Abstract
Nucleoporin 98 (Nup98), a glycine-leucine-phenylalanine-glycine (GLFG) amino acid repeat-containing nucleoporin, plays a critical part in nuclear trafficking. Injection of antibodies to Nup98 into the nucleus blocks the export of most RNAs. Nup98 contains binding sites for several transport factors; however, the mechanism by which this nucleoporin functions has remained unclear. Multiple subcellular localizations have been suggested for Nup98. Here we show that Nup98 is indeed found both at the nuclear pore complex and within the nucleus. Inside the nucleus, Nup98 associates with a novel nuclear structure that we term the GLFG body because the GLFG domain of Nup98 is required for targeting to this structure. Photobleaching of green fluorescent protein-Nup98 in living cells reveals that Nup98 is mobile and moves between these different localizations. The rate of recovery after photobleaching indicates that Nup98 interacts with other, less mobile, components in the nucleoplasm. Strikingly, given the previous link to nuclear export, the mobility of Nup98 within the nucleus and at the pore is dependent on ongoing transcription by RNA polymerases I and II. These data give rise to a model in which Nup98 aids in direction of RNAs to the nuclear pore and provide the first potential mechanism for the role of a mobile nucleoporin.
INTRODUCTION
The nuclear pore complex is a massive structure that conducts all traffic between the nucleus and cytoplasm (reviewed by Ohno et al., 1998; Gorlich and Kutay, 1999; Nakielny and Dreyfuss, 1999; Ryan and Wente, 2000; Vasu and Forbes, 2001). The pore has been studied intensely at the structural level in both yeast and vertebrate systems (Stoffler et al., 1999a; Allen et al., 2000). Although the vertebrate pore is larger and thought to contain a greater number of constituent proteins, the pores of both yeast and vertebrates share a similar structural organization. The central mass of the nuclear pore displays eightfold symmetry around a central axis perpendicular to the nuclear envelope. Two distinct sets of fibers extend out from the cytoplasmic and nuclear faces of the pore. On the nuclear side, the fibers are joined together at their distal ends by a ring to form the nuclear basket of the pore. Additionally, the nuclear basket of both yeast and vertebrate pores has associated filaments that extend for considerable distances into the nuclear interior and may serve to direct transport cargoes to and/or from the pore.
A subset of the nuclear pore complex proteins (nucleoporins or Nups) each contain a domain with multiple, nontandem repeats of the amino acid sequences FG, FXFG, or GLFG (glycine-leucine-phenylalanine-glycine). These domains provide docking sites for a family of nuclear transport signal receptor proteins (known as importins, exportins, and transportin, or collectively as karyopherins). The repeat domain nucleoporins are found in multiple substructures of the nuclear pore and thus are thought to facilitate the movement of receptors between the nucleus and the cytoplasm (Mattaj and Englmeier, 1998; Nakielny and Dreyfuss, 1999; Hood and Silver, 2000). The affinity of repeat domains for transport receptor-cargo complexes varies at different sites in the pore; an import receptor-cargo complex binds most tightly to Nup153, which is found on the nuclear side of the pore (Shah et al., 1998; Ben-Efraim and Gerace, 2001). Conversely, Nup214 on the cytoplasmic fibers has the highest affinity for a nuclear export complex (Fornerod et al., 1997). These observations have led to one model in which cargo-receptor complexes cross the pore by progressively binding to higher affinity sites (Ben-Efraim and Gerace, 2001). Alternatively, a second model suggests that translocation through the nuclear pore is an essentially stochastic process, with those cargo-receptor complexes that do reach the opposite side of the pore finding one high-affinity–binding site, Nup153 or Nup214, and then being rapidly disassembled to release the cargo (Rout et al., 2000; Ribbeck and Gorlich, 2001).
At its terminal binding site, the transport complex is disassembled by the GTPase, Ran. Import cargo complexes are disassembled by Ran-GTP, whereas export cargo complexes are stabilized by Ran-GTP and disassembled after GTP hydrolysis. To facilitate the directionality of traffic, Ran is maintained in the GTP-bound state within the nucleus and in the GDP-bound state in the cytoplasm (reviewed by Gorlich and Kutay, 1999; Azuma and Dasso, 2000; Conti and Izaurralde, 2001).
A surprising feature of current models is their depiction of the pore complex as a static structure, essentially a scaffold for the binding sites on which transport receptors traffic. An important question is whether the pore itself contributes to transport other than by providing the proper arrangement of receptor-binding sites. Observations of conformational changes of the nuclear pore in response to elevated calcium levels (Kiseleva et al., 1996; Stoffler et al., 1999b; Goldberg et al., 2000), Balbiani ring granule binding to the basket (Kiseleva et al., 1996), or after injection of Ran-GTP (Goldberg et al., 2000) suggest that the nuclear pore may undergo dynamic structural changes coupled to trafficking. A second indication of a more active role is the observation that some nucleoporin move on and off the pore (Daigle et al., 2001), or may themselves translocate through the pore, but precisely how this would be linked to substrate trafficking is as yet unclear (Nakielny et al., 1999; Zolotukhin and Felber, 1999; Dilworth et al., 2001).
How transport through the pore is linked to the nuclear interior is also unclear. The pore-associated protein Tpr has been shown to form filaments that extend from the nuclear pore basket into the nucleoplasm (Cordes et al., 1993). In yeast, two proteins, MLP1p and MLP2p, are believed to form similar filaments (Strambio-de-Castillia et al., 1999). Such filaments could direct imported proteins from the pore to sites in the nuclear interior; alternatively, they could aid in targeting export cargo to the pore. The possibility of filaments directing transport cargo to or from the pore is especially intriguing in light of recent protein mobility measurements within the nucleus (Kruhlak et al., 2000; Phair and Misteli, 2000). Nuclear proteins move through the nucleoplasm far more slowly than predicted by diffusion, whereas a nonnuclear protein such as green fluorescent protein (GFP), when artificially directed to the nucleus, moves at nearly the predicted diffusion rate. The slowed mobility is believed to result from repeated transient interactions between nuclear proteins and other, more immobile nuclear components (Misteli, 2001; Pederson, 2001).
We have focused on the function of one nucleoporin, Nup98, as a means to address the workings of the nuclear pore. Nup98 is the only known vertebrate member of the GLFG repeat domain family (Powers et al., 1995; Radu et al., 1995). In yeast, this family comprises five nucleoporins, three of which, Nups100, 116, and 145, are most closely related to Nup98 and function primarily in nuclear export (Wente and Blobel, 1993, 1994; Fabre et al., 1994; Bailer et al., 1998). Similarly, Nup98 seems to play its primary role in nuclear export. We have previously shown that injection of Nup98-specific antibodies blocks export of most RNAs while leaving protein import unaltered (Powers et al., 1997). In keeping with this, Nup98 binds in vitro to multiple export factors, including RaeI/Gle2, TAP, and CRM (Neville et al., 1997; Pritchard et al., 1999; Bachi et al., 2000; Strasser et al., 2000; Strawn et al., 2001). However, Nup98 can also bind to several import receptors in vitro and may contribute in some way to nuclear import (Fontoura et al., 2000). At the nuclear pore complex, Nup98 was localized by electron microscopy to the basket on the nuclear face of the pore (Radu et al., 1995). A fraction of Nup98 is likely to be within the nucleus, based both on our original immunofluorescence localization in Xenopus cells (Powers et al., 1995) and on the observation that overexpression of Rev can cause relocalization of Nup98 to nucleoli (Zolotukhin and Felber, 1999). Additionally, Nup98 was recently shown to bind in vitro to the nuclear filament protein Tpr (Fontoura et al., 2001).
To address the potential mechanism of action of Nup98, we have applied a powerful new approach for the study of nuclear trafficking: photobleaching of a fluorescently tagged nucleoporin in living cells. We initially carried out a careful analysis of the localization of Nup98 within the nucleus using a combination of immunofluorescence and electron microscopy. Both Nup98 and GFP-Nup98 are localized not only at the nuclear pore complex but also in the nuclear interior, where Nup98 can associate with a novel nuclear body. We used photobleaching of GFP-Nup98 in live cells to address Nup98 mobility and function. We present data indicating that Nup98 moves between the nuclear interior and the pore, as well as between the nucleus and the cytoplasm. Most importantly, through the use of specific inhibitors, we show that the mobility of Nup98 is coupled to ongoing transcription of RNA.
MATERIALS AND METHODS
DNA Constructs
The full-length, hemagglutinin-tagged human Nup98 cDNA was a gift from Dr. Julian Borrow. An EcoRI site was inserted at the N terminus by polymerase chain reaction (PCR) and the Nup98-coding sequence was ligated in-frame into pEGFP-C1 (CLONTECH Laboratories, Palo Alto, CA) to generate GFP-Nup98. The N terminus (amino acids 1–225), GLFG domain (221–504), and C terminus (506–920) of Nup98 were each amplified by PCR and inserted into pEGFP-C3 (CLONTECH). All constructs produced by PCR were sequenced for confirmation.
Cell Culture
XL177 cells were a gift from Dr. Rebecca Heald and were grown at room temperature in 70% Leibovitz L-15 medium supplemented with 15% fetal calf serum (FCS), 100 μg/ml penicillin, and 100 U/ml streptomycin. HeLa cells were grown in high glucose DMEM (Life Technologies, Rockville, MD) supplemented with 10% FCS, 2 mM glutamine, 100 μg/ml penicillin, and 100 U/ml streptomycin. tsBN2 cells were a gift from Dr. Ian Macara and were maintained in the same medium as HeLa cells but at 33.5°C. For temperature shifts, tsBN2 cells were placed at 39°C for 4 h before imaging. For inhibitor studies, leptomycin B (a gift from Dr. Minoru Yoshida), 5,6-dichloro-1-β-d-ribofuranosylbenzimidazole (DRB; Calbiochem La Jolla, CA), and actinomycin D (actinomycin C1; Roche, Indianapolis, IA) were used at concentrations of 0.1, 75, and 5 μg/ml, respectively. For transient transfections, fugene 6 (Roche) was used according to the manufacturer's instructions. To estimate the relative levels of expression for GFP-Nup98 and endogenous Nup98, transfections were scored by fluorescence for percentage of transfected cells, then lysed, and immunoblotted with antibody to the C terminus of human Nup98 (anti-hNup98; amino acids 506–920; 1:1500). The relative signal intensities were corrected for percentage of transfection to arrive at an estimate of relative protein amounts.
