Abstract
Phosphorus (P) is a macronutrient necessary for plant growth and development. Inorganic phosphate (Pi) deficiency modulates the signaling pathway of the phytohormone jasmonate in Arabidopsis thaliana, but the underlying molecular mechanism currently remains elusive. Here, we confirmed that jasmonate signaling was enhanced under low Pi conditions, and the CORONATINE INSENSITIVE1 (COI1)-mediated pathway is critical for this process. A mechanistic investigation revealed that several JASMONATE ZIM-DOMAIN (JAZ) repressors physically interacted with the Pi signaling-related core transcription factors PHOSPHATE STARVATION RESPONSE1 (PHR1), PHR1-LIKE2 (PHL2), and PHL3. Phenotypic analyses showed that PHR1 and its homologs positively regulated jasmonate-induced anthocyanin accumulation and root growth inhibition. PHR1 stimulated the expression of several jasmonate-responsive genes, whereas JAZ proteins interfered with its transcriptional function. Furthermore, PHR1 physically associated with the basic helix–loop–helix (bHLH) transcription factors MYC2, MYC3, and MYC4. Genetic analyses and biochemical assays indicated that PHR1 and MYC2 synergistically increased the transcription of downstream jasmonate-responsive genes and enhanced the responses to jasmonate. Collectively, our study reveals the crucial regulatory roles of PHR1 in modulating jasmonate responses and provides a mechanistic understanding of how PHR1 functions together with JAZ and MYC2 to maintain the appropriate level of jasmonate signaling under conditions of Pi deficiency.
PHOSPHATE STARVATION RESPONSE1 (PHR1) functions with JASMONATE ZIM-DOMAIN (JAZ) repressors and MYC2 to maintain the appropriate level of jasmonate signaling under conditions of phosphate deficiency.
Introduction
Phosphorus (P) is a macronutrient essential for various biological processes in plants. Plants take up P from the soil as inorganic phosphate (Pi; Raghothama 1999; Nussaume et al. 2011). Although P is abundant in the soil, its effective utilization is limited by its fixation (e.g. by metals in the soil), low diffusion rate, and conversion to organic phosphate by microorganisms (Raghothama 1999; Veneklaas et al. 2012; López-Arredondo et al. 2014). In response to Pi deficiency, plants have evolved various developmental, physiological, and biochemical adaptations (Péret et al. 2011; Zhang et al. 2014; Castrillo et al. 2017). Pi deficiency-induced changes in Arabidopsis thaliana mainly include remodeling of the root system architecture (i.e. inhibition of root growth), decreased photosynthesis, and the accumulation of anthocyanins and starch (Yuan and Liu 2008; López-Arredondo et al. 2014; Crombez et al. 2019; Liu et al. 2022).
Mechanistic investigations revealed the sophisticated signaling cascade networks underlying plant responses to Pi conditions (Thibaud et al. 2010; Yang and Finnegan 2010; Wild et al. 2016; Dong et al. 2017; Puga et al. 2017; Ham et al. 2018; Sega and Pacak 2019; He et al. 2020). The SPX (SYG1/PHO81/XPR1) domain proteins act as repressors of several downstream transcription factors in mediating Pi starvation responses (Lv et al. 2014; Wang et al. 2014; Ueda et al. 2020; Yang et al. 2022). Under conditions of Pi sufficiency, SPX1 and SPX2 interact with the MYB-CC-type transcription factor PHOSPHATE STARVATION RESPONSE1 (PHR1), which prevents PHR1 from binding to and activating the promoters of PHOSPHATE STARVATION-INDUCED (PSI) genes (Wykoff et al. 1999; Rubio et al. 2001; Wang et al. 2018; He et al. 2020; Ried et al. 2021; Paz-Ares et al. 2022). Under conditions of Pi deficiency, the physical associations of SPX1 and SPX2 with PHR1 in the nucleus are diminished (Lv et al. 2014). The released PHR1 subsequently binds to the PSI promoters and upregulates their expression, thereby promoting Pi uptake and utilization by plants (Bustos et al. 2010; Lv et al. 2014). In addition, SPX proteins also associate with other critical regulators, such as the nitrate sensor NITRATE TRANSPORTER1.1B (NRT1.1B) and the key transcription factor NIN-LIKE PROTEIN3 (NLP3), to integrate Pi and nitrogen (N) signaling networks in plants (Hu et al. 2019; Hu and Chu 2020; Yang et al. 2022).
The PHR1 transcription factor and its close homolog PHR1-LIKE (PHL) are crucial activators of the Pi signaling pathway in Arabidopsis (Rubio et al. 2001; Müller et al. 2015; Sun et al. 2016; Wang et al. 2018). A loss-of-function mutation of PHR1 leads to decreases in PSI expression and cell Pi content as well as impaired anthocyanin accumulation. Moreover, the plant biomass and shoot-to-root growth ratio are significantly lower for the phr1 mutant than for the wild-type control (Rubio et al. 2001). In contrast, the overexpression of PHR1 results in the increased accumulation of Pi in cells and plant tolerance to Pi deficiency (Nilsson et al. 2007). PHR1 and its PHL homologs directly modulate the expression of PHOSPHATE1 (PHO1), PHOSPHATE TRANSPORTER1 (PHT1), PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1), and RIBONUCLEASE1 (RNS1) and regulate Pi uptake and redistribution (Martín et al. 2000; Rubio et al. 2001; Bari et al. 2006; Bustos et al. 2010). Among these genes, PHO1 is an important Pi transporter, mainly involved in the transfer of Pi from roots to shoots (Hamburger et al. 2002; Stefanovic et al. 2007; Arpat et al. 2012; Wege et al. 2016). PHT1, PHF1, and RNS1 contribute to the translocation of Pi within plant cells (Nilsson et al. 2007; Nussaume et al. 2011; Guo et al. 2015; Sun et al. 2016). PHR1 also upregulates the expression of the FLAVANONE 3-HYDROXYLASE (F3'H) and LEUCOANTHOCYANIDIN DIOXYGENASE (LDOX) genes to mediate the synthesis of anthocyanins under low Pi conditions (Liu et al. 2022).
Furthermore, PHR1 acts as a crucial node that is modulated by several key transcription factors involved in light and phytohormone signaling pathways. For example, the essential regulators of phytochrome signaling, FAR-RED ELONGATED HYPOCOTYL3 (FHY3) and ELONGATED HYPOCOTYL5 (HY5), as well as the crucial transcription factor of ethylene signaling ETHYLENE-INSENSITIVE3 (EIN3), directly regulate PHR1 expression as part of the molecular mechanisms underlying the regulatory effects of light and ethylene on Pi responses (Liu et al. 2017; Sega and Pacak 2019). The transcription factors AUXIN RESPONSE FACTOR7 (ARF7) and ARF19, which participate in the auxin signaling pathway, mediate plant root growth by binding to the PHR1 promoter and modulating its expression (Huang et al. 2018; Sega and Pacak 2019). Although there has been substantial progress in elucidating the Pi signaling network in recent years, the effects of Pi deficiency and PHR1 on endogenous phytohormone signaling and the associated molecular mechanisms remain elusive.
The phytohormone jasmonate is a fatty acid compound ubiquitous in the plant kingdom and crucial for various physiological processes, including anthocyanin accumulation, primary root development, trichome formation, male fertility, and stress responses (Chini et al. 2016; Hu et al. 2017; Guo et al. 2018; Howe et al. 2018; Zhou et al. 2019; Wasternack 2020; Cao et al. 2022; Han et al. 2023a). Jasmonate is perceived by the receptor CORONATINE INSENSITIVE1 (COI1), which is an F-box protein that interacts with Arabidopsis SKP1-like1 (ASK1) and ASK2, Cullin, and RING-BOX1 (Rbx1) to form the SCFCOI1 complex (Xie et al. 1998; Xu et al. 2002; Chini et al. 2007; Thines et al. 2007; Yan et al. 2009). When jasmonoyl–isoleucine concentrations increase in plants, JASMONATE ZIM-DOMAIN (JAZ) proteins, critical repressors of jasmonate signaling, are degraded via the SCFCOI1–26S proteasome pathway, triggering jasmonate signaling (Thines et al. 2007; Sheard et al. 2010; Yan et al. 2013; Wu et al. 2020; Li et al. 2021a; Hu et al. 2023).
The JAZ repressors negatively regulate jasmonate signaling by suppressing the functions of transcription factors from multiple families (Fernández-Calvo et al. 2011; Kazan and Manners 2013; Chini et al. 2016; Mao et al. 2017). The basic helix–loop–helix (bHLH) transcription factor MYC2 was the first JAZ-binding factor to be identified. It was subsequently revealed to target a large proportion of jasmonate-responsive genes and regulate diverse jasmonate-mediated physiological processes (Boter et al. 2004; Schweizer et al. 2013; Wang et al. 2019a; Zander et al. 2020; Zhai et al. 2020). MYC2 and its homologous MYC3 activate various jasmonate responses through a large transcription factor network (Zander et al. 2020). Additionally, JAZ repressors also mediate the crosstalk between jasmonate signaling and other phytohormone signaling pathways by targeting some essential components, such as EIN3, ABSCISIC ACID INSENSITIVE5 (ABI5), and DELLA proteins (Song et al. 2011; Zhu et al. 2011; Hu et al. 2013; Qi et al. 2014; Boter et al. 2015; Zhai et al. 2015; Mei et al. 2022). Although the jasmonate signaling network has been relatively thoroughly investigated, most studies focused on the interactions between endogenous signals. Hence, it is largely unclear whether external signals are integrated with jasmonate signaling to regulate physiological processes in plants.
Previous studies have highlighted a connection between the Pi signaling pathway and jasmonate signaling pathway (Morcuende et al. 2007; Ribot et al. 2008; Khan et al. 2016; Zhao et al. 2018; Kong et al. 2021; Pandey et al. 2021). For instance, rice (Oryza sativa) OsPHR2 directly mediates the transcription of OsJAZ11 and OsMYC2 and modulates immune defense and Pi deficiency responses, respectively (Kong et al. 2021; Pandey et al. 2021). In Arabidopsis, the synthesis and signaling of jasmonate were induced by Pi deficiency in both roots and shoots (Morcuende et al. 2007; Aparicio-Fabre et al. 2013; Wang et al. 2014; Khan et al. 2016). Consistently, Pi starvation leads to increased plant tolerance to insect herbivory (Khan et al. 2016). Nevertheless, the exact regulatory mechanisms underlying how jasmonate signaling is enhanced under low Pi conditions remain to be further elucidated.
In this study, we used molecular and genetic approaches to reveal the biological functions of PHR1 in the jasmonate signaling pathway and to clarify how Pi deficiency cooperates with endogenous jasmonate signaling to mediate physiological processes in plants. We initially confirmed that Pi deficiency activates jasmonate-related responses in Arabidopsis, including anthocyanin accumulation, decreased primary root growth, and increased expression of jasmonate-responsive genes. We also observed that the COI1-mediated pathway is critical for Pi deficiency-stimulated jasmonate signaling. Further analyses indicated that several JAZ repressors interact with PHR1, PHL2, and PHL3 in yeast and plants. Phenotypic analyses revealed that PHR1 and its PHL homologs have redundant positive effects on jasmonate-induced responses. Compared with the wild-type control, the phr1 phl2, phr1 phl3, and phl2 phl3 double mutants and the phr1 phl2 phl3 triple mutant were less sensitive to jasmonate, whereas transgenic seedlings overexpressing PHR1, PHL2, or PHL3 were more sensitive to jasmonate. PHR1 stimulates the expression of several jasmonate-responsive genes, whereas JAZ proteins interfere with the transcriptional function of PHR1. Genetic analyses indicated that the overexpression of PHR1 suppresses the reduced jasmonate sensitivity phenotype of JAZ1-accumulating plants. Additionally, PHR1 interacts with the transcription factors MYC2, MYC3, and MYC4. Furthermore, PHR1 and MYC2 function coordinately in the jasmonate signaling pathway to activate downstream target genes. Taken together, our results suggest that PHR1, the core transcription factor of Pi signaling, positively regulates jasmonate-mediated anthocyanin accumulation and root growth inhibition. The findings of this study provide a mechanistic understanding of how jasmonate signaling is enhanced under Pi-deficient conditions.
Results
Pi deficiency activates jasmonate signaling, and the COI1-mediated pathway is critical for this process
Previous studies have shown that Pi deficiency upregulates the expression of several jasmonate-responsive genes (Morcuende et al. 2007; Wang et al. 2014; Khan et al. 2016). To verify whether Pi deficiency promotes plant responses to jasmonate, we treated wild-type seedlings with methyl jasmonate (MeJA) on modified half-strength Murashige and Skoog (MS) medium containing different concentrations of Pi. The seedlings grown under Pi-sufficient conditions accumulated more anthocyanins and had shorter primary roots in the presence of MeJA compared to the mock-treated seedlings (Supplemental Fig. S1, A to C). Moreover, the MeJA-induced anthocyanin accumulation and root growth inhibition were further enhanced under Pi-deficient conditions. We also compared the changes in anthocyanin content and root length response to MeJA at different Pi concentrations, which indicated that these differences were greater under Pi-deficient conditions (Supplemental Fig. S1, D and E). These observations suggest that Pi deficiency promotes plant responses to jasmonate.
To further validate these results, we performed a reverse transcription quantitative PCR (RT-qPCR) analysis to examine the expression of several well-characterized jasmonate-responsive genes. These genes included anthocyanin synthesis-related gene LDOX and the jasmonate-induced ALLENE OXIDE SYNTHASE (AOS) and LIPOXYGENASE2 (LOX2) genes. As shown in Supplemental Fig. S1F, LDOX, AOS, and LOX2 were more highly expressed under Pi-deficient conditions than under Pi-sufficient conditions upon MeJA treatment. Together, these findings show that Pi deficiency stimulates jasmonate-induced anthocyanin accumulation and root growth inhibition.
To investigate the molecular basis underlying Pi deficiency-activated jasmonate signaling, we tested whether the crucial components of the endogenous jasmonate signaling pathway modulate these processes. The F-box protein COI1 is the receptor of jasmonate and positively regulates jasmonate responses (Liu et al. 2004; Ren et al. 2005; Yan et al. 2009, 2013; Wasternack 2020). Two leaky loss-of-function COI1 mutants, coi1-2 and coi1-16, did not differ in anthocyanin content under both Pi-sufficient and Pi-deficient conditions in the presence of MeJA, although in both conditions, the anthocyanin content of two mutants was much lower when compared to the wild-type control (Supplemental Fig. S2, A and B). Moreover, coi1-2 and coi1-6 had longer roots when compared to the wild type regardless of Pi concentrations, as in both conditions, the root length of the mutants was increased (Supplemental Fig. S2, A and C). JAZ proteins are vital repressors of jasmonate signaling (Chini et al. 2007; Thines et al. 2007; Sheard et al. 2010). jazQ quintuple mutants (lacking five JAZ repressors; Campos et al. 2016), similar to the wild-type controls, had a significant increase in anthocyanin level under Pi-deficient conditions when compared to the Pi-sufficient conditions (Supplemental Fig. S2, A and B). In both conditions, the jazQ mutants had greater anthocyanin levels than the wild-type controls (Supplemental Fig. S2, A and B). In addition, the root length of jazQ was reduced when compared to the wild type under both Pi conditions in response to MeJA (Supplemental Fig. S2, A and C). Subsequent data analyses showed that the coi1-2 and coi1-16 were less sensitive to changes in Pi conditions when comparing anthocyanin levels while the jazQ mutant remained responsive (Supplemental Fig. S2D). Unlikely, in the root length assays, all mutant and wild-type lines analyzed were responsive to changes in Pi conditions (Supplemental Fig. S2E). Nevertheless, the changes in root elongation upon MeJA treatment were less extensive in the coi1-2 and coi1-16 mutants under both Pi conditions compared with those of wild type and jazQ (Supplemental Fig. S2E). Considered together, these results suggest that the COI1-mediated endogenous pathway promotes Pi deficiency-stimulated jasmonate responses.
JAZ repressors physically interact with PHR1, PHL2, and PHL3
JAZ proteins negatively regulate jasmonate signaling by interacting with and inhibiting downstream transcription factors (Kazan and Manners 2013; Howe et al. 2018). Moreover, JAZ repressors function together with several transcriptional modulators to integrate jasmonate with other signaling pathways (Hou et al. 2010; Zhou et al. 2019; Mei et al. 2022). PHR1 is a core stimulator of Arabidopsis Pi starvation-dependent responses (Rubio et al. 2001; Bari et al. 2006; Lv et al. 2014; Wang et al. 2018). Because JAZ repressors are implicated in Pi deficiency-enhanced jasmonate signaling, we queried whether JAZ also physically associate with PHR1.
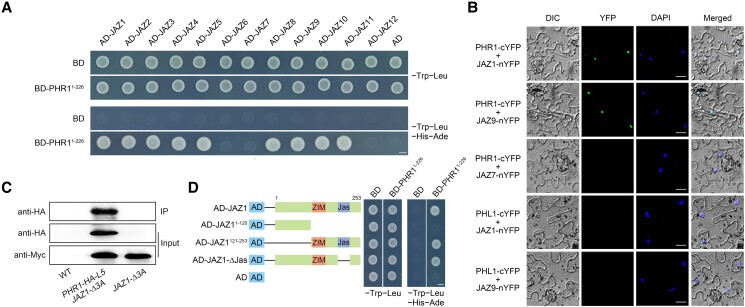
To verify this possibility, we used the yeast two-hybrid (Y2H) system to analyze the possible interactions between JAZ and PHR1. For these analyses, sequences encoding the full-length JAZ proteins were ligated with the sequence encoding the Gal4 activation domain in the prey vector (AD-JAZ), whereas the sequence encoding the N-terminal region of PHR1 (amino acids 1 to 226) was fused to the sequence encoding the Gal4 DNA-binding domain of the bait vector (BD-PHR11–226). As shown in Fig. 1A, PHR1 interacted with JAZ1, JAZ2, JAZ3, JAZ4, JAZ5, JAZ8, JAZ9, JAZ10, and JAZ11 in yeast. We also analyzed the possible physical associations between JAZ and PHL (PHL1 to PHL4). PHL2 and PHL3 were observed to interact with JAZ4, JAZ6, JAZ8, and JAZ9 in yeast (Supplemental Fig. S3). No interaction was detected between JAZ and PHL1 or PHL4 (Supplemental Fig. S3).
