half-lives of over an hour yet remain in the nucleus. They are not subjected to nonsense mediated mRNA decay (NMD) and many of the detained introns are evolutionarily conserved (Boutz et al., 2015). In plants, intron retention isoforms are also not degraded by NMD, and this is mainly due to their nuclear retention that make them unavailable to this degradation machinery (Kalyna et al., 2012; Leviatan et al., 2013; Göhring et al., 2014; Fuchs et al., 2021). A remarkable extreme scenario is that of human anucleate platelets, where splicing of specific introns of accumulated precursor mRNAs occurs in response to cellular activation in an environment that lacks any nuclear regulation or de novo transcription (Denis et al., 2005; Schwertz et al., 2006). Interestingly, though intron retention is generally scientifically neglected in animals, data from more than 40 diverse human and mouse cell and tissue types shows that intron retention affects transcripts from most genes (Braunschweig et al., 2014). Moreover, the authors of the study claim that “these retained introns act widely to functionally “tune” transcriptomes by further reducing the expression of relatively low abundance transcripts that often lack physiological relevance to the cells and tissues they are detected” (Braunschweig et al., 2014). Generally speaking, the presence of an intron in an mRNA seems to be linked to fine-tuning of gene expression in plants (as well as in animals). It is relevant to mention that plant introns are short and, hence, less prone to contain regulatory sequences or signals as those for cleavage and polyadenylation, while mammalian introns are much longer, which increases the chances to find these kinds of regulatory sequences or signals, and reduces the probability of finding the whole intron retained in a transcript.
Introns, and transcripts that retain introns, may have more than the mere passive functions associated with nuclear retention or detention that can be resumed by post-transcriptional splicing. On the one hand, intron lariats (see Figure 1), one of the final products of splicing, were associated with microRNA (miRNA) biogenesis. Using an RNA debranching enzyme mutant in Arabidopsis (dbr1), Li and colleagues showed that the accumulation of intron lariats reduces miRNA accumulation. This seems to be linked to these lariats’ secondary structure, which resembles a primary miRNA that can act as a decoy that associates with the DCL1/HYL1 dicing complex, sequestering it and preventing this complex from processing proper miRNAs (Li et al., 2016). Although the evidence was obtained from a weak mutant allele of dbr1 (the null mutant is embryo lethal in both plants and animals) and from the overexpression of particular lariats, both of which are artifactual molecular phenotypes, the results indicate that if there is such a condition that increases the amounts of any lariat, or of several lariats, then these would interfere with miRNA biogenesis, thereby impacting different plant responses. On the other hand, nuclear retention of transcripts was recently shown in human cells as a mechanism that might support robust mRNA concentration homeostasis. This is based on negative regulation of RNA polymerase II (RNAPII) transcription by the accumulation of nuclear mRNAs (Berry et al., 2022). If this mechanism is conserved in plants, then the accumulation of transcripts (those mRNAs that retain introns) in the nuclear compartment may be another way, directly and actively, to globally regulate gene expression.
In addition to the general function(s) of introns, they can offer a significative benefit under certain conditions, as systematically removing all known introns from budding yeast (Saccharomyces cerevisiae) genes, renders organisms that poorly adapt to starvation conditions regardless of host-gene function (Parenteau et al., 2019). Similarly, a less known role of introns, most conspicuously studied in Arabidopsis, is the enhancement of gene expression. This process generally involves introns located near the 5′-end of the genes associated with conserved sequences that may have a function at the DNA level (Parra et al., 2011; Rose et al., 2011). More interestingly, in some cases intron-mediated enhancement of gene expression requires a functional intron (i.e. the splicing itself). This direct effect of splicing-competent introns on gene expression is a bona fide case of intron-mediated enhancement and provides evidence of crosstalk between splicing and transcription (Moabbi et al., 2012; Dwyer et al., 2021). Moreover, as exon junction complexes deposited on spliced mRNAs enhance translation, the splicing affects the output of a gene at different levels (Lee et al., 2009). Finally, I would like to draw the attention toward a poorly considered and less evaluated option. This is the possibility that a transcript that is retained in the nucleus could be acting as a regulatory long noncoding RNA (lncRNA) that, as an example, titrates a splicing factor or any other RNA binding protein. This possibility is partially supported by recent findings in animal cells showing that upon ultraviolet (UV) irradiation there is an increase of a noncoding transcript of a gene (ASCC3) that counteracts the function of the protein-coding isoform (Williamson et al., 2017).
Therefore, introns are clearly advantageous, and this is most likely related to the addition or synergy between different effects: they are good places for recombination at the DNA level; they have different roles in regulating gene expression, some that are passive and others (far more interesting) that could be active; and they are fundamental to the phenomenon of alternative splicing. This process leads to a substantial expansion of the transcriptome as well as potential proteome diversification and may be an important component of organismal responses and adaptation to the environment (Staiger and Brown, 2013).
The blue pill or the red pill? The fates of splicing alternatives
Now that we understand the process and ramifications of splicing, we can start arguing about its alternatives. Alternative splicing means exactly that: to have options to the process of recognition of intronic (and exonic) sequences, or in other words, the spliceosome might recognize the same region as an intron in some conditions or as part of an exon in others. The spliceosome can eliminate a particular sequence in response to a stimulus, or it could retain that region in the mature RNA molecule in other cases. The first part of this review focused on one kind of alternative splicing event, intron retention. This is, in fact, a very special member of much more heterogeneous set, as we will see next.
