Summary
Background
Matricellular proteins comprising matrisome and adhesome are responsible for structure integrity and interactions between cells in the tumour microenvironment of breast cancer. Changes in the gene expression of matrisome and adhesome augment metastasis. Since inflammatory breast cancer (IBC) is characterized by high metastatic behaviour. Herein, we compared the gene expression profile of matrisome and adhesome in non-IBC and IBC in fresh tissue and ex vivo patient-derived explants (PDEs) and we also compared the secretory inflammatory mediators of PDEs in non-IBC and IBC to identify secretory cytokines participate in cross-talk between cells via interactions with matrisome and adhisome.
Methods
Fifty patients (31 non-IBC and 19 IBC) were enrolled in the present study. To test their validation in clinical studies, PDEs were cultured as an ex vivo model. Gene expression and cytokine array were used to identify candidate genes and cytokines contributing to metastasis in the examined fresh tissues and PDEs. Bioinformatics analysis was applied on identified differentially expressed genes using GeneMANIA and Metascape gene annotation and analysis resource to identify pathways involved in IBC metastasis.
Results
Normal and cancer fresh tissues and PDEs of IBC were characterized by overexpression of CDH1 and MMP14 and downregulation of CTNNA1 and TIMP1 compared with non-IBC. The secretome of IBC cancer PDEs is characterized by significantly high expression of interleukin 6 and monocyte chemoattractant protein-1 (CCL2) compared with non-IBC.
Conclusion
Genes expressed by adhisome and matrisome play a significant role in IBC metastasis and should be considered novel target therapy.
Introduction
Breast cancer incidence is increasing globally in the past decade with high mortality rate in low-income countries.1 Inflammatory breast cancer (IBC) is the highly aggressive and metastatic subtype of breast cancer. The median age of patients diagnosed with IBC is ∼5.25 years younger compared with non-IBC patients with an overall survival rate less than 4 years.2 Moreover, in the Middle East and northern Africa, IBC prevalence reached 10% compared with other parts of the world.3 Thus, the aggressiveness of IBC warrants a collective effort to better understand its distinctive biology, which will help in developing novel targeted therapies.
Many genomic studies were conducted to discover novel biological targets in IBC but have not led to the successful discovery of distinct mechanisms.4 However, mechanisms of IBC carcinogenesis, formation of tumor emboli and upregulated signalling pathways found to be linked with TME cellular interactions via matrisome, adhesome and cytokinome.5,6,7
Patient-derived explants (PDEs) representing the ex vivo culture of segments for the freshly resected tumour tissue preserve matrisome and adhesome organization that retains the histological features of original tumour. PDE models mimic the TME without any destruction of fresh samples obtained during surgery and are used now to study cancer progression and to assess drug responses.8 Herein, we compared genomic profile of matrisome and adhesome in fresh tissue and PDEs of non-IBC and IBC patients. Furthermore, we cultured PDEs for 48 h to test their viability and their secretory molecules, and confirm ability to preserve phenotype and genotype similar to the excised fresh tissue to be used as preclinical model for drug testing.
Materials and methods
Patients’ recruitment and selection
This study was approved by the ethics committee of Ain Shams University (IRB#00006379) and all participants signed a consent form. Diagnosis of IBC and non-IBC was performed as described before.9,10 Fifty breast cancer patients were participated in this study and grouped into non-IBC (n = 31) and IBC (n = 19).
Collection of fresh tissue and preparation of PDEs
Fresh tissues were divided into two parts once excised from patients during Modified Radical Mastectomy (MRM) part preserved in RNAlater (ThermoFisher Scientific, MA, USA) and part used in preparation of PDEs. The PDEs were prepared from tissue as described by Sineh Sepehr et al.11 with slight modifications. In brief, excised cancer tissues were placed in the lab into a transferring medium (Dulbecco’s Modified Eagle Medium [DMEM] with 1% penicillin/streptomycin antibiotic solution) (Lonza, Basel, Switzerland), and the adipose tissues, blood vessels and muscular tissues surrounding the cancer mass were removed during dissection. Normal breast tissues were homogenized from glandular tissues (mostly breast ducts surrounded by connective, fibrous and adipose tissues). Subsequently, PDEs of normal and cancer tissues were washed with DMEM/F-12 supplemented with 10% foetal bovine serum (FBS), 100 IU/ml penicillin, 100 μg/ml streptomycin and 2% l-glutamine. Each tissue sample was placed in a 700-μL growth medium (Lonza) in a Falcon organ culture dish (BD Biosciences, NJ, USA) and incubated for 48 h at 37°C in a humidified CO2 incubator for further studies. It should be noted that the size of the excised tissue and cell viability are appropriate for the preparation of PDEs.
Cell viability and proliferation assay
To test whether culturing of PDEs in growth media will affect cell viability and proliferation, we used MTT [3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyl-2H-tetrazolium bromide] as we described before.12 PDE specimens were mechanically dissociated using a scalpel, transferred to a DMEM solution containing 20 mg/ml Gibco collagenase I (ThermoFisher Scientific) and incubated at 37°C in a humidified CO2 incubator. PDE specimens dissociated into single cells were washed with phosphate-buffered saline and cell pellets were collected by centrifugation at 1500 rpm for 5 min. Isolated cells were seeded at a density of 4.0 × 103 cells in 100 μl DMEM/F-12 culture medium per well (96 wells plate) and incubated for 48 h at 37°C in a humidified CO2 incubator. After that, 10 μl of 5 mg/ml MTT was added to each well along with a serum-free medium and incubated for 4 h at 37°C. Then MTT was removed and 100 μl dimethyl sulfoxide was added to each well. The absorbance was measured at 570 nm wavelength.
