Abstract
The yeast SIR2 gene and many of its homologs have been identified as NAD+-dependent histone deacetylases. To get a broader view of the relationship between the histone deacetylase activity of Sir2p and its in vivo functions we have mutated eight highly conserved residues in the core domain of SIR2. These mutations have a range of effects on the ability of Sir2p to deacetylate histones in vitro and to silence genes at the telomeres and HM loci. Interestingly, there is not a direct correlation between the in vitro and in vivo effects in some of these mutations. We also show that the histone deacetylase activity of Sir2p is necessary for the proper localiztion of the SIR complex to the telomeres.
INTRODUCTION
SIR2 affects processes as widespread as silencing (Rine and Herskowitz, 1987), recombination (Gottlieb and Esposito, 1989), DNA repair (Tsukamoto et al., 1997), and aging (Kaeberlein et al., 1999). SIR2 was first identified with the other SIR genes in a screen for mutations that derepress the silent mating type loci (HM) (Rine and Herskowitz, 1987) and later shown to silence markers located in the telomeres (Aparicio et al., 1991). SIR2 is also the only SIR gene that is necessary for silencing at the rDNA (Bryk et al., 1997; Smith and Boeke, 1997).
Clues to the functions of SIR2 and the other SIR genes first came from genetic studies that linked HM silencing to chromatin. Mutations in the N-terminal tail of histone H3 and H4 can derepress silencing at HM and the telomeres (Kayne et al., 1988; Park and Szostak, 1990; Aparicio et al., 1991; Thompson et al., 1994). In addition, the histones in silenced regions of the genome are hypoacetylated in an SIR-dependent manner (Braunstein et al., 1993, 1996). These observations emphasize an important connection between the SIR genes, the deacetylated state of silent chromatin, and the ability to repress. SIR2 appears to play an especially important role in this regard because its overexpression leads to decreases in global acetylation levels of histones (Braunstein et al., 1993).
The first insight for an enzymatic activity came when CobB, a bacterial homolog to SIR2, was shown to be involved in transferring a phospho-ribose moiety from nicotinate mononucleotide to 5,6-dimethylbenzimidazole (Tsang and Escalante-Semerena, 1998). It was then discovered that the human homolog to SIR2 could use NAD+ to ADP-ribosylate protein (Frye, 1999) and that yeast SIR2 could ADP-ribosylate histones, albeit inefficiently (Tanny et al., 1999). Sir2p was then also shown to deacetylate histone tails in an NAD+-dependent manner in a robust manner (Imai et al., 2000). Related studies showed that the protein product of HST2, a yeast gene that has sequence homology to SIR2, and CobB were also capable of deacetylating histones in an NAD+-dependent manner (Landry et al., 2000b; Smith et al., 2000).
Recent investigations into the mechanism of the histone deacetylation activity of Sir2p suggest that it is linked to the hydrolysis of NAD+ (Landry et al., 2000a; Tanner et al., 2000; Tanny and Moazed, 2001). These two activities are coupled in a reaction that transfers the acetyl group from histones to the ADP-ribose from NAD+ and forms an O-acetyl-ADP-ribose moiety. It is currently speculated that the deacetylation reaction requires the energy yielded from the hydrolysis of NAD+ (Tanny and Moazed, 2001) or that the O-acetyl-ADP-ribose moiety itself could have a signaling function in the cell (Tanner et al., 2000).
NAD+ and SIR2 are important regulators of yeast aging. Deletions of SIR2 decrease yeast life span, whereas overexpressing SIR2 leads to an extension (Kaeberlein et al., 1999). SIR2 is also necessary for the increase in life span in caloric restricted yeast and NAD+ levels are crucial for this increase (Lin et al., 2000). Interestingly, SIR2 is also an important regulator of aging in nematodes. When the Caenorhabditis elegans homolog of SIR2 is overexpressed, the worm's life span is increased by as much as 50% (Tissenbaum and Guarente, 2001).
To understand this gene that affects so many processes, we have mutated SIR2 and investigated its effects in vitro and in vivo. Previous studies have shown that mutations in SIR2 can affect both the enzymatic activity and its silencing phenotypes (Tanny et al., 1999; Imai et al., 2000). Other studies have shown that mutations outside the most conserved part of SIR2 create locus-specific alleles that silence either the telomeres or the rDNA but not both (Cuperus et al., 2000). In this study, we mutated residues in the highly conserved core domain of SIR2 to try to better understand the relationship between the in vivo phenotypes of SIR2 and its in vitro enzymatic activity.
MATERIALS AND METHODS
Yeast Strains, Plasmids, and Antibodies
Yeast strains are listed in Table 1. pRS305-SIR2, an integrating plasmid that contains sir2 driven by its native promoter, was used. Mutant SIR2 genes were also cloned into these vectors. SIR2 and mutant sir2 strains were generated by cutting pRS305-SIR2 within the LEU2 gene at an AflII site and integrated using standard yeast transformation protocols. SIR2 or mutant sir2 cloned into the pET28a vector was used for the production of recombinant protein. The hemagglutinin (HA) tagging of SIR4 was done with the pSF323-SIR4–3XHA vector (a gift from Steve Bell), which integrates a tagged version of SIR4 into the native SIR4 locus. Rabbit antibody to Sir2p and Sir3p have been previously described (Mills et al., 1999; Imai et al., 2000). The 12CA5 antibody to the HA epitope was obtained from Covance and the acetylated histone H3 and acetylated histone H4 were obtained from Upstate Biotechnology (Lake Placid, NY).
Table 1.
