Abstract
Arena3Dweb is an interactive web tool that visualizes multi-layered networks in 3D space. In this update, Arena3Dweb supports directed networks as well as up to nine different types of connections between pairs of nodes with the use of Bézier curves. It comes with different color schemes (light/gray/dark mode), custom channel coloring, four node clustering algorithms which one can run on-the-fly, visualization in VR mode and predefined layer layouts (zig-zag, star and cube). This update also includes enhanced navigation controls (mouse orbit controls, layer dragging and layer/node selection), while its newly developed API allows integration with external applications as well as saving and loading of sessions in JSON format. Finally, a dedicated Cytoscape app has been developed, through which users can automatically send their 2D networks from Cytoscape to Arena3Dweb for 3D multi-layer visualization. Arena3Dweb is accessible at http://arena3d.pavlopouloslab.info or http://arena3d.org
INTRODUCTION
Network biology has become a major domain in modern biology that captures associations among biomedical terms and entities such as organisms, diseases, genes, proteins, drugs and processes (1,2). To be able to view these networks, many visualization tools have been proposed (3,4). While Cytoscape (5,6), Gephi (7), Tulip (8) and Pajek (9) are few of the most widely used applications, latest efforts focus on addressing more targeted problems. NORMA (10,11) for example focuses on distinguishing annotated groups in a network using innovative layout strategies. OmicsNet (12) is developed to support multi-omics integration and network visual analytics. Graphia (13) comes with advanced layout algorithms to visualize a network in 3D, and STRING (14) uses an in-house graph visualizer to support multi-edged connections (different channels of information) between two nodes (proteins) (15).
Arena3Dweb (16), the web-based successor to Arena3D (17,18), is the first, fully interactive 3D web tool for processing, analyzing and visualizing multilayered networks. It allows advanced 3D visualizations to be created and navigated by providing a rich collection of layout algorithms, which one can apply on a selected set of layers individually or in combination, the ability to color layers, nodes and edges, and options to highlight and resize nodes based on topological features.
Here, we present an updated version of Arena3Dweb, which goes far beyond the aforementioned features. It now supports directed networks to allow visualization of regulatory networks, multi-edge connections to allow multiple types of edges between the same two nodes, and intra/ inter-layer node clustering. The new version also features alternative color themes including a light mode suitable for designing figures for print, and enhanced navigation virtual reality (VR) functionality for viewing large networks in 3D. Finally, we provide import/export functions in JSON format and an Application Programming Interface (API) for integration with other applications such as connecting Cytoscape with Cytoscape with Arena3Dweb through the new Arena3D Cytoscape app.
UI/UX ENHANCEMENT
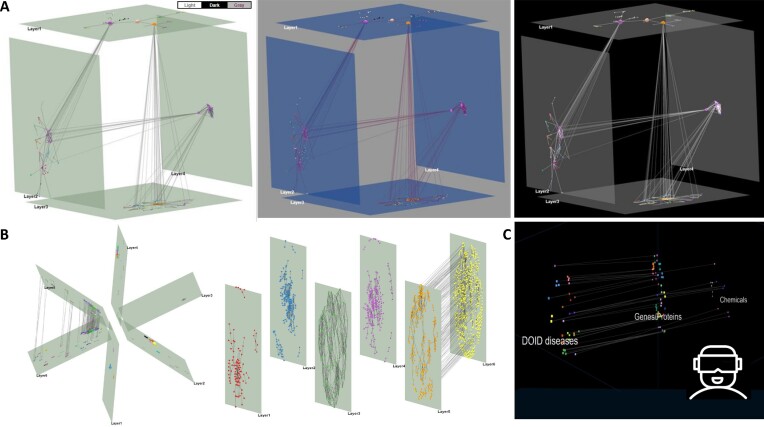
In this version, Arena3Dweb comes with major aesthetic improvements. The tool now offers three different color themes including light, gray and dark mode (Figure 1A). The light mode in particular enables users to produce publication-ready figures with a white background more easily, without the need to tweak the respective scene, layer and edge colors manually. In addition, several predefined layer layouts are also offered. To this end, initial 3D layer setups are produced automatically and include a zig-zag, a star and a cube layout (Figure 1B). In the star layout, a virtual sphere of 360° is equally divided by the total number of layers whereas in the cube layout, each cube can contain up to six layers. In the case of more than 6 layers, additional cubes are created and placed next to each other.
Figure 1.
Updated viewing options. (A) Three default themes for a cube multi-layered network including (from left to right) a light, a gray and a dark mode option. (B) A star predefined layout (left) and a zig-zag layer layout (right) of six layers. (C) A VR view for a network with three layers (disease, proteins and chemicals).
