Abstract
The dynamics of gene expression in crop grains has typically been investigated at the transcriptional level. However, this approach neglects translational regulation, a widespread mechanism that rapidly modulates gene expression to increase the plasticity of organisms. Here, we performed ribosome profiling and polysome profiling to obtain a comprehensive translatome data set of developing bread wheat (Triticum aestivum) grains. We further investigated the genome-wide translational dynamics during grain development, revealing that the translation of many functional genes is modulated in a stage-specific manner. The unbalanced translation between subgenomes is pervasive, which increases the expression flexibility of allohexaploid wheat. In addition, we uncovered widespread previously unannotated translation events, including upstream open reading frames (uORFs), downstream open reading frames (dORFs), and open reading frames (ORFs) in long noncoding RNAs, and characterized the temporal expression dynamics of small ORFs. We demonstrated that uORFs act as cis-regulatory elements that can repress or even enhance the translation of mRNAs. Gene translation may be combinatorially modulated by uORFs, dORFs, and microRNAs. In summary, our study presents a translatomic resource that provides a comprehensive and detailed overview of the translational regulation in developing bread wheat grains. This resource will facilitate future crop improvements for optimal yield and quality.
A unique translatomic resource provides insight into the translational control of gene expression during bread wheat grain development, facilitating crop improvement for optimal yield and quality.
IN A NUTSHELL.
Background: Bread wheat (Triticum aestivum) is one of the most widely cultivated cereal crops worldwide. The genetic code of DNA is first transcribed to produce mRNAs, which are then translated to produce proteins. This process is regulated at each step. Translational regulation is a widespread mechanism that rapidly modulates gene expression to maintain growth and development. Our current understanding of gene regulation in wheat grains focuses on the transcriptional level, neglecting the translational level.
Question: How is gene expression regulated at the translational level in developing wheat grains? What functional proteins are involved in grain development?
Findings: We used 2 approaches, namely, ribosome profiling and polysome profiling, to obtain a unique translatome data set of developing bread wheat grains. The translation of many functional genes is modulated in a stage-specific manner. The divergence of the translational regulation between the wheat subgenomes is pervasive, which increases the expression flexibility of allohexaploid wheat. Widespread open reading frames (ORFs) are actively expressed in wheat grains. Upstream open reading frames (uORFs) that function as translational regulatory elements can repress or even activate the translation of mRNAs. Gene translation may be combinatorially modulated by uORFs, downstream ORFs, and microRNAs.
Next steps: Our work presents valuable translatomic resources for understanding the translational control of gene expression during wheat grain development. Further studies will explore functional translational regulatory elements to improve wheat yield and quality.
Introduction
Cereals are the most widely cultivated crops worldwide (Hickey et al. 2019). Their grains or seeds are used as food sources, accounting for most of the calories consumed by humans worldwide. The importance of cereals has driven research interest in genetic improvement strategies to maximize the yield and quality of the grains, which requires a comprehensive understanding of the gene expression dynamics affecting traits such as grain shape and quality (Borrill et al. 2019; Hickey et al. 2019). Due to the reduced cost and advanced technologies of high-throughput sequencing (Stark et al. 2019), many researchers have studied the regulation of gene expression in bread wheat at the transcriptional level (Ramirez-Gonzalez et al. 2018; Chen et al. 2023). A series of functional genes involved in grain development and their transcriptional regulatory networks in crops were identified (Wang et al. 2010; Zhan et al. 2015; Li and Li 2016; Xiang et al. 2019; Yi et al. 2019). In one such study, for example, transcriptome profiling was performed at 7 stages of embryo development, 2 endosperm stages, and one pericarp stage using polyploid wheats (Triticum spp.) and their diploid ancestors (Xiang et al. 2019). A temporal transcriptome of early maize (Zea mays) seed development was utilized to construct transcriptional regulatory networks (Yi et al. 2019). The most recent investigations into the regulation of gene expression are focused on the transcriptional level, whereas our understanding of translational regulation in crop grains is sparse.
Gene expression in organisms is modulated at multiple levels, from transcription to protein production, in which translation is the biological process that decodes genetic information into functional proteins (Reddy et al. 2013; Brar and Weissman 2015; Ingolia 2016). Multiple approaches have been developed for translatome exploration (Brar and Weissman 2015; Zhao et al. 2019). Translating ribosome affinity purification incorporating FLAG-tagged RIBOSOMAL PROTEIN L18 (RPL18) into ribosomes was developed for the quantification of mRNA translation in a specific tissue or organ (Zanetti et al. 2005; Juntawong et al. 2014; Reynoso et al. 2019; Traubenik et al. 2020; Yoo et al. 2020). Polysome profiling (Poly-seq) has been used to perform genome-wide profiling of translation. In this technique, polysome-associated mRNA is isolated through differential centrifugation and fractionation and is further quantified using high-throughput sequencing (Bunnik et al. 2013; Juntawong et al. 2014; Bai et al. 2017; Zhang et al. 2017). In addition, ribosome profiling (Ribo-seq) has been utilized to obtain ribosome-protected mRNA fragments (Juntawong et al. 2014; Lei et al. 2015; Hsu et al. 2016; Bazin et al. 2017; Chotewutmontri and Barkan 2021; Zhu et al. 2021a), which enables the quantification of translation control and the positions of the translating ribosomes on a genome-wide scale. The ribosome reads the information, one codon (3 nucleotides) at a time. Ribosome profiling has been utilized to uncover unannotated translation events, such as upstream open reading frames (uORFs) and small ORFs (sORFs) encoding small peptides (Juntawong et al. 2014; Lei et al. 2015; Hsu et al. 2016; Bazin et al. 2017; Zhang et al. 2018a, 2021; van Heesch et al. 2019; Wu et al. 2019; Chen et al. 2020a; Patraquim et al. 2020). Recent studies found that variation in the uORFs causes allelic diversity in protein abundance in maize (Gage et al. 2022) and soybean (Glycine max) (Guo et al. 2022); thus, these experimental approaches can provide great opportunities for the identification of translated ORFs and the quantification of translational control in crop grains on a genome-wide scale.
In plants, some studies have investigated the translational dynamics of genes in diverse aspects of development, growth, and environmental adaptation, including photomorphogenesis, C4 photosynthesis, the circadian clock, seed germination, and the responses to hypoxia stress, drought stress, phosphorus deficiency, and flooding (Liu et al. 2013; Juntawong et al. 2014; Lei et al. 2015; Bai et al. 2017; Bazin et al. 2017; Srivastava et al. 2018; Reynoso et al. 2019; Chung et al. 2020; Bonnot and Nagel 2021; Chotewutmontri and Barkan 2021). The identification of translated ORFs in plants has been investigated in Arabidopsis (Arabidopsis thaliana), tomato (Solanum lycopersicum), and maize (Juntawong et al. 2014; Hsu et al. 2016; Wu et al. 2019; Liang et al. 2021). Most studies use a single type of translation investigation method. Ribosome profiling allows the determination of ribosome positioning on mRNA at a single-nucleotide resolution and the identification of translated ORFs, but it is far less sensitive in detecting less-abundant mRNAs (Ingolia et al. 2009; Kronja et al. 2014; Gandin et al. 2016) and has relatively lower read alignment accuracy due to the short length of the sequencing reads. Polysome profiling can reveal the translationally active fraction of an mRNA (Kronja et al. 2014) and produce longer sequencing reads. These reports highlight the power of combining ribosome profiling and polysome profiling when studying the translatome, especially in allohexaploid wheat.
Bread wheat (Triticum aestivum, 2n = 6x = 42, BBAADD), which originated from 2 rounds of polyploidization, provides about 20% of the calories consumed by humans worldwide (International Wheat Genome Sequencing Consortium et al. 2018; Laugerotte et al. 2022). At the transcriptional level, a large number of studies in allohexaploid wheat have revealed that stage-specific transcriptional expression and subgenome asymmetry are common (Liu et al. 2015; Ramirez-Gonzalez et al. 2018; Takahagi et al. 2018; Xiang et al. 2019), and homoeolog expression bias may affect phenotypes (He et al. 2022); however, the translational dynamics of genes during allohexaploid wheat grain development remain unknown. Here, we combined ribosome profiling and polysome profiling to construct a grain translatome data set for bread wheat and investigate translational control during grain development. We uncovered extensive stage-specific and subgenome-divergent translational regulation of mRNAs at a genome-wide scale. In addition, we uncovered actively translated ORFs, including uORFs in the 5′ untranslated regions (UTRs), downstream ORFs (dORFs) in the 3′ UTRs, and ORFs in long noncoding RNAs (lncRNAs), and characterized their expression dynamics. The combinatorial control of gene expression at the translational level by uORFs, dORFs, and microRNAs (miRNAs) was also revealed. This study sheds light on the regulation of gene expression, unannotated ORFs, and subgenome asymmetry at the post-transcriptional level in polyploid crops.
Results
A wheat grain time series translatome data set based on ribosome profiling and polysome profiling
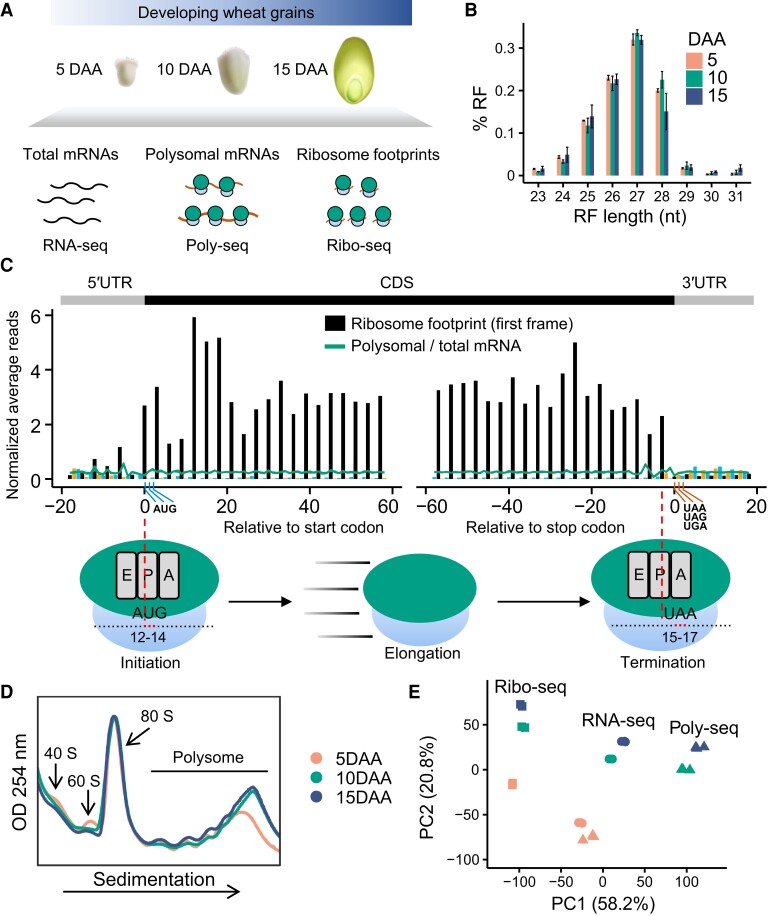
To characterize the translational dynamics of genes during grain development at a genome-wide scale, we performed next-generation sequencing on the total mRNAs (RNA-seq), polysomal mRNAs (Poly-seq), and ribosome-protected mRNA fragments (Ribo-seq) of wheat grains at successive developmental stages, including 5 d after anthesis (DAA), 10 DAA, and 15 DAA (Fig. 1A; Supplemental Data Set 1). On average, the Illumina sequencing platform yielded 76.9 million and 75.7 million reads from each replicate of the total and polysomal mRNA samples, respectively, of which 89.1% and 90.3% could be mapped to the bread wheat Chinese Spring reference genome IWGSC RefSeq v2.1 (International Wheat Genome Sequencing Consortium et al. 2018; Zhu et al. 2021b). In addition, around 60.0 million ribosome footprint (RF) reads per replicate were generated for the ribosome profiling samples. After filtering out fragments of housekeeping RNAs (ribosomal [rRNA], transfer [tRNA], and small nucleolar RNA [snoRNA]), nearly 78.6% of the RF reads could be mapped to the wheat reference genome. The high mapping rates emphasize the high quality of our data sets.
Figure 1.
Polysome profiling and ribosome profiling expose translational dynamics during grain development. A) A schematic overview of the experimental approach. The wheat grains from 3 developmental stages were used for the translatome investigation. B) Size distribution of the ribosome footprint (RF). Mean and standard error are shown. C) Metagene analysis of the RF reads near the annotated translation start and stop sites. The 27-nucleotide RF reads and the combination of replicates are shown. The density of reads at each position was normalized across the density of the surrounding reads. Bars show RFs, and lines show total and polysomal mRNAs. The predicted position of the ribosome's peptidyl (P) site of RF reads relative to the CDS start and stop codons is shown. The position of the RF reads is indicated by its 12th nucleotide within each footprint. The x-axis represents the relative distance of each RF reads to the start codon or the stop codon. The 0 on the x-axis represents that the 12th nucleotide of the RF reads was mapped to the 1st nucleotide of the start codon. The red, blue, and green bars represent the RF reads mapped to the 1st (expected), 2nd, and 3rd reading frames, respectively. The inferred peptidyl (P) site (nucleotides 12 to 14) in the start codon and the acceptor (A) site (nucleotides 15 to 17) in the stop codon are illustrated. D) Polysome profiling was analyzed using a sucrose gradient sedimentation with 3 replicates using grain sampled at 5, 10, and 15 d after anthesis (DAA), and the optical density and a wavelength of 254 nm (OD254; arbitrary units) was measured for the 3 grain developmental stages at the times indicated. E) Principal component analysis (PCA) of the RNA-seq, Poly-seq, and Ribo-seq data from the 3 grain development stages using the 10,000 most variable genes.