Immunofluorescence
For immunofluorescence experiments, cells were fixed in 4% paraformaldehyde for 10 min and permeabilized with 0.2% TritonX-100 on ice for 10 min. Samples were blocked with 3% bovine serum albumin plus 0.02% TritonX-100 in phosphate-buffered saline (PBS; block) for 30 min, incubated with primary antibody diluted in block for 30 min, and washed four times in 1.5% bovine serum albumin plus 0.02% Triton X-100 in PBS. Fluorescent secondary antibody diluted in block was applied for 30 min, and cells were washed as described above. The dye H33258 (Hoechst; 1 μg/ml; Calbiochem) was included in the penultimate wash step to visualize DNA. Coverslips were mounted with Vectashield (Vector Laboratories, Burlingame, CA). The following antibodies used: anti-xNup98 (1:50; Powers et al., 1995), anti-hNup98, (1:3000), anti-PML (mAb 5E10, 1:10; Santa Cruz Biotech, Santa Cruz, CA), anti-SC35 (1:1000; Accurate Chemical, Westbury, NY), anti-p80 coilin (antibody R288, 1:400; Andrade et al., 1993), anti-Ran (1:1000; Transduction Laboratories, San Diego, CA), goat anti-rabbit rhodamine isothiocyanate (1:800; Jackson Immunoresearch, West Grove, PA), and goat anti-mouse rhodamine isothiocyanate (1:500; Jackson Immunoresearch).
Still images were captured using either a BX60 microscope (Olympus, Tokyo, Japan) with an 8-bit camera (Dage-MTI, Michigan City, IN) and IP Lab software (Scanalytics, Fairfax, VA) or an LSM 510 confocal microscope (Zeiss, Thornwood, NY). For deconvolution microscopy, stacks of images were deconvolved using HazeBuster software from Vaytek (Fairfield, IA), running a nearest-neighbor algorithm to remove out-of-field fluorescence.
Live Cell Imaging
For live cell imaging, cells were grown and transfected in LabTek II-chambered coverslips (Nalgene, Rochester, NY). Before imaging, the medium was removed and replaced with DMEM supplemented with 20% FCS and 25 mM HEPES, pH 7.0, and the chambers were sealed with petroleum jelly to prevent evaporation of the medium. Imaging was performed using a Zeiss LSM510 confocal microscope. An ASI 400 air stream incubator (Nevtek, Burnsville, VA) was used to maintain the cells at 37°C during imaging.
Fluorescence Recovery after Photobleaching (FRAP) and Fluorescence Loss in Photobleaching (FLIP) Analyses
For qualitative FRAP analysis, cells expressing GFP-tagged proteins were identified on the Zeiss LSM 510 confocal microscope using the 488-nm line of a Kr/Ar laser operating at 75% laser power and 1.1% transmission (imaging intensity). An area of the selected cell was outlined for bleaching. Four imaging scans of the whole cell were performed, and then the selected area was bleached using 25 bleaching iterations with 75% laser power and 100% transmittance (bleaching intensity). After the photobleach, scans were taken at imaging intensity every 1.5 s for 2 min or until the fluorescence intensity reached a plateau.
To quantitate the recovery of GFP-Nup98 or GFP-GLFG in nuclear bodies, bleaching was performed as described above except that the pinhole was opened to maximum diameter to provide a deeper Z-axis. For quantitation of the faster recovery of GFP or GFP-Nup98 in the nucleoplasm, cells were imaged using an ROI (region of interest) zoom that allows for faster image acquisition. The cells were imaged 10 times before photobleaching and 90 times afterward with a scan speed of 250 ms. All measurements were corrected for background fluorescence, as determined by measuring the average noncellular fluorescence in the sample. To correct for the loss of fluorescence during the bleach pulse and during imaging, the total cellular fluorescence was measured in each scan, and fluorescence in the region of interest was normalized to the change in cellular fluorescence using the following equation: Inorm = T0It/TtI0, where Inorm is the normalized intensity of the region of interest, T0 is the cellular fluorescence before photobleach, Tt is the cellular fluorescence at time t, I0 is the intensity at the region of interest before photobleach, and It is the intensity in the region of interest at time t (Phair and Misteli, 2000). To graph the data, the first scan after photobleaching was set as 0 s, and the normalized intensity at each time point multiplied by 100 was plotted as the percentage of initial fluorescence. The data points were then fitted with a nonlinear curve using Origin 6.1 software (OriginLab, Northampton, MA). The half-time of recovery was the time at which the fluorescence had recovered to an intensity halfway between the postbleach intensity and the maximal recovery. The percentage difference between recovery to 100% of the prebleached fluorescence intensity and the maximal recovery was determined to be the immobile fraction.
For FLIP analyses, a region of the cytoplasm was selected and bleached 10 times, followed by two imaging scans. This pattern was repeated for 100 iterations, and the nuclear fluorescence was measured over time. To graph fluorescence lost in photobleaching, the background-corrected fluorescence at the time before the initial photobleach was set as 100%. The corrected fluorescence at each time point after bleaching was divided by the prebleach intensity and multiplied by 100 to give the percentage of initial fluorescence.
For videos, scans were collected as described for the appropriate procedure, false colored, and ordered in IP Lab and exported as QuickTime (Apple Computer, Cupertino, CA) movies.
Electron Microscopy
XL177 cells on coverslips were fixed in 4% paraformaldehyde and 0.2% glutaraldehyde and washed with 0.1 M phosphate buffer, pH 7.4 (PB), followed by PB containing 0.1% sodium borohydride to inactivate residual aldehyde groups. The cells were then permeabilized with PB containing 0.05% Triton X-100 for 20 min at room temperature and washed with PB. Blocking solution was PBS, pH 7.4, containing 4% normal goat serum (NGS). After blocking, cells were incubated with affinity-purified rabbit anti-xNup98 antibody (1:50) in PBS containing 1% NGS. After six washes with PBS, cells were incubated with a biotinylated goat anti-rabbit secondary antibody (1:200) in PBS and 1% NGS. After washing, cells were incubated in avidin-biotin complex (Vector ABC Elite, Vector Laboratories). Immunoreactivity was visualized by incubation in 0.05% 3–3′-diaminobenzidine tetrahydrochloride (DAB; Sigma, St. Louis, MO) and 0.003% hydrogen peroxide in 0.05 M Tris, pH 7, for 2 min. Cells were washed, postfixed with 2.5% glutaraldehyde in PB, washed again, fixed with 0.5% osmium tetroxide for 15 min, dehydrated, and embedded in Eponate 12 resin (Ted Pella, Redding, CA) for sectioning.
For immunogold detection, the above protocol was followed with the following changes. The blocking solution contained 0.1% cold water fish skin gelatin (Aurion, Wageningen, The Netherlands) in addition to NGS. The cells were incubated overnight at 4°C with the secondary antibody, an ultrasmall gold-conjugated Fab2 fragment of goat anti-rabbit IgG (Aurion) diluted 1:100 in PBS and 1% NGS. The cells were washed with PBS and postfixed with 2.5% glutaraldehyde in PB for 2 h. Silver enhancement was performed with R-gent SE-EM solution (Aurion) according to manufacturer's instructions.
RESULTS
Nup98 Is Targeted to the Nucleoplasm and Nuclear Bodies, in Addition to the Nuclear Pore
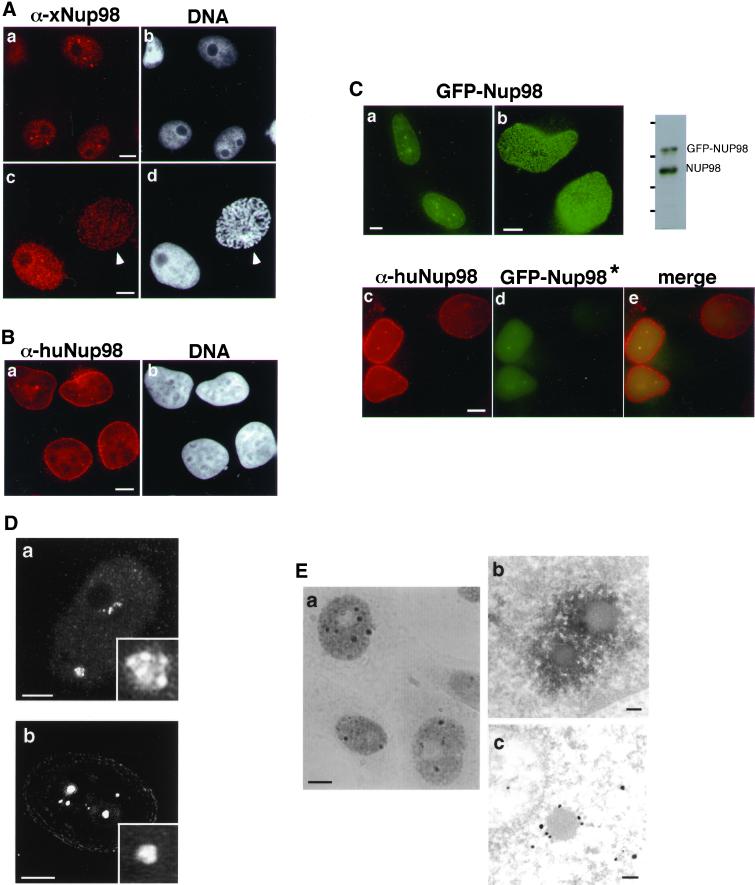
The localization of the nucleoporin Nup98 has been controversial. In Xenopus cells using an affinity-purified antibody raised against xNup98 isolated from Xenopus egg extracts, we have consistently observed Nup98 within the nucleus, both in the nucleoplasm and in intensely fluorescent dots (Figure 1A, a; Powers et al., 1995). Another group reported that Nup98 is primarily at the nuclear pore complex based on both immunofluorescence and immunoelectron microscopy (Radu et al., 1995). Under some circumstances, such as during prophase of the cell cycle, our antibody did indeed reveal a nuclear rim stain (Figure 1A, c and d; Powers et al., 1995). These localization patterns suggested to us that a substantial pool of Nup98 within the nucleus could be masking the population of Nup98 at the nuclear rim. This intranuclear population might be better recognized by some antibodies than by others or be lost from the nucleus under some fixation conditions. We raised an antibody against the C-terminal domain of human Nup98 and investigated Nup98 localization in human cells. In HeLa cells, we more readily observed a nuclear rim stain; additionally, we detected nucleoplasmic staining and intranuclear dots (Figure 1B). Although the intranuclear dots were not as prominently stained as in Xenopus cells, they were observed in approximately one-third of HeLa cells. When a fusion of GFP to the amino terminus of human Nup98 was transfected into either HeLa or Cos7 cells, the live cells exhibited a punctate nuclear rim fluorescence and also reproduced the diffuse nucleoplasmic stain and the fluorescent intranuclear dots (Figure 1C, a and b). An immunoblot of the transfected cells demonstrated that GFP-Nup98 was not greatly overexpressed in these cells; from the relative signal intensities and extent of transfection, we estimate the average level of GFP-Nup98 per cell to be no more than double that of endogenous Nup98 (Figure 1C). As further demonstration that the GFP-Nup98 localization is representative of endogenous protein, fluorescent colocalization established that the same intranuclear dots contain both endogenous Nup98 and transfected GFP-Nup98. To avoid recognition of the GFP-Nup98 protein by the anti–C-terminal antibody used to detect the endogenous Nup98 protein, for this experiment, we transfected GFP fused to only the GLFG domain of Nup98 (GFP-Nup98*) because this fusion is localized to the intranuclear dots, although not to the nuclear pore (Figure 1C, c–e; see also Figure 4B). Additionally, we have found that in vivo, like the endogenous Nup98, GFP-Nup98 is able to bind and coprecipitate its partner protein, RaeI/Gle2 (Griffis and Powers, unpublished results). Taking all of these data together, we conclude that Nup98 is found both at the nuclear envelope and within the nucleus and that this localization is accurately represented by GFP-Nup98.
