Abstract
Small RNAs (sRNAs) associate with ARGONAUTE (AGO) proteins forming effector complexes with key roles in gene regulation and defense responses against molecular parasites. In multicellular eukaryotes, extensive duplication and diversification of RNA interference (RNAi) components have resulted in intricate pathways for epigenetic control of gene expression. The unicellular alga Chlamydomonas reinhardtii also has a complex RNAi machinery, including 3 AGOs and 3 DICER-like proteins. However, little is known about the biogenesis and function of most endogenous sRNAs. We demonstrate here that Chlamydomonas contains uncommonly long (>26 nt) sRNAs that associate preferentially with AGO1. Somewhat reminiscent of animal PIWI-interacting RNAs, these >26 nt sRNAs are derived from moderately repetitive genomic clusters and their biogenesis is DICER-independent. Interestingly, the sequences generating these >26-nt sRNAs have been conserved and amplified in several Chlamydomonas species. Moreover, expression of these longer sRNAs increases substantially under nitrogen or sulfur deprivation, concurrently with the downregulation of predicted target transcripts. We hypothesize that the transposon-like sequences from which >26-nt sRNAs are produced might have been ancestrally targeted for silencing by the RNAi machinery but, during evolution, certain sRNAs might have fortuitously acquired endogenous target genes and become integrated into gene regulatory networks.
IN A NUTSHELL.
Background: Noncoding RNAs are not translated into proteins. However, many play key roles in various biological processes. In eukaryotes, small RNAs (sRNAs) can bind to matching messenger RNAs (mRNAs) and trigger their degradation or shut down their translation into proteins, a phenomenon called RNA interference. These sRNAs associate with ARGONAUTE (AGO) proteins and regulate the expression of protein-coding genes. Other kinds of sRNAs help fight viruses and transposons. Generally, all these types of sRNAs are produced by cleaving longer double-stranded RNAs via Dicer ribonucleases. Indeed, in many organisms, diverse Dicer-dependent sRNAs affect growth, development, and disease tolerance.
Question: We wanted to characterize the sRNAs functioning in the single-cell alga Chlamydomonas reinhardtii. This green alga lives in many different environments throughout the world and has become a valuable model for biological research.
Findings: C. reinhardtii contains a class of sRNAs (greater than 26 nucleotides in length) that associates preferentially with a specific AGO protein named AGO1. These sRNAs are atypical since they do not require Dicer enzymes for their production. They are derived from moderately repetitive, transposon-like sequences conserved in related Chlamydomonas species. Interestingly, the abundance of these sRNAs increased substantially in cells subject to nitrogen or sulfur deprivation, simultaneously with the downregulation of predicted target mRNAs. We postulate that this class of sRNAs might have a role in algal tolerance to nutritional stress.
Next steps: We would like to find out how these sRNAs may help the alga cope with nitrogen or sulfur deprivation and the molecular mechanism(s) by which they may regulate gene expression. This information may be helpful for improving nutrient use efficiency in crops.
Introduction
Small silencing RNAs (sRNAs) play important roles in the regulation of gene expression, heterochromatin formation, DNA methylation, maintenance of genome stability, intercellular communication, transposon repression, and/or defense against viruses in a wide range of eukaryotes (Ghildiyal and Zamore 2009; Borges and Martienssen 2015; Wendte and Pikaard 2017; Yu et al. 2017; Bartel 2018; Ozata et al. 2019; Chen and Rechavi 2022). Endogenous sRNAs are generally 20 to 35 nucleotides (nt) in length and associate with members of the ARGONAUTE (AGO) family of proteins, forming effector complexes (Ghildiyal and Zamore 2009; Swarts et al. 2014; Borges and Martienssen 2015; Wendte and Pikaard 2017; Yu et al. 2017; Bartel 2018; Ozata et al. 2019; Chen and Rechavi 2022; Iwakawa and Tomari 2022). These effector complexes then recognize target sequences by complementarity with the guide sRNAs and trigger post-transcriptional or transcriptional gene silencing by diverse molecular mechanisms. Beyond these defining features, many different classes of sRNAs have been described in a broad spectrum of eukaryotes (Katiyar-Agarwal et al. 2007; Ghildiyal and Zamore 2009; Axtell 2013; Borges and Martienssen 2015; Yu et al. 2017; Bartel 2018; Hardcastle et al. 2018; Ozata et al. 2019; Feng et al. 2020; Lunardon et al. 2020; Müller et al. 2020; Rzeszutek and Betlej 2020; Wu et al. 2020; Alves and Nogueira 2021; Chen et al. 2021; Reshetnyak et al. 2021; Baldrich et al. 2022; Chen and Rechavi 2022; Haase 2022).
In most species, endogenous sRNAs derived from dsRNA precursors processed by DICER-like (DCL) endonucleases can be grouped into 2 main classes: microRNAs (miRNAs) and small interfering RNAs (siRNAs; Ghildiyal and Zamore 2009; Axtell 2013; Borges and Martienssen 2015; Yu et al. 2017; Bartel 2018; Lunardon et al. 2020; Müller et al. 2020; Chen et al. 2021). miRNAs are typically 20 to 22 nt in length, processed from imperfectly paired stem-loop regions of ssRNA precursors, and regulate gene expression through mRNA degradation and/or translational repression (Ghildiyal and Zamore 2009; Axtell 2013; Borges and Martienssen 2015; Yu et al. 2017; Bartel 2018). siRNAs are produced from near-perfectly complementary dsRNAs of various origins, participate in post-transcriptional or transcriptional gene silencing, and are grouped into multiple subclasses (Ghildiyal and Zamore 2009; Axtell 2013; Borges and Martienssen 2015; Lunardon et al. 2020; Chen et al. 2021; Baldrich et al. 2022; Chen and Rechavi 2022). In angiosperms, the most abundant siRNA subclass, ∼24-nt heterochromatic siRNAs, is involved in the canonical RNA-directed DNA methylation pathway primarily targeting transposons and other repeats (Axtell 2013; Borges and Martienssen 2015; Lunardon et al. 2020; Chen et al. 2021; Baldrich et al. 2022; Chen and Rechavi 2022).
Another major siRNA subclass comprises secondary siRNAs, including trans-acting siRNAs, phased siRNAs, and epigenetically activated siRNAs, which may silence genes and/or transposons. Their biogenesis is generally triggered by miRNA-directed cleavage of a transcript, which is then converted to dsRNA by an RNA-dependent RNA polymerase and processed by DCL proteins into siRNAs, often in a phased pattern (Axtell 2013; Borges and Martienssen 2015; Lunardon et al. 2020; Chen et al. 2021; Baldrich et al. 2022; Chen and Rechavi 2022). Several additional siRNA subtypes (e.g. natural antisense transcript-derived siRNAs, hairpin-derived siRNAs, DNA double-strand break-induced sRNAs, and bacteria-induced long siRNAs) have also been described in land plants, particularly in model species such as Arabidopsis (Arabidopsis thaliana), and the boundaries among the siRNA classes are becoming quite diffuse (Katiyar-Agarwal et al. 2007; Axtell 2013; Borges and Martienssen 2015; Hardcastle et al. 2018; Lunardon et al. 2020; Rzeszutek and Betlej 2020; Wu et al. 2020; Alves and Nogueira 2021; Reshetnyak et al. 2021; Chen and Rechavi 2022). Metazoans have an additional large class of longer sRNAs (∼22 to 35 nt) termed PIWI-interacting RNAs (piRNAs). piRNAs are distinct from siRNAs since they derive from ssRNAs, do not require DCL proteins for their processing, and bind to PIWI (P-element Induced Wimpy Testis) proteins, an evolutionarily distinct clade of AGO proteins (Ghildiyal and Zamore 2009; Swarts et al. 2014; Ozata et al. 2019; Haase 2022).
sRNA-mediated silencing is generally accepted to have evolved as a defense mechanism against viruses and transposable elements, before being co-opted to regulate the expression of endogenous genes (Cerutti and Casas-Mollano 2006; Shabalina and Koonin 2008). Extensive duplication and specialization of proteins involved in sRNA biogenesis and/or effector functions have contributed to pathway diversification (Ghildiyal and Zamore 2009; Swarts et al. 2014; Borges and Martienssen 2015; Lee and Carroll 2018; Wang et al. 2021; Chen and Rechavi 2022); and the sorting of sRNAs into specific AGOs/effector complexes ultimately determines their biological function(s) (Ghildiyal and Zamore 2009; Czech and Hannon 2011; Borges and Martienssen 2015; Iwakawa and Tomari 2022).
In land plants, the structure of sRNA duplex precursors (such as thermodynamic asymmetry), the presence and location of mismatches and bulges, as well as the identity of the 5′-terminal nucleotide of sRNAs affect their loading onto individual AGOs and the choice of guide strand (Mi et al. 2008; Takeda et al. 2008; Havecker et al. 2010; Czech and Hannon 2011; Zhu et al. 2011; Frank et al. 2012; Endo et al. 2013; Zhang et al. 2014; Borges and Martienssen 2015; Iwakawa and Tomari 2022). For instance, Arabidopsis has 10 AGOs, with AGO1 and AGO10 preferentially binding sRNAs with a 5′ uracil and AGO5 showing a bias for 5′ cytosine, whereas AGO2, AGO4, AGO6, and AGO9 prefer sRNAs with 5′ adenine (Mi et al. 2008; Takeda et al. 2008; Havecker et al. 2010; Czech and Hannon 2011; Frank et al. 2012; Borges and Martienssen 2015). The resulting large variety of sRNA-directed effector complexes participate in distinct, yet often intertwined, regulatory pathways that influence development, responses to abiotic and biotic stresses, reproduction, and genome reprogramming (Ghildiyal and Zamore 2009; Czech and Hannon 2011; Borges and Martienssen 2015; Lee and Carroll 2018; Chen and Rechavi 2022). Moreover, new sRNA classes and new members of existing classes continue to be discovered, sometimes highlighting the evolution of lineage-specific regulatory mechanisms.
The green unicellular alga Chlamydomonas (Chlamydomonas reinhardtii) is a classical reference organism for studying photosynthesis, chloroplast biology, cell cycle control, and cilia structure and function (Salomé and Merchant 2019). This alga was the first unicellular organism in which miRNAs were described, and deep sequencing of short RNAs revealed that it contains a wide variety of endogenous sRNAs (Molnár et al. 2007; Zhao et al. 2007; Voshall et al. 2015; Müller et al. 2020). Chlamydomonas also encodes a complex RNA interference (RNAi) machinery consisting of 3 DCL proteins and 3 AGOs but, like most metazoans, lacks a canonical RNA-dependent RNA polymerase (Casas-Mollano et al. 2008; Voshall et al. 2015; Valli et al. 2016; Chung et al. 2019). In addition, distinct from land plants, Chlamydomonas miRNAs appear to use a metazoan-like seed matching rule to identify their target transcripts but, distinct from animals, the binding sites are predominantly located within the mRNA coding sequences rather than in the 3′ untranslated regions (UTRs; Yamasaki et al. 2013; Chung et al. 2017; Iwakawa and Tomari 2017). Nonetheless, the role(s) of most endogenous sRNAs and core RNAi components, particularly with regards to pathway specialization, remains to be explored in Chlamydomonas. The highly similar AGO2 and AGO3 proteins (∼90% amino acid identity) bind preferentially to 20 to 22-nt sRNAs (Voshall et al. 2015; Chung et al. 2019) and AGO3 has been shown to associate with most bona fide miRNAs and mediate target transcript cleavage and/or translational repression (Voshall et al. 2015; Yamasaki et al. 2016; Chung et al. 2019). However, the biological function(s) of AGO1 remains largely uncharacterized. To gain further insights into the evolution of sRNAs and RNAi-related pathways in eukaryotes, particularly in unicellular photosynthetic organisms, we characterized here a unique class of >26-nt sRNAs that associate preferentially with AGO1 in Chlamydomonas.
