Abstract
Brassinosteroid (BR), a growth-promoting phytohormone, regulates many plant growth processes including cell development. However, the mechanism by which BR regulates fiber growth is poorly understood. Cotton (Gossypium hirsutum) fibers are an ideal single-cell model in which to study cell elongation due to their length. Here we report that BR controls cotton fiber elongation by modulating very-long-chain fatty acid (VLCFA) biosynthesis. BR deficiency reduces the expression of 3-ketoacyl-CoA synthases (GhKCSs), the rate-limiting enzymes involved in VLCFA biosynthesis, leading to lower saturated VLCFA contents in pagoda1 (pag1) mutant fibers. In vitro ovule culture experiments show that BR acts upstream of VLCFAs. Silencing of BRI1-EMS-SUPPRESOR 1.4 (GhBES1.4), encoding a master transcription factor of the BR signaling pathway, significantly reduces fiber length, whereas GhBES1.4 overexpression produces longer fibers. GhBES1.4 regulates endogenous VLCFA contents and directly binds to BR RESPONSE ELEMENTS (BRREs) in the GhKCS10_At promoter region, which in turn regulates GhKCS10_At expression to increase endogenous VLCFA contents. GhKCS10_At overexpression promotes cotton fiber elongation, whereas GhKCS10_At silencing inhibits cotton fiber growth, supporting a positive regulatory role for GhKCS10_At in fiber elongation. Overall, these results uncover a mechanism of fiber elongation through crosstalk between BR and VLCFAs at the single-cell level.
Brassinosteroid signaling modulates the expression of 3-ketoacyl-CoA synthases, the rate-limiting enzymes in very long-chain fatty acid biosynthesis, thus regulating cotton fiber elongation.
IN A NUTSHELL.
Background: Cotton (Gossypium hirsutum) produces natural fibers for textile production. Cotton fibers are single-celled epidermal seed trichomes that are excellent models to study plant cell development. In recent years, brassinosteroid (BR) plant hormones and very-long-chain fatty acids (VLCFAs) have been reported to promote fiber elongation. However, the molecular mechanism by which BRs and VLCFAs regulate fiber elongation remains largely unknown. Therefore, we explored the mechanisms and relationship between BRs and VLCFAs in regulating fiber elongation.
Question: How do BRs regulate fiber cell elongation? What is the relationship between BRs and VLCFAs in regulating fiber elongation?
Findings: BR deficiency reduced the expression of 3-ketoacyl-CoA synthase genes (GhKCSs), which encode the rate-limiting enzymes involved in VLCFA biosynthesis, leading to lower saturated VLCFA contents. In vitro cultured ovules showed that BR acts upstream of VLCFAs. GhBES1.4, a master transcription factor of BR signaling, increased VLCFA contents and fiber elongation. DNA binding assays revealed that GhBES1.4 directly binds to the promoter of GhKCS10_At to induce its expression, thereby promoting fiber elongation. GhKCS10_At regulated the biosynthesis of endogenous VLCFAs and acts as a positive regulator of fiber development. These results indicated that BRs regulate cotton fiber elongation by modulating VLCFA biosynthesis.
Next steps: Since BRs regulate fiber elongation through modulating VLCFA biosynthesis, how VLCFAs work as a signal molecule in regulating fiber development is a fascinating question for future research.
Introduction
Cotton (Gossypium hirsutum) fibers are single-celled seed trichomes, which can elongate up to 65 mm and thus provide a unique system for studying cell elongation. Cotton fiber formation initiates from the ovule surface on the day of anthesis and then undergoes rapid elongation, generally from 5 to 20 DPA (days post anthesis), during which the final fiber length is determined, a key parameter of fibers for the textile industry.
Cotton fibers elongate through a unique tip-biased, diffuse growth mode regulated through a complex mechanism (Yu et al. 2019). In vitro ovule cultures have demonstrated that plant hormones are critical regulators of fiber development, among which, brassinosteroid (BR), auxin, gibberellin acid, and ethylene serve as positive regulators of fiber development. Conversely, cytokinin and abscisic acid inhibit fiber development (Kim and Triplett 2001; Shi et al. 2006; Yang et al. 2014). Transcriptome analysis and research on transgenic plant lines have identified several genes in plant hormone biosynthesis and signaling pathways involved in fiber development. In addition to plant hormones, reactive oxygen species and saturated very-long-chain fatty acids (VLCFAs) have also been shown to promote fiber elongation (Qin et al. 2007a; Qin and Zhu 2011; Tang et al. 2014; Yang et al. 2020). However, the potential relationship between various hormones and metabolites remains unclear.
VLCFAs, with characteristically fully saturated unbranched hydrocarbon chains (>20 carbons), play crucial roles in plant structure, physiology, and signaling. VLCFAs are synthesized in the endoplasmic reticulum through 4 successive reactions catalyzed by 4 different enzymes, among which 3-ketoacyl-CoA synthases (KCSs) are the rate-limiting enzyme and likely to determine the substrate and tissue specificity in fatty acid elongation (Millar and Kunst 1997; Haslam and Kunst 2013). VLCFAs serve as the precursors for sphingolipids and glycolipids, which are essential cell membrane lipids and participate in intercellular communication. VLCFAs and their derivatives have been generally recognized for their roles as cell membrane components or in seed storage. However, an increasing number of studies show that VLCFAs also function as signaling molecules to mediate various biological processes. VLCFAs affect membrane structure and functional dynamics during cell division, and act as signals in response to biotic or abiotic stresses (De Bigault Du Granrut and Cacas 2016).
VLCFAs are synthesized in the epidermis and send signals to internal tissues to restrict cell proliferation (Nobusawa et al. 2013). Analysis of a kcs1 mutant indicated that VLCFAs also function as signal molecules between cell layers to mediate regeneration in Arabidopsis thaliana (Shang et al. 2016; Trinh et al. 2019). Disruption of FIDDLEHEAD/KCS10 results in fewer trichomes, indicating that VLCFAs regulate protoderm cell differentiation into trichomes (Yephremov et al. 1999). VLCFAs produced by KCS6 function as signaling molecules to activate water transfer from papilla cells to pollen during pollen hydration (Zhan et al. 2018). VLCFAs also promote the expression of ACC SYNTHASE1 (ACS1) (a key enzyme of ethylene biosynthesis) and elevated ethylene induces aerenchyma formation in the rice (Oryza sativa) root cortex (Yamauchi et al. 2015). Comparative transcriptomic and genomic analyses revealed a critical role for VLCFAs in fiber development (Liu et al. 2015; Zhang et al. 2015). In particular, C24:0 promotes cotton fiber elongation by activating the expression of ACC OXIDASES (GhACOs), which encode enzymes required for ethylene biosynthesis (Qin et al. 2007a). These studies indicate that VLCFAs may function upstream of the ethylene pathway and contribute to fiber development, although the upstream regulators of VLCFAs are still unknown.
BRs, a group of polyhydroxylated steroidal phytohormones, are essential components in the regulation of diverse processes in plant development (Clouse and Sasse 1998). Characterization of BR biosynthesis and signaling mutants has shown that BR is one of the primary phytohormones responsible for promoting plant growth (Szekeres et al. 1996; Kang et al. 2001). BR signaling is transduced from the cell surface by the receptor BR INSENSITIVE1 (BRI1) to BRI1-EMS-SUPPRESSOR1 (BES1) and BRASSINAZOLE-RESISTANT1 (BZR1) family transcription factors (TFs), which control BR-regulated gene expression and function directly in BR promotion of plant growth. BES1/BZR family TFs have a highly conserved DNA binding domain with a basic helix-loop-helix (bHLH)-like motif. These TFs regulate target gene expression by binding to promoter sequences containing E-box (CANNTG, including G-box CACGTG) or BR-response elements (BRRE; CGTGC/TG) (Wang et al. 2002; He et al. 2005; Chen et al. 2019; Nolan et al. 2020; Liu et al. 2022; Li et al. 2023).
Specific concentrations of BR promote the initiation and elongation of cotton fibers in ovule cultures in vitro (Sun et al. 2005; Li et al. 2022). The upregulation of BR biosynthesis genes during fiber elongation implies that BR contributes to fiber development (Shi et al. 2006). Fiber length is significantly reduced in BR-deficient cotton mutants with suppression of the BR biosynthesis gene STEROID 5α-REDUCTASE DE-ETOLIATED2 (GhDET2). By contrast, overexpression of BR biosynthesis or signaling genes results in longer fibers (Yang et al. 2014; Zhou et al. 2015; Zhao et al. 2018). Recently, the bHLH TF FIBER-RELATED PROTEIN1 GhFP1 was found to promote fiber elongation by modulating BR biosynthesis and signaling (Liu et al. 2020). Another bHLH TF, FIBER-RELATED PROTEIN2 (GhFP2), antagonistically regulates cotton fiber elongation together with ACTIVATORS FOR CELL ELONGATION1 (GhACE1) (Lu et al. 2022). Despite these discoveries, the mechanism through which BR signaling regulates fiber development has remained elusive.
Findings in this study show that BR regulates cotton fiber elongation by modulating VLCFA biosynthesis. We found that BR deficiency inhibits fiber elongation by reducing the expression of GhKCSs and VLCFA contents, while BR functions upstream of VLCFAs to regulate fiber elongation in vitro. BR signaling regulates fiber elongation through the master TF GhBES1. Subsequent DNA binding and reporter assays indicate that GhBES1.4 directly binds to a BRRE in the GhKCS10_At promoter region to activate its expression, increasing VLCFA accumulation, and ultimately promoting fiber elongation. Taken together, these results illustrate how crosstalk between BR and VLCFAs regulates cell elongation at the single-cell level and depicts a blueprint to cultivate a high-quality variety by modulating the signaling between BR and VLCFAs.
Results
BR deficiency affects endogenous VLCFA biosynthesis
Our previous transcriptome analysis revealed that genes involved in VLCFA biosynthesis are downregulated in the BR-deficient short fiber mutant, pag1 and fiber length decreased from 28.12 ± 0.63 mm in ZM24 plants to 24.64 ± 0.61 mm in pag1 mutant cotton (Fig. 1A) (Yang et al. 2014). To explore whether BR affects VLCFA biosynthesis, we examined the GhKCSs in the upland cotton (G. hirsutum) genome. GhKCSs encode essential VLCFA biosynthesis pathway enzymes and we identified 57 GhKCSs in upland cotton, including 50 FATTY ACID ELONGATION1 (FAE1)-type GhKCSs and 7 ELONGASE (ELO)-type GhKCSs in the ZM24 genome (G. hirsutum_ZM24_ICR) (Supplemental Fig. S1).
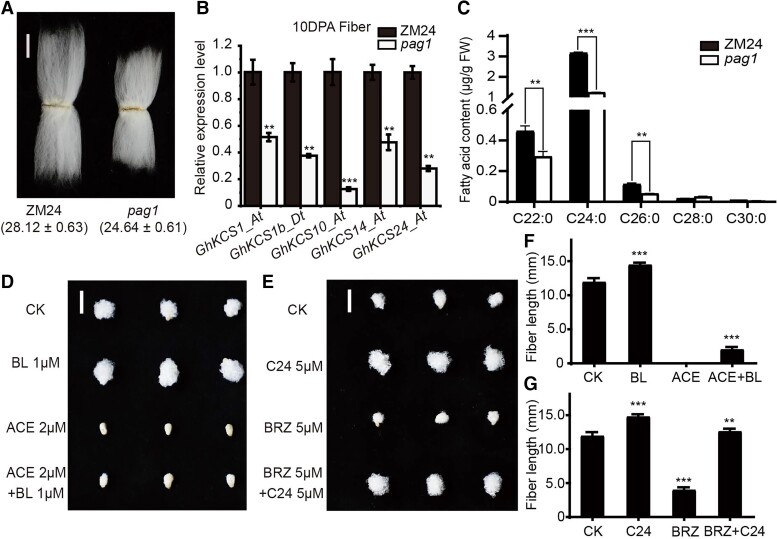
Figure 1.
