ABSTRACT
We assessed the level of evidence for the presence of new periodontal pathogens by (i) comparing the occurrence of non-classical periodontal taxa between healthy vs. periodontitis patients (Association study); (ii) assessing the modifications in the prevalence and levels of these species after treatments (Elimination study). In the Association study, we compared the prevalence and levels of 39 novel bacterial species between periodontally healthy and periodontitis patients. In the Elimination study, we analyzed samples from periodontitis patients assigned to receive scaling and root planing alone or with metronidazole+ amoxicillin TID/ 14 days. Levels of 79 bacterial species (39 novel and 40 classic) were assessed at baseline, 3 and 12 months post-therapy. All samples were analyzed using Checkerboard DNA-DNA hybridization. Out of the 39 novel species evaluated, eight were categorized as having strong and four as having moderate association with periodontitis. Our findings suggest strong evidence supporting Lancefieldella rimae, Cronobacter sakazakii, Pluralibacter gergoviae, Enterococcus faecalis, Eubacterium limosum, Filifactor alocis, Haemophilus influenzae, and Staphylococcus warneri, and moderate evidence supporting Escherichia coli, Fusobacterium necrophorum, Spiroplasma ixodetis, and Staphylococcus aureus as periodontal pathogens. These findings contribute to a better understanding of the etiology of periodontitis and may guide future diagnostic and interventional studies.
KEYWORDS: Periodontitis, etiology, new pathogens, periodontal treatment, oral microbiology, Microbiota
Introduction
The occurrence and progression of periodontitis involve a complex interaction between the periodontal microbiome and the immune system [1,2]. As a disease associated with polymicrobial dysbiosis, effective prevention, and management of periodontitis is dependent on the identification of microorganisms associated with its onset and progression.
Socransky et al. [3] used the Checkerboard DNA-DNA hybridization technique to evaluate the composition of subgingival biofilm samples (n = 13,261) from 160 volunteers with periodontitis and 25 with periodontal health. The authors described three microbial complexes (green, purple, and yellow) associated with periodontal health and two groups of microorganisms (red and orange) strongly associated with clinical signs of disease. The red complex was composed of Porphyromonas gingivalis, Tannerella forsythia, and Treponema denticola while the orange complex congregated ten species from different genera, such as Fusobacterium, Prevotella, and Campylobacter. Importantly, a subsequent study demonstrated that four Actinomyces species were also closely related to periodontal health [4]. These studies have greatly contributed to increasing knowledge of the periodontal microbial ecology. Furthermore, the 40 bacterial species from these microbial complexes have been successfully used as biological markers in studies testing the effects of periodontal therapies on reestablishing a health-associated microbiome [5,6,7–11,12,13–15].
As growing knowledge on the oral microbiome composition emerges, especially through next-generation sequencing (NGS), other species have been suggested as possible bacterial pathogens [15,16,17–19,20,21]. A systematic review pointed out that bacteria from the phyla Bacteroidetes, Firmicutes, Proteobacteria, Saccharibacteria, Spirochaetes, and Synergistetes might be related to periodontal disease onset and progression [21].
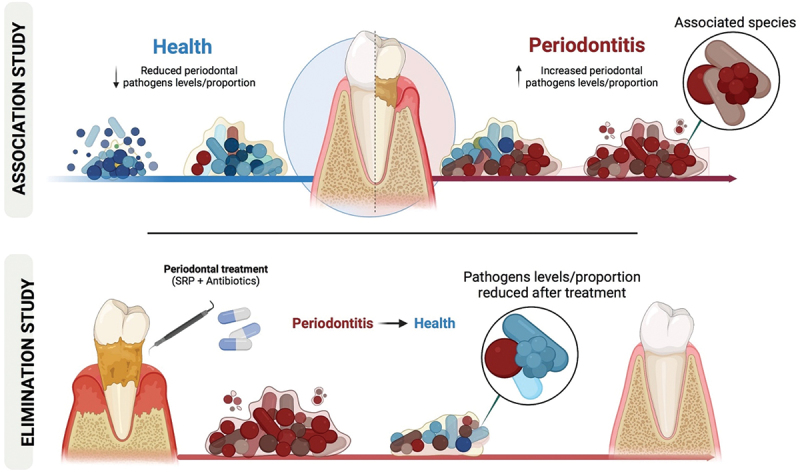
The main criteria used to associate a certain microorganism with the etiology of infection are the postulates established by [22]. In the context of periodontal infections, the postulates modified by Socransky [23], have been largely used to establish a true periodontal pathogen, as follows: 1) Association with disease implies increased proportions or levels of the microorganism at sites of disease and reduced proportions or levels (or absence) in healthy sites or sites with other forms of disease; 2) Elimination of the organism is a critical test of its effect during disease by its elimination or reduction leading to disease suppression; 3) A pathogen should lead to an increased or decreased host immune response; 4) Animal models should replicate what is observed in human disease; 5) A putative microorganism should have particular pathogenic mechanisms.
Socransky [23] suggested that the most important studies to associate a microorganism with a given pathology are the Association and Elimination studies (postulates 1 and 2). Several Association studies have evaluated the possible involvement of newly identified species in the etiology of periodontitis, either by targeted or open-ended microbiological techniques, as shown by previous studies [15,21]. However, most studies evaluated few biofilm samples. In addition, Elimination studies aiming to determine changes in the levels or proportions of these species after periodontal treatment are scarce [15]. Combining Association and Elimination studies may help to identify new species involved in the etiology of periodontitis and understand the ecology of the oral cavity. Hence, we determined the level of evidence for novel periodontal pathogens by (i) comparing the occurrence of 39 novel bacterial species in healthy and periodontitis patients (Association study) and (ii) assessing their behavior after periodontal treatment (Elimination study).
Materials and methods
This investigation included two different sections: (i) an Association and (ii) an Elimination study. The Association study aimed at comparing the prevalence and levels of 39 novel bacterial species between periodontally healthy and diseased patients. The Elimination study was designed as a randomized controlled trial to test changes in the prevalence and levels of these species after treatment.
Study population and inclusion and exclusion criteria: Association and Elimination Studies
Patients with stages III and IV, grades B and C, generalized periodontitis, and periodontally healthy individuals were selected from the Periodontal Clinic at Guarulhos University (Guarulhos, SP, Brazil). Informed consent was provided by each patient after a thorough explanation of the risks and benefits of their participation in the study. The study was approved by the human subjects ethics board of Guarulhos University (Protocol 437.155).
Inclusion criteria for patients with periodontitis were: at least 6 teeth with≥1 interproximal non-contiguous sites with probing depth (PD) and clinical attachment level (CAL) ≥5 mm; at least 15 teeth; ≥30% of sites with bleeding on probing (BoP) and PD and CAL≥4 mm; ≥30 years old.
Inclusion criteria for periodontally healthy patients were: ≥15 teeth; no site with PD and CAL>3 mm; <10% of sites with BoP; ≥30 years old.
Exclusion criteria for all patients were: previous periodontal treatment; smokers or former smokers (≤5 years); pregnancy or lactation; any systemic disease that can affect the pathogenesis of periodontal diseases need for antibiotic prophylaxis; current orthodontic treatment; use of antibiotics in the previous 6 months; allergy to metronidazole (MTZ) or penicillin for patients with periodontitis.
Clinical assessment: association and Elimination Studies
Two examiners participated in a calibration exercise to assess intra- and inter-examiner variability for PD and CAL measurements. Calibration was carried out according to Araujo et al. [24] and the standard error (SE) of measurement was calculated. The average level of intra- and inter-examiner agreement for the categorical variables was>93% (Kappa test). Clinical measurements were performed at baseline for periodontally healthy individuals and periodontitis patients (Association study), and at 3 and 12 months for patients with periodontitis (Elimination study). Mean CAL, PD, and the percentage of sites with plaque, gingival bleeding, BoP and suppuration (SUP) were assessed at 6 sites/teeth using a North Carolina periodontal probe (PCPUNC-BR 15 Hu-Friedy, Rio de Janeiro, RJ, Brazil).
Microbial assessment: association and Elimination Studies
For patients with periodontitis, 9 subgingival biofilm samples per patient were collected at baseline, and at 3 and 12 months post-treatment, 3 from each of the following categories of sites: shallow (PD ≤3 mm), moderate (PD = 4–6 mm), and deep (PD ≥7 mm). For healthy patients, 9 randomly biofilm samples were collected from sites with PD ≤3 mm. After supragingival biofilm and calculus removal, subgingival plaque samples were collected with periodontal curettes (Gracey mini-five 11–12) and kept in pre-coded sterile microtubes with 150 μL TE buffer (10 mM Tris-HCL) (Invitrogen Life Technologies, Carlsbad, CA, USA). Then, 100 μL of 0.5 M NaOH solution was added. The microtubes were kept frozen (−20ºC) until the biofilm samples were analyzed using Checkerboard DNA-DNA hybridization [9,25,26].
