Abstract
The taxonomy of viruses is developed and overseen by the International Committee on Taxonomy of Viruses (ICTV), which scrutinizes, approves and ratifies taxonomic proposals, and maintains a list of virus taxa with approved names (https://ictv.global). The ICTV has approximately 180 members who vote by simple majority. Taxon-specific Study Groups established by the ICTV have a combined membership of over 600 scientists from the wider virology community; they provide comprehensive expertise across the range of known viruses and are major contributors to the creation and evaluation of taxonomic proposals. Proposals can be submitted by anyone and will be considered by the ICTV irrespective of Study Group support. Thus, virus taxonomy is developed from within the virology community and realized by a democratic decision-making process. The ICTV upholds the distinction between a virus or replicating genetic element as a physical entity and the taxon category to which it is assigned. This is reflected by the nomenclature of the virus species taxon, which is now mandated by the ICTV to be in a binomial format (genus + species epithet) and is typographically distinct from the names of viruses. Classification of viruses below the rank of species (such as, genotypes or strains) is not within the remit of the ICTV. This article, authored by the ICTV Executive Committee, explains the principles of virus taxonomy and the organization, function, processes and resources of the ICTV, with the aim of encouraging greater understanding and interaction among the wider virology community.
Keywords: virus taxonomy, virus classification, virus nomenclature, International Committee on Taxonomy of Viruses, ICTV
ICTV organization
First established in 1966 as the International Committee on Nomenclature of Viruses (ICNV) and changing its name to the International Committee on Taxonomy of Viruses (ICTV) in 1975, the ICTV oversees the classification and taxonomic nomenclature of viruses and some other mobile genetic elements, including satellite nucleic acids, viriforms and viroids [1]. The ICTV is a committee of the Virology Division of the International Union of Microbiological Societies (IUMS), and ICTV-authorized classification and nomenclature serve as the official reference point for virus taxonomy (see Glossary for definitions). There is a distinction between the ICTV taxonomy and other ways of classifying viruses, such as the Baltimore classification [2, 3] or the grouping of viruses into non-phylogenetic assemblages (such as “arboviruses” or “mycoviruses”) based on phenotypic characteristics. Thus, while there can be multiple classification systems, there is only one approved virus taxonomy [4]. ICTV affairs are governed by its Statutes (https://ictv.global/statutes), and the rules of virus taxonomy are explained in the International Code of Virus Classification and Nomenclature (ICVCN) (https://ictv.global/code).
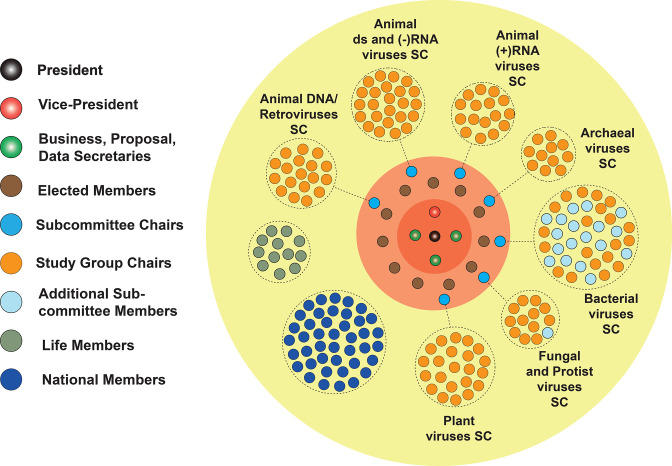
The ICTV comprises an Executive Committee (EC) of 23 members, about 40 National Members (representing virologists in different countries), 11 Life Members, the chairs of over 120 taxon-specific Study Groups, and additional Subcommittee members (Fig. 1). Study Group chairs are selected by the Subcommittee chairs and subsequently invite Study Group members to contribute their expertise relating to a particular virus taxon or discipline (such as virology, structural biology, bioinformatics, evolutionary biology or medicine). The ICTV members’ election procedures, tenure and duties are described in detail in the ICTV Statutes. The decisions of the ICTV EC and ICTV are made by majority voting. The ICTV facilitates the implementation of virus taxonomy by authorizing changes in response to submitted taxonomic proposals (TaxoProps). This process contrasts with the classification systems of animals and plants, in which the recognition and naming of species are traditionally based on valid publication and specimen deposition, with priority being given to the first description of a taxon. All members of the ICTV EC and Study Groups are volunteers and do not receive any financial remuneration for their work.
Fig. 1.
The organization of the ICTV. ICTV members (yellow circle) include the Officers (dark orange circle), Elected Members including Subcommittee chairs (light orange circle), Life Members, National Members and Study Group chairs. The Bacterial Viruses Subcommittee and Fungal and Protist Virus Subcommittee also include ICTV members who are not Study Group chairs. The Officers, Elected Members, and Life Members are elected by a vote of the full ICTV membership. National Members are nominated by Member Societies of the Virology Division of the International Union of Microbiological Societies (IUMS). Members of Virus Subcommittees are appointed by the Virus Subcommittee chairs (who are themselves appointed from the Elected members by the Executive Committee) and include all Study Group chairs. An unlimited number of Study Group members are assembled by the Study Group chairs for their expertise on viruses of a particular taxon but are not part of the ICTV. The names of the many hundreds of Study Group Members are published on the ICTV website to recognize their valuable contribution to virus taxonomy (https://ictv.global/sc/dna, https://ictv.global/sc/rna-m, https://ictv.global/sc/rnas, https://ictv.global/sc/archaeal, https://ictv.global/sc/bacterial, https://ictv.global/sc/fungal-protist, https://ictv.global/sc/plants).