BR affects endogenous VLCFA contents in cotton fiber. A) The phenotype and statistics of the length of mature fiber of ZM24 and pag1 cotton. Bar = 1 cm. B) The expression levels of 5 GhKCS genes in 10 DPA fibers of pag1 and ZM24. Histone3 was used as the internal control. The error bars represent SD for 3 independent experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01, ***P < 0.001). C) Fatty acid contents in 10 DPA fibers of pag1 and ZM24. The error bars represent SD for 3 independent experiments. The “FW” means fresh weight. All data reported here were obtained from 3 independent experiments with 30 fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01, ***P < 0.001). D) Phenotypes of ZM24 ovules (collected at 1 DPA) cultured for 14 d in medium without or with BL (1 μM) or with ACE (2 μM) or with both BL (1 μM) and ACE (2 μM). The “CK” means ZM24. Bar = 1 cm. E) Phenotypes of ZM24 ovules (collected at 1 DPA) cultured for 14 d in medium without or with C24 (5 μM) or with BRZ (5 μM) or with both C24 (5 μM) and BRZ (5 μM). Bar = 1 cm. F) The fiber lengths measured at the end of the 14-d culture period in (D). The error bars represent the SD in 10 different fibers of CK, BL (1 μM), ACE (2 μM), and BL (1 μM) + ACE (2 μM), respectively. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (***P < 0.001 by t-test). G) The fiber lengths measured at the end of the 14-d culture period in (E). The error bars represent the SD in 10 different fibers of CK, C24 (5 μM), BRZ (5 μM), and C24 (5 μM) + BRZ (5 μM), respectively. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01, ***P < 0.001 by t-test).
After renaming these GhKCS genes according to their chromosomal order (Supplemental Table S1), we focused 35 GhKCSs that were preferentially expressed in fibers (Supplemental Fig. S2). Among them, transcriptome analysis showed that 5 GhKCSs were significantly downregulated in 10 DPA fiber of pag1 (Supplemental Fig. S3; Supplemental Table S2). Reverse transcription-quantitative PCR (RT-qPCR) analysis also confirmed these results (Fig. 1B). Among these, GhKCS10_At expression was significantly decreased by 87.72% (Fig. 1B). In contrast, treatment with 1 μM brassinolide (BL) resulted in upregulation of these 5 GhKCSs in ZM24 fibers (Supplemental Fig. S4). In parallel with the above findings, GhKCS10_At showed approximately 3-fold increased expression after BL treatment (Supplemental Fig. S4). These results indicated that GhKCS10_At gene expression is induced by BR.
To determine whether BR deficiency affects endogenous VLCFA contents, we quantified VLCFAs in pag1 and ZM24 fibers (Fig. 1C). Except for C28:0 and C30:0, all saturated VLCFA levels were lower in pag1 10 DPA fibers compared with that in ZM24 cotton plants (Fig. 1C), which is consistent with GhKCSs expression data. More specifically, C22:0, C24:0, and C26:0 contents as much as 35.9%, 60.8%, and 52.6% lower, respectively, in 10 DPA pag1 fibers were compared with their levels in ZM24 (Fig. 1C). Taken together, these results suggested that VLCFA contents increased with increasing BR in fibers.
C24:0 reverses the inhibitory effect of BR deficiency on fiber growth
Exogenous application of BR and VLCFAs reportedly promotes fiber elongation (Qin et al. 2007a), while their respective inhibitors, BRZ (brassinazole) and ACE (acetochlor), inhibit fiber elongation. After confirming that the inhibitory effects of BRZ and ACE could be reversed by BR and C24:0 application, respectively (Supplemental Figs. S5 and S6), cultured ZM24 ovules were exposed to different concentrations of exogenous BR, BRZ, C24:0, or ACE to determine their respective effects on fiber development and fiber-related gene expression (Supplemental Figs. S5 and S6). The results showed that the optimal concentration of each compound for inducing longer or shorter fibers was consistent with those reported in previous studies (Shi et al. 2006; Qin et al. 2007a).
Furthermore, exogenous BR and VLCFAs both induced higher expression of known fiber elongation-related genes, such as GhACS6 (ACC SYNTHASE6), GhACO1 (ACC OXIDASES1), and GhEXP1 (EXPANSIN1) (Supplemental Figs. S5 and S6), while the addition of 5 μM BRZ or 2 μM ACE in ZM24 ovule culture medium resulted in severe inhibition of fiber elongation. Notably, the addition of 5 μM C24:0 to ovule culture medium containing 5 μM BRZ could overcome this inhibition, resulting in an approximately 3-fold increase in fiber length after 14 d of culture compared with ovules treated with BRZ alone (Fig. 1, E and G). On the contrary, the inhibitory effects of ACE on fiber elongation were not reversed by the addition of BR (Fig. 1, D and F). Furthermore, fiber elongation in pag1 cotton plants was partially restored by exposure to C24:0 (Supplemental Fig. S7). These results implied that BR acts upstream of VLCFAs in regulating cotton fiber elongation.
GhBES1.4, the master TFs of BR signaling, positively regulates fiber elongation
BES1 family genes function as central regulators of the BR signaling pathway by controlling the expression of thousands of downstream BR response genes (Wang et al. 2002; Yin et al. 2002), with 22 GhBES1 genes identified in upland cotton (Liu et al. 2018). Our previous expression analysis demonstrated that GhBES1.4 was preferentially expressed in rapidly elongating fibers and induced by BR treatment (Liu et al. 2018). Further heterologous transformation experiments showed that A. thaliana plants overexpressing GhBES1.4 exhibited an excessive BR phenotype, such as curled leaves with long and bent petioles (Supplemental Fig. S8). These results indicated that GhBES1.4 functions in BR signaling.
To explore the function of GhBES1.4 in cotton fiber elongation, we first constructed GhBES1.4 RNA interference (RNAi) transgenic plants. We performed RT-qPCR to assess the expression level of GhBES1.4 in the 10 DPA fibers of all 3 observed T3GhBES1.4-RNAi lines. The expression level of GhBES1.4 was more than 5-fold lower compared with their expression in ZM24 (Fig. 2D). The fiber length of GhBES1.4-RNAi lines was significantly reduced by 11.76% to 12.43% than that of ZM24 (Fig. 2, A and B). To further confirm the role of GhBES1.4 in fiber development, a GhBES1.4 expression vector driven by the cauliflower mosaic virus (CaMV) 35S promoter was transformed into ZM24 cotton. RT-qPCR analysis confirmed that GhBES1.4 expression was substantially enhanced in 10 DPA fibers of all 3 T3GhBES1.4 overexpression (OE) lines (Fig. 2C). The mature fiber length of GhBES1.4-OE lines was 8.09% to 8.57% longer than that of ZM24 (Fig. 2, A and B).
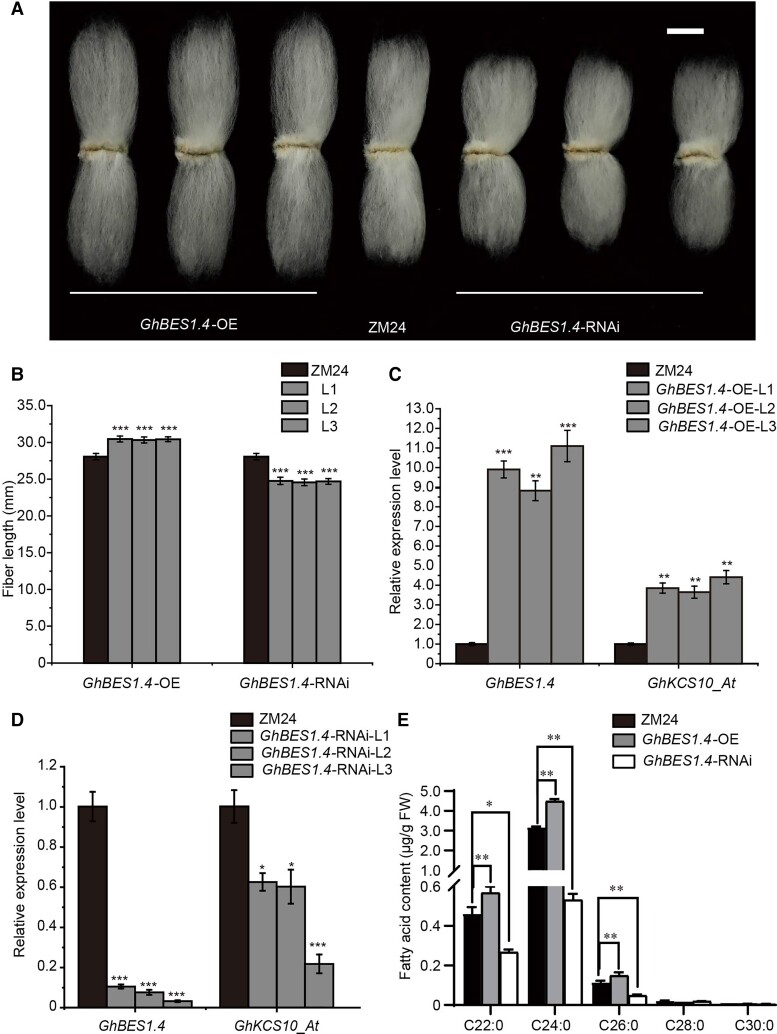
Figure 2.
GhBES1.4 positively regulates fiber elongation. A) The phenotypes of GhBES1.4-OE/GhBES1.4-RNAi and ZM24 mature fibers. Bar = 1 cm. B) Fiber lengths of GhBES1.4-OE/GhBES1.4-RNAi and ZM24 mature fibers. The error bars represent the SD in 30 different mature fibers of GhBES1.4-OE/GhBES1.4-RNAi and ZM24, respectively. All data reported here were obtained from 3 independent experiments with 30 mature fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (***P < 0.001 by t-test). C) The expression level of GhBES1.4 and GhKCS10_At in GhBES1.4-OE and ZM24 10 DPA fibers. Histone3 was used as the internal control. The error bars represent SD for 3 independent experiments. The expression level in the ZM24 sample was set to 1. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01, ***P < 0.001 by t-test). D) The expression levels of GhBES1.4 and GhKCS10_At in GhBES1.4-RNAi and ZM24 10 DPA fibers. Histone3 was used as the internal control. The error bars represent SD for 3 independent experiments. The expression level in the ZM24 sample was set to 1. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (*P < 0.05, ***P < 0.001 by t-test). E) Fatty acid contents of GhBES1.4-OE and GhBES1.4-RNAi fibers at 10 DPA. The error bars represent SD for 3 independent experiments. The “FW” means fresh weight. All data reported here were obtained from 3 independent experiments with 30 different 10 DPA fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (*P < 0.05, **P < 0.01 by t-test).
We also performed in vitro ovule culture of GhBES1.4-OE and GhBES1.4-RNAi transgenic cotton. BL application could increase fiber length by 24.38% and 0.83% that of ZM24 and GhBES1.4-RNAi lines, respectively, compared with mock treatment, suggesting that silencing GhBES1.4 reduced sensitivity to BL (Supplemental Fig. S9). By contrast, BRZ application resulted in 65.69% and 2.82% respective decreases in ZM24 and GhBES1.4-OE plants compared with that in the mock treatment controls, indicating that GhBES1.4-OE cotton plants were less sensitive to BRZ (Supplemental Fig. S9). These results indicated that GhBES1.4 participates in the BR signaling pathway and acts as a positive regulator of cotton fiber elongation.