In addition to the layouts, object manipulation has been significantly enhanced in this version. Raycasting via the three.js package has been implemented for easier layer and node selection with the mouse. Nodes change color on mouse hover as a visual cue while hovered layers are highlighted in red. Layer and node selection through double-click is also offered, while layers can be dragged with the mouse on the 3D scene as an alternative to the navigation control action buttons. The scene orbit controls (middle-click drag) have also been enhanced, allowing for smoother 3D scene rotations compared to the old version of the tool.
Finally, Arena3Dweb now allows network exploration in VR mode (Figure 1C). VR views are static and can be accessed via a VR headset or a mobile phone with a gyroscope. For better clarity, layer floors are disabled while layer labels always face the user's camera. Notably, the VR view is always offered in a new tab and visualizes the running view of Arena3Dweb at any time point. For consistency, the objects’ coordinate system is always adjusted in the VR view. We note that the VR functionality is only accessible through the web application and not through the local version of Arena3Dweb, since all required VR files are served internally through the API (see section ‘Interoperability’).
MULTIPLE EDGE CHANNELS AND DIRECTIONS
In biology, two entities can often be linked with more than one type of connection. In the STRING database, for example, two genes can be co-expressed, co-mentioned in literature or bind to each other. In a signal transduction network, a petri-net or a pathway, signal tracing as well as up- and down-stream analysis can be monitored by following directed paths visualized by arrows rather than plain lines. Therefore, the support of directed and multi-edged graphs is of great importance.
In this version of Arena3Dweb, we address both issues by allowing arrows and up to nine different types of connections between two nodes. In the case of multi-edged networks, each information channel must be labeled in the Arena3Dweb input file and the corresponding edges are visualized as Bézier curves with distinct colors. In case of directed graphs, two intra- or inter-layer nodes can be connected via straight or curved arrows. The default channel colors are slightly adjusted to each theme but can be manually changed via color palettes at any time, while arrow sizes and curvature can be adjusted via dedicated sliders.
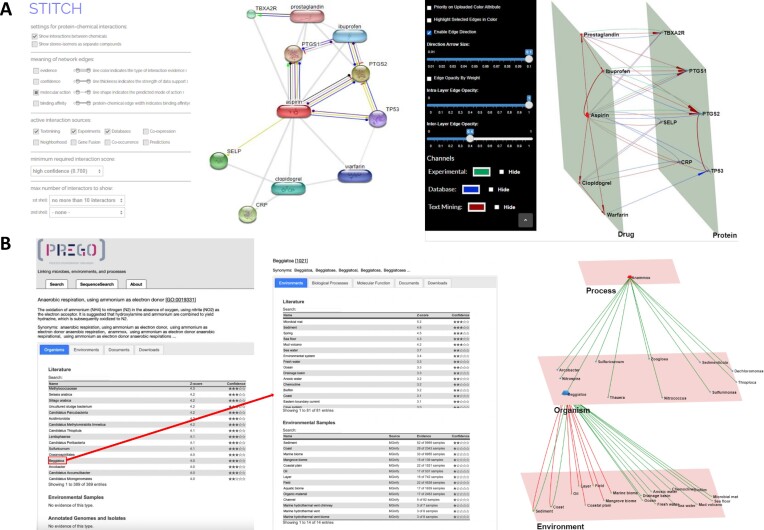
To demonstrate how Arena3Dweb handles both directed and multi-edged connections, we provide two use cases. In the first case, we queried the STITCH (19) database for ‘aspirin’ and filtered its interactions by only keeping the high-confidence ones (score > 0.7). We allow up to ten interactors for the channels: ‘Experimental’, ‘Database’ and ‘Text Mining’, depicted in green, blue and red, respectively (Figure 2A). Drug and protein nodes are separated into different layers.
Figure 2.
Multi-channel use cases. (A) A protein–chemical interaction network generated by STITCH. Left: Aspirin compound with its top ten interactors and molecular interaction channels is shown. Right: The same network in Arena3Dweb format with three channels of information: ‘Experimental’, ‘Database’ and ‘Text-Mining''. Interactions are colored in green, blue and red edges respectively. The network consists of two layers, Drugs and Proteins. (B) PREGO’s Process, Organism and Environment association tables. Left: The associations of the ‘anaerobic ammonium oxidation’ process with organisms. Middle: The associations of the genus Beggiatoa with the corresponding Environments and their respective channel sources (‘Literature’ (text-mining), ‘Environmental Samples’). Right: The multilayer and multi-edge Arena3Dweb simultaneous visualization of these two distinct tables maintaining the source channel information (‘Literature’ (text-mining) shown via green edges, while ‘Environmental Sample’ derived-ones shown in red).