Genomic probes were prepared for 39 ”novel” pathogens not yet been proven to be related to the etiology or progression of periodontitis. The selection of these potential pathogens was performed as follows:
Species that have been suggested as possible periodontal pathogens by Pérez-Chaparro et al. [21]. The authors reported that 34 newly identified bacterial species could be considered periodontal pathogens. As 22 of these are cultivable species, they could be analyzed by Checkerboard DNA-DNA hybridization. We obtained DNA from 9 of these 22 cultivable species and designed their probes. However, 3 of these bacterial species (Eubacterium saphenum, Selenomonas sputigena, and Acinetobacter baumannii) were eventually excluded because of nonfunctioning DNA probes. Thus, 6 of these bacterial species were evaluated in the present study: Dialister pneumosintes, Enterococcus faecalis, Escherichia coli, Eubacterium brachy, Filifactor alocis, and Porphyromonas endodontalis.
Bacterial species of medical importance and/or suggested by Dr. Sigmund Socransky and Dr. Flavia Teles (advisory expert opinion): seven species (Lancefieldella rimae, Haemophilus influenzae, Klebsiella pneumoniae, Prevotella oris, Rothia dentocariosa, Staphylococcus aureus, and Staphylococcus epidermidis) that had already been associated with the etiology of periodontitis in at least one previous publication were selected [15]. The other 26 microorganisms were selected for being pointed out as bacterial species of medical importance or related to extra-oral infections. Streptococcus pneumoniae was only included in the Elimination study for being identified in healthy volunteers only [27]. Therefore, 39 species were evaluated in the Association study and 40 species were assessed in the Elimination study.
Checkerboard DNA-DNA hybridization
Microbial analyses were performed using Checkerboard DNA-DNA Hybridization according to Socransky and Haffajee [25], and Soares et al. [26] (Suppl. materials 1).
Periodontal treatment: elimination study
After biofilm samples were collected, patients with periodontitis were randomly assigned to receive scaling and root planing (SRP) with or without systemic antibiotics. The control group received SRP+ placebos TID for 14 days, and the test group (MTZ+AMX) received SRP+ MTZ (400 mg) + amoxicillin (AMX, 500 mg), TID for 14 days. This was a double-blind, placebo-controlled interventional study. Microbiological monitoring of the 39 new periodontal pathogens and 40 classic periodontal species was conducted at baseline and at 3 and 12 months after therapy. SRP was performed in 4 to 6 appointments within 14 days by a trained periodontist using an ultrasonic device and curette. The antibiotic or placebo regimens began on the first SRP session. Patients were engaged in supportive periodontal therapy every 3 months, including supragingival plaque and calculus removal, subgingival scaling of sites with PD ≥4 mm, and oral hygiene instructions.
Statistical analysis
Association study
Mean CAL and PD and mean percentage of sites with plaque, gingival bleeding, BoP, and suppuration were recorded for each patient and within each group. The bacterial levels and prevalence of the ‘new’ bacterial species were recorded from biofilm samples of patients with periodontal health and disease The mean levels (x105) of each species were recorded per site, per patient, and per group. The same procedures were applied for microbiological data recorded from shallow sites (i.e. PD ≤3 mm) of both groups. The percentage of sites and patients colonized by each species was recorded. The differences between periodontally healthy and periodontitis groups were assessed by the Mann-Whitney test. The chi-square test was used to compare gender and the frequency of patients colonized by each bacterial species. Statistical significance was established as 5%. Microbiological analyses were performed with and without adjusting for multiple comparisons. A p-value of<0.00125 was applied for the adjusted comparisons, as proposed by Socransky et al. [28].
Elimination study
The ideal sample size to assure adequate power for this study was based on a difference of at least 4 sites with PD ≥5 mm [29] between groups and a standard deviation of 5 sites with PD ≥5 mm [9]. Considering a significance level of 5%, 20 subjects per group would be necessary to provide a power of 80%. Mean CAL and PD, the percentage of sites with gingival bleeding, plaque, BoP, and suppuration, and mean number of sites with PD ≥5 mm were recorded per patient and within the groups. Intragroup differences (from baseline to 1-year post-treatment) were assessed using the Wilcoxon test. Thedifferences between treatment groups at each time point were assessed by the Mann-Whitney test. The percentage of patients achieving the clinical endpoint for treatment (≤4 sites PD ≥5 mm [29]; 1-year post-therapy was evaluated using the Chi-square test.
Microbiological data from subgingival biofilm samples of both therapeutic groups were expressed as bacterial loads (levels/count), prevalence, and proportion. The mean levels (x105) and prevalence of each new species were recorded per site, per patient, and per group at each time point. Intragroup differences (baseline, 3 months, and 1 year) for these microbiological parameters were assessed by the Friedman test. Differences between treatment groups at each time point were assessed by the Mann-Whitney test. The mean proportions of the microbial complexes [3] between baseline and 1 year within each treatment group were compared using the Wilcoxon test, while the mean proportions of the microbial complexes between groups at each time point were compared by the Mann-Whitney test. Statistical significance was established as 5%. Microbiological analyses were performed with and without adjustment for multiple comparisons. A p-value of<0.00125 was considered for multiple comparisons, as proposed by Socransky et al. [28].
Classification of potential “new” periodontal pathogens
Results from the Association and the Elimination studies were grouped, and bacterial species were categorized into 2 levels of evidence for the status of ‘periodontal pathogen’ according to the following criteria:
Strong evidence: when the species was significantly elevated in periodontitis patients when compared to periodontally healthy individuals considering the following 5 parameters: % of patients colonized (i); % of colonized sites considering all sites (ii) or only shallow sites (iii); mean bacterial counts considering all sites (iv) or only shallow sites (v). Besides that, the species should be reduced after treatment according to the following parameters: percentage of colonized sites (i) and mean levels (ii), by at least one of the treatment protocols. For the analysis of ‘shallow sites’, adjusted and non-adjusted statistical significance for multiple comparisons were considered, whereas for the other parameters only adjusted significances were considered.
Moderate evidence: when the species was elevated in periodontitis patients when compared to healthy individuals for at least 4 of the 5 abovementioned parameters, considering adjusted and non-adjusted statistical significances. Besides that, the species should be reduced for the following parameters: percentage of colonized sites (i) and mean levels (ii), by at least one of the treatment protocols.
Results
Association Study
Demographic and clinical baseline characteristics of healthy and periodontitis groups are presented in Table 1. All clinical parameters were significantly higher forpatients with periodontitis (n = 40) than for periodontally healthy (n = 17) individuals (p < 0.05).
Table 1.
Association study: Clinical and demographic parameters of patients at baseline.
| |
Groups |
|
|
|---|---|---|---|
| Healthy (n = 17) |
Periodontitis (n = 40) |
||
| Variables | Mean ± SD | Mean ± SD | p-value |
| Gender (F/M) | 10/7 | 18/12 | >0.05# |
| Age (years) | 40.6 ± 4.1 | 45.6 ± 8.1 | 0.5674* |
| PD (mm) | 1.96 ± 0.6 | 3.82 ± 0.71 | 0.0007* |
| CAL (mm) | 1.04 ± 0.5 | 4.23 ± 0.93 | 0.0099* |
| sites with: | |||
| Visible plaque (%) | 34.5 ± 9.9 | 84.1 ± 12.3 | 0.0103* |
| Gingival bleeding (%) | 4.5 ± 2.9 | 29.2 ± 11.1 | 0.0029* |
| BOP (%) | 6.1 ± 3.2 | 81.1 ± 14.1 | 0.0004* |
| SUP (%) | 0.0 ± 0.0 | 2.5 ± 3.2 | 0.0000* |
SD, Standard deviation; PD, Probing depth; CAL, Clinical attachment level; BOP, Bleeding on probing; SUP, Suppuration. *Mann-Whitney test. # Chi-square test.
Table 2 depicts the 17 bacterial species that colonized a higher percentage of subjects with periodontitis than with periodontal health (p < 0.05). The mean percentage of all sites colonized by the 39 bacterial species evaluated is presented in Figure 1. When all sites were analyzed (Figure 1a), 22 species were observed in a significantly higher percentage of sites in patients with periodontitis than in periodontally healthy individuals (p < 0.05). When only shallow sites were considered (Figure 1b), E. faecalis, Staphylococcus warneri, and Cronobacter sakazakii colonized a higher percentage of sites of patients with periodontitis (adjusting for multiple comparisons). Eleven species (L. rimae, Pluralibacter gergoviae, Enterococcus faecium, E. coli, Eubacterium limosum, F. alocis, Fusobacterium necrophorum, H. influenzae, Serratia marcescens, Spiroplasma ixodetis, and S. aureus) were detected in a higher percentage of sites and Aggregatibacter aphrophilus in a lower percentage of sites of periodontitis patients than in healthy individuals, when the non-adjusted analysis was pondered.