GhBES1.4 alters VLCFA contents and regulates the expression of GhKCSs
To explore the relationship between GhBES1.4 and VLCFAs, we analyzed the VLCFA composition in GhBES1.4-OE and GhBES1.4-RNAi transgenic cotton plants. In GhBES1.4-OE plants, contents of the saturated VLCFAs C22:0, C24:0, and C26:0 were significantly higher, by 22.7%, 41.5%, and 29.9%, respectively, compared with those in ZM24 (Fig. 2E). Alternatively, C22:0, C24:0, and C26:0 in GhBES1.4 silenced plants were significantly lower than those of ZM24, especially C24:0, which was reduced by 83.3% (Fig. 2E).
Subsequently, we performed RNA-seq analysis to identify differentially expressed genes in 10 DPA fibers between ZM24 and GhBES1.4-OE plants. Fatty acid biosynthesis genes were significantly upregulated in GhBES1.4-OE fibers, which was consistent with their higher VLCFAs contents. In addition, the expression of ethylene-, cell wall-, and cytoskeleton-related genes was also significantly increased in GhBES1.4-OE fibers (Supplemental Fig. S10; Supplemental Table S3).
Examination of the expression levels of the 5 above-mentioned GhKCSs by RT-qPCR showed that all 5 GhKCSs were downregulated in 10 DPA fibers of GhBES1.4-RNAi lines, but upregulated in fibers of GhBES1.4-OE lines compared with that in ZM24 fibers (Fig. 2, C and D; Supplemental Fig. S11). Notably, changes in GhKCS10_At expression were the most obvious, displaying an approximately 2.5-fold increase in GhBES1.4-OE plants and an approximately 80% decrease in GhBES1.4 silenced plants relative to its expression in ZM24 (Fig. 2, C and D), indicating the key role of GhKCS10_At in the GhBES1.4 affecting VLCFA biosynthesis in cotton fiber.
GhKCS10_At is a functional elongase in VLCFA biosynthesis
In order to explore its biological function in VLCFA biosynthesis, GhKCS10_At was first transformed into the CER6 (CUT1) dwarf Arabidopsis mutant cut1. The average plant height of transgenic cut1-GhKCS10_At-OE lines was approximately twice that of cut1 mutant plants (Fig. 3, A and B), close to that of wild-type (WT) Arabidopsis. RT-qPCR assays confirmed that GhKCS10_At was highly expressed in the transgenic lines (Fig. 3C). These results indicated that heterologous expression of GhKCS10_At could rescue the dwarf phenotype of the cut1 mutant in Arabidopsis.
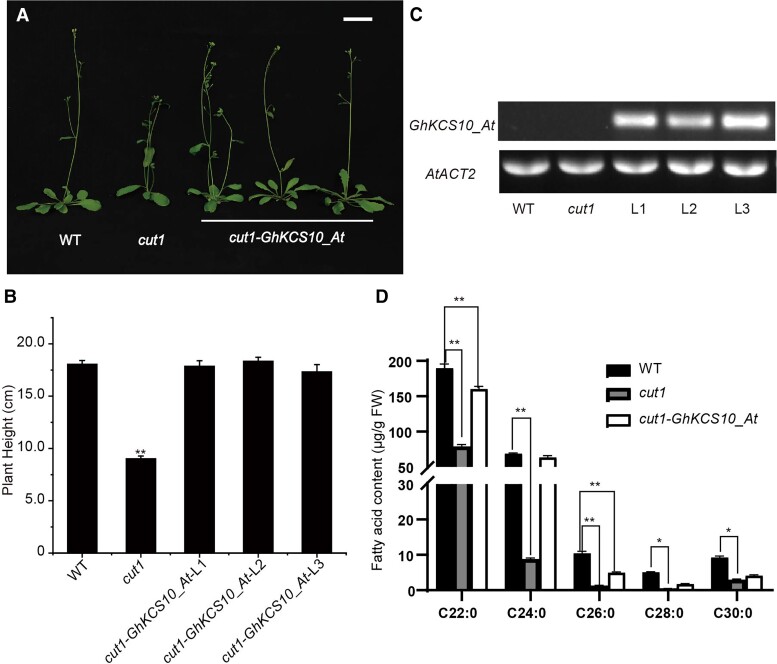
Figure 3.
GhKCS10_At complementary cut1 phenotype and fatty acid contents. A) Overexpression GhKCS10_At in cut1 mutant in Arabidopsis. Bar = 1 cm. B) Plant height of GhKCS10_At after transfer into cut1 mutant. The error bars represent the SD in 25 different aerial parts in WT, cut1, and cut1-GhKCS10_At transgenic lines, respectively. All data reported here were obtained from 3 independent experiments with 30 aerial parts measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the WT were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01 by t-test). C) Detection of the expression in GhKCS10_At-OE in cut1 mutant. AtACT2 was used as an internal control. D) The fatty acid content of GhKCS10_At after transfer into cut1 mutant. The error bars represent SD for 3 independent experiments. The “FW” means fresh weight. All data reported here were obtained from 3 independent experiments with 30 aerial parts measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the WT were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01 by t-test).
To explore whether this altered phenotype was linked to changes in saturated VLCFA contents, we analyzed fatty acid composition in aerial tissues of WT, cut1, and cut1-GhKCS10_At transgenic Arabidopsis. Consistent with the known function of cut1, cut1 plants contained significantly lower VLCFA levels (from C22:0 to C30:0) than those of WT, while overexpression of GhKCS10_At in cut1 resulted in significantly greater VLCFA accumulation (from C22:0 to C26:0). Moreover, treatment with C24:0 rescued the cut1 dwarf phenotype (Supplemental Fig. S12). Correspondingly, C24:0 contents in GhKCS10_At-OE lines increased approximately 7.8-fold compared with that in cut1 mutant plants (Fig. 3D). These cumulative results verified that GhKCS10_At encodes a functional VLCFA elongase.
GhKCS10_At is a positive regulator of fiber development
To investigate the role of GhKCS10_At in fiber development, we constructed GhKCS10_At-RNAi transgenic cotton plants. The expression of GhKCS10_At was checked by RT-qPCR, which showed that its expression was significantly lower in the 3 independent T3 RNAi lines than that in ZM24 (Fig. 4D). Additionally, GhKCS10_At-RNAi plants had shorter mature fibers compared with those of ZM24 (Fig. 4, A and B), an approximate 8.25% to 9.43% decrease, suggesting a role in fiber development.
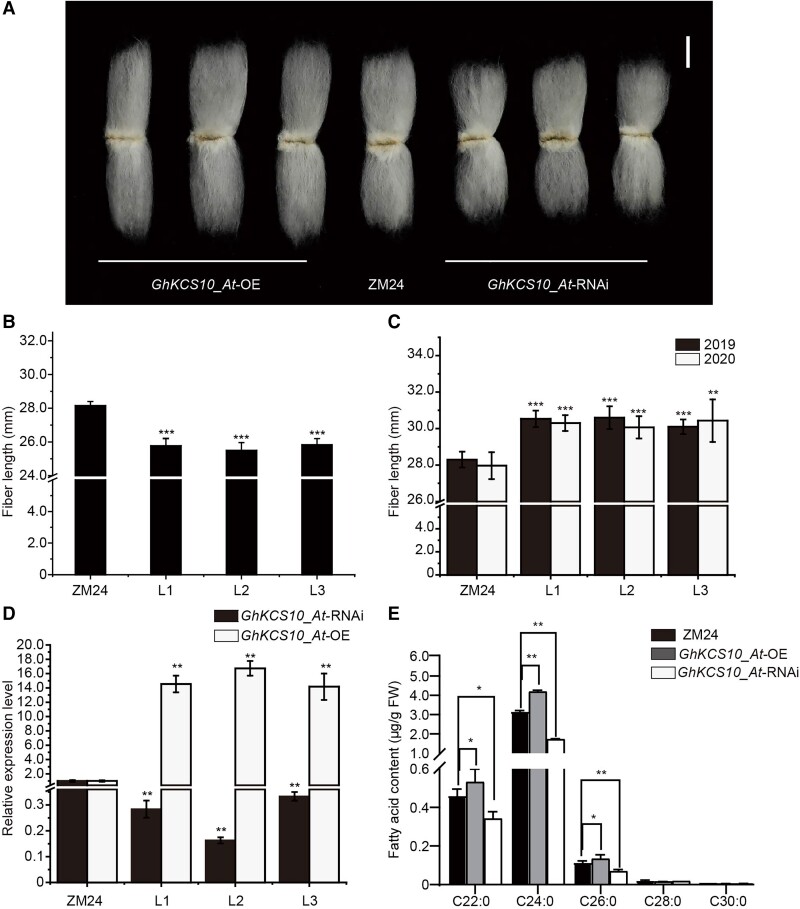
Figure 4.
GhKCS10_At overexpression affects fiber elongation and fatty acid contents. A) The phenotypes of GhKCS10_At-OE/GhKCS10_At-RNAi and ZM24 mature fibers. Bar = 1 cm. B) Determination of mature fiber length of GhKCS10_At-RNAi cotton. The error bars represent the SD in 30 different mature fibers of GhKCS10_At-RNAi and ZM24, respectively. All data reported here were obtained from 3 independent experiments with 30 mature fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (***P < 0.001 by t-test). C) Determination of mature fiber length of GhKCS10_At overexpressed cotton in 2019 and 2020. The error bars represent the SD in 30 different mature fibers of GhKCS10_At-OE and ZM24, respectively. All data reported here were obtained from 3 independent experiments with 30 mature fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01, ***P < 0.001 by t-test). D) Detection of GhKCS10_At expression in GhKCS10_At-OE lines and GhKCS10_At-RNAi lines. Histone3 was used as the internal control. The error bars represent SD for 3 independent experiments. The expression level in the ZM24 sample was set to 1. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01 by t-test). E) Fatty acid contents in 10 DPA fibers of GhKCS10_At-OE, GhKCS10_At-RNAi, and ZM24 plants. The error bars represent SD for 3 independent experiments. All data reported here were obtained from 3 independent experiments with 30 different 10 DPA fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (*P < 0.05, **P < 0.01 by t-test).
To further confirm GhKCS10_At function, we transformed the GhKCS10_At cDNA sequence driven by the CaMV 35S promoter into upland cotton ZM24 and selected 3 independent homozygous lines with elevated GhKCS10_At expression in fiber tissues for subsequent analyses (Fig. 4D). In 2 yr of field trials, mature GhKCS10_At-OE cotton fibers were 6.36% to 8.81% longer than ZM24 cotton fibers (Fig. 4, A and C). Moreover, the fiber strength in GhKCS10_At-OE lines was also significantly increased over that of ZM24 fibers (Supplemental Table S4). GhKCS10_At-OE lines also produced larger bolls, with higher fiber yields per 50 mature bolls compared with ZM24. Despite a lower lint percentage, lint yield was significantly higher (by up to 37%) in the GhKCS10_At-OE lines compared with WT (Supplemental Fig. S13; Supplemental Table S5).
Aligning well with increased GhKCS10_At activity, our results showed that saturated VLCFA levels for C22:0, C24:0, and C26:0 were significantly increased by 15.1%, 32.1%, and 16.9%, respectively, in GhKCS10_At-OE plants compared with their contents in ZM24 (Fig. 4E). Alternatively, C22:0, C24:0, and C26:0 levels in plants silenced for GhKCS10_At plants were significantly lower than those in ZM24, especially C24:0 which reduced by 45.6% (Fig. 4E). These results indicated that GhKCS10_At modulated the biosynthesis of endogenous VLCFAs and could, therefore, act as a positive regulator of fiber development.
In vitro ovule culture experiments showed that fiber length was longer in GhKCS10_At-OE lines than in ZM24 after 14 d of culture, whereas fibers of GhKCS10_At-RNAi lines were shorter than those of ZM24 (Supplemental Fig. S14) consistent with the mature fiber phenotype observed above (Fig. 4A). Application of BL resulted in 24.38% and 7.95% increases in fiber length of ZM24 and GhKCS10_At-RNAi plants, respectively, compared with the mock treatment, suggesting that silencing GhKCS10_At reduced sensitivity to BL. Conversely, compared with mock-treated plants, fiber length decreased by 65.69% and 2.14%, respectively, in ZM24 and GhKCS10_At-OE plants following BRZ application, suggesting that GhKCS10_At overexpression could decrease sensitivity to BRZ (Supplemental Fig. S14). Taken together, the above results suggested that GhKCS10_At plays a positive role in BR-mediated regulation of cotton fiber elongation.