We investigated the RF features that are strongly associated with the translation mechanism in organisms (Ingolia 2016; Calviello and Ohler 2017) to examine the quality of the Ribo-seq libraries. We found that the RF size was about 27 nucleotides for the 3 grain developmental stages (Fig. 1B), which is similar to the footprint reported for Arabidopsis (Juntawong et al. 2014; Hsu et al. 2016), tomato (Wu et al. 2019), and maize (Lei et al. 2015; Reynoso et al. 2019). Importantly, the distribution of RFs from wheat grains exhibited strong 3-nucleotide periodicity (Fig. 1C; Supplemental Fig. S1), a typical characteristic of actively translating ribosomes that shift down the mRNA by 3 nucleotides at a time. The RF metagene analysis showed that the peptidyl (P) sites within the wheat grain ribosomes are located between nucleotides 12 and 14 and the acceptor (A) site is located between nucleotides 15 and 17 for the 27-nucleotide RFs (Fig. 1C). In addition, in comparison with the RNA-seq and Poly-seq reads, the Ribo-seq reads were mainly mapped to the annotated coding region and were sparse in the UTR (Supplemental Fig. S2). The replicates of the 3 types of libraries were highly correlated (R > 0.94; Supplemental Fig. S3), with replicates clustering together as expected.
We performed a gene expression quantification analysis (Supplemental Data Set 2) for all libraries. We took the average fragments per kilobase of exon per million mapped fragment (FPKM) values as the abundance of the associated genes. The read count was normalized by sequencing depth with a sample-specific size factor implemented in DESeq2 (Love et al. 2014). The total mRNAs were positively correlated with the RFs (R = 0.78 on average) and polysomal mRNA (R = 0.91 on average) (Supplemental Fig. S4). In addition, we performed a principal component analysis (PCA) for all samples and found that PC1 (explaining 58.2% of the variation) separated the samples into RNA-seq (total mRNA/transcription), Poly-seq (polysomal mRNA/translation), and Ribo-seq (RF/translation) groups, while PC2 (20.8% of the variation) separated the developmental process (Fig. 1E).
We analyzed the polysome profiles of diverse developmental stages and found that changes in polysome profiles occur throughout all developmental phases (Fig. 1D), which is consistent with the result of the PCA. The Poly-seq data are most appropriate for the quantification of transcription abundance because Poly-seq reads have high mapping accuracy, while the Ribo-seq data are best used to uncover the positions of the ORFs. Our data sets are suitable for the investigation of translational regulation during wheat grain development.
Extensive stage-specific translational regulation of gene expression during grain development
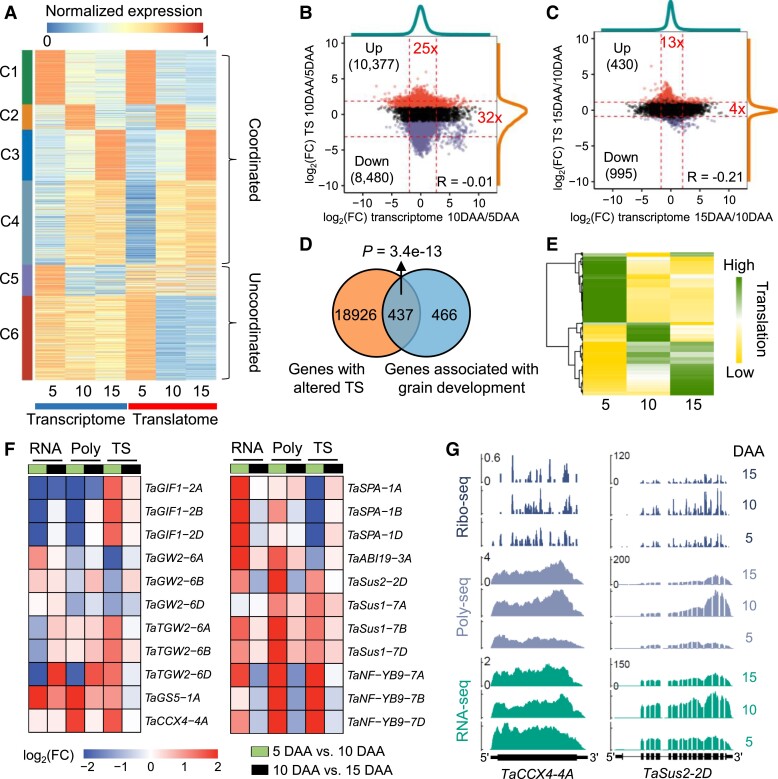
To gain a comprehensive understanding of the transcriptional and translational dynamics during the grain developmental process, we identified a collection of 33,815 differentially expressed genes (DEGs), accounting for 64.1% of expressed genes, across the 3 types of libraries representing diverse developmental stages (Supplemental Data Set 3). This high total number of DEGs suggests that large numbers of genes are involved in the regulation of grain development, which is consistent with a previous grain transcriptome study (Xiang et al. 2019). In addition, more than half of the DEGs in the Ribo-seq data sets could also be detected in the RNA-seq or Poly-seq data sets (Supplemental Fig. S5), indicating high correlations among the 3 types of libraries.
We further performed a K-means coexpression analysis for the transcript abundance of the DEGs, resulting in 6 coexpression clusters with a range from 2,537 to 8,418 DEGs (Fig. 2A; Supplemental Data Set 4). The expression patterns of clusters 5 and 6 were not consistent between the transcriptome and the translatome, implying that translational regulation may have altered the expression of some genes. Furthermore, we acquired 857 differentially expressed proteins (DEPs; Supplemental Data Set 5) from a public proteomic data set of the wheat cv. Cranbrook grains (Daba et al. 2020) and found that the translatomic data can recover more DEPs than the transcriptomic data (Supplemental Fig. S6).
Figure 2.
Translational dynamics of gene expression during grain development. A) Coexpression of differentially expressed genes (DEGs) at the transcriptional and translational levels at 3 different grain development stages (5, 10, and 15 d after anthesis). For the transcriptome and translatome, the expression values (FPKMs) were normalized by dividing by the maximum FPKM at the transcriptional or translational level. The K-means method was employed to identify coexpression clusters. B and C) Scatterplots showing the fold changes of the translation state (TS) and transcriptional expression levels during grain development. The transitions from 5 d after anthesis (DAA) to 10 DAA B) and from 10 to 15 DAA C) were analyzed. Dashed lines represent the 2.5 and 97.5 percentiles. The fold changes of the mRNA abundance ratio B) and TS ratio C) between the 2.5 and 97.5 percentiles are indicated in red text, respectively. Red and purple points indicate genes with upregulated and downregulated TSs, respectively. D) The Venn diagram shows that most of the genes involved in grain development have an altered TS. The genes involved in grain development were taken from the literature (Yao et al. 2018; Chen et al. 2020b). P-value, hypergeometric test. E) Translational expression patterns of 437 grain development–related genes with altered TS. The Z-scores of the expression levels of each gene at the translational level were visualized. The high Z-score on the gradient scale represents the high expression levels at the translational level. F) Expression profiles of 9 functional genes. Differential expression and translation analyses were performed. G) Total mRNA (RNA-seq), polysomal mRNA (Poly-seq), and ribosome footprint (Ribo-seq) read coverage on key genes regulating wheat grain development and filling. The read coverage was normalized by the million reads mapped to nuclear coding sequences.
The translation state (TS; calculated as the ratio of polysomal mRNA abundance to that of total RNA) was used to measure the utilization efficiency of mRNA for protein synthesis. To reduce the count errors from short reads mapping, we used the Poly-seq and RNA-seq data to calculate TSs of mRNAs. We identified a total of 19,363 genes with altered TS to investigate the dynamics of translational regulation (Supplemental Data Set 6) and observed more genes with altered TS and a greater change in gene abundance at the transition from 5 to 10 DAA. In detail, at the comparison of 5 to 10 DAA, 95% of genes spanned a 25-fold range in the transcript abundance ratio and a larger range (32-fold) in the TS ratio (Fig. 2B). The reverse situation was observed at the transition from 10 to 15 DAA (that is, the change in the transcript abundance ratio is larger than that in the TS ratio) (Fig. 2C).
We next aimed to determine the biological processes in which the differentially translated genes might be involved and thus performed a Gene Ontology (GO) enrichment analysis. The results revealed a significant enrichment in categories related to starch biosynthesis (Supplemental Fig. S7). Based on the literature and public resources (Yao et al. 2018; Chen et al. 2020b), we explored 903 genes associated with grain development (Supplemental Data Set 2) and found that almost half of them have altered TS (P = 3.4e−13, hypergeometric test) (Fig. 2D). An expression analysis revealed that these functional genes with a changed TS may be involved in diverse grain developmental stages (Fig. 2E). We viewed the expression patterns of several well-known genes (Fig. 2F). During the grain developmental process, GRAIN INCOMPLETE FILLING 1 (GIF1), known to control grain filling and yield (Wang et al. 2008), had an increased TS. GRAIN WIDTH 2 (GW2) (Song et al. 2007) and THOUSAND GRAIN WEIGHT 2 (TGW2) (Ruan et al. 2020), the homologs of which negatively regulate grain width and weight in rice (Oryza sativa), also had altered TSs. A previous study showed that higher GRAIN SIZE 5 (GS5) abundance is correlated with larger rice grain size (Li et al. 2011), and we found that GS5 had an increased TS in the 10-DAA grains. CATION CALCIUM EXCHANGER 4 (CCX4), a gene that controls the salicylic acid signaling pathway and compensates for cell enlargement (Fujikura et al. 2020), had an increased TS (Fig. 2G). In addition, we also examined the translational regulation of the starch biosynthesis–related genes. The sucrose synthase–related gene SUCROSE SYNTHASE 1 (Sus1) (Baroja-Fernandez et al. 2012) had an increased TS in the 5-DAA grain, and Sus2 had an increased TS in the 10-DAA and 15-DAA grains (Fig. 2G). NUCLEAR FACTOR Y B9 (NF-YB9), involving in starch biosynthesis during cereal endosperm development (Niu et al. 2021), had an increased TS in the 10-DAA grain. STORAGE PROTEIN ACTIVATOR (SPA), encoding a bZIP transcription factor that is negatively correlated with prolamin and starch accumulation in wheat grain, had a decreased TS. ABSCISIC ACID INSENSITIVE 19 (ABI19), encoding a grain filling–initiation regulator (Yang et al. 2021a), had a decreased TS in the 10-DAA grain (Fig. 2F). These observations demonstrate that functional genes are under extensive translational regulation during grain development.
Subgenome asymmetry of translational regulation increases expression divergence between the wheat homoeologs
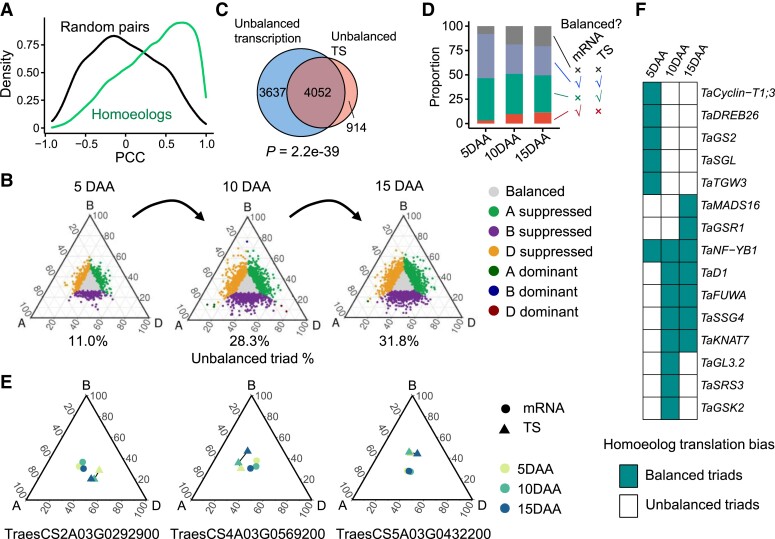
Hexaploid wheat that has undergone 2 rounds of polyploidization displays an unbalanced transcriptional expression between homoeologs (Ramirez-Gonzalez et al. 2018; Xiang et al. 2019; He et al. 2022); however, the divergence of the translational regulation between the allohexaploid wheat subgenomes remains unknown. We calculated TSs of mRNAs using the Poly-seq and RNA-seq data. Given that homoeologs originate from orthologs that are then aggregated into a single genome through allopolyploidization (Glover et al. 2016), we expected and observed a high correlation of TS values between the homoeolog pairs (Fig. 3A).
Figure 3.