ICTV taxonomy
The nature and origins of viruses have engaged virologists for decades [5, 6]. Viruses are biological entities that replicate and have co-evolved intimately with their cellular hosts for billions of years. The classification and naming of viruses have an essential role in clinical, agricultural, educational and regulatory fields.
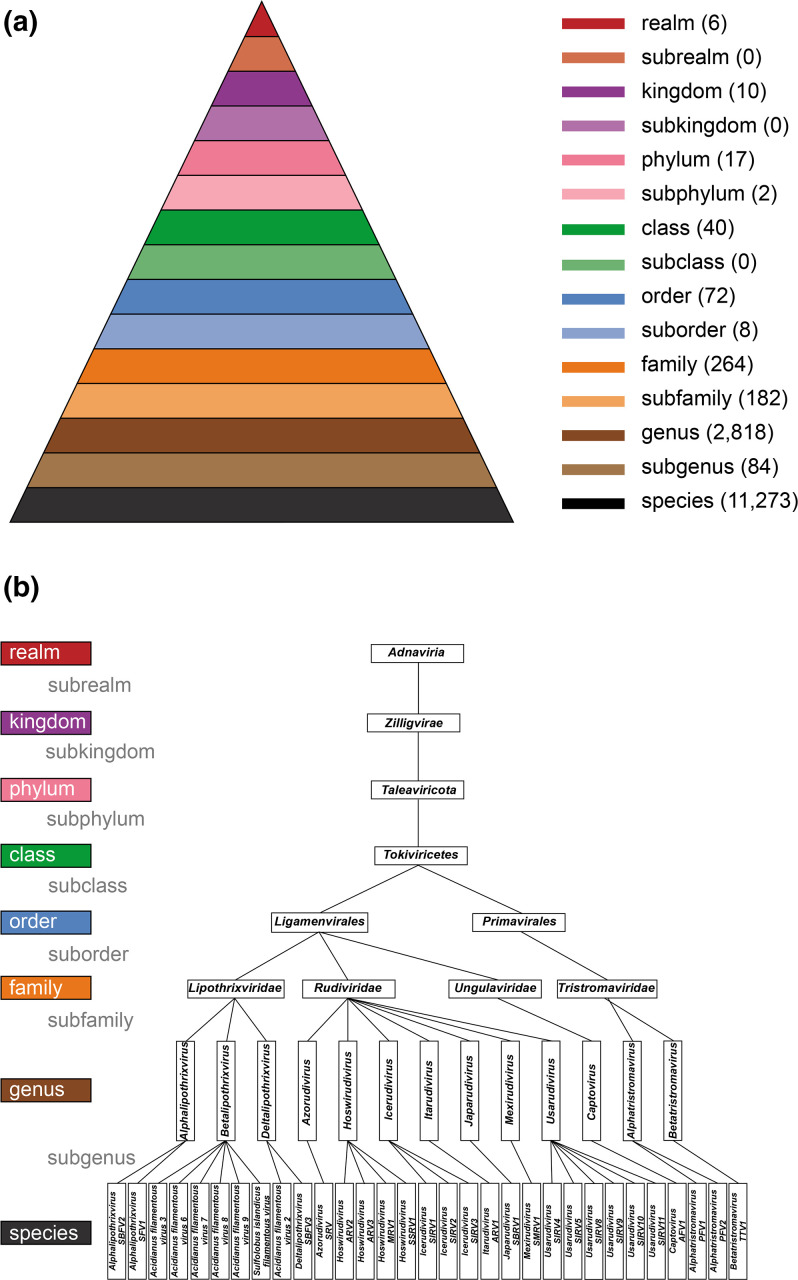
Viruses are classified into taxa that are placed into hierarchical ranks; the primary ranks are named realm, kingdom, phylum, class, order, family, genus and species [7]. Additionally, secondary ranks are available between the primary ranks (e.g. subrealm, subkingdom and subphylum), but there is no rank below that of species (Fig. 2a). This 15-rank taxonomy is designed to accommodate the entire spectrum of genetic diversity and evolutionary relationships in the virosphere and is reminiscent of the Linnaean taxonomic system for cellular organisms. The guiding principle for current virus taxonomy is that taxonomic assignments should be congruent with inferred virus evolutionary histories; i.e. taxa should be monophyletic [4], albeit with the caveat that virus genomes can have complex reticulate evolutionary histories. Phenotypic and ecological properties of viruses may be used to inform the boundaries between ranks, but all taxa should be monophyletic and their relationships based on evolutionary analysis of appropriate genomic characters. At present, 80 % of 11 273 virus species taxa are assigned to a family taxon and 94 % of species taxa have been assigned to one of the six currently established realms. Assignment to ranks other than genus and species is not mandatory.
Fig. 2.
The ICTV taxonomic rank hierarchy. (a) The ICTV-authorized 15-rank hierarchal taxonomy of viruses as ratified in 2023. Taxonomic ranks are shown with an idealized distribution pattern of taxa; actual numbers of taxa are shown in brackets. When the ranks are described as a hierarchy, the species rank is often referred to as the lowest rank and the realm rank as the highest rank. When the ranks are used as phylogenetic terms, the realm rank may be described as basal, and the species rank as apical. (b) Taxonomic structure of the realm Adnaviria [45]. Species taxon names are as currently assigned, but will be revised to binomial format in 2023.