GhKCS10_At is a direct target of GhBES1.4
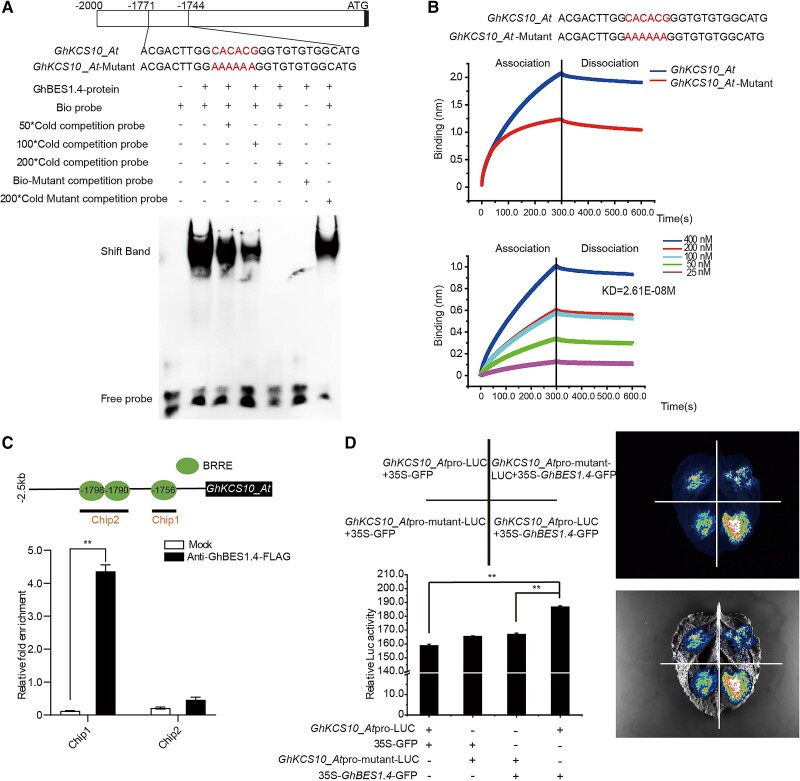
Our above finding that GhKCS10_At expression was significantly increased by GhBES1.4-OE and reduced by GhBES1.4 silencing (Fig. 2C) implied that GhKCS10_At might be directly regulated by GhBES1.4. To further verify this, we first analyzed the GhKCS10_At promoter sequence and found 3 GhBES1 binding motifs (BRREs) at positions −1,756, −1,790, and −1,798 bp relative to the predicted transcriptional start site (Supplemental Fig. S15). Electrophoretic mobility shift assays (EMSA) investigating whether GhBES1.4 could indeed bind to these recognition elements in the GhKCS10_At promoter revealed that the GhBES1.4 protein directly bound to 28 bp DNA probes for the −1771 to −1744 region containing the CACACG element (−1,756 bp), while the addition of unlabeled probes significantly decreased GhBES1.4 protein binding to the labeled probes (Fig. 5A). In addition, further EMSA also showed that GhBES1.4 could not bind the other 2 BRREs at −1,790 and −1,798 bp, respectively (Supplemental Fig. S16A). Furthermore, GhBES1.4 could not bind to a BRRE (−1,756 bp) harboring a mutated BRRE motif with the CACACG replaced by AAAAAA, indicating that GhBES1.4 specifically bound the BRRE (−1,756 bp) in the GhKCS10_At promoter (Fig. 5A). To further confirm that GhBES1.4 interacted with the GhKCS10_At promoter, we measured their binding affinity in real time using biolayer interferometry (BLI). The binding ability of GhBES1.4 to the GhKCS10_At promoter fragment gradually declined with decreasing concentration (Fig. 5B). In addition, the binding affinity of the BRRE (−1,756 bp) mutant in the GhKCS10_At promoter with GhBES1.4 protein decreased (Fig. 5B).
Figure 5.
GhKCS10_at is a direct target of GhBES1.4. A) EMSA in vitro showing GhBES1.4 protein binds to the GhKCS10_At promoter. B) Real-time binding analysis of GhBES1.4 protein to GhKCS10_At promoter. C) Chip analysis confirmed GhBES1.4 protein is directly bound to the GhKCS10_At promoter through the BRRE element (−1,756 bp). D) Luc activity analysis of the interaction of GhBES1.4 and GhKCS10_At through the BRRE element (−1,756 bp). Quantification of relevant Luc activities. The error bars represent the SD in 30 different injected tobacco leaves for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the control group (GhKCS10pro-LUC + 35S-GFP) were determined using one-way ANOVA analysis of variance software combined with Student's t-test (**P < 0.01).
To examine whether GhBES1.4 activated the transcription of GhKCS10_At in vivo, we performed chromatin immunoprecipitation (ChIP) coupled with qPCR analysis to detect GhBES1.4 binding at the GhKCS10_At promoter region. ChIP-qPCR assays with GhBES1.4-FLAG antibodies in GhBES1.4-OE transgenic cotton confirmed that the binding enrichment of GhBES1.4 to the BRRE (−1,756 bp) in the GhKCS10_At promoter (Fig. 5C). We then fused 2,000-bp upstream promoter sequences of GhKCS10_At and a promoter carrying the mutated −1,756-bp BRRE were each fused to the Luc reporter gene to construct GhKCS10_Atpro-Luc and GhKCS10_Atpro-mutant-Luc, respectively. Co-expression of GhKCS10_Atpro-Luc with CaMV 35S:GhBES1.4 showed obviously stronger Luc activity than that of GhKCS10_Atpro-mutant-Luc co-expression with CaMV 35S:GhBES1.4 (Fig. 5D), indicating that GhBES1.4 could activate transcription of the Luc reporter gene driven by the GhKCS10_At promoter. Reporter fusions with the other 2 mutated BRRE, at −1,790 and −1,798 bp, were generated and co-injected with CaMV 35S:GhBES1.4 into Nicotiana benthamiana leaves. The results showed that mutations in these 2 elements did not affect the transcriptional activation of GhKCS10_At by GhBES1.4, thus proving that these 2 elements were not the binding elements of GhBES1.4 (Supplemental Fig. S16, D and F).
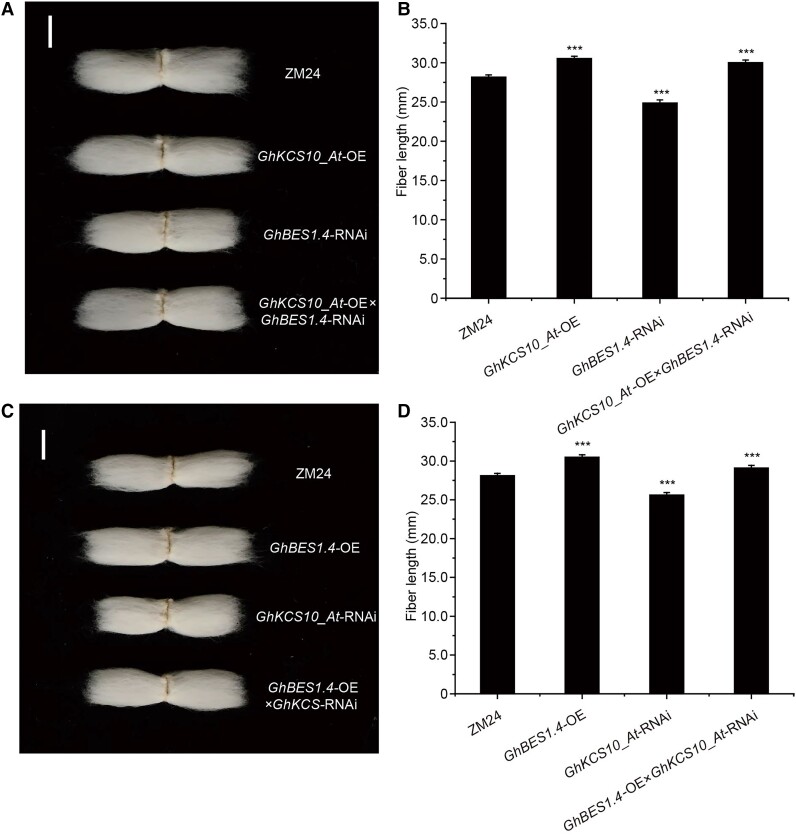
To test the genetic relationship between GhBES1.4 and GhKCS10_At, we crossed 2 transgenic cotton plants to generate GhKCS10_At-OE × GhBES1.4-RNAi and GhBES1.4-OE × GhKCS10_At-RNAi hybrid progeny. Phenotypic analysis showed that GhKCS10_At-OE lines showed an 8.39% increase, while GhBES1.4-RNAi showed 11.74% decreased fiber length than ZM24 fiber; however, GhKCS10_At-OE × GhBES1.4-RNAi lines showed 6.56% increased fiber length when compared with ZM24 fiber indicating that GhKCS10_At-OE rescued the short fiber phenotype of GhBES1.4-RNAi plants (Fig. 6, A and B). Similarly, GhBES1.4-OE lines showed an 8.48% increase while GhKCS10_At-RNAi showed 8.91% deceased fiber length than ZM24 fiber; however, GhBES1.4-OE × GhKCS10_At-RNAi lines showed 3.53% increased fiber length when compared with ZM24 fiber indicating that GhKCS10_At-RNAi could suppress the elongated fiber phenotype of GhBES1.4-OE plants (Fig. 6, C and D). Taken together, these genetic and physiological data suggested that GhBES1.4 promotes fiber elongation by regulating GhKCS10_At-mediated VLCFAs synthesis in cotton (Fig. 7).
Figure 6.
The mature fiber phenotypes of GhKCS10_At × GhBES1.4 by hybridization. A) Phenotypes of GhKCS10_At-OE, GhBES1.4-RNAi, GhKCS10_At-OE × GhBES1.4-RNAi, and ZM24 mature fibers. Bar = 1 cm. B) Determination of mature fiber length of GhKCS10_At-OE, GhBES1.4-RNAi, GhKCS10_At-OE × GhBES1.4-RNAi, and ZM24 cotton. The error bars represent the SD in 30 different mature fibers of GhKCS10_At-OE, GhBES1.4-RNAi, GhKCS10_At-OE × GhBES1.4-RNAi, and ZM24, respectively. All data reported here were obtained from 3 independent experiments with 30 mature fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (***P < 0.001 by t-test). C) Phenotypes of GhBES1.4-OE, GhKCS10_At-RNAi, GhBES1.4-OE × GhKCS10_At-RNAi, and ZM24 mature fibers. Bar = 1 cm. D) Determination of mature fiber length of GhBES1.4-OE, GhKCS10_At-RNAi, GhBES1.4-OE × GhKCS10_At-RNAi, and ZM24 cotton. The error bars represent the SD in 30 different mature fibers of GhBES1.4-OE, GhKCS10_At-RNAi, GhBES1.4-OE × GhKCS10_At-RNAi, and ZM24, respectively. All data reported here were obtained from 3 independent experiments with 30 mature fibers measured for each treatment and are presented as mean ± SD of triplicate experiments. Significant differences compared with the ZM24 were determined using one-way ANOVA analysis of variance software combined with Student's t-test (***P < 0.01 by t-test).