Table 2.
Species colonizing a statistically significantly higher percentage of patients with periodontitis than periodontally healthy individuals.
| |
Groups |
|
|
|---|---|---|---|
| Microrganismos | Healthy (n = 17) | Periodontitis (n = 40) | p-value |
| Actinomyces meyeri | 76.5 | 97.5 | 0.0242** |
| Lancefieldella rimae | 23.5 | 80.0 | 0.0002* |
| Cronobacter sakazakii | 58.8 | 100 | <0.0001* |
| Enterobacter cloacae | 41.2 | 80.0 | 0.0060** |
| Enterobacter gergoviae | 5.9 | 67.5 | <0.0001* |
| Enterococcus faecallis | 58.8 | 97.5 | <0.0001* |
| Escherichia coli | 82.4 | 100 | 0.0200** |
| Eubacterium limosum | 0.0 | 82.5 | <0.0001* |
| Filifactor alocis | 47.1 | 90.0 | 0.0010* |
| Fusobacterium necrophorum | 52.9 | 90.0 | 0.0035** |
| Haemophilus influenzae | 0.0 | 75.0 | <0.0001* |
| Spiroplasma ixodetis | 41.2 | 90.0 | 0.0020** |
| Staphylococcus aureus | 58.8 | 95.0 | 0.0018** |
| Staphylococcus warneri | 70.6 | 100 | <0.0001* |
| Streptococcus pyogenes | 82.4 | 100 | 0.0232** |
| Veillonella díspar | 64.7 | 97.5 | 0.0019** |
| Vibrio nereis | 47.1 | 87.5 | 0.0023** |
p < 0.05 means statistically significant differences between groups by the Chi-square test, with (*) and without (**) adjustments for multiple comparisons.
Figure 1.
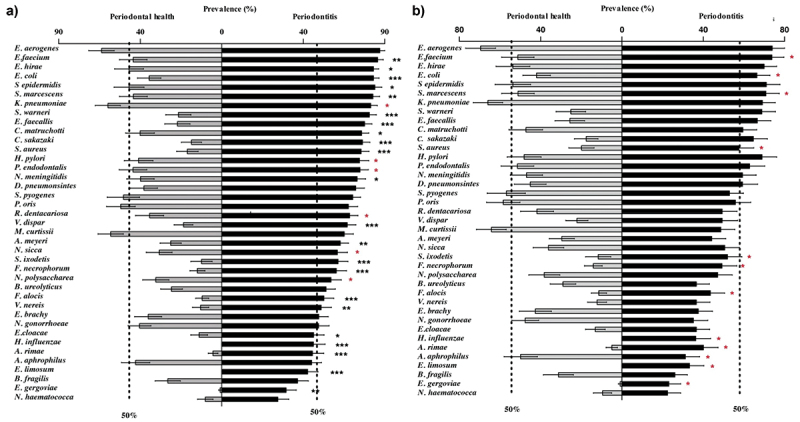
Mean percentage of all evaluated sites (a) and of shallow sites (b) colonized by the 39 ‘new’ bacterial species in periodontal health and disease. The species are presented in descending order according to their prevalence in the periodontitis group. (*p < 0.05, Mann-Whitney test). (* in black) indicates statistical significance adjusted for multiple comparisons; (* in red) indicates unadjusted statistical significance.
The levels of 30 species (25 after adjustments for multiple comparisons) were higher in periodontitis than in healthy patients considering all sites (p < 0.05). The levels of 15 species (2 after adjustments for multiple comparisons) were higher in periodontitis compared to healthy patients considering the shallow sites only (Figure 2).
Figure 2.
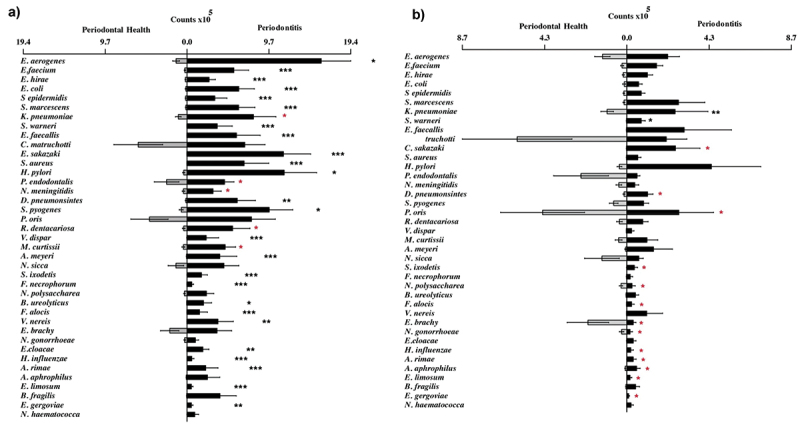
Mean counts of the 39 ‘new’ bacterial species in periodontal health and disease of all evaluated sites (a) and in shallow sites (b). The species are listed in descending order according to the average percentage of colonized sites in the periodontitis group (Figure 1). The statistical significance of differences between groups was assessed using the Mann-Whitney test (p < 0.05). (* in black) indicates statistical significance adjusted for multiple comparisons; (* in red) indicates unadjusted statistical significance.
Elimination Study
Table 3 presents the demographic characteristics and baseline clinical parameters of the groups, before and after periodontal therapy. Clinical parameters did not differ between treatment groups at baseline (p > 0.05). At 1-year post-therapy, improved clinical parameters were observed for both groups. However, the group that received SRP + MTZ + AMX showed more striking clinical benefits than the placebo group (p < 0.05). The mean number of sites with PD ≥5 mm 1 year after periodontal therapy was 14.5 ± 10.2 for the placebo group and 4.1 ± 4.8 for the SRP+MTZ+AMX group (p < 0.05, adjusted for baseline values). 70% of patients in the test group achieved the clinical endpoint for periodontal treatment (≤4 sites with PD ≥5 mm [29]; as opposed to 10% of those in the placebo group.
Table 3.
Elimination study: Demographic characteristics and clinical parameters of both treatment groups, before and after periodontal therapy.
| Treatment groups |
||||
|---|---|---|---|---|
| Variables | Time | SRP (n = 20) | SRP+ MTZ + AMX (n = 20) | p-value |
| Gender (F/M) | Baseline | 16/4 | 12/8 | 0.300# |
| Age (Mean ± SD, years) | Baseline | 44.8 ± 8.9 | 46.2 ± 8.9 | 0.473* |
| PD (Mean ± SD, mm) | Baseline | 3.88 ± 0.64 | 3.70 ± 0.66 | 0.514* |
| CAL (Mean ± SD, mm) | Baseline | 4.25 ± 0.74 | 4.15 ± 0.89 | 0.375* |
| Mean number (± SD) of sites with PD ≥5 mm | Baseline | 38.8 ± 21.6 A | 33.0 ± 20.0 A | 0.089* |
| 1 year | 14.5 ± 10.2 B, a | 4.1 ± 4.8 B, b | <0.0001* | |
| Number and % of patients achieving the clinical endpoint for treatment, i.e. < 4 sites PD > 5 mm [29]. | 1 year | 2 (10.0%)a | 14 (70.0%)b | <0.0001# |
The statistical significance of differences between groups at baseline and at 1-year post-treatment was assessed by the Mann-Whitney (*) test or the Chi-square test (#). Different lowercase letters indicate significant differences between groups, p < 0.05.
The statistical significance of differences within each group for the mean number of sites with PD ≥5 mm (between baseline and 1 year) was assessed by the Wilcoxon test. Different capital letters indicate significant differences between time points, p < 0.05.
SRP, scaling and root planing; MTZ, metronidazole; AMX, amoxicillin; PD, probing depth, CAL, clinical attachment level; SD: Standard deviation.
Figure 3 displays the effects of therapies (Elimination study) on the mean number of sites colonized by the ‘new’ species at 3 months and 1 year, and the comparison between groups between baseline and 1 year. For the analysis adjusted for multiple comparisons, SRP significantly reduced 3 species (H. influenzae, N. meningitidis, and P. endodontalis), and SRP+MTZ+AMX, 17 species (Campylobacter ureolyticus, C. sakazakii, E. cloacae, Enteroccocus hirae, F. necrophorum, H. influenzae, N. meningitidis, N. polysaccharea, Neisseria sicca, P. endodontalis, R. dentocariosa, S. marcescens, S. ixodetis, S. aureus, S. epidermidis, S. warneri, and S. pneumoniae). When the groups were compared at 1 year, patients in the SRP+MTZ+AMX presented fewer sites colonized by 9 species (L. rimae, C. sakazakii, E. hirae, F. alocis, N. gonorrhoeae, N. polysaccharea, S. aureus, V. nereis, and E. cloacae) than those in the SRP group. However, these differences were only observed for the non-adjusted comparisons.