Yeast strains
| Strain | Genotype |
|---|---|
| W303R5-Δsir2 | MATa ade2-1 leu2-3,112 can1-100 trp 1-1 ura3-52 his3-11,15 RAD5 rDNA-ADE2 sir2∷TRP1 |
| W303RT | MATa ade2-1 leu2-3,112 can1-100 trp 1-1 ura3-52 his3-11,15 rad5-535 TEL VIIL-URA3 rDNA-ADE2 |
| W303RT-Δsir2 | W303RT sir2∷TRP1 |
| W303R5-SIR2-wt | W303R5-Δsir2 leu2∷LEU2-SIR2 |
| W303R5-Δsir2-pRS305 | W303R5-Δsir2 leu2∷LEU2 |
| W303R5-sir2T261A | W303R5-Δsir2 leu2∷LEU2-sir2T261A |
| W303R5-sir2G270A | W303R5-Δsir2 leu2∷LEU2-sir2G270A |
| W303R5-sir2I271A | W303R5-Δsir2 leu2∷LEU2-sir2I271A |
| W303R5-sir2F274A | W303R5-Δsir2 leu2∷LEU2-sir2F274A |
| W303R5-sir2R275A | W303R5-Δsir2 leu2∷LEU2-sir2R275A |
| W303R5-sir2N345A | W303R5-Δsir2 leu2∷LEU2-sir2N345A |
| W303R5-sir2D347A | W303R5-Δsir2 leu2∷LEU2-sir2D347A |
| W303R5-sir2H364A | W303R5-Δsir2 leu2∷LEU2-sir2H364A |
| W303RT-SIR2-wt | W303RT-Δsir2 leu2∷LEU2-SIR2 |
| W303RT-Δsir2-pRS305 | W303RT-Δsir2 leu2∷LEU2 |
| W303RT-sir2T261A | W303RT-Δsir2 leu2∷LEU2-sir2T261A |
| W303RT-sir2G270A | W303RT-Δsir2 leu2∷LEU2-sir2G270A |
| W303RT-sir2I271A | W303RT-Δsir2 leu2∷LEU2-sir2I271A |
| W303RT-sir2F274A | W303RT-Δsir2 leu2∷LEU2-sir2F274A |
| W303RT-sir2R275A | W303RT-Δsir2 leu2∷LEU2-sir2R275A |
| W303RT-sir2N345A | W303RT-Δsir2 leu2∷LEU2-sir2N345A |
| W303RT-sir2D347A | W303RT-Δsir2 leu2∷LEU2-sir2D347A |
| W303RT-sir2H364A | W303RT-Δsir2 leu2∷LEU2-sir2H364A |
| W303R5-SIR2-wt-HASIR4 | W303R5-SIR2-wt sir4∷URA3-SIR4HA |
| W303R5-Δsir2-HASIR4 | W303R5-Δsir2-pRS305 sir4∷URA3-SIR4HA |
| W303R5-sir2T261A-HASIR4 | W303R5-sir2T261A sir4∷URA3-SIR4HA |
| W303R5-sir2R275A-HASIR4 | W303R5-sir2R275A sir4∷URA3-SIR4HA |
| W303R5-sir2D347A-HASIR4 | W303R5-sir2D347A sir4∷URA3-SIR4HA |
| W303RT-SIR2/SIR2 | W303RT-leu2∷LEU2-SIR2 |
| W303RT-SIR2/sir2T261A | W303RT-leu2∷LEU2-sir2T261A |
| W303RT-SIR2/sir2G270A | W303RT-leu2∷LEU2-sir2G270A |
| W303RT-SIR2/sir2I271A | W303RT-leu2∷LEU2-sir2I271A |
| W303RT-SIR2/sir2F274A | W303RT-leu2∷LEU2-sir2F274A |
| W303RT-SIR2/sir2R275A | W303RT-leu2∷LEU2-sir2R275A |
| W303RT-SIR2/sir2N345A | W303RT-leu2∷LEU2-sir2N345A |
| W303RT-SIR2/sir2D347A | W303RT-leu2∷LEU2-sir2D347A |
| W303RT-SIR2/sir2H364A | W303RT-leu2∷LEU2-sir2H364A |
| Cky20 | MATα arg1 tsm11 |
Site-directed Mutagenesis of SIR2
Site-directed mutants were generated in pRS305-SIR2 as per Imai et al. (2000) and subcloned into pET-28a (Imai et al., 2000). The mutations were sequenced to ensure that the mutagenesis was successful. Expression of Sir2p in yeast was monitored by Western blot analysis of whole cell extracts probed with anti-Sir2p antibody.
Purification of Recombinant Protein and Enzymatic Assays
Six-his–tagged recombinant Sir2p and mutant Sir2p were purified from BL21 bacteria that overexpressed the gene on a pET28a plasmid as described previously (Imai et al., 2000). ADP-ribosylation of histones was detected as described previously (Imai et al., 2000). Histone deacetylation activity was measured using a peptide corresponding to the N-terminal tail of histone H4 (SGRGKGGKGLGKGGAKRHRC) labeled with tritiated acetate by using the histone deacetylase assay kit from Upstate Biotechnology. The assay was performed by incubating 2 μg of recombinant protein with the labeled peptide in 1 mM NAD+ overnight. Ethyl acetate was then used to separate acetyl groups freed by the reaction from those still bound to the peptide. Deaceytlation activity was then measured by counting the free tritiated acetate in a scintillation counter. Histone deacetylation assays were also measured by performing the reaction for 1 h and running the reaction products on a high-performance liquid chromatography (HPLC) as previously described (Imai et al., 2000).
Silencing and rDNA Recombination Assays
To test silencing at the telomeres, 10-fold dilutions of the derivatives of W303RT were spotted on media containing 5-fluoroorotic acid (5-FOA). To assay for HM silencing, W303R derivatives were patched onto YPD with the tester strain CKy20 and after overnight growth were replica plated to minimal media with no supplemented amino acids. rDNA recombination rates were measured as in Kaeberlein et al. (1999).
Immunoprecipitation of HA-Sir4 and Sir2
Whole cell extracts were prepared from cells grown in 100 ml of YPD to an OD of 1.0 (Strahl-Bolsinger et al., 1997). Extract (200 μl) was diluted to 500 μl with lysis buffer to which 3 μl of anti-HA antibody was added and incubated at 4°C overnight. Protein A beads were then added and further incubated at 4°C for 1 h. The beads were washed three times with lysis buffer and then boiled in 60 μl of SDS running buffer. Ten microliters was run on a 7.5% PAGE gel for Western blotting analysis.
Chromatin Immunoprecipitation
Yeast were grown in 100 ml of YPD to and OD of 1.0. Immunoprecipitation of cross-linked extract was performed essentially as described (Strahl-Bolsinger et al., 1997), by using 2.5 μl of anti-SIR3 polyclonal antibody or 5.0 μl of anti-SIR2 polyclonal, anti-acetylated histone H3 antibody, or anti-aceytlated histone H4 antibody. Polymerase chain reaction (PCR) analysis of immunoprecipitated DNA was performed in 50-μl reaction volumes by using 1:25, 1:75, and 1:225 of the total immunoprecipitated DNA. PCR reaction conditions were as described using the following primers: TEL-300.fwd, GGATATGTCAAAATTGGATACGCTTATG; TEL-300.rev, CTATAGTTGATTATAGATCCTCAATGATC; TEL-3000.fwd, TGATTCTGCTTTATCTACTTGCGTTTC; TEL-3000.rev, AGAGTAACCATAGCTATTTACAATAGG; XV-internal2.fwd, GTAGTTCGTTAGGTATGGACATTGATTTGGCC; and XV-internal2.rev, AAATGAA-ATGTATTGGGGCCTAGGTTCGCA. Slot blot analysis was performed by blotting 10 μl of immunoprecipitated (IP) DNA or 5 μl of input DNA to a Zeta-Probe membrane by using a Bio-Rad slot blot apparatus. The blot was then probed with a 32P-labeled DNA fragment corresponding to the 5S rDNA sequence.
RESULTS
Mutations in Core Domain of SIR2 Affect Enzymatic Activity
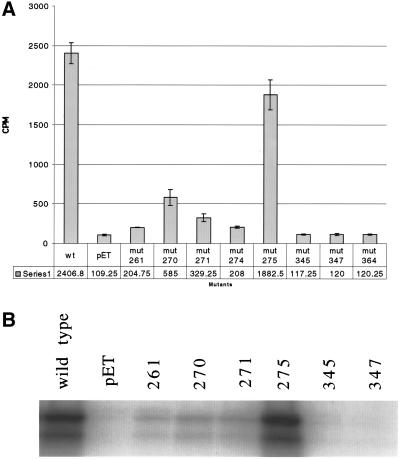
We chose eight amino acid residues that are absolutely conserved between yeast SIR2, mouse SIR2alpha, and CobB for mutational analysis (Figure 1a). We mutated the following residues in the yeast SIR2 gene to alanine: Thr-261, Gly-270, Ile-271, Phe-274, Arg-275, Asn-345, Asp-347, and His-364. Each mutant was expressed in bacteria and purified over a Ni2+-NTA column (Figure 1b).