Expression bias of homoeologs at the translational level. A) Pearson correlation coefficients (PCCs) of the translation state (TS) between homoeolog triads (green) and random (black) gene pairs. B) Ternary plots showing the relative TSs of triads in grains at 5 d after anthesis (DAA), 10, and 15 DAA. Each point corresponds to a gene triad with an A, B, and D coordinate. The color indicates the homoeolog bias pattern. The bottom percentage is the proportion of unbalanced homoeolog triads. The unbalanced group consists of 6 other groups of triads, excluding the balanced triads. C) Overlap between the triads with unbalanced transcriptional expressions and the triads with unbalanced TSs. P-value, hypergeometric test. Triads were combined for the 3 developmental stages. D) Summary of the homoeolog expression bias patterns at the transcriptional and translational levels in 5-, 10-, and 15-DAA grains. The ordinate shows the proportion of each triad group. The right panel indicates whether homoeolog triads in each group have balanced mRNA abundance or TE. E) Several examples show the unbalanced TSs between the homoeolog triads that are balanced at the transcriptional level. The relative TS of each triad was calculated. F) Unbalanced translation of triads of the well-known genes involved in grain development. Each block represents a homoeolog triad.
We next analyzed a total of 10,124 stably expressed homoeolog triads (Supplemental Data Set 7) and found that 11.0% (5 DAA) to 31.8% (15 DAA) of the triads have an unbalanced TS between the subgenomes (Fig. 3B). The total number of unbalanced triads increased with grain development, suggesting that the change in the translational dynamics of homoeologs is related to grain development processes. A significant overlap between the triads with unbalanced transcription and the triads with unbalanced TS was found (Supplemental Fig. S8; Fig. 3C), suggesting that the triads with unbalanced transcription also tend to have unbalanced translational regulations. We discovered that nearly half of the triads have the same transcriptional and translational expression patterns across all stages (Fig. 3D). Notably, 1,575 stage-specific triads have balanced transcriptional expressions but unbalanced translational expressions. The profiling of several individual homoeolog triads showed different homoeolog bias patterns between the transcriptional and translational levels (Fig. 3E). In homoeolog triads with unbalanced TS, some genes involved in mRNA transport, rRNA processing, lipid homeostasis, and starch catabolic process were overrepresented (Supplemental Fig. S9). Many stage-specific enrichments were found, such as DNA repair, RNA splicing, and chaperone-mediated protein folding. We observed an unbalanced TS for the homoeolog triads of some well-known genes, such as Cyclin-T1;3 (Qi et al. 2012), GS2 (Hu et al. 2015), GRAIN LENGTH 3.2 (GL3.2) (Xu et al. 2015), SHORT GRAIN LENGTH (SGL) (Wu et al. 2014), NF-YB1 (Bai et al. 2016), and SUBSTANDARD STARCH GRAIN 4 (SSG4) (Matsushima et al. 2014) (Fig. 3F). These observations demonstrate that subgenome asymmetry for translational regulation is pervasive during grain development.
Sequence length, GC content, and normalized minimal free energy alter the translation of mRNAs
The mRNA sequence features may play important roles in translational regulation, modulating RNA translation, and protein abundance (Kawaguchi and Bailey-Serres 2005; Juntawong et al. 2014; Lei et al. 2015; Traubenik et al. 2020). To identify sequence features associated with translational regulation in wheat grains, we compared the sequence length, GC content, and normalized minimal free energy (NMFE) of the coding sequence (CDS), 5′ UTR, and 3′ UTR regions of the mRNAs (Supplemental Fig. S10). According to their TS, the mRNAs were classified into 3 groups: high TSs, middle TSs, and low TSs (see Materials and methods). The results showed that the well-translated mRNAs generally have extreme CDS, 5′ UTR, and 3′ UTR sequence lengths compared with the poorly translated mRNAs (Supplemental Fig. S10A–C). Furthermore, we observed that the mRNAs with a high GC content and low NMFE in the 5′ UTR are generally poorly translated. In contrast, the mRNAs with a low GC content and high NMFE in the 3′ UTR and CDS are generally poorly translated. The above findings were also previously observed in maize (Lei et al. 2015), highlighting similar sequence features affecting mRNA translation in crops. In addition, the dynamic triads have significantly higher (P < 0.01, Wilcoxon test) coefficients of variation (CVs) among homoeologs than the stable triads in terms of sequence length, GC content, and NMFE of CDS, 5′ UTR, and 3′ UTR (Supplemental Fig. S10, D–F), suggesting that these sequence features may contribute to unbalanced translation between homoeologs.
Identification and characterization of active annotated and unannotated ORFs at a genome-wide scale
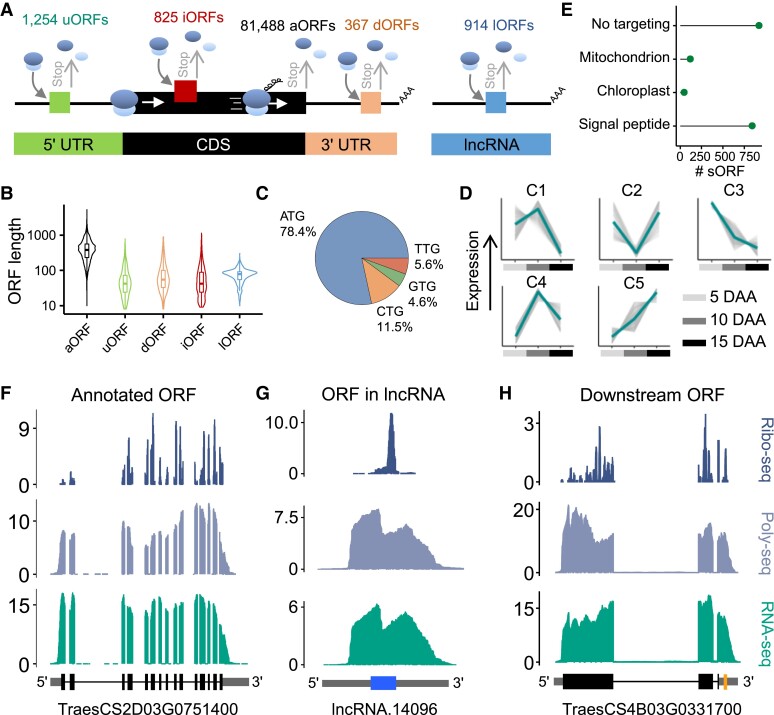
To catalog the translated sequences in the wheat grain, we identified genome-wide ORFs using the Ribo-seq data based on the annotated transcripts in the reference genome and the newly assembled transcripts (Fig. 4A). According to previously described methods (Zhao et al. 2018; Tian et al. 2019), we assembled lncRNAs in the wheat grains using the sequencing data sets generated in this study. We then identified a total of 81,488 annotated ORFs (aORFs) within protein-coding genes in the reference genome. In addition, we discovered 3,360 unannotated ORFs potentially encoding peptides (Supplemental Data Set 8), including 1,254 uORFs in the 5′ UTRs, 367 dORFs in the 3′ UTRs, 825 internal ORFs (iORFs) in annotated CDSs but in a different reading frame, and 914 ORFs in the lncRNAs (lORFs), the transcripts of which lacked prior protein-coding annotations. We found that most of the unannotated ORFs have a shorter sequence than the aORFs in CDSs (Fig. 4B), and 21.6% of the unannotated ORFs had non-AUG start codons (Fig. 4C). The Ribo-seq reads mapping to the unannotated ORFs exhibited the expected strong 3-nucleotide periodicity of positions (Supplemental Fig. S11), indicating that we had identified active unannotated ORFs on a genome-wide scale.
Figure 4.
Genome-wide identification and characterization of actively translated open reading frames (ORFs). A) Summary of the ORFs identified, including the upstream ORFs (uORFs), annotated ORFs (aORFs), downstream ORFs (dORFs), internal ORFs (iORFs), and ORFs in lncRNAs (lORFs). B) Size distribution of the identified ORFs. The lengths were normalized. C) Start-codon usage of the identified ORFs. Numbers within the plots represent the percentages of ORFs with different start-codon usages. D) Translational expression patterns of the identified short ORFs (sORFs), which contain fewer than 300 nucleotides. Expression values were scaled to Z-scores. The K-means method was used to construct coexpression clusters of sORFs. The thick line in the middle represents mean. E) Predicted subcellular localization of the proteins encoded by sORFs. The TargetP software (Almagro Armenteros et al. 2019) was used here. F–H) Coverage of RNA-seq, Poly-seq, and Ribo-seq in examples of an aORF F), lORF G), and dORF H). Read coverage was normalized by the million reads mapped to nuclear coding sequences. The gene model of the aORF is well supported by the sequencing reads F). In long noncoding RNA (lncRNA), the sequencing reads support the expression of a lORF G). Sequencing reads in aORFs provided strong support for dORFs H).
Small peptides, encoded by sORFs, play fundamental roles in plant growth, development, and environmental adaptation. We identified a total of 1,883 sORFs (Supplemental Data Set 9) from the newly identified uORFs, dORFs, lORFs, and iORFs. To characterize the expression dynamics of these sORFs at the translational level, we performed a coexpression analysis using the K-means method and obtained 5 sORF coexpression clusters representing different expression patterns (Fig. 4D). For instance, genes in cluster C3 might have important roles in the early development of the grain because of their higher abundance in the 5-DAA grain. A further subcellular localization analysis predicted 636 ORFs harboring signal peptides, 28 ORFs harboring chloroplast transit peptides, and 76 ORFs harboring mitochondrial transit peptides (Fig. 4E). These findings highlight that the identified sORFs may be functionally expressed.
Our translatome data sets can be used to improve the genome annotation of the complex allohexaploid wheat genome. The profiling of several individual transcripts showed that both the aORFs (Fig. 4F) and the newly identified ORFs (Fig. 4, G and H) have evidence of ribosome and polysome reads within the coding regions.
uORFs act as regulatory elements to influence the translation efficiency of mRNAs
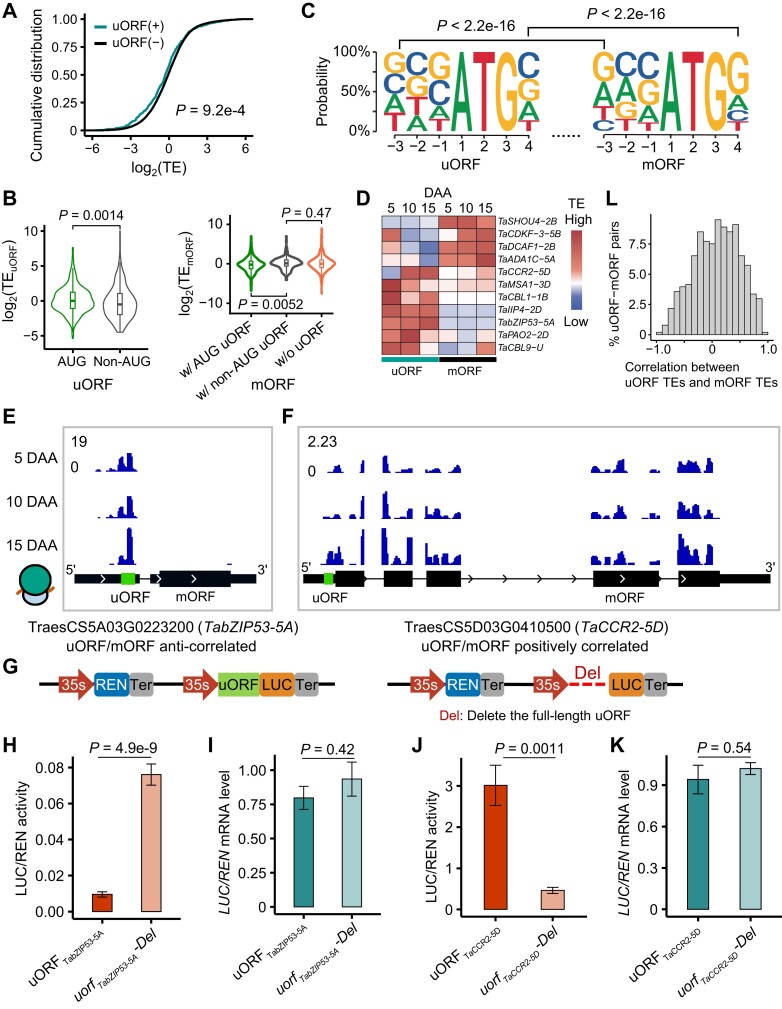
Some uORFs can affect the translation of the main ORFs (mORFs) in plants (Juntawong et al. 2014; Lei et al. 2015; Wu et al. 2019), but their roles in bread wheat remain unknown. To investigate the effect of uORFs on gene translation, we calculated the translation efficiencies (TEs) of ORFs using the RNA-seq and Ribo-seq data. As expected, mRNAs with uORFs have lower TEs than mRNAs without uORFs (Fig. 5A). Furthermore, we found that uORFs with AUG start codons have higher TEs than uORFs with non-AUG start codons and that the mRNAs with AUG uORFs have lower TEs than the mRNAs with non-AUG uORFs (Fig. 5B), implying that AUG-start uORFs may have a stronger suppressive effect on mORF abundance.
Figure 5.