Results
AGO1-associated sRNAs
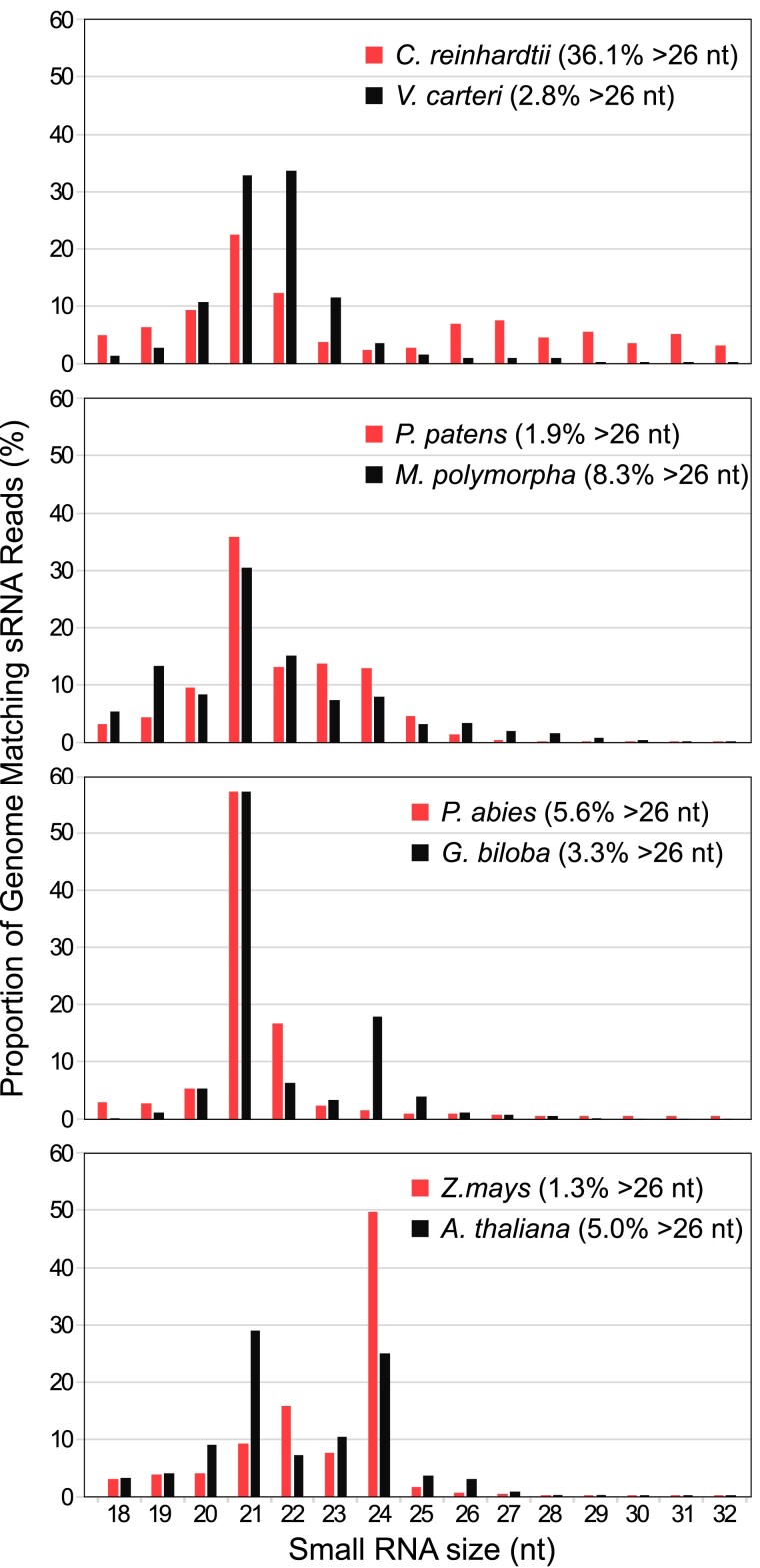
Deep sequencing of total sRNA libraries from vegetative Chlamydomonas cells has previously revealed that this unicellular organism contains a complex array of sRNAs, including miRNAs and a variety of endogenous sRNAs (Molnár et al. 2007; Zhao et al. 2007; Voshall et al. 2015; Müller et al. 2020). Notably, more than 36% of the sequenced sRNAs were longer than 26 nt (Fig. 1). By contrast, the abundance of >26-nt sRNAs is fairly low in diverse libraries from land plants (Fig. 1; Axtell 2013; Hardcastle et al. 2018; Feng et al. 2020; Lunardon et al. 2020; Chen et al. 2021; Baldrich et al. 2022), including mosses (e.g. Physcomitrium patens), liverworts (e.g. Marchantia polymorpha), gymnosperms (e.g. Norway spruce [Picea abies] and gingko [Ginkgo biloba]), and angiosperms (e.g. Arabidopsis and maize [Zea mays]). sRNAs longer than 26 nt are also rare in Volvox carteri (Fig. 1; Li et al. 2014; Dueck et al. 2016), a green alga belonging to the same family, Volvocaceae, as Chlamydomonas (Craig et al. 2021). However, we acknowledge that the detection of >26-nt sRNAs in published libraries may be affected by certain experimental procedures, such as RNA size selection during library construction, and we cannot rule out the existence of longer sRNAs in other photosynthetic eukaryotes. Also, at present, little is known about the biogenesis and function of the >26-nt sRNAs detected in Chlamydomonas.
Figure 1.
Size distribution of genome-mapped total sRNAs in green algae and land plants. References for the analyzed sRNA libraries are indicated in parenthesis. nt, nucleotide. Green algal species: C. reinhardtii (Valli et al. 2016) and V. carteri (Li et al. 2014). Land plant species: P. patens (Xia et al. 2016), M. polymorpha (Lin et al. 2016), P. abies (Wilkinson et al. 2021), G. biloba (Zhang et al. 2015), Z. mays (Liu et al. 2014), and A. thaliana (Schalk et al. 2017).
Similarly, the role(s) of AGO1, the most abundant AGO in Chlamydomonas (Chung et al. 2019), is not known. Phylogenetic analyses indicated that Chlamydomonas AGO1 and AGO2/AGO3 belong to different clades, possibly having diverged in a common ancestor of the Chlorophyceae algal class (Supplemental Fig. S1). However, putative orthologs of AGO1 appear to have been lost in some lineages of the Chlorophyceae (including V. carteri; Supplemental Fig. S1). From a functional perspective, Chlamydomonas AGO1 contains the 4 conserved domains typical of eukaryotic AGOs, namely the N-terminal, PAZ, MID, and PIWI domains (Casas-Mollano et al. 2008; Chung et al. 2019). Moreover, the PIWI domain includes the RNase H-like active site (Iwakawa and Tomari 2022), with putative functional residues in the catalytic tetrad, suggesting that AGO1 has the capability to cleave target transcripts (Chung et al. 2019).
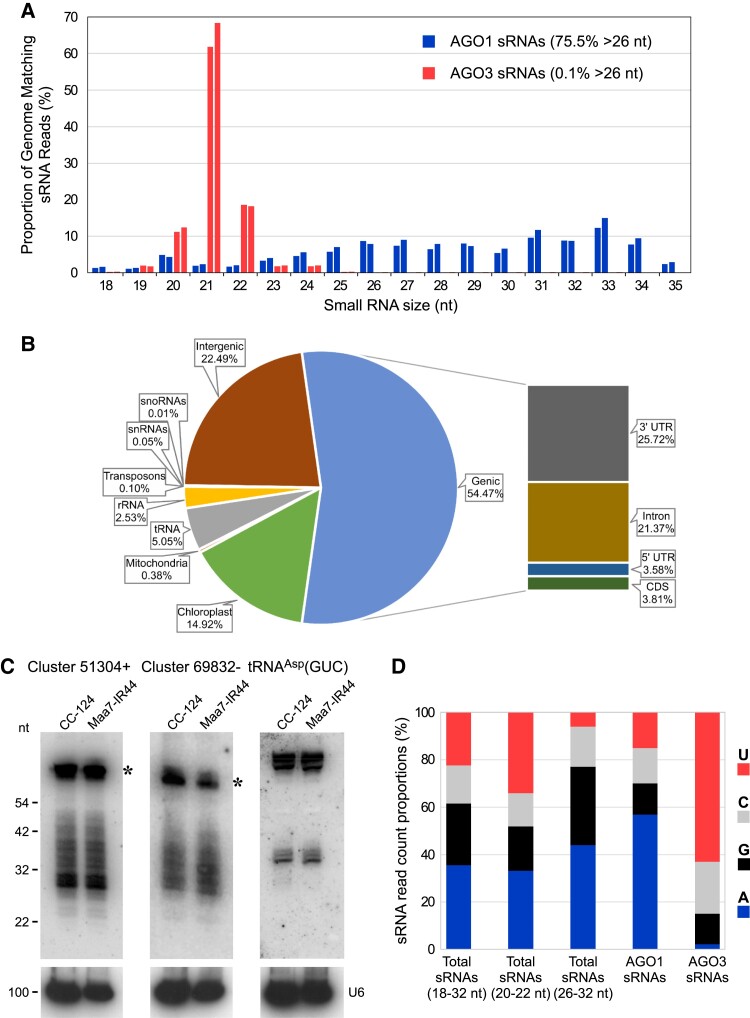
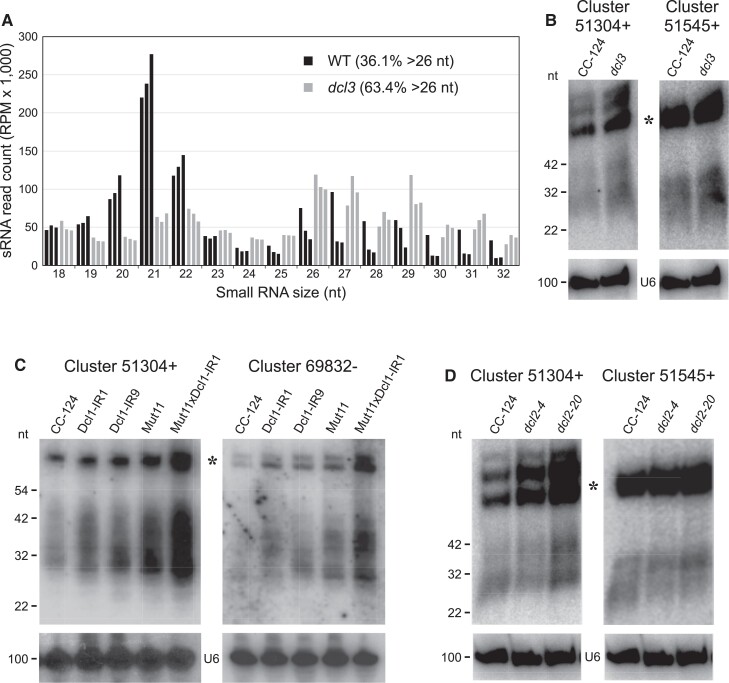
As previously described for AGO3 (Voshall et al. 2015), we introduced a transgene encoding FLAG-tagged AGO1 into the Chlamydomonas Maa7-IR44 strain, which exhibits resistance to 5-fluoroindole due to RNAi of MAA7 (METHYL ANTHRANILIC ACID 7; Ma et al. 2013). We then isolated AGO1 and its associated sRNAs by co-immunoprecipitation with an anti-FLAG antibody and generated 2 independent sRNA libraries for deep sequencing. Analysis of the obtained genome-matching reads revealed that >75% of the AGO1-associated sRNAs were longer than 26 nts (Fig. 2A). By contrast, as already reported (Voshall et al. 2015; Chung et al. 2019), AGO3 is associated preferentially with sRNAs 20 to 22 nt in length, with only 0.1% of the reads being longer than 26 nt (Fig. 2A). Redundant sRNAs associated with AGO1 predominantly matched introns (∼21.4%) and 3′ UTRs (∼25.7%) of predicted protein-coding genes as well as intergenic regions (∼22.5%) in the nuclear genome (Fig. 2B). AGO1 also bound to abundant transfer RNA (tRNA) fragments derived from genes encoded in the nuclear (∼5.1%) or chloroplast (∼14.9%) genomes (Fig. 2B). We verified the existence of a subset of AGO1-associated >26-nt sRNAs by RNA gel blot analyses (Fig. 2C), which also confirmed the occurrence of sRNA size variants ranging from 26 to almost 42 nt in length. tRNA-derived fragments were instead more precisely defined in size (Fig. 2C).
Figure 2.
AGO1-associated sRNAs in C. reinhardtii. A) Size distribution of genome-mapped AGO1-associated sRNAs (blue) and AGO3-associated sRNAs (red). Data from replicate sRNA libraries are shown as bars of the same color. B) Abundance (as % of all genome-mapped reads) of AGO1-associated redundant sRNAs matching sequences of distinct functional categories. The chloroplast fraction corresponds to tRNAs present in the chloroplast genome. C) RNA gel blot analyses of sRNAs isolated from the indicated strains and detected with probes specific for the most abundant >26-nt sRNAs of Clusters 51304+ or 69832− (Supplemental Table S3) or tRNA fragments derived from nuclear tRNAAsp(GUC). The same filters were reprobed with the U6 small nuclear RNA sequence (U6) as a control for equivalent loading of the lanes. The asterisks indicate putative RNA precursors of the >26-nt sRNAs. CC-124, wild-type strain; Maa7-IR44, CC-124 transformed with an inverted repeat (IR) transgene designed to induce RNAi of MAA7 (encoding tryptophan synthase β-subunit). D) 5′-terminal nucleotide preference of genome-mapped total sRNAs (of the indicated size fractions), AGO1-associated sRNAs and AGO3-associated sRNAs.
We also examined the 5′-terminal nucleotide bias in total sRNA libraries from wild-type Chlamydomonas strains, which presumably include sRNAs associated with AGO1, AGO2, and AGO3 as well as some unbound sRNAs (Fig. 2D). Relative to the total sRNA population (i.e. sRNAs of 18 to 32 nt in length), the fraction 26 to 32 nt in length was enriched in sRNAs starting with A, whereas the fraction 20 to 22 nt in length was enriched in sRNAs starting with U (Fig. 2D). Consistent with these findings and as an indication of binding specificity, 57% of redundant AGO1-associated sRNAs (mainly >26 nt in length) had an A at the 5′-most position (Fig. 2D), whereas most AGO3-bound sRNAs had a U as the 5′ nucleotide (Voshall et al. 2015).
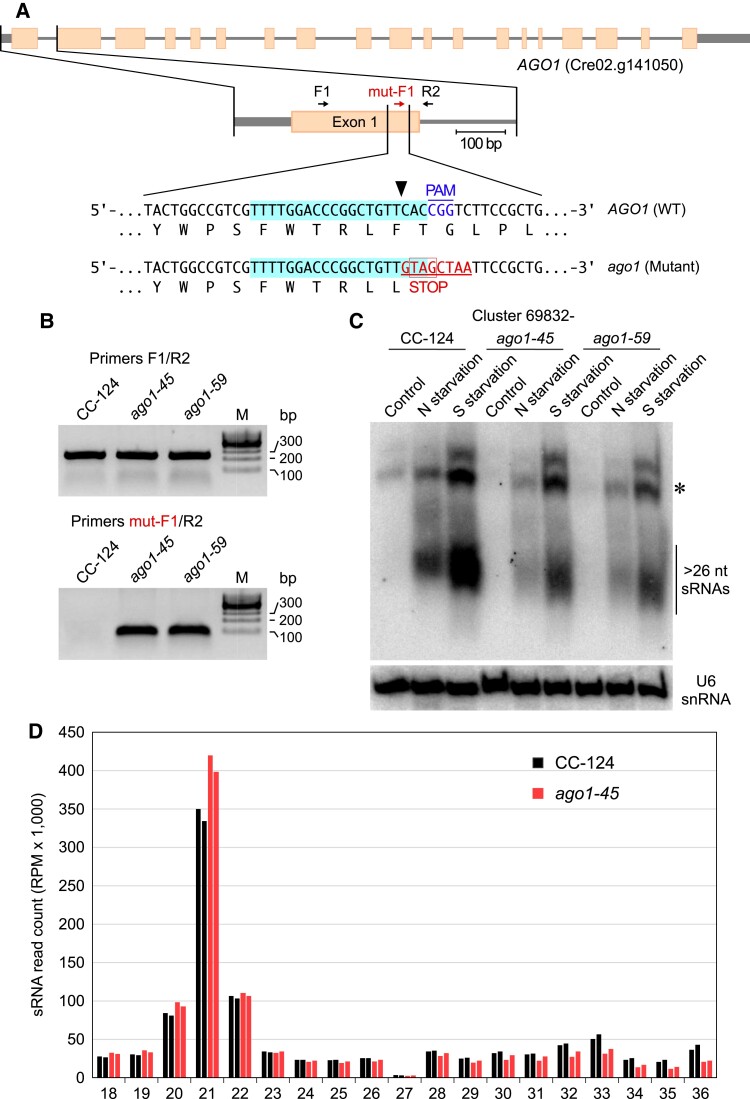
To validate the preferential association of Chlamydomonas >26-nt sRNAs with native AGO1, we used clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated nuclease 9 (Cas9)-mediated genome editing (Akella et al. 2021) to generate knockout mutants of the AGO1 gene. To this end, we electroporated cells of the wild-type strain CC-124 with both a ribonucleoprotein (RNP) complex, consisting of Cas9 and a guide RNA targeting exon 1 of AGO1, and a single-stranded oligodeoxynucleotide (ssODN), with a mutated DNA sequence overlapping the Cas9 cleavage site. Precise repair of the DNA double-strand break caused by the Cas9 RNP complex, using the ssODN as a homologous template, would substitute 8 bp within exon 1 of AGO1 (Fig. 3A), introducing an in-frame stop codon that destroys the Cas9 protospacer adjacent motif (PAM).