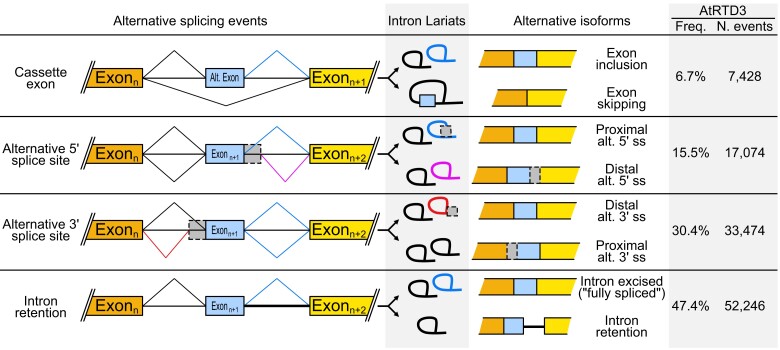
The alternative splicing mechanism can be viewed as the differential recognition and selection of 5′ ss and 3′ ss by the spliceosome (Kornblihtt et al., 2013; Vaquero-Garcia et al., 2016; Dent et al., 2021). Considering this, we can build a simple list of all possible types of alternative splicing events (Figure 2). These are: (1) cassette exons, leading to exon inclusion or skipping, (2) alternative 5′ ss, that increase or reduce the length of an exon in its 3′ end, (3) alternative 3′ ss, that increase or reduce the length of an exon in its 5′ end, and (4) the above-mentioned case of intron retention, the complete retention of an intron in the mature RNA. However, as you may guess, there are different combinations of all these options and there are also particular sub-types with specific features. As shown in Figure 2, when considering internal alternative splicing events, intron retention is the most prevalent, with almost 50% of relative frequency. It is followed by alternative 3′ ss, then alternative 5′ ss, and finally exon skipping, according to AtRTD3, the most comprehensive dataset of Arabidopsis (Zhang et al., 2022). Moreover, this curated transcriptome recognizes 40,929 total genes, and 27,199 that are multi-exonic (with at least one intron). Considering this, almost half of the total annotated genes (nearly 50%), and about 75% of the multi-exonic genes, have more than one isoform. However, these numbers also include alternative transcription start sites (TSS) and polyadenylation sites. If considering only alternative splicing events, then the frequency is similar to what was previously reported for Arabidopsis, around 60% (Marquez et al., 2012; Zhang et al., 2017). These numbers indicate that alternative splicing events are common in this model organism. In other plant species, reports indicate that a frequency greater than 60% or 80% of intron-containing genes undergo alternative splicing in soybean (Glycine max) and maize (Zea mays), around 50% in rice, and 24% in wheat (Triticum aestivum L.). In all the analyzed cases, intron retention events are the most conspicuous (see Li et al., 2021 for further details). Hence, there are some linked emerging questions: Are the different alternative splicing isoforms coding for different proteins with distinctive domains or functions? If not, what are the fates or possible functions of the different mRNA isoforms?
Figure 2.
Major types of alternative splicing events and their frequency in Arabidopsis. Alternative splicing events can be divided into four different general categories: (1) cassette exons are alternative splicing events where an exon is included/excluded in/from the mature RNA, (2) alternative (Alt.) 5′ ss corresponds to the differential recognition and usage of different alternative 5′ ss giving place to mature RNAs that have a longer/shorter exon (in its 3′ end), (3) similar to the latter, alternative 3′ ss corresponds to the differential usage of 3′ ss in competition giving isoforms with a longer/shorter exon (in the 5′ end of the corresponding exon in this case), and (4) intron retention is the permanence of a region that can be identified as an intron and excised in other isoforms, it is a lack of recognition and/or usage of the involved 5′ and 3′ ss. As the fundamental unit of splicing is the intron, the figure includes the ^ to visualize the recognized and used alternative splice sites that results in each isoform and the corresponding lariats. Alternative splicing isoforms and common labels are depicted, and the frequency and number of each event according to the AtRTD3 transcriptome are shown (Zhang et al., 2022).
Let us try to answer these questions by using the most complete and highly curated dataset available for plants, the Arabidopsis transcriptome AtRTD3 (Zhang et al., 2022). If intron retention events are 47.40% of total internal alternative splicing events, then the remanent 52.60% is constituted by cassette exons and alternative 5′ and 3′ ss. The vast majority of the alternative splicing events fall into the coding sequence and they may consequently have an impact on the proteome (Zhang et al., 2017, 2022; Marquez et al., 2012). On the other hand, 17.4% of the multi-exon protein-coding genes are predicted to generate NMD-targeted isoforms (Kalyna et al., 2012; Drechsel et al., 2013). Since introns often have termination codons and favor the retention of the transcripts that contain them in the nucleus, we could ignore them as contributors to protein diversity. Removing these important events and considering NMD, we can deduce that only about 40% of the total alternative splicing events could have an impact on the proteome. In this sense, Yu and colleagues combined uniquely aligned reads from transcriptome and translatome to ensure the comprehensive and reliable detection of alternative splicing events that could be translated. Using stringent parameters for intron identification, they found between 22,541 and 22,896 total genes, with 17,652–17,670 (77%–78%) that are intron-containing. Depending on the pipeline used for transcript identification, between 34.9% and 40.1% of the alternative splicing events were found associated with polysomes (Yu et al., 2016). It is important to mention that these results are somewhat misleading, as the authors did not actually evaluate whether different isoforms of a same gene are translatable. It could be the case that only a minor proportion of the polysome-associated events is linked to individual transcriptional units, whilst the vast majority of the associated events are actually represented in the translatome by the reference isoforms, that are commonly coding and used to build the annotated gene models. As deduced above, although intron retention is the most abundant alternative splicing type, it is also more likely to be excluded from the translatome (Yu et al., 2016). Importantly, our deduction relies on a flawed assumption, that all introns are the same. In fact, different introns can confer distinct properties to the transcripts in which they are embedded.