Figure 1.
Physical interactions of JAZ repressors with PHR1, PHL2, and PHL3. A) Y2H assay analyses. Protein interactions were indicated by the ability of cells to grow on dropout medium lacking Leu, Trp, His, and Ade and containing 20 mM 3-aminotriazole after a 2-d incubation. pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. Bars = 2.5 mm. B) BiFC analyses. The fluorescence detected in the nucleus of transformed Nicotiana benthamiana cells co-expressing JAZ1-nYFP (or JAZ9-nYFP) with PHR1-cYFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Pro35S). No signal was observed in the negative controls where PHR1-cYFP and JAZ7-nYFP or PHL1-cYFP and JAZ1-nYFP (or JAZ9-nYFP) were co-expressed. Nuclei are indicated by DAPI staining. Bars = 15 µm. C) CoIP assays. Total proteins were extracted from 8-d-old transgenic Arabidopsis seedlings simultaneously overexpressing PHR1 and JAZ1 (PHR1-HA-L5 JAZ1-Δ3A) under the control of Pro35S. 3Myc-fused JAZ1 was immunoprecipitated using an anti-Myc antibody (1:250) and the co-immunoprecipitated PHR1-HA protein was detected using an anti-HA antibody (1:10,000). Protein input for 3Myc-fused JAZ1 in the immunoprecipitated complexes was also tested and is displayed. Experiments were repeated three times, with similar results. IP, immunoprecipitation. D) Y2H assay showing that the ZIM domain of JAZ1 is responsible for the interaction with PHR1. Left: diagram of the full-length and truncated JAZ1 constructs with specific deletions. Right: interactions are indicated by the ability of yeast cells to grow on the dropout medium lacking Leu, Trp, His, and Ade and containing 20 mM 3-aminotriazole after a 2-d incubation. BD and AD were used as negative controls. Bars = 2.5 mm.
To further confirm the interactions between PHR1 and JAZ proteins, we performed bimolecular fluorescence complementation (BiFC) assays in Nicotiana benthamiana. The sequence encoding the N-terminal fragment of yellow fluorescent protein (nYFP) under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Pro35S) was fused with the sequence encoding JAZ1, JAZ7, or JAZ9 to produce JAZ1-nYFP, JAZ7-nYFP, and JAZ9-nYFP. Next, sequences encoding PHR1 and PHL1 were ligated to the sequences encoding the C-terminal fragment of YFP (cYFP) to generate PHR1-cYFP and PHL1-cYFP. When JAZ1-nYFP or JAZ9-nYFP was co-expressed with PHR1-cYFP in N. benthamiana leaves, strong YFP fluorescence was detected in the nucleus of transformed cells stained with 4′,6-diamidino-2-phenylindole (DAPI; Fig. 1B; Supplemental Fig. S4A). However, fluorescence was undetectable in the negative controls in which JAZ7-nYFP was co-expressed with PHR1-cYFP or when JAZ1-nYFP or JAZ9-nYFP was co-expressed with PHL1-cYFP (Fig. 1B; Supplemental Fig. S4A). In addition to BiFC assays, co-immunoprecipitation (CoIP) assays provided further in vivo evidence of the association between JAZ1 and PHR1 in transgenic Arabidopsis plants simultaneously overexpressing JAZ1 and PHR1 (PHR1-HA-L5 JAZ1-Δ3A). These plants were developed by introducing PHR1 overexpression into JAZ1-Δ3A plants (transgenic plants overexpressing JAZ1 with deletion of Jas-encoding domain under the control of Pro35S; Han et al. 2018; Fig. 1C). To further characterize which JAZ1 protein region is required for the interaction with PHR1, we performed a directed Y2H analysis, which indicated the regions containing the ZIM domain of JAZ1 interacted with PHR1 in yeast (Fig. 1D). Based on these experiments, we conclude that PHR1 physically interacts with several JAZ proteins.
Disruption of PHR1, PHL2, and PHL3 attenuates jasmonate-induced anthocyanin accumulation and root growth inhibition
Having demonstrated that JAZ repressors physically interact with PHR1, PHL2, and PHL3, we investigated whether these transcription factors modulate jasmonate responses. To assess this possibility, we initially analyzed the transcription of PHR1 and its close homologs (PHL1 to PHL4) in wild-type seedlings with or without MeJA treatment. Similar with JAZ1 expression (as a positive control), their expression levels increased significantly in the MeJA-treated seedlings (Supplemental Fig. S5), suggesting that jasmonate triggers the transcription of PHR1 and its close homologs. To validate whether PHR and PHL respond to jasmonate, we compared the phenotypes of the wild-type control and the loss-of-function phr1, phl2, and phl3 single mutants upon MeJA treatment on a medium with different concentrations of Pi. The anthocyanin contents and root lengths were similar between the phr1, phl2, and phl3 single mutants and the wild-type control in the presence of 30 µM MeJA (Supplemental Fig. S6). Accordingly, mutations to PHR1, PHL2, or PHL3 alone have little effect on jasmonate responses.
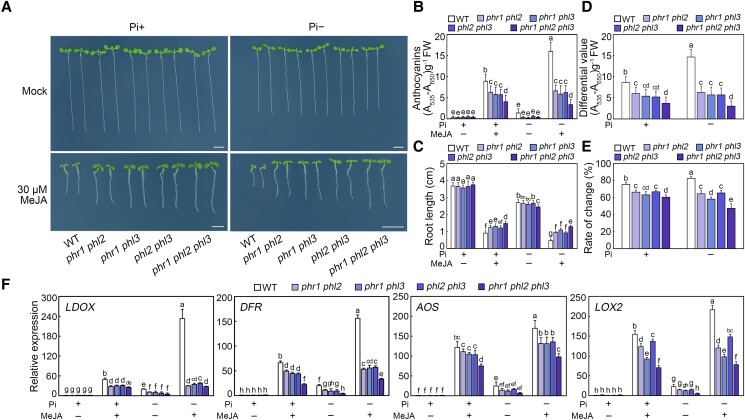
Because PHR1, PHL2, and PHL3 have redundant functions in plant responses to Pi deficiency (Rubio et al. 2001; Guo et al. 2015; Müller et al. 2015; Sun et al. 2016; Wang et al. 2018), we speculated that they may have partially overlapping roles in mediating jasmonate signaling. To test this hypothesis, we constructed phr1 phl2, phr1 phl3, and phl2 phl3 double mutants and the phr1 phl2 phl3 triple mutant through genetic crossing. Next, we sowed the seeds of these double and triple mutants on a medium with or without 30 µM MeJA for an analysis of the resulting seedlings in terms of their anthocyanin accumulation and primary root growth. In response to the MeJA treatment, the anthocyanin contents were lower in these mutants, especially the triple mutant, than in the wild-type plants under both Pi-sufficient and Pi-deficient conditions (Fig. 2, A and B). The roots of these mutants grown on 30 µM MeJA-containing medium were significantly longer than those of wild-type plants (Fig. 2, A and C); however, in the absence of MeJA, the root lengths were similar between the mutants and the wild-type control. Moreover, in the presence of MeJA, the Pi deficiency-induced changes in anthocyanin contents and root lengths were reduced in these mutants than in the wild-type seedlings (Fig. 2, D and E). To confirm these observations, we examined the expression of several jasmonate-responsive genes [i.e. LDOX, DIHYDROFLAVONOL 4-REDUCTASE (DFR), AOS, and LOX2] in double and triple mutants treated with MeJA under different Pi concentrations. The transcript levels of these genes were lower in the double mutants than in the wild-type seedlings following the MeJA treatment at different Pi conditions, but they were even lower in the triple mutant (Fig. 2F). Hence, PHR1, PHL2, and PHL3 may positively modulate jasmonate-induced anthocyanin accumulation and root growth inhibition.
Figure 2.
Mutation of PHR1, PHL2, and PHL3 attenuated jasmonate-induced anthocyanin accumulation and root growth inhibition. A) Phenotypes of 8-d-old wild-type (WT), phr1 phl2, phr1 phl3, phl2 phl3, and phr1 phl2 phl3 seedlings grown on modified half-strength Murashige and Skoog (MS) medium containing different concentrations of Pi with or without (Mock) 30 µM methyl jasmonate (MeJA). In the absence of MeJA (mock), an equal volume of 10% (v/v) ethanol was added. The media containing 0.65 mM or 1 µM KH2PO4 were used as the Pi-sufficient (Pi+) and Pi-deficient (Pi−) media, respectively. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). Bars = 5 mm. B) Anthocyanin contents in 8-d-old WT, phr1 phl2, phr1 phl3, phl2 phl3, and phr1 phl2 phl3 seedlings grown on Pi+ or Pi− media with or without 30 µM MeJA. Experiments were performed eight times with more than 100 seedlings for each sample per replicate. Data are means ± standard deviation (Sd) from eight independent experiments (n = 8 replicates). FW, fresh weight. C) Root length of 8-d-old WT, phr1 phl2, phr1 phl3, phl2 phl3, and phr1 phl2 phl3 seedlings grown on Pi+ or Pi− media with or without 30 µM MeJA. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). The root length of 20 representative seedlings was measured for each sample per replicate by using a vernier caliper. Data are means ± Sd (n = 20 representative plants). D) Anthocyanin content changes in 8-d-old WT, phr1 phl2, phr1 phl3, phl2 phl3, and phr1 phl2 phl3 seedlings induced by 30 µM MeJA. Data represent the anthocyanin content differences of seedlings grown on Pi+ or Pi− media. The differential value on the y axis represents the difference between the anthocyanin content of seedlings treated with 30 µM MeJA and that of mock-treated seedlings. The data used to calculate these difference values are from panel B. Data are means ± Sd from eight independent experiments (n = 8 times). E) Percentage of root length changes in 8-d-old WT, phr1 phl2, phr1 phl3, phl2 phl3, and phr1 phl2 phl3 seedlings induced by 30 µM MeJA. Data represent the percentages of root length changes of seedlings grown on Pi+ or Pi− media. The percentages represent the differences of root length with or without 30 µM MeJA treatment divided by root length without MeJA exposure. The data used to calculate percentage are from panel C. Data are means ± Sd (n = 20 representative plants). F) RT-qPCR analyses of LDOX, DFR, AOS, and LOX2 expression levels in WT, phr1 phl2, phr1 phl3, phl2 phl3, and phr1 phl2 phl3 seedlings. For LDOX and DFR, total RNA was extracted from 8-d-old seedlings (more than 60 seedlings for each sample per replicate) grown on Pi+ or Pi− media with or without 30 µM MeJA. For AOS and LOX2, total RNA was extracted from 8-d-old seedlings (more than 60 seedlings for each sample per replicate grown on Pi+ or Pi− media) which were treated with or without 100 μM MeJA for 6 h. Data are means ± Sd from five independent experiments (n = 5 times). Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by a two-way ANOVA using Tukey's HSD test.
In addition to the PHR1 and PHLs, we considered whether the Pi transport-related proteins PHF1, RNS1, and PHT1 also contribute to jasmonate signaling. To examine this possibility, we analyzed the deletion mutants phf1-1 and rns1 as well as the pht1;1 pht1;5 double mutant regarding their anthocyanin contents and root growth on modified half-strength MS medium with or without MeJA. There were no significant differences in the anthocyanin accumulation and root growth between these mutants and the wild-type seedlings (Supplemental Fig. S7). Thus, these Pi transport-related proteins may not participate in the modulation of jasmonate signaling in plants.
An earlier study showed that endogenous jasmonate content was elevated in wild-type seedlings under Pi-deficient conditions (Khan et al. 2016). This observation prompted us to further investigate whether PHR1 and its PHL homologs mediate the biosynthesis of jasmonate. To test this possibility, we measured jasmonate content in roots of wild-type and phr1 phl2 phl3 mutant seedlings under both Pi-sufficient and Pi-deficient conditions with or without exogenous MeJA treatment. As shown in Supplemental Fig. S8, the phr1 phl2 phl3 mutants accumulated less jasmonate in roots compared with the wild-type plants under Pi-deficient conditions without MeJA addition. However, the jasmonate levels in wild-type roots were similar with those in phr1 phl2 phl3 mutant roots when 30 μM MeJA was applied, regardless of Pi conditions (Supplemental Fig. S8). These findings suggest that PHR1 and its PHL homologs positively affect jasmonate synthesis under Pi-deficient conditions in the absence of MeJA, whereas they exert little effect on jasmonate accumulation in response to MeJA exposure.
Transgenic seedlings overexpressing PHR1, PHL2, or PHL3 exhibit enhanced jasmonate-mediated anthocyanin accumulation and root growth inhibition
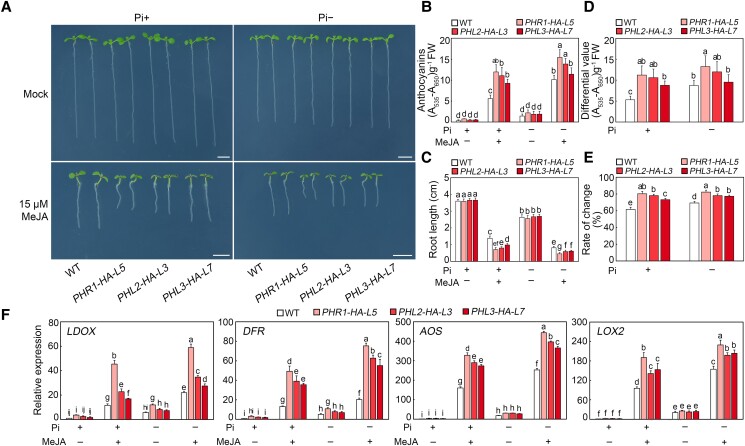
To further reveal the PHR1 role in jasmonate signaling, we generated transgenic plants overexpressing PHR1 under the control of Pro35S. The RT-qPCR analysis showed that PHR1 was most highly expressed in the PHR1-HA-L5 and PHR1-HA-L8 transgenic plants (Supplemental Fig. S9). We subsequently examined the anthocyanin accumulation and root elongation of the T5 progeny of PHR1-HA-L5 and PHR1-HA-L8 seedlings grown on modified half-strength MS medium with different concentrations of Pi upon MeJA treatment. The PHR1-HA-L5 and PHR1-HA-L8 seedlings accumulated more anthocyanins and had shorter roots than the wild-type control in the presence of MeJA under both Pi-sufficient and Pi-deficient conditions (Fig. 3, A to C; Supplemental Fig. S10, A and B). The statistical analysis of the data indicated that the PHR1-HA-L5 and PHR1-HA-L8 plants were more sensitive to MeJA than the wild-type plants in terms of the jasmonate-induced changes to anthocyanin contents and root elongation at all Pi concentrations (Fig. 3, D and E; Supplemental Fig. S10, C and D). Having demonstrated the partial functional redundancy of PHR1, PHL2, and PHL3 in the regulation of jasmonate signaling, we speculated whether the overexpression of PHL2 or PHL3 also promotes jasmonate responses. To test this possibility, we further generated PHL2- or PHL3-overexpressing transgenic plants (PHL2-HA-L3 and PHL3-HA-L7; Supplemental Fig. S9) and analyzed their phenotypes. Similar to PHR1-HA-L5 plants, PHL2-HA-L3 and PHL3-HA-L7 seedlings were also more responsive to jasmonate (Fig. 3, A to D).
Figure 3.
Overexpression of PHR1, PHL2, or PHL3 enhances jasmonate-induced anthocyanin accumulation and root growth inhibition. A) Phenotypes of 8-d-old wild-type (WT), PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 seedlings grown on modified half-strength Murashige and Skoog (MS) medium containing different concentrations of Pi with or without (Mock) 15 µM methyl jasmonate (MeJA). In the absence of MeJA (mock), an equal volume of 10% (v/v) ethanol was added. The media with 0.65 mM and 1 µM KH2PO4 were used as the Pi-sufficient (Pi+) and Pi-deficient (Pi−) media, respectively. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). Bars = 5 mm. B) Anthocyanin contents in 8-d-old WT, PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed eight times with more than 100 seedlings for each sample per replicate. Data are means ± Sd from eight independent experiments (n = 8 times). FW, fresh weight. C) Root length of 8-d-old WT, PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). The root length of 20 representative seedlings was measured for each sample per replicate. Data are means ± Sd (n = 20 representative plants). D) Anthocyanin content changes in 8-d-old WT, PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 seedlings induced by 15 µM MeJA. Data represent the anthocyanin content differences of seedlings grown on Pi+ or Pi− media. The differences mean the anthocyanins of seedlings with 15 µM MeJA treatment minus those of seedlings without MeJA treatment. The data used to calculate these difference values are from panel B. Data are means ± Sd from eight independent experiments (n = 8 times). E) Percentage of root length changes in 8-d-old WT, PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 seedlings induced by 15 µM MeJA. Data represent the percentages of root length changes of seedlings grown on Pi+ or Pi− media. The percentages mean the differences of root length with or without 15 µM MeJA treatment divided by root length without MeJA exposure. The data used to calculate percentage are from panel C. Data are means ± Sd (n = 20 representative plants). F) RT-qPCR analyses of LDOX, DFR, AOS, and LOX2 expression levels in WT, PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 seedlings. For LDOX and DFR, total RNA was extracted from 8-d-old seedlings (more than 60 seedlings for each sample per replicate) grown on Pi+ or Pi− media with or without 15 µM MeJA. For AOS and LOX2, total RNA was extracted from 8-d-old seedlings (more than 60 seedlings for each sample per replicate grown on Pi+ or Pi− media) which were treated with or without 100 μM MeJA for 6 h. Data are means ± Sd from five independent experiments (n = 5 times). Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by a two-way ANOVA using Tukey's HSD test.
To substantiate these observations, we analyzed the expression of several jasmonate-responsive genes, including LDOX, DFR, AOS, and LOX2 in PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 plants treated with MeJA at different Pi conditions. These genes were significantly more highly expressed upon MeJA treatment in the PHR1-HA-L5, PHL2-HA-L3, and PHL3-HA-L7 plants than in the wild-type plants, and their MeJA-induced expression was enhanced by Pi deficiency (Fig. 3F). Therefore, the overexpression of PHR1, PHL2, or PHL3 renders the plants more responsive to jasmonate. Overall, these findings further demonstrate that PHR1, PHL2, and PHL3 positively regulate jasmonate-induced anthocyanin accumulation and root growth inhibition.