Figure 3.
Abundance of >26-nt sRNAs in AGO1 edited mutants. A) Schematic diagram of the AGO1 gene showing the Cas9 target region in exon 1. Short arrows indicate primers used for PCR analysis. In the wild-type sequence (top sequence), the target region is highlighted in light blue. The Cas9 cleavage site is indicated by a black arrowhead and the PAM is shown in blue font. Homology-directed repair of the DNA double-strand break, using an electroporated ssODN as template, introduces 8-bp changes (underlined red font) into the genome, including an in-frame stop codon (bottom sequence). B) The AGO1 target region was amplified by PCR with primers F1/R2 (annealing outside the edited sequence) or mut-F1/R2 (with one primer annealing exclusively to the edited sequence). The panels show representative inverted contrast images of PCR products resolved by agarose gel electrophoresis. CC-124, wild-type strain; ago1-45 and ago1-59, edited mutants with a disrupted AGO1 gene in the CC-124 background. C) RNA gel blot analysis of sRNAs isolated from the indicated strains and detected with probes specific for the most abundant >26-nt sRNAs of Clusters 51304+ or 69832−. The same membranes were reprobed with the U6 small nuclear RNA sequence as a control for equivalent loading of the lanes. The asterisks indicate putative RNA precursors of the >26-nt sRNAs. Examined cells were cultured under various nutritional conditions: Control, nutrient-replete medium; N starvation, nitrogen deprivation for 24 h; and S starvation, sulfur deprivation for 24 h. D) Size distribution of genome-mapped total sRNAs from wild type (CC-124) or ago1 mutant (ago1-45) strains. Data from replicate sRNA libraries are shown as bars of the same color.
We identified 2 (independently generated) precisely edited mutants by PCR analysis of selected Chlamydomonas colonies, using a primer that anneals exclusively to the altered sequence (Fig. 3, A and B). Sequencing of a 235-bp PCR product (Fig. 3A, primers F1/R2), overlapping the mutated site, revealed no additional, unintended changes to the DNA sequence of AGO1 exon 1. In both edited mutants, AGO1 transcript abundance was substantially reduced, presumably because of nonsense-mediated mRNA decay triggered by a premature termination codon (Supplemental Fig. S2A).
In these mutants, impaired formation of AGO1 effector complexes is expected to result in destabilization of the associated sRNAs, although some sRNAs may persist by alternative association with the intact AGO2/AGO3 proteins. Indeed, RNA gel blot analyses showed that the abundance of all tested >26-nt sRNAs is lower in the ago1 knockout mutants in comparison to the wild-type strain (Fig. 3C; Supplemental Fig. S2, B and C). Deep sequencing of total sRNA libraries from the parental and the ago1-45 Chlamydomonas strains also revealed that, based on normalized read counts, the ago1 mutant shows a relative decrease in the >26-nt sRNAs, concomitantly with a relative increase in 20 to 21-nt sRNAs (Fig. 3D; Supplemental Fig. S2D). These results demonstrate that Chlamydomonas contains a unique class of >26-nt sRNAs, with a bias for adenine as the starting nucleotide, that is bound preferentially by AGO1.
Genomic loci generating AGO1-associated >26-nt sRNAs
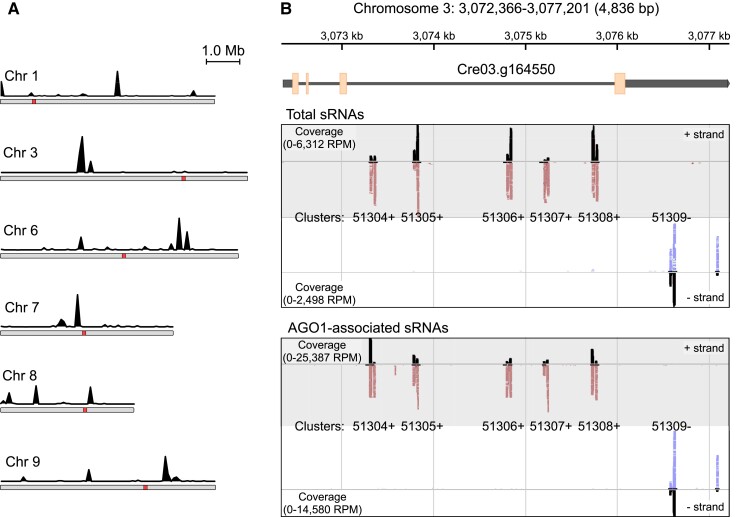
Abundant >26-nt sRNAs (excluding tRNA fragments) mapped to a limited number of genomic loci in chromosomes 1, 3, 6, 7, 8, and 9 of Chlamydomonas (Fig. 4A). For cataloging purposes, we annotated adjacent reads mapping to the nuclear genome no more than 100 bp apart, regardless of a strand, as belonging to the same sRNA cluster (Supplemental Fig. S3). However, the sequences of most of these sRNA clusters were partly similar to each other (Table 1; Supplemental Fig. S3), corresponding to moderately repetitive elements.
Figure 4.
AGO1-associated >26-nt sRNAs originate from clustered genomic loci. A) Location of genomic loci producing AGO1-associated >26-nt sRNAs on Chlamydomonas chromosomes. The black traces indicate the number of AGO1-associated long sRNAs (from normalized libraries) mapped in sliding 10-kb windows across the Chlamydomonas chromosomes. Red squares represent putative centromeres (Craig et al. 2021). B) Genome browser view of the clusters matching to the Cre03.g164550 gene, encoding a homolog of the conserved eukaryotic FRA2/BolA-like protein 2. The upper panel shows the chromosome 3 coordinates and a diagram of the Cre03.g164550 gene with exons (orange boxes), introns (thin gray lines), and UTRs (thick gray lines). The middle panel shows sRNAs from total sRNA libraries mapping to the sense strand (upper) or the antisense strand (lower). Individual mapped sRNAs are shown as red marks (mapped on + strand) or blue marks (mapped on − strand) along with coverage in black (indicated as RPM). The bottom panel shows AGO1-associated >26-nt sRNAs mapping to the sense strand (upper) or the antisense strand (lower).
Table 1.
AGO1-associated >26-nt sRNAs and miRNAs in Chlamydomonas
| Cluster/miRNAa | sRNA sequenceb | Chrc | Wild typed | dcl3 d | AGO1 IPd | AGO3 IPd |
|---|---|---|---|---|---|---|
| 3142−, 3151− | AUGGGUCCGAUCGGGAAGCUUUAUC* | 1 | 2,489 | 4,801 | 23,221 | 7 |
| 3152− | AUGGGUCCGGCCGGGAAGCUUUAUC* | 1 | 71 | 115 | 130 | 0 |
| 51198+ | GAGGUCCGACAGCGAGAGGGUUA* | 3 | 960 | 1,558 | 982 | 0 |
| 51293−, 51295−, 51304+, 51308+, 51309−, 78707− | AUGGGUCCGACCGGGAAGCUUUAUC* | 3, 9 | 8,803 | 10,666 | 77,472 | 142 |
| 51294−, 51305+, 51306+ | AUGGGUCCGAACGGGAAGCUUUAUC* | 3 | 3,111 | 4,532 | 13,680 | 14 |
| 51307+ | AGGGUCCGAACGGCAAGCUUUAUC* | 3 | 124 | 352 | 2,231 | 6 |
| 51545+ | ACGAGGUCCGACCGUAGAGGUUUAC* | 3 | 785 | 1393 | 1,528 | 10 |
| 66035+ | AUGGGCCGAACGGGCAGGUUUAUC* | 6 | 259 | 560 | 3,394 | 2 |
| 69832− | AGGGUCCGAACGCGAAGCUUUAUC* | 7 | 300 | 464 | 12,166 | 2 |
| 74058+ | AUGGUCCGCCGGGGAAGCGUUAUC* | 8 | 135 | 226 | 396 | 1 |
| 74059+ | AUGGUCCGCCCUGGAAGCGUUAUCAAG* | 8 | 74 | 98 | 123 | 0 |
| miR9897-5p | UACCGGGCGUGGGGAGGGCAGG | 10 | 169 | 1 | 18 | 44,247 |
| miR9897-3p | UUACGGCUCCUUCUUAUCGGC | 10 | 168 | 9 | 15 | 16,083 |
| miR1157-3p | UUCAGGUAGCGGGACCAGGUG | 12 | 42 | 0 | 5 | 6,431 |
| miR1169-3p | UGUGGAUGUUGCUUGCUGGAU | 16 | 11 | 0 | 6 | 1,860 |
| miR1162-3p | UGUUGUAGUAGUUUAGCCCUGC | 6 | 210 | 1 | 52 | 15,085 |
| miR1147-3p | UUCAGGUAGCGGGACCAGGUG | 12 | 42 | 0 | 5 | 6,431 |
| miR1153-5p | UGGGCCAUCGUAUUACUAUCAG | 4 | 20 | 0 | 15 | 18,557 |
| miR910 | AGCAGCGUCGGGCUCGACCGC | 14 | 689 | 4 | 4 | 31 |
The encoding strand for each cluster is indicated as + or − (C. reinhardtii genome v5.5).
The 5′ end sequence for the predominant >26-nt sRNAs in the examined clusters is followed by an asterisk denoting length and sequence variation at their 3′ ends (see Supplemental Fig. S3).
Chromosome where the cluster or miRNA locus is located.
Normalized read counts (as reads per million) are shown for sRNAs from total sRNA libraries in the wild-type or dcl3 mutant strains and from libraries of co-immunoprecipitated sRNAs associated with AGO1 or AGO3.
The loci producing >26-nt sRNAs also tended to form superclusters at certain genomic locations. As an example, the Chlamydomonas Cre03.g164550 gene encodes a homolog of the conserved eukaryotic FRA2 (Fe repressor of activation-2)/BolA-like protein 2, which in both fungi and mammals plays an essential role in trafficking (2Fe–2S) clusters to certain enzymes and transcription factors that control iron metabolism (Rey et al. 2019; Talib and Outten 2021). The third intron of this gene is ∼3 kb in length and contains 5 clusters producing AGO1-associated sRNAs in the sense orientation (Fig. 4B, clusters 51304+, 51305+, 51306+, 51307+, and 51308+). An additional cluster is located in the 3′ UTR of the same gene in the antisense orientation (Fig. 4B, cluster 51309−).
The structure of the FRA2/BolA-like protein 2 gene, with 4 exons and 3 introns, is conserved in algae of the Trebouxiophyceae (e.g. Coccomyxa subellipsoidea) and Chlorophyceae (e.g. Chlamydomonas) classes, which diverged 750 to 850 million years ago (Leliaert et al. 2012). However, in most algal species, the third intron is relatively short, as in Tetradesmus obliquus, V. carteri, and Edaphochlamys debaryana within the Chlorophyceae (Supplemental Fig. S4). By contrast, this third intron has experienced substantial size expansion in Chlamydomonas and its close relatives Chlamydomonas incerta and Chlamydomonas schloesseri (Supplemental Fig. S4), when compared to the more distant relative E. debaryana (for details on the phylogenetic relationships see Craig et al. 2021). Interestingly, the three Chlamydomonas species contain conserved sequences homologous to the AGO1-associated >26-nt sRNAs in the third intron and 3′ UTR of their FRA2/BolA-like protein 2 genes (Fig. 4B; Supplemental Figs S5 and S6); moreover, these sequences have been amplified so that the size of the third intron has increased in parallel with the number of sRNA-producing clusters (1 in C. schloesseri, 3 in C. incerta, and 5 in C. reinhardtii at a threshold of >70% nucleotide identity).
We also examined the existence of sRNAs homologous to those from clusters 51304+, 51545+, and 69832− in E. debaryana and C. incerta by RNA gel blot analysis (Supplemental Fig. S7, A, C, and D). Using probes specific for the most abundant reads of each cluster, we detected >26-nt sRNAs in C. reinhardtii and its close relative C. incerta. Moreover, changes in steady-state levels of these related sRNAs were conserved in the two species in response to several nutrient limitation conditions (see below), (Supplemental Fig. S7, A, C, and D). Reprobing the same membrane with an oligonucleotide hybridizing to a C. reinhardtii miRNA, miR912 (21 nt in length), substantiated the notion that the detected sRNAs are indeed larger in size than canonical miRNAs and also showed that miR912 is not conserved in C. incerta (Supplemental Fig. S7B). The E. debaryana genome does not appear to harbor loci with homology to the examined C. reinhardtii sRNAs; we also did not observe obvious hybridization signals to RNA isolated from this species in RNA gel blots (Supplemental Fig. S7). Thus, although the origin of the repetitive genomic loci producing these >26-nt sRNAs remains unknown, some of these sequences have been conserved and amplified within core Chlamydomonas species. Moreover, their expression under various nutritional limitation conditions also appears to be conserved.