Figure 3.
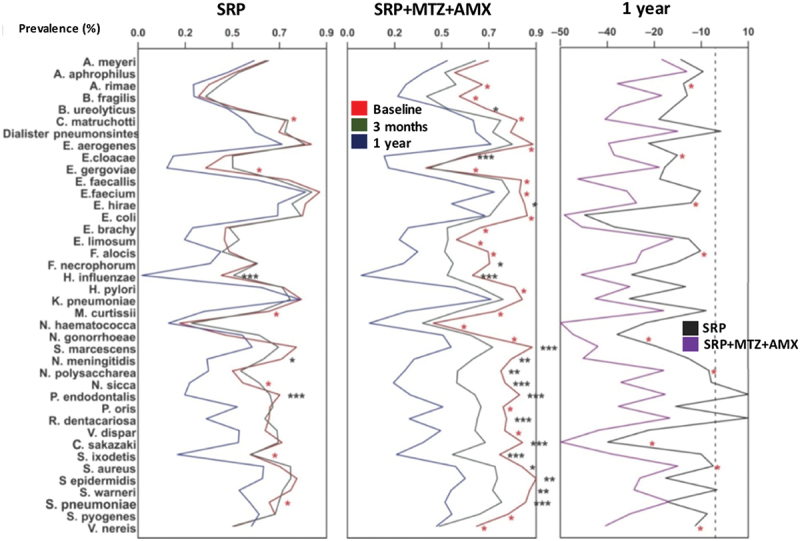
Average of periodontal sites colonized (prevalence) by the new pathogens evaluated for the treatment group SRP or SRP+MTZ+AMX at baseline, after 3 months and 1 year. Comparison between treatment groups after 1 year. SRP – Scaling root planing; MTZ – Metronidazole; AMX – Amoxicillin. The statistical significance of differences between groups was assessed using the Mann-Whitney or Friedman test (p < 0.05). (* in black) indicates statistical significance adjusted for multiple comparisons; (* in red) indicates unadjusted statistical significance.
The effects of treatments on the individual mean levels of the 40 classic bacterial species from the microbial complexes [3] are presented in Figure 4. Both treatments reduced the individual levels of pathogens from the red and orange complexes and led to an increase in several host-compatible species. Nonetheless, the reduction in counts of the three red complex pathogens was more striking in the antibiotic group. The mean proportions of the microbial complexes [3]at baseline and 1 year are presented in Figure 5. This analysis includes the 40 bacterial species from the traditional Checkerboard DNA-DNA hybridization panel [26]. The proportions of orange and red complexes were significantly reduced by both treatments and the Actinomyces species increased (p < 0.05). At 1 year, patients treated with adjunctive MTZ + AMX had lower proportions of red complex (3%) and higher proportions of Actinomyces (36%), in comparison with those treated with SRP-only (9% and 26%, respectively). As a comparison, the mean proportions of all bacterial species evaluated in this study are presented in Supplementary Figure 1. Some classical and putative periodontal pathogens were among the species present in the highest proportions, such as T. forsythia, P. gingivalis, and several Fusobacterium species.
Figure 4.
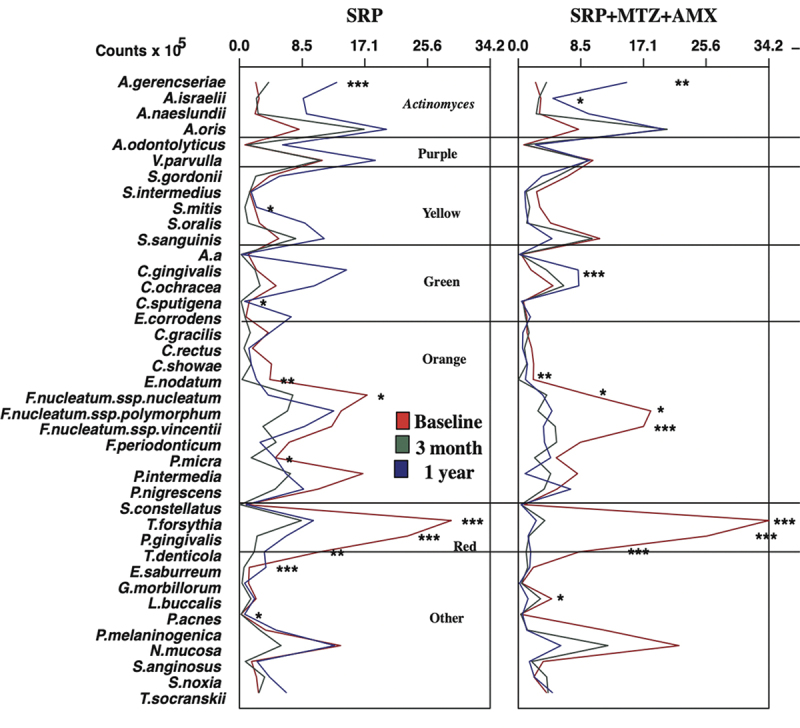
Mean levels of the classic 40 bacterial species 3Mean levels of the 40 classic bacterial species from the microbial complexes [3] at baseline, 3 months and 1 year. SRP – Scaling root planning; MTZ – Metronidazole; AMX – Amoxicillin. The statistical significance of differences within eacg treatment group over time was assessed using the Friedmantest (p < 0.05) (* in black) indicates statistical significance adjusted for multiple comparisons; (*in red) indicates unadjusted statistical significance.
Figure 5.
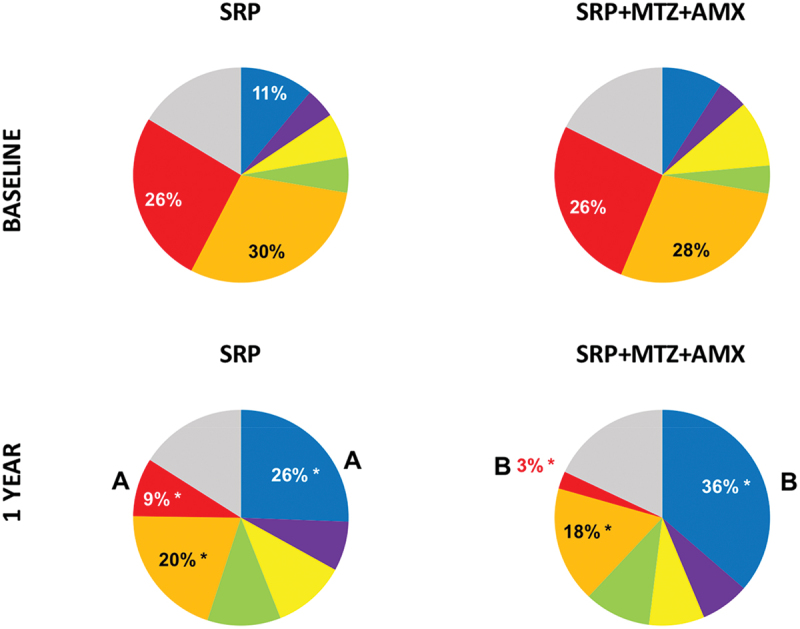
Mean proportions of the microbial species described by Socransky et al. [3] in the two treatment groups at baseline and 1 year. The significance of differences between groups was determined by the Mann-Whitney test (different letters = p < 0.05), and within each group (between the two time points) by the Wilcoxon test (*p < .05).
Classification of potential “new” periodontal pathogens
Table 4 Species categorized in the two levels of evidence – Strong or Moderate – for the status of ‘periodontal pathogen’. Eight species were included in the Strong evidence category (L. rimae, C. sakazakii, E gergoviae, E. faecalis, E. limosum, F. alocis, H. influenzae, and S. warneri) while four species were categorized as Moderate evidence (E. coli, F. necrophorum, S. ixodetis, and S. aureus). Table 5 presents the summary of key findings from Association and Elimination studies used to determine the levels of evidence.
Table 4.