Figure 1.
Mutagenesis of the core domain of SIR2. (A) Eight amino acid residues that are conserved between bacterial CobB, Saccharomyces SIR2, and mouse SIR2α were mutated to alanine in ySIR2. They correspond to the following residues: Thr-261, Gly-270, Ile-271, Phe-274, Arg-275, Asn-345, Asp-347, and His-364. (B) rSir2p from E. coli was purified over a Ni+ column. The purified protein was run on a polyacrylamide gel to test for purity and stability.
To test the effect of these mutations on the NAD+-dependent histone deacetylase activity of Sir2p, we performed a histone deacetylation assay in the presence of NAD+ by incubating the recombinant proteins with an H4 peptide with tritiated acetyl groups and counting the amount of tritium that was freed in the reaction (Figure 2a). The mutations fall into three different categories based on their ability to deacetylate. The first category consisting of mutations in the following residues: Asn-345, Asp-347, and His-364, shows complete loss of histone deacetylation activity of Sir2p. The second category, Arg-275, shows almost no loss of histone deacetylation activity, releasing ∼1900 cpm of acetate compared with wild type's 2400 cpm. The third category, including the most N-terminal core domain mutants Thr-261, Gly-270, Ile-271, and Phe-274, shows drastic decrease in the histone deacetylation activity, yet not a total loss of it, with activities ranging from 7 to 20% the level of wild type.
Figure 2.
Analysis of Sir2p's enzymatic activities. (A) Tritiated H4 peptide was used to measure the efficiency of the mutants in an NAD+-dependent deacetylation reaction. The peptide was incubated with 1 mM NAD+ and 2 μg of recombinant protein. The graph measures deacetylation activity by counting the amount of tritiated acetate released from the peptide. (B) Capability of the mutant rSir2 proteins to ribosylate histone H3 was measured by incubating recombinant enzyme with [32P]NAD+ and calf histone H3. The products were run out on a polyacrylamide gel and the gel was exposed to film to see whether the histones incorporated label from the [32P]NAD+.
We also investigated the ADP-ribosyl transferase activity of a subset of the mutants (261, 270, 271, 275, 345, and 347) (Figure 2b). Again, the mutations appear to fall into three different categories. The most C-terminal mutations (345 and 347) have no noticeable level of activity. Mutation 275 is as robust as wild type. The N-terminal mutations (261, 270, and 271) appear to have levels of activity that are dramatically weakened, but higher than the nonactivity of mutants 345 and 347 or the empty vector control. The effect of mutations on ADP-ribosylation correlates well with their effects on deacetylation.
Mutations Affect In Vivo Functions of SIR2
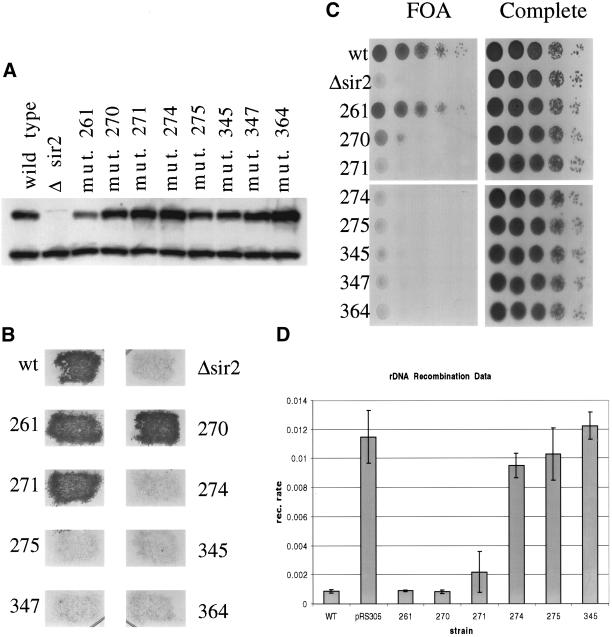
We integrated wild-type SIR2, empty vector, and each of the mutants into a Δsir2 strain. We performed Western blots on yeast whole cell extract and saw that each of the mutants expresses at levels comparable to the wild-type allele, suggesting that none of the mutations alters the stability of the protein in vivo (Figure 3a).
Figure 3.
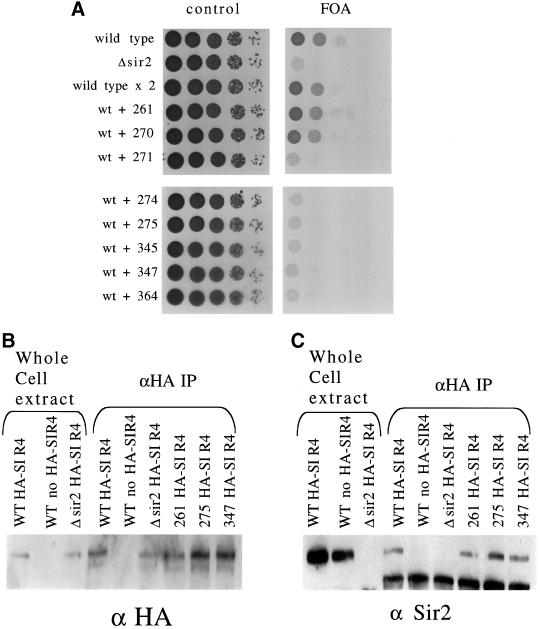
Testing sir2 mutant phenotypes in vivo. (A) Whole cell protein extracts were made from each of the indicated strains. Whole cell extract (15 μg) was run on a polyacrylamide gel and blotted to a polyvinylidene difluoride membrane for Western blot analysis. The blot was probed with anti-Sir2p to measure the level of Sir2p in each of the mutant strains. The upper band is the Sir2p band. (B) To investigate silencing at the HMLα locus, the mutant strains were mated with a mating tester strain and grown on selective media to select for diploids. (C) To test for telomere silencing, SIR2 was mutated in a strain background with the URA3 marker at the end of telomere VII. Each mutant strain was tested for its ability to silence the marker by growing on media containing 5-FOA (a substrate that is toxic to yeast expressing URA3). (D) rDNA recombination rates were measured in a strain background with the ADE2 marker located in the rDNA array. The rate of marker loss was measured for each mutant by counting the number of colonies that lost the marker in the first generation after plating.
To test silencing at the HM loci, we crossed each mutant with a mating tester strain to see whether they could successfully mate (Figure 3b). Wild type, and mutants 261, 270, and 271 formed diploids, whereas the other strains failed to mate, suggesting that mutants 261, 270, and 271 silence the HM loci, whereas mutant 274, 275, 345, 347, and 364 are unable to silence it.