Taxonomy is a dynamic science that develops as analytical methods improve and new data become available. For example, there are six realms, each of which is inferred to represent a unique monophyletic evolutionary origin of its constituent viruses. However, recent evidence suggests that the number of independent and ancient evolutionary origins of viruses may be larger [8], implying that revisions to the current taxonomy, especially at the highest ranks, are inevitable. A functioning virus taxonomy must also recognize and rectify incorrect classifications as new information becomes available. For example, from 1975 onwards the genus Rubivirus was included in the family Togaviridae along with members of the genus Alphavirus. However, rubiviruses (including rubella virus) differ considerably from alphaviruses such as Semliki Forest virus and chikungunya virus in their mode of transmission, virion structure and sequence relationships. Thus, in 2019, the genus was moved to a newly created family, Matonaviridae, which now includes both human and animal viruses [9].
The ICTV is charged with the development of a universal taxonomy that can be applied to all viruses irrespective of their hosts, whether they have been isolated and characterized phenotypically or if they have been identified only via sequences of nucleic acids in the metagenomic (or metatranscriptomic) analysis of viromes or environmental samples [10].
The term “universal” does not mean that all viruses must be classified and named using a single approach or placed within a single tree. Nor does it mean that the ranking of taxa for different virus lineages must necessarily represent equivalent degrees of genetic divergence. The problems of “ranking” in natural classification systems are well known [11] and they apply equally well to virus classification [12]. There is also no requirement for all taxonomic ranks to be used. For example, in the realm Adnaviria, sub-ranks are not used (Fig. 2b), and there are many other instances in which several primary and secondary ranks have not yet been assigned (Table 1).
Table 1.
Examples of virus taxonomy
|
RANK |
Taxonomy |
||||||||
|---|---|---|---|---|---|---|---|---|---|
|
realm |
Adnaviria |
Duplodnaviria |
Monodnaviria |
Riboviria |
Riboviria |
Ribozyviria |
Varidnaviria |
||
|
kingdom |
Zilligvirae |
Heunggongvirae |
Sangervirae |
Orthornavirae |
Orthornavirae |
Bamfordvirae |
|||
|
phylum |
Taleaviricota |
Uroviricota |
Phixviricota |
Lenarviricota |
Pisuviricota |
Preplasmiviricota |
|||
|
class |
Tokiviricetes |
Caudoviricetes |
Malgrandaviricetes |
Leviviricetes |
Pisoniviricetes |
Polintoviricetes |
Naldaviricetes |
||
|
order |
Ligamenvirales |
Crassvirales |
Petitvirales |
Nidovirales |
Orthopolintovirales |
Lefavirales |
|||
|
suborder |
Arnidovirineae |
||||||||
|
family |
Lipothrixviridae |
Crevaviridae |
Microviridae |
Arteriviridae |
Kolmioviridae |
Adintoviridae |
Baculoviridae |
Ampullaviridae |
|
|
subfamily |
Coarsevirinae |
Bullavirinae |
Variarterivirinae |
||||||
|
genus |
Alphalipothrixvirus |
Junduvirus |
Gequatrovirus |
Hohglivirus |
Betaarterivirus |
Daazvirus |
Alphadintovirus |
Alphabaculovirus |
Bottigliavirus |
|
subgenus |
Chibartevirus |
||||||||
|
species |
Alphalipothrixvirus SBFV2 |
Junduvirus communis |
Gequatrovirus talmos |
Hohglivirus simiicola |
Betaarterivirus sheoin |
Daazvirus cynopis |
Alphadintovirus mayetiola |
Alphabaculovirus ardigrammae |
Bottigliavirus ABV |
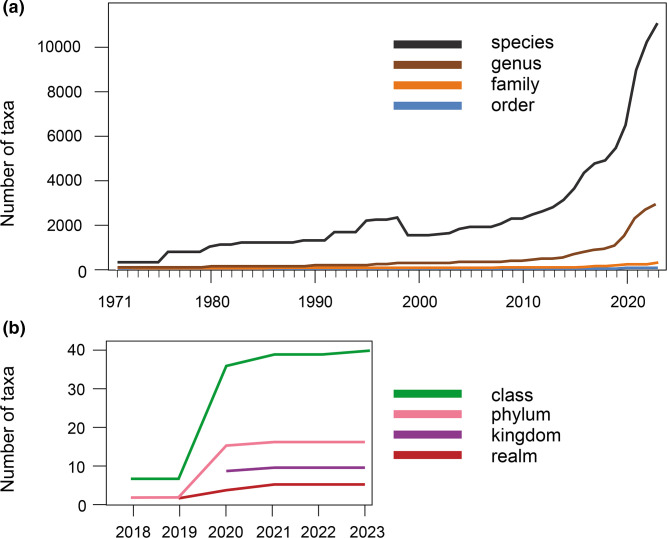
A description of the many methods that can be used to infer the evolutionary histories of viruses and support classification decisions is beyond the scope of this article but has been summarized elsewhere [4]. These may focus on virus genomes, hallmark genes or virus-encoded proteins, and range from the simple calculation of pairwise nucleotide distances to the comparison and classification of virus protein structures [13, 14]. As conflicting evolutionary signals can be provided as a result of recombination, e.g. from the acquisition of genes from other viruses or hosts, different approaches have been developed to disentangle alternative evolutionary histories [15, 16]. The decision regarding which methods are used is left to the authors of taxonomic proposals, usually, but not always, the taxon-specific Study Groups that include experts with domain-specific knowledge. Again, this emphasizes the community-based origin of virus taxonomy. The current taxonomy is available as the Master Species List (MSL) at https://ictv.global/msl and can be searched using the taxonomy browser at https://ictv.global/taxonomy. The number of taxa in each of the primary taxonomic ranks from 1971 onwards is shown in Fig. 3.