Classification of potential ‘New’ periodontal pathogens according to the level of association with periodontitis.
| Species | ATCC Strain | |
|---|---|---|
| Strong | Lancefieldella rimae | 49626 |
| Cronobacter sakazakii | 12868 | |
| Enterobacter gergoviae | 33028 | |
| Enterococcus faecallis | 29212 | |
| Eubacterium limosum | 8486 | |
| Filifactor alocis | 35896 | |
| Haemophilus influenzae | 33533 | |
| Staphylococcus warneri | 27836 | |
| Moderate | Escherichia coli | 10799 |
| Fusobacterium necrophorum | 25286 | |
| Spiroplasma ixodetis | 33835 | |
| Staphylococcus aureus | 33591 |
Strong evidence: when the species was significantly elevated in periodontitis patients when compared to periodontally healthy individuals considering the following 5 parameters: % of patients colonized (i); % of colonized sites considering all sites (ii) or only shallow sites (iii); mean bacterial counts inconsidering all sites (iv) or only shallow sites (v). Besides that, the species should be reduced aftertreatment according to the following parameters: percentage of colonized sites (i) and mean levels (ii), by at least one of the treatment protocols. For the analysis of ‘shallow sites’, adjusted and non-adjusted statistical significance for multiple comparisons were considered, whereas for the other parameters only adjusted significances were considered.
*For the analysis of ‘shallow sites’, adjusted and non-adjusted statistical significance for multiple comparisons were considered, whereas for the other parameters only adjusted significances were considered.
Moderate evidence: when the species was elevated in periodontitis patients when compared to healthy individuals for at least 4 of the 5 abovementioned parameters, considering adjusted and non-adjusted statistical significances. Besides that, the species should be reduced for the following parameters: percentage of colonized sites (i) and mean levels (ii), by at least one of the treatment protocols.
Table 5.
Summary of key findings from Association and Elimination studies.
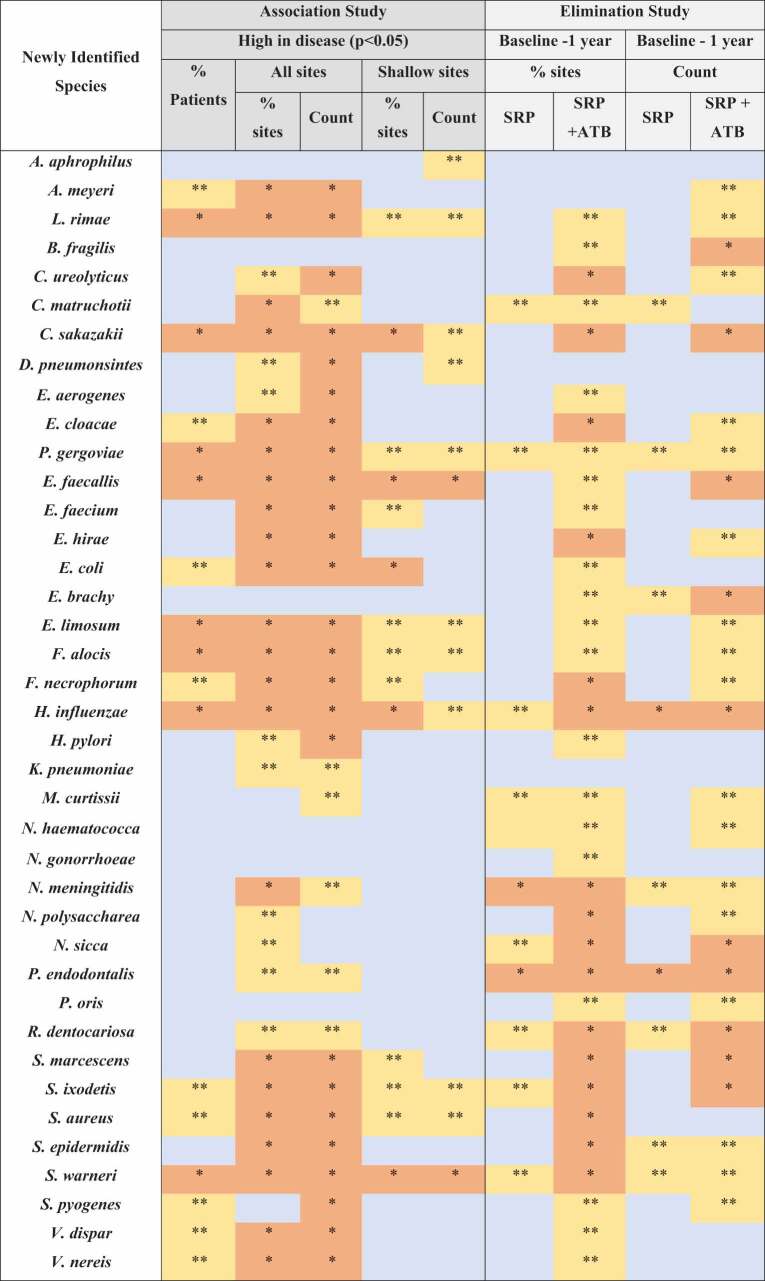 |
Adjusted statistical significance for multiple comparisons  ; Unadjusted statistical significance
; Unadjusted statistical significance  ; No difference
; No difference  % sites: percentage of sites colonized; % patients: percentage of patients colonized; Baseline −1 year: bacterial species significantly reduced between baseline and 1-year post-therapies; SRP: scaling.
% sites: percentage of sites colonized; % patients: percentage of patients colonized; Baseline −1 year: bacterial species significantly reduced between baseline and 1-year post-therapies; SRP: scaling.
Discussion
The data of this study suggested the association of 12 new bacterial species with the etiopathogenesis of periodontitis, with strong or moderate evidence. The bacterial species that showed strong evidence to be potential novel pathogens were L. rimae, C. sakazakii, P. gergoviae, E. faecalis, E. limosum, F. alocis, H. influenzae, and S. warneri; and those with moderate evidence were E. coli, F. necrophorum, S. ixodetis, and S. aureus.
Ecological studies have revealed that levels and proportions of a microorganism in periodontal pockets are more relevant to understand their role in health and disease than their mere presence/absence [4,30,31]. Thus, it is noteworthy that among the eight species showing strong association with periodontitis, three - C. sakazakii, E. faecalis, and S. warneri - were detected at higher levels and colonized a higher percentage of sites in patients with periodontitis than in healthy ones, when ‘all sites’ and only ‘shallow sites’ were evaluated. In addition, they were found at very low levels and prevalence in healthy volunteers [32].
Sakazakii is a gram-negative rod that belongs to the Enterobacteriaceae family and is not part of the normal microbiota of the human/animal oral cavity or gastrointestinal tract. This microorganism is recognized as an opportunistic pathogen of food origin and has been related to cases of neonatal meningitis [33,34]. In periodontics, C. sakazakii has been found in the subgingival biofilm of HIV-positive patients with necrotizing periodontitis [35]. However, this species had not yet been associated with periodontitis in systemically healthy patients [15,21]. Therefore, this is new information that deserves further assessment.
Warneri is a coagulase-negative Staphylococcus (CNS) commonly present in human mucous membranes and epithelial microbiota. This microorganism has been considered a potential new pathogen in non-oral severe infections [36,37]; but has rarely been studied in periodontology. The epidemiology and the pathogenesis of S. warneri are still unclear [38,39]. Colombo et al. [40] detected this species in subgingival biofilm samples in patients refractory to conventional periodontal treatment. These authors identified other bacterial species not commonly found in the oral cavity, including A. baumannii, Gemella haemolysans, Pseudomonas aeruginosa, and E. faecalis – a species also strongly associated with periodontitis in the current study.
Faecalis is a common commensal microorganism of the human gastrointestinal tract but may be an important opportunistic pathogen in other parts of the body. It is the third most frequent pathogen isolated from bacteremia, causing most of the postoperative infections in intensive care units [41,42]. Regarding oral infections, this species has been associated with cases of root canal treatment failure [43–48]. Several Association studies showed a relationship between E. faecalis and periodontitis in Brazilian patients [40,49,50], which could be an example of geographical specificity. Chidambar et al. [51] also reported a higher prevalence of E. faecalis in subgingival plaque samples of Indian patients with periodontitis. Those studies corroborate historical data showing subgingival E. faecalis occurring in 1% of early-onset periodontitis patients and in approximately 5% of adults with periodontitis [52]. While E. faecalis were suggested to populate periodontal pockets as superinfecting organisms, this theory has not been proven. In any case scenario, in heavily infected patients, this microorganism could contribute to periodontal breakdown [52]. Interestingly, this species has not been commonly detected in studies using NGS techniques [21,15].
Two other species worth mentioning are S. aureus and E. coli, both classified as Moderate evidence. They followed a similar pattern to C. sakazaki, E. faecalis, and S. warneri but were less robust since some of the significance was only reached in the non-adjusted analyses.
Aureus secretes several toxins which are related to respiratory infections [53]. One of the pathogenic mechanisms used by this microorganism is the production of virulence factors that effectively alter specific target cell functions [54]. S. aureus has been observed in the oral cavity, mainly infecting root canals [55]. Souto et al. [49] found S. aureus elevated in patients with chronic periodontitis. Conversely, da Silva-Boghossian et al. [50] demonstrated that the frequency and counts of S. aureus in subgingival biofilm did not differ from sites with chronic periodontitis and periodontal health. Furthermore, other investigations have reported that S. aureus can be found in the subgingival biofilm regardless of the patient’s periodontal condition [56,57]. More recently, the role of S. aureus on polymicrobial peri-implant infections has been investigated [58,59]. Hence, additional studies evaluating S. aureus isolated from healthy and diseased teeth and implants are needed to clarify these controversial findings.