Up to this point in the review, introns have been considered as one big category containing all those RNA sequences or regions that can be recognized and excised by the spliceosome, though some of these can also be retained in certain conditions. However, not all introns are the same. Besides their capacity of being recognized and excised by the spliceosome, introns can have completely different features and provoke different fates for the RNAs that contain them. Intron retention, defined as the permanence of an intron in an otherwise processed transcript, can be developmentally or physiologically regulated, constituting a relevant layer in gene expression control (Boutz et al., 2015; Jacob and Smith, 2017). The effect of a retained intron in a transcript is a direct consequence of its relative location within the mature RNA and of its specific features (mainly associated with the cis-elements constituting the intron).
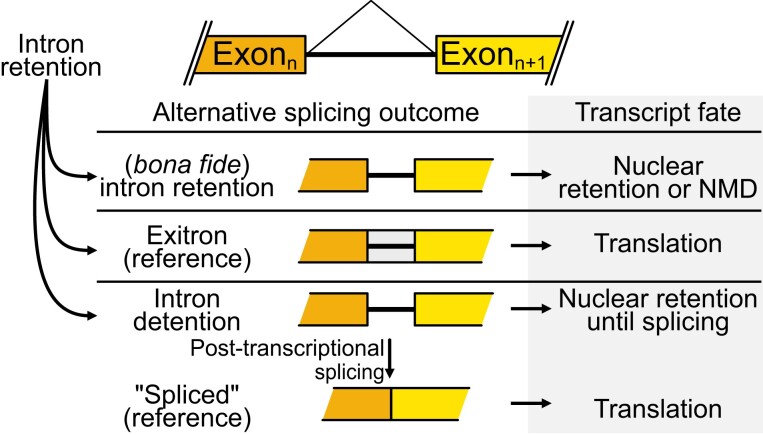
Regarding the relative location, an intron in the UTR or in the coding sequence will lead to different fates for the retention variants. Interestingly, introns at 5′ UTRs can modulate the output of a gene by intron-mediated enhancement of gene expression (Parra et al., 2011). Intron-mediated enhancement of gene expression may reflect a fundamental feature of eukaryotic gene expression since it has been observed in diverse phyla, including plants. As explained above, since intron sequences can have an effect at the DNA or at the RNA level, Rose and colleagues used sequences from introns that stimulate gene expression in both sense and anti-sense orientations. Both orientations were stimulating, suggesting that the enhancement occurs at the DNA level (Parra et al., 2011; Rose et al., 2011). This favors the idea of an enhancer sequence present in these introns. However, intron-mediated enhancement of expression is often related to the splicing of the introns involved (Moabbi et al., 2012). Hence, modulating the splicing/retention of those introns is a way to control the total output of the gene. More broadly, introns in the 5′ UTRs can also insert an upstream open reading frame (uORF), or other structural features with consequences for translational efficiency (Jacob and Smith, 2017). Intron retention events in the 3′ UTR can introduce cis elements that affect the stability or translational efficiency of the corresponding mRNA. Moreover, if the intron in the 3′ UTR is more than ∼55 nt downstream the natural stop codon, while its excision could lead to NMD, its retention can stabilize the mRNA (Bicknell et al., 2012). On the other hand, if the intron is in the coding sequence, then it is key to consider its particular properties. As anticipated above, many introns contain stop or termination codons, hence their retention can generate transcripts with premature termination codons (PTCs). The presence of PTCs is often coupled to cytoplasmic degradation of the transcripts via the NMD pathway (Kalyna et al., 2012; Drechsel et al., 2013; Lykke-Andersen and Jensen, 2015). As shown in Figure 3, transcripts with retained introns can also be stored in the nucleus to be post-transcriptionally spliced in response to appropriate signals, giving place to functional mRNAs that are consequently exported and translated (Boothby et al., 2013; Mauger et al., 2016; Jia et al., 2020). On the other hand, these transcripts can be kept in the nucleus and degraded in this compartment afterwards (Pendleton et al., 2018), being dead ends or, similarly, they can be post-transcriptionally spliced to give rise to fully processed transcripts containing PTCs that are then subjected to NMD, as another way to remove them from the nucleus but actively degrade them (Boutz et al., 2015). In all these cases, intron retention serves as a diversion. This detour can be temporary, as in the case of intron detention events where splicing resumes upon a signal; or absolute, as in the case of those transcripts with retained introns that are degraded in the nucleus or by cytosolic NMD. In both cases, intron retention would be regulating the final transcriptional output of functional variants (i.e. translated) of a gene (Figure 3).