PHR1 directly stimulates the expression of several jasmonate-responsive genes, whereas JAZ proteins interfere with the transcriptional function of PHR1
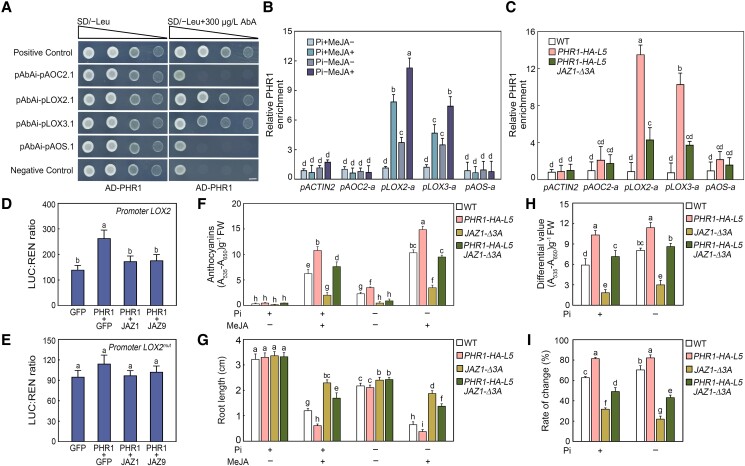
Having ascertained that PHR1 positively mediates jasmonate signaling in plants, we further analyzed whether PHR1 directly regulates the expression of downstream genes responding to jasmonate. More specifically, we initially conducted yeast one-hybrid (Y1H) analyses to assess whether PHR1 binds the promoter regions of several jasmonate-inducible genes, such as LOX2, LOX3, AOS, and ALLENE OXIDE CYCLASE2 (AOC2). Previous studies revealed that PHR1 recognizes the PHR1-binding sequence (P1BS) cis-element (GNATATNC) in the promoters of PSI genes (Rubio et al. 2001; Wu et al. 2013; Puga et al. 2014; Guo et al. 2015; Ruan et al. 2015). The putative P1BS element fragments in the LOX2, LOX3, AOS, and AOC2 promoters were cloned into the pAbAi vector to generate pAbAi-pLOX2.1, pAbAi-pLOX3.1, pAbAi-pAOS.1, and pAbAi-pAOC2.1, respectively (Fig. 4A; Supplemental Fig. S11). We inserted the full-length PHR1 coding sequence into pGADT7 to produce the AD-PHR1 construct. The results based on Y1H analyses showed that PHR1 interacted with the LOX2 and LOX3 promoter regions (pLOX2.1 and pLOX3.1) in yeast cells (Fig. 4A; Supplemental Fig. S11). To verify these findings, we performed chromatin immunoprecipitation (ChIP) assays by using PHR1-HA-L5 seedlings grown under both Pi-sufficient and Pi-deficient conditions with or without MeJA treatment. As shown in Fig. 4B, PHR1 was enriched at the LOX2 and LOX3 promoter regions (pLOX2-a and pLOX3-a; Supplemental Table S1) under Pi-deficient conditions. Moreover, the enrichment of PHR1 on the LOX2 and LOX3 promoters was increased upon MeJA treatment (Fig. 4B). These results suggested that PHR1 directly associates with the promoters of LOX2 and LOX3 in vivo under Pi-deficient conditions, and this association is responsive to jasmonate.
Figure 4.
PHR1 directly stimulates the expression of several jasmonate-responsive genes, whereas JAZ proteins interfere with the transcriptional function of PHR1. A) Yeast one-hybrid analyses of the binding of PHR1 to the LOX2 and LOX3 promoters in yeast cells. The empty triangle represents the range of yeast concentrations from the dilutions 100 (OD600 = 0.8) to 10−3. pGADT7-p53 + pAbAi-p53 was used as a positive control, and pGADT7 + pAbAi-p53 was used as a negative control. Bars = 2.5 mm. B and C) ChIP-qPCR analyses of the enrichment of PHR1 in the AOC2 (pAOC2-a), LOX2 (pLOX2-a), LOX3 (pLOX3-a), and AOS (pAOS-a) promoters. Eight-d-old PHR1-HA-L5 seedlings grown on half-strength Murashige and Skoog (MS) medium with different concentrations of Pi were treated with or without 100 µM methyl jasmonate (MeJA) for 1 h (B). The media with 0.65 mM and 1 µM KH2PO4 were used as the Pi-sufficient (Pi+) and Pi-deficient (Pi−) media, respectively. Eight-d-old wild-type (WT), PHR1-HA-L5, and PHR1-HA-L5 JAZ1-Δ3A seedlings grown on Pi− media were treated with 100 µM MeJA for 1 h (C). More than 50 seedlings for each sample were pooled for ChIP assays using an anti-HA antibody. qPCR data from the ChIP assays with the ACTIN2 untranslated region sequence (pACTIN2) as a negative control. Data are means ± Sd from three independent biological replicates. D) Transient dual-luciferase reporter assays indicating that PHR1 promotes the transcription of LOX2, whereas the JAZ repressors interfere with the transcriptional activation of LOX2 by PHR1. Data are means ± Sd from three independent biological replicates. E) Transient dual-luciferase reporter assays showing that the P1BS cis-element of the LOX2 promoter is crucial for the regulation by PHR1. The reporter construct comprised a mutant version (the putative P1BS element was mutated) of LOX2 promoter (promoter LOX2mut) fused to the LUC gene. Data are means ± Sd from three independent biological replicates. F) Anthocyanin contents in 7-d-old WT, PHR1-HA-L5, JAZ1-Δ3A, and PHR1-HA-L5 JAZ1-Δ3A seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed eight times with more than 100 seedlings for each sample per replicate. Data are means ± Sd from eight independent experiments (n = 8 times). FW, fresh weight. G) Root length of 7-d-old WT, PHR1-HA-L5, JAZ1-Δ3A, and PHR1-HA-L5 JAZ1-Δ3A seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). The root length of 20 representative seedlings was measured for each sample per replicate. Data are means ± Sd (n = 20 representative plants). H) Anthocyanin content changes in 7-d-old WT, PHR1-HA-L5, JAZ1-Δ3A, and PHR1-HA-L5 JAZ1-Δ3A seedlings induced by 15 µM MeJA. Data represent the anthocyanin content differences of seedlings grown on Pi+ or Pi− media. The differences mean the anthocyanins of seedlings with 15 µM MeJA treatment minus those of seedlings without MeJA treatment. The data used to calculate these difference values are from panel F. Data are means ± Sd from eight independent experiments (n = 8 times). I) Percentage of root length changes in 7-d-old WT, PHR1-HA-L5, JAZ1-Δ3A, and PHR1-HA-L5 JAZ1-Δ3A seedlings induced by 15 µM MeJA. Data represent the percentages of root length changes of seedlings grown on Pi+ or Pi− media. The percentages mean the differences of root length with or without 15 µM MeJA treatment divided by root length without MeJA exposure. The data used to calculate percentage are from panel G. Data are means ± Sd (n = 20 representative plants). Bars with different letters are significantly different from each other (P < 0.05). Data shown in panels D and E were analyzed by a one-way ANOVA, and others were analyzed by a two-way ANOVA using Tukey's HSD test.
Because JAZ proteins physically interact with PHR1 (Fig. 1), we investigated the modulatory effects of JAZ proteins on transcriptional functions of PHR1. To test whether JAZ1 affects the binding of PHR1 to the promoter regions of LOX2 and LOX3 (pLOX2-a and pLOX3-a), we conducted ChIP analyses by using PHR1-HA-L5 JAZ1-Δ3A plants grown under Pi-deficient conditions with MeJA treatment. As shown in Fig. 4C, ChIP assays revealed that the enrichment of PHR1 on the LOX2 and LOX3 promoters was reduced in PHR1-HA-L5 JAZ1-Δ3A compared with PHR1-HA-L5 plants. This observation suggests that JAZ proteins interfere with the binding of PHR1 to promoter regions of downstream target genes. To further analyze the regulation of JAZ proteins on PHR1, we conducted dual-luciferase (LUC) reporter assays using Arabidopsis wild-type mesophyll protoplasts (Yoo et al. 2007). The effector constructs contained PHR1, JAZ1, JAZ9, or GFP under the control of Pro35S, whereas the reporter construct comprised a native or mutant version of LOX2 promoter fused to the LUC gene (Supplemental Fig. S11). For the mutant form of LOX2 promoter (named LOX2mut promoter), the putative P1BS element was mutated. Compared with the effect of GFP alone, the co-expression of PHR1 and GFP substantially increased the expression of LUC driven by the LOX2 promoter (Fig. 4D). Additionally, the LUC expression level was lower when PHR1 and JAZ1 were co-expressed than when PHR1 and GFP were co-expressed (Fig. 4D). Similar assay results were obtained when JAZ9 was co-expressed with PHR1 (Fig. 4D). However, co-expression of PHR1 with GFP or JAZ proteins had little effect on LOX2 transcription when the P1BS element was mutated (Fig. 4E). These results suggest that the P1BS element is crucial for the regulation of PHR1 on LOX2 expression. Taken together, these results suggest that PHR1 is a transcriptional activator of the jasmonate-responsive LOX2 gene, but its function is attenuated by JAZ repressors.
To further dissect the relationship between JAZ and PHR1, we tested whether PHR1 overexpression could partially restore the phenotype of JAZ1-Δ3A plants. Similar with the coi1-2 and coi1-16 mutants, the observed decrease in anthocyanin accumulation and the increase in the primary root length suggested that JAZ1-Δ3A seedlings were less sensitive to jasmonate under both Pi-sufficient and Pi-deficient conditions (Fig. 4, F and G). Following the MeJA treatment, the PHR1-HA-L5 JAZ1-Δ3A plants were in between the PHR1-HA-L5 and JAZ1-Δ3A plants in terms of their anthocyanin content and root growth (Fig. 4, F and G). Further statistical analyses of the data indicated that the overexpression of PHR1 partially rescued the reduced MeJA sensitivity of the JAZ1-Δ3A plants (Fig. 4, H and I). Based on these findings, we concluded that PHR1 positively mediates jasmonate signaling in a process that is compromised by JAZ proteins.
MYC transcription factors physically associate with PHR1 and promote Pi deficiency-induced jasmonate signaling
The MYC transcription factors are the most extensively characterized JAZ-binding factors that positively regulate multiple jasmonate-related processes (Dombrecht et al. 2007; Chen et al. 2012; Qi et al. 2015; Liu et al. 2019; You et al. 2019). Considering that PHR1 also interacts with JAZ repressors, we wondered whether PHR1 physically associates with the MYC factors. To investigate this possibility, we constructed prey vectors to produce full-length MYC2, MYC3, or MYC4 fused to the Gal4 activation domain (AD-MYC2, AD-MYC3, and AD-MYC4). The Y2H results showed that PHR1 interacts with MYC2, MYC3, and MYC4 in yeast (Fig. 5A). We also identified the PHR1 domain required for the associations with the MYC factors. More specifically, PHR1 was divided into the N-terminal fragment (amino acid residues 1 to 226), C-terminal fragment (including the MYB and CC domains; amino acid residues 219 to 410), and the fragment containing only the CC domain (amino acid residues 293 to 410) (Fig. 5A). The assay results indicated the N-terminal region of PHR1 is essential for the interactions with MYC (Fig. 5A). To further characterize the MYC2 domain involved in the interaction, we divided MYC2 into the N-terminal fragment (including the TAD domain; amino acid residues 1 to 188), the mid-terminal fragment (amino acid residues 189 to 445), and the C-terminal fragment (including the bHLH domain; amino acid residues 396 to 924). The subsequent analysis demonstrated that the N-terminal and mid-terminal fragments of MYC2 interact with PHR1 (Fig. 5B).
Figure 5.
MYC transcription factors physically associate with PHR1. A) Mapping the PHR1-interacting domain of MYC2, MYC3, and MYC4 according to a Y2H assay. Interactions are indicated by the ability of cells to grow on dropout medium lacking Leu, Trp, His, and Ade and containing 20 mM 3-aminotriazole after a 2-d incubation. pGBKT7 (BD) and pGADT7 (AD) were used as negative controls. Bars = 2.5 mm. B) Mapping the MYC2-interacting domain of PHR1 according to a Y2H assay. Interactions are indicated by the ability of cells to grow on dropout medium lacking Leu, Trp, His, and Ade and containing 20 mM 3-aminotriazole after a 2-d incubation. BD and AD were used as negative controls. Bars = 2.5 mm. C) BiFC analyses. The fluorescence detected in the nucleus of transformed N. benthamiana cells co-expressing MYC2-nYFP (or MYC3-nYFP) with PHR1-cYFP or PHR11–226-cYFP under the control of the cauliflower mosaic virus (CaMV) 35S promoter (Pro35S). No signal was observed in the negative controls where PHR1-cYFP (or PHR11–226-cYFP) and MYC2396–624-nYFP or PHR1293–410-cYFP and MYC2-nYFP co-expressed. Nuclei are indicated by DAPI staining. Bars = 15 µm. D) CoIP assays. Total proteins were extracted from 8-d-old transgenic Arabidopsis seedlings simultaneously overexpressing PHR1 and MYC2 (PHR1-HA-L5 MYC2-4Myc) under the control of Pro35S. 4Myc-fused MYC2 was immunoprecipitated using an anti-Myc antibody (1:250) and the co-immunoprecipitated PHR1-HA protein was detected using an anti-HA antibody (1:10,000). Protein input for 4Myc-fused MYC2 in the immunoprecipitated complexes was also detected and is shown. Experiments were repeated three times with similar results. IP, immunoprecipitation.
To verify the interactions between PHR1 and MYC factors in plant cells, we performed BiFC assays. Sequences encoding truncated PHR1 proteins were fused to the YFP C-terminal fragment and expressed under the control of Pro35S to generate PHR11–226-cYFP and PHR1293–410-cYFP. Next, the full-length coding sequences of MYC2 and MYC3 as well as the sequence encoding the C-terminal fragment of MYC2 were ligated to the sequence encoding the N-terminal fragment of YFP (nYFP) to generate MYC2-nYFP, MYC3-nYFP, and MYC2396–624-nYFP. When PHR1-cYFP or PHR11–226-cYFP was co-expressed with MYC2-nYFP in N. benthamiana, strong YFP fluorescence was observed in the nucleus of transformed cells stained with DAPI (Fig. 5C; Supplemental Fig. S4B). The same result was obtained for N. benthamiana cells in which PHR1-cYFP and MYC3-nYFP were co-expressed. Fluorescence was undetectable in the negative controls (Fig. 5C; Supplemental Fig. S4B). These results reflect the interactions between PHR1 and MYC factors in the plant cell nucleus. The CoIP assays further provided in vivo evidence of the association between PHR1 and MYC2 in transgenic plants simultaneously overexpressing PHR1 and MYC2 (PHR1-HA-L5 MYC2-4Myc; Fig. 5D), which were derived from a cross between the PHR1-HA-L5 with transgenic plants overexpressing MYC2 under the control of Pro35S (MYC2-4Myc; Chen et al. 2011, 2012; Zhai et al. 2013). Collectively, these findings show that MYC factors interact with PHR1 in plant cells.
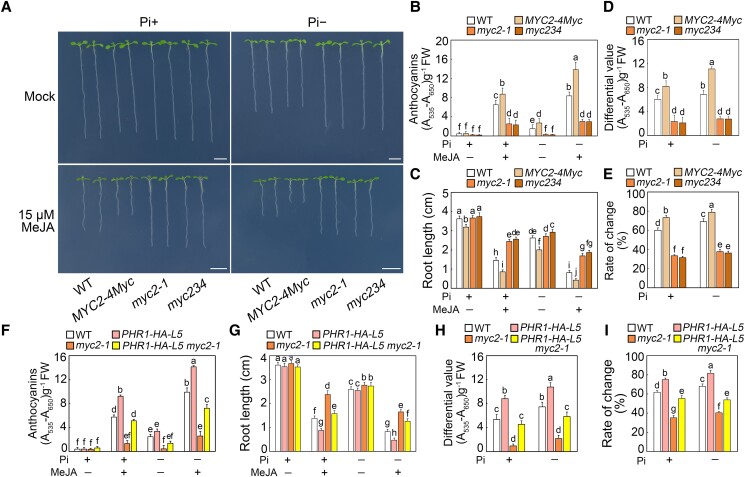
Because MYC factors physically associate with PHR1, we investigated whether they are involved in Pi deficiency-activated jasmonate signaling. We examined the phenotypes of MYC2-4Myc, the loss-of-function mutant myc2-1, and the triple mutant myc2 myc3 myc4 (myc234) on medium supplemented with MeJA and different concentrations of Pi. On the medium containing MeJA, the jasmonate-sensitive responses of MYC2-4Myc plants were increased under Pi-deficient conditions. In contrast, the myc2-1 and myc234 had lower anthocyanin levels and longer primary roots, regardless of the Pi concentration (Fig. 6, A to C). Compared with the Pi-sufficient conditions, the changes in the anthocyanin content and root elongation were significantly greater in the MYC2-4Myc plants under Pi-deficient conditions, but significantly less extensive in the myc2-1 and myc234 mutants at all Pi concentrations (Fig. 6, D and E). These results indicate MYC factors promote Pi deficiency-induced jasmonate signaling.
Figure 6.