Human extracellular matrix and adhesion molecules polymerase chain reaction array
Total RNA was purified from fresh tissue and PDEs using QIAzol lysis reagent (Qiagen, Hilden, Germany). The total RNA concentration was measured by Multiskan SkyHigh microplate spectrophotometer (ThermoFisher Scientific) and RNA integrity was tested by separating the RNA on a 1% standard agarose gel and examining the ribosomal RNA bands. One microgram of RNA was transcribed into complementary DNA (cDNA) using a High-Capacity cDNA Reverse Transcription Kit (ThermoFisher Scientific). To study the gene expression profiles of the extracellular matrix (ECM) and cell adhesion molecules, we used RT2 ECM and adhesion molecule polymerase chain reaction (PCR) array (Qiagen) as we recently described.13 Amplification specificity was verified using melting curve analysis. Data were analysed using the Qiagen Gene globe web tool (https://geneglobe.qiagen.com/analyze/) after normalization to ACTB, B2M, GAPDH, HPRT1 and RPLP0 housekeeping genes.
Human cytokine antibody array
Cytokines profiling of the secretome of IBC versus non-IBC PDEs was characterized quantitatively using the human cytokine array c3 kit (RayBiotech Life, GA, USA) as we described before.14 The relative density of each cytokine was analysed using densitometric methods in ImageJ software National Institutes of Health (NIH) (Bethesda, MD, USA).15,16
Enrichment analysis of differentially expressed genes
Enrichment analysis of differentially expressed genes (DEGs) was analysed using GeneMANIA (http://genemania.org/) and Metascape (http://metascape.org/gp/index.html#/main/step1) online tools. Gene expression profiling for the studied samples was genotyped by clustering analysis and heat maps allying the values of the expressed genes and samples were created.13 Protein–protein interactions (PPIs) among input DEGs and neighbouring genes were extracted from the PPI data source and formed a PPI network. The molecular complex detection (MCODE) algorithm was then applied to this network to identify neighbourhoods where proteins are densely connected.17–20
Statistical analysis
The statistical package of the Social Sciences software, version 22.0 was used for data analysis. The data were input as the mean ± SD. Shapiro–Wilk test was used to prove the normality of data distribution. Chi-square and t-tests were used for normally distributed data, whereas the Mann–Whitney U-test was used for data that were not normally distributed. The P-values < 0.05 were considered statistically significant.
Results
Clinical and pathological characterization of breast cancer patients
Table 1 describes the patients’ clinicopathological properties. IBC patients were characterized by significantly larger tumour size (P = 0.01) and increased lymph node metastasis (P = 0.0001) than non-IBC patients.
Table 1.
Clinical and pathological characterization of non-IBC versus IBC patients
| Characteristic | Non-IBC | IBC | P-value |
|---|---|---|---|
| (N = 31) | (N = 19) | ||
| Age (year) | |||
| Range | 29–80 | 39–69 | 0.384a |
| Mean ± SD | 49.8 ± 11.3 | 54.3 ± 10.3 | |
| Tumour size (cm) | |||
| Mean ± SD | 3.7 ± 1.9 | 4.5 ± 2.9 | 0.01*b |
| ≤4 | 21 (67.7%) | 6 (31.6%) | |
| >4 | 10 (32.3%) | 13 (68.4%) | |
| Tumour grade | |||
| G1 | 1 (3.2%) | 1 (5.3%) | 0.413b |
| G2 | 22 (71%) | 16 (84.2%) | |
| G3 | 8 (25.8%) | 2 (10.2%) | |
| Axillary lymph node metastasis | |||
| Negative | 21 (67.7%) | 1 (5.3%) | 0.0001*b |
| Positive | 10 (32.3%) | 18 (94.7%) | |
| Lymphovascular invasion | |||
| Negative | 19 (61.3%) | 10 (52.6%) | 0.378b |
| Positive | 12 (38.7%) | 9 (47.4%) | |
| ER | |||
| Negative | 8 (25.8%) | 5 (26.3%) | 0.731b |
| Positive | 23 (74.2%) | 14 (73.7%) | |
| PR | |||
| Negative | 9 (29%) | 11 (57.9%) | 0.111b |
| Positive | 22 (71%) | 8 (42.1%) | |
| Her-2 | |||
| Negative | 23 (74.2%) | 13 (68.4%) | 0.726b |
| Positive | 8 (25.8%) | 6 (31.6%) |
Data are reported as means ± SD.
Student’s t-test.
Chi-square test.
Significant P-values (P < 0.05).