In the second example, we queried the PREGO knowledge base (20) for the ‘anaerobic ammonium oxidation’ biological process (GO:0019331, anammox) and explored the associated microorganism taxa. The top 11 organisms (at the genus level) co-mentioned in the scientific literature with anammox were extracted (Figure 2B,left). For illustration purposes (Figure 2B, middle and right), the genus ‘Beggiatoa’ was selected to show its associated environment types, including the top 12 environments from the Literature (text-mining) channel (edges indicated in green) along with the top 8 environments from the Environmental Sample channel (edges indicated in red).
In addition, using PREGO’s knowledge base, we compiled a multilayer and multi-edged network based on saline environments from environment ontology terms (Supplementary Figure 1). The network contains associations from 12 saline environments with 557 microbes, 114 biological processes and 73 molecular functions as extracted from PREGO’s Literature (text mining), Environmental Samples (experiments) and Annotated Genomes (knowledge) channels. After filtering the highest scored associations, we ended up with a network consisting of 4 layers, 758 nodes and 904 edges in total (see https://github.com/lab42open-team/prego_use_case for the methodology). This network is orders of magnitude larger compared to the previous one, hence visualization and clustering is key to investigate the network. Cytoscape 2D view is useful to locate the key nodes, in terms of high degree and interconnection of big clusters, and the disconnected components of the network. The 3D visualization of Arena3Dweb makes it intuitive and quick to identify some global properties; the network is highly clustered and most microorganisms are unique to each environment term. Also, the associations between microorganisms and environments are inferred from multiple PREGO channels whereas associations from environments with the other layers are derived from the text mining channel. This is displayed without the need of color cues. The edges colors and spatial orientation clear the clutter of overlapping edges which is inevitable in large 2D networks.
CLUSTERING
Arena3Dweb now supports four main clustering algorithms offered by the igraph (21) library in combination with various global and local layouts. These are:
Louvain (22): It is a multi-level modularity optimization algorithm for finding community structures and based on the modularity measure and a hierarchical approach.
Walktrap (23): It performs random walks to detect densely connected neighborhoods. It is based on the assumption that random walks tend to restrict themselves in the same community.
Fast-Greedy (24): It optimizes a modularity score to identify densely connected communities.
Label propagation (25): It runs on a nearly linear time and tries to label the nodes with unique labels and update them by majority voting in the neighborhood of the node.
Users may apply any of these algorithms for a set of selected layers and channels to run them separately (per layer) or in combination (across layers). In case a selected subnetwork of layers and nodes cannot form a graph (e.g. layer with a single node), a relevant warning is shown. Otherwise, upon clustering, the grouped nodes are placed on their respective layer according to the local layout selected from those offered by the igraph library. These layouts vary from simple ones like circle, grid, star, random to more advanced force-directed ones such as Fruchterman–Reingold (26), Distributed Recursive (Graph) Layout (27), Multidimensional scaling (28), Kamada–Kawai (29), Large Graph Layout (LGL) and Graphopt or hierarchical-based ones like Reingold-Tilford (30) and Sugiyama. A more detailed description of these algorithms can be found in our previous Arena3Dweb article (16). The resulting clusters and their node members are shown in the ‘View Data’ panel. This panel offers additional downloadable data tables such as the network edge list as well as topological metrics data (Degree, Betweenness Centrality and Clustering Coefficient) if the respective algorithms have been run through the ‘Layer Selection & Layouts’ panel. Moreover, we show an additional table regarding the selected-only edges from the network, offered through right-clicking on any node and choosing a neighbor-selecting option.
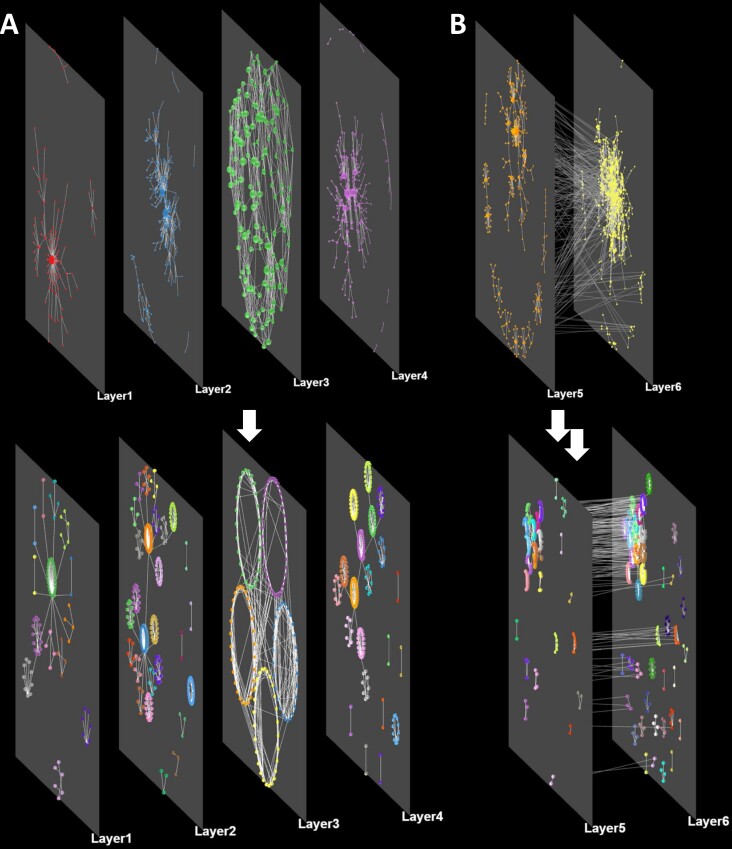
To minimize the cluster overlaps, Arena3Dweb follows NORMA’s layout strategy named ‘Super nodes’ to visualize the clustered groups (11). To this end, a virtual super node is created to represent each cluster whereas the links among these super nodes (clusters) originate from initial node connections. After applying a global layout of preference, all of the initial nodes will be placed around their respective virtual super-nodes according to a user-selected local layout. Figure 3 demonstrates a visual example of the described procedure.