Figure 1.
Nup98 shows a substantial intranuclear localization in human and Xenopus cells. (A) Immunofluorescence using antibodies raised against xNup98 revealed brightly staining structures as well as a diffuse intranuclear stain at interphase in XL177 cells (a). At prophase, a population of Nup98 at the nuclear rim is revealed (c, prophase cell indicated by arrowhead). b and d are stained with Hoechst. (B) Endogenous human Nup98 in live HeLa cells generates a nuclear rim stain with intranuclear foci and diffuse nucleoplasmic signal (a). b is stained with Hoechst. (C) The same localization pattern was observed in HeLa cells expressing GFP-Nup98. a is a section through the midplane of the nucleus, and b is focused on the surface. To the right is an immunoblot of transfected cells probed with antibodies raised against human Nup98. Molecular mass markers correspond to 184, 121, 86, and 68 kDa. In the bottom panels, GFP-Nup98* (d) is localized to the same intranuclear foci as the endogenous protein (c). Note that to avoid cross-reactivity with the antibody, this construct contains only the GLFG domain of Nup98, and thus signals for targeting to the nuclear pore are absent in this GFP protein. (D) Deconvolution microscopy of the endogenous xNup98 in XL177 cells shows that the intranuclear structures are clusters of smaller bodies (a and inset). The GFP-Nup98 bodies in HeLa cells appear similarly lobular (b and inset). (E) In XL177 cells, endogenous Nup98 localized using DAB and visualized by phase contrast (a) recapitulates the Nup98 localization pattern seen with immunofluorescence. Transmission electron microscopy analysis using either DAB (b) or immunogold (c) confirms that the bodies seen at the light level are in fact clusters of smaller structures. Scale bars, 5 μm in light micrographs and 100 nm in electron micrographs.
Figure 4.
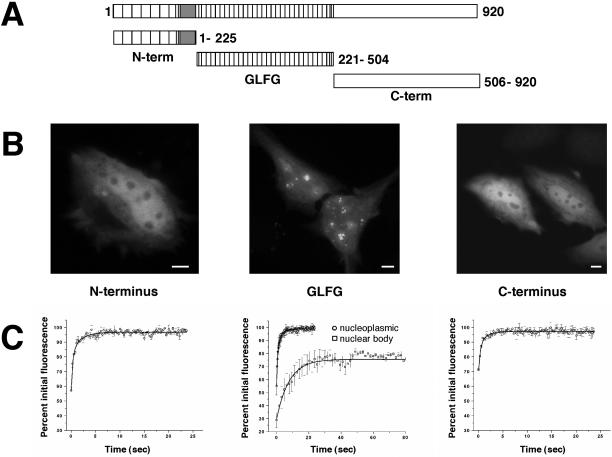
The GLFG domain of Nup98 targets to intranuclear bodies. (A) The domain organization of human Nup98 (920 amino acids) is shown. Each of the depicted domains was N terminally fused to GFP and transfected into HeLa cells. (B) The localization of each of these GFP-domain constructs in HeLa cells showed that only the GLFG domain associates with nuclear bodies. Scale bars, 5 μm. (C) FRAP analysis was carried out on each domain within the nucleoplasm following the same protocol used to quantitate the recovery of Nup98 in the nucleoplasm. The average recovery for five independent experiments is plotted (open circles). In the middle graph, the open squares represent the average recovery after photobleaching of the GFP-GLFG nuclear bodies. Note the difference in the X-axis values between GLFG and N- and C-terminal domain graphs.
To further investigate the nuclear organization of Nup98, we utilized deconvolution microscopy (Figure 1D; Wang, 1998; McNally et al., 1999). This analysis of endogenous Nup98 in Xenopus cells revealed that the dots observed within the nucleus were actually composed of a cluster of smaller structures, which gave them an overall irregular, rather than round, surface (Figure 1D, a). When we imaged GFP-Nup98 transiently expressed in HeLa cells, a similar cluster-like structure was observed (Figure 1D, b), suggesting that GFP-Nup98 is incorporated into an equivalent structure or body.
The fine structure of the Nup98-containing bodies was further probed by electron microscopy. Xenopus cells were stained for Nup98 by immunohistochemistry using DAB, which produced a pattern identical to that seen in immunofluorescence (Figure 1E, a), and enabled identification of the Nup98 bodies in the electron microscope. DAB was consistently deposited around smooth, spherical bodies of ∼0.2 μm in diameter (Figure 1E, b). Immunogold staining with anti-Nup98 labeled the same spherical structures (Figure 1E, c). In some sections, it was possible to see more than one spherical structure, as would be expected if the structures were clustered. In these cases, one body has a diameter of ∼0.2 μm, whereas the other appears smaller, because the section does not pass through its center.
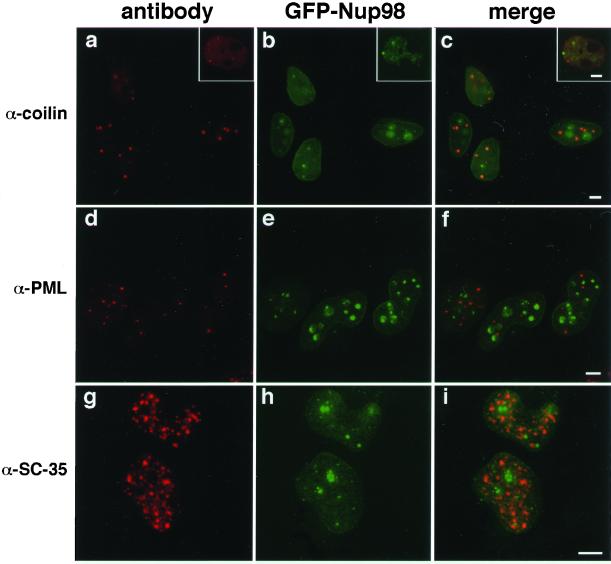
To address the nature of these nuclear bodies, we performed colocalizations with antibodies to components of known nuclear bodies (reviewed by Matera, 1999). Because very few of these antibody markers cross-react with the homologous Xenopus proteins, colocalization experiments were generally performed in HeLa cells transfected with GFP-Nup98 (Figure 2). As before, GFP-Nup98 was found at the nuclear envelope and also in the nuclear bodies. The Nup98 bodies did not colocalize with p80 coilin (Figure 2, c), a component of the Cajal bodies (coiled bodies; Gall, 2000), nor did the Nup98 structures correspond to either PML/ND10 bodies (Figure 2, f) or nuclear splicing speckles (Figure 2, i). An antibody to Xenopus coilin was available and, when used for colocalization, indicated that endogenous xNup98 did not associate with the Cajal bodies (Figure 2, a–c, insets). Based on these results, we conclude that the nuclear bodies in which we detect Nup98 are most probably novel structures.
Figure 2.
The intranuclear Nup98 bodies are novel structures. HeLa cells expressing GFP-Nup98 were stained for markers of other, previously characterized nuclear bodies. GFP-Nup98–expressing cells (b) were stained with anti-p80 coilin (a), and the two images were merged in c. The insets show the same staining in Xenopus cells. GFP-Nup98–expressing cells (e) were stained to visualize PML bodies using either mAb 5E10 (d) or an antibody to SUMO (Griffis and Powers, unpublished results), and the images were merged in f. Splicing factor speckles were visualized with mAb SC-35 (g) in GFP-Nup98–expressing cells (h), and the results are shown merged in i. Scale bars, 5 μm.