MYC transcription factors are involved in Pi deficiency-promoted jasmonate signaling. A) Phenotypes of 8-d-old wild-type (WT), MYC2-4Myc, myc2-1, and myc2 myc3 myc4 (myc234) seedlings grown on modified half-strength Murashige and Skoog (MS) medium containing different concentrations of Pi with or without (Mock) 15 µM methyl jasmonate (MeJA). In the absence of MeJA (mock), an equal volume of 10% (v/v) ethanol was added. The media with 0.65 mM and 1 µM KH2PO4 were used as the Pi-sufficient (Pi+) and Pi-deficient (Pi−) media, respectively. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). Bars = 5 mm. B) Anthocyanin contents in 8-d-old WT, MYC2-4Myc, myc2-1, and myc234 seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed eight times with more than 100 seedlings for each sample per replicate. Data are means ± Sd from eight independent experiments (n = 8 times). FW, fresh weight. C) Root length of 8-d-old WT, MYC2-4Myc, myc2-1, and myc234 seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). The root length of 20 representative seedlings was measured for each sample per replicate. Data are means ± Sd (n = 20 representative plants). D) Anthocyanin content changes in 8-d-old WT, MYC2-4Myc, myc2-1, and myc234 seedlings induced by 15 µM MeJA. Data represent the anthocyanin content differences of seedlings grown on Pi+ or Pi− media. The differences mean the anthocyanins of seedlings with 15 µM MeJA treatment minus those of seedlings without MeJA treatment. The data used to calculate these difference values are from panel B. Data are means ± Sd from eight independent experiments (n = 8 times). E) Percentage of root length changes in 8-d-old WT, MYC2-4Myc, myc2-1, and myc234 seedlings induced by 15 µM MeJA. Data represent the percentages of root length changes of seedlings grown on Pi+ or Pi− media. The percentages mean the differences of root length with or without 15 µM MeJA treatment divided by root length without MeJA exposure. The data used to calculate percentage are from panel C. Data are means ± Sd (n = 20 representative plants). F) Anthocyanin contents in 8-d-old WT, PHR1-HA-L5, myc2-1, and PHR1-HA-L5 myc2-1 seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed eight times with more than 100 seedlings for each sample per replicate. Data are means ± Sd from eight independent experiments (n = 8 times). FW, fresh weight. G) Root length of 8-d-old WT, PHR1-HA-L5, myc2-1, and PHR1-HA-L5 myc2-1 seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). The root length of 20 representative seedlings was measured for each sample per replicate. Data are means ± Sd (n = 20 representative plants). H) Anthocyanin content changes in 8-d-old WT, PHR1-HA-L5, myc2-1, and PHR1-HA-L5 myc2-1 seedlings induced by 15 µM MeJA. Data represent the anthocyanin content differences of seedlings grown on Pi+ or Pi− media. The differences mean the anthocyanins of seedlings with 15 µM MeJA treatment minus those of seedlings without MeJA treatment. The data used to calculate these difference values are from panel F. Data are means ± Sd from eight independent experiments (n = 8 times). I) Percentage of root length changes in 8-d-old WT, PHR1-HA-L5, myc2-1, and PHR1-HA-L5 myc2-1 seedlings induced by 15 µM MeJA. Data represent the percentages of root length changes of seedlings grown on Pi+ or Pi− media. The percentages mean the differences of root length with or without 15 µM MeJA treatment divided by root length without MeJA exposure. The data used to calculate percentage are from panel G. Data are means ± Sd (n = 20 representative plants). Bars with different letters are significantly different from each other (P < 0.05). Data were analyzed by a two-way ANOVA using Tukey's HSD test.
PHR1 functions synergistically with MYC2 in jasmonate signaling, whereas JAZ1 inhibits their transcriptional functions and physical interaction
In this study, we showed that the myc2-1 mutant accumulated less anthocyanins and had longer primary roots than the wild-type plants upon jasmonate treatment (Fig. 6, A to E), which is in accordance with the findings of earlier studies (Lorenzo et al. 2004; Niu et al. 2011). Because of the confirmed physical interaction between PHR1 and MYC2, both of which mediate Pi deficiency-induced jasmonate signaling, we examined the possible genetic relationship between PHR1 and MYC2 by assessing whether the overexpression of PHR1 could rescue the phenotype of myc2-1 plants. We crossed PHR1-HA-L5 with myc2-1 to generate PHR1-HA-L5 myc2-1 plants, which were then treated with MeJA for phenotypic analyses. Consistent with the above-mentioned results, PHR1-HA-L5 was more sensitive to jasmonate, whereas myc2-1 was less sensitive, in terms of their anthocyanin contents and root growth on medium with MeJA (Fig. 6, F and G). However, PHR1-HA-L5 myc2-1 seedlings had higher anthocyanin contents and more inhibited primary root growth than the myc2-1 mutant plants, regardless of the Pi concentration (Fig. 6, F and G). Further statistical analyses suggested that the overexpression of PHR1 compensates for the reduced sensitivity of myc2-1 to MeJA (Fig. 6, H and I). To confirm this phenotypic observation, we quantitatively analyzed the expression of the jasmonate-responsive LDOX, DFR, AOS, and LOX2 genes in MeJA-treated PHR1-HA-L5, myc2-1, and PHR1-HA-L5 myc2-1 seedlings. Relative transcript levels of these genes were lower in myc2-1 than in PHR1-HA-L5 myc2-1 seedlings (Supplemental Fig. S12). Accordingly, overaccumulation of PHR1 partially suppressed the phenotype of myc2-1 in response to jasmonate.
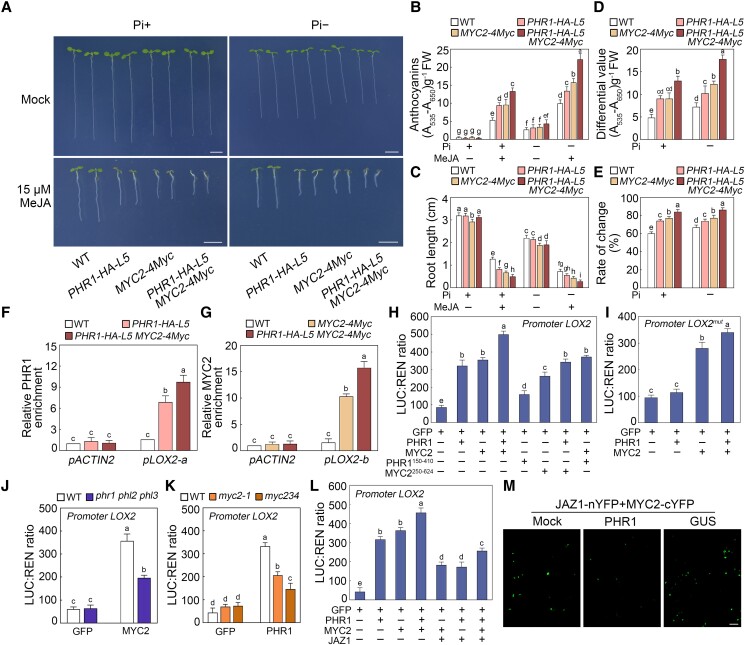
Based on the genetic association between PHR1 and MYC2, we speculated whether PHR1 works in concert with MYC2 in the jasmonate signaling pathway. We further examined the PHR1-HA-L5 MYC2-4Myc phenotype in response to MeJA treatment. As expected, at different Pi concentrations, the jasmonate response of PHR1-HA-L5 MYC2-4Myc plants was more robust than that of PHR1-HA-L5 and MYC2-4Myc plants (i.e. a much higher anthocyanin content and significantly inhibited primary root growth) (Fig. 7, A to C). Notably, the data suggested that PHR1-HA-L5 MYC2-4Myc plants were more responsive to jasmonate under Pi-deficient conditions than under Pi-sufficient conditions (Fig. 7, D and E). These findings support the idea that PHR1 works synergistically with MYC2 in the Pi deficiency-promoted jasmonate signaling pathway.
Figure 7.
PHR1 functions synergistically with MYC2 in jasmonate signaling. A) Phenotypes of 7-d-old wild-type (WT), PHR1-HA-L5, MYC2-4Myc, and PHR1-HA-L5 MYC2-4Myc seedlings grown on modified half-strength Murashige and Skoog (MS) medium containing different concentrations of Pi with or without (Mock) 15 µM methyl jasmonate (MeJA). In the absence of MeJA (mock), an equal volume of 10% (v/v) ethanol was added. The media with 0.65 mM and 1 µM KH2PO4 were used as the Pi-sufficient (Pi+) and Pi-deficient (Pi−) media, respectively. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). Bars = 5 mm. B) Anthocyanin contents in 7-d-old WT, PHR1-HA-L5, MYC2-4Myc, and PHR1-HA-L5 MYC2-4Myc seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed eight times with more than 100 seedlings for each sample per replicate. Data are means ± Sd from eight independent experiments (n = 8 times). FW, fresh weight. C) Root length of 7-d-old WT, PHR1-HA-L5, MYC2-4Myc, and PHR1-HA-L5 MYC2-4Myc seedlings grown on Pi+ or Pi− media with or without 15 µM MeJA. Experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). The root length of 20 representative seedlings was measured for each sample per replicate. Data are means ± Sd (n = 20 representative plants). D) Anthocyanin content changes in 7-d-old WT, PHR1-HA-L5, MYC2-4Myc, and PHR1-HA-L5 MYC2-4Myc seedlings induced by 15 µM MeJA. Data represent the anthocyanin content differences of seedlings grown on Pi+ or Pi− media. The differences mean the anthocyanins of seedlings with 15 µM MeJA treatment minus those of seedlings without MeJA treatment. The data used to calculate these difference values are from panel B. Data are means ± Sd from eight independent experiments (n = 8 times). E) Percentage of root length changes in 7-d-old WT, PHR1-HA-L5, MYC2-4Myc, and PHR1-HA-L5 MYC2-4Myc seedlings induced by 15 µM MeJA. Data represent the percentages of root length changes of seedlings grown on Pi+ or Pi− media. The percentages mean the differences of root length with or without 15 µM MeJA treatment divided by root length without MeJA exposure. The data used to calculate percentage are from panel C. Data are means ± Sd (n = 20 representative plants). F) ChIP-qPCR analyses of the enrichment of PHR1 in the LOX2 (pLOX2-a) promoter after the enhancement by MYC2. Eight-d-old WT, PHR1-HA-L5, and PHR1-HA-L5 MYC2-4Myc seedlings grown on Pi− media after treatment with 100 µM MeJA for 1 h were used in ChIP assays. More than 50 seedlings for each sample were pooled for ChIP assays using an anti-HA antibody. qPCR data from the ChIP assays with the ACTIN2 untranslated region sequence (pACTIN2) were used as a negative control. Data are means ± Sd from three independent biological replicates. G) ChIP-qPCR analyses of the enrichment of MYC2 to the LOX2 (pLOX2-b) promoter after the enhancement by PHR1. Eight-d-old WT, MYC2-4Myc, and PHR1-HA-L5 MYC2-4Myc seedlings grown on Pi− media after treatment with 100 µM MeJA for 1 h were used in ChIP assays. More than 50 seedlings for each sample were pooled for ChIP assays using an anti-Myc antibody. qPCR data from the ChIP assays with the ACTIN2 untranslated region sequence (pACTIN2) as a negative control. Data are means ± Sd from three independent biological replicates. H and I) Transient dual-luciferase reporter assays indicating that PHR1 acts synergistically with MYC2 to promote the transcription of LOX2. The physical interaction between PHR1 and MYC2 (H) and the P1BS element of LOX2 (I) exerts effects on PHR1–MYC2 co-activation. Data are means ± Sd from three independent biological replicates. J) Transient dual-luciferase reporter assays indicating that the activation of LOX2 promoter by MYC2 decreases in the phr1 phl2 phl3 mutant. Data are means ± Sd from three independent biological replicates. K) Transient dual-luciferase reporter assays indicating that the activation of LOX2 promoter by PHR1 decreases in the myc2-1 and myc234 mutants. Data are means ± Sd from three independent biological replicates. L) Transient dual-luciferase reporter assays indicating that transcriptional activation of LOX2 by PHR1 in concert with MYC2 is repressed by JAZ1. Data are means ± Sd from three independent biological replicates. Bars with different letters are significantly different from each other (P < 0.05). Data shown in panels H, I, and L were analyzed by a one-way ANOVA, and others were analyzed by a two-way ANOVA using Tukey's HSD test. M) BiFC analyses showing that PHR1 diminishes the interaction between JAZ1 and MYC2. As a negative control, GUS (β-glucuronidase) was co-expressed with JAZ1-nYFP and MYC2-cYFP. Fluorescence was detected 48 h after co-expression of JAZ1-nYFP + MYC2-cYFP (mock), PHR1 + JAZ1-nYFP + MYC2-cYFP (PHR1), or GUS + JAZ1-nYFP + MYC2-cYFP (GUS). Bars = 15 µm.
To elucidate the biochemical mechanism underlying the synergistic effects of PHR1 and MYC2, we analyzed whether PHR1 and MYC2 co-activate the expression of downstream genes. We initially conducted ChIP assays to analyze the enrichment of these two factors at the LOX2 promoter in the PHR1-HA-L5 MYC2-4Myc seedlings under Pi-deficient conditions with MeJA treatment. More PHR1 and MYC2 were enriched at the LOX2 promoter regions (pLOX2-a and pLOX2-b, respectively; Supplemental Tables S1 and S2) when they were both present than when only one was present (Fig. 7, F and G). To explain whether this outcome is caused by altered MYC2 expression level in the transgenic lines, we compared the MYC2 expression in MYC2-4Myc and PHR1-HA-L5 MYC2-4Myc seedlings. As shown in Supplemental Fig. S13A, the expression level of MYC2 was similar in both transgenic lines. We also found no significant changes in MYC2 expression in phr1 phl2 phl3 and PHR1-HA-L5, further supporting that PHR1 does not affect the expression of MYC2 (Supplemental Fig. S13B). These results imply that PHR1 and MYC2 reciprocally enhance each other's enrichment on the promoter regions of downstream target genes.
Next, we used the dual-luciferase reporter assay to determine the effects of PHR1 and MYC2 on LOX2 transcription in wild-type mesophyll protoplasts (Yoo et al. 2007). LUC expression driven by the LOX2 promoter was much higher when PHR1 and MYC2 were co-expressed than when PHR1 and GFP or MYC2 and GFP were co-expressed (Fig. 7H). To further investigate whether the protein interaction of PHR1–MYC2 is necessary for the regulation, we constructed effectors including a truncated PHR1 or MYC2 protein, disrupting the physical association of PHR1 with MYC2 (Supplemental Fig. S11). One effector contained a mutant version of PHR1 with the N-terminal 149 amino acids deleted (PHR1150–410) (Supplemental Fig. S11). The other effector included a truncated MYC2 fragment removing N-terminal 249 amino acids (MYC2250–624) (Supplemental Fig. S11). As shown in Fig. 7H, LOX2 promoter-driven LUC expression was much lower when co-expressing PHR1 with MYC2250–624 or co-expressing PHR1150–410 with MYC2 than when co-expressing PHR1 with MYC2. These results suggest that PHR1 and MYC2 function cooperatively to activate the transcription of target genes, and their physical interaction is essential for this regulatory relationship.
Then, we also analyzed the influence of the P1BS element on the PHR1–MYC2 co-activation of LOX2. When the P1BS element in the LOX2 promoter was mutated, MYC2 co-expression with GFP still increased LUC expression in wild-type protoplasts (Fig. 7I). Moreover, co-expression of MYC2 with PHR1 also moderately upregulated LUC expression driven by the LOX2mut promoter compared with co-expression of MYC2 with GFP in protoplasts of wild type (Fig. 7I). To assess whether functional defects of PHR1 would affect the transcriptional function of MYC2, we examined the ability of MYC2 to activate LOX2 transcription in phr1 phl2 phl3 protoplasts (Fig. 7J). We observed that LOX2 promoter-driven LUC activity was lower in MYC2-expressing phr1 phl2 phl3 protoplasts than in wild-type protoplasts (Fig. 7J). Similarly, LOX2 promoter-driven LUC activity was decreased in PHR1-expressing myc2-1 and myc234 protoplasts than in wild-type protoplasts (Fig. 7K). Collectively, these results support the notion that PHR1 and MYC2 function synergistically to positively regulate the expression of target genes.
Considering the interactions of JAZ with PHR1 and MYC2, we further performed the dual-luciferase reporter assays to investigate the regulatory effect of JAZ1 on these two factors in wild-type mesophyll protoplasts (Yoo et al. 2007). As shown in Fig. 7L, co-expression of JAZ1 with PHR1 and/or MYC2 downregulated LOX2 promoter-driven LUC activity compared with the expression of PHR1 and/or MYC2 in protoplasts of wild type. These results suggest that JAZ1 interferes with the transcriptional functions of PHR1 and MYC2. To further uncover the regulatory relationship among JAZ1, PHR1, and MYC2, we conducted the BiFC assays to detect the interaction between JAZ1 and MYC2 with or without PHR1. When PHR1 was co-expressed with JAZ1-nYFP and MYC2-cYFP, the YFP fluorescence signal was dramatically reduced in leaves of N. benthamiana (Fig. 7M; Supplemental Fig. S14). As a negative control, when β-glucuronidase (GUS) was co-expressed with JAZ1-nYFP and MYC2-cYFP, the YFP fluorescence intensity was not obviously changed (Fig. 7M; Supplemental Fig. S14). These observations show that PHR1 competes with JAZ1 to bind MYC2.
Discussion
The phytohormone jasmonate is a critical signaling compound that regulates diverse physiological processes, such as root growth inhibition, anthocyanin accumulation, and stress responses (Wasternack and Hause 2013; Huang et al. 2017; Zhang et al. 2017; Guo et al. 2018; Howe et al. 2018). The jasmonate signaling pathway is involved in a complex network that includes its crosstalk with other phytohormone signaling pathways (Spoel et al. 2003, 2007; Navarro et al. 2008; Cheng et al. 2009; Grant and Jones 2009; Sun et al. 2009; Zhou et al. 2019). Although the jasmonate signaling network has been studied, it is still largely unclear whether crucial environmental signals and jasmonate work together to modulate physiological processes in plants. P is an essential macronutrient for plant growth and development (Marschner 1995; Raghothama 1999; Nussaume et al. 2011). Plants exposed to Pi deficiency produce local signals that lead to changes in the root system architecture to enhance Pi uptake (Péret et al. 2011; Zhang et al. 2014; Crombez et al. 2019). Plants have evolved complex and sophisticated signaling cascades that mitigate the effects of Pi deficiency (Duan et al. 2008; Kant et al. 2011; Lin et al. 2013; Lei et al. 2016). Numerous studies have indicated that phytohormones modulate the Pi-deficient responses of plants (Franco-Zorrilla et al. 2005; Perez-Torres et al. 2008; Chacón-López et al. 2011; Mayzlish-Gati et al. 2012; Kumar et al. 2015; Liu et al. 2017). Interestingly, previous studies found that Pi deficiency induces jasmonate synthesis and signaling (Morcuende et al. 2007; Aparicio-Fabre et al. 2013; Wang et al. 2014; Khan et al. 2016). Consistently, we confirmed that Pi deficiency activates jasmonate signaling-related responses in Arabidopsis thaliana, including anthocyanin accumulation, root growth inhibition, and the activation of genes involved in jasmonate synthesis and signaling (Supplemental Fig. S1, A to F). A phenotypic analysis revealed that the COI1 mutants coi1-2 and coi1-16 are less sensitive to Pi deficiency-activated jasmonate signaling, suggesting that the COI1-mediated endogenous pathway is crucial for Pi deficiency-induced jasmonate responses (Supplemental Fig. S2, A to E).