Upstream open reading frames (uORFs) alter the translation efficiency (TE) of mRNAs in wheat. A) Cumulative distributions of the TEs of mRNAs with uORFs. A 2-sided Wilcoxon test was performed. B) TE of uORFs with and without an AUG start codon and their downstream mORFs. The left panel shows TEs of AUG uORFs and non-AUG uORFs. The right panel shows TEs of mORFs with and without an AUG uORF. The “w/o uORF” group represents 1,000 randomly selected mORFs without uORFs. Two-sided Wilcoxon tests were performed. C) Kozak sequences of uORFs and main ORFs (mORFs). A chi-square test was performed. D) Several genes with uORFs alter the mORF TE. The TEs of the uORFs and mORFs across developmental stages are anticorrelated. E and F) Coverage of Ribo-seq reads in TabZIP53-5AE) and TaCCR2-5DF). Read coverage was normalized by the million reads mapped to nuclear coding sequences. In the region upstream of the mORF, the sequencing reads support the expression of uORFs. Green boxes, uORFs. The base sequences and the length of the uORFs (uORFTabZIP53-5A, uORFTaCBL1-1B) can be found in Supplemental Data Set 10. G) Schematic of the dual-luciferase system used to investigate the effect of uORFs on the translation of mORFs. 35S, cauliflower mosaic virus 35S promoter. REN, Renilla reniformis luciferase; LUC, firefly luciferase. Del indicates the deletion of a uORF. H–K) The effect of the different cassettes on the LUC/REN activity H and J) and LUC/REN mRNA level I and K) associated with the mORF in the dual-luciferase reporter system. P-values were calculated using 2-sided Student's t-test. uORFTabZIP53-5A and uORFTaCCR2-5 were derived from TabZIP53-5A and TaCCR2-5D, respectively. H and I) TabZIP53-5A. J and K) TaCCR2-5D. Mean and standard error are shown. L) A frequency distribution histogram of the correlation between the TEs of uORFs and those of their mORFs. TEs of uORFs and their mORFs were calculated, respectively. All replicates from 3 developmental stages were used. The Spearman correlation coefficients were used to measure the correlation.
Kozak sequences are important for start codon recognition and translation initiation in organisms (Kozak 1987). We observed an expected well-conserved Kozak sequence feature at the positions of −3 (A/G) and 4 (G) around the AUG start codons of the aORFs (Supplemental Fig. S12) and mORFs, but not uORFs (Fig. 5C). A further GO enrichment analysis of the mRNAs with uORFs showed that these mRNAs may be involved in photosynthesis, rhythmic processes, and protein autophosphorylation (Supplemental Fig. S13A). In addition, we found that the mRNAs with uORFs have lower TEs than their homologous genes with similar functional annotations and without uORFs (Supplemental Fig. S13B), suggesting that some homologous genes with similar functions may have different translation efficiencies due to the variation of the uORF. Some uORFs are negatively correlated with the translation of mORFs, such as in CALCINEURIN B-LIKE PROTEIN 9 (CBL9) (Pandey et al. 2004), ILA1-INTERACTING PROTEIN 4 (IIP4) (Zhang et al. 2018c), SHOU4 (Polko et al. 2018), and DDB1-CUL4 ASSOCIATED FACTOR 1 (DCAF1) (Zhang et al. 2008) (Fig. 5D), suggesting that the uORF has a repressive effect.
Over evolutionary time, conserved proteins could be functionally important. To investigate the evolutionary history of uORFs, we acquired 4 eudicot genomes and 4 monocot genomes and identified the evolutionarily conserved peptide uORF (CPuORF) (Supplemental Data Set 8). With a whole-genome search for uORF homologs, we identified 134 angiosperm-conserved CPuORFs, which may represent the uORFs encoding functionally important proteins (Supplemental Fig. S14). In addition, we found 58 Poaceae-specific CPuORFs in all 5 Poaceae genomes (Supplemental Fig. S14), which may contribute to Poaceae evolution. There is not a statistically significant difference between the TEs of the mRNAs with CPuORFs and those with non-CP uORFs (Supplemental Fig. S13C). Interestingly, 317 homoeolog triads have present–absent variations (PAVs) of uORFs among the 3 subgenomes, which might contribute to homoeolog translation bias. In addition, we found that the uORFs of BASIC LEUCINE ZIPPER 53 (bZIP53) and genes encoding the ultraviolet-B receptor UVR8 have different RF coverage between homoeologs (Supplemental Fig. S15). In summary, these observations highlight the potential role of uORFs in wheat evolution.
To further verify the effect of uORFs on the translation of the mORFs, we measured the expression of ORFs in Nicotiana benthamiana leaves using a dual-luciferase (LUC) system by calculating the ratio of LUC activity to the control Renilla reniformis luciferase (REN) activity (Hellens et al. 2005). We selected 2 active uORFs with Ribo-seq reads for further experiments (Fig. 5E; Supplemental Fig. S16A). These uORFs are located in the 5′ UTR region of the genes TabZIP53-5A and TaCBL1-1B. In an expression cassette driven by the 35S promoter, cassettes with only the complete uORFs or deleted uORFs were constructed upstream of the LUC-coding region. A second 35S promoter–driven cassette expressing REN as an internal vector control was also included in the resulting constructs (Fig. 5G). Compared with cassettes with the intact uORFs, cassettes with the deleted uORFs consistently generated substantially higher LUC/REN activity levels for TabZIP53-5A (Fig. 5H) and TaCBL1-1B (Supplemental Fig. S16B), with fold increases of approximately 7.0 and 3.5, respectively, while reverse transcription quantitative PCR (RT-qPCR) assays revealed that the LUC/REN mRNA ratios of transcripts did not differ significantly (Fig. 5I; Supplemental Fig. S16C). These results demonstrated that these 2 uORFs suppress the translation of the downstream mORFs with no effect on transcription.
Furthermore, we identified a uORF located in the 5′ UTR of CINNAMOYl-COA REDUCTASE 2 on chromosome 5D (TaCCR2-5D) (Fig. 5F), which displayed a positive correlation between the translation of the uORF and the mORF, and studied it to identify whether uORFs can improve the translation of mORFs. Compared with the mutant uorfTaCCR2-5D-Del, uORFTaCCR2-5D had a higher level of LUC/REN activity (Fig. 5J) and a comparable LUC/REN mRNA level (Fig. 5K), showing that the uORF enhances the translation of the TaCCR2-5D mORF. In addition, we analyzed the correlation between uORF TEs and mORF TEs and observed an overall mildly positive correlation between uORF TEs and mORF TEs, rather than an anticorrelation (Fig. 5L), which was also observed in a previous study in human hearts (van Heesch et al. 2019). These findings suggest that some uORFs can enhance the translation of mORFs, but through an unknown mechanism.
Combinatorial regulation of mRNA translation by uORFs, dORFs, and miRNAs
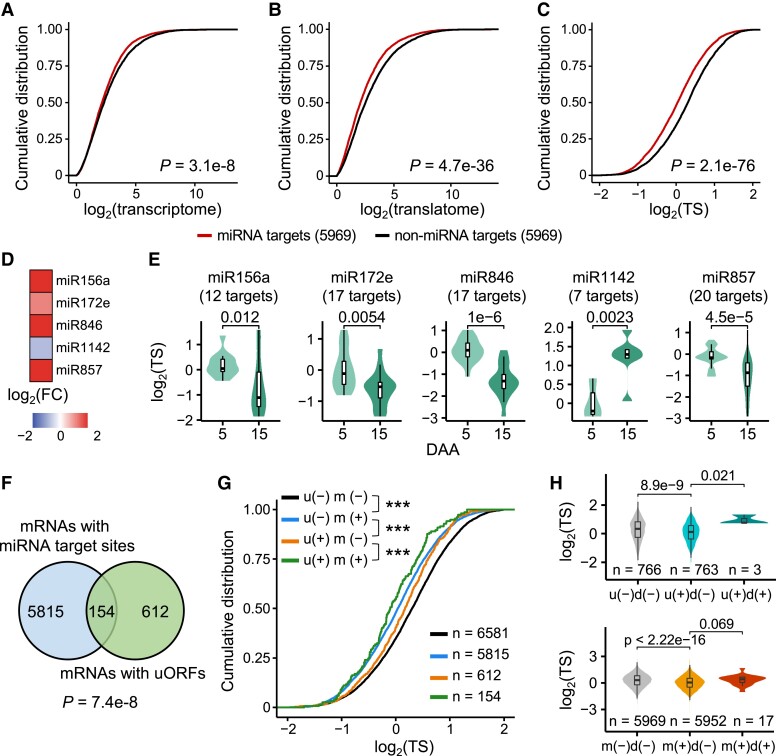
MicroRNAs (miRNAs) can pair with target sites to downregulate gene expression by degrading the resulting mRNAs or suppressing their translation (Axtell 2013; Rogers and Chen 2013; Axtell and Meyers 2018; Song et al. 2019); the effects of miRNAs on gene translation in crops require further study. To determine the effect of miRNAs on gene expression in wheat, we first predicted the miRNA-mediated translational regulation relationships on a genome-wide scale and obtained a set of 14,441 putative miRNA targets. The TSs based on the RNA-seq and Poly-seq data were used here. The miRNA targets have lower transcriptional expression levels than the mRNAs without miRNA target sites (Fig. 6A). Consistent with the previous report that miRNAs negatively regulate gene expression through translational repression, more drastic differences were observed when we analyzed the transcript abundance of miRNA targets at the translational level (Fig. 6, B and C). The mRNAs with miRNA target sites had lower levels of translational expression (Fig. 6B) and lower TSs (Fig. 6C), highlighting that miRNAs might downregulate gene expression by suppressing translation. In addition, we obtained a public small RNA sequencing data set of 5- and 15-DAA grains (Meng et al. 2013) to investigate miRNA expression levels. We analyzed several examples of miRNAs whose expression is associated with reduced translation of their downstream targets (Fig. 6, D and E). The miRNAs, including miR156, miR172, miR846, miR1142, and miR857, have changed expression levels during grain development (Fig. 6D). A further analysis shows that the patterns of abundance change of the miRNAs were opposite to those of the TSs of their predicted targets, suggesting that these miRNAs can repress the translation of the predicted targets (Fig. 6E). In addition, we analyzed the subgenome variations in miRNA target sites and found that the dynamic triad group (23.3%) has a slightly higher proportion (Chi-squared test P = 0.10; see Materials and methods) of miRNA target site PAVs than the stable triad group (19.5%).
Figure 6.
Combinatorial regulation of gene expression at the translational level by upstream open reading frames (uORFs), downstream ORFs (dORFs), and microRNAs (miRNAs). A–C) Cumulative distributions of the transcriptional level (A), translational level (B), and translation state (TS) (C) of the miRNA target genes and non-miRNA target genes. The different colored lines represent mRNAs with or without miRNA target sites. Two-sided Wilcoxon tests were performed. Only expressed genes with a FPKM > 1 were retained. A random set of the gene without miRNA target sites was used as a control. D) The fold changes in expression levels of several miRNAs during grain development. The comparison of 15-DAA grains to 5-DAA grains was shown. E) The TS distributions of the targets of several miRNAs. The predicted targets with miRNA-mediated translation repression were shown. Two-sided Wilcoxon tests were performed. F) A Venn diagram illustrates the cooccurrence of miRNAs and uORFs in gene models. A hypergeometric test was performed. G) The TS distributions of the “u−m−,” “u−m+,” “u+m−,” and “u+m+” mRNAs. “u−,” genes without an uORF; “u+,” genes with an uORF; “m−,” non-miRNA target genes; “m+,” miRNA target genes. A random set of the gene without uORFs or miRNA target sites was used as a control. Two-sided Wilcoxon tests were performed. ***P < 0.001. H) dORFs enhance the TSs of mRNAs with uORFs or microRNA target sites. “d−”, genes without a dORF. “d+”, genes with a dORF. The random set of the gene without specific features was used as a control.
Given that both miRNAs and uORFs can downregulate gene expression at the translational level, we investigated the locations of the miRNA target sites and uORFs and found a high cooccurrence ratio in mRNA (Fig. 6F), suggesting that uORFs and miRNAs tend to coregulate shared mRNAs. To further examine the combinatorial effect of miRNAs and uORFs on gene expression at the translational level, we divided genes into 4 groups: “u−m−,” “u+m−,” “u−m+,” and “u+m+,” by the presence or absence of uORFs (u+ or u−, respectively) or miRNA target sites (m+ or m−, respectively). In comparison with the other mRNA sets, the “u−m−” mRNAs had higher TSs, as expected (Fig. 6G). Compared with the TSs of the “u−m−” mRNAs, both the “u+m−” mRNAs and the “u−m+” mRNAs had a reduced TS. The “u+m+” mRNAs had the lowest TSs.
We found that dORFs can also enhance the translation of the mRNAs (Supplemental Fig. S17), and thus, we hypothesized that dORFs may weaken the translation inhibition by uORFs or miRNAs. We investigated the location of the dORFs but did not observe a significant cooccurrence between the dORFs and the miRNAs or uORFs (Supplemental Fig. S18). In comparison with the TS of the “u+d−” mRNAs and the “m+d−” mRNAs, the “u+d+” mRNAs and the “m+d+” mRNAs had higher TSs (Fig. 6H). These observations reveal that uORFs and miRNAs may tend to combinatorially repress gene translation, while the presence of dORFs may enhance the translation of mRNAs to some extent.
Discussion
Translational regulation can modulate gene expression during plant growth and environmental adaptation. Many researchers have studied gene regulation at the transcriptional level in crop grains (Pfeifer et al. 2014; Xiang et al. 2019), while a global understanding of the control of translation is still lacking. Multiple approaches, such as Ribo-seq and Poly-seq, have been developed for translatome investigation (Ingolia 2016; Zhao et al. 2019). Here, we performed Ribo-seq, Poly-seq, and RNA-seq to examine the transcriptome and translatome of developing grains in the important food crop wheat.