Biogenesis of AGO1-associated >26-nt sRNAs
To assess whether the >26-nt sRNAs show any evidence of specific processing, we aligned the sRNAs within each cluster to their corresponding genomic sequences and evaluated the precision with which their 5′ and 3′ ends had been generated as well as the possible folding of precursor RNAs into secondary structures. In most cases, we predicted a putative precursor with a fold-back hairpin secondary structure, ∼60 to 80 nt in length, by using RNAfold from the Vienna RNA package (Lorenz et al. 2011; Fig. 5, cluster 51304+, and Supplemental Fig. S3). Moreover, RNA gel blot analyses suggested the existence of easily detectable RNA precursors of the predicted length (Figs 2C and 3C, asterisks).
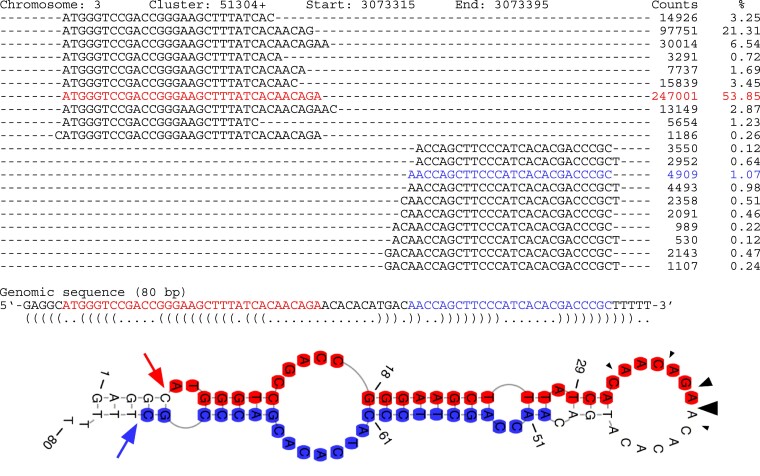
Figure 5.
Predicted precursor RNA structure for cluster 51304+. Sequences of AGO1-associated >26-nt sRNAs were aligned to the corresponding genomic sequence and their frequency in the libraries is shown as raw counts. The secondary structure of the matching genomic sequence was predicted with RNAfold (Lorenz et al. 2011) and depicted at the bottom. The most abundant >26-nt sRNAs present in the 5′ arm or the 3′ arm of the predicted hairpin are indicated in red and blue, respectively. Fairly precise putative cleavage sites on the left side of the depicted hairpin are indicated by red and blue arrows. More imprecise putative cleavage sites in the terminal loop of the depicted hairpin are indicated by black arrowheads.
The 5′ end of the >26-nt sRNAs encoded in the 5′ arm of the predicted hairpin and the 3′ end of the >26-nt sRNAs encoded in the 3′ arm of the hairpin appeared to be quite precisely processed, as evidenced in both strand ends on the left side of the secondary structure diagrams (Fig. 5; Supplemental Fig. S3). However, in very few instances, the precursor sequences formed a stable, perfectly base-paired dsRNA stem at the putative cleavage sites (Supplemental Fig. S3), making it unlikely that they were processed by RNase III enzymes such as DCL (Vermeulen et al. 2005; Macrae et al. 2006; Song and Rossi 2017). Additionally, the processing of the right ends of the hairpin secondary structures (i.e. 3′ end of >26-nt sRNAs encoded in the 5′ arm of the hairpin and 5′ end of >26-nt sRNAs encoded in the 3′ arm of the hairpin) appeared to be rather imprecise and the putative cleavage site(s) often corresponded to predicted ssRNA regions. Also, we cannot rule out that some of the apparent specificity in processing 5′ ends of >26-nt sRNAs may reflect the predominant stabilization of sequences starting with adenine because of the AGO1 binding preference.
Although the secondary structure of predicted >26-nt sRNA precursors suggested that they are not processed by DCLs, we tested more directly whether the biogenesis of the AGO1-associated sRNAs was dependent on these RNase III endonucleases. As already mentioned, Chlamydomonas encodes 3 DCL proteins, of which DCL2 and DCL3 are fairly similar to each other (Casas-Mollano et al. 2008; Valli et al. 2016). Examination of sRNA libraries from a Chlamydomonas dcl3 null mutant (Valli et al. 2016) revealed that lack of DCL3 function did not affect the production of the >26-nt sRNAs preferentially associated with AGO1 (Fig. 6A; Table 1). However, as already described (Valli et al. 2016), the accumulation of 20 to 22-nt sRNA (including most miRNAs) was substantially lower in this mutant (Fig. 6A; Table 1). RNA gel blot analysis indicated that the steady-state level of >26-nt sRNAs (and of their putative precursors) is slightly increased in a dcl3 mutant relative to the wild-type strain (Fig. 6B).
Figure 6.
The biogenesis of AGO1-associated >26-nt sRNAs is independent of DCL1, DCL2, and DCL3. A) Size distribution of genome-mapped total sRNAs from the wild-type strain (black) and the dcl3 null mutant (gray). Data from replicate sRNA libraries are shown as bars of the same color. RPM, reads per million. B) RNA gel blot analysis of sRNAs isolated from the wild-type (CC-124) or dcl3 mutant (dcl3) strains and detected with probes specific for the most abundant >26-nt sRNAs of Clusters 51304+ or 51545+ (Supplemental Table S3). The same filter was reprobed with the U6 small nuclear RNA sequence (U6) as a control for equivalent loading of the lanes. The asterisk indicates putative RNA precursors of the >26-nt sRNAs. C) RNA gel blot analysis of sRNAs isolated from the indicated strains and detected with probes specific for the most abundant >26-nt sRNAs of Clusters 51304+ or 69832− (Supplemental Table S3). CC-124, wild-type strain; Dcl1-IR1 or Dcl1-IR9, CC-124 transformed with an IR transgene designed to induce RNAi of DCL1 (Casas-Mollano et al. 2008); Mut11, mutant defective in a core subunit of H3K4 methyltransferase complexes required for chromatin-mediated silencing (van Dijk et al. 2005); Mut11xDcl1-IR1, Mut11 transformed with an IR transgene designed to induce RNAi of DCL1 (Casas-Mollano et al. 2008). D) RNA gel blot analysis of sRNAs isolated from the wild-type (CC-124) or dcl2 mutant (dcl2-4 and dcl2-20) strains and detected with probes specific for the most abundant >26-nt sRNAs of Clusters 51304+ or 51545+.
To examine the role of DCL2 in the biogenesis of the >26-nt sRNAs associated with AGO1, we generated Cas9-edited mutants by using a strategy similar to the one described for AGO1 disruption (Supplemental Fig. S8A). We identified 2 (independently generated) edited mutants, one containing the designed premature stop codon in DCL2 exon 1, accompanied by a downstream insertion of part of the homologous repair template, and one displaying a 2-bp deletion and insertion of the repair template (in antisense orientation) at the Cas9 cleavage site (Supplemental Fig. S8, B and C). In RNA gel blot analyses of the dcl2 knockout mutant strains, as with Chlamydomonas dcl3 mutants, the abundance of the >26-nt sRNAs was not affected or somewhat higher than in the wild-type strain (Fig. 6D).
We previously generated Chlamydomonas strains where DCL1 expression was suppressed by RNAi in a wild-type background (CC-124) and in a mutant (Mut11) defective in chromatin-mediated transcriptional silencing (Casas-Mollano et al. 2008). We used these strains to demonstrate that DCL1 plays a role in transposon repression, particularly when chromatin-mediated silencing was compromised (Casas-Mollano et al. 2008). RNA gel blot analyses of the same strains indicated that production of the AGO1-associated >26-nt sRNAs is not affected by DCL1 suppression (Fig. 6C). On the contrary, the abundance of both the >26-nt sRNAs and their putative precursors increased in the DCL1 RNAi lines, and even more so in the strain that is also deficient in chromatin-mediated transcriptional silencing (Fig. 6C).
These results suggest that the biogenesis of the >26-nt sRNAs is independent of Chlamydomonas DCL1, DCL2, and DCL3. Compensatory functions between any of the DCL proteins seem unlikely since all these enzymes appear to antagonize to some degree (admittedly small in some cases) the production of >26-nt sRNAs and their putative precursors (Fig. 6). Moreover, as already mentioned, the predicted precursor hairpin RNAs do not seem to be cleaved at perfectly base-paired dsRNA regions, as would be required for DICER-mediated processing (Vermeulen et al. 2005; Macrae et al. 2006; Song and Rossi 2017). Thus, our observations strongly suggest that the Chlamydomonas >26-nt sRNAs associated with AGO1 have distinct biogenesis from that of canonical miRNAs and siRNAs, although the actual molecular mechanism(s) remains elusive.
Potential role(s) of AGO1-associated >26-nt sRNAs
Patterns of adaptive evolution have been observed in animal RNAi-pathway genes involved in the control of transposable elements and/or viruses, such as those coding for PIWI proteins and a subset of insect AGOs including orthologs of Drosophila melanogaster AGO2 (Wynant et al. 2017; Palmer et al. 2018; Ozata et al. 2019). Their accelerated evolution has been hypothesized to reflect an “evolutionary arms race” with the rapidly evolving molecular parasites they are targeting (Wynant et al. 2017; Palmer et al. 2018). Directional selective pressure can be identified by testing for an elevated rate of nonsynonymous nucleotide substitutions (Ka) relative to synonymous substitutions (Ks) within the coding sequence of a protein (Hurst 2002; Wynant et al. 2017). Using tree-based methods for the calculation of Ka/Ks ratios of Chlamydomonas AGO genes, we observed substantially elevated values for the AGO1 and AGO2/AGO3 genes in comparison to the housekeeping ACTIN gene (Supplemental Fig. S9). The Ka/Ks ratios for Chlamydomonas AGOs, particularly AGO1, were similar to those of the fast-evolving animal RNAi-pathway genes (Wynant et al. 2017), suggesting that AGO1 might likewise play a role in defense responses against transposons and/or viruses.
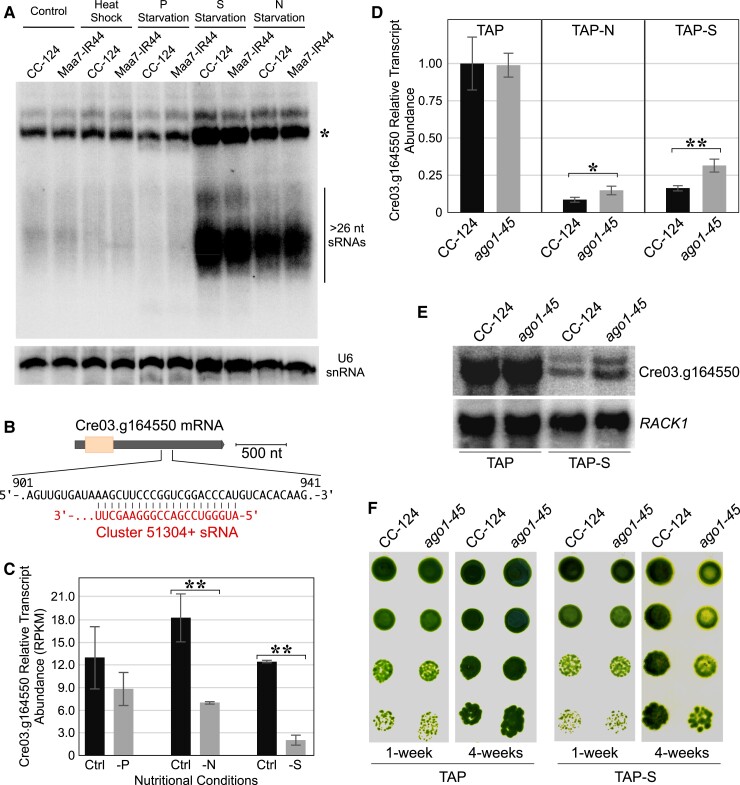
As described above, AGO1 associates predominantly with uncommonly long sRNAs derived from moderately repetitive sequences and, in principle, it might also have a role in regulating the expression of endogenous genes using the associated >26-nt sRNAs as guides. To obtain insight into the possible function(s) of these sRNAs, we examined the expression of the abundant sequences corresponding to cluster 51304+ under a variety of stress conditions. Interestingly, in RNA gel blot analyses of Chlamydomonas, the levels of putative precursor and mature sRNAs from cluster 51304+ increased markedly under sulfur or nitrogen deprivation, in comparison to those of cells grown in nutrient-replete medium (Fig. 7A). We observed a similar response to nutrient deprivation in Chlamydomonas and its close relative C. incerta for the >26-nt sRNAs corresponding to clusters 51304+, 51545+, and 69832− (Supplemental Fig. S7, A, C, and D). Examination of total sRNA libraries generated from Chlamydomonas cells cultured under nutrient-replete or nitrogen-deprived conditions also revealed an increased relative abundance of a subset of the AGO1-associated >26-nt sRNAs when nitrogen was omitted from the medium (Supplemental Table S1, clusters 51198+, 51293−, 51295−, 51304+, 51308+, 51309+, 78707−, and 66035+).
Figure 7.
The abundance of AGO1-associated >26-nt sRNAs increases under certain nutritional deprivation conditions, concurrently with the downregulation of the Cre03.g164550 transcript. A) RNA gel blot analysis of >26-nt sRNAs from cluster 51304+ in Chlamydomonas cells cultured under the denoted conditions. Control, nutrient-replete standard laboratory conditions; Heat shock, 42 °C for 20 min; P starvation, phosphorus deprivation for 24 h; S starvation, sulfur deprivation for 24 h; N starvation, nitrogen deprivation for 24 h. sRNAs were detected with a probe specific for the most abundant >26-nt sRNAs of cluster 51304+ (Supplemental Table S3). The same filter was reprobed with the U6 small nuclear RNA sequence as a control for equivalent loading of the lanes. The asterisk indicates putative RNA precursors of the >26-nt sRNAs. CC-124, wild-type strain; Maa7-IR44, CC-124 transformed with an IR transgene designed to induce RNAi of MAA7 (encoding tryptophan synthase β-subunit). B) Schematic diagram of the binding site of Cluster 51304+ sRNAs in the 3′ UTR of the predicted target Cre03.g164550. C) Relative Cre03.g164550 mRNA levels in wild-type Chlamydomonas grown under the indicated nutritional conditions. RPKM, reads per kilobase per million determined from RNA-seq data. Samples indicated with double asterisks are significantly different (P < 0.01). D) Relative Cre03.g164550 transcript abundance examined by RT-qPCR in cells grown under the indicated nutritional conditions. Values are means ± standard deviation (Sd) of 3 independent experiments and normalized to those in CC-124 grown in standard TAP medium. Samples indicated with asterisks are significantly different (**P < 0.01; *P < 0.05) in a 2 tailed Student's t-test. ago1-45, edited mutant with a disrupted AGO1 gene in the CC-124 background. E) RNA gel blot analysis of Cre03.g164550 transcript levels. The same filter was reprobed with the coding sequence of RACK1 (encoding RECEPTOR FOR ACTIVATED C KINASE 1) as a control for similar loading of the lanes. F) Growth and survival of the indicated strains on nutrient replete TAP medium (TAP) or on the same medium lacking sulfur (TAP–S). The images show serially diluted cells in spots, after 1 and 4 wk of incubation under dim lights.