Coli is a bacterium that normally colonizes human and some animal intestines, but in some cases can cause infections, such as urinary and intestinal infections [60,61]. Like E. faecalis, this species has been associated with the etiology of periodontitis mostly in studies evaluating Brazilian patients [49]. Therefore, this may be another example of a specific periodontal pathogen of the Brazilian population that deserves further investigation.
The fact that P. endondontalis did not show any association with periodontitis in the present study was unexpected. This species has been considered as a possible periodontal pathogen in studies using nested PCR [62], Sanger sequencing [63], or pyrosequencing [19,20]. A similar situation occurred with D. pneumosintes, which also showed no specific association with periodontitis but had already been associated with periodontitis in previous investigations [16,21,62]. On the other hand, F. alocis, included in our study in the Strong evidence category, had already been suggested as a potential periodontal pathogen in several previous studies evaluating the microbiota of adults [16,19,20,63,64] and young patients with periodontitis [64; 65]. This species has also been related to xinfected root canals [66] and peri-implantitis [67].
The results of the Elimination study confirm data from previous studies suggesting that both treatments lead to improvements in periodontal clinical parameters, but when MTZ + AMX are used in combination with SRP these improvements are more robust [68–73]. The results of the 40 bacterial species from the traditional Checkerboard DNA-DNA hybridization panel underpin the clinical results and confirm the value of these species as biological markers for evaluating the effectiveness of different periodontal therapies [5,9,11,14,26] Patients treated with adjunctive MTZ+AMX showed a more beneficial biofilm composition even 1 year after antibiotic intake. This indicates that this treatment protocol is more effective than mechanical treatment alone to prevent biofilm resilience. In microbial ecology, resilience is the capacity of an ecosystem to deal with perturbation without shifting to an alternative state [74]. Most importantly, along with the benefits of these therapies, especially of the SRP+ MTZ + AMX, in the clinical parameters and the classic bacterial species, the Elimination study confirms the effect of these therapies in reducing the levels and prevalence of species that were considered as possible new periodontal pathogens. All 12 species categorized as Strong or Moderate evidence were significantly reduced by SRP+MTZ+AMX, while only 4 of them (P. gergoviae, H. influenzae, S warneri, and S. ixodetis) were significantly reduced by SRP only.
NGS analysis has increased our knowledge about bacterial communities due to a broad detection of species, including as-yet-uncultured taxa [75]. Although studies using NGS techniques are encouraged, the 40 species of the Checkerboard DNA-DNA hybridization panel continue to be consistent markers for oral dysbiosis and homeostasis. Additionally, the Checkerboard test allows the quantification of individual species, data not provided by sequencing techniques. Importantly, up until now, OMICS knowledge has not changed how we treat patients in daily practice [15]. Also, studies using meta-transcriptomic analysis to assess the metabolic functions and the virulence factors expressed by these potential novel pathogens would be fundamental to broadening our vision of the complex functionality of the oral microbiome. Thus, future standardized, large, multi-center case-control studies using multiple sequencing techniques (e.g. 16S rRNA gene sequencing, metagenomic, transcriptomics, proteomics, and metabolomics) must be encouraged.
The main limitations of this study are the low number of healthy patients evaluated and the close-ended microbial test used, which precludes the identification of uncultivated organisms. The main strength is to be the first study to combine an Association and an Elimination study and to evaluate a high number of species to weigh the evidence for their potential as new periodontal pathogens. These two experimental designs are considered the most important level of evidence when establishing a causal relationship between a microorganism and a particular infection [23].
The evaluation of the ‘new’ candidate pathogens in deep and shallow sites is also considered a strength of the study design. Previous Association studies have determined that species considered true pathogens, such as the red complex microorganisms, and newly identified pathogens are found at higher levels in deep sites of volunteers with periodontitis [3,15,76]. Even so, a continuous debate in the periodontal literature questions whether higher levels/prevalence of a microorganism in deep pockets of patients with periodontitis would configure a causal association with periodontitis. It has been speculated that this might be due to an overgrowth of these species, favored by inflammatory environmental stimuli in the periodontal pocket, such as the absence of oxygen and the large availability of nutrients for the growth of these pathogens (e.g., metals, amino acids, and peptides) [77–79]. Hence, the strategy used in our study of determining the presence of microorganisms also in shallow sites (PD ≤3 mm) helps to establish a causal relationship between their presence and the onset of infection.
Nevertheless, the question of whether periodontal disease onset is triggered by the new potential pathogens identified in this study (isolated or in combination) is extremely difficult to answer in humans. This is true even for classic pathogens in prospective clinical study designs including longitudinal repeated sampling of the same patients. Those limitations strengthen the rationale for asking such questions experimentally using germ-free mouse models, for example. In such models, each species can be introduced separately or in combination and their individual and combined effects on periodontal onset and/or progression may be assessed [80].
Another point of consideration relates to the functionality of these potential new pathogens, such as the ‘pathobionts’ concept [79]. According to this theory,under conditions of disrupted homeostasis, commensal microorganisms have the potential to cause deregulated inflammation and disease . This might explain why healthy patients are colonized with some of these newly identified pathogens without developing disease [81].
Regarding the statistical analysis, it is important to highlight that concerning the shallow site data, most of the statistically significant differences were observed only by the unadjusted analysis. This was expected since the number of samples is reduced to one-third when only shallow sites are considered. Similarly, for the Elimination study, the periodontitis group (n = 40) was divided into two groups of 20 volunteers, which also reduces the power of the study and may make it difficult to obtain statistical significance when adjusting for multiple comparisons. Therefore, when categorizing species according to the level of evidence for periodontal pathogen status, it was important to consider the results obtained in the adjusted and unadjusted analyses for shallow site data and the Elimination study.
Conclusions
Our findings suggest strong evidence supporting L. rimae, C. sakazakii, P. gergoviae, E. faecalis, E. limosum, F. alocis, H. influenzae, and S. warneri, and moderate evidence supporting E. coli, F. necrophorum, S. ixodetis, and S. aureus as periodontal pathogens. Together, these findings contribute to a better understanding of the etiology of periodontitis and may guide future diagnostic and interventional studies
Supplementary Material
Funding Statement
The author(s) reported there is no funding associated with the work featured in this article.
Supplementary Material
Supplemental data for this article can be accessed online at https://doi.org/10.1080/20002297.2023.2213111
Disclosure statement
No potential conflict of interest was reported by the authors.
Sources of funding statement
The authors acknowledge the Brazilian Coordination for the Improvement of Higher Education Personnel (CAPES) (Brazil), the National Council for Scientific and Technological Development (CNPq, Brazil) (Brazil) and Latin American Oral Health Association (LAOHA) (Brazil) for supporting some researchers involved in this study.