Nup98 Is Dynamically Associated with Nuclear Bodies and with the Nuclear Pore
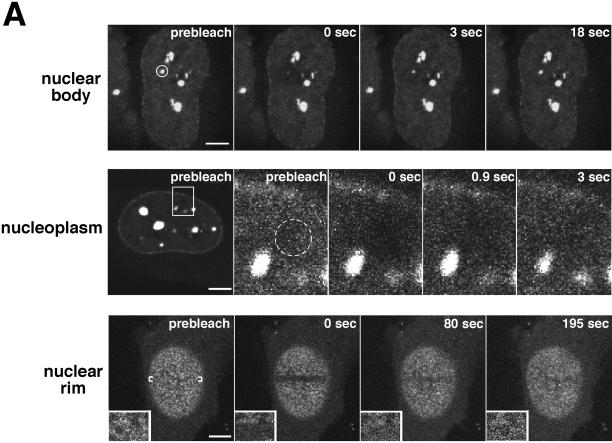
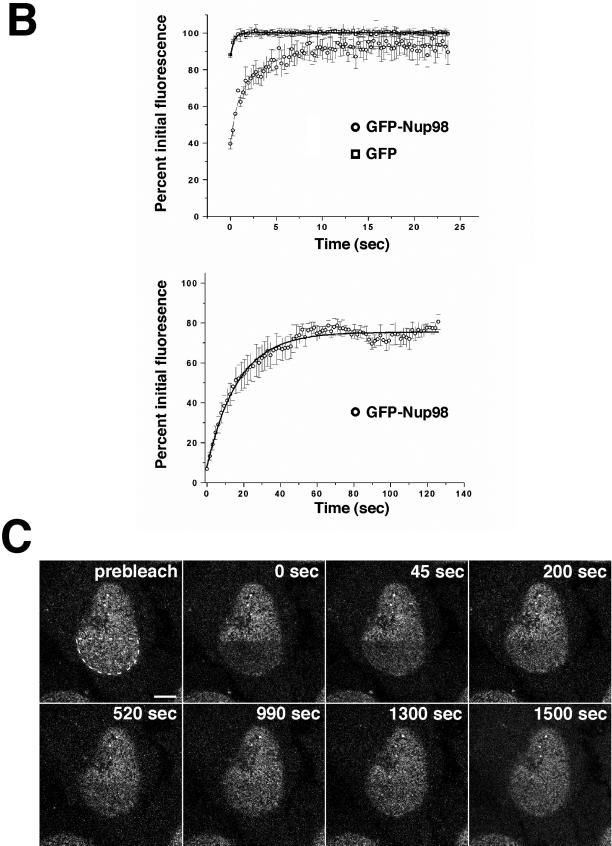
Because we observed Nup98 in multiple compartments of the nucleus, it was important to understand whether this protein can readily move within and between these different nuclear domains. To address this, we took advantage of the GFP-Nup98-expressing cells and the technique of FRAP (reviewed by Lippincott-Schwartz et al., 2001) to measure the mobility of Nup98 in the nucleus. The top row of panels in Figure 3A depicts photobleaching of an Nup98-containing intranuclear body. Images were collected every 1.5 s for a total of 2 min after bleaching. In the middle row are images from a photobleach and recovery of Nup98 in the nucleoplasm. In this experiment, the area indicated in the first was imaged as shown in succeeding panels. After the bleach, images were taken every 150 ms for 25 s to monitor recovery. It is evident that Nup98 moves dynamically in and between these compartments; recovery is observed within seconds, with maximum recovery after ∼20 s for the nuclear bodies and after 3–4 s for the nucleoplasmic protein. Recovery of fluorescence in the nuclear bodies represents exchange of bleached for unbleached protein; over long time courses we never observed that the bodies increased in size as unbleached protein became associated. For comparison, a similar analysis was performed on cells expressing free GFP, which recovered nearly instantaneously in the nucleoplasmic bleach protocol (Griffis and Powers, unpublished results).
Figure 3.
Nup98 is mobile within the nucleoplasm and dynamically associates with both the nuclear bodies and the nuclear pore complex. (A) In the top row, the indicated area containing a single nuclear body was photobleached and then imaged every 1.5 s for 2 min. To improve the time resolution for analyzing Nup98 recovery in the nucleoplasm, the inset area indicated in the first panel of the middle row was enlarged and imaged as a region of interest. The area circled in the prebleach panel was then photobleached and imaged every 150 ms for 20 s. The bottom row represents a tangential section through the nuclear envelope. The indicated area of the nuclear rim was photobleached and then imaged every 1.5 s for a total of 200 s. Insets show close-ups of an area near the edge of the bleach zone. (B) FRAP was quantitated. Values were background corrected and adjusted for bleaching as described in MATERIALS AND METHODS. The top graph represents the average recovery of fluorescence of GFP-Nup98 and free GFP after photobleaching of the nucleoplasm of transfected cells. The bottom graph shows the recovery of GFP-Nup98 in intranuclear bodies after photobleaching. Each graph represents the average of five independent experiments. Error bars depict the SEM. (C) The focal plane was set at the surface of the nuclear envelope, and the region indicated in the first panel was photobleached. The bleached area was then imaged every 20 s for 25 min, and fluorescence recovery was determined. Scale bars, 5 μm. A, top and bottom are presented as videos in the supplementary material.
Fluorescence quantitation was performed to more precisely compare the relative half-times of recovery. Background fluorescence was subtracted from each measurement, and values were corrected for the small amount of bleaching that results from repetitive imaging (see MATERIALS AND METHODS). The corrected values were then normalized to the prebleach fluorescence to obtain the percentage of initial fluorescence. From the data in Figure 3B, we were able to calculate the half-time of recovery for Nup98 in different locations. In the nucleoplasm, GFP-Nup98 recovers with a half-time of 1.16 s. In contrast, GFP has a half-time that is shorter than 200 ms; we were unable to obtain significant bleaching because of the very rapid recovery of the bleached area. As has been previously described for the RNA-splicing factor, ASF, the reduced mobility of a nuclear protein fused to GFP relative to that of GFP alone most likely reflects transient interactions between the nuclear protein and other nuclear components (Kruhlak et al., 2000). In contrast, GFP, which has no nuclear function, would not be expected to interact with nuclear components and thus should exhibit unrestricted movement. Nup98 fluorescence in the nuclear bodies recovers with a half-time of 11.8 s and exhibits an immobile fraction of ∼25%. The nuclear bodies could be subjected to multiple rounds of FRAP, and each time showed similar kinetics of recovery and the same immobile fraction (Griffis and Powers, unpublished results), demonstrating that the bleach was complete and did not cause short-term damage to the cell.
The data above indicated that Nup98 could rapidly associate with and disassociate from one structure, the intranuclear body. We next asked whether Nup98 interacts dynamically with the nuclear pore complex. The bottom row in Figure 3A illustrates FRAP performed on the nuclear envelope of a GFP-Nup98–expressing cell. Insets in each panel contain a magnification of a small region of the nuclear envelope that crosses the bleach boundary. After the bleach, GFP-Nup98 fluorescence returns to the nuclear rim, an indication that Nup98 does in fact move on and off of the pore complex. Because the minimal optical slice of 0.3 μm is greater than the thickness of the pore, it was not possible to absolutely exclude recovery of some photobleached protein within the nucleoplasm from our quantitative measurements. Therefore, we could not accurately measure a half-life of Nup98 recovery at the nuclear pore; however, it is clear that this recovery is not as rapid as recovery after photobleaching of the intranuclear bodies (in Figure 3A, compare top and bottom). We also performed this experiment in the presence of cycloheximide and found that recovery was not further slowed (Griffis and Powers, unpublished results). Thus, the recovery at the pore did not depend on new protein synthesis.
Recovery of GFP-Nup98 fluorescence at the nuclear rim could occur by one of two mechanisms. Unbleached GFP-Nup98 from the nucleoplasm could exchange for the bleached GFP-Nup98 on the nuclear pore complex. Alternatively, nuclear pore complexes could be sufficiently mobile in the nuclear envelope that unbleached pores would move into the region previously occupied by pores containing photobleached Nup98. These two mechanisms would lead to very different predictions about the pattern of recovery after photobleach of a large region of the nuclear envelope. To distinguish between these two models, we performed a photobleach of one-half the area of the nuclear envelope and watched the subsequent recovery of fluorescence (Figure 3C). The GFP-Nup98 signal did not return as a gradually shrinking bleached circle, as would be expected if nuclear pores were moving through the envelope from all directions around the bleached area. Rather, the signal increased simultaneously throughout the bleached area along with a concomitant drop in fluorescence within the unbleached region of the nuclear envelope. Our results are in agreement with, and supported by, the recent elegant demonstration by Ellenberg and colleagues that the nuclear pores are not mobile within the plane of the nuclear envelope (Daigle et al., 2001). Thus, we conclude that Nup98 moves continuously on and off the nuclear pore.
The GLFG Domain Targets Nup98 to the Intranuclear Bodies
Like many nucleoporins, Nup98 has a well-defined domain organization. To determine which region(s) of the protein was required for the localization pattern and dynamics that we observed, we fused GFP individually at the N terminus of: 1) the N-terminal domain including the Gle2/Rae1-binding sequences, 2) the GLFG repeat domain, and 3) the C-terminal domain, which contains the putative nucleoporin RNA-binding motif (Figure 4). We determined the localization and the recovery after photobleaching for each domain. Strikingly, although each of these fusion proteins was at least partially localized to the nucleus, only the GLFG repeat domain associated with intranuclear bodies (Figure 4B). None of the individual domains showed a preferential localization to the nuclear pores, a finding we have also seen with in vitro assembled nuclear pores (Smith and Powers, unpublished results). FRAP experiments were performed to assess the mobility of each of the domains. We found that the N- and C-terminal fusions recover rapidly after photobleaching of the nucleoplasm (half-time of 0.38 and 0.37 s, respectively), although they recover significantly slower than does GFP (Figure 3). In the nucleoplasm, the GLFG domain fusion consistently had a slightly longer half-time of recovery (0.54 s). When the nuclear bodies were photobleached, the GLFG domain showed a time course of recovery closer to that of the full-length GFP-Nup98 (half-time of 5.6 s). We conclude that, although each of these domains participates in some interactions with other nuclear proteins, it is the GLFG repeat domain that targets Nup98 to intranuclear bodies; consequently, we have referred to these as GLFG bodies.