Figure 3.
Layout and clustering combinations. (A) A network consisting of four different layers (no inter-layer edges) before and after applying the Louvain clustering algorithm per layer. A force-directed global layout and a circular local layout have been selected. (B) A network consisting of two interconnected layers before and after clustering with the same parameters as before.
INTEGRATION WITH OTHER TOOLS
Interoperability
In this version, Arena3Dweb comes with its own API and a slightly updated input file format to allow multiple channels as well as a different import/export format for loading and saving sessions. Starting from the latter, Arena3Dweb can save and load the running visualization in a JSON format. More specifically, the JSON object contains information regarding the: (i) scene object rotation, translation, scaling and background color, (ii) layer names, positions, rotations, scales, colors and widths, (iii) node colors, respective layers, positions, scales, colors and optional accompanying metadata, (iv) edge source and target nodes, opacities, colors, optional channels and (v), an optional global label color as well as an optional flag for enabling edge direction.
The JSON format is suitable for calling Arena3Dweb from external applications and services. For this purpose, a dedicated API route has been implemented in Node.js. This API call handles POST requests of an Arena3Dweb JSON object, as described in the paragraph above (also see Ηelp page, ‘API’) and opens the generated network object in an Arena3Dweb viewer. For any missing parameters in the imported object, the tool provides default values where applicable.
To demonstrate the API’s usability we provide two examples. Through the API utilization, Arena3Dweb can be called from Darling (31), a text-mining disease-oriented software that produces 2D networks among tagged biomedical terms of ten different categories. In Darling, a user can query for diseases, chemicals, proteins, tissues or publications and then construct networks that link the relevant extracted biological entities based on literature co-mentions. In Arena3Dweb, the various biomedical entities will be placed on different layers according to their type (one layer per type). Similarly, through the recently published functional and literature enrichment analysis tool named Flame (32), two-layered Arena3Dweb networks can be produced, one layer corresponding to the terms of a user-selected enriched category and a second layer containing the related genes/proteins. Two examples demonstrating the aforementioned APIs, are provided in Supplementary Figure 2.
Integration with cytoscape
Networks from Cytoscape can be sent to Arena3Dweb through a dedicated Cytoscape app called Arena3DwebApp. This app is implemented in Java and provides a simple interface, where the user can configure how the Cytoscape network will be transferred to Arena3Dweb. The most important setting is to choose which node attribute in Cytoscape should be used to define the layers; this can be any numeric or string value that defines up to 18 different non-overlapping groups. Arena3DwebApp retains much of the styling of the network from Cytoscape, extracting the currently displayed color, size and coordinates of each node as well as the directionality, color, thickness, and transparency of each edge. It also transfers the network-wide node label and network background colors. The user can choose which column to use for the node description and URL that can be seen in Arena3Dweb as additional node information (on node right-click). If there are nodes that do not participate in any named layer, they are added to a layer named ‘unassigned’ by default, but the user can choose to not import them in Arena3Dweb. The app generates a JSON file that is automatically sent to Arena3Dweb and gets displayed in the user's default web browser. If users want to share the layered network or open it later, they can export the JSON file from Cytoscape and import it in Arena3Dweb.