References
- [1].Duran-Pinedo AE, Chen T, Teles R, et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. Isme J. 2014;8(8):1659–15. DOI: 10.1038/ismej.2014.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Lamont RJ, Hajishengallis G.. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol Med. 2015;21(3):172–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Socransky SS, Haffajee AD, Cugini MA, et al. Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. [DOI] [PubMed] [Google Scholar]
- [4].Socransky SS, Haffajee AD. Dental biofilms: difficult therapeutic targets. Periodontol 2000. 2002;28:2–55. [DOI] [PubMed] [Google Scholar]
- [5].Carvalho LH, DAvila GB, Leão A, et al. Scaling and root planing, systemic metronidazole and professional plaque removal in the treatment of chronic periodontitis in a Brazilian population II–microbiological results. J Clin Periodontol. 2005;32:406–411. [DOI] [PubMed] [Google Scholar]
- [6].Haffajee AD, Patel M, Socransky SS. Microbiological changes associated with four different periodontal therapies for the treatment of chronic periodontitis. Oral Microbiol Immunol. 2008;23:148–157. [DOI] [PubMed] [Google Scholar]
- [7].Matarazzo F, Figueiredo LC, Cruz SE, et al. Clinical and microbiological benefits of systemic metronidazole and amoxicillin in the treatment of smokers with chronic periodontitis: a randomized placebo-controlled study. J Clin Periodontol. 2008;35:885–896. [DOI] [PubMed] [Google Scholar]
- [8].Feres M, Gursky LC, Faveri M, et al. Clinical and microbiological benefits of strict supragingival plaque control as part of the active phase of periodontal therapy. J Clin Periodontol. 2009;36:857–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Mestnik MJ, Feres M, Figueiredo LC, et al. Short-term benefits of the adjunctive use of metronidazole plus amoxicillin in the microbial profile and in the clinical parameters of subjects with generalized aggressive periodontitis. J Clin Periodontol. 2010;37(4):353–365. [DOI] [PubMed] [Google Scholar]
- [10].Sampaio E, Rocha M, Figueiredo LC, et al. Clinical and microbiological effects of azithromycin in the treatment of generalized chronic periodontitis: a randomized placebo-controlled clinical trial. J Clin Periodontol. 2011;38:838–846. [DOI] [PubMed] [Google Scholar]
- [11].Socransky SS, Haffajee AD, Teles R, et al. Effect of periodontal therapy on the subgingival microbiota over a 2-year monitoring period. I. Overall effect and kinetics of change. J Clin Periodontol. 2013;40(8):771–780. DOI: 10.1111/jcpe.12117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Faveri M, Rebello A, de Oliveira Dias R, et al. Clinical and microbiological effects of adjunctive metronidazole plus amoxicillin in the treatment of generalized chronic periodontitis: smokers versus non-smokers. J Periodontol. 2014;85(4):581–591. [DOI] [PubMed] [Google Scholar]
- [13].Tamashiro NS, Duarte PM, Miranda TS, et al. Amoxicillin plus metronidazole therapy for Patients with Periodontitis and Type 2 Diabetes: a 2-year Randomized Controlled Trial. J Dent Res. 2016;95(7):829–836. DOI: 10.1177/0022034516639274 [DOI] [PubMed] [Google Scholar]
- [14].Feres M, Figueiredo LC, Soares GM, et al. Systemic antibiotics in the treatment of periodontitis. Periodontol 2000. 2015;67(1):131–186. [DOI] [PubMed] [Google Scholar]
- [15].Feres M, Retamal-Valdes B, Gonçalves C, et al. Did Omics change periodontal therapy? Periodontol 2000. 2021;85(1):182–209. [DOI] [PubMed] [Google Scholar]
- [16].Kumar PS, Griffen AL, Moeschberger ML, et al. Identification of candidate periodontal pathogens and beneficial species by quantitative 16s clonal analysis. J Clin Microbiol. 2005;43:3944–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Matarazzo F, Ribeiro AC, Feres M, et al. Diversity and quantitative analysis of Archaea in aggressive periodontitis and periodontally healthy subjects. J Clin Periodontol. 2011;38(7):621–627. [DOI] [PubMed] [Google Scholar]
- [18].Teles FRF, Teles RP, Siegelin Y, et al. RNA-oligonucleotide quantification technique (ROQT) for the enumeration of uncultivated bacterial species in subgingival biofilms. Oral Microbiol. 2011;26:127–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Griffen AL, Beall CJ, Campbell JH, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. Isme J. 2012;6:176–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Abusleme L, Dupuy AK, Dutza NN, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. Isme J. 2013;7:1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Pérez-Chaparro PJ, Gonçalves C, Figueiredo LC, et al. Newly identified pathogens associated with periodontitis: a systematic review. J Dent Res. 2014;93(9):846–858. DOI: 10.1177/0022034514542468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Koch R. Die Aetiologie der Tuberculose. Berliner klin Wochenschr. 1882;19:221–230. [Google Scholar]
- [23].Socransky SS. Criteria for the infectious agents in dental caries and periodontal disease. J Clin Periodontol. 1979;6:16–21. [DOI] [PubMed] [Google Scholar]
- [24].Araujo MW, Hovey KM, Benedek JR, et al. Reproducibility of probing depth measurement using a constant-force electronic probe: analysis of inter and intra-examiner variability. J Periodontol. 2003;74:1736–1740. [DOI] [PubMed] [Google Scholar]
- [25].Socransky SS, Smith C, Martin L, et al. ”Checkerboard“ DNA-DNA hybridization. Biotechniques. 1994;17:788–792. [PubMed] [Google Scholar]
- [26].Soares GM, Mendes JA, Silva MP, et al. Metronidazole alone or with amoxicillin as adjuncts to non-surgical treatment of chronic periodontitis: a secondary analysis of microbiological results from a randomized clinical trial. J Clin Periodontol. 2014;41(4):366–376. DOI: 10.1111/jcpe.12217 [DOI] [PubMed] [Google Scholar]
- [27].Szafranski SP, Wos-Oxley M, Vilchez-Vargas R, et al. High-resolution taxonomic profiling of the subgingival microbiome for biomarker discovery and periodontitis diagnosis. Appl Environ Microbiol. 2015;81(3):1047–1058. DOI: 10.1128/AEM.03534-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Socransky SS, Haffajee AD, Smith C, et al. Relation of counts of microbial species to clinical status at the sample sites. J Clin Periodontol. 1991;18:766–775. [DOI] [PubMed] [Google Scholar]
- [29].Feres M, Retamal-Valdes B, Faveri M, et al. Proposal of a clinical endpoint for periodontal trials: the treat-to-target approach. J Int Acad Periodontol. 2020;22(2):41–53. [PubMed] [Google Scholar]
- [30].Teles RP, Haffajee AD, Socransky SS. Microbiological goals of periodontal therapy. Periodontol 2000. 2006;42:180–218. [DOI] [PubMed] [Google Scholar]
- [31].Haffajee AD, Teles RP, Socransky SS. The effect of periodontal therapy on the composition of the subgingival microbiota. Periodontol 2000. 2006;42:219–258. [DOI] [PubMed] [Google Scholar]
- [32].Haffajee AD, Cugini MA, Dibart S, et al. The effect of SRP on the clinical and/microbiological parameters of periodontal diseases. J Clin Periodontol. 1997;24:324–334. [DOI] [PubMed] [Google Scholar]
- [33].Masood N, Moore K, Farbos A, et al. Draft genome sequence of a meningitic isolate of Cronobacter sakazakii clonal complex 4, strain 8399. Genome Announc. 2013;1. DOI: 10.1128/genomeA.00833-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Hunter CJ, Bean JF. Cronobacter: an emerging opportunistic pathogen associated with neonatal meningitis, sepsis and necrotizing enterocolitis. J Perinatol. 2013;33:581–585. [DOI] [PubMed] [Google Scholar]
- [35].Gaetti-Jardim Júnior E, Nakano V, Wahasugui TC, et al. Occurrence of yeasts, enterococci and other enteric bacteria in subgingival biofilm of HIV-positive patients with chronic gingivitis and necrotizing periodontitis. Braz J Microbiol. 2008;39:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Pinheiro-Hubinger L, Moraes Riboli DF, Abraão LM, et al. Coagulase-negative staphylococci clones are widely distributed in the hospital and community. Pathogens. 2021. Jun 23;10(7):792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Center KJ, Reboli AC, Hubler R, et al. Decreased vancomycin susceptibility of coagulase-negative staphylococci in a neonatal intensive care unit: evidence of spread of Staphylococcus warneri. J Clin Microbiol. 2003. Oct;41(10):4660–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Campoccia D, Montanaro L, Visai L, et al. Characterization of 26 Staphylococcus warneri isolates from orthopedic infections. Int J Artif Organs. 2010;33:575–581. [DOI] [PubMed] [Google Scholar]
- [39].Kini GD, Patel K, Parris AR, et al. An unusual presentation of endocarditis caused by Staphylococcus warneri. Open Microbiol J. 2010;4:103–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Colombo AP, Teles RP, Torres MC, et al. Subgingival microbiota of Brazilian subjects with untreated chronic periodontitis. J Periodontol. 2002:360–369. DOI: 10.1902/jop.2002.73.4.360 [DOI] [PubMed] [Google Scholar]