Figure 3.
Game of introns. During splicing, introns are excised; however, some introns are retained and are present in the otherwise completely processed and mature RNA. Depending on the features of the retained intron, and on the characteristics of retention (definitive or temporary), the alternative splicing outcome has different fates. Exitrons are reference isoforms that are exported and translated as the (not-so-appropriately-called) “fully spliced”, the commonly coding and fully processed isoforms. Canonical intron retention, where the introns likely include stop codons in their sequence, leads to nuclear retention or to NMD in case of export to the cytoplasm. Intron detention is a specific case of intron retention that is temporary, as the splicing resumes (post-transcriptionally) after a signal or activation of the cell. See the text for further details.
Can we think of an active function for intron retention isoforms? Although this alternative splicing outcome seems to have mostly passive functions, it could also give rise to a hitherto unexplored possibility, the generation of isoforms that have a regulatory role, as such long noncoding RNAs (Schmitz et al., 2017; and although not a regular case of intron retention, see also Williamson et al., 2017). However, intron retention variants could also actively affect translation efficiency and mRNA stability, and furthermore, they can directly contribute to proteome diversity. In this sense, there is a category of introns that have features making them more like exons than other introns; these are known as exitrons, as named by Yamile Marquez, Andrea Barta, Maria Kalyna, and collaborators when originally characterized; first as cryptic introns and then as exonic introns (Marquez et al., 2012, 2015). As the authors nicely described, exitrons constitute intra-exonic regions that, when retained, never introduce any stop codons as these are fully coding sequences. In this sense, exitrons are always part of the reference isoforms. Among the many specific features of these exitrons, like the higher GC content compared to other introns and their full-coding capacity, of fundamental relevance is a lack of sequence determinants that could cause their nuclear retention. Captivatingly, their name actually includes this fundamental property, as transcripts with exitrons are exported out of the nucleus. So, even though exitrons were detected as a subset of retained introns, called cryptic introns in the first report, they are clearly distinguishable from “canonical” introns. About half of the 1,002 Arabidopsis and 923 human exitrons have sizes of multiples of 3 nt, so they keep the translation reading frame either excised or retained. Moreover, their alternative inclusion/excision affects the presence of protein domains, disordered regions, sites of various PTMs, signal peptides, etc. clearly contributing to protein diversity. Consistently with these exonic features, those exitrons that can be evolutionarily compared with ancestral states, showing that they originated from alternative exons (Marquez et al., 2015).
The spliceosome recognizes introns. That is its job, and it is not an easy task in some conditions. This process (like alternative splicing) occurs mostly cotranscriptionally (Beyer and Osheim, 1988; Kornblihtt et al., 2013; Zhu et al., 2020). However, some introns are not efficiently or rapidly spliced, and they remain in the otherwise completely processed transcript (Ner-Gaon et al., 2004; Braunschweig et al., 2014; Boutz et al., 2015; Jia et al., 2020). As explained several times in this text, these transcripts are defined as intron retention isoforms (Jacob and Smith, 2017), however, this definition encompasses several different types of introns that can be retained. Braunschweig, Irimia, Blencowe, and colleagues proposed the existence of three different types of introns, based on their particular features/sequence, types A–C: “Type A are ancestral introns flanked by constitutive exons, Type B arose by “intronization” of ancestral exonic sequence, and Type C are located adjacent to one or more alternative exons that may or may not be conserved between species.” (Braunschweig et al., 2014). Interestingly, Irimia characterized type B introns in Caenorhabditis elegans by their intra-exonic location, high GC content, shortest length, weakest splice sites, and highest retention (Irimia et al., 2008). These are all features shared with exitrons (Marquez et al., 2012, 2015); in fact, type B introns and exitrons are most likely the same type of intron. Among these features underlies the difference that determines the efficient export of transcripts containing exitrons from the nucleus, in contrast to that of other intron retention isoforms. The coding capacity could be of relevance, however it is more likely that their nuclear export or retention is regulated by the 5′ and 3′ ss sequences (Palazzo and Lee, 2018). Interestingly, Legrain and Rosbash concluded in 1989 that early acting factors of the spliceosome interact with the 5′ ss and the branch-point sequence to commit the precursor to splicing, thereby preventing its transport to the cytoplasm (Legrain and Rosbash, 1989).