In human (Homo sapiens) AGO2, the conserved helix 7 and residue I365 have been reported to shape the miRNA seed region for rapid target RNA recognition (Klum et al. 2018; Gebert and MacRae 2019). These features (particularly the equivalent isoleucine and nearby residues) are conserved in Chlamydomonas AGO1 (Chung et al. 2019), consistent with a similar guide targeting mode as canonical AGO proteins. Thus, we used the web server psRNATarget (Dai et al. 2018), with near default parameters (i.e. following miRNA-like pairing rules), to identify target mRNAs hybridizing to the first 21 nucleotides of the abundant >26-nt sRNAs derived from the 5′ arm of cluster 51304+ (Fig. 5). We note that this sequence is conserved in several additional >26-nt sRNA clusters present in the Chlamydomonas genome (Table 1; Supplemental Fig. S3, clusters 51293−, 51295−, 51304+, 51308+, 51309+, and 78707−) as well as in genomic loci of the closely related species C. incerta and C. schloesseri. Thirty-three putative targets were predicted using this approach (Supplemental Data Set S1), including Cre03.g164550, which contains the 51304+ cluster within its third intron and complementary sequences (in antisense orientation) within its 3′ UTR (Fig. 7B; Supplemental Fig. S10). The predicted targets appear to play a role in diverse cellular functions rather than being confined to specific cellular or metabolic pathways (Supplemental Data Set S1).
We then examined publicly available transcriptome sequencing (RNA-seq) data sets collected from wild-type Chlamydomonas cultured under nutrient-replete or under phosphorus-, sulfur-, or nitrogen-deprived conditions (Ngan et al. 2015), to assess the expression level of the predicted targets of sRNAs from cluster 51304+ (Supplemental Data Set S2). With the caveat that some target predictions may represent false positives or the predicted binding sites may not be accessible for interaction with a guide sRNA, 45.5% (15 out of 33) of the predicted target transcripts were downregulated under sulfur deprivation and 42.4% (14 out of 33) of the predicted target transcripts were downregulated under nitrogen deprivation (Supplemental Data Set S2). Additionally, 10 out of the 33 predicted targets (30.3%) were downregulated under both nutritional deprivation conditions, suggesting that they are controlled by a similar mechanism(s) (Supplemental Data Set S2). Of note, Cre03.g164550 transcript abundance was significantly downregulated under sulfur or nitrogen deprivation (Fig. 7C; Supplemental Data Set S2), coinciding with increased abundance of the cluster 51304+ >26-nt sRNAs (Fig. 7A).
In the AGO1-edited mutants, which showed diminished steady-state levels of >26-nt sRNAs under all examined trophic conditions (Fig. 3C; Supplemental Fig. S2, B–D), the abundance of Cre03.g164550 transcripts increased moderately under sulfur or nitrogen deprivation relative to the wild-type strain (Fig. 7, D and E). Two additional predicted targets, Cre01.g036950 (encoding Cobalamin 5′-phosphate synthase) and Cre06.g278097 (encoding a putative mRNA export factor; Supplemental Data Set S2), also displayed somewhat enhanced steady-state mRNA levels in the ago1-45 mutant strain under nutritional stress (Supplemental Fig. S11, A and B). However, despite repeated attempts, we were unable to detect cleavage of any of these predicted target transcripts, by a modified 5′ rapid amplification of cDNA ends (5′ RACE) protocol (Yamasaki et al. 2013; Voshall et al. 2015), within the putative >26-nt sRNA binding elements. Thus, our observations suggest that AGO1-associated >26-nt sRNAs might target specific genes for silencing under sulfur or nitrogen limitation, but the actual mechanism(s) remains to be explored. Additionally, AGO1 mutant strains became chlorotic faster and/or more acutely than the wild type under sulfur or nitrogen deprivation (Fig. 7F; Supplemental Fig. 11C), suggesting a role for Chlamydomonas AGO1 (and, presumably, its associated sRNAs) in tolerance to nutritional stresses.
Discussion
RNAi, mediated by small noncoding RNAs, is a highly conserved mechanism influencing gene regulation, genome stability as well as defense responses against genomic parasites (Ghildiyal and Zamore 2009; Borges and Martienssen 2015; Wendte and Pikaard 2017; Yu et al. 2017; Bartel 2018; Lee and Carroll 2018; Ozata et al. 2019; Chen and Rechavi 2022). In multicellular eukaryotes, such as land plants, animals, and fungi, extensive duplication and diversification of RNAi machinery components have resulted in complex, partly overlapping pathways for epigenetic regulation (Ghildiyal and Zamore 2009; Czech and Hannon 2011; Borges and Martienssen 2015; Lee and Carroll 2018; Wang et al. 2021). Duplicated RNAi components appear to allow the evolution of new gene regulatory mechanisms that use distinct sRNAs as sequence-specific determinants, as well as more effective strategies to counteract the action of invading viruses and transposable elements. By contrast, many unicellular eukaryotes appear to have entirely lost the RNAi machinery or have retained only a basic set of RNAi components (Cerutti and Casas-Mollano 2006; Casas-Mollano et al. 2008). Intriguingly, the alga Chlamydomonas has an RNAi machinery much more complex than might be expected for a unicellular organism, consisting of 3 AGOs, 3 DCLs, and a diverse assortment of sRNAs (Molnár et al. 2007; Zhao et al. 2007; Casas-Mollano et al. 2008; Voshall et al. 2015; Valli et al. 2016; Chung et al. 2019; Müller et al. 2020). Of the core RNAi proteins, Chlamydomonas DCL3 and AGO3 have been implicated in the biogenesis and activity of many (all) miRNAs (Voshall et al. 2015; Valli et al. 2016; Yamasaki et al. 2016; Chung et al. 2019). Their closely related paralogous genes, DCL2 and AGO2, are expressed at relatively low levels and AGO2 binds to ∼21-nt siRNAs of unknown function (Chung et al. 2019). By contrast, the more divergent paralogs DCL1 and AGO1 have not been characterized in detail (Casas-Mollano et al. 2008). Likewise, the biogenesis and function(s) of most endogenous sRNAs in Chlamydomonas, including a unique, abundant class of >26-nt sRNAs (Fig. 1), remain to be elucidated.
As described here, AGO1 is associated predominantly with these >26-nt sRNAs, having a bias for adenine as the 5′-terminal nucleotide (Figs 2 and 3). The most abundant AGO1-associated sRNAs were generated from a limited number of genomic loci (Fig. 4; Table 1), corresponding to moderately repetitive sequences that match either intergenic regions or introns/3′ UTRs of predicted protein-coding genes (Fig. 2B). These features are in agreement with several characteristics of the recently annotated locus class 4, uncovered by using data-driven machine-learning approaches to systematically classify sRNA loci in Chlamydomonas (Müller et al. 2020). Moreover, the sequences producing >26-nt sRNAs, as well as their expression, were conserved in closely related Chlamydomonas species (Supplemental Figs S4–S7). The >26-nt sRNAs are likely processed from ∼60 to 80-nt precursor RNAs that can fold into hairpin secondary structures (Fig. 5; Supplemental Fig. S3). However, most putative precursor RNAs do not appear to be cleaved at perfectly base-paired dsRNA regions (Supplemental Fig. S3) and >26-nt sRNA biogenesis was independent of DCL1, DCL2, and DCL3 (Fig. 6; Table 1).
The Chlamydomonas >26-nt sRNAs share some features with animal piRNAs (Ghildiyal and Zamore 2009; Ozata et al. 2019; Haase 2022) such as a longer size than canonical miRNAs/siRNAs, being produced from genomic clusters, and DICER-independent biogenesis. However, piRNAs are processed from ssRNAs without obvious secondary structures and they associate with PIWI proteins (Ghildiyal and Zamore 2009; Ozata et al. 2019; Haase 2022), which are absent from plant and green algal species (Cerutti and Casas-Mollano 2006; Swarts et al. 2014). In land plants, most sRNAs are shorter than ∼26 nts (Axtell 2013; Borges and Martienssen 2015; Hardcastle et al. 2018; Feng et al. 2020; Lunardon et al. 2020; Chen et al. 2021; Baldrich et al. 2022) and, to our knowledge, sRNAs with similar characteristics to the Chlamydomonas >26-nt sRNAs have not been described. Thus, the Chlamydomonas >26-nt sRNAs appear to add to a growing list of atypical sRNAs with unique biogenesis pathways and/or functions (Rzeszutek and Betlej 2020; Wu et al. 2020; Alves and Nogueira 2021; Meseguer 2021; Reshetnyak et al. 2021).
Based on its fairly high Ka/Ks ratio (Supplemental Fig. S9), similar to that of PIWIs and certain animal AGOs involved in the control of viruses and/or transposons (Wynant et al. 2017; Palmer et al. 2018; Ozata et al. 2019), we hypothesize that Chlamydomonas AGO1 may play a role in defense responses against molecular parasites. We previously implicated Chlamydomonas DCL1 in transposon silencing (Casas-Mollano et al. 2008); and, intriguingly, DCL1 and AGO1 might possibly be functionally linked since they are encoded by adjacent, divergently transcribed genes (an arrangement conserved in the genomes of the related species C. incerta and C. schloesseri; Casas-Mollano et al. 2008). However, as already described (Jeong et al. 2002; Casas-Mollano et al. 2008; Kim et al. 2015), multiple, partly redundant epigenetic mechanisms are involved in preventing transposon mobilization in Chlamydomonas. In cells grown under standard laboratory conditions, transposons appear to be primarily repressed by a chromatin-based mechanism(s), and only when this system is defective can the involvement of RNAi as a transposon silencing mechanism be fully appreciated (Jeong et al. 2002; Casas-Mollano et al. 2008; Kim et al. 2015).
AGO1 might also have a role in regulating the expression of endogenous genes, presumably using the associated >26-nt sRNAs as guides. Interestingly, we observed that the abundance of several AGO1-associated sRNAs was markedly enhanced under certain nutritional stresses, such as nitrogen or sulfur deprivation (Fig. 7A; Supplemental Fig. S7 and Table S1). Under these same conditions, a substantial fraction of putative target transcripts, identified by sequence complementarity to the most abundant reads from cluster 51304+, displayed a reduction in their mRNA levels in wild-type Chlamydomonas (Supplemental Data Set S2). One of the downregulated transcripts corresponded to the FRA2/BolA-like protein 2 gene (Fig. 7C), which hosts several >26-nt sRNA clusters within its third intron but also contains complementary target sequences within its 3′ UTR (Fig. 4B; Supplemental Fig. S10). In the AGO1-edited mutants, the transcript abundance of the FRA2/BolA-like protein 2 gene increased moderately under sulfur or nitrogen deprivation relative to wild-type levels (Fig. 7, D and E). Similar changes in transcript abundance, comparing ago1 mutants to the wild-type strain, were detected for other predicted target genes, namely Cre01.g036950 (encoding Cobalamin 5′-phosphate synthase) and Cre06.g278097 (encoding a putative mRNA export factor; Supplemental Fig. S11, A and B). Additionally, ago1 mutant strains displayed an accelerated and/or more severe chlorotic phenotype under sulfur or nitrogen deprivation (Fig. 7F; Supplemental Fig. 11C). These observations are consistent with the regulation of certain genes, in response to nutritional stresses, by an effector complex comprised of AGO1 and its associated sRNAs, but the actual molecular mechanism(s) is not obvious.
Curiously, some of the conserved cluster sequences producing >26-nt sRNAs, although not matching known transposable elements, have been amplified in closely related Chlamydomonas species (Supplemental Figs S4–S6). Also, since DCL mutations result in somewhat enhanced >26-nt sRNA steady-state levels in Chlamydomonas (Fig. 6), the RNAi machinery appears to antagonize the production of >26-nt sRNAs and of their putative RNA precursors, as expected for a defense mechanism against genomic parasites. Thus, although the origin of the sequences encoding these sRNAs remains elusive, in several respects they appear to have transposon-like behavior. Within this context, it was recently reported that sequences of diverse giant viruses can integrate into the nuclear genomes of several Chlamydomonas species (Moniruzzaman et al. 2022). We surmise that the transposon-like sequences generating >26-nt sRNAs might have been ancestrally targeted for silencing by AGO1 effector complexes. Yet, in the course of evolution, some of the >26-nt sRNAs may have fortuitously acquired endogenous target genes, conferring a selective advantage in gene regulation and leading to the fixation of the encoding clusters.
At their core, eukaryotic RNAi systems consist of a short guide RNA, allowing for sequence-specific target recognition, and an effector protein of the AGO family, which mediates downstream effects with varying outcomes (Burroughs et al. 2014; Swarts et al. 2014; Dexheimer and Cochella 2020). The guide sRNAs identified in earlier studies were derived from dsRNA precursors processed by RNAse III type DICER endonucleases (Ghildiyal and Zamore 2009; Burroughs et al. 2014; Borges and Martienssen 2015; Yu et al. 2017; Bartel 2018; Chen and Rechavi 2022). However, it is now becoming apparent that guide sRNAs can be generated by a variety of alternative pathways (Ghildiyal and Zamore 2009; Yang and Lai 2010; Burroughs et al. 2014; Wendte and Pikaard 2017; Bartel 2018; Alves and Nogueira 2021; Haase 2022). The functional role of AGOs also appears to have changed during evolution, from relatively simple host-defense proteins against viruses and transposons to key players in complex multiprotein regulatory pathways in multicellular organisms (Shabalina and Koonin 2008; Ghildiyal and Zamore 2009; Burroughs et al. 2014; Swarts et al. 2014; Borges and Martienssen 2015; Lee and Carroll 2018; Dexheimer and Cochella 2020; Chen and Rechavi 2022). Based on our observations, it is tempting to speculate that the minimal RNAi (protein) machinery might consist of a single AGO acting in association with sRNAs generated by a DICER-independent mechanism(s).