Fig. 3.
The number of taxa in primary ranks. (a) The number of taxa in the species, genus, family, and order ranks, 1971–2023. The drop in species taxa numbers in 1999 was due to some closely related viruses being consolidated into single species taxa at the same time as the systematic introduction of virus species taxa. (b) The number of taxa in the class, phylum, kingdom and realm ranks, 2018–2023. Note that the MSL in which these changes occurred generally has the previous calendar year as an identifier, indicating the year of the EC meeting at which the taxonomic changes were proposed.
ICTV processes
Taxonomic proposals (TaxoProps)
The process used to create or abolish taxa, to change taxon names, or to change the position of taxa in the taxonomic hierarchy begins with the submission of a TaxoProp to the ICTV. Although most proposals are submitted by taxon-specific Study Groups, which are tasked by the ICTV EC with the classification of viruses within specific families or other high-rank taxa, anyone can debate and submit a proposal for consideration. There are several websites that collate information on particular groups of viruses, including viruses that have yet to be classified (e.g. www.picornaviridae.com, sites.google.com/site/adenoseq). TaxoProps are submitted to the Subcommittee chair for the relevant group of viruses (https://ictv.global/sc); i.e. animal DNA viruses and retroviruses, animal dsRNA and (-)RNA viruses, animal (+)RNA viruses, archaeal viruses, bacterial viruses, fungal and protist viruses, and plant viruses (Fig. 1). Subcommittee chairs may propose revisions to prepare TaxoProps for consideration by the EC. If the taxa within the proposals span multiple hosts or virus groups, proposals may be submitted to multiple Subcommittee chairs. TaxoProps are evaluated once a year; the deadline for submission is typically in May/June, with the initial review at the EC meeting in July/August. Proposals that are not approved by the EC are either rejected or returned to the authors for amendment before reconsideration.
When a TaxoProp is approved by the EC, it will enter a ratification vote by the full ICTV membership (Fig. 1); this vote typically takes place early in the year following the EC meeting at which the proposal was accepted. If ratified, the now accepted taxonomic changes will be published online in the MSL and Virus Metadata Resource (VMR) (see below). At all stages of the process, each TaxoProp is posted publicly for comment (https://ictv.global/files/proposals/pending); ratified TaxoProps are archived at https://ictv.global/files/proposals/approved. TaxoProp templates and instructions can be downloaded from https://ictv.global/taxonomy/templates.
TaxoProps take the form of Microsoft Word documents summarizing the evidence for the proposed taxonomic changes or additions, and Excel spreadsheets itemizing any new taxa, as well as changes to existing taxonomic names or their place in the taxonomic hierarchy. TaxoProps must propose taxon names, provide supporting evidence for proposed taxonomic placements, and in the case of new species taxa, must provide nucleotide sequence information via GenBank accession numbers of an exemplar virus isolate. All TaxoProps must be based on the availability of at least one near-complete genome sequence of an exemplar virus. The ICTV does not classify viruses characterized only by partial genome sequences. Particular care must be taken with the accuracy of the Excel spreadsheets, because the process of correcting even an incorrectly spelled taxon name can be lengthy and may require submission of a corrected TaxoProp.
One of the most important elements of a TaxoProp is the evidence that supports the assignment of a group of viruses into a taxon and the placement of that taxon in a ranking hierarchy that is built upon the inclusion principle (see Glossary). Thus, a proposal to classify a group of viruses for the first time must be clear about the criteria used to assign viruses to a taxon at the species rank and assign species to higher-rank taxa. At a minimum, species taxa must be assigned to a genus and, optionally, other higher-rank taxa. TaxoProps normally include a phylogenetic analysis of relevant nucleotide or amino-acid sequences, or present alternative evidence justifying monophyly. If the viruses are to be classified in an existing taxonomy, the established assignment criteria must be used or a convincing scientific argument to change these criteria must be presented.
Orthography
In virus taxonomy, the names of all taxa are written in italics. Names of taxa placed in ranks above species must end with a mandated suffix, as outlined in Table 2. After consultation [17], discussion by the EC, and a ratification vote, species taxon names are required to be written in a binomial format that is comprised of the genus name followed by a space and then a species epithet. Species epithets can be in any form as long as they are a single word made of letters from the Latin alphabet and numbers only. Species epithets may be Latinized [18]; guidance on the format of non-Latinized epithets is available at https://ictv.global/taxonomy/templates. The binomial format requirement is mandated for all new species taxon names and is being introduced retrospectively for existing names, with a deadline of the EC meeting of 2023.
Table 2.