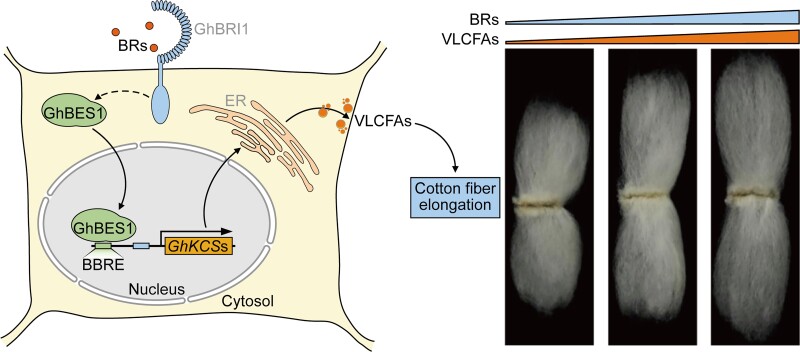
Figure 7.
. A model for the regulation of cotton fiber elongation through the crosstalk between BR and VLCFA. In the presence of BR, the key TF in the BR signaling pathway, GhBES1, moves to the nucleus and directly binds to the BRRE of GhKCSs to activate its expression. High expression of GhKCSs, encoding the rate-limiting enzyme of VLCFA biosynthesis, increases endogenous VLCFAs contents, which promotes fiber elongation.
Discussion
Plant cell elongation is regulated by the external environment as well as endogenous hormone signaling. BRs were initially discovered due to their function in cell elongation (Wang et al. 2012). Cotton fibers are extremely elongated single cells, and the BR signaling pathway plays a vital role in the regulation of fiber elongation. Until now, the regulatory mechanism of fiber cell elongation mediated by BR has remained obscure (Lu et al. 2022). In this study, we explored the mechanism of BR-mediated regulation of fiber elongation by genetic, physiological, biochemical, and transcriptome analyses. Our results demonstrated that BR could modulate VLCFA biosynthesis to regulate cotton fiber elongation. More specifically, the BR signaling GhBES1.4 TF modulates the expression of GhKCS10_At via directly binding to the BRRE in its promoter region, which in turn regulates cotton fiber elongation by increasing VLCFA biosynthesis (Fig. 7). This study thus uncovers a mechanism in which crosstalk between BR and VLCFAs underpins control of cotton fiber elongation.
BES1, the core component of BR, regulates fiber cell elongation in cotton
A predominant effect of BR signaling is to promote cell elongation. Among the isolated BR signaling components, BES1 family proteins are master TFs governing BR-regulated gene expression. Reduced BES1 family gene expression in rice produces semi-dwarf plants and decreased BR sensitivity (Zhang et al. 2014). Furthermore, a hextuple Arabidopsis mutant disrupted for all BES1 family genes displays a severe dwarf phenotype highly similar to the BR receptor null mutant and has blocked expression of BR-regulated genes, illustrating the indispensable role of BES1 family genes in BR signaling (Bai et al. 2007; Qiao et al. 2017; Chen et al. 2019).
Cotton fiber is one of the longest plant cells, which provide a unique system for studying cell elongation. Suppression of GhBES1.4 in cotton inhibits fiber cell elongation, whereas GhBES1.4-OE significantly enhances fiber length (Fig. 2). GhBES1s would modulate the expression of a number of downstream target genes involved in BR regulating fiber elongation (Lee et al. 2010; Bajwa et al. 2015; Li et al. 2018). Thus, despite cotton fiber being a specifically elongated single cell, BR regulates its elongation through a conserved mechanism in which GhBES1s have a central role.
Arabidopsis and rice only have 6 and 4 BES1 family genes, respectively (Bai et al. 2007; Chen et al. 2019). However, gene duplication events in allotetraploid cotton have resulted in 22 GhBES1 TFs in the upland cotton genome, 6 of which were found to be preferentially expressed in fibers during the rapid elongation stage (Liu et al. 2018). The GhBES1.4-RNAi lines showed shortened fiber phenotype; however, the fiber length reduction is not as severe as that caused by BR deficiency (Fig. 1A), probably due to functional redundancy. Further research is necessary to dissect how these GhBES1 genes cooperatively regulate fiber development.
VLCFAs regulate fiber elongation in a variety of ways
VLCFAs are well known to regulate plant development. For instance, the VLCFA-deficient mutants, pas2-1 and cut1, exhibit dwarf phenotypes, indicating that VLCFAs are required for cell elongation (Qin et al. 2007a; Zhu et al. 2020). Transcriptome analysis indicated that VLCFA biosynthesis genes participate in fiber development, and subsequent in vitro assays confirmed that VLCFAs function directly in fiber elongation (Qin et al. 2007a). Population genetic analysis showed that VLCFA biosynthesis genes (KCS, KCR, and ECR) are positively selected for fiber improvement (Zhang et al. 2015). Our work revealed that suppression of the VLCFA biosynthesis gene, GhKCS10_At, hindered fiber elongation. By contrast, GhKCS10_At overexpression increases VLCFA contents and results in longer fibers (Fig. 4). These stable transgenic cotton lines thus provide direct evidence supporting VLCFA regulation of fiber elongation. KCSs specifically act on saturated fatty acyl-CoA substrates in plants (Blacklock and Jaworski 2006). In previous reports, GhCER6 was shown to catalyze the synthesis of C26:0 in yeast cells (Qin et al. 2007b). Here, we found that C22:0, C24:0, and C26:0 contents in ZM24 cotton plants overexpressing GhKCS10_At are significantly higher than those in ZM24, especially C24:0 content was increased by 32.1% (Fig. 4E). We, therefore, speculate that GhKCS10_At mainly contributes to elongating VLCFAs ranging from C22:0 to C24:0, although further experimental verification is needed to test this possibility.
VLCFAs have been shown to act as signal molecules in regulating a variety of developmental processes in plants. For example, VLCFAs can protect plants from hypoxia-induced damage by regulating the ethylene signaling pathway (Xie et al. 2015), and VLCFAs also promote ethylene biosynthesis inducing aerenchyma formation in the rice root cortex (Yamauchi et al. 2015). VLCFAs have been shown to modulate phytohormone signaling, especially auxin and ethylene, to regulate fiber elongation (Batsale et al. 2021). Actually, VLCFAs activate ethylene biosynthesis in fibers (Qin et al. 2007a). In the current study, ethylene content increased in 10 DPA fibers of GhKCS10_At-OE plants, but decreased in corresponding fibers of GhKCS10_At-RNAi plants (Supplemental Fig. S17). These findings support the idea that VLCFAs regulate fiber elongation, at least in part, by modulating endogenous ethylene content. In addition, we found that both VLCFA and ethylene levels were substantially decreased in 10 DPA fibers of the BR-deficient mutant pag1 (Fig. 1C; Supplemental Fig. S17), suggesting that VLCFAs may serve as the point at which BR- and ethylene-responsive regulatory signals are coordinated to determine fiber length (Fig. 7). This is an interesting phenomenon, as fiber length plays a significant role in determining cotton fiber quality and fabric properties.
It also warrants mention that lipid raft activity in the cell membrane is higher during fiber elongation and lower during secondary wall synthesis, suggesting that VLCFA-containing sphingolipids may promote membrane stabilization by enhancing lipid raft activity (Xu et al. 2021). VLCFAs are essential components of the vacuole membrane. The central vacuole, which imposes turgor pressure indispensable for fiber elongation, takes up 98% of the elongating fiber cell volume (Hu et al. 2019). So, we should not exclude the possibility that VLCFAs work as membrane constituents. Despite these findings, the exact mechanism of how VLCFAs affect fiber development requires further research. Since BR, VLCFAs, and ethylene promote fiber cell elongation, exploring the relationship between them will likely reveal the regulatory mechanisms responsible for fiber cell development.
BR modulates VLCFA levels to affect different processes
BR is known to regulate various developmental processes in plants via crosstalk with other phytohormones and signal molecules at multiple levels (Planas-Riverola et al. 2019; Nolan et al. 2020; Kour et al. 2021). In this study, we demonstrate that BR regulates VLCFA biosynthesis during cotton fiber development. Firstly, in vitro application of C24:0 effectively reversed the inhibitory effect of BR inhibitor BRZ on fiber growth (Fig. 1E). Nevertheless, BR did not weaken the inhibitory effect of ACE (Fig. 1D). Secondly, the expression of 5 GhKCSs was downregulated in fibers of both BR-deficient mutants pag1 and GhBES1.4 silenced plants (Fig. 1B, Supplemental Fig. S11), whereas BL treatment induced their transcriptional upregulation (Supplemental Fig. S4). Thirdly, VLCFA contents were significantly reduced in pag1 and GhBES1.4-silenced fibers (Figs. 1C and 2E), while significantly elevated in GhBES1.4-OE plants (Fig. 2E). Furthermore, in vitro and in vivo protein and DNA interaction analyses indicated that GhBES1.4 directly activates the expression of GhKCS10_At (Fig. 5).
In addition to GhKCS10_At, BES1-specific recognition elements have been identified in the promoter of 12 other GhKCSs (Supplemental Fig. S15), suggesting that these GhKCSs may be the potential targets of GhBES1s. In fact, GhKCS14_At and GhKCS1b_Dt were upregulated in GhBES1.4-OE lines (Supplemental Fig. S11), indicating that they might also be the direct target of GhBES1.4. This finding also implies that different GhBES1s may target a variety of GhKCSs to regulate fiber elongation.
In Arabidopsis, KCS1 is an early BR response gene. AtMYB30, a known direct target of AtBES1, regulates BR-induced gene expression, and AtMYB30 can directly bind the KCS1 promoter to activate its expression (Li et al. 2009). Chip-chip data have shown that KCS1 should be a direct target gene of BZR1 (Lisso et al. 2005; Sun et al. 2010), so BES1 and BZR1 simultaneously regulate the expression of KCS1 to regulate VLCFA biosynthesis. BR regulates VLCFA biosynthesis at multiple levels. Unlike regulating fiber elongation in cotton, BR modulates VLCFA synthesis to activate hypersensitive cell death response in Arabidopsis (Raffaele et al. 2008). Although crosstalk between BR and VLCFAs may be ubiquitous in plants, the regulatory effects on various biological functions may diverge among different plant species. Another fascinating question is which other processes are affected by BR-mediated variation VLCFA levels.
Materials and methods
Plant materials and growth conditions
G. hirsutum, cultivar ZM24, cotton plants were planted on the mixed substrate of nutrient soil (v/v = 1:1) and grown in a greenhouse with 16-h light/8-h dark at 30 °C. The lighting was provided by a LED flat panel with an intensity of 80,000 lux. On the day of anthesis, the flowers and bolls were marked; the day of anthesis was treated as 0 DPA. WT A. thaliana ecotype Landsberg erecta (Ler-0) was used in this study. The mutant line cut1 (SALK_CS6242) was obtained from the Arabidopsis Biological Resource Center (ABRC) (https://abrc.osu.edu/). Arabidopsis mutant seeds were kept at 4 °C for 2 d in the dark prior to germination. Seeds were surface sterilized with sodium hypochlorite (NaOCl), followed by washing 5 times with sterile water. Germination was carried out in MS medium under aseptic conditions. After 2 wk, the plants were transferred to soil and grown in a greenhouse with 16-h light/8-h dark at 23 °C. For cut1 Arabidopsis treated with BL and C24:0, seed germination was carried out in MS medium supplemented with 5 μM L−1 C24:0 or 1 μM L−1 BL. After 2 wk, the plants were transferred to soil and grown in a greenhouse. The cut1 plants received 50 mL of 5 μM L−1 C24:0 or 1 μM L−1 BL supplementation daily from the beginning on the day of bolting for each pot.
Phylogenetic analysis and expression profile of GhKCS family genes
Arabidopsis KCS gene sequences were downloaded from TAIR10 (https://www.arabidopsis.org/) and used as queries for genome-wide identification of KCS genes in G. hirsutum (G. hirsutum_ZM24_ICR). To analyze evolutionary relationships, full-length protein sequences were aligned and used to construct a phylogenetic tree using the neighbour joining method (1,000 Bootstrap Replication, 95% Site Coverage Cutoff) in MEGA7.0 (Supplemental Files S1 and S2) (Kumar et al. 2016). The high-throughput G. hirsutum ZM24 transcriptome sequencing data was used to investigate the GhKCSs gene expression patterns in fiber tissues. The heatmap of GhKCSs gene expression patterns in cotton fiber (5, 10, 20, and 25 DPA) was constructed using TBtools software (Chen et al. 2020).