Intron retention is the trickiest type of alternative splicing event. It is completely different from the other categories in the sense that there are no splice sites in competition. It is just an intron singing, Should I stay or should I go?. When considering cassette exons, alternative 5′ ss or alternative 3′ ss, there are always two or more splice sites that compete for the recognition by the spliceosome. This competition leads to the selection of the strongest, in terms of sequence, or the most favored one according to other regulators, such as RNAPII transcription elongation, chromatin modifications, and splicing factors (Black, 2003; Kornblihtt et al., 2013). Interestingly, when considering intron retention, there is only one 5′ ss and one 3′ ss to consider, but the mechanism is such that in some cases these splice sites are recognized, and the intron is correctly excised, while in others there is no recognition of these sites, and the intron remains in the mature RNA. Hence, we need to consider splicing efficiency, spliceosome efficacy, recognition of the different sites versus usage, and the temporal dynamics of splicing and export, as being of key relevance to the general cotranscriptionality of the splicing and the possibility of posttranscriptional processing of some introns. Some introns may be retained until the RNA is completely degraded, meaning it is the final state of that molecule. This is the case with bona fide intron retention; but it is also the case of exitrons, as transcripts with exitrons are actively translated and then degraded as any other coding mRNA of the cell. Other introns can be excised post-transcriptionally, in response to a signal or a change in the cell. In this case the intron retention variant can be considered as an intermediate. Thus, it can be important to determine whether an intron is retained as an outcome of splicing or as an intermediate and regulated step in the processing of a transcript. Novel approaches and technologies might help us to clearly distinguish these different pathways and fates of introns to gain further knowledge of their relevance in genomes and in gene expression regulation.
Conclusion and future perspectives
One fundamental take-home message of this review should be the importance of clear definitions of exons and introns. Although we can guess, based on sequence, if a particular region of a precursor RNA is an exon or an intron (i.e. 5′ ss, 3′ ss, GC richness, coding capacity, etc.), the spliceosome knows better. Whatever the sequence, if it is excised by this complex machinery, it is indeed an intron, and the regions upstream and downstream of the excised segment are exons. Importantly, all the internal exons (i.e. from the second till the penultimate) are defined by splicing; however, the terminal exons are different. The first exon is defined by the transcription start site (TSS) on the 5′ end, and by splicing in its 3′ end; while the last exon is defined by splicing at its 5′ end, and by cleavage and polyadenylation at its 3′ end. Interestingly, a region that is recognized and excised as an intron in one isoform of a gene can be part of the mature mRNA of another isoform of the same gene. This is the basis of alternative splicing. Transcripts with different termini that arise from alternative TSSs or alternative polyadenylation sites should not be confused as alternative splicing isoforms.
The alternative splicing process generates different transcripts from the same gene. It is normally accepted that this results in an expansion of the proteome, however, the contribution of the different alternative splicing isoforms to protein isoforms is still elusive in plants, and also in animal systems. A study in moss (Physcomitrium patens) suggests that alternative splicing does not substantially modify the proteome (Fesenko et al., 2017), as only 85 isoform-specific peptides, representing 25 differentially alternatively spliced genes, were found in this organism. Another study, using Arabidopsis, allows the authors to conclude that the low numbers of alternatively spliced events that can be confirmed using proteomics datasets are the result of a relatively low depth of sampling in the experiments (Severing et al., 2011). Interestingly, using a TRAP-seq (translating ribosome affinity purification followed by sequencing) approach, Tian and colleagues identified domain-specific alternative splicing events, reflecting differential contribution of selected alternative splicing events during shoot domain specification in Arabidopsis. Precisely, they identified 751 genes whose isoforms show domain-specific enrichment in the translatome data (Tian et al., 2019). Similarly, an evaluation of polysome-bound mRNAs estimated that 35% of the alternative splicing events are most likely translated (Yu et al., 2016). Although intron retention is the most represented event in the transcriptome, its frequency in the translatome is substantially reduced. Of course, these numbers must be considered in light of all the different potential features and fates of introns discussed above. When analyzing the different possibilities, exitrons are among the events that significantly contribute to the proteome (Yu et al., 2016; Marquez et al., 2015), while the more canonical events of intron retention contribute much less. Similar to exitrons, cassette exons and alternative 5′ ss and 3′ ss events that are expressed and identified in the transcriptome, are mostly present in the translatome (Yu et al., 2016).
Intron retention is the most conspicuous type of alternative splicing in plants, and is also relatively common, although often ignored, in animals. As explained above, aside from the case of exitrons, intron retention does not contribute to the proteome, begging the question of why it persists. This review is an attempt to answer these questions, although it raises others, including the key question of whether a retained intron is a dead end or a yet-to-be-processed region in an mRNA. Deep sequencing methods that analyze intron lariat populations under different conditions could be used to explore this question. Since splicing and alternative splicing are all about introns and their recognition, analyzing the intron lariats is arguably the best way to study the regulation of these processes (Zhang et al., 2019). In fact, intron lariats together with their mature mRNAs are the actual products of splicing (see Figure 1), hence they could be directly and accurately linked to changes in splicing decisions. This kind of perspective, together with the new technologies for long-read sequencing, could lead to a new revolution in our understanding of alternative splicing, its outcomes, and its functions. In the coming years it could be exciting to explore if coding genes that undergo alternative splicing, could give rise to isoforms that lack coding capacities and act as regulatory long noncoding RNAs (Williamson et al., 2017), blurring the lines between different subfields of RNA biology.
Acknowledgments
To Dante and Gri, eternal gratitude and love. I would like to thank Maria Kalyna for her kind support, careful reading, and helpful criticisms and Andrea Barta for her mentoring and help throughout this process (and always). Gracias Messi et al. por la tercera. This work was partially supported by the funding agencies and grants listed below, but it was possible mainly thanks to the Robin Hood of science, Alexandra Elbakyan. I would also like to thank all the wonderful people at IFIBYNE for generating such a great atmosphere to learn, work, and progress, not only to the scientific community but also to all the people that make our work possible with their contributions. Special thanks to my group for giving me the trust and joy of doing research together. I would like to apologize to all those colleagues whose works could not be cited owing to space constraints. Let us make science great again by making it more accessible and open.