Mandated suffixes for virus taxon names
|
Rank |
Suffix |
|---|---|
|
realm |
…viria |
|
subrealm |
…vira |
|
kingdom |
…virae |
|
subkingdom |
…virites |
|
phylum |
…viricota |
|
subphylum |
…viricotina |
|
class |
…viricetes |
|
subclass |
…viricetidae |
|
order |
…virales |
|
suborder |
…virineae |
|
family |
…viridae |
|
subfamily |
…virinae |
|
genus |
…virus |
|
subgenus |
…virus |
|
species |
not applicable |
ICTV resources
Master Species List and Virus Metadata Resource
The ICTV maintains a list of all virus taxa and their history, called the Master Species List (MSL). The MSL is updated each year following the ratification vote and is available from the ICTV website (https://ictv.global/msl). A complementary list, called the Virus Metadata Resource (VMR), provides information about at least one virus assigned to each species taxon, including the virus name, an accession number for all or part of the virus genome, the nature of the virus genome (DNA or RNA, single- or double-stranded and positive- or negative-sense), and the natural host (plant/animal/fungi/protist/bacteria or archaea), if known. This means that, by searching the VMR, it is possible to link virus names to the corresponding species and higher taxa and to link virus species taxa with the genome sequence of an exemplar virus (https://ictv.global/help/vmr/find_species_name). The VMR is updated several times a year and is available as a downloadable spreadsheet (https://ictv.global/vmr).
The MSL and VMR also form the basis of the virus section of the National Center for Biotechnology Information (NCBI) Taxonomy Database [19]. However, implementation of the new taxonomy releases into the NCBI database is not automatic, and short delays are expected each year. Thus, the most recent MSL and VMR databases should be used to confirm virus taxonomic assignments.
The ICTV website
The ICTV website provides access to the current taxonomy database in online and downloadable formats and maintains a complete history of virus taxa dating back to the first release in 1971 (https://ictv.global/taxonomy/history). The site also provides information about the ICTV, recent news on virus taxonomy and forums for general and taxonomic discussions.
The ICTV Report and Profiles
The ICTV Report is published online as a continuously updated open-access publication (https://ictv.global/report). It strives to be a comprehensive description of all virus taxa and includes extensive information on virion structure, genome structure and replication, biology and phylogeny. Chapters typically refer to a particular virus family and are written by ICTV Study Groups and edited by members of the EC. In addition, summaries of the individual ICTV Report chapters are published in a special section of the Journal of General Virology, called “ICTV Virus Taxonomy Profiles” (http://www.microbiologyresearch.org/content/ictv-virus-taxonomy-profiles). These Profiles provide concise summaries of the classification, structure and properties of members of the virus taxa included in the Report chapter. The Profiles are indexed in PubMed (for example, [20, 21]) and can be used to cite the corresponding chapters of the ICTV Report, even when the content of the Report has been subsequently updated.
Virology Division News
The Virology Division News (VDN) section of Archives of Virology is currently an important freely accessible vehicle for communicating ICTV news to virologists and others (https://www.springer.com/journal/705/updates/18970520). For example, previous VDN reviews and articles have encompassed the development of virus taxonomy, including official taxonomic changes [22], the substance of taxonomic proposals under consideration by the ICTV [23] and informal suggestions for discussion [17].
Virus classification and nomenclature below the rank of species
Classification
The remit of the ICTV only extends to the assignment, placement and naming of taxa at and above the species rank. No general guidance can be given about classification below the species rank because criteria differ across groups of viruses. For example, viruses assigned to the same species taxon may be grouped into different serotypes or genotypes based upon cross-neutralisation or genetic divergence of their nucleotide or encoded amino-acid sequences. Examples include the numerous (sero)types of enteroviruses within the species Enterovirus A (family Picornaviridae) or (geno)types of avian hepatitis E viruses (species Avihepevirus magniiecur, family Hepeviridae). Other viruses belonging to a single species taxon have been grouped using other biological characteristics, such as disease associations, host range or virion structure. In some cases, there may be a need to group viruses that are not congruent with evolutionary relationships, and these classifications may include viruses from several species taxa. This is often the case when the groupings have relevance for disease, bioregulation or biology. However, these alternative classifications cannot be regarded as virus taxonomies.
Nomenclature
An issue separate from classification below the level of the species taxon is the names given to viruses as physical entities or the diseases they produce. As an example of the use of these different kinds of names, rubella virus (the virus name), the etiological agent of rubella or German measles (the disease names), is a member of the species Rubivirus rubellae (the species taxon name) in the family Matonaviridae (the family taxon name). Virus names are not regulated by the ICTV [1, 24] and are usually those used in the first description of the virus, either from a publication or a nucleotide accession entry. To help distinguish virus names from taxon names (especially the species taxon name), virus names should be written without italics. They should also be written without capital letters except for proper nouns that are virus name components. In addition, it is preferable for virus names to be distinct from virus species taxon names to help avoid category confusion, something that has been particularly widespread for plant and bacterial viruses. The confusion of virus and species taxon names should become less acute following the mandatory use of binomial virus species taxon names, but the advice for naming viruses is still valid. The use of place names or personal names as parts of virus names should be carefully considered before being adopted, as the chosen name may not be unique or the association with a virus may be unwelcome [25].