Expression pattern of GhKCSs in pag1 and BL treatment
Fiber tissues at 10 DPA of pag1 and ZM24 were taken as samples. For BL treatment, 1 μM L−1 BL was added to the Beasley and Ting (BT) medium for the ZM24 ovule. Ovule culture was conducted at 30 °C in the dark. After collection, plant tissues were frozen immediately in liquid nitrogen and RT-qPCR was used to detect the expression of GhKCSs.
In vitro ovule culture
Ovules of 1 DPA ZM24 cotton plants were sterilized with 75% anhydrous ethanol for 5 min followed by soaking in 95% anhydrous ethanol for 2 to 3 s and washing 5 times with distilled water. The sterilized ovules were taken out and kept in a liquid BT medium. Next, BL (0.1, 1, and 5 μM L−1), BRZ (0.1, 1, and 5 μM L−1), and VLCFAs inhibitor ACE (0.2, 2, and 20 μM L−1) were both dissolved in anhydrous ethanol and placed in the BT medium. Different concentrations of C24:0 (1, 5, and 10 μM L−1) were dissolved in methyl tertiary butyl ether and added to the BT medium (Qin et al. 2007a). For the C24:0 restored pag1 ovules experiment, pag1 and ZM24 ovules were cultured for 14 d in the dark at 30 °C. For the C24:0 reversed BRZ inhibiting fiber experiment, ZM24 ovules were cultured for 14 d in the dark at 30 °C and fiber length was measured. For the BL, BRZ, C24:0, and ACE treated ovules experiment, the ZM24 ovules were cultured for 14 d in the dark at 30 °C and fiber length was measured.
Phenotypic analysis for cotton fibers
The lengths of 10 DPA fibers were measured with a ruler. GhBES1.4-OE/GhBES1.4-RNAi transgenic cotton fibers were collected in Anyang in 2019. GhKCS10_At-OE transgenic cotton fibers were collected in Anyang in 2019 and 2020. GhKCS10_At-RNAi transgenic cotton fibers were collected in Anyang in 2021. All mature fibers (15 g) were collected from the bolls at a similar position on plants. The mature fiber samples were then analyzed at the Cotton Fiber Quality Inspection and Testing Center, Chinese Ministry of Agriculture (Anyang, Henan, China) for quality measurements.
RNA isolation and RT-qPCR analysis
For cotton samples, total RNA was extracted from fibers at specified time points. For Arabidopsis samples, total RNA was extracted from seedlings at 38 d after transplanting into soil. Total RNA was extracted using an RNA prep Pure Plant Kit (TIANGEN, Beijing, China). First-strand cDNA was reverse-transcribed using a PrimeScript RT reagent kit (Takara, Dalian, China). For the RT-qPCR assay, Premix Ex Taq II (Takara) was used along with the LightCycler 480 system (Roche Diagnostics, Mannheim, Germany). The reaction parameters of the RT-qPCR assay were as follows: 95 °C for 10 min, followed by 40 cycles of 95 °C for 10 s and 60 °C for 30 s. Specific primers for GhKCS1_At, GhKCS10_At, GhKCS14_At, GhKCS24_At, and GhKCS1b_Dt were designed from the 5-UTR region. The accession numbers of the identified genes and primers for the RT-qPCR analysis are given the Supporting Information (Supplemental Table S6). Cotton histone3 (GenBank accession number AF024716) and AtACT2 (GenBank accession number AT3G18780) were used as internal controls in PCR experiments. The relative expression of each target gene was calculated by the equation Y = 10 (CtGhhistone3−CtGhgene)/3 × 100%, and the detailed experimental method refers to previous reports (Li et al. 2005). Each RT-qPCR was performed in triplicates, and the results were calculated as the mean and standard deviation.
Vector construction and Arabidopsis transformation
The GhKCS10_At and GhBES1.4 coding sequences were retrieved from CottonGen (https://www.cottongen.org/). Genes were amplified from the ZM24 cDNA library using gene-specific primers listed in Supplemental Table S6 and constructed into a pCAMBIA2300 vector driven by the CaMV 35S promoter. Notably, overexpression of the GhBES1.4 coding sequence contains a point mutation modification designed by PrimerX (http://www.bioinformatics.org/primerx/cgi-bin/DNA_1.cgi) (Liu et al. 2018). For the GhBES1.4 and GhKCS10_At-RNAi vector, the GhBES1.4 and GhKCS10_At specific fragment was cloned from the ZM24 cDNA library using gene-specific primers listed in Supplemental Table S6 and constructed into a pBI121 vector driven by the CaMV 35S promoter. The GhKCS10_At and GhBES1.4 vectors were introduced into Agrobacterium tumefaciens strain GV3101 to transform Arabidopsis via the floral dip method (Zhang et al. 2006). The transgenic plants were selected on a half-strength MS medium containing 50 mg L−1 kanamycin.
Cotton transformation
All overexpression and RNAi constructs were introduced into the Agrobacterium strain LBA4404. ZM24 seeds were sterilized and cultured in a chamber without light for 6 d at 30 °C (Ge et al. 2022). The cotton transformation was performed using hypocotyl segments as explants. The explants were immersed in an A. tumefaciens suspension after they were cut into 5-mm segments. After callus induction, proliferation, embryogenic callus induction, embryo differentiation, and plantlet regeneration, the putative transgenic plants were grown in the greenhouse. The successful transformation and integrity of the transgene were confirmed via PCR analysis of genomic DNA with the 35S-F/GhKCS10_At-R, 35S-F/GhBES1.4-RNAi-R, GhBES1.4-RNAi-F/intron-R, and GhKCS10_At-RNAi-F/intron-R primers in Supplemental Table S6. RT-qPCR was used to detect GhKCS10_At and GhBES1.4 expression levels in transgenic lines, and cotton histone3 was used as an internal control (Supplemental Table S6).
Fatty acid extraction and gas chromatography-mass spectrometry analysis
Before fatty acid extraction, 0.2 g of fiber (10 DPA) and Arabidopsis aerial parts tissues were placed in a mortar and ground to a fine powder with a pestle under cryogenic conditions using liquid nitrogen. The fibers were immersed in 5% concentrated sulfuric acid/methanol (v/v). A total of 10 μL butylated hydroxytoluene methanol was added to the samples followed by vortex mixing for 1 min. Next, the samples were incubated for 30 min at 90 °C and placed at room temperature for 10 min. Then 2 mL of n-hexane and 10 mL of saturated NaCl solution were added. After vortexing for 1 min, the supernatant was taken after allowing to settle for 15 min for analysis. A standard fatty acid methyl ester mixture (Supelco 37 Component FAME Mix, USA) was used to identify the fatty acid methyl esters. Fatty acid content was analyzed by HP-5 capillary chromatography column (30 m × 0.25 mm ×0.25 μm, Agilent, USA) coupled to a gas chromatography (GC) system (Agilent 7890-5977, USA), which was connected to the National Institute of Standards and Technology databases. In this study, the external standard method was used to measure the fatty acid content (Dursun and Güler 2021). The fatty acid content of each sample is expressed as μg/g fresh weight (FW).
Ethylene measurements
The amount of ethylene produced was measured by a gas chromatograph (GC-2010 Plus; Shimadzu) equipped with a Flame Ionization Detector (FID) and 30-m HP-PLOT column (RXT-1), the sensitivity of the GC-FID was 0.01 ppm. For measurement of ethylene (ET) production in cotton fiber development, approximately 5.0 g freshly collected 10 DPA fibers of ZM24, GhKCS10_At-OE, GhKCS10_At-RNAi, and pag1 were placed in 50-mL glass flasks in darkness at 30 °C for 48 h, and the released gas was collected with a needle via the drain method, and 100 μL gas sample was analyzed using GC-FID. All measurements were recorded from 6 replicates, and the final assessment results are calculated per gram per hour (nL g−1 FW h−1) (Li et al. 2020).
Electrophoretic mobility shift assay
The EMSA was performed using an EMSA kit (Thermo Fisher Scientific, Shanghai, China) according to a previously described method (Hou et al. 2014). The promoter fragments of GhKCS10_At containing the native and mutant BRRE (−1,756 bp) were labeled with biotin on both ends of the probe. The promoter fragment of GhKCS10_At containing the native BRRE (−1,798 and −1,790 bp) was labeled with biotin on both ends of the probe. GhBES1.4 was constructed in the pET30a vector and induced with IPTG at 37 °C for 4 h. Non-labeled probes were used as cold competitors.
BLI binding measurements
We used the Octet RED96 system (ForteBio) to measure DNA–protein binding kinetics by streptavidin-coated biosensors. The biotin-labeled probe on the GhKCS10_At promoter was used as described above. First, the GhBES1.4 protein interacted with the DNA-immobilized sensor surface in the dilution buffer for 3 min. Sensors were then washed before monitoring protein association for 5 min. Next, PBS (10%)/Tween (0.5%) buffers were used to elute the DNA–protein compound on the sensor surface for 10 min. Dissociation curves were obtained by maintaining this set-up for 15 min and transferring the sensors back to tubes. The data were fitted to a 1:1 binding mode to calculate the association rate constants. The graphics were drawn using OriginPro2022 software (https://www.originlab.com/).
ChIP assays
ChIP assays were performed as described previously (Oh et al. 2009). The 10 DPA fibers were cross-linked with 1% (v/v) formaldehyde and sonicated to obtain 200 to 500 bp fragments from cell nuclei. Afterwards, GhBES1.4-FLAG antibodies (Beyotime, China, catalog:AF519) were used to immunoprecipitate the DNA fragments. DNA fragments pulled down without any antibodies were set as control. To ensure the authenticity of ChIP data, the input sample was analyzed with each primer set listed in Supplemental Table S6. The enrichment degrees were analyzed by RT-qPCR analysis using SYBR Premix Ex Taq (Takara) in a LightCycler 480 system (Roche Diagnostics, Mannheim, Germany).
Transient expression assay
The 2,000 bp native and mutant promoter regions of GhKCS10_At were constructed into the vector pGWB435 containing a luciferase reporter gene to construct the GhKCS10_Atpro-Luc and GhKCS10_Atpro-mutant-Luc, respectively. GhBES1.4 was cloned into CAMBIA2300 to generate the CaMV 35S: GhBES1.4, while CaMV 35S: GFP was used as an empty vector. Agrobacterium strains were allowed to infiltrate into tobacco leaves. The tobacco leaves were placed in the dark for 12 h, followed by 48 h in the growth chamber at 23 °C. The tobacco leaves were sprayed with luciferin and kept out of the light for 10 min. A low-light cooled charge-coupled device was used to take photos (Vilber NEWTON7.0 Bio). Quantitative analysis was done using a Tanon 5200 Multi chemiluminescent imaging system.
Statistical analysis
All data were analyzed using one-way ANOVA or a two-tailed Student's t-test with software Microsoft Excel. The values are represented as mean ± SD. Statistical data are provided as Supplemental Data Set 1.