Funding
This work was supported by the ANPCyT (Agencia Nacional de Promoción Científica y Tecnológica, Argentina, PICTs: 2017-1343, 2019-01690 and 2020-02865). E.P. is a career researcher from CONICET.
References
- Akinyi MV, Frilander MJ (2021) At the intersection of major and minor spliceosomes: crosstalk mechanisms and their impact on gene expression. Front Genet 12: 700744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berget SM, Moore C, Sharp PA (1977) Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc Natl Acad Sci USA 74(8): 3171–3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry S, Müller M, Rai A, Pelkmans L (2022) Feedback from nuclear RNA on transcription promotes robust RNA concentration homeostasis in human cells. Cell Syst 13(6): 454–470.e15 [DOI] [PubMed] [Google Scholar]
- Beyer AL, Osheim YN (1988) Splice site selection, rate of splicing, and alternative splicing on nascent transcripts. Genes Dev 2(6): 754–765 [DOI] [PubMed] [Google Scholar]
- Bicknell AA, Cenik C, Chua HN, Roth FP, Moore MJ (2012) Introns in UTRs: why we should stop ignoring them. Bioessays 34(12): 1025–1034 [DOI] [PubMed] [Google Scholar]
- Black DL (2003) Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72(1): 291–336 [DOI] [PubMed] [Google Scholar]
- Boothby TC, Zipper RS, van der Weele CM, Wolniak SM (2013) Removal of retained introns regulates translation in the rapidly developing gametophyte of Marsilea vestita. Dev Cell 24(5): 517–529 [DOI] [PubMed] [Google Scholar]
- Boutz PL, Bhutkar A, Sharp PA (2015) Detained introns are a novel, widespread class of post-transcriptionally spliced introns. Genes Dev 29(1): 63–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunschweig U, Barbosa-Morais NL, Pan Q, Nachman EN, Alipanahi B, Gonatopoulos-Pournatzis T, Frey B, Irimia M, Blencowe BJ (2014) Widespread intron retention in mammals functionally tunes transcriptomes. Genome Res 24(11): 1774–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buccitelli C, Selbach M (2020) mRNAs, proteins and the emerging principles of gene expression control. Nat Rev Genet 21(10): 630–644 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T (1991) Intron phylogeny: a new hypothesis. Trends Genet 7(1): 145–148 [PubMed] [Google Scholar]
- Chow LT, Gelinas RE, Broker TR, Roberts RJ (1977) An amazing sequence arrangement at the 5′ ends of adenovirus 2 messenger RNA. Cell 12(1): 1–8 [DOI] [PubMed] [Google Scholar]
- Collins L, Penny D (2005) Complex spliceosomal organization ancestral to extant eukaryotes. Mol Biol Evol 22(4): 1053–1066 [DOI] [PubMed] [Google Scholar]
- Denis MM, Tolley ND, Bunting M, Schwertz H, Jiang H, Lindemann S, Yost CC, Rubner FJ, Albertine KH, Swoboda KJ, et al. (2005) Escaping the nuclear confines: signal-dependent pre-mRNA splicing in anucleate platelets. Cell 122(3): 379–391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent CI, Singh S, Mukherjee S, Mishra S, Sarwade RD, Shamaya N, Loo KP, Harrison P, Sureshkumar S, Powell D, et al. (2021) Quantifying splice-site usage: a simple yet powerful approach to analyze splicing. NAR Genom Bioinform 3(2): lqab041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drechsel G, Kahles A, Kesarwani AK, Stauffer E, Behr J, Drewe P, Rätsch G, Wachter A (2013) Nonsense-mediated decay of alternative precursor mRNA splicing variants is a major determinant of the Arabidopsis steady state transcriptome. Plant Cell 25(10): 3726–3742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer K, Agarwal N, Pile L, Ansari A (2021) Gene architecture facilitates intron-mediated enhancement of transcription. Front Mol Biosci 8: 669004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagundes NJR, Bisso-Machado R, Figueiredo PICC, Varal M, Zani ALS (2022) What we talk about when we talk about “junk DNA”. Genome Biol Evol 14(5): evac055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fesenko I, Khazigaleeva R, Kirov I, Kniazev A, Glushenko O, Babalyan K, Arapidi G, Shashkova T, Butenko I, Zgoda V, et al. (2017) Alternative splicing shapes transcriptome but not proteome diversity in Physcomitrella patens. Sci Rep 7(1): 2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs A, Riegler S, Ayatollahi Z, Cavallari N, Giono LE, Nimeth BA, Mutanwad KV, Schweighofer A, Lucyshyn D, Barta A, et al. (2021) Targeting alternative splicing by RNAi: from the differential impact on splice variants to triggering artificial pre-mRNA splicing. Nucleic Acids Res 49(2): 1133–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring NH, Roignant J-Y (2021) Anything but ordinary—emerging splicing mechanisms in eukaryotic gene regulation. Trends Genet 37(4): 355–372 [DOI] [PubMed] [Google Scholar]
- Gilbert W (1978) Why genes in pieces? Nature 271(5645): 501–501 [DOI] [PubMed] [Google Scholar]