Another subsidiary issue concerns the naming of variants or strains of the same virus. There is no established convention for this within the field of virology and the ICTV has no remit in this matter. For some viruses, isolates are named according to a defined pattern. For example, for influenza A viruses (species Alphainfluenzavirus influenzae, family Orthomyxoviridae), the isolate designation form is type/host (if not human)/country/strain number/year (haemagglutinin/neuraminidase type)—giving “influenza A virus, isolate A/swine/Italy/81220/2017(H1N1)”. Such names may simplify the computational analysis of virus sequences, but they can also become cumbersome. For bacterial viruses, a common naming pattern takes the form “Acinetobacter phage Apostate” or the formulaic “Acinetobacter phage vB_AbaM_Apostate”, consisting of the virus host genus followed by “phage” and a unique phage name, which can take the form “vB” (virus of bacteria), “Aba” (abbreviated host genus and species name), “M” (virus morphotype, indicating myovirus in this case), followed by the phage common name or identifier which should be unique [26, 27].
Conclusions
In the last few years, virus taxonomy has changed from a specialist niche to an interdisciplinary field that encompasses virology, bioinformatics, evolutionary biology and, occasionally, philosophical considerations on the meaning of taxonomy, particularly when inferring deeper, less compelling, evolutionary relationships [6, 28–33] or the nature of virus species and the associated principles of categorization [34–36]. This change of scope has largely been due to the introduction of high-throughput sequencing technology [37, 38], recognition of the true diversity of the virosphere [8, 39–41] and progress in computational phylogenetics [42–44]. The next decade of virus taxonomy will be very challenging and requires a continuing effort to unravel the genetic make-up and evolution of virus genomes, as these are shaped by biological and ecological mechanisms. The goal of this effort, which is supported by the ICTV, is to translate this knowledge into a taxonomy that helps us make more sense of the natural world.
Glossary
Classification. The rational process of assigning viruses to variously defined groups.
Hallmark genes. A group of orthologous genes or gene sets, that are evolutionary conserved and used in the construction of a hierarchical taxonomy.
Inclusion principle. Viruses assigned to a lower taxon based on certain virus characteristics must also have the characteristics needed for classification into a higher-ranked taxon that includes the lower taxon.
Monophyletic. A monophyletic taxon is defined as one that includes a single most recent common ancestor of a group of viruses and all of its descendants.
Nomenclature. The assignment of names to taxa.
Phylogenetics (of viruses). The inference of evolutionary history and relationships among or within groups of viruses.
Polyphyletic. A polyphyletic taxon is defined as one that groups together the descendants of more than one most recent common ancestor.
Ranking. In biological classification, the relative level of a group of organisms (a taxon) in an ancestral or hereditary hierarchy.
Reassortment (or pseudorecombination). The exchange of one or more genetic segments that occurs in viruses with segmented genomes.
Recombination. The combining of genetic information between nucleic acid molecules from divergent genomes. Recombination (involving the homologous or non-homologous exchange of genetic material) occurs within a virus genome, between divergent co-infecting viruses or between virus and host (leading to gene acquisition).
Taxon. A monophyletic category that includes viruses sharing certain characteristics related by descent from a common ancestor. The commonly accepted correct plural of “taxon” is the fake Greek “taxa”.
Virome. The assemblage of viruses found associated with a particular ecosystem, organism or community of organisms.
Virosphere. The pool of viruses in all hosts and all environments on planet Earth.
Virion. Extracellular infectious virus particles that are capable of initiating a virus replication cycle.
Virus. A replicative non-cellular entity that is obliged to depend on a host cell for replication.
Virus taxonomy. The classification of viruses into taxa and taxon nomenclature.
Funding information
D.B.S. is supported by a grant from the Microbiology Society. B.E.D. is supported by the European Research Council (ERC) Consolidator grant 865694: DiversiPHI, the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany ´s Excellence Strategy – EXC 2051 – Project-ID 390713860, the Alexander von Humboldt Foundation in the context of an Alexander von Humboldt-Professorship founded by German Federal Ministry of Education and Research, and the European Union’s Horizon 2020 research and innovation programme, under the Marie Skłodowska-Curie Actions Innovative Training Networks grant agreement no. 955 974 (VIROINF). E.J.L. – this publication was supported by the National Institutes of Health (NIH) National Institute of Allergy And Infectious Diseases (NIAID), under Award Number U24AI162625. This work was also supported in part through the Laulima Government Solutions, LLC, prime contract with the NIH NIAID, under Contract No. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under Contract No. HHSN272201800013C. A.R.M. is a Program Director at the U.S. National Science Foundation (NSF); the statements and opinions expressed herein are made in a personal capacity and do not constitute endorsement by NSF or the government of the United States. H.M.O. was supported by the University of Helsinki and Academy of Finland funding for FINStruct, part of Biocenter Finland and Instruct-ERIC. D.L.R. is funded by the MRC (MC_UU_12014/12). S.S. acknowledges support from the Mississippi Agricultural and Forestry Experiment Station (MAFES), USDA-ARS project 58-6066-9-033 and the National Institute of Food and Agriculture, US Department of Agriculture, Hatch Project, under Accession Number 1021 494. The content and views contained in this document are solely the responsibility of the authors and should not be interpreted as necessarily representing the official policies or views, either expressed or implied, of the U.S. Department of Health and Human Services, the NIH, or the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products or organizations imply endorsement by the U.S. Government. Furthermore, the opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the U.S. Centers for Disease Control and Prevention or the U.S. Government.
Acknowledgements
The authors thank Anya Crane (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, MD, USA) for critically editing the manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: EC, executive committee; ICTV, International Committee on Taxonomy of Viruses; IUMS, International Union of Microbiological Societies; MSL, master species list; NCBI, National Center for Biotechnology Information; TaxoProp, taxonomic proposal; VDN, Archives of Virology’s Virology Division News; VMR, virus metadata resource.