Growth media supplemented with FBS do not alter cell viability to non-IBC and IBC ex vivo normal and cancer PDEs
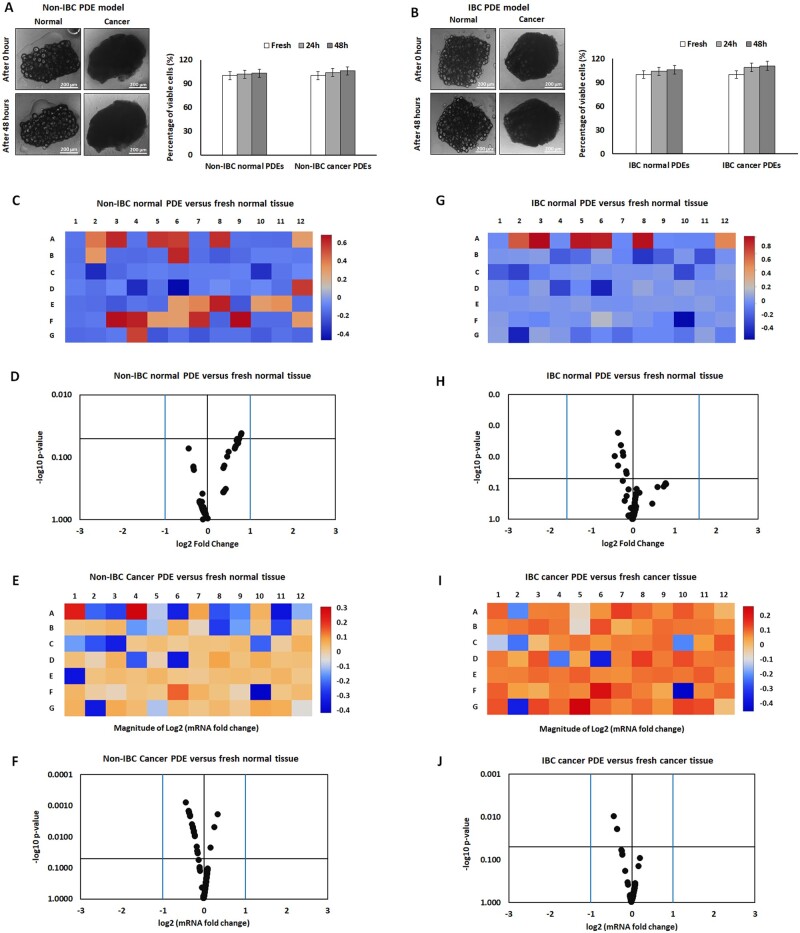
The results of the MTT assay showed no significant changes in cell viability of IBC normal and cancer PDEs seeded in growth media supplemented with nutrients and FBS (Figure 1A and B).
Figure 1.
Morphological and gene expression comparison between fresh normal and cancer tissues and their autologous PDEs collected from non-IBC and IBC patients. (A and B) Microscopic imaging and bars showed no significant changes in cell viability of fresh normal and cancer tissues and their autologous PDEs collected from non-IBC and IBC patients. (C–J) Heat maps and volcano diagrams showed no significant differences in the expression level of matrisome and adhisome genes in non-IBC and IBC fresh normal and cancer versus their autologous PDEs collected from non-IBC and IBC patients. P-values were calculated using Student’s t-test, where (P < 0.05) represented significance.
Alteration in the expression of matrisome and adhisome genes of fresh cancer tissues compared with cancer ex vivo PDEs in non-IBC and IBC patients
Quantitative PCR array results showed nonsignificant alterations in the level of expression of matrisome and adhisome genes between fresh normal and cancer tissues and their autologous PDEs after 48 h in both non-IBC and IBC patients (Figure 1C and J).
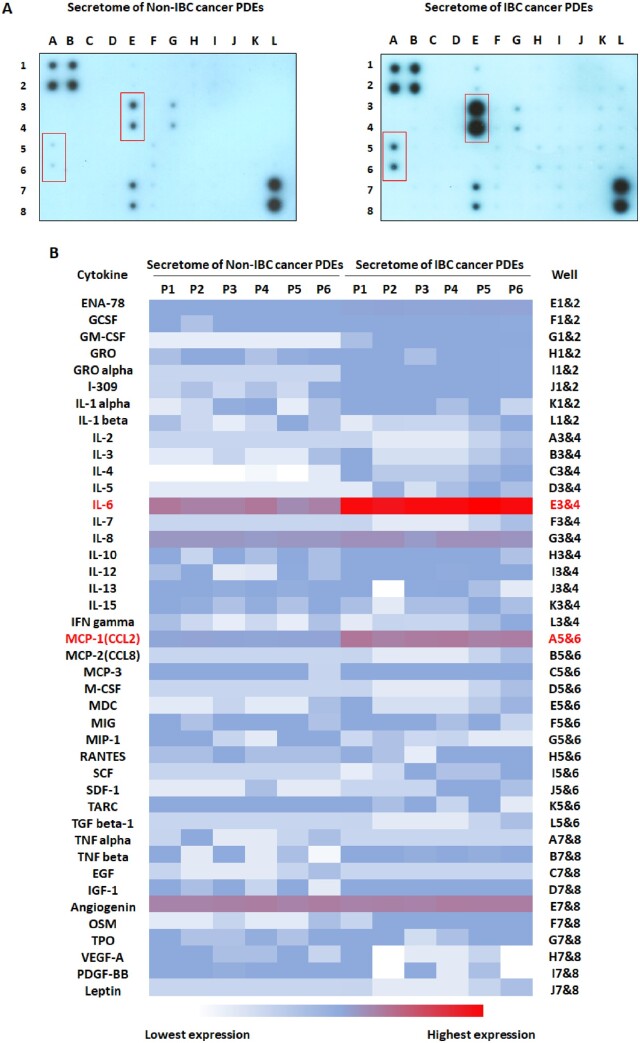
Profiling of cytokines secreted by non-IBC and IBC cancer PDEs
Culture media containing PDEs secretome was profiled by cytokine array. Results showed that the secretome of IBC cancer PDEs (n = 19) compared with non-IBC (n = 31) is characterized by significantly high expression levels of interleukin 6 (IL-6) (P = 0.01) and monocyte chemoattractant protein-1 (MCP-1/CCL2) (P = 0.04) (Figure 2).
Figure 2.
Profiling of cytokines in the secretome of non-IBC and IBC cancer PDEs. (A) Cytokine array detected differently expressed cytokines in the conditioned media of non-IBC and IBC cancer PDEs. (B) The heat map showed differently expressed cytokines in the conditioned media of non-IBC and IBC cancer PDEs. P-values were calculated using Student’s t-test, where (P < 0.05) represented significance.