Nup98 Moves between the Nucleus and the Cytoplasm
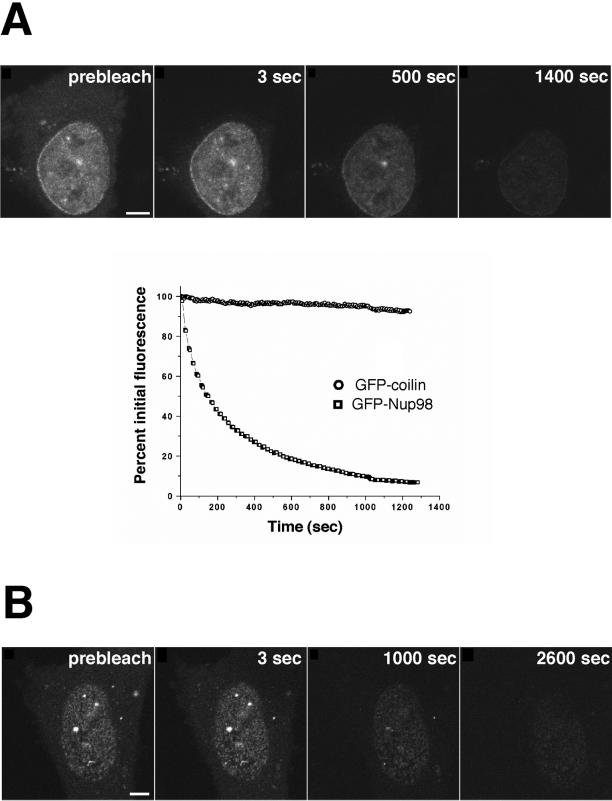
FLIP (reviewed by White and Stelzer, 1999; Lippincott-Schwartz et al., 2001) provides a powerful approach to assess whether distinct cellular compartments are in equilibrium. To determine whether nuclear or nuclear pore complex-associated Nup98 could exchange with the very small population of Nup98 in the cytoplasm, we repetitively bleached a region of the cytoplasm and asked whether GFP-Nup98 fluorescence decreased in the nucleus. For comparison, we monitored the effect of the same bleach protocol on GFP-coilin, which has been reported to shuttle between the nucleus and cytoplasm in Xenopus oocytes (Bellini and Gall, 1999). Strikingly, the FLIP experiments revealed that Nup98 does move between the nucleus and the cytoplasm (Figure 5A, top). GFP-Nup98 fluorescence was reduced to background levels in ∼20 min. To our surprise, we observed that human coilin did not shuttle at an appreciable rate in either HeLa or Cos7 cells (Figure 5B; Griffis and Powers, unpublished results). This may reflect a difference between human and Xenopus coilin or, alternatively, a distinction between oocytes and somatic cells. However, the fact that we did not observe nucleocytoplasmic shuttling made GFP-coilin an ideal control for the effects of nonspecific photobleaching during the FLIP experiment. Less than 8% of the GFP-coilin signal was lost from the nucleus during the time course of this experiment (Figure 5A).
Figure 5.
FLIP reveals that Nup98 can shuttle between the nucleus and cytoplasm. (A) An area of cytoplasm was repeatedly bleached, and the loss of fluorescence from the nucleus was measured over time. As a control, a cell expressing GFP-coilin was repeatedly bleached in the cytoplasm, and the loss of nuclear fluorescence was measured. As the graph indicates, only 8% of the nuclear GFP-coilin was photobleached over the full time course of the experiment. In contrast, GFP-Nup98 fluorescence decreased >94% during the same time course. The data points plotted are not averaged but are representative of 10 independent experiments. (B) The FLIP protocol was modified for measuring the loss of fluorescence of GFP-Nup98 at nuclear pores. The focal plane is set at the surface of the nuclear envelope and, to reduce photobleaching during imaging, fewer scans were taken over a longer time by inserting a 5-s time delay between images. Scale bar, 5 μm. A and B are included as videos in the supplementary material.
The possibility existed that a mobile population of GFP-Nup98 might move between the nucleoplasm and cytoplasm but not assemble into the nuclear pore complex. To test this, we carried out FLIP as described above but monitored fluorescence at the nuclear rim. Again, we found that GFP-Nup98 fluorescence was lost with repetitive bleaching of the cytoplasm (Figure 5A, bottom). Photobleaching of GFP-Nup98 at the nuclear pore occurred with a time course of ∼40 min. We conclude from this that the mobile population of GFP-Nup98 is in equilibrium with the population at the nuclear pore complex.
Nup98 Trafficking Is Independent of the Ran Gradient
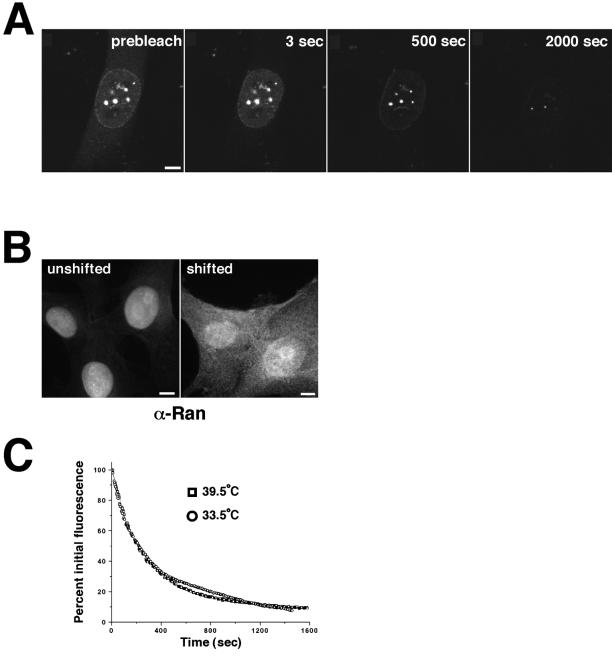
The trafficking of most, but not all, nuclear proteins is dependent on the small GTPase, Ran, and GTP. However, a growing number of substrates is proving to be exceptions to this rule. The export of spliced mRNA, unlike that of tRNA and snRNA, does not require nuclear Ran GTP (Clouse et al., 2001). RCC1, the Ran exchange factor required to generate the GTP-bound form of Ran, can be imported by two distinct transport pathways, one of which is independent of both energy and the Ran gradient (Nemergut and Macara, 2000). The importin β family of transport receptors, when not carrying cargo, can traffic through the pore independently of Ran (Kose et al., 1999). We asked whether the trafficking of Nup98 between cellular compartments requires an existing Ran gradient. GFP-Nup98 was expressed in tsBN2 cells, which carry a temperature sensitive mutation in the RCC1 gene. These cells were then shifted to the nonpermissive temperature for 4 h to remove RCC1 and cause Ran to accumulate in the GDP-bound form. Under these conditions, the steady-state localization of GFP-Nup98 was not changed (Figure 6A, prebleach). Photobleaching of the cytoplasm in an FLIP experiment indicated that GFP-Nup98 continued to traffic between the nuclear and cytoplasmic compartments (Figure 6A). At the same time point, Ran was monitored by immunofluorescence and found to have substantially relocalized to the cytoplasm, an indicator of loss of the Ran gradient (Nemergut and Macara, 2000; Figure 6B). The time course of GFP-Nup98 FLIP was compared in temperature-shifted and unshifted tsBN2 cells and found to be indistinguishable (Figure 6C). We conclude that, like some other components of the transport machinery, Nup98 can traffic through the nuclear pore independently of Ran.
Figure 6.
Nup98 shuttling does not require the Ran gradient. (A) tsBN2 cells were transfected with GFP-Nup98, and, before imaging, the cells were shifted to the nonpermissive temperature (39°C) for 4 h. FLIP was then carried out as described above. (B) To confirm that the temperature shift had resulted in loss of the Ran gradient, Ran was localized by immunofluorescence in tsBN2 cells either growing at 33.5°C (unshifted) or shifted to 39°C for 4 h. The shifted cells that lacked a functional RCC1 protein could not sustain the nuclear Ran gradient and thus did not maintain nuclear localization of Ran. Scale bars, 5μm. (C) Loss of GFP-Nup98 fluorescence was quantitated in both temperature-shifted and unshifted cells. The graphs are not averaged but are representative of five independent experiments. A is presented as a video in the supplementary material.
The Mobility of Nup98 in the Nucleus Is Sensitive to Inhibitors of Transcription
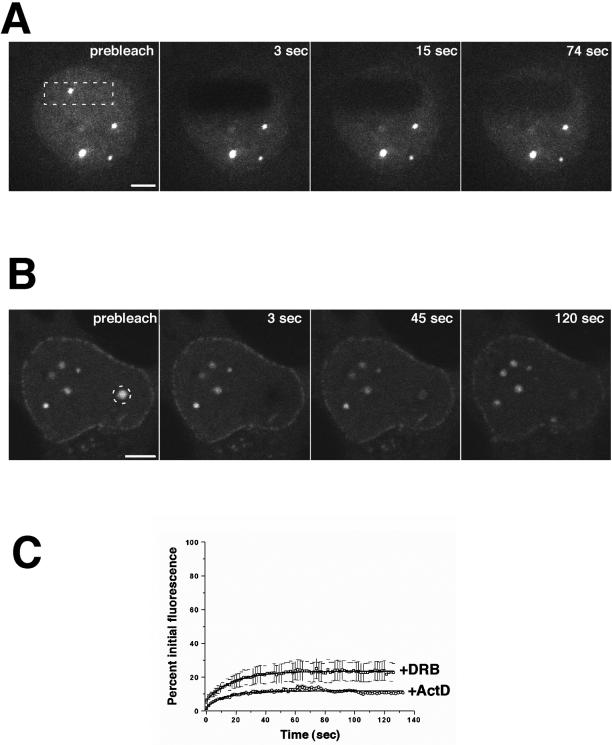
The results presented above suggested that Nup98 is a remarkably mobile nucleoporin, both within the nucleus and between the nucleus and the cytoplasm. However, these experiments could not reveal the functional significance of Nup98 trafficking. To begin to assess the physiological role of this mobile nucleoporin, we treated GFP-Nup98–expressing cells with chemical inhibitors of a variety of processes. Our previous work had indicated a role for Nup98 in nuclear RNA export (Powers et al., 1997). To determine whether there was a link between RNA export and the mobility of Nup98, we initially treated cells with actinomycin D to inhibit transcription by RNA polymerases I and II and carried out FRAP as described above. Remarkably, we observed that after actinomycin D treatment there was virtually no recovery after photobleaching in the nucleus (Figures 7, A and C). This loss of mobility was observed for GFP-Nup98 in both the nucleoplasm and the nuclear bodies. This was not a generalized inhibition of all mobility within the nucleus; FRAP analysis was conducted on both GFP-ASF and GFP-fibrillarin after actinomycin D treatment, and no decrease in their recovery was observed (Griffis and Powers, unpublished results). We also found loss of fluorescence recovery when GFP-Nup98 was photobleached at the nuclear pore complexes (Griffis and Powers, unpublished results). In contrast to these nuclear results, the small amount of GFP-Nup98 that is present in the cytoplasm remained fully mobile after photobleaching. When the cytoplasm was repetitively bleached in an FLIP experiment in the presence of transcription inhibitors, there was no significant loss of nuclear fluorescence (Griffis and Powers, unpublished results). Thus, in the absence of transcription, Nup98 was no longer in equilibrium between the nuclear and cytoplasmic compartments.