Accession numbers
The RNA-seq data related to this research were deposited at NCBI SRA, which can be found under the following accession numbers: SRR1695191 (5 DPA fiber), SRR1695192 (10 DPA fiber), SRR1695193 (20 DPA fiber), SRR1695194 (25 DPA fiber). The gene sequence data from this article can be found in the Cotton Functional Genomics Database (https://cottonfgd.net/) under the following accession numbers: GhBES1.4, Ghicr24_A05G201500.1; GhACS6, Ghicr24_A12G253200.1; GhACS7, Ghicr24_D05G029800.1; GhACO1, Ghicr24_D05G173700.1; GhACO2, Ghicr24_A06G178700.1; GhACO3, Ghicr24_D08G054500.1; GhACO4, Ghicr24_A07G092100.1; GhEXP1, Ghicr24_D10G120700.1; GhTUB1, Ghicr24_A08G201600.1; GhACT1, Ghicr24_D11G059000.1; GhPP2A, Ghicr24_D01G134900.1. The gene sequence data from this article can be found in the TAIR10 (https://www.arabidopsis.org/) under the following accession numbers: AtACO2, AT1G63280; AtACO4, AT1G05010; AtCPD, AT5G05690; AtDWF4, AT3G50660; AtPRE5, AT3G28857; AtEXP8, AT2G40610.
Supplementary Material
Acknowledgments
We thank Prof. Jiahe Wu for constructive comments.
Contributor Information
Zuoren Yang, Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, 450001 Henan, China; State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China; Western Agricultural Research Center, Chinese Academy of Agricultural Sciences, Changji, 831100 Xinjiang, China.
Zhao Liu, Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, 450001 Henan, China; State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China.
Xiaoyang Ge, Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, 450001 Henan, China; State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China.
Lili Lu, State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China.
Wenqiang Qin, State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China.
Ghulam Qanmber, Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, 450001 Henan, China; State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China.
Le Liu, State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China.
Zhi Wang, Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, 450001 Henan, China; State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China.
Fuguang Li, Zhengzhou Research Base, State Key Laboratory of Cotton Biology, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, 450001 Henan, China; State Key Laboratory of Cotton Biology, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, 455000 Henan, China; Western Agricultural Research Center, Chinese Academy of Agricultural Sciences, Changji, 831100 Xinjiang, China.
Author contributions
F.G.L. and Z.R.Y. conceived and designed the research. Z.R.Y., Z.L., X.Y.G., L.L.L., G.Q., and L.L. performed the experiments. W.Q.Q. contributed to materials and phenotyping. Z.L., W.Q.Q., and Z.W. analyzed the data. Z.R.Y., Z.L., and F.G.L. wrote the paper.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Evolutionary tree analysis of GhKCS genes in upland cotton.
Supplemental Figure S2. Heatmap representation of the expression of GhKCS in ZM24 fiber.
Supplemental Figure S3. Heatmap representation of the expression of GhKCS in ZM24 and pag1 fiber.
Supplemental Figure S4. GhKCS gene expression patterns after BL treatment.
Supplemental Figure S5. Effects of exogenously applied BL and BRZ on fiber cell elongation.
Supplemental Figure S6. Effects of exogenously applied C24 and ACE on fiber cell elongation.
Supplemental Figure S7. C24 restores the inhibition fiber phenotype of pag1 and ZM24.
Supplemental Figure S8. Plants overexpressing GhBES1.4 showed BR response phenotype.
Supplemental Figure S9. GhBES1.4 transgenic cotton showed response to BR.
Supplemental Figure S10. Heatmap representation of the expression from DEGs in ZM24 and GhBES1.4-OE fiber.
Supplemental Figure S11. Expression analysis of GhKCS genes in GhBES1.4-RNAi/OE cotton.
Supplemental Figure S12. Phenotypes of C24 and BL exogenously applied to cut1.
Supplemental Figure S13. Phenotypes of GhKCS10_At-OE and ZM24 in 15 DPA and mature fibers.
Supplemental Figure S14. GhKCS10_At-RNAi transgenic cotton is insensitive to BR.
Supplemental Figure S15. Analysis of cis-acting elements of GhKCS gene promoter.
Supplemental Figure S16. GhKCS10_At is a site-specific target of GhBES1.4.
Supplemental Figure S17. Ethylene production in cultured ZM24, GhKCS10_At-OE, GhKCS10_At-RNAi, and pag1 10 DPA fiber.
Supplemental Table S1. The information of 57 GhKCS genes.
Supplemental Table S2. The comparative analysis of GhKCSs gene expression levels of ZM24 and pag1 mutant in 10-DPA fiber.
Supplemental Table S3. Representative genes in GhBES1.4-overexpressing cotton fiber.
Supplemental Table S4. The fiber quality of GhKCS10_At overexpression lines and ZM24 in 2019 and 2020 growing in Anyang, China.
Supplemental Table S5. Comparisons of 50 bolls’ yield and lint yield of GhKCS10_At expression lines and ZM24 in 2020.
Supplemental Table S6. Primer sequences used in this study.
Supplemental Data Set S1. Summary of statistical analyses.
Supplemental File S1. Multiple protein sequence alignment used to generate the phylogenetic tree shown in Supplemental Figure S1.
Supplemental File S2. Newick file format of the phylogenetic tree shown in Supplemental Figure S1.
Funding
This work was funded by the National Key R&D Program of China (2022YFF1001400); National Natural Science Foundation of China (No. 31971987 and No. 31621005); Natural Science Foundation of Henan (No. 212300410093); and Natural Science Foundation of Xinjiang Uygur Autonomous Region (No. 2022D01E08).
Data availability
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplemental data.
References
- Bai M-Y, Zhang L-Y, Gampala SS, Zhu S-W, Song W-Y, Chong K, Wang Z-Y. Functions of OsBZR1 and 14-3-3 proteins in brassinosteroid signaling in rice. Proc Nat Acad Sci. 2007:104(34):13839–13844. 10.1073/pnas.0706386104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajwa KS, Shahid AA, Rao AQ, Bashir A, Aftab A, Husnain T. Stable transformation and expression of GhEXPA8 fiber expansin gene to improve fiber length and micronaire value in cotton. Front Plant Sci. 2015:6(1):838. 10.3389/fpls.2015.00838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batsale M, Bahammou D, Fouillen L, Mongrand S, Joubes J, Domergue F. Biosynthesis and functions of very-long-chain fatty acids in the responses of plants to abiotic and biotic stresses. Cells. 2021:10(6):1284. 10.3390/cells10061284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blacklock BJ, Jaworski JG. Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem Biophys Res Commun. 2006:346(2):583–590. 10.1016/j.bbrc.2006.05.162 [DOI] [PubMed] [Google Scholar]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant. 2020:13(8):1194–1202. 10.1016/j.molp.2020.06.009 [DOI] [PubMed] [Google Scholar]
- Chen L-G, Gao Z, Zhao Z, Liu X, Li Y, Zhang Y, Liu X, Sun Y, Tang W. BZR1 Family transcription factors function redundantly and indispensably in BR signaling but exhibit BRI1-independent function in regulating anther development in Arabidopsis. Mol Plant. 2019:12(10):1408–1415. 10.1016/j.molp.2019.06.006 [DOI] [PubMed] [Google Scholar]
- Clouse SD, Sasse JM. Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Phys. 1998:49(1):427–451. 10.1146/annurev.arplant.49.1.427 [DOI] [PubMed] [Google Scholar]
- De Bigault Du Granrut A, Cacas JL. How very-long-chain fatty acids could signal stressful conditions in plants? Front Plant Sci. 2016:7(1):1490. 10.3389/fpls.2016.01490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dursun A, Güler Z. Internal of external standard techniques for quantification of free fatty acids (FFAs) in raw milk and kefir samples. Sci Study Res Chem Chem Eng Biotechnol Food Ind. 2021:22(2):125–139. [Google Scholar]
- Ge X, Xu J, Yang Z, Yang X, Wang Y, Chen Y, Wang P, Li F. Efficient genotype-independent cotton genetic transformation and genome editing. J Integr Plant Biol. 2022. 10.1111/jipb.13427 [DOI] [PubMed] [Google Scholar]
- Haslam TM, Kunst L. Extending the story of very-long-chain fatty acid elongation. Plant Sci. 2013:210:93–107. 10.1016/j.plantsci.2013.05.008 [DOI] [PubMed] [Google Scholar]
- He JX, Gendron JM, Sun Y, Gampala SS, Gendron N, Sun CQ, Wang ZY. BZR1 Is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science. 2005:307(5715):1634–1638. 10.1126/science.1107580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Zhou J, Liu C, Liu L, Shen L, Yu H. Nuclear factor Y-mediated H3K27me3 demethylation of the SOC1 locus orchestrates flowering responses of Arabidopsis. Nat Commun. 2014:5(1):1–14. 10.1038/ncomms5601 [DOI] [PubMed] [Google Scholar]
- Hu Y, Chen J, Fang L, Zhang Z, Ma W, Niu Y, Ju L, Deng J, Zhao T, Lian J, et al. Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat Genet. 2019:51(4):739–748. 10.1038/s41588-019-0371-5 [DOI] [PubMed] [Google Scholar]
- Kang JG, Yun J, Kim DH, Chung KS, Fujioka S, Kim JI, Dae HW, Yoshida S, Takatsuto S, Song PS, et al. Light and brassinosteroid signals are integrated via a dark-induced small G protein in etiolated seedling growth. Cell. 2001:105(5):625–636. 10.1016/S0092-8674(01)00370-1 [DOI] [PubMed] [Google Scholar]
- Kim HJ, Triplett BA. Cotton fiber growth in planta and in vitro. Models for plant cell elongation and cell wall biogenesis. Plant Physiol. 2001:127(4):1361–1366. 10.1104/pp.010724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kour J, Kohli SK, Khanna K, Bakshi P, Sharma P, Singh AD, Ibrahim M, Devi K, Sharma N, Ohri P. Brassinosteroid signaling, crosstalk and, physiological functions in plants under heavy metal stress. Front Plant Sci. 2021:12:608061. 10.3389/fpls.2021.608061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016:33(7):1870–1874. 10.1093/molbev/msw054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Burns TH, Light G, Sun Y, Fokar M, Kasukabe Y, Fujisawa K, Maekawa Y, Allen RD. Xyloglucan endotransglycosylase/hydrolase genes in cotton and their role in fiber elongation. Planta. 2010:232(5):1191–1205. 10.1007/s00425-010-1246-2 [DOI] [PubMed] [Google Scholar]
- Li L, Yu XF, Thompson A, Guo M, Yoshida S, Asami T, Chory J, Yin Y. Arabidopsis MYB30 is a direct target of BES1 and cooperates with BES1 to regulate brassinosteroid-induced gene expression. Plant J. 2009:58(2):275–286. 10.1111/j.1365-313X.2008.03778.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Xing K, Qanmber G, Chen G, Liu L, Guo M, Hou Y, Lu L, Qu L, Liu Z. GhBES1 mediates brassinosteroid regulation of leaf size by activating expression of GhEXO2 in cotton (Gossypium hirsutum). Plant Mol Biol. 2023:111(1–2):89–106. 10.1007/s11103-022-01313-5 [DOI] [PubMed] [Google Scholar]
- Li S, Zhu B, Pirrello J, Xu C, Zhang B, Bouzayen M, Chen K, Grierson D. Roles of RIN and ethylene in tomato fruit ripening and ripening-associated traits. New Phytol. 2020:226(2):460–475. 10.1111/nph.16362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Liu W, Ren Z, Wang X, Liu J, Yang Z, Zhao J, Pei X, Liu Y, He K. Glucose regulates cotton fiber elongation by interacting with brassinosteroid. J Exp Bot. 2022:73(3):711–726. 10.1093/jxb/erab451 [DOI] [PubMed] [Google Scholar]