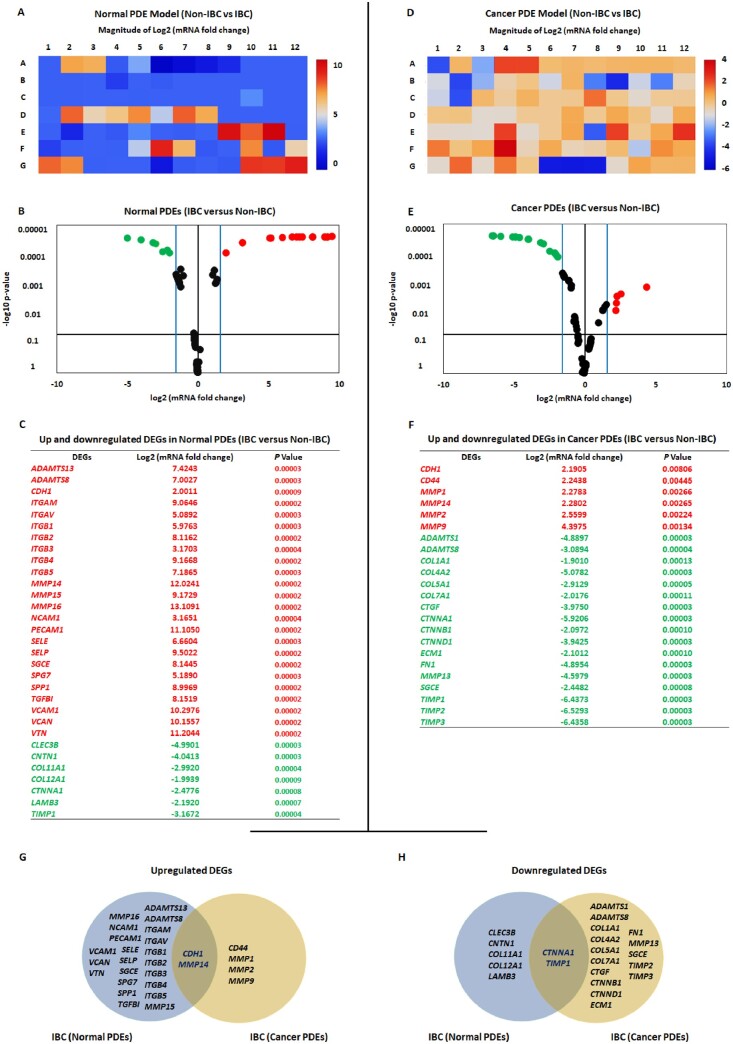
Differential expression of matrisome and adhesome genes in normal and cancer IBC PDEs compared with non-IBC PDEs
Considering genes showing fold change more than 3, the IBC-PDEs of normal tissues showed overexpression of 7 matrisome and 17 adhesome genes compared with non-IBC. Conversely, three matrisome and four adhisome genes were downregulated in IBC compared with non-IBC PDEs of normal tissues (Figure 3A–C). Cancer tissues of IBC-PDEs were characterized by overexpression of four matrisome and two adhesomes genes compared with non-IBC. The down-regulated genes were 11 matrisomes and 6 adhisomes in IBC compared with non-IBC PDEs of cancer tissues (Figure 3D–F). Comparing non-IBC with IBC DEGs, significantly upregulated genes in IBC were CDH1 and MMP14 (Figure 3G) and downregulated genes were CTNNA1 and TIMP1 (Figure 3H).
Figure 3.
Differentially expressed matrisome and adhisome genes in normal and cancer PDEs of IBC compared with non-IBC. (A and B) Heat map and volcano diagram showing significantly upregulated DEGs in IBC compared with non-IBC normal PDEs. (C) The table showing the Log2 mRNA fold change and P-values of significantly up- and downregulated DEGs in IBC compared with non-IBC normal PDEs. (D and E) Heat map and volcano diagram showing significantly upregulated DEGs in IBC compared with non-IBC cancer PDEs. (F) The table showing the Log2 mRNA fold change and P-values of significantly up- and downregulated DEGs in IBC compared with non-IBC cancer PDEs. (G and H) Venn diagrams show the overlap between up- and downregulated DEGs in normal and cancer IBC compared with non-IBC PDEs. Data are plotted as the mean ± SD. P-values were calculated using a t-test, with significance set at P < 0.05.