To investigate silencing at the telomeres, we used a strain in which the URA3 marker has been integrated into the telomere at the left arm of chromosome VII. When SIR2 is functional, the URA3 gene is repressed and the strain can grow on media containing 5-FOA, a toxic substrate to cells that express the URA3 gene. We measured telomere silencing in the sir2 mutant strains by serial dilution of the mutants onto 5-FOA media (Figure 3c). The mutants fall into three categories based on their ability to silence telomeres. The first class, mutants 271, 274, 275, 345, 347, and 364, is incapable of silencing the URA3 marker. The second class, mutant 261, grows as well as wild type, suggesting no defect in telomere silencing. The third class, mutant 270, shows very limited but noticeable growth on 5-FOA, suggesting it can partially silence the marker.
SIR2 suppresses recombination at the rDNA and silences markers integrated there. To investigate how mutations affect the role of SIR2 at the rDNA, we measured the recombination rate in a strain with the ADE2 marker in the rDNA array by counting half-sectoring colonies (Figure 3d). Mutants 274, 275, and 345 have a rate of marker loss comparable to the sir2 disrupted strain, indicating they have lost the ability to suppress rDNA recombination. Mutants 261 and 270 have low rates of marker loss comparable to wild type. Finally, mutant 271 is intermediate, with twice the rate of marker loss as wild type, but 5 times lower than a sir2 deletion.
Overall, the mutants fall into three phenotypic categories. The first class, which only contains mutant 261, shows the same phenotypes as the wild-type allele. The second class, mutants 270 and 271, shows some ability to silence as well as wild type in some assays, yet they are either partially or completely defective in silencing at other loci. The final class, mutants 274, 275, 345, 347, and 364, is incapable of silencing at any locus tested.
Interestingly, there is not perfect correlation between the in vivo and in vitro effects for some of the mutants. There are, in particular, two puzzling mutants, mutant 261 and mutant 275. Mutant 261 has a weak enzymatic activity, yet appears to be as strong as wild type in all of its in vivo phenotypes. Alternatively, mutant 275 has near wild-type levels of activity in vitro, but is totally defective in vivo. To test whether the SIR2 homologous HST genes were able to help mutant 261 silence, we knocked out the genes HST1, HST2, HST3, and HST4 individually in a mutant 261 background but saw no effect on the ability of mutant 261 to silence (our unpublished data).
Acetylation State of Histones In Vivo
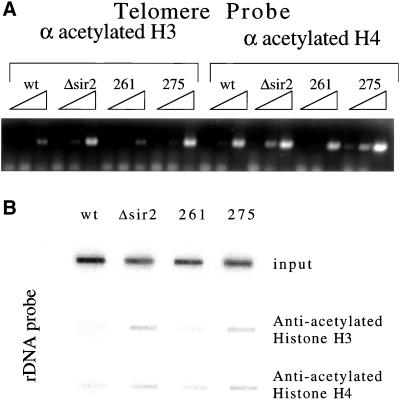
Because mutant 261 showed low activity in vitro despite behaving like wild type in vivo and mutant 275 had the opposite effect, we determined the acetylation state of silent chromatin by chromatin immunoprecipitation (ChIP). After cross-linking, we used both anti-acetylated H3 and anti-acetylated H4 to pull down the acetylated histones that had been cross-linked to DNA. After reversing the cross-linking we probed for telomere sequences by using primers that specifically recognize the telomere on the right arm of chromosome VI (Figure 4a). In strains with a wild-type allele of SIR2, very little telomeric sequence is pulled down compared with a sir2 disruption. This result was expected, because regions that are silenced are hypoacetylated in a SIR-dependent manner (Braunstein et al. 1993, 1996). The telomeres in mutant 261 are hypoacetylated much like wild type, whereas telomeres in mutant 275 is hyperacetylated much like the sir2 disruption.
Figure 4.
Chromatin IP by using anti-acetylated histone antibody to measure histone acetylation at silent loci. (A) Chromatin IP was used to investigate the acetylation state of the histones at the telomeres in mutants 261 and 275. Threefold dilutions of anti-acetylated H3 and anti-acetylated H4 IP DNA were probed for telomeres by using PCR primers corresponding to the telomere on the right arm of chromosome VI. (B) IP DNA described above was blotted to a membrane by using a slot blot apparatus. The DNA was then probed for rDNA sequences to test the level of acetylation of histone H3 and H4 at the rDNA in the mutant strains.
The effect of SIR2 on the acetylation state of histones at the rDNA has not been examined in yeast. We used chromatin IP as described above to see what effect SIR2 and the mutants have at the rDNA. Using slot blots, we saw that histones do indeed show an increase in acetylation at the rDNA when SIR2 is deleted (Figure 4b). When we looked at the mutants, we saw that mutant 261 was hypoacetylated like wild type, whereas mutant 275 is hyperacetylated like Δsir2, much like what we saw at the telomeres. Thus, for these unusual mutants, the histone acetylation state correlates with the observed degree of silencing.
Enzymatic Analysis of Mutant 261 and 275
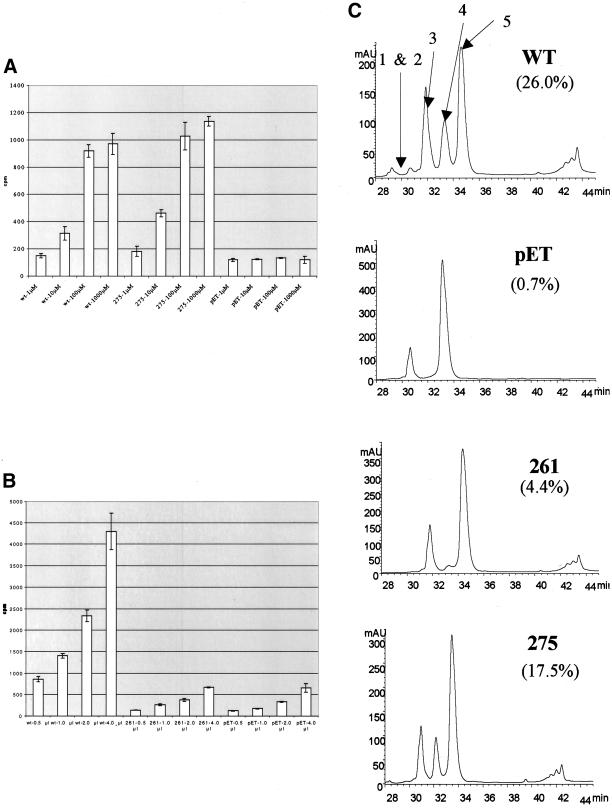
To see whether mutant 275 affected the ability of Sir2p to bind and use NAD, we repeated the histone deacetylation reaction under varying concentrations of NAD+ (Figure 5a). As expected, when we used wild-type Sir2p and increased the NAD+ concentration from 1 μM to 1 mM, an increase in the efficiency of deacetylation was observed. Interestingly, the efficiency of the reaction does not change much between 100 μM and 1 mM concentrations of NAD. The effect of varying NAD+ concentration on mutant 275 very closely paralleled the effect it had on wild-type Sir2p. This suggests that mutant 275 does not strongly affect NAD+ binding.
Figure 5.