The 3 highly similar subgenomes of bread wheat mean that the short Ribo-seq reads may be incorrectly mapped; however, we combined Ribo-seq and Poly-seq reads to maximize the power of our translatome investigation by examining both the ORF positions and expression levels. We used Poly-seq reads (paired-end 150-bp reads) to perform gene expression quantification and translation analysis and used the Ribo-seq reads mainly to annotate new translation events and their genome positions. We also observed that most of the DEGs in the Ribo-seq data sets could be detected in RNA-seq or Poly-seq data sets, and we further experimentally validated the Ribo-seq-based expression patterns of several uORFs. The significant 3-nucleotide periodicity further verified the quality of the translatome data set. The present translatome resource represents a comprehensive translation investigation of developing crop grains.
Multiple wheat genome assemblies are available (International Wheat Genome Sequencing Consortium et al. 2018; Walkowiak et al. 2020), but the current gene annotations in assemblies are generally automatically produced by universal pipelines and result in many unannotated and mis-annotated events. The translated ORFs are also yet to be investigated in wheat. Using high-quality translatome data sets, we unveiled a large number of previously unannotated translation events, including uORFs, dORFs, iORFs, and lORFs (i.e. ORFs in lncRNAs), and provided a resource for the validation of aORFs in CDSs. We found that 21.6% of unannotated ORFs use non-AUG start codons, which can be further investigated using translation inhibitors and translation factor mutants (Kearse and Wilusz 2017). Similarly, we found that uORFs with a non-AUG start codon have lower TE. We revealed that many sORFs have stage-specific expression patterns and possess the predicted signal and mitochondrial transit peptides, suggesting that these sORFs may play important roles during grain development.
However, several limitations are also present when exploring Ribo-seq data in polyploid wheat. Accurately mapping short Ribo-seq reads to the genome has always been, and continues to be, a challenge for the polyploid plant community. Putative ORFs predicted by Ribo-seq data require further independent experimental validation. Some uORFs and dORFs in the wheat genome remain unidentified because about 39% of the high-confidence genes still lack annotated UTRs, and the current tools for translated ORF detection, including RiboTaper (Calviello et al. 2016) and RiboCode (Xiao et al. 2018), are based on annotated transcript information. Future genomic studies should therefore focus on improving genome annotation.
Our study provides a valuable opportunity to investigate translational regulation during grain development. We found that the expression of some genes is stage specific and regulated at the translational level during wheat grain development. Many well-known genes, such as TabZIP53, TaGIF1, TaGW2, TaGS5, TaSPA, TaABI19, TaSus2, TaSus1, and TaNF-YB9, have an altered TS throughout the developmental process. We found that the bZIP53 CPuORF is conserved in angiosperms and is homologous to the uORFs of S1 class bZIPs. The S1 class bZIPs control amino acid and sugar metabolism (Rahmani et al. 2009; van der Horst et al. 2020). Increasing sucrose levels induces ribosome stalling near the stop codon of the CPuORF, thereby inhibiting bZIP11 protein production (Wiese et al. 2004; van der Horst et al. 2020). We also performed a phylogenetic analysis for 260 wheat bZIPs and found that several homologs of TabZIP53-5A have uORFs and that uORFs of 3 S1-group bZIPs are conserved in the 3 wheat subgenomes (Supplemental Fig. S19). The absence of reads on the TabZIP53-5A mORF suggested that the sucrose content was high in developing wheat grains.
Interestingly, our study revealed that, contrary to expectations, some uORFs are positively associated with gene translation in wheat, which has also been shown in human hearts (van Heesch et al. 2019). We further used a dual-luciferase system to verify this finding. Furthermore, the translation of genes may be combinatorially repressed by uORFs and miRNAs, although the presence of dORFs may weaken this inhibition. Notably, even good prediction tools are not foolproof, as miRNA-target complementarity, underpinned by central matches, is not the sole dictator of the mRNA silencing outcome (Li et al. 2014). Overall, these observations reveal that extensive translational regulation increases the flexibility of gene expression during the grain developmental process.
An allopolyploid genome necessitates the orchestration of complex intergenomic gene expression across multiple aspects, from transcription to protein production (Coate et al. 2014; Pfeifer et al. 2014; Yang et al. 2021b). A previous study produced a unique polysome profiling data set of tetraploid wheat seedlings and revealed that the RNA structure can modulate translation asymmetry between subgenomes of tetraploid wheat (Yang et al. 2021b). Allohexaploid wheat (Triticum aestivum, BBAADD) has undergone 2 rounds of polyploidization and has a nonbalanced expression between homoeologs at the transcriptional level (Ramirez-Gonzalez et al. 2018). Our study provides a comprehensive resource for investigating the diverse translational regulations between the allohexaploid wheat subgenomes. We found that many homoeolog triads have unbalanced TSs across the subgenomes, and their unbalanced translation regulation is stage specific. We also found that the triads with unbalanced transcription tend to have unbalanced translational regulation. One possible explanation is the presence of variable variant sites between the subgenomes. The presence of unbalanced TSs between the homoeologs of many well-known genes suggests that the unbalanced translational regulation between homoeologs is a crucial step in modulating gene expression during grain development. The analyses of the variations in uORFs, miRNA target sites, sequence length, GC content, and NMFE among homoeologs have shed light on the possible mechanisms causing unbalanced translational regulation. These observations provide insights into the unbalanced gene expression between subgenomes.
Our research used Ribo-seq, Poly-seq, and RNA-seq to create a valuable resource for the study of translational regulation and actively translated ORFs in developing wheat grains. These findings also explain the expression divergence between subgenomes at the post-transcriptional level. Overall, our study highlights the importance of translational regulation in modulating gene expression during grain development in crops.
Materials and methods
Plant materials and growth conditions
Triticum aestivum cv. Chinese Spring was utilized in this investigation. The sterilized seeds were soaked in water for 3 d in the dark at 4°C, after which the seedlings were grown in a growth chamber at 22°C/18°C (day/night), 16 h light/8 h dark, with a light intensity of 3,000 lux (Master GreenPower CG T 400W E40; Philips), and 60% relative humidity. The grain samples were sampled at 5, 10, and 15 d after anthesis (DAA) and were harvested from different plants grown in the growth chamber. For each sample, developing seeds from 5 different plants were mixed to represent one biological replicates, and at least 2 or 3 biological replicates were used. All samples were immediately frozen in liquid nitrogen and stored at −80°C until required.
Ribosome profiling assays
After the grains at the 3 phases of development were pulverized into fine powder in liquid nitrogen, 0.3 g of the ground grains was lysed in 2 mL extraction buffer containing 50 mM Tris–HCl (pH 7.5), 200 mM KCl, 15 mM MgCl2, 1% (v/v) Triton X-100, 20 mM β-mercaptoethanol, 1 U/μL DNase I, and 50 mg mL−1 cycloheximide. After 10 min in an ice bath, the cell debris was removed by centrifugation at 20,000 × g for 10 min at 4°C. The lysates with an A260 value of 2 were used to digest at the condition of room temperature for 2 h with adding 15 μL RNase I (Invitrogen, cat. no. AM2294) and then stopped by adding 20 μL RNase inhibitor (Invitrogen, cat. no. AM2694). The sample was immediately transferred into a MicroSpin S-400 column to enrich the RNA-ribosome complex (monosomes). We then utilized an improved hybridization capture technology to remove virtually all the cytoplasmic (nuclear-encoded) and chloroplast ribosomal RNA (rRNA) using Ribo-Zero Magnetic Kit (MRZSR116; Madison, WI, USA).
Preparation of total and polysomal RNA
The ground grains (0.2 g) were lysed in 0.4 mL extraction buffer containing 200 mM Tris–HCl (pH 9.0), 200 mM KCl, 35 mM MgCl2, 2 mM EGTA, 1% (v/v) Triton X-100, 1% (v/v) Tween 20, 2% (v/v) polyoxyethylene, 2.5 mg heparin, 5 mM DTT, 100 μg mL−1 chloramphenicol, and 100 μg mL−1 cycloheximide. After 10 min in an ice bath, the cell debris was removed by centrifugation at 13,200 × g for 15 min at 4°C. The supernatants were centrifuged for 2 h at 4°C at 41,000 rpm (SW55Ti rotor in a Beckman L-100XP ultracentrifuge) in a density gradient of 15% to 60% sucrose. Gradient Profiler (BioComp Instruments) was used to separate the various sucrose components after a speed reduction without braking and to measure their optical densities at 254 nm. Fourteen 350-mL fractions were collected automatically and used for the subsequent extraction of polysome-bound RNA. For polysome-RNA isolation, the fractions marked as “polysome” in Fig. 1, D were pooled and then extracted using TRIzol Reagent (Thermo Fisher Scientific, Waltham, MA, USA). Total RNA was extracted from grains using TRIzol Reagent. We then used Hieff NGS MaxUp II Dual-Mode mRNA Library Prep Kit for Illumina (cat no.12300; Yeasen, Shanghai, China) which used beads coupling of Oligo (dT) to isolate intact poly (A)+ RNA from previously isolated total RNA.
Library construction and sequencing
The total and polysomal mRNA samples were used to prepare RNA-seq libraries using the Illumina standard mRNA-seq library preparation kit, following the manufacturer's protocol. The RNA-seq and Poly-seq libraries were sequenced to generate 150 bp paired-end reads on the Illumina NovaSeq 6000 platform. Ribosome-protected mRNA was used to prepare a library with the NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs, Ipswich, MA, USA) in accordance with the manufacturer's instructions. After PCR amplification, the samples were used for quality control and deep sequencing with an Illumina NovaSeq 6000 platform.
Reads mapping and processing
All RNA-seq and Poly-seq reads were processed and filtered using fastp (Chen et al. 2018), with the default parameters. The high-quality reads were mapped to the bread wheat Chinese Spring reference genome IWGSC RefSeq v2.1 (International Wheat Genome Sequencing Consortium et al. 2018; Zhu et al. 2021) using STAR (Dobin et al. 2013). Only uniquely mapped reads were used for the subsequent analysis. In addition, all high-quality Ribo-seq reads with a length of 18 to 40 nucleotides were retained. The rRNA, tRNA, and snoRNA sequences in wheat were downloaded from Ensembl Plants (Cunningham et al. 2022), and the remaining Ribo-seq reads were aligned against these sequences using bowtie2 (Langmead and Salzberg 2012) to produce the unaligned reads. The unaligned reads were mapped to the wheat genome using STAR, allowing up to 2 mismatches and a maximum of 50 multimapping positions. One alignment was randomly selected for the multimapping reads over all possible best alignments, as described in a previous study (Hsu et al. 2016). These options can improve the read count for individual transcripts (Hsu et al. 2016). FeatureCounts (Liao et al. 2014) with the default parameters was used to count the reads mapped to each feature. Expression quantification was followed by read normalization and size factor estimation using DESeq2 (Love et al. 2014). The FPKM values were used to measure the expression levels of the genes.
The RF length distribution, 3-nucleotide periodicity, and metagene analysis were performed in R. For the 3-nucleotide periodicity analysis, each read was assigned a relative distance to the start codon and end codon, and the cumulative read depth at each relative distance was obtained. The relative phasing of each site was calculated according to the previously described method (Lei et al. 2015).
To avoid bias from low-expression gene models, only those gene models with a mean FPKM > 1 were considered to be expressed genes. The expression levels from the biological replicates were combined. The Pearson correlation coefficients between samples were calculated using the cor.test function in R. A PCA analysis was performed using the prcomp function in R and visualized using ggplot2. The normalized expression values (log2 (FPKM + 1)) were used for the correlation analysis, and the Z-scores were used for the PCA analysis.
Identification of wheat lncRNAs
Wheat grains from multiple developmental stages were pooled to construct strand-specific RNA-seq libraries using dUTP-based methods. The libraries were sequenced to generate 150-bp paired-end reads on the Illumina NovaSeq platform. A strict identification pipeline was used to discover lncRNAs in wheat, as previously described (Zhao et al. 2018). For each biological replicate, the transcripts were first assembled using StringTie (Pertea et al. 2015). StringTie merge was utilized to merge the assembled transcripts from all replicates into a final, unified set of transcripts. Only the transcripts with class codes “u,” “x,” and “i” from Cuffcompare (Trapnell et al. 2012) were retained. The transcripts with an FPKM > 1 and a length of more than 200 bp were used for further analysis. Taking the transcripts as the query sequence and the Swiss-Prot protein sequences as the database, the transcripts with protein-coding potential were first removed using the BLASTX (Altschul et al. 1990) program with an E value cutoff of 1e−3. The coding-potential calculator (Kong et al. 2007) was further utilized to remove the transcripts with protein-coding potential. The retained transcripts were considered potential lncRNAs.
Differential expression analysis
To perform a DEG analysis, the nonnormalized read counts were used as inputs for the R package DESeq2 (Love et al. 2014). The genes with a false discovery rate (FDR) of <0.01 and a fold change >1 were considered to be DEGs. In addition, the nonnormalized read counts were used as inputs for the software RiboDiff (Zhong et al. 2017) to perform a differential TS analysis. The genes with an FDR <0.05 and a fold change > 1 were considered to have differential TSs. The expression levels of all genes across samples were scaled to the Z-scores. The R function kmeans was then used to generate coexpression clusters. The R package pheatmap was used for the visualization.
Functional enrichment analysis
The function descriptions and homologs of genes were acquired from Triticeae-GeneTribe (TGT) (Chen et al. 2020b). The GO enrichment analysis was performed using TGT, and terms with an FDR <0.05 were considered significant. All expressed genes were used as a background gene set. In addition, the grain development gene enrichment analysis was based on the hypergeometric test. The genes associated with grain development were acquired from the funRiceGenes database (Yao et al. 2018). The functional genes were also acquired from the WheatOmics database (Ma et al. 2021).