Jasmonate signaling involves profound transcriptional reprogramming of cytogenetic programs associated with complex interactions between positive and negative regulators (e.g. COI1 receptor and JAZ repressors). Additionally, JAZ repressors modulate jasmonate signaling by physically interacting with downstream transcription factors (Fonseca et al. 2009; Kazan and Manners 2013; Huang et al. 2017; Ju et al. 2019; Pan et al. 2020). In the current study, the PHR1 transcription factor and its homologs PHL2 and PHL3 were found to interact with several JAZ proteins (Fig. 1, A to D; Supplemental Fig. S3), implying that PHR1, PHL2, and PHL3 may be critical regulators of the jasmonate signaling pathway. PHR1 is a core modulator of the response to Pi deficiency (Rubio et al. 2001; Sun et al. 2016; Wang et al. 2018; Guo et al. 2021; Navarro et al. 2021). Several crucial transcription factors were demonstrated to mediate the expression of PHR1 (Liu et al. 2017; Huang et al. 2018; Sega and Pacak 2019). Moreover, PHR1 entry into the nucleus and its transcriptional function are mainly suppressed by SPX proteins (Lv et al. 2014; Puga et al. 2014; Osorio et al. 2019). Nevertheless, the reports on the physical associations between PHR1 and other regulatory proteins are very limited. Because PHR1, PHL2, and PHL3 interact with JAZ repressors (Fig. 1, A to D), we further analyzed their functions related to jasmonate signaling. Based on the phenotypic results (Figs. 2, A to F and 3, A to F; Supplemental Fig. S10), we concluded that PHR1, PHL2, and PHL3 positively regulate jasmonate-induced anthocyanin accumulation and root growth inhibition. Biochemical analyses showed that PHR1 binds the promoters of several jasmonate-responsive genes to increase their transcription (Fig. 4, A to E). Furthermore, we revealed that JAZ proteins repress the transcriptional functions of PHR1 in the jasmonate signaling pathway (Fig. 4, C to E). By examining the phenotypes of transgenic plants simultaneously overexpressing PHR1 and JAZ1, we detected that the reduced sensitivity of JAZ1-Δ3A plants to jasmonate was partially attenuated by the PHR1 protein (Fig. 4, F to I). These results show that PHR1 acts together with JAZ repressors to integrate jasmonate signaling and Pi signaling pathways through direct protein–protein interactions.
Although PHR1-HA-L5 mimicked the phenotypes of PHL2-HA-L3 and PHL3-HA-L7 upon MeJA treatment, the performances of PHR1-HA-L5 and PHL2-HA-L3 or PHL3-HA-L7 were significantly different (Fig. 3, A to F). One possible explanation for the phenotypic discrepancies is that the PHR1 levels were higher in PHR1-HA-L5 than those of PHL2 and PHL3 in PHL2-HA-L3 and PHL3-HA-L7, respectively (Supplemental Fig. S15A). It is also possible that PHR1 exerts relatively greater regulatory effects on downstream genes compared with PHL2 and PHL3. Consistent with this notion, LOX2 promoter-driven LUC expression was higher in PHR1-expressing wild-type protoplasts than in PHL2- or PHL3-expressing protoplasts (Supplemental Fig. S15B). The expression of PHR1 and its close homologs (PHL1 to PHL4) was upregulated in wild-type seedlings by exogenous application of MeJA (Supplemental Fig. S3). Considering the direct linkage between the JAZ repressors and PHR1 protein, jasmonate signaling may have a dual regulatory function on PHR1. Similarly, recent research has revealed that the root hair-related bHLH genes ROOT HAIR DEFECTIVE 6 (RHD6), RHD6 LIKE1 (RSL1), RSL2, RSL4, and RSL5 were upregulated in MeJA-treated wild-type plants (Han et al. 2020). Meanwhile, JAZ repressors bind to and interfere with RHD6 and RSL1 transcription factors, thereby mediating root hair development (Han et al. 2020).
Earlier studies have shown that jasmonate triggers the transcription of MYC2 and that the JAZ repressors directly target MYC2 involved in a variety of jasmonate-mediated physiological processes (Chung et al. 2008; Katsir et al. 2008; Wang et al. 2017; Liu et al. 2019). In addition, Qi et al. (2011) demonstrated that JAZ interact with transcription factors such as MYB75, GLABLA1 (GL1), TRANSPARENT TESTA8 (TT8), and GLABLA3 (GL3) to regulate anthocyanin accumulation and trichome initiation. Moreover, the transcriptional levels of those factors are induced by jasmonate (Qi et al. 2011). Based on the findings, we hypothesize that jasmonate’s dual regulatory function on downstream transcription factors is an adaptive approach for maintaining proper jasmonate signaling and ensuring optimal plant survival in specific physiological contexts like Pi deficiency.
Previous works confirmed that PHR1 is a central modulator of Pi signaling and mediates the expression of PSI genes including PHT1, PHF1, and RNS1, which encode Pi transporters (Rubio et al. 2001; Guo et al. 2015; Sun et al. 2016; Wang et al. 2018). Among them, PHT1 is the most intensively studied Pi transporter, which transports Pi from the endoplasmic reticulum through vesicles to the plasma membrane, and this process is required for the function of PHF1 (Mudge et al. 2002; Shin et al. 2004; González et al. 2005; Bayle et al. 2011; Nussaume et al. 2011). The ribonuclease RNS1 has been reported to be involved in the recirculation and reactivation of Pi (Bariola et al. 1994; Duan et al. 2008). We further analyzed whether these Pi transporters are also involved in regulating jasmonate signaling. By observing the jasmonate responses of loss-of-function phf1-1, rns1, and pht1;1 pht1;5 mutants, we found that the above-mentioned transporters are not involved in the jasmonate signaling pathway (Supplemental Fig. S7, A and B). Interestingly, it has been demonstrated that PHO1, a transporter involved in Pi transfer from root to shoot, contributes to jasmonate synthesis and signal transduction (Hamburger et al. 2002; Stefanovic et al. 2007; Arpat et al. 2012; Khan et al. 2016; Wege et al. 2016). In the Pi-deficient mutant pho1, jasmonate synthesis was enhanced and jasmonate signaling pathway was activated, as well as plants accumulated more anthocyanins, compared with the wild-type control (Khan et al. 2016). Ribot et al. (2008) found that application of the jasmonate precursor 12-oxo-phytodienoic acid (OPDA), but not MeJA, increased the expression of AtPHO1;H10. These observations suggest that the linkage of Pi transport and jasmonate signaling is sophisticated and complex, and that the precise regulatory relationship needs to be further investigated in depth. In addition to the Pi transporters, SPX proteins are widely reported to be the main repressors of PHR1 and its homologs (Lv et al. 2014; Wang et al. 2014, 2018). In recent years, SPX proteins have been reported to integrate signaling pathways for more efficient utilization of N and Pi in plants (Medici et al. 2019; Hu et al. 2019, 2020; Ueda et al. 2020; Yang et al. 2022). However, it is currently unknown whether SPX help the jasmonate signaling pathway in modulating plant growth and development.
The MYC transcription factors MYC2, MYC3, and MYC4 are the most comprehensively studied JAZ-interacting proteins that mediate a subset of jasmonate processes (e.g. inhibition of root elongation and defense responses) (Lorenzo et al. 2004; Dombrecht et al. 2007; Qi et al. 2015; Liu et al. 2019; Wang et al. 2019a). In this study, we found that PHR1 physically associated with MYC transcription factors to form protein complexes in plant cells (Fig. 5, A to D). Further phenotypic analyses revealed that MYC transcription factors are likely essential for Pi deficiency-activated jasmonate signaling (Fig. 6, A to E). Considering that both PHR1 and MYC positively regulate the jasmonate signaling pathway, we clarified their genetic and biochemical regulatory relationships (Figs. 6, F to I and 7, A to M). We found that PHR1 and MYC2 synergistically upregulate the expression of several downstream jasmonate-responsive genes, thereby co-activating Pi deficiency-mediated jasmonate signaling (Fig. 7, A to K). Nevertheless, PHR1 may also be involved in mediating jasmonate signaling through other key regulators, such as the close homologs of MYC2. Consistently, overexpression of PHR1 partially rescued the less sensitive phenotype of myc2-1 in response to MeJA (Fig. 6, F to I; Supplemental Fig. S12). Transient transactivation assays revealed that LOX2 promoter-driven LUC expression was lower in PHR1-expressing protoplasts of the myc234 triple mutant than in protoplasts of the myc2-1 single mutant (Fig. 7K). Moreover, other transcription factors downstream of JAZ, such as the bHLH subgroup IIId factors MYB75, EIN3, and CONSTANS (CO), are also engaged in jasmonate-mediated anthocyanin accumulation and primary root growth inhibition (Song et al. 2013, 2014; Nakata et al. 2013; Sasaki-Sekimoto et al. 2013; Han et al. 2023b; Fonseca et al. 2014; Serrano-Bueno et al. 2022). The possible connections between PHR1 and those transcription factors also need to be clarified in future investigations.
In the agricultural production, plants (crops) are less efficient in their uptake of Pi fertilizers (Raghothama 1999; Veneklaas et al. 2012; López-Arredondo et al. 2014). Plants have evolved a range of physiological or developmental strategies to synchronize internal biological processes with surrounding environmental Pi deficiency (Zhang et al. 2014; Castrillo et al. 2017; Crombez et al. 2019). The finding that Pi deficiency amplifies jasmonate signaling suggests that environmental and phytohormonal signals coordinate to establish an appropriate balance among development or stress signaling pathways so that growth and stress tolerance are optimized for the prevailing conditions (Jia et al. 2022). Specifically, plants accumulate more anthocyanins under low Pi conditions in the presence of jasmonate (Supplemental Fig. S1; He et al. 2020; Liu et al. 2022; Song et al. 2022). The accumulation of anthocyanins prevents overexcitation and photosynthetic damage associated with Pi starvation (Gould et al. 2018; Liu et al. 2022). In addition, anthocyanins scavenge reactive oxygen species produced under stressful conditions (Zhang et al. 2013; Yan et al. 2020), which also allows plants to adapt to Pi-limiting conditions and survive.
Another co-regulation of Pi signaling pathway and jasmonate signaling pathway is to remodel the root structure of plants, such as total root length, root branching, and root hairs (Jiang et al. 2007; López-Arredondo et al. 2014; Crombez et al. 2019). This strategy is a way for plants to establish a specific developmental program in response to perceived environmental stimuli that maximize an organ’s expansion and elongation capacity. Remodeling of root structure may facilitate nutrient (i.e. Pi) acquisition with minimal energy cost. In addition, Pi deficiency stimulates jasmonate signaling to enhance resistance of plants against pathogens and insect herbivory (Khan et al. 2016; Wang et al. 2019b; Kong et al. 2021; Li et al. 2021b; Tang et al. 2022). The integration of Pi signaling pathway and jasmonate signaling pathway also provides profound insights into the regulation of trade-offs between plant growth and defense. Plant genetic engineering should appreciate this regulatory crosstalk and the exact molecular mechanisms that underpin it to improve the efficiency of Pi utilization.
To further elucidate the molecular basis of the Pi deficiency-activated jasmonate signaling in Arabidopsis, we propose the following simplified model involving JAZ–PHR1–MYC2 (Fig. 8). Under Pi-deficient conditions, PHR1 is expressed and the encoding protein is modified and activated (Miura et al. 2005; Bari et al. 2006; Nilsson et al. 2007; Lei et al. 2016; Sun et al. 2016; He et al. 2020). When the jasmonate concentration increases, the receptor COI1 perceives jasmonate and targets JAZ proteins for degradation via the SCFCOI1–26S proteasome pathway (Chini et al. 2007; Thines et al. 2007; Sheard et al. 2010; Yan et al. 2013). The degradation of JAZ repressors enables MYC2 and PHR1 to form a protein complex that positively regulates jasmonate-induced anthocyanin accumulation and root growth inhibition (Fig. 8). PHR1 may also modulate jasmonate signaling through other critical regulators, such as two MYC2 homologs (MYC3 and MYC4). Our results reveal critical roles of PHR1 in modulating jasmonate responses and provide mechanistic insights into how jasmonate signaling is fine-tuned under Pi-deficient conditions.
Figure 8.

Working model for Pi deficiency-activated jasmonate signaling. Under Pi-deficient conditions, PHR1 is expressed and the encoding protein is modified and activated. When the jasmonate concentration increases, the receptor COI1 perceives jasmonate and targets JAZ proteins for degradation via the SCFCOI1–26S proteasome pathway. The degradation of JAZ repressors enables MYC2 and PHR1 to form a protein complex that positively regulates jasmonate-induced anthocyanin accumulation and root growth inhibition.
Materials and methods
Materials and plant growth conditions
The phytohormone MeJA was purchased from Sigma-Aldrich. Common chemicals were obtained from Shanghai Sangon (Shanghai, China), and Taq DNA polymerases were purchased from Takara Biotechnology (Dalian, China). The anti-Myc (Sigma-Aldrich, catalog no. M4439) and anti-HA (Sigma-Aldrich, catalog no. H9658) antibodies used in this study were purchased from Sigma-Aldrich. The wild-type and mutant Arabidopsis thaliana plants are in the Columbia (Col-0) genetic background. The mutant or transgenic plants coi1-2 (Xu et al. 2002), coi1-16 (Pan et al. 2020), jazQ (Campos et al. 2016), myc2-1, myc2 myc3 myc4 (Fernández-Calvo et al. 2011), JAZ1-Δ3A (Han et al. 2018), and MYC2-4Myc (Chen et al. 2012) were described previously. The phr1 (SALK_067629C), phl2 (SALK_114420C), phl3 (SALK_113627C), phf1-1 (SALK_037068C), rns1 (SALK_087165C), pht1;1 (SALK_088586C), and pht1;5 (SALK_138009C) mutants were obtained from The Arabidopsis Biological Resource Center at Ohio State University (http://abrc.osu.edu). The double mutants phr1 phl2, phr1 phl3, phl2 phl3, and pht1;1 pht1;5 as well as the triple mutant phr1 phl2 phl3 were generated via genetic crosses using standard techniques. To generate PHR1-HA, PHL2-HA, and PHL3-HA transgenic plants, the full-length PHR1, PHL2, and PHL3 cDNA sequences were inserted into the binary vector pOCA30 in the sense orientation for the subsequent expression under the control of Pro35S (Hu et al. 2013).
Arabidopsis seeds were surface-sterilized for 12 min in 20% (v/v) bleach and then sown on modified half-strength MS medium and kept at 4 °C for 3 d pre-germination. The media containing 0.65 mM or 1 µM KH2PO4 were used as the Pi-sufficient (Pi+) and Pi-deficient (Pi−) media, respectively. For the MeJA treatment, MeJA was dissolved in ethanol to produce a 10 mM stock solution. In the absence of MeJA (mock), an equal volume of 10% (v/v) ethanol was added. Arabidopsis seedlings were grown on medium with or without different concentrations of MeJA. Arabidopsis and N. benthamiana plants were incubated in an artificial growth chamber at 22 °C with a 16-h light (100 µE m−2 s−1, white fluorescent bulbs, full wavelength)/8-h dark photoperiod.
Anthocyanin content and primary root length measurements
To measure the anthocyanin content, Arabidopsis seedlings were grown on modified half-strength MS (Pi+, Pi−, MeJA+, or MeJA−) medium before measuring the anthocyanin content as previously described (Qi et al. 2011). The anthocyanin content was expressed as (A535 − A650) per gram fresh weight. All experiments were performed eight times with more than 100 seedlings for each sample per replicate. Data are means ± standard deviation (Sd) from eight independent experiments (n = 8 replicates).
To measure the primary root length, seedlings were grown vertically on modified half-strength MS (Pi+, Pi−, MeJA+, or MeJA−) medium before measuring the root length. All experiments were performed four times with similar results by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate). The root length of 20 representative seedlings was measured for each sample per replicate by using a vernier caliper (Supplemental Data Set S1). Data are means ± Sd (n = 20 representative plants).
RNA extraction and RT-qPCR
Total RNA was extracted from seedlings (more than 60 seedlings for each sample) treated with different concentrations of Pi and MeJA using the TRIzol reagent (Invitrogen). The RT-qPCR analysis was conducted as described by Han et al. (2018). Briefly, 1.0 μg DNase-treated RNA was reverse transcribed in a 20-μL reaction volume containing oligo-(dT)19 primer and Moloney murine leukemia virus reverse transcriptase (Fermentas, Hanover, MD, USA). The cDNA was diluted 1:1 prior to use. A 1.0-μL aliquot of the cDNA solution was used for the RT-qPCR analysis, which was performed using the SYBR Premix Ex Taq kit (Takara Biotechnology) and the LightCycler 480 real-time PCR system (Roche). The RT-qPCR analyses were completed using five biological replicates by analyzing different batches of seedlings (more than 60 seedlings for each sample per replicate), each with three technical replicates. Changes in the expression of the target gene were calculated using the 2–ΔΔCt method relative to the expression of ACTIN2 (AT3G18780). The gene-specific RT-qPCR primers are listed in Supplemental Table S3.