References
- 1.Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, et al. 50 years of the International Committee on Taxonomy of Viruses: progress and prospects. Arch Virol. 2017;162:1441–1446. doi: 10.1007/s00705-016-3215-y. [DOI] [PubMed] [Google Scholar]
- 2.Baltimore D. Expression of animal virus genomes. Bacteriol Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koonin EV, Krupovic M, Agol VI. The Baltimore classification of viruses 50 years later: how does it stand in the light of virus evolution? Microbiol Mol Biol Rev. 2021;85:e0005321. doi: 10.1128/MMBR.00053-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Simmonds P, Adriaenssens EM, Zerbini FM, Abrescia NGA, Aiewsakun P, et al. Four principles to establish a universal virus taxonomy. PLoS Biol. 2023;21:e3001922. doi: 10.1371/journal.pbio.3001922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harris HMB, Hill C. A place for viruses on the Tree of Life. Front Microbiol. 2020;11:604048. doi: 10.3389/fmicb.2020.604048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koonin EV, Dolja VV, Krupovic M. The logic of virus evolution. Cell Host Microbe. 2022;30:917–929. doi: 10.1016/j.chom.2022.06.008. [DOI] [PubMed] [Google Scholar]
- 7.International Committee on Taxonomy of Viruses Executive Committee The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat Microbiol. 2020;5:668–674. doi: 10.1038/s41564-020-0709-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koonin EV, Krupovic M, Dolja VV. The global virome: How much diversity and how many independent origins? Environ Microbiol. 2023;25:40–44. doi: 10.1111/1462-2920.16207. [DOI] [PubMed] [Google Scholar]
- 9.Mankertz A, Chen M-H, Goldberg TL, Hübschen JM, Pfaff F, et al. ICTV Virus Taxonomy Profile: Matonaviridae 2022. J Gen Virol. 2022;103:001817. doi: 10.1099/jgv.0.001817. [DOI] [PubMed] [Google Scholar]
- 10.Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, et al. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol. 2017;15:161–168. doi: 10.1038/nrmicro.2016.177. [DOI] [PubMed] [Google Scholar]
- 11.van der Gulik PTS, Hoff WD, Speijer D. Renewing Linnaean taxonomy: a proposal to restructure the highest levels of the Natural System. Biol Rev Camb Philos Soc. 2023;98:584–602. doi: 10.1111/brv.12920. [DOI] [PubMed] [Google Scholar]
- 12.Aiewsakun P, Simmonds P. The genomic underpinnings of eukaryotic virus taxonomy: creating a sequence-based framework for family-level virus classification. Microbiome. 2018;6:38. doi: 10.1186/s40168-018-0422-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bao Y, Chetvernin V, Tatusova T. Improvements to pairwise sequence comparison (PASC): a genome-based web tool for virus classification. Arch Virol. 2014;159:3293–3304. doi: 10.1007/s00705-014-2197-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mönttinen HAM, Ravantti JJ, Poranen MM. Structure unveils relationships between RNA virus polymerases. Viruses. 2021;13:313. doi: 10.3390/v13020313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pérez-Losada M, Arenas M, Galán JC, Palero F, González-Candelas F. Recombination in viruses: mechanisms, methods of study, and evolutionary consequences. Infect Genet Evol. 2015;30:296–307. doi: 10.1016/j.meegid.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Irwin NAT, Pittis AA, Richards TA, Keeling PJ. Systematic evaluation of horizontal gene transfer between eukaryotes and viruses. Nat Microbiol. 2022;7:327–336. doi: 10.1038/s41564-021-01026-3. [DOI] [PubMed] [Google Scholar]
- 17.Siddell SG, Walker PJ, Lefkowitz EJ, Mushegian AR, Dutilh BE, et al. Binomial nomenclature for virus species: a consultation. Arch Virol. 2020;165:519–525. doi: 10.1007/s00705-019-04477-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Postler TS, Rubino L, Adriaenssens EM, Dutilh BE, Harrach B, et al. Guidance for creating individual and batch latinized binomial virus species names. J Gen Virol. 2022;103:001800. doi: 10.1099/jgv.0.001800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoch CL, Ciufo S, Domrachev M, Hotton CL, Kannan S, et al. NCBI Taxonomy: a comprehensive update on curation, resources and tools. Database. 2020;2020:baaa062. doi: 10.1093/database/baaa062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gatherer D, Depledge DP, Hartley CA, Szpara ML, Vaz PK, et al. ICTV Virus Taxonomy Profile: Herpesviridae 2021. J Gen Virol. 2021;102 doi: 10.1099/jgv.0.001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Benkő M, Aoki K, Arnberg N, Davison AJ, Echavarría M, et al. ICTV Virus Taxonomy Profile: Adenoviridae 2022. J Gen Virol. 2022;103 doi: 10.1099/jgv.0.001721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuhn JH, Adkins S, Alkhovsky SV, Avšič-Županc T, Ayllón MA, et al. 2022 taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales . Arch Virol. 2022;167:2857–2906. doi: 10.1007/s00705-022-05546-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pénzes JJ, Söderlund-Venermo M, Canuti M, Eis-Hübinger AM, Hughes J, et al. Reorganizing the family Parvoviridae: a revised taxonomy independent of the canonical approach based on host association. Arch Virol. 2020;165:2133–2146. doi: 10.1007/s00705-020-04632-4. [DOI] [PubMed] [Google Scholar]