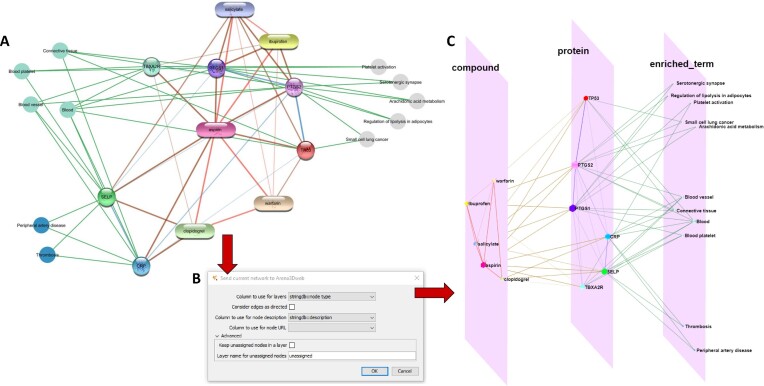
To illustrate the interoperability between Cytoscape and Arena3Dweb, we used stringApp v2.0 (33) and Arena3DwebApp in combination from Cytoscape. Specifically, we used the ‘STITCH: protein/compound query’ function of stringApp to search for the compound ‘aspirin’ with a confidence score cutoff of 0.7 and up to ten additional interactors (compounds or human proteins). We then used stringApp to retrieve functional enrichment for the proteins interacting with aspirin and added all enriched diseases, tissues and KEGG pathways to the network (two, four, and five, respectively). The resulting network in Cytoscape is shown in Figure 4A. To transfer the network to Arena3Dweb, we opened the Arena3DwebApp dialog box shown in Figure 4B. We chose to use the column ‘stringdb::node type’ to define the layers in Arena3Dweb, which means that chemical compounds will be placed in one layer, proteins in a second, and enriched terms in a third. We selected to not consider edges as directed, since STRING networks are undirected, and that the column ‘stringdb::description’ should be used for node descriptions. Finally, we submitted this three-layer network to Arena3Dweb for further 3D manipulations (Figure 4C). In the enriched_term layer, we show the three categories of enriched terms from STRING in three separate neighborhoods; KEGG pathways on top, tissues in the middle and diseases on the bottom.
Figure 4.
From Cytoscape to Arena3Dweb. (A) An aspirin chemical-protein interaction network along with enriched terms including diseases, tissues and KEGG pathways generated through stringApp in Cytoscape. Enriched disease nodes are colored red, tissues green and pathways gray. (B) The Arena3DwebApp dialogue window, where users are prompted to choose the node column that will indicate the various layers in Arena3Dweb. Other options include edge directionality, columns for node descriptions and URLs and how to handle nodes without layer information. (C) The generated Arena3Dweb network in three layers; compounds, proteins and enriched terms. Enriched pathways are placed on top, tissues in the middle and diseases on the bottom of the layer. The chemical compound interactions channel is colored red, compound-protein channel in brown, protein-protein in blue and protein participation in the various enriched terms in green. The node coloring scheme is in agreement with the initial STRING network in Cytoscape.
SUMMARY
Arena3Dweb is an innovative network analysis tool dedicated to the visualization of multilayered networks in 3D. Its main purpose is to visualize heterogeneous information of higher complexity in meaningful and appealing ways and simultaneously allow users to capture publication-ready, story-telling figures. Arena3Dweb retains an interdisciplinary character and is able to serve case-studies from various fields. For example, Arena3Dweb visualizations could potentially be applied to study and interpret multi-layered neural networks, following the information flux from the input towards the output layer. Even though deep learning models could be difficult to illustrate due to their unknown number of hidden layers and nodes, visible neural networks (VNNs) where input-output translation is based on prior knowledge (34,35), could be visualized and further explored in 3D.
Future directions will include time series analyses and major integration with: (i) biomedical repositories for automated data extraction and information retrieval (e.g. PPI databases (36)) (ii) biomedical text-mining applications (e.g. STRING (14), BioTextQuest (37), STITCH (19), EXTRACT (38), OnTheFly (39), UniRed (40), PREGO (20)) and (iii) functional enrichment analysis tools (e.g. g:Profiler (41), aGOtool (42), DAVID (43), Panther (44)). To this end, integration with Cytoscape addresses many of these challenges, as several functionalities are offered through the available Cytoscape apps, whose results can be exported and visualized with Arena3Dweb directly through the Arena3Dweb Cytoscape app.
DATA AVAILABILITY
Arena3Dweb is an open source application. Its code can be downloaded from the GitHub repository: https://github.com/PavlopoulosLab/Arena3Dweb. Downloadable examples for both ‘upload’ and ‘import’ Arena3Dweb options can be found in the GitHub repository in the Arena3Dweb/www/data/ folder, as well as in the tool's help panel (Help/Examples). Permanent DOIs: Arena3Dweb (10.5281/zenodo.7943901), Arena3Dweb_CytoscapeApp (10.5281/zenodo.7943908).
The Docker image is available at: https://hub.docker.com/r/pavlopouloslab/arena3dweb.
The service is available online at http://arena3d.pavlopouloslab.info or http://arena3d.org.
The API is accessible through https://bib.fleming.gr/bib/api/arena3dweb.
The Arena3Dweb Cytoscape plugin is available at: https://apps.cytoscape.org/apps/arena3DwebApp.