Y2H assays
To analyze the physical interactions of PHR1 and its homologs with JAZ proteins or MYC transcription factors, the full-length PHR1 and PHL coding sequences were cloned into pGBKT7 to produce the bait constructs (BD-PHR1 and BD-PHL), whereas the full-length JAZ and MYC coding sequences were inserted into pGADT7 to generate the prey constructs (AD-JAZ and AD-MYC). To identify the specific protein regions responsible for the interactions, multiple sequences encoding truncated forms of PHR1 were inserted into pGBKT7, whereas sequences encoding truncated forms of JAZ1 or MYC2 were incorporated into pGADT7. Y2H assays were performed as previously described (Yang et al. 2021). Briefly, yeast (Saccharomyces cerevisiae) strain AH109 cells were co-transformed with the bait and prey constructs. Protein interactions were indicated by the ability of the transformed cells to grow on a dropout medium lacking Leucine (Leu), Tryptophan (Trp), Histidine (His), and Adenine (Ade) and containing 20 mM 3-aminotriazole after a 2-d incubation. The primers used for cloning are listed in Supplemental Data Set S2.
BiFC assays
The cDNA sequences encoding the N-terminus (173 amino acids) of YFP (nYFP) and the C-terminus (64 amino acids) of YFP (cYFP) were amplified by PCR and inserted into separate pFGC5941 plasmids to produce pFGC-nYFP and pFGC-cYFP, respectively (Kim et al. 2008). The sequences encoding full-length or truncated PHR1 and full-length PHL1 and MYC2 were inserted into pFGC-cYFP to produce the following fusion proteins: PHR1-cYFP, PHR11–226-cYFP, PHR1293–410-cYFP, PHL1-cYFP, and MYC2-cYFP. Similarly, sequences encoding full-length JAZ1, JAZ7, or JAZ9 were inserted into pFGC-nYFP to produce the following fusion proteins: JAZ1-nYFP, JAZ7-nYFP, and JAZ9-nYFP. Sequences encoding full-length or truncated MYC2 and full-length MYC3 were fused with the sequence encoding nYFP to generate the following proteins: MYC2-nYFP, MYC2396–624-nYFP, and MYC3-nYFP. The resulting recombinant plasmids were inserted into Agrobacterium tumefaciens (strain EHA105) cells for the infiltration of N. benthamiana leaves as previously described (Hu et al. 2019). The experiments were performed at least three times using different batches of plants. For each biological replicate, more than 12 N. benthamiana plants were infiltrated and more than 600 cells were examined. The leaves were analyzed at 48 h post-infiltration. Specifically, YFP and DAPI fluorescent signals were detected using a confocal laser scanning microscope (Olympus Fluoview FV1000, Tokyo, Japan). For DAPI staining, infected leaves were stained with DAPI solution (10 mM) for 5 min before observation. The YFP signals were imaged with excitation at 488 nm (the intensity was 24%, and the gains were 1), and the emission signal was collected between 510 and 530 nm. The DAPI signals were imaged with excitation at 405 nm (the intensity was 15%, and the gains were 1), and the emission signal was collected between 420 and 440 nm. The primers used for cloning are listed in Supplemental Data Set S2.
CoIP assays
To verify the PHR1–JAZ1 and PHR1–MYC2 interactions, we extracted proteins from 8-d-old transgenic Arabidopsis plants simultaneously overexpressing PHR1 and JAZ1 (PHR1-HA-L5 JAZ1-Δ3A) or PHR1 and MYC2 (PHR1-HA-L5 MYC2-4Myc) under the control of Pro35S. The protein extraction was performed using extraction buffer containing 50 mM Tris–HCl (pH 7.4), 1 mM EDTA, 150 mM NaCl, 10% (v/v) glycerol, 0.1% (v/v) Triton X-100, 1 mM PMSF, and 1× Roche Protease Inhibitor Cocktail. Immunoprecipitation experiments were performed using Protein A/G Plus agarose beads (Santa Cruz Biotechnology, catalog no. D1217) following the manufacturer's protocol. Briefly, cell lysates were pre-cleared using the Protein A/G Plus agarose beads and then incubated with the anti-Myc antibody (1:250) and the Protein A/G Plus agarose beads at 4 °C overnight in the extraction buffer. The agarose beads were washed three times with the extraction buffer and then the co-immunoprecipitated protein was detected by immunoblotting using an anti-HA antibody (1:10,000). The primers used for cloning are listed in Supplemental Data Set S2.
Y1H assays
Y1H assays were performed using the Matchmaker Yeast One-Hybrid System Kit (Clontech) according to the manufacturer's instructions. The full-length PHR1 coding sequence was inserted into pGADT7 to produce the AD-PHR1 construct. The putative promoter fragments of AOS, LOX2, LOX3, and AOC2 were cloned into the pAbAi vector to generate pAbAi-pAOS.1, pAbAi-pLOX2.1, pAbAi-pLOX3.1, and pAbAi-pAOC2.1, respectively, which were linearized using BstBI and then inserted into Y1HGold yeast cells. The transformed cells were grown on the SD/−Ura medium in plates for 3 d. Next, AD-PHR1 was then inserted into the cells harboring pAbAi-pAOS.1, pAbAi-pLOX2.1, pAbAi-pLOX3.1, and pAbAi-pAOC2.1 and then selected on the SD/−Leu medium in plates. Co-transformed cells were cultured on the SD/−Leu medium containing aureobasidin A (AbA: 300 µg/L) in plates for 3 d. Positive clones were obtained for several yeast cell concentrations from the dilutions 100 (OD600 = 0.8) to 10−3. The primers used for cloning are listed in Supplemental Data Set S2.
ChIP assays
The ChIP assays were performed essentially as previously described (Mukhopadhyay et al. 2008; Jiang et al. 2014). Briefly, the wild-type, PHR1-HA-L5, PHR1-HA-L5 JAZ1-Δ3A, MYC2-4Myc, and PHR1-HA-L5 MYC2-4Myc seedlings were treated with 1% formaldehyde (cross-linking treatment) and then their chromatin was isolated. The anti-HA and anti-Myc antibodies (1:1,000) were used to immunoprecipitate the protein–DNA complexes. The precipitated DNA was purified using a commercial PCR purification kit (Qiagen). To quantitatively analyze the PHR1–DNA (target promoters) and MYC2–DNA binding, the RT-qPCR analysis was performed as previously described (Mukhopadhyay et al. 2008), with the ACTIN2 3′ untranslated region sequence as the endogenous control. Relative enrichment was calculated in terms of the DNA binding rate. The analyses were completed using data of three biological replicates by analyzing different batches of seedlings (more than 50 seedlings for each sample per replicate). The primers used for the ChIP assays are listed in Supplemental Tables S1 and S2.
Transient transcriptional activation assays
Full-length PHR1, JAZ1, JAZ9, PHL2, PHL3, MYC2, and GFP, and truncated PHR1150–410 and MYC2250–624 coding sequences were amplified by PCR and cloned into separate pGreenII 62-SK vectors for the subsequent expression under the control of the CaMV 35S promoter (i.e. as effectors) (Fig. 6A). The putative promoter sequences of LOX2 and LOX2mut (the putative PHR1-recognized P1BS element “GTATATAC” of the LOX2 promoter was mutated to “TCCGCGGA”) were amplified by PCR and inserted into separate pGreenII 0800-LUC vectors (i.e. as reporters) (Hellens et al. 2005). Different combinations of the recombinant plasmids were used to transform Arabidopsis leaf mesophyll protoplasts as previously described (Sheen 2001). Transfected cells were cultured for 16 to 18 h before the relative LUC activity was analyzed using the Dual-Luciferase Reporter Assay system (Promega, Madison, WI, USA), which measures the activities of firefly LUC and the internal control Renilla reniformis LUC (REN). The primers used for cloning are listed in Supplemental Data Set S2.
Statistical analysis
The effects of interactions between Pi and MeJA on anthocyanin accumulation, primary root elongation, and expression levels of several jasmonate-induced genes in seedlings were tested. All analyses of multifactorial variance were performed using the generalized linear model procedure in SPSS for Windows. The model results showed that the interactive effect between Pi and MeJA was significant (P < 0.05) or highly significant (P < 0.01) (Supplemental Data Set S3). Based on the experiments, statistical analysis was further performed by Student's t test or one-way or two-way ANOVA using Tukey's honest significant difference (HSD) as a post hoc test. The results are shown in Supplemental Data Set S4.
Accession numbers
Arabidopsis Genome Initiative numbers for the genes discussed in this article are as follows: COI1, AT2G39940; JAZ1, AT1G19180; JAZ2, AT1G74950; JAZ3, AT3G17860; JAZ4, AT1G48500; JAZ5, AT1G17380; JAZ6, AT1G72450; JAZ7, AT2G34600; JAZ8, AT1G30135; JAZ9, AT1G70700; JAZ10, AT5G13220; JAZ11, AT3G43440; JAZ12, AT5G20900; MYC2, AT1G32640; MYC3, AT5G46760; MYC4, AT4G17880; PHR1, AT4G28610; PHL1, AT5G29000; PHL2, AT3G24120; PHL3, AT4G13640; PHL4, AT2G20400; AOC2, AT3G25770; AOS, AT5G42650; LOX2, AT3G45140; LOX3, AT1G17420; PHF1, AT3G52190; RNS1, AT2G02990; PHT1;1, AT5G43350; PHT1;5, AT2G32830; and ACTIN2, AT3G18780.
Supplementary Material
Acknowledgments
We thank D. Xie (Tsinghua University, China), G.A. Howe (Michigan State University), R. Solano (Campus Universidad Autónoma), X. Chen (Institute of Plant Physiology and Ecology, Shanghai Institutes for Biological Sciences), C. Li (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences), and Z. Chen (Purdue University) for sharing research materials. We thank J. Wu and J. Qi (Kunming Institute of Botany, Chinese Academy of Sciences) for measuring the jasmonate content. We also thank the Central Laboratory of Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences for technical supports.
Contributor Information
Kunrong He, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Jiancan Du, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Xiao Han, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Huiqiong Li, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Mengyi Kui, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Juping Zhang, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; College of Life Sciences, University of Chinese Academy of Sciences, Beijing 100049, China.
Zhichong Huang, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Qiantang Fu, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Yanjuan Jiang, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China; State Key Laboratory for Conservation and Utilization of Bio-Resources in Yunnan, School of Life Sciences, Yunnan University, Kunming 650091, China.
Yanru Hu, CAS Key Laboratory of Tropical Plant Resources and Sustainable Use, Xishuangbanna Tropical Botanical Garden, Chinese Academy of Sciences, Kunming, Yunnan 650223, China.
Author Contributions
Y.H. and K.H. designed this study. K.H., J.D., X.H., H.L., K.M., J.Z., Z.H., Q.F., Y.J., and Y.H. performed experiments or interpreted data. Y.H. and K.H. interpreted data and wrote the article. All authors read and approved the final article.
Supplemental data
The following materials are available in the online version of this article.
Supplemental Figure S1. Pi deficiency enhances jasmonate signaling.
Supplemental Figure S2. The COI1-mediated pathway is essential for Pi deficiency-stimulated jasmonate signaling.
Supplemental Figure S3. Y2H analyses of the interactions between JAZ repressors and PHL2 or PHL3.
Supplemental Figure S4. Percentage of transformed cells with YFP fluorescence in the BiFC assays presented in Figures 1B and 5C.
Supplemental Figure S5. Jasmonate promotes the transcription of PHR1 and its homologs PHL1, PHL2, PHL3, and PHL4 in plants.
Supplemental Figure S6. Jasmonate responses of phr1, phl2, and phl3 single mutants.
Supplemental Figure S7. Jasmonate responses of Pi transport-related mutants.
Supplemental Figure S8. The jasmonate content in roots of wild-type and phr1 phl2 phl3 mutant seedlings under different Pi concentrations with or without MeJA treatment.
Supplemental Figure S9. RT-qPCR analysis of PHR1, PHL2, and PHL3 expression in transgenic overexpression lines.
Supplemental Figure S10. Sensitivity of PHR1-HA-L5 and PHR1-HA-L8 seedlings to jasmonate.
Supplemental Figure S11. Schematic of the AOC2, LOX2, LOX3, and AOS promoter fragments used in the Y1H assays and the effectors and reporters used in the transient transactivation assays.
Supplemental Figure S12. RT-qPCR analyses of LDOX, DFR, AOS, and LOX2 expression levels in WT, PHR1-HA-L5, myc2-1, and PHR1-HA-L5 myc2-1.
Supplemental Figure S13. RT-qPCR analyses of MYC2 expression levels in wild type, MYC2-4Myc, PHR1-HA-L5 MYC2-4Myc, phr1 phl2 phl3, and PHR1-HA-L5.
Supplemental Figure S14. Quantitative analysis of YFP fluorescence intensity in Figure 7M.
Supplemental Figure S15. The accumulation of PHR1, PHL2, and PHL3 in transgenic plants and the regulatory effects of PHR1, PHL2, and PHL3 on LOX2 expression.
Supplemental Table S1. Information for detecting PHR1 binding on AOC2, LOX2, LOX3, and AOS promoter sequences.
Supplemental Table S2. Information for detecting MYC2 binding on the LOX2 promoter sequence.
Supplemental Table S3. Primers used for RT-qPCR analyses.
Supplemental Data Set S1. Supplementary data for root length measurements.
Supplemental Data Set S2. Primers used for generating various clones.
Supplemental Data Set S3. Analyses of the interactive effects between Pi, MeJA and genotype.
Supplemental Data Set S4. ANOVA and Student's t-test results.
Funding
This work was supported by the National Natural Science Foundation of China (31922009, 31870259, and 32270613), the Applied Basic Research Project of Yunnan Province (202305AS350010, 202001AV070009, 202001AT070118, and 202101AW070005), the CAS “Light of West China” Program (to X.H.), the Youth Innovation Promotion Association of the of Chinese Academy of Sciences (Y201973 and 2022399), and the Yunnan High Level Talents Special Support Plan (YNWR-QNBJ-2018-075).
Data availability
All data supporting the findings of this study are available within the article and its supplementary materials.