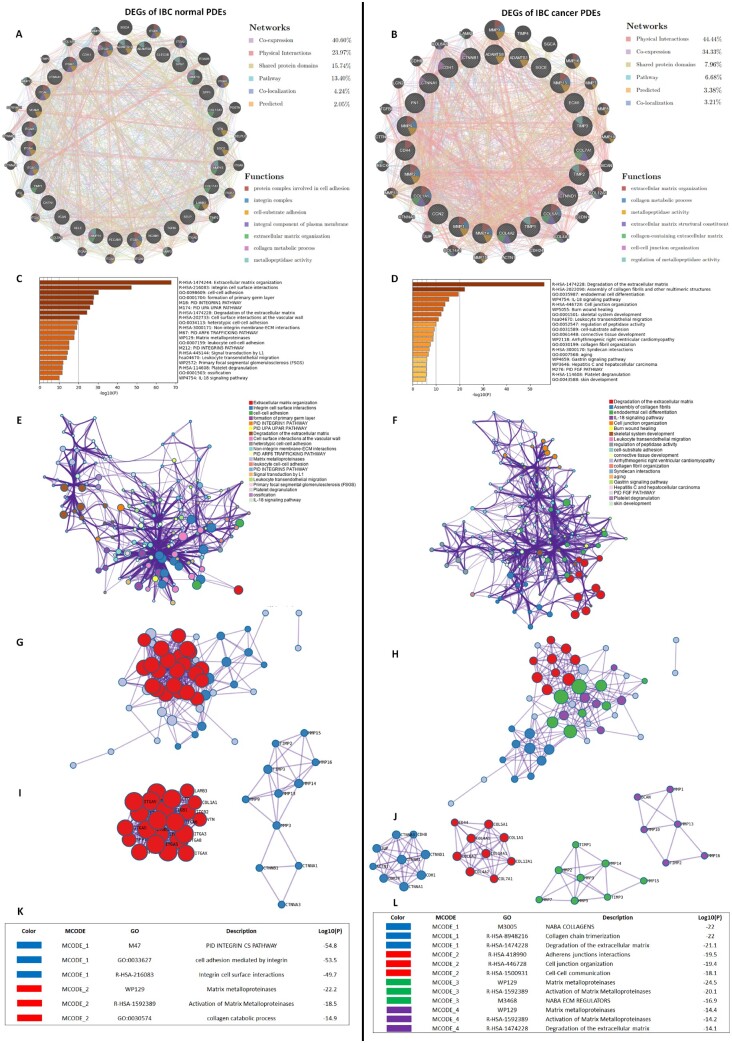
Functional enrichment analysis and PPI network construction
Through GeneMANIA analysis, we identified the top 25 neighbouring genes with the highest frequency association with differential expressed matrisome- and adhisome-related genes in IBC normal and cancer PDEs compared with non-IBC. GeneMANIA Network for IBC normal PDEs was based on 40.6% co-expression, 23.97% physical interactions and 15.74% shared protein domains, while for IBC cancer PDEs was based on 44.44% physical interactions, 34.33% co-expression and 7.96% shared protein domains (Figure 4A and B). The functions of these matrisome- and adhisome-related genes and their neighbouring genes were predicted using Metascape. The top pathway enrichment analysis for normal IBC-PDEs represented pathways including ECM organization, integrin cell surface interactions, cell–cell adhesion and matrix metalloproteinases (Figure 4C). The top pathway enrichment analysis for IBC-PDEs represented pathways associated with IBC progression and metastasis including degradation of the ECM, assembly of collagen fibrils and other multimeric structures, IL-18 signalling pathway and cell junction organization (Figure 4D). Moreover, to better understand the relationship between Cell Adhesion and ECM-related genes and their neighbouring genes, and IBC, we then performed a Metascape PPI enrichment analysis. The PPI network and MCODE components are shown in (Figure 4E–L). Data showed that the biological functions of matrisome- and adhisome-related genes and their neighbouring genes in normal IBC PDEs are mainly enriched in PID integrin CS pathway, cell adhesion mediated by integrin, integrin cell surface interactions matrix metalloproteinases (MMPs) and collagen catabolic process, while for cancer IBC-PDEs are mainly enriched in collagen chain trimerization, degradation of the ECM, cell junction organization, cell–cell communication, activation of MMPs and NABA ECM regulators.
Figure 4.
Gene enrichment analysis for the differentially expressed matrisome and adhisome genes in normal and cancer PDEs of IBC compared with non-IBC. (A and B) Gene–gene interaction network of upregulated and downregulated cell adhesion and ECM DEGs and their neighbouring genes in normal and cancer PDEs, respectively. Each node represents a gene and the size of which represents the strength of the interaction. (C and D) Bar graphs of enriched terms across input gene lists, coloured by P-values. (E and F) Network of enriched terms coloured by P-values, where terms containing more genes tend to have a more significant P-values. (G–J) Protein–protein interaction networks and MCODE components identified in the gene lists. (K and L) The tables showed the functional analysis of MCODEs components.
Discussion
Matricellular proteins composed of matrisome and adhisome are key regulatory partners in cancer development playing essential role in the tissue remodelling and metastasis.21 In breast cancer development, matrisome and adhisome contribute to cell growth, biological turn over, cell division, differentiation, epithelial mesenchymal transition, motility, migration and metastasis.22 Tumour bed cavity and wound healing after tumour excision are hubs of tremendous changes in the gene expression of matrisome, adhisome and inflammatory mediators triggering recurrence and metastasis.23 Although there are different studies discussing the role of matrisome and adhisome in non-IBC poor prognosis, their role in IBC progression is poorly understood. In the present study, we first compared the genomic profile of matrisome and adhisome in fresh normal and carcinoma tissues of non-IBC and IBC patients. Since ex vivo PDEs represent a culture model that resembles in vivo patients’ anatomy preserved by the expression of matrisome and adhisome proteins, we also tested whether the tissue preparation and culture of PDEs will preserve its phenotype and genotype properties via compering matrisome and adhisome gene expression of PDEs cultured for 48 h with their autologous primary patients’ fresh tissue. Indeed, the cross talk between cells in the TME is governed to great extent by cytokines; herein, we also profiled PDEs-secreted cytokines, chemokines and growth factors to identify candidate molecules that might modulate the expression of matrisome and adhisome proteins. Using integrated bioinformatic approaches, we analysed the results and identified the key matrisome, adhisome and secretory cytokines that are crucial for IBC cancer development, matrix remodelling, disease recurrence and metastasis compared with non-IBC.
In continuity to our previous research,13,24 herein we utilized quantitative real-time PCR array to demonstrate the differences between matrisome and adhisome of fresh normal and cancer tissues and PDEs in non-IBC and IBC patients. The DEGs analysis showed that IBC normal, cancer tissues and PDEs are characterized by upregulation of CDH1 and MMP-14 and downregulation of CTNNA1 and TIMP1 compared with non-IBC normal and cancer PDEs. CDH1 or N-cadherin is associated with EMT, cancer metastasis and poor prognosis in different malignancies including breast cancer.25 The transmembrane proteases MMP-14 is considered as target therapy since it plays significant role in the degradation of ECM proteins, breast cancer motility invasion and metastasis of triple-negative breast cancer.26 Our previous study showed that IBC tissues overexpressed MMP-14 and correlated with MMP-2 and -9 suggesting MMPs role in IBC progression.27 Thus, both CDH1 and MMP-14 play significant role in cancer metastasis. In addition, bioinformatic analysis employing MCODE identified the module of collagen chain trimerization that implies collagen solubility and degradation of ECM proteins in PPI as main properties of IBC cancer fresh tissue and PDEs (Figure 4L). Indeed, solubility of collagen and degradation of ECM proteins are essential for cancer cell motility, invasion and metastasis.