Sequence feature analysis
For the sequence feature analysis, the transcript with the longest coding sequence was used to represent a gene. The genes were grouped according to their TS, resulting in 3 groups: “high TS” (TS > 2), “middle TS” (0.5 < TS < 2), and “low TS” (TS < 0.5). The sequence length and GC content calculations were performed using in-house scripts. RNAfold was used to calculate the NMFE, representing the sequence stability of the secondary structure. We obtained 2 triad groups, namely, the stable triad group with balanced TS and the dynamic triad group with unbalanced TS. CVs of sequence features among homoeologs were calculated according to the formula and represent the extent of sequence variation.
Detection and analysis of actively translated ORFs
The wheat reference genome IWGSC RefSeq v2.1 and the identified lncRNAs were combined to facilitate the identification of the genome-wide unannotated translated ORFs. For the ORF discovery analysis, the offset parameters (i.e. the inferred P-site position for each footprint length) were first obtained through a metagene analysis. For the 25-, 26-, 27-, and 28-nucleotide footprints, the inferred offset values were 9, 10, 11, and 12, respectively. The merged bam files were utilized for ORF discovery using RiboCode (Xiao et al. 2018) and the defined offset parameters. The alternative start codons, such as CTG, GTG, and TTG, were used. The minimum size was set at 8 amino acids. The uORFs or dORFs that overlapped with annotated CDSs were discarded, and the remaining ORFs were further classified into 5 types: uORFs, dORFs, aORFs, iORFs, and lORFs. The ORFs with fewer than 300 nucleotides were considered to be sORFs, potentially encoding small peptides. The TargetP software (Almagro Armenteros et al. 2019) was used to determine the subcellular location of proteins with the plant model.
Translation efficiency analysis
Because allohexaploid wheat has 3 similar subgenomes, we combined Poly-seq and Ribo-seq to investigate the translational landscape. To reduce the errors from short reads mapping, we mainly used the Poly-seq and RNA-seq data to calculate TSs of mRNAs using the formula FPKMPoly-seq/FPKMRNA-seq. To improve the accuracy of the analyses, genes with an FPKM > 1 were retained for further analysis. In addition, we used the Ribo-seq data to investigate TEs of unannotated sORFs because Ribo-seq provides unique opportunities for investigating the translation of sORFs. The formula is FPKMRibo-seq/FPKMRNA-seq. The Spearman correlation coefficient between the TEs of the uORFs and CDSs was calculated using the R function cor.
Evolutionary analysis of uORFs
Genome assemblies and annotations of Triticum aestivum, Hordeum vulgare, Brachypodium distachyon, Oryza sativa, Zea mays, Solanum lycopersicum, Vitis vinifera, Glycine max, and Arabidopsis thaliana were obtained from Ensembl Plants (http://plants.ensembl.org/) and TGT. TransDecoder was used to predict all potential ORFs with a minimum length of 8 amino acids. We then searched for homologous sequences of uORFs using BLASTP with all the predicted ORFs in the other 8 genomes as the database. Results were filtered to retain only the best hit (an identity of >50% and an e-value of <0.01). Only the complete ORFs from start to stop codon were retained.
Phylogenetic analysis
To perform the phylogenetic analysis of wheat bZIPs, we first identified potential bZIPs using PlantTFdb (Jin et al. 2017) and iTAK (Zheng et al. 2016). The intersections between the genes identified by PlantTFdb and those by iTAK were considered the high-confidence bZIP genes. The transcript with the longest coding sequence is used to represent a gene. The protein sequences were acquired from the IWGSC RefSeq v2.1 annotation and were then aligned using MUSCLE (Edgar 2004). trimAL (Capella-Gutierrez et al. 2009) was further used to remove poor aligned regions. The phylogenetic tree was constructed in MEGA-X (Kumar et al. 2018) using the neighbor-joining method with 10,000 bootstrap replicates. The evolutionary distances were computed using the Poisson correction method. The tree file can be found at https://chenym1.github.io/data_resources/.
Translation state bias analysis across homoeolog triads
Using a previously reported method as a reference (Ramirez-Gonzalez et al. 2018), the relative TS and homoeolog TS bias categories were defined. The homoeolog triads were acquired from the TGT database. To avoid bias from low-expression gene models, only the genes with an FPKM sum > 2 and a transcription FPKM minimum above 0.5 were retained. The relative TS (RTS) was calculated as follows:
| (1) |
where A, B, and D represent the genes corresponding to the A, B, and D homoeologs in the triad. The relative TS of each homoeolog per triad was visualized using the R package ggtern (Hamilton and Ferry 2018).
Identification and analysis of miRNA targets
The expressed miRNAs in wheat grains were acquired from the previous studies (Meng et al. 2013; Sun et al. 2014; Li et al. 2015). The expression levels of miRNAs in the 5-DAA and 15-DAA grains were acquired from a public sRNA sequencing data set (Meng et al. 2013). In addition, psRNATarget (Dai et al. 2018), a widely used software for predicting miRNA targets in plants, was used to identify potential miRNA targets using the miRNA sequences and the IWGSC RefSeq v2.1 annotation as the input. Scoring schema V2 (2017) in psRNATarget was used, and the central mismatch range of 10 to 11 nucleotides, leading to translational inhibition, was used to identify potential miRNA-mediated translational regulations. Potential miRNA targets with only central mismatches were used for subsequent analyses. We considered a mRNA to be translationally repressed by miRNA if it has at least one miRNA target site causing translation repression. Chi-squared test was used to determine the significance of miRNA-induced homoeolog translation bias.
Dual-luciferase reporter system and reverse transcription quantitative PCR
The wild-type and mutant (deleted) forms of each gene's uORF sequence were cloned separately into a pGreenII0800-LUC vector and expressed in Nicotiana benthamiana leaf cells by Agrobacterium tumefaciens–mediated gene transformation (Hellens et al. 2005). About 2 d after infiltration, the activities of firefly luciferase (LUC) and the internal control Renilla luciferase (REN) were detected separately using dual-luciferase assay reagents (Promega, Madison, Wisconsin, USA) following the manufacturer's instructions. The ratios of the LUC/REN activities and LUC/REN mRNA levels were calculated as described previously (Xu Greene et al. 2017; Xu Yuan et al. 2017; Zhang et al. 2018b). All primers used in this study are listed in Supplemental Data Set 10. Total RNA was extracted from N. benthamiana leaves using TRIzol Reagent (Thermo Fisher Scientific), and cDNAs were generated using the HiScript Q RT SuperMix (Vazyme Biotech, Nanjing, China). RT-qPCR was conducted using AceQ qPCR SYBR Green Master Mix (Vazyme Biotech) on a CFX96 real-time PCR machine (Bio-Rad Laboratories, Hercules, CA, USA). RT-qPCR was performed as technical triplicates per sample. Three biological replicates were performed and similar results were obtained; the figure shows the results of one replicate. All primers used in this study are listed in Supplemental Data Set 10. Significance was calculated by t-test. The detailed statistical information is shown in Supplemental Data Set 11.
Accession numbers
Accession numbers of the genes which are discussed in the main text are provided in Supplemental Data Set 12.
Supplementary Material
Acknowledgments
We thank Zhen Qin and Xuejun Tian for the technical support, Shuchen Deng for figure preparation, and Yutong Li for language polishing. We thank the anonymous reviewers for their useful comments and suggestions to improve the manuscript.
Contributor Information
Yiwen Guo, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Yongming Chen, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Yongfa Wang, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Xiaojia Wu, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Xiaoyu Zhang, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Weiwei Mao, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Hongjian Yu, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Kai Guo, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Jin Xu, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Liang Ma, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Weilong Guo, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Zhaorong Hu, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Mingming Xin, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Yingyin Yao, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Zhongfu Ni, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Qixin Sun, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Huiru Peng, Frontiers Science Center for Molecular Design Breeding, Key Laboratory of Crop Heterosis and Utilization, Beijing Key Laboratory of Crop Genetic Improvement, China Agricultural University, Beijing 100193, China.
Author contributions
H.P., Z.N., and Q.S. supervised and conceived this work. Y.G. performed ribosome profiling and polysome profiling experiments. Y.G., Y.W., X.W., X.Z., W.M., H.Y., K.G., J.X., and L.M. collected plant materials. Y.C. and Y.G. performed analysis. Y.C., Y.G., W.G., Z.H., M.X., Y.Y., Z.N., Q.S., and H.P. interpreted data. Y.C. and Y.G. wrote the manuscript, and H.P. revised it. All authors read and approved the final manuscript.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1 . Metagene analysis of the RF reads at different lengths near the annotated translation start and stop sites.
Supplemental Figure S2 . Read distribution across various transcript features.
Supplemental Figure S3 . Correlations of the gene expression levels between biological replicates.
Supplemental Figure S4 . Correlations of the gene expression levels between the transcriptome and translatome.
Supplemental Figure S5 . DEGs in developing wheat grains at the transcriptional and translational levels.
Supplemental Figure S6 . Comparison of the translatome data set to a public proteomic data set.
Supplemental Figure S7 . GO enrichment analysis for the genes with altered translation state.
Supplemental Figure S8 . Overlap between the triads with unbalanced transcription and the triads with unbalanced TS in the different developmental stages.
Supplemental Figure S9 . GO enrichment for the unbalanced homoeolog triads at the translational level.
Supplemental Figure S10 . Sequence features of mRNAs change their TSs.
Supplemental Figure S11 . The 3-nucleotide periodicity of the RF reads at different lengths in the identified unannotated ORFs.
Supplemental Figure S12 . Kozak sequences of the aORFs.
Supplemental Figure S13 . The function analysis and effect on mRNA translation of uORFs.
Supplemental Figure S14 . Evolutionary dynamics of uORFs across angiosperms.
Supplemental Figure S15 . RF coverage on 2 homoeolog triads with uORFs.
Supplemental Figure S16 . The uORF suppresses the translation of the TaCBL1-1B mORF.
Supplemental Figure S17 . dORFs influence the translation states of mRNAs in wheat.
Supplemental Figure S18 . miRNAs and dORFs and uORFs and dORFs overlap in the gene models
Supplemental Figure S19 . uORFs and phylogenetic analysis of wheat bZIP genes.
Supplemental Data Set 1 . Total number of reads and aligned reads per sample.
Supplemental Data Set 2 . Expression levels of genes.
Supplemental Data Set 3 . List of differentially expressed genes.
Supplemental Data Set 4 . Coexpression clusters of differentially expressed genes.
Supplemental Data Set 5 . List of differentially expressed proteins.
Supplemental Data Set 6 . List of the genes with differential translation states.
Supplemental Data Set 7 . Translation state and RNA abundance of homoeologs in wheat.
Supplemental Data Set 8 . List of unannotated ORFs encoding peptides.
Supplemental Data Set 9 . Expression levels and subcellular localizations of small ORFs.
Supplemental Data Set 10 . List of primers and base sequences of upstream ORFs.
Supplemental Data Set 11 . Results of statistical analysis.
Supplemental Data Set 12 . Symbol names of the genes mentioned in this study.
Funding
This work was supported by the National Natural Science Foundation of China (Grant no. 31971942 and 31991210).
Data availability
Sequencing data sets are available at the NCBI Sequencing Read Archive (SRA), BioProject ID PRJNA871371. Tracks for all the translatomic data are available through the genome browser (http://wheat.cau.edu.cn/wheat_grain_translatome_browser/).