References
- Aparicio-Fabre R, Guillén G, Loredo M, Arellano J, Valdés-López O, Ramírez M, Íñiguez LP, Panzeri D, Castiglioni B, Cremonesi P, et al. . Common bean (Phaseolus vulgaris L.) PvTIFY orchestrates global changes in transcript profile response to jasmonate and phosphorus deficiency. BMC Plant Biol. 2013:13(1):26. 10.1186/1471-2229-13-26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arpat AB, Magliano P, Wege S, Rouached H, Stefanovic A, Poirier Y. Functional expression of PHO1 to the Golgi and trans-Golgi network and its role in export of inorganic phosphate. Plant J. 2012:71(3):479–491. 10.1111/j.1365-313X.2012.05004.x [DOI] [PubMed] [Google Scholar]
- Bari R, Datt Pant B, Stitt M, Scheible WR. PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 2006:141(3):988–999. 10.1104/pp.106.079707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bariola PA, Howard CJ, Taylor CB, Verburg MT, Jaglan VD, Green PJ. The Arabidopsis ribonuclease gene RNS1 is tightly controlled in response to phosphate limitation. Plant J. 1994:6(5):673–685. 10.1046/j.1365-313X.1994.6050673.x [DOI] [PubMed] [Google Scholar]
- Bayle V, Arrighi JF, Creff A, Nespoulous C, Vialaret J, Rossignol M, Gonzalez E, Paz-Ares J, Nussaume L. Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell. 2011:23(4):1523–1535. 10.1105/tpc.110.081067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Golz JF, Giménez-Ibañez S, Fernandez-Barbero G, Franco-Zorrilla JM, Solano R. FILAMENTOUS FLOWER is a direct target of JAZ3 and modulates responses to jasmonate. Plant Cell. 2015:27(11):3160–3174. 10.1105/tpc.15.00220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boter M, Ruíz-Rivero O, Abdeen A, Prat S. Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 2004:18(13):1577–1591. 10.1101/gad.297704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos R, Castrillo G, Linhares F, Puga MI, Rubio V, Pérez-Pérez J, Solano R, Leyva A, Paz-Ares J. A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 2010:6(9):e1001102. 10.1371/journal.pgen.1001102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campos ML, Yoshida Y, Major IT, de Oliveira Ferreira D, Weraduwage SM, Froehlich JE, Johnson BF, Kramer DM, Jander G, Sharkey TD, et al. . Rewiring of jasmonate and phytochrome B signalling uncouples plant growth-defense tradeoffs. Nat Commun. 2016:7(1):12570. 10.1038/ncomms12570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Liu L, Ma K, Wang W, Lv H, Gao M, Wang X, Zhang X, Ren S, Zhang N, et al. . The jasmonate-induced bHLH gene SlJIG functions in terpene biosynthesis and resistance to insects and fungus. J Integr Plant Biol. 2022:64(5):1102–1115. 10.1111/jipb.13248 [DOI] [PubMed] [Google Scholar]
- Castrillo G, Teixeira PJPL, Paredes SH, Law TF, de Lorenzo L, Feltcher ME, Finkel OM, Breakfield NW, Mieczkowski P, Jones CD, et al. . Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017:543(7646):513–518. 10.1038/nature21417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacón-López A, Ibarra-Laclette E, Sánchez-Calderón L, Gutiérrez-Alanis D, Herrera-Estrella L. Global expression pattern comparison between low phosphorus insensitive 4 and WT Arabidopsis reveals an important role of reactive oxygen species and jasmonic acid in the root tip response to phosphate starvation. Plant Signal Behav. 2011:6(3):382–392. 10.4161/psb.6.3.14160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen R, Jiang H, Li L, Zhai Q, Qi L, Zhou W, Liu X, Li H, Zheng W, Sun J, et al. . The Arabidopsis mediator subunit MED25 differentially regulates jasmonate and abscisic acid signaling through interacting with the MYC2 and ABI5 transcription factors. Plant Cell. 2012:24(7):2898–2916. 10.1105/tpc.112.098277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q, Zhou W, Qi L, Xu L, Wang B, Chen R, Jiang H, Qi J, et al. . The basic helix-loop-helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell. 2011:23(9):3335–3352. 10.1105/tpc.111.089870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Song S, Xiao L, Soo HM, Cheng Z, Xie D, Peng J. Gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 2009:5(3):e1000440. 10.1371/journal.pgen.1000440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, Adie B, Chico JM, Lorenzo O, García-Casado G, López-Vidriero I, Lozano FM, Ponce MR, et al. . The JAZ family of repressors is the missing link in jasmonate signaling. Nature. 2007:448(7154):666–671. 10.1038/nature06006 [DOI] [PubMed] [Google Scholar]
- Chini A, Gimenez-Ibanez S, Goossens A, Solano R. Redundancy and specificity in jasmonate signalling. Curr Opin Plant Biol. 2016:33:147–156. 10.1016/j.pbi.2016.07.005 [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiol. 2008:146(3):952–964. 10.1104/pp.107.115691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crombez H, Motte H, Beeckman T. Tackling plant phosphate starvation by the roots. Dev Cell. 2019:48(5):599–615. 10.1016/j.devcel.2019.01.002 [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, et al. . MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell. 2007:19(7):2225–2245. 10.1105/tpc.106.048017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J, Pineros MA, Li X, Yang H, Liu Y, Murphy AS, Kochian LV, Liu D. An Arabidopsis ABC transporter mediates phosphate deficiency-induced remodeling of root architecture by modulating iron homeostasis in roots. Mol Plant. 2017:10(2):244–259. 10.1016/j.molp.2016.11.001 [DOI] [PubMed] [Google Scholar]
- Duan K, Yi K, Dang L, Huang H, Wu W, Wu P. Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 2008:54(6):965–975. 10.1111/j.1365-313X.2008.03460.x [DOI] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, Chico J-M, Gimenez-Ibanez S, Geerinck J, Eeckhout D, Schweizer F, Godoy M, Franco-Zorrilla JM, et al. . The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell. 2011:23(2):701–715. 10.1105/tpc.110.080788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Chico JM, Solano R. The jasmonate pathway: the ligand, the receptor and the core signaling module. Curr Opin Plant Biol. 2009:12(5):539–547. 10.1016/j.pbi.2009.07.013 [DOI] [PubMed] [Google Scholar]
- Fonseca S, Fernández-Calvo P, Fernández GM, Díez-Díaz M, Gimenez-Ibanez S, López-Vidriero I, Godoy M, Fernández-Barbero G, Van Leene J, De Jaeger G, et al. . bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS One. 2014:9(1):e86182. 10.1371/journal.pone.0086182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Martín AC, Leyva A, Paz-Ares J. Interaction between phosphate-starvation, sugar, and cytokinin signaling in Arabidopsis and the roles of cytokinin receptors CRE1/AHK4 and AHK3. Plant Physiol. 2005:138(2):847–857. 10.1104/pp.105.060517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- González E, Solano R, Rubio V, Leyva A, Paz-Ares J. PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell. 2005:17(12):3500–3512. 10.1105/tpc.105.036640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould KS, Jay-Allemand C, Logan BA, Baissac Y, Bidel LPR. When are foliar anthocyanins useful to plants? Re-evaluation of the photoprotection hypothesis using Arabidopsis thaliana mutants that differ in anthocyanin accumulation. Environ Exp Bot. 2018:154:11–22. 10.1016/j.envexpbot.2018.02.006 [DOI] [Google Scholar]
- Grant MR, Jones JD. Hormone (dis)harmony moulds plant health and disease. Science. 2009:324(5928):750–752. 10.1126/science.1173771 [DOI] [PubMed] [Google Scholar]
- Guo Q, Major IT, Howe GA. Resolution of growth-defense conflict: mechanistic insights from jasmonate signaling. Curr Opin Plant Biol. 2018:44:72–81. 10.1016/j.pbi.2018.02.009 [DOI] [PubMed] [Google Scholar]
- Guo M, Ruan W, Li C, Huang F, Zeng M, Liu Y, Yu Y, Ding X, Wu Y, Wu Z, et al. . Integrative comparison of the role of the PHOSPHATE RESPONSE1 subfamily in phosphate signaling and homeostasis in rice. Plant Physiol. 2015:168(4):1762–1776. 10.1104/pp.15.00736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M, Ruan W, Zhang Y, Zhang Y, Wang X, Guo Z, Wang L, Zhou T, Paz-Ares J, Yi K. A reciprocal inhibitory module for Pi and iron signaling. Mol Plant. 2021:15(1):138–150. 10.1016/j.molp.2021.09.011 [DOI] [PubMed] [Google Scholar]
- Ham B-K, Chen J, Yan Y, Lucas WJ. Insights into plant phosphate sensing and signaling. Curr Opin Biotechnol. 2018:49:1–9. 10.1016/j.copbio.2017.07.005 [DOI] [PubMed] [Google Scholar]
- Hamburger D, Rezzonico E, MacDonald-Comber Petétot J, Somerville C, Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002:14(4):889–902. 10.1105/tpc.000745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Hu Y, Zhang G, Jiang Y, Chen X, Yu D. Jasmonate negatively regulates stomatal development in Arabidopsis cotyledons. Plant Physiol. 2018:176(4):2871–2885. 10.1104/pp.17.00444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Kui M, He K, Yang M, Du J, Jiang Y, Hu Y. Jasmonate-regulated root growth inhibition and root hair elongation. J Exp Bot. 2023a:74(4):1176–1185. 10.1093/jxb/erac441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Kui M, Xu T, Ye J, Du J, Yang M, Jiang Y, Hu Y. CO interacts with JAZ repressors and bHLH subgroup IIId factors to negatively regulate jasmonate signaling in Arabidopsis seedlings. Plant Cell. 2023b:35(2):852–873. 10.1093/plcell/koac331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Zhang M, Yang M, Hu Y. Arabidopsis JAZ proteins interact with and suppress RHD6 transcription factor to regulate jasmonate-stimulated root hair development. Plant Cell. 2020:32(4):1049–1062. 10.1105/tpc.19.00617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Zhang X, Li L, Sun Z, Li J, Chen X, Hong G. SPX4 interacts with both PHR1 and PAP1 to regulate critical steps in phosphorus-status-dependent anthocyanin biosynthesis. New Phytol. 2020:230(1):205–217. 10.1111/nph.17139 [DOI] [PubMed] [Google Scholar]
- Hellens RP, Allan AC, Friel EN, Bolitho K, Grafton K, Templeton MD, Karunairetnam S, Gleave AP, Laing WA. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods. 2005:1(1):13. 10.1186/1746-4811-1-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou X, Yen L, Lee C, Xia K, Yan Y, Yu H. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Dev Cell. 2010:19(6):884–894. 10.1016/j.devcel.2010.10.024 [DOI] [PubMed] [Google Scholar]
- Howe GA, Major IT, Koo AJ. Modularity in jasmonate signaling for multistress resilience. Annu Rev Plant Biol. 2018:69(1):387–415. 10.1146/annurev-arplant-042817-040047 [DOI] [PubMed] [Google Scholar]
- Hu B, Chu C. Nitrogen-phosphorus interplay: old story with molecular tale. New Phytol. 2020:225(4):1455–1460. 10.1111/nph.16102 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang Y, Han X, Wang H, Pan J, Yu D. Jasmonate regulates leaf senescence and tolerance to cold stress: crosstalk with other phytohormones. J Exp Bot. 2017:68(6):1361–1369. 10.1093/jxb/erx004 [DOI] [PubMed] [Google Scholar]
- Hu B, Jiang Z, Wang W, Qiu Y, Zhang Z, Liu Y, Li A, Gao X, Liu L, Qian Y, et al. . Nitrate-NRT1.1B-SPX4 cascade integrates nitrogen and phosphorus signalling networks in plants. Nat Plants. 2019:5(4):401–413. 10.1038/s41477-019-0384-1 [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D. Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis. Plant Cell. 2013:25(8):2907–2924. 10.1105/tpc.113.112631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu S, Yu K, Yan J, Shan X, Xie D. Jasmonate perception: ligand-receptor interaction, regulation, and evolution. Mol Plant. 2023:16(1):23–42. 10.1016/j.molp.2022.08.011 [DOI] [PubMed] [Google Scholar]
- Huang H, Liu B, Liu L, Song S. Jasmonate action in plant growth and development. J Exp Bot. 2017:68(6):1349–1359. 10.1093/jxb/erw495 [DOI] [PubMed] [Google Scholar]
- Huang K-L, Ma G-J, Zhang M-L, Xiong H, Wu H, Zhao C-Z, Liu C-S, Jia H-X, Chen L, Kjorven JO, et al. . The ARF7 and ARF19 transcription factors positively regulate PHOSPHATE STARVATION RESPONSE1 in Arabidopsis roots. Plant Physiol. 2018:178(1):413–427. 10.1104/pp.17.01713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia Z, Giehl RFH, von Wirén N. Nutrient-hormone relations: driving root plasticity in plants. Mol Plant. 2022:15(1):86–103. 10.1016/j.molp.2021.12.004 [DOI] [PubMed] [Google Scholar]
- Jiang C, Gao X, Liao L, Harberd NP, Fu X. Phosphate starvation root architecture and anthocyanin accumulation responses are modulated by the Gibberellin-DELLA signaling pathway in Arabidopsis. Plant Physiol. 2007:145(4):1460–1470. 10.1104/pp.107.103788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Liang G, Yang S, Yu D. Arabidopsis WRKY57 functions as node of convergence for jasmonic acid and auxin-mediated signaling in jasmonic acid-induced leaf senescence. Plant Cell. 2014:26(1):230–245. 10.1105/tpc.113.117838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju L, Jing Y, Shi P, Liu J, Chen J, Yan J, Chu J, Chen K, Sun J. JAZ proteins modulate seed germination through interacting with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019:223(1):246–260. 10.1111/nph.15757 [DOI] [PubMed] [Google Scholar]
- Kant S, Peng M, Rothstein SJ. Genetic regulation by NLA and microRNA827 for maintaining nitrate-dependent phosphate homeostasis in Arabidopsis. PLoS Genet. 2011:7(3):e1002021. 10.1371/journal.pgen.1002021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Chung HS, Koo AJ, Howe GA. Jasmonate signaling: a conserved mechanism of hormone sensing. Curr Opin Plant Biol. 2008:11(4):428–435. 10.1016/j.pbi.2008.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. MYC2: the master in action. Mol Plant. 2013:6(3):686–703. 10.1093/mp/sss128 [DOI] [PubMed] [Google Scholar]
- Khan GA, Vogiatzaki E, Glauser G, Poirier Y. Phosphate deficiency induces the jasmonate pathway and enhances resistance to insect herbivory. Plant Physiol. 2016:171(1):632–644. 10.1104/pp.16.00278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KC, Lai Z, Fan B, Chen Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell. 2008:20(9):2357–2371. 10.1105/tpc.107.055566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y, Wang G, Chen X, Li L, Zhang X, Chen S, He Y, Hong G. OsPHR2 modulates phosphate starvation-induced OsMYC2 signalling and resistance to Xanthomonas oryzae pv. oryzae. Plant Cell Environ. 2021:44(10):3432–3444. 10.1111/pce.14078 [DOI] [PubMed] [Google Scholar]
- Kumar M, Pandya-Kumar N, Kapulnik Y, Koltai H. Strigolactone signaling in root development and phosphate starvation. Plant Signal Behav. 2015:10(7):e1045174. 10.1080/15592324.2015.1045174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei K-J, Lin Y-M, Ren J, Bai L, Miao Y-C, An G-Y, Song C-P. Modulation of the phosphate-deficient responses by MicroRNA156 and its targeted SQUAMOSA PROMOTER BINDING PROTEIN-LIKE 3 in Arabidopsis. Plant Cell Physiol. 2016:57(1):192–203. 10.1093/pcp/pcv197 [DOI] [PubMed] [Google Scholar]
- Li Y, Li S, Du R, Wang J, Li H, Xie D, Yan J. Isoleucine enhances plant resistance against Botrytis cinerea via jasmonate signaling pathway. Front. Plant Sci. 2021b:12:628328. 10.3389/fpls.2021.628328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Yu G, Cao C, Liu P. Metabolism, signaling, and transport of jasmonates. Plant Commun. 2021a:2(5):100231. 10.1016/j.xplc.2021.100231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin W-Y, Huang T-K, Chiou T-J. Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell. 2013:25(10):4061–4074. 10.1105/tpc.113.116012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Du M, Deng L, Shen J, Fang M, Chen Q, Lu Y, Wang Q, Li C, Zhai Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell. 2019:31(1):106–127. 10.1105/tpc.18.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F, Ni W, Griffith ME, Huang Z, Chang C, Peng W, Ma H, Xie D. The ASK1 and ASK2 genes are essential for Arabidopsis early development. Plant Cell. 2004:16(1):5–20. 10.1105/tpc.017772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Wu X, Wang E, Liu Y, Wang Y, Zheng Q, Han Y, Chen Z, Zhang Y. PHR1 positively regulates phosphate starvation-induced anthocyanin accumulation through direct upregulation of genes F3'h and LDOX in Arabidopsis. Planta. 2022:256(2):42. 10.1007/s00425-022-03952-w [DOI] [PubMed] [Google Scholar]
- Liu Y, Xie Y, Wang H, Ma X, Yao W, Wang H. Light and ethylene coordinately regulate the phosphate starvation response through transcriptional regulation of PHOSPHATE STARVATION SPONSE1. Plant Cell. 2017:29(9):2269–2284. 10.1105/tpc.17.00268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Arredondo DL, Leyva-González MA, González-Morales SI, López-Bucio J, Herrera-Estrella L. Phosphate nutrition: improving low-phosphate tolerance in crops. Annu Rev Plant Biol. 2014:65(1):95–123. 10.1146/annurev-arplant-050213-035949 [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell. 2004:16(7):1938–1950. 10.1105/tpc.022319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Q, Zhong Y, Wang Y, Wang Z, Zhang L, Shi J, Wu Z, Liu Y, Mao C, Yi K, et al. . SPX4 negatively regulates phosphate signaling and homeostasis through its interaction with PHR2 in rice. Plant Cell. 2014:26(4):1586–1597. 10.1105/tpc.114.123208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao Y-B, Liu Y-Q, Chen D-Y, Chen F-Y, Fang X, Hong G-J, Wang L-J, Wang J-W, Chen X-Y. Jasmonate response decay and defense metabolite accumulation contributes to age-regulated dynamics of plant insect resistance. Nat Commun. 2017:8(1):13925. 10.1038/ncomms13925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marschner. Mineral nutrition of higher plants. J Ecol. 1995:76:681–861. 10.1146/annurev.pp.31.060180.001323 [DOI] [Google Scholar]
- Martín AC, del Pozo JC, Iglesias J, Rubio V, Solano R, de La Peña A, Leyva A, Paz-Ares J. Influence of cytokinins on the expression of phosphate starvation responsive genes in Arabidopsis. Plant J. 2000:24(5):559–567. 10.1046/j.1365-313x.2000.00893.x [DOI] [PubMed] [Google Scholar]
- Mayzlish-Gati E, De-Cuyper C, Goormachtig S, Beeckman T, Vuylsteke M, Brewer PB, Beveridge CA, Yermiyahu U, Kaplan Y, Enzer Y, et al. . Strigolactones are involved in root response to low phosphate conditions in Arabidopsis. Plant Physiol. 2012:160(3):1329–1341. 10.1104/pp.112.202358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medici A, Szponarski W, Dangeville P, Safi A, Dissanayake IM, Saenchai C, Emanuel A, Rubio V, Lacombe B, Ruffel S, et al. . Identification of molecular integrators shows that nitrogen actively controls the phosphate starvation response in plants. Plant Cell. 2019:31(5):1171–1184. 10.1105/tpc.18.00656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei S, Zhang M, Ye J, Du J, Jiang Y, Hu Y. Auxin contributes to jasmonate-mediated regulation of abscisic acid signaling during seed germination in Arabidopsis. Plant Cell. 2023:35(3):1110–1133. 10.1093/plcell/koac362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miura K, Rus A, Sharkhuu A, Yokoi S, Karthikeyan AS, Raghothama KG, Baek D, Koo YD, Jin JB, Bressan RA, et al. . The Arabidopsis SUMO E3 ligase SIZ1 controls phosphate deficiency responses. Proc Natl Acad Sci U S A. 2005:102(21):7760–7765. 10.1073/pnas.0500778102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morcuende R, Bari R, Gibon Y, Zheng W, Pant BD, Bläsing O, Usadel B, Czechowski T, Udvardi MK, Stitt M, et al. . Genome-wide reprogramming of metabolism and regulatory networks of Arabidopsis in response to phosphorus. Plant Cell Environ. 2007:30(1):85–112. 10.1111/j.1365-3040.2006.01608.x [DOI] [PubMed] [Google Scholar]