Figure 7.
Nup98 mobility is sensitive to inhibitors of transcription. (A) HeLa cells expressing GFP-Nup98 were treated with actinomycin D for 4 h before FRAP analysis. The area outlined was bleached, and fluorescence recovery was measured every 1.5 s for 2 min. (B) HeLa cells expressing GFP-Nup98 were treated with DRB for 4 h before FRAP. The area outlined was bleached, and the fluorescence recovery was measured every 1.5 s for 2 min. Scale bars, 5 μm. (C) Quantitation of the recovery of Nup98 nuclear bodies shows that the mobility of Nup98 is severely reduced by transcriptional inhibitors. Each data point is the average of five independent experiments. Error bars depict the SEM. ActD, actinomycin D. A and B are presented in video form on the CD.
Treatment of cells with a second inhibitor of transcription, DRB, produced the same loss of Nup98 mobility, both within the nucleus and at the nuclear pore (Figure 7, B and C). Again, this is not a generalized effect on nuclear proteins; as previously reported (Kruhlak et al., 2000), we found no decrease in GFP-ASF mobility after DRB treatment. Importantly, the inhibitory effect on Nup98 was specific to transcription-blocking agents. Treatment of cells with leptomycin B, a potent inhibitor of protein export via the CRM-dependent pathway, had no effect on Nup98 mobility (Griffis and Powers, unpublished results). Likewise, cycloheximide, an inhibitor of protein synthesis, aphidicolin, an inhibitor of DNA replication, verapamil, an inhibitor of the multidrug transporter, and A23187, a calcium ionophore, had no effects (Griffis and Powers, unpublished results). Because DRB acts via inhibition of RNA polymerase II phosphorylation, we asked whether other kinase inhibitors might produce a similar decrease in Nup98 mobility. We found that neither staurosporine, a broad-spectrum kinase inhibitor, nor roscovitine, a cdk inhibitor, caused any alteration in Nup98 mobility in any cellular compartment (Griffis and Powers, unpublished results). We conclude from these inhibitor studies that Nup98 mobility is strongly coupled to ongoing transcription, especially by RNA polymerases I and II. Thus, Nup98 is a nucleoporin that may play an important role by linking RNA transcription to RNA export via the nuclear pores.
DISCUSSION
Through a combination of immunofluorescence and immunoelectron microscopy, we have demonstrated that the nucleoporin Nup98 is localized both within the nucleus and at the nuclear pore. Within the nucleus, Nup98 is in equilibrium between the nucleoplasm and a novel nuclear structure, the GLFG body. Further, through photobleaching studies in live cells, we have shown that Nup98 moves dynamically between the nuclear interior and the nuclear pore complex and that this movement is dependent on ongoing transcription. Additionally, Nup98 moves between the nucleus and the cytoplasm. Although movement of Nup98 between these compartments is sensitive to inhibitors of transcription, the mobility of Nup98 in the cytoplasm is not affected. Taken together, these results indicate that Nup98, which we have previously shown to contribute to RNA export, may provide a functional connection between RNA transcription and export.
Several years ago, we first reported the localization of a fraction of Nup98 to the nuclear interior (Powers et al., 1995). At that time, it was unclear precisely how a nucleoporin might function other than at the pore complex, although Nup98-depleted nuclei had major defects in nuclear organization and DNA replication. Since then, other nuclear pore-associated proteins with at least partial nucleoplasmic distributions have been described. Ranbp3 is a Ran GTP-binding protein of the Ranbp1 family and is found in the nucleoplasm (Mueller et al., 1998). Rip/Rab is an HIV Rev-binding protein with FG repeat motifs (Bogerd et al., 1995; Fritz et al., 1995; Stutz et al., 1995). Nup50 is an FG repeat-containing nucleoporin that interacts with Nup153 (Guan et al., 2000). Both of these proteins are found in the nucleoplasm and at the nuclear pore complex. Rae1/Gle2 is an RNA export factor that binds to Nup98 at the pore and is also seen both inside the nucleus and in the cytoplasm (Murphy et al., 1996; Bharathi et al., 1997; Pritchard et al., 1999). Perhaps most significantly, Tpr was found to form fibers that extend into the nucleus from the nuclear pore complex (Cordes et al., 1997; Zimowska et al., 1997). Fontoura et al. (2001) recently reported that Tpr can interact with Nup98 in vitro. Intriguingly, in cultured Drosophila cells, Tpr can also be seen by immunofluorescence microscopy in bodies within the nucleus (Zimowska et al., 1997).
What are the intranuclear GLFG bodies? They are clearly distinct from the well characterized Cajal bodies, PML/ND10 bodies, and splicing factor speckles based on both staining with marker antibodies and electron microscopic structure. The 0.2 μm diameter spheres most closely resemble the simple nuclear bodies previously noted in electron microscopy studies, sometimes seen as multiple associated bodies (reviewed by Brasch and Ochs, 1992). However, novel bodies have been emerging as more proteins are shown to associate with nuclear foci (Pombo et al., 1998; Nayler et al., 2000). Most such nuclear bodies appear to function as storage or recycling sites for transcription and processing factors (Matera, 1999). In at least one case, however, active transcription is associated with a type of nuclear body (Pombo et al., 1998). However, we have found that the active, phosphorylated form of RNA polymerase II is not concentrated in the GLFG bodies (Griffis and Powers, unpublished results). It is possible that the GLFG bodies serve to store Nup98 and perhaps other factors involved in RNA export.
How is Nup98 targeted to the nuclear GLFG bodies? It is clear that the GLFG repeat domain is responsible for directing Nup98 to these structures. This could occur through interaction with an endogenous factor or it is possible that the formation of bodies is initiated by self-interaction between multiple Nup98 proteins. Such a self-interaction has been proposed for the initiation of Cajal body formation by p80 coilin (Hebert and Matera, 2000). Gel exclusion chromatography indicates that during mitosis Nup98 exists in a complex of ∼450 kDa (Macaulay et al., 1995; Matsuoka et al., 1999). The stoichiometry of this complex is unknown. However, because the only other known member is the 42-kDa protein, RaeI/Gle2, it is possible that Nup98 does in fact associate with itself to form a multimeric complex. Our preliminary colocalization experiments did not detect Tpr in the GLFG bodies, but a careful analysis to exclude the presence of Tpr should entail the use of antibodies to multiple domains of this large protein, because some regions may not be accessible when incorporated into a structure. Similarly, we did not find other nucleoporins (Nup214, Nup153, p62) or transport factors (importin α, importin β, CRM, TAP) in the GLFG bodies (Griffis and Powers, unpublished results). Identification of the full composition of the Nup98 bodies may await purification of these structures and characterization of protein components.
When observing GFP-Nup98–expressing cells over long periods of time (8–10 h), the GLFG bodies do not move far from their original location within the nucleus; they appear to be tethered in one region (Griffis, Altan, Lippincott-Schwartz, and Powers, unpublished observation). Such limited mobility over time has been similarly reported for splicing factor bodies (Eils et al., 2000; Kruhlak et al., 2000) and coiled bodies (Platani et al., 2000). This is distinctly different from the artifactual foci observed after transfection with fluorescent histone deacetylase, which move rapidly around the nucleus over a short time span (Kruhlak et al., 2000). Cajal bodies, although less mobile than the histone deacetylase, appear to be more motile than either GLFG or splicing factor bodies (Platani et al., 2000). Over time, the GLFG bodies do not vary appreciably in size or intensity, even without cycloheximide treatment. In this respect, they are similar to Cajal bodies, which have an established size and change only by fusion or fission (Platani et al., 2000). Nup98 trafficks in and out of the GLFG bodies with an immobile fraction of only 25%. The dynamic association of constituent proteins is also characteristic of the known nuclear compartments or bodies (reviewed by Matera, 1999; Misteli, 2001).
Why is Nup98 not freely mobile in the nucleus? In FRAP experiments similar to those presented here, other nuclear protein-GFP fusions have also proven to have significantly longer recovery times after photobleaching than does free GFP. It is proposed that nuclear proteins, even when localized in the nucleoplasm rather than in a nuclear structure, undergo continuous transient interactions with other nuclear proteins that may be relatively less mobile (Misteli, 2001; Pederson, 2001). Such interactions would not occur with the nonphysiological, nuclear GFP. It is not surprising that Nup98 might interact with other proteins in the nucleoplasm. In particular, the GLFG repeat domain of Nup98 binds to soluble nuclear export factors such as TAP and CRM (Neville et al., 1997; Bachi et al., 2000; Strasser et al., 2000; Strawn et al., 2001). The N- and C-terminal domains are inherently slightly more mobile than the repeat domain, suggesting that they may participate in fewer, or perhaps more transient, protein interactions within the nucleus. However, all three domains of Nup98 are significantly less mobile than free GFP. A sequence within the N-terminal domain of Nup98 constitutes a binding site for the RNA export factor, RaeI/Gle2 (Pritchard et al., 1999), and the C-terminal domain could interact with as yet unidentified proteins. Nup98 also binds to Tpr, a component of nuclear filaments, although the domain of Nup98 involved in this interaction is as yet undetermined (Fontoura et al., 2001).
We found that the transcription inhibitor actinomycin D dramatically reduced both the mobility of Nup98 within the nucleus and the movement of Nup98 between nuclear and cytoplasmic compartments. At the concentrations used here, actinomycin D is a potent inhibitor of both RNA polymerases (Pol) I and II. Our previous antibody injection experiments indicated a role for Nup98 in nuclear export of both Pol I and Pol II transcripts (Powers et al., 1997). It is interesting to note that such injection experiments could not distinguish where the antibody was impairing the function of Nup98, in the nucleoplasm, at the nuclear pore, or possibly at both locations. DRB blocks only Pol II transcription; it is not known to inhibit rRNA transcription by Pol I. We observed a slightly less complete inhibition of mobility by DRB, which may reflect the continuing production of some export substrates. When individual domains of Nup98 were examined, it was the GLFG domain alone whose mobility was sensitive to actinomycin D (Griffis and Powers, unpublished results). Strikingly, the GLFG domain has been proposed to interact with the transcription apparatus; fusion of this domain to a DNA-binding motif results in aberrant activation of transcription (Kasper et al., 1999). Although the precise mechanism of transcriptional activation is as yet unclear, cellular transformation and transcriptional activation by the fusion protein correlate with the ability of the GLFG repeat domain to bind in vitro to the transcriptional coactivators, CREB-binding protein and p300. Thus, it is the GLFG repeat domain that links Nup98 to transcription.