- [41].Vebø HC, Snipen L, Nes IF, et al. The transcriptome of the nosocomial pathogen Enterococcus faecalis V583 reveals adaptive responses to growth in blood. PLoS ONE. 2009;4:e7660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vu J, Carvalho J. Enterococcus: review of its physiology, pathogenesis, diseases and the challenges it poses for clinical microbiology. Front Biol. 2011;6:357–366. [Google Scholar]
- [43].Stuart CH, Schwartz S, Beeson TJ, et al. Enterococcus faecalis: its role in root canal treatment failure and current concepts in retreatment. J Endod. 2006;32:93–98. [DOI] [PubMed] [Google Scholar]
- [44].Wang Q-Q, Zhang C-F, Chu C-H, et al. Prevalence of Enterococcus faecalis in saliva and filled root canals of teeth associated with apical periodontitis. J Oral Sci. 2012;4:19–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Murad CF, Sassone LM, Faveri M, et al. Microbial diversity in persistent root canal infections investigated by checkerboard DNA-DNA hybridization. J Endod. 2014;40(7):899–906. [DOI] [PubMed] [Google Scholar]
- [46].Prada I, Micó-Muñoz P, Giner-Lluesma T, et al. Influence of microbiology on endodontic failure. Literature Review Med Oral Patol Oral Cir Bucal. 2019;24(3):e364–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Alghamdi F, Shakir M. The influence of Enterococcus faecalis as a dental root canal pathogen on endodontic treatment: a systematic review. Cureus. 2020. Mar 13;12(3):e7257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Barbosa-Ribeiro M, Arruda-Vasconcelos R, Louzada LM, et al. Microbiological analysis of endodontically treated teeth with apical periodontitis before and after endodontic retreatment. Clin Oral Investig. 2021;25(4):2017–2027. [DOI] [PubMed] [Google Scholar]
- [49].Souto R, Andrade AF, Uzeda M, et al. Prevalence of “non-oral” pathogenic bacteria in subgingival biofilm of subjects with chronic periodontitis. Braz J Microbiol. 2006;37:208–215. [Google Scholar]
- [50].da Silva-Boghossian CM, Do Souto RM, Luiz RR, et al. Association of red complex, A. actinomycetemcomitans, and non-oral bacteria with periodontal diseases. Arch Oral Biol. 2011;56(9):899–906. [DOI] [PubMed] [Google Scholar]
- [51].Chidambar CK, Shankar SM, Raghu P, et al. Detection of Enterococcus faecalis in subgingival biofilms of healthy, gingivitis, and chronic periodontitis subjects. J Indian Soc Periodontol. 2019;23(5):416–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Rams TE, Feik D, Young V, et al. Enterococci in human periodontitis. Oral Microbiol Immunol. 1992. Aug;7(4):249–252. [DOI] [PubMed] [Google Scholar]
- [53].Valour F, Chebib N, Gillet Y, et al. Infections broncho-pulmonaires à Staphylococcus aureus. Rev Pneumol Clin. 2013;69:368–382. [DOI] [PubMed] [Google Scholar]
- [54].Hu DL, Nakane A. Mechanisms of staphylococcal enterotoxin-induced emesis. Eur J Pharmacol. 2014;722:95–107. [DOI] [PubMed]
- [55].Wyman TP, Dowden WE, Langeland K. Staphylococcus aureus isolation from a clinically nonexposed root canal. J Endod. 1978;4:122–128. [DOI] [PubMed] [Google Scholar]
- [56].Koukos G, Sakellari D, Arsenakis M, et al. Prevalence of Staphylococcus aureus and methicillin resistant Staphylococcus aureus (MRSA) in the oral cavity. Arch Oral Biol. 2015;60(9):1410–1415. [DOI] [PubMed] [Google Scholar]
- [57].Zhuang LF, Watt RM, Mattheos N, et al. Periodontal and peri-implant microbiota in patients with healthy and inflamed periodontal and peri-implant tissues. Clin Oral Implants Res. 2016;27(1):13–21. [DOI] [PubMed] [Google Scholar]
- [58].Tambone E, Marchetti A, Ceresa C, et al. Counter-acting Candida albicans-Staphylococcus aureus mixed biofilm on titanium implants using microbial biosurfactants. Polymers. 2021. Jul 23;13(15):2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Pérez-Tanoira R, Fernández-Arias M, Potel C, et al. Silver nanoparticles produced by laser ablation and re-irradiation are effective preventing peri-implantitis multispecies biofilm formation. Int J Mol Sci. 2022. Oct 10;23(19):12027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Hannan TJ, Mysorekar IU, Chen SL, et al. LeuX tRNA-dependent and -independent mechanisms of Escherichia coli pathogenesis in acute cystitis. Mol Microbiol. 2008;67(1):116–128. DOI: 10.1111/j.1365-2958.2007.06025.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Hodges K, Hecht G. Bacterial infections of the small intestine. Curr Opin Gastroenterol. 2013;29:159–163. [DOI] [PubMed] [Google Scholar]
- [62].Mayanagi G, Sato T, Shimauchi H, et al. Detection frequency of periodontitis-associated bacteria by polymerase chain reaction in subgingival and supragingival plaque of periodontitis and healthy subjects. Oral Microbiol Immunol. 2004;19:379–385. [DOI] [PubMed] [Google Scholar]
- [63].Kumar PS, Griffen AL, Barton J, et al. New bacterial species associated with chronic periodontitis. J Dent Res. 2003;82:338–344. [DOI] [PubMed] [Google Scholar]
- [64].Schlafer S, Riep B, Griffen AL, et al. Filifactor alocis involvement in periodontal biofilms. BMC Microbiol. 2010;10:10–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Fine DH, Markowitz K, Fairlie K, et al. A consortium of Aggregatibacter actinomycetemcomitans, Streptococcus parasanguinis, and Filifactor alocis is present in sites prior to bone loss in a longitudinal study of localized aggressive periodontitis. J Clin Microbiol. 2013;51:2850–2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Gomes BP, Jacinto RC, Pinheiro ET, et al. Molecular analysis of Filifactor alocis, Tannerella forsythia, and Treponema denticola associated with primary endodontic infections and failed endodontic treatment. J Endod. 2006;32:937–940. [DOI] [PubMed] [Google Scholar]
- [67].da Silva ES, Feres M, Figueiredo LC, et al. Microbiological diversity of peri-implantitis biofilm by Sanger sequencing. Clin Oral Implants Res. 2014;25(10):1192–1199. [DOI] [PubMed] [Google Scholar]
- [68].Feres M, Soares GM, Mendes JA, et al. Metronidazole alone or with amoxicillin as adjuncts to non-surgical treatment of chronic periodontitis: a 1-year double-blinded, placebo-controlled, randomized clinical trial. J Clin Periodontol. 2012;39(12):1149–1158. [DOI] [PubMed] [Google Scholar]
- [69].Mestnik MJ, Feres M, Figueiredo LC, et al. The effects of adjunctive metronidazole plus amoxicillin in the treatment of generalized aggressive periodontitis: a 1-year double-blinded, placebo-controlled, randomized clinical trial. J Clin Periodontol. 2012;39(10):955–961. DOI: 10.1111/j.1600-051X.2012.01932.x [DOI] [PubMed] [Google Scholar]
- [70].Goodson JM, Haffajee AD, Socransky SS, et al. Control of periodontal infections: a randomized controlled trial I. The primary outcome attachment gain and pocket depth reduction at treated sites. J Clin Periodontol. 2012;39:525–536. [DOI] [PubMed] [Google Scholar]
- [71].Sgolastra F, Gatto R, Petrucci A, et al. Effectiveness of systemic amoxicillin/metronidazole as adjunctive therapy to scaling and root planing in the treatment of chronic periodontitis: a systematic review and meta-analysis. J Periodontol. 2012;83:1257–1269. [DOI] [PubMed] [Google Scholar]
- [72].Zandbergen D, Slot DE, Cobb CM, et al. The clinical effect of scaling and root planing and the concomitant administration of systemic amoxicillin and metronidazole: a systematic review. J Periodontol. 2012;84(3):332–351. [DOI] [PubMed] [Google Scholar]
- [73].Teughels W, Feres M, Oud V, et al. Adjunctive effect of systemic antimicrobials in periodontitis therapy: a systematic review and meta-analysis. J Clin Periodontol. 2020;47:257–281. [DOI] [PubMed] [Google Scholar]
- [74].Rosier BT, Marsh PD, Mira A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J Dent Res. 2018;97:371–380. [DOI] [PubMed] [Google Scholar]
- [75].Varoni EM, Bavarian R, Robledo-Sierra J, et al. World Workshop on Oral Medicine VII: targeting the microbiome for oral medicine specialists-Part 1. A methodological guide. Oral Dis. 2019. Jun;25(Suppl 1):12–27. [DOI] [PubMed] [Google Scholar]
- [76].Pérez-Chaparro PJ, McCulloch JA, Mamizuka EM, et al. Do different probing depths exhibit striking differences in microbial profiles? J Clin Periodontol. 2018;45(1):26–37. DOI: 10.1111/jcpe.12811 [DOI] [PubMed] [Google Scholar]
- [77].Hajishengallis G, Liang S, Payne MA, et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011. Nov 17;10(5):497–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Hajishengallis G, Darveau RP, Curtis MA. The keystone-pathogen hypothesis. Nat Rev Microbiol. 2012. Oct;10(10):717–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Hajishengallis G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 2014. JanOct;35(1):3–11. [Epub 2013 23]. DOI: 10.1016/j.it.2013.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Louail R, Florin F, Bernard S, et al. Invasion of intestinal cells by Staphylococcus warneri, a member of the human gut microbiota. Gut Pathog. 2023. Jan 27;15(1):4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Herrera D, Contreras A, Gamonal J, et al. Subgingival microbial profiles in chronic periodontitis patients from Chile, Colombia and Spain. J Clin Periodontol. 2008;35:106–113. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