- Mudge SR, Rae AL, Diatloff E, Smith FW. Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 2002:31(3):341–353. 10.1046/j.1365-313X.2002.01356.x [DOI] [PubMed] [Google Scholar]
- Mukhopadhyay A, Deplancke B, Walhout AJM, Tissenbaum HA. Chromatin immunoprecipitation (ChIP) coupled to detection by quantitative real-time PCR to study transcription factor binding to DNA in Caenorhabditis elegans. Nat Protoc. 2008:3(4):698–709. 10.1038/nprot.2008.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller J, Toev T, Heisters M, Teller J, Moore KL, Hause G, Dinesh DC, Bürstenbinder K, Abel S. Iron-dependent callose deposition adjusts root meristem maintenance to phosphate availability. Dev Cell. 2015:33(2):216–230. 10.1016/j.devcel.2015.02.007 [DOI] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. Plant Cell. 2013:25(5):1641–1656. 10.1105/tpc.113.111112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signaling. Curr Biol. 2008:18(9):650–655. 10.1016/j.cub.2008.03.060 [DOI] [PubMed] [Google Scholar]
- Navarro C, Mateo-Elizalde C, Mohan TC, Sánchez-Bermejo E, Urrutia O, Fernández-Muñiz MN, García-Mina JM, Muñoz R, Paz-Ares J, Castrillo G, et al. . Arsenite provides a selective signal that coordinates arsenate uptake and detoxification through the regulation of PHR1 stability in Arabidopsis. Mol Plant. 2021:14(9):1489–1507. 10.1016/j.molp.2021.05.020 [DOI] [PubMed] [Google Scholar]
- Nilsson L, Müller R, Nielsen TH. Increased expression of the MYB-related transcription factor, PHR1, leads to enhanced phosphate uptake in Arabidopsis thaliana. Plant Cell Environ. 2007:30(12):1499–1512. 10.1111/j.1365-3040.2007.01734.x [DOI] [PubMed] [Google Scholar]
- Niu YJ, Figueroa P, Browse J. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J Exp Bot. 2011:62(6):2143–2154. 10.1093/jxb/erq408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nussaume L, Kanno S, Javot H, Marin E, Pochon N, Ayadi A, Nakanishi TM, Thibaud MC. Phosphate import in plants: focus on the PHT1 transporters. Front Plant Sci. 2011:2:83. 10.3389/fpls.2011.00083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osorio MB, Ng S, Berkowitz O, De Clercq I, Mao C, Shou H, Whelan J, Jost R. SPX4 acts on PHR1-dependent and -independent regulation of shoot phosphorus status in Arabidopsis. Plant Physiol. 2019:181(1):332–352. 10.1104/pp.18.00594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J, Hu Y, Wang H, Guo Q, Chen Y, Howe GA, Yu D. Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell. 2020:32(12):3846–3865. 10.1105/tpc.19.00838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey BK, Verma L, Prusty A, Singh AP, Bennett MJ, Tyagi AK, Giri J, Mehra P. OsJAZ11 regulates phosphate starvation responses in rice. Planta. 2021:254(1):8. 10.1007/s00425-021-03657-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paz-Ares J, Puga MI, Rojas-Triana M, Martinez-Hevia I, Diaz S, Poza-Carrion C, Minambres M, Leyva A. Plant adaptation to low phosphorus availability: core signaling, crosstalks, and applied implications. Mol Plant. 2022:15(1):104–124. 10.1016/j.molp.2021.12.005 [DOI] [PubMed] [Google Scholar]
- Péret B, Clément M, Nussaume L, Desnos T. Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 2011:16(8):442–450. 10.1016/j.tplants.2011.05.006 [DOI] [PubMed] [Google Scholar]
- Perez-Torres CA, López-Bucio J, Cruz-Ramírez A, Ibarra-Laclette E, Dharmasiri S, Estelle M, Herrera-Estrella L. Phosphate availability alters lateral root development in Arabidopsis by modulating auxin sensitivity via a mechanism involving the TIR1 auxin receptor. Plant Cell. 2008:20(12):3258–3272. 10.1105/tpc.108.058719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Mateos I, Charukesi R, Wang Z, Franco-Zorrilla JM, de Lorenzo L, Irigoyen ML, Masiero S, Bustos R, Rodríguez J, et al. . SPX1 is a phosphate-dependent inhibitor of Phosphate Starvation Response 1 in Arabidopsis. Proc Natl Acad Sci U S A. 2014:111(41):14947–14952. 10.1073/pnas.1404654111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puga MI, Rojas-Triana M, de Lorenzo L, Leyva A, Rubio V, Paz-Ares J. Novel signals in the regulation of Pi starvation responses in plants: facts and promises. Curr Opin Plant Biol. 2017:39:40–49. 10.1016/j.pbi.2017.05.007 [DOI] [PubMed] [Google Scholar]
- Qi T, Huang H, Song S, Xie D. Regulation of jasmonate-mediated stamen development and seed production by a bHLH-MYB complex in Arabidopsis. Plant Cell. 2015:27(6):1620–1633. 10.1105/tpc.15.00116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell. 2014:26(3):1118–1133. 10.1105/tpc.113.121731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. The jasmonate-ZIM-domain proteins interact with the WD-repeat/bHLH/MYB complexes to regulate jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell. 2011:23(5):1795–1814. 10.1105/tpc.111.083261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghothama KG. Phosphate acquisition. Annu Rev Plant Physiol Plant Mol Biol. 1999:50(1):665–693. 10.1146/annurev.arplant.50.1.665 [DOI] [PubMed] [Google Scholar]
- Ren C, Pan J, Peng W, Genschik P, Hobbie L, Hellmann H, Estelle M, Gao B, Peng J, Sun C, et al. . Point mutations in Arabidopsis Cullin1 reveal its essential role in jasmonate response. Plant J. 2005:42(4):514–524. 10.1111/j.1365-313X.2005.02394.x [DOI] [PubMed] [Google Scholar]
- Ribot C, Zimmerli C, Farmer EE, Reymond P, Poirier Y. Induction of the Arabidopsis PHO1; H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiol. 2008:147(2):696–706. 10.1104/pp.108.119321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ried MK, Wild R, Zhu J, Pipercevic J, Sturm K, Broger L, Harmel RK, Abriata LA, Hothorn LA, Fiedler D, et al. . Inositol pyrophosphates promote the interaction of SPX domains with the coiled-coil motif of PHR transcription factors to regulate plant phosphate homeostasis. Nat Commun. 2021:12(1):384. 10.1038/s41467-020-20681-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan W, Guo M, Cai L, Hu H, Li C, Liu Y, Wu Z, Mao C, Yi K, Wu P, et al. . Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol Biol. 2015:87(4-5):429–440. 10.1007/s11103-015-0289-y [DOI] [PubMed] [Google Scholar]
- Rubio V, Linhares F, Solano R, Martín AC, Iglesias J, Leyva A, Paz-Ares J. A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 2001:15(16):2122–2133. 10.1101/gad.204401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiol. 2013:163(1):291–304. 10.1104/pp.113.220129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweizer F, Fernández-Calvo P, Zander M, Diez-Diaz M, Fonseca S, Glauser G, Lewsey MG, Ecker JR, Solano R, Reymond P. Arabidopsis basic helix-loop-helix transcription factors MYC2, MYC3, and MYC4 regulate glucosinolate biosynthesis, insect performance, and feeding behavior. Plant Cell. 2013:25(8):3117–3132. 10.1105/tpc.113.115139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sega P, Pacak A. Plant PHR transcription factors: put on a map. Genes (Basel). 2019:10(12):1018. 10.3390/genes10121018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano-Bueno G, de Los Reyes P, Chini A, Ferreras-Garrucho G, Sánchez de Medina-Hernández V, Boter M, Solano R, Valverde F. Regulation of floral senescence in Arabidopsis by coordinated action of CONSTANS and jasmonate signaling. Mol Plant. 2022:15(11):1710–1724. 10.1016/j.molp.2022.09.017 [DOI] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, Withers J, Ben-Nissan G, Hinds TR, Kobayashi Y, Hsu F-F, Sharon M, Browse J, et al. . Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature. 2010:468(7322):400–405. 10.1038/nature09430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheen J. Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 2001:127(4):1466–1475. 10.1104/pp.010820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin H, Shin H-S, Dewbre GR, Harrison MJ. Phosphate transport in Arabidopsis: Pht1; 1 and Pht1; 4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 2004:39(4):629–642. 10.1111/j.1365-313X.2004.02161.x [DOI] [PubMed] [Google Scholar]
- Song S, Liu B, Song J, Pang S, Song T, Gao S, Zhang Y, Huang H, Qi T. A molecular framework for signaling crosstalk between jasmonate and ethylene in anthocyanin biosynthesis, trichome development, and defenses against insect herbivores in Arabidopsis. J Integr Plant Biol. 2022:64(9):1770–1788. 10.1111/jipb.13319 [DOI] [PubMed] [Google Scholar]
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genet. 2013:9(7):e1003653. 10.1371/journal.pgen.1003653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. Plant Cell. 2011:23(3):1000–1013. 10.1105/tpc.111.083089 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Wasternack C, Xie D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr Opin Plant Biol. 2014:21:112–119. 10.1016/j.pbi.2014.07.005 [DOI] [PubMed] [Google Scholar]
- Spoel SH, Johnson JS, Dong X. Regulation of tradeoffs between plant defenses against pathogens with different lifestyles. Proc Natl Acad Sci U S A. 2007:104(47):18842–18847. 10.1073/pnas.0708139104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux J-P, Brown R, Kazan K, et al. . NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003:15(3):760–770. 10.1105/tpc.009159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanovic A, Ribot C, Rouached H, Wang Y, Chong J, Belbahri L, Delessert S, Poirier Y. Members of the PHO1 gene family show limited functional redundancy in phosphate transfer to the shoot, and are regulated by phosphate deficiency via distinct pathways. Plant J. 2007:50(6):982–994. 10.1111/j.1365-313X.2007.03108.x [DOI] [PubMed] [Google Scholar]
- Sun L, Song L, Zhang Y, Zheng Z, Liu D. Arabidopsis PHL2 and PHR1 act redundantly as the key components of the central regulatory system controlling transcriptional responses to phosphate starvation. Plant Physiol. 2016:170(1):499–514. 10.1104/pp.15.01336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Xu Y, Ye S, Jiang H, Chen Q, Liu F, Zhou W, Chen R, Li X, Tietz O, et al. . Arabidopsis ASA1 is important for jasmonate-mediated regulation of auxin biosynthesis and transport during lateral root formation. Plant Cell. 2009:21(5):1495–1511. 10.1105/tpc.108.064303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J, Wu D, Li X, Wang L, Xu L, Zhang Y, Xu F, Liu H, Xie Q, Dai S, et al. . Plant immunity suppression via PHR1-RALF-FERONIA shapes the root microbiome to alleviate phosphate starvation. EMBO J. 2022:41(6):109102. 10.15252/embj.2021109102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibaud MC, Arrighi JF, Bayle V, Chiarenza S, Creff A, Bustos R, Paz-Ares J, Poirier Y, Nussaume L. Dissection of local and systemic transcriptional responses to phosphate starvation in Arabidopsis. Plant J. 2010:64(5):775–789. 10.1111/j.1365-313X.2010.04375.x [DOI] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature. 2007:448(7154):661–665. 10.1038/nature05960 [DOI] [PubMed] [Google Scholar]
- Ueda Y, Kiba T, Yanagisawa S. Nitrate-inducible NIGT1 proteins modulate phosphate uptake and starvation signalling via transcriptional regulation of SPX genes. Plant J. 2020:102(3):448–466. 10.1111/tpj.14637 [DOI] [PubMed] [Google Scholar]
- Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelock CE, Plaxton WC, Price CA, Scheible W-R, Shane MW, White PJ, et al. . Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol. 2012:195(2):306–320. 10.1111/j.1469-8137.2012.04190.x [DOI] [PubMed] [Google Scholar]
- Wang F, Deng M, Xu J, Zhu X, Mao C. Molecular mechanisms of phosphate transport and signaling in higher plants. Semin Cell Dev Biol. 2018:74:114–122. 10.1016/j.semcdb.2017.06.013 [DOI] [PubMed] [Google Scholar]
- Wang H, Li S, Li Y, Xu Y, Wang Y, Zhang R, Sun W, Chen Q, Wang X-J, Li C, et al. . MED25 connects enhancer-promoter looping and MYC2-dependent activation of jasmonate signaling. Nat Plants. 2019a:5(6):616–625. 10.1038/s41477-019-0441-9 [DOI] [PubMed] [Google Scholar]
- Wang H, Li Y, Pan J, Lou D, Hu Y, Yu D. The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol Plant. 2017:10(11):1461–1464. 10.1016/j.molp.2017.08.007 [DOI] [PubMed] [Google Scholar]
- Wang J, Wu D, Wang Y, Xie D. Jasmonate action in plant defense against insects. J Exp Bot. 2019b:70(13):3391–3400. 10.1093/jxb/erz174 [DOI] [PubMed] [Google Scholar]
- Wang H, Xu Q, Kong Y-H, Chen Y, Duan J-Y, Wu W-H, Chen Y-F. Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1; 1 expression in response to phosphate starvation. Plant Physiol. 2014:164(4):2020–2029. 10.1104/pp.113.235077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. Determination of sex by jasmonate. J Integr Plant Biol. 2020:62(2):162–164. 10.1111/jipb.12840 [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. Ann Bot. 2013:111(6):1021–1058. 10.1093/aob/mct067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wege S, Khan GA, Jung J-Y, Vogiatzaki E, Pradervand S, Aller I, Meyer AJ, Poirier Y. The EXS domain of PHO1 participates in the response of shoots to phosphate deficiency via a root-to-shoot signal. Plant Physiol. 2016:170(1):385–400. 10.1104/pp.15.00975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild R, Gerasimaite R, Jung J-Y, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, et al. . Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science. 2016:352(6288):986–990. 10.1126/science.aad9858 [DOI] [PubMed] [Google Scholar]
- Wu F, Deng L, Zhai Q, Zhao J, Chen Q, Li C. Mediator subunit MED25 couples alternative splicing of JAZ genes with fine-tuning of jasmonate signaling. Plant Cell. 2020:32(2):429–448. 10.1105/tpc.19.00583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu P, Shou H, Xu G, Lian X. Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol. 2013:16(2):205–212. 10.1016/j.pbi.2013.03.002 [DOI] [PubMed] [Google Scholar]
- Wykoff DD, Grossman AR, Weeks DP, Usuda H, Shimogawara K. Psr1, a nuclear localized protein that regulates phosphorus metabolism in Chlamydomonas. Proc Natl Acad Sci U S A. 1999:96(26):15336–15341. 10.1073/pnas.96.26.15336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie D-X, Feys BF, James S, Nieto-Rostro M, Turner JG. COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science. 1998:280(5366):1091–1094. 10.1126/science.280.5366.1091 [DOI] [PubMed] [Google Scholar]
- Xu L, Liu F, Lechner E, Genschik P, Crosby WL, Ma H, Peng W, Huang D, Xie D. The SCF(COI1) ubiqutin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell. 2002:14(8):1919–1935. 10.1105/tpc.003368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan S, Chen N, Huang Z, Li D, Zhi J, Yu B, Liu X, Cao B, Qiu Z. Anthocyanin fruit encodes an R2R3-MYB transcription factor, SlAN2-like, activating the transcription of SlMYBATV to fine-tune anthocyanin content in tomato fruit. New Phytol. 2020:225(5):2048–2063. 10.1111/nph.16272 [DOI] [PubMed] [Google Scholar]
- Yan J, Li H, Li S, Yao R, Deng H, Xie Q, Xie D. The Arabidopsis F-box protein CORONATINE INSENSITIVE1 is stabilized by SCFCOI1 and degraded via the 26S proteasome pathway. Plant Cell. 2013:25(2):486–498. 10.1105/tpc.112.105486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, Bai Z, Zhang W, Qi T, Cheng Z, Peng W, Luo H, Nan F, et al. . The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell. 2009:21(8):2220–2236. 10.1105/tpc.109.065730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang XJ, Finnegan PM. Regulation of phosphate starvation responses in higher plants. Ann Bot. 2010:105(4):513–526. 10.1093/aob/mcq015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Han X, Yang J, Jiang Y, Hu Y. The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell. 2021:33(9):3022–3041. 10.1093/plcell/koab168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Zhao X, Chen Y, Li G, Li X, Xia M, Sun Z, Chen Y, Li Y, Yao L, et al. . Identification, structural, and expression analyses of SPX genes in giant duckweed (Spirodela polyrhiza) reveals its role in response to low phosphorus and nitrogen stresses. Cells. 2022:11(7):1167. 10.3390/cells11071167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo S-D, Cho Y-H, Sheen J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat Protoc. 2007:2(7):1565–1572. 10.1038/nprot.2007.199 [DOI] [PubMed] [Google Scholar]
- You Y, Zhai Q, An C, Li C. LEUNIG_HOMOLOG mediates MYC2-dependent transcriptional activation in cooperation with the coactivators HAC1 and MED25. Plant Cell. 2019:31(9):2187–2205. 10.1105/tpc.19.00115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu D. Signaling components involved in plant responses to phosphate starvation. J Integr Plant Biol. 2008:50(7):849–859. 10.1111/j.1744-7909.2008.00709.x [DOI] [PubMed] [Google Scholar]
- Zander M, Lewsey MG, Clark NM, Yin L, Bartlett A, Saldierna GJP, Hann E, Langford AE, Jow B, Wise A, et al. . Integrated multi-omics framework of the plant response to jasmonic acid. Nat Plants. 2020:6(3):290–302. 10.1038/s41477-020-0605-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Deng L, Li C. Mediator subunit MED25: at the nexus of jasmonate signaling. Curr Opin Plant Biol. 2020:57:78–86. 10.1016/j.pbi.2020.06.006 [DOI] [PubMed] [Google Scholar]
- Zhai Q, Yan L, Tan D, Chen R, Sun J, Gao L, Dong M-Q, Wang Y, Li C. Phosphorylation-coupled proteolysis of the transcription factor MYC2 is important for jasmonate-signaled plant immunity. PLoS Genet. 2013:9(4):e1003422. 10.1371/journal.pgen.1003422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. Plant Cell. 2015:27(10):2814–2828. 10.1105/tpc.15.00619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, De Stefano R, Schoonbeek H-J, Magusin A, Pagliarani C, Wellner N, Hill L, Orzaez D, Granell A, et al. . Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr Biol. 2013:23(12):1094–1100. 10.1016/j.cub.2013.04.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Liao H, Lucas WJ. Molecular mechanisms underlying phosphate sensing, signaling, and adaptation in plants. J Integr Plant Biol. 2014:56(3):192–220. 10.1111/jipb.12163 [DOI] [PubMed] [Google Scholar]
- Zhang L, Zhang F, Melotto M, Yao J, He SY. Jasmonate signaling and manipulation by pathogens and insects. J Exp Bot. 2017:68(6):1371–1385. 10.1093/jxb/erw478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P, Wang L, Yin H. Transcriptional responses to phosphate starvation in Brachypodium distachyon roots. Plant Physiol Biochem. 2018:122:113–120. 10.1016/j.plaphy.2017.11.010 [DOI] [PubMed] [Google Scholar]
- Zhou W, Lozano-Torres JL, Blilou I, Zhang X, Zhai Q, Smant G, Li C, Scheres B. A jasmonate signaling network activates root stem cells and promotes regeneration. Cell. 2019:177(4):942–956. 10.1016/j.cell.2019.03.006 [DOI] [PubMed] [Google Scholar]
- Zhu Z, An F, Feng Y, Li P, Xue L, A M, Jiang Z, Kim J-M, To TK, Li W, et al. . Derepression of ethylene-stabilized transcription factors (EIN3/EIL1) mediates jasmonate and ethylene signaling synergy in Arabidopsis. Proc Natl Acad Sci USA. 2011:108(30):12539–12544. 10.1073/pnas.1103959108 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data supporting the findings of this study are available within the article and its supplementary materials.