- Li X-B, Fan X-P, Wang X-L, Cai L, Yang W-C. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell. 2005:17(3):859–875. 10.1105/tpc.104.029629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Wang NN, Wang Y, Liu D, Gao Y, Li L, Li XB. The cotton XLIM protein (Gh XLIM 6) is required for fiber development via maintaining dynamic F-actin cytoskeleton and modulating cellulose biosynthesis. Plant J. 2018:96(6):1269–1282. 10.1111/tpj.14108 [DOI] [PubMed] [Google Scholar]
- Lisso J, Steinhauser D, Altmann T, Kopka J, Müssig C. Identification of brassinosteroid-related genes by means of transcript co-response analyses. Nucl Acids Res. 2005:33(8):2685–2696. 10.1093/nar/gki566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu GJ, Xiao GH, Liu NJ, Liu D, Chen PS, Qin YM, Zhu YX. Targeted lipidomics studies reveal that linolenic acid promotes cotton fiber elongation by activating phosphatidylinositol and phosphatidylinositol monophosphate biosynthesis. Mol Plant. 2015:8(6):911–921. 10.1016/j.molp.2015.02.010 [DOI] [PubMed] [Google Scholar]
- Liu L, Chen G, Li S, Gu Y, Lu L, Qanmber G, Mendu V, Liu Z, Li F, Yang Z. A brassinosteroid transcriptional regulatory network participates in regulating fiber elongation in cotton. Plant Physiol. 2022. 10.1093/plphys/kiac590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Qanmber G, Lu L, Qin W, Liu J, Li J, Ma S, Yang Z, Yang Z. Genome-wide analysis of BES1 genes in Gossypium revealed their evolutionary conserved roles in brassinosteroid signaling. Sci China Life Sci. 2018:61(12):1566–1582. 10.1007/s11427-018-9412-x [DOI] [PubMed] [Google Scholar]
- Liu ZH, Chen Y, Wang NN, Chen YH, Wei N, Lu R, Li Y, Li XB. A basic helix-loop-helix protein (GhFP1) promotes fibre elongation of cotton (Gossypium hirsutum) by modulating brassinosteroid biosynthesis and signalling. New Phytol. 2020:225(6):2439–2452. 10.1111/nph.16301 [DOI] [PubMed] [Google Scholar]
- Lu R, Li Y, Zhang J, Wang Y, Zhang J, Li Y, Zheng Y, Li XB. The bHLH/HLH transcription factors GhFP2 and GhACE1 antagonistically regulate fiber elongation in cotton. Plant Physiol. 2022:189(2):628–643. 10.1093/plphys/kiac088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar AA, Kunst L. Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 1997:12(1):121–131. 10.1046/j.1365-313X.1997.12010121.x [DOI] [PubMed] [Google Scholar]
- Nobusawa T, Okushima Y, Nagata N, Kojima M, Sakakibara H, Umeda M. Synthesis of very-long-chain fatty acids in the epidermis controls plant organ growth by restricting cell proliferation. PLoS Biol. 2013:11(4):e1001531. 10.1371/journal.pbio.1001531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolan TM, Vukašinović N, Liu D, Russinova E, Yin Y. Brassinosteroids: multidimensional regulators of plant growth, development, and stress responses. Plant Cell. 2020:32(2):295–318. 10.1105/tpc.19.00335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh E, Kang H, Yamaguchi S, Park J, Lee D, Kamiya Y, Choi G. Genome-wide analysis of genes targeted by PHYTOCHROME INTERACTING FACTOR 3-LIKE5 during seed germination in Arabidopsis. Plant Cell. 2009:21(2):403–419. 10.1105/tpc.108.064691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planas-Riverola A, Gupta A, Betegón-Putze I, Bosch N, Ibañes M, Caño-Delgado AI. Brassinosteroid signaling in plant development and adaptation to stress. Development. 2019:146(5):dev151894. 10.1242/dev.151894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiao S, Sun S, Wang L, Wu Z, Li C, Li X, Wang T, Leng L, Tian W, Lu T. The RLA1/SMOS1 transcription factor functions with OsBZR1 to regulate brassinosteroid signaling and rice architecture. Plant Cell. 2017:29(2):292–309. 10.1105/tpc.16.00611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y-M, Hu C-Y, Pang Y, Kastaniotis AJ, Hiltunen JK, Zhu Y-X. Saturated very-long-chain fatty acids promote cotton fiber and Arabidopsis cell elongation by activating ethylene biosynthesis. Plant Cell. 2007a:19(11):3692–3704. 10.1105/tpc.107.054437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin YM, Pujol FM, Hu CY, Feng JX, Kastaniotis AJ, Hiltunen JK, Zhu YX. Genetic and biochemical studies in yeast reveal that the cotton fibre-specific GhCER6 gene functions in fatty acid elongation. J Exp Bot. 2007b:58(3):473–481. 10.1093/jxb/erl218 [DOI] [PubMed] [Google Scholar]
- Qin YM, Zhu YX. How cotton fibers elongate: a tale of linear cell-growth mode. Curr Opin Plant Biol. 2011:14(1):106–111. 10.1016/j.pbi.2010.09.010 [DOI] [PubMed] [Google Scholar]
- Raffaele S, Vailleau F, Leger A, Joubes J, Miersch O, Huard C, Blee E, Mongrand S, Domergue F, Roby D. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell. 2008:20(3):752–767. 10.1105/tpc.107.054858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang B, Xu C, Zhang X, Cao H, Xin W, Hu Y. Very-long-chain fatty acids restrict regeneration capacity by confining pericycle competence for callus formation in Arabidopsis. Proc Natl Acad Sci USA. 2016:113(18):5101–5106. 10.1073/pnas.1522466113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y-H, Zhu S-W, Mao X-Z, Feng J-X, Qin Y-M, Zhang L, Cheng J, Wei L-P, Wang Z-Y, Zhu Y-X. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006:18(3):651–664. 10.1105/tpc.105.040303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Fan X-Y, Cao D-M, Tang W, He K, Zhu J-Y, He J-X, Bai M-Y, Zhu S, Oh E. Integration of brassinosteroid signal transduction with the transcription network for plant growth regulation in Arabidopsis. Dev Cell. 2010:19(5):765–777. 10.1016/j.devcel.2010.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Veerabomma S, Abdel-Mageed HA, Fokar M, Asami T, Yoshida S, Allen RD. Brassinosteroid regulates fiber development on cultured cotton ovules. Cell Physiol. 2005:46(8):1384–1391. 10.1093/pcp/pci150 [DOI] [PubMed] [Google Scholar]
- Szekeres M, Nemeth K, Koncz-Kalman Z, Mathur J, Kauschmann A, Altmann T, Redei GP, Nagy F, Schell J, Koncz C. Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell. 1996:85(2):171–182. 10.1016/S0092-8674(00)81094-6 [DOI] [PubMed] [Google Scholar]
- Tang W, Tu L, Yang X, Tan J, Deng F, Hao J, Guo K, Lindsey K, Zhang X. The calcium sensor GhCaM7 promotes cotton fiber elongation by modulating reactive oxygen species (ROS) production. New Phytol. 2014:202(2):509–520. 10.1111/nph.12676 [DOI] [PubMed] [Google Scholar]
- Trinh DC, Lavenus J, Goh T, Boutte Y, Drogue Q, Vaissayre V, Tellier F, Lucas M, Voss U, Gantet P, et al. PUCHI Regulates very long chain fatty acid biosynthesis during lateral root and callus formation. Proc Natl Acad Sci USA. 2019:116(28):14325–14330. 10.1073/pnas.1906300116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z-Y, Bai M-Y, Oh E, Zhu J-Y. Brassinosteroid signaling network and regulation of photomorphogenesis. Annu Rev Genet. 2012:46(1):701–724. 10.1146/annurev-genet-102209-163450 [DOI] [PubMed] [Google Scholar]
- Wang Z-Y, Nakano T, Gendron J, He J, Chen M, Vafeados D, Yang Y, Fujioka S, Yoshida S, Asami T. Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell. 2002:2(4):505–513. 10.1016/S1534-5807(02)00153-3 [DOI] [PubMed] [Google Scholar]
- Xie L-J, Chen Q-F, Chen M-X, Yu L-J, Huang L, Chen L, Wang F-Z, Xia F-N, Zhu T-R, Wu J-X. Unsaturation of very-long-chain ceramides protects plant from hypoxia-induced damages by modulating ethylene signaling in Arabidopsis. Plos Genet. 2015:11(3):e1005143. 10.1371/journal.pgen.1005143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Chen Q, Huang L, Luo M. Advances about the roles of membranes in cotton fiber development. Membranes. 2021:11(7):471. 10.3390/membranes11070471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi T, Shiono K, Nagano M, Fukazawa A, Ando M, Takamure I, Mori H, Nishizawa NK, Kawai-Yamada M, Tsutsumi N. Ethylene biosynthesis is promoted by very-long-chain fatty acids during lysigenous aerenchyma formation in rice roots. Plant Physiol. 2015:169(1):180–193. 10.1104/pp.15.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Qanmber G, Wang Z, Yang Z, Li F. Gossypium genomics: trends, scope, and utilization for cotton improvement. Trends Plant Sci. 2020:25(5):488–500. 10.1016/j.tplants.2019.12.011 [DOI] [PubMed] [Google Scholar]
- Yang Z, Zhang C, Yang X, Liu K, Wu Z, Zhang X, Zheng W, Xun Q, Liu C, Lu L. PAG1, A cotton brassinosteroid catabolism gene, modulates fiber elongation. New Phytol. 2014:203(2):437–448. 10.1111/nph.12824 [DOI] [PubMed] [Google Scholar]
- Yephremov A, Wisman E, Huijser P, Huijser C, Wellesen K, Saedler H. Characterization of the FIDDLEHEAD gene of Arabidopsis reveals a link between adhesion response and cell differentiation in the epidermis. Plant Cell. 1999:11(11):2187–2201. 10.1105/tpc.11.11.2187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin Y, Wang Z-Y, Mora-Garcia S, Li J, Yoshida S, Asami T, Chory J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell. 2002:109(2):181–191. 10.1016/S0092-8674(02)00721-3 [DOI] [PubMed] [Google Scholar]
- Yu Y, Wu S, Nowak J, Wang G, Han L, Feng Z, Mendrinna A, Ma Y, Wang H, Zhang X, et al. Live-cell imaging of the cytoskeleton in elongating cotton fibres. Nat Plants. 2019:5(5):498–504. 10.1038/s41477-019-0418-8 [DOI] [PubMed] [Google Scholar]
- Zhan H, Xiong H, Wang S, Yang ZN. Anther endothecium-derived very-long-chain fatty acids facilitate pollen hydration in Arabidopsis. Mol Plant. 2018:11(8):1101–1104. 10.1016/j.molp.2018.05.002 [DOI] [PubMed] [Google Scholar]
- Zhang C, Bai M-Y, Chong K. Brassinosteroid-mediated regulation of agronomic traits in rice. Plant Cell Rep. 2014:33(5):683–696. 10.1007/s00299-014-1578-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang TZ, Hu Y, Jiang WK, Fang L, Guan XY, Chen JD, Zhang JB, Saski CA, Scheffler BE, Stelly DM, et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat Biotechnol. 2015:33(5):531–U252. 10.1038/nbt.3207 [DOI] [PubMed] [Google Scholar]
- Zhang X, Henriques R, Lin S-S, Niu Q-W, Chua N-H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protocols. 2006:1(2):641–646. 10.1038/nprot.2006.97 [DOI] [PubMed] [Google Scholar]
- Zhao B, Cao JF, Hu GJ, Chen ZW, Wang LY, Shangguan XX, Wang LJ, Mao YB, Zhang TZ, Wendel JF, et al. Core cis-element variation confers subgenome-biased expression of a transcription factor that functions in cotton fiber elongation. New Phytol. 2018:218(3):1061–1075. 10.1111/nph.15063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Zhang ZT, Li M, Wei XZ, Li XJ, Li BY, Li XB. Cotton (Gossypium hirsutum) 14-3-3 proteins participate in regulation of fibre initiation and elongation by modulating brassinosteroid signalling. Plant Biotechnol J. 2015:13(2):269–280. 10.1111/pbi.12275 [DOI] [PubMed] [Google Scholar]
- Zhu X, Tellier F, Gu Y, Li S. Disruption of very-long-chain-fatty acid synthesis has an impact on the dynamics of cellulose synthase in Arabidopsis thaliana. Plants (Basel). 2020:9(11):1599. 10.3390/plants9111599 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplemental data.