Herein, PDEs of IBC are characterized by high secretion of IL-6 and MCP-1. The IL-6 stimulates cancer cell proliferation via antiapoptotic response.28,29 Also, IL-6 induces expression of MMP-14 which in turn enhances motility, invasion and metastasis of cancer cells. This mechanism modulated by IL-6 down regulation to p5330 IL-6 also increases metastatic properties of melanoma via phosphorylation of STAT3 which in turn upregulates TWIST and CDH1 (N-cadherin).31 Similarly, MCP-1 which is secreted by IBC-PDEs found to induce expression of MMPs family.32 MCP-1 expression within the TME by the non-tumour stromal cells promotes the lung metastasis in T41 murine breast cancer cells.33 In addition, IL-6, like MCP-1, is expressed by mesenchymal stem cells which help to promote the migration and metastasis of breast cancer cells, and considered as one of the factors of tumour progression and metastasis.34
In contrast, gene expression arrays showed that CTNNA1 and TIMP1 were downregulated in IBC cancer tissue and PDEs. CTNNA1 induces the expression of E-cadherin,35 which is a hall mark of IBC highly expressed in IBC tumour emboli and associated with IBC poor prognosis.36 Down regulated TIMP1 detected in IBC known to enhance metastatic potential of cancer cells.37
Bioinformatic results using gene enrichment analysis identified the neighbouring genes, function and pathways of matrisome and adhisome DEGs. The pathway enrichment and PPI network analysis of DEGs and their neighbouring genes (Figure 4K) showed strong association mainly with PID integrin CS pathway, cell adhesion mediated by integrin, integrin cell surface interactions, MMPs and collagen catabolic process in IBC normal PDEs, while for cancer IBC PDEs are mainly enriched in collagen chain trimerization, degradation of the ECM, cell junction organization, cell–cell communication, MMPs and NABA ECM regulators (Figure 4L). Our results agree with previous studies,21,38–40 that used bioinformatic analysis to identify involvement of matrisome and adhisome in IBC progression; however, herein we used freshly resected tissues from breast cancer patients cultured as ex vivo model that retains biological, histological and anatomical features of the original tissues to identify key adhisome and matrisome and cytokinome proteins that may contribute to IBC metastasis.
Conclusion
Genes expressed by adhisome and matrisome play a significant role in IBC metastasis. The result of the present study highlights the complexity of matricellular gene expression in IBC carcinoma tissues and their implications for clinical trial. We also presented PDEs ex vivo model to be used for studying live cell interactions and identify therapeutic targets. Further studies with large samples size are warranted to validate PDEs in understanding pathobiology of breast cancer disease (non-IBC and IBC) and investigate the mechanisms that undergo disease recurrence and metastatic potential after therapeutic intervention.
Ethics approval
The study protocol was approved by the Institutional Review Board (IRB#00006379), Faculty of Medicine, Ain Shams University, Egypt. Before participation, all patients signed consent forms, including approval for publication of the study results.
Acknowledgments
This work was conducted in the Cancer Biology Research Laboratory, Faculty of Science, Cairo University, Giza, Egypt, and part of this work was conducted at Galala University, Galala City, Suez, Egypt. Special thanks to Egyptian breast cancer patients who participated in the present study.
Contributor Information
Alshaimaa Tarek, From the Department of Zoology, Faculty of Science, Cairo University, Giza 12613, Egypt.
Hossam Taha Mohamed, From the Department of Zoology, Faculty of Science, Cairo University, Giza 12613, Egypt; Faculty of Biotechnology, October University for Modern Sciences and Arts, Giza 12451, Egypt.
Aya Ali El-Sharkawy, From the Department of Zoology, Faculty of Science, Cairo University, Giza 12613, Egypt.
Shrouk Khalaf El-Sayed, From the Department of Zoology, Faculty of Science, Cairo University, Giza 12613, Egypt.
Jon Mark Hirshon, School of Medicine, University of Maryland, Baltimore, MD 21201, USA.
Wendy A Woodward, Morgan Welch Inflammatory Breast Cancer Research Program and Clinic, Department of Radiation Oncology, The University of Texas MD Anderson Cancer Center, Houston, TX 77030, USA.
Mohamed El-Shinawi, Faculty of Medicine, Galala University, Suez 43511, Egypt; Department of General Surgery, Faculty of Medicine, Ain Shams University, Cairo 11566, Egypt.
Mona Mostafa Mohamed, From the Department of Zoology, Faculty of Science, Cairo University, Giza 12613, Egypt.
Data availability
All supporting data are included within the main article and its supplementary file. All data are available upon request and can be accessed by contacting the authors.
Author contributions
Alshaimaa Tarek (Formal analysis [equal], Methodology [equal]), Hossam Taha Mohamed (Formal analysis [equal], Methodology [equal], Validation [lead], Writing—review and editing [equal]), Aya Ali El-Sharkawy (Methodology [equal]), Shrouk Khalaf El-Sayed (Formal analysis [equal]), Jon Mark Hirshon (Project administration [equal], Resources [equal]), Wendy A. Woodward (Supervision [supporting]), Mohamed El-Shinawi (Resources [equal], Supervision [equal]), Mona Mostafa Mohamed (Conceptualization [lead], Project administration [lead], Resources [equal], Supervision [lead], Validation [equal], Writing—original draft [lead], Writing—review and editing [equal]).