Histone deacetylation activity of mutants under different NAD+ and peptide concentrations. (A) Histone deacetylation activity of mutant 275 along with wild-type Sir2p and no enzyme control was measured using tritiated H4 peptide that was incubated with enzyme and concentrations of NAD+ ranging from 1 μM to 1 mM. Deacetylation activity was measured by counting the amount of free acetate removed during the reaction. (B) Histone deacetylation activity of mutant 261 along with wild-type Sir2p and no enzyme was measured while the amount of tritiated peptide was varied from 0.5 to 4 μl. (C) HPLC chromatograms showing absorbance at 220 nm of products of deacetylation assays with yeast Sir2p, pET (no recombinant protein), mutant 261, and mutant 275 after a 1-h reaction. The efficiencies of the reactions are calculated as a fraction of the areas under peaks 1, 2, and 4 compared with the area under all of the peaks.
Mutant 261 severely diminishes the histone deacetylation activity of Sir2p. It is possible that loss of activity is the result of poor substrate binding and that in vivo a higher histone concentration than what is used in vitro could explain why the mutant is capable of deacetylating histones in vivo. To test this possibility we used increasing amounts of peptide to see whether the efficiency of the histone deacetylation activity of mutant 261 increased (Figure 5b). Wild-type Sir2p shows a proportional increase in release the of acetyl groups as the peptide concentration is increased. The defect in the 261 mutant was not rescued by higher peptide concentrations, suggesting that mutant 261 affects the efficiency of the enzymatic activity and not the efficiency of histone binding.
The histone deacetylation assays were done overnight raising the possibility that either mutant 275 had a slow initial rate, but over time was able to catch up to wild-type Sir2p. To test this we measured histone deacetylation rates after 1-h reactions by incubating an acetylated histone H3 peptide with enzyme and NAD+ and then running the reaction products on an HPLC apparatus comparable to what was done in Imai et al. (2000). When wild-type Sir2p is compared with no protein, three novel peaks appear in the wild-type reaction that correspond to deacetylated products (Figure 5c, peaks 1, 2, and 4). When mutant 261 was used in the reaction a very small peak appears that corresponds to a deacetylated peptide. This indicates that like in the overnight reaction, mutant 261 has very weak activity when the reaction is carried out for shorter periods (4.4 vs. 26.0% for wild type). When mutant 275 was used in the reaction, the deacetylated peaks were almost as prominent as they were when wild-type Sir2p was used (17.5 vs. 26.0%). This indicates that 275 is not defective in deacetylating under shorter reaction times and its in vivo defect is unlikely to be a result of an initial rate defect.
Mutations Effects on Aging
In light of the importance that SIR2 plays in yeast life span, we wanted to know what effect some of these mutations might have on aging. The life spans of mutants 261, 270, and 275 were measured in strains with HMLα deleted. Mutants 261 and 270 had life spans comparable to wild type, whereas mutant 275 had a life span comparable to Δsir2 (Figure 6a). HMLα was deleted so that a/α effects would not play a role; a/α strains show a shortened life span (Kaeberlein et al., 1999). Thus, mutants 261 and 270 do not affect life span, whereas mutant 275 does.
Figure 6.
Life span of mutant sir2 strains. (A) In an HMLα-deleted background, the life spans of mutant 261 and 270 are the same as wild type, whereas mutant 275 is comparable to Δsir2. The mean life spans are as follows: wild type, 20.7; Δsir2, 11.9; mutant 261, 20.8; mutant 270, 21.4; and mutant 275, 11.9. (B) In an HML + background, the life spans of mutants 261 and 270 are intermediate. The mean life spans are as follows: wild type, 24.5; Δsir2, 11.3; mutant 261, 19.9; and mutant 270, 18.9.
We also measured the life span of strains that were not deleted at HMLα. Mutants 261 and 275 had an intermediate life span, shorter than wild type, but not as short as Δsir2 (Figure 5b). This suggests that these mutants have an intermediate effect on life span that is HMLα dependent. This could be the result of only partial silencing of MATα expression from HML, i.e., mutants 261 and 270 silence HML strongly enough to allow mating, but allow enough expression of MATα to shorten life span. This is also the only phenotype seen for mutant 261 that differs from wild-type SIR2, suggesting that mutant 261 is a slight hypomorph, which can only be detected with the most sensitive assays.
Mutations Are Dominant Negative and Do Not Affect Formation of a Sir2/Sir4 Complex
Several labs have observed that mutations in SIR2 can be dominant negative (Sherman et al., 1999; Tanny et al., 1999; Cuperus et al., 2000). This led us to test whether our mutations were dominant negative. We integrated each mutant into a wild-type strain with URA3 at TEL-VIIL so that each strain would have both a wild-type and mutant copy of SIR2 present. We spotted the cells on 5-FOA media to test their ability to silence the telomere (Figure 7a). Mutants 271, 274, 275, 345, 347, and 364 abolished silencing at telomeres by the wild-type copy of SIR2. This is interesting and suggests that even though these mutants fail to silence, they are probably forming a complex with Sir2p-binding proteins, thereby poisoning the wild-type activity.
Figure 7.
Dominance of mutations and coimmunoprecipitation of SIR2 and SIR4. (A) W303-ARUT strains with a wild-type and mutant copy of SIR2 were spotted onto control synthetic media and 5-FOA plates to test whether the mutations had a dominant effect on telomere silencing. (B) Whole cell protein extract from selected mutant strains (wild type, Δsir2, mutant 261, mutant 275, and mutant 347) with an HA-tagged SIR4 gene was immunoprecipitated with antibodies against the HA epitope. The IP extract along with whole cell extract was run on a polyacrylamide gel and blotted to a polyvinylidene difluoride membrane so that Western blotting analysis could be performed. The membrane was probed using antibody to the HA epitope to measure the efficiency of the pull down. (C) Polyvinylidene difluoride membrane is reprobed with antibody recognizing Sir2p to measure the ability of the mutant Sir2ps to interact with Sir4p in vivo.
It is known that Sir2p and Sir4p interact directly and coimmunoprecipitate from whole cell extracts (Moazed et al., 1997; Strahl-Bolsinger et al., 1997). To test whether these mutants form the Sir complex, we tagged the native SIR4 gene with HA in SIR2 wild-type, Δsir2, mutant 261, mutant 275, and mutant 347 strains. We then isolated whole cell extract from these strains and used anti-HA antibody to pull down Sir4p. HA-Sir4p was precipitated from all of the strains as seen by Western blot (Figure 7b). We then reprobed the Western to see whether Sir2p coprecipitated (Figure 7c). Sir2p coprecipitated from wild-type extract but not from extracts with untagged SIR4. Moreover, Sir2p was coprecipitated with Sir4p in all three mutant extracts. This suggests that all mutant Sir2p tested, including mutant 275 assemble with Sir4p.