References
- Almagro Armenteros JJ, Salvatore M, Emanuelsson O, Winther O, von Heijne G, Elofsson A, Nielsen H. Detecting sequence signals in targeting peptides using deep learning. Life Sci Alliance. 2019:2(5):e201900429. 10.26508/lsa.201900429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990:215(3):403–410. 10.1016/S0022-2836(05)80360-2 [DOI] [PubMed] [Google Scholar]
- Axtell MJ. Classification and comparison of small RNAs from plants. Annu Rev Plant Biol. 2013:64(1):137–159. 10.1146/annurev-arplant-050312-120043 [DOI] [PubMed] [Google Scholar]
- Axtell MJ, Meyers BC. Revisiting criteria for plant microRNA annotation in the era of big data. Plant Cell. 2018:30(2):272–284. 10.1105/tpc.17.00851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai AN, Lu XD, Li DQ, Liu JX, Liu CM. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016:26(3):384–388. 10.1038/cr.2015.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai B, Peviani A, van der Horst S, Gamm M, Snel B, Bentsink L, Hanson J. Extensive translational regulation during seed germination revealed by polysomal profiling. New Phytol. 2017:214(1):233–244. 10.1111/nph.14355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baroja-Fernandez E, Munoz FJ, Li J, Bahaji A, Almagro G, Montero M, Etxeberria E, Hidalgo M, Sesma MT, Pozueta-Romero J. Sucrose synthase activity in the sus1/sus2/sus3/sus4 Arabidopsis mutant is sufficient to support normal cellulose and starch production. Proc Natl Acad Sci USA. 2012:109(1):321–326. 10.1073/pnas.1117099109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazin J, Baerenfaller K, Gosai SJ, Gregory BD, Crespi M, Bailey-Serres J. Global analysis of ribosome-associated noncoding RNAs unveils new modes of translational regulation. Proc Natl Acad Sci USA. 2017:114(46):E10018–E10027. 10.1073/pnas.1708433114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonnot T, Nagel DH. Time of the day prioritizes the pool of translating mRNAs in response to heat stress. Plant Cell. 2021:33(7):2164–2182. 10.1093/plcell/koab113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borrill P, Harrington SA, Uauy C. Applying the latest advances in genomics and phenomics for trait discovery in polyploid wheat. Plant J. 2019:97(1):56–72. 10.1111/tpj.14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brar GA, Weissman JS. Ribosome profiling reveals the what, when, where and how of protein synthesis. Nat Rev Mol Cell Biol. 2015:16(11):651–664. 10.1038/nrm4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunnik EM, Chung D-WD, Hamilton M, Ponts N, Saraf A, Prudhomme J, Florens L, Le Roch KG. Polysome profiling reveals translational control of gene expression in the human malaria parasite Plasmodium falciparum. Genome Biol. 2013:14(11):R128–R118. 10.1186/gb-2013-14-11-r128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calviello L, Mukherjee N, Wyler E, Zauber H, Hirsekorn A, Selbach M, Landthaler M, Obermayer B, Ohler U. Detecting actively translated open reading frames in ribosome profiling data. Nat Methods. 2016:13(2):165–170. 10.1038/nmeth.3688 [DOI] [PubMed] [Google Scholar]
- Calviello L, Ohler U. Beyond Read-Counts: Ribo-seq data analysis to understand the functions of the transcriptome. Trends Genet. 2017:33(10):728–744. 10.1016/j.tig.2017.08.003 [DOI] [PubMed] [Google Scholar]
- Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009:25(15):1972–1973. 10.1093/bioinformatics/btp348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Brunner AD, Cogan JZ, Nunez JK, Fields AP, Adamson B, Itzhak DN, Li JY, Mann M, Leonetti MD, et al. Pervasive functional translation of noncanonical human open reading frames. Science. 2020a:367(6482):1140–1146. 10.1126/science.aay0262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Guo Y, Guan P, Wang Y, Wang X, Wang Z, Qin Z, Ma S, Xin M, Hu Z, et al. A wheat integrative regulatory network from large-scale complementary functional datasets enables trait-associated gene discovery for crop improvement. Mol Plant. 2023:16(2):393–414. 10.1016/j.molp.2022.12.019 [DOI] [PubMed] [Google Scholar]
- Chen Y, Song W, Xie X, Wang Z, Guan P, Peng H, Jiao Y, Ni Z, Sun Q, Guo W. A collinearity-incorporating homology inference strategy for connecting emerging assemblies in the Triticeae tribe as a pilot practice in the plant pangenomic era. Mol Plant. 2020b:13(12):1694–1708. 10.1016/j.molp.2020.09.019 [DOI] [PubMed] [Google Scholar]
- Chen SF, Zhou YQ, Chen YR, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chotewutmontri P, Barkan A. Ribosome profiling elucidates differential gene expression in bundle sheath and mesophyll cells in maize. Plant Physiol. 2021:187(1):59–72. 10.1093/plphys/kiab272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung BYW, Balcerowicz M, Di Antonio M, Jaeger KE, Geng F, Franaszek K, Marriott P, Brierley I, Firth AE, Wigge PA. An RNA thermoswitch regulates daytime growth in Arabidopsis. Nat Plants. 2020:6(5):522–532. 10.1038/s41477-020-0633-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coate JE, Bar H, Doyle JJ. Extensive translational regulation of gene expression in an allopolyploid (Glycine dolichocarpa). Plant Cell. 2014:26(1):136–150. 10.1105/tpc.113.119966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham F, Allen JE, Allen J, Alvarez-Jarreta J, Amode MR, Armean IM, Austine-Orimoloye O, Azov AG, Barnes I, Bennett R, et al. Ensembl 2022. Nucleic Acids Res. 2022:50(D1):D988–D995. 10.1093/nar/gkab1049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daba SD, Liu X, Aryal U, Mohammadi M. A proteomic analysis of grain yield-related traits in wheat. AoB Plants. 2020:12(5):plaa042. 10.1093/aobpla/plaa042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Zhuang Z, Zhao PX. psRNATarget: a plant small RNA target analysis server (2017 release). Nucleic Acids Res. 2018:46(W1):W49–W54. 10.1093/nar/gky316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013:29(1):15–21. 10.1093/bioinformatics/bts635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004:32(5):1792–1797. 10.1093/nar/gkh340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikura U, Ezaki K, Horiguchi G, Seo M, Kanno Y, Kamiya Y, Lenhard M, Tsukaya H. Suppression of class I compensated cell enlargement by xs2 mutation is mediated by salicylic acid signaling. PLoS Genet. 2020:16(6):e1008873. 10.1371/journal.pgen.1008873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gage JL, Mali S, McLoughlin F, Khaipho-Burch M, Monier B, Bailey-Serres J, Vierstra RD, Buckler ES. Variation in upstream open reading frames contributes to allelic diversity in maize protein abundance. Proc Natl Acad Sci USA. 2022:119(14):e2112516119. 10.1073/pnas.2112516119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gandin V, Masvidal L, Hulea L, Gravel SP, Cargnello M, McLaughlan S, Cai Y, Balanathan P, Morita M, Rajakumar A, et al. nanoCAGE reveals 5′ UTR features that define specific modes of translation of functionally related MTOR-sensitive mRNAs. Genome Res. 2016:26(5):636–648. 10.1101/gr.197566.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover NM, Redestig H, Dessimoz C. Homoeologs: what are they and how do we infer them? Trends Plant Sci. 2016:21(7):609–621. 10.1016/j.tplants.2016.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Z, Cao H, Zhao J, Bai S, Peng W, Li J, Sun L, Chen L, Lin Z, Shi C, et al. A natural uORF variant confers phosphorus acquisition diversity in soybean. Nat Commun. 2022:13(1):3796. 10.1038/s41467-022-31555-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton NE, Ferry M. ggtern: ternary diagrams using ggplot2. J Stat Softw. 2018:87(Code Snippet 3):1–17. 10.18637/jss.v087.c03 [DOI] [Google Scholar]
- He F, Wang W, Rutter WB, Jordan KW, Ren J, Taagen E, DeWitt N, Sehgal D, Sukumaran S, Dreisigacker S, et al. Genomic variants affecting homoeologous gene expression dosage contribute to agronomic trait variation in allopolyploid wheat. Nat Commun. 2022:13(1):826. 10.1038/s41467-022-28453-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005:1(1):13. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hickey LT, Hafeez AN, Robinson H, Jackson SA, Leal-Bertioli SCM, Tester M, Gao C, Godwin ID, Hayes BJ, Wulff BBH. Breeding crops to feed 10 billion. Nat Biotechnol. 2019:37(7):744–754. 10.1038/s41587-019-0152-9 [DOI] [PubMed] [Google Scholar]
- Hsu PY, Calviello L, Wu HL, Li FW, Rothfels CJ, Ohler U, Benfey PN. Super-resolution ribosome profiling reveals unannotated translation events in Arabidopsis. Proc Natl Acad Sci USA. 2016:113(45):E7126–E7135. 10.1073/pnas.1614788113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu J, Wang Y, Fang Y, Zeng L, Xu J, Yu H, Shi Z, Pan J, Zhang D, Kang S, et al. A rare allele of GS2 enhances grain size and grain yield in rice. Mol Plant. 2015:8(10):1455–1465. 10.1016/j.molp.2015.07.002 [DOI] [PubMed] [Google Scholar]
- Ingolia NT. Ribosome footprint profiling of translation throughout the genome. Cell. 2016:165(1):22–33. 10.1016/j.cell.2016.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009:324(5924):218–223. 10.1126/science.1168978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Wheat Genome Sequencing Consortium, Appels R, Eversole K, Stein N, Feuillet C, Keller B, Rogers J, Pozniak CJ, Choulet F, Distelfeld A, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science. 2018:361(6403):eaar7191. 10.1126/science.aar7191 [DOI] [PubMed] [Google Scholar]
- Jin J, Tian F, Yang DC, Meng YQ, Kong L, Luo J, Gao G. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017:45(D1):D1040–D1045. 10.1093/nar/gkw982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juntawong P, Girke T, Bazin J, Bailey-Serres J. Translational dynamics revealed by genome-wide profiling of ribosome footprints in Arabidopsis. Proc Natl Acad Sci USA. 2014:111(1):E203–E212. 10.1073/pnas.1317811111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi R, Bailey-Serres J. mRNA sequence features that contribute to translational regulation in Arabidopsis. Nucleic Acids Res. 2005:33(3):955–965. 10.1093/nar/gki240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse MG, Wilusz JE. Non-AUG translation: a new start for protein synthesis in eukaryotes. Genes Dev. 2017:31(17):1717–1731. 10.1101/gad.305250.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007:35(suppl_2):W345–W349. 10.1093/nar/gkm391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. An analysis of 5′-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987:15(20):8125–8148. 10.1093/nar/15.20.8125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronja I, Yuan B, Eichhorn SW, Dzeyk K, Krijgsveld J, Bartel DP, Orr-Weaver TL. Widespread changes in the posttranscriptional landscape at the Drosophila oocyte-to-embryo transition. Cell Rep. 2014:7(5):1495–1508. 10.1016/j.celrep.2014.05.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. 2018:35(6):1547. 10.1093/molbev/msy096 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012:9(4):357–359. 10.1038/nmeth.1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laugerotte J, Baumann U, Sourdille P. Genetic control of compatibility in crosses between wheat and its wild or cultivated relatives. Plant Biotechnol J. 2022:20(5):812–832. 10.1111/pbi.13784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei L, Shi J, Chen J, Zhang M, Sun S, Xie S, Li X, Zeng B, Peng L, Hauck A, et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015:84(6):1206–1218. 10.1111/tpj.13073 [DOI] [PubMed] [Google Scholar]
- Li Y, Fan C, Xing Y, Jiang Y, Luo L, Sun L, Shao D, Xu C, Li X, Xiao J, et al. Natural variation in GS5 plays an important role in regulating grain size and yield in rice. Nat Genet. 2011:43(12):1266–1269. 10.1038/ng.977 [DOI] [PubMed] [Google Scholar]
- Li N, Li Y. Signaling pathways of seed size control in plants. Curr Opin Plant Biol. 2016:33:23–32. 10.1016/j.pbi.2016.05.008 [DOI] [PubMed] [Google Scholar]
- Li T, Ma L, Geng Y, Hao C, Chen X, Zhang X. Small RNA and degradome sequencing reveal complex roles of miRNAs and their targets in developing wheat grains. PLoS One. 2015:10(10):e0139658. 10.1371/journal.pone.0139658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Reichel M, Millar AA. Determinants beyond both complementarity and cleavage govern microR159 efficacy in Arabidopsis. PLoS Genet. 2014:10(3):e1004232. 10.1371/journal.pgen.1004232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang Y, Zhu W, Chen S, Qian J, Li L. Genome-wide identification and characterization of small peptides in maize. Front Plant Sci. 2021:12:695439. 10.3389/fpls.2021.695439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014:30(7):923–930. 10.1093/bioinformatics/btt656 [DOI] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, Wu SH. Translational landscape of photomorphogenic Arabidopsis. Plant Cell. 2013:25(10):3699–3710. 10.1105/tpc.113.114769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Xin M, Qin J, Peng H, Ni Z, Yao Y, Sun Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015:15(1):152. 10.1186/s12870-015-0511-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014:15(12):550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma S, Wang M, Wu J, Guo W, Chen Y, Li G, Wang Y, Shi W, Xia G, Fu D, et al. WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol Plant. 2021:14(12):1965–1968. 10.1016/j.molp.2021.10.006 [DOI] [PubMed] [Google Scholar]
- Matsushima R, Maekawa M, Kusano M, Kondo H, Fujita N, Kawagoe Y, Sakamoto W. Amyloplast-localized SUBSTANDARD STARCH GRAIN4 protein influences the size of starch grains in rice endosperm. Plant Physiol. 2014:164(2):623–636. 10.1104/pp.113.229591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng F, Liu H, Wang K, Liu L, Wang S, Zhao Y, Yin J, Li Y. Development-associated microRNAs in grains of wheat (Triticum aestivum L.). BMC Plant Biol. 2013:13(1):140. 10.1186/1471-2229-13-140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu B, Zhang Z, Zhang J, Zhou Y, Chen C. The rice LEC1-like transcription factor OsNF-YB9 interacts with SPK, an endosperm-specific sucrose synthase protein kinase, and functions in seed development. Plant J. 2021:106(5):1233–1246. 10.1111/tpj.15230 [DOI] [PubMed] [Google Scholar]