In keeping with our findings, it was previously suggested that Nup98 and Nup214 might be mobile components of the nuclear pore complex (Zolotukhin and Felber, 1999). Interestingly, the consequences of actinomycin D treatment in that report were strikingly different from our results; in contrast to the loss of Nup98 mobility that we observe, Zolotukhin and Felber reported that Nup98 relocalized to the cytoplasm, as has been seen with other RNA-binding proteins such as hnRNP A1. This distinct difference in outcome may possibly result from a difference in cell lines. The HeLa-derived cell line used in the experiments of Zolotukhin and Felber constitutively expressed the HIV Tat protein, a regulator of HIV transcriptional elongation that has also been suggested to interact with transcriptional coactivators CREB-binding protein and p300 (Hottiger and Nabel, 1998; Vo and Goodman, 2001). Zolotukhin and Felber did not observe relocalization of Nup98 in Xenopus cells after actinomycin D treatment, nor, in similar experiments, did we (Smith and Powers, unpublished data). It would be interesting to test the mobility of GFP-Nup98 in the HeLa-tat cells.
In summary, we have shown that the GLFG nucleoporin, Nup98, is a dynamic component of the nuclear pore complex. Nup98 is mobile within the nucleoplasm and associates both with a novel structure, the GLFG body, and with the nuclear pore complex. The data presented here, along with our previous studies showing a role for Nup98 in nuclear export, are consistent with a model in which Nup98 interacts with the pore-associated filament network to facilitate export of mRNA, snRNA, and rRNA. Nup98 could transiently bind to or progress along this network through a mechanism that is coupled to ongoing RNA transcription. Thus, one role for a mobile nucleoporin may be the direction of export substrates to the pore. This may be a common function of the mobile nucleoporins, or there may be as yet undetermined functions contributed by other dynamic components of the nuclear pore complex.
Supplementary Material
ACKNOWLEDGMENTS
We sincerely thank our colleagues who provided us with reagents. We thank Dr. Joe Gall for anticoilin, Dr. Tom Mistelli for the GFP-SF2 and GFP-fibrillarin constructs, Drs. Michael Hebert and Greg Matera for the human GFP-coilin, Dr. Julian Borrow for the human Nup98 clone, Dr. Rebecca Heald for XL177 cells, Dr. Ian Macara for the tsBN2 cell line, and Dr. Minoru Yoshida for leptomycin B. We are grateful to Dawn Greenside, Ashley Smith, and Melanie Lester for excellent technical assistance, and to Hong Yi of the Emory Neuroscience Microscopy Facility for expert assistance with electron microscopy. We thank Dr. Paula Vertino for essential advice, Dr. Keith Berland for helpful discussions, and Drs. Katharine Ullman, Win Sale, and Barry Shur for comments on the manuscript. This work was supported by a grant from the National Institutes of Health to M.A.P. (GM-59975). E.R.G. is a predoctoral trainee of the National Institutes of Health.
Abbreviations used:
- DAB
3–3′-diaminobenzidine tetrahydrochloride
- DRB
5,6-dichloro-1-β-d-ribofuranosylbenzimidazole
- FCS
fetal calf serum
- FLIP
fluorescence loss in photobleaching
- FRAP
fluorescence recovery after photobleaching
- GLFG
glycine-leucine-phenylalanine-glycine
- GFP
green fluorescent protein
- NGS
normal goat serum
- Nup
nucleoporin
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
Footnotes
Online version of this article contains video material for some figures. Online version available at www.molbiolcell.org.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–11–0538. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–11–0538.
REFERENCES
- Allen TD, Cronshaw JM, Bagley S, Kiseleva E, Goldberg MW. The nuclear pore complex: mediator of translocation between nucleus and cytoplasm. J Cell Sci. 2000;113:1651–1659. doi: 10.1242/jcs.113.10.1651. [DOI] [PubMed] [Google Scholar]
- Andrade LE, Tan EM, Chan EK. Immunocytochemical analysis of the coiled body in the cell cycle and during cell proliferation. Proc Natl Acad Sci USA. 1993;90:1947–1951. doi: 10.1073/pnas.90.5.1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azuma Y, Dasso M. The role of Ran in nuclear function. Curr Opin Cell Biol. 2000;12:302–307. doi: 10.1016/s0955-0674(00)00093-4. [DOI] [PubMed] [Google Scholar]
- Bachi A, Braun IC, Rodrigues JP, Pante N, Ribbeck K, von Kobbe C, Kutay U, Wilm M, Gorlich D, Carmo-Fonseca M, Izaurralde E. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Nup 116p and Nup 100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. EMBO J. 1998;17:1107–1119. doi: 10.1093/emboj/17.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellini M, Gall JG. Coilin shuttles between the nucleus and cytoplasm in Xenopus oocytes. Mol Biol Cell. 1999;10:3425–3434. doi: 10.1091/mbc.10.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben-Efraim I, Gerace L. Gradient of increasing affinity of importin beta for nucleoporins along the pathway of nuclear import. J Cell Biol. 2001;152:411–417. doi: 10.1083/jcb.152.2.411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bharathi A, Ghosh A, Whalen WA, Yoon JH, Pu R, Dasso M, Dhar R. The human RAE1 gene is a functional homologue of Schizosaccharomyces pombe rae1 gene involved in nuclear export of Poly(A)+ RNA. Gene. 1997;198:251–258. doi: 10.1016/s0378-1119(97)00322-3. [DOI] [PubMed] [Google Scholar]
- Bogerd HP, Fridell RA, Madore S, Cullen BR. Identification of a novel cellular cofactor for the Rev/Rex class of retroviral regulatory proteins. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- Brasch K, Ochs RL. Nuclear bodies (NBs): a newly “rediscovered”organelle. Exp Cell Res. 1992;202:211–223. doi: 10.1016/0014-4827(92)90068-j. [DOI] [PubMed] [Google Scholar]
- Clouse KN, Luo MJ, Zhou Z, Reed R. A Ran-independent pathway for export of spliced mRNA. Nat Cell Biol. 2001;3:97–99. doi: 10.1038/35050625. [DOI] [PubMed] [Google Scholar]
- Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Kohler A, Stuurman N, van Driel R, Franke WW. Intranuclear filaments containing a nuclear pore complex protein. J Cell Biol. 1993;123:1333–1344. doi: 10.1083/jcb.123.6.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes VC, Reidenbach S, Rackwitz HR, Franke WW. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments. J Cell Biol. 1997;136:515–529. doi: 10.1083/jcb.136.3.515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigle N, Beaudouin J, Hartnell L, Imreh G, Hallberg E, Lippincott-Schwartz J, Ellenberg J. Nuclear pore complexes form immobile networks and have a very low turnover in live mammalian cells. J Cell Biol. 2001;154:71–84. doi: 10.1083/jcb.200101089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dilworth DJ, Suprapto A, Padovan JC, Chait BT, Wozniak RW, Rout MP, Aitchison JD. Nup2p dynamically associates with the distal regions of the yeast nuclear pore complex. J Cell Biol. 2001;153:1465–1478. doi: 10.1083/jcb.153.7.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eils R, Gerlich D, Tvarusko W, Spector DL, Misteli T. Quantitative imaging of pre-mRNA splicing factors in living cells. Mol Biol Cell. 2000;11:413–418. doi: 10.1091/mbc.11.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabre E, Boelens WC, Wimmer C, Mattaj I, Hurt EC. Nup145p is required for nuclear export of mRNA and binds homopolymeric RNA in vitro via a novel conserved motif. Cell. 1994;78:275–289. doi: 10.1016/0092-8674(94)90297-6. [DOI] [PubMed] [Google Scholar]
- Fontoura BM, Blobel G, Yaseen NR. The nucleoporin Nup98 is a site for GDP/GTP exchange on ran and termination of karyopherin beta 2-mediated nuclear import. J Biol Chem. 2000;275:31289–31296. doi: 10.1074/jbc.M004651200. [DOI] [PubMed] [Google Scholar]
- Fontoura BM, Dales S, Blobel G, Zhong H. The nucleoporin Nup98 associates with the intranuclear filamentous protein network of TPR. Proc Natl Acad Sci USA. 2001;98:3208–3213. doi: 10.1073/pnas.061014698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti KG, Fransen J, Grosveld G. The human homologue of yeast CRM1 is in a dynamic subcomplex with CAN/Nup214 and a novel nuclear pore component Nup88. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fritz CC, Zapp ML, Green MR. A human nucleoporin-like protein that specifically interacts with HIV Rev. Nature. 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- Gall JG. Cajal bodies: the first 100 years. Annu Rev Cell Dev Biol. 2000;16:273–300. doi: 10.1146/annurev.cellbio.16.1.273. [DOI] [PubMed] [Google Scholar]
- Goldberg MW, Rutherford SA, Hughes M, Cotter LA, Bagley S, Kiseleva E, Allen TD, Clarke PR. Ran alters nuclear pore complex conformation. J Mol Biol. 2000;300:519–529. doi: 10.1006/jmbi.2000.3891. [DOI] [PubMed] [Google Scholar]
- Gorlich D, Kutay U. Transport between the cell nucleus and the cytoplasm. Annu Rev Cell Dev Biol. 1999;15:607–660. doi: 10.1146/annurev.cellbio.15.1.607. [DOI] [PubMed] [Google Scholar]
- Guan T, Kehlenbach RH, Schirmer EC, Kehlenbach A, Fan F, Clurman BE, Arnheim N, Gerace L. Nup50, a nucleoplasmically oriented nucleoporin with a role in nuclear protein export. Mol Cell Biol. 2000;20:5619–5630. doi: 10.1128/mcb.20.15.5619-5630.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert MD, Matera AG. Self-association of coilin reveals a common theme in nuclear body localization. Mol Biol Cell. 2000;11:4159–4171. doi: 10.1091/mbc.11.12.4159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hood JK, Silver PA. Diverse nuclear transport pathways regulate cell proliferation and oncogenesis. Biochim Biophys Acta. 2000;1471:M31–41. doi: 10.1016/s0304-419x(00)00018-4. [DOI] [PubMed] [Google Scholar]