Mutations Affect Ability of Sir Complex to Localize to Telomeres
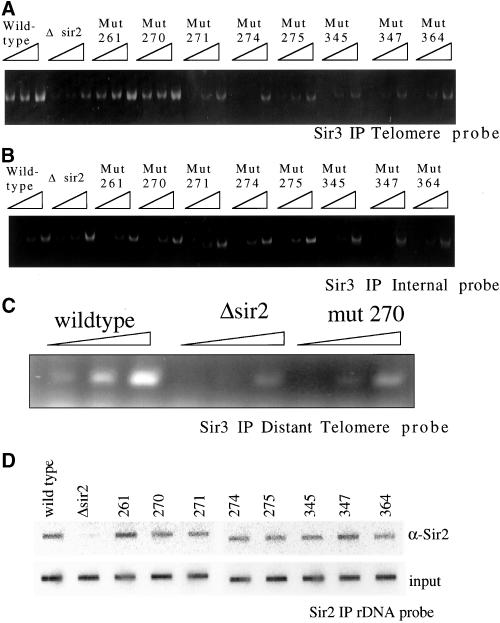
Although the mutants appear to form the Sir complex, it is possible they fail to localize to the telomeres or other silenced loci. To investigate this possibility we immunoprecipitated with anti-Sir3 antibody from extracts prepared after cross-linking and probed the coprecipitated DNA by using PCR with primers that recognize sequences 300 base pairs from the end of the right arm of chromosome VI (Figure 8a). As expected, a wild-type strain supported immunoprecipitation of the telomeric sequence, whereas a Δsir2 strain did not. Extracts from the sir2 mutants 261 and 270 gave a comparable signal to wild type. However, in the remaining mutants: 271, 274, 275, 345, 347, and 364, the telomere signal was absent, indicating Sir3p does not localize to the telomeres. We observed the same pattern by using anti-Sir2 antibody (our unpublished data). As a control for specificity and the amount of DNA present in the IP sample, we reprobed the immunoprecipitated DNA with primers corresponding to internal sequences and saw the same low level of pull down from all of the extracts (Figure 8b). These localization data show a strong correlation between the silencing activities at telomeres and the ability of the Sir complex to localize properly in the mutant strains. Because mutant 270 only partially silences the telomeres, we checked more distal regions of the telomere by probing sequences that are 3000 base pairs from the end (Figure 8c). Mutant 270 localizes much more poorly than wild type at this distal site, suggesting that this Sir mutant complex is weakened in its ability to further polymerize along the telomeres.
Figure 8.
Chromatin IP of Sir complex. Chromatin immunoprecipitation was used to investigate the ability of the Sir complex to localize to the telomeres and rDNA in the sir2 mutant strains. (A) Cross-linked chromatin was immunoprecipitated with antibodies to Sir3p. To test for localization of the Sir complex at the telomeres, the DNA pulled down was probed for telomere sequences by using PCR primers corresponding to 300 base pairs from the end of the telomere at the right arm of chromosome VI. Threefold dilutions of the immunoprecipitated DNA were used in the PCR reaction. (B) Pulled down DNA was probed for sequences corresponding to an internal nonsilenced location on chromosome XII. (C) Pulled down DNA from wild type, Δsir2, and mutant 270 was probed for more distal telomere sequences by using PCR primers that recognize sequences that are 3000 base pairs from the end of the telomere at the right arm of chromosome VI. (D) Cross-linked chromatin pulled down with anti-Sir2p was blotted to a membrane by using a slot blotting apparatus and probed for rDNA sequences.
At the rDNA, Sir2p interacts with a novel set of proteins termed the RENT complex, whereas the other SIR genes do not have a direct effect on the rDNA (Shou et al., 1999; Straight et al., 1999). We determined whether Sir2p was localizing to the rDNA in the mutant strains. The DNA from anti-Sir2 ChIPs was blotted to a filter and then probed for rDNA sequences (Figure 8d). As expected DNA from a wild-type strain contained rDNA sequences, whereas DNA from a sir2 disruption did not. All of the mutants localized to the rDNA, even though their ability to elicit hypoacetylation of histones and repress recombination varied at that site. This is not too unexpected because it has previously been shown that a mutation in residue 364 still localizes to the rDNA (Tanny et al., 1999); however, it is interesting that this phenomenon holds up for a wide range of core domain mutants. This finding contrasts to telomeres where there is a strong correlation between the silenced state and the localization of the Sir proteins.
DISCUSSION
In this article, we investigated the effect of core domain mutations on the in vitro enzymatic activity and in vivo phenotypes of SIR2. A simple model would have predicted that mutations that affect the histone deacetylase activity of Sir2p would also affect the ability of SIR2 to silence, and furthermore, that the most severe mutations would show the strongest desilencing phenotypes. Although most mutations do indeed affect the in vivo and in vitro functions of SIR2 in similar ways, two of the mutations showed unexpected differences between in vivo and in vitro activity. A summary of the mutations and their effects in vivo and in vitro is listed in Table 2.
Table 2.
Summary of mutant phenotypes
| Mutant | ADP-Ribose activity (% of wt) | Deacetylase activitya | HM silencing | Telomere silencing | rDNA recombinations rate (%) | ChIP at telomeres |
|---|---|---|---|---|---|---|
| wt | 100 | 100 | + | + | 0.1 | + |
| Δsir2 | N/A | N/A | − | − | 1.2 | − |
| Thr-261 | 4 | 5 | + | + | 0.1 | + |
| Gly-270 | 7 | 21 | + | ± | 0.1 | ± |
| Ile-271 | 8 | 9 | + | − | 0.2 | − |
| Phe-274 | ND | 9 | − | − | 0.9 | − |
| Arg-275 | 100 | 78 | − | − | 1.0 | − |
| Asn-345 | 0 | 0.3 | − | − | 1.2 | − |
| Asp-347 | 0 | 0.4 | − | − | ND | − |
| His-364 | ND | 0.4 | − | − | ND | − |
N/A, not applicable; ND, not determined; wt, wild type.
As a percentage of wild-type minus the pET negative control.
Correlations between Mutations' Effects on Enzymatic Activity and Silencing Phenotypes
When we examined the effects of the mutations on the enzymatic activity of Sir2p they fell into three categories: totally inactive, partially active, and fully active. The first class of mutants had three members: mutants 345, 347, and 364. None of these mutants were capable of silencing in any of the in vivo assays tested, even though they properly formed complexes and properly localized to some chromosomal loci. The results with these mutants seem to correlate with what was observed in Tanny et al. (1999) who also mutated residue 364 and made many of the same observations with it that we do herein.
Only mutant 275 showed full enzymatic activity. Surprisingly, this mutant was incapable of silencing in any of the in vivo assays. The mutant presumably has a subtler defect that only manifests itself in vivo.
The other class of mutants, 261, 270, 271, and 274, partially disrupts the enzymatic activity of Sir2p, showing anywhere from 5 to 20% of wild-type levels of histone deacetylase activity. Of these, only mutant 274 is completely inactive in all of the in vivo assays tested. Mutants 270 and 271 may not silence at wild-type levels because their enzymatic activity is reduced by 5- to 10-fold. Mutant 261, however, is exceptional because it has even lower enzymatic activity than mutants 270 and 271, yet it has no noticeable silencing defects, although it does have a shorter life span that appears to be the result of a failure to fully silence the HM loci
Paradoxes of Mutants 261 and 275
The lack of a direct correlation between the histone deacetylation activity and the silencing phenotypes of mutants 261 and 275 suggests that the role of SIR2 is more complex than that solely of a histone deacetylase. One explanation would be the in vitro histone deacetylation activity of SIR2 is not crucial for silencing. Instead, perhaps in vivo the relevant substrate for the deacetylation activity of Sir2p is something other than histones. By this scenario, mutant 261 would weaken activity for histones but not for the other substrate, whereas mutant 275 would have the opposite effect. Lending support to this possibility is the observation that Hst2p appears to provide most of the NAD+-dependent histone deacetylase activity in cell extracts (Smith et al., 2000). Also, new evidence has suggested that SIR2 homologs can deacetylate proteins other than histones (Sutton et al., 2001; Imai, Park, and Guarente, unpublished data). However, hypoacetylation of histones at silent loci is dependent upon SIR2, which suggests that histones are the Sir2p substrate. Also, Sir2p preferentially deacetylates residue 16 of histone H4, the residue that plays the most important role in silencing (Imai et al., 2000; Tanny and Moazed, 2001).