Supplementary Material
Notes
Present address: Georgios A. Pavlopoulos, Institute for Fundamental Biomedical Research, BSRC “Alexander Fleming”, 34 Fleming Street, Vari 16672, Greece.
Contributor Information
Maria Kokoli, Institute for Fundamental Biomedical Research, BSRC “Alexander Fleming”, Vari, 16672, Greece.
Evangelos Karatzas, Institute for Fundamental Biomedical Research, BSRC “Alexander Fleming”, Vari, 16672, Greece.
Fotis A Baltoumas, Institute for Fundamental Biomedical Research, BSRC “Alexander Fleming”, Vari, 16672, Greece.
Reinhard Schneider, University of Luxembourg, Luxembourg Centre for Systems Biomedicine, Bioinformatics Core, Esch-sur-Alzette, Luxembourg.
Evangelos Pafilis, Institute of Marine Biology, Biotechnology and Aquaculture (IMBBC), Hellenic Centre for Marine Research (HCMR), Former U.S. Base of Gournes, Heraklion 71003, Greece.
Savvas Paragkamian, Institute of Marine Biology, Biotechnology and Aquaculture (IMBBC), Hellenic Centre for Marine Research (HCMR), Former U.S. Base of Gournes, Heraklion 71003, Greece; Department of Biology, University of Crete, Voutes University Campus, P.O. Box 2208, 70013 Heraklion, Crete, Greece.
Nadezhda T Doncheva, Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, Copenhagen N DK-2200, Denmark.
Lars Juhl Jensen, Novo Nordisk Foundation Center for Protein Research, University of Copenhagen, Copenhagen N DK-2200, Denmark.
Georgios A Pavlopoulos, Institute for Fundamental Biomedical Research, BSRC “Alexander Fleming”, Vari, 16672, Greece; Center of New Biotechnologies & Precision Medicine, Medical School, National and Kapodistrian University of Athens, Athens 11527, Greece.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
Hellenic Foundation for Research and Innovation (to H.F.R.I.) under the ‘First Call for H.F.R.I. Research Projects to support faculty members and researchers and the procurement of high-cost research equipment grant’ [1855-BOLOGNA]; G.A.P. was also supported by the project ‘The Greek Research Infrastructure for Personalized Medicine (pMedGR)’ [MIS 5002802], which is implemented under the Action ‘Reinforcement of the Research and Innovation Infrastructure’, funded by the Operational Program ‘Competitiveness, Entrepreneurship and Innovation’ [NSRF 2014-2020]; Greece and the European Union (European Regional Development Fund); F.B. was supported by the Fondatión Sante; N.T.D. and L.J.J. were funded by the Novo Nordisk Foundation [NNF14CC0001]; S.P. was supported by the Hellenic Foundation for Research and Innovation (HFRI) under the 3rd Call for HFRI PhD Fellowships [Fellowship number: 5726].
Conflict of interest statement. All authors have read and approved the manuscript and declare no conflict of interest.
REFERENCES
- 1. Koutrouli M., Karatzas E., Paez-Espino D., Pavlopoulos G.A.. A guide to conquer the biological network era using graph theory. Front. Bioeng Biotechnol. 2020; 8:34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baltoumas F.A., Zafeiropoulou S., Karatzas E., Koutrouli M., Thanati F., Voutsadaki K., Gkonta M., Hotova J., Kasionis I., Hatzis P.et al.. Biomolecule and bioentity interaction databases in systems biology: a comprehensive review. Biomolecules. 2021; 11:1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Gehlenborg N., O’Donoghue S.I., Baliga N.S., Goesmann A., Hibbs M.A., Kitano H., Kohlbacher O., Neuweger H., Schneider R., Tenenbaum D.et al.. Visualization of omics data for systems biology. Nat. Methods. 2010; 7:S56–S68. [DOI] [PubMed] [Google Scholar]
- 4. Pavlopoulos G.A., Wegener A.-L., Schneider R.. A survey of visualization tools for biological network analysis. BioData Min. 2008; 1:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T.. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Saito R., Smoot M.E., Ono K., Ruscheinski J., Wang P.-L., Lotia S., Pico A.R., Bader G.D., Ideker T.. A travel guide to Cytoscape plugins. Nat. Methods. 2012; 9:1069–1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bastian M., Heymann S., Jacomy M.. Gephi: an open source software for exploring and manipulating networks. Proceedings of the international AAAI conference on web and social media. 2009; 3:361–362. [Google Scholar]
- 8. Auber D., Archambault D., Bourqui R., Delest M., Dubois J., Lambert A., Mary P., Mathiaut M., Melançon G., Pinaud B.et al.. Alhajj R., Rokne J.. Tulip 5. Encyclopedia of Social Network Analysis and Mining. 2017; NY: Springer New York; 1–28. [Google Scholar]