Funding
This work was supported by Fogarty International Center, National Institutes of Health (NIH) Grant Number R21TW010952 (PI: J.M.H. and Co-PIs: M.M.M., W.A.W. and M.E.-S.), Academy of Scientific Research and Technology (ASRT), Egypt, APPLE Call Grant Number 10142021205540 (PI: M.M.M. and Co-PI: M.E.-S.) and the Cairo University Scientific Research Sector (M.M.M.).
Conflict of interests
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- 1. Kashyap D, Pal D, Sharma R, Garg VK, Goel N, Koundal D, et al Global increase in breast cancer incidence: risk factors and preventive measures. BioMed Res Int 2022; 2022:9605439. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 2. Hance KW, Anderson WF, Devesa SS, Young HA, Levine PH.. Trends in inflammatory breast carcinoma incidence and survival: the surveillance, epidemiology, and end results program at the National Cancer Institute. J Natl Cancer Inst 2005; 97:966–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Boussen H, Bouzaiene H, Ben Hassouna J, Dhiab T, Khomsi F, Benna F, et al Inflammatory breast cancer in Tunisia: epidemiological and clinical trends. Cancer 2010; 116:2730–5. [DOI] [PubMed] [Google Scholar]
- 4. Lim B, Woodward WA, Wang X, Reuben JM, Ueno NT.. Inflammatory breast cancer biology: the tumour microenvironment is key. Nat Rev Cancer 2018; 18:485–99. [DOI] [PubMed] [Google Scholar]
- 5. Ross JS, Ali SM, Wang K, Khaira D, Palma NA, Chmielecki J, et al Comprehensive genomic profiling of inflammatory breast cancer cases reveals a high frequency of clinically relevant genomic alterations. Breast Cancer Res Treat 2015; 154:155–62. [DOI] [PubMed] [Google Scholar]
- 6. Morrow RJ, Etemadi N, Yeo B, Ernst M.. Challenging a misnomer? The role of inflammatory pathways in inflammatory breast cancer. Mediat Inflamm 2017;2017:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Arora J, Sauer SJ, Tarpley M, Vermeulen P, Rypens C, Van Laere S, et al Inflammatory breast cancer tumor emboli express high levels of anti-apoptotic proteins: use of a quantitative high content and high-throughput 3D IBC spheroid assay to identify targeting strategies. Oncotarget 2017; 8:25848–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Powley IR, Patel M, Miles G, Pringle H, Howells L, Thomas A, et al Patient-derived explants (PDEs) as a powerful preclinical platform for anti-cancer drug and biomarker discovery. Br J Cancer 2020; 122:735–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nouh MA, Mohamed MM, El-Shinawi M, Shaalan MA, Cavallo-Medved D, Khaled HM, et al Cathepsin B: a potential prognostic marker for inflammatory breast cancer. J Transl Med 2011; 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. El-Shinawi M, Mohamed HT, El-Ghonaimy EA, Tantawy M, Younis A, Schneider RJ, et al Human cytomegalovirus infection enhances NF-kappaB/p65 signaling in inflammatory breast cancer patients. PLoS ONE 2013; 8:e55755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sineh Sepehr K, Razavi A, Saeidi M, Mossahebi-Mohammadi M, Abdollahpour-Alitappeh M, Hashemi SM.. Development of a novel explant culture method for the isolation of mesenchymal stem cells from human breast tumor. J Immunoass Immunochem 2018; 39:207–17. [DOI] [PubMed] [Google Scholar]
- 12. Mohamed MM, Cavallo-Medved D, Rudy D, Anbalagan A, Moin K, Sloane BF.. Interleukin-6 increases expression and secretion of cathepsin B by breast tumor-associated monocytes. Cell Physiol Biochem 2010; 25:315–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mohamed HT, El-Sharkawy AA, El-Shinawi M, Schneider RJ, Mohamed MM.. Inflammatory breast cancer: the secretome of HCMV(+) tumor-associated macrophages enhances proliferation, invasion, colony formation, and expression of cancer stem cell markers. Front Oncol 2022; 12:899622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ibrahim AS, El-Shinawi M, Sabet S, Ibrahim SA, Mohamed MM.. Role of adipose tissue-derived cytokines in the progression of inflammatory breast cancer in patients with obesity. Lipids Health Dis 2022; 21:67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Mohamed MM. Conditioned media stimulate fibronectin expression and spreading of inflammatory breast cancer cells in three-dimensional culture: a mechanism mediated by IL-8 signaling pathway. Cell Commun Signal 2012; 10:3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ahmed S, Mohamed HT, El-Husseiny N, El Mahdy MM, Safwat G, Diab AA, et al IL-8 secreted by tumor associated macrophages contribute to lapatinib resistance in HER2-positive locally advanced breast cancer via activation of Src/STAT3/ERK1/2-mediated EGFR signaling. Biochim Biophys Acta Mol Cell Res 2021; 1868:118995. [DOI] [PubMed] [Google Scholar]