If SIR2 is deacetylating histones in vivo then how can we explain the effects of mutants 261 and 275? Mutant 275 may create a functional defect unrelated to its enzymatic activity such as an inability to interact with other proteins. However, Sir2p and Sir4p coimmunoprecipitated in mutant 275 strains, suggesting that the mutation did not disrupt that crucial interaction. It does not, however, rule out the possibility that another protein interaction is disrupted by mutant 275. In mutant 261, it is possible that the mutation causes a folding defect in Escherichia coli that does not exist in yeast. Purification of the mutant from yeast extract, however, showed that it was still a poor histone deacetylase, making that explanation unlikely (Armstrong, unpublished data). It is possible that the limited activity that mutant 261 possesses is adequate to silence in vivo but this would seem unlikely in light of the results with mutants 270, 271, and 275. Another reason that could explain both mutants might be a difference in how they interact with chromatin as opposed to histones. Mutant 275 is a very effective deacetylase with histone peptides as the substrate, but perhaps it is unable to deacetylate nucleosomes in the full chromatin context that is seen in the cell. Alternatively, perhaps mutant 261 is far more efficient at using full chromatin as the substrate.
SIR2 Mutations and Their Effects on Localization
We were also curious about what effect the mutations would have on where the Sir complex localized. Mutants that failed to silence at the telomeres also failed to localize there. This included mutant 275 along with the mutations that render the enzyme inactive. Is the mutants' failure to silence a result of their failure to localize? This could explain why mutant 275 does not silence. However, all of the mutants localize to the rDNA regardless of their ability to affect rDNA recombination. If mutant 275 did not silence merely because it failed to localize then one would expect it to suppress rDNA recombination, yet it does not.
Conversely, it seems plausible that the failure of these mutants to deacetylate could lead to their inability to localize to the telomeres. Mutants 345, 347, and 364 strongly support this idea because they do not deacetylate histones in vitro and they fail to localize to the telomeres. If telomere localization were independent of the acetylation state then these mutants should localize properly. The failure of the Sir complex in mutant 275 to localize to the telomeres could also be a by-product of the mutant's failure to deacetylate in vivo. These observations imply a model in which Sir2p first deacetylates the histones and then the Sir complex tightly binds to the telomeres, forming a fully functional silencing complex. This fits observations showing that Sir3p and Sir4p are capable of binding to the tails of histone H4 (Hecht et al., 1995). Others have suggested that a silenced state can be achieved in part by the binding of the Sir proteins to the unacetylated histone tail to create heterochromatin (Wu and Grunstein, 2000). These localization data in connection with previous observations suggest that a silencing complex is formed in the following way. The telomere binding protein Rap1p binds to the telomere and initially recruits the Sir complex. Sir2p then deacetylates histones in the region, allowing the Sir complex to bind to the histone tails and create silenced chromatin. The deacetylation of nearby histones by Sir2p may also be responsible for the polymerization of the Sir complex along the telomere. Even if histone deacetylation is a crucial step for the proper localization of the Sir complex to the telomeres, it is not necessary for Sir2p to localize to the rDNA.
Structure of Sir2
The crystal structure of the Archaeglobus fulfidus homolog to SIR2 bound to NAD+ has recently been solved, allowing us to compare our mutants to the crystal structure (Min et al., 2001). There are four major parts to the structure: a Rossman fold made up of six parallel strands and six helices, a loop that appears to be the most structurally flexible part of the protein, a helical domain, and a Zn binding domain. NAD+ binds in a pocket between the helical domain and the Rossman fold and flanked by the loop.
Mutant 261 is on the border between the first part of the Rossman fold and the loop. Mutants 345 and 347 fall in the border region between the second part of the Rossman fold and the Zinc binding domain. Mutant 364 falls in the middle of the Zinc binding domain. Mutants 270, 271, 274, and 275 all fall within the loop. Mutant 275's location is interesting because it corresponds to residue Arg33 in A. fulfidus, which we see form a hydrogen bond with NAD+ in one of their crystals, suggesting an important role in NAD+ binding. If mutant 275 were crucial for NAD+ binding then one would expect this mutation to destroy the enzymatic activity, but it does not. On the other hand, in the monoclinic crystal we do not see the residue binding to NAD+, challenging the importance of that residue in NAD+ binding. It is interesting to note that mutants 261 and 275 both lie in or near the loop, suggesting that learning more about this loop region could reveal additional clues about the function of SIR2.
ACKNOWLEDGMENTS
We thank members of the Guarente lab for helpful comments throughout the work and E. Ford for helpful comments on the manuscript. This work was funded by a National Institutes of Health predoctoral grant to C.A. and M.K.; a grant from The Human Frontier Science Program Organization to S.I.; and grants from the National Institutes of Health, Seaver Foundation, Ellison Medical Foundation, and Howard and Linda Stern Fund to L.G.
Footnotes
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.01–10–0482. Article and publication date are at www.molbiolcell.org/cgi/doi/10.1091/mbc.01–10–0482.