- Göhring J, Jacak J, Barta A (2014) Imaging of endogenous messenger RNA splice variants in living cells reveals nuclear retention of transcripts inaccessible to nonsense-mediated decay in Arabidopsis. Plant Cell 26(2): 754–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Roy SW (2014) Origin of spliceosomal introns and alternative splicing. Cold Spring Harb Perspect Biol 6(6): a016071–a016071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irimia M, Rukov JL, Penny D, Vinther J, Garcia-Fernandez J, Roy SW (2008) Origin of introns by ‘intronization’ of exonic sequences. Trends Genet 24(8): 378–381 [DOI] [PubMed] [Google Scholar]
- Jacob AG, Smith CWJ (2017) Intron retention as a component of regulated gene expression programs. Hum Genet 136(9): 1043–1057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia J, Long Y, Zhang H, Li Z, Liu Z, Zhao Y, Lu D, Jin X, Deng X, Xia R, et al. (2020) Post-transcriptional splicing of nascent RNA contributes to widespread intron retention in plants. Nat Plants 6(7): 780–788 [DOI] [PubMed] [Google Scholar]
- Jo B-S, Choi SS (2015) Introns: the functional benefits of introns in genomes. Genomics Inform 13(4): 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyna M, Simpson CG, Syed NH, Lewandowska D, Marquez Y, Kusenda B, Marshall J, Fuller J, Cardle L, McNicol J, et al. (2012) Alternative splicing and nonsense-mediated decay modulate expression of important regulatory genes in Arabidopsis. Nucleic Acids Res 40(6): 2454–2469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keren H, Lev-Maor G, Ast G (2010) Alternative splicing and evolution: diversification, exon definition and function. Nat Rev Genet 11(5): 345–355 [DOI] [PubMed] [Google Scholar]
- Kornblihtt A (2021) No, no Está Bien. Una pasión argentina por la ciencia (y por el arte y la política) (Penguin Random House Grupo Editorial Argentina), Está mal [Google Scholar]
- Kornblihtt AR, Schor IE, Alló M, Dujardin G, Petrillo E, Muñoz MJ (2013) Alternative splicing: a pivotal step between eukaryotic transcription and translation. Nat Rev Mol Cell Biol 14(3): 153–165 [DOI] [PubMed] [Google Scholar]
- Larue GE, Eliáš M, Roy SW (2021) Expansion and transformation of the minor spliceosomal system in the slime mold Physarum polycephalum. Curr Biol 31(14): 3125–3131.e4 [DOI] [PubMed] [Google Scholar]
- Lee HC, Choe J, Chi S-G, Kim YK (2009) Exon junction complex enhances translation of spliced mRNAs at multiple steps. Biochem Biophys Res Commun 384(3): 334–340 [DOI] [PubMed] [Google Scholar]
- Legrain P, Rosbash M (1989) Some cis- and trans-acting mutants for splicing target pre-mRNA to the cytoplasm. Cell 57(4): 573–583 [DOI] [PubMed] [Google Scholar]
- Leviatan N, Alkan N, Leshkowitz D, Fluhr R (2013) Genome-Wide survey of cold stress regulated alternative splicing in Arabidopsis thaliana with tiling microarray. PLOS ONE 8(6): e66511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li A, Shen W, Ye N, Wang G, Zhang J (2021) Global survey of alternative splicing in rice by direct RNA sequencing during reproductive development: landscape and genetic regulation. Rice (N Y) 14(1): 75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Wang S, Cheng J, Su C, Zhong S, Liu Q, Fang Y, Yu Y, Lv H, Zheng Y, et al. (2016) Intron lariat RNA inhibits MicroRNA biogenesis by sequestering the dicing complex in Arabidopsis. PLoS Genet 12(11): e1006422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Jensen TH (2015) Nonsense-mediated mRNA decay: an intricate machinery that shapes transcriptomes. Nat Rev Mol Cell Biol 16(11): 665–677 [DOI] [PubMed] [Google Scholar]
- Marquez Y, Brown JWS, Simpson C, Barta A, Kalyna M (2012) Transcriptome survey reveals increased complexity of the alternative splicing landscape in Arabidopsis. Genome Res 22(6): 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquez Y, Höpfler M, Ayatollahi Z, Barta A, Kalyna M (2015) Unmasking alternative splicing inside protein-coding exons defines exitrons and their role in proteome plasticity. Genome Res 25(7): 995–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin W, Koonin EV (2006) Introns and the origin of nucleus–cytosol compartmentalization. Nature 440(7080): 41–45 [DOI] [PubMed] [Google Scholar]
- Mauger O, Lemoine F, Scheiffele P (2016) Targeted intron retention and excision for rapid gene regulation in response to neuronal activity. Neuron 92(6): 1266–1278 [DOI] [PubMed] [Google Scholar]
- Moabbi AM, Agarwal N, El Kaderi B, Ansari A (2012) Role for gene looping in intron-mediated enhancement of transcription. Proc Natl Acad Sci USA 109(22): 8505–8510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39(6): 877–885 [DOI] [PubMed] [Google Scholar]