Material and methods
C. reinhardtii strains, mutants, and culture conditions
Strain Maa7-IR44, containing an inverted repeat transgene designed to silence the MAA7 gene (encoding Tryptophan Synthase β-subunit), was previously described (Ma et al. 2013). Mut11 is an insertional mutant defective in a core subunit of histone H3 lysine 4 methyltransferase complexes, which is required for chromatin-mediated silencing (Jeong et al. 2002; van Dijk et al. 2005). Chlamydomonas strains where DCL1 expression was suppressed by transgenic RNAi in the wild-type background (CC-124) or in the Mut11 mutant background were also described (Casas-Mollano et al. 2008). For the isolation of AGO1-associated sRNAs, we cloned a sequence encoding the FLAG tag (Einhauer and Jungbauer 2001; Voshall et al. 2015) in-frame and upstream of the Chlamydomonas AGO1 (Cre02.g141050) coding sequence. This construct was placed under the control of PsaD regulatory sequences and transformed into Maa7-IR44 by electroporation (Yamano et al. 2013). The PsaD regulatory sequences allow reliable expression with reduced incidence of gene silencing of cDNA transgenes in Chlamydomonas (Fischer and Rochaix 2001).
Unless noted otherwise, Chlamydomonas or other algal species were routinely grown in tris-acetate phosphate (TAP) medium (Harris 2009). For nitrogen deprivation analyses, cells initially grown in nutrient-replete TAP medium to the middle of the logarithmic phase (OD750 ∼0.3 to 0.4) were collected by centrifugation (2,000 × g for 8 min), washed twice, and resuspended at a density of ∼1.0 × 106 cells mL−1 in TAP medium or TAP medium lacking nitrogen (i.e. lacking ammonium chloride; Harris 2009; Voshall et al. 2017). After 24 h of incubation under continuous illumination (∼150 μmol m−2 s−1 photosynthetic photon flux density; fluorescent cool white light bulbs), cells were harvested and immediately frozen in liquid nitrogen for total RNA isolation. A similar protocol was used for the analysis of phosphate- or sulfur-deprived cells, following prior specifications (Moseley et al. 2009; González-Ballester et al. 2010; Voshall et al. 2017). For heat-shock treatments, mid-log phase cells in TAP medium were incubated in a water bath at 42 °C for 20 min. For phenotypic analyses, cells grown to logarithmic phase in TAP medium were serially diluted, spotted on agar plates of the appropriate medium (see figure legends), and incubated for 35 d under dim lights (Ma et al. 2013; Voshall et al. 2017).
AGO1 and DCL2 gene editing
Following a previously described protocol (Akella et al. 2021), cells of the wild-type CC-124 strain were electroporated with an in vitro assembled Cas9 RNP, targeting either AGO1 exon 1 or DCL2 exon 1, together with a corresponding modified ssODN, to serve as a template for homology-directed DNA repair. After a 48-h recovery period, electroporated cells were spread on TAP agar plates containing oxyfluorfen (Akella et al. 2021). Surviving colonies were screened by PCR amplification (Cao et al. 2009) of the AGO1 or the DCL2 target site, using a primer (AGO1-mut-F1 or DCL2-mut-F1) that anneals exclusively to the edited sequence. The oligonucleotides and primers used for these experiments are listed in Supplemental Table S3.
Isolation of AGO1-associated sRNAs, library preparation, and sequencing
FLAG-tagged AGO1 was affinity-purified from cell lysates as previously described (van Dijk et al. 2005; Voshall et al. 2015). RNAs associated with AGO1 were purified with TRI reagent (Molecular Research Center) and contaminant DNA was removed by DNase I treatment (Ibrahim et al. 2010; Ma et al. 2013). Small cDNA libraries (2 independent replicates) were prepared with a small RNA v1.5 sample prep kit (Illumina), following the manufacturer's protocol, and sequenced on a Hiseq1500 instrument (Illumina). All sequencing data were deposited into the NCBI Sequence Read Archive (SRA) under BioProject PRJNA765360.
Purification of total sRNAs, library preparation, and sequencing
Total RNA was purified from wild-type (CC-124) or ago1-45 mutant cells, grown to the middle of the logarithmic phase, by using TRI reagent (Molecular Research Center). Contaminating genomic DNA was removed with DNase I treatment (Ibrahim et al. 2010; Ma et al. 2013). Small cDNA libraries (2 independent replicates from each strain) were prepared with a NEXTFLEX Small RNA-Seq v3 kit (Perkin Elmer), which includes randomized primer adapter technology to reduce ligation bias. The libraries were sequenced on a NextSeq550 platform (Illumina). All sequencing data were deposited into the NCBI Sequence Read Archive (SRA) under BioProject PRJNA765360.
sRNA mapping and profiling
Adaptor-trimmed reads were mapped, by using Bowtie2 (version 2.2.9) with default parameters (Langmead and Salzberg 2012), to the unmasked C. reinhardtii genome v5.5 downloaded from Phytozome version 12.1 (https://phytozome.jgi.doe.gov/pz/portal.html; Goodstein et al. 2012). Mapped reads were filtered to remove those shorter than 18 nts or aligning to gaps or mismatches. The population of reads that mapped perfectly to the genome was then profiled based on length. Genome-mapped reads were also classified as matching the chloroplast or the mitochondrial genomes, ribosomal RNAs (rRNAs), small nuclear or nucleolar RNAs (snRNAs or snoRNAs), transfer RNAs (tRNAs), or transposable elements. All sequences except for transposable elements were taken from Genbank (accession numbers BK000554.2 and NC_001638.1 for the chloroplast and mitochondrial genomes, respectively). Transposon sequences were taken from Repbase (Kapitonov and Jurka 2008). After filtering the above sequences, the remaining mapped reads were further classified based on their match to structural categories in the annotated Chlamydomonas nuclear genome (Creinhardtii_281_v5.5.gene.gff3), such as intergenic or genic regions, the latter partitioned into 5′ UTRs, exons, introns, or 3′ UTRs. The expression level in reads per million (RPM) for each mapped sRNA was determined by the formula: RPM = [(106C)/N], where C is the number of mapped reads corresponding to an individual sRNA sequence in the library and N is the total number of mapped reads in the library (Voshall et al. 2015). For comparative analyses, publicly available small RNA sequencing data from wild-type C. reinhardtii and a dcl3 null mutant (Valli et al. 2016) were retrieved from the NCBI Sequence Read Archive (BioProject PRJEB10672). AGO3-associated small RNA sequencing data (Voshall et al. 2015, 2017) were also retrieved from NCBI SRA (BioProjects PRJNA271880 and PRJNA303189).
Genome clustering of AGO1-associated sRNAs and secondary structure prediction of putative RNA precursors
After removing reads that mapped to known noncoding RNA categories, transposons, the chloroplast, or the mitochondrial genomes, the remaining sRNA reads were clustered by genomic location such that there were no more than 100 nts between adjacent reads, regardless of the strand, in a given cluster. The reads from both strands in the same genomic location were placed in the same cluster. Then the genomic sequence for each strand of a cluster was folded using version 2.3.1 of RNAfold from the Vienna RNA package (Lorenz et al. 2011), to assess whether putative precursor RNAs for the sequenced sRNAs may have secondary structures. Clusters were also manually examined to assess the processing accuracy of the 5′ and 3′ ends of the predominant read(s), the frequency of the predominant read(s), and the extent of complementarity (i.e. dsRNA stem formation) between the 2 arms of predicted hairpins.
Analysis of sRNA size distribution in green algae and land plants
Publicly available total small RNA sequencing data corresponding to whole cells or whole organisms (in nonreproductive phases) were retrieved from the NCBI SRA for the following species: C. reinhardtii (E-MTAB-3851), V. carteri (SRR1029443), P. patens (SRR1842137), M. polymorpha (SRR2179617), P. abies (SRR14056790), G. biloba (SRR1658901), Z. mays (SRR895785), and A. thaliana (SRR4124967). The 3′ end adaptor sequences were trimmed using the fastx toolkit (http://hannonlab.cshl.edu/fastx_toolkit/), and the reads were then mapped to the respective reference genomes as described above. Genome-mapped redundant reads, longer than 17 nts, were then profiled based on their size.
Prediction of Chlamydomonas >26-nt sRNA targets
Putative >26-nt sRNA target transcripts were predicted using psRNATarget schemaV2 (2017 release; Dai et al. 2018). Default parameters were used except for the expectation parameter, which was set to 4 (instead of the default 5) to increase stringency. The transcript library used in the search was “cDNA library Phytozome 11, 281_v5.5.” Putative functions of the predicted targets were evaluated by using the annotations of Chlamydomonas genes [if available through Phytozome v. 12.1, (Goodstein et al. 2012)] as well as conserved protein domains.
Differential expression analysis of predicted >26-nt sRNA targets under various nutritional conditions
Publicly available poly(A) mRNA sequencing data collected from wild-type Chlamydomonas grown under different nutritional conditions were retrieved from the NCBI SRA for the following treatments: phosphorus deprivation (SRR1216592, SRR1216593, SRR1216594, SRR1216595), sulfur deprivation (SRR1521728, SRR1521729, SRR1521742, SRR1521743; Ngan et al. 2015) and nitrogen deprivation (SRR1521680, SRR1521684, SRR1521722, SRR1521723; Ngan et al. 2015). Standard RNA-seq analysis tools, Tophat and Cuffdiff, were used to compare gene expression under nutrient-replete or nutrient-deprived conditions (Trapnell et al. 2009). Transcript abundance was analyzed as Reads Per Kilobase of transcript per Million mapped reads (RPKM), which normalizes read counts based on both transcript length and total number of reads, using the formula: RPKM = [(109C)/(NL)], where C is the number of reads mapped to each transcript, N is the total number of mapped reads in the library, and L is the transcript length in nucleotides (Mortazavi et al. 2008).
Phylogenetic analysis
AGO protein sequences from Viridiplantae were obtained from the NCBI database and the Phycocosm algal portal (https://phycocosm.jgi.doe.gov/phycocosm/home). Polypeptides homologous to AGO-PIWI were identified by BLAST or PSI-BLAST searches of protein and/or translated genomic DNA sequences (see Supplemental Fig. S1 for their accession numbers). The phylogenetic analysis was carried out using the pipeline implemented in the Phylogeny.fr server (Dereeper et al. 2008). Sequence alignment of the full-length proteins was performed with MUSCLE with default parameters (BLOSUM62; maximum number of iterations = 16; Edgar 2004). Blocks of conserved, aligned sequences were selected with Gblocks with default parameters (Talavera et al. 2007), resulting in 221 aligned positions in 10 selected blocks. Maximum likelihood phylogeny reconstruction was performed with PhyML (Guindon et al. 2010) and bootstrapping statistical test for branch support with a WAG amino acid substitution model (gamma shape parameter: 1.380; number of categories: 4; proportion of invariant: 0.032; bootstrapped data sets: 500; Dereeper et al. 2008). Visualization of the phylogeny was done using TreeDyn (Chevenet et al. 2006).
Calculation of Ka/Ks ratios
Orthologous candidates for the AGO1 and AGO2/AGO3 genes (Supplemental Table S2) were identified from C. reinhardtii and the closely related species C. incerta, C. schloesseri, E. debaryana, and Chlamydomonas eustigma, based on the phylogeny shown in Supplemental Fig. S1. Multiple sequence alignment of each orthologous protein set was performed using the E-INS-i algorithm of MAFFT (Katoh et al. 2019). The nucleotide sequences coding for the AGO1 and AGO2/AGO3 proteins were aligned based on the protein alignments using TranslatorX (Abascal et al. 2010). The ratios of nonsynonymous (Ka) to synonymous (Ks) nucleotide substitution rates among these coding sequences were estimated by using the Ka/Ks Calculation tool available at the Norwegian Bioinformatics platform (http://services.cbu.uib.no/tools/kaks). It uses the methods found in Liberles (2001) and Siltberg and Liberles (2002). For each data set, the coding sequence alignment produced using MAFFT was provided, and the maximum likelihood phylogenies (with the discrete Zhang substitution matrix option) were reconstructed. The boxplot of the results was generated using R (https://www.R-project.org/).
RNA analyses
Total RNA was purified with TRI reagent (Molecular Research Center), in accordance with the manufacturer's instructions (Ibrahim et al. 2010; Ma et al. 2013), from C. reinhardtii, C. incerta, or E. debaryana cells grown under different trophic conditions. For RNA gel blot analyses of sRNAs, total RNA samples were resolved in 15% polyacrylamide/7 M urea gels and electroblotted to Hybond-XL membranes (GE Healthcare; Ibrahim et al. 2010; Ma et al. 2013). Blots were hybridized with 32P-labeled oligo-DNA probes using a High-Efficiency Hybridization System (Molecular Research Center) at 40 °C for 48 h (Ibrahim et al. 2010; Ma et al. 2013). Specific sRNAs were detected by hybridization with DNA oligonucleotides labeled at their 5′ termini with [γ-32P]ATP and T4 Polynucleotide Kinase (New England Biolabs) as previously described (Ibrahim et al. 2010; Ma et al. 2013). The oligonucleotides used as hybridization probes are listed in Supplemental Table S3.
For reverse-transcription quantitative real-time PCR (RT-qPCR), total RNA from CC-124 and ago1 mutant strains was purified and treated with DNase I (Ambion, Austin, TX, USA) to remove any contaminating genomic DNA as previously reported (Ibrahim et al. 2010; Ma et al. 2013). Reverse transcription was performed as previously described (Carninci et al. 1998), but using Superscript III (Invitrogen). The synthesized first-strand cDNA was then used as a template in end-point PCR or qPCR reactions (Sambrook and Russell 2001). DNA fragments were amplified and quantified with an RT2 SYBR Green/Fluorescein qPCR mastermix (Qiagen) on an iCycler Real-Time PCR Detection System (Bio-Rad). The ACTIN and RACK1 (Cre06.g278222, initially described as CBLP) transcript levels were examined for normalization purposes. All primers used for end-point PCR and qPCR are listed in Supplemental Table S3.