REFERENCES
- Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Rose AB, Holmes SG, Allis CD, Broach JR. Transcriptional silencing in yeast is associated with reduced nucleosome acetylation. Genes Dev. 1993;7:592–604. doi: 10.1101/gad.7.4.592. [DOI] [PubMed] [Google Scholar]
- Braunstein M, Sobel RE, Allis CD, Turner BM, Broach JR. Efficient transcriptional silencing in Saccharomyces cerevisiae requires a heterochromatin histone acetylation pattern. Mol Cell Biol. 1996;16:4349–56. doi: 10.1128/mcb.16.8.4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryk M, Banerjee M, Murphy M, Knudsen KE, Garfinkel DJ, Curcio MJ. Transcriptional silencing of Ty1 elements in the RDN1 locus of yeast. Genes Dev. 1997;11:255–269. doi: 10.1101/gad.11.2.255. [DOI] [PubMed] [Google Scholar]
- Cuperus G, Shafaatian R, Shore D. Locus specificity determinants in the multifunctional yeast silencing protein Sir2. EMBO J. 2000;19:2641–2651. doi: 10.1093/emboj/19.11.2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye RA. Characterization of five human cDNAs with homology to the yeast SIR2 gene: Sir2-like proteins (sirtuins) metabolize NAD and may have protein ADP-ribosyltransferase activity. Biochem Biophys Res Commun. 1999;260:273–279. doi: 10.1006/bbrc.1999.0897. [DOI] [PubMed] [Google Scholar]
- Gottlieb S, Esposito RE. A new role for a yeast transcriptional silencer gene, SIR2, in regulation of recombination in ribosomal DNA. Cell. 1989;56:771–776. doi: 10.1016/0092-8674(89)90681-8. [DOI] [PubMed] [Google Scholar]
- Hecht A, Laroche T, Strahl-Bolsinger S, Gasser SM, Grunstein M. Histone H3 and H4 N-termini interact with SIR3 and SIR4 proteins: a molecular model for the formation of heterochromatin in yeast. Cell. 1995;80:583–592. doi: 10.1016/0092-8674(95)90512-x. [DOI] [PubMed] [Google Scholar]
- Imai S, Armstrong CM, Kaeberlein M, Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- Kaeberlein M, McVey M, Guarente L. The SIR2/3/4 complex and SIR2 alone promote longevity in Saccharomyces cerevisiae by two different mechanisms. Genes Dev. 1999;13:2570–2580. doi: 10.1101/gad.13.19.2570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kayne PS, Kim UJ, Han M, Mullen JR, Yoshizaki F, Grunstein M. Extremely conserved histone H4 N terminus is dispensable for growth but essential for repressing the silent mating loci in yeast. Cell. 1988;55:27–39. doi: 10.1016/0092-8674(88)90006-2. [DOI] [PubMed] [Google Scholar]
- Landry J, Slama JT, Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem Biophys Res Commun. 2000a;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- Landry J, Sutton A, Tafrov ST, Heller RC, Stebbins J, Pillus L, Sternglanz R. The silencing protein SIR2 and its homologs are NAD-dependent protein deacetylases. Proc Natl Acad Sci USA. 2000b;97:5807–5811. doi: 10.1073/pnas.110148297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin SJ, Defossez PA, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- Mills KD, Sinclair DA, Guarente L. MEC1-dependent redistribution of the Sir3 silencing protein from telomeres to DNA double-strand breaks. Cell. 1999;97:609–620. doi: 10.1016/s0092-8674(00)80772-2. [DOI] [PubMed] [Google Scholar]
- Min J, Landry J, Sternglanz R, Xu RM. Crystal structure of a SIR2 homolog-NAD complex. Cell. 2001;105:269–279. doi: 10.1016/s0092-8674(01)00317-8. [DOI] [PubMed] [Google Scholar]
- Moazed D, Kistler A, Axelrod A, Rine J, Johnson AD. Silent information regulator protein complexes in Saccharomyces cerevisiae: a SIR2/SIR4 complex and evidence for a regulatory domain in SIR4 that inhibits its interaction with SIR3. Proc Natl Acad Sci USA. 1997;94:2186–2191. doi: 10.1073/pnas.94.6.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park EC, Szostak JW. Point mutations in the yeast histone H4 gene prevent silencing of the silent mating type locus HML. Mol Cell Biol. 1990;10:4932–4934. doi: 10.1128/mcb.10.9.4932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman JM, Stone EM, Freeman-Cook LL, Brachmann CB, Boeke JD, Pillus L. The conserved core of a human SIR2 homologue functions in yeast silencing. Mol Biol Cell. 1999;10:3045–3059. doi: 10.1091/mbc.10.9.3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shou W, Seol JH, Shevchenko A, Baskerville C, Moazed D, Chen ZW, Jang J, Shevchenko A, Charbonneau H, Deshaies RJ. Exit from mitosis is triggered by Tem1-dependent release of the protein phosphatase Cdc14 from nucleolar RENT complex. Cell. 1999;97:233–244. doi: 10.1016/s0092-8674(00)80733-3. [DOI] [PubMed] [Google Scholar]
- Smith JS, Boeke JD. An unusual form of transcriptional silencing in yeast ribosomal DNA. Genes Dev. 1997;11:241–54. doi: 10.1101/gad.11.2.241. [DOI] [PubMed] [Google Scholar]
- Smith JS, Brachmann CB, Celic I, Kenna MA, Muhammad S, Starai VJ, Avalos JL, Escalante-Semerena JC, Grubmeyer C, Wolberger C, Boeke JD. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc Natl Acad Sci USA. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strahl-Bolsinger S, Hecht A, Luo K, Grunstein M. SIR2 and SIR4 interactions differ in core and extended telomeric heterochromatin in yeast. Genes Dev. 1997;11:83–93. doi: 10.1101/gad.11.1.83. [DOI] [PubMed] [Google Scholar]
- Straight AF, Shou W, Dowd GJ, Turck CW, Deshaies RJ, Johnson AD, Moazed D. Net1, a Sir2-associated nucleolar protein required for rDNA silencing and nucleolar integrity. Cell. 1999;97:245–256. doi: 10.1016/s0092-8674(00)80734-5. [DOI] [PubMed] [Google Scholar]
- Sutton A, Heller RC, Landry J, Choy JS, Sirko A, Sternglanz R. A novel form of transcriptional silencing by sum1–1 requires hst1 and the origin recognition complex. Mol Cell Biol. 2001;21:3514–3522. doi: 10.1128/MCB.21.10.3514-3522.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanner KG, Landry J, Sternglanz R, Denu JM. Silent information regulator 2 family of NAD-dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc Natl Acad Sci USA. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanny JC, Dowd GJ, Huang J, Hilz H, Moazed D. An enzymatic activity in the yeast Sir2 protein that is essential for gene silencing. Cell. 1999;99:735–745. doi: 10.1016/s0092-8674(00)81671-2. [DOI] [PubMed] [Google Scholar]
- Tanny JC, Moazed D. Coupling of histone deacetylation to NAD breakdown by the yeast silencing protein Sir2: evidence for acetyl transfer from substrate to an NAD breakdown product. Proc Natl Acad Sci USA. 2001;98:415–420. doi: 10.1073/pnas.031563798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JS, Ling X, Grunstein M. Histone H3 amino terminus is required for telomeric and silent mating locus repression in yeast. Nature. 1994;369:245–247. doi: 10.1038/369245a0. [DOI] [PubMed] [Google Scholar]
- Tissenbaum HA, Guarente L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature. 2001;410:227–230. doi: 10.1038/35065638. [DOI] [PubMed] [Google Scholar]
- Tsang AW, Escalante-Semerena JC. CobB, a new member of the SIR2 family of eucaryotic regulatory proteins, is required to compensate for the lack of nicotinate mononucleotide: 5,6-dimethylbenzimidazole phosphoribosyltransferase activity in cobT mutants during cobalamin biosynthesis in Salmonella typhimurium LT2. J Biol Chem. 1998;273:31788–31794. doi: 10.1074/jbc.273.48.31788. [DOI] [PubMed] [Google Scholar]
- Tsukamoto Y, Kato J, Ikeda H. Silencing factors participate in DNA repair and recombination in Saccharomyces cerevisiae. Nature. 1997;388:900–903. doi: 10.1038/42288. [DOI] [PubMed] [Google Scholar]
- Wu J, Grunstein M. 25 Years after the nucleosome model: chromatin modifications. Trends Biochem Sci. 2000;25:619–623. doi: 10.1016/s0968-0004(00)01718-7. [DOI] [PubMed] [Google Scholar]