- 17. Wang T, Zhang Y, Bai J, Xue Y, Peng Q.. MMP1 and MMP9 are potential prognostic biomarkers and targets for uveal melanoma. BMC Cancer 2021; 21:1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li M, He M, Xu F, Guan Y, Tian J, Wan Z, et al Abnormal expression and the significant prognostic value of aquaporins in clear cell renal cell carcinoma. PLoS ONE 2022; 17:e0264553-e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jin Y, Lin Q, Fei H, Xue L, Li L, Xi Q, et al Bioinformatics analysis of potential therapeutic targets and prognostic biomarkers amid CXC chemokines in ovarian carcinoma microenvironment. J Oncol 2021; 2021:8859554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ota A, Hyon S-H, Sumi S, Matsumura K.. Gene expression analysis of human induced pluripotent stem cells cryopreserved by vitrification using StemCell Keep. Biochem Biophys Rep 2021; 28:101172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Winkler J, Abisoye-Ogunniyan A, Metcalf KJ, Werb Z.. Concepts of extracellular matrix remodelling in tumour progression and metastasis. Nat Commun 2020; 11:5120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhao Y, Zheng X, Zheng Y, Chen Y, Fei W, Wang F, et al Extracellular matrix: emerging roles and potential therapeutic targets for breast cancer. Front Oncol 2021; 11:650453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang Y, Xu H, Zhu B, Qiu Z, Lin Z.. Systematic identification of the key candidate genes in breast cancer stroma. Cell Mol Biol Lett 2018; 23:44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mohamed HT, Kamel G, El-Husseiny N, El-Sharkawy AA, El-Sherif AA, El-Shinawi M, et al Synchrotron Fourier-transform infrared microspectroscopy: characterization of in vitro polarized tumor-associated macrophages stimulated by the secretome of inflammatory and non-inflammatory breast cancer cells. Biochim Biophys Acta Mol Cell Res 2023; 1870:119367. [DOI] [PubMed] [Google Scholar]
- 25. Cao ZQ, Wang Z, Leng P.. Aberrant N-cadherin expression in cancer. Biomed Pharmacother 2019; 118:109320. [DOI] [PubMed] [Google Scholar]
- 26. Ling B, Watt K, Banerjee S, Newsted D, Truesdell P, Adams J, et al A novel immunotherapy targeting MMP-14 limits hypoxia, immune suppression and metastasis in triple-negative breast cancer models. Oncotarget 2017; 8:58372–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Al-Raawi D, Abu-El-Zahab H, El-Shinawi M, Mohamed MM.. Membrane type-1 matrix metalloproteinase (MT1-MMP) correlates with the expression and activation of matrix metalloproteinase-2 (MMP-2) in inflammatory breast cancer. Int J Clin Exp Med 2011; 4:265–75. [PMC free article] [PubMed] [Google Scholar]
- 28. Kurebayashi J. Regulation of interleukin-6 secretion from breast cancer cells and its clinical implications. Breast Cancer (Tokyo, Japan) 2000; 7:124–9. [DOI] [PubMed] [Google Scholar]
- 29. Knüpfer H, Preiss R.. Significance of interleukin-6 (IL-6) in breast cancer (review). Breast Cancer Res Treat 2007; 102:129–35. [DOI] [PubMed] [Google Scholar]
- 30. Cathcart JM, Banach A, Liu A, Chen J, Goligorsky M, Cao J.. Interleukin-6 increases matrix metalloproteinase-14 (MMP-14) levels via down-regulation of p53 to drive cancer progression. Oncotarget 2016; 7:61107–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Na YR, Lee JS, Lee SJ, Seok SH.. Interleukin-6-induced Twist and N-cadherin enhance melanoma cell metastasis. Melanoma Res 2013; 23:434–43. [DOI] [PubMed] [Google Scholar]
- 32. Liu J-F, Chen P-C, Chang T-M, Hou C-H.. Monocyte chemoattractant protein-1 promotes cancer cell migration via c-Raf/MAPK/AP-1 pathway and MMP-9 production in osteosarcoma. J Exp Clin Cancer Res 2020; 39:254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yoshimura T, Howard OM, Ito T, Kuwabara M, Matsukawa A, Chen K, et al Monocyte chemoattractant protein-1/CCL2 produced by stromal cells promotes lung metastasis of 4T1 murine breast cancer cells. PLoS ONE 2013; 8:e58791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. De Luca A, Lamura L, Gallo M, Maffia V, Normanno N.. Mesenchymal stem cell-derived interleukin-6 and vascular endothelial growth factor promote breast cancer cell migration. J Cell Biochem 2012; 113:3363–70. [DOI] [PubMed] [Google Scholar]
- 35. Chi Q, Xu H, Song D, Wang Z, Wang Z, Ma G.. α-E-catenin (CTNNA1) inhibits cell proliferation, invasion and EMT of bladder cancer. Cancer Manag Res 2020; 12:12747–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Colpaert CG, Vermeulen PB, Benoy I, Soubry A, van Roy F, van Beest P, et al Inflammatory breast cancer shows angiogenesis with high endothelial proliferation rate and strong E-cadherin expression. Br J Cancer 2003; 88:718–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schelter F, Halbgewachs B, Bäumler P, Neu C, Görlach A, Schrötzlmair F, et al Tissue inhibitor of metalloproteinases-1-induced scattered liver metastasis is mediated by hypoxia-inducible factor-1α. Clin Exp Metastasis 2011; 28:91–9. [DOI] [PubMed] [Google Scholar]
- 38. Yousefi H, Vatanmakanian M, Mahdiannasser M, Mashouri L, Alahari NV, Monjezi MR, et al Understanding the role of integrins in breast cancer invasion, metastasis, angiogenesis, and drug resistance. Oncogene 2021; 40:1043–63. [DOI] [PubMed] [Google Scholar]
- 39. Quintero-Fabián S, Arreola R, Becerril-Villanueva E, Torres-Romero JC, Arana-Argáez V, Lara-Riegos J, et al. Role of Matrix Metalloproteinases in Angiogenesis and Cancer, Front Oncol2019; 9:1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Jena MK, Janjanam J.. Role of extracellular matrix in breast cancer development: a brief update. F1000Research 2018; 7:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All supporting data are included within the main article and its supplementary file. All data are available upon request and can be accessed by contacting the authors.