- Hottiger MO, Nabel GJ. Interaction of human immunodeficiency virus type 1 Tat with the transcriptional coactivators p300 and CREB binding protein. J Virol. 1998;72:8252–8256. doi: 10.1128/jvi.72.10.8252-8256.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasper LH, Brindle PK, Schnabel CA, Pritchard CE, Cleary ML, van Deursen JM. CREB binding protein interacts with nucleoporin-specific FG repeats that activate transcription and mediate NUP98-HOXA9 oncogenicity. Mol Cell Biol. 1999;19:764–776. doi: 10.1128/mcb.19.1.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiseleva E, Goldberg M, Daneholt B, Allen TD. RNP export is mediated by structural reorganization of the nuclear basket. J Mol Biol. 1996;260:304–311. doi: 10.1006/jmbi.1996.0401. [DOI] [PubMed] [Google Scholar]
- Kose S, Imamoto N, Tachibana T, Yoshida M, Yoneda Y. Beta-subunit of nuclear pore-targeting complex (importin-beta) can be exported from the nucleus in a Ran-independent manner. J Biol Chem. 1999;274:3946–3952. doi: 10.1074/jbc.274.7.3946. [DOI] [PubMed] [Google Scholar]
- Kruhlak MJ, Lever MA, Fischle W, Verdin E, Bazett-Jones DP, Hendzel MJ. Reduced mobility of the alternate splicing factor (ASF) through the nucleoplasm and steady state speckle compartments. J Cell Biol. 2000;150:41–51. doi: 10.1083/jcb.150.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippincott-Schwartz J, Snapp E, Kenworthy A. Studying protein dynamics in living cells. Nat Rev Mol Cell Biol. 2001;2:444–456. doi: 10.1038/35073068. [DOI] [PubMed] [Google Scholar]
- Macaulay C, Meier E, Forbes DJ. Differential mitotic phosphorylation of proteins of the nuclear pore complex. J Biol Chem. 1995;270:254–262. doi: 10.1074/jbc.270.1.254. [DOI] [PubMed] [Google Scholar]
- Matera AG. Nuclear bodies: multifaceted subdomains of the interchromatin space. Trends Cell Biol. 1999;9:302–309. doi: 10.1016/s0962-8924(99)01606-2. [DOI] [PubMed] [Google Scholar]
- Matsuoka Y, Takagi M, Ban T, Miyazaki M, Yamamoto T, Kondo Y, Yoneda Y. Identification and characterization of nuclear pore subcomplexes in mitotic extract of human somatic cells. Biochem Biophys Res Commun. 1999;254:417–423. doi: 10.1006/bbrc.1998.9953. [DOI] [PubMed] [Google Scholar]
- Mattaj IW, Englmeier L. Nucleocytoplasmic transport: the soluble phase. Annu Rev Biochem. 1998;67:265–306. doi: 10.1146/annurev.biochem.67.1.265. [DOI] [PubMed] [Google Scholar]
- McNally JG, Karpova T, Cooper J, Conchello JA. Three-dimensional imaging by deconvolution microscopy. Methods. 1999;19:373–385. doi: 10.1006/meth.1999.0873. [DOI] [PubMed] [Google Scholar]
- Misteli T. Protein dynamics: implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- Mueller L, Cordes VC, Bischoff FR, Ponstingl H. Human RanBP3, a group of nuclear RanGTP binding proteins. FEBS Lett. 1998;427:330–336. doi: 10.1016/s0014-5793(98)00459-1. [DOI] [PubMed] [Google Scholar]
- Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of thte Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Mol Biol Cell. 1996;7:1921–1937. doi: 10.1091/mbc.7.12.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakielny S, Dreyfuss G. Transport of proteins and RNAs in and out of the nucleus. Cell. 1999;99:677–690. doi: 10.1016/s0092-8674(00)81666-9. [DOI] [PubMed] [Google Scholar]
- Nakielny S, Shaikh S, Burke B, Dreyfuss G. Nup153 is an M9-containing mobile nucleoporin with a novel Ran-binding domain. EMBO J. 1999;18:1982–1995. doi: 10.1093/emboj/18.7.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayler O, Hartmann AM, Stamm S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J Cell Biol. 2000;150:949–962. doi: 10.1083/jcb.150.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemergut ME, Macara IG. Nuclear import of the ran exchange factor, RCC1, is mediated by at least two distinct mechanisms. J Cell Biol. 2000;149:835–850. doi: 10.1083/jcb.149.4.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neville M, Stutz F, Lee L, Davis LI, Rosbash M. The importin-beta family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- Ohno M, Fornerod M, Mattaj IW. Nucleocytoplasmic transport: the last 200 nanometers. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- Pederson T. Protein mobility within the nucleus: what are the right moves? Cell. 2001;104:635–638. doi: 10.1016/s0092-8674(01)00258-6. [DOI] [PubMed] [Google Scholar]
- Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- Platani M, Goldberg I, Swedlow JR, Lamond AI. In vivo analysis of Cajal body movement, separation, and joining in live human cells. J Cell Biol. 2000;151:1561–1574. doi: 10.1083/jcb.151.7.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pombo A, Cuello P, Schul W, Yoon JB, Roeder RG, Cook PR, Murphy S. Regional and temporal specialization in the nucleus: a transcriptionally-active nuclear domain rich in PTF, Oct1 and PIKA antigens associates with specific chromosomes early in the cell cycle. EMBO J. 1998;17:1768–1778. doi: 10.1093/emboj/17.6.1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J Cell Biol. 1997;136:241–250. doi: 10.1083/jcb.136.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers MA, Macaulay C, Masiarz FR, Forbes DJ. Reconstituted nuclei depleted of a vertebrate GLFG nuclear pore protein, p97, import but are defective in nuclear growth and replication. J Cell Biol. 1995;128:721–736. doi: 10.1083/jcb.128.5.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard CE, Fornerod M, Kasper LH, van Deursen JM. RAE1 is a shuttling mRNA export factor that binds to a GLEBS-like NUP98 motif at the nuclear pore complex through multiple domains. J Cell Biol. 1999;145:237–254. doi: 10.1083/jcb.145.2.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81:215–222. doi: 10.1016/0092-8674(95)90331-3. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Gorlich D. Kinetic analysis of translocation through nuclear pore complexes. EMBO J. 2001;20:1320–1330. doi: 10.1093/emboj/20.6.1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. J Cell Biol. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan KJ, Wente SR. The nuclear pore complex: a protein machine bridging the nucleus and cytoplasm. Curr Opin Cell Biol. 2000;12:361–371. doi: 10.1016/s0955-0674(00)00101-0. [DOI] [PubMed] [Google Scholar]
- Shah S, Tugendreich S, Forbes D. Major binding sites for the nuclear import receptor are the internal nucleoporin Nup153 and the adjacent nuclear filament protein Tpr. J Cell Biol. 1998;141:31–49. doi: 10.1083/jcb.141.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffler D, Fahrenkrog B, Aebi U. The nuclear pore complex: from molecular architecture to functional dynamics. Curr Opin Cell Biol. 1999a;11:391–401. doi: 10.1016/S0955-0674(99)80055-6. [DOI] [PubMed] [Google Scholar]
- Stoffler D, Goldie KN, Feja B, Aebi U. Calcium-mediated structural changes of native nuclear pore complexes monitored by time-lapse atomic force microscopy. J Mol Biol. 1999b;287:741–752. doi: 10.1006/jmbi.1999.2637. [DOI] [PubMed] [Google Scholar]
- Strambio-de-Castillia C, Blobel G, Rout MP. Proteins connecting the nuclear pore complex with the nuclear interior. J Cell Biol. 1999;144:839–855. doi: 10.1083/jcb.144.5.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strasser K, Bassler J, Hurt E. Binding of the Mex67p/Mtr2p heterodimer to FXFG, GLFG, and FG repeat nucleoporins is essential for nuclear mRNA export. J Cell Biol. 2000;150:695–706. doi: 10.1083/jcb.150.4.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strawn LA, Shen T, Wente SR. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. J Biol Chem. 2001;276:6445–6452. doi: 10.1074/jbc.M008311200. [DOI] [PubMed] [Google Scholar]
- Stutz F, Neville M, Rosbash M. Identification of a novel nuclear pore-associated protein as a functional target of the HIV-1 Rev protein in yeast. Cell. 1995;82:495–506. doi: 10.1016/0092-8674(95)90438-7. [DOI] [PubMed] [Google Scholar]
- Vasu SK, Forbes DJ. Nuclear pores and nuclear assembly. Curr Opin Cell Biol. 2001;13:363–375. doi: 10.1016/s0955-0674(00)00221-0. [DOI] [PubMed] [Google Scholar]
- Vo N, Goodman RH. CREB-binding protein and p300 in transcriptional regulation. J Biol Chem. 2001;276:13505–13508. doi: 10.1074/jbc.R000025200. [DOI] [PubMed] [Google Scholar]
- Wang YL. Digital deconvolution of fluorescence images for biologists. Methods Cell Biol. 1998;56:305–315. doi: 10.1016/s0091-679x(08)60432-x. [DOI] [PubMed] [Google Scholar]
- Wente SR, Blobel G. A temperature-sensitive NUP116 null mutant forms a nuclear envelope seal over the yeast nuclear pore complex thereby blocking nucleocytoplasmic transport. J Cell Biol. 1993;123:275–284. doi: 10.1083/jcb.123.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wente SR, Blobel G. NUP145 encodes a novel yeast glycine-leucine-phenylalanine-glycine (GLFG) nucleoporin required for nuclear envelope structure. J Cell Biol. 1994;125:955–969. doi: 10.1083/jcb.125.5.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J, Stelzer E. Photobleaching GFP reveals protein dynamics inside live cells. Trends Cell Biol. 1999;9:61–65. doi: 10.1016/s0962-8924(98)01433-0. [DOI] [PubMed] [Google Scholar]
- Zimowska G, Aris JP, Paddy MR. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes. J Cell Sci. 1997;110:927–944. doi: 10.1242/jcs.110.8.927. [DOI] [PubMed] [Google Scholar]
- Zolotukhin AS, Felber BK. Nucleoporins nup98 and nup214 participate in nuclear export of human immunodeficiency virus type 1 Rev. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.