- Pandey GK, Cheong YH, Kim KN, Grant JJ, Li L, Hung W, D’Angelo C, Weinl S, Kudla J, Luan S. The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell. 2004:16(7):1912–1924. 10.1105/tpc.021311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patraquim P, Mumtaz MAS, Pueyo JI, Aspden JL, Couso JP. Developmental regulation of canonical and small ORF translation from mRNAs. Genome Biol. 2020:21(1):128. 10.1186/s13059-020-02011-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015:33(3):290–295. 10.1038/nbt.3122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeifer M, Kugler KG, Sandve SR, Zhan B, Rudi H, Hvidsten TR, International Wheat Genome Sequencing Consortium, Mayer KF, Olsen OA. Genome interplay in the grain transcriptome of hexaploid bread wheat. Science. 2014:345(6194):1250091. 10.1126/science.1250091 [DOI] [PubMed] [Google Scholar]
- Polko JK, Barnes WJ, Voiniciuc C, Doctor S, Steinwand B, Hill JL Jr, Tien M, Pauly M, Anderson CT, Kieber JJ. SHOU4 proteins regulate trafficking of cellulose synthase complexes to the plasma membrane. Curr Biol. 2018:28(19):3174–3182.e6. 10.1016/j.cub.2018.07.076 [DOI] [PubMed] [Google Scholar]
- Qi P, Lin YS, Song XJ, Shen JB, Huang W, Shan JX, Zhu MZ, Jiang L, Gao JP, Lin HX. The novel quantitative trait locus GL3.1 controls rice grain size and yield by regulating cyclin-T1; 3. Cell Res. 2012:22(12):1666–1680. 10.1038/cr.2012.151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmani F, Hummel M, Schuurmans J, Wiese-Klinkenberg A, Smeekens S, Hanson J. Sucrose control of translation mediated by an upstream open reading frame-encoded peptide. Plant Physiol. 2009:150(3):1356–1367. 10.1104/pp.109.136036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Gonzalez RH, Borrill P, Lang D, Harrington SA, Brinton J, Venturini L, Davey M, Jacobs J, van Ex F, Pasha A, et al. The transcriptional landscape of polyploid wheat. Science. 2018:361(6403):eaar6089. 10.1126/science.aar6089 [DOI] [PubMed] [Google Scholar]
- Reddy AS, Marquez Y, Kalyna M, Barta A. Complexity of the alternative splicing landscape in plants. Plant Cell. 2013:25(10):3657–3683. 10.1105/tpc.113.117523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynoso MA, Kajala K, Bajic M, West DA, Pauluzzi G, Yao AI, Hatch K, Zumstein K, Woodhouse M, Rodriguez-Medina J, et al. Evolutionary flexibility in flooding response circuitry in angiosperms. Science. 2019:365(6459):1291–1295. 10.1126/science.aax8862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers K, Chen X. Biogenesis, turnover, and mode of action of plant microRNAs. Plant Cell. 2013:25(7):2383–2399. 10.1105/tpc.113.113159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan B, Shang L, Zhang B, Hu J, Wang Y, Lin H, Zhang A, Liu C, Peng Y, Zhu L, et al. Natural variation in the promoter of TGW2 determines grain width and weight in rice. New Phytol. 2020:227(2):629–640. 10.1111/nph.16540 [DOI] [PubMed] [Google Scholar]
- Song XJ, Huang W, Shi M, Zhu MZ, Lin HX. A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat Genet. 2007:39(5):623–630. 10.1038/ng2014 [DOI] [PubMed] [Google Scholar]
- Song X, Li Y, Cao X, Qi Y. MicroRNAs and their regulatory roles in plant-environment interactions. Annu Rev Plant Biol. 2019:70(1):489–525. 10.1146/annurev-arplant-050718-100334 [DOI] [PubMed] [Google Scholar]
- Srivastava AK, Lu Y, Zinta G, Lang Z, Zhu JK. UTR-dependent control of gene expression in plants. Trends Plant Sci. 2018:23(3):248–259. 10.1016/j.tplants.2017.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark R, Grzelak M, Hadfield J. RNA Sequencing: the teenage years. Nat Rev Genet. 2019:20(11):631–656. 10.1038/s41576-019-0150-2 [DOI] [PubMed] [Google Scholar]
- Sun F, Guo G, Du J, Guo W, Peng H, Ni Z, Sun Q, Yao Y. Whole-genome discovery of miRNAs and their targets in wheat (Triticum aestivum L.). BMC Plant Biol. 2014:14(1):142. 10.1186/1471-2229-14-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahagi K, Inoue K, Mochida K. Gene co-expression network analysis suggests the existence of transcriptional modules containing a high proportion of transcriptionally differentiated homoeologs in hexaploid wheat. Front Plant Sci. 2018:9:1163. 10.3389/fpls.2018.01163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian C, Wang Y, Yu H, He J, Wang J, Shi B, Du Q, Provart NJ, Meyerowitz EM, Jiao Y. A gene expression map of shoot domains reveals regulatory mechanisms. Nat Commun. 2019:10(1):141. 10.1038/s41467-018-08083-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 2012:7(3):562–578. 10.1038/nprot.2012.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traubenik S, Reynoso MA, Hobecker K, Lancia M, Hummel M, Rosen B, Town C, Bailey-Serres J, Blanco F, Zanetti ME. Reprogramming of root cells during nitrogen-fixing symbiosis involves dynamic polysome association of coding and noncoding RNAs. Plant Cell. 2020:32(2):352–373. 10.1105/tpc.19.00647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Horst S, Filipovska T, Hanson J, Smeekens S. Metabolite control of translation by conserved peptide uORFs: the ribosome as a metabolite multisensor. Plant Physiol. 2020:182(1):110–122. 10.1104/pp.19.00940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Heesch S, Witte F, Schneider-Lunitz V, Schulz JF, Adami E, Faber AB, Kirchner M, Maatz H, Blachut S, Sandmann CL, et al. The translational landscape of the human heart. Cell. 2019:178(1):242–260.e29. 10.1016/j.cell.2019.05.010 [DOI] [PubMed] [Google Scholar]
- Walkowiak S, Gao LL, Monat C, Haberer G, Kassa MT, Brinton J, Ramirez-Gonzalez RH, Kolodziej MC, Delorean E, Thambugala D, et al. Multiple wheat genomes reveal global variation in modern breeding. Nature. 2020:588(7837):277–283. 10.1038/s41586-020-2961-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang E, Wang J, Zhu X, Hao W, Wang L, Li Q, Zhang L, He W, Lu B, Lin H, et al. Control of rice grain-filling and yield by a gene with a potential signature of domestication. Nat Genet. 2008:40(11):1370–1374. 10.1038/ng.220 [DOI] [PubMed] [Google Scholar]
- Wang L, Xie W, Chen Y, Tang W, Yang J, Ye R, Liu L, Lin Y, Xu C, Xiao J, et al. A dynamic gene expression atlas covering the entire life cycle of rice. Plant J. 2010:61(5):752–766. 10.1111/j.1365-313X.2009.04100.x [DOI] [PubMed] [Google Scholar]
- Wiese A, Elzinga N, Wobbes B, Smeekens S. A conserved upstream open reading frame mediates sucrose-induced repression of translation. Plant Cell. 2004:16(7):1717–1729. 10.1105/tpc.019349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T, Shen Y, Zheng M, Yang C, Chen Y, Feng Z, Liu X, Liu S, Chen Z, Lei C, et al. Gene SGL, encoding a kinesin-like protein with transactivation activity, is involved in grain length and plant height in rice. Plant Cell Rep. 2014:33(2):235–244. 10.1007/s00299-013-1524-0 [DOI] [PubMed] [Google Scholar]
- Wu HL, Song G, Walley JW, Hsu PY. The tomato translational landscape revealed by transcriptome assembly and ribosome profiling. Plant Physiol. 2019:181(1):367–380. 10.1104/pp.19.00541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang D, Quilichini TD, Liu Z, Gao P, Pan Y, Li Q, Nilsen KT, Venglat P, Esteban E, Pasha A, et al. The transcriptional landscape of polyploid wheats and their diploid ancestors during embryogenesis and grain development. Plant Cell. 2019:31(12):2888–2911. 10.1105/tpc.19.00397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Z, Huang R, Xing X, Chen Y, Deng H, Yang X. De novo annotation and characterization of the translatome with ribosome profiling data. Nucleic Acids Res. 2018:46(10):e61. 10.1093/nar/gky179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F, Fang J, Ou S, Gao S, Zhang F, Du L, Xiao Y, Wang H, Sun X, Chu J, et al. Variations in CYP78A13 coding region influence grain size and yield in rice. Plant Cell Environ. 2015:38(4):800–811. 10.1111/pce.12452 [DOI] [PubMed] [Google Scholar]
- Xu G, Greene GH, Yoo H, Liu L, Marques J, Motley J, Dong X. Global translational reprogramming is a fundamental layer of immune regulation in plants. Nature. 2017:545(7655):487–490. 10.1038/nature22371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu G, Yuan M, Ai C, Liu L, Zhuang E, Karapetyan S, Wang S, Dong X. uORF-mediated translation allows engineered plant disease resistance without fitness costs. Nature. 2017:545(7655):491–494. 10.1038/nature22372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Guo L, Ji C, Wang H, Wang J, Zheng X, Xiao Q, Wu Y. The B3 domain-containing transcription factor ZmABI19 coordinates expression of key factors required for maize seed development and grain filling. Plant Cell. 2021a:33(1):104–128. 10.1093/plcell/koaa008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Yu H, Sun W, Ding L, Li J, Cheema J, Ramirez-Gonzalez R, Zhao X, Martín AC, Lu F, et al. Wheat in vivo RNA structure landscape reveals a prevalent role of RNA structure in modulating translational subgenome expression asymmetry. Genome Biol. 2021b:22(1):326. 10.1186/s13059-021-02549-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao W, Li G, Yu Y, Ouyang Y. Funricegenes dataset for comprehensive understanding and application of rice functional genes. Gigascience. 2018:7(1):1–9. 10.1093/gigascience/gix119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F, Gu W, Chen J, Song N, Gao X, Zhang X, Zhou Y, Ma X, Song W, Zhao H, et al. High temporal-resolution transcriptome landscape of early maize seed development. Plant Cell. 2019:31(5):974–992. 10.1105/tpc.18.00961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H, Greene GH, Yuan M, Xu G, Burton D, Liu L, Marques J, Dong X. Translational regulation of metabolic dynamics during effector-triggered immunity. Mol Plant. 2020:13(1):88–98. 10.1016/j.molp.2019.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanetti ME, Chang IF, Gong F, Galbraith DW, Bailey-Serres J. Immunopurification of polyribosomal complexes of Arabidopsis for global analysis of gene expression. Plant Physiol. 2005:138(2):624–635. 10.1104/pp.105.059477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhan J, Thakare D, Ma C, Lloyd A, Nixon NM, Arakaki AM, Burnett WJ, Logan KO, Wang D, Wang X, et al. RNA Sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell. 2015:27(3):513–531. 10.1105/tpc.114.135657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Dou S, He F, Luo J, Wei L, Lu J. Genome-wide maps of ribosomal occupancy provide insights into adaptive evolution and regulatory roles of uORFs during Drosophila development. PLoS Biol. 2018a:16(7): e2003903. 10.1371/journal.pbio.2003903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Feng S, Chen F, Chen H, Wang J, McCall C, Xiong Y, Deng XW. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell. 2008:20(6):1437–1455. 10.1105/tpc.108.058891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Liu X, Gaikwad K, Kou X, Wang F, Tian X, Xin M, Ni Z, Sun Q, Peng H, et al. Mutations in eIF5B confer thermosensitive and pleiotropic phenotypes via translation defects in Arabidopsis thaliana. Plant Cell. 2017:29(8):1952–1969. 10.1105/tpc.16.00808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Si X, Ji X, Fan R, Liu J, Chen K, Wang D, Gao C. Genome editing of upstream open reading frames enables translational control in plants. Nat Biotechnol. 2018b:36(9):894–898. 10.1038/nbt.4202 [DOI] [PubMed] [Google Scholar]
- Zhang H, Wang Y, Wu X, Tang X, Wu C, Lu J. Determinants of genome-wide distribution and evolution of uORFs in eukaryotes. Nat Commun. 2021:12(1):1076. 10.1038/s41467-021-21394-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Xu Z, Cao S, Chen K, Li S, Liu X, Gao C, Zhang B, Zhou Y. An uncanonical CCCH-tandem zinc-finger protein represses secondary wall synthesis and controls mechanical strength in rice. Mol Plant. 2018. c:11(1):163–174. 10.1016/j.molp.2017.11.004 [DOI] [PubMed] [Google Scholar]
- Zhao X, Li J, Lian B, Gu H, Li Y, Qi Y. Global identification of Arabidopsis lncRNAs reveals the regulation of MAF4 by a natural antisense RNA. Nat Commun. 2018:9(1):5056. 10.1038/s41467-018-07500-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Qin B, Nikolay R, Spahn CMT, Zhang G. Translatomics: the global view of translation. Int J Mol Sci. 2019:20(1):212. 10.3390/ijms20010212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Jiao C, Sun H, Rosli HG, Pombo MA, Zhang P, Banf M, Dai X, Martin GB, Giovannoni JJ, et al. iTAK: a program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol Plant. 2016:9(12):1667–1670. 10.1016/j.molp.2016.09.014 [DOI] [PubMed] [Google Scholar]
- Zhong Y, Karaletsos T, Drewe P, Sreedharan VT, Kuo D, Singh K, Wendel HG, Ratsch G. RiboDiff: detecting changes of mRNA translation efficiency from ribosome footprints. Bioinformatics. 2017:33(1):139–141. 10.1093/bioinformatics/btw585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu T, Wang L, Rimbert H, Rodriguez JC, Deal KR, De Oliveira R, Choulet F, Keeble-Gagnere G, Tibbits J, Rogers J, et al. Optical maps refine the bread wheat Triticum aestivum cv. Chinese Spring genome assembly. Plant J. 2021b:107(1):303–314. 10.1111/tpj.15289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Xu J, Chen S, Chen J, Liang Y, Zhang C, Li Q, Lai J, Li L. Large-scale translatome profiling annotates the functional genome and reveals the key role of genic 3′ untranslated regions in translatomic variation in plants. Plant Commun. 2021a:2(4):100181. 10.1016/j.xplc.2021.100181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data sets are available at the NCBI Sequencing Read Archive (SRA), BioProject ID PRJNA871371. Tracks for all the translatomic data are available through the genome browser (http://wheat.cau.edu.cn/wheat_grain_translatome_browser/).