- 9. Mrvar A., Batagelj V.. Analysis and visualization of large networks with program package Pajek. Complex Adaptive Syst. Model. 2016; 4:6. [Google Scholar]
- 10. Koutrouli M., Karatzas E., Papanikolopoulou K., Pavlopoulos G.A.. NORMA: the network makeup artist - a web tool for network annotation visualization. Genomics Proteomics Bioinformatics. 2021; 3:578–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Karatzas E., Koutrouli M., Baltoumas F.A., Papanikolopoulou K., Bouyioukos C., Pavlopoulos G.A.. The network makeup artist (NORMA-2.0): distinguishing annotated groups in a network using innovative layout strategies. Bioinformatics Adv. 2022; 2:vbac036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou G., Pang Z., Lu Y., Ewald J., Xia J.. OmicsNet 2.0: a web-based platform for multi-omics integration and network visual analytics. Nucleic Acids Res. 2022; 50:W527–W533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Freeman T.C., Horsewell S., Patir A., Harling-Lee J., Regan T., Shih B.B., Prendergast J., Hume D.A., Angus T.. Graphia: a platform for the graph-based visualisation and analysis of complex data. Bioinformatics. 2020; 18:e1010310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M., Santos A., Doncheva N.T., Roth A., Bork P.et al.. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017; 45:D362–D368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Koutrouli M., Hatzis P., Pavlopoulos G.. Exploring Networks in the STRING and Reactome Database. Reference Module in Biomedical Sciences. 2020; Elsevier. [Google Scholar]
- 16. Karatzas E., Baltoumas F.A., Panayiotou N.A., Schneider R., Pavlopoulos G.A.. Arena3Dweb: interactive 3D visualization of multilayered networks. Nucleic Acids Res. 2021; 49:W36–W45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pavlopoulos G.A., O’Donoghue S.I., Satagopam V.P., Soldatos T.G., Pafilis E., Schneider R.. Arena3D: visualization of biological networks in 3D. BMC Syst. Biol. 2008; 2:104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Secrier M., Pavlopoulos G.A., Aerts J., Schneider R.. Arena3D: visualizing time-driven phenotypic differences in biological systems. BMC Bioinf. 2012; 13:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Szklarczyk D., Santos A., von Mering C., Jensen L.J., Bork P., Kuhn M.. STITCH 5: augmenting protein-chemical interaction networks with tissue and affinity data. Nucleic Acids Res. 2016; 44:D380–D384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zafeiropoulos H., Paragkamian S., Ninidakis S., Pavlopoulos G.A., Jensen L.J., Pafilis E.. PREGO: a literature and data-mining resource to associate microorganisms, biological processes, and environment types. Microorganisms. 2022; 10:293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Csardi G., Nepusz T.. The igraph software package for complex network research. Inter Journal. 2006; 1695:1–9. [Google Scholar]
- 22. Blondel V.D., Guillaume J.-L., Lambiotte R., Lefebvre E.. Fast unfolding of communities in large networks. J. Stat. Mech. 2008; 2008:P10008. [Google Scholar]
- 23. Walkowiak K. Yolum P., Güngör T., Gürgen F., Özturan C.. A Unified Approach to Survivability of Connection-Oriented Networks. Computer and Information Sciences - ISCIS 2005, Lecture Notes in Computer Science. 2005; 3733:Berlin, Heidelberg: Springer; 3–12. [Google Scholar]
- 24. Clauset A., Newman M.E.J., Moore C.. Finding community structure in very large networks. Phys. Rev. E. 2004; 70:066111. [DOI] [PubMed] [Google Scholar]
- 25. Raghavan U.N., Albert R., Kumara S.. Near linear time algorithm to detect community structures in large-scale networks. Phys. Rev. E. 2007; 76:036106. [DOI] [PubMed] [Google Scholar]
- 26. Fruchterman T.M.J., Reingold E.M.. Graph drawing by force-directed placement. Software: Pract. Exp. 1991; 21:1129–1164. [Google Scholar]
- 27. Martin S.B., Brown W.M., Klavans R., Boyack K.W. DrL: distributed recursive (graph) layout. SAND Reports. 2008; 10.11578/dc.20210416.20. [DOI] [Google Scholar]
- 28. Cox T.F., Cox M.A.A.. Multidimensional Scaling. 2001; 2nd edn.Boca Raton: Chapman & Hall/CRC. [Google Scholar]
- 29. Kamada T., Kawai S.. An algorithm for drawing general undirected graphs. Inform. Process. Lett. 1989; 31:7–15. [Google Scholar]
- 30. Reingold E.M., Tilford J.S.. Tidier Drawings of Trees. IEEE Transactions on Software Engineering. 1981; SE-7:223–228. [Google Scholar]