Accession numbers
All sequencing data generated in this study have been deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject PRJNA765360. Accession numbers of public data sets analyzed in this work are listed in Materials and methods under the appropriate sections. Gene identifiers (Phytozome v. 12.1): Cre02.g141050 (AGO1); Cre04.g214250 (AGO2); Cre16.g689647 (AGO3); Cre02.g141000 (DCL1); Cre16.g684715 (DCL2); Cre07.g345900 (DCL3); Cre03.g164550 (FRA2/BolA-like protein 2); Cre01.g036950 (CBA1); Cre06.g278097 (RAE1); Cre13.g603700 (ACTIN); Cre06.g278222 (RACK1).
Supplementary Material
Acknowledgments
We are thankful to members of the Cerutti and Moriyama laboratories for helpful technical suggestions on RNA gel analyses of sRNAs.
Contributor Information
Yingshan Li, School of Biological Sciences and Center for Plant Science Innovation, University of Nebraska-Lincoln, Nebraska-Lincoln, NE 68588-0666, USA.
Eun-Jeong Kim, Department of Life Science, Chung-Ang University, Seoul 06974, Korea.
Adam Voshall, School of Biological Sciences and Center for Plant Science Innovation, University of Nebraska-Lincoln, Nebraska-Lincoln, NE 68588-0666, USA; Division of Genetics and Genomics, Boston Children's Hospital and Harvard Medical School, Boston, MA 02115, USA.
Etsuko N Moriyama, School of Biological Sciences and Center for Plant Science Innovation, University of Nebraska-Lincoln, Nebraska-Lincoln, NE 68588-0666, USA.
Heriberto Cerutti, School of Biological Sciences and Center for Plant Science Innovation, University of Nebraska-Lincoln, Nebraska-Lincoln, NE 68588-0666, USA.
Author contributions
E.M. and H.C. conceived and designed the project. Y.L., E.J.K., and A.V. performed the experiments. Y.L., A.V., E.M., and H.C. implemented software and analyzed the data. Y.L., E.J.K., E.M., and H.C. wrote and edited the manuscript. All authors read and approved the final article.
Supplemental data
Supplemental Figure S1 . Maximum likelihood phylogeny among AGO homologs from Viridiplantae.
Supplemental Figure S2 . Abundance of >26-nt sRNAs in wild-type and ago1 mutant strains of C. reinhardtii.
Supplemental Figure S3 . Secondary structure plots of predicted precursor RNAs for clusters of AGO1-associated >26-nt sRNAs.
Supplemental Figure S4 . Comparison of the structure of FRA2/BolA-like protein 2 gene orthologs in algae of the Trebouxiophyceae and Chlorophyceae classes.
Supplemental Figure S5 . VISTA plot of nucleotide conservation of the C. incerta FRA2/BolA-like protein 2 gene (g5696.t1) relative to its C. reinhardtii ortholog.
Supplemental Figure S6 . VISTA plot of nucleotide conservation of the C. schloesseri FRA2/BolA-like protein 2 gene (g16856.t1) relative to its C. reinhardtii ortholog.
Supplemental Figure S7 . sRNAs longer than 26 nt in C. reinhardtii, C. incerta, and E. debaryana.
Supplemental Figure S8 . Disruption of the DCL2 gene by CRISPR/Cas9-mediated genome editing in C. reinhardtii.
Supplemental Figure S9 . Ratios of nonsynonymous (Ka) to synonymous (Ks) substitution rates calculated from ACTIN, AGO1, and AGO2/AGO3 coding sequences from C. reinhardtii and 4 closely related algal species.
Supplemental Figure S10 . Binding site of Cluster 51304+ sRNAs on the 3′ UTR of the Cre03.g164550 (FRA2/BolA-like protein 2) transcript.
Supplemental Figure S11 . Expression of predicted target genes of Cluster 51304+ sRNAs in ago1 mutant strains under nutrient deprivation.
Supplemental Table S1 . Abundance of AGO1-associated >26-nt sRNAs and miRNAs in C. reinhardtii cultured under nutrient-replete or nitrogen-deprived conditions.
Supplemental Table S2 . List of accession numbers for the coding sequences used for the Ka/Ks calculation.
Supplemental Table S3 . Oligonucleotides and primers used in this study.
Supplemental Data Set S1 . Predicted targets of the most abundant AGO1-associated >26-nt sRNAs from cluster 51304+.
Supplemental Data Set S2 . Differential expression analysis of predicted target transcripts of cluster 51304+ sRNAs under sulfur, nitrogen, or phosphorus deprivation.
Supplemental Data Set S3 . Summary of statistical analyses.
Funding
This work was supported in part by grants from the National Science Foundation (MCB 1616863 and MCB 2131783) and the Gordon and Betty Moore Foundation (Award No. 4968.01) to H.C.
References
- Abascal F, Zardoya R, Telford MJ. Translatorx: multiple alignment of nucleotide sequences guided by amino acid translations. Nucleic Acids Res. 2010:38(Suppl_2):W7–W13. 10.1093/nar/gkq291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akella S, Ma X, Bacova R, Harmer ZP, Kolackova M, Wen X, Wright DA, Spalding MH, Weeks DP, Cerutti H. Co-targeting strategy for precise, scarless gene editing with CRISPR/Cas9 and donor ssODNs in Chlamydomonas. Plant Physiol. 2021:187(4):2637–2655. 10.1093/plphys/kiab418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves CS, Nogueira FTS. Plant small RNA world growing bigger: tRNA-derived fragments, longstanding players in regulatory processes. Front Mol Biosci. 2021:8:638911. 10.3389/fmolb.2021.638911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013:64(1):137–159. 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- Baldrich P, Bélanger S, Kong S, Pokhrel S, Tamim S, Teng C, Schiebout C, Gurazada SG, Gupta P, Patel P, et al. The evolutionary history of small RNAs in Solanaceae. Plant Physiol. 2022:189:644–665. 10.1093/plphys/kiac089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. Metazoan microRNAs. Cell. 2018:173(1):20–51. 10.1016/j.cell.2018.03.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges F, Martienssen RA. The expanding world of small RNAs in plants. Nat Rev Mol Cell Biol. 2015:16(12):727–741. 10.1038/nrm4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burroughs AM, Ando Y, Aravind L. New perspectives on the diversification of the RNA interference system: insights from comparative genomics and small RNA sequencing. Wiley Interdiscip Rev RNA. 2014:5(2):141–181. 10.1002/wrna.1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M, Fu Y, Guo Y, Pan J. Chlamydomonas (Chlorophyceae) colony PCR. Protoplasma. 2009:235(1–4):107–110. 10.1007/s00709-009-0036-9 [DOI] [PubMed] [Google Scholar]
- Carninci P, Nishiyama Y, Westover A, Itoh M, Nagaoka S, Sasaki N, Okazaki Y, Muramatsu M, Hayashizaki Y. Thermostabilization and thermoactivation of thermolabile enzymes by trehalose and its application for the synthesis of full length cDNA. Proc Natl Acad Sci U S A. 1998:95(2):520–524. 10.1073/pnas.95.2.520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas-Mollano JA, Rohr J, Kim EJ, Balassa E, van Dijk K, Cerutti H. Diversification of the core RNA interference machinery in Chlamydomonas reinhardtii and the role of DCL1 in transposon silencing. Genetics. 2008:179(1):69–81. 10.1534/genetics.107.086546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti H, Casas-Mollano JA. On the origin and functions of RNA-mediated silencing: from protists to man. Curr Genet. 2006:50(2):81–99. 10.1007/s00294-006-0078-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li J, Feng J, Liu B, Feng L, Yu X, Li G, Zhai J, Meyers BC, Xia R. sRNAanno—a database repository of uniformly annotated small RNAs in plants. Hortic Res. 2021:8(1):45. 10.1038/s41438-021-00480-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Rechavi O. Plant and animal small RNA communications between cells and organisms. Nat Rev Mol Cell Biol. 2022:23(3):185–203. 10.1038/s41580-021-00425-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevenet F, Brun C, Bañuls AL, Jacq B, Christen R. Treedyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformatics. 2006:7(1):439. 10.1186/1471-2105-7-439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Deery MJ, Groen AJ, Howard J, Baulcombe DC. Endogenous miRNA in the green alga Chlamydomonas regulates gene expression through CDS-targeting. Nat Plants. 2017:3(10):787–794. 10.1038/s41477-017-0024-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BY, Valli A, Deery MJ, Navarro FJ, Brown K, Hnatova S, Howard J, Molnar A, Baulcombe DC. Distinct roles of Argonaute in the green alga Chlamydomonas reveal evolutionary conserved mode of miRNA-mediated gene expression. Sci Rep. 2019:9(1):11091. 10.1038/s41598-019-47415-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig RJ, Hasan AR, Ness RW, Keightley PD. Comparative genomics of Chlamydomonas. Plant Cell. 2021:33(4):1016–1041. 10.1093/plcell/koab026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech B, Hannon GJ. Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet. 2011:12(1):19–31. 10.1038/nrg2916 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018:46(W1):W49–W54. 10.1093/nar/gky316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dereeper A., Guignon V., Blanc G., Audic S., Buffet S., Chevenet F., Dufayard JF., Guindon S., Lefort V., Lescot M., et al. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008:36(Web Server):W465–W469. 10.1093/nar/gkn180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dexheimer PJ, Cochella L. MicroRNAs: from mechanism to organism. Front Cell Dev Biol. 2020:8:409. 10.3389/fcell.2020.00409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dueck A, Evers M, Henz SR, Unger K, Eichner N, Merkl R, Berezikov E, Engelmann JC, Weigel D, Wenzl S, et al. Gene silencing pathways found in the green alga Volvox carteri reveal insights into evolution and origins of small RNA systems in plants. BMC Genomics. 2016:17(1):853. 10.1186/s12864-016-3202-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004:32(5):1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Einhauer A, Jungbauer A. The FLAG peptide, a versatile fusion tag for the purification of recombinant proteins. J Biochem Biophys Methods. 2001:49(1–3):455–465. 10.1016/S0165-022X(01)00213-5 [DOI] [PubMed] [Google Scholar]
- Endo Y, Iwakawa HO, Tomari Y. Arabidopsis ARGONAUTE7 selects miR390 through multiple checkpoints during RISC assembly. EMBO Rep. 2013:14(7):652–658. 10.1038/embor.2013.73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng L, Zhang F, Zhang H, Zhao Y, Meyers BC, Zhai J. An online database for exploring over 2,000 Arabidopsis small RNA libraries. Plant Physiol. 2020:182(2):685–691. 10.1104/pp.19.00959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer N, Rochaix JD. The flanking regions of PsaD drive efficient gene expression in the nucleus of the green alga Chlamydomonas reinhardtii. Mol Genet Genomics. 2001:265(5):888–894. 10.1007/s004380100485 [DOI] [PubMed] [Google Scholar]
- Frank F, Hauver J, Sonenberg N, Nagar B. Arabidopsis Argonaute MID domains use their nucleotide specificity loop to sort small RNAs. EMBO J. 2012:31(17):3588–3595. 10.1038/emboj.2012.204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert LFR, MacRae IJ. Regulation of microRNA function in animals. Nat Rev Mol Cell Biol. 2019:20(1):21–37. 10.1038/s41580-018-0045-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat Rev Genet. 2009:10(2):94–108. 10.1038/nrg2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González-Ballester D, Casero D, Cokus S, Pellegrini M, Merchant SS, Grossman AR. RNA-seq analysis of sulfur-deprived Chlamydomonas cells reveals aspects of acclimation critical for cell survival. Plant Cell. 2010:22(6):2058–2084. 10.1105/tpc.109.071167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodstein DM, Shu S, Howson R, Neupane R, Hayes RD, Fazo J, Mitros T, Dirks W, Hellsten U, Putnam N, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012:40(D1):D1178–D1186. 10.1093/nar/gkr944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010:59(3):307–321. 10.1093/sysbio/syq010 [DOI] [PubMed] [Google Scholar]
- Haase AD. An introduction to PIWI-interacting RNAs (piRNAs) in the context of metazoan small RNA silencing pathways. RNA Biol. 2022:19(1):1094–1102. 10.1080/15476286.2022.2132359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardcastle TJ, Müller SY, Baulcombe DC. Towards annotating the plant epigenome: the Arabidopsis thaliana small RNA locus map. Sci Rep. 2018:8(1):6338. 10.1038/s41598-018-24515-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris EH. The Chlamydomonas sourcebook: introduction to Chlamydomonas and its laboratory use. San Diego: (CA: ): Academic Press; 2009. [Google Scholar]
- Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC. The Arabidopsis RNA-directed DNA methylation Argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell. 2010:22(2):321–334. 10.1105/tpc.109.072199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurst LD. The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 2002:18(9):486–487. 10.1016/S0168-9525(02)02722-1 [DOI] [PubMed] [Google Scholar]
- Ibrahim F, Rymarquis LA, Kim E-J, Becker J, Balassa E, Green PJ, Cerutti H. Uridylation of mature miRNAs and siRNAs by the MUT68 nucleotidyltransferase promotes their degradation in Chlamydomonas. Proc Natl Acad Sci U S A. 2010:107(8):3906–3911. 10.1073/pnas.0912632107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. Silencing messages in a unique way. Nat Plants. 2017:3(10):769–770. 10.1038/s41477-017-0028-2 [DOI] [PubMed] [Google Scholar]
- Iwakawa HO, Tomari Y. Life of RISC: formation, action, and degradation of RNA-induced silencing complex. Mol Cell. 2022:82(1):30–43. 10.1016/j.molcel.2021.11.026 [DOI] [PubMed] [Google Scholar]
- Jeong BR, Wu-Scharf D, Zhang C, Cerutti H. Suppressors of transcriptional transgenic silencing in Chlamydomonas are sensitive to DNA-damaging agents and reactivate transposable elements. Proc Natl Acad Sci U S A. 2002:99(2):1076–1081. 10.1073/pnas.022392999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapitonov VV, Jurka J. A universal classification of eukaryotic transposable elements implemented in Repbase. Nat Rev Genet. 2008:9(5):411–412. 10.1038/nrg2165-c1 [DOI] [PubMed] [Google Scholar]