- 24.Calisher CH, Briese T, Brister JR, Charrel RN, Dürrwald R, et al. Strengthening the interaction of the Virology Community with the International Committee on Taxonomy of Viruses (ICTV) by linking virus names and their abbreviations to virus species. Syst Biol. 2019;68:828–839. doi: 10.1093/sysbio/syy087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zerbini FM, Siddell SG, Mushegian AR, Walker PJ, Lefkowitz EJ, et al. Differentiating between viruses and virus species by writing their names correctly. Arch Virol. 2022;167:1231–1234. doi: 10.1007/s00705-021-05323-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adriaenssens E, Brister JR. How to name and classify your phage: an informal guide. Viruses. 2017;9:70. doi: 10.3390/v9040070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kropinski AM, Prangishvili D, Lavigne R. Position paper: the creation of a rational scheme for the nomenclature of viruses of Bacteria and Archaea. Environ Microbiol. 2009;11:2775–2777. doi: 10.1111/j.1462-2920.2009.01970.x. [DOI] [PubMed] [Google Scholar]
- 28.Gorbalenya AE, Lauber C, Siddell S. Taxonomy of viruses. Reference Module in Biomedical Sciences. 2019 [Google Scholar]
- 29.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, et al. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev. 2020;84:e00061-19. doi: 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krupovic M, Makarova KS, Koonin EV. Cellular homologs of the double jelly-roll major capsid proteins clarify the origins of an ancient virus kingdom. Proc Natl Acad Sci. 2022;119:e2120620119. doi: 10.1073/pnas.2120620119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kuhn JH. In: Encyclopedia of Virology. 4th. Bamford DH, Zuckerman M, editors. Oxford: Academic Press; 2021. Virus taxonomy; pp. 28–37. edn. [Google Scholar]
- 32.Nasir A, Romero-Severson E, Claverie J-M. Investigating the concept and origin of viruses. Trends Microbiol. 2020;28:959–967. doi: 10.1016/j.tim.2020.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner D, Kropinski AM, Adriaenssens EM. A roadmap for genome-based phage taxonomy. Viruses. 2021;13:506. doi: 10.3390/v13030506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van Regenmortel MHV. The species problem in virology. Adv Virus Res. 2018;100:1–18. doi: 10.1016/bs.aivir.2017.10.008. [DOI] [PubMed] [Google Scholar]
- 35.Gibbs AJ, Gibbs MJ. A broader definition of “the virus species.”. Arch Virol. 2006;151:1419–1422. doi: 10.1007/s00705-006-0775-2. [DOI] [PubMed] [Google Scholar]
- 36.Simmonds P. A clash of ideas - the varying uses of the “species” term in virology and their utility for classifying viruses in metagenomic datasets. J Gen Virol. 2018;99:277–287. doi: 10.1099/jgv.0.001010. [DOI] [PubMed] [Google Scholar]
- 37.Cobbin JC, Charon J, Harvey E, Holmes EC, Mahar JE. Current challenges to virus discovery by meta-transcriptomics. Curr Opin Virol. 2021;51:48–55. doi: 10.1016/j.coviro.2021.09.007. [DOI] [PubMed] [Google Scholar]
- 38.Dutilh BE, Varsani A, Tong Y, Simmonds P, Sabanadzovic S, et al. Perspective on taxonomic classification of uncultivated viruses. Curr Opin Virol. 2021;51:207–215. doi: 10.1016/j.coviro.2021.10.011. [DOI] [PubMed] [Google Scholar]
- 39.Dominguez-Huerta G, Wainaina JM, Zayed AA, Culley AI, Kuhn JH, et al. The RNA virosphere: How big and diverse is it? Environ Microbiol. 2023;25:209–215. doi: 10.1111/1462-2920.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Neri U, Wolf YI, Roux S, Camargo AP, Lee B, et al. Expansion of the global RNA virome reveals diverse clades of bacteriophages. Cell. 2022;185:4023–4037. doi: 10.1016/j.cell.2022.08.023. [DOI] [PubMed] [Google Scholar]
- 41.Zayed AA, Wainaina JM, Dominguez-Huerta G, Pelletier E, Guo J, et al. Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science. 2022;376:156–162. doi: 10.1126/science.abm5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC, Taylor J, Lin V, Altman T, Barbera P, et al. Petabase-scale sequence alignment catalyses viral discovery. Nature. 2022;602:142–147. doi: 10.1038/s41586-021-04332-2. [DOI] [PubMed] [Google Scholar]
- 43.Gorbalenya AE, Lauber C. Bioinformatics of virus taxonomy: foundations and tools for developing sequence-based hierarchical classification. Curr Opin Virol. 2022;52:48–56. doi: 10.1016/j.coviro.2021.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Lauber C, Seitz S. Opportunities and challenges of data-driven virus discovery. Biomolecules. 2022;12:1073. doi: 10.3390/biom12081073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krupovic M, Kuhn JH, Wang F, Baquero DP, Dolja VV, et al. Adnaviria: a new realm for archaeal filamentous viruses with linear a-form double-stranded DNA genomes. J Virol. 2021;95:e0067321. doi: 10.1128/JVI.00673-21. [DOI] [PMC free article] [PubMed] [Google Scholar]