- Palazzo AF, Lee ES (2018) Sequence determinants for nuclear retention and cytoplasmic export of mRNAs and lncRNAs. Front Genet 9: 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenteau J, Maignon L, Berthoumieux M, Catala M, Gagnon V, Abou Elela S (2019) Introns are mediators of cell response to starvation. Nature 565(7741): 612–617 [DOI] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Rose AB, Korf I (2011) Comparative and functional analysis of intron-mediated enhancement signals reveals conserved features among plants. Nucleic Acids Res 39(13): 5328–5337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendleton KE, Park S-K, Hunter OV, Bresson SM, Conrad NK (2018) Balance between MAT2A intron detention and splicing is determined cotranscriptionally. RNA 24(6): 778–786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poverennaya IV, Roytberg MA (2020) Spliceosomal introns: features, functions, and evolution. Biochemistry (Mosc) 85(7): 725–734 [DOI] [PubMed] [Google Scholar]
- Rogozin IB, Carmel L, Csuros M, Koonin EV (2012) Origin and evolution of spliceosomal introns. Biol Direct 7(1): 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose AB, Emami S, Bradnam K, Korf I (2011) Evidence for a DNA-based mechanism of intron-mediated enhancement. Front Plant Sci 2: 98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz U, Pinello N, Jia F, Alasmari S, Ritchie W, Keightley M-C, Shini S, Lieschke GJ, Wong JJ-L, Rasko JEJ (2017) Intron retention enhances gene regulatory complexity in vertebrates. Genome Biol 18(1): 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwertz H, Tolley ND, Foulks JM, Denis MM, Risenmay BW, Buerke M, Tilley RE, Rondina MT, Harris EM, Kraiss LW, et al. (2006) Signal-dependent splicing of tissue factor pre-mRNA modulates the thrombogenecity of human platelets. J Exp Med 203(11): 2433–2440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Severing EI, van Dijk AD, van Ham RC (2011) Assessing the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana using proteomics data. BMC Plant Biol 11(1): 82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shalgi R, Hurt JA, Lindquist S, Burge CB (2014) Widespread inhibition of posttranscriptional splicing shapes the cellular transcriptome following heat shock. Cell Rep 7(5): 1362–1370 [DOI] [PubMed] [Google Scholar]
- Smathers CM, Robart AR (2019) The mechanism of splicing as told by group II introns: ancestors of the spliceosome. Biochimica Biophys Acta – Gene Regul Mech 1862(11–12): 194390. [DOI] [PubMed] [Google Scholar]
- Staiger D, Brown JWS (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25(10): 3640–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wang Y, Yu H, He J, Wang J, Shi B, Du Q, Provart NJ, Meyerowitz EM, Jiao Y (2019) A gene expression map of shoot domains reveals regulatory mechanisms. Nat Commun 10(1): 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaquero-Garcia J, Barrera A, Gazzara MR, González-Vallinas J, Lahens NF, Hogenesch JB, Lynch KW, Barash Y (2016) A new view of transcriptome complexity and regulation through the lens of local splicing variations. Elife 5: e11752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- William Roy S, Gilbert W (2006) The evolution of spliceosomal introns: patterns, puzzles and progress. Nat Rev Genet 7(3): 211–221 [DOI] [PubMed] [Google Scholar]
- Williamson L, Saponaro M, Boeing S, East P, Mitter R, Kantidakis T, Kelly GP, Lobley A, Walker J, Spencer-Dene B, et al. (2017) UV irradiation induces a non-coding RNA that functionally opposes the protein encoded by the same gene. Cell 168(5): 843–855.e13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H, Tian C, Yu Y, Jiao Y (2016) Transcriptome survey of the contribution of alternative splicing to proteome diversity in Arabidopsis thaliana. Mol Plant 9(5): 749–752 [DOI] [PubMed] [Google Scholar]
- Zhang R, Calixto C, Marquez Y, Venhuizen P, Tzioutziou NA, Guo W, Spensley M, Entizne JC, Lewandowska D, ten Have S, et al. (2017) A high quality Arabidopsis transcriptome for accurate transcript-level analysis of alternative splicing. Nucleic Acids Res 45(9): 5061–5073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Kuo R, Coulter M, Calixto CPG, Entizne JC, Guo W, Marquez Y, Milne L, Riegler S, Matsui A, et al. (2022) A high-resolution single-molecule sequencing-based Arabidopsis transcriptome using novel methods of iso-seq analysis. Genome Biol 23(1): 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Zhang Y, Wang T, Li Z, Cheng J, Ge H, Tang Q, Chen K, Liu L, Lu C, et al. (2019) A comprehensive map of intron branchpoints and lariat RNAs in plants. Plant Cell 31(5): 956–973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu D, Mao F, Tian Y, Lin X, Gu L, Gu H, Qu L, Wu Y, Wu Z (2020) The features and regulation of co-transcriptional splicing in Arabidopsis. Mol Plant 13(2): 278–294 [DOI] [PubMed] [Google Scholar]
- Zhu L, Zhang Y, Zhang W, Yang S, Chen J-Q, Tian D (2009) Patterns of exon-intron architecture variation of genes in eukaryotic genomes. BMC Genomics 10(1): 47. [DOI] [PMC free article] [PubMed] [Google Scholar]