- Katiyar-Agarwal S, Gao S, Vivian-Smith A, Jin H. A novel class of bacteria-induced small RNAs in Arabidopsis. Genes Dev. 2007:21(23):3123–3134. 10.1101/gad.1595107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform. 2019:20(4):1160–1166. 10.1093/bib/bbx108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Ma X, Cerutti H. Gene silencing in microalgae: mechanisms and biological roles. Bioresour Technol. 2015:184:23–32. 10.1016/j.biortech.2014.10.119 [DOI] [PubMed] [Google Scholar]
- Klum SM, Chandradoss SD, Schirle NT, Joo C, MacRae IJ. Helix-7 in Argonaute2 shapes the microRNA seed region for rapid target recognition. EMBO J. 2018:37(1):75–88. 10.15252/embj.201796474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012:9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CH, Carroll BJ. Evolution and diversification of small RNA pathways in flowering plants. Plant Cell Physiol. 2018:59:2169–2187. 10.1093/pcp/pcy167 [DOI] [PubMed] [Google Scholar]
- Leliaert F, Smith DR, Moreau H, Herron MD, Verbruggen H, Delwiche CF, De Clerck O. Phylogeny and molecular evolution of the green Algae. CRC Crit Rev Plant Sci. 2012:31(1):1–46. 10.1080/07352689.2011.615705 [DOI] [Google Scholar]
- Li J, Wu Y, Qi Y. MicroRNAs in a multicellular green alga Volvox carteri. Sci China Life Sci. 2014:57(1):36–45. 10.1007/s11427-013-4580-3 [DOI] [PubMed] [Google Scholar]
- Liberles DA. Evaluation of methods for determination of a reconstructed history of gene sequence evolution. Mol Biol Evol. 2001:18(11):2040–2047. 10.1093/oxfordjournals.molbev.a003745 [DOI] [PubMed] [Google Scholar]
- Lin PC, Lu CW, Shen BN, Lee GZ, Bowman JL, Arteaga-Vazquez MA, Liu LYD, Hong SF, Lo CF, Su GM, et al. Identification of miRNAs and their targets in the liverwort Marchantia polymorpha by integrating RNA-seq and degradome analyses. Plant Cell Physiol. 2016:57(2):339–358. 10.1093/pcp/pcw020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Qin C, Chen Z, Zuo T, Yang X, Zhou H, Xu M, Cao S, Shen Y, Lin H, et al. Identification of miRNAs and their target genes in developing maize ears by combined small RNA and degradome sequencing. BMC Genomics. 2014:15(1):25. 10.1186/1471-2164-15-25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz R, Bernhart SH, Höner zu Siederdissen C, Tafer H, Flamm C, Stadler PF, Hofacker IL. Vienna RNA package 2.0. Algorithms Mol Biol. 2011:6(1):26. 10.1186/1748-7188-6-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunardon A, Johnson NR, Hagerott E, Phifer T, Polydore S, Coruh C, Axtell MJ. Integrated annotations and analyses of small RNA-producing loci from 47 diverse plants. Genome Res. 2020:30(3):497–513. 10.1101/gr.256750.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Kim EJ, Kook I, Ma F, Voshall A, Moriyama E, Cerutti H. Small interfering RNA-mediated translation repression alters ribosome sensitivity to inhibition by cycloheximide in Chlamydomonas reinhardtii. Plant Cell. 2013:25(3):985–998. 10.1105/tpc.113.109256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macrae IJ, Li F, Zhou K, Cande WZ, Doudna JA. Structure of Dicer and mechanistic implications for RNAi. Cold Spring Harb Symp Quant Biol. 2006:71(0):73–80. 10.1101/sqb.2006.71.042 [DOI] [PubMed] [Google Scholar]
- Meseguer S. MicroRNAs and tRNA-derived small fragments: key messengers in nuclear-mitochondrial communication. Front Mol Biosci. 2021:8:643575. 10.3389/fmolb.2021.643575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi S, Cai T, Hu Y, Chen Y, Hodges E, Ni F, Wu L, Li S, Zhou H, Long C, et al. Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell. 2008:133(1):116–127. 10.1016/j.cell.2008.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molnár A, Schwach F, Studholme DJ, Thuenemann EC, Baulcombe DC. miRNAs control gene expression in the single-cell alga Chlamydomonas reinhardtii. Nature. 2007:447(7148):1126–1129. 10.1038/nature05903 [DOI] [PubMed] [Google Scholar]
- Moniruzzaman M, Erazo-Garcia MP, Aylward FO. Endogenous giant viruses contribute to intraspecies genomic variability in the model green alga Chlamydomonas reinhardtii. Virus Evol. 2022:8(2):veac102. 10.1093/ve/veac102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008:5(7):621–628. 10.1038/nmeth.1226 [DOI] [PubMed] [Google Scholar]
- Moseley JL, Gonzalez-Ballester D, Pootakham W, Bailey S, Grossman AR. Genetic interactions between regulators of Chlamydomonas phosphorus and sulfur deprivation responses. Genetics. 2009:181(3):889–905. 10.1534/genetics.108.099382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller SY, Matthews NE, Valli AA, Baulcombe DC. The small RNA locus map for Chlamydomonas reinhardtii. PLoS One. 2020:15(11):e0242516. 10.1371/journal.pone.0242516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ngan CY, Wong CH, Choi C, Yoshinaga Y, Louie K, Jia J, Chen C, Bowen B, Cheng H, Leonelli L, et al. Lineage-specific chromatin signatures reveal a regulator of lipid metabolism in microalgae. Nat Plants. 2015:1(8):15107. 10.1038/nplants.2015.107 [DOI] [PubMed] [Google Scholar]
- Ozata DM, Gainetdinov I, Zoch A, O'Carroll D, Zamore PD. PIWI-interacting RNAs: small RNAs with big functions. Nat Rev Genet. 2019:20(2):89–108. 10.1038/s41576-018-0073-3 [DOI] [PubMed] [Google Scholar]
- Palmer WH, Hadfield JD, Obbard DJ. RNA-interference pathways display high rates of adaptive protein evolution in multiple invertebrates. Genetics. 2018:208(4):1585–1599. 10.1534/genetics.117.300567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reshetnyak G, Jacobs JM, Auguy F, Sciallano C, Claude L, Medina C, Perez-Quintero AL, Comte A, Thomas E, Bogdanove A, et al. An atypical class of non-coding small RNAs is produced in rice leaves upon bacterial infection. Sci Rep. 2021:11(1):24141. 10.1038/s41598-021-03391-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey P, Taupin-Broggini M, Couturier J, Vignols F, Rouhier N. Is there a role for glutaredoxins and BOLAs in the perception of the cellular iron status in plants? Front Plant Sci. 2019:10:712. 10.3389/fpls.2019.00712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzeszutek I, Betlej G. The role of small noncoding RNA in DNA double-strand break repair. Int J Mol Sci. 2020:21(21):8039. 10.3390/ijms21218039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Merchant SS. A series of fortunate events: introducing Chlamydomonas as a reference organism. Plant Cell. 2019:31(8):1682–1707. 10.1105/tpc.18.00952 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Russell DW. Molecular cloning—a laboratory manual. Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schalk C, Cognat V, Graindorge S, Vincent T, Voinnet O, Molinier J. Small RNA-mediated repair of UV-induced DNA lesions by the DNA DAMAGE-BINDING PROTEIN 2 and ARGONAUTE 1. Proc Natl Acad Sci U S A. 2017:114(14):E2965–E2974. 10.1073/pnas.1618834114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabalina SA, Koonin EV. Origins and evolution of eukaryotic RNA interference. Trends Ecol Evol. 2008:23(10):578–587. 10.1016/j.tree.2008.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siltberg J, Liberles DA. A simple covarion-based approach to analyse nucleotide substitution rates. J Evol Biol. 2002:15(4):588–594. 10.1046/j.1420-9101.2002.00416.x [DOI] [Google Scholar]
- Song MS, Rossi JJ. Molecular mechanisms of Dicer: endonuclease and enzymatic activity. Biochem J. 2017:474(10):1603–1618. 10.1042/BCJ20160759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swarts DC, Makarova K, Wang Y, Nakanishi K, Ketting RF, Koonin EV, Patel DJ, van der Oost J. The evolutionary journey of Argonaute proteins. Nat Struct Mol Biol. 2014:21(9):743–753. 10.1038/nsmb.2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda A, Iwasaki S, Watanabe T, Utsumi M, Watanabe Y. The mechanism selecting the guide strand from small RNA duplexes is different among Argonaute proteins. Plant Cell Physiol. 2008:49(4):493–500. 10.1093/pcp/pcn043 [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J, Kjer K, Page R, Sullivan J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007:56(4):564–577. 10.1080/10635150701472164 [DOI] [PubMed] [Google Scholar]
- Talib EA, Outten CE. Iron-sulfur cluster biogenesis, trafficking, and signaling: roles for CGFS glutaredoxins and BolA proteins. Biochim Biophys Acta Mol Cell Res. 2021:1868(1):118847. 10.1016/j.bbamcr.2020.118847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. Tophat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009:25(9):1105–1111. 10.1093/bioinformatics/btp120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valli AA, Santos BACM, Hnatova S, Bassett AR, Molnar A, Chung BY, Baulcombe DC. Most microRNAs in the single-cell alga Chlamydomonas reinhardtii are produced by Dicer-like 3-mediated cleavage of introns and untranslated regions of coding RNAs. Genome Res. 2016:26(4):519–529. 10.1101/gr.199703.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk K, Marley KE, Jeong BR, Xu J, Hesson J, Cerny RL, Waterborg JH, Cerutti H. Monomethyl histone H3 lysine 4 as an epigenetic mark for silenced euchromatin in Chlamydomonas. Plant Cell. 2005:17(9):2439–2453. 10.1105/tpc.105.034165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermeulen A, Behlen L, Reynolds A, Wolfson A, Marshall WS, Karpilow J, Khvorova A. The contributions of dsRNA structure to Dicer specificity and efficiency. RNA. 2005:11(5):674–682. 10.1261/rna.7272305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voshall A, Kim EJ, Ma X, Moriyama EN, Cerutti H. Identification of AGO3-associated miRNAs and computational prediction of their targets in the green alga Chlamydomonas reinhardtii. Genetics. 2015:200(1):105–121. 10.1534/genetics.115.174797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voshall A, Kim EJ, Ma X, Yamasaki T, Moriyama EN, Cerutti H. miRNAs in the alga Chlamydomonas reinhardtii are not phylogenetically conserved and play a limited role in responses to nutrient deprivation. Sci Rep. 2017:7(1):5462. 10.1038/s41598-017-05561-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Liang H, Xu Y, Li L, Wang H, Sahu DN, Petersen M, Melkonian M, Sahu SK, Liu H. Genome-wide analyses across Viridiplantae reveal the origin and diversification of small RNA pathway-related genes. Commun Biol. 2021:4(1):412. 10.1038/s42003-021-01933-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendte JM, Pikaard CS. The RNAs of RNA-directed DNA methylation. Biochim Biophys Acta Gene Regul Mech. 2017:1860(1):140–148. 10.1016/j.bbagrm.2016.08.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson SW, Vivian-Smith A, Krokene P, Mageroy MH. The microRNA response associated with methyl jasmonate-induced resistance in Norway spruce bark. Plant Gene. 2021:27:100301. 10.1016/j.plgene.2021.100301 [DOI] [Google Scholar]
- Wu H, Li B, Iwakawa HO, Pan Y, Tang X, Ling-hu Q, Liu Y, Sheng S, Feng L, Zhang H, et al. Plant 22-nt siRNAs mediate translational repression and stress adaptation. Nature. 2020:581(7806):89–93. 10.1038/s41586-020-2231-y [DOI] [PubMed] [Google Scholar]
- Wynant N, Santos D, Vanden Broeck J. The evolution of animal Argonautes: evidence for the absence of antiviral AGO Argonautes in vertebrates. Sci Rep. 2017:7(1):9230. 10.1038/s41598-017-08043-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia J, Wang X, Perroud PF, He Y, Quatrano R, Zhang W. Endogenous small-noncoding RNAs and potential functions in desiccation tolerance in Physcomitrella patens. Sci Rep. 2016:6(1):30118. 10.1038/srep30118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamano T, Iguchi H, Fukuzawa H. Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J Biosci Bioeng. 2013:115(6):691–694. 10.1016/j.jbiosc.2012.12.020 [DOI] [PubMed] [Google Scholar]
- Yamasaki T, Kim EJ, Cerutti H, Ohama T. Argonaute3 is a key player in miRNA-mediated target cleavage and translational repression in Chlamydomonas. Plant J. 2016:85(2):258–268. 10.1111/tpj.13107 [DOI] [PubMed] [Google Scholar]
- Yamasaki T, Voshall A, Kim EJ, Moriyama E, Cerutti H, Ohama T. Complementarity to an miRNA seed region is sufficient to induce moderate repression of a target transcript in the unicellular green alga Chlamydomonas reinhardtii. Plant J. 2013:76(6):1045–1056. 10.1111/tpj.12354 [DOI] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle. 2010:9(22):4455–4460. 10.4161/cc.9.22.13958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Y, Jia T, Chen X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 2017:216(4):1002–1017. 10.1111/nph.14834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Q, Li J, Sang Y, Xing S, Wu Q, Liu X. Identification and characterization of microRNAs in Ginkgo biloba var. epiphylla Mak. PLoS One. 2015:10:e0127184. 10.1371/journal.pone.0127184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Niu D, Carbonell A, Wang A, Lee A, Tun V, Wang Z, Carrington JC, Chang CA, Jin H, et al. ARGONAUTE PIWI domain and microRNA duplex structure regulate small RNA sorting in Arabidopsis. Nat Commun. 2014:5(1):5468. 10.1038/ncomms6468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Li G, Mi S, Li S, Hannon GJ, Wang XJ, Qi Y. A complex system of small RNAs in the unicellular green alga Chlamydomonas reinhardtii. Genes Dev. 2007:21(10):1190–1203. 10.1101/gad.1543507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu H, Hu F, Wang R, Zhou X, Sze SH, Liou L, Barefoot A, Dickman M, Zhang X. Arabidopsis Argonaute10 specifically sequesters miR166/165 to regulate shoot apical meristem development. Cell. 2011:145(2):242–256. 10.1016/j.cell.2011.03.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.