- 31. Karatzas E., Baltoumas F.A., Kasionis I., Sanoudou D., Eliopoulos A.G., Theodosiou T., Iliopoulos I., Pavlopoulos G.A.. Darling: a web application for detecting disease-related biomedical entity associations with literature mining. Biomolecules. 2022; 12:520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Thanati F., Karatzas E., Baltoumas F.A., Stravopodis D.J., Eliopoulos A.G., Pavlopoulos G.A.. FLAME: a web tool for functional and literature enrichment analysis of multiple gene lists. Biology (Basel). 2021; 10:665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Doncheva N.T., Morris J.H., Holze H., Kirsch R., Nastou K.C., Cuesta-Astroz Y., Rattei T., Szklarczyk D., von Mering C., Jensen L.J.. Cytoscape stringApp 2.0: analysis and visualization of heterogeneous biological networks. J. Proteome Res. 2022; 22:637–646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kuenzi B.M., Park J., Fong S.H., Sanchez K.S., Lee J., Kreisberg J.F., Ma J., Ideker T.. Predicting drug response and synergy using a deep learning model of human cancer cells. Cancer Cell. 2020; 38:672–684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ma J., Yu M.K., Fong S., Ono K., Sage E., Demchak B., Sharan R., Ideker T.. Using deep learning to model the hierarchical structure and function of a cell. Nat. Methods. 2018; 15:290–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bajpai A.K., Davuluri S., Tiwary K., Narayanan S., Oguru S., Basavaraju K., Dayalan D., Thirumurugan K., Acharya K.K.. Systematic comparison of the protein-protein interaction databases from a user's perspective. J. Biomed. Inform. 2020; 103:103380. [DOI] [PubMed] [Google Scholar]
- 37. Papanikolaou N., Pavlopoulos G.A., Pafilis E., Theodosiou T., Schneider R., Satagopam V.P., Ouzounis C.A., Eliopoulos A.G., Promponas V.J., Iliopoulos I.. BioTextQuest(+): a knowledge integration platform for literature mining and concept discovery. Bioinformatics. 2014; 30:3249–3256. [DOI] [PubMed] [Google Scholar]
- 38. Pafilis E., Buttigieg P.L., Ferrell B., Pereira E., Schnetzer J., Arvanitidis C., Jensen L.J.. EXTRACT: interactive extraction of environment metadata and term suggestion for metagenomic sample annotation. Database (Oxford). 2016; 2016:baw005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Baltoumas F.A., Zafeiropoulou S., Karatzas E., Paragkamian S., Thanati F., Iliopoulos I., Eliopoulos A.G., Schneider R., Jensen L.J., Pafilis E.et al.. OnTheFly 2.0: a text-mining web application for automated biomedical entity recognition, document annotation, network and functional enrichment analysis Bioinformatics. NAR Genom Bioinform. 2021; 3:lqab090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Theodosiou T., Papanikolaou N., Savvaki M., Bonetto G., Maxouri S., Fakoureli E., Eliopoulos A.G., Tavernarakis N., Amoutzias G.D., Pavlopoulos G.A.et al.. UniProt-Related Documents (UniReD): assisting wet lab biologists in their quest on finding novel counterparts in a protein network. NAR Genom. Bioinform. 2020; 2:lqaa005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Raudvere U., Kolberg L., Kuzmin I., Arak T., Adler P., Peterson H., Vilo J.. g:profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019; 47:W191–W198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Schölz C., Lyon D., Refsgaard J.C., Jensen L.J., Choudhary C., Weinert B.T.. Avoiding abundance bias in the functional annotation of post-translationally modified proteins. Nat. Methods. 2015; 12:1003–1004. [DOI] [PubMed] [Google Scholar]
- 43. Sherman B.T., Hao M., Qiu J., Jiao X., Baseler M.W., Lane H.C., Imamichi T., Chang W.. DAVID: a web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022; 50:W216–W221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Mi H., Muruganujan A., Casagrande J.T., Thomas P.D.. Large-scale gene function analysis with the PANTHER classification system. Nat. Protoc. 2013; 8:1551–1566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Arena3Dweb is an open source application. Its code can be downloaded from the GitHub repository: https://github.com/PavlopoulosLab/Arena3Dweb. Downloadable examples for both ‘upload’ and ‘import’ Arena3Dweb options can be found in the GitHub repository in the Arena3Dweb/www/data/ folder, as well as in the tool's help panel (Help/Examples). Permanent DOIs: Arena3Dweb (10.5281/zenodo.7943901), Arena3Dweb_CytoscapeApp (10.5281/zenodo.7943908).
The Docker image is available at: https://hub.docker.com/r/pavlopouloslab/arena3dweb.
The service is available online at http://arena3d.pavlopouloslab.info or http://arena3d.org.
The API is accessible through https://bib.fleming.gr/bib/api/arena3dweb.
The Arena3Dweb Cytoscape plugin is available at: https://apps.cytoscape.org/apps/